Abstract
Plasmacytoid dendritic cells (pDCs) are an immune subset devoted to the production of high amounts of type 1 interferons in response to viral infections. Whereas conventional dendritic cells (cDCs) originate mostly from a common dendritic cell progenitor (CDP), pDCs have been shown to develop from both CDPs and common lymphoid progenitors. Here, we found that pDCs developed predominantly from IL-7R+ lymphoid progenitor cells. Expression of SiglecH and Ly6D defined pDC lineage commitment along the lymphoid branch. Transcriptional characterization of SiglecH+Ly6D+ precursors indicated that pDC development requires high expression of the transcription factor IRF8, whereas pDC identity relies on TCF4. RNA sequencing of IL-7R+ lymphoid and CDP-derived pDCs mirrored the heterogeneity of mature pDCs observed in single-cell analysis. Both mature pDC subsets are able to secrete type 1 interferons, but only myeloid-derived pDCs share with cDCs their ability to process and present antigen.
Dendritic cells (DCs) are a specialized immune subset dedicated to sensing pathogens and inducing the appropriate immune response1. Under steady-state conditions, DCs can be subdivided into cDCs and pDCs2–5. cDCs are specialized in antigen uptake and presentation to naïve T cells and can be further subdivided into cDC1 and cDC2, expressing the transcription factors IRF8 and IRF4, respectively6–8. pDCs are a distinct lineage dedicated to the production of high amounts of type 1 interferons in response to viral infections9–11. Development of DCs occurs in the bone marrow (BM) and requires a complex transcriptional network, in which progressive lineage specification gradually and hierarchically limits and excludes alternative fates4,8. A CDP able to give rise to both cDCs and pDCs has been described in the BM12,13. Furthermore, cDCs and pDCs share not only their dependency on the cytokine FLT3L but also the expression of several transcription factors, thus suggesting common regulatory networks14–16.
Immediate precursors with exclusive differentiation potential have been identified for both cDC subsets17. Although the molecular mechanism defining cDC1 lineage specification has been dissected17, cDC2 commitment is still unclear4. Multiple pathways appear to converge into a phenotypically homogenous but transcriptionally heterogeneous cDC2 lineage4,5,18,19. In addition, pDC development seems to be ‘promiscuous’, because both CDPs and common lymphoid progenitors can give rise to pDCs13,16,20,21. Two distinct pDC progenitors have been characterized22,23. Within the BM, CCR9–MHC-IIlo pDCs have been characterized as the immediate precursors of CCR9+ mature pDCs22,23, although these pDC precursors already express mature markers and appear functional. pDCs have also been reported to arise mostly from CD135+CD115– CD127– precursor cells. Despite their greater pDC potential than CDPs, these progenitors maintain the ability to generate cDCs, thus suggesting that they are either heterogeneous or still uncommitted in nature22–24.
Molecularly, pDC development and identity depend on the expression of the transcription factor TCF4 (also referred to as E2-2)25,26. TCF4 deficiency is prenatally lethal, and haploinsufficiency in humans results in Pitt–Hopkins syndrome, which is characterized by impaired pDC development, thus indicating a conserved requirement of TCF4 across species25. Whereas commitment to pDCs is regulated by the expression of TCF4, development into cDC1 depends on the expression of the transcriptional repressor ID2, which specifically inhibits TCF4 and therefore pDC lineage commitment26. The branching of these two DC subsets at the CDP stage is determined by the balance of ID2 and TCF4. In particular, the long isoform of TCF4 (TCF4L) in complex with the transcription factor CBFA2T3 (also referred to as MTG16) induces pDC-target genes while repressing Id2 and therefore cDC1 commitment27,28. The zinc-finger transcription factor ZEB2 has also been shown to be involved in the regulation of early DC development29,30. Zeb2 deficiency results in decreases in pDCs and cDC2, and in an increase of cDC1, whereas its overexpression leads to slightly decreased cDC1 and unaltered numbers of cDC2 and pDCs29. According to these observations, and the elevated expression of Id2 observed in Zeb2−/− mice29, ZEB2 has been suggested to potentially repress Id2, which is required for cDC1 commitment. However, the unaltered pDC pool in Itgax-cre × R26-Zeb2Tg/Tg mouse progeny indicates that an active lineage commitment involving a more complex, TCF4-dependent transcriptional network that goes beyond the previously supposed cDC1–pDC dichotomy is required during pDC development. Furthermore, these results may also suggest a dual origin of pDCs, in which the requirement for TCF4 and ZEB2 is lineage and stage specific. The complete absence of pDCs in the progeny of Zeb2fl/fl crossed to Mx1-cre30 mice, and their partial decrease in Itgax-cre29, supports either a CDP-independent origin of pDCs or an incomplete deletion of Zeb2 in CDPs.
A prerequisite for determining the molecular mechanisms involved in lineage specification is to define the developmental stage at which pDC commitment occurs and to identify the pDC-committed precursor (pre-pDC) with exclusive lineage potential. Given the complexity of the transcriptional interactions occurring at different stages during pDC and cDC commitment and the possible developmental convergence from lymphoid and myeloid lineages into a single phenotypically consistent population, we decided to perform an in-depth exploration of the paths leading to pDC differentiation. Here, we found that pDCs developed mostly from IL-7R+ lymphoid precursor cells (IL-7R+ LPs) and identified a bona fide committed pre-pDC with exclusive lineage potential within the IL-7R-expressing pool. In addition, we characterized the transcriptional landscape of pDC development from IL-7R+ LPs to mature pDCs. Transcriptionally, the pre-pDC precursors identified here showed high expression of IRF8 before the acquisition of pDC identity and functionality, which was gained only after the expression of TCF4. Finally, we showed, through single-cell analysis, that despite developmental convergence to a phenotypically similar population, lymphoid- and myeloid-derived mature pDCs are transcriptionally and functionally heterogeneous.
Results
pDCs develop primarily from IL-7R+ LPs
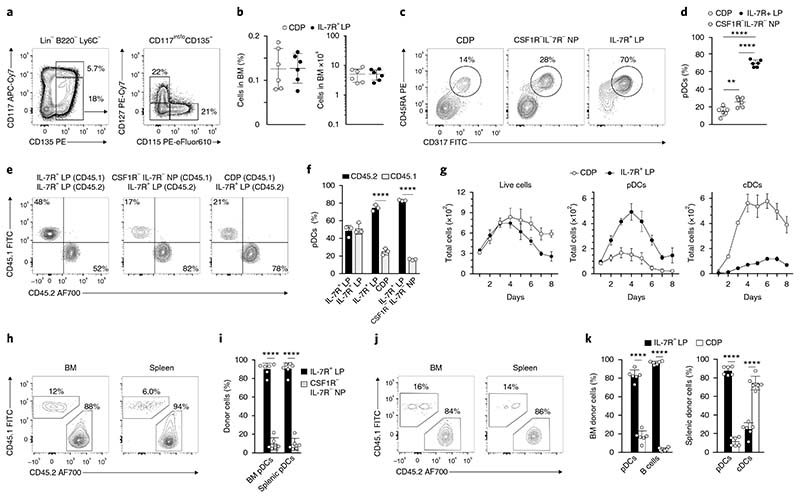
Both CDPs and common lymphoid progenitors are able to generate pDCs in vitro, but the independent contributions of these two subsets to the mature pDC pool in vivo are unclear12,13,16,21. All DCs, including cDCs and pDCs, originate from Lin–B220–Ly6C–CD117int/loCD135+ hematopoietic progenitor cells. Within this subset, the expression of CD115 (CSF1R) and CD127 (IL-7R) allowed for the identification of three populations: CD115+CD127– cells (CDPs hereafter), CD115–CD127+ cells (IL-7R+ LPs hereafter) and CD115–CD127– cells (CSF1R–IL-7R– NPs hereafter) (Fig. 1a). The frequency and abundance of CDPs and IL-7R+ LPs were similar in the BM of wild-type mice under steady-state conditions (Fig. 1b). To understand the cDC and pDC potential of each BM progenitor subset, we cultured them in the presence of FLT3L and assessed the development of CD45RA+CD317+ pDCs, CD11c+MHC-IIhi cDCs, CD11c+MHC-IIhiCD11b+ cDC2 and CD11c+MHC-IIhi CD24+ cDC1, unless otherwise specified. IL-7R+ LPs generated approximately fivefold more pDCs than did CDPs, which predominantly gave rise to cDCs, and more than threefold more pDCs than did CSF1R–IL-7R– NPs (Fig. 1c,d and Supplementary Fig. 1b). Because IL-7R+ LPs represent approximately 0.12% of total BM cells and include approximately 0.04% Sca1+ common lymphoid progenitors31, which are progenitors of B cells, we assessed the ability of CDPs, IL-7R+ LPs and CSF1R–IL-7R– NPs to develop into CD19+ B cells by culturing them under B cell–polarizing conditions in the presence of FLT3L and OP9 stromal cells (Methods). Only IL-7R+ LPs developed into CD19+ B cells and remained the most efficient population at generating pDCs under these B cell–permissive conditions (Supplementary Fig. 1a,c).
Fig. 1. pDCs develop primarily from IL-7R+ lymphoid progenitors.
a,b, All subsets are pregated on Lin–B220–Ly6C–CD117int/loCD135+. Shown are the gating strategy (a) of CDPs (CD115+CD127–), IL-7R–M-CSFR– NPs (CD115–CD127–) and IL-7R+ LPs (CD115–CD127+), and the frequency (b) of the indicated progenitors in the BM of C57BL/6 mice (n = 6; each dot represents a mouse, and thin lines represent the mean ± s.d.). c,d, Sort-purified CDPs, IL-7R–MCSFR– NPs and IL-7R+ LPs were cultured for 4 d in the presence of FLT3L. pDC output was determined according to the expression of CD317 (Bst2) and CD45RA (c) and is shown as percentage output (d) (n = 6; each dot represents a mouse, and thin lines represent the mean ± s.d.). e–k, Sort-purified progenitors isolated from CD45.1 and CD45.2 mice were cocultured (e,f) for 4 d in the presence of FLT3L. Shown are two-color histograms for the expression of CD45.1 and CD45.2 pregated on CD45RA+CD317+ pDCs (e) and percentage output (f) (n = 3; each dot represents a mouse, and thin lines represent the mean ± s.d.). g, IL-7R+ LPs and CDPs were cultured in competitive settings in a 1:1 ratio. Shown is the total, pDC and cDC output over 8 d of culture (n = 3 mice; thin lines represent the mean ± s.e.m.). h–k, BM and splenic pDC output was determined 4 d after intravenous co-transfer of CD45.2-positive IL-7R+ LPs in competition with CD45.1-positive IL-7R–M-CSFR– NPs (h,i) or CDPs (j,k). Shown are two-color histograms for the expression of CD45.1 and CD45.2 pregated on CD45RA+CD317+ pDCs (h,j) (n = 6 mice). Shown are percentage donor-derived BM pDCs and B cells and splenic pDCs and cDCs, as indicated (i,k) (n = 6; each dot represents a mouse, and thin lines represent the mean ± s.d.). Statistical analysis was done with one-way ANOVA with Tukey post-test (d,f) or two-tailed Student’s t test (i,k). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To examine the pDC, cDC and B cell potential of these progenitor subsets under competitive conditions, we isolated IL-7R+ LPs from the BM of CD45.2 mice and cocultured them with CDPs or CSF1R– IL-7R– NPs from CD45.1 congenic mice (Supplementary Fig. 1i). Regardless of the presence or absence of stromal cells, IL-7R+ LPs had significantly greater pDC potential than did CDPs or CSF1R– IL-7R– NPs at all analyzed time points (Fig. 1e,f and Supplementary Fig. 1g) and were the only progenitors able to differentiate into CD19+ B cells (Supplementary Fig. 1f,h). To exclude differential proliferative capacity and the possibility that these progenitor subsets might be distinct developmental stages of one another, we performed a time-course analysis (Fig. 1g). The total cell output from CDPs and IL-7R+ LPs was comparable and peaked at day 4. However, IL-7R+ LP–derived pDCs outnumbered those derived from CDPs at every time point (Supplementary Fig. 1d). In contrast, cDCs were mostly CDP derived (Fig. 1g and Supplementary Fig. 1e). After day 4 of culture, the total cellular output from CDPs was higher than that of IL-7R+ LPs (Fig. 1g), thus suggesting that the ability of IL-7R+ LPs to generate progeny decreased, that the IL-7R+ LP–derived cells had diminished survival ability in vitro, or a combination of both. To discriminate among these possibilities, we performed the same experiment, using the proliferation tracer CellTrace Violet. We detected no major differences in the proliferation rates of CDP- or IL-7R+ LP–derived cells (data not shown), thus suggesting that in vitro–generated pDCs have lower survival ability than in vitro–generated cDCs, independently of their origin, in line with previous reports32.
To assess the pDC, cDC and B cell in vivo potential of these progenitors, we co-transferred IL-7R+ LPs isolated from BM in a 1:1 ratio with congenic CSF1R–IL-7R– NPs (Fig. 1h, i) or CDPs (Fig. 1j,k and Supplementary Fig. 1l) into sublethally irradiated mice and analyzed the BM and spleen of the recipient mice by flow cytometry 4 d after transfer (Fig. 1h–k). IL-7R+ LPs generated a 5- to 15-fold-higher output of SiglecH+CD317+ pDCs than CSF1R–IL-7R– NPs or CDPs in both tissues (Fig. 1h–k). Donor-derived CD19+ B cells were detected only in the BM and were exclusively IL-7R+ LP derived (Fig. 1k and Supplementary Fig. 1l), whereas cDCs, which were recovered only in the spleen, were 80% CDP derived (Fig. 1k and Supplementary Fig. 1l). Early uncommitted Lin–c-kithi BM progenitors, when co-injected, had equal potential to generate SiglecH+CD317+ BM and splenic pDCs, thus suggesting that both congenic strains had equal pDC reconstitution potential (Supplementary Fig. 1j–k). Collectively, mature BM and splenic pDCs differentiate in vitro and in vivo predominantly from IL-7R-expressing BM progenitors and not from CDPs or CSF1R–IL-7R– NPs.
SiglecH+Ly6D+ IL-7R+ LPs have exclusive pDC potential
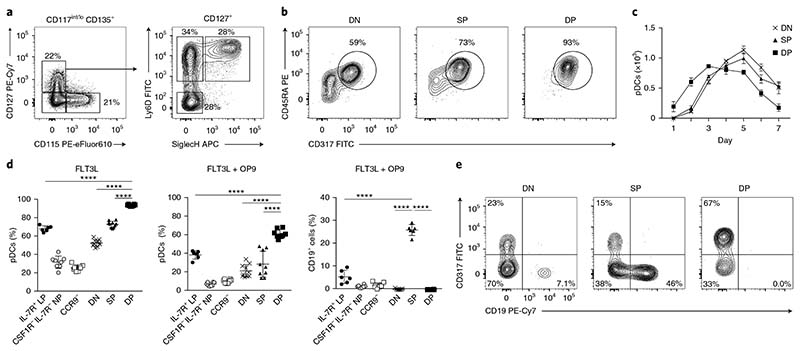
We then further investigated the pDC and B cell potential within the IL-7R+ LPs. Staining for SiglecH and Ly6D allowed us to subdivide IL-7R+ LPs into three fractions with relatively equal distribution: SiglecH–Ly6D– (double negative, DN), SiglecH–Ly6D+ (single positive, SP) and SiglecH+Ly6D+ (double positive, DP) (Fig. 2a and Supplementary Fig. 2a). All three subsets showed a high differentiation potential into CD317+CD45RA+ pDCs that increased from DN to SP to DP (Fig. 2b). Furthermore, DP cells had almost exclusive pDC lineage potential, showing a pDC commitment of more than 90% (Fig. 2b). To understand the developmental relationship among DN, SP and DP progenitors, we sorted and performed a time-course analysis examining the pDC output over 7 d of culture. All three subsets had a similar total output of mature pDCs (Fig. 2c). However, DP progenitors developed into mature pDCs faster, peaking at day 3 (Fig. 2c), thus suggesting that DP progenitors may be a more mature subset. In vitro, the pDC developmental ability of DP progenitors was also largely superior to those of CDPs and the previously reported pDC precursors CSF1R–IL-7R– NPs23 and CD11c+CD317+CCR9– cells (CCR9– progenitors hereafter)22,24 (Supplementary Fig. 2b–d). The percentage of CD317–CD45RA– non-pDCs that developed in FLT3L-treated cultures in vitro was approximately 40% for DN cells and 30% for SP cells, and was limited to approximately 5% for DP progenitors (Fig. 2b and Supplementary Fig. 2e). Most of these CD317–CD45RA– cells expressed cDC markers such as CD172, CD11b, CD24 and MHC-II. In comparison, CDPs generated approximately 50% cDC1 and 35% cDC2, and showed a pDC output of approximately 15% (Fig. 2d and Supplementary Fig. 2c–e). When we exposed DN, SP and DP cells, CSF1R–IL-7R– NPs, CCR9– progenitors and CDPs to B cell–polarizing conditions (FLT3L and OP9 stromal cells; Supplementary Fig. 2f–j), CSF1R–IL-7R– NPs, CCR9– progenitors and CDPs had no CD19+ B cell potential, IL-7R+ LPs and SP had CD19+ B cell potential, and DP progenitors were unable to develop into CD19+ B cells and maintained a high pDC output (Fig. 2d–e and Supplementary Fig. 2f,g,i). SiglecH+Ly6D+IL-7R+ LPs are committed to the pDC lineage: they showed almost exclusive pDC potential in the presence of FLT3L and had no ability to differentiate into B cells when cultured under B cell–polarizing conditions in the presence of OP9 stromal cells.
Fig. 2. SiglecH+Ly6D+IL-7R+ LPs have exclusive pDC potential.
a, IL-7R+ LPs, as defined in Fig. 1a, were further subdivided on the basis of expression of SiglecH and Ly6D. Shown is the gating strategy for DN, Ly6D SP, and SiglecH and Ly6D DP progenitors (n = 6 mice). b–e, Sort-purified IL-7R–M-CSFR– NPs, IL-7R+ LPs, CCR9– (CD317+ B220+CD11c+CCR9–), DN, SP and DP precursors were cultured in the presence of FLT3L. Two-color histograms for the expression of CD45RA and CD317 (b) and the percentage of mature pDCs (d) are shown for day 4 of culture (n = 6 mice, with 1 or 2 technical replicates. Each dot represents a sample, and thin lines represent the mean ± s.d.). c, Total pDCs generated over 7 d of culture from sort-purified DN, SP and DP precursors (n = 3 mice; thin lines represent the mean ± s.e.m.). d,e, The same progenitors cultured for 4 d in the presence of FLT3L and OP9 stromal cells. Shown are percentages of mature pDCs and B cells (d) and two-color histograms for the expression of CD317+ pDCs and CD19+ B cells (e) (n = 6 mice with 1 or 2 technical replicates; each dot represents a sample, and thin lines represent the mean ± s.d.). Statistical analysis was done with one-way ANOVA with Tukey post-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
SiglecH+Ly6D+ DP cells are bona fide pDC progenitors
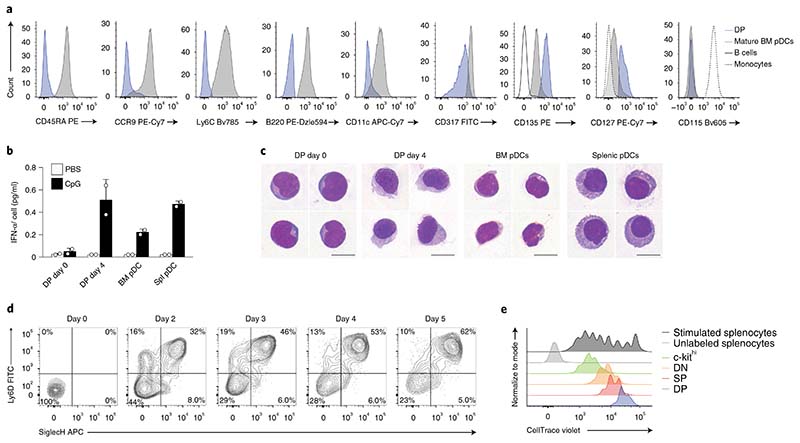
Because SiglecH and Ly6D33 are expressed on mature pDCs, we phenotypically and functionally compared DP progenitors directly with freshly isolated BM and splenic mature pDCs. We detected higher expression of CD127, CD135, CCR9, CD45RA, Ly6C, B220, CD11c, MHC-II, Sca1 and CD317 on mature SiglecH+B220+Ly6D+Ly6C+ pDCs than on DP progenitors (Fig. 3a and Supplementary Fig. 3a). Further, DP progenitors did not express CCR9 (Fig. 3a), thus indicating that pDC progenitors reside within the CCR9– compartment22. In Zbtb46gfp/+ mice, Zbtb46-GFP or CD115, which are cDC-specific markers, were not detected on either DP cells or mature pDCs. (Fig. 3a and Supplementary Fig. 3a). Stimulation with CpG-A induced the production of the cytokine IFN-α by mature BM and splenic pDCs, but not by DP progenitors, which acquired this ability after 4 d in culture (Fig. 3b). Similarly, SP and DN progenitors produced type 1 IFN only after 4 d, upon maturation (Supplementary Fig. 3b). Morphologic maturation, as assessed by Giemsa staining, was achieved by DN, SP and DP progenitors after 4 d in culture (Fig. 3c and Supplementary Fig. 3c).
Fig. 3. SiglecH+Ly6D+ DP cells are bona fide pDC progenitors.
a, Representative single-color histograms for the indicated surface markers expressed by DP pre-pDCs (blue), mature pDCs (gray), B cells (black) and monocytes (broken line) (n = 3 independent experiments). b, DP pre-pDC progenitors were cultured for 4 d in the presence of FLT3L. IFN-α was measured in the supernatants collected on day 0 and day 4, or on freshly plated mature pDCs 16 h after stimulation with CpG-A, as indicated (n = 3 independent experiments, with one representative experiment shown; each dot represents a technical replicate, and thin lines represent the mean ± s.d.). Spl, splenic. c, May–Gruenwald staining of sort-purified mature BM, splenic pDCs and DP progenitors at day 0 and after 4 d of culture (n = 4 representative images taken from 3 independent experiments; scale bars, 10 μ m). d, DN progenitors, as defined in Fig. 2a, were cultured in the presence of FLT3L for 5 d. Shown are two-color histograms for the expression of Ly6D and SiglecH analyzed at the indicated time points (n = 3 independent experiments, with representative data from one experiment shown). e, DP (blue), SP (red), DN (orange) and c-kithi (Lin–B220–Ly6C–CD117hi) (green) progenitors were cultured for 4 d in the presence of FLT3L. Proliferation was assessed through CellTrace Violet dilution. Unlabeled (light gray) or labeled (dark gray) splenocytes were stimulated with anti-CD3 and anti-CD28 (n = 3 independent experiments, with representative data from one experiment shown). Statistical analysis was done with one-way ANOVA with Tukey post-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We next tested whether the DN, SP and DP maturation stages could be recapitulated in vitro. A time-course analysis of cell-sorted DN progenitors showed progressive accumulation of SP and DP cells, which were detectable in culture at day 2 (Fig. 3d). Mature CD45RA+SiglecH+ pDCs developed from DP progenitors after 2 d of culture, whereas SP progenitors initially upregulated SiglecH at day 1 and transitioned into mature cells from day 3 (Supplementary Fig. 3d,e). Similarly, analysis of proliferation showed that DP cells required fewer divisions than SP and DN cells to develop into mature pDCs (Fig. 3e), thereby indicating progressive maturation from DN via SP to DP status. Thus, DP progenitors acquire the expression of lineage-specific markers, the morphology and the ability to produce IFN-α characteristic of mature splenic pDCs after two cell divisions.
Stage-specific transcriptional signatures define pDC commitment
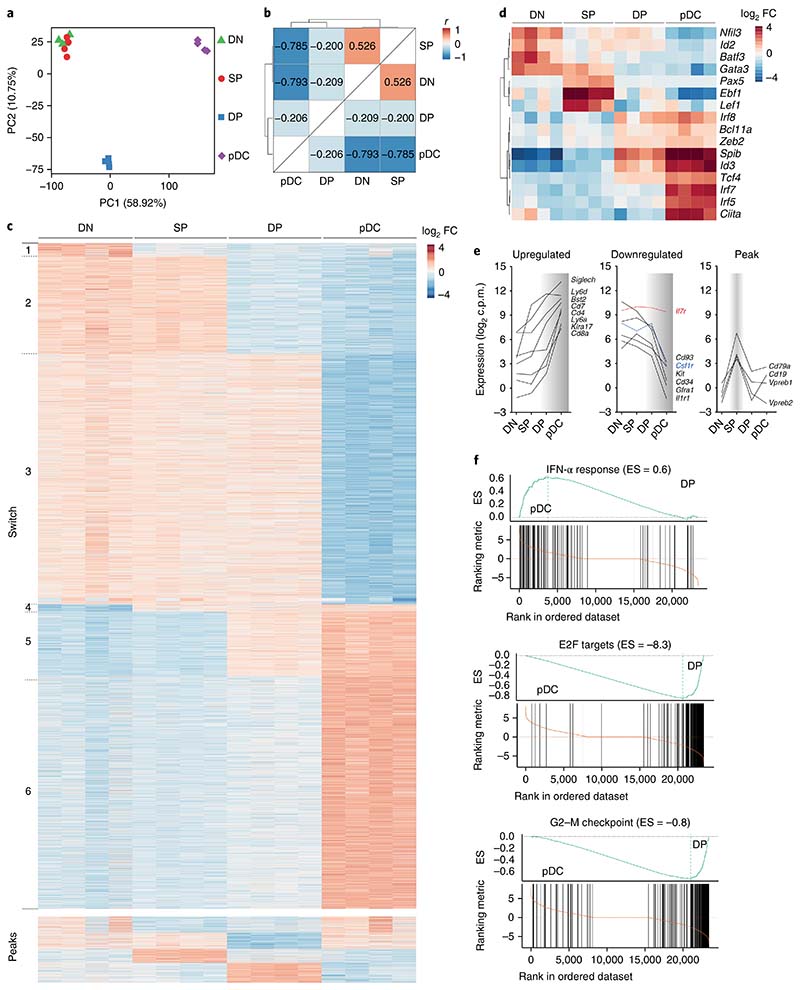
We next sought to define the transcriptional signature that recapitulates the commitment to pDCs. We performed RNA sequencing on DN, SP and DP progenitors isolated from wild-type BM and compared their transcriptional landscapes with that of B220+SiglecH+Ly6C+Ly6D+ mature BM pDCs. Principal component analysis (PCA) showed individual segregation of mature pDCs and DP progenitors: whereas DN cells and SP cells clustered together and partially shared their transcriptomes, DP cells were a distinct subset also different from mature pDCs (Fig. 4a). Hierarchical clustering of the subsets on the basis of Pearson’s correlation coefficient confirmed the results obtained by PCA (Fig. 4b), in which DP cells were transcriptionally closely related to mature pDCs.
Fig. 4. Stage-specific transcriptional signatures define pDC commitment.
a–f, Bulk RNA sequencing was performed on cell-sorted DN, SP and DP progenitors, and mature pDCs (Methods). a,b, PCA (a) and hierarchical clustering (b) of the progenitor subsets and mature pDCs on the basis of Pearson’s correlation coefficient calculated on the 25% of genes with the highest variance (calculated as interquartile range). PC, principal component. c, Heat map based on a developmental-stage model from DN to SP to DP and to mature pDCs, generated on selected genes (fold change (FC) > 1 and P < 0.05; Methods). Genes were ordered in switches and peaks according to their expression profiles for the different subsets. d, Correlation heat map for selected transcription factors. e, Specific surface markers, plotted according to their expression profiles (upregulated, downregulated or peak) across the indicated developmental stages from DN to mature pDCs. Highlighted are the transcript levels as c.p.m. for Il7r (red) and Csf1r (blue). f, Gene set enrichment analysis, performed on the Molecular Signature Database (MSigDb v5.2) comparing the enrichment score (ES) for DP progenitors with mature pDCs, as depicted (IFN-α response, P < 0.00358; E2F targets, P < 0.00146; G2–M checkpoint, P < 0.000946). RNA was collected from sort-purified subsets in n = 4 independent experiments. For each experiment, all progenitors were obtained from one mouse.
To better understand the dynamics of pDC commitment, we generated a heat map based on genes uniquely expressed at the DN stage (switch 1) and at the DN and SP stages (switch 2) through the mature pDC stage (switch 6) (Fig. 4c). Transcripts were also distributed according to shared expression patterns across two cell subsets, defined as peaks (Fig. 4c and Supplementary Table 1). The resulting developmental-expression heat map reflected the transcriptional landscape for pDC commitment and was used to evaluate switch-specific transcription factors and surface receptors (Fig. 4d,e and Supplementary Fig. 4a,b). Spib and Irf7 were highly expressed only in mature pDCs (Fig. 4d), a result consistent with the inability of DN, SP and DP progenitors to produce IFN-α and with their requirement at later stages of development. The SP stage, which is the only one permissive for B cell development, was marked by the expression of B cell–specific transcription-factor transcripts, such as Pax5 and Ebf1 (Fig. 4d) as well as surface-receptor transcripts, such as Cd19, Vpreb1 and Vpreb2, and Cd79a (Fig. 4e). Transcripts encoding transcription factors known to be essential during pDC development, particularly Tcf4, Irf8, Zeb2 and Bcl11a, were expressed at the DP stage and were further upregulated after maturation (Fig. 4d), thus suggesting that pDC lineage specification was achieved at the DP stage. Importantly, whereas the Il7r transcript as well as IL-7R protein are expressed during development and on mature pDCs, the Csf1r transcript was detected on all progenitors, in the absence of the protein at all stages (Fig. 4e and Fig. 2a). This result provides a potential explanation for the apparent conflict of lineage tracing in mice, in which pDCs were labeled in both Il7rcre (ref. 34) and Csf1rcre mice35.
We next searched for differentially expressed genes between DP cells and all the other analyzed subsets (Supplementary Fig. 4d–f and Supplementary Table 1). Gene set enrichment analysis between DP progenitors and mature pDCs identified changes in three major pathways: E2F targets, the G2–M checkpoint and IFN-α production, thus suggesting that maturation was achieved through the downregulation of cell-cycle-associated genes and the upregulation of genes mediating functional properties (Fig. 4f, Supplementary Fig. 4c and Supplementary Table 2). Collectively, these data reveal that pDC lineage specification is already transcriptionally established at the DP stage.
Expression of IRF8 marks pDC lineage commitment on SP cells
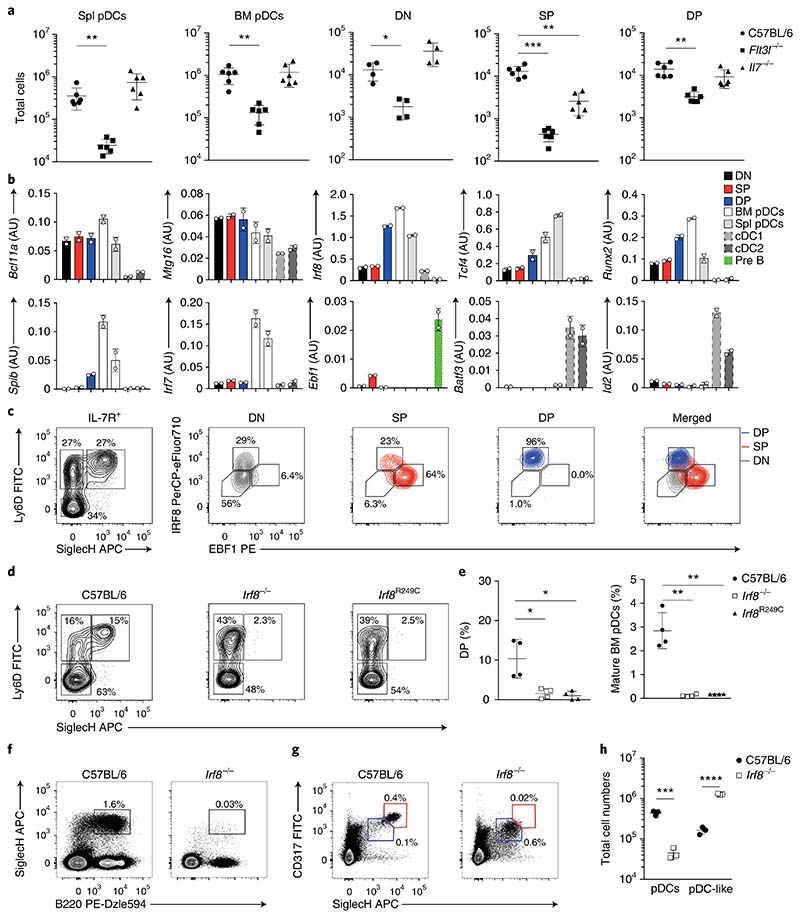
Given the sustained expression of CD135 and CD127 at all stages of pDC development and on mature pDCs, we examined the requirement of the corresponding ligands FLT3L and IL-7 during commitment. Flt3l−/− mice showed impaired pDC development across all stages, with approximately tenfold-fewer total DN progenitors than those in wild-type control mice (Fig. 5a and Supplementary Fig. 5a,e). Il7−/− mice, compared with wild-type controls, had unaltered numbers of DN and DP progenitors, as well as mature pDCs, but markedly lower numbers of SP progenitors and mature B cells (Fig. 5a and Supplementary Fig. 5a,e,f). This result correlated with the greater B cell–specific developmental and transcriptional bias of SP precursors and suggested that SP cells were heterogeneous and already committed to either the B or the pDC lineage. Quantitative PCR analysis validated the stage-specific expression of several transcription factors important for B, pDC and cDC development4,8,36 (Fig. 5b and Supplementary Fig. 5b). Transcripts for Spib and Irf7 were low or absent on progenitors but were induced in mature pDCs, whereas the expression of Irf8, Tcf4 and Runx2 was already established at the DP progenitor stage and further increased in mature pDCs (Fig. 5b). Notably, Ebf1 expression was confined within the SP subset, in agreement with the exclusive ability of SP progenitors to generate B cells (Fig. 5b). To address whether SP progenitors were heterogeneous, we examined the expression of EBF1 and IRF8 on DN, SP and DP progenitors in wild-type, Il7−/− and Flt3l−/− mice. Indeed, SP progenitors could be split into EBF1+IRF8int and EBF1–IRF8hi cells, whereas DP cells were exclusively EBF1–IRF8hi (Fig. 5c). Furthermore, induction of EBF1, but not IRF8 expression, was compromised at the SP stage in Il7−/− mice compared with littermate controls, in agreement with fewer mature B cells37 (Supplementary Fig. 5c,d).
Fig. 5. Expression of IRF8 marks pDC lineage commitment on SP cells.
a, Total mature BM and splenic pDCs and DN, SP and DP progenitors in wild-type, Flt3l−/− and Il7−/− mice, as indicated (n = 6 independent experiments; each symbol represents a mouse, and thin lines represent the mean ± s.d.). b, Expression of selected genes (arbitrary units (AU) relative to β-actin) on sort-purified DN, SP and DP BM progenitors, gated as in Fig. 2a, mature BM and splenic pDCs (SiglecH+CD317+), cDC1 (CD11c+MHCIIhiCD24+XCR1+) and cDC2 (CD11c+MHCIIhiCD11b+Sirp-α+) were analyzed for the expression of selected genes (n = 3 independent experiments, with one representative experiment shown; each dot represents a technical replicate, and thin lines represent the mean ± s.d.). c, Expression of IRF8 and EBF1, determined on DN (black), SP (red) and DP (blue) progenitors (n = 6 mice; representative experiment shown). d,e, DN, SP and DP subsets and mature pDCs were analyzed in Irf8−/− and BXH2 (IRF8R249C) mice. Shown are representative two-color histograms for the expression of Ly6D and SiglecH (d) and the percentage DP pre-pDCs and mature BM pDCs (e) (n = 4; each symbol represents a mouse, and thin lines represent the mean ± s.d.). f–h, Splenic and BM pDCs analyzed in C57BL/6 and Irf8−/− mice. Shown are representative two-color histograms for the expression of SiglecH and B220 on BM cells (f) and CD317 and SiglecH on splenocytes (g). CD317hiSiglecHhi pDCs (red) and CD317intSiglecHint pDC-like cells (blue), gated as in g, were quantified (h) (n = 3; each symbol represents a mouse, and thin lines represent the mean ± s.d.). Statistical analysis was done with two-tailed Student’s t test (b,e,h). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Whereas FLT3L deficiency resulted in a partial decrease in DP progenitors, Irf8−/− or Irf8R249C mutant mice (Methods), in which an R249C mutation prevents the interaction with partner transcription factors such as PU.1, IRF2 and SpiB38, completely lacked DP cells6,39 (Fig. 5d, e). SP cells accumulated in both Irf8−/− and Irf8R249C mice, a result indicative of a developmental block at the transition from SP to DP cells (Fig. 5d). In agreement with this finding, Irf8−/− mice lacked mature SiglecH+B220+ pDCs in the BM and SiglecHhiCD317hi in the spleen (Fig. 5e–h). However, a population of SiglecHintCD317int pDC-like cells was detected and was even found to be elevated in the Irf8−/− spleens (Fig. 5g,h). These results indicate that SP cells can be subdivided into IRF8hi, IL-7-independent pDC-committed and EBF1+, IL-7-dependent B cell–committed progenitors.
IRF8 and EBF1 define pDC and B cell lineage dichotomy
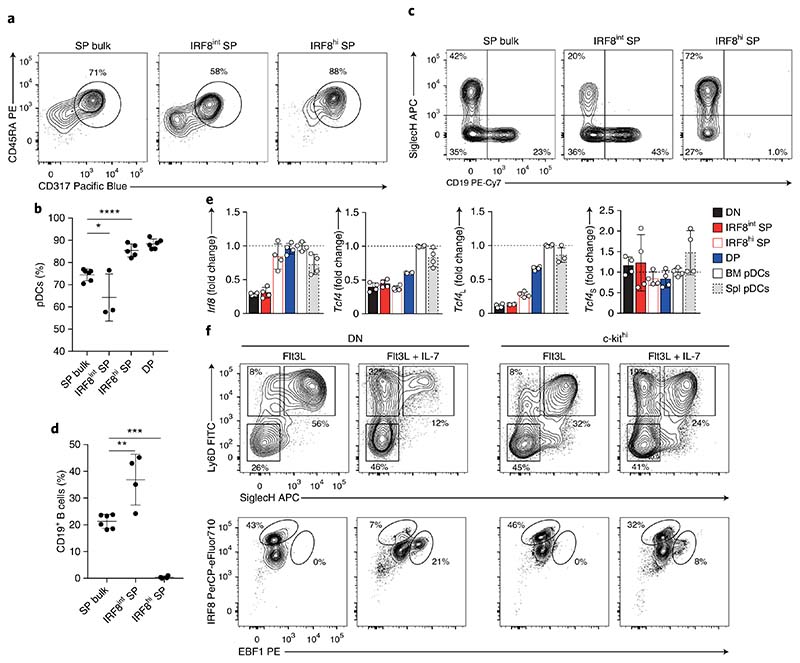
To dissect the heterogeneity of the SP compartment, we used IRF8-eGFP and EBF1-hCD2 reporter mice, which express a 3′ IRES-GFP and an IRES-human CD2, respectively, thus allowing us to trace the genes and sort the expressing subsets. SP cells from IRF8-eGFP or EBF1-hCD2 mice could be sorted into IRF8-GFPint (IRF8int SP) or IRF8-GFPhi (IRF8hi SP) and into EBF1-hCD2– (EBF1− SP) and EBF1-hCD2+ SP (EBF1+ SP) cells (Supplementary Fig. 6a,c). We then assessed the pDC- and B cell–differentiation potential of each SP subset in vitro, as described above. IRF8hi SP progenitors had almost exclusive pDC output and could not differentiate into B cells (Fig. 6a–d and Supplementary Fig. 6b,d), thus suggesting that the B cell potential was lost concomitant with the induction of high IRF8 expression at the SP precursor stage. Similarly, EBF1– SP progenitors did not differentiate into B cells on OP9 stromal cells under B cell–polarizing conditions (Supplementary Fig. 6e–h), thus suggesting that EBF1 expression at the SP stage is necessary to promote B cell lineage commitment.
Fig. 6. IRF8 and EBF1 define pDC and B cell lineage dichotomy.
a–d, Progenitors from C57BL/6 (bulk) and Irf8egfp reporter mice, as indicated and gated as in Supplementary Fig. 6a, were sort-purified and analyzed for pDC and B cell potential. Specifically, DP and unfractionated SP (SP bulk) and SP progenitors expressing intermediate (IRF8int SP) or high (IRF8hi SP) levels of IRF8 were sort-purified and cultured for 4 d in the presence of FLT3L (a,b) or FLT3L and OP9 stromal cells (c,d), as indicated. Shown are two-color histograms and percentages of CD45RA+CD317+ pDCs (a,b) and SiglecH–CD19+ (c,d) (n = 3 independent experiments with 1 or 2 technical replicates; each symbol represents a sample, and thin lines represent the mean ± s.d.). e, Expression of Irf8, Tcf4 and its long (Tcf4L) or short (Tcf4S) isoforms, quantified in DN, SP IRF8int, SP IRF8hi, DP, BM and splenic pDCs from IRF8-eGFP reporter mice. Shown are the expression levels indicated as a ratio to BM pDCs (n = 2 independent experiments with 2 technical replicates; data shown as mean ± s.d). f, DN and c-kithi progenitors were cultured for 5 d in the presence of FLT3L with or without IL-7 as indicated. Shown are two-color histograms for the expression of Ly6D/SiglecH (top) and IRF8/EBF1 (bottom) (n = 3 independent experiments, with one representative experiment shown). Statistical analysis was done with one-way ANOVA with Tukey post-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Expression of TCF4, specifically its long isoform (Tcf4L), is required for pDC development, because pDCs do not develop in its absence25,27. RT–qPCR analysis indicated comparable expression of Tcf4L in DP cells and mature BM and splenic pDCs (Fig. 6e). The expression of Irf8 in IRF8 hi SP cells was comparable to that observed in DP cells and mature pDCs, whereas the expression of Tcf4L in IRF8hi SP cells was low, and similar to that in DN cells (Fig. 6e), thus suggesting that the expression of IRF8 but not TCF4L mirrored the acquisition of pDC-lineage specification.
BM niches are likely to influence progenitor lineage choice through the availability of cytokines and other cues. We simulated the contact and the exposure to the BM stromal environment by using a Transwell culture system, in which SP cells and DP cells were cultured in the presence of FLT3L, either in direct contact with OP9 stromal cells or exposed to soluble factors released by OP9 stromal cells. Differentiation of SP and DP cells toward CD317+ pDCs was significantly inhibited by direct contact with OP9 stromal cells, whereas differentiation of SP cells toward CD19+ B cells required direct contact (Supplementary Fig. 6i–l). To understand the role played by polarizing cytokines in pDC and B cell lineage specification, we examined the induction of IRF8 and EBF1 in uncommitted DN and c-kithi progenitors exposed to FLT3L and IL-7. Exposure of DN and c-kithi precursors to FLT3L resulted in a strong induction of IRF8 expression as well as accumulation of DP progenitors after 5 d of culture (Fig. 6f). The addition of IL-7 promoted the accumulation of SP progenitors and induced EBF1 expression in the absence of OP9 stromal cells (Fig. 6f). This result indicates that lineage specification toward pDCs or B cells occurs in SiglecH–Ly6D+ SP cells and is defined by the mutually exclusive, high expression of IRF8 or EBF1, which was in turn governed by the exposure to FLT3L or IL-7 and influenced by contact with stromal cells.
Single-cell analysis elucidates pDC heterogeneity
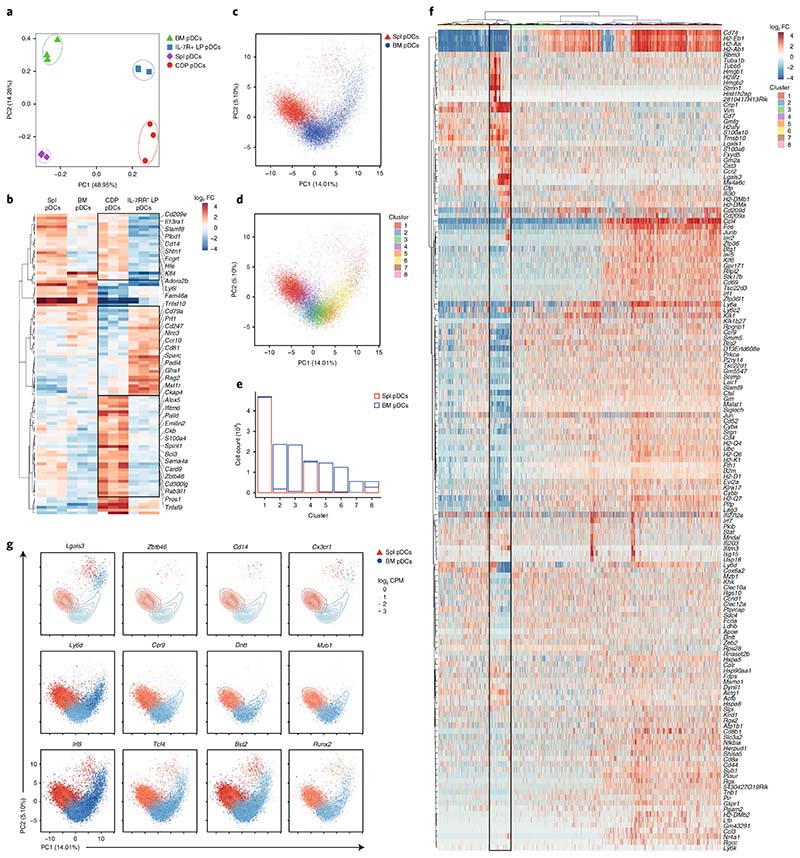
To understand how tissue imprinting as well as ontogeny might influence the transcriptional landscape of pDCs, we performed bulk as well as single-cell RNA sequencing. Bulk RNA sequencing was done on ex vivo–isolated mature pDCs from the BM and spleen, and on in vitro–generated CD317+SiglecH+ pDCs from IL-7R+ LPs and CDPs. A high correlation coefficient ranging from 0.8 to 0.95 was obtained across all pDC samples analyzed (Supplementary Fig. 7a). Through PCA, we were able to highlight differences related to in vitro–generated versus ex vivo–isolated pDCs (principal component 1, 49%), and differences associated with tissue imprinting, in splenic versus BM pDCs (principal component 2, 14%) (Fig. 7a). Further analysis of the sample was performed after filtering for the 25% most variable genes. By focusing on differences related to their ontogeny and using a stringent cutoff (log2 fold change > 2) we identified 107 genes differentially expressed in CDP- and IL-7R+ LP–derived pDCs (Fig. 7b and Supplementary Fig. 7c,d). Importantly, whereas pDC-related transcripts, such as Irf8, Siglech, Tcf4 and Bst2, showed high expression, most of the genes expressed differentially between CDP and IL-7R+ LP–derived pDCs, such as Rag2 and Cd14, were expressed at low levels (Supplementary Fig. 7b).
Fig. 7. Single-cell analysis elucidates pDC heterogeneity.
a,b, Bulk RNA sequencing on mature BM and splenic pDCs, and on IL-7R+ LP– and CDP-derived pDCs, performed as described in Methods. a, PCA performed on the 25% most variable genes. b, Heat map showing relative expression for differentially expressed genes (log2 fold change > 2.0) from IL-7R+ LPs versus CDP-derived pDCs. c–g, Single-cell RNA sequencing, performed as described in Methods, on sort-purified BM and splenic pDCs. c,d, PCA based on the 148 hypervariable genes (biological variation > 0.1 and false discovery rate < 0.05). Colors indicate the tissue of origin (c) or the identified clusters (d). e, Number of cells identified for each cluster in the BM and spleen. f, Heat map for 148 hypervariable genes across all 14,744 cells. At top, colors indicate the identified clusters as in d. g, Expression of the indicated genes from individual BM (blue) and splenic (red) pDCs. The size of each dot corresponds to the relative expression of a given gene for each cell. The contour lines indicate the density of the BM (blue) and splenic (red) cells in the PCA space. Cells for bulk and single-cell RNA sequencing were harvested from n = 3 mice in 3 independent experiments.
In line with their ontogeny, IL-7R+ LP–derived pDCs were enriched in expression of lymphoid-associated genes, such as Rag2 and Cd79, whereas CDP-derived pDCs expressed several cDC- and myeloid-related genes, such as Zbtb46, Cd14 and Klf4 (Fig. 7b). To validate the ontogeny-based heterogeneity of mature pDCs, we performed single-cell RNA sequencing on approximately 8,000 BM and 7,000 splenic CD317+SiglecH+ pDCs, detecting approximately 2,000 genes per cell. PCA based on 148 hypervariable genes confirmed tissue-specific identity at the single-cell level. Each cell was plotted in two-dimensional PCA space, and whereas splenic pDCs split into two discrete subgroups, BM pDCs spread along a diagonal over principal components 1 and 2 (Fig. 7c). Clustering analysis (Methods) identified eight clusters (Fig. 7d–f), with splenic pDCs splitting into three major clusters (1, 4 and 8), and BM pDCs splitting into six clusters (2, 3, 5, 6, 7 and 8) (Fig. 7e and Supplementary Fig. 7e). The relative and absolute frequency of cells belonging to each cluster showed almost exclusive tissue specificity for each cluster except for cluster 8, which was equally represented in both tissues and accounted for approximately 5% of the total pDC population (Fig. 7e and Supplementary Fig. 7e). A pairwise comparison of the differentially expressed genes (absolute log2 fold change > 1.5) across all clusters showed limited differences across clusters 1–6, and only clusters 7 and 8 diverged from the other clusters (Supplementary Fig. 7f). The numbers of detected genes per cluster and cell-cycle-associated genes indicated that BM cluster 7 was actively cycling and was probably an immediate precursor of cluster 8 (Supplementary Fig. 7g,h). Transcripts associated with cluster 8, i.e., Lgals3, Zbtb46 and Cd14, were reminiscent of the CDP-derived pDCs. Clusters 1 to 6 showed expression of genes previously identified in IL-7R+ LP–derived pDCs (Fig. 7b,g and Supplementary Table 3), i.e., Ly6d, Ccr9 and Dntt. All clusters expressed high amounts of pDC-specific transcripts, such as Tcf4, Irf8 and Bst2 (Fig. 7g), thus ruling out contamination of cluster 8 with other myeloid lineages. As such, single-cell analysis revealed heterogeneity of the pDC compartment and validated the segregation of lymphoid and myeloid signatures in two distinct subsets: the conventional pDCs and cluster 8, referred to as pDC-like cells.
The functional heterogeneity of pDCs is developmentally encoded
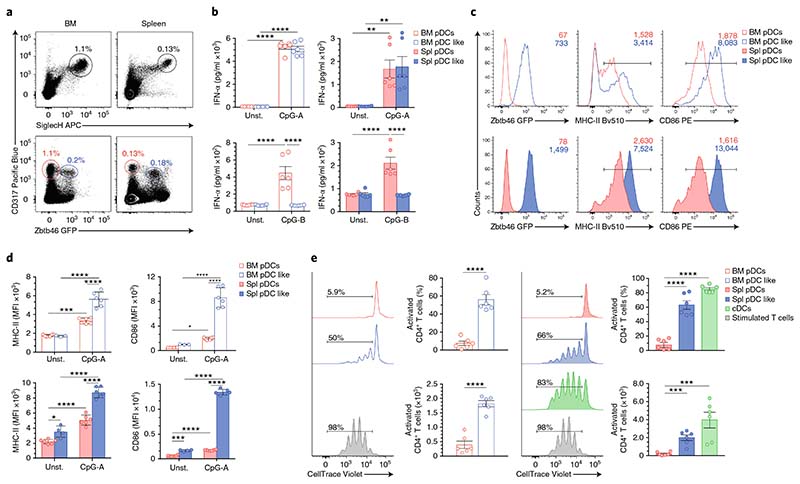
A prerequisite for performing a functional analysis of pDC-like cells was the identification of specific markers and the establishment of a gating strategy, which would enable us to sort and directly compare pDCs and pDC-like cells. We used Zbtb46gfp/wt mice to sort Zbtb46-GFP–SiglecHhiCD317+ pDCs and Zbtb46-GFP+SiglecHintCD317+ pDC-like cells (Fig. 8a and Supplementary Fig. 8a). We extended our phenotypic analysis of pDCs and pDC-like cells by comparing the transcript expression and, when possible, the protein expression of several pDC- and cDC-related genes. Both subsets expressed similar levels of pDC- and cDC-specific transcripts, such as Siglech, Bst2, Ly6c, H2-Aa, Itgax, Runx2 and Irf8. In addition, the protein expression of SiglecH, Bst2, MHC-II, CD11c and Ly6C was comparable between the two subsets, and only slight differences were detetcted (Supplementary Fig. 8b–f). However, CX3CR1 was expressed exclusively on pDC-like cells (Supplementary Fig. 8d,e), a result reminiscent of nonconventional CX3CR1+CD8α+ DCs previously identified40, with the exception that pDC-like cells were CD8α– (data not shown). Both subsets produced IFN-α when stimulated with CpG-A (Fig. 8b). However, unlike conventional pDCs, pDC-like cells did not respond to CpG-B stimulation (Fig. 8b), thus suggesting different signal regulation. Furthermore, when stimulated with CpG-A, pDC-like cells had higher surface expression of MHC-II and the co-stimulatory molecule CD86 than did conventional pDCs (Fig. 8c,d). Importantly, in contrast to conventional pDCs, pDC-like cells from BM and spleen were efficient at taking up, processing and presenting protein and induced strong proliferation of OT2 T cells, at levels comparable to those observed in cDCs41 (Fig. 8e) and consistent with their expression of cDC transcripts. In summary, pDC-like cells were phenotypically and transcriptionally similar to conventional pDCs but also exhibited several cDC features, including efficient antigen processing and presentation.
Fig. 8. Functional heterogeneity of pDCs is developmentally encoded.
a–e, BM and splenic pDCs and pDC-like cells, analyzed in Zbtb46gfp/+ mice. a, Two-color histograms for the expression of CD317/SiglecH and CD317/Zbtb46-GFP on BM and splenic pDCs pregated on CD11c+MHCII– cells. BM and splenic pDCs are labeled in red, and pDC-like cells are marked in blue (n = 6 mice, with one representative experiment shown). b, IFN-α, measured from BM and splenic pDCs and pDC-like cells stimulated for 16 h with CpG-A or CpG-B, or left unstimulated (unst.), as indicated (n = 3 independent experiments with 2 technical replicates; each symbol represents a sample, and thin lines represent the mean ± s.e.m.). Expression of Zbtb46-GFP, MHC-II and CD86, determined in BM and splenic pDCs (red) and pDC-like cells (blue) 16 h after CpG-A stimulation. c,d Representative single-color histograms with mean fluorescence intensity (MFI) (c) and the compiled values from BM and splenic cells (d) (n = 3 independent experiments with 2 technical replicates; each symbol represents a sample, and thin lines represent the mean ± s.d.). e, Proliferation of OT-II T cells induced by sort-purified BM and splenic pDCs (red), pDC-like cells (blue) and splenic cDCs (green) (Methods). Cells were cultured in the presence of CpG-A, lipopolysaccharide and OVA-protein for 4 d. T cells stimulated with anti-CD3 and anti-CD28 (gray) were used as controls (n = 3 independent experiments with 2 technical replicates; each symbol represents a sample, and thin lines represent the mean ± s.e.m.). Statistical analysis was done with two-way ANOVA with Tukey post-test (b,d,e). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Discussion
Here we demonstrated that pDC development predominantly occurred from IL-7R+ LPs rather than from CDPs or the previously identified CSF1R–IL-7R– NPs2,23. Within the IL-7R+ LPs, Ly6D+SiglecH+ DP precursors gave rise almost exclusively to pDCs when cultured in the presence of FLT3L, thus suggesting that they might represent committed pre-pDCs that depend on the expression of IRF8 and are only two divisions away from maturity. Whereas Ly6D–SiglecH– DN progenitors were still uncommitted and showed the broadest lineage potential, Ly6D+SiglecH– SP cells were able to differentiate into pDCs and B cells, depending on the expression of IRF8 or EBF1, respectively. High expression of IRF8 within the SP cells determined loss of B cell potential and pDC lineage specification, which already occured in the absence of TCF4L, thus suggesting that the amount of IRF8 is key for pDC commitment.
How IRF8 is induced and regulated during pDC development remains an open question. Proliferation may be a key factor determining accumulation or dilution rates, as has been suggested for PU.1 during macrophage versus B cell differentiation42. BM niches rich in IL-7 would promote high proliferation of precursors, maintain low levels of IRF8 and allow for the induction of Pax5 by EBF1, thus leading to efficient B cell differentiation. Progressive expansion of the progenitor pool would also separate distal cells from the IL-7-rich BM niches, thereby limiting their proliferation and consequently leading to the accumulation of a specific IRF8 threshold promoting pDC development. A transcriptional mechanism may conceivably result in autoregulatory induction of IRF8 acting during pDC lineage specification, similarly to the one described for cDC1 (ref. 17). Beside the induction of IRF8 at the SP stage, transition to DP cells was characterized by the progressive expression of lineage-specific genes, including TCF4L, IRF7 and SpiB, concomitant with the downregulation of cell-cycle-associated genes. Further studies will be necessary to characterize the intrinsic and extrinsic players acting in the context of pDC development, given that different mechanisms are likely to shape differentiation along the lymphoid and the myeloid branch. Although the levels of ID2 and TCF4 were shown to be critical for the commitment toward cDC1 and pDCs along the myeloid differentiation pathway, repression of ID2 and cDC1 commitment may not be necessary along the lymphoid, IL-7R+ LP–derived developmental pathway, in which B cell potential instead must be prevented.
Neither DP cells nor conventional type 1 IFN–producing pDCs develop in mice lacking IRF8 or carrying the IRF8R249C mutation. In these mice, we observed the expansion of an alternative type of pDCs, which were unable to produce type 1 IFNs in response to CpG-B39, a pDC-defining hallmark9–11, and which expressed several features reminiscent of cDCs, including the ability to process and present antigens to T cells39. Nevertheless, caution is necessary when interpreting data from knockout mice. Although most pDCs arose from IL-7R+ LPs, they also developed from myeloid CDPs. In agreement with the greater contribution of lymphocyte progenitors than CDPs to the mature pDC pool, pDCs have a slower turnover rate than cDCs. Approximately 10% of mature pDCs are replaced within 2 d, a rate similar to that of T cells, whereas the turnover of cDCs is much faster, such that approximately 50% of the mature pool is replaced within the same timeframe43. These results support either a different number of cell divisions necessary for pDCs and cDCs to acquire maturity from CDPs, or, as is more likely and is supported by our data, for a different ontogeny of most pDCs and cDCs. Further supporting a major lymphoid developmental path of pDCs, a novel computational fate-mapping analysis performed on hematopoietic cells (FateID) revealed the presence of a common early progenitor shared by B cells and pDCs44.
The dual origin of pDCs may suggest a heterogeneous pool of mature cells able to perform the variety of functions ascribed to pDCs45. Our data on CDP- and IL-7R+ LP–derived pDCs as well as single-cell RNA sequencing highlighted this heterogeneity, revealing at least two subsets of mature pDCs: conventional pDCs and a small subset of pDC-like cells. pDC-like cells, which account for approximately 5–10% of the mature pDCs in the BM and spleen, were characterized by the concomitant expression of pDC-specific and cDC-associated transcripts. A small subset of peripheral blood mononuclear cells that combine features of pDCs and cDCs has recently also been identified in humans46,47. The use of Zbtb46gfp (ref. 48) mice was key in enabling us to identify and purify pDCs and pDC-like cells to perform a direct comparison of the two pDC subsets. Beyond phenotypic differences, conventional pDCs and pDC-like cells were functionally distinct. Although both subsets secreted type 1 IFNs in response to CpG-A stimulation, pDC-like cells were unable to do so when stimulated with CpG-B. Similarly to cDCs, pDC-like cells were better than conventional pDCs at antigen processing and presentation. This feature, which is atypical for pDCs41, may explain the ability of mature pDCs, including pDC-like cells, to induce antitumor responses in clinical trials on patients with melanoma49. In summary, our investigation of the developmental trajectory of pDCs led to the identification and characterization of a novel subset of antigen-presenting cells, pDC-like cells, which share transcriptional and functional features with both pDCs and cDCs.
Methods
Mice
All animals were bred and maintained in a specific-pathogen-free animal facility according to institutional guidelines (Veterinäramt BS, license number 2786_26606). Mice of the following genotypes were purchased from Jackson Laboratories: C57BL/6 J, CD11c-cre50, IRF8R249C (ref. 38) and Irf8f/f (ref. 51). Irf8−/− mice were generated by crossing Irf8f/f mice with CMV-Cre mice52. Il7−/− (ref. 53), Flt3l−/− (ref. 54), Ebf1hCD2 (ref. 55), Irf8egfp (ref. 56) and Zbtb46egfp48 were bred in house. For BM chimera experiments, B6.SJL57 mice were purchased from Jackson Laboratories. Unless otherwise indicated, experiments used sex- and age-matched littermates between 6 and 14 weeks of age.
Progenitor-cell harvest
BM was collected from femurs, tibia and pelvic bones. Bones were fragmented with a mortar and pestle, and debris was removed by filtration through a 70-μ m strainer. Red blood cells were lysed with ACK lysis buffer. Cells were counted, then stained for analysis or cell sorting. c-kithi progenitors were identified as Lin−B220−Ly6C−CD16/32−CD117hiCD135+ cells; CDPs were identified as Lin−CD16/32−B220−Ly6C−CD117int/lo CD135+CD115+CD127− cells13; pDC progenitors were gated as Lin−CD16/32−B220−Ly6C−CD117int/loCD135+CD115–CD127− (CSF1R–IL-7R–NPs)23 and CD317+B220+CD11c+CCR9− (CCR9–) cells22; IL-7R+ LPs were identified as Lin−CD16/32−B220−Ly6C−CD117int/loCD135+CD115−CD127+ cells; DN, SP and DP progenitors were included within the IL-7R+ LP gate and were defined by the expression of SiglecH, and Ly6D, as indicated in Fig. 2a (lineage markers CD3, CD19, Ter119, CD105 and NK1.1). The following gating strategy was used for bulk RNA sequencing: mature BM pDCs were gated as Lin–CD11c+SiglecH+ Ly6C+Ly6D+, whereas mature splenic pDCs were characterized and sorted as Lin–SiglecH+Bst2+Ly6C+. For single-cell RNA sequencing, pDCs from the BM and spleen were sorted as Lin–CD11c+BST2+SiglecH+. For cell sorting, a BD FACSAria II instrument with a custom built-in violet laser was used. Cells were sorted into PBS supplemented with 0.5% BSA and 2.5 mM EDTA. Cell purities of at least 95% were confirmed by post-sort analysis.
In vivo transfer
3 × 104 to 6 × 104 sort-purified CD45.1 or CD45.2 progenitors were co-injected intravenously at a 1:1 ratio into sublethally irradiated CD45.1/2 mice. The reconstitution ability of progenitors was assessed 4 d after injection, through flow cytometry.
Cell culture
5 × 102 to 5 × 103 sort-purified progenitors were cultured for 4 d in IMDM (Gibco) supplemented with 10% FCS (MPbio). To induce pDC or cDC development, cells were cultured in the presence of 100 ng/ml recombinant hFLT3L. To induce B cell differentiation, cells were cultured on irradiated OP9 stromal cells in the presence of 100 ng/ml recombinant hFLT3L (Peprotech), as previously described58,59. In some experiments, as indicated, 100 ng/ml recombinant mIL-7 was added. For Transwell experiments (Corning), 5.6 × 103 irradiated OP9 stromal cells were plated in the lower compartment, and 2 × 103 sort-purified progenitors were cultured either over stromal cells (lower chamber) or in the upper chamber.
Type 1 IFN ELISA
2 × 104 mature pDCs and 2 × 103 progenitor cells were sort-purified and stimulated with CpG-A 2216 (6 μ g/ml) or CpG-B 1826 (6 μ g/ml) for 16 h at day 0 or after 4 d of culture. Supernatants were analyzed with a Mouse IFN alpha Platinum ELISA Kit (eBioscience).
May–Gruenwald Giemsa staining
Cytospins of 5 × 103 sort-purified cells were stained with May–Gruenwald Giemsa (Sigma Aldrich), according to the manufacturer’s instructions. Slides were air-dried and sealed with Eukit quick-hardening mounting medium (Sigma Aldrich), and images were taken with a Leica DMI 4000 microscope.
In vitro proliferation assays
Total splenocytes or sort-purified progenitors were stained with CellTrace Violet (Invitrogen), according to the manufacturer’s guidelines. Total splenocytes were cultured on dishes coated with anti-CD3 and anti-CD28 (0.5 mg/ml) and were used as a positive control. Dilution of the cell dye was determined by flow cytometry.
OT-II proliferation assays
2 × 103 sort-purified conventional pDCs, pDC-like cells and splenic cDCs were co-cultured with 1 × 104 labeled OT-II CD4+ T cells in the presence of OVA protein (1 μ g/ml) and stimulated with LPS and CpG-A (both at 6 μ g/ml) or left unstimulated. OT-II T cells were labeled with CellTrace Violet (Thermo Fisher Scientific) according to the manufacturer’s instructions. Proliferation rates were measured after 4 d in culture. As controls, labeled and unlabeled OT-II T cells were cultured on anti-CD3 (1 µ g/ml) and anti-CD28 (0.5 µ g/ml) precoated wells.
Antibodies and flow cytometry
Cells were stained as previously described18 with the antibodies listed in the Supplementary Information Note. Cells were analyzed on a BD LSR Fortessa instrument, and data were analyzed with FlowJo X software (TreeStar).
Intracellular cytokine staining
For intracellular cytokine staining, cells were surface stained and subsequently fixed and permeabilized with a BD Cytofix/Cytoperm Kit.
Quantitative PCR
RNA of sort-purified progenitors was extracted, and cDNA was generated as previously described18. A KAPA SYBR Fast universal qPCR kit (KapaBiosystems) was used, and samples were run on an Applied Biosystems StepOnePlus qPCR machine. Primers are listed in the Supplementary Information Note.
Statistical analysis
Analysis of all data was done with paired two-tailed Student’s t test or one-way ANOVA with Tukey post-test with a 95% confidence interval (Prism, GraphPad Software). P < 0.05 was considered significant. *0.01 < P < 0.05; **0.001 < P < 0.01; ***P < 0.001; ****P < 0.0001.
RNA-seq analysis
Bulk RNA-seq
Total RNA was isolated from cells with an Ambion RNAqueous Micro Kit. RNA quality was assessed with a Fragment Analyzer. cDNA was prepared with a SMART-Seq v4 Ultra Low Input RNA Kit (Clontech). RNA libraries were prepared with a Nextera XT DNA Library Preparation Kit (Illumina). Indexed cDNA libraries were sequenced on an Illumina HiSeq 2500 machine and Illumina NexSeq 500. The sequence quality of the obtained single end reads (SR51) was assessed with the FastQC tool (version 0.11.3). Reads were mapped to the mouse genome assembly, version mm10 (http://genome.ucsc.edu/), with RNA-STAR (version 2.5.2a)60 with default parameters. As an exception, reporting for multimappers comprised only one hit in the final alignment files (outSAMmultNmax = 1) and filtering reads without evidence in the spliced junction table (outFilterType = ”BySJout”). All subsequent gene expression data analysis was done in the R software package (R Foundation for Statistical Computing). With RefSeq mRNA coordinates from UCSC (http://genome.ucsc.edu/, downloaded in December 2015) and the qCount function from the QuasR package (version 1.16.0)61, we quantified gene expression as the number of reads that started within any annotated exon of a gene. The differentially expressed genes were identified with the Generalized Linear Model (GLM) framework in the edgeR package (version 3.18.1)62. Factors indicating mouse IDs were included in the model as covariates. Genes with a false discovery rate < 0.05 and a minimum log2 FC of 1 were considered differentially expressed genes. Gene set enrichment analysis was performed with the function ‘camera’ from the edgeR package and with all gene sets from the Molecular Signature Database (MSigDB v5.2). We considered only sets containing more than ten genes and used later implementations of camera, in which correlations of genes within gene sets were set to a fixed value of 0.01. Data were corrected for batch effects (mouse ID) for visualization purposes. A linear model with a single factor indicating mouse ID was fitted to the log-transformed data (The ‘voom’ function from the edgeR package was used to transform the data, and the lmFit function from the limma package (version 3.32.10) was used to fit the model.) The residuals from this model were used for PCA and heat-map figures. The developmental heat map was generated as follows: the samples were first ordered according to their developmental stages, and all possible combinations of peak/switch models were fitted to the expression of individual genes with the GLM framework in the edgeR package, with mouse ID used as a covariate. For each gene, the best-fitting model was selected, and the gene was assigned to the corresponding category. All genes with a false discovery rate < 0.05 and a FC > 1 were selected and plotted in Fig. 4c and are listed in Supplementary Table 1. Subsequently, the generated list was filtered for transcription-factor log2 FC > 1.5 (TF, GO 0003677) and cell-surface-marker log2 FC > 3 (CS, GO:0009986), both filtered for log2 CPM > 1 in at least one set of replicates, and log2 FC > 1.5 values were individually plotted (Supplementary Fig. 4c,d). Each identified set of genes was tested for an enrichment in biological processes (package GO.db version 3.4.1) with the hypergeometric test implemented in the GOstats package (version 2.42.0).
Single-cell RNA-seq
BM and splenic pDCs were sorted from three mice and counted with a Countess II FL instrument (Life Technologies). 3,000 cells from each sample were loaded on a 10 × Genomics Chromium Single Cell Controller. Single-cell capture and cDNA and library preparation were performed with a Single Cell 3′ v2 Reagent Kit (10 × Genomics) according to the manufacturer’s instructions. Sequencing libraries were loaded on an Illumina NextSeq 500 instrument with high-output 75-cycle kits and paired-end sequenced with the following read lengths: read 1, 26 cycles; read 2, 8 cycles; read 3, 58 cycles. Single-cell sequencing files (basecalls) were processed with the Cell Ranger Single Cell Software Suite (version 2.0.0) to perform quality control, sample demultiplexing, barcode processing and single-cell 3′ gene counting (https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome/). Samples were first demultiplexed and then were aligned to the UCSC mouse (mm10) transcriptome and genome with ‘cellranger’ with default parameters for all six samples. UMI were counted with ‘cellranger count’. Samples were merged with the ‘cellranger aggregate’ procedure without downscaling. Further analysis was performed in R (version 3.4.0) with the scran (1.4.5) and scatter (1.4.0) packages by following the Bioconductor workflow (for version 3.5). Cells with log library sizes (or log total features) more than three median absolute deviations (MADs) below the median log library size (or log total features) were filtered out. Similarly, cells with a proportion of reads mapping to the mitochondrial genome more than three MADs above the median percentage of reads mapping to the mitochondrial genome were removed. Low-abundance genes with average log2 CPM. counts < 0.0023 were filtered out. This threshold was estimated from the distribution of average log2 CPM. counts after fitting two normal distributions (assuming two populations of genes: not expressed – background, and expressed) with Mclust function (mclust version 5.3) and choosing a threshold of P = 0.05 for background population. Expression values of 11,753 genes for 14,744 cells were kept. The raw UMI counts were normalized with the size factors estimated from pools of cells to avoid dominance of zeros in the matrix63. A mean-dependent trend was fitted to the variances of the log expression values of endogenous genes to distinguish between genuine biological variability and technical noise, under the assumption that most genes are not differentially expressed across cells, and their variance is mainly technical (trendVar function with ‘loess’ trend and span of 0.01 to better fit the sparse data). Afterward, the fitted technical noise was subtracted, the genes were sorted on the basis of the biological components of their variance, and those with a variance larger than 0.1 were used for clustering of cells and PCA (n = 148). The clustering of cells into putative subpopulations was done on log expression values (hierarchical clustering on the Euclidean distances between cells, with Ward’s criterion to minimize the total variance within each cluster). The clusters of cells were identified by applying a dynamic tree cut64, which resulted in eight putative subpopulations. Afterward, the marker genes specific for each cluster were identified with the findMarkers function (scran package), which fits a linear model to the log-transformed expression values for each gene with the imma framework65. The expression profiles of individual clusters were also compared in a pairwise analysis (P values in those analyses were considered only as ranks, because the same data were used for cluster identification and statistical testing).
Supplementary Material
Reporting Summary.
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Acknowledgements
We thank A. G. Rolink (DBM University of Basel), K. M. Murphy (Washington University in St. Louis), and P. Tsapogas (DBM University of Basel) for sharing reagents, expertise and discussions; B. Lambrecht and M. Guilliams (VIB-UGent Center for Inflammation Research) for sharing data and reagents; M. Busslinger (Research Institute of Molecular Pathology IMP, Vienna) for kindly providing us with Ebf1hCD2 reporter mice; D. Schreiner and C. King for constant input and critical reading of the manuscript; D. Labes (DBM University of Basel), K. Eschbach and C. Beisel (Department of Biosystems Science and Engineering (D-BSSE) University of Zurich, Basel) for excellent technical support; the DBM-Microscopy Core Facility; A. Brülhart and all the animal caretakers of WRO1060; and C. Cannavo for IT support. This study was supported by SNF project number PP00P3_150714 and by the Novartis Foundation for medical-biological research n.16A052. This work is dedicated to the memory of Ton Rolink.
Footnotes
Author contributions
P.R.F. and R.T. designed the project, performed experiments, interpreted the data and wrote the manuscript. L.A.-S. and R.I. analyzed the RNA-sequencing data and contributed to writing the paper. A.E. and G.E.G.-R. performed experiments. All authors agreed to the submission of the manuscript for publication.
Competing interests
The authors declare no competing interests.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Methods
Methods, including statements of data availability and any associated accession codes and references, are available at https://doi.org/10.1038/s41590-018-0136-9.
Data availability
The RNA-seq data generated in the current study are available in the Gene Expression Omnibus database under accession code GSE114315.
References
- 1.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 2.Guilliams M, et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol. 2014;14:571–578. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colonna M, Trinchieri G, Liu YJ. Plasmacytoid dendritic cells in immunity. Nat Immunol. 2004;5:1219–1226. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- 4.Murphy TL, et al. Transcriptional control of dendritic cell development. Annu Rev Immunol. 2016;34:93–119. doi: 10.1146/annurev-immunol-032713-120204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mildner A, Jung S. Development and function of dendritic cell subsets. Immunity. 2014;40:642–656. doi: 10.1016/j.immuni.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 6.Tamura T, et al. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki S, et al. Critical roles of interferon regulatory factor 4 in CD11bhighCD8alpha– dendritic cell development. Proc Natl Acad Sci USA. 2004;101:8981–8986. doi: 10.1073/pnas.0402139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belz GT, Nutt SL. Transcriptional programming of the dendritic cell network. Nat Rev Immunol. 2012;12:101–113. doi: 10.1038/nri3149. [DOI] [PubMed] [Google Scholar]
- 9.Perussia B, Fanning V, Trinchieri G. A leukocyte subset bearing HLA-DR antigens is responsible for in vitro alpha interferon production in response to viruses. Nat Immun Cell Growth Regul. 1985;4:120–137. [PubMed] [Google Scholar]
- 10.Siegal FP, et al. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 11.Cella M, et al. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat Med. 1999;5:919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- 12.Naik SH, et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- 13.Liu K, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karsunky H, Merad M, Cozzio A, Weissman IL, Manz MG. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J Exp Med. 2003;198:305–313. doi: 10.1084/jem.20030323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Amico A, Wu L. The early progenitors of mouse dendritic cells and plasmacytoid predendritic cells are within the bone marrow hemopoietic precursors expressing Flt3. J Exp Med. 2003;198:293–303. doi: 10.1084/jem.20030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sathe P, Vremec D, Wu L, Corcoran L, Shortman K. Convergent differentiation: myeloid and lymphoid pathways to murine plasmacytoid dendritic cells. Blood. 2013;121:11–19. doi: 10.1182/blood-2012-02-413336. [DOI] [PubMed] [Google Scholar]
- 17.Grajales-Reyes GE, et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α+ conventional DC clonogenic progenitor. Nat Immunol. 2015;16:708–717. doi: 10.1038/ni.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tussiwand R, et al. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity. 2015;42:916–928. doi: 10.1016/j.immuni.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller JC, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13:888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shigematsu H, et al. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity. 2004;21:43–53. doi: 10.1016/j.immuni.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Pelayo R, et al. Derivation of 2 categories of plasmacytoid dendritic cells in murine bone marrow. Blood. 2005;105:4407–4415. doi: 10.1182/blood-2004-07-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlitzer A, et al. Identification of CCR9– murine plasmacytoid DC precursors with plasticity to differentiate into conventional DCs. Blood. 2011;117:6562–6570. doi: 10.1182/blood-2010-12-326678. [DOI] [PubMed] [Google Scholar]
- 23.Onai N, et al. A clonogenic progenitor with prominent plasmacytoid dendritic cell developmental potential. Immunity. 2013;38:943–957. doi: 10.1016/j.immuni.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Schlitzer A, et al. Tissue-specific differentiation of a circulating CCR9– pDC-like common dendritic cell precursor. Blood. 2012;119:6063–6071. doi: 10.1182/blood-2012-03-418400. [DOI] [PubMed] [Google Scholar]
- 25.Cisse B, et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh HS, Cisse B, Bunin A, Lewis KL, Reizis B. Continuous expression of the transcription factor e2-2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity. 2010;33:905–916. doi: 10.1016/j.immuni.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grajkowska LT, et al. Isoform-specific expression and feedback regulation of E protein TCF4 control dendritic cell lineage specification. Immunity. 2017;46:65–77. doi: 10.1016/j.immuni.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosh HS, et al. ETO family protein Mtg16 regulates the balance of dendritic cell subsets by repressing Id2. J Exp Med. 2014;211:1623–1635. doi: 10.1084/jem.20132121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott CL, et al. The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J Exp Med. 2016;213:897–911. doi: 10.1084/jem.20151715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X, et al. Transcription factor Zeb2 regulates commitment to plasmacytoid dendritic cell and monocyte fate. Proc Natl Acad Sci USA. 2016;113:14775–14780. doi: 10.1073/pnas.1611408114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 32.Swiecki M, et al. Type I interferon negatively controls plasmacytoid dendritic cell numbers in vivo. J Exp Med. 2011;208:2367–2374. doi: 10.1084/jem.20110654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blasius AL, et al. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J Immunol. 2006;177:3260–3265. doi: 10.4049/jimmunol.177.5.3260. [DOI] [PubMed] [Google Scholar]
- 34.Schlenner SM, et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 35.Loschko J, et al. Inducible targeting of cDCs and their subsets in vivo. J Immunol Methods. 2016;434:32–38. doi: 10.1016/j.jim.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 37.Kikuchi K, Lai AY, Hsu CL, Kondo M. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J Exp Med. 2005;201:1197–1203. doi: 10.1084/jem.20050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tailor P, Tamura T, Morse HC, III, Ozato K. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood. 2008;111:1942–1945. doi: 10.1182/blood-2007-07-100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sichien D, et al. IRF8 transcription factor controls survival and function of terminally differentiated conventional and plasmacytoid dendritic cells, respectively. Immunity. 2016;45:626–640. doi: 10.1016/j.immuni.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 40.Bar-On L, et al. CX3CR1+ CD8alpha+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proc Natl Acad Sci USA. 2010;107:14745–14750. doi: 10.1073/pnas.1001562107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krug A, et al. Interferon-producing cells fail to induce proliferation of naive T cells but can promote expansion and T helper 1 differentiation of antigen-experienced unpolarized T cells. J Exp Med. 2003;197:899–906. doi: 10.1084/jem.20021091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kueh HY, Champhekar A, Nutt SL, Elowitz MB, Rothenberg EV. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science. 2013;341:670–673. doi: 10.1126/science.1240831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen M, Huang L, Shabier Z, Wang J. Regulation of the lifespan in dendritic cell subsets. Mol Immunol. 2007;44:2558–2565. doi: 10.1016/j.molimm.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herman JS, Sagar, Grün D. FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat Methods. 2018;15:379–386. doi: 10.1038/nmeth.4662. [DOI] [PubMed] [Google Scholar]
- 45.Swiecki M, Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol. 2015;15:471–485. doi: 10.1038/nri3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.See P, et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science. 2017;356:eaag3009. doi: 10.1126/science.aag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villani AC, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Satpathy AT, et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med. 2012;209:1135–1152. doi: 10.1084/jem.20120030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tel J, et al. Natural human plasmacytoid dendritic cells induce antigen-specific T-cell responses in melanoma patients. Cancer Res. 2013;73:1063–1075. doi: 10.1158/0008-5472.CAN-12-2583. [DOI] [PubMed] [Google Scholar]
- 50.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J Exp Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng J, et al. IFN regulatory factor 8 restricts the size of the marginal zone and follicular B cell pools. J Immunol. 2011;186:1458–1466. doi: 10.4049/jimmunol.1001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Freeden-Jeffry U, et al. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKenna HJ, et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–3497. [PubMed] [Google Scholar]
- 55.Vilagos B, et al. Essential role of EBF1 in the generation and function of distinct mature B cell types. J Exp Med. 2012;209:775–792. doi: 10.1084/jem.20112422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H, et al. IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood. 2008;112:4028–4038. doi: 10.1182/blood-2008-01-129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Janowska-Wieczorek A, et al. Platelet-derived microparticles bind to hematopoietic stem/progenitor cells and enhance their engraftment. Blood. 2001;98:3143–3149. doi: 10.1182/blood.v98.10.3143. [DOI] [PubMed] [Google Scholar]
- 58.Nakano T, Kodama H, Honjo T. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science. 1994;265:1098–1101. doi: 10.1126/science.8066449. [DOI] [PubMed] [Google Scholar]
- 59.Balciunaite G, Ceredig R, Massa S, Rolink AGA. A B220+ CD117+ CD19- hematopoietic progenitor with potent lymphoid and myeloid developmental potential. Eur J Immunol. 2005;35:2019–2030. doi: 10.1002/eji.200526318. [DOI] [PubMed] [Google Scholar]
- 60.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gaidatzis D, Lerch A, Hahne F, Stadler MB. QuasR: quantification and annotation of short reads in R. Bioinformatics. 2015;31:1130–1132. doi: 10.1093/bioinformatics/btu781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lun AT, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 65.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated in the current study are available in the Gene Expression Omnibus database under accession code GSE114315.