Abstract
HIV-1 Gag metamorphoses inside each virion from an immature lattice that forms during viral production to a mature capsid that drives infection. Here we show that the immature lattice is required to concentrate the cellular metabolite inositol hexakisphosphate (IP6) into virions to catalyse mature capsid assembly. Disabling HIV-1’s ability to enrich IP6 does not prevent immature lattice formation or production of the virus. However, without sufficient IP6 molecules inside each virion, HIV-1 can no longer build a stable capsid and fails to become infectious. IP6 cannot be replaced by other inositol phosphate (IP) molecules, as substitution with other IPs profoundly slows mature assembly kinetics and results in virions with gross morphological defects. Our results demonstrate that whilst HIV-1 can become independent of IP6 for immature assembly, it remains dependent upon the metabolite for mature capsid formation.
Introduction
IP6 has recently been identified as a co-factor in HIV-1 replication[1, 2], but whether HIV-1 can become independent of this metabolite is unknown. Genetic knockout (KO) of kinases Inositol polyphosphate multikinase (IPMK) or Inositol-pentakisphosphate 2-kinase (IPPK), which catalyse IP6 biosynthesis, modestly decreases virus particle production and infectivity[3–5]. However, HIV-1 packages IP5 into virions instead of IP6 in IPPK KO cells[3], illustrating that without knowing how efficiently HIV-1 utilises different inositol phosphate (IP) species and what these IP levels are in different cell lines, any dependence of the virus on IP6 is difficult to unpick. IP6 levels have been measured at 22 – 44 μM in human tissues[6] and in rat tissues between 11 – 16 μM[7]. The minimum concentration of IP6 required for HIV-1 assembly is unknown, but the metabolite is actively enriched into virions during assembly[3]. This enrichment is mediated by the binding of IP6 to two lysine rings – formed by residues 158 and 227 of the CA domain of Gag – within hexamers of the immature Gag lattice [3, 8]. IP6 occupies the centre of each hexamer, coordinating the two rings and promoting immature VLP assembly by stabilizing the immature Gag lattice [1]. Mutation of either K158 or K227 prevents IP6 enrichment, decreases virion production and results in a profound loss of infectivity[3]. Importantly, loss of IP6 enrichment is a causative factor in the replication defects exhibited by K158 and K227 mutants, as passaging either mutant leads to the acquisition of second-site mutations, such as T8I in the spacer peptide 1 (SP1) domain of Gag, which concomitantly restore IP6 incorporation, particle production and infectivity[8]. These second-site mutations act by stabilizing the immature Gag lattice.
Once HIV-1 virions have budded, they undergo a maturation step in which the immature Gag lattice is cleaved by the viral protease and the newly released capsid protein (CA) assembles into a mature capsid made up of hexamers and 12 pentamers (referred to collectively as capsomers). The capsomers of the mature capsid lattice also have two charged rings (formed by R18 and K25) that bind IP6[1–3, 9]. IP6 binding stabilises the capsomers and promotes capsid assembly and stability in an analogous manner to IP6 binding to Gag hexamers in the immature lattice[1, 2]. The fact that reduced IP6 incorporation in K158 or K227 mutants decreases virus infectivity as well as particle production highlights that IP6 is important for making stable capsids. However, it is not possible to rule out indirect effects as these mutants have altered immature lattice stability, which is known to affect Gag processing and maturation[8, 10, 11]. Importantly, the IP6 binding site in mature capsomers was first identified as the electrostatic pore through which nucleotides are imported to drive reverse transcription and protect newly synthesised DNA from sensors and nucleases[12]. Thus, the charged rings in mature capsomers have multiple roles that have not been disentangled.
Whilst previous reports establish that IP6 is an important HIV-1 cofactor, it remains unclear if it is essential for HIV-1 replication and if so whether it is necessary for assembly of the immature Gag lattice or the mature capsid, or both. Here we show that HIV-1 can become independent of IP6 at the level of immature Gag lattice assembly, Gag processing and virion production. However, preventing enrichment of IP6 into virions, or reducing the concentration of both IP5 and IP6 in producer cells below a threshold level, leads to aberrant virion morphologies and dramatically reduces HIV-1 infectivity. These results suggest that the presence of lysine rings in the immature lattice are a viral adaptation to ensure IP6 enrichment into virions, rather than IP6 being essential to build an immature HIV-1 Gag lattice. We propose that HIV-1 is dependent upon IP6 because of its requirement in mature capsid formation, where it is necessary to build capsomers and stabilise the strongly charged pores necessary for nucleotide import.
Results
Immature VLPs assemble at 100-fold lower IP6 concentrations than mature capsids
IP6 has been shown to promote the assembly of both immature VLPs and mature capsids[1]. We sought to quantify the relative dependence of these two assembly reactions on IP6. The kinetics of HIV-1 capsid assembly can be studied by monitoring the increase in light scattering during oligomerisation[13]. Assembly results in the formation of structures of different sizes and shapes; these include, in the presence of IP6, either immature-like VLPs or mature-like capsids depending on the reaction conditions[1, 14]. We measured the in vitro assembly kinetics of either DMA-CANC protein [comprising part of the matrix (MA) domain and all of the CA and nucleocapsid (NC) domains] or CA. The maximum assembly value (Amax), given by maximal light scattering at a range of IP6 concentrations, was used to calculate the effective concentration needed for half-maximal assembly (EC50). Fitting the data for mature CA assembly gives an EC50MA for WT HIV-1 of > 200 μM (Supplementary Figure 1A&B). Negative stain electron microscopy (EM) was used to confirm that the assembled particles were morphologically consistent with HIV-1 capsids (Supplementary Figure 1C). We repeated these experiments using DMA-CANC in the presence of RNA to obtain an estimate of the IP6 concentration needed for immature assembly. Immature assembly proceeded with sigmoidal assembly kinetics (Supplementary Figure 1D), as expected for viral capsids or spherical polymers like immature VLPs[15]. The fitted value for half-maximal immature assembly (EC50IM) was ~ 3 μM IP6 (Supplementary Figure 1E). Again, negative stain EM was used to confirm that assemblies were as expected for immature VLPs (Supplementary Figure 1F). However, assembly is a multistep process with a complex dependence on the concentration of capsid protein and ligands, which will be distributed between free monomer, nucleating structures, assembly intermediates and full VLPs [15]. Thus, we stress that the fitted values for IP6 concentration dependence are only indicative of the relative values necessary for immature vs mature particle formation. Nevertheless, these highlight that immature VLP assembly occurs at IP6 concentrations ~ 100-fold below that required for mature VLP assembly and are in agreement with previous end-point data[1, 14]. As the cellular concentration of IP6 is between 22-44 μM [6], this means that while immature assembly can occur efficiently in producer cells, HIV-1 must have a mechanism to enrich IP6 into virions for mature capsids to form post-budding. Moreover, as capsid assembly requires much higher IP6 concentrations, it is likely to be much more sensitive than immature assembly to a reduction in cellular IP6 bioavailability.
Altering IP composition in producer cells impairs HIV-1 particle production, Gag processing and infectivity
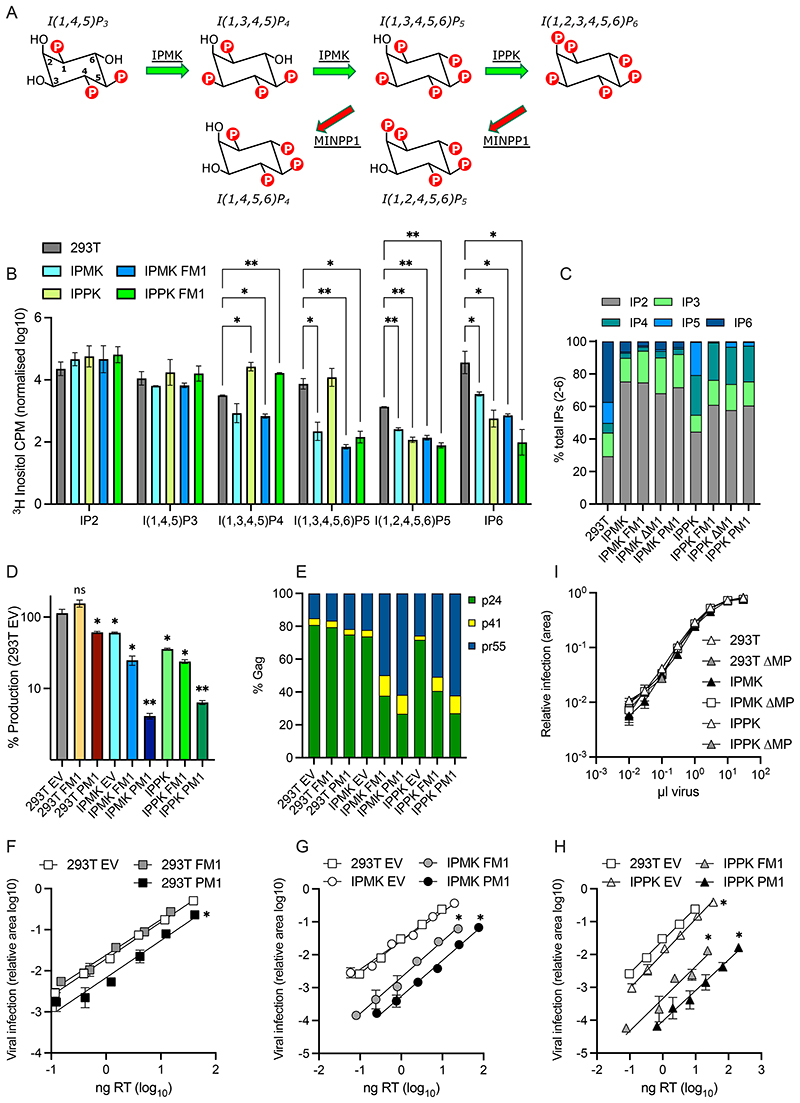
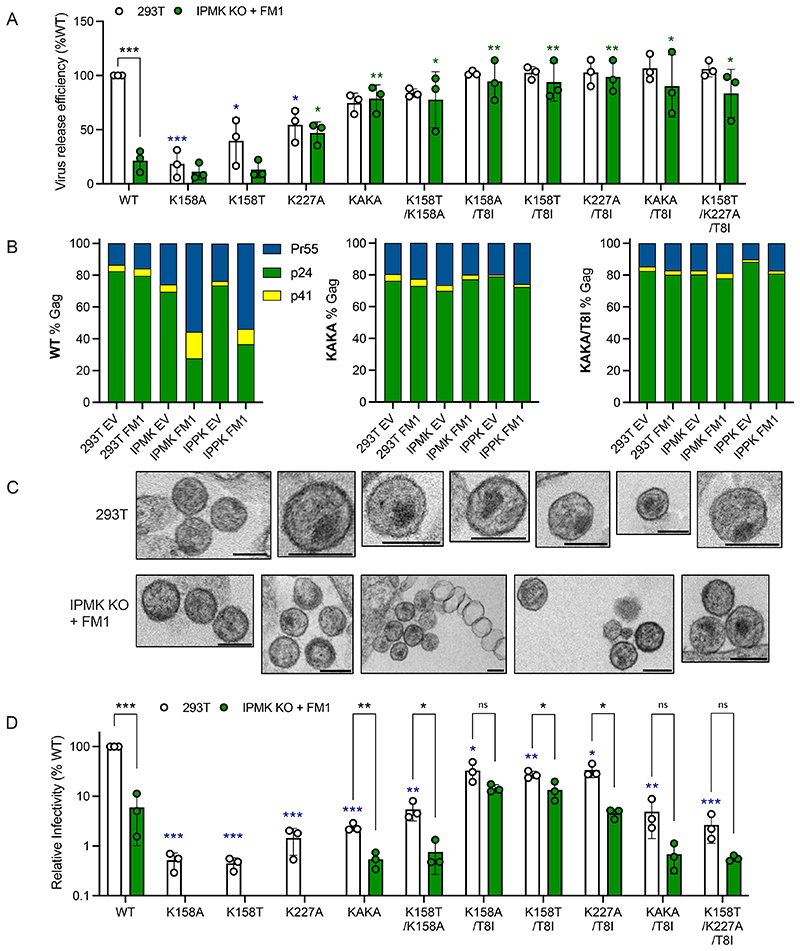
To directly link IP availability to specific aspects of HIV-1 assembly and infectivity, we manipulated IP levels in producer and target cells. Key roles in IP6 biosynthesis have been identified for the multikinase IPMK, which synthesises IP4 and IP5, and the kinase IPPK, which phosphorylates IP5 to IP6 (Figure 1A). Steady-state cellular levels of IP5 and IP6 are also regulated by the phosphatase Minpp1 [16], which hydrolyses 3’ phosphates (Figure 1A). Previously, we have shown that knockout of IPMK reduces but does not eliminate IP6, whilst in IPPK KOs IP6 levels are reduced but IP5 is unaffected and is incorporated into virions instead[3]. Over-expression of the phosphatase Minpp1 in IPPK KOs has been reported to reduce HIV-1 infectivity but how this alters the relative proportion of different IP species was not determined[4]. We therefore overexpressed a full-length construct of Minpp1 (FM1) in WT, IPMK KO and IPPK KO cells supplemented with tritiated inositol and used SAX-HPLC fractionation to determine their IP profile. We observed that Minpp1 (FM1) drove a dramatic reduction in I(1,3,4,5,6)P5 levels in IPPK KO cells and decreased IP6 in both kinase KOs (Figure 1B). We used the relative changes in IP levels to calculate the probable concentrations in the different cell lines, based on published quantitative values from 293T cells (Supplementary Table 1). There were marked differences between the cell lines, with IP4 levels substantially elevated in IPPK KOs + Minpp1 but not in IPMK KO + Minpp1 cells. The different IP profiles are most apparent when plotting the relative abundance of each species as a fraction of the total (Figure 1C). Indeed, IP4 levels in IPPK KO + Minpp1 cells are > 100-fold higher than the levels of IP5 and IP6. This is significant as production of HIV-1 has been shown to be reduced in IPPK KO + Minpp1 cells [4], yet IP4 can catalyse assembly of immature VLPs in vitro[1].
Figure 1. Altering IP composition in producer cells changes HIV-1 particle production, Gag processing and infectivity.
(A) Simplified biosynthetic scheme for IP6. (B-C) 293Ts, CRISPR KOs for IPMK or IPPK[3], or KOs over-expressing full-length (FM1), plasma-membrane targeted (PM1) or cytosol-targeted (DM1) Minpp1 were grown in tritiated inositol. IP species were extracted and separated by SAX-HPLC as previously described[31]. (B) The counts per minute (CPM) of each inositol species normalised to total lipid is shown. Error bars depict mean CPM ± SEM from at least two independent experiments. An unpaired t test against 293Ts was used for statistical analysis of IPs in each cell line and only significant differences indicated (P = 0.05 (*), < 0.005 (**)). (C) The proportion of each IP species as a fraction of the total IP2-IP6 concentration. (D) HIV-1 production, as measured by RT activity, expressed as a percentage of virus production in 293T cells. An unpaired t test against 293T EV was used for statistical analysis and significant differences indicated (P = 0.05 (*), < 0.005 (**)). (E) Gag cleavage efficiency in purified virions, calculated as the percentage of p24 (CA), p41 and Pr55Gag. (F-H) Infectivity of viruses from (D) plotted against quantity of input virus. Error bars depict the SEM from three independent experiments. Nonlinear regression was used to compare Y-intercepts against 293T EV and significant differences indicated (P < 0.05 (*)). (I) Infection of 293Ts, CRISPR KOs for IPMK or IPPK, or KOs over-expressing cytosol-targeted (ΔM1) Minpp1 by HIV-1. Error bars depict the SEM from three independent experiments.
Despite the impact of FM1 expression on IP5 and IP6 levels in kinase KO cells, there was only a small reduction in HIV-1 production (Figure 1D). We considered that this may be because Minpp1 is localized to the endoplasmic reticulum (ER), whereas HIV-1 buds from the plasma membrane and should thus be impacted most by plasma membrane IP concentrations. We therefore designed a Minpp1 variant that was re-localised to the plasma membrane (PM1) by removing the signal peptide (residues 1 – 28) and ER retention sequence (C-terminal residues 484 onwards), which results in cytoplasmic Minpp1 expression[16]; this modified Minpp1 was fused to a construct encoding GFP and the first 15 residues of Gnai2, which contain motifs that direct N-myristoylation and S-palmitoylation [17]. As expected, we observed little difference in the total cell IP profile of PM1-expressing cells compared to unmodified Minpp1 (Figure 1C). However, compared to cells expressing FM1, there was a dramatic impact on HIV-1 production in both IPMK and IPPK KOs and an exacerbated Gag processing defect in which there was an accumulation of Pr55Gag (Figure 1D&E, Supplementary Figure 2). Expression of PM1 also potentiated the infectivity defect of viruses produced in IPPK and IPMK KO cells (Figure 1F-H). Most notably, PM1 was sufficient to reduce the infectivity of viruses produced in WT 293T cells (Figure 1F). This result is in contrast to previous data showing that Minpp1 overexpression in WT 293Ts does not alter the production of infectious HIV-1[4] and highlights that HIV-1 likely incorporates IP6 at the plasma membrane during immature Gag lattice assembly and viral budding. Our results agree with data showing that viruses produced in Minpp1 over-expressing cells give reduced infection [4] but reveal this is because of independent defects in both production and infectivity.
Next, we investigated whether IP6 is important in target cells. Previous studies have tested this in the context of IPMK and IPPK KOs[3, 5] but these cells retain combined IP5 and IP6 levels of > 5 μM (Supplementary Table 1), or by overexpressing a full-length Minpp1 that is localised to the ER[4]. Because our data suggest that local IP6 concentrations are important, we modified our PM1 construct to remove the Gnai2 sequence and re-localise Minpp1 to the cytoplasm. Overexpression of this construct, denoted DM1, led to reduced IP5 and IP6 levels in IPMK and IPPK KO cells (Figure 1C). We infected each cell line with HIV-1 produced in unmodified 293T cells. Remarkably, despite IPMK KO + DM1 and IPPK + DM1 cells having ~ 100-fold lower levels of IP5 and IP6 there was no impact on HIV-1 infection (Figure 1I). This further supports that HIV-1 infection is not dependent upon IP6 in target cells[3–5].
HIV-1 particles produced in IP5/IP6-depleted cells display aberrant morphology and lack a condensed capsid
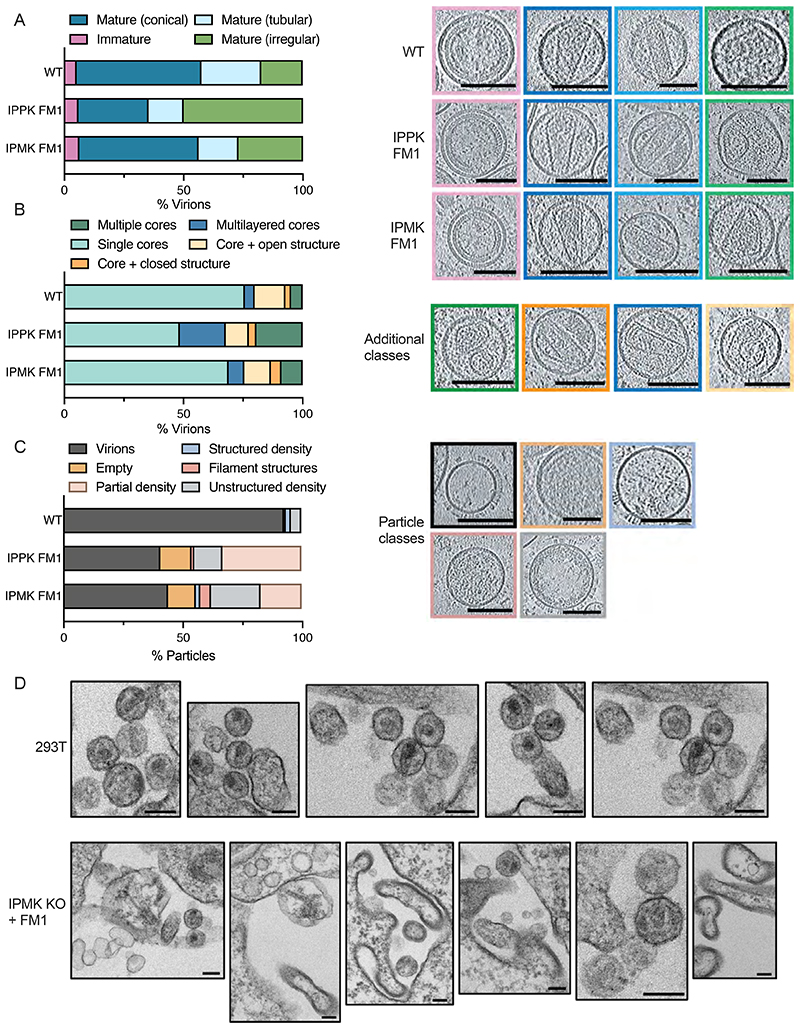
To investigate how a lack of IP5 and IP6 in producer cells might alter HIV-1 infectivity, we used cryoET to analyse particles produced in kinase KO cells over-expressing Minpp1 (FM1). Surprisingly, classifying those particles that possessed Gag structures revealed only a mild reduction in the proportion that had mature conical cores in the case of virions produced in IPPK KO + FM1 cells and no reduction in those from IPMK KO + FM1 cells (Figure 2A). Further classification revealed the increased presence of additional structures in virions from either KO cell line when compared to those from WT cells, which may be due to decreased lattice stability in the presence of reduced IP6 (Figure 2B). However, the magnitude of the morphology defects revealed by cryoET does not explain the severe infectivity defect of virions produced in the KO + FM1 cells. We noted that there were far fewer virions on grids prepared from KO + FM1 cells than expected based on their quantification by RT, suggesting the presence of particles that contain viral proteins but no visible Gag ultrastructure. In addition, virions from KO + FM1 cells were more likely to be empty or contain only partial density (Figure 2C).
Figure 2. HIV-1 particles produced in IP5/IP6-depleted cells display aberrant morphology and lack a condensed capsid.
(A-C) Cryo-ET on indicated HIV-1 mutants produced in IPMK or IPPK KO cells with Minpp1 overexpression. Tilt series were collected and reconstructions performed to assess capsid morphology. A total of 131 WT, 34 IPPK + FM1 and 48 IPMK + FM1 particles were analysed. (A) Virions were classified into the indicated categories: Immature (pink), Mature Conical (dark blue), Mature Tubular (light blue), Mature Irregular (green). Slices through representative tomograms of the virions are shown together with quantification. Scale bars, 100 nm. (B) Virions with mature lattices were further subdivided into: Multiple Cores (green), Single Cores (cyan), Cores with additional closed structure (orange), Cores with additional open structure (light orange), Multilayered Cores (blue). Slices through example tomograms of the virions are shown together with quantification. Scale bars, 100 nm. (C) All particles that were categorized as VSV-G positive (as indicated by the spikes on the surface of the particles) on the grid but did not contain a clear assembled lattice were categorized into: Virions (black), Partial density (gray), Empty (orange), Structured density (blue), Filament structures (dark pink), Partial density (light pink). Slices through representative tomograms of the virions are shown together with quantification. Scale bars, 100 nm. (D) Thin-section electron-microscopy of HIV-1 virions produced in 293T cells or IPMK KO cells over-expressing Minpp1. Scale bar = 120 nm.
To better understand the nature of the aberrant or empty particles produced by IPMK KO + FM1 cells, we used transmission EM (TEM) to examine viral assembly and budding. We observed profound morphological differences in the particles produced in these cells compared to WT 293T cells (Figure 2D). There was an abundance of large, irregular particles and few spherical VLPs with electron-dense material indicative of an assembled Gag or capsid lattice. Taken together, the TEM data suggest that lack of IP6, or substitution of IP6 for smaller IPs, results in the production of aberrant particles lacking an organised capsid structure.
Immature particles assemble more slowly with smaller IPs, are less stable and display altered Gag processing relative to particles assembled in the presence of IP5 or IP6
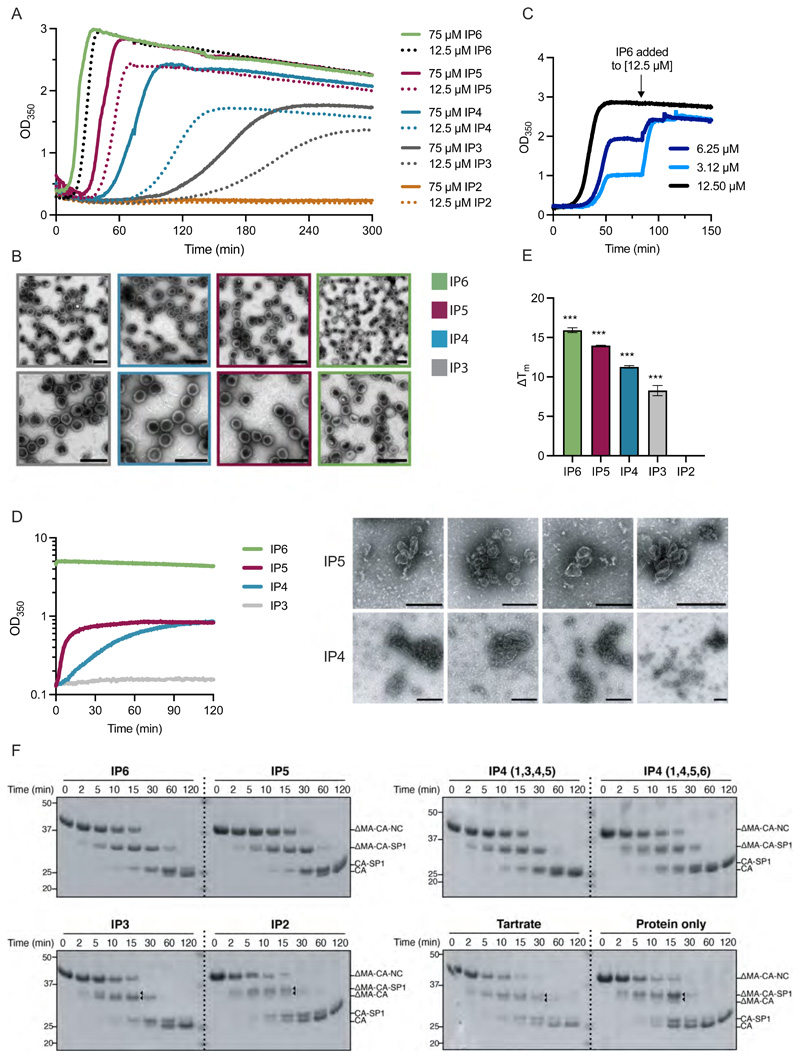
Next, we compared the ability of different IPs to promote in vitro assembly of immature and mature particles. At an IP concentration of 50 μM, IP5 and IP6, but not IP2-4, promoted immature assembly, with IP6 promoting substantially faster assembly with a smaller time lag (Supplementary Figure 3A). Increasing IP concentration (Supplementary Figure 3B-D), DMA-CANC concentration (Supplementary Figure 3E-G) or temperature to 37 °C (Figure 3A&B) allowed assembly using IP3 to IP6, albeit with decreased kinetics and efficiency. Increasing the stoichiometry of 1 IP6 per immature hexamer did not alter assembly appreciably (Figure 3A), while sub-stoichiometric ratios (e.g. 0.25:1) gave reduced assembly that could be recovered by restoring a 1:1 ratio (Figure 3C). In contrast to IP6, increasing the concentration of IP3 and IP4 beyond a 1:1 stoichiometry increased both the rate and efficiency of assembly (Figure 3A), likely because they have a lower affinity, therefore, a higher concentration is needed to achieve binding. Indeed, assembly in the presence of IP3 at 12.5 μM (1:1 ratio) plateaued prematurely and could only be restored by adding 12.5 μM IP6 (Supplementary Figure 3H) or 500 μM IP3 (Supplementary Figure 3I). Thus, a stoichiometry of 1 IP molecule per 1 immature hexamer is sufficient for optimal immature particle assembly if the affinity is sufficient to drive the equilibrium to full occupancy. Next, we compared the ability of different IPs to promote assembly of mature particles. We observed that even at high IP concentration (2.5 mM), only IP6 promoted rapid mature particle assembly (Figure 3D). IP5 and IP4 promoted some assembly, but with little evidence of regular conical capsids in micrographs. Taken together, the data indicate that mature assembly is significantly more impacted than immature assembly when forced to utilise smaller IPs
Figure 3. Immature particles assemble more slowly with smaller IPs, are less stable and have altered Gag processing relative to particles assembled in the presence of IP5 or IP6.
(A) In vitro assembly of immature particles using recombinant DMA-CANC protein. 7.5 μM RNA was added to 75 μM DMA-CANC and assembly monitored through light scattering changes at 350 nm at 37 °C. Indicated IPs were added at equimolar DMA-CANC concentration or 12.5 μM, to achieve a stoichiometry with immature hexamers of 6:1 or 1:1, respectively. Representative of two experiments. (B) EM images of negative-stained samples of the final assembly reactions shown in (A). Scale bars are 200 nm. (C) In vitro assembly reaction of immature particles as in (A) except IP6 was added at stoichiometric ratios with respect to immature hexamers of 1:1 (12.5 μM), 0.5:1 (6.25 μM) or 0.25:1 (3.12 μM). Once assembly had plateaued, additional IP6 was added to achieve 1:1 stoichiometry, resulting in similar final yields by light scattering. Representative of two experiments. (D) In vitro assembly of mature particles using 150μM recombinant CA protein and 2.5 mM of the indicated IP. EM images of negative-stained samples of the final assembly reactions are shown to the right. Size bars are 200 nm. Representative of at least three experiments. (E) Thermostability of in vitro assembled particles with 7.5 μM RNA, 75 μM ΔMA-CANC and 12.5 μM IP was measured by differential scanning fluorimetry (DSF). The change in melt temperature (ΔTm) was calculated with respect to the thermostability of unassembled ΔMA-CANC protein. An unpaired t test against unassembled ΔMA-CANC was used for statistical analysis and significant differences indicated (P < 0.0005 (***)). (F) In vitro assembled particles as in (E) but with an additional condition including 375 μM Tartrate rather than IP and unassembled DMA-CANC were incubated with recombinant HIV-1 protease for the indicated times and analysed by SDS PAGE and western blot. The probable cleavage products, based on size, are indicated. Triangles point to additional cleavage products that are not present in particles assembled with IP6.
Usage of different IP molecules would be predicted to alter not just assembly kinetics but also the stability of the lattice once formed. To test this, we measured the thermal stability of immature particles assembled with different IPs. Samples from completed assembly reactions carried out as shown in Figure 3A were analysed by nanoscale differential scanning fluorimetry (nanoDSF) and the melt temperature (TM) compared to unassembled starting material. Immature particles assembled in the presence of IP3 and IP4 were less stable than those assembled in the presence of IP5 or IP6 (Figure 3E). A limitation of this approach is that assembled material is not homogenous; however, there is a clear correlation between the efficiency of assembly and thermostability when using different IPs. Reduced stability may contribute to the altered Gag processing observed in virions produced by IPMK or IPPK KO cells over-expressing Minpp1. To test this, we added purified HIV-1 protease to immature particles that had been assembled in vitro with different IPs and monitored the cleavage reaction over time. We observed a distinctive processing sequence for IP6-assembled particles, closely matching that within authentic HIV-1 virions, beginning with NC cleavage and ending with SP1 liberation from CA (Figure 3F & Supplementary Figure 4). Processing of particles assembled in the presence of IP5 or IP6 was very similar, consistent with IP5 being capable of functionally replacing IP6[3]. However, particles assembled with smaller IPs, most noticeably with IP3 or IP2, or tartrate, displayed subtle differences in processing.
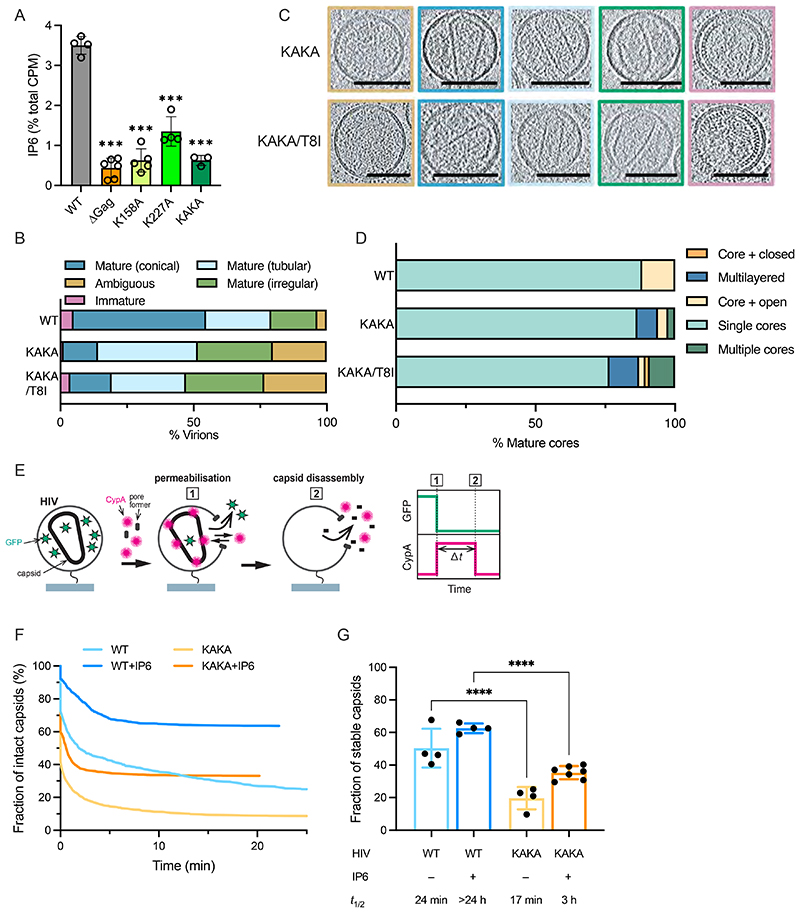
HIV-1 can become independent of IP6 for immature particle assembly
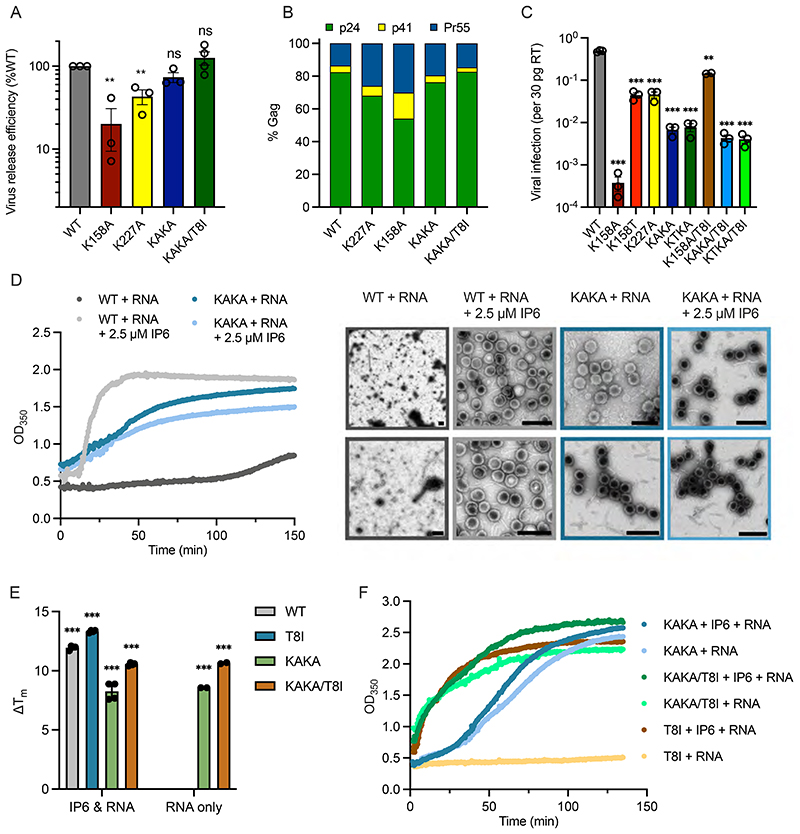
The marked preference of mature assembly for IP6 and the fact that capsids require a concentration of several hundred μM IP6, well above cellular levels, highlights the need for HIV-1 to enrich IP6 into virions. As mentioned, HIV-1 recruits IP6 into virions using two lysine rings that are formed by K158 and K227 at the centre of immature hexamers (Supplementary Figure 5)[8]. We therefore investigated how virus production, maturation and infection are affected by mutation of these rings (K158A/K227A or ‘KAKA’). Surprisingly, virus production was largely unaffected by simultaneously mutating both lysines to alanine and was not significantly increased by inclusion of the stabilising mutation SP1-T8I (Figure 4A). Analysis of Gag processing revealed that whilst the individual lysine mutants K227A and K158A exhibit mild cleavage defects, the double mutant KAKA displayed similar cleavage efficiency as WT virus (Figure 4B & Supplementary Figure 6). In contrast to the limited effect on virus production and maturation, the KAKA mutant displayed markedly reduced infectivity, with a ~100-fold lower infectivity than WT (Figure 4C). K158A and K227A mutants can be rescued via the second-site mutation SP1-T8I, which increases infectivity by promoting immature hexamer stability and restoring IP6 enrichment[8]. In contrast, addition of SP1-T8I to KAKA had no impact on infectivity (Figure 4C). We created an additional mutant K158T/K227A (‘KTKA’), which introduces a threonine instead of an alanine at position 158 and increases stability of the single K158 mutant, presumably by improving hydrophobic packing [8]. The KTKA mutant had a similar infectivity defect as the KAKA variant and the addition of SP1-T8I again had no impact on infectivity. The inability of either SP1-T8I or K158T to improve infectivity is likely because with both lysine rings missing, there is no way to recruit IP6 into the assembling immature Gag lattice no matter how stable immature hexamers become.
Figure 4. HIV-1 can become independent of IP6 for immature particle assembly.
(A) Virus release efficiency of Gag mutants, calculated as the percentage of particle-associated p24 (CA) as a fraction of total (cell- + particle-associated Gag) normalised to WT virus. Error bars depict the SEM from at least three independent experiments. An unpaired t test against WT was used for statistical analysis and significant differences indicated [P < 0.005 (**)]. (B) Gag cleavage efficiency in purified virions, calculated as the percentage of p24 (CA), p41 and Pr55Gag. (C) Infectivity of Gag mutants normalised to the quantity of input virus [per 30 pg of reverse transcriptase (RT)]. Error bars depict the SEM from three independent experiments. An unpaired t test against WT was used for statistical analysis and significant differences indicated [P = 0.005 (**), < 0.0005 (***)]. (D) In vitro assembly of immature particles using recombinant DMA-CANC protein [comprising capsid (CA) and nucleocapsid (NC) domains]. 7.5 μM RNA was added to 75 μM ΔMA-CANC and assembly monitored through light scattering changes at 350 nm. EM images of negative stained samples of the final assembly reactions. Scale bars are 200 nm. Representative of two experiments. (E) The thermostability of in vitro assembled particles was measured by differential scanning fluorimetry and expressed as a change in melt temperature (Tm) compared to unassembled ΔMA-CANC. An unpaired t test against unassembled ΔMA-CANC was used for statistical analysis and significant differences indicated [P < 0.0005 (***)]. (F) Kinetics of immature particle assembly of ΔMA-CANC mutants from (E), using 10 μM RNA and 100 μM CANC. Representative of at least two experiments.
Next, we tested the ability of KAKA to assemble immature VLPs in vitro. Importantly, we found that whilst WT assembly required both IP6 and RNA, RNA alone was sufficient to drive assembly of the KAKA mutant (Figure 4D). Moreover, these KAKA VLPs were similar in size to WT VLPs assembled with IP6. Addition of IP6 had no effect on KAKA assembly, consistent with the two lysines being necessary to bind the metabolite. However, the KAKA mutant does not assemble quite as rapidly as WT, in contrast to previous data [18]. We also compared the thermal stability of immature VLPs (Figure 4E). When WT ΔMA-CANC assembles in the presence of IP6 and RNA, we observed a > 10 °C increase in melting temperature (Tm) relative to unassembled protein (Figure 4E). There was no change in WT Tm upon addition of RNA alone (in the absence of IP6), consistent with a lack of assembly under this condition (Figure 4D). The Tm of KAKA DMA-CANC increased by ~8 °C after assembly with RNA and there was no further increase in Tm for assembly reactions also containing IP6 (Figure 4E). Consistent with its ability to stabilise immature hexamers, adding the SP1-T8I mutant to WT and KAKA resulted in an increase in Tm and accelerated assembly kinetics. However, WT/T8I was still depended IP6 for assembly whereas KAKA/T8I was not (Figure 4F). We conclude that an immature lattice can form without IP6 with close to WT efficiency and stability if K158 and K227 are not present.
HIV-1 virions lacking enriched IP6 assemble mature capsids with low efficiency and which have reduced stability
The inability of IP6 to promote immature KAKA Gag assembly suggested that this mutant may be unable to enrich IP6 into budding HIV-1 virions. To test this, we produced virus particles in cells supplemented with tritiated inositol and used SAX-HPLC to fractionate the inositol phosphates from purified virions, as described previously[3, 8]. WT HIV-1 Gag was responsible for increasing IP6 abundance in virions by ~ 8-fold (Figure 5A). In contrast, KAKA lost the ability to enrich IP6, displaying similar levels to those of the single K158A mutant but lower than those of K227A (Figure 5A). The ability of the KAKA mutant to undergo immature particle assembly, followed by efficient Gag cleavage, suggested that its reduced infectivity may be due to defective mature core formation as a consequence of reduced IP6 in budded virions. We investigated this by using cryo-electron tomography (cryoET) to examine the structures of capsids in purified virions. We found that KAKA virions had fewer conical capsids than WT (Figure 5B&C), but did not display an over-assembly phenotype characterised by frequent additional aberrant structures (Figure 5D) as seen with K158 virions [8]. Both infectivity and the formation of single morphologically normal cores can be rescued in K158 through the second-site compensating mutation SP1-T8I [8], but the addition of SP1-T8I had little effect on KAKA (Figure 5B-D). This is consistent with the in vitro assembly data and suggests that the KAKA mutant is defective in mature but not immature lattice formation.
Figure 5. HIV-1 virions lacking enriched IP6 assemble mature capsids with low efficiency and which have reduced stability.
(A) Viruses produced in cells supplemented with tritiated inositol were purified and inositol phosphate species extracted and fractioned by SAX-HPLC. Data were analysed as previously described[8] and the counts per minute (CPM) of IP6 shown as a fraction of total CPM in the sample. Error bars depict mean CPM ± SEM from at least two independent experiments. An unpaired t test against WT was used for statistical analysis and significant differences indicated [P < 0.0005 (***)]. (B-D) Cryo-ET on indicated HIV-1 mutants. Tilt-series were collected and reconstructions performed to assess capsid morphology. A total of 163 KAKA and 187 KAKA/T8I particles were analysed. (B) Virions produced by the indicated Gag mutants were classified into the indicated categories: Ambiguous (orange), Immature (pink), Mature Conical (dark blue), Mature Tubular (light blue), Mature Irregular (green). (C) Slices through tomograms show representative examples of the viral morphologies in (B). Scale bars, 100 nm. (D) Virions with mature lattices were further subdivided into: Multiple Cores (green), Single Cores (cyan), Cores with additional closed structure (orange), Cores with additional open structure (light orange), Multilayered Cores (blue). (E) Schematic diagram of a viral particle in the kinetic TIRF assay detecting capsid uncoating. HIV particles are loaded with low levels of EGFP using a cleavable fusion protein with EGFP and Vpr. These EGFP-loaded HIV particles are immobilised then permeabilised in the presence of AF568-labelled CypA. Fluorescence traces are recorded at the locations of individual HIV particles by TIRF microscopy. Permeabilisation of the viral membrane (step 1) with a pore-forming protein leads to loss of the EGFP signal and concomitant binding of AF647-CypA molecules to the capsid. Note that any capsid internalized EGFP is not detected above background. Capsid uncoating (step 2) is detected as the loss of the AF647-CypA signal and capsid lifetime is calculated as the time difference (Δt) between permeabilization and uncoating. (F-G) Capsid survival curves (G) constructed from the lifetimes of all particles in the field of view reveal an unstable subpopulation that decays away within the first few minutes (no or short-lived AF647-CypA signal with a half-life of < 2 min) and a stable subpopulation with slow uncoating kinetics (long-lived AF647-CypA signal). The fraction of stable capsids for WT and KAKA mutant in the absence and presence of IP6 is shown (G), with the calculated half-life of the stable fraction obtained by fitting of survival curves (F) in the presence or absence of 100 μM IP6. A one-way ANOVA was used for statistical analysis and significant differences indicated [P < 0.0001 (****)].
Previously we have used real-time TIRF microscopy to show that IP6 is required to keep the capsid stable once the protection of the viral membrane is lost [2]. Next, we tested the stability of KAKA capsids that have formed in the absence of enriched IP6 using a similar approach. Briefly, purified virions loaded with low levels of EGFP as a fluid phase marker are immobilised then permeabilised in the presence of AF647-CypA (Figure 5E). Permeabilisation is recorded by the loss of GPF signal, while capsid lifetime is measured by the time taken for loss of the AF647-CypA which ‘paints’ the capsid lattice (Figure 5E). Survival analysis of all particles (Figure 5F) reveals a subpopulation showing no or short-lived AF647-CypA signals, attributed to particles with no mature capsid lattice or one that rapidly collapses[19]. For simplicity, these are classified as ‘unstable’. The proportion of virions with stable capsids can therefore be determined (Figure 5G), while the survival curves can be fitted to obtain a half-life for stability (Figure 5G). Comparison of WT and KAKA virions revealed a ~2-fold decrease in the fraction of detectable capsid lattice. Moreover, these detectable KAKA capsid lattices had a shorter half-life (Figure 5G). As shown previously, maintaining the presence of IP6 through addition to the buffer extends the half-life of WT capsid stability beyond meaningful measurement (> 24 hours) [2]. In contrast, IP6 only led to a partial rescue of KAKA capsid stability, with most capsids falling apart with a half-life of 3 hours (Figure 5F). Taken together with the cryoET data, this shows that KAKA forms fewer mature capsids and those that form are highly unstable. Importantly, KAKA capsids are not rescued through the addition of exogenously added IP6. This suggests that KAKA capsids cannot easily obtain IP6 from target cells upon viral membrane fusion, explaining this mutants poor infectivity.
Simultaneously reducing cellular IP6 and the ability of HIV-1 to enrich it into virions amplifies infectivity defects
While the KAKA mutant reduces the ability of HIV-1 to enrich IP6 into virions, it likely does not prevent passive IP6 incorporation into virus particles. We therefore investigated how HIV-1 is impacted when the cellular pool of IP6 is reduced simultaneously with its ability to enrich IP6 into budding virions. Measuring virus release efficiency, we observed that removing both lysine rings in the immature lattice partially rescues the production defect in IPMK KO + FM1 cells (Figure 6A). In contrast, virus production remained low when only a single lysine, K227 or K158, was mutated. The KAKA mutation also rescued Gag processing in IP6-depleted cells; irrespective of kinase KO or over-expression of Minpp1, KAKA achieved close to WT levels of Gag cleavage (Figure 6B & Supplementary Figure 7). This correlates with the ability of KAKA, but not single-lysine mutants, to assemble a stable immature lattice independently of IP6. Consistent with this interpretation, KAKA was sufficient to reverse the profound morphological defect of VLPs produced by IPMK KO + FM1 cells (Figure 6C). This suggests that the defective morphology of virions budded from IPMK KO + FM1 cells is a result of problems associated with the immature lattice. Making the assembly of the immature lattice IP6-independent, via KAKA mutation, restores the production, correct particle morphology and Gag processing of viruses produced in IP6-depleted cells. Importantly, the combination of IP6 depletion and lysine mutation decreased infection additively (Figure 6D). While we were unable to measure IP6 levels in virions produced under these conditions, this is consistent with both perturbations combining to decrease IP6 levels in budded virions further than either alone.
Figure 6. Simultaneously reducing cellular IP6 and the ability of HIV-1 to enrich it into virions amplifies infectivity defects.
(A) Virus release efficiency of Gag mutants from either 293T cells or IPMK KOs overexpressing FM1, calculated as the percentage of particle-associated p24 (CA) as a fraction of total (cell- + particle-associated Gag) normalised to WT virus. Error bars depict the SEM from at least three independent experiments. An unpaired T test was used for statistical analysis and only significant differences indicated [P = 0.05 (*), = 0.005 (**), < 0.0005 (***)]. Blue asterisks refer to significant differences to WT virus produced in 293Ts, whilst green asterisks refer to significant differences to WT virus produced in IPMK KO + FM1 cells. (B) Gag cleavage efficiency of WT, KAKA and KAKA/T8I purified virions, calculated as the relative amount of p24 (CA), p41 and Pr55Gag as a percentage of total Gag. (C) Thin-section electron-microscopy of KAKA mutant HIV-1 virions produced in 293T cells or IPMK KO cells expressing FM1. Scale bar = 120 nm. (D) Infectivity of Gag mutants normalised to the quantity of input virus as determined by RT assay and relative to WT HIV-1. Error bars depict the SEM from three independent experiments. An unpaired T test was used for statistical analysis and significant differences indicated [P = 0.05 (*), = 0.005 (**), < 0.0005 (***)]. Blue asterisks refer to significant differences from WT virus infection of 293Ts.
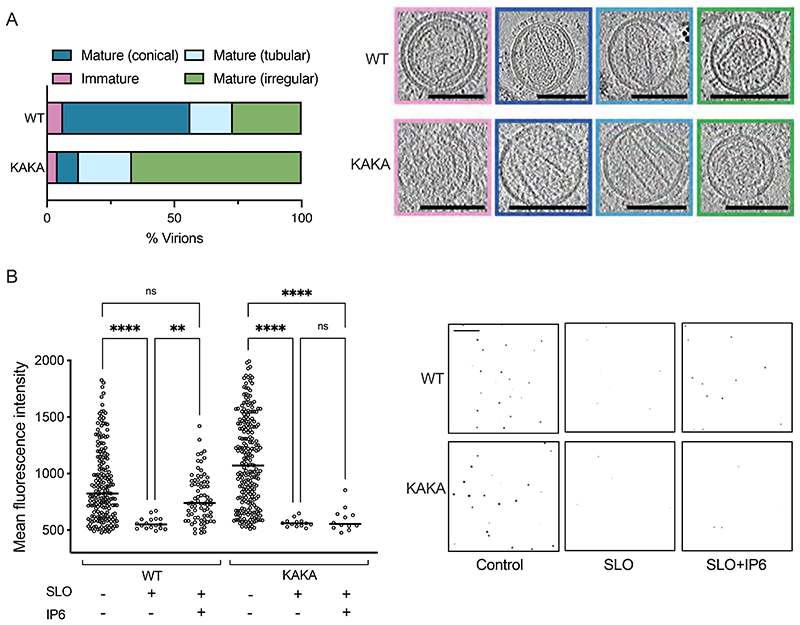
To determine the combined impact of low IP6 availability and no enrichment mechanism on HIV-1 capsid formation, we collected tomograms on KAKA virions produced in IPMK KO + FM1 cells. Of 42 reconstructed particles, only 24 could be unambiguously defined as virions and of these only 2 contained conical capsids (Figure 7A). In contrast, ~50% of WT virions had conical capsids. This confirms that IP6 incorporation into virions is important to build a mature capsid. Next, we measured the stability of those few IP6-deficient capsids that are able to form using TIRF microscopy. To perform a comparison with KAKA virions produced in IPMK KO + FM1 cells, we used a modified protocol in which virions were first classified as mature or immature by their p24 fluorescence intensity (Supplementary Figure 8A&B). Comparison of WT virions with the protease-inactivating mutant D25A, which cannot undergo maturation[20], was used to establish the fluorescence of immature virions (Supplementary Figure 8B). Next, WT virions were pre-incubated for 30 minutes with the pore-forming protein Streptolysin O (SLO), which permeabilises the HIV-1 membrane and allows the measurement of capsid stability[21], before VSV-G and p24 antibody staining. VSV-G staining of nonpermeabilized control samples confirmed SLO activity, since the antibody (Abcam, ab1874) targets an internal epitope only exposed upon pore-formation or membrane permeabilization (Supplementary Figure 8C). Meanwhile, VSV-G staining of permeabilised samples confirmed that pore-formation does not lead to destruction or loss of virions (Supplementary Figure 8D). In contrast to the VSV-G signal, we observed a significant loss of capsid fluorescence upon SLO treatment, consistent with published data showing that without exogenous IP6 the capsid collapses and p24 protein dissociates from virions in < 10 minutes [2, 19] (Figure 7B). As expected however, and consistent with previous data[2], there was no significant loss of capsid fluorescence in the presence of exogenous IP6. A similar reduction in capsid fluorescence was observed upon SLO treatment of KAKA virions produced in IPMK KO + FM1 cells. Importantly, however, and in contrast to WT HIV-1, the addition of exogenous IP6 did not preserve KAKA capsid fluorescence upon SLO permeabilization (Figure 7B). These results are consistent with previous TIRF data showing that K158A, which like KAKA does not package IP6 into virions, collapses soon after virion permeabilization and before it can be stabilised by exogenously added IP6[8]. Only restoration of IP6 packaging through the addition of second-site mutation SP1-T8I rescues WT-like capsid stability in the case of K158A. Taken together, the cryoET and TIRF data suggest that the infectivity defect of KAKA virions, which is exacerbated by production in IP6-depleted cells, is caused by a combination of reduced capsid formation and extreme capsid instability. These results also reinforce why IP6 in the target cell does not rescue incoming IP6-deficient KAKA capsids – because they collapse too quickly to recruit the metabolite.
Figure 7. KAKA virions produced in IP6-low cells have fewer and less-stable capsids.
(A) Cryo-ET comparison of KAKA and WT virus produced in IPMK KO + FM1 cells. Tilt-series were collected and reconstructions performed to assess capsid morphology. A total of 48 WT and 24 KAKA particles were analysed. Virions were classified into the indicated categories: Immature (pink), Mature Conical (dark blue), Mature Tubular (light blue), Mature Irregular (green). Slices through representative tomograms of the virions are shown together with quantification. Scale bars, 100 nm. (B) TIRF microscopy on WT virions produced in 293T cells and KAKA mutants produced in IPMK KO + FM1 cells. Virions were adhered to Ibidi slides and treated with SLO in the presence or absence of IP6. Samples were fixed, permeabilised and labelled with VSV-G and p24 antibody. Virions from three independent images for each condition were analysed for mean fluorescence object intensity above threshold (see Supplementary Figure 8). A one-way ANOVA was used for statistical analysis and significant differences indicated [P = 0.01 (**), < 0.0001 (****)]. Right panel shows representative images of virions used in the analysis. Scale = 20μm.
Discussion
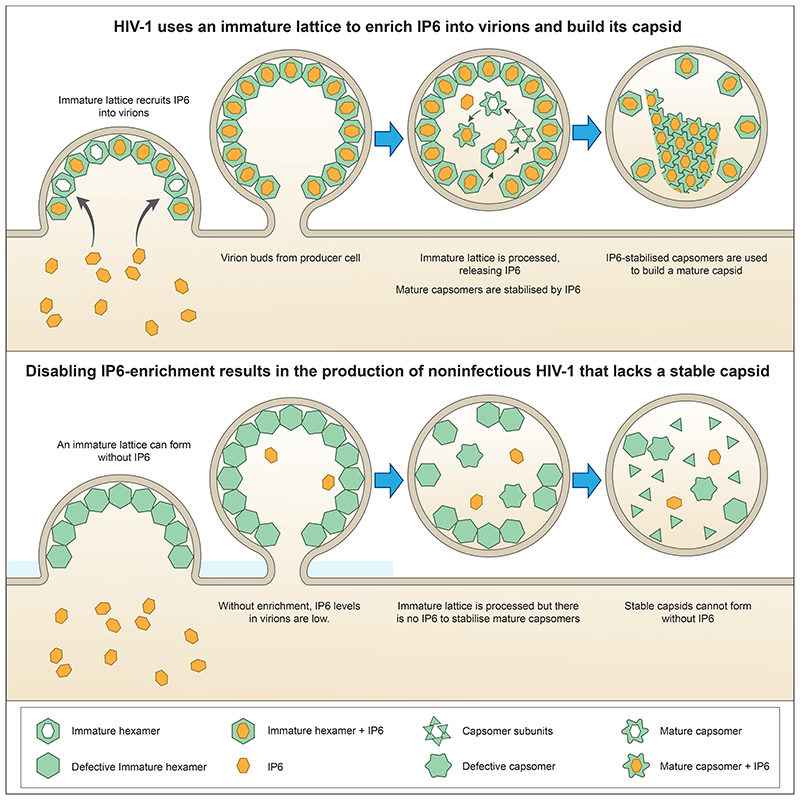
IP6 is important for both production and infectivity[2] and promotes the assembly of both immature VLPs and mature capsids[22]. Surprisingly, our data show that HIV-1 immature assembly and production can become independent of IP6 simply by mutating the two IP6-binding residues in Gag. However, these mutants remain critically dependent upon IP6 for mature capsid formation and infectivity (Figure 8). These data support a hypothesis in which the immature lattice has evolved to bind IP6 in order to enrich the metabolite into virions, so that once budded from the cell there are sufficient IP6 molecules inside each virion to build an infection-competent mature capsid. Disabling this IP6 enrichment mechanism does not prevent efficient production of virions from producer cells, but if the immature lattice does not enrich IP6 into virions then stable mature capsids do not form and infectivity is largely abolished (Figure 8).
Figure 8. The HIV-1 immature lattice enriches IP6 into virions to catalyse mature capsid assembly.
The immature lattice does not intrinsically need IP6 for assembly but instead acts as a ‘net’ to capture IP6 from producer cells and enrich it into virions. Virions that have budded from the cell undergo maturation during which the immature lattice is cleaved by the viral protease. This results in the liberation of CA protein from Gag and the release of IP6 from its binding site in the immature Gag lattice. The newly freed IP6 promotes the assembly of CA into capsomers (predominately hexamers but also pentamers), which are used to construct the conical capsid characteristic of mature HIV-1 particles. Because maturation happens inside virions, separated from the cell, if there are insufficient IP6 molecules packaged into virions then stable mature capsids cannot form and the resulting HIV-1 particles fail to become infectious.
Comparing the in vitro assembly kinetics of immature VLPs and mature capsids highlights that while the former requires single-digit μM concentrations of IP6, the latter requires several hundred μM. These figures are significant because the cellular IP6 concentration is 22 - 44 μM, below the concentration required to stimulate mature lattice assembly. We also find that while immature assembly occurs with similar kinetics in the presence of IP5 or IP4, or slightly slower with IP3, mature assembly kinetics are substantially attenuated when using anything other than IP6. This highlights the profound IP6 dependence of mature capsid assembly. Tritiated inositol measurements suggest there may be 300 - 800 IP6 molecules per virion[2] (assuming 1000 – 1500 CA molecules per particle), or 1.2 – 3.2 IP6 molecules per hexamer in a fully assembled capsid. This is consistent with there being two known binding sites per hexamer provided by R18[1, 2] and K25[23]. If IP6 is not enriched into virions then there will only be enough molecules to stabilise 10 - 30 % of capsomers. Our cryoET data reveal that KAKA virions do form some capsids, despite IP6 concentrations being well below the EC50MA. This may be driven by the extremely high CA concentration inside each viral particle. However, TIRF microscopy data suggest that many of these capsids immediately collapse upon viral membrane permeabilization, coincident with the sudden drop in CA concentration. The inability to prevent this collapse with exogenous IP6 may explain why capsids that form in virions lacking sufficient IP6, such as KAKA, cannot simply sequester it from the target cell during infection.
Importantly, IP6 sequestration by the immature lattice is the only way to ensure there is sufficient IP6 inside virions to fully saturate the mature capsid. This is because IP6 precipitates above 49 μM in the presence of physiological bivalent cation concentrations [24]. This solubility limit has important implications for IP6-driven maturation. There are two broad models for maturation: disassembly/reassembly or partially displacive transition. These models postulate, respectively, either that mature assembly only starts once Gag has been fully cleaved or begins as soon as p24 CA protein is liberated at the edge of the immature lattice[25]. We suggest that a disassembly and reassembly model is unlikely because IP6 would precipitate within the virion, resulting in a concentration too low to support capsid formation and too few IP6 molecules to stabilise all hexamers in the lattice. A partially displacive model, as has been proposed[26–29], seems more likely in the context of IP6-driven assembly.
Previous experiments have tested the dependence of HIV-1 on IP6 by knocking out biosynthetic IP kinases[3–5]. However, knockout of IPMK or IPPK results in modest phenotypes that are hard to interpret without knowing their quantitative impact on cellular IP levels. Correlating HIV-1 production and infectivity phenotypes with the IP profiles of different cell lines has provided several important insights into the virus’ dependence on these metabolites. As shown in detail here, IPMK and IPPK KOs are not equivalent and result in very different IP profiles: IPMK KOs have 10-fold more IP6 than IPPK KOs, whilst in IPPK KOs IP5 has become the predominant species. This is because IPPK KO IP6 levels are reduced ~ 100-fold to ~ 0.44 μM while IP4 and IP5(OH) levels have increased 5 - 10-fold and ~2-fold respectively (Supplementary Table 1). Meanwhile, IP6 is only reduced in IPMK KOs by 6-fold, to ~5 μM, but IP5(OH) is also decreased 30-fold. Although in both IPMK and IPPK KOs, the total concentration of IP5 + IP6 exceeds what is required to promote immature assembly, only viruses produced in IPPKs have an infectivity defect (Figure 1G&H). This highlights that IP5 cannot fully substitute for IP6 and correlates with in vitro measurements showing mature assembly with IP5 is less efficient[1] and slower (Figure 3D). Over-expressing the phosphatase Minpp1 substantially reduces the levels of IP5 in IPPK KOs and IP6 in IPMK KOs. In IPPKs, Minpp1 overexpression also results in a substantial increase in IP4 to levels 100-fold higher than those of IP5 and IP6. The fact that HIV-1 from IPPK KO/Minpp1 has a production and infectivity defect that is at least as severe as virus from IPMK KO/Minpp1 cells suggests that IP4 cannot functionally replace IP6. In vitro data shows that IP4 allows immature assembly but fails to promote efficient mature assembly, making this latter step the likely point of sensitivity. Targeting Minpp1 expression to the plasma membrane further exacerbates these phenotypes. It also reduces infection by viruses produced in wild-type cells, in contrast to previously published data where Minpp1 was overexpressed but not plasma membrane targeted[4]. These results suggest that IP6 is recruited at the plasma membrane, consistent with this being the site of immature lattice assembly and IP6 binding hexameric but not monomeric Gag. Finally, the IP profiles of different cell lines demonstrate that HIV-1 fitness is most severely affected when combined IP5 and IP6 concentrations drop below the EC50IM for immature assembly. This is likely because at sub-μM IP5 or IP6 concentrations, HIV-1 can no longer actively enrich these metabolites into virions, which is catastrophic for the maturation of infectious virions.
Taken together, our data show that HIV-1 immature assembly and virion production, but not the maturation of infectious particles, can become independent of IP6 with just two mutations (KAKA). Whether there are equivalent mutations in the mature capsid that could allow HIV-1 to become fully IP6-independent remains to be determined. A natural equivalent of the HIV-1 KAKA mutant exists in the form of the alpharetrovirus RSV, which does not require IP6 for immature assembly[30]. Interestingly, RSV requires a substantially lower IP6 concentration for mature capsid assembly than HIV-1. It has been proposed that this may be because IP6 is incorporated into the forming RSV capsid before membrane scission[30]. Alternatively, RSV mature capsomers may be intrinsically more stable than those of HIV-1 and may have acquired the ability to utilise lower IP6 concentrations or other polyanions for their assembly. Perhaps HIV-1 mutants that increase capsid stability could also allow the virus to utilise lower IP6 levels and obviate the need for IP6 enrichment by the immature lattice. However, we propose that as long as mature HIV-1 capsomers retain a positively charged ring, R18, for nucleotide import and reverse transcription IPs will remain essential to build and stabilise functional HIV-1 capsids.
Materials & Methods
Cells and Plasmids
293T CRL-3216 cells were purchased from ATCC. All cells are regularly tested and are mycoplasma free. HEK293T and HeLa cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin (GIBCO) at 37°C with 5% CO2). Replication deficient VSV-G pseudotyped HIV-1 virions were produced in HEK293T cells using the packaging plasmid pMDG2, which encodes VSV-G envelope (Addgene plasmid # 12259), pNL4-3-derived pCRV GagPol (HIV-1 clade B)[32], and pCSGW[33] as described previously[34]. Mutagenesis of CA was performed using the QuickChange method (Stratagene) against pCRV GagPol. The HIV-1 clade B infectious molecular clone pNL4-3 was used for all passage and virus release experiments. Mutant constructs were generated with the NEB Q5 site directed mutagenesis kit (NEB E0554).
Virus Production & Infection Experiments
Viruses were produced from 2.5 x 106 cells in a 10cm dish or 5 x 105 cells per well of a 6-well plate, plated the day before. Transfection mixtures were made using 200μl OptiMem (GIBCO), 1μg pMDG2, 1.5μg pCSGW, 1μg pCRV GagPol and 12μl FuGENE6 (Promega). Mixtures were incubated at room temp for 15 min and then added in entirety to 10cm dishes or 60μl added to a well of a 6-well plate. Viral supernatants were harvested 48hr posttransfection and filtered through a 0.45 μm filter and stored at -80C. For infection experiments with 293T, cells were seeded at 0.75x104 cells per well into 96-well plates and left to adhere overnight. Indicated amounts of virus were added, and the plates were scanned every 8 h for up to 72 h in an IncuCyte!(Satorius) to identify GFP-expressing cells.
Virus Quantification
The level of RT enzyme was quantified using qRT-PCR as described previously with slight alterations[35]. In brief, 5 μl of viral supernatant was mixed with 5 μl lysis buffer (0.25% Triton X-100, 50 mM KCl, 100 mM Tris-HCl (pH 7.4), 40% glycerol) and 0.1 μl RNase Inhibitor and incubated for 10 min at room temperature before diluting to 100 μl with nuclease-free water. 2 μl of lysate was added to 5 μl TaqMan Fast Universal PCR Mix, 0.1 μl MS2 RNA, 0.05 μl RNase Inhibitor and 0.5 μl MS2 primer mix, to a final volume of 10μl. The reaction was run on an ABI StepOnePlus Real Time PCR System (Life Technologies), with additional reverse transcription step (42°C 20 min).
Purification and analysis of inositol phosphates
This was carried out as previously described[8]. Briefly, virus particles were produced in 293T cells pre-cultured in inositol-free media supplemented with [3H] inositol (5 μCi/ml). Particles were concentrated by ultracentrifugation and inositol phosphates extracted and analysed by HPLC following a published protocol[31]. Cells or pelleted virions labelled with [3H] inositol were resuspended in 200μl of extraction solution (1M Perchloric acid, 5mM EDTA) and incubated on ice or 100 °C for 10 mins. Inositol phosphates were resolved by strong anion exchange chromatography Sax-HPLC on a Partisphere SAX 4.6 °— 125 mm column (Hichrom). Fractions (1 ml) were collected and analyzed by scintillation counting after adding 4 ml of Ultima-Flo AP cocktail (Perkin Elmer, 6013599). The counts per minute (CPM) of IP6 were normalized to total lipids.
Virus Release
Virus release assays were performed as described previously[36]. Briefly, HEK293T cells were transfected with 2 μg of pNL4-3 WT or mutant plasmids in 6-well plates. 0.25 μg of FM1 or empty vector was used in co-transfections. Linear polyethylenimine (1mg/ml) was used as the transfection reagent. At 48 h post-transfection, viral supernatants were filtered and pelleted by ultracentrifugation at 4°C. Virus pellets and remaining cells were lysed and probed for Gag via western blot. HIV-Ig (NIH AIDS Reagent Program Cat. #3957) was used as the primary antibody for Gag detection and an anti-human IgG HRP-tagged antibody (Sigma Cat. # GENA933) was used as the secondary antibody. Supersignal West Pico Plus (Thermo 34578) was used as the chemiluminescent substrate. Imaging and band quantification were performed using the Sapphire Biomolecular Imager and Azure Spot analysis software (Azure Biosystems). Virus release was calculated using the following formula: virus p24/(virus p24 + cell p24 + cell Pr55).
Western Blotting
Samples were run on 4-12% Bis Tris gels and transferred onto nitrocellulose membranes using iBlot (Life Technologies) and detected by ECL or by Li-COR for quantification. Anti-HIV-1 p24 (183-H12-5C) was obtained from the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Anti-HIV-1 p24 Monoclonal (183-H12-5C) (Cat# 3537) from Dr. Bruce Chesebro and Kathy Wehrly[37, 38], loading control COX IV (P/N 926-42214) was obtained from Li-Cor Biosciences. For virus release, samples were subjected to SDS-PAGE (4-20%), then transferred to a polyvinylidene fluoride (PVDF) membrane (Immobilon, Millipore) via semi-dry transfer (Bio-Rad Trans-Blot Turbo). The membrane was blocked for 1 h with 5% non-fat milk in Trisbuffered saline + 0.05% Tween 20 detergent (TBST) and incubated overnight at 4°C with anti-HIV-1 IgG. The membrane was then washed with TBST and incubated for 2 h with anti-human horseradish peroxidase-conjugated secondary antibody and washed again. SuperSignal West Pico PLUS (Thermo Scientific) was used to reveal protein bands.
Jess capillary protein detection system
Samples were run following manufacturers protocol. Shortly, protein standard is run in each capillary and in the presence of specific human antibodies these serve as primary antibodies that are then detected with anti-human HRP secondary antibody. Bio-Techne software Compass was used to quantify antibody titres in the samples.
Virus particle production for tomography
Virus-like particles were produced in HEK293T as described above. Supernatants were harvested and passed through a 0.45 μm filter followed by a 0.22μm filter. The particles were concentrated by ultracentrifugation over a 20 % (wt/vol) sucrose cushion (2 h at 28,000 rpm in a Beckman SW32 rotor; Beckman Coulter Life Sciences). The pellet was resuspended in PBS and incubated at 4°C overnight to allow full resuspension.
Cryo-Tomography
Virus-like particles were produced in HEK293T as described above. Supernatants were harvested and passed through a 0.45 μm filter followed by a 0.22-μm filter. The particles were concentrated by ultracentrifugation over a 20% (wt/vol) sucrose cushion (2 h at 28,000 rpm in a Beckman SW32 rotor; Beckman Coulter Life Sciences). The pellet was resuspended in PBS. 10-nm-diameter colloidal gold beads were added to the purified HIV-1 mutants. 4 μl sample-gold suspension was applied to a glow discharged C-Flat 2/2 3C (20 mA, 40 s). Grids were blotted and plunge-frozen in liquid ethane with a FEI Vitrobot Mark II at 15 °C and 100% humidity. Tomographic tilt series of KAKA and WT were acquired between -40° and +40°with increments of 3°, on a TF2 Tecnai F20 transmission electron microscope equipped with a Falcon III Direct Electron detector at 200 kV using Serial-EM under low-dose conditions at a magnification of 50000x and a defocus between -3 μm and -6 μm. Tomography of the IPMK and IPPK mutants was performed on a FEI Titan Krios transmission electron microscope at 300 kV equipped with a Gatan K2 summit direct electron detector and a Gatan Quantum energy-filter (GIF). Tilt series were acquired between -60° and +60° with increments of 3° using a dose symmetric scheme using Serial-EM[39]. Images were collected at a magnification of 33000x with 10 frames per tilt and a total dose of ∼120 e-/Å2 across all of the tilts. Frames were aligned in SerialEM with a final pixel size of 3.667 Å per pixel in the unbinned image stacks. Tomograms were reconstructed using IMOD (4.9)[40]. The alignment of 2D projection images of the tilt series was performed using gold beads as fiducial markers, tomograms were reconstructed by back projection.
Transmission Electron Microscopy
293T cells (WT or IPMK KO) were transfected with WT or mutant pNL4-3 along with an empty vector (WT cells) or Minpp1 FM1 (IPMK KO cells). Transfections were performed with 1mg/ml linear polyethyleneimine (PEI) in 6-well plates seeded the previous day with 6 x 106 cells. Fixation of cells, preparation of samples, and transmission EM were performed as previously described[36].
Protein production and purification
The Capsid proteins were expressed in E.coli C41 cells for 4 h at 37°C, lysed in lysis buffer (50 mM Tris-HCl (pH 8.0), 200 mM NaCl, 20% BugBuster, Protease inhibitor tablets, 1 mM DTT) and centrifuged (24 000 rpm, 1h). The supernatant was precipitated with 25% ammoniumsulphate (wt/vol) followed by centrifugation (13 000 rpm, 20 min). The precipitated CA was resuspended and dialysed against 50 mM MES (pH 6.0), 20 mM NaCl, 1mM DTT. The CA protein was further purified via a cation-exchange column with a gradient from 20mM -1M NaCl followed by size exclusion chromatography with Tris pH 8.0, 20 mM NaCl, 1mM DTT, concentrated and snap frozen.
The ΔMA-CANC protein was expressed as described previously with a few alterations [1]. Briefly, ΔMA-CANC was expressed in E.coli C41 cells for 4 h at 25°C, lysed in lysis buffer (50 mM Tris-HCl pH 7.4, 500 mM NaCl, 1 mM DTT, 10 μM ZnCl2, 20% BugBuster, Protease Inhibitor Tablets). 0.1% PEI (v/v) was added and the lysate was stirred for 10 min. The lysate then was centrifuged (24 000 rpm, 1h) and the supernatant was precipitated in 25% ammonium sulphate, followed by centrifugation (13 000 rpm, 20 min). The precipitated protein was resuspended in dialysis buffer (50 mM Tris-HCl pH 7.4, 40 mM NaCl, 1mM DTT, 10 μM ZnCl2), dialysed into the same buffer and applied to an anion-exchange column. The flow-through was precipitated with 0.1% PEI, followed by centrifugation and the supernatant was precipitated with 25% ammonium sulphate. The precipitate was resuspended in dialysis buffer and applied to a cation-exchange column and eluted with a gradient of 40 mM -1M NaCl. The protein was concentrated and snap-frozen.
The HIV-1 protease was purified as previously described[41]. Briefly, protease was expressed in E. coli BL21(DE3) cells for 4 h at 37°C. Cells were harvested by centrifugation, resuspended in PR buffer (20 mM Tris [pH 8.0], 0.1 M NaCl, 5 mM imidazole, 1 mM β-mercaptoethanol) supplemented with protease inhibitor tablets (Roche). After centrifugation the pellet containing inclusion bodies was washed twice with 30 ml of PR buffer supplemented with 2 M urea and 1% Triton X-100 and once with water with subsequent centrifugation. The pellet was resuspended in PR buffer supplemented with 8 M urea and incubated overnight. The protein was purified on Ni (NTA) resin (Qiagen) and diluted with 20 mM Tris-HCl (pH 7.9), 100 mM NaCl, 5 mM imidazole, and 8 M urea. PR was refolded by stepwise dialysis against a solution containing 20 mM Tris-HCl (pH 7.9), 100 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10% glycerol, and 1 mM DTT, with gradually decreasing urea concentrations from 8 to 0 M. Finally, refolded protein was concentrated and snap frozen.
Turbidity Assays
CA proteins were dialysed against 50mM MES (pH 6.0), 40 mM NaCl, 1mM DTT. CA proteins at a final concentration of 25-100 μM were mixed with IP6 at 25°C (final concentration 50 μM-2 mM). ΔMA-CANC proteins were diluted into assembly buffer to the indicated concentrations (20 mM Tris [pH 7.5], 140 mM KCl, 10 mM NaCl, 5 mM MgCl2, 10 mM TCEP). Assembly of 75μM CA was initiated by adding 7.5 μM ssRNA GT25 and IP6, IP5, IP4, IP3 or IP2 at the indicated concentrations (SiChem). The apparent increase in absorbance reflecting increased light scattering (OD350) was measured using a PHERAstar FSX Plate reader (BMG Labtech) in 384-well plate with shaking between each measurement at 25°C or 37°C. The concentration of IP6 needed for half-maximal assembly was obtained by fitting the maximum endpoint light scattering (OD350) at different IP6 concentrations to the equation: Y=Amin+(Xh)*(Amax-Amin)/(EC50MAh + Xh); where X=concentration of IP6, Y=light scattering, Amin and Amax is the minimum and maximum assembly, EC50MA is the effective concentration for half-maximal mature assembly and h is the Hill slope.
Negative stain
4 μl of sample from the assembly assay was put onto a glow discharged carbon coated grid (Cu, 300 mesh, Electron Microscopy Services), washed and stained with 2% Uranyl-acetate. Micrographs were taken at room temperature on a Tencai Spirit (FEI) operated at an accelerated voltage of 120 keV and Gatan 2k × 2 k CCD camera. Images were collected with a total dose of ~30 e-/A° 2 and a defocus of 1–3 μm.
Cleavage Assays
100 μM ΔMA-CANC was assembled with ssRNA and 50 μM IP6 at 37C for 2h. The protein was diluted 1:2 with cleavage buffer (20 mM [MES] [pH 6.0], 140 mM KCl, 10 mM NaCl, 5 mM MgCl2, 10 mM TCEP) and incubated for 1h at 37C. Protease was added to the assembly mixture at a 1:50 ratio to Gag and incubated at 25°C. Samples were taken at the indicated time points the reaction was stopped with NuPAGE® LDS Sample Buffer (Invitrogen) to stop the reaction, and then subjected to NuPAGE Novex 4-12% Bis-Tris gel (Invitrogen) for cleavage products analysis and visualized by Coomassie blue staining.
Nanoscale Differential Scanning Fluorimetry (NanoDSF)
DSF measurements were performed using a Prometheus NT.48 (NanoTemper Technologies) over a temperature range of 20-95°C using a ramp rate of 2.5°C / min. Samples were taken from assembly reactions under the conditions described in the main text or using 75 μM ΔMA-CANC assembled with ssRNA with and without 50 μM IP6.
Statistical Analysis
Unless otherwise indicated, statistical analyses were Student’s t-tests and performed using GraphPad Prism 9 software (GraphPad). Error bars depict the mean +/-SEM unless indicated otherwise.
CypA-paint TIRF imaging
HEK293T cells were transfected using PEI with a mixture of the plasmids pCRV1 Gag-Pol, pCSGW and pEGFP-PS-Vpr (molar ratio of 1:1.7:2.1) to produce GFP-loaded HIV particles lacking envelope protein. The plasmid pEGFP-PS-Vpr encodes a fusion protein consisting of EGFP, a protease site cleaved by HIV protease and Vpr. This fusion protein is packaged into HIV particles and processed during maturation to release EGFP as a solution phase marker for viral particles. The medium was exchanged 18 h post transfection and the virus-containing medium was collected 72 hours post transfection and centrifuged (2100 x g, 20 min, 4 °C) to remove cells. The viral particles were then biotinylated using EZ-Link Sulfo-NHS-LC-LC-Biotin and purified by size exclusion chromatography.
TIRF microscopy was carried out following the previously published method[19, 42]. Biotinylated viral particles were captured onto coverslips adhered to microfluidic flow cells cast from PDMS and imaged using a custom built TIRF microscope with an ASI-RAMM frame (Applied Scientific Instrumentation), a Nikon 100 x CFI Apochromat TIRF (1.49 NA) oil immersion objective and NicoLase laser system. Immobilised virions were treated with imaging buffer containing 200 nM PFO, to permeabilize the lipid envelope, and labelled CypA (0.5 – 1 μM), to detect the capsid. Dual-colour TIRF images were then acquired with a frequency of 1 frame/6 s using lasers with a 20 ms exposure time for excitation of the EGFP solution phase marker (488 nm) and Alexa Fluor 647-CypA (647 nm) and an Andor iXon 888 EMCCD camera for detection. Single-virion fluorescence traces were extracted from the TIRF image stacks using the JIM Immobilized Microscopy analysis package (https://github.com/lilbutsa/JIM-Immobilized-Microscopy-Suite) and analysed in MATLAB (The MathWorks, Inc). Briefly, the duration of the CypA signal was extracted from fluorescence traces by step-fitting using change point analysis. Capsid stability was quantified as the time difference between acquisition of Alexa Fluor 647-CypA upon permeabilization and loss of fluorescence upon capsid uncoating.
Anti-p24 TIRF imaging
Lentiviruses were produced as described in ‘Virus Production & Infection Experiments’ section but in 2% FBS to decrease cell debris. Supernatants were filtered and analysed by RT qPCR to normalise particle numbers used for experiments. 8-well glass bottom Ibidi-dishes were prepared by covering with Poly-L-lysine (Sigma P4707) for 1 h and washing with PBS. Virions were added to wells for 1 h at dilutions adjusted for RT. After washing, samples were fixed with 4 % formaldehyde (Thermo Scientific 28908) for 20 min and permeabilised with 0.1 % Triton-X100 for 5 min. Capsids were labelled with p24 antibody (mix of Mab EF7 and 38-96k) and VSV-G antibody (Abcam, ab1874) followed by secondary Alexa Fluor 647 against mouse IgG1 and 405 against rabbit. Streptolysin O (SLO, Sigma, S5265) was diluted in 50 μl PBS and 2 mM TCEP to obtain ~ 5 μM stock solution. Bound virions were treated with 100 nM SLO in the presence or absence of 100 μM IP6 (Sigma, 593648) for 30 min at 37°C. After washing with PBS, samples were fixed and processed as above. All images were acquired using a Nikon TIRF inverted microscope with a 100x/1.49NA oil-immersion objective, a 1.5x intermediate magnification and Prime95B sCMOS camera from Photometrics resulting into a 74nm pixel size. All laser powers were kept identical during the imaging. Image analysis was performed in Fiji, where images were median filtered and background subtracted. An intensity threshold was used to create a mask and a watershed step allowed separation of touching particles. The threshold was adjusted to select particles of interest. In particular, the WT/KAKA analysis was performed with particles whose intensity was > 330, to exclude virions that may not have undergone maturation (based on D25A protease mutant viruses). Finally, ROI were filtered by area within 5 to 500 pixels and mean fluorescence intensities measured in the original image. Graphs were made using GraphPad Prism.
Supplementary Material
Footnotes
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Study was conceived by NR, AK, DLM, AS, EOF & LCJ. Manuscript was written by LCJ with contributions from all authors. Experiments were performed by NR, AK, DLM, AA, AS, KMRF. Analysis was carried out by all authors.
Acknowledgements
This work was supported by the MRC (UK; U105181010), a Wellcome Trust Investigator Award (200594/Z/16/Z), a Wellcome Trust Collaborator Award (214344/A/18/Z) and the NHMRC (APP1182212 to TB). Research in the Freed laboratory is supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health. AK was supported in part by an Intramural AIDS Research Fellowship. AS is supported by Medical Research Council UK grant MR/T028904/1. We are grateful to the MRC-LMB Electron Microscopy Facility for access and support of electron microscopy sample preparation and data collection, MRC-LMB Light Microscopy facility, in particular Jerome Boulanger, for help with TIRF image analysis and MRC-LMB Visual Aids department. We thank Vaibhav Shah for help with particle production and TIRF assays at UNSW.
References
- 1.Dick RA, Zadrozny KK, Xu C, Schur FKM, Lyddon TD, Ricana CL, Wagner JM, Perilla JR, Ganser-Pornillos BK, Johnson MC, Pornillos O, et al. Inositol phosphates are assembly co-factors for HIV-1. Nature. 2018;560(7719):509–512. doi: 10.1038/s41586-018-0396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mallery DL, Marquez CL, McEwan WA, Dickson CF, Jacques DA, Anandapadamanaban M, Bichel K, Towers GJ, Saiardi A, Bocking T, James LC. IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis. Elife. 2018;7 doi: 10.7554/eLife.35335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mallery DL, Faysal KMR, Kleinpeter A, Wilson MSC, Vaysburd M, Fletcher AJ, Novikova M, Bocking T, Freed EO, Saiardi A, James LC. Cellular IP6 Levels Limit HIV Production while Viruses that Cannot Efficiently Package IP6 Are Attenuated for Infection and Replication. Cell Rep. 2019;29(12):3983–3996.:e4. doi: 10.1016/j.celrep.2019.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ricana CL, Lyddon TD, Dick RA, Johnson MC. Primate lentiviruses require Inositol hexakisphosphate (IP6) or inositol pentakisphosphate (IP5) for the production of viral particles. PLoS Pathog. 2020;16(8):e1008646. doi: 10.1371/journal.ppat.1008646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sowd GA, Aiken C. Inositol phosphates promote HIV-1 assembly and maturation to facilitate viral spread in human CD4+ T cells. PLoS Pathog. 2021;17(1):e1009190. doi: 10.1371/journal.ppat.1009190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu D, Wilson MS, Eisenbeis VB, Harmel RK, Riemer E, Haas TM, Wittwer C, Jork N, Gu C, Shears SB, Schaaf G, et al. Analysis of inositol phosphate metabolism by capillary electrophoresis electrospray ionization mass spectrometry. Nat Commun. 2020;11(1):6035. doi: 10.1038/s41467-020-19928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Letcher AJ, Schell MJ, Irvine RF. Do mammals make all their own inositol hexakisphosphate? Biochem J. 2008;416(2):263–70. doi: 10.1042/BJ20081417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mallery DL, Kleinpeter AB, Renner N, Faysal KMR, Novikova M, Kiss L, Wilson MSC, Ahsan B, Ke Z, Briggs JAG, Saiardi A, et al. A stable immature lattice packages IP6 for HIV capsid maturation. Sci Adv. 2021;7(11) doi: 10.1126/sciadv.abe4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu C, Fischer DK, Rankovic S, Li W, Dick R, Runge B, Zadorozhnyi R, Ahn J, Aiken C, Polenova T, Engelman AN, et al. Permeability of the HIV-1 capsid to metabolites modulates viral DNA synthesis. bioRxiv. 2020:2020.04.30.071217. doi: 10.1371/journal.pbio.3001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adamson CS, Ablan SD, Boeras I, Goila-Gaur R, Soheilian F, Nagashima K, Li F, Salzwedel K, Sakalian M, Wild CT, Freed EO. In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (Bevirimat) J Virol. 2006;80(22):10957–71. doi: 10.1128/JVI.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waki K, Durell SR, Soheilian F, Nagashima K, Butler SL, Freed EO. Structural and functional insights into the HIV-1 maturation inhibitor binding pocket. PLoS Pathog. 2012;8(11):e1002997. doi: 10.1371/journal.ppat.1002997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacques DA, McEwan WA, Hilditch L, Price AJ, Towers GJ, James LC. HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nature. 2016;536(7616):349–353. doi: 10.1038/nature19098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanman J, Sexton J, Sakalian M, Prevelige PE., Jr Kinetic analysis of the role of intersubunit interactions in human immunodeficiency virus type 1 capsid protein assembly in vitro. J Virol. 2002;76(14):6900–8. doi: 10.1128/JVI.76.14.6900-6908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kucharska I, Ding P, Zadrozny KK, Dick RA, Summers MF, Ganser-Pornillos BK, Pornillos O. Biochemical Reconstitution of HIV-1 Assembly and Maturation. J Virol. 2020;94(5) doi: 10.1128/JVI.01844-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Endres D, Zlotnick A. Model-based analysis of assembly kinetics for virus capsids or other spherical polymers. Biophys J. 2002;83(2):1217–30. doi: 10.1016/S0006-3495(02)75245-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chi H, Yang X, Kingsley PD, O’Keefe RJ, Puzas JE, Rosier RN, Shears SB, Reynolds PR. Targeted deletion of Minpp1 provides new insight into the activity of multiple inositol polyphosphate phosphatase in vivo. Mol Cell Biol. 2000;20(17):6496–507. doi: 10.1128/mcb.20.17.6496-6507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clift D, McEwan WA, Labzin LI, Konieczny V, Mogessie B, James LC, Schuh M. A Method for the Acute and Rapid Degradation of Endogenous Proteins. Cell. 2017;171(7):1692–1706.:e18. doi: 10.1016/j.cell.2017.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dostalkova A, Kaufman F, Krizova I, Vokata B, Ruml T, Rumlova M. In Vitro Quantification of the Effects of IP6 and Other Small Polyanions on Immature HIV-1 Particle Assembly and Core Stability. J Virol. 2020;94(20) doi: 10.1128/JVI.00991-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marquez CL, Lau D, Walsh J, Shah V, McGuinness C, Wong A, Aggarwal A, Parker MW, Jacques DA, Turville S, Bocking T. Kinetics of HIV-1 capsid uncoating revealed by single-molecule analysis. Elife. 2018;7 doi: 10.7554/eLife.34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loeb DD, Swanstrom R, Everitt L, Manchester M, Stamper SE, Hutchison CA., 3rd Complete mutagenesis of the HIV-1 protease. Nature. 1989;340(6232):397–400. doi: 10.1038/340397a0. [DOI] [PubMed] [Google Scholar]
- 21.Marquez CL, Lau D, Walsh J, Faysal KMR, Parker MW, Turville SG, Bocking T. Fluorescence Microscopy Assay to Measure HIV-1 Capsid Uncoating Kinetics in vitro. Bio Protoc. 2019;9(13):e3297. doi: 10.21769/BioProtoc.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dick RA, Mallery DL, Vogt VM, James LC. IP6 Regulation of HIV Capsid Assembly, Stability, and Uncoating. Viruses. 2018;10(11) doi: 10.3390/v10110640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renner N, Mallery DL, Faysal KMR, Peng W, Jacques DA, Bocking T, James LC. A lysine ring in HIV capsid pores coordinates IP6 to drive mature capsid assembly. PLoS Pathog. 2021;17(2):e1009164. doi: 10.1371/journal.ppat.1009164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veiga N, Torres J, Dominguez S, Mederos A, Irvine RF, Diaz A, Kremer C. The behaviour of myo-inositol hexakisphosphate in the presence of magnesium(II) and calcium(II): protein-free soluble InsP6 is limited to 49 microM under cytosolic/nuclear conditions. J Inorg Biochem. 2006;100(11):1800–10. doi: 10.1016/j.jinorgbio.2006.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan A, Pak AJ, Morado DR, Voth GA, Briggs JAG. Immature HIV-1 assembles from Gag dimers leaving partial hexamers at lattice edges as potential substrates for proteolytic maturation. Proc Natl Acad Sci U S A. 2021;118(3) doi: 10.1073/pnas.2020054118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ganser-Pornillos BK, Yeager M, Pornillos O. Assembly and architecture of HIV. Adv Exp Med Biol. 2012;726:441–65. doi: 10.1007/978-1-4614-0980-9_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller PW, Huang RK, England MR, Waki K, Cheng N, Heymann JB, Craven RC, Freed EO, Steven AC. A two-pronged structural analysis of retroviral maturation indicates that core formation proceeds by a disassembly-reassembly pathway rather than a displacive transition. J Virol. 2013;87(24):13655–64. doi: 10.1128/JVI.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ning J, Erdemci-Tandogan G, Yufenyuy EL, Wagner J, Himes BA, Zhao G, Aiken C, Zandi R, Zhang P. In vitro protease cleavage and computer simulations reveal the HIV-1 capsid maturation pathway. Nat Commun. 2016;7:13689. doi: 10.1038/ncomms13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schur FK, Hagen WJ, Rumlova M, Ruml T, Muller B, Krausslich HG, Briggs JA. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 A resolution. Nature. 2015;517(7535):505–8. doi: 10.1038/nature13838. [DOI] [PubMed] [Google Scholar]
- 30.Obr M, Ricana CL, Nikulin N, Feathers JR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA. Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer. Nat Commun. 2021;12(1):3226. doi: 10.1038/s41467-021-23506-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azevedo C, Saiardi A. Extraction and analysis of soluble inositol polyphosphates from yeast. Nat Protoc. 2006;1(5):2416–22. doi: 10.1038/nprot.2006.337. [DOI] [PubMed] [Google Scholar]
- 32.Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. Journal of virology. 2004;78(21):12058–61. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272(5259):263–7. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 34.Price AJ, Jacques DA, McEwan WA, Fletcher AJ, Essig S, Chin JW, Halambage UD, Aiken C, James LC. Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly. PLoS pathogens. 2014;10(10):e1004459. doi: 10.1371/journal.ppat.1004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vermeire J, Naessens E, Vanderstraeten H, Landi A, Iannucci V, Van Nuffel A, Taghon T, Pizzato M, Verhasselt B. Quantification of reverse transcriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PLoS One. 2012;7(12):e50859. doi: 10.1371/journal.pone.0050859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waheed AA, Ono A, Freed EO. Methods for the study of HIV-1 assembly. Methods Mol Biol. 2009;485:163–84. doi: 10.1007/978-1-59745-170-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toohey K, Wehrly K, Nishio J, Perryman S, Chesebro B. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology. 1995;213(1):70–9. doi: 10.1006/viro.1995.1547. [DOI] [PubMed] [Google Scholar]
- 38.Wehrly K, Chesebro B. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods. 1997;12(4):288–93. doi: 10.1006/meth.1997.0481. [DOI] [PubMed] [Google Scholar]
- 39.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–6. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen HL, Nguyen TT, Vu QT, Le HT, Pham Y, Trinh PL, Bui TP, Phan TN. An efficient procedure for the expression and purification of HIV-1 protease from inclusion bodies. Protein Expr Purif. 2015;116:59–65. doi: 10.1016/j.pep.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 42.Márquez CL, Lau D, Walsh J, Rifat Faysal K, Parker MW, Turville SG, Böcking T. Fluorescence Microscopy Assay to Measure HIV-1 Capsid Uncoating Kinetics in vitro. Bio-protocol. 2019;9(13):e3297. doi: 10.21769/BioProtoc.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.