Abstract
The prolactin receptor (PRLR) signals predominantly through the JAK2-STAT5 pathway regulating multiple physiological functions relating to fertility, lactation, and metabolism. However, the molecular pathology and role of PRLR mutations and signalling are incompletely defined, with progress hampered by a lack of reported disease-associated PRLR variants. To date, two common germline PRLR variants are reported to demonstrate constitutive activity, with one, Ile146Leu, overrepresented in benign breast disease, while a rare activating variant, Asn492Ile, is reported to be associated with an increased incidence of prolactinoma. In contrast, an inactivating germline heterozygous PRLR variant (His188Arg) was reported in a kindred with hyperprolactinaemia, while an inactivating compound heterozygous PRLR variant (Pro269Leu/Arg171Stop) was identified in an individual with hyperprolactinaemia and agalactia. We hypothesised that additional rare germline PRLR variants, identified from large-scale sequencing projects (ExAC and GnomAD), may be associated with altered in vitro PRLR signalling activity. We therefore evaluated >300 previously uncharacterised non-synonymous, germline PRLR variants and selected 10 variants for in vitro analysis based on protein prediction algorithms, proximity to known functional domains and structural modelling. Five variants, including extracellular and intracellular domain variants, were associated with altered responses when compared to the wild-type receptor. These altered responses included loss- and gain-of-function activities related to STAT5 signalling, Akt and FOXO1 activity, as well as cell viability and apoptosis. These studies provide further insight into PRLR structure–function and indicate that rare germline PRLR variants may have diverse modulating effects on PRLR signalling, although the pathophysiologic relevance of such alterations remains to be defined.
Keywords: hyperprolactinaemia, prolactinoma, proliferation, JAK-STAT5
Introduction
The prolactin receptor (PRLR), and its ligand, the hormone prolactin (PRL) are reported to have diverse roles that include induction and maintenance of lactation in the peripartum and postpartum periods (Ben-Jonathan et al. 2008), parental behaviour, immune function, reproduction and metabolic functions such as pregnancy-related increases in β-cell mass, and regulation of lipid content and body temperature (Freemark et al. 2002, Schuff et al. 2002, Viengchareun et al. 2004, Viengchareun et al. 2008, Huang et al. 2009, Banerjee et al. 2016, Smiley et al. 2022). PRL binds to the PRLR, a class I cytokine receptor, which is functionally active as a homodimer (Gadd & Clevenger 2006, Qazi et al. 2006, Brooks et al. 2014). Each mature PRLR monomer has a multi-domain structure consisting of a highly conserved ligand-binding extracellular domain (ECD, 1–210), a transmembrane α-helix (TM, residues 211–234), and an intracellular domain (ICD, residues 235–598). Structural analysis of the ECD has revealed two subdomains, designated D1 and D2, which are important in ligand binding and subsequent receptor activation (Svensson et al. 2008, Broutin et al. 2010, van Agthoven et al. 2010, Rao & Brooks 2011, Brooks 2012), and the WSxWS motif, which acts as a molecular switch during activation (Broutin et al. 2010, van Agthoven et al. 2010, Dagil et al. 2012).
The ICD interacts with the Janus kinase-2 (JAK2) protein that associates with a highly conserved structural motif named Box 1 (residues 243–251) and potentially, with a second motif, Box 2 (residues 287–296) within the ICD (Lebrun et al. 1995). Moreover, recent structural studies have revealed that both PRLR and the related growth hormone receptor (GHR) harbour conserved regions that interact with lipids, referred to as lipid-interacting domains or LIDs within the inner plasma membrane leaflet thereby allowing a greater surface area and potentially, simultaneous interaction with multiple signalling kinases (Haxholm et al. 2015, Bugge et al. 2016). Hormone binding to the ECD activates conformational changes within the TMD and ICD, allowing separation of the ICDs and initiating phosphorylation cascades downstream of JAK2 (Brown et al. 2005, Brooks et al. 2014, Haxholm et al. 2015, Bugge et al. 2016). JAK2 activates complex signalling pathways, predominantly via interaction with the signal activator of transcription 5 (STAT5) pathway (Brooks 2012), and also by the phosphatidylinositol 3-kinase (PI3K)/Akt and mitogen-activated protein kinase pathways (Fresno Vara et al. 2000, Amaral et al. 2004, Brooks 2012). These signal pathways lead to transcription of target genes that regulate proliferation, differentiation, and cell survival (Fresno Vara et al. 2000, Amaral et al. 2004, Brooks 2012). Despite these findings, the function of individual residues in receptor activation and signal transduction is poorly understood.
A number of human studies have highlighted residues that are important for receptor function and subsequent signal transduction. The PRLR variants Ile76Val and Ile146Leu were reported to be gain-of-function variants with constitutive activity that occur in 15% of women with breast fibroadenomas (Bogorad et al. 2008, Courtillot et al. 2010). However, more recent studies did not detect such correlations (Glasow et al. 2001, Vaclavicek et al. 2006, Lee et al. 2007, Nyante et al. 2011) or marked changes in signalling activity (Bernard et al. 2016, Gorvin et al. 2018b), although the Ile146 residue has been shown to be important for receptor folding and stability (Dagil et al. 2012, Zhang et al. 2015). A loss-of-function pathogenic germline PRLR variant (His188Arg), which affected a highly conserved His188 residue within the D2 domain that is important for hormone binding, was described in a family with hyperprolactinaemia (Kulkarni et al. 2010, Newey et al. 2013). Subsequently, an individual with hyperprolactinaemia and agalactia was reported with germline compound heterozygous nonsense (Arg171Stop) and missense (Pro269Leu) PRLR variants (Kobayashi et al. 2018), and recently, a germline Asn492Ile PRLR variant that increases receptor activity via the PI3K-Akt pathway was reported to be associated with a higher incidence of prolactinoma (Gorvin et al. 2018b).
Both the Ile146Leu and Asn492Ile variants are present in recently described population databases such as the Exome Aggregation Consortium (ExAC) and the Genome Aggregation Database (GnomAD) (Karczewski et al. 2020), and we hypothesised that a further examination of these population-based databases could yield important structural and functional insights for individual PRLR residues and provide information on activation of specific signalling pathways. Indeed, similar studies have previously identified residues within the adaptor protein-2 sigma subunit that are important for calcium homeostasis (Gorvin et al. 2018a) and residues in α-N-acetylglucosaminidase that contribute to the rare lysosomal storage disease Sanfilippo type-B (Clark et al. 2018). We therefore examined the ExAC/GnomAD datasets with the aim of identifying missense coding variants in the PRLR, which could be characterised by their functional consequences.
Materials and methods
Protein sequence alignment and three-dimensional modelling of PRLR structure
The population frequencies of germline non-synonymous PRLR single nucleotide variants (SNVs) were evaluated using ExAC and Genome Aggregation Databases (both datasets (ExAC and GnomAD v.2.1) now reported at GnomAD (https://gnomad.broadinstitute.org/) (Karczewski et al. 2020)). SIFT, MutationTaster, Polyphen-2, and REVEL were used to predict the effect of amino acid substitutions (Kumar et al. 2009, Adzhubei et al. 2010, Schwarz et al. 2014, Ioannidis et al. 2016). Amino acid conservation was examined in PRLR mammalian orthologs using ClustalW2(Larkin et al. 2007). The crystal structure of the two chains of the rat PRLR extracellular domain in complex with PRL (Protein Data Bank (PDB) accession code 3NPZ) and the NMR structure of the human PRLR ECD D2 domain (PDB:2LFG) (van Agthoven et al. 2010, Dagil et al. 2012) were used to predict the effect of ECD variants on PRLR structure. The NMR structure of the single-pass transmembrane domain of PRLR (PDB:2N7I) (Bugge et al. 2016) was used to predict the structural effect of TMD variants. Figures were prepared using the PyMOL Molecular Graphics System (Schrödinger, New York, NY, USA).
Cell culture and transfection
PRLR variants were introduced into the wild-type (WT) pdEYFP-PRLR construct by site-directed mutagenesis using the Quikchange Lightning Kit (Agilent Technologies) and gene-specific primers (Sigma) and were confirmed as previously described (Newey et al. 2013). Expression constructs were transiently transfected into HEK293 cells and were maintained in DMEM-Glutamax media (Gibco) with 10% fetal bovine serum (Gibco) at 37ºC, 5% CO2, using Lipofectamine 2000 (LifeTechnologies), as described (Newey et al. 2013), and functional studies were performed using poly-l-lysine-treated plates. Western blot analysis was used to assess the expression of transfected PRLR and endogenous α-tubulin as a loading control, using anti-PRLR (1:1000, Santa Cruz Biotechnology) and anti-α-tubulin (1:1000, Abcam) antibodies, as described (Newey et al. 2013).
Confocal microscopy
Confocal imaging was performed as previously described (Gorvin et al. 2018b). Cells were plated in six-well plates containing poly-l-lysine-treated coverslips and cultured at 37°C. Cells were transiently transfected with 1000 ng of either WT or variant PRLR expression constructs. After 24 h, cells were fixed in 4% paraformaldehyde/PBS (Sigma-Aldrich), permeabilised in 1% Triton-X100/PBS (Thermo Scientific), and immunostained with primary anti-PRLR (1:200, Santa Cruz Biotechnology) and secondary antibody Alexa Fluor-488 (1:1000, Molecular Probes). Cells were mounted in Prolong Gold Antifade reagent (Invitrogen). Images were captured using a Zeiss LSM780 confocal microscope with a Plan-Apochromat x63/1.2/water DIC objective. An argon laser (488 nm) was used to excite Alexa Fluor-488.
AlphaScreen SureFire assays
AlphaScreen assays were performed as previously described (Newey et al. 2013, Gorvin et al. 2018b). Cells were transiently transfected in 48-well plates with 200 ng of either WT or variant PRLR vectors. After 30 h, cells were incubated in serum-free media for 12 h prior to treatment with human recombinant PRL (PromoCell, Heidelberg, Germany) for 20 min at concentrations ranging from 0 to 1000 ng/mL. Cells were lysed in Surefire lysis buffer, and AlphaScreen Surefire pSTAT5 or pAkt assays (PerkinElmer) were performed according to manufacturer’s instructions (Binder et al. 2008). The fluorescence signal was measured using a PHERAstar FS microplate reader (BMG Labtech, Aylesbury, UK). A minimum of four independently transfected replicates were used for each construct within each experiment, and each experiment was performed on four to five separate occasions with different cell passages. Data were plotted as fold-change responses relative to the response at 0 ng/mL in cells expressing the WT PRLR expression construct. Statistical analyses were performed using two-way ANOVA with Dunnett’s or Tukey’s multiple-comparison tests for pSTAT5 studies and by one-way ANOVA with Sidak’s multiple-comparison tests for pAkt studies
Luciferase reporter assays
The Forkhead box O1 (FOXO1) promoter region was PCR amplified from human genomic DNA using previously described primers (Essaghir et al. 2009) and cloned into the pGL4.10 vector (Promega). The sequence of the insert was confirmed by Sanger DNA sequencing (Source Bioscience, Nottingham, UK). The pGL4.10 vector containing the cytokine-inducible SH2-containing protein (CISH) reporter has been described previously (Newey et al. 2013). HEK293 cells were transiently co-transfected in 24-well plates with 100 ng of pGL4.10-CISH reporter gene construct, 10 ng of PRL (renilla) control vector, and 100 ng of WT or variant PRLR vectors. Following transfection, cells were incubated in serum-free media overnight. Cells were then treated with 0–500 ng/mL PRL for 24 h in serum-free media. Cells were lysed and assayed for luciferase activity using a Turner Biosystems (Promega) or Centro LB960 (Berthold Technologies, Harpenden, Hertfordshire, UK) luminometer and the Dual-Luciferase Reporter assay system (Promega). The firefly luciferase activity was adjusted for Renilla luciferase activity (Firefly/Renilla ratio) and ratios were expressed as a fold-change relative to cells treated with 0 ng/mL of PRL within each group. A minimum of four independently transfected replicates were performed in each experiment, and each experiment was performed on four to seven separate occasions with different cell passages. Statistical analysis was performed by two-way ANOVA with Sidak’s or Dunnett’s multiple-comparisons test for CISH and by Kruskal–Wallis with Dunn’s test or one-way ANOVA with Dunnett’s test for FOXO1.
Cell viability assay
Cells were plated in 96-well plates and transfected with 50 ng of WT or variant PRLR per well. Following 24 h, cells were treated with 200 ng/mL of PRL and cell viability was assessed 96 h later using the CellTiter Blue kit (Promega) (Gorvin et al. 2018b). The cell count for day 1 (i.e. time 0 before PRL was added) was set as 1 and each cell count was expressed relative to this original cell count. Plates were read on a PHERAstar FS microplate reader (BMG Labtech). A minimum of four independently transfected replicates were performed in each experiment, and each experiment was performed on four separate occasions with different cell passages. Statistical analysis was performed by one-way ANOVA with Dunnett’s test or Kruskal–Wallis with Dunn’s test.
Apoptosis assay
Cells were plated in 96-well plates and transfected with 50 ng of WT or variant PRLR vectors per well. Following 24 h, cells were treated with 0 ng/mL or 200 ng/mL of PRL and apoptosis was assessed at 0 and 96 h post-PRL treatment using the Caspase-Glo-3/7 kit (Promega) (Gorvin et al. 2018b). Plates were read on a Centro LB960 luminometer. A minimum of four independently transfected replicates were performed in each experiment, and each experiment was performed on four to six separate occasions with different cell passages. Statistical analysis was performed by one-way ANOVA with Dunnett’s multiple comparisons test.
Statistics
The number of experimental replicates denoted by n is indicated in each figure legend. Data were plotted and statistical analyses were performed in Graphpad Prism 7. Normality tests (Shapiro–Wilk or D’Agostino–Pearson) were performed on all datasets to determine whether parametric or non-parametric tests were appropriate. A P-value of < 0.05 was considered statistically significant. A minimum of four independently transfected replicates was performed in all cell-based assays, and each experiment was performed on separate occasions with different passages of cells. Specific details of each test are outlined in the figure legends and within the relevant methods section.
Results
Identification of non-synonymous PRLR variants and their predicted effects on protein function
An analysis of the ExAC (v1.0) and GnomAD (v2.1.1) databases was performed to identify non-synonymous, missense PRLR variants in the full-length, membrane-expressed (i.e. excluding the 24 amino acid signal peptide) protein. These analyses revealed 310 non-synonymous missense PRLR variants comprising 85 ECD variants, 10 TMD variants, and 215 ICD variants. The distribution of variants between the ECD, TMD, and ICD was 27.4, 3.2, and 69.4%, respectively. This was significantly different from that expected based on the size of each region (P < 0.05, χ2-test), with fewer variants observed in the ECD than expected (37.7%), indicating this region may be less tolerant to variation.
The predicted deleteriousness/pathogenicity of each PRLR variant was determined by assessing their population frequency; their effect on protein function using four online prediction software packages (SIFT, Polyphen-2, MutationTaster, and REVEL (Kumar et al. 2009, Adzhubei et al. 2013, Schwarz et al. 2014, Ioannidis et al. 2016)) and the evolutionary conservation of each residue in mammalian species. Following exclusion of variants that have previously been functionally expressed (e.g. Gly57Ser, Ile76Val, Ile146Leu, Gly376Gln, Asn492Ile, and Glu554Gln (Bogorad et al. 2008, Newey et al. 2013, Bernard et al. 2016, Gorvin et al. 2018b)), examination of the minor allele frequencies (MAF) of all the remaining variants revealed them to be rare (defined as a MAF of <1% (Agarwala et al. 2013)). Variants that were predicted benign or tolerated by all the prediction programs, and those that affected residues that were conserved in fewer than two mammalian orthologues, were excluded from further analyses. Thus, 42 ECD, 5 TMD, and 93 ICD variants (Table 1) were predicted to be potentially deleterious/pathogenic by these criteria, indicating that they may have functional consequences on PRLR signalling.
Table 1.
PRLR rare variants predicted to be potentially deleterious/pathogenic in the GnomAD database.
| Amino acid change | Residue | Protein predictionsa | Evolutionary conservationb | Predicted structural effectc | ||||
|---|---|---|---|---|---|---|---|---|
| SIFT | Polyphen | MutationTaster | REVEL | |||||
| ECD D1 | Pro>Leu | 3 (27) | Tolerated | Probably damaging | Disease causing | Benign | 4 | No change |
| Pro>His | 3 (27) | Deleterious | Probably damaging | Polymorphism | Benign | 4 | No change | |
| Lys>Asn | 6 (30) | Deleterious | Probably damaging | Disease causing | Benign | 4 | Loss of contact with Asp91 | |
| Pro>Ser | 7 (31) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | Gains contact with Val79 | |
| Arg>His | 13 (37) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | No change | |
| Glu>Lys | 18 (42) | Deleterious | Probably damaging | Disease causing | Benign | 4 | No change | |
| Thr>Ile | 21 (45) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | No change | |
| Gly>Arg | 30 (54) | Deleterious | Probably damaging | Disease causing | Benign | 4 | Gains contact with Asn83 | |
| Gly>Arg | 31 (55) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | No change | |
| Thr>Ser | 34 (58) | Deleterious | Benign | Disease causing | Benign | 4 | No change | |
| His>Arg | 49 (73) | Deleterious | Possibly damaging | Polymorphism | Benign | 2 | No change | |
| Cys>Tyr | 51 (75) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | Gains contact with Phe63 | |
| Gly>Asp | 58 (82) | Deleterious | Probably damaging | Disease causing | Benign | 4 | No change | |
| Asn>Ser | 60 (84) | Deleterious | Possibly damaging | Disease causing | Benign | 4 | No change | |
| Ile>Met | 76 (100) | Tolerated | Possibly damaging | Polymorphism | Benign | 3 | No change | |
| Gln>His | 84 (108) | Deleterious | Possibly damaging | Disease causing | Benign | 3 | No change | |
| Met>Ile | 85 (109) | Tolerated | Probably damaging | Disease causing | Benign | 4 | No change | |
| Ser>Arg | 88 (112) | Deleterious | Possibly damaging | Polymorphism | Benign | 4 | Gains contact with Ser87 and loses contact with Pro4 | |
| Val>Leu | 95 (119) | Deleterious | Possibly damaging | Disease causing | Benign | 4 | No change | |
| Asp>Glu | 96 (120) | Tolerated | Probably damaging | Disease causing | Benign | 3 | No change | |
| Val>Met | 97 (121) | Deleterious | Probably damaging | Disease causing | Benign | 4 | No change | |
| Tyr>His | 99 (123) | Deleterious | Probably damaging | Disease causing | Benign | 4 | Loses contact with PRL | |
| Ile>Val | 100 (124) | Deleterious | Probably damaging | Disease causing | Benign | 4 | No change | |
| ECD D2 | Leu>Met | 109 (133) | Deleterious | Possibly damaging | Polymorphism | Benign | 4 | No change |
| Pro>Ala | 129 (153) | Deleterious | Possibly damaging | Disease causing | Likely benign | 4 | No change | |
| Pro>Leu | 129 (153) | Deleterious | Possibly damaging | Disease causing | Disease causing | 4 | No change | |
| Thr>Met | 141 (165) | Deleterious | Benign | Disease causing | Benign | 3 | Loses contact with Asp187 | |
| Tyr>Asn | 144 (168) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | No change | |
| Glu>Asp | 145 (169) | Deleterious | Probably damaging | Disease causing | Benign | 4 | Loses 3 of 4 contacts with Arg183, gains contact with Ile146 | |
| Arg>Gln | 147 (171) | Tolerated | Benign | Disease causing | Benign | 4 | No change | |
| Glu>Lys | 151 (175) | Deleterious | Benign | Disease causing | Benign | 3 | No change | |
| Glu>Lys | 155 (179) | Deleterious | Possibly damaging | Disease causing | Benign | 3 | Loses contact with Lys114 on opposite PRLR protomer | |
| Trp>Leu | 156 (180) | Deleterious | Possibly damaging | Disease causing | Benign | 4 | No change | |
| Ile>Val | 169 (193) | Tolerated | Benign | Disease causing | Likely benign | 2 | No change | |
| Leu>Val | 172 (196) | Tolerated | Benign | Disease causing | Benign | 4 | No change | |
| Gly>Glu | 175 (199) | Deleterious | Probably damaging | Disease causing | Benign | 4 | No change | |
| Leu>Phe | 179 (203) | Tolerated | Benign | Disease causing | Benign | 4 | No change | |
| Val>Ile | 180 (204) | Tolerated | Benign | Disease causing | Benign | 4 | No change | |
| Arg>His | 183 (207) | Tolerated | Probably damaging | Disease causing | Benign | 4 | Loses 2 of 4 contacts with Glu145; loses contact with Ala193 of WSxWS motif | |
| Asp>Glu | 187 (211) | Deleterious | Possibly damaging | Disease causing | Benign | 4 | Loses contact with His188 | |
| Gln>Arg | 201 (225) | Deleterious | Benign | Polymorphism | Benign | 2 | No change | |
| Asp>Asn | 205 (229) | Tolerated | Benign | Disease causing | Benign | 4 | No change | |
| TMD | Ala>Gly | 222 (246) | Deleterious | Benign | Disease causing | Benign | 4 | No change |
| Ile>Thr | 227 (251) | Tolerated | Possibly damaging | Disease causing | Disease causing | 4 | No change | |
| Trp>Cys | 230 (254) | Deleterious | Possibly damaging | Disease causing | Disease causing | 4 | No change | |
| Val>Ala | 232 (256) | Deleterious | Benign | Disease causing | Disease causing | 4 | No change | |
| Val>Met | 232 (256) | Tolerated | Benign | Possibly damaging | Benign | 4 | No change | |
| ICD | Lys>Arg | 235 (259) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | Unknown |
| Gly>Ser | 236 (260) | Tolerated | Possibly damaging | Disease causing | Benign | 4 | LID1, Box 1 | |
| Met>Leu | 239 (263) | Tolerated | Benign | Possibly damaging | Benign | 4 | LID1, Box 1 | |
| Cys>Arg | 242 (266) | Tolerated | Probably damaging | Disease causing | Disease causing | 4 | LID1, Box 1 | |
| Ile>Val | 243 (267) | Tolerated | Benign | Disease causing | Likely benign | 4 | LID1, Box 1 | |
| Pro>Leu | 245 (269) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | LID1, Box 1 | |
| Pro>Ser | 246 (270) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | LID1, Box 1 | |
| Lys>Asn | 251 (275) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | LID1, Box 1 | |
| Lys>Arg | 253 (277) | Tolerated | Probably damaging | Disease causing | Benign | 4 | LID1 | |
| Lys>Glu | 253 (277) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | LID1 | |
| Phe>Ser | 255 (279) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | LID1 | |
| Asp>Ala | 256 (280) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | LID1 | |
| Leu>Trp | 260 (284) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | LID1 | |
| Gly>Ser | 263 (287) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | LID1 | |
| Gly>Asp | 263 (287) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | LID1 | |
| Lys>Gln | 264 (288) | Deleterious | Probably damaging | Disease causing | Benign | 4 | LID1 | |
| Ser>Cys | 265 (289) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | LID1 | |
| Glu>Gln | 266 (290) | Deleterious | Probably damaging | Disease causing | Benign | 4 | LID1 | |
| Ser>Thr | 270 (294) | Tolerated | Benign | Disease causing | Benign | 3 | LID1 | |
| Ser>Arg | 270 (294) | Deleterious | Benign | Disease causing | Benign | 3 | LID1 | |
| Ala>Val | 271 (295) | Deleterious | Possibly damaging | Disease causing | Benign | 4 | LID1 | |
| Leu>Ser | 272 (296) | Deleterious | Possibly damaging | Disease causing | Disease causing | 4 | LID1 | |
| Gly>Val | 273 (297) | Tolerated | Probably damaging | Disease causing | Benign | 3 | LID1 | |
| Asp>Tyr | 276 (300) | Deleterious | Possibly damaging | Disease causing | Benign | 4 | LID1 | |
| Pro>His | 278 (302) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | LID1 | |
| Ser>Phe | 281 (305) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | LID1 | |
| Ser>Tyr | 281 (305) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | LID1 | |
| Asp>Asn | 285 (309) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | LID1 | |
| Asp>Glu | 285 (309) | Tolerated | Benign | Disease causing | Likely benign | 4 | LID1 | |
| Val>Ile | 293 (317) | Tolerated | Possibly damaging | Disease causing | Benign | 4 | LID1, Box 2 | |
| Asp>Glu | 294 (318) | Tolerated | Possibly damaging | Polymorphism | Benign | 4 | LID1, Box 2 | |
| Asp>Asn | 298 (322) | Deleterious | Benign | Disease causing | Benign | 4 | LID1 | |
| Leu>Ile | 301 (325) | Deleterious | Possibly damaging | Disease causing | Benign | 4 | ||
| Met>Ile | 302 (326) | Tolerated | Benign | Possibly damaging | Benign | 4 | ||
| Pro>Arg | 316 (340) | Deleterious | Possibly damaging | Polymorphism | Benign | 4 | ||
| Asp>Tyr | 320 (344) | Deleterious | Probably damaging | Polymorphism | Benign | 4 | ||
| Arg>Gln | 327 (351) | Tolerated | Benign | Disease causing | Benign | 2 | Degradation motif | |
| Arg>Pro | 327 (351) | Deleterious | Probably damaging | Disease causing | Disease causing | 2 | Degradation motif | |
| Arg>Trp | 327 (351) | Deleterious | Probably damaging | Disease causing | Disease causing | 2 | Degradation motif | |
| Asp>Asn | 331 (355) | Deleterious | Probably damaging | Disease causing | Benign | 4 | ||
| Pro>Arg | 333 (357) | Deleterious | Possibly damaging | Disease causing | Benign | 2 | ||
| Ser>Phe | 334 (358) | Tolerated | Probably damaging | Disease causing | Benign | 4 | ||
| Cys>Tyr | 340 (364) | Tolerated | Benign | Disease causing | Benign | 4 | ||
| Glu>Lys | 342 (366) | Deleterious | Possibly damaging | Dsease causing | Benign | 4 | ||
| Pro>Thr | 353 (377) | Tolerated | Probably damaging | Polymorphism | Benign | 4 | LID2 | |
| Glu>Gly | 376 (400) | Deleterious | Possibly damaging | Polymorphism | Benign | 2 | LID2 | |
| Tyr>Cys | 381 (405) | Tolerated | Benign | Disease causing | Benign | 2 | LID2 | |
| Gly>Glu | 386 (410) | Tolerated | Possibly damaging | Polymorphism | Benign | 2 | ||
| Gln>Glu | 396 (420) | Deleterious | Probably damaging | Polymorphism | Benign | 2 | ||
| Arg>Ile | 403 (427) | Deleterious | Possibly damaging | Polymorphism | Benign | 4 | ||
| Asp>Val | 411 (435) | Deleterious | Possibly damaging | Disease causing | Disease causing | 4 | ||
| Asp>Tyr | 411 (435) | Deleterious | Possibly damaging | Polymorphism | Disease causing | 4 | ||
| Cys>Arg | 413 (437) | Tolerated | Possibly damaging | Polymorphism | Benign | 4 | ||
| Cys>Tyr | 413 (437) | Tolerated | Possibly damaging | Polymorphism | Benign | 4 | ||
| Thr>Pro | 425 (449) | Tolerated | Possibly damaging | Polymorphism | Likely benign | 2 | ||
| Asp>Asn | 463 (487) | Tolerated | Benign | Disease causing | Benign | 2 | ||
| Gln>His | 472 (496) | Tolerated | Possibly damaging | Polymorphism | Benign | 4 | ||
| Lys>Glu | 481 (505) | Tolerated | Possibly damaging | Polymorphism | Benign | 3 | ||
| Asp>Asn | 484 (508) | Deleterious | Possibly damaging | Disease causing | Disease causing | 3 | ||
| Val>Met | 486 (510) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | ||
| Glu>Ala | 487 (511) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | ||
| Glu>Lys | 487 (511) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | ||
| Ile>Phe | 488 (512) | Deleterious | Probably damaging | Disease causing | Benign | 4 | ||
| Ile>Thr | 488 (512) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | ||
| His>Tyr | 489 (513) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | ||
| Gly>Val | 495 (519) | Deleterious | Probably damaging | Disease causing | Benign | 4 | ||
| Leu>Phe | 497 (521) | Deleterious | Probably damaging | Polymorphism | Benign | 4 | ||
| Leu>Arg | 500 (524) | Deleterious | Possibly damaging | Polymorphism | Disease causing | 2 | ||
| Pro>Gln | 501 (525) | Deleterious | Probably damaging | Disease causing | Benign | 4 | ||
| Pro>Thr | 501 (525) | Tolerated | Possibly damaging | Disease causing | Benign | 4 | ||
| Pro>Leu | 501 (525) | Tolerated | Benign | Disease causing | Benign | 4 | ||
| Arg>Thr | 504 (528) | Deleterious | Benign | Polymorphism | Benign | 2 | ||
| Lys>Asn | 520 (544) | Deleterious | Probably damaging | Disease causing | Benign | 4 | ||
| Glu>Lys | 521 (545) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | ||
| Lys>Glu | 524 (548) | Deleterious | Possibly damaging | Disease causing | Disease causing | 4 | ||
| Asp>His | 530 (554) | Deleterious | Probably damaging | Disease causing | Benign | 4 | ||
| Asp>Gly | 530 (554) | Tolerated | Possibly damaging | Disease causing | Benign | 4 | ||
| Asn>Lys | 531 (555) | Deleterious | Benign | Polymorphism | Benign | 3 | ||
| Asn>Ser | 532 (556) | Tolerated | Benign | Disease causing | Benign | 4 | ||
| Leu>Val | 534 (558) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | ||
| Val>Met | 535 (559) | Deleterious | Probably damaging | Disease causing | Disease causing | 4 | ||
| Asp>His | 539 (563) | Deleterious | Probably damaging | Polymorphism | Benign | 3 | ||
| Ala>Asp | 542 (566) | Tolerated | Possibly damaging | Polymorphism | Benign | 4 | ||
| Glu>Lys | 549 (573) | Deleterious | Possibly damaging | Polymorphism | Benign | 4 | LID3 | |
| Glu>Gly | 549 (573) | Deleterious | Possibly damaging | Polymorphism | Benign | 4 | LID3 | |
| Ala>Asp | 552 (576) | Deleterious | Benign | Polymorphism | Disease causing | 4 | LID3 | |
| Lys>Arg | 553 (577) | Tolerated | Possibly damaging | Polymorphism | Benign | 4 | LID3 | |
| Asn>Lys | 562 (586) | Tolerated | Possibly damaging | Polymorphism | Benign | 2 | LID3 | |
| Asn>Tyr | 562 (586) | Deleterious | Probably damaging | Polymorphism | Benign | 2 | LID3 | |
| Leu>Pro | 580 (604) | Tolerated | Probably damaging | Disease causing | Disease causing | 3 | LID3 | |
| Asp>Tyr | 586 (610) | Deleterious | Probably damaging | Disease causing | Disease causing | 3 | LID3 | |
| Asp>Gly | 589 (613) | Deleterious | Probably damaging | Disease causing | Disease causing | 3 | LID3 | |
| Pro>Arg | 590 (614) | Deleterious | Probably damaging | Disease causing | Disease causing | 3 | LID3 | |
| Cys>Arg | 592 (616) | Tolerated | Possibly damaging | Polymorphism | Disease causing | 3 | LID3 | |
aProtein predictions were made using SIFT, Polyphen-2, MutationTaster, and REVEL; bEvolutionary conservation was based on the PRLR ortholog sequence in four species (cow, dog, mouse, and rat) in comparison to the human protein. The score shows the number of species (out of four) in which the residue is conserved; cStructural analyses were performed using the reported crystal structure of the ligand-bound homodimeric ECD and the NMR structure of the D2 domain (van Agthoven et al. 2010, Dagil et al. 2012) (Figs. 1, 2 and 3). The ten PRLR variants shown in bold are those that were functionally characterised.
Structural characterisation of the ECD PRLR variants
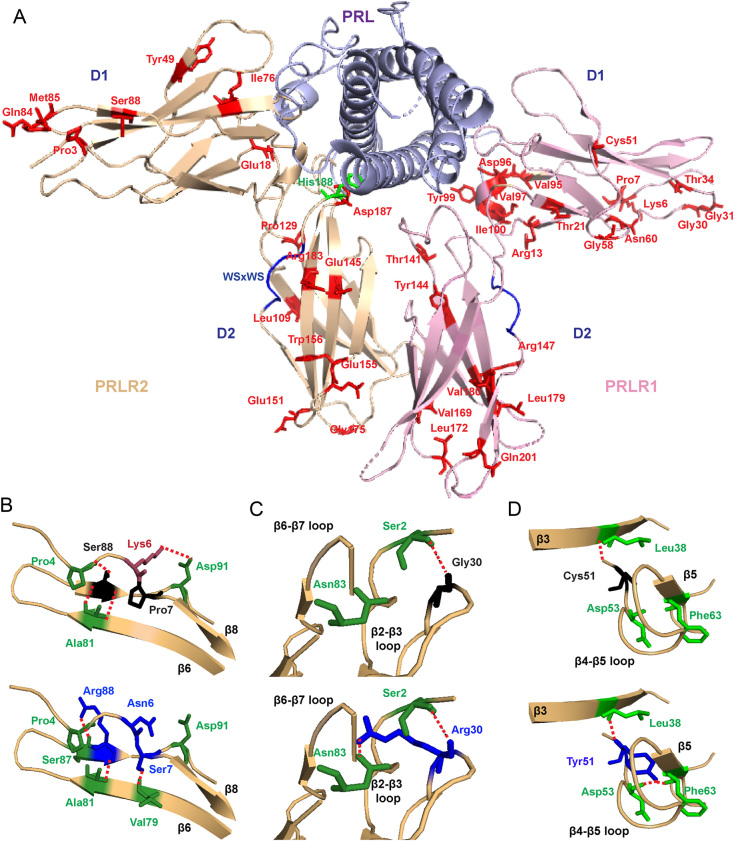
Three-dimensional modelling using the crystal structure of the PRL-bound homodimeric ECD and the NMR structure of the ligand-free D2 domain (van Agthoven et al. 2010, Dagil et al. 2012) was performed to determine the locations of the ECD PRLR variants and predict their effects on structural integrity. This revealed that 23 of the total 42 variants were within the D1 lobe and the remaining 19 variants were within the D2 lobe, of which 11 variants (6 in D1 and 5 in D2) were predicted to have structural effects on the PRLR protein (Table 1, Fig. 1A). Three variants (Pro7Ser, Gly30Arg, Cys51Tyr) are predicted to gain contacts with adjacent residues, six variants (Lys6Asn, Tyr99His, Thr141Met, Glu155Lys, Arg183His, Asp187Glu) are predicted to lose contacts with adjacent residues, and two variants (Ser88Arg, Glu145Asp) are predicted to both lose and gain contacts with adjacent PRLR residues (Fig. 1B, C, D and 2, Table 1). These predicted changes may affect ECD flexibility and disrupt PRLR activation.
Figure 1.
Location of the PRLR ECD variants and structural characterisation of rare variants located in D1. (A) Crystal structure showing the two monomers (PRLR1 in pink and PRLR2 in light brown) of the rat PRLR extracellular domain (ECD) in complex with the PRL hormone (purple) (PDB 3NPZ (van Agthoven et al. 2010)). Each PRLR ECD monomer is comprised of two subdomains, designated D1 (residues 1–101) and D2 (residues 109–210), which are important in ligand binding and subsequent receptor activation (Svensson et al. 2008, Broutin et al. 2010, van Agthoven et al. 2010, Rao and Brooks 2011, Brooks 2012). Side chains of the potential deleterious variant residues from ExAC and GnomAD are shown in red. The His188 residue that is mutated in hyperprolactinaemia is shown in green. Residue 49 is labelled as Tyr in the rat structure, and in humans, this is His49. Residue 169 is labelled as Val in the rat structure, and in human, this is Ile169. Rare variants are shown in only one monomer. (B) Lys6, Pro7, and Ser88 are located in close proximity within the β6-β7-β8 region (Top). The variant Asn6 loses contact with Asp91 on the β8 strand, Ser7 gains contact with Val79 on the β6 strand, and Arg88 loses contact with Pro4 and gains a new contact with the adjacent Ser87 (all blue, bottom). (C) The Gly30 residue, located in the β2-β3 loop, forms a polar contact with the Ser2 residue (Top). When mutated to Arg30 (blue, bottom), the longer side chain is able to retain this contact and form a new hydrogen bond with Asn83 in the β6-β7 loop. (D) The Cys51 residue is located in the β4-β5 loop. Mutation in Tyr51 (blue, bottom) adds a more bulky residue within this tightly packed region, which is predicted to result in gain of two contacts with Asp53 and Phe63 of the β4-β5 loop.
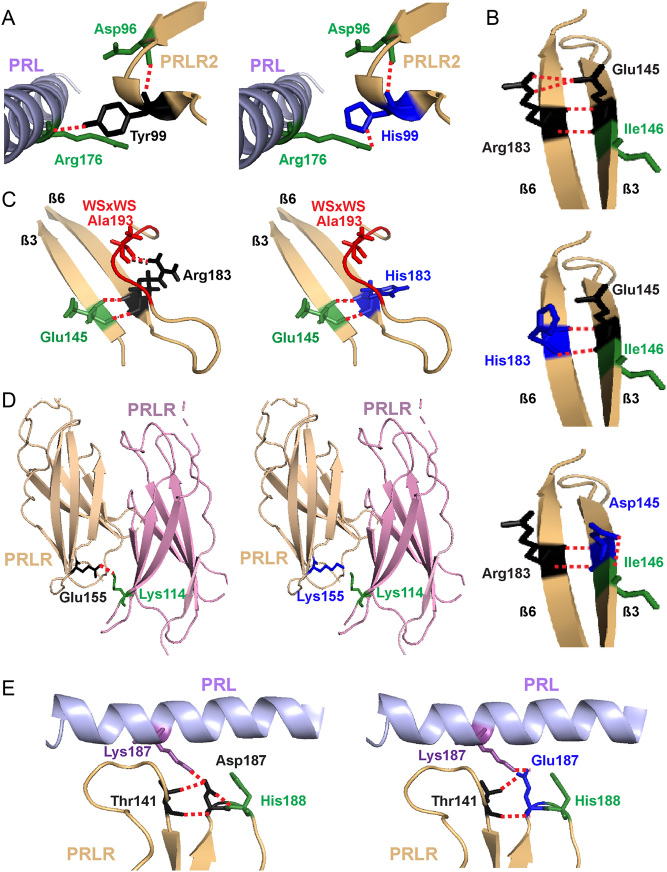
Figure 2.
Structural characterisation of rare variants located in the C-terminal D1 and D2 of PRLR ECD. (A) The Tyr99 residue is located close to the PRL-binding site with the PRLR. The hydroxyl group of the wild-type Tyr99 contacts Arg176 on the PRL protein (left). Mutation to His99 (blue, right) predicts retention of the contact, but at a more distal site, further away from the PRL α-helix and therefore is likely to increase the distance between the PRL and PRLR molecules which may affect binding and activation. (B) The Glu145 and Arg183 PRLR residues are located in adjacent β-strands and form polar contacts with each other. Mutation to His183 (middle) and Asp145 (bottom) is predicted to disrupt two of these contacts. Additionally, Asp145 is predicted to lose contact with the neighbouring Ile146, a residue previously demonstrated to be important for PRLR folding and stability (Dagil et al. 2012, Zhang et al. 2015). (C) The His183 also loses contact with Ala193, which forms part of the highly conserved WSxWS motif (red). (D) The wild-type Glu155 forms a contact with Lys114 on the opposite PRLR protomer. The Lys155 variant loses this contact and may disrupt homodimeric structural stability. (E) The wild-type Asp187 residue lies close to the PRL binding site and forms a contact with the His188 residue of PRLR, which has a critical role in ligand binding (Kulkarni et al. 2010) (Left). Mutation to Glu187 (blue, right) leads to loss of this contact, which may affect activation of the PRLR protein.
Several variants were predicted to affect important structural components in the C-terminal region of D1 and in D2 of the PRLR ECD (Fig. 2). The Tyr99His variant, in D1, is predicted to retain contact with the PRL ligand but at a more distal site (i.e further away from the PRL α-helix) and therefore is likely to increase the distance between the PRL and PRLR molecules, which may affect agonist binding or PRLR activation (Fig. 2A, Table 1). Two D2 variant residues, His183 and Asp145, are predicted to disrupt contacts between the wild-type Arg183 and Glu145 residues. The Glu145 and Arg183 PRLR residues are located in adjacent β-strands and form four direct contacts (Fig. 2B). The variants Asp145 and His183 are predicted to disrupt two of these contacts and may increase the flexibility of the D2 lobe (Fig. 2B). Additionally, the Asp145 variant is predicted to lose contact with the neighbouring Ile146, a residue previously demonstrated to be important for PRLR folding and stability (Dagil et al. 2012, Zhang et al. 2015) (Fig. 2B); while His183 loses contact with Ala193, a residue that forms part of the highly-conserved WSxWS motif (Fig. 2C). Arg183 is one of five highly conserved residues (Ile146, Glu151, Glu155, Tyr178, Arg183) that together with residues of the Trp-Arg ladder undergo conformational changes to switch PRLR to an active state (Dagil et al. 2012). Rare variants were identified to affect two more of these five residues (Glu151Lys, Glu155Lys) (Table 1). Moreover, the variant Lys155 loses contact with Lys114 on the adjacent PRLR protomer, which may affect receptor dimerisation and protein stability (Fig. 2D, Table 1). The Asp187Glu variant lies close to the PRL binding site and is predicted to lose contact with the adjacent His188 residue, which has a critical role in ligand binding (Kulkarni et al. 2010) and is mutated in some individuals with hyperprolactinaemia (Newey et al. 2013) (Fig. 2E, Table 1). Thus, the PRLR D2 domain mutations are predicted to affect PRL binding, flexibility of the PRLR structure, and PRLR activation.
Structural characterisation of the TMD and ICD PRLR variants
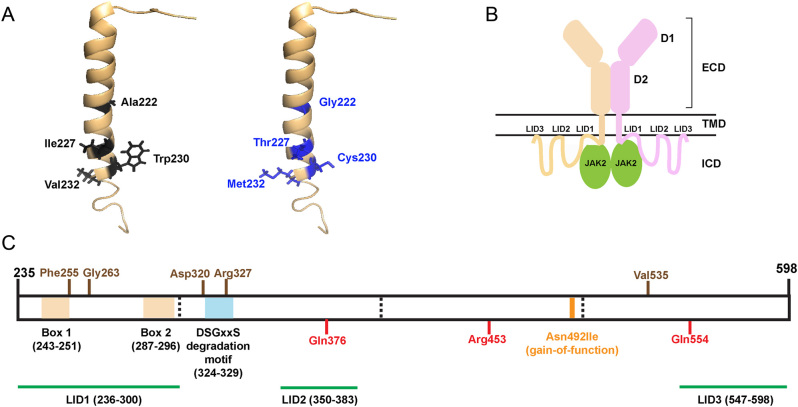
The NMR structure of the single-pass transmembrane domain of the PRLR (PDB:2N7I (Bugge et al. 2016)) was used to study the structural effects of the five highly conserved TMD PRLR variants (Ala222Gly, Ile227Thr, Trp230Cys,Val232Ala, Val232Met) that are predicted to be potentially deleterious/pathogenic (Fig. 3A). Three-dimensional modelling did not predict any changes in interactions between the TMD located PRLR variants. However, the side chains of each amino acid project into the plasma membrane bilayer, and therefore, the variant residues may affect interactions with lipids, which cannot be predicted using the current structural models. In addition to the TMD, plasma membrane interactions also occur with three regions of the ICD (LID1–3) (Bugge et al. 2016) (Fig. 3B). About 45 PRLR rare variants affect residues within these LID regions, 31 in LID1, 3 in LID2, and 11 in LID3 (Table 1). The ICD contains two other structural features, Box 1 and 2, the binding sites for JAK2 (Lebrun et al. 1995) and a conserved degradation motif, DSGxxS (located at residues 324–329) (Plotnikov et al. 2009) (Fig. 3C). Seven rare variants were identified in residues within Box 1, and two variants within Box 2, while three variants within the same residue were identified in the degradation motif.
Figure 3.
Structural characterisation of the PRLR variants located in the TMD and ICD. (A) A single α-helix forms the transmembrane domain (TMD) of the PRLR. Five rare PRLR variants (Ala222Gly, Ile227Thr, Trp230Cys, Val232Ala, Val232Met) are located within the TMD, and four of these are present within the published NMR structure of the PRLR TMD (PDB:2N7I (Bugge et al. 2016)). The four WT residues (black) are located at the cytoplasmic end of the TMD and each forms backbone contacts with adjacent residues within the α-helix. The mutant residues (blue) are not predicted to affect these backbone contacts. (B) Cartoon depicting the PRLR structure with the two monomers shown in brown and pink. The extracellular domain (ECD) contains two domains (D1 and D2) and is connected to the intracellular domain (ICD) via the TMD. The ICD is predicted to interact with the plasma membrane in at least three regions known as lipid-interacting domains (LID1-3). The ICD also interacts with the JAK2 proteins that activate signalling downstream of the PRLR. (C) Cartoon showing the known functional domains of the PRLR ICD, with the amino acid residues (236–300, 243–251, 324–329, 350–383, and 547–598) involved shown in parentheses. The LIDs are shown in green. Two regions, Box 1 and Box 2 (brown-shaded regions), are binding sites for JAK2 (Lebrun et al. 1995), and the DSGxxS region (blue-shaded region) acts as a degradation motif (Plotnikov et al. 2009). The PRLR residues investigated in this study are shown in brown above the cartoon and residues investigated in previous studies are shown in red below the cartoon (Bernard et al. 2016, Gorvin et al. 2018a). The location of the Asn492Ile gain-of-function PRLR variant that is associated with prolactinoma is indicated in orange.
Functional analysis of the PRLR rare variants
Based on their predicted effects on pathogenicity, evolutionary conservation, and structure, we chose to assess ten PRLR rare variants. Five ECD variants (Tyr99His, Glu145Asp, Glu155Lys, Arg183His, Asp187Gly) (Fig. 2A, B, C, D and E) were selected including ones within the PRL-binding region, in the homodimeric interface and close to the WSxWS motif and five ICD variants including, two residues within LID1 located between Box 1 and 2 (Phe255Ser, Gly263Asp); two residues close to or within the degradation motif (Asp320Tyr, Arg327Gln); and one distal rare variant close to LID3 (Val535Met) (Fig. 3C).
Effect of rare variants on PRLR expression
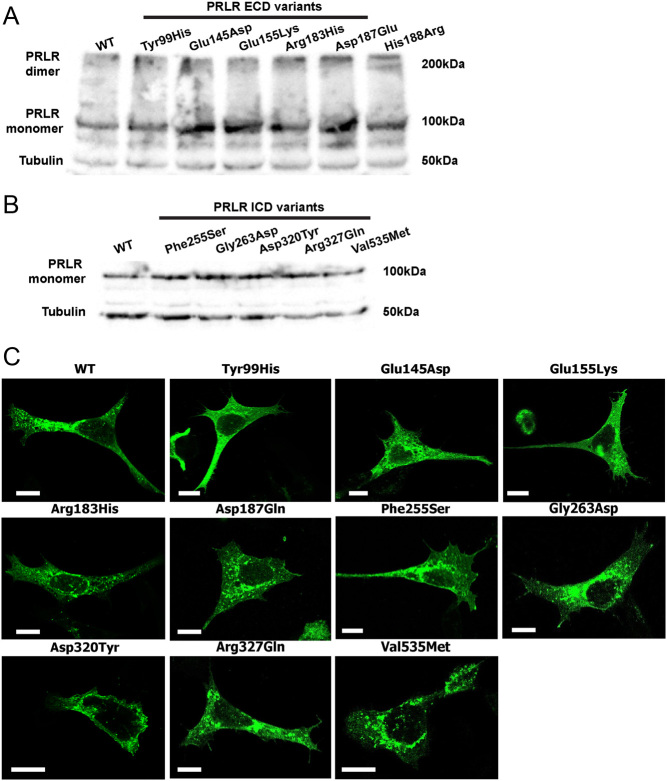
Initially, PRLR protein expression was assessed by Western blot analyses in HEK293 cells transiently transfected with WT or variant PRLR (Fig. 4A and B). The PRLR protein was expressed at equivalent levels in cells transfected with the five ECD and five ICD rare variants and WT PRLR constructs (Fig. 4A and B), indicating it is unlikely that any of the variants affect protein folding. Although structural analysis had predicted that the Glu155Lys variant may affect contacts between the two PRLR protomers, concentrations of PRLR dimers were similar in cells transfected with WT or the ECD variants (Fig. 4A). Therefore, the Glu155Lys variant may not have a major impact on homodimer stability.
Figure 4.
Expression of the PRLR ECD and ICD variants. Western blot analyses of HEK293 cells expressing: (A) PRLR ECD rare variants and (B) PRLR ICD rare variants. Lysates show approximately equal expression levels of PRLR in cells transfected with each rare variant and the WT PRLR. Tubulin was used as a loading control. (C) Confocal microscopy images of the PRLR WT and variant proteins in transfected HEK293 cells. Bar indicates 10 µm.
The cellular expression of each of the PRLR variants was then assessed using confocal microscopy. As previously observed, the WT PRLR protein is located within the cytoplasm and at the plasma membrane (Gorvin et al. 2018b). A similar expression pattern was observed for the ten PRLR rare variants (Fig. 4C). Therefore, it is unlikely that the variant residues affect protein expression or trafficking of the PRLR to the cell surface.
Effect of the PRLR rare variants on STAT5 signalling
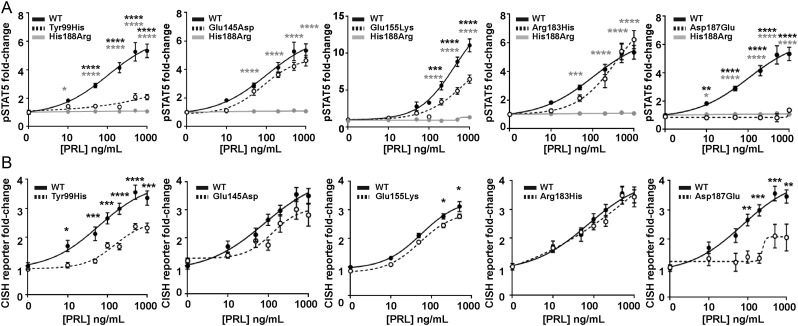
Previous studies have demonstrated PRLR to predominantly signal via the STAT5 pathway (Brooks 2012) and we therefore assessed this pathway by measuring immediate signalling by phospho-STAT5 (pSTAT5) and later downstream effects on transcription by measuring the STAT5 target gene CISH. The effects on PRLR signalling were assessed together with that of the His188Arg mutant PRLR that has been reported to result in a loss of function in association with familial hyperprolactinaemia (Newey et al. 2013, Bernard et al. 2016). We first assessed the ECD variants and demonstrated that increasing concentrations of PRL led to an increase in pSTAT5 and CISH luciferase reporter in a similar dose-dependent manner in cells expressing the PRLR variants Glu145Asp and Arg183His and cells expressing WT PRLR (Fig. 5A and B). However, responses were significantly reduced in cells expressing the PRLR ECD variants Tyr99His, Glu155Lys, and Asp187Glu, when compared to those expressing WT PRLR (Fig. 5A and B, Table 2).
Figure 5.
Functional characterisation of the JAK-STAT signalling pathway by PRLR ECD variants. (A) pSTAT5 responses following prolactin (PRL) treatment in cells expressing wild-type (WT), mutant His188Arg, or ECD variants Tyr99His, Glu145Asp, Glu155Lys, Arg183His, Asp187Glu. PRL-induced pSTAT5 production was abolished in His188Arg- and Asp187Glu-expressing cells and significantly reduced in Tyr99His- and Glu155Lys-expressing cells compared to cells expressing WT. (B) CISH luciferase reporter activity in cells transfected with WT or the five ECD variant PRLRs. CISH reporter activity was significantly reduced in Tyr99His-, Glu155Lys-, and Asp187Glu-expressing cells compared to WT cells. Mean from four to five independent assays for all panels. Statistical analyses show comparisons between WT and the five ECD PRLR variants (black) and WT and the His188Arg mutant (grey) by two-way ANOVA with Dunnett’s or Sidak’s multiple comparisons tests. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05.
Table 2.
Summary of effects of rare variants on PRLR function.
| PRLR variant | Predicted pathogenicity | STAT5 signalling | PI3K/ Akt signalling | Cell viability | Apoptosis | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Prediction programsa | Evolutionary conservationb | Structural predictions | pSTAT5 | CISH | pAkt | FOXO1 | ||||
| WT | 0 | N/A | N/A | ++ | ++ | ++ | ++ | ++ | ++ | |
| His188Arg | 4 | 4 | Affects PRL binding | - | - | - | - | ++ | +++ | |
| ECD |
Tyr99His | 3 | 4 | Affects PRL binding | + | + | ++ | ++ | ++ | ++ |
| ECD | Glu145Asp | 3 | 4 | Loss and gain of contacts | ++ | ++ | ++ | ++ | ++ | ++ |
| ECD | Glu155Lys | 3 | 3 | Disrupts homodimer region | + | + | ++ | + | ++ | ++ |
| ECD | Arg183His | 2 | 4 | Loses contact with WSxWS motif | ++ | ++ | ++ | ++ | ++ | ++ |
| ECD | Asp187Glu | 3 | 4 | Loses contact with His188 | - | + | + | ++ | ++ | ++ |
| ICD |
Phe255Ser | 4 | 4 | LID1, close to JAK2 binding site | - | + | ++ | ++ | +++ | ++ |
| ICD | Gly263Asp | 4 | 4 | LID1 | ++ | ++ | ++ | ++ | ++ | ++ |
| ICD | Asp320Tyr | 2 | 4 | Unknown | ++ | ++ | ++ | ++ | ++ | ++ |
| ICD | Arg327Gln | 1 | 2 | Degradation motif | +++ | +++ | +* | ++ | +++ | ++ |
| ICD | Val535Met | 4 | 4 | Unknown | ++ | ++ | ++ | ++ | ++ | ++ |
aA score out of four based on protein predictions using SIFT, Polyphen-2, MutationTaster, and REVEL is given. If the variant was predicted to be probably damaging or damaging/pathogenic, it was classified as affected; bEvolutionary conservation was based on the PRLR ortholog sequence in four species (cow, dog, mouse, and rat) in comparison to the human protein. The score shows the number of species in which the residue is conserved out of 4; +++Gain of function; ++Normal; +Impaired; -Abolished.;*Arg327Gln has significantly increased basal Akt activity, which likely accounts for the impaired PRL-induced activity.
N/A, not applicable.
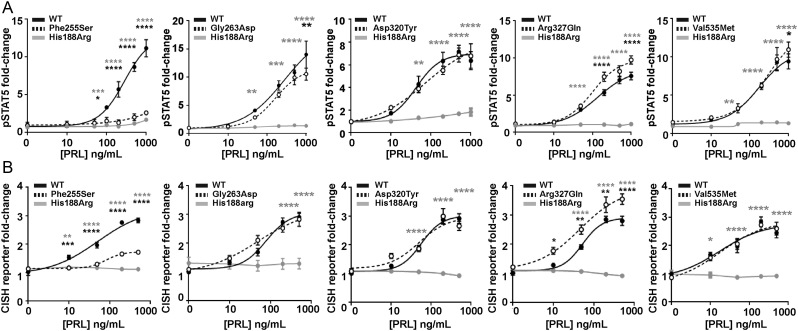
The effects of the five PRLR ICD rare variants were next assessed on STAT5 signalling. Increasing concentrations of PRL led to an increase in pSTAT5 and CISH luciferase reporter in a similar dose-dependent manner in cells expressing the PRLR rare variants Gly263Asp, Asp320Tyr, and Val535Met and cells expressing WT PRLR (Fig. 6A and B). In contrast, the Phe255Ser rare ICD PRLR variant significantly reduced pSTAT5 and CISH reporter responses, while the Arg327Gln variant had significantly elevated pSTAT5 and CISH reporter responses, when compared to WT-expressing cells (Fig. 6A and B, Table 2). There was consistently no response in cells expressing the His188Arg-mutant protein to increasing concentrations of PRL (Fig. 6, Table 2).
Figure 6.
Functional characterisation of the JAK2-STAT5 signalling pathway by PRLR ICD variants. (A) pSTAT5 responses following prolactin (PRL) treatment in cells expressing wild-type (WT), mutant His188Arg, or ICD variants Phe255Ser, Gly263Asp, Asp320Tyr, Arg327Gln, Val535Met. PRL-induced pSTAT5 production was abolished in His188Arg expressing cells, significantly reduced in Phe255Ser and significantly increased in Arg327Gln expressing cells when compared to WT cells. Additionally, pSTAT5 was reduced in Gly263Asp- and Val535Met-expressing cells when treated with high (1000 ng/mL) PRL. (B) CISH luciferase reporter activity in cells transfected with WT, mutant His188Arg, or the five ICD variant PRLRs. CISH reporter activity was abolished in His188Arg-expressing cells, significantly reduced in Phe255Ser and significantly increased in Arg327Gln-expressing cells when compared to WT cells. Mean from four to five independent assays for all panels. Statistical analyses show comparisons between WT and the five ICD PRLR variants (black) and WT and the His188Arg mutant (grey) by two-way ANOVA with Dunnett’s or Tukey’s multiple comparisons tests. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05.
Effect of the PRLR rare variants on Akt signalling
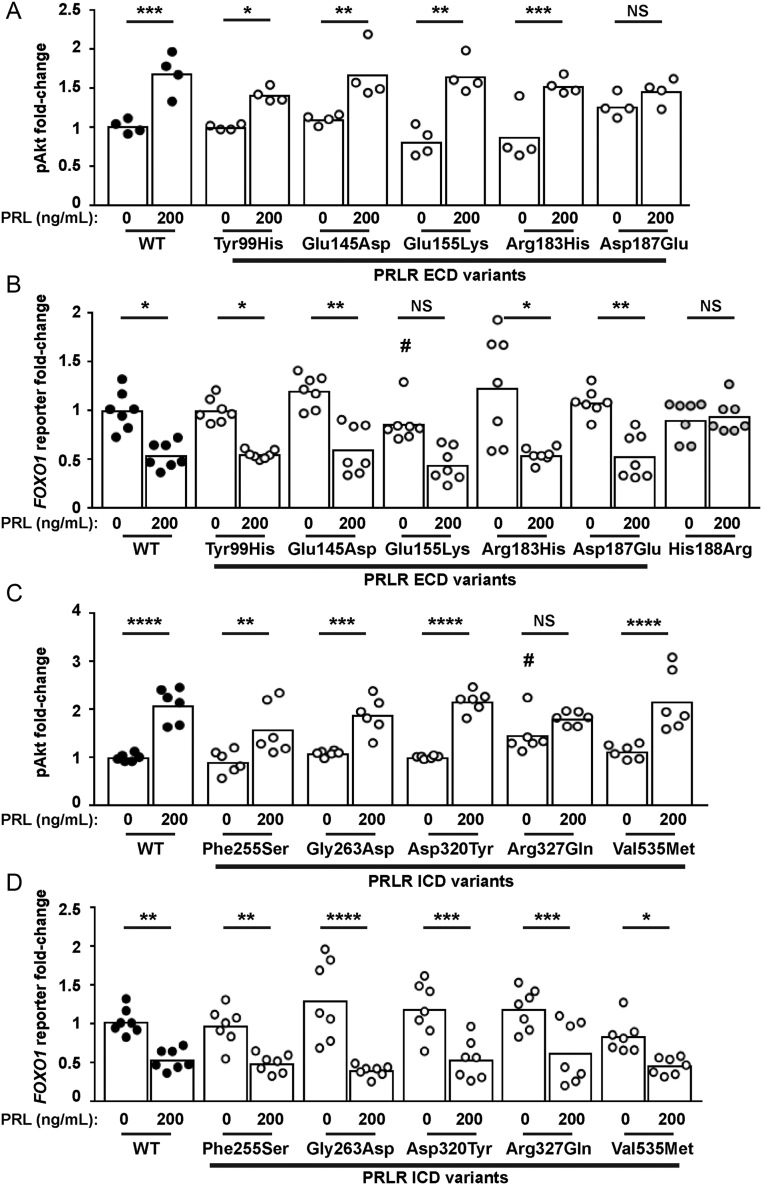
PRLR can also signal by the Akt pathway, and we have previously demonstrated that some PRLR rare variants affect signalling by this pathway (Gorvin et al. 2018b). To assess the effects of the PRLR variants on Akt signalling, we investigated PRL-induced responses of phospho-Akt by AlphaScreen analysis and luciferase reporter activity by the Akt-target gene FOXO1 (Essaghir et al. 2009). Exposure of four of the PRLR ECD variants (Tyr99His, Glu145Asp, Glu155Lys, Arg183His) to 200 ng/mL PRL led to an increase in pAkt activity, which was not significantly different from that observed in WT PRLR expressing cells (Fig. 7A, Table 2). However, cells expressing the Asp187Glu rare variant were unable to induce increases in p-Akt in response to 200 ng/mL PRL (Fig. 7A).
Figure 7.
Functional characterisation of the Akt signalling pathway by PRLR rare variants. (A) pAkt responses following PRL treatment in cells transfected with wild-type (WT) or the five extracellular domain (ECD) variant PRLRs. Asp187Glu had impaired pAkt responses when compared to WT-expressing cells. (B) FOXO1 luciferase reporter activity in cells transfected with WT, mutant His188Arg, or the five ECD variant PRLRs. Addition of 200 ng/mL PRL reduces FOXO1 luciferase activity in WT and four variant cell lines. However, no response was observed in His188Arg cells and Glu155Lys had reduced basal FOXO1 activity and impaired PRL-induced responses. (C) pAkt responses following PRL treatment in cells transfected with WT or the five intracellular domain (ICD) variant PRLRs. Arg327Gln-expressing cells had elevated basal pAkt activity. (D) FOXO1 luciferase activity in cells transfected with WT or the five ICD variant PRLRs. Addition of 200 ng/mL prolactin to WT and variant PRLRs reduces FOXO1 luciferase activity similarly in all cell lines. Data in all panels were expressed relative to WT cells treated with 0 ng/mL PRL. Mean from 4 to 6 independent assays for pAkt assays. Mean (panel D) or median (panel B) from seven independent assays for luciferase reporter assays. Statistical analyses performed by one-way ANOVA with Sidak’s or Dunnett’s multiple comparisons tests for panels A, C, and D. Statistical analysis performed by Kruskal–Wallis test with Dunn’s multiple comparisons tests for panel B. Comparisons show 0 vs 200 nM PRL (asterisks) or between WT and variant (#). ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, NS, not significant.
Akt phosphorylates FOXO proteins, resulting in their exclusion from the nucleus and subsequent degradation. Thus, PRL activation of the PRLR, which induces increases in Akt signalling, will reduce FOXO1 transcription (Essaghir et al. 2009). Assessment of FOXO1 luciferase activity in cells expressing the WT or PRLR ECD variants showed that all cells could reduce FOXO1 luciferase reporter activity in response to 200 ng/mL PRL (Fig. 7B). However, cells expressing the Glu155Lys variant had lower basal expression of FOXO1 luciferase activity (Fig. 7B). In contrast, the His188Arg mutant, which has previously been shown to reduce pAkt activity (Gorvin et al. 2018b), was unable to reduce FOXO1 luciferase reporter activity following exposure to 200 ng/mL PRL (Fig. 7B), consistent with impaired pAkt activity.
Assessment of the ICD PRLR rare variants showed that four variants (Phe255Ser, Gly263Asp, Asp320Tyr, Val535Met) had similar PRL-induced pAkt responses to WT PRLR-expressing cells (Fig. 7C, Table 2). However, the Arg327Gln variant did not increase pAkt in response to 200 ng/mL PRL. This may have been a consequence of constitutively high basal pAkt concentrations in Arg327Gln-expressing cells (Fig. 7C, Table 2). None of the five ICD variants had a significant effect on PRL-induced FOXO1 responses compared to the wild-type PRLR (Fig. 7D).
Effect of the PRLR rare variants on cell viability and apoptosis
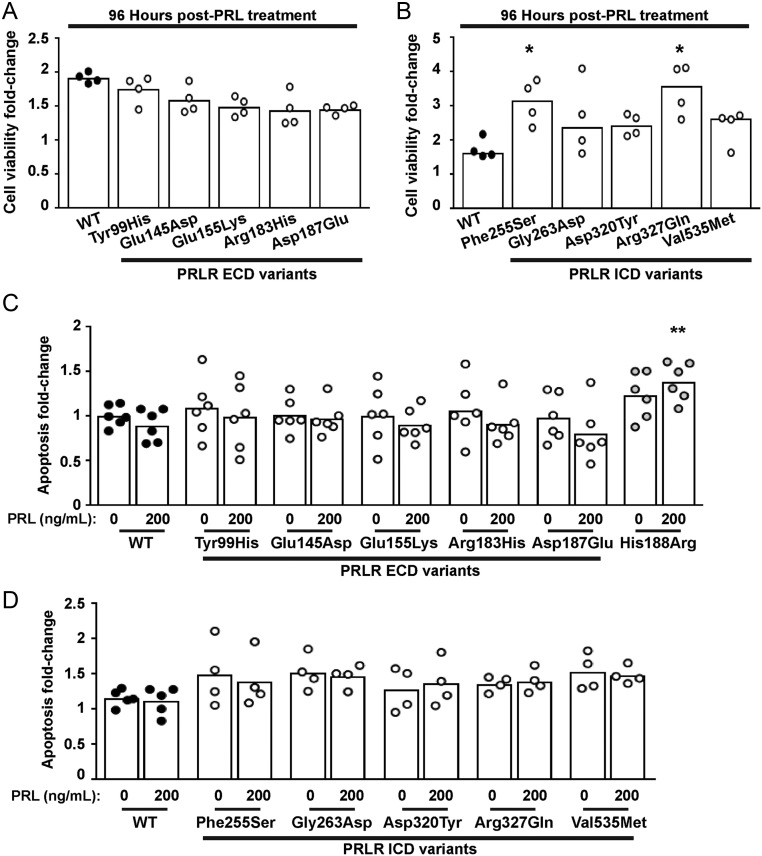
Both the STAT5 and Akt signalling pathways lead to transcription of target genes that regulate proliferation and cell survival (Fresno Vara et al. 2000, Amaral et al. 2004, Brooks 2012), and previous studies have demonstrated that the Asn492Ile PRLR variant increases proliferation, while the His188Arg mutation increases apoptosis (Gorvin et al. 2018b). We therefore assessed the effect of the ten PRLR rare variants on cell viability using the CellTiter Blue assay and on apoptosis using a Caspase-Glo-3/7 assay (Gorvin et al. 2018b). This demonstrated that all five ECD variants (Tyr99His, Glu145Asp, Glu155Lys, Arg183His, Asp187Glu) and three ICD variants (Gly263Asp, Asp320Tyr Val535Met) had a similar effect on cell viability when compared to cells expressing WT PRLR, following exposure to 200 ng/mL PRL for 96 h (Fig. 8A and B, Table 2). The ICD Phe255Ser and Arg327Gln PRLR variants were associated with significantly increased numbers of viable cells after 96 h of PRL treatment. Assessment of apoptosis was performed in WT and variant PRLR expressing cells after 96 h of exposure to 200 ng/mL PRL. The ECD His188Arg loss-of-function mutation increased apoptosis in cells treated with 200 ng/mL PRL (Fig. 8C), consistent with our previous report (Gorvin et al. 2018b). However, none of the other ECD (Fig. 8C) or ICD (Fig. 8D) PRLR variants had a significant effect on apoptosis. Therefore, both loss-of-function and gain-of-function mutations in the PRLR increased the number of viable cells, although none of these investigated rare variants affected apoptosis.
Figure 8.
Effect of the PRLR rare variants on cell viability and apoptosis. (A-B) Effect of PRL (200 ng/mL) on viability in cells expressing WT, or (A) the extracellular domain (ECD) or (B) the intracellular domain (ICD) variant PRLRs. Cell viability was increased in cells expressing the Phe255Ser and Arg327Gln variant PRLRs at 96 h post-treatment with PRL, when compared to WT cells. (C-D) Effect of PRL (200 ng/mL) on apoptosis in cells expressing WT, mutant His188Arg, or (C) the ECD or (D) the ICD variant PRLRs. Each point shows one biological replicate (derived from the mean of four technical replicates) performed on independent occasions. Statistical analyses performed by one-way ANOVA with Dunnett’s multiple comparisons tests for panels A, C, and D. Statistical analysis performed by Kruskal–Wallis test with Dunn’s multiple comparisons tests for panel B. Comparisons show WT vs variant (asterisks). ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05.
Discussion
Evaluating the clinical significance of rare coding variants within genes associated with Mendelian disorders and complex traits represents a significant challenge, and consequently, a range of in silico methods have been developed to facilitate the identification of potentially deleterious variants resulting in altered protein function. Our analysis of PRLR variants demonstrated that in silico tools could not accurately predict those that affected PRLR function (Table 2). This is consistent with previous studies that have shown that algorithms are only 65–80% accurate in predicting known disease variants (Thusberg et al. 2011) as pathogenic and often, over-predict missense changes as deleterious, while they are unreliable in predicting variants with milder effects (Choi et al. 2012). As such, the American College of Medical Genetics and Genomics (ACMG) recommends that protein prediction software is not used as the sole source of information to make clinical decisions (Richards et al. 2015). For the PRLR, only one variant, Phe255Ser, was correctly predicted deleterious in all four in silico methods (Table 1 and 2); while the gain-of-function Arg327Gln variant was predicted benign in three of four tools examined. This is consistent with previous studies of missense variants in the ABCC8, GCK, and KCNJ11 genes that showed SIFT and Polyphen to be better at predicting inactivating than gain-of-function mutations (Flanagan et al. 2010). Furthermore, most in silico tools use the evolutionary conservation of the affected residue as a parameter to predict deleteriousness/pathogenicity (Richards et al. 2015). Such reliance on evolutionary conservation may be poorly predictive for PRLR variants, as the receptor has a specific role in lactation, and thus a lack of conservation with non-lactating species may be unimportant and could account for the poor predictive capability of in silico tools for PRLR.
Our functional studies identified five of ten PRLR germline variants that were associated with altered signalling, and these variants were all located in regions of the PRLR that have known receptor functions (Table 2). Thus, the three ECD variants that reduced PRLR function were predicted to affect ligand binding, receptor activation, and homodimerisation, while the ICD variants are located close to the JAK2-binding site and a known degradation motif (Fig. 1, 2 and 3). Examination of the ExAc/GnomAD databases identified significantly fewer ECD variants than predicted to occur in a region of this size and that there are more singleton variants (i.e. those identified in a single individual), which have previously been reported to have a higher probability of being functionally damaging and typically have occurred recently in evolutionary terms (Tennessen et al. 2012). These findings indicate that the ECD may be less tolerant of genetic variation, due to its critical roles in ligand binding and receptor activation, and that identification of variants in known functional domains is a reasonable predictor of possible pathogenicity. It is of note that several PRLR variants from other regions had similar responses to WT PRLR or were associated with, at most, modest effects on receptor function that may only be identified at supra-physiological concentrations of PRL (e.g. pSTAT5 responses for Gly263Asp and Val535Met).
The Glu155Lys variant is predicted to disrupt a contact formed across the homodimer interface and was associated with a partial loss of function for both STAT5 and pAkt signalling pathways (Fig. 2, 5 and 7, Table 2). However, analysis of protein expression in Glu155Lys-expressing cells showed no discernible difference in PRLR dimer or monomer concentrations (Fig. 4), indicating that loss of this homodimeric interaction is not sufficient to impair dimer formation. However, the partial loss of function associated with this Glu155Lys variant indicates that this residue may have an important role in facilitating conformational changes across the dimer interface that are necessary for receptor activation. The Glu155 residue lies in proximity to the WSXWS motif, a highly conserved motif of cytokine receptors, that holds the receptor in an ‘off-state’, until ligand interaction occurs, inducing formation of a Trp-Arg ladder to activate the receptor (Dagil et al. 2012). The Glu155Lys variant may disrupt these conformational changes, with a consequent reduction in signalling as observed in functional studies (Table 2). The elucidation of the full-length PRLR structure in the active and inactive states could help resolve whether the Glu155Lys variant has such functional effects.
Within the ICD, the Phe255 residue is located in the region between the two JAK2-binding sites, within a series of residues that are unnecessary for JAK2 phosphorylation but critical for downstream transcription of the beta-casein reporter gene (Lebrun et al. 1995, Ali & Ali 1998). We therefore hypothesise that Phe255 is involved in interaction with downstream signalling partners of PRLR that are necessary for pSTAT5 signalling but not for pAkt signalling, which was unaffected by the variant (Fig. 6 and 7, Table 2). Interestingly, the Phe255Ser variant was associated with increased numbers of viable cells, which we have previously observed for the gain-of-function Asn492Ile ICD variant (Gorvin et al. 2018b), and showed for the activating Arg327Gln variant in this study (Fig. 8, Table 2). Thus, both inactivating and activating ICD PRLR variants may be associated with increased cell survival and proliferation. It is likely that increased cell survival by the three receptor variants involves different mechanisms. The Asn492Ile variant had no effect on PRL-induced pSTAT5 signalling but increased Akt signalling, which could be rectified by treatment with a PI3K inhibitor (Gorvin et al. 2018b). Thus, it is likely this variant increases cell survival by activating the Akt-PI3K signalling pathway. The Arg327Gln variant does not increase PRL-induced Akt signalling but has constitutive Akt activity (Fig. 7, Table 2), which may contribute to increased numbers of viable cells. Additionally, the Arg327Gln variant significantly increases STAT5 signalling, which has previously been shown to promote mammary cell proliferation (Iavnilovitch et al. 2002). Thus, proliferation may be increased by both enhanced PRL-induced STAT5 signalling and constitutive Akt signalling. Furthermore, the Arg327 residue lies within the PRLR degradation motif (Plotnikov et al. 2009) and the Arg327Gln variant may impair receptor degradation. However, Western blot analyses did not show enhanced protein expression, indicating that Arg327Gln is unlikely to affect protein turnover. The mechanism by which the Phe255Ser variant increases cell viability is unknown. It is possible that this residue enhances binding of negative regulators of PRLR signalling, such as the suppressors of cytokine signalling (SOCS) proteins (Tomic et al. 1999), or may activate signalling pathways that are yet to be identified. Examination of proteins that regulate the cell cycle, including expression of cyclin D1 and transcription factors such as c-Myc, which are controlled by prolactin-mediated JAK-STAT and Akt signalling (Brockman et al. 2002, Acosta et al. 2003), could provide more insights into the different effects of PRLR variants on cell survival. The use of pathway-specific inhibitors, such as the Akt1/2 inhibitor previously used to examine the prolactinoma-associated Asn492Ile PRLR variant, may be required to further elucidate these mechanisms. Moreover, studies of additional PRLR ICD variants within the LID1 region may identify other residues with similar effects on signalling and proliferation.
Although these studies identified several inactivating PRLR variants, some activity was retained by all the variants, in contrast to the hyperprolactinaemia-associated His188Arg variant (Newey et al. 2013), which abolishes signalling. This is in keeping with the observation that the His188 residue occurs within the high-affinity ligand-binding interface and has a functional role in ligand binding and receptor activation (Kulkarni et al. 2010). The retention of some signalling activity may also explain why only the His188Arg variant is associated with enhanced apoptosis (Gorvin et al. 2018b). It is unclear whether the partial loss-of-activity associated with the PRLR variants examined in this study would affect PRLR physiological activities, as all the variants have been identified in the heterozygous state. Previous in vitro studies of the Pro269Leu PRLR variant identified in a compound heterozygote individual with hyperprolactinaemia showed that the variant impaired STAT5 phosphorylation but had no effect on STAT5 signalling when expressed with WT PRLR (Kobayashi et al. 2018). Thus, it is possible that the ten variants characterised in this study may have minimal effect on PRLR function in heterozygous individuals, unless expressed with other PRLR variants. Further investigation of the inactivating and activating PRLR variants in large well-characterised populations is required to determine their physiological consequences.
This study and our previous analysis of PRLR variants associated with prolactinoma demonstrated that both JAK-STAT and Akt signalling can be impaired by genetic variants of the receptor (Gorvin et al. 2018a). The PRLR has also been described to activate other signalling pathways including Ras–Raf-mediated mitogen-activated protein kinase (MAPK) signalling (Bole-Feysot et al. 1998) and can activate Src family kinases independently of JAK2 phosphorylation (Fresno Vara et al. 2000) to increase focal-adhesion kinase/MAPK and PI3K-Akt signalling, upregulate c-Myc and cyclin d1 mRNA expression, enhance cell proliferation, and accelerate receptor internalisation (Acosta et al. 2003, Piazza et al. 2009). It is possible that the PRLR variants studied in this manuscript may also affect these other signalling pathways and the observed effects on Akt could be mediated by JAK2-independent Src signalling. Src family kinases and MAPK signalling proteins are expressed in HEK293 cells (Della Rocca et al. 1997), and future studies of PRLR variants could expand the screening pipeline to include examination of these proteins.
In summary, these studies give further insight into PRLR structure–function and highlight that rare PRLR variants are associated with alterations in receptor signalling. Future studies of rare coding variants will require a combination of molecular, in vivo, and epidemiological approaches to appropriately classify the significance of such variants.
Declaration of interest
The authors declare that there is no conflict of interest.
Funding
This work was supported by the United Kingdom Medical Research Council (MRC) programme grant G1000467/2010 (RVT); Chief Scientist Office, UK Fellowship SCAF/15/01 (PJN); start-up funds from the University of Birmingham and COMPARE (CMG); a Wellcome Trust Senior Investigator Award (RVT); National Institute for Health Research (NIHR) Oxford Biomedical Research Centre Programme (RVT); and NIHR Senior Investigator Award (RVT).
References
- Acosta JJ, Munoz RM, Gonzalez L, Subtil-Rodriguez A, Dominguez-Caceres MA, Garcia-Martinez JM, Calcabrini A, Lazaro-Trueba I, Martin-Perez J.2003Src mediates prolactin-dependent proliferation of T47D and MCF7 cells via the activation of focal adhesion kinase/ERK1/2 and phosphatidylinositol 3-kinase pathways. Molecular Endocrinology 172268–2282. ( 10.1210/me.2002-0422) [DOI] [PubMed] [Google Scholar]
- Adzhubei I, Jordan DM, Sunyaev SR.2013Predicting functional effect of human missense mutations using PolyPhen-2. Current Protocols in Human Genetics 761–7. ( 10.1002/0471142905.hg0720s76) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR.2010A method and server for predicting damaging missense mutations. Nature Methods 7248–249. ( 10.1038/nmeth0410-248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwala V, Flannick J, Sunyaev SGoT2D Consortium & Altshuler D.2013Evaluating empirical bounds on complex disease genetic architecture. Nature Genetics 451418–1427. ( 10.1038/ng.2804) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, Ali S.1998Prolactin receptor regulates Stat5 tyrosine phosphorylation and nuclear translocation by two separate pathways. Journal of Biological Chemistry 2737709–7716. ( 10.1074/jbc.273.13.7709) [DOI] [PubMed] [Google Scholar]
- Amaral ME, Cunha DA, Anhe GF, Ueno M, Carneiro EM, Velloso LA, Bordin S, Boschero AC.2004Participation of prolactin receptors and phosphatidylinositol 3-kinase and MAP kinase pathways in the increase in pancreatic islet mass and sensitivity to glucose during pregnancy. Journal of Endocrinology 183469–476. ( 10.1677/joe.1.05547) [DOI] [PubMed] [Google Scholar]
- Banerjee RR, Cyphert HA, Walker EM, Chakravarthy H, Peiris H, Gu X, Liu Y, Conrad E, Goodrich L, Stein RWet al. 2016Gestational diabetes mellitus from inactivation of prolactin receptor and MafB in islet beta-cells. Diabetes 652331–2341. ( 10.2337/db15-1527) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Jonathan N, LaPensee CR, LaPensee EW.2008What can we learn from rodents about prolactin in humans? Endocrine Reviews 291–41. ( 10.1210/er.2007-0017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard V, Bouilly J, Beau I, Broutin I, Chanson P, Young J, Binart N.2016Germline prolactin receptor mutation is not a major cause of sporadic prolactinoma in humans. Neuroendocrinology 103738–745. ( 10.1159/000442981) [DOI] [PubMed] [Google Scholar]
- Binder C, Lafayette A, Archibeque I, Sun Y, Plewa C, Sinclair A, Emkey R.2008Optimization and utilization of the SureFire phospho-STAT5 assay for a cell-based screening campaign. Assay and Drug Development Technologies 627–37. ( 10.1089/adt.2007.111) [DOI] [PubMed] [Google Scholar]
- Bogorad RL, Courtillot C, Mestayer C, Bernichtein S, Harutyunyan L, Jomain JB, Bachelot A, Kuttenn F, Kelly PA, Goffin Vet al. 2008Identification of a gain-of-function mutation of the prolactin receptor in women with benign breast tumors. PNAS 10514533–14538. ( 10.1073/pnas.0800685105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bole-Feysot C, Goffin V, Edery M, Binart N, Kelly PA.1998Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocrine Reviews 19225–268. ( 10.1210/edrv.19.3.0334) [DOI] [PubMed] [Google Scholar]
- Brockman JL, Schroeder MD, Schuler LA.2002PRL activates the cyclin D1 promoter via the Jak2/Stat pathway. Molecular Endocrinology 16774–784. ( 10.1210/mend.16.4.0817) [DOI] [PubMed] [Google Scholar]
- Brooks AJ, Dai W, O'Mara ML, Abankwa D, Chhabra Y, Pelekanos RA, Gardon O, Tunny KA, Blucher KM, Morton CJet al. 2014Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science 344 1249783. ( 10.1126/science.1249783) [DOI] [PubMed] [Google Scholar]
- Brooks CL.2012Molecular mechanisms of prolactin and its receptor. Endocrine Reviews 33504–525. ( 10.1210/er.2011-1040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broutin I, Jomain JB, Tallet E, van Agthoven J, Raynal B, Hoos S, Kragelund BB, Kelly PA, Ducruix A, England Pet al. 2010Crystal structure of an affinity-matured prolactin complexed to its dimerized receptor reveals the topology of hormone binding site 2. Journal of Biological Chemistry 2858422–8433. ( 10.1074/jbc.M109.089128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RJ, Adams JJ, Pelekanos RA, Wan Y, McKinstry WJ, Palethorpe K, Seeber RM, Monks TA, Eidne KA, Parker MWet al. 2005Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nature Structural and Molecular Biology 12814–821. ( 10.1038/nsmb977) [DOI] [PubMed] [Google Scholar]
- Bugge K, Papaleo E, Haxholm GW, Hopper JT, Robinson CV, Olsen JG, Lindorff-Larsen K, Kragelund BB.2016A combined computational and structural model of the full-length human prolactin receptor. Nature Communications 7 11578. ( 10.1038/ncomms11578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP.2012Predicting the functional effect of amino acid substitutions and indels. PLoS One 7 e46688. ( 10.1371/journal.pone.0046688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark WT, Yu GK, Aoyagi-Scharber M, LeBowitz JH.2018Utilizing ExAC to assess the hidden contribution of variants of unknown significance to SanFilippo Type B incidence. PLoS One 13 e0200008. ( 10.1371/journal.pone.0200008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtillot C, Chakhtoura Z, Bogorad R, Genestie C, Bernichtein S, Badachi Y, Janaud G, Akakpo JP, Bachelot A, Kuttenn Fet al. 2010Characterization of two constitutively active prolactin receptor variants in a cohort of 95 women with multiple breast fibroadenomas. Journal of Clinical Endocrinology and Metabolism 95271–279. ( 10.1210/jc.2009-1494) [DOI] [PubMed] [Google Scholar]
- Dagil R, Knudsen MJ, Olsen JG, O'Shea C, Franzmann M, Goffin V, Teilum K, Breinholt J, Kragelund BB.2012The WSXWS motif in cytokine receptors is a molecular switch involved in receptor activation: insight from structures of the prolactin receptor. Structure 20270–282. ( 10.1016/j.str.2011.12.010) [DOI] [PubMed] [Google Scholar]
- Della Rocca GJ, van Biesen T, Daaka Y, Luttrell DK, Luttrell LM, Lefkowitz RJ.1997Ras-dependent mitogen-activated protein kinase activation by G protein-coupled receptors. Convergence of Gi- and Gq-mediated pathways on calcium/calmodulin, Pyk2, and Src kinase. Journal of Biological Chemistry 27219125–19132. ( 10.1074/jbc.272.31.19125) [DOI] [PubMed] [Google Scholar]
- Essaghir A, Dif N, Marbehant CY, Coffer PJ, Demoulin JB.2009The transcription of FOXO genes is stimulated by FOXO3 and repressed by growth factors. Journal of Biological Chemistry 28410334–10342. ( 10.1074/jbc.M808848200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan SE, Patch AM, Ellard S.2010Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genetic Testing and Molecular Biomarkers 14533–537. ( 10.1089/gtmb.2010.0036) [DOI] [PubMed] [Google Scholar]
- Freemark M, Avril I, Fleenor D, Driscoll P, Petro A, Opara E, Kendall W, Oden J, Bridges S, Binart Net al. 2002Targeted deletion of the PRL receptor: effects on islet development, insulin production, and glucose tolerance. Endocrinology 1431378–1385. ( 10.1210/endo.143.4.8722) [DOI] [PubMed] [Google Scholar]
- Fresno Vara JA, Carretero MV, Geronimo H, Ballmer-Hofer K, Martin-Perez J.2000Stimulation of c-Src by prolactin is independent of Jak2. Biochemical Journal 34517–24. ( 10.1042/0264-6021:3450017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadd SL, Clevenger CV.2006Ligand-independent dimerization of the human prolactin receptor isoforms: functional implications. Molecular Endocrinology 202734–2746. ( 10.1210/me.2006-0114) [DOI] [PubMed] [Google Scholar]
- Glasow A, Horn LC, Taymans SE, Stratakis CA, Kelly PA, Kohler U, Gillespie J, Vonderhaar BK, Bornstein SR.2001Mutational analysis of the PRL receptor gene in human breast tumors with differential PRL receptor protein expression. Journal of Clinical Endocrinology and Metabolism 863826–3832. ( 10.1210/jcem.86.8.7753) [DOI] [PubMed] [Google Scholar]
- Gorvin CM, Metpally R, Stokes VJ, Hannan FM, Krishnamurthy SB, Overton JD, Reid JG, Breitwieser GE, Thakker RV.2018aLarge-scale exome datasets reveal a new class of adaptor-related protein complex 2 sigma subunit (AP2sigma) mutations, located at the interface with the AP2 alpha subunit, that impair calcium-sensing receptor signalling. Human Molecular Genetics 27901–911. ( 10.1093/hmg/ddy010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorvin CM, Newey PJ, Rogers A, Stokes V, Neville MJ, Ntali G, Lees P, Morrison PJ, Singhellakis PN, Malandrinou FCet al. 2018bAssociation of prolactin receptor (PRLR) variants with prolactinomas. Human Molecular Genetics 281023–1037. ( 10.1093/hmg/ddy396) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxholm GW, Nikolajsen LF, Olsen JG, Fredsted J, Larsen FH, Goffin V, Pedersen SF, Brooks AJ, Waters MJ, Kragelund BB.2015Intrinsically disordered cytoplasmic domains of two cytokine receptors mediate conserved interactions with membranes. Biochemical Journal 468495–506. ( 10.1042/BJ20141243) [DOI] [PubMed] [Google Scholar]
- Huang C, Snider F, Cross JC.2009Prolactin receptor is required for normal glucose homeostasis and modulation of beta-cell mass during pregnancy. Endocrinology 1501618–1626. ( 10.1210/en.2008-1003) [DOI] [PubMed] [Google Scholar]
- Iavnilovitch E, Groner B, Barash I.2002Overexpression and forced activation of stat5 in mammary gland of transgenic mice promotes cellular proliferation, enhances differentiation, and delays postlactational apoptosis. Molecular Cancer Research 132–47. (available at: https://aacrjournals.org/mcr/article/1/1/32/232305/Overexpression-and-Forced-Activation-of-Stat5-in) [PubMed] [Google Scholar]
- Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, Musolf A, Li Q, Holzinger E, Karyadi Det al. 2016REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. American Journal of Human Genetics 99877–885. ( 10.1016/j.ajhg.2016.08.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DPet al. 2020The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581434–443. ( 10.1038/s41586-020-2308-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Usui H, Tanaka H, Shozu M.2018Variant prolactin receptor in agalactia and hyperprolactinemia. New England Journal of Medicine 3792230–2236. ( 10.1056/NEJMoa1805171) [DOI] [PubMed] [Google Scholar]
- Kulkarni MV, Tettamanzi MC, Murphy JW, Keeler C, Myszka DG, Chayen NE, Lolis EJ, Hodsdon ME.2010Two independent histidines, one in human prolactin and one in its receptor, are critical for pH-dependent receptor recognition and activation. Journal of Biological Chemistry 28538524–38533. ( 10.1074/jbc.M110.172072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC.2009Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols 41073–1081. ( 10.1038/nprot.2009.86) [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez Ret al. 2007Clustal W and Clustal X version 2.0. Bioinformatics 232947–2948. ( 10.1093/bioinformatics/btm404) [DOI] [PubMed] [Google Scholar]
- Lebrun JJ, Ali S, Goffin V, Ullrich A, Kelly PA.1995A single phosphotyrosine residue of the prolactin receptor is responsible for activation of gene transcription. PNAS 924031–4035. ( 10.1073/pnas.92.9.4031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SA, Haiman CA, Burtt NP, Pooler LC, Cheng I, Kolonel LN, Pike MC, Altshuler D, Hirschhorn JN, Henderson BEet al. 2007A comprehensive analysis of common genetic variation in prolactin (PRL) and PRL receptor (PRLR) genes in relation to plasma prolactin levels and breast cancer risk: the multiethnic cohort. BMC Medical Genetics 8 72. ( 10.1186/1471-2350-8-72) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newey PJ, Gorvin CM, Cleland SJ, Willberg CB, Bridge M, Azharuddin M, Drummond RS, van der Merwe PA, Klenerman P, Bountra Cet al. 2013Mutant prolactin receptor and familial hyperprolactinemia. New England Journal of Medicine 3692012–2020. ( 10.1056/NEJMoa1307557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyante SJ, Faupel-Badger JM, Sherman ME, Pfeiffer RM, Gaudet MM, Falk RT, Andaya AA, Lissowska J, Brinton LA, Peplonska Bet al. 2011Genetic variation in PRL and PRLR, and relationships with serum prolactin levels and breast cancer risk: results from a population-based case-control study in Poland. Breast Cancer Research 13 R42. ( 10.1186/bcr2864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza TM, Lu JC, Carver KC, Schuler LA.2009SRC family kinases accelerate prolactin receptor internalization, modulating trafficking and signaling in breast cancer cells. Molecular Endocrinology 23202–212. ( 10.1210/me.2008-0341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov A, Varghese B, Tran TH, Liu C, Rui H, Fuchs SY.2009Impaired turnover of prolactin receptor contributes to transformation of human breast cells. Cancer Research 693165–3172. ( 10.1158/0008-5472.CAN-08-4033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qazi AM, Tsai-Morris CH, Dufau ML.2006Ligand-independent homo- and heterodimerization of human prolactin receptor variants: inhibitory action of the short forms by heterodimerization. Molecular Endocrinology 201912–1923. ( 10.1210/me.2005-0291) [DOI] [PubMed] [Google Scholar]
- Rao GV, Brooks CL.2011Functional epitopes for site 1 of human prolactin. Biochemistry 501347–1358. ( 10.1021/bi101838s) [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector Eet al. 2015Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine 17405–424. ( 10.1038/gim.2015.30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuff KG, Hentges ST, Kelly MA, Binart N, Kelly PA, Iuvone PM, Asa SL, Low MJ.2002Lack of prolactin receptor signaling in mice results in lactotroph proliferation and prolactinomas by dopamine-dependent and -independent mechanisms. Journal of Clinical Investigation 110973–981. ( 10.1172/JCI15912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D.2014MutationTaster2: mutation prediction for the deep-sequencing age. Nature Methods 11361–362. ( 10.1038/nmeth.2890) [DOI] [PubMed] [Google Scholar]
- Smiley KO, Brown RSE, Grattan DR.2022Prolactin action is necessary for parental behavior in male mice. Journal of Neuroscience 428308–8327. ( 10.1523/JNEUROSCI.0558-22.2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson LA, Bondensgaard K, Norskov-Lauritsen L, Christensen L, Becker P, Andersen MD, Maltesen MJ, Rand KD, Breinholt J.2008Crystal structure of a prolactin receptor antagonist bound to the extracellular domain of the prolactin receptor. Journal of Biological Chemistry 28319085–19094. ( 10.1074/jbc.M801202200) [DOI] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun Get al. 2012Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 33764–69. ( 10.1126/science.1219240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thusberg J, Olatubosun A, Vihinen M.2011Performance of mutation pathogenicity prediction methods on missense variants. Human Mutation 32358–368. ( 10.1002/humu.21445) [DOI] [PubMed] [Google Scholar]
- Tomic S, Chughtai N, Ali S.1999SOCS-1, -2, -3: selective targets and functions downstream of the prolactin receptor. Molecular and Cellular Endocrinology 15845–54. ( 10.1016/s0303-7207(9900180-x) [DOI] [PubMed] [Google Scholar]
- Vaclavicek A, Hemminki K, Bartram CR, Wagner K, Wappenschmidt B, Meindl A, Schmutzler RK, Klaes R, Untch M, Burwinkel Bet al. 2006Association of prolactin and its receptor gene regions with familial breast cancer. Journal of Clinical Endocrinology and Metabolism 911513–1519. ( 10.1210/jc.2005-1899) [DOI] [PubMed] [Google Scholar]
- van Agthoven J, Zhang C, Tallet E, Raynal B, Hoos S, Baron B, England P, Goffin V, Broutin I.2010Structural characterization of the stem-stem dimerization interface between prolactin receptor chains complexed with the natural hormone. Journal of Molecular Biology 404112–126. ( 10.1016/j.jmb.2010.09.036) [DOI] [PubMed] [Google Scholar]
- Viengchareun S, Bouzinba-Segard H, Laigneau JP, Zennaro MC, Kelly PA, Bado A, Lombes M, Binart N.2004Prolactin potentiates insulin-stimulated leptin expression and release from differentiated brown adipocytes. Journal of Molecular Endocrinology 33679–691. ( 10.1677/jme.1.01563) [DOI] [PubMed] [Google Scholar]
- Viengchareun S, Servel N, Feve B, Freemark M, Lombes M, Binart N.2008Prolactin receptor signaling is essential for perinatal brown adipocyte function: a role for insulin-like growth factor-2. PLoS One 3 e1535. ( 10.1371/journal.pone.0001535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Cherifi I, Nygaard M, Haxholm GW, Bogorad RL, Bernadet M, England P, Broutin I, Kragelund BB, Guidotti JEet al. 2015Residue 146 regulates prolactin receptor folding, basal activity and ligand-responsiveness: potential implications in breast tumorigenesis. Molecular and Cellular Endocrinology 401173–188. ( 10.1016/j.mce.2014.12.006) [DOI] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a