Orho-Melander et al. recently reported that lower low-density lipoprotein cholesterol (LDLC) as predicted by the T-allele of the variant rs12916 in HMGCR was associated with a decreased risk of developing breast cancer [odds ratio (OR) = 0.89; 95% confidence interval (CI): 0.82–0.96].1 Their analysis was embedded in a wider Mendelian randomization (MR) study performed using genotype data from a prospective cohort of 26,589 individuals that included 16,022 women and 1176 incident breast cancer cases. HMGCR encodes 3-hydroxy-3-methylglutaryl-coenzyme A reductase, the enzyme inhibited by statins. The T-allele of rs12916 is associated with reduced HMGCR expression and therefore, in principle, its associations should be analogous to the effects of lifelong statin administration starting at birth.2 The MR study of Orho-Melander et al. also found that a genome-wide LDLC score based on 32 independent LDLC-associated single nucleotide polymorphisms (SNPs) was not associated with breast cancer. In light of this finding, they suggested that the protective association of the rs12916 T-allele with breast cancer may be the result of a distinct non-LDLC-based mechanism that is regulated by rs12916 and HMGCR.
Orho-Melander et al. found that rs12916 was not associated with body mass index (BMI) in their study (P = 0.29). However, we noted that this SNP has previously been found to be associated with BMI by a genome-wide association meta-analysis of 315,585 adults (Table 1).3 This pleiotropic BMI-increasing association of the LDLC-lowering T-allele of rs12916 is noteworthy because it has been found by another large MR study by Guo et al. that higher genetically-predicted adult BMI is associated with reduced risk of estrogen receptor (ER)-positive and ER-negative breast cancer.4 We hypothesized that either (a) HMGCR rs12916 T-allele via this pleiotropic association with increased BMI, rather than its LDLC-lowering association, is responsible for the reported association with breast cancer protection or (b) BMI is a confounder of the association between LDLC (as predicted by genetic variation in/near HMGCR) and breast cancer (Figure 1). The latter possibility is supported by the association between BMI and breast cancer reported in the MR study by Guo et al. and by the association between higher BMI and lower LDLC reported in an MR study by Holmes et al.4,5
Table 1. Associations between individual SNPs in the HMGCR region and LDLC, BMI, and breast cancer.a.
| SNP | Allele | LDLC | BMI | ER-pos BC | ER-neg BC | Overall BC | ER-pos/ER-neg | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | P | Beta | P | Beta | P | Beta | P | Beta | P | HetP | ||
| rs10066707 | G | -0.0497 | 3.0E-19 | 0.0097 | 0.02 | -0.0112 | 0.15 | -0.0113 | 0.34 | -0.0136 | 0.04 | 0.99 |
| rs12916 | T | -0.0733 | 7.8E-78 | 0.0182 | 7.3E-09 | -0.0136 | 0.07 | -0.0105 | 0.36 | -0.0113 | 0.07 | 0.82 |
| rs17238484 | G | -0.0627 | 1.4E-21 | -- | -- | -0.0237 | 0.01 | -0.0151 | 0.26 | -0.0156 | 0.03 | 0.59 |
| rs2006760 | C | -0.0533 | 1.7E-13 | 0.006 | 0.27 | -0.0161 | 0.08 | -0.0052 | 0.71 | -0.0135 | 0.08 | 0.52 |
| rs2303152 | G | -0.0423 | 1.0E-09 | 0.0158 | 2.8E-03 | -0.0222 | 0.07 | -0.0005 | 0.98 | -0.0077 | 0.45 | 0.33 |
| rs5909 | G | -0.0617 | 4.9E-13 | -0.0008 | 0.90 | 0.0066 | 0.59 | 0.0057 | 0.76 | -0.0029 | 0.78 | 0.97 |
| rs2112347b | T | -0.0443 | 4.4E-30 | 0.0261 | 6.2E-17 | -0.0154 | 0.04 | -0.0343 | 3.3E-03 | -0.0183 | 4.4E-03 | 0.18 |
Obtained from the data sets in references 3,10, and 11.
rs2112347 is the SNP most strongly associated with BMI in the HMGCR region in the BMI genetic association meta-analysis.3
Beta coefficients are signed with reference to the allele shown.
Abbreviations: BC: breast cancer; ER-pos: estrogen receptor positive; ER-neg: estrogen receptor negative; LDLC: low density lipoprotein cholesterol; BMI: body mass index, Het P: P-value from Cochran's Q test for heterogeneity.
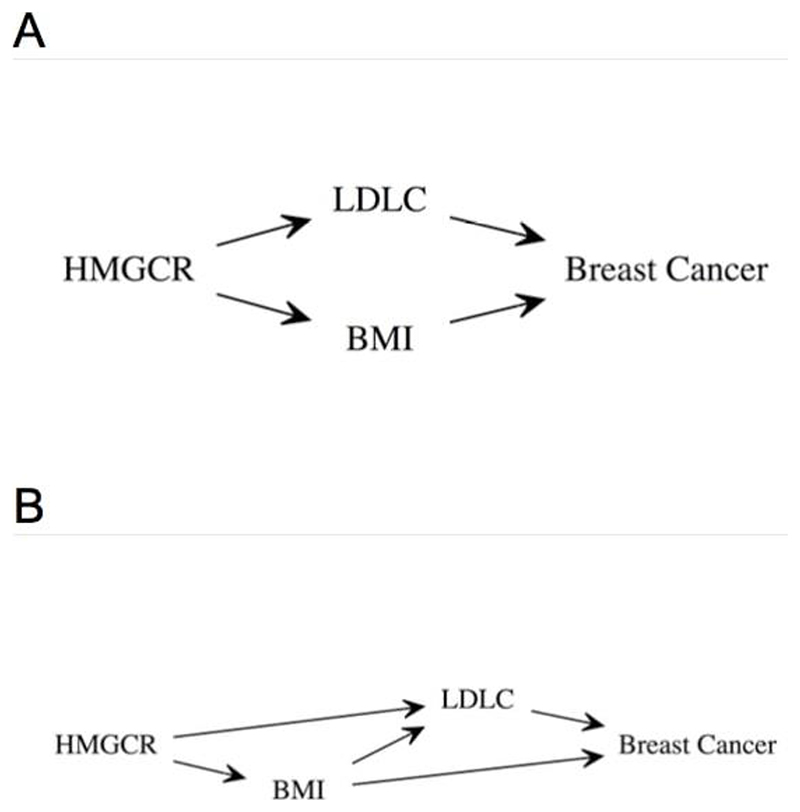
Figure 1.
Directed acyclic graphs (DAGs) showing the hypothesized relationships between HMGCR single nucleotide polymorphisms, low-density lipoprotein cholesterol (LDLC), body mass index (BMI), and breast cancer. DAG (A) illustrates a pleiotropic hypothesis while DAG (B) illustrates a hypothesis where BMI confounds the association between LDLC and breast cancer that was identified by Orho-Melander et al.
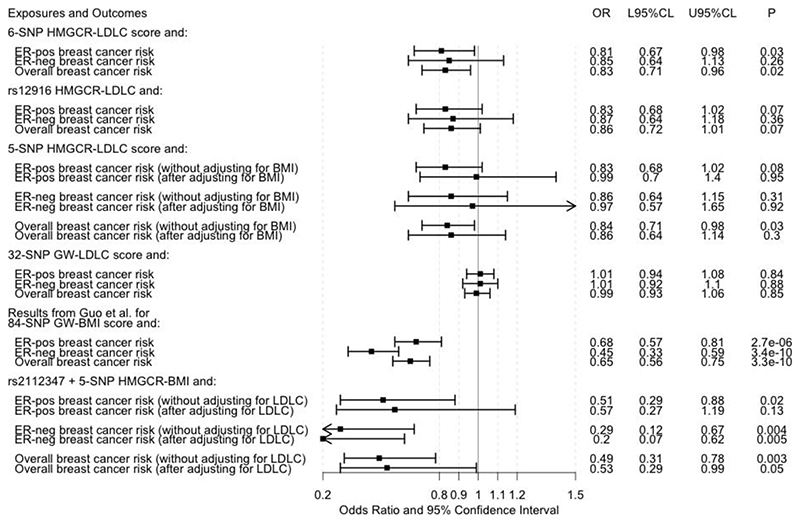
We tested our hypotheses using a two-sample Mendelian randomization study with a multivariable approach.6,7 While multivariable MR cannot distinguish between the pleiotropic and confounding models described (Figure 1), it can provide an estimate of the direct association between LDLC (as predicted by HMGCR SNPs) and breast cancer. It is not possible to perform a multivariable MR analysis using only one SNP as a genetic instrumental variable and so we constructed a multi-SNP instrumental variable for this purpose. Six SNPs in or within 100 kb of HMGCR (including rs12916) have previously been used to construct a genetic instrumental variable for LDLC lowering via inhibition of HMGCR in studies that have compared this genetically-predicted association with the actual effect of statins on cardiovascular outcomes observed in clinical trials.8,9 Forward stepwise-regression models based on individual-level data have shown that each of these SNPs has a strong and independent association with circulating LDLC level.8 We extracted summary results for these SNPs (Table 1) from publicly available genetic association meta-analysis data sets for LDLC (maximum n = 168,357 individuals),10 BMI (max. n = 315,585 individuals),3 ER-positive breast cancer risk (69,501 cases/105,974 controls),11 ER-negative breast cancer risk (21,468 cases/105,974 controls),11 and overall breast cancer risk (122,977 cases/105,974 controls).11 The overall breast cancer risk data set included breast cancer cases whose ER-status was unknown, in addition to all ER-positive and ER-negative cases included in the two ER-specific data sets. One of the six SNPs, rs17238484, had not been mapped in the BMI genetic association meta-analysis.3 For four of the five SNPs with BMI data available, the LDLC-lowering allele was the increasing BMI-associated allele and associated with a protection against breast cancer (Table 1). We used a random-effect linear regression model weighted by the inverse of the variance of the SNP associations with breast cancer for MR analysis.12,13 We obtained the genetic correlations between the six SNPs using the ‘TwoSampleMR’ R package and accounted for these genetic correlations in our MR analyses using the weighted generalized linear regression method implemented in the ‘MendelianRandomization’ R package.13,14 Univariable analysis confirmed that lower LDLC as predicted by all six HMGCR SNPs put together was associated with reduced ER-positive and overall breast cancer risk (Figure 2). For the corresponding ER-negative breast cancer risk analysis, while the confidence interval included one, the point estimate of the OR was consistent in magnitude and direction with the ER-positive and overall breast cancer associations (Figure 2). We also obtained similar point estimates when using rs12916 alone as a genetic instrumental variable to test the association between LDLC and breast cancer risk by the Wald ratio method (Figure 2).15 However, multivariable Mendelian randomization analysis, wherein standardized regression coefficients from the genotype-BMI association analysis were included as a covariate in the model,7 attenuated the protective association with breast cancer risk of lower LDLC as collectively predicted by the five HMGCR SNPs that had both LDLC and BMI data available (Figure 2). The shift in OR towards the null after adjusting for the association with BMI was evident for ER-positive breast cancer (from 0.83 to 0.99) and ER-negative breast cancer (from 0.86 to 0.97). The was no heterogeneity in the HMGCR SNP associations by ER-subtype (Table 1). On balance, the multivariable MR analyses suggested that the pleiotropic associations with BMI might mediate the association between LDLC-lowering alleles in HMGCR and reduced breast cancer risk.
Figure 2.
Forest plot of the results of Mendelian randomization analyses. The odds ratios (ORs) presented for breast cancer risk are per 1-standard deviation (~1 mmol/L) decrease in low-density lipoprotein cholesterol (LDLC) or per 1-standard deviation (~4.65 kg/m2) increase in adult body mass index (BMI) except for the ORs from Guo et al.,4 which are scaled per 5 kg/m2 increase in adult BMI. The estrogen receptor (ER)-positive (ER-pos), ER-negative (ER-neg), and overall breast cancer results from Guo et al.4 were based on analyses of 69,556, 49,770, and 88,807 women, respectively, and this data set overlapped entirely with the data set in reference 11. Other abbreviations: L95%CL: lower 95% confidence limit; U95%CL: upper 95% confidence limit; SNP: single nucleotide polymorphism; GW: genome-wide.
We confirmed that the 32-SNP genome-wide LDLC score from Orho-Melander et al. was not associated with breast cancer risk in our data sets (using beta coefficients for these SNPs from the 2013 Global Lipid Genetics Consortium data set10 and without adjustment for BMI; Figure 2). We also used the SNP that had the strongest association with BMI in the HMGCR genomic region,3 rs2112347 which is 321 kb from HMGCR and has a correlation (r2)of 0.37 with rs12916 in European-ancestry populations from the 1000 Genomes Project,16 in conjunction with the five HMGCR SNPs to create a HMGCR-BMI score. Higher BMI based on this score was associated with lower risk of ER-positive, ER-negative, and overall breast cancer in univariable MR analysis and in multivariable MR analysis (adjusted for LDLC associations at the six SNPs) for ER-negative and overall breast cancer (Figure 2). Taken together with the negative association between BMI genetically-predicted by 84 SNPs across the genome and breast cancer risk identified by Guo et al. (Figure 2),4 these findings lend additional support to BMI, rather than LDLC, being a more likely mechanism underlying the potential protective association of genetic inhibition of HMGCR with breast cancer risk that was identified by Orho-Melander et al.
Acknowledgements
This work builds on multiple, publicly available data sets. The low-density lipoprotein cholesterol association data were obtained from the Global Lipid Genetics Consortium (GLGC). The body mass index association data were obtained from the Genetic Investigation of ANthropometric Traits (GIANT) Consortium. The breast cancer genetic association data were obtained from the Breast Cancer Association Consortium (BCAC). We are grateful to each of the three consortia.
Footnotes
Data sets:
Global Lipid Genetics Consortium (GLGC):
http://csg.sph.umich.edu/abecasis/public/lipids2013/
Genetic Investigation of ANthropometric Traits (GIANT) consortium:
Breast Cancer Association Consortium (BCAC): http://bcac.ccge.medschl.cam.ac.uk/bcacdata/oncoarray/gwas-icogs-and-oncoarray-summary-results/
References
- 1.Orho-Melander M, Hindy G, Borgquist S, et al. Blood lipid genetic scores, the HMGCR gene and cancer risk: a Mendelian randomization study. Int J Epidemiol. 2017 Nov 20; doi: 10.1093/ije/dyx237. [DOI] [PubMed] [Google Scholar]
- 2.Swerdlow DI, Preiss D, Kuchenbaecker KB, et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet. 2015 Jan 24;385(9965):351–361. doi: 10.1016/S0140-6736(14)61183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015 Feb 12;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo Y, Warren Andersen S, Shu X-O, et al. Genetically Predicted Body Mass Index and Breast Cancer Risk: Mendelian Randomization Analyses of Data from 145,000 Women of European Descent. PLoS Med. 2016 Aug;13(8):e1002105. doi: 10.1371/journal.pmed.1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmes MV, Lange LA, Palmer T, et al. Causal effects of body mass index on cardiometabolic traits and events: a Mendelian randomization analysis. Am J Hum Genet. 2014 Feb 6;94(2):198–208. doi: 10.1016/j.ajhg.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015 Feb 15;181(4):251–260. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgess S, Dudbridge F, Thompson SG. Re: ‘Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects’. Am J Epidemiol. 2015 Feb 15;181(4):290–291. doi: 10.1093/aje/kwv017. [DOI] [PubMed] [Google Scholar]
- 8.Ference BA, Robinson JG, Brook RD, et al. Variation in PCSK9 and HMGCR and Risk of Cardiovascular Disease and Diabetes. N Engl J Med. 2016;375(22):2144–2153. doi: 10.1056/NEJMoa1604304. [DOI] [PubMed] [Google Scholar]
- 9.Ference BA, Kastelein JJP, Ginsberg HN, et al. Association of Genetic Variants Related to CETP Inhibitors and Statins With Lipoprotein Levels and Cardiovascular Risk. JAMA. 2017;318(10):947–956. doi: 10.1001/jama.2017.11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willer CJ, Schmidt EM, Sengupta S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013 Nov;45(11):1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michailidou K, Lindström S, Dennis J, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017 Nov 2;551(7678):92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013 Nov;37(7):658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017 Dec 1;46(6):1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016 May 20;35(11):1880–1906. doi: 10.1002/sim.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife. 2018 May 30;7 doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015 Nov 1;31(21):3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]