Abstract
Peptides are biopolymers, typically consisting of 2–50 amino acids. They are biologically produced by the cellular ribosomal machinery or by non-ribosomal enzymes and, sometimes, other dedicated ligases. Peptides are arranged as linear chains or cycles, and include post-translational modifications, unusual amino acids and stabilizing motifs. Their structure and molecular size render them a unique chemical space, between small molecules and larger proteins. Peptides have important physiological functions as intrinsic signalling molecules, such as neuropeptides and peptide hormones, for cellular or interspecies communication, as toxins to catch prey or as defence molecules to fend off enemies and microorganisms. Clinically, they are gaining popularity as biomarkers or innovative therapeutics; to date there are more than 60 peptide drugs approved and more than 150 in clinical development. The emerging field of peptidomics comprises the comprehensive qualitative and quantitative analysis of the suite of peptides in a biological sample (endogenously produced, or exogenously administered as drugs). Peptidomics employs techniques of genomics, modern proteomics, state-of-the-art analytical chemistry and innovative computational biology, with a specialized set of tools. The complex biological matrices and often low abundance of analytes typically examined in peptidomics experiments require optimized sample preparation and isolation, including in silico analysis. This Primer covers the combination of techniques and workflows needed for peptide discovery and characterization and provides an overview of various biological and clinical applications of peptidomics.
Introduction
Peptides occur in all three domains of life. Their native functions comprise acting as peptide hormones for cellular signalling, as secretory peptides for interspecies communication and interaction, as predatory peptide toxins and as defence peptides against microorganisms, viruses and herbivores1. Peptides are important molecules that play a fundamental role in human physiology and pathology. In addition, natural sources including animals, plants, fungi and microorganisms provide rich sources of biologically active peptides1,2. Research has focused on investigating and understanding the biological function of peptides and their potential as disease biomarkers or therapeutic lead compounds and drugs3,4.
Peptides are molecular entities of amino acids linked via amide bonds rendering the peptide backbone or peptide chain. Although the size discrimination between a large peptide and a small protein is rather arbitrary, the scientific community typically refers to a peptide containing between 2 and 50 amino acids. Most peptides are encoded by DNA or RNA and, hence, are products of translation, transcription and the cellular protein manufacturing machinery. Typically, they contain many and often complex post-translational modifications, including side-chain modifications, disulfide bond formation, individual residue isomerization and, sometimes, head-to-tail or side-chain cyclizations. Non-ribosomal enzymatically synthesized peptides (typically produced by bacteria and fungi) further increase the variety of peptide species by incorporating non-proteinogenic building blocks and a set of additional tailoring reactions. Overall, naturally occurring peptides range between simple linear and often heavily modified molecules, which occupy a large and unique chemical space in between small organic molecules and larger proteins5.
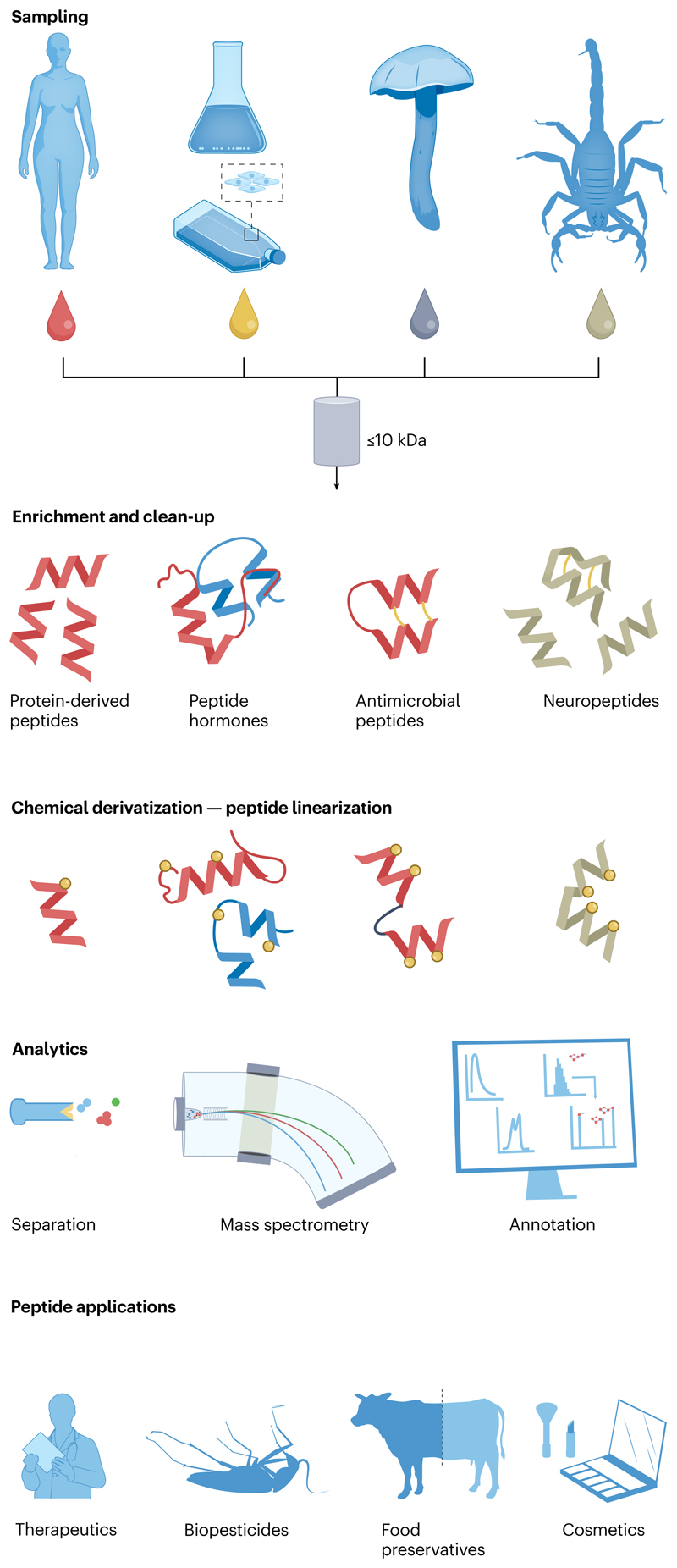
The highly diverse and easily modified peptides from nature are among the most interesting molecules. Peptides can be obtained from various sources and can range from structurally simple to highly functionalized and complex structures, giving them a plethora of biological features (Fig. 1). Although differences in the extent of modification might appear extreme, they could still exert the same biological function. Owing to their relatively small size and chemical potential, peptides can possess valuable chemical information for the design of novel drugs6.
Fig. 1. Peptides, from analysis to application.
The sources of peptides are vast, and many serve diverse biological roles. However, peptides are often difficult to detect without proper enrichment or clean-up strategies for use in downstream applications. Subsequent modification or derivatization of the peptides may be required to elucidate their sequence or structural features, which is essential for further design or use of discovered peptides in medical or industrial applications
The term peptidomics was introduced to define a strategy for the direct measurement and structural characterization of endogenous peptides in biological systems in a high-throughput manner, with robust and unprecedented sensitivity7–9. Since then, peptidomics became a fast developing and progressing multidisciplinary field that combines state-of-the-art separation techniques including liquid chromatography, modern mass spectrometry technologies, innovative bioinformatics and statistics for qualitative and quantitative analysis of peptides relevant to fundamental biology and human health sciences. Although the field of peptidomics resorts to various technologies that have been developed for proteomics, analytical chemistry and genomics, it is the unique sample preparation and specialized combination of these tools and methods that make peptidomics a unique discipline and research field.
Peptidomics refers to a system-level study of a set of analytes with the aim to describe the number and identity as well as the relative or absolute levels of peptides. As with other biological omics fields, peptidomics has a significant overlap with genomics and proteomics. Bioinformatic approaches have been utilized to discover genes encoding for peptides and proteins the peptide product could have originated from, for potential use as novel medicinal products or as clinical biomarkers. With more newly discovered peptides, as the algorithms used become more powerful, bioinformatics and high-resolution mass spectrometry are now shaping the future of biological sciences.
Peptides can have various applications, ranging from food preservatives to therapeutic agents in humans (Fig. 1). However, the use of peptides goes even beyond that, as peptides can act as catalysts, improving the yields and enantioselectivity of chemical synthesis10. Highly efficient and selective catalysis was originally considered to be the domain of proteins, but peptides can be modified to catalyse simple chemical reactions. Biomaterials are now a trending topic, where sustainability and recyclability will be a requirement for future industries11,12. Peptides are accessible and chemically diverse oligomers, making them a popular choice for research and development, with ample application outside biological sciences. However, the discovery and characterization of new peptides lays the foundation for their future utilization.
This Primer aims to define and describe peptidomics technologies, tools and workflows for identification and analysis of endogenous peptides in a qualitative and quantitative manner. The field has rapidly advanced in the past three decades with the development of computational and mass spectrometry-based techniques. Remaining analytical challenges and pending questions in peptidome analysis are discussed for selected applications. The Primer covers various in silico and peptide analysis methodologies including de novo sequencing, high-throughput and automated peptidomics workflows, peptide imaging and quantitative mass spectrometry approaches. The application of peptidomics in biology, in drug discovery and in the clinic to identify novel biomarkers and understand disease mechanisms are discussed. Lastly, reproducibility and data deposition are discussed, followed by current limitations and the outlook regarding ongoing developments in the field of peptidomics.
Experimentation
Modern peptidomics workflows encompass the analysis of genetic information, characterization of peptides and computational processing of the data. Although this multilevel analysis allows for more comprehensive read-outs, a single-step analysis also qualifies to be counted into the field of peptidomics. Genetic information is accessed by peptide precursor mining or metabolic network analysis. Working at the peptide level can be subdivided into several steps, including sample preparation and clean-up, mass spectrometric analysis and data evaluation or integration.
Sample preparation
Multiple workflows have been described that proved useful for analysis of various peptide entities. State-of-the-art peptidomics workflows are amenable to complex samples, as they are obtained from plant, microbial cell or animal/human tissue extractions. However, sample processing steps are usually implemented to enrich analytes over matrix compounds and to concentrate low-abundance analytes. This Primer describes common pipelines; several of these methods can be modularly combined for workflows suitable to address the needs of any specific research questions. Approaches are optimized by trial and error to obtain the desired outcomes.
Sample harvest, cell lysis and extraction
Depending on the source of peptides, there are several harvest and extraction procedures available. A common problem during sample preparation is the degradation of peptides — especially low-abundance peptides, for example by cellular proteases. For instance, neuropeptides and peptide hormones are biosynthesized as larger precursor proteins; they are converted into their bioactive form by processing enzymes that cleave their precursor into several final active messengers or hormones13. As a neuropeptide’s biological message is transient, active peptides become inactivated by proteases14. Optimizing sampling approaches is important to counteract rapid postmortem/post-harvest degradation. To increase the yield of active peptides for analysis, degrading enzymes can be inactivated by heating tissue/cells, or by addition of protease inhibitors or chaotropic agents15,16. Extended heat treatments should be avoided as peptides are prone to chemical reactions, such as hydrolysis and oxidation. Processing/storing samples at low temperature helps avoid peptide degradation/modification. These steps are less crucial for stabilized peptides, such as plant defence peptides or venom-derived peptide toxins. For efficient extraction of peptide analytes, the cells/tissue must be broken up by cell lysis. For instance, cells derived from cell cultures or from collagen-treated tissues can be lysed with hypotonic buffers followed by ultra-sonication to disintegrate the phospholipid bilayers to release intracellular material17. Throughout these steps, a buffer amenable to dissolving the peptides should be used, which can be aqueous or organic depending on the nature of the sample. Obviously, the heat treatment of biological samples for protease inactivation will already be the first step to induce lysis. The lysis of microorganisms or plants requires specialized protocols18. For instance, to extract peptides from plants, the source material is mechanically ground or pulverized (using a grinder/shredder or liquid N2 and mortar/pestle); the peptides are then extracted by use of organic solvents in combination with alcohols/aqueous buffers19. The organic solvent used to remove chlorophyll not only helps break down cellular membranes to release the peptides into the buffer but also inhibits any unwanted protease degradation. Generally, detergent-based lysis using cationic, zwitterionic or anionic detergents is not compatible with mass spectrometry (but there are alternative methods available20). The efficient and repeatable peptide extraction from biological matrices is key for further sample processing, and to achieve maximum concentration of the peptide analytes.
Enrichment and clean-up strategies
After extraction, crude biological samples contain a low abundance of peptides, as they also contain salts, lipids, proteins and carbohydrates21, which makes purification steps necessary. The complexity of this matrix background (molecules other than the peptides of interest) can impair the ability of mass spectrometry to identify the peptides of interest. Although less complicated sample preparation techniques are needed for peptidomics compared with proteomics, several clean-up techniques are effective for enriching the peptides of interest prior to instrumental analysis22. Solid-phase extraction (SPE) is commonly applied for sample concentration and desalting, as a rapid, stand-alone tool. This is especially important if the samples are being analysed directly by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry, a soft ionization technique, as high salt concentrations suppress analyte–matrix crystallization and ionization. For reversed-phase SPE there are many different types of cartridges (ranging in size, volume, chemistry, vendor) available. They can be used for single or multi-channel sample processing using vacuum manifold systems, which can handle up to 24 cartridges simultaneously, or in 96/384-well format, utilizing SPE containing plates for automated sample clean-up. For rapid clean-up of small sample volumes, ZipTips or alternative pipette tip SPE devices have become useful23. Alternative but low-resolution (and sometimes lowrecovery) methods for sample clean-up are molecular weight cut-off or nanofiltration devices and classical sample precipitation techniques. A more efficient method for sample clean-up is liquid chromatography. This technique can be used offline or coupled directly to mass spectrometry. It is compatible with numerous separation chemistries. One example is food-derived peptides, a large class of bioactive molecules usually ranging in size from 2 to 20 amino acids. They are generated from enzymatic food hydrolysates during digestion or fermentation. The isolation of peptides from the hydrolysate matrix is achieved by a combination of ultrafiltration and chromatographic separation techniques, such as reversed-phase, ion exchange and size exclusion chromatography24. The isolated peptides are further analysed to the amino acid level using amino acid quantitation, Edman degradation or mass spectrometry-based de novo sequencing.
Chemical derivatization and peptide labelling
For certain peptidomics applications, such as discovery, isolation and analysis of cysteine-rich peptides, chemical derivatization of cysteine residues may be beneficial. Disulfide bridged or cysteine knot peptides are chemically reduced to the sulfhydryl-containing compound with mild reduction reagents and, then, immediately converted into stable derivatives. Thiol-containing reductants are β-mercaptoethanol, glutathione and dithiothreitol (DTT) supporting sulfhydryl deprotonation under basic conditions. Phosphine-based reagents, for example tris(2-carboxyethyl)phosphine (TCEP), are considered more stable than thiols and are amenable to all pH conditions25. The cysteine sulfhydryl group derivatization makes further use of a few standard reagents, preferably halogenated acetate derivatives (for example, iodoacetamide, iodoacetic acid), N-substituted maleimides (for example, N-ethyl maleimide), 4-vinylpyridine or thiosulfonate reagents (for example, methyl-methane thiosulfonate)26,27. Iodoacetamide has become a standard working horse for proteomics and peptidomics applications. Derivatization was also explored for peptide quantification via cysteine derivatization. For instance, owing to its chromophore, 5,5-dithiobis(2-nitrobenzoate) (DTNB) provides labelled peptides that can be quantitatively analysed by HPLC–UV detectors. Other labelling approaches established in the peptidomics field support the detection and quantitation or comparative analysis of peptidome samples.
Labelling of peptides with stable isotopes is a common strategy in peptidomics to increase sensitivity and detection and to enable quantitative analysis. Labelling of peptides of interest with stable isotopes in biological models can be achieved by metabolic, enzymatic and chemical strategies. Metabolic labelling introduces stable isotopes through metabolic incorporation in vivo, often via diet or culture media, and thus leads to long experimental procedures. Several chemistry-based labelling protocols have been implemented that enable stable isotope tagging of peptides in vitro28–32. There are several commercial tags available, for example tandem mass tags (TMT)33 and isobaric tags for relative and absolute quantitation (iTRAQ)34; there are also many lowcost alternatives available, including N,N-dimethyl leucine (DiLeu)31, deuterium isobaric amine reactive tag (DiART)35 and 10-plex isobaric tags (IBT)36.
Peptide separation
Crude cell or tissue extracts typically present high chemical complexity and a large concentration range of diverse compounds. As peptides may exist in solution as charged molecules with differing degrees of hydrophobicity, they are amenable to several separation technologies. Although reversed-phase liquid chromatography has been widely used, size exclusion, ion exchange and mixed-mode applications are gaining attraction with continuous development of new sorbent materials37. Chromatographic separation systems are either directly hyphenated to mass spectrometry via the electrospray ionization (ESI) interface or performed offline38. Reversed-phase chromatography promises high peak capacity and excellent resolution. Particles in use are modified silica gels with alkyl group substitution, with pore sizes typically ranging from 110 to 300 Å to facilitate adsorptive analyte–stationary phase interactions. The alkyl chain length is the major determinant for overall hydrophobicity of the material and the retention power for analytes. Octadecyl (C18), octyl (C8) or butyl (C4) alkylated silica materials are the most common. The selectivity for analytes and their peak shape can further be improved by end-capping, cross-linking or other modifications. For instance, trimethylsilyl or polar groups are used for fine-tuning selectivity, resolution and retention capacity of the stationary phase. The optimal choice of reversed-phase stationary material depends on the structural and chemical diversity of peptide analytes. For example, C8 material commonly enables better separation of basic and neutral molecules (for example, tryptic peptides) under acidic conditions. The mobile phases in reversed-phase applications are aqueous and organic solvents, usually methanol or acetonitrile with acidic modifiers. Trifluoroacetic acid (TFA) is a strong ion-pairing counter-ion providing very reliable and reproducible peak shapes and overall separations39. TFA is unfortunately not well compatible with the ESI technique commonly applied in liquid chromatography–mass spectrometry (LC-MS) systems. Here, formic acid is used instead and ammonium formate or ammonium acetate can be used as the mass spectrometry-compatible buffer system.
More specialized applications such as affinity-based and immobilized-metal affinity chromatography are used to enrich or isolate phosphopeptides from tissue extracts, whereas hydrophilic liquid interaction chromatography is utilized for separation of glycopeptides40. For mass and volume-limited samples, capillary electrophoresis can be used as a front-end separation method. One advantage of capillary electrophoresis is that it is compatible with one to two orders of magnitude smaller sample volumes than liquid chromatography systems, and therefore is the best choice for cellular and subcellular peptidomics.
The most recent addition to peptide separation approaches is ion mobility spectrometry (IMS) that sorts and separates gas-phase ions according to their 3D shapes. Placing an IMS module between the source and the mass analyser increases ion utilization efficiency, improves sensitivity and specificity of detection, and broadens the dynamic range. Because of the introduction of a new range of IMS instruments, this approach is rapidly gaining application in peptide characterization and quantitation.
Glossary.
Fourier transform ion cyclotron resonance mass spectrometers
(FTICR-MS). High-resolution mass analysers that trap ions in a cyclotron radius by applying a fixed magnetic field and an oscillating electronic field. As the ions rotate, an interferogram signal is recorded by electrodes and the useful mass spectrum is extracted with a Fourier transformation.
Hyphenated front-end separation platforms
Platforms that separate the analytes online before they enter the mass spectrometers. Techniques include, but are not limited to, liquid chromatography, gas chromatography, ion mobility spectrometry (IMS), solidphase extraction (SPE) and capillary electrophoresis.
Ion mobility spectrometry
(IMS). An analytical technique that sorts and separates gas-phase ions based on their mobility in a carrier buffer gas under the influence of an electrical field, which is related to the conformation and 3D shapes of the molecules.
Multiple reaction monitoring
A type of analysis for tandem mass spectrometers providing capabilities for quantitation of analytes. Pre-defined precursor ions (m/z) are selected by the first mass analyser and submitted to a fragmentation, and the selected product ion m/z signals are detected by the second mass analyser.
Peptide dereplication
Refers to the identification of known peptides in a sample by comparing mass spectrometric data with a library. The identification can be obtained by comparison of m/z mass signals, including the isotopologue intensities and pattern of isotopologues, giving information on the chemical composition as well as on tandem mass spectrometry (MS/MS) fragmentation spectra match with library data.
Peptide spectrum match
(PSM). A scoring function in which the mass spectrum of a peptide is compared with a theoretical peptide sequence to determine the probability of the measured peptide matching the theoretical peptide.
Post-source decay
A type of fragmentation technique that applies when metastable ions spontaneously decompose in the drift region between the ion source and reflectron.
Short open reading frames
(sORFs). Open reading frames that occur throughout the genome and usually comprise <100 codons. They are a possible source for peptides with biological relevance.
Mass spectrometry technologies
Mass spectrometry instrumentation
There are various different mass spectrometry systems available, which can be categorized by the ionization technique, resolution of the mass analyser, single or tandem set-up of the system or mass analyser type. The main soft ionization techniques are MALDI41,42 and ESI38. Fourier transform ion cyclotron resonance mass spectrometers (FTICR-MS) are high-resolution instruments suitable for peptide mapping and characterization via accurate mass matching and mass spectrometry imaging (MSI) studies. High-speed mass spectrometry systems, typically equipped with a quadrupole, time-of-flight (TOF) or ion trap mass analyser, are useful for quantitative analysis43,44. Most of these systems have the capacity to perform tandem mass spectrometry and fragmentation experiments in the MS/MS or MS/MS/MS mode (such as ion trap and orbitrap systems)45,46. Peptide fragmentation uses different techniques, such as post-source decay, high-energy collision-induced dissociation or electron transfer dissociation methods47–49. Peptidomics studies utilize several mass analysers of various types, such as TOF, quadrupole, ion trap and, more recently due to its high resolving power, orbitrap23 or ion cyclotron resonance analysers.
Tandem mass spectrometry workflows
Shotgun peptidomics is based on high-throughput automated sequencing and identification of endogenous peptides representative of a biological sample. Sequencing is realized by implementation of tandem mass spectrometry (MS/MS)45 with two main protocols: data-dependent and data-independent acquisition (DDA and DIA, respectively). The benefit of DDA is highquality MS/MS spectra resulting from a user-specified number of the most intense precursor ions in a given chromatographic time frame subjected to fragmentation. Identification of low-abundance ions is facilitated by dynamic exclusion of previously sampled precursor ions from MS/MS. Given the limited number of precursors sampled within a duty cycle, the resolution of hyphenated front-end separation platforms plays a crucial role in the complexity of the mass spectrometry spectrum and, respectively, in the degree of sample characterization via MS/MS. In DIA-based analysis, all precursor ions from the mass spectrometry survey scan are selected for MS/MS via stepping broad isolation windows across the entire m/z range. Implementation of DIA improves reproducibility across samples, which in turn reduces missing values and greatly improves the quantitative accuracy of peptidomics assay. Although useful for quantitation of peptides, the main limitation of DIA is dependence on reference spectral libraries, typically generated via DDA analysis of additional samples. Current efforts are channelled towards development of library-free DIA approaches50–52.
Mass spectrometry imaging
The development of MALDI MSI maps the spatial locations and distribution patterns of the biomolecules in tissue samples53. Currently, MALDI MSI remains the most common method for spatial mapping of lipids, metabolites and peptides/proteins. Other than MALDI MSI, secondary ion mass spectrometry or SIMS-MSI54, desorption ESI MSI55 and scanning microprobe MALDI MSI56, surface-assisted laser desorption/ionization mass spectrometry57 and nanostructure imaging mass spectrometry58 are also used to examine the localization of proteins/peptides. Advantages and disadvantages of the different ionization techniques have been reviewed elsewhere59. Unlike immunohistochemistry, radio-immunoassays and fluorescence microscopy that require extensive sample preparation and prior knowledge of the target analytes60–62, MSI involves relatively simple sample preparation and enables the localization of hundreds to thousands of different analytes on a tissue slice in a single experiment63. To localize the peptide, the tissue of interest needs to be properly prepared. After dissection it is usually fresh frozen or embedded in gelatine, sodium carboxymethyl cellulose or paraffin. Formalin-fixed paraffin-embedding is a special embedding technique that can preserve the specimen under room temperature for more than a decade64,65. Although optimal cutting temperature (OCT) compound is a common tissue-embedding solution, the high concentration of polyethylene glycol (PEG) in the OCT compound can affect the analyte signals and, thus, OCT compound is not recommended for tissue embedding in MSI experiments66. Cleaning procedures to remove interferring embedding substances are needed before using OCT compound or formalin-fixed paraffin-embedded tissue for MSI experiments67–69. For endogenous peptide MSI experiments, sample preparation requires fewer steps; before imaging, fresh frozen or embedded tissues are sectioned into 5–20 μm slices70. Before imaging, background salts and lipids need to be removed from the tissue for which various washing techniques such as organic solvents have been evaluated71. Washing is also adaptable for peptide imaging experiments, but may increase the loss of hydrophilic peptides72 and cause analyte diffusion73. Optimization of the washing procedure for formalin-fixed paraffin-embedded tissue sections shows signal enhancement for neuropeptide imaging74. Tissue slices with or without a washing procedure can then be analysed by imaging instruments (excluding MALDI MSI). For MALDI MSI, matrix application is needed as the last step before instrumental analysis. The matrix choice, concentration and application method are important for signal intensity and resolution and need to be carefully selected75,76. The resulting images from the MSI experiment can be analysed using various software choices depending on the instrument used.
In silico peptide mining
Peptides of diverse origins serve various roles in nature, and therefore may bear various modifications. The modifications pose a particular difficulty for peptide analytics and characterization, and the taxonomic origin (Fig. 1) requires adapted/tailored research strategies. In the post-genomic era, scientists have access to databases77–82 and tools to address these issues (Table 1). Most publicly available ab initio gene and protein sequence data are annotated by programs such as GeneMark83 or Prodigal84. These are robust platforms but come with their limitations as they do not always annotate short open reading frames (sORFs) where peptides are often found. For that purpose, specialized tools exist with their own rule sets for in silico peptide mining. They can be specialized to predict multiple post-translational enzymes, amino acid substrates or biosynthetic tailoring of non-ribosomal peptides, assisting the researcher in annotating possible peptide modifications. In the case of eukaryotic organisms, bioactive peptides may originate from sORFs or derive from breakdown products from other enzymes58,85,86. For eukaryotic genetically encoded peptides, there are tools such as SPADA87, MiPepid88, DeepCPP89 or rAMPage90 (Table 1 and Supplementary Table 1) if genomic or transcriptomic data are available. For protein-derived bioactive peptides, PeptideLocator86 can be used on protein sequences. Alternatively, comparative genomics can be helpful with tools such as CoGe91 or EDGAR92 (Table 1), which may be used when searching conserved homologues for validation; for a summary of databases and software tools, refer to Supplementary Table 1.
Table 1. Selected tools for peptide mining.
| Tool | Organism | Data type | Description |
|---|---|---|---|
| Sequence annotation | |||
| SPADA | Plant | DNA | sORF annotation |
| MiPepid | Eukaryotic | DNA | sORF annotation |
| rAMPage | Eukaryotic | RNA | Antimicrobial peptide detection |
| DeepCPP | Eukaryotic | RNA | sORF annotation |
| antiSmash | Bacterial/fungal/plant | DNA | Gene cluster annotation |
| RODEO | Bacterial | Amino acid | Gene neighbourhood analysis |
| BAGEL | Bacterial | DNA | Gene cluster annotation |
| DeepBGC | Bacterial | DNA | Gene cluster annotation |
| Comparative genomics | |||
| EvoMining | Bacterial/fungal/plant | Amino acid | Phylogenetic gene cluster search |
| CoGE | Eukaryotic/prokaryotic | DNA | Genome comparison |
| EDGAR | Eukaryotic/prokaryotic | DNA | Genome comparison |
| Product prediction | |||
| SANDPUMA | Bacterial | Amino acid | A-domain specificity prediction |
| Peptidelocator | Eukaryotic | Amino acid | Prediction of bioactive peptides derived from enzyme degradation |
| Multi-omics | |||
| Nerpa | Bacterial/fungal | DNA/mass spectrometry | Maps NRPs back to their respective gene clusters |
| BioCAT | Bacterial/fungal | DNA/mass spectrometry | Maps NRPs back to their respective gene clusters |
| DeepRiPP | Bacterial/fungal | DNA/mass spectrometry | Structure elucidation of RiPPs from mass spectra and sequence data |
| PoGo | Eukaryotic | DNA/mass spectrometry | Peptidogenetic tool, mapping peptides to the genomic loci |
| MetaMiner | Bacterial | DNA/mass spectrometry | Large-scale screening platform for microbial peptides |
In silico peptide mining is aided by a myriad of interesting tools from sequence annotation, comparative genomics, product predictions and multi-omics approaches. However, a tool needs to be matched with its proper use, as the genes and genomic architecture of plants, animals, bacteria or fungi are not the same, and will therefore often need detection rules, dedicated to each clade of life. The detection and discovery of potential peptides can be done at each level of sequence data from genomic (DNA) to transcriptomic (RNA) or by further investigation into proteins (amino acid) for their degradation products, or neighbouring genes may be bioactive peptides or involved in bioactive peptide processing. Furthermore, comparison of sequence data with mass spectra has led to the development of robust multi-omics platforms to aid researchers in high-throughput peptidomics. A-domain, adenylation domain; NRP, non-ribosomally synthesized peptide; RiPP, ribosomally synthesized and post-translationally modified peptide; sORF, short open reading frame.
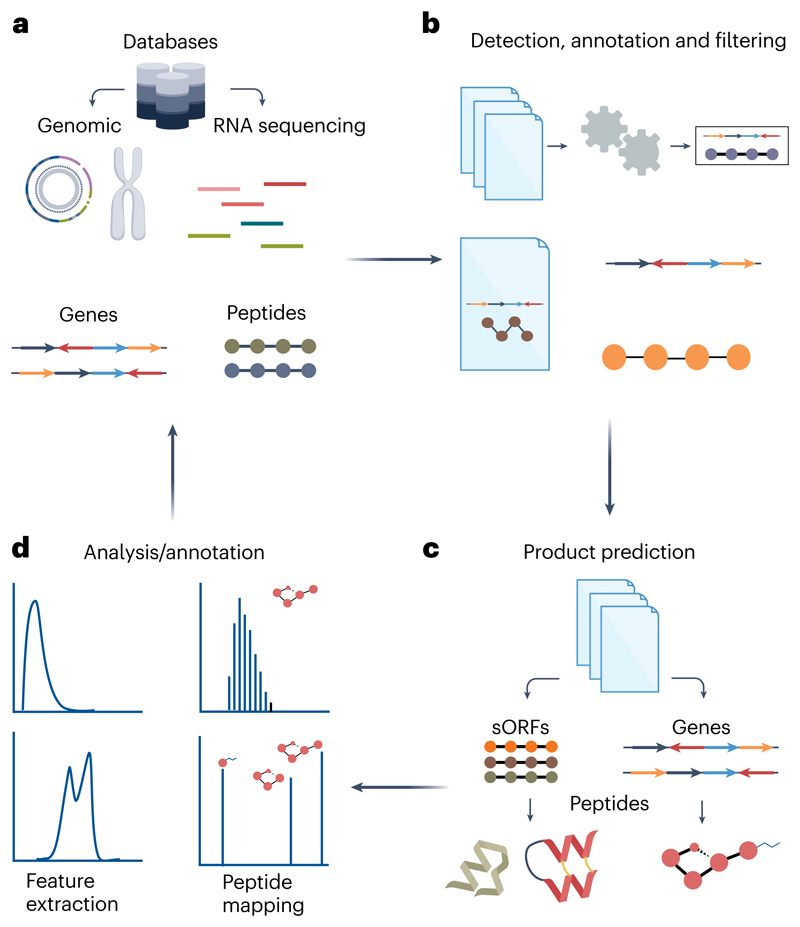
The biosynthetic genes of ribosomally synthesized and posttranslationally modified peptides (RiPPs) and non-ribosomally synthesized peptides (NRPs) from bacteria and fungi93–95 are commonly encoded in biosynthetic gene clusters and require programs specialized in biosynthetic gene cluster detection, such as antiSMASH95 and DeepBGC96 (Tables 1 and2). These tools can be complemented by phylogenetic genome mining using EvoMining97 for the discovery of homologous gene clusters. There are even more specialized tools for NRPs, such as SANDPUMA98, for the prediction of the substrates of the adenylation domains (A-domains). Furthermore, there are other mining tools for RIPP analysis, such as BAGEL99 or RODEO100 for detection and classification of RiPPs from the genome or DeepRiPP101 for classification, structure prediction and spectral assignment (Tables 1 and2). These programs use genomic or transcriptomic data to discover potential peptides. Alternatively, tools such as PoGo102 map detected ribosomally synthesized peptides from LC-MS data to the genome. Recently, a retro-biosynthetic tool for NRPs has been developed103, which allowed non-ribosomal peptides to be mapped back to their respective gene clusters by tools such as Nerpa104 and BioCAT105. The process of discovering novel peptides is an iterative process of in silico and laboratory work, where new discoveries constantly feed the expanding databases, allowing for more precise and detailed tools to be developed (Fig. 2).
Table 2. List of important databases and tools.
| Databases and software | Use case | Data available/input data | Key features |
|---|---|---|---|
| Database | |||
| NCBI | Biological sequences | DNA, RNA, amino acid | Repository of various biological sequences |
| Dictionary of Natural Products a | Physicochemical data | Solubility, UV–Vis | Extensive database resource of natural products and their physiochemical properties |
| Software for in silico annotation | |||
| antiSMASH | RiPPs and NRPs | DNA | Rule-based cluster detection |
| SANDPUMA | NRPs | DNA | A-domain specificity prediction |
| DeepBGC | RiPPs and NRPs | DNA | Gene cluster detection |
| Software for peptidogenetic pipelines | |||
| DeepRiPP | RiPPs | DNA (open reading frame) | Classification, processing and spectral matching |
| Software for mass spectrometry analysis | |||
| DEREPLICATOR+ | RiPPs and NRPs | LC-MS/MS | data Natural product identification from mass spectrometry spectra (GNPS framework) |
| MS-FINDER | Mass spectrometry data analysis | EI-MS, GC-MS, MS/MS | Formula predictions, fragment annotations and structure elucidation |
| Software for MSIa | |||
| MSiReader | Mass spectrometry data analysis | MSI | MSI platform for analysis |
| SCiLS Lab a | Mass spectrometry data analysis | MSI | MSI platform |
| ImageQuest a | Mass spectrometry data analysis | MSI | MSI platform |
| High Definition Imaging a | Mass spectrometry data analysis | MSI | MSI platform |
| msiQuant | Mass spectrometry data analysis | MSI | MSI platform for analysis |
Various resources can aid in peptidomics and one of the main resources most researchers start with are biological sequence databases, to gather genomic (DNA), transcriptomic (RNA), proteomic (amino acid) or other data that may be relevant to their research. These data types can be further complemented by specialized software or other physiochemical or spectral data to make more accurate predictions or annotations of peptides that may be present in the sample. Databases and further tools are continued in Supplementary Table 1. A-domain, adenylation domain; EI-MS, electron ionization–mass spectrometry; GC-MS, gas chromatography–mass spectrometry; LC-MS, liquid chromatography–mass spectrometry; MSI, mass spectrometry imaging; MS-MS, tandem mass spectrometry; NRP, non-ribosomally synthesized peptide; RiPP, ribosomally synthesized and post-translationally modified peptide.
Commercial platforms, may be subject to licensing charges.
Fig. 2. In silico peptide mining.
General workflow of how multi-omics approaches may be applied to peptidomics. a, Novel peptide candidates are often discovered in publicly available data sets, from genomics, transcriptomics or proteins. b, FASTA files are fed into the annotation platforms, where they utilize their own detection rule sets to predict the likelihood of true peptide candidates. c, Peptides derived from ribosomal transcription may undergo extensive modification, or non-ribosomal proteins may be responsible for peptide biosynthesis; as such, product prediction tools have been made to aid in novel peptide discovery. d, Finally, candidates validated by chemical analysis will be deposited in databases, increasing our collective knowledge and data amount to further improve algorithms and bioinformatics tools. sORF, short open reading frame.
Results
The comprehensive sequence identification of peptides includes the full assignment of amino acids in the correct sequence orientation, which is usually determined by the encoding genes and/or the ribosomal protein translational machinery of a cell. The identification and the site of post-translational modifications or tailoring reactions are an additional challenge to allow full assignment of the native peptide. This section provides an overview on the workflows and frameworks that have been implemented to analyse different sets of peptide analytes using peptide mapping and de novo sequencing, automated LC-MS/MS workflows and MSI (Fig. 3).
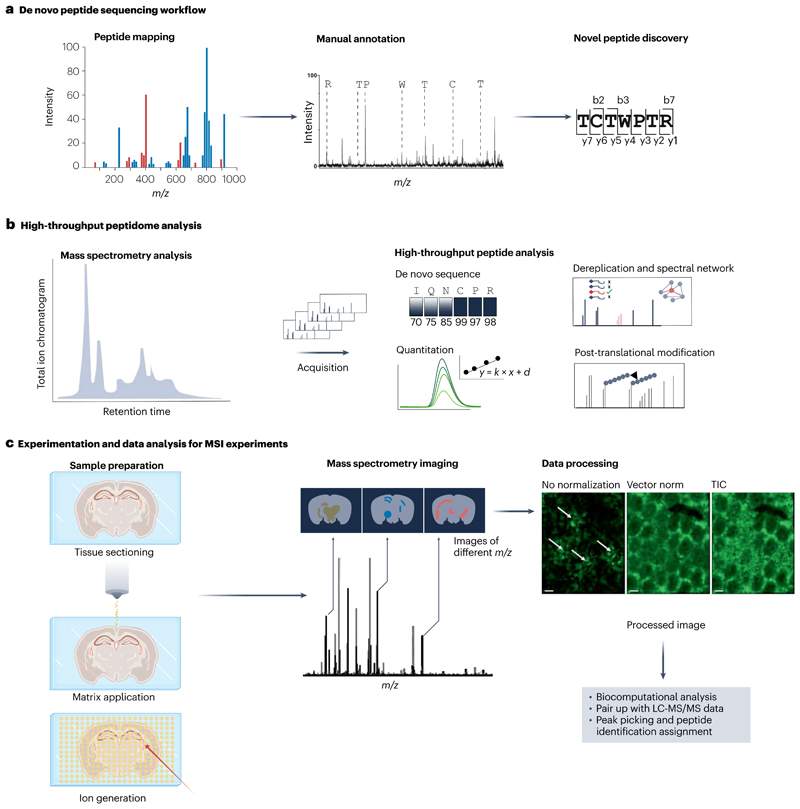
Fig. 3. Overview of common peptidomics workflows.
a, De novo peptide sequencing is utilized in peptide discovery research. Purified peptides are analysed with matrix-assisted laser desorption/ionization time-of-flight/time-offlight mass spectrometry (MALDI-TOF/TOF-MS) or tandem mass spectrometry (MS/MS) systems. b, High-throughput peptidome analysis is often employed for automated sequence analysis, dereplication and/or spectral network analysis, quantitation and analysis for post-translational modifications. These analyses are usually paired with high-throughput analytical systems such as high-resolution liquid chromatography–tandem mass spectrometry (LC-MS/MS). Data acquisition generates MS/MS spectra that can be analysed with de novo sequencing algorithms or used for identification of peptides with a database search. Quantitation of peptides can be achieved at the level of the mass spectrometry peak as well as with multiple reaction monitoring acquisition. Post-translational modification analysis can be performed for tryptic peptides or for endogenous peptides using a peptide fragment spectrum for database search. c, Experimental protocol and data analysis procedure for mass spectrometry imaging (MSI). After sample preparation of tissue slices (for example, mouse brain tissue), the matrix is applied and co-registered with a tissue image to precisely define sample position in the mass spectrometer. The data acquisition over the entire raster generates a summed total ion chromatogram (TIC) spectrum. To visualize m/z maps, each m/z signal can be selected to show extracted ion chromatograms. The data can be processed (normalization, smoothing, data compression) for various biocomputational analysis. Scale bars, 200 μm. Part c data processing images reprinted from ref. 133, Springer Nature Limited.
Peptide mapping and de novo sequencing
Despite the use of coupled LC-MS methods, for certain applications, such as complex peptide analytes, an offline workflow can be beneficial. For such applications, MALDI-TOF/TOF-MS41 is often the method of choice as it results in spectra with singly charged ions, for example [M + H]+, suitable for manual peptide annotation and de novo sequencing (Fig. 3a).
Peptide dereplication and peptide mapping
Peptide dereplication is commonly applied for rapid pre-screening of peptide libraries, for example peptide natural products in extracted samples from microbial or plant origin106. Dereplication can be achieved at different levels, for example by matching experimental m/z signals (mass spectrometry) or spectral MS/MS data to libraries. Peptide m/z signals provide valuable information for peptide content mapping by comparison of those experimentally determined with calculated molecular masses in databases. HRMS data further enable comparison of isotopologue intensities with theoretical data-based intensities as well as prediction of chemical sum formula, which can provide a further layer of evidence for the matched library hit. Despite the lack of comprehensive databases targeting natural product/microbial-derived peptides, they are listed in common natural product databases, such as the Dictionary of Natural Products (Taylor & Francis) or Antibase107 (others are under development), or specialized databases such as Bactibase dedicated to subsets or subclasses of peptides (Table 2 and Supplementary Table 1). The search runs are performed by using computational-aided tools for assignment and further evaluation. DEREPLICATOR+108 (Table 2), SEQUEST109 and MZmine3 have emerged as helpful tools. For example, the MZmine3 engine offers an all-in-one solution for peak picking, chromatographic peak detection, peak identification and quantitation, and data processing/visualization functionalities110. Using high-resolution mass spectrometers, such as FTICR-MS or orbitrap devices, it has become possible to derive molecular formulas of peptides based on their determined accurate mass. Common tools for molecular formula determination based on high-resolution spectra are pacMass111, MS-FINDER112 (Table 2) or MetaSpace113. Moreover, spectral library search approaches are also advancing into the peptidomics field as, besides customized in-house spectral databases, commercial (for example, mzCloud) or open access (such as NIST) spectral libraries are readily available for mapping experiments. A combination of database dereplication, highresolution mass detection and spectral database annotation is state of the art for peptide mapping approaches today. Several manufacturers of mass spectrometers offer software solutions, such as Metaboscape (Bruker Daltonics) or Compound Discoverer (Thermo Fisher Scientific), to perform the identification of peptides using one or several of the described methods in one package. The annotation of branched or cyclic peptides was addressed by CycloBranch2 (refs. 114,115) or the VarQuest algorithm using spectral networks116. As a general limitation, a comprehensive peptide database including high-resolution accurate mass, sequence information, fragmentation spectra is still not available to the research field to date.
De novo sequencing
MALDI-TOF/TOF systems are capable of postsource decay fragmentation. Owing to metastable decay, the system offers less efficient fragmentation than collision-induced dissociation or electron transfer dissociation methods. This fragmentation technology provides signal intensities for specific fragment ions, which are useful for manual de novo sequencing applications117 (Fig. 3a). De novo sequencing examines characteristic mass shifts among the fragment ions to reconstruct ion series indicating non-random amino acid combinations, thus allowing for the detection of novel sequences118. De novo interpretation of fragmentation spectra is still the best choice to derive sequence information of highly functionalized peptides (for example, peptides with post-translational modifications and/or non-natural amino acids). The post-source decay fragmentation approach is useful for studying single to few peptides without much need for further automation. High-throughput analysis and combinations together with other omics techniques made a software-assisted solution for de novo interpretation of MS/MS spectra indispensable to handle these large data sets. Bioinformatic tools, such as PEAKS119, are algorithms for de novo peptide/protein sequencing and allow combinatorial database annotation. An important consideration of de novo sequencing is handling of false positive assignments. Deep learning methods have significantly improved the power of de novo sequencing methods, allowing ≥97% sequence coverage120. Although the expected sequence coverages with and without the assistance of databases improved over the past two decades, the algorithms to assign amino acid sequences de novo to experimental MS/MS data are still a source of variation. As these algorithms can substantially differ from each other, for example in their cut-offs for profitability probing or in other decision-making processes, their output for one spectrum can be conflicting with obvious false assignments. To account for this remaining challenge of software-assisted analysis, de novo sequence work should be carefully validated using known or reference peptides whenever possible. The quality of de novo data may also depend on instrument performance, spectra quality, peptide fragmentation efficiency, presence of post-translational modifications, the abundance of precursor ions or sample size69,121,122.
Peptide databases and identification workflows
Regardless of the acquisition method (DDA or DIA), the output of a typical LC-MS/MS experiment comprises hundreds of thousands of peptide fragmentation spectra correlated to chromatographic retention time and precursor ion mass (Fig. 3b). Interpretation of such spectral data typically relies on querying them against in silico predicted theoretical spectra from protein sequences found in a species proteome database (Table 2 and Supplementary Table 1). More effective, however, is a de novo sequence tag approach that infers the peptide sequence directly from characteristic mass shifts between peptide fragment ions, and then matches the tag to proteins in the database. The advantage of de novo sequencing is its ability to discover peptides outside the proteome database, as unassigned tags can be searched against expressed sequence tag (EST) repositories or mass spectrometry databases of homologous species from other studies. With either matching algorithm, experimental data fit to matched proteins are statistically evaluated for probability and the false discovery rate (FDR).
Peptide identification from DDA spectra is typically done by ranking the probability of non-random fit of peak patterns in the MS/MS spectrum into certain amino acid sequences using mass spectrometry vendor-specific analysis tools or the universal format software PEAKS7. Annotation of post-translational modifications is typically included with either option. It is important to recognize that popular proteomics tools MaxQuant123 and Mascot124 are not suitable for native peptide identifications as they rely on in silico spectral libraries of theoretical peptides potentially originating from enzymatic cleavages of proteins in a database; when the enzyme is not specified, dramatic expansion of search space overwhelms computational resources
Owing to a conceptually different fragmentation approach in DIA experiments, alternative software is needed for identification and quantitation of peptides, for example Skyline125, DIA-NN50, OpenSWATH126, Spectronaut127 and DIA-Umpire51. Statistical analysis of DIA measured peptides can be performed with the output result files using Excel, R programming, Python and Perseus123 (Table 2 and Supplementary Table 1).
Quantitative peptidomics
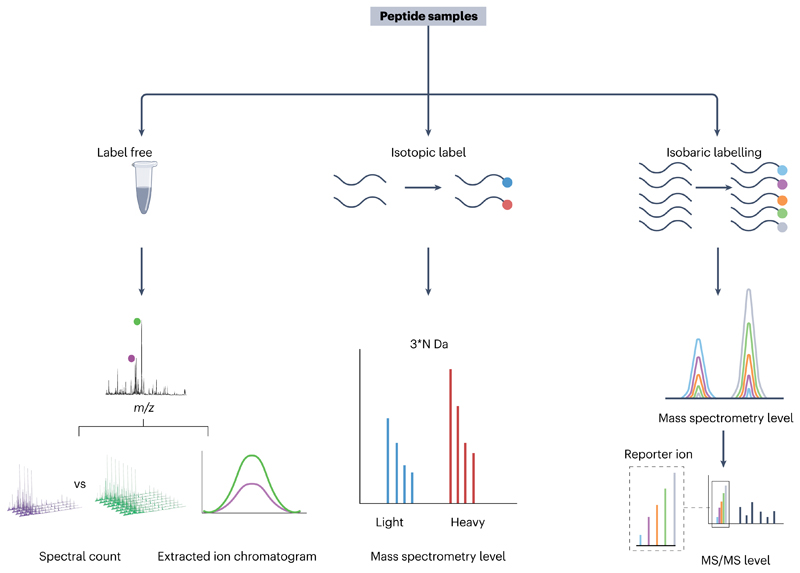
The application of mass spectrometry-based peptide quantitation is rapidly growing in clinical, applied and basic research. Traditionally, in bottom-up proteomics, the mass spectrometry quantitation approach is based on comparison of protein levels via summation of measurements from several encoded tryptic peptides. In peptidomics, however, individual bioactive peptides may have independent levels in relation to pathological or experimental conditions, even if originating from the same protein or prohormone. Therefore, endogenous peptides that are subject to peptidomics investigation will be quantified individually at the peptide level, not the protein level. With that difference in mind, practical strategies for quantitative peptidomics are similar to those widely used in bottom-up proteomics and include stable isotope or chemical labelling and label-free methods28–32 (Fig. 4). An advantage of the label-free quantitation approach is its low cost and simplicity of sample preparation. The two commonly used label-free quantitation techniques are based on the signal intensity (using extracted ion chromatograms) and spectral counting128. Both methods can be used for relative and absolute quantitation. For absolute quantitation, a peptide standard that is similar to the target peptide can be added43, or ideally a synthetic stable isotope-labelled internal standard for each peptide of interest is used, for example AQUA46 peptide. In silico algorithm-based methods may also be used to achieve absolute quantitation and assess the actual concentration of the target peptide in one sample58,129,130. Additionally, multiple reaction monitoring can be used in targeted peptide quantitation. Multiple reaction monitoring focuses on selected fragment ions for peptides of interest and allows the detection of low-abundance peptides44.
Fig. 4. Quantitation methods for peptidomics.
Techniques include label-free, isotopic labelling and isobaric labelling quantitation. Label-free quantitation (left panel) is based on the mass spectrometry level and can be achieved through either spectral count or extracted ion chromatograms; labelled strategies can be achieved at both the mass spectrometry and tandem mass spectrometry (MS/MS) levels. Samples labelled with an isotopic labelling strategy (middle panel) can be quantified through the mass defect on the mass spectrometry level, whereas isobaric-labelled samples (right panel) are quantified on the MS/MS level. Bottom panel reprinted with permission from ref. 165, Wiley.
The advantage of the stable isotope labelling strategy is multiplexing and throughput, although often at high cost. The quantitation of labelled peptides is based on the mass spectrometry signal intensity, and the complexity of analysis increases with the size of data sets. Conceptually different in vitro strategies involve isobaric labels131 that produce identical mass shifts on the mass spectrometry level, but generate distinct reporter ions associated with different labelling channels during peptide fragmentation in MS/MS events131. We refer the readers to another article for more extensive discussion on isobaric labelling131. Raw files obtained from both label-free and labelling strategies can be analysed through the software mentioned in the previous section and detailed in Table 2 and Supplementary Table 1.
Mass spectrometry imaging and spatial patterning
MSI is a complementary method to IHC staining60, radio-immunoassays61 and fluorescence microscopy62, to yield images of the target analyte in relatively high throughput53. Figure 3c demonstrates the experimental procedure and data processing steps for MSI experiments. Distribution of a list of the target m/z can be visualized in one experiment from the same tissue slice. Although MSI provides direct qualitative results for the target m/z, quantitative analysis can also be done with the aid of commercial and open access software. To relatively quantify the desired m/z for a certain peptide, direct comparison can be done among the different tissue regions or different tissue slices63. Normalization is usually completed pre and post processing132. The software is equipped with normalization tools, using the total ion chromatogram (TIC), vector norm through root mean square, median and noise133. Similar to LC-MS, isotopic labelling may be transformed for MSI relative quantitation134. Absolute quantitation is a relatively untapped area, and such quantitation can be performed by LC-MS/MS or by adding the calibrant to the solvent stream in desorption ESI MSI experiments135,136. Another way to perform absolute quantitation is to create a calibration curve by spotting the standards of interest onto a different tissue section that is adjacent to the analysed tissue sections137.
Various software tools (Table 2 and Supplementary Table 1) have been developed for MSI visualization and analysis, including MSiReader138, SCiLS Lab, ImageQuest, High Definition Imaging, MassImager139 and msiQuant140. Several statistical analyses can be performed using some of these software packages, including analysis of variance, principal component analysis and partial least squares coupled to discriminant analysis (PCA-DA and PLS-DA, respectively), and receiver operator characteristic curve analysis for biomarker tests63. Subsequent statistical and classification analysis can also be done using machine learning and in silico algorithm-based software. MALDI MSI data are usually paired with LC-MS/MS data for specific peak assignment and peptide identification. We refer the readers to a more thorough review article63 for a discussion on MSI.
Applications
Function, diversity and evolution in biology
Microbial peptide biosynthesis
Some peptide species from bacteria and fungi are distinct from peptides commonly found in higher organisms. These peptides, according to their more specialized biosynthetic mechanisms, can be categorized into two groups: RiPPs and NRPs. RiPPs are encoded as precursor peptides in the genome and undergo post-translational modifications beyond those commonly known from animal/plant-derived peptides and proteins141. RiPPs are subcategorized by their characteristic post-translational modifications, which include thioethers, heterocycles, amino acid side-chain functionalizations and various side-chain cross links. Recently, examples such as the lipolanthines emerged where RiPP biosynthesis is combined with other biosynthesis pathways, such as fatty acid biosynthesis of polyketide synthesis (PKS)142.
By contrast, NRPs are commonly synthesized by modular multidomain synthetases that can incorporate non-proteinogenic amino acids and other substrates. Modules for peptide assembly consist of a basic domain set: A-domains responsible for amino acid/substrate activation and selectivity; a peptidyl-carrier protein domain; and a condensation domain (C-domain) forming the amide bonds. A thioesterase domain (TE-domain) releases a linear or cyclic peptide. Additional domains may be interspersed to perform epimerizations (E-domains), oxidations/reductions (Ox/Red-domains), cyclizations (Cy-domains) or other modifications143. Tailoring of the peptide substrate may occur to an extent that, sometimes, a peptide structure is hardly recognized in the final product, for example in the β-lactams (precursor peptide l-aminoadipoyl-l-Cysd-Val). The structural diversity of NRPs is even further extended by mixing biosynthetic functions with PKS, rendering lipopeptides or even stronger morphed PKS-NRP-like structures, which probably mark the borders of the peptidomics field144. Although, recently, genome mining considerably facilitated the discovery of new classes and types of microbial peptides derived from RiPP and NRP biosynthesis, the field still is in its infancy, as estimates consider that only 2.1% of the global prokaryotic taxa are represented in sequenced genomes145.
Venomics
Venomous animals (including snakes, spiders, scorpions, amphibians, snails and even platypuses) have evolved multiple times throughout evolution, and many venom cocktails are rich in peptides146–148. In fact, the diversity of venom peptides is unprecedented: their estimated number exceeds millions149. Peptidomics has provided detailed insights into the diversification of venom. The venom content has also evolved, based on the targeted prey or predators to defend against. In research on the predation of cone snails Conus marmoreus and Conus geographus, their conotoxins and the defence stings showed remarkably different venoms. Whereas defensive venoms are localized in the proximal duct, the predation venoms are in the distal duct of the venom gland150,151. Predatory venoms evolved to incapacitate or kill the prey with high selectivity, whereas the defensive venoms had little to no activity against their prey but contained high amounts of paralytic peptides (conotoxins) acting on mammalian ion channels. Scorpions have also been studied extensively for this purpose. Their venom components differ concerning defensive and predatory behaviours; they might sting multiple times, but the venom composition may vary in response to the threat of the animal152,153. These examples underline the requirement for appropriate extraction methods when working with venomous animals.
Invertebrate neuropeptide discovery
There is an overwhelming diversity of (neuro)peptidomics studies on invertebrates, including the classic model species in insects154–156, molluscs157–160, worms, crustaceans161–165 and cnidarians166–168. Multiple approaches proved effective for discovery and characterization of neuropeptides in invertebrates. With growing numbers of sequenced genomes or transcriptomes, the bioinformatics annotation of prohormones and the prediction of endogenous putative peptides facilitates peptidomics studies169. The characterization of structurally simple peptides can be achieved by matching the mass spectrometry-detected peptide masses to theoretical masses of peptides predicted from a protein/prohormone sequence while accounting for possible post-translational modifications170. This is an effective approach suitable for single-cell peptidomics171, and thus allowing for the discovery of chemical messengers in well-defined neuronal circuits172 that control physiological functions and behaviour, for which invertebrates are extremely well suited due to the simplicity of their organization and conservation of signalling molecules along the evolution tree. For characterization of the peptidome of an organism with no genomic information, shotgun peptidomics on a larger tissue sample works best with mass spectrometry data annotated using the protein database of a related species (homology search)173. Specifically, hybrid bioinformatics approaches for interpretation of the MS/MS data match experimental de novo sequence tags to a database of related species proteins while accounting for potential single point mutations174. A more targeted analysis aimed at peptide-level validation of selected gene expression and/or function in specific cells or tissue often involves a multi-omics workflow175 or gene cloning followed by gene expression mapping to guide tissue or cell sampling for mass spectrometry analysis176–179. Elegant multi-platform studies combining shotgun peptidomics for peptide library construction followed by MSI of tissue sections have led to mapping of putative bioactive peptides in nervous system tissue sections under different biological paradigms, as well as the exploration of cellular heterogeneity and organelle peptide complements. Implementation of mass spectrometry-based, peptidecentred workflows has accelerated the discovery of enzyme-derived d-amino acid-containing peptides (DAACPs) of high physiological importance in invertebrates176,180,181. DAACPs typically co-exist with all-l-amino acid-containing peptide counterparts in tissue extracts and can be fractionated via RP-HPLC and can be validated via trapped IMS and MS/MS182,183. An alternative discovery pipeline based on enzymatic screening, separation and amino acid analysis is greatly enhanced by MS/MS184–186.
Human and mammalian neuropeptide discovery
Peptidergic systems are abundant ligand–receptor signalling systems in mammals and are of high interest to further understand human signalling networks. There are numerous peptide hormones in the body but their receptor targets, many of them from the G-protein-coupled receptor family, and the physiological role of these systems remain elusive. Peptidomics assisted the deorphanization process of peptide/protein target signalling networks in the past. Many neuropeptides and their endogenous receptors were discovered with bioinformatics tools, as the human genome sequence data enabled large-scale analysis. Conceptionally, bioinformatics has limitations to detect precursor splicing, secretion signal sequences and post-translational modifications. Peptidomics approaches were applied in neuropeptide discovery but have limited sensitivity for the detection, and the identification is often knowledge-based using pre-defined search libraries187. Recent developments highlight a robust analytic framework for extracting, analysing and identifying endogenous peptides188. Integrated computational and experimental approaches have been powerful tools for peptide–orphan GPCR pairing189,190. Human peptide ligands of orphan receptors were predicted based on common sequence motifs, for example secretion sequences and conserved regions in their encoding precursors. The identified peptides can be chemically synthesized for testing in vitro for activation of receptor systems. These deciphered systems revealed new peptide ligands for the GPR1, GPR15, GPR55, GPR68 and BB3 receptors189. The human neuropeptide discovery field is hugely significant to understand physiological process and pathophysiological conditions, providing clinical opportunities for new therapies of brain disorders. For example, combinatorial workflows to enable large-scale mass spectrometry-based peptidomics for drug discovery or integrated workflows to decipher signalling systems provided significant contributions188,189. Overall, there is a need for in-depth studies of the human neuropeptidome, which is still at the frontiers stage compared with other omics technologies.
Drug discovery
From venoms to drugs
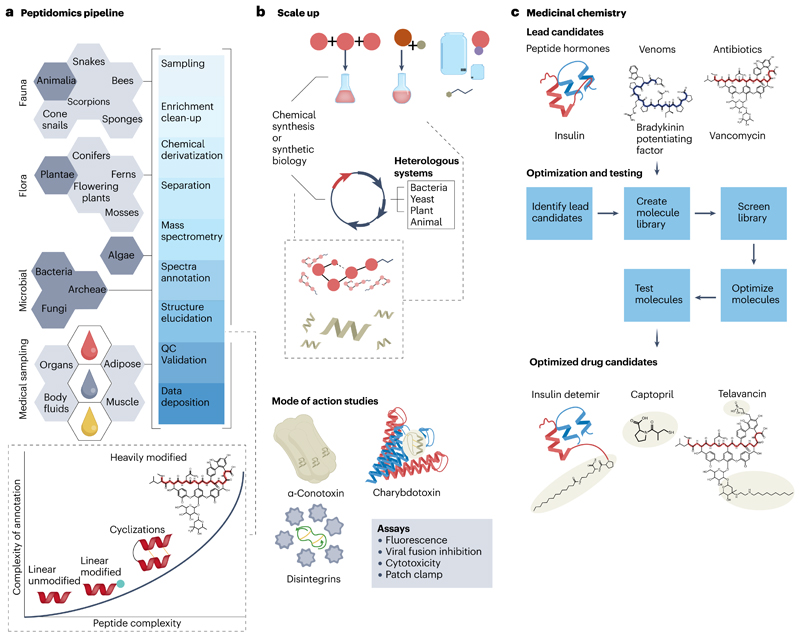
Historically, the discovery of drugs has been accomplished by natural observation, followed by trial and error experimentation with various plants and animal extracts191,192. Traditional knowledge, passed on by generations, became the pillar of drug discovery as medical sciences were established and methods were developed to validate the bioactivity193. Modern drug discovery can be split into two phases: compound screening at the molecular targets (Fig. 5a,b) and medicinal chemistry efforts (Fig. 5b,c) to improve the pharmacological properties194. Peptidomics has played a crucial role in the discovery of peptide drugs and peptide-derived drugs. A prominent example is the bradykinin potentiating peptide isolated from the Brazilian pit viper Bothrops jararaca; the initial peptide isolated was the template for developing a small-molecule peptidomimetic resulting in the angiotensin converting enzyme inhibitor captopril, a hypertension medication195 (Fig. 5c).
Fig. 5. From nature to medicine.
a, Generalized scheme of peptide sources and the peptidomics analysis pipeline. During structural elucidation, the complexity of spectral annotation will increase with the structural complexity of the peptides (for example, by post-translational modifications such as disulfide bond formation, cyclization). b, After peptide analysis, desired products are scaled up by chemical synthesis or synthetic biology (heterologous expression in various host organisms) for mode of action studies and pharmacological analysis. c, Optimization of the lead peptides (for example, insulin, bradykinin potentiating factor or vancomycin) by medicinal chemistry. Optimization is a multi-step process, including pharmacophore analysis, chemical synthesis, molecule library preparation and screening, structure–activity studies and pharmacology to generate optimized peptide or peptidomimetic drug candidates (for example, insulin detemir, captopril or telavancin) for applications in medicine. Structural alterations during lead to drug optimization are highlighted by pale yellow ovals (peptide backbones are coloured blue and red).
Owing to their neurotoxic effects, peptide analgesics are known to be common components of venoms. Peptidomics analysis has shed light on the composition and structures of these highly complex peptide mixtures. Therefore, it has been possible to isolate and identify peptides from various animals such as scorpions196, cnidarians197 and cone snails198. One such example, the cone snail conotoxin MVIIA is an N-type channel blocker. The synthetic version, marketed as Ziconotide (Prialt), is used to treat chronic pain199. Venom peptides also target peptide hormone systems. Remarkably, cone snails have weaponized peptide hormones by generating fish insulin analogues, releasing them from their venom glands and sending their prey into hypoglycaemic shock200. Peptidomics was the key technology to isolate and identify the activity-bearing peptides. The last example worth mentioning is the Gila monster (lizard) peptide exendin 4. It is a long-acting GLP1 mimetic, which led to the development of the GLP1 antagonist exenatide201 (Fig. 5c) preceding liraglutide and semaglutide as peptide drugs in the same therapeutic area202. GLP1 and GIP are endogenous peptide hormones, which amplify the response of glucose-induced insulin secretion. Hence, these venom-derived and hormone-derived peptides, identified with the help of peptidomics, are being used clinically as antidiabetic drugs.
Antimicrobial peptides and derivatives
Although possibly not regarded as antimicrobial peptides in the classical sense, microbial peptides and derivatives have been used for decades as highly successful, mostly anti-infective drugs. Examples of marketed NRP-derived peptide drugs, which all show sophisticated mechanisms of action, are vancomycin203, daptomycin204, bleomycin205 (anticancer), cyclosporin206 (immunosuppressant) and even β-lactams. Among RiPPs, nisin (E234) is the frequently mentioned example serving as a food preservative207. Antimicrobial peptides in a narrower sense include defensins, which kickstarted the discovery of peptides belonging to the innate immune system208. Antimicrobial peptides originate from multicellular organisms (fungi, animals and plants) and range between 10 and 120 amino acids in size. The assignment to this family is very broad, preferably with an overall positive charge and cross-linked by disulfide bridges209, but other structural types are known. Antimicrobial peptides often affect cells by rupturing the lipid layer, pore formation or inhibiting cell-wall synthesis205. Recent peptide discoveries with impressive antibacterial activity against relevant Gram-negative bacteria include albicidin (NRP)210 and darobactin (RiPP)211, which may contribute to the much-needed demand for new antibiotics.
Body fluid peptidomics — novel antiviral drug candidates
Although it is established that peptides of the innate immune system possess antibacterial and antifungal properties, these peptides also possess antiviral or anticancer properties6,212,213. These functions are frequent in biology and are termed moonlighting activities. The features that improve antiviral properties correlate to the cationic charge and the peptide’s hydrophobicity. The peptides directly bind to the viral particles themselves, preventing viral fusion with the host cell. These peptides display characteristics to prevent or to encounter viral infections6,214 and further development may eventually lead to future drugs.
Clinical applications
Biomarker discovery
Mass spectrometry has made significant contributions to identify and validate potential peptide biomarkers. By comparing healthy and diseased tissues or body fluids, differential display of the endogenous peptides can indicate potential biomarkers or a therapeutic target for a disease. The development of improved mass spectrometry techniques enabled the discovery of low-abundance peptides in clinical samples, especially for peptides from biofluids. Many studies utilized sensitive mass spectrometry tools to investigate the urinary peptidome for kidney-related diseases215. The urinary peptidome was investigated with capillary electrophoresis–TOF-MS and further verified with capillary electrophoresis–FTICR-MS, where 273 peptides were identified to be associated with advanced chronic kidney disease216, which later proved to be biomarkers for chronic kidney disease progression and diabetic nephropathy217,218. Instead of the traditionally used positron emission tomography technique219, researchers have also leveraged MALDI MSI to study diseases with analyte localization information. Exemplarily, the spatial progression of amyloid aggregates for Alzheimer disease was investigated through multimodal MALDI MSI220.
Targeted characterization and quantitation of peptides with specific post-translational modifications can also be highly valuable. Among the diverse modifications, glycosylation has attracted increased interest221, due to its close association with neurodegenerative disorders222, cancer223 and autoimmune diseases224. The in-depth characterization and quantitation of glycosylated peptides remain challenging due to their low abundance in vivo and high chemical complexity and structural diversity221. Many mass spectrometryrelated methodologies for glycosylated peptide detection have been reported, including separation and enrichment of the glycopeptides during sample preparation, enhanced fragmentation techniques (for example, collision-induced dissociation, electron transfer dissociation, EThCD)47–49 and DIA-MS139,225. As an example, a targeted mass spectrometry approach was employed with oxonium ion-triggered EThCD to achieve the first large-scale discovery of O-glycosylation on signalling peptides in human and mouse pancreatic islets226.
Pathophysiology/physiology: mechanism of disease and treatment
Since the discovery of insulin, neuropeptides and other peptide hormones have been regarded as an important class of chemical regulators broadly involved in mediating numerous physiological functions227. Various animal models relevant to human physiology and pathophysiology provided an opportunity to link neurochemical changes to behavioural output and extrapolate findings to humans. Mass spectrometry played a significant role in the discovery and characterization of signalling peptides in evolutionary conserved pathways governing homeostasis225, pain228, complex behaviour229, learning and memory230, and ageing231,232, to name a few. High-throughput LC-MSpowered inquiries of animal peptidomes provided molecular links between native peptide dynamic states and environmental factors, nutrition, disease and behaviour233. As an example of a tour de force discovery effort, a label-free LC-MS approach was employed to identify and measure neuropeptide levels in a murine migraine model234. From 1,500 neuropeptides screened, 16 were linked to migraine and/or opioid-induced hyperalgesia234. To focus on secreted peptides, peptidomic analysis can be performed on synaptoneurosomes, dense core vesicles235 or captured single-cell releasates36 to probe neuropeptidome dynamics236, synaptic dysfunction and brain neurodegeneration. A more elegant but difficult approach is an in vivo measurement of secreted peptides via microdialysis coupled to mass spectrometry platforms for identification237. Another way of gaining insights into intercellular communication is selective in vitro analysis of secreted, physiologically relevant endogenous peptides released from neuronal networks in response to physiological stimulation, which can be achieved via microfluidic devices238. Microfluidics integration with mass spectrometry provides capabilities for molecular structural characterization and label-free and absolute quantitation of peptides239.
Reproducibility and data deposition
The field of peptidomics deals with highly variable sources of peptides, requiring various extraction methods, clean-up, derivatization and different approaches for analysis. If automated methods are used to assist the analysis, researchers will need to report the FDR. The FDR is a statistical method for determining the rate at which type 1 errors occur in null hypothesis testing. The FDR provides the global confidence of the data set, in contrast to the P value of a peptide spectrum match (PSM) which refers to the percentage likelihood of incorrect assignment. For the FDR estimation, the decoy database is the null hypothesis. Accordingly, the FDR is the number of hits from the null hypothesis (decoy) divided by the number of total hits, providing a global confidence of the data set. Although the P value only accounts for a single PSM and the FDR for the global data set, methods to exclude certain PSMs are termed controlling procedures. The simplest would be the q value, often interpreted as the minimum posterior probability of the null hypothesis or the FDR, which means the FDR and the α threshold are the same. Then, if set at 1%, all PSMs with P ≥ 0.01 will be rejected240. This method may not be sufficient, as many algorithms try to improve the FDR along different parameters using the posterior error probability, which can depend on the length, charge and modifications241. Other approaches to controlling the FDR may include using P values, covariates, z scores or the family-wise error rate (FWER)242. Reporting how the FDR is controlled is important to any omics approach, along with the null hypothesis of the experiment. This allows the user to answer questions such as whether the incorrect PSMs are truly incorrect if post-translational modifications prevent correct assignment or whether peptides in the experiment are present in the database file. As such, the availability of annotated peptide sequences is an invaluable resource for researchers, and they are encouraged to deposit them in relevant databases.
For reproducibility and traceability, discovered peptides and their modifications should be deposited in open access databases. The repositories of the National Center for Biotechnology Information (NCBI)78, the European Molecular Biology Laboratory European Bioinformatics Institute (EMBL-EBI)243 and the DNA Data Bank of Japan (DDBJ)244 share access to data between themselves under the FAIR (Findability, Accessibility, Interoperability and Reusability) guidelines245. The EMBL-EBI has three portals for submission relevant to peptidomics research: SPIN (Edman degradation or manual interpretation of MS/MS spectra)246, ENA (nucleotide translations of protein-level data)82 and PRIDE247 (for sequences identified using search engines). Raw and unprocessed mass spectrometry data can be deposited along with processed protein and peptide output files. The use of a vendor data file format is less common. Instead, open source mzXML (or similar) is standard. The data collection should also contain meta files, such as sample preparation protocols and device settings, as well as information on the sample origin (cell type, tissue). The data repositories have detailed requirements and provide a unique identifier to connect publications with the deposit data files247. Specifically, in the case of microbial RiPPs and NRPs (and their corresponding gene clusters), the resource MiBIG relies on very simple inputs of the GenBank accession for all genes in the gene cluster and the SMILES for the compound discovered81.
Limitations and optimizations
Peptide degradation
Sampling that truly reflects the in vivo state of the tissue is imperative for finding potential biomarkers and regulators, but this is not entirely possible. Native peptides in freshly isolated biological samples are subject to a multitude of interferences including biased sampling, variable sample stability and fast degradation that make measurement and identification of endogenous peptides more challenging relative to traditional bottom-up proteomics. Enzymatic degradation of ubiquitous proteins is particularly detrimental to mass spectrometry analysis, as degradation products fall into a typical peptide mass range, thus obscuring the detection of native peptides typically present in much lower amounts in tissue extracts or biological fluids. To prevent enzymatic protein degradation during tissue sampling, several tissue stabilization approaches have been implemented248, with heat stabilization being one of the most effective methods of sample preparation249. Heat stabilization arrests the ex vivo peptidase activity, thereby conserving the chemical composition of the sample250.
Biological variation versus sensitivity
Biological systems involve a complex interplay of the organism and its environment, and much of that context is lost in the laboratory setting. Sample collection and culturing of microorganisms, for example, only covers a tiny fraction of the strains that were present in the original sample as cultivation conditions are not known for many strains. However, peptide extraction and analysis without propagation may reveal novel peptides or keep some biosynthetic gene clusters active251,252. The same applies for clinical settings: every individual has their own genetics, lifestyle and risk factors that affect health. For instance, where routine clinical diagnostics did not suffice, a novel multi-omics approach managed to identify Bacteroides vulgatus proteases as a novel risk factor for ulcerative colitis by adding metapeptidomic data to their analysis, which has often been ignored in clinical practice253. The use of multi-omics approaches in conjunction with peptidomics will help address the biological variability and improve the sensitivity by proper sampling and analysis. This might become important, for instance, in personalized medicine applications.
Analysis, algorithms and big data
Nowadays, there are several analysis tools available (Tables 1 and 2 and Supplementary Table 1). Peptide assignment and annotation from genomic data are straightforward if genome data of the organisms of interest are available. However, in silico-derived and mass spectrometry-based data sets are not necessarily identical in routine analysis254. These discrepancies are due to ambiguities, such as technical shortcomings in mass spectrometric detection and data processing or because of false positive peptide assignment and annotation. For instance, for certain organisms, such as bacteria and fungi, it can be challenging, due to the small reading frames and/or massive biosynthetic modifications. Furthermore, in silico workflows are biased towards well-known examples and prone to propagating bias255. Continuous research on such peptides aids training of the algorithms to improve peptide identifications in the future95,96,99,100. Analysis can become cumbersome if access to genomic or transcriptomic data is limited. As sequencing costs continue to decrease and sequencing power continuously increases, more genome data will become available in the future. Furthermore, other sources of peptides are breakdown products of proteins, for example the opioid peptides and haemorphins derived from haemoglobin58. This is being addressed by deep learning algorithms being developed to detect bioactive peptides in protein sequences86.
A special consideration is required for RiPPs and NRPs from bacterial and fungal sources. Previously, classical screening approaches were based on the taxonomic characterization of the strain and the subsequent workflow of analytical and assay techniques. Nowadays, massive bioinformatic data from genome sequences, however, made genome mining the dominating technique: genes or gene clusters are analysed by predictive tools for their putative function, which are subsequently validated experimentally. Precedence of structural and functional data eases assignment to biosynthetic classes. Particularly, in the RiPP field, the increasing availability of DNA sequences in the databases led to a massive boost in the discovery of putative but also new structures. NRPs, where the amino acid sequence is not encoded in the mRNA sequence, are a special case. To predict the potential product of these synthetases, the A-domain specificities98,256 of subdomains involved in substrate recruitment are used. However, this procedure may fail for new amino acid motifs or for complex-type synthetases if the co-linearity rule is violated. Thus, sequence data on the synthetases alone often do not suffice to predict the structure of the searched peptide2. This can be overcome with an increase in known NRP structures and their biosynthetic gene clusters. Although most commercial platforms can do an excellent job on properly linearized and derivatized peptides, recently algorithms have been developed for more complex peptides of microbial origins to be annotated directly from the spectra101,257. Machine learning258 and spectral networks have been explored to identify and assign the chemical nature of peptide natural products259.
Outlook
Method and instrumentation developments
Rapidly evolving mass spectrometry instrumentation opens new opportunities for in-depth interrogation of peptidomes even in smallvolume samples. The latest two-step methodology integrated an ion mobility with TOF or orbitrap mass analysers, leading to unprecedented sensitivity and the highest quality of peptide sequencing. Implementing ion mobility as an additional dimension of separation resulted in improved peptide identification rates, enhanced peptide coverage and greater confidence of post-translational modification assignments260. Even unique post-translational modifications such as isomerization that were difficult to deduce by other mass spectrometry methods due to a lack of characteristic mass shifts now can be unambiguously measured and validated. This technology may attract more attention in the peptidomics field soon. The ion mobility separation can be further combined with MSI to enable the investigation of spatially resolved peptidomics in a high-throughput manner with enhanced chemical information.
Data deposition, open source, unity
Standardization of data deposition and annotation has improved in the past few years with centralized databases such as the NCBI78, the EMBL243 and UniProt79 (Table 2). However, valuable data are still stored in various ‘in-house’ or decentralized databases. Servers such as Bactibase (bacteriocins)261, ConoServer (conotoxins)80, CyBase (circular peptides)262 or the PeptideAtlas (peptide spectra)263 use their own accession codes and aim to solve pre-existing problems. The generation and deposition of data should always be a mutual goal of researchers to make sure that the information generated does not fade away as, eventually, websites will be archived, and information lost. As was the case for ArachnoServer264, a database for spider venoms, which no longer responds to connection requests, whereas copies of its content remain on UniProt cross-referenced databases. Data deposition does not overwrite the utility of these databases, as many of them also come equipped with various tools or query-specific types of data. The best workarounds currently available are those of MiBIG81 and the VEu-PathDB project77. They connect data from the NCBI, the EMBL or other sources to their specific applications. Researchers are encouraged to make this the standard practice, to deposit biological sequence data with the major repositories (NCBI, EMBL, DDBJ), fetch data through their services and focus on adding layers relevant to their field onto that information. The VeuPathDB project has several resources for researchers, and MiBIG is a centralized resource used by most tools in natural product discovery. Even if the tools may be lost to the aeons, researchers are encouraged to deposit their codes on publicly available servers if tools are to be discontinued (for example, Github, Bitbucket, SourceForge). For future prospects, the Protein Data Bank (PDB)265 has potentially the most important feature of any database, the option to deposit unpublished protein structures. This will hopefully become more standard practice where researchers can deposit unpublished experimental data under certain guidelines, as this would deal with the most significant loss of scientific data, and bring them out of the cupboards and into the Big Data landscape.
Bioinformatics, systems biology and artificial intelligence
Current bioinformatics approaches have been used successfully to identify new genes with machine learning. The machine learning methods primarily relied on hidden Markov models, support vector machines or random forest algorithms that laid the foundation for most bioinformatic approaches today266. Currently, deep learning algorithms267 are becoming more frequent, as they have the advantage of being able to learn more complex features than their predecessor. Deep learning has been successfully implemented in the discovery of new genes and peptides89, but possibly its most impressive feat is the accurate prediction of protein structures74,268. As the algorithms continue to improve along with access to graphics processing units to train neural networks, researchers in all fields, even with relatively little experience in programming, will be able to make use of the power of deep learning for their research. With the coming improvements from bioinformatics, bioanalytics and the omics fields, the discipline of systems biology aims to harness all levels of data it can, to understand further how each of these fields may work together269. Systems biology approaches have been applied in the field of metabolomics by generating genome-scale metabolic models270. These approaches are commonly used for production optimization, and industry relies on them to improve yields from fermentation271,272. The field shows promise in combining biological data for clinical applications and could facilitate the transition into personalized medicine94.
Biological sciences are in a major transition into the big data landscape, where a lot of the focus has been on genomics, transcriptomics, proteomics and metabolomics. Peptidomics emerges as a bridge connecting proteomics and metabolomics, bridging the functions between proteins and small molecules. With the advances in deep learning and artificial intelligence, the biochemical space made available by peptides can be better exploited, and novel peptidomimetics can be developed for medicine or industry. Spanning from novel therapeutics to peptide-assisted catalysts10, the field of peptidomics has just started to show a tiny portion of its tremendous potential.
Supplementary Material
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s43586-023-00205-2.
Acknowledgements
Work in the laboratory of C.W.G. has been supported by the Austrian Science Fund (FWF) through projects P32109 and ZK 81B. The work of A.S. and R.D.S. was funded by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) under Germany’s Excellence Strategy — EXC 2008-390540038 (UniSysCat) and RTG 2473 ‘Bioactive Peptides’. The work of L.L. is supported by the National Science Foundation (NSF) (CHE-2108223) and National Institutes of Health (NIH) through grants (R01DK071801, R01 AG078794 and RF1AG052324). The work of J.V.S. is supported by the National Institute on Drug Abuse under Award No. P30DA018310 and the National Institute of Neurological Disorders and Stroke (NINDS) through R01NS031609.
Footnotes
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
The authors declare no competing interests.
Peer review information Nature Reviews Methods Primers thanks Vivian Hook, Dong-Woo Lee and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
antiSMASH: https://antismash.secondarymetabolites.org
DeepBGC: https://github.com/Merck/deepbgc
DeepRiPP: http://deepripp.magarveylab.ca
DEREPLICATOR+: https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp
Dictionary of Natural Products: https://dnp.chemnetbase.com/faces/chemical/ChemicalSearch.xhtml
High Definition Imaging: https://www.waters.com/waters/en_US/High-Definition-Imaging-(HDI)-Software/nav.htm?cid=134833914&locale=en_US
ImageQuest: https://www.thermofisher.com/order/catalog/product/10137985
MS-FINDER: http://prime.psc.riken.jp/compms/msfinder/main.html
MSiReader: https://msireader.com/
msiQuant: https://ms-imaging.org/paquan/
NCBI: https://www.ncbi.nlm.nih.gov
SANDPUMA: https://bitbucket.org/chevrm/sandpuma/src/master/
SCiLS Lab: https://www.bruker.com/en/products-and-solutions/mass-spectrometry/ms-software/scils-lab.html
UniProt: https://www.uniprot.org
References
- 1.Gruber CW, Muttenthaler M, Freissmuth M. Ligand-based peptide design and combinatorial peptide libraries to target G protein-coupled receptors. Curr Pharm Des. 2010;16:3071–3088. doi: 10.2174/138161210793292474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dang T, Süssmuth RD. Bioactive peptide natural products as lead structures for medicinal use. Acc Chem Res. 2017;50:1566–1576. doi: 10.1021/acs.accounts.7b00159. [DOI] [PubMed] [Google Scholar]
- 3.Fosgerau K, Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discov Today. 2015;20:122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Muttenthaler M, King GF, Adams DJ, Alewood PF. Trends in peptide drug discovery. Nat Rev Drug Discov. 2021;20:309–325. doi: 10.1038/s41573-020-00135-8. [This comprehensive review discusses the importance of peptides as drug leads and innovative therapeutics.] [DOI] [PubMed] [Google Scholar]
- 5.Craik DJ, Fairlie DP, Liras S, Price D. The future of peptide-based drugs. Chem Biol Drug Des. 2013;81:136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 6.Munch J, Standker L, Forssmann WG, Kirchhoff F. Discovery of modulators of HIV-1 infection from the human peptidome. Nat Rev Microbiol. 2014;12:715–722. doi: 10.1038/nrmicro3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baggerman G, et al. Peptidomics. J Chromatogr B. 2004;803:3–16. doi: 10.1016/j.jchromb.2003.07.019. [DOI] [PubMed] [Google Scholar]
- 8.Schrader M, Schulz-Knappe P, Fricker LD. Historical perspective of peptidomics. EuPA Open Proteom. 2014;3:171–182. [Google Scholar]
- 9.Schulz-Knappe P, et al. Peptidomics: the comprehensive analysis of peptides in complex biological mixtures. Comb Chem High TScr. 2001;4:207–217. doi: 10.2174/1386207013331246. [DOI] [PubMed] [Google Scholar]
- 10.Metrano AJ, et al. Asymmetric catalysis mediated by synthetic peptides, version 2.0: expansion of scope and mechanisms. Chem Rev. 2020;120:11479–11615. doi: 10.1021/acs.chemrev.0c00523. [This review article discusses peptide-assisted asymmetric synthesis reactions and recent advances in the field.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collier JH, Segura T. Evolving the use of peptides as components of biomaterials. Biomaterials. 2011;32:4198–4204. doi: 10.1016/j.biomaterials.2011.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agnieray H, Glasson JL, Chen Q, Kaur M, Domigan LJ. Recent developments in sustainably sourced protein-based biomaterials. Biochem Soc Trans. 2021;49:953–964. doi: 10.1042/BST20200896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malandrino N, Smith RJ. In: Principles of Endocrinology and Hormone Action. Belfiore A, LeRoith D, editors. Springer International; 2018. pp. 29–42. [Google Scholar]
- 14.Yi J, Warunek D, Craft D. Degradation and stabilization of peptide hormones in human blood specimens. PLoS ONE. 2015;10:e0134427. doi: 10.1371/journal.pone.0134427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svensson M, et al. Heat stabilization of the tissue proteome: a new technology for improved proteomics. J Proteome Res. 2009;8:974–981. doi: 10.1021/pr8006446. [DOI] [PubMed] [Google Scholar]
- 16.Yang N, Anapindi KDB, Romanova EV, Rubakhin SS, Sweedler JV. Improved identification and quantitation of mature endogenous peptides in the rodent hypothalamus using a rapid conductive sample heating system. Analyst. 2017;142:4476–4485. doi: 10.1039/c7an01358b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feist P, Hummon AB. Proteomic challenges: sample preparation techniques for microgram-quantity protein analysis from biological samples. Int J Mol Sci. 2015;16:3537–3563. doi: 10.3390/ijms16023537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison ST. Bacterial cell disruption: a key unit operation in the recovery of intracellular products. Biotechnol Adv. 1991;9:217–240. doi: 10.1016/0734-9750(91)90005-g. [DOI] [PubMed] [Google Scholar]
- 19.Koehbach J, et al. Cyclotide discovery in Gentianales revisited — identification and characterization of cyclic cystine-knot peptides and their phylogenetic distribution in Rubiaceae plants. Biopolymers. 2013;100:438–452. doi: 10.1002/bip.22328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen EI, Cociorva D, Norris JL, Yates JR. 3rd Optimization of mass spectrometry-compatible surfactants for shotgun proteomics. J Proteome Res. 2007;6:2529–2538. doi: 10.1021/pr060682a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panuwet P, et al. Biological matrix effects in quantitative tandem mass spectrometrybased analytical methods: advancing biomonitoring. Crit Rev Anal Chem. 2016;46:93–105. doi: 10.1080/10408347.2014.980775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finoulst I, Pinkse M, Van Dongen W, Verhaert P. Sample preparation techniques for the untargeted LC-MS-based discovery of peptides in complex biological matrices. J Biomed Biotechnol. 2011;2011:245291. doi: 10.1155/2011/245291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tubaon RM, Haddad PR, Quirino JP. Sample clean-up strategies for ESI mass spectrometry applications in bottom-up proteomics: trends from 2012 to 2016. Proteomics. 2017;17:1700011. doi: 10.1002/pmic.201700011. [DOI] [PubMed] [Google Scholar]
- 24.Sosalagere C, Adesegun Kehinde B, Sharma P. Isolation and functionalities of bioactive peptides from fruits and vegetables: a reviews. Food Chem. 2022;366:130494. doi: 10.1016/j.foodchem.2021.130494. [DOI] [PubMed] [Google Scholar]
- 25.Mthembu SN, Sharma A, Albericio F, de la Torre BG. Breaking a couple: disulfide reducing agents. Chembiochem. 2020;21:1947–1954. doi: 10.1002/cbic.202000092. [DOI] [PubMed] [Google Scholar]
- 26.Hellinger R, et al. Importance of the cyclic cystine knot structural motif for immunosuppressive effects of cyclotides. ACS Chem Biol. 2021;16:2373–2386. doi: 10.1021/acschembio.1c00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai PL, Chen SF, Huang SY. Mass spectrometry-based strategies for protein disulfide bond identification. Rev Anal Chem. 2013;32:257–268. [Google Scholar]
- 28.Han DK, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat Biotechnol. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Proteolytic 18O labeling for comparative proteomics: model studies with two serotypes of adenovirus. Anal Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 30.Hsu J-L, Huang S-Y, Chow N-H, Chen S-H. Stable-isotope dimethyl labeling for quantitative proteomics. Anal Chem. 2003;75:6843–6852. doi: 10.1021/ac0348625. [DOI] [PubMed] [Google Scholar]
- 31.Greer T, Lietz CB, Xiang F, Li L. Novel isotopic N,N-dimethyl leucine (iDiLeu) reagents enable absolute quantification of peptides and proteins using a standard curve approach. J Am Soc Mass Spectrom. 2014;26:107–119. doi: 10.1007/s13361-014-1012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeSouza LV, et al. Multiple reaction monitoring of mTRAQ-labeled peptides enables absolute quantification of endogenous levels of a potential cancer marker in cancerous and normal endometrial tissues. J Proteome Res. 2008;7:3525–3534. doi: 10.1021/pr800312m. [DOI] [PubMed] [Google Scholar]
- 33.Thompson A, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 34.Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7:340–350. doi: 10.1002/pmic.200600422. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Wang Y, Li S. Deuterium isobaric amine-reactive tags for quantitative proteomics. Anal Chem. 2010;82:7588–7595. doi: 10.1021/ac101306x. [DOI] [PubMed] [Google Scholar]
- 36.Atkins N, Jr, et al. Functional peptidomics: stimulus- and time-of-day-specific peptide release in the mammalian circadian clock. ACS Chem Neurosci. 2018;9:2001–2008. doi: 10.1021/acschemneuro.8b00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gedela S, Medicherla NR. Chromatographic techniques for the separation of peptides: application to proteomics. Chromatographia. 2007;65:511–518. [Google Scholar]
- 38.Udeshi ND, Compton PD, Shabanowitz J, Hunt DF, Rose KL. Methods for analyzing peptides and proteins on a chromatographic timescale by electron-transfer dissociation mass spectrometry. Nat Protoc. 2008;3:1709–1717. doi: 10.1038/nprot.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahoney WC, Hermodson MA. Separation of large denatured peptides by reverse phase high performance liquid chromatography. Trifluoroacetic acid as a peptide solvent. J Biol Chem. 1980;255:11199–11203. [PubMed] [Google Scholar]
- 40.Yoshida T. Peptide separation by hydrophilic-interaction chromatography: a review. J Biochem Biophys Meth. 2004;60:265–280. doi: 10.1016/j.jbbm.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 41.Hillenkamp F, Karas M. Mass spectrometry of peptides and proteins by matrix-assisted ultraviolet laser desorption/ionization. Meth Enzymol. 1990;193:280–295. doi: 10.1016/0076-6879(90)93420-p. [DOI] [PubMed] [Google Scholar]
- 42.Dreisewerd K. The desorption process in MALDI. Chem Rev. 2003;103:395–426. doi: 10.1021/cr010375i. [DOI] [PubMed] [Google Scholar]
- 43.Dong X, et al. A LC-MS/MS method to monitor the concentration of HYD-PEP06, a RGD-modified Endostar mimetic peptide in rat blood. J Chromatogr B. 2018;1092:296–305. doi: 10.1016/j.jchromb.2018.05.042. [DOI] [PubMed] [Google Scholar]
- 44.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ludwig C, et al. Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial. Mol Syst Biol. 2018;14:e8126. doi: 10.15252/msb.20178126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Follmann R, Goldsmith CJ, Stein W. Spatial distribution of intermingling pools of projection neurons with distinct targets: a 3D analysis of the commissural ganglia in Cancer borealis. J Comp Neurol. 2017;525:1827–1843. doi: 10.1002/cne.24161. [DOI] [PubMed] [Google Scholar]
- 48.Mechref Y. Use of CID/ETD mass spectrometry to analyze glycopeptides. Curr Protoc Protein Sci. 2012;68:12.11.1-12.11.11. doi: 10.1002/0471140864.ps1211s68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riley NM, Malaker SA, Driessen MD, Bertozzi CR. Optimal dissociation methods differ for N-and O-glycopeptides. J Proteome Res. 2020;19:3286–3301. doi: 10.1021/acs.jproteome.0c00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Demichev V, Messner CB, Vernardis SI, Lilley KS, Ralser M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat Methods. 2020;17:41–44. doi: 10.1038/s41592-019-0638-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsou C-C, et al. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat Methods. 2015;12:258–264. doi: 10.1038/nmeth.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinitcyn P, et al. MaxDIA enables library-based and library-free data-independent acquisition proteomics. Nat Biotechnol. 2021 doi: 10.1038/s41587-021-00968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 54.Tyler BJ, Rayal G, Castner DG. Multivariate analysis strategies for processing ToF-SIMS images of biomaterials. Biomaterials. 2007;28:2412–2423. doi: 10.1016/j.biomaterials.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eberlin LS, et al. Desorption electrospray ionization then MALDI mass spectrometry imaging of lipid and protein distributions in single tissue sections. Anal Chem. 2011;83:8366–8371. doi: 10.1021/ac202016x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bouschen W, Spengler B. Artifacts of MALDI sample preparation investigated by high-resolution scanning microprobe matrix-assisted laser desorption/ionization (SMALDI) imaging mass spectrometry. Int J Mass Spectrom. 2007;266:129–137. [Google Scholar]
- 57.Iakab S-A, et al. SALDI-MS and SERS multimodal imaging: one nanostructured substrate to rule them both. Anal Chem. 2022;94:2785–2793. doi: 10.1021/acs.analchem.1c04118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ali A, Baby B, Soman SS, Vijayan R. Molecular insights into the interaction of hemorphin and its targets. Sci Rep. 2019;9:14747. doi: 10.1038/s41598-019-50619-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rocha B, Ruiz-Romero C, Blanco FJ. Mass spectrometry imaging: a novel technology in rheumatology. Nat Rev Rheumatol. 2017;13:52–63. doi: 10.1038/nrrheum.2016.184. [DOI] [PubMed] [Google Scholar]
- 60.Ramos-Vara J. Technical aspects of immunohistochemistry. Vet Pathol. 2005;42:405–426. doi: 10.1354/vp.42-4-405. [DOI] [PubMed] [Google Scholar]
- 61.Skelley D, Brown L, Besch P. Radioimmunoassay. Clin Chem. 1973;19:146–186. [PubMed] [Google Scholar]
- 62.Lichtman JW, Conchello J-A. Fluorescence microscopy. Nat Methods. 2005;2:910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 63.Buchberger AR, DeLaney K, Johnson J, Li L. Mass spectrometry imaging: a review of emerging advancements and future insights. Anal Chem. 2018;90:240. doi: 10.1021/acs.analchem.7b04733. [This comprehensive review discusses various aspects of MSI, spanning from sample preparation and mass spectrometry instrumentation to data analysis and diverse applications.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lemaire R, et al. Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. J Proteome Res. 2007;6:1295–1305. doi: 10.1021/pr060549i. [DOI] [PubMed] [Google Scholar]
- 65.Kokkat TJ, et al. Archived formalin-fixed paraffin-embedded (FFPE) blocks: a valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv Biobank. 2013;11:101–106. doi: 10.1089/bio.2012.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ren Y, et al. Reagents for isobaric labeling peptides in quantitative proteomics. Anal Chem. 2018;90:12366–12371. doi: 10.1021/acs.analchem.8b00321. [DOI] [PubMed] [Google Scholar]
- 67.Truong JX, et al. Removal of optimal cutting temperature (OCT) compound from embedded tissue for MALDI imaging of lipids. Anal Bioanal Chem. 2021;413:2695–2708. doi: 10.1007/s00216-020-03128-z. [DOI] [PubMed] [Google Scholar]
- 68.Tian Y, Bova GS, Zhang H. Quantitative glycoproteomic analysis of optimal cutting temperature-embedded frozen tissues identifying glycoproteins associated with aggressive prostate cancer. Anal Chem. 2011;83:7013–7019. doi: 10.1021/ac200815q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bogdanow B, Zauber H, Selbach M. Systematic errors in peptide and protein identification and quantification by modified peptides. Mol Cell Proteom. 2016;15:2791–2801. doi: 10.1074/mcp.M115.055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwartz SA, Reyzer ML, Caprioli RM. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J Mass Spectrom. 2003;38:699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- 71.Lemaire R, et al. MALDI-MS direct tissue analysis of proteins: improving signal sensitivity using organic treatments. Anal Chem. 2006;78:7145–7153. doi: 10.1021/ac060565z. [DOI] [PubMed] [Google Scholar]
- 72.Buchberger AR, Sauer CS, Vu NQ, DeLaney K, Li L. Temporal study of the perturbation of crustacean neuropeptides due to severe hypoxia using 4-plex reductive dimethylation. J Proteome Res. 2020;19:1548–1555. doi: 10.1021/acs.jproteome.9b00787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaletaç BK, et al. Sample preparation issues for tissue imaging by imaging MS. Proteomics. 2009;9:2622–2633. doi: 10.1002/pmic.200800364. [DOI] [PubMed] [Google Scholar]
- 74.Jumper J, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leopold J, Popkova Y, Engel KM, Schiller J. Recent developments of useful MALDI matrices for the mass spectrometric characterization of lipids. Biomolecules. 2018;8:173. doi: 10.3390/biom8040173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.DeLaney K, et al. Mass spectrometry quantification, localization, and discovery of feeding-related neuropeptides in cancer borealis. ACS Chem Neurosci. 2021;12:782–798. doi: 10.1021/acschemneuro.1c00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amos B, et al. VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res. 2022;50:D898–D911. doi: 10.1093/nar/gkab929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sayers EW, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022;50:D20–D26. doi: 10.1093/nar/gkab1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.The UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaas Q, Yu R, Jin AH, Dutertre S, Craik DJ. ConoServer: updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 2012;40:D325–D330. doi: 10.1093/nar/gkr886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kautsar SA, et al. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2020;48:D454–D458. doi: 10.1093/nar/gkz882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leinonen R, et al. The European Nucleotide Archive. Nucleic Acids Res. 2011;39:D28–D31. doi: 10.1093/nar/gkq967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33:W451–W454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hyatt D, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hazarika RR, et al. ARA-PEPs: a repository of putative sORF-encoded peptides in Arabidopsis thaliana. BMC Bioinformatics. 2017;18:37. doi: 10.1186/s12859-016-1458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mooney C, Haslam NJ, Holton TA, Pollastri G, Shields DC. PeptideLocator: prediction of bioactive peptides in protein sequences. Bioinformatics. 2013;29:1120–1126. doi: 10.1093/bioinformatics/btt103. [DOI] [PubMed] [Google Scholar]
- 87.Zhou P, et al. Detecting small plant peptides using SPADA (Small Peptide Alignment Discovery Application) BMC Bioinformatics. 2013;14:335. doi: 10.1186/1471-2105-14-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhu M, Gribskov M. MiPepid: microPeptide identification tool using machine learning. BMC Bioinformatics. 2019;20:559. doi: 10.1186/s12859-019-3033-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Y, Jia C, Fullwood MJ, Kwoh CK. DeepCPP: a deep neural network based on nucleotide bias information and minimum distribution similarity feature selection for RNA coding potential prediction. Brief Bioinform. 2021;22:2073–2084. doi: 10.1093/bib/bbaa039. [DOI] [PubMed] [Google Scholar]
- 90.Lin D, et al. Mining amphibian and insect transcriptomes for antimicrobial peptide sequences with rAMPage. Antibiotics. 2022;11:952. doi: 10.3390/antibiotics11070952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lyons E, Freeling M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008;53:661–673. doi: 10.1111/j.1365-313X.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- 92.Dieckmann MA, et al. EDGAR3.0: comparative genomics and phylogenomics on a scalable infrastructure. Nucleic Acids Res. 2021;49:W185–W192. doi: 10.1093/nar/gkab341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Medema MH, Fischbach MA. Computational approaches to natural product discovery. Nat Chem Biol. 2015;11:639–648. doi: 10.1038/nchembio.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weber T, Kim HU. The secondary metabolite bioinformatics portal: computational tools to facilitate synthetic biology of secondary metabolite production. Synth Syst Biotechnol. 2016;1:69–79. doi: 10.1016/j.synbio.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blin K, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [This work presents the most significant genome mining platform for natural products, covering a wide range of compounds, and is a recommended read for anyone interested in natural product research.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hannigan GD, et al. A deep learning genome-mining strategy for biosynthetic gene cluster prediction. Nucleic Acids Res. 2019;47:e110. doi: 10.1093/nar/gkz654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sélem-Mojica N, Aguilar C, Gutiérrez-García K, Martínez-Guerrero CE, Barona-Gómez F. EvoMining reveals the origin and fate of natural product biosynthetic enzymes. Microb Genom. 2019 doi: 10.1099/mgen.0000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chevrette MG, Aicheler F, Kohlbacher O, Currie CR, Medema MH. SANDPUMA ensemble predictions of nonribosomal peptide chemistry reveal biosynthetic diversity across Actinobacteria. Bioinformatics. 2017;33:3202–3210. doi: 10.1093/bioinformatics/btx400. [This work discusses how SANDPUMA has aided NRP discovery and continues to provide valuable predictions for researchers involved in NRP research.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van Heel AJ, et al. BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018;46:W278–W281. doi: 10.1093/nar/gky383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ramesh S, et al. Bioinformatics-guided expansion and discovery of graspetides. ACS Chem Biol. 2021;16:2787–2797. doi: 10.1021/acschembio.1c00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Merwin NJ, et al. DeepRiPP integrates multiomics data to automate discovery of novel ribosomally synthesized natural products. Proc Natl Acad Sci USA. 2020;117:371–380. doi: 10.1073/pnas.1901493116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schlaffner CN, Pirklbauer GJ, Bender A, Choudhary JS. Fast, quantitative and variant enabled mapping of peptides to genomes. Cell Syst. 2017;5:152–156.:e4. doi: 10.1016/j.cels.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ricart E, et al. rBAN: retro-biosynthetic analysis of nonribosomal peptides. J Cheminform. 2019;11:13. doi: 10.1186/s13321-019-0335-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kunyavskaya O, et al. Nerpa: a tool for discovering biosynthetic gene clusters of bacterial nonribosomal peptides. Metabolites. 2021 doi: 10.3390/metabo11100693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Konanov DN, Krivonos DV, Ilina EN, Babenko VV. BioCAT: search for biosynthetic gene clusters producing nonribosomal peptides with known structure. Comput Struct Biotechnol J. 2022;20:1218–1226. doi: 10.1016/j.csbj.2022.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grundemann C, Koehbach J, Huber R, Gruber CW. Do plant cyclotides have potential as immunosuppressant peptides. J Nat Prod. 2012;75:167–174. doi: 10.1021/np200722w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van Santen JA, et al. The Natural Products Atlas: an open access knowledge base for microbial natural products discovery. ACS Cent Sci. 2019;5:1824–1833. doi: 10.1021/acscentsci.9b00806. [This work discusses how the Natural Products Atlas provides valuable information, visualization and validation of discovered compounds.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mohimani H, et al. Dereplication of microbial metabolites through database search of mass spectra. Nat Commun. 2018;9:4035. doi: 10.1038/s41467-018-06082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Diament BJ, Noble WS. Faster SEQUEST searching for peptide identification from tandem mass spectra. J Proteome Res. 2011;10:3871–3879. doi: 10.1021/pr101196n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Claesen J, Valkenborg D, Burzykowski T. De novo prediction of the elemental composition of peptides and proteins based on a single mass. J Mass Spectrom. 2020;55:e4367. doi: 10.1002/jms.4367. [DOI] [PubMed] [Google Scholar]
- 112.Lai Z, et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat Methods. 2018;15:53–56. doi: 10.1038/nmeth.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Palmer A, et al. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat Methods. 2017;14:57–60. doi: 10.1038/nmeth.4072. [DOI] [PubMed] [Google Scholar]
- 114.Novak J, Skriba A, Havlicek V. CycloBranch 2: molecular formula annotations applied to imzML data sets in bimodal fusion and LC-MS data files. Anal Chem. 2020;92:6844–6849. doi: 10.1021/acs.analchem.0c00170. [DOI] [PubMed] [Google Scholar]
- 115.Ricart E, Pupin M, Muller M, Lisacek F. Automatic annotation and dereplication of tandem mass spectra of peptidic natural products. Anal Chem. 2020;92:15862–15871. doi: 10.1021/acs.analchem.0c03208. [DOI] [PubMed] [Google Scholar]
- 116.Gurevich A, et al. Increased diversity of peptidic natural products revealed by modification-tolerant database search of mass spectra. Nat Microbiol. 2018;3:319–327. doi: 10.1038/s41564-017-0094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Seidler J, Zinn N, Boehm ME, Lehmann WD. De novo sequencing of peptides by MS/MS. Proteomics. 2010;10:634–649. doi: 10.1002/pmic.200900459. [DOI] [PubMed] [Google Scholar]
- 118.Yang H, Chi H, Zeng W-F, Zhou W-J, He S-M. pNovo 3: precise de novo peptide sequencing using a learning-to-rank framework. Bioinformatics. 2019;35:i183–i190. doi: 10.1093/bioinformatics/btz366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tran NH, et al. Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry. Nat Methods. 2019;16:63–66. doi: 10.1038/s41592-018-0260-3. [DOI] [PubMed] [Google Scholar]
- 120.Tran NH, Zhang X, Xin L, Shan B, Li M. De novo peptide sequencing by deep learning. Proc Natl Acad Sci USA. 2017;114:8247–8252. doi: 10.1073/pnas.1705691114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Elias JE, Gygi SP. Target-decoy search strategy for mass spectrometry-based proteomics. Meth Mol Biol. 2010;604:55–71. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Reiter L, et al. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol Cell Proteom. 2009;8:2405–2417. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 124.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 125.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Röst HL, et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat Biotechnol. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 127.Bruderer R, et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol Cell Proteom. 2015;14:1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Neilson KA, et al. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11:535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]
- 129.Chang C, et al. LFAQ: toward unbiased label-free absolute protein quantification by predicting peptide quantitative factors. Anal Chem. 2018;91:1335–1343. doi: 10.1021/acs.analchem.8b03267. [DOI] [PubMed] [Google Scholar]
- 130.Ishihama Y, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteom. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 131.Sivanich MK, Gu TJ, Tabang DN, Li L. Recent advances in isobaric labeling and applications in quantitative proteomics. Proteomics. 2022;22:e2100256. doi: 10.1002/pmic.202100256. [This critical review article discusses isobaric labelling strategies for quantitative proteomics and peptidomics applications as well as current limitations and future outlooks.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fonville JM, et al. Robust data processing and normalization strategy for MALDI mass spectrometric imaging. Anal Chem. 2012;84:1310–1319. doi: 10.1021/ac201767g. [DOI] [PubMed] [Google Scholar]
- 133.Deininger S-O, et al. Normalization in MALDI-TOF imaging datasets of proteins: practical considerations. Anal Bioanal Chem. 2011;401:167–181. doi: 10.1007/s00216-011-4929-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Källback P, Shariatgorji M, Nilsson A, Andrén PE. Novel mass spectrometry imaging software assisting labeled normalization and quantitation of drugs and neuropeptides directly in tissue sections. J Proteom. 2012;75:4941–4951. doi: 10.1016/j.jprot.2012.07.034. [DOI] [PubMed] [Google Scholar]
- 135.Shariatgorji M, et al. Direct targeted quantitative molecular imaging of neurotransmitters in brain tissue sections. Neuron. 2014;84:697–707. doi: 10.1016/j.neuron.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 136.Lanekoff I, Thomas M, Laskin J. Shotgun approach for quantitative imaging of phospholipids using nanospray desorption electrospray ionization mass spectrometry. Anal Chem. 2014;86:1872–1880. doi: 10.1021/ac403931r. [DOI] [PubMed] [Google Scholar]
- 137.Hansen HT, Janfelt C. Aspects of quantitation in mass spectrometry imaging investigated on cryo-sections of spiked tissue homogenates. Anal Chem. 2016;88:11513–11520. doi: 10.1021/acs.analchem.6b02711. [DOI] [PubMed] [Google Scholar]
- 138.Robichaud G, Garrard KP, Barry JA, Muddiman DC. MSiReader: an open-source interface to view and analyze high resolving power MS imaging files on Matlab platform. J Am Soc Mass Spectrom. 2013;24:718–721. doi: 10.1007/s13361-013-0607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Alexander J, Oliphant A, Wilcockson DC, Webster SG. Functional identification and characterization of the diuretic hormone 31 (DH31) signaling system in the green shore crab, Carcinus maenas. Front Neurosci. 2018;12:454. doi: 10.3389/fnins.2018.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Källback P, Nilsson A, Shariatgorji M, Andrén PE. msIQuant — quantitation software for mass spectrometry imaging enabling fast access, visualization, and analysis of large data sets. Anal Chem. 2016;88:4346–4353. doi: 10.1021/acs.analchem.5b04603. [DOI] [PubMed] [Google Scholar]
- 141.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Product Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [This comprehensive review introduces the reader to RiPPs, from classification to biosynthesis and bioactivity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wiebach V, et al. The anti-staphylococcal lipolanthines are ribosomally synthesized lipopeptides. Nat Chem Biol. 2018;14:652–654. doi: 10.1038/s41589-018-0068-6. [This research article discusses a novel type of anti-staphylococcal RiPP, utilizing a short peptide conjugated with a lipid moiety.] [DOI] [PubMed] [Google Scholar]
- 143.Sussmuth RD, Mainz A. Nonribosomal peptide synthesis-principles and prospects. Angew Chem Int Ed. 2017;56:3770–3821. doi: 10.1002/anie.201609079. [This comprehensive review about NRPs explains biosynthesis, structures and bioactivity or NRPs.] [DOI] [PubMed] [Google Scholar]
- 144.Tang S, et al. Discovery and characterization of a PKS-NRPS hybrid in Aspergillus terreus by genome mining. J Nat Prod. 2020;83:473–480. doi: 10.1021/acs.jnatprod.9b01140. [DOI] [PubMed] [Google Scholar]
- 145.Zhang Z, Wang J, Wang J, Wang J, Li Y. Estimate of the sequenced proportion of the global prokaryotic genome. Microbiome. 2020 doi: 10.1186/s40168-020-00903-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.V V, et al. Venom peptides — a comprehensive translational, perspective in pain management. Curr Res Toxicol. 2021;2:329–340. doi: 10.1016/j.crtox.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.King GF, Hardy MC. Spider-venom peptides: structure, pharmacology, and potential for control of insect pests. Annu Rev Entomol. 2013;58:475–496. doi: 10.1146/annurev-ento-120811-153650. [DOI] [PubMed] [Google Scholar]
- 148.Munawar A, Ali SA, Akrem A, Betzel C. Snake venom peptides: tools of biodiscovery. Toxins. 2018 doi: 10.3390/toxins10110474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.King GF. Venoms as a platform for human drugs: translating toxins into therapeutics. Expert Opin Biol Ther. 2011;11:1469–1484. doi: 10.1517/14712598.2011.621940. [DOI] [PubMed] [Google Scholar]
- 150.Dutertre S, et al. Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat Commun. 2014;5:3521. doi: 10.1038/ncomms4521. [This research article investigates the diferences between the defensive and predatory venoms of cone snails.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Prashanth JR, Dutertre S, Lewis RJ. In: Evolution of Venomous Animals and Their Toxins Ch. Malhotra A, editor. Vol. 18. Springer; 2017. pp. 105–123. [Google Scholar]
- 152.Coelho P, Kaliontzopoulou A, Rasko M, Meijden A, Portugal S. A 'striking' relationship: scorpion defensive behaviour and its relation to morphology and performance. Funct Ecol. 2017;31:1390–1404. [This work presents a fascinating investigation into the diferent methods of the defensive behaviours of scorpions, measuring both the speed and frequency of stings in response to stimuli.] [Google Scholar]
- 153.Nisani Z, Hayes WK. Defensive stinging by Parabuthus transvaalicus scorpions: risk assessment and venom metering. Anim Behav. 2011;81:627–633. [Google Scholar]
- 154.Diesner M, Predel R, Neupert S. Neuropeptide mapping of dimmed cells of adult Drosophila brain. J Am Soc Mass Spectrom. 2018;29:890–902. doi: 10.1007/s13361-017-1870-1. [DOI] [PubMed] [Google Scholar]
- 155.Habenstein J, et al. Transcriptomic, peptidomic, and mass spectrometry imaging analysis of the brain in the ant Cataglyphis nodus. J Neurochem. 2021;158:391–412. doi: 10.1111/jnc.15346. [DOI] [PubMed] [Google Scholar]
- 156.Zeng H, et al. Genomics- and peptidomics-based discovery of conserved and novel neuropeptides in the American cockroach. J Proteome Res. 2021;20:1217–1228. doi: 10.1021/acs.jproteome.0c00596. [DOI] [PubMed] [Google Scholar]
- 157.El Filali Z, Van Minnen J, Liu WK, Smit AB, Li KW. Peptidomics analysis of neuropeptides involved in copulatory behavior of the mollusk Lymnaea stagnalis. J Proteome Res. 2006;5:1611–1617. doi: 10.1021/pr060014p. [DOI] [PubMed] [Google Scholar]
- 158.Parmar BS, et al. Identification of non-canonical translation products in C elegans using tandem mass spectrometry. Front Genet. 2021;12:728900. doi: 10.3389/fgene.2021.728900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Van Bael S, et al. A Caenorhabditis elegans mass spectrometric resource for neuropeptidomics. J Am Soc Mass Spectrom. 2018;29:879–889. doi: 10.1007/s13361-017-1856-z. [DOI] [PubMed] [Google Scholar]
- 160.Wood EA, et al. Neuropeptide localization in Lymnaea stagnalis: from the central nervous system to subcellular compartments. Front Mol Neurosci. 2021;14:670303. doi: 10.3389/fnmol.2021.670303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.DeLaney K, Buchberger A, Li L. Identification, quantitation, and imaging of the crustacean peptidome. Methods Mol Biol. 2018;1719:247–269. doi: 10.1007/978-1-4939-7537-2_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.DeLaney K, Li L. Capillary electrophoresis coupled to MALDI mass spectrometry imaging with large volume sample stacking injection for improved coverage of C borealis neuropeptidome. Analyst. 2019;145:61–69. doi: 10.1039/c9an01883b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu Y, Li G, Li L. Targeted top-down mass spectrometry for the characterization and tissue-specific functional discovery of crustacean hyperglycemic hormones (CHH) and CHH precursor-related peptides in response to low pH stress. J Am Soc Mass Spectrom. 2021;32:1352–1360. doi: 10.1021/jasms.0c00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Xu LL, et al. Major shrimp allergen peptidomics signatures and potential biomarkers of heat processing. Food Chem. 2022;382:132567. doi: 10.1016/j.foodchem.2022.132567. [DOI] [PubMed] [Google Scholar]
- 165.Phetsanthad A, et al. Recent advances in mass spectrometry analysis of neuropeptides Mass. Spectrom Rev. 2021;42:706–750. doi: 10.1002/mas.21734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fujisawa T, Hayakawa E. Peptide signaling in Hydra. Int J Dev Biol. 2012;56:543–550. doi: 10.1387/ijdb.113477tf. [DOI] [PubMed] [Google Scholar]
- 167.Monroe EB, et al. Exploring the sea urchin neuropeptide landscape by mass spectrometry. J Am Soc Mass Spectrom. 2018;29:923–934. doi: 10.1007/s13361-018-1898-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Takahashi T. Neuropeptides and epitheliopeptides: structural and functional diversity in an ancestral metazoan Hydra. Protein Pept Lett. 2013;20:671–680. doi: 10.2174/0929866511320060006. [DOI] [PubMed] [Google Scholar]
- 169.Southey BR, Romanova EV, Rodriguez-Zas SL, Sweedler JV. Bioinformatics for prohormone and neuropeptide discovery. Methods Mol Biol. 2018;1719:71–96. doi: 10.1007/978-1-4939-7537-2_5. [This methodological article describes a pipeline for annotation of neuropeptide prohormones from genomic assemblies using freely available public toolsets and databases.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hu CK, et al. Identification of prohormones and pituitary neuropeptides in the African cichlid, Astatotilapia burtoni. BMC Genomics. 2016;17:660. doi: 10.1186/s12864-016-2914-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chan-Andersen PC, Romanova EV, Rubakhin SS, Sweedler JV. Profiling 26,000 Aplysia californica neurons by single cell mass spectrometry reveals neuronal populations with distinct neuropeptide profiles. J Biol Chem. 2022;298:102254. doi: 10.1016/j.jbc.2022.102254. [This work presents an elegant mass spectrometry-based approach for robust categorization of large cell populations based on a single-cell neuropeptide profile.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jiménez CR, et al. Peptidomics of a single identified neuron reveals diversity of multiple neuropeptides with convergent actions on cellular excitability. J Neurosci. 2006;26:518–529. doi: 10.1523/JNEUROSCI.2566-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Green DJ, et al. cAMP, Ca2+, pH and NO regulate H-like cation channels that underlie feeding and locomotion in the predatory sea slug Pleurobranchaea californica. ACS Chem Neurosci. 2018;9:1986–1993. doi: 10.1021/acschemneuro.8b00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Han Y, Ma B, Zhang K. SPIDER: software for protein identification from sequence tags with de novo sequencing error. J Bioinform Comput Biol. 2005;3:697–716. doi: 10.1142/s0219720005001247. [DOI] [PubMed] [Google Scholar]
- 175.Romanova EV, Aerts JT, Croushore CA, Sweedler JV. Small-volume analysis of cell-cell signaling molecules in the brain. Neuropsychopharmacology. 2014;39:50–64. doi: 10.1038/npp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bai L, et al. Characterization of GdFFD, a D-amino acid-containing neuropeptide that functions as an extrinsic modulator of the Aplysia feeding circuit. J Biol Chem. 2013;288:32837–32851. doi: 10.1074/jbc.M113.486670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Checco JW, et al. Aplysia allatotropin-related peptide and its newly identified D-amino acid-containing epimer both activate a receptor and a neuronal target. J Biol Chem. 2018;293:16862–16873. doi: 10.1074/jbc.RA118.004367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Romanova EV, et al. Urotensin II in invertebrates: from structure to function in Aplysia californica. PLoS ONE. 2012;7:e48764. doi: 10.1371/journal.pone.0048764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhang G, et al. Newly identified Aplysia SPTR-gene family-derived peptides: localization and function. ACS Chem Neurosci. 2018;9:2041–2053. doi: 10.1021/acschemneuro.7b00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mast DH, Checco JW, Sweedler JV. Differential post-translational amino acid isomerization found among neuropeptides in Aplysia californica. ACS Chem Biol. 2020;15:272–281. doi: 10.1021/acschembio.9b00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mast DH, Checco JW, Sweedler JV. Advancing D-amino acid-containing peptide discovery in the metazoan. Biochim Biophys Acta Proteins Proteom. 2021;1869:140553. doi: 10.1016/j.bbapap.2020.140553. [This review discusses the prevalence of enzyme-derived DAACPs among animals, physiological consequences of peptide isomerization and analytical methods for structural characterization/discovery of DAACPs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lambeth TR, Julian RR. Differentiation of peptide isomers and epimers by radical-directed dissociation. Methods Enzymol. 2019;626:67–87. doi: 10.1016/bs.mie.2019.06.020. [DOI] [PubMed] [Google Scholar]
- 183.Mast DH, Liao HW, Romanova EV, Sweedler JV. Analysis of peptide stereochemistry in single cells by capillary electrophoresis-trapped ion mobility spectrometry mass spectrometry. Anal Chem. 2021;93:6205–6213. doi: 10.1021/acs.analchem.1c00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Checco JW, et al. Molecular and physiological characterization of a receptor for D-amino acid-containing neuropeptides. ACS Chem Biol. 2018;13:1343–1352. doi: 10.1021/acschembio.8b00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Livnat I, et al. A D-amino acid-containing neuropeptide discovery funnel. Anal Chem. 2016;88:11868–11876. doi: 10.1021/acs.analchem.6b03658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yussif BM, Checco JW. Evaluation of endogenous peptide stereochemistry using liquid chromatography-mass spectrometry-based spiking experiments. Methods Enzymol. 2022;663:205–234. doi: 10.1016/bs.mie.2021.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Jiang L, et al. A quantitative proteome map of the human body. Cell. 2020;183:269–283.:e19. doi: 10.1016/j.cell.2020.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Secher A, et al. Analytic framework for peptidomics applied to large-scale neuropeptide identification. Nat Commun. 2016;7:11436. doi: 10.1038/ncomms11436. [This article introduces a comprehensive analytical workflow for large-scale mammalian peptidomics studies, detailing procedures ranging from sample preparation to data analysis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Foster SR, et al. Discovery of human signaling systems: pairing peptides to G protein-coupled receptors. Cell. 2019;179:895–908.:e21. doi: 10.1016/j.cell.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hauser AS, Gloriam DE, Brauner-Osborne H, Foster SR. Novel approaches leading towards peptide GPCR de-orphanisation. Br J Pharmacol. 2020;177:961–968. doi: 10.1111/bph.14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Scarpa A. Pre-scientific medicines: their extent and value. Soc Sci Med A Med Psychol Med Sociol. 1981;15:317–326. doi: 10.1016/0271-7123(81)90061-4. [DOI] [PubMed] [Google Scholar]
- 192.Pina AS, Hussain A, Roque ACA. In: Ligand-Macromolecular Interactions in Drug Discovery: Methods and Protocols. Roque ACA, editor. Humana; 2010. pp. 3–12. [Google Scholar]
- 193.Heinrich M. Ethnobotany and its role in drug development. Phytother Res. 2000;14:479–488. doi: 10.1002/1099-1573(200011)14:7<479::aid-ptr958>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 194.Campbell IB, Macdonald SJF, Procopiou PA. Medicinal chemistry in drug discovery in Big Pharma: past, present and future. Drug Discov Today. 2018;23:219–234. doi: 10.1016/j.drudis.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 195.Camargo ACM, Ianzer D, Guerreiro JR, Serrano SMT. Bradykinin-potentiating peptides: beyond captopril. Toxicon. 2012;59:516–523. doi: 10.1016/j.toxicon.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 196.Cesa-Luna C, et al. Structural characterization of scorpion peptides and their bactericidal activity against clinical isolates of multidrug-resistant bacteria. PLoS ONE. 2019;14:e0222438. doi: 10.1371/journal.pone.0222438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Jouiaei M, et al. Ancient venom systems: a review on Cnidaria toxins. Toxins. 2015;7:2251–2271. doi: 10.3390/toxins7062251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jin AH, et al. Conotoxins: chemistry and biology. Chem Rev. 2019;119:11510–11549. doi: 10.1021/acs.chemrev.9b00207. [This review article on conotoxins explains the chemistry and biology behind their function by using 3D structural models, thus providing a deeper understanding of the topic.] [DOI] [PubMed] [Google Scholar]
- 199.McGivern JG. Ziconotide: a review of its pharmacology and use in the treatment of pain. Neuropsychiatr Dis Treat. 2007;3:69–85. doi: 10.2147/nedt.2007.3.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Safavi-Hemami H, et al. Specialized insulin is used for chemical warfare by fish-hunting cone snails. Proc Natl Acad Sci USA. 2015;112:1743–1748. doi: 10.1073/pnas.1423857112. [This article is interesting for researchers involved in peptide hormone research, discussing the weaponization of peptide hormones by animals.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Furman BL. The development of Byetta (exenatide) from the venom of the Gila monster as an anti-diabetic agent. Toxicon. 2012;59:464–471. doi: 10.1016/j.toxicon.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 202.Muller TD, Bluher M, Tschop MH, DiMarchi RD. Anti-obesity drug discovery: advances and challenges. Nat Rev Drug Discov. 2022;21:201–223. doi: 10.1038/s41573-021-00337-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Rubinstein E, Keynan Y. Vancomycin revisited — 60 years later. Front Public Health. 2014 doi: 10.3389/fpubh.2014.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Heidary M, et al. Daptomycin. J Antimicrob Chemother. 2018;73:1–11. doi: 10.1093/jac/dkx349. [DOI] [PubMed] [Google Scholar]
- 205.Felnagle EA, et al. Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol Pharm. 2008;5:191–211. doi: 10.1021/mp700137g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Flores C, Fouquet G, Moura IC, Maciel TT, Hermine O. Lessons to learn from low-dose cyclosporin-a: a new approach for unexpected clinical applications. Front Immunol. 2019 doi: 10.3389/fimmu.2019.00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Additives E, et al. Safety of nisin (E 234) as a food additive in the light of new toxicological data and the proposed extension of use. EFSA J. 2017;15:e05063. doi: 10.2903/j.efsa.2017.5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Nakatsuji T, Gallo RL. Antimicrobial peptides: old molecules with new ideas. J Invest Dermatol. 2012;132:887–895. doi: 10.1038/jid.2011.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lei J, et al. The antimicrobial peptides and their potential clinical applications. Am J Transl Res. 2012;11:3919. [PMC free article] [PubMed] [Google Scholar]
- 210.Zborovsky L, et al. Improvement of the antimicrobial potency, pharmacokinetic and pharmacodynamic properties of albicidin by incorporation of nitrogen atoms. Chem Sci. 2021;12:14606–14617. doi: 10.1039/d1sc04019g. [This work is an example of how medicinal chemistry can be used to improve the bioactive qualities of peptides.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Imai Y, et al. A new antibiotic selectively kills Gram-negative pathogens. Nature. 2019;576:459–464. doi: 10.1038/s41586-019-1791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Vilas Boas LCP, Campos ML, Berlanda RLA, de Carvalho Neves N, Franco OL. Antiviral peptides as promising therapeutic drugs. Cell Mol Life Sci. 2019;76:3525–3542. doi: 10.1007/s00018-019-03138-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Bosso M, Ständker L, Kirchhoff F, Münch J. Exploiting the human peptidome for novel antimicrobial and anticancer agents Bioorg. Med Chem. 2018;26:2719–2726. doi: 10.1016/j.bmc.2017.10.038. [DOI] [PubMed] [Google Scholar]
- 214.Kuroki A, Tay J, Lee GH, Yang YY. Broad-spectrum antiviral peptides and polymers. Adv Healthc Mater. 2021;10:e2101113. doi: 10.1002/adhm.202101113. [DOI] [PubMed] [Google Scholar]
- 215.Klein J, Bascands J-L, Mischak H, Schanstra JP. The role of urinary peptidomics in kidney disease research. Kidney Int. 2016;89:539–545. doi: 10.1016/j.kint.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 216.Good DM, et al. Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol Cell Proteom. 2010;9:2424–2437. doi: 10.1074/mcp.M110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Argiles A, et al. CKD273, a new proteomics classifier assessing CKD and its prognosis. PLoS ONE. 2013;8:e62837. doi: 10.1371/journal.pone.0062837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Roscioni S, et al. A urinary peptide biomarker set predicts worsening of albuminuria in type 2 diabetes mellitus. Diabetologia. 2013;56:259–267. doi: 10.1007/s00125-012-2755-2. [DOI] [PubMed] [Google Scholar]
- 219.Nakamura A, et al. High performance plasma amyloid-β biomarkers for Alzheimer's disease. Nature. 2018;554:249–254. doi: 10.1038/nature25456. [DOI] [PubMed] [Google Scholar]
- 220.Kaya I, Zetterberg H, Blennow K, Hanrieder JR. Shedding light on the molecular pathology of amyloid plaques in transgenic Alzheimer's disease mice using multimodal MALDI imaging mass spectrometry. ACS Chem Neurosci. 2018;9:1802–1817. doi: 10.1021/acschemneuro.8b00121. [DOI] [PubMed] [Google Scholar]
- 221.Reily C, Stewart TJ, Renfrow MB, Novak J. Glycosylation in health and disease. Nat Rev Nephrol. 2019;15:346–366. doi: 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Chen Z, et al. In-depth site-specific analysis of N-glycoproteome in human cerebrospinal fluid and glycosylation landscape changes in Alzheimer's disease. Mol Cell Proteom. 2021;20:100081. doi: 10.1016/j.mcpro.2021.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 224.Li Q, et al. Site-specific glycosylation quantitation of 50 serum glycoproteins enhanced by predictive glycopeptidomics for improved disease biomarker discovery. Anal Chem. 2019;91:5433–5445. doi: 10.1021/acs.analchem.9b00776. [DOI] [PubMed] [Google Scholar]
- 225.Alim FZD, et al. Seasonal adaptations of the hypothalamo-neurohypophyseal system of the dromedary camel. PLoS ONE. 2019;14:e0216679. doi: 10.1371/journal.pone.0216679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Yu Q, et al. Targeted mass spectrometry approach enabled discovery of O-glycosylated insulin and related signaling peptides in mouse and human pancreatic islets. Anal Chem. 2017;89:9184–9191. doi: 10.1021/acs.analchem.7b01926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Anapindi KDB, Romanova EV, Checco JW, Sweedler JV. Mass spectrometry approaches empowering neuropeptide discovery and therapeutics. Pharmacol Rev. 2022;74:662–679. doi: 10.1124/pharmrev.121.000423. [This review article discusses the historical, current and future states of neuropeptidomics with mass spectrometry and their implications for therapeutic strategies in neurological disorders.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Tillmaand EG, et al. Peptidomics and secretomics of the mammalian peripheral sensory-motor system. J Am Soc Mass Spectrom. 2015;26:2051–2061. doi: 10.1007/s13361-015-1256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Ramachandran S, et al. A conserved neuropeptide system links head and body motor circuits to enable adaptive behavior. eLife. 2021 doi: 10.7554/eLife.71747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Van Damme S, et al. Neuromodulatory pathways in learning and memory: lessons from invertebrates. J Neuroendocrinol. 2021;33:e12911. doi: 10.1111/jne.12911. [DOI] [PubMed] [Google Scholar]
- 231.Greenwood MP, et al. The effects of aging on biosynthetic processes in the rat hypothalamic osmoregulatory neuroendocrine system. Neurobiol Aging. 2018;65:178–191. doi: 10.1016/j.neurobiolaging.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Pan F, et al. Peptidome analysis reveals the involvement of endogenous peptides in mouse pancreatic dysfunction with aging. J Cell Physiol. 2019;234:14090–14099. doi: 10.1002/jcp.28098. [DOI] [PubMed] [Google Scholar]
- 233.Hook V, Lietz CB, Podvin S, Cajka T, Fiehn O. Diversity of neuropeptide cell-cell signaling molecules generated by proteolytic processing revealed by neuropeptidomics mass spectrometry. J Am Soc Mass Spectrom. 2018;29:807–816. doi: 10.1007/s13361-018-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Anapindi KDB, et al. PACAP and other neuropeptide targets link chronic migraine and opioid-induced hyperalgesia in mouse models. Mol Cell Proteom. 2019;18:2447–2458. doi: 10.1074/mcp.RA119.001767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Jiang Z, et al. Differential neuropeptidomes of dense core secretory vesicles (DCSV) produced at intravesicular and extracellular pH conditions by proteolytic processing. ACS Chem Neurosci. 2021;12:2385–2398. doi: 10.1021/acschemneuro.1c00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Podvin S, et al. Dysregulation of neuropeptide and tau peptide signatures in human Alzheimer's disease brain. ACS Chem Neurosci. 2022;13:1992–2005. doi: 10.1021/acschemneuro.2c00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Al-Hasani R, et al. In vivo detection of optically-evoked opioid peptide release. eLife. 2018 doi: 10.7554/eLife.36520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Vitorino R, Guedes S, Costa JPD, Kasicka V. Microfluidics for peptidomics, proteomics, and cell analysis. Nanomaterials. 2021 doi: 10.3390/nano11051118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ong TH, Tillmaand EG, Makurath M, Rubakhin SS, Sweedler JV. Mass spectrometry-based characterization of endogenous peptides and metabolites in small volume samples. Biochim Biophys Acta. 2015;1854:732–740. doi: 10.1016/j.bbapap.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Burger T. Gentle introduction to the statistical foundations of false discovery rate in quantitative proteomics. J Proteome Res. 2018;17:12–22. doi: 10.1021/acs.jproteome.7b00170. [This work is a worthwhile introduction to the statistics behind FDRs, highly recommended for all researchers working in proteomics or peptidomics.] [DOI] [PubMed] [Google Scholar]
- 241.Käll L, Storey JD, MacCoss MJ, Noble WS. Posterior error probabilities and false discovery rates: two sides of the same coin. J Proteome Res. 2008;7:40–44. doi: 10.1021/pr700739d. [DOI] [PubMed] [Google Scholar]
- 242.Korthauer K, et al. A practical guide to methods controlling false discoveries in computational biology. Genome Biol. 2019;20:118. doi: 10.1186/s13059-019-1716-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Kanz C, et al. The EMBL nucleotide sequence database. Nucleic Acids Res. 2005;33:D29–D33. doi: 10.1093/nar/gki098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Fukuda A, Kodama Y, Mashima J, Fujisawa T, Ogasawara O. DDBJ update: streamlining submission and access of human data. Nucleic Acids Res. 2021;49:D71–D75. doi: 10.1093/nar/gkaa982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wilkinson MD, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [This work on the FAIR Guiding Principles is an essential read for all researchers as data management will become more important as data continue to be generated worldwide.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Pichler K, Warner K, Magrane M, UniProt C. SPIN: submitting sequences determined at protein level to UniProt. Curr Protoc Bioinformatics. 2018;62:e52. doi: 10.1002/cpbi.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Ternent T, et al. How to submit MS proteomics data to ProteomeXchange via the PRIDE database. Proteomics. 2014;14:2233–2241. doi: 10.1002/pmic.201400120. [DOI] [PubMed] [Google Scholar]
- 248.Segerstrom L, Gustavsson J, Nylander I. Minimizing postsampling degradation of peptides by a thermal benchtop tissue stabilization method. Biopreserv Biobank. 2016;14:172–179. doi: 10.1089/bio.2015.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Fridjonsdottir E, Nilsson A, Wadensten H, Andren PE. Brain tissue sample stabilization and extraction strategies for neuropeptidomics. Methods Mol Biol. 2018;1719:41–49. doi: 10.1007/978-1-4939-7537-2_2. [DOI] [PubMed] [Google Scholar]
- 250.Stingl C, Soderquist M, Karlsson O, Boren M, Luider TM. Uncovering effects of ex vivo protease activity during proteomics and peptidomics sample extraction in rat brain tissue by oxygen-18 labeling. J Proteome Res. 2014;13:2807–2817. doi: 10.1021/pr401232e. [DOI] [PubMed] [Google Scholar]
- 251.Katz M, Hover BM, Brady SF. Culture-independent discovery of natural products from soil metagenomes. J Ind Microbiol Biotechnol. 2016;43:129–141. doi: 10.1007/s10295-015-1706-6. [DOI] [PubMed] [Google Scholar]
- 252.Reher R, et al. Native metabolomics identifies the rivulariapeptolide family of protease inhibitors. Nat Commun. 2022;13:4619. doi: 10.1038/s41467-022-32016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Mills RH, et al. Multi-omics analyses of the ulcerative colitis gut microbiome link Bacteroides vulgatus proteases with disease severity. Nat Microbiol. 2022;7:262–276. doi: 10.1038/s41564-021-01050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Hellinger R, et al. Peptidomics of circular cysteine-rich plant peptides: analysis of the diversity of cyclotides from viola tricolor by transcriptome and proteome mining. J Proteome Res. 2015;14:4851–4862. doi: 10.1021/acs.jproteome.5b00681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Haynes WA, Tomczak A, Khatri P. Gene annotation bias impedes biomedical research. Sci Rep. 2018;8:1362. doi: 10.1038/s41598-018-19333-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Flissi A, et al. Norine: update of the nonribosomal peptide resource. Nucleic Acids Res. 2020;48:D465–D469. doi: 10.1093/nar/gkz1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Wang M, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Saldivar-Gonzalez FI, Aldas-Bulos VD, Medina-Franco JL, Plisson F. Natural product drug discovery in the artificial intelligence era. Chem Sci. 2022;13:1526–1546. doi: 10.1039/d1sc04471k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Mohimani H, et al. Dereplication of peptidic natural products through database search of mass spectra. Nat Chem Biol. 2017;13:30–37. doi: 10.1038/nchembio.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Jeanne Dit Fouque K, et al. Fast and effective ion mobility-mass spectrometry separation of D-amino-acid-containing peptides. Anal Chem. 2017;89:11787–11794. doi: 10.1021/acs.analchem.7b03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Hammami R, Zouhir A, Le Lay C, Ben Hamida J, Fliss I. BACTIBASE second release: a database and tool platform for bacteriocin characterization. BMC Microbiol. 2010;10:22. doi: 10.1186/1471-2180-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wang CK, Kaas Q, Chiche L, Craik DJ. CyBase: a database of cyclic protein sequences and structures, with applications in protein discovery and engineering. Nucleic Acids Res. 2008;36:D206–D210. doi: 10.1093/nar/gkm953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Deutsch EW. The PeptideAtlas Project. Methods Mol Biol. 2010;604:285–296. doi: 10.1007/978-1-60761-444-9_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Pineda SS, et al. ArachnoServer 3.0: an online resource for automated discovery, analysis and annotation of spider toxins. Bioinformatics. 2018;34:1074–1076. doi: 10.1093/bioinformatics/btx661. [DOI] [PubMed] [Google Scholar]
- 265.wwPDB consortium. Protein Data Bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019;47:D520–D528. doi: 10.1093/nar/gky949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Larranaga P, et al. Machine learning in bioinformatics. Brief Bioinform. 2006;7:86–112. doi: 10.1093/bib/bbk007. [This interesting review discusses the machine learning methods that got bioinformatics to where it is today.] [DOI] [PubMed] [Google Scholar]
- 267.Min S, Lee B, Yoon S. Deep learning in bioinformatics. Brief Bioinform. 2017;18:851–869. doi: 10.1093/bib/bbw068. [This article describes the use, applications and architecture of deep learning networks, providing the readers with insight into the direction that bioinformatics is heading in the next decade.] [DOI] [PubMed] [Google Scholar]
- 268.Baek M, et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Breitling R. What is systems biology? Front Physiol. 2010;1:9. doi: 10.3389/fphys.2010.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Heirendt L, et al. Creation and analysis of biochemical, constraint-based models using the COBRA Toolbox v.3.0. Nat Protoc. 2019;14:639–702. doi: 10.1038/s41596-018-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Mitra S, Dhar R, Sen R. Designer bacterial cell factories for improved production of commercially valuable non-ribosomal peptides. Biotechnol Adv. 2022;60:108023. doi: 10.1016/j.biotechadv.2022.108023. [DOI] [PubMed] [Google Scholar]
- 272.Helmy M, Smith D, Selvarajoo K. Systems biology approaches integrated with artificial intelligence for optimized metabolic engineering. Metab Eng Commun. 2020;11:e00149. doi: 10.1016/j.mec.2020.e00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.