Abstract
Background
Obesity and type 2 diabetes (T2D) are correlated risk factors for chronic kidney disease (CKD).
Methods
Using summary data from GIANT, DIAGRAM and CKDGen, we examined causality and directionality of the association between obesity and kidney function. Bi-directional two-sample Mendelian randomization (MR) estimated the total causal effects of body mass index (BMI) and waist-to-hip ratio (WHR) on kidney function, and vice versa. Effects of adverse obesity and T2D were examined by stratifying BMI variants by their association with WHR and T2D. Multi-variable MR estimated the direct causal effects of BMI and WHR on kidney function. The inverse variance weighted random-effects MR for Europeans was the main analysis, accompanied by several sensitivity MR analyses.
Results
One standard deviation (SD≈4.8 kg/m2) genetically higher BMI was associated with decreased estimated glomerular filtration rate (β=-0.032 [95% confidence intervals:-0.036,-0.027] log[eGFR], P=1x10-43), increased blood urea nitrogen (β=0.010 [0.005,0.015] log[BUN], P=3x10-6), increased urinary albumin-to-creatinine ratio (β=0.199 [0.067,0.332] log[UACR], P=0.003) in individuals with diabetes, and increased risk of microalbuminuria (OR=1.15 [1.04-1.28], P=0.009) and CKD (1.13 [1.07-1.19], P= 3x10-6). Corresponding estimates for WHR and for trans-ethnic populations were overall similar. The associations were driven by adverse obesity, and for microalbuminuria additionally by T2D. While genetically high BMI, unlike WHR, was directly associated with eGFR, BUN and CKD, the pathway to albuminuria was likely through T2D. Genetically predicted kidney function was not associated with BMI or WHR.
Conclusions
Genetically high BMI is associated with impaired kidney function, driven by adverse obesity, and for albuminuria additionally by T2D.
Keywords: Mendelian Randomization Analysis, Body Mass Index, Waist-Hip Ratio, Obesity, Diabetes Mellitus, Type 2, Kidney Function Tests, Glomerular Filtration Rate, Blood Urea Nitrogen, Albuminuria, Renal Insufficiency, Chronic
Introduction
Numerous observational studies have linked high body mass index (BMI) with impaired kidney function (1–7). This association is of immense importance since the prevalences of both obesity and kidney disease are very high and increasing globally (8, 9), while obesity is potentially reversible and preventable.
Chronic kidney disease (CKD) is defined as estimated glomerular filtration rate (eGFR) below 60 ml/min/1.73 m2 and/or kidney damage, often ascertained as albuminuria, i.e., increased albumin excretion in urine (10). A recent meta-analysis in more than 5 million individuals showed that BMI was independently associated with eGFR decline (1). Interestingly, in this meta-analysis, increasing BMI was a stronger risk factor for eGFR decline in the general population than in individuals with preexisting CKD and diabetes. In contrast, in a Norwegian general population study of 1261 middle aged individuals without diabetes, cardiovascular, or kidney disease, neither BMI nor waist-to-hip ratio (WHR) was associated with rapid annual eGFR decline (11). Previous studies in individuals with type 2 diabetes (T2D) and obesity have shown that compared to usual care, bariatric surgery was associated with lower incidence of CKD, suggesting that weight loss may have beneficial effects on kidney function (12, 13). However, the beneficial effects may be limited to short-term effects in individuals with T2D and obesity, and possibly mediated through either weight loss and/or improvement of T2D.
Thus, whether the association between obesity and kidney function is causal, i.e., whether weight loss directly can improve or delay the decline of kidney function in the general population is presently uncertain.
While observational studies are prone to reverse causation and confounding, using the Mendelian randomization (MR) approach circumvents both and allows for causal inference. This approach uses the genetic variants (typically single nucleotide polymorphisms [SNPs]) associated with life-long exposure of interest (e.g., BMI), to assess the magnitude of the effect of genetically predicted exposure on outcome (e.g., kidney function). Since genotype is established at conception (Mendel’s law of segregation), the possibility of reverse causation is minimized. Likewise, potential confounders are evenly distributed across genotype (Mendel’s law of independent assortment), thus mimicking the random distribution of confounders by randomization in randomized controlled trials. Recent MR studies have supported a causal role for BMI and waist circumference in kidney function (14–16). Hence, the purpose of our study was to expand on current evidence by performing a bi-directional two-sample MR on the association between obesity measures (BMI, WHR and WHR adjusted for BMI [WHRadjBMI]) and all outcomes related to kidney function and damage that were available (eGFR, blood urea nitrogen [BUN], annual eGFR decline, urinary albumin-to-creatinine ratio [UACR], microalbuminuria and CKD) by increasing statistical power via inclusion of larger population samples and additional genetic instruments. Compared to previous MR studies, the novelty of our study includes: 1) an increased number of outcomes related to kidney function and damage, 2) increased sample size for eGFR and BUN, 3) ten-fold increased number of SNPs for BMI, 4) examination of the effect of favorable and adverse obesity as well as unexpected fat distribution (i.e., WHRadjBMI) on kidney function outcomes, 5) testing the directionality by MR Steiger and Steiger filtering (see Methods), and by using eGFR and UACR as exposures for obesity outcomes, and 6) conducting multivariable MR (MVMR) with BMI, WHR and T2D as exposures for outcomes related to kidney function and damage. For simplicity, these outcomes are onwards termed “kidney function outcomes”.
Methods
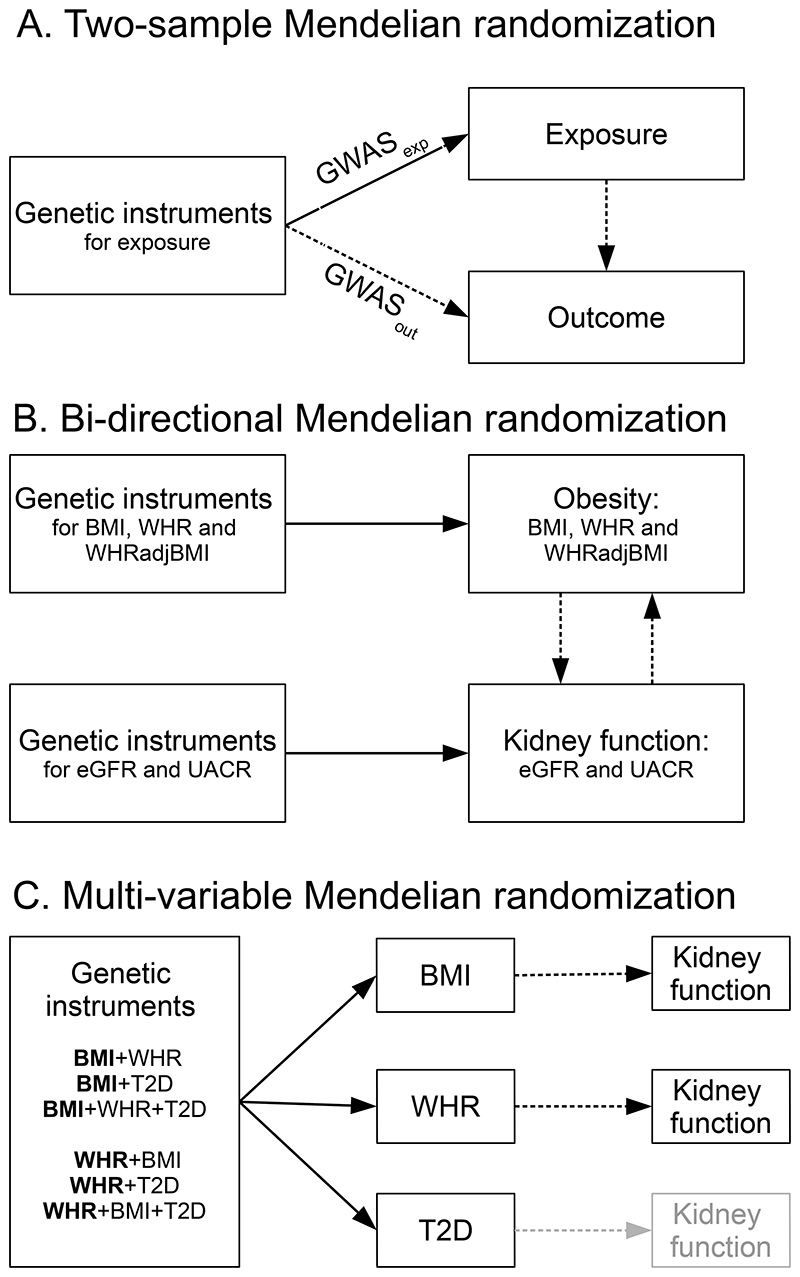
Study design is illustrated in Figure 1 and described in detail in the online Supplemental Methods.
Figure 1. Schematic diagrams illustrating the study design.
All genetic instruments were single nucleotide polymorphisms (SNPs).
A) The two-sample Mendelian randomization (MR) approach uses summary data from two different population samples, provided by large genome-wide association studies (GWASs) consortia to assess causality between the exposure and the outcome. Summary data (β coefficients and standard errors) for the SNP-exposure (GWASexp) and SNP-outcome (GWASout) associations for this study were from the GIANT Consortium for obesity (body mass index [BMI], waist-to-hip ratio [WHR] and WHR adjusted for BMI [WHRadjBMI]) and CKDGen for kidney function (estimated glomerular filtration rate [eGFR], blood urea nitrogen, annual eGFR decline, urinary albumin-to-creatinine ratio [UACR], microalbuminuria and chronic kidney disease). Supplemental Tables 1 shows an overview of the studies included.
B) The bi-directional MR approach uses genetic instruments for both exposure and outcome to evaluate whether the “exposure” causes the “outcome” or vice versa. We identified genetic instruments for obesity (BMI, WHR and WHRadjBMI) and kidney function (estimated glomerular filtration rate [eGFR] and urinary albumin-to-creatinine ratio [UACR]). Supplemental Tables 2 shows the identification of genetic instruments.
C) While MR estimates the total causal effects using a single exposure at a time, the multi-variable MR estimates the direct causal effects of each exposure using multiple exposures simultaneously. In this study, we examined the direct causal effects of BMI and WHR on each kidney function outcome, while accounting for each other and/or type 2 diabetes (T2D). Hence the bold font denotes the three models for BMI and WHR each (Supplemental Tables 11), whereas the direct effects of T2D were not examined (shown in light grey). Summary data for T2D were from the DIAGRAM consortium.
Exposures: Identification of genetic instruments
Details on identification of SNPs used as genetic instruments are shown in online Supplemental Table 2. We identified genetic instruments from published studies reporting top independent SNPs in main tables, supplementary tables or links to online repositories (Supplemental Methods).
In populations of European ancestry (EA), we identified 941 SNPs for BMI (17), 316 SNPs for WHR (18), and 346 SNPs for WHRadjBMI (18) from the GIANT and UK Biobank combined, and 77 SNPs for BMI from the GIANT alone (BMIGIANT SNPs, supplementary analyses) (19). Since some SNPs for WHRadjBMI exhibited sexual dimorphisms, i.e., had significantly different effects between the two sexes (18), we excluded the dimorphic SNPs from the main analyses (but included in the online Supplemental Methods).
From the CKDGen and UK Biobank combined, we identified 173 SNPs associated with eGFRcrea, and additionally validated by being associated with eGFRcysC and inversely associated with BUN, hence named eGFRval SNPs (20). From the CKDGen Consortium alone, we identified 126 SNPs for eGFRcrea (validated by being inversely associated with BUN) (21) and 61 SNPs for UACR (online Supplemental Table 1) (22).
For MVMR analyses, in addition to 941 SNPs for BMI and 316 SNPs for WHR, we also identified 246 SNPs associated with T2D (23).
Outcomes
In EA populations, kidney function outcomes were: eGFRcysC, eGFRcreaCKD-EPI, BUN, annual eGFRcreaMDRD decline, UACR, microalbuminuria and CKD. Furthermore, annual eGFRcreaMDRD decline was stratified by CKD (yes/no), and UACR by diabetes (yes/no). In supplementary analyses, we examined eGFRcreaMDRD stratified by diabetes. Obesity outcomes were BMI, WHR and WHRadjBMI.
Statistical analysis
Analyses were performed in R version 4.0.4 using TwoSampleMR and MendelianRandomization packages.
We excluded SNPs in linkage disequilibrium within a 10 Mb window and R2 cut-off at 0.01 (PLINK clumping method included in the TwoSampleMR package).
For each SNP, the effect allele was defined as the allele associated with increased level of the relevant exposure (BMI, WHR, WHRadjBMI, eGFRval, eGFRcrea, and UACR). For all outcomes, we extracted summary data for the relevant exposure SNPs, and aligned the effect to the effect allele (data harmonization). This excluded inconsistent and palindromic SNPs with effect allele frequencies close to 50%.
The main overall causal estimates were assessed by the inverse variance weighting multiplicative random effects (IVW-RE) meta-analysis applied across the individual instrumental estimates and their standard errors (24). We used the random-effects rather than fixed-effects meta-analysis because it accounts for the heterogeneity of the individual causal estimates (assessed by the I2 index, range: 0%-100%, increases with increasing heterogeneity) (25). Because the inverse variance weighting method requires that all SNPs are valid genetic instruments, we also performed sensitivity analyses with different assumptions: MR-PRESSO (MR Pleiotropy RESidual Sum and Outlier), weighted median (WM) and MR-Egger regression analyses. MR-PRESSO detects and corrects for horizontal pleiotropy by excluding “outliers”, thus narrowing the confidence intervals. WM provides reliable estimates if at least 50% of the weight comes from valid genetic instruments (26), and MR-Egger in cases where even fewer than 50% of the genetic variants are valid instruments, thus broadening the confidence intervals (27). The directional pleiotropy was tested by the MR-Egger intercept test and “NO Measurement Error” (NOME) violation was quantified by the I2GX (range: 0%-100%, decreases with increasing NOME violation) (27). The causal direction was assessed by MR Steiger as true if the exposure likely causes the outcome, or false if the exposure unlikely causes the outcome. Additional MR analyses were performed after Steiger filtering, i.e. after excluding SNPs that explained more of the variation in outcomes than the exposures (28).
A Bonferroni correction was used to control for false positive findings due to multiple testing, and with four exposures (BMI, WHR, eGFR and UACR) and up to 14 outcomes, a P<0.003 (0.05/18) was considered significant.
To reduce the risk of population stratification, we restricted the main analyses to EA populations.
In order to investigate the potential causal effects of adverse and favorable obesity, we stratified SNPs associated with BMI (by their association with WHR) into: adverse obesity (when BMI increasing alleles were nominally associated [P<0.05] with increased WHR, 78% of all BMI SNPs), favorable obesity (when BMI increasing alleles were nominally associated with decreased WHR, 3% of all SNPs), and WHR indifferent obesity (when BMI increasing alleles were not associated [p≥0.05] with WHR, 19% of all SNPs).
We employed the same approach to examine whether the potential causal effects of BMI on kidney function were driven by SNPs associated with T2D. BMI SNPs were stratified into: T2D concordant (when BMI increasing alleles were nominally associated with increased risk of T2D, 49% of all SNPs), T2D discordant (when BMI increasing alleles were nominally associated with decreased risk of T2D, 5% of all SNPs), and T2D indifferent (when BMI increasing alleles were not associated with risk of T2D, 46% of all SNPs).
Finally, since MR estimates the total causal effects, we additionally performed multi-variable MR (MVMR) in order to assess the direct causal effects of BMI and WHR (29). We performed three MVMR analyses for both exposures, taking each other and/or T2D into account (for BMI: +WHR, +T2D and +WHR+T2D; for WHR: +BMI, +T2D and +BMI+T2D, Figure 1C).
Results
Total causal effects of obesity on eGFR and BUN
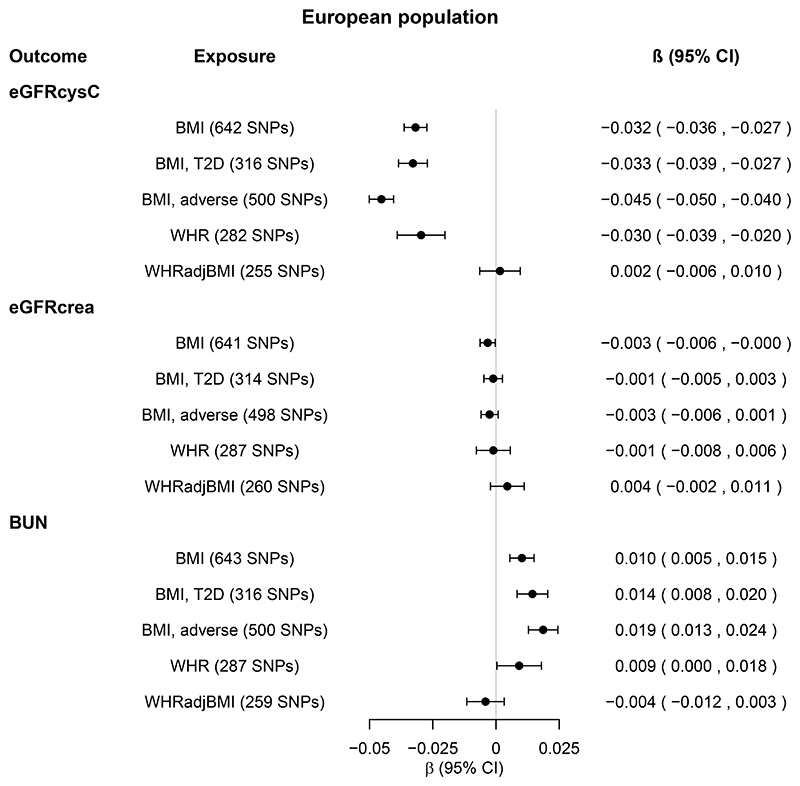
In EA populations, one standard deviation (SD≈4.8 kg/m2) of genetically higher BMI was associated with 0.032 log[eGFRcysC] decrease (Figure 2, β=−0.032 [95% confidence intervals: -0.036, -0.027], P=1x10-43) and 0.010 log[BUN] increase (Figure 2, β= 0.010 [0.005, 0.015], P=3x10-5). The effect was driven by SNPs associated with adverse obesity, and for BUN additionally by SNPs associated with T2D (Figure 2, online Supplemental Tables 3-6). These results were supported by supplementary analyses, including sensitivity MR, MR Steiger, Steiger filtering and trans-ethnic analyses (online Supplemental Tables 3-6, online Supplemental Figure 1).
Figure 2. Total causal effects of obesity on eGFR and BUN in the European population.
Estimates (β coefficients and 95% confidence intervals [CIs]) are from the inverse variance weighted random-effects Mendelian randomization analysis, and expressed in log units per standard deviation increase in the relevant exposure.
Obesity exposures from the GIANT Consortium were BMI (body mass index, N=795,624), WHR (waist-to-hip ratio, N=697,702), and WHRadjBMI (WHR adjusted for BMI, N=694,469). For each obesity exposure, the number of single nucleotide polymorphisms (SNPs) included in the analysis is shown in parenthesis.
BMI, T2D and BMI, adverse (obesity) refer to BMI SNPs restricted to SNPs nominally associated with type 2 diabetes (T2D) and WHR, respectively. For details, see Methods. Kidney function outcomes from the CKDGen Consortium and UK Biobank combined were eGFRcysC (estimated glomerular filtration rate [eGFR] based on serum cystatin C, N=460,826), eGFRcrea (eGFR based on serum creatinine using the CKD-EPI equation, N=1,004,040) and BUN (blood urea nitrogen, N=679,531). Sensitivity MR analyses are shown in Supplemental Tables 3-4.
In contrast, genetically high BMI was associated with neither eGFRcreaCKD-EPI (Figure 2) nor eGFRcreaMDRD (online Supplemental Table 3).
While the estimates for genetically high WHR were similar to those for BMI, genetically high WHRadjBMI was associated with neither eGFR nor BUN (Figure 2 and online Supplemental Tables 3-6).
Total causal effects of obesity on annual eGFR decline
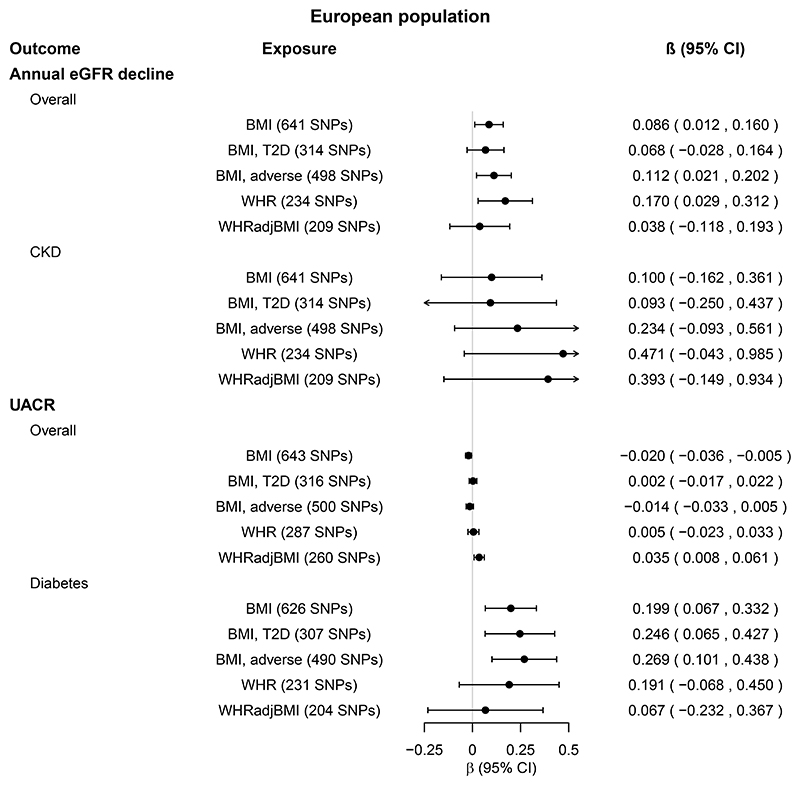
Genetically high BMI, WHR and WHRadjBMI were not associated with annual eGFRcreaMDRD decline after Bonferroni correction (Figure 3, online Supplemental Tables 3-4). However, genetically high BMI and WHR were nominally associated with increased annual eGFRcreaMDRD decline in the overall population (online Supplemental Tables 3-4). The effect was driven by SNPs associated with adverse obesity (Figure 3, online Supplemental Tables 3-4). The estimates were larger in the CKD population, but only nominally significant in BMIGIANT SNPs in IVW-RE and MR-PRESSO, but not other sensitivity MR analyses nor MR Steiger (online Supplemental Tables 3-4).
Figure 3. Total causal effects of obesity on eGFR decline and UACR in the European population.
Estimates (β coefficients and 95% confidence intervals [CIs]) are from the inverse variance weighted random-effects Mendelian randomization analysis, and expressed in log units per standard deviation increase in the relevant exposure.
Obesity exposures from the GIANT Consortium were BMI (body mass index, N=795,624), WHR (waist-to-hip ratio, N=697,702), and WHRadjBMI (WHR adjusted for BMI, N=694,469). For each obesity exposure, the number of single nucleotide polymorphisms (SNPs) included in the analysis is shown in parenthesis.
BMI, T2D and BMI, adverse (obesity) refer to BMI SNPs restricted to SNPs nominally associated with type 2 diabetes (T2D) and WHR, respectively. For details, see Methods. Kidney function outcomes from the CKDGen Consortium were annual eGFR (estimated glomerular filtration rate) decline (based on serum creatinine levels and calculated by the MDRD equation), available in overall population and subpopulation with chronic kidney disease (CKD, NCKDcases=3338 cases, NCKDcontrols=39,653, Noverall=43,008), and UACR (urinary albumin-to-creatinine ratio), available in overall population and subpopulation with diabetes (NDM=11,529, Noverall=118,460). Sensitivity MR analyses are shown in Supplemental Tables 3-4.
We were unable to examine annual eGFR decline in trans-ethnic populations due to lack of GWAS on this outcome in trans-ethnic populations.
Total causal effects of obesity on UACR
In EA populations, one SD of genetically higher BMI was associated with 0.199 log[UACR] increase in individuals with diabetes (β =0.199 [0.067,0.332], P=0.003, Figure 3, online Supplemental Tables 3-4), but not in the overall population. This was supported by BMIGIANT SNPs, MR-PRESSO and trans-ethnic, but not other sensitivity MR analyses. (online Supplemental Figure 2, online Supplemental Tables 3-6). The effect was driven by SNPs associated with adverse obesity, as well as T2D (online Supplemental Tables 34). MR Steiger results were contradictory: false direction (exposure unlikely causes the outcome) in the EA and true direction (exposure likely causes the outcome) in the trans-ethnic populations (online Supplemental Tables 3-6). In contrast, genetically high BMI and WHRadjBMI were nominally associated with decreased and increased UACR in the overall EA, but not trans-ethnic populations (P=0.01, Figure 3, online Supplemental Figure 2).
Genetically high WHR and WHRadjBMI were not robustly associated with UACR in individuals with diabetes, nor in the overall population (Figure 3, online Supplemental Figure 2, online Supplemental Tables 3-6).
Total causal effects of obesity on microalbuminuria and CKD
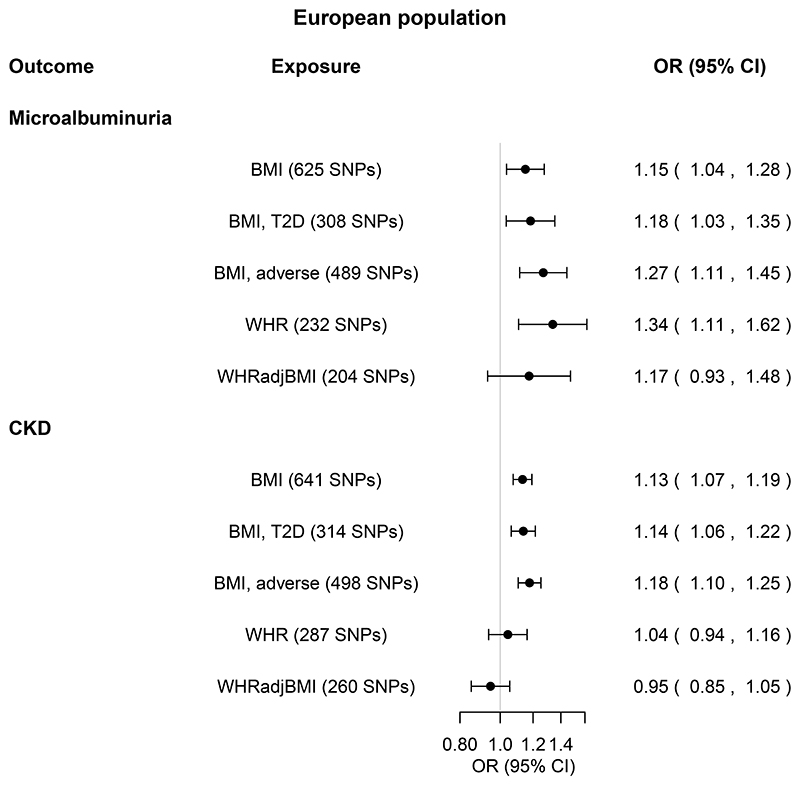
In EA populations, one SD of genetically higher BMI was associated with an odds ratio of 1.15 (1.04-1.28, P=0.01) for microalbuminuria and 1.13 (1.07-1.19, P=3x10-6) for CKD (Figure 4, online Supplemental Tables 3-4). These results were supported by MR-PRESSO and MR Steiger, but not by other MR sensitivity analyses nor Steiger filtering (Figure 4, online Supplemental Tables 3-4). The effect was driven by SNPs associated with adverse obesity, and for microalbuminuria additionally by SNPs associated with T2D (Figure 4, online Supplemental Tables 3-4).
Figure 4. Total causal effects of obesity on microalbuminuria and CKD in the European population.
Estimates are from the inverse variance weighted random-effects Mendelian randomization analysis, and expressed as odds ratios (ORs) and 95% confidence intervals (CIs).
Obesity exposures from the GIANT Consortium were BMI (body mass index, N=795,624), WHR (waist-to-hip ratio, N=697,702), and WHRadjBMI (WHR adjusted for BMI, N=694,469). For each obesity exposure, the number of single nucleotide polymorphisms (SNPs) included in the analysis is shown in parenthesis.
BMI, T2D and BMI, adverse (obesity) refer to BMI SNPs restricted to SNPs nominally associated with type 2 diabetes (T2D) and WHR, respectively. For details, see Methods. Kidney function outcomes from the CKDGen Consortium were microalbuminuria (defined as urinary albumin-to-creatinine ratio above 25 mg/g in women and 17 mg/g in men, Ncases=5996, Ncontrols=48,140), and CKD (chronic kidney disease, defined as estimated glomerular filtration rate below 60 ml/min/1.73 m2, NCKDcases=41,395 cases,
Ncontrols=439,303 controls). Sensitivity MR analyses are shown in Supplemental Tables 34.
There was no robust association between genetically high WHR and WHRadjBMI and microalbuminuria and CKD (Figure 4, online Supplemental Tables 3-6, online Supplemental Figure 3).
Total causal effects of eGFR and UACR on obesity
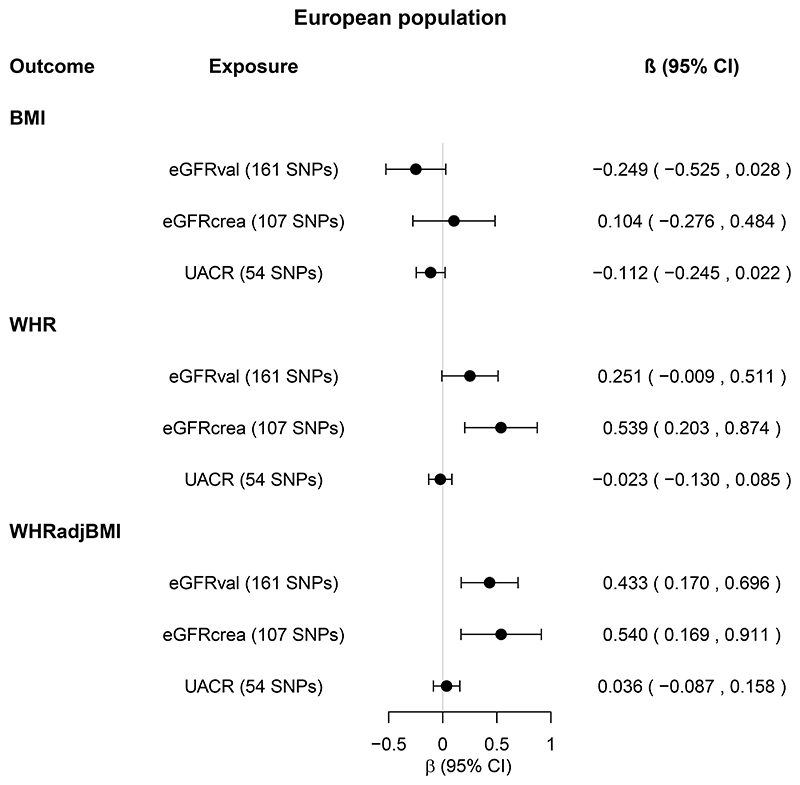
Genetically high eGFR and UACR were not associated with BMI and WHR (Figure 5, online Supplemental Figure 4, online Supplemental Tables 7-10). After Bonferroni correction, only genetically high eGFRval was associated with WHRadjBMI (Figure 5, online Supplemental Figure 4, online Supplemental Tables 7-10). This association was supported by MR-PRESSO and MR Steiger, but not other sensitivity MR analyses. Steiger filtering reduced the number of SNPs by >50%, and somewhat attenuated the association (online Supplemental Tables 7-8).
Figure 5. Total causal effects of kidney function on obesity in the European population.
Estimates (β coefficients and 95% confidence intervals [CIs]) are from the inverse variance weighted random-effects Mendelian randomization analysis, and expressed in log units per standard deviation increase in the relevant exposure.
Kidney function exposures from the CKDGen Consortium were eGFRval (estimated glomerular filtration rate based on creatinine and calculated by CKD-EPI equation, and validated by being associated with cystatin C and inversely associated with blood urea nitrogen [BUN]), eGFRcrea (eGFR based on creatinine and calculated by CKD-EPI equation, and validated by being inversely associated with BUN, N=567,460-1,004,040) and UACR (urinary albumin-to-creatinine ratio, N=547,361). For each kidney function exposure, the number of single nucleotide polymorphisms (SNPs) included in the analysis is shown in parenthesis.
Obesity outcomes from the GIANT Consortium and UK Biobank combined were BMI (body mass index, N=795,624), WHR (waist-to-hip ratio, N=697,702), and WHRadjBMI (WHR adjusted for BMI, N=694,469). Sensitivity MR analyses are shown in Supplemental Tables 7-8.
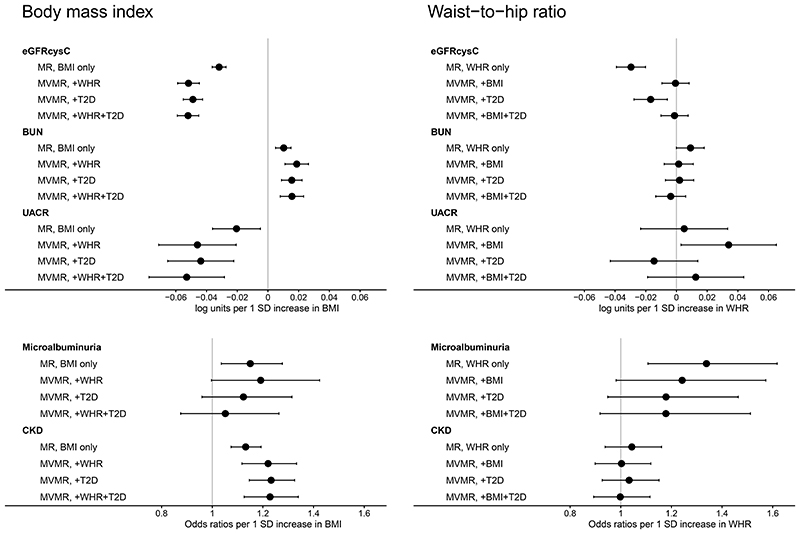
Multivariable MR (MVMR): Direct causal effects of obesity on kidney function
Genetically high BMI was associated with decreased eGFRcysC, increased BUN and increased risk of CKD when WHR and/or T2D were accounted for (Figure 6, online Supplemental Tables 11). In fact, the direct causal effects were larger than the total causal effects. Meanwhile, genetically high WHR was not associated with eGFRcysC, BUN nor CKD when BMI was accounted for (Figure 6, online Supplemental Tables 11).
Figure 6. Direct causal effects of obesity on kidney function in the European population.
Estimates (and corresponding 95% confidence intervals) are from the inverse variance weighted random-effects Mendelian randomization (MR) analyses, and expressed in log units per standard deviation (SD) increase in exposure for continuous outcomes, and odds ratios for binary outcomes.
MR approach estimated the total causal effects of body mass index (BMI) and waist-to-hip ratio (WHR) on kidney function outcomes (shown in Supplemental Table 3).
Multivariable MR (MVMR) based on the inverse-variance weighted method estimated the direct causal effects of BMI and WHR on each kidney function outcome, while accounting for each other and/or type 2 diabetes (T2D).
eGFRcysC: estimated glomerular filtration rate based on serum cystatin C. BUN: blood urea nitrogen. UACR: urinary albumin-to-creatinine ratio. CKD: chronic kidney disease.
Genetically high BMI was directly associated with decreased UACR, when WHR and/or T2D were accounted for (Figure 6, Supplemental Table 11). Meanwhile, genetically high WHR was directly associated with increased UACR when we accounted for BMI (in agreement with MR estimates for WHRadjBMI, Figure 3), but not when we accounted for T2D (Figure 6, online Supplemental Tables 11).
Genetically high BMI and WHR were both associated with increased risk of microalbuminuria. Although the direct causal estimates were imprecise (large confidence intervals), both the total and the direct causal effects were larger for genetically high WHR than BMI (Figure 6, online Supplemental Tables 11).
Discussion
This large two-sample MR study demonstrated that genetically high BMI is associated with impaired kidney function, and not vice versa. Genetically predicted high BMI was associated with decreased eGFRcysC, increased BUN, increased UACR in individuals with diabetes, and increased risk of microalbuminuria and CKD. These associations were driven by adverse obesity, and for albuminuria also by T2D. While genetically high BMI, unlike WHR, was directly associated with eGFRcysC, BUN and CKD, the pathway to albuminuria was likely through T2D.
Interestingly, genetically high BMI was only associated with decreased eGFR based on serum cystatin C, but not serum creatinine measures. Current evidence supports that eGFRcysC may be a better biomarker for kidney function than eGFRcrea (20, 30). Indeed, estimates of GFR based on creatinine may overestimate kidney function in individuals with relatively low muscle mass, whereas cystatin C based estimates are unaffected, which may explain the discrepancy between the two methods, particularly in an elderly population (31).
In agreement with a previous MR study, we also found that genetically high BMI was associated with decreased eGFRcysC and increased BUN (14). In that study, Xu et al showed that a one SD increase in genetically predicted BMI (NSNPs=72, NGIANT=339,224) was associated with log[eGFRcysC] decrease of 0.038 in one-sample (NUKBB=303,373) and 0.047 in two-sample MR analysis (NCKDGen=33,152). Indeed, since we used summary statistics for eGFRcysC based on CKDGen and UK Biobank cohort combined (NCKDGen+UKBB=460,826), our estimates were very similar (β=-0.032) to the previous one-sample, and somewhat lower than the previous two-sample MR analysis. Likewise, our findings of 0.010 log[BUN] increase per one SD increase in genetically predicted BMI, were somewhat lower than previously reported by Xu et al: 0.020 for one-sample (NUKBB=314,731) and 0.032 for two-sample (NCKDGen=243,031) (14). This is likely explained by our two-to three-fold larger sample size, as we used summary statistics for BUN based on CKDGen and UK Biobank cohort combined (NCKDGen+UKBB=679,531) and a 10fold increased number of SNPs used as instruments (643 versus 74 SNPs). Indeed, when we performed replication using CKDGen alone, our estimates were 0.030 using 74 SNPs and 0.010 using 643 SNPs.
Since annual eGFR decline examined in this study was based on eGFRcrea and relatively small sample size (particularly for the CKD subpopulation), this study calls for re-evaluation with future larger meta-GWAS for annual eGFRcysC decline (stratified by CKD at baseline). Indeed, we cannot completely exclude that genetically high BMI and WHR may be associated with annual eGFR decline, particularly in the subpopulation with CKD. Accordingly, in a trans-ethnic US population of 2489 elderly (70-79 years old) individuals, observationally high BMI was associated with eGFRcysC decline (30% decrease in eGFRcysC during a median follow-up of 9 years) as well as incident CKD. (4) Meanwhile, in the large meta-analysis demonstrating an association between observationally high BMI and eGFR decline, eGFR decline was defined as eGFRcreaCKD-EPI decline>40%, eGFRcreaCKD-EPI <10 mL/minute/1.73 m2, or initiation of kidney replacement therapy, i.e., a binary outcome comparable to severe CKD, but not annual eGFR decline (1).
In a study of more than 400,000 individuals from the UK Biobank, representing the general population, genetically high BMI and WHR adjusted for BMI were independently associated with increased albuminuria (7). In our study, genetically high BMI was associated with increased risk of microalbuminuria, and increased UACR in the population with diabetes only. Together with the fact that the association with microalbuminuria was driven by BMI SNPs associated with T2D, this suggests that the pathway from obesity to albuminuria may be mediated through T2D.
Previous MR studies have shown varying effect sizes for genetically high BMI on CKD with ORs between 1.16 and 1.78 (14–16). This is likely explained by the differences in numbers of SNPs used and sample sizes.
Interestingly, while we found no support for an association between genetically high WHR in CKD, a very recent study of 281,228 individuals form the UK Biobank found that one SD of genetically higher WHR was associated with an OR of 1.29 for CKD (16). Possible explanations for the discrepancy include differences in 1) MR methods (sex-combined two-sample MR on summary level data versus sex-specific genetic risk score on individual level data), 2) SNP independency criteria (R2<0.01 separated by 10 Mb versus R2<0.1 separated by 1 Mb, and thus slightly different number of SNPs: 287 versus 394), and 3) sample sizes (480,698 versus 281,228).
The directionality of the association between genetically high BMI and kidney function in this study agrees with previous MR studies (14, 15). Nevertheless, we cannot completely exclude the possibility that genetically high eGFR may be associated with increased WHR and WHRadjBMI. Because these associations were not confirmed by UACR (supplementing eGFR as an exposure for kidney function), they likely represent chance findings due to multiple testing.
Our finding that genetically high BMI was directly associated with decreased eGFR and increased risk of CKD is in line with previous MR findings showing that the effect of glycemic traits on CKD was weak, suggesting that T2D may have glucose-independent mechanisms (such as obesity, hypertension and dyslipidemia) to influence CKD (15). One major strength of this study is the consistent associations for obesity (genetically high BMI and WHR), and the consistent lack of associations for genetically unexpected fat distribution (WHRadjBMI) across many different kidney function outcomes. Our study was unlikely to suffer from weak instrument bias since we only included SNPs with sufficient instrument strength (F>10) associated with their respective outcomes at GWAS significance level (P<5x10−8), and excluded SNPs in linkage disequilibrium. We employed two different SNP sets for BMI (77 BMIGIANT and 941 BMIGIANT+UKBB in EA populations) and eGFR (173 eGFRval and 126 eGFRcrea in EA populations, and 188 eGFRval and 147 eGFRcrea in trans-ethnic populations), showing largely consistent results, thus further validating these SNP sets as genetic instruments for BMI and eGFR. Although the main analyses were restricted to EA, the results were largely replicated in trans-ethnic populations. However, as Europeans comprised the majority of trans-ethnic populations, there is a need to explore whether our findings apply to individuals of other ancestries as well. Another strength is the large sample sizes, particularly for eGFRcysC, eGFRcreaCKD-EPI and BUN, used for the first time in the present MR study. Although pleiotropy is impossible to rule out completely, we addressed it by 1) performing sensitivity MR analyses (MR-PRESSO, weighted median and MR Egger) with different assumptions regarding pleiotropy, and 2) stratifying BMI SNPs according to their association with WHR and T2D. One important limitation of our two-sample MR study based on summary data is inability to investigate a potentially nonlinear association between obesity (genetically high BMI and WHR) and kidney function. Another potential limitation is the substantial overlap of the participants in the GWASs included in the two-sample MR, which could have biased our estimates (32). However, any bias would likely be minimal (32), and a recent simulation study has shown that two-sample MR methods can be safely used for one-sample MR performed within large cohorts, as was the case in this study (33). Unfortunately, this does not apply to the MVMR approach, and having set the pairwise covariances between SNP-exposure associations to zero (assuming independent samples) even though the samples were largely overlapping may have biased our results. Nevertheless, this potential bias was unlikely substantial since our MVMR estimates were in agreement with the results from our stratification analyses (genetic instruments for BMI stratified by association with WHR and T2D).
In conclusion, this large two-sample MR study showed that genetically high BMI is associated with impaired kidney function, and not vice versa. The effect was particularly pronounced in adverse obesity (genetically high BMI and high WHR). While BMI was directly associated with eGFR, BUN and CKD, the pathway to microalbuminuria was likely through T2D. These findings expand the previously published MR reports.
Supplementary Material
Acknowledgements
The authors thank the GIANT, DIAGRAM and CKDGen consortia for providing the summary statistics data used in this study.
Grant support
KJS and TWW were supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Project-ID 387509280 – SFB 1350 (subproject C6, Principal Investigator: Iris M Heid).
ADK is funded by an unrestricted grant from Novo Nordisk.
Abbreviations
- BMI
body mass index
- BUN
blood urea nitrogen
- CI
confidence intervals
- CKD
chronic kidney disease
- CKD-EPI
Chronic Kidney Disease Epidemiology Collaboration
- CKDGen
CKD Genetics
- DIAGRAM
DIAbetes Genetics Replication And Meta-analysis
- EA
European ancestry
- eGFR
estimated glomerular filtration rate
- eGFRcrea
eGFR based on serum creatinine
- eGFRcysC
eGFR based on serum cystatin C
- GIANT
Genetic Investigation of ANthropometric Traits
- GWAS
genome-wide association study
- IVW-RE
inverse variance weighted random-effects
- MR
Mendelian randomization
- MR-PRESSO
MR Pleiotropy RESidual Sum and Outlier
- MVMR
Multi-variable MR
- NOME
NO Measurement Error
- OR
odds ratio
- SD
standard deviation
- SNP
single nucleotide polymorphism
- T2D
type 2 diabetes
- UACR
urinary albumin-to-creatinine ratio
- WHR
waist-to-hip ratio
- WHRadjBMI
WHR adjusted for BMI
- WM
weighted median
- β
beta-coefficient from a regression analysis
Footnotes
Human Genes
None described.
Disclosures
The authors declare no known competing financial interests or personal relationships that could have appeared to influence this study.
Contributor Information
Alisa D. Kjaergaard, Email: alisa.kjaergaard@auh.rm.dk, Steno Diabetes Center Aarhus, Aarhus University Hospital, Aarhus, Denmark.
Alexander Teumer, Email: ateumer@uni-greifswald.de, Institute for Community Medicine, University Medicine Greifswald, Greifswald, Germany, and DZHK (German Center for Cardiovascular Research), partner site Greifswald, Greifswald, Germany.
Daniel R. Witte, Email: daniel.witte@ph.au.dk, Steno Diabetes Center Aarhus, Aarhus University Hospital, Aarhus, Denmark, and Department of Public Health, Aarhus University, Aarhus, Denmark.
Kira-Julia Stanzick, Email: Kira-Julia.Stanzick@klinik.uni-regensburg.de, Department of Genetic Epidemiology, University of Regensburg, Regensburg, Germany.
Thomas W. Winkler, Email: Thomas.Winkler@klinik.uni-regensburg.de, Department of Genetic Epidemiology, University of Regensburg, Regensburg, Germany.
Stephen Burgess, Email: sb452@medschl.cam.ac.uk, MRC Biostatistics Unit, University of Cambridge, Cambridge, and Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge.
Christina Ellervik, Email: christina.ellervik@childrens.harvard.edu, Department of Clinical Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, DK-2200, Denmark; Department of Data and Development, Sorø, Region Zealand, Denmark; Department of Pathology, Harvard Medical School, Boston, MA-02215, USA; and Department of Laboratory Medicine, Boston Children’s Hospital, Boston, MA-02215, USA.
References
- 1.Chang AR, Grams ME, Ballew SH, Bilo H, Correa A, Evans M, et al. Adiposity and risk of decline in glomerular filtration rate: meta-analysis of individual participant data in a global consortium. Bmj. 2019;364:k5301. doi: 10.1136/bmj.k5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garofalo C, Borrelli S, Minutolo R, Chiodini P, De Nicola L, Conte G. A systematic review and meta-analysis suggests obesity predicts onset of chronic kidney disease in the general population. Kidney Int. 2017;91(5):1224–35. doi: 10.1016/j.kint.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 3.Herrington WG, Smith M, Bankhead C, Matsushita K, Stevens S, Holt T, et al. Bodymass index and risk of advanced chronic kidney disease: Prospective analyses from a primary care cohort of 1.4 million adults in England. PLoS One. 2017;12(3):e0173515. doi: 10.1371/journal.pone.0173515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madero M, Katz R, Murphy R, Newman A, Patel K, Ix J, et al. Comparison between Different Measures of Body Fat with Kidney Function Decline and Incident CKD. Clin J Am Soc Nephrol. 2017;12(6):893–903. doi: 10.2215/CJN.07010716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vivante A, Golan E, Tzur D, Leiba A, Tirosh A, Skorecki K, et al. Body mass index in 1.2 million adolescents and risk for end-stage renal disease. Arch Intern Med. 2012;172(21):1644–50. doi: 10.1001/2013.jamainternmed.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu H, Kuja-Halkola R, Chen X, Magnusson PKE, Svensson P, Carrero JJ. Higher body mass index is associated with incident diabetes and chronic kidney disease independent of genetic confounding. Kidney Int. 2019;95(5):1225–33. doi: 10.1016/j.kint.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 7.Zhu P, Lewington S, Haynes R, Emberson J, Landray MJ, Cherney D, et al. Cross-sectional associations between central and general adiposity with albuminuria: observations from 400,000 people in UK Biobank. Int J Obes (Lond) 2020;44(11):2256–66. doi: 10.1038/s41366-020-0642-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill NR, Fatoba ST, Oke JL, Hirst JA, O’Callaghan CA, Lasserson DS, et al. Global Prevalence of Chronic Kidney Disease-A Systematic Review and Meta-Analysis. PLoS One. 2016;11(7):e0158765. doi: 10.1371/journal.pone.0158765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blüher M. Obesity: global epidemiology and pathogenesis. Nature Reviews Endocrinology. 2019;15(5):288–98. doi: 10.1038/s41574-019-0176-8. [DOI] [PubMed] [Google Scholar]
- 10.Stevens PE, Levin A. Evaluation and management of chronic kidney disease: synopsis of the kidney disease: improving global outcomes 2012 clinical practice guideline. Ann Intern Med. 2013;158(11):825–30. doi: 10.7326/0003-4819-158-11-201306040-00007. [DOI] [PubMed] [Google Scholar]
- 11.Stefansson VTN, Schei J, Solbu MD, Jenssen TG, Melsom T, Eriksen BO. Metabolic syndrome but not obesity measures are risk factors for accelerated age-related glomerular filtration rate decline in the general population. Kidney Int. 2018;93(5):1183–90. doi: 10.1016/j.kint.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 12.Liakopoulos V, Franzén S, Svensson A-M, Sattar N, Miftaraj M, Björck S, et al. Renal and Cardiovascular Outcomes After Weight Loss From Gastric Bypass Surgery in Type 2 Diabetes: Cardiorenal Risk Reductions Exceed Atherosclerotic Benefits. Diabetes Care. 2020;43(6):1276–84. doi: 10.2337/dc19-1703. [DOI] [PubMed] [Google Scholar]
- 13.O’Brien R, Johnson E, Haneuse S, Coleman KJ, O’Connor PJ, Fisher DP, et al. Microvascular Outcomes in Patients With Diabetes After Bariatric Surgery Versus Usual Care: A Matched Cohort Study. Ann Intern Med. 2018;169(5):300–10. doi: 10.7326/M17-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X, Eales JM, Jiang X, Sanderson E, Scannali D, Morris AP, et al. Obesity as a cause of kidney disease-insights from Mendelian randomisation studies. medRxiv. 2020:2020.09.13.20155234 [Google Scholar]
- 15.Zheng J, Zhang Y, Rasheed H, Walker V, Sugawara Y, Li J, et al. Trans-ethnic Mendelian randomization study reveals causal relationships between cardio-metabolic factors and chronic kidney disease. medRxiv. 2020:2020.09.04.20188284. doi: 10.1093/ije/dyab203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu P, Herrington WG, Haynes R, Emberson J, Landray MJ, Sudlow CLM, et al. Conventional and Genetic Evidence on the Association between Adiposity and CKD. J Am Soc Nephrol. 2021;32(1):127–37. doi: 10.1681/ASN.2020050679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, et al. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum Mol Genet. 2018;27(20):3641–9. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pulit SL, Stoneman C, Morris AP, Wood AR, Glastonbury CA, Tyrrell J, et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet. 2019;28(1):166–74. doi: 10.1093/hmg/ddy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;5187(538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanzick K-J, Li Y, Gorski M, Wuttke M, Pattaro C, Köttgen A, et al. Discovery and prioritization of variants and genes for kidney function in >1.2 million individuals. bioRxiv. 2020:2020.09.04.283713. doi: 10.1038/s41467-021-24491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wuttke M, Li Y, Li M, Sieber KB, Feitosa MF, Gorski M, et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet. 2019;51(6):957–72. doi: 10.1038/s41588-019-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teumer A, Li Y, Ghasemi S, Prins BP, Wuttke M, Hermle T, et al. Genome-wide association meta-analyses and fine-mapping elucidate pathways influencing albuminuria. Nat Commun. 2019;10(1):4130. doi: 10.1038/s41467-019-11576-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505–13. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartwig FP, Davies NM, Hemani G, Davey Smith G. Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol. 2016;45(6):1717–26. doi: 10.1093/ije/dyx028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greco MF, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med. 2015;34(21):2926–40. doi: 10.1002/sim.6522. [DOI] [PubMed] [Google Scholar]
- 26.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40(4):304–14. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan NA, Thompson JR. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int J Epidemiol. 2016;45(6):1961–74. doi: 10.1093/ije/dyw220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13(11):e1007081. doi: 10.1371/journal.pgen.1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanderson E, Davey Smith G, Windmeijer F, Bowden J. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int J Epidemiol. 2019;48(3):713–27. doi: 10.1093/ije/dyy262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lees JS, Welsh CE, Celis-Morales CA, Mackay D, Lewsey J, Gray SR, et al. Glomerular filtration rate by differing measures, albuminuria and prediction of cardiovascular disease, mortality and end-stage kidney disease. Nat Med. 2019;25(11):1753–60. doi: 10.1038/s41591-019-0627-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Husain SA, Willey JZ, Park Moon Y, Elkind MSV, Sacco RL, Wolf M, et al. Creatinine-versus cystatin C-based renal function assessment in the Northern Manhattan Study. PLoS One. 2018;13(11):e0206839. doi: 10.1371/journal.pone.0206839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol. 2016;40(7):597–608. doi: 10.1002/gepi.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minelli C, Fabiola Del Greco M, van der Plaat DA, Bowden J, Sheehan NA, Thompson J. The use of two-sample methods for Mendelian randomization analyses on single large datasets. bioRxiv. 2020:2020.05.07.082206. doi: 10.1093/ije/dyab084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.