Abstract
Dynamic measurements of molecular machines can provide invaluable insights into their mechanism but have been challenging in living cells. Here, we developed live-cell tracking of single fluorophores with nanometer spatial and millisecond temporal resolution in two and three dimensions using the recently introduced super-resolution technique MINFLUX. Using this approach, we resolved the precise stepping motion of the motor protein Kinesin-1 as it walks on microtubules in living cells. Nanoscopic tracking of motors walking on the microtubules of fixed cells also enabled us to resolve the architecture of the microtubule cytoskeleton with protofilament resolution.
Molecular machines drive many processes essential for life. For example, members of the myosin, kinesin, and dynein families are ATP-driven processive molecular motors that recognize the intrinsic structural polarity of cytoskeletal filaments to drive directed movement important for cellular processes like intracellular transport and cell division (1–3) Over the past decades, the microtubule plus-end directed motor kinesin-1 has served as a key model both for the understanding of motor dynamics and for the development of improved single-molecule methods, such as optical tweezers and single particle tracking (4–7) These studies have revealed that kinesin-1 moves in a hand-over-hand manner, in which each step along the microtubule encompasses a 16 nm displacement of the N-terminal motor domain, which leads to an 8 nm displacement of the C-terminal cargo binding domain (Fig. 1A). Despite the large success of existing single-molecule techniques in studying purified motors in well-controlled in vitro reconstitution experiments, performing such experiments in the living cell has proven challenging: Large labels required for optical tweezers typically bind multiple motors in undefined ways and single fluorophores do not provide sufficient spatial and temporal resolution to resolve the fast stepping behavior of motors under physiological ATP concentrations. As a workaround, bright nanoparticles taken up via endocytosis into transport vesicles have been used as proxies to study motor dynamics in living cells (8, 9), but in such experiments, the identity and copy numbers of the motors that drive transport are unknown. As such, the stepping dynamics of specific motors inside living cells has remained experimentally inaccessible.
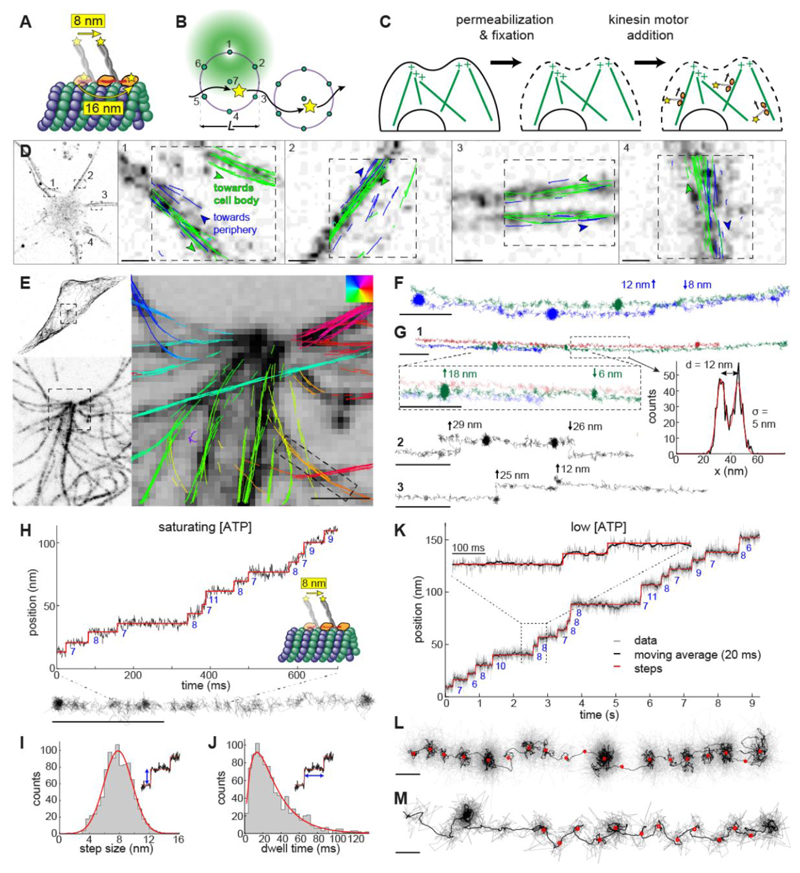
Figure 1. MINFLUX tracking of kinesin-1 in fixed cells.
(A) Kinesin-1 walks on microtubules (MTs) in a hand-over-hand manner. The apparent step size is 8 nm when the label is attached to the C-terminal tail domain and 16 nm when it is attached to the N-terminal motor domain. (B) 2D MINFLUX tracking of a single molecule. A donut beam probes seven positions around a fluorophore to determine its location with nanometer precision. The scan pattern is iteratively centered on the fluorophore during tracking. (C) Motor-PAINT approach to track kinesin in fixed cells. Cells are first permeabilized to extract cell contents and then gently fixed to preserve MTs. Purified fluorescently labeled kinesins (DmKHC (1-421)-SNAP-tag-6xHis) are added and tracked as they walk towards the plus-ends of the MTs (movie S1). (D – M) MINFLUX motor-PAINT in fixed cells. (D) Confocal images of a neuron and overlaid kinesin trajectories in four neurites. Most of the neurites show kinesin trajectories in both directions, i.e. towards (green) and away (blue) from the soma, as expected for dendrites. (E) Confocal microscopy images of GFP-α-tubulin in U2OS cells showing what appears to be a centrosome and overlaid kinesin trajectories with color-coded walking directions. (F) Tracks as indicated in (E) show side-stepping. (G) Tracks as close as 12 nm are clearly resolved and display kinesin switching laterally between neighboring MTs or protofilaments (movie S2). (H) Representative track and the corresponding time versus position plot at saturating ATP concentrations (>1 mM, here 6 mM), showing 8 nm walking steps. (I) Histogram of step size at saturating ATP concentrations from 7 experiments, 49 tracks, and 956 steps and Gaussian fit (7.8 nm ± 2.7 (std) ± 0.09 nm (sem); red line). (J) Dwell time histogram and fit with a convolution of two exponential functions (average dwell time of 30.8 ms; red line). (K, L and movie S3) A representative track at low ATP concentrations (10 μΜ) and a corresponding time versus position, raw data (gray), and 20 ms running mean (black), clearly showing 8 nm walking steps. (M, fig. S2A, E and movie S4) A representative track showing a zigzag trajectory, indicating an asymmetric arrangement of the label within a kinesin molecule (see fig. S3 for additional examples). Scale bars: 10 nm (L, M), 100 nm (F, G, H), 1 μm (D, E).
MINFLUX, a super-resolution microscopy technology (10), holds great potential to overcome these limitations. By efficiently utilizing the limited photon budget of single fluorophores, MINFLUX enables high spatial resolution for imaging (10–12) and temporal resolution for fluorophore tracking (13, 14). In MINFLUX tracking, a donut-shaped excitation beam is scanned around a single fluorophore (Fig. 1B). From the intensities measured at specific positions, the coordinate of the fluorophore is calculated and the scanning pattern is re-centered on this position before the next iteration. Keeping the fluorophore close to the dark center of the beam results in a high localization precision and minimizes photobleaching. In a companion paper, MINFLUX is used to dissect the conformational stages of kinesin-1 stepping on in vitro polymerized microtubules (15). We now show this technique can enable the high-resolution tracking of fast molecular motors in living cells.
Optimization of MINFLUX for motor protein tracking
To establish MINFLUX tracking of molecular motors in living cells, we first optimized the workflow on single molecules and fluorescent beads in vitro to maximize precision and speed (fig. S1A-C) and achieved a localization precision of ≈ 2 nm with a sub-millisecond temporal resolution. We then optimized kinesin tracking using motor-PAINT (16). Here, cells are permeabilized and fixed, before fluorescently labeled kinesin motors are added that walk along microtubules towards the plus-end (Fig. 1C and movie S1). Compared to high-resolution in vitro assays that use large beads as labels (5, 17), we used small fluorescent tags that reduced the linkage error to around 3 nm (as predicted by Alphafold2, (18, 19)), comparable to the system resolution. Motor-PAINT is less challenging than live-cell tracking as it allows us to precisely control the concentration of kinesin motors and, importantly, their speed via the ATP concentration. Using MINFLUX motor-PAINT, we were able to reconstruct cellular microtubules with a precision of ≈ 2 nm (Fig. 1D-E, table S1). Additionally, the directionality of kinesin reveals the orientation of the microtubules. Compared to standard motor-PAINT with a widefield microscope, the use of MINFLUX improved the localization precision 5-fold, the temporal resolution 50-fold, and the number of localizations per track by more than one order of magnitude. In neurons, this allowed us to better resolve individual microtubules inside dendrites compared to our earlier motor-PAINT study (Fig. 1D)(16). In human osteosarcoma (U2OS) cells, we could resolve individual trajectories of the purified motors in the crowded area around the centrosome with near protofilament resolution (Fig. 1E, F). Tracks, just 12 nm apart, were easily resolvable and we regularly observed side-stepping between different protofilaments (Fig. 1F, G and movie S2). These side steps often occurred after stalling events, suggesting that motors were circumventing obstructions, i.e. microtubule-associated proteins (MAPs) that became fixed to the microtubules, or microtubule defects from the fixation process.
A closer inspection of individual tracks showed clusters of localizations that correspond to the 8 nm steps of the labeled C-terminus. Indeed, these steps become obvious when plotting the position of the motor along the microtubule over time (Fig. 1H). This allowed us to quantify the precise step size and dwell time distributions under saturating (physiological) ATP concentrations (Fig. 1I, J). From 956 steps in 49 tracks, we measured a step size of 7.8 ± 2.7 (standard deviation, std) ± 0.09 (standard error of the mean, sem) nm and an average dwell time of 30.8 ms. To investigate the stepping behavior in greater detail, we reduced the ATP concentration to slow down the motors (20), resulting in a similar step size but a reduced rate constant for ATP binding (fig. S2A, E, table S1, S2). Under these conditions, we could measure hundreds to thousands of localizations per step (Fig. 1K, L and movie S3). Averaging over the coordinates allowed us to calculate the position of each step with sub-angstrom precision (sem). We note that currently, the accuracy of the measurements is not limited by the detected photons, but by the stability of the sample and microscope, as well as the offset of the label from the microtubule binding site. Interestingly, we noticed that 58 % of the tracks displayed zigzag motion with every other step displaced perpendicular to the track center by, on average, 3.6 nm (Fig. 1M, fig. S3A, C and movie S4). This motion is likely due to an asymmetric positioning of the fluorophore with respect to the two motor domains imaged in a top view (fig. S3D) (21), demonstrating that MINFLUX can reveal intricate details of the conformational dynamics of individual motor proteins.
Application of MINFLUX/motor-PAINT in living cells
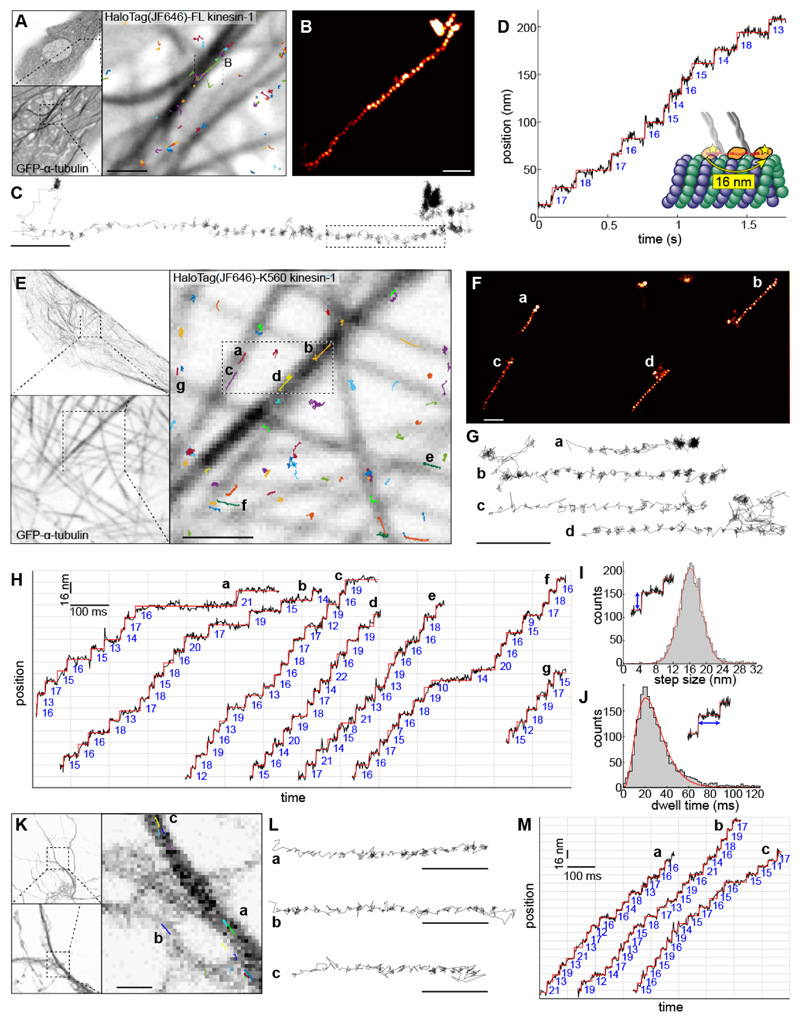
We next attempted MINFLUX tracking of kinesin in living cells. To this end, we expressed HaloTag-kinesin-1 (full-length) in U2OS cells (movie S5) and labeled at most a single motor domain per dimer by addition of the dye JF646 at very low concentrations. Individual tracks clearly revealed the 16 nm steps of the motor domains (Fig. 2A-D, movie S6). On average, we found a step size of 15.7 ± 3.8 (std) ± 0.25 (sem) nm and an average dwell time of 46.8 ms (fig. S2B, F). We also observed tracks with frequent switching between microtubules, back-slipping potentially due to multiple competing motors (movie S7), and, unlike in motor-PAINT, tracks without clear steps (fig. S4). The latter potentially stem from kinesins that are attached to dynamic microtubules or cargoes driven by other motors and are thus passively dragged along. However, the number of tracks we could acquire was low, likely due to kinesins assuming an autoinhibited form with only a low fraction in the processive state (22, 23). To increase the throughput, we used the truncated kinesin-1 variant K560, in which cargo binding and auto-inhibition are removed, and treated cells with Taxol to increase the number of stabilized microtubules preferred by kinesin-1 (movie S8) (24). With these changes, we could readily observe multiple tracks in a single field of view (Fig. 2E-H and movie S9). This allowed us to measure precise in vivo step size and dwell time distributions from 2887 steps in 330 tracks (Fig. 2I, J). This revealed that, while the average step size of 16.2 ± 3.8 (std) ± 0.07 (sem) nm was similar to full-length kinesin, the average dwell time of 27.5 ms was much shorter, consistent with the higher speeds that we observed with K560 (table S2). To test whether our approach could be extended to more complex and sensitive cell types, we examined kinesin-1 dynamics in the axons of live primary mouse cortical neurons (Fig. 2K-M), which critically depend on motor-driven transport. Here, we could clearly quantify the 16 nm stepping dynamics of kinesin-1 without Taxol treatment (step size = 15.7 ± 3.7 (std) ± 0.21 (sem) nm, average dwell time 29.2 ms, fig. S2C, G), demonstrating that MINFLUX reveals conformational dynamics of individual motor proteins in complex cellular systems.
Figure 2. MINFLUX tracking of kinesin-1 in live cells.
(A - D) Tracking of full-length kinesin-1 labeled N-terminally with a HaloTag bound to JF646 in live U2OS cells. (A) Confocal images of GFP-α-tubulin in untreated live U2OS cells, and overlaid full-length human kinesin-1 trajectories. (B) A kinesin-1 track where the localizations are rendered as a super-resolution image, in the region indicated in (A). (C) A line plot connecting each localization. (D) Time versus position plot of the highlighted portion of the track in (C), showing steps of 16 nm. (E - J) Tracking of truncated kinesin-1 (HaloTag-K560) in Taxol-treated live U2OS cells. (E) Confocal images of GFP-α-tubulin and overlaid kinesin-1 tracks. The tracks indicated in (E) rendered as a super-resolution image (F), and line plots connecting each localization (G and movie S9), showing clear walking steps (localization precision: 2 nm; temporal resolution: 1 ms). (H) Time versus position plots of representative kinesin-1 tracks as indicated in (E), showing clear 16 nm stepwise movements. (I) Step size histogram (161 experiments, 330 tracks, and 2887 steps) and a Gaussian fit (16.2 ± 3.8 (std) ± 0.07 (sem) nm). (J) Dwell time histogram, fit with a convolution of four exponential functions (average dwell time of 27.5 ms; red line). (K - M) Tracking of kinesin-1 (HaloTag-K560) in untreated live primary mouse cortical neurons. (K) Confocal images of GFP-α-tubulin and overlaid kinesin-1 tracks. (L) Representative tracks corresponding to those indicated in (K) as line plots and (M) time versus position plots, showing 16 nm stepwise movements (see fig. S2C, G for step size and dwell time histograms). Scale bars: 100 nm (B, C, F, G, L), 1 μm (A, E, K).
Three-dimensional tracking in live cells
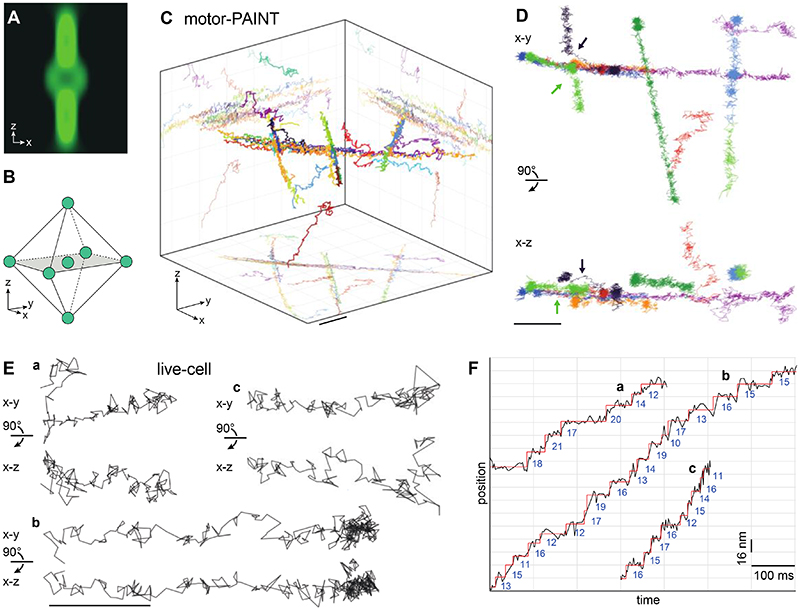
Because most biological structures extend into three dimensions, only 3D tracking can capture the true dynamics and avoid projection artifacts that limit the accuracy in 2D tracking. Unfortunately, standard single-particle tracking provides, at best, poor z resolution (25). MINFLUX has been used to image cellular structures in 3D (11), but for tracking it has so far remained limited to 2D. We therefore adapted MINFLUX for 3D tracking by scanning a 3D donut beam in 3D (Fig. 3A, B). We achieved a localization precision of 2.5 nm, 3.1 nm, and 3.9 nm in the x,y,z-directions, respectively (fig. S1D). When used with motor-PAINT, we could resolve many tracks on crossing microtubules (Fig. 3C) including jumps between microtubules (arrow in Fig. 3D). Importantly, we could also establish 3D tracking in live cells with a similar spatial and temporal resolution (3.9 nm and 3.0 ms, table S1), allowing us to resolve the 16 nm steps of kinesin in 3D (Fig. 3E, F, fig. S2D, H, and movie S10). When we investigated these trajectories in the cross-sectional views, we found dynamic movements along the z-axis including side steps and vertical trajectories. 3D tracking allowed us to extract accurate step sizes from a strongly inclined trajectory (average 15.1 nm), which, when analyzed in 2D, showed a bias towards smaller step sizes (average 9.0 nm) (fig. S5). Thus, MINFLUX tracking opens the possibility to quantify the precise 3D dynamics of molecular machines in living cells.
Figure 3. 3D MINFLUX tracking of kinesin-1.
(A, B) 3D MINFLUX tracking. (A) A 3D donut beam probes the intensity at (B) seven three-dimensionally distributed positions around a fluorophore. (C, D) 3D tracking with motor-PAINT in fixed U2OS cells. (C) 3D rendering of kinesin-1 tracks at crossing MTs with a volumetric size of 1.2 μm × 1.2 μm × 1 μm. (D) Selected tracks from (C) in top and side views, including ascending and descending trajectories, and two trajectories in which motors switch MTs (arrows). (E, F) 3D tracking in live cells. (E) Representative kinesin-1 tracks in live U2OS cells in top and side views, showing stepwise movements both in the x-y plane and along the z-axis (see fig. S2D, H for histograms, fig. S9 confocal overview images and movie S10). (F) Position versus time plots of the tracks from (E), showing 16 nm steps. Scale bars: 100 nm (E), 200 nm (C, D).
Discussion
Here we established MINFLUX tracking of kinesin-1 with nanometer spatial and sub-millisecond temporal resolution, and demonstrated that we can directly resolve steps of individual motors in live cells. In contrast to recent in vitro MINFLUX measurements (15), we did not observe clear 4 nm sub-steps of kinesin in motor-PAINT, likely because of insufficient spatial resolution. However, in live cell experiments with our N-terminally labelled motor, we could occasionally observe 8 nm sub-steps (fig. S6). This encouraged us to also image C-terminally labeled kinesin-1 in living cells. Here, the faster dynamics and higher background fluorescence made the measurements challenging, but we could observe tracks with the expected 8 nm step size (fig. S7). We saw that these C-terminally labelled motors move with a slightly higher average velocity (table S2), an effect which could be the result of motor domain labeling, an important consideration for future experiments.
Our study paves the way for investigating how stepping kinetics of motors in cells are modulated by the presence or absence of different MAPs or cargo adaptors (26). Such measurements could help to explain the observed discrepancy of kinesin-1 stepping behavior, such as dwell time, stalling and side-stepping, in motor-PAINT (in vitro-like) and live cells (presence of regulatory MAPs such as MAP7, higher salt concentrations, etc.) (table S1, 2). One parameter that was consistent across studies is the average step size, despite the fact that in live cells, the motion of the microtubules themselves could contribute to the measured step size. As expected, the average step size was ≈16 nm, both in U2OS cells (which have some microtubule sliding (27)) and in neuronal axons (where microtubules are largely immobile (28)) (table S1). The impact of microtubule movement is thus likely to be minimal at the time scale of kinesin steps.
MINFLUX tracking is not limited to kinesin-1, but can be used to study the precise motion of any protein in living cells with high spatio-temporal resolution and minimal perturbance due to its compatibility with single-fluorophore labels (see fig. S8 for tracking of Myosin-V). In the future, developing MINFLUX to simultaneously track two colors will enable monitoring of the relative 3D positions of labeled protein domains with nanometer spatial and sub-millisecond temporal resolution. Such measurements of conformational changes of molecular machines in their native environment will provide important insights into their function and regulation.
Supplementary Material
One-sentence summary:
We developed an approach to measure nanoscale conformational dynamics of individual motor proteins in living cells.
Acknowledgements
We thank Koki Watanabe and Klara I. Jansen for generating full-length wild type Myosin-V and K560-HaloTag constructs, respectively, Abberior Instruments and specifically Roman Schmidt for MINFLUX technical support, Franziska Fichtner, Angelica Pacheco, Ulf Matti, and Lucia Perez for their help in sample preparations, and the EMBL Imaging Centre for access to the MINFLUX instrument. JF646 and JFX646 HaloTag ligand was a kind gift of Luke Lavis, HHMI Janelia Research Campus.
Funding
This work was supported by H2020 Marie Sklodowska-Curie Actions (RobMin, No. 101031734 to T.D., EMBL ARISE fellowship, No. 945405 to S.L., EIPOD4 program, No. 847543 to J.H.), the European Research Council (grant no. ERC CoG-724489 to J.R. and CoG-819219 to L.C.K.) and the European Molecular Biology Laboratory (T.D., E.S., C.H., L.S., J.H., S.S., P.H., S.L, V.C., K.N., and J.R.).
Footnotes
Author contributions:
J.R. conceived the project. L.S., P.H., and V.C. designed and generated constructs. M.K.I. purified proteins for motor-PAINT. J.H. cultured neurons. T.D., M.K.I., E.S., C.H., L. S., J.H. prepared samples. T.D., T.W., and S.S. established MINFLUX tracking protocols. T.D. and S.L. acquired MINFLUX tracking data. E.S. and C.H. acquired fluorescence images. J.R. wrote analysis software. T.D., M.K.I., E.S., C.H., L.C.K., and J.R. analyzed the data. K.N., L.C.K., J.R. supervised the research. T.D., M.K.I, E.S., C.H., J.H., P.H., L.C.K., and J.R. wrote the manuscript with input from all authors.
Competing interests:
The authors declare that they have no competing interests.
Data and materials availability
All MINFLUX tracking data is available at BioStudies https://www.ebi.ac.uk/biostudies/studies/S-BIAD608.
References
- 1.Howard J, Hudspeth AJ, Vale RD. Movement of microtubules by single kinesin molecules. Nature. 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- 2.Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 3.Shingyoji C, Higuchi H, Yoshimura M, Katayama E, Yanagida T. Dynein arms are oscillating force generators. Nature. 1998;393:711–714. doi: 10.1038/31520. [DOI] [PubMed] [Google Scholar]
- 4.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 5.Sudhakar S, Abdosamadi MK, Jachowski TJ, Bugiel M, Jannasch A, Schäffer E. Germanium nanospheres for ultraresolution picotensiometry of kinesin motors. Science. 2021;371:eabd9944. doi: 10.1126/science.abd9944. [DOI] [PubMed] [Google Scholar]
- 6.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin Walks Hand-Over-Hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 7.Stepp WL, Ökten Z. Resolving kinesin stepping: one head at a time. Life Science Alliance. 2019;2 doi: 10.26508/lsa.201900456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng CS, Zhang Y, Liu Q, Marti GE, Huang Y-WA, Südhof TC, Cui B, Chu S. 2022 doi: 10.1038/s41589-024-01694-2. https://www.biorxiv.org/content/10.1101/2022.01.05.475120v1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nan X, Sims PA, Xie XS. Organelle Tracking in a Living Cell with Microsecond Time Resolution and Nanometer Spatial Precision. ChemPhysChem. 2008;9:707–712. doi: 10.1002/cphc.200700839. [DOI] [PubMed] [Google Scholar]
- 10.Balzarotti F, Eilers Y, Gwosch KC, Gynnâ AH, Westphal V, Stefani FD, Elf J, Hell SW. Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science. 2017;355:606–612. doi: 10.1126/science.aak9913. [DOI] [PubMed] [Google Scholar]
- 11.Gwosch KC, Pape JK, Balzarotti F, Hoess P, Ellenberg J, Ries J, Hell SW. MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells. Nat Methods. 2020;17:217–224. doi: 10.1038/s41592-019-0688-0. [DOI] [PubMed] [Google Scholar]
- 12.Pape JK, Stephan T, Balzarotti F, Büchner R, Lange F, Riedel D, Jakobs S, Hell SW. Multicolor 3D MINFLUX nanoscopy of mitochondrial MICOS proteins. Proc Natl Acad Sci USA. 2020;117:20607–20614. doi: 10.1073/pnas.2009364117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eilers Y, Ta H, Gwosch KC, Balzarotti F, Hell SW. MINFLUX monitors rapid molecular jumps with superior spatiotemporal resolution. Proc Natl Acad Sci USA. 2018;115:6117–6122. doi: 10.1073/pnas.1801672115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt R, Weihs T, Wurm CA, Jansen I, Rehman J, Sahl SJ, Hell SW. MINFLUX nanometer-scale 3D imaging and microsecond-range tracking on a common fluorescence microscope. Nat Communications. 2021;12:1478. doi: 10.1038/s41467-021-21652-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolff JO, Scheiderer L, Engelhardt T, Engelhardt J, Matthias J, Hell SW. 2022. if now published, omit the URL and provide only a standard reference https://www.biorxiv.org/content/10.1101/2022.07.25.501426v1.
- 16.Tas RP, Chazeau A, Cloin BMC, Lambers MLA, Hoogenraad CC, Kapitein LC. Differentiation between Oppositely Oriented Microtubules Controls Polarized Neuronal Transport. Neuron. 2017;96:1264–1271.:e5. doi: 10.1016/j.neuron.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isojima H, Iino R, Niitani Y, Noji H, Tomishige M. Direct observation of intermediate states during the stepping motion of kinesin-1. Nat Chem Biol. 2016;12:290–297. doi: 10.1038/nchembio.2028. [DOI] [PubMed] [Google Scholar]
- 18.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Židek A, Potapenko A, Bridgland A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnitzer MJ, Block SM. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 21.Liu D, Liu X, Shang Z, Sindelar CV. Structural basis of cooperativity in kinesin revealed by 3D reconstruction of a two-head-bound state on microtubules. eLife. 2017;6:e24490. doi: 10.7554/eLife.24490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coy DL, Hancock WO, Wagenbach M, Howard J. Kinesin’s tail domain is an inhibitory regulator of the motor domain. Nat Cell Biol. 1999;1:288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- 23.Friedman DS, Vale RD. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat Cell Biol. 1999;1:293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- 24.Cai D, McEwen DP, Martens JR, Meyhofer E, Verhey KJ. Single Molecule Imaging Reveals Differences in Microtubule Track Selection Between Kinesin Motors. PLOSBiology. 2009;7:e1000216. doi: 10.1371/journal.pbio.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrecka J, Ortega Arroyo J, Takagi Y, de Wit G, Fineberg A, MacKinnon L, Young G, Sellers JR, Kukura P. Structural dynamics of myosin 5 during processive motion revealed by interferometric scattering microscopy. eLife. 2015;4:e05413. doi: 10.7554/eLife.05413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hooikaas PJ, Martin M, Mühlethaler T, Kuijntjes G-J, Peeters CAE, Katrukha EA, Ferrari L, Stucchi R, Verhagen DGF, van Riel WE, Grigoriev I, et al. MAP7 family proteins regulate kinesin-1 recruitment and activation. Journal of Cell Biology. 2019;218:1298–1318. doi: 10.1083/jcb.201808065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansen KI, Burute M, Kapitein LC. 2021 https://www.biorxiv.org/content/10.1101/2021.06.23.449589v1 . [Google Scholar]
- 28.Burute M, Jansen KI, Mihajlovic M, Vermonden T, Kapitein LC. Local changes in microtubule network mobility instruct neuronal polarization and axon specification. Science Advances. 2022;8:eabo2343. doi: 10.1126/sciadv.abo2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beyer HM, Gonschorek P, Samodelov SL, Meier M, Weber W, Zurbriggen MD. AQUA Cloning: A Versatile and Simple Enzyme-Free Cloning Approach. PLOS ONE. 2015;10:e0137652. doi: 10.1371/journal.pone.0137652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gumy LF, Katrukha EA, Grigoriev I, Jaarsma D, Kapitein LC, Akhmanova A, Hoogenraad CC. MAP2 Defines a Pre-axonal Filtering Zone to Regulate KIF1-versus KIF5-Dependent Cargo Transport in Sensory Neurons. Neuron. 2017;94:347–362.:e7. doi: 10.1016/j.neuron.2017.03.046. [DOI] [PubMed] [Google Scholar]
- 31.Thevathasan JV, Kahnwald M, Cieśliński K, Hoess P, Peneti SK, Reitberger M, Heid D, Kasuba KC, Hoerner SJ, Li Y, Wu Y-L, et al. Nuclear pores as versatile reference standards for quantitative superresolution microscopy. Nat Methods. 2019;16:1045–1053. doi: 10.1038/s41592-019-0574-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mora-Bermúdez F, Gerlich D, Ellenberg J. Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase. Nat Cell Biol. 2007;9:822–831. doi: 10.1038/ncb1606. [DOI] [PubMed] [Google Scholar]
- 33.Diekmann R, Kahnwald M, Schoenit A, Deschamps J, Matti U, Ries J. Optimizing imaging speed and excitation intensity for single-molecule localization microscopy. Nat Methods. 2020;17:909–912. doi: 10.1038/s41592-020-0918-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer Control of Microscopes Using pManager. Current Protocols in Molecular Biology. 2010;92:14.20.1-14.20.17. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deschamps J, Ries J. EMU: reconfigurable graphical user interfaces for Micro-Manager. BMC Bioinformatics. 2020;21:456. doi: 10.1186/s12859-020-03727-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ries J. SMAP: a modular super-resolution microscopy analysis platform for SMLM data. Nat Methods. 2020;17:870–872. doi: 10.1038/s41592-020-0938-1. [DOI] [PubMed] [Google Scholar]
- 37.Loeff L, Kerssemakers JWJ, Joo C, Dekker C. AutoStepfinder: A fast and automated step detection method for single-molecule analysis. Patterns. 2021;2:100256. doi: 10.1016/j.patter.2021.100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All MINFLUX tracking data is available at BioStudies https://www.ebi.ac.uk/biostudies/studies/S-BIAD608.