Abstract
Unlike early transcriptional responses to mitogens, later events are less well-characterized. Here, we identified delayed down-regulated genes (DDGs) in mammary cells after prolonged treatment with epidermal growth factor (EGF). The expression of these DDGs was low in mammary tumors and correlated with prognosis. The proteins encoded by several DDGs directly bind to and inactivate oncoproteins and might therefore act as tumor suppressors. The transcription factor teashirt zinc finger homeobox 2 (TSHZ2) is encoded by a DDG, and we found that overexpression of TSHZ2 inhibited tumor growth and metastasis and accelerated mammary gland development in mice. Although the gene TSHZ2 localizes to a locus (20q13.2) that is frequently amplified in breast cancer, we found that hypermethylation of its promoter correlated with down-regulation of TSHZ2 expression in patients. Yeast two-hybrid screens and protein-fragment complementation assays in mammalian cells indicated that TSHZ2 nucleated a multiprotein complex containing PRC1/Ase1, cyclin B1, and additional proteins that regulate cytokinesis. TSHZ2 increased the inhibitory phosphorylation of PRC1, a key driver of mitosis, mediated by cyclin-dependent kinases. Furthermore, similar to the tumor suppressive transcription factor p53, TSHZ2 inhibited transcription from the PRC1 promoter. By recognizing DDGs as a distinct group in the transcriptional response to EGF, our findings uncover a group of tumor suppressors and reveal a role for TSHZ2 in cell cycle regulation.
Introduction
While cancer almost invariably initiates when mutations start accumulating in individual cells, due to external or internal mechanisms (1), the simple concept assuming that tumor progression depends solely on the mutations and intrinsic properties of cancer cells has given way to more complex paradigms in which progression depends on metabolic and other interactions between tumors and the host (2, 3). For example, the tumor microenvironment profoundly influences not only cell growth but also angiogenesis and dissemination, as well as suppresses immune responses (4, 5). The cross-talk between tumor cells and their stroma is mediated by both insoluble molecules, such as components of the extracellular matrix, and soluble molecules, such as metabolites and peptides. Soluble polypeptides mediating paracrine (heterotypic) signaling between epithelial and stromal cells critically regulate the proliferative and metastatic behavior of cancer cells. For example, colony-stimulating factor 1 produced by mammary carcinoma cells promotes expression of the epidermal growth factor (EGF) by macrophages, and this enhances matrix invasion by carcinoma cells (6).
Resolving the logic of gene expression programs stimulated in context of paracrine and autocrine modes of cell regulation might disclose mechanisms relevant to the interaction between tumors and their stroma. Our previous studies portrayed a wave-like gene regulatory program, which initiates by rapid disappearance of 24 microRNAs, followed by an abrupt rise of immediate early genes (IEGs) (7). Concurrent with the fall of IEGs, a larger group—the delayed early genes (DEGs), which encode for phosphatases and other inhibitors (8)—is up-regulated, and its own fall overlaps the rise or fall of the final wave of late-response genes. The delayed wave persists and determines long-term phenotype acquisition. Here, we set out to investigate these late-wave genes, especially a persistently down-regulated group. Assuming that this group normally arrests cell growth, we focused therein on a prototypic gene, teashirt zing finger homeobox 2 (TSHZ2), the insect paralog of which patterns the trunk and midgut (9). TSHZ2 is commonly down-regulated in breast cancer, although it maps to an amplicon. This, we found here, may result from targeted promoter hypermethylation, which is frequent among patients with breast cancer. Upon further investigation of TSHZ2, our findings uncovered a new mechanism of cell cycle regulation that is altered in patients with breast cancer.
Results
Multiple genes undergoing down-regulation after mitogenic stimulation are lowly expressed in tumors and this correlates with shorter survival of patients with breast cancer
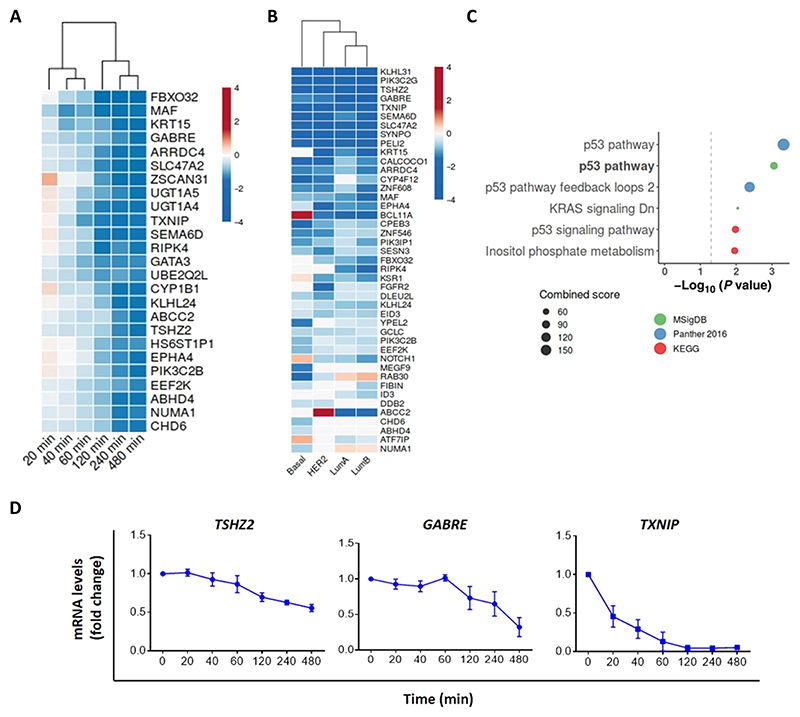
To interrogate cellular functions of the late wave of EGF-responsive genes, we focused on a group of delayed down-regulated genes (DDGs) (10). About 100 genes were persistently repressed after stimulation of mammary cells with EGF (table S1). This group was subjected to ranking according to statistical significance of down-regulation by EGF, and the top 25 are shown, including TSHZ2 (Fig. 1A). Next, we analyzed expression of all DDGs in the TCGA (The Cancer Genome Atlas) database of patients with breast cancer (n = 838, including 86 normal breast samples) and classified them by their molecular subtype. Thereafter, we computed differential expression using DESeq2, to select those genes that were significantly down-regulated in malignant tissue, as compared to the adjacent normal tissue. This analysis identified 43 genes that were down-regulated in two or more molecular subtypes (Fig. 1B). Using these 43 genes as seeds for enrichment analysis that used three different collections of annotated gene sets [the Molecular Signatures Database (MsigDB), Panther, and Kyoto Encyclopedia of Genes and Genomes (KEGG)] revealed that all three tools converged on the “P53 pathway” as the most significantly represented (Fig. 1C). The gene encoding p53 is the most frequently mutated tumor suppressor gene in human cancers, with deletions and point mutations seen in nearly half of all tumors (11). Therefore, the identification of the p53 pathway as the most enriched group in the cohort of genes down-regulated after stimulation with a mitogen is in line with growth suppressive functions of TSHZ2.
Fig. 1. Several delayed down-regulated genes are lowly expressed in mammary tumors, relative to peritumor regions.
(A) Heatmap of top 25 genes that were consistently down-regulated in MCF10A cells after 240- to 480-min-long exposure to EGF. Data are from GSE24391. (B) The ratio of gene expression in tumors versus expression in the respective peritumors of the four subtypes of breast cancer was analyzed using TCGA data. The heatmap presents the DDGs lowly expressed in at least one subtype of breast cancer. (C) Pathway overrepresentation analysis using MSigDB, Panther 2016, and KEGG databases. The group of 43 genes from (B) was used. Shown are the most significant pathways (P < 0.05). Note that “KRAS signaling Dn” refers to genes down-regulated by KRAS activation. (D) Real-time qPCR using mRNA isolated from EGF-stimulated MCF10 cells to assess transcript levels of three DDGs. Shown are fold changes relative to basal levels (means ± SEM of three independent experiments). GAPDH transcripts were used for normalization.
Next, we used real-time quantitative polymerase chain reaction (RT-qPCR) to verify persistent down-regulation of three representative DDGs: TSHZ2, GABRE, and TXNIP (Fig. 1D). To examine association between DDGs and prognosis of patients with breast cancer, we used the Kaplan-Meier (KM) plotter database (12). As predicted, high expression levels of the majority of genes associated with longer relapse-free survival of patients (exemplified by nine shown here; fig. S1). These observations motivated us to study indepth one to two DDGs. As a prelude, we considered previous experimental data linking specific DDGs to cancer and summarized the most relevant genes (table S2). Unexpectedly, the list included several putative or established suppressors of tumor growth. For example, FBXO32 encodes an E3 ubiquitin ligase that suppresses breast cancer by sorting for degradation the Krüppel-like factor 4 (KLF4), an oncogenic transcription factor (13). Another, SEMA6D, encodes a repulsive axon guidance factor playing roles in promoting survival of patients with breast cancer (14). These observations proposed that some DDGs function as inducers of growth arrest or as tumor suppressor genes. In summary, a subset of genes that undergo down-regulation in response to mitogenic stimulation was found to be associated with growth arrest. Correspondingly, this group is lowly expressed in tumors relative to adjacent tissues, and high expression in tumors predicts relatively longer survival in patients.
EGF induces down-regulation of the thioredoxin-interacting protein, which binds AKT and predicts survival of patients with breast cancer
One DDG-encoded protein we identified was thioredoxin-interacting protein (TXNIP), which binds to and inhibits thioredoxin and, hence, modulates the cellular redox state (15). First, we confirmed EGF- and time-dependent down-regulation of the 50-kDa TXNIP protein (fig. S2A). Studies performed using TXNIP-ablated fibroblasts revealed physical binding with and inhibition of the kinase AKT (16). Because EGF can stimulate AKT, these findings raised the possibility that down-regulation of TXNIP can enhance activation of AKT in cancer cells stimulated by stromal growth factors. Hence, we next examined one aspect of this model in mammary cells, namely, physical interactions between TXNIP and AKT. To verify interactions and colocalization, we applied the proximity ligation assay (PLA), which uses oligonucleotides attached to antibodies against two proteins of interest (17). Accordingly, we applied antibodies to the endogenous forms of AKT and TXNIP and observed strong protein-protein interactions in mammary cells (fig. S2B).
Per one report, TXNIP expression in breast cancer is inversely correlated to the abundance of the EGF family receptor HER2 (human EGFR 2) as well as associated with better overall patient survival (18). These attributes are consistent with the identification of TXNIP as a DDG. To extend the analysis and specifically refer to disease subtypes devoid of the estrogen receptor (ER), we applied the KM plotter web tool. The resulting curves present the overall survival probability of patients with ER-positive tumors versus ER-negative tumors (fig. S2C). These analyses confirmed statistically significant correlations between high TXNIP and longer patient survival time, but ER status may not affect the prognostic impact of TXNIP. In conclusion, TXNIP emerges as a prototype DDG: The respective protein undergoes down-regulation in response to mitogenic stimulation, and it directly interacts with oncogenic proteins like AKT and thioredoxin.
Reduced expression of TSHZ2 characterizes invasive carcinomas and predicts shorter patient survival
Unlike the well-studied functions of TXNIP, only two previous studies implicated TSHZ2 in cancer (19, 20). Hence, we made use of this DDG to test the hypothesis arguing that a fraction of the DDGs encodes yet uncharacterized tumor suppressors. TSHZ2 is a nuclear protein acting as a transcriptional repressor of zinc finger protein GLI1, in the presence of the corepressor CTBP2 (19). To confirm EGF-induced down-regulation of the corresponding transcript, we isolated RNA from the immortalized 184A1 cell line, which was established from a normal mammary tissue. Cells were prestimulated with EGF for different time intervals, before isolating RNA and RT-qPCR analysis (fig. S3A). In line with the DDG assignment, we observed delayed, time-dependent down-regulation of TSHZ2 transcripts. Similarly, immunoblotting of cell extracts confirmed partial down-regulation of the respective protein (fig. S3B).
Next, we examined the abundance of TSHZ2 in breast cancer. To begin with, we surveyed a collection of 83 human mammary cell lines (Gene Expression Omnibus: GSE73526), including four un-transformed cells and 79 cancer lines derived from all subtypes (fig. S3C). Three of the four untransformed lines exhibited detectable expression of TSHZ2, whereas only one of the four transformed lines did. Next, we compared the abundance of TSHZ2 transcripts in invasive ductal carcinomas from patients with breast cancer versus that in the mammary epithelium of otherwise healthy donors. We examined two datasets that collectively comprised 1556 invasive tumors and 144 control tissues and found a substantial decrease in TSHZ2 abundance in the invasive tumors (fig. S3D). That down-regulation of this homeobox transcription factor confers that survival benefits was apparent from the patient survival curves (fig. S3E). A small but statistically significant separation of the curves supported the notion that high abundance of TSHZ2 contributes to longer patient survival. Thus, our findings collectively indicate that reduced expression of TSHZ2 characterizes EGF-stimulated mammary cells and invasive ductal carcinomas and predicts shorter survival of patients with breast cancer.
Despite amplification of the corresponding locus, promoter hypermethylation suppresses TSHZ2 expression in mammary tumors
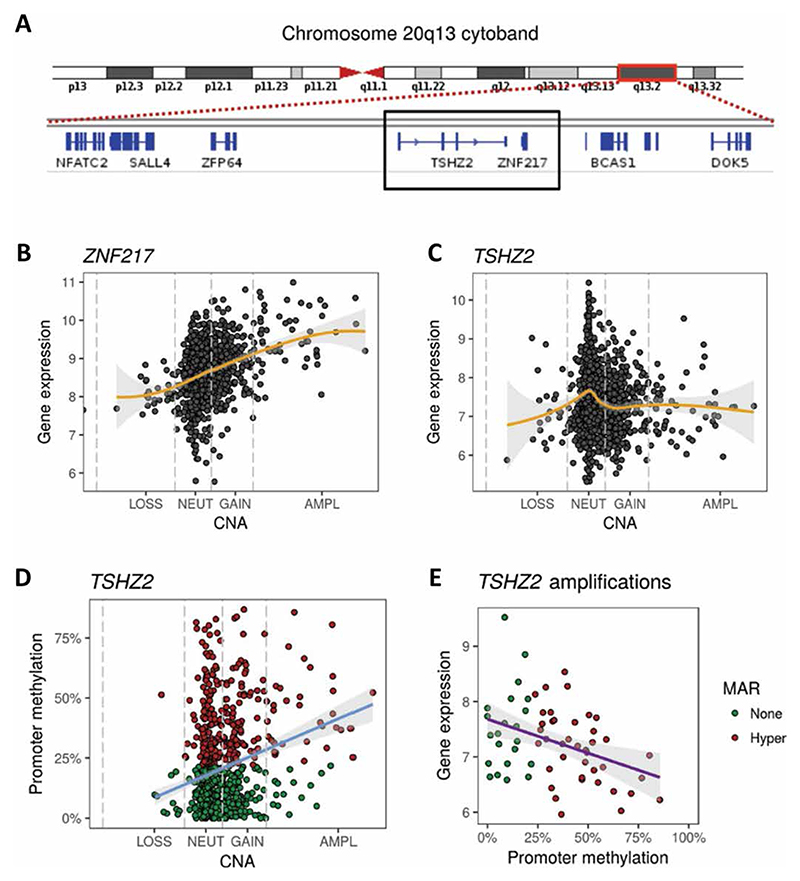
Comparative genomic hybridization analyses and other approaches revealed that breast cancer is driven largely by genomic copy number aberrations (CNAs) rather than point mutations or indels (21). A well-characterized gain of chromosome 20q sequences, which includes the locus of TSHZ2 (20q13.2) (Fig. 2A), has been identified in breast cancer (22, 23), as well as in colorectal and other tumors. The association between gain of an about 0.5-Mb region centered at TSHZ2 and 20q13.2, along with the observed frequent down-regulation of TSHZ2 in tumors, raised the question how an amplicon resident is frequently down-regulated in mammary tumors, despite the regional numerical gain. The likely target of the gain is ZNF217, which flanks TSHZ2, and similarly encodes a zinc finger repressor amplified not only in breast but also in ovarian tumors (24). We posited that down-regulation of TSHZ2 is driven by targeted methylation of the TSHZ2 promoter in a fraction of breast tumors. To begin addressing this model, we analyzed CNAs in a set of 1482 breast tumors and found that 62 tumors had four or more copies of ZNF217, which is defined as an amplification. Next, we examined relationships between expression of ZNF217 and CNA. ZNF217 displayed the expected increase in expression, such that breast tumors with the amplification exhibited significantly higher expression of ZNF217 (Fig. 2B). Although all 62 tumors displayed an amplification of the adjacent gene (Fig. 2B), they did not show higher expression of TSHZ2 (Fig. 2C). In line with the proposed model, examining the promoter of TSHZ2 revealed that breast tumors with the amplification had higher average promoter methylation (Fig. 2D). About 64% of tumors with amplifications had significant, 3.5-fold enrichment in promoter hypermethylation. Exploring the regulatory role of TSHZ2 promoter methylation revealed that hypermethylation was associated with down-regulation of the gene in tumors with amplifications (Fig. 2E). In summary, the 20q13.2 amplicon of breast cancer encodes two zinc finger transcription factors: ZNF217, which is oncogenic and frequently amplified in mammary tumors, and the tumor suppressive gene TSHZ2, which is frequently down-regulated in breast cancer. Our data indicate that this configuration of gene coamplification is permitted by virtue of targeted hypermethylation of the TSHZ2 promoter.
Fig. 2. The anticipated effects of copy number amplification of the TZHZ2 locus are buffered by promoter hypermethylation.
(A) Shown is the cytoband corresponding to 20q13. The centromere (red) and chromosome arms are shown. The red rectangle is enlarged, and the respective genes (blue) are indicated. The black rectangle highlights TSHZ2 and the adjacent gene, ZNF217. (B and C) Relationships between expression levels of ZNF217 (B) or TSHZ2 (C) and copy number alteration (CNA) for 1482 breast tumors. The gold line represents the LOESS smoothed curve (95% confidence interval; shaded in gray). Note that 62 tumors had four or more copies of ZNF217, which is defined as an amplification. (D) Relationship between TSHZ2 promoter methylation and CNA for 1482 breast tumors. The blue line represents the linear regression slope. LOSS, loss of copy number; NEUT, neutral copy number; GAIN, gain of copy number; AMPL, amplification of copy number. P = 1.4 × 10-6 by Fisher’s exact test. (E) Relationships between TSHZ2 expression and promoter methylation for tumors with amplification (copies ≥ 4). The purple line represents the linear regression slope (95% confidence interval, shaded gray; P = 0.0024). MAR, methylation altered region.
In MCF10A normal mammary cells, EGF-induced downregulation of TSHZ2 is mediated by the EGFR-to-MEK pathway and controls cell cycle progression
The response of mammary cells to EGF comprises simultaneous activation of multiple signaling pathways. To identify the pathway responsible for activation-induced down-regulation of TSHZ2, we preincubated MCF10A mammary cells with various inhibitors, each blocked a specific signaling route. Thereafter, cells were stimulated with EGF, and levels of TSHZ2 mRNA and protein were determined. The results (fig. S4, A and B) confirmed a requirement for EGF receptor (EGFR) activation, because an EGFR-specific kinase inhibitor blocked the down-regulation of TSHZ2. Of the other inhibitors used, a MEK (MAPK/ERK kinase)–specific drug better inhibited ligand-induced down-regulation of both mRNA and protein corresponding to TSHZ2. Hence, we concluded that EGF-stimulated MEK-to-ERK signaling is essential for inhibiting TSHZ2.
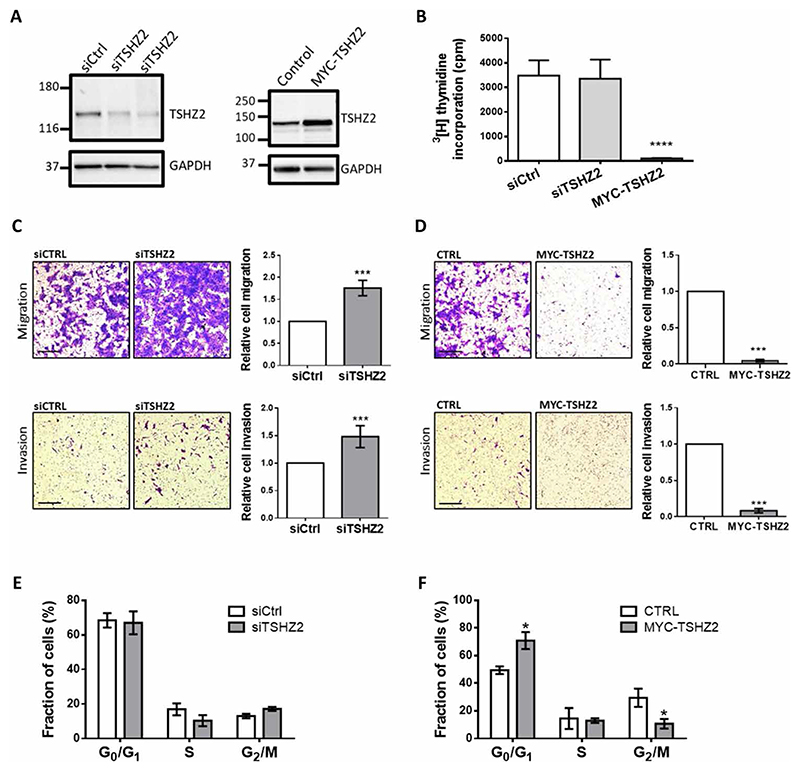
Because EGFR activation and downstream signaling to MEK and ERK control multiple mRNAs, we next applied loss- and gain-of-function strategies to singly address the functional consequences of TSHZ2 down-regulation. MCF10A cells were transfected with either small interfering RNA (siRNA) oligonucleotides specific to TSHZ2 or with an expression vector encoding a MYC-tagged TSHZ2. Immunoblotting analyses of cell extracts confirmed the expected changes in TSHZ2 protein levels (Fig. 3A). Next, we measured the incorporation of radioactive thymidine into DNA. The results (Fig. 3B) indicated that overexpression of TSHZ2 inhibited incorporation of the radionucleoside into DNA, but partial down-regulation of TSHZ2 induced only minor effects. A recent study concluded that motility markers of mammary cancer significantly better associate with patient survival than proliferation markers (25). To measure effects on cell migration, we plated MC-F10A cells in the upper compartment of transwell trays and allowed them to migrate for 21 hours through an intervening filter. Thereafter, cells that migrated across the filter were photographed. A similar protocol examined invasion across tissue barriers by coating the intervening filters with a preparation of extracellular matrix. The results of both assays reflected strong inhibitory effects of TSHZ2 on cellular motility (Fig. 3, C and D), but this might also incorporate effects on cell proliferation. Yet, another assay determined cell cycle distributions of the genetically manipulated MCF10A cells. Flow cytometry of cells prelabeled with bromodeoxyuridine (BrdU) quantified cells at the G0/G1 phase versus the S or G2/M phases (Fig. 3, E and F). While no consistent effects of knocking down TSHZ2 were detectable, we noted clear effects of the overexpressed transcription factor: Significantly, more cells were found at the G0/G1 phase, and correspondingly fewer cells were captured at the G2/M phase. In conclusion, by activating the EGFR-to-MEK pathway, mammary cells stimulated with EGF down-regulate TSHZ2 levels; this arrests cell growth and retards their ability to migrate and invade across extracellular matrix barriers.
Fig. 3. TSHZ2 abundance controls DNA synthesis, cell cycle progression, and motility of normal mammary cells.
(A) Cell extracts from MCF10A cells pretransfected with siTSHZ2 (or control siRNAs), or with a plasmid encoding a MYC-tagged TSHZ2, were analyzed 48 hours later using immunoblots. (B) Cells from (A) were plated, and 12 hours later, the media were replaced with media containing 3H-thymidine. Acid-precipitable radioactivity was determined 48 hours later and presented as averages + SEM (triplicates). One-way analysis of variance (ANOVA) with Dunnett’s multiple comparison test: ****P < 0.0001. (C and D) MCF10A cells pretransfected with either siRNAs or a MYC-tagged TSHZ2 plasmid were seeded in migration or invasion chambers. After 21 hours, migrating or invading cells were stained. Photos of five fields were quantified and presented as means ± SEM of four independent experiments. Scale bars, 100 μm. Unpaired t test with Welch’s correction: ***P < 0.001. (E and F) Cell cycle analysis, using flow cytometry of MCF10 cells transfected with siTSHZ2, control siRNA, or MYC-TSHZ2 plasmid (untransfected cells, Ctrl) and labeled with BrdU. Shown are means ± SEM of three experiments. Multiple t-test (two-tailed): *P < 0.05.
Overexpression of TSHZ2 unveils inhibitory roles in tumor growth and metastasis
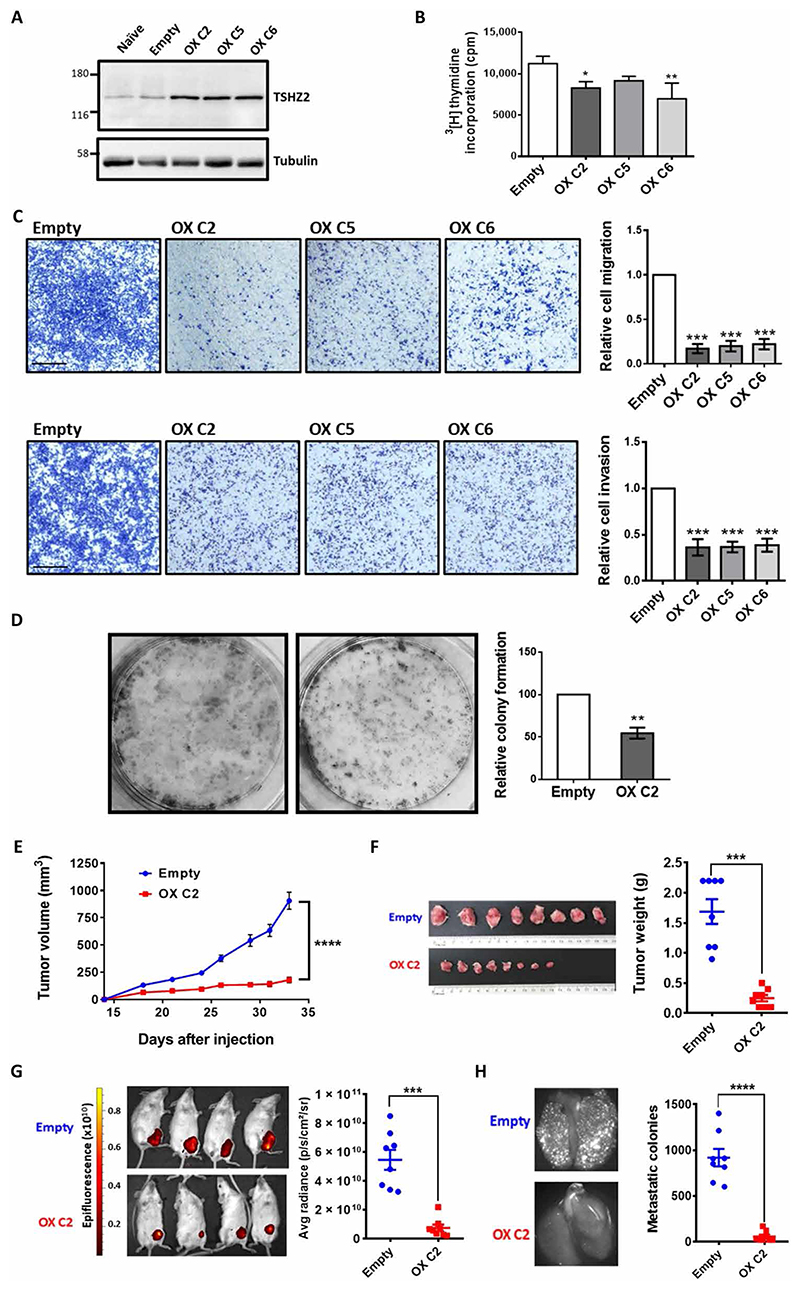
MDA-MB-231 cells expressing very low levels of TSHZ2 (20) constitute a suitable model of breast cancer, because they are highly invasive in vitro, and, in animal models, they spontaneously metastasize. Hence, we established derivatives that inducibly (or stably) overexpress TSHZ2. Cells expressing TSHZ2 under control of the tetracycline-inducible expression system (iTSHZ2 cells) were exposed to increasing concentrations of the inducer (doxycycline). Immunoblotting of cell extracts confirmed inducibility of the ectopic TSHZ2 (fig. S5A), and accordingly, viability assays indicated that the addition of doxycycline was followed by reduced viability (fig. S5B). In analogy to observations made with MCF10A cells, adding doxycycline to the growth medium of iTSHZ2 cells was sufficient to inhibit both cell migration (fig. S5C) and cell invasion (fig. S5D), but control cells did not detectably respond to the inducer. To confirm these observations, we established another derivative of MDA-MB-231 cells, which stably overexpressed TSHZ2 fused to the Discosoma red fluorescent protein (DsRed; Fig. 4A). Subsequent tests, which examined proliferation (Fig. 4B), migration and invasion (Fig. 4C), as well as colony formation (Fig. 4D), confirmed that the clones we established displayed reduced activities in vitro.
Fig. 4. Overexpression of TSHZ2 inhibits growth, motility, and colony formation in vitro, as well as metastasis and tumor growth in animals.
(A) Three MDA-MB-231 single-cell clones pretransfected with a DsRed-TSHZ2 plasmid were analyzed using immunoblotting. (B) Radioactive thymidine incorporation assays were performed with the indicated TSHZ2-overexpressing clones, in triplicates. (C) Migration and invasion assays were performed with MDA-MB-231 clones (6 × 104 cells), in triplicates. Scale bars, 500 μm. (D) Cells (2 × 103) stably overexpressing either TSHZ2 (C2 clone), or an empty plasmid, were incubated for 7 to 10 days. Colonies were stained and four fields were quantified (n = 5). (E and F) C2 or control MDA-MB-231 cells (2.5 × 106) were injected into the subaxillary fat pad of female SCID mice (n = 8 per group). Tumor sizes were measured twice weekly (E), and excised tumors and their weights were assessed (F). (G and H) Primary control tumors or tumors overexpressing DsRed-TSHZ2 were detected non-invasively. Mice were euthanized and the lungs were removed to visualize micro-metastases. Representative fluorescence images of lungs are shown. Data are means ± SEM. One-way ANOVA with Dunnett’s multiple comparison test (B and C), two-tailed t test (D), two-way ANOVA with Bonferroni’s multiple comparisons test (E), and by unpaired t test with Welch’s correction (F, G, and H): *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
For in vivo tests, we implanted the overexpressing C2 clone in subaxillary mammary fat pads of female severe combined immunodeficient (SCID) mice (eight mice per group). Palpable tumors emerged after a few days and their sizes were measured twice weekly. The results (Fig. 4E) indicated that overexpression of TSHZ2 retarded the rate of tumorigenic growth. Two additional tests confirmed this conclusion: (i) The average weights of the excised tumors displayed similarly large differences (Fig. 4F), and (ii) noninvasive imaging of DsRed fluorescence, which we recorded in anesthetized mice, reflected large differences between the two groups (Fig. 4G). To follow metastasis, we harvested all lungs at the end of the experiment and used a stereomicroscope equipped with a fluorescence illuminator. As expected, data quantification indicated that TSHZ2 overexpression inhibited metastasis from the mammary fat pad to lungs (Fig. 4H). Because metastasis might be affected as an indirect consequence of primary tumor growth, we injected red fluorescent protein–labeled MDA-MB-231 into the mammary fat pad of NOG (NOD/Shi-scid/IL-2Rγnull) mice. Once tumors reached about 250 mm3 (fig. S5E), they were resected. Control animals (Dox−) were fed with water containing sucrose, and the experimental group was treated with doxycycline and sucrose (Dox+). After 4 weeks of treatment, all animals were sacrificed. As shown, unlike untreated animals, the group treated with doxycycline showed marked reductions in lung metastasis (fig. S5F). These observations imply a direct impact of TSHZ2 on the metastatic potential of breast cancer cells, independent of the effects exerted on primary tumor growth. In conclusion, when overexpressed in highly tumorigenic breast cancer cells, the homeobox transcription factor acted as a suppressor of all attributes of cancer cells we tested, including tumorigenic growth in an orthotopic animal model and metastasis to the lung.
Loss-of-function strategies using shRNAs or Tshz2 ablation validated involvement in tumor growth and metastasis
The consistently weaker impact of TSHZ2 knockdown using siRNAs (Fig. 3), as opposed to the stronger influence of overexpression, could be due to a threshold effect or to the cell line we used. To circumvent such issues, we used shRNAs and HCC70 human triple negative breast cancer cells. First, we established by means of viral transduction several sublines, each stably and singly expressing a different shRNA specific to TSHZ2. Immunoblotting of the corresponding extracts confirmed variable but consistently lowered levels of the TSHZ2 protein (fig. S6A). Next, we performed a series of assays that compared control cells and two shTSHZ2 clones: assays assessing cell viability (fig. S6B), migration and invasion (fig. S6, C and D), and colony formation (fig. S6E). In all four assays, the shTSHZ2 clones displayed enhanced activities relative to shControl cells, confirming inhibition of both proliferation and motility by TSHZ2.
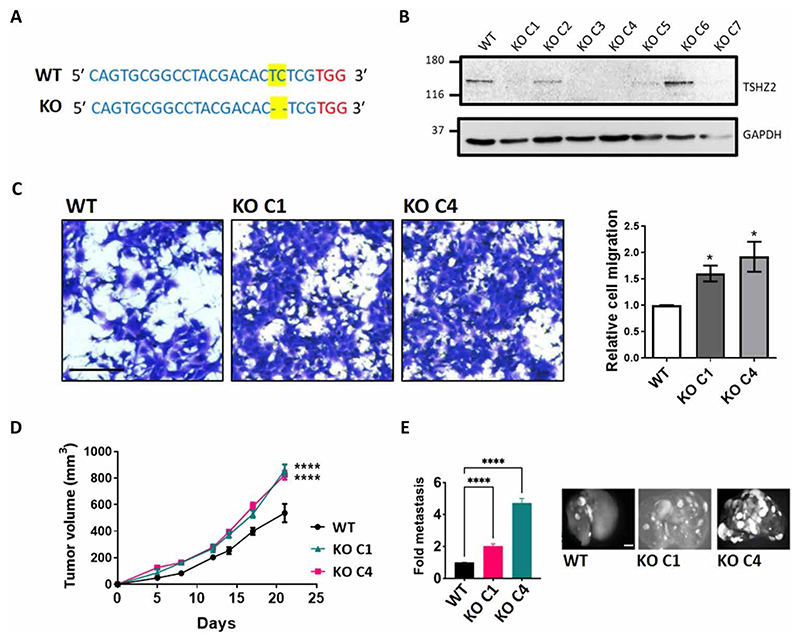
As an alternative loss-of-function strategy, we used the CRISPR-Cas9 technology to ablate expression of murine Tshz2, as described (26). After selection of cell clones under puromycin and confirming deletion of two bases (Fig. 5A), we subjected both clones to analysis in vitro and in animals. The results confirmed gene ablation (Fig. 5B) and indicated that this accelerated migration of 4T1 clones (Fig. 5, B and C). Furthermore, the resulting 4T1 clones exhibited moderately enhanced tumor growth in immunocompetent BALB/c mice (Fig. 5D) and strongly enhanced metastasis in the tail vein assay (Fig. 5E). In summary, the combination of gain- and loss-of-function approaches, which we applied in vitro and in animals, confirmed the ability of TSHZ2 to suppress attributes of malignancy (in vitro), as well as tumor progression in both immunocompetent and immune-compromised animal models.
Fig. 5. Tshz2 knockout enhances migration in vitro and both metastasis and tumor growth in animals.
(A) The CRISPR-Cas9 technology was used to ablate expression of Tshz2 in murine mammary 4T1 cells. Cell clones were selected under puromycin and assayed using DNA sequencing to confirm deletion of a pair of DNA bases (yellow), next to the chosen PAM sequence (red). (B) Whole extracts derived from the parental [wild-type (WT)] and KO clones were analyzed using immunoblotting. (C) Migration assays were performed with parental cells and two selected clones (KO C1 and KO C4). Scale bar, 200 μm (n = 3). (D) WT, KO C1, or KO C4 4T1 cells (1 × 106) were injected into the subaxillary fat pad of female BALB/c mice (n = 6, per group). Tumor volumes were measured once every 3 days until day 22. (E) WT, KO C1, or KO C4 4T1 cells (2 × 105) labeled with GFP were injected into the tail vein of female BALB/c mice (n = 6 per group), and animals were followed for 4 weeks. After sacrificing the mice, the lungs were excised and photographed. The bar graph shows the quantification of the micro-metastatic tumor nodules. Representative images of lungs are shown. Scale bar, 0.5 cm. Data are means + SEM. One-way ANOVA with Dunnett’s multiple comparison test (C) or two-way ANOVA with Dunnett’s multiple comparison test (D and E): *P < 0.05 and ****P < 0.0001.
TSHZ2 overexpression in the mammary gland of mice increases ductal branching and bud maturation
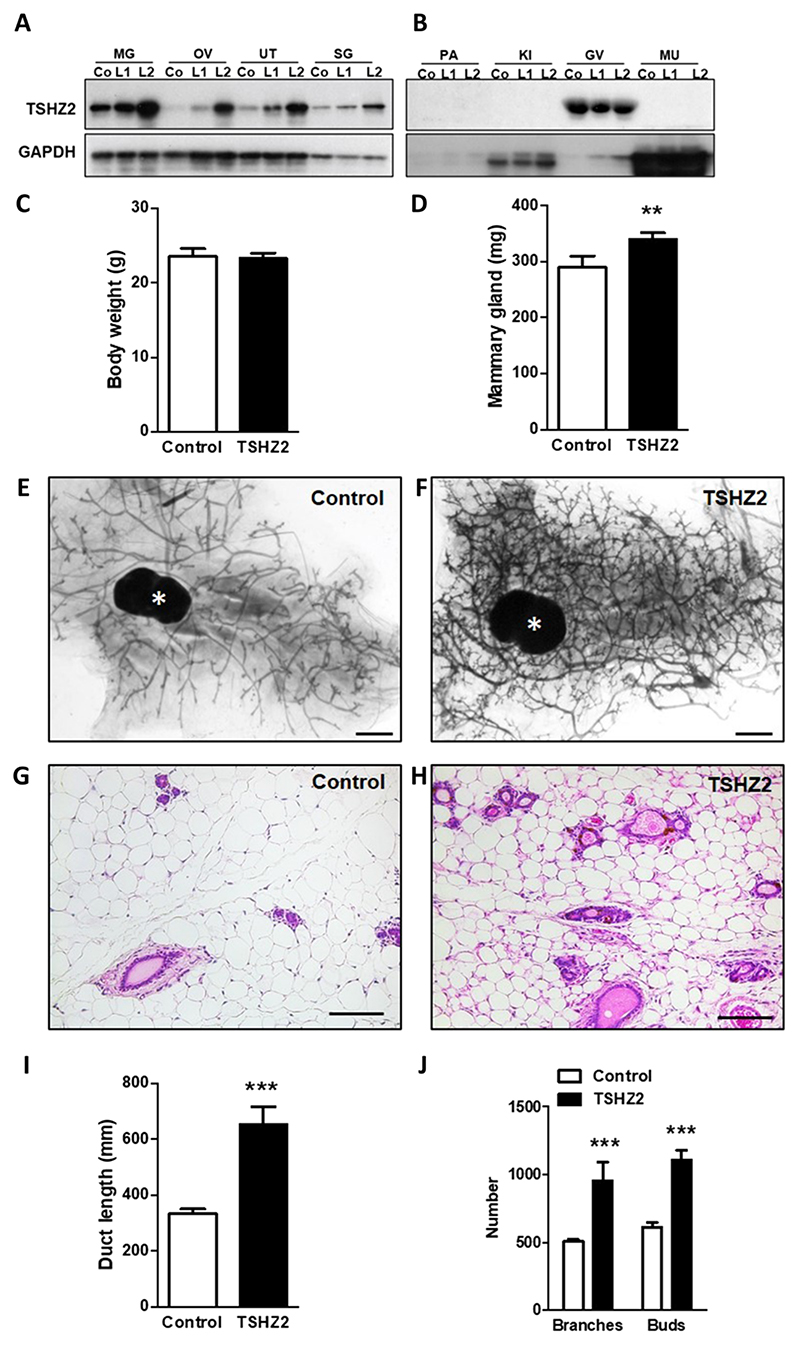
The normal function of organ-specific tumor suppressor genes, such as breast cancer type 1 susceptibility gene (BRCA1), might relate to essential functions in development. For example, BRCA1 controls differentiation of ductal luminal progenitors to mature luminal cells (27, 28). Because Drosophila teashirt (tsh) acts as a region-specific homeotic gene that specifies trunk identity (29), we investigated developmental functions in the murine mammary gland. To this end, we placed the murine Tshz2 cDNA downstream to the mouse mammary tumor virus (MMTV) promoter, essentially as previously described (30). Next, the plasmid was linearized and injected into zygotes of C57BL/6N mice. Two independent mouse lines (L1 and L2) expressing exogenous TSHZ2 in the mammary gland were established. Both lines showed the same phenotype, but all results presented herein relate to line L2.
First, we collected different organs from control animals and from the two transgenic lines. Analysis of protein extracts using immunoblotting detected the recombinant protein in organs that normally display MMTV promoter activity (Fig. 6A): the mammary gland, ovary, uterus, and the salivary gland. Consistently, organs with no MMTV promoter activity, such as pancreas, kidney, and muscle, displayed no expression of the ectopic protein and also no endogenous Tshz2, but the glandula vesicularis (murine seminal vesicle) showed high endogenous expression (Fig. 6B). The transgenic mice were morphologically similar to control mice, and their average body weights were almost identical (Fig. 6C). However, the transgenic mammary glands showed significantly increased weights (Fig. 6D). Whole mounts of mammary glands from a 6-month-old virgin female MMTV-Tshz2 mouse and a control littermate are shown (Fig. 6, E and F), along with the respective hematoxylin and eosin staining patterns (Fig. 6, G and H). Compared to the control mice, the transgenic mammary glands showed richer branching patterns and more visible buds. Morphometric analysis of the mammary glands from four mice of each group confirmed association between the transgene and significantly longer ducts (Fig. 6I), as well as more abundant branches and buds (Fig. 6J).
Fig. 6. TSHZ2 overexpression regulates growth rates of ducts and buds of the mouse mammary gland.
(A and B) Organs obtained from control animals (Co) and from two MMTV-Tshz2 mouse lines (L1 and L2) were analyzed using immunoblots. Shown are the organs with (A) and without (B) MMTV promoter activity. MG, mammary gland; OV, ovary; UT, uterus; SG, salivary gland; PA, pancreas; KI, kidney; GV, germinal epithelium of the seminal vesicle; MU, muscle. (C and D) Body and mammary gland weights of 6-month-old virgin female MMTV-Tshz2 mice and control littermates. Shown are means + SD of four mice per group. (E and F) Representative whole mounts of mammary glands from a female MMTV-Tshz2 mouse and a control littermate. Asterisks mark lymph nodes of the inguinal mammary gland. Scale bars, 1 mm. (G and H) Representative hematoxylin and eosin staining photos of mammary glands from 6-month-old virgin female MMTV-TSHZ2 mouse and a control littermate. Scale bars, 0.1 mm. (I and J) Morphometric analysis of mammary glands from female MMTV-TSHZ2 mice and control littermates (n = 4 mice each). Duct lengths and number of branches and buds were determined. Shown are means + SD. Two-tailed Student’s t tests: **P < 0.01 and ***P < 0.001.
In conclusion, overexpression of TSHZ2 in the mammary gland appears to enhance gland maturation, as reflected by ductal branching and bud maturation, but loss-of-function evidence would be needed to firmly establish roles in differentiation. Several EGFR ligands, including EGF and TGF-α, are involved in mammary gland development and partially share phenotypes with the TSHZ2-overexpressing mice. For example, mammary transplants overexpressing amphiregulin developed into hyperplastic lobules (31), whereas loss of amphiregulin stunted ductal outgrowth during puberty (32). Yet, another EGFR ligand, betacellulin, indirectly regulates the mammary gland by modulating prolactin release from the pituitary gland (33).
Yeast two-hybrid screens unveil physical interactions between TSHZ2 and a multiprotein complex containing the oncogenic protein regulator of cytokinesis, PRC1
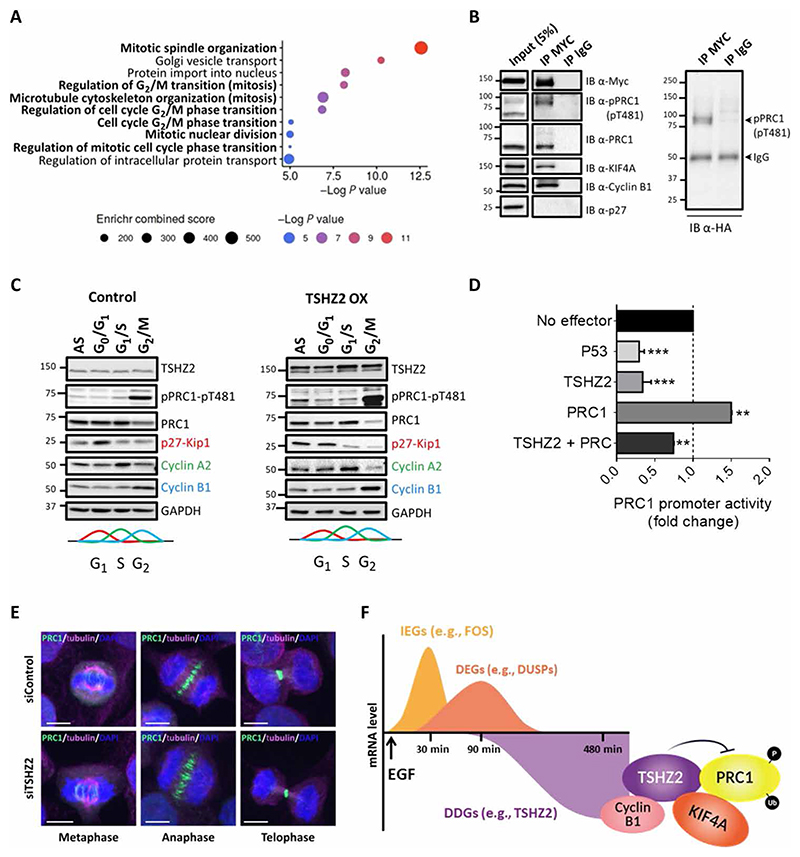
Presumably, DDGs like TSHZ2 execute growth arrest and tumor suppression by means of inactivating specific oncoproteins. Consistent with this model, listing the known functions of DDGs (table S2) identified candidate oncoproteins. For example, TXNIP directly inactivates both AKT and thioredoxin (34), and two other DDGs encode for the phosphatidylinositol-3-kinase–interacting protein 1 and FBXO32 (F-box protein 32), which respectively inhibits the PI3K/AKT/mTOR pathway (35) or sorts KLF4 for degradation (13). This commonality raised the possibility that in addition to DNA binding, TSHZ2 might bind with specific oncoproteins. To exhaustively identify direct binders, we performed yeast two-hybrid screens. The screens used the nucleotide sequence corresponding to TSHZ2 (amino acids 36 to 1034), which was cloned next to LexA and used as a bait to screen a random-primed human breast cancer cDNA library, screened as previously described (36). Overall, the screen identified 82 potential TSHZ2 interactors (table S3 and data file S1). Despite their multiplicity, Gene Ontology (GO) enrichment analysis found relatively narrow overrepresentation of GO terms, the majority were relevant to mitosis, G2/M transition, and spindle organization (Fig. 7A). Hence, we focused on the top three putative interactors (fig. S7A): (i) PRC1, a microtubule-associated protein playing key roles in organizing antiparallel microtubules in the central spindle during cytokinesis (37); (ii) KIF4A, a kinesin implicated in the regulation of chromosome segregation during mitotic cell division (38); and (iii) alpha-actinin-1 (ACTN1), a member of the alpha-actinin family known to cross-link actin filaments at the actomyosin ring that mediates abscission (39, 40). Aberrant expression of PRC1 predicts recurrence in men with prostate cancer (41), contributes to drug resistance and poor prognosis in ovarian cancer (42), and enhances liver and lung oncogenesis (43, 44). Hence, we examined the possibility that PRC1 can act as an oncogenic counterpart of TSHZ2.
Fig. 7. Yeast two-hybrid and other analyses unveil involvement of TSHZ2 in cell cycle regulation by binding with PRC1.
(A) Biological process enrichment analysis of TSHZ2-interacting proteins identified using the yeast two-hybrid screens. Gene Ontology (GO) enrichment was used. Size and color of dots refer to a combined enrichment score and an adjusted P value. (B) HEK293T cells pretransfected with plasmids encoding MYC-TSHZ2 and HA-ubiquitin were extracted 48 hours after transfection. Cleared extracts were subjected to immunoprecipitation (IP) using a normal mouse immunoglobulin or an anti-MYC antibody, and a fraction (5%) was separately analyzed (input). After protein transfer to filters, we applied immunoblotting (IB) with an anti-HA antibody (right) or with antibodies against the indicated proteins (left). (C) HEK293 cells were transfected with a DsRed-Empty plasmid (control) or with DsRed-TSHZ2 plasmid (TSHZ2 OX). After 24 hours, the cells were treated as follows: G0/G1, 17 hours on serum starvation plus 1 hour with fetal bovine serum; G1/S, 18 hours with 2 mM thymidine; G2/M, 18 hours with nocodazole (100 ng/ml). Asynchronized cells (AS) were used as control. Protein extracts were immunoblotted using antibodies against PRC1 and the phosphorylated form (pThr481). Antibodies against p27, cyclin A2, and cyclin B1 were used to assess growth arrest. (D) A luciferase reporter plasmid corresponding to the promoter of PRC1 was transfected into HEK293 cells along with the indicated vectors, and luminescence signals were determined 48 hours later. Data are averages ± SEM from three independent experiments. **P < 0.01 and ***P < 0.001. (E) HeLa cells preseeded in round bottom glasses were transfected with siTSHZ2 (or control siRNAs). Forty-eight hours later, cells were prepared for immunofluorescence analyses using antibodies against PRC1 (green) and tubulin (magenta). DNA was stained with DAPI (blue). The slides were viewed in a spinning disc microscope. Representative data (in B, C, and E) are of three experiments. (F) Summary model. A burst of immediate early genes (IEGs) rapidly follows stimulation of mammary cells with mitogens like EGF. Another set of genes, called delayed early genes (DEGs) undergo delayed activation, and like the IEGs, they rapidly rise and fall. Last, about 2.5 hours after stimulation, a third set of genes, the delayed down-regulated genes (DDGs), begin to fall. TSHZ2 is encoded by one of these DDGs. This transcription factor physically interacts with several mitotic proteins, including cyclin B1, KI-F4A, and PRC1 and inhibits PRC1. As we report herein, both promoter inactivation and enhancement of CDK1-mediated phosphorylation are mechanisms by which TSHZ2 inhibits PRC1 (which is also inhibited by the tumor suppressor p53). As also demonstrated herein, TSHZ2 may be down-regulated in cancer cells through promoter methylation or constitutive growth factor stimulation. FOS, proto-oncogene c-FOS; DUSPs, dual specificity phosphatases.
Two different approaches validated direct interactions between TSHZ2 and each of the three top candidates: (i) Immunofluorescence confirmed colocalization of each pair when ectopically expressed in human embryonic kidney (HEK) 293T cells (fig. S7B), and (ii) protein-fragment complementation assays (45) detected direct interactions (fig. S7C). The former approach made use of a DsRed-TSHZ2 fusion protein and the corresponding three green fluorescent protein (GFP) fusions, whereas the other used Gaussia luciferase (Gluc) split into two fragments, amino-terminal (Gluc1) and carboxyl-terminal (Gluc2). Gluc2 was fused to TSHZ2, whereas PRC1, KIF4A, and ACTN1 were separately fused upstream to Gluc1. As summarized (fig. S7C), all three Gluc1 fusion proteins showed highly significant luciferase activities only when cells were cotransfected with the TSHZ2-Gluc2 plasmid. The immunofluorescence assays revealed punctate nuclear localization of TSHZ2, which we addressed in HeLa cells pretransfected with a plasmid encoding a DsRed-TSHZ2 fusion protein (fig. S7D). This analysis confirmed that TSHZ2 is localized to nuclei, and it is confined to punctate structures. Because increasing evidence shows that subnuclear compartmentalization of regulatory factors, such as splicing factor compartments and chromatin domains (46, 47), plays key roles in nuclear function, we examined whether TSHZ2, the chromatin, and histone markers are sequestered within the same subnuclear structures. While TSHZ2 was found to be colocalized with the most extensively studied heterochromatin protein, HP1a, we detected no colocalization with a marker of the euchromatin, H3K4me3. Likewise, we detected only minimal colocalization with gamma-H2AX, a marker of damaged DNA.
We note that previous studies demonstrated physical interactions between KIF4A and PRC1 (48), and our screens identified not only PRC1 and KIF4A but also cyclin B1 as a ligand of TSHZ2 (table S3). To test this latter interaction, along with formation of a ternary PRC1-TSHZ2-KIF4A complex, we performed coimmunoprecipitation assays, which validated the existence of three binary complexes: TSHZ2-PRC1, TSHZ2-KIF4A, and TSHZ2-cyclin B1 (Fig. 7B). In summary, the presented studies identified a putative heterochromatin complex involving TSHZ2 and physical partners involved in regulating mitosis, the central spindle and the contractile ring. Collectively, our results implicated TSHZ2 in cytokinesis, a hitherto unknown function of this zinc finger homeobox protein.
TSHZ2 inhibits PRC1 by means of increasing inhibitory phosphorylation and, in similarity to p53, by means of decreasing transcription
PRC1 undergoes both phosphorylation and ubiquitination, and its expression level peaks during the S and G2/M phases (49). While roles for ubiquitination are less clear, PRC1 contains two CDK1 phosphorylation sites, Thr470 and Thr481, modification of which is important for mitotic suppression of microtubule bundling (50). Probing TSHZ2 immunoprecipitates indicated that the phosphorylated and ubiquitinated form(s) of PRC1 physically associates with TSHZ2 (Fig. 7B). To gain cell cycle phase resolution, we arrested TSHZ2-overexpressing HEK293T cells at G0/G1, G1/S, and G2/M. As expected, the effects we observed implicated TSHZ2 in the late phases: nocodazole, an inhibitor of microtubule polymerization known to arrest cells at G2/M, increased Thr481 phosphorylation and decreased PRC1 levels, especially when TSHZ2 was overexpressed (Fig. 7C). Concomitantly, TSHZ2 overexpression associated with increased and decreased expression of cyclins B1 and A2, respectively.
Presumably, TSHZ2 overexpression destabilizes PRC1 in a mechanism involving the anaphase promoting complex (51), or it might block transcription from the respective promoter. Note that another tumor suppressor, p53, directly binds with and inhibits the promoter of PRC1 (52). To address similar relations, we used a luciferase reporter corresponding to PRC1’s promoter. When introduced into HEK293T cells, together with a TSHZ2 plasmid, we observed strong promoter inhibition, similar to the effect of a p53 plasmid (Fig. 7D). Chromatin immunoprecipitation (ChIP) analyses confirmed direct binding of TSHZ2 with PRC1’s promoter (fig. S8A). Despite promoter regulation, we detected no effect of siTSHZ2 on PRC1’s localization or spindle formation (Fig. 7E). Together, our observations predicted reciprocal relationships between the levels of TZHZ2 and PRC1. Several lines of evidence supported this model: Overexpression of TSHZ2 in MDA-MB-231 cells decreased PRC1’s transcript and protein, whereas TSHZ2 knockdown oppositely regulated PRC1 (fig. S8, B and C). Likewise, analysis of MCF10A cells prestimulated with EGF detected delayed up-regulation of PRC1, which paralleled down-regulation of TSHZ2 (fig. S8D). These relationships further predicted negative correlation between TSHZ2 and PRC1 levels in human mammary tumors. This prediction was confirmed by analyzing data from 1992 patients with breast cancer [Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) dataset; fig. S8E]. In conclusion, expression levels of PRC1 are normally repressed at the promoter level by two tumor suppressors, TSHZ2 and p53, and this mechanism might explain the observed reciprocal interactions between PRC1, an oncoprotein, and TSHZ2.
In addition to the herein described role for TSHZ2 in EGFR signaling, according to a previous report, this repressor inhibits GLI1, a transcription factor transmitting signals generated by another pathway, the Hedgehog-Patched route, which culminates in increased expression of AEBP1 and CXCR4 (19). To examine this model in our experimental system, we isolated RNA from TSHZ2-overexpressing MDA-MB-231 cells (OX C2). Using primers specific to TSHZ2, CXCR4, and AEBP1, we found that high expression of TSHZ2 significantly reduced transcript levels of both CXCR4 and AEBP1 (fig. S8F), in line with the possibility that TSHZ2 integrates inputs relayed by two families of growth factors, EGF and Hedgehog (53).
In summary, we report a large group of putative tumor suppressors, which undergo delayed down-regulation in response to mitogenic stimulation. Here, we focused on two DDGs: TSHZ2 and TXNIP. TSHZ2 localizes to a chromosomal site frequently amplified in mammary tu-mors, but we found that hypermethylation of the respective promoter permits frequent down-regulation in patients with breast cancer. As befits a bona fide tumor suppressor, overexpression of TSHZ2 inhibited the Hedgehog pathway, reduced invasive features in vitro, as well as retarded tumor growth and metastasis in animals. While aiming at putative oncogenic partners, we identified >80 TSHZ2’s binders. PRC1, an oncogenic binder, bundles microtubules and controls a late checkpoint of the cell cycle. In response to EGF, PRC1 undergoes transcriptional up-regulation, which is negated by binding of two tumor suppressors, p53 and TSHZ2, to PRC1’s promoter. As we demonstrate herein, TSHZ2 might inhibit PRC1 also by means of a nontranscriptional mechanism involving phosphorylation by CDK1 (Fig. 7F). Thus, by identifying a previously unrecognized group of genes, the DDGs, we propose a new mechanism of oncogenesis: CNAs, promoter methylation, and mitogen-induced down-regulation of tumor suppressors like TSHZ2 deinactivate a diverse group of oncoproteins, such as PRC1 and additional regulators of the cell cycle.
Discussion
A strictly regulated temporal cascade of viral gene expression, which can be divided into three classes termed immediate-early, early, and late, is initiated during infection of mammalian cells by herpesviruses (54). An analogously interdependent cascade of gene expression is initiated when growth factors activate target cells in development or during postnatal life (55, 56). The first genes activated, the IEGs, typically encode oncogenic transcription factors. Another set, the DEGs, encoding negatively acting players, peaks about 2 hours after the stimulus (8). Last, about 2 hours after stimulation, a third set of genes begins to change to reach steady-state levels 4 to 8 hours later. Here, we studied EGF-responsive genes and focused on the less studied, DDGs. About 100 genes were persistently repressed after simulation of mammary cells with EGF. This group was subjected to two tests aimed at validating relevance to breast cancer. The first ranked DDGs by the difference in transcript abundance in tumors relative to the peri-tumor tissue, whereas the other divided patients with breast cancer to two groups (high and low expressors) and compared patient relapse times. The most important finding that emerged from the gene list is a clear association between the selected DDGs and genes promoting growth arrest or tumor suppression. The selected genes showed no preferential expression in a particular breast cancer subtype, which implies that their collective down-regulation may underlay processes common to all subtypes, such as regulation of metabolism or the cell cycle.
The DDG-encoded TSHZ2 protein is a zinc finger repressor of transcription. It has previously been suggested that TSHZ2 forms a complex with the C-terminal binding protein 2 (CtBP2) and GLI1, the main transcription factor of the Hedgehog pathway (19). Conceivably, growth factors dynamically regulate TSHZ2’s abundance in development of the mammary gland and in cancer, thereby enable noncanonical activation of GLI1’s and Hedgehog’s target genes. This model is consistent with the reported synergistic crosstalk between the EGFR pathway and Hedgehog signaling (53). Similarly, we predict that additional DDG-encoded proteins physically inhibit oncogenic complexes. It is notable that each of the two DDGs we studied directly binds two oncogenic proteins: AKT and thioredoxin, in the case of TXNIP (16), and both PRC1 and a complex containing CtBP2 and GLI1 (19) in the case of TSHZ2. Direct interactions between TSHZ2 and four proteins involved in spindle assembly and mitosis, PRC1, KIF4A, cyclin B1, and alpha-actinin, are reported herein. PRC1 acts as a microtubule-binding protein directly involved in bundling antiparallel microtubules (57). Accordingly, down-regulation of either PRC1 or KIF4A disorganized the midzone spindle comprising antiparallel interdigitating microtubules, which assemble between separating chromosomes (48). Because the midzone is required for maintenance of overall spindle architecture, we infer that down-regulation of TSHZ2 after stimulation with EGF serves as an essential component of the mitogenic process. In line with this, PRC1’s promoter is inhibited by another tumor suppressor, p53 (52). However, unlike TSHZ2, p53 also regulates an earlier checkpoint of the cell cycle, the restriction (R) point, crossing of which is recognized as the major step regulated by mitogens (58). In conclusion, by studying TSHZ2, one out of a large group of DDGs, we unveiled an unanticipated link between growth factors and cytokinesis. Predictably, tracking additional genes of the newly recognized DDG group will uncover more pairs of tumor suppressors and oncoproteins controlling developmentally essential hubs, which might be hijacked by cancer.
Materials And Methods
Chemicals and cell culture
Unless indicated, chemicals were from Sigma-Aldrich. Cell lines were purchased from the American Type Culture Collection. MCF10A cells were grown in Dulbecco’s modified Eagle’s medium (DMEM)/F12 supplemented with antibiotics, insulin (10 μg/ml), cholera toxin (0.1 μg/ml), hydrocortisone (0.5 μg/ml), heat-inactivated horse serum (5%, v/v), and EGF (10 ng/ml). The 184A1 cells were grown in mammary epithelial cell growth medium (MEGM) (Lonza, Basel, Switzerland) mixed 1:1 with DMEM/F12, supplemented with insulin, EGF, and hydrocortisone. MDA-MB-231, HCC70, and 4T1 cell lines were cultured in RPMI with 10% serum. HEK293T and HeLa cells were cultured in DMEM containing fetal bovine serum (10%). All cell lines were incubated at 37°C and 5% CO2. For time series experiments, cells were starved overnight in DMEM/F12 or MEGM without additives (starvation medium), and EGF was added to a final concentration of 10 ng/ml. All inhibitors (from MedChem Express; afatinib, PD0325901, everolimus, BEZ235, AKT-VIII, and BAY 11-7085) were dissolved in dimethyl sulfoxide. For cell cycle synchronization, HEK293T cells were arrested in different phases as follows: serum withdrawal for 17 hours followed by addition of serum for 1 hour (G0/G1), 2 mM thymidine (G1/S, 18 hours), or nocodazole (100 ng/ml) (G2/M, 18 hours).
siRNAs, plasmids, and transfection
siRNA-TSHZ2 (ON-TARGETplus; Dharmacon) or siRNA-CTRL (ON-TARGETplus Non-targeting; Dharmacon) was transfected into cells using the Dharmafect-1 reagent. pCMV-MYC-TSHZ2 and pDsRed-TSHZ2 were gifts from K. Kasai (Aichi Medical University School of Medicine). pEGFP-PRC1 was a gift from K. Hui (National Cancer Centre, Singapore). pEGFP-KIF4A was a gift from K. Samejima (University of Edinburgh). pEGFP-ACTN1 (#11908) and pSpCas9(BB)-2A-Puro (PX459) V2.0 (#62988) were from Addgene. Transient transfections were performed using Lipofectamine 2000 Reagent (Thermo Fisher Scientific), or jetPEI (Polyplus-transfection). The lentiviral vector pLV[Tet]-EGFP-TRE3G>hTSHZ2 (Vector-Builder, ID: VB180711-1040ara) was used for inducible overexpression of TSHZ2 (iTSHZ2), and the pHIV-Luc-ZsGreen (Addgene, #39196) was used to generate GFP-labeled cells. Viral particles encoding iTSHZ2, ZsGreen plasmid, or shRNAs targeting TSHZ2 (Sigma-Aldrich) were produced in HEK293T cells using the vectors psPAX2 (Addgene, #12260) and pMD2.G (Addgene #12259). HCC70, MDA-MB-231, and 4T1 cells were infected and selected under puromycin (2 μg/ml) or sorted using FACSAria Fusion instrument (BD Bioscience). Inducible overexpression experiments used doxycycline (2 μg/ml). For stable overexpression of TSHZ2, we used pDsRed-TSHZ2 (Takara, #632466), along with Amaxa Cell Line Nucleofector Kit V (Lonza).
Generation of 4T1-GFP knockout cells
The mouse Tshz2 gene was mutated using CRISPR-Cas9 technology. The web tool http://crispor.tefor.net/ was used to design guide RNA (gRNA) complementary to mTshz2. DNA templates for gRNA synthesis were generated by annealing of two single-stranded DNA oligonucleotides (see table S4). The gRNA was cloned to generate pSpCas9(Tshz2)-2A-Puro plasmid. 4T1 cells were transiently transfected with the sequencing-confirmed plasmid and selected under puromycin (2 μg/ml). Control cells were generated using an empty plasmid. Clones were screened for the presence of the deletion by restriction enzyme digestion using AleI and T7 endonuclease. Positive clones were sequenced to detect the mutation. Identified knockout (KO) cell clones were confirmed by Western blot analysis.
Mouse xenograft models
All protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the Weizmann Institute of Science. MDA-MB-231 cells stably expressing DsRed-TSHZ2, the inducible-TSHZ2 vector, or an empty plasmid were inoculated subcutaneously in the mammary fat pad of 6-week-old female SCID (Envigo, Israel) or NOG/NSG (NOD scid gamma mice) (the Jackson Laboratory, USA) mice. 4T1-GFP (KO and wild-type) cells were injected into the sub-axillary fat pad or tail vein of female Balb/C mice (6 weeks old; Envigo, Israel). Tumor growth was monitored once every 3 days with a caliper to measure width (W) and length (L), and tumor volume was calculated according to the formula: 3.14X(W2XL)/6. Mice were anesthetized with ketamine and xylazine, and primary tumors were detected noninvasively using an in vivo optical imaging system (IVIS 100, Xenogen Corp.). Living Image Software was used for sequential fluorescent image acquisition and superimposition of photographic images of mice and color-coded fluorescent images. Mice were euthanized when tumor size reached 800 to 1000 mm3. Thereafter, tumors were removed and weighed, and lungs were extracted and analyzed for micro-metastases using a Zoom Stereo Microscope SZX-RFL-2 (Olympus). The ImageJ software was used for image analysis. Primary tumors of animals injected with inducible TSHZ2-expressing cells were surgically excised under sterile conditions. Once the tumors were removed, the drinking water of the Dox+ group was supplemented with doxycycline (3 μg/ml) and sucrose (5 mg/ml).
Generation of MMTV-TSHZ2 mice
All animal experiments were approved by the institutional committee on animal care and carried out in accordance with the German Animal Protection Law, with permission from the responsible veterinary authority (Az.:55.2-1-54-2532-182-14). Mice were maintained in the C57BL/6N background (Janvier Labs, France) and housed under specific pathogen–free conditions in a closed barrier facility. Animals had free access to water and a standard rodent diet (V1534, Ssniff, Soest, Germany). Pronuclear microinjection was used to generate two independent mouse lines. The sequence of Tshz2 was amplified from a murine cDNA pool using the listed primers (table S4). The cDNA was ligated into the EcoRI site of the MMTV-SV40-Bssk vector, downstream of the MMTV promoter. The plasmid was linearized and later injected into zygotes of C57BL/6N mice.
RNA extraction and RT-qPCR
RNA isolation was performed using the RNeasy kit (QIAGEN). For cDNA synthesis, we used the qScript cDNA synthesis kit (Quantabio). RT-qPCR analysis was performed using Fast SYBR Green Master Mix (Thermo Fisher Scientific) and specific primers (listed in table S4). Transcripts encoding β-2 microglobulin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used for normalization.
Cell proliferation, migration, invasion, and colony formation assays
The assays used to measure proliferation {MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] and radioactive thymidine incorporation}, migration, invasion (Boyden chamber assays), and colony formation were applied as previously described (59).
Cell cycle analysis
Pretransfected cells were incubated for 60 min with BrdU (10 μM) and then washed, harvested, and fixed in ethanol (at 4°C). Thereafter, cells were incubated in a denaturation solution (2 N HCl and 0.5% Triton X-100; 30 min), followed by a neutralization solution [0.1 M sodium borate (pH 8.5); 30 min]. BrdU incorporated into newly synthesized DNA was then assayed using a fluorescein isothiocyanate–conjugated anti-BrdU antibody (see table S5). Total DNA content was determined by using propidium iodide solution supplemented with ribonuclease A (RNase A). Cell cycle distribution was detected by flow cytometry on a BD FACSAria Fusion instrument (BD Bioscience) and analyzed using the FACSDiva v8.0.1 Software.
Whole-mount mammary gland preparation
For whole-mount preparations, the right inguinal mammary gland complex was mounted on slides, fixed overnight, stained with carmine alum, defatted, clarified, and mounted with EUKITT, as described earlier (33). The left inguinal mammary complex was fixed in 4% paraformaldehyde (PFA) and stained with hematoxylin and eosin. Morphometric analyses were performed on whole-mount mammary gland preparations from four virgin female control mice (control) and four MMTV-TSHZ2 mice, at 6 months of age, using the Leica Application Suite software.
Yeast two-hybrid screens
The screens, which were outsourced to Hybrigenics Services (www.hybrigenics-services.com/), used the coding sequence for human TSHZ2 (amino acids 36 to 1034), which was PCR-amplified and cloned into the pB27 (N-LexA-bait-C fusion) vector. The construct was used as a bait to screen a random-primed human breast epithelial cancer cDNA library. Overall, 93 million interactions were screened as previously described (36). Two hundred seventy-five His+ colonies were selected in a medium lacking tryptophan, leucine, and histidine. The prey fragments of the positive clones were amplified and sequenced at their 5′ and 3′ junctions. The resulting sequences were used to identify the corresponding interacting proteins in the GenBank database (National Center for Biotechnology Information). A confidence score was attributed to each interaction as described (60). The tool “Enrichr” (61) was used to perform GO enrichment analysis from the TSHZ2-interacting proteins.
Immunofluorescence analyses and PLA
Cells fixed with 4% PFA for 15 min, permeabilized for 10 min with phosphate-buffered saline/Triton X-100 (0.2%), and blocked for 1 hour (in 1% bovine serum albumin) were hybridized with primary antibodies overnight. On the next day, cells were incubated with secondary fluorescent-tagged antibodies and stained with 4′,6-diamidino-2-phenylindole (DAPI) for 15 min. For PLA, the cells were processed using the Duolink In Situ Red Starter Kit (Mouse/Rabbit, Sigma-Aldrich). Samples were examined using a spinning-disc confocal microscope (Zeiss).
Coimmunoprecipitation and immunoblotting
Transfected HEK293T cells were harvested and lysed in radio-immunoprecipitation assay buffer containing protease and phosphatase inhibitors. The lysates (0.5 mg) were subjected to coimmunoprecipitation using Pierce Protein A/G Magnetic Beads (Thermo Fisher Scientific) according to the manufacturer’s instructions. Antibodies against MYC tag and control [immunoglobulin G (IgG)] were used. Proteins were eluted into 1× SDS gel loading buffer before being subjected to electrophoresis and immunoblotting. Tissues were homogenized in Laemmli extraction buffer, and the protein content was estimated using the bicinchoninic acid protein assay. For immunoblotting, cleared lysates (from cells or tissues) were resolved by electrophoresis and transferred to nitrocellulose membranes (GE Healthcare). Membranes were probed with the primary antibodies at 4°C overnight (see table S5) and incubated with horseradish peroxidase–conjugated secondary antibodies. Bands were detected using the enhanced chemiluminescence reagent (Bio-Rad).
Luciferase reporter and protein complementation assays
Luciferase plasmid containing the promoter region of PRC1 (pmGL3-PRC1) was a gift from W. Seol (Wonkwang University). Plasmids encoding TSHZ2, PRC1, and p53 were used as effectors. Luciferase-reporter and protein complementation assays were applied essentially as previously described (59). Briefly, expression vectors encoding TSHZ2 and the candidate interacting proteins fused to N- and C-terminal fragments of Gluc plasmid were constructed using specific primers (see table S4). The constructs were either transiently transfected (individually) or cotransfected into HEK293T cells, and 16 hours after transfection, bioluminescence luciferase assays were used to detect luciferase activity.
Chromatin immunoprecipitation qPCR
Cell fixation, ChIP, and qPCR were performed as described (62). ChIP assays were carried out using about 10 million HEK293T cells per sample. Forty-eight hours after transfection, cells were cross-linked for 10 min in 1% formaldehyde at 37°C. This reaction was subsequently quenched in 2.5 M glycine for 5 min. Chromatin from formaldehyde-fixed cells was fragmented to a size range of 200 to 700 bases using a sonicator. Solubilized chromatin was immunoprecipitated with anti-MYC and control (IgG) antibodies overnight at 4°C. Antibody-chromatin complexes were pulled down with Protein G Dynabeads (Life Technologies), washed, and then eluted. After cross-link reversal, RNase A, and proteinase K treatment, immunoprecipitated DNA was extracted with AMP Pure beads (Beckman Coulter). The DNA region of interest was detected using RT-qPCR and the listed primers (table S4).
Identification of DDGs
Our previously published dataset (GSE24391) of EGF-responsive genes was used (10). Differential expression analysis was performed using Limma (63). First, we selected genes that were down-regulated at least –1 LFC (log2 fold change P < 0.05) for each time point. There-after, we selected all mRNAs that showed persistent down-regulation in at least two consecutive time points. Genes were ranked according to significance of down-regulation by EGF at the two last time points (240 and 480 min). RNA sequencing expression data for breast tumors and adjacent tissue were obtained from TCGA. Differential expression analysis was performed on the TCGA Breast Invasive Carcinoma RNAseqV2 dataset using the DESeq2 (64). Gene expression graphs were created using R version 3.6.2. The pathway overrepresentation analysis was performed using MSigDB (www.gsea-msigdb.org/gsea/index.jsp), Panther 2016 (www.pantherdb.org/), and KEGG (www.genome.jp/kegg/) databases.
Analyses of DNA methylation and clinical datasets
TSHZ2 expression was analyzed using the KM plotter dataset and a set of 1980 patients with breast cancer established by the METABRIC (65–67). Patient survival models were fitted using packages in R. To evaluate the independent roles of DNA methylation and of CNA in explaining gene expression, as well as to make an informed choice for the PFMR (principal functional methylation region) per gene, we applied a novel algorithm called spatially coordinated CpG sites within the RRBS (reduced representation bisulphite sequencing) universe in breast cancer. This algorithm transforms RRBS data into distinct regions comprising multiple CpG sites coordinated in their methylation behaviors, beyond sites identified as DMRs (differentially methylated regions). For each region-gene combination, gene expression (normalized log2 intensities) was treated as the independent variable, and both DNA methylation (M value) and CNA (segmented mean log2 ratios) served as continuous co-variates. Only tumors displaying matched gene expression, CNA (single-nucleotide polymorphism 6.0 array), and DNA methylation data were selected for this analysis. A total of 15,968 genes with gene expression (microarray), CNA (array), and DNA methylation data were used.
Statistical analysis
All statistical tests were performed using GraphPad Prism 8.0.1. The methods for statistical test, fitting, and plot are described in the corresponding figure legends. All representative experiments shown were repeated three or more times. P values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank K. Kasai, K. M. Hui, K. Samejima, W. Seol, and M. Oren for reagents.
Funding
This work was performed at the Marvin Tanner Laboratory for Research on Cancer. Y.Y. is the incumbent of the Harold and Zelda Goldenberg Professorial Chair in Molecular Cell Biology. C.C. received the Weston Visiting Professorship from the Weizmann Institute of Science. Y.Y. received funds from the Israel Science Foundation (ISF), the Israel Cancer Research Fund (ICRF), the European Research Council (ERC), and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (AMRF). C.C. was supported with funding from Cancer Research UK (Caldas Core Grant A16942 and CRUK Cambridge Institute Core Grant A29580) and an ERC Advanced Grant from the European Union’s Horizon 2020 research and innovation programme (ERC-2015-AdG-694620).
Footnotes
Author contributions: M.L.U. and Y.Y. designed the experiments and wrote the manuscript. M.L.U. and M.D. established the cell and animal models. M.L.U., M.D., N.B.N., Y.H., L.K., A.S., I.M., S.G., I.V., S.L., and L.K. performed the experiments. M.L.U., R.N.B., and D.D.-G. performed bioinformatic analyses of clinical data. Y.T., I.C., O.M.R., and C.C. reviewed the manuscript.
Competing interests: C.C. is a member of AstraZeneca’s iMED External Science Panel and of Illumina’s Scientific Advisory Board. In addition, he received in the past research grants (not related to this work) from AstraZeneca, Genentech, Roche, and Servier. All other authors declare that they have no competing interests.
Data and materials availability
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References
- 1.Tomasetti C, Vogelstein B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu M, Polyak K. Microenvironmental regulation of cancer development. Curr Opin Genet Dev. 2008;18:27–34. doi: 10.1016/j.gde.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vitale I, Manic G, Coussens LM, Kroemer G, Galluzzi L. Macrophages and metabolism in the tumor microenvironment. Cell Metab. 2019;30:36–50. doi: 10.1016/j.cmet.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.David CJ, Massagué J. Contextual determinants of TGF b action in development, immunity and cancer. Nat Rev Mol Cell Biol. 2018;19:419–435. doi: 10.1038/s41580-018-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, Stanley ER, Segall JE, Condeelis JS. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65:5278–5283. doi: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- 7.Avraham R, Sas-Chen A, Manor O, Steinfeld I, Shalgi R, Tarcic G, Bossel N, Zeisel A, Amit I, Zwang Y, Enerly E, et al. EGF decreases the abundance of microRNAs that restrain oncogenic transcription factors. Sci Signal. 2010;3:ra43. doi: 10.1126/scisignal.2000876. [DOI] [PubMed] [Google Scholar]
- 8.Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, Amariglio N, et al. A module of negative feedback regulators defines growth factor signaling. Nat Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- 9.Burglin TR, Affolter M. Homeodomain proteins: An update. Chromosoma. 2016;125:497–521. doi: 10.1007/s00412-015-0543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeisel A, Kostler WJ, Molotski N, Tsai JM, Krauthgamer R, Jacob-Hirsch J, Rechavi G, Soen Y, Jung S, Yarden Y, Domany E. Coupled pre-mRNA and mRNA dynamics unveil operational strategies underlying transcriptional responses to stimuli. Mol Syst Biol. 2011;7:529. doi: 10.1038/msb.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 13.Zhou H, Liu Y, Zhu R, Ding F, Wan Y, Li Y, Liu Z. FBXO32 suppresses breast cancer tumorigenesis through targeting KLF4 to proteasomal degradation. Oncogene. 2017;36:3312–3321. doi: 10.1038/onc.2016.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen D, Li Y, Wang L, Jiao K. SEMA6D expression and patient survival in breast invasive carcinoma. Int Breast J Cancer. 2015;2015:539721. doi: 10.1155/2015/539721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elgort MG, O’Shea JM, Jiang Y, Ayer DE. Transcriptional and translational downregulation of thioredoxin interacting protein is required for metabolic reprogramming during G1. Genes Cancer. 2010;1:893–907. doi: 10.1177/1947601910389604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huy H, Song HY, Kim MJ, Kim WS, Kim DO, Byun JE, Lee J, Park YJ, Kim TD, Yoon SR, Choi EJ, et al. TXNIP regulates AKT-mediated cellular senescence by direct interaction under glucose-mediated metabolic stress. Aging Cell. 2018;17:e12836. doi: 10.1111/acel.12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius K-J, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson L-G, Landegren U. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 18.Nie W, Huang W, Zhang W, Xu J, Song W, Wang Y, Zhu A, Luo J, Huang G, Wang Y, Guan X. TXNIP interaction with the Her-1/2 pathway contributes to overall survival in breast cancer. Oncotarget. 2015;6:3003–3012. doi: 10.18632/oncotarget.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riku M, Inaguma S, Ito H, Tsunoda T, Ikeda H, Kasai K. Down-regulation of the zinc-finger homeobox protein TSHZ2 releases GLI1 from the nuclear repressor complex to restore its transcriptional activity during mammary tumorigenesis. Oncotarget. 2016;7:5690–5701. doi: 10.18632/oncotarget.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto M, Cid E, Bru S, Yamamoto F. Rare and frequent promoter methylation, respectively, of TSHZ2 and 3 genes that are both downregulated in expression in breast and prostate cancers. PLOS ONE. 2011;6:e17149. doi: 10.1371/journal.pone.0017149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dawson SJ, Rueda OM, Aparicio S, Caldas C. A new genome-driven integrated classification of breast cancer and its implications. EMBO J. 2013;32:617–628. doi: 10.1038/emboj.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kallioniemi A, Kallioniemi OP, Piper J, Tanner M, Stokke T, Chen L, Smith HS, Pinkel D, Gray JW, Waldman FM. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci USA. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 24.Quinlan KG, Verger A, Yaswen P, Crossley M. Amplification of zinc finger gene 217 (ZNF217) and cancer: When good fingers go bad. Biochim Biophys Acta. 2007;1775:333–340. doi: 10.1016/j.bbcan.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Nair NU, Das A, Rogkoti V-M, Fokkelman M, Marcotte R, de Jong CG, Koedoot E, Lee JS, Meilijson I, Hannenhalli S, Neel BG, et al. Migration rather than proliferation transcriptomic signatures are strongly associated with breast cancer patient survival. Sci Rep. 2019;9:10989. doi: 10.1038/s41598-019-47440-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiang H-C, Zhang X, Li J, Zhao X, Chen J, Wang HT-H, Jatoi I, Brenner A, Hu Y, Li R. BRCA1-associated R-loop affects transcription and differentiation in breast luminal epithelial cells. Nucleic Acids Res. 2019;47:5086–5099. doi: 10.1093/nar/gkz262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiang HC, Zhang X, Zhao X, Zhang C, Chen J, Garza P, Smith S, Ludwig T, Baer RJ, Li R, Hu Y. Gene-specific genetic complementation between Brca1 and Cobra1 during mouse mammary gland development. Sci Rep. 2018;8:2731. doi: 10.1038/s41598-018-21044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manfroid I, Caubit X, Kerridge S, Fasano L. Three putative murine Teashirt orthologues specify trunk structures in Drosophila in the same way as the Drosophila teashirt gene. Development. 2004;131:1065–1073. doi: 10.1242/dev.00977. [DOI] [PubMed] [Google Scholar]
- 30.Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 31.Kenney NJ, Smith GH, Rosenberg K, Cutler ML, Dickson RB. Induction of ductal morphogenesis and lobular hyperplasia by amphiregulin in the mouse mammary gland. Cell Growth Differ. 1996;7:1769–1781. [PubMed] [Google Scholar]
- 32.Luetteke NC, Qiu TH, Fenton SE, Troyer KL, Riedel RF, Chang A, Lee DC. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development. 1999;126:2739–2750. doi: 10.1242/dev.126.12.2739. [DOI] [PubMed] [Google Scholar]
- 33.Dahlhoff M, Blutke A, Wanke R, Wolf E, Schneider MR. In vivo evidence for epidermal growth factor receptor (EGFR)-mediated release of prolactin from the pituitary gland. J Biol Chem. 2011;286:39297–39306. doi: 10.1074/jbc.M111.243493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park JW, Lee SH, Woo GH, Kwon HJ, Kim DY. Downregulation of TXNIP leads to high proliferative activity and estrogen-dependent cell growth in breast cancer. Biochem Biophys Res Commun. 2018;498:566–572. doi: 10.1016/j.bbrc.2018.03.020. [DOI] [PubMed] [Google Scholar]
- 35.Lee K, Kitagawa M, Liao PJ, Virshup DM, Lee SH. A Ras-LSD1 axis activates PI3K signaling through PIK3IP1 suppression. Oncogenesis. 2020;9:2. doi: 10.1038/s41389-019-0185-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen PA, Field CM, Mitchison TJ. Prc1E and Kif4A control microtubule organization within and between large Xenopus egg asters. Mol Biol Cell. 2018;29:304–316. doi: 10.1091/mbc.E17-09-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazumdar M, Sundareshan S, Misteli T. Human chromokinesin KIF4A functions in chromosome condensation and segregation. Cell J Biol. 2004;166:613–620. doi: 10.1083/jcb.200401142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy ACH, Young PW. The actinin family of actin cross-linking proteins-a genetic perspective. Cell Biosci. 2015;5:49. doi: 10.1186/s13578-015-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sjöblom B, Salmazo A, Djinovi K, Carugo ć. α-actinin structure and regulation. Cell Mol Life Sci. 2008;65:2688–2701. doi: 10.1007/s00018-008-8080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo H-W, Chen Q-B, Wan YP, Chen G-X, Zhuo Y-J, Cai Z-D, Luo Z, Han Z-D, Liang Y-X, Zhong W-D. Protein regulator of cytokinesis 1 overexpression predicts biochemical recurrence in men with prostate cancer. Biomed Pharmacother. 2016;78:116–120. doi: 10.1016/j.biopha.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 42.Bu H, Li Y, Jin C, Yu H, Wang X, Chen J, Wang Y, Ma Y, Zhang Y, Kong B. Overexpression of PRC1 indicates a poor prognosis in ovarian cancer. Int J Oncol. 2020;56:685–696. doi: 10.3892/ijo.2020.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, Rajasekaran M, Xia H, Zhang X, Kong SN, Sekar K, Seshachalam VP, Deivasigamani A, Goh BKP, Ooi LL, Hong W, et al. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the Wnt/b-catenin signalling pathway. Gut. 2016;65:1522–1534. doi: 10.1136/gutjnl-2015-310625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanselmann S, Wolter P, Malkmus J, Gaubatz S. The microtubule-associated protein PRC1 is a potential therapeutic target for lung cancer. Oncotarget. 2018;9:4985–4997. doi: 10.18632/oncotarget.23577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov. 2007;6:569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- 46.Littmann M, Goldberg T, Seitz S, Boden M, Rost B. Detailed prediction of protein sub-nuclear localization. BMC Bioinformatics. 2019;20:205. doi: 10.1186/s12859-019-2790-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carmo-Fonseca M. The contribution of nuclear compartmentalization to gene regulation. Cell. 2002;108:513–521. doi: 10.1016/s0092-8674(02)00650-5. [DOI] [PubMed] [Google Scholar]
- 48.Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 2004;23:3237–3248. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.She ZY, Wei YL, Lin Y, Li YL, Lu MH. Mechanisms of the Ase1/PRC1/MAP65 family in central spindle assembly. Biol Rev Camb Philos Soc. 2019;94:2033–2048. doi: 10.1111/brv.12547. [DOI] [PubMed] [Google Scholar]
- 50.Mollinari C, Kleman JP, Jiang W, Schoehn G, Hunter T, Margolis RL. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. Cell J Biol. 2002;157:1175–1186. doi: 10.1083/jcb.200111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Juang Y-L, Huang J, Peters J-M, McLaughlin ME, Tai C-Y, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- 52.Li C, Lin M, Liu J. Identification of PRC1 as the p53 target gene uncovers a novel function of p53 in the regulation of cytokinesis. Oncogene. 2004;23:9336–9347. doi: 10.1038/sj.onc.1208114. [DOI] [PubMed] [Google Scholar]
- 53.Mangelberger D, Kern D, Loipetzberger A, Eberl M, Aberger F. Cooperative Hedgehog-EGFR signaling. Front Biosci (Landmark Ed) 2012;17:90–99. doi: 10.2741/3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Everett RD. The regulation of transcription of viral and cellular genes by herpesvirus immediate-early gene products (review) Anticancer Res. 1987;7:589–604. [PubMed] [Google Scholar]
- 55.Feldman ME, Yarden Y. Steering tumor progression through the transcriptional response to growth factors and stroma. FEBS Lett. 2014;588:2407–2414. doi: 10.1016/j.febslet.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Avraham R, Yarden Y. Feedback regulation of EGFR signalling: Decision making by early and delayed loops. Nat Rev Mol Cell Bio. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 57.Kellogg EH, Howes S, Ti SC, Ramirez-Aportela E, Kapoor TM, Chacon P, Nogales E. Near-atomic cryo-EM structure of PRC1 bound to the microtubule. Proc Natl Acad Sci USA. 2016;113:9430–9439. doi: 10.1073/pnas.1609903113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zwang Y, Sas-Chen A, Drier Y, Shay T, Avraham R, Lauriola M, Shema E, Lidor-Nili E, Jacob-Hirsch J, Amariglio N, Lu Y, et al. Two phases of mitogenic signaling unveil roles for p53 and EGR1 in elimination of inconsistent growth signals. Mol Cell. 2011;42:524–535. doi: 10.1016/j.molcel.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Srivastava S, Nataraj NB, Sekar A, Ghosh S, Bornstein C, Drago-Garcia D, Roth L, Romaniello D, Marrocco I, David E, Gilad Y, et al. ETS proteins bind with glucocorticoid receptors: Relevance for treatment of Ewing sarcoma. Cell Rep. 2019;29:104–117.:e4. doi: 10.1016/j.celrep.2019.08.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, Jacq B, et al. Protein interaction mapping: A Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blecher-Gonen R, Barnett-Itzhaki Z, Jaitin D, Amann-Zalcenstein D, Lara-Astiaso D, Amit I. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat Protoc. 2013;8:539–554. doi: 10.1038/nprot.2013.023. [DOI] [PubMed] [Google Scholar]
- 63.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA, Chin SF, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 66.Pereira B, Chin S-F, Rueda OM, Vollan H-KM, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut S-J, Tsui DWY, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ali HR, Rueda OM, Chin S-F, Curtis C, Dunning MJ, Aparicio SA, Caldas C. Genome-driven integrated classification of breast cancer validated in over 7,500 samples. Genome Biol. 2014;15:431. doi: 10.1186/s13059-014-0431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Gräf S, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chou J-L, Su H-Y, Chen L-Y, Liao Y-P, Hartman-Frey C, Lai Y-H, Yang H-W, Deatherage DE, Kuo C-T, Huang Y-W, Yan PS, et al. Promoter hypermethylation of FBXO32, a novel TGF-b/SMAD4 target gene and tumor suppressor, is associated with poor prognosis in human ovarian cancer. Lab Invest. 2010;90:414–425. doi: 10.1038/labinvest.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Folgueira MAAK, Brentani H, Katayama MLH, Patrão DFC, Carraro DM, Mourão Netto M, Barbosa EM, Caldeira JRF, Abreu APS, Lyra EC, Kaiano JHL, et al. Gene expression profiling of clinical stages II and III breast cancer. Braz J Med Biol Res. 2006;39:1101–1113. doi: 10.1590/s0100-879x2006000800013. [DOI] [PubMed] [Google Scholar]
- 71.Blain SW, Montalvo E, Massague J. Differential interaction of the cyclin-dependent kinase (Cdk) inhibitor p27Kip1 with cyclin A-Cdk2 and cyclin D2-Cdk4. J Biol Chem. 1997;272:25863–25872. doi: 10.1074/jbc.272.41.25863. [DOI] [PubMed] [Google Scholar]
- 72.He X, Zhu Z, Johnson C, Stoops J, Eaker AE, Bowen W, DeFrances MC. PIK3IP1, a negative regulator of PI3K, suppresses the development of hepatocellular carcinoma. Cancer Res. 2008;68:5591–5598. doi: 10.1158/0008-5472.CAN-08-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kristensen H, Haldrup C, Strand S, Mundbjerg K, Mortensen MM, Thorsen K, Ostenfeld MS, Wild PJ, Arsov C, Goering W, Visakorpi T, et al. Hypermethylation of the GABRE~miR-452~miR-224 promoter in prostate cancer predicts biochemical recurrence after radical prostatectomy. Clin Cancer Res. 2014;20:2169–2181. doi: 10.1158/1078-0432.CCR-13-2642. [DOI] [PubMed] [Google Scholar]
- 74.Oh JH, Rhyu MG, Jung SH, Choi SW, Kim SI, Hong SJ. Slow overmethylation of housekeeping genes in the body mucosa is associated with the risk for gastric cancer. Cancer Prev Res. 2014;7:585–595. doi: 10.1158/1940-6207.CAPR-13-0320. [DOI] [PubMed] [Google Scholar]
- 75.Chen JL-Y, Merl D, Peterson CW, Wu J, Liu PY, Yin H, Muoio DM, Ayer DE, West M, Chi J-T. Lactic acidosis triggers starvation response with paradoxical induction of TXNIP through MondoA. PLOS Genet. 2010;6:e1001093. doi: 10.1371/journal.pgen.1001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu Q, Liu Y, Zheng X, Zhu Q, Shen Z, Wang H, He H, Lin N, Jiang H, Yu L, Zeng S. Histone H3 lysine 4 trimethylation, lysine 27 trimethylation, and lysine 27 acetylation contribute to the transcriptional repression of solute carrier family 47 member 2 in renal cell carcinoma. Drug Metab Dispos. 2017;45:109–117. doi: 10.1124/dmd.116.073734. [DOI] [PubMed] [Google Scholar]
- 77.Stefely JA, Zhang Y, Freiberger EC, Kwiecien NW, Thomas HE, Davis AM, Lowry ND, Vincent CE, Shishkova E, Clark NA, Medvedovic M, et al. Mass spectrometry proteomics reveals a function for mammalian CALCOCO1 in MTOR-regulated selective autophagy. Autophagy. 2020;16:2219–2237. doi: 10.1080/15548627.2020.1719746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu G, Xiong D, Liu Z, Li Y, Chen K, Tang H. Long noncoding RNA LINC00052 inhibits colorectal cancer metastasis by sponging microRNA-574-5p to modulate CALCOCO1 expression. J Cell Biochem. 2019;120:17258–17272. doi: 10.1002/jcb.28988. [DOI] [PubMed] [Google Scholar]
- 79.Cengiz B, Yumrutas O, Bozgeyik E, Borazan E, Igci YZ, Bozgeyik I, Oztuzcu S. Differential expression of the UGT1A family of genes in stomach cancer tissues. Tumour Biol. 2015;36:5831–5837. doi: 10.1007/s13277-015-3253-1. [DOI] [PubMed] [Google Scholar]
- 80.Morrison JA, Pike LA, Sams SB, Sharma V, Zhou Q, Severson JJ, Tan AC, Wood WM, Haugen BR. Thioredoxin interacting protein (TXNIP) is a novel tumor suppressor in thyroid cancer. Mol Cancer. 2014;13:62. doi: 10.1186/1476-4598-13-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y, He W, Zhang S. Seeking for correlative genes and signaling pathways with bone metastasis from breast cancer by integrated analysis. Front Oncol. 2019;9:138. doi: 10.3389/fonc.2019.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Heim D, Cornils K, Schulze K, Fehse B, Lohse AW, Brummendorf TH, Wege H. Retroviral insertional mutagenesis in telomerase-immortalized hepatocytes identifies RIPK4 as novel tumor suppressor in human hepatocarcinogenesis. Oncogene. 2015;34:364–372. doi: 10.1038/onc.2013.551. [DOI] [PubMed] [Google Scholar]
- 83.Si W, Huang W, Zheng Y, Yang Y, Liu X, Shan L, Zhou X, Wang Y, Su D, Gao J, Yan R, et al. Dysfunction of the reciprocal feedback loop between GATA3-and ZEB2-nucleated repression programs contributes to breast cancer metastasis. Cancer Cell. 2015;27:822–836. doi: 10.1016/j.ccell.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 84.Saintigny P, Peng S, Zhang L, Sen B, Wistuba II, Lippman SM, Girard L, Minna JD, Heymach JV, Johnson FM. Global evaluation of Eph receptors and ephrins in lung adenocarcinomas identifies EphA4 as an inhibitor of cell migration and invasion. Mol Cancer Ther. 2012;11:2021–2032. doi: 10.1158/1535-7163.MCT-12-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun Y, Qian J, Lu M, Xu H. Lower and reduced expression of EphA4 is associated with advanced TNM stage, lymph node metastasis, and poor survival in breast carcinoma. Pathol Int. 2016;66:506–510. doi: 10.1111/pin.12444. [DOI] [PubMed] [Google Scholar]
- 86.Xiao M, Xie J, Wu Y, Wang G, Qi X, Liu Z, Wang Y, Wang X, Hoque A, Oakhill J, Proud CG, et al. The eEF2 kinase-induced STAT3 inactivation inhibits lung cancer cell proliferation by phosphorylation of PKM2. Cell Commun Signal. 2020;18:25. doi: 10.1186/s12964-020-0528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Simpson CD, Hurren R, Kasimer D, MacLean N, Eberhard Y, Ketela T, Moffat J, Schimmer AD. A genome wide shRNA screen identifies a/b hydrolase domain containing 4 (ABHD4) as a novel regulator of anoikis resistance. Apoptosis. 2012;17:666–678. doi: 10.1007/s10495-012-0723-4. [DOI] [PubMed] [Google Scholar]
- 88.Salvador Moreno N, Liu J, Haas KM, Parker LL, Chakraborty C, Kron SJ, Hodges K, Miller LD, Langefeld C, Robinson PJ, Lelievre SA, et al. The nuclear structural protein NuMA is a negative regulator of 53BP1 in DNA double-strand break repair. Nucleic Acids Res. 2019;47:2703–2715. doi: 10.1093/nar/gkz138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ohata H, Miyazaki M, Otomo R, Matsushima-Hibiya Y, Otsubo C, Nagase T, Arakawa H, Yokota J, Nakagama H, Taya Y, Enari M. NuMA is required for the selective induction of p53 target genes. Mol Cell Biol. 2013;33:2447–2457. doi: 10.1128/MCB.01221-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.