Abstract
Transcriptional networks are crucial to integrate various internal and external signals into optimal responses during plant growth and development. Primary root vasculature patterning and proliferation are controlled by a network centred around the basic Helix-Loop-Helix transcription factor complex formed by TARGET OF MONOPTEROS 5 (TMO5) and LONESOME HIGHWAY (LHW), which control cell proliferation and division orientation by modulating cytokinin response and other downstream factors. Despite recent progress, many aspects of the TMO5/LHW pathway are not fully understood. In particular, the upstream regulators of TMO5/LHW activity remain unknown. Here, using a forward genetic approach to identify new factors of the TMO5/LHW pathway, we discovered a novel function of the MYB-type transcription factor MYB12. MYB12 physically interacts with TMO5 and dampens the TMO5/LHW-mediated induction of direct target gene expression as well as the periclinal/radial cell divisions. The expression of MYB12 is activated by the cytokinin response, downstream of TMO5/LHW, resulting in a novel MYB12-mediated negative feedback loop that restricts TMO5/LHW activity to ensure optimal cell proliferation rates during root vascular development.
Keywords: Arabidopsis thaliana, cytokinin, EMS screen, root development, transcription factors, vascular development
Introduction
Transcription factors (TFs) play a crucial role in controlling virtually all developmental processes in eukaryotes by regulating the expression of specific subsets of target genes. TFs do not typically act alone but are embedded in complex transcriptional networks, which modulate their activity to ensure optimal transcriptional output in response to various environmental and developmental signals. Transcriptional networks often rely on feedback regulation, where a TF promotes the expression of its own activator (positive feedback) or repressor (negative feedback), respectively (Ohashi-Ito and Fukuda, 2020).
During vascular development in the plant embryo and primary root apical meristem, the heterodimer complex formed by the basic Helix-Loop-Helix (bHLH) TFs TARGET OF MONOPTEROS 5 (TMO5) and LONESOME HIGHWAY (LHW) controls vascular cell proliferation leading to radial expansion of the vascular bundle (De Rybel et al., 2014; De Rybel et al., 2013; Ohashi-Ito and Bergmann, 2007; Ohashi-Ito et al., 2013; Ohashi-Ito et al., 2014). The TMO5/LHW dimer is active in xylem cells, where it directly activates the expression of LONELY GUY 3 (LOG3), LOG4 and BETA GLUCOSIDASE 44 (BGLU44), encoding key enzymes in the biosynthesis and deconjugation of cytokinin (De Rybel et al., 2014; Kurakawa et al., 2007; Kuroha et al., 2009; Ohashi-Ito et al., 2014; Yang et al., 2021). This leads to a local increase of cytokinin, which is thought to diffuse to the neighbouring procambium cells (De Rybel et al., 2014; Ohashi-Ito et al., 2014) and trigger the expression of members of the DNA-BINDING WITH ONE FINGER (DOF) type TF family (Miyashima et al., 2019; Smet et al., 2019). These DOF-type TFs in turn lead to a switch in division plane orientation from anticlinal to periclinal and radial in specific subsets of procambium and phloem pole cells, depending on the DOF family member. The actual molecular mechanisms are however not yet fully explored (Otero et al., 2022). The activity of the TMO5/LHW complex is negatively regulated by members of the SUPPRESSOR OF ACAULIS51-LIKE (SACL) subclade of bHLH TFs (Katayama et al., 2015; Vera-Sirera et al., 2015). Similarly to TMO5, SACLs physically interact with LHW. By competing with TMO5 for LHW binding, the SACLs reduce the amount of functional TMO5/LHW complexes, and thus dampen the activity of the pathway (Katayama et al., 2015; Vera-Sirera et al., 2015). As SACL genes are themselves downstream targets of TMO5/LHW, they constitute a typical negative feedback loop (Katayama et al., 2015; Vera-Sirera et al., 2015).
Besides forming bHLH homo- or heterodimers, bHLH proteins have also been shown to directly interact with other proteins such as MYB-type TFs, which can enhance or supress their transcriptional activity (Carretero-Paulet et al., 2010; Cui et al., 2021; Feller et al., 2011; Zhao et al., 2008). MYB TFs are defined by their highly conserved DNA-binding MYB-domain that contains up to four α-helical “R” repeats (Du et al., 2009; Ogata et al., 1996). The class (R1, R2 or R3, depending on their similarity to c-Myb R repeats) and number of the R repeats are the basis of MYB protein classification (Dubos et al., 2010). Most plant MYBs belong to the R2R3-MYB subfamily (Stracke et al., 2001), which is involved in a plethora of processes including phenylpropanoid biosynthesis (Liu et al., 2015), development of tissues and organs (Lee and Schiefelbein, 1999; Oppenheimer et al., 1991) and hormonal responses (Jin and Martin, 1999). Exemplary bHLH-MYB interactions take place during epidermal cell fate specification. The formation of trichomes and root hairs depends on the assembly of different heterotrimeric bHLH/WD40/MYB complexes. In addition to the WD40 protein TRANSPARENT TESTA GLABRA 1 (TTG1), the core bHLH proteins GLABRA 3 (GL3) or ENHANCER OF GLABRA 3 (EGL3) interact with the R2R3 MYB proteins WEREWOLF (WER) or GLABRA 1 (GL1), forming an active transcriptional complex that promotes root hair or trichome formation, respectively. Alternatively, the recruitment of CAPRICE (CPC), TRIPTYCHON (TRY) or ENHANCER OF TRY AND CPC 1, single-repeat R3 MYBs that lack the C-terminal transcriptional activation domain and compete with the active R2R3 MYBs for bHLH binding, results in the formation of a transcriptionally inactive complex that prevents trichome/root hair formation (Kirik et al., 2004; Ramsay and Glover, 2005; Tominaga-Wada et al., 2017; Wada et al., 1997). The single-repeat R3 MYBs are downstream targets of the active MYB/bHLH/WD40 complex, and at the same time its non-cell autonomous inhibitors. The bHLH and MYB TFs thus constitute a negative feedback loop that lies at the core of epidermal cell type specification and patterning (Song et al., 2015; Wang et al., 2008). A similar bHLH/MYB/WD40 complex controls the expression of a core enzyme in the proanthocyanin biosynthetic pathway (Appelhagen et al., 2011; Xu et al., 2015; Xu et al., 2013). As such, interactions between MYB and bHLH TFs are key to various developmental processes.
The closely related R2R3 MYB proteins MYB11, MYB12 and MYB111 promote the expression of genes encoding key flavonol biosynthetic enzymes (Mehrtens et al., 2005; Stracke et al., 2007; Stracke et al., 2010; Stracke et al., 2017). Flavonols are a subgroup of flavonoids, besides the red to purple anthocyanins and brown proanthocyanidins (Lepiniec et al., 2006; Winkel-Shirley, 2001). Flavonoids convey color to fruits and seeds and aid in abiotic stress response (Wang et al., 2016). MYB11, MYB12 and MYB111 induce flavonol biosynthesis at different developmental stages, depending on their distinct expression patterns: While MYB12 is mostly active in roots, MYB11 acts in meristematic tissues and MYB111 functions in the hypocotyl and cotyledons (Stracke et al., 2007). The genes encoding flavonol biosynthesis enzymes CHALCONE SYNTHASE (CHS), CHALCONE FLAVANONE ISOMERASE (CHI), FLAVANONE 3’-HYDROXYLASE (F3’H), and FLAVONOL SYNTHASE (FLS) catalyse consecutive steps of flavonol production (Forkmann and Martens, 2001) and are regulated by MYB TFs via the MYB recognition element in their promoter regions. CHS and FLS are directly transcriptionally activated by MYB12 (Mehrtens et al., 2005). Consequently, the levels of the flavonols kaempferol and quercetin are decreased in the myb12 mutant, while MYB12 overexpression leads to increased flavonol levels (Mehrtens et al., 2005).
Here, we discover a novel role of MYB12 as a negative regulator of the TMO5/LHW pathway during vascular proliferation. MYB12 is a downstream target of TMO5/LHW; interacts with TMO5 and represses TMO5/LHW transcriptional activity, thus constituting a negative feedback loop in the regulation of vascular development. Our work highlights the importance of bHLH-MYB interactions in multiple developmental processes; and demonstrates concomitant activator and repressor functions of the same TF in different transcriptional network contexts.
Materials and Methods
Plant material and growth conditions
Seedlings were grown at 22°C under continuous light on ½ Murashige and Skoog (MS) medium without 1% sucrose, after seeds were stratified for 24h-48h. For dexamethasone (DEX) treatment, 10 μM DEX (Sigma-Aldrich) was added to the growth medium from a 10 mM DMSO stock solution; seedlings were either germinated on DEX-containing medium or transferred from MS medium at the indicated time point. For the CK sensitivity assay, seedlings were germinated on 10μM 6-benzylaminopurine (6-BAP; Duchefa)-containing medium. For the CK treatment of pMYB12::nGFP/GUS, 5-day old seedlings were incubated in liquid ½ MS supplemented with 10μM 6-BAP for 6h.The AGI identifiers for the genes used in this manuscript are as followed: TMO5 (AT3G25710), LHW (AT2G27230), MYB11 (AT3G62610), MYB12 (AT2G47460), MYB111 (AT5G49330), LOG4 (AT3G53450), GH10 (AT4G38650) and ARR5 (AT3G48100). The following mutant and transgenic lines were described previously: myb12-1f (Mehrtens et al., 2005); myb11 myb12-1f myb111 (myb triple) (Stracke et al., 2007); pRPS5A::TMO5:GR x pRPS5A::LHW:GR (dGR) (Smet et al., 2019); pLOG4::n3GFP; pTMO5::nGFP/GUS (De Rybel et al., 2014). The lines ins4/lhw-8 and hyp2/myb12-2 were generated in the dGR background by EMS mutagenesis (see below). The lines pGH10::n3GFP, pRPS5A::MYB12, pRPS5A::MYB12 hyp2, pMYB12::nGFP-GUS and pMYB12::gMYB12:sYFP were obtained by transforming the respective expression clones into Col-0 or hyp2 by the floral dip method (Clough and Bent, 1998). The pLOG4::n3GFP and pGH10::n3GFP were introduced into the dGR and pRPS5A::MYB12 hyp2/myb12-2 dGR backgrounds by genetic crossing and analysed in F1 generation seedlings.
EMS mutagenesis and screening
The dGR line (Smet et al., 2019) was used for the EMS mutagenesis. Approximately 10,000 seeds were incubated shaking in water overnight. The water was replaced with 15 ml of 0.05 % Triton X-100. After mixing well, the seeds were incubated for 5 min in this solution then twice washed with water. The seeds were mutagenized by treatment with 30 mM EMS in 0.1 M phosphate buffer (pH 7.5) for 6-7 hours. Afterwards, the EMS solution was removed, and mutagenesis was stopped by adding 0.1 M Na2S2O3 for 5 min five times. The Na2S2O3 was washed away with water seven times. These seeds were afterwards stratified in 0.1% agarose overnight. Approximately 50 seeds were sown together in a pot per pool. A total of 228 pools was maintained. For each pool, about 1,000 M2 seeds were initially screened on 10 μM DEX containing ½ MS media, leading to a selection of 260 mutants from 110 pools. Next, the root length and root width of one-week-old M3 seedlings was measured in both mock (DMSO) and 10 μM DEX. Changes in root length and meristem width were measured upon DEX treatment and compared to a Col-0 and dGR control.
Mapping causal mutation of EMS mutants
Selected EMS mutants were backcrossed with the parental dGR line, and one-week-old BC1F2 seedlings with the desired phenotype were collected for DNA extraction. DNA was extracted using hexadecyltrimethylammoniumbromide (CTAB) extraction buffer (0.1 M Tris pH7.5, 0.7M NaCl, 0.01 M EDTA and 0.03 M CTAB) and afterwards isolated using chloroform:isoamylalcohol (24:1) and isopropanol. RNA was degraded by RNase treatment between the chloroform and isopropanol isolation steps. The bulked genomic DNA was sequenced by using the Illumina NextSeq 500 system. For the library preparation, an insert size of 400-500 bp was used. Paired end sequencing was performed, with a read length of 2x150 bp length and 50x coverage. Potential causal mutations were selected using the SHORE map analysis tool (Schneeberger et al., 2009).
Molecular cloning
The promoters and coding sequences were PCR-amplified using a high-fidelity polymerase (primers used are shown in Table S3). All constructs were made by MultiSite Gateway cloning (Karimi et al., 2002). Promoter regions were amplified from genomic DNA and introduced into the pDONRP4P1R vector. The coding sequences were amplified from genomic DNA or root cDNA and introduced into the pDONR221 vector. All entry clones were sequence verified before further steps. The MYB12 promoter was subcloned into pmK7S*nF14mGW destination vector. The construct was transformed in Col-0 and dGR via Agrobacterium-mediated flower dipping (Clough and Bent, 1998).
Root phenotyping
For root length measurements, one-week-old roots were scanned on a flatbed scanner and root length was measured in FIJI (Schindelin et al., 2012) with the integrated NEURONJ plugin (https://imagescience.org/meijering/software/neuronj/) (Meijering et al., 2004). Root width of one-week-old seedlings were measured by dissecting the roots and mounting them in clearing agent (60 % lactic acid, 20 % glycerol and 20 % H2O). Width of the root tips was measured at the beginning of the elongation zone for all roots in FIJI. Imaging of differentiated primary xylem vessels was performed on one-week-old roots mounted in the clearing agent described above. GUS staining of pMYB12::GUS was performed as described previously (De Rybel et al., 2010). The slides were imaged using a Differential Interference contrast (DIC) microscope (Olympus BX51) equipped with a Nikon camera and image captured with ToupView software.
Statistics and visualization of the data
All boxplots and statistical analysis were generated and performed within R studio (R Core Team, 2020). In these plots, the boxes indicate the descriptive statistics with the interquartile range (IQR) with the central black line marking the median, and the 25th and 75th percentile of the data, the whiskers extend to minima and maxima within 1.5 IQR of the 25th and 75th percentiles, and outliers are indicated as single empty circles. Additionally, the means (blue rhombus) and a measure of their precision (blue standard error bars) as well as the compact letter display (blue letters) are displayed. The ‘n’ represents the number of data points, which are plotted as the grey dots. For the continuous variables, root width and root length, one-way or two-way analysis of variance (ANOVA) with post-hoc Tukey testing at 5% level of significance was performed. Significances asterisks: * = p-value < 0.05; ** = p-value < 0.01; *** = p-value < 0.001. A generalized linear model with log link function has been fitted to count data (e.g. number of cell files). The lower-case letters (compact letter display) associated to the mean and its precision within the boxplots, indicate which groups with any common letter are not significantly different, determined by pairwise comparisons of estimated marginal means testing with p-value adjustment. P-values have been adjusted for multiple testing by the Bonferroni method, if not otherwise specified in Table S1.
Confocal imaging and processing
Transcriptional and translational fluorescent reporter lines were imaged on a Leica SP8 confocal microscope with a 40x NA 1.1 water immersion objective. Seedlings were mounted in propidium iodide (PI); GFP and sYFP reporter lines were excited at 488, resp. 514 nm and detected at 500-535, resp. 515-550 nm; PI was detected at 600-700 nm. For the vascular cell file number measurements in root tips, one-week-old seedlings were fixed and stained using the mPS-PI protocol and imaged using the Leica SP2 or SP8 confocal microscopes as described previously (Arents et al., 2022; Truernit et al., 2008). For the vascular cell file number of the late primary root and secondary root, 4-day, 14-day or 21-day old seedlings were used respectively. Roots were sampled 0,5 cm below the hypocotyl and vibratome cross-sections were made as described previously (Arents et al., 2022). Cell walls were stained with calcofluor and lignin in xylem vessels was stained with basic fuchsin as described before (Ursache et al., 2018). Cell counting was done using the cell counter plugin in FIJI (Schindelin et al., 2012). The vascular bundle cell number quantifications included the pericycle cell layer, except if mentioned otherwise. The analysis of CK treated pMYB12::nGFP/GUS was the sum of 10 Z-slices and measured Integrated density of the whole image using FIJI (Schindelin et al., 2012). All figures were assembled and processed using Inkscape and Adobe Illustrator.
RNA isolation and qRT-PCR
For dGR induction, plants were grown on ½ MS (1% agar) for 5 days before transferring to either mock or 10 μM DEX for 2h. For CK treatment, 5-day-old seedlings were transferred to medium containing 10 μM 6-benzylaminopurine (6-BAP; Duchefa) from a 10 mM DMSO stock solution. All samples were ground in liquid nitrogen and RNA was extracted using RNA isolation protocol for non-fiberous tissue by the RNA Tissue Miniprep System (Promega). cDNA synthesis was done using 1μg of total RNA with the qScriptTM cDNA Supermix kit (Quanta BioSciences). The qRT-PCR primers were designed by Universal Probe Library Design Center (Roche) (Table S3). The qRT-PCR was performed using UBC and EEF as reference genes on a Roche Lightcycler 480 device (Roche Molecular Systems Inc.) with SYBR Green I Master kit (Roche). The gene expression analysis was done using qBase v3.2 software (Biogazelle, Zwijnaarde, Belgium - www.qbaseplus.com).
DNA extraction and genotyping
Genomic DNA was isolated using the CTAB extraction method. The T-DNA mutants (myb11/SALK077068 and myb111/GK291D01) were genotyped using PCR based method (Table S3). The myb12-1f mutant (Mehrtens et al., 2005) was genotyped using cleaved amplified polymorphic sequence (CAPS). An amplicon of 547 bp was amplified (using primers described in Table S3), and was cut by using HphI restriction. The wild type allele was cut into two bands of 399 bp and 148 bp, while the mutant remained uncut.
Tobacco infiltration and co-immunoprecipitation
Nicotiana benthamiana leaves (3 to 4 week old) were infiltrated with a mixture of 4 C58C1Rif (pMP90) Agrobacterium strains containing the following constructs: p35S::P19, p35S::TMO5:3xHA, p35S::LL1:3xHA and pUBI10-XVE-p35Sminimal:MYB12:GFP. Leaves were harvested and immediately snap frozen by immersion in liquid nitrogen and stored at -70°C until further processing. Frozen leaves were ground with pestle and mortar until a fine powder was obtained. The ground tissue was then dissolved in extraction buffer (50mM Tris – 300mM NaCl – 5mM DTT – 1mM PMSF – pH 7.4 supplemented with proteinase inhibitor tablets 1 per 50ml of buffer) and sonicated (3x15sec on, 3x 15sec off). For each gram of plant material, 3ml of extraction buffer was used. After sonication the sample was kept on ice for 15 min and centrifuged for 2x15 min at 20.000 rcf. 100μl of anti-GFP magnetic beads (Chromotek – catalogue number: gtm-20) were added to the supernatant and incubated for 2h at 4°C on a rotating wheel. After incubation, the beads were removed from the sample and thoroughly washed with extraction buffer. Elution was performed by adding 25μl boiling sample buffer and boiling for another 10 min. Upon SDS-PAGE and Western blotting, detection was performed with 1/5000 anti-HA-HRP antibodies (Abcam, catalogue number ab1190).
Yeast-2-Hybrid (Y2H) analysis
The MYB12, TMO5 and LHW coding sequences were cloned into pDEST22 (Prey: GAL4AD-x Yeast selection marker: TRP1) and pDEST32 (Bait: GAL4DB-y Yeast selection marker: LEU2). These plasmids were transformed into Saccharomyces cerevisiae strain AH109 (Clontech). At least 3 independent yeast transformants were checked for each pairwise interaction according to (Cuellar et al., 2013) with minor modifications: the protein-protein interactions were validated with undiluted overnight yeast culture droplets manually pipetted on selective SD Base-Leu/-Trp/-His and grown for 3-4 days at 30°C before imaging.
Knock sideways
The knock sideways (KSD) assay was performed as described previously (Winkler et al., 2021). Briefly, N. benthamiana leaves were transiently co-transformed with the constructs p35S::TMO5-EGFP-FKBP, p35S::MITO-FRB, and pG1090::XVE>>MYB12-TagBFP2 or p35S::TagBFP2 as a negative control. After 2 days, the transformed leaves were infiltrated with 1 μM rapamycin or H2O mock control and images were acquired 2-4 h thereafter on a Leica SP8X or Zeiss LSM880 confocal microscope in line sequential scanning mode. The pG1090::XVE>>MYB12-TagBFP2 construct was originally intended for estradiol-inducible expression, but turned out very leaky in expression in the N. benthamiana system and was thus used for constitutive expression instead.
Results
A mutant screen identifies modulators of TMO5/LHW activity
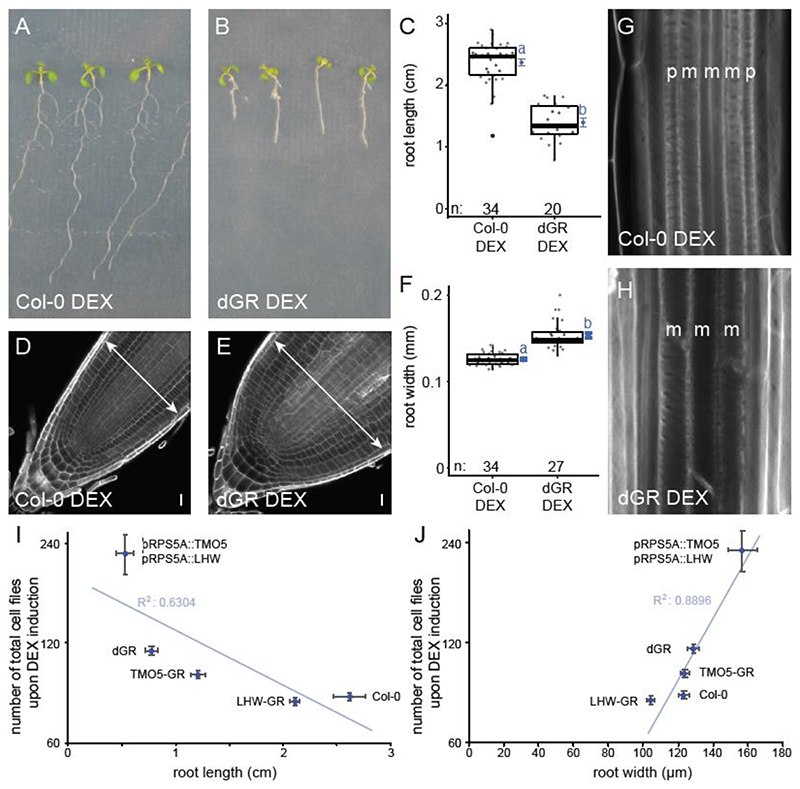
In order to identify novel regulators of TMO5/LHW activity leading to vascular proliferation via control of oriented cell divisions, we designed an EMS-based forward genetic screen in the previously described dexamethasone (DEX)-inducible pRPS5A::TMO5:GR x pRPS5A::LHW:GR double misexpression line (double GR or dGR) in Col-0 background (Smet et al., 2019). Upon exogenous DEX treatment, root apical meristem width is increased in this line due to the ectopic periclinal and radial cell divisions (De Rybel et al., 2013), protoxylem differentiation is inhibited due to increased cytokinin levels (De Rybel et al., 2014) and additionally, primary root length is reduced (De Rybel et al., 2013) (Fig. 1A-H, Table S1). We reasoned those mutations in positive/negative regulators of the TMO5/LHW pathway would suppress/enhance these dGR phenotypes. Although the TMO5/LHW activity was previously shown by a detailed quantification of the vascular cell file number (Arents et al., 2022; Wendrich et al., 2020; Yang et al., 2021), such experiments are labour intensive and require fixed samples, making them incompatible with high-throughput screening. We thus first evaluated whether root length and meristem width could serve as reliable read-outs for TMO5/LHW activity and hence cell proliferation capacity by plotting the root length or root width parameters against the total number of quantified cell files in multiple transgenic lines with increasing levels of TMO5/LHW heterodimer activity (Col-0, pRPS5A::LHW:GR, pRPS5A::TMO5:GR, the inducible dGR line and a constitutive double TMO5/LHW misexpression line). We observed a clear inverse correlation between root length and TMO5/LHW activity and a positive correlation between root width and TMO5/LHW activity (Fig. 1I-J, Table S1). These results suggest that root length and width can serve as reliable proxies for the number of cell file number and thus TMO5/LHW activity.
Figure 1. Root phenotype of Col-0 and the dGR line on induced (DEX) media.
(A-B) 1- week-old Col-0 (A) and dGR (B) plants grown on 10 μM DEX. (C) Boxplot of root length of Col-0 and dGR plants grown for 5 days on 10 μM DEX. (D-E) Col-0 (D) and dGR (E) root tips grown on 10 μM DEX. Arrows are highlighting root meristem width. (F) Boxplot of root width of Col-0 and dGR plants grown on 10 μM DEX. Black lines indicates the median and grey boxes indicate data ranges. The mean and its precision are plotted as the blue rhombus with blue SE bars. Blue lower-case letters (compact letter display) at these means in C, F not sharing a letter indicate significantly different groups as determined by one-way ANOVA with post-hoc Tukey testing at 5% significance level. The n marks the number of datapoints for each sample. (G-H) The vascular differentiation phenotype of Col-0 (G) and dGR (H) plants grown on 10 μM DEX. The p and m indicate protoxylem and metaxylem strands respectively. Root width of Col-0 and dGR plants grown for 5 days on 10 μM DEX (n ≥ 20). (I-J) 1-week old seedlings grown on 10 μM DEX (n ≥ 10), were used to plot the number of total cell files in the root meristem against the root length (I) or root width (J). Error bars indicate standard error. Scale bars in D-E indicate 10 μm.
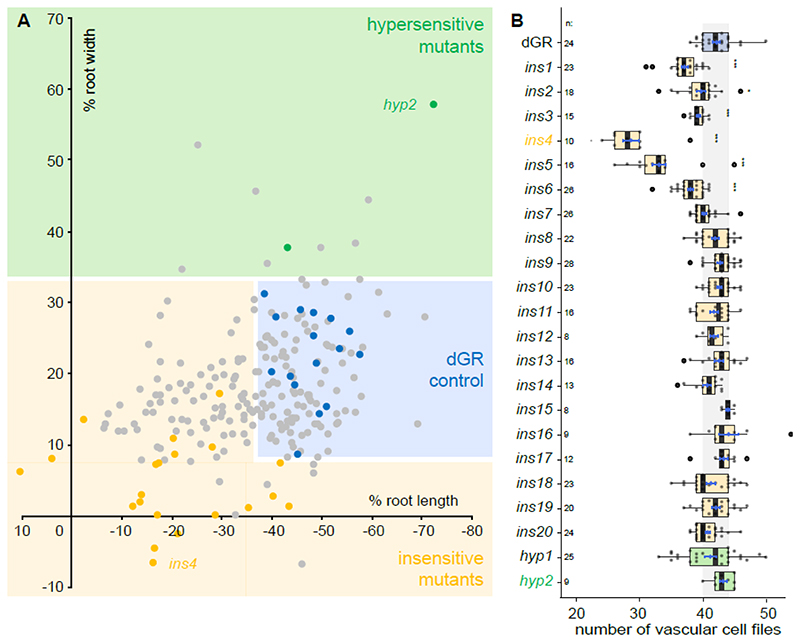
Having established the screening strategy, we performed EMS mutagenesis of dGR seeds and screened 228 pools of EMS mutagenized M2 dGR seedlings for alterations in root length upon DEX induction. This first round of selection yielded 310 candidate mutants from 110 pools, of which 260 produced viable M3 seeds. In total, 50 albino plants were observed among these 228 pools of mutants, suggesting that the EMS mutagenesis was successful (Micol-Ponce et al., 2014). In the M3 generation, we quantified both root length and root meristem width of the 260 candidate mutants (Fig. 2A), resulting in 20 validated mutants with reduced responses (insensitive 1-20, ins1-20) and 2 mutants showing hypersensitive responses (hypersensitive 1-2, hyp1-2) (Fig. S1-3, Table S1). The insensitive mutants are defined as not showing a change in root length and/or root meristem width upon TMO5/LHW induction (dGR on DEX) compared to the non-induced control (dGR on mock). The hypersensitive mutants are defined as having an even more pronounced change in root length and/or root meristem width upon TMO5/LHW induction (dGR on DEX) compared to the non-induced control (dGR on mock). We next performed a detailed quantification of the vascular cell file number as the read-out of TMO5/LHW activity used previously (De Rybel et al., 2014; De Rybel et al., 2013; Ohashi-Ito and Bergmann, 2007; Ohashi-Ito et al., 2013; Ohashi-Ito et al., 2014). Notably, 5 insensitive mutants already showed a significantly reduced number of vascular cell files in mock conditions compared to the non-induced control (dGR) (Fig. 2B, Table S1), suggesting that these mutants might inherently have differential TMO5/LHW activity and further confirming that our multi-step screening procedure using root length and width as proxies was successful. A segregation analysis further showed that the observed phenotypes in ins2 and ins7 could not be explained by a recessive mutation at a single locus (Table S2). These mutants were therefore excluded from further analysis. We finally focussed our attention on the mutants with the most pronounced phenotype in each category: ins4 and hyp2 (Fig. 2B, Fig. S1, Table S1), and mapped the causal mutations by next generation sequencing.
Figure 2. Overview of obtained EMS mutants.
(A) A total overview of all 260 primary selected EMS mutants and several parental dGR lines, plotted for their sensitivity of root length changes against the sensitivity of root width changes relative to mock treatment of the same genotype. Thus, the percentage represent mock vs DEX treatment of each mutant or parental dGR line. Data from the EMS screening was used. Dots in the blue box represent EMS mutants behaving similar to parental dGR control and dots in the yellow box represent mutants that behave insensitive to dGR response (significant longer and/or thinner roots) compared to the dGR parental line, while in the green box mutants behave hypersensitive to dGR induction. Yellow and green dots represent the 22 selected EMS mutants, the yellow dots represent the ins mutants and green dots the hyp mutants. Grey dots represent other EMS mutants selected from the primary screen and blue dots represent non-mutagenized parental dGR. Each data point was compared with a dGR parental control grown on the same plate. This internal control explains why some non-significant EMS mutant lines (grey dots) are further away from the parental dGR (blue dots) then some significant EMS mutant lines (yellow or green dots). For each data point the average was used from 10 biological repeats. (B) Overview vascular cell files phenotype in candidate mutants. Counts of vascular cell files in the root meristem of 1-week-old dGR (blue), ins (yellow) and hyp (green) seedlings. All genotypes described contain the dGR constructs. Black lines indicate the median and boxes indicate data ranges. The mean and its precision are plotted as the blue rhombus with blue SE bars. The n is above the genotype labels indicates number of datapoints for each genotype. Pairwise comparisons with Dunnett method p-value adjustment, was performed to evaluate significant differences between a mutant’s and dGR number of vascular cell files (Table S1). Significances asterisks: * = p-value < 0.05; ** = p-value < 0.01; *** = p-value < 0.001.
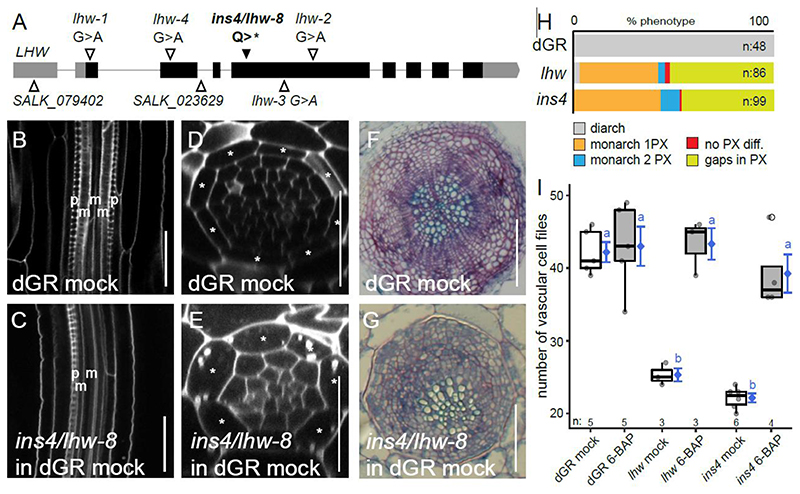
A strong lhw allele is causal to the ins4 phenotype
The insensitive ins4 mutant (in dGR background) showed a strong reduction in the number of vascular cell files under mock condition and a repression of the increased root thickness upon DEX treatment (Fig. 2B, Fig. S1, Table S1). Indeed, upon TMO5/LHW induction in ins4, the number of vascular cell files increased compared to mock, but the response of ins4 on DEX did not differ from dGR on mock (Fig S4A-B, Table S1). Sequencing and SHORE map analysis (Schneeberger et al., 2009) focusing on the reduced vascular cell files phenotype under mock conditions revealed that ins4 carried a premature stop codon in LHW (Fig. 3A) which was confirmed using Sanger sequencing. Similar to the published lhw mutant alleles (De Rybel et al., 2013; Ohashi-Ito and Bergmann, 2007; Ohashi-Ito et al., 2013; Parizot et al., 2008), the ins4 mutant showed a monarch vascular architecture in the primary root meristem, resulting in an off-centre xylem bundle during secondary growth (Fig. 3B-H, Table S1). The number of vascular cell files could also be rescued by exogenous cytokinin application (Fig. 3I, Table S1) as was shown before to bypass the TMO5/LHW dependent cytokinin biosynthesis (De Rybel et al., 2014). It is counter intuitive that dGR induction could not rescue the ins4 mutant while ectopic cytokinin treatment could, as both treatments converge on increased CK levels. Thus, we explored LOG4 expression levels upon dGR induction in the ins4 mutant background and found these were not changed to the extent seen in the dGR control (Fig. S4C). This suggests that within the ins4 mutant background, dGR is incapable of increasing cytokinin levels, explaining the discrepancy between the dGR induction and cytokinin treatment experiments in ins4. It is however unclear whether this is due to the truncated LHW protein or caused by another independent mutation. Taken together, the mapping and phenotypic characterization show that ins4 is a novel, strong lhw allele. We thus termed ins4 as lhw-8. Although ins4/lhw-8 itself does not provide new insight into the regulation of TMO5/LHW activity, it further confirms that our screening and mapping set-up was successful to uncover causative EMS mutations.
Figure 3. The insensitive mutant ins4 is a novel lhw allele.
(A) Alleles of lhw mutants with ins4/lhw-8 having a point mutation, resulting in a premature stop codon in exon (black bar) 4 of LHW. (B-C) Longitudinal view of the root vascular tissue shown for dGR (B) and ins4/lhw-8 (dGR) (C). (D-E) Optical cross-section through the root meristem of dGR (D) and ins4/lhw-8 (dGR) (E) show smaller vascular cylinder for ins4/lhw-8 (dGR). (F-G) Secondary growth phenotype can be observed in sections of dGR (F) and ins4/lhw-8 (dGR) (G) through the hypocotyl of 3-week-old seedlings. Scale bars in B-E are 25 μm and in F-G 100 μm. (H) The frequency of xylem differentiation (diff.) phenotype plotted for dGR, lhw and ins4 (dGR). The asterisks mark the endodermis cells in D-E, ‘p’ an ‘m’ represent protoxylem and metaxylem cell files in B-C. (I) The number of vascular cell files of 1-week-old seedlings treated with cytokinin (6-BAP). Black lines indicate mean values and grey/white boxes indicate data ranges. n marks the number of datapoints for each sample. The mean and its precision are plotted as the blue rhombus with blue SE bars. Count data samples were compared pairwise based estimated means of a generalized linear model with log link function. Common blue lower-case letters (compact letter display) at these means indicate non-significantly different groups as determined by the pairwise comparisons (Table S1).
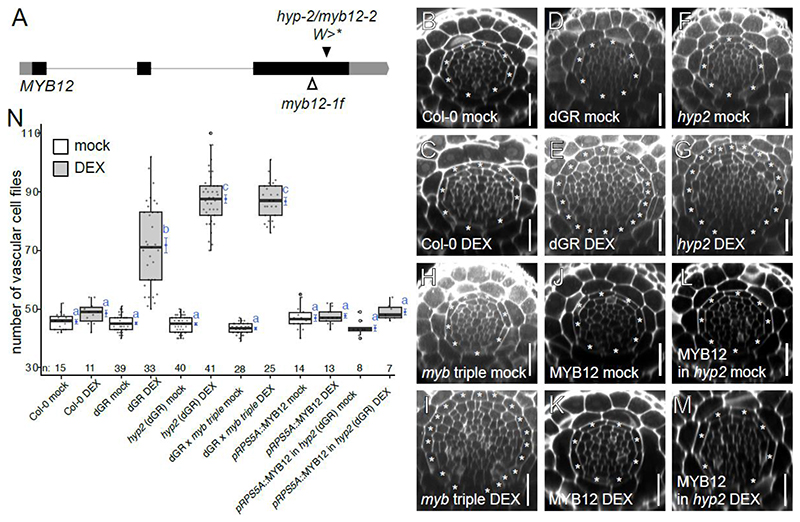
hyp2 is a novel myb12 allele
At the other side of the selected mutant spectrum, the recessive hyp2 mutant showed no clear phenotype under normal growth conditions, but a strong hypersensitive response upon DEX treatment (Fig. 2B, Fig. 4N, Fig. S1, Fig. S5-7, Table S1). SHORE map analysis (Schneeberger et al., 2009) identified an early stop codon in the gene encoding the R2R3 transcription factor MYB12 (Fig. 4A). To confirm the causality of the MYB12 mutation for the observed dGR hypersensitive phenotype, we first crossed the previously published myb11 myb12-1f myb111 triple mutant (Stracke et al., 2007) into our dGR parental line. The triple mutant was chosen as at least the closely related myb111 is also dGR inducible (Figure S8C) (Smet et al., 2019). A hypersensitive response comparable to hyp2 was detected in the myb11 myb12-1f myb111 dGR mutant (Fig. 4B-I, N, Table S1). This triple mutant also did not show an aberrant phenotype under mock conditions in the Col-0 control background (Fig. 4B, H, N, Table S1). Collectively, this data also suggests that MYB12 acts as a repressor of ectopically induced TMO5/LHW vascular proliferation. To further test this hypothesis, we introduced a construct driving the MYB12 coding sequence from the strong meristematic RPS5A promoter (Weijers et al., 2001) in the hyp2 mutant background. pRPS5A::MYB12 lines with increased MYB12 levels showed mild repression of the number of vascular cell files already in mock conditions (Fig. S8A-B, Table S1). Upon DEX treatment, the pRPS5A::MYB12 construct strongly repressed the TMO5/LHW-induced vascular cell proliferation (Fig. 4J-N, Table S1). Taken together, hyp2 is a novel mutant allele of MYB12, which we designated as myb12-2. Our initial results hint towards a new function for this TF and suggest that MYB12 might act as a negative regulator of the TMO5/LHW pathway.
Figure 4. The hyp2 is hypersensitive to dGR response and MYB12 acts as a repressor for TMO5/LHW activity.
(A) MYB12 gene marked with known myb12-1f transposon insertion site and hyp2/myb12-2 point mutation site, which results in premature stop codon. (B-M) Representative root meristem cross-sections of Col-0 on mock (B), Col-0 on DEX (C), dGR on mock (D), dGR on DEX (E), hyp2/myb12-2 (dGR) on mock (F), hyp2/myb12-2 (dGR) on DEX (G), myb11 myb12-1f myb111 triple mutant (referred to as myb triple) (dGR) on mock (H), myb triple (dGR) on DEX (I), pRPS5A::MYB12 on mock (J), pRPS5A::MYB12 on DEX (K), pRPS5A::MYB12 (in myb12-2 (dGR)) line on mock (L) and on DEX (M). The asterisks mark the endodermis cells and counted vascular cell file number are within this cell type. Scale bars are 25 μm. (N) Boxplot plotting the vascular cell file number. Black lines indicates mean values and grey/white boxes indicate data ranges. n marks the number of datapoints for each sample. The mean and it precision are plotted as the blue rhombus with blue SE bars. Count data samples were compared pairwise based estimated means of a generalized linear model with log link function. No shared blue lower-case letters (compact letter display) at these means indicate significantly different groups as determined by the pairwise comparisons (Table S1).
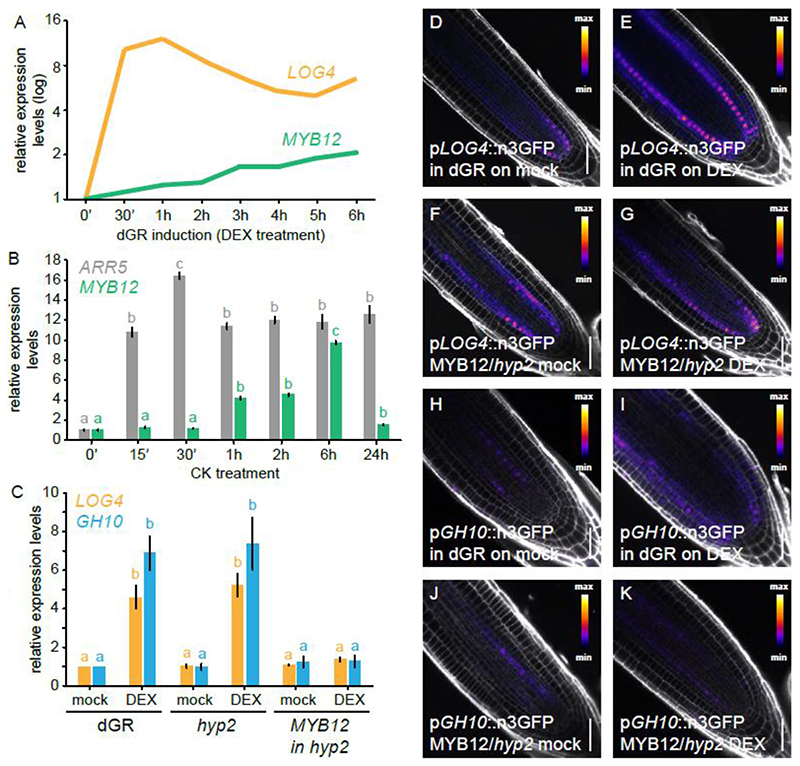
We previously found that MYB12 is transcriptionally upregulated upon TMO5/LHW induction in the dGR line (Smet et al., 2019) (Fig. 5A). We thus introduced the previously described MYB12 transcriptional reporter (Stracke et al., 2007) into the dGR line and observed increased activity of the MYB12 promoter in the root differentiation zone upon dGR induction (Fig. S9A-B). Similar increase in expression levels upon TMO5/LHW induction in the dGR background was observed in newly generated pMYB12::nGFP/GUS and pMYB12::gMYB12:sYFP reporter lines based on the full genomic fragment of MYB12 including the introns and a longer promoter sequence (Fig. S9C-F). Given the slow induction kinetics compared to direct TMO5/LHW target genes such as LOG4 (Fig. 5A) (De Rybel et al., 2014; Ohashi-Ito et al., 2014), we hypothesized that the induction of MYB12 is likely indirect and possibly triggered by cytokinin signalling downstream of TMO5/LHW (De Rybel et al., 2014; Ohashi-Ito et al., 2014). Indeed, we found MYB12 to be cytokinin inducible by qRT-PCR analysis (Fig. 5B, Table S1), confirming previous reports (Brenner and Schmulling, 2012). Analysis of the newly generated pMYB12::nGFP/GUS reporter line further revealed that ectopic cytokinin treatment results in higher activity of the MYB12 promoter (Fig. S9G-I). These results suggest that MYB12 might be part of a negative feedback loop where TMO5/LHW, via increased cytokinin signalling, activates its own repressor to modulate vascular proliferation rates.
Figure 5. MYB12 acts downstream of TMO5/LHW-mediated CK production and represses TMO5/LHW targets.
(A) Relative expression levels LOG4 and MYB12 genes over different DEX treatment durations on dGR line derived from microarray data described in Smet et al 2019 (Smet et al., 2019), with 0h DEX expression levels set to 1. (B) Relative expression levels of the CK-inducible A-type ARR5 and MYB12 in a time course experiment following cytokinin treatment. (C) Relative expression of LOG4 and GH10 in 5-days-old seedlings of dGR, hyp2/myb12-2 (dGR) and pRPS5A::MYB12 in hyp2/myb12-2 (dGR) where TMO5/LHW activity was induced for 2h on mock or DEX. Common lower-case letters in B, C indicate non-significantly different groups as determined by respectively one-way and two-way ANOVA with post-hoc Tukey testing. Error bars are standard errors. (D-G) Expression of pLOG4::n3GFP in F1 5-days-old seedlings in dGR background (D-E) and pRPS5A::MYB12 in hyp2/myb12-2 (dGR) (F-G) background after 24h on mock (D,F) or DEX (E,G). Expression of pGH10::n3GFP in F1 5-days-old seedlings in dGR background (H-I) and pRPS5A::MYB12 in hyp2/myb12-2 (dGR) background (J-K) after 24h on mock (H,J) or DEX (I,K). Scale bars are 50 μm.
MYB12 represses TMO5/LHW transcriptional activity
One possible way how MYB12 could repress the TMO5/LHW activity downstream of the cytokinin response would be to alter the cytokinin response itself. To test this hypothesis, we analysed the inhibition of root length caused by increasing concentrations of exogenously applied cytokinin in myb12 mutants. No major differences in cytokinin sensitivity were observed between either myb12 allele and their respective control lines under mock conditions (Col-0 for myb12-1f and dGR for hyp2/myb12-2) (Fig. S11, Table S1), suggesting the repression of TMO5/LHW activity does not act at the level of cytokinin signalling or perception. Next, we tested possible repression at the level of the activity of the TMO5/LHW heterodimer itself by analysing the expression levels of direct TMO5/LHW target genes in the hyp2/myb12-2 and pRPS5A::MYB12 hyp2/myb12-2 dGR lines in comparison to the dGR control. The expression levels of the direct target genes LOG4 and GH10 can be used as molecular read-out of TMO5/LHW activity (De Rybel et al., 2014; Ohashi-Ito et al., 2014; Vera-Sirera et al., 2015). Upon DEX treatment, relative expression levels of LOG4 and GH10 were induced in control (dGR) and hyp2/myb12-2 in dGR backgrounds (Fig. 5C, Table S1). In the pRPS5A::MYB12 hyp2/myb12-2 dGR line, however, no induction in LOG4 and GH10 expression was observed (Fig. 5C, Table S1), suggesting that MYB12 might directly inhibit TMO5/LHW activity. To verify these results, we next introduced the transcriptional reporter of LOG4 (De Rybel et al., 2014) and a newly generated reporter for GH10 into the pRPS5A::MYB12 hyp2/myb12-2 dGR line and the parental dGR line as control. Both the pLOG4::n3GFP and pGH10::n3GFP transcriptional reporters showed a clear induction in expression strength and ectopic expression upon DEX treatment in dGR/+ background compared to a mock DMSO treatment (Fig. 5D-E, H-I). This induction was repressed in the pRPS5A::MYB12/+ hyp2/+ dGR/+ background (Fig. 5F-G, J-K); confirming the qRT-PCR results (Fig. 5C, Table S1). Taken together, these results suggest that MYB12 represses TMO5/LHW activity by inhibiting direct target gene expression. Importantly, MYB12 does not contain a characteristic EAR motif associated with transcriptional repressors (Kagale and Rozwadowski, 2011; Liu et al., 2015) and directly activates transcription of the CHS and FLS genes (Mehrtens et al., 2005). This shows that MYB12 is thus not a typical transcriptional repressor, but represses TMO5/LHW transcriptional activity in another way.
MYB12 non-competitively binds to TMO5
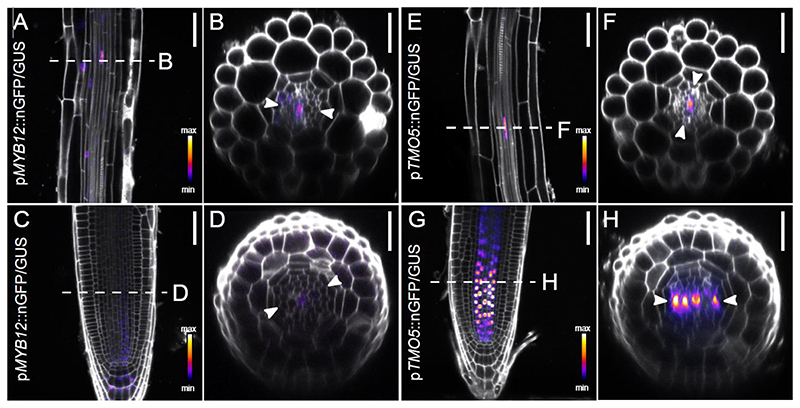
TMO5/LHW activity is known to be repressed by the SACL bHLH proteins, which compete with TMO5 for binding to LHW and thus reduce the amount of active TMO5/LHW dimers (Katayama et al., 2015; Vera-Sirera et al., 2015). Given the well documented interactions between MYB and bHLH TFs (Carretero-Paulet et al., 2010; Cui et al., 2021; Feller et al., 2011; Zhao et al., 2008), we hypothesized that MYB12 function might involve direct binding to the TMO5/LHW complex (Katayama et al., 2015; Vera-Sirera et al., 2015). Firstly, if MYB12 were to bind the TMO5/LHW complex, it would need to be present in the same cells. As single-cell transcriptomics (Wendrich et al., 2020) suggested a broader MYB12 meristematic expression domain than revealed by the existing MYB12 transcriptional reporter (Stracke et al., 2007) (Fig. S9a, Fig. S10A), we speculated that additional regulatory elements might be present in the MYB12 coding region. To this end, we more closely examined our newly generated reporter lines (Fig. 6A-D, Fig. S10B). In agreement with previous findings (Stracke et al., 2007; Struk et al., 2022), the expression was the strongest in most cells from the elongation zone onwards, including xylem cells, and in the lateral root cap. Nonetheless, our genomic fusion containing introns and a longer promoter region revealed additional MYB12 expression also in epidermis, cortex, and importantly, xylem cells of the meristematic zone (Fig. S10B), confirming the recently published single cell transcriptomic data (Wendrich et al, 2020) (Fig. S10A). It is thus conceivable that MYB12 could directly interact with the TMO5/LHW complex, expressed in xylem cells throughout the root meristem as driven from their endogenous promoters (Fig. 6, S9-10) (De Rybel et al., 2013).
Figure 6. MYB12 and TMO5 have overlapping expression patterns in root tissues.
(A-B) Expression pattern of 1-week-old pMYB12::nGFP/GUS in root elongation zone and (C-D) root meristem. (E-F) Expression pattern of 1-week-old pTMO5::nGFP/GUS in root elongation zone and (E-F) and (C-D) root meristem. The dashed line marks location optical cross section was made. Arrowheads indicate xylem axis in optical cross sections. Scale bars are 50 μm (A,C,E,G) and 25 μm (B,D,H,F).
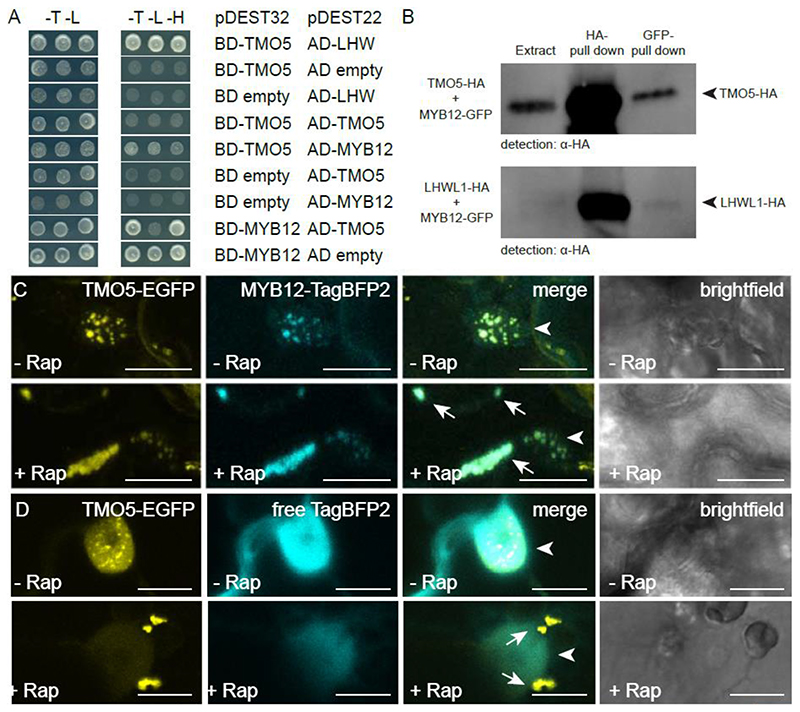
We therefore next tested the capacity of MYB12 to directly interact with TMO5 and/or LHW. Yeast-2-Hybrid (Y2H) analysis showed that MYB12 is able to bind to TMO5 (Fig. 7A). Binding of MYB12 to LHW could not be evaluated due to auto-activation of LHW-BD and MYB12-BD in the yeast system (Fig. S12). This could be caused by the innate transcriptional activity of LHW and MYB12. We next performed co-immunoprecipitation and could confirm the interaction between TMO5-HA and MYB12-GFP. Additionally, a putative interaction between MYB12-GFP and LHW-LIKE1-HA was found (Fig. 7B, S13). To provide additional confirmation of these interactions in planta using an independent system, we took advantage of the recently developed rapamycin-dependent knock sideways assay in transiently transformed N. benthamiana leaves (Winkler et al., 2021). This assay is based on the ability of FKBP and FRB protein domains to solely dimerize in presence of the drug rapamycin (Belshaw et al., 1996). In control conditions, we observed that simultaneous infiltration of plasmids carrying TMO5-GFP-FKB, MYB12-TagBFP2 and a mitochondria-targeted FRB resulted in nuclear localization of the TMO5 and MYB12 fusions, as expected from transcription factors (Fig. 7C). In the presence of rapamycin, TMO5-GFP-FKBP bound to mito-FRB and delocalized to the mitochondria (Fig. 7D). Together with TMO5-GFP-FKBP, MYB12-TagBFP2, but not free TagBFP2, co-translocalized towards the mitochondria (Fig. 7C-D), indicating that TMO5 interacted with MYB12. Taken together, these experiments show that TMO5, and perhaps LHW homologs, can directly interact in vivo and in planta with MYB12. This corresponds with previous findings where MYB transcription factors are part of the bHLH/MYB/WD40 complex (Ramsay and Glover, 2005).
Figure 7. MYB12 binds to TMO5 in yeast and tobacco leaves.
(A) Yeast-two-hybrid assay with pDEST22 (prey) or pDEST32 (bait) constructs containing fusion proteins of the MYB12 and TMO5 coupled to respectively, the activator (AD) or binding domain (BD). The yeast colonies are representative colonies of 24 independent yeast transformants per prey and bait pair. The empty pDEST22 or empty pDEST32 plasmids were used to check for auto-activation. Transformed yeast grown on the selective -Trp/-Leu (-T -L) medium and interaction verifying - Trp/-Leu/-His (-T -L -H) medium. (B) co-immunoprecipitation of TMO5-HA and LHW-LIKE1-HA with MYB12-GFP (full blot is shown in Figure S13). (C-D) Knock-sideways with Mito-FRB, TMO5-EGFP-FKGP and MYB12-TagBFP2 (C) or free TagBFP2 (D) as control in absence or presence of rapamycin. Arrows indicate the aggregated mitochondria and arrowheads indicate the nucleus. (n ≥10 for C and n ≥20 for D) Scale bars are 20 μm in C and 10 μm in D.
Altogether, using forward genetics, we have identified the R2R3 MYB transcription factor MYB12 as a novel regulator of the TMO5/LHW pathway during root vascular proliferation. MYB12 directly interacts with TMO5 and represses the activity of the TMO5/LHW complex at the level of direct target gene expression. MYB12 itself is a downstream target gene of the TMO5/LHW pathway, thus constituting a negative feedback loop which contributes to fine tuning the activity of the TMO5/LHW complex during vascular development in the root meristem.
Discussion
The patterning and proliferation of the vascular bundle during primary root growth relies on a complex regulatory network of transcriptional, hormonal and other signals (De Rybel et al., 2016). The key heterodimeric bHLH transcription factor complex, TMO5/LHW, promotes cytokinin biosynthesis via promoting the expression of LOG3, LOG4 and BGLU44 in the xylem cells (De Rybel et al., 2014; Ohashi-Ito et al., 2014; Yang et al., 2021). This locally produced cytokinin is thought to act as a mobile signal that coordinates the radial growth and correct patterning of the vascular bundle (Wybouw and De Rybel, 2019). In this study, we have taken a forward genetic approach to find new regulators of the TMO5/LHW pathway and discovered a novel function of the previously described transcription factor MYB12. Our data revealed that myb12 mutants are hypersensitive to the gain-of-function phenotypes caused by TMO5/LHW misexpression, while MYB12 misexpression represses vascular proliferation by inhibiting the transcriptional activation of direct TMO5/LHW targets genes. Moreover, MYB12 is transcriptionally activated by the cytokinin response downstream of TMO5/LHW, and MYB12 directly interacts with TMO5 and possibly LHW homologs. All these findings indicate that MYB12 acts as a repressor of the TMO5/LHW transcriptional pathway, while at the same time being its downstream target. Hence, we have found a novel negative feedback loop regulating the TMO5/LHW transcriptional network via the action of MYB12.
This negative feedback loop is reminiscent of the previously described regulation of the TMO5/LHW pathway by the SACL genes. Nonetheless, there are several key differences between the MYB12- and SACL-mediated negative feedback loops. Firstly, MYB12 appears to be a secondary TMO5/LHW target induced indirectly by the downstream cytokinin response, while the SACL genes are direct targets of TMO5/LHW (Katayama et al., 2015; Vera-Sirera et al., 2015). This would suggest that the MYB12-mediated negative feedback is slower in comparison to the SACL loop, which might be important for spatiotemporal fine tuning TMO5/LHW activity. Furthermore, the cytokinin response levels are affected by numerous factors other than TMO5/LHW (Kieber and Schaller, 2018). Thus, the cytokinin-inducible MYB12 can, unlike the SACL proteins, help optimize vascular proliferation rates by integrating the TMO5/LHW activity with other developmental signals. In support of the SACL- and MYB12-mediated negative feedback loops acting on different spatiotemporal scales, SACL and MYB12 have very distinct expression patterns. SACLs are co-expressed with TMO5 and LHW in all xylem cells in the root meristem zone (Vera-Sirera et al., 2015). MYB12 is most prominently expressed in older root tissues from the differentiation zone onwards and in the late meristematic xylem cells, consistent with providing slower and more indirect feedback. However, the SACL and MYB12 regulatory loops do not seem to be mutual exclusive, as myb12 mutants are hypersensitive towards increased TMO5/LHW activity in the root meristem. Unfortunately, despite clear inhibitory effects on vascular proliferation in both SACL and MYB12 gain-of-function lines, a lack of prominent aberrant phenotypes in the respective loss-of-function mutants makes it difficult to dissect the exact function of these genes during vascular development. This further emphasizes the pronounced genetic redundancy operating in plant development, especially during the control of such vital processes like vascular tissue patterning.
We have shown that MYB12 directly interacts with TMO5, and likely also LHW homologs and inhibits the transcriptional activation of direct TMO5/LHW target genes. Nonetheless, the exact molecular mechanism of MYB12 action remains unclear. MYB12 does not contain an EAR or TLLLFR motif typical for MYB TF repressors (Kagale and Rozwadowski, 2011; Ma and Constabel, 2019). Additionally, MYB12 lacks the bHLH-binding motif present in other known bHLH-interacting MYB TFs (Wang and Chen, 2014; Zimmermann et al., 2004), and it functions as a bona fide transcriptional activator (Forkmann and Martens, 2001; Mehrtens et al., 2005).
Therefore, the MYB12-mediated inhibition of TMO5/LHW activity must depend on another molecular mechanism. In one scenario, MYB12 might act as a passive repressor by preventing TMO5/LHW interaction with DNA and/or recruitment of the RNA polymerase II complex (Kazan, 2006; Krogan and Long, 2009). Another and more likely possibility is that, rather than acting as a conventional repressor, MYB12 might redirect TMO5/LHW activity away from LOG4, GH10 and other genes involved in vascular proliferation, and contribute to activating different TMO5/LHW target genes instead. This explanation would fit best with the previously described function of MYB12 as a classical transcriptional activator of several genes in the flavonoid biosynthesis pathway (Forkmann and Martens, 2001; Mehrtens et al., 2005). Target gene specificity has previously been associated with the MYB TFs in heteromeric bHLH-MYB transcriptional complexes (Ramsay and Glover, 2005). TMO5-LIKE 1 (T5L1), a close homolog of TMO5, is able to promote ectopic xylem differentiation in addition to its role in promoting radial growth (Katayama et al., 2015); The same bHLH TF thus functions in two very different developmental processes that require the activation of completely different gene sets. It is conceivable that such alternative functionalities of bHLH TFs could be achieved by interactions with different MYBs. In such a scenario, the TMO5/LHW complex would recruit an unknown MYB TF to promote the expression of genes required for vascular proliferation, while the alternative recruitment of MYB12 would lead to the activation of different target genes. To take this speculation even further, the dual roles of MYB12 in flavonol biosynthesis (Forkmann and Martens, 2001; Mehrtens et al., 2005) and vascular proliferation (this study) could then be explained by alternative interactions with TMO5 and an unknown bHLH TF needed for MYB12-mediated induction of the CHS and FLS flavonol biosynthesis genes. Further investigations into the precise molecular mechanisms responsible for MYB12 as well as other related MYB TFs action will be needed to shed light on these intriguing open questions and hypotheses.
What is the biological meaning of the same transcription factor MYB12 being involved in flavonol biosynthesis as well as vascular proliferation is another open question arising from our study. Interestingly, the bHLH TF TRANSPARENT TESTA 8 (TT8) has been previously implied in flavonoid biosynthesis (Nesi et al., 2000) and trichome development (Maes et al., 2008), indicating that dual functions in different metabolic and developmental pathways might be a common feature of multiple transcription factors from different families (Zhang et al., 2017). This might reflect the need of certain metabolic changes for a specific developmental process. For example, trichomes are rich in biotic stress defence compounds which include flavonoids (Karabourniotis et al., 2020). Utilizing TT8 to control both trichome development and flavonoid biosynthesis might thus aid in coordinating the two processes. Likewise, the transition from vascular proliferation to differentiation might involve so far unappreciated metabolic changes in addition to the decline of TMO5/LHW activity, both hypothetically controlled by MYB12. Alternatively, dampening the TMO5/LHW pathway while promoting flavonoid biosynthesis might contribute to the balance between growth and defence processes. Different stresses often lead to increased reactive oxygen species levels, which can be mitigated by flavonoid antioxidant activity (Wang et al., 2016). In such conditions, attenuating the TMO5/LHW-mediated radial growth in favour of flavonoid biosynthesis by the increased MYB12 levels could be important for optimal resource allocation.
In summary, we have uncovered a novel role of the transcription factor MYB12 as a negative regulator of the TMO5/LHW pathway during vascular proliferation. The MYB12-mediated negative feedback loop is distinct from the modus operandi of the previously described SACL proteins in both molecular mechanism and spatiotemporal dynamics, showing that TMO5/LHW activity is being controlled using multiple distinct mechanisms. The full molecular details of MYB12 mode of action, as well as the biological meaning of its dual functions in vascular development and flavonoid biosynthesis, remain exciting challenges for future investigations. Our work establishes that a bona fide transcriptional activator can function as a repressor in a different transcriptional network. Furthermore, our results show that functional interactions between bHLH and MYB transcription factors are involved in multiple unrelated transcriptional networks, highlighting them as a powerful and possibly underappreciated developmental module.
Supplementary Material
Highlight.
A forward genetics screen identifies MYB12 as a repressor of TMO5/LHW activity which ensures optimal cell proliferation rates during root vascular development.
Footnotes
Author Contributions:
B.D.R. conceived the project; B.D.R., M.G., D.V.D., B.W. and H.E.A. designed experiments and analysed data; B.W., H.E.A., B.Y., J.N., W.S., M.V. and M.G. performed experiments; ; M.M., X.L. and I.D.C. contributed to revising the manuscript; B.D.R. and M.G. supervised the project and wrote the paper with input of all authors.
Conflict of interest:
The authors declare no conflict of interest.
Contributor Information
Brecht Wybouw, Email: brechtwybouw@helsmki.fi.
Helena E. Arents, Email: helena.arents@gmail.com.
Baojun Yang, Email: bayan@psb.vib-ugent.be.
Jonah Nolf, Email: jonol@psb.vib-ugent.be.
Wouter Smet, Email: wouter.smet@kaust.edu.sa.
Michael Vandorpe, Email: midor@psb.vib-ugent.be.
Max Minne, Email: max.minne@psb.vib-ugent.be.
Xiaopeng Luo, Email: xiaopeng.luo@psb.vib-ugent.be.
Inge De Clercq, Email: inge.declercq@psb.vib-ugent.be.
Daniёl Van Damme, Email: dadam@psb.vib-ugent.be.
Matouš Glanc, Email: magla@psb.vib-ugent.be.
Acknowledgements
The authors thank Ralf Stracke and Sofie Goormachtig for sharing the myb11myb12-1f,myb111 and myb12-f mutant seeds, respectively; Davy Opdenacker for help with the EMS mutagenesis and Frederik Coppens for help with the SHOREmap analysis. B.D.R. would like to thank T.G.A. and A.P.M. for helpful discussions completely unrelated to this work.
Funding
This work was funded by The Research Foundation - Flanders (FWO; Odysseus II G0D0515N and post-doc fellowship 1215820N); the Netherlands Organization for Scientific Research (NWO; VIDI 864.13.00); the European Research Council (ERC Starting Grant TORPEDO; 714055); the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 885979 “DIVISION BELL”; EMBO (long term fellowship ALTF 1005-2019); and ERC consolidator Grant T-REX; 682436 to DVD; ERC Starting Grant COSI; 949808 to I.D.C and the China Scholarship Council (PhD fellowship 201706910099) to X.L..
Data availability
All data supporting the findings of this study and respective statistics are available within the paper in Table S1. Materials and resources of this study are available from the corresponding author upon reasonable request.
References
- Appelhagen I, Jahns O, Bartelniewoehner L, Sagasser M, Weisshaar B, Stracke R. Leucoanthocyanidin Dioxygenase in Arabidopsis thaliana: characterization of mutant alleles and regulation by MYB-BHLH-TTG1 transcription factor complexes. Gene. 2011;484:61–68. doi: 10.1016/j.gene.2011.05.031. [DOI] [PubMed] [Google Scholar]
- Arents HE, Eswaran G, Glanc M, Mahonen AP, De Rybel B. Means to Quantify Vascular Cell File Numbers in Different Tissues. Methods in Molecular Biology. 2022;2382:155–179. doi: 10.1007/978-1-0716-1744-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw PJ, Ho SN, Crabtree GR, Schreiber SL. Controlling protein association and subcellular localization with a synthetic ligand that induces heterodimerization of proteins. Proceedings of the National Academy of Sciences U S A. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Schmulling T. Transcript profiling of cytokinin action in Arabidopsis roots and shoots discovers largely similar but also organ-specific responses. BMC Plant Biology. 2012;12:112. doi: 10.1186/1471-2229-12-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martinez-Garcia JF, Bilbao-Castro JR, Robertson DL. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiology. 2010;153:1398–1412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. the Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cuellar AP, Pauwels L, De Clercq R, Goossens A. Yeast two-hybrid analysis of jasmonate signaling proteins. Methods in Molecular Biology. 2013;1011:173–185. doi: 10.1007/978-1-62703-414-2_14. [DOI] [PubMed] [Google Scholar]
- Cui D, Zhao S, Xu H, Allan AC, Zhang X, Fan L, Chen L, Su J, Shu Q, Li K. The interaction of MYB, bHLH and WD40 transcription factors in red pear (Pyrus pyrifolia) peel. Plant Molecular Biology. 2021;106:407–417. doi: 10.1007/s11103-021-01160-w. [DOI] [PubMed] [Google Scholar]
- De Rybel B, Adibi M, Breda AS, Wendrich JR, Smit ME, Novak O, Yamaguchi N, Yoshida S, Van Isterdael G, Palovaara J, Nijsse B, et al. Plant development. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science. 2014;345:1255215. doi: 10.1126/science.1255215. [DOI] [PubMed] [Google Scholar]
- De Rybel B, Mahonen AP, Helariutta Y, Weijers D. Plant vascular development: from early specification to differentiation. Nature Reviews Molecular Cell Biology. 2016;17:30–40. doi: 10.1038/nrm.2015.6. [DOI] [PubMed] [Google Scholar]
- De Rybel B, Moller B, Yoshida S, Grabowicz I, Barbier de Reuille P, Boeren S, Smith RS, Borst JW, Weijers D. A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Developmental Cell. 2013;24:426–437. doi: 10.1016/j.devcel.2012.12.013. [DOI] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, Moller B, et al. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Current Biology. 2010;20:1697–1706. doi: 10.1016/j.cub.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Du H, Zhang L, Liu L, Tang XF, Yang WJ, Wu YM, Huang YB, Tang YX. Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry (Moscow) 2009;74:1–11. doi: 10.1134/s0006297909010015. [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends in Plant Science. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. the Plant Journal. 2011;66:94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- Forkmann G, Martens S. Metabolic engineering and applications of flavonoids. Current Opinion in Biotechnology. 2001;12:155–160. doi: 10.1016/s0958-1669(00)00192-0. [DOI] [PubMed] [Google Scholar]
- Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Molecular Biology. 1999;41:577–585. doi: 10.1023/a:1006319732410. [DOI] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K. EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics. 2011;6:141–146. doi: 10.4161/epi.6.2.13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabourniotis G, Liakopoulos G, Nikolopoulos D, Bresta P. Protective and defensive roles of non-glandular trichomes against multiple stresses: structure-function coordination. Journal of Forestry Research. 2020;31:1–12. [Google Scholar]
- Karimi M, Inze D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Katayama H, Iwamoto K, Kariya Y, Asakawa T, Kan T, Fukuda H, Ohashi-Ito K. A Negative Feedback Loop Controlling bHLH Complexes Is Involved in Vascular Cell Division and Differentiation in the Root Apical Meristem. Current Biology. 2015;25:3144–3150. doi: 10.1016/j.cub.2015.10.051. [DOI] [PubMed] [Google Scholar]
- Kazan K. Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends in Plant Science. 2006;11:109–112. doi: 10.1016/j.tplants.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE. Cytokinin signaling in plant development. Development. 2018;145 doi: 10.1242/dev.149344. [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Huelskamp M, Schiefelbein J. The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Developmental Biology. 2004;268:506–513. doi: 10.1016/j.ydbio.2003.12.037. [DOI] [PubMed] [Google Scholar]
- Krogan NT, Long JA. Why so repressed? Turning off transcription during plant growth and development. Current Opinion in Plant Biology. 2009;12:628–636. doi: 10.1016/j.pbi.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- Kuroha T, Tokunaga H, Kojima M, Ueda N, Ishida T, Nagawa S, Fukuda H, Sugimoto K, Sakakibara H. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell. 2009;21:3152–3169. doi: 10.1105/tpc.109.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/s0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M. Genetics and biochemistry of seed flavonoids. Annual Review of Plant Biology. 2006;57:405–430. doi: 10.1146/annurev.arplant.57.032905.105252. [DOI] [PubMed] [Google Scholar]
- Liu J, Osbourn A, Ma P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Molecular Plant. 2015;8:689–708. doi: 10.1016/j.molp.2015.03.012. [DOI] [PubMed] [Google Scholar]
- Ma D, Constabel CP. MYB Repressors as Regulators of Phenylpropanoid Metabolism in Plants. Trends in Plant Science. 2019;24:275–289. doi: 10.1016/j.tplants.2018.12.003. [DOI] [PubMed] [Google Scholar]
- Maes L, Inze D, Goossens A. Functional specialization of the TRANSPARENT TESTA GLABRA1 network allows differential hormonal control of laminal and marginal trichome initiation in Arabidopsis rosette leaves. Plant Physiology. 2008;148:1453–1464. doi: 10.1104/pp.108.125385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiology. 2005;138:1083–1096. doi: 10.1104/pp.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry part A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Micol-Ponce R, Aguilera V, Ponce MR. A genetic screen for suppressors of a hypomorphic allele of Arabidopsis ARGONAUTE1. Scientific Reports. 2014;4:5533. doi: 10.1038/srep05533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S, Roszak P, Sevilem I, Toyokura K, Blob B, Heo JO, Mellor N, Help-Rinta-Rahko H, Otero S, Smet W, Boekschoten M, et al. Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature. 2019;565:490–494. doi: 10.1038/s41586-018-0839-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata K, Kanei-Ishii C, Sasaki M, Hatanaka H, Nagadoi A, Enari M, Nakamura H, Nishimura Y, Ishii S, Sarai A. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nature Structural Molecular Biology. 1996;3:178–187. doi: 10.1038/nsb0296-178. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC. Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development. 2007;134:2959–2968. doi: 10.1242/dev.006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Fukuda H. Transcriptional networks regulating root vascular development. Current Opinion in Plant Biology. 2020;57:118–123. doi: 10.1016/j.pbi.2020.08.004. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Matsukawa M, Fukuda H. An atypical bHLH transcription factor regulates early xylem development downstream of auxin. Plant and Cell Physiology. 2013;54:398–405. doi: 10.1093/pcp/pct013. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Saegusa M, Iwamoto K, Oda Y, Katayama H, Kojima M, Sakakibara H, Fukuda H. A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Current Biology. 2014;24:2053–2058. doi: 10.1016/j.cub.2014.07.050. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell. 1991;67:483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- Otero S, Gildea I, Roszak P, Lu Y, Di Vittori V, Bourdon M, Kalmbach L, Blob B, Heo JO, Peruzzo F, Laux T, et al. A root phloem pole cell atlas reveals common transcriptional states in protophloem-adjacent cells. Nature Plants. 2022;8:954–970. doi: 10.1038/s41477-022-01178-y. [DOI] [PubMed] [Google Scholar]
- Parizot B, Laplaze L, Ricaud L, Boucheron-Dubuisson E, Bayle V, Bonke M, De Smet I, Poethig SR, Helariutta Y, Haseloff J, Chriqui D, et al. Diarch symmetry of the vascular bundle in Arabidopsis root encompasses the pericycle and is reflected in distich lateral root initiation. Plant Physiology. 2008;146:140–148. doi: 10.1104/pp.107.107870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. RStudio | Open source professional software for data science teams-RStudio. 2020. Available at: https://www.rstudio.com/RStudio.
- Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends in Plant Science. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jorgensen JE, Weigel D, Andersen SU. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nature Methods. 2009;6:550–551. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- Smet W, Sevilem I, de Luis Balaguer MA, Wybouw B, Mor E, Miyashima S, Blob B, Roszak P, Jacobs TB, Boekschoten M, Hooiveld G, et al. DOF2.1 Controls Cytokinin-Dependent Vascular Cell Proliferation Downstream of TMO5/LHW. Current Biology. 2019;29:520–529.:e526. doi: 10.1016/j.cub.2018.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SK, Kwak SH, Chang SC, Schiefelbein J, Lee MM. WEREWOLF and ENHANCER of GLABRA3 are interdependent regulators of the spatial expression pattern of GLABRA2 in Arabidopsis. Biochemical and Biophysical Research Communications. 2015;467:94–100. doi: 10.1016/j.bbrc.2015.09.115. [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. the Plant Journal. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Fernie AR, Weisshaar B. Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12-and MYB111-independent flavonol glycoside accumulation. New Phytologist. 2010;188:985–1000. doi: 10.1111/j.1469-8137.2010.03421.x. [DOI] [PubMed] [Google Scholar]
- Stracke R, Turgut-Kara N, Weisshaar B. The AtMYB12 activation domain maps to a short C-terminal region of the transcription factor. Zeitschrift für Naturforschung C. 2017;72:251–257. doi: 10.1515/znc-2016-0221. [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Current Opinion in Plant Biology. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Struk S, Braem L, Matthys C, Walton A, Vangheluwe N, Van Praet S, Jiang L, Baster P, De Cuyper C, Boyer FD, Stes E, et al. Transcriptional Analysis in the Arabidopsis Roots Reveals New Regulators that Link rac-GR24 Treatment with Changes in Flavonol Accumulation, Root Hair Elongation and Lateral Root Density. Plant and Cell Physiology. 2022;63:104–119. doi: 10.1093/pcp/pcab149. [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R, Kurata T, Wada T. Localization of ENHANCER OF TRY AND CPC1 protein in Arabidopsis root epidermis. Journal of Plant Physiology. 2017;214:48–52. doi: 10.1016/j.jplph.2017.04.001. [DOI] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthelemy J, Palauqui JC. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell. 2008;20:1494–1503. doi: 10.1105/tpc.107.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursache R, Andersen TG, Marhavy P, Geldner N. A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. the Plant Journal. 2018;93:399–412. doi: 10.1111/tpj.13784. [DOI] [PubMed] [Google Scholar]
- Vera-Sirera F, De Rybel B, Urbez C, Kouklas E, Pesquera M, Alvarez-Mahecha JC, Minguet EG, Tuominen H, Carbonell J, Borst JW, Weijers D, et al. A bHLH-Based Feedback Loop Restricts Vascular Cell Proliferation in Plants. Developmental Cell. 2015;35:432–443. doi: 10.1016/j.devcel.2015.10.022. [DOI] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science. 1997;277:1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- Wang F, Kong W, Wong G, Fu L, Peng R, Li Z, Yao Q. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Molecular Genetics and Genomics volume. 2016;291:1545–1559. doi: 10.1007/s00438-016-1203-2. [DOI] [PubMed] [Google Scholar]
- Wang S, Chen JG. Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Frontiers in Plant Science. 2014;5:133. doi: 10.3389/fpls.2014.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Hubbard L, Chang Y, Guo J, Schiefelbein J, Chen JG. Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biology. 2008;8:81. doi: 10.1186/1471-2229-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Franke-van Dijk M, Vencken RJ, Quint A, Hooykaas P, Offringa R. An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development. 2001;128:4289–4299. doi: 10.1242/dev.128.21.4289. [DOI] [PubMed] [Google Scholar]
- Wendrich JR, Yang B, Vandamme N, Verstaen K, Smet W, Van de Velde C, Minne M, Wybouw B, Mor E, Arents HE, Nolf J, et al. Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science. 2020;370 doi: 10.1126/science.aay4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J, Mylle E, De Meyer A, Pavie B, Merchie J, Grones P, Van Damme DL. Visualizing protein-protein interactions in plants by rapamycin-dependent delocalization. Plant Cell. 2021;33:1101–1117. doi: 10.1093/plcell/koab004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wybouw B, De Rybel B. Cytokinin-A Developing Story. Trends in Plant Science. 2019;24:177–185. doi: 10.1016/j.tplants.2018.10.012. [DOI] [PubMed] [Google Scholar]
- Xu W, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends in Plant Science. 2015;20:176–185. doi: 10.1016/j.tplants.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Xu W, Grain D, Le Gourrierec J, Harscoet E, Berger A, Jauvion V, Scagnelli A, Berger N, Bidzinski P, Kelemen Z, Salsac F, et al. Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytologist. 2013;198:59–70. doi: 10.1111/nph.12142. [DOI] [PubMed] [Google Scholar]
- Yang B, Minne M, Brunoni F, Plackova L, Petrik I, Sun Y, Nolf J, Smet W, Verstaen K, Wendrich JR, Eekhout T, et al. Non-cell autonomous and spatiotemporal signalling from a tissue organizer orchestrates root vascular development. Nature Plants. 2021;7:1485–1494. doi: 10.1038/s41477-021-01017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yperman K, Wang J, Eeckhout D, Winkler J, Vu LD, Vandorpe M, Grones P, Mylle E, Kraus M, Merceron R, Nolf J, et al. Molecular architecture of the endocytic TPLATE complex. Science Advances. 2021;7 doi: 10.1126/sciadv.abe7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ivanova A, Vandepoele K, Radomiljac J, Van de Velde J, Berkowitz O, Willems P, Xu Y, Ng S, Van Aken O, Duncan O, et al. The Transcription Factor MYB29 Is a Regulator of ALTERNATIVE OXIDASE1a. Plant Physiology. 2017;173:1824–1843. doi: 10.1104/pp.16.01494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development. 2008;135:1991–1999. doi: 10.1242/dev.016873. [DOI] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. the Plant Journal. 2004;40:22–34. doi: 10.1111/j.1365-313X.2004.02183.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study and respective statistics are available within the paper in Table S1. Materials and resources of this study are available from the corresponding author upon reasonable request.