Abstract
Pathogenic and non-pathogenic fungi synthesize glycosphingolipids, which have a crucial role in growth and viability. Glycosphingolipids also contribute to fungal-associated pathogenesis. The opportunistic yeast pathogen Candida albicans synthesizes phospholipomannan (PLM), which is a glycosphingolipid of the mannosylinositol phosphorylceramide family. Through its lipid and glycan moieties, PLM contributes to the initial recognition of the yeast, causing immune system disorder and persistent fungal disease through activation of host signaling pathways. The lipid moiety of PLM activates the deregulation signaling pathway involved in yeast phagocytosis whereas its glycan moiety, composed of β-1,2 mannosides (β-Mans), participates to inflammatory processes through a mechanism involving Galectin-3. Biosynthesis of PLM β-Mans involves two β-1,2 mannosyltransferases (Bmts) that initiate (Bmt5) and elongate (Bmt6) the glycan chains. After generation of double bmtsΔ mutants, we show that Bmt5 has redundant activity with Bmt2, which can replace Bmt5 in bmt5Δ mutant. We also report that PLM is located in the inner layer of the yeast cell wall. PLM seems to be not essential for systemic infection of the yeast. However, defect of PLM β-mannosylation increases resistance of C. albicans to inhibitors of β-glucans and chitin synthesis, highlighting a role of PLM in cell wall homeostasis.
Keywords: Glycosphingolipid; β-1,2 mannosyltransferases; yeast; cell wall; virulence
1. Introduction
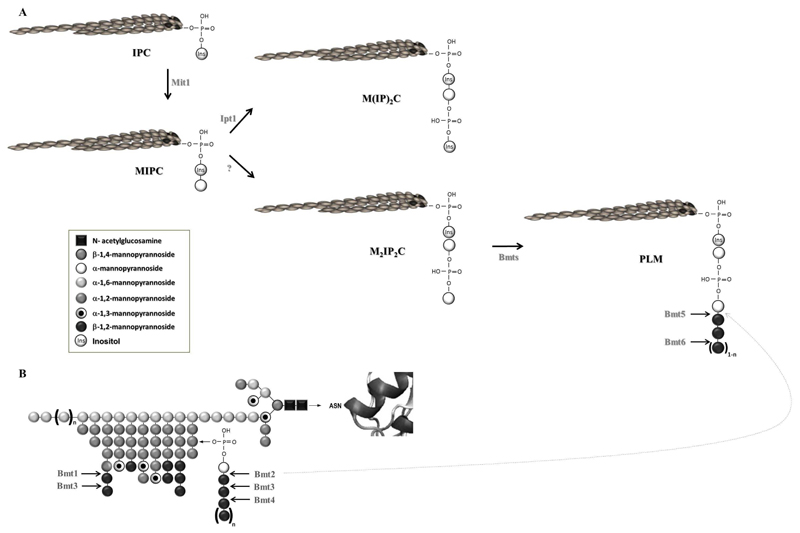
Yeast sphingolipids are essential molecules for proper yeast growth and viability. In addition to be primary structural components of cell membranes together with glycerophospholipids and sterols, they act as second messengers in signal transduction pathways to mediate activities as diverse as growth regulation, responses to stress and cell wall integrity [1,2]. Sphingolipids synthesis leading to the production of complex sphingolipids from the mannosylinositol phosphorylceramide family and the enzymes involved have been well characterized in the baker’s yeast Saccharomyces cerevisiae [2]. The opportunistic yeast pathogen Candida albicans also sequentially synthesizes inositol phosphorylceramide (IPC), mannose inositol phosphorylceramide (MIPC) and mannosyl-diinositol-phosphorylceramide (M(IP)2C), respectively (Figure 1, [3–5]). MIPC and M(IP)2C are secreted into the plasma membrane, whereas non glycosylated IPC is transported to a vacuole.
Figure 1. Biosynthesis of C. albicans glycosphingolipids from the MIPC family and mannosylation pathways of C. albicans proteins.
(A) The C. albicans M(IP)2C and PLM biosynthesis pathways are schematized. (B) The structure of C. albicans N-linked glycans are represented.
The arrows indicate the function of the enzymes.
The dashed arrow represents the redundant activity of Bmt2 in bm5Δ mutant as highlighted in the present study.
Another complex glycosphingolipid named phospholipomannan (PLM) has been identified in C. albicans. Like M(IP)2C, PLM is an end-product of the sphingolipid pathway (Figure 1A, [3,6]). M(IP)2C and PLM synthesis differ after the MIPC step. MIPC from C. albicans can be phosphomannosylated rather than linked to phosphoryl-inositol. Phosphomannosylated MIPC (M2IP2C) is further β-1,2 mannosylated to form PLM. β-1,2 mannosides (β-Mans) addition confers to PLM marked hydrophilic properties, which favor its transfer from the plasma membrane to the cell wall [7,8].
The role of PLM in yeast biology is still unknown but PLM has been considered as a factor that contributes to C. albicans virulence [9]. Lipid and glycan moieties of PLM modulate the signaling pathways linked to the survival of C. albicans in macrophages and favor interaction with soluble galectin-3 to amplify inflammatory response, respectively [9–11]. PLM is all the more considered essential for C. albicans pathogenicity since it was shown that this complex glycosphingolipid is shed in contact with host cells [12]. Location of PLM in the yeast cell wall needs nevertheless to be further investigated.
β-Mans are also present on the other cell wall glycoconjugates: phosphopeptidomannan (PPM) [13,14] and mannoproteins (MPs) [15] (Figure 1B). β-mannosylation of cell wall glycoconjugates involves a family of β-1,2 mannosyltransferases (Bmts) with relative strict substrate and step of mannosylation specificities (Figure 1, [6]). Bmts1-4 add β-mannose on PPM and MPs [16] whereas Bmts5-6 act on PLM [17]. Compensation of the loss of Bmt5 activity in bmt5Δ mutant highlighted the probable action of another Bmt to initiate β-mannosylation on PLM in this mutant. This observation prompted us to study further PLM β-mannosylation and to determine its impact on cell wall homeostasis and C.albicans systemic infection. Location of PLM in the yeast cell wall was also investigated.
2. Materials and methods
2.1. Fungal strains, preculture and growth conditions
All strains used in this study are listed in Table 1. Pre-cultured cells were obtained by growing yeast cells overnight at 37°C in YPD broth (1% yeast extract, 2% Bactopeptone, 2% glucose) with continuous shaking at 150 rpm. The cells were washed twice and suspended in distilled water prior to inoculation in appropriate media.
Table 1. C. albicans strains used in this study.
| Strain | Parental strain | Genotype | Reference |
|---|---|---|---|
| CAI-4+CIp10 | CAI-4 | RPS1::rps1 ::CIp10-URA3 | Courjol et al., submitted |
| bmt1 Δ+CIp10 | bmt1Δ | As bmt1Δ but bmi1Δ::dp1200 bmi1Δ::dp1200. RPS1/rps1 Δ::CIp10-URA3 | Courjol et al., submitted |
| bmt2Δ+CIp10 | bmt2Δ | As bmt2Δ but bmi2Δ::dp1200 bmi2Δ::dp1200. RPS1/rps1 Δ::CIp10-URA3 | This study |
| bmt5Δ+CIp10 | bmt5Δ | As bmt5Δ but bmi5Δ::dp1200 bmi5Δ::dp1200. RPS1/rps1 Δ::CIp10-URA3 | This study |
| bmt1Δ/bmt2Δ | bmt2Δ | As CAI-4 but bmt1Δ::dp1200/bmt1Δ::dp1200.bmt2Δ::dp1200/bmt2Δ::dp1200. RPS1/rps1Δ::CIp10-URA3 | This study |
| bmt2Δ/bmt5Δ | bmt2Δ | As CAI-4 but bmt2Δ::dp1200/bmt2Δ::dp1200,bmt5Δ::dp1200/bmt5Δ::dp1200, RPS1/rps1Δ::CIp10-URA3 | This study |
Yeast cells were inoculated into YPD broth at a density of 106 cells ml-1 and grown at 28°C or 37°C, pH 2.0 or 6.0, with continuous shaking at 150 rpm until the optical density at 600 nm of the culture reached 1.0 (approximately 2x107 cells ml-1).
Spheroplasts were prepared as previously described [18]. Briefly, exponential-phase yeast cells were washed in 0.1M EDTA pH 7.5, suspended in 0.1M EDTA pH 9.0 containing 0.3 M β-mercaptoethanol and incubated at 24°C for 15 min. Cells were pelleted by centrifugation at 2000g and suspended in 1.0M sorbitol, 0.lM EDTA pH 7.5 containing Zymolyase 20T (ICN) (80 units per g wet weight of yeast cells). After 30 min at 37°C, cells were washed in 1.0M sorbitol, 0.lM EDTA pH 7.5. The SDS lysis test was performed to monitor the efficiency of spheroplast formation.
For animal experiments and macrophage stimulation, cells were grown on YPD at 28°C for 16h.
2.2. Gene disruption
All mutants were generated in the CAI-4 background using the mini ura blaster method as previously described [19]. Region of pDDB57 plasmid containing URA3 selection marker is amplified by PCR with primers listed in Table 2 (BMT knockout Fwd/BMT knockout Rev) containing 20 and 80 bases homologous to the plasmid and to the gene of interest, respectively. The PCR fragment was used to transform CAI-4 by the lithium acetate method described previously [20]. Clones formed on YNB (0,67% Yeast Nitrogen Based, 2% glucose, 2% agar) plates were spread onto 5-fluoroorotic acid plates to recycle selection URA3 marker. A second round of transformation was performed to delete the second allele by the same method. Double mutants were generated after two additional rounds of transformation to delete the two alleles of the second gene. Homologous integration of the cassette was checked by Southern blot with probes obtained with primers (probe Fwd/ probe Rev) listed in Table 2. To give the corresponding controls, linearized CIp10 alone was transformed into CAI-4 and the different mutants. A single integration of these plasmids at the RPS1 locus in the same allele was confirmed by Southern blot analysis with probes obtained with primers (probe Fwd/ probe Rev) listed in Table 2.
Table 2. C. albicans primers used in this study.
| Primers | Séquences |
|---|---|
| BMT1 knockout Fwd | CTAAAAAAGGTAAACGAACAATTATATTCCCAAATAATTTCAATCATGTTCATGATCATAAAGGTTCT TATATGATGAAAGTTTTCCCAGTCACGACGTT |
| BMT1 knockout Rev | TCTTTTTCAATTGTCCAAGATGAAATACCATTGGGGATAATTAAATTATAATGTTCACATAAACCTTT ATCTAAATACCATGTGGAATTGTGAGCGGATA |
| BMT2 knockout Fwd | CCTTCAGCCCTGTTTGGAAAAGTGTACAAAATAGGCACAAAGTTAAACTTTACACTACTTGCCCTTTG CTTACTTTTGGCATTTTCCCAGTCACGACGTT |
| BMT2 knockout Rev | TAAACCCATAATCATTCAATTCTAATAATTTCAGGTTTGGATCATCAAATAGTATTGACTTTAATAAT CCTTTGATATGATGTGGAATTGTGAGCGGATA |
| BMT5 knockout Fwd | GCAGTACCGATTTGCCCCAAAGTCAATATTCACATTTGTGTTTCTATGTTTTGTTGCAATAGTTGTCAT AATATCCACATCTTTTCCCAGTCACGACGTT |
| BMT5 knockout Rev | CTGTTTGTTTGCAATAAATATTCAGATAATATCGACTTTAGTATTCCCTTCATGTGTATAATATCAACT GTAGAATCAGATGTGGAATTGTGAGCGGATA |
| probe BMT1 Fwd | ATACAATCATTTAGTCATCAA |
| probe BMT1 Rev | ATACTGGGATAGGGGCGATT |
| probe BMT2 Fwd | GAGAAATGTGGCTGTGGTGA |
| probe BMT2 Rev | TGTTTTTCGGGACCGTATGT |
| probe BMT5 Fwd | GACTCGCCGTTATTGGACAT |
| probe BMT5 Rev | ATTGGCACACCAAAATCCAT |
| probe URA3 Fwd | GCCTCACCAGTAGCACAACGATTA |
| probe URA3 Rev | GCATTCCAACCAGCATCTCTATACC |
2.3. Lectin and monoclonal antibody
Biotinylated concanavalin A (Con A) specific for terminal α-D-mannosyl and α-D-glucosyl residues was purchased from Sigma. Monoclonal antibody (mAb) 5B2 (rat-mouse immunoglobulin M [IgM]) is specific for β-Mans with a mannobiose as minimal epitope [21–23].
2.4. Whole-cell protein extraction, SDS-PAGE separation and western blot analysis
Cells were washed twice with 50 mM Tris buffer (pH 8.0). Cells were suspended in lysis buffer (50 mM Tris [pH 8.0], 10 mM EDTA, protease inhibitors [protease inhibitor cocktail setIV; Calbiochem]), lysed and homogenized by vortexing with acid-washed glass beads (0.5 mm; Sigma) for 30 min at 4°C. Lysates were then incubated 15 min at 100°C to solubilized PPM and chilled 5min on ice. Loading buffer was added to equal amount of each sample in order to have the following final concentrations: 2% SDS, 50mM Tris pH 6.8, 10% glycerol and 2% β-mercaptoethanol. Samples were boiled 5 min and centrifuged 5 min at 10000g. Supernatants were analyzed by SDS–polyacrylamide gel electrophoresis [PAGE; [24]] on 5–20% acrylamide gel slabs. Membranes were probed, as previously described [15], with mAb 5B2 diluted 1:2000 and then incubated with a 1:2000 dilution of alkaline phosphatase-conjugated anti-rat IgM. Enzyme activity was detected with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (Promega) diluted in 0.1 M Tris (pH 9.5), 0.15 M NaCl, 5 mM MgCl2.
For lectin staining, membranes were incubated with biotinylated ConA diluted 1:1000 and then incubated with HRP-labelled Streptavidin (AbCam). Peroxidase activity was detected with diamidobenzidine (SIGMAFAST DAB; Sigma).
2.5. Phylogenetic analysis
The phylogenetic tree of Bmts was calculated using clustalW2 Multiple Sequence Alignment tool (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The resulting tree was generated using clustalW2 Phylogeny tool and the Unweighted Pair Group Method with Arithmetic Mean (UPGMA), distance correction and GAPs exclusion (http://www.ebi.ac.uk/Tools/phylogeny/clustalw2_phylogeny/). The tree is based on the entire amino acid sequence of Bmts.
2.6. PPM extraction and Fluorophore Assisted Carbohydrate Electrophoresis analyses of β-Mans released from PPM
PPM from cells grown in YPD at 28°C or 37°C was extracted by autoclaving in citrate buffer at pH 7.0 [25]. Briefly, cell pellets were suspended in 20 mM citrate buffer and autoclaved at 125°C for 90 min. Suspensions were harvested, and Fehling’s solution was added to the supernatant to precipitate PPM. The PPM was then washed in methanol/acetic acid (8/1) and dried in a Speed Vac concentrator. Sugar concentrations were estimated by the sulfuric-phenol colorimetric method [26]. One μg of PPM was hydrolyzed in 20 mM HCl for 1 h at 100°C to release β-Mans from the phosphomannosides. After neutralization, hydrolysates were then dried and tagged with 0.15 M 8-amino-naphthalene-1,3,6-trisulfonate (ANTS) and 1 M sodium cyanoborohydride for 16 h at 37°C [27]. The ANTS-labelled oligomannosides were dried and resuspended in glycerol/water (1/4) prior to separation on 25-30% (w/v) acrylamide gels. Oligomannosides purified from PPM phosphomannosides and separated by gel filtration chromatography, were also tagged with ANTS and used as carbohydrate standards. Gels were visualized with a Gel Doc 2000 image analysis apparatus from Biorad equipped with a 365 nm UV-transilluminator.
2.7. Indirect immunofluorescence assays
Yeast cells were fixed with formalin and coated on 10 wells slides (Thermoscientific). Wells were washed with PBS-0.5%BSA, blocked with PBS containing 5% BSA and incubated 1h at 37°C with monoclonal antibody or biotinylated ConA, all diluted 1:200 in PBS. Wells were washed with PBS-0.5%BSA and incubated with a 1:100 dilution of FITC-conjugated anti-rat IgM or FITC-conjugated streptavidin, all containing 0.002% of Evans blue counterstain. After wells were washed, slides were mounted and visualized by fluorescent microscopy.
2.8. Phenotypic characterization of C. albicans bmtsΔ mutants
To determine the sensitivity to chemical and antifungal agents, 2μl of serial 1:10 dilutions of an overnight culture were spotted onto YPD agar plates containing 100μg/ml Congo red, 100μg/ml caspofungin, 100μg/ml calcofluor, 1 and 5 mM H2O2, 0.01% SDS, 1M NaCl and 50mM CaCl2. Plates were incubated at 30°C and 37°C for 1 to 3 days.
2.9. Animal experiments
All murine experiments were performed according to protocols approved by the Minister of superior education and of the Research and in accordance with the European legal and institutional guidelines (2010/63/UE) for the care and use of laboratory animals. All animals had free access to food and water throughout the experiments.
Yeast cells were harvested, washed twice with sterile physiological saline solution and suspended in sterile physiological saline solution to produce inocula of 2.105 cfu in 100 μl. For each C. albicans strain tested, six female BALB/c mice (8-10 weeks old – average weight: 18.8+/-0.7g) were injected intravenously with 100 μl of cell suspension into the lateral tail vein. Mice were monitored daily and humanely killed when they showed signs of distress or were unable to freely reach food and water.
The results shown are from three independent experiments. Survival data were analyzed by the Kaplan Meier survival analysis.
P values < 0.05 were considered significant.
All statistical analyses were performed with IBM SPSS Statistics Software.
2.10. Stimulation of macrophages
Cells of the mouse macrophage-like J774 lineage (ECAC 85011428), derived from a tumor of a female BALB/c mouse were used. Adherent cells were cultured at 37°C in an atmosphere containing 5% CO2 in DMEM (Life technologies) supplemented with 10% heat-inactivated fetal calf serum (Valbiotech), 5 mM L-glutamine, 100 μg/ml streptomycin, and 50 μg/ml penicillin (Life Technologies). Before use, the cells were gently scraped off with a rubber policeman and plated onto 24-wells tissue culture dishes. Yeast cells were harvested, washed twice with PBS and resuspended at the desired cell concentration. J774 cells were incubated with C. albicans yeast cells for 4 h at 37°C at a yeast:macrophage ratio of 1:1, 5:1, or 10:1.Cell-free supernatants were collected and stored at −80°C. The concentration of TNF-α was determined by enzyme-linked immunosorbent assay according to the manufacturer's instructions (R&D Systems).
Dose-dependent evaluations were compared by ANOVA or the Student's t test for simple comparisons. P < 0.05 was considered to indicate statistical significance.
3. Results and discussion
3.1. Bmt2 and Bmt5 have redundant activity on PLM
Following the screening of bmtΔ mutants for PLM β-mannosylation, we reported that Bmt5 and Bmt6 add the first and the third β-mannose, respectively, on this glycosphingolipid (Figure 1A, [17]). These two Bmts are specific for PLM as bmt5Δ and bmt6Δ mutants don’t display impairment of PPM and MPs β-mannosylation. Furthermore, no other Bmt replaces Bmt1, Bmt2, Bmt3 and Bmt4 on PPM and MPs in the corresponding bmtΔ single mutants (Figure 1B, [16]). When grown at 37°C, bmt5Δ mutant still contains some β-Mans with high polymerization degrees [17]. As this mutant displays no impairement of sphingolipids biosynthesis, the data suggest that another Bmt shares redundant activity with Bmt5. In the present study, we analyzed if the different Bmts specific for PPM and MPs could replace Bmt5.
C. albicans is a versatile microorganism that is able to survive and grow in different environments displaying distinct pressures. Environmental adaptation has a clear impact on the yeast cell wall and the different glycosylation mechanisms {Hall, 2015 #47}. Like the classical yeast culture medium (YPD, Sabouraud…), 28°C and pH 2.0 have no human physiological relevance but they are relevant culture parameters to analyze β-1,2 mannosylation. By contrast with PLM β-mannosylation that has almost a constitutive expression, PPM and MPs β-mannosylation is regulated by temperature and pH [28,29]. PPM and MPs β-mannosylation is optimal at 28°C, whereas it is inhibited at pH2.0. In the present study, we analyzed PLM β-mannosylation of bmt5Δ mutant and its parental strain in growth conditions favoring (28°C, pH6.0) or inhibiting (37°C, pH2.0) β-mannosylation of PPM and MPs.
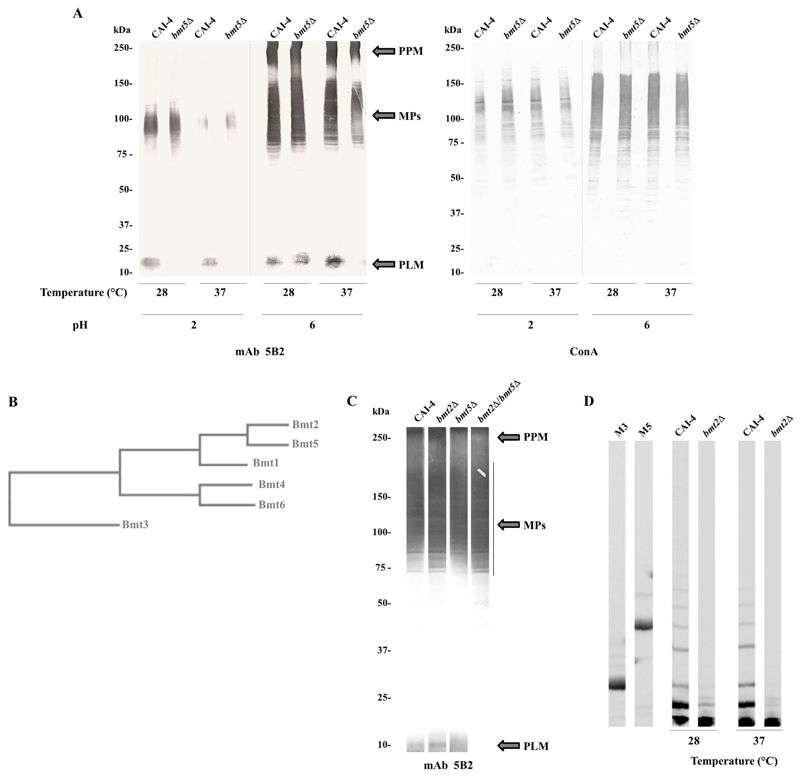
Western blot analysis of whole cell extracts with anti-β-Mans mAb 5B2 confirmed that only PLM β-1,2 mannosylation is affected in bmt5Δ mutant compared to the parental strain in a same growth condition (Figure 2A). As expected, reactivity of mAb 5B2 to bmt5Δ PLM was dramatically reduced when cells where grown at 37°C and at pH6.0. However, when bmt5Δ mutant and its parental strain were grown at 28°C and at pH 6.0, their PLM were almost equally β-1,2 mannosylated. This observation suggests that overlapping Bmt5 activity is rather regulated than partial. This was confirmed when cells were grown at acidic pH (pH2.0). In this condition, whatever the growth temperature, no β-Mans was added on bmt5Δ PLM compared to parental PLM (Figure 2A). ConA staining of the different extracts shows that α-mannosylation of the parental and mutant strains was comparable in the different growth conditions tested. Thus, the loss of Bmt5 activity in bmt5Δ mutant is compensated by the activity of another Bmt whose expression is enhanced at 28°C and repressed at pH2.
Figure 2. Analysis of β-1,2 mannosylation of C. albicans glycoconjugates and cluster analysis of Bmts.
(A) Western blots of whole-cell extracts from the parental strain CAI-4 and bmt5Δ mutant grown at 28°C or 37°C, pH 2.0 or 6.0, were stained with anti-β-Mans mAb 5B2 and lectin ConA, specific for terminal α-mannoside.
PPM, MPs and PLM are indicated.
(B) Phylogenetic tree of C. albicans PPM and PLM specific Bmts. This tree, based on the entire enzyme sequences, illustrates the relatedness of Bmts and reveals a cluster of Bmts according to the β-mannosylation step and common acceptor.
(C) Western blot of whole-cell extracts from the parental strain CAI-4 and bmt2Δ, bmt5Δ and bmt2Δ/bmt5Δ mutants grown at 28°C, pH 6.0 was stained with anti-β-Mans mAb 5B2. PPM, MPs and PLM are indicated.
(D) Analysis of β-Mans released from purified PPM of the parental strain CAI-4 and bmt5Δ mutant. Oligomannosides released by acid hydrolysis from purified PPMs were analyzed by FACE. Two carbohydrate standards were used to evaluate the monomer number in the oligomannoside chains: M3, β-mannotriose; M5, β-mannopentaose.
Cluster analysis of the six PPM and PLM specific Bmts classify these enzymes into groups depending on their specificities (Figure 2B): their acceptor and their involvement in the β-mannosylation steps (Figure 1). From the enzymes adding the first β-mannose, Bmt2 and Bmt5 display the strongest similarity with 49.09 sequence identity. These two enzymes have the same acceptor, a phosphomannose, but on different glycoconjugates: PPM and MPs for Bmt2 and PLM for Bmt5. As Bmt2 activity is optimal at 28°C and at pH6 [28,29], we hypothesized that it could replace Bmt5 in bmt5Δ mutant.
We generated a strain lacking the two alleles of the genes coding for Bmt2 and Bmt5. Western blot analysis with mAb5B2 was performed on whole cell extracts of the resulting strain (bmt2Δ/bmt5Δ mutant), bmt2Δ and bmt5Δ mutants and their parental strain, CAI-4. These strains were grown at 28°C and at pH 6.0 to favor Bmt5 overlapping activity in strains deprived of this enzyme. As expected, PLM of bmt2Δ mutant was β-mannosylated like the PLM of the parental strain (Figure 2C). PLM of bmt5Δ mutant displays a weak β-mannosylation defect, whereas no β-Mans could be detected on PLM of bmt2Δ/bmt5Δ mutant. This data reveals that Bmt2 has strict specificity of acceptor but not of glycoconjugate. As already suggested [16], analysis of PPM phosphomannosides from bmt2Δ mutant and its parental strain by fluorescent assisted carbohydrate electrophoresis shows that PPM phosphomannose of bmt2Δ mutant is not β-mannosylated, whatever the growth conditions (Figure 2D). Loss of Bmt2 activity in bmt2Δ mutant is not compensated by Bmt5 or any other enzyme. Spatial conformation of the glycoconjugate and not only the acceptor certainly determines Bmt5 specificity. It is assumed that Bmt5 can add both the first and the second β-Mans to PLM glycan [17] such as Mnt1 and Mnt2 that are able to add both the first and the second α-1,2 Mans to mannoprotein [30]. Bmt2 has not such activity on PPM but we cannot rule out that it might add the second β-mannose on PLM in bmt5Δmutant. Bmt3, which adds the second β-mannose on PPM and MPs (Figure 1B) could also be involved. Interestingly, Bmt6 is the only Bmt that can add the third β-mannose and the following ones on PLM [17]. This suggests that initiation of PLM β-mannosylation might be essential for the yeast.
3.2. Location of PLM in the cell wall
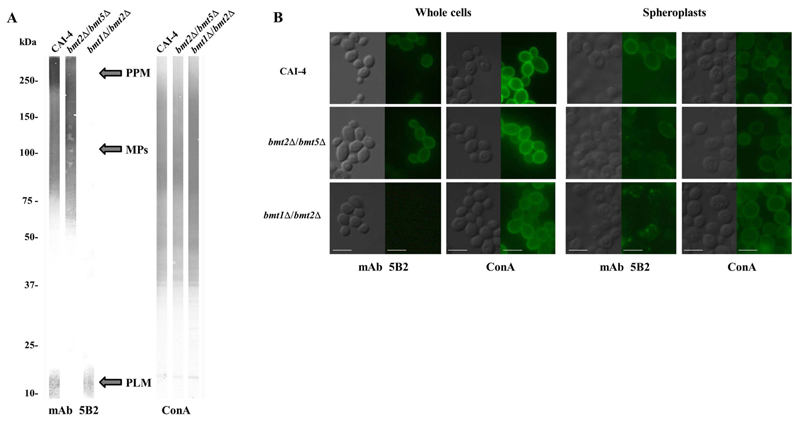
Anti-β-Mans antibodies have been helpful tools to characterize β-1,2 mannosylated glycoconjugates [15,31], to identify the different Bmts activities and specificities [16,17] and to localize β-Mans at the cell wall surface [7,32]. By contrast to SDS-PAGE, native conditions prevent differentiation between the different glycoconjugates that carry β-Mans. The relative strict specificities of Bmts give new prospects to target specific β-mannosylated glycoconjugates in the cell wall. To analyze the location of PLM in C. albicans cell wall, we generated a strain, which does not initiate β-Mans synthesis on PPM and MPs. The two alleles of the genes coding for Bmt1 and Bmt2, which add the first β-mannose on the PPM/MPs α1,2 mannosides and phosphomannosides, respectively (Figure 1B), were deleted. Western blot analysis of whole cell extracts with mAb5B2 confirmed that bmt1Δ/bmt2Δ mutant has β-Mans epitopes only on PLM whereas bmt2Δ/bmt5Δ mutant carry them only on PPM and MPs (Figure 3A). As expected, the parental strain expressed β-Mans epitopes on PPM, MPs and PLM. All the strains displayed the same amount of α-mannosides.
Figure 3. Analysis of β-1,2 oligomannoside epitopes on C. albicans glycoconjugates and at the surface of yeast cells.
(A) Western blots of whole-cell extracts from the parental strain CAI-4, bmt2Δ/bmt5Δ mutant and bmt1Δ/bmt2Δ mutant grown at 28°C, pH 6.0 were stained with anti-β-Mans mAb 5B2 and lectin ConA, specific for terminal α-mannoside.
PPM, MPs and PLM are indicated.
(B) Indirect immunofluorescence assays were performed on intact yeast cells and spheroplasts from bmt2Δ/bmt5Δ and bmt1Δ/bmt2Δ mutants and their parental strain, CAI-4 using anti-β-Mans mAb 5B2 and lectin ConA. Evan’s blue was used as counterstain to decrease the nonspecific background fluorescence. For each strain, a micrograph of phase contrast and a fluorescence micrograph with the mentioned antibody or lectin are shown.
Scale bars = 5 μm
As previously reported [28], immunofluorescence assay of parental strain with mAb 5B2 showed a great variability in the surface expression of β-Mans from cell to cell (Figure 3B). Cells were either highly labelled (around 70%) or unlabeled. A similar distribution of β-Mans at cell surface of bmt2Δ/bmt5Δ mutant was observed (Figure 3B). All cells of parental strain and bmt2Δ/bmt5Δ mutant were stained by the lectin ConA, showing that they all express α-mannosides on their surface. Immunofluorescence assay with ConA revealed that all cells of bmt1Δ/bmt2Δ mutant were also decorated with α-Mans but none of them expressed β-Mans on their surface (Figure 3B). Same results were obtained when cells were grown in different conditions (data not shown), revealing that PLM, whose β-Mans are not detected at the cell surface, is certainly not a cell surface molecule. To detect inner wall molecules, we generated spheroplasts prior to the immunofluorescence assay. Treatment of cells with zymolyase, a β-1,3 glucanase, was monitored to obtain yeast with inner cell wall layers. Immunofluorescence assay with ConA confirmed that the three strains have remaining cell wall as they all have α-Mans uniformly exposed, even if there is a weaker amount of α-Mans expressed at the spheroplasts surface than at the whole cells surface (Figure 3B). Parental strain and bmt2Δ/bmt5Δ mutant spheroplasts expressed weak amount of β-Mans epitopes with patches on the surface. The bmt1Δ/bmt2Δ mutant only displayed a patchy distribution of β-Mans epitopes, confirming the inner location of concentrated PLM in the cell wall and probably also in the plasma membrane like MIPC and M(IP)2C [33]. Location of β-Mans in the plasma membrane, surely through PLM, of C. albicans has been reported some decades ago [18]. Contribution of PLM to surface expression of β-Mans has been hypothesized [7], which would enable PLM to directly interact with host receptors and modulate the host response [34]. Our present study together with previous ones [28,29] clearly demonstrate that this molecule is certainly located in the inner layer of the cell wall. Location of PLM within the deeper cell wall layers does not prevent it from interacting with host cells. PLM was indeed reported to be shed in contact with host cells through a mechanism not yet explored/elucidated [12]. Furthermore, we reported in a recent study [10] that phagocytosis favors yeast cells to activate a macrophage pro-inflammatory response. PLM could be released or exposed at the cell surface during the phagocytosis process and then could modulate the macrophage inflammatory response.
3.3. Inhibition of PLM β-1,2 mannosylation enhances resistance to inhibitors of β-glucan synthesis
Function of PLM during Candida-Host interaction has been well documented with its lipid core being involved in macrophage apoptosis [11] and β-Mans being important for the inflammatory response [9,10]. The function of PLM in Candida biology is unknown even if it certainly shares functions of other complex glycosphingolipid such as MIPC and M(IP)2C. The different bmtΔ mutants were tested for their susceptibility to different chemical agents inducing osmotic stress (NaCl), oxidative stress (H2O2), calcium homeostasis (CaCl2) and cell integrity (SDS) impairments and disruption of cell wall homeostasis (Calcofluor white, Congo red and Caspofungin). None of the strains were sensitive or resistant to NaCl, H2O2, SDS and CaCl2, suggesting that β-1,2 mannosylation of PLM is not required for osmotic, oxidative and calcium homeostasis of C. albicans (data not shown). Interestingly, by contrast with IPC and MIPC, M(IP)2C also does not affect calcium homeostasis of Candida cells [35].
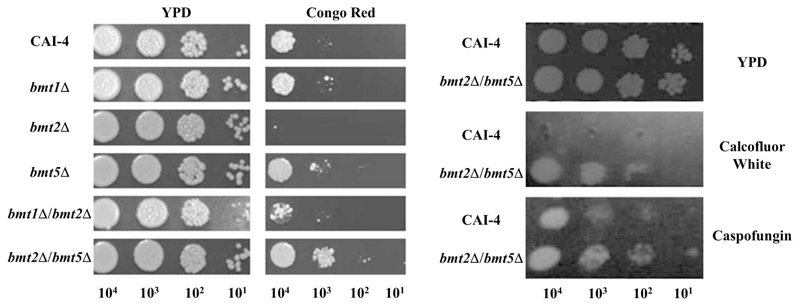
Surprisingly, two mutant strains, bmt5Δ and bmt2Δ/bmt5Δ, display different level of resistance to Congo red, which is an anionic dye believed to inhibit chitin and β-glucan synthases after binding to nascent β-1,3 glucan or chitin fibers (Figure 4A). This resistance seems to be linked to β-mannosylation of PLM as bmt1Δ and bmt1Δ/bmt2Δ are as sensitive as the parental strain and bmt2Δ is more sensitive than the parental strain. Furthermore, bmt2Δ/bmt5Δ mutant is more resistant than bmt5Δ mutant, surely because Bmt2 replaces Bmt5 in that mutant. Involvement of PLM β-mannosylation in the resistance to chitin and β-glucan synthesis inhibitor was surprising but further confirmed after incubation of bmt2Δ/bmt5Δ and its parental strain with Calcofluor White, an anionic dye with the same property than Congo red, and an antifungal drug, caspofungin, which is an echinocandin that inhibits specifically the β-1,3 glucan synthase FKS1. The double mutant was more resistant to Calcofluor White and caspofungin (Figure 4B). We can hypothesize that non β-1,2 mannosylated PLM accumulates in the plasma membrane and disturbs the sphingolipids homeostasis of the plasma membrane. As a result, the interaction between caspofungin and Fks1 could be blocked, conferring resistance to this echinocandin. Modulation of echinocandin-Fks by sphingolipids has been observed in the pathogenic yeast C. glabrata [36]. Resistance to Congo red and Calcofluor White certainly involves regulation of β-glucan and chitin synthases activity, which could be partly dependent of plasma membrane PLM. Interestingly, non-β-mannosylated PLM does not impair polyene-ergosterol complex as no resistance of bmt2Δ/bmt5Δ to amphotericin B, an antifungal polyene, was observed (data not shown). A better knowledge of the mechanism of action of these two anionic dyes would enable us to draw conclusions. We believe nevertheless that resistance to these compounds is linked to accumulation of non-β-1,2 mannosylated PLM (M2IP2C) in the plasma membrane.
Figure 4. Sensitivity of C. albicans bmtΔ mutants and parental strain to inhibitors of β-glucans synthesis.
Serial 10-fold dilution of CAI-4 (parental strain) and bmt1Δ, bmt2Δ, bmt5Δ, bmt1Δ/bmt2Δ and bmt2Δ/bmt5Δ mutants were inoculated on YPD agar medium or YPD agar medium containing 100μg/mL Congo Red (A) or 100μg/ml of either Calcofluor White or Caspofongin (B).
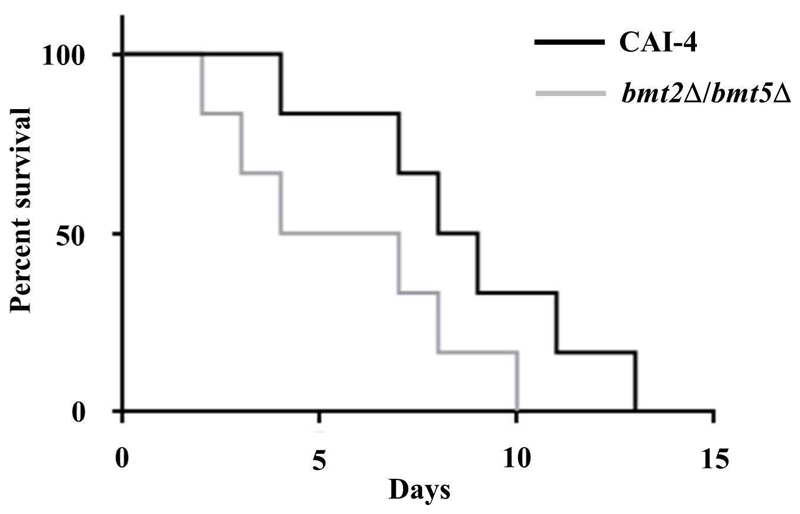
3.4. Inhibition of PLM β-1,2 mannosylation has no impact on C. albicans systemic infection
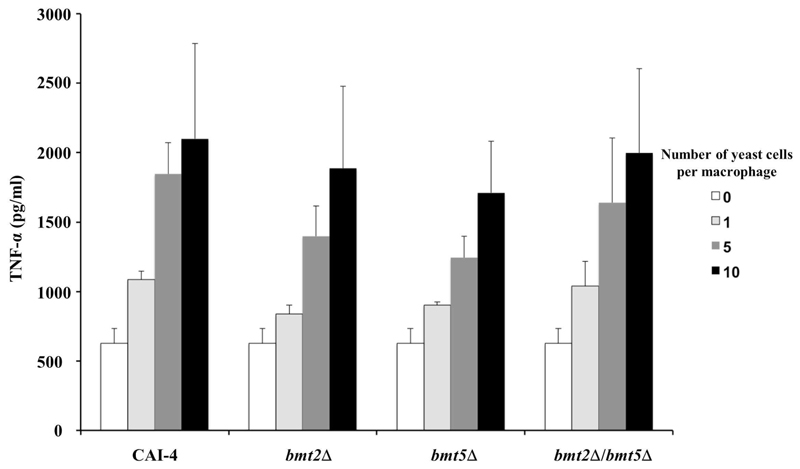
The mutant lacking b-1,2 mannosylated PLM and its parental strain were tested in a murine model of disseminated candidiasis. No significant difference was observed between the survival rates of the two strains (Figure 5). Mice infected with bmt2Δ/bmt5Δ mutant even seemed to die slightly faster than mice receiving the parental strain. This result suggests that β-mannosylation of PLM is not essential for C. albicans systemic infection. Knowing the probable role of PLM, through its lipid and glycan moieties, for regulating the host immune response to favor survival of the fungus, we can hypothesize that its lipid part is involved during systemic infection whereas its β-Mans are certainly more important during the invasion processes. Reduce virulence of mit1Δ mutant, which lacks the MIPC synthase (Figure 1A), in a same model strengthens our hypothesis by highlighting the role of the sphingolipids (MIPC, M(IP)2C and M2IP2C) in C. albicans systemic infection. We were puzzled to show that PLM β-1,2 mannosylation is dispensable for immune recognition of C. albicans by macrophages. Cells of the murine macrophage lineage responded with equal amount of TNF-α when stimulated with bmt2Δ, bmt5Δ and bmt2Δ/bmt5Δ mutants and their reference strain (Figure 6). C. albicans expresses different PAMPs able to induce inflammatory signals through recognition by distinct PRRs. These signals certainly compensate loss of PLM stimulus through its β-Mans. Furthermore, bmt2Δ/bmt5Δ mutant expresses β-Mans on cell surface molecules (MPs and PPM) (Fig.3B), which might also stimulate macrophages. The role of the PLM precursor (M2IP2C) needs to be analyzed. It is then crucial to characterize the enzyme(s) responsible for the phosphomannosylation of MIPC to generate cells expressing only MIPC and M(IP)2C. This strain will be definitively a useful tool to better characterize the role of PLM in C. albicans biology and virulence.
Figure 5. Virulence assay.
Cumulative mortality of mice injected with 2.105 yeast cells from the parental strain CAI-4 and bmt2Δ/bmt5Δ mutant.
The results shown are from three independent experiments.
Figure 6. Cytokine production by J774 macrophage cells in response to live C. albicans bmtΔ mutants.
J774 cells were incubated with either CAI-4 (parental strain) or bmt2Δ, bmt5Δ or bmt2Δ/bmt5Δ mutants at a yeast:macrophage ratio of 1:1, 5:1, or 10:1. Production of TNF-α was measured in cell-free supernatants after 4 h at 37°C. Values shown are the means ± standard error of three independent experiments performed in triplicate.
4. Conclusion
Different studies have highlighted the role of glycolipids, including sphingolipids, in the development of fungi and fungal-induced pathogenesis [1,37–40]. C. albicans, a prominent opportunistic fungal pathogen in developed countries, uses the biosynthetic pathway of complex sphingolipids from the MIPC family to express a glycosphingolipid named PLM with an unusual linear β-Mans [8]. PLM β-mannosylation certainly favors its transfer from the plasma membrane to the lower layer of the cell wall and surely has an impact on sphingolipids distribution and location, promoting chitin and β-glucan homeostasis. This glycosylation step is important for C. albicans as Bmt redundant activity was highlighted for the initiation of β-Mans synthesis. If PLM β-1,2 mannosylation is not essential for the yeast systemic infection, we believe that it is involved in other steps of C. albicans infection. It could account, at least in part, for regulating the host immune response to favor survival of the fungus [9]. The knowledge of the PLM biosynthesis pathway will help to further identify selective inhibitors, which may open novel therapeutic perspectives able to prevent invasive candidiasis in high-risk patients [41].
Acknowledgements
This work was supported by the “Agence Nationale de la Recherche” [ANR-09-MIE-031-01]; the European project “AllFun” from the 7thFramework programme-Health [260338] and the “College Doctoral Lille Nord de France” [Ouverture Internationale des Etudes et de la Formation Doctorale en Région Nord-Pas de Calais].
We gratefully acknowledge Prof. A. P. Mitchell (Carnegie Mellon University, Pittsburgh, USA), Prof. A. J. P. Brown (Department of Molecular and Cell Biology, Aberdeen, UK) and Prof. R. Robert (GEIHP, Univesrité d’Angers, France) for providing pDDB57 and CIp10 plasmids and mAb 5B2, respectively.
In memorandum to Pierre-André Trinel who discovered and characterized phospholipomannan from Candida albicans
References
- 1.Dickson RC. Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast. Journal of lipid research. 2008;49:909–921. doi: 10.1194/jlr.R800003-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dickson RC, Sumanasekera C, Lester RL. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Progress in lipid research. 2006;45:447–465. doi: 10.1016/j.plipres.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Mille C, Janbon G, Delplace F, Ibata-Ombetta S, Gaillardin C, Strecker G, Jouault T, Trinel PA, Poulain D. Inactivation of CaMIT1 inhibits Candida albicans phospholipomannan beta-mannosylation, reduces virulence, and alters cell wall protein beta-mannosylation. The Journal of biological chemistry. 2004;279:47952–47960. doi: 10.1074/jbc.M405534200. [DOI] [PubMed] [Google Scholar]
- 4.Prasad T, Saini P, Gaur NA, Vishwakarma RA, Khan LA, Haq QM, Prasad R. Functional analysis of CaIPT1, a sphingolipid biosynthetic gene involved in multidrug resistance and morphogenesis of Candida albicans. Antimicrobial agents and chemotherapy. 2005;49:3442–3452. doi: 10.1128/AAC.49.8.3442-3452.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong W, Jeffries MW, Georgopapadakou NH. Inhibition of inositol phosphorylceramide synthase by aureobasidin A in Candida and Aspergillus species. Antimicrobial agents and chemotherapy. 2000;44:651–653. doi: 10.1128/aac.44.3.651-653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fabre E, Hurtaux T, Fradin C. Mannosylation of fungal glycoconjugates in the Golgi apparatus. Current opinion in microbiology. 2014;20:103–110. doi: 10.1016/j.mib.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Poulain D, Slomianny C, Jouault T, Gomez JM, Trinel PA. Contribution of phospholipomannan to the surface expression of beta-1,2-oligomannosides in Candida albicans and its presence in cell wall extracts. Infection and immunity. 2002;70:4323–4328. doi: 10.1128/IAI.70.8.4323-4328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trinel PA, Maes E, Zanetta JP, Delplace F, Coddeville B, Jouault T, Strecker G, Poulain D. Candida albicans phospholipomannan, a new member of the fungal mannose inositol phosphoceramide family. The Journal of biological chemistry. 2002;277:37260–37271. doi: 10.1074/jbc.M202295200. [DOI] [PubMed] [Google Scholar]
- 9.Fradin C, Bernardes ES, Jouault T. Candida albicans phospholipomannan: a sweet spot for controlling host response/inflammation. Seminars in immunopathology. 2015;37:123–130. doi: 10.1007/s00281-014-0461-5. [DOI] [PubMed] [Google Scholar]
- 10.Devillers A, Courjol F, Fradin C, Coste A, Poulain D, Pipy B, Bernardes ES, Jouault T. Deficient beta-mannosylation of Candida albicans phospholipomannan affects the proinflammatory response in macrophages. PloS one. 2013;8:e84771. doi: 10.1371/journal.pone.0084771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibata-Ombetta S, Idziorek T, Trinel PA, Poulain D, Jouault T. Candida albicans phospholipomannan promotes survival of phagocytosed yeasts through modulation of bad phosphorylation and macrophage apoptosis. The Journal of biological chemistry. 2003;278:13086–13093. doi: 10.1074/jbc.M210680200. [DOI] [PubMed] [Google Scholar]
- 12.Jouault T, Fradin C, Trinel PA, Bernigaud A, Poulain D. Early signal transduction induced by Candida albicans in macrophages through shedding of a glycolipid. The Journal of infectious diseases. 1998;178:792–802. doi: 10.1086/515361. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi H, Shibata N, Suzuki S. Evidence for oligomannosyl residues containing both beta-1,2 and alpha-1,2 linkages as a serotype A-specific epitope(s) in mannans of Candida albicans. Infection and immunity. 1992;60:2106–2109. doi: 10.1128/iai.60.5.2106-2109.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibata N, Ichikawa T, Tojo M, Takahashi M, Ito N, Okubo Y, Suzuki S. Immunochemical study on the mannans of Candida albicans NIH A-207, NIH B-792, and J-1012 strains prepared by fractional precipitation with cetyltrimethylammonium bromide. Archives of biochemistry and biophysics. 1985;243:338–348. doi: 10.1016/0003-9861(85)90511-9. [DOI] [PubMed] [Google Scholar]
- 15.Fradin C, Slomianny MC, Mille C, Masset A, Robert R, Sendid B, Ernst JF, Michalski JC, Poulain D. Beta-1,2 oligomannose adhesin epitopes are widely distributed over the different families of Candida albicans cell wall mannoproteins and are associated through both N- and O-glycosylation processes. Infection and immunity. 2008;76:4509–4517. doi: 10.1128/IAI.00368-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mille C, Bobrowicz P, Trinel PA, Li H, Maes E, Guerardel Y, Fradin C, Martinez-Esparza M, Davidson RC, Janbon G, Poulain D, et al. Identification of a new family of genes involved in beta-1,2-mannosylation of glycans in Pichia pastoris and Candida albicans. The Journal of biological chemistry. 2008;283:9724–9736. doi: 10.1074/jbc.M708825200. [DOI] [PubMed] [Google Scholar]
- 17.Mille C, Fradin C, Delplace F, Trinel PA, Masset A, Francois N, Coddeville B, Bobrowicz P, Jouault T, Guerardel Y, Wildt S, et al. Members 5 and 6 of the Candida albicans BMT family encode enzymes acting specifically on beta-mannosylation of the phospholipomannan cell-wall glycosphingolipid. Glycobiology. 2012;22:1332–1342. doi: 10.1093/glycob/cws097. [DOI] [PubMed] [Google Scholar]
- 18.Li RK, Cutler JE. A cell surface/plasma membrane antigen of Candida albicans. Journal of general microbiology. 1991;137:455–464. doi: 10.1099/00221287-137-3-455. [DOI] [PubMed] [Google Scholar]
- 19.Wilson RB, Davis D, Enloe BM, Mitchell AP. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast. 2000;16:65–70. doi: 10.1002/(SICI)1097-0061(20000115)16:1<65::AID-YEA508>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 20.Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrobial agents and chemotherapy. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elguezabal N, Maza JL, Moragues MD, Ponton J. Monoclonal antibody-mediated inhibition of adhesion of Candida albicans and Candida dubliniensis to human epithelial cells. European journal of oral sciences. 2009;117:474–478. doi: 10.1111/j.1600-0722.2009.00642.x. [DOI] [PubMed] [Google Scholar]
- 22.Elguezabal N, Maza JL, Ponton J. Inhibition of adherence of Candida albicans and Candida dubliniensis to a resin composite restorative dental material by salivary secretory IgA and monoclonal antibodies. Oral diseases. 2004;10:81–86. doi: 10.1046/j.1354-523x.2003.00969.x. [DOI] [PubMed] [Google Scholar]
- 23.Trinel PA, Faille C, Jacquinot PM, Cailliez JC, Poulain D. Mapping of Candida albicans oligomannosidic epitopes by using monoclonal antibodies. Infection and immunity. 1992;60:3845–3851. doi: 10.1128/iai.60.9.3845-3851.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Faille C, Wieruszeski JM, Michalski JC, Poulain D, Strecker G. Complete 1H-and 13C-resonance assignments for D-mannooligosaccharides of the beta-D-(1-->2)-linked series released from the phosphopeptidomannan of Candida albicans VW.32 (serotype A) Carbohydrate research. 1992;236:17–27. doi: 10.1016/0008-6215(92)85004-j. [DOI] [PubMed] [Google Scholar]
- 26.Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith F. A colorimetric method for the determination of sugars. Nature. 1951;168:167. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- 27.Goins TL, Cutler JE. Relative abundance of oligosaccharides in Candida species as determined by fluorophore-assisted carbohydrate electrophoresis. Journal of clinical microbiology. 2000;38:2862–2869. doi: 10.1128/jcm.38.8.2862-2869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trinel PA, Cantelli C, Bernigaud A, Jouault T, Poulain D. Evidence for different mannosylation processes involved in the association of beta-1,2-linked oligomannosidic epitopes in Candida albicans mannan and phospholipomannan. Microbiology. 1996;142(Pt 8):2263–2270. doi: 10.1099/13500872-142-8-2263. [DOI] [PubMed] [Google Scholar]
- 29.Trinel PA, Jouault T, Cutler JE, Poulain D. Beta-1,2-mannosylation of Candida albicans mannoproteins and glycolipids differs with growth temperature and serotype. Infection and immunity. 2002;70:5274–5278. doi: 10.1128/IAI.70.9.5274-5278.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munro CA, Bates S, Buurman ET, Hughes HB, Maccallum DM, Bertram G, Atrih A, Ferguson MA, Bain JM, Brand A, Hamilton S, et al. Mnt1p and Mnt2p of Candida albicans are partially redundant alpha-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. The Journal of biological chemistry. 2005;280:1051–1060. doi: 10.1074/jbc.M411413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trinel PA, Borg-von-Zepelin M, Lepage G, Jouault T, Mackenzie D, Poulain D. Isolation and preliminary characterization of the 14- to 18-kilodalton Candida albicans antigen as a phospholipomannan containing beta-1,2-linked oligomannosides. Infection and immunity. 1993;61:4398–4405. doi: 10.1128/iai.61.10.4398-4405.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li RK, Cutler JE. Chemical definition of an epitope/adhesin molecule on Candida albicans. The Journal of biological chemistry. 1993;268:18293–18299. [PubMed] [Google Scholar]
- 33.Malathi K, Higaki K, Tinkelenberg AH, Balderes DA, Almanzar-Paramio D, Wilcox LJ, Erdeniz N, Redican F, Padamsee M, Liu Y, Khan S, et al. Mutagenesis of the putative sterol-sensing domain of yeast Niemann Pick C-related protein reveals a primordial role in subcellular sphingolipid distribution. The Journal of cell biology. 2004;164:547–556. doi: 10.1083/jcb.200310046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jouault T, Sarazin A, Martinez-Esparza M, Fradin C, Sendid B, Poulain D. Host responses to a versatile commensal: PAMPs and PRRs interplay leading to tolerance or infection by Candida albicans. Cellular microbiology. 2009;11:1007–1015. doi: 10.1111/j.1462-5822.2009.01318.x. [DOI] [PubMed] [Google Scholar]
- 35.Leber A, Fischer P, Schneiter R, Kohlwein SD, Daum G. The yeast mic2 mutant is defective in the formation of mannosyl-diinositolphosphorylceramide. FEBS letters. 1997;411:211–214. doi: 10.1016/s0014-5793(97)00692-3. [DOI] [PubMed] [Google Scholar]
- 36.Healey KR, Katiyar SK, Raj S, Edlind TD. CRS-MIS in Candida glabrata: sphingolipids modulate echinocandin-Fks interaction. Molecular microbiology. 2012;86:303–313. doi: 10.1111/j.1365-2958.2012.08194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerantola V, Guillas I, Roubaty C, Vionnet C, Uldry D, Knudsen J, Conzelmann A. Aureobasidin A arrests growth of yeast cells through both ceramide intoxication and deprivation of essential inositolphosphorylceramides. Molecular microbiology. 2009;71:1523–1537. doi: 10.1111/j.1365-2958.2009.06628.x. [DOI] [PubMed] [Google Scholar]
- 38.Garcia J, Sims KJ, Schwacke JH, Del Poeta M. Biochemical systems analysis of signaling pathways to understand fungal pathogenicity. Methods in molecular biology. 2011;734:173–200. doi: 10.1007/978-1-61779-086-7_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shea JM, Kechichian TB, Luberto C, Del Poeta M. The cryptococcal enzyme inositol phosphosphingolipid-phospholipase C confers resistance to the antifungal effects of macrophages and promotes fungal dissemination to the central nervous system. Infection and immunity. 2006;74:5977–5988. doi: 10.1128/IAI.00768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki E, Tanaka AK, Toledo MS, Levery SB, Straus AH, Takahashi HK. Trypanosomatid and fungal glycolipids and sphingolipids as infectivity factors and potential targets for development of new therapeutic strategies. Biochimica et biophysica acta. 2008;1780:362–369. doi: 10.1016/j.bbagen.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 41.Courjol F, Jouault T, Fradin C. In: Antifungals: From Genomics to Resistance and the Development of Novel Agents. Coste AT, Vandeputte P, editors. Caister Academic Press; Norfolk, UK: 2015. Modulation of the Host Response to Control Invasive Fungal Infections; pp. 237–266. [Google Scholar]