Abstract
Purpose
With the development of PARP inhibitors for treatment of cancer patients with an altered BRCA1 or BRCA2 gene, there is an urgent need to ensure that there are appropriate strategies for identifying mutation carriers whilst balancing the increased demand for and cost of cancer genetics services. To date, the majority of mutation prediction tools have been developed in women of European descent where the age and cancer-subtype distributions are different from that in Asian women.
Methods
In this study, we built a new model (ARiCa: Asian Risk Calculator) for estimating the likelihood of carrying a pathogenic variant in BRCA1 or BRCA2 gene, using germline BRCA genetic testing results in a cross-sectional population-based study of 8,162 Asian breast cancer patients. We compared the model performance to existing mutation prediction models. The models were evaluated for discrimination and calibration.
Results
ARiCa included age of diagnosis, ethnicity, bilateral breast cancer, tumour biomarkers, and family history of breast cancer or ovarian cancer as predictors. The inclusion of tumour grade improved significantly the model performance. The full model was calibrated (Hosmer-Lemeshow p-value=0.614) and discriminated well between BRCA and non-BRCA pathogenic variant carriers (Area Under Receiver Operating Curve 0.80, 95% Confidence Interval=0.75-0.84). Addition of grade to the existing clinical genetic testing criteria targeting breast cancer patients below 45 years reduced the proportion of patients referred for genetic counselling and testing from 37% to 33% (p-value=0.003), thereby improving the overall efficacy.
Conclusion
Population-specific customisation of mutation prediction models and clinical genetic testing criteria improved the accuracy of BRCA mutation prediction in Asian patients.
Introduction
Germline genetic testing for BRCA1 and BRCA2 (BRCA) has enabled risk management for individuals at elevated cancer risk, and with the advent of PARP inhibitor treatment, enabled treatment selection with improved outcomes.1 In high resource countries, clinical genetics services are well established and patients are referred for germline BRCA genetic testing using criteria based on age of onset of cancer, breast cancer histology, and cancer family history.2,3
Similar genetic testing criteria have been incorporated into clinical practice guidelines in Asian countries, but these pose significant resource challenges, particularly in low- and middle-income Asian countries with limited clinical genetics services.4 Notably, because of the shifting reproductive and behavioural patterns, the incidence in many Asian countries have doubled or tripled in the past 40 years.5 This dramatic increase in incidence in younger generations means that the mean age of diagnosis for breast cancer in Asian women is approximately 10 years younger than that in European women,6,7 thus, a higher proportion of breast cancer patients fulfil clinical genetic testing criteria for referral, exacerbating the challenges in access to genetic services in Asian countries.4
In European or North American populations, models have been developed for predicting the likelihood of carrying germline BRCA pathogenic variants (PVs), mostly using data of breast cancer patients ascertained through genetic clinics, and these models are well-calibrated for these populations.8,9,10,11,12,13,14,15 Evaluation of such models in high-risk breast cancer patients of Asian descent living in North America16 or in Asia17,18,19,20,21 showed that these models underestimated the proportion of BRCA PVs carriers, especially for BRCA2 PVs carriers16,17,20,21 and for breast cancer patients with no family history of breast cancer.17,18 Recently, a BRCA carrier prediction algorithm (KOHCal) was developed in South Korea, based on high-risk breast cancer patients and was found to have better discrimination and calibration in South Koreans than models built on women of European descent.22
With the approval of PARP inhibitors for treatment of breast and ovarian cancer patients with BRCA PVs, there is an urgent need to determine the performance of these models in diverse populations. To date, no studies have evaluated the performance of clinical genetic testing criteria or developed a BRCA carrier prediction model in a population-based study of Asian breast cancer patients. In this study, we evaluated the performance of existing BRCA carrier prediction models, developed a new prediction model, and customised clinical genetic testing criteria in a population-based study of 8,162 Asian breast cancer patients from Malaysia and Singapore unselected for age of diagnosis and family history of cancer.
Methods
Study population
The study participants were women diagnosed clinically with breast cancer (invasive and non-invasive) who were recruited in the Malaysian Breast Cancer Genetic (MyBrCa) study23 and the Singapore Breast Cancer Cohort (SGBCC) study. Cases were recruited from two hospitals in Malaysia and six hospitals in Singapore. Germline DNA were sequenced in two batches, using targeted sequencing panels described previously.24 Carriers of pathogenic variants in non-BRCA genes were treated as non-carriers.
Statistical analyses
Existing BRCA carrier prediction models
Three existing BRCA carrier prediction models were evaluated in this study; two empirical models (PENNII, KOHCal) and a genetic risk model BOADICEA 5.0.25 Model performance was determined based on model calibration, assessed using Hosmer-Lemeshow (HL) test, and discrimination, assessed using area under receiver operating curve (AUC).
Development and validation of population-specific BRCA carrier prediction model
The study sample was randomly split into training and validation sets, comprising 70% and 30% of the samples, respectively. Candidate predictors of BRCA PV included age of breast cancer diagnosis, ethnicity, bilateral breast cancer, pathological features, and family history of breast or ovarian cancer (Supplementary Table 1). Missing data in the training set were imputed using multiple imputation by chained equations, whilst missing data in the validation set were imputed using single or multiple imputation by chained equations, under the missing at random (MAR) assumption.26,27 Given that multiple imputation generates more than one imputed dataset, results for single imputation in the validation dataset is presented in the main Figures and Tables, with results of multiple imputation included in Supplementary data where relevant. Additional sensitivity test was performed to ensure that the validation test results are comparable after single and multiple imputations. BRCA carrier prediction models were built based on logistic regression method using the training set. Model calibration and discrimination were evaluated in the validation set using HL test and AUC, respectively. The optimal carrier probability threshold for genetic testing was chosen based on the intersection of sensitivity and specificity curves.28
Customisation and evaluation of existing clinical criteria for germline BRCA genetic testing
Modified Clinical Criteria (MCC) were developed starting with the MCGplus Criteria2 by considering combinations of age of diagnosis of proband in 5 year intervals, with and without considering grade, resulting in a total of 96 different categories. The efficacy of MCC was evaluated in the validation set based on detection ratio (number of patients to be screened to detect one carrier).
All the data were analysed using Stata version 13.0 (Stata Corp., College Station., Texas, USA) and a p-value < 0.05 (two-tailed) was deemed to be statistically significant. See Supplemental Methods for more details.
Results
Study population characteristics
In this cross-sectional population-based study of 8,162 breast cancer patients, 323 (4.0%) had germline BRCA1 or BRCA2 PVs (Supplementary Table 1). The majority of patients were Chinese (75.4%), with a mean age of diagnosis of 52.3 years (SD=10.77). Compared to Chinese women, Indian women had a higher proportion of HR- and TNBC breast cancer cases, whereas Malay women had higher proportions of HER2+ and Luminal B breast cancer cases. There was a higher proportion of carriers amongst Indian and Malay women. Whilst the tumour characteristics tested were significantly associated with BRCA1 status, these were also associated with BRCA2 status, with the exception of ER, PR, HR, and TNBC (Supplementary Table 2).
Development and validation of population-specific BRCA carrier prediction model
Prediction models were developed using 5,714 breast cancer cases (228 BRCA carriers) and validated using 2,448 cases (95 BRCA carriers) (Supplementary Fig 1). Collinearity tests showed that ER, PR, HER2, TNBC, HR, and immune-histochemical subtypes were correlated (correlation coefficients, r>0.40). Hence, six combinations of tumour biomarkers along with the remaining predictors were considered in the analyses: (a) TNBC, (b) ER, (c) ER and HER2, (d) HR and HER2, (e) HER2, and (f) immune-histochemical subtypes. The best-performing model was selected based on the highest AUC and the lowest non-significant HL score in the validation set (Supplementary Table 3). Model (a) (AUC=0.86, HL=2.63) and Model (e) (AUC=0.75, HL=10.89) were the best-performing models for BRCA1 and BRCA2 PVs carrier status, whereas Model (c) was the best-performing model for overall BRCA (AUC=0.80, HL=5.43). The predictive performance of Model (c) by mutation-type were similar to the respective best-performing models (BRCA1: Model (c) versus Model (a) – AUC (HL): 0.86 (4.33) versus 0.86 (2.63); BRCA2: Model (c) versus Model (e) – AUC (HL): 0.75 (12.15) versus 0.75 (10.89)). Analyses after multiple imputation showed similar results. Hence, Model (c), subsequently referred to as ARiCa (Asian Risk Calculator), was selected as the final model for predicting overall BRCA PVs carrier status. We evaluated the performance of ARiCa by ethnicity and found that the model had high discriminatory power and well-calibrated across ethnic groups (Supplementary Table 4).
In ARiCa, younger age of diagnosis, Indian ethnicity, bilateral breast cancer, ER-negativity, HER2-negativity, higher grade, and presence of first degree family history of breast or ovarian cancer were associated with overall BRCA PVs carrier status (Table 1). These variables were also associated with BRCA1 PVs carrier status except grade, whereas BRCA2 was only associated with younger age, HER2-negativity, higher grade and first degree family history of breast cancer. Notably, both Malay and Indian ethnicities were associated with higher odds of being BRCA1 PVs carriers compared to Chinese ethnicity.
Table 1. Multivariable regression of ARiCa.
| Variable | Category |
BRCA vs Non-BRCA n=5,714) |
BRCA1 vs Non-BRCA1 (n=5,714) |
BRCA2 vs Non-BRCA2 (n=5,714) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Odds Ratio |
95% CI | P- value |
Odds Ratio |
95% CI | P- value |
Odds Ratio |
95% CI | P- value |
|||||
| Age* | 0.94 | 0.93 | 0.96 | <0.001 | 0.93 | 0.91 | 0.95 | <0.001 | 0.95 | 0.94 | 0.97 | <0.001 | |
| Ethnicity | Chinese | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| Malay | 1.26 | 0.87 | 1.82 | 0.220 | 1.86 | 1.05 | 3.29 | 0.034 | 0.98 | 0.61 | 1.57 | 0.920 | |
| Indian | 2.06 | 1.37 | 3.09 | <0.001 | 3.20 | 1.75 | 5.82 | <0.001 | 1.35 | 0.77 | 2.36 | 0.295 | |
| Other | 1.50 | 0.45 | 4.96 | 0.511 | 1.79 | 0.23 | 13.91 | 0.58 | 1.37 | 0.33 | 5.77 | 0.667 | |
| Bilateral | Unilateral | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| Contralateral | 2.56 | 1.57 | 4.17 | <0.001 | 4.31 | 2.15 | 8.61 | <0.001 | 1.56 | 0.80 | 3.06 | 0.195 | |
| Ipsilateral | 1.21 | 0.46 | 3.21 | 0.689 | 1.70 | 0.45 | 6.49 | 0.437 | 0.86 | 0.20 | 3.59 | 0.833 | |
| ER | Er+ | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| ER- | 1.62 | 1.17 | 2.24 | 0.004 | 5.59 | 3.15 | 9.92 | <0.001 | 0.71 | 0.45 | 1.10 | 0.126 | |
| HER2 | HER2+ | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| HER2- | 2.35 | 1.59 | 3.48 | <0.001 | 3.11 | 1.61 | 6.01 | 0.001 | 1.84 | 1.13 | 3.00 | 0.015 | |
| Grade | One | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| Two | 3.18 | 1.38 | 7.32 | 0.006 | 2.27 | 0.41 | 12.49 | 0.346 | 3.62 | 1.39 | 9.39 | 0.008 | |
| Three | 4.02 | 1.72 | 9.42 | 0.001 | 2.91 | 0.53 | 16.02 | 0.219 | 4.34 | 1.62 | 11.66 | 0.004 | |
| FHBC | No | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| Yes | 3.01 | 2.23 | 4.07 | <0.001 | 3.48 | 2.13 | 5.69 | <0.001 | 2.56 | 1.77 | 3.71 | <0.001 | |
| FHOC | No | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| Yes | 4.57 | 2.51 | 8.30 | <0.001 | 7.95 | 3.63 | 17.41 | <0.001 | 1.93 | 0.75 | 4.92 | 0.170 | |
Sample: 5714 MyBrCa and SGBCC breast cancer patients in multiply imputed training set.
Abbreviations: Bilateral, Bilateral Breast Cancer; FHBC, First Degree Family History for Breast Cancer; FHOC, First Degree Family History for Ovarian Cancer; 95% CI, 95% Confidence Interval.
Age of diagnosis for breast cancer of proband.
We determined the optimal carrier probability threshold for ARiCa in the validation set as the intercept of sensitivity and specificity (Supplementary Fig 2). At the optimal threshold, corresponding to a mutation prevalence of 4%, 31% (95%CI=29-33) of breast cancer patients would require germline BRCA genetic testing and 71% (95%CI=61-80) of BRCA PVs carriers would be identified (Supplementary Table 5). Performance of ARiCa were consistent across imputed validation sets after multiple imputation.
Comparison of BRCA carrier prediction models
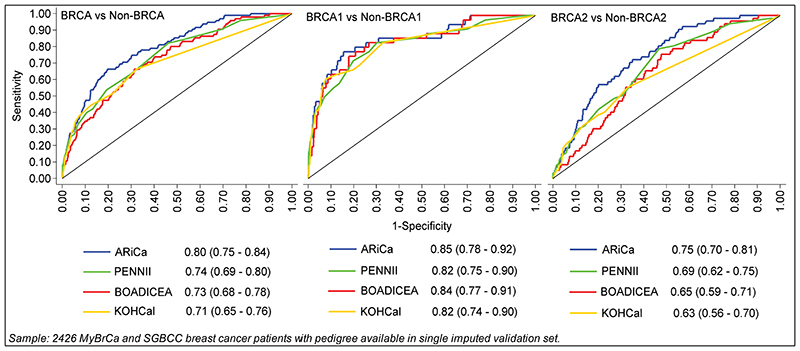
We compared the performance of ARiCa with models which have been developed in other populations using data for 2,426 patients from the validation set for whom data are available for variables required in all considered models. For overall BRCA, ARiCa had the highest AUC (0.80), followed by PENNII (0.74), BOADICEA (0.73), and KOHCal (0.71) (Fig 1). The AUCs for BRCA1 were similar across models, but ARiCa had significantly better discriminatory ability than PENNII, BOADICEA, and KOHCal for BRCA2 (0.75, 0.69, 0.65, and 0.63 respectively).
Fig. 1. ROC curves of BRCA carrier prediction models.
Sample: 2426 MyBrCa and SGBCC breast cancer patients with pedigree available in single imputed validation set.
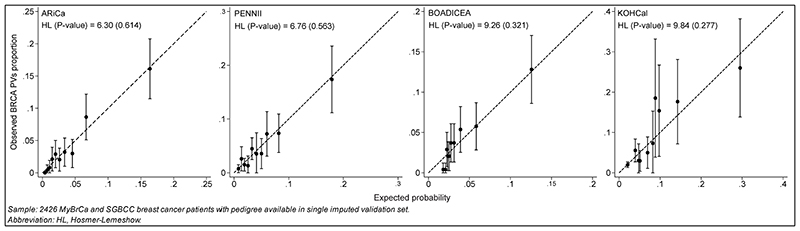
All models were well-calibrated; ARiCa had the lowest HL for overall BRCA (Fig 2). There was no significant difference between the observed proportion and expected probability for BRCA1 and BRCA2 PVs carriers and a majority were distributed close to the bisector (Supplementary Fig 3).
Fig. 2. Observed proportion and expected probability of BRCA carrier prediction models.
Sample: 2426 MyBrCa and SGBCC breast cancer patients with pedigree available in single imputed validation set.
Abbreviation: HL, Hosmer-Lemeshow.
We compared the efficacy measures (sensitivity, specificity) at the optimal and the conventional 10% and 20% thresholds.17,21,29 All models had poor sensitivity at the 10% and 20%, so we focused on the lower optimal thresholds (Table 2).17 At the respective optimal thresholds for each model, the sensitivities of all models for overall BRCA were 63-71% and the specificities were 67-71%. Whilst all models achieved a sensitivity of 83% for BRCA1, KOHCal (56%), BOADICEA (56%), and PENNII (51%) had lower sensitivity for BRCA2 than ARiCa (66%). ARiCa achieved relatively high sensitivity and specificity for overall BRCA (71%, 71%), BRCA1 (83%, 70%), and BRCA2 (66%, 70%) at the optimal threshold.
Table 2. Performance of BRCA carrier prediction models at different thresholds.
| Threshold (%) |
Model |
BRCA vs Non-BRCA (n=2,426) |
BRCA1 vs Non-BRCA1 (n=2,426) |
BRCA2 vs Non-BRCA2 (n=2,426) |
|||
|---|---|---|---|---|---|---|---|
| Sensitivity (%) |
Specificity (%) |
Sensitivity (%) |
Specificity (%) |
Sensitivity (%) |
Specificity (%) |
||
| 4.0 * | ARiCa | 71 | 71 | 83 | 70 | 66 | 70 |
| 8.0 * | PENNII | 63 | 70 | 83 | 70 | 51 | 69 |
| 2.2 * | BOADICEA | 66 | 67 | 83 | 67 | 56 | 67 |
| 4.0 * | KOHCal | 66 | 68 | 83 | 67 | 56 | 67 |
| 10.0 ** | ARiCa | 34 | 94 | 58 | 93 | 19 | 93 |
| PENNII | 60 | 74 | 78 | 73 | 49 | 73 | |
| BOADICEA | 22 | 95 | 44 | 95 | 8 | 95 | |
| KOHCal | 44 | 88 | 64 | 87 | 32 | 87 | |
| 20.0 ** | ARiCa | 15 | 99 | 25 | 98 | 8 | 98 |
| PENNII | 15 | 99 | 25 | 98 | 8 | 98 | |
| BOADICEA | 7 | 99 | 14 | 99 | 3 | 99 | |
| KOHCal | 22 | 97 | 42 | 97 | 10 | 96 | |
Sample: 2426 MyBrCa and SGBCC breast cancer patients with pedigree available in single imputed validation set.
Optimal threshold.
Conventional threshold.
Customisation and evaluation of existing clinical criteria for germline BRCA genetic testing
We evaluated the NCCN and Mainstreaming Cancer Genetics (UK) clinical genetic testing criteria in the validation set. Applying the NCCN and MCG Criteria would lead to 37% and 39% being referred with 72% and 69% of BRCA PVs carriers identified, respectively. Addition of family history variables to the MCG Criteria (MCGPlus) increased the screening rate from 39% to 49% and improved the detection rate from 69% to 81% (data not shown). Whilst the expanded NCCN Criteria detected the highest detection rate (96%), more than three-quarters of breast cancer patients (88%) would need to be screened.
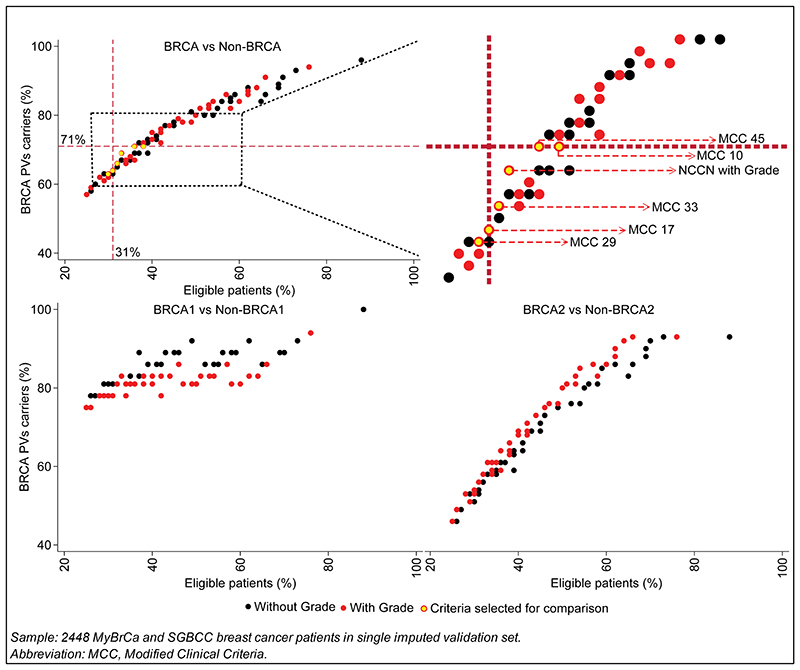
Given that patients in this study had a younger age of diagnosis for breast cancer than those in the Western populations and grade was a significant predictor of BRCA PVs carrier status, we customised MCGplus Criteria by considering several combinations of age of diagnosis for breast cancer and higher-grade breast cancer (grade 2 or 3) to improve the overall efficacy. Applying 96 Modified Clinical Criteria (48 MCC with grade and 48 MCC without grade) (Fig 3), we found that the detection rates for BRCA1 (78-92%) were higher than for BRCA2 (46-93%). Notably, at similar detection rate, the addition of grade resulted in reduction in screening rate of 1% to 10% (average=4%) for overall BRCA. Similarly, the addition of grade resulted in reduction in screening rate of 3% to 12% (average=5%) for existing clinical genetic testing criteria. There was no difference in reduction rates between BRCA1 and BRCA2.
Fig. 3. Eligible patients and BRCA PVs carriers detected for clinical criteria with and without grade.
Sample: 2448 MyBrCa and SGBCC breast cancer patients in single imputed validation set. Abbreviation: MCC, Modified Clinical Criteria.
We identified 3 clinical criteria categories (MCC 17, 29, 33) from Fig 3 with similar screening rates to ARiCa for overall BRCA (screening rate=31%) (Supplementary Table 5). These criteria had identical criteria for grade 2 or 3 breast cancer (≤40) and bilateral breast cancer (≤60), but they had different thresholds for age of diagnosis of proband with TNBC and family history of breast or ovarian cancer (Table 3).
Table 3. Evaluation of clinical criteria with grade.
| Criteria | BC + Grade* | TNBC* | Bilateral* | OC | FHBC* | FHOC* | Eligible patients (%)** |
BRCA PVs carriers (%)** |
Detection ratio |
|---|---|---|---|---|---|---|---|---|---|
| MCC 29 | ≤40 | ≤45 | ≤60 | 30.0 | 63.0 | 12 : 1 | |||
| MCC 17 | ≤40 | ≤60 | ≤60 | ≤60 | ≤60 | 31.0 | 64.0 | 13 : 1 | |
| MCC 33 | ≤40 | ≤50 | ≤60 | 32.0 | 66.0 | 12 : 1 | |||
| NCCN with grade | ≤45 | ≤60 | 46-50 | 46-50 | 33.0 | 69.0 | 12 : 1 | ||
| MCC 45 | ≤40 | ≤60 | 38.0 | 71.0 | 14 : 1 | ||||
| MCC 10 | ≤45 | ≤50 | ≤60 | ≤60 | ≤60 | 36.0 | 71.0 | 13 : 1 | |
Sample: 2448 MyBrCa and SGBCC breast cancer patients in single imputed validation set.
Abbreviations: MCC, Modified Clinical Criteria; BC, Breast Cancer of proband; TNBC, Triple Negative Breast Cancer; Bilateral, Bilateral Breast Cancer; OC, Ovarian Cancer; FHBC, one or more first degree relatives with Breast Cancer; FHOC, one or more first degree relatives with Ovarian Cancer.
Age of diagnosis for breast cancer of proband.
Fulfilled at least one criterion.
We also identified 3 clinical criteria categories (NCCN with grade, MCC 10, 45) from Fig 3 with similar detection rates to ARiCa for overall BRCA (detection rate=71%) (Supplementary Table 5). These criteria had different combinations of age of diagnosis of proband with grade 2 or 3 breast cancer, TNBC, bilateral breast cancer, and family history of breast or ovarian cancer (Table 3).
All 6 modified criteria resulted in lower detection ratios, when compared to existing clinical genetic testing criteria (Expanded NCCN=24:1; MCGplus=15:1; MCG=14:1; NCCN=13:1) (Table 3). NCCN with grade outperformed the 5 modified criteria by achieving a higher detection rate (69%) at the lowest detection ratio (12:1). Nonetheless, all 6 modified criteria still underperformed ARiCa. For instance, MCC (17, 29, 33) had lower detection rates of 63-66% compared to 71% for ARiCa. Similarly, NCCN with grade and MCC (10, 45) had higher screening rate of 33-38% compared to 31% for ARiCa (Table 3).
Discussion
Whilst germline BRCA1 or BRCA2 PVs testing has an established role in risk management, this is increasingly relevant in the selection of therapy.1 We showed that logistic regression models built based on a large Asian population-based study of breast cancer patients, unselected for age of diagnosis and family history, outperformed the genetic risk model (BOADICEA) developed using data on European-ancestry populations and the empirical models (PENNII, KOHCal) developed using breast cancer patients with early onset or familial breast cancer. The Modified Clinical Criteria (MCC) customised to the Asian breast cancer patients in combination with presence of grade were more efficient than existing clinical genetic testing criteria.
In multivariable regression analyses, we found that the risk factors significantly associated with BRCA PVs carrier status in this study were consistent with previously published findings from Asian countries, including younger age of diagnosis, bilateral breast cancer, ER-negative status, HER2-negative status, higher grade, and presence of first degree family history of breast cancer or ovarian cancer.20,22,30,31,32,33,34,35,36,37,38
We found that all the BRCA carrier prediction models and the Modified Clinical Criteria (MCC) were more sensitive (sensitivity/detection rate) and accurate (discrimination) for BRCA1 than BRCA2, which is likely to be driven by the stronger association between BRCA1 and the ER-negative status, TNBC subtype, and ovarian cancer family history.39,40,41,42,43 Indeed, several studies have previously demonstrated that the use of pathologic characteristics, namely ER and TNBC improved the sensitivity and discrimination for BRCA1 when selecting individuals for germline genetic testing in high-risk breast cancer patients.44,45,46,47,48
All three existing models tested performed similarly in our study population (AUC=0.71-0.74) as previously reported in other Asian populations (AUC=0.69-0.76), but the AUCs were lower than those reported in women of European descent, especially for BOADICEA (AUC=0.77).13,17,22,29 BOADICEA had lower discriminatory ability for overall BRCA and BRCA2, consistent with the observation that BOADICEA performed better at lower thresholds because it underestimated carrier probability of Asian breast cancer patients with the lowest sensitivity at conventional thresholds, particularly those with germline BRCA2 PVs.17 Nevertheless, BOADICEA outperformed PENNII for BRCA1.17 These observations are not surprising. Given that BOADICEA is a genetic risk model, it relies on population-specific parameters for breast cancer incidences, PV frequencies, and tumour-pathology distributions as input parameters. Customisation of BOADICEA using population-specific parameters and addition of tumour grade (a clear predictor of carrier status in our analysis) could substantially improve its discrimination.
In terms of calibration, models built on Asian populations had better calibration for BRCA2, whilst models built on women of European descent had better calibration for BRCA1. Notably, this was evident in KOHCal and BOADICEA that appeared to be calibrated with the lowest HL for BRCA2 and BRCA1, respectively. A possible explanation could be due to the variation in mutation prevalence. Whilst BRCA2 mutations are more common than BRCA1 mutations in Asian, it is the opposite in many European populations.17
Notwithstanding BRCA carrier prediction models have good discrimination, there are challenges in their implementation in resource constrained settings. Clinical genetic testing criteria are likely to continue as a mainstay for referral of patients for genetic counselling and testing. In our evaluation, NCCN with grade had a sensitivity of 69% at a screening rate of 33%. This is marginally better than the existing NCCN and MCG criteria where at similar detection rates of 69-72%, about 37-39% patients would be referred for genetic counselling and testing. It is possible that this improvement is because of the age threshold for TNBC (≤60 vs no age restriction), bilateral breast cancer (46-50 vs ≤60), and first degree family history of breast cancer (46-50 vs no age restriction), but the inclusion of grade is also an important consideration. Indeed, higher grade was identified as a strong predictor not only for BRCA1 PVs carriers but also for BRCA2 PVs carriers.42 Previous studies have shown that inclusion of grade can improve the sensitivity and discrimination of germline BRCA PVs prediction in high-risk breast cancer patients.44,45 Given that BRCA1 (91%) and BRCA2 (89%) PVs carriers were of higher grade than in non-carriers (76%), future improvements in BRCA carrier prediction tools could include grade.45,49
Limitations and strengths
The validation sample was relatively small with only 95 BRCA PVs carriers. Future independent studies should aim to assess the models developed here. The analysis was also restricted to BRCA1 or BRCA2, but gene-panels that include additional susceptibility genes are now widely used, which include additional genes (e.g., PALB2) that may be relevant in informing treatment.3 However, the present sample size is too small to allow the prediction of carrying PVs in other genes. Although grade was identified as a potential variable to include in risk prediction models, it is noteworthy that quality assurance may be required for this and other variables in order to ensure model accuracy. Finally, whilst ARiCa was shown to perform equally well across different ethnic groups in Malaysia and Singapore, studies in other Asian populations are needed to evaluate its utility in these populations.
Despite the limitations, this is the first study to develop a logistic regression BRCA carrier prediction model and customise clinical genetic testing criteria for use in mainstream germline BRCA genetic testing based on unselected sample of breast cancer patients in South East Asia.
Conclusion
With the advent of germline genetic testing for treatment selection, more women may consider genetic testing as part of their treatment plans. Given that Asian women have a younger age of diagnosis for breast cancer and different distribution of breast cancer subtypes compared to women of European descent, population-specific customisation of BRCA carrier prediction tools is important to enable more accurate BRCA mutation prediction in diverse populations.
Supplementary Material
Context Summary.
Key objective
Increasing breast cancer incidence and limited resources pose a significant challenge to genetic counselling and testing in many low- and middle-income countries in Asia. Whilst existing mutation prediction models underestimate proportion of carriers in Asian women, a logistic regression model was developed and validated based on South East Asian breast cancer patients unselected for age of diagnosis and family history of cancer to estimate the likelihood of carrying a pathogenic variant in BRCA1 or BRCA2 gene, called ARiCa (Asian Risk Calculator).
Knowledge generated
ARiCa outperformed existing mutation prediction models. Discrimination of mutation prediction model and efficacy of clinical genetic testing criteria were significantly enhanced by the inclusion of tumour grade.
Relevance
Population-specific customisation of mutation prediction tools is important to enable more accurate BRCA mutation prediction in diverse populations for referral of breast cancer patients for genetic counselling and testing who may benefit from the selection of therapy.
Acknowledgements
MyBrCa thanks study participants and all research staff at Cancer Research Malaysia, University Malaya, and Sime Darby Medical Centre who assisted in recruitment and interviews (including, Meow Keong Thong, Daphne SC Lee, Sheau-Yee Lee, Sze-Yee Phuah, Kah-Nyin Lai, Shao Yan Lau, Shivaani Mariapun, Siu Wan Wong, Heamanthaa Padmanabhan, Hanani Che Halim, Leelavathy Krishnan) for their contributions and commitment to this study. For SGBCC, we want to thank the program manager Jenny Liu, clinical research coordinators/research assistants Siew-Li Tan, Siok-Hoon Yeo, Ting-Ting Koh, Amanda Ong, Jin-Yee Lee, Michelle Mok, Ying-Jia Chew, Jing-Jing Hong, and Hui-Min Lau for their contributions in recruitment, Yen-Shing Yeoh for data preparation, and Alexis J. Khng for processing the DNA samples. We also want to thank all the participants’ support to SGBCC.
Financial Support
Supported in whole, or in part, by Wellcome Trust (Grant No.: v203477/Z/16/Z). For the purpose of open access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission. The Malaysian Breast Cancer Genetic Study was established using funds from the Malaysian Ministry of Science, and the Malaysian Ministry of Higher Education High Impact Research Grant (Grant No.: UM.C/HIR/MOHE/06), and additional funding was received from Yayasan Sime Darby, Yayasan PETRONAS, Estee Lauder Group of Companies, Khind Starfish Foundation, and other donors of Cancer Research Malaysia. SGBCC was supported by the National Research Foundation Singapore (Grant No.: NRF-NRFF2017-02, awarded to J.L.), NUS start-up Grant, National University Cancer Institute Singapore (NCIS) Centre Grant (Grant No.: NMRC/CG/NCIS/2010, NMRC/CG/012/2013, CGAug16M005), Breast Cancer Prevention Programme (BCPP), Asian Breast Cancer Research Fund, and the NMRC Clinician Scientist Award (SI Category; Grant No.: NMRC/CSA-SI/0015/2017) and the Breast Cancer Screening and Prevention Programme Grant (Grant No.: NUHSRO/2020/121/BCSPP/LOA). A.C.A. is supported by Cancer Research UK (Grant No.: C12292/A20861, PPRPGM-Nov20\100002).
Footnotes
Conception and design:
Boon Hong Ang, Weang Kee Ho, Pui Yoke Kwan, Douglas F. Easton, Antonis C. Antoniou, Soo Hwang Teo
Financial support:
Soo Hwang Teo, Douglas F. Easton, Antonis C. Antoniou, Weang Kee Ho, Mikael Hartman, Jingmei Li, Nur Aishah Mohd Taib
Administrative support:
Eldarina Wijaya, Siti Norhidayu Hasan, Tiara Hassan, Mei-Chee Tai
Provision of study materials or patients:
Nur Aishah Mohd Taib, Cheng Har Yip, Mikael Hartman, Swee Ho Lim, Ern Yu Tan, Benita K. T. Tan, Su-Ming Tan, Veronique K. M. Tan, Jingmei Li
Collection and assembly of data:
Sook Yee Yoon, Tiara Hassan, Pei Sze Ng, Siti Norhidayu Hasan, Joanna M. C. Lim, Mei-Chee Tai, Peh Joo Ho, Alexis J. Khng, Jamie Allen, Alison M. Dunning
Data analysis and interpretation:
Boon Hong Ang, Weang Kee Ho, Eldarina Wijaya, Andrew Lee
Manuscript writing:
All authors
Final approval of manuscript:
All authors
Accountable for all aspects of the work:
All authors The authors confirm that they have no competing financial interests.
References
- 1.Tung NM, Garber JE. BRCA 1/2 testing: therapeutic implications for breast cancer management. Br J Cancer. 2018;119:141. doi: 10.1038/s41416-018-0127-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kemp Z, Turnbull A, Yost S, et al. Evaluation of cancer-based criteria for use in mainstream BRCA1 and BRCA2 genetic testing in patients with breast cancer. JAMA Netw Open. 2019;2:e194428. doi: 10.1001/jamanetworkopen.2019.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yadav S, Hu C, Hart SN, et al. Evaluation of Germline Genetic Testing Criteria in a Hospital-Based Series of Women with Breast Cancer. J Clin Oncol. 2020;38:1409. doi: 10.1200/JCO.19.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakamura S, Kwong A, Kim SW, et al. Current status of the management of hereditary breast and ovarian cancer in Asia: first report by the Asian BRCA consortium. Public Health Genomics. 2016;19:53–60. doi: 10.1159/000441714. [DOI] [PubMed] [Google Scholar]
- 5.Boyle P, Levin B. World cancer report 2008. IARC Press, International Agency for Research on Cancer; 2008. [Google Scholar]
- 6.Lin CH, Yap YS, Lee KH, et al. Contrasting epidemiology and clinicopathology of female breast cancer in Asians vs the US population. J Natl Cancer Inst. 2019;111:1298–1306. doi: 10.1093/jnci/djz090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song QK, Li J, Huang R, et al. Age of diagnosis of breast cancer in china: almost 10 years earlier than in the United States and the European union. Asian Pac J Cancer Prev. 2014;15:10021–10025. doi: 10.7314/apjcp.2014.15.22.10021. [DOI] [PubMed] [Google Scholar]
- 8.Apicella C, Andrews L, Hodgson SV, et al. Log odds of carrying an Ancestral Mutation in BRCA1 or BRCA2for a Defined personal and family history in an Ashkenazi Jewish woman (LAMBDA) Breast Cancer Res. 2003;5:R206. doi: 10.1186/bcr644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berry DA, Iversen ES, Jr, Gudbjartsson DF, et al. BRCAPRO validation, sensitivity of genetic testing of BRCA1/BRCA2 and prevalence of other breast cancer susceptibility genes. J Clin Oncol. 2002;20:2701–2712. doi: 10.1200/JCO.2002.05.121. [DOI] [PubMed] [Google Scholar]
- 10.Couch FJ, DeShano ML, Blackwood MA, et al. BRCA1 mutations in women attending clinics that evaluate the risk of breast cancer. N Engl J Med. 1997;336:1409–1415. doi: 10.1056/NEJM199705153362002. [DOI] [PubMed] [Google Scholar]
- 11.de la Hoya M, Osorio A, Godino J, et al. Association between BRCA1 and BRCA2 mutations and cancer phenotype in Spanish breast/ovarian cancer families: implications for genetic testing. Int J Cancer. 2002;97:466–471. doi: 10.1002/ijc.1627. [DOI] [PubMed] [Google Scholar]
- 12.Frank TS, Deffenbaugh AM, Reid JE, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–1490. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 13.Lindor NM, Johnson KJ, Harve H, et al. Predicting BRCA1 and BRCA2 gene mutation carriers: comparison of PENN II model to previous study. Fam Cancer. 2010;9:495–502. doi: 10.1007/s10689-010-9348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shattuck-Eidens D, Oliphant A, McClure M, et al. BRCA1 sequence analysis in women at high risk for susceptibility mutations: risk factor analysis and implications for genetic testing. JAMA. 1997;278:1242–1250. [PubMed] [Google Scholar]
- 15.Vahteristo P, Eerola H, Tamminen A, et al. A probability model for predicting BRCA1 and BRCA2 mutations in breast and breast-ovarian cancer families. Br J Cancer. 2001;84:704–708. doi: 10.1054/bjoc.2000.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurian AW, Gong GD, Chun NM, et al. Performance of BRCA1/2 mutation prediction models in Asian Americans. J Clin Oncol. 2008;26:4752. doi: 10.1200/JCO.2008.16.8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung FH, Wang YA, Jian JW, et al. Evaluating BRCA mutation risk predictive models in a Chinese cohort in Taiwan. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-46707-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang E, Park SK, Yang JJ, et al. Accuracy of BRCA1/2 mutation prediction models in Korean breast cancer patients. Breast Cancer Res Treat. 2012;134:1189–1197. doi: 10.1007/s10549-012-2022-8. [DOI] [PubMed] [Google Scholar]
- 19.Kim H, Choi DH. Distribution of BRCA1 and BRCA2 mutations in Asian patients with breast cancer. J Breast Cancer. 2013;16:357–365. doi: 10.4048/jbc.2013.16.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thirthagiri E, Lee SY, Kang P, et al. Evaluation of BRCA1 and BRCA2 mutations and risk-prediction models in a typical Asian country (Malaysia) with a relatively low incidence of breast cancer. Breast Cancer Res. 2008;10:R59. doi: 10.1186/bcr2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwong A, Wong CH, Suen DT, et al. Accuracy of BRCA1/2 mutation prediction models for different ethnicities and genders: experience in a southern Chinese cohort. World J Surg. 2012;36:702–713. doi: 10.1007/s00268-011-1406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang E, Park SK, Lee JW, et al. KOHBRA BRCA risk calculator (KOHCal): a model for predicting BRCA1 and BRCA2 mutations in Korean breast cancer patients. J Hum Genet. 2016;61:365–371. doi: 10.1038/jhg.2015.164. [DOI] [PubMed] [Google Scholar]
- 23.Tan MM, Ho WK, Yoon SY, et al. A case-control study of breast cancer risk factors in 7,663 women in Malaysia. PloS One. 2018;13:e0203469. doi: 10.1371/journal.pone.0203469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorling, et al. Breast cancer risk genes: association analysis of rare coding variants in 34 genes in 60,466 cases and 53,461 controls. N Engl J Med. 2021 [Google Scholar]
- 25.Lee A, Mavaddat N, Wilcox AN, et al. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet Med. 2019;21:1708–1718. doi: 10.1038/s41436-018-0406-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howlader N, Cronin KA, Kurian AW, et al. Differences in breast cancer survival by molecular subtypes in the United States. Cancer Epidemiol Biomarkers Prev. 2018;27:619–626. doi: 10.1158/1055-9965.EPI-17-0627. [DOI] [PubMed] [Google Scholar]
- 27.Stuart EA, Azur M, Frangakis C, et al. Multiple imputation with large data sets: a case study of the Children’s Mental Health Initiative. Am J Epidemiol. 2009;169:1133–1139. doi: 10.1093/aje/kwp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosmer DW, Lemeshow SL. Applied logistic regression. ed 2. A Wiley-Interscience; New York: 2000. [Google Scholar]
- 29.Antoniou AC, Hardy R, Walker L, et al. Predicting the likelihood of carrying a BRCA1 or BRCA2 mutation: validation of BOADICEA, BRCAPRO, IBIS, Myriad and the Manchester scoring system using data from UK genetics clinics. J Med Genet. 2008;45:425–431. doi: 10.1136/jmg.2007.056556. [DOI] [PubMed] [Google Scholar]
- 30.Kwong A, Wong LP, Wong HN, et al. Clinical and pathological characteristics of Chinese patients with BRCA related breast cancer. Hugo J. 2009;3:63–76. doi: 10.1007/s11568-010-9136-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Ide Y, Inuzuka M, et al. BRCA1/BRCA2 mutations in Japanese women with ductal carcinoma in situ. Mol Genet Genomic Med. 2019;7:e493. doi: 10.1002/mgg3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noh JM, Han BK, Choi DH, et al. Association between BRCA mutation status, pathological findings, and magnetic resonance imaging features in patients with breast cancer at risk for the mutation. J Breast Cancer. 2013;16:308–314. doi: 10.4048/jbc.2013.16.3.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong ES, Shekar S, Chan CH, et al. Predictive factors for BRCA1 and BRCA2 genetic testing in an Asian clinic-based population. PloS One. 2015;10:e0134408. doi: 10.1371/journal.pone.0134408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yip CH, Taib NA, Choo WY, et al. Clinical and pathologic differences between BRCA1-, BRCA2-, and non-BRCA-associated breast cancers in a multiracial developing country. World J Surg. 2009;33:2077–2081. doi: 10.1007/s00268-009-0146-8. [DOI] [PubMed] [Google Scholar]
- 35.Yu JH, Lee JW, Son BH, et al. Characteristics of BRCA1/2 mutation-positive breast cancers in Korea: a comparison study based on multicenter data and the Korean Breast Cancer Registry. J Breast Cancer. 2014;17:129–135. doi: 10.4048/jbc.2014.17.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang XR, Devi BC, Sung H, et al. Prevalence and spectrum of germline rare variants in BRCA1/2 and PALB2 among breast cancer cases in Sarawak, Malaysia. Breast Cancer Res Treat. 2017;165:687–97. doi: 10.1007/s10549-017-4356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwong A, Shin VY, Ho JC, et al. Comprehensive spectrum of BRCA1 and BRCA2 deleterious mutations in breast cancer in Asian countries. J Med Genet. 2016;5:15–23. doi: 10.1136/jmedgenet-2015-103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wen WX, Allen J, Lai KN, et al. Inherited mutations in BRCA1 and BRCA2 in an unselected multiethnic cohort of Asian patients with breast cancer and healthy controls from Malaysia. J Med Genet. 2018;55:97–103. doi: 10.1136/jmedgenet-2017-104947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H, Wu J, Zhang Z, et al. Association between BRCA status and triple-negative breast cancer: a meta-analysis. Front Pharmacol. 2018;9:909. doi: 10.3389/fphar.2018.00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lakhani SR, Van De Vijver MJ, Jacquemier J, et al. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20:2310–2318. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- 41.Tung N, Wang Y, Collins LC, et al. Estrogen receptor positive breast cancers in BRCA1 mutation carriers: clinical risk factors and pathologic features. Breast Cancer Res. 2010;12:R12. doi: 10.1186/bcr2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spurdle AB, Couch FJ, Parsons MT, et al. Refined histopathological predictors of BRCA1 and BRCA2 mutation status: a large-scale analysis of breast cancer characteristics from the BCAC, CIMBA, and ENIGMA consortia. Breast Cancer Res. 2014;16:1–6. doi: 10.1186/s13058-014-0474-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ford D, Easton DF, Stratton M, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am J Hum Genet. 1998;62:676–89. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farshid G, Balleine RL, Cummings M, et al. Morphology of breast cancer as a means of triage of patients for BRCA1 genetic testing. Am J Surg Pathol. 2006;30:1357–1366. doi: 10.1097/01.pas.0000213273.22844.1a. [DOI] [PubMed] [Google Scholar]
- 45.James PA, Doherty R, Harris M, et al. Optimal selection of individuals for BRCA mutation testing: a comparison of available methods. J Clin Oncol. 2006;24:707–715. doi: 10.1200/JCO.2005.01.9737. [DOI] [PubMed] [Google Scholar]
- 46.Lidereau R, Eisinger F, Champème MH, et al. Major improvement in the efficacy of BRCA1 mutation screening using morphoclinical features of breast cancer. Cancer Res. 2000;60:1206–1210. [PubMed] [Google Scholar]
- 47.Phuah SY, Looi LM, Hassan N, et al. Triple-negative breast cancer and PTEN (phosphatase and tensin homologue) loss are predictors of BRCA1 germline mutations in women with early-onset and familial breast cancer, but not in women with isolated late-onset breast cancer. Breast Cancer Res. 2012;14:R142. doi: 10.1186/bcr3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robertson L, Hanson H, Seal S, et al. BRCA1 testing should be offered to individuals with triple-negative breast cancer diagnosed below 50 years. Br J Cancer. 2012;106:1234–1238. doi: 10.1038/bjc.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stratton MR. Pathology of familial breast cancer: differences between breast cancers in carriers of BRCA1 or BRCA2 mutations and sporadic cases. The Lancet. 1997;349:1505–1510. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.