Summary
Protective immunity relies on the interplay of innate and adaptive immune cells with complementary and redundant functions. Innate lymphoid cells (ILCs) have recently emerged as tissue-resident, innate mirror images of the T cell system with which they share lineage-specifying transcription factors and effector machinery1. Located at barrier surfaces, ILCs are among the first responders against invading pathogens and might thus determine the outcome of the immune response2. However, it has been impossible until now to dissect the unique contributions of ILCs to protective immunity due to limitations in specifically targeting ILC subsets. Thus, all of the available data have either been generated in mice lacking the adaptive immune system or with tools that also affect other immune cell subsets. In addition, it was proposed that ILCs might be dispensable for a proper immune response because other immune cells could compensate for their absence3–7. Here, we report the generation of a new mouse model based on the Nmurl promoter as a driver for simultaneous expression of Cre recombinase and green fluorescent protein (GFP), which allows gene targeting in ILC2s without affecting other innate and adaptive immune cells. By removing Id2 and Gata-3 using Cre-mediated gene deletion in Nmur1-expressing cells, we have generated mice with selective and specific deficiency in ILC2s. ILC2-deficient mice have decreased eosinophil counts in steady state and are unable to recruit eosinophils in the airways in models of allergic asthma. Further, ILC2-deficient mice fail to mount an appropriate immune and epithelial type 2 response resulting in a profound defect in worm expulsion and a non-protective type 3 immune response. In total, our data establish non-redundant functions for ILC2s in the presence of adaptive immune cells at steady state and during diseases and argue for a multilayered organization of the immune system based on a spatiotemporal division of labor.
Results
A novel mouse model based on Nmur1 P2A-iCre-T2A-eGFP selectively labels ILC2s
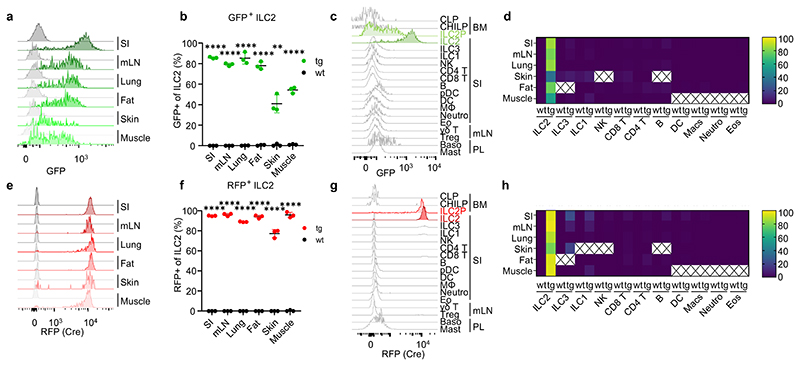
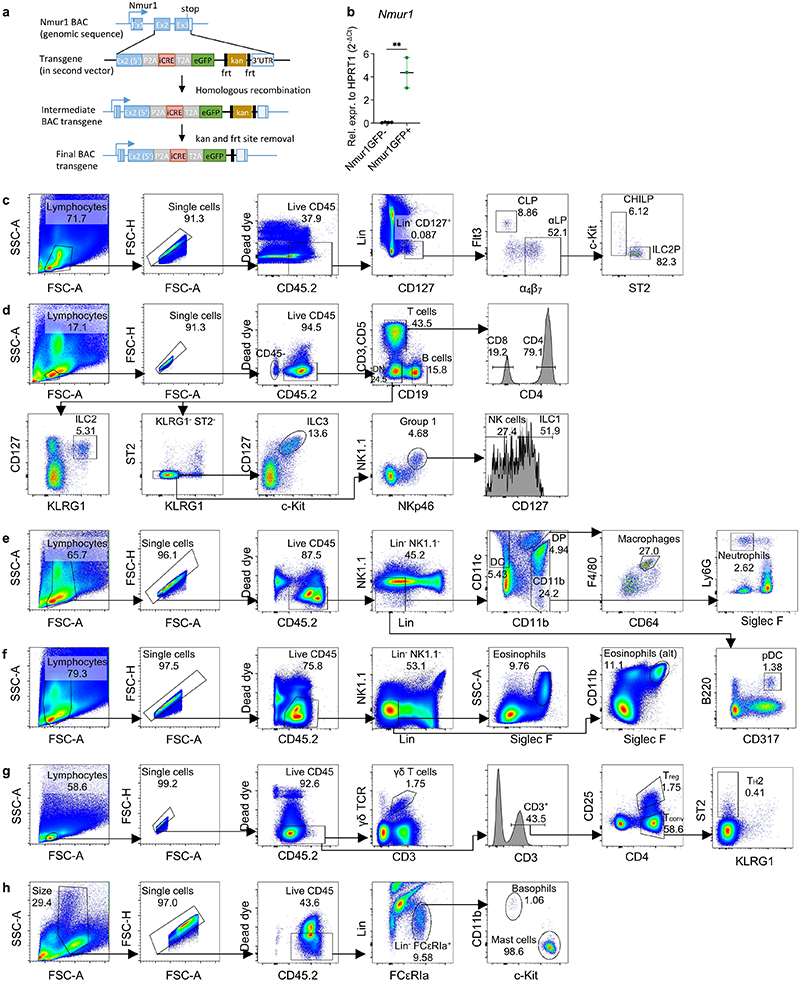
While many transcription factors and effector cytokines are shared between ILCs and other immune cells, the neuropeptide receptor Neuromedin U receptor 1 (Nmur1) was reported to be selectively expressed by ILC2s8–11. We have generated a BAC-transgenic mouse, in which a P2A-iCre-T2A-eGFP cassette was inserted in exon 2 and 3 of the Nmur1 gene (referred to as Nmur1cre-T2A-GFP mice) (Extended Data Fig.1a). Indeed, Nmur1 GFP reflected the endogenous expression since Nmur1 was highly expressed in GFP+ but not in GFP- cells (Extended Data Fig.1b). Consistent with previous data8–11, Nmur1 GFP was detected in ILC2s from all tissues investigated (Fig. 1a, b). Further, Nmur1 GFP was not observed during lymphocyte development in common lymphoid progenitors (CLP)12 or common helper-like ILC progenitors (CHILP)13,14 but upregulated in ILC2 progenitors15,16 in the bone marrow albeit to lower levels than in mature ILC2s (Fig. 1c, Extended Data Fig. 1c). Nmur1 GFP was selectively expressed in the ILC2 lineage but was not detected in other immune cells investigated, including Natural Killer (NK) cells, ILC1s, ILC3s, CD4+ and CD8+ T cells, B cells, macrophages, plasmacytoid (p) dendritic cells (DCs) and conventional (c)DCs, neutrophils, eosinophils, γδ T cells, basophils, mast cells, T helper 2 (TH2) cells, regulatory T cells (Treg) cells and CD45- cells (Fig. 1c,d; Extended Data Fig. 1d-h, Extended Data Fig. 2a-e). To monitor Cre activity, we next crossed Nmur1Cre-T2A-GFP mice to Rosa26Rfp/Rfp mice, which carry a floxed stop codon in front of the red fluorescent protein (RFP) open reading frame, thus preventing RFP production. Similar to the data obtained for GFP expression, RFP was detected in ILC2s among tissues, whereas in other cell types investigated, only a negligible fraction expressed RFP (Fig. 1e-h; Extended Data Fig. 2f-q). To investigate if Nmur1 expression is induced in cell types that do not express Nmur1 at steady state, we infected Nmur1Cre-T2A-GFP Rosa26Rfp/+ mice with the worm Nippostrongylus brasiliensis (N. brasiliensis), a strong inducer of type 2 inflammation. Similar to the expression pattern measured at steady state, Nmur1 GFP and RFP expression was enriched in ILC2s but not in other cell types after worm infection and lung inflammation (Extended Data Fig. 2 r-t), indicating that Nmur1Cre-T2A-GFP could be used for selective gene targeting in models of type 2 inflammation.
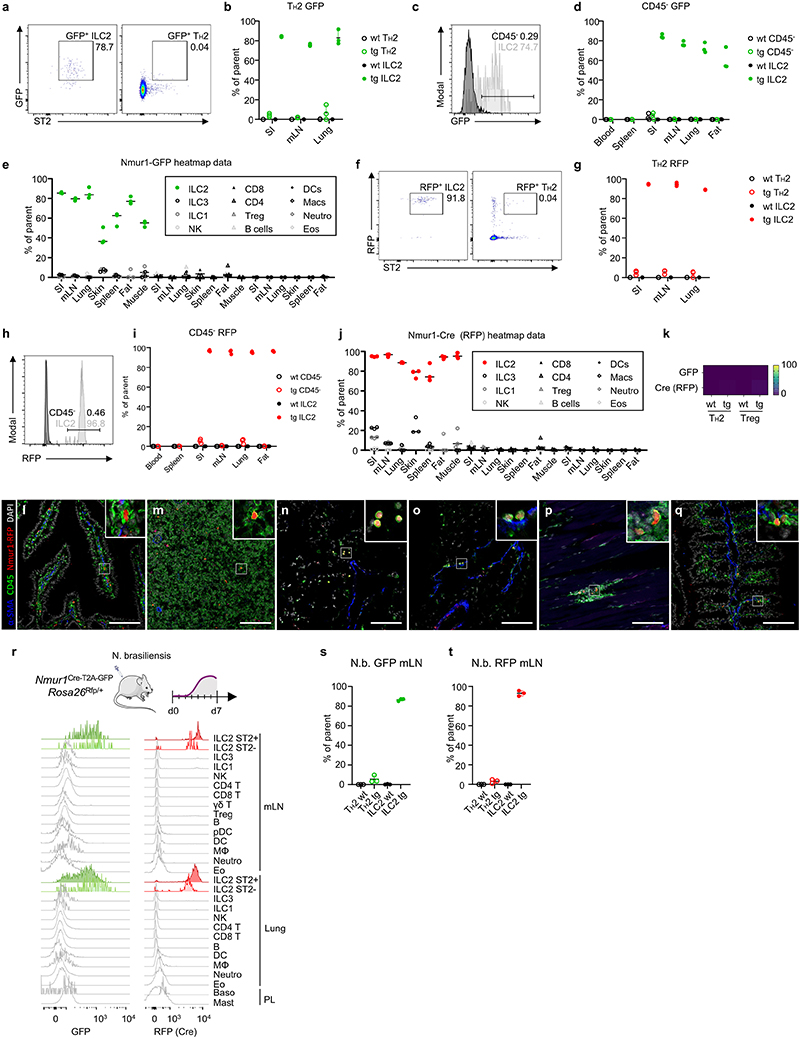
Figure 1. NmurlCre-T2A-GFP faithfully labels ILC2s.
a, Flow cytometric histograms of GFP expression in ILC2s of Nmur1Cre-T2A-GFP (green) and C57BL/6 WT mice (gray) across organs. ILC2s were gated on live CD45+ Lin- (CD3, CD5, CD19) CD127+ and KLRG1+ or ST2+. b, Quantification of ILC2s expressing GFP as in (a). c, Flow cytometric histograms of GFP expression in Nmur1Cre-T2A-GFP mice across immune cell populations in the steady state. d, Heatmap of flow-cytometric quantification of GFP-expressing cells, across immune cell populations and organs. Values represent percentage of positive cells of parental gate, median of three mice. X, not determined. e, Flow cytometric histograms of Cre expression, measured by RFP fluorescence in Nmur1Cre-T2A-GFP x Rosa26Rfp/RfP F1 offspring, in ILC2s across organs in the steady state. f, Quantification of ILC2s expressing RFP as in (e). g, Flow cytometric histograms of Cre (RFP) expression across immune cell populations in the steady state. h, Heatmap of flow-cytometric quantification of Cre-expressing cells, across immune cell populations and organs. SI small intestine, mLN mesenteric lymph nodes, BM bone marrow, PL peritoneal lavage. Each symbol represents data from one mouse, data are representative of two experiments with three mice per group. Mean +/- SD, Student’s t-test, ** p<0.01, *** p<0.001, **** p<0.0001.
Nmur1Cre-T2A-GFP Id2flox/flox and Nmur1Cre-T2A-GFP Gata3flox/flox mice selectively lack ILC2s
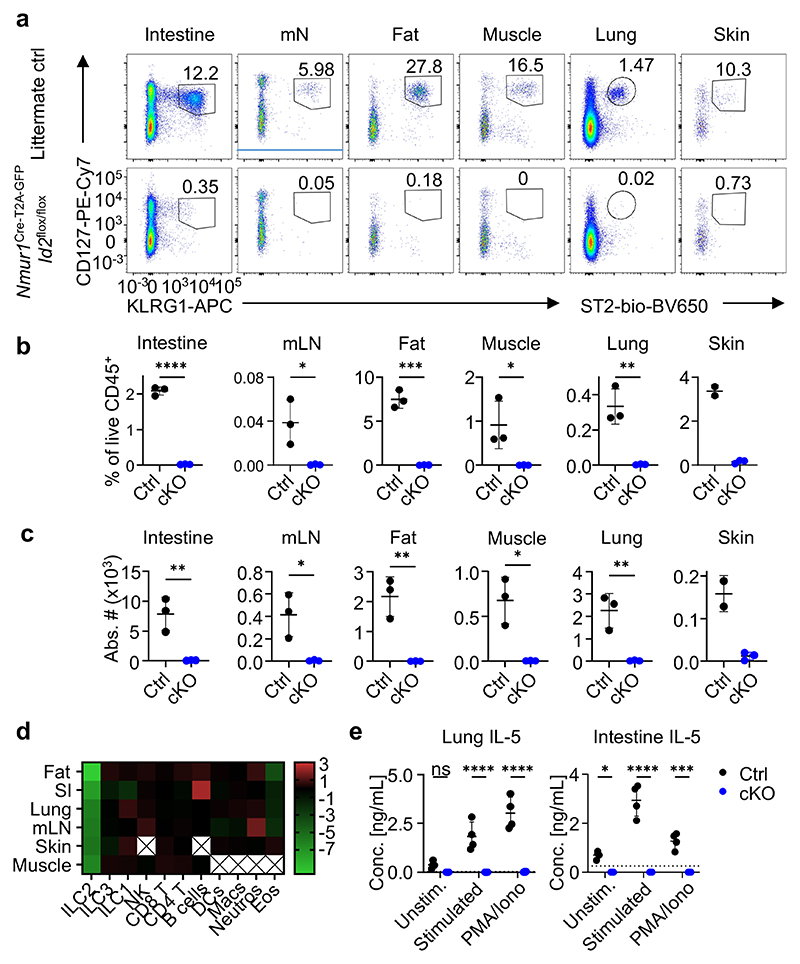
We aimed to generate ILC2-deficient mice by breeding Nmur1Cre-T2A-GFP mice with floxed alleles for Id2 or Gata3, both essential transcriptional regulators for the development and maintenance of ILC2s15,17–19. Indeed, Nmur1Cre-T2A-GFP Id2flox/flox and Nmur1Cre-T2A-GFP Gata3flox/flox mice lacked ILC2s in the small intestine, mesenteric lymph node, adipose tissue, muscle, lung, and skin (Fig. 2a-c; Extended Data Fig.3a,b). We did not detect a reduction in other cell types investigated including NK cells, ILC1s, ILC3s, CD4+ and CD8+ T cells, B cells, macrophages, DCs, neutrophils, TH2 cells, and Treg cells except eosinophils, which will be discussed in the next section (Fig. 2d; Extended Data Fig.3c-e).
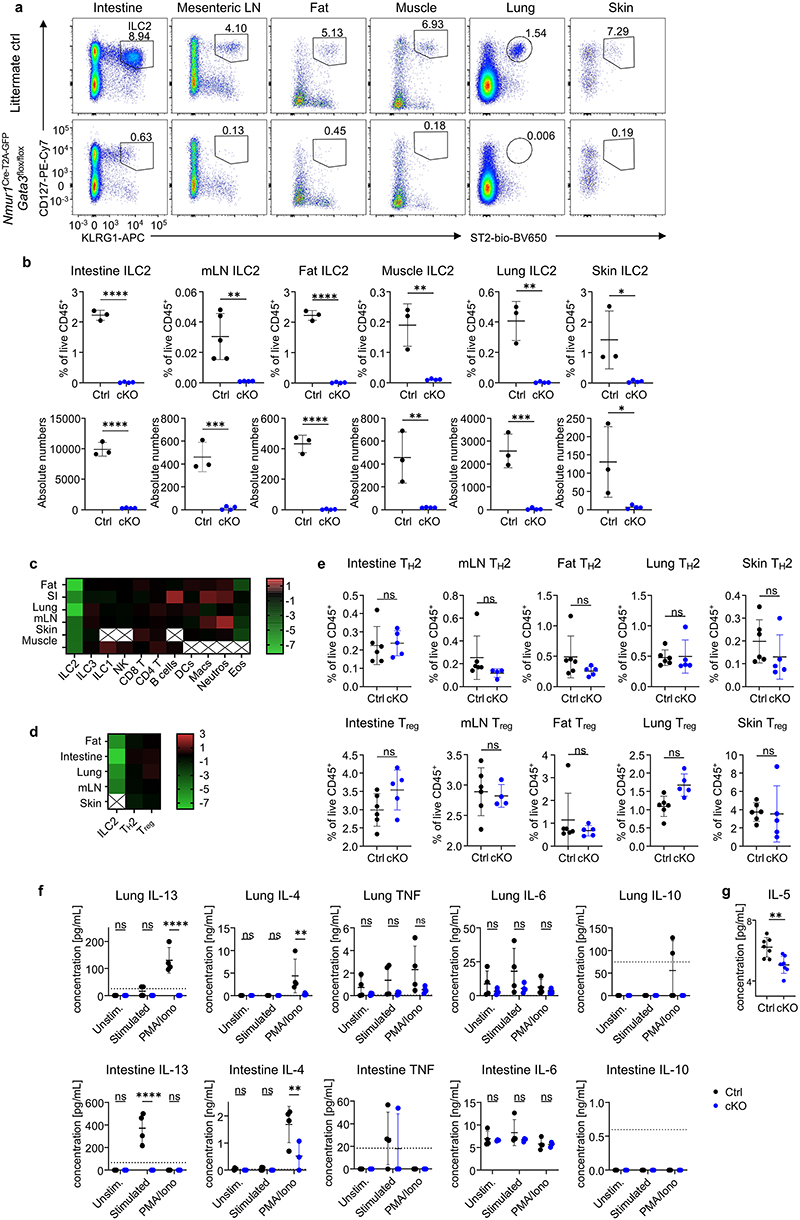
Figure 2. NmurlCre-T2A-GFP Id2flox/flox mice lack ILC2s in all tissues investigated.
a, Flow-cytometric plots of ILC2s gated from live CD45+ Lin- (CD3, CD5, CD19) cells as CD127+ KLRG1+ or CD127+ ST2+ across organs of Nmur1Cre-T2A-GFP Id2flox/floxmice (cKO) and littermate controls (Ctrl). Numbers denote percentage of cells inside the gate. b,c, Quantification of (a), Student’s t-test. d, Heatmap displaying Log2-fold changes of the median cell frequencies among live CD45+ cells across different populations and organs of Nmur1Cre-T2A-GFP Id2flox/flox mice, compared to littermate controls. X, not determined. e, IL-5 secretion measured in the supernatant of cultured immune cells isolated from lung or intestine of Nmur1Cre-T2A-GFP Id2flox/flox mice and littermate controls, stimulated with IL-2, IL-7, IL-25 and IL-33 or PMA/Ionomycin for 8 h. Each symbol represents data from one mouse, mean +/- SD, two-way ANOVA with Šídák’s multiple comparison tests, data are representative of two independent experiments with 3-4 mice per group. ns not significant, *p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. Littermate controls for a-d were Id2flox/+ and Nmur1Cre-T2A-GFP Id2flox/+combined.
ILC2s mediate immune responses to a large degree by rapid secretion of type 2 cytokines after activation, such as interleukin (IL)-5 and IL-13. To investigate if the immediate type 2 cytokine response is affected in Nmur1Cre-T2A-GFP Id2flox/flox mice, we cultured bulk purified leukocytes and either stimulated them in vitro with medium containing cytokines known to bolster type 2 cytokine production, such as IL-2, IL-7, IL-25 and IL-33, or with phorbol 12-myristate 13-acetate (PMA)/Ionomycin and measured cytokines in the supernatant. High levels of IL-5 and IL-13 were measured in the supernatant of control cells, whereas IL-5 and IL-13 were not detectable above baseline in samples from Nmur1Cre-T2A-GFP Id2flox/flox mice (Fig. 2e; Extended Data Fig.3f). We also observed a smaller but significant decrease in IL-4 production but only after PMA/Ionomycin stimulation. We did not detect any significant reduction in secretion of IL-6, IL-10 and TNF, which may be produced by non-ILC2s under these experimental conditions (Extended Data Fig.3f). Consistent with the in vitro data, we detected reduced IL-5 concentration in the serum of Nmur1Cre-T2A-GFP Id2flox/flox mice using a highly sensitive bead-based immunoassay (Extended Data Fig. 3g). These data confirm the absence of ILC2s and impaired IL-5 and IL-13 production in Nmur1Cre-T2A-GFP Id2flox/flox mice. We concluded from these experiments that Nmur1Cre-T2A-GFP Gata3flox/flox and Nmur1Cre-T2A-GFP Id2flox/flox mice specifically lack ILC2s and are a suitable model to investigate ILC2 function in vivo.
ILC2s are essential for eosinophils and type 2 inflammation
We found reduced eosinophil numbers in ILC2-deficient mice in steady state in many organs, including bone marrow, blood, spleen, intestine, mesenteric lymph node, adipose tissue, lung, and skin (Fig. 3a,b; Extended Data Fig.4a). Since eosinophils do not express Nmur1, our gene targeting strategy excludes direct effects on eosinophils (Fig. 1c,g). Thus, the reduction in eosinophil numbers has to be a consequence of the lack of ILC2s in tissues. To test whether the hematopoietic compartment or non-hematopoietic compartment is required for proper eosinophil development, we generated bone marrow chimeras. To this end, we used lethally irradiated Nmur1Cre-T2A-GFP Id2flox/flox (KO) or Id2flox/flox (WT) mice as hosts and reconstituted them with either KO or WT bone marrow resulting in four experimental groups, which had a KO or WT hematopoietic, non-hematopoietic or both compartments. Selectively, the Nmur1Cre-T2A-GFP Id2flox/flox hematopoietic compartment regulated eosinophil numbers (Fig. 3c; Extended Data Fig. 4b). Further, a clear correlation between ILC2 and eosinophil numbers in the tissue of the bone marrow chimeras is supporting the notion that ILC2s regulate eosinophils in tissues (Fig. 3d). ILC2s were reported to be a dominant source of IL-5 in tissues20. In line, ILC2-deficient mice showed decreased IL-5 levels at steady state and failed to secrete IL-5 upon stimulation (Fig. 2e; Extended Data Fig.3g). Furthermore, ILC2-deficient mice phenocopy the effects on eosinophils reported in IL-5-deficient mice (Extended Data Fig.4c)21.
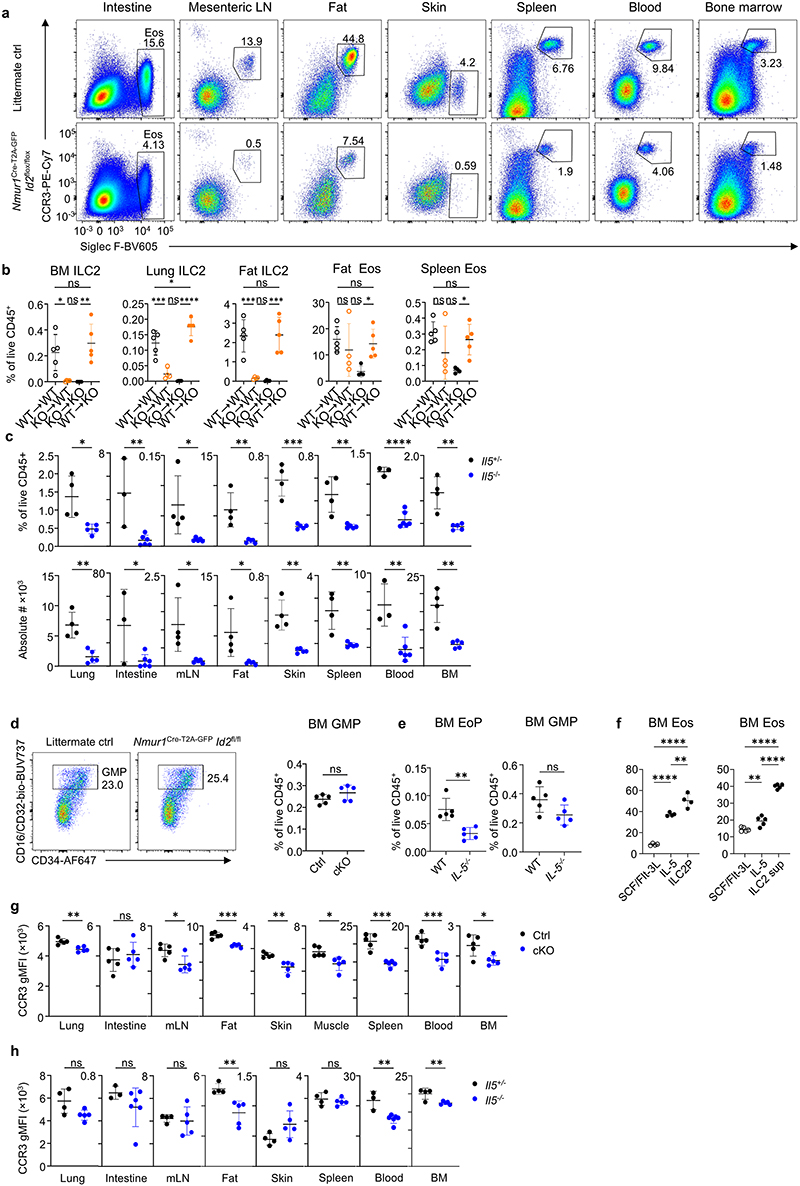
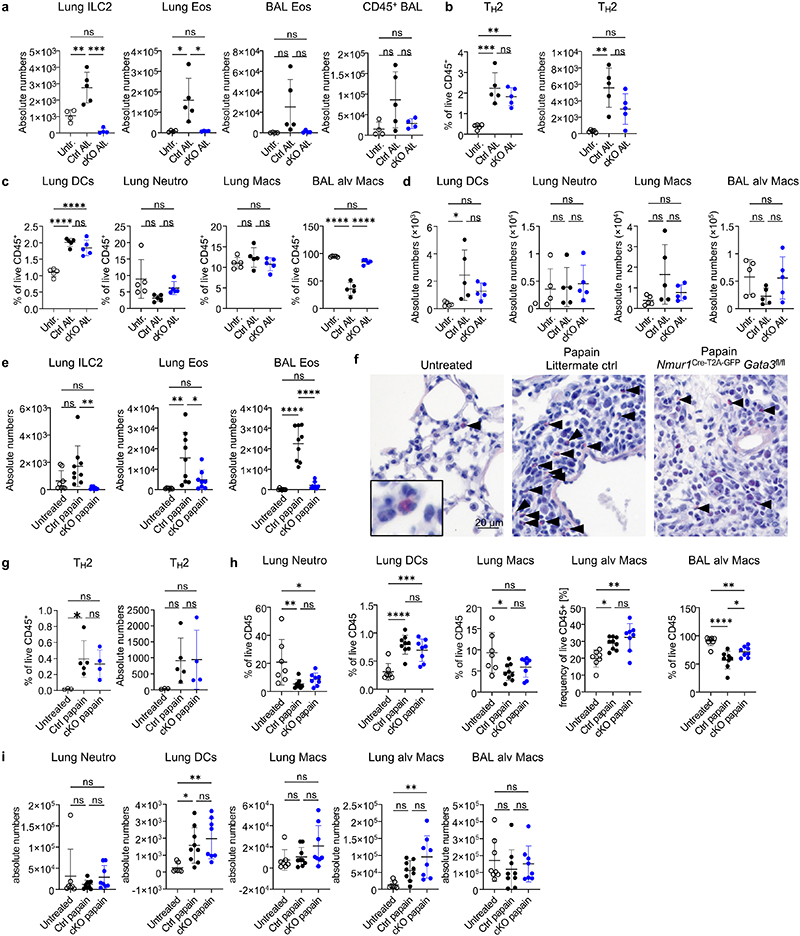
Figure 3. ILC2-deficient mice have reduced eosinophil counts and fail to develop eosinophilia in allergic asthma models.
a, Lung eosinophils in untreated Nmur1Cre-T2A-GFP Id2flox/flox and littermate controls gated from live CD45+Lin- (CD3, CD5, CD19, Ly6G) cells using Siglec F and CCR3. b, Percentage and absolute numbers of eosinophils across organs. c, Eosinophil numbers in bone marrow chimeras of Nmur1Cre-T2A-GFP Id2flox/flox (KO) and Id2flox/flox (WT) recipients transplanted with bone marrow of KO or WT donors. d, Pearsons correlation and linear regression of ILC2s and eosinophils frequencies. R2, coefficient of determination; p, P value. e, Eosinophil-lineage-committed progenitor (EoP) gated from live CD45+ Lin- (CD3, CD5, CD19, Gr1, B220) Sca1- CD34+ and relative quantification in bone marrow of Nmur1Cre-T2A-GFP Id2flox/flox mice compared to littermate controls. f, UMAP displaying cell population clusters of scRNA-sequenced lung myeloid (blue shades) and epithelial (red shades) cells of untreated Nmur1Cre-T2A-GFP Id2flox/flox mice and littermate controls (Id2flox/+ or Id2flox/flox). g, Violin plots of differentially expressed genes in populations marked by triangles in Extended Data figure 5 e-g. h-j, Lung eosinophils of Nmur1Cre-T2A-GFP Id2flox/flox mice and littermate controls, 7 (h,i) or 17 (j) days after allergy induction by intranasal application of Alternaria alternata (h), papain (i), or house dust mite (j) and percentage of the indicated cell subsets. Untreated littermate controls in h and j were Id2flox/flox, papain-treated littermate controls were Id2flox/+ or Id2flox/flox or Nmur1Cre-T2A-GFP Id2flox/+). BAL, bronchoalveolar lavage. Data are representative of the following number of independent experiments and mice per group: four experiments with 3-5 mice (a,b), two experiments with 4-5 mice(c,) 3-5 (e), 3-5 mice (h,j) or pooled from two experiments with 3-4 mice (i). Mean +/- SD, Student’s t-test (b,e) or one-way ANOVA with Tukey’s multiple comparisons (c, h-j), ns not significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
A more detailed analysis of eosinophil development in the bone marrow revealed that eosinophil precursors were diminished in ILC2-deficient mice whereas granulocyte/macrophage progenitors (GMPs) were detected in comparable frequencies in both groups (Fig. 3e; Extended Data Fig. 4d). Similar results were obtained form Il5-/- mice (Extended Data Fig.4e). Supernatant from stimulated ILC2s or co-culture with ILC2 precursors promoted eosinophil differentiation from myeloid progenitors indicating that ILC2s control early eosinophil development (Extended Data Fig. 4f). Moreover, mature eosinophils of Nmur1Cre-T2A-GFP Id2flox/flox and of Il5-/- mice had reduced expression of the eotaxin receptor CCR3 in many tissues suggesting further effects of ILC2s on eosinophil maturation and recruitment to tissues (Extended Data Fig. 4g,h). Collectively, our data demonstrate that ILC2s are required for the development and maturation of eosinophils at steady state.
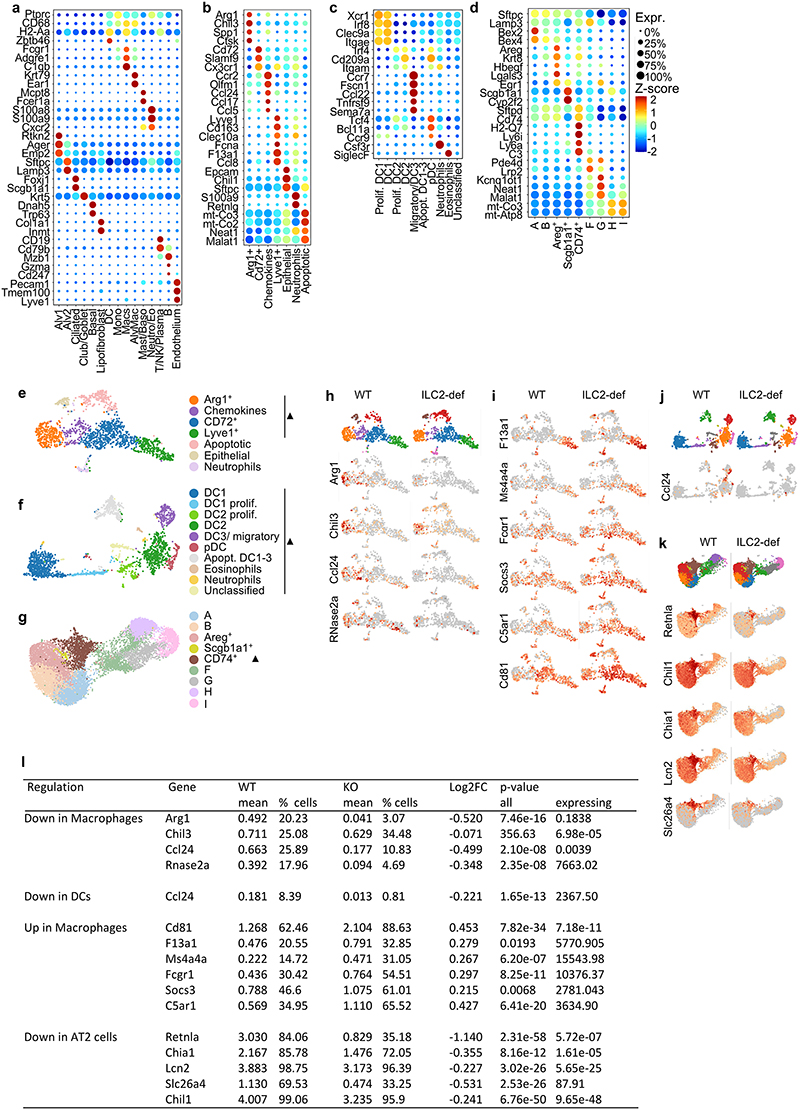
To explore how ILC2s mediate immunoregulation in tissues, we performed single-cell sequencing of myeloid cells and epithelial cells from the lung of ILC2-deficient and control mice. We obtained clustering and annotated myeloid and epithelial cell subsets according to signature genes for these populations (Fig. 3f,g; Extended Data Fig. 5a-l). In total, the absence of ILC2s in the lung affected the expression of immunoregulatory genes linked to type 2 immunity in both myeloid and epithelial subsets. For example, we found diminished expression of Ccl24 (eotaxin-2) in macrophages and dendritic cells, indicating that ILC2s could also mediate effects on eosinophil recruitment indirectly via activation of other cells secreting eotaxins. Further, ILC2-deficiency altered gene expression in macrophages leading to reduced expression of genes associated with an M2 macrophage phenotype, such as Arg1, Ccl24 and Chil3 (YM1) (Fig.3g; Extended Data Fig. 5e-l). Moreover, we detected downregulation of transcripts linked to type 2 inflammation, such as Retnla (Relm-α), Chia1 (AMCase) and Slc26a4 in type 2 pneumocytes (Fig.3g; Extended Data Fig. 5e-l). Altogether, these data indicate that ILC2s maintain multiple signaling pathways with hematopoietic and non-hematopoietic cells in the lung to orchestrate type 2 inflammation.
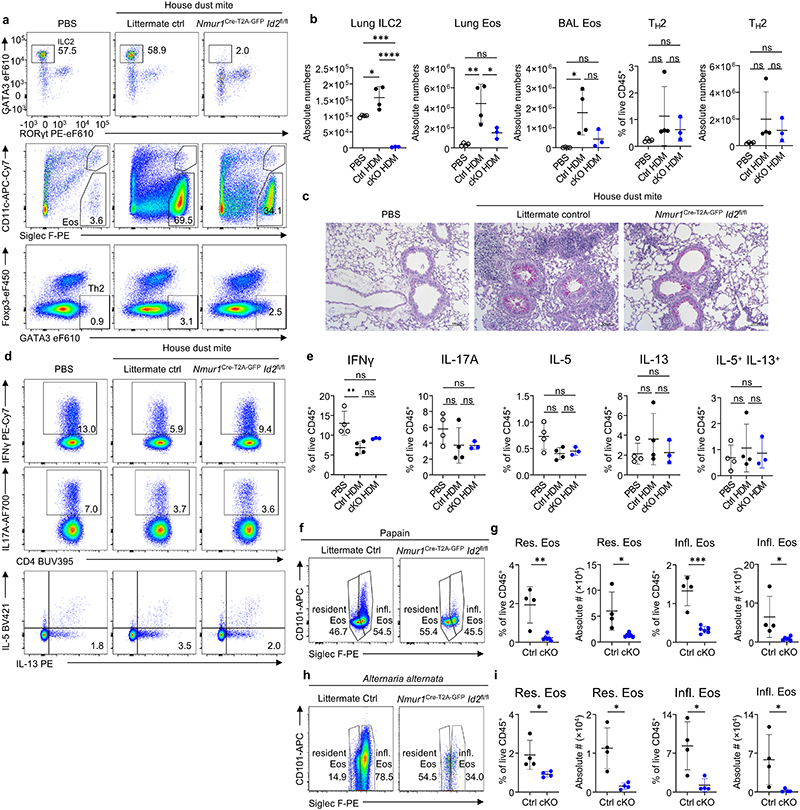
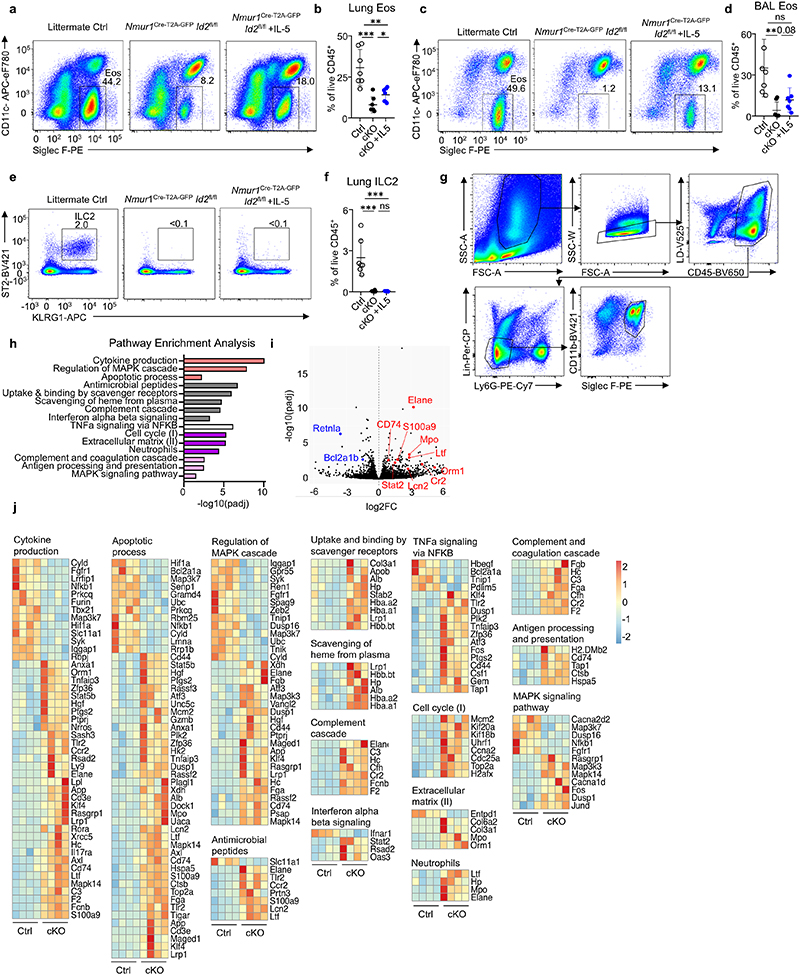
Considering the effects of ILC2-deficiency on immune homeostasis in the lung at steady state, we aimed to investigate how ILC2s regulate type 2 inflammation in mouse models of allergic asthma. To this end, we provoked allergic lung inflammation by administration of fungal extract of Alternaria alternata, the protease allergen papain or house dust mite (HDM) extract in ILC2-deficient and control mice. Allergen treatment resulted in strong eosinophilia in bronchoalveolar lavage (BAL) and lung tissue in control mice. However, ILC2-deficient mice lacked eosinophilia, with eosinophil numbers remaining at steady state levels in the BAL and lung despite comparable proportions of TH2 cells (Fig. 3 h-j; Extended Data Fig. 6; Extended Data Fig. 7a-e). Further, ILC2-deficiency resulted in a significant reduction of both resident Siglec Flow CD101- and inflammatory Siglec Fhigh CD101+ eosinophils (Extended Data Fig. 7f-i). Injection of recombinant IL-5 in ILC2-deficient mice resulted in a partial rescue of eosinophil numbers indicating that ILC2-derived IL-5 contributed to eosinophil recruitment in allergic lung inflammation (Extended Data Fig. 8a-f).
To explore the signaling pathways in eosinophils controlled by ILC2s, we performed bulk RNA-sequencing of eosinophils from papain-treated mice deficient or sufficient in ILC2s. Pathway analysis based on differentially regulated genes revealed that eosinophils from ILC2-deficient mice have enrichment in pathways, such as neutrophils, antimicrobial peptides, binding and uptake of ligands by scavenger receptors, complement cascade and antigen processing and presentation (Extended Data Fig. 8g-j). Upregulation of neutrophil primary granule proteins was reported during eosinophil maturation22. Therefore, these data suggest that eosinophils fail to terminally mature in the absence of ILC2s and express neutrophil effector genes, such as Elane, Lcn2, Mpo and Ltf. We detected additional alterations in pathways such as interferon-, TNF- and MAPK-signaling, cell cycle, apoptosis and cytokine production indicating that the lack of ILC2-derived signals changes multiple basic biological processes in eosinophils associated with proliferation, maturation, activation and survival (Extended Data Fig. 8g-j). Together these data provide evidence for a non-redundant role of ILC2s in allergic lung inflammation and expose that the eosinophil response is strictly dependent on the presence of ILC2s in the tissue at steady state and during type 2 inflammation.
ILC2-deficient mice are highly susceptible to worm infections
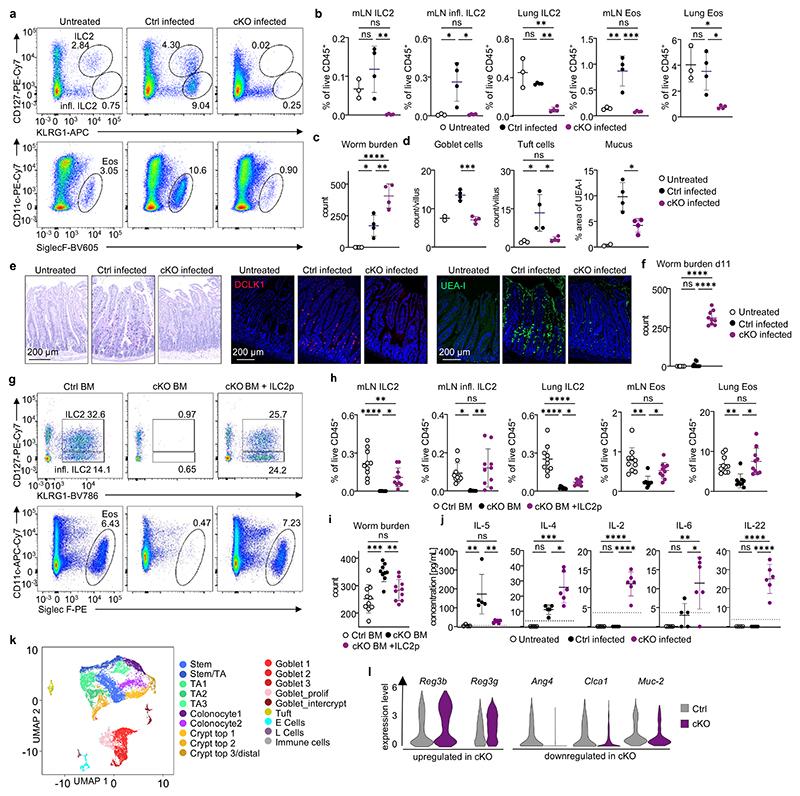
To test whether ILC2s play a non-redundant role in resistance to helminth infection in lymphoreplete mice, we infected ILC2-deficient mice and control mice with N. brasiliensis. As expected, natural and inflammatory ILC2s remained undetectable following worm infection in Nmur1Cre-T2A-GFP Id2flox/flox and Nmur1Cre-T2A-GFP Gata3flox/flox mice but not in control mice (Fig. 4a,b, Extended Data Fig. 9d,e). Consistent with our data in steady state and in allergic asthma models, the eosinophil influx in tissues was totally abrogated in ILC2-deficient mice, with eosinophil frequencies not exceeding those of naïve mice (Fig. 4a,b, Extended Data Fig. 9d,e) whereas TH2 frequencies were comparable to infected control (Extended Data Fig. 9a).-We further measured higher neutrophil counts in the lung of ILC2-deficient mice, suggesting a stronger unresolved inflammation (Extended Data Fig. 9b,c). Both Nmur1Cre-T2A-GFP Id2flox/flox and Nmur1Cre-T2A-GFP Gata3flox/flox mice were more susceptible to N. brasiliensis infection compared to their littermate controls measured by higher worm burden in the intestine on day 7 after infection (Fig. 4c; Extended Data Fig. 9f). In addition, ILC2-deficient mice failed to trigger an epithelial response to control the worm infection characterized by the lack of goblet- and tuft cell hyperplasia and consequently decreased mucus production as a sign of the impaired type 2 immune response (Fig. 4d,e; Extended Data Fig. 9g,h). The high susceptibility of ILC2-deficient mice to N. brasiliensis infection persisted at day 11 post-infection when control mice had already expelled the worm. At that time point, the intestine of ILC2-deficient mice was still filled with worms and the type 2 immune response was altered (Fig. 4f; Extended Data Fig. 9i-m). We concluded from these experiments that ILC2s are not only strictly required for the initial worm control and induction of type 2 inflammation but also for worm expulsion.
Figure 4. ILC2-deficient mice are susceptible to N. brasiliensis infection.
a,b Natural and inflammatory ILC2s and eosinophils in the mLN of untreated WT animals and infected Nmur1Cre-T2A-GFP Id2flox/flox ILC2-deficient mice (cKO) and littermate controls (Ctrl). b, Relative ILC2- and eosinophil numbers in mLN and lung. c, Worm burden in untreated WT animals and infected Nmur1Cre-T2A-GFP Id2flox/flox ILC2-deficient mice or littermate controls. d, Goblet- and tuft cell numbers per villus and quantification of UEA-I+ pixels. e, Periodic acid-Schiff staining of murine small intestine seven days after infection (left). Immunofluorescence micrographs of murine small intestine, tuft cells (Dclk1, red, middle) mucus (UEA-I, green, right) and DAPI (blue). Data in a-e are representative of two independent experiments with 3-4 mice per group. f, Worm burden as in (c), 11 days after infection. g-i, Infection of BM chimeras with Nippostrongylus brasiliensis. C57BL/6 recipients were transplanted with bone marrow of Id2flox/flox (Ctrl), Nmur1Cre-T2A-GFP Id2flox/flox (cKO) donors, or bone marrow of Nmur1Cre-T2A-GFP Id2flox/flox(cKO) donors supplemented with sorted ILC2 precursors from C57BL/6 donors (cKO + ILC2p). g, ILC2s and eosinophils in the mLN. h, ILC2s and eosinophils in mLN and lung. i, Worm burden 7 days after infection. Data are representative of two independent experiments with 3-6 mice per group. j, Serum cytokines 7 days after infection as in a-e. Dotted lines indicate detection limits. a-j One-way ANOVA with Tukey’s multiple comparisons. ns not significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. k, UMAP displaying cell population clusters of scRNA-sequencing of colon epithelium from N.b. infected Nmur1Cre-T2A-GFP Id2flox/flox mice and littermate controls. l, Differentially expressed genes in colonic epithelial cells of infected Nmur1 Cre-T2A-GFP Id2flox/flox mice (cKO) compared to littermate controls (Ctrl) in clusters expressing these genes (see extended data 10). Untreated littermate controls in (a-l) were C57BL/6, N. b.-infected littermate controls were Id2flox/+ or Nmur1Cre-T2A-GFP Id2flox/+).
To more directly scrutinize if ILC2s are the cell type mediating anti-helminth immunity, we aimed to reconstitute ILC2s in Nmur1Cre-T2A-GFP Id2flox/flox mice. To this end, we reconstituted lethally irradiated B6 WT mice with Id2flox/flox or Nmur1Cre-T2A-GFP Id2flox/flox bone marrow and included a group, in which Nmur1Cre-T2A-GFP Id2flox/flox bone marrow was supplemented with sort-purified ILC2ps15. Mice reconstituted with Nmur1Cre-T2A-GFP Id2flox/flox bone marrow failed to control N. brasiliensis infection compared to mice reconstituted with Id2flox/flox bone marrow. However, mice reconstituted with Nmur1Cre-T2A-GFP Id2flox/flox bone marrow supplemented with sort-purified ILC2ps showed an increased type 2 immune response and comparable resistance to worm infection as in Id2flox/flox chimeras (Fig. 4g-i; Extended Data Fig. 9n-r). Altogether, these data provide strong evidence for a non-redundant role of ILC2s in triggering the type 2 response and resistance to worm infection.
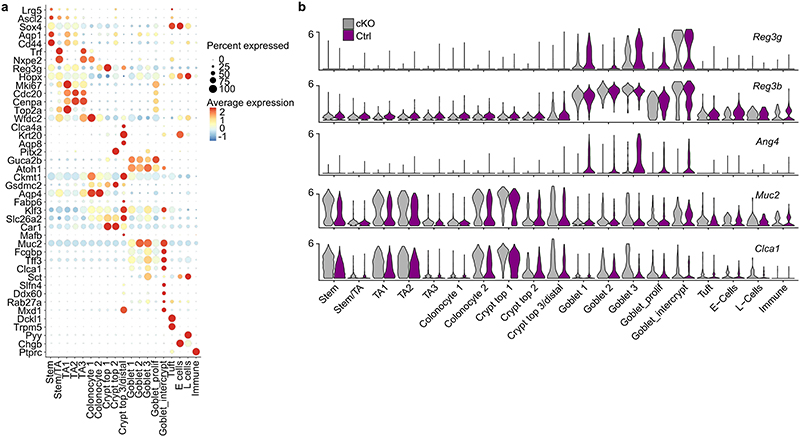
Consistent with a defective ILC2 response, IL-5 levels in the serum were significantly reduced, whereas other cytokines not associated with type 2 immunity, such as IL-2, IL-6 and IL-22 were upregulated in ILC2-deficient mice (Fig. 4j). Interestingly, these cytokines are linked to type 3 immune responses and RORγt+lymphocytes, including ILC3s23–26, which protect the epithelial barrier from bacterial, viral and fungal infections via secretion of antimicrobial peptides in epithelial cells, such as Reg3β and Reg3γ regulated by IL-22 (Fig. 4j; Extended Data Fig. 9s)27–29. Single-cell sequencing revealed that Reg3β and Reg3γ were upregulated in the epithelium of ILC2-deficient mice, whereas genes such as Ang4, Clca1 and Muc2 associated with worm expulsion, goblet cells and type 2 immunity were downregulated (Fig. 4k,l; Extended Data Fig. 10a,b)30. Collectively, these data indicate that ILC2-dependent defects are critical for proper type 2 immunity at mucosal barriers and important for resistance against helminth infections. Lack of ILC2s resulted not only in a defective myeloid and epithelial immune response but also, due to the missing counter-regulation of type 3 immunity, in an exaggerated, frustrated type 3 immune response characterized by increased expression of IL-22 and epithelial response genes.
Research of more than a decade has witnessed the emergence of ILCs as a complementary innate system to T cells with evolutionary conserved transcriptional programs. Given the overlap in immune effector molecules between ILCs and T cells, it is tempting to scrutinize if the lack of ILCs could be compensated by other hematopoietic cells resulting in measurable but redundant functions of ILCs in tissues. Undeniably a substantial amount of work investigating ILC function was carried out in lymphopenic mice lacking the adaptive immune system and the results obtained there could overestimate the importance of ILCs in lymphoreplete mice. However, using a novel mouse model for genetic ablation of ILC2s, this study now clearly describes non-redundant functions of ILC2s. ILC2s are essential for eosinophil homeostasis and lack of ILC2s results in an attenuated type 2 immune response with functional relevance in resistance to helminth infection and allergic asthma, making ILC2s a suitable target for therapeutic intervention.
Methods
Mouse strains
C57BL/6 mice (Mus musculus) were purchased from Janvier. Nmur1Cre-T2A-GFP, Id2flox/flox 31, Gata3flox/flox 32, Il5-/-20, and Rosa26Rfp/Rfp33 on a C57BL/6 background were bred locally at Charité. To generate Nmur1Cre-T2A-GFP mice, a P2A-iCre-T2A-eGFP-SV40pA cassette followed by a kanamycin resistance cassette surrounded by two Frt sites was inserted in BAC RP23-403I3 in exon 2 and 3 of Nmur1 via homologous recombination. The kanamycin cassette was removed by Flp mediated recombination. A LoxP site and a LoxP511 site contained in the BAC were also removed via homologous recombination. The modified BAC was injected in C57BL/6 fertilized eggs and two founders were obtained with the help of Cyagen. Sex and age-matched male and female animals, usually aged 7-14 weeks, were used for experiments. Id2flox/flox mice were used as littermate controls for Nmur1Cre-T2A-GFP Id2flox/flox animals, and Gata3flox/flox mice were used as littermate controls for Nmur1Cre-T2A-GFP Gata3flox/flox animals if not otherwise indicated.-We did not use blinding or randomization to assign animals to experimental groups. The sample size of experimental groups was not determined by statistical methods. All animal experiments were approved and are in accordance with the local animal care committees (LAGeSo Berlin, Institutional Animal Care and Use Committee at Weill Cornell Medicine).
Cell Isolation
Small intestine or colon was removed, cleaned from remaining fat tissue and washed in ice-cold PBS. Peyer’s patches were eliminated, small intestine was opened longitudinally and washed in ice-cold PBS. Dissociation of epithelial cells was performed by incubation on a shaker at 37°C in HBSS (Sigma-Aldrich) containing 10 mM Hepes (Gibco) and 5 mM EDTA (Roboklon) two times for 15 min. After each step, samples were vortexed and the epithelial fraction discarded. Afterwards, remaining tissue was chopped into small pieces and enzymatic digestion was performed using dispase (0.5 U/ml; Corning), collagenase D (0.5 mg/ml; Roche) and DNaseI (100 μg/ml; Sigma-Aldrich). Leukocytes were further enriched by Percoll gradient centrifugation (GE Healthcare). Lungs were chopped and incubated in the enzyme cocktail described above for 40 min on a shaker at 37°C. The remaining tissues were mashed with a syringe plunger and single cell suspensions were filtered through a 70 μm cell strainer. Leukocytes were then further enriched by Percoll gradient centrifugation. Mesenteric lymph nodes and spleens were chopped and incubated in RPMI 1640 medium (Gibco) supplemented with 1% BSA (Sigma-Aldrich), collagenase II (1 mg/ml; Sigma-Aldrich) and DNaseI (100 μg/ml) for 20 min on a shaker at 37°C. Afterwards, cells were dissociated using a pasteur pipette, and filtered through a 70 μm cell strainer. Epididymal white adipose tissue was removed and incubated in the same digestion buffer for 45 min on a shaker at 37°C. After incubation, cells were dissociated using a Pasteur pipette, filtered through a 70 μm cell strainer, spun down and the adipocyte layer was aspirated. Muscle tissue was treated similar to mesenteric lymph node and spleen but with double the concentration of DNaseI and collagenase II, 30 min incubation time on a shaker at 37°C and Percoll gradient purification. For skin preparation, ears were removed, split and put dermal side down in DMEM supplemented with Liberase TL (0.5 mg/ml; Roche) at 37°C for 1.5 h. The tissue was then mashed through a 70 μm cell strainer and the leukocytes further enriched by Percoll gradient centrifugation.
Flow cytometry and cell sorting
Dead cells were routinely excluded with Fixable Aqua Dead Cell Stain or SYTOX Blue Dead Cell Stain (Thermo Fisher Scientific). Single cell suspensions were incubated on ice with anti-CD16/CD32 antibody and the following conjugated antibodies in PBS (Ca2+ and Mg2+-free, Sigma-Aldrich). If indicated, lineage-positive cells were excluded by staining for CD3ε (145-2C11 or 500A2), CD5 (53-7.3), CD19 (1D3 or 6D5), FcεRI (Mar-1) and Ly6G (1A8). For surface staining the following antibodies were used: KLRG1 (2F1 or MAFA), CD45 (30-F11) or CD45.2 (104), CD4 (GK1.5 and RM4-5), Sca-1 (D7), c-Kit (2B8), CD127 (A7R34), ST2 (RMST2-33), CD90 (30H12), NK1.1 (PK136), NKp46 (29A1.4), CCR6 (29-2L17), CD11b (M1/70), CD11c (N418), Siglec F (E50-2440), F4/80 (BM8), CD64 (X54-5/7.1), CD317 (927), B220 (RA3-6B2), CCR3 (J073E5), Flt3 (A2F10), α4β7 integrin (DATK32), PD-1 (29F.1A12), TCRγδ (GL3), CD25 (PCG1.5), CD103 (2E7), CD101 (REA301, Miltenyi), Gr1 (RB6-8C5), CD16/CD34 (93), IL-5Rα (T21), CD34 (RAM34), Foxp3 (FJK-16s), GATA3 (TWAJ), T-bet (4B10), RORγt (B2D), IFN-γ (XMG1.2), IL17A (eBio17B7), IL-5 (TRFK5), IL-13 (eBio13A) and IL-22 (IL22JOP). All antibodies used in flow cytometry were purchased from eBioscience, Biolegend or BD Biosciences if not otherwise indicated. All flow cytometry experiments were acquired using a custom configuration Fortessa flow cytometer and the FACS Diva software (BD Biosciences) and were analyzed with FlowJo V9.9.3 or V10.6.2 software (TreeStar) or sort-purified by using a custom configuration FACSAria cell sorter (BD Biosciences).
Quantitative real-time PCR
Sorted cells were homogenized in Trizol (Thermo Fisher Scientific) and stored at -80°C. RNA was extracted with chloroform and RNA concentration was determined using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific). Reverse transcription of total RNA was performed using the High Capacity cDNA Reverse Transcription kit according to the protocol provided by the manufacturer (Thermo Fisher Scientific). Reaction was detected on a QuantStudio 5 Real-Time PCR (Thermo Fisher Scientific) using Taqman Gene Expression Assay (Applied Biosystems) with Nmur1(Mm00515885_m1). Gene expression was normalized to the housekeeping gene Hprt1(Mm00446968_m1).
Helminth infection and allergic asthma induction
Third-stage larvae (L3) of N. brasiliensis were purified with a Baermann apparatus. After washing three times in PBS, larvae were counted and 500 purified larvae were injected subcutaneously in PBS. Mice were killed, organs were analyzed and worm burden was determined in the small intestine 7 or 11 days post infection.
For bone marrow chimeras, recipient mice were preconditioned with the Sulfatrim diet (200 mg trimethoprim and 40 mg sulfamethoxazole) for a week and irradiated with 11 Gy from an X-ray irradiator (Rad Source Technologies) in two rounds of 5.5 Gy irradiation with a 2-hour break between the doses. The irradiated mice were rested for at least 2 hours before reconstitution. For the ILC2 progenitor (ILC2p) add-back bone marrow chimera mice, full bone marrow (BM) cells were collected from the femur, tibia, and pelvis bones from Id2flox/flox mice, Nmur1Cre-T2A-GFP Id2flox/flox mice by flushing. For ILC2p cells, BM cells harvested from B6 WT mice were crudely enriched by using Dynabeads™ Untouched™ Mouse CD4 Cells Kit (Thermo Scientific) and FACS sorted for CD45+Lin− Sca1+CD127+CD25+ ILC2p cells (Lin:CD3, CD5, CD19, FceRIa, NK1.1, CD19, and Ly6G). The irradiated recipient mice received either 2x106 Id2flox/flox BM cells, 2x106, Nmur1Cre-T2A-GFP Id2flox/flox BM cells, or 2x106 Nmur1Cre-T2A-GFP Id2flox/flox BM cells together with 5x104 ILC2p cells intravenously. The recipient mice were reconstituted for at least 8 weeks before analysis. During the reconstitution, the recipient mice were maintained on the Sulfatrim diet for two weeks, followed by 6 weeks on the regular diet.
For allergic asthma induction, 30 μg of Papain (Roche) or 10 μg Alternaria Alternata extract (Greer Laboratories Inc.) in PBS were administered intranasally on three consecutive days. Mice were killed seven days after initial administration, organs were collected and analyzed. In some experiments recombinant murine IL-5 (250 ng, R&D Systems) in 200 μl PBS or 200 μl PBS were injected i.p. every other day.
House dust mite extract-induced model of allergic airway inflammation
Mice were sensitized to allergic responses by intranasal administration of house dust mite (HDM) extract (Der p 1, Greer Laboratories Inc.)34. Briefly, mice were anesthetized with isoflurane (Isothesia, Henry Schein Animal Health) and sensitized by 25 μg (protein content) in 25 μl of PBS for 3 consecutive days (days 0, 1, 2), and challenged after two weeks with 5 μg (protein content) in 25 μl of PBS for 3 days (days 14, 15, 16)34. Mice were killed one day after the last challenge for analysis.
Generation of bone marrow chimeras
Mice were irradiated with 11 Gy split in two doses of 5.5 Gy Gammacell 40 (MDS Nordion) and a 4 h break between the two cycles. Afterwards mice were reconstituted with full bone marrow (2x106 cells) from Nmur1Cre-T2A-GFP Id2flox/flox mice or Id2flox/flox mice. Baytril (2.5%) was delivered 1 week before and 2 weeks after bone marrow transplantation and mice were used for experiments 8 weeks after transplantation.
Cytokine measurement
Bulk purified immune cells were incubated in DMEM with high glucose supplemented with 10% FCS, 10 mM Hepes, 1 mM sodium pyruvate, non-essential amino acids, 80 μM 2-Mercaptoethanol, 2 mM Glutamine, 100 U/ml Penicillin and 100 μg/ml Streptomycin (all from Gibco) in 96-well U-bottom microtiter plates (Nunc) for 8 h at 37°C and 5% CO2. If indicated, the culture was supplemented with a cocktail of IL-2, IL-7, IL-25, IL-33 (Biolegend, 100 ng/ml, each) or phorbol 12-myristate 13-acetate (PMA, 1 μg/ml) and ionomycin (Invitrogen, 0.5 μg/ml).
Cytokine concentration in culture supernatant or serum samples were determined by using the LEGENDplex murine Th Cytokine Panel V03 and LEGENDplex murine TH2 Panel V03 multiplex beads-based assay (Biolegend) or the BD Cytometric Bead Array Mouse IL-5 Enhanced Sensitivity Flex Set (BD Biosciences) according to the manufacture’s protocol. Samples were recorded on a custom configuration Fortessa flow cytometer and the FACS Diva software (BD Biosciences) and the flow cytometry data files were analyzed using the Legendplex cloud-based analysis software suite (Biolegend).
In vitro bone marrow cell culture
Bone marrow (BM) cells were isolated from femurs, tibia and pelvis of C57BL/6 mice. BM cells were cultured in vitro in RPMI 1640 medium (Gibco) supplemented with 20% FBS (Sigma-Aldrich), 1% penicillin-streptomycin, 2 mM of L-glutamine, 25 mM of HEPES and 1% nonessential amino acids, 1 mM sodium pyruvate (all from Gibco), and 0.05 mM 2-ME (Sigma-Aldrich). Stem cell factor (SCF; 100 ng/ml; PeproTech) and FLT3 ligand (FLT3-L; 100 ng/ml: PeproTech) were added to the culture. After 3 days, BM cells were cultured in 96-well plate (100,000 cells each well) in 200 μL with one of the following conditions: SCF (100 ng/mL) and FLT3-L (100 ng/mL), IL-5 (10 ng/mL; Biolegend), ILC2 progenitors or supernatant from cytokine-stimulated ILC2s. ILC2 progenitors were sort-purified from bone marrow of C57BL/6N mice as lineage-CD127+Sca1+CD25+ST2+ cells and stimulated with a cytokine cocktail (IL-33, IL-25, IL-7; 100 ng/mL, respectively). Sorted single and viable lung ILC2s defined as lineage-CD90+CD127+CD25+ST2+ cells were cultured for 10 days in the presence of IL-2, IL-7, IL-25 and IL-33 (each at 10 ng/mL).
Bulk RNA-seq analysis
Eosinophils were sort-purified as Sytox blue- CD45+ Lineage (CD3, CD5, CD19, Ly6G)-CD11b+Siglec F+ from the lung of Nmur1Cre-T2A-GFP Id2flox/flox and control mice on day 7 after papain administration. Cells were sorted into RLT Plus Lysis Buffer supplemented with 40 mM DTT and RNA was isolated using the RNeasy Plus Micro kit (Qiagen) according to the protocol provided by the manufacturer. RNA-seq libraries were prepared by the MDC BIMSB Core Bioinformatic Facility using the Ovation® SoLo RNA-Seq Systems (Tecan Genomics) or theSMARTer Stranded Total RNA-Seq Kit – Pico (Takara). Sequencing was performed on a NovaSeq 6000 (Illumina), yielding 100 bp single-end reads. RNA-Seq reads were mapped to the mouse genome (GRCm38.p5) with STAR35 version 2.7.3a using default parameters. Reads were assigned to genes with FeatureCounts36 with the following parameters: -t exon -g gene_id, gene annotation - Gencode GRCm38/M12. The differential expression was carried out with DESeq2 version 1.22.137 using default parameters. We kept genes that have at least 5 reads in at least 3 samples.
Single cell RNA- sequencing of lung myeloid and epithelial cells
Briefly, lung and trachea were chopped and digested for 30 minutes with DNaseI (100 μg/ml; Sigma-Aldrich), Liberase TM (0.5 mg/ml; Roche) and 2 μg/ml actinomycin D at 37°C on a shaker. Afterwards, cells were dissociated using a pasteur pipette, and mashed through a 70 μm cell strainer. Cells were counted after trypan blue exclusion and the same number of cells from four mice per group were pooled. Myeloid cells were sort-purified using live CD45+ (CD11c and/or CD11b)+, epithelial cells were sorted as live CD45-/bEpCAM+.
Library preparation and sequencing
Single-cell sequencing was performed as previously described in Ferreira-Gomes et al. 202138. Briefly, myeloid cells from Nmur1Cre-T2A-GFP Id2flox/floxmice were pooled with epithelial cells from littermate controls and vice versa, and applied to the 10X Genomics workflow for cell capturing and scRNA gene expression (GEX) library preparation using the Chromium Single Cell 5’ v2 Library & Gel Bead Kit (10x Genomics). Final GEX libraries were obtained after fragmentation, adapter ligation, and final Index PCR using the Single Index Kit TT Set A. Qubit HS DNA assay kit (Life Technologies) was used for library quantification and fragment sizes were determined using the Fragment Analyzer with the HS NGS Fragment Kit (1–6000bp) (Agilent). Sequencing was performed on a NextSeq2000 device (Illumina) using a NextSeq 1000/2000 P2 reagent (200 cycles) with the recommended sequencing conditions for 5’ GEX libraries (read1: 26nt, read2: 90nt, index1: 10nt, index2: 10).
Single-cell transcriptome
Raw sequence reads were processed using cellranger (version 5.0.0), including the default detection of intact cells. Mkfastq and count were used in default parameter settings for demultiplexing and quantification of gene expression. Refdata-cellranger- mm10-1.2.0 was used as reference. The number of expected cells was set to 3000.
The cellranger output was analyzed in R (version 4.0.3) using the Seurat package (version 4.0.2). The single cell transcriptomic profiles for 12000 KO cells and 12000 WT cells were integrated, normalized, variable genes were detected and a uniform manifold approximation and projection (UMAP) was performed in default parameter settings using FindIntegrationAnchors, IntegrateData, FindVariableGenes, RunPCA and RunUMAP with 30 principle components. Expression values are represented as ln (10,000 * UMIsGene)/UMIsTotal +1). Similar clusters were identified using shared nearest neighbor (SNN) modularity optimization, SNN resolutions ranging from 0.1 to 1.0 in 0.1 increments were computed, or gating was performed manually using the Loupe Browser (10X Genomics). Subsequently, clusters were annotated by projection of indicated genes on the UMAP to assign different cell types. Signature genes were identified using FindAllMarkers in default parameter settings. Heatmaps are based on z-transformed expression values for genes with significant differences to means in different clusters as judged by a Bonferroni corrected P-value (Wilcoxon rank sum Test) below 0.01 and a minimal absolute fold-change to the mean of log2(1.3).
Single cell RNA-sequencing of colon epithelial cells
Cell isolation
For isolation of the intestinal epithelium for sort-purification, the intestine was prepared and cleaned as described above. The intestine was incubated in PBS supplemented with 30 mM EDTA (Roboklon) and 1.5 mM DTT (Sigma-Aldrich) for 20 min on ice. Next, the tissue was transferred to prewarmed PBS/30 mM EDTA and incubated on a shaker at 37°C for 10 minutes and vortexed afterwards to remove the epithelium. The epithelium was washed in PBS/10% FCS and afterwards incubated in HBSS (Sigma-Aldrich) supplemented with dispase (0.5 U/ml; Corning) on a shaker at 37°C for 10 minutes and another 5 minutes after addition of FCS (1% final concentration) and DNaseI (100 μg/ml; Sigma-Aldrich) and filtered through a 70 μm and a 40 μm cell strainer.
For single-cell sequencing, the epithelium was sort-purified (Sytox blue- CD45-/lowEpCam+) from the large intestine of Nmur1Cre-T2A-GFP Id2flox/floxand control mice on day 7 after N. brasiliensis infection.
Library preparation and sequencing
The purified cDNA was amplified using 11 cycles of PCR and then used for 3’ gene expression (GEX) library preparation. Through fragmentation, adapter ligation and index PCR the final libraries were obtained. The quality of single cell 3’ GEX was assessed by Qubit quantification and Tapestation analysis (HS D1000, Agilent). The sequencing was performed on a NovaSeq6000 (Illumina) with a S1 xp flow cell, paired-end sequencing with 400mln reads per library and otherwise the recommended sequencing conditions for 3’ GEX (read1: 28nt, read2: 90nt, index i7: 10nt, index i5: 10nt). ScRNA-Seq data alignment and gene expression quantification was carried out with Cellranger (version-5.0.0), using as a reference refdata-gex-mm10-2020-A mouse genome. Data were analyzed with R (version 4.0.2) Seurat package (version 4.0.1)39. All analyses were performed using a filtered feature barcode matrix. We excluded genes expressed in less than 3 cells and cells expressing less than 200 genes or more than 6000 genes, as well as cells with more than 20% mitochondrial reads. Counts were normalized with “LogNormalize” method and applied a scale factor of 10.000. Variable features were identified with “FindVariableFeatures” (using “vst” method). 2 Seurat objects (wild type and mutant) were integrated using “SelectIntegrationFeatures”, “FindIntegrationAnchors” and “IntegrateData” functions. Data were scaled and centered with “ScaleData” function and dimensionality reduction was done using PCA (30 PCs selected). Further dimensionality reduction was used for data visualization using UMAP algorithm. The “FindClusters” function, utilizing shared nearest neighbor modularity optimization-based algorithm was used with resolution 0.5 to identify 18 clusters. The differential expression per cluster was carried out with “FindMarkers” functions default parameters.
Histology and immunofluorescence microscopy
For eosinophil visualization, lungs were injected with 4% PFA and fixed for at least 16 h at 4°C. Sections from paraffin-embedded tissue were stained by modified Sirius Red stain and counter-stained with hematoxylin40. Intestine was fixed in 4% PFA at 4°C until tissue embedding. Sections from paraffin-embedded tissues were stained with periodic-acid-Schiff (PAS).
For immunofluorescence imaging of Nmur1Cre-T2A-GFP Rosa26Rfp/+ cells, organs were fixed in 1% PFA at 4°C over night, and immersed in 30% sucrose until tissues sank to the bottom. Tissues were frozen in Tissue-tek (Sakura), 7 μm cryosections were obtained on a cryostat (Leica), permeabilized with 0.5% Triton-X-100 in PBS and blocked with 2% FCS in PBS. Tissues were counterstained with antibodies against mouse-anti-human/mouse alpha smooth muscle actin coupled to Alexa Fluor 488 (1A4, Thermo Fisher Scientific), rat-anti-mouse CD45 (30F11, BD Bioscience) and subsequently goat-anti-rat coupled to Alexa Fluor 647 (Thermo Fisher Scientific). Images were acquired on a Zeiss LSM780 confocal microscope.
Small intestine from N. brasiliensis-infected animals was fixed was fixed in 4% PFA at 4°C until tissue embedding. Paraffin-embedded sections were de-paraffinized and rehydrated. Sections were permeabilized with 0.5% Triton-X in PBS and blocked with PBS 0.5% Triton X-100 and 10% serum and stained. The following antibodies were used: rabbit anti-Dclk1 (Abcam) followed by donkey anti-rabbit antibody coupled to Alexa Fluor 555 (Thermo Fisher Scientific), UEA-I-Fluorescein (Vector Laboratories) and nuclei were counterstained with DAPI (Thermo Fisher Scientific). Images were captured on a Zeiss Axio Observer 7 microscope and analysed with Zen software (Zeiss). For goblet and tuft cell numbers, three representative villi were counted on five independent images per mouse using ImageJ. For measuring UEA-I+ pixels, images were loaded into ImageJ and channels were split. The green (UEA-I) channel was used for the analysis and the threshold was set manually to create binary images. The UEA-I+ area was calculated relative to the total tissue area including the mucus layer on the image.
Extended Data
Extended data 1 (related to Figure 1). Gating strategy for flow cytometry.
a, Generation of genetic construct of the Nmur1Cre-T2A-GFP BAC transgene. 70th bp of exon 2 to 29th bp downstream of the TGA stop codon of exon 3 was replaced by a cassette containing iCre and eGFP linked with P2A and T2A. (86,388,138-86,386,296) were replaced through homologous recombination. The FRT-flanked kanamycin selection cassette was deleted after FLP-mediated recombination. Removal of LoxP and LoxP511 sites on BAC vector backbone. b, Expression of the Nmur1 gene in GFP+and GFP’ cells purified by FACsorting from the intestinal lamina propria. c, Gating strategy of bone marrow precursors. Lineage: CD3, CD5, CD19, Ly6G, FcεRIa. d, Gating strategy of intestinal lymphoid populations. e, Gating strategy of intestinal myeloid populations. Lineage: CD3, CD5, CD19, FcεRIa. f, Gating strategy of intestinal eosinophils, 2 alternative gates shown. Lineage: CD3, CD5, CD19, Ly6G. g, Gating strategy of γδ T cells, regulatory T cells (Treg) and TH2 cells from the mesenteric lymph node. h, Gating strategy of mast cells and basophils from the peritoneal lavage. Lineage: CD3, CD5, CD19, Ly6G
Extended data 2 (related to Figure 1). Nmur1Cre-T2A-GFP specifically labels ILC2s in steady state and during type 2 inflammation.
a, Flow-cytometric plots of GFP and ST2 in lung ILC2s (left) and TH2 cells (right) of Nmur1Cre-T2A-GFP mice. Numbers denote percentage of cells inside the gate. b, GFP expression in TH2 cells and ILC2s of Nmur1Cre-T2A-GFP mice, quantification of (a). c, GFP expression in mLN CD45- cells (black) and ILC2s (gray). d, Quantification of (c). e, Quantification of GFP expression across organs and cell types as displayed in the heatmap of Fig. 1d. f, RFP and ST2 in lung ILC2s (left) and TH2 cells (right) of Nmur1Cre-T2A-GFP x Rosa26Rfp/Rfp F1 offspring. Numbers denote percentage of cells inside the gate. g, RFP expression in TH2 cells and ILC2s, quantification of (f). h, RFP expression in mLN CD45- cells (black) and ILC2s (gray) of Nmur1Cre-T2A-GFP x Rosa26Rfp/Rfp F1 offspring. i, Quantification of (h). j, RFP expression across organs and cell types as displayed in the heatmap of Fig. 1h. k, Heatmap of flow-cytometric quantification of GFP and RFP expression in TH2 and Treg cells. Values represent median percentage of positive cells of parental gate, n=3 mice per group. l-q, Immunofluorescence micrographs of RFP expression in Nmur1 Cre-T2A-GFP xRosa26Rlp/Rlp F1 offspring stained for alpha smooth muscle actin (α-SMA), CD45 and DAPI in the small intestine (l), mesenteric lymph nodes (m), lung (n), mesenteric fat (o), femoral muscle (p) and large intestine (q). r, GFP and Cre (RFP) expression seven days after Nippostrongylus brasiliensis (N.b.) helminth infection across immune cell populations in Nmur1Cre-T2A-GFP x Rosa26Rfp/Rfp F1 offspring. s,t, GFP (s) and RFP (t) expression in Nmur1Cre-T2A-GFP Rosa26Rfp/+ mice, seven days after infection with N.b.. Data in (r-t) are representative of two independent experiments with 3 mice per group.
Extended Data 3, (related to Figure 2). Nmur1Cre-T2A-GFPGata3flox/flox and Nmur1Cre-t2A-GFP id2flox/flox mice lack ILC2s in all tissues investigated.
a, Flow-cytometric plots of ILC2s gated from live CD45+ Lin- (CD3, CD5, CD19) cells as CD127+ KLRG1+ or CD127+ ST2+ across organs of Nmur1Cre-T2A-GFP Gata3flox/flox mice and littermate controls. Numbers denote percentage of cells inside the gate. b, Quantification of (a). Mean +/- SD, Student’s t-test. c-d, Heatmaps displaying Log2-fold changes of the median cell frequencies among live CD45+ cells across different populations and organs of Nmur1Cre-T2A-GFP Gata3flox/flox (c) and Nmur1Cre-T2A-GFP Id2flox/flox (d) mice, compared to the respective littermate controls. Results are representative for 2 independent experiments with 3-4 mice per group. Littermate controls for (a-c) were Gata3flox/+ or Nmur1Cre-T2A-GFP Gata3flox/+, for (d) Id2flox/+ or Nmur1Cre-T2A-GFP Id2flox/+. e, Quantification of relative THh2 (top) and Treg (bottom) numbers in Nmur1Cre-T2A-GFP Id2flox/flox mice and Id2flox/flox littermate controls. Results are representative for 3 independent experiments with 3-6 mice per group. Mean +/- SD, Student’s t-test. f, Cytokine secretion by cultured Nmur1Cre-T2A-GFP Id2flox/flox immune cells isolated from lung and intestine, stimulated with IL-2, IL-7, IL-25 and IL-33 or PMA/Ionomycin for 8 h, measured in the supernatant by cytometric bead assay. Mean +/-SD, two-way ANOVA with Šídák’s multiple comparison tests, data are representative of two independent experiments with 3-4 mice per group. g, Serum IL-5 in untreated Id2flox/flox and Nmur1Cre-T2A-GFP Id2flox/flox animals as determined by the BD Cytometric Bead Array Mouse IL-5 Enhanced Sensitivity Flex Set, Student’s t-test. ns not significant, *p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. (a-g) One symbol represents data from one mouse.
Extended Data 4 (related to Figure 3). ILC2s regulate eosinophils in tissue.
Analysis of eosinophils across organs in the steady state. a, Eosinophils were gated from live CD45+ Lin- (CD3, CD5, CD19, Ly6G) cells using Siglec F and CCR3, across organs of Nmur1Cre-T2A-GFP Id2flox/flox mice and littermate controls. b, Relative ILC2 and eosinophil numbers in bone marrow chimera of Nmur1Cre-T2A-GFP Id2flox/flox (KO) and Id2flox/flox (WT) recipients transplanted with bone marrow of Nmur1Cre-T2A-GFPId2flox/flox (KO) or Id2flox/flox (WT) donors. c, Quantification of eosinophils in IL5-/- mice and respective IL5+/- littermate controls. d, Flow-cytometric plots of granulocyte/macrophage progenitor (GMP) gated from live CD45+ Lin- (CD3, CD5, CD19, Gr1, B220) Sca-1- c-Kithi cells as CD34+CD16/CD32hi and relative quantification in bone marrow of Nmur1Cre-T2A-GFP Id2flox/flox mice and Id2flox/flox littermate controls. e, Quantification of relative Eosinophil precursors (EoP) and GMPs in the bone marrow of IL-5-/- compared to WT controls. f, Flow-cytometric quantification of relative numbers of bone marrow-derived eosinophils gated from live CD45+ Lin- (CD3, CD5, CD19) CD11b+ cells as SSChi and Siglec F+. BM cells were cultured with either SCF and FLT3L, IL-5, ILC2P or supernatant from ILC2s for 10 days. g,h Mean fluorescence intensity of CCR3 on eosinophils in Nmur1Cre-T2A-GFP Id2flox/flox (g) and Il5-/- (h) mice and respective littermate controls. b-e, g-h One symbol represents data from one mouse (b-e, g-h) or one well (f), mean +/- SD, c-e, g-h Student s t-Test. b, f Oneway ANOVA with Tukey’s multiple comparisons, ns not significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
Extended Data 5 (related to Figure 3). ILC2 deletion regulates myeloid and epithelial populations in the lung.
a, Bubble plot of marker genes of global cell populations in scRNA sequencing of myeloid and epithelial cells in the steady state lung of Nmur1Cre-T2A-GFP Id2flox/flox mice and littermates. b, Marker genes of subclusters of interstitial macrophages. c, Marker genes of DC subclusters. d, Marker genes in subclusters of alveolar type 2 epithelial cells. e-g, Subclusters in e macrophages, f dendritic cells and g alveolar type 2 epithelial cells. h, Macrophage UMAPs color-coded gray-to-red with genes downregulated in Macrophages of Nmur1Cre-T2A-GFP Id2foxl/flox mice. i, Macrophage UMAPs color-coded with genes upregulated in macrophages of Nmur1Cre-T2A-GFP Id2flox/flox mice. j, DC UMAP color-coded by Ccl24 expression. k, UMAPs of alveolar type 2 epithelial cells color-coded with genes downregulated in alveolar type 2 epithelial cells in Nmur1Cre-T2A-GFP Id2flox/flox mice. l, Statistical evaluation of genes displayed in f-h. Cells from four mice per group were pooled. Littermate controls for (a-l) were Id2flox/flox or Id2flox/+.
Extended Data 6 (related to Figure 3). ILC2s regulate myeloid cells in type-2 inflammatory allergy models of the lung.
a-d, Flow-cytometric quantification of cell population in Nmur1Cre-T2A-GFP Id2flox/flox mice and littermate controls sensitized with A. alternata extract for three consecutive days, analyzed 7 days after the first dose. a, absolute numbers of lung ILC2s, lung an BAL eosinophils and total CD45+ cells in the BAL. b, Relative and absolute numbers of TH2 cells in the lung. c,d, relative (c) and absolute (d) numbers of myeloid populations from the lung interstitium, and alveolar macrophages from the BAL. Data in a-d are representative of two independent experiments with 4-6 animals per group. e-i, Analysis of Nmur1Cre-T2A-GFPId2flox/flox mice and littermate controls sensitized with papain for three consecutive days, analyzed 7 days after the first dose. e, Absolute numbers of ILC2s in the lung interstitium, and eosinophils in the lung interstitium and BAL. f, Eosinophils are visualized by modified Sirius red stain in the lung. g, Relative and absolute numbers of TH2 cells in the lung. h,i, relative (h) and absolute (i) numbers of myeloid populations from the lung interstitium, and alveolar macrophages from the BAL. Data in e and g-i are pooled from two independent experiments with 4-5 mice per group. Untreated controls for (e-i) were Id2flox/flox, papain-treated littermate controls were Id2flox/flox, Id2flox/+ or Nmur1Cre-T2A-GFP Id2flox/+. One symbol represents data from one mouse, mean+/- SD, Oneway ANOVA with Tukey’s multiple comparison tests, ns: not significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. BAL, bronchoalveolar lavage.
Extended Data 7 (related to Figure 3). ILC2s regulate myeloid cells in type-2 inflammatory allergy models of the lung.
a-e, Analysis of Nmur1Cre-T2A-GFP Id2flox/flox mice (cKO) and littermate controls (Ctrl) sensitized and challenged with house dust mite. a, Lung ILC2s (top), BAL eosinophils (middle) and Lung TH2 cells (bottom) were gated from live CD45+ Lin- (CD11b-CD11c-FcεR1a-B220-CD3-5-) cells. The second gate in the eosinophil plots marks alveolar macrophages. b, Quantification of (a). c, Periodic-acid-Schiff staining of lungs. d, Gating of cytokine-expressing T helper cells isolated from the lung. e, Quantification of (d). f-j, Resident and inflammatory eosinophils from the lungs of papain- and A. alternata-treated mice. f, Flow-cytometric plots showing resident and inflammatory eosinophils from lung after Papain treatment of Nmur1Cre-T2A-GFP Id2flox/flox mice (cKO) and littermate controls (Ctrl). g, Relative and absolute numbers of resident and inflammatory eosinophils. h, Flow-cytometric plots showing resident and inflammatory eosinophils from lung after Altenaria alternata treatment of Nmur1Cre-T2A-GFP Id2flox/flox mice (cKO) and littermate controls (Ctrl). i, Relative and absolute numbers of resident and inflammatory eosinophils. Data are representative for two independent experiments with 3-5 mice per group (f-i). Symbols represent data from one mouse, mean +/- SD, One-way ANOVA with Tukey’s multiple comparisons (b-e) or Student’s t-test (g,i), ns not significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
Extended Data 8 (related to Figure 3). Eosinophils are altered in ILC2-deficient mice.
a-f, Flow-cytometric analysis of eosinophils and ILC2s in the BAL and lung interstitium of Id2flox/flox, Nmur1Cre-T2A-GFP Id2flox/flox mice treated with either PBS or recombinant IL-5, seven days after allergy induction by intranasal application of Alternaria alternata extract. a,b Eosinophils in the lung interstitium, c,d Eosinophils in the BAL, e,f ILC2s in the lung interstitium. Symbols represent data from one mouse, data are pooled from two independent experiments. Mean +/- SD, Student’s t-test, ns not significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. g Flow-cytometric plots showing the gating strategy for the analysis and sorting of eosinophils from lung after Papain treatment of Nmur1Cre-T2A-GFP Id2flox/flox mice (cKO) and littermate controls (Ctrl). Data are representative of one experiment with 5 mice per group. h, Pathways from tmod gene set enrichment analysis (GSEA) of bulk RNA sequencing from eosinophils of 4 Nmur1Cre-T2A-GFP Id2flox/flox mice vs 4 WT control animals. Bars are color-coded according to Databases: Gene Ontology (GO, red), Reactome (gray), Hallmark (white), tmod (purple), KEGG (pink). Genes differentially expressed in eosinophils after papain treatment of Nmur1Cre-T2A-GFP Id2flox/flox mice (cKO) and littermate controls (Ctrl) were used for the analysis. i, Volcano plot showing differentially expressed genes of interest in eosinophils after papain treatment of Nmur1Cre-T2A-GFP Id2flox/flox mice (cKO) and littermate controls (Ctrl) j, Heatmaps of genes differentially expressed in eosinophils after papain treatment of Nmur1Cre-T2A-GFP Id2flox/flox mice (cKO) and littermate controls (Ctrl). Each heatmap corresponds to one pathway shown in (h).
Extended Data 9 (related to Figure 4). ILC2-deficient mice are highly susceptible to N. brasiliensis infection.
Nmur1Cre-T2A-GFP Id2flox/flox mice (cKO) and littermate controls (Ctrl) were analyzed 7 (a-c) or 11 days (i-m) after infection with N.b., alongside untreated WT controls. a, TH2 cells. b,c, Neutrophils in the lung. Littermate controls for (a-c) were Id2flox/+ or Nmur1Cre-T2A-GFP Id2flox/+. d-h, Nmur1iCre-GFP Gata3flox/flox mice (cKO) and littermate controls (Ctrl) 7 days after N. b. infection alongside untreated WT controls. d-e, ILC2s (top) and eosinophils (bottom) in the mLN (d) or mLN and lung (e). f, Worm burden g, Goblet- and tuft cells per villus, mucus area quantified from UEA-I stain, all as shown in (h). h, left: Periodic acid-Schiff staining; middle: intestinal tuft cell staining with Dclk1 (red), DAPI (blue); right: intestinal mucus stained with UEA-I (green), DAPI (blue). Littermate controls for (d-h) were Gata3flox/+. i,j, ILC2s and eosinophils in the mLN (i) or mLN and lung (j) 11 days after infection. k, TH2 cells. l, Goblet- and tuft cells per villus, mucus area quantified from UEA-I stain as shown in (m). m, intestine as shown in (h), 11 days after infection.
n-r Infection of BM chimeras of C57BL/6 recipients transplanted with bone marrow of Id2flox/flox(Ctrl), Nmur1Cre-T2A-GFP Id2flox/flox (cKO) donors, or bone marrow of cKO donors supplemented with sorted ILC2 precursors (cKO + ILC2p) from C57BL/6 donors, with N.b..n, Periodic acid-Schiff staining of small intestine seven days after N. b. infection. o, Goblet cells per villus. p,q, Neutrophils in the lung. r, TH2 cells. s, ILC3s, LTi-like ILC3s and NCR+ ILC3s 7 days after N.b. infection of Nmur1Cre-T2A-GFP Id2flox/flox mice (cKO) and littermate controls (Ctrl). (a-s) One symbol represents data from one mouse, one-way ANOVA with Tukey’s multiple comparisons, ns not significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
Extended Data 10 (related to Figure 4). Gene expression in epithelial cells of ILC2-deficient mice during N. brasiliensis infection.
a, Heatmap showing scaled expression of marker genes for colonic cell populations in scRNA Seq Data shown in Fig 4m. b, Violin plots showing normalized count distributions of differentially expressed genes Reg3b, Reg3g, Muc-2, Clca1 and Ang4 in colonic epithelial cells between infected NmurlCre-T2A-GFPId2 flox/flox mice (cKO) and littermate controls (Ctrl) in all clusters.
Acknowledgments
We thank C. Braeuning, T. Borodina, T. Conrad and the other members of the Genomics Platform at MDC Berlin-Mitte for carrying out the RNA sequencing experiments, A. Branco and M. Fernandes of the Benjamin Franklin Flow Cytometry Facility for cell sorting, J. Fehling (Rosa26Rfp/RfP), A. Lasorella (Id2flox/flox) and M. Busslinger (Gata3fl/fl) for providing mice, L. Paul for technical assistance, I. Mattiola for discussion and M. Witkowski for reading the manuscript. This work was supported by grants from the European Research Council Starting Grant (ERCEA; 803087 to CSNK), the German Research Foundation (DFG; Project-ID 259373024 – CRC/TRR 167, FOR2599 project 5, B05 – CRC/TRR 241, - KL 2963/5-2, SPP1937 - KL 2963/2-1 and KL 2963/3-1 to CSNK), the Swiss National Science foundation (Grant-ID: 184425 to MJ), the Leibnitz Association (Leibnitz Collaborative Excellence, TargArt) to C.R. and M.F.M, the Crohn’s and Colitis Foundation Research Fellowship Award (Award #527125 to A.M.T; #851136 to M.A.), the WCM Department of Pediatrics Junior Faculty Pilot Award, and The Jill Roberts Center Pilot Award for Research in IBD (to A.M.T.); the US National Institutes of Health (DK126871, AI151599, AI095466, AI095608, AI142213, AR070116, AI172027, DK132244 to D.A.), LEO foundation, Cure for IBD, Jill Roberts Institute, the Sanders Family, the Rosanne H. Silbermann Foundation (all to D.A.). and the state of Berlin and the “European Regional Development Fund” to M.F.M. (ERDF 2014-2020, EFRE 1.8/11, 563 Deutsches Rheuma-Forschungszentrum).
Footnotes
Authorship contributions
K.J.J., P.M.T., M.O.J., H.Y., M.A. X.G. and C.S.N.K. carried out most experiments and analyzed the data. S.B., V.S-T, P.L., A.P. Z.A.R, A.S., A.M.T., C.C, C.S., S.F. and A.A.K helped performing the experiments. A.I., D.B., F.F.H. G.G., P.D. and M-F.M. performed RNA-seq analysis. C.U.D., C.R., A.D. and D.A. provided crucial input and tools for the study. K.J.J. and C.S.N.K. conceived the project and wrote the manuscript with input from all co-authors.
Declaration of interests
D.A. has contributed to scientific advisory boards at Pfizer, Takeda, FARE, and the KRF. The other authors declare no competing interests.
Statistical analysis
Data is plotted showing the mean +/- standard deviation. P values of data sets were determined by unpaired two-tailed Student’s t-test, ordinary one-way ANOVA with Tukey’s multiple comparisons test, or two-way ANOVA with Šidák’s multiple comparisons test, with 95% confidence interval. Normal distribution was assumed. To test correlation between ILC2 and eosinophil numbers, squared Pearson correlation coefficients (R2) were calculated; P-values were computed in a two-tailed analysis with a 95% confidence interval._Before mentioned statistical tests were performed with Graph Pad Prism V9 software (GraphPad Software, Inc.). (*p <0.05; **p <0.01; ***p <0.001; ****p <0.0001 and n.s., not significant). Genes regulated in scRNAseq of the Lung were tested using Mann-Whitney test with Bonferroni correction.
Additional information
Correspondence and requests for materials should be addressed to Christoph Klose (christoph.klose@charite.de).
Data availability
The RNA-seq data are deposited in the Gene Expression Omnibus (GEO) repository database under the accession numbers GSE212011 and GSE212940. Source data are provided with this manuscript.
Code availability
Data was analyzed using the standard Seurat 4.0.1 or 4.0.2 pipeline, or with the stated variations. User scripts will be shared upon request.
References
- 1.Vivier E, et al. Innate Lymphoid Cells: 10 Years On. Cell. 2018;174:1054–1066. doi: 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 2.Klose CS, Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nature immunology. 2016;17:765–774. doi: 10.1038/ni.3489. [DOI] [PubMed] [Google Scholar]
- 3.Vely F, et al. Evidence of innate lymphoid cell redundancy in humans. Nature immunology. 2016;17:1291–1299. doi: 10.1038/ni.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rankin LC, et al. Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nature immunology. 2016;17:179–186. doi: 10.1038/ni.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colonna M. Innate Lymphoid Cells: Diversity, Plasticity, and Unique Functions in Immunity. Immunity. 2018;48:1104–1117. doi: 10.1016/j.immuni.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonnenberg GF, Hepworth MR. Functional interactions between innate lymphoid cells and adaptive immunity. Nature reviews Immunology. 2019;19:599–613. doi: 10.1038/s41577-019-0194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orange JS, et al. Comment on: Evidence of innate lymphoid cell redundancy in humans. Nature immunology. 2018;19:788–789. doi: 10.1038/s41590-018-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klose CSN, et al. The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature. 2017;549:282–286. doi: 10.1038/nature23676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallrapp A, et al. The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature. 2017;549:351–356. doi: 10.1038/nature24029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardoso V, et al. Neuronal regulation of type 2 innate lymphoid cells via neuromedin U. Nature. 2017;549:277–281. doi: 10.1038/nature23469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinette ML, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nature immunology. 2015;16:306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 13.Klose CSN, et al. Differentiation of Type 1 ILCs from a Common Progenitor to All Helper-like Innate Lymphoid Cell Lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 14.Constantinides MG, et al. PLZF expression maps the early stages of ILC1 lineage development. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:5123–5128. doi: 10.1073/pnas.1423244112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoyler T, et al. The Transcription Factor GATA-3 Controls Cell Fate and Maintenance of Type 2 Innate Lymphoid Cells. Immunity. 2012;37:634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halim TY, et al. Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity. 2012;37:463–474. doi: 10.1016/j.immuni.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Moro K, et al. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. 2010;463:540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- 18.Furusawa J, et al. Critical role of p38 and GATA3 in natural helper cell function. J Immunol. 2013;191:1818–1826. doi: 10.4049/jimmunol.1300379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein Wolterink RG, et al. Essential, dose-dependent role for the transcription factor Gata3 in the development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10240–10245. doi: 10.1073/pnas.1217158110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nussbaum JC, et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 2013;502:245–248. doi: 10.1038/nature12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kopf M, et al. IL-5-deficient mice have a developmental defect in CD5+ B-1 cells and lack eosinophilia but have normal antibody and cytotoxic T cell responses. Immunity. 1996;4:15–24. doi: 10.1016/s1074-7613(00)80294-0. [DOI] [PubMed] [Google Scholar]
- 22.Kim K, et al. Terminally Differentiating Eosinophils Express Neutrophil Primary Granule Proteins as well as Eosinophil-specific Granule Proteins in a Temporal Manner. Immune Netw. 2017;17:410–423. doi: 10.4110/in.2017.17.6.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou L, et al. Innate lymphoid cells support regulatory T cells in the intestine through interleukin-2. Nature. 2019;568:405–409. doi: 10.1038/s41586-019-1082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cupedo T, et al. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nature immunology. 2009;10:66–74. doi: 10.1038/ni.1668. [DOI] [PubMed] [Google Scholar]
- 25.Cella M, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takatori H, et al. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. The Journal of experimental medicine. 2009;206:35–41. doi: 10.1084/jem.20072713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanos SL, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nature immunology. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satoh-Takayama N, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nature medicine. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 30.Harris NL, Loke P. Recent Advances in Type-2-Cell-Mediated Immunity: Insights from Helminth Infection. Immunity. 2017;47:1024–1036. doi: 10.1016/j.immuni.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Niola F, et al. Id proteins synchronize stemness and anchorage to the niche of neural stem cells. Nat Cell Biol. 2012;14:477–487. doi: 10.1038/ncb2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grote D, Souabni A, Busslinger M, Bouchard M. Pax 2/8-regulated Gata 3 expression is necessary for morphogenesis and guidance of the nephric duct in the developing kidney. Development. 2006;133:53–61. doi: 10.1242/dev.02184. doi:dev.02184 [pii] [DOI] [PubMed] [Google Scholar]
- 33.Luche H, Weber O, Nageswara Rao T, Blum C, Fehling HJ. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. European journal of immunology. 2007;37:43–53. doi: 10.1002/eji.200636745. [DOI] [PubMed] [Google Scholar]
- 34.Halim TYF, et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 40:425–35. doi: 10.1016/j.immuni.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 37.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferreira-Gomes M, et al. SARS-CoV-2 in severe COVID-19 induces a TGF-beta-dominated chronic immune response that does not target itself. Nature communications. 2021;12:1961. doi: 10.1038/s41467-021-22210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stuart T, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.:e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyerholz DK, Griffin MA, Castilow EM, Varga SM. Comparison of histochemical methods for murine eosinophil detection in an RSV vaccine-enhanced inflammation model. Toxicol Pathol. 2009;37:249–255. doi: 10.1177/0192623308329342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The RNA-seq data are deposited in the Gene Expression Omnibus (GEO) repository database under the accession numbers GSE212011 and GSE212940. Source data are provided with this manuscript.
Data was analyzed using the standard Seurat 4.0.1 or 4.0.2 pipeline, or with the stated variations. User scripts will be shared upon request.