Introductory Paragraph
Mammalian telomeres protect chromosome ends from aberrant DNA repair1. TRF2, a component of the telomere-specific Shelterin protein complex, facilitates end protection through sequestration of the terminal telomere repeat sequence within a lariat t-loop structure2,3. Deleting Trf2 in somatic cells abolishes t-loop formation, which coincides with telomere deprotection, chromosome end-to-end fusions, and inviability3–9. In contrast, we establish here that TRF2 is largely dispensable for telomere protection in mouse pluripotent embryonic and epiblast stem cells. Embryonic stem cell (ESC) telomeres devoid of TRF2 instead activate an attenuated telomeric DNA Damage Response (DDR) without accompanying telomere fusions and propagate for multiple generations. Induction of telomere dysfunction in ESCs, consistent with somatic Trf2 deletion, only occurs following removal of the entire Shelterin complex. Consistent with TRF2 being largely dispensable for telomere protection specifically during early embryonic development, cells exiting pluripotency rapidly switch to TRF2-dependent end protection, and Trf2-null embryos arrest prior to implantation with evidence of strong DDR signaling and apoptosis specifically in the non-pluripotent compartment. Finally, we show that ESCs form t-loops independently of TRF2, revealing why TRF2 is dispensable for end protection during pluripotency. Collectively, these data establish that telomere protection is solved by distinct mechanisms in pluripotent and somatic tissues.
Keywords: telomeres, telomere end protection, TRF2, stem cells, pluripotency, t-loops, shelterin, DNA repair
Results
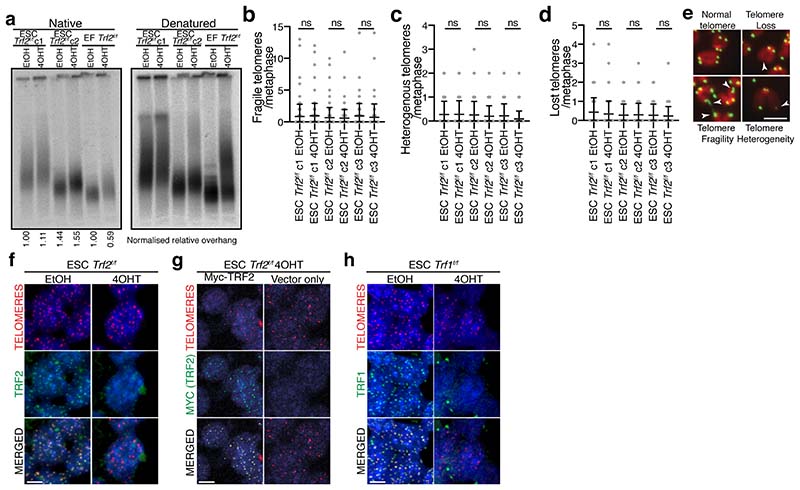
It is assumed that mechanisms of telomere protection are conserved in somatic and stem cells, yet evidence is currently lacking. To investigate this directly, we derived isogenic Trf2f/f ERT2Cre pluripotent mouse ESCs and somatic mouse Ear Fibroblasts (EFs). TRF2, the central effector of telomere end protection in somatic cells3–9, is rapidly lost in both cell types following 4-Hydroxytamoxifen (4OHT) treatment (Fig. 1a). As reported previously, TRF2 loss in EFs induces robust telomere end-to-end fusions, accumulation of cells with 4N DNA content, and cell death (Fig. 1b-d, Extended Data Fig. 1a-g)6,7. In contrast, TRF2 loss in three independent ESC clones did not induce telomere end-to-end fusions either 4 or 10 days following 4OHT treatment (Fig. 1-c and Extended Data Fig. 1h-k). Furthermore, ESCs lacking TRF2 proliferated for many generations with only a minor reduction in growth rate, and showed no cell cycle alteration or changes in expression of pluripotency markers OCT4 and NANOG (Fig. 1a and Extended Data Fig. 1a-g)10. ESCs lacking TRF2 also retained a normal telomere 3’ G-overhang, and did not display telomere fragility, heterogeneous telomere length, telomere loss, or telomere sister chromatid exchanges (Fig. 1e-f and Extended Data Fig. 2a-e). ESC telomeres without TRF2 therefore replicate normally, are not fused, and do not undergo recombination.
Figure 1. TRF2 is largely dispensable for telomere end protection and cell viability in ESCs.
(a) Western blot of whole cell extracts from the indicated cells 96 hours after EtOH or 4OHT treatment. (b) Mitotic chromosome spreads from the indicated cells 96 h after treatment with 4OHT or EtOH. The DNA is stained with DAPI (red) and the telomeric DNA by fluorescent in situ hybridization (FISH, green). The mean percentage of fused chromosome ends from (c) is indicated. (c) Quantification of telomere fusions from the experiment shown in (b). Clonal lines are denoted c1-c3 (mean ± s.d., ≥70 spreads/condition over 3 independent experiments, oneway ANOVA.) (d) Flow cytometry determination of DNA content for the indicated cells 96 hours after 4OHT or EtOH treatment (n=3, ≥10,000 cells/condition). (e-f) Images and quantification of telomere-sister chromatid exchange (T-SCE) analysis in the indicated cells 96 h after treatment with EtOH or 4OHT, or 12 h after induction of TRF1-FOK1 expression. (mean ± s.d., ≥70 spreads/condition over 3 independent experiments, one-way ANOVA). (g) 53BP1 TIFs in Trf2f/f ESCs or EFs 96 h after treatment with 4OHT or EtOH. 53BP1 was detected by immunofluorescence (IF, green), the telomeric DNA with FISH (red), and DNA with DAPI (blue). (h) Quantification of (g) (mean ± s.d., ≥300 cells/condition over 4 independent experiments, one-way ANOVA). (i) Western blots of whole cell extracts from the indicated cells 96 hours after EtOH or 4OHT treatment. Where indicated, IR represents 2 h post-treatment with 2 Gy IR, and UV represents 2 h post-treatment with 10 J/m2 UV. For all panels: Scale bar = 5 μm; ns = not significant; **** p ≤ 0.0001.
Prompted by this observation, we interrogated DDR activation at EF and ESC telomeres using SILAC-PICh (Proteomics of Isolated Chromatin Loci)11, immunofluorescence, and western blotting. We detected broadly similar telomeric proteomes and all Shelterin components at ESC and EF telomeres in cells expressing TRF2 (Extended Data Fig. 2f-h,3a-d). Following TRF2 loss, DDR markers γH2AX and 53BP1 were readily detected at EF telomeres in Telomere Dysfunction Induced Foci (TIFs) (Fig. 1g-h, Extended Data Fig. 3e-f)12. Notably, significantly fewer Trf2-null ESC telomeres co-localized with 53BP1 and/or γH2AX, and many ESCs lacking TRF2 displayed no TIFs (Fig. 1-h, Extended Data Fig. 3e-i). Additionally, while TRF2 loss in EFs induced robust ATM, CHK2, and p53 phosphorylation, TRF2 loss in ESCs conferred only weak ATM and CHK2 phosphorylation, and p53 activation was absent (Fig. 1i). ESCs lacking TRF2 therefore exhibit a quantitatively attenuated telomeric DDR.
Despite these distinct responses to Trf2-deletion, both ESCs and EFs robustly activate ATM, CHK2, CHK1 and p53 in response to Ionizing Radiation (IR) or Ultraviolet light (UV) (Fig. 1i). ESCs also respond to IR-induced DSBs with rapid focal accumulation of γH2AX and 53BP1 and transient G2/M arrest, indicating that DDR signaling remains intact (Extended Data Fig. 4a-c). Moreover, both ESCs and EFs rapidly repair IR-induced breaks, which is impaired in both cells by addition of a DNA-PKcs inhibitor that blocks non-homologous end joining (NHEJ, Extended Data Fig. 4d-e), the DSB repair pathway that drives somatic telomere fusions following Trf2 deletion. Furthermore, preventing NHEJ through genetic deletion of Ku70 or DNA Ligase IV (LigIV) induces severe IR-sensitivity in ESCs and neither prolonged G1 arrest nor induction of ATM signaling with IR were sufficient to induce telomere fusions in Trf2-null ESCs (Extended Data Fig. 4f-h,5a-e). Collectively, these data show that ATM and NHEJ are functional in ESCs and the lack of a robust response to TRF2 deletion is neither due to an attenuated DDR, DNA repair capacity, nor the short G1 phase present in this cell type.
The unexpected dispensability of TRF2 in ESCs led us to investigate other Shelterin components. In somatic cells, TPP1 mediates recruitment of POT1 to telomeric G-overhangs to suppress RPA loading and ATR kinase activation13. Previously, TPP1-depletion was shown to induce NHEJ at fusion-resistant but Trf2-null telomeres7. TPP1 depletion in ESCs induced a moderate 53BP1 TIF response that was augmented by TRF2 co-depletion (Extended Data Fig. 6a-d). Loss of both TRF2 and TPP1 from ESCs, but not either protein alone, induced moderate telomere fusions, CHK2 activation, and G2/M arrest without affecting OCT4 levels (Extended Data Fig. 6a,e-g). This is consistent with the reported protective somatic functions of TPP1. In somatic cells TRF1 facilitates telomere replication14. In agreement, Trf1 deletion in ESCs induced telomere replication stress, which manifests as telomere fragility and telomere loss, 53BP1 TIF formation, telomeric RPA-pSer33 recruitment, phosphorylation of the replication stress markers CHK1-pSer345 and γH2AX, and reduced proliferation (Fig. 2a-d, Extended Data Fig. 6h-k). TRF1 thus facilitates ESC telomere replication consistent with the protein’s somatic activity, indicating that TRF2 is distinct in being largely dispensable for ESC telomere function.
Figure 2. The Shelterin complex is required for telomere protection in ESCs.
(a) Western blot of whole cell extracts 72 h after EtOH or 4OHT treatment. UV indicates 2 h post-treatment with 10 J/m2 UV. (b) Mitotic chromosome spreads 72 h after 4OHT or EtOH treatment. DNA is stained with DAPI (red) and the telomeres by FISH (green). Fragile telomeres are marked with an asterisk, telomere loss with an arrowhead. (c-d) Quantification of telomere fragility (c) and loss (d) from (b)_(unpaired two-tailed t-test). (e) Western blot of whole cell extracts from indicated ESC cultures 72 h after EtOH or 4OHT treatment. (f) Mitotic chromosome spreads from indicated ESCs 72 h after 4OHT or EtOH treatment, stained as in (b). (g) Quantitation of telomere fusions in (f) (one-way ANOVA). (h) 53BP1 TIFs in ESCs 36 h after 4OHT or EtOH treatment. 53BP1 (green) was detected by IF, telomeric DNA by FISH (red), DNA with DAPI (blue). (i) Quantification of (h) (mean ± s.d. ≥300 cells/condition over 3 independent experiments, one-way ANOVA). (j) Western blot of whole cell extracts from non-targeting control (NTC) or Atm-Knockout (Atm-KO) Trf1f/f Trf2f/f ESCs 48 h or 72 h after EtOH or 4OHT treatment (*non-specific band). (k) Telomere fusions in NTC and Atm-KO Trf1f/f Trf2f/f ESCs ± ATR inhibitor (ATRi) 72 h after EtOH or 4OHT treatment (one-way ANOVA). (l, m) Telomere fusions in NTC, Ku70-KO (l), or DNA Ligase VI-Knockout (LigIV-KO) (m) Trf1f/f Trf2f/f ESCs 72 h after EtOH or 4OHT treatment. Clonal lines are denoted c1 and c2 (one-way ANOVA). For all panels: Scale bar = 5 μm; ns = not significant;, ** p=0.0011, **** p ≤ 0.0001. Mean telomere fusion percentage shown in (b) and (f). For (c, d, g, k-m) mean ± s.d., ≥70 spreads/condition over 3 independent experiments.
We next examined the consequence of removing the entire Shelterin complex from ESC telomeres by co-deleting Trf1 and Trf215. The resulting Shelterin-free ESCs displayed extensive telomere fusions, prominent 53BP1 TIF, robust CHK2 activation, G2/M arrest, and cell death 72 hours after 4OHT administration (Fig. 2e-i, Extended Data Fig. 7a-d). Furthermore, 72 h after 4OHT treatment, ESCs lacking both TRF1 and TRF2, but not TRF1 or TRF2 alone, had lost expression of the pluripotency markers NANOG and OCT4 (Fig. 2e). Since excessive IR-induced DNA damage also promotes differentiation concomitant with cell death (Extended Fig. 7e), we infer that robust DDR and/or multicentric chromosomes induced by Shelterin-free telomeres are incompatible with pluripotency.
To understand the temporal relationship between telomere dysfunction and pluripotency loss we performed timed experiments following Trf1 and Trf2 co-deletion in ESCs. Following gene knockout with 4OHT, we observed a strong TIF response (24-36 hours) that preceded telomere fusions and CHK2 activation (36-48 h), and apoptosis (48-72 h) (Fig. 2h-j, Extended Data Fig. 8a-c). Notably, the TIF response, CHK2 activation, and telomere fusions all preceded the loss of OCT4 and NANOG (48-72 hours). Pluripotency loss is therefore not the driver of telomere dysfunction or fusions in Shelterin-free ESCs. Furthermore, these data indicate ESCs possess the ability to fuse telomeres if left unprotected (Fig. 2f-g). Shelterin-free ESC telomeres accumulated γH2AX and 53BP1 TIFs similar to levels observed in Shelterin free MEFs and significantly elevated relative to ESCs lacking TRF1 or TRF2 (Fig. 2h-I, Extended Data Fig. 8d-g). Additionally, Shelterin-free, but not Trf2-null, ESCs arrested in G2/M (Extended Data Fig. 7d). Telomere fragility, however, was comparable between Trf1-null and Shelterin-free ESCs (Extended Data Fig. 8h).
ESCs lacking Shelterin exhibit both telomere fusions, typically signaled via ATM, and telomere replication stress, typically signaled via ATR7,14. In agreement, both ATM-pSer1987 and RPA-pSer33 co-localize with Shelterin free ESC telomeres (Extended Data Fig. 9a-h)7,15. We generated Trf1f/f Trf2f/f Atm-knockout ESC clones and observed that Atm deletion, but not pharmacological ATR inhibition, significantly reduced CHK2 activation, apoptosis and telomere fusions in Shelterin-free ESCs (Fig. 2j,k). This is consistent with Shelterin free MEFs and suggests that whilst both ATM and ATR could trigger telomere fusions at dysfunctional ESC telomeres, ATM plays a more prominent role (Extended Data Fig. 10a-c). Likewise, genetic deletion of Ku70 or LigIV significantly reduced telomere fusions in Shelterin-free ESCs, indicating that fusions in Shelterin-free ESCs are pre-dominantly NHEJ driven (Fig. 2l,m, Extended Data Fig. 10d-g). ATM signaling and NHEJ are thus primarily responsible for telomere fusions in Shelterin-free ESCs, consistent with Trf2-null somatic cells7. This reaffirms the functionality of ATM signaling and NHEJ repair in ESCs. We propose that ESC telomeres protect chromosome ends from ATM and NHEJ-dependent fusion through a Shelterin-dependent mechanism predominantly independent of TRF2.
Since ESCs progress through canonical developmental stages before lineage specification and differentiation16, we next probed when in development the transition from largely TRF2-independent to strictly TRF2-dependent end protection occurs. We derived Epiblast Stem Cells (EpiSC) from E6.5 embryos, representing the primed pluripotent state in development, distinct from naïve pluripotent E3.5-derived ESCs17,18. Consistent with ESCs, Trf2 deletion did not affect the expression of EpiSC pluripotent markers19, conferred a minor TIF response, (relative to somatic cells) and did not induce substantial telomere fusions, robust CHK2 activation, or apoptosis (Fig. 3a-d, Extended Data Fig. 11a-i). We conclude that TRF2 is largely dispensable for end protection in cultured pluripotent embryonic stem cells.
Figure 3. TRF2-dependent chromosome end protection begins at pluripotency exit.
(a) Western blots of whole cell extracts from the indicated cells 96 h after EtOH or 4OHT treatment. (b) Mitotic chromosome spreads from Trf2f/f EpiSCs 96 h after 4OHT or EtOH treatment. DNA is stained with DAPI (red), telomeres with FISH (green). (c) Quantitation of telomere fusions from (b) (mean ± s.d., ≥80 spreads/condition over 3 independent experiments, one-way ANOVA). (d) 53BP1 TIFs in Trf2f/f EpiSCs and Trf2f/f EFs 96 h after 4OHT or EtOH treatment. (mean ± s.d., ≥300 cells/condition over 3 independent experiments, one-way ANOVA). (e) Differentiation protocol used in (f-m). (f) Growth curve of Trf2f/f ESCs differentiated as shown in (e) (n=3 biologically independent experiments, mean ± s.e.m.). (g) Brightfield images depicting Trf2f/f ESCs differentiated as in (e). (h) Mitotic chromosome spreads from Trf2f/f ESCs differentiated as shown in (e) and stained as in (b). (i) quantitation of telomere fusions from (h) (mean ± s.d. ≥60 spreads/condition over 3 independent experiments, unpaired two-tailed t-test). (j) 53BP1 TIFs in Trf2f/f ESCs and Trf2f/f ESCs differentiated as in (e), 96 h after EtOH or 4OHT treatment. 53BP1 (green) was detected by IF, telomeric DNA by FISH (red) and DNA with DAPI (blue). (k) Quantification of (j) (mean ± s.d., ≥300 cells/condition over 4 independent experiments, one-way ANOVA). (l) 53BP1 TIFs in non-targeting control (NTC) and Atm-knockout (Atm-KO) Trf2f/f ESCs differentiated as shown in (e) (mean ± s.d. n=3, ≥300 spreads/condition, unpaired two-tailed t-test). (m) Telomere fusions from NTC and Atm-KO Trf2f/f ESCs differentiated as in (e) (mean ± s.d., ≥ 60 spreads/condition over 3 independent experiments, unpaired two-tailed t-test). For b,h,j Scale bar = 5 μm. For all panels: **** p ≤ 0.0001. Mean telomere fusion percentages are shown in (b) and (h).
ESCs can be differentiated in vitro through an EpiSC-like state into neural progenitors through culture in FGF4-containing media20 (Fig. 3e, Extended Data Fig. 11j-n). Trf2 deletion did not affect changes in gene expression accompanying differentiation, nor the ability of ESCs to acquire an EpiSC-like state (Extended Data Fig. 11o-r). However, we failed to obtain Trf2-null cells after day three of in vitro differentiation, the timepoint at which control cells begin to adopt a neural identity, consistent with Trf2-null cells dying as they exit pluripotency (Fig. 3f-g)20. In agreement, upon exit from pluripotency, in vitro differentiated Trf2-deleted ESCs displayed a striking induction of 53BP1 TIFs, telomere fusions, CHK2 activation, and apoptosis (Fig. 3h-k, Extended Data Fig. 11s). Consistent with observations in somatic cells, Atm-knockout rescued the TIFs, telomere fusions, and CHK2 activation in FGF4-media cultured Trf2-null ESCs without affecting differentiation (Fig. 3l-m, Extended Data Fig. 11t-v). Collectively, these data demonstrate TRF2 becomes essential for end protection and cellular viability upon somatic differentiation.
To determine the impact of TRF2 loss in vivo, we bred Trf2+/- mice and obtained embryos at various developmental stages. The expected mendelian ratios of Trf2-/- embryos were observed at E3.5 but were significantly underrepresented at E6.5 (Fig. 4a). To examine when Trf2-/- embryos become compromised, we obtained fertilized oocytes from Trf2+/- crossings at E0.5 and visualized development using EmbryoScope live imaging. Trf2-/- embryos were capable of initiating blastocyst formation with normal frequency and developmental rate before the blastocysts became severely compromised during expansion and subsequent hatching (Fig. 4b-d). ESCs represent a subset of NANOG-expressing pluripotent cells in blastocysts. Immunostaining revealed that Trf2-/- E3.5 embryos accumulate large numbers of TIFs, which are markedly reduced in the NANOG-positive cells, mirroring what is observed in cultured Trf2-null pluripotent cells (Fig. 1g,h and 4e,f). Trf2-/- E3.5 embryos also display increased cells with pan-nuclear γH2AX staining with blebbing, indicative of apoptosis, which are specifically NANOG-negative (Extended Data Fig. 12a-c). Trf2-null embryos are therefore compromised in their ability to maintain blastocyst formation and continue development, possibly through cell death in the non-pluripotent compartment. The relatively low level of DDR signaling and lack of apoptosis in the embryonic NANOG-positive cells from TRF2-null embryos is consistent with TRF2 being dispensable for telomere end protection in the pluripotent compartment of the developing embryo.
Figure 4. TRF2-null embryos are compromised as expanding blastocysts.
(a) Genotypes of embryos obtained at various developmental stages after crossing Trf2+/- mice. Count = number of embryos with each genotype. (b) EmbryoScope images of developing Trf2+/- and Trf2-/- embryos obtained at E0.5. (c) Time at which Trf2+/+, Trf2+/-, and Trf2-/- embryos shown in (b) reach key developmental stages in EmbryoScope culture. Number of embryos of each genotype analysed is shown (mean ± s.d.). (d) The percentage of embryos from the experiment in (b-c) that reach each developmental stage. (e) IF-FISH analysis of γH2AX TIFs and NANOG within embryos of the indicated genotypes. γH2AX (green) and NANOG (magenta) were detected by IF, telomeric DNA by FISH (red) and DNA with DAPI (blue). Whole embryo maximum intensity projection is shown on the left, and a maximum intensity projection restricted to the pluripotent Inner Cell Mass (ICM) is on the right. (f) Quantification of γH2AX TIFs in NANOG-positive and NANOG-negative cells within embryos of indicated genotypes from (e). Each data point represents the mean number of TIFs across all NANOG-positive or Nanog-negative cells within each embryo. Number of embryos of each genotype analysed is shown (two-way ANOVA, **** p ≤ 0.0001, mean ± s.d.).
While TRF2 is dispensable for most end protective functions in ESCs, Trf2 deletion confers weak CHK2 activation and low levels of TIFs in these cells (Fig. 1g-i). We observed this weak DDR induced by Trf2 deletion in ESCs is dependent on Atm (Fig. 5a-c, Extended Data Fig. 13a-b). To explore this further, we complemented Trf2-null ESCs using wild type (WT), mutant, or hybrid TRF2 alleles that impact DDR signaling (Extended Data Fig. 13c-d). TRFcT lacks the TRFH domain from TRF2 that suppresses ATM signaling, while TRFcH and TRF2-DiDDR both lack the TRF2 iDDR domain that suppresses DDR signaling downstream of ATM activation21. Both WT TRF2 and TRFcT, but neither TRFcH nor TRF2-DiDDR, complemented the subtle TIF induction following TRF2 deletion in ESCs (Fig. 5d-e, Extended Data Fig. 13d-e). The TRF2-iDDR domain therefore suppresses ATM signaling at ESC telomeres. However, the TRFH domain, which was previously implicated in t-loop formation, was dispensable3. This prompted us to investigate t-loops in ESCs.
Figure 5. T-loop formation occurs independently of TRF2 in ESCs.
(a) Western blots of whole cell extracts from non-targeting control (NTC) and Atm-Knockout (Atm-KO) Trf2f/f ESCs 96 h after EtOH or 4OHT treatment (* non-specific band). Clonal lines are denoted c1 and c2. (b) 53BP1 TIFs in NTC and Atm-KO Trf2f/f ESCs 96 h after treatment with 4OHT or EtOH. 53BP1 was detected by IF (green), telomeric DNA by FISH (red) and DNA with DAPI (blue). Scale bar = 5 μm. (c) Quantification of (b). (d) 53BP1 TIFs in Trf2f/f ESCs stably expressing Myc-tagged mouse WT TRF2 (TRF2) or the indicated mutant or hybrid TRF2 allele, 96 h after EtOH or 4OHT treatment. Samples were stained as in (b), scale bar = 5 μm. (e) Quantification of (d). (f) T-loops and linear telomeres from ESCs imaged by Airyscan super-resolution microscopy. Scale bar = 2 μm. (g) Quantitation of t-loops in Trf2f/f MEFs and ESCs 96 h after EtOH or 4OHT treatment. Data are exclusive of ambiguous telomere molecules (one-way ANOVA, data are mean ± SEM, minimum four experiments scoring ≥ 2804 molecules per replicate). (h) Measurement of the loop portion of t-loops from the data in in (g) (mean ± s.d.). For all panels: ns = not significant, **** p ≤ 0.0001. For (c, e), mean ± s.d., ≥300 cells/condition over 3 independent experiments, one-way ANOVA.
Super-resolution Airyscan microscopy of the somatic macromolecular telomere structure previously identified that telomere ATM activation occurs concomitantly with t-loop unfolding and exposure of the linear chromosome end3. Using the same methods, MEFs and ESCs expressing TRF2 were found to possess t-loops with similar frequency (33.1 % vs 32.0%) and size (Fig. 5f-h, Extended Data Fig. 13f-g). Strikingly, however, unlike somatic cells, the percentage of t-loops in ESCs was unaffected by Trf2 deletion (Fig. 5g). In accordance, ESCs lacking TRF2 retain a normal G-overhang (Extended Data Fig. 2a). ESC telomeres can therefore form t-loops independently of TRF2.
In conclusion, our data reveal that TRF2, the central mediator of telomere protection in somatic cells, is largely dispensable for telomere protection in pluripotent cells during early embryonic development. ESC telomeres lacking TRF2 activate a mild ATM-dependent DDR but do not undergo NHEJ, despite the ATM and NHEJ pathways being functional in ESCs. Upon differentiation this unique attribute of stem cells is lost and TRF2 assumes its full role in end protection. Surprisingly, we show that t-loops form independently of TRF2 in ESCs. The retention of end protection in the presence of t-loops, but absence of TRF2, confirms a long-suspected dogma that t-loops are a key mediator of telomere end protection irrespectively of how they form. We anticipate the presence of a developmental switch upon exit from pluripotency that transitions t-loops from forming independently of TRF2 to being reliant on TRF2 for their formation/stabilisation. How this developmental switch is controlled, how t-loops form and are stabilized without TRF2, whether this alternative mechanism occurs in other contexts, and why it has evolved are open questions for investigation.
Methods
Cell culture procedures
SV40-LT-immortalized Trf2f/f Rosa26-ERT2Cre mouse embryonic fibroblasts (MEFs) were a kind gift of Eros Lazzerini Denchi. Trf1f/f Trf2f/f p53-/- mouse embryonic fibroblasts (MEFs) were a kind gift of Titia de Lange. SV40-LT-immortalised Trf2f/f Rosa26-ERT2Cre mouse ear fibroblasts (EFs) were obtained from adult mice of the same genotype and immortalised with SV40-LT. DR4 MEF feeder cells were obtained from DR4 mouse embryos and irradiated with 20 Gy at passage 2 to induce senescence. MEFs and EFs were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10 % fetal bovine serum (FBS, Invitrogen), L-glutamine, and penicillin-streptomycin (Gibco). HEK 293FT cells (Invitrogen) were grown in DMEM with 10% FBS. Trf2f/f Rosa26-ERT2Cre mouse embryonic stem cells (ESCs) were obtained from embryos of the same genotype by standard methods22. Trf1+/+ Trf2f/f Rosa26-ERT2Cre, Trf1f/f Trf2+/+ Rosa26-ERT2Cre and Trf1f/f Trf2f/f Rosa26-ERT2Cre ESCs were isolated from embryos obtained by crossing of the individually targeted Trf1f/f Rosa26-ERT2Cre and Trf2f/f Rosa26-ERT2Cre mice. Genotypes were determined by Transnetyx Inc. using real time PCR with allele-specific probes. ESCs were maintained in ESM (Knockout DMEM, 10% batch-tested ESM FBS, 1x Penicillin-streptomycin-glutamine, 1x Glutamax, 1x Non-essential Amino Acids (all Gibco) and 240 U/ml LIF (ESGRO, Millipore)) or Knockout Serum Replacement (KOSR) media (DMEM, 15 % Knockout Serum Replacement, 1x Non-essential amino acids, 1x Glutamax, 1x Penicillin-Streptomycin-Glutamine (all Gibco) and 240 U/ml LIF (Millipore)) on DR4 MEF feeder cells. ESCs were adapted onto Matrigel for minimum of 2 passages for experiments. For growth curves, a set number of cells were seeded at each passage and cells counted at the next passage 2 or 3 days later and ESC growth curves were performed with cells grown on DR4 feeders. Deletion of floxed alleles in Rosa26-ERT2Cre EFs, MEFs and ESCs was performed by 16 h treatment with 100 nM 4-hydroxy tamoxifen (4OHT) and successful targeting was checked via genotyping with validated primers and/or western blot at the indicated time points. An equal amount of EtOH was used as a vehicle control. For deletion of floxed alleles in MEF Trf1f/f Trf2f/f p53-/- cells, cells were infected with AdCRE (Vector biolabs 1045) and AdGFP (Vector biolabs, 1060) viruses with an MOI of 100 as described previously9. Where shown, brightfield images of cultured cells were taken using an Olympus GKX41 microscope and GXCapture software. Cells were regularly tested for mycoplasma.
Creation of TRF2 alleles and cell line generation
Production of lentivirial supernatants and transductions of Trf2f/f Rosa26-ERT2Cre ESCs were done essentially as described earlier9. Trf2f/f Rosa26-ERT2Cre ESCs were infected with pLVEF1a-IRES-Puromycin lentiviruses expressing empty vector, mouse Myc-tagged TRF2 or TRF2 mutants. TRFcT and TRFcH mutants were a kind gift of Eros Lazzenrini Denchi. TRF2-DiDDR was obtained from wild type TRF2 using Q5® Site-Directed Mutagenesis Kit and the following primers - TGCTTTGGGCTTCTTCTC and GTTCAGGCACCAGGTGAA. These Trf2 alleles were cloned into the pLV-EF1a-IRES-puromycin lentivirus vector which was a kind gift of Dr Tobias Meyer. Transduced ESCs were selected with puromycin (0.2 μg/ml) for 3 days.
Generation of CRISPR Cas9 knockouts
ESCs were co-transfected with a PiggyBac Transposase (SystemsBio) and PiggyBac EF1α-MCS-IRES-Neo vector modified to contain mammalian adapted Cas9-GFP23. Cells were selected with G418 (100 μg/ml) for 3 days and single clones were selected and grown. Successful Cas9 activity was tested as described24. ESCs expressing functional Cas9 were transduced with lentiguide-puro (a kind gift of Feng Zhang) containing Non-targeting Control (NTC) (GTATTACTGATATTGGTGGG), Atm (TGCAAGATACACATGAATCG), Ku70 (GAAGACGTAGTACTCATGGT) or DNA Ligase IV (LigIV - GCACAACGTCACCACAGATC) gRNAs and selection performed with puromycin (0.2 ug/ml) for 3 days. gRNA sequences were obtained from the Brie library or previous studies25,26. Single cells were seeded, clones were picked after outgrowth for 10 days and tested for ATM, Ku70 or LigIV expression via western blotting after 10 days. Clones showing no ATM, Ku70 or LigIV expression were selected as Atm-Knockout, Ku70-knockout or LigIV-knockout clones for experiments.
Derivation and culture of Epiblast Stem Cells
Epiblast stem cells (EpiSC) were derived and cultured as described previously17. Briefly, embryos were obtained from crossing of Trf2f/+ Rosa26-ERT2Cre mice. Mouse epiblast was dissected from individual mouse embryos at day E6.5 and plated in 4-well dishes (Nunc) coated with DR4 feeder cells in Chemically Defined Media (0.5x F12 NUT Mix, 0.5x Iscoves IDMEM with Glutamine, 0.5 % BSA, 0.1 % Chemically defined lipid concentrate, 1x Penicillin-Streptomycin (all Gibco), 400 uM Monothioglycerol, 7 ug/ml Insulin (both Sigma), 15 ug/ml Transferin, 20 ng/ml Activin (both Roche), 12 ng/ml FGF4 (R&D Systems)). Colonies of epiblast stem cells were picked from the first outgrowth on day 7 and propagated on DR4 feeder cells. Cells were passaged every 2/3 days in CDM using Collagenase Type IV (Invitrogen) with media being changed every day. Genotyping was performed using established primers and a Trf2f/f Rosa26-ERT2Cre epiSC clone was selected for experiments. This clone was validated by western blotting for TRF2 96 h after 4OHT treatment and by qPCR for known EpiSC markers OCT4, NANOG, SOX2, FGF4 and OTX2.
Directed differentiation of ESCs into neuromesoderm
ESCs were maintained on feeder cells in ESM with passaging every 2 days. 24 h prior to differentiation protocol, cells were treated with 100 nM 4OHT or equal volume of EtOH as a vehicle control for 16 h. Cells were trypsinised, resuspended and “panned” to remove feeders. Cells were washed with N2B27 medium (1x DMEM/F12, 1x Neurobasal medium, 0.08% BSA, 1x N2 Plus Supplement, 1x B27 serum free Supplement, 1x Penicillin-Strepomycine-Glutamine, 77 nM β-mercaptoethanol (all Gibco)) and seeded at 1x104 cells/cm2 on gelatin coated Corning dishes in N2B27 + 10 ng/ml FGF4 (R&D Systems). Media was replaced with N2B27 + 10 ng/ml FGF4 at 48 h after seeding and changed into N2B27 without growth factors at 72 h after seeding (Differentiation Day 3) to stimulate midbrain neural differentiation. For growth curves, duplicate dishes of cells seeded as above were trypsinised and counted at the indicated time points after seeding three times. Successful differentiation was checked by RT-qPCR for ESC, EpiSC and neural markers in cells harvested at day 2, 3, 4 and 5 of the differentiation protocol as outlined in Figure 4e. ESCs were seeded onto gelatin in ESM at the same time as being seeded for neural differentiation and these cells were harvested to provide the ESC controls for these experiments.
Flow Cytometry
Cells were trypsinised and fixed in 70% ethanol. Cells were then resuspended in an RNase A (20 mg/ml) and propidium iodide (50 mg/ml) solution, passed through a 70 mm cell strainer and the cell cycle distribution of the cells analyzed by flow cytometry, using a 610/20 gate. Gating and analysis were performed manually using FlowJo v10 (FlowJo).
Cell cycle synchronization
ESCs were synchronized in G1 by Mimosine block. Briefly, cells were treated with 250 nM Mimosine (Sigma-Aldrich) for 16 hours, washed three times and then released in fresh ESM. Synchronisation and release were confirmed by flow cytometry as outlined above.
Cell lysis, Western blotting, DNA Damaging and Drug treatments
Cells were harvested and resuspended in ice-cold RIPA buffer (1x EDTA-free Complete protease inhibitor cocktail (Roche), 1x PhosSTOP phosphatase inhibitor cocktail (Roche)) and lysed for 30 minutes on ice. Cell lysates were briefly vortexed, spun and the soluble protein fractions were collected after centrifugation at 16000 x g for 10 minutes at 4°C. Western blotting analysis was performed by standard methods, as described previously9, using the following antibodies: gH2AX, 05-636; ATM-pSer1987, 05-740; Chk2, 05-649 - all Millipore; Myc, 9E10 - Santa Cruz Biotechnologies; TRF2, D1Y5D; Chk1, 2360; Chk1-pSer345 133D3; Cleaved Caspase 3, 9661; P53, 1C12; P53-pSer15, 9284 – all Cell Signalling Technology; ATM, A1106; Flag, M2 – both Sigma Aldrich; DNA Ligase IV, ab80514; Histone H3, ab10799; Ku70, Ab2624; Nanog, ab80892; Oct4, ab19857; TRF1, ab10579; Vinculin, ab11194 – all Abcam and secondary antibodies: Swine Anti-Rabbit Immunoglobulins P0399 and Goat Anti-Mouse Immunoglobulins P0447 – both Dako. For inhibition of ATR, VE821 (SelleckChem), was used at a final concentration of 500 nM for 36 hours. DNA-PKcs was inhibited by NU7441 (SelleckChem) at a final concentration of 2 uM added 30 min prior to IR treatment. An equal amount of EtOH or DMSO was used as a vehicle control. When indicated, cells were exposed to ionizing radiation (IR) in plates in a Cs-137 irradiator and media changed 5 min after. UV treatment was performed by washing cells with PBS, removing PBS and exposing to a UVC source for a total of 10 J/m2 UV, before addition of fresh media.
IR Sensitivity assays
16000 cells of each genotype were seeded onto Matrigel-coated 96 well plates in ESM and immediately irradiated in a Cs-137 irradiator with 0–2.4 Gy of IR. Cells were then grown for 4 days with regular media changes. Cell survival and proliferation was then measured with a using CellTiter-Glo (Promega) according to manufacturer instructions. Luminescence was read at all wavelengths using a CLARIOStar (BMGLabtech).
Inducible TRF1-FOK1 expression
Flag-TRF1-FOK1 was a kind gift from Roger Greenberg. Trf2f/f ESCs were co-transfected with a PiggyBac Transposase (SystemsBio) and PiggyBac EF1α-MCS-IRES-Neo vector modified to inducibly express flag-TRF1-FOK1. Cells were selected with G418 (100 μg/ml) for 3 days and single clones were selected and grown. Clones expressing Flag-TRF1-FOK1 were screened using IF for Flag and a clone with rapid and uniform expression upon doxycycline treatment was selected. These cells were treated with 10 ng doxycycline for 12 h for experiments. Expression and telomeric localisation of Flag-TRF1-FOK1 was confirmed via western blotting and IF-FISH using a Flag antibody (M2, Sigma).
siRNA treatment and siRNA oligos
Transfections with siRNA oligos were performed using Lipofectamine RNAiMax (Thermo Fisher Scientific). Briefly, ESCs at density of 2.0x105 cells/well were transfected in a 6-well plate with 20 pmol non-targeting (D-001810-10, SMARTpool ON-TARGETplus, Dharmacon, sequences: UGGUUUACAUGUCGACUAA, UGGUUUACAUGUUGUGUGA, UGGUUUACAUGUUUUCUGA, UGGUUUACAUGUUUUCCUA) or TPP1 siRNA (L-057987-01, SMARTpool ON-TARGETplus, Dharmacon – sequences: CCAGAGAGGCAUCGUGAUA, CCAGAGAGGCAUCGUGAUA, GAAAGUGGUGCCAGCGUUA, AGUCAGAAGGCUCGGGGAA). 16 h after transfection the medium was exchanged. 72 h post-transfection the cells were harvested and the levels of TPP1 expression were assessed by qPCR using validated Quantitect Primers (Qiagen) as detailed below.
RealTime(RT)-qPCR
RNA was first isolated using the RNeasy Mini Kit (QIAGEN) and then reverse transcribed using the High-Capacity RNA-to-cDNA kit (Thermo Fisher Scientific) according to the manufacturers’ instructions. RT-qPCR was performed using the SsoAdvanced Universal Supermix (Biorad) with Quantitect primers (TPP1 only) or previously validated primers20,27. Primer sequences available upon request.
Telomere FISH and CO-FISH of metaphase cells
Telomeric Peptide Nucleic Acid Fluorescence In Situ Hybridisation (PNA FISH) on cytogenetic chromosome spreads was performed largely as described before28. Briefly, cells were treated with 0.2 μg/ml of colcemid for 120 minutes to arrest cells in metaphase. Trypsinized cells were incubated in 3:1 H20:DMEM for 10 min, fixed with cold methanol:acetic acid (3:1), and spread on glass slides. Slides were rehydrated in PBS for 5 minutes, fixed in 4% formaldehyde for 2 minutes, treated with 1 mg/ml of pepsin for 10 minutes at 37 °C, and fixed in 4% formaldehyde for 5 minutes. Slides were dehydrated in 70%, 85%, and 100% (v/v) ethanol for 15 minutes each and air-dried. Metaphase chromosome spreads were hybridized with telomeric TelCCy5 PNA probe (Panagene) for 90 seconds at 80C, incubated at room temperature for 2 h, then washed twice in washing buffer (70% formamide, 10mM Tris-HCl pH 7.2), and thrice in 2XSSC, with DAPI in the second wash. Slides were rinsed in H2O and mounted using ProLong Gold antifade with DAPI (Life Technologies). For CO-FISH, cells were treated with 10 uM BrdU for 18 h prior to harvesting as above and CO-FISH was performed essentially as described29 using Alexa 488-TelG and TelC-Cy5 PNA probes (Panagene). Slides were mounted using ProLong Gold antifade with DAPI (Life Technologies). Chromosome images and telomere signals were captured using Zeiss AxioImager M1 microscope equipped with an Orca Spark camera (Hammamatsu) controlled by Micromanager software (ImageJ) using a 63x objective and imported to ImageJ for manual analysis.
Indirect Immunofluorescence (IF) and IF-FISH
For 53BP1, gH2AX and Myc IFs, cells were grown on #1.5 glass coverslips, washed with PBS and fixed with ice cold methanol on ice for 10 minutes then washed thrice with PBS. For RPA-pSer33; ATM-pSer1987, TRF1 and TRF2IFs, cells were washed with ice cold PBS, extracted for 5 min on ice in CSK buffer (10 mM PIPES, pH 6.8, 100mM NaCl, 300 mM sucrose, 1mM MgCl2, 1 mM EGTA, 1mM DTT) containing 0.5% Triton X-100, fixed for 10 min with 4% formaldehyde at room temperature and then washed thrice with PBS. Cells were blocked with ADB (Antibody Dilution Buffer; 10% normal goat serum, 0.1% Triton X-100, 0.1% saponin in PBS) for 30 min, incubated with primary antibody (diluted in ADB) for 1 hour at room temperature (53BP1 A272-300, Bethyl; gH2AX, 05-636, Millipore; Myc, 9E10, SCBT; RPA-pSer33, A300-246A, Bethyl; ATM-pSer1987, 05-740, Millipore; TRF1, TRF12-A, ADI; TRF2, D1Y5D, CST), washed three times with PBS and then counterstained with Alexa Fluor secondary antibodies (A11001, A11003, A11008, A11010, Invitrogen) diluted in ADB for 1 hour at room temperature. Cells were washed three times with 1X PBS, then mounted with Prolong Gold mounting agent supplemented with DAPI (Life Technologies). For IF-FISH, cells were treated as for IF until washing of secondary antibodies. After washing, cells were fixed again with 2% (w/v) formaldehyde in PBS for 20 min at room temperature and then washed twice with PBS. Samples were incubated in 2XSSC at 72 degrees for 40 min, then in 0.1 M NaOH for 10 min, washed in PBS and dehydrated in 70%, 85%, and 100% (v/v) ethanol for 5 minutes each and then air-dried. Dry coverslips were hybridized with a telomeric TelC-Cy5 PNA probe (Panagene) in hybridizing solution (70% formamide, 0.5% blocking reagent (Roche), 10mM Tris-HCl pH 7.2) for 90 s at 80C followed by 2 hours at room temperature and washed thrice in 2XSSC for 10 min at 42C, with DAPI in the second wash. The coverslips were mounted onto glass slides with Prolong Gold mounting agent supplemented with DAPI. Images were acquired using a Zeiss Upright 710 confocal microscope with a 40X objective, and Zen software. Following acquisition, images were imported into Imaris for manual quantitation and figure generation.
Crossing of Trf2+/- mice and IF-FISH of embryos
Trf2+/- mice were crossed and embryos harvested by flushing at E3.5 or from uterine dissection at E6.5 and E7.5. Embryos were washed in PBS, lysed (10 mM Tris pH 8.5, 50 mM KCl, 0.01% gelatin, 300 μg/ml proteinase K) and genotyped using validated primers6. Separately, Trf2+/- mice were crossed and fertilised oocytes were obtained from the infundibulum at E0.5. Mouse embryos were cultured in drops of pre-equilibrated Global medium (LGGG-20; CooperSurgical inc.) supplemented with 10% HSA (ART-3001-5;CooperSurgical inc.) and overlaid with mineral oil (Origio; ART-4008-5P). Zygote stage mouse embryos were incubated at 37 C and 5.5% CO2 in an EmbryoScope+ time-lapse incubator (Vitrolife). Embryos were imaged for the next 5 days, then lysed and DNA harvested for genotyping as above. Prior to genotyping, images were analysed to determine whether individual embryos reached specific stereotypical developmental states and the times after placement in the embryoscope at which these states were obtained. Trf2 genotyping of these embryos was performed as above. For IF-FISH, Blastocysts were fixed with 4% PFA for 15 min at RT followed by permeabilization with PBS 0.25% Triton for 20min. Embryos were blocked in 10% Fetal Bovine Serum at 4°C then incubated with primary antibodies (gH2AX, 05-636 Millipore; NANOG, RCAB003P, 2B Scientific) for 2h at RT in blocking buffer. After washing in PBS/0.1% Triton, embryos were incubated with secondary antibodies in blocking buffer for 1h at RT. Embryos were then fixed with 2% PFA for 10 min and subjected to FISH with telomere probe as above, with slight modifications: embryos were equilibrated with increased concentration of hybridisation buffer in PBS (25%, 50%, 75%, 90%, 100%). Then, embryos were incubated at 80°C for 15 min in hybridisation buffer and left overnight at 4°C, before washing in FISH washing buffer and storage in PBS/DAPI Prolong Antifade. Stained embryos were imaged using an Olympus FV1000 confocal microscope with 40x magnification using a sufficient z-stacks to capture the entire embryo and images exported into ImageJ for analysis. Representative images shown were obtained using a maximal intensity projection of part of the z-stack to avoid having excessive overlapping cells in image.
Airyscan super-resolution imaging and Super-resolution microscopy scoring criteria
Sample preparation for super-resolution microscopy, cross-linking efficiency determination and Airyscan imaging were performed as described previously3,8. For analysis, post capture and processing, images were exported to ImageJ as TIF images with maintained scales. Images were quantified manually with researchers blinded to the experimental conditions. Telomere molecules were scored if they had a traceable telomere contour of ≥ 1 μm and contained no gaps ≥ 500 nm in telomere staining. Molecules were classified as t-loops when an individual molecule consisting of a closed loop structure with a single attached tail could be discerned. Individual molecules that depicted two visible ends, containing no loops or branched structures were classified as linear. All molecules that did not conform to the looped or linear definition were classified as ambiguous. Densely packed areas of coverslips with overlapping telomere molecules were not scored. Each loop and linear molecule were measured for contour length using the ImageJ trace function.
In-gel analysis of single-stranded telomeric DNA
Mouse telomeric overhang and telomeric restriction fragment patterns were analysed 96 h after EtOH or 4OHT treatment by in-gel hybridization with a γ-32P-ATP end-labelled [AACCCT]4 probe, as previously described6. ImageJ software was used to quantify the single-stranded telomere overhang signals and the signal from total telomeric DNA in the same lane in the denatured gel.
Pulse-field Gel Electrophoresis analysis of DNA DSBs
After treatment with 20 Gy IR from an Cs-137 irradiator, media was changed. At the indicated times after treatment, 106 cells were cast in 1 % agarose into plug moulds. After lysis and washing as previously described30, DNA was separated with a CHEF-DR III (Biorad) system and visualised with EtBr staining using a Biorad GelDoc XR+ imager. DSB fragments and total DNA intensity were calculated for each lane with ImageJ software and DSBs normalised to total DNA for each lane.
Proteomics of Isolated Chromatin (PICh), mass spectrometric analyses and protein identification
ESCs were grown for 8-14 population doublings in either SILAC DMEM R0K0 or R10K8 (Geminibio) supplemented with 15% of SILAC FBS (Geminibio), NEAA, Glutamine and P/S and amplified for PICh on matrigel-coated plates. 96 h after treatment with EtOH or 4OHT, cells were crosslinked in 3.6% formaldehyde 30 min before PICh protocol which was performed as described29 with minor modifications: 25x15cm plates of ESCs were used per condition. Each condition was treated separately until sonication in 50mM Tris pH8, 10mM EDTA, 200mM NaCl, 1% SDS, and subsequent desalting. Equal amounts of chromatin (OD260) were mixed (EtOH R0K0 with 4OHT R10K8 and EtOH R10K8 with 4OHT R0K0), hybridised with 5 μl of 100uM 2’-F RNA telomere probe (Desthiobiotin-108 Carbons-5-(UUAGGG)7.5) and pulled-down with streptavidin beads. EF were grown for 8-14 population doublings in either SILAC DMEM R0K0 or R10K8 (Geminibio) supplemented with 10% of SILAC FBS (Geminibio) and amplified for PICh. 72 h after treatment with EtOH or 4OHT, cells were crosslinked in 3.6% formaldehyde for 30 min before PICh protocol was performed as above. Short NuPAGE gels were run for 1cm prior to reduction, alkylation and overnight ingel trypsin digestion. Peptides were analyzed by nano-liquid chromatography tandem mass spectrometry (nano-LC-MS/MS) using an Ultimate 3000 nano-HLPC coupled to a Lumos Tribrid Orbitrap (Thermo Scientific) as described31. Normalised SILAC ratios were generated using Maxquant to search the raw data against the Uniprot mus musculus reference proteome (UP000000589 https://www.uniprot.org/proteomes/UP000000589) at a false detection rate of 1% and visualised with Perseus.
Statistics and Reproducibility
Statistical analyses were performed using GraphPad PRISM version 8.0 software (GraphPad Inc.). Statistical significance of data was assessed by 2-tailed t-test or one-way ANOVA unless noted otherwise. Data represent mean ±s.e.m. or ±s.d. as indicated in figure legends. P > 0.05 was considered not significant. For all experiments where data is displayed as a dot plot, all values from each repeat of the experiment are displayed together. The mean ± s.d. displayed are the mean and s.d. from all data points across all 3 experiments and are not taken from the average values from each experiment. For all experiments shown, similar results were obtained from three biologically independent experiments, with the following exceptions: similar results were obtained from 2 biologically independent repeats of the experiments shown in Extended Data Figures 4a-b, 5d-c, 9a-b, 9e-h and 11b-f; similar results were obtained from 4 biologically independent repeats of the experiments shown in Figures 1g-h and 3j-k and Extended Data Figures 6e-f, 9c-d and 11g-h.
Extended Data
Extended Data Figure 1. TRF2-null ESCs do not undergo telomere fusions and show only a mild growth defect.
(a-b) Growth curves of Trf2f/f ESCs and EFs after treatment with 4OHT or EtOH. Cells were counted and re-seeded every 2 or 3 days (mean ± s.d., n=3 biologically independent experiments). (c) Brightfield images of the indicated cells 7 days after treatment with EtOH or 4OHT. (d) Western blot of whole cell extracts from Trf2f/f ESCs at the indicated times after treatment with EtOH or 4OHT. (e) Flow cytometry determination of DNA content for the indicated cells 96 hours after 4OHT or EtOH treatment (≥10,000 cells/condition, n=3 independent experiments). (f) Western blot of whole cell extracts from indicated cells 96 h after treatment with EtOH or 4OHT. IR indicates 2 h post-treatment with 20 Gy IR. (g) Gating strategy for flow cytometry determination of DNA content. Example shown is Trf2f/f EF EtOH sample from Fig.1d. Similar strategies to gate live cells and singlets were applied for all flow cytometry experiments. (h) Western blot of whole cell extracts from three independently generated Trf2f/f ESC clones 96 h after treatment with EtOH or 4OHT. (i) Mitotic chromosome spreads from Trf2f/f ESCs 10 days after treatment with 4OHT or EtOH. The DNA is stained with DAPI (red) and the telomeres by FISH (green), with the mean percentage of telomere fusions from (j) indicated. Scale bar = 5 μm (j) Quantification of the experiment shown in (i) (mean ± s.d., ≥ 60 spreads/condition across 3 independent experiments, unpaired two-tailed t-test, ns = not significant). (k) Western blot of whole cell extracts from Trf2f/f ESCs 10 days after treatment with EtOH or 4OHT.
Extended Data Figure 2. Telomeres in TRF2-null ESCs are phenotypically normal.
(a) In-gel hybridization assay for single stranded telomeric G-overhang DNA after Trf2 deletion. Left: TelC signals under the native condition. Right: same gel re-hybridized after in situ denaturation of the DNA. Overhang signals (bottom) were normalized to the total telomeric signals (right) and compared to Trf2f/f EFs treated with EtOH, which was given an arbitrary value of 1 in each of three independent experiments. The mean of these normalised values is shown for each condition (n=3 biologically independent experiments, representative example shown). (b-d) Quantification of telomere fragility (b), heterogeneity (c) and loss (d) from mitotic chromosome spreads shown in Fig.1b (mean ± s.d., ≥70 spreads/condition across 3 independent experiments, unpaired two-tailed t-tests) (e) Representative examples of the phenotypes quantified in (b-d) with relevant abnormalities marked with arrows. The DNA is stained with DAPI (red) and the telomeres by FISH (green). Scale bar = 5 μm (f) IF-FISH analysis to detect endogenous TRF2 in Trf2f/f ESCs 96 h after treatment with 4OHT or EtOH. TRF2 was detected by IF (green), telomeric DNA with FISH (red), and the DNA with DAPI (blue). (g) IF-FISH analysis to detect exogenous Myc-TRF2 in Trf2f/f ESCs stably expressing Myc-TRF2 or a vector-only control 96 h after treatment with 4OHT. Myc-TRF2 was detected by IF (green), telomeric DNA with FISH (red), and the DNA with DAPI (blue). (h) IF-FISH analysis to detect TRF1 in Trf1f/f ESCs 48 h after treatment with 4OHT or EtOH. TRF1 was detected by IF (green), telomeric DNA with FISH (red), and the DNA with DAPI (blue). For (fh) the scale bar = 5 μm. For all panels: ns = not significant.
Extended Data Figure 3. The telomeric DDR is attenuated in ESCs lacking TRF2.
(a) Cartoon depicting the SILAC-PICh workflow used in (b, c). (b-c) Results of SILAC-PICh-Mass Spectrometry of Trf2f/f ESCs (b) and Trf2f/f EFs (c) 96 h after 4OHT or EtOH treatment. Data are shown as the fold change of heavy/light ratio for each protein in duplicate experiments for which opposite labelling with heavy/light isotopes was used. Shelterin subunits and select DNA repair factors are indicated in the plot. NA indicates protein was not detected in that experiment. (d) Venn diagram showing overlap of the proteins identified by SILAC-PICh in Wild Type Trf2f/f ESCs and Trf2f/f EFs (EtOH conditions). (e) IF-FISH analysis to detect γH2AX TIFs in Trf2f/f ESCs and Trf2f/f EFs 96 h after treatment with 4OHT or EtOH. γH2AX was detected by IF (red), telomeric DNA with FISH (white), and the DNA with DAPI (blue). Scale bar = 5 μm. (f) Quantification of the experiment shown in (e) (mean ± s.d., ≥ 300 cells/condition across 3 independent experiments, one-way ANOVA). (g) Quantification of the experiment shown in (e) (each dot represents percentage of cells with >3 TIFs in each of 3 independent experiments analysing ≥100 cells/condition per experiment, bars represent mean ± s.d. of these values, one-way ANOVA) (h) Quantification from experiment shown in Fig. 1g (each dot represents percentage of cells with >3 TIFs in each of 4 independent experiments analysing ≥100 cells/condition per experiment, bars represent mean ± s.d. of these values, one-way ANOVA). (i) Quantification of IF-FISH to detect 53BP1 TIFs in the indicated cells, 96 hours after treatment with EtOH or 4OHT. 53BP1 was detected by IF (green), telomeric DNA with FISH (red) and DNA with DAPI (blue). c1, c2 and c3 indicate 3 independently derived clones. (mean ± s.d., ≥ 200 cells/condition across 3 independent experiments, one-way ANOVA). For all panels: **** p ≤ 0.0001.
Extended Data Figure 4. The DNA Damage Response and NHEJ are functional in ESCs.
(a) IF to detect γH2AX (green) and 53BP1 (red) in untreated Trf2f/f ESCs and Trf2f/f EFs or 1 h after treatment with 0.5 Gy IR. DNA stained with DAPI (blue), scale bar = 5 μm. (b) Quantification from experiment shown in (a) (each dot represents percentage of cells with >4 foci in each of 2 independent experiments analysing ≥100 cells/condition per experiment, bars represent mean ± s.d. of these values, one-way ANOVA). (c) Flow cytometry determination of DNA content for the indicated cells at the indicated times after IR treatment, 96 h after treatment with EtOH or 4OHT (n=3 independent experiments, ≥10,000 cells/condition in each experiment). (d) DNA from Trf2f/f ESCs and Trf2f/f MEFs was harvested, subjected to pulsed-field gel electrophoresis and stained with EtBr at the indicated times after irradiation with 20 Gy. The band of DNA resulting from double strand breaks (DSBs) is indicated. (e) Quantitation of the experiment in (d). The DNA band resulting from DNA DSBs was quantified and normalised to the unbroken DNA. The level of DNA resulting from DSBs was subsequently compared to the sample harvested 10 minutes after IR from that condition, which was normalized as 100% DSBs remaining (mean ± s.d. n=3 biologically independent experiments). (f) Survival of non-targeting control (NTC), Ku70 knockout (Ku70-KO), or DNA Ligase IV knockout (LigIV-KO) Trf1f/f Trf2f/f ESCs after exposure to the indicated doses of Ionising Radiation as measured using CellTitreGlo, 4 days after IR treatment (n=3 biologically independent experiments, mean ± s.e.m). Individual clonal knockout lines are denoted c1 and c2. (g-h) Western blot of whole cell extracts from the cells used in (f). * indicates non-specific band. For all panels: ns = not significant, **** p ≤ 0.0001.
Extended Data Figure 5. The absence of telomere fusions in TRF2-null ESCs is not the result of a weak DDR or a short G1 phase.
(a) 96 h after treatment with EtOH or 4OHT, Trf2f/f ESCs were grown asynchronously or treated for 16 h with Mimosine to induce G1 arrest. DNA content from these cells was determined using flow cytometry at 0 h, 4 h and 9 h after release from Mimosine. (b) Mitotic chromosome spreads from asynchronous cells and cells released from Mimosine block for 9 h as in (a). The DNA was stained with DAPI (red) and the telomeric DNA with FISH (green). The mean percentage of fused chromosomes from (c) is indicated. (c) Quantification from experiment shown in (a, b) (mean ± s.d., ≥ 70 spreads/condition across 3 independent experiments, unpaired two-tailed t-test). (d) Mitotic chromosome spreads from Trf2f/f ESCs 96 h after treatment with 4OHT and 24 h after mock treatment or treatment with 2 Gy IR. Samples are stained as in (b) and the mean percentage of telomere fusions from (e) is indicated. (e) Quantification of experiment shown in (d) (mean ± s.d., ≥ 50 spreads/condition across 2 independent experiments, unpaired two-tailed t-test). For all panels: Scale bar = 5 μm; ns = not significant.
Extended Data Figure 6. TPP1 and TRF1 function are conserved in ESC and somatic tissues.
(a) Western blot of whole cell extracts from indicated cells 96 h after EtOH or 4OHT treatment, and 72 h after siRNA transfection. (b) TPP1 expression in the indicated cells analysed by RT-qPCR. Data were normalized to the ESC Trf2f/f EtOH Control siRNA (siCon) condition, which was arbitrarily assigned a value of 1 (mean ± s.d., n=3 biologically independent experiments, one-way ANOVA). (c) Quantification of 53BP1 TIFs from cells shown in (d) (mean ± s.d., ≥ 300 cells/condition across 3 independent experiments, one-way ANOVA, ** p = 0.0013). (d) 53BP1 TIFs in the indicated cells. 53BP1 was detected by IF (green), telomeres by FISH (red) and DNA with DAPI (blue). (e) Mitotic chromosome spreads from indicated cells 96 h after 4OHT or EtOH treatment, and 72 h after siRNA transfection. Telomeres stained with FISH (green) and the DNA with DAPI (red). The mean percentage of fused telomeres from (f) is shown. (f) Quantification of telomere fusions from (e) (mean ± s.d., 90 spreads/condition across 4 independent experiments, one-way ANOVA). (g) Flow cytometry determination of DNA content for the indicated cells 96 hours after 4OHT or EtOH treatment (n=3, ≥ 10,000 cells/condition). (h) Telomeric RPA-pSer33 foci in Trf1f/f ESCs 72 h after treatment with EtOH or 4OHT. RPA-pSer33 was detected by IF (green), telomeres by FISH (red), and DNA with DAPI (blue). (i) Quantification of the experiment in (h) (mean ± s.d., ≥ 300 cells/condition across 3 independent experiments, unpaired two-tailed t-test). (j) 53BP1 TIFs in Trf1f/f ESCs 72 h after treatment with 4OHT or EtOH. Samples are stained as in (d). (k) Quantification of (j) (mean ± s.d., ≥ 300 cells/condition across 3 independent experiments, unpaired two-tailed t-test). For all panels: Scale bar = 5 μm; ns = not significant, **** p ≤ 0.0001.
Extended Data Figure 7. ESCs require Shelterin for chromosome end protection.
(a) Equal numbers of the indicated cells were seeded and treated with EtOH or 4OHT. The cells were counted at 72 h and the number of cells in each condition normalized to the ESC Trf1+/+ Trf2f/f EtOH condition, which was given an arbitrary value of 1 (mean ± s.d., n=3 biologically independent experiments). (b) Flow cytometry determination of DNA content for the indicated cells 48 or 72 hours after 4OHT or EtOH treatment (n=3, ≥ 10,000 cells/condition). (c) 53BP1 TIFs in the indicated cells 72 h after treatment with 4OHT or EtOH. 53BP1 was detected by IF (green), the telomeres with FISH (red) and DNA with DAPI (blue). Scale bar = 5 μm. (d) Quantification of the experiment shown in (c) (mean ± s.d. ≥ 300 cells/condition across 3 independent experiments, one-way ANOVA, **** p ≤ 0.0001) (e) Western blot of whole cell extracts from Trf2f/f ESCs at indicated times after treatment with 2 Gy or 20 Gy IR.
Extended Data Figure 8. Shelterin-free ESCs activate a full DNA Damage Response equivalent to that in Shelterin-free somatic cells.
(a) Western blotting of whole cell extracts from Trf1f/f Trf2f/f ESCs at indicated times after EtOH or 4OHT treatment. (b,c) Quantification of telomere fusions (b) or γH2AX TIFs (c) in Trf1f/f Trf2f/f ESCs at the indicated times after 4OHT or EtOH treatment (mean ± s.d., ≥70 mitotic spreads/condition for (b) or, ≥300 cells/condition for (c), each across 3 independent experiments, one-way ANOVA). (d) 53BP1 TIFs in the indicated cells, 48 h after EtOH or 4OHT treatment or infection with AdGFP or AdCRE. 53BP1 was detected by IF (green), telomeres by FISH (red) and DNA with DAPI (blue). (e) Quantification of the experiment shown in (d) (mean ± s.d., ≥ 200 cells/condition across 2 independent experiments, one-way ANOVA). (f) Quantification of γH2AX TIFs in the indicated cells 48 after treatment with EtOH or 4OHT or infection with AdGFP or AdCRE (mean ± s.d., ≥ 200 cells/condition across 3 independent experiments, one-way ANOVA). (g) Western blotting of whole cell extracts from Trf1f/f Trf2f/f ESCs and Trf1f/f Trf2f/f MEFs at the indicated times after EtOH or 4OHT treatment or infection with AdGFP or AdCRE. (h) Quantification of telomere fragility from mitotic chromosome spreads shown in Fig.2f, 72 h after EtOH or 4OHT treatment (mean ± s.d., ≥70 spreads/condition examined over 3 experiments, one-way ANOVA) (i) γH2AX TIFs in the indicated cells 36 h after EtOH or 4OHT treatment. γH2AX was detected by IF (green) telomeric DNA with FISH (red), and the DNA with DAPI (blue). (j) Quantification of the experiment shown in (i) (mean ± s.d., ≥ 300 cells/condition across 3 independent experiments, one-way ANOVA). For all panels: the scale bar = 5 μm; ns = not significant; **** p ≤ 0.0001.
Extended Data Figure 9. Shelterin free ESC and MEF telomeres are substrates for both ATM and ATR kinases.
(a) IF-FISH analysis to detect telomeric ATM-pSer1987 foci in CSK-extracted Trf1f/f Trf2f/f ESCs 36 h after treatment with 4OHT or EtOH. ATM-pSer1987 was detected by IF (green), telomeres by FISH (red), and DNA with DAPI (blue). (b) Quantification from experiment shown in (a) (mean ± s.d., n=2 independent experiments, ≥100 cells/condition per experiment, unpaired two-tailed t-test). (c) IF-FISH analysis to detect telomeric RPA-pSer33 foci in CSK-extracted Trf1f/f Trf2f/f ESCs 36 h after treatment with 4OHT or EtOH. RPA-pSer33 (green) was detected by IF, telomeres by FISH (red), DNA with DAPI (blue). (d) Quantification from experiment shown in (c) (mean ± s.d., n=4 independent experiments, ≥100 cells/condition per experiment, unpaired two-tailed t-test). (e) IF-FISH analysis to detect telomeric ATM-pSer1987 foci in CSK-extracted Trf1f/f Trf2f/f MEFs 48 h after infection with AdGFP or AdCRE. ATM-pSer1987 was detected by IF (green), telomeres by FISH (red), and DNA with DAPI (blue). (f) Quantification from experiment shown in (e) (mean ± s.d., n=2 independent experiments, ≥150 cells/condition per experiment, unpaired two-tailed t-test). (g) IF-FISH analysis to detect telomeric RPA-pSer33 foci in CSK-extracted Trf1f/f Trf2f/f MEFs 48 h after infection with AdGFP or AdCRE. RPA-pSer33 was detected by IF (green), telomeres by FISH (red), and DNA with DAPI (blue). (h) Quantification from experiment shown in (g) (mean ± s.d., n=2 independent experiments, ≥150 cells/condition per experiment, unpaired two-tailed t-test). For all panels: scale bar = 5 μm; **** p ≤ 0.0001. For (b, d, f, g), each dot represents value from each independent experiment, bars represent mean ± s.d., of these values.
Extended Data Figure 10. Telomere fusions in Shelterin-free ESCs are primarily driven by ATM signaling and NHEJ.
(a) Mitotic chromosome spreads from the indicated cells 72 h after treatment with 4OHT or EtOH, mock treated or treated with an ATR inhibitor (ATRi). The DNA is stained with DAPI (red) and the telomeres by FISH (green). The mean percentage of fused chromosome ends from Fig. 2k is noted. (b) Western blotting of whole cell lysates from the indicated cells, 72 h after infection with AdGFP or AdCRE. * indicates non-specific band (c) Quantification of telomere fusions in the indicated cells 72 h after infection with AdGFP or AdCRE. Cells were either mock treated or treated with an ATRi (n≥50 spreads/condition analysed over 2 biologically independent experiments, one-way ANOVA. ns=not significant; **** p ≤ 0.0001). (d) Western blotting of whole cell lysates from Ku70-Knockout (Ku70-KO) or non-targeting control (NTC) Trf1f/f Trf2f/f ESCs 72 h after treatment with EtOH or 4OHT. (e) Mitotic chromosome spreads from the indicated cells 72 h after treatment with 4OHT or EtOH. Cells were stained as in (a) and the mean percentage of fused chromosome ends from Fig. 2l is shown. (f) Western blotting of whole cell lysates from DNA Ligase IV-Knockout (LigIV-KO) or NTC Trf1f/f Trf2f/f ESCs 72 h after treatment with EtOH or 4OHT. * indicates non-specific band (g) Mitotic chromosome spreads from the indicated cells 72 h after treatment with 4OHT or EtOH. Cells were stained as in (a). The mean percentage of chromosome ends fused from Fig 2m is noted. For all panels: Scale bar = 5 μm
Extended Data Figure 11. TRF2 is required for telomere protection and viability upon loss of pluripotency.
(a) Brightfield images of Trf2f/f EpiSCs 96 hours after EtOH or 4OHT treatment. (b-f) Gene expression in the indicated cells analysed by RT-qPCR 96 h after EtOH or 4OHT treatment. Data were normalized to expression in the ESC Trf2f/f EtOH sample, which was arbitrarily assigned a value of 1 for each gene in each experiment. (mean ± s.d., n=2 biologically independent experiments). (g, h) γH2AX TIF images (g) and quantitation (h), 96 h hours after EtOH or 4OHT treatment (mean ± s.d. ≥300 cells across 4 independent experiments, unpaired two-tailed t-test, **** p ≤ 0.0001). γH2AX was detected by IF (green), telomeres by FISH (red), and DNA with DAPI (blue). (i) 53BP1 TIFs in Trf2f/f EpiSCs 96 h after 4OHT or EtOH treatment. 53BP1 was detected by IF (green), telomeric DNA by FISH (red) and the DNA with DAPI. (jn) Gene expression in the indicated cells analysed by RT-qPCR from 2-5 days after initiating differentiation. Data were normalized as described for (b-f). “Diff Day x” refers to cells on day “x” of the differentiation protocol (mean ± s.d., n=3 biologically independent experiments). (or) Gene expression in the indicated cells analysed by RT-qPCR. Data were normalized as described in (b-f) (mean ± s.d., n=3 biologically independent experiments). (s) Western blots of whole cell extracts from Trf2f/f ESCs differentiated as in Fig. 3e. (t, u) Nanog and Fgf5 gene expression in the indicated cells analysed by RT-qPCR 96 hours after treatment with EtOH or 4OHT. Data were normalized as described for (b-f) (mean ± s.d., n=3) (v) Western blot of whole cell extracts from the indicated cells 96 h after treatment with EtOH or 4OHT. * indicates non-specific band. For h, i: Scale bar = 5 μm.
Extended Data Figure 12. TRF2 is required for blastocyst development prior to implantation.
(a) Quantification of NANOG-positive and NANOG-negative cells in each of the embryos assessed in Fig. 5e, f (mean ± s.d., one-way ANOVA, ns = not significant). (b) Maximal projection of z-stacks from Trf2-/- embryos displaying pan-nuclear γH2AX-positive cells which are highlighted with arrows. γH2AX (green) and NANOG (magenta) were detected by IF, telomeres by FISH (red), DNA with DAPI (blue). (c) Quantification of percentage of cells within an embryo showing pan-nuclear γH2AX staining from experiment shown in (b) (mean ± s.d., one-way ANOVA).
Extended Data Figure 13. TRF2 is not required for t-loop formation in ESCs.
(a) IF-FISH analysis to detect telomeric ATM-pSer1987 foci in Trf2f/f ESCs 96 h after treatment with 4OHT or EtOH. ATM-pSer1987 was detected by IF (green), telomeres by FISH (red), DNA with DAPI (blue). (b) Quantification from experiment shown in (a) (mean ± s.d., ≥300 cells across 3 independent experiments, unpaired two-tailed t-test). (c) Schematic representation of TRF1 and the TRF2 variants used in this study21. (d) Western blots of whole cell extracts from Trf2f/f ESCs expressing WT, mutant, or hybrid TRF2 alleles 96 hours after treatment with EtOH or 4OHT. (e) IF-FISH detection of γH2AX TIFs in the indicated cells 96 h after treatment with 4OHT or EtOH. γH2AX was detected by IF (green), telomeric DNA by FISH (red), and DNA with DAPI (blue). (f) Quantification of the experiment shown in (e) (mean ± s.d., ≥300 cells across 3 independent experiments, two-tailed t-test) (g) Quantitation (upper panel) of the cross-linking efficiency test (bottom panel) in Trf2f/f ESCs 96 h after treatment with EtOH or 4OHT (mean ± s.e.m., n=3 biologically independent experiments, unpaired two-tailed t-test). (h) Measurement of linear and t-loop molecules shown in Fig. 5f (mean ± s.e.m., n=3 biological replicates scoring ≥ 2804 molecules per replicate). T-loop measurements are a sum of the loop and tail portions of the molecule. For (a) and (e), scale bar = 5 μm. For all panels: ns = not significant, **** p ≤ 0.0001.
Acknowledgements
We thank members of the Boulton and Cesare labs for suggestions and discussions and for critical reading of the manuscript. We thank Scott Page and the Australian Cancer Research Foundation Telomere Analysis Centre at the Children’s Medical Research Institute (CMRI) for microscopy infrastructure. We thank the Crick BRF and GEMs for support with animal experiments. We thank Titia de Lange for advice on G-overhang analysis. The work in the Boulton lab is supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC0010048), the UK Medical Research Council (FC0010048), and the Wellcome Trust (FC0010048); a European Research Council (ERC) Advanced Investigator Grant (TelMetab); and Wellcome Trust Senior Investigator and Collaborative Grants. The laboratory of A.J.C. is supported by grants from the Australian NHMRC (1162886 and 1185870) and philanthropy from the Neil and Norma Hill Foundation.
Footnotes
Author Contribution
P.R., P.M. and S.J.B. conceived the study; P.R. generated all cell lines and performed the large majority of experiments in this manuscript, including all those not listed below; V.B. A.M. and P.R. performed EmbryoScope and embryo IF-FISH experiments; G.R.K. adapted stem cell culture for t-loop visualization, D.V-L. performed all AiryScan imaging and analysis, P.M and S.H. performed PICh-MS analysis. P.R., D.V.L., V.B., G.R.K., A.M., S.H., P.M., A.J.C. and S.J.B. analysed data; P.R., A.J.C, and S.J.B. wrote the manuscript with editorial input from D.V.L., P.M., J.B., and K.K.N.
Author Information
All data are archived at the Francis Crick Institute or CMRI. The authors declare no competing interests. Requests for materials should be addressed to simon.boulton@crick.ac.uk.
Data Availability
The datasets generated during and/or analysed during the current study are either included alongside the manuscript and are available from the corresponding author on reasonable request. Data were searched against a recent copy of the uniprot mus musculus reference proteome UP000000589 https://www.uniprot.org/proteomes/UP000000589. For gel source data, see Supplementary Figure 1.
References
- 1.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annual review of genetics. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 2.Griffith JDC, C L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian Telomeres End in a Large Duplex Loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 3.Van Ly D, et al. Telomere Loop Dynamics in Chromosome End Protection. Mol Cell. 2018;71:510–525.:e516. doi: 10.1016/j.molcel.2018.06.025. [DOI] [PubMed] [Google Scholar]
- 4.van Steensel B, S A, de Lange T. TRF2 Protects Human Telomeres from End-to-End Fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 5.Karlseder J, B D, Dai Y, Hardy S, de Lange T. p53- and ATM-Dependent Apoptosis Induced by Telomeres Lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 6.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 7.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 8.Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155:345–356. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarek G, et al. CDK phosphorylation of TRF2 controls t-loop dynamics during the cell cycle. Nature. 2019;575:523–527. doi: 10.1038/s41586-019-1744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 11.Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman JR, et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takai Kaori K, Kibe T, Donigian Jill R, Frescas D, de Lange T. Telomere Protection by TPP1/POT1 Requires Tethering to TIN2. Molecular Cell. 2011;44:647–659. doi: 10.1016/j.molcel.2011.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science (New York N.Y) 2012;336:593–597. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takaoka K, Hamada H. Cell fate decisions and axis determination in the early mouse embryo. Development. 2012;139:3–14. doi: 10.1242/dev.060095. [DOI] [PubMed] [Google Scholar]
- 17.Brons IG, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 18.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 19.Acampora D, Di Giovannantonio LG, Simeone A. Otx2 is an intrinsic determinant of the embryonic stem cell state and is required for transition to a stable epiblast stem cell condition. Development. 2013;140:43–55. doi: 10.1242/dev.085290. [DOI] [PubMed] [Google Scholar]
- 20.Gouti M, et al. In vitro generation of neuromesodermal progenitors reveals distinct roles for wnt signalling in the specification of spinal cord and paraxial mesoderm identity. PLoS Biol. 2014;12:e1001937. doi: 10.1371/journal.pbio.1001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamoto K, et al. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013;494:502–505. doi: 10.1038/nature11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding H, et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 23.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tzelepis K, et al. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Rep. 2016;17:1193–1205. doi: 10.1016/j.celrep.2016.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zelensky AN, Schimmel J, Kool H, Kanaar R, Tijsterman M. Inactivation of Pol theta and C-NHEJ eliminates off-target integration of exogenous DNA. Nat Commun. 2017;8:66. doi: 10.1038/s41467-017-00124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wamaitha SE, et al. Gata6 potently initiates reprograming of pluripotent and differentiated cells to extraembryonic endoderm stem cells. Genes Dev. 2015;29:1239–1255. doi: 10.1101/gad.257071.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 29.Panier S, et al. SLX4IP Antagonizes Promiscuous BLM Activity during ALT Maintenance. Mol Cell. 2019;76:27–43.:e11. doi: 10.1016/j.molcel.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meltser V, Ben-Yehoyada M, Reuven N, Shaul Y. c-Abl downregulates the slow phase of double-strand break repair. Cell Death Dis. 2010;1:e20. doi: 10.1038/cddis.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellelli R, et al. Polepsilon Instability Drives Replication Stress, Abnormal Development, and Tumorigenesis. Mol Cell. 2018;70:707–721.:e707. doi: 10.1016/j.molcel.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analysed during the current study are either included alongside the manuscript and are available from the corresponding author on reasonable request. Data were searched against a recent copy of the uniprot mus musculus reference proteome UP000000589 https://www.uniprot.org/proteomes/UP000000589. For gel source data, see Supplementary Figure 1.