Abstract
Background
Pediatric disorders include a range of highly genetically heterogeneous conditions amenable to genome-wide diagnostic approaches. Finding a molecular diagnosis is challenging but can have profound lifelong benefits.
Methods
The Deciphering Developmental Disorders (DDD) study recruited >13,500 families with severe, likely monogenic, difficult-to-diagnose developmental disorders from 24 regional genetics services around the UK and Ireland. We collected standardised phenotype data and performed exome sequencing and microarray analysis to investigate novel genetic causes. We developed an iterative variant analysis pipeline, reporting candidate variants to clinical teams for validation, diagnostic interpretation and communication to families. We performed multiple regression analyses evaluating factors affecting probability of diagnosis.
Results
We reported ~1 candidate variant per parent-offspring trio and ~2.5 variants per singleton proband. Using clinical and computational approaches to variant classification, we achieved a diagnosis in ~41% (5502 probands), of whom ~76% have a pathogenic de novo variant. Another 22% have variants of uncertain significance in genes robustly linked with monogenic developmental disorders. Recruitment as a parent-offspring trio had the largest impact on chance of diagnosis (OR=4.70). Probands who were extremely premature (OR=0.39), had in-utero exposure to antiepileptic medications (OR=0.44), or whose mothers had diabetes (OR=0.52) were less likely to be diagnosed, as were those of African ancestry (OR=0.51).
Conclusions
The DDD study shows multimodal analysis of genome-wide data has good diagnostic power, even after prior attempts at diagnosis.
(Funded by Wellcome Trust and others.)
Introduction
Genomic sequencing has made extraordinary strides towards identifying novel molecular causes for rare monogenic disorders, and is becoming increasingly available in diagnostic clinics throughout the world.1,2 Pediatrics has particularly benefited from the use of high-throughput next generation sequencing technologies, partly because of the high clinical need and potential for lifelong impact of diagnosis and treatment.3 In addition, the early presentation of severe disease makes genetic diagnosis more tractable as causal variants are largely absent from control datasets.4
Progress in pediatric rare disease genomics has been spearheaded by numerous diagnostic research groups across the world.5,6 One of the first studies to combine large-scale genomic research with individual patient feedback was the Deciphering Developmental Disorders (DDD) study,7–9 with >13,500 families with exome sequence and microarray data and rich clinical phenotypes recorded by >200 clinicians across the UK and Ireland. Here we describe analytical strategies developed over a decade by the DDD study to identify and classify thousands of new molecular diagnoses, and report factors affecting the probability of receiving a diagnosis.
Methods
Study Overview
The DDD study was granted UK Research Ethics Committee (REC) approval by the Cambridge South REC (10/H0305/83) and Republic of Ireland REC (GEN/284/12). A multicentre research collaboration was set up with all 24 Regional Genetics Services, and a management committee (comprising clinicians, scientists and a bioethicist) was created to provide ongoing ethical oversight (Table 1). In addition to genomic and data scientists, a social scientist was employed to do ethics research.10
Table 1. Ethical considerations in the DDD study.
The DDD study depended crucially upon integration of ethics into decision-making and collaboration-building, both upfront and throughout the project, allowing important ethical questions to be identified and ethical policies to be developed through a consensual process.
| Ethical Domain | Key Issues | Resolution within DDD Study |
|---|---|---|
| Building and maintaining partnerships at clinical-research interface |
|
|
| Recruitment, consent, capacity, and eligibility |
|
|
| Sample inclusion, collection, and verification |
|
|
| Sharing clinically- relevant variants |
|
|
| Sharing genome-wide variants |
|
|
| Managing withdrawal |
|
|
Cohort
13,610 cases (88% as parent-offspring trios) were ascertained and recruited between April 2011-2015 by consultant clinical geneticists, facilitated by research nurses/genetic counsellors. Families gave informed consent for participation. Eligibility criteria included any of the following: neurodevelopmental disorders; congenital anomalies; abnormal growth parameters (single parameter >4SD or two or more parameters >3SD above the mean); dysmorphic features; unusual behavioural phenotypes; and genetic disorders with a significant impact for which the molecular basis was unknown. The study was initially limited to probands <16 years at the date of recruitment, but this age limit was later removed (except in Scotland). Most probands had previously undergone clinical chromosomal microarray (85%) and/or single gene testing (53%) but remained undiagnosed. Probands were assigned pseudonymised IDs and basic clinical information, quantitative growth data, developmental milestones and Human Phenotype Ontology (HPO)11 terms were recorded for all participants via a bespoke standardised interface in DECIPHER.12
Genomic Analyses
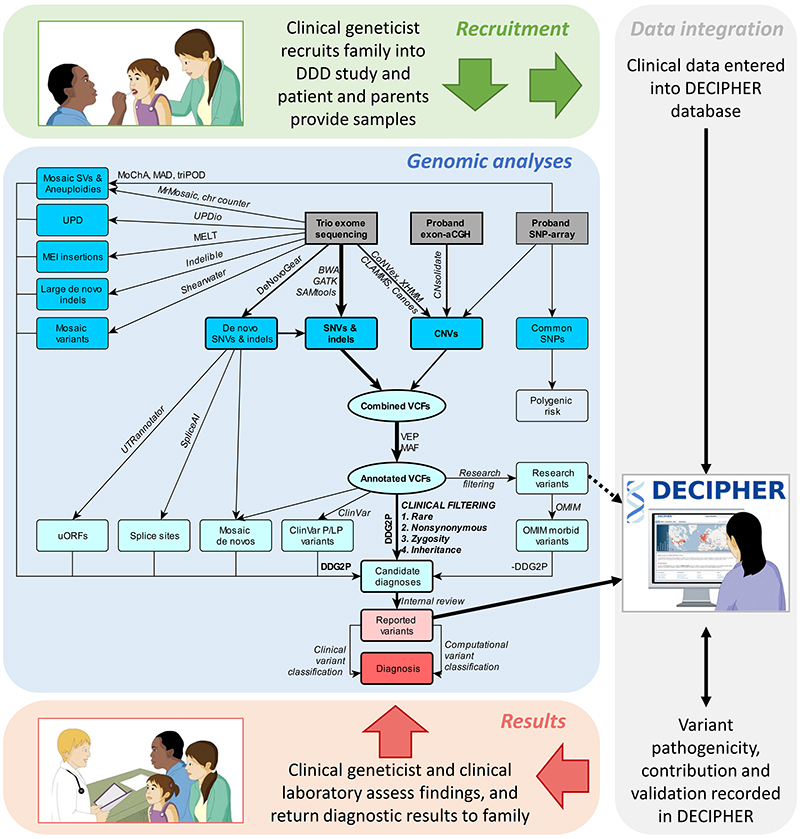
Detailed assay protocols13,14 and variant filtering pipelines7,15 have been described elsewhere (Supplementary Information). Briefly, three independent genomic assays were performed: exome sequencing (ES) of complete family trios and singleton probands (i.e. non-trios); exon-array comparative genomic hybridisation (aCGH) of probands; and genome-wide SNP-genotyping of probands. Assay designs remained largely unchanged throughout the study. Datasets were processed in batches, and multiple different algorithms were used to detect and annotate sequence and structural variants (Figure 1). The inheritance status of variants in the proband were determined by comparison with parental data16. For clinical reporting, we selected high-quality, rare, non-synonymous variants overlapping genes in the Developmental Disorders Gene2Phenotype database (DDG2P)17 with appropriate zygosity and inheritance (where available). We augmented this pipeline with additional analyses to find missing likely causal variants, including ClinVar pathogenic/likely pathogenic variants,18 and de novo variants that were mosaic,19 created upstream open reading frames,20 affected splicing,21 or were coding insertions/deletions of intermediate size22 or caused by mobile element insertions23 or mosaic chromosomal alterations.24
Figure 1. Overview of DDD variant detection and filtering pipelines.
Physician-patient interactions within the DDD study were supported by the DECIPHER database, including recruitment, barcoded sample collection and phenotyping at the start, and variant reporting, diagnostic interpretation and discussion of results at the end. Genomic assays are shown in grey boxes, variants in blue boxes, variant subsets in light blue circles, and reported and diagnostic variants in red boxes; variant callers and analytical processes are annotated on arrows (further detail and references in Supplementary Information). Once candidate variants were deposited into DECIPHER, clinical judgement was used to assess whether a patient’s phenotype fitted with the genotype prior to returning confirmed diagnoses to families. Diagrams were taken from www.ddduk.org. aCGH = array comparative genomic hybridisation; CNVs = copy number variants; DDG2P = Developmental Disorders Gene2Phenotype database; indels = insertions/deletions; MAF = minor allele frequency; MEI = mobile element insertion; OMIM = Online Mendelian Inheritance in Man database; P/LP = pathogenic/likely pathogenic (variants in the ClinVar database); SNP = single nucleotide polymorphism; SNVs = single nucleotide variants; SVs = structural variants; UPD = uniparental disomy; uORFs = upstream open reading frames; VEP = Variant Effect Predictor; VCFs = variant call files.
Defining a Diagnosis
Candidate diagnostic variants identified bioinformatically were reviewed by a central clinical review panel to evaluate analytical and clinical validity prior to reporting in batches to regional genetics teams via DECIPHER (April 2014-2022, Figure 2). The referring clinician then evaluated the reported variant(s), requested diagnostic laboratory confirmation where required, and communicated diagnoses to the family. At the time of writing, clinical classifications of variant pathogenicity (benign/ likely benign/ uncertain/ likely pathogenic/ pathogenic) and contribution to the phenotype (full/ partial/ unknown/ none) were recorded in DECIPHER for 84% of variants. These were supplemented by automated predictions for variant classification criteria (BA1, BS1, BP4, BP7, PVS1, PS1, PS2, PP3 and PM2) based on published guidelines from the American College of Medical Genetics and Genomics (ACMG) and Association of Molecular Pathologists25 and UK Association of Clinical Genetic Scientists (ACGS)26. A provisional variant classification was calculated using a log-additive Bayesian framework described elsewhere27 (Supplementary Information). Variants with a posterior probability of >0.9 were classified as likely pathogenic and pathogenic >0.99, or <0.1 as likely benign and benign <0.001. For genes with ≥10 pathogenic/likely pathogenic variants, computational phenotype matching was performed using IMPROVE-DD,28 applying the same Bayesian framework to combine variant classifications and gene-disease models; phenotype-based likelihoods were scaled appropriately and used at the evidence equivalent of “Strong”.27 Probands were categorised as “diagnosed” if ≥1 variant(s) or ≥2 compound heterozygous variants were annotated as pathogenic/likely pathogenic by either the proband’s referring clinician and/or the predicted classification, and the contribution to phenotype was not clinically annotated as “none”. Factors influencing the chance of receiving a diagnosis (based on clinical annotation only) were investigated using multivariable logistic regression with Bonferroni correction to account for multiple hypothesis testing (Supplementary Information).
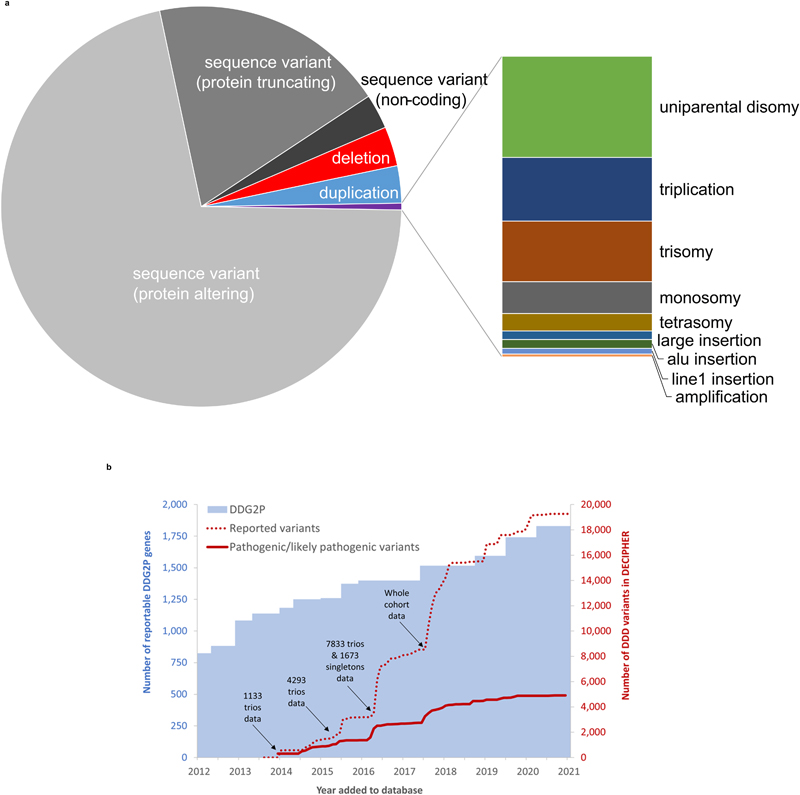
Figure 2. Summary of DDD variant deposition into DECIPHER.
(a) Variant classes reported into DECIPHER. Sequence variants were detected using ES and included variants <100bp in DDG2P genes; structural variants range from >100bp to whole chromosomes and were detected using microarrays and ES. (b) Changes in DDG2P and number of variants reported and annotated as pathogenic/likely pathogenic with time. Gene-disease entities were added to the DDG2P database following curation of the literature by consultant clinical geneticists or burden analyses within the DDD study. Participants were sequenced and analysed in batches based on recruitment date, sample receipt and family trio status. Variant filtering was repeated over the course of the study to enable evaluation of novel variants and variants in newly included genes. As a result of this iterative variant filtering strategy, some probands were evaluated up to six times and all were evaluated at least twice (see Figure S3). Following evaluation, variants were deposited into DECIPHER, usually in batches, for evaluation by clinical teams. Clinical annotation of pathogenicity was not immediate up deposition, but once annotated, the vast majority (97%) of variants did not change their annotation. Blue bars = cumulative number of reportable DDG2P genes; red dotted line = cumulative number of total DDD variants deposited into DECIPHER; red continuous line = cumulative number of clinically annotated pathogenic/likely pathogenic DDD variants in DECIPHER.
Results
Cohort Characteristics
The DDD study includes 13,449 probands (9,859 in parent-offspring trios) with severe, previously undiagnosed developmental disorders with ES, exon-aCGH and SNP genotyping data, recruited across the UK and Ireland with a median recruitment per centre of 216 probands per million population (range=69-588). The median age was 7 years (range=0-63) at recruitment for probands and 31 years (range=15-70 at the proband’s birth) for parents; 42% of probands were female and 16% were non-European (Table S1). A median of 6 HPO terms (range=1-36) were recorded per proband, including 65% with global developmental delay/intellectual disability, and 72% of probands were the only affected member of their family.
Genetic Findings
To date, 19,285 potentially pathogenic sequence and structural variants have been identified in DDD probands and returned to referring clinicians through up to six rounds of iterative re-analysis and batch reporting, involving 18 different variant detection algorithms (Figure 1 and Table S2).7,15 The majority of variants were identified using a clinically-curated database of 1,840 DD-associated genes (DDG2P)17 which was updated at a rate of approximately 100 genes/year through literature curation and cohort-wide enrichment analyses (including 60 novel DD-associated genes identified by DDD; Figure 2);5,13,14,29,30 44% of reported variants were in genes added to DDG2P after the first round of reporting in 2014. The majority of reported variants were single nucleotide variants and small insertions/deletions detected using ES data (71% protein-altering, 19% protein-truncating, 3% non-coding variants), while structural variants were identified through a combination of microarray and ES analyses (6% copy number variants, 1% other structural variants; Figure 2). On average, one variant was reported per trio versus 2.5 per singleton proband (Figure S1), and probands with non-European ancestry had more variants reported (Figure S2). Each new round of analysis resulted in approximately one additional variant being reported for every six trios (Figure S3). Consistent with similar studies,31 de novo variants in the proband and variants inherited from a mosaic parent (i.e. post-zygotic parental de novo variants) in dominant genes provided the highest diagnostic yield, with 79% of reported variants clinically classified as pathogenic/likely pathogenic; in contrast, 32% of variants in autosomal recessive conditions, 23% maternally inherited on the X-chromosome, 11% in autosomal dominant conditions inherited from an affected parent or with unknown inheritance were clinically classified as pathogenic/likely pathogenic (Figure S4).
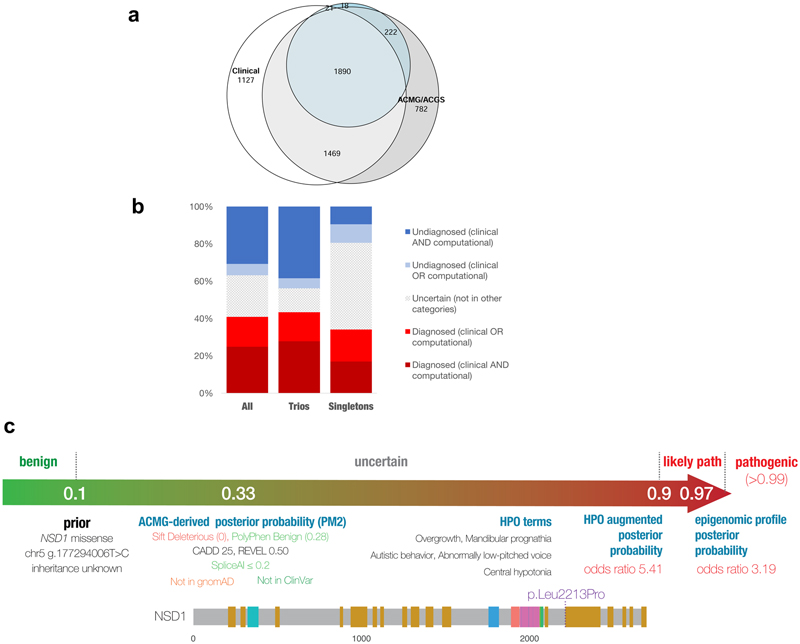
There was a high rate of concordance between clinical and predicted classifications of variant pathogenicity and benignity (N=4425; sensitivity=99.5%, specificity=85.0%, PPV=96.5%, NPV=97.9%; Figure S6).25–27 Discrepancies (N=149; 3%) were due to false positive variant calls, incorrect clinical classifications (e.g. atypical disease presentations) or inappropriate ACMG/ACGS criteria assignment (e.g. incorrect disease mechanisms). Based on concordance between clinical and predicted classifications of variant pathogenicity, we estimate that a minimum of 25% of probands (N=3347) are diagnosed, which rises to 32% (N=4363) for predicted only, 33% (N=4484) for clinical only, and 41% (N=5502) for either clinical or predicted (Figure 3). Of those who were diagnosed by clinical assertion: 76% of those in family trios (N=2750/3599) have a pathogenic de novo variant (Supplementary Information); 13% (N=561) are partially diagnosed, and a further 3% (N=121) have two or more different genetic diagnoses potentially resulting in a composite phenotype, which rises to 7% (N=359) including computational predictions.32 Although 30% (N=4021) have no reported variants, the rest of the cohort have either likely benign variants or variants of uncertain significance, of which 0.7% (N=99) have a predicted Bayesian posterior probability of pathogenicity 0.8-0.9. High rates of concordance were also seen in the subset of variants for which we were able to derive a phenotype-based gene-disease model using IMPROVE- DD,28 and a further 18 variants of uncertain significance were raised to likely pathogenic based on the individual’s phenotype.
Figure 3. Summary of diagnoses in the DDD study.
(a) Venn diagram showing overlap of diagnoses based on clinical assertion (white) versus predicted ACMG/ACGS variant classifications (grey), augmented with phenotype-based IMPROVE-DD gene-disease models (blue); figure created using eulerr. (b) Diagnostic ranges in trios and singleton probands, based on clinical and/or predicted variant classifications. (c) Example of computational Bayesian variant classification, incorporating genotypic and phenotypic data in a DDD proband: only PM2 could be applied to the missense variant, resulting in an uncertain classification, but the proband’s phenotype was consistent with the IMPROVE-DD model for NSD1, allowing the variant to be upgraded to likely pathogenic; additional data (e.g. epigenomic profiling)40 was used to further increase the robustness of the diagnosis of Sotos syndrome.
Factors Influencing Diagnostic Rate
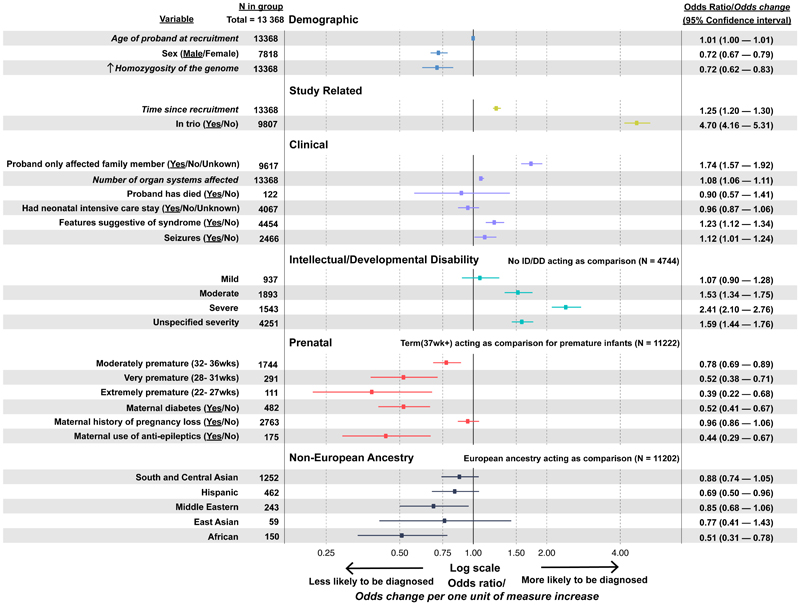
We performed multiple logistic regression to investigate how demographic, clinical, phenotypic, prenatal and ancestral factors affected the chance of receiving a clinical diagnosis from the DDD study (Figure 4). The model explained ~14% of the variance. Being recruited in a parent-offspring trio had the largest impact on the chance of being diagnosed (OR: 4.70, 95% CI: 4.16-5.31). Other factors significantly increasing the chance of diagnosis included: having severe intellectual/ developmental delay (OR: 2.41, 95% CI: 2.10-2.76); time since recruitment (increased odds of diagnosis: 1.25 per additional year, 95% CI: 1.20-1.30); being the only affected family member (OR: 1.74, 95% CI: 1.57-1.92) or having fewer affected first-degree relatives (Figure S7); having features suggestive of a syndrome (OR: 1.23, 95% CI: 1.12-1.34); and having more organ systems affected (increased odds of diagnosis: 1.08 per additional organ system, 95% CI: 1.06-1.11). Probands born prematurely (OR: 0.73, 95% CI: 0.64-0.82), or who had in utero exposure to antiepileptic medications (OR: 0.44, 95% CI: 0.29-0.67) or whose mothers had diabetes (OR: 0.52, 95% CI: 0.41-0.67) were less likely to have a genetic diagnosis. Male sex (OR: 0.72, 95% CI: 0.67-0.79) also reduced the odds of getting a diagnosis, as did increasing homozygosity due to consanguinity (decreased odds of diagnosis: 0.72 for each increase equivalent to the offspring of first cousins, 95% CI: 0.62-0.83). Probands with African ancestry had a lower diagnostic rate than those with other ancestries (OR: 0.51, 95% CI: 0.31-0.78), which was driven by fewer diagnoses in singleton probands (Figure S8).
Figure 4. Factors influencing the probability of being diagnosed.
Odds associated with being fully or partially diagnosed by the DDD study (based on clinician assertions of variant pathogenicity) are shown for covariates included in a multivariable logistic regression, adjusted for recruitment centre and number of variants reported in DECIPHER. Odds ratios are presented for binary and categorical variables. For quantitative variables (italics), odds change per one unit of measure of increase are presented. P-values and 95% confidence intervals are also shown; underline in variable column = outcome variable plotted, with N referring to the number of probands in this group. See Supplementary Information for further analysis of the number of affected first-degree relatives (Figure S6) and ancestry (Figure S7 and S8
Discussion
The DDD study has identified and communicated molecular diagnoses to thousands of families across the UK and Ireland affected by severe, previously undiagnosed developmental disorders. Despite excellent provision of clinical genetics and genetic testing services across the UK, these probands show how a genome-driven approach combined with detailed phenotyping can improve diagnostic yield over the previous standard of care. Our analysis highlights the value of using diverse and agnostic variant detection methodologies, combined with stringent variant filtering rules and repeated, iterative variant analysis and classification to enable new diagnoses to be made from existing data.15
The high burden of pathogenic de novo variants and current diagnostic yield of around 41% is consistent with similar studies.33 Our analysis supports clinical intuition about the likelihood of establishing a molecular diagnosis in developmental disorders (e.g. availability of parental genotype data, as well as sex, ethnicity and phenotypic severity) and moves towards quantifying the expectation of making such a diagnosis. The work also highlights groups with lower diagnostic rates in our cohort (e.g. non-trios, families with multiple affected members, and those with non-European ancestry or high consanguinity), and reinforces the imperative to increase research participation for under-represented groups. Probands of African ancestry had a particularly low diagnostic rate, partly caused by the lack of ancestry-matched controls to estimate allele frequency and partly by being less likely to be recruited as a trio. Excluding cohort-specific factors, our multivariable logistic regression model predicts that probands in the top decile of probability of being diagnosed have a diagnostic rate of 52% versus 16% in the bottom decile. We hypothesise that the lower diagnostic rate found in probands with certain prenatal risk factors reflects a larger role for environmental influences in these individuals. Prematurity34, maternal diabetes35 and in utero exposure to antiepileptic medications36 are known risk factors for developmental disorders. Further exploration is needed to better understand the relative contributions and interplay of genetic/environmental influences in this cohort.
The genetic architecture of developmental disorders is heterogeneous; although the large burden of highly-penetrant de novo variants facilitates both diagnosis and large-scale gene-disease discovery,5 the number of composite and partial diagnoses suggests that many individuals are likely to have multiple contributing factors, including rare and common incompletely penetrant genetic variants and non-genetic causes. Under a liability threshold model of disease,37 probands who have a substantial environmental contribution may require less severe or even no large-effect genetic variants to develop a neurodevelopmental disorder. Nonetheless, statistical burden analyses suggest that many more diagnoses remain in protein-coding genes than in non-coding elements,38 which will likely be identified through novel DD-associated gene discovery (especially for dominant disorders), evaluation of incompletely penetrant variants, and functional assays to improve interpretation of existing candidate variants. Ultimately, clinical interpretation remains indispensable for determining the relevance of genomic findings for an individual patient.
The DDD study pioneered a hybrid clinical-research approach, requiring development of new methodologies to facilitate both large-scale analysis and individual variant feedback, which has since become standard practice in genomic medicine. The study primarily recruited infants and children and hence pioneered a conservative approach to individual variant feedback that focussed on diagnosis,7–9 whilst exploring attitudes to communicating incidental findings39 that influenced subsequent approaches.1 A large network of expert clinician-researchers and the integration of ethics at a high level throughout the project lifecycle served to both facilitate collaboration and enable real-time ethical issues to be openly and responsibly addressed (Table 1). To date, in addition to making thousands of new diagnoses for patients, the DDD study has resulted in >270 publications (https://www.ddduk.org/publications.html), identified around 60 new disorders and enabled >350 genotype/phenotype-specific projects led by clinician-researchers across all 24 recruitment sites. DECIPHER12 was another key component of the DDD study, enabling nationwide recruitment, systematic phenotyping, individual feedback, variant interpretation and data sharing. DECIPHER is a live online platform enabling reported variants to be re-evaluated with current data (e.g. gene-disease associations, population frequencies, co-located variants reported in ClinVar, DECIPHER or publications) each time a patient is reviewed in the clinic, thus facilitating new opportunities for diagnosis as knowledge grows.
Although many of our conclusions are widely applicable across a range of rare diseases, the generalisability is limited by a number of factors. Recruitment of families following clinician-led differential diagnosis and routine diagnostic testing (karyotyping, aCGH and targeted single gene testing) resulted in a cohort likely depleted of clinically recognisable syndromes, large pedigrees with segregating pathogenic variants, recessive conditions in consanguineous families, and large structural variants. These biases will reduce the estimated diagnostic yield relative to first-tier testing, and skew the factors affecting getting a diagnosis. The diagnostic yield in DDD therefore represents a conservative estimate with higher yields anticipated if genomic sequencing had been offered as a first-line investigation. Our genotyping approach (ES and microarrays) did not assay most non-coding variants and could not detect all complex structural variants or tissue-specific mosaicism, and our analytical approach was insensitive to incomplete penetrance. Furthermore, the study was not funded to capture longitudinal phenotype data, evaluate parental phenotypes in detail, record the impact of diagnosis on subsequent clinical management of families, or comprehensively assess social or environmental contributions to developmental disorders – all of which, in retrospect, would have enhanced the project. Finally, despite the large cohort size, due to the enormous genetic and phenotypic heterogeneity, we often had insufficient probands (particularly across different ancestries) with the same ultra-rare condition to enable confident variant interpretation, highlighting the need to aggregate phenotype information and structured electronic health data across cohorts internationally to improve variant interpretation.
That being said, through its pioneering genomic analysis of a large clinical cohort using a hybrid clinical-research model, the DDD study shows how the fusion of clinical expertise, genomic science and bioinformatics can drive diagnosis and discovery in families for whom standard clinically-driven diagnostic approaches have failed.
Supplementary Material
Acknowledgements
The authors are deeply indebted to the patients and families involved in the DDD study, as well as the UK NHS and Irish HSE genetics services and the many scientists who have worked on DDD data at the Wellcome Sanger Institute (Supplementary Information). The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network. The study uses DECIPHER (https://www.deciphergenomics.org), which is funded by Wellcome [grant number WT206194]. PC was supported by an NIHR Academic Clinical Fellowship.
Funding
The DDD study presents independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1009-003], a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Sanger Institute [grant number WT098051]. DECIPHER is funded by Wellcome [WT223718/Z/21/Z]. The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
Data Availability
A Supplementary Table of diagnoses per gene is provided. Individual-level datasets are available under managed access for research into developmental disorders via the European Genome-phenome Archive (EGAS00001000775). Individual pathogenic/likely pathogenic variants are openly accessible with phenotypes via DECIPHER, and are provided in a Supplementary File in the European Genome-Phenome Archive.
References
- 1.100,000 Genomes Project Pilot Investigators. Smedley D, Smith KR, et al. 100,000 Genomes Pilot on Rare-Disease Diagnosis in Health Care - Preliminary Report. N Engl J Med. 2021;385(20):1868–80. doi: 10.1056/NEJMoa2035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams DR, Eng CM. Next-Generation Sequencing to Diagnose Suspected Genetic Disorders. N Engl J Med. 2018;379(14):1353–62. doi: 10.1056/NEJMra1711801. [DOI] [PubMed] [Google Scholar]
- 3.Lalonde E, Rentas S, Lin F, Dulik MC, Skraban CM, Spinner NB. Genomic diagnosis for pediatric disorders: revolution and evolution. Front Pediatr. 2020;8:373. doi: 10.3389/fped.2020.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett CA, Petrovski S, Oliver KL, Berkovic SF. ExACtly zero or once: A clinically helpful guide to assessing genetic variants in mild epilepsies. Neurol Genet. 2017;3(4):e163. doi: 10.1212/NXG.0000000000000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplanis J, Samocha KE, Wiel L, et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature. 2020;586(7831):757–62. doi: 10.1038/s41586-020-2832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaulieu CL, Majewski J, Schwartzentruber J, et al. FORGE Canada Consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am J Hum Genet. 2014;94(6):809–17. doi: 10.1016/j.ajhg.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright CF, Fitzgerald TW, Jones WD, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385(9975):1305–14. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright CF, Hurles ME, Firth HV. Principle of proportionality in genomic data sharing. Nat Rev Genet. 2016;17(1):1–2. doi: 10.1038/nrg.2015.5. [DOI] [PubMed] [Google Scholar]
- 9.Wright CF, Middleton A, Barrett JC, et al. Returning genome sequences to research participants: Policy and practice. [version 1; peer review: 2 approved] Wellcome Open Res. 2017;2:15. doi: 10.12688/wellcomeopenres.10942.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Middleton A, Parker M, Wright CF, Bragin E, Hurles ME. DDD Study. Empirical research on the ethics of genomic research. Am J Med Genet A. 2013;161A(8):2099–101. doi: 10.1002/ajmg.a.36067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Köhler S, Doelken SC, Mungall CJ, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42(Database issue):D966-74. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firth HV, Richards SM, Bevan AP, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84(4):524–33. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519(7542):223–8. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542(7642):433–8. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright CF, McRae JF, Clayton S, et al. Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families withdevelopmental disorders. Genet Med. 2018;20(10):1216–23. doi: 10.1038/gim.2017.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramu A, Noordam MJ, Schwartz RS, et al. DeNovoGear: de novo indel and point mutation discovery and phasing. Nat Methods. 2013;10(10):985–7. doi: 10.1038/nmeth.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thormann A, Halachev M, McLaren W, et al. Flexible and scalable diagnostic filtering of genomic variants using G2P with Ensembl VEP. Nat Commun. 2019;10(1):2373. doi: 10.1038/s41467-019-10016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright CF, Eberhardt RY, Constantinou P, Hurles ME, FitzPatrick DR, Firth HV. Evaluating variants classified as pathogenic in ClinVar in the DDD Study. Genet Med. 2020 doi: 10.1038/s41436-020-01021-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright CF, Prigmore E, Rajan D, et al. Clinically-relevant postzygotic mosaicism in parents and children with developmental disorders in trio exome sequencing data. Nat Commun. 2019;10(1):2985. doi: 10.1038/s41467-019-11059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright CF, Quaife NM, Ramos-Hernández L, et al. Non-coding region variants upstream of MEF2C cause severe developmental disorder through three distinct loss- of-function mechanisms. Am J Hum Genet. 2021;108(6):1083–94. doi: 10.1016/j.ajhg.2021.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lord J, Gallone G, Short PJ, et al. Pathogenicity and selective constraint on variation near splice sites. Genome Res. 2019;29(2):159–70. doi: 10.1101/gr.238444.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardner EJ, Sifrim A, Lindsay SJ, et al. Detecting cryptic clinically relevant structural variation in exome-sequencing data increases diagnostic yield for developmental disorders. Am J Hum Genet. 2021;108(11):2186–94. doi: 10.1016/j.ajhg.2021.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardner EJ, Prigmore E, Gallone G, et al. Contribution of retrotransposition to developmental disorders. Nat Commun. 2019;10(1):4630. doi: 10.1038/s41467-019-12520-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eberhardt RY, Wright CF, FitzPatrick DR, Hurles ME, Firth HV. Detection of mosaic chromosomal alterations in children with severe developmental disorders recruited to the DDD study. medRxiv. 2022 [Google Scholar]
- 25.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellard S, Baple E, Callaway A, Berry I, Forrester N Clare. ACGS Best Practice Guidelines for Variant Classification in Rare Disease 2020. 2020. [Google Scholar]
- 27.Tavtigian SV, Greenblatt MS, Harrison SM, et al. Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet Med. 2018;20(9):1054–60. doi: 10.1038/gim.2017.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aitken S, Firth HV, Wright CF, Hurles ME, FitzPatrick DR, Semple CA. IMPROVE-DD: Integrating Multiple Phenotype Resources Optimises Variant Evaluation in genetically determined Developmental Disorders. HGG Adv. 2023;4(1):100162. doi: 10.1016/j.xhgg.2022.100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin HC, Jones WD, McIntyre R, et al. Quantifying the contribution of recessive coding variation to developmental disorders. Science. 2018;362(6419):1161–4. doi: 10.1126/science.aar6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akawi N, McRae J, Ansari M, et al. Discovery of four recessive developmental disorders using probabilistic genotype and phenotype matching among 4,125 families. Nat Genet. 2015;47(11):1363–9. doi: 10.1038/ng.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vissers LELM, de Ligt J, Gilissen C, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42(12):1109–12. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 32.Posey JE, Harel T, Liu P, et al. Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation. N Engl J Med. 2017;376(1):21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srivastava S, Love-Nichols JA, Dies KA, et al. Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet Med. 2019;21(11):2413–21. doi: 10.1038/s41436-019-0554-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blencowe H, Lee ACC, Cousens S, et al. Preterm birth-associated neurodevelopmental impairment estimates at regional and global levels for 2010. Pediatr Res. 2013;74(Suppl 1):17–34. doi: 10.1038/pr.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen S, Zhao S, Dalman C, Karlsson H, Gardner R. Association of maternal diabetes with neurodevelopmental disorders: autism spectrum disorders, attention-deficit/hyperactivity disorder and intellectual disability. Int J Epidemiol. 2021;50(2):459–74. doi: 10.1093/ije/dyaa212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coste J, Blotiere P-O, Miranda S, et al. Risk of early neurodevelopmental disorders associated with in utero exposure to valproate and other antiepileptic drugs: a nationwide cohort study in France. Sci Rep. 2020;10(1):17362. doi: 10.1038/s41598-020-74409-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walsh R, Tadros R, Bezzina CR. When genetic burden reaches threshold. Eur Heart J. 2020;41(39):3849–55. doi: 10.1093/eurheartj/ehaa269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Short PJ, McRae JF, Gallone G, et al. De novo mutations in regulatory elements in neurodevelopmental disorders. Nature. 2018;555(7698):611–6. doi: 10.1038/nature25983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Middleton A, Morley KI, Bragin E, et al. Attitudes of nearly 7000 health professionals, genomic researchers and publics toward the return of incidental results from sequencing research. Eur J Hum Genet. 2016;24(1):21–9. doi: 10.1038/ejhg.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choufani S, Cytrynbaum C, Chung BHY, et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat Commun. 2015;6:10207. doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A Supplementary Table of diagnoses per gene is provided. Individual-level datasets are available under managed access for research into developmental disorders via the European Genome-phenome Archive (EGAS00001000775). Individual pathogenic/likely pathogenic variants are openly accessible with phenotypes via DECIPHER, and are provided in a Supplementary File in the European Genome-Phenome Archive.