Abstract
We aimed to investigate potential causal associations between serum 25-hydroxyvitamin D [25(OH)D] levels and incidence of lung cancer overall and histologic types.
We performed a Mendelian randomization (MR) analysis using a prospective cohort study in Norway, including 54580 individuals and 676 incident lung cancer cases. A 25(OH)D allele score was generated based on vitamin D-increasing alleles of rs2282679, rs12785878 and rs10741657. Hazard ratios (HRs) with 95% confidence intervals (CIs) for incidence of lung cancer and histologic types were estimated in relation to the allele score. Inverse-variance weighted method using summarized data of individual single-nucleotide polymorphisms was applied to calculate the MR estimates.
The allele score accounted for 3.4% of the variation in serum 25(OH)D levels. There was no association between the allele score and lung cancer incidence overall, with HR being 0.99 (95% CI 0.93 to 1.06) per allele score. A 25 nmol/L increase in genetically determined 25(OH)D level was not associated with the incidence of lung cancer overall (MR estimate HR 0.96, 95% CI 0.54 to 1.69) or any histologic type.
MR analysis did not suggest causal association between 25(OH)D levels and risk of lung cancer overall or histologic types in this population-based cohort study.
Keywords: histologic types, lung cancer, Mendelian randomization, serum 25-hydroxyvitamin D [25(OH)D], single-nucleotide polymorphisms, vitamin D
Introduction
Vitamin D has been suggested to have a number of anti-carcinogenic potentials, such as stimulating differentiation, inducing apoptosis, and inhibiting invasion and metastasis [1, 2]. Epidemiological studies of the associations between circulating vitamin D and various cancers have shown inconsistent results [3, 4]. Lung cancer has been the most common cancer type for several decades worldwide, and it is also the most deadly cancer [5]. The main histologic types of lung cancer are small cell lung cancer (SCLC), adenocarcinoma and squamous cell carcinoma [6]. Adenocarcinoma is the most common histologic type of lung cancer in many countries [7]. Unlike SCLC and squamous cell carcinoma, the association between smoking and adenocarcinoma is much weaker [8]. Thus, identifying other risk factors than tobacco smoking is necessary for further prevention of lung cancer overall and certain histologic types.
Two meta-analyses of observational cohort studies suggested an inverse association between serum vitamin D and risk of lung cancer overall [9, 10]. However, conventional observational epidemiological studies have limited capability to identify causal associations due to potential bias from confounding and reverse causation [11]. Although well-designed prospective cohort studies can reduce the possibility of reverse causation, residual and unmeasured confounding is inevitable in observational studies. Mendelian randomization (MR) studies, however, have been suggested to be able to overcome these limitations and to help make causal inferences of modifiable risk factors on health-related outcomes, provided that the crucial assumptions are satisfied [11, 12].
Therefore, we performed a MR study using three single-nucleotide polymorphisms (SNPs) as instrumental variables for serum 25-hydroxyvitamin D [25(OH)D], the primary circulating form of vitamin D, to explore potential causal associations of serum 25(OH)D levels with incidence of lung cancer overall and different histologic types in a population-based prospective cohort.
Material and methods
Study population and data linkage
The Nord-Trøndelag Health Study (HUNT) is a large population-based health study in Norway consisting of three separate surveys: HUNT1 (1984–1986), HUNT2 (1995–1997) and HUNT3 (2006–2008). The current study was based on data from HUNT2, in which 65227 subjects aged ≥20 years living in the county of Nord-Trøndelag participated (response rate 70%). All participants completed a general questionnaire including questions on health, lifestyle and socio-economic status. Blood samples were drawn and body weight and height were measured at a clinical examination. The HUNT Research Center received updated information about deaths of all causes and emigration of the HUNT participants from the Norwegian National Registry in which the dates of such events were recorded for all people living in Norway.
Using the unique 11-digit personal identification number of all residents in Norway, the data on HUNT2 participants were linked with data from the Cancer Registry of Norway [13]. The ICD-10 (Tenth Revision of the International Statistical Classification of Diseases and Related Health Problems) topography codes C33-C34 were used to identify incident lung cancer cases among the HUNT2 study participants. Histologic types of lung cancer were classified according to the International Classification of Diseases of Oncology (ICD-O) [14]. The participants in HUNT2 were followed from the date of participation to the date of lung cancer diagnosis, death, emigration, or end of follow-up (December 31, 2014), whichever occurred first.
We excluded subjects who reported ever cancer (n=2400) in the HUNT2 questionnaire at baseline, lung cancer cases diagnosed before the participation date (n=13) in the HUNT2 study and subjects who did not have information on genotype (n=8234), leaving 54580 subjects in the analysis cohort. Moreover, a 10% random sample (n=6613) of the HUNT2 participants was selected as a subcohort for serum 25(OH)D measurement. After further excluding individuals without serum and genotype information, 5546 individuals remained in the analysis subcohort.
Measurement and standardization of serum 25(OH)D levels
Serum 25(OH)D level is widely recognized as the best available proxy measure for body vitamin D status [15, 16]. Serum 25(OH)D levels were measured at the HUNT Biobank using LIAISON 25-OH Vitamin D TOTAL (DiaSorin, Saluggia, Italy), a fully automated, antibody-based, chemiluminescence assay. The detection range of the assay is 10–375 nmol/L. Because seasonal fluctuations in 25(OH)D levels were expected due to the high- latitude geographical position of Norway, a cosinor model based on month of blood draw was used to calculate season-standardized 25(OH)D level (nmol/L) that represents the annual average value of 25(OH)D for each subject [17]. The season-standardized 25(OH)D was calculated using the package cosinor (version 1.1) in R (version 3.4.2).
Genotyping and imputation of SNPs and allele score as instrumental variables
DNA was isolated from blood samples collected in HUNT2 and stored at the HUNT biobank. Genotyping was performed using Illumina HumanCoreExome arrays as described elsewhere [18]. Imputation was performed on samples of recent European ancestry using Minimac3 (v2.0.1, http://genome.sph.umich.edu/wiki/Minimac3) [19] from a merged reference panel constructed from the Haplotype Reference Consortium panel (release version 1.1) [20] and a local reference panel based on 2201 whole-genome sequenced HUNT participants [21]. In total 3 SNPs located in or near genes for vitamin D synthesis and metabolism were selected as instrumental variables for serum 25(OH)D based on two widely-cited GWAS studies [22, 23]: rs2282679 (GC), rs12785878 (NADSYN1/DHCR7), rs10741657 (CYP2R1). Information on rs6013897 that was included in Wang et al. [23] and its proxy SNPs was not available in the HUNT study, this SNP, however, showed the weakest effect on 25(OH)D level [23, 24]. The effect allele [25(OH)D increasing allele] was coded as 1 and the other allele was coded as 0 (rs2282679: T=1; rs12785878: T=1; and rs10741657: A=1). A 25(OH)D allele score, that was a sum of the number of effect alleles of rs2282679, rs12785878 and rs10741657, was generated to increase the statistical power of the analyses [25]. The R2 values for linkage disequilibrium between these 3 SNPs were calculated [26].
Statistical analyses
Linear regression was applied to calculate the F-statistic and R2 value between SNPs or the allele score and season-standardized 25(OH)D levels. Values of F-statistic greater than 10 suggest that the SNPs or allele score are valid instrumental variables [11]. Linear regression was used to estimate the associations between the allele score and continuous covariates in order to test the assumption that the instrumental variables were not associated with potential confounders for the association between serum 25(OH)D and lung cancer; logistic regression was used in corresponding analyses of binary covariates. To test if there was a causal association between serum 25(OH)D and risk of lung cancer, we used Cox proportional hazards regression to calculate hazard ratios (HRs) with 95% confidence intervals (CIs) for the incidence of lung cancer overall or histologic types in relation to the allele score. Age was used as the time scale in the models. The proportional hazards assumption was satisfied for all SNPs and the allele score. In analyses estimating the risk of a specific histologic type, all other subtypes were censored at the date of diagnosis.
To calculate Mendelian randomization estimates of serum 25(OH)D on lung cancer risk, we generated summarized data of coefficients and standard errors from linear regression of individual SNPs on season-standardized 25(OH)D levels in the subcohort (n=5546), as well as coefficients [ln(hazard ratio)] and standard errors from Cox regression of individual SNPs on risk of lung cancer overall or a histologic type in the cohort (n=54580). Inverse-variance weighted (IVW) and median-based methods were used for the summarized data to calculate MR estimates of serum 25(OH)D for lung cancer overall and histologic types [27]. An IVW estimate of the causal effect combines the ratio estimates using each genetic variant in a fixed-effect meta-analysis model [28]. To test for pleiotropy we used MR-Egger to calculate the intercept and 95% CIs [29]. Additionally, we tested for heterogeneity between SNPs using I2 and Cochran’s Q statistic. To test the robustness of our findings, we performed a two-sample MR as sensitivity analysis using summarized data of SNPs–25(OH)D association derived from a previous consortium study (n≈35000) [24].
Analyses with summarized data of individual SNPs were carried out using the package MendelianRandomization (version 0.2.2) in R (version 3.4.2). All other statistical analyses were performed with Stata/SE 14.2 (College Station, TX, USA).
Ethics
The study was approved by the Norwegian Regional Committees for Medical and Health Research Ethics. All participants gave their informed consent on participation in HUNT, linkage to previous HUNT surveys and specific registries.
Results
During a median follow up of 18 years, a total of 676 incident lung cancer cases were diagnosed among the 54580 cohort participants. Table 1 shows the distribution of baseline characteristics in the cohort (n=54580) and subcohort (n=5546) of the HUNT2 study. In general, the distribution of baseline characteristics was similar between the cohort and subcohort. Supplementary table 1 presents the characteristics of SNPs included in the 25(OH)D allele score in the HUNT2 study. There was no evidence of departure from the Hardy-Weinberg equilibrium for the 3 SNPs. The allele frequency was in line with that of the 1000 Genomes Phase 3 data. The R2 values for linkage disequilibrium between the 3 SNPs were <0.1.
Table 1. Distribution of baseline characteristics in the cohort and subcohort of the HUNT2 study, 1995–1997.
| Cohort | Subcohort* | |
|---|---|---|
| Number of subjects | 54580 | 5546 |
| Age (years) | 49.2±16.6 | 49.1±16.6 |
| Season-standardized 25(OH)D level (nmol/L) | - | 48.3±17.1 |
| Lung cancer cases | 676 | 77 |
| Sex, % (women/men) | 52.7/47.3 | 52.8/47.2 |
| Smoking, % (never/ever/unknown) | 42.3/55.8/1.9 | 42.5/55.4/2.1 |
| Family history of cancer, % (no/yes) | 74.5/25.5 | 74.1/25.9 |
| Education (years), % (<10/≥10/unknown) | 33.6/61.8/4.6 | 32.9/62.5/4.6 |
| Economic difficulties, % (no/yes/unknown) | 49.8/21.8/28.3 | 51.0/21.1/27.9 |
| Body mass index (BMI, kg/m2), % (<25/≥25/unknown) | 39.8/59.4/0.7 | 40.6/58.8/0.6 |
| Physical activity, % (inactive/active/unknown) | 21.4/48.8/29.9 | 21.1/49.0/29.8 |
| Alcohol consumption (times/month), % (never/≥1/unknown) | 33.6/58.0/8.4 | 32.9/59.0/8.1 |
| Chronic bronchitis, % (no/yes/unknown) | 94.6/3.4/2.0 | 95.0/3.4/1.7 |
25(OH)D: 25-hydroxyvitamin D; HUNT2: The Nord-Trøndelag Health Study Survey 2
Data are given as percentage of subjects or mean ± standard deviation
Those with measured serum 25(OH)D levels and genotype information
F-statistics and R2 values between SNPs/the allele score and season-standardized 25(OH)D levels are presented in Table 2. The SNP rs2282679 had the highest F-statistic and R2 value among the 3 SNPs, showing 4.0 nmol/L increase in 25(OH)D per effect allele. The 25(OH)D allele score had a F-statistic of 197 and accounted for 3.4% of the variation of serum 25(OH)D levels. The associations between the allele score and the potential confounders are presented in Supplementary table 2. In general, taking account of multiple testing, there were no clear associations observed.
Table 2. Coefficient, F-statistic and R2 value of linear regression between SNP/allele score and season-standardized 25(OH)D level (nmol/L) in subcohort of the HUNT2 study (n=5546).
| SNP | Coefficient* | 95 CI | P value | F-statistic | R2 |
|---|---|---|---|---|---|
| rs2282679 | 4.00 | (3.30 to 4.71) | 1.63×10-28 | 124 | 0.022 |
| rs12785878 | 1.92 | (1.26 to 2.59) | 1.36×10-08 | 32 | 0.006 |
| rs10741657 | 2.48 | (1.84 to 3.13) | 6.98×10-14 | 56 | 0.010 |
| Allele score† | 2.74 | (2.36 to 3.12) | 5.1536×10-44 | 197 | 0.034 |
25(OH)D: 25-hydroxyvitamin D; CI: confidence interval; HUNT2: The Nord-Trøndelag Health Study Survey 2; SNP: Single-Nucleotide
Coefficient: change of season-standardized 25(OH)D (nmol/L) per effect allele or per allele score
The 25(OH)D allele score is sum of the number of effect alleles of rs2282679, rs12785878 and rs10741657
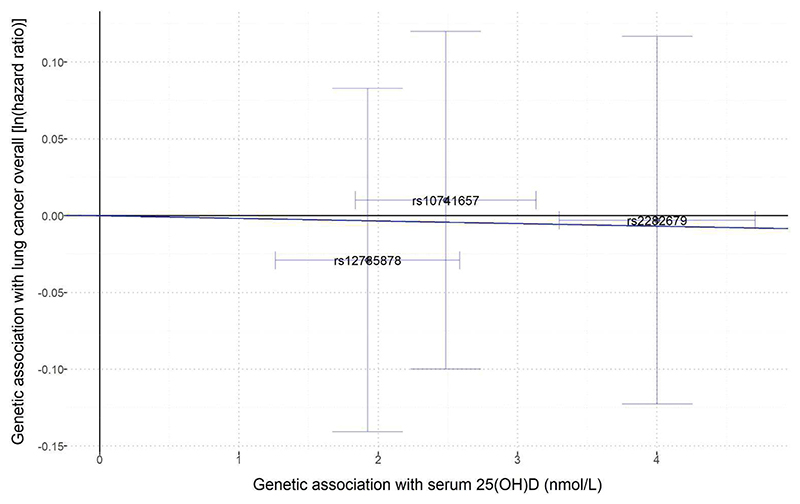
Table 3 shows that the 25(OH)D allele score was not associated with the incidence of lung cancer overall, with HR being 0.99 (95% CI 0.93 to 1.06) per allele score. There was no clear association between the allele score and risk of any histologic type of lung cancer. Based on MR estimates using either IVW method or weighted median method, there was little evidence that genetically determined season-standardized 25(OH)D was associated with risk of lung cancer overall or any histologic type (Table 4 and Figure 1). Using the IVW method, the MR estimate HR for lung cancer overall was 0.96 (95% CI 0.54 to 1.69) per 25 nmol/L increase in the genetically determined 25(OH)D level.
Table 3. The associations between the 25(OH)D allele score and risk of lung cancer overall and histologic types in the HUNT2 study (n=54580).
| Number of cases | Rate (100000 PY* | HR† | 95% CI | P value | |
|---|---|---|---|---|---|
| Lung cancer overall | 676 | 73.8 | 0.99 | (0.93 to 1.06) | 0.83 |
| SCLC | 90 | 9.8 | 0.92 | (0.77 to 1.10) | 0.35 |
| Adenocarcinoma | 189 | 20.6 | 0.92 | (0.82 to 1.04) | 0.20 |
| Squamous cell carcinoma | 141 | 15.4 | 0.98 | (0.85 to 1.14) | 0.82 |
| Other/unknown subtypes | 256# | 27.9 | 1.08 | (0.97 to 1.21) | 0.14 |
25(OH)D: 25-hydroxyvitamin D; CI: confidence interval; HR: hazard ratio; HUNT2: The Nord-Trøndelag Health Study Survey 2; PY: person-years; SCLC: small cell lung cancer
Time at risk for outcomes: 916480 person-years
Per vitamin D-increasing allele score
Among which 87 cases were other subtypes
Table 4. Mendelian randomization estimates of the associations between a 25 nmol/L increase in genetically determined season-standardized 25(OH)D and risk of lung cancer overall and histologic types in the HUNT2 study (n=54580).
| IVW method | Weighted median method | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Outcome | MR estimate HR (95% CI) | P value | I2 (95% CI) | P value of Q statistic | MR estimate (95% CI) | P value | |||
| Lung cancer overall | 0.96 | (0.54 to 1.69) | 0.88 | 0.00 | (0.00 to 0.24) | 0.87 | 1.00 | (0.54 to 1.85) | 0.99 |
| SCLC | 0.58 | (0.12 to 2.69) | 0.48 | 0.45 | (0.00 to 0.84) | 0.16 | 0.82 | (0.14 to 4.76) | 0.82 |
| Adenocarcinoma | 0.54 | (0.18 to 1.57) | 0.26 | 0.00 | (0.00 to 0.62) | 0.76 | 0.61 | (0.19 to 1.95) | 0.41 |
| Squamous cell carcinoma | 0.64 | (0.19 to 2.17) | 0.47 | 0.08 | (0.00 to 0.90) | 0.34 | 0.64 | (0.17 to 2.44) | 0.52 |
| Other/unknown subtypes | 2.22 | (0.86 to 5.75) | 0.10 | 0.00 | (0.00 to 0.72) | 0.69 | 2.69 | (0.94 to 7.71) | 0.07 |
25(OH)D: 25-hydroxyvitamin D; CI: confidence interval; HR: hazard ratio; IVW: inverse-variance weighted; MR: Mendelian randomization; SCLC: small cell lung cancer
Figure 1.
Effect of single-nucleotide polymorphisms (SNPs)-determined serum 25(OH)D on the risk of lung cancer overall and histologic types in the HUNT2 study (n=54580). The ln(hazard ratio) for risk of outcome of each SNP (Y-axis) was plotted against each SNP’s effect on increase of serum 25(OH)D (nmol/L) (X-axis). Each trend line, derived from the inverse-variance weighted method and set through the origin of the axes arbitrarily, represents ln(HR) for the risk of lung cancer or histologic type per nmol/L increase in genetically determined serum 25(OH)D. Vertical and horizontal lines around points show 95% confidence intervals for each SNP. Outcomes: A) Lung cancer overall; B) Small cell lung cancer: C) Andenocarcinoma; D) Squamous cell carcinoma; E) Other/unknown subtypes.
As shown in Table 4, I2 and Cochran’s Q statistic showed no evidence for heterogeneity between the SNPs (I2 0.00, 95% CI 0.00 to 0.24, P value was 0.87 for lung cancer overall). The P value of the intercept by MR-Egger method was 0.79 (intercept -0.03, 95% CIs -0.25 to 0.19) for lung cancer overall, suggesting no substantial pleiotropic effect of these SNPs (Table 5).
Table 5. MR-Egger pleiotropy test of associations between a 25 nmol/L increase in genetically determined season-standardized 25(OH)D and risk of lung cancer overall and histologic types in the HUNT2 study (n=54580).
| Outcome | MR-Egger method | ||
|---|---|---|---|
| Intercept (95% CI) | P value | ||
| Lung cancer overall | -0.03 | (-0.25 to 0.72) | 0.79 |
| SCLC | -0.27 | (-1.27 to 0.72) | 0.59 |
| Adenocarcinoma | -0.14 | (-0.55 to 0.27) | 0.50 |
| Squamous cell carcinoma | 0.36 | (-0.12 to 0.84) | 0.14 |
| Other/unknown subtypes | -0.09 | (-0.45 to 0.28) | 0.64 |
25(OH)D: 25-hydroxyvitamin D; CI: confidence interval; MR: Mendelian randomization; SCLC: small cell lung cancer
MR estimates of a 10% increase in genetically determined 25(OH)D level with risks of lung cancer and histologic types in a two-sample MR are presented in Supplementary tables 3-4 and Supplementary figure 1 as sensitivity analyses. All estimates in the two-sample MR were similar to those derived from the primary analyses.
Discussion
Main findings
In this MR analysis of a population-based prospective cohort study including 54580 subjects, we found no substantial evidence of a causal association of serum 25(OH)D level with the incidence of lung cancer overall, SCLC, adenocarcinoma or squamous cell carcinoma.
Comparison with other studies
The finding of the current MR study is inconsistent with the conclusion from two meta- analyses of observational studies [9, 10]. The results of the meta-analyses may be largely driven by the inclusion of a large cohort study showing an inverse association between 25(OH)D levels and incidence of lung cancer [30], whereas others showed no association [9, 10]. The current study is also inconsistent with results from our own observational study from the same cohort showing that lower 25(OH)D levels were associated with a lower risk of adenocarcinoma, particularly in obese individuals [31]. The present MR analysis conformed to our speculation that residual confounding by adiposity or adiposity related factors could have biased the observational results [31].
Few MR studies have explored the potential causal association between circulating vitamin D levels and lung cancer risk. Our findings are consistent with that of the study by Dimitrakopoulou et al. who used summarized data of a consortium (TRICL-ILCCO) including a large number of lung cancer cases (n=12537) [32]. Having assumed sufficient statistical power to detect moderate effects, they found no causal association between circulating vitamin D concentration and risk of lung cancer and certain histologic types (adenocarcinoma and squamous cell carcinoma) [32]. Although the conclusions are the same, our study differs from the study by Dimitrakopoulou et al. in study design. Our study investigated the association in a homogeneous population-based prospective cohort study with a long follow-up duration, while TRICL-ILCCO mainly consisted of case-control studies from different geographical areas and ethnicities [33]. Selection bias is more likely in case-control studies than in prospective cohort studies, and survivor bias in MR studies has recently been discussed as a methodological issue [34]. Nevertheless, we need to note that the causal effect estimates in MR studies generally reflect a life-time risk, regardless of the follow-up time.
Strengths and limitations
The current study is one of the first MR analyses using a long-term prospective population-based study to investigate the associations of serum 25(OH)D levels with the risk of lung cancer and histologic types. Information about diagnosis of lung cancer at the Cancer Registry of Norway is nearly complete and reasonably accurate [35]. However, there were many cases with unknown subtypes, resulting in limited statistical power in the analyses of histologic types.
Compared with observational studies, MR studies are not vulnerable to reverse causation and unmeasured confounding when the assumptions of MR studies are satisfied. We performed both one-sample MR in which we have measured serum 25(OH)D levels in a reasonably large subcohort and two-sample MR as a sensitivity analysis. Even though two- sample MR is getting common with the access to MR Base [36], one-sample MR still has its advantages, such as testing the important assumptions of MR directly. F-statistics and R2 values from the regression of SNPs/allele score on 25(OH)D levels indicated sufficient strength of the instrumental variables of the exposure in the current study. The variation of 25(OH)D explained by the 3 SNPs used in the present study was larger than that explained by 4 SNPs in the study by Vimaleswaran et al (3.4% vs. 1.9%) [24]. We could also investigate the associations between SNPs or the allele score with a broad range of measured and reported characteristics at baseline. Even though the instruments may still be associated with unmeasured confounding, they were not associated with important confounders such as smoking and socio-economic status in HUNT2. The last important assumption in MR is that the instrument (SNPs or allele score) should be associated with the outcome of interest (lung cancer) only via the exposure (circulating vitamin D levels). We found no violation of this essential assumption according to MR-Egger tests, but fewer genetic instruments may have a relatively low power to detect horizontal pleiotropy [29].
This study had several potential limitations. Non-participation in HUNT2 was about 30% of the population. As participants in the HUNT studies were shown to be healthier than non-participants, our findings might differ to some degree from the true situation in the general Norwegian population [37]. The applied MR analysis was based on the assumption that the exposure-outcome relationship is linear and in dose-response [11]. We were not able to investigate the non-linear association in MR due to the lack of methods for binary outcomes [38], whereas many of the reported associations between circulating 25(OH)D levels and health outcomes were non-linear [31, 39]. The sample size of this study was likely insufficient to reveal a weak to moderate effect of vitamin D on lung cancer risk based on the wide confidence intervals of the MR estimates, but the sample size of our cohort seemed adequate to detect risk factors with large effects on lung cancer, such as smoking (Supplementary table 5). Besides, our results were consistent with the null findings of the aforementioned MR study that reported a sufficient study power of a case-control design [32]. Nevertheless, consortia consisting of data from European population-based prospective studies with long follow-up duration are warranted to further investigate the causality of vitamin D on lung cancer in MR analysis. MR studies are also called for in Asia and the Middle East where populations are reported to have lower vitamin D levels than populations from Europe [40]. In addition, results from ongoing large clinical trials [41, 42] are expected to clarify the causal association of vitamin D with cancer and other adverse outcomes, especially in individuals with low vitamin D status prior to intervention.
Conclusions
In summary, Mendelian randomization analysis indicated that serum 25(OH)D levels were not causally associated with the risk of lung cancer overall or histologic types in a population-based prospective cohort study.
Supplementary Material
Acknowledgments
The Nord-Trøndelag Health Study (HUNT) is a collaboration between the HUNT Research Centre (Faculty of Medicine and Health Sciences, NTNU, Norwegian University of Science and Technology), the Nord-Trøndelag County Council, and the Norwegian Institute of Public Health. The authors especially thank the HUNT Research Centre laboratory personnel for the measurement of serum 25(OH)D levels.
Funding
YQS and this work were supported by The Norwegian Cancer Society (project ID 5769155-2015) and The Research Council of Norway “Gaveforsterkning”. BMB was supported by a Research grant (#46055500-10) from The Liaison Committee for education, research and innovation in Central Norway. The K.G. Jebsen Center for Genetic Epidemiology is financed by Stiftelsen Kristian Gerhard Jebsen, Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology (NTNU) and Central Norway Regional Health Authority.
Footnotes
Disclaimer
The study has used data from the Cancer Registry of Norway. The interpretation and reporting of these data are the sole responsibility of the authors, and no endorsement by the Cancer Registry of Norway is intended nor should be inferred.
Contributors
YQS, YC and XMM contributed to the study design. XMM contributed to data collection. YQS, BMB, CB, SJL, SB and XMM contributed to statistical analyses of MR. YQS conducted statistical analyses, interpreted results and wrote the initial draft of the manuscript. BMB, CB, SJL, SB, FS, YC, TILN, PRR and XMM participated in the data interpretation and helped to write the final draft of the manuscript.
Competing interests
No
References
- 1.Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nat Rev Cancer. 2007;7(9):684–700. doi: 10.1038/nrc2196. [DOI] [PubMed] [Google Scholar]
- 2.Feldman D, Krishnan AV, Swami S, Giovannucci E, Feldman BJ. The role of vitamin D in reducing cancer risk and progression. Nat Rev Cancer. 2014;14(5):342–357. doi: 10.1038/nrc3691. [DOI] [PubMed] [Google Scholar]
- 3.Giovannucci E, Liu Y, Rimm EB, Hollis BW, Fuchs CS, Stampfer MJ, Willett WC. Prospective study of predictors of vitamin D status and cancer incidence and mortality in men. J Natl Cancer Inst. 2006;98(7):451–459. doi: 10.1093/jnci/djj101. [DOI] [PubMed] [Google Scholar]
- 4.Jacobs ET, Kohler LN, Kunihiro AG, Jurutka PW. Vitamin D and Colorectal, Breast, and Prostate Cancers: A Review of the Epidemiological Evidence. J Cancer. 2016;7(3):232–240. doi: 10.7150/jca.13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.GLOBOCAN. Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. 2012. Available from: http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx.
- 6.Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. Lyon, France: IARC Press; 2004. [Google Scholar]
- 7.Cheng TY, Cramb SM, Baade PD, Youlden DR, Nwogu C, Reid ME. The International Epidemiology of Lung Cancer: Latest Trends, Disparities, and Tumor Characteristics. J Thorac Oncol. 2016;11(10):1653–1671. doi: 10.1016/j.jtho.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee PN, Forey BA, Coombs KJ. Systematic review with meta-analysis of the epidemiological evidence in the 1900s relating smoking to lung cancer. BMC Cancer. 2012;12:385. doi: 10.1186/1471-2407-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Dong Y, Lu C, Wang Y, Peng L, Jiang M, Tang Y, Zhao Q. Meta-analysis of the correlation between vitamin D and lung cancer risk and outcomes. Oncotarget. 2017;8(46):81040–81051. doi: 10.18632/oncotarget.18766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Wang S, Che X, Li X. Vitamin D and lung cancer risk: a comprehensive review and meta-analysis. Cell Physiol Biochem. 2015;36(1):299–305. doi: 10.1159/000374072. [DOI] [PubMed] [Google Scholar]
- 11.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27(8):1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 12.Lawlor DA. Commentary: Two-sample Mendelian randomization: opportunities and challenges. International journal of epidemiology. 2016;45(3):908–915. doi: 10.1093/ije/dyw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Registry of Norway. Available from: https://www.kreftregisteret.no/
- 14.International classification of diseases for oncology (ICD-O) 3rd. World Health Organization; 2013. [Google Scholar]
- 15.Holick MF. Vitamin D deficiency. N Engl J Med. 2007;357(3):266–281. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- 16.Zerwekh JE. Blood biomarkers of vitamin D status. Am J Clin Nutr. 2008;87(4):1087s–1091s. doi: 10.1093/ajcn/87.4.1087S. [DOI] [PubMed] [Google Scholar]
- 17.Degerud E, Hoff R, Nygard O, Strand E, Nilsen DW, Nordrehaug JE, Midttun O, Ueland PM, de Vogel S, Dierkes J. Cosinor modelling of seasonal variation in 25-hydroxyvitamin D concentrations in cardiovascular patients in Norway. Eur J Clin Nutr. 2016;70(4):517–522. doi: 10.1038/ejcn.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira MA, Vonk JM, Baurecht H, Marenholz I, Tian C, Hoffman JD, Helmer Q, Tillander A, Ullemar V, van Dongen J, Lu Y, et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet. 2017 doi: 10.1038/ng.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou W, Fritsche LG, Das S, Zhang H, Nielsen JB, Holmen OL, Chen J, Lin M, Elvestad MB, Hveem K, Abecasis GR, et al. Improving power of association tests using multiple sets of imputed genotypes from distributed reference panels. Genet Epidemiol. 2017;41(8):744–755. doi: 10.1002/gepi.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, Gallicchio L, Jacobs EJ, Ascherio A, Helzlsouer K, Jacobs KB, Li Q, et al. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. 2010;19(13):2739–2745. doi: 10.1093/hmg/ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, Kiel DP, Streeten EA, Ohlsson C, Koller DL, Peltonen L, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376(9736):180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vimaleswaran KS, Berry DJ, Lu C, Tikkanen E, Pilz S, Hiraki LT, Cooper JD, Dastani Z, Li R, Houston DK, Wood AR, et al. Causal relationship between obesity and vitamin D status: bi-directional Mendelian randomization analysis of multiple cohorts. PLoS Med. 2013;10(2):e1001383. doi: 10.1371/journal.pmed.1001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. International journal of epidemiology. 2013;42(4):1134–1144. doi: 10.1093/ije/dyt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. International journal of epidemiology. 2017 doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. International journal of epidemiology. 2015;44(2):512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Afzal S, Bojesen SE, Nordestgaard BG. Low plasma 25-hydroxyvitamin D and risk of tobacco-related cancer. Clin Chem. 2013;59(5):771–780. doi: 10.1373/clinchem.2012.201939. [DOI] [PubMed] [Google Scholar]
- 31.Sun YQ, Langhammer A, Wu C, Skorpen F, Chen Y, Nilsen TIL, Romundstad PR, Mai XM. Associations of serum 25-hydroxyvitamin D level with incidence of lung cancer and histologic types in Norwegian adults: a case-cohort analysis of the HUNT study. Eur J Epidemiol. 2017 doi: 10.1007/s10654-017-0324-1. [DOI] [PubMed] [Google Scholar]
- 32.Dimitrakopoulou VI, Tsilidis KK, Haycock PC, Dimou NL, Al-Dabhani K, Martin RM, Lewis SJ, Gunter MJ, Mondul A, Shui IM, Theodoratou E, et al. Circulating vitamin D concentration and risk of seven cancers: Mendelian randomisation study. BMJ (Clinical researched) 2017;359:j4761. doi: 10.1136/bmj.j4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The International Lung Cancer Consortium (ILCCO) Availablefrom: http://ilcco.iarc.fr/
- 34.Vansteelandt S, Dukes O, Martinussen T. Survivor bias in Mendelian randomization analysis. Biostatistics. 2017 doi: 10.1093/biostatistics/kxx050. [DOI] [PubMed] [Google Scholar]
- 35.Larsen IK, Smastuen M, Johannesen TB, Langmark F, Parkin DM, Bray F, Moller B. Data quality at the Cancer Registry of Norway: an overview of comparability, completeness, validity and timeliness. Eur J Cancer. 2009;45(7):1218–1231. doi: 10.1016/j.ejca.2008.10.037. [DOI] [PubMed] [Google Scholar]
- 36.Hemani G, Zheng J, Wade KH, Laurin C, Elsworth B, Burgess S, Bowden J, Langdon R, Tan V, Yarmolinsky J, Shihab HA, et al. MR-Base: a platform for systematic causal inference across the phenome using billions of genetic associations. [cited 2017];bioRxiv beta, the preprint server for biology. 2017 Available from: https://www.biorxiv.org/content/early/2016/12/16/078972. [Google Scholar]
- 37.Langhammer A, Krokstad S, Romundstad P, Heggland J, Holmen J. The HUNT study: participation is associated with survival and depends on socioeconomic status, diseases and symptoms. BMC Med Res Methodol. 2012;12:143. doi: 10.1186/1471-2288-12-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staley JR, Burgess S. Semiparametric methods for estimation of a nonlinear exposure-outcome relationship using instrumental variables with application to Mendelian randomization. Genet Epidemiol. 2017;41(4):341–352. doi: 10.1002/gepi.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun YQ, Langhammer A, Skorpen F, Chen Y, Mai XM. Serum 25-hydroxyvitamin D level, chronic diseases and all-cause mortality in a population-based prospective cohort: the HUNT Study, Norway. BMJ Open. 2017;7(6):e017256. doi: 10.1136/bmjopen-2017-017256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Schoor NM, Lips P. Worldwide vitamin D status. Best Pract Res Clin Endocrinol Metab. 2011;25(4):671–680. doi: 10.1016/j.beem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 41.Kupferschmidt K. Uncertain verdict as vitamin D goes on trial. Science. 2012;337(6101):1476–1478. doi: 10.1126/science.337.6101.1476. [DOI] [PubMed] [Google Scholar]
- 42.Pilz S, Rutters F, Dekker JM. Disease prevention: vitamin D trials. Science. 2012;338(6109):883. doi: 10.1126/science.338.6109.883-c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.