Abstract
Ribosomes are produced in large quantities during oogenesis and stored in the egg. However, the egg and early embryo are translationally repressed1–4. Using mass-spectrometry and cryo-EM analyses of ribosomes isolated from zebrafish and Xenopus eggs and embryos, we provide molecular evidence that ribosomes transition from a dormant to an active state during the first hours of embryogenesis. Dormant ribosomes are associated with four conserved factors that form two modules, consisting of Habp4-eEF2 and of Dap1b/Dap-eIF5a. Both modules occupy functionally important sites and act together to stabilize ribosomes and repress translation. Dap1b/Dapl1 is a newly discovered translational inhibitor that stably inserts into the polypeptide exit tunnel. Addition of recombinant zebrafish Dap1b protein is sufficient to block translation and reconstitute the dormant egg ribosome state in a mammalian translation extract in vitro. Thus, a developmentally programmed, conserved ribosome state plays a key role in ribosome storage and translational repression in the egg.
Introduction
Ribosomes are amongst the most abundant macromolecular complexes stored in the quiescent egg5,6. These maternally provided ribosomes (in short: maternal ribosomes) are essential for progression through embryogenesis as they are required for the translation of maternal and zygotic transcripts7–9 (Fig. 1a). Although the overall number of ribosomes per embryo remains constant throughout early embryogenesis, prior studies from multiple organisms indicate that translational activity increases in the embryo, suggesting that translation is repressed in the mature egg1–4,10,11. Several mechanisms, including shortening of mRNA polyadenine tails12 and interference with the formation of the translational initiation factor eIF4F13, have been implicated in dampening translation in the egg and early embryo; however, direct evidence for a contribution of the ribosome itself is missing.
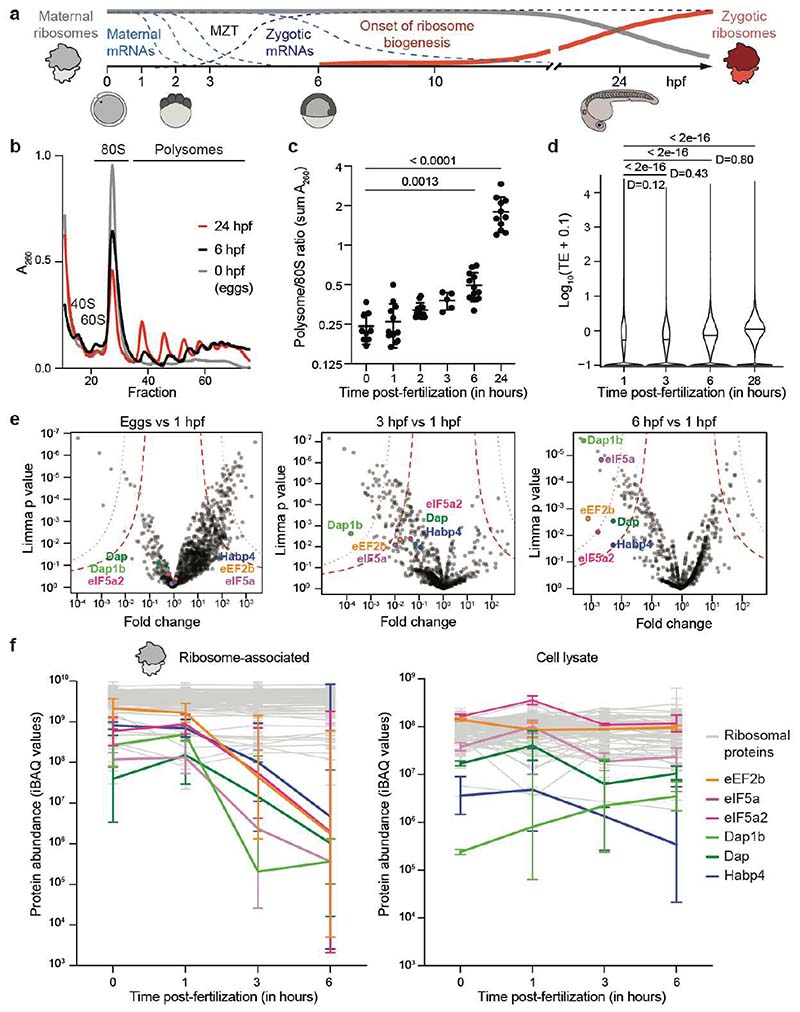
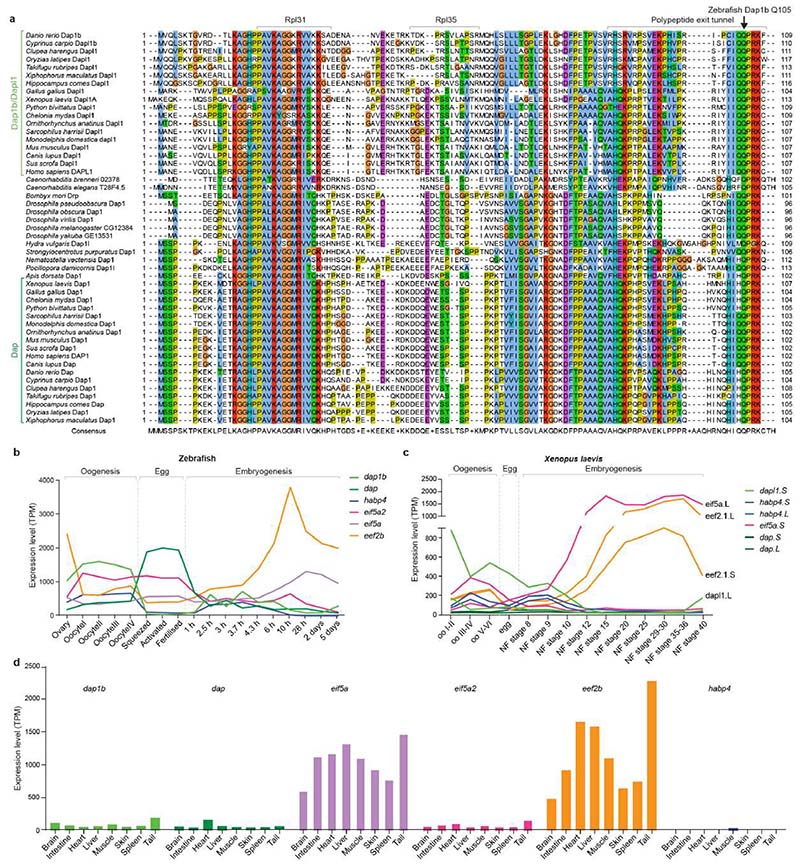
Fig. 1. Translation increase during zebrafish embryogenesis anti-correlates with the presence of ribosome-bound factors.
a, Schematic of the maternal-to-zygotic transition (MZT). Clearance of maternal mRNAs is coordinated with the activation of transcription during the first hours post-fertilization (hpf). In zebrafish, replacement of maternal by zygotic ribosomes takes several days. b, Representative zebrafish polysome profiles. A260, absorbance at 260 nm. c, Quantification of polysome-to-monosome ratios (0 hpf: n = 9; 1 hpf: n = 13; 2 hpf: n = 13, 3 hpf: n = 5; 6 hpf: n = 12; 24 hpf: n = 11). Significance was determined with Kruskal-Wallis and Dunn’s two-sided test. Data are mean ± standard deviation (SD). d, Violin plots showing the distribution of the median translation efficiency (TE) during embryogenesis. Significance was assessed by the two-sided Wilcoxon pairwise test and effect size was estimated by Cohen’s D58 (< 0.2: “negligible”; [0.2-0.5]: “small”; [0.5-0.8]: “medium”; > 0.8: “large”). e, Volcano plots based on mass spectrometry data showing fold enrichments of proteins in the ribosome fraction of 1 hpf embryos compared to eggs (left), 3 hpf (middle) and 6 hpf (right) embryos (n = 3 for each time-point). All significantly enriched or depleted proteins are listed in Supplementary Table 1. Permutation-based false discovery rates (FDRs) are displayed as dotted (FDR < 0.01) and dashed (FDR < 0.05) lines. f, Abundance changes of a subset of factors relative to ribosomal proteins in the ribosome-associated proteome (left) and cell lysate (right) (n = 3). Abundances are reported as iBAQ values. 74 ribosomal proteins are plotted in grey. Error bars correspond to geometric SD. For c, e and f, n are biologically independent samples.
Translationally inactive ribosomes have been observed in pro- and eukaryotes in specific cellular contexts14. Under stress conditions, prokaryotic 70S ribosomes dimerize to form 100S particles via the association of HPF and RMF in Gram-negative bacteria15, or LHPF in Gram-positive bacteria16. In eukaryotes, several factors have been reported to associate with inactive ribosomes upon sporulation (i.e. MDF1 and MDF2 in microsporidia17) and nutrient deprivation. The latter include SERBP1 (Stm1 in yeast)18–21, IFRD218 and CCDC124 (Lso2 in yeast)22. Apart from the inhibition of ribosomes upon induction of endogenously encoded factors, prokaryotic ribosomes are targeted by eukaryotic proline-rich antimicrobial peptides (PrAMPs) like Bac723 and Apidaecins24,25, which are known to inhibit translation by inserting into the polypeptide exit tunnel of bacterial ribosomes.
Studies from the 1970s with sea urchin eggs indicated the presence of inhibitory proteins which were thought to be associated with maternal ribosomes26–28, yet the relevance and molecular identity of these factors remained unclear. How ribosomes are stored in an inactive state for extended amounts of time in the mature egg, and whether regulation of the ribosome itself contributes to translational repression in the egg and the subsequent increase in translational activity during embryogenesis remains thus unknown.
Translation is repressed in zebrafish eggs
To investigate the timing of translational activation during the egg-to-embryo transition in vertebrates, we examined the translational status of zebrafish embryos at different stages of development (Fig. 1a). Specifically, we used polysome gradients to uncover the fraction of ribosomes present as individual subunits (40S and 60S), monosomes (80S) and polysomes. In the absence of triggers of ribosome stalling, an increase in the polysome fraction generally indicates increased active translation29. We found that ribosomes in the egg and in the 1-hour post fertilization (hpf) embryo were almost exclusively present as monosomes, whereas the polysome fraction started to increase from 3-6 hpf onwards (Fig. 1b, c). As an orthogonal approach, we calculated translational efficiency (TE) values based on ribosome-protected mRNA fragments over the course of zebrafish embryogenesis, using published ribosome profiling and RNA-Seq data sets30,31. We observed that translational efficiency increases over the course of embryogenesis, with a clear increase at 6 hpf (Fig. 1d). Together, these data suggest that, similarly to other vertebrates1–4,10,11, most ribosomes present in the zebrafish egg and early embryo are not engaged in active translation yet transition to a translationally active state during embryogenesis.
Identification of dormant ribosome factors
To uncover factors that regulate maternal ribosome activity during the first hours after fertilization, we purified crude ribosomes (monosomes and polysomes) from zebrafish eggs and embryos for mass-spectrometry (MS) analysis. While the relative abundances of core ribosomal proteins were similar at all time points, we identified four evolutionarily conserved factors, namely eIF5a/eIF5a2, eEF2b, Habp4 and the paralogs Dap1b/Dap that were specifically associated with ribosomes in zebrafish eggs and 1 hpf embryos, but depleted from ribosomes in 3 hpf and 6 hpf embryos (Fig. 1e, f; Supplementary Table 1). The levels of these ribosome-associated factors remained relatively stable in total embryo lysates over the first 6 hours of development (Fig. 1f), suggesting that they were released from the ribosome but not immediately degraded. Importantly, we found that the same set of factors were enriched in ribosomes purified from Xenopus eggs versus 24 hpf embryos (a developmental stage equivalent to zebrafish 6 hpf embryos) (Extended Data Fig. 1a). Thus, the association of these four factors with ribosomes in the mature egg is conserved in zebrafish and Xenopus and correlates with suppressed translation.
Structure of the dormant egg ribosome
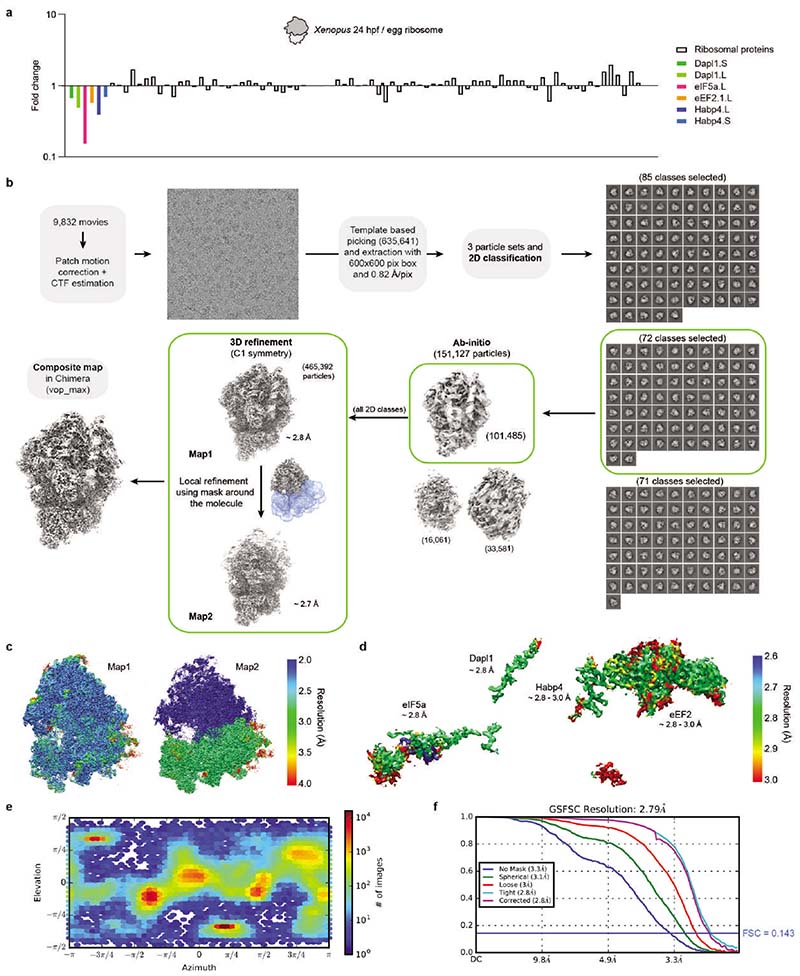
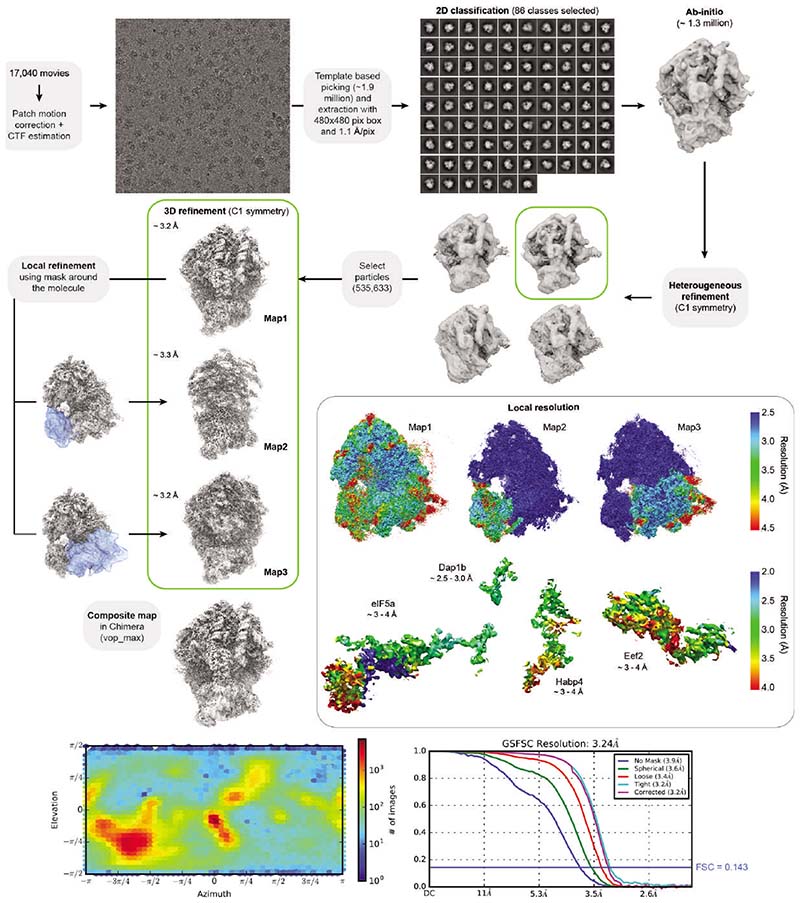
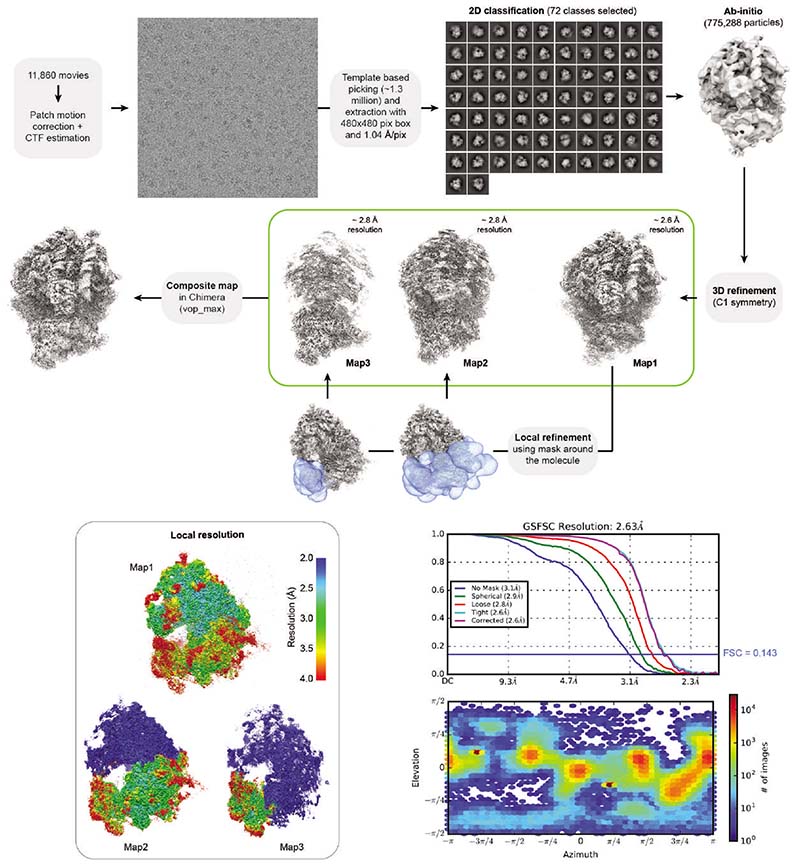
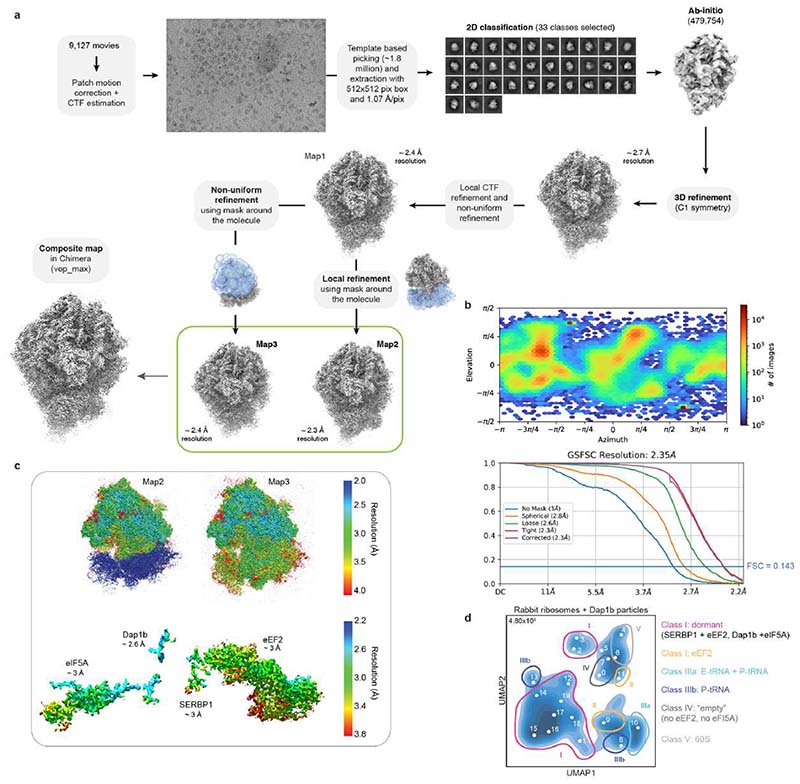
To elucidate how these factors establish a translationally repressed ribosome state, we determined the structures of maternal ribosomes isolated from 1 hpf zebrafish embryos (Fig. 2a) and Xenopus eggs (Fig. 2b) by electron cryo-microscopy (cryo-EM) at 3.2 and 2.8 Å average resolution, respectively (Supplementary Table 2; Extended Data Figs. 1b-f and 2). We were able to assign densities for all four factors in ribosomes from both zebrafish and Xenopus (Fig. 2c; Extended Data Fig. 3a-d). In parallel, we performed cryo-EM analysis of ribosomes isolated from 6 hpf zebrafish embryos (2.6 Å average resolution; Extended Data Figs. 3e and 4; Supplementary Table 2). In agreement with the MS data, ribosomes from 6 hpf zebrafish embryos showed low density levels for eEF2b and no densities for eIF5a/eIF5a2, Habp4 and Dap1b/Dap (Extended Data Figs. 3e and 4).
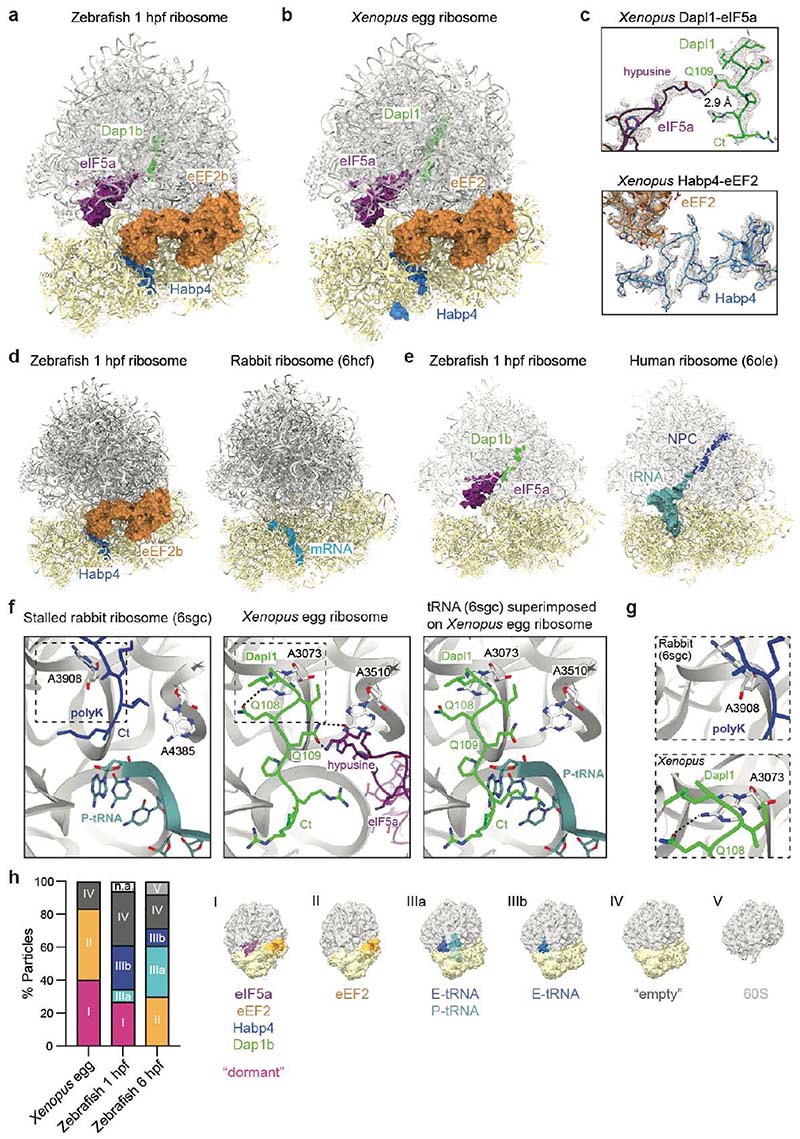
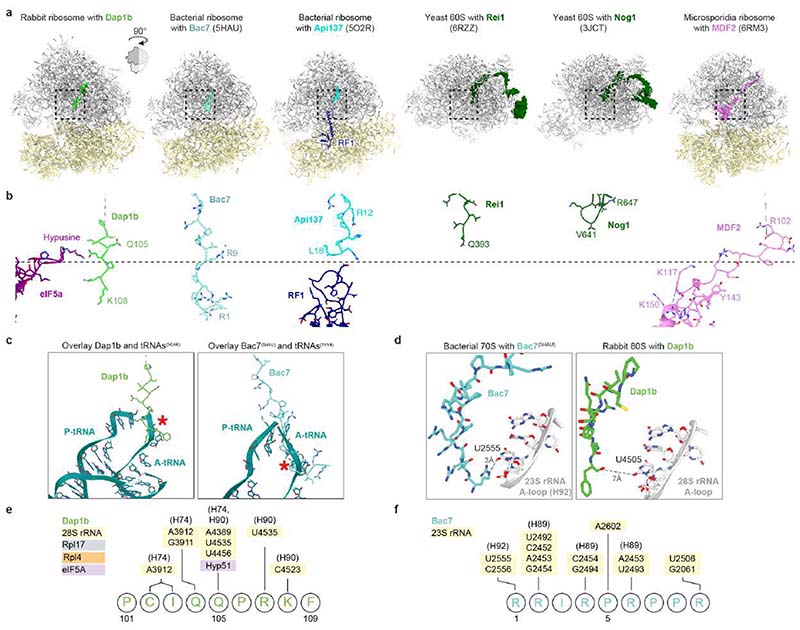
Fig. 2. A conserved set of factors blocks functionally important sites of the egg ribosome.
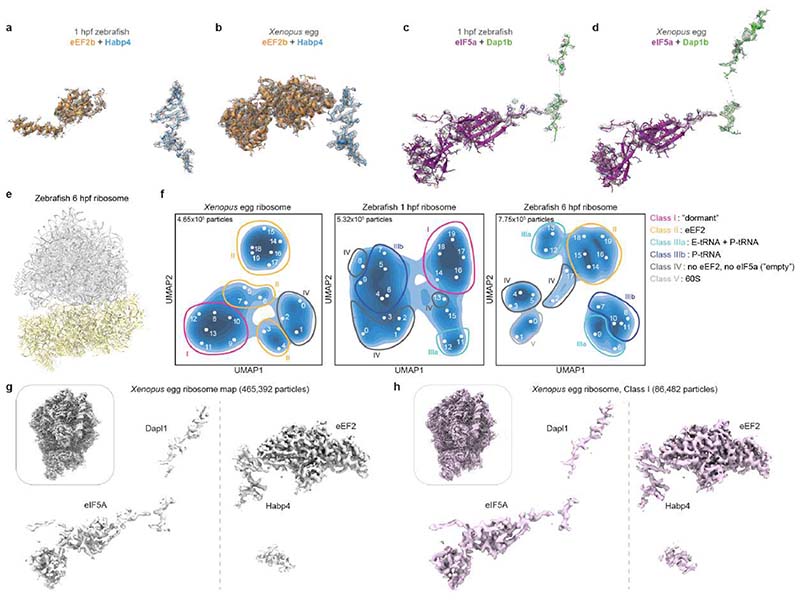
a-b, Overview of the dormant ribosome structure from 1 hpf zebrafish (a) and Xenopus egg (b). Ribosome-associated factors are shown as surface representations; eEF2b and eIF5a correspond to PDB-6MTE18 and PDB-5DAT. c, Density maps (in mesh) of the two modules of the Xenopus dormant ribosome. d, Habp4 and eEF2b (6MTE18) are shown as surface representations in the zebrafish dormant ribosome (left). A rabbit ribosome stalled on an mRNA (6HCF59) is shown for comparison (right). e, Clipping of the zebrafish dormant ribosome shows eIF5a (5DAT) and Dap1b bound within the polypeptide exit tunnel (left). The structure of a stalled human ribosome (6OLE60) containing a nascent polypeptide chain (NPC) and a P/E-tRNA is shown for comparison (right). f, Comparison of the peptidyl-transferase center of a rabbit 80S ribosome stalled with a poly-Lysine NPC and a P/E-tRNA (6SGC61, left) and of the Xenopus dormant ribosome (middle). For comparison, the P/E-tRNA (6SGC61), absent in dormant ribosomes, was superimposed onto the Xenopus egg ribosome (right; eIF5A is hidden). Critical amino acids and 28S rRNA nucleotides are labeled; interactions are depicted with dashed lines. Boxed areas are shown at higher magnification in g. g, A3073 of Xenopus 28S rRNA (bottom; equivalent to A3908 of rabbit 28S, top) displays a different conformation when interacting with Gln108 of Dapl1. h, Distribution of ribosomal particles among the classes obtained with cryoDRGN48. Representative filtered maps of the major classes are shown (Extended Data Fig. 3g, h; Supplementary Table 3; n.a refers to “non-assigned” particles). Ct, C-terminus.
eIF5a and eEF2 are known essential translation factors. eIF5a normally promotes translation elongation and termination, particularly upon ribosome stalling at specific amino acid sequence contexts32,33. We found that eIF5a/eIF5a2 binds between the exit (E) and peptidyl (P) site of the ribosome (Fig. 2a, b), as previously reported for a stalled ribosome34. eEF2 mediates ribosomal translocation and transiently interacts with the tRNA-mRNA complex at the aminoacyl (A) site of the ribosome35,36. The eEF2 interaction at the A site has been reported to be stabilized in inactive ribosomes by Stm1/SERBP1 (proteins of the Habp4 family)18,37, but not by Habp4 itself. Indeed, we found Habp4 bound to the mRNA entry channel of zebrafish 1 hpf and Xenopus egg ribosomes at the same position as previously reported for its mammalian homolog SERBP118,20,38, sequestering eEF2 at the A site and blocking the interaction sites of the tRNA-mRNA complex on the ribosome (Fig. 2d; Extended Data Fig. 3a, b).
Death Associated Protein 1b (Dap1b)/Dapl1 (in Xenopus Dapl1.L/Dapl1.S; in mammals also known as EEDA39,40) and Dap are conserved proteins across animals (Extended Data Fig. 5a). In mammals, Dapl1 has been linked to epithelial differentiation and proliferation39–41, and Dap (in mammals Dap1) has been described as a positive regulator of programmed cell death42 and a negative regulator of autophagy43. Strikingly, we found density in the polypeptide exit tunnel (PET) of zebrafish 1 hpf and Xenopus egg ribosomes that we could attribute to the conserved C-terminus of Dap1b/Dapl1 or Dap (Fig. 2e; Extended Data Fig. 3c, d). Dap1b was ~10-fold more abundant than Dap in ribosomes from 1 hpf zebrafish embryos according to our MS analysis (Fig. 1f), and only Dapl1 was identified by MS in ribosomes from Xenopus eggs (Extended Data Fig. 1a). Thus, we modeled the C-terminus of zebrafish Dap1b and Xenopus Dapl1, which we found to extend by five amino acids beyond the position that is occupied by the C-terminal amino acid residue of a nascent polypeptide chain (Fig. 2c, f). The fifth-last invariant amino acid in Dap1b/Dapl1 (Gln105 in zebrafish, Gln109 in Xenopus; Extended Data Fig. 5a) occupies the same position as the C-terminal residue of a nascent peptide chain38 and forms a hydrogen bond with the hypusine residue of eIF5a (Fig. 2f), which is essential for eIF5a function44,45. Moreover, we observe additional hydrogen-bond interactions between the immediately adjacent Glutamine in Dap1b/Dapl1 (Gln104 in zebrafish, Gln108 in Xenopus) with the 28S rRNA (Fig. 2f, g). Interestingly, the interaction of this Glutamine with adenine 3073 in Xenopus (A3168 in zebrafish) restricts this adenine to a conformation distinct from previously reported ribosomal structures (Fig. 2g). Moreover, Dap1b/Dapl1’s path differs from other proteins that have been previously found to be inserted into the PET of mature or precursor large ribosomal subunits, including antimicrobial peptides (e.g. Bac723 and Api13724,25), biogenesis factors (e.g. Rei1 and Nog146,47) and the dormancy factor MDF217 (Extended Data Fig. 6). Our structural analyses suggest that Dap1b/Dapl1’s specific interactions with eIF5a and the 28S rRNA contribute to Dap1b/Dapl1’s binding to the PET of a fully assembled eukaryotic 80S ribosome, thus making them unique among all ribosome-associated factors characterized so far.
All four factors bind to the same ribosome
To determine whether the four factors are bound to the same ribosomal particles, we performed a 3D variability analysis using cryoDRGN48. Each dataset’s latent representation showed distinct clusters when visualized in UMAPs (Extended Data Fig. 3f). A detailed analysis of the particles in these clusters revealed six major classes based on the presence or absence of eEF2, eIF5a, and tRNAs (Supplementary Table 3). In contrast to actively translating ribosomes, which show mutually exclusive binding of eIF5a and eEF218,32,49, our analysis revealed that 40.2% and 26.9% of ribosomes in Xenopus eggs and in 1 hpf zebrafish embryos, respectively, were simultaneously bound by eIF5a and eEF2 (Fig. 2h, class I particles). The reconstruction of a high-resolution map from Xenopus egg’s class I ribosomes revealed that Dapl1 and Habp4 are also present (Fig. 2h; Extended Data Fig. 3g, h). Given that class I ribosomal particles were exclusively identified in eggs and early embryos, which are characterized by a low translational activity (Fig. 1b-d), we will refer to these ribosomes as "dormant". Other ribosomal particle classes, including class II particles containing only eEF2, class III particles containing tRNAs, and “empty” class IV particles lacking eEF2, eIF5a, and tRNAs were identified to varying levels in all data sets (Fig. 2h; Supplementary Table 3). We hypothesize that “empty” class IV particles may correspond to ribosomes which have lost ribosome-associated factors or tRNAs during the purification since they do not cluster together in the UMAPs (Extended Data Fig. 3f). Together, our results reveal a novel ribosome state in which four factors (Habp4, eEF2, Dap1b/Dap and eIF5a) bind to the same ribosome.
Crosslinking of dormant ribosomes
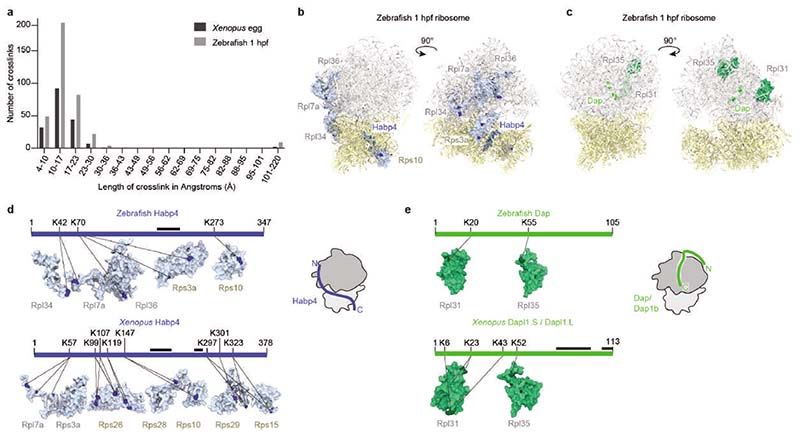
While we could assign Cryo-EM densities for small amino acid stretches within Habp4 and Dap1b/Dapl1 and Dap proteins, the majority of the polypeptide chains of these factors are not visible in our ribosome structures. To determine the path of these novel ribosome-associated proteins within the ribosome, we performed crosslinking MS with the primary amine crosslinker disuccinimidyl sulfoxide (DSSO), using ribosomes purified from 1 hpf zebrafish embryos and Xenopus eggs (Extended Data Fig. 7, Supplementary Table 4). We obtained more than 1000 crosslinked peptides for each ribosome sample, and 95% (zebrafish) and 90% (Xenopus) of all mapped crosslinks had Cα-Cα distances below the expected maximum crosslinking length of 23 Å50 (Extended Data Fig. 7a). We found crosslinks of the N-terminus of Habp4, which was not visible in cryo-EM densities, with small and large subunit ribosomal proteins, including Rpl7a (eL8) and Rps3a (eS1), in dormant ribosomes from both zebrafish and Xenopus (Extended Data Fig. 7b, d; Supplementary Table 4). Crosslinking MS analyses of Dap1b/Dapl1 and Dap showed that their N-terminal regions are proximal to the polypeptide exit site, which is consistent with insertion of their C-termini into the PET. In particular, we found crosslinks between a highly conserved N-terminal region of Dapl1/Dap and Rpl31 (eL31), and between the central region of Dapl1/Dap and Rpl35 (uL29) (Extended Data Fig. 7c, e; Supplementary Table 4), indicating that both N- and C-terminal regions of Dap1b/Dapl1 and Dap are in close proximity to the ribosome.
In vivo role of dormant ribosomes
Zebrafish and Xenopus habp4, dap1b/dapl1, dap and eif5a2 mRNAs are highly expressed during oogenesis, whereas eif5a and eef2 transcripts are abundant in all tissues (Extended Data Fig. 5bd). Moreover, Habp4 family proteins have been linked to ribosome stabilization during nutrient deprivation in yeast (Stm1)19 and to non-translating ribosomes in yeast19,51, Drosophila (Vig2)38 and mammals (SERBP1)18,21,22,38. Based on these observations, we hypothesized that Habp4 and Dap1b/Dapl1/Dap play key roles in the prolonged ribosomal storage and/or translational repression in the egg. To test this hypothesis, we used CRISPR/Cas9-based mutagenesis to generate individual (habp4-/-), double (dap-/-, dap1b-/-) and triple mutant (dap-/-, dap1b-/-, habp4-/-) zebrafish lines (Supplementary Fig. 1a-c).
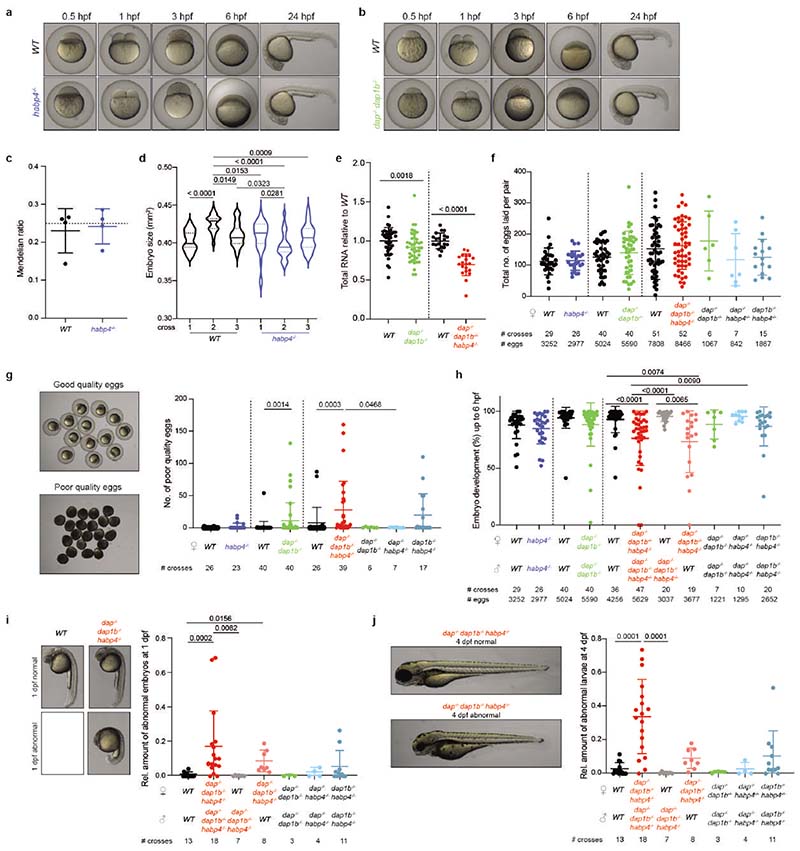
Habp4-/- mutants were viable and showed normal egg clutch sizes, fertility, embryo development and survival rates (Extended Data Fig. 8a, c, d, f-h). However, 1-3 hpf embryos derived from habp4-/- parents contained 30% fewer monosomes as well as about 30% less total RNA (80-90% of the total cellular RNA is estimated to be ribosomal RNA52) compared to their wild-type siblings (Fig. 3a-c). Importantly, this molecular phenotype could be rescued by oocyte-specific transgenic expression of Habp4 (Fig. 3a; Supplementary Fig. 1d). In contrast to the reduction in ribosomes observed in habp4-/- embryos, no significant difference was observed in polysome levels at 1 hpf in habp4-/- compared to wild-type embryos (Fig. 3c; Supplementary Fig. 1k). These findings demonstrate that Habp4 functions in stabilizing ribosomes. Moreover, the lack of an overt developmental phenotype in habp4-/- embryos suggests that ribosomes are stored in excess in the zebrafish egg given that a reduction in ribosome levels had previously been shown to cause developmental defects (e.g. minute phenotypes in Drosophila53) and to alter cell differentiation (e.g. in embryonic stem cells54 and tumor formation in adult zebrafish55).
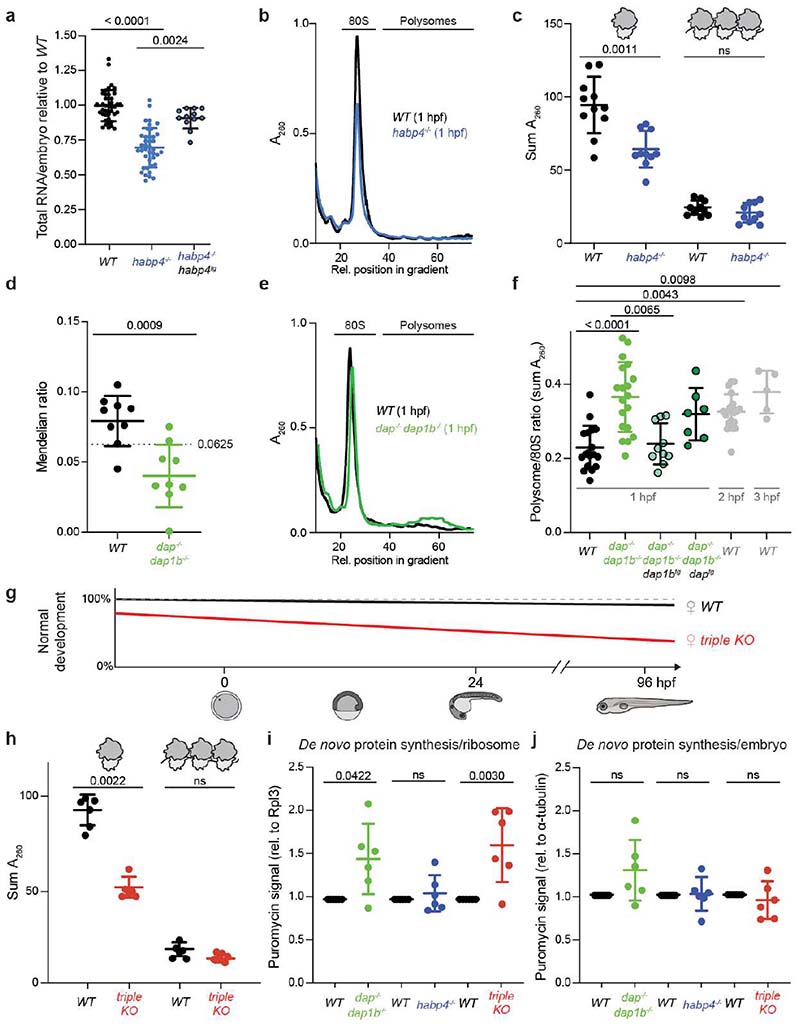
Fig. 3. Habp4 stabilizes monosomes, while Dap1b/Dap represses translation in embryos.
a, RNA isolated from wild-type (WT), habp4-/- and transgenic habp4-/- embryos expressing Habp4 (habp4-/-, habp4tg) (WT: n = 40; habp4-/-: n = 40; habp4-/-, habp4tg: n = 12). b, Representative polysome profiles from WT and habp4-/- embryos at 1 hour post-fertilization (hpf). A260, absorbance at 260 nm. c, Quantification of monosomes and polysomes from 1 hpf embryos (WT: n = 11; habp4-/-: n = 10). d, Mendelian ratio analysis of fin-clipped adult fish. Adult dap-/-, dap1b-/- and WT fish are expected at 6.25% from a dap+/-, dap1b+/- incross (n = 9 independent crosses; heterozygous combinations are not shown; total number of fish genotyped: 1029). e, Representative polysome profiles from 1 hpf WT and dap-/-, dap1b-/- embryos. f, Quantification of polysome-to-monosome ratios of 1 hpf embryos derived from WT, dap-/-, dap1b-/-, and transgenic dap-/-, dap1b-/- expressing Dap or Dap1b (WT, 1 hpf: n = 18; dap-/-, dap1b-/-: n = 19; dap-/-, dap1b-/-, daptg: n = 7; dap-/-, dap1b-/-, dap1btg: n = 10; WT, 2 hpf: n = 21; WT, 3 hpf: n = 5). g, Scheme of the percentage of WT and dap-/-, dap1b-/-, habp4-/- (triple KO) embryos that developed into larvae (Extended Data Fig. 8g-j). h, Quantification of monosomes and polysomes from 1 hpf embryos (n = 6 per genotype; representative profile in Supplementary Fig. 1j). i-j, Quantification of protein synthesis rates normalized to Rpl3 (i) or alpha-Tubulin (j) (n = 6 per genotype; Supplementary Fig. 1l; for uncropped images of membranes see Supplementary Fig. 2c). For a, c, f and h-j, n are biologically independent samples. In a, c, d, f and h-j, data are scatter dot plots with mean ± standard deviation (SD); ns, not significant. Statistical analysis was performed with Kruskal-Wallis followed by Dunn’s two-sided tests (a, f), two-sided Mann-Whitney (c, d, h), and one-way ANOVA followed by one-sided Tukey’s multiple comparisons test (i, j).
Adult dap-/-, dap1b-/- fish were obtained from a double heterozygous parent in-cross (dap+/-, dap1b+/-) at sub-Mendelian ratios compared to wild-type siblings (Fig. 3d), indicating a fitness defect. Though surviving dap-/-, dap1b-/- adults appeared morphologically normal and showed normal egg clutch sizes, fertility, and embryo development and survival rates (Extended Data Fig. 8b, f-j), their progeny displayed a significantly higher polysome-to-monosome ratio at 1 hpf compared to 1 hpf wild-type embryos (Fig. 3e, f). This ratio was similar to that observed for 2 hpf and 3 hpf wild-type embryos (Fig. 3f). Transgenic overexpression of Dap1b in dap-/-, dap1b-/- embryos was sufficient to rescue the higher polysome-to-monosome ratio observed in these mutants (Fig. 3f; Supplementary Fig. 1e-i), pointing to a role for Dap1b in translational inhibition. Importantly, dap-/-, dap1b-/- embryos did not show differences in monosome levels (Supplementary Fig. 1i), which suggests that, in contrast to Habp4, Dap1b and Dap are not important for ribosome stabilization in the egg.
Given that Habp4 and Dap/Dap1b associate with the same ribosomes and contribute to their dormancy, we generated triple knockout (KO) (dap-/-, dap1b-/-, habp4-/-) mutants to assess the physiological importance of the dormant egg ribosome state. Importantly, while eggs and embryos derived from habp4-/- and dap-/-, dap1b-/- mutants developed without overt morphological abnormalities, lack of all three proteins in adult triple KO females resulted in impaired oogenesis and embryogenesis (Fig. 3g). On average 30% of the eggs laid by triple KO females were of poor quality (Extended Data Fig. 8g) and, of the remaining 70%, only 74% (93% for wild-type eggs) were fertilized and showed proper early embryo development (Extended Data Fig. 8h). Moreover, an increased fraction of embryos from triple KO fish showed severe abnormalities by 1 and 4 days post fertilization, including abnormal head and eye development (Extended Data Fig. 8i, j). Taking into consideration all oogenesis and embryogenesis defects, less than 50% of eggs from triple KO fish developed into phenotypically normal larvae (Fig. 3g). Notably, these defects are due to the lack of dormancy factors in the egg (maternal phenotype) since these phenotypes were only observed in embryos derived from crosses with triple KO females (Extended Data Fig. 8h-j). Analysis of rRNA and monosome content in developing triple KO embryos revealed a reduction in the amount of ribosomes compared to wild type (Fig. 3h; Extended Data Fig. 8e), thus corroborating the important role of these factors in storing translationally repressed ribosomes in the egg.
To assess the in vivo roles of Dap1b/Dap and Habp4 in translation, we used puromycin incorporation as a direct readout of protein synthesis in early embryos. Consistent with our polysome profiling experiments, we observed increased translation per ribosome (normalized to ribosome content (Rpl3)) in dap-/-, dap1b-/- and triple KO, but not in habp4-/- embryos (Fig. 3i). Importantly, while dap-/-, dap1b-/- embryos also show a trend towards more active translation per embryo (normalized to alpha-Tubulin), translation levels per embryo in habp4-/- and triple KO samples resemble wild type (Fig. 3j). Together, our in vivo data suggests that Habp4 and Dap/Dap1b may play distinct roles in ribosome dormancy, with Habp4 contributing to ribosome stability and Dap/Dap1b to translational repression. However, a role for Habp4 in translational repression can at this stage not be ruled out.
Dap1b represses translation in vitro
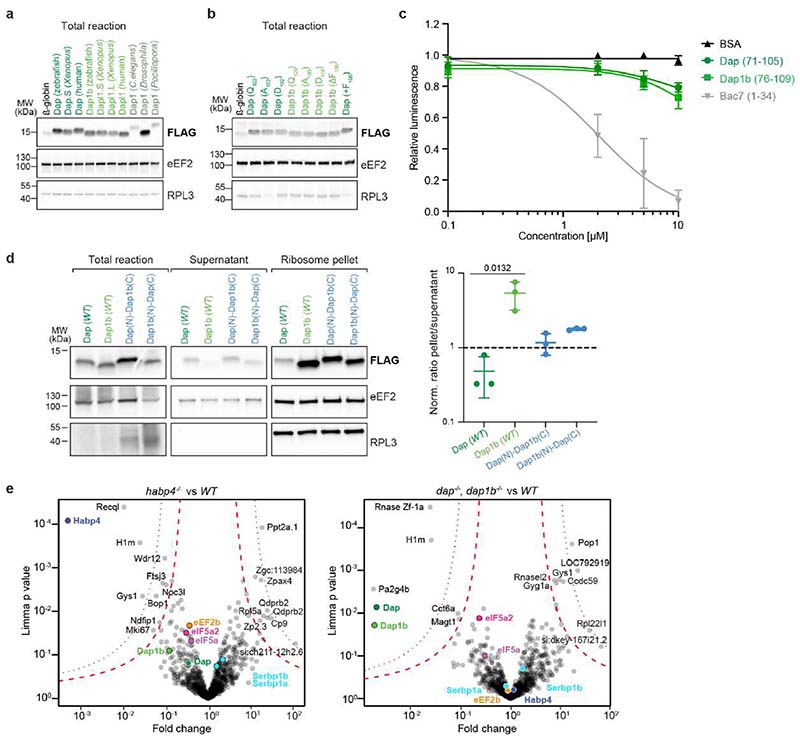
To directly assess Dap1b/Dapl1’s and Dap’s function in translational repression independently of other layers of translational regulation present in vivo, we performed in vitro translation assays in rabbit reticulocyte lysate (RRL) in the presence of recombinant Dap and Dap1b proteins (Fig. 4a). Dap and Dap1b/Dapl1 are small (~15 kDa), unstructured proteins that are rich in basic amino acids and prolines (15%). While invertebrates contain only one ortholog, vertebrates have evolved two paralogs (Extended Data Fig. 5a). Despite displaying less than 50% overall sequence identity, all Dap1b/Dapl1 and Dap proteins share two highly conserved motifs at the N- and C-terminus, which we found proximal to Rpl31 (Extended Data Fig. 5a and 7c, e) and within the PET (Fig. 2e, f), respectively. Whereas recombinant zebrafish Dap or the negative control BSA did not affect the translational activity of RRL, recombinant zebrafish Dap1b repressed the translation of a luciferase mRNA to a similar extent as the antimicrobial peptide Bac7 (Fig. 4b), which is known to bind to the PET and inhibit translation23. To investigate whether the observed difference in repressive activity between zebrafish Dap and Dap1b was generalizable to other Dap and Dap1b/Dapl1 proteins, we assessed ribosomal binding of in vitro translated flag-tagged dap and dap1b/dapl1 mRNAs from various species (Fig. 4a). Consistent with a general difference in repressive activity between Dap and Dap1b in vitro, Dap showed a weaker affinity for rabbit ribosomes than Dap1b/Dapl1 from three different vertebrate species (Fig. 4c, d; Extended Data Fig. 9a). Analysis of ancestral Dap1 proteins from invertebrates revealed that Dap1 from C. elegans and from the coral Pocillopora had an affinity for rabbit ribosomes in between that observed for vertebrate Dap and Dap1b proteins, while Dap from D. melanogaster did not bind to rabbit ribosomes (Fig. 4c, d; Extended Data Fig. 9a).
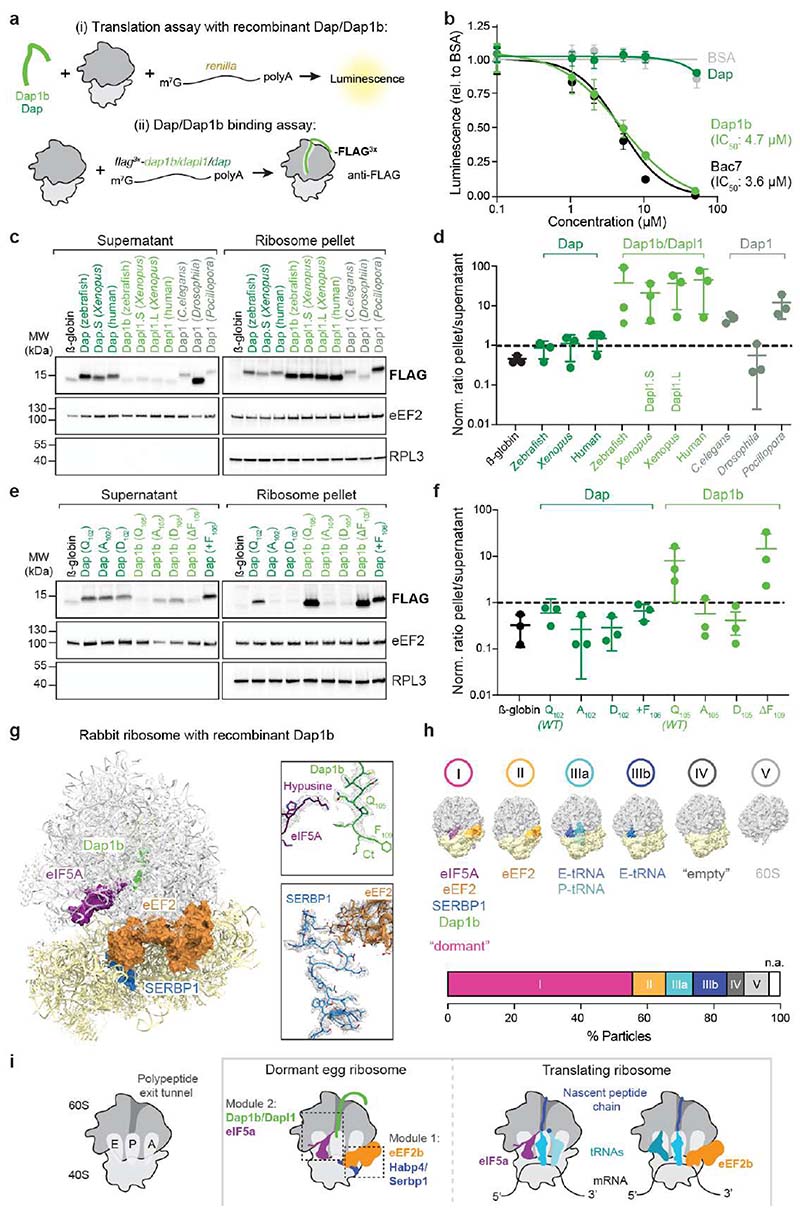
Fig. 4. Dap1b/Dapl1 binding to mammalian ribosomes blocks translation and reconstitutes the egg-like dormant ribosome state.
a, Scheme of the assays performed in rabbit reticulocyte lysate (RRL). (Top) Recombinant zebrafish Dap or Dap1b and in vitro synthesized renilla mRNA were added to RRL. Translation was assessed by measuring Luciferase activity. (Bottom) In vitro synthesized mRNAs encoding FLAG-tagged proteins were translated in RRL. Ribosomes were pelleted and proteins were quantified by Western blot. b, IC50 analyses of Dap1b and Dap (a, top). Bac7 and BSA were used as controls (n = 4; plot shows mean ± standard deviation (SD)). c, Ribosome binding assays of in vitro translated proteins (a, bottom). β-globin was used as control. eEF2 and RPL3 were used as a readout of fractionation and for normalization. Total reaction is shown in Extended Data Fig. 9a. d, Quantification of the blots shown in c. Values are normalized to loading (eEF2 or RPL3) and levels of the FLAG-tagged factors (n = 3). e, Ribosome binding assays with β-globin (control), wild-type and mutant versions of zebrafish Dap and Dap1b. Total reaction is shown in Extended Data Fig. 9b. f, Quantification of the blots shown in e (n = 3). g, Structure of the rabbit ribosome with zebrafish Dap1b. Ribosome-associated factors are shown as surface representations. Densities (in mesh) are shown on the right. Critical amino acids are indicated. Ct, C-terminus. h, Distribution of ribosomal particles from RRL among the classes obtained with cryoDRGN48. Representative filtered maps of the major classes (top) with superimposed colored densities for eEF2, eIF5A and tRNAs (Supplementary Table 3; n.a refers to “non-assigned” particles). i, Scheme depicting the main features of the dormant ribosomes identified in this study. (Left) Cartoon of important functional sites of the ribosome. (Middle) Dormant ribosomes are associated with four factors. (Right) Translating ribosomes. For b, d and f, n are independent experiments. In d and f, data are scatter plots with mean ± SD, and statistical analysis was performed with Kruskal-Wallis followed by Dunn’s two-sided test. Uncropped images of membranes shown in c and e are provided in Supplementary Fig. 2a, b.
We noticed that all rabbit ribosome-binding vertebrate Dap1b/Dapl1 proteins as well as the ancestral Pocillopora and C. elegans Dap1 proteins contain at least one additional residue at the C-terminus compared to non-binding vertebrate and Drosophila Dap proteins (Extended Data Fig. 5a). However, extending zebrafish Dap or shortening zebrafish Dap1b by one amino acid did not alter their ribosome binding capability (Fig. 4e, f; Extended Data Fig. 9b). Instead, mutating the conserved, hypusine-interacting Gln105 of zebrafish Dap1b (as well as the corresponding Gln102 of zebrafish Dap) abolished binding of Dap1b (and Dap) to the ribosome (Fig. 4e, f; Extended Data Fig. 9b). Interestingly, while the invariant Gln105 (Xenopus Gln109) is therefore key in stabilizing Dap1b binding to the ribosome, Dap1b’s C-terminus (76-109) alone was not sufficient to repress translation (Extended Data Fig. 9c). This is in contrast to the similarly short peptide Bac7, suggesting that both N- and C-terminal regions of Dap1b are important for its insertion into the PET. In agreement with this conclusion, chimeric proteins formed by Dap’s N-terminus and Dap1b’s C-terminus (and vice versa) bound less efficiently to the ribosome than Dap1b, but more efficiently than Dap (Extended Data Fig. 9d). Together, these analyses revealed that Dap1b/Dapl1 but not Dap efficiently binds to mammalian ribosomes in vitro and is sufficient to inhibit translation.
Reconstitution of ribosome dormancy
To assess whether Dap1b was inserted into the PET of mammalian ribosomes, and whether Dap1b addition to translational extracts indeed reconstitutes the dormant ribosome state, we performed cryo-EM on ribosomes isolated from an in vitro translation extract supplemented with 20 µM of recombinant zebrafish Dap1b and obtained a map at 2.3 Å average resolution (Fig. 4g; Extended Data Fig. 10a, b; Supplementary Table 2). Notably, we were able to assign densities not only for Dap1b in the PET, but also for the other dormant ribosome factors, namely eIF5A, eEF2, and the Habp4 homolog SERBP1 (Fig. 4g; Extended Data Fig. 10c), which are present in RRL18. A 3D classification of these ribosomal particles using cryoDRGN48 revealed that, after addition of zebrafish Dap1b, 56% of particles were in a dormant state similar to ribosomes from 1 hpf zebrafish embryos and Xenopus eggs (Fig. 4h (class I); Extended Data Fig. 10d; Supplementary Table 3). While several cryo-EM ribosome structures isolated from RRL have been published, none of them has reported the co-existence of eIF5A and eEF218,36, which is consistent with Dap1b being causally linked to this dormant ribosome state. Although Dap1b is required for generating the dormant ribosome state, absence of Dap1b in zebrafish embryos resulted only in a decreased ribosome association of eIF5A but not of eEF2 and Habp4 (Extended Data Fig. 9e). In contrast, ribosomes isolated from habp4-/- embryos showed decreased levels of all three proteins (Extended Data Fig. 9e), suggesting that Habp4 is required for binding of the other dormancy factors. While this reduction did not reach significance, the extent of the effect could be masked by two Habp4 paralogs, Serbp1a and Serbp1b, which are associated with ribosomes from habp4-/- embryos (Extended Data Fig. 9e) and might in part compensate for the loss of Habp4 in these embryos.
Discussion
Together, our results support a novel, dormant ribosome state present in the mature egg in which four functionally important sites of the ribosome, namely the A site and the E/P site, the mRNA channel and the PET are occupied by conserved factors (Fig. 4i). Based on direct protein interactions, these factors are organized in two modules: the Habp4-eEF2 module and the Dap1b/Dap-eIF5a module. Both in vivo and in vitro data revealed Dap1b as a novel translational repressor. While Habp4 binding to the mRNA-entry channel is also incompatible with translation, our in vivo data points to a main function for Habp4 in stabilizing ribosomes rather than in repressing translation. It is of note that two Habp4 paralogs, namely Serbp1a and Serbp1b, are associated with ribosomes at 1 hpf and might contribute to translational repression in habp4-/- mutants. Overall, both modules are important for establishing ribosome dormancy since loss of Dap, Dap1b and Habp4 in triple KO embryos results in a reduced amount of dormant ribosomes and strong developmental phenotypes. Establishing ribosome dormancy during oogenesis might thus be important for the increase in translation upon fertilization, which we speculate to be achieved via the release of dormant factors from the ribosome. Alternatively, dormant ribosomes might have additional roles in protecting the essential translational factors eIF5a and eEF2, which have been shown to be prone to aggregate with aging56, and/or serve as a source of pyrimidines to maintain nucleotide homeostasis in the embryo (as proposed for C. elegans57).
While ribosome storage is remarkably different in the egg compared to any other so-far described ‘inactive’ ribosome state in somatic cells, it is intriguing to speculate that similar mechanisms may be used at other stages in the lifetime of animals that require prolonged periods of low metabolic activity (e.g. the diapause in C. elegans). The mechanism we describe here, namely having functionally important sites of the ribosome occupied by evolutionarily conserved factors, two of which are needed for resumption of translation later on (Fig. 4i), appears ideally suited to accomplish energy-preserving long-term storage of ribosomes in a variety of developmentally programmed and metabolically induced contexts.
Methods
Zebrafish and Xenopus lines and husbandry
Zebrafish (Danio rerio) were raised according to standard protocols (28°C; 14/10 hour light/dark cycle). TLAB fish, generated by crossing zebrafish AB with the natural variant TL (Tupfel Longfin), served as wild-type zebrafish for all experiments. Zebrafish dap, dap1b and habp4 mutants were generated as part of this study and are described below. Wild-type Xenopus laevis were obtained from NASCO (USA) and maintained in the IMP animal facility. All fish and Xenopus experiments were conducted according to Austrian and European guidelines for animal research and approved by local Austrian authorities (animal protocols for work with zebrafish: GZ 342445/2016/12 and MA 58-221180-2021-16; animal protocols for work with Xenopus: BMWFW-66.006/0012-WF/II/3b/2014, BMWFW-66.006/0003-WF/V/3b/2016).
Generation of knockout and transgenic fish lines
Zebrafish dap, dap1b and habp4 mutants were generated by Cas9-mediated mutagenesis. A pool of two guide-RNAs (sgRNAs) targeting the first exon of either dap (chr24:22071216-22103555), dap1b (chr9:52378482-52386733) or habp4/zgc:103482 (chr21:20396891-20402554) were generated according to published protocols62 (see Supplementary Table 5 for oligos used for generating sgRNAs). Cas9 protein and sgRNA pools were co-injected into one-cell stage TLAB embryos. Putative founders were outcrossed to TLAB, and germline mutations were identified by a size difference in PCR amplicons (see Supplementary Table 5 for primers used for genotyping). Embryos from founder fish were raised to adulthood. Homozygous knockout fish were generated by crossing heterozygous fish. To generate dap-/-, dap1b-/- double mutants, dap+/- and dap1b+/- fish were crossed, and double heterozygous mutants were identified by genotyping and raised to adulthood. Double homozygous mutants (dap-/-, dap1b-/-) and wild-type (dap+/+, dap1b+/+) siblings were obtained from in-crossing double heterozygous mutants. To generate dap-/-, dap1b-/-, habp4-/- triple mutants, dap+/-, dap1b+/- and habp4+/- fish were crossed, and triple heterozygous mutants were identified by genotyping and raised to adulthood. Triple homozygous mutants (dap-/-, dap1b-/-, habp4-/-) and double mutants (dap+/+, dap1b-/-, habp4-/-; dap-/-, dap1b+/+, habp4-/-; dap-/-, dap1b-/-, habp4+/+) and wild-type siblings were obtained from in-crossing double heterozygous mutants. To generate N-terminally tagged 3xFlag-sfGFP-Dap or 3xFlag-sfGFP-Dap1b transgenic fish lines, the respective ORF was PCR amplified from zebrafish cDNA and cloned via the Gibson assembly method63 and BamHI/EcoRV sites into a vector for Tol2-based transgenesis containing the actb2 promoter, a Xenopus globin 5’-UTR, an SV40 3’UTR and an Ampicillin resistance gene (final constructs: Tol2 - actb2: globin 5’UTR - 3xflag-sfgfp-dap - SV40 3’UTR; Tol2 - actb2: globin 5’UTR - 3xflag-sfgfp-dap1b - SV40 3’UTR).
Plasmids (15 pg) were injected together with 35 pg Tol2 mRNA into the cell of 1-cell stage TLAB embryos. Putative founders were crossed to the dap-/-, dap1b-/- line, and transgenic offspring were identified by GFP expression and genotyped for dap and dap1b mutations at 8 weeks. Adult fish expressing the transgene and heterozygous for dap and dap1b were incrossed. Larvae from this cross were screened at 2 days post-fertilization for GFP expression and genotyped for dap and dap1b mutations at 8 weeks.
To generate a C-terminally tagged Habp4-3xFlag transgenic fish line, the respective ORF lacking the STOP codon was PCR amplified from zebrafish cDNA (RNA from multiple embryonic stages was pooled) and cloned via the Gibson assembly method63 and XmaI/NheI sites into a vector for Tol2-based transgenesis containing the zp3 promoter, a Xenopus globin 5’-UTR, an SV40 3’UTR, a cmlc2:GFP as transgenesis marker and an Ampicillin resistance gene (final constructs: Tol2 - zp3: globin 5’UTR - habp4-3xflag - SV40 3’UTR, cmlc2:gfp). The plasmid was injected into embryos as described above. Putative founders were crossed to habp4+/- fish and transgenic offspring were identified by screening for cmcl2-driven heart-specific expression of GFP at 30 hpf.
Phenotypic characterization
Adult fish were crossed at least twice prior to phenotypic characterization. Females and males were kept at equal numbers per tank. To quantify egg quality, individual male and female fish were set up together in breeding cages the night before mating. Wild-type and mutant sibling females were crossed to wild-type male fish. Fish were put together by removing the divider and allowing fish to mate for approximately 60 minutes. Eggs were collected from individual breeding cages and moved to E3 medium with 0.1% methylene blue at 28°C. Eggs were scored under a dissection microscope. Eggs were considered as “good quality” if they were normal-looking one-cell stage embryos (clear separation of yolk and cell; normal egg activation and chorion swelling; non-opaque appearance). Eggs were considered as “poor quality” if they were opaque, deformed, and no clear cell or chorion elevation was visible. Pictures of early embryo development were taken at 0.5, 1, 2, 3, 6 and 24 hours post-fertilization (hpf) on a ZEISS Stemi 508 stereo microscope with camera (2x magnification, FlyCapture2 software). To calculate the fertilization rate, the number of fertilized eggs was divided by the number of “good quality” eggs. Clutch size was calculated as the sum of “good quality” eggs (fertilized and unfertilized). The size of the embryos was measured as area (mm2) at 6 hpf in Fiji. Successful and normal embryonic development was assessed at 6 hpf. Abnormal looking embryos were removed from the dish. Survival rate was calculated by dividing the number of larvae at 24 hpf by the number of fertilized eggs. Moreover, the numbers of normal and abnormal looking larvae were counted at 4 days post-fertilization. Statistical analysis was performed in GraphPad Prism v8.0.2.
Polysome gradients
Wild-type and mutant embryos were dechorionated with pronase (1 mg/mL). 200 dechorionated embryos per genotype were lysed in 550 μL of lysis buffer (20 mM Tris-Cl pH 7.5, 30 mM MgCl2, 100 mM NaCl, 0.25% Igepal-630 (v/v), 100 μg/mL cycloheximide, 0.5 mM dithiothreitol (DTT), and 1 mg/mL heparin). Embryo lysates were incubated for 10 min on ice and centrifuged for 10 min at 4°C with 21,000 xg. 200 μL of clarified lysates were loaded onto a continuous 10-50% (w/v) sucrose gradient prepared in TMS buffer (20 mM Tris-Cl pH 7.5, 5 mM MgCl2, 140 mM NaCl). Gradients were centrifuged in a SW40 Ti rotor (Beckman) at 4 °C and 35,000 rpm for 165 min. Polysome gradients were analyzed using a gradient station (BioComp) coupled to a Model Triax™ Flow Cell detector (FC-2). The precise location of the fractions along the UV-tracing was monitored using the Gradient profiler v1.25 (BioComp) software. Polysome gradients were normalized prior to quantification for baseline differences. The area under the curve was calculated for the defined fractions. Polysome-to-monosome ratio was calculated by dividing the value of the polysome fraction by the obtained value of the 80S fraction.
RNA isolation
Total RNA was isolated from 10 homogenized zebrafish embryos per sample using the RNeasy Mini Kit (Qiagen). RNA concentration was measured on a spectrophotometer (DeNovix DS-11 FX+). Mutant RNA concentration was normalized to wild-type RNA concentration for each experiment.
Analysis of translation efficiency
To calculate translational efficiencies over the time-course of embryogenesis, published PolyA+ RNASeq data31 (GSE32898) and ribosome profiling data30 (GSE46512) was pre-processed according to standard bioinformatic procedures. For ribosome profiling data (ribo-seq), 3’ adaptor sequences were removed using bbduk from bbmap v38.26, and trimmed reads were mapped to a set of rRNAs and abundant sequences downloaded from the Illumina iGenome using bowtie2. The remaining reads were aligned to the zebrafish reference genome GRCz10 using tophat2. For RNA-seq analysis, adaptor sequences were removed with bbduk from bbmap v38.26, and abundant reads that mapped to abundant sequences using bowtie2 were removed, similar to ribo-seq reads. Remaining reads were mapped with tophat2 to the zebrafish transcriptome using the GRCz10 genome assembly and the Ensembl 82 transcriptome release.
For ribo-seq and RNA-seq, FPKM values for the coding sequence (CDS) were calculated for transcripts with an annotated start and stop codon. FPKM values were defined as read counts over CDS/(CDS length * total read count) * 109. Translational efficiencies were obtained by dividing FPKM of ribo-seq by FPKM of RNA-seq. TE values + 0.1 were used for plotting in R 4.0.2 (1 hpf: n = 35975; 3 hpf: n = 36872; 6 hpf: n = 39916; 28 hpf: n = 41690 quantified transcripts). Statistical significance was assessed by the two-sided Wilcoxon pairwise test, and effect size was estimated by Cohen’s D58 (< 0.2: “negligible”; [0.2-0.5]: “small”; [0.5-0.8]: “medium”; > 0.8: “large”).
Sample collection for ribosome isolation
The evening prior to the zebrafish egg or embryo collections, male and female zebrafish were separated in breeding cages. To collect mature, un-activated eggs, female zebrafish were anesthetized in the morning using 0.1% (w/v) Tricaine (25x stock solution in dH2O, buffered to pH 7-7.5 with 1 M Tris pH 9.0). After being gently dried on a paper towel, the female was transferred to a dry Petri dish, and eggs were carefully expelled from the female by applying mild pressure on the fish belly with a finger and stroking from anterior to posterior. The eggs were separated from the female using a small paintbrush, and the female was transferred back to the breeding cage filled with fish water for recovery. To prevent activation, eggs were kept in sorting medium (Leibovitz’s medium, 0.5% BSA, pH 9)64 at room temperature (RT). Zebrafish embryos were collected from regular matings. In the case of zebrafish embryos (1,000 embryos per sample for mass spectrometry (MS) and electron cryo-microscopy (Cryo-EM), and 5,000 embryos per sample for crosslinking-MS), eggs were collected in Petri dishes with blue water, which consist of fish water, 0.025% (v/v) Instant Ocean salts (Aquarium Systems, 218035), and 0.0001% (v/v) methylene blue (Sigma-Aldrich, M9140), pH 7. Embryos were incubated at 28°C, collected at the desired time points (1 hpf and 6 hpf), and incubated with 1 mg/mL of pronase for 5 min at RT for dechorionation.
To obtain samples from Xenopus, egg collection and in vitro fertilization were performed following a previously described protocol65. In brief, sexually matured wild-type females were primed with 50 IU of pregnant mare serum gonadotropin (PMSG, ProSpec, HOR-272) one week before the experiments. In the evening before egg collection, PMSG-primed females were injected with 500 IU of human chorionic gonadotropin (hCG, Sigma-Aldrich, CG10). Freshly laid eggs were collected and chemically dejellied in 2% (w/v) L-cysteine/1xMMR (Marc’s Modified Ringer) pH 7.8 before ribosome extraction. For the 24 hpf sample, freshly laid eggs were in vitro fertilized with wild-type sperm and dejellied in 2% (w/v) L-cysteine/0.1xMMR pH 7.8 at the 1-cell stage. Embryos were allowed to develop at RT for 24 h before ribosome extraction.
Ribosome isolation
Samples were washed in 2 volumes of lysis buffer, containing 20 mM HEPES-KOH pH 7.4, 150 mM KCl, 10 mM MgCl2, 0.5 mM DTT, rRNasin (Promega), 0.25% RNaseOUT (v/v) (ThermoFisher), 0.25% SUPERaseIN (v/v) (ThermoFisher), 0.25% Igepal (v/v), and cOmpleteTM-EDTA-free protease inhibitor (Roche). Embryos were lysed in 2 mL (for 1,000 embryos) or 5 mL (for 5,000 embryos) of lysis buffer using a pre-cooled Dounce homogenizer. Following incubation on ice for 5 min, the supernatant was cleared by centrifugation at 21,000 xg for 10 min at 4°C. Ribosome isolation was adapted from66. Briefly, the clarified supernatant was loaded onto a 30% (w/v) sucrose cushion prepared in buffer A (20 mM Tris-Cl pH 7.5, 2 mM Mg(OAc)2 and 150 mM KCl) and centrifuged at 116,000 xg for 5 h at 4°C in a TL100.3 rotor (Beckman). The ribosome pellet was resuspended by orbital shaking (1 h at 4°C) in buffer B, consisting of 20 mM Tris-Cl pH 7.5, 6 mM Mg(OAc)2, 150 mM KCl, 6.8% sucrose (w/v), 1 mM DTT, 0.25% RNasin Plus (v/v) (ThermoFisher), and 0.25% RNase Inhibitor (v/v), (Promega). Resuspended ribosomes were loaded onto a 15-30% (w/v) sucrose gradient prepared in buffer C (100 mM KCl, 5 mM Mg(OAc)2, 20 mM HEPES-KOH pH 7.6, 1 mM DTT, and 10 mM NH4Cl) and centrifuged for 16 h at 4°C with 18,000 rpm in an SW60Ti rotor (Beckman). Gradients were hand-fractionated and ribosome concentrations were determined by absorbance at 260 nm (A260) and SDS-PAGE.
To isolate ribosomes from rabbit reticulocyte lysates (RRL) (Green Hectares; composition and preparation of the lysate are described in the section “In vitro translation assays” below), RRL was supplemented with 20 µM recombinant zebrafish Dap1b protein and incubated for 1 h at 30°C. A one-step centrifugation of 1 mL of in vitro translation reaction was performed over 3 mL of a 30% (w/v) sucrose cushion prepared in RNC buffer (50 mM HEPES-KOH pH 7.4, 100 mM KOAc, 5 mM Mg(OAc)2, and 1 mM DTT) in a TLA100.3 rotor (Beckman) at 116,000 xg for 5 h. The ribosome pellet was resuspended in 120 mL of RNC buffer.
For MS and cryo-EM, sucrose was removed from the gradient fractions by loading the resuspended ribosome solution on a Zeba™ Spin Desalting Columns (7K MWCO).
To analyze ribosome composition by Western blot, 1000 embryos or larvae were used per condition. Total lysate samples were collected from the cell free extract after the initial centrifugation step. The cytoplasmic fraction overlaying the sucrose cushion was collected after the ultracentrifugation step. Pelleted ribosomes after the first ultracentrifugation step were directly resuspended in 200 µL 1x Laemmli buffer. All collected fractions were frozen in liquid nitrogen.
Digest of total cell lysates and ribosomes for mass spectrometry (MS)
Total cell lysates and isolated ribosomes from zebrafish eggs and embryos and from Xenopus eggs were either directly denatured in 8 M urea in 100 mM ammoniumbicarbonate (ABC) buffer or acetone precipitated before being dissolved in 8 M urea in 100 mM ABC. DTT was added to a final concentration of 10 mM and the sample was incubated 1 h at 37°C. Alkylation was performed by adding iodoacetamide (IAA) to a final concentration of 20 mM and incubating for 30 min at RT in the dark. The reaction was quenched by addition of 5 mM DTT and incubated again 30 min at RT.
Samples were diluted to 6 M urea with 100 mM ABC followed by addition of Lys-C (FUJIFILM Wako Pure Chemical Corporation) at a ratio of 1:100 and incubation at 37°C for 2 h. The samples were diluted to 2 M urea with 100 mM ABC. Trypsin (Promega, Trypsin Gold) was added at a ratio of 1:100 and incubated at 37°C overnight. 500 ng of each sample were analyzed by liquid chromatography-mass spectrometry (LC-MS/MS).
Crosslinking of purified ribosomes and digest for mass spectrometry (MS)
For elution of zebrafish 1 hpf and Xenopus egg ribosome fractions used in crosslinking-MS experiments, buffer C was adjusted to 20 mM HEPES-KOH pH 7.6, 100 mM KCl, and 5 mM Mg(OAc)2. 0.5 mM of disuccinimidyl sulfoxide (DSSO) from a 5 mM stock solution (in dimethyl sulfoxide, DMSO) was added to 10 µL of 500 nM ribosomes and incubated for 45 min at RT. The reaction was quenched by adding Tris-Cl pH 7.5 to a final concentration of 100 mM and incubated for 15 min at RT. The band pattern of the crosslinking reaction was analyzed by SDS-PAGE. DSSO-crosslinked ribosomes were denatured in 8 M urea, 100 mM ABC followed by reduction, alkylation and proteolytic digest using Lys-C and trypsin (both added 1:20) as described above. Digests were acidified to 1% TFA and desalted using Oasis HLB Sorbent (Oasis HLB Microelution plate, Waters) according to the manufacturer’s description. Peptides were eluted with 2 x 30 μL 70% acetonitrile (ACN), 0.1% TFA and the sample was concentrated under reduced pressure to 20 μL. The samples were supplemented with 5% DMSO. To enrich for crosslinked peptides, the samples were fractionated by size-exclusion chromatography (SEC) on a TSKgel SuperSW2000 column (300 mm × 4.5 mm × 4 μm, Tosoh Bioscience), which was operated at 200 μL/min in 30% ACN, 0.1% TFA. Fractions were collected every minute, and ACN was removed under reduced pressure. DMSO was again added to 5%.
LC-MS/MS
Generated peptides were analyzed on a nano-reversed phase HPLC (RSLC nano system, ThermoFisher) coupled to a Q Exactive HF-X mass spectrometer (ThermoFisher), equipped with a Nanospray Flex ion source (ThermoFisher).
Peptides were loaded onto a trap column (PepMap C18, 5 mm × 300 μm ID, 5 μm particles, 100 Å pore size) at a flow rate of 25 μL/min using 0.1% TFA as mobile phase. After 10 min, the trap column was switched in line with the analytical column (PepMap C18, 500 mm × 75 μm ID, 2 μm, 100 Å, ThermoFisher), which was operated at a flow rate of 230 nl/min at 30°C. For separation a solvent gradient was applied, starting with 98% buffer A (0.1% formic acid in water) and 2% buffer B (80% ACN, 0.1% formic acid), followed by an increase to 35% buffer B over the next 180 or 360 min for digested lysates and ribosomes. For unfractionated and SEC-enriched crosslinked samples, a gradient to 40% buffer B in 180 or 120 min, respectively, was used. This was followed by a steep gradient to 90% buffer B in 5 min, staying there for five min and decreasing to 2% buffer B in another 5 min.
For the digested lysates and ribosomes, the mass spectrometer was operated in data-dependent mode, using a full scan (m/z range 375-1500, resolution of 60,000, target value 1E6) followed by MS/MS scans of the 10 most abundant ions. MS/MS spectra were acquired using a normalized collision energy of 28, isolation width of 1.4 m/z, resolution of 30,000, maximum fill time of 105 ms and a target value of 1E5. Precursor ions selected for fragmentation (excluding charge state 1, 7, 8, >8) were put on a dynamic exclusion list for 60 s. Additionally, the minimum AGC target was set to 5E3 and intensity threshold was calculated to be 4.8E4. The peptide match feature was set to preferred, and the exclude isotopes feature was enabled.
For crosslinked samples, the mass spectrometer was operated in data-dependent mode, using a full scan (m/z range 350-1600, resolution of 120,000 and a target value of 1E6) followed by MS/MS scans of the 15 most abundant ions. MS/MS spectra were acquired using a stepped normalized collision energy of 27+/-6, isolation width of 1 m/z, resolution of 30,000, maximum fill time of 150 ms and a target value of 5E4. Precursor ions selected for fragmentation (excluding charge state 1, 2, >7) were put on a dynamic exclusion list for 30 s. Additionally, the minimum AGC target was set to 5E3, and intensity threshold was calculated to be 3.3E4. The peptide match feature was set to preferred, and the exclude isotopes feature was enabled.
Analysis of MS data
For peptide identification, the RAW-files were loaded into Proteome Discoverer (v2.5.0.402, ThermoFisher). All hereby created MS/MS spectra were searched using MSAmanda v2.5.0.16129, Engine v2.0.0.1612967. For the first step search, the RAW-files were searched against a combined zebrafish sequence database (Danio_rerio GRCz11) and Uniprot downloaded on 21st March 2019 (58,524 sequences; 34,079,443 residues), and against the Xenopus laevis Uniprot proteome UP000186698. All RAW-files were supplemented with common contaminants using the following search parameters: (i) the peptide mass tolerance was set to ±5 ppm and the fragment mass tolerance to ±8 ppm, and (ii) the maximal number of missed cleavages was set to 2 using tryptic enzymatic specificity. The result was filtered to 1% FDR on protein level using Percolator algorithm68 integrated in Proteome Discoverer. A sub-database was generated for further processing. For the second step, the RAW-files were searched against the created sub-database. Oxidation on methionine, deamidation on asparagine and glutamine, phosphorylation on serine, threonine and tyrosine; iodoacetamide derivative on cysteine, beta-methylthiolation on cysteine and carbamylation on lysine were set as variable modifications. Monoisotopic masses were searched within unrestricted protein masses for tryptic enzymatic specificity. The peptide mass tolerance was set to ±5 ppm and the fragment mass tolerance to ±8 ppm. The maximal number of missed cleavages was set to 2. The result was filtered to 1% FDR on peptide level using Percolator algorithm integrated in Proteome Discoverer. The localization of the post-translational modification sites within the peptides was performed with the tool ptmRS, based on the tool phosphoRS69. Peptide areas have been quantified using in-house-developed tool apQuant70. Proteins were quantified by summing unique and razor peptides and applying the intensity-based absolute quantification (iBAQ) calculation. Normalization of protein abundances was performed on the sum of areas of all identified ribosomal proteins. Statistical significance of differentially expressed proteins was determined using limma71.
Analysis of crosslinking-MS data
Data analysis was performed within Proteome Discoverer v2.5.0.402 using MS Annika (v1.0.18345)72. The workflow tree consisted of the MS Annika Detector node (MS tolerance 10 ppm, crosslink modification: DSSO +185.004 Da at lysine and at protein N-termini, diagnostic ions: 138.0911; 155.179; 170.0634; 187.0900, crosslink modification addition: 18.010565, doublet pair selection in combined mode) followed by MS Annika Search (full tryptic digest, 5/10 ppm peptide/fragment mass tolerance, maximal 3 missed cleavages, carbamidomethyl +57.021 Da at cysteine as static and oxidation +15.995 Da at methionine as dynamic modification) and completed with MS Annika Validator (1% FDR cutoff at CSM and crosslink level, separate Intra/Inter-link FDR set to false). The search was performed against a database generated from a MS analysis of a digested non-crosslinked zebrafish (containing 1,092 proteins) or Xenopus (containing 1,173 proteins) ribosome sample, respectively. To generate these specific databases the two non-crosslinked samples were searched using MS Amanda against the zebrafish Uniprot reference proteome UP000000437 (downloaded 2020-09-15, 46,847 sequences, 24,556,292 residues) or Xenopus laevis Uniprot proteome UP000186698 (downloaded 2020-01-07, 43,235 sequences, 19,251,456 residues), both supplemented with common contaminants. Search parameters were set as described for time course experiments but using 5/10 ppm peptide/fragment mass tolerance and maximum 3 missed cleavages. Carbamidomethylation on cysteine was set as a fixed modification, oxidation on methionine, deamidation on asparagine and glutamine and acetylation on the protein N-terminus were set as variable modifications.
To validate the quality of the chosen search strategy, an additional crosslink search was performed using equal settings but a database file containing sequences aligned to the dormant ribosome structures of Xenopus and zebrafish. The resulting crosslinks at 1% FDR were plotted onto the structure using ChimeraX v1.173, and the distance between connected amino acids (Cα-Cα distance) was measured. Crosslinks including Cα residues missing in the structure file were removed, yielding 360 and 178 crosslinks aligned to the zebrafish and Xenopus structures, respectively.
DNA constructs and mRNA synthesis
dap, dap1b, habp4, renilla luciferase and zebrafish β-globin constructs and dap/dap1b mutant and chimeric versions for in vitro translation in rabbit reticulocyte lysate (RRL) (Green Hectares) were cloned into pCDNA3.1 by Gibson assembly63. Zebrafish coding sequences were amplified from cDNA. Dap, dapl1 and dap1 coding sequences from other species were ordered as Gblocks from IDT (amino acid sequences are shown in Extended Data Fig. 5a). Capped mRNAs of all cloned constructs were synthesized with the Sp6 mMessage Machine kit (Ambion) according to the manufacturer’s protocol, using linearized plasmids as templates.
Recombinant protein expression and purifications
Zebrafish dap1b and dap coding sequences were codon optimized for E. coli, cloned into a vector providing an N-terminal 6xHis tag followed by a 3C protease-cleavage site, and transformed into BL21(DE3) E. coli cells. For Dap1b expression, cells were grown in LB at 37°C until reaching an OD600 of 0.7, induced with 0.5 mM IPTG, and grown overnight at 18°C. For Dap expression, bacterial cultures were grown on terrific broth medium supplemented with 1.5% (w/v) lactose for 1 h at 37°C, and for 23 h at 18°C. Cells were pelleted, resuspended in lysis buffer (50 mM Tris-Cl pH 8.0, 300 mM NaCl, cOmplete Protease Inhibitor Cocktail [Merck]) and lysed by sonication (19 mm probe, 4 min: pulse on 1 s, pulse off 2 s, 60% amplitude). The intracellular soluble fraction was isolated by centrifugation at 20,000 xg, 30 min, 4°C. Imidazole was added to a final concentration of 20 mM. The lysate was loaded onto a 5 mL HisTrap FF column (Merck), washed with buffer A (50 mM Tris-Cl pH 8.0, 300 mM NaCl), and eluted with buffer A supplemented with 500 mM imidazole. 1 mg of 3C protease was added to the elution fraction during dialysis overnight against buffer A. The cleaved protein was loaded onto a 5 mL HisTrap FF column (Merck); the flow-through, containing the cleaved protein, was concentrated (Vivaspin 20, MWCO 5 kDa) and loaded onto a Superdex 75 26/60 column (GE Life Sciences) equilibrated with SEC buffer (20 mM HEPES-KOH pH 7.5, 150 mM KCl).
Analysis of de novo translation
De novo translation in zebrafish embryos was measured using incorporation of puromycin. 180 µM puromycin (approximate final concentration 2 µM) was injected into 1-cell stage embryos. Embryos were incubated for 30 min, during which they were manually dechorionated. Following the 30 minute incubation period, embryos were manually deyolked (de-capped). Cell caps were immediately transferred into 1x Laemmli sample buffer and snap-frozen in liquid nitrogen. Each individual replicate consisted of embryos derived from a single pair of wild-type fish and a single pair of mutant fish of the respective genotype. Only two pairs (one wild-type and one mutant pair) were allowed to mate at the same time, and the exact time point of the mating onset was recorded for each pair and sample processing was adapted accordingly. For each sample 10 embryos were lysed in 50 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, 12.5 mM EDTA containing cOmplete Protease Inhibitor Cocktail (Roche, 11836170001). Embryos treated with 100 µg/ml cycloheximide 10 min prior to injection with puromycin were used as a negative control. Puromycin incorporation was assessed by Western blotting (see below).
In vitro translation (IVT) assays
Rabbit reticulocyte lysate (RRL; Green Hectares) was optimized for translation as previously described74,75. The final reaction contained 35% (v/v) RRL, 20 mM HEPES-KOH pH 7.4, 10 mM KOH, 50 mM KOAc, 1 mM ATP, 1 mM GTP, 12 mM creatine phosphate, 0.1 mg/mL tRNAs (Merck), 40 μg/mL creatine kinase, 2 mM MgCl2, 1 mM reduced glutathione, 0.3 mM spermidine, and 40 μM of each of the 20 amino acids. 1 μg of mRNA was added to a total volume of 30 μL of RRL. To compare translation of different factors, equimolar amounts of mRNA were added per reaction. Reactions were performed at 30°C for 60 min (unless stated otherwise). To assess ribosome binding, 250 μL of the reaction was loaded onto a 30% (w/v) sucrose cushion in ribosome-stabilizing RNC buffer (50 mM HEPES-KOH pH 7.4, 100 mM KOAc, 5 mM Mg(OAc)2, and 1 mM DTT), and centrifuged for 5 h with 116,000 xg at 4°C in a TLA100.3 rotor (Beckman). Binding to the different fractions was assessed by Western blot.
To analyze the sufficiency of recombinant zebrafish Dap1b and Dap proteins and C-terminal peptides to repress translation, different concentrations of recombinant proteins or synthetic peptides in protein buffer (20 mM HEPES-KOH pH 7.4, 150 mM KCl) and 1 μg renilla luciferase mRNA were added to RRL to a total volume of 25 μL. Bac7(1-35) synthetic peptide (1-RRIRPRPPRLPRPRPRPLPFPRPGPRPIPRPLPFP-35) was used as a positive control. BSA served as negative control. Reactions were performed at 30°C for 90 min. Renilla luciferase activity (Promega, E1960) was measured in duplicates on a Synergy H1 plate reader (BioTek), using the software Gen5 (BioTek Instruments, Inc.). Samples were normalized to buffer control containing 1 μg of renilla luciferase mRNA. Relative IC50 was calculated in GraphPad Prism v8.0.2 using nonlinear regression and the equation [Inhibitor] vs. response - Variable slope (four parameters).
Western blotting and quantification
Western blotting was performed following standard protocols. In brief, protein samples were boiled for 5 min in 1x Laemmli sample buffer and separated by SDS-PAGE using Mini-PROTEAN® TGXTM Precast Protein Gels or 10–20% Mini-PROTEAN® Tris-Tricine Gels (Bio-Rad). Blotting was performed onto a nitrocellulose membrane (GE Healthcare) using a wet-blot system (BioRad). The following antibodies were used: anti-FLAG (mouse, 1:1000, Sigma-Aldrich F1804), anti-eEF2 (rabbit, 1:1000, Proteintech, 20107-1-AP), anti-Puromycin (mouse, 1:20000, Sigma-Aldrich, MABE343), anti-alpha-Tubulin (mouse, 1:10000, Sigma-Aldrich, T6074), anti-RPL3 (rabbit, 1:1000, GeneTex, GTX124464), F(ab’)2 anti-rabbit IgG (H+L)-HRPO (goat, 1:10000, 111-036-045, Dianova), and F(ab’)2 anti-mouse IgG (H+L)-HRPO (goat, 1:10000, 115-036-062, Dianova). The chemiluminescent signal was quantified using ImageJ 1.8.0_172. To quantify ribosome binding of FLAG-tagged proteins after in vitro translation, signal intensities were simultaneously measured for FLAG, RPL3 and eEF2 for all three membranes (total reaction, supernatant, ribosome pellet). The ratio between the FLAG signal in the supernatant and the ribosome pellet was normalized to the total amount translated (FLAG signal in the total reaction) and to sample loading (RPL3 and eEF2 signals). To quantify de novo translation in wild-type and mutant embryos, signal intensities were measured for Puromycin, RPL3 and alpha-Tubulin. The Puromycin signal was normalized to the RPL3 (per ribosomal content) or alpha-Tubulin (per embryo) signal. Transgenic expression of tagged Dap and Dap1b as well as changes in ribosome binding during development were determined by measuring the signal intensity for FLAG, RPL3, alpha-Tubulin and eEF2 for all three membranes (total reaction, supernatant, ribosome pellet). The relative amount in each fraction was determined by normalization of the FLAG signal in the supernatant and in the ribosome pellet fraction to the total amount expressed at the respective stage and the amount loaded. Raw images of uncropped blots are shown in Supplementary Fig. 2.
Grid preparation for cryo-EM
A 2 nm thick continuous carbon film was produced on an Auto306 high vacuum evaporator (Boc Edwards). This film was floated onto Quantifoil Cu 3.5/1, 200 mesh grids, and grids were dried. Grids were glow-discharged for 1 min in a SCD005 sputter coater (Bal-Tec) at ~20 mA. 4 µL of sample (at a concentration of 200 ng/µL of RNA, which corresponds to ~ 100 nM ribosomes) were applied to the grid and incubated for 60 s at 70% humidity and 4°C in a Leica EM GP. Subsequently, grids were blotted for 2 s using the proximity sensor and plunge-frozen in liquid ethane at –180°C.
Cryo-EM data collection
All grids were screened on a Glacios TEM (ThermoFisher) to check for particle distribution and grid quality; data were recorded with EPU2. Grids that passed the evaluation were recorded on a Titan Krios microscope equipped either with a Falcon 3EC or a K3 detector, using the software SerialEM v3.8. A pixel size of about 1 Å/px was chosen, and micrographs with a total dose of about 40 e/Å2 fractionated in 39 frames were collected with a target defocus between –1 and –2.5 µm.
Electron microscopy data processing
Most data processing was performed in Cryosparc version 2.16.076. Micrographs were motion corrected and dose weighted using Patch motion correction. CTF (Contrast Transfer Function) parameters were determined using Patch CTF. About 500 particles were manually selected and 2D classified to create templates. Automated particle picking was performed using these templates. Picking was manually inspected, and a threshold that excludes low signal-to-noise and high signal-to-dirt false positive picks and minimizes the false negative picks was chosen. The remaining particles were extracted with a box size between 440 and 512 and subjected to 2D classification. Classes that showed clear ribosomal densities were selected and subjected to heterogeneous abinitio model generation. Subsequently, the remaining particles were 3D refined to generate a consensus model. Afterwards, optics groups were assigned to the individual particles depending on the applied beam shift during recording. Global and local CTF refinements were then performed, and all particles were refined using non-uniform refinement. To generate masks for local refinements, the density was filtered to 20 Å, and sub-densities of the entire 40S subunit and the head region were generated using the Volume eraser tool in UCSF Chimera 1.13.177. These densities were binarized, extended by 7 pixels and another 7 pixels as soft edge. Locally refined maps using the corresponding masks were obtained in Cryosparc, and composite maps were generated with the vop maximum command in UCSF Chimera 1.13.177.
To analyze particle dynamics and heterogeneity, the consensus refined particles were subjected to CryoDRGN analysis48. Due to the large pixel box size of our data sets, the dimensions of the ribosome particles were reduced to allow for feasible computation. The particles were down-sampled to a box size of 128 or 256, and two iterative rounds of the CryoDRGN VAE were trained using a network with 3 decoder and 3 encoder layers with 1024 as outer dimensions and a bottleneck latent dimension of 8. The resulting z values were classified using kmeans clustering and gaussian mixture models and plotted using a UMAP approach. Particles belonging to class I (containing all four factors) from the Xenopus egg ribosome dataset were selected from the originally refined particles in CryoDRGN’s Jupyter notebook and subjected to an additional round of 3D refinement. Only particles located at the center of the cluster were taken in order to obtain a map as free from heterogeneity as possible. The final maps were polished with deepEMhancer78.
Model building and refinement
The molecular models of the 1 hpf and 6 hpf ribosomes from zebrafish were built using PDB-4UG066 as an initial model for core ribosomal proteins and rRNA. In the case of the 1 hpf zebrafish ribosome, additional factors were modeled from previously published structures: eIF5A from PDB-5DAT, and eEF2 and Habp4 (SERBP1 paralog) from PDB-6MTE18. All chains were first rigid body-fitted in Coot 0.8.979 and manually mutated to the zebrafish sequences, taking MS data from purified zebrafish ribosomes (protein identity and abundance) into consideration. In the case of Habp4, another protein (Serbp1a, Uniprot ID F1Q5Q3) could also fit into the 1 hpf ribosome density. Given that Habp4 was more abundant than Serbp1a in 1 hpf zebrafish ribosomes according to MS, Habp4 was modeled.
The dormant ribosome structure from Xenopus eggs was built based on the 60S subunit of the 1 hpf zebrafish ribosome, and the 40S subunit of the 6 hpf zebrafish ribosome, given that both 40S subunits showed the same swiveled conformation. Protein and rRNA molecules from zebrafish were mutated to the corresponding sequences in Xenopus, taking into consideration the MS data from purified Xenopus ribosomes. Dapl1.S was built de novo in Coot and then used as a template to build Dap1b in the zebrafish 1 hpf ribosome. Although two Dap proteins (Dapl1.S and Dapl1.L in Xenopus, and Dap1b and Dap in zebrafish) could fit into the corresponding densities, Dapl1.S and Dap1b were modeled according to gene expression data (Extended Data Fig. 5b-d) and protein abundance (measured by MS; Fig. 1f and Extended Data Fig. 1a).
The rabbit ribosome model with recombinant zebrafish Dap1b was built using 6MTE18 as an initial model for core ribosomal proteins, rRNA, eEF2 and SERBP1. eIF5A was modeled using PDB-5DAT as reference. All models were manually adjusted to fit the observed density in Coot 0.8.979 and real-space-refined using Phenix 1.17.180. The 40S subunit of the rabbit ribosome was only submitted to rigid body fitting in Phenix. Working files were saved in PDB and the final models were converted to mmCIF using PDB_extract81. Cryo-EM densities and models were visualized using UCSF ChimeraX version 0.9173. Structure alignments were performed using the command mmaker in ChimeraX.
Reporting summary
Further information on research design, statistics and reproducibility is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. Mass-spectrometry and cryo-EM analysis of Xenopus egg ribosomes.
a, Fold change of ribosome-associated factors and core ribosomal proteins after comparing mass spectrometry data of purified ribosomes from unfertilized Xenopus eggs and 24 hpf larvae (stage 14) (n = 1 experiment). b, Processing pipeline of the Xenopus egg ribosome. All steps were done in Cryosparc v3.2.0. Maps are shown in grey, masks in blue. c-d, Maps showing the local resolution of Map1 and Map2 (c), and of the four factors associated with Xenopus egg ribosomes (d). Note that the resolution of the large subunit in Map2 (in c) is 0 (shown in blue) since this region was outside the mask used for obtaining this map (see b). e, Orientation distribution plot for all particles contributing to Map1. f, Gold-Standard Fourier Shell Correlation (GSFSC) of Map1.
Extended Data Fig. 2. Processing pipeline of the 1 hpf zebrafish ribosome.
All steps were done in Cryosparc v3.2.0. Maps are shown in grey, masks in blue. The orientation distribution plot for all particles contributing to Map1 and the Gold-Standard Fourier Shell Correlation (GSFSC) of the respective map is shown on the bottom. Local resolution maps were calculated for Map1, Map2, Map3, and for the four ribosome-associated factors. Note that in the box with local resolution maps, the resolution of the large subunit in Map2 and Map3, and of the small subunit’s head in Map3 is 0 (shown in blue) since these regions were outside the masks used for generating these maps.
Extended Data Fig. 3. Characterization of the dormant ribosome state in zebrafish 1 hpf and Xenopus egg ribosomes.
a-d, Densities of the two modules, namely Habp4-eEF2b/eEF2 (a-b), and Dap1b/Dapl1-eIF5a (c-d), that are characteristic for dormant ribosomes in zebrafish 1 hpf embryos and Xenopus eggs. e, Overview of the ribosome structure isolated from 6 hpf zebrafish embryos lacking the specific egg ribosome-associated factors. f, Latent space representations of ribosomal particles from Xenopus eggs (left), 1 hpf zebrafish embryos (middle) and 6 hpf zebrafish embryos (right) as UMAP embeddings after training a cryoDRGN latent variable model. Classes are depicted with circles in Roman numbers, map volumes are indicated with Arabic numbers. Total particle numbers are shown on the top left of each graph. g, Densities of ribosome-associated factors from Xenopus eggs. An overview of the map is shown on the top left. h, Densities of ribosome-associated factors in a map reconstructed from class I particles from Xenopus eggs obtained with cryoDRGN. An overview of the map is shown on the top-left.
Extended Data Fig. 4. Processing pipeline of the 6 hpf zebrafish ribosome.
All steps were done in Cryosparc v3.2.0. Maps are shown in grey, masks in blue. The orientation distribution plot for all particles contributing to Map1 and the Gold-Standard Fourier Shell Correlation (GSFSC) of the respective map is shown on the bottom-right. Local resolution maps were calculated for Map1, Map2 and Map3. Note that in the box with local resolution maps, the resolution of the large subunit in Map2 and Map3, and of the small subunit’s body in Map3 is 0 (shown in blue) since these regions were outside the masks used for generating these maps.
Extended Data Fig. 5. Sequence conservation of the Dap/Dap1b/Dapl1 protein family and RNA expression of ribosome-associated factors.
a, Protein sequence alignment of the Dap/Dap1b/Dapl1 protein family illustrates conserved motifs. Vertebrates have two paralogs, namely Dap1b/Dapl1 and Dap. Invertebrates only encode one homolog (Dap1) that clusters in between Dap1b/Dapl1 and Dap proteins. b, Zebrafish mRNA expression levels (PolyA+ RNA-seq31,83) of eif5a/eif5a2 (purple), eef2 (orange), dap1b/dap (green) and habp4 (blue) during oogenesis and embryogenesis. c, Xenopus mRNA expression levels of all paralogs of eif5a, eef2, dap, dapl1 and habp4 derived from riboMinus-seq data84. d, mRNA expression levels of zebrafish eif5a/eif5a2, eef2, dap1b/dap and habp4 in adult tissues85. TPM, transcripts per million.
Extended Data Fig. 6. Structural comparison of Dap1b and other factors that insert into the polypeptide exit tunnel (PET).
a, Structures of ribosomes with proteins and peptides inserted into the PET. From left to right: zebrafish Dap1b inserted into the rabbit ribosome, Bac7 (5HAU86), Api137 (5O2R87), Rei1 (6RZZ88), Nog1 (3JCT89) and MDF2 (6RM317). Models were clipped to have a better view of the PET. Boxed areas (dashed boxes) are shown at higher magnification in b. b, Detail of the peptidyl-transferase center (PTC) of the ribosomes shown in a. The dashed line indicates the position of the PTC. All previously known factors use different mechanisms than Dap1b to achieve their functions: Bac7 interacts with the ribosomal A-loop via its N-terminus (see below) to block translation initiation, Api137 interacts with the release factors RF1 or RF2 to block termination of bacterial ribosomes, Rei1 and Nog1 insert into the PET of 60S subunits during ribosome biogenesis and their C-termini do not extend beyond the PTC, and MDF2’s C-terminus interferes with P-tRNA and eIF5A binding to establish dormancy in the microsporidian ribosome. c, Superimposition of Dap1b (left) and Bac7 (right) with eukaryotic (5GAK34) and prokaryotic (1VY490) A- and P-tRNAs, respectively. Red asterisks denote clashes of Dap1b and Bac7 with the A-tRNA. d, Dap1b’s C-terminus does not interact with the A-loop (right), in contrast to Bac7 (left). Dashed lines mark distances between Arg1 of Bac7 and Phe109 of Dap1b with a conserved uracil of the A-loop. e, Scheme of the interactions of Dap1b’s C-terminus within the ribosome. Dap1b interacts with helix 74 (H74) and H90 of the 28S rRNA. f, Scheme of the interactions of Bac7’s N-terminus within the ribosome. Bac7 interacts with H89 and the A-loop (H92).
Extended Data Fig. 7. Crosslinking and mass spectrometry (MS) analysis of ribosomes from 1 hpf zebrafish embryos and Xenopus eggs.
a, Cα-Cα distance distribution of the DSSO-induced crosslinks identified in Xenopus egg and zebrafish 1 hpf ribosomes. b-c, Proteins crosslinked to zebrafish Habp4 (b) or Dap (c) are shown on the 1 hpf zebrafish ribosome as surface representations, with crosslinked residues depicted in darker color. d, Crosslinking mapping of Habp4 (shown as a scheme; modeled regions highlighted with a black line) to proteins of the zebrafish embryo (top) and Xenopus egg (bottom) ribosome. Crosslinked proteins are shown as surface representations, with crosslinked residues depicted in dark blue. A cartoon (right) shows a model of Habp4 on the ribosome. e, Crosslinking mapping of zebrafish Dap (top) and Xenopus Dapl1 (bottom; for details, see d).
Extended Data Fig. 8. Characterization of single (habp4-/-), double, and triple (dap-/-, dap1b-/-, habp4-/-) zebrafish mutants.
a-b, Early embryo development of habp4-/- (a) and dap-/ -, dap1b-/- mutants (b) compared to wildtype (WT). c, Mendelian ratio analysis of fin-clips from adult fish obtained from heterozygous habp4+/- parents (25% expected to be habp4-/-; n = 4 for both genotypes). d, Size of WT and habp4-/- embryos at 6 hours post-fertilization (hpf) (WT-1: n = 31; WT-2: n = 22; WT-3: n = 35; habp4-/--1: n = 36; habp4-/--2: n = 24; habp4-/--3: n = 33). e, Total RNA of 1-3 hpf embryos (WT: n = 42; dap-/-, dap1b-/-: n = 41; WT and triple KO: n = 20 for both genotypes). f, Number of eggs laid by single, double, and triple KO females compared to matching WT. g, Representative images (left) and quantification (right) of poor quality eggs. h, Percentage of embryos from single, double, and triple KO mutant pairs displaying normal embryo development until 6 hpf compared to matching WT pairs. i-j, Relative number of embryos and larvae (in relation to the embryos that developed normally up to 6 hpf; see h) that showed abnormalities at 1 (i) and 4 (j) days post-fertilization (dpf). Example images are shown on the left. Data in c and e-j are represented as scatter dot plots with means ± standard deviation (SD). Data in d are represented as a violin plot with median and quartiles. In f-h, dotted vertical lines indicate separate experiments. Significance was determined using Kruskal-Wallis and Dunn’s two-sided test (d, f-j; for more than 2 sample group comparisons) or Mann-Whitney test (c, and in e-h for pairwise comparisons between habp4-/- or dap-/-, dap1b-/- versus WT). For c and f, n are independent crosses; for d, n are individual embryos; for e, n are biologically independent samples. #, number of crosses.
Extended Data Fig. 9. Recombinant Dap1b binds to the polypeptide exit tunnel of rabbit ribosomes.
a-b, Western blot of the total in vitro translation reaction shown in Fig. 4c (a) and Fig. 4e (b). Uncropped images of membranes are provided in Supplementary Fig. 2a, b. c, Translation activity assays (Fig. 4a top) of renilla luciferase mRNA upon addition of increasing concentrations of C-terminal Dap and Dap1b peptides. BSA and N-terminal Bac7 are used as negative and positive controls, respectively (n = 4 biologically independent samples). Dots represent means and error bars are standard deviation (SD). d, Ribosome binding assays (Fig. 4a bottom) of in vitro translated FLAG-tagged Dap-Dap1b chimeras compared to full-length (WT) Dap and Dap1b. A representative Western blot from a single experiment is shown on the left; quantification of three independent experiments is shown on the right. Data are represented as scatter dot plots with means ± standard deviation (SD. Significance was assessed with Kruskal-Wallis followed by Dunn’s two-sided test. Uncropped images of membranes are provided in Supplementary Fig. 2d. e, Mass spectrometry data of ribosomes isolated from habp4-/- and WT (left), and from dap-/-, dap1b-/- and WT (right) embryos at 1 hpf, represented as volcano plots (n = 3 independent experiments). Permutation-based false discovery rates (FDRs) are displayed as dotted (FDR < 0.01) and dashed (FDR < 0.05) lines.
Extended Data Fig. 10. Processing pipeline of the rabbit ribosome with recombinant zebrafish Dap1b.
a, Processing pipeline for obtaining an 80S density map of the rabbit ribosome with zebrafish Dap1b. b, Orientation distribution plot (top) and Gold-Standard Fourier Shell Correlation (GSFSC; bottom) of Map1. c, Local resolution maps calculated for Map2 and Map3. Densities and local resolutions of eIF5A, Dap1b, eEF2 and SERBP1 are shown on the bottom. Note that the resolution of the small subunit in Map2 is 0 (shown in blue) since this region was outside the mask used for generating this map. d, Latent space representation of particles from rabbit ribosomes with zebrafish Dap1b as a UMAP embedding after training a cryoDRGN latent variable model. Classes are depicted with circles in Roman numbers, map volumes are indicated with Arabic numbers. Total particle number is shown on the top left of the graph.
Supplementary Material
Acknowledgements
We thank L.E. Cabrera-Quio and M. Novatchkova for ribosome profiling data analysis; K.R. Gert for providing non-activated zebrafish eggs; the Mass Spectrometry Facility at the Vienna BioCenter Core Facilities GmbH (VBCF), in particular S. Opravil for the processing of our samples, and R. Imre and G. Dürnberger for their help with data analysis; the VBCF Protein Technologies facility, in particular David Drechsel and A. Lehner, for the expression and purification of recombinant Dap1b and Dap proteins; M. Madalinski for synthesizing the Bac7, Dap(71-105) and Dap1b(76-109) peptides; A. Schleiffer for his help in the phylogenetic analysis of Dap and Dap1b/Dapl1; the VBCF Electron Microscopy Facility for the support and maintenance of the facilities; H. Stark (MPI BPC) for providing additional Krios measurement time; the IMP animal facility personnel, in particular F. Ecker, J. König and D. Sunjic for their excellent care of fish and Xenopus; M. Binner, J. Kiraly, T. Mylenko and A. Bandura for their help with genotyping; L. Leiendecker and the entire Obenauf lab for discussions and sharing reagents; E. Tanaka and T. Clausen for funding T.-Y.L. and A.M., respectively; the entire Pauli group for valuable discussions on the project; the VBC RNA Salon and the RNA Deco-SFB community for providing useful feedback and suggestions; A. Andersen (Life Science Editors), C. Plaschka, A. Stark, J. Brennecke, A. Carter, F. Peske, U. Hohmann, E. Calo, A. Shah and A. Blaha for critical reading and valuable feedback on the manuscript. This work was supported by the Institute of Molecular Pathology (IMP), which receives institutional funding from Boehringer Ingelheim and the Austrian Research Promotion Agency (Headquarter grant FFG-852936), and funding from the FWF START program (Y 1031-B28 to A.P.), the Human Frontier Science Program (HFSP) Career Development Award (CDA00066/2015 to A.P.), the SFB RNA-Deco (project number F 80 to A.P.) and EMBO-YIP funds (to A.P.). L.L.-O. was supported by an SNF Early Postdoc Mobility fellowship (P2GEP3_191204), an EMBO long-term fellowship (ALTF 1165-2019) and an MSCA-IF-EF-SE (890218). A.C. was supported by an MSCA-IF-EF-SE and a VIP2 postdoctoral fellowship. We acknowledge the cryo-electron microscopy facility CEITEC MU of CIISB, Instruct-CZ Centre, supported by MEYS CR (LM2018127), and Diamond Light Source for granting us access and support at the cryo-EM facilities at the UK’s national Electron Bio-imaging Centre (eBIC) under proposal EM BI25222, funded by the Wellcome Trust, MRC and BBSRC. For the purpose of Open Access, the authors have applied a CC BY public copyright license to any Author Accepted Manuscript (AAM) version arising from this submission.
Footnotes
Author contributions. F.L., L.L.-O., D.H. and A.P. conceived the study; F.L. performed most experiments with the help of C.P., further supported by J. R. and A. C.; L.L.-O. obtained the final cryo-EM maps and modeled all ribosomes with contributions of F.L., A.M. and D.H. and help from S.K.; I.G. prepared and screened grids, collected EM data, and together with F.L. and D.H., contributed to the initial cryo-EM data processing with help from K.B.; M.M. and E.R. performed and analyzed the crosslinking sample preparation, and K.M. supervised mass-spectrometry experiments and analyzed the crosslinking samples; T.-Y.L. provided Xenopus eggs and 24 hpf embryos; D.H. performed the cryoDRGN analysis and F.L. and L.L-O. compared the resulting maps; A.P. and D.H. supervised the project; L.L.-O., F.L., and A.P. wrote the manuscript with input from all authors.
Competing interests. The authors declare no competing interests.
Data availability
Cryo-EM maps and molecular models generated in this study have been deposited in the Protein Data Bank (PDB) with IDs 7OYA (zebrafish 1 hpf), 7OYB (zebrafish 6 hpf), 7OYC (Xenopus eggs) and 7OYD (rabbit ribosome with zebrafish Dap1b), and in the Electron Microscopy Data Bank with accession codes EMD-13111 (zebrafish 1 hpf), EMD-13112 (zebrafish 6 hpf), EMD-13113 (Xenopus egg ribosome) and EMD-13114 (rabbit ribosome with zebrafish Dap1b). Raw micrographs, particle stacks and CryoDRGN models were deposited at the EMPIAR Database (EMPIAR-11274). Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE82 partner repository with the dataset identifier PXD026866. Source data are provided with this paper.
Previously published structures used in this study are PDB-1VY4, PDB-3JCT, PDB-4UG0, PDB-5DAT, PDB-5GAK, PDB-5HAU, PDB-5O2R, PDB-6HCF, PDB-6MTE, PDB-6OLE, PDB-6SGC, PDB-6RM3 and PDB-6RZZ. The ribosome profiling data and RNA-seq data were published previously and are accessible at Gene Expression Omnibus (GEO) with accession numbers GSE4651230 (ribosome profiling), GSE3290031 (subseries GSE32898) and GSE14711283 (RNA-seq).
Code availability
No custom codes were used for this study.
References
- 1.Woodland HR. Changes in the polysome content of developing Xenopus laevis embryos. Developmental Biology. 1974;40:90–101. doi: 10.1016/0012-1606(74)90111-0. [DOI] [PubMed] [Google Scholar]
- 2.Brandis JW, Raff RA. Translation of oogenetic mRNA in sea urchin eggs and early embryos. Demonstration of a change in translational efficiency following fertilization. Dev Biol. 1978;67:99–113. doi: 10.1016/0012-1606(78)90303-2. [DOI] [PubMed] [Google Scholar]
- 3.Kronja I, et al. Widespread Changes in the Posttranscriptional Landscape at the Drosophila Oocyte-to-Embryo Transition. Cell Reports. 2014;7:1495–1508. doi: 10.1016/j.celrep.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachvarova R, De Leon V. Stored and polysomal ribosomes of mouse ova. Developmental Biology. 1977;58:248–254. doi: 10.1016/0012-1606(77)90090-2. [DOI] [PubMed] [Google Scholar]
- 5.Burkholder GD, Comings DE, Okada TA. A storage form of ribosomes in mouse oocytes. Experimental Cell Research. 1971;69:361–371. doi: 10.1016/0014-4827(71)90236-9. [DOI] [PubMed] [Google Scholar]
- 6.Alberts B, et al. Molecular Biology of the Cell. 4th edition. 2002. Eggs. [Google Scholar]
- 7.Locati MD, et al. Linking maternal and somatic 5S rRNA types with different sequence-specific non-LTR retrotransposons. RNA. 2017;23:446–456. doi: 10.1261/rna.059642.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Locati MD, et al. Expression of distinct maternal and somatic 5.8S, 18S, and 28S rRNA types during zebrafish development. RNA. 2017;23:1188–1199. doi: 10.1261/rna.061515.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cenik ES, et al. Maternal Ribosomes Are Sufficient for Tissue Diversification during Embryonic Development in C. elegans. Developmental Cell. 2019;48:811–826.:e6. doi: 10.1016/j.devcel.2019.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danilchik MV, Hille MB. Sea urchin egg and embryo ribosomes: differences in translational activity in a cell-free system. Dev Biol. 1981;84:291–298. doi: 10.1016/0012-1606(81)90397-3. [DOI] [PubMed] [Google Scholar]
- 11.Chassé H, Boulben S, Cormier P, Morales J. Translational Control of Canonical and Non-Canonical Translation Initiation Factors at the Sea Urchin Egg to Embryo Transition. International Journal of Molecular Sciences. 2019;20:626. doi: 10.3390/ijms20030626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stebbins-Boaz B, Cao Q, de Moor CH, , Mendez R, Richter JD. Maskin Is a CPEB-Associated Factor that Transiently Interacts with eIF-4E. Molecular Cell. 1999;4:1017–1027. doi: 10.1016/s1097-2765(00)80230-0. [DOI] [PubMed] [Google Scholar]
- 14.Smith PR, Pandit SC, Loerch S, Campbell ZT. The space between notes: emerging roles for translationally silent ribosomes. Trends in Biochemical Sciences. 2022 doi: 10.1016/j.tibs.2022.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beckert B, et al. Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1. Nat Microbiol. 2018;3:1115–1121. doi: 10.1038/s41564-018-0237-0. [DOI] [PubMed] [Google Scholar]
- 16.Beckert B, et al. Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. EMBO J. 2017;36:2061–2072. doi: 10.15252/embj.201696189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barandun J, Hunziker M, Vossbrinck CR, Klinge S. Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome. Nat Microbiol. 2019;4:1798–1804. doi: 10.1038/s41564-019-0514-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown A, Baird MR, Yip MC, Murray J, Shao S. Structures of translationally inactive mammalian ribosomes. eLife. 2018;7:e40486. doi: 10.7554/eLife.40486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Dyke N, Baby J, Van Dyke MW. Stm1p, a Ribosome-associated Protein, is Important for Protein Synthesis in Saccharomyces cerevisiae under Nutritional Stress Conditions. Journal of Molecular Biology. 2006;358:1023–1031. doi: 10.1016/j.jmb.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Smith PR, et al. Functionally distinct roles for eEF2K in the control of ribosome availability and p-body abundance. Nat Commun. 2021;12:6789. doi: 10.1038/s41467-021-27160-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shetty S, Hofstetter J, Battaglioni S, Ritz D, Hall MN. TORC1 phosphorylates and inhibits the ribosome preservation factor Stm1 to activate dormant ribosomes. 2022:2022.08.08.503151. doi: 10.1101/2022.08.08.503151. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wells JN, et al. Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes. PLOS Biology. 2020;18:e3000780. doi: 10.1371/journal.pbio.3000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seefeldt AC, et al. Structure of the mammalian antimicrobial peptide Bac7(1–16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Res. 2016;44:2429–2438. doi: 10.1093/nar/gkv1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casteels P, Ampe C, Jacobs F, Vaeck M, Tempst P. Apidaecins: antibacterial peptides from honeybees. EMBO J. 1989;8:2387–2391. doi: 10.1002/j.1460-2075.1989.tb08368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krizsan A, Prahl C, Goldbach T, Knappe D, Hoffmann R. Short Proline-Rich Antimicrobial Peptides Inhibit Either the Bacterial 70S Ribosome or the Assembly of its Large 50S Subunit. Chembiochem. 2015;16:2304–2308. doi: 10.1002/cbic.201500375. [DOI] [PubMed] [Google Scholar]
- 26.Metafora S, Felicetti L, Gambino R. The Mechanism of Protein Synthesis Activation After Fertilization of Sea Urchin Eggs. PNAS. 1971;68:600–604. doi: 10.1073/pnas.68.3.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gambino R, Metafora S, Felicetti L, Raisman J. Properties of the ribosomal salt wash from unfertilized and fertilized sea urchin eggs and its effect on natural mRNA translation. Biochimica et Biophysica Acta (BBA) - Nucleic Acids and Protein Synthesis. 1973;312:377–391. doi: 10.1016/0005-2787(73)90382-1. [DOI] [PubMed] [Google Scholar]
- 28.Hille MB. Inhibitor of protein synthesis isolated from ribosomes of unfertilised eggs and embryos of sea urchins. Nature. 1974;249:556–558. doi: 10.1038/249556a0. [DOI] [PubMed] [Google Scholar]
- 29.Chassé H, Boulben S, Costache V, Cormier P, Morales J. Analysis of translation using polysome profiling. Nucleic Acids Research. 2017;45:e15. doi: 10.1093/nar/gkw907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chew G-L, et al. Ribosome profiling reveals resemblance between long non-coding RNAs and 5 leaders of coding RNAs. Development. 2013;140:2828–2834. doi: 10.1242/dev.098343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pauli A, et al. Toddler: An Embryonic Signal That Promotes Cell Movement via Apelin Receptors. Science. 2014;343 doi: 10.1126/science.1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutierrez E, et al. eIF5A Promotes Translation of Polyproline Motifs. Molecular Cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuller AP, Wu CC-C, Dever TE, Buskirk AR, Green R. eIF5A Functions Globally in Translation Elongation and Termination. Mol Cell. 2017;66:194–205.:e5. doi: 10.1016/j.molcel.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt C, et al. Structure of the hypusinylated eukaryotic translation factor eIF-5A bound to the ribosome. Nucleic Acids Research. 2016;44:1944–1951. doi: 10.1093/nar/gkv1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 36.Flis J, et al. tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis. Cell Reports. 2018;25:2676–2688.:e7. doi: 10.1016/j.celrep.2018.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayashi H, et al. Tight interaction of eEF2 in the presence of Stm1 on ribosome. J Biochem. 2018;163:177–185. doi: 10.1093/jb/mvx070. [DOI] [PubMed] [Google Scholar]
- 38.Anger AM, et al. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 39.Sun L, Ryan DG, Zhou M, Sun T-T, Lavker RM. EEDA: a protein associated with an early stage of stratified epithelial differentiation. J Cell Physiol. 2006;206:103–111. doi: 10.1002/jcp.20433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma X, et al. Regulation of cell proliferation in the retinal pigment epithelium: Differential regulation of the death-associated protein like-1 DAPL1 by alternative MITF splice forms. Pigment Cell & Melanoma Research. 2018;31:411–422. doi: 10.1111/pcmr.12676. [DOI] [PubMed] [Google Scholar]
- 41.Ma X, et al. DAPL1, a susceptibility locus for age-related macular degeneration, acts as a novel suppressor of cell proliferation in the retinal pigment epithelium. Hum Mol Genet. 2017;26:1612–1621. doi: 10.1093/hmg/ddx063. [DOI] [PubMed] [Google Scholar]
- 42.Deiss LP, Feinstein E, Berissi H, Cohen O, Kimchi A. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 1995;9:15–30. doi: 10.1101/gad.9.1.15. [DOI] [PubMed] [Google Scholar]
- 43.Koren I, Reem E, Kimchi A. DAP1, a Novel Substrate of mTOR, Negatively Regulates Autophagy. Current Biology. 2010;20:1093–1098. doi: 10.1016/j.cub.2010.04.041. [DOI] [PubMed] [Google Scholar]
- 44.Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park MH, Nishimura K, Zanelli CF, Valentini SR. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010;38:491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greber BJ, Boehringer D, Montellese C, Ban N. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat Struct Mol Biol. 2012;19:1228–1233. doi: 10.1038/nsmb.2425. [DOI] [PubMed] [Google Scholar]
- 47.Klingauf-Nerurkar P, et al. The GTPase Nog1 co-ordinates the assembly, maturation and quality control of distant ribosomal functional centers. eLife. 2020;9:e52474. doi: 10.7554/eLife.52474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong ED, Bepler T, Berger B, Davis JH. CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Nat Methods. 2021;18:176–185. doi: 10.1038/s41592-020-01049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rossi D, et al. Evidence for a Negative Cooperativity between eIF5A and eEF2 on Binding to the Ribosome. PLoS One. 2016;11:e0154205. doi: 10.1371/journal.pone.0154205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kao A, et al. Development of a Novel Cross-linking Strategy for Fast and Accurate Identification of Cross-linked Peptides of Protein Complexes. Mol Cell Proteomics. 2011;10:M110.002212. doi: 10.1074/mcp.M110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balagopal V, Parker R. Stm1 modulates translation after 80S formation in Saccharomyces cerevisiae. RNA. 2011;17:835–842. doi: 10.1261/rna.2677311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blobel G, Potter VR. Studies on free and membrane-bound ribosomes in rat liver: I. Distribution as related to total cellular RNA. Journal of Molecular Biology. 1967;26:279–292. doi: 10.1016/0022-2836(67)90297-5. [DOI] [PubMed] [Google Scholar]
- 53.Marygold SJ, et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007;8:R216. doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fortier S, MacRae T, Bilodeau M, Sargeant T, Sauvageau G. Haploinsufficiency screen highlights two distinct groups of ribosomal protein genes essential for embryonic stem cell fate. PNAS. 2015;112:2127–2132. doi: 10.1073/pnas.1418845112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amsterdam A, et al. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vecchi G, et al. Proteome-wide observation of the phenomenon of life on the edge of solubility. PNAS. 2020;117:1015–1020. doi: 10.1073/pnas.1910444117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y, et al. Autophagy-dependent ribosomal RNA degradation is essential for maintaining nucleotide homeostasis during C. elegans development. eLife. 2018;7:e36588. doi: 10.7554/eLife.36588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cohen J. Statistical Power Analysis for the Behavioral Sciences. Routledge; 1988. [DOI] [Google Scholar]
- 59.Juszkiewicz S, et al. ZNF598 Is a Quality Control Sensor of Collided Ribosomes. Molecular Cell. 2018;72:469–481.:e7. doi: 10.1016/j.molcel.2018.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li W, et al. Structural basis for selective stalling of human ribosome nascent chain complexes by a drug-like molecule. Nat Struct Mol Biol. 2019;26:501–509. doi: 10.1038/s41594-019-0236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chandrasekaran V, et al. Mechanism of ribosome stalling during translation of a poly(A) tail. Nat Struct Mol Biol. 2019;26:1132–1140. doi: 10.1038/s41594-019-0331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gagnon JA, et al. Efficient Mutagenesis by Cas9 Protein-Mediated Oligonucleotide Insertion and Large-Scale Assessment of Single-Guide RNAs. PLOS ONE. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 64.Nair S, Lindeman RE, Pelegri F. In vitro oocyte culture-based manipulation of zebrafish maternal genes. Dev Dyn. 2013;242:44–52. doi: 10.1002/dvdy.23894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sive HL, Grainger RM, Harland RM. Early Development of Xenopus laevis. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 66.Khatter H, et al. Purification, characterization and crystallization of the human 80S ribosome. Nucleic Acids Res. 2014;42:e49. doi: 10.1093/nar/gkt1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dorfer V, et al. MS Amanda, a Universal Identification Algorithm Optimized for High Accuracy Tandem Mass Spectra. J Proteome Res. 2014;13:3679–3684. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 69.Taus T, et al. Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res. 2011;10:5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- 70.Doblmann J, et al. apQuant: Accurate Label-Free Quantification by Quality Filtering. J Proteome Res. 2019;18:535–541. doi: 10.1021/acs.jproteome.8b00113. [DOI] [PubMed] [Google Scholar]
- 71.Smyth GK. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Springer; 2005. limma: Linear Models for Microarray Data; pp. 397–420. [DOI] [Google Scholar]
- 72.Pirklbauer GJ, et al. MS Annika: A New Cross-Linking Search Engine. J Proteome Res. 2021;20:2560–2569. doi: 10.1021/acs.jproteome.0c01000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goddard TD, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sharma A, Mariappan M, Appathurai S, Hegde RS. In vitro dissection of protein translocation into the mammalian endoplasmic reticulum. Methods Mol Biol. 2010;619:339–363. doi: 10.1007/978-1-60327-412-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng Q, Shao S. In vitro reconstitution of translational arrest pathways. Methods. 2018;137:20–36. doi: 10.1016/j.ymeth.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 76.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 77.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 78.Sanchez-Garcia R, et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun Biol. 2021;4:1–8. doi: 10.1038/s42003-021-02399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 80.Liebschner D, et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang H, et al. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2004;60:1833–1839. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- 82.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cabrera-Quio LE, Schleiffer A, Mechtler K, Pauli A. Zebrafish Ski7 tunes RNA levels during the oocyteto-embryo transition. PLOS Genetics. 2021;17:e1009390. doi: 10.1371/journal.pgen.1009390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Session AM, et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:336–343. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fujihara Y, et al. The conserved fertility factor SPACA4/Bouncer has divergent modes of action in vertebrate fertilization. Proceedings of the National Academy of Sciences. 2021;118:e2108777118. doi: 10.1073/pnas.2108777118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gagnon MG, et al. Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition. Nucleic Acids Res. 2016;44:2439–2450. doi: 10.1093/nar/gkw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Florin T, et al. An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome. Nat Struct Mol Biol. 2017;24:752–757. doi: 10.1038/nsmb.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kargas V, et al. Mechanism of completion of peptidyltransferase centre assembly in eukaryotes. eLife. 2019;8:e44904. doi: 10.7554/eLife.44904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu S, et al. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature. 2016;534:133–137. doi: 10.1038/nature17942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Polikanov YS, Steitz TA, Innis CA. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat Struct Mol Biol. 2014;21:787–793. doi: 10.1038/nsmb.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps and molecular models generated in this study have been deposited in the Protein Data Bank (PDB) with IDs 7OYA (zebrafish 1 hpf), 7OYB (zebrafish 6 hpf), 7OYC (Xenopus eggs) and 7OYD (rabbit ribosome with zebrafish Dap1b), and in the Electron Microscopy Data Bank with accession codes EMD-13111 (zebrafish 1 hpf), EMD-13112 (zebrafish 6 hpf), EMD-13113 (Xenopus egg ribosome) and EMD-13114 (rabbit ribosome with zebrafish Dap1b). Raw micrographs, particle stacks and CryoDRGN models were deposited at the EMPIAR Database (EMPIAR-11274). Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE82 partner repository with the dataset identifier PXD026866. Source data are provided with this paper.
Previously published structures used in this study are PDB-1VY4, PDB-3JCT, PDB-4UG0, PDB-5DAT, PDB-5GAK, PDB-5HAU, PDB-5O2R, PDB-6HCF, PDB-6MTE, PDB-6OLE, PDB-6SGC, PDB-6RM3 and PDB-6RZZ. The ribosome profiling data and RNA-seq data were published previously and are accessible at Gene Expression Omnibus (GEO) with accession numbers GSE4651230 (ribosome profiling), GSE3290031 (subseries GSE32898) and GSE14711283 (RNA-seq).
No custom codes were used for this study.