Abstract
Atypical chemokine receptor sub-type 3 (ACKR3), a chemokine receptor, couples selectively to β-arrestins (βarrs) but not to G proteins despite having seven transmembrane (7TM) helix architecture. Yen et al. present cryogenic-electron microscopy (cryoEM) structures of agonist-bound ACKR3, elucidating a distinct chemokine-binding mechanism, and offering a structural template to probe the transducer-coupling bias at this receptor.
Chemokines (see Glossary) are small proteins secreted by immune cells that activate integral membrane receptors present in the plasma membrane known as chemokine receptors [1]. These receptors typically belong to the large family of G protein-coupled receptors (GPCRs), which are characterized by their 7TM architecture, coupling to heterotrimeric G proteins, GPCR kinases (GRKs), and multifunctional proteins known as βarrs [1]. Che-mokine receptors are critically involved in the functioning and regulation of our immune response, including inflammatory mechanisms in a pathophysiological context [1]. Interestingly, however, some chemokine receptors – such as the CXC-type chemokine receptor 7 (CXCR7, also known as ACKR3) – fail to activate G pro-teins but still maintain a robust interaction with βarrs [2–4]. These arrestin-coupled receptors (ACRs) represent natural examples of intrinsically biased 7TM receptors (7TMRs) [5]. Understanding the molecular mechanisms of their activation and inherent transducer-coupling bias re-mains an important direction in the broader context of 7TMR activation, signaling, and downregulation. Yen et al. present structural snapshots of agonist-bound ACKR3 using cryo-EM [6] which provide a frame-work to analyze chemokine interaction and activation-dependent conformational changes in this receptor.
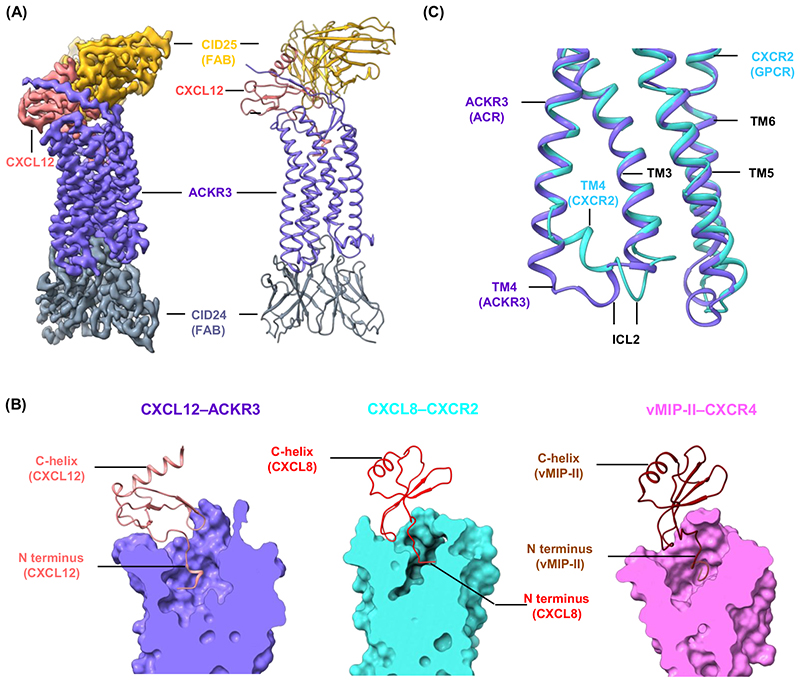
Considering the small size of ACKR3, Yen et al. first generated and characterized antibody fragments (FABs) targeted against the extracellular and intracellular surface of the receptor, and utilized these FABs, either together or individually, to determine the structure of ACKR3 using cryo-EM [6](Figure 1A). In these structures, the receptor is occupied by either its natural chemokine agonist, CXCL12, a modified version of CXCL12 referred to as CXCL12LRHQ, or a small-molecule agonist, CCX662. So, what does the structure of ACKR3 tell us about the chemokine recognition by the receptor? The binding interface of chemokines on chemokine receptors is typically believed to follow a two-site model involving the N terminus of the receptor [known as chemokine recognition site 1 (CRS1)] and the extracellular face of the transmembrane bundle [known as chemokine recognition site 2 (CRS2)] [7]. The binding of CXCL12 to ACKR3 exhibits overall a similar pattern where the β1-strand of CXCL12 interfaces with the N terminus of ACKR3, while the C terminus of CXCL12 inserts deeper into the receptor core sitting in proximity with multiple residues from various trans-membrane helices (TMs) and ECL2 (Figure 1B). Interestingly, however, the positioning of CXCL12 on the receptor differs significantly from the previously visualized chemokine–chemokine receptor complexes. For example, a significant rotation of CXCL12 on ACKR3 is observed in comparison with the binding mode of other CXC chemokines such as CXCL8 in complex with CXCR2 and a viral chemokine analog vMIP-II with CXCR4 (Figure 1B). This distinctive binding pose of CXCL12 does not arise due to the stabilizing FAB (CID25), as the structure of CXCL12–ACKR3 – even in the absence of FAB – shows an almost identical binding orientation [6].
Figure 1. Chemokine recognition and activation of atypical chemokine receptor subtype 3 (ACKR3).
(A) Overall structural snapshot of CXCL12–ACKR3 in the presence of two stabilizing antigen-binding fragments of antibodies (FABs) (CID25 and CID24). (B) Distinct binding pose of CXCL12 to ACKR3 compared with CXCL8 and viral macrophage inflammatory protein II (vMIP-II) on CXCR2 and CXCR4, respectively. (C) Transmembrane helix 4 (TM4) in ACKR3 is an extended conformation at the cytoplasmic end while it exhibits a kink in the CXCL8-bound CXCR2 structure. These structural snapshots are designed based on PDB ID 7SK3, 6LFO, and 4RWS for ACKR3, CXCR2, and CXCR4, respectively. Abbreviations: ACR, arrestin-coupled receptor; GPCR, G protein-coupled receptor.
The overall arrangement of TMs in ACKR3 is comparable to other active GPCR structures, including that of CXCR2 in complex with heterotrimeric G proteins. Some of the key motifs and microswitches such as the NPXXY in TM7 and the PIF signature encompassing TM3, 5, and 6 exhibit active-like conformation, which is also accompanied by an outward movement of TM6 [6]. These conformational changes do not appear to be influenced by the binding of FAB (CID24) on the cytoplasmic side of the receptor as they are also reflected in the structure determined without it. These active-like features observed in ACKR3 may explain its constitutive trafficking and association with βarr2 that has been reported previously in some cell types [8].
Does structural visualization of ACKR3 elucidate the mechanism of its transducer-coupling bias compared to other prototypical chemokine receptors? There are some interesting insights and testable hypotheses into this aspect that can be drawn from the structures. For example, the cytoplasmic end of TM4 in ACKR3 shows an extended conformation and lacks a kink that was observed in the previously determined structure of CXCR2 (Figure 1C). This kink in CXCR2 presumably helps ICL2 position in a manner that allows the docking of the N-terminal helix of Gαi to the receptor, and the extended TM4 in ACKR3 may interfere with the optimal positioning of Gαi. It should be noted, however, that the potential collision with Gαi due to extended TM4 in ACKR3 is likely to be rather small con-sidering a much larger binding interface of Gαi, as observed for other GPCRs, and therefore it may not be the sole reason for the lack of Gαi coupling to ACKR3. As the interaction with βarrs is driven primarily by receptor phosphorylation, the active-like features observed in ACKR3 may be sufficient for GRK engagement and subsequent recruitment of βarrs.
So, what are the key questions that remain unanswered? First, while the structure offers a putative hypothesis for the lack of G protein coupling to ACKR3, establishing the precise molecular mechanism requires additional experimentation. Considering the dynamic nature of receptor–G protein interaction, it is plausible that a direct answer will emerge from studies focused on local conformational changes in the receptor, especially at the interface of cytoplasmic ends of TM helices and the connecting intracellular loops [9]. Moreover, considering that ACKR3 is a βarr-biased 7TMR, the molecular basis of its interaction with βarrs and how it differs from prototypical GPCRs also remain central and unanswered questions. The interaction of GPCRs with βarrs typically involves a biphasic mechanism involving the phosphorylated C terminus of the receptor (tail interaction) and the activated transmembrane bundle (core interaction) [10]. While the structural snapshots do not preclude the possibility of core interaction, it is tempting to speculate that ACKR3 may engage βarrs primarily through the tail interaction, considering that the core interaction facilitates the termination of G protein coupling. However, this hypothesis remains to be tested experimentally using biophysical and structural approaches, including direct visualization of ACKR3–βarr complexes. While some chemokine receptors exhibit a broad promiscuity in terms of chemokine binding, ACKR3 recognizes only two chemokines – namely, CXCL12 and CXCL11 – and therefore it would be interesting to also visualize the binding mode of CXCL11 to ACKR3.
Finally, there are several additional examples of βarr-biased 7TMRs including a non-chemokine receptor: the complement C5a receptor subtype 2 (C5aR2) [5]. Future studies focusing on deciphering the structural features of these additional receptors are essential to develop a holistic framework of intrinsic bias at 7TMRs with direct implications for biased agonism at GPCRs in general.
In summary, the work by Yen et al. provides direct structural visualization of CXCL12–ACKR3 interaction, and a distinct binding pattern of CXCL12 compared to other chemokines offers a potential structural template to probe the implications of distinct binding modalities on downstream functional responses. However, the quest to understand the structural basis of intrinsic bias exhibited by ACKR3 and other similar βarr-biased receptors remains very much open.
Acknowledgments
The research program in the laboratory of A.K.S. is supported by the Senior Fellowship of DBT Wellcome Trust India Alliance (IA/S/20/1/504916), and Science and Engineering Research Board (SERB) (SPR/2020/000408 and IPA/2020/000405), and Indian Council for Medical Research (F.NO.52/15/2020/BIO/BMS).
Glossary
- Arrestin-coupled receptors (ACRs)
seven transmembrane receptors (7TMRs) which selectively engage βarrs without any measurable activation of G proteins such as ACKR3 (CXCR7).
- Biased agonism
the ability of an agonist to preferentially engage one signal transducer over another and activate the corresponding downstream signaling, such as G protein versus βarrs.
- Chemokines
small proteins secreted by the immune cells that mediate cellular migration.
- FABs
antigen-binding fragments of antibodies consisting of the variable heavy and light chain domains with a part of the constant domain without the Fc region.
- Intrinsic bias
the property of certain 7TMRs to selectively recruit and signal through a specific signal transducer even in response to activation by its natural (endogenous) agonist.
Footnotes
Declaration of interests
The authors declare no conflicts of interest.
References
- 1.Allen Sj, et al. Chemokine: receptor structure, interactions, and antagonism. Annu Rev Immunol. 2007;25:787–820. doi: 10.1146/annurev.immunol.24.021605.090529. [DOI] [PubMed] [Google Scholar]
- 2.Rajagopal S, et al. Beta-arrestin-but not G protein-mediated signaling by the "decoy" receptor CXCR7. Proc Natl Acad Sci U S A. 2010;107:628–632. doi: 10.1073/pnas.0912852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarma YHP, et al. Molecular insights into intrinsic transducer-coupling bias in the CXCR4-CXCR7 system. bioRxiv. 2022 doi: 10.1101/2022.06.06.494935. Published online June 7 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachelerie F, et al. New nomenclature for atypical chemokine receptors. Nat Immunol. 2014;15:207–208. doi: 10.1038/ni.2812. [DOI] [PubMed] [Google Scholar]
- 5.Pandey S, et al. Intrinsic bias at non-canonical, beta-arrestin-coupled seven transmembrane receptors. Mol Cell. 2021;81:e4611. doi: 10.1016/j.molcel.2021.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yen Yc, et al. Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias. Sci Adv. 2022;8:eabn8063. doi: 10.1126/sciadv.abn8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scholten Dj, et al. Pharmacological modulation of che-mokine receptor function. Br J Pharmacol. 2012;165:1617–1643. doi: 10.1111/j.1476-5381.2011.01551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luker Ke, et al. Constitutive and chemokine-dependent internalization and recycling of CXCR7 in breast cancer cells to degrade chemokine ligands. Oncogene. 2010;29:4599–4610. doi: 10.1038/onc.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleist Ab, et al. Conformational selection guides beta-arrestin recruitment at a biased G protein-coupled receptor. Science. 2022;377:222–228. doi: 10.1126/science.abj4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maharana J, et al. Emerging structural insights into GPCR-beta-arrestin interaction and functional outcomes. Curr Opin Struct Biol. 2022;75:102406. doi: 10.1016/j.sbi.2022.102406. [DOI] [PMC free article] [PubMed] [Google Scholar]