Abstract
Species belonging to the order Ranunculales have attracted much attention because of their phylogenetic position as a sister group to all other eudicot lineages and their ability to produce unique yet diverse benzylisoquinoline alkaloids (BIAs). The Papaveraceae family in Ranunculales is often used as a model system for studying BIA biosynthesis. Here, we report the chromosome-level genome assembly of Corydalis tomentella, a species of Fumarioideae—one of the two subfamilies of Papaveraceae. Based on the comparisons of sequenced Ranunculalean species, we present clear evidence of a shared whole-genome duplication (WGD) event that has occurred before the divergence of Ranunculales but after its divergence from other eudicot lineages. The C. tomentella genome enabled us to integrate isotopic labelling and comparative genomics to reconstruct the BIA biosynthetic pathway for both sanguinarine biosynthesis shared by papaveraceous species and the cavidine biosynthesis specific to Corydalis. Also, our comparative analysis revealed that gene duplications, especially tandem gene duplications, underlie the diversification of BIA biosynthetic pathways in Ranunculales. In particular, tandemly duplicated berberine bridge enzyme-like genes appear to be involved in cavidine biosynthesis. In conclusion, our study of the C. tomentella genome provides important insights into the occurrence of WGDs during the early evolution of eudicots as well as into the evolution of BIA biosynthesis in Ranunculales.
Keywords: benzylisoquinoline alkaloids, Papaveraceae, Corydalis tomentella, whole-genome duplication, evolution
Introduction
Within eudicots, the order Ranunculales forms a sister group to all other extant eudicot lineages (Group, 2016), holding an important phylogenetic position for understanding the evolution and diversification of eudicots. In addition, Ranunculales is an order of significant pharmaceutical importance because of its unique biosynthesis of benzylisoquinoline alkaloid (BIA) compounds. To date, more than 2,500 BIAs have been identified in species of the Ranunculaceae, Papaveraceae, Berberidaceae, and Menispermaceae families (Liscombe et al., 2005; Hao, 2018). Many well-known BIAs have medicinal value, such as the antitumor activity of noscapine from Papaver somniferum; analgesic activity of morphine and codeine from P. somniferum; and antibacterial activity of sanguinarine, berberine, and palmatine from most Papaveraceae species (Shamma, 2012).
To study BIA biosynthesis, opium poppy (P. somniferum) has long served as a model system. The complete biosynthetic pathways of some well-known BIAs, including morphine, noscapine, and sanguinarine, have been decoded using the opium poppy system (Hagel and Facchini, 2010; Winzer et al., 2012; Liu et al., 2017). In addition, the P. somniferum genome has revealed the critical roles of gene duplications and gene clusters in the evolution of BIA genes and alkaloid production (Guo et al., 2018; Li et al., 2020a). However, the focus on opium poppy can only offer a limited view of the evolution of BIA biosynthetic pathways in Papaveraceae. In fact, the poppy family Papaveraceae comprises two subfamilies, namely Papaveroideae and Fumarioideae, which diverged from each other around 87.4 million years ago (Valtuena et al., 2012). Although noscapine, morphine, and codeine production is unique to certain species of the Papaver genus of Papaveroideae, species in both Papaveroideae and Fumarioideae can biosynthesize sanguinarine (Li et al., 2020b), and its pathway is still unclear. In addition, Fumarioideae species have evolved certain independent BIA biosynthetic pathways, which differ from the ones known in Papaveroideae (Xu et al., 2021), offering a system to understand the diversification of BIA biosynthetic pathways along with the divergence of lineages. Furthermore, the sequenced Ranunculalean genomes of P. somniferum (Guo et al., 2018), Eschscholzia californica (Hori et al., 2018), and Macleaya cordata (Liu et al., 2017) are all from the subfamily Papaveroideae. Hence, genome sequencing of species from the subfamily Fumarioideae of Papaveraceae is imperative to study the evolution of BIA biosynthesis in Ranunculales and Papaveraceae.
The insights gained by analyzing the diversification of BIA biosynthesis in Papaveraceae may not only shed light on the evolution of plant secondary metabolic pathways but also assist enzymatic and metabolic engineering for industrial BIA biosynthesis. Specifically, although the microbial production of several BIAs has been successfully implemented (Minami et al., 2008; Galanie et al., 2015; Li et al., 2018; Courdavault et al., 2020), the current yield cannot meet the requirements of industrial production; therefore, potential BIA biosynthetic genes in the species of Papaveraceae, specifically Fumarioideae, may be important to assist enzymatic and metabolic engineering for industrial BIA biosynthesis.
Here, we report the genome of Corydalis tomentella, which is the first genome of the subfamily Fumarioideae of the poppy family. The subfamily Fumarioideae of Papaveraceae comprises over 570 species in 20 genera (Perez-Gutierrez et al., 2012). Corydalis is the largest genus within Fumarioideae, with 465 species native to the Northern Hemisphere and South Africa. Approximately 80% Corydalis species are distributed in China, mainly in the Hengduan Mountains and the Qinghai-Tibet Plateau. Species of the genus occur at various altitudes and in diverse habitats, such as forest margins, wet meadows, wastelands, rock crevices, and even dry and rocky limestone cliffs. Corydalis has undergone reticulate evolution and intensive differentiation (Jiang et al., 2018; Ren et al., 2019), resulting in remarkable variability in morphology, even within the same species, which hinders species identification. The Corydalis genome will provide critical insights for the use of medicinal resources and study of herbgenomics (Xin et al., 2019; Xu et al., 2020).
Similar to other species in the poppy family, Corydalis produces sanguinarine, in addition to a class of anti-inflammatory BIA compounds called cavidines (e.g., cavidine, apocavidine, dehydroapocavidine, and dehydrocavidine) (Bhakuni and Chaturvedi, 1983). As rich sources of these unique BIA compounds, some Corydalis species are widely used in traditional Chinese medicine because of their antibacterial, antiviral, and anticancer activities (Zhang et al., 2016; Liu et al., 2019; Tian et al., 2020). For instance, the tuber of Corydalis yanhusuo and the whole plant of Corydalis bungeana have been noted in the Chinese Pharmacopoeia for their medicinal usage in invigorating blood circulation and analgesic effects (Committee;, 2015). Furthermore, the dry herbs of C. tomentella and C. saxicola are used in traditional Chinese medicine to alleviate fever and hepatitis.
The sequenced C. tomentella genome is crucial for comprehensively investigating the biosynthetic pathways of sanguinarine and other BIAs through comparisons between the two subfamilies of Papaveraceae as well as between the Papaveraceae and other Ranunculalean species. In addition, the C. tomentella genome allows us to examine the origin of cavidine biosynthetic pathway in the Corydalis lineage. Here, using isotope tracking, metabolite profiling, and gene expression analyses, we dissected the biosynthetic pathways of active BIAs in C. tomentella. Furthermore, we identified the gene clusters and tandem duplication events involved in the divergent evolution of BIA biosynthesis through comparative genomics.
Results
Genome assembly and annotation
Genome of the medicinal plant C. tomentella exhibits low heterozygosity (about 0.3%). The genome size is 258.56 Mb based on the frequency distribution of 21 k-mer (Figure S1) based on 33.66 Gb Illumina sequencing reads (Table S1-2). We generated 27.64 Gb (~107 × coverage) raw data using the third-generation Sequel sequencing platform (Table S3). The N50 length of filtered subreads (4,077,968) was 9.63 Kb. The error-corrected raw reads were pre-assembled into seed sequence using hierarchical genome assembly, and the seed reads from different hierarchical clusters with an average length of 14.37 Kb were further assembled and polished using FALCON. The polished assembly contained 1,321 contigs with the contig N50 length of 2.36 Mb. After removing heterozygous contigs using Redundans, the final assembly comprised 1,022 contigs; the genome size was 248.9 Mb, the N50 length was 2.52 Mb, covering 96.26% of the estimated nuclear genome size; the longest contig was 9.83Mb; the GC content was 36.79%. Then, a library of Chromosome conformation capture techniques (Hi-C) was constructed, and 39.67 Gb sequencing reads were produced for scaffolding. A total of 953 contigs, covering 248.6 Mb (99.9%) of the assembled genome, were anchored to eight pseudochromosomes (2n=16) (Figure 1A, Figure S2, Table S4-5). Moreover, 36 Gb RNA-Seq data from roots, stems, leaves, and flowers of C. tomentella were aligned to the assembled genome at an average mapping rate of 95.34% (Table S2). De novo assembled transcripts from the RNA-Seq reads were aligned to the C. tomentella genome, and 82.65% reads aligned with at least 90% coverage and 90% identity. In addition, 1,334 (97.67%) of the 1,375 embryophyta single-copy orthologs from BUSCO were identified ‘as complete’ in the C. tomentella genome (Simao et al., 2015), suggesting that the genome assembly was of high quality (Table S4).
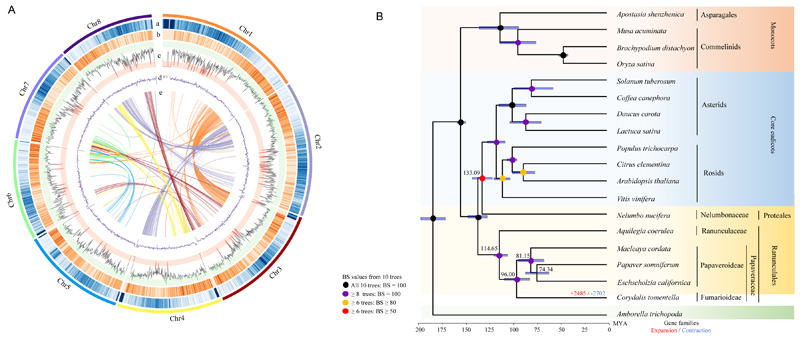
Figure 1. Genomic features and phylogenetic position of C. tomentella.
A. Chromosome level assembly of the C. tomentella genome. (a) transposable elements in 100 kb windows, (b) gene density in 100 kb windows, (c) gene expression level estimated from read counts per million mapped reads in 100 kb windows, (d) GC content in 100 kb windows, and (e) synteny blocks of paralogous sequences of C. tomentella genome. B. A phylogenetic tree showing the topology and divergence times for 19 plant species. The different circles signify the bootstrap support (BS) values from 10 phylogenetic trees based on the different gene sets. Numbers at the branch leading to C. tomentella indicate the expansion and contraction of gene families. Blue bars at the internodes indicate 95% confidence intervals of estimated divergence time.
Approximately 44.24% (110,107,533 bp) of the C. tomentella genome was annotated as transposable elements (TEs) (see Materials and Methods and Table S6), similar to that in the Macleaya cordata genome (43.5%) also from Papaveraceae. Of the TEs, 19.37% were long terminal repeat (LTR) retrotransposons. A total of 81,630 LTR elements were identified, of which 22,370 (6.05%) belonged to the Gypsy superfamily and 13,905 (3.99%) to the Copia superfamily. A total of 136,330 simple sequence repeats (SSRs) were annotated (see Materials and Methods), providing valuable molecular markers for future genetic diversity studies of C. tomentella (Table S7). We confidently predicted 37,808 protein-coding genes by integrating ab initio gene predictions, homologous protein searches, and de novo assembled transcripts from the RNA-Seq reads (see Materials and Methods). Complete orthologs were identified for 94.8% of the embryophyta BUSCOs, indicating that the predicted protein-coding genes were largely complete (Table S4).
Phylogenomic analysis and dating
We used OrthoFinder to delineate orthologous groups of proteomes from 19 angiosperms and obtained 21,299 orthologous groups covering 497,631 genes. Here, we selected 10 different gene sets, with 78 single-copy gene families, and 120, 259, 350, 424, 540, 693, 749, 850, and 1,714 low-copy gene families, to infer a high-confidence species phylogeny using Amborella trichopoda as an outgroup (see Materials and Methods). The final phylogenetic relationships of the candidate species using the different gene sets were consistent with the Angiosperm Phylogeny Group IV botanical classification system. The phylogenetic analysis, as expected, revealed that C. tomentella from Fumarioideae is sister to all sequenced Papaveroideae species, including M. cordata, P. somniferum, and E. californica (Figure 1B). Molecular dating using nucleotide sequences of the 78 single-copy genes and four fossil age calibrations indicated that the two subfamilies of Papaveraceae, that is Fumarioideae and Papaveroideae, diverged approximately 96.00 million years ago (MYA), with a 95% confidence interval (CI) of 82.65 to 109.09 MYA (see Materials and Methods). The split between Papaveraceae and Ranunculaceae was dated to approximately 114.65 MYA, with a 95% CI of 106.15 to 120.28 MYA.
Whole-genome duplications (WGDs) in the Ranunculales
Intragenomic colinearity analysis revealed remnants of one WGD event in the C. tomentella genome (Figure S3). Intergenomic co-linearity analyses between C. tomentella and Vitis vinifera, Aquilegia coerulea, and Nelumbo nucifera supported the identified WGD event. For instance, two paralogous segments in the C. tomentella genome correspond to three, two, and two orthologous regions in the V. vinifera, A. coerulea, and N. nucifera genomes, respectively (Figure S4). Moreover, the identified WGD event was supported by chromosome printing of the C. tomentella genome with the ancestral eudicot karyotype (AEK) pre-γ chromosomes (Murat et al., 2017) (Figure 2A). In addition, distributions of synonymous substitutions per synonymous site (KS) for all paralogous genes and for paralogous genes in the collinear regions of C. tomentella showed a clear peak at KS ≈ 1.04, indicative of a WGD event (Figure 2B and Figure S5). Similarly, previously sequenced genomes of Ranunculales species, including A. coerulea, M. cordata, and E. californica, showed a signature peak for a WGD event in their genomes, albeit with different KS peak values (Figure S5), while the P. somniferum genome showed two KS peaks, including a recent major peak (KS ≈ 0.1) and a more ancient minor peak (KS ≈ 1.5) (Guo et al., 2018).
Figure 2. Synteny and KS analysis of the C. tomentella WGD.
A. Synteny comparison between the C. tomentella genome and ancestral eudicot karyotype (AEK) pre-γ chromosomes. Two paralogous blocks in C. tomentella chromosomes are shown in shared colors but different filling. B. KS distributions of anchor pairs for the paralogous genes of C. tomentella, M. cordata, A. coerulea, and V. vinifera, and for the one-to-one orthologs between C. tomentella and M. cordata, A. coerulea, and V. vinifera, respectively. C. Phylogeny of C. tomentella and other species with branch lengths in KS units. Different blocks represent the inferring of WGD or WGT events based on KS values of paralogs.
The various KS peak values for the WGD signatures of different published Ranunculalean genomes sparked a debate on whether Ranunculalean species share an ancient Ranunculalean-specific WGD (Guo et al., 2018), or whether the observed WGD is shared by all eudicots and contributes to the hexaploidization event (referred to as gamma) that all core eudicots share (Aköz and Nordberg, 2019). Analysis of the A. coerulea genome suggested a model of hybridization for the origin of core eudicots, including a pan-eudicot WGD (tetraploidization) followed by a hybridization forming the hexaploid common ancestor of the core eudicots (Aköz and Nordberg, 2019). However, the proposed scenario contradicts the results of the analysis of the Nelumbo genome and other Ranunculalean genomes, in which the identified WGDs were inferred to have occurred after the divergence of the core eudicots and Nelumbo (Ming et al. 2013; Shi and Chen, 2020) or even after the divergence of Ranunculales (Guo et al., 2018). Interestingly, comparing with the peak in the KS age distribution of one-to-one orthologs between C. tomentella and V. vitis revealed a smaller KS value of peak identified in the C. tomentella genome (i.e., younger age) but a larger value of peak identified in the A. coerulea genome (i.e., older age, Figure 2B), seemingly reflecting different WGD scenarios.
To resolve the various KS values for the WGD peaks in different Ranunculalean species, we argue that these differences are due largely to the variability in substitution rates among the Ranunculalean lineages. Indeed, comparison of one-to-one orthologous KS distributions between V. vinifera and sequenced Ranunculalean species (C. tomentella, M. cordata, P. somniferum, E. californica, and A. coerulea) revealed some differences in orthologous KS peaks, suggesting that the studied Ranunculaclean species have evolved at different substitution rates (Figure S6). Indeed, because all orthologous KS peaks reflect the divergence between Ranunculales and core eudicots (represented by Vitis), these lineages should have similar KS values if the substitution rates are similar. Therefore, crude comparisons of paralogous and orthologous KS distributions, regardless of the differences in substation rates, do not necessarily reflect the exact timing of WGD events.
To more carefully date the identified WGD events in the Ranunculalean genomes, considering the various substitution rates observed for different Ranunculales species, we used the 78 single-copy gene families to infer branch lengths in KS units on the species phylogeny (see Materials and Methods). The investigated Ranunculalean species presented different branch lengths following divergence, which further supports the various substitution rates of these lineages (Figure 2C). By mapping the different KS values for WGD peaks identified in the Ranunculalean and Protealean species on the phylogeny (see Materials and Methods), we inferred that the ancient WGD in P. somniferum and the WGDs in C. tomentella, M. cordata, and A. coerulea occurred on the stem branch of Ranunculales around the same time, suggesting a shared WGD event among all Ranunculalean species. Notably, based on our results, the California poppy (E. californica) genome has undergone two WGD events—an ancient event shared by all Ranunculalean species and a recent lineage-specific WGD event—similar to the poppy (P. somniferum) genome (Figure 2C). However, because of the more recent WGDs and the higher substitution rate of the sequenced Ranunculalean species (Figure S6), the ancient Ranunculalean WGD event only has a (very) vague remnant in the paralogous KS distributions of E. californica and P. somniferum.
In addition, both the WGD in Nelumbo and the hexaploidization in Vitis are assumed to have occurred after their divergence from Ranunculales (Figure 2C). Hence, our results support a scenario describing independent paleopolyploidizations after the divergence of Ranunculales, Proteales, and core eudicots. Nevertheless, the branch leading to the divergence between Proteales and the core eudicots is extremely short, with a KS of 0.0034, while the WGD events in Vitis and Ranunculales are close to the early speciation events of eudicots (Figure 2C) Therefore, we cannot completely rule out other scenarios involving more complicated hybridizations (Kellogg, 2016) or lineage-specific rediploidization (Robertson et al., 2017).
BIA accumulation in C. tomentella
BIAs are enriched in different C. tomentella tissues. The contents of different BIAs in roots, stems, leaves, and flowers were spectrophotometrically quantified. Obvious chromatographic peaks were observed at retention times of 25.74, 36.33, and 40.01 min, which corresponded to dehydrocheilanthifoline, coptisine, and dehydrocavidine, respectively (Figure S7 – 9). The key intermediate compound, dehydrocheilanthifoline, shows relatively higher accumulation in flowers than in other organs. Specific accumulation of coptisine in flowers was detected, suggesting the involvement of flower-specific oxidoreductases in coptisine biosynthesis. The biomarker compound dehydrocavidine was mainly accumulated in the roots, stems, and flowers, exhibiting the highest content among all tested alkaloids of C. tomentella (Figure S7).
Cavidines, including dehydroapocavidine and dehydrocavidine, are exclusively present in Corydalis species. We employed an integrative strategy to investigate cavidine biosynthesis in C. tomentella. First, to identify metabolites involved in cavidine biosynthesis in C. tomentella, we employed the isotopic tracer method by feeding the 13C6 (benzene-ring)-labeled tyrosine into the culture medium of C. tomentella. Using liquid chromatography-mass spectrometry (LC-MS), we identified 21 metabolites involved in BIA biosynthesis in C. tomentella (Figure S10 – 11). According to a known BIA biosynthetic process (Figure 3), we missed two metabolites, namely 4-hydroxyphenylpyruvate and tetrahydropalmatine. Given the detection of their derivates, we assume that they are present in C. tomentella but were probably missed due to their extremely low accumulation.
Figure 3.
The proposed biosynthetic pathway of BIAs in C. tomentella. The compounds, labeled by the ring-13C6, were identified by LC/MS. M+6 and M+12 represent one labeled ring and two labeled rings, respectively. The brown lines represent the scoulerine biosynthesis, conserved upstream biosynthesis of BIAs in Ranunculales species, the green lines show the biosynthesis of columbamine and palmatine, also accumulated in many Ranunculales species, the blue lines reflect the specific accumulation of cavidines in Corydalis, and the red line indicates the sanguinarine biosynthesis in most Papaveraceae species. TYDC: Tyrosine decarboxylase, TYR: tyramine 3-hydroxylase, TAT tyrosine aminotransferase, HPPDC 4-hydroxyphenylpyruvate decarboxylase, NCS: norcoclaurine synthase, 6OMT: norcoclaurine 6-O-methyltransferase, CNMT: coclaurine N-methyltransferase, NMCH: N-methylcoclaurine 3’-hydroxylase, 4’OMT: 3’-hydroxy-N-methylcoclaurine 4’-O-methyltransferase, BBE: berberine bridge enzyme, 9OMT: scoulerine 9-O-methyltransferase, TNMT: tetrahydroprotoberberine N-methyltransferase, MSH: methylstylopine hydroxylase, P6H: protopine 6-hydroxylase, OMT: O-methyltransferase, CMT: C-methyltransferase, BBEL: berberine bridge enzyme-like. The red triangles represent undetected compounds via LC-MS analysis.
Next, to identify the BIA genes involved in cavidine biosynthesis, we used BIA genes in the poppy genome to identify the ones in the upstream of BIA biosynthesis in C. tomentella; most of the BIA biosynthetic genes, except those involved in Corydalis-specific cavidine biosynthesis, have been identified in the P. somniferum genome (see the brown and green flows in Figure 3). We then compared the BIA genes from A. coerulea, C. tomentella, M. cordata, and P. somniferum in Ranuncluales and N. nucifera in Proteales (Table S8), to understand the evolution of BIAs in Ranunculales and aid the identification of cavidine-specific BIA genes in C. tomentella. By integrating chemical compound structural analyses and comparative genomics, we propose a potential biosynthetic pathway of the cavidines in C. tomentella (see the blue flow Figure 3).
Comparative genomics of BIA genes upstream of cavidine biosynthesis
Starting from L-tyrosine, (S)-Norcoclaurine (NOR) is a central intermediate metabolite of BIA biosynthesis, which is present in all analyzed Ranunculalean species as well as in N. nucifera (Figure 4A). NOR is synthesized from two L-tyrosine products, dopamine and 4-hydroxyphenylacetadehyde, by the action of norcoclaurine synthase (NCS); the genes encoding NCS are conserved in all the Ranunculalean species and N. nucifera. Ranunculalean species harbor more NCS genes than N. nucifera (four in N. nucifera versus nine in C. tomentella to 58 in P. somniferum; Table S8). Interestingly, a gene cluster including seven NCS genes, which has originated through tandem duplication, was identified in the C. tomentella genome, and collinearity analysis revealed the corresponding syntenic blocks, including five NCS genes in M. cordata, one NCS gene in A. coerulea, and one NCS gene in N. nucifera, suggesting tandem duplications played a key role in the expansion of NCS genes following the divergence of Ranunculales and Proteales (Figure S12, S13).
Figure 4. Tandem gene duplications of CtBBEL involved in the biosynthesis of cavidines and coptisine.
A. The distributions of NOR (norcoclaurine), RET (reticuline), CHE (cheilanthifoline), SAN (sanguinarine), DHCH (dehydrocheilanthifoline), CAV (cavidine), DHCA (dehydrocavidine), STY (stylopine), COP (coptisine), MOR (morphine), and NOS (noscapine) in N. nucifera, A. coerulea, M. cordata, P. somniferum, and C. tomentella. The collinearity analysis identified syntenic blocks containing the tandem duplications of BBEL genes in C. tomentella. Two syntenic blocks in N. nucifera and P. somniferum, respectively, originated from their lineage-specific WGD event. The hollow blocks with different colors, which were linked by dotted lines, represent the syntenic blocks of non-BIA genes among candidate species. B. Phylogenetic tree constructed for BBEL genes from the syntenic blocks of M. cordata, P. somniferum, and C. tomentella with the CtBBE gene as outgroup. C. Gene expression profile of CtBBEL genes from the syntenic block and CtBBE gene in different tissues of C. tomentella, including root, flower, stem, and leaf. D. In vivo catalytic assays of CtBBEL8, CtBBEL31, and CtBBEL22 in Sf9 insect cells using cheilanthifoline, cavidine, and stylopine as substrates, respectively. E. Proposed biosynthetic pathway of cavidines and coptisine from cheilanthifoline in C. tomentella. The red full lines represent the identified steps in this study.
After NOR, a serial of methyltransferases, including 6-O-methyltransferase (6OMT), coclaurine N-methyltransferase (CNMT), N-methylcoclaurine hydroxylase (NMCH), and 4-O-methyltransferase (4’OMT), catalyze the biosynthesis of (S)-reticuline (RET). Gene expression profiling analysis revealed that the RET biosynthetic homologs of NCS, 6OMT, CNMT, NMCH, and 4’OMT were highly expressed in the roots, stems, leaves, and flowers of C. tomentella (Figure S12, Table S9). All studied Ranunculalean species can synthesize reticuline, whereas N. nucifera cannot. Consistently, our phylogenetic analysis showed that the orthologs encoding 6OMT, CNMT, and NMCH are present in both Ranunculales and Proteales, while the orthologs of 4’OMT are unique to the former order (Figure S14 – 16). In P. somniferum, C. tomentella, and M. cordata genomes, the homologous of 6OMT, CNMT, NMCH and 4’OMT are present at different locations (Figure S12). However, they appear in more or less similar genomic contexts as the MRCA of Ranunculales or even that of eudicots, as we observed collinear blocks for 6OMT among C. tomentella, M. cordata, and A. coerulea (Figure S14); for CNMT among C. tomentella, M. cordata, P. somniferum, A. coerulea, and N. nucifera (Figure S15); and for NMCH among C. tomentella, M. cordata, P. somniferum, and A. coerulea (Figure S14). Sequence alignments using Ranunculalean WGD-derived paralogs showed that the TYDC and TAT genes in P. somniferum as well as CNMT in C. tomentella, A. coerulea, and M. cordata were duplicated and underwent Ranunculalean WGD event.
Cheilanthifoline (CHE) biosynthesis involves the berberine bridge enzyme (BBE) for the conversion of RET to into (S)-scoulerine, which is further catalyzed by CYP719 members, such as CYP719A14 and CYP719A2, to (S)-cheilanthifoline and (S)-stylopine (Figure 3). Both (S)-cheilanthifoline and (S)-stylopine are the common intermediates of sanguinarine, and cheilanthifoline is present in all studied Ranunculalean species (Figure 4A). Four CYP719 subfamily genes were identified in the C. tomentella genome (Table S8). Furthermore, we detected collinear regions containing CYP719 genes encoding cheilanthifoline synthase (CYP719A14 or CHS) and stylopine synthase (CYP719A2 or STS) among the C. tomentella, M. cordata, and P. somniferum genomes, suggesting that the biosynthesis of the two common intermediates is conserved in Papaveraceae (Figure S17). Interestingly, the CHS and STS genes are adjacent in Papaveraceae genomes, indicating that they were formed through a tandem duplication event. These results, together with phylogenetic findings, indicated that tandem duplication must have occurred before the divergence of Ranunculales. Further, although A. coerulea possesses seven paralogs of CHS genes, it does not harbor the orthologs of STS related to sanguinarine biosynthesis. Similarly, N. nucifera lacks cheilanthifoline and stylopine because of the absence of CYP719A members (Table S8).
Sanguinarine is widely distributed in all the Papaveraceae species (Figrue 4A), and two CYP450 members from the CYP82N subfamily, encoding (S)-N-methylstylopine hydroxylase (MSH) and (S)-protopine-6-hydroxylase (P6H), form a critical step catalyzing the production of sanguinarine, which have been reported in both P. somniferum and M. cordata (Liu et al., 2017). A gene cluster including one SDR gene, one BBE-like (BBEL) gene, three OMT genes, 12 CYP450 genes, and one 2OGD gene was identified in the C. tomentella genome; among these, the CYP450 cluster contained the MSH and NMCH genes (Figure S12). Genome synteny analysis revealed that the gene cluster in C. tomentella well aligned with the gene cluster in M. cordata (Figure S18), suggesting that the upstream biosynthetic genes of sanguinarine are conserved in Papaveraceae. In addition, MSH and P6H were created by gene duplications before the divergence of Papaveraceae (Figure S17); therefore, N. nucifera and A. coerulea lack these genes (Table S8), consistent with the specific emergence of sanguinarine biosynthesis in Papaveraceae.
Finally, noscapine biosynthesis is unique to Papaver (Figure 4A). Specifically, in P. somniferum, CYP82Y1, CYP82X2, and CYP82X1 catalyze the formation of hydroxy products of (S)-N-methylcanadine. The opium poppy genome harbors 11 CYP82X/Y genes (Table S8), which are sister to the MSH (CYP82N) genes. In the phylogenetic tree, a duplication event before the divergence of Papaveraceae formed two clades of these genes (Figure S19). In the opium poppy genome, PsCYP82N4 (PsMSH), PsCYP82X1, PsCYP82X2, PsCYP82Y1, and PsCYP82Y2 are located in a 569 Kb gene cluster (Figure S19). Collinearity analysis of this CYP82 gene cluster revealed that the two collinearity regions in C. tomentella and M. cordata genomes only contain the orthologous of CYP82N4 and have lost the orthologs of PsCYP82X1, PsCYP82X2, PsCYP82Y1, and PsCYP82Y2 genes related to noscopine biosynthesis. In addition, these syntenic blocks in N. nucifera and A. coerulea genomes are completely lost (Table S8). These results indicate that only one of the duplicates was retained in the Papaver lineage, followed by a series of lineage-specific gene duplications (Figure S19).
Origin of cavidine biosynthesis in Corydalis
BBEL enzymes, a subgroup of the superfamily of FAD-linked oxidases, are conserved in bacteria, fungi, and plants (Daniel et al., 2017). The BBE and BBEL genes catalyze two- and four-electron oxidation steps in the BIA biosynthesis, such as the conversion of the central intermediate RET to (S)-scoulerine in all Ranunculaleans, and the conversion of (S)-tetrahydrocolumbamin to columbamine in Papaver (Figure 3). Therefore, we speculate that in cavidine biosynthesis, the four-electron oxidation steps from (S)-cheilanthifoline to dehydrocheilanthifoline, from apocavidine to dehydroapocavidine, from cavidine to dehydrocavidine, and from (S)-stylopine to coptisine are catalyzed by BBEL enzymes (Figure 3). Respectively 16, 17, 34, 42, and 43 BBEL genes were identified in the N. nucifera, A. coerulea, C. tomentella, M. cordata, and P. somniferum genomes (Table S8), suggesting the expansion of BBEL genes in Papaveraceae species.
Additionally, we found the largest cluster of 25 Corydalis BBEL genes originating from tandem duplications among all the compared genomes (Figure S12). Collinearity analysis of the orthologous regions of this cluster among the N. nucifera, A. coerulea, M. cordata, and P. somniferum genomes revealed that the N. nucifera and A. coerulea genomes lack BBEL genes, while the M. cordata and P. somniferum genomes harbor only four BBEL genes in collinear regions (Figure 4A). The expression profiles of the BBEL genes in the cluster were also in consistent with the accumulation of coptisine and cavidines in C. tomentella, with the specific expression of CtBBEL22 in the flower and of CtBBEL31 in all the investigated tissues (Figure 4C). In the phylogenetic tree, CtBBEL8 and CtBBEL31 clustered on the Corydalis-specific BBEL branch (Figure 4B), and the expression of these two genes in all tested tissues (FPKM per sample > 5) was relatively higher than that of other genes from the tandem gene cluster of Corydalis after its divergence with P. somniferum.
To further confirm the catalytic activities of BBEL enzymes in cavidine and coptisine biosynthesis, we successfully expressed three BBEL proteins (CtBBEL8, CtBBEL22, and CtBBEL31) from C. tomentella in Sf9 insect cells (see Materials and Methods). In vivo assays using tetrahydrocolumbamin, tetrahydropalmatine, cheilanthifoline, cavidine, and stylopine as substrates revealed that the candidate BBEL genes catalyzed the four-electron oxidation of all the tested BIAs (Figure 4D, Figure S20 – 22). In addition, ultra-performance liquid chromatography (UPLC) and LC-MS analysis revealed that CtBBEL22 likely catalyzes the conversion of stylopine to coptisine, while CtBBEL8 and CtBBEL31 likely catalyze the conversion of cheilanthifoline to dehydrocheilanthifoline and of cavidine to dehydrocavidine (Figure 4E, Figure S21 – 22). The distinct substrate specificities of these BBEL genes might depend on the methylenedioxy bridge of BIA substrates. As a negative control, we tested whether BBEL genes from other species, but closely related to the Corydalis BBEL genes in the cluster genome, could catalyze the oxidation of cheilanthifoline and cavidine. Briefly, we expressed BwSTOX from Berberis wilsoniae in Sf9 insect cells. Although BwSTOX can transform tetrahydropalmatine into palmatine (Gesell et al., 2011), it did not act on cheilanthifoline and cavidine in our analysis (Figure S21), suggesting that the specific biosynthesis of dehydrocheilanthifoline and dehydrocavidine is closely correlated with the unique tandem duplication of BBEL genes in the Corydalis genus after its divergence from P. somniferum and M. cordata.
Stylopine and coptisine are conserved in most Papaveraceae species, and the CtBBEL22 branch that originated before the divergence of C. tomentella, M. cordata and P. somniferum may be related to coptisine biosynthesis. Together, collinearity and phylogenetic analyses revealed that the 25 BBEL genes in C. tomentella were first formed by tandem duplications before the divergence of Papaveraceae, followed by additional tandem duplications in C. tomentella after its divergence from P. somniferum and M. cordata (Figure 4B). BBEL21, BBEL22, BBEL23, BBEL24, and BBEL25 in C. tomentella are the orthologs of conserved PS0410460 and PS0515260 in P. somniferum as well as of BVC80_1779g11 and BVC80_1779g18 in M. cordata, which are more likely to be involved in coptisine biosynthesis from stylopine. In addition, BBEL8 and BBEL31 have exclusively evolved in C. tomentella through tandem duplication. Hence, we conclude that the dramatic expansion of BBEL genes in the C. tomentella genome may be related to the appearance of cavidines and coptisine in Corydalis, which further obtained the function of catalyzing the four-electron oxidation steps in the biosynthesis of specific BIAs.
Discussion
The four lineages of eudicots, Ranunculales, Proteales (Sabiaceae), Trochodendrales, as well as Buxales, all share deeper common ancestors with the core eudicots than do the species within the core eudicots (Group, 2016). The C. tomentella genome sequence reported herein provides a valuable genomic resource for Papaveraceae in Ranunculales. Many Papaveraceae species, such as P. somniferum, M. cordata, and C. tomentella, produce common or species-specific BIAs, which have important medical and economic value. The genome of C. tomentella, as the first sequenced species of Fumarioideae, offers a crucial reference for studying the early evolution of eudicots and elucidating the diversification of BIA biosynthesis in Ranunculales. In particular, we uncovered once common WGD event that was likely shared by all species of Ranunculaleans after its divergence from other eudicot lineages. Furthermore, the analyzed Ranunculalean species present variable synonymous substitution rates, leading to various KS values of peaks representing the shared WGD event.
Genome mining can effectively promote discovery of natural product biosynthetic pathways and facilitate their characterization. Our analysis of the BIA biosynthetic pathway revealed genes involved in the biosynthesis of common or lineage-specific BIAs in Ranunculales and highlighted the importance of gene duplications, especially tandem gene duplications, and loss in the diversification of plant secondary metabolic pathways. Cavidine biosynthesis in Corydalis is likely shaped by the unique expansion of BBEL genes via tandem gene duplication. Similarly, the sanguinarine and noscapine biosynthetic genes have probably originated through a duplication event that occurred before the divergence of Papaveraceae. Interestingly, only one duplicate was retained and the other was lost in most Papaveraceae lineages, except the genus Papaver. In most Papaveraceae species, the retained duplicate evolved and gave rise to the CYP82N subfamily, which contains MSH and P6H that are crucial for sanguinarine biosynthesis. Meanwhile, in Papaver, the retained duplicate underwent further tandem duplication and gave rise to the CYP82X/Y subfamily, which contains key enzymes involved in the Papaver-specific biosynthesis of noscapine. The lack of 4’OMT, CHS (CYP719A14), STS (CYP719A2), and CYP82N/X/Y members in Proteobacteria mainly contribute to the complete loss of BIA compounds. In contrast, tandem duplications and loss of crucial genes related to BIA biosynthesis are important drivers of the specification and diversity of BIA accumulation in Ranunculales.
Cavidines, including cavidine, apocavidine, dehydroapocavidine, and dehydrocavidine, have mainly been isolated from Corydalis species. Cavidines are compounds with a conserved methyl moiety at the C-14 position and methylenedioxy groups at the C-9 and C-10 positions in the skeleton structure of protoberberine alkaloids. According to the 13C6 (benzene ring)-labeled BIAs and the known BIA biosynthetic pathway, we assume that cavidines are the downstream products of cheilanthifoline under the catalytic activity of CMT or OMT. In addition, sinactine is the only reported O-methylation product of cheilanthifoline, synthesized through the action of an hitherto uncharacterized enzyme (Beaudoin and Facchini, 2014). In a previous study, sinactine was isolated from the genus Corydalis (Bhakuni and Chaturvedi, 1983); however, we could not identify the fragment ions of sinactine in our 13C6 (benzene ring)-labeled datasets of LC-MS in Corydalis tomentella, suggesting the trace level of sinactine. Moreover, 14-C-methylated cavidines and dehydrocheilanthifoline were abundant in Corydalis tomentella. Therefore, we believe that C-methylation likely precedes O-methylation during cavidine biosynthesis. The biosynthesis of dehydrocavidine and dehydroapocavidine in Corydalis is likely shaped by the unique expansion of BBEL genes via tandem gene duplication. In our study, the BBEL8/31 could both accept cheilanthifoline and cavidine as substrates, suggesting that both enzymes are promiscuous regarding the C-14 methylation pattern of the substrates. However, the substrate specificities or promiscuity of BBEL enzymes need more evidence, which will provide important insight in the findings and synthesis of diverse BIA compounds with two- and four-electron oxidation.
Materials and Methods
Sample collection and genomic survey
C. tomentella was identified using DNA barcoding (Ren et al., 2019). For genomic DNA extraction, a C. tomentella plant was collected from the Chongqing City in Nanchuan District (29°N, 107°E), China. Two libraries of 250 and 500 insert DNAs were constructed, and 2 × 125 paired-end sequencing was performed using the Illumina HiSeq 4000 platform. Illumina reads were used to estimate the genome size of C. tomentella based on the distribution of 21 K-mers. For transcriptome sequencing, different tissues of C. tomentella, including roots, stems, leaves, and flowers, were collected.
SMRT sequencing and genome assembly
The high-molecular-weight (HMG) gDNA of C. tomentella was extracted following the methods of megabase-size DNA preparation (Zhang et al., 2012). HMG DNA was used to prepare 20 kb templates with the BluePippin size selection system (PAC20KB/BLF7510). Libraries were constructed, and SMRT sequencing of seven cells was performed on the Sequel platform. Raw data were filtered to remove adapters and low-quality reads through SMRT Portal analysis.
The C. tomentella genome was assembled de novo using HGAP (v4) with default parameters, including error correction, pre-assembled reads, assembly to contig, and genome polishing (https://www.pacb.com/products-and-services/analytical-software/smrt-analysis/). Heterozygous contigs were removed from the draft assembly using Redundans with paired-end and SMRT sequencing data (Pryszcz and Gabaldon, 2016). The Embryophya odb 10 dataset was selected to estimate the completeness of the Corydalis tomentella genome assembly using BUSCO (v. 4) (Simao et al., 2015).
To improve genome assembly, young leaves of C. tomentella were collected for Hi-C library construction and paired-end sequencing. Cross-linked and lysed cells were digested using the HindIII restriction enzyme. Paired-end reads were mapped to the draft assembly, and the misjoins of the chimeric contigs were split and corrected using the 3D de novo assembly pipeline (Dudchenko et al., 2017). The corrected contigs were anchored to pseudo-chromosomes using the ALLHiC pipeline (Zhang et al., 2019).
Genome annotation and gene expression analysis
Structural repeat annotation of the C. tomentella genome was performed using the RepeatModeler (v1.0.9) package (http://www.repeatmasker.org/RepeatModeler/). Two de novo repeat finding programs, namely RECON and RepeatScout, were employed to identify and classify the repeat elements in the C. tomentella genome. Moreover, Repbase (v. 21.12) was used, and consensus classification of TEs was performed according to the default parameters. Finally, RepeatMasker (v. 4.0.6) was used to calculate and mask TE sequences (http://www.repeatmasker.org/).
Transcriptomic data from different tissues were assembled de novo using Trinity (v. 2.2.0) (Grabherr et al., 2011). Putative protein-coding genes were ab initio predicted using the MAKER (v 2.31.9) annotation pipeline with the following parameters (Cantarel et al., 2008). A. thaliana was selected as the gene prediction species model in AUGUSTUS. Unigenes and protein sequences predicted based on the RNA-Seq data of C. tomentella and the annotated protein sequences from A. thaliana and M. cordata were subjected to combined EST and proteomic analysis with BLAST and Exonerate alignment. The completeness of genome annotation was estimated using BUSCO (v. 4) (Simao et al., 2015).
RNA-Seq data from the roots, stems, leaves, and flowers of C. tomentella were filtered using Trimmomatic (v0.39) (Bolger et al., 2014). Clean reads from different tissues were separately aligned to the C. tomentella genome using HISAT2 (v2.2.0) with default parameters. CUFFLINKS (v. 2.2.1) (Trapnell et al., 2012) was used to calculate the FPKM values of annotated genes.
Phylogenetic tree construction and phylogenomic dating
OrthoFinder (v. 2.2.7) (Emms and Kelly, 2019) was used to identify the orthogroups and orthologs using the default parameters. The orthologs were obtained from eight core eudicots [Coffea canephora (Denoeud et al., 2014), Lactuca sativa (Reyes-Chin-Wo et al., 2017), A. thaliana (Arabidopsis Genome, 2000), Populus trichocarpa (Tuskan et al., 2006), Solanum tuberosum (Tomato Genome, 2012), Citrus clementina (Wu et al., 2014), Daucus carota (Garcia-Mas and Rodriguez-Concepcion, 2016), and V. vinifera (Jaillon et al., 2007)], six sister lineages to core eudicots [N. nucifera (Ming et al., 2013), A. coerulea (Filiault et al., 2018), C. tomentella, M. cordata (Liu et al., 2017), P. somniferum (Guo et al., 2018), and E. californica (Hori et al., 2018)], four monocots (Apostasia shenzhenica (Zhang et al., 2017), Brachypodium distachyon (International Brachypodium, 2010), Musa acuminata (D'Hont et al., 2012), and Oryza sativa (Goff et al., 2002)], and the sister lineage to all other extant angiosperms [A. trichopoda (Amborella Genome, 2013)]. Single- and low-copy genes in 19 vascular plants were concatenated, and the sequences were aligned and trimmed using MAFFT (v. 6.240) (Katoh and Standley, 2013) and trimAI (v. 1.2) (Capella-Gutierrez et al., 2009), respectively. A species tree was constructed using RAxML (v. 8.2.9) (Stamatakis, 2006) based on nucleotide sequences with the GTR+GAMMA model and protein sequences with the JTT+GAMMA model. The evolutionary timescale was analyzed using the MCMCtree of the PAML package (Yang, 2007) with 500,000 iterations and 150 sample frequencies, following 500,000 iterations as burn-in. The divergence time of species was estimated based on the following fossil-based age constraints: B. distachyon and O. sativa divergence time of 42–52 MYA; P. trichocarpa and C. elementina divergence time of 98–117 MYA; monocot and eudicot divergence time of 130 MYA with a hard boundary; and A. trichopoda and other angiosperms divergence time of 173 – 199 MYA. CAFÉ (v. 3.1) was used to predict gene family evolution, including gene expansion and contraction (De Bie et al. 2006). The gene families with rapid expansion were annotated using the InterProScan tool (http://www.ebi.ac.uk/interpro/).
Identification of WGD events
KS-based age distributions for paralogous genes of the sister lineages to core eudicots (N. nucifera, A. coerulea, C. tomentella, M. cordata, P. somniferum, and E. californica) and V. vinifera were constructed using the “wgd” pipeline (Zwaenepoel and Van de Peer, 2019). Briefly, the paralogous genes for each species were identified using the Diamond (v. 0.9.18.119) (Buchfink et al., 2015) sequence similarity search tool, with an e-value cutoff of 1e−10. The paralogous gene families were clustered using the Markov Cluster Algorithm (MCL) (Enright et al., 2002), with an inflation factor of 2.0. Each paralogous gene family was aligned using MAFFT (v7.453) (Katoh and Standley, 2013), with default parameters. The KS values of all gene pairs within a gene family were estimated using the CODEML program in the PAML package (v4.9) (Yang, 2007). KS estimates were subsequently node-weighted to correct for redundancy, and phylogenetic trees were constructed for all families using FastTree (v2.1.7) (Price et al., 2009). The KS age distributions of paralogous genes in all tested species are shown as gray bars in Figure S5. The intragenomic collinear segments and the corresponding anchor pairs for each genome were identified using i-ADHoRe (v. 3.0) (Proost et al., 2012). The KS distribution of paralogous genes located in the collinear segments was estimated using the CODEML program of the PAML package (v. 4.9) (Yang, 2007). The KS age distribution of anchor pairs for each genome is shown as black bars in Figure S7.
The KS-based age distributions of orthologs in tested species were constructed using the “wgd” pipeline, as described above. Orthologs between species were selected according to the best hits of Diamond (v. 0.9.18.119) (Buchfink et al., 2015). The KS values of all orthologs or anchor pairs from i-ADHoRe (v. 3.0) (Proost et al., 2012) were estimated using the CODEML program in the PAML package (v. 4.9) (Yang, 2007). To trace the WGD events identified in Ranunculales, anchor-pair KS distributions between C. tomentella and A. coerulea, M. cordata, and V. vinifera were identified. To confirm the different substitution rates in the sister lineages of core eudicots, the KS distributions of orthologs between V. vinifera and N. nucifera, A. coerulea, C. tomentella, M. cordata, P. somniferum, and E. californica were compared one-to-one.
The different substitution rates of sister lineages to core eudicots led to variability in the placement of (likely the same) WGD events. To further correctly place the WGD events, single-copy genes from the OrthoFinder (v. 2.2.7) (Emms and Kelly, 2019) analysis of all tested species were used to calculate the branch lengths of phylogenies in the KS unit using the CODEML program of the PAML package (v. 4.9) (Yang, 2007) with the free-ratio model. To map the identified peaks of KS distributions to the phylogeny (in the KS unit), half of the KS peak values were placed from the tip toward the root of the phylogeny, with the assumption that duplicated genes in each genome evolved with similar substitution rates after a WGD event.
Identification of BIAs in different tissues of Corydalis tomentella
Different tissues of Corydalis tomentella, including roots, stems, leves and flowers, collected from the same cultivated plant in the Chongqing City in Nanchuan District, China, were dried at 45°C and ground to a fine powder (20 mesh). To isolate potential metabolites, 100 mg of dried powder, which was weighed accurately and dissolved in 25 mL of mobile phase, was extracted using ultrasound (100 kHz) at 25°C for 30 min. The same solvent was added to compensate for the weight lost during extraction. The extract filtered through a 0.22 μm membrane for HPLC analysis.
The Waters 2695 (Milford, USA) liquid chromatograph equipped with a quaternary pump solvent management system, an online degasser, an autosampler, a DAD detector, and the Xbridge BEH C18 (4.6 mm × 250 mm, 5 μm) column was used. The mobile phase was composed of acetonitrile (A) and water (0.2% phosphoric acid, 20 mmol·L-1 potassium dihydrogen phosphate, and 10 mmol·L-1 triethylamine), using the gradient elution of 15–22% A at 0–10 min, 22% A at 10–40 min, and 22–15% A at 40–50 min. The flow rate of the mobile phase was set to 0.8 mL·min-1. The injection volume for each sample was 10 μL.
Isotopic labelling of BIA biosynthesis and liquid chromatography quadrupole time-of-flight mass spectrometry (LC-Q-TOF-MS) analysis
C. tomentella plants were collected and cultured in a greenhouse at 25°C for 7 days. The plants were then cultured in water with or without isotopically labeled [ring-13C6]-tyrosine (4 mg·mL-1) for a week. The unlabeled and labeled samples with three replicates were dried at 45°C and ground to a fine powder (20 mesh). The dried powder (100 mg) was accurately weighed, dissolved in 2 mL of methanol, and extracted using ultrasound (100 Hz) at room temperature for 60 min. After centrifugation at 4,500 rpm for 10 min, the supernatant was filtered through a 0.22 μm membrane and used for LC–MS analysis.
Chromatographic separation was performed on the Agilent 1290 instrument using the Agilent Zorbax SB C18 (100 × 2.1 mm,3.5 μm) column. The mobile phase was composed of A (water with 0.1% formic acid) and B (methanol with 0.1% formic acid), using the gradient elution of 10–20% B at 0–3 min, 20–25% B at 3–13 min, 25–30% B at 13–23 min, and 30–40% B at 23–33 min. The flow rate of the mobile phase was set at 0.3 mL·min-1, and the column temperature was maintained at 35°C. The sample injection volume was 5 μL. UPLC was coupled to Q-TOF-MS (6550A, Agilent). Data were acquired in the positive ionization and auto-MS/MS modes (m/z 50-500). Spray parameters were set as follows: gas temperature, 200°C; gas flow, 16 L·min-1; nebulizer, 50 psi; sheath gas temperature, 350°C; sheath gas flow, 12 L·min-1; Vcap voltage, 3,500 V; nozzle voltage, 500 V; and collision energy, 20eV. Agilent PCDL Manager was used to establish the Corydalis tomentella alkaloid database, and Agilent MassHunter Profinder was used to analyze and extract isotopologues (match tolerance mass ± 20 ppm, RT ± 10 min).
Heterologous expression of CtBBELs and activity assay
BBEL genes were expressed using the Bac-to-Bac system. BBEL genes (CtBBEL8, CtBBEL22, and CtBBEL31) from C. tomentella and BwSTOX from B. wilsoniae were cloned into the pFastBacDual vector, and the pFastBacDual-BBELs were transformed into the competent E. coli strain DH10Bac. Recombinant bacmid DNA was used for the transfection of Sf9 insect cells at a density of 2 × 106 cells·mL-1. After 24 h at 27°C for infection, the alkaloid standards, including tetrahydrocolumbamin, tetrahydropalmatine, stylopin, cheilanthifoline, and cavidine, were respectively added to the medium for additional 36 h incubation. The cultures were further extracted using 1 volume butanol and dried in vacuum. The dried substances were resuspended in methyl alcohol for HPLC and LC-MS analyses.
Supplementary Material
Acknowledgements
This work was supported by National Key R&D Program of China (2019YFC1711100), CAMS Innovation Fund for Medical Sciences (CIFMS) (Grant No. 2021-I2M-1-029), National Science and Technology Major Project for “Significant New Drugs Development” (2019ZX09201005-006-003), and the State Scholarship Fund (CSC no. 201808110110). ZX acknowledges Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team). ZL is funded by a postdoctoral fellowship from the Special Research Fund of Ghent University (BOFPDO2018001701). YVdP acknowledges funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (Grant No. 833522).
Footnotes
Author contributions
Z.X. and J.S. designed and coordinated the study. F.R. supplied the plant materials. Z.X. assembled and annotated the genome. Z.X. and Z.L. conducted WGD analysis. R.G., Z.W., J.Z., X.M., X.P., and T.X. performed the experiments and analyzed the data. Z.X., Z.L., R.G., T.Z., W.S., S.R., Y.V.d.P., S.C., and J.S. wrote and revised the manuscript.
Conflict of interest statement
The authors declare no conflict of interest.
Data availability
The raw data of genome and transcriptome sequencing reported in this paper have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number CRA003885 that are publicly accessible at http://bigd.big.ac.cn/gsa. The assembled genome and gene structures have been deposited in the Genome Warehouse in National Genomics Data Center under the accession number GWHAORS00000000 with the BioProject ID (PRJCA003323) and the BioSample ID (SAMC231736), which is publicly accessible at https://bigd.big.ac.cn/gwh.
References
- Amborella Genome P. The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome I. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Beaudoin GA, Facchini PJ. Benzylisoquinoline alkaloid biosynthesis in opium poppy. Planta. 2014;240:19–32. doi: 10.1007/s00425-014-2056-8. [DOI] [PubMed] [Google Scholar]
- Bhakuni DS, Chaturvedi R. The alkaloids of Corydalis meifolia. J Nat Prod. 1983;46:320–324. doi: 10.1021/np50027a004. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, Holt C, Sanchez Alvarado A, Yandell M. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome research. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee CP. Pharmacopoeia of the People’s Republic of China. China Beijing: China Medical Science and Technology Press; 2015. [Google Scholar]
- Courdavault V, O'Connor SE, Oudin A, Besseau S, Papon N. Towards the Microbial Production of Plant-Derived Anticancer Drugs. Trends Cancer. 2020;6:444–448. doi: 10.1016/j.trecan.2020.02.004. [DOI] [PubMed] [Google Scholar]
- D'Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, Da Silva C, et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488:213–217. doi: 10.1038/nature11241. [DOI] [PubMed] [Google Scholar]
- Daniel B, Konrad B, Toplak M, Lahham M, Messenlehner J, Winkler A, Macheroux P. The family of berberine bridge enzyme-like enzymes: A treasure-trove of oxidative reactions. Arch Biochem Biophys. 2017;632:88–103. doi: 10.1016/j.abb.2017.06.023. [DOI] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M, Zheng C, Alberti A, Anthony F, Aprea G, Aury JM, et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 2014;345:1181–1184. doi: 10.1126/science.1255274. [DOI] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, Aiden EL. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356:92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic acids research. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiault DL, Ballerini ES, Mandakova T, Akoz G, Derieg NJ, Schmutz J, Jenkins J, Grimwood J, Shu S, Hayes RD, Hellsten U, et al. The Aquilegia genome provides insight into adaptive radiation and reveals an extraordinarily polymorphic chromosome with a unique history. Elife. 2018;7 doi: 10.7554/eLife.36426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanie S, Thodey K, Trenchard IJ, Filsinger Interrante M, Smolke CD. Complete biosynthesis of opioids in yeast. Science. 2015;349:1095–1100. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mas J, Rodriguez-Concepcion M. The carrot genome sequence brings colors out of the dark. Nature genetics. 2016;48:589–590. doi: 10.1038/ng.3574. [DOI] [PubMed] [Google Scholar]
- 21.Gesell A, Chavez ML, Kramell R, Piotrowski M, Macheroux P, Kutchan TM. Heterologous expression of two FAD-dependent oxidases with (S)-tetrahydroprotoberberine oxidase activity from Arge mone mexicana and Berberis wilsoniae in insect cells. Planta. 2011;233:1185–1197. doi: 10.1007/s00425-011-1357-4. [DOI] [PubMed] [Google Scholar]
- 22.Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 23.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Group, T.A.P. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society. 2016;181:1–20. [Google Scholar]
- 25.Guo L, Winzer T, Yang X, Li Y, Ning Z, He Z, Teodor R, Lu Y, Bowser TA, Graham IA, Ye K. The opium poppy genome and morphinan production. Science. 2018;362:343–347. doi: 10.1126/science.aat4096. [DOI] [PubMed] [Google Scholar]
- 26.Hagel JM, Facchini PJ. Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat Chem Biol. 2010;6:273–275. doi: 10.1038/nchembio.317. [DOI] [PubMed] [Google Scholar]
- 27.Hao D. Ranunculales medicinal plants: biodiversity, chemodiversity and pharmacotherapy. Academic Press; 2018. [Google Scholar]
- 28.Hori K, Yamada Y, Purwanto R, Minakuchi Y, Toyoda A, Hirakawa H, Sato F. Mining of the Uncharacterized Cytochrome P450 Genes Involved in Alkaloid Biosynthesis in California Poppy Using a Draft Genome Sequence. Plant Cell Physiol. 2018;59:222–233. doi: 10.1093/pcp/pcx210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.International Brachypodium, I. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- 30.Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jiang L, Li M, Zhao F, Chu S, Zha L, Xu T, Peng H, Zhang W. Molecular Identification and Taxonomic Implication of Herbal Species in Genus Corydalis (Papaveraceae) Molecules. 2018;23 doi: 10.3390/molecules23061393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA. Has the connection between polyploidy and diversification actually been tested? Current Opinion in Plant Biology. 2016;30:25–32. doi: 10.1016/j.pbi.2016.01.002. [DOI] [PubMed] [Google Scholar]
- Li Q, Ramasamy S, Singh P, Hagel JM, Dunemann SM, Chen X, Chen R, Yu L, Tucker JE, Facchini PJ, Yeaman S. Gene clustering and copy number variation in alkaloid metabolic pathways of opium poppy. Nature communications. 2020a;11:1190. doi: 10.1038/s41467-020-15040-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Winzer T, He Z, Graham IA. Over 100 Million Years of Enzyme Evolution Underpinning the Production of Morphine in the Papaveraceae Family of Flowering Plants. Plant Commun. 2020b;1:100029. doi: 10.1016/j.xplc.2020.100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li S, Thodey K, Trenchard I, Cravens A, Smolke CD. Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:E3922-E3931. doi: 10.1073/pnas.1721469115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscombe DK, MacLeod BP, Loukanina N, Nandi OI, Facchini PJ. Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry. 2005;66:2501–2520. doi: 10.1016/j.phytochem.2005.04.044. [DOI] [PubMed] [Google Scholar]
- Liu X, Zheng H, Lu R, Huang H, Zhu H, Yin C, Mo Y, Wu J, Liu X, Deng M, Li D, et al. Intervening Effects of Total Alkaloids of Corydalis saxicola Bunting on Rats With Antibiotic-Induced Gut Microbiota Dysbiosis Based on 16S rRNA Gene Sequencing and Untargeted Metabolomics Analyses. Front Microbiol. 2019;10:1151. doi: 10.3389/fmicb.2019.01151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Liu Y, Huang P, Ma Y, Qing Z, Tang Q, Cao H, Cheng P, Zheng Y, Yuan Z, Zhou Y, et al. The Genome of Medicinal Plant Macleaya cordata Provides New Insights into Benzylisoquinoline Alkaloids Metabolism. Molecular plant. 2017;10:975–989. doi: 10.1016/j.molp.2017.05.007. [DOI] [PubMed] [Google Scholar]
- Minami H, Kim JS, Ikezawa N, Takemura T, Katayama T, Kumagai H, Sato F. Microbial production of plant benzylisoquinoline alkaloids. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7393–7398. doi: 10.1073/pnas.0802981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, VanBuren R, Liu Y, Yang M, Han Y, Li LT, van der Klink JJ, Zhang Q, Kim MJ, Schatz MC, Campbell M, et al. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn) Genome Biol. 2013;14:R41. doi: 10.1186/gb-2013-14-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat F, Armero A, Pont C, Klopp C, Salse J. Reconstructing the genome of the most recent common ancestor of flowering plants. Nature genetics. 2017;49:490–496. doi: 10.1038/ng.3813. [DOI] [PubMed] [Google Scholar]
- Perez-Gutierrez MA, Romero-Garcia AT, Salinas MJ, Blanca G, Fernandez MC, Suarez-Santiago VN. Phylogeny of the tribe Fumarieae (Papaveraceae s.l.) based on chloroplast and nuclear DNA sequences: evolutionary and biogeographic implications. Am J Bot. 2012;99:517–528. doi: 10.3732/ajb.1100374. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S, Fostier J, De Witte D, Dhoedt B, Demeester P, Van de Peer Y, Vandepoele K. i-ADHoRe 3.0--fast and sensitive detection of genomic homology in extremely large data sets. Nucleic acids research. 2012;40:e11. doi: 10.1093/nar/gkr955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryszcz LP, Gabaldon T. Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic acids research. 2016;44:e113. doi: 10.1093/nar/gkw294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren FM, Wang YW, Xu ZC, Li Y, Xin TY, Zhou JG, Qi YD, Wei XP, Yao H, Song JY. DNA barcoding of Corydalis, the most taxonomically complicated genus of Papaveraceae. Ecol Evol. 2019;9:1934–1945. doi: 10.1002/ece3.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Chin-Wo S, Wang Z, Yang X, Kozik A, Arikit S, Song C, Xia L, Froenicke L, Lavelle DO, Truco MJ, Xia R, et al. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nature communications. 2017;8:14953. doi: 10.1038/ncomms14953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson FM, Gundappa MK, Grammes F, Hvidsten TR, Redmond AK, Lien S, Martin SAM, Holland PWH, Sandve SR, Macqueen DJ. Lineage-specific rediploidization is a mechanism to explain time-lags between genome duplication and evolutionary diversification. Genome Biol. 2017;18:111. doi: 10.1186/s13059-017-1241-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamma M. The isoquinoline alkaloids chemistry and pharmacology. Elsevier; 2012. [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Tian B, Tian M, Huang SM. Advances in phytochemical and modern pharmacological research of Rhizoma Corydalis. Pharm Biol. 2020;58:265–275. doi: 10.1080/13880209.2020.1741651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome C. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Valtuena FJ, Preston CD, Kadereit JW. Phylogeography of a Tertiary relict plant, Meconopsis cambrica (Papaveraceae), implies the existence of northern refugia for a temperate herb. Mol Ecol. 2012;21:1423–1437. doi: 10.1111/j.1365-294X.2012.05473.x. [DOI] [PubMed] [Google Scholar]
- Winzer T, Gazda V, He Z, Kaminski F, Kern M, Larson TR, Li Y, Meade F, Teodor R, Vaistij FE, Walker C, et al. A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science. 2012;336:1704–1708. doi: 10.1126/science.1220757. [DOI] [PubMed] [Google Scholar]
- Wu GA, Prochnik S, Jenkins J, Salse J, Hellsten U, Murat F, Perrier X, Ruiz M, Scalabrin S, Terol J, Takita MA, et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nature biotechnology. 2014;32:656–662. doi: 10.1038/nbt.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin T, Zhang Y, Pu X, Gao R, Xu Z, Song J. Trends in herbgenomics. Sci China Life Sci. 2019;62:288–308. doi: 10.1007/s11427-018-9352-7. [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Valtuena FJ, Preston CD, Kadereit JW. Phylogeography of a Tertiary relict plant, Meconopsis cambrica (Papaveraceae), implies the existence of northern refugia for a temperate herb. Mol Ecol. 2012;21:1423–1437. doi: 10.1111/j.1365-294X.2012.05473.x. [DOI] [PubMed] [Google Scholar]
- Winzer T, Gazda V, He Z, Kaminski F, Kern M, Larson TR, Li Y, Meade F, Teodor R, Vaistij FE, Walker C, Bowser TA, Graham IA. A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science. 2012;336:1704–1708. doi: 10.1126/science.1220757. [DOI] [PubMed] [Google Scholar]
- Wu GA, Prochnik S, Jenkins J, Salse J, Hellsten U, Murat F, Perrier X, Ruiz M, Scalabrin S, Terol J, Takita MA, et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nature biotechnology. 2014;32:656–662. doi: 10.1038/nbt.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin T, Zhang Y, Pu X, Gao R, Xu Z, Song J. Trends in herbgenomics. Sci China Life Sci. 2019;62:288–308. doi: 10.1007/s11427-018-9352-7. [DOI] [PubMed] [Google Scholar]
- Xu D, Lin H, Tang Y, Huang L, Xu J, Nian S, Zhao Y. Integration of full-length transcriptomics and targeted metabolomics to identify benzylisoquinoline alkaloid biosynthetic genes in Corydalis yanhusuo. Hortic Res. 2021;8:16. doi: 10.1038/s41438-020-00450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Gao R, Pu X, Xu R, Wang J, Zheng S, Zeng Y, Chen J, He C, Song J. Comparative Genome Analysis of Scutellaria baicalensis and Scutellaria barbata Reveals the Evolution of Active Flavonoid Biosynthesis. Genomics Proteomics Bioinformatics. 2020;18:230–240. doi: 10.1016/j.gpb.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang B, Huang R, Hua J, Liang H, Pan Y, Dai L, Liang D, Wang H. Antitumor lignanamides from the aerial parts of Corydalis saxicola. Phytomedicine : international journal of phytotherapy and phytopharmacology. 2016;23:1599–1609. doi: 10.1016/j.phymed.2016.09.006. [DOI] [PubMed] [Google Scholar]
- Zhang GQ, Liu KW, Li Z, Lohaus R, Hsiao YY, Niu SC, Wang JY, Lin YC, Xu Q, Chen LJ, Yoshida K, et al. The Apostasia genome and the evolution of orchids. Nature. 2017;549:379–383. doi: 10.1038/nature23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zhang Y, Scheuring CF, Wu CC, Dong JJ, Zhang HB. Preparation of megabase-sized DNA from a variety of organisms using the nuclei method for advanced genomics research. Nature protocols. 2012;7:467–478. doi: 10.1038/nprot.2011.455. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang S, Zhao Q, Ming R, Tang H. Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nature plants. 2019;5:833–845. doi: 10.1038/s41477-019-0487-8. [DOI] [PubMed] [Google Scholar]
- Zwaenepoel A, Van de Peer Y. wgd-simple command line tools for the analysis of ancient whole-genome duplications. Bioinformatics. 2019;35:2153–2155. doi: 10.1093/bioinformatics/bty915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of genome and transcriptome sequencing reported in this paper have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number CRA003885 that are publicly accessible at http://bigd.big.ac.cn/gsa. The assembled genome and gene structures have been deposited in the Genome Warehouse in National Genomics Data Center under the accession number GWHAORS00000000 with the BioProject ID (PRJCA003323) and the BioSample ID (SAMC231736), which is publicly accessible at https://bigd.big.ac.cn/gwh.