Abstract
β-arrestin plays a key role in G protein-coupled receptor (GPCR) signaling and desensitization. Despite recent structural advances, the mechanisms that govern receptor-β-arrestin interactions at the plasma membrane of living cells remain elusive. Here, we combine single-molecule microscopy with molecular dynamics simulations to dissect the complex sequence of events involved in β-arrestin interactions with both receptors and the lipid bilayer. Unexpectedly, our results reveal that β-arrestin spontaneously inserts into the lipid bilayer and transiently interacts with receptors via lateral diffusion on the plasma membrane. Moreover, they indicate that, following receptor interaction, the plasma membrane stabilizes β-arrestin in a longer-lived, membrane-bound state, allowing it to diffuse to clathrin-coated pits separately from the activating receptor. These results expand our current understanding of β-arrestin function at the plasma membrane, revealing a critical role for β-arrestin preassociation with the lipid bilayer in facilitating its interactions with receptors and subsequent activation.
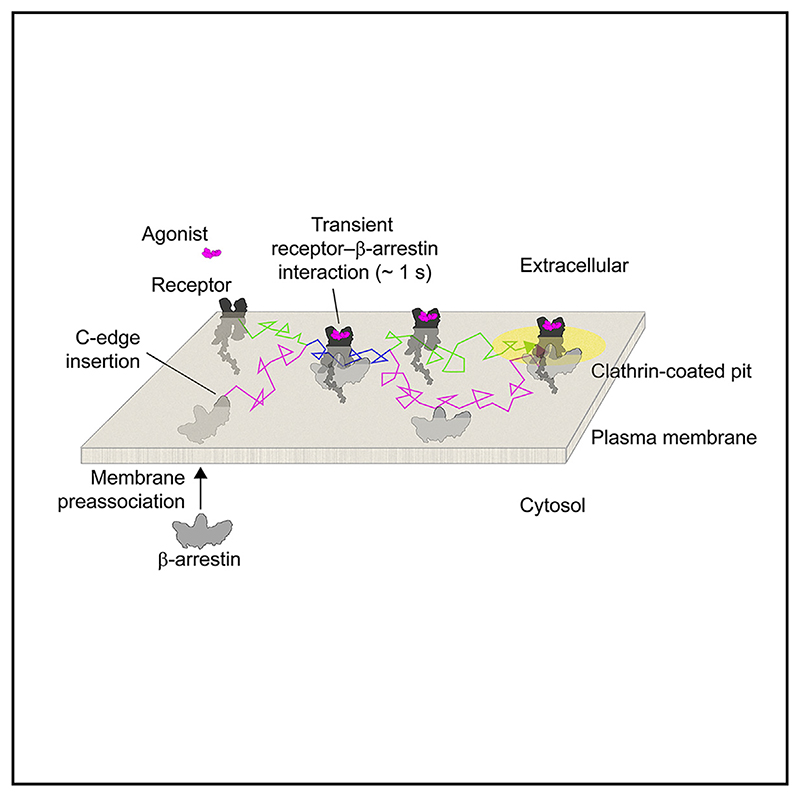
Graphic abstract.
Introduction
G protein-coupled receptors (GPCRs) are implicated in virtually every physiological process and are major drug targets.1,2 Following agonist-mediated GPCR activation and phosphorylation by G protein-coupled-receptor kinases (GRKs), β-arrestins translocate from the cytosol to bind agonist-occupied, phosphorylated receptors on the plasma membrane. There are four arrestins—two visual arrestins (also known as arrestin 1 and 4), β-arrestin 1 (βArr1) (arrestin 2), and β-arrestin 2 (βArr2) (arrestin 3).
By interacting with the receptor core, β-arrestins mediate rapid signal desensitization.3 In addition, β-arrestins trigger receptor internalization via interaction with the adaptor protein 2 (AP2) and clathrin heavy chain.4 Moreover, β-arrestins have been proposed to mediate “non-classical” G protein-independent effects,5 providing a mechanism for “biased” signaling.6 This diversity of functions has been linked to multiple conformations in receptor-arrestin complexes revealed by structural and biophysical studies with purified proteins.7–17 Furthermore, recent findings of prolonged β-arrestin activation18–20 suggest that β-arrestin signaling might be more complex than previously thought. However, how this complexity operates on the plasma membrane of living cells remains largely unknown.
Here, we combined an innovative multicolor single-molecule microscopy approach21,22 with molecular dynamics (MD) simulations to dissect the sequence of events in receptor-β-arrestin interactions at the plasma membrane of living cells with ~20 nm spatial and 30 ms temporal resolution.21,22 Our results reveal that β-arrestin binds directly to the lipid bilayer, allowing it to transiently interact with receptors via lateral diffusion, and reaches clathrin-coated pits (CCPs) separately from the activating receptor.
Results
Single-molecule imaging reveals spontaneous membrane translocation and lateral diffusion of βArr2
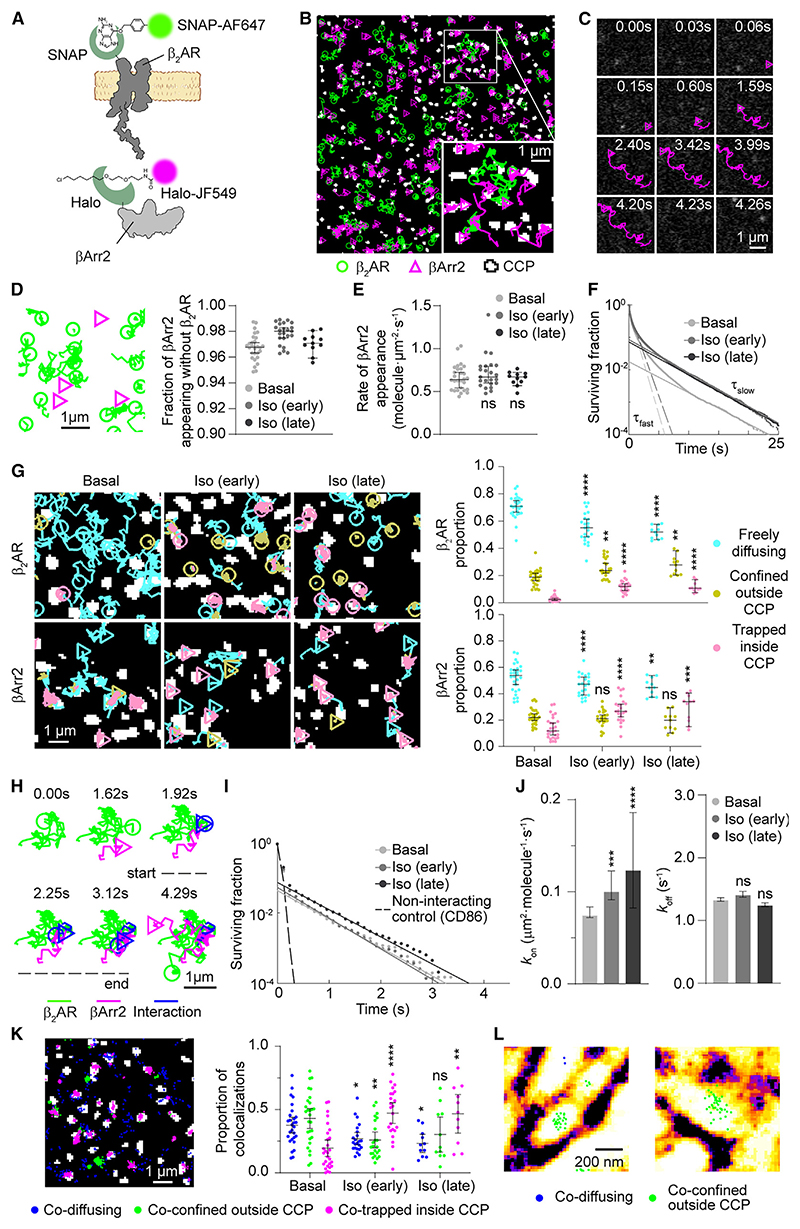
As a main model, we chose βArr2 and the β2-adrenergic receptor (β2AR), a prototypical family-A GPCR that regulates numerous physiological processes.23,24 To investigate the behavior of individual βArr2 and β2AR molecules on the plasma membrane of living cells, we labeled them with two distinct organic fluorophores via fusion of Halo25 and SNAP26 tags to their C and N termini, respectively (Figure 1A). Both constructs were transiently expressed at low physiological levels in Chinese hamster ovary (CHO) cells, which have no detectable β1AR/β2AR.27 Both βArr2-Halo and SNAP-β2AR constructs are functional; βArr2-Halo binds receptors and mediates receptor internalization to a similar extent as wild-type (WT) βArr2, and co-localizes with internalized receptors in endosomes (Figures S1A–S1D; see STAR Methods for details). CHO cells were then labeled with saturating concentrations of both organic fluorophores (Figure S1E) and were simultaneously imaged by fast multicolor total internal reflection fluorescence (TIRF) microscopy combined with single-particle tracking22 (Figure 1B; Videos S1 and S2); unspecific labeling was <1% (Figure S1F). We additionally visualized CCPs by cotransfection of GFP-labeled clathrin light chain. Data were acquired both under basal conditions and after early (2–7 min) and late (8–15 min) stimulation with the β-adrenergic full agonist isoproterenol (Iso). An excess of 5.8 million individual molecular trajectories were acquired and analyzed in this study. Numbers of trajectories and particle densities in the analyzed groups are given in Table S1.
Figure 1. Single-molecule imaging of β2AR and βArr2.
(A) Labeling strategy.
(B) Representative single-molecule results.
(C) Representative trajectory of a βArr2 molecule appearing and transiently diffusing on the plasma membrane.
(D) Sites of βArr2 appearance on the plasma membrane.
(E) Rates of βArr2 appearance on the plasma membrane.
(F) Survival curves of βArr2 molecules at the plasma membrane.
(G) Diffusivity states of β2AR and βArr2 molecules.
(H) Transient single-molecule co-localization event between β2AR and βArr2 on the plasma membrane.
(I) Survival curves of β2AR-βArr2 interactions, based on deconvolution of apparent co-localization times. CD86 was used as a non-interacting control.
(J) Estimated kon and koff of β2AR-βArr2 interactions.
(K) Representative spatial map (left) and overall distributions (right) of β2AR-βArr2 co-localization events in cells stimulated with isoproterenol (10 μM; late), color-coded based on the diffusivity states of the involved molecules.
(L) β2AR-βArr2 single-molecule co-localizations over super-resolved (SRRF) image of actin filaments.
Data are median ± 95% confidence interval. Differences in (G), (J) (kon), and (K) are statistically significant by Kruskal-Wallis test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 versus basal by t test with Bonferroni correction. ns, statistically not significant.
The results revealed that βArr2 molecules stochastically translocate from the cytosol to the plasma membrane, resulting in their sudden appearance in the TIRF field (Figure 1C). Unexpectedly, the newly translocated βArr2 molecules often diffused on the plasma membrane for multiple frames before disappearing (Figure 1C). Since photobleaching occurred on a much longer timescale (τ ~ 52 s) (Figure S1G), their disappearance was mostly due to membrane dissociation. Of note, the majority (>95%) of detected βArr2 molecules appeared at membrane sites that were not occupied by β2ARs (Figure 1D). Although we observed accumulation of βArr2 molecules on the plasma membrane after isoproterenol stimulation (Figure S1H), the rate of their appearance on the membrane was remarkably similar between basal and stimulated conditions (Figure 1E). We therefore investigated the membrane residency time of βArr2 molecules. Under basal conditions, the corresponding survival curve had one larger, fast component (τfast = 0.58 s) and one smaller, slow component (τslow = 4.53 s) (Figure 1F). We hypothesized that these might correspond to inactive β-arrestins that only briefly interact with the plasma membrane and “active” β-arrestins stabilized at the plasma membrane, respectively. Isoproterenol caused a 4.1-/4.8-fold increase in the magnitude of the second component (i.e., the number of slowly dissociating βArr2 molecules), without affecting the τ values of either component (τfast = 0.65/0.64 s and τslow = 4.26/4.21 s for early/late stimulation) (Figure 1F).
Both β2AR and βArr2 molecules showed heterogeneous diffusion and alternated phases of confinement and diffusion as revealed by a spatial confinement analysis28 (Figure 1G). Isoproterenol stimulation caused ~4- and ~2-fold increases in the frequency of β2AR and βArr2 molecules trapped in CCPs and a modest increase (~1.4-fold) in that of β2ARs confined outside CCPs (Figure 1G). The survival curves of the corresponding states had a fast and a slow component. Isoproterenol increased the relative amplitude of the slow component (τ ~ 3.5 s) of βArr2 trapped inside CCPs. As expected, no agonist-dependent changes were observed in the absence of β2AR (Figures S1I and S1J).
These results suggest that βArr2 spontaneously associates with the plasma membrane, and its agonist-dependent membrane accumulation is mainly due to an increase in the fraction of slowly dissociating βArr2 molecules.
Initial β2AR-βArr2 interactions at the plasma membrane are highly dynamic and occur via lateral diffusion
We then asked how and for how long β2ARs and βArr2 interact at the plasma membrane of living cells. Surprisingly, we found that most new single-molecule β2AR-βArr2 co-localizations were highly transient and often involved laterally diffusing β2AR and βArr2 molecules (Figure 1H). To estimate the frequency and duration of the underlying interactions, we applied our previously developed method based on deconvolution of apparent colocalization times with those of random co-localizations22 (Figure 1I), which we estimated by replacing β2AR with the unrelated integral membrane protein CD8622 expressed at comparable levels (0.53 ± 0.12 molecules · μm–2). We estimated that β2AR-βArr2 interactions that lead to the formation of true complexes, i.e., excluding unproductive collisions,22 occurred with an association rate constant (kon) of 0.075 μm2 · molecule–1 · s–1 (95% confidence interval: 0.072–0.083) and lasted on average only ~0.7 s (dissociation rate koff = 1.34 s–1; 95% confidence interval: 1.31–1.37) in the absence of agonist (Figure 1J)—this is considerably shorter than the average lifetime on the plasma membrane of the slowly dissociating component of βArr2 molecules (τ ~4.5 s) (Figure 1F). Isoproterenol stimulation caused ~1.3- and ~1.7-fold increases in kon at early and late time points, respectively, while causing no significant changes in koff (Figure 1J). Similar interaction times (~0.6 s) were observed between β2AR and a previously used βArr2 construct tagged with Halo N-terminally (Halo-βArr2) instead of C-terminally29 (Figure S1K).
Under basal conditions, β2AR-βArr2 co-localizations mainly involved freely diffusing molecules (37%) or molecules confined outside CCPs (47%) (Figure 1K), which could be explained by co-trapping in nanodomains defined by the actin cytoskeleton (Figure 1L) similar to receptor-G protein interactions.22 As expected, isoproterenol stimulation increased (~3-fold) the proportion of single-molecule co-localization events between trapped molecules in CCPs (Figure 1K).
These results revealed that the initial β2AR-βArr2 interactions mostly occur via lateral diffusion, are highly transient, and are mainly controlled by kon.
Receptors with varying affinity for βArr2 show different association rates but similar dissociation rates
Seminal studies identified two main classes of GPCRs based on their interactions with β-arrestins and trafficking properties.30,31 Class A GPCRs, such as the β2AR, bind β-arrestins relatively weakly and appear to dissociate from β-arrestins during internalization. In contrast, class B GPCRs have been suggested to bind β-arrestins more strongly and co-internalize with receptors in endosomes where they co-localize for extended periods of time.31 Typical examples of class B GPCRs are the vasopressin V2 receptor (V2R) or the β2V2R chimeric receptor,8,30 which carries the C-tail of the V2R fused to the β2AR core.
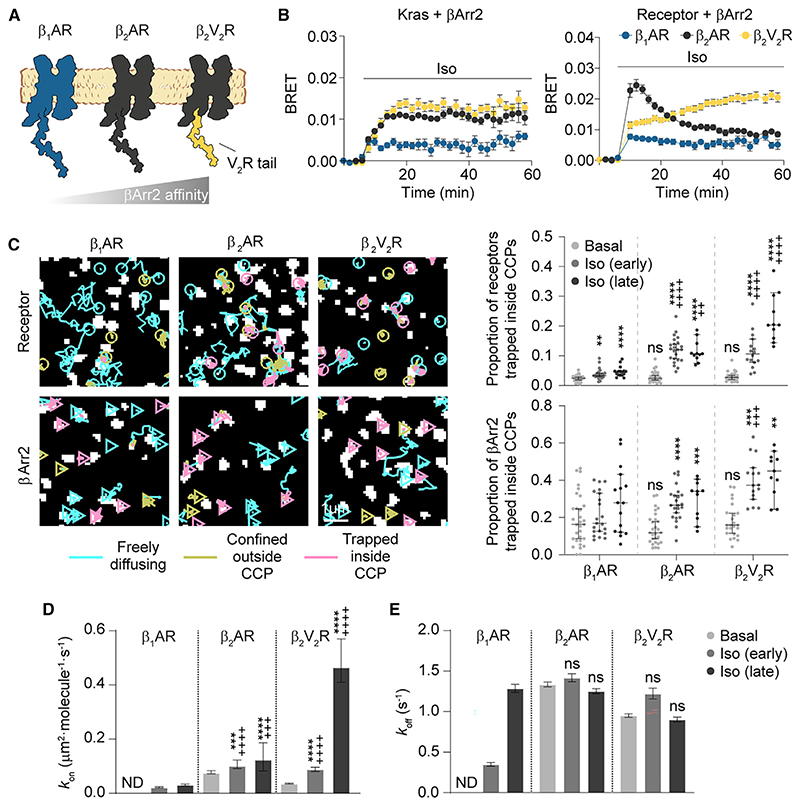
To explore possible differences among receptors, we compared the β2AR with two additional GPCRs—the β1AR, which has an even weaker interaction with βArr2 than β2AR, and the β2V2R, widely used as a model of strong βArr2 association8,10 (Figure 2A). All receptors showed similar lateral diffusion on the plasma membrane (Figures S2A–S2C). Real-time bioluminescence resonance energy transfer (BRET) measurements (Figure 2B) showed relatively weaker βArr2 plasma membrane translocation in the case of the β1AR and persistent, strong βArr2 interaction with β2V2R at late time points, as expected. Single-molecule imaging revealed a stronger accumulation of β2V2R than β2AR in CCPs after isoproterenol, with only a minor increase for β1AR (Figures 2C and S2B). βArr2 accumulation in CCPs followed the same order of the receptors, albeit with higher basal and stimulated levels (Figures 2C and S2B), indicating that receptor and β-arrestin accumulation are not stoichiometric.
Figure 2. Affinity for receptor C-tail governs βArr2 interaction with receptors and plasma membrane behavior.
(A) Schematic of the investigated receptors.
(B) Kinetics of βArr2 recruitment to the plasma membrane (Kras) and receptor upon isoproterenol (10 μM) stimulation monitored by BRET.
(C) Diffusivity states of receptor and βArr2 molecules. Trajectories (left) are after isoproterenol stimulation (10 μM; late).
(D and E) Estimated kon (D) and koff (E) values for β1AR, β2AR, and β2V2R-βArr2 interactions.
Data are mean ± SEM in (B) and median ± 95% confidence interval in (C)–(E). Results in (C) and (D) are statistically significant by Kruskal-Wallis test. **p < 0.01, ***p < 0.001, ****p < 0.0001 versus corresponding basal condition, and ++p<0.01, +++p < 0.001, ++++p < 0.0001 versus corresponding β1AR condition by t test with Bonferroni correction. ns, statistically not significant.
Basal β1AR-βArr2 interactions estimated by deconvolution were undistinguishable from those with CD86 (Figure 2D), consistent with the low basal affinity of β1AR for βArr2. In contrast, we detected basal β2V2R-βArr2 interactions (kon = 0.035 μm2 · molecule–1 ·s–1; 95% confidence interval: 0.034–0.038) as for β2AR. Isoproterenol increased kon for all three receptors (β1AR ND; β2AR ~1.7-fold; β2V2R ~13-fold; Figure 2D), in good agreement with their βArr2 affinity. Relatively smaller differences were observed among koff values (Figure 2E), with estimated average interaction times ~1 s for all conditions, except for β1AR after early stimulation (~2.9 s).
As for β2AR, basal co-localization between β1AR or β2V2R and βArr2 molecules mainly involved free/confined molecules outside CCPs, with a variable increase inside CCPs after isoproterenol stimulation (Figure S2D).
These results further support the notion that initial receptor-β-arrestin interactions are mainly controlled by kon and are short lived, even in the case of the strongly interacting β2V2R.
Sequence of events
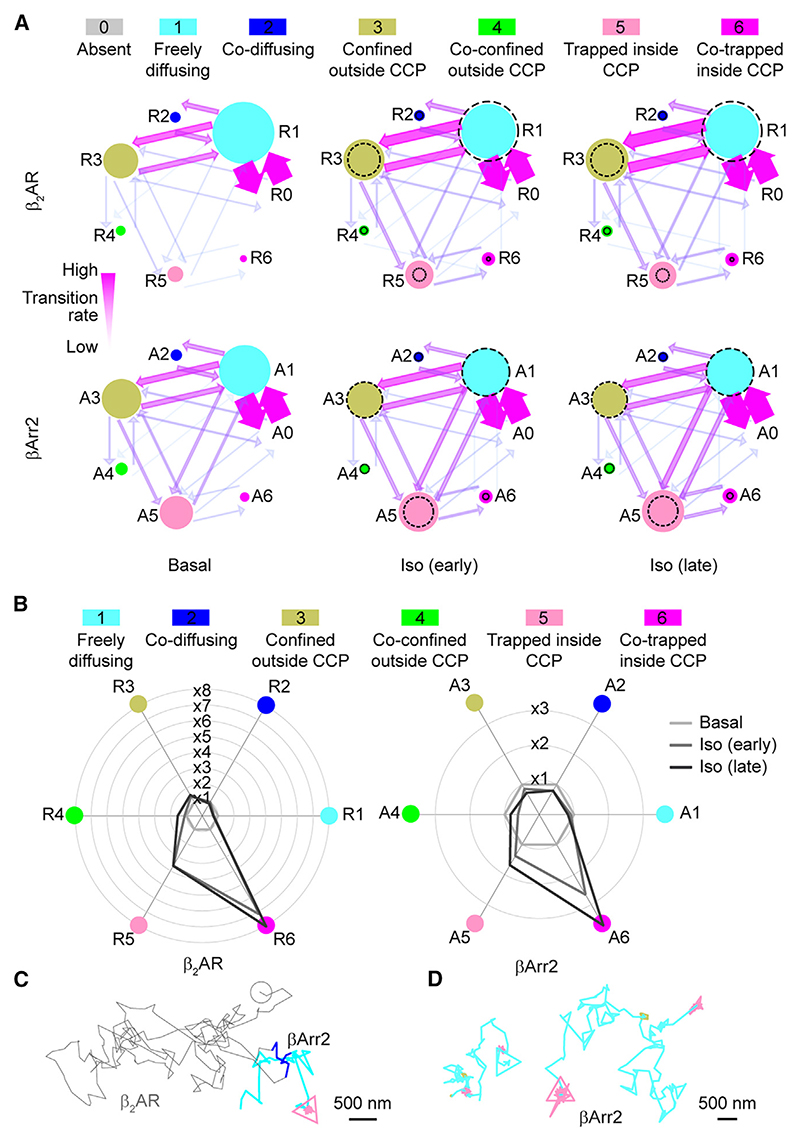
To further dissect the sequence of events in β2AR-βArr2 interactions, we assigned to each molecule at each frame a state (R1–6 and A1–6 for receptor and β-arrestin, respectively) defined by their motion (free/confined), mutual co-localization (present/absent), and trapping at CCPs (present/absent). A dummy state (R0/A0) was added to represent molecules before/after their appearance/disappearance from the plasma membrane (Figure 3A; Table S2). This information was used to build Markov chains describing receptor and β-arrestin state occupancies and transitions (Figure 3A; Table S3). Under basal conditions, β2ARs were prevalently exchanging between free diffusion (R1) and confinement outside CCPs (R3), with a small fraction trapped in CCPs alone (R5) (Figure 3A). Only a minor fraction was co-diffusing with βArr2 molecules (R2). βArr2 showed a similar pattern, albeit with relatively higher trapping in CCPs (A5) (Figure 3A). Isoproterenol stimulation increased trapping alone in CCPs by 3.6- and 1.7-fold, for β2AR and βArr2, respectively (Figure 3B). Unexpectedly, the transition from co-diffusion (R2, A2) to co-trapping in CCPs (R6, A6) was remarkably low under all conditions (Figure 3A). Instead, the main transition leading to co-trapping in CCPs was from the states corresponding to either molecule trapped alone in CCPs (R5, A5) (Figure 3A).
Figure 3. Sequence of events in β2AR-βArr2 interactions.
(A) Simplified representation of the results of the Markov chain analysis. Dashed circles, corresponding basal occupancies. See Table S3 for full transition probabilities.
(B) Changes in single-molecule state occupancies induced by isoproterenol (10 μM) stimulation. Data are normalized to the corresponding basal levels.
(C) Example of a βArr2 molecule undergoing a transient interaction with a β2AR to then reach a CCP without an accompanying receptor.
(D) Examples of βArr2 molecules visiting multiple CCPs via lateral diffusion.
See also Figures S3 and S4 and Table S5.
We then focused on the sequence of states preceding/following the first time β2AR and βArr2 molecules co-localized (Figures S3A and S3B). A2 (co-diffusion) was preceded in 84% of the cases by A1 (βArr2 diffusing alone) and only in 16% by A0 (absent βArr2), corresponding to βArr2 translocation from the cytosol. No relevant changes were observed after stimulation. Importantly, A2 was immediately followed by A1 in 89% of the cases, indicating that most βArr2 molecules continued to diffuse on the plasma membrane alone after co-diffusing with a receptor. Unexpectedly, the state corresponding to βArr2 co-trapped with β2AR in CCPs (A6) was mainly preceded by βArr2 trapped in CCPs alone (A5), which in turn was often preceded by βArr2 diffusing on the plasma membrane alone (A1), and we only rarely observed co-diffusing β2AR and βArr2 molecules (R2, A2) reaching CCPs together (<1.5%) (Figures S3C and S3D). A small proportion (11%) of co-trapping in CCPs (A6) was preceded by co-confinement just outside CCPs (A4), possibly due to cytoskeletal barriers increasing local βArr2/β2AR concentrations.
Remarkably, we also directly observed βArr2 molecules diffusing on the plasma membrane after transient β2AR-βArr2 co-localization until they reached and became trapped in a CCP alone (Figure 3C). Moreover, we observed βArr2 molecules visiting multiple CCPs via lateral diffusion (Figure 3D), indicating that βArr2 trapping at CCPs is reversible.
Of note, although β2AR and βArr2 mostly diffused to CCPs independently, β2AR and βArr2 molecules eventually became co-trapped in CCPs, consistent with the well-known role of β-arrestin in mediating β2AR CCP accumulation and internalization.32–34 The requirement of β-arrestin for receptor trapping and accumulation in CCPs was further confirmed by experiments in βArr1/2 CRISPR-Cas9 knockout CHO cells, in which βArr2 re-expression was required for β2AR CCP trapping (Figure S1L).
Similar results, albeit with some quantitative differences, were observed for β1AR and β2V2R (Figures S4A and S4B). Remarkably, even in the case of the strongly interacting β2V2R after isoproterenol stimulation, the majority of βArr2 molecules (95% and 91% at early and late stimulation time points, respectively) diffused to CCPs alone.
These results indicate that receptor and β-arrestin molecules mostly reach CCPs separately via lateral diffusion and not via co-diffusion as long-lived complexes.
βArr2 spontaneously inserts into the lipid bilayer
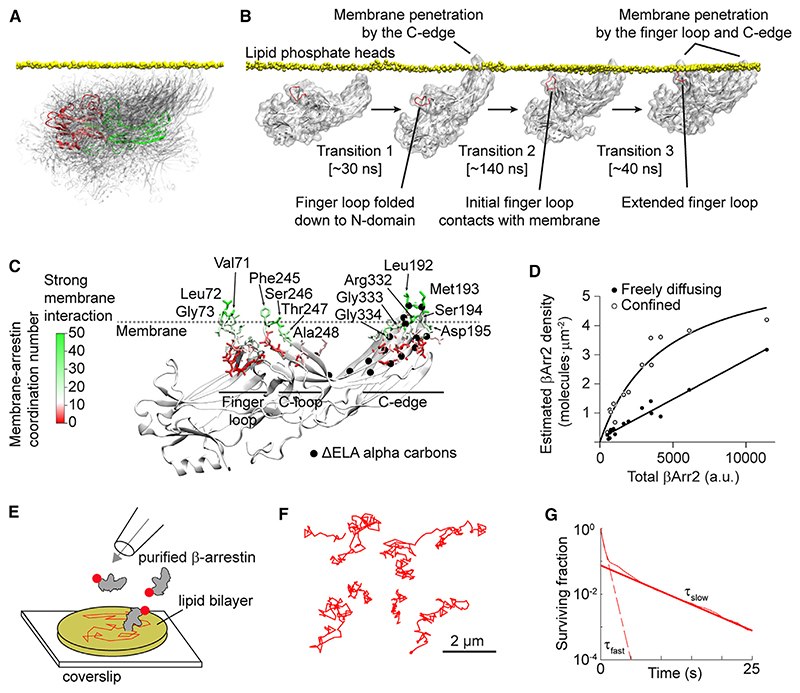
Based on our results and recent structural and biophysical data,15,17,35 we hypothesized that the β-arrestin molecules seen spontaneously translocating to the plasma membrane and laterally diffusing without a receptor might be directly bound to the lipid bilayer. To further explore this hypothesis, we performed MD simulations (40 × 60 ns) starting with βArr2 close to the plasma membrane. As expected, βArr2 in solution assumed a wide range of positions relative to the plasma membrane (Figure 4A). Remarkably, in 4 out of 40 simulations, we observed spontaneous insertion of the βArr2 C-edge into the lipid bilayer (Figure 4B; Table S4). In additional simulations (20 × 200 ns), starting from βArr2 with the C-edge inserted into the lipid bilayer, this was surprisingly followed at times (3/20 simulations) by membrane penetration of the finger loop (Figure 4B), a key β-arrestin region involved in interaction with the receptor core.8,13 Based on extended MD simulations (3 × 1 μs), starting from the fully membrane-anchored conformation obtained in the previous simulations, we further refined a major predicted lipid anchoring site in the C-edge (Leu192, Met193, Ser194, Asp195, Arg332, Gly333, Gly334), as well as two additional sites in the finger loop (Val71, Leu72, Gly73) and C-loop (Phe245, Ser246, Thr247, Ala248) (Figure 4C).
Figure 4. Spontaneous β-arrestin insertion into the lipid bilayer.
(A) Superposition of βArr2 conformations sampled in solution in MD simulations. A selected conformation is highlighted to show the positions of the N- (red) and C- (green) domains.
(B) Linked molecular dynamics (MD) simulations showing spontaneous insertion of the βArr2 C-edge into the lipid bilayer followed by a conformational rearrangement of the finger loop and its penetration into the bilayer.
(C) Extended MD simulations (3 μs accumulated time) of membrane-bound βArr2. The results are shown on a representative structure of fully membrane-inserted βArr2 obtained in the simulations. Color indicates the lipid coordination numbers of βArr2 residues. Main interacting residues (lipid coordination number > 20) are labeled.
(D) Densities of freely diffusing βArr2 molecules on the plasma membrane of cells in which βArr2 expression was varied ~25-fold.
(E) Schematic of the in vitro reconstitution experiments with purified β-arrestin and supported lipid bilayers.
(F) Representative single-molecule trajectories showing lateral diffusion of purified β-arrestin in supported lipid bilayers.
(G) Survival curve of β-arrestin molecules on supported lipid bilayers.
See also Table S5.
In support of our hypothesis, single-molecule experiments in which βArr2 expression was varied ~25-fold showed no saturation of freely diffusing βArr2 molecules on the plasma membrane at high βArr2 expression, consistent with their binding to high abundance sites such as membrane lipids. In contrast, saturation was observed for confined βArr2 molecules, used as a control (Figure 4D).
To provide further evidence for direct binding of β-arrestin to the lipid bilayer, we resorted to a reconstituted system consisting of fluorescently labeled, purified β-arrestin and supported lipid bilayers obtained from giant unilamellar vesicles (GUVs) (Figure 4E). Importantly, also in supported lipid bilayers, we observed spontaneous insertion and lateral diffusion of β-arrestin molecules (Figure 4F). Similar to living cells (Figure 1F), the residency times of β-arrestin molecules on the supported lipid bilayers had a fast and a slow component (τfast = 0.55 s; τslow = 5.56 s) (Figure 4G). Although the relative amplitudes of the two components differed between living cells and lipid bilayers, which could be explained by the far more complex organization of living cells, it is remarkable that their average lifetimes were very similar.
These findings provided strong evidence that β-arrestin can spontaneously insert into lipid bilayers and remain associated with them for several seconds.
βArr2 membrane preassociation drives receptor-β-arrestin interactions
To further test the underlying mechanisms and functional consequences, we took advantage of a well-characterized βArr2 mutant (ΔPIP2) in which the basic amino acids that form a high affinity phosphatidylinositol 4,5-bisphosphate (PIP2) binding site are mutated to glutamine (K233Q/R237Q/K251Q).36,37 A second extended lipid anchoring deficient mutant (ΔELA) (R189Q/F191E/L192S/M193G/T226S/K227E/T228S/K230Q/K231E/K233Q/R237E/K251Q/K325Q/K327Q/V329S/V330D/R332E) (Figure 4C) was designed based on the available structures and our MD simulations to additionally interfere with C-edge lipid interactions. MD simulations of the ΔELA mutant predicted that the substitutions should not alter βArr2 overall folding (data not shown). Furthermore, the ΔELA mutant could be activated in vitro by a phosphopeptide corresponding to the C-terminal region of the V2R, as shown by immunoprecipitation with Fab30 (Figure S1M), a synthetic antibody fragment that selectively recognizes an active conformation in βArr1/2.7,11
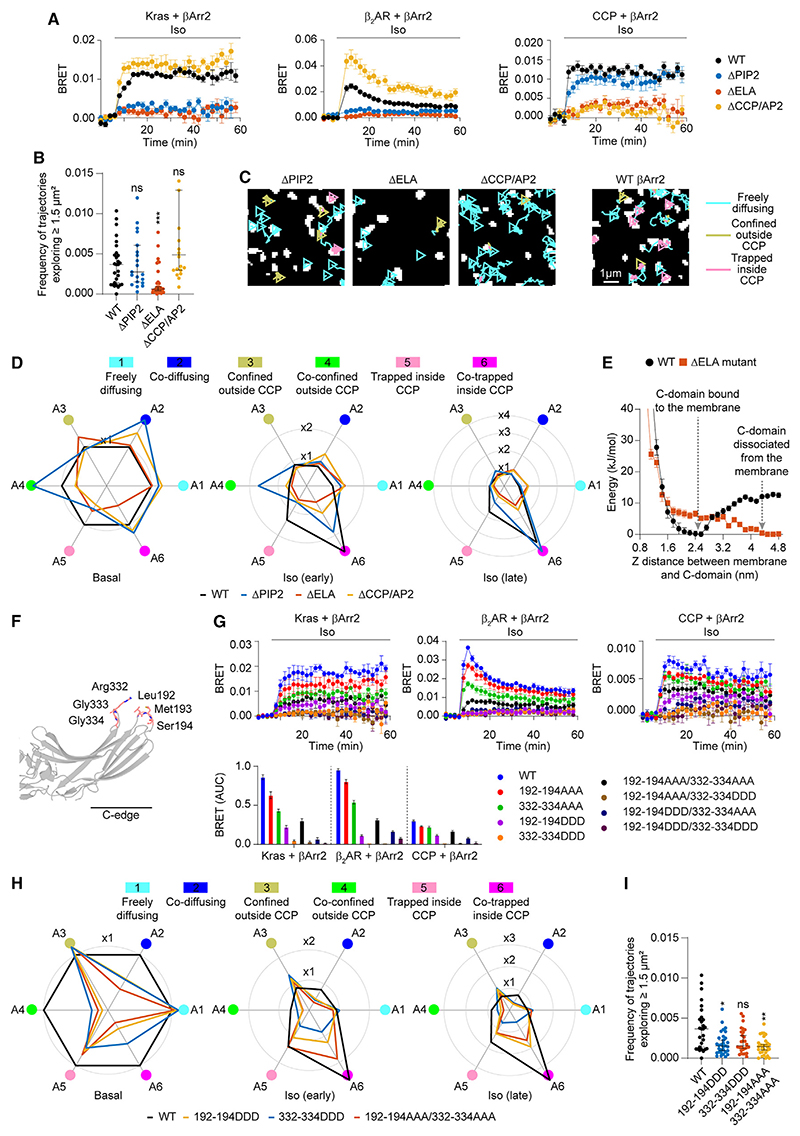
Mutating the PIP2 binding site alone (ΔPIP2) interfered with agonist-dependent increases in βArr2 binding to β2AR as well as to the plasma membrane and slowed down its accumulation at CCPs (Figure 5A), consistent with a role of PIP2 in facilitating receptor-β-arrestin interactions.16,38 However, it did not alter the basal frequency of βArr2 molecules exploring space via lateral diffusion (Figures 5B and 5C). In contrast, the ΔELA mutant was not only largely defective in agonist-dependent translocation and CCP accumulation (Figures 5A and 5D) but also in its ability to preassociate with and diffuse laterally on the plasma membrane (Figures 5B and 5C). These results were further supported by metadynamics MD simulations with WT βArr2, which revealed a low energy well at a 2.5-nm distance from the lipid bilayer, corresponding to βArr2 with the C-edge inserted in the bilayer, which was lost in the case of the ΔELA mutant (Figure 5E).
Figure 5. Functional consequences of βArr2 membrane preassociation.
(A) Kinetics of βArr2 mutant(ΔPIP2, ΔELA, ΔCCP/AP2) recruitment to the plasma membrane (Kras; left), β2AR(middle), or CCPs(right) upon isoproterenol (10 μM) stimulation monitored by BRET.
(B) Propensity of βArr2 mutants to explore the plasma membrane.
(C) Diffusivity states of βArr2 mutants. Shown are representative trajectories after stimulation with isoproterenol (10 μM; late).
(D) Changes in single-molecule state occupancies induced by isoproterenol (1 0 μM) stimulation. Data are normalized to Halo-tagged WT βArr2 with SNAP-tagged WT β2AR basal.
(E) Well-tempered metadynamics simulations comparing the ΔELA mutant and WT βArr2. Shown are the free-energy landscapes as the proteins are pulled out of the membrane, using as collective variable the distance between the C-domain and the plasma membrane.
(F) Positions of the targeted mutations introduced in the C-edge of βArr2.
(G) Kinetics of targeted C-edge mutant recruitment to the plasma membrane (Kras; left), β2AR (middle), or CCPs (right) upon isoproterenol (10 μM) stimulation monitored by BRET.
(H) Changes in single-molecule state occupancies of targeted C-edge mutants induced by isoproterenol (10 μM) stimulation. Data are normalized to Halo-tagged WT βArr2 with SNAP-tagged WT β2AR basal.
(I) Propensity of targeted C-edge mutants to explore the plasma membrane. Shown are the relative frequencies of molecules exploring ≥1.5 μm2 in unstimulated cells.
Halo-tagged WT βArr2 is included in (A)-(D) and (G)-(I) for comparison. Data are mean ± SEM in (A) and (G) and median ± 95% confidence interval in (B) and (I). Differences in (B) and (I) are statistically significant by Kruskal-Wallis test. *p < 0.05, **p < 0.01, ***p < 0.001 versus Halo-tagged WT by t test with Bonferroni correction.
Prompted by these results, we designed targeted mutations in the C-edge to more selectively impair C-edge-mediated βArr2 membrane anchoring. We generated eight constructs, in which we mutated two separate groups of three aa each in two C-edge loops (Leu192/Met193/Ser194 and Arg332/Gly333/Gly334) that were predicted to interact with the lipid bilayer in our MD simulations (Figure 5F). Further MD simulations suggested that these mutations might be sufficient to destabilize C-edge membrane binding (Table S4). All eight mutants were then tested in BRET experiments in the presence of β2AR. Substituting all six aa to alanine (192-194AAA/332-334AAA) or mutating only one of the two groups to aspartate (192-194DDD or 332-334DDD) largely impaired agonist-dependent βArr2 membrane recruitment, β2AR interaction, and accumulation in CCPs (Figure 5G). These three mutants were then further investigated by single-molecule microscopy, which showed that they could not be efficiently activated by agonist stimulation (Figure 5H) and were defective in membrane exploration via lateral diffusion (Figure 5I). In spite of this, all three mutants could be activated in vitro by the V2R C-tail phosphopeptide (Figure S1M).
Our hypothesis was further supported by experiments with a clathrin/AP2 binding-deficient βArr2 mutant (ΔCCP/AP2), which, instead of accumulating in CCPs, continued diffusing laterally on the plasma membrane after isoproterenol stimulation (Figures 5A-5D). This also ruled out the possibility that the laterally diffusing βArr2 molecules might be bound to the plasma membrane via clathrin/AP2.
These results suggest that βArr2 binds directly to the plasma membrane with major contribution of the identified C-edge loops, whereas the known PIP2 binding site appears dispensable. Moreover, they indicate that βArr2 membrane preassociation is required for efficient receptor interaction and CCP accumulation.
Role of core and C-tail interactions
Binding of β-arrestin to receptors has been shown to involve two distinct interactions (Figure 6A): a first one between a polar core in β-arrestin N-domain and the receptor phosphorylated C-tail and a second one between β-arrestin finger loop and the receptor core. Both interactions have been reported to participate in receptor-β-arrestin binding, albeit with variable contribution among receptors.8,13,19,39–42
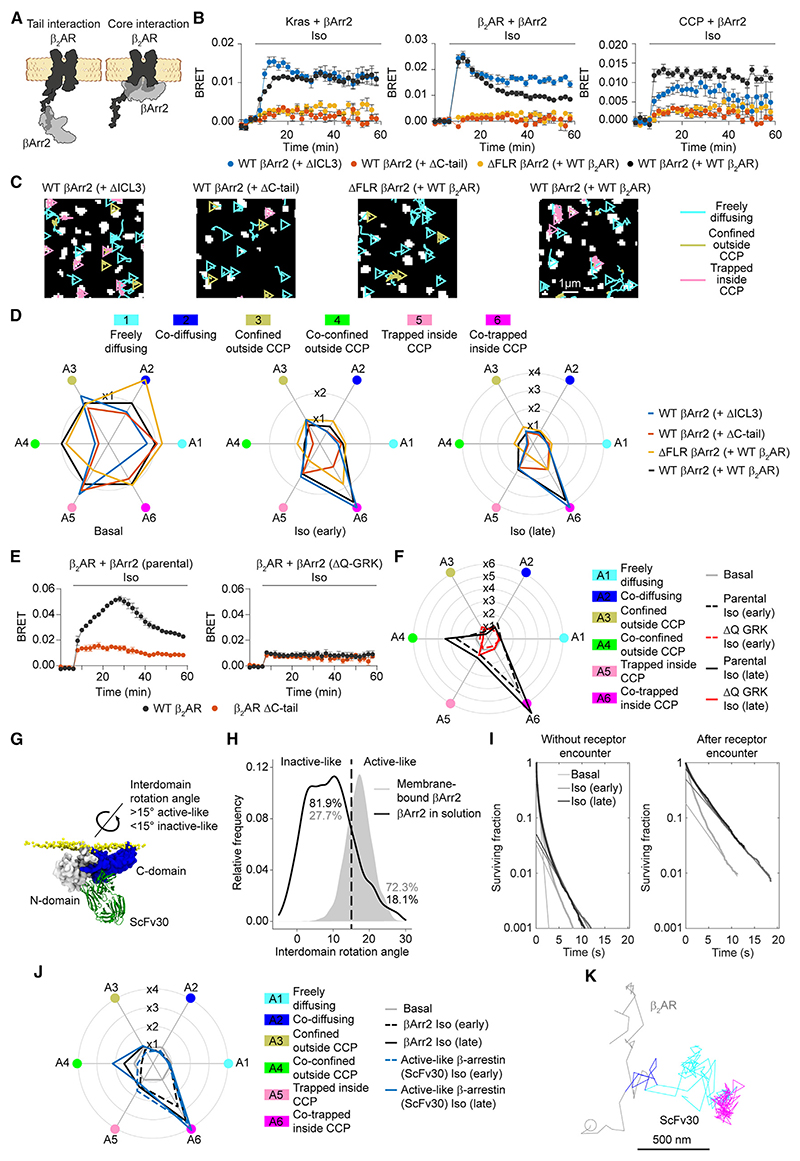
Figure 6. Mechanisms of βArr2 activation and stabilization at the plasma membrane.
(A) Schematic of C-tail and core receptor-arrestin interactions.
(B) Kinetics of βArr2 recruitment to the plasma membrane (Kras; left), β2AR (middle), or CCPs (right) upon isoproterenol (10 μM) stimulation for the indicated construct combinations monitored by BRET.
(C) βArr2 diffusivity states in cells expressing the same construct combinations. Shown are representative trajectories after stimulation with isoproterenol (10 μM; late). The results with SNAP-tagged WT β2AR and Halo-tagged WT βArr2 are included for comparison.
(D) Corresponding changes in single-molecule state occupancies induced by isoproterenol (10 μM) stimulation. Data are normalized to Halo-tagged WT βArr2 with SNAP-tagged WT β2AR basal, included for comparison.
(E) Kinetics of βArr2 recruitment to β2AR and β2AR ΔC-tail upon isoproterenol (10 μM) stimulation in parental (left) and ΔQ-GRK KO cells (right) monitored by BRET.
(F) Corresponding changes in single-molecule state occupancies induced by isoproterenol (10 μM) stimulation.
(G) Recognition of active-like membrane-bound βArr2 by Fab30/ScFv30. The structural model was obtained by aligning the membrane-bound βArr2 conformation obtained in MD simulations to the crystal structure of the active βArr1-Fab30 complex (PDB: 4JQI).
(H) MD simulations comparing βArr2 conformations in solution and bound to the lipid bilayer.
(I) Survival curves at the plasma membrane of βArr2 molecules without receptor encounter and after receptor encounter.
(J) Radar plot obtained from single-molecule experiments comparing the behavior of ScFv30, recognizing active-like β-arrestin, and total βArr2.
(K) Example of an active-like β-arrestin molecule, visualized with ScFv30, undergoing transient interaction with a β2AR molecule (blue) to then diffuse away alone (cyan) and meet another receptor in a CCP (magenta).
Data are mean ± SEM in (B) and (E).
See also Figures S5, S6, and S7 and Table S5.
To further investigate their contribution with our model receptors, we examined receptor constructs carrying a deletion in either the third intracellular loop (ΔICL3) or C-tail (ΔC-tail). Using bimane fluorescence spectroscopy analyses, the ΔICL3 deletion has been previously demonstrated to virtually completely abolish βArr1 core interaction with the β2V2R and V2R, while retaining comparable βArr1 binding to the phosphorylated receptor C-tails.11,14,43 To further validate this approach, we performed BRET experiments comparing βArr1 and βArr2 recruitment to WT and ΔICL3 β2V2R, which confirmed that the ΔICL3 mutant does not impair agonist-dependent recruitment of either βArr1 or βArr2 to β2V2R—if anything, it makes it faster (Figure S5A). In contrast, the DC-tail mutation largely prevented βArr1/2 recruitment to β2V2R (Figure S5A), consistent with previous ensemble measurements.14 When evaluated in single-molecule experiments, the ΔC-tail but not the ΔICL3 mutation largely interfered with agonist-induced β2V2R-βArr1/2 interactions and their accumulation in CCPs (Figure S5B). While βArr1 and 2 behaved in an overall similar manner, βArr1 showed stronger agonist-dependent changes and co-confinement outside CCPs. Similar results were obtained for the β2AR with βArr2, where the ΔC-tail but not the ΔICL3 mutation hampered agonist-dependent receptor-β-arrestin interactions and CCP accumulation both in BRET and single-molecule experiments, with co-trapping in CCPs being particularly affected (Figures 6B-6D, S6A, and S6B). These results indicate that at least in the case of these receptors, the presence of an intact C-tail is required for efficient receptor-β-arrestin interactions and their co-trapping in CCPs.
Furthermore, we searched for receptors with a documented relevant contribution of the core interaction, which include the V2R and parathyroid hormone receptor (PTHR).29 In BRET experiments, ΔC-tail mutants of both PTHR and V2R retained partial βArr2 recruitment upon agonist stimulation, although at substantially reduced levels compared with the full-length receptors (Figure S7A). The stronger recruitment to full-length receptors was largely, albeit not completely, due to GRK-dependent C-tail phosphorylation, as indicated by experiments in CRISPR-Cas9 edited human embryonic kidney (HEK) cells lacking GRK 2/3/5/6 expression (ΔQ-GRK)29 (Figure S7A). The residual recruitment could be explained by phosphorylation by other kinases or by a contribution of the unphosphorylated C-tail.
Motivated by these results, we performed additional single-molecule experiments on the V2R (Figure S7B). Similar to β1AR, β2AR, and β2V2R, the interactions of full-length V2R with βArr2 mainly involved preassociated, laterally diffusing βArr2 molecules (Figure S4C). Also in this case, agonist stimulation mainly increased kon and the interactions were short lived, lasting ~1.2 and ~2.3 s at early and late time points after stimulation, respectively (Figures S7C and S7D). Single-molecule experiments showed agonist-dependent increases in V2R ΔC-tail-βArr2 accumulation in CCPs, albeit at substantially lower levels than with full-length V2R (Figure S7B). We were also able to detect a small number of single-molecule V2R ΔC-tail-βArr2 interactions (Figure S7E), which were too small to estimate association/dissociation rate constants. Analogous experiments with β2AR in ΔQ-GRK cells indicated that GRK-dependent phosphorylation is required for efficient β2AR-βArr2 interactions and their co-trapping in CCPs (Figures 6E and 6F).
We then introduced a βArr2 mutant carrying a deletion in the finger loop (ΔFLR), which has been shown to interfere with β-arrestin core interaction.13 Although the ΔFLR mutant was capable of binding to the plasma membrane and exploring space via lateral diffusion under basal conditions (frequency of molecules exploring ≥1.5 μm2 ~0.007), it was largely deficient in agonist-dependent translocation to the plasma membrane, β2AR interaction, and accumulation in CCPs (Figures 6B-6D, S6A, and S6B), in striking contrast to the ΔICL3 mutant.
These results indicate that at least for the four tested receptors, the C-tail interaction appears required for efficient β-arrestin interaction and receptor-β-arrestin co-trapping in CCPs. Moreover, they suggest that an intact finger loop is required for βArr2 activation and accumulation in CCPs.
Lipid interactions stabilize βArr2 in a longer-lived state at the plasma membrane
Our MD simulations (Figures 4B and 4C) suggested a previously unrecognized potential interaction with the plasma membrane of the finger loop, whose integrity appears required for efficient β-arrestin activation and accumulation in CCPs. We therefore hypothesized that the interaction with the plasma membrane and possible extension of the finger loop might help stabilize β-arrestin in an “active-like” conformation, where by active-like we refer to a set of conformations that resemble those of β-arrestin in complex with an active receptor and that are characterized by a key inter-domain rotation between its N- and C-domains (inactive-like < 15°, active-like > 15°)44 (Figure 6G).
To explore this possibility, we compared the results of the previous extended MD simulations of βArr2 on the plasma membrane with those of an additional set of 3 × 1 μs simulations of βArr2 in solution, starting in both cases from the conformation that βArr2 spontaneously adopted when it was fully inserted (finger loop, C-loop, and C-edge) in the lipid bilayer (Figure 4B). Monitoring the inter-domain rotation angle as a proxy for βArr2 activation, we confirmed that both membrane-bound and cytosolic βArr2 rapidly sample a wide range of conformational states. Importantly, we observed that once placed in solution, βArr2 rapidly reverts to spending most of its time (81.9%) in the inactive-like state, whereas if associated with the lipid bilayer, it continues to mainly explore the active-like state (72.3%) (Figure 6H).
Based on these simulations, we further hypothesized that transient receptor-β-arrestin interactions may promote the stabilization of β-arrestin on the plasma membrane. To verify this hypothesis, we used our single-molecule data to compute the membrane residency times of βArr2 molecules after transient receptor encounter, compared with the entire residency time of βArr2 molecules that did not encounter a receptor (Figure 6I). Under basal condition, the curve corresponding to βArr2 molecules that do not encounter receptors is mainly characterized by a fast component (τfast ~ 0.4 s) and a second, much smaller slow component (τslow ~ 2 s), which could be attributed to partial, spontaneous βArr2 activation. Remarkably, this second component is larger and longer-lived (τslow ~ 4.3 s) after transient receptor encounter, and its magnitude is further increased by ~4-fold after agonist stimulation, consistent with our hypothesis.
To further test this hypothesis, we took advantage of an intra-body based on ScFv30, a single-chain version of Fab30, that selectively recognizes the active-like rotation in βArr1/28,43,45 (Figure 6G). BRET experiments in βArr1/2 CRISPR-Cas9 knockout cells, in which we re-expressed βArr2, confirmed the ability of ScFv30 to recognize active-like βArr2, as shown by its plasma membrane translocation following isoproterenol stimulation, which did not occur in control knockout cells (Figure S6C). Single-molecule imaging revealed striking similarities between the behavior of ScFv30 and βArr2, including the presence of laterally diffusing ScFv30 molecules with characteristics similar to those of βArr2 and capable of reaching and becoming trapped in CCPs without an accompanying receptor (Figures 6J, 6K, and S6D-S6G). Of note, receptor-ScFv30 interactions lasted on average ~1–2 s (dissociation rate koff = 0.43/1.00 s–1; 95% confidence interval: 0.36–0.47/0.96–1.05 for early/late), which is shorter than the average membrane residency time of slowly dissociating βArr2 molecules (~4.5 s). Moreover, the slow component of the survival curve of ScFv30 on the plasma membrane had decays (τslow = 5.1, 5.4, and 5.0 s, for basal, early, and late, respectively) similar to those observed for βArr2 (Figure S6H). These results suggest that ScFv30 binding does not substantially prolong the lifetime of active-like β-arrestin at the plasma membrane. This view is further supported by the fact that ScFv30 overexpression did not substantially modify the survival curve of βArr2 on the plasma membrane (Figure S6I).
These results provide further evidence that transient interaction with an active receptor catalyzes the conversion of β-arrestin into a longer-lived state at the plasma membrane, allowing it to diffuse to CCPs independently of the activating receptor.
Discussion
According to the current model, which is largely based on ensemble measurements, β-arrestin is assumed to translocate from the cytosol to directly bind an active receptor on the plasma membrane and remain bound to the same receptor at least until they reach a CCP together. In contrast, our single-molecule results reveal an unexpected scenario whereby β-arrestin spontaneously preassociates with the plasma membrane, allowing it to explore space via lateral diffusion and to undergo highly transient interactions with receptors that lead to β-arrestin activation. Importantly, this prolongs the duration of β-arrestin at the plasma membrane, allowing it to reach CCPs independently of the initial, short-lived receptor-β-arrestin complexes.
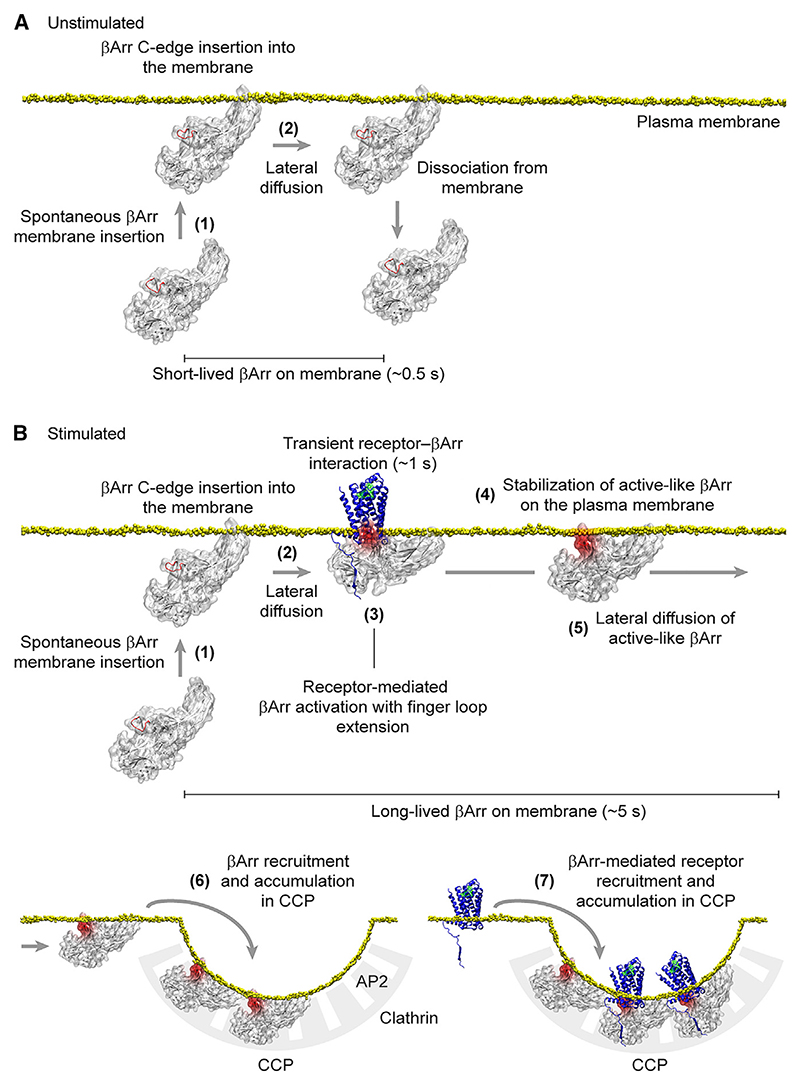
Based on our detailed single-molecule measurements, we propose a revised multistep model for receptor-β-arrestin interactions: (1) β-arrestin spontaneously inserts into the plasma membrane via its C-edge, (2) laterally diffuses on the plasma membrane, (3) transiently (~1 s) interacts with an active receptor via lateral diffusion and becomes activated with extension of the finger loop, (4) is stabilized in a membrane-bound, apparently active-like, conformation with possible involvement of the finger loop, (5) diffuses to CCPs separately from the activating receptor, (6) becomes trapped in CCPs via its interactions with clathrin and AP2, and (7) mediates receptor trapping and accumulation in CCPs (Figure 7).
Figure 7. Proposed model of receptor-β-arrestin interactions.
(A) Behavior of β-arrestin at the plasma membrane under unstimulated conditions. Inactive β-arrestin in the cytosol spontaneously binds to the plasma membrane via insertion of the C-edge into the lipid bilayer (1), allowing it to explore space via lateral diffusion (2). Most β-arrestin molecules remain on the plasma membrane for a short time before dissociating and returning to the cytosol.
(B) Behavior of β-arrestin at the plasma membrane in the presence of a stimulated receptor. After spontaneous insertion into the plasma membrane (1), β-arrestin reaches the receptor via lateral diffusion (2). Transient interaction with the receptor catalyzes β-arrestin activation, including β-arrestin inter-domain rotation and extension of the finger loop (3). Following dissociation from the receptor, the interaction of the extended finger loop with the lipid bilayer likely contributes to stabilizing β-arrestin in a membrane-bound, active-like conformation (4). This causes β-arrestin molecules to stay longer and accumulate on the plasma membrane, allowing them to reach CCPs vial lateral diffusion separately from the activating receptors (5). The increase in the number of active β-arrestin molecules and time they spend diffusing on the plasma membrane leads their recruitment and accumulation in CCPs via interaction with AP2 and clathrin (6). β-arrestin molecules tethered to CCPs bind receptors diffusing on the plasma membrane, also causing their recruitment and accumulation in CCPs (7).
A first key finding of our study is that β-arrestin spontaneously interacts with the lipid bilayer. This reveals a previously unexpected role of the lipid bilayer in facilitating receptor-β-arrestin interactions, which likely occurs via at least three separate mechanisms. First, β-arrestin preassociation with the lipid bilayer via its C-edge restricts β-arrestin in an overall orientation relative to the plasma membrane that resembles the orientation in receptor-β-arrestin complexes. Second, the preassociation of β-arrestin with the plasma membrane increases its local concentration close to the receptors. Third, the switch from 3D diffusion in the cytosol to 2D diffusion on the plasma membrane can reduce the time required to reach a receptor.46,47 Whereas C-edge lipid interactions were previously proposed to contribute to stabilizing the fully engaged visual arrestin-rhodopsin complex,35 our results on β-arrestin reveal an unanticipated role for β-arrestin membrane association in facilitating receptor-β-arrestin interactions and keeping β-arrestin bound to the plasma membrane after transient receptor interaction.
Another key finding of our study is that the initial receptor-β-arrestin interactions on the plasma membrane, i.e., before they both accumulate in CCPs, are highly dynamic. Whereas the exact duration of these key interactions was previously unknown, receptor-β-arrestin complexes were widely assumed to be sufficiently stable to allow receptors and β-arrestins to reach CCPs together (class A receptors) or even to co-internalize and to remain associated in endosomes (class B receptors).30,31,48 In contrast, our single-molecule results reveal that at least for the receptors investigated in this study, the initial receptor-β-arrestin interactions are highly transient, lasting on average only ~1 s. This is also true for β2V2R and V2R, which have been previously reported to co-internalize and to remain co-localized with β-arrestin in endosomes.30 Our findings have a number of important implications. First, they mean that most receptors and β-arrestins must diffuse to CCPs separately. Moreover, they imply that a typical receptor meets multiple β-arrestin molecules during its time on the plasma membrane, permitting a much more dynamic regulation of GPCR signaling than previously thought. This likely occurs at least 2 times—a first one when a receptor and a β-arrestin molecule transiently interact, leading to β-arrestin activation, and a second, crucial one when receptors and β-arrestins meet again in CCPs. Once receptors and β-arrestins are in CCPs, the presence of clathrin, AP2, PIP2, and their high local concentrations likely contribute to further stabilizing receptor-β-arrestin complexes so that they can be efficiently internalized, in line with a wealth of previous observations.32–34
Our finding that the initial receptor-β-arrestin interactions are mainly controlled by their association rate (kon) indicates that, similar to receptor-G protein interactions,22,49 they are not diffusion-limited but rather controlled by conformational changes. This is common for protein-protein interactions that involve large conformational changes like those involved in the formation of receptor-β-arrestin complexes and is essentially the only way to regulate the speed of catalytic reactions, which is typical for signaling proteins.22,50–52 Moreover, our data are in good agreement with previous fluorescence resonance energy transfer (FRET) measurements, suggesting that receptor phosphorylation increases kon for receptor-β-arrestin interactions.53
Several studies have investigated the relative contribution of C-tail and core interactions.8,13,19,39–42 Whereas both interactions have been suggested to independently trigger β-arrestin activation with potentially synergistic effects,20 their relative contribution appears to differ among receptors. For many receptors, the presence of an intact C-tail and GRK-dependent phosphorylation appear required for efficient β-arrestin binding.7,8,11,14,29,54,55 Our results with four model GPCRs are overall in line with this view. Importantly, also in the case of the class B V2R, where the core contribution appears relatively larger and where we can detect single-molecule interactions in the absence of the C-tail, we show that the initial β-arrestin interactions involve its preassociation with the plasma membrane and are short lived, indicating that our proposed mechanism is likely shared independently of the relative contribution of core and C-tail interactions.
Moreover, our study identified the C-edge as the critical region for β-arrestin membrane anchoring. This appears to occur spontaneously as suggested by our MD simulations and results with supported lipid bilayers, albeit at a low rate in unstimulated cells. In contrast, the interaction with an active receptor likely catalyzes this transition, leading to a substantial increase in the amount of active β-arrestin on the plasma membrane. This is consistent with the finding that interactions of proximal phosphate groups in the receptor C-tail can trigger the release of an ionic lock that keeps the finger loop in its inactive conformation.56 This in turn could allow the finger loop to interact with the plasma membrane, stabilizing β-arrestin in a membrane-bound, active-like conformation, capable of reaching CCPs alone to mediate GPCR internalization and non-classical signaling. This view is supported by our finding that agonist stimulation increases the duration of β-arrestin at the plasma membrane and is consistent with previous FRET results indicating that β-arrestin conformational changes last longer than direct receptor interaction after transient receptor activation.18 Since we estimate that active-like β-arrestin dissociates from the plasma membrane much slower than inactive β-arrestin, this emerges as the main reason why β-arrestin accumulates on the plasma membrane after agonist stimulation as opposed to the formation of rather stable, long-lived receptor-β-arrestin complexes, as previously thought.
At the same time, our results indicate that the known β-arrestin PIP2 binding site,36,37 although dispensable for membrane anchoring, plays an important role in facilitating receptor-β-arrestin interactions, consistent with the recent finding of a PIP2 bridge in the structure of the neurotensin receptor 1-βArr1 complex.16 A key role of PIP2 is further supported by a recent study demonstrating that PIP2 can promote an active conformation in β-arrestin and stabilize receptor-β-arrestin complexes.38
These insights were only possible thanks to our single-molecule approach, which allowed us to directly observe the entire life of individual β-arrestin molecules on the plasma membrane of living cells. While being consistent with a wealth of published results, our data explain how the average behaviors of receptor and β-arrestin populations emerge from previously unrecognized, highly dynamic interactions among individual receptors, β-arrestins, and the lipid bilayer.
Altogether, our findings redefine the current model of receptor-β-arrestin interactions by revealing a critical role of β-arrestin binding to the lipid bilayer for efficient β-arrestin interaction with receptors and for accumulation on the plasma membrane.
Limitations of the study
Whereas our findings of β-arrestin preassociation and lateral diffusion should be relevant independently of the receptor under investigation, qualitative and/or quantitative differences in receptor-β-arrestin interactions might exist among the approximately 800 GPCRs encoded in the human genome.
While our experiments show that the Halo-tagged βArr2 construct used in this study is functional and is essentially undistinguishable from untagged WT βArr2 in its ability to bind β2AR and promote its internalization, like for any other modification we cannot completely rule out that the Halo tag might have more subtle consequences on βArr2 function.
Although we did not observe major effects of Fab30/ScFv30 on the lifetime of βArr on the plasma membrane, we cannot completely rule out that it might increase the stability of βArr on the plasma membrane.
Due to technical limitations, it is currently not possible to compute the full process of β-arrestin binding to the plasma membrane, receptor-mediated activation, and membrane unbinding in MD simulations. Therefore, our MD results cannot be used to make quantitative predictions about the relative amount of active/inactive β-arrestin in the cytosol versus on the plasma membrane at equilibrium. Please also note that the βArr2 model used in our MD simulations does not include the flexible distal C-tail of βArr2, which is not resolved in the available structures. We therefore cannot rule out a role for this region in modulating βArr2 interactions with the plasma membrane.
Star*Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| βArr2 polyclonal antibody | ThermoFisher | Cat#PA1-732; RRID: AB_2060256 |
| GAPDH monoclonal antibody | ThermoFisher | Cat#AM4300; RRID: AB_2536381 |
| Peroxidase AffiniPure goat anti-rabbit IgG | Jackson ImmunoResearch Labs | Cat#111-035-144; RRID: AB_2307391 |
| Peroxidase AffiniPure goat anti-mouse IgG | Jackson ImmunoResearch Labs | Cat#115-035-003; RRID: AB_10015289 |
| Bacterial and virus strains | ||
| E. coli BL21(DE3) cells | New England Biolabs | Cat#C2527I |
| Chemicals, peptides, and recombinant proteins | ||
| Lipofectamine 2000 | ThermoFisher | Cat#11668019 |
| Polyethylenimine | Generon | Cat#23966-1 |
| ScFv30 (single-chain version of Fab30) | Shukla et al.7 | N/A |
| Phosphopeptide corresponding to the C-terminal region of the V2R (V2Rpp) | Shukla et al.7 | N/A |
| Furimazine | Promega | Cat#N1120 |
| Isoproterenol hydrocholoride | Tocris | Cat#1747 |
| Arginine-vasopressin | Sigma | Cat#V9879 |
| Parathyroid hormone 1-34 | Bachem | Cat#4011474 |
| QuickExtract DNA Solution | Cambio | Cat#QE0905T |
| Lipofectamine CRISPRMAX | ThermoFisher | Cat#CMAX00008 |
| Alt-R S.p. Cas9 Nuclease | Integrated DNA Technologies | Cat#1081058 |
| HaloTag R110Direct | Promega | Cat#G322A |
| SNAP-Surface Alexa Fluor 647 | New England Biolabs | Cat#S9136S |
| HaloTag Janelia Fluor 549 | Promega | Cat#GA1110 |
| Alexa Fluor 647 C2 Maleimide | Invitrogen | Cat#A20347 |
| Deposited data | ||
| Results of the unbiased MD simulations | This paper | GPCRmd: https://submission.gpcrmd.org/dynadb/publications/1493/ |
| Experimental models: Cell lines | ||
| CHO-K1 cells | ATCC | ATCC-CCL-61 |
| HEK293T cells | ATCC | ATCC-CRL3216 |
| HEK293 βArr1/2 CRISPR/Cas9 KO cells | Schrage et al.57 and O’Hayre et al.58 | N/A |
| HEK293 parental and ΔQ-GRK CRISPR/Cas9 KO cells | Drube et al.29 | N/A |
| CHO-K1 βArr1/2 CRISPR/Cas9 KO cells | This paper | N/A |
| Oligonucleotides | ||
| crRNA targeting βArr1: 5’-ATCGACCTCGTGGACCCTG | Integrated DNA | N/A |
| TGGG-3’ | Technologies | |
| crRNA targeting βArr2: 5’-GCGCGACTTTGTAGACCAC | Integrated DNA | N/A |
| CTGG-3’ | Technologies | |
| Software and algorithms | ||
| MATLAB 2018b | MathWorks | N/A |
| Prism 9.4.1 | GraphPad | N/A |
| Fiji ImageJ | N/A | https://imagej.net/software/fiji |
| Andor Solis 4.31.30023.0 | Andor | N/A |
| Metamorph 7.10.5.476 | Molecular Devices | N/A |
| Zen Black 2012 | Zeiss | N/A |
| PhERAstar Multi-user Reader Control and MARS Data Analysis Software | BMG Labtech | N/A |
| CHOPCHOP web toolbox | Labun et al.59 | https://chopchop.cbu.uib.no |
| MATLAB analysis scripts | This paper | Zenodo: https://doi.org/10.5281/zenodo.7682343; GitHub: https://github.com/CalebiroLab/sm_bArr |
| Other | ||
| Pierce protein L agarose | ThermoFisher | Cat#20510 |
| PVDF membrane | Immobilon-P | Cat#IPVH00010 |
| TetraSpeck fluorescent beads | ThermoFisher | Cat#T7279 |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Davide Calebiro (d.calebiro@bham.ac.uk).
Materials availability
Plasmids generated in this study are available from the lead contact. This study did not generate new unique reagents.
Data and code availability
The raw single-molecule microscopy data reported in this study cannot be deposited in a public repository because of their large size. To request access, contact the lead contact. The results of the unbiased MD simulations have been deposited at GPCRmd and are publicly available as of the date of publication. The accession number is listed in the key resources table.
All original code has been deposited at Zenodo and is publicly available as of the date of publication. The DOI is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental Model and Subject Details
Cell culture and transfection
Chinese hamster ovary K1 (CHO-K1) cells (ATCC) were cultured in phenol red-free Dulbecco’s modified Eagle’s medium (DMEM)/F12, supplemented with 10% FBS, 100 U/ml penicillin and 0.1 mg/ml streptomycin at 37 °C, 5% CO2.
Human embryonic kidney 293T (HEK293T) (ATCC), βArr1/2 CRISPR KO HEK293 (kindly provided by Asuka Inoue),57,58 ΔQ-GRK CRISPR KO HEK293 and parental HEK293 cells29 were cultured in DMEM, supplemented with 10% FBS, 100 U/ml penicillin and 0.1 mg/ml streptomycin at 37 °C, 5% CO2.
For single-molecule microscopy experiments, cells were seeded onto ultraclean 25-mm round glass coverslips at a density of 3 x 105 cells per well. On the next day, they were transfected using Lipofectamine 2000, following the manufacturer’s protocol (ThermoFisher). Cells were labeled and imaged by single-molecule microscopy 3.5-4 hours after transfection to obtain low physiological expression levels.22,27
For BRET experiments, cells were seeded at a density of 7 x 105 (HEK293T and βArr1/2 CRISPR KO HEK293) and 1 x 106 (ΔQ-GRK CRISPR KO HEK293 and parental HEK293) cells per well in a 6-well plate and, on the next day, transfected with Lipofectamine 2000, following the manufacturer’s protocol. After 24 hours, they were resuspended in FluoroBrite phenol red-free DMEM medium supplemented with 4 mM L-glutamine and 5% FBS and replated into poly-D-lysine-coated 96-well white polystyrene Nunc microplates (Sigma) at a density of 1 x 105 cells per well. Forty-eight hours post transfection, the medium was replaced with Hank’s balanced salt solution (HBSS), supplemented with 10 mM HEPES, and containing 100 nM HaloTag R110Direct (Promega) and incubated for 1 h at 37 °C for labeling, followed by addition of 10 μM furimazine (Promega).
For β-arrestin-Fab30 binding experiments, 7 x 105 HEK293T cells were seeded in 15-cm Petri dishes and, upon reaching 80% confluency, transfected with 40 μg DNA and 120 μg 25 kDa linear polyethylenimine (PEI) (Polysciences). Cells were harvested 48 hours post-transfection in phosphate buffered saline (PBS) supplemented with 2 mM ethylenediaminetetraacetic acid (EDTA), resuspended and collected by centrifugation at 1,000 x g for 5 min at 4 °C.
For measurements of β-arrestin expression levels, HEK293T cells were seeded at a density of 1 x 105 per well in a 6-well plate and transfected at 80% confluency with PEI. Cells were harvested two days post-transfection in PBS supplemented with 2 mM EDTA, resuspended and centrifuged at 1,000 x g for 5 min at 4 °C. Cell pellets were resuspended in loading buffer containing 50 mMTris-HCl pH 6.8, 1% sodium dodecyl sulfate (SDS), 10% glycerol, 0.02% bromophenol blue, 2 mM dithiothreitol (DTT).
Cells were routinely tested for mycoplasma contamination.
Generation of βArr1/2 CRISPR-Cas9 KO CHO-K1 cells
βArr1 and 2 were sequentially knocked out in CHO-K1 cells using the Alt CRISPR-Cas9 System (Integrated DNA Technologies). Guide sequences (5’-ATCGACCTCGTGGACCCTGTGGG-3’ and 5’-GCGCGACTTTGTAGACCACCTGG-3’) targeting exons 3 of βArr1 and βArr2, respectively, were designed using the CHOPCHOP web toolbox.59 The guide sequences were synthesized as CRISPR RNAs (crRNAs), which were annealed to a trans-activating CRISPR RNA (tracrRNA) to form a single guide RNA (sgRNA) for each gene. Knockout of βArr2 was carried out by incubating the βArr2 sgRNA with Cas9 protein to form a functional RNP complex, which was then transfected into CHO-K1 cells using the Lipofectamine CRISPRMAX reagent (ThermoFisher), according to the manufacturer’s instructions. After 48 hours, the cells were diluted and seeded in 96-well plates at an average density of 0.5 cells/well to generate single-cell clones. Once confluent, genomic DNA (gDNA) was extracted from each clone using the QuickExtract DNA Solution (Cambio) and the gDNA used in PCR reactions with the following βArr2-specific primers: 5’-GTCTTCAAGAAGTCGAGCCCTA-3’ and 5’-GAATTCCTTCTTCTTCCTGCCT-3’. The resulting PCR fragments were sequenced, and clones containing indels in both βArr2 alleles were kept. One clone was selected and used to knockout βArr1 using the same procedure as for βArr2. The resulting clones were screened for the presence of indels in βArr1 using the following βArr1-specific PCR primers: 5’-GCTCCCTGCCTAGTT CAGAGTA-3’ and 5’-TATTCTGCAGTGTACCTGGTGG-3’. Knockout of both βArr1 and 2 was verified by Western blotting using a rabbit polyclonal antibody recognizing βArr1 and 2. One clone was selected and used in subsequent experiments.
Method Details
Molecular biology
Plasmids encoding N-terminally SNAP-tagged human β1AR (SNAP-β1AR), β2AR (SNAP-β2AR) and CD86 (SNAP-CD86) were reported previously.27 The functionality of the SNAP-β2AR construct has been extensively validated in previous studies.27 The βArr2-Halo construct showed strong agonist-stimulated recruitment to β2AR, plasma membrane and CCPs in BRET measurements (Figure S1A). Moreover, it behaved very similarly to wild-type βArr2 in experiments in βArr1/2 CRISPR/Cas9 knockout cells comparing their binding to the β2AR (Figure S1B) or ability to mediate β2AR internalization (Figure S1C). In addition, it showed the typical pattern of rapid recruitment to the plasma membrane and rapid co-internalization with the V2R in endosomes, as seen with GFP-tagged βArr2, which has been extensively used in previous studies on β-arrestin-dependent receptor internalization and trafficking30,31,60,61 (Figure S1D). A plasmid encoding N-terminally SNAP-tagged human β2AR carrying the V2R C-tail (SNAP-β2V2R) was generated by replacing the C-tail in the SNAP-β2AR construct with that of V2R (ARGRTPPSLGPQDESCTTASSSLAKDTSS). Plasmids encoding N-terminally SNAP-tagged β2AR with a deletion in the third intracellular loop (SNAP-β2AR ΔICL3) or lacking the entire C-tail (SNAP-β2AR ΔC-tail) and N-terminally SNAP-tagged β2V2R with a deletion in the third intracellular loop (SNAP-β2V2R ΔICL3) were generated by PCR using standard procedures. A plasmid encoding N-terminally SNAP-tagged V2R was generated by gene synthesis (Twist Bioscience). A plasmid encoding N-terminally SNAP-tagged V2R carrying the ΔC-tail deletion was generated by PCR. Plasmids encoding bovine βArr2 tagged C-terminally with Halo (βArr2-Halo) or GFP (βArr2-GFP) were generated by replacing CFP with the Halo tag or GFP, respectively, in a previously described βArr2-CFP construct.18 A plasmid encoding C-terminally Halo-tagged bovine βArr1 (βArr1-Halo) was generated by replacing βArr2 with βArr1 in the previous construct. A plasmid encoding C-terminally Halo-tagged bovine βArr2 carrying mutations interfering with binding to both clathrin (L373A/I374A/F376A) and AP2 (R393A/R395A)62,63 (βArr2 ΔCCPAP2-Halo) was generated by PCR mutagenesis. Similar procedures were used to generate plasmids encoding C-terminally Halo-tagged bovine βArr2 with a deletion of the finger loop (YGREDLDVLGLSFRK) (βArr2 ΔFLR-Halo)13 or carrying mutations (K233Q/R237Q/K251Q) that interfere with PIP2 binding (βArr2 ΔPIP2-Halo).36,37 An additional βArr2-Halo construct carrying a panel of mutations (R189Q/F191E/L192S/M193G/T226S/K227E/T228S/K230Q/K231E/K233Q/R237E/K251Q/K325Q/K327Q/V329S/V330D/R332E) designed to prevent plasma membrane interactions (βArr2 ΔELA-Halo) was generated by PCR mutagenesis of the βArr2 ΔPIP2-Halo construct, followed by Gibson assembly.64 Plasmids encoding C-terminally Halo-tagged βArr2 carrying the 192-194AAA, 332-334AAA, 192-194DDD, 332-334DDD, 192-194AAA/332-334AAA, 192-194AAA/332-334DDD, 192-194DDD/332-334AAA, or 192-194DDD/332-334DDD mutations were generated by Gibson assembly, using the βArr2-Halo construct as a template. The ΔELA, 192-194AAA/332-334AAA, 192-194DDD, and 332-334DDD mutant scan be activated in vitro by a phosphopeptide corresponding to the C-terminal region of the V2R (Figure S1M). A plasmid encoding N-terminally Halo-tagged βArr2 was previously described.29 All βArr2-Halo constructs used in this study had roughly similar expression levels in Western blot analyses (Figure S1N).
A plasmid encoding human β2AR containing NanoLuciferase (Nluc) fused to its C-terminus (β2AR-Nluc) was previously described.65 Plasmids encoding β1AR, β2V2R, β2AR ΔICL3 and β2AR ΔC-tail with Nluc fused to their C-termini were cloned from the β2AR-Nluc construct by replacing the β2AR coding sequence with those of the corresponding receptor constructs. Plasmids encoding V2R, V2R ΔC-tail, PTHR, PTHR ΔC-tail and β2V2R ΔICL3 with Nluc fused to their C-termini were generated by gene synthesis (Twist Bioscience). Plasmids encoding K-Ras with Nluc or YFP fused to its N-terminus were kindly provided by Kevin Pfleger.66 A plasmid encoding N-terminally GFP-tagged clathrin light chain (GFP-CCP) was kindly provided by Emanuele Cocucci and Tom Kirchhausen.67 A plasmid encoding N-terminally CFP-tagged clathrin light chain (CFP-CCP) was cloned by replacing GFP in GFP-CCP using PCR and Gibson assembly. A plasmid encoding Lifeact-YFP was generated by replacing GFP with YFP in a previously described Lifeact-GFP construct (kindly provided by Antje Gohla).22 A plasmid encoding N-terminally Nluc-tagged clathrin light chain was obtained by gene synthesis (Twist Bioscience). A plasmid encoding C-terminally Halo-tagged ScFv30 (ScFv30-Halo) was generated by replacing the YFP sequence with Halo in a previously described ScFv30-YFP construct.68,69
BRET measurements
BRET measurements between Nluc fused to Kras, receptors or clathrin-light chain and Halo-tagged βArr1/2 or ScFv30, labeled with R110, were performed at 37 °C using a PHERAstar Microplate Reader (BMG Labtech) with a dual-luminescence readout BRET1 plus filter (460-490 nm band-pass, 520-550 nm long-pass). Following 4 baseline measurements, the cells were treated with vehicle or the indicated agonist concentration and measured for an additional hour. BRET acceptor/donor ratios were calculated separately for each well. The results were normalized to the baseline values and those obtained with vehicle. Measurements were performed in triplicate readouts.
β-arrestin–Fab30 binding assay
Each cell pellet was suspended in 150 μl assay buffer (150 mM NaCl, 20 mM HEPES pH 7.4), homogenized using a Dounce homogenizer and centrifuged at 21,000 x g for 1 h at 4 °C. The supernatant (120 μl) was transferred to a new tube, followed by addition of 60 μg Fab30 and 3.6 nmol of a phosphopeptide corresponding to the C-terminal region of the V2R (V2Rpp peptide). After 1 h incubation at room temperature under gentle rotation to allow complex formation, the sample was added to 50 μl protein L agarose (ThermoFisher) equilibrated with assay buffer, followed by further incubation for 3 h at room temperature under gentle rotation. Each supernatant was transferred to a new tube and the agarose resin was washed three times with 500 μl assay buffer. Equal fractions of the initial complex (i), supernatant (s/n) and agarose resin resuspended in 150 μl assay buffer (r) were mixed with 5x loading buffer (250 mM Tris-HCl pH 6.8, 5% SDS, 50% glycerol, 10 mM DTT, 0.1% bromophenol blue).
Western blotting
Lysates were incubated for 5 min at 95 °C and centrifuged at 20,000 x g for 1 min at room temperature (RT). The supernatants were separated by electrophoresis on a 10% SDS polyacrylamide gel and transferred to a PVDF membrane (Millipore). The membrane was blocked with TBS (10 mM Tris-HCl pH 7.4, 100 mM NaCl) supplemented with 0.1% Tween and 5% skim milk powder for 1 h at RT and incubated with a rabbit βArr2 polyclonal antibody (1:10,000) or a mouse monoclonal GAPDH antibody (1:10,000) overnight at 4 °C, followed by incubation with an anti-rabbit or anti-mouse HRP-conjugate secondary antibody (1:20,000) in a buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.2% NP-40, 0.2% bovine serum albumin (BSA). The membranes were detected with the Amersham ECL Prime Western Blotting Detection reagent (Cytiva), according to the manufacturer’s protocol and imaged on a Chemi Doc system (Bio-Rad).
β-arrestin purification and labeling
A minimal cysteine mutant of bovine β-arrestin 1 carrying an N-terminal GST-tag separated by a thrombin-cleavage site and cloned in the pGEX4T3 expression vector was used.11 Two additional amino acids, Ala-Cys, were introduced at the C-terminus to allow site-specific labeling and the construct was confirmed by DNA sequencing. GST-β-arrestin was expressed in E. coli BL21(DE3) cells. A starter culture grown in LB broth supplemented with 100 μg/ml ampicillin at 37 °C to an A600 of 0.6 was used to inoculate 2 liters of Terrific Broth supplemented with 100 μg/ml ampicillin, which was also grown at 37 °C. When A600 reached 0.6-0.8, the cultures were equilibrated to 18 °C, and expression was induced with 50 μM Isopropyl β-D-1-thiogalactopyranoside for 16 h. The cells were harvested in PBS, resuspended in 180 ml of cold lysis buffer (25 mM Tris-HCl pH 8.5, 150 mM NaCl, 1 mM phenylmethanesulfonyl fluoride, 2 mM benzamidine hydrochloride, 1 mM EDTA, 5% glycerol, 2 mM DTT) with the addition of 10 μl Benzonase nuclease, lysed by sonication and cleared by ultracentrifugation at 100,000 g at 4 °C for 40 min. All following steps were done at 4 °C unless otherwise stated. The cleared lysate was filtered through a 0.22-μm syringe filter, applied to 20 ml of Glutathione Sepharose 4B resin, pre-equilibrated in wash buffer (50 mM Tris-HCl pH 8.5, 150 mM NaCl) and incubated overnight under gentle rotation. The Sepharose suspension was spun at 1,000 g for 15 min, the supernatant decanted and the resin transferred to a glass chromatography column filled with wash buffer. The resin was washed with 20 column volumes (CV) of high-salt wash buffer (50 mM Tris-HCl pH 8.5, 1 M NaCl), and 10 CV of wash buffer. GST-β-arrestin was eluted in 1 CV fractions of elution buffer (50 mM Tris-HCl pH 8.5, 150 mM NaCl, 2 mM DTT, 20 mM glutathione), the protein content was estimated by Pierce BCA Protein Assay, and the fractions containing proteins pooled and concentrated to 5 ml. The buffer was adjusted to 350 mM NaCl and 0.02% n-dodecyl β-D-maltoside (DDM), thrombin protease (250 U) was added, and cleavage was allowed for 2 h at room temperature before it was stopped by the addition of 2 mM benzamidine hydrochloride. The solution was concentrated to 600 μl and β-arrestin was isolated on a Superdex 200 Increase 10/300 GL column (Cytiva) equilibrated in 25 mM Tris-HCl pH 8.5, 350 mM NaCl, 0.02% DDM at room temperature. Peak fractions were pooled, concentrated to 250 μl, and the pH was adjusted by the addition of 40 mM Tris-HCl pH 6.8. Disulphide bridges were reduced by the addition of 0.8 μM Tris(2-carboxyethyl)phosphine hydrochloride. β-arrestin was labeled by incubation with 0.8 μM Alexa Fluor™ 647 C2 Maleimide for 2 h at room temperature in the dark, followed by polishing on a Superdex 200 Increase 10/300 GL column as described above. Peak fractions were pooled and aliquots flash-frozen in liquid nitrogen and stored at -80 °C.
Giant unilamellar vesicle preparation
Giant unilamellar vesicles (GUVs) were obtained by electroformation. A total of 7 μl of di-oleoyl-phosphatidylcholine lipid solution (10 mg/ml in chloroform) was added to indium tin oxide-coated glass slides to form lipid films on the conductive surfaces. Once the lipid films were dry, a chamber with 0.3-mm gap was assembled and filled with a 200 mM sucrose solution. The assembled chamber was then placed at 50°C and the following conditions were applied for GUV electroformation: 11 Hz, 1V alternating electric current for 2 h.
Experiments with supported lipid bilayers
Custom imaging chambers were assembled as previously described.70 Briefly, 0.75-mm diameter inlet/outlet holes were drilled in glass microscopy slides at a distance of 3–4 mm from the edges to create a flow channel. Coverslips (VWR, 24 x 40 mm) were cleaned by sequential sonication in chloroform and 5 M NaOH solution, rinsed in distilled water and allowed to dry. To assemble the flow chambers, double-sided Scotch tape was sandwiched between a slide and a coverslip and the edges were sealed with an epoxy glue (5-Minute Epoxy, Thorlabs), resulting in the formation of flow channels connected to the inlet/outlet holes. The chambers were rinsed with 200 μl PBS. A total of 20 μl of the GUV suspension was mixed with 0.2 μl of a lipophilic dye solution (5 μM 3,3’-dioctadecyloxacarbocyanine perchlorate in N,N-dimethylformamide) and loaded into the chambers, followed by incubation for 1 h at room temperature in the dark to allow for the GUVs to break through osmotic shock and form lipid bilayers. Afterwards, the chambers were rinsed with 200 μl PBS, followed by 200 μl PBS containing 0.1% BSA. Purified β-arrestin was suspended to a concentration of 1 nM in an oxygen-scavenging buffer (10 mM Tris-HCl pH 7.4, 50 mM NaCl, 0.1% BSA, 1% D-glucose, 2 mM Trolox, 25 U/ml glucose oxidase, 250 U/ml catalase) and loaded into a chamber, which was immediately sealed and imaged by single-molecule microscopy as described above for live cells.
Live cell protein labeling for single-molecule microscopy
Cells were labeled with a combination of 1 μM SNAP-Surface Alexa Fluor 647 (AF647, cell impermeable, New England Biolabs) and 1 μM HaloTag Janelia 549 (JF549, cell permeable, Promega) in complete culture medium for 20 min at 37 °C. These concentrations were selected based on titration experiments to obtain saturation labeling of both SNAP- and Halo-tagged proteins (Figure S1C), which results in ~90% and >70% labeling efficiencies for extracellular and intracellular tags as determined by single-molecule microscopy22,27 and competition labeling experiments.71 Cells were then washed three times with complete culture medium, allowing 5 min incubation between washes. Non-specific labeling was <1% (Figure S1D).
Single-molecule microscopy
Single-molecule microscopy experiments were performed using total internal reflection fluorescence (TIRF) illumination on a custom system (assembled by CAIRN Research) based on an Eclipse Ti2 microscope (Nikon) equipped with a 100x oil-immersion objective (SR HP APO TIRF NA 1.49, Nikon), 405, 488, 561, and 637 nm diode lasers (Coherent, Obis), an iLas2 TIRF illuminator (Gataca Systems), quadruple band excitation and dichroic filters, a quadruple beam splitter, 1.5x tube lens, four EMCCD cameras (iXon Ultra 897, Andor), hardware focus stabilization, and a temperature-controlled enclosure. The sample and objective were maintained at 37 °C throughout the experiments. Coverslips were mounted in a microscopy chamber filled with HBSS supplemented with 10 mM HEPES, pH 7.5. A reduced oxygen environment (2-4% O2) was provided in the imaging chamber to decrease photobleaching without increasing cytotoxicity using a mixture of nitrogen and air and a home-built gas mixing and humidifying system as previously described.72 The oxygen concentration in the imaging solution was measured in real-time using a needle-type oxygen sensor connected to an OXY-1 microsensor (OXY-1 ST PreSens). Multi-color single-molecule image sequences were acquired simultaneously on the four synchronized EMCCDs at a rate of one image every 30 ms. Only individual cells with comparable expression levels of both the receptor and β-arrestin constructs used in this study were selected for single-molecule analyses, resulting in similar densities within the compared groups. Details about the number of trajectories and the densities of both receptor and β-arrestin molecules in all groups are given in Table S1.
HILO microscopy
Highly inclined and laminated optical sheet (HILO) microscopy was performed on the same TIRF system as for single-molecule microscopy using a subcritical incident illumination angle. Image sequences were acquired simultaneously on two of the four synchronized EMCCDs at a rate of one image every 60 s.
Single-particle tracking
Automated single-particle detection and tracking were performed with the u-track software73 and the obtained trajectories were further analyzed using custom algorithms in MATLAB environment as previously described.22 Image sequences from different channels were registered against each other using a linear piecewise transformation, based on reference points taken with multi-color fluorescent beads (100 nm, TetraSpeck).22 The inter-channel localization precision after coordinate registration was ~20 nm.
Single-molecule interaction analysis and estimation of kon/koff values
The frequency and duration of receptor–βArr2 interactions were estimated using our previously described method based on deconvolution of the distribution of single-molecule colocalization times with the one expected for random colocalizations.22 The distribution for random colocalizations was estimated in cells co-transfected with βArr2-Halo and SNAP-tagged CD86, a non-related membrane protein that does not interact with βArr2 and has diffusion characteristics comparable to those of the investigated receptors.
Single-molecule interactions and microscopic kon/koff values were estimated as previously described.22 Briefly, for each particle in channel 1 at each frame, all particles in channel 2 falling within a defined search radius R0 were identified as colocalizing.
For each colocalization event, the starting and terminating frame was obtained. A Monte Carlo approach was used to link colocalization events that were prematurely terminated due to uncertainty in the assignment of trajectory segments after a splitting event. Data obtained in cells expressing SNAP-CD86 labeled with SNAP-AF647, βArr2-Halo labeled with Halo-JF549 and unlabeled wildtype β2AR were used to estimate the frequency and duration of random colocalizations.
The distributions of true interaction times were estimated by deconvolving the observed distributions of colocalization times with that obtained for the non-interacting control pair (SNAP-CD86 and βArr2-Halo).
Of note, it is sufficient for two molecules to be detected at a distance higher than R0 just for one frame to interrupt a colocalization. Therefore, to avoid premature termination of the colocalizations one needs to choose R0 to be higher than the localization error (~20 nm). In our case, we chose R0 = 150 nm, which corresponds to a probability of correctly detecting a true interaction that lasts n frames of 0.9998n (e.g. ~0.98 after 100 frames). A key feature of our method is that, since the same search radius is used for the test and control conditions, the resulting deconvolved distribution is largely insensitive to the choice of R0. This allows the reliable detection and estimation of the duration of true interactions provided that they last longer than ~280 ms on average.22
To estimate dissociation rate constant (koff) values, normalized relaxation curves obtained from the distributions of true interaction times were fitted to an exponential decay function:
| (Equation 1) |
where Fr(t) is the fraction of surviving interactions at time t, Ftrue (≤1) is the fraction of true interactions at t = 0, and kloss is a correction factor accounting for premature termination of the interactions due to photobleaching or potential particle loss at detection or tracking. kloss was estimated based on simulated image sequences of randomly diffusing, non-dissociating particles with characteristics (particle densities, point spread functions, diffusion coefficients, background levels, signal-to-noise ratios, fluorophore bleaching rates) matching the experimental ones.
Association rate constant (kon) values are related to Ftrue and the rate of new colocalizations per unit of area d[C]ρ/dt by the following equation:
| (Equation 2) |
where []ρ denotes density and [R]ρ and [A]ρ are the densities of free receptor and β-arrestin molecules, which can in turn be derived by subtracting the estimated density of receptor-β-arrestin complexes ([RA]ρ) from the total densities measured in channels 1 [Ch1]ρ and 2 [Ch2]ρ, respectively.
Finally, [RA]ρ can be deduced based on the balance between association and dissociation rates at equilibrium using the formula:
| (Equation 3) |
These relationships allowed us to estimate kon from measured observables using to the following formula:
| (Equation 4) |
kon was estimated separately at each frame and the given values were obtained by averaging over the analyzed frames.
TAMSD analysis
The time-averaged mean squared displacement (TAMSD)74,75 of individual trajectories was computed as previously described.22 TAMSD data were fitted to the equation describing the ensemble averaged TAMSD for an ergodic process:
| (Equation 5) |
where Δ indicates lag time, α is the anomalous diffusion exponent and σerr is a constant offset accounting for localization error. Only trajectories lasting at least 100 frames were analyzed. Data corresponding to the first 10 lag time points were used for the fitting.
For the analysis of sub-trajectories, all trajectory segments lasting at least 50 frames were included. Segments characterized by free diffusion were fitted using Equation 5. Segments characterized by confinement/trapping were fitted using the equation describing the diffusion of a molecule inside a harmonic potential76:
| (Equation 6) |
where is the squared distance from the center of the trap at the beginning of the diffusive process and λ is the inverse of the mean reversion time. From this, the approximate confinement/trap diameter (d) was deduced as. Similar distributions of d were observed under basal and stimulated conditions for both β2AR and βArr2.
Space exploration analysis
To evaluate the tendency of individual molecules to laterally diffuse on the plasma membrane, we developed an algorithm to estimate their explored area. Because trajectories are fundamentally one dimensional, albeit embedded in a two-dimensional space, there is no unique way to estimate their explored area. The approach we used takes in account the position uncertainty of the trajectory coordinates and the random nature of molecular displacements. First, for each localization along the trajectory we draw a disk of radius , where is the average diffusion coefficient for βArr2 and Δt = 30 ms is the time between 2 frames, which accounts for the uncertainty in coordinate localization and arrestin diffusion between frames. Then, for each pair of consecutive localizations, we compute the convex hull that contains their associated disks. Finally, the total area explored is calculated as the area of the union of all convex hulls computed along the trajectory. With this approach, trajectories of molecules that are either immobile or stay on the membrane for a single frame result in an explored area of ~0.05 μm2, which corresponds the area chosen for the disk. The figures report the proportion of trajectories exploring ≥ 1.5 μm2, which on average corresponds to ~30 frames of free lateral diffusion for .
CCP detection
CCP detection was performed by applying a frame-by-frame binary mask to GFP-CCP image sequences. Image sequences were preprocessed to remove local background and enhance contrast using a bottom and top-hat filter with a disk-shaped structuring element of 11-pixel diameter (‘imtophat’ and ‘imbothat’ functions in MATLAB). A Kalman filter with low gain (0.1) was applied to reduce noise. The image sequences were then deconvolved with the theoretical PSF of the system using the Lucy-Richardson algorithm.77,78 Binary masks corresponding to CCPs were obtained by thresholding the image sequences with a value corresponding to the mean plus 1 standard deviation of each frame. Only pixels persisting at least 45 consecutive frames (= 1.5 s) were included in the final CCP masks.
Spatial confinement analysis
A spatial confinement analysis was used to identify trajectory segments characterized by confinement/trapping, using our recently described algorithm.28 Briefly, for each trajectory, we computed a recurrence matrix (Mij) containing information about the distance between each pair of points in the trajectory:
| (Equation 7) |
where i and j run over each step of the trajectory, x is the position of the particle, and λ is the test length scale of the analysis. Mij approaches 1 if the difference between xi and xj is smaller than λ. To minimize the effects of localization error and possible misdetected outliers, Mij was smoothened by local averaging and thresholded to obtain a binary matrix (B), where Bij = 1 if Mij > e–1 or zero otherwise. Trapped portions of the trajectory appear in B as square blocks of ones along its diagonal. To detect these blocks, three quantities were calculated, i.e. block time (t|(n)), neighbouring time (t⊥(n)) and persistence time (t∥(n)), from which we computed the invariant quantity v(n):
| (Equation 8) |
In the idealized case of a perfect squared box of ones it is easy to verify that v(n) = 1. In practice, blocks are never perfect squares, and we use a cut-off of v(n) = 3/4 to identify blocks corresponding to potential confined trajectory segments. A statistical test based on the probability of detecting a larger block by chance for a particle with 2D fractional Brownian motion and P value = 0.05 is then applied to decide if the detected block is a confinement event.
Markov chain analysis
A Markov chain analysis was used to estimate the relative occupancies and forward transition probabilities. Each molecule at each frame was labeled with four binary numbers describing the presence/absence of spatial confinement, CCP localization, colocalization with either a β-arrestin molecule (in case of a receptor) or a receptor (in case of β-arrestin), and confinement of the colocalizing partner. Considering all possible combinations and discarding the physically irrelevant ones, such as freely diffusing molecules on a CCP or colocalization between a freely diffusing and a confined molecule, we obtained a set of 6 unique states (R1–6 and A1–6 for receptor and β-arrestin, respectively) plus a dummy state (R0/A0) corresponding to molecules before or after their detection on the plasma membrane (Table S2). R0/A0 was introduced to take into account the movements of molecules between the cytosol and the plasma membrane (in the case of β-arrestin), diffusion between the basal and apical membrane (not visible in TIRF), internalization as well as disappearance due to fluorophore photobleaching or particle loss at tracking. Only states lasting at least 10 frames (300 ms) and their transitions were considered. This cutoff removes ~95% of random colocalizations, while retaining most (~86%) true interactions. Since these removed interactions are only a minor fraction and are very short lived (typically ≪ 300 ms), this has negligible impacts on the results.
For each cell, we computed the transition rates rij, i.e. the average number of forward transitions per frame from state i to state j with i,j ∈ [0,1,..,6], except for the case where i = 0 and j = 0 which cannot be estimated. Then the values obtained in each cell were combined by computing a pondered average 〈rij〉, with weights corresponding to the plasma membrane densities of the considered molecule (receptor or β-arrestin) to compensate for cell-to-cell variability.
The results are shown using a simplified visual representation, where each state is represented by a circle with area proportional to its relative occupancy, calculated for each state i as the sum of the transition rates 〈rij〉 where j ∈ [0,1,..,6] divided by the sum of all transition rates, and each arrow from i to j (where i ≠ j) represents the outward transition rates. To highlight the most relevant transitions while also showing smaller contributions, the width and color of the arrows were chosen to scale with the logarithm of 〈rij〉. Outward transition rates smaller than 1/10,000 of the sum of all outward transition rates are not shown. Note that this representation is different from the usual representation of Markov chains, where, for each state i, the transition rates are normalized by the sum of all the forward transitions departing from the same state, which corresponds to transition probabilities per frame. Transition probability matrices of the Markov chain analyses performed in this study are given in Table S3.
History plots
Plots displaying all the observed sequences of events before and after a given target state were generated in several steps. First, we collected all the trajectories that contained the target state lasting for at least 10 frames (300 ms). Then, for each trajectory, we separately gathered the h states preceding and following the target state. The information was used to build two separate tree graphs (past and future), where the branches represent all observed sequences of events. Taking advantage of graph theory, we assigned a branch number to each history in either graph using the following formula:
| (Equation 9) |
Where i runs over all states in the sequence (from the target state i = 0 to the farthest state from present to be analyzed for i = h) with ni being the state number of the state i and N the number of distinct states (here N = 7).
Finally, histories were sorted according to their branch number. Graphic representations were obtained by stacking all histories, each represented by a thin horizontal line, color coded according to the contained states.
Super-resolution radial fluctuation imaging
Super-resolved Lifeact images were generated from image sequences using the super-resolution radial fluctuations (SRRF) algorithm implemented in the NanoJ-SRRF ImageJ plugin.79 In overlaid images, the coordinates of single-molecule trajectories and CCPs binary masks were rescaled to match the higher resolution of SRRF images.
Unbiased molecular dynamics (MD) simulations
The initial conformation of βArr2 was modelled with MODELLER80,81 using the structure of βArr1 in complex with the muscarinic M2 receptor (PDB code: 6U1N).17 This template was selected as the C-edge is resolved in a conformation that exposes hydrophobic residues towards the membrane interface more favorably than other existing βArr1 complexes (6PWC, 6TKO), which is expected to promote membrane interaction.35 The inactive conformation of the finger loop was obtained from the inactive structure of βArr2 (PDB code: 3P2D).82 Of note, our model did not include the distal disordered C-tail of β-arrestin, which is not resolved in the available structures.
To study spontaneous interaction with the plasma membrane, βArr2 was placed in proximity to the lipid bilayer. To mimic the experimental conditions as much as possible, we have taken into account the membrane composition of CHO cells83 and incorporated the five most abundant membrane components into our simulation setup: 10% cholesterol, 38% palmitoyl-oleoyl-phosphatidylcholine, 28% dioleoyl-phosphatidylcholine, and 24% dioleoyl-phosphatidylethanolamine.
Membrane building and system solvation were done using the CHARMM-GUI server.84,85 The ionic strength of the system was kept at 0.15 M using NaCl ions.
Simulations were performed with the ACEMD3 package.86 Parameters for system components were obtained from CHARMM36m87 and CHARMM36 force fields.88,89 The simulation data are made available via the GPCRmd platform.90
All the simulated systems were first relaxed during 50 ns of simulations under constant pressure and temperature (NPT) with a time step of 2 fs, with harmonic constraints applied to the protein backbone. The temperature was maintained at 310 K using the Langevin thermostat91 and pressure was kept at 1 bar using the Berendsen barostat.92 The equilibration run was followed by production runs in conditions of constant volume and temperature (NVT) with a 4-fs time step. No constraints were applied during this stage. In all simulations, we used van der Waals and short-range electrostatic interactions with a cut-off of 9 Å and the particle mesh Ewald method93 for long-range electrostatic interactions. The resulting simulation frames were analyzed using VMD94 and tools available within, as well as in-house scripts.
To study spontaneous plasma membrane insertion of βArr2, we carried out 40 x production runs of 60 ns each, starting with βArr2 in the vicinity of the membrane, which resulted in C-edge insertion in 4/40 cases. These simulations were further expanded by performing additional 20 x 200 ns production runs, starting from a representative conformation of C-edge-anchored β-arrestin obtained from the previous simulations. These simulations revealed the additional penetration of the finger loop in 3/20 cases.
To further study the conformations explored by membrane-anchored βArr2, we carried out a separate set of 3 x 1 μs production runs, starting from the fully membrane-anchored conformation obtained in the previous simulations. We compared the results with those of an additional set of 3 x 1 μs production runs starting from the same βArr2 conformation observed when it is fully membrane-anchored but, this time, with βArr2 placed in solution.
To generate the ΔELA mutant (R189Q/F191E/L192S/M193G/T226S/K227E/T228S/K230Q/K231E/K233Q/R237E/K251Q/K325Q/K327Q/V329S/V330D/R332E), as well as the three targeted C-edge mutants (L192D/M193D/S194D; R332D/G333D/G334D; L192A/M193A/S194A/R332A/G333A/G334A), the involved modifications were introduced within the structure of the fully membrane-anchored βArr2. Of note, our computational experiments do not allow us to reach equilibrium of arrestin binding/unbinding due to current limitations in simulating such long timescales.
Membrane unbinding of the C-edge was defined based on the z-coordinate of the CA atom of L192. Its average z-coordinate for wild-type βArr2 is 117 Å, which corresponds to stable membrane anchoring. Membrane unbinding is reported in our system when the z-coordinate drops below 105 Å.
Metadynamics
Due to the high complexity and huge degrees of freedom of the arrestin–membrane ensemble, the simulation time required to obtain a converged energetic estimate of the membrane (un)binding process would exceed our computational capabilities. To overcome this limitation, we simulated the membrane (un)binding of the C-domain of βArr2 (P176-P347) alone, with or without the investigated mutations. Even though these simulations do not include the whole arrestin-membrane ensemble, they provide valuable insights into the effects of mutations on the free energy associated with membrane (un)binding.
All well-tempered metadynamics simulations were performed using ACEMD3.3 with the plumed 2.6.1 plugin.
The collective variable (CV) used to bias the simulations was the distance (z1-z2) between the mean position of phosphorous atoms (z1 coordinate) in the lower leaflet of the membrane (cytosolic surface) and the geometric center of the Ca positions in the C-domain of βArr2 (z2 coordinate). The choice of this collective variable ensures sampling of only the βArr2 (un)binding event, while excluding the movements of βArr2 along the xy plane of the membrane. An upper wall for the CV was set at a distance of 7.5 nm with a force constant of K = 500 KJ · mol-1 · nm-2, aiming to avoid the escape of the C-domain into the bulk solvent and, thus, helping the convergence of the metadynamics simulations and reducing the computational burden.
The starting structure for the metadynamics simulations had the C-domain bound to the membrane and was obtained by equilibrating the C-domain for 50 ns under NPT conditions. The same general parameters (time step, thermostat, etc.) of the unbiased MD simulations were used. Metadynamics simulations were performed using an initial bias height of 1 KJ·mol-1, a width of 0.5, a bias factor of 16 and the rate of Gaussian deposition set to 500 steps. We performed 2 independent well-tempered metadynamics simulations for both wild-type and mutant C-domain until they reached convergence.
To check for convergence, we performed a block analysis of the histogram of the CV, which showed (for both simulations) a hyperbolic-like graph, suggesting that the system had converged at 1.8 μs. To compute the free energy, we followed the protocol described by Bussi and Laio95 for well-tempered metadynamics, and we estimated the error on the free energy by using the previous block analysis.
Analysis of contacts in MD simulations
To quantify the interactions, we monitored the distance from the membrane of reference amino acids in the finger loop (A61-D79), C-loop (A240-Q251) and C-edge (T188-H199, N224-E231, S329-V336), using the following rational switching function:
| (Equation 10) |
where R0 is 0.5 nm and r is the distance between the geometric center of the sidechain of each amino acid (Hα1 and Hα2 for glycine) and all the atoms composing the membrane. The main reason for using the geometric center of each sidechain instead of the position of the contained amino acids is to normalize for the differences in the number of sidechain atoms between amino acids. At the same time, we ensure that the sidechain orientation is relevant for the total number of contacts.
The coordination number N for each amino acid is defined as the sum over all the atoms of the membrane in the simulation according to the following expression:
| (Equation 11) |
By using this approach, we can quantify the coordination number for each of the residues studied, which is directly related to the strength of the interaction between βArr2 and the lipid bilayer. The coordination number is calculated for each residue in each frame for the three accumulated microseconds to posteriorly calculate its mean value for the selected finger loop, C-loop and the C-edge residues. To simplify the comparison between wild-type and mutant βArr2, total coordination numbers for the finger loop, C-loop and C-edge were calculated by summing up the coordination numbers of each of the residues in each region.
Inter-domain rotation angle
The rotation angle between βArr2 N- and C-domains was computed as previously described.44 The utilized scripts were kindly provided by Naomi Latoracca.20 To evaluate the ability of Fab30/ScFv30 to recognize the observed active-like conformation of the fully membrane engaged βArr2, we compared the conformations obtained in MD simulations with that of the structure of βArr1 in complex with Fab30 and a fully phosphorylated 29-amino-acid C-terminal peptide derived from the V2R (PDB code: 4JQI).7
Quantification and Statistical Analysis
Statistical analyses were performed using MATLAB (version 2018b). Differences between three or more groups were assessed by a non-parametric Kruskal-Wallis test followed by unpaired two-tailed t test with Bonferroni correction. Differences were considered significant for P < 0.05. Single-molecule data were analyzed by automated scripts with no user intervention during the analysis. Statistical parameters are reported in the figure legends.
Supplementary Material
Highlights.
b-arrestin spontaneously preassociates with the plasma membrane via its C-edge
Preassociated b-arrestin interacts with receptors via lateral diffusion
Receptor-b-arrestin interactions are short lived and trigger b-arrestin activation
Active b-arrestin and receptors reach clathrin-coated pits separately via diffusion
ACKNOWLEDGMENTS
This study was supported by a Wellcome Trust Senior Research Fellowship (212313/Z/18/Z to D.C.). Research in the laboratory of A.K.S. is supported by a Senior Fellowship of the DBT/Wellcome Trust India Alliance (IA/S/20/1/504916). Research in the laboratory of S.J.H. is supported by the MRC (grant no. MR/W016176/1). J.S. is supported by the Instituto de Salud Carlos III FEDER (PI18/00094) and the ERA-NET NEURON & Ministry of Economy, Industry, and Competitiveness (AC18/00030). A.K.S. is a EMBO Young Investigator and Joy Gill Chair Professor. J. Grimes, Z.K., Y.L., T.M., S.L.O’B., T.M.S., B.M.-L., C.H., J.S., and D.C. participate in the European COST Action CA18133 (ERNEST). We thank Chris Tate for discussions during the initial phase of this study.
Footnotes
Author Contributions: D.C. conceived the study. J. Grimes, Z.K., Y.L., T.M., and S.L.O’B. performed the single-molecule and BRET experiments and analyzed the data. Y.L. and D.C. developed the mathematical analyses and wrote the software. T.M.S. and B.M.-L. performed the MD simulations. D.C. and Z.K. supervised the single-molecule experiments. S.J.H. and D.C. supervised the BRET experiments. J.S. supervised the MD simulations. M.B. and A.K.S. generated the Fab30/ScFv30 probe and provided support with Fab30/ScFv30 experiments. T.M. purified β-arrestin for the experiments with supported lipid bilayers. M.M. and D.M.O. prepared the GUVs and provided support with the experiments with supported lipid bilayers. J. Goulding assisted with single-molecule microscopy experiments. J.D. and C.H. generated the ΔQ-GRK cell line and provided support with the experiments investigating the role of GRK-dependent phosphorylation. R.M. generated the CRISPR-Cas9 CHO-K1 line. D.C., J. Grimes, Z.K., and Y.L. wrote the manuscript with contributions from J.S., T.M.S., and B.M.-L. All authors discussed the results and edited the manuscript.
Declaration of Interests:
The authors declare no competing interests.
Inclusion and Diversity:
We support inclusive, diverse and equitable conduct of research.
References
- 1.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Hauser AS, Attwood MM, Rask-Andersen M, Schioth HB, Glo-riam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017;16:829–842. doi: 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. β-arrestin: a protein that regulates β-adrenergic receptor function. Science. 1990;248:1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- 4.Pierce KL, Lefkowitz RJ. Classical and new roles of β-ar-restins in the regulation of G-protein-coupled receptors. Nat Rev Neuro-sci. 2001;2:727–733. doi: 10.1038/35094577. [DOI] [PubMed] [Google Scholar]
- 5.Daaka Y, Luttrell LM, Ahn S, Della Rocca GJ, Ferguson SS, Caron MG, Lefkowitz RJ. Essential role for G protein-coupled receptor endocytosis in the activation of mitogen-activated protein kinase. J Biol Chem. 1998;273:685–688. doi: 10.1074/jbc.273.2.685. [DOI] [PubMed] [Google Scholar]
- 6.Reiter E, Ahn S, Shukla AK, Lefkowitz RJ. Molecular mechanism of β-arrestin-biased agonism at seven-transmembrane receptors. Annu Rev Pharmacol Toxicol. 2012;52:179–197. doi: 10.1146/annurev.pharmtox.010909.105800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shukla AK, Manglik A, Kruse AC, Xiao K, Reis RI, Tseng WC, Staus DP, Hilger D, Uysal S, Huang LY, et al. Structure of active β-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature. 2013;497:137–141. doi: 10.1038/nature12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shukla AK, Westfield GH, Xiao K, Reis RI, Huang LY, Tripathi-Shukla P, Qian J, Li S, Blanc A, Oleskie AN, et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, Barty A, White TA, Yefanov O, Han GW, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomsen ARB, Plouffe B, Cahill TJ, 3rd, Shukla AK, Tarrasch JT, Dosey AM, Kahsai AW, Strachan RT, Pani B, Mahoney JP, et al. GPCR-G protein-β-arrestin super-complex mediates sustained G protein signaling. Cell. 2016;166:907–919. doi: 10.1016/j.cell.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumari P, Srivastava A, Banerjee R, Ghosh E, Gupta P, Ranjan R, Chen X, Gupta B, Gupta C, Jaiman D, et al. Functional competence of a partially engaged GPCR-β-arrestin complex. Nat Commun. 2016;7:13416. doi: 10.1038/ncomms13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou XE, He Y, de Waal PW, Gao X, Kang Y, Van Eps N, Yin Y, Pal K, Goswami D, White TA, et al. Identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors. Cell. 2017;170:457–469.:e13. doi: 10.1016/j.cell.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cahill TJ, 3rd, Thomsen AR, Tarrasch JT, Plouffe B, Nguyen AH, Yang F, Huang LY, Kahsai AW, Bassoni DL, Gavino BJ, et al. Distinct conformations of GPCR-β-arrestin complexes mediate desensitization, signaling, and endocytosis. Proc Natl Acad Sci USA. 2017;114:2562–2567. doi: 10.1073/pnas.1701529114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumari P, Srivastava A, Ghosh E, Ranjan R, Dogra S, Yadav PN, Shukla AK. Core engagement with β-arrestin is dispensable for agonist-induced vasopressin receptor endocytosis and ERK activation. Mol Biol Cell. 2017;28:1003–1010. doi: 10.1091/mbc.E16-12-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y, Warne T, Nehme R, Pandey S, Dwivedi-Agnihotri H, Chatur-vedi M, Edwards PC, Garcia-Nafria J, Leslie AGW, Shukla AK, et al. Molecular basis of β-arrestin coupling to formoterol-bound β1-adrenoceptor. Nature. 2020;583:862–866. doi: 10.1038/s41586-020-2419-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang W, Masureel M, Qu Q, Janetzko J, Inoue A, Kato HE, Robertson MJ, Nguyen KC, Glenn JS, Skiniotis G, et al. Structure of the neurotensin receptor 1 in complex with β-arrestin 1. Nature. 2020;579:303–308. doi: 10.1038/s41586-020-1953-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staus DP, Hu H, Robertson MJ, Kleinhenz ALW, Wingler LM, Capel WD, Latorraca NR, Lefkowitz RJ, Skiniotis G. Structure of the M2 muscarinic receptor-β-arrestin complex in a lipid nanodisc. Nature. 2020;579:297–302. doi: 10.1038/s41586-020-1954-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nuber S, Zabel U, Lorenz K, Nuber A, Milligan G, Tobin AB, Lohse MJ, Hoffmann C. β-arrestin biosensors reveal a rapid, receptor-dependent activation/deactivation cycle. Nature. 2016;531:661–664. doi: 10.1038/nature17198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichel K, Julli D, Barsi-Rhyne B, Latorraca NR, Masureel M, Sibar-ita JB, Dror RO, von Zastrow M. Catalytic activation of β-arrestin by GPCRs. Nature. 2018;557:381–386. doi: 10.1038/s41586-018-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latorraca NR, Wang JK, Bauer B, Townshend RJL, Hollingsworth SA, Olivieri JE, Xu HE, Sommer ME, Dror RO. Molecular mechanism of GPCR-mediated arrestin activation. Nature. 2018;557:452–456. doi: 10.1038/s41586-018-0077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calebiro D, Sungkaworn T. Single-molecule imaging of GPCR interactions. Trends Pharmacol Sci. 2018;39:109–122. doi: 10.1016/j.tips.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 22.Sungkaworn T, Jobin ML, Burnecki K, Weron A, Lohse MJ, Calebiro D. Single-molecule imaging reveals receptor-G protein interactions at cell surface hot spots. Nature. 2017;550:543–547. doi: 10.1038/nature24264. [DOI] [PubMed] [Google Scholar]
- 23.Lohse MJ, Engelhardt S, Eschenhagen T. What is the role of β-adrenergic signaling in heart failure? Circ Res. 2003;93:896–906. doi: 10.1161/01.RES.0000102042.83024.CA. [DOI] [PubMed] [Google Scholar]
- 24.Zhang P, Mende U. Regulators of G-protein signaling in the heart and their potential as therapeutic targets. Circ Res. 2011;109:320–333. doi: 10.1161/CIRCRESAHA.110.231423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 26.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 27.Calebiro D, Rieken F, Wagner J, Sungkaworn T, Zabel U, Borzi A, Cocucci E, Zurn A, Lohse MJ. Single-molecule analysis of fluorescently labeled G-protein-coupled receptors reveals complexes with distinct dynamics and organization. Proc Natl Acad Sci USA. 2013;110:743–748. doi: 10.1073/pnas.1205798110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanoiselee Y, Grimes J, Koszegi Z, Calebiro D. Detecting transient trapping from a single trajectory: A structural approach. Entropy (Basel) 2021;23:1044. doi: 10.3390/e23081044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drube J, Haider RS, Matthees ESF, Reichel M, Zeiner J, Fritzwanker S, Ziegler C, Barz S, Klement L, Filor J, et al. GPCR kinase knockout cells reveal the impact of individual GRKs on arrestin binding and GPCR regulation. Nat Commun. 2022;13:540. doi: 10.1038/s41467-022-28152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oakley RH, Laporte SA, Holt JA, Barak LS, Caron MG. Association of β-arrestin with G protein-coupled receptors during clathrin-mediated endocytosis dictates the profile of receptor resensitization. J Biol Chem. 1999;274:32248–32257. doi: 10.1074/jbc.274.45.32248. [DOI] [PubMed] [Google Scholar]
- 31.Oakley RH, Laporte SA, Holt JA, Caron MG, Barak LS. Differential affinities of visual arrestin, β arrestin1, and β arrestin2 forG protein-coupled receptors delineate two major classes of receptors. J Biol Chem. 2000;275:17201–17210. doi: 10.1074/jbc.M910348199. [DOI] [PubMed] [Google Scholar]
- 32.Ferguson SS, Downey WE, 3rd, Colapietro AM, Barak LS, Menard L, Caron MG. Role of β-arrestin in mediating agonist-promoted G protein-coupled receptor internalization. Science. 1996;271:363–366. doi: 10.1126/science.271.5247.363. [DOI] [PubMed] [Google Scholar]
- 33.Goodman OB, Jr, Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL. β-arrestin acts as a clathrin adaptor in endocytosis of the β2-adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- 34.Kang DS, Tian X, Benovic JL. Role of β-arrestins and arrestin domain-containing proteins in G protein-coupled receptor trafficking. Curr Opin Cell Biol. 2014;27:63–71. doi: 10.1016/j.ceb.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lally CC, Bauer B, Selent J, Sommer ME. C-edge loops of arrestin function as a membrane anchor. Nat Commun. 2017;8:14258. doi: 10.1038/ncomms14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaidarov I, Krupnick JG, Falck JR, Benovic JL, Keen JH. Arrestin function in G protein-coupled receptor endocytosis requires phosphoinositide binding. EMBO J. 1999;18:871–881. doi: 10.1093/emboj/18.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milano SK, Pace HC, Kim YM, Brenner C, Benovic JL. Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry. 2002;41:3321–3328. doi: 10.1021/bi015905j. [DOI] [PubMed] [Google Scholar]
- 38.Janetzko J, Kise R, Barsi-Rhyne B, Siepe DH, Heydenreich FM, Kawakami K, Masureel M, Maeda S, Garcia KC, von Zastrow M, et al. Membrane phosphoinositides regulate GPCR-β-arrestin complex assembly and dynamics. Cell. 2022;185:4560–4573.:e19. doi: 10.1016/j.cell.2022.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurevich VV, Gurevich EV. The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol Ther. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gimenez LE, Kook S, Vishnivetskiy SA, Ahmed MR, Gurevich EV, Gurevich VV. Role of receptor-attached phosphates in binding of visual and non-visual arrestins to G protein-coupled receptors. J Biol Chem. 2012;287:9028–9040. doi: 10.1074/jbc.M111.311803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gimenez LE, Vishnivetskiy SA, Gurevich VV. Targeting individual GPCRs with redesigned nonvisual arrestins. Handb Exp Pharmacol. 2014;219:153–170. doi: 10.1007/978-3-642-41199-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seyedabadi M, Gharghabi M, Gurevich EV, Gurevich VV. Receptor-arrestin interactions: the GPCR perspective. Biomolecules. 2021;11:218. doi: 10.3390/biom11020218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh E, Dwivedi H, Baidya M, Srivastava A, Kumari P, Stepniew-ski T, Kim HR, Lee MH, van Gastel J, Chaturvedi M, et al. Conformational sensors and domain swapping reveal structural and functional differences between β-arrestin isoforms. Cell Rep. 2019;28:3287–3299.:e6. doi: 10.1016/j.celrep.2019.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dwivedi-Agnihotri H, Chaturvedi M, Baidya M, Stepniewski TM, Pandey S, Maharana J, Srivastava A, Caengprasath N, Hanyaloglu AC, Selent J, et al. Distinct phosphorylation sites in a prototypical GPCR differently orchestrate β-arrestin interaction, trafficking, and signaling. Sci Adv. 2020;6:abb8368. doi: 10.1126/sciadv.abb8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosh E, Srivastava A, Baidya M, Kumari P, Dwivedi H, Nidhi K, Ranjan R, Dogra S, Koide A, Yadav PN, et al. A synthetic intrabody-based selective and generic inhibitor of GPCR endocytosis. Nat Nanotechnol. 2017;12:1190–1198. doi: 10.1038/nnano.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benichou O, Grebenkov D, Levitz P, Loverdo C, Voituriez R. Optimal reaction time for surface-mediated diffusion. Phys Rev Lett. 2010;105:150606. doi: 10.1103/PhysRevLett.105.150606. [DOI] [PubMed] [Google Scholar]
- 47.Benichou O, Grebenkov DS, Levitz PE, Loverdo C, Voituriez R. Mean first-passage time of surface-mediated diffusion in spherical domains. J Stat Phys. 2011;142:657–685. doi: 10.1007/s10955-011-0138-6. [DOI] [Google Scholar]
- 48.Oakley RH, Laporte SA, Holt JA, Barak LS, Caron MG. Molecular determinants underlying the formation of stable intracellular G protein-coupled receptor-β-arrestin complexes after receptor endocytosis. J Biol Chem. 2001;276:19452–19460. doi: 10.1074/jbc.M101450200. [DOI] [PubMed] [Google Scholar]
- 49.Calebiro D, Koszegi Z, Lanoiselee Y, Miljus T, O’Brien SL. G protein-coupled receptor-G protein interactions: a single-molecule perspective. Physiol Rev. 2021;101:857–906. doi: 10.1152/physrev.00021.2020. [DOI] [PubMed] [Google Scholar]
- 50.Schreiber G, Haran G, Zhou HX. Fundamental aspects of protein-protein association kinetics. Chem Rev. 2009;109:839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prakash MK. Insights on the role of (dis)orderfrom protein-protein interaction linear free-energy relationships. J Am Chem Soc. 2011;133:9976–9979. doi: 10.1021/ja201500z. [DOI] [PubMed] [Google Scholar]
- 52.Kastritis PL, Bonvin AM. On the binding affinity of macromolecular interactions: daring to ask why proteins interact. J R Soc Interface. 2013;10:20120835. doi: 10.1098/rsif.2012.0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krasel C, Bunemann M, Lorenz K, Lohse MJ. β-arrestin binding to the β2-adrenergic receptor requires both receptor phosphorylation and receptor activation. J Biol Chem. 2005;280:9528–9535. doi: 10.1074/jbc.M413078200. [DOI] [PubMed] [Google Scholar]
- 54.Xiao K, Shenoy SK, Nobles K, Lefkowitz RJ. Activationdependent conformational changes in β-arrestin 2. J Biol Chem. 2004;279:55744–55753. doi: 10.1074/jbc.M409785200. [DOI] [PubMed] [Google Scholar]
- 55.Nobles KN, Guan Z, Xiao K, Oas TG, Lefkowitz RJ. The active conformation of β-arrestin1: direct evidence for the phosphate sensor in the N-domain and conformational differences in the active states of β-arrestins1 and-2. J Biol Chem. 2007;282:21370–21381. doi: 10.1074/jbc.M611483200. [DOI] [PubMed] [Google Scholar]
- 56.Sente A, Peer R, Srivastava A, Baidya M, Lesk AM, Balaji S, Shukla AK, Babu MM, Flock T. Molecular mechanism of modulating arrestin conformation by GPCR phosphorylation. Nat Struct Mol Biol. 2018;25:538–545. doi: 10.1038/s41594-018-0071-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schrage R, Schmitz AL, Gaffal E, Annala S, Kehraus S, Wenzel D, Bullesbach KM, Bald T, Inoue A, Shinjo Y, et al. The experimental power of FR900359 to study Gq-regulated biological processes. Nat Commun. 2015;6:10156. doi: 10.1038/ncomms10156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O’Hayre M, Eichel K, Avino S, Zhao X, Steffen DJ, Feng X, Kawakami K, Aoki J, Messer K, Sunahara R, et al. Genetic evidence that β-arrestins are dispensable for the initiation of β2-adrenergic receptor signaling to ERK. Sci Signal. 2017;10:eaal3395. doi: 10.1126/scisig-nal.aal3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barak LS, Ferguson SS, Zhang J, Caron MG. A β-ar-restin/green fluorescent protein biosensor for detecting G protein-coupled receptor activation. J Biol Chem. 1997;272:27497–27500. doi: 10.1074/jbc.272.44.27497. [DOI] [PubMed] [Google Scholar]
- 61.Zhang J, Barak LS, Anborgh PH, Laporte SA, Caron MG, Ferguson SS. Cellular trafficking of G protein-coupled receptor/β-ar-restin endocytic complexes. J Biol Chem. 1999;274:10999–11006. doi: 10.1074/jbc.274.16.10999. [DOI] [PubMed] [Google Scholar]
- 62.Laporte SA, Oakley RH, Zhang J, Holt JA, Ferguson SS, Caron MG, Barak LS. The β2-adrenergic receptor/β-arrestin complex recruits the clathrin adaptor AP-2 during endocytosis. Proc Natl Acad Sci USA. 1999;96:3712–3717. doi: 10.1073/pnas.96.7.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laporte SA, Oakley RH, Holt JA, Barak LS, Caron MG. The interaction of β-arrestin with the AP-2 adaptor is required for the clustering of β2-adrenergic receptor into clathrin-coated pits. J Biol Chem. 2000;275:23120–23126. doi: 10.1074/jbc.M002581200. [DOI] [PubMed] [Google Scholar]
- 64.Heydenreich FM, Miljus T, Jaussi R, Benoit R, Milić D, Veprint-sev DB. High-throughput mutagenesis using a two-fragment PCR approach. Sci Rep. 2017;7:6787. doi: 10.1038/s41598-017-07010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kilpatrick LE, Alcobia DC, White CW, Peach CJ, Glenn JR, Zimmerman K, Kondrashov A, Pfleger KDG, Ohana RF, Robers MB, et al. Complex formation between VEGFR2 and the β2-adrenocep-tor. Cell Chem Biol. 2019;26:830–841.:e9. doi: 10.1016/j.chembiol.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White CW, Vanyai HK, See HB, Johnstone EKM, Pfleger KDG. Using nanoBRET and CRISPR/Cas9 to monitor proximity to a genome-edited protein in real-time. Sci Rep. 2017;7:3187. doi: 10.1038/s41598-017-03486-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cocucci E, Aguet F, Boulant S, Kirchhausen T. The first five seconds in the life of a clathrin-coated pit. Cell. 2012;150:495–507. doi: 10.1016/j.cell.2012.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Min K, Yoon HJ, Park JY, Baidya M, Dwivedi-Agnihotri H, Maha-rana J, Chaturvedi M, Chung KY, Shukla AK, Lee HH. Crystal structure of β-arrestin 2 in complex with CXCR7 phosphopeptide. Structure. 2020;28:1014–1023.:e4. doi: 10.1016/j.str.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pandey S, Kumari P, Baidya M, Kise R, Cao Y, Dwivedi-Agnihotri H, Banerjee R, Li XX, Cui CS, Lee JD, et al. Intrinsic bias at non-canonical, β-arrestin-coupled seven transmembrane receptors. Mol Cell. 2021;81:4605–4621.:e11. doi: 10.1016/j.molcel.2021.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jain A, Liu R, Xiang YK, Ha T. Single-molecule pull-down for studying protein interactions. Nat Protoc. 2012;7:445–452. doi: 10.1038/nprot.2011.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Willy NM, Colombo F, Huber S, Smith AC, Norton EG, Kural C, Cocucci E. CALM supports clathrin-coated vesicle completion upon membrane tension increase. Proc Natl Acad Sci USA. 2021;118:e2010438118. doi: 10.1073/pnas.2010438118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsunoyama TA, Watanabe Y, Goto J, Naito K, Kasai RS, Suzuki KGN, Fujiwara TK, Kusumi A. Super-long single-molecule tracking reveals dynamic-anchorage-induced integrin function. Nat Chem Biol. 2018;14:497–506. doi: 10.1038/s41589-018-0032-5. [DOI] [PubMed] [Google Scholar]
- 73.Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kepten E, Weron A, Sikora G, Burnecki K, Garini Y. Guidelines for the fitting of anomalous diffusion mean square displacement graphs from single particle tracking experiments. PLoS One. 2015;10:e0117722. doi: 10.1371/journal.pone.0117722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lanoiselee Y, Sikora G, Grzesiek A, Grebenkov DS, Wyłoman-ska A. Optimal parameters for anomalous-diffusion-exponent estimation from noisy data. Phys Rev E. 2018;98:062139. doi: 10.1103/PhysRevE.98.062139. [DOI] [Google Scholar]
- 76.Cherstvy AG, Thapa S, Mardoukhi Y, Chechkin AV, Metzler R. Time averages and their statistical variation for the Ornstein-Uhlenbeck process: role of initial particle distributions and relaxation to stationarity. Phys Rev E. 2018;98:022134. doi: 10.1103/PhysRevE.98.022134. [DOI] [PubMed] [Google Scholar]
- 77.Richardson WH. Bayesian-based iterative method of image restoration. J Opt Soc Am. 1972;62:55–59. doi: 10.1364/JOSA.62.000055. [DOI] [Google Scholar]
- 78.Lucy LB. An iterative technique for the rectification of observed distributions. Astron J. 1974;79:745. doi: 10.1086/111605. [DOI] [Google Scholar]
- 79.Gustafsson N, Culley S, Ashdown G, Owen DM, Pereira PM, Henriques R. Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations. Nat Commun. 2016;7:12471. doi: 10.1038/ncomms12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 81.Fiser A, Do RK, Sali A. Modeling of loops in protein structures. Prot Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhan X, Gimenez LE, Gurevich VV, Spiller BW. Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual subtypes. J Mol Biol. 2011;406:467–478. doi: 10.1016/j.jmb.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Symons JL, Cho KJ, Chang JT, Du G, Waxham MN, Hancock JF, Levental I, Levental KR. Lipidomic atlas of mammalian cell membranes reveals hierarchical variation induced by culture conditions, subcellular membranes, and cell lineages. Soft Matter. 2021;17:288–297. doi: 10.1039/d0sm00404a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 85.Wu EL, Cheng X, Jo S, Rui H, Song KC, Davila-Contreras EM, Qi Y, Lee J, Monje-Galvan V, Venable RM, et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J Comput Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harvey MJ, Giupponi G, Fabritiis GD. ACEMD: accelerating BioMolecular Dynamics in the microsecond time scale. J Chem Theor Comput. 2009;5:1632–1639. doi: 10.1021/ct9000685. [DOI] [PubMed] [Google Scholar]
- 87.Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmuller H, MacKerell AD., Jr CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr, Pastor RW. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, Macker-ell AD. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J Chem Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rodríguez-Espigares I, Torrens-Fontanals M, Tiemann JKS, Aranda-García D, Ramírez-Anguita JM, Stepniewski TM, Worp N, Varela-Rial A, Morales-Pastor A, Medel-Lacruz B, et al. GPCRmd uncovers the dynamics of the 3D-GPCRome. Nat Methods. 2020;17:777–787. doi: 10.1038/s41592-020-0884-y. [DOI] [PubMed] [Google Scholar]
- 91.Grest GS, Kremer K. Molecular dynamics simulation for polymers in the presence of a heat bath. Phys Rev A Gen Phys. 1986;33:3628–3631. doi: 10.1103/physreva.33.3628. [DOI] [PubMed] [Google Scholar]
- 92.Eslami H, Mozaffari F, Moghadasi J, Muller-Plathe F. Molecular dynamics simulation of confined fluids in isosurface-isothermal-isobaric ensemble. J Chem Phys. 2008;129:194702. doi: 10.1063/1.3009844. [DOI] [PubMed] [Google Scholar]
- 93.Darden T, York D, Pedersen L. Particle mesh Ewald: an Nolog(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 94.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 95.Bussi G, Laio A. Using metadynamics to explore complex free-energy landscapes. Nat Rev Phys. 2020;2:200–212. doi: 10.1038/s42254-020-0153-0. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw single-molecule microscopy data reported in this study cannot be deposited in a public repository because of their large size. To request access, contact the lead contact. The results of the unbiased MD simulations have been deposited at GPCRmd and are publicly available as of the date of publication. The accession number is listed in the key resources table.
All original code has been deposited at Zenodo and is publicly available as of the date of publication. The DOI is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.