Summary
In many developing tissues, the patterns of gene expression that assign cell fate are organized by graded secreted signals. Cis-regulatory elements (CREs) interpret these signals to control gene expression, but how this is accomplished remains poorly understood. In the neural tube, a gradient of the morphogen sonic hedgehog (Shh) patterns neural progenitors. We identify two distinct ways in which CREs translate graded Shh into differential gene expression in mouse neural progenitors. In most progenitors, a common set of CREs control gene activity by integrating cell-type-specific inputs. By contrast, the most ventral progenitors use a unique set of CREs, established by the pioneer factor FOXA2. This parallels the role of FOXA2 in endoderm, where FOXA2 binds some of the same sites. Together, the data identify distinct cis-regulatory strategies for the interpretation of morphogen signaling and raise the possibility of an evolutionarily conserved role for FOXA2 across tissues.
Graphic abstract.
Introduction
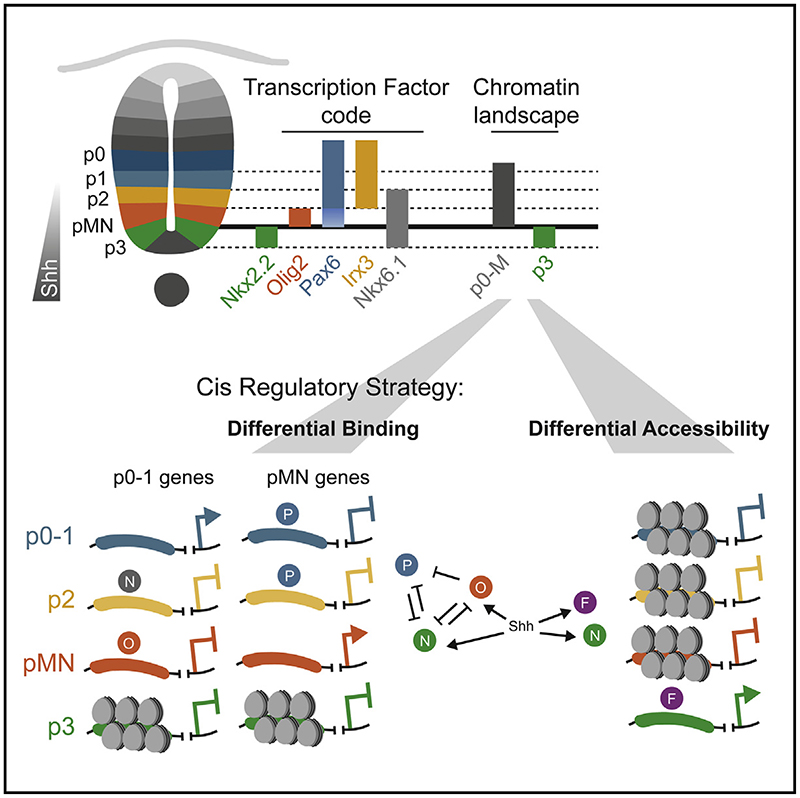
During development signaling cues direct cell fate decisions. Precise spatiotemporal gene expression is essential for this process. Cis-regulatory elements (CREs) integrate inputs from tissue and cell-type-specific transcription factors (TFs), as well as signaling effectors, to direct gene expression.1 This suggests a straightforward mechanism in which distinct signals produce different cell types by activating different transcriptional effectors that bind to different CREs. However, in many tissues, a single signal directs multiple, alternative cell fates.2 Two strategies can be envisioned for how CREs interpret a single signal to define multiple cell fates. Different CREs could function in different cell types. In this strategy—differential accessibility—the availability of an element is the principal determinant of cell identity. In an alternative strategy—differential binding—the same CREs could be used in all cell types in the tissue and the configuration of proteins bound to the elements determines cell fate. An implication of these two strategies is that choosing between two fates will require chromatin remodeling in the case of the differential availability of CREs but can happen without remodeling in the case of different protein configurations at the CREs.
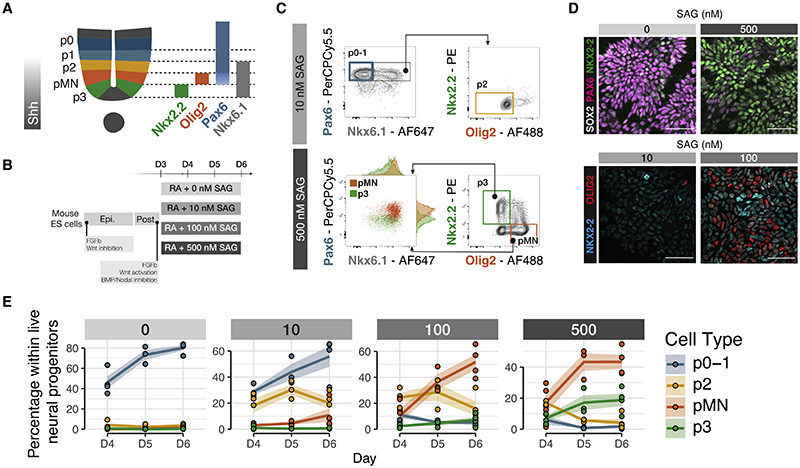
A well-characterized example of a signal controlling multiple cell types is the morphogen sonic hedgehog (Shh). Shh controls the pattern of neuronal subtype generation in ventral regions of the developing neural tube.3 Shh, initially secreted from the notochord and later from the floor plate (FP),4,5 forms a ventral to dorsal gradient in the neural tube and establishes a set of molecularly distinct domains that occupy characteristic positions along the dorsal-ventral axis. Progenitors in each domain differentiate into distinct classes of post-mitotic neurons. This organization is necessary for the subsequent assembly of the locomotor and sensory circuits.6,7 The progenitor domains are identifiable by characteristic gene expression programs (Figure 1A). NKX2.2 is expressed in p3 progenitors, the most ventral domain;8 OLIG2 is expressed in adjacent motor neuron progenitors (pMNs);9 PAX6 is expressed in more dorsal progenitor domains, including p0, p1, and p2 domains, whereas lower expression is detected in pMN;10 NKX6.1 is expressed in, p3, pM, and p2 domains.11
Figure 1. A stem cell model of ventral neural tube progenitors.
(A) Schematic of ventral spinal cord progenitors and the markers used for the combinatorial multi-color flow cytometry and sorting strategies.
(B) Schematic of the protocol for the differentiation of mouse ES cells to generate ventral neural progenitors following equivalent signaling cues to embryonic development.
(C) Representative flow cytometry plots of the gating strategies used for both analysis and sorting of NPs p0-1, p2, pMN, and p3. Cells were gated as NPs by excluding dead cells and selecting SOX2+.
(D) Representative immunohistochemistry of ES cells differentiated for 6 days show expression PAX6 when exposed to 0 nM SAG and NKX2.2 if exposed to 500 nM. At 100 nM SAG, both OLIG2 or NKX2.2 are detected compared with little or no signal at 10 nM SAG. Scale bars, 50 μm.
(E) Proportion of NPs at each SAG concentration shows generation of higher proportions of more ventral cell types at increasing SAG concentrations. Dots are individual samples. n = 4 biological replicates for each SAG concentration. Line represents the average. Shaded areas, SEM.
SAG, smoothened agonist.
See also Figure S1.
The pattern of progenitor gene expression is determined by a gene regulatory network (GRN). Broadly expressed tissue-wide activators, such as SOX2, promote the transcriptional programs of multiple progenitor domains.12–14 Concurrently, a set of TFs encoding Groucho/TLE-dependent transcriptional repressors are differentially expressed in distinct progenitor domains.15 Pairs of these TFs, expressed in adjacent domains, cross-repress each other and form a network of transcriptional repression that selects a single progenitor identity by repressing all other cell fates.16–18 The transcriptional effectors of Shh signaling, the GLI (Gli1, Gli2, and Gli3) proteins provide the spatially polarized input that initiates the patterning process.19–21 The combination of the positive and negative inputs generated by this network establish and position the discrete boundaries of gene expression domains in response to graded Shh signaling.11,22,23
Although the genetic architecture of the GRN is well established, how cell fate choice is implemented through CREs is less clear. Enhancer elements that drive expression of domain-specific TFs have been identified.13,14 The presence of these cell-type-specific enhancer elements could support the differential accessibility strategy in which different CREs are available in different cell types. However, many of these CREs are bound by SOX2, the Shh effector GLI1 and the repressive TFs expressed in alternative cell types, including NKX2.2, OLIG2, and NKX6.1.13,16,17 This suggests that the same CREs are employed in different cell types and the composition of TFs at these CREs determines activity: the differential binding strategy.
To distinguish between these two regulatory control strategies, we made use of neural progenitors (NPs) differentiated from embryonic stem (ES) cells.24,25 We reasoned chromatin accessibility was the best approach to capture the global landscape of functional regulatory regions. We therefore needed a cell-type-specific accessibility assay, in a system without cell surface markers. Here, we developed an ATAC-seq work-flow—crosslinked and TF-sorted ATAC-seq (CaTS-ATAC) — based on intracellular flow sorting that allowed the assay of chromatin accessibility of NP cell types. We found that most NPs shared a common set of CREs. These elements are bound by the known repressive TFs, indicating that these cell fate decisions are made by changing the composition of proteins bound to the elements. By contrast, the most ventral NP cell type, p3, has a distinct regulatory program. We identify the role of FOXA2 in driving this chromatin remodeling and show that it is required for p3 specification. This is reminiscent of the pioneering role of FOXA2 in endoderm lineages, raising the intriguing possibility that it represents a remnant of a GRN co-opted from this germlayer or a shared evolutionary origin for these cell types.
Results
A stem cell model of ventral neural tube progenitors
To generate NPs of different dorsoventral identities, we made use of an established protocol for the directed differentiation of mouse ES cells.24,25 To mimic graded Shh signaling, we used different concentrations, ranging from 0 to 500 nM of the Shh agonist SAG (smoothened agonist) (Figures 1A and 1B). We observed a dose response as measured by Shh target genes Gli1 and Ptch1 (Figure S1A). Using markers for specific progenitor domains,26 we assayed cell identity with single-cell resolution (Figures 1C and 1D). Each SAG condition generated a mixture of two or three NP types. Using the combinatorial expression of TFs, we quantified the proportions of different cell types for each concentration by intracellular staining followed by flow cytometry (Figure 1C). This showed enrichment of the expected cell types. The highest concentration (500 nM SAG) produced the highest percentage of the most ventral domain, p3, characterized by the expression of NKX2.2; pMN, which express the TF OLIG2, were produced at 500 and 100 nM SAG; p2 progenitors, which express NKX6.1 and PAX6, were produced at 100 and 10 nM. PAX6 expression in the absence of NKX6.1 identifies p0 and p1 progenitors, and these were produced at 0 and 10 nM SAG. The expected combinatorial expression of TFs was also observed: All p3 and pMN cells also expressed NKX6.1 (Figure 1E). Moreover, we observed a lower level of PAX6 in pMN cells compared with p3 cells, consistent with in vivo data (Figure S1B).
The different SAG conditions also generated the expected neuronal subtypes after the addition of Notch inhibitor dibenzazepine (DBZ), which induces neuronal differentiation (Figure S1C). Overall, the data indicate that the ES-derived model of neural tube patterning ventral of progenitors faithfully mimics the in vivo response of NPs to Shh.
CaTS-ATAC identifies cell-type-specific regulatory program
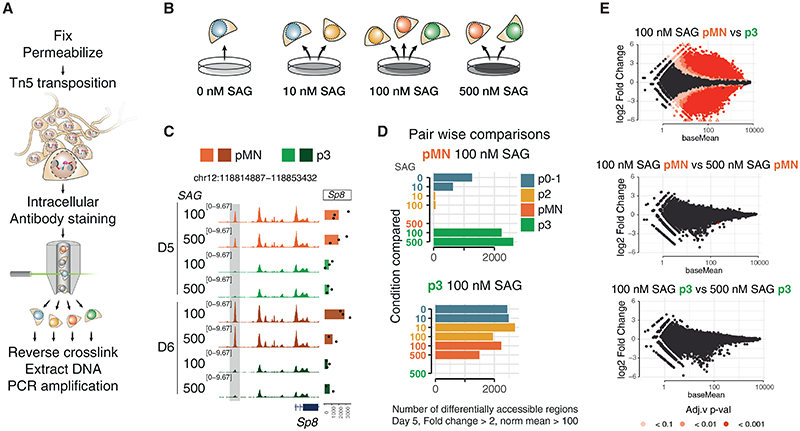
To distinguish whether the gradient of Shh signaling is interpreted via differential accessibility or differential binding, we reasoned we needed global chromatin accessibility information with cell-type specificity. ATAC-seq is conventionally performed in live, permeabilized cells. However, the cell-type-specific markers that distinguish NPs are intracellular and therefore cannot be used on live cells. Based on previous work,27 we devised a strategy—CaTS-ATAC—to perform ATAC-seq on formaldehyde fixed cells. This involved intracellularly staining and flow cytometry sorting of cells followed by bulk ATAC library preparation (Figure 2A; STAR Methods).
Figure 2. Chromatin accessibility reflects cell-type identity independent of SAG concentration.
(A) Schematic of CaTS-ATAC, a strategy for cell-type-specific ATAC-seq based on intracellular markers developed for this study.
(B) Cell types analyzed from each SAG concentration.
(C) Representative genome coverage plot of a differentially accessible region and expression of the nearby gene, Sp8, shows accessibility is consistent for each cell type regardless of the SAG concentration from which it was generated.
(D) Quantification of differentially accessible regions between the indicated sample and all other samples at day 5 of differentiation shows no significant differences with the same cell type generated from a different SAG concentration. Thresholds used: absolute fold change > 2, basemean > 100.
(E) MA-plot (log2-fold change versus base mean) for the indicated comparisons show large numbers of differentially accessible elements between different cell types generated under the same SAG concentration, but not between the same cell type generated in different SAG concentrations. n = 3 biological replicates. See also Figure S2.
We assessed the overall quality of the method by comparing it to ATAC-seq from live cells using the Omni-ATAC protocol.28 To this end, we sorted Olig2-expressing cells using a previously developed reporter cell line25 and compared it with CaTS-ATAC pMN samples, both from 100nM SAG at day 5 of differentiation. CaTS-ATAC data were of high quality as assessed by the insert size distribution, proportion of mitochondria DNA, and FRiP (Fraction of Reads in Peaks) score (Figures S2A–S2C). There was some underrepresentation of the smallest size fragments, which we assume is due to the reverse crosslinking steps, as noted by others29 (Figure S2A). The correlation between samples from the same method was high (R2 > 0.94) (Figures S2D and S2E) and a bit lower between methods (R2 > 0.72 for any pair wise comparison) (Figure S2F). We note that the samples will be slightly different as the reporter-sorted cells will include newly differentiated MNs and some OLIG2+ NKX2.2+ double positive cells, which are excluded from out CaTS-ATAC pMN. Overall, we were satisfied with the quality assessments and used standard ATAC QC metrics for all further samples.
We performed CaTS-ATAC-seq over a 4-day time course following the addition of different concentrations of SAG. For each condition, we sorted the two or three predominant cell types. This allows us to compare the regulatory landscape of the same cell type originating at different SAG concentrations as well as different cell types within the same dish (Figure 2B).
To explore this dataset, we first examined the correlation between samples based on accessibility at transcriptional start sites (TSSs) or distal elements (not overlapping with TSS). As previously reported in other systems,30 accessibility of distal elements exhibited greater cell-type specificity and higher dynamic range than promoter elements (Figures S2G and S2H). This confirms our previous observation that ATAC-seq can capture the regulatory landscape of NPs,31 and it can do so with cell type specificity.
Cell-type-specific accessibility configuration is independent of Shh agonist concentration
We first investigated the effect of SAG concentration on cell type-specific chromatin accessibility. To do this, we compared global accessibility changes between different NP types generated under the same SAG concentration and the same NP type generated under different SAG concentrations.
We found that the accessibility profiles between the same NP type isolated from different SAG concentrations were indistinguishable (Figures 2C–2E). Differential accessibility analysis using DESeq2 revealed no significant differentially accessible peaks between pMN derived from different SAG concentrations (Figures 2D and 2E) nor between p3 derived from different SAG concentrations (Figures 2D and 2E). By contrast, over 2,000 differentially accessible peaks were identified between pMN and p3 generated under the same SAG concentration (thresholds: fold change over 2, FDR < 0.01 and base mean > 100 normalized counts) (Figure 2D). Similarly, p0-1 or p2 arising from different concentrations had few if any differentially accessible elements (Figure S2I, arrowhead), whereas almost 500 elements were highly differentially accessible between these two NP types generated from the same SAG concentration (Figure S2I).
Thus, each cell type, identified by marker gene expression, acquired a specific chromatin landscape, irrespective of the agonist concentration to which they were exposed. This indicates the GRN mechanism of morphogen interpretation, that converts graded Shh input into distinct cell identities, establishes not only the discrete transcriptional identities of NPs but also their chromatin landscapes. Thus the differential chromatin accessibility induced by different levels of Shh signaling reflect cell-type-specific identity and are a product of the GRN that patterns progenitors.
A shared chromatin landscape for p0-1, p2, and pMN
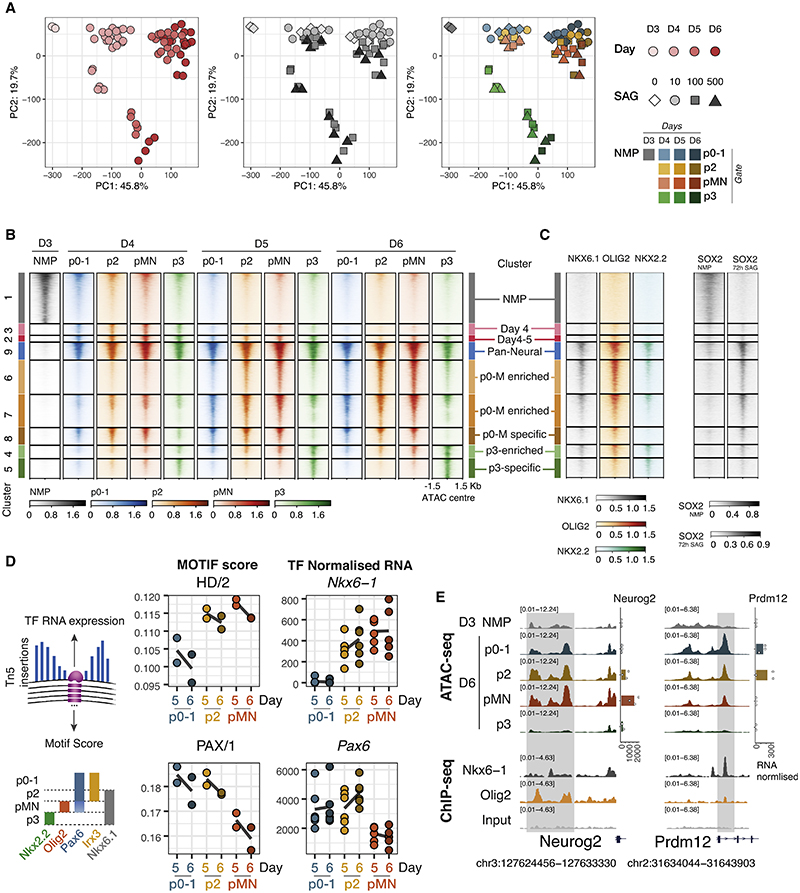
To examine the global accessibility dynamics, we performed principal component analysis (top 30,000 elements, Figure 3A) and clustered differentially expressed elements across all conditions using k-means (Figure 3B; STAR Methods). The main change across all data is the global decommissioning of the neuromesodermal progenitor (NMP) program and establishment of the NP program, which occurs between day 3 and day 4 of differentiation (Figures 3A and 3B). This involves closing a large proportion of elements open in D3 NMPs (Figure 3B, cluster 1 “NMP”), as well as opening elements in NPs (Figure 3B, cluster 3 “day 4,” cluster 2 “days 4 and 5,” cluster 9 “pan-neural”). This is expected as cells adopt neural identity. Consistent with this, SOX2 ChIP-seq from NMPs32 is enriched in cluster 1 NMP elements, and SOX2 ChIP-seq from neural cells13 is enriched in neural clusters (9, 6, 7, 8, 4, and 5) (Figure 3C).
Figure 3. Two regulatory landscapes underlie the Shh response of neural progenitors.
(A) Dimensionality reduction (principal component analysis) based on the most variable 30,000 consensus elements shows two different behaviors: a shared one for p0-1, p2, pMN samples, and a different one for p3 samples, regardless of SAG concentration. Each symbol represents a sample colored by differentiation day, SAG concentration, or cell type as indicated in the legend. n = 3 biological replicates (independent differentiations).
(B) Heatmap showing ATAC-seq coverage for elements differentially accessible between any two cell types or time points with the same dynamics, grouped in clusters (see STAR Methods) shows decommissioning of the NMP program (cluster 1), a shared pan-neuronal cluster (cluster 4) and two behaviors in cell type-specific accessibility, a shared for p0-1, p2, and pMN samples (clusters 6–8), and a unique program for p3 (clusters 4 and 5). Elements ordered by average accessibility.
(C) Heatmap of ChIP-seq coverage for the same elements for NKX6.1, OLIG2, and NKX2.216 performed in neural embryoid bodies treated with SAG shows binding to both pan-neuronal and cell-type-specific elements, and SOX2 from either NMP stage32 or SAG-treated neural embryoid bodies13 correlates with accessibility in either NMP or neural progenitors, respectively.
(D) Footprinting scores (using TOBIAS, see STAR Methods) for motifs with highly variable scores between p0-1, p2, and pMN at days 5 and 6, and normalized RNA counts for the most correlated TF within the motif archetype. The motifs for cell-type-specific TFs show expected footprinting differences.
(E) Example loci that are accessible across p0-pMN but were not accessible at day 3 and do not became accessible in p3 NPs. ChIP-seq for NKX6.1 and OLIG216 shown for the same elements. Normalized expression for nearby genes Neurog2 and Prdm12 shown. Points are individual samples; bars represent average expression.
See also Figure S3.
From day 4 onward, as cells adopt one of the ventral NP fates, the analysis revealed that pMN, p2 and p0-1 were remarkably similar (Figures 3A and 3B). This is surprising as these progenitors are molecularly distinct and give rise to functional different neurons: pMNs, characterized by the expression of OLIG2 generate motor neurons, whereas the p2 domain expresses IRX3 and generates V2 interneurons. RNA expression of marker genes from paired RNA-seq samples confirmed the expected identity of these cell types (Figure S3A). NP p3 were, however, distinct in their chromatin accessibility profile (Figures 3A and 3B).
The markers that define NP domains are repressive TFs and are directly involved in cell-type specification. OLIG2 is expressed in the pMN and represses p3 and p2 fate. NKX6.1 is expressed in p2, pMN and p3 and represses p0-p1. Binding data (ChIP-seq) from these TFs16 showed both NKX6.1 and OLIG2 bind regions of open chromatin accessible across all the panneural (cluster 9) and regions shared by pMN, p2, and p0-1 (p0-M enriched/specific clusters 6, 7, 8) (Figures 3C, S3B, and S3C). Since these TFs are only expressed in a subset of the cell types, this suggests that the same elements are open across multiple cell types and occupied by different TFs in different NP subtypes.
To explore gene regulation differences between p0-p1, p2, and pMN NPs, we turned to footprinting. This computational approach uses transposase insertion sites to identify motifs that are under-transposed within open chromatin and are thus likely to be protected by a bound protein. We used TOBIAS33 with motifs from three databases,34–36 grouping motifs into published archetypes based on position weight matrix clustering of motifs.37 The analysis revealed homeodomain (HD) and PAX motifs among those most differentially scoring between pMN, p2, and p0-1 at day 5 and day 6 (Figure 3D). From all the TFs associated with these motifs, the genes Nkx6-1 and Pax6 had RNA expression levels most consistent with the footprint score for HD and PAX, respectively (Figure 3D). In addition, we found the Ebox motif to be among the most variable footprints between these NPs (Figure S3D). The high footprint score in pMN is expected as these NP express Olig2. However, this is not the only Ebox binding TF expressed in these cell types that could explain this footprint. Neurog2 and Neurod1 both bind the same motif and are expressed in p2 and pMN, potentially explaining the Ebox footprint score in p2 NPs. This highlights the challenges in assigning footprints to specific TFs. Overall, the differential footprints found between p0-1, p2, and pMN supports the differential binding strategy, in which a common set of open chromatin regions is shared across cell types and different protein composition of the CREs determine gene activity (Figure 3E).
FOXA2 establishes the p3 regulatory landscape
By contrast to the other NPs, p3 cells had a distinct global accessibility profile, consistent with the differential accessibility strategy (Figures 3A and 3B). Accessibility was apparent at a unique set of open chromatin regions not shared with any other NP (“p3-specific” cluster 5), and at a set of elements highly enriched in this cell type (“p3-enriched” cluster 4). These differences are evident in the second principal component of the PCA where p3 cells are clearly distinct from all other ventral progenitors (Figure 3A).
Analysis of ChIP-seq for the p3-specific TF NKX2.2 revealed NKX2.2 was enriched in the p3-enriched (cluster 4) and p3-specific clusters (cluster 5) (Figures 3C and S3B). However, NKX2.2 was not specific, and it also bound the elements shared across all cell types (pan-neural cluster 9) and, to a lesser extent, the p0-pMN-enriched clusters (clusters 6 and 7) (Figures 3C, S3B, and S3C). Thus, the expression of NKX2.2 did not appear to explain the distinct chromatin landscape of p3. What then drives this distinct chromatin remodeling?
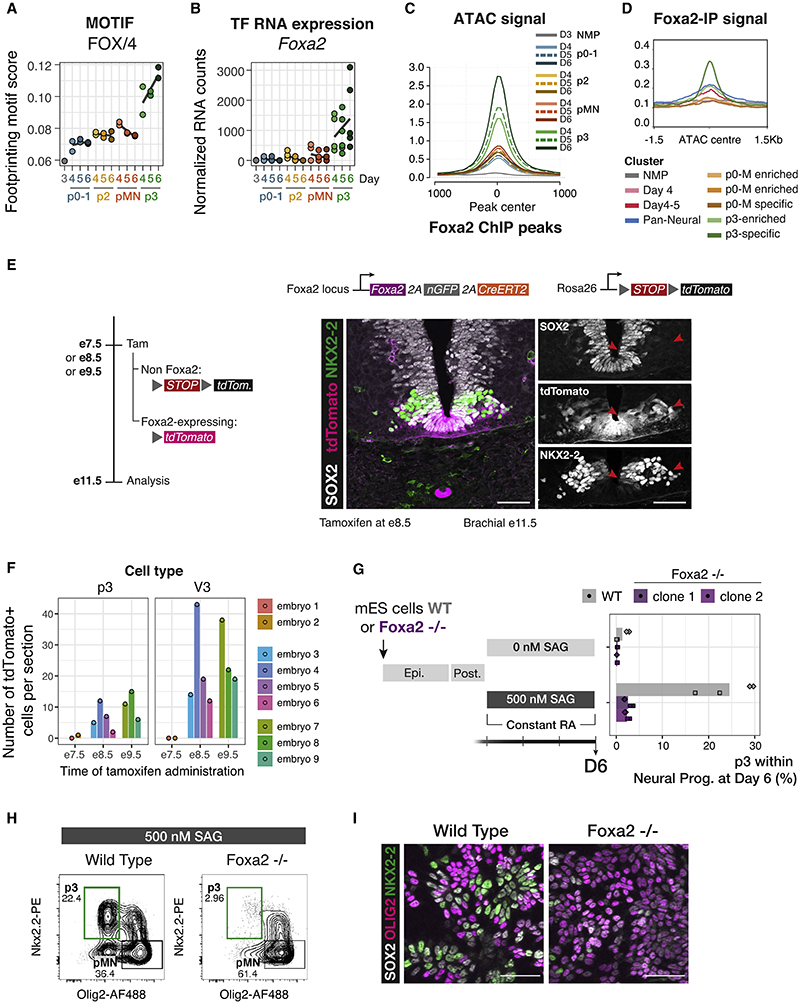
To explore which TFs made p3 NP different from other NPs, we compared footprinting scores across all conditions. The strongest footprint distinguishing p3 from other NPs was predicted to correspond to FOXA2 (Figures 4A, 4B, and S4C). This was intriguing as FOXA2 is known to be expressed during FP development.38–40 FOXA2 responds to Shh,41 and it induces Shh expression,42 thus generating a new signaling center in the ventral neural tube.
Figure 4. FOXA2 drives the p3-specific chromatin accessibility program.
(A) Footprinting score for the FOX motif is highest in p3 samples.
(B) Foxa2 expression in p3 NPs suggests it is the most likely candidate to drive the footprinting signal.
(C) Average ATAC-seq accessibility at FOXA2 ChIP-seq peaks13 in the indicated samples shows these regions are highly accessible in p3 NPs.
(D) Normalized FOXA2 ChIP-seq coverage showing accessibility in p3-specific elements from the groups of ATAC-seq elements in the indicated clusters from Figure 3B.
(E) Genetic lineage tracing indicates that cells that expressed Foxa2 at E8.5 (tamoxifen administration) have contributed to the p3 progenitor and V3 neuronal cell types by E11.5 (red arrows).
(F) Quantifications of p3 and V3 cells expressing tdTomato in embryos collected after tamoxifen administration at the indicated times. Biological replicates: n = 2 for E7.5, n = 4 for E8.5, n = 3 for E9.5.
(G) Foxa2−/− ES cells fail to generate p3 NPs when exposed to 500 nM SAG.
(H) Representative flow cytometry plots of the quantifications in (G) showing a marked reduction in p3 NPs from Foxa2−/− ES cells compared with wild type. Cells are gated for SOX2+ live neural progenitors.
(I) Representative immunohistochemistry staining for SOX2, OLIG2 and NKX2,2 showing reduced number of cells expressing NKX2.2 in Foxa2−/− mutant cells at day 6 of differentiation treated with 500 nM SAG.
Scale bars, 50 μm.
See also Figure S4.
To exclude the possibility that the cells isolated as p3 NPs (SOX2+ NKX2.2+ OLIG2–) contained FP cells, we devised culture conditions that promoted expression of mature FP markers (Arx, Shh) in which Foxa2 is expressed at high levels. We compared our spinal cord (SC) differentiation with FP differentiation and observed little expression of FP markers in SC conditions even at the last time point sorted, D6 (Figure S4A). Moreover, the expression of FP markers even at later time points was very inefficient in SC conditions, suggesting that the FOXA2 footprint is not the result of FP induction (Figures S4A and S4B).
We next analyzed published FOXA2 ChIP-seq generated from neuralized embryoid bodies treated with SAG.13 This revealed FOXA2 binding only to p3-specific peaks (Figure 4D). Consequently, we observed open chromatin in p3 NP samples in the regions bound by FOXA2 (Figure 4C). This contrasts with ChIP-seq of other NP TFs, which showed accessibility across all NP cell types (Figure S3C). Together, the data prompt the hypothesis that FOXA2 is driving the unique chromatin accessibility profile we observe in p3 cells.
In vivo p3 cells have a history of FOXA2 expression
If FOXA2 plays a role in generating the p3 chromatin accessibility profile, it should be expressed in p3 cells. In vivo NKX2.2 and FOXA2 are co-expressed at early stages of neural tube development,43,44 but these FOXA2 expressing cells have been thought to mark the future FP.45 To determine whether cells that express FOXA2 also contribute to the p3 domain, we took advantage of genetic lineage tracing in mouse embryos. Foxa2-driven CreERT2 expression46 induced recombination of a fluorescent reporter when tamoxifen was administered to pregnant mice at E8.5 or E9.5. Embryos analyzed at E11.5 showed expression of the reporter in both p3 (as identified by SOX2 and NKX2.2 expression) and V3 neurons (identified as laterally located cells expressing NKX2.2 alone) (Figures 4E and 4F). When tamoxifen was administered at E7.5, zero and one p3 cells were detected in two embryos, consistent with de novo Foxa2 expression induced by ventral Shh signaling (Figure 4F). This demonstrates that p3 cells and their neuronal progeny express Foxa2 during their history and supports the hypothesis that FOXA2 expression establishes a unique chromatin landscape in p3.
FOXA2 is required for p3 cell fate specification
Our results are consistent with the hypothesis that FOXA2 drives extensive remodeling of the chromatin landscape in p3 cells, which in turn is essential for p3 identity. To test this hypothesis, we used genome engineering to create a FOXA2 knockout mouse ES cell line. Foxa2−/− cells showed a marked reduction in the proportion of p3 cells (25% in wild type to 2% in the mutants) under 500 nM SAG (Figures 4G–4I).
Cells co-expressing NKX2.2 and OLIG2 were still present in Foxa2−/− (Figure 4H). This indicates FOXA2 is not directly required for the expression of NKX2.2. Instead, we hypothesize that FOXA2-driven remodeling is required in order for NKX2.2 to exert its repressive activity on OLIG2 and establish the p3 domain. Consistent with this, we observed binding of FOXA2 at the same locations as NKX2.2 (Figure S4D).
FOXA2 expression substitutes for Shh signaling early in p3 specification
FOXA2 is expressed in NPs closest to the source of Shh, and it is only co-expressed with NKX2.2 at early stages of neural tube development. To ask whether early exposure to Shh signaling is necessary to establish a p3 identity, we took advantage of our in vitro NP differentiation protocol to investigate the timing requirements for FOXA2 expression and Shh signaling in p3 specification. To this end, we exposed cells to 0 nM SAG for 24 h before changing regime to 500 nM SAG. Compared with a constant exposure to 500 nM SAG, cells exposed to delayed SAG are greatly impaired in their generation of p3 NPs (Figures S5A and S5B). This was not due to reduced Shh transduction (Figure S5B). The reduction in p3 specification was reminiscent of the outcome of exposing Foxa2−/− NPs to 500 nM SAG (Figures 4G and 4H) as we also saw NPs co-expressing NKX2.2 and OLIG2 (Figure S5B). When cells were exposed to SAG from day 3, a proportion of them show low but detectable levels of FOXA2 after 24 h. In the delayed SAG addition conditions, however, cells did not upregulate FOXA2 in the equivalent time frame (Figure S5D).
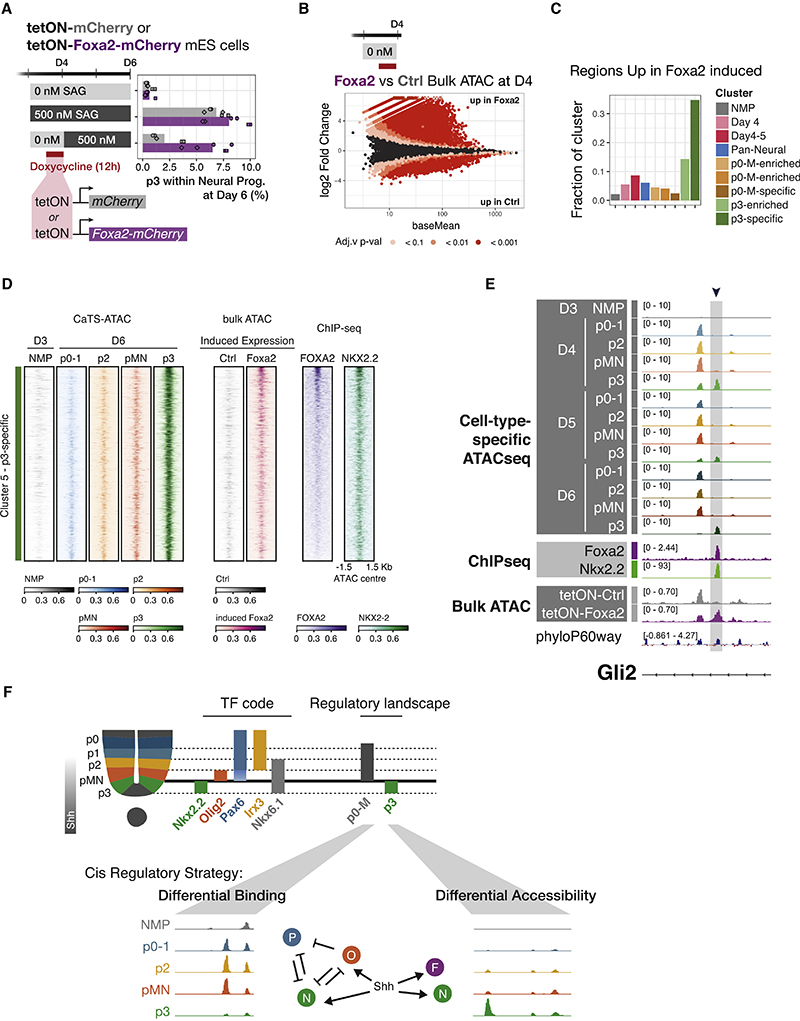
This regime provided the opportunity to test whether FOXA2 expression within the initial 24 h of NP differentiation was sufficient to rescue p3 generation. We used doxycycline inducible lines that expressed either mCherry or a Foxa2-mCherry fusion under the control of the doxycycline-responsive promoter tetON. Consistent with prior experiments, overexpression of mCherry alone in this “delayed SAG” regime showed a reduction in p3 generation at D6 (Figure 5A). By contrast, enforced expression of Foxa2 for 12 h during the initial 24 h of neural differentiation rescued p3 generation to levels comparable with those observed with constant 500 nM SAG (Figure 5A).
Figure 5. FOXA2 can replace Shh in early p3 specification where it opens p3-specific regulatory elements.
(A) A delayed SAG regime greatly reduces p3 generation in control cells (mCherry). A 12 h overexpression of tetON-Foxa2-mCherry rescues p3 generation. n = 3 biological replicates (independent differentiations), and 2 independent samples from each differentiation, for each condition.
(B) Differential accessibility in Foxa2 forced expression versus control mCherry shows FOXA2 predominantly opens elements. n = 3 biological replicates (independent differentiations).
(C) Overlap of differentially upregulated regions from (B) with ATAC clusters (Figure 3B) reveals a large fraction of the p3-specific cluster is opened upon Foxa2 overexpression.
(D) Heatmap showing CaTS-ATAC-seq coverage over all sites in the p3-specific cluster 5, bulk ATAC-seq coverage after induced expression of control or FOXA2, as well as ChIP-seq coverage for FOXA2 and NKX2.2. Regions are ordered by FOXA2 ChIP-signal and depict higher accessibility when FOXA2 (but not control) is induced, concomitant with increased binding of FOXA2 and NKX2.2 in the same regions.
(E) Example element in a Gli2 intron with p3-specific chromatin accessibility, FOXA2 and NKX2.2 binding that gains accessibility upon Foxa2 overexpression but not control.
(F) Diagram describing the two regulatory landscapes that underlie the molecular identity of ventral neural progenitors, a differential binding strategy is used to distinguish p0-1, p2, pMN, whereas p3 employ a differential accessibility strategy.
See also Figure S5.
These data indicate that high Shh at the onset of SC progenitor specification is required to induce Foxa2. This expression of FOXA2 is sufficient to confer competence for Shh signaling to specify p3 identity.
FOXA2 is sufficient to remodel the chromatin landscape
To test whether FOXA2 is responsible for the early p3-specific chromatin remodeling, we took advantage of the tetON cell lines. We performed bulk ATAC-seq at D4, which is 24 h after the induction of neural identity and 12 h after doxycycline exposure (Figure 5B). Both control and FOXA2-overexpressing conditions showed accessibility across the previously defined NMP and pan-neuronal clusters (Figure S5F). Although neither cell line had been exposed to SAG, FOXA2-expressing cells displayed opened chromatin across the p3-specific cluster (Figure S5F).
Consistent with its known role as a pioneer factor,47 FOXA2 overexpression resulted in chromatin opening (Figure 5B). Over 30% of the regions in the p3-specific cluster overlapped with regions more accessible in FOXA2-overexpressing cells (Figure 5C). Increased accessibility in cluster 5 (p3-specific) regions in response to induced FOXA2 is observed together with binding of p3-specific TF NKX2-2, with areas of highest binding also sharing FOXA2 binding (Figure 5D). An example of this behavior is shown for a Gli2 intron (Figure 5E), as well as three additional loci (Figure S5G). These results support a model in which FOXA2 opens chromatin regions required for the p3 fate. Thus, Shh induction of Foxa2 at early neural developmental stages is required to configure the p3-specific response of the cells to Shh. Cell fate choice between p0-1, p2, and pMN employ a differential binding strategy, whereas p3 rely on differential accessibility mediated by pioneer factor FOXA2 (Figure 5F).
A common lineage-pioneering role for FOXA2 across germ layers
FOXA TFs are required for the development of endoderm-derived lineages, pancreas, liver, and lung, where they play partially redundant roles.48–53 Although many TFs play roles in multiple tissues, they are generally thought to act via different enhancers in a combinatorial fashion with other tissue-specific TFs.54,55 Pioneer TFs are proposed to act by opening up compacted chromatin, thus allowing other TFs to bind.56 Hence, a given pioneer TF could have access to the same binding sites independent of the cellular context. However, the binding of pioneer TFs has been shown to depend on epigenetic features.57 Indeed, cell-type-specific co-factors direct cell-type-specific binding of FOXA2 in endogenously expressing cell lines and in FOXA2 overexpressing cell lines.58
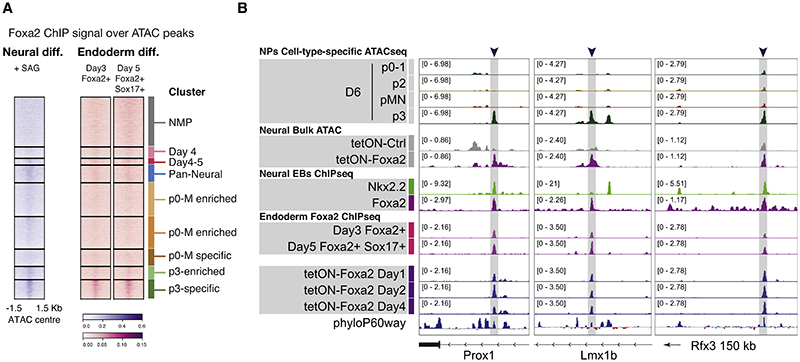
We therefore asked how much of FOXA2’s pioneering activity is tissue specific between endoderm and neural p3 progenitors. We examined the binding of FOXA2 in cellular differentiation models of endoderm at day 3 and day 550 and asked whether endoderm FOXA2 was bound to any of the SC differentiation clusters. To our surprise, we found FOXA2 also bound to the p3-specific cluster of elements in endoderm (cluster 5) (Figure 6A). This included genes such as Prox1, expressed in both lineages;59,60 Lmx1b, required for specification of hindbrain p3-derived serotonergic neurons61,62 and for in vitro pancreatic islet generation;63 and Rfx3, also required for pancreatic endocrine differentiation64 and expressed in the ventral neural tube65 (Figure 6B). This argues for a functional role in both tissues for at least some of these targets. Moreover, many of these sites were either opened in NPs upon brief overexpression of Foxa2 (Figures 6B and S5F) or occupied by overexpressed FOXA2 in endoderm differentiation (Figure 6B).
Figure 6. A common regulatory role of FOXA2 in ventral neural and endoderm lineages.
(A) Heatmap of ChIP-seq coverage for neural FOXA2 and endoderm FOXA2 at two differentiation time points50 show binding for both in the p3-specific accessibility cluster.
(B) Potentially functionally relevant target genes expressed and/or required in both tissues, with p3-specific accessibility, opened in response to FOXA2 overexpression, bound by FOXA2 in neural and endoderm, including overexpression of FOXA2.
See also Figure S6.
Although endoderm and NPs also have tissue-specific FOXA2 binding, these data raise the possibility that the two tissues share a core set of targets. Mechanistically, this could be mediated by the classical pioneer role of FOXA2 in this subset of targets or by the presence of common co-factors in both tissues. Intriguingly, it could offer evidence of an evolutionarily conserved GRN.
Discussion
Here, we identify two cis-regulatory strategies that direct cell fate choice in developing NPs. One strategy—differential binding — relies on a common regulatory landscape. The different composition of TFs at these CREs dictates differential gene expression and cell fate decisions. The second strategy—differential accessibility—relies on cell-type-specific chromatin remodeling (Figure 5F). This is the case for p3 NPs, which have a unique set of accessible elements that distinguish them from all other NPs. We show that FOXA2, expressed early in response to Shh, is responsible for the remodeling that opens p3-specific elements. Many of these elements then bind the p3-specific TF, NKX2.2. Intriguingly, a subset of the elements bound and opened by FOXA2 in NPs are also bound by FOXA2 during endoderm differentiation, where it plays a parallel role opening elements later required for specific endoderm lineages.51 The identification of these two regulatory strategies establishes a framework for classifying the genomic implementation of cell fate-decision mechanisms.
To assay cell-type-specific chromatin accessibility, we combined intracellular flow cytometry with methods developed to measure protein levels at single-cell resolution27 that produced high quality chromatin accessibility data for specific cell types.29
The method is applicable to other tissues, in vitro and in vivo, and will be of particular use in cases where fluorescent reporters are not readily available or when the identification of the desired cell types depends on the intersectional expression of multiple markers. Moreover, the approach could be extended to isolate cells based on other intracellular characteristics, such as post-translationally modified signaling effectors.
A common set of pan-neural elements, not accessible prior to neural induction, was apparent in all NPs. These elements were enriched for binding of TFs involved in the neural tube GRN, including SOX2, NKX6.1, NKX2.2, and OLIG2. The existence of this set of elements is reminiscent of the idea of morphogenetic fields—discrete, modular units of embryonic development66 that display a coordinated response to patterning signals.66 Comparison of chromatin accessibility in the same NP cell type generated using different levels of Shh signaling revealed that cell identity, rather than morphogen concentration, correlated with chromatin accessibility pattern. This indicates that the morphogen interpretation mechanism that converts graded Shh input into distinct cell identities establishes not only the discrete transcriptional identities of NPs but also their chromatin landscapes.
The shared chromatin landscape of the majority of ventral NPs suggests a differential binding strategy governs gene expression selection for these cells. This is consistent with the “selection by exclusion” mechanism that has been proposed previously to explain the specification of progenitor subtype identity by graded Shh signaling.16,17 In this view, the positive and negative transcriptional inputs supplied by the GRN are integrated at CREs associated with target genes, which are accessible in NPs, regardless of whether the target gene is active. Thus, cell fate choice does not depend on major chromatin remodeling but instead is determined by the composition of transcriptional effectors bound. These findings challenge the validity of approaches typically used to infer gene regulation and cell identity from genomic data, which equate open chromatin with gene activity and cell fate. Our data demonstrate that a major regulatory strategy—“differential binding”—violates this assumption and indicate the necessity of developing more nuanced methods.
By contrast, the differential accessibility strategy appears to dictate p3 identity. These NPs are distinguished by accessibility at a distinct subset of CREs that depend on FOXA2. Despite the transience of FOXA2 expression in p3 NPs,67 accessibility at the CREs opened by FOXA2 persists and are bound by the p3-specific TF NKX2.2. This is similar to the observation that the presence of FOXA is not required to maintain stable epigenetic states in liver cells.68 The marked changes in chromatin landscape initiated by FOXA2 thus represents an example of epigenetic memory and demonstrate how the pioneering activity of a TF rewires a GRN to specify a specific cell type.
The different regulatory strategies have important implications for cellular reprogramming. Cell types sharing the same landscape (differential binding) would be relatively plastic, enabling transitions between the different fates by altering the TF configuration on already available CREs. Consequently, the barrier to reprogram cells between such fates would be lower. On the other hand, the differential accessibility strategy involves chromatin remodeling to facilitate the binding of TFs previously unable to bind specific CREs. The expression of the end point TFs alone might not be sufficient for cell fate acquisition if the elements these TFs need to bind are inaccessible. This might explain the inefficiency or incompleteness of some transgene-driven reprogramming approaches that use only the TFs expressed in the final state of the target cell type.69 Understanding and recapitulating the epigenetic trajectory cells follow to reach the desired endpoint might improve such differentiation protocols.
Our results are consistent with prior genetic studies showing the p3 NPs are the only NPs that require activating GLI proteins for their specification.19,70 The demonstration that FOXA2 expression can substitute for Shh signaling during the early steps of p3 specification indicates that the requirement for activating GLI proteins is for the expression of FOXA2. This requirement restricts p3 induction to the most ventral progenitors in the neural tube. The expression of FOXA2 and NKX2.2 in p3 NPs also appears to contribute to establishing p3 identity by modulating the cell intrinsic response to Shh.43 NKX2.2 promotes Foxa2 via a mechanism involving Gli3 repression43 and FOXA2 binds and opens an intronic region in the gene encoding Gli2 (Figure 5D). Together, these data suggest that the interpretation of Shh signaling by p3 NPs is distinct from the adjacent p0-pMN region. This distinction involves remodeling the regulatory landscape of p3 NPs.
FOXA2 has been proposed to act as a pioneer factor in the endoderm, where it remodels chromatin to allow the binding of other TFs.51,56 Although TFs often have roles in multiple tissues, they are usually thought to act in a combinatorial fashion with other TFs via different elements in different tissues.54,55,58 The observation that FOXA2 binds to p3 NP-specific CREs in endodermal cells raises the possibility these two cell types share some aspects of their regulatory control. Consistent with this, pancreas specification, an endoderm derivative, involves many TFs that are also expressed in the ventral neural tube, specifically in the p3 domain.71 The potential shared control by FOXA2 of functionally relevant genes in endoderm and neural tissue raises the question of how this might have arisen. The role of FOXA proteins in endoderm development is evolutionarily conserved72 and predates the origin of bilaterians. One possibility therefore is the co-option of an endoderm FOXA2 regulatory program into NPs in an ancestor of vertebrates. Alternatively, it could be evidence of convergent evolution or of a common evolutionary origin for endoderm and neural cells. This latter possibility would be supported by recent work suggesting that parts of the digestive system in the cnidaria Nematostella are of ectodermal origin73 and that a common progenitor generates neurons and secretory cells in this species.74,75 Thus, the shared regulatory features seen in vertebrate endoderm and neural cells might be a vestige of a common evolutionary origin of these cell types. A deeper understanding of the GRNs in neural tissue and endoderm will be needed to explore this possibility. More generally, it will be important to determine whether other tissues use similar cis-regulatory strategies to specify distinct cell types.
Limitations of the study
In this study, we used Foxa2 knockout ES cell lines to show a requirement for Foxa2 in NPs. Whether there is a similar requirement for FOXA2 in vivo is not known because the removal of Foxa2 in vivo disrupts axis formation and is embryonic lethal at an earlier time point.76,77 Additionally, the previously published ChIP-seq data used in this study was generated from a different directed neural differentiation protocol, and there could be differences in binding of the ventral TFs as a result. Finally, although we demonstrate a good correlation between data produced by CaTS-ATAC and conventional ATAC protocols that use live sorted cells, there could be limitations introduced by the paraformaldehyde fixation necessary for CaTS-ATAC. Paraformaldehyde disrupts TF binding to mitotic chromatin, and this might result in a loss of any cell cycle phase specific signal.
Star★Methods
Key Resources Table.
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Sox2-V450 | BD Biosciences | Cat#561610; clone O30-678 |
| Pax6-PerCPCy5.5 | BD Biosciences | Cat#562388; clone 018-1330 |
| Nkx6.1-AlexaFluor647 | BD Biosciences | Cat#563338; clone R11-560 |
| Goat anti-Olig2 | R&D | Cat#AF2418 |
| Nkx2.2-PE | BD Biosciences | Cat#564730; clone 74.5A5 |
| Tubb3-Biotin | BD Biosciences | Cat#560394; TUJ1 |
| Foxa2-AlexaFluor488 | SC Biotechnology | Cat#sc-374376 AF488; clone H-4 |
| Goat anti-Sox2 | R&D | Cat#AF2018 |
| Rat anti-Sox2 | Invitrogen | Cat#14-9811-80, clone Btjce |
| Mouse anti-Nkx2.2 | BD Biosciences | Cat#564731, clone 74.5A5 |
| Rabbit anti-Olig2 | Merck Millipore | Cat#AB9610 |
| Rabbit anti-Arx | Kind gift from J.Chelly | N/A |
| Goat anti-Foxa2 | R&D | Cat#AF2400 |
| Rabbit anti-Pax6 | Merck Millipore | Cat#AB2237 |
| Donkey anti-rat 405 | Biotium | Cat#20419 |
| Donkey anti-mouse AF488 | Invitrogen | Cat#A21202 |
| Donkey anti-goat AF488 | Invitrogen | Cat#A11055 |
| Donkey anti-rabbit AF488 | Invitrogen | Cat#A21206 |
| Strep-APC-Cy7 | BioLegend | Cat#405208 |
| Donkey anti-mouse AF568 | Invitrogen | Cat#A10037 |
| Donkey anti-goat AF568 | Invitrogen | Cat#A11057 |
| Donkey anti-mouse AF647 | Invitrogen | Cat#A31571 |
| Donkey anti-goat AF647 | Invitrogen | Cat#A21447 |
| Bacterial and virus strains | ||
| BL21-Gold(DE3) | Agilent | Cat#230132 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant Mouse LIF Protein | Merck | Cat#ESG1107 |
| Dulbecco's Modified Eagle Medium (DMEM) - Knock OUT |
Gibco | Cat#10829-018 |
| ES Foetal Bovine Serum (FBS) | Pan Biotech | Cat#P30-2602 |
| Penicillin/Streptomycin | Gibco | Cat#15140122 |
| L-Glutamine | Gibco | Cat#25030024 |
| Non-essential amino acids | Gibco | Cat#11140-035 |
| 2-mercaptoethanol | Gibco | Cat#21985-023 |
| 0.05% trypsin-EDTA | Gibco | Cat#25300054 |
| Gelatin | Gibco | Cat#G1393-100ML |
| Advanced DMEM-F12 | Gibco | Cat#21331-020 |
| Neurobasal medium | Gibco | Cat#A35829-01 |
| N2 | Gibco | Cat#17502001 |
| B27 | Gibco | Cat#17504001 |
| Bovine Serum Albumin solution | Merck | Cat#A7979-50ML |
| Matrigel | Corning | Cat#356231 |
| bFGF | R&D | Cat#100-18B |
| LGK | Cayman Chemical Company | Cat#1.800.364.9897 |
| CHIR99021 | Axon Medchem | Cat#1386 |
| SB-431542 | Tocris | Cat#S0400 |
| DMH1 | Adooq Bioscience | Cat#A12820 |
| Retinoic Acid (RA) | Sigma | Cat#R2625 |
| Smoothened Agonist (SAG) | Merck | Cat#566660-5mg |
| DBZ | Biotechne | Cat#4489 |
| Doxycycline | Sigma-Aldrich | Cat#D9891 |
| Tamoxifen | Sigma-Aldrich | Cat#T5648-5G |
| Lipofectamine 2000 | Invitrogen | Cat#11668030 |
| Hygromycin-B | Thermo Fisher | Cat#10687010 |
| Puromycin | Thermo Fisher | Cat#A1113802 |
| LIVE/DEADTM Fixable Dead Cell Stain Near-IR fluorescent reactive dye |
ThermoScientific | Cat#L34976 |
| Phosphate Buffer Saline (PBS) | Gibco | Cat#14190-094 |
| Accutase | Gibco | Cat#00-4555-56 |
| Paraformaldehyde (PFA) | ThermoScientific | Cat#28908 |
| Triton-X100 | VWR Chemicals | Cat#28817.295 |
| Superscript III reverse transcriptase | Invitrogen | Cat#18080-051 |
| SYBR Green PCR assay | ThermoFisher Scientific | Cat#A25742 |
| ProLong Gold antifade reagent | Invitrogen | Cat#P36930 |
| Igepal CA-630 | Sigma | Cat#I8896-100ML |
| Tween-20 | Sigma | Cat#P2287-500ML |
| Digitonin | Invitrogen | Cat#BN2006 |
| Proteinase K | Ambion | Cat#AM2546 |
| 2X PCR Master Mix NEB | NEB | Cat#M0541S |
| AMPureXP beads | Beckman Coulter | Cat#A63882 |
| Ni-Sepharose column | Cytiva | Cat#17526801 |
| RNasin Plus | Promega | Cat#N2615 |
| Critical commercial assays | ||
| Zymo DNA concentrator kit | Zymo | Cat#D4013 |
| RecoverAll RNA/DNA Isolation Kit | Thermo Fisher Scientific | Cat#AM1975 |
| SMART-Seq HT kit | Takara | Cat#634437 |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat#FC-131-1096 |
| TDE tagmentation enzyme | Illumina | Cat#15027865 |
| 2X TD tagmentation buffer | Illumina | Cat#15027866 |
| RNeasy Qiagen kit with DNAse digest | QIAGEN | Cat#74106 |
| Mouse Embryonic Stem Cell Nucleofector™ Kit |
Lonza | Cat#VPH-1001 |
| Deposited data | ||
| CaTS-ATAC, live Omni-ATAC, RNA-seq and bulk ATAC-seq |
This study | GSE204921 |
| Experimental models: Cell lines | ||
| Mus musculus (Male): HM1 | Doetschman et al.78 | N/A |
| Mus musculus (Male): HM1-Foxa2-/- | This study | N/A |
|
Mus musculus (Male): HM1-TetON-Foxa2- mCherry |
This study | N/A |
|
Mus musculus (Male): HM1-TetON- mCherry |
This study | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: Foxa2nEGFP-CreERT2/+ | Imuta et al.46 | MGI:5490029 |
| Mouse: R26tdTomatoAi14/tdTomatoAi14 (Gt(ROSA)26Sortm14(CAG-tdTomato) Hze |
Madisen et al.79 | MGI:3809524 |
| Oligonucleotides | ||
| qPCR primers used in this study, see Table S1 |
N/A | |
| Primers for cloning of Foxa2 guides into pX458, see Table S1 |
This study | N/A |
| Oligonucleotides for Tn5 assembly, see Table S1 |
Picelli et al.80 | N/A |
| Oligonucleotides for CaTS-ATAC and ATAC-seq library, see Table S1 |
This study | N/A |
| Recombinant DNA | ||
| pX458 vector | Ran et al.81 | Addgene plasmid 48138 |
| PB-tetOn-destination-PGK-hygro | Stuart et al.82 | N/A |
| PB-CAG-rtTA3-PGK-puro | Stuart et al.82 | N/A |
| Software and algorithms | ||
| nf-core atacseq pipeline | https://nf-co.re/atacseq | https://doi.org/10.5281/zenodo.2634132 |
| DESeq2 | Love et al.83 | https://doi.org/10.18129/B9.bioc.DESeq2 |
| nf-core chipseq pipeline | https://nf-co.re/chipseq | https://doi.org/10.5281/zenodo.3240506 |
| TOBIAS footprinting tools | Bentsen et al.33 | https://doi.org/10.1038/s41467-020-18035-1 |
| nf-core rnaseq pipeline | https://nf-co.re/rnaseq | https://doi.org/10.5281/zenodo.1400710 |
| R | R Core Team | https://www.R-project.org/ |
| Fiji | Schindelin et al.84 | https://imagej.net/software/fiji/ |
| Other | ||
| Superfrost Plus slides | Thermo Scientific | Cat#10149870 |
| 22-mm-by-50-mm no. 1.5 thickness coverslips |
VWR | Cat#631-0138 |
| 6-well plates | Corning | Cat#353046 |
| LoBind 1.5 ml Eppendorf tubes | Eppendorf | Cat#Z666548 |
| DNA High Sensitivity DNA Kit | Agilent | Part No 5067-4626 |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, James Briscoe (james.briscoe@crick.ac.uk).
Materials availability
Murine ES cell lines generated in this study area available upon request.
Experimental Model And Subject Details
Lineage tracing of Foxa2-expressing cells
Foxa2-2a-nEGFP-2a-CreERT2/+;R26Tomato Ai14/ Tomato Ai14 embryos were obtained from time matings. Mouse lines Foxa2nEGFP-CreERT2/+ (MGI:5490029)46 and R26tdTomatoAi14/tdTomatoAi14 (Gt(ROSA)26Sortm14(CAG-tdTomato) Hze (MGI:3809524)79 were backcrossed and maintained in a C57BL6 background. Induction of recombination was achieved by tamoxifen administration by oral gavage at 0.08 mg/body weight.
All animal procedures were performed in accordance with the Animal (Scientific Procedures) Act 1986 under the UK Home Office project licenses PP8527846 and PF59163DB. Animals were housed under a 12-h light, 12-h dark cycle at the Francis Crick Institute animal facility. Animals were housed in singly-ventilated cages. The relative humidity was kept at 45 to 65%. Mouse rooms and cages were kept at a temperature range of 20-24°C. Animals had 24 h access to RO water and autoclaved pelleted food. Caging, bedding, nesting materials and enrichments were autoclaved prior to use and cages are changed routinely. A maximum of 5 adult mice were housed per cage if they were over 20 g. Animals were monitored visually daily for health concerns and once a week a full health check was carried out during a discretionary cage change
Cell lines
WT experiments were performed with the mouse embryonic stem cell line HM1.78 All cell lines were maintained at 37°C with 5% carbon dioxide (CO2).
Generation of Foxa2-/- ES cell lines
Generation of Foxa2 knockout cell lines was performed by electroporating two different guides cloned into the pX458 vector81 with the AMAXA nucleofector kit (Lonza Cat no. VPH-1001). Cells were sorted for GFP as single cells one day after electroporation. Clonal cell lines were genotyped and two clones were used. Both carried the same deletion.
Generation of tetON-Foxa2 and control ES cell lines
PiggyBac (PB)-tetOn-destination-PGK-hygro and PB-CAG-rtTA3-PGK-puro vectors were used from Stuart et al.82 Gateway cloning (Invitrogen) was used to insert the coding sequence for mCherry or FoxA2:mCherry fusion protein into the tetOn destination vector. Stable transgenic HM1 mouse ES cell lines were generated by transfection as follows: 1 μg PB-tetOn expression vector, 1 μg PB-CAG-rtTA3 vector, 1 μg non-integrating PBase transposase vector and 2 μl Lipofectamine-2000 (Invitrogen) were incubated for 20 min in 100 μl DMEMF12 (Gibco) at room temperature, then applied to 300,000 cells/6well in 1.6 ml medium for 18 h. Selection was applied to transfectants for at least 5 passages prior to use (150 μg/ml hygromycin-B (ThermoFisher) and 1 μg/ml puromycin [ThermoFisher]). Transfection and selection were performed in feeder-free 2iLIF culture conditions as described in Stuart et al.,82 then adapted back to feeders and ES cell medium regime as described below for at least 4 passages prior to experiments.
Method Details
Cell culture and neural progenitor differentiation
Mouse ES cell lines were maintained in ES cell medium (Dulbecco’s Modified Eagle Medium (DMEM) Knock Out (Gibco, Cat No. 10829-018) supplemented with 10% Foetal Bovine Serum (Pan Biotech, Cat. No. P30-2602), Penicillin/Streptomycin (Gibco, Cat No. 15140122), 2mM L-Glutamine (Gibco, Cat No. 25030024), 2mM Non-essential amino acids (Gibco, Cat No. 11140-035), and 0.1 mM 2-mercaptoethanol (Gibco, Cat. No. 21985-023)) with 1000 U/ml LIF (Chemicon, Int ESG1107) on mitotically inactive primary mouse embryo fibroblasts (feeders). For spinal cord neural differentiation, ES cells were dissociated using 0.05% trypsin-EDTA (Gibco, Cat No. 25300054) and then plated onto 0.1% gelatinised (Gibco Cat no. G1393-100ML) tissue culture plates for three successive periods of 15 mins to remove feeders. Differentiations were carried out in N2B27 media (Advanced DMEM - F12 (Gibco, Cat. No. 21331-020) and Neurobasal medium (Gibco, Cat. No. A35829-01) (1:1), supplemented with 1xN2 (Gibco Cat no. 17502001), 1xB27 (Gibco Cat no. 17504001), 2 mM L-glutamine (Gibco, Cat No. 25030024), 40 μg/ml BSA (Sigma-Aldrich, Cat No. A7979-50ML), and 0.1 mM 2-mercaptoethanol) with the indicated additives. Cells were plated onto 6-well plates (Corning, Cat. No. 353046) precoated in Matrigel (Corning, Cat. No. 356231) diluted 1/50 in Advanced DMEM - F12 at a density of 20,000 cells per 35mm well in 1.5 ml. The media was supplemented on the different days as follows: Day 0 to Day 2, 10ng/ml bFGF (R&D, Cat. No. 100-18B) and 5 μM LGK (Cayman Chemical Company, Cat. No. 1.800.364.9897); Day 2 to Day 3 for 20 h, 10 ng/ml bFGF, 5 μM CHIR99021 (Axon Medchem, Cat. No. 1386), 10 μM SB-431542 (Tocris, Cat. No. S0400), and 2μM DMH1 (Adooq Bioscience, Cat. No. A12820); from Day 3 onwards, 100 nM RA (Sigma, Cat. No. R2625) and either 0 nM, 10 nM, 100 nM or 500 nM SAG (Merck, Cat. No. 566660-5mg). The “delayed SAG” regime consisted of 100 nM RA and 0 nM SAG from Day 3 to Day 4, followed by a constant concentration of RA and increased SAG to 500 nM. Induction of the tetON system was achieved by supplementing the media with 1 μg/ml Doxycycline (Sigma-Aldrich, Cat. No. D9891) for the second 12h of Day 4. Floor plate conditions were achieved by addition of 500 nM SAG on Day 3 without any addition of RA.
Flow cytometry of intracellular markers
Sample collection
1 μl/ml of LIVE/DEADTM Fixable Dead Cell Stain Near-IR fluorescent reactive dye (ThermoScientific, Cat. No. L34976) was added to cells in culture and incubated at 37°C for 30 mins. Cells were washed twice with Phosphate Buffer Saline (PBS) (Gibco, Cat. No. 14190-094), and were dissociated using 0.5ml Accutase (Gibco, Cat. No. 00-4555-56) per well of a 6 well plate incubated 5 min at 37°C. Cell were collected, centrifuged at 400g for 4 min and resuspended in 100μl of 4% paraformaldehyde (PFA) (ThermoScientific, Cat. No. 28908). PFA fixation was carried out for 10 min at room temperature. Cells were washed in PBS and resuspended in 500μl PBS + 0.5% BSA.
Staining
1 million cells were stained for flow cytometry analysis in LoBind 1.5 ml Eppendorf tubes. Pellets were resuspended in 0.1ml of PBS + 0.5% BSA and 0.1% Triton-X100 (VWR Chemicals, Cat No. 28817.295) and were incubated on ice for 10 mins. Cells were then incubated with primary antibodies or directly conjugated antibodies diluted in the same buffer for 1.5 h protected from light at room temperature. When necessary, secondary antibodies were incubated under the same conditions for 40 min. Cells were washed in PBS + 0.5% BSA and 0.1% Triton-X100, and resuspended in 300μl of PBS + 0.5% BSA for analysis on a BD Fortessa analyzer (Becton Dickinson). The antibody panels used were as follows: “D-V progenitors”, Sox2-V450 (1:100), Pax6-PerCPCy5.5 (1:100), Nkx6.1-AlexaFluor647 (1:100), Goat Olig2 unconjugated (1:400), Nkx2.2-PE (1:100), and Tubb3-Biotin (1:800) followed by secondaries donkey anti-goat AlexaFluor488 (1:1000) and Strep-APC-Cy7 (1:20,000); “mCherry inducible”, Sox2-V450 (1:100), Pax6-PerCPCy5.5 (1:100), Goat Olig2 unconjugated (1:400), Nkx2.2-AlexaFluor647(1:100), secondaries donkey anti-goat AlexaFluor488 (1:1000); “D-V with Foxa2”, Sox2-V450 (1:100), Pax6-PerCPCy5.5 (1:100), Goat Olig2 unconjugated (1:400), Nkx2.2-PE (1:100) and Foxa2-AlexaFluor488 (1:100), with secondary donkey anti-goat AlexaFluor647 (1:1000). Antibody details in listed in the key resources table.
RNA extraction, cDNA preparation, and qPCR analysis
Wells were washed with PBS and lysed in 350 μl of RLT buffer (QIAGEN, Cat. No. 1015762) per 35mm well. After cells were mixed, they were collected in a 2 ml RNase-free Eppendorf tube and stored at -20°C. RNA extraction was performed using RNeasy Qiagen kit with DNAse digest (QIAGEN, Cat. No. 74106) as per manufacturer’s protocol. cDNA was synthesized from 1ug of RNA using Superscript III reverse transcriptase (Invitrogen 18080-051) with random hexamers, and qRT-PCR analyses were performed by QuantStudio 12K Flex Real-Time PCR system (ThermoFisher Scientific) using SYBR Green PCR assay (ThermoFisher Scientific, Cat. No. A25742). All experiments were performed in technical duplicates, biological duplicates or triplicates for each time point analysed. Expression values were normalized against β-actin. Primer sequences in Table S1.
Immunofluorescence staining of cell differentiations
Cells for immunofluorescence staining were cultured in matrigel-coated glass slides in 12-well plates with all the volumes adjusted to achieve the same cell density. Wells were washed with PBS and fixed in 4% PFA at 4°C for exactly 15 mins. Glass slides were transferred to a new 12-well plate for staining. Blocking was performed for 10 mins with PBS + 1% BSA + 0.1% Triton-X100 at room temperature, cells were incubated in PBS + 1% BSA + 0.1% Triton-X100 with primary antibodies overnight at 4°C, with secondary antibodies at room temperature for 1hr, and mounted with ProLong Gold antifade reagent (Invitrogen, Cat. No. P36930). Fluorescent images were taken with Leica SP8 confocal microscope or an Apotome microscope.
The following primary antibodies were used: goat anti-Sox2 (1:500), mouse anti-Nkx2.2 (1:1000), rabbit anti-Olig2 (1:500), rabbit anti-Pax6 (1:1000), rat anti-Sox2 (1:1000), goat anti-Foxa2 (1:500), rabbit anti-Arx (1:1000). Corresponding donkey-raised secondaries were used. Antibody details listed in the key resources table.
Generation of mouse sections and immunostainings
Mouse spinal cord sections staining were performed as previously described.85 Mouse embryos from timed pregnant females were collected and fixed in 4% PFA for 1.5h at 4°C, washed in PBS, and transferred to 15% sucrose in phosphate buffer overnight at 4°C. Embryos were subsequently embedded in gelatin solution (7.5% gelatin, 15% sucrose in phosphate buffer) and snap-frozen in isopentane on dry ice. Transverse cryosections (thickness: 14 mm) were cut using a Leica CM3050S cryostat (Leica Microsystems Limited, Milton Keynes, UK) and placed on Superfrost Plus slides (Thermo Scientific, Cat. No. 10149870). Slides were stored at −80°C until ready to be processed for immunohistochemistry. After immunohistochemistry, 22-mm–by–50-mm no. 1.5 thickness coverslips (VWR, Cat. No. 631-0138) were mounted onto the sections using ProLong Gold antifade reagent. Immunohistochemistry was performed in the same way as for cell culture slides using the same antibodies against Sox2 and Nkx2.2.
Quantification of lineage tracing from mouse sections
Images acquired on a Leica SP8 40X objective were segmented using the custom scripts described in https://github.com/tiagu/ Nucleus. The DAPI signal was used to segment nuclei and signal intensity for all other channels calculated for each nuclei. The raw data and code to reproduce the quantifications of tdTomato p3 or V3 cells can be found in https://github.com/MJDelas/tdTom_LinTracing. In brief, a threshold was found for the intensities of SOX2, NKX2.2 and tdTomato. Cells expressing SOX2 and NKX2.2 were classified as p3. Cells expressing only NKX2.2 were classified as V3. Position was used as a quality control step.
Because of background binding by a mouse raised antibody in mouse sections, NKX2.2 signal emanating from cells far from the p3 domain were excluded from the analysis. The number of either p3 or V3 expressing tdTomato per section were plotted.
CaTS-ATAC (Crosslinked and TF-Sorted ATAC-seq)
Sample collection
Cell type specific ATAC-seq based on intracellular markers was carried out from HM1 differentiations at the timepoints and SAG concentrations indicated. Dishes were washed with PBS and dissociated with 0.5ml Accutase (Gibco, Cat. No. 00-4555-56) per well of a 6 well plate incubated 5 min at 37°C. Cells were collected in Eppendor LoBind tubes (Eppendorf cat. #Z666548), the wells were rinsed with 1ml PBS and the samples were spun at 400 g for 4 min at room temperature. Samples were resuspended in 1μl/ml of LIVE/DEAD™ Fixable Dead Cell Stain Near-IR fluorescent reactive dye as per manufacturer’s instructions. After 30 min incubation on ice protected from light the samples were spun again and resuspended in 300 μl of PBS, followed by addition of 100 μl of PFA to achieve 1% PFA. Fixation was carried out for 15 min with rotation at 4°C. Fixation was quenched with 25 μl of 2M Glycine for 5 min at 4°C with rotation, spun and resuspended in PBS with 0.5% BSA. Cells were counted and 1 million cells were transferred to a new LoBind Eppendorf tube.
Transposition
Tn5 transposition was carried out as previously described28 with some modifications. Briefly, samples were resuspended in 0.5 ml of RSB buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2) supplemented with 0.1% Igepal CA-630 (Sigma, Cat. No. I8896-100ML), 0.1% Tween-20 (Sigma, Cat. No. P2287-500ML), and 0.01% digitonin (Invitrogen, Cat No. BN2006), and incubated on ice for 3 min. 1 ml of RSB with 0.1% Tween-20 was then added and samples were spun for 10 min at 2000 g and 4°C. The samples were then resuspended in 1 ml of ATAC mix(2X TDE buffer (Illumina), 50 μl TDE (Illumina), 0.01% digitonin, 0.1% Tween-20 and 330 μl of PBS). Transposition was carried out at 37°C with 1000 rpm shaking for exactly 30 min. The reaction was stopped with 30 μl of EDTA, spun at 2000g for 5 min at 4°C and resuspended in 100 μl of PBS with 0.5% BSA and 0.1% Triton X-100.
FACS
Flow cytometry staining was carried out as described in “Flow cytometry of intracellular markers”. Samples processed in a BD Fusion cell sorter (Beckton Dickinson). 15,000 cells of each desired population were sorted in to 0.5ml of PBS with 0.5% BSA. Sorted cells were spun for 10 min at 3000 g and 4°C and resuspended in Reverse crosslink buffer (50mM Tris-HCl ph8, 0.5% Tween-20, 0.5% Igepal CA-630) supplemented with 1 μl of 20 mg/ml Proteinase K (Ambion, Cat. No. AM2546) and incubated overnight at 65°C shaking at 300 rpm.
DNA isolation
The following day samples were spun down to collect condensation and DNA was isolated using the DNA clean up & Concentrator kit (Zymo, Cat. No. D4013) as per manufacturer’s instructions. DNA was eluted in 21 μl of DNAse-free water. DNA was stored at -20°C until ready to be processed.
Library generation
Transposed DNA was first amplified with 2X PCR Master Mix NEB (Cat. No. M0541S), and 1 μl of constant forward primer and indexed reversed primer (Table S1) (both at 25 μM) in a total reaction volume of 50 ul. The program was as follows: 5 min at 72°C, 30 s at 98°C, followed by 7 cycles of 30 s at 98°C, 30 s at 63 °C and 60 s at 72°C, with a final extension of 5 min at 72°C. DNA was cleaned up with 1.8x volumes of AMPureXP beads (Beckman Coulter, Cat. No. A63882) and eluted in 18 μl of EB buffer. qPCR was carried out to determine the number of extra cycles using 2 μl of the amplified DNA in technical duplicates. Each 20 μl reaction contained 20 μl of 2X SYBR Green PCR assay, using PowerUP SYBR Green Master Mix (Thermo Fisher, Cat. No. A25742) and 2.5 μl of each primer (at 25 μM each). The qPCR program was as follows: initial activation for 2 min at 50°C, 30 s at 98°C, followed by 40 cycles of 10 s at 98°C, 30 s at 63°C and 60 s at 72°C. The number of additional cycles was calculated as 1/4 of the maximum amplification.86 The second amplification was done using 12.5 μl of amplified DNA, 2X PCR Master Mix NEB and 2.5 μl of each primer (at 25 uM) in a total volume of 50 ul. The program was the same as the first amplification without the initial extension and for the calculated number of cycles. Finally, re-amplified DNA was cleaned up using 1.8x volumes AMPureXP beads and eluted in 30 ul. Samples were quantified in the QuBit, size profiles examined in the Bioanalyzer using the DNA High Sensitivity DNA Kit (Agilent, Part No. 5067-4626). Samples were pooled and sequenced in a NovaSeq by Novogene (Cambridge, UK).
Bulk ATAC-seq
Tn5 production
Methods were adapted from Hennig et al.87 as follows: The pETM11-Sumo3-Tn5E54K,L372P plasmid was obtained from EMBL-Heidelberg and transformed into Escherichia coli BL21 (DE3) Gold cells (Agilent). Bacterial cultures were grown at 30°C to a density of OD600=0.6-0.8 and protein expression was induced with 0.5 mM isopropyl-β-D-thiogalactoside (IPTG). Prior to induction, cultures were cooled and maintained at 20°C for protein expression overnight. Cell pellets were harvested by centrifugation for 10 m at 4000 rpm and then resuspended in lysis buffer (50 mM Tris-HCl pH 8.0, 20mM Imidazole, 0.5 M NaCl, 10% Glycerol, 1mM TCEP, 1U/ml Benzonase, 1 tablet/50ml Protease inhibitor tablets (cOmplete, Roche) and lysed with sonication. The cell lysate was then centrifuged for 30 m at 80,000 x g. The supernatant was collected and applied onto a 5ml Ni-Sepharose column (Cytiva). The column was washed with 10 Column Volumes wash buffer A (50mM Tris-HCl pH 8.0, 20 mM Imidazole, 0.5 M NaCl,10% Glycerol, 1mM TCEP). Bound proteins were eluted with buffer A containing 300mM Imidazole. Fractions containing His-Sumo3-Tn5E54K,L372P were pooled and incubated overnight at 4°C with SenP2 protease to remove the fusion tag. Next day, the sample was diluted six times and applied onto a 5ml Heparin HP column pre-equilibrated in buffer B (50mM Tris-HCl pH 8.0, 0.05 M NaCI, 1mM TCEP). Bound Tn5 protein was eluted with a linear gradient to 100% buffer B containing 1M NaCl, concentrated and loaded onto a Superdex 200 Increase 10/300 GL column (Cytiva) equilibrated in buffer C (50 mM Tris pH 8.0, 150 mM NaCl, 1 mM TCEP and 5% glycerol). Fractions corresponding to the Tn5 peak were pooled, concentrated to 10μM and flash frozen in liquid nitrogen.
Tn5 assembly
Dilution and oligo assembly of Tn5 was performed as previously described.28,88 In brief: purified protein was diluted to 4 μM in Dilution Buffer (50 mM Tris, 100 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.1% NP-40, and 50% glycerol). Two independent oligo mixes were assembled (Mix A, Mix B), each with 5 μl of 100 μM Tn5MEREV oligo, 5 μl of 100 μM of either Tn5_1 (Mix A) orTn5_2_ME_comp (Mix B), and 40ml of nuclease-free water by incubating each mix in a PCR thermocycler as follows: 95°C for 3 m, 65°C for 3 m and ramp down to 24°C at a rate of -1C/min. Once annealed, Mix A and Mix B were combined with 100 μl glycerol to create a 5 μM, 50% glycerol adaptor mix. Equal parts of diluted Tn5 transposase and adaptor mix were mixed and incubated at 25°C for 60 min. For oligo sequence see Table S1
Transposition and library generation
Wells were washed and cells were dissociated using 0.5 ml Accutase per well of a 6 well plate incubated 5 min at 37°C. Cells were washed in PBS, and 50,000 cells per condition and replicates were used for ATAC-seq following established protocols28,89 (for sequences used see Table S1) and as described for CaTS-ATAC. After transposition, DNA was directly purified and used for library preparation (as described for CaTS-ATAC).
Crosslinked and TF-Sorted RNA-seq
Samples for CaTS-RNAseq were collected in the same way as for CaTS-ATAC but all buffers were supplemented with 1 μL/ml of RNasin Plus (Promega, Cat. No. N2615). After fixation and quenching, samples were stained for intracellular TFs, also as described in “Flow cytometry of intracellular markers” with RNasin-supplemented buffers.
Cells were sorted into 0.5 ml PBS with 0.5% BSA and 1 μl/ml of RNasin Plus. RNA extraction from fixed samples were performed with the RecoverAll RNA/DNA Isolation Kit (Thermo Fisher Scientific, Cat. No. AM1975) with the modifications described in Amamoto et al.90 In brief, sorted cells were spun down at 3000 g for 7 min at 4°C and as much supernatant as possible was removed (leaving 40-50 μl). 100 μl of Digestion Buffer previously mixed with 4 μl of Protease mix was added to each sample and resuspended. The tubes were incubated at 50°C for 3 h and then stored at -80°C. RNA isolation was performed as per manufacturer’s instructions, eluting in 17 μl of Nuclease-free water.
Due to low quality and limiting amounts, the maximum amount (10.5 μl) was used for library preparation using the SMART-Seq HT kit (Takara, Cat. No. 634437) followed by Nextera XT DNA Library Preparation Kit (Illumina, Cat. No. FC-131-1096).
ATAC-seq processing
Data was processed using the nf-core atacseq pipeline (https://nf-co.re/atacseq) with the following options: –genome mm10–macs_fdr 0.00001 –skip_diff_analysis –min_reps_consensus 2 -r 1.2.0. Peaks were further filtered out if they didn’t pass qval < 0.00001 in at least 1 sample (qval generated from MACS peak calling in each sample).
ATAC-seq differential expression analysis
Read counts within the consensus intervals generated by featureCounts were used as input for DESeq2.83 Principal Component Analysis was performed using the top 30000 most variable elements and coloured by different sample metadata.
To assess the cell type specific accessibility for the same NP cell types at different SAG concentrations, all by all differential expression was performed for all conditions at day 5. Number of differentially accessible intervals plotted (fold change > 2, basemean > 100) as a bar graph, and individual elements are plotted color-coded by their p-adjusted values for specific pairwise comparisons.
Selection of differentially accessible elements across cell types and timepoint was performed by pairwise DESeq2 analysis between any two cell types within a timepoint and any two timepoints for the same cell type. Elements that fulfilled padj < 0.01 & abs(log2FoldChange) > 2 & baseMean > 100 were selected for subsequent clustering.
ATAC element clusters by kmeans
Varianced stabilized transformed data generated using DESeq2 were used as input to identify clusters of elements with the same dynamics. Clustering was performed using kmeans with a high number of centers, 30, and subsequently re-grouping clusters of very similar dynamics using hclust and target of 9 final clusters. This was chosen as independent iterations resulted in reproducible clusters and dynamics.
Analysis of published ChIP-seq data
Processing of published ChIP-seq datasets was for NKX6.1, NKX2.2, OLIG2,16SOX2 and FOXA213from neural embryoids with SAG were processing using the nf-core chipseq pipeline as follows: nextflow run nf-core/chipseq –input design.csv –single_end –genome mm10 -profile crick -r 1.1.0.
FOXA2 from endoderm differentiations50 were process with the nf-core atacseq pipeline due to lack of input samples. nextflow run nf-core/atacseq –input design.csv –single_end –mito_name false –genome mm10 -profile crick -r 1.1.0.
Footprinting analysis
BAM files for merged replicates were used as input for TOBIAS ATACorrect,33 followed by TOBIAS Footprint. TOBIAS BINDetect was run on all samples combined using the following the JASPAR2018, HOCOMOCO and Taipale databases.34–36
Motifs that were amongst the top 5% highest absolute fold change or 5% smallest pvalues in each pair-wise comparison were selected. Motifs were grouped in archetypes.37 The most variable archetypes for the desired comparison (either p0-1, p2, pM at day 5 and 6, or all conditions) were selected and RNA expression for each TF that is associated with the archetype was compared to the motif score for that archetype. This correlation analysis between motif score and RNA expression was used to find candidate TFs driving the chromatin changes observed.
RNA-seq data processing
Data was processed using the nf-core rnaseq pipeline with the following options: –star_index ’star_ercc_mm10_genome’ –gtf’mm10.refGene_wERCC.gtf’ –fc_group_features_type gene_id –fc_extra_attributes gene_id -r 1.4.2. Gene counts were obtained from featureCounts with the option ignoreDup=TRUE to try and remove PCR duplicates. Samples were excluded from downstream analysis based on a number of QC criteria particularly applicable to this type of low input samples from PFA-fixed cells including: too small proportion of the library being of mouse origin (excess spike-in representation), large number of overrepresented sequences, and loss of dynamic range in spike-in quantification.
Quantification And Statistical Analysis
Details for bioinformatic data analysis and software used are detailed in each method section. No additional statistical analyses were conducted. All code has been deposited at https://github.com/MJDelas/Neural_DV_ATAC, including parameters and versions for all software used. Figure legends contain number of replicates and details of data plotted (mean, s.e.m).
Supplementary Material
In brief.
Delás et al. provide evidence that spinal cord neural progenitors use two different chromatin regulatory strategies to control differential gene expression in response to the morphogen Shh. This highlights a conserved role of the pioneer factor FOXA2 between endoderm and neural cells, suggesting potential evolutionary relationships between these cell types.
Highlight.
CaTS-ATAC reveals chromatin accessibility from intracellular marker cell sorting
Neural progenitors employ two chromatin regulatory strategies in response to Shh
Most progenitors share accessibility across the same regulatory elements
FOXA2 mediates a “differential accessibility” strategy in the most ventral progenitors
Acknowledgments
We are grateful to Andreas Sagner, Vicki Metzis, and Teresa Rayon and members of the Briscoe lab for advice and reagents. We thank Tiago Rito for nuclei segmentation of images. We thank Anne Grapin-Botton and Uli Technau for critical reading of the manuscript. We thank the Crick Science Technology Platforms, in particular, Vangelis Christodoulou from the Structural Biology STP for production of Tn5 enzyme, Flow Cytometry, Advanced Sequencing, Advanced Light Microscopy, BRF and Advanced Computing. This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK, the UK Medical Research Council and Wellcome Trust (all under FC001051). Work in the Briscoe lab is funded by the European Research Council under European Union (EU) Horizon 2020 research and innovation program grant 742138. This work was also funded by the Wellcome Trust (220379/D/20/Z). H.T.S. is supported by a Sir Henry Wellcome postdoctoral fellowship.
Declarations
Supplemental Information
Supplemental information can be found online at https://doi.org/10.1016/j.devcel.2022.11.016.
Author Contributions
M.J.D. and J.B. conceived the project, interpreted the data, and wrote the manuscript. M.J.D. designed and performed experimentsand also performed bioinformatic analysis and data analysis. C.M.K., T.F., M.D., and I.Z. performed experiments. H.T.S., E.C., and E.M.T. generated and characterized the HM1-tetON cell lines. K.I. designed and assisted with embryology experiments.
Declaration Of Interests
The authors declare no competing interests.
Data and code availability
Sequencing data (CaTS-ATAC, live Omni-ATAC, RNA-seq and bulk ATAC-seq) have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Accession number for the SuperSeries is GSE204921.
Microscopy data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at https://github.com/MJDelas/Neural_DV_ATAC and is publicly available as of the date of publication. Custom code for lineage tracing quantification are available at https://github.com/MJDelas/tdTom_LinTracing.
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- 1.Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 2.Rogers KW, Schier AF. Morphogen gradients: from generation to interpretation. Annu Rev Cell Dev Biol. 2011;27:377–407. doi: 10.1146/annurev-cellbio-092910-154148. [DOI] [PubMed] [Google Scholar]
- 3.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 4.Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 5.Roelink H, Augsburger A, Heemskerk J, Korzh V, Norlin S, Ruiz i Altaba AR, Tanabe Y, Placzek M, Edlund T, Jessell TM. Floor plate and motor neuron induction by vhh-1, a vertebrate ho-molog of hedgehog expressed by the notochord. Cell. 1994;76:761–775. doi: 10.1016/0092-8674(94)90514-2. [DOI] [PubMed] [Google Scholar]
- 6.Balaskas N, Abbott LF, Jessell TM, Ng D. Positional strategies for connection specificity and synaptic organization in spinal sensory-motor circuits. Neuron. 2019;102:1143–1156.:e4. doi: 10.1016/j.neuron.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bikoff JB, Gabitto MI, Rivard AF, Drobac E, Machado TA, Miri A, Brenner-Morton S, Famojure E, Diaz C, Alvarez FJ, et al. Spinal inhibitory interneuron diversity delineates variant motor microcircuits. Cell. 2016;165:207–219. doi: 10.1016/j.cell.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briscoe J, Sussel L, Serup P, Hartigan-O’Connor D, Jessell TM, Rubenstein JL, Ericson J. Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature. 1999;398:622–627. doi: 10.1038/19315. [DOI] [PubMed] [Google Scholar]
- 9.Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 10.Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- 11.Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodo-main protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 12.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 13.Peterson KA, Nishi Y, Ma W, Vedenko A, Shokri L, Zhang X, McFarlane M, Baizabal J-M, Junker JP, van Oudenaarden A, et al. Neural-specific Sox2 input and differential Gli-binding affinity provide context and positional information in Shh-directed neural patterning. Genes Dev. 2012;26:2802–2816. doi: 10.1101/gad.207142.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oosterveen T, Kurdija S, Alekseenko Z, Uhde CW, Bergsland M, Sandberg M, Andersson E, Dias JM, Muhr J, Ericson J. Mechanistic differences in the transcriptional interpretation of local and long-range Shh morphogen signaling. Dev Cell. 2012;23:1006–1019. doi: 10.1016/j.devcel.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 15.Muhr J, Andersson E, Persson M, Jessell TM, Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell. 2001;104:861–873. doi: 10.1016/s0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- 16.Nishi Y, Zhang X, Jeong J, Peterson KA, Vedenko A, Bulyk ML, Hide WA, McMahon AP. A direct fate exclusion mechanism by Sonic hedgehog-regulated transcriptional repressors. Development. 2015;142:3286–3293. doi: 10.1242/dev.124636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kutejova E, Sasai N, Shah A, Gouti M, Briscoe J. Neural progenitors adopt specific identities by directly repressing all alternative progenitor transcriptional programs. Dev Cell. 2016;36:639–653. doi: 10.1016/j.devcel.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- 19.Persson M, Stamataki D, te Welscher PT, Andersson E, Bose J, Rüther U, Ericson J, Briscoe J. Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes Dev. 2002;16:2865–2878. doi: 10.1101/gad.243402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamataki D, Ulloa F, Tsoni SV, Mynett A, Briscoe J. A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes Dev. 2005;19:626–641. doi: 10.1101/gad.325905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hui CC, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- 22.Balaskas N, Ribeiro A, Panovska J, Dessaud E, Sasai N, Page KM, Briscoe J, Ribes V. Gene regulatory logic for reading the sonic hedgehog signaling gradient in the vertebrate neural tube. Cell. 2012;148:273–284. doi: 10.1016/j.cell.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen M, Page KM, Perez-Carrasco R, Barnes CP, Briscoe J. A theoretical framework for the regulation of Shh morphogen-controlled gene expression. Development. 2014;141:3868–3878. doi: 10.1242/dev.112573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gouti M, Tsakiridis A, Wymeersch FJ, Huang Y, Kleinjung J, Wilson V, Briscoe J. In vitrogeneration of neuromesodermal progenitors reveals distinct roles for wnt signalling in the specification of spinal cord and paraxial mesoderm identity. PLOS Biol. 2014;12:e1001937. doi: 10.1371/journal.pbio.1001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sagner A, Gaber ZB, Delile J, Kong JH, Rousso DL, Pearson CA, Weicksel SE, Melchionda M, Mousavy Gharavy SNM, Briscoe J, et al. Olig2 and Hes regulatory dynamics during motor neuron differentiation revealed by single cell transcriptomics. PLoS Biol. 2018;16:e2003127. doi: 10.1371/journal.pbio.2003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delias MJ, Briscoe J. Repressive interactions in gene regulatory networks: when you have no other choice. Curr Top Dev Biol. 2020;139:239–266. doi: 10.1016/bs.ctdb.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, Litzenburger UM, Wei Y, Schep AN, LaGory EL, Choudhry H, Giaccia AJ, Greenleaf WJ, Chang HY. Joint single-cell DNA accessibility and protein epitope profiling reveals environmental regulation of epigenomic heterogeneity. Nat Commun. 2018;9:4590. doi: 10.1038/s41467-018-07115-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baskar R, Chen AF, Favaro P, Reynolds W, Mueller F, Borges L, Jiang S, Park HS, Kool ET, Greenleaf WJ, et al. Integrating transcription-factor abundance with chromatin accessibility in human erythroid lineage commitment. Cell Rep Methods. 2022;2:100188. doi: 10.1016/j.crmeth.2022.100188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metzis V, Steinhauser S, Pakanavicius E, Gouti M, Stamataki D, Ivanovitch K, Watson T, Rayon T, Mousavy Gharavy SNM, Lovell-Badge R, et al. Nervous system regionalization entails axial allocation before neural differentiation. Cell. 2018;175:1105–1118.:e17. doi: 10.1016/j.cell.2018.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blassberg R, Patel H, Watson T, Gouti M, Metzis V, Delás MJ, Briscoe J. Sox2 levels regulate the chromatin occupancy of WNT mediators in epiblast progenitors responsible for vertebrate body formation. Nat Cell Biol. 2022;24:633–644. doi: 10.1038/s41556-022-00910-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bentsen M, Goymann P, Schultheis H, Klee K, Petrova A, Wiegandt R, Fust A, Preussner J, Kuenne C, Braun T, et al. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat Commun. 2020;11:4267. doi: 10.1038/s41467-020-18035-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Chèneby J, Kulkarni SR, Tan G, et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2017;46:gkx1126. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kulakovskiy IV, Vorontsov IE, Yevshin IS, Sharipov RN, Fedorova AD, Rumynskiy EI, Medvedeva YA, Magana-Mora A, Bajic VB, Papatsenko DA, et al. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via largescale ChIP-Seq analysis. Nucleic Acids Res. 2017;46:gkx1106. doi: 10.1093/nar/gkx1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. DNA-binding specificities ofhuman transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 37.Vierstra J, Lazar J, Sandstrom R, Halow J, Lee K, Bates D, Diegel M, Dunn D, Neri F, Haugen E, et al. Global reference mapping of human transcription factor footprints. Nature. 2020;583:729–736. doi: 10.1038/s41586-020-2528-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasaki H, Hogan BL. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development. 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- 39.Ruizi Altaba AR, Prezioso VR, Darnell JE, Jessell TM. Sequential expression of HNF-3ß and HNF-3α by embryonic organizing centers: the dorsal lip/node, notochord and floor plate. Mech Dev. 1993;44:91–108. doi: 10.1016/0925-4773(93)90060-B. [DOI] [PubMed] [Google Scholar]
- 40.Ang SL, Wierda A, Wong D, Stevens KA, Cascio S, Rossant J, Zaret KS. The formation and maintenance of the definitive endoderm lineage in the mouse: involvement of HNF3/forkhead proteins. Development. 1993;119:1301–1315. doi: 10.1242/dev.119.4.1301. [DOI] [PubMed] [Google Scholar]
- 41.Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding sitefor Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 42.Epstein DJ, McMahon AP, Joyner AL. Regionalization of Sonic hedgehog transcription along the anteroposterior axis of the mouse central nervous system is regulated by Hnf3-dependent and -independent mechanisms. Development. 1999;126:281–292. doi: 10.1242/dev.126.2.281. [DOI] [PubMed] [Google Scholar]
- 43.Lek M, Dias JM, Marklund U, Uhde CW, Kurdija S, Lei Q, Sussel L, Rubenstein JL, Matise MP, Arnold H-H, et al. A homeodo-main feedback circuit underlies step-function interpretation of a Shh morphogen gradient during ventral neural patterning. Development. 2010;137:4051–4060. doi: 10.1242/dev.054288. [DOI] [PubMed] [Google Scholar]
- 44.Ribes V, Balaskas N, Sasai N, Cruz C, Dessaud E, Cayuso J, Tozer S, Yang LL, Novitch B, Marti E, et al. Distinct Sonic Hedgehog signaling dynamics specify floor plate and ventral neuronal progenitors in thevertebrate neural tube. Genes Dev. 2010;24:1186–1200. doi: 10.1101/gad.559910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho G, Lim Y, Cho I-T, Simonet JC, Golden JA. Arx together with FoxA2, regulates Shh floor plate expression. Dev Biol. 2014;393:137–148. doi: 10.1016/j.ydbio.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imuta Y, Kiyonari H, Jang CW, Behringer RR, Sasaki H. Generation of knock-in mice that express nuclear enhanced green fluorescent protein and tamoxifen-inducible Cre recombinase in the notochord from Foxa2and Tloci. Genesis. 2013;51:210–218. doi: 10.1002/dvg.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) andGATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 48.Wang A, Yue F, Li Y, Xie R, Harper T, Patel NA, Muth K, Palmer J, Qiu Y, Wang J, et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermallineage intermediates. Cell Stem Cell. 2015;16:386–399. doi: 10.1016/j.stem.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee K, Cho H, Rickert RW, Li QV, Pulecio J, Leslie CS, Huangfu D. FOXA2 is required for enhancer priming during pancreatic differentiation. Cell Rep. 2019;28:382–393.:e7. doi: 10.1016/j.celrep.2019.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cernilogar FM, HasenÖder S, Wang Z, Scheibner K, Burtscher I, Sterr M, Smialowski P, Groh S, Evenroed IM, Gilfillan GD, et al. Pre-marked chromatin and transcription factor co-binding shape the pioneering activity of Foxa2. Nucleic Acids Res. 2019;47:9069–9086. doi: 10.1093/nar/gkz627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geusz RJ, Wang A, Lam DK, Vinckier NK, Alysandratos K-D, Roberts DA, Wang J, Kefalopoulou S, Ramirez A, Qiu Y, et al. Sequence logic at enhancers governs a dual mechanism of endodermal organ fate induction by FOXA pioneer factors. Nat Commun. 2021;12:6636. doi: 10.1038/s41467-021-26950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wan H, Dingle S, Xu Y, Besnard V, Kaestner KH, Ang S-L, Wert S, Stahlman MT, Whitsett JA. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J Biol Chem. 2005;280:13809–13816. doi: 10.1074/jbc.M414122200. [DOI] [PubMed] [Google Scholar]
- 53.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 54.Meredith DM, Borromeo MD, Deering TG, Casey BH, Savage TK, Mayer PR, Hoang C, Tung K-C, Kumar M, Shen C, et al. Program Specificity for Ptf1a in Pancreas versus Neural Tube Development Correlates with Distinct Collaborating Cofactors and chromatin Accessibility. Mol Cell Biol. 2013;33:3166–3179. doi: 10.1128/MCB.00364-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EEM. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
- 56.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Donaghey J, Thakurela S, Charlton J, Chen JS, Smith ZD, Gu H, Pop R, Clement K, Stamenova EK, Karnik R, et al. Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat Genet. 2018;50:250–258. doi: 10.1038/s41588-017-0034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burke Z, Oliver G. Prox1 is an early specific marker for the developing liver and pancreas in the mammalian foregut endoderm. Mech Dev. 2002;118:147–155. doi: 10.1016/s0925-4773(02)00240-x. [DOI] [PubMed] [Google Scholar]
- 60.Kaltezioti V, Antoniou D, Stergiopoulos A, Rozani I, Rohrer H, Politis PK. Prox1 regulates Olig2 expression to modulate binary fate decisions in spinal cord neurons. J Neurosci. 2014;34:15816–15831. doi: 10.1523/JNEUROSCI.1865-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng L, Chen C-L, Luo P, Tan M, Qiu M, Johnson R, Ma Q. Lmx1b, Pet-1, and Nkx2.2 coordinately specify serotonergic neurotransmitter phenotype. J Neurosci. 2003;23:9961–9967. doi: 10.1523/JNEUROSCI.23-31-09961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ding Y-Q, Marklund U, Yuan W, Yin J, Wegman L, Ericson J, Deneris E, Johnson RL, Chen Z-F. Lmx1b is essential for the development of serotonergic neurons. Nat Neurosci. 2003;6:933–938. doi: 10.1038/nn1104. [DOI] [PubMed] [Google Scholar]
- 63.Alvarez-Dominguez JR, Donaghey J, Rasouli N, Kenty JHR, Helman A, Charlton J, Straubhaar JR, Meissner A, Melton DA. Circadian entrainment triggers maturation ofhuman in vitro is-lets. Cell Stem Cell. 2020;26:108–122.:e10. doi: 10.1016/j.stem.2019.11.011. [DOI] [PubMed] [Google Scholar]
- 64.Ait-Lounis A, Baas D, Barras E, Benadiba C, Charollais A, Nlend Nlend RN, LiÈgeois D, Meda P, Durand B, Reith W. Novel function of the ciliogenic transcription factor RFX3 in development of the endocrine pancreas. Diabetes. 2007;56:950–959. doi: 10.2337/db06-1187. [DOI] [PubMed] [Google Scholar]
- 65.Cruz C, Ribes V, Kutejova E, Cayuso J, Lawson V, Norris D, Stevens J, Davey M, Blight K, Bangs F, et al. Foxj1 regulates floor plate cilia architecture and modifies the response of cells to sonic hedgehog signalling. Development. 2010;137:4271–4282. doi: 10.1242/dev.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gilbert SF, Opitz JM, Raff RA. Resynthesizing evolutionary and developmental Biology. Dev Biol. 1996;173:357–372. doi: 10.1006/dbio.1996.0032. [DOI] [PubMed] [Google Scholar]
- 67.Wang H, Lei Q, Oosterveen T, Ericson J, Matise MP. Tcf/Lef repressors differentially regulate Shh-Gli target gene activation thresholds to generate progenitor patterning in the developing CNS. Development. 2011;138:3711–3721. doi: 10.1242/dev.068270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reizel Y, Morgan A, Gao L, Schug J, Mukherjee S, García MF, Donahue G, Baur JA, Zaret KS, Kaestner KH. FoxA-dependent demethylation of DNA initiates epigenetic memory of cellular identity. Dev Cell. 2021;56:602–612.:e4. doi: 10.1016/j.devcel.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nashun B, Hill PW, Hajkova P. Reprogramming of cell fate: epigenetic memory and the erasure of memories past. EMBO J. 2015;34:1296–1308. doi: 10.15252/embj.201490649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Litingtung Y, Chiang C. Specification of ventral neuron types is mediated by an antagonistic interaction between Shhand Gli3. Nat Neurosci. 2000;3:979–985. doi: 10.1038/79916. [DOI] [PubMed] [Google Scholar]
- 71.Larsen HL, Grapin-Botton A. The molecular and morphogenetic basis of pancreas organogenesis. Semin Cell Dev Biol. 2017;66:51–68. doi: 10.1016/j.semcdb.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 72.Stainier DYR. A glimpse into the molecular entrails of endoderm formation. Genes Dev. 2002;16:893–907. doi: 10.1101/gad.974902. [DOI] [PubMed] [Google Scholar]
- 73.Steinmetz PRH, Aman A, Kraus JEM, Technau U. Gutlike ectodermal tissue in a sea anemone challenges germ layer homology. Nat Ecol Evol. 2017;1:1535–1542. doi: 10.1038/s41559-017-0285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.TourniÈre O, Gahan JM, Busengdal H, Bartsch N, Rentzsch F. Insm1-expressing neurons and secretory cells develop from a common pool of progenitors in the sea anemone Nematostella vectensis. Sci Adv. 2022;8:eabi7109. doi: 10.1126/sciadv.abi7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Steger J, Cole AG, Denner A, Lebedeva T, Genikhovich G, Ries A, Reischl R, Taudes E, Lassnig M, Technau U. Single-cell transcriptomics identifies conserved regulators of neuroglandular lineages. Cell Rep. 2022;40:111370. doi: 10.1016/j.celrep.2022.111370. [DOI] [PubMed] [Google Scholar]
- 76.Weinstein DC, Ruiz i Altaba AR, Chen WS, Hoodless P, Prezioso VR, Jessell TM, Darnell JE. Thewinged-helixtranscription factor HNF-3ß is required for notochord development in the mouse embryo. Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- 77.Ang S-L, Rossant J. HNF-3ß is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- 78.Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, Smithies O. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 79.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Picelli S, BjÖrklund AK, Reinius B, Sagasser S, Winberg G, Sandberg R. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014;24:2033–2040. doi: 10.1101/gr.177881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stuart HT, Stirparo GG, Lohoff T, Bates LE, Kinoshita M, Lim CY, Sousa EJ, Maskalenka K, Radzisheuskaya A, Malcolm AA, et al. Distinct molecular trajectories converge to induce naive pluripotency. Cell Stem Cell. 2019;25:388–406.:e8. doi: 10.1016/j.stem.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rayon T, Stamataki D, Perez-Carrasco R, Garcia-Perez L, Barrington C, Melchionda M, Exelby K, Lazaro J, Tybulewicz VLJ, Fisher EMC, et al. Species-specific pace ofdevelopment is associated with differences in protein stability. Science. 2020;369:eaba7667. doi: 10.1126/science.aba7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hennig BP, Velten L, Racke I, Tu CS, Thoms M, Rybin V, Besir H, Remans K, Steinmetz LM. Large-scale low-cost NGS library preparation using a robust Tn5 purification and tagmentation protocol. G3. 2018;8:79–89. doi: 10.1534/g3.117.300257. Bethesda. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma S, Zhang B, LaFave LM, Earl AS, Chiang Z, Hu Y, Ding J, Brack A, Kartha VK, Tay T, et al. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell. 2020;183:1103–1116.:e20. doi: 10.1016/j.cell.2020.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yoshida H, Lareau CA, Ramirez RN, Rose SA, Maier B, Wroblewska A, Desland F, Chudnovskiy A, Mortha A, Dominguez C, et al. The cis-regulatory atlas of the mouse immune system. Cell. 2019;176:897–912.:e20. doi: 10.1016/j.cell.2018.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Amamoto R, Garcia MD, West ER, Choi J, Lapan SW, Lane EA, et al. Probe-Seq enables transcriptional profiling of specific cell types from heterogeneoustissue by RNA-based isolation. eLife. 2019;8:e51452. doi: 10.7554/eLife.51452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data (CaTS-ATAC, live Omni-ATAC, RNA-seq and bulk ATAC-seq) have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Accession number for the SuperSeries is GSE204921.
Microscopy data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at https://github.com/MJDelas/Neural_DV_ATAC and is publicly available as of the date of publication. Custom code for lineage tracing quantification are available at https://github.com/MJDelas/tdTom_LinTracing.
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.