Abstract
Purpose
The DNA Damage Immune Response (DDIR) assay was developed in breast cancer (BC) based on biology associated with deficiencies in homologous recombination and Fanconi Anemia (HR/FA) pathways. A positive DDIR call identifies patients likely to respond to platinum-based chemotherapies in breast and oesophageal cancers. In colorectal cancer (CRC) there is currently no biomarker to predict response to oxaliplatin. We tested the ability of the DDIR assay to predict response to oxaliplatin-based chemotherapy in CRC and characterised the biology in DDIR-positive CRC.
Methods
Samples and clinical data were assessed according to DDIR status from patients who received either 5FU or FOLFOX within the FOCUS trial (n=361, stage 4), or neo-adjuvant FOLFOX in the FOxTROT trial (n=97, stage 2/3). Whole transcriptome, mutation and immunohistochemistry data of these samples were used to interrogate the biology of DDIR in CRC.
Results
Contrary to our hypothesis, DDIR negative patients displayed a trend towards improved outcome for oxaliplatin-based chemotherapy compared to DDIR positive patients. DDIR positivity was associated with Microsatellite Instability (MSI) and Colorectal Molecular Subtype 1 (CMS1). Refinement of the DDIR signature, based on overlapping interferon-related chemokine signalling associated with DDIR positivity across CRC and BC cohorts, further confirmed that the DDIR assay did not have predictive value for oxaliplatin-based chemotherapy in CRC.
Conclusions
DDIR positivity does not predict improved response following oxaliplatin treatment in CRC. However, data presented here suggests the potential of the DDIR assay in identifying immune-rich tumours that may benefit from immune checkpoint blockade, beyond current use of MSI status.
Keywords: Colorectal cancer, DNA damage response, immune-oncology, bioinformatics, molecular pathology
Introduction
Colorectal cancer (CRC) is the fourth most common cancer and the second most common cause of cancer related death in the UK (1). CRC diagnostic classification relies on the WHO classification and the tumour-node-metastasis (TNM) staging system. While histological assessment provides valuable prognostic information, it cannot identify specific patient subgroups within tumour type, grade or clinical stage that respond best to chemotherapy. Despite advances in treatment regimens, 5-year overall survival (OS) rates in the unresectable metastatic setting remain at 10% (2). In patients with stage III or histologically high-risk stage II tumours, recurrence is seen in 45% and 16% of patients respectively, following surgery and adjuvant 5-FU based chemotherapy (2). The addition of oxaliplatin to 5-FU based regimens has led to a 20% risk reduction in OS following surgery for patients with stage III CRC (3–5). However chronic peripheral neuropathy occurs in ~50% of patients exposed to oxaliplatin (6), and there is no clinically-validated test available to predict oxaliplatin response. Therefore, a significant proportion of patients may endure distressing side effects from this treatment with no clinical benefit (7). This highlights the need for the development of improved predictive tools to guide treatment decision making and ultimately improve patient outcomes (8).
Numerous models suggest that conventional chemotherapy elicits high levels of DNA damage and DNA strand breaks in highly proliferative cancer cells that can either prime them for cell death, or tip already primed cells into apoptosis (9). The efficacy of chemotherapy in cancer cells is often compromised due to dysfunctional damage detection or cell death mechanisms, allowing cell survival (9). Certain chemotherapeutic agents target vulnerabilities inherent in tumours with defective DNA damage repair machinery, leading to neoplastic cell death. In CRC, the most common defective DNA damage repair mechanism occurs in tumours with microsatellite instability (MSI), characterised by defects in DNA mismatch repair. MSI tumours account for ~15% of stage II/III CRC and ~4% of stage IV patients, and are largely characterised by hypermutation, an increase in cancer-specific neoantigen production, high immune infiltration, and a favourable prognosis in earlier stages (10,11). Interestingly, in the recent FOxTROT neoadjuvant colon cancer chemotherapy clinical trial, this immune-rich MSI subgroup, defined by loss of MMR, specifically failed to gain a clear significant benefit from oxaliplatin-based neoadjuvant therapy (7). The DNA damage immune response (DDIR) signature, which comprises a 44-gene transcriptional signature based on loss of the Fanconi anemia/BRCA (FA/BRCA) DNA damage response pathway, was previously developed in breast cancer (BC), where it demonstrated clinical utility for the identification of patients with a good response to anthracycline and/or cyclophosphamide-based neoadjuvant chemotherapy (12,13). DDIR-positive tumours (exhibiting defective DNA damage repair) are characterised by an inflammatory tumour microenvironment (TME), upregulation of interferon signalling genes and high lymphocytic infiltration. Additional studies in BC indicated that DDIR-positive tumours have increased levels of CXCL10 and enhanced signalling through the cGAS/STING pathway (14).
Given these predictive findings, the Stratification in COloRecTal cancer (S:CORT) consortium (15) hypothesised that the DDIR signature would be predictive of oxaliplatin benefit in CRC, based on its ability to predict benefit from DNA-damaging therapy in BC. In this study we tested the ability of the DDIR signature to identify patients that may respond to oxaliplatin-based chemotherapy in both metastatic and neoadjuvant CRC settings, employing transcriptional profiling and bioinformatic analysis of subsets of samples from the FOCUS (first-line metastatic, n=391) and FOxTROT (first-line neoadjuvant, n=97 randomised controlled trials. We ascertained if DDIR-positivity was associated with improved outcomes in metastatic CRC patients treated with FOLFOX compared to 5FUFA alone (bolus and infusional 5-FU and folinic acid on the modified de Gramont schedule), and in patients with localised disease treated with FOLFOX in the neo-adjuvant setting. We also performed a series of analyses to comprehensively characterise the underlying biology of DDIR subtypes in CRC compared to BC.
Materials and Methods
As part of the MRC Stratified Medicine in Colorectal Cancer Consortium (S:CORT) (15), tumour biospecimens with associated clinical trial data were identified for exploration of potential stratifiers for oxaliplatin treatment. The randomised MRC FOCUS trial was selected for exploration in the metastatic setting and the FOxTROT trial was selected for exploration of short course FOLFOX in the neoadjuvant setting. The studies were performed in accordance with the Declaration of Helsinki. All subjects provided written informed consent for further research on their samples at the time of consent to the clinical trials. Both the original clinical trials (FOCUS Ref: 79877428; FOxTROT 07/SO703/57) and the studies reported here (S:CORT ref 15/EE/0241) were approved by the National Research Ethics Service in the UK.
FOCUS Trial
FOCUS was a large UK-based randomised controlled trial comparing different strategies of sequential or combination therapies of 5FUFA (bolus and infusion 5-FU with folinic acid) with or without oxaliplatin or irinotecan as first- or second-line therapies in patients with newly-diagnosed advanced CRC (16). A total of 2135 patients were recruited between 2000-03 and randomised between three strategies of first- or second-line combination therapy. Control strategy: First-line 5FUFA alone, followed by single-agent irinotecan; second strategy: first-line 5FUFA alone, followed by second-line combination chemotherapy; third strategy: combination chemotherapy in first line treatment. Within the two research strategies, the combination regimen was an additional randomisation: either 5FUFA plus oxaliplatin (FOLFOX), or 5FUFA plus irinotecan (FOLFIRI). For the DDIR analysis, samples from patients with colonic primaries from a biobank of archival diagnostic tissue were selected from consenting patients in the relevant arms where a randomised comparison could be made between first-line 5FUFA alone or in combination with oxaliplatin (85mg/m2 two-weekly) (Supplementary Figure 1A). 385 samples were obtained from 371 primary resections, 8 primary biopsies, 6 metastatic samples (3 liver, 2 nodal and 1 lung). The primary outcome for FOCUS was overall survival (OS), but data were also available for progression-free survival (PFS) and objective response rate (ORR).
FOxTROT Trial
FOxTROT was an international randomised trial (1052 patients) which has reported its main finding (7). Patients were eligible if they had been diagnosed with locally advanced colon cancer (CC) without evidence of distance metastasis and with surgical resection of the primary tumour planned. Patients were randomised into one of three chemotherapy groups:
Group A: Patients had 6-weeks pre-surgery chemotherapy (oxaliplatin with either 5FUFA or capecitabine) and 18-weeks chemotherapy that commenced 4-8 weeks after surgical resection of the tumour.
Group B: Patients had no pre-surgery chemotherapy but had 24-weeks chemotherapy (OxMdG or OxCap) after their surgical resection.
Group C: For patients who were RAS wild-type on baseline biopsy and randomised to neo-adjuvant chemotherapy, the option of a secondary randomisation between panitumumab or not, for the 6 weeks prior to surgery.
For patients randomised into Group A, FOxTROT provided an opportunity to measure DDIR in the tissue biopsy in a subset at baseline and determine whether DDIR was predictive of response to neo-adjuvant OxMdG therapy prior to resection surgery, excluding patients in Group C and those with complete response (Supplementary Figure 1B).
Gene Expression Profiling
All the archival formalin-fixed paraffin-embedded (FFPE) tumour tissue samples were tested at Almac’s Diagnostic CLIA Laboratories. Samples were reviewed and tumour material identified on an adjacent H&E stained slide for microdissection. Total RNA was extracted from two sequential 5μm sections using the Roche High Pure FFPE Extraction Kit (Roche Life Sciences, Penzberg, Germany) and amplified using the NuGen Ovation FFPE Amplification System v3 (NuGen San Carlos, California, USA). The amplified product was hybridised to the Almac Diagnostics XCEL array (Almac, Craigavon, UK), a cDNA microarray-based technology optimised for archival FFPE tissue, and analysed using the Affymetrix Genechip 3000 7G scanner (Affymetrix, Santa Clara, California, USA) as previously described (12). Microarray data were quality checked (see Supplementary methods) then pre-processed where raw CEL files underwent the Robust Multiarray Average (RMA) normalisation for the Almac Diagnostic XCEL array with the affy package (v1.56.0) (17). Gene expression profiles from a total of 391 samples from FOCUS and 97 samples from FOxTROT were made available.
For the biological analysis, a subset of gene expression profiles from n=361 primary tumour resection samples from FOCUS were used (exclusions detailed in supplementary Figure 1A) and n=97 pre-treatment biopsy samples from FOxTROT (exclusions detailed in supplementary Figure 1B). Probes were annotated using annotation file “Xcel Annotations, CSV format, Release 36” available for download from (http://www.affymetrix.com/support/technical/byproduct.affx?product=xcel), and then collapsed to their corresponding genes using WGCNA package (version 1.68), based on the probe with highest average value for each gene (18). For comparative analysis between BC and CRC, TRASNBIG BC cohort (19) containing gene expression profiles for 198 fresh frozen samples from patients with node-negative T1-T2 (≤5cm) breast performed on Affymetrix Human Genome U133A array was downloaded from Gene Omnibus Expression (GEO; www.ncbi.nlm.nih.gov/geo/) (accession number ‘GSE7390’).
DDIR Signature
A total of 484 clinical samples (391 from FOCUS and 97 from FOxTROT) had DDIR signature scores calculated and predefined cut-points applied. The pre-defined threshold of 0.1094 was optimised in an independent technical study of 260 CRC samples whereby the optimal threshold was detected at the score where the sensitivity and specificity meant a joint maximum to accurately detect the DDIR-positive subgroup as defined in hierarchical clustering (Personal communication Almac Diagnostics). The threshold was then applied independently to the validation cohorts, dichotomising patients as DDIR-positive (>0.1094) or DDIR-negative (≤0.1094).
TRANSBIG BC cohort (19) used in the original study had information available on predetermined DDIR threshold of 0.37 along with DDIR continuous score (12), that was used on our analysis.
Consensus Molecular Subtyping and CRC Intrinsic Subtyping
To obtain CMS calls, genes with multiple probesets were collapsed by mean and the CMSclassifier package was used (20). Classification by random forest with the default posterior probability of 0.5 showed a higher frequency of unclassified samples compared to the original publication (20). To derive calls with comparable frequencies, single sample predictor calls were computed after row-centring the expression data. Final CMS calls were generated when there was a match between both methods without applying any cut-off. To obtain CRIS calls, probesets with the highest average levels for each gene were selected and the CRISclassifier package was used (21). Samples with a Benjamini-Hochberg-corrected False Discovery Rate (BH.FDR) > 0.2 were left unclassified as originally reported (21).
Mutational Analysis
Mutation data was generated by DNA target capture (SureSelect, Agilent) spanning all coding exons of 80 CRC driver genes (listed in Supplementary Methods) followed by next generation sequencing (Illumina). Variant calling was performed with Caveman for point mutations and Pindel for indel mutations. Driver mutations in KRAS, NRAS, PIK3CA and TP53 were considered for binary classification (e.g. depending on whether genes are dominant/recessive, mutations reported as recurrent or an internal curated list) based on frequency and relevance. BRAF was classified as mutated only with a V600E mutation. Tumours showing more than two mutations in n=123 MSI markers within the panel were classified as MSI, otherwise as MSS. The FOxTROT cohort showed a high failure rate (55/97 missing data, 57%) due to lack of enough tissue in small biopsies after RNA profiling. Therefore, MSI classification form additional FOxTROT tumours were derived with a RNA signature (22). Two borderline tumours were not classified.
Gene Set Enrichment Analysis (GSEA)
GSEA was performed in the three cohorts to investigate biological pathways associated with DDIR (23,24), using Hallmarks gene set collection (h.all.v6.2.symbols.gmt [Hallmarks]) from Molecular Signature Database (MSigDB) (25,26). GSEA version 19.0.26 was accessed from the GenePattern cloud server web interface: https://cloud.genepattern.org. All default parameters were utilised, with the exception of ‘collapse dataset’ which was set to ‘FALSE’, as the probes were collapsed to their genes a priori, and the random seed was stated to be ‘40218336’. Normal enrichment score (NES) and false discovery rate (FDR) values were noted for each gene set within the two phenotypic (DDIR) groups, where FDR q-value below 25% was justified to be a significant gene set.
Microenvironment Cell Population Analysis
The MCPcounter (version MCPcounter_1.1.0) R package was downloaded from GitHub (https://github.com/ebecht/MCPcounter), and was used to generate MCP estimation scores for ten stromal and immune cell infiltrates from the transcriptomic data of the three cohorts (27). Estimates were compared between DDIR-positive and DDIR-negative to determine their stromal/immune content, and the differences in cellular composition between the cancer types.
Differential Gene Expression and Pathway Analysis
Partek Genomics Suite (PGS) version 6.6 was utilised to perform ANOVA analysis to identify differentially expressed genes with FDR of < 0.05, and fold change (FC) adjusted to 1.5 for FOCUS and FOxTROT cohorts; for TRANSBIG due to the large number of differentially expressed genes, FC value was increased to 2.5. Differentially expressed genes were assessed using Ingenuity Pathway Analysis (IPA - 49932394) to examine any significant biological pathways associated with DDIR subtypes. All parameters were set to default.
Statistical Analysis
Statistical analyses were conducted according to pre-specified statistical analysis plans that were agreed prior to inspection of any DDIR-stratified outcome data. All clinical-related analyses for Objective response rate, progression-free-survival and overall survival were performed using Stat version 15.0 (Stata Corporation, Texas City, USA) or R (version 3.4.1). Further detailed statistical analysis on FOCUS and FOxTROT cohort is available in Supplementary Methods.
All statistical analyses undertaken for further biological exploration, including Pearson’s Correlation Coefficient, Fisher’s exact test, Student’s t-test, Wilcoxon rank sum test, Kruskal-Wallis rank sum test, and one-way ANOVA followed by Tukey’s Honest Significance Difference test were performed to generate p-values for statistical significance using R stats package in R (version 3.4.0) and RStudio (version 1.1383). In addition to base R packages, ggplot2 R package (version 3.2.1) with other supporting packages, including cowplot (version 0.9.4), ggpubr (version 0.2.3) and grid (version 3.4.0) were used for graphical visualisation.
Data and Script Availability
FOCUS and FOxTROT gene expression dataset and clinicopathological information are provided from S:CORT (https://www.s-cort.org/contact), with transcriptional data, mutation data (for KRAS, NRAS, PIK3CA, BRAF and TP53) and MSI call available on GEO under reference GSE156915. All scripts required to reproduce figures in this manuscript are available from corresponding author on request or from www.dunne-lab.com.
Results
Case selection from FOCUS metastatic CRC clinical trial
A total of n=391 patients were available for DDIR analysis from the FOCUS trial. Following exclusion of rectal cancer cases and prioritisation of resected tissue to ensure there was sufficient tumour tissue for molecular analyses, n=310 from the 5FU alone group and n=81 in the 5FU+oxaliplatin group were used for outcome analyses (Supplementary Table S1). Assessment of baseline characteristics of patients excluded from the DDIR analysis compared to those included in the DDIR analysis revealed that there were no other obvious selection biases between the groups (Supplementary Table S1, Supplementary figure S1). A total of 76/391 patients were classified as DDIR positive (Supplementary Figure S2), generating a prevalence of 19% [95% CI 16-24] overall, with a reasonable balance between the randomised groups of 63 (20%) versus 13 (16%) in the 5FU and 5FU+oxaliplain groups respectively, (Chi-squared p-value for difference=0.39; Supplementary Table S1).
The overall prevalence of DDIR was lower than anticipated when compared with data from other cohorts of patients with CRC (28) and other disease indications (12,13,29) but was similar to the technical study of 260 metastatic CRC used to set the threshold for DDIR positivity (Personal communication Almacgroup).
Survival analyses according to DDIR status in the FOCUS trial
During the course of follow-up between 16th May 2000 and 18th October 2006, there were a total of 383 PFS events (357 during the first 15 months) and 342 OS events. During the first 12-weeks of first-line chemotherapy, there were 157 (40%) complete or partial responders and 234 (60%) stable or progressive disease non-responders. A comparison between randomised groups, without stratification for DDIR, confirmed the anticipated treatment effect of oxaliplatin; PFS adjusted HR (95% CI) = 0.63 (0.48, 0.81), p=0.001 and ORR adjusted OR (95% CI) = 4.07 (2.37, 7.01), p<0.001 (Supplementary figure S3).
In the FOCUS control arm, we identified no prognostic effect of DDIR status for patients with metastatic colon cancer treated with first line 5FU alone, either on OS (Unadjusted HR (95% CI) = 0.95 (0.71, 1.28), p = 0.73, Test of proportional hazards: χ2 = 1.42 on 1 d.f., p=0.20, Supplementary Figure S2b), or on PFS (Adjusted HR = 1.11 (95% CI 0.79 – 1.54), p = 0.55). This result remained non-significant when adjusted for clinical variables, CMS status and other molecular variables.
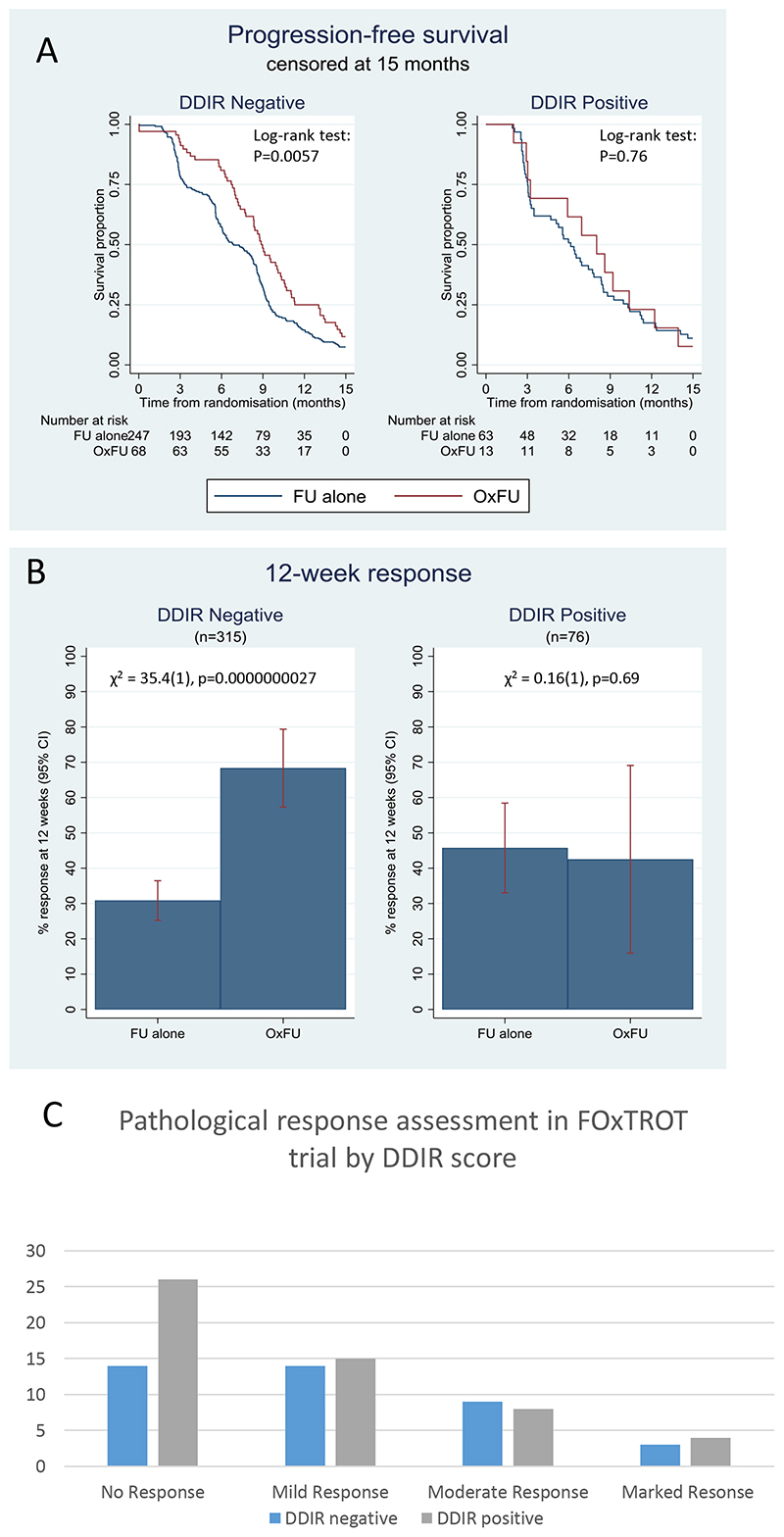
Using fully adjusted models, we next explored the predictive effects of DDIR for all outcomes, with PFS at 15 months as the primary outcome (Figure 1A). Contrary to the expectation that DDIR-positive patients would derive the most benefit from oxaliplatin, DDIR-negative patients appeared to respond more frequently to FOLFOX (ratio of odds ratios for ORR = 0.15 (95% CI 0.04 – 0.65), test for interaction p = 0.011; Table 1, Figure 1B). Although this inverted direction of effect was the same for the survival outcomes, the tests for interaction were non-significant (Table 1).
Figure 1. Clinical outcomes in patients randomised to FUFA or to OxFU in FOCUS trial by DDIR score.
A) Progression free survival (to 15 months) B) Overall response rate (ORR) C. Pathological response assessment in resected primary following 6 weeks oxaliplatin based chemotherapy in FOxTROT trial by DDIR score.
Table 1. Statistical outcomes to oxaliplatin based therapy by DDIR status in 1. FOCUS trial and 2. FoxTROT trial sample sets.
| DDIR negative (81%) | DOIR positive (19%) | |||||
|---|---|---|---|---|---|---|
| Outcome (FOCUS) | HR or OR for OxFU vs 5FU alone | (95% Cl) pvalue | HR or OR for OxFU vs 5FU alone | (95% Cl) P-value | interaction HR or OR | (95% Cl) p value |
| PTS (1S months) | 0.59 | (0.44, 080) P=0 001 | 0 85 | (0.45.1.62) P=0.63 | 1.43 | (0.70, 2,92) P=0.32 |
| PIS (full) | 0.58 | (0.43,076) p<0.001 | 1.00 | (0.54,1.87) P=0.99 | 1.73 | (0.87, 3.43) P=0.12 |
| OS (Full) | 0.58 | (0.65, 1.18) P-0.38 | 1-26 | (0.65, 2.46 P-0.50 | 1.44 | (0.69, 3.01)P-0.34 |
| ORR | 5 64 | (3.01. 10.56) p<0.001 | 0.86 | (0.23. 3.16) p=0.82 | 0.15 | (0.04,0-65) p=0.011 |
| DDIR negative (41%) | DDIR positive (59%) | |||||
|---|---|---|---|---|---|---|
| Outcome (FoxTrot) ORR | N | % | N | % | Unadjusted oriknal rogression | (95% Cl) p-value |
| excel | 14 | 35% | 26 | 49% | 0.62 | (0.29,133) P=0.128 |
| Mild Response | 14 | 35% | 15 | 28% | ||
| Moderate Response | 9 | 23% | 8 | 15% | ||
| Marked Response | 3 | 7% | 4 | 8% | ||
Case selection and survival analyses according to DDIR in the FOxTROT neoadjuvant CRC clinical trial
Following these analyses in the metastatic setting, we next assessed the clinical utility of the DDIR in the CRC neoadjuvant setting. A total of 97 patients who received neoadjuvant FOLFOX were selected from Group A of the FOxTROT dataset. Patients were excluded if they withdrew from the trial, if they did not receive neo-adjuvant chemotherapy or if they received OxCap prior to surgery. Additionally, no patients with complete pathological response were forwarded to S:CORT for analysis. These selections led to a somewhat biased subset compared to the main study with less responders, less MSI and more KRAS wildtype tumours (Supplementary Table 2). Of these 97 patients, 4 had no associated response data, leaving a total of 93 patients who were included in the final analysis. There were a total of 40 non-responders, 29 mild-responders, 17 moderate responders and 7 marked responders. The DDIR threshold was set at the same value defined in the FOCUS cohort, resulting in 57% DDIR positive patients, which was considerably higher than the 19% seen in the metastatic FOCUS dataset (Supplementary Figure S2c). Using ordinal regression across the 4 response groups, there were marginally better responses in the DDIR-negative group (Figure 1C), but this was not statistically significant using unadjusted ordinal regression OR = 0.62 [95% CI 0.29 – 1.33], p=0.218 (Table 1). After adjustment for age, sex, pT-stage, pN-stage, primary tumour location, MSI and RAS status, the coefficient reduced slightly to 0.55 [95% CI 0.21-1.39], p=0.205. Employing DDIR as a continuous variable, the unadjusted OR for response was 0.19 [95% CI 0.02-1.79], p=0.148. When adjusted for age, sex, T-stage, N-stage, left/right, MSI and RAS status the OR reduced to 0.11 [95% CI 0.01-1.66], p=0.110 (Supplementary Table S2).
Given these counter-intuitive findings, we next set out to investigate if there was a biological explanation for this potentially inverted and inconsistent effect between previous breast cohorts and our CRC trial cohorts.
Association between DDIR and colorectal cancer subtypes
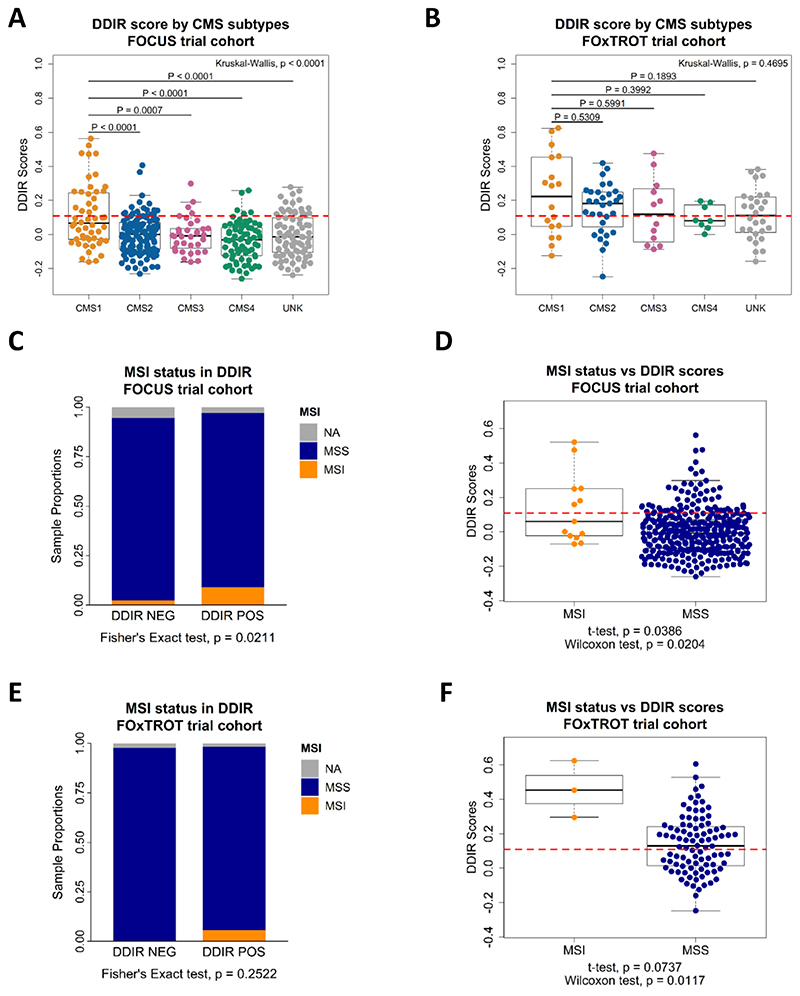
Investigation into the biological relevance of DDIR signature led to the comparison against CRC Consensus Molecular Subtypes (CMS) which is largely based on histological (stroma and immune) features (20). In the FOCUS cohort, immune-rich CMS1 tumours are significantly associated with increased DDIR scores when compared to all other CMS subtypes (Figure 2A; Kruskal-Wallis, p < 0.0001). Despite CMS1 tumours having a significantly higher proportion of DDIR-positive tumours compared to the other subtypes (Supplementary Figure 6A; Fisher’s exact test, p = 0.0002), given the low prevalence of DDIR-positivity across the whole cohort, 68% of CMS1 subtypes are below the DDIR threshold (Figure 2A). Of note, there are proportionally more CMS4 tumours within DDIR-negative classification in the FOCUS cohort (Supplementary Figure 6A). In pre-treatment biopsies from the smaller FOxTROT cohort, CMS1 tumours show a non-significant trend towards DDIR positivity (Figure 2B; Kruskal-Wallis, p = 0.4695, and Supplementary Figure 6B; Fisher’s exact test, p = 0.4879). Additionally, we also examined DDIR on Colorectal Cancer Intrinsic Subtypes (CRIS) that represents CRC tumour-intrinsic (epithelial) biology (21). Contrary to CMS, no significant association between the CRIS subtypes and DDIR-positive or DDIR-negative tumours in both the FOCUS and FOxTROT cohort was found (Supplementary Figures 6C-F). These findings suggest that, in CRC, DDIR-positivity is primarily associated with (and potentially influenced by) CMS-related tumour microenvironment (TME) factors, such as differences in stromal/immune infiltrates, rather than epithelial-derived intrinsic factors.
Figure 2. Consensus molecular subtypes (CMS) and CRC intrinsic subtypes (CRIS) in association with DDIR in adjuvant FOCUS and neoadjuvant FOxTROT clinical trial cohorts.
A) Distribution of CMS samples against DDIR score in FOCUS and B) FOxTROT cohort, shown with DDIR threshold value at 0.1094 (red dash line). Statistics: Kruskal-Wallis rank sum test for global p-value, and Tukey’s HSD test following one-way ANOVA for comparison between two groups. C) Proportion of MSI/MSS CRCs in the FOCUS cohort comparing DDIR positive and DDIR negative, and D) number of MSI/MSS CRCs in the FOCUS cohort samples against DDIR continuous score. E) Proportion of MSI/MSS CRCs in the FOxTROT cohort comparing DDIR-positive and DDIR-negative, and F) number of MSI/MSS CRCs in the FOxTROT cohort samples against DDIR continuous score. Statistics: Pearson’s Coefficient Correlation, Fisher’s exact test, Student’s t-test and Wilcoxon rank sum test.
Originally, DDIR signature was developed based on defective DNA damage response and repair machinery of Homologous Recombination (HR) and Fanconi Anaemia (FA) in breast cancer (12). However, there is limited evidence on their role in CRC tumorigenesis (30). Thus, we explored the relationship between HR/FA and DDIR in CRC cohorts and made comparison against TRANSBIG BC cohort which was used in the development of the DDIR signature. Our investigation suggested that within CRC, these pathways do not show any association with DDIR, contrary to that in BC (see Supplementary Results; Supplementary Figure 4). Microsatellite instability (MSI), a result of defective DNA mismatch repair mechanisms, defines a proportion of CRC patients associated with high tumour mutational burden, leading to development of immune-responsive TME. Despite the limited number of MSI tumours in the metastatic FOCUS CRC cohort (n=13), we observe that MSI tumours contain a significantly higher proportion of DDIR-positives (Figure 2C; Fisher’s exact test, p = 0.0211). However, DDIR-positivity is not a biomarker of MSI status, as only 46% of MSI tumours are DDIR-positive (6 out of 13) while the majority of DDIR-positive tumours overall are MSS (Figure 2D; MSI/DDIR+ n=6, MSS/DDIR+ n=59). In the FOxTROT cohort, MSI trends observed are in line with the larger FOCUS cohort (Figure 2E; Fisher’s exact test, p = 0.2522, and Figure 2F; Student’s t-test, p = 0.0737), but this result cannot be used to confirm the FOCUS findings due to small (n=3) MSI sample size (Figure 2F). Furthermore, while MSI tumours collectively contain higher mutational burden than MSS as expected, mutational burden is not associated with DDIR-positivity in either of the CRC cohorts (Supplementary Figure 6G; Student’s t-test, p = 0.1279 and Supplementary Figure 6H; Student’s t-test, p = 0.4534).
Enhanced immune-related signalling pathways define DDIR-positive tumours
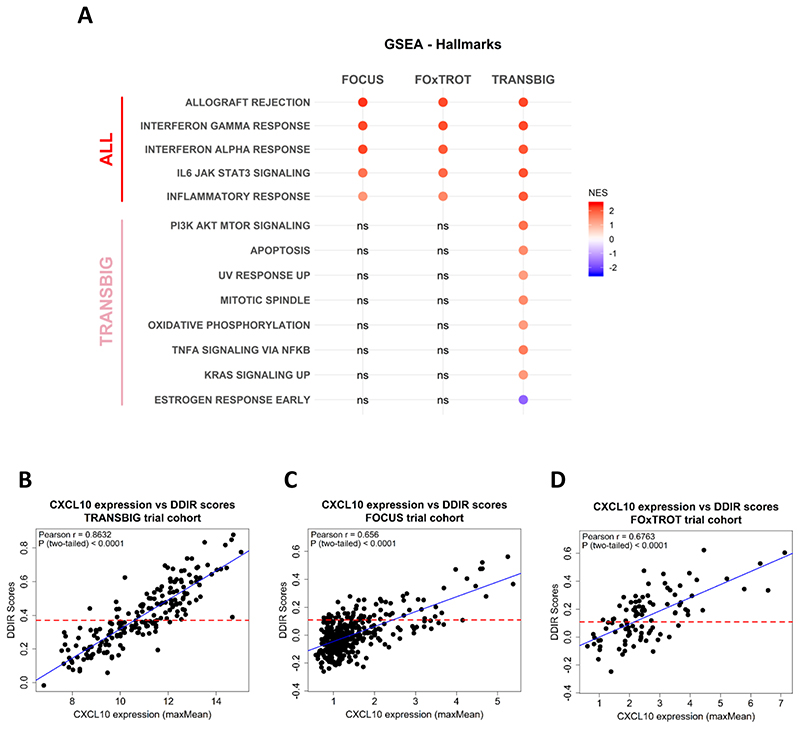
To further characterise the biological functions and pathways associated with DDIR, we performed GSEA, using the “Hallmark” collection, to compare DDIR-positive and DDIR-negative tumours in FOCUS and FOxTROT CRC cohorts, compared to the same analyses in the TRANSBIG BC cohort. GSEA between DDIR-positive and DDIR-negative tumours generated different numbers of significant Hallmarks genesets in each cohorts (Supplementary Figure 7). However, in general, between the three cohorts five common significantly-enriched genesets in DDIR-positive CRC and BC tumours were identified, namely allograft rejection, IL6/JAK/STAT3 signalling, inflammatory response, interferon-α response and interferon-γ response (Figure 3A; FDR q-value < 0.25), suggesting that a common immune and/or inflammatory-like signalling defines DDIR-positivity, regardless of the cancer type. Interestingly, we also observe eight unique gene sets that are only associated with DDIR in BC and not in CRC (Figure 3A).
Figure 3. Inflammatory and immune response-related pathways are elevated in DDIR positive tumours.
A) Gene set enrichment analysis on the two CRC cohorts (FOCUS and FOxTROT) and a BC cohort (TRANSBIG) identifies five common pathways associated with DDIR positive tumours in both cancer types; Benjamini-Hochberg False Discovery Rate (FDR) < 0.25 considered significant, Normalised Enrichment Score (NES) bar (DDIR POS > 0, DDIR NEG < 0). B) Expression of CXCL10 correlated with DDIR scores in TRANSBIG, C) FOCUS, and D) FOxTROT cohort, displayed with line of best fit (blue).
Previous studies of DDIR signalling in BC have highlighted increased levels of the interferon gamma-induced chemokine CXCL10 gene/protein expression in DDIR-positive tumour cells, leading to lymphocytic trafficking into the tumour (14). Here, we showed that CXCL10 expression has a strong positive (>6) correlation with DDIR scores in both BC and CRC cohorts (Figure 3B, 3C and 3D). Additionally, it was previously demonstrated that DDIR-positivity in BC was specifically associated with activation of cGAS/STING/TBK1 innate immune response axis (14). This, however, was not found to be the case in CRC (see Supplementary Results).
DDIR-defined tumour microenvironment reflects immune-rich colorectal subtype
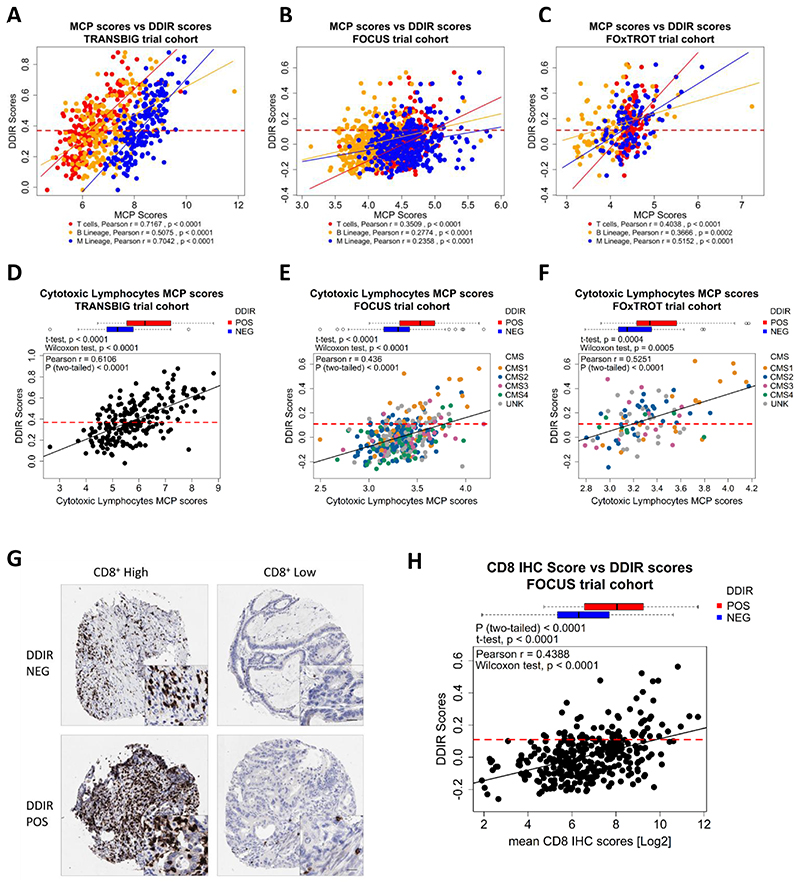
We tested the association between immune/stromal composition, based on gene expression profiles using microenvironment cell population (MCP) analysis, where we identified consistent correlations between DDIR scores and T cell, B cell and monocytic immune lineages, confirming an increase in lymphocytic infiltration in DDIR-positive BC (Figure 4A; Pearson r; T cells = 0.7167, B Lineage = 0.5075, Monocytic Lineage = 0.7042). While we also observe correlative trends in both CRC cohorts (Figure 4B; Pearson r; T cells = 0.3509, B Lineage = 0.2774, Monocytic Lineage = 0.2358 and Figure 4C; Pearson r; T cells = 0.4038 and Monocytic Lineage = 0.5152 and B Lineage, r = 0.3666), these correlations were not as strong as those observed in BC. Moreover, cytotoxic lymphocytes scores also demonstrate a positive correlation with DDIR using both a positive versus negative categorical (Figure 4D; Student’s t-test, p < 0.0001) or DDIR continuous score (Figure 4D; Pearson r = 0.6106) in the TRANSBIG BC cohort. Similar, albeit weaker, correlations were observed in both FOCUS (Figure 4E: Student’s t-test, p < 0.0001; Pearson r = 0.436) and FOxTROT (Figure 4F: Student’s t-test, p = 0.0004; Pearson r = 0.5251) CRC cohorts using the MCP-derived cytotoxic lymphocyte scores. Incorporation of CMS in the CRC analyses demonstrated the association between CMS1, lymphocytic infiltration and increased DDIR score. Levels of cytotoxic CD8+ T-lymphocytic infiltration were further assessed in situ in the FOCUS cohort by IHC (Figure 4G), where a significant association between CD8 IHC scores and DDIR score was observed, in line with MCP assessments in these tumours (Figure 4H: Student’s t-test, p < 0.0001; Pearson r = 0.4388). Conversely, fibroblast levels and CMS4 subtypes were negatively correlated with DDIR score in the FOCUS cohort (Supplementary Figure 8A and 8B; t-test, p = 0.0109; Pearson r = -0.1597), while no association was noted in FOxTROT cohort (Supplementary Figure 8C and 4D: t-test, p = 0.9984; Pearson r = 0.0291).
Figure 4. Increased immune infiltrates highly correlates with DDIR positivity.
A) MCP scores of three immune infiltrates – T cells (red), B lineage (yellow) and monocytic lineage (blue) – correlated against DDIR scores with line of best fit for each immune infiltrates for TRANSBIG, B) FOCUS, and C) FOxTROT cohort.; shown DDIR threshold value at 0.37 for BC and 0.1094 for two CRC cohorts (red dash line). D) Cytotoxic lymphocytes MCP scores correlated with DDIR score in TRANSBIG, E) with overlay of CMS in FOCUS, and F) FOxTROT cohort; shown DDIR threshold value at 0.37 for BC and 0.1094 for two CRC cohorts (red dash line). G) Immunohistochemistry (IHC) images of DDIR negative and DDIR positive tumours stained with CD8+ marker in FOCUS cohort (x10; inset x40, 20μm bar). H) Comparison of average CD8+ log-transformed scores from IHC analysis between DDIR positive (red) and DDIR negative (blue) shown in boxplot above scatterplot examining correlation with DDIR continuous score; line of best fit (black) and DDIR threshold value at 0.1094 (red dash line). Statistics: Student’s t-test, Wilcoxon rank sum test and Pearson’s Coefficient Correlation.
Overlapping interferon-responsive biology in DDIR-positive CRC and BC
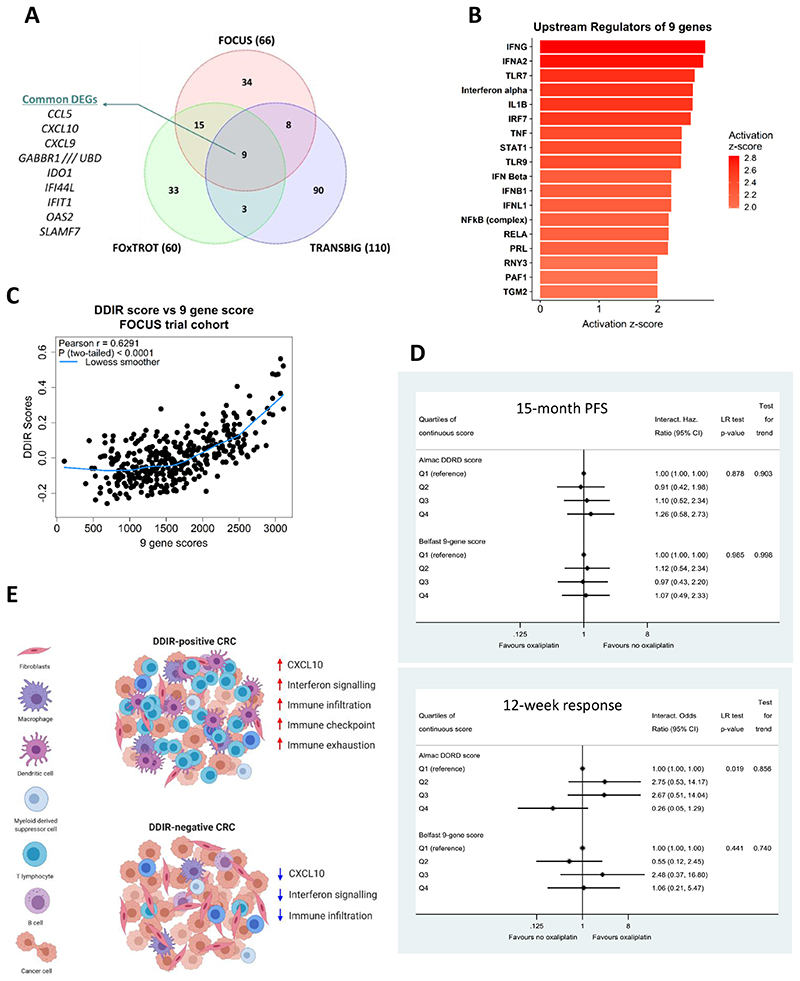
Next, we set out to identify overlapping individual differentially expressed genes between DDIR subtypes in both BC and CRC. Differential gene expression analysis comparing DDIR-positive and DDIR-negative tumours identified 66 and 60 differentially expressed genes in FOCUS and FOxTROT cohorts respectively (FDR < 0.05, FC = 1.5; Figure 5A). We observed 975 differential genes between DDIR-positive and negative tumours in the BC cohort compared to CRC; thus, in order to limit these analyses to a similar sized gene list for the TRANSBIG cohort, we increased the FC for analysis, identifying 110 differentially expressed genes (FDR < 0.05, FC = 2.5; Figure 5A). Comparison of gene lists from the three cohorts identified nine genes that are consistently upregulated in DDIR-positive tumours in both cancer types (Figure 5A). This list contained members of chemokines family, including two genes (CXCL10 and IDO1) that are part of the 44-gene DDIR signature. Using these nine differentially expressed genes common in all three cohorts, pathway analysis was performed, which revealed 18 potential upstream regulators of conserved biology contributing to DDIR-positivity across CRC and BC, including key regulators of inflammatory and interferon-related signalling; such as IFN-alpha, IFN-gamma, STAT1 and the NFkB complex (Figure 5B and Supplementary Figure 9A).
Figure 5. Differential gene expression analysis identifies distinct and conserved DDIR biology across BC and CRC.
A) Venn diagram of differentially expressed genes between DDIR positive and DDIR negative in three cohorts shows nine common genes, including chemokines such as CCL5 and CXCL10. B). Ingenuity Pathway Analysis (IPA) was used to identify potential elevated/activated upstream regulators of the conserved 9 genes identified in (A). C) Correlation and distribution of DDIR compared to a sum cumulative score generated from the 9 gene overlap in (A). D) 15-month PFS (top) and 12-week objective response rate (bottom) comparing the Almac DDIR score and the modified 9-gene score. Estimates adjusted for WHO PS, left vs right-sided, liver resection, number of mets, source and age of sample, CMS, KRAS, BRAF, PIK3CA, TP53, MSI, imputed (N=361). E) Diagram displaying DDIR-positive and DDIR-negative specific tumour microenvironment and upregulation of biological features such as CXCL10 expression in CRC. DDIR-positive CRCs are riddled with immune infiltrates responding to inflammatory/interferon signalling leading to ‘inflamed’ TME. On the contrary, DDIR-negative CRCs are immune ‘cold’ with low level of CXCL10, interferon signalling and overall low immune cells.
Using these nine consensus DDIR-related genes to generate an unweighted cumulative score, we observed a strong positive correlation between this new overlapping ranked sum score and the original DDIR score (Figure 5C; Pearson r = 0.6291, p < 0.0001). In line with this overlap, we also observed similar correlative trends for both CMS and MSI (Supplementary Figure 9B and 9C), with the nine gene score as observed with the original DDIR score (Figure 2). Finally, a Cox regression model (for PFS) and a logistic regression model (for response) were fitted with main effects for oxaliplatin and for each of three quartiles of Almac DDIR or 9-gene score relative to Q1 (reference), and interactions between oxaliplatin and the three quartiles (Figure 5D). As with the response and outcomes analyses using the original DDIR score, this overlapping nine gene score fails to predict a benefit for the addition of oxaliplatin to 5FU in the FOCUS trial. Importantly, however, this new refined CRC DDIR signature removes the trend for increased response to oxaliplatin observed in the DDIR-negative group in the original DDIR.
Discussion
The original characterisation of the DDIR signature demonstrated its predictive value as a biomarker for platinum-based chemotherapy treatment in BC, and subsequently oesophageal adenocarcinoma (OAC) (12,29). In the initial BC study, the biology underpinning DDIR was based on dysfunctional DNA damage response and repair machinery regulated via the HR and FA/BRCA pathways, which is targeted by some chemotherapies as a mode of action (31). The multi-disciplinary S:CORT consortium (15) was established to identify and test new molecular stratification methods to predict CRC response to treatments, through the discovery of new and/or validation of existing molecular biomarker-based assays. In this study, we tested the clinical utility of the 44-gene DDIR signature from archival FFPE tumour tissue profiled at Almac’s Diagnostic CLIA Laboratories as previously described, to predict response to the addition of oxaliplatin to 5-FU-based chemotherapy in both metastatic CRC (FOCUS cohort) and neoadjuvant CRC (FOxTROT) clinical trial settings. Accompanying this clinical assessment, we utilised the molecular and histological data generated to further interrogate the biological signalling associated with CRC-specific DDIR positivity in contrast to BC.
DDIR-positivity was observed in 19% of primary tumours from stage IV FOCUS cohort and 57% of primary tumour biopsy material from stage II/III FOxTROT cohort. A previous study of DDIR-positivity in CRC reported a 35% incidence in a predominantly (94%) non-metastatic population (28). This was comparable to findings in BC (34%) (12) and OAC (24%) (29). Differing DDIR rates in our study could be credited to the cancer stage or other (molecular) criteria used for patient selection in the original trials. Patients with localised disease, as in the neo-adjuvant FOxTROT study, have a higher proportion of tumours with immune infiltration (32), a factor associated with DDIR-positivity in BC and OAC, and also with MSI and CMS1 tumours in CRC. Similarly, the reduction in DDIR-positivity to ~20% in metastatic disease is consistent with a lower relative proportion of patients with MSI in metastatic disease, which falls from ~20% in localised CC in ~4% in mCRC, as in the FOCUS cohort.
MSI is the most notable feature in CRC displaying defective DNA damage response and repair via mismatch repair (MMR) system (30). MSI and CMS1 are closely linked together with high tumour mutation burden, overproduction of tumour-specific neoantigens, increased immune infiltration and show favourable clinical outcome in early stage disease (20). Given their high levels of immune infiltration and mutation burden, these tumours have responded well to checkpoint blockade immune-oncology (IO) treatments (33). There is a strong association of DDIR status with CMS1, MSI status (28) (Figure 2) in FOCUS cohort, and a similar trend is observed in FOxTROT cohort, given its small sample size (Figure 2), reflecting the observed clinical utility of immunotherapeutic interventions in this molecular subtype (34,35). However, our findings do not validate the correlation between DDIR and mutational burden in the FOCUS cohort observed in the CRC threshold development abstract (28), likely due to the difference in disease stage (FOCUS as mCRC) and mutational panel sequencing methods used with S:CORT.
Contrary to our primary hypothesis, it was noted that response to the addition of oxaliplatin to 5FUFA was more likely to benefit DDIR-negative patients in both FOCUS and FOxTROT cohorts rather than DDIR-positive patients. While this was only statistically significant in terms of response in the metastatic FOCUS trial setting (ratio of odds ratios for ORR = 0.15, test for interaction p = 0.011), the trend was consistent across all endpoints in both cohorts examined. However, the refinement of DDIR gene signature to only 9-genes signature through our analysis showed no additional benefit from oxaliplatin for either DDIR-positive or DDIR-negative patients (Figure 5). The original and subsequent DDIR study in BC with the South Western Oncology Group (13) demonstrated improved response to anthracycline and/or cyclophosphamide-based neoadjuvant and adjuvant chemotherapy in DDIR-positive patients. Similarly, in OAC, DDIR-positivity was predictive of improved response to cisplatin-containing chemotherapy (29). Oxaliplatin is known to differ in its mechanism of cytotoxicity compared to cisplatin and may have more complex mode of action in CRC (36).
Although we show no additional interaction between DDIR-positivity and oxaliplatin treatment, biologically, our study highlights promising immunotherapeutic opportunities among DDIR-positive CRC patients, beyond the use of general immune infiltration or MSI status. DDIR-positivity may have value in identifying additional subsets of MSS CRC patients who exhibit high tumour mutational burden and/or high TME activity, who have the potential to respond to immune checkpoint blockade such as PD-L1 inhibition (35,42,43). The search for biomarkers to distinguish immune “cold” tumours (that display limited response to IO) from immune “hot” tumours (that respond to IO) has gained traction in recent years. Our findings indicate that in CRC, although DDIR-positivity is associated with increased levels of both innate and cytotoxic infiltration, likely to be driven by interferon-related signalling, the immune system is in an “exhausted” state and unable to efficiently clear these tumours, due to the concurrent expression of checkpoints such as IDO1 and PD-L1 (CD274) (Figure 6E). These findings may also provide an explanation for the non-correlation of DDIR with oxaliplatin-based chemotherapy response, as induction of immune tolerance is a common response pattern to inflammation in the gut and tumour-associated inflammation (as seen in DDIR positive tumours) that leads to a predominantly immune suppressive milieu, which is further reinforced by additional chemotherapy-related inflammatory signalling. Indeed, MSI tumours are largely non-responsive to chemotherapy, as has been demonstrated recently in the neoadjuvant FOxTROT trial (7), as are immune-rich/MSI tumours when assessed in other non-trial adjuvant cohorts (44). Very recent trial data reported 100% response rate in early-stage MSI CC, including 60% pathological complete response, to neoadjuvant IO treatment (combined CTLA-4 and PD1 blockade) (45). Results from that study also indicate that only 27% of MSS tumours displayed any response. Importantly, however, these data confirmed the predictive nature of CD8+ T cell infiltration for IO response in MSS tumours; a phenotype associated with the biology underpinning DDIR-positivity in MSS CRC presented in this study, supporting clinical testing of DDIR as a predictive assay to select MSS patients in this setting.
The approach adopted in our study highlights the clinical utility and high success rates associated with molecular profiling of FFPE material (Supplementary Table 1), even in tissue-limited pre-treatment diagnostic biopsy material used to guide treatment decisions in the neoadjuvant setting, as in FOxTROT. The TRANSBIG data used in the original DDIR study poses a potential limitation on our BC analysis due to the platform employed in the original analysis (Affymetrix Human Genome U133A Array) not being identical to the one used for the transcriptional profiling in the CRC cohorts, which was the Almac XCEL array. To ensure cross-platform comparison for DDIR was not confounding our study, Almac have classified DDIR according to their diagnostic assay on all cohorts tested.
In summary, our study shows that, in contrast to BC and OAC, DDIR does not predict improved response or survival to oxaliplatin treatment. We have identified the underlying biology of the signalling associated with DDIR in CRC that could effect the outcome. While we identify significant overlap in DDIR signalling across BC and CRC, particularly immune-related TME signalling, we also highlight that signalling associated with both HR/BRCA and STING pathways is not significantly associated with DDIR in CRC. Overall, our data supports further testing of the utility of the DDIR signature in selecting patients who may respond to IO-based therapy.
Supplementary Material
Translational relevance.
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide, with around 1.3 million cases diagnosed each year. Efforts to develop biomarkers of prognosis and response to chemotherapy in CRC have resulted in stratification systems based on components of the tumour microenvironment (TME), highlighting the importance of characterising both molecular and pathological features. The DNA Damage Immune Response (DDIR) transcriptional assay was developed as a predictive biomarker for identifying breast cancer (BC) patients that benefit from DNA-damaging chemotherapy, based on signalling associated with defective homologous recombination DNA repair. Here we show that the DDIR signature does not predict outcomes from oxaliplatin based chemotherapy for localised or metatastic CRC patients in clinical trials. We show that although this predictive assay identifies tumours enriched for defects in the DNA mismatch repair machinery, it primarily identifies immune-rich, albeit exhausted, CRC tumours with competent repair signalling that may respond to immune checkpoint blockade.
Acknowledgements
WE are grateful to all the patients and their families who participated in the FOCUS and FOxTROT clinical trials and gave consent to further research on their samples. We are also grateful to the Trial Management Groups and Trial Steering Committees for FOCUS and FOxTROT trials who allowed this work to proceed. This work was originally led by Professor Paddy Johnston from Queen’s University Belfast. Sadly, soon after the project commenced Paddy passed away and we would like to dedicate this work to him.
Funding
The stratification in colorectal cancer consortium (S:CORT) is funded by a UK Medical Research Council (MRC) Stratified Medicine Consortium programme grant (grant ref MR/M016587/1) and co-funded by Cancer Research-UK. Brown, Fisher and Kaplan are partially funded by an MRC Core funding grant for the MRC Clinical Trials Unit at UCL (grant code 12023/20). This work was supported by a Cancer Research UK programme grant (Dunne, Longley, Johnston; C212/A13721).Sample collection for FOxTROT was funded by Yorkshire Cancer Research.
Footnotes
Conflicts of Interest
In-depth clinical and biological exploration of DNA Damage Immune Response (DDIR) as a biomarker for oxaliplatin use in colorectal cancer
Malla S Nothing to declare
Fisher DJ Nothing to declare
Domingo E Nothing to declare
Blake A Nothing to declare
Hassanieh S Nothing to declare
Redmond K Nothing to declare
Richman SD Nothing to declare
Youdell M Nothing to declare
Walker SM Employment: Almac Diagnostics. Patents, Royalties, Other Intellectual Property: Named inventor on Almac Diagnostics patents”
Logan GE Employment: Almac Diagnostic Services
Chatzpili K Nothing to declare
Amirkhah R Nothing to declare
Humphries M Nothing to declare
Craig S Nothing to declare
McDermott U Nothing to declare
Seymour M Nothing to declare
Morton D Nothing to declare
West NP consultancies from Eisai and Adlai Nortye
Quirke P PQ: has received research funding from Roche, Amgen, Halio, Intragen, Genomic Health, GeneFirst, Adlai Nordlyte, Leica and acted as a consultant or speaker for Roche, Bayer, Amgen and Merck Serono. He is a National Institute for Health Senior Investigator.
Salto-Tellez M Advisory boards and/or speaking engagement honoraria from: Roche, MSD, Pfizer, Astra Zeneca, BMS. Research Funding from: Phillips & Roche
Kennedy R Employment: Almac Diagnostics. Patents, Royalties, Other Intellectual Property: Named inventor on Almac Diagnostics patents. Honoraria: AstraZeneca, Tesaro
Johnston P PJ had stock and ownership interests in Almac Diagnostics before his death
Tomlinson I Nothing to declare
Koelzer V invited speaker on behalf of Indica labs outside the submitted work”
Campo L Nothing to declare
Kaplan R Nothing to declare
Longley D Nothing to declare
Lawler M ML has received honoraria from Pfizer, EMD Serono and Roche for presentations unrelated to this work
Maughan TS Honoraria: Astrazeneca, Array. Reseach funding from Almac as a partner in S:CORT consortium
Brown LC Nothing to declare
Dunne PD Nothing to declare
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018 doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Research UK. Bowel Cancer Statistics. 2018. [cited 2019 May 28]. [Internet]. Available from: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bowel-cancer.
- 3.André T, Boni C, Mounedji-Boudiaf L, Navarro M, Tabernero J, Hickish T, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004 doi: 10.1056/NEJMoa032709. [DOI] [PubMed] [Google Scholar]
- 4.Kuebler JP, Wieand HS, O’Connell MJ, Smith RE, Colangelo LH, Yothers G, et al. Oxaliplatin combined with weekly bolus fluorouracil and leucovorin as surgical adjuvant chemotherapy for stage II and III colon cancer: Results from NSABP C-07. J Clin Oncol. 2007 doi: 10.1200/JCO.2006.08.2974. [DOI] [PubMed] [Google Scholar]
- 5.Haller DG, Tabernero J, Maroun J, De Braud F, Price T, Van Cutsem E, et al. Capecitabine plus oxaliplatin compared with fluorouracil and folinic acid as adjuvant therapy for stage III colon cancer. J Clin Oncol. 2011 doi: 10.1200/JCO.2010.33.6297. [DOI] [PubMed] [Google Scholar]
- 6.Seretny M, Currie GL, Sena ES, Ramnarine S, Grant R, Macleod MR, et al. Incidence, prevalence, and predictors of chemotherapy-induced peripheral neuropathy: A systematic review and meta-analysis. Pain. 2014 doi: 10.1016/j.pain.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 7.Seymour MT, Morton D. FOxTROT: an international randomised controlled trial in 1052 patients (pts) evaluating neoadjuvant chemotherapy (NAC) for colon cancer. J Clin Oncol. 2019 May 20;37(15_suppl):3504–3504. [Google Scholar]
- 8.Lawler M, Alsina D, Adams RA, Anderson AS, Brown G, Fearnhead NS, et al. Critical research gaps and recommendations to inform research prioritisation for more effective prevention and improved outcomes in colorectal cancer. Gut. 2018 Jan 1;67(1):179–193. doi: 10.1136/gutjnl-2017-315333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8(3):193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 10.Ward R, Meagher A, Tomlinson I, O’Connor T, Norrie M, Wu R, et al. Microsatellite instability and the clinicopathological features of sporadic colorectal cancer. Gut. 2001;48(6):821–9. doi: 10.1136/gut.48.6.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boland CR, Goel A. Microsatellite Instability in Colorectal Cancer. Gastroenterology. 2010 May;138(6):2073–2087.:e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulligan JM, Hill LA, Deharo S, Irwin G, Boyle D, Keating KE, et al. Identification and Validation of an Anthracycline/Cyclophosphamide–Based Chemotherapy Response Assay in Breast Cancer. JNCI J Natl Cancer Inst. 2014 Jan;106(1):235–7. doi: 10.1093/jnci/djt335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma P, Barlow WE, Godwin AK, Parkes EE, Knight LA, Walker SM, et al. Validation of the DNA damage immune response signature in patients with triple-negative breast cancer from the SWOG 9313c trial. J Clin Oncol. 2019 doi: 10.1200/JCO.19.00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkes EE, Walker SM, Taggart LE, McCabe N, Knight LA, Wilkinson R, et al. Activation of STING-dependent innate immune signaling by s-phase-specific DNA damage in breast cancer. J Natl Cancer Inst. 2017 doi: 10.1093/jnci/djw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawler M, Kaplan R, Wilson RH, Maughan T. Changing the Paradigm—Multistage Multiarm Randomized Trials and Stratified Cancer Medicine. Oncologist. 2015 Aug 12;20(8):849–51. doi: 10.1634/theoncologist.2015-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seymour MT, Maughan TS, Ledermann JA, Topham C, James R, Gwyther SJ, et al. Different strategies of sequential and combination chemotherapy for patients with poor prognosis advanced colorectal cancer (MRC FOCUS): a randomised controlled trial. Lancet. 2007 Jul;370(9582):143–52. doi: 10.1016/S0140-6736(07)61087-3. [DOI] [PubMed] [Google Scholar]
- 17.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy - Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004 doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 18.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, et al. Strong Time Dependence of the 76-Gene Prognostic Signature for Node-Negative Breast Cancer Patients in the TRANSBIG Multicenter Independent Validation Series. Clin Cancer Res. 2007;13(11):3207–14. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 20.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015 Nov 12;21(11):1350–6. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isella C, Brundu F, Bellomo SE, Galimi F, Zanella E, Porporato R, et al. Selective analysis of cancer-cell intrinsic transcriptional traits defines novel clinically relevant subtypes of colorectal cancer. Nat Commun. 2017 May;8:15107. doi: 10.1038/ncomms15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian S, Roepman P, Popovici V, Michaut M, Majewski I, Salazar R, et al. A robust genomic signature for the detection of colorectal cancer patients with microsatellite instability phenotype and high mutation frequency. J Pathol. 2012 doi: 10.1002/path.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003 doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian A, Subramanian A, Tamayo P, Tamayo P, Mootha VK, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015 doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016 Dec 20;17(1):218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsantoulis P, Hill LA, Walker SM, Wirapati P, Graham DM, Wilson RH, et al. Association of a specific innate immune response to DNA damage with DNA repair deficient colorectal cancers. J Clin Oncol. 2016 May 20;34(15_suppl):3035-3035 [Google Scholar]
- 29.Turkington RC, Knight LA, Blayney JK, Secrier M, Douglas R, Parkes EE, et al. Immune activation by DNA damage predicts response to chemotherapy and survival in oesophageal adenocarcinoma. Gut. 2019:1–10. doi: 10.1136/gutjnl-2018-317624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muzny DM, Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chartron E, Theillet C, Guiu S, Jacot W. Targeting homologous repair deficiency in breast and ovarian cancers: Biological pathways, preclinical and clinical data. Crit Rev Oncol Hematol. 2019 March;133:58–73. doi: 10.1016/j.critrevonc.2018.10.012. 2018. [DOI] [PubMed] [Google Scholar]
- 32.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science (80-) 2006 doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 33.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science (80-) 2017 Jul 28;357(6349):409–13. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gkekas I, Novotny JAN, Pecen L, Strigård K, Palmqvist R, Gunnarsson ULF. Microsatellite Instability as a Prognostic Factor in Stage II Colon Cancer Patients, a Meta-Analysis of Published Literature. 2017;6574:6563–74. doi: 10.21873/anticanres.12113. [DOI] [PubMed] [Google Scholar]
- 35.Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):1–14. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruno PM, Liu Y, Park GY, Murai J, Koch CE, Eisen TJ, et al. A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress. Nat Med. 2017 doi: 10.1038/nm.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018;23(1):239–254.:e6. doi: 10.1016/j.celrep.2018.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dietlein F, Thelen L, Reinhardt HC. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet. 2014;30(8):326–39. doi: 10.1016/j.tig.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Esteban-Jurado C, Franch-Expósito S, Muñoz J, Ocaña T, Carballal S, López-Cerón M, et al. The Fanconi anemia DNA damage repair pathway in the spotlight for germline predisposition to colorectal cancer. Eur J Hum Genet. 2016;24(10):1501–5. doi: 10.1038/ejhg.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.An X, Zhu Y, Zheng T, Wang G, Zhang M, Li J, et al. An Analysis of the Expression and Association with Immune Cell Infiltration of the cGAS/STING Pathway in Pan-Cancer. Mol Ther - Nucleic Acids. 2019 Mar;14:80–9. doi: 10.1016/j.omtn.2018.11.003. March. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Overman M, Repair M. Where We Stand With Immunotherapy in Colorectal Cancer : Toxicity Management. ASCO Educ B; 2018. pp. 239–47. [DOI] [PubMed] [Google Scholar]
- 43.Goodman AM, Sokol ES, Frampton GM, Lippman SM, Kurzrock R. Microsatellite-stable tumors with high mutational burden benefit from immunotherapy. Cancer Immunol Res. 2019;7(10):1570–3. doi: 10.1158/2326-6066.CIR-19-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunne PD, Alderdice M, O’Reilly PG, Roddy AC, McCorry AMB, Richman S, et al. Cancer-cell intrinsic gene expression signatures overcome intratumoural heterogeneity bias in colorectal cancer patient classification. Nat Commun. 2017 doi: 10.1038/ncomms15657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chalabi M, Fanchi LF, Van den Berg JG, Beets GL, Lopez-Yurda M, Aalbers AG, et al. Ann Oncol. Elsevier Enhanced Reader; 2018. Neoadjuvant ipilimumab plus nivolumab in early stage colon cancer. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.