Abstract
Background
Although a number of health outcomes such as cardiovascular diseases, metabolic-related outcomes, neurological disorders, pregnancy outcomes and cancers have been identified in relation to B vitamins, evidence is of uneven quality and volume, and there is uncertainty about putative causal relationships.
Objectives
To explore the effects of B vitamins and homocysteine on a wide range of health outcomes based on a large biorepository linking biological samples and electronic medical records.
Methods
First, we performed a phenome-wide association study (PheWAS) to investigate associations of genetically predicted plasma concentrations (genetic component of the circulating concentrations) of folate, vitamin B6, vitamin B12 and their metabolite homocysteine with a wide range of disease outcomes (including both prevalent and incident events) among 385,917 individuals in the UK Biobank. Second, two-sample Mendelian randomization (MR) analysis was used to replicate any observed associations and detect causality. We considered MR p <0.05 as significant for replication. Third, dose-response, mediation and bioinformatics analyses were carried out to examine any non-linear trends and to disentangle the underlying mediating biological mechanisms for the identified associations.
Results
In total 1,117 phenotypes were tested in each PheWAS analysis. After multiple correction, 32 phenotypic associations of B vitamins and homocysteine were identified. Two-sample MR analysis supported that three of them were causal, including associations of higher plasma vitamin B6 with lower risk of calculus of kidney (OR: 0.64; 95% CI: 0.42, 0.97; P=0.033); higher homocysteine concentration with higher risk of hypercholesterolemia (OR: 1.28, 95% CI: 1.04, 1.56; P=0.018) and chronic kidney disease (OR: 1.32, 95% CI: 1.06, 1.63; P=0.012). Significant nonlinear dose-response relationships were observed for the associations of folate with anemia, vitamin B12 with vitamin B-complex deficiencies, anemia and cholelithiasis, and homocysteine with cerebrovascular disease.
Conclusions
This study provides strong evidence for the associations of B vitamins and homocysteine with endocrine/metabolic and genitourinary disorders.
Keywords: B vitamins, Homocysteine, PheWAS, Mendelian randomization
Introduction
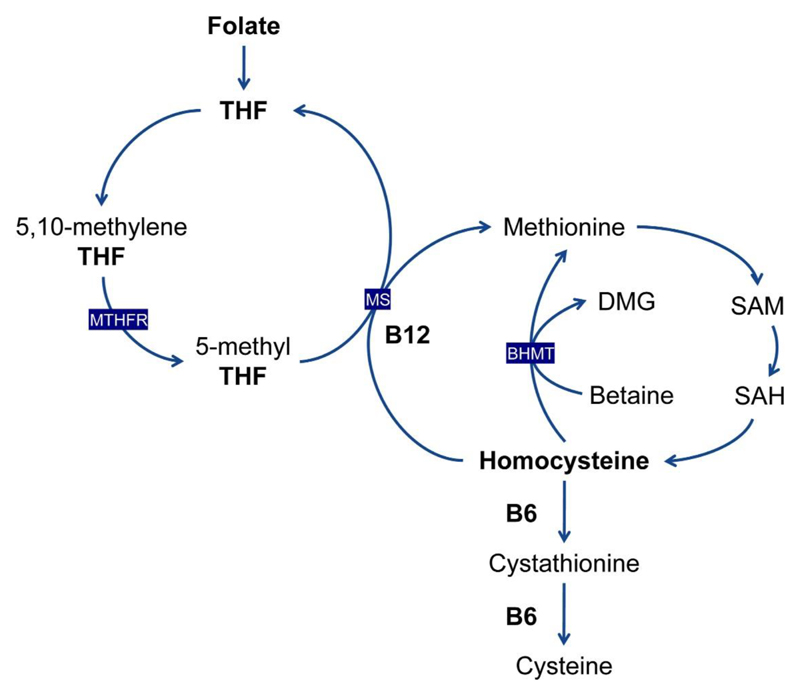
The B vitamins, including folate (B9), vitamin B6, vitamin B12, have been investigated to have effects on many health and disease outcomes. In a recent umbrella review of meta-analyses of observational studies and of randomized clinical trials (RCTs), folate (dietary intake, dietary supplementary and plasma concentration) has been linked to more than 100 unique health outcomes (1). In addition, vitamin B6 and B12 (dietary intake, dietary supplementary and plasma concentration) have been identified as associated with the risk of cardiovascular disease, metabolic-related outcomes, neurological disorders, pregnancy outcomes and several cancers (2–4). One-carbon metabolism is a process in which folate transfers one-carbon units to support a wide range of biological processes including DNA synthesis and methylation (5). Vitamin B6 and B12 interact with folate as methyl donors within this network, and deficiency of either folate, B6 or B12 can lead to increased circulating concentration of the related metabolite, homocysteine (Figure 1) (5), which has been suggested as an independent risk factor for cardiovascular disease (6–8).
Figure 1. Interrelation between folate, vitamin B6, vitamin B12 and homocysteine metabolism.
Folate is reduced to THF. A methyl group is then transferred to THF, forming 5,10-methylene-THF. 5,10-methylene-THF can be reduced by MTHFR to 5-methyl-THF. The methyl group of 5-methyl-THF is transferred to homocysteine by MS, generating methionine and regenerating THF. Vitamin B12 is a cofactor for MS. Methionine can also be generated independently of folate and B12, by the action of BHMT, which transfers a methyl group from betaine to homocysteine. Methionine is then activated to form SAM, which serves as a universal methyl donor for numerous reactions. SAH is one of the products of these methylation reactions and is subsequently hydrolyzed to generate homocysteine. Homocysteine is used either to regenerate methionine, or it is converted to cystathionine and then cysteine. BHMT, betaine homocysteine methyltransferase; DMG, dimethylglycine; MS, methionine synthase; MTHFR, methylenetetrahydrofolate reductase; SAH, S-adenosyl homocysteine; SAM, S-adenosyl methionine; THF, tetrahydrofolate.
Based on a study of elderly twins, genetic polymorphisms account for a substantial component of the heritability of B vitamin concentrations, with estimates of 56%, 59% and 66% for folate, vitamin B12 and homocysteine, respectively (9). Thus, identification of genetic variants that affect circulating levels of B vitamins can give insights into the interplay of diet, genetics and human health. Genome-wide association studies (GWASs) of serum folate, vitamin B6, vitamin B12 and homocysteine have identified and replicated several single nucleotide polymorphisms (SNPs) associated with these biomarkers (10–12). The relevant genetic variants can be used as instruments to predict biomarker concentrations and examination of their associations with disease outcomes can help strengthen causal inference (13).
With the recently increased availability of dense electronic health records (EHRs), phenomes related to a participant’s health conditions, lifestyle and environmental exposures can be characterized (14). By linking large biorepositories containing human DNA samples and EHRs, we can comprehensively evaluate associations between genetically predicted biomarkers and a wide range of phenotypes using the phenome-wide association study design (PheWAS) (15). PheWAS has been introduced as an approach to replicate associations previously identified in observational studies and detect novel genotype-phenotype associations (16). In this study, we aim to explore the phenotypic effects of B vitamins and homocysteine under the phenome-wide association framework, using genetic proxies in the UK Biobank.
Methods
Study design
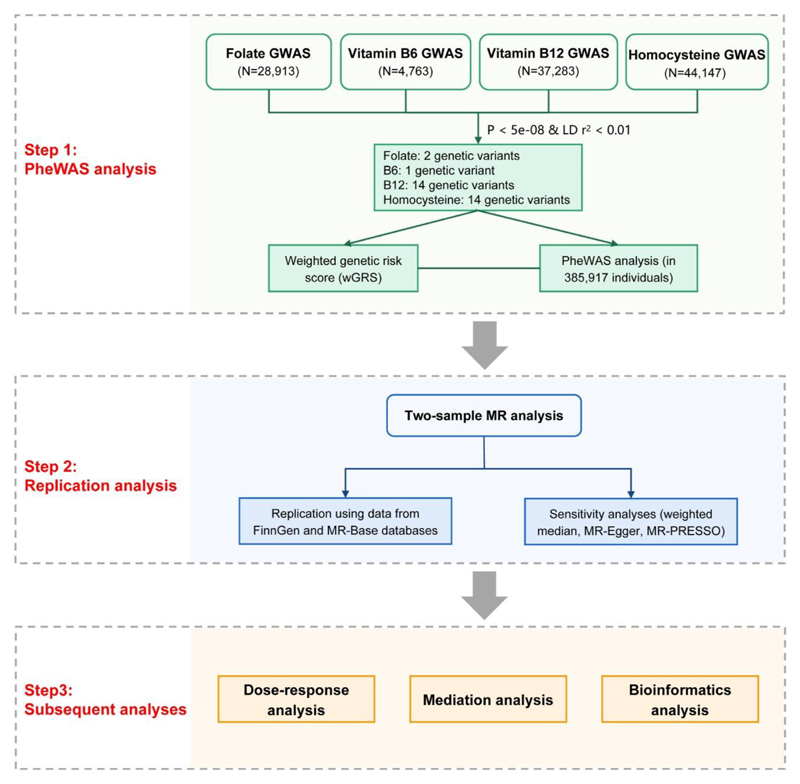
The UK Biobank genetic data contains genotypes for 488,366 participants. After quality control, a total of 385,917 individuals were retained for the subsequent analyses including PheWAS, MR, dose-response, and mediation analyses. We first performed a PheWAS analysis to investigate associations between genetic proxies of four biomarkers (folate, vitamin B6, vitamin B12 and homocysteine) and a wide range of disease outcomes. Associations with false discovery rate (FDR) less than 0.05 were considered significant. Second, the observed significant associations were replicated and regarded as successful with a threshold of Mendelian randomization (MR) p <0.05. Third, dose-response, mediation and bioinformatics analyses were conducted to characterize and quantify the associations between the biomarkers and the associated outcomes. The study design is presented in Figure 2.
Figure 2. Flowchart for the study design.
Step 1, PheWAS analysis was performed for each biomarker to investigate associations with a wide range of disease outcomes; Step 2, the observed significant associations were replicated using several MR approaches (IVW, weighted median, MR-Egger and MR-PRESSO); Step 3, dose-response, mediation and bioinformatics analyses were conducted to quantify and characterize the associations between the biomarkers and the associated outcomes.
UK Biobank
The UK Biobank is a large-scale, population-based prospective cohort study, which recruited 502,490 adult participants aged between 40 and 69 years in 2006-2010 and combined extensive measurement of baseline data and genotype data with linked wide-ranged health outcomes (17). 613,944 SNPs were genotyped by the Affymetrix UK BiLEVE Axiom array and Affymetrix UK Biobank Axiom array, and 30,798,054 SNPs were imputed based on a merged reference panel of the Haplotype Reference Consortium and the UK10K haplotype resources. A variety of health outcomes from three main different types of national medical records (e.g., in-patient hospital episode records, cancer registry and death registry) were incorporated into the UK Biobank to follow up the disease diagnosis, cancer occurrence, and causes of death among the enrolled participants. For quality control of genotype data, a list of field variables was made available by the UK Biobank to indicate the genotype quality, population structure, and genetic relatedness. In order to minimize the influence of diverse population structure within UK Biobank, quality control of samples was also conducted and those that were identified as outliers with high heterozygosity or with high missing rate, sex mismatch or putative aneuploidy in sex chromosome, non-European ancestry, or individuals with relatedness were excluded from the analysis. More details on the quality control of UK Biobank samples are given in Supplementary Figure 1.
Weighted genetic risk score
To generate weighted genetic risk scores (wGRSs) for plasma folate, vitamin B6, vitamin B12 and homocysteine, we applied a two-sample study design (exposures and outcomes are measured in non-overlapping populations) by retrieving GWAS summary data from previous published studies, aiming to increase the statistical power by incorporating multiple sources from the same ancestry. SNPs associated with plasma folate and vitamin B12 were identified from GWASs with 37,341 and 45,576 individuals of Icelandic and Danish populations (10). SNPs associated with plasma vitamin B6 and homocysteine were identified in GWASs of 4,763 and 44,147 individuals of European descent, respectively (11, 12). We selected variants that were associated with biomarkers at genome-wide significance (P < 5x10-8) and clumped them using a linkage disequilibrium (LD) threshold of r2 < 0.01 according to the European reference panel of the 1000 Genomes project. As a result, we selected two SNPs for folate, one SNP for vitamin B6, 14 SNPs for vitamin B12 and 14 SNPs for homocysteine from the external GWAS summary data, which explained 0.55% of variance for folate, 0.65% of variance for vitamin B6, 5.59% of variance for vitamin B12, and 3.22% of variance for homocysteine, respectively (Supplementary Table 1). We also calculated the F-statistic of each instrument and no weak instruments were identified (F-statistic > 10). For individuals in the UK Biobank, a wGRS for each biomarker was then calculated by adding up the number of biomarker-increasing alleles for each SNP weighted for the reported effect size of the variant.
Phenome framework and PheWAS analysis
The ontology of the phenome was defined based on the International Classification of Diseases (ICD) codes in the EHRs. Records date back to 1997 for England, 1998 for Wales and 1981 for Scotland. All of the current UK Biobank linked English and most Welsh hospital data are coded in ICD10. However, because the collection of Scottish data collection began in 1981, the Scottish data collected prior to 1997 are coded in ICD-9, and small number of Welsh records are coded with ICD-9. Thus, we included both ICD-9 and ICD-10 codes in our PheWAS analysis. Individual ICD codes could not be directly used to define the phenome, as they represent specific sub-phenotypes of a similar set of outcomes, instead of independent phenotypes. To account for the correlations between ICD codes, we define the phenome framework using the PheCODE schema that combines one or more related ICD codes into distinct outcome groups (18). For a given phenotype, the case group included patients recorded as having the specific phecode that most closely related to the etiology of the disease, and the control group was defined based on the absence of the phecode. Participants with a disease code that was related to one of the examined case group were also excluded from the control group (19). As suggested by a simulation study, outcomes with more than 200 cases were included in the analysis (20). In the PheWAS analysis, multivariable logistic regression was used to explore associations between wGRS and phecodes with adjustment for multiple covariates including sex, age, assessment center, and the first 10 genetic principal components (PCs). We also conducted sex-stratified analysis by dividing the population into male and female subgroups and re-conducted PheWAS analysis in each subgroup. The Cochran Q test and the I square metric were calculated to detect potential heterogeneity among subgroups. The Benjamini-Hochberg method was applied for each biomarker to account for multiple testing and associations with FDR <0.05 were considered statistically significant (21). The PheWAS analysis was implemented by using the “PheWAS” (19) package (R version 4.0.3).
MR analysis
To replicate and inform potential causality for biomarker-outcome associations identified in the PheWAS analysis, we carried out two-sample MR analysis in other independent European populations derived from the OpenGWAS database (22). In addition, we also examined the causal relationships between the biomarkers and some intermediate phenotypes including total cholesterol (TC), triglycerides (TG), high-density lipoprotein (HDL) cholesterol, low-density lipoprotein (LDL) cholesterol, cystatin C and creatinine. We applied the inverse variance weighted (IVW) approach as the main statistical method where at least two exposure SNPs were available, and the causal estimates were calculated by meta-analyzing SNP-specific Wald ratio estimates based on the random-effects inverse variance method that weights each ratio by its standard error (23). Where one exposure SNP was available for analysis, we used the Wald ratio method. In addition, three sensitivity analyses, including the weighted median, MR-Egger and MR-PRESSO, were applied to detect horizontal pleiotropic effects in the causal estimates when performing multivariable MR analysis (23–25). Under the assumption that the association of each genetic variant with the exposure is independent of the pleiotropic effect of the variant (not via the exposure), the MR-Egger regression can be used to detect the bias due to directional horizontal pleiotropy when conducting MR analysis using multiple genetic instruments (23). The weighted median model generates consistent estimates of causal effects if at least half of the weights come from valid SNPs (24). The MR-PRESSO method can detect outlying SNPs and provide causal estimates after removal of possible outliers under the assumption that the used SNPs are valid (25). The odds ratios (ORs) and corresponding 95% confidence intervals (CIs) of outcomes were scaled to one-standard deviation (SD) increase in genetically predicted circulating concentrations of B vitamins and homocysteine. The association with a P < 0.05 was deemed significant in the two-sample MR analysis for replication. All tests were conducted using the “TwoSampleMR” (22) and “MR-PRESSO” (25) packages in R Software 4.0.3.
Dose-response analysis
For significant PheWAS associations, we first conducted linear regression to test for any potential linear dose-response relationship, however, no significant linear association was identified. Then, we performed non-linear dose-response analysis using a restricted cubic spline function with five knots located at the 5th, 25th, 50th, 75th, and 95th percentiles of genetically predicted biomarker concentrations (26). Multivariable logistic regression models were adopted to estimate ORs with 95% CIs for the risk of outcomes, adjusted for age, sex, assessment center, and the first 10 PCs. Prediction regression implemented in “rms” R package (27) was applied to test for trends by assigning the median value for each category and modelling this variable as a continuous variable, and a P < 0.05 stands for the significance of non-linearity. A dose-response curve was used to present the dose-response relationship between the biomarker and the risk of outcome.
Mediation analysis
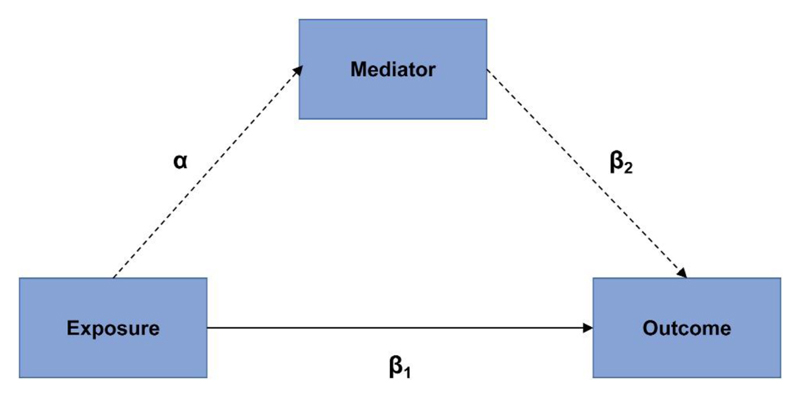
Given that B vitamins, including folate and vitamins B6 and B12, are important for the metabolism of homocysteine, abnormal levels of any of these B vitamins can lead to a change in homocysteine concentration. Therefore, to further explore any pleiotropic association derived from the correlations between B vitamins and homocysteine, we performed mediation analysis by using wGRSs as proxies for blood biomarker levels considering that data for B vitamin supplements in UKB are dietary estimates and only cover one-fifth of the population. We used genetically predicted plasma biomarker levels as the exposure and performed the mediation analysis with the “mediation” R package (28). The package performs causal mediation analysis (CMA) under the assumption of sequential ignorability, which implies that there is no unmeasured confounding of the exposure-mediator, exposure-outcome and mediator-outcome relationships while conditioning on covariates. More details regarding the rationale of mediation analysis can be found in Figure 3. The analysis reports an average causal mediation effect (ACME) that is transmitted via mediator to the outcome and an average direct effect (ADE) that is explained by the exposure as well as the proportion of explained variance by the mediator. This approach was applied to disentangle the underlying mediating pathophysiological processes.
Figure 3. Schematic diagram illustrating the rationale of mediation analysis.
Dotted lines (αβ2) refer to the indirect effect of the exposure on the identified outcome through the mediator. Solid line (β1) is the direct effect of the exposure on the identified outcome. The assumption under mediation analysis is that 1) the relationship between the exposure and the outcome is entirely mediated by the mediator; 2) the exposure is truly associated with the mediator; and 3) the exposure is not associated with any confounder of the relationship between the mediator and the outcome.
Bioinformatics analysis
For outcomes validated in MR analysis, subsequent bioinformatics analysis was carried out to explore the underlying biological mechanisms. First, to identify whether the instrumental variants exert effects through regulating the expression of located genes, we conducted expression quantitative trait loci (eQTL) analysis based on the GTEx v8 release (https://gtexportal.org/home/), which consists of normalized gene expression data in whole blood tissue. Briefly, the cis-eQTL mapping window was defined as 1 MB up- and down-stream of the transcription start site. The effect sizes were defined as the slopes of the linear regression computed as the effect of the alternate allele relative to the reference allele (29). Then, to explore the interactions between biomarker-associated genes and outcome-associated genes, we used STRING (https://string-db.org/) and Cytoscape (https://cytoscape.org/) to construct a protein-protein interaction (PPI) network using the instrumental SNPs located genes and the top ten associated genes of biomarker and outcome (30, 31). The top 10 associated genes of biomarker and outcome were searched through the GeneCards website (https://www.genecards.org/) (32). The biological processes in which these genes are involved were also explored by performing pathway enrichment analysis. The Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome were used as sources (33, 34). FDR correction was used for multiple testing and an adjusted P < 0.05 was regarded as a cutoff threshold (21).
Results
UK Biobank participants
In this study, we used the UK Biobank genetic data of 488,366 participants. Genetic quality control was done centrally by UK Biobank. A total of 385,917 unrelated European individuals were included in our final analysis, consisting of 177,690 males (46.0% of the cohort) and 208,227 females (54.0% of the cohort). Basic characteristics including socio-demographic information and biomarker concentrations are present in Table 1.
Table 1. Baseline characteristics of participants in the UK Biobank.
| Baseline characteristic | All participants (n=385917) |
|---|---|
| Sex, n (%) | |
| Female | 208227 (54.0) |
| Male | 177690 (46.0) |
| Age (years), mean (sd) | 56.7 (8.0) |
| BMI (kg/m2), mean (sd) | 27.4 (4.8) |
| Townsend deprivation index, mean (sd) | -1.5 (3.0) |
| Smoking status, n (%) | |
| Current | 40039 (10.4) |
| Former | 136651 (35.4) |
| Never | 207296 (53.7) |
| Unknown | 1931 (0.5) |
| Blood pressure (mmHg), mean (sd) | |
| Diastolic blood pressure | 82.2(10.7) |
| Systolic blood pressure | 139.8 (19.7) |
| Biomarker concentration (mmol/L), mean (sd) | |
| LDL-cholesterol | 3.6 (0.9) |
| HDL-cholesterol | 1.5 (0.4) |
| Total cholesterol | 5.7 (1.1) |
| Triglycerides | 1.7 (1.0) |
PheWAS associations
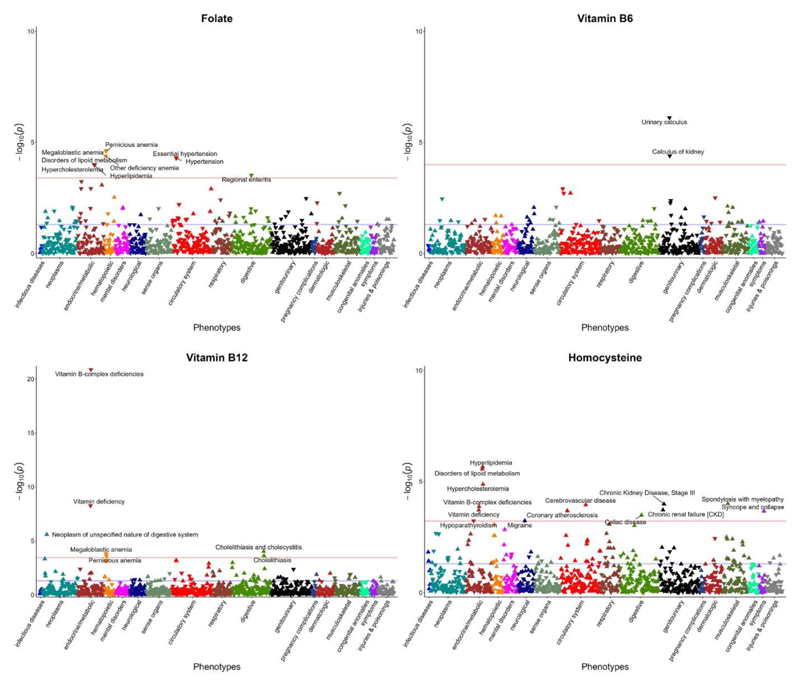
The phenome defined by PheCODE schema consisted of 1,804 distinct phecodes. After filtering out the phecodes with less than 200 cases, PheWAS analysis was performed for 1,117 phecodes that could be classified into 17 broadly related disease categories (Supplementary Table 2). Findings from PheWAS analysis indicated that higher genetically predicted plasma folate was significantly associated with lower risk of megaloblastic anemia (OR: 0.91; 95% CI: 0.87, 0.95; P=2.50×10-5), essential hypertension (OR: 0.98; 95% CI: 0.98, 0.99; P=5.01×10-5), lipid metabolism disorders including hypercholesterolemia (OR: 0.98; 95% CI: 0.97, 0.99; P=9.92×10-5) and hyperlipidemia (OR: 0.98; 95% CI: 0.97, 0.99; P=1.06×10-4), and regional enteritis (OR: 0.92; 95% CI: 0.89, 0.96; P=3.02×10-4) (Table 2 and Figure 4). Higher blood vitamin B6 was inversely associated with the risk of urinary calculus (OR: 0.95; 95% CI: 0.93, 0.97; P=7.72×10-7) and calculus of kidney (OR: 0.94; 95% CI: 0.92, 0.97; P=4.03×10-5). Higher vitamin B12 concentration was related to lower risk of vitamin B-complex deficiencies (OR: 0.47; 95% CI: 0.40, 0.55; P=1.36×10-21) and megaloblastic anemia (OR: 0.73; 95% CI: 0.62, 0.86; P=1.53×10-4), and may increase the risk of neoplasm of digestive system (OR: 1.50; 95% CI: 1.26, 1.77; P=2.54×10-6) and cholelithiasis and cholecystitis (OR: 1.10; 95% CI: 1.05, 1.16; P=9.36×10-5). As expected, higher circulating homocysteine was positively associated with the risk of vitamin B-complex deficiencies (OR: 1.57; 95% CI: 1.25, 1.98; P=1.33×10-4). This served as positive control for the genetic instrument of homocysteine. Additionally, higher homocysteine was related to higher risk of lipid metabolism disorders including hyperlipidemia (OR: 1.14; 95% CI: 1.08, 1.20; P=2.17×10-6) and hypercholesterolemia (OR: 1.13; 95% CI: 1.07, 1.19; P=1.34×10-5), spondylosis with myelopathy (OR: 3.27; 95% CI: 1.80, 5.95; P=9.99×10-5), kidney disease including chronic kidney disease (OR: 1.29; 95% CI: 1.13, 1.46; P=1.04×10-4) and renal failure (OR: 1.21; 95% CI: 1.09, 1.34; P=1.90×10-4), circulatory disease primarily cerebrovascular disease (OR: 1.21; 95% CI: 1.10, 1.33; P=1.12×10-4) and coronary atherosclerosis (OR: 1.14; 95% CI: 1.06, 1.22; P=2.07×10-4), celiac disease (OR: 1.50; 95% CI: 1.20, 1.87; P=3.27×10-4), syncope and collapse (OR: 1.19; 95% CI: 1.09, 1.31; P=2.17×10-4), celiac disease (OR: 1.50; 95% CI: 1.20, 1.87; P=3.27×10-4), and migraine (OR: 1.32; 95% CI: 1.12, 1.54; P=5.90×10-4). Sex-stratified analysis revealed considerable heterogeneity in the associations of vitamin B12 with vitamin deficiency (Pheterogeneity=0.022, I2=81%) among males and females. More specifically, per 1SD high blood vitamin B12 was associated with vitamin deficiency in males only (OR: 0.64; 95% CI: 0.55, 0.76; P=6.50×10-8) (Supplementary Table 3).
Table 2. Significant PheWAS associations by using weighted GRS in the UK Biobank (n = 385917).
| Biomarker | Phecode | Description | Group | Cases | Participants | OR (95%CI) | P-value | FDR adjusted p-value |
|---|---|---|---|---|---|---|---|---|
| Folate | 281.11 | Pernicious anemia | hematopoietic | 1144 | 355132 | 0.88 (0.83, 0.93) | 2.37E-05 | 1.13E-02 |
| 281.1 | Megaloblastic anemia | hematopoietic | 1862 | 355850 | 0.91 (0.87, 0.95) | 2.50E-05 | 1.13E-02 | |
| 281 | Other deficiency anemia | hematopoietic | 1972 | 355960 | 0.91 (0.87, 0.95) | 4.19E-05 | 1.13E-02 | |
| 401.1 | Essential hypertension | circulatory system | 109289 | 385661 | 0.98 (0.98, 0.99) | 5.01E-05 | 1.13E-02 | |
| 401 | Hypertension | circulatory system | 109545 | 385917 | 0.98 (0.98, 0.99) | 5.06E-05 | 1.13E-02 | |
| 272 | Disorders of lipid metabolism | endocrine/metabolic | 54333 | 385917 | 0.98 (0.97, 0.99) | 9.79E-05 | 1.48E-02 | |
| 272.11 | Hypercholesterolemia | endocrine/metabolic | 50143 | 381727 | 0.98 (0.97, 0.99) | 9.92E-05 | 1.48E-02 | |
| 272.1 | Hyperlipidemia | endocrine/metabolic | 54099 | 385683 | 0.98 (0.97, 0.99) | 1.06E-04 | 1.48E-02 | |
| 555.1 | Regional enteritis | digestive | 2166 | 315062 | 0.92 (0.89, 0.96) | 3.02E-04 | 3.74E-02 | |
| Vitamin B6 | 594 | Urinary calculus | genitourinary | 8724 | 385126 | 0.95 (0.93, 0.97) | 7.72E-07 | 8.62E-04 |
| 594.1 | Calculus of kidney | genitourinary | 4748 | 381150 | 0.94 (0.92, 0.97) | 4.03E-05 | 2.25E-02 | |
| Vitamin B12 | 261.2 | Vitamin B-complex deficiencies | endocrine/metabolic | 2246 | 380355 | 0.47 (0.40, 0.55) | 1.36E-21 | 1.52E-18 |
| 261 | Vitamin deficiency | endocrine/metabolic | 5055 | 383164 | 0.74 (0.67, 0.82) | 5.51E-09 | 3.08E-06 | |
| 158 | Neoplasm of unspecified nature of digestive system | neoplasms | 1540 | 362470 | 1.50 (1.26, 1.77) | 2.54E-06 | 9.44E-04 | |
| 574 | Cholelithiasis and cholecystitis | digestive | 20567 | 383100 | 1.10 (1.05, 1.16) | 9.36E-05 | 2.61E-02 | |
| 281.1 | Megaloblastic anemia | hematopoietic | 1862 | 355850 | 0.73 (0.62, 0.86) | 1.53E-04 | 3.42E-02 | |
| 574.1 | Cholelithiasis | digestive | 18324 | 380857 | 1.10 (1.05, 1.16) | 2.37E-04 | 4.01E-02 | |
| 281.11 | Pernicious anemia | hematopoietic | 1144 | 355132 | 0.68 (0.55, 0.83) | 2.51E-04 | 4.01E-02 | |
| Homocysteine | 272.1 | Hyperlipidemia | endocrine/metabolic | 54099 | 385683 | 1.14 (1.08, 1.20) | 2.17E-06 | 1.53E-03 |
| 272 | Disorders of lipid metabolism | endocrine/metabolic | 54333 | 385917 | 1.14 (1.08, 1.20) | 2.74E-06 | 1.53E-03 | |
| 272.11 | Hypercholesterolemia | endocrine/metabolic | 50143 | 381727 | 1.13 (1.07, 1.19) | 1.34E-05 | 4.98E-03 | |
| 721.2 | Spondylosis with myelopathy | musculoskeletal | 335 | 374722 | 3.27 (1.80, 5.95) | 9.99E-05 | 2.08E-02 | |
| 585.33 | Chronic Kidney Disease, Stage III | genitourinary | 7753 | 362882 | 1.29 (1.13, 1.46) | 1.04E-04 | 2.08E-02 | |
| 433 | Cerebrovascular disease | circulatory system | 14058 | 384206 | 1.21 (1.10, 1.33) | 1.12E-04 | 2.08E-02 | |
| 261.2 | Vitamin B-complex deficiencies | endocrine/metabolic | 2246 | 380355 | 1.57 (1.25, 1.98) | 1.33E-04 | 2.12E-02 | |
| 585.3 | Chronic renal failure [CKD] | genitourinary | 12777 | 367906 | 1.21 (1.09, 1.34) | 1.90E-04 | 2.20E-02 | |
| 261 | Vitamin deficiency | endocrine/metabolic | 5055 | 383164 | 1.34 (1.15, 1.57) | 1.92E-04 | 2.20E-02 | |
| 411.4 | Coronary atherosclerosis | circulatory system | 28433 | 369752 | 1.14 (1.06, 1.22) | 2.07E-04 | 2.20E-02 | |
| 788 | Syncope and collapse | symptoms | 14378 | 385917 | 1.19 (1.09, 1.31) | 2.17E-04 | 2.20E-02 | |
| 557.1 | Celiac disease | digestive | 2470 | 315366 | 1.50 (1.20, 1.87) | 3.27E-04 | 3.05E-02 | |
| 252.2 | Hypoparathyroidism | endocrine/metabolic | 355 | 379815 | 0.36 (0.20, 0.64) | 5.89E-04 | 4.71E-02 | |
| 340 | Migraine | neurological | 5026 | 376143 | 1.32 (1.12, 1.54) | 5.90E-04 | 4.71E-02 |
Figure 4. Plots for PheWAS associations of folate, vitamin B6, vitamin B12 and homocysteine (n=385917).
The x axis represents distinct phenotypic groups by using different colors, the y axis represents the p value for the phenotypic associations.
MR analysis
The MR IVW analysis revealed that genetically proxied high vitamin B6 was associated with lower risk of calculus of kidney (OR: 0.64; 95% CI: 0.42, 0.97; P=0.033) (Table 3). For homocysteine, genetically proxied high concentration was associated with a reduction in HDL cholesterol levels (OR: 0.95; 95% CI: 0.90, 1.00; P=0.041, Pheterogeneity=0.021, PMR-Egger=0.311) and higher risk of hypercholesterolemia (OR: 1.28; 95% CI: 1.04, 1.56; P=0.018, Pheterogeneity=0.046, PMR-Egger=0.695). In addition, genetically predicted high homocysteine was related to elevated creatinine levels (OR: 1.11; 95% CI: 1.01, 1.21; P=0.027, Pheterogeneity<0.001, PMR-Egger=0.437) and higher risk of chronic kidney disease (OR: 1.32; 95% CI: 1.06, 1.63: P=0.012, Pheterogeneity=0.276, PMR-Egger=0.784). All MR estimates for associations identified by PheWAS are presented in Supplementary Table 4.
Table 3. Replication of MR effect estimates in OpenGWAS database.
| Exposure | Outcome & data source | MR method | Beta | SE | OR (95%CI) | P-value1 | Intercept2 | P-value2 | Cases | Participants |
|---|---|---|---|---|---|---|---|---|---|---|
| Vitamin B6 | Calculus of kidney and ureter | |||||||||
| UKBB | PheWAS | -0.058 | 0.014 | 0.94 (0.92, 0.97) | 4.03E-05 | - | - | 4748 | 381150 | |
| OpenGWAS: finn-N14_CALCUKIDUR | Wald ratio3 | -0.453 | 0.213 | 0.64 (0.42, 0.97) | 0.033 | - | - | 3856 | 176613 | |
| Homocysteine | HDL cholesterol levels | |||||||||
| OpenGWAS: ieu-4844 | IVW | -0.055 | 0.027 | 0.95 (0.90, 1.00) | 0.041 | - | - | - | 77409 | |
| Weighted median4 | -0.061 | 0.027 | 0.94 (0.89, 0.99) | 0.022 | - | - | ||||
| MR-Egger4 | -0.065 | 0.061 | 0.94 (0.83, 1.06) | 0.311 | 0.001 | 0.853 | ||||
| MR-PRESSO4 | -0.059 | 0.026 | 0.94 (0.90, 0.99) | 0.044 | - | - | ||||
| Hypercholesterolemia | ||||||||||
| UKBB | PheWAS | 0.122 | 0.028 | 1.13 (1.07, 1.19) | 1.34E-05 | - | - | 50143 | 381727 | |
| OpenGWAS: finn-E4_HYPERCHOL | IVW | 0.245 | 0.103 | 1.28 (1.04, 1.56) | 0.018 | - | - | 6840 | 167301 | |
| Weighted median | 0.210 | 0.106 | 1.23 (1.00, 1.52) | 0.047 | - | - | ||||
| MR-Egger | 0.093 | 0.230 | 1.10 (0.70, 1.72) | 0.695 | 0.013 | 0.471 | ||||
| MR-PRESSO | 0.245 | 0.103 | 1.28 (1.04, 1.56) | 0.035 | - | - | ||||
| Creatinine levels | ||||||||||
| OpenGWAS: met-Creatinine | IVW | 0.103 | 0.046 | 1.11 (1.01, 1.21) | 0.027 | - | - | - | 110058 | |
| Weighted median | 0.013 | 0.022 | 1.01 (0.97, 1.06) | 0.553 | - | - | ||||
| MR-Egger | 0.087 | 0.108 | 1.09 (0.88, 1.35) | 0.437 | 0.001 | 0.875 | ||||
| MR-PRESSO | 0.071 | 0.031 | 1.07 (1.01, 1.14) | 0.490 | - | - | ||||
| Chronic kidney disease | ||||||||||
| UKBB | PheWAS | 0.252 | 0.065 | 1.29 (1.13, 1.46) | 1.04E-04 | - | - | 7753 | 362882 | |
| OpenGWAS: ebi-GCST008026 | IVW | 0.276 | 0.110 | 1.32 (1.06, 1.63) | 0.012 | - | - | 1533 | 20920 | |
| Weighted median | 0.134 | 0.149 | 1.14 (0.85, 1.53) | 0.368 | - | - | ||||
| MR-Egger | 0.070 | 0.250 | 1.07 (0.66, 1.75) | 0.784 | 0.017 | 0.376 | ||||
| MR-PRESSO | 0.276 | 0.110 | 1.32 (1.06, 1.63) | 0.026 | - | - | ||||
The p value of the MR effect estimates from different methods.
The Egger regression interceopt and horizontal pleiotropy p value of MR-Egger analysis.
Wald ratio method was applied to examine MR estimates for vitamin B6 as only one SNP was used as instrumental variable for vitamin B6.
These three sensitivity methods were applied to detect any pleiotropic effect when performing multivariable MR analysis.
Dose-response analysis
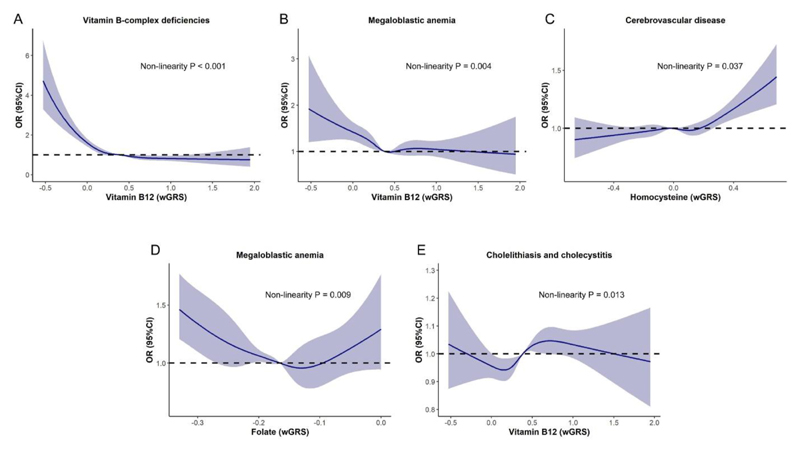
The dose-response results showed that the risk of vitamin B-complex deficiencies (P<0.001) and megaloblastic anemia (P=0.004) reduced when vitamin B12 levels increase, while the estimated ORs of developing cerebrovascular disease increased in response to the increase of homocysteine (P=0.037) (Figure 5A-C). In addition, significant U-shaped dose-response relationships were identified for the associations of folate with megaloblastic anemia (P=0.009), and vitamin B12 with cholelithiasis and cholecystitis (P=0.013) (Figure 5D, 5E).
Figure 5. Dose-response relationships between genetically predicted biomarker concentrations and the risk of outcomes identified by PheWAS (n=385917).
(A) Non-linear relationship between genetically predicted vitamin B12 concentration and vitamin-B complex deficiencies risk; (B) Non-linear relationship between genetically predicted vitamin B12 concentration and megaloblastic anemia risk; (C) Non-linear relationship between genetically predicted homocysteine concentration and cerebrovascular disease risk; (D) Non-linear relationship between genetically predicted folate concentration and megaloblastic anemia risk; (E) Non-linear relationship between genetically predicted vitamin B12 concentration and cholelithiasis and cholecystitis risk.
Mediation analysis
Considering the limited variance of folate and vitamin B6 explained by the selected genetic instruments, we only examined the mediation effects of homocysteine on the associations between vitamin B12 and the identified outcomes. The results showed that homocysteine is a potential mediator in the associations of vitamin B12 with vitamin deficiency (ACME=-1.20×10-4, P<0.001, Supplementary Table 5), vitamin B-complex deficiencies (ACME=-9.01×10-5, P<0.001), megaloblastic anemia (ACME=-6.45×10-5, P<0.001) and pernicious anemia (ACME=-5.19×10-5, P=0.020), but the mediation only accounts for 3.4%, 2.2%, 4.2% and 4.3% of the total effects, respectively.
Bioinformatics analysis
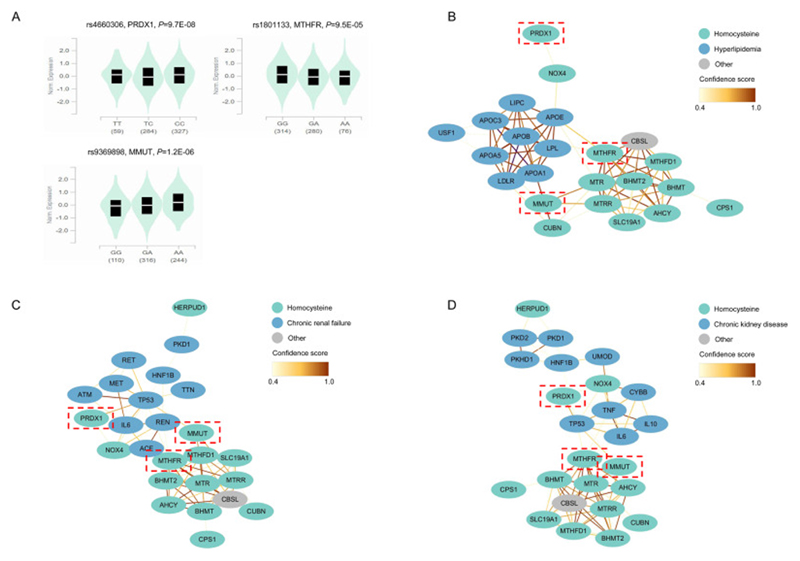
Using gene expression data from GTEx, we found that three homocysteine-associated genetic instruments including rs4660306, rs1801133 and rs9369898 were associated with the expression of PRDX1, MTHFR and MMUT (Figure 6A). The protein-protein interaction analysis identified interactions between the homocysteine-associated genes (MTHFR, MMUT but not PRDX1) and hyperlipidemia-associated genes (APOB, APOE, APOA5) (Figure 6B). Pathway analysis revealed that these genes were significantly enriched in biological processes related to cholesterol metabolism, plasma lipoprotein remodeling, assembly of active lipoprotein lipase and hepatic triacylglycerol lipase complexes (Supplementary Table 6), which play an important role in lipid metabolism, thus alterations in these pathways may lead to the development of hyperlipidemia. The protein-protein interaction results also revealed interactions between the homocysteine-associated genes (PRDX1, MTHFR, MMUT) and kidney disease-associated genes (TP53, TNF, IL-6, REN, ACE, ALB) (Figure 6C-D), which are aggregated in the renin-angiotensin system, biological oxidations, and immune-related signaling (Supplementary Table 6). These pathways are biologically relevant to the development and progression of chronic kidney diseases. These data suggest possible mechanisms of action that may underlie the observed PheWAS associations between homocysteine and hyperlipidemia and chronic kidney disease.
Figure 6. Bioinformatics analysis for homocysteine-associated instrumental variants.
(A) eQTL violin plots of the associations between homocysteine-associated SNPs and expression of located genes; (B) Diagram of the interactions between homocysteine-associated genes and hyperlipidemia-associated genes; (C) Diagram of the interactions between homocysteine-associated genes and chronic renal failure-associated genes; (D) Diagram of the interactions between homocysteine-associated genes and chronic kidney disease-associated genes.
Discussion
In this study, we comprehensively investigated the effects of B vitamins (folate, vitamin B6, vitamin B12) and homocysteine on a wide range of disease outcomes. A total of 32 pairs of genotype-phenotype associations were identified, resulting in 25 unique health outcomes belonging to several disease groups including hematopoietic (e.g., megaloblastic anemia), endocrine/metabolic (e.g., hyperlipidemia), circulatory (e.g., hypertension), genitourinary (e.g., chronic kidney disease), digestive (e.g., cholelithiasis and cholecystitis), musculoskeletal (e.g., spondylosis with myelopathy), neurological disorders (e.g. migraine), neoplasm (neoplasm of digestive system), and symptoms (e.g., syncope and collapse). Two-sample MR analysis further confirmed potential causal effects of B vitamins and homocysteine on calculus of kidney, hypercholesterolemia and chronic kidney disease. When incorporating evidence from published MR studies, the present study replicated the associations homocysteine and lipid metabolism disorders, and identified a potentially new causal association between vitamin B6 and calculus of kidney, and homocysteine and chronic kidney disease. Our findings provide evidence for further investigation on the clinical relevance of B vitamins and homocysteine with the identified disease outcomes.
Our PheWAS and MR analyses consistently demonstrated that genetically proxied high blood vitamin B6 was significantly associated with decreased risk of urinary calculus and calculus of kidney. Studies on vitamin B6 in relation to calculus are scarce and conflicting. Previous cohort studies found that the intake of vitamin B6 was not correlated with the risk of kidney stone formation among males in the Health Professionals Follow-up Study (HPFS), while large dose of vitamin B6 seemed to be protective among females in the Nurses’ Health Study (NHS) (35, 36). A recent prospective study conducted in the same cohorts with larger number of incident events (6,308 incident kidney stones) and additional follow-up time (3,108,264 person-years) showed that there was no association between vitamin B6 intake and the risk of kidney stones in either males or females (37). The present study confirmed the previous findings of a lack of association between vitamin B6 and the risk of kidney stones among females and identified that higher vitamin B6 appeared to exert a protective effect on calculus in males. In addition, our study adopted a MR approach to mitigate the impacts of residual confounding and reverse causality, and provided further supportive evidence showing that high plasma vitamin B6 was associated with lower risk of calculus of kidney. From a biological perspective, it may be related to the role of vitamin B6 in reducing urinary excretion of oxalate and thus the risk of developing calcium oxalate kidney stones (38).
For vitamin B12, this study found that higher genetically proxied plasma vitamin B12 was significantly associated with decreased risk of vitamin B-complex deficiencies and megaloblastic anemia in a dose-response manner, consistent with its known causal relationship with megaloblastic anemia (39, 40). The mediation analysis revealed that homocysteine may mediate the associations of vitamin B12 with vitamin deficiency and anemia with small effects. A possible explanation may be that low vitamin B12 results in elevated homocysteine. As one of the major pathways of homocysteine metabolism, re-methylation of homocysteine to methionine, requires vitamin B12 as the essential cofactor (41), vitamin B12 deficiency inhibits homocysteine to be methylated and thus leading to the accumulation of homocysteine, which also exerted positive effects on the development of vitamin deficiency and anemia observed in our study.
Homocysteine, a by-product of the one-carbon metabolism, has been previously associated with lipid metabolism (42, 43). In the current study, we found that higher genetically predicted plasma homocysteine was associated with a reduction in HDL cholesterol levels and increased risk of hypercholesterolemia. In addition, we conducted an overview of MR studies on the health effects of homocysteine and identified that genetically predicted high homocysteine has been reported to be related to lower HDL cholesterol and higher oxidized LDL cholesterol (44), providing support for our findings. As homocysteine is reported to reduce the concentration of HDL cholesterol in plasma by inhibiting the hepatic synthesis of apoA-I (45, 46), this is a possible mechanism linking homocysteine to the development of hypercholesterolemia. In addition, the present study revealed significant causal associations of higher genetically proxied homocysteine concentration with elevated creatinine levels (a biomarker for the evaluation of kidney function) and increased risk of chronic kidney disease. Consistently, a recent MR study revealed that higher blood homocysteine decreased estimated glomerular filtration rate (eGFR) and thus reduced kidney function (47), which may partly explain the associations between homocysteine and chronic kidney disease. Moreover, we identified that the underlying potential biological mechanisms may be related to the effects of homocysteine-associated genetic variants on the expression of genes within important pathways (e.g., biological oxidations and immune-related signaling) that are involved in the development of chronic kidney disease (48).
Folate status during pregnancy is essential for adequate fetal development and for the long-term health of the individual. Low maternal folate status may induce adverse birth/pregnancy outcomes including neural tube defects (NTD) (49, 50). Therefore, several countries (i.e. Canada and the US) have implemented public policies to fortify foods with folic acid since 1998, which resulted in the decrease of NTD prevalence (51, 52). However, due to limited data on pregnancy-related outcomes in the UK Biobank, we did not identify any relevant outcome associated with the instrumental variable for genetically predicted folate.
The present study has several strengths and limitations. We applied a rigorous study design, starting with a PheWAS analysis, based on which we investigated the relationship of genetically predicted concentrations of B vitamins and homocysteine with a wide spectrum of phenotypes. We applied additional analyses including MR, dose-response analysis, mediation analysis and pathway enrichment analysis to further elucidate the identified associations. The study also has some limitations. Our study was confined to individuals of European ancestry in order to minimize population structure bias, which hinders the generalizability of our findings to other populations. Moreover, considering that participants in the UK Biobank are exclusively from the UK, our results may not be applicable to the wider Caucasian population. Another limitation is the small variance of exposure explained by the genetic instruments, which may lead to inadequate statistical power in detecting phenotypic associations and underestimated association and mediation effects. In addition, we did not use the sex specific GWAS SNPs as instrumental variables when performing stratified analysis due to lack of sex specific GWAS data. Finally, we only incorporated EHR data for case ascertainment, and survival bias may occur since the medical records start prior to the time of cohort participation. More advanced criteria should be developed to help improve the coverage and validity of case definition for future PheWAS studies.
Conclusions
This study suggests robust associations between genetically predicted B vitamins and homocysteine concentrations with a group of disease outcomes, including hematopoietic, metabolic, circulatory, genitourinary, digestive, musculoskeletal, neurological disorders, neoplasm, and symptoms. In particular, abnormal levels of B vitamins and homocysteine could be associated with calculus of kidney, hypercholesterolemia, and chronic kidney disease. The biomarker-associated genetic variants exert their effects on the expression of genes that play important role in the development of the outcomes, which may be potential biological mechanisms underlying the identified associations.
Supplementary Material
Acknowledgements
We thank UK Biobank for their help in providing the data.
The authors’ responsibilities were as follows: ET, JL and XL conceived and designed the study; LW, ET and XL designed the methodology; LW conducted data analysis, made figures, and drafted the manuscript; All authors advised on statistical analyses and made critical revisions of the manuscript for important intellectual content. All authors have read and approved the final version of the manuscript.
Abbreviations
- ACME
average causal mediation effect
- ADE
average direct effect
- CMA
causal mediation analysis
- EHR
electronic health record
- eQTL
expression quantitative trait loci
- FDR
false discovery rate
- GWAS
genome-wide association study
- HDL
high-density lipoprotein
- ICD
International Classification of Diseases
- IVW
inverse variance weighted
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LDL
low-density lipoprotein
- MR
Mendelian randomization
- PheWAS
phenome-wide association study
- PPI
protein protein interaction
- RCT
randomized clinical trial
- SNP
single nucleotide polymorphism
- TC
total cholesterol
- TG
triglycerides
- wGRS
weighted genetic risk score
Footnotes
Conflict of interest and funding disclosure: All authors declare no competing interest. LW is supported by a Darwin Trust PhD studentship. ET is supported by a Cancer Research UK Career Development Fellowship (C31250/A22804). This work was in part supported by the Canadian Institutes of Health Research (CIHR) - Funding Reference Number: 175263.
Data availability
This research has been conducted using the UK Biobank Resource under Application Number 10775. Data are available from the UK Biobank (https://www.ukbiobank.ac.uk/) for researchers who meet the criteria and gain approvals to access the research database from the UK Biobank.
References
- 1.Bo Y, Zhu Y, Tao Y, Li X, Zhai D, Bu Y, Wan Z, Wang L, Wang Y, Yu Z. Association Between Folate and Health Outcomes: An Umbrella Review of Meta-Analyses. Front Public Health. 2020;8:550753. doi: 10.3389/fpubh.2020.550753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green R, Allen LH, Bjorke-Monsen AL, Brito A, Gueant JL, Miller JW, Molloy AM, Nexo E, Stabler S, Toh BH, et al. Vitamin B12 deficiency. Nat Rev Dis Primers. 2017;3:17040. doi: 10.1038/nrdp.2017.40. [DOI] [PubMed] [Google Scholar]
- 3.Peterson CT, Rodionov DA, Osterman AL, Peterson SN. B Vitamins and Their Role in Immune Regulation and Cancer. Nutrients. 2020;12(11) doi: 10.3390/nu12113380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith AD, Refsum H. Homocysteine, B Vitamins, and Cognitive Impairment. Annu Rev Nutr. 2016;36:211–39. doi: 10.1146/annurev-nutr-071715-050947. [DOI] [PubMed] [Google Scholar]
- 5.Ducker GS, Rabinowitz JD. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017;25(1):27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jakubowski H. Homocysteine Modification in Protein Structure/Function and Human Disease. Physiol Rev. 2019;99(1):555–604. doi: 10.1152/physrev.00003.2018. [DOI] [PubMed] [Google Scholar]
- 7.Spence JD, Yi Q, Hankey GJ. B vitamins in stroke prevention: time to reconsider. Lancet Neurol. 2017;16(9):750–60. doi: 10.1016/S1474-4422(17)30180-1. [DOI] [PubMed] [Google Scholar]
- 8.Jardine MJ, Kang A, Zoungas S, Navaneethan SD, Ninomiya T, Nigwekar SU, Gallagher MP, Cass A, Strippoli G, Perkovic V. The effect of folic acid based homocysteine lowering on cardiovascular events in people with kidney disease: systematic review and meta-analysis. BMJ. 2012;344:e3533. doi: 10.1136/bmj.e3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nilsson SE, Read S, Berg S, Johansson B. Heritabilities for fifteen routine biochemical values: findings in 215 Swedish twin pairs 82 years of age or older. Scand J Clin Lab Invest. 2009;69(5):562–9. doi: 10.1080/00365510902814646. [DOI] [PubMed] [Google Scholar]
- 10.Grarup N, Sulem P, Sandholt CH, Thorleifsson G, Ahluwalia TS, Steinthorsdottir V, Bjarnason H, Gudbjartsson DF, Magnusson OT, Sparso T, et al. Genetic architecture of vitamin B12 and folate levels uncovered applying deeply sequenced large datasets. PLoS Genet. 2013;9(6):e1003530. doi: 10.1371/journal.pgen.1003530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazra A, Kraft P, Lazarus R, Chen C, Chanock SJ, Jacques P, Selhub J, Hunter DJ. Genome-wide significant predictors of metabolites in the one-carbon metabolism pathway. Hum Mol Genet. 2009;18(23):4677–87. doi: 10.1093/hmg/ddp428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Meurs JB, Pare G, Schwartz SM, Hazra A, Tanaka T, Vermeulen SH, Cotlarciuc I, Yuan X, Malarstig A, Bandinelli S, et al. Common genetic loci influencing plasma homocysteine concentrations and their effect on risk of coronary artery disease. Am J Clin Nutr. 2013;98(3):668–76. doi: 10.3945/ajcn.112.044545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emdin CA, Khera AV, Kathiresan S. Mendelian Randomization. JAMA. 2017;318(19):1925–6. doi: 10.1001/jama.2017.17219. [DOI] [PubMed] [Google Scholar]
- 14.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012;13(6):395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 15.Bush WS, Oetjens MT, Crawford DC. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat Rev Genet. 2016;17(3):129–45. doi: 10.1038/nrg.2015.36. [DOI] [PubMed] [Google Scholar]
- 16.Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, Field JR, Pulley JM, Ramirez AH, Bowton E, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–10. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–9. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26(9):1205–10. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll RJ, Bastarache L, Denny JC. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30(16):2375–6. doi: 10.1093/bioinformatics/btu197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verma A, Bradford Y, Dudek S, Lucas AM, Verma SS, Pendergrass SA, Ritchie MD. A simulation study investigating power estimates in phenome-wide association studies. BMC Bioinformatics. 2018;19(1):120. doi: 10.1186/s12859-018-2135-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate-a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. [Google Scholar]
- 22.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018:7. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40(4):304–14. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–8. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desquilbet L, Mariotti F. Dose-response analyses using restricted cubic spline functions in public health research. Stat Med. 2010;29(9):1037–57. doi: 10.1002/sim.3841. [DOI] [PubMed] [Google Scholar]
- 27.Harrell FE, Jr, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA. 1982;247(18):2543–6. [PubMed] [Google Scholar]
- 28.Zhang Z, Zheng C, Kim C, Van Poucke S, Lin S, Lan P. Causal mediation analysis in the context of clinical research. Ann Transl Med. 2016;4(21):425. doi: 10.21037/atm.2016.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D12. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 2016;54:1301–13. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillespie M, Jassal B, Stephan R, Milacic M, Rothfels K, Senff-Ribeiro A, Griss J, Sevilla C, Matthews L, Gong C, et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022;50(D1):D687–D92. doi: 10.1093/nar/gkab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curhan GC, Willett WC, Speizer FE, Stampfer MJ. Intake of vitamins B6 and C and the risk of kidney stones in women. J Am Soc Nephrol. 1999;10(4):840–5. doi: 10.1681/ASN.V104840. [DOI] [PubMed] [Google Scholar]
- 36.Curhan GC, Willett WC, Rimm EB, Stampfer MJ. A prospective study of the intake of vitamins C and B6, and the risk of kidney stones in men. J Urol. 1996;155(6):1847–51. [PubMed] [Google Scholar]
- 37.Ferraro PM, Taylor EN, Gambaro G, Curhan GC. Vitamin B6 intake and the risk of incident kidney stones. Urolithiasis. 2018;46(3):265–70. doi: 10.1007/s00240-017-0999-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curhan GC, Taylor EN. 24-h uric acid excretion and the risk of kidney stones. Kidney Int. 2008;73(4):489–96. doi: 10.1038/sj.ki.5002708. [DOI] [PubMed] [Google Scholar]
- 39.Stabler SP. Clinical practice. Vitamin B12 deficiency. N Engl J Med. 2013;368(2):149–60. doi: 10.1056/NEJMcp1113996. [DOI] [PubMed] [Google Scholar]
- 40.Green R. Vitamin B12 deficiency from the perspective of a practicing hematologist. Blood. 2017;129(19):2603–11. doi: 10.1182/blood-2016-10-569186. [DOI] [PubMed] [Google Scholar]
- 41.Sukumar N, Saravanan P. Investigating vitamin B12 deficiency. BMJ. 2019;365:11865. doi: 10.1136/bmj.11865. [DOI] [PubMed] [Google Scholar]
- 42.Elshorbagy AK, Nurk E, Gjesdal CG, Tell GS, Ueland PM, Nygard O, Tverdal A, Vollset SE, Refsum H. Homocysteine, cysteine, and body composition in the Hordaland Homocysteine Study: does cysteine link amino acid and lipid metabolism? Am J Clin Nutr. 2008;88(3):738–46. doi: 10.1093/ajcn/88.3.738. [DOI] [PubMed] [Google Scholar]
- 43.Vanizor Kural B, Orem A, Cimsit G, Uydu HA, Yandi YE, Alver A. Plasma homocysteine and its relationships with atherothrombotic markers in psoriatic patients. Clin Chim Acta. 2003;332(1-2):23–30. doi: 10.1016/s0009-8981(03)00082-2. [DOI] [PubMed] [Google Scholar]
- 44.Zhao JV, Schooling CM. Homocysteine-reducing B vitamins and ischemic heart disease: a separate-sample Mendelian randomization analysis. Eur J Clin Nutr. 2017;71(2):267–73. doi: 10.1038/ejcn.2016.246. [DOI] [PubMed] [Google Scholar]
- 45.Barter PJ, Rye KA. Homocysteine and cardiovascular disease: is HDL the link? Circ Res. 2006;99(6):565–6. doi: 10.1161/01.RES.0000243583.39694.1f. [DOI] [PubMed] [Google Scholar]
- 46.Devlin AM, Lentz SR. ApoA-I: a missing link between homocysteine and lipid metabolism? Circ Res. 2006;98(4):431–3. doi: 10.1161/01.RES.0000214406.87060.e0. [DOI] [PubMed] [Google Scholar]
- 47.Park S, Lee S, Kim Y, Cho S, Kim K, Kim YC, Han SS, Lee H, Lee JP, Joo KW, et al. Causal Effects of Homocysteine, Folate, and Cobalamin on Kidney Function: A Mendelian Randomization Study. Nutrients. 2021;13(3) doi: 10.3390/nu13030906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalantar-Zadeh K, Jafar TH, Nitsch D, Neuen BL, Perkovic V. Chronic kidney disease. Lancet. 2021;398(10302):786–802. doi: 10.1016/S0140-6736(21)00519-5. [DOI] [PubMed] [Google Scholar]
- 49.Blom HJ, Shaw GM, den Heijer M, Finnell RH. Neural tube defects and folate: case far from closed. Nat Rev Neurosci. 2006;7(9):724–31. doi: 10.1038/nrn1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crider KS, Devine O, Hao L, Dowling NF, Li S, Molloy AM, Li Z, Zhu J, Berry RJ. Population red blood cell folate concentrations for prevention of neural tube defects: Bayesian model. BMJ. 2014;349:g4554. doi: 10.1136/bmj.g4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dietary Reference Intakes for Thiamin, Riboflavin, Niacin, Vitamin B6, Folate, Vitamin B12, Pantothenic Acid, Biotin, and Choline. Washington (DC): 1998. Edtion ed. [PubMed] [Google Scholar]
- 52.Garrett GS, Bailey LB. A public health approach for preventing neural tube defects: folic acid fortification and beyond. Ann N Y Acad Sci. 2018;1414(1):47–58. doi: 10.1111/nyas.13579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This research has been conducted using the UK Biobank Resource under Application Number 10775. Data are available from the UK Biobank (https://www.ukbiobank.ac.uk/) for researchers who meet the criteria and gain approvals to access the research database from the UK Biobank.