Abstract
Linear and disturbed flow differentially regulate gene expression, with disturbed flow priming endothelial cells for a pro-inflammatory, atheroprone expression profile and phenotype. Here, we investigated the role of the transmembrane protein neuropilin-1 (NRP1) in endothelial cells (ECs) exposed to flow using cultured ECs, mice with an endothelium-specific knockout of NRP1, and a mouse model of atherosclerosis. We demonstrated that NRP1 was a constituent of adherens junctions that interacted with VE-cadherin and promoted its association with p120 catenin, stabilizing adherens junctions and inducing cytoskeletal remodeling in alignment with the direction of flow. We also showed that NRP1 interacted with transforming growth factor-β (TGF-β) receptor II (TGFBR2) and reduced the plasma membrane localization of TGFBR2 and TGF-β signaling. NRP1 knockdown increased the abundance of pro-inflammatory cytokines and adhesion molecules, resulting in increased leukocyte rolling and atherosclerotic plaque size. These findings describe a role for NRP1 in promoting endothelial function and reveal a mechanism by which NRP1 reduction in ECs may contribute to vascular disease by modulating adherens junction signaling and promoting TGF-β signaling and inflammation.
Introduction
Endothelial cell (EC) junctions are essential to vascular integrity and determine the selective semi-permeable property of the endothelial barrier, allowing nutrient exchange and leukocyte extravasation from the circulation and into the surrounding tissues (1). Accordingly, inflammatory cytokines activate signaling pathways that weaken intercellular junctions and induce gaps at contact points, allowing immune cells to move across the endothelium (2). In cardiovascular disease, endothelial activation drives the expression of pro-inflammatory cytokines and pro-inflammatory adhesion molecules which promote permeability, leukocytes trans-endothelial migration and inflammation (3, 4).
Several lines of evidence indicate that adherens junctions play a critical role in modulating the barrier function of the endothelium and EC signaling (4, 5). In ECs, vascular endothelial-cadherin (VE-cadherin) is the main constituent of adherens junctions and its cadherin repeats in the extracellular domain promote intercellular VE-cadherin homophilic interactions between neighboring ECs (6–8). The intracellular domain of VE-cadherin associates with adaptor proteins of the armadillo family such as β-catenin and p120 catenin, which couple the adherens junction complex to the actin cytoskeleton. (8). The interaction of p120 catenin with the juxtamembrane domain of VE-cadherin also regulates VE-cadherin availability at junctional adhesion sites (9, 10) by preventing clathrin-mediated endocytosis (11). Accordingly, inhibition of the interaction between p120 catenin and VE-cadherin reduces VE-cadherin levels and destabilizes adherens junctions, decreasing endothelial barrier function (12). Adherens junction stability is modulated by molecular mechanisms controlling VE-cadherin availability at junctional adhesion sites but also by post-translational modification of VE-cadherin, with phosphorylation of Tyr685 and Tyr731 residues regulating vascular permeability and leukocyte diapedesis (12), respectively.
In addition to determining the tightness of the endothelial monolayer, adherens junctions modulate cell signaling pathways in ECs. Junctional clustered VE-cadherin interacts with transforming growth factor-β (TGF-β) type II and type I transmembrane serine-threonine kinase receptors and induces TGF-β receptor activation and localization in signaling puncta, thus promoting TGF-β signaling (13). In addition, VE-cadherin is part of a junctional mechanosensing complex together with platelet endothelial cell adhesion molecule-1 (PECAM-1) and vascular endothelial growth factor receptor-2 (VEGFR2), which function as a mechanical force transmitter and transduce intracellular signaling, respectively (14, 15). Endothelial dysfunction and increased permeability occur in atherogenesis, causing the accumulation of lipoproteins and immune cells in the subendothelial region of sites of atherosclerosis (16). In mice, the endothelium at sites of atherosclerotic plaques presents with disorganized, discontinued VE-cadherin junctions compared to healthy endothelium (17). Moreover, a high cholesterol diet increases the degradation of VE-cadherin and endothelial permeability in a manner dependent on m-calpain-mediated cleavage of VE-cadherin (18). Treatment with a function-blocking antibody inhibiting VE-cadherin clustering at adherens junctions reduces endothelium-dependent dilatation in arteries in response to acetylcholine (19, 20), supporting a role of VE-cadherin in regulating EC function.
Neuropilin-1 (NRP1) is a transmembrane protein expressed by several cell types including ECs (21, 22). Originally identified as an adhesion molecule (23), NRP1 has been extensively studied as a VEGF and Semaphorin co-receptor in ECs and neurons (24, 25). Global and endothelium-specific NRP1 knockout mouse mutants die in utero with abnormal yolk sac and neuronal vascularization (26–28), consistent with defects in VEGF signaling (29–31). However, NRP1-dependent VEGF signaling is not essential for vascular development (32), indicating that NRP1 regulates angiogenesis by integrating VEGF-dependent and -independent pathways (29–31), including extracellular-matrix-mediated signaling (33, 34), and by suppressing endothelial TGF-β signaling in sprouting vessels (32).
NRP1 promotes vascular permeability in a VEGF/VEGFR2 dependent mouse model of choroidal neovascularization akin to age-related macular degeneration through a mechanism requiring the NRP1 cytoplasmic domain and activation of Src family kinases and ABL1 (35). NRP1 can also promote vascular permeability independently of VEGFR2 activation (36). NRP1 binding to VEGF-A, R-x-x-R carboxyl-terminal motif-containing peptides (CendR) (36), or a NRP1-targeting ligand-blocking antibody promotes lateral NRP1 delocalization at cell-cell contacts and increased permeability in a manner dependent on NRP1 cytoplasmic domain but independent of VEGFR2 activity (36). This finding suggests a direct role of NRP1 in modulating cell-cell junction-mediated signaling with NRP1 playing a role in flow-induced signaling upstream of the junctional complex in ECs. Accordingly, PlexinD1 associates with NRP1 and VEGFR2 to form a mechano-sensing complex that activates early signaling in response to shear stress including VEGFR2 phosphorylation and its association with the tyrosine kinase Src (37).
NRP1 also acts as a co-receptor for TGF-β and interacts with TGF-β type II and type I transmembrane serine-threonine kinase receptors with an affinity increased by TGF-β binding (38). In agreement, in T-lymphocytes and breast cancer cells, NRP1 promotes TGF-β-mediated SMAD2/3 phosphorylation and signaling (38, 39). However, in ECs, NRP1 promotes angiogenesis and tip cell specification by inhibiting SMAD2/3 activation downstream of TGF-β and bone morphogenic protein 9 (BMP9). The molecular mechanism by which NRP1 suppresses TGF-β signaling in ECs is not completely understood but it requires the extracellular and transmembrane domain of NRP1 but not the cytoplasmic domain (32).
In this study, we showed that NRP1 interacted with VE-cadherin in a shear stress-dependent manner and that NRP1 promotes VE-cadherin interaction with p120 catenin, thus promoting adherens junction stability. Accordingly, knockdown of NRP1 or p120 catenin in ECs exposed to laminar flow induced adherens junction destabilization and cytoskeleton remodeling. The destabilization of adherens junctions caused by the knockdown of NRP1 or p120 catenin increased TGFBR2 plasma membrane localization and increased SMAD2/3 phosphorylation, thus activating TGF-β signaling. We further showed that NRP1 prevented activation in ECs exposed to atheroprotective laminar flow and that genetic depletion of NRP1 in the mouse endothelium increased leukocyte rolling and atherosclerosis plaque deposition in ApoE knockout hyperlipidemic mice. Together, our data reveal that NRP1 promotes vascular function by modulating adherens junction signaling and suppressing TGF-β-dependent signaling and endothelial activation, thereby promoting endothelial anti-inflammatory properties.
Results
Shear stress modulates NRP1 levels which promotes flow-induced gene expression in ECs
Under normal physiological conditions, ECs are exposed to shear forces generated by the flow of blood. Thus, to investigate the role of NRP1 in endothelial function in an in vitro model simulating the physiological environment of blood vessels, primary human umbilical vein endothelial cells (HUVECs) were exposed to shear stress using an Ibidi Quad microfluidic device. HUVECs were exposed to either high unidirectional shear stress (laminar; 20 dynes/cm2), low bidirectional shear stress (oscillatory; 5 dynes/cm2; 1 Hz) or cultured in the absence of flow (static). The high unidirectional laminar shear stress mimics the atheroprotective unidirectional flow occurring in atheroprotected regions of the coronary arteries and the descending aorta whereas low bidirectional shear stress simulates the pro-inflammatory and pro-atherogenic multidirectional disturbed flow of atheroprone regions such as that occurring at sites of bifurcation and the aortic arch (40–42). Phalloidin staining of filamentous actin (F-actin) confirmed that HUVECs exposed to laminar flow (Fig. 1A, single-headed white arrow) aligned in the direction of flow whereas HUVECs exposed to oscillatory flow (Fig. 1A, double-headed white arrow) or cultured in the absence of flow lacked a specific directional orientation (Fig. 1A). These data are consistent with previous reports showing alignment of ECs with high-shear laminar flow (43–46) and indicate that the Ibidi system effectively generates laminar flow. Immunostaining indicated that HUVECs exposed to laminar flow had higher NRP1 levels than those cultured under static conditions (Fig. 1A, B) or exposed to oscillatory flow (Fig. 1A, B). These data indicate that exposure to physiological, atheroprotective shear stress increases NRP1 levels in ECs.
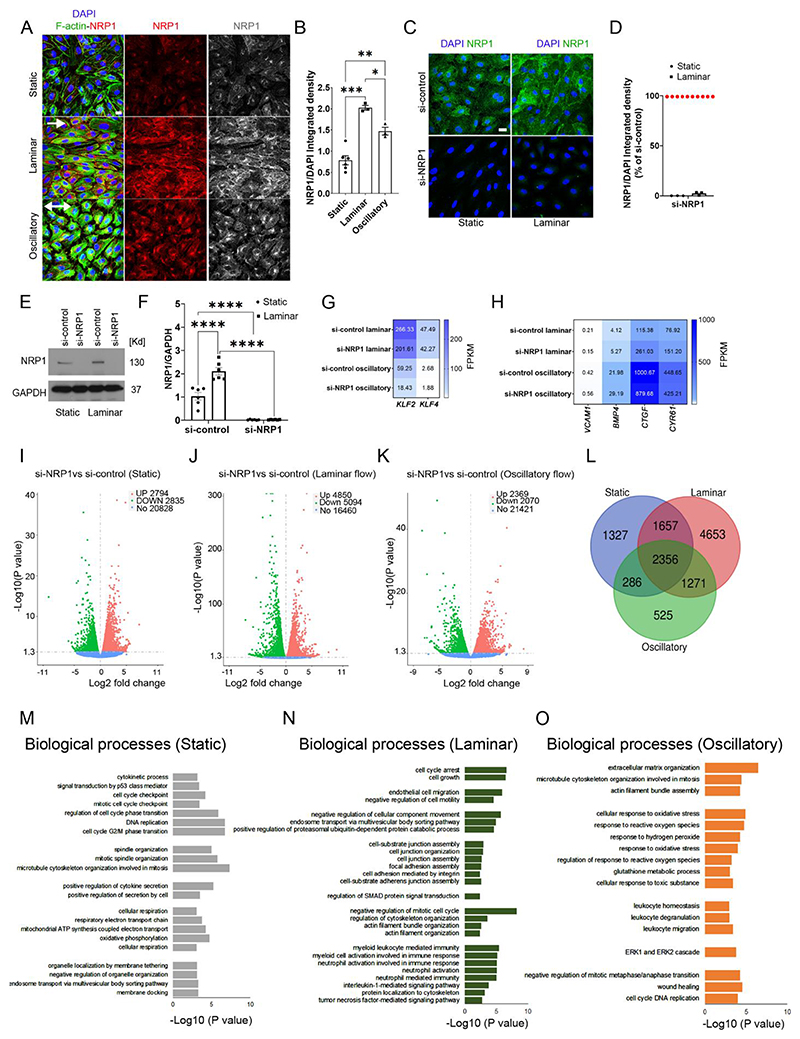
Fig. 1. NRP1 regulates flow-induced gene expression in endothelial cells.
(A) HUVECs exposed to laminar or oscillatory flow or cultured under static conditions for 24 hours were stained for NRP1 (red and grey), F-actin (green) and DAPI (blue). Scale bar =20 μm. White arrows show the direction of flow. (B) NRP1 integrated density was measured in optical z-stacks and normalized to DAPI. Data are presented as means ± SEM. N=3 (laminar or oscillatory) or 6 (static) biological replicates per group from 3 independent experiments. *p < 0.05, **p < 0.005, ***p < 0.001 by one-way ANOVA. (C) HUVECs transfected for 72 hours with si-control or si-NRP1 and exposed to laminar flow for 24 hours or cultured under static condition were stained for NRP1 (green) and DAPI (blue). Scale bar = 20 μm. (D) NRP1 integrated density was measured in optical z-stacks, normalized to DAPI and expressed as percentage of si-control. Data are presented as means ± SEM. N = 3 biological replicates per group. (E) Representative immunoblotting for NRP1 and GAPDH of HUVECs transfected for 72 hours with si-control or si-NRP1 and exposed to static or laminar flow for 24 hours. (F) Quantification of NRP1 signal normalized to GAPDH signal expressed as fold change of static si-control. Data are presented as means ± SEM. N = 6 biological replicates per group. **p < 0.005, ***p < 0.001 by two-way ANOVA. (G, H) Heat-map of gene expression in HUVECs transfected for 72 hours with si-control or si-NRP1 exposed to laminar or oscillatory flow showing fragments per kilobase of transcript per million mapped reads (FPKM). (I-K) Volcano plots of RNA-seq transcriptomic data displaying significantly differentially expressed genes in si-NRP1-transfected HUVECs relative to si-control-transfected HUVECs cultured under static conditions (I), exposed to laminar flow (J) or oscillatory flow (K) for 24 hours. (L) Venn diagram of RNA-seq transcriptome data displaying the number of differentially expressed genes between si-control and si-NRP1 common or specific to static, laminar or oscillatory conditions. (M-O) GO analysis of biological processes differentially modulated in HUVECs transfected with si-control or si-NRP1 for 72 hours and cultured under static conditions (M) or laminar flow (N) or oscillatory flow (O) for 24 hours. For (G) to (O), N = 3 biological replicates per group.
Next, we investigated whether NRP1 has a role in modulating gene expression in response to flow by performing a full transcriptomic profile using next-generation RNA sequencing of HUVECs depleted of NRP1 using a previously validated siRNA (si-NRP1) approach (33–35, 47). First, we confirmed effective NRP1 downregulation in HUVECs cultured under static or flow conditions by immunostaining (Fig. 1C, D), immunoblotting (Fig. 1E, F) and RT-qPCR analyses (Fig. S1A). NRP1 mRNA expression was similar in HUVECs cultured under static conditions or exposed to high-shear laminar flow (Fig. S1A), suggesting that the increase in NRP1 protein levels (Fig. 1A,B; Fig. 1E,F) was likely due to enhanced protein stability. We compared the transcriptomic profiles of HUVECs transfected with siNRP1 or a non-targeting siRNA (si-control) and exposed to laminar or oscillatory flow. RNAseq and RT-qPCR analyses showed that the atherogenic genes encoding Kruppel-like factor (KLF) 2 and KLF4 were similarly expressed in si-control and si-NRP1 HUVECs exposed to atheroprotective laminar flow and expressed at lower levels in cells exposed to atherogenic oscillatory flow (Fig. 1G; Fig. S1B). Both si-NRP1 and si-control treated HUVECs showed a similar increase in the expression of genes upregulated by disturbed, atherogenic flow, such as those encoding the pro-inflammatory vascular cell adhesion molecule 1 (VCAM1), bone morphogenic protein 4 (BMP4), connective tissue growth factor (CTGF) and cysteine-rich angiogenic inducer 61 (CYR61) (48) (Fig. 1H). These data indicate that shear stress generated by the Ibidi system effectively induces a mechanosensory response in HUVECs and that exposure to different flow patterns for 24 hours induces a similar mechanosensor-dependent transcriptional response in control transfected HUVECs and those depleted of NRP1.
We identified genes with differential expression under the different flow patterns to provide insight into the role of NRP1 in the transcriptional response to shear stress. Under static conditions, NRP1 knockdown upregulated 2794 genes and downregulated 2835 genes (Fig. 1I). In response to laminar flow, NRP1 knockdown upregulated 4850 genes and downregulated 5094 genes (Fig. 1J). In HUVECs exposed to oscillatory flow, NRP1 knockdown upregulated 2369 genes and downregulated 2070 genes (Fig. 1K). Comparison of the differentially expressed genes modulated by NRP1 knockdown in each condition showed that 1327 genes were exclusively modulated under static conditions, 525 in oscillatory flow and 4653 in laminar flow conditions (Fig. 1L; Data File S1). Gene Ontology (GO) analysis revealed that NRP1 knockdown affected genes with functions related to cytoskeletal remodeling and cell proliferation in cells cultured under static and both flow conditions (Fig 1M-O), in agreement with previous reports using ECs cultured under static conditions (33, 47). Furthermore, in cells cultured under static conditions, NRP1 knockdown affected genes in pathways involved in mitochondrial respiration and organelle organization by membrane tethering (Fig. 1M). In cells exposed to atheroprotective laminar flow, NRP1 knockdown affected genes in pathways involved in cell-matrix adhesion, cell junctions, adherens junction, leukocyte and neutrophil-mediated immune responses and interleukin-1 mediated signaling (Fig. 1N). In HUVECs exposed to pro-inflammatory, atherogenic oscillatory flow, GO analysis showed that NRP1 knockdown affected genes in pathways involved in leukocyte homeostasis and oxidative stress (Fig. 1O). Together, these data indicate that although NRP1 is dispensable for sensing shear stress and activating a flow response, it modulates flow-dependent transcriptional changes involved in cell adhesion, proliferation, oxidative stress and inflammation independently of typical flow-induced genes such as KLF2 and KLF4 (Fig 1I-O; Data File 1).
NRP1 forms a complex with VE-cadherin and promotes adherens junction stability in ECs exposed to high-shear laminar flow
Because GO analysis revealed that NRP1 control pathways regulating cell-cell contact and cellular junctions (Fig. 1N), because NRP1 regulates endothelial permeability (1, 12), and because pro-permeability factors modulate NRP1 junctional localization (36), we decided to explore whether NRP1 plays a role in regulating adherens junctions in HUVECs exposed to laminar or oscillatory flow. F-actin staining showed that control HUVECs exposed to laminar flow aligned with the direction of flow (Fig. 2A), whereas VE-cadherin staining showed a continuous signal across the plasma membranes of neighboring cells (Fig. 2A). NRP1 knockdown in HUVECs exposed to laminar flow induced a rounded cellular morphology with abundant cortical actin (Fig. 2A) and a loss of cell-cell contacts, as indicated by discontinuous VE-cadherin staining and by the appearance of gaps between adjacent cells (Fig. 2A, red arrows; Fig. 2B). The integrated density for VE-cadherin was not changed, suggesting that NRP1 downregulation altered VE-cadherin organization without affecting VE-cadherins levels (Fig. 2A, C). Exposure of HUVECs treated with si-control or si-NRP1 to oscillatory flow induced a rounded morphology and the formation of intercellular gaps between adjacent cells (Fig. 2A, B). NRP1 knockdown did not affect VE-cadherin immunostaining levels (Fig. 2A, C). These data suggest that laminar flow promotes adherens junction stability, whereas oscillatory flow has a destabilizing effect on cell-cell contacts. In addition, these data suggest that NRP1 is required to promote adherens junction stability in response to flow. To further examine the relationship between NRP1 and adherens junction stability we performed a gain-of-function experiment in HUVECs with endogenous NRP1 knockdown that were reconstituted with mouse NRP1 (mNRP1) (Fig. 2D-G, Fig. S2A,B). The expression of mNRP1 rescued the intercellular gap defect (Fig. 2D, white asterisk) observed between adjacent cells with endogenous NRP1 knockdown (Fig. 2D-G), thus confirming the role of NRP1 in regulating adherens junctions.
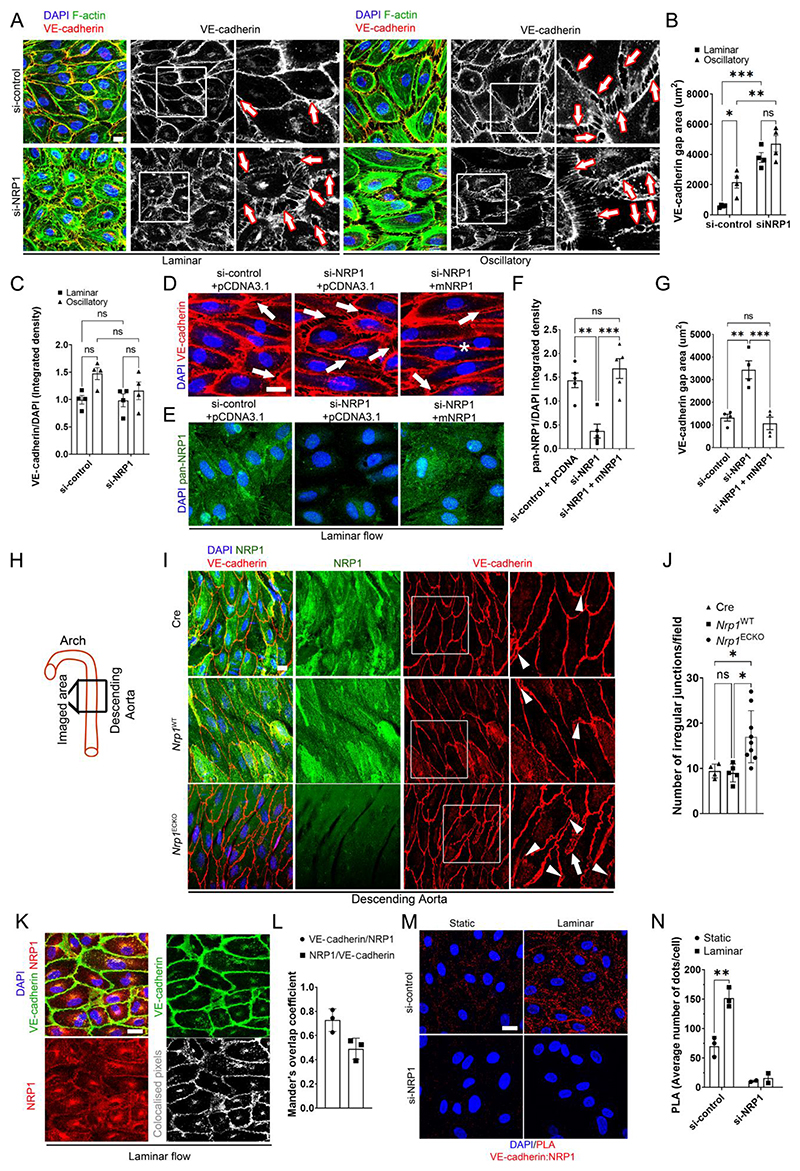
Fig. 2. NRP1 regulates adherence junctions by VE-cadherin.
(A) HUVECs transfected with si-control or si-NRP1 for 72 hours were exposed to laminar or oscillatory flow for 24 hours and stained for VE-cadherin (red and grey), F-actin (green) and DAPI (blue). Scale bar = 20 μm. White squares indicate the areas shown in higher magnification. Arrows indicate representative gaps between adjacent cells. (B) VE-cadherin staining gap (μm2) between neighboring cells measured in optical z-stacks. Data are presented as means ± SEM. N = 4 biological replicates per group. ns = non-significant, *p < 0.05, **p < 0.005, ***p < 0.001 by two-way ANOVA. (C) VE-cadherin integrated density measured in optical z-stacks normalized to DAPI. Data are presented as means ± SEM. N = 4 biological replicates per group. ns = non-significant by two-way ANOVA. (D and E) HUVECs transfected with si-control or si-NRP1 and pCDNA3.1 empty vector or pCDNA3.1 encoding WT mouse NRP1 for 72 hours and exposed to laminar flow for 24 hours were stained for VE-cadherin (red) (D) or with a pan-NRP1 antibody (green) (E) and counterstained with DAPI (blue). Scale bar = 20 μm. Arrows indicate representative gaps between adjacent cells. Asterisk indicates normal cellcell junction. (F) Quantification of NRP1 integrated density measured in optical z-stacks and normalized to DAPI. Data are presented as means ± SEM. N = 4 biological replicates per group. **p < 0.005, ***p < 0.001 by one-way ANOVA. (G) VE-cadherin gap (μm2) between adjacent cells measured in optical z-stacks Data are presented as means ± SEM. N = 4 biological replicates per group. **p < 0.005, ***p < 0.001 by one-way ANOVA. (H) Schematic indicating the aortic regions analyzed in (I) and (J). (I) C57BL/6 mice carrying two WT NRP1 alleles expressing Cdh5(PAC)-iCreERT2 (Cre), Nrp1fl/fl (Nrp1WT) or Nrp1fl/fl;Cdh5(PAC)-iCreERT2 (Nrp1ECKO) littermates injected daily with tamoxifen (12.5mg/kg) for 5 days at 4 weeks of age. Aortae were collected after 4 weeks and immunostained for NRP1 (green), VE-cadherin (red), DAPI (blue); Scale bar = 20 μm. White squares indicate the areas shown in higher magnification. White arrowheads indicate VE-cadherin discontinuous irregular patterns. White arrows show finger-like protrusions. (J) Number of irregular VE-cadherin junctions per field in optical z-stacks. Data are presented as means ± SD. N > 4 mice per group. *p < 0.05 by one-way ANOVA. (K) HUVECs subjected to laminar flow for 24 hours were stained for NRP1 (red), VE-cadherin (green) and DAPI (blue). Grey scale image represents NRP1/VE-cadherin co-localized pixels. Scale bar = 20 μm. (L) Mander’s coefficient of VE-cadherin and NRP1 colocalization. Data are presented as means ± SD. N = 3 biological replicates per group. (M) PLAs (red) for NRP1 and VE-cadherin in HUVECs transfected with si-control or si-NRP1 for 72 hours and subjected to static or laminar flow for 24 hours. Cells were counterstained with DAPI (blue). Scale bar = 20 μm. (N) Average PLA signal per cell measured in a minimum of 80 cells per experiment in 3 experiments for si-control and 2 experiments for si-NRP1. Data are presented as means ± SD. *p < 0.05 by paired t-test).
To further investigate the role of NRP1 in adherens junction stability in ECs, we analyzed VE-cadherin organization in analogous areas of whole-mounted descending aortae (Fig. 2H) isolated from tamoxifen-inducible endothelium-specific NRP1 knockout (Nrp1ECKO), NRP1-expressing control littermates carrying Nrp1 floxed alleles (Nrp1WT), and mutants carrying Nrp1 wild-type alleles and expressing tamoxifen-inducible endothelium-specific Cre (Cre). Nrp1ECKO mice showed reduced NRP1 levels in the endothelium compared to Nrp1WT or Cre control mutants, confirming the efficacy of the Nrp1 gene deletion strategy (Fig. 2I). ECs of aortae isolated from Nrp1WT and Cre mutants showed continuous VE-cadherin staining at the junctions, with few regions showing discontinuous or irregular patterns, and a consistent alignment with the blood flow direction (Fig. 2I, J). In contrast, VE-cadherin staining in Nrp1ECKO mice showed a statistically significant increase in irregular junctions (Fig. 2I, J), with gaps between adjacent cells (Fig. 2I, arrowheads) and finger-like protrusions (Fig. 2I, arrows), consistent with our in vitro observations (Fig. 2A-C). Together, these data indicate that NRP1 promotes adherens junction stability in ECs exposed to high-shear laminar flow.
Because knockdown of NRP1 affects VE-cadherin organization and adherens junction stability, we hypothesized that NRP1 was part of the multi-protein complex forming adherens junctions. Co-localization analysis of HUVECs exposed to laminar flow showed that NRP1 and VE-cadherin colocalized at the plasma membrane (Fig. 2K, L), in line with previous observations in ECs cultured in the absence of flow (35, 36). We performed proximity ligation assays (PLAs), which generate a fluorescent signal when target proteins are within 40 nm of each other, to investigate whether NRP1 and VE-cadherin formed a complex and whether this complex was modulated by laminar flow. PLA analysis showed that NRP1 and VE-cadherin formed a complex in the absence of flow and that laminar flow significantly increased this interaction (Fig. 2M, N). Moreover, the PLA signal was reduced in HUVECs with NRP1 knockdown, confirming the specificity of the assay. Together, these data show that NRP1 is part of the adherens junctions and that exposure to laminar shear stress increases the interaction of NRP1 and VE-cadherin.
NRP1 forms a complex with p120 catenin and VE-cadherin and promotes the interaction of p120 catenin and VE-cadherin to stabilize adherens junctions
To further investigate the interaction between NRP1 and VE-cadherin, we performed a HRP–based proximity-labelling assay that results in the biotinylation of proteins within a 20 nm radius (49) (Fig. 3A) in HUVECs exposed to laminar flow. Western blotting demonstrated that the streptavidin pulldowns of biotinylated proteins contained endogenous NRP1, NRP1-HRP and VE-cadherin but not the intracellular proteins GAPDH, β-tubulin or transferrin receptor (CD71), a plasma membrane protein (Fig. 3B-D). The streptavidin pulldowns also contained p120 catenin, which interacts with the intracellular juxtamembrane domain of VE-cadherin to promote adherens junction stability by preventing VE-cadherin endocytosis (5, 10) (Fig. 3B-D). These data indicate that the three proteins form a protein complex.
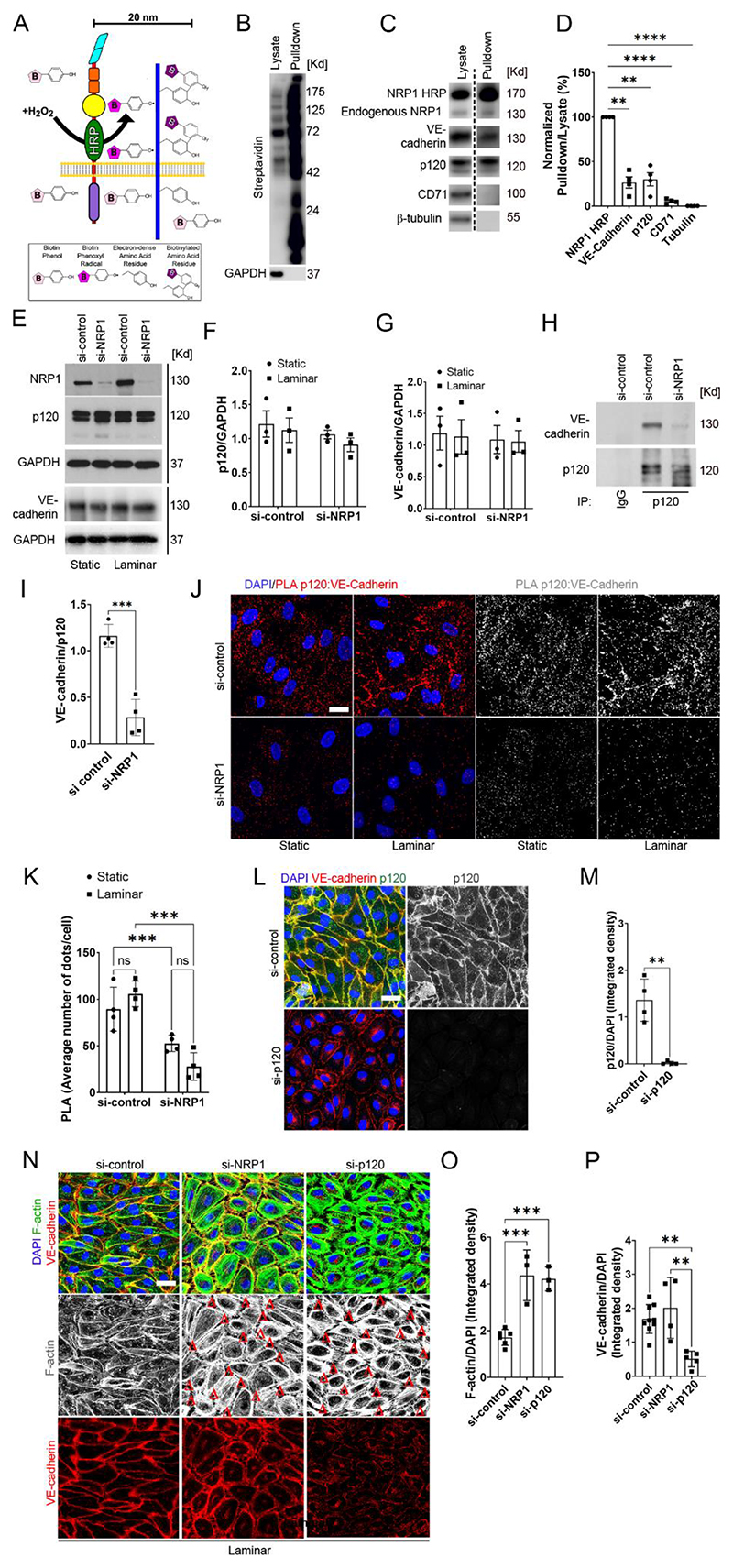
Fig. 3. Loss of NRP1 reduces the interaction between p120 catenin cadherin and VE-cadherin, resulting in adherens junction and cytoskeletal disruption.
A) Schematic illustrating the HRP–based proximity-labelling in which an HRP-containing protein exposed to hydrogen peroxide for 1 minute oxidizes fluid-phase–fed biotin tyramide, resulting in the biotinylation of proteins within a 20 nm radius. (B and C) Biotinylated proteins were pulled down from lysates with streptravidin beads, and pulldown samples and lysates were analysed by immunoblotting for the indicated proteins (dashed line indicates images are from different exposure times). (D) Quantification of pulled down proteins normalized to pulled down NRP1-HRP (100%) and tubulin (0%). Data are presented as means ± SEM. N = 4 biological replicates per group. **p<0.009; ****p<0.0001 by repeated measure one-way ANOVA with Dunnett post hoc test. (E to G) Representative immunoblotting (E) for NRP1, p120 catenin, VE-Cadherin and GAPDH in HUVECs transfected for 72 hours with si-control or si-NRP1 and exposed to laminar flow or static conditions for 24 hours. Densitometry analysis of p120 catenin (F) and VE-cadherin (G) relative to GAPDH. Data are presented as means ± SEM. N = 3 biological replicates per group. Two-way ANOVA. (H) Endogenous VE-cadherin and p120 catenin were co-immunoprecipitated from lysates of HUVECs cultured under static conditions and transfected with control or NRP1 siRNAs for 72 hours. p120 catenin or control IgG immunoprecipitates were immunoblotted for VE-cadherin and p120 catenin. (I) Densitometry analysis of endogenous co-immunoprecipitated VE-cadherin normalized to immunoprecipitated p120 catenin. Data are presented as means ± SEM. N = 4 biological replicates per group. ***p < 0.001 by paired t-test. (J) PLA (red, grey) for p120 catenin:VE-cadherin in HUVECs transfected with si-control or si-NRP1 for 72 hours, exposed to static or laminar flow conditions for 24 hours and counterstained with DAPI (blue). Scale bar = 20 μm. (K) Average PLA signal per cell was measured in a minimum of 80 cells per experiment from 4 independent experiments. Data are presented as means ± SD. **p < 0.001 by two-way ANOVA. (L) Staining for VE-cadherin (red), p120 catenin (green and gray) of HUVECs transfected for 72 hours with si-control or si-p120 catenin and exposed to laminar flow for 24 hours. Scale bar = 20 μm. (M) Quantification of p120 catenin integrated density in optical z-stacks normalized to DAPI. Data are presented as means ± SD. N = 4 biological replicates per group. **p < 0.001 by t-test. (N) HUVECs transfected for 72 hours with si-control, si-NRP1 or si-p120 catenin were subjected to laminar flow for 24 hours and stained for VE-cadherin (red), F-actin (green and grey) and DAPI (blue). Red Δs indicate gaps between adjacent cells. Scale bar = 20 μm. (O) Quantification of F-actin integrated density in optical z-stacks normalized to DAPI. Data are presented as means ± SD. N ≥ 3 biological replicates per group. ns = non-significant, ***p < 0.0001 by one-way ANOVA. (P) Quantification of VE-cadherin integrated density in optical z-stacks and normalized to DAPI. Data are presented as means ± SD. N ≥ 4 biological replicates per group. ns = non-significant, **p < 0.005 by one-way ANOVA.
To investigate the mechanism by which NRP1 regulates the stability of adherens junctions in ECs exposed to laminar flow, we investigated whether NRP1 modulates the interaction of VE-cadherin with p120 catenin. Immunoblotting (Fig. 3E) and immunostaining (Fig. S2C) analyses of ECs exposed to laminar flow showed that NRP1 knockdown did not affect p120 catenin and VE-cadherin levels (Fig. 3E-G). Next, we investigated whether NRP1 knockdown affected the ability of p120 catenin to interact with VE-cadherin. Co-immunoprecipitation analysis performed in HUVECs cultured under static conditions showed that NRP1 knockdown reduced the interaction of VE-cadherin with p120 catenin (Fig. 3H, I). Accordingly, PLA analysis showed that in control HUVECs, VE-cadherin and p120 catenin formed a complex under both static and laminar flow conditions and that NRP1 knockdown reduced the PLA signal compared to control cells under both static and flow conditions (Fig. 3J, K). Together, these data demonstrate that NRP1 promotes the interaction between p120 catenin and VE-cadherin (Fig. 3H-K).
We next investigated whether p120 catenin or NRP1 knockdown similarly affected adherens junctions and the actin cytoskeleton. NRP1 or p120 catenin knockdown (Fig. 3L, M; Fig. S3A-C) induced a rounded cell morphology characterized by abundant cortical actin and an increase in F-actin staining compared to control cells (Fig. 3N, O). In HUVECs depleted of either p120 catenin or NRP1, F-actin staining exhibited gaps between adjacent cells (Fig. 3N, red deltas) suggesting that NRP1 and p120 catenin contributed to a similar extent to the stability of adherens junctions and regulate actin cytoskeleton dynamics. HUVECs with p120 catenin knockdown, but not with NRP1 knockdown, showed a significant decrease in VE-cadherin levels (Fig. 3P), in agreement with a role for p120 catenin in preventing VE-cadherin endocytosis (Fig. 2A, C; Fig. 3E, G, N, P; Fig. S2C). These data suggest that in the absence of NRP1, the residual interaction between p120 catenin and VE-cadherin maintains VE-cadherin protein levels but is insufficient to stabilize the junctions between adjacent cells. Another possibility is that NRP1 knockdown reduces or delays the trafficking of VE-cadherin into endosomes and subsequent VE-cadherin degradation, as previously shown for VEGFR2 (50) and as suggested by changes in endosomal trafficking pathways revealed by GO analysis (Fig. 1M, N). Together, these data demonstrate that NRP1 forms a protein complex with VE-cadherin and that NRP1 promotes p120 catenin interaction with VE-cadherin, increasing the stability of adherens junctions, thus promoting cell-cell contact and regulating actin cytoskeleton remodeling.
Destabilizing adherens junctions induces TGF-β membrane localization and TGF-β signaling
VE-cadherin associates with TGFβR1 and TGFβR2 receptors and modulates TGF-β downstream signaling by promoting TGF-β localization in puncta on the plasma membrane in ECs cultured under static conditions (13). NRP1 promotes TGF-β signaling in breast cancer cells (38) whereas it suppresses TGF-β pathways in ECs (32). GO analysis suggested a role for NRP1 in regulating TGF-β-dependent SMAD pathways in HUVECs exposed to laminar flow (Fig. 1N). Thus, we hypothesized that NRP1 may suppress VE-cadherin-dependent TGF-β signaling in ECs exposed to flow. Co-staining showed that NRP1 and TGFBR2 localized at the plasma membrane and in intracellular compartments as expected (Fig. 4A) and that TGFBR2 knockdown reduced TGFBR2 protein levels, confirming the specificity of the anti-TGFBR2 antibody (Fig. 4A; Fig. S3D-F). HUVECs with knockdown of either NRP1 or p120 catenin showed marked TGFBR2 localization at the plasma membrane (Fig. 4A; Fig. S3F). Accordingly, we showed that NRP1 knockdown increased the co-localization of TGFBR2 and p120 catenin in HUVECs exposed to laminar flow (Fig. S4A, B) without altering TGFBR2 protein levels (Fig. S4A, C, D, E). These data indicate that NRP1 inhibited TGFBR2 plasma membrane localization and suggest that by promoting adherens junction stability, NRP1 and p120 catenin suppressed TGFBR2-dependent signaling in ECs exposed to flow. We therefore assessed the phosphorylation status of the TGF-β downstream effectors SMAD2/3. Immunoblotting analysis showed that knockdown of NRP1 or p120 catenin increased the phosphorylation of SMAD2/3 (pSMAD2/3) compared to control cells (Fig. 4B, C, Fig. S5), indicating that the plasma membrane localization of TGFBR2 correlated with enhanced TGF-β signaling. The inner curvature of aortic arches is exposed to turbulent flow, resulting in low laminar shear stress, which induces TGF-β signaling in ECs (51). Whole-mount co-staining for NRP1 and pSMAD2/3 in aortic arches (Fig. 4D) showed a statistically significant increase in SMAD2/3 phosphorylation in the endothelium of Nrp1ECKO compared to Nrp1WT littermates (Fig. 4E, F). Moreover, aortic ECs of Nrp1WT and Nrp1ECKO showed a cobblestone morphology, with zipper-like VE-cadherin junctions (Fig. 4E) as expected in vascular regions subjected to disturbed flow (52). Together, these data suggest that by promoting adherens junction stability, NRP1 suppresses TGF-β downstream signaling in ECs exposed to flow.
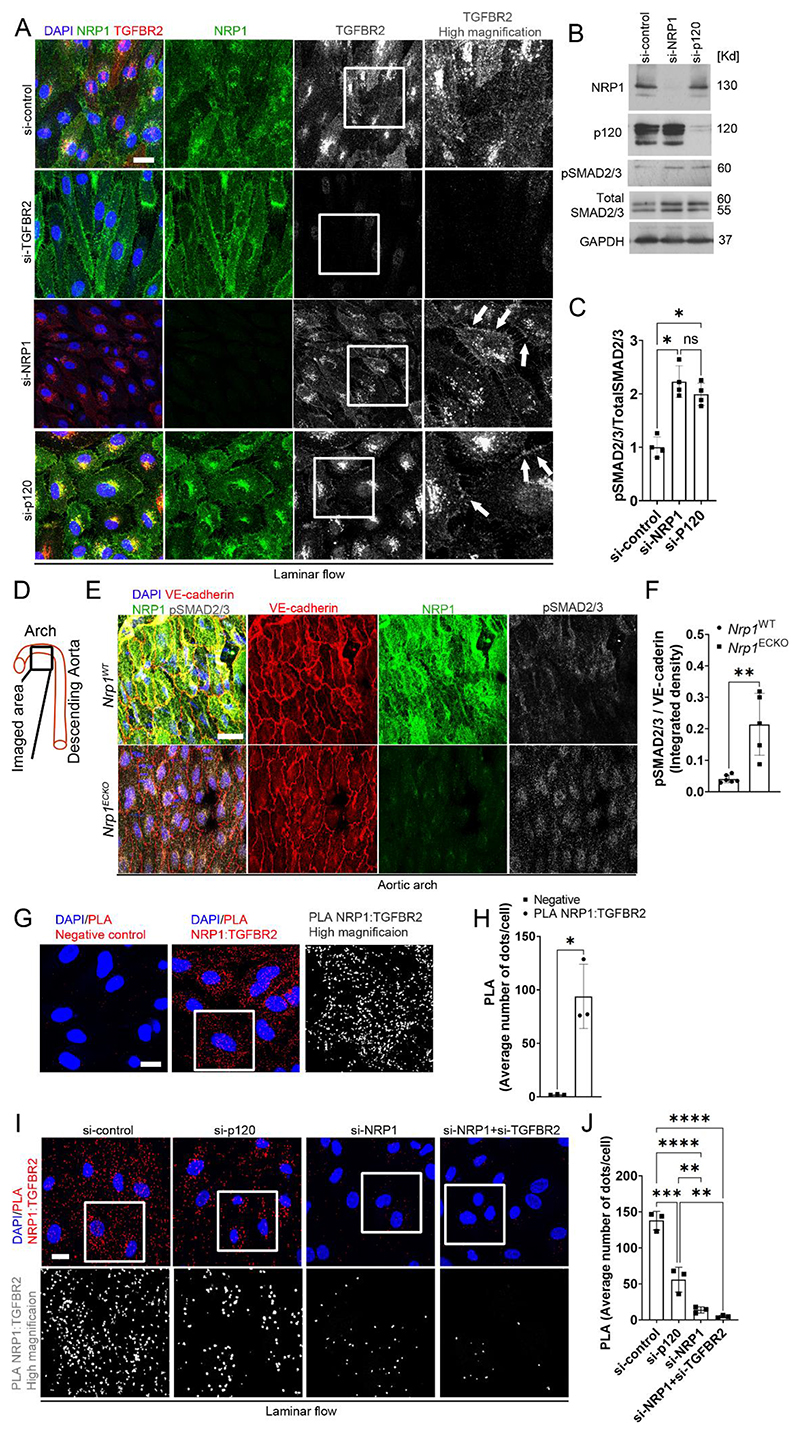
Fig. 4. NRP1 and p120 catenin cooperate to suppress TGF-β signaling.
(A) HUVECs transfected for 72 hours with si-control, si-NRP1, si-p120 catenin or si-TGFBR2 and exposed to laminar flow for 24 hours were stained for TGFBR2 (red and grey), NRP1 (green) and DAPI (blue). Scale bar = 20 μm. White squares indicate the areas shown in higher magnification. White arrows show TGFBR2 plasma membrane enrichment. N = 3 biological replicates per group. (B) Representative immunoblotting for NRP1, p120 catenin, pSMAD2/3, total SMAD2/3 and GAPDH of HUVECs transfected for 72 hours with si-control, si-NRP1 or si-p120 catenin. (C) Quantification of SMAD2/3 phosphorylation relative to total SMAD2/3 expressed as fold change compared to si-control. Data are presented as means ± SD. N = 4 biological replicates per group. ns = non-significant, *p < 0.05 by one-way ANOVA. (D) Schematic indicating the region in the aortic arch analyzed in (E) and (F). (E) Aortic arches of Nrp1WT or Nrp1ECKO mice injected daily with tamoxifen (12.5mg/kg) for 5 days at 4 weeks of age, collected after 4 weeks and immunostained for NRP1 (green), VE-cadherin (red), pSMAD2/3 (grey) and DAPI (blue). Scale bar = 20 μm. (F) Quantification of pSMAD2/3 integrated density relative to VE-cadherin. Data are presented as means ± SD. N ≥ 5 mice per group. **p < 0.005 by unpaired t-test. (G) PLA (red, grey) for NRP1:TGFBR2 in HUVECs cultured under static conditions. Scale bar = 20 μm. White square shows the location of enhanced magnification of PLA shown in grey scale in the righthand side panel. (H) Average PLA signal per cell measured in a minimum of 80 cells per experiment from 3 independent experiments. Data are presented as means ± SD. *p < 0.05 by paired t-test. (I) PLA (red, grey) for NRP1 :TGFBR2 in HUVECs transfected for 72 hours with si-control, si-NRP1, si-p120 catenin or co-transfected with si-NRP1 and si-TGFBR2, exposed to laminar flow for 24 hours and counterstained with DAPI (blue). Scale bar = 20 μm. White square shows the location of enhanced magnification of PLA shown in grey scale in the bottom panels. (J) Average PLA signal was measured in a minimum of 80 cells per experiment from 3 independent experiments. Data are presented as means ± SD. **p < 0.005, ***p < 0.001, ****p < 0.0001 by one-way ANOVA.
To further investigate the mechanism by which NRP1 regulates TGF-β signaling, we investigated whether NRP1 forms a complex with TGFBR2 in ECs. PLAs showed that NRP1 and TGFBR2 formed a complex in HUVECs cultured under static conditions (Fig. 4G, H), in line with previous observations in other cell types and in cell-free assays (38). We next investigated whether junction stability affected the interaction between NRP1 and TGFBR2. PLAs showed that HUVECs with p120 catenin knockdown exhibited a significant reduction in the interaction of TGFBR2 and NRP1 compared to control HUVECs (Fig. 4I, J). In addition, HUVECs with NRP1 knockdown alone or in combination with TGFBR2 knockdown showed a significant reduction in PLA signals between TGFBR2 and NRP1, confirming the specificity of the PLA assay (Fig. 4I, J). Together, these data indicate that under laminar flow, adherens junction stability promotes NRP1 and TGFBR2 interaction which reduces TGFBR2 plasma membrane localization and inhibits TGF-β signaling.
NRP1 suppresses atherosclerosis and endothelial activation
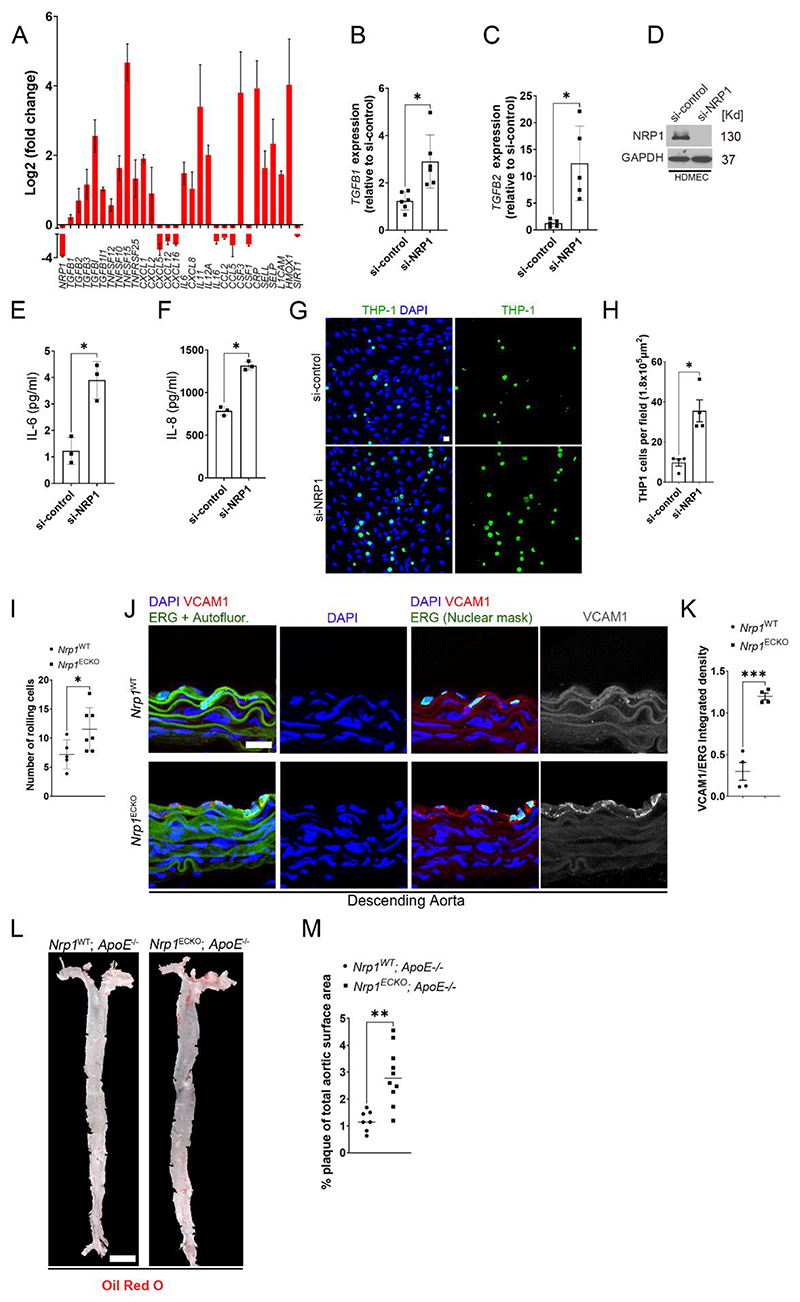
Endothelial TGF-β signaling drives vascular inflammation and atherosclerosis and inhibition of TGF-β pathways reduces inflammation, vascular permeability and atherosclerosis progression (53). Thus, we analyzed the differential expression of genes with a role in atherogenesis and vascular inflammation. NRP1 knockdown induced the expression of mRNAs encoding TGF-β isoforms, TNF superfamily members, C-reactive protein (CRP) and pro-inflammatory cytokines belonging to the interleukin family such as IL6, IL11 and IL12 (Fig. 5A). Furthermore, NRP1 knockdown cells showed increased expression of mRNAs encoding the pro-inflammatory neutrophil chemokines CXCL1 and CXCL8 and reduced expression of chemokines with an inhibitory role in atherogenesis such as CXCL5, CXCL12 and CXCL16 (54, 55) (Fig. 5A). Also, NRP1 knockdown increased the expression of mRNAs encoding pro-inflammatory adhesion molecules that regulate leukocyte adhesion to sites of inflammation such as P-selectin (SELP) and L-selectin (SELL) (Fig. 5A). We used RT-qPCR analysis to confirm the transcriptional regulation of some of the differentially expressed genes such as TGFB1 and TGFB2 in HUVECs (Fig. 5B, C) and measured the release of IL-6 and IL-8 in human microvascular endothelial cells (HDMECs) (Fig. 5D-F).
Fig. 5. NRP1 suppresses inflammatory pathways and atherosclerosis.
(A) Fold changes (Log2) in gene expression of HUVECs transfected with si-NRP1 for 72 hours and exposed to laminar flow for 24 hours relative to HUVECs transfected with si-control under the same experimental conditions. Data are presented as means ± SD. N = 3 biological replicates per group. (B) TGFB1 gene expression in HUVECs transfected for 72 hours with si-control or si-NRP1 and cultured under static conditions, expressed as fold change relative to si-control. Data are presented as means ± SD. N = 6 biological replicates per group. *p < 0.05 by paired t-test. (C) TGFB2 gene expression was measured in HUVECs transfected for 72 hours with si-control or si-NRP1 and cultured under static conditions. TGF-β2 gene expression was expressed as fold change relative to si-control. Data are presented as means ± SD. N = 5 biological replicates per group. *p < 0.05 by paired t-test. (D) Representative immunoblotting for NRP1 and GAPDH of HDMECs transfected for 72 hours with si-control or si-NRP1 and cultured under static conditions. (E and F) Quantification by ELISA of IL-6 (E) and IL-8 (F) secretion in HDMECs transfected for 72 hours with si-control or si-NRP1. Data are presented as means ± SD. N = 3 biological replicates per group. *p < 0.05 by t-test. (G) THP-1 cells labelled with Calcein-AM 1μM (green) adhering to a confluent monolayer of HUVECs transfected with si-control or si-NRP1 for 72 hours and counterstained with DAPI (blue). (H) Number of adhering THP-1 cells per field. Data are presented as means ± SEM. N = 3 biological replicates per group. *p < 0.05 by paired t-test. (I) Rolling leukocytes in the postcapillary venules of the mesentery of Nrp1WT (n=5) or Nrp1ECKO (n=7) mice injected daily with tamoxifen (12.5mg/kg) for 5 days at 4 weeks of age and imaged after 2 weeks. Data are presented as means ± SD. *p < 0.05 by unpaired t-test. (J) Rings of descending aortae from Nrp1WT or Nrp1ECKO mice injected daily with tamoxifen (12.5mg/kg) for 5 days at 4 weeks of age and immunostained after 4 weeks for ERG (green), VCAM1 (red and grey) and counterstained with DAPI (blue). Scale bar = 20 μm. ImageJ software was used to mask the autofluorescence in the green channel to reveal ERG-DAPI double positive ECs. (K) Quantification of VCAM1 integrated density relative to ERG. Data are presented as means ± SD. N = 4 mice per group. ***p < 0.001 by paired t-test. (L) En-face aortae from Nrp1WT;ApoE-/- or Nrp1ECKO;ApoE-/- mice, injected daily with tamoxifen (12.5mg/kg) for 5 days at 4 weeks and stained 18 weeks of age with Oil-Red-O (Red). Red staining shows plaque deposition on the inner wall of aortae. Scale bar = 2.1 mm (M) Quantification of plaque deposition was expressed as percentage of total aortic surface coverage. Data are presented as means ± SD. N ≥ 7 mice per group. **p < 0.005 by t-test.
We investigated whether these transcriptional changes resulted in endothelial activation and increased interaction with immune cells. We found that more THP-1 leukemia monocytic cells (56) adhered to monolayers formed by HUVECs with NRP1 knockdown than to control HUVEC monolayers (Fig 5G, H). We sought to determine whether deletion of NRP1 in the endothelium increased the leukocyte-endothelium interaction in vivo. Consistent with the in vitro data, intravital microscopy revealed a significant increase in leukocyte rolling in the postcapillary venules of the mesentery in Nrp1ECKO mice compared to those of Nrp1WT littermates (Fig. 5I). We assessed whether deletion of NRP1 up-regulated VCAM1, an immunoglobulin-like adhesion molecule expressed by activated endothelial cells that promotes monocyte adhesion and plays a key role in atherosclerosis (57, 58). Immunostaining for VCAM1 and the endothelial marker ERG (59) showed that Nrp1ECKO aortic rings from descending aortas showed significantly increased endothelial VCAM1 expression compared to those from Nrp1WT littermates (Fig. 5J,K). The lack of change in VCAM1 gene expression in HUVECs with NRP1 knockdown (Fig. 1H) suggests that long-term deletion of NRP1 is required to increase VCAM1 expression. Together, these data indicate that NRP1 suppresses a pro-inflammatory phenotype in endothelial cells, preventing endothelial cell activation and the interaction of leukocytes with the vascular endothelium.
Endothelial activation and dysfunction occur at an early stage of atherosclerosis and contribute to disease progression leading to a chronic inflammatory environment at sites of atheroma formation. Because NRP1 suppresses TGF-β signaling and endothelial activation (Fig. 4A-F; Fig. 5A-K), we hypothesized that NRP1 has atheroprotective functions. Thus, we crossed Nrp1 endothelium-specific inducible knockout mice with ApoE knockout mutants (60) and measured the total atherosclerotic plaque area within the aorta in chow-fed mice, 14 weeks after tamoxifen injection to achieve CRE activation. Whereas Nrp1WT; ApoE-/- mutants showed few Oil-red-O positive lesions and small plaque areas in the aorta, Nrp1ECKO;ApoE-/- mice showed statistically significant increases in Oil-red-O staining, indicating an increase in plaque area (Fig. 5L, M). These data suggest that NRP1 plays a tonic repressive role in the endothelium to prevent EC activation, thereby reducing atherosclerotic plaque growth.
Discussion
Endothelial NRP1 is a key regulator of developmental angiogenesis. In this context, NRP1 regulates sprouting angiogenesis by integrating growth factor- and extracellular matrix component-mediated signaling (61). In the vasculature of adult mice, NRP1 regulates physiological and pathological VEGF- and Semaphorin 3A-induced permeability (35, 36, 62), but its broader role in maintaining EC function and consequently vascular homeostasis is less defined. Shear stress is a key regulator of physiological and pathological aspects of endothelial function, with physiological unidirectional shear stress in atheroprotected regions of the vasculature suppressing pro-inflammatory endothelial activation and leukocyte recruitment in response to inflammatory stimuli (63, 64). Conversely, low multidirectional shear stress experienced by ECs in atheroprone regions induces leukocyte migration into the sub-intimal space by driving oxidative stress, reduced endothelial nitric oxide synthase (eNOS) expression and upregulation of inflammatory cytokines and adhesion molecules (63, 64). Whereas NRP1 is part of a mechanosensing complex that regulates VEGFR2 phosphorylation during an acute response to shear stress (37), the role of NRP1 in regulating longer-term shear stress-induced responses such as alignment to flow, junctional remodeling and flow-mediated gene transcription remained to be explored. Here we showed that exposure to shear stress for 24 hours increased NRP1 levels (Fig. 1A, B) and that NRP1 is a component of adherens junctions (Fig. 2M,N; Fig. 3A-J) required to stabilize cell-cell junctions and regulate adherens junction signaling under protective laminar flow conditions. Our data also showed that ECs with or without NRP1 similarly modulated the transcription of flow-responsive genes such as KLF2 (65) and KLF4 (66, 67) (Fig. 1G; Fig. S1B), suggesting that NRP1 is not required for sensing shear stress in long-term flow conditions. However, transcriptomic analysis showed that NRP1 regulated the transcriptomic profile of ECs exposed to different flow patterns and affected pathways involved in cell-cell adhesion, inflammation, immune response, oxidative stress and TGF-β signaling, which are all involved in modulating endothelial function (Fig. 1I, O).
Arterial endothelial ECs overlying atherosclerotic plaques in ApoE-/- mice show disrupted junctional morphology with chaotic small membrane extensions and finger-like projections between neighboring ECs and paracellular gaps, consistent with the increased vascular permeability, leukocyte adhesion and transmigration characteristic of atherogenesis (68). Our data revealed that NRP1 knockdown or knockout in endothelial cells increased leukocyte adhesion and rolling (Fig. 5G-I) and induced irregular junctions in ECs exposed to laminar flow and in those lining the descending aorta (Fig. 2A-J), suggesting that NRP1 is required for promoting the atheroprotective signal of laminar unidirectional flow. Accordingly, we showed that endothelium-specific deletion of NRP1 in a hypercholesterolemic mouse model resulted in increased atherosclerotic plaque growth (Fig. 5L,M). Because mice with endothelium-specific NRP1 knockout do not have defects in lipid absorption and metabolism (69), our data suggest that by regulating adherens junctions and the transcription of inflammatory genes involved in EC activation, NRP1 suppresses vascular inflammation (Fig. 5A-K) and atherogenesis (Fig. 5L, M; Fig. 6 A and B).
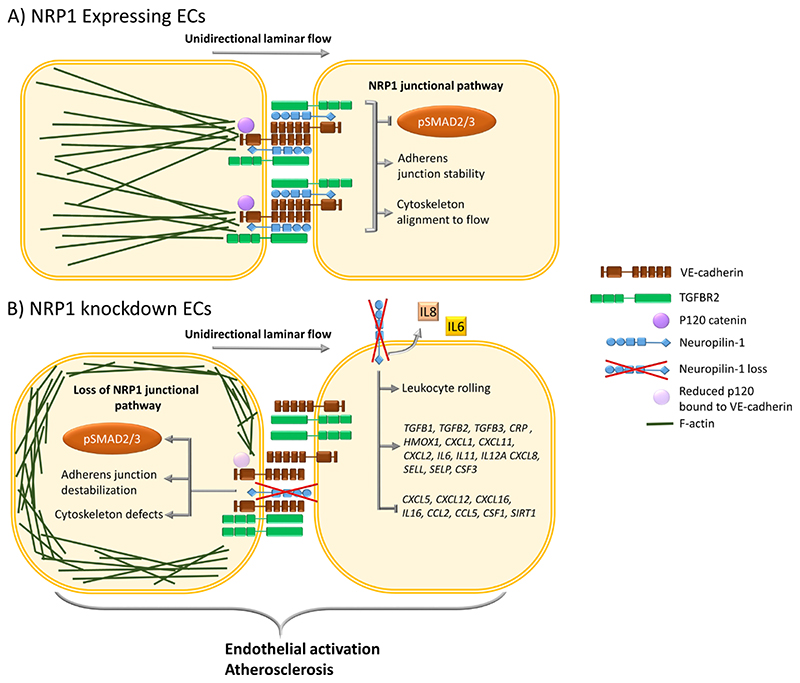
Fig. 6. Schematic representation of the effect of NRP1 on the TGFBR2-VE-cadherin pathway in regulating adherens junctions and inflammatory responses.
(A) In ECs, NRP1 interacts with VE-cadherin and TGFBR2, reducing TGF-β signaling and stabilizing adherens junctions. NRP1 promotes VE-cadherin interaction with p120 catenin, leading to optimal coupling of the actin cytoskeleton with the adherens junction complex and to alignment of the cytoskeleton to the direction of flow. (B) In the absence of NRP1, the interaction of NRP1 with VE-cadherin is lost and that of VE-cadherin with p120 catenin is reduced, leading to cytoskeleton remodeling and abundant cortical actin and resulting in a lack of cytoskeleton alignment to the flow direction. Furthermore, in ECs lacking NRP1, adherens junction destabilization increases the plasma membrane localization of TGFBR2, resulting in the downstream phosphorylation of SMAD2/3. Loss of NRP1-dependent signaling pathways results in EC activation, leading to increases in the expression of genes encoding proinflammatory cytokines, chemokines, adhesion molecules, secretion of IL6 and IL8, leukocyte rolling, and plaque formation in an atherosclerosis mouse model.
Our finding that NRP1 formed a complex with VE-cadherin (Fig. 2M, N; Fig. 3B-D) and stabilized adherens junctions in ECs exposed to atheroprotective flow (Fig. 2A,G) provides insight into the role of NRP1 in these subcellular structures and suggests that this function promotes atheroprotective signaling and prevents atherosclerosis. We provided mechanistic data concerning the regulation of adherens junction plasticity with potential implications for the understanding of the role of NRP1 in controlling vascular permeability. NRP1 re-localizes at cell-cell contacts upon binding of VEGF or CendR tetramers, an event that coincides with the induction of vascular leakage in a manner requiring the NRP1 cytoplasmic domain but independently of VEGFR2 expression and activity (36). Thus, this work suggests that in addition to promoting VEGFA-induced vascular permeability by acting as a VEGF co-receptor and enhancer of VEGFR2-dependent signaling (35), NRP1 autonomously regulates the remodeling of endothelial junctions. Our findings that NRP1 interacted with VE-cadherin (Fig. 2M,N; Fig. 3B-D) to promote adherens junction stability supports the published work showing a role for NRP1 in regulating permeability and raises the possibility that binding of VEGF or CendR peptides could interfere with the ability of NRP1 to bind VE-cadherin, thus directly affecting junctional stability and consequently permeability.
We propose that in addition to binding to VE-cadherin, NRP1 promotes the interaction of VE-cadherin with p120 catenin (Fig. 3H, K). In addition to regulating VE-cadherin availability at cell-cell contact sites by inhibiting VE-cadherin endocytosis (70), p120 catenin interacts with p120 catenin-p190RhoGAP and regulates actin cytoskeleton remodeling through Rac-dependent antagonism of RhoA in fibroblasts, with p120 catenin knockdown enhancing RhoA activity and actin stress fibers (71). Our data showed that knockdown of p120 catenin or NRP1 resulted in disorganized adherens junctions and accumulation of cortical stress fibers (Fig. 3N-P). These data suggest that by interacting with VE-cadherin and promoting the binding of p120 catenin to VE-cadherin, NRP1 provides a spatial cue to coordinate RhoGTPase-dependent actin remodeling and junctional stability. These data are consistent with previous evidence showing that NRP1 regulates fibronectin-induced actin cytoskeleton, migration (33) and the activity of the small GTPase CDC42 in ECs (34).
In a sepsis-induced lung injury mouse model, inflammatory stimuli attenuate p120 catenin expression. Conversely, deficiency of p120 catenin in the lung vasculature in mice enhances LPS-induced increases in TNFα, IL6 and neutrophil adhesion to pulmonary ECs and transmigration (72). Because knockdown of NRP1 or p120 catenin induced similar cytoskeletal changes and junctional gaps (Fig. 3N-P), it could be argued that by interacting with VE-cadherin and by promoting the interaction of p120 catenin and VE-cadherin, NRP1 suppresses the pro-inflammatory phenotype, which was reflected in the enhanced leukocyte adhesion (Fig. 5G-I), endothelial activation (Fig. 5J, K) and atherogenesis in NRP1 endothelial knockout mouse mutants (Fig. 5L-M). However, the lack of effect of NRP1 knockdown on p120 catenin and VE-cadherin abundance (Fig. 3E, G, N, P), combined with the decrease in VE-cadherin protein levels induced by p120 catenin knockdown (Fig. 3N, P), indicates that alternative NRP1-dependent pathways, such as those controlling oxidative stress and endothelial cell senescence (47), may contribute to endothelial activation and transcriptional changes.
NRP1 suppresses the TGF-β pathway in ECs (32) through an unknown molecular mechanism. Here, we confirmed that NRP1 suppressed TGF-β signaling in ECs by showing that NRP1 knockdown or genetic deletion in the endothelium increased SMAD2/3 phosphorylation in cultured cells and in mouse aortic endothelium (Fig. 4B-F). We further demonstrated that NRP1 and TGFBR2 formed a complex in ECs exposed to laminar flow (Fig. 4I, J) and that p120 catenin knockdown reduced this interaction, leading to increased SMAD2/3 phosphorylation, comparable to that following NRP1 depletion (Fig. 4B,C). Because NRP1 interacted with VE-cadherin and TGFBR2 and because p120 catenin knockdown increased TGFBR2 plasma membrane localization (Fig. 4A) and reduced the interaction of NRP1 with TGFBR2 (Fig. 4 I,J), our data suggest that adherens junction stability plays a role in suppressing TGFβ signaling through a mechanism requiring NRP1 interaction with VE-cadherin and TGFBR2 (Fig. 6, A and B). VE-cadherin junctional clustering recruits and promotes the assembly of TGFBRs to enhance TGF-β signaling (1). Thus, our data suggest that by interacting with VE-cadherin and TGFBR2, NRP1 suppresses VE-cadherin-dependent TGF-β signaling. Furthermore, endothelial TGF-β signaling drives vascular inflammation and atherosclerosis (53). We propose that NRP1 regulates atheroprotective signals which modulates transcription of inflammatory genes and EC activation, adherens junction stability and VE-cadherin-dependent TGF-β signaling. Inhibition or reduction of endothelial NRP1 in ECs may contribute to the progression of vascular disease.
Materials and Methods
Cell culture and transfection
HUVECs from pooled donors (Lonza, UK; cat#: C2519A) were cultured on 0.5 % gelatin (diluted from 2 % stock; Scientific Laboratory Supplies, UK; cat#: G1393-100) in EGM2 media with supplements (Lonza, UK; cat#: CC-3162). HDMECs were cultured on 0.5 % gelatin in MV2 media with supplements (Promocell, UK; cat#: C-22120). THP-1 cells were cultured in RPMI-1640 supplemented with 10% FBS (Labtech, UK); 2mM L-glutamine (Thermo Fisher Scientific, UK); 100 U/ml Penicillin-Streptomycin (Thermo Fisher Scientific, UK); 50 μM 2-mercaptoethanol (Merck, UK). For siRNA transfection, HUVECs and HDMECs were cultured for up to six passages and transfected with Lipofectamine RNAiMAX (Thermo Fisher Scientific, UK). Media was replaced 24 hours after transfection. The following siRNAs were used to transfect cells: SMARTPool SiRNA targeting NRP1 (Dharmacon, USA; cat#: L-019484-00-0020) sequence 1, CGAUAAAUGUGGCGAUACU (sense) and AGUAUCGCCACAUUUAUCG (antisense); sequence 2, GGACAGAGACUGCAAGUAU (sense) and: AUACUUGCAGUCUCUGUCC (antisense); sequence 3, GUAUACGGUUGCAAGAUAA (sense) and UUAUCUUGCAACCGUAUAC (antisense); sequence 4, AAGACUGGAUCACCAUAAA (sense) and UUUAUGGUGAUCCAGUCUU (antisens); SMARTPool SiRNA targeting TGFBR2 (Dharmacon, USA; cat#: L-003930-00-0005) sequence 1, CAACAACGGUGCAGUCAAG (sense) and CUUGACUGCACCGUUGUUG (antisense); sequence 2, GACGAGAACAUAACACUAG (sense) and CUAGUGUUAUGUUCUCGUC (antisense); sequence 3, GAAAUGACAUCUCGCUGUA (sense) and UACAGCGAGAUGUCAUUUC (antisense); sequence 4, CCAAUAUCCUCGUGAAGAA (sense) and UUCUUCACGAGGAUAUUGG (antisense); siRNA targeting TGFBR2 siRNA#2 (Silencer validated siRNA AM51331, ID#388), GGAAGUCUGUGUGGCUGU (sense) and UACAGCCACACAGACUUCC (antisense); SMARTPool SiRNA targeting p120 catenin (Dharmacon, USA; cat#: L-012572-00-0005) sequence 1, GGAAUGUGAUGGUUUAGUU (sense) and AACUAAACCAUCACAUUCC (antisense); sequence 2, UAGCUGACCUCCUGACUAA (sense) and UUAGUCAGGAGGUCAGCUA (antisense); sequence 3, GGACCUUACUGAAGUUAUU (sense) and AAUAACUUCAGUAAGGUCC (antisense); GAGUGAAGCUCGCCGGAAA (sense) and UUUCCGGCGAGCUUCACUC (antisense); siRNA targeting p120 catenin siRNA#2 (Silencer Pre-designed siRNA AM16704 ID#145776), CCUUACUGAAGUUAUUACC (sense) and GGUAAUAACUUCAGUAAGG (antisense); and Silencer® negative control siRNA (Life Technology, UK; cat#: AM4635). To induce NRP1 overexpression, HUVECs were co-transfected with control siRNA or siRNA targeting NRP1 and pCDNA3.1 encoding WT mouse NRP1(33) or pCDNA3.1 empty vector using Dharmafect Duo (Horizon, UK) following the manufacturer’s instructions. For the HRP–based proximity-labelling assay, the NRP1 HRP1 construct was synthesized by Geneart Gene Synthesis (Thermo Fisher Scientific, UK) and subcloned into the pLNT SFFV plasmid. Lentiviral particles were generated by transfecting Lenti-X 293T (Takara Bio, Saint-Germain-en-Laye, France) with the NRP1 HRP pLNT-SFFV plasmid alongside pGagPol, pMDVSVG. HUVECs were transduced with viral particles overnight.
Ibidi Pump System Quad
Untreated or siRNA-transfected HUVECs were seeded at a density of 2 x 105 per 100 μL in Ibidi slides (Thistle Scientific Ltd, UK; cat#: single-channel: IB-80178; multi-channel: IB-80606) precoated with 1% gelatin at 37°C at 5% CO2 for 1 hour. Cells were allowed to adhere and reach confluency for 24 hours before being connected to the fluidic unit. In parallel, the fluidic units of Ibidi Pump System Quad (Thistle Scientific Ltd, UK; cat#: 10962) were assembled in sterile conditions and loaded with EGM2 media. The system was allowed to equilibrate for 24 hours at 37°C at 5% CO2. The following day, slides were connected to the perfusion sets and the desired shear stress values were applied for 24hours (software Pump control v.1.5.4 by Ibidi v.1.5.4). HUVECs were exposed to different levels of shear stress depending on the experiments: laminar, 20 dynes/cm2 unidirectional; oscillatory, 5 dynes/cm2 1Hz (46, 73). At the end of the experiment, cells were washed with PBS (Fisher Scientific UK Ltd, UK; cat#: 11530546), fixed for 30 minutes at room temperature with warm 4% paraformaldehyde (Sigma-Aldrich-Merk, UK; cat#: P6148-500G) and washed in PBS before being stored in PBS at 4°C.
RNA extraction and RT-qPCR analysis
mRNA was collected using the Monarch® Total RNA Miniprep Kit (New England BioLabs, UK; cat#: T2010S) and cDNA was prepared using LunaScript® RT SuperMix Kit (New England BioLabs, UK; cat#: E3010L). RT-qPCR analysis was performed in an AriaMx thermocycler (Agilent, UK) using SYBR Green PCR mastermix (Fisher Scientific, UK; cat#: A25918) following the manufacturer’s instructions. The following oligonucleotide primers were used: human NRP1, GAAAAATCGAATGCTGAT (forward) and AATCCGGGGGACTTTATCAC (reverse); human GAPDH: CAAGGTCATCCATGACAACTTTG (forward) and GGGCCATCCACAGTCTTCTG (reverse); human TGFB2, GCTTACACTGTCCCTGCTGC (forward) and TTAGCAGGAGATGTGGGGTC (reverse); human TGFB1, CTTCCAGCCGAGGTCCTT (forward) and CCCTGGACACCAACTATTGC (reverse); human KLF2, TTGCAGTGGTAGGGCTTCTC (forward) and ACTCACACCTGCAGCTACGC (reverse). Data were analyzed with Agilent Aria 1.71 software (Agilent, US).
RNA sequencing
HUVECs transfected with siRNA targeting NRP1 or with a non-targeting siRNA were cultured under static conditions or exposed to laminar or oscillatory flow for 24 hours. RNAs from 3 independent experiments were isolated using the protocol described above and transcriptomic analysis performed by Novogene, UK. Sequencing libraries were generated using NEBNext® Ultra TM RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X) or by sonication with Diagenode bioruptor Pico to break RNA strands. First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H). Second strand cDNA synthesis was performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends by exonuclease or polymerase. After adenylation of 3’ ends of DNA fragments, NEBNext Adaptors with hairpin loop structure were ligated to prepare for hybridization. The library fragments were purified with AMPure XP system (Beckman Coulter, Beverly,USA). 3 μl USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 minutes followed by 5 minutes at 95 °C before PCR, which was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using PE Cluster Kit cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina platform and paired-end reads were generated. Paired-end clean reads were aligned to the reference genome using the Spliced Transcripts Alignment to a Reference (STAR) software. The read numbers mapped for each gene was counted by FeatureCounts and reads per kilobase of exon model per million mapped reads (RPKM) of each gene was calculated based on the length of the gene and read counts mapped to this gene. Differential expression analysis was performed using DESeq2 R package. The resulting P values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate (FDR). Genes with an adjusted P value < 0.05 found by DESeq2 were assigned as differentially expressed. Gene Ontology enrichment analysis of differentially expressed genes was implemented by the clusterProfiler R package. Gene Ontology terms with corrected P value less than 0.05 were considered significantly enriched by differential expressed genes.
Immunoblotting
Cells were lysed in 150 mM NaCl, 50 mM Tris-HCl pH 7.4 (MERCK, UK; cat#: T5941-500G), 50 mM Glycerophosphate (MERCK, UK), 1 % Tween-20 (MERCK, UK; cat#: P1379-100ML), 0.2 % Igepal©CA-630 (MERCK, UK; cat#: I8896-50ML) in the presence of protease inhibitor cocktail 2 (MERCK, UK; cat#: P8340-1ML) and phosphatase inhibitor cocktail 2 (MERCK, UK; cat#: P5726-1ML). Protein concentration was calculated using PierceTM BCA Protein Assay Kit (Thermo Fisher Scientific, UK; cat#23227), according to the manufacturing guide lines. 35μg of protein lysate were prepared in Laemmli buffer 1 X, denatured for 5 minutes at 95°C, separated by SDS-PAGE and transferred to nitrocellulose membrane (Whatman, USA; cat#: 10600007). Nitrocellulose membranes were immunoblotted with the following primary antibodies: anti-human-GAPDH (rabbit) (Sigma-Aldrich, UK; G9545-100UL); anti-human-NRP1 (rabbit) (Cell Signaling Technology Europe BV, UK; cat#: 3725S); anti-human-pSMAD2/3 (rabbit) (Cell Signaling Technology Europe BV, UK; cat#: 8828S); antihuman -Total SMAD2/3 (rabbit) (Cell Signaling Technology Europe BV, UK; cat#: 8685S); anti-human-p120 catenin (mouse) (BD Bioscience, UK; cat#: 610133). HRP conjugated secondary antibodies were used for chemiluminescence detection with ECL prime (Cytiva-AmerhamUK; cat#: RPN2232) and protein levels were quantified by densitometry with ImageJ (NIH, Bethesda US) and normalized to GAPDH.
For the HRP-proximity labelling assay, PVDF membranes were utilized in place of nitrocellulose. In addition to the previously listed antibodies, the following primary antibodies were used: streptavidin polyclonal HRP (Thermo Fisher Scientific, UK cat#: 21140), anti-human-VE-Cadherin (goat) (Santa Cruz Biotechnology, USA, cat#: sc-6458), anti-human-CD71 (mouse) (Santa Cruz Biotechnology, USA, cat#:sc-65882), anti-recombinant-GAPDH (rabbit) (Abcam, UK cat#: ab181603), anti-human-β-Tubulin (Sigma-Aldrich, UK, cat#: T4026). NRP1, VE-Cadherin, p120 catenin and CD71 were labelled with HRP conjugated secondary antibodies (Agilent, USA, cat#: anti-mouse P0447, anti-rabbit P0448, anti-goat P0449). Chemiluminescent detection was performed with Radiance Plus (Azure Biosystems, USA, cat#: AC2103). GAPDH and β-tubulin were multiplexed with other target proteins and detected with infrared dyes (IR Dye, LI-COR, USA, anti-mouse (goat)-680 cat#: 926-68070; anti-mouse (donkey)-800 cat#: 926-32212, anti-rabbit (goat)-680 cat#: 926-68071.) Blots were imaged using an Azure C600 instrument (Azure Biosystems, USA) and protein levels quantified with ImageStudio Lite (LI-COR, USA).
Co-immunoprecipitation analysis
HUVECs transfected with si-control or si-NRP1 and cultured under static conditions were lysed in 50 mM Tris, pH 8.0, 50 mM KCl, 1% (vol/vol) IGEPAL in the presence of protease inhibitor cocktail 2 and phosphatase inhibitor cocktail. 1.5 mg of protein was incubated with 3 μg goat anti-human VE-Cadherin (R&D #AF938) or control goat IgG (R&D # AB-108-C) and then with 30 μl magnetic protein G Dynabeads (Life Technologies) at 4°C overnight. Beads were collected with a Dynabead magnet, washed three times with lysis buffer on a rotating wheel at 4°C for 5 min, and resuspended in 50 μl Laemmli sample buffer for SDS-PAGE and immunoblotting.
ELISA
HDMECs were transfected with si-control and si-NRP1 for 72 hours. Media was changed 24 hours after transfection and cells were cultured for an additional 48 hours. Cell culture supernatants were collected and analyzed with human IL-6 and IL-8 DuoSet ELISA kit following the manufacturer’s instructions (R&D System, UK; cat# DY208). Results were expressed as means of three independent experiments with triplicate samples.
Immunofluorescence
For immunostaining experiments, HUVECs exposed to flow or cultured under static conditions were fixed in 4% paraformaldehyde in PBS for 30 minutes, washed with PBS, permeabilized in PBS 0.25% Triton X-100 for 3 minutes, washed with PBS, blocked for 30 minutes in PBS 0.1% BSA and incubated overnight at 4°C with the following primary antibodies: anti-human-NRP1 (R&D Systems, USA; Mab, clone 446921; cat#: MAB3870); anti-human-VE-cadherin (R&D Systems, USA; cat#: AF938); anti-human-p120 catenin (BD Bioscience, USA; cat#: 610133); anti-human-TGFBR2 (R&D Systems, USA; cat#:AF-241-NA). Primary antibodies were detected by incubating cells with appropriate combinations of the following secondary antibodies for 1 hour at room temperature: donkey anti-goat 488 (Invitrogen, UK; cat#: A1105); donkey anti-goat 555 (Invitrogen, UK; cat#: A21432); donkey anti-goat 647 (Invitrogen, UK; cat#: A21447); donkey anti-rabbit 488 (Invitrogen, UK; cat#: A21206); donkey anti-rabbit 555 (Invitrogen, UK; cat#: A31572); donkey anti-rabbit 647 (Invitrogen, UK; cat#: A31573); donkey anti-mouse 488 (Invitrogen, UK; cat#: A21202); donkey anti-mouse 555 (Invitrogen, UK; cat#: A31570); donkey anti-mouse 647 (Invitrogen, UK; cat#: A31571). Nuclei were counterstained with DAPI (MERCK, UK cat#: D9564-10MG). Filamentous actin was stained with DyLightTM 488 Phalloidin (Cell signalling technology, UK; cat#: 129355). Samples were washed with PBS, post-fixed in 4% paraformaldehyde for 5 minutes, washed with PBS, mounted with mounting media (Fisher Scientific, UK; cat#: 10662815) and imaged using an LSM 880 confocal microscope equipped with an Apochromat 63X 1.4 NA oil objective (Zeiss, Germany). Maximal projection of optical z-stacks were acquired with an LSM 880 confocal microscope and quantified using the threshold method with ImageJ (Bethesda, US). Pixel area, integrated density and mean intensity were normalized to DAPI. For each experiment, randomly selected areas were imaged. A minimum of three pictures per experimental conditions were acquired and analyzed and values were averaged for a single biological replicate or experiment.
Co-localization analysis
HUVECs were co-stained for VE-cadherin and NRP1 or TGFBR2 and p120 catenin, counterstained with DAPI and imaged with a LSM880 confocal microscope with a plan apochromat 63X 1.4 NA oil objective (Zeiss, Germany). Manders’ coefficient was calculated by analyzing maximal intensity projections of optical z-stacks with the JACoP ImageJ plugin (74). The analysis was performed on a minimum of 3 images and values were averaged for a single biological replicate or experiment.
HRP-based proximity-labelling assay
After 24 hours under flow (as above), HUVECs were incubated for 30 minutes with 500 μM biotin tyramide (Iris Biotech, Marktredwitz, Germany) under flow. Cells were removed from flow and the Ibidi chamber was flushed once and basal media supplemented with 1 mM H2O2 was added for a total exposure time of 1 minute. The labelling reaction was quenched by three washes with PBS containing 10 mM sodium azide, 10 mM ascorbate, 5 mM Trolox. Cells were lysed in RIPA buffer (Sigma-Aldrich, UK), supplemented with Protease Inhibitor Cocktail (Sigma-Aldrich, UK) and 10 mM sodium azide (to inhibit HRP activity). Biotinylated content was pulled down with PierceHigh Capacity Streptavidin Agarose (Thermo Fisher Scientific, UK). Beads were washed three times with lysis buffer before lysate was added. Beads were rotated with lysate overnight at 4°C and washed three times with either PBS supplemented with 10 mM sodium azide and protease inhibitor cocktail. Biotinylated content was released from beads for immunoblotting by boiling for 5 minutes in 1x SDS-PAGE sample buffer(Thermofisher) with 5% β-mercaptoethanol.
Proximity Ligation Assay (PLA)
HUVECs exposed to shear stress or cultured under static conditions were grown on gelatinized Ibidi chambers (Labtek, Sigma-Aldrich, UK). Cells were fixed with 4% paraformaldehyde and PLAs were performed following the manufacturer’s instructions (Duolink, MERCK, UK). PLAs were performed using the following combinations of primary antibodies: (i) mouse anti-human-NRP1 and goat anti-human-VE-cadherin; (ii) mouse anti-human-p120 catenin and goat anti-human-VE-cadherin; (iii) mouse anti-human-NRP1 and goat anti-human TGFBR2. Cells were counterstained with DAPI, mounted with Vectashield (Vector Laboratories, USA) and imaged with a LSM880 confocal microscope with a plan apochromat 63X 1.4 NA oil objective (Zeiss, Germany). PLAs were analyzed in z-stack projections with the Analyze Particle function of ImageJ (NIH, Bethesda, USA). A minimum of three images per experimental condition were captured to acquire PLA signals for a total of minimum 80 cells and values were averaged for a single biological replicate or experiment.
Monocyte adhesion assay
HUVECs were transfected with non-targeting or NRP1-targeting siRNAs. 24 hours after transfection cells were seeded on coverslips coated with fibronectin (10μg/ml) (Merck, UK Cat.# F1141-1MG) and allowed to reach confluency. THP-1 cells were labelled with Calcein-AM (1μM) (Cat.# 425201; Bioloegend, UK) and 5x105 labelled THP-1 incubated with the confluent HUVECs for 60 minutes. Cells were washed with PBS, fixed with 4% paraformaldehyde, counterstained with DAPI and imaged with a LSM880 confocal microscope with a plan apochromat 20X objexctive. Double-positive DAPI/fluorescein THP-1 cells per field were quantified in a minimum of 3 images per condition and values were averaged for a single biological replicate or experiment.
Animal studies
Experiments were conducted according to the Animals (Scientific Procedures) Act 1986, United Kingdom, approved by the United Kingdom Home Office guidelines. We used C57BL/6 mice carrying two floxed conditional null Nrp1 alleles (75) lacking or expressing a Cdh5(PAC)-iCreERT2 (76) (Nrp1WT = Nrp1fl/fl; Nrp1ECKO = Nrp1fl/fl; Cdh5(PAC)-iCreERT2). In some experiments, we used C57BL/6 mice carrying two WT NRP1 alleles expressing Cdh5(PAC)-iCreERT2 (Cre). For the atherosclerosis experiments we crossed Nrp1fl/fl; Cdh5(PAC)-iCreERT2 mice with C57BL/6 ApoE+/- mice (60) to generate Nrp1fl/fl; ApoE-/- Cdh5(PAC)-iCreERT2 (Nrp1ECKO; ApoE-/-) and Nrp1fl/fl; ApoE-/- (Nrp1WT; ApoE-/-) mice. For tamoxifen-induction of Cre-mediated recombination, 0.25 mg of tamoxifen (Sigma Aldrich, UK) dissolved in peanut oil to 2.5 mg/mL in 100μl total volume was administered for 5 days to 4 week old Nrp1ECKO and Nrp1WT littermates by intraperitoneal injection. 4 weeks later, animals were perfusion fixed and the entire aorta was isolated. For the atherosclerosis experiments, Nrp1ECKO; ApoE-/- and Nrp1WT; ApoE-/-mice received 1 daily injection for a total of 3 additional injections after 4 weeks. Animals were perfusion fixed and aortae were isolated when mice were 18 weeks old and stained with Oil-red-O to visualize atherosclerotic lesions.
Genotyping
Ear biopsies were collected on 2 week old pups. Biopsies were digested for 30 minutes at 95°C in 25 mM NaOH. Digestion was inhibited with an equal volume of 40 mM Tris-HCl. DNA samples were centrifuged at 300g for 10 minutes. 1 μL of the supernatant was used to prepare the PCR Red BioMix (Scientific Laboratory Supplies Ltd, UK; cat#: BIO25006), The following primers were used for genotyping: Nrp1, AAGGAGTGGCACAGCATCTT (forward) and TCACACCCAAACTTCCTTCC (reverse); ApoE, GCCTAGCCGAGGGAGAGCCG (forward), GCCGCCCCGACTGCATCT (reverse for ApoE-/-), and TGTGACTTGGGAGCTCTGCAGC (reverse for ApoE+/+); Cdh5(PAC)-iCreERT2, GCCTGCATTACCGGTCGATGCAACGA (forward) and GTGGCAGATGGCGCGGCAACACCATT (reverse). PCR products were run in 1 % Agarose gel containing GelRed® Nucleic acid gel stain (Insight Biotechnology, UK; cat#: 41003-T) and visualized using a FluorCheE (ProteinSimple, US).
En-face aorta and aortic ring immunostaining
Mice were perfused with PBS by the left ventricle, then with 4% paraformaldehyde in PBS at physiological pressure to fix the vascular tree. The whole aortae were dissected, cleaned of fat, permeabilized in 0.25% Triton X-100 in PBS for 5 minutes, blocked for 1 hour in 3% BSA in PBS-T 0.05%, and incubated overnight at 4°C with the following primary antibodies: anti-mouse-NRP1 (R&D Systems, USA; cat#: AF566); anti-mouse-VE-cadherin (ABCAM, UK; cat#: ab91064); anti-pSMAD2/3 (Cell Signaling Technology Europe BV, UK; cat#: 8828S). Depending on the experiment, primary antibodies were detected with donkey anti-goat 488; donkey anti-goat 647; donkey Anti-Rabbit 488; donkey anti-rabbit 647; goat anti-rat 555 (Invitrogen, UK; cat#: 21434). DAPI was used to counterstain the nuclei. Samples were washed with PBS, post-fixed in 4% paraformaldehyde for 5 minutes, washed with PBS, and mounted with mounting media (Fisher Scientific, UK; cat#: 10662815). Z-stacks of equivalent regions within the descending aorta or the aortic arch were acquired with an LSM 880 confocal microscope equipped with a plan apochromat 63X 1.4 NA oil objective. Maximal intensity projections were generated with ImageJ (NIH, Bethesda, USA) and staining was normalized to DAPI.
Mice were perfused with PBS by the left ventricle, then with 4 % paraformaldehyde in PBS. The whole aortae were dissected and cleaned of fat. Descending aortae were cryoprotected by overnight incubation in 15% and then 30% sucrose at 4°C. Tissues were embedded in OCT cryo-microtomy embedding medium (Cellpath, UK) and cryo-sectioned into sections of 15 μm thickness. Sections were air-dried, permeabilized, and immunostained as described above. Anti-ERG antibody (Abcam, UK; Cat# ab92513) was used for aortic ring immunostaining.
Quantification of VE-cadherin junctions in vitro and in vivo
Following VE-cadherin staining of HUVECs exposed to laminar or oscillatory flow, gaps between adjacent cells were analyzed using ImageJ. Regions of interest (ROI) of 800 x 800 pixels from a z-stacks tile-scans acquired with an LSM 880 confocal microscope were randomly selected. For each experiment, 9 random selections were analyzed per experimental condition and values were averaged for a single biological replicate or experiment. Gaps were highlighted with the magic wand tool in ImageJ and pixel area measured. Quantification of irregular junction in the aortic endothelium was performed on maximal intensity projections of z-stacks of equivalent regions of descending aortae. Maximal intensity z-projections were generated using ImageJ (Bethesda, US). Irregular junctions consisting of protrusions and of areas of surface VE-cadherin discontinuity outside the individual cellular outline were ROI marked and counted for each picture. A minimum two images were analyzed per aorta and values averaged to represent a single biological replicate or experiment (77).
Oil red O staining
Mice were perfusion fixed with 4% paraformaldehyde. Aortae were dissected, cleaned of fat, washed with MilliQ water and 60% isopropyl alcohol, and incubated for 20 minutes at room temperature in Oil red O working solution (60 % of 0.5 % Oil-Red-O (Sigma-Aldrich, UK; cat#: O0625-25G) in isopropyl alcohol and 40 % of 1 % aqueous dextrin (Sigma-Aldrich, UK; cat#: D2256) in MilliQ water). Aortae were washed once with 60% isopropyl alcohol, MilliQ water and at least 3 times in PBS. Images of en-face staining were acquired with a digital camera (Samsung 16 MP, f/1.9, 27mm wide, AF) and the total area covered by plaque was quantified using ImageJ.
Intravital microscopy of the mesentery
Mice of 5 weeks of age were anaesthetized with ketamine (150 mg/kg) and xylazine (7.5 mg/kg). Body temperature was maintained at 37 °C. An incision was made through the skin and the peritoneal wall. The mouse was positioned on the viewing stage of an inverted microscope and the mesentery was gently exteriorized. Warm Krebs buffer [132 mmol/L NaCl, 4.7 mmol/L KCl, 1.2 mmol/L MgSO4, 17.9 mmol/L NaHCO3, and 2.0 mmol/L CaCl2; gassed with 5% CO2 and 95% N2] was constantly released onto the mesentery at a rate of 2 mL/min. Venules of 20-40 μm in diameter and 100 μm in length were imaged with a 40X water immersion objective. The mesentery was observed with a bright field microscope and leukocyte rolling was measured by manual counting in 3 different measurements of 1 minute per venule analyzed, with 3 different venules analyzed per animal. Thus, a total of 9 measurements from each animal were acquired and values averaged to represent a single n value.
Statistical analysis
Statistical analyses were performed with GraphPad (Prism) or Office Excel (Microsoft). Data shown are representative of at least three experiments (unless otherwise stated) and are expressed as the mean± standard error of the mean (SEM) or standard deviation (SD). For in vivo experiments, group sizes were determined using power calculation based on our experience of characterizing murine phenotypes. Animals were randomized using identification numbers and experiments were performed blind to animal genotype and treatment group. Statistical significance was determined by Student’s t-test, one-way ANOVA or two-way ANOVA as indicated in the figure legends for each experiment. One-way or two-way analysis of variance ANOVA was followed by post hoc analysis using a Sidak multiple-comparison test. All statistical tests were two sided.
Supplementary Material
One-Sentence Summary.
The barrier function and quiescence of the endothelium are supported by transmembrane protein NRP1.
Editor’s Summary.
Maintaining a quiescent endothelium
Atherosclerosis causes disturbed blood flow that activates inflammatory pathways in endothelial cells, which attracts leukocytes to plaques and exacerbates disease progression. Bosseboeuf et al. investigated the role of the transmembrane protein NRP1 in the response of the endothelium to flow. Under normal flow patterns, NRP1 stabilized protein complexes at cell-cell junctions called adherens junctions and suppressed endothelial inflammation. In mice, NRP1 deficiency was associated with greater numbers of rolling leukocytes on endothelial cells in vitro and in vivo and larger plaque sizes in a model of atherosclerosis. Thus, NRP1 maintains the endothelium in a quiescent state and may limit inflammation in the endothelium under the disturbed blood flow patterns that are characteristic of atherosclerosis.
Acknowledgments
We thank Professor Adrian Hobbs for the help with the animal experiments. We thank Dr. Neil Dufton for his help with the staining protocols of animal tissues. We would like to thank the staff of the Biological Resources Units at Imperial College London and Queen Mary University London, particularly Reiss Browning (QMUL) and Mason Arnold (QMUL) for their help with mouse husbandry. We are grateful to Steven Rothery at the Facility Imaging by Light Microscopy at Imperial College London and to Professor Paul Chapple for his help with confocal imaging at QMUL. We thank Professor Dorian Haskard for the help with the experimental plan for the atherosclerosis experiments. We thank Professor Christiana Ruhrberg (UCL) for support during the development of the project. We thank Dr Christina Warboys and Dr Tasilenia Lampropoulou for the discussion on flow-dependent signaling in endothelial cells. We thank Elias Raimondi for the help with the immunofluorescence analysis. We thank Dr Stavroula Kanoni for providing a statistical review of the manuscript.
Funding
This work was funded by British Heart Foundation (BHF) fellowship FS/16/22/32045 to CR, QMUL startup funding grant code MCP1105B to CR, SGUL startup funding grant code 12526-25 to AC. The confocal facility was supported by Barts and the London Charity grant MGU0293.
Footnotes
Author contributions:
EB and CR performed and analyzed immunoblots, qPCR, immunostaining and oil red O staining. EB managed the breeding of the mouse colonies, performed genotyping and contributed to all in vivo experiments. AC performed and analyzed ELISAs and analyzed the RNAseq data. AC and CR performed and analyzed PLAs. DV and RR performed and analyzed qPCR data. RK and CR performed and analyzed leukocyte adhesion by performing intravital microscopy of the mesentery. TPM and TDN performed and analyzed the HRP labeling proximity assay. EB, AC and CR wrote the manuscript. EB, CR, AC, AA, AR and JM reviewed and edited the article. CR, AC, AA, AR, JM designed and supervised the research.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability
The RNA-seq data have been deposited in Annotare 2.0 ArrayExpress accession: E-MTAB-12191. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References
- 1.Dejana E, Orsenigo F, Lampugnani MG. The role of adherens junctions and VE-cadherin in the control of vascular permeability. J Cell Sci. 2008;121:2115–2122. doi: 10.1242/jcs.017897. [DOI] [PubMed] [Google Scholar]
- 2.Muller WA. How endothelial cells regulate transmigration of leukocytes in the inflammatory response. Am J Pathol. 2014;184:886–896. doi: 10.1016/j.ajpath.2013.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nourshargh S, Alon R. Leukocyte migration into inflamed tissues. Immunity. 2014;41:694–707. doi: 10.1016/j.immuni.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Muller WA. Leukocyte-endothelial-cell interactions in leukocyte transmigration and the inflammatory response. Trends Immunol. 2003;24:327–334. doi: 10.1016/s1471-4906(03)00117-0. [DOI] [PubMed] [Google Scholar]
- 5.Lampugnani MG, et al. VE-cadherin regulates endothelial actin activating Rac and increasing membrane association of Tiam. Mol Biol Cell. 2002;13:1175–1189. doi: 10.1091/mbc.01-07-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boggon TJ, et al. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 7.Setyawati MI, et al. Titanium dioxide nanomaterials cause endothelial cell leakiness by disrupting the homophilic interaction of VE-cadherin. Nat Commun. 2013;4:1673. doi: 10.1038/ncomms2655. [DOI] [PubMed] [Google Scholar]
- 8.Gavard J. Endothelial permeability and VE-cadherin: a wacky comradeship. Cell Adh Migr. 2014;8:158–164. doi: 10.4161/cam.29026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao K, et al. Cellular levels of p120 catenin function as a set point for cadherin expression levels in microvascular endothelial cells. J Cell Biol. 2003;163:535–545. doi: 10.1083/jcb.200306001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao K, et al. p120-Catenin regulates clathrin-dependent endocytosis of VE-cadherin. Mol Biol Cell. 2005;16:5141–5151. doi: 10.1091/mbc.E05-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wessel F, et al. Leukocyte extravasation and vascular permeability are each controlled in vivo by different tyrosine residues of VE-cadherin. Nat Immunol. 2014;15:223–230. doi: 10.1038/ni.2824. [DOI] [PubMed] [Google Scholar]
- 13.Rudini N, et al. VE-cadherin is a critical endothelial regulator of TGF-beta signalling. EMBO J. 2008;27:993–1004. doi: 10.1038/emboj.2008.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shay-Salit A, et al. VEGF receptor 2 and the adherens junction as a mechanical transducer in vascular endothelial cells. Proc Natl Acad Sci U S A. 2002;99:9462–9467. doi: 10.1073/pnas.142224299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tzima E, et al. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–431. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- 16.Davignon J, Ganz P. Role of endothelial dysfunction in atherosclerosis. Circulation. 2004;109:III27–32. doi: 10.1161/01.CIR.0000131515.03336.f8. [DOI] [PubMed] [Google Scholar]
- 17.Beldman TJ, et al. Nanoparticle-Aided Characterization of Arterial Endothelial Architecture during Atherosclerosis Progression and Metabolic Therapy. ACS Nano. 2019;13:13759–13774. doi: 10.1021/acsnano.8b08875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyazaki T, et al. m-Calpain induction in vascular endothelial cells on human and mouse atheromas and its roles in VE-cadherin disorganization and atherosclerosis. Circulation. 2011;124:2522–2532. doi: 10.1161/CIRCULATIONAHA.111.021675. [DOI] [PubMed] [Google Scholar]
- 19.Flavahan NA. In Development-A New Paradigm for Understanding Vascular Disease. J Cardiovasc Pharmacol. 2017;69:248–263. doi: 10.1097/FJC.0000000000000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flavahan S, Chang F, Flavahan NA. Local renin-angiotensin system mediates endothelial dilator dysfunction in aging arteries. Am J Physiol Heart Circ Physiol. 2016;311:H849–854. doi: 10.1152/ajpheart.00422.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pellet-Many C, Frankel P, Jia H, Zachary I. Neuropilins: structure, function and role in disease. Biochem J. 2008;411:211–226. doi: 10.1042/BJ20071639. [DOI] [PubMed] [Google Scholar]
- 22.Soker S, Takashima S, Miao HQ, Neufeld G, Klagsbrun M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell. 1998;92:735–745. doi: 10.1016/s0092-8674(00)81402-6. [DOI] [PubMed] [Google Scholar]
- 23.Takagi S, et al. Expression of a cell adhesion molecule, neuropilin, in the developing chick nervous system. Dev Biol. 1995;170:207–222. doi: 10.1006/dbio.1995.1208. [DOI] [PubMed] [Google Scholar]
- 24.Lampropoulou A, Ruhrberg C. Neuropilin regulation of angiogenesis. Biochem Soc Trans. 2014;42:1623–1628. doi: 10.1042/BST20140244. [DOI] [PubMed] [Google Scholar]
- 25.Schwarz Q, Ruhrberg C. Neuropilin, you gotta let me know: should I stay or should I go? Cell Adh Migr. 2010;4:61–66. doi: 10.4161/cam.4.1.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouvree K, et al. Semaphorin3A, Neuropilin-1, and PlexinA1 are required for lymphatic valve formation. Circ Res. 2012;111:437–445. doi: 10.1161/CIRCRESAHA.112.269316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ochsenbein AM, Karaman S, Jurisic G, Detmar M. The role of neuropilin-1/semaphorin 3A signaling in lymphatic vessel development and maturation. Adv Anat Embryol Cell Biol. 2014;214:143–152. doi: 10.1007/978-3-7091-1646-3_11. [DOI] [PubMed] [Google Scholar]
- 28.Jurisic G, et al. An unexpected role of semaphorin3a-neuropilin-1 signaling in lymphatic vessel maturation and valve formation. Circ Res. 2012;111:426–436. doi: 10.1161/CIRCRESAHA.112.269399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan Q, et al. Blocking neuropilin-1 function has an additive effect with anti-VEGF to inhibit tumor growth. Cancer Cell. 2007;11:53–67. doi: 10.1016/j.ccr.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 30.Fantin A, et al. Neuropilin 1 (NRP1) hypomorphism combined with defective VEGF-A binding reveals novel roles for NRP1 in developmental and pathological angiogenesis. Development. 2014;141:556–562. doi: 10.1242/dev.103028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gelfand MV, et al. Neuropilin-1 functions as a VEGFR2 co-receptor to guide developmental angiogenesis independent of ligand binding. Elife. 2014;3:e03720. doi: 10.7554/eLife.03720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aspalter IM, et al. Alk1 and Alk5 inhibition by Nrp1 controls vascular sprouting downstream of Notch. Nat Commun. 2015;6:7264. doi: 10.1038/ncomms8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raimondi C, et al. Imatinib inhibits VEGF-independent angiogenesis by targeting neuropilin 1-dependent ABL1 activation in endothelial cells. J Exp Med. 2014;211:1167–1183. doi: 10.1084/jem.20132330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fantin A, et al. NRP1 Regulates CDC42 Activation to Promote Filopodia Formation in Endothelial Tip Cells. Cell Rep. 2015;11:1577–1590. doi: 10.1016/j.celrep.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fantin A, et al. VEGF165-induced vascular permeability requires NRP1 for ABL-mediated SRC family kinase activation. J Exp Med. 2017;214:1049–1064. doi: 10.1084/jem.20160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth L, et al. Neuropilin-1 mediates vascular permeability independently of vascular endothelial growth factor receptor-2 activation. Sci Signal. 2016;9:ra42. doi: 10.1126/scisignal.aad3812. [DOI] [PubMed] [Google Scholar]
- 37.Mehta V, et al. The guidance receptor plexin D1 is a mechanosensor in endothelial cells. Nature. 2020;578:290–295. doi: 10.1038/s41586-020-1979-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glinka Y, Stoilova S, Mohammed N, Prud’homme GJ. Neuropilin-1 exerts co-receptor function for TGF-beta-1 on the membrane of cancer cells and enhances responses to both latent and active TGF-beta. Carcinogenesis. 2011;32:613–621. doi: 10.1093/carcin/bgq281. [DOI] [PubMed] [Google Scholar]
- 39.Glinka Y, Prud’homme GJ. Neuropilin-1 is a receptor for transforming growth factor beta-1, activates its latent form, and promotes regulatory T cell activity. J Leukoc Biol. 2008;84:302–310. doi: 10.1189/jlb.0208090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davies PF. Hemodynamic shear stress and the endothelium in cardiovascular pathophysiology. Nature clinical practice Cardiovascular medicine. 2009;6:16–26. doi: 10.1038/ncpcardio1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Passerini AG, et al. Coexisting proinflammatory and antioxidative endothelial transcription profiles in a disturbed flow region of the adult porcine aorta. Proc Natl Acad Sci U S A. 2004;101:2482–2487. doi: 10.1073/pnas.0305938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brooks AR, Lelkes PI, Rubanyi GM. Gene expression profiling of human aortic endothelial cells exposed to disturbed flow and steady laminar flow. Physiological genomics. 2002;9:27–41. doi: 10.1152/physiolgenomics.00075.2001. [DOI] [PubMed] [Google Scholar]
- 43.Kroon J, et al. Flow-induced endothelial cell alignment requires the RhoGEF Trio as a scaffold protein to polarize active Rac1 distribution. Mol Biol Cell. 2017;28:1745–1753. doi: 10.1091/mbc.E16-06-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C, Baker BM, Chen CS, Schwartz MA. Endothelial cell sensing of flow direction. Arterioscler Thromb Vasc Biol. 2013;33:2130–2136. doi: 10.1161/ATVBAHA.113.301826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Traub O, Berk BC. Laminar shear stress: mechanisms by which endothelial cells transduce an atheroprotective force. Arterioscler Thromb Vasc Biol. 1998;18:677–685. doi: 10.1161/01.atv.18.5.677. [DOI] [PubMed] [Google Scholar]
- 46.Malek AM, Alper SL, Izumo S. Hemodynamic shear stress and its role in atherosclerosis. JAMA. 1999;282:2035–2042. doi: 10.1001/jama.282.21.2035. [DOI] [PubMed] [Google Scholar]
- 47.Issitt T, et al. Neuropilin-1 Controls Endothelial Homeostasis by Regulating Mitochondrial Function and Iron-Dependent Oxidative Stress. iScience. 2019;11:205–223. doi: 10.1016/j.isci.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakajima H, Mochizuki N. Flow pattern-dependent endothelial cell responses through transcriptional regulation. Cell Cycle. 2017;16:1893–1901. doi: 10.1080/15384101.2017.1364324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kostelnik KB, et al. Dynamic trafficking and turnover of JAM-C is essential for endothelial cell migration. PLoS Biol. 2019;17:e3000554. doi: 10.1371/journal.pbio.3000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lanahan A, et al. The neuropilin 1 cytoplasmic domain is required for VEGF-A-dependent arteriogenesis. Dev Cell. 2013;25:156–168. doi: 10.1016/j.devcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi YX, et al. PDGF-BB and TGF-{beta}1 on cross-talk between endothelial and smooth muscle cells in vascular remodeling induced by low shear stress. Proc Natl Acad Sci U S A. 2011;108:1908–1913. doi: 10.1073/pnas.1019219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giannotta M, Trani M, Dejana E. VE-cadherin and endothelial adherens junctions: active guardians of vascular integrity. Dev Cell. 2013;26:441–454. doi: 10.1016/j.devcel.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 53.Chen PY, et al. Endothelial TGF-beta signalling drives vascular inflammation and atherosclerosis. Nat Metab. 2019;1:912–926. doi: 10.1038/s42255-019-0102-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramji DP, Davies TS. Cytokines in atherosclerosis: Key players in all stages of disease and promising therapeutic targets. Cytokine Growth Factor Rev. 2015;26:673–685. doi: 10.1016/j.cytogfr.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zernecke A, Weber C. Chemokines in atherosclerosis: proceedings resumed. Arterioscler Thromb Vasc Biol. 2014;34:742–750. doi: 10.1161/ATVBAHA.113.301655. [DOI] [PubMed] [Google Scholar]
- 56.Kaplanski G, et al. Thrombin-activated human endothelial cells support monocyte adhesion in vitro following expression of intercellular adhesion molecule-1 (ICAM-1; CD54) and vascular cell adhesion molecule-1 (VCAM-1; CD106) Blood. 1998;92:1259–1267. [PubMed] [Google Scholar]
- 57.Cybulsky MI, et al. A major role for VCAM-1 but not ICAM-1 in early atherosclerosis. J Clin Invest. 2001;107:1255–1262. doi: 10.1172/JCI11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park JG, et al. Evaluation of VCAM-1 antibodies as therapeutic agent for atherosclerosis in apolipoprotein E-deficient mice. Atherosclerosis. 2013;226:356–363. doi: 10.1016/j.atherosclerosis.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 59.Birdsey GM, et al. The endothelial transcription factor ERG promotes vascular stability and growth through Wnt/beta-catenin signaling. Dev Cell. 2015;32:82–96. doi: 10.1016/j.devcel.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang SH, Reddick RL, Piedrahita JA, Maeda N. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science. 1992;258:468–471. doi: 10.1126/science.1411543. [DOI] [PubMed] [Google Scholar]
- 61.Raimondi C, Brash JT, Fantin A, Ruhrberg C. NRP1 function and targeting in neurovascular development and eye disease. Prog Retin Eye Res. 2016;52:64–83. doi: 10.1016/j.preteyeres.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Acevedo LM, Barillas S, Weis SM, Gothert JR, Cheresh DA. Semaphorin 3A suppresses VEGF-mediated angiogenesis yet acts as a vascular permeability factor. Blood. 2008;111:2674–2680. doi: 10.1182/blood-2007-08-110205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cunningham KS, Gotlieb AI. The role of shear stress in the pathogenesis of atherosclerosis. Lab Invest. 2005;85:9–23. doi: 10.1038/labinvest.3700215. [DOI] [PubMed] [Google Scholar]
- 64.Souilhol C, et al. Endothelial responses to shear stress in atherosclerosis: a novel role for developmental genes. Nat Rev Cardiol. 2020;17:52–63. doi: 10.1038/s41569-019-0239-5. [DOI] [PubMed] [Google Scholar]
- 65.Wang N, et al. Shear stress regulation of Kruppel-like factor 2 expression is flow pattern-specific. Biochem Biophys Res Commun. 2006;341:1244–1251. doi: 10.1016/j.bbrc.2006.01.089. [DOI] [PubMed] [Google Scholar]
- 66.McCormick SM, et al. DNA microarray reveals changes in gene expression of shear stressed human umbilical vein endothelial cells. Proc Natl Acad Sci U S A. 2001;98:8955–8960. doi: 10.1073/pnas.171259298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hamik A, et al. Kruppel-like factor 4 regulates endothelial inflammation. J Biol Chem. 2007;282:13769–13779. doi: 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
- 68.Kluza E, et al. Diverse ultrastructural landscape of atherosclerotic endothelium. Atherosclerosis. 2021;339:35–45. doi: 10.1016/j.atherosclerosis.2021.11.017. [DOI] [PubMed] [Google Scholar]
- 69.Zhang F, et al. Lacteal junction zippering protects against diet-induced obesity. Science. 2018;361:599–603. doi: 10.1126/science.aap9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chiasson CM, Wittich KB, Vincent PA, Faundez V, Kowalczyk AP. p120-catenin inhibits VE-cadherin internalization through a Rho-independent mechanism. Mol Biol Cell. 2009;20:1970–1980. doi: 10.1091/mbc.E08-07-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wildenberg GA, et al. p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell. 2006;127:1027–1039. doi: 10.1016/j.cell.2006.09.046. [DOI] [PubMed] [Google Scholar]
- 72.Wang YL, et al. Innate immune function of the adherens junction protein p120-catenin in endothelial response to endotoxin. J Immunol. 2011;186:3180–3187. doi: 10.4049/jimmunol.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vion AC, et al. Endothelial Cell Orientation and Polarity Are Controlled by Shear Stress and VEGF Through Distinct Signaling Pathways. Front Physiol. 2020;11:623–769. doi: 10.3389/fphys.2020.623769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 75.Gu C, et al. Neuropilin-1 conveys semaphorin and VEGF signaling during neural and cardiovascular development. Dev Cell. 2003;5:45–57. doi: 10.1016/s1534-5807(03)00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y, et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature. 2010;465:483–486. doi: 10.1038/nature09002. [DOI] [PubMed] [Google Scholar]
- 77.Seebach J, Cao J, Schnittler HJ. Quantitative dynamics of VE-cadherin at endothelial cell junctions at a glance: basic requirements and current concepts. Discoveries (Craiova) 2016;4:e63. doi: 10.15190/d.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited in Annotare 2.0 ArrayExpress accession: E-MTAB-12191. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.