Abstract
Although the plant kingdom provides an enormous diversity of metabolites with potentially beneficial applications for humankind, a large fraction of these metabolites and their biosynthetic pathways remains unknown. Resolving metabolite structures and their biosynthetic pathways is key to gaining biological understanding and to allow metabolic engineering. In order to retrieve novel biosynthetic genes involved in specialized metabolism, we developed a novel untargeted system-wide method in Arabidopsis thaliana, subjecting qualitative metabolic traits to a genome-wide association study (designated as Qualitative Trait GWAS or QT-GWAS), along with the more conventional metabolite GWAS (mGWAS) that considers the quantitative variation of metabolites. As proof of the validity of the QT-GWAS and mGWAS, 23 and 15 of the retrieved associations were supported by previous research. Furthermore, seven gene-metabolite associations retrieved by QT-GWAS were confirmed in this study through reverse genetics combined with metabolomics and/or in vitro enzyme assays. As such, we established that CYTOCHROME P450 706A5 (CYP706A5) is involved in the biosynthesis of chroman derivatives, UGT76C3 is able to hexosylate guanine in vitro and in planta, and SULFOTRANSFERASE 202B1 (SULT202B1) catalyzes the sulfation of neolignans in vitro.
Introduction
Plant metabolism entails a complex network of biochemical pathways, in which enzymes perform conversions that connect a wide variety of structurally diverse metabolites. The diversity and abundance of metabolites can be considered as a phenotypic output of the genome (Contrepois et al., 2016). The plant metabolome can be classified into the primary and secondary/specialized metabolome. The former describes the collection of metabolites involved in processes essential for growth and development, whereas the latter comprises a specialized assortment of metabolites that allow the proper functioning and adaptation of an organism in a particular environment. Many metabolites in the specialized metabolome offer a use for humankind. They can lead to the development of medicines (Bishayee and Sethi, 2016; Desborough and Keeling, 2017), or commercially valuable products such as cosmetics, dyes, and oils (Loza-Tavera, 1999; Shin et al., 2013; Rose et al., 2018; Góral and Wojciechowski, 2020). They can also contribute attractive properties to food, improve feed quality (Ricachenevsky et al., 2019), and present agricultural benefits such as enhanced biotic (Fürstenberg-Hägg et al., 2013; Arbona and Gomez-Cadenas, 2016; Chowański et al., 2016) and abiotic stress tolerance (Varela et al., 2016; Peng et al., 2017), enhanced yield (Steenackers et al., 2019), or improved interaction with beneficial organisms such as pollinators (Dudareva et al., 2013) or micro-organisms (Bouwmeester et al., 2019). Exploring the genetic mechanisms controlling the abundances of such metabolites is therefore crucial for both fundamental biological understanding and the engineering of plants to enhance metabolic traits.
Because of the vast amount of both metabolites and the genes underlying their biosynthesis, systems-wide tools are often employed to unravel biosynthetic pathways of specialized metabolites (Desmet et al., 2021a). By exploiting natural genomic variation, genome-wide association studies (GWAS) permit the search for associations of genomic markers, such as single-nucleotide polymorphisms (SNPs), with phenotypic traits of interest. In case the traits are metabolite abundances, this procedure is referred to as metabolite GWAS (mGWAS). mGWAS has become a valuable tool for uncovering new and uncharacterized genes/pathways using either targeted or untargeted metabolomics. For example, in Arabidopsis thaliana (Arabidopsis), mGWAS led to the discovery of genes involved in biosynthetic pathways of both primary (Strauch et al., 2015; Wu et al., 2016; Angelovici et al., 2017) and specialized metabolism (Chan et al., 2010; Routaboul et al., 2012; Li et al., 2014). An mGWAS study on Arabidopsis employed under different environmental conditions led to a list of 70 candidate genes putatively involved in the metabolism of several metabolic classes, of which five genes were experimentally validated by means of reverse genetics (Wu et al., 2018). mGWAS has been successfully used to discover genes controlling natural variation of metabolite abundances in crop plants such as rice (Chen et al., 2014; Dong et al., 2015; Matsuda et al., 2015; Chen et al., 2016; Brotman et al., 2021), maize (Wen et al., 2014; Wen et al., 2015; Chen et al., 2016; Baseggio et al., 2020), wheat (Chen et al., 2020; Shi et al., 2020) and tomato (Nunes-Nesi et al., 2019; Alseekh et al., 2020; Tohge et al., 2020).
In mGWAS, the number of loci that significantly contribute to the variation in metabolite abundance is often lower for specialized metabolites than for primary metabolites (Fernie and Tohge, 2017). Because of the highly polygenic basis underlying primary metabolism, loci determining variation in primary metabolism are often of relatively small effect (<30% for a given locus) (Schauer et al., 2008; Wen et al., 2015; Wu et al., 2016; Knoch et al., 2017), whereas for specialized metabolism, loci with larger effect sizes (>30%) can often be identified (Chen et al., 2014; Wen et al., 2014; Luo, 2015), suggesting that the abundance of such specialized metabolites is predominantly affected by only a small number of major genes. Many specialized metabolites are decorated derivatives of common core structures (Morreel et al., 2014; Wang et al., 2019). Because many of the genes involved in this variety of decorations are conserved only among the most closely related species, specialized metabolism pathways are likely the result of neo-functionalizations following tandem gene duplications (Moghe and Last, 2015). This allows the creation of novel specialized metabolites of which the abundance is affected mostly by those specific duplicated genes. Furthermore, specialized metabolic pathways tend to be less subjected to natural selection compared with primary metabolic pathways. In primary metabolism, a major selective pressure is generally enforced by the large metabolic flux requirements inherent to the crucial role of primary metabolites. This high selective pressure ensures high precision of metabolic conversions and catalytic efficiency of enzymes in primary metabolism (Mukherjee et al., 2015). Genes underlying primary metabolism therefore tend to be highly conserved across the plant kingdom (Weng, 2014). In contrast, the selective pressure on specialized metabolism drives the variation in secondary metabolites rather than flux increases(Mukherjee et al., 2015). For example, when a particular specialized metabolite is no longer required for a specific subpopulation of a certain species to thrive (e.g., when the species enters a new niche), this metabolite could completely disappear in that subpopulation as a result of the accumulation of mutations in biosynthetic genes for that specialized metabolite. Such metabolites then represent distinct features in a natural population, only occurring in specific accessions (Sotelo-Silveira et al., 2015). The presence/absence of such a metabolite can be considered as a qualitative rather than a quantitative trait. The absence of a qualitative metabolic trait can be expected to show monogenic inheritance, considering that a knock-out mutation in one of the biosynthetic genes, will result in the loss of that metabolite. Such inheritance facilitates the discovery of the underlying genes (Kushalappa and Gunnaiah, 2013). In addition, because of their major effect, it is reasonable to hypothesize that, when subjecting qualitative metabolic traits to genetic analysis, biosynthetic genes can readily be identified. For example, surveying leaf sinapate esterprofiles from 96 Arabidopsis accessions demonstrated that the Pna-10 accession, which accumulates sinapoylglucose instead of sinapoylmalate, is a natural deletion mutant of SNG1 encoding sinapoylglucose:malate sinapoyltransferase (Li et al., 2010). Similarly, a novel flavonol phenylacyltransferase gene (FPT2) has been identified based on a targeted study of saiginols present only in a subset of the analyzed accessions (Tohge et al., 2016). The absence of flavonol 3-O-gentiobioside 7-O-rhamnoside in 42 out of 81 analyzed Arabidopsis accessions led to the identification of an acyl-glucose-dependent glucosyltransferase (BGLU6) (Ishihara et al., 2016). This rather limited number of examples illustrates that analyses of qualitative metabolic traits (metabolic traits that are absent in part of the studied population) can pinpoint enzyme-encoding genes that underlie these metabolic conversions. Here, we performed an untargeted association analysis of 4,479 qualitative metabolic traits (Qualitative Trait GWAS or QT-GWAS) and 1,147 quantitative metabolic traits (mGWAS) obtained from liquid-chromatography–mass spectrometry (LC-MS) chromatograms with 250K SNPs of 183 Arabidopsisaccessions. Both methods enriched for genes labelled with “metabolic process” related GOterms, but QT-GWAS did so more significantly. When focussing on associations involving characterised metabolites, we found 30 valid associations retrieved by the QT-GWAS of which 23 were supported by previous research and seven (involving the three genes UGT76C3, CYP706A5 and SULT202B1) were newly confirmed in this study. Our results show that through an untargeted QT-GWAS, valid gene–metabolite associations can be retrieved at the level of enzyme-encoding genes involved in metabolic conversions, and can retrieve new associations not found by mGWAS. This is, to our knowledge, the first time an untargeted GWAS approach has been combined with qualitative metabolic traits.
Results
LC–MS based metabolic profiles
An LC–MS analysis was performed on methanol extracts prepared from 14-day-old seedlings belonging to 183 accessions (Supplemental Table 1). A total of 5,082 metabolic features (peaks) was detected across all chromatograms (Supplemental Data Set 1), of which 603 were detected in all accessions. The other 4,479 features that were below the detection limit (ion intensity of 500 in this study) in at least 1 of the 183 accessions were considered as qualitative traits. Features with an average abundance above 500 and that were present in at least 100 accessions (to reduce the false-positive rate resulting from too-small sample sizes and to ensure a continuous distribution of quantitative traits across the used population), were selected as quantitative traits (Wu et al., 2018). Based on the applied filtering, 1,147 of the 5,082 features were selected as quantitative traits, 702 of which could be identified as both qualitative and quantitative traits in this study (Figure 1).
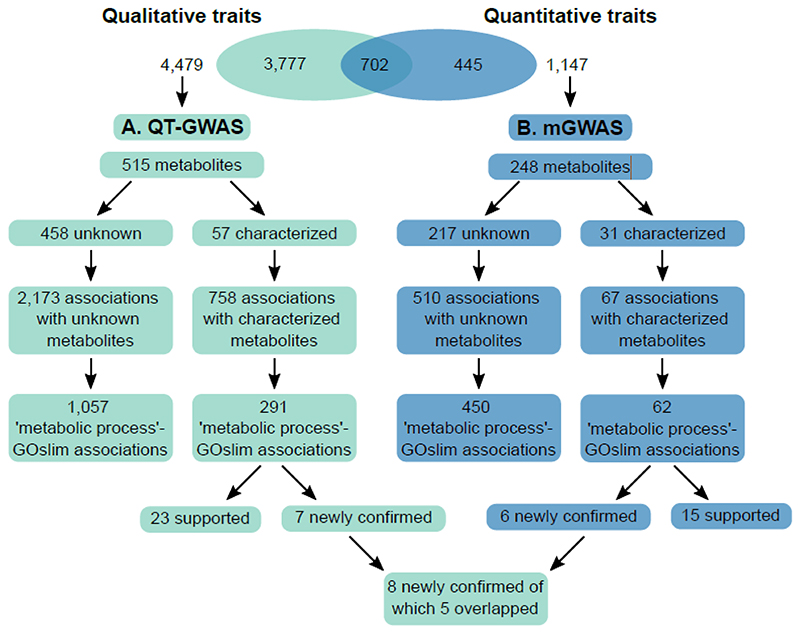
Figure 1. Schematic overview of the qualitative and quantitative traits and their recorded associations.
A, 4,479 qualitative metabolic traits were used as input for the QT-GWAS. Associations were retrieved for 709 qualitative traits, estimated to correspond to 515 metabolites, of which 57 were characterized and 458 remained unknown. 2,173 associations were retrieved for the unknown metabolites, of which 1,057 involved loci encoding enzymes labeled with ‘metabolic process’-related GOslim categories. The 57 characterized metabolites were involved in 758 associations. In this dataset, 291 loci contained genes labeled with ‘metabolic process’-related GOslim categories. At least 23 of these associations were confirmed or supported by previous research and seven (involving three loci) were newly confirmed in this study. B, 1,147 quantitative traits were used as input for the mGWAS. Associations were retrieved for 288 of estimated to correspond to 248 metabolites. Of these metabolites, 31 were characterized and 217 remained unknown. The unknowns were involved in 510 associations of which 450 encompassed loci containing genes labeled with ‘metabolic process’-related GOslim categories. The 31 characterized metabolites were involved in 67 associations. In this dataset, 62 loci contained genes labeled with ‘metabolic process’-related GOslim categories, of which at least 15 were confirmed or supported by previous research and six (involving 3 loci) were newly confirmed in this study. In total, eight new associations were confirmed in this study of which five overlapped between the two methods.
Associations between qualitative metabolic traits and SNP data
SNP data were used from a previously published 250K SNP data set (Horton et al., 2012) for the 183 selected Arabidopsis accessions. SNPs that were monomorphic in all accessions were removed from the data set. Significant associations of qualitative features with SNPs were retrieved by a Fisher’s exact test using a P-value <10-6 threshold. Of the 4,479 qualitative features, 709 features (16%) were involved in 53,464 associations. After a filtering step to remove (i) redundant associations between a particular feature and SNPs that are located within a 20-kb window (10 kb up- and 10 kb downstream of the most significant SNP) based on the average linkage disequilibrium (LD) size in Arabidopsis (Kim et al., 2007), and (ii) redundant associations between a particular SNP and features representing the same metabolite [most metabolites are represented by multiple features in LC-MS (Mahieu et al., 2016)], 515 features (hereafter referred to as metabolites) remained that were involved in 2,931 associations (Figure 1A, Supplemental Data Set 2). MS/MS spectra were recorded for 140 of the 515 metabolites, of which 57 were tentatively characterized (Supplemental Table 2) using the DynLib spectral database (Desmet et al., 2021b). The metabolites were characterized as glucosinolates (13 metabolites), (neo)lignans/oligolignols (11, of which 5 were found to be sulfated); flavonoids (6), jasmonates (5), organic acids (4), aromatic polyketides (3), phenylpropanoids (3), benzenoids/coumarins (2), lipids (2), purines (2), an amino acid (1), and others (5).
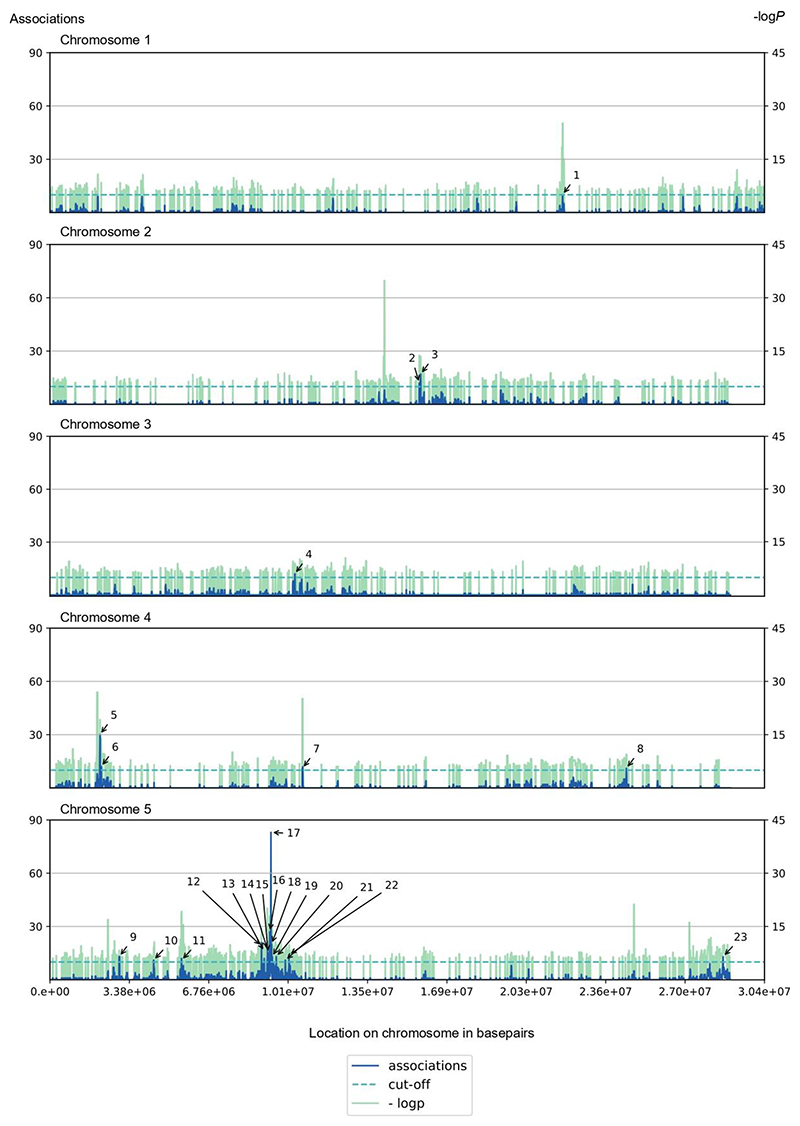
From the 515 metabolites, 397 (77%) were associated with one locus and 118 (23%) with multiple loci (Figure 2). Here a locus is defined as the 10-kb upstream and 10-kb downstream region of the associated SNP based on the average size of LD in Arabidopsis. Considering loci with 10 or more associated metabolites (threshold based on the observed elbow in Supplemental Figure 1) as pleiotropic loci, 23 such loci (L1-23) of the 2931 loci were found of which 1, 2, 1, 4, and 15 loci mapped on chromosomes 1 to 5, respectively (Figure 3, Supplemental Table 3). The pleiotropic locus L17 showed 83 associations, the highest number of associations of all loci in this data set, of which seven involved characterized glucosinolates, known to be affected by the METHYLTHIOALKYLMALATE 1 (MAM1), MAM2 and MAM3 genes within this locus (Textor et al., 2004).
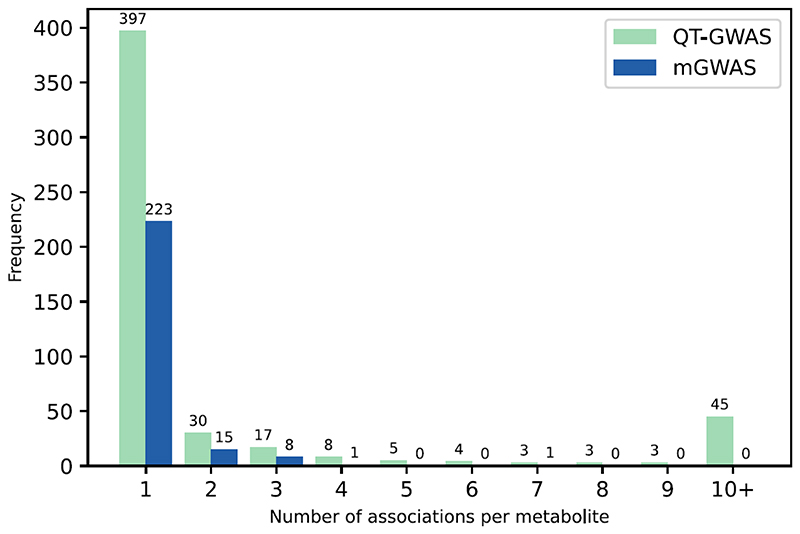
Figure 2. Distribution plot of the number of associations per metabolite retrieved for the 515 metabolites in the QT-GWAS (blue) and the 248 in the mGWAS (green).
Of the 515 metabolites associated in QT-GWAS, 397 show an association with one locus, 118 with two or more loci. In mGWAS, 223 metabolites showed an association with one locus and 25 to multiple loci.
Figure 3. Overview of the pleiotropic loci (> 10 associations) obtained with the QT-GWAS.
For each locus, the number of associations is represented in dark blue, the negative logarithmic P-value with base 10 (-logP) of the association in green. The dashed line represents the cut-off of ten associations to qualify as a pleiotropic locus. Pleiotropic loci are numbered according to Supplemental Table S3. A locus is defined as the region 10-kb upstream and 10-kb downstream of the most significant SNP (lead SNP) associated with a particular metabolite.
Associations between quantitative metabolic traits and SNP data
Next, we investigated the continuous variation of metabolic features in our data set. For the 1,147 quantitative features, the average abundance across the five replicates of each accession was calculated, and these averages were then subjected to a quantitative mGWAS following the EMMAX procedure (Kang et al., 2010). A genomic locus was considered as significantly associated with a particular feature, when at least 1 SNP in that locus showed a P-value <10-6 (the most significant SNP is then defined as the lead SNP) and at least an additional 5 of 40 SNPs in a region upstream and downstream of the lead SNP showed a P-value ≤10-3. When such associations were retrieved, an associated locus was defined as the window of SNPs still in LD (showing a significant association) with the lead SNP. In this way, 288 quantitative traits (of which 71 were also defined as qualitative traits), estimated to correspond to 248 metabolites after feature grouping, were associated with 577 loci (Figure 1, Supplemental Data Set 3). Of the 248 metabolites, 31 could be characterized, all of which were also present in the QT-GWAS data set. The characterized metabolites contained glucosinolates (10), jasmonates (5), (neo)lignans/oligolignols (4), organic acids (3), flavonoids (2), phenylpropanoids (2), purines (2), aromatic polyketides (2), and an amino acid (1). Of the quantitative traits, 223 (90%) had only one significant association, whereas 25 (10%) showed multiple associations (Figure 2).
For the mGWAS, six pleiotropic loci could be identified (involved in associations with ten or more metabolites; Supplemental Figure 2, Supplemental Table 4). Two of the six pleiotropic loci were shared with the QT-GWAS (L5 and L17). The four other pleiotropic loci (L24-27) were all situated on chromosome 5, surrounding the pleiotropic MAM locus (L17). Both L5 and L17 showed associations with glucosinolates and are both known to contribute to natural variation in glucosinolates (Kliebenstein et al., 2001; Kroymann et al., 2001). Possibly, the association of various glucosinolates to L24-27 could be explained by the existence of an extended LD block surrounding the MAM locus in L17 (Chan et al., 2010), resulting in SNPs, which are further away from the MAM locus, to still be associated with glucosinolates. A detailed overview of the loci and corresponding associated characterized metabolites can be found in Supplemental Table 4.
Gene Ontology term enrichment and overlap analysis
To investigate whether the loci associated with the metabolic traits were enriched in genes involved in metabolic processes, a GOterm enrichment was performed for both approaches, using the PANTHER webtool. For the qualitative approach, 103 GOterms in the ‘biological process’ category were significantly enriched (Bonferroni corrected P-value <0.05), not counting the unclassified category. Of these, twenty two GOterms (21%), related to various metabolic processes (Supplemental Table 5). For the quantitative approach, 25 GOterms were significantly enriched, of which five involved metabolic processes (20%). Of these 25 GOterms, all but one (‘seed development’ – GO:0048316) were also enriched in the QT-GWAS (Supplemental Table 6). Consequently, all GOterms involving metabolic processes that were enriched in the mGWAS were also enriched in the QT-GWAS. Notably, all of these metabolic GOterms were more significantly enriched in the QT-GWAS, although the fold-changes of the enrichments were of similar size (Supplemental Table 7).
Of the 702 traits that were selected as both qualitative and quantitative metabolites, 71 traits yielded associations for both QT-GWAS and mGWAS. QT-GWAS and mGWAS retrieved 2556 and 529 unique genes for these 71 traits. Of these genes, 167 were retrieved by both methods. The overlap coefficient (Szymkiewicz–Simpson coefficient; the ratio of the intersection of both gene sets and the size of the smallest gene set, here the mGWAS genes) is 0.32. This indicates that only 32% of the genes retrieved by mGWAS (the smallest set) overlapped with the genes retrieved by QT-GWAS, suggesting that the QT-GWAS is able to retrieve new associations not found by mGWAS.
Proof of concept
Knowing the structure of a metabolite facilitates the search for candidate enzyme-encoding genes involved in its biosynthesis (e.g., glycosylated metabolites associated with genes encoding glycosyltransferases) – in many cases not even requiring full metabolite annotation if key functional groups can be identified. Therefore, all associations in the filtered QT-GWAS data set involving the 57 characterized metabolites were investigated (758 associations out of 2,931 associations, Figure 1; Supplemental Data Set 4). In order to pinpoint enzyme-encoding genes involved in metabolic processes, the QT-GWAS data set was further filtered for genes labeled with GOslim categories (high level summaries of related GOterms) related to metabolic processes. In this way, 291 associations with 34 metabolites remained. Of this selection, at least 23 associations involving 21 of the 34 metabolites were supported by published research (Table 1, Supplemental Results), demonstrating that the QT-GWAS approach allows to pinpoint enzyme-encoding genes affecting the biosynthesis of the associated metabolites. Similarly, in the mGWAS data set, 67 associations out of the 577 involved the 31 characterized metabolites. After filtering for genes labeled with GOslims related to metabolic processes, 62 associations remained involving the 31 metabolites (Supplemental Data Set 5). Of these 62 associations, at least 15 associations involving 14 of the 31 characterized metabolites were supported by published research (Table 1, Supplemental Results). Twelve of the supported associations overlapped between QT-GWAS and mGWAS.
Table 1. Overview of associations supported by literature and newly confirmed associations for QT-GWAS, mGWAS, and ASRgwas (population structure correction for qualitative metabolites).
“No.” refers to the corresponding metabolite number as used in the text and Supplemental Table S2. “NA” indicates that no association was recorded for the corresponding method. FA, ferulic acid; SA, sinapic acid, ISF, detected as in-source fragment.
| Associations confirmed by previous research | |||||
| DAAR1 | acetyl-(iso)leucine | 18 | 1.10 x 10-27 | 1.86 x 10-26 | 7.41 x 10-11 |
| MAM1/2/3 | 3-methylsulfinylpropyl glucosinolate | 1 | 1.58 x 10-8 | NA | 0.019 |
| 3-oxathiolesulfinylpropyl glucosinolate | 2 | NA | 3.03 x 10-7 | NA | |
| butyl glucosinolate | 6 | 4.03 x 10-8 | 2.17 x 10-9 | 0.022 | |
| 3-methylsulfinylbutyl glucosinolate | 7 | 1.48 x 10-12 | NA | 1.85 x 10-6 | |
| methylbutenyl glucosinolate | 8 | NA | 2.86 x 10-7 | NA | |
| glutathionylated methylsulfinylhexyl glucosinolate | 15 | 3.47 x 10-13 | NA | 1.54 x 10-5 | |
| sulfohydroxyimidazolone-G(8-O-4)FA | 17 | 3.56 x 10-11 | NA | 2.17 x 10-3 | |
| dihydroxynonenyl glucosinolate | 20 | NA | 9.64 x 10-7 | NA | |
| phenylpropyl glucosinolate | 23 | 2.24 x 10-17 | NA | 6.06 x 10-5 | |
| 4-benzoyloxybutyl glucosinolate | 40 | 1.92 x 10-10 | 8.67 x 10-18 | 8.55 x 10-9 | |
| 7-methylthioheptyl glucosinolate | 47 | 1.38 x 10-8 | 2.69 x 10-7 | 4.80 x 10-4 | |
| AOP3 | 3-oxathiolesulfinylpropyl glucosinolate | 2 | 8.14 x 10-12 | 8.92 x 10-7 | 5.95 x 10-5 |
| 4-O-sulfo-G(8-O-4)FA (isomer) | 29 | 2.48 x 10-7 | NA | 9.24 x 10-6 | |
| GSL-OH/2OG | glutathionylated methylsulfinylhexyl glucosinolate | 15 | 3.14 x 10-8 | NA | 9.00 x 10-5 |
| sinapoyl-(4-O-hexosylsinapoyl)-hexose | 34 | 9.30 x 10-9 | NA | 8.78 x 10-4 | |
| sinapoyl hexose(8-8)Sred | 42 | 4.13 x 10-9 | NA | 5.01 x 10-5 | |
| BGLU6 | quercetin-3-O-hexosyl(1->6)hexoside-7-O-deoxyhexoside | 19 | 5.72 x 10-23 | 5.80 x 10-10 | 3.60 x 10-4 |
| kaempherol-3-O-hexosyl(1->6)hexoside-7-O-deoxyhexoside | 25 | 1.49 x 10-24 | 9.13 x 10-9 | 2.09 x 10-11 | |
| isorhamnetin-3-O-hexosyl(1->6)hexoside-7-O-deoxyhexoside | 27 | 3.50 x 10-25 | NA | 3.32 x 10-11 | |
| Associations supported by previous research | |||||
| GH3 | 12-hydroxy-4,5-didehydrojasmonoyl glutamine hexoside | 24 | 1.50 x 10-15 | 4.62 x 10-19 | 2.71 x 10-11 |
| 12-hydroxy-4,5-didehydrojasmonoyl glutamine hexoside (isomer) | 26 | 2.34 x 10-19 | 2.19 x 10-14 | 4.18 x 10-5 | |
| 12-hydroxy-4,5-didehydrojasmonoyl glutamine hexoside (isomer) | 30 | 1.25 x 10-15 | 1.94 x 10-17 | 2.79 x 10-8 | |
| 12-hydroxy-4,5-didehydrojasmonoyl glutamine | 45 | 6.04 x 10-20 | 4.68 x 10-9 | 4.54 x 10-5 | |
| malonyl (2-hydroxy-4,5-didehydrojasmonoyl glutamine) | 46 | 1.06 x 10-12 | 1.65 x 10-10 | 8.19 x 10-5 | |
| SCPL13/8/9 | sinapoyl-(4-O-hexosylsinapoyl)-hexose | 34 | 3.96 x 10-9 | NA | 7.00 x 10-4 |
| Newly confirmed associations | |||||
| CYP706A5 | 6-hydroxy-2-methoxy-2-(2’-propanone-C-hexoside)-chroman | 37 | 1.12 x 10-23 | NA | 2.88 x 10-11 |
| 6-hydroxy-2-methoxy-2-(pentane-2’,4’-dione-5’-C-hexoside)-chroman | 48 | 7.12 x 10-26 | 7.41 x 10-13 | 5.11 x 10-12 | |
| 6-hydroxy-2-methoxy-2-(pentane-2’,4’-dione-5’-C-hexoside)-chroman (isomer) | 49 | 3.44 x 10-23 | 1.03 x 10-17 | 1.02 x 10-12 | |
| UGT76C3 | guanine (benzoyl) sulfohexoside | 22 | 3.25 x 10-14 | 1.37 x 10-14 | 5.11 x 10-4 |
| guanine (benzoyl) hexoside | 28 | 1.17 x 10-17 | 1.02 x 10-7 | 2.25 x 10-9 | |
| SULT202B1 | sulfo-G(8-O-4)FA | 21 | 7.37 x 10-8 | 2.55 x 10-12 | 6.07 x 10-7 |
| sulfo-G(8-O-4)FA (isomer) | 29 | NA | 1.40 x 10-10 | NA | |
| sulfo-G(8-O-4)SA | 44 | 4.92 x 10-7 | NA | 3.43 x 10-5 | |
Retrieving new candidate genes involved in metabolic pathways
Guided by structural information of the associated metabolites, seven associations involving three genes were selected for validation through comparative metabolome profiling of knockout mutants and/or in vitro enzyme assays. Of these seven validated associations, five were also retrieved by the mGWAS.
CYP7065A, a gene involved in chroman biosynthesis
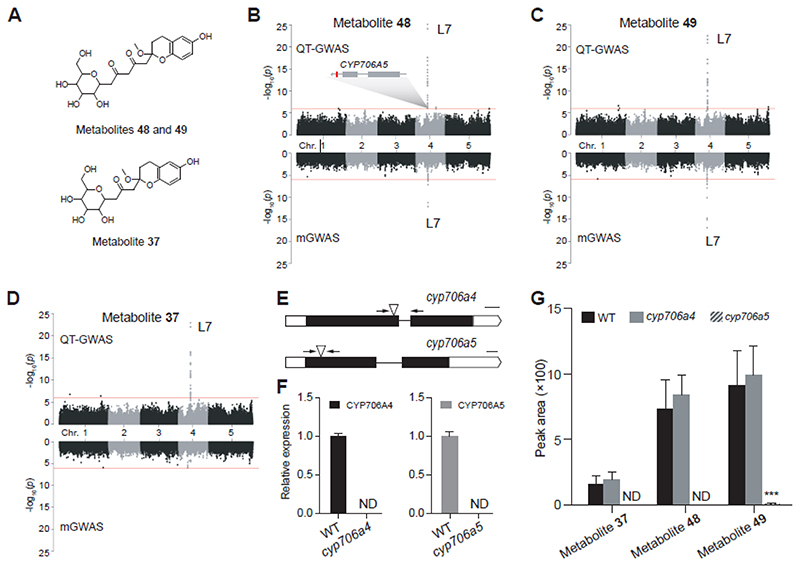
Chroman derivatives are heterocyclic compounds sharing a chroman (benzodihydropyran) backbone. Chromans constitute various important plant metabolites such as tocopherols, known for their vitamin E activity. In addition, the chroman skeletal structure occurs in specialized metabolites such as flavonoids (Jiang et al., 2020). In the characterized qualitative data set, the second most significant association (P-value = 7.12 x 10-26) in the QT-GWAS with characterized metabolites, was found between 6-hydroxy-2-methoxy-2-(pentane-2’,4’-dione-5’-C-hexoside)-chroman (metabolite 48) and SNP 4_7310453 located in the 3’ UTR of AT4G12310 (Supplemental Data Set 4, Figure 4A and 4B). This gene encodes a cytochrome P450 monooxygenase (CYP706A5) and was also identified as part of a pleiotropic locus (locus L7, Figure 3, Supplemental Table 3). Two neighboring genes, AT4G12300 and AT4G12320, also encode CYP706A proteins (CYP706A4 and CYP706A6, respectively). Furthermore, the locus contains two copper amine oxidase genes (AT4G12280 and AT4G12290). However, these two genes do not show expression at the seedling stage according to external datasets and were therefore excluded as candidate genes to be involved in the metabolism of metabolite 48. According to the ATTED-II database (Obayashi et al., 2022), CYP706A5 is strongly co-expressed with CYP706A6, suggesting they may be involved in the same metabolic pathway. Via QT-GWAS, also the abundances of an isomer of metabolite 48, i.e. metabolite 49, as well as of 6-hydroxy-2-methoxy-2-(2’-propanone-C-hexoside)-chroman (metabolite 37, Figure 4A), were associated with SNPs located in locus L7 (SNP 4_7309739, P-value =3.44 x 10-23 and SNP 4_7310453, P-value =1.12 x 10-23, respectively; Figure 4C and 4D). In addition, metabolites 48 and 49 were also associated with SNP 4_7310453 located in L7 via mGWAS (Table 1, Supplemental Data Set 5). No significant association was found for metabolite 37 via mGWAS.Q-Q plots for metabolites 48, 49 and 37 show a tail reflecting the small P-values of significantly associated SNPs and the SNPs in LD, as can be expected (Supplemental Figure 9). Nevertheless, the plots suggest an inflation of the P-values for QT-GWAS which could be the result of population structure. In conclusion, CYP706A4, A5 and A6 were considered as candidate genes underlying the variance in abundance of metabolites 48, 49 and 37.
Figure 4. Overview of the CYP706A locus and its associated metabolites.
A: putative structure of associated metabolites 48, 49 and 37. B, C, D: Manhattan plots of metabolites 48, 49 and 37 respectively, for QT-GWAS (upper panel) and mGWAS (lower panel): the x-axis represents the location of the recorded SNPs on the genome, the y-axis represents the negative logarithmic P-value for the association of each SNP to the respective metabolites. E. Schematic representation of CYP706A4 and CYP706A5; intronic regions are represented by a line, exonic regions are indicated in black and UTR regions in white; T-DNA insertion locations of the mutant lines are indicated by the triangles; Scale bar indicates 100 basepair. F. Relative expression of CYP706A4 and CYP706A5 as determined by RT-qPCR, primers are indicated by the arrows. G. Comparative metabolite profiling shows a significant reduction in the abundance of metabolites 37, 48 and 49 in cyp706a5 uniquely. Data are presented as mean ± standard deviation, n = 5; ND, not detected. *** P-value <0.001
In order to confirm the effect of L7 on the abundances of the chroman derivatives 37, 48 and 49, one homozygous T-DNA insertion mutant was obtained for CYP706A4 (cyp706a4; Figure 4E) and one for CYP706A5 (cyp706a5; Figure 4E). No T-DNA insertion mutant was available for CYP706A6. The reduction in expression of CYP706A4 and CYP706A5 was confirmed by RT-qPCR in the corresponding mutant lines (Figure 4F), while the expression of the three other CYP706A genes was not altered (Supplemental Figure 3). Comparative metabolite profiling of seedling metabolic extracts from each of the homozygous mutants revealed significant reductions for metabolites 37, 48 and 39 in cyp706a5, but not in cyp706a4 (Figure 4G, Supplemental Data Set 6-7). These results support the hypothesis that the CYP706A5 is involved in the production of chromans 37, 48 and 49, as predicted by the QT-GWAS approach.
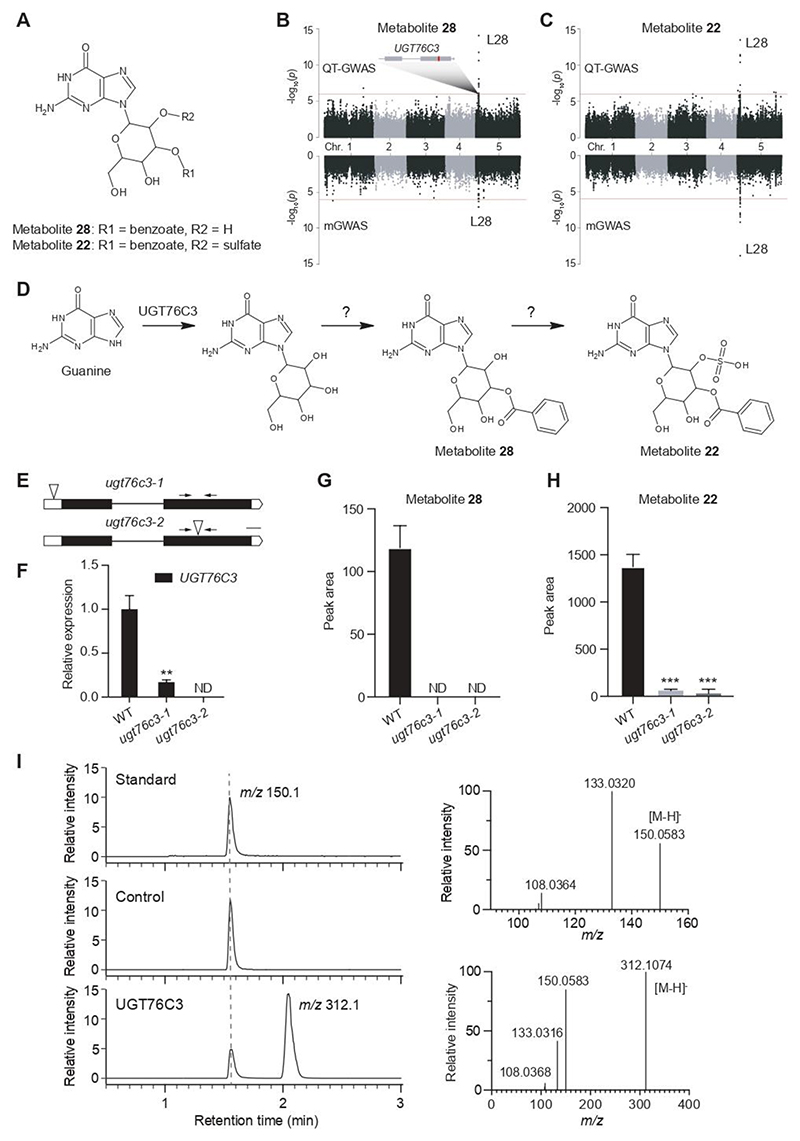
UGT67C3, a gene involved in guanine glycosylation
Guanine is one of the five essential nucleobases that make up part of the building blocks of nucleic acids and are ubiquitous in all known life forms. Nucleobases such as guanine can be taken up from the environment (Girke et al., 2014) or can be released from nucleotides or nucleic acids (Barbado et al., 2018). Little is known about guanine biosynthesis and metabolism, especially in plants. Based on MS/MS spectral data, two metabolites were characterized as guanine hexoside derivatives (metabolites 28 and 22; Figure 5A). In the QT-GWAS data set, the variations in both guanine hexoside pools were significantly associated with SNP 5_1776009 located in L28 on chromosome 5 (Figure 5B and 5C). This locus contains seven enzyme-encoding genes: DEOXYHYPUSINE SYNTHASE (DHS, AT5G05920), GUANYLYL CYCLASE 1 (GC1, AT5G05930), and five UDP-GLYCOSYLTRANSFERASE genes (UGT76C1-5). DHS is involved in hypusine synthesis from peptidyl-lysine, whereasGC1 is described to catalyze the formation of guanosine 3’,5’-cyclic monophosphate (cGMP)from guanosine5’-triphosphate (GTP) (Ludidi and Gehring, 2003; Duguay et al., 2007). UGT76C1 (AT5G05870) and UGT76C2 (AT5G05860) are described cytokinin UGTs (Šmehilová et al., 2016). UGT76C4 (AT5G05880) and UGT76C5 (AT5G05890) are known nicotinate UGTs (Wang et al., 2011; Li et al., 2015). The function of UGT76C3 (AT5G05900) has not been reported yet. In addition, the most significantly associated SNP is located in the coding sequence of UGT76C3, implying UGT76C3 is the most likely candidate glycosyltransferase involved in the biosynthesis of the guanine hexoside derivatives. Analogous to the chroman derivatives, Q-Q plots for metabolites 22 and 28 show a tail reflecting the small P-values of significantly associated SNPs and the SNPs in LD, and indicate some inflation of the P-values for QT-GWAS (Supplemental Figure 10). The associations of L28 with metabolites 28 and 22 were also retrieved in the mGWAS (Table 1).
Figure 5. Overview of the UGT76C locus and its associated metabolites.
A: putative structure of associated metabolites 28 and 22. The sugar moiety is N-linked, but its exact position and that of the substituents R1 and R2 are unknown. B and C: Manhattan plots of metabolites 28 and 22 respectively, for QT-GWAS (upper panel) and mGWAS (lower panel): the x-axis represents the location of the recorded SNPs on the genome, the y-axis represents the negative logarithmic P-value for the association of each SNP to the respective metabolites. A schematic representation of the candidate gene (UGT76C3) in L28 is shown and the most significantly associated SNP is marked in red within the gene. D. Proposed pathway for the biosynthesis of metabolites 28 and 22 in which UGT76C3 catalyzes the first step, namely the hexosylation of guanine. E. Schematic representation of UGT76C3; intronic regions are represented by a line, exonic regions are indicated in black and UTR regions in white; insertion locations of the mutant lines are indicated by the triangles. F. Relative expression of UGT76C3 as determined by RT-qPCR, primers are indicated by the arrows. G and H. Comparative metabolite profiling shows a significant reduction of metabolites 28 and 22 in both mutant lines. Data are presented as mean ± SD, n = 5; ND, not detected. ***P-value < 0.001, **0.001 < P-value < 0.01. I. In vitro enzyme assays show the conversion of guanine to guanine glucoside in the UGT76C3 reaction exclusively (left panel). GST-tagged RFP was used as negative control. MS/MS spectral data confirmed the characterization of m/z 150.1 as guanine (right upper panel) and m/z 312.1 as guanine glucoside (right lower panel).
To independently confirm the association between the UGT76C3 candidate gene and metabolites 22 and 28, two homozygous T-DNA insertion lines were obtained for UGT76C3 (ugt76c3-1 and ugt76c3-2, Figure 5E). The reduced UGT76C3 expression in the ugt76c3 mutant lines was confirmed by RT-qPCR (Figure 5F), whereas the expression of the other UGT76C genes was not altered (Supplemental Figure 4). Comparative metabolite profiling of ugt76c3-1 ugt76c3-2 versus WT seedling samples showed that metabolite 28 was completely absent in both mutants and metabolite 22 was significantly decreased in both mutants (ANOVA; PFDR <0.05) (Figure 5G and 5H, Supplemental Data Set 8-9). Root tissues of both mutant lines and WT plants were subjected to metabolic profiling, given the relatively high expression of UGT76C3 in roots (based on external datasets and RT-qPCR, Supplemental Figure 4). These results were in agreement with the seedling metabolic profiling (Supplemental Data Set 10-11). These results support that UGT76C3 is causal for the natural variation in the abundance of the associated guanine hexoside derivatives in Arabidopsis seedlings and suggest that the glycosylation of guanine, the first step of the proposed metabolic pathway, is catalyzed by UGT76C3 (Figure 5D).
To further investigate whether UGT76C3 catalyzes the glycosylation of guanine as suggested (Figure 5D), UGT76C3 was expressed in E. coli and purified as GST-tagged UGT76C3 recombinant protein (GST-UGT76C3) for enzymatic assays. Guanine was incubated with GST-UGT76C3 in the presence of UDP-glucose as a sugar donor. In contrast to the negative control, the GST-UGT76C3 reaction product gave rise to one peak at m/z 312.1, which was consistent with the theoretical monoisotopic mass of the guanine glucoside [M-H]- ion (C11H14N5O6-; 312.09441 Da) (Figure5I). The MS/MS fragmentation spectrum further showed a neutral loss of 162Da, corresponding to the loss of a hexose (minus H2O) moiety, indicating that the glucose moiety and guanine are N-linked, not C-linked. These observations demonstrate that UGT76C3 can catalyze the N-glycosylation of guanine in vitro.
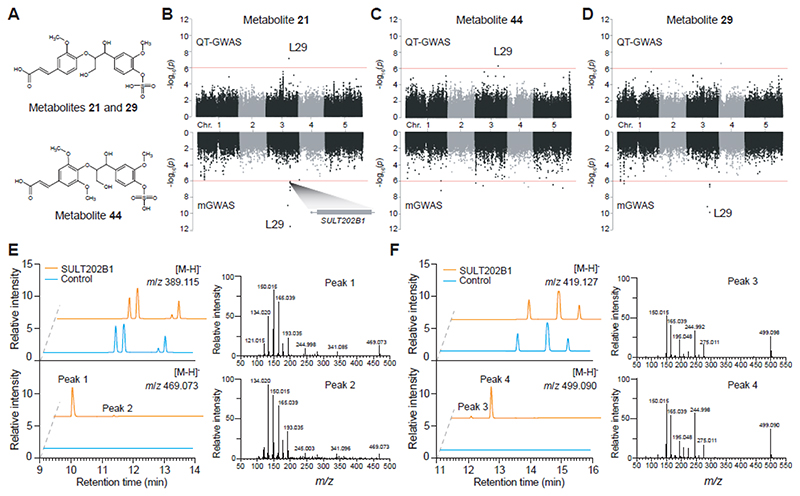
SULT202B1, a gene involved in neolignan sulfation
Sulfation of flavonoids has been reported in various studies (Teles et al., 2018) and several underlying genes have been reported in Arabidopsis (Klein and Papenbrock, 2004; Hashiguchi et al., 2014). However, genes responsible for the sulfation of neolignans remain unknown. Three sulfated neolignans, sulfo-G(8-O-4)FA (G stands for coniferyl alcohol and FA for ferulic acid, metabolite 21) and its isomer (metabolite 29), and sulfo-G(8-O-4)SA (SA stands for sinapic acid, metabolite 44) were characterized in this study (Figure 6A). In the QT-GWAS data set, metabolites 21 (SNP 3_16490051; P-value = 7.37 x 10-8, Figure 6B) and 44 (SNP 3_16487258;P-value=4.92 x 10-7, Figure 6C), were associated with locus L29 that contains four genes: a nitrate transporter (NRT2.6; AT3G45060), a sulfotransferase (SULT202B1; AT3G45070) and two nucleoside triphosphate hydrolases (AT3G45080 and AT3G45090). The sulfotransferase SULT202B1 has been reported to operate on flavonoids (Gidda and Varin, 2006; Hashiguchi et al., 2013). The associations of metabolites 21 and 44 with L29 suggest that SULT202B1 could potentially act upon neolignans as well. Alternatively, the association could be the result of competition between neolignans and flavonoids for conjugation. Q-Q plots for metabolites 21 and 44 show a less profound tail, compared to the chroman and guanin derivatives under investigation as a result of the less significant associations (Supplemental Figure 11).In the mGWAS, an association between locus L29 and metabolites 21 and 29 (Figure 6, B and D) could be retrieved, but not with metabolite 44, even though it was selected as quantitative trait as well.
Figure 6. Overview of the SULT202B1 locus and its associated metabolites.
A. Putative structure of associated metabolites 21, 29 and 44. The exact position of the sulfate group could not be determined based on MS/MS spectral data. B, C and D: Manhattan plots of metabolites 21, 44 and 29 respectively, for QT-GWAS (upper panel) and mGWAS (lower panel): the x-axis represents the location of the recorded SNPs on the genome, the y-axis represents the negative logarithmic P-value for the association of each SNP to the respective metabolites. The candidate gene (SULT202B1) in L29 is shown. E and F: Chromatograms of in vitro enzymatic reactions of SULT202B1 with chemically synthesized G(8-O-4)FA and G(8-O-4)SA, respectively. GST-tagged RFP was used as negative control. MS/MS spectral data confirmed the characterization of two m/z 469.073 products (peaks 1 and 2) as sulfo-G(8-O-4)FA and two m/z 499.090 products (peaks 3 and 4) as sulfo-G(8-O-4)SA.
To test whether SULT202B1 is active on G(8-O-4)FA or G(8-O-4)SA as predicted, recombinant GST-tagged SULT202B1 was produced in E. coli and purified for in vitro enzyme assays with chemically synthesized G(8-O-4)FA and G(8-O-4)SA (Supplemental Methods). G(8-O-4)FA and G(8-O-4)SA were tested as potential substrates (see methods), whereas the flavonol galangin was used as a positive control. As previously reported, SULT202B1 displayed activity towards galangin (Gidda and Varin, 2006). When incubated with G(8-O-4)FA, the SULT202B1 reaction product gave rise to two peaks exhibiting an m/z value and fragmentation spectrum corresponding to sulfo-G(8-O-4)FA (Figure 6E). Similarly, two sulfo-G(8-O-4)SA peaks were observed when G(8-O-4)SA was incubated with SULT202B1 (Figure 6F). These observations illustrate that SULT202B1 can catalyze the sulfation of G(8-O-4)FA and G(8-O-4)SA in vitro.
We assessed the effect of population structure correction for qualitative metabolites whose associations were supported by previous research or validated in this study through ASRgwas (Table 1). As expected, for all tested traits, the associations retrieved by QT-GWAS were less significantly detected by ASRgwas. Notably, 19 out of the 30 validated associations retrieved by QT-GWAS were no longer deemed significant by ASRgwas (p-values > 10-6) even though they have been experimentally validated in this study or were supported by previous research. These results suggest population structure correction is in some cases too conservative and could overlook the identification of real associations, as also noted by (Klasen et al., 2016). We visualized the potential influence of population structure on the associations retrieved by QT-GWAS, through Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP, (McInnes et al., 2020)) (Supplemental Figure 5, Supplemental Results, Supplemental Methods).
Discussion
QT-GWAS and mGWAS both retrieve loci involved in the biosynthesis of specialized metabolites and are partially complementary
Several studies have illustrated that targeted genetic analyses of qualitative metabolic traits, only present in a subset of a population of accessions, can efficiently pinpoint biosynthetic genes (Li et al., 2010; Ishihara et al., 2016; Tohge et al., 2016). Such discoveries are highly important both for fundamental knowledge and to open up possibilities towards metabolic engineering. By performing an untargeted association analysis through Fisher’s exact tests of 4,479 qualitative metabolic traits and 250K SNPs in Arabidopsis, we retrieved 2,931 significant associations with 515 metabolites (11% of all qualitative traits defined here). In comparison, mGWAS yielded 577 associations involving 248 (22% of 1147) quantitative metabolites. Considering that QT-GWAS and mGWAS picked up at least 23 and 15 literature-supported associations, respectively, both mGWAS and QT-GWAS methods are valid approaches to retrieve associations involved in the biosynthesis of specialized metabolites. The complementarity of both methods is illustrated by the observation that only twelve of the supported associations overlapped between both methods. Of the eight novel associations confirmed in this study, only five were retrieved by both methods.
The complementarity between both approaches could be explained by the fact that the QT-GWAS differentiates between genotypes in which the trait is absent and those expressing the trait, whereas mGWAS narrows associations down to those genotypes affecting the trait quantitatively rather than qualitatively. Nevertheless, the complementarity could also in part be explained by the different selection criteria for qualitative and quantitative traits. However, for the traits that were selected both as quantitative and qualitative traits, the two methods yielded different results in agreement with the different statistical strategies of both methods (only 32% of the genes overlapped based on the Szymkiewicz–Simpson coefficient). For example, QT-GWAS was able to retrieve a correct association between 6-hydroxy-2-methoxy-2-(2’-propanone-C-hexoside)-chroman (metabolite 37) and CYP705A6 and between sulfo-G(8-O-4)SA (metabolite 44) and SULT202B1, which were not retrieved by mGWAS at the current threshold, even though both metabolites were also selected as quantitative traits for the mGWAS. QT-GWAS was able to retrieve 11 associations that were supported by previous research that were not retrieved by mGWAS, despite the fact that the involved metabolites were used as input in both methods. Analogously, mGWAS yielded three literature-supported associations that were not found by QT-GWAS. The complementarity could (in part) be the result of the exclusion of rare alleles (minor allele frequency <5%) from mGWAS. Such alleles were not excluded from the QT-GWAS.
An enrichment for ‘metabolic process’-related GOterms was observed for both the mGWAS and the QT-GWAS. The QT-GWAS showed a more significant enrichment for all 5 ‘metabolic process’-related GOterms that were enriched in the mGWAS, and also for 17 additional ‘metabolic process’-related GOterms that were not enriched in the mGWAS. These results further support that the novel QT-GWAS is indeed able to retrieve genes involved in metabolism.
Novel enzyme–metabolite associations
In order to further demonstrate the validity of the QT-GWAS approach, candidate genes of unknown function were selected from genomic loci associated with characterized metabolites and subjected to reverse genetics. Comparative metabolic profiling of the T-DNA insertion mutant in candidate CYP706A5 versus the WT showed that the abundance of the associated chroman derivatives 37, 48 and 49 were significantly reduced in cyp706a5, indicating that CYP706A5 is involved in the biosynthesis of these metabolites. Furthermore, these metabolites were not differential in cyp706a4, indicating that the functions of CYP706A4 and CYP706A5 are not redundant. Metabolites 37, 48 and 49 have previously been reported to be highly abundant in leaf vacuoles of Arabidopsis (Dima et al., 2015). Moreover, the putative structures of the associated metabolites 37, 48 and 49 resemble polyketide synthase derailment products, because of their putative polyketide like nature (Yamaguchi et al., 1999; Jiang et al., 2006; Lim et al., 2016). Most polyketide synthases are promiscuous in the number of elongation steps, causing some polyketide intermediates to derail, resulting in heterocyclic truncation products (Lim et al., 2016). Although the involvement of CYP706A5 in the production of metabolites 37, 48, and 49 was demonstrated by reverse genetics, the identity of its in vivo substrate remains unclear.
Similarly, the metabolite profiling of T-DNA insertion lines in UGT76C3, located in L28, confirmed the involvement of the UGT in the production of metabolites 22 and 28, as predicted by both the QT-GWAS and mGWAS. The biosynthesis of metabolites 22 and 28 could potentially start from the hexosylation of guanine by UGT76C3 (Figure 5D). This hypothesis is supported by the in vitro enzyme assay, which illustrates that UGT76C3 is indeed capable of catalyzing the glycosylation of guanine in vitro. This is, to our knowledge, the first time that guanine glycosylation has been reported in plants.Here, guanine (benzoyl) hexoside (metabolite 28) and guanine (benzoyl)sulfo-hexoside (metabolite 22)were detected, instead of guanine hexoside. A possible reason is that guanine hexoside is rapidly conjugated with benzoate and/or sulfate. This may also explain why guanine hexoside has not been reported yet in other organisms. The guanine (benzoyl) hexoside and guanine (benzoyl) sulfo-hexoside could possibly be storage molecules, as molecules are frequently hexosylated for storage in the vacuole (Dima et al., 2015; Le Roy et al., 2016; Desmet et al., 2021b). Because UGT76C3 is highly expressed in roots, it could play a role in storage of excess guanine. Guanine has been shown to be taken up from the environment (Girke et al., 2014), but could also play a role as a nitrogen storage form in nitrogen-poor or -fluctuating environments (Mojzeš et al., 2020).
The QT-GWAS and mGWAS suggested that in addition to flavonoids, SULT202B1 could also catalyze the sulfation of neolignans, based on the observed associations with metabolites 21, 29 and 44. This hypothesis was confirmed by in vitro enzyme assays. This is, to our knowledge, the first report of an enzyme capable of catalyzing the sulfation of neolignans in vitro. The sulfation of neolignans could serve to improve their solubility in water, which could possibly alter their sequestration and/or transport to the vacuole or cell wall (Routaboul et al., 2012). Alternatively, sulfation of specialized metabolites, such as neolignans, could be the result of an adaptation to a specific environment. For example, a strong correlation was observed between the occurrence of sulfated flavonoids and plants growing near aquatic environments rich in mineral salts. Hence, the binding of sulfate to flavonoids and neolignans could be a mechanism for deactivation of excessive inorganic sulfate (Li et al., 2014).
QT-GWAS and mGWAS generate valuable databases
The QT-GWAS and mGWAS data sets retrieved a total of 26 literature-supported associations. Furthermore eight novel associations were confirmed here. These results illustrate that both methods lead to valuable gene–metabolite associations. In the QT-GWAS data set, 1,057 associations involved unknown metabolites and contained genes categorized under GOslims categories related to metabolic processes (Figure 1). In the list of QT-GWAS associations with characterized metabolites, at least 30 of the 291 associations (10%) involving metabolic genes were validated (23 through previous research and seven in this study). Extrapolating this percentage to the unknown associations, at least another 106 associations are promising leads. Analogously, in the mGWAS data set, 450 associations involved unknown metabolites and contained genes labeled with GOslims related to metabolic processes. In the mGWAS associations with characterized metabolites, 21 of the 62 associations (34%) involving metabolic genes were validated (15 through previous research and six in this study). Extrapolating this percentage to the unknown associations, at least 153 associations are predicted as promising leads. The 1,057 and 450 ‘metabolic process’-related loci involving unknown metabolites in QT-GWAS and mGWAS, respectively, can be mined further for additional candidate genes, which could be subjected to a combination of reverse genetics and metabolite profiling to investigate whether the abundance of the associated unknown metabolite is affected in the mutant. The identity of the potential substrate can be obtained through (partial) metabolite characterization, in case MS/MS spectral fragmentation data allows elucidation, isotope labeling (Simpson et al., 2021) or through purification and NMR analysis of the unknown metabolite.
Methods
Plant growth and experimental setup
Seeds of 183 A. thaliana accessions (Supplemental Table 1) were sown on MS-agar plates (0.5X Murashige and Skoog medium, 0.5% (w/v) sucrose, 1.2% (w/v) plant tissue agar), and stratified at 4°C for five days. Based on a randomize block design, 14-day-old seedlings (five replicates per accession) were then transferred to 96-well plates (MultiScreen, Merck Millipore) with two seedlings per well in 1 mL liquid MS-medium (0.5X MS medium, 0.5% (w/v) sucrose). The 96-well plates were placed in a growth chamber for 12 days (21°C, 16 h light/8 h dark with 120 μEs-1m-2). For metabolite extraction, the growth medium was removed by vacuum filtration and samples were quenched with 1 mL dry-ice-cold methanol. After heating the samples to 70°C for 15 min, the methanol extract was collected in a receiver plate using a vacuum manifold. Subsequently, 500 μL of the collected extract was cleaned with a Sep-Pak C18 96-well plate (Waters), dried in a vacuum concentrator and suspended in 50 μL ultrapure water. The 250K SNP data was obtained from from the Gregor Mendel Institute (available on github: https://github.com/Gregor-Mendel-Institute/atpolydb/tree/master/250k_snp_data).
All T-DNA insertion lines in this study were obtained from the SALK collection through the Nottingham Arabidopsis Stock Centre and homozygous mutants were identified by PCR amplification using T-DNA and gene specific primers (Supplemental Table 11). The Columbia-0 (Col-0) accession was used as WT.
Metabolite profiling and data processing
To obtain the metabolic profiles of all 183 A. thaliana accessions, each replicate (10 μL injected) was analyzed via negative ionization mode using a ultrahigh performance liquid chromatography (UHPLC) system hyphenated to electrospray ionization (ESI)-quadrupole-time of flight (QTOF)-MS (Acquity UPLC system coupled to a Synapt High Definition MS, Waters Corporation, Manchester, UK, Supplemental Methods). T-DNA insertion lines were analyzed on a Vion QTOF-MS (Waters Corporation, Manchester, UK, Supplemental Methods).
Peak grouping on the recorded LC–MS peaks was performed following Morreel et al. (2014) to estimate the number of detected metabolites. Statistical tests were performed in R version 4.0.0 (R Core Team, 2020). Data were transformed as described previously (Desmet et al., 2021b). Significant differences in the feature abundances between the T-DNA mutant lines and the WT were obtained following a one-way analysis of variance (ANOVA; lm() function) followed by Tukey honestly significant difference [HSD; TukeyHSD()] post hoc tests (α=0.05). The ANOVA model P-values were subjected to a false discovery rate [FDR, PFDR<0.05; p.adjust()] correction.
MS-based structural elucidation
In addition to MS/MS spectra recorded via QTOF-MS, MSn spectra were generated via reversed-phase UHPLC-ESI-Fourier transform ion cyclotron resonance (FTICR)-MS (Accela UHPLC system coupled to an LTQ FT Ultra, Thermo Scientific, Bremen, Germany) using the same separation conditions as mentioned for the QTOF-MS-based analyses. MS settings were as previously described (Desmet et al., 2021b). The MS/MS and MSn spectra mutually aligned and interpreted using RDynLib (Desmet et al., 2021b). Spectral interpretation was further assisted using CSI:FingerID (Dührkop et al., 2015) and competitive fragmentation modeling (CFM)-ID (Allen et al., 2014), and by applying previously published MS fragmentation pathways (Morreel et al., 2006; Morreel et al., 2010a; Morreel et al., 2010b; Desmet et al., 2021b) (Supplemental Methods).
GWAS of qualitative traits (QT-GWAS)
LC–MS processed features were converted to binary traits and subjected to Fisher exact tests using R version 4.0.0 [R Core Team, 2020; see “Qualitative GWAS.R available on Github”]. Whenever a feature was absent in more than half of the replicates for a particular accession (3 or more out of 5), the feature was considered absent in that accession. Some 4,479 features remained for the QT-GWAS (after removing features either present or absent in all 183 accessions). Using the 250K SNP data set, cosegregation analysis between the presence/absence of a feature and the presence/absence of a SNP was performed with Fisher exact tests [fisher.test(); -log10(P-value) >6] (see “Qualitative GWAS.R”). To accommodate for multiple hypotheses testing a P-value threshold of P-value < 10-6 was selected to define associations as significant.All scripts are made available at Github (https://github.com/mabro7766/QT). Manhattan and Q-Q plots were generated with the “qqman” package in R (Turner, 2014).
GWAS of quantitative traits (mGWAS)
mGWAS was performed on metabolite abundances measured in the 183 selected A. thaliana accessions. m/z signals with an average abundance lower than the detection limit of 500 were put to missing. The analysis was limited to those metabolites where at least 100 accessions had a value above the detection limit. Only SNPs were considered with a minor allele fraction (MAF) of at least 0.05. The association study was performed analogous to Kang et al. (2010), see Supplemental Methods for detailed descriptions. To accommodate for multiple hypotheses testing correction a P-value threshold of P-value < 10-6 was selected to define associations as significant. All scripts are made available at Github (https://github.com/mabro7766/mGWAS).
Comparative GOterm analysis and overlap coefficient
Gene Ontology data was obtained from TAIR (https://www.arabidopsis.org/). A GOterm enrichment was performed using the PANTHER webtool (http://pantherdb.org/) for both QT-GWAS and mGWAS. The inputted genes included duplicates, [genes occurring in more than one association were included in the enrichment as many times as they showed associations (13,795 genes for the QT-GWAS and 3,535 genes for the mGWAS)]. Enrichment was assessed versus all Arabidopsis genes present in the database through Bonferroni-corrected Fisher’s exact tests. The overlap coefficient (Szymkiewicz–Simpson) was calculated as ratio of the intersection and the size of the mGWAS gene set (equation 1).
Gene expression analysis via RT-qPCR and external datasets
Shoot or root tissues of 2-week-old Arabidopsis seedlings were collected and immediately frozen in liquid nitrogen. Six plants were pooled per biological replicate and three replicates were harvested per line. All plant materials were disrupted in 2-mL Eppendorf tubes using a Retsch MM300 mill (20 Hz, 3-mm bead). Total RNA was extracted using ReliaPrep RNA Tissue Miniprep System (Promega). A total of 1 μg RNA was used as a template for cDNA synthesis using the qScript cDNA SuperMix (Quantabio). RT-qPCRs were performed using SYBR® Green Mix (Roche)in the Lightcycler® 480 System (Roche). Arabidopsis UBIQUITIN CONJUGATION ENZYME 9 (UBC9, AT4G27960) and UBC21 (AT5G25760) genes were used as reference genes. Gene expression of AT4G12280, AT4G12290, AT4G12300, AT4G12310, AT4G12320, and AT5G05900 was checked via eFP browser (Winter et al., 2007).
Enzymatic assays
In vitro enzymatic assays of UGT76C3 were performed as described (Peng et al., 2017) enzymatic assays of SULT202B1 were conducted as described (Hashiguchi et al., 2013).Recombinant GST-tagged red fluorescent protein (RFP) was used as negative control. For recombinant protein expression and purification see Supplemental Methods.
LC–MS analyses of the reaction mixtures were performed on an Acquity UPLC system coupled to a Synapt-XS high resolution MS (Waters Corporation, Manchester, UK). Ten μL reaction product was injected and analyzed as described for “Metabolite profiling and data processing” with the following altered parameter settings: desolvation temperature was set to 550°C, the desolvation gas flow was set to 800L/h and transfer collision energy was set to 4 V.
Supplementary Material
Supplemental Information is available at Molecular Plant Online.
Short summary.
A novel genome-wide association method to uncover biosynthetic loci underlying qualitative metabolic traits (designated as QT-GWAS) was developed and performed alongside a conventional quantitative metabolite GWAS (mGWAS). At least 26 of the associations found were supported by previous research and 8 associations involving three metabolic enzyme-encoding genes (CYP706A5, UGT76C3 and SULT202B1) were newly confirmed, illustrating the power of the novel QT-GWAS.
Acknowledgments
We would like to thank dr. engr. Joost Keurentjes for providing the Arabidopsis accessions and Marnik Vuylsteke (Gnomixx) for analyzing the data with the R packages ASRgwas and ASReml-R. We would like to thank Annick Bleys for critically reading the manuscript. No conflict of interest declared.
Funding
M.B. is indebted for funding to ERC-Advanced Grant 2019 POPMET and FWO-SB predoctoral fellowship (1S38920N), M.P. is indebted for funding to MSCA (CHORPATH – 897918), R.H. is indebted for funding to OMICS@VIB Marie Curie COFUND fellowship. I.E.H. is indebted for funding to FWO-SB predoctoral fellowship (1S04020N), EMBO research grant (STF-8658) and iBOF (Next-BIOREF, 01IB4220). We also thank the Bijzonder Onderzoeksfonds-Zware Apparatuur of Ghent University for the Fourier transform ion cyclotron resonance mass spectrometer (174PZA05) and the Hercules program of Ghent University for the Synapt QTOF High Definition MS (Grant AUGE/014). V.I.T. and J.R. were funded by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-SC0018409). W.B. is indebted for funding by Stanford University’s Global Climate and Energy Project‘ Towards New Degradable Lignin Types’ and by the ERC-Advanced Grant POPMET.
Footnotes
Author Contributions
M.B., K.M. & W.B. designed the research; M.B., M.P., R.H., C.D. & I.E.H. performed experiments; M.B., K.M., V.S., & Y.S. developed scripts for data acquisition; V.I.T. and J.R. produced authentic synthetic compounds, M.B., M.P. & K.M. performed data analysis; M.B., M.P., K.M. & W.B. wrote the manuscript that all authors edited and approved.
References
- Allen F, Pon A, Wilson M, Greiner R, Wishart D. CFM-ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acids Res. 2014;42:W94–W99. doi: 10.1093/nar/gku436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseekh S, Ofner I, Liu Z, Osorio S, Vallarino J, Last RL, Zamir D, Tohge T, Fernie AR. Quantitative trait loci analysis of seed-specialized metabolites reveals seed-specific flavonols and differential regulation of glycoalkaloid content in tomato. Plant J. 2020;103:2007–2024. doi: 10.1111/tpj.14879. [DOI] [PubMed] [Google Scholar]
- Angelovici R, Batushansky A, Deason N, Gonzalez-Jorge S, Gore MA, Fait A, DellaPenna D. Network-guided GWAS improves identification of genes affecting free amino acids. Plant Physiol. 2017;173:872–886. doi: 10.1104/pp.16.01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbona V, Gomez-Cadenas A. Metabolomics of disease resistance in crops. Curr Issues Mol Biol. 2016;19:13–30. [PubMed] [Google Scholar]
- Barbado C, Córdoba-Cañero D, Ariza RR, Roldán-Arjona T. Nonenzymatic release of N7-methylguanine channels repair of abasic sites into an AP endonuclease-independent pathway inArabidopsis. Proc Natl Acad Sci USA. 2018;115:E916–E924. doi: 10.1073/pnas.1719497115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baseggio M, Murray M, Magallanes - Lundback M, Kaczmar N, Chamness J, Buckler ES, Smith ME, DellaPenna D, Tracy WF, Gore MA. Natural variation for carotenoids in fresh kernels is controlled by uncommon variants in sweet corn. Plant Genome. 2020;13:e20008. doi: 10.1002/tpg2.20008. [DOI] [PubMed] [Google Scholar]
- Bishayee A, Sethi G. Bioactive natural products in cancer prevention and therapy: progress and promise. Semin Cancer Biol. 2016;40-41:1–3. doi: 10.1016/j.semcancer.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Bouwmeester H, Schuurink RC, Bleeker PM, Schiestl F. The role of volatiles in plant communication. Plant J. 2019;100:892–907. doi: 10.1111/tpj.14496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotman Y, Llorente-Wiegand C, Oyong G, Badoni S, Misra G, Anacleto R, Parween S, Pasion E, Tiozon RN, Jr, Anonuevo JJ, et al. The genetics underlying metabolic signatures in a brown rice diversity panel and their vital role in human nutrition. Plant J. 2021;106:507–525. doi: 10.1111/tpj.15182. [DOI] [PubMed] [Google Scholar]
- Chan EKF, Rowe HC, Kliebenstein DJ. Understanding the evolution of defense metabolites in Arabidopsis thaliana using genome-wide association mapping. Genetics. 2010;185:991–1007. doi: 10.1534/genetics.109.108522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Hu X, Shi T, Yin H, Sun D, Hao Y, Xia X, Luo J, Fernie AR, He Z, et al. Metabolite-based genome-wide association study enables dissection of the flavonoid decoration pathway of wheat kernels. Plant Biotechnol J. 2020;18:1722–1735. doi: 10.1111/pbi.13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Gao Y, Xie W, Gong L, Lu K, Wang W, Li Y, Liu X, Zhang H, Dong H, et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat Genet. 2014;46:714–721. doi: 10.1038/ng.3007. [DOI] [PubMed] [Google Scholar]
- Chen W, Wang W, Peng M, Gong L, Gao Y, Wan J, Wang S, Shi L, Zhou B, Li Z, et al. Comparative and parallel genome-wide association studies for metabolic and agronomic traits in cereals. Nat Commun. 2016;7:12767. doi: 10.1038/ncomms12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowański S, Adamski Z, Marciniak P, Rosiński G, Büyükgüzel E, Büyükgüzel K, Falabella P, Scrano L, Ventrella E, Lelario F, et al. A review of bioinsecticidal activity of solanaceae alkaloids. Toxins. 2016;8:60. doi: 10.3390/toxins8030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contrepois K, Liang L, Snyder M. Can metabolic profiles be used as a phenotypic readout of the genome to enhance precision medicine? Clin Chem. 2016;62:676–678. doi: 10.1373/clinchem.2015.251181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desborough MJR, Keeling DM. The aspirin story – from willow to wonder drug. Br J Haematol. 2017;177:674–683. doi: 10.1111/bjh.14520. [DOI] [PubMed] [Google Scholar]
- Desmet S, Brouckaert M, Boerjan W, Morreel K. Seeing the forest for the trees: retrieving plant secondary biochemical pathways from metabolome networks. Comp Struct Biotechnol J. 2021a;19:72–85. doi: 10.1016/j.csbj.2020.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet S, Saeys Y, Verstaen K, Dauwe R, Kim H, Niculaes C, Fukushima A, Goeminne G, Vanholme R, Ralph J, et al. Maize specialized metabolome networks reveal organ-preferential mixed glycosides. Comp Struct Biotechnol J. 2021b;19:1127–1144. doi: 10.1016/j.csbj.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dima O, Morreel K, Vanholme B, Kim H, Ralph J, Boerjan W. Small glycosylated lignin oligomers are stored in Arabidopsis leaf vacuoles. Plant Cell. 2015;27:695–710. doi: 10.1105/tpc.114.134643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Gao Y, Chen W, Wang W, Gong L, Liu X, Luo J. Spatiotemporal distribution of phenolamides and the genetics of natural variation of hydroxycinnamoyl spermidine in rice. Mol Plant. 2015;8:111–121. doi: 10.1016/j.molp.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Dudareva N, Klempien A, Muhlemann JK, Kaplan I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013;198:16–32. doi: 10.1111/nph.12145. [DOI] [PubMed] [Google Scholar]
- Duguay J, Jamal S, Liu Z, Wang T-W, Thompson JE. Leaf-specific suppression of deoxyhypusine synthase in Arabidopsis thaliana enhances growth without negative pleiotropic effects. J Plant Physiol. 2007;164:408–420. doi: 10.1016/j.jplph.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Dührkop K, Shen HB, Meusel M, Rousu J, Böcker S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc Natl Acad Sci USA. 2015;112:12580–12585. doi: 10.1073/pnas.1509788112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Tohge T. The genetics of plant metabolism. Annu Rev Genet. 2017;51:287–310. doi: 10.1146/annurev-genet-120116-024640. [DOI] [PubMed] [Google Scholar]
- Fürstenberg-Hägg J, Zagrobelny M, Bak S. Plant defense against insect herbivores. Int J Mol Sci. 2013;14:10242–10297. doi: 10.3390/ijms140510242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda SK, Varin L. Biochemical and molecular characterization of flavonoid 7-sulfotransferase from Arabidopsis thaliana. Plant Physiol Biochem. 2006;44:628–636. doi: 10.1016/j.plaphy.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Girke C, Daumann M, Niopek-Witz S, Möhlmann T. Nucleobase and nucleoside transport and integration into plant metabolism. Front Plant Sci. 2014;5:443. doi: 10.3389/fpls.2014.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Góral I, Wojciechowski K. Surface activity and foaming properties of saponin-rich plants extracts. Adv Colloid Interface Sci. 2020;279:102145. doi: 10.1016/j.cis.2020.102145. [DOI] [PubMed] [Google Scholar]
- Hashiguchi T, Sakakibara Y, Hara Y, Shimohira T, Kurogi K, Akashi R, Liu M-C, Suiko M. Identification and characterization of a novel kaempferol sulfotransferase from Arabidopsis thaliana. Biochem Biophys Res Commun. 2013;434:829–835. doi: 10.1016/j.bbrc.2013.04.022. [DOI] [PubMed] [Google Scholar]
- Hashiguchi T, Sakakibara Y, Shimohira T, Kurogi K, Yamasaki M, Nishiyama K, Akashi R, Liu M-C, Suiko M. Identification of a novel flavonoid glycoside sulfotransferase in Arabidopsis thaliana. J Biochem. 2014;155:91–97. doi: 10.1093/jb/mvt102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton MW, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjálmsson BJ, et al. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet. 2012;44:212–216. doi: 10.1038/ng.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H, Tohge T, Viehöver P, Fernie AR, Weisshaar B, Stracke R. Natural variation in flavonol accumulation in Arabidopsis is determined by the flavonol glucosyltransferase BGLU6. J Exp Bot. 2016;67:1505–1517. doi: 10.1093/jxb/erv546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Schommer CK, Kim SY, Suh DY. Cloning and characterization of chalcone synthase from the moss, Physcomitrella patens. Phytochemistry. 2006;67:2531–2540. doi: 10.1016/j.phytochem.2006.09.030. [DOI] [PubMed] [Google Scholar]
- Jiang S, Su S, Chen M, Peng F, Zhou Q, Liu T, Liu L, Xue W. Antibacterial activities of novel dithiocarbamate-containing 4H-chromen-4-one derivatives. J Agric Food Chem. 2020;68:5641–5647. doi: 10.1021/acs.jafc.0c01652. [DOI] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong S-y, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M. Recombination and linkage disequilibrium inArabidopsis thaliana. Nat Genet. 2007;39:1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]
- Klasen JR, Barbez E, Meier L, Meinshausen N, Bühlmann P, Koornneef M, Busch W, Schneeberger K. A multi-marker association method for genome-wide association studies without the need for population structure correction. Nature communications. 2016;7:13299. doi: 10.1038/ncomms13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M, Papenbrock J. The multi-protein family of Arabidopsis sulphotransferases and their relatives in other plant species. J Exp Bot. 2004;55:1809–1820. doi: 10.1093/jxb/erh183. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T. Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 2001;126:811–825. doi: 10.1104/pp.126.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoch D, Riewe D, Meyer RC, Boudichevskaia A, Schmidt R, Altmann T. Genetic dissection of metabolite variation in Arabidopsis seeds: evidence for mQTL hotspots and a master regulatory locus of seed metabolism. J Exp Bot. 2017;68:1655–1667. doi: 10.1093/jxb/erx049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J, Textor S, Tokuhisa JG, Falk KL, Bartram S, Gershenzon J, Mitchell-Olds T. A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol. 2001;127:1077–1088. [PMC free article] [PubMed] [Google Scholar]
- Kushalappa AC, Gunnaiah R. Metabolo-proteomics to discover plant biotic stress resistance genes. Trends Plant Sci. 2013;18:522–531. doi: 10.1016/j.tplants.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Le Roy J, Huss B, Creach A, Hawkins S, Neutelings G. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front Plant Sci. 2016;7:735. doi: 10.3389/fpls.2016.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang F, Chang Y, Zhao T, Schranz ME, Wang G. Nicotinate O-glucosylation is an evolutionarily metabolic trait important for seed germination under stress conditions in Arabidopsis thaliana. Plant Cell. 2015;27:1907–1924. doi: 10.1105/tpc.15.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Bergelson J, Chapple C. The ARABIDOPSIS accession Pna-10 is a naturally occurring sng1 deletion mutant. Mol Plant. 2010;3:91–100. doi: 10.1093/mp/ssp090. [DOI] [PubMed] [Google Scholar]
- Li X, Svedin E, Mo H, Atwell S, Dilkes BP, Chapple C. Exploiting natural variation of secondary metabolism identifies a gene controlling the glycosylation diversity of dihydroxybenzoic acidsin Arabidopsis thaliana. Genetics. 2014;198:1267–1276. doi: 10.1534/genetics.114.168690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YP, Go MK, Yew WS. Exploiting the biosynthetic potential of type III polyketide synthases. Molecules. 2016;21:806. doi: 10.3390/molecules21060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza-Tavera H. Monoterpenes in essential oils. Biosynthesis and properties. Adv Exp Med Biol. 1999;464:49–62. doi: 10.1007/978-1-4615-4729-7_5. [DOI] [PubMed] [Google Scholar]
- Ludidi N, Gehring C. Identification of a novel protein with guanylyl cyclase activity in Arabidopsis thaliana. J Biol Chem. 2003;278:6490–6494. doi: 10.1074/jbc.M210983200. [DOI] [PubMed] [Google Scholar]
- Luo J. Metabolite-based genome-wide association studies in plants. Curr Opin Plant Biol. 2015;24:31–38. doi: 10.1016/j.pbi.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Mahieu NG, Spalding JL, Gelman SJ, Patti GJ. Defining and detecting complex peak relationships in mass spectral data: the Mz.unity algorithm. Anal Chem. 2016;88:9037–9046. doi: 10.1021/acs.analchem.6b01702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Nakabayashi R, Yang Z, Okazaki Y, Yonemaru J-i, Ebana K, Yano M, Saito K. Metabolome-genome-wide association study dissects genetic architecture for generating natural variation in rice secondary metabolism. Plant J. 2015;81:13–23. doi: 10.1111/tpj.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, Melville J. Umap: Uniform manifold approximation and projection for dimension reduction. 2020. arXiv:1802.03426 [Google Scholar]
- Moghe GD, Last RL. Something old, something new: conserved enzymes and the evolution of novelty in plant specialized metabolism. Plant Physiol. 2015;169:1512–1523. doi: 10.1104/pp.15.00994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojzeš P, Gao L, Ismagulova T, Pilátová J, Moudříková S, Gorelova O, Solovchenko A, Nedbal L, Salih A. Guanine, a high-capacity and rapid-turnover nitrogen reserve in microalgal cells. Proc Natl Acad Sci USA. 2020;117:32722–32730. doi: 10.1073/pnas.2005460117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K, Dima O, Kim H, Lu F, Niculaes C, Vanholme R, Dauwe R, Goeminne G, Inzé D, Messens E, et al. Mass spectrometry-based sequencing of lignin oligomers. Plant Physiol. 2010a;153:1464–1478. doi: 10.1104/pp.110.156489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K, Goeminne G, Storme V, Sterck L, Ralph J, Coppieters W, Breyne P, Steenackers M, Georges M, Messens E, et al. Genetical metabolomics of flavonoid biosynthesis in Populus a case study. Plant J. 2006;47:224–237. doi: 10.1111/j.1365-313X.2006.02786.x. [DOI] [PubMed] [Google Scholar]
- Morreel K, Kim H, Lu F, Dima O, Akiyama T, Vanholme R, Niculaes C, Goeminne G, Inzé D, Messens E, et al. Mass spectrometry-based fragmentation as an identification tool in lignomics. Anal Chem. 2010b;82:8095–8105. doi: 10.1021/ac100968g. [DOI] [PubMed] [Google Scholar]
- Morreel K, Saeys Y, Dima O, Lu F, Van de Peer Y, Vanholme R, Ralph J, Vanholme B, Boerjan W. Systematic structural characterization of metabolites in Arabidopsis via candidate substrate-product pair networks. Plant Cell. 2014;26:929–945. doi: 10.1105/tpc.113.122242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D, Mukherjee A, Ghosh TC. Evolutionary rate heterogeneity of primary and secondary metabolic pathway genes inArabidopsis thaliana. Genome Biol Evol. 2015;8:17–28. doi: 10.1093/gbe/evv217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Alseekh S, de Oliveira Silva FM, Omranian N, Lichtenstein G, Mirnezhad M, González RRR, Sabio y Garcia J, Conte M, Leiss KA, et al. Identification and characterization of metabolite quantitative trait loci in tomato leaves and comparison with those reported for fruits and seeds. Metabolomics. 2019;15:46. doi: 10.1007/s11306-019-1503-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Hibara H, Kagaya Y, Aoki Y, Kinoshita K. ATTED-II v11: a plant gene coexpression database using a sample balancing technique by subagging of principal components. Plant Cell Physiol. 2022;63:869–881. doi: 10.1093/pcp/pcac041. [DOI] [PubMed] [Google Scholar]
- Peng M, Shahzad R, Gul A, Subthain H, Shen S, Lei L, Zheng Z, Zhou J, Lu D, Wang S, et al. Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. Nat Commun. 2017;8 doi: 10.1038/s41467-017-02168-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: a language and environment for statistical computing. Foundation for Statistical Computing; Vienna, Austria: 2020. http://www.R-project.org/ [Google Scholar]
- Ricachenevsky FK, Vasconcelos MW, Shou H, Johnson AAT, Sperotto RA. Editorial: Improving the nutritional content and quality of crops: promises, achievements, and future challenges. Front Plant Sci. 2019;10:738. doi: 10.3389/fpls.2019.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PM, Cantrill V, Benohoud M, Tidder A, Rayner CM, Blackburn RS. Application of anthocyanins from blackcurrant (Ribes nigrum L.) fruit waste as renewable hair dyes. J Agric Food Chem. 2018;66:6790–6798. doi: 10.1021/acs.jafc.8b01044. [DOI] [PubMed] [Google Scholar]
- Routaboul J-M, Dubos C, Beck G, Marquis C, Bidzinski P, Loudet O, Lepiniec L. Metabolite profiling and quantitative genetics of natural variation for flavonoids inArabidopsis. J Exp Bot. 2012;63:3749–3764. doi: 10.1093/jxb/ers067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Balbo I, Steinfath M, Repsilber D, Selbig J, Pleban T, Zamir D, Fernie AR. Mode of inheritance of primary metabolic traits in tomato. Plant Cell. 2008;20:509–523. doi: 10.1105/tpc.107.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Zhu A, Jia J, Hu X, Chen J, Liu W, Ren X, Sun D, Fernie AR, Cui F, et al. Metabolomics analysis and metabolite‐agronomic trait associations using kernels of wheatTriticum aestivum recombinant inbred lines. Plant J. 2020;103:279–292. doi: 10.1111/tpj.14727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SW, Jung E, Kim S, Kim J-H, Kim E-G, Lee J, Park D. Antagonizing effects and mechanisms of afzelin against UVB-induced cell damage. PLoS ONE. 2013;8:e61971. doi: 10.1371/journal.pone.0061971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JP, Wunderlich C, Li X, Svedin E, Dilkes B, Chapple C. Metabolic source isotopic pair labeling and genome-wide association are complementary tools for the identification of metabolite–gene associations in plants. The Plant Cell. 2021;33:492–510. doi: 10.1093/plcell/koaa046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmehilová M, Dobrůšková J, Novák O, Takáč T, Galuszka P. Cytokinin-specific glycosyltransferases possess different roles in cytokinin homeostasis maintenance. Front Plant Sci. 2016;7:1264. doi: 10.3389/fpls.2016.01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotelo-Silveira M, Chauvin A-L, Marsch-Martinez N, Winkler R, de Folter S. Metabolic fingerprinting of Arabidopsis thaliana accessions. Front Plant Sci. 2015;6:365. doi: 10.3389/fpls.2015.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenackers W, El Houari I, Baekelandt A, Witvrouw K, Dhondt S, Leroux O, Gonzalez N, Corneillie S, Cesarino I, Inzé D, et al. cis-Cinnamic acid is a natural plant growth-promoting compound. J Exp Bot. 2019;70:6293–6304. doi: 10.1093/jxb/erz392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch RC, Svedin E, Dilkes B, Chapple C, Li X. Discovery of a novel amino acid racemase through exploration of natural variation inArabidopsis thaliana. Proc Natl Acad Sci USA. 2015;112:11726–11731. doi: 10.1073/pnas.1503272112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles YCF, Souza MSR, de Souza MDFV. Sulphated flavonoids: biosynthesis, structures, and biological activities. Molecules. 2018;23:480. doi: 10.3390/molecules23020480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Textor S, Bartram S, Kroymann J, Falk KL, Hick A, Pickett JA, Gershenzon J. Biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana: recombinant expression and characterization of methylthioalkylmalate synthase, the condensing enzyme of the chain-elongation cycle. Planta. 2004;218:1026–1035. doi: 10.1007/s00425-003-1184-3. [DOI] [PubMed] [Google Scholar]
- Tohge T, Scossa F, Wendenburg R, Frasse P, Balbo I, Watanabe M, Alseekh S, Jadhav SS, Delfin JC, Lohse M, et al. Exploiting natural variation in tomato to define pathway structure and metabolic regulation of fruit polyphenolics in the lycopersicum complex. Mol Plant. 2020;13:1027–1046. doi: 10.1016/j.molp.2020.04.004. [DOI] [PubMed] [Google Scholar]
- Tohge T, Wendenburg R, Ishihara H, Nakabayashi R, Watanabe M, Sulpice R, Hoefgen R, Takayama H, Saito K, Stitt M, et al. Characterization of a recently evolved flavonol-phenylacyltransferase gene provides signatures of natural light selection in Brassicaceae. Nat Commun. 2016;7:12399. doi: 10.1038/ncomms12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SD. qqman: an R package for visualizing GWAS results using QQ and manhattan plots. Biorxiv. 2014 doi: 10.1101/005165. [DOI] [Google Scholar]
- Varela MC, Arslan I, Reginato MA, Cenzano AM, Luna MV. Phenolic compounds as indicators of drought resistance in shrubs from Patagonian shrublands (Argentina) Plant Physiol Biochem. 2016;104:81–91. doi: 10.1016/j.plaphy.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Wang J, Ma X-M, Kojima M, Sakakibara H, Hou B-K. N-glucosyltransferase UGT76C2 is involved in cytokinin homeostasis and cytokinin response in Arabidopsis thaliana. Plant Cell Physiol. 2011;52:2200–2213. doi: 10.1093/pcp/pcr152. [DOI] [PubMed] [Google Scholar]
- Wang S, Alseekh S, Fernie AR, Luo J. The structure and function of major plant metabolite modifications. Mol Plant. 2019;12:899–919. doi: 10.1016/j.molp.2019.06.001. [DOI] [PubMed] [Google Scholar]
- Wen W, Li D, Li X, Gao Y, Li W, Li H, Liu J, Liu H, Chen W, Luo J, et al. Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat Commun. 2014;5:3438. doi: 10.1038/ncomms4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Li K, Alseekh S, Omranian N, Zhao L, Zhou Y, Xiao Y, Jin M, Yang N, Liu H, et al. Genetic determinants of the network of primary metabolism and their relationships to plant performance in a maize recombinant inbred line population. Plant Cell. 2015;27:1839–1856. doi: 10.1105/tpc.15.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J-K. The evolutionary paths towards complexity: a metabolic perspective. New Phytol. 2014;201:1141–1149. doi: 10.1111/nph.12416. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Alseekh S, Cuadros-Inostroza Á, Fusari CM, Mutwil M, Kooke R, Keurentjes JB, Fernie AR, Willmitzer L, Brotman Y. Combined use of genome-wide association data and correlation networks unravels key regulators of primary metabolism in Arabidopsis thaliana. PLoS Genet. 2016;12:e1006363. doi: 10.1371/journal.pgen.1006363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tohge T, Cuadros-Inostroza Á, Tong H, Tenenboim H, Kooke R, Méret M, Keurentjes JB, Nikoloski Z, Fernie AR, et al. Mapping the Arabidopsis metabolic landscape by untargeted metabolomics at different environmental conditions. Mol Plant. 2018;11:118–134. doi: 10.1016/j.molp.2017.08.012. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Kurosaki F, Suh DY, Sankawa U, Nishioka M, Akiyama T, Shibuya M, Ebizuka Y. Cross-reaction of chalcone synthase and stilbene synthase overexpressed in Escherichia coli. FEBS Lett. 1999;460:457–461. doi: 10.1016/s0014-5793(99)01403-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.