Abstract
Hypoxic adaptation pathways, essential for Candida albicans pathogenesis, are tied to its transition from a commensal to a pathogen. Herein, we identify a WW domaincontaining protein, Ifu5, as a determinant of hypoxic adaptation that also impacts normoxic responses in this fungus. Ifu5 activity supports glycosylation homeostasis via the Cek1 mitogen-activated protein kinase-dependent up-regulation of PMT1, under normoxia. Transcriptome analysis of ifu5Δ/Δ under normoxia shows a significant up-regulation of the hypoxic regulator EFG1 and EFG1-dependent genes. We demonstrate physical interaction between Ifu5 by virtue of its WW domain and Efg1 that represses EFG1 expression under normoxia. This interaction is lost under hypoxic growth conditions, relieving EFG1 repression. Hypoxic adaptation processes such as filamentation and biofilm formation are affected in ifu5Δ/Δ cells revealing the role of Ifu5 in hypoxic signalling and modulating pathogenicity traits of C. albicans under varied oxygen conditions. Additionally, the WW domain of Ifu5 facilitates its role in hypoxic adaptation, revealing the importance of this domain in providing a platform to integrate various cellular processes. These data forge a relationship between Efg1 and Ifu5 that fosters the role of Ifu5 in hypoxic adaptation thus illuminating novel strategies to undermine the growth of C. albicans.
Keywords: Biofilm, cell wall integrity, hypoxia, Efg1, hyphal morphogenesis, WW domain
1. Introduction
The ability of Candida albicans, a commensal pathogen, to adapt to environmental challenges while it colonises and infects the human host, is critical for its survival. One such challenge is to be able to adapt to a variety of oxygen concentrations that this pathogen may experience at various stages of establishing an infection in the mammalian host. The response of C. albicans to hypoxic microenvironment is crucial for various virulence traits, starting from infection (commensal-to-pathogen transition) to biofilm formation (Ernst & Tielker, 2009; Grahl, Shepardson, Chung, & Cramer, 2012). Hypoxic responses are also critical for C. albicans to ensure its occupancy as a commensal in hypoxic niches such as the lower gastrointestinal tract, in its human host (Pierce & Kumamoto, 2012).
A body of studies has identified distinct transcription factors and regulatory circuits that function to facilitate the adaptation of C. albicans exclusively under hypoxia and differ from those required under normoxia. In this fungus during hypoxia, genes involved in ergosterol and fatty acid biosynthesis, iron metabolism, cell wall regulation, glycolysis, and fermentation are up-regulated, whereas respiratory genes are repressed (Chang, Bien, Lee, Espenshade, & Kwon-Chung, 2007; Desai, van Wijlick, Kurtz, Juchimiuk, & Ernst, 2015; Setiadi, Doedt, Cottier, Noffz, & Ernst, 2006). Whereas, the transcription factor Upc2 directs an increase in the ergosterol biosynthesis transcripts (Synnott, Guida, Mulhern-Haughey, Higgins, & Butler, 2010), the regulators Tye7 and Efg1 activate the glycolytic and fermentation genes and unstaurated fatty acid metabolism, respectively during hypoxia (Askew et al., 2009; Bonhomme et al., 2011; Setiadi et al., 2006). The pleiotropic virulence regulator Efg1 is considered central for hypoxic adaptation in this fungus as Efg1 not only contributes to the regulation of half of all hypoxia-responsive genes but also prevents the expression of normoxia associated genes (Setiadi et al., 2006; Stichternoth & Ernst, 2009). Thus, compromising the function of the key hypoxia-regulated genes or regulators results in attenuated virulence in murine models of fungal infection (Askew et al., 2009; Desai et al., 2015).
The link between C. albicans ability to adapt to hypoxia with its pathogenesis is reinforced by the observation that hypoxic conditions enable C. albicans to establish a successful infection by inducing the masking of cell wall β-glucan. As a consequence, C. albicans is able to impede its clearance by evading recognition by the polymorphonuclear leukocytes, pointing to the contribution of hypoxic niches in enhancing virulence (Lopes et al., 2018; Pradhan et al., 2018). Additionally, the ability of this fungal pathogen to undergo the bud-to-hyphae transition and form biofilms in response to hypoxia also permits this pathogen to colonise hypoxic niches in the human host. Both these processes are regulated by Efg1 wherein although Efg1 promotes hypoxic filamentation and biofilm formation at 37°C, it inhibits hypoxic hypha formation during growth on agar specifically at temperatures ≤35°C (Desai et al., 2015; Setiadi et al., 2006). The temperature-dependent inhibition on hypoxic hypha formation requires the concerted action of Efg1, Bcr1, Brg1, and Ace2 transcription factors (Desai et al., 2015). Furthermore, transcriptional induction of genes such as those involved in glucose metabolism, sulphur metabolism, peroxisomal functions, and iron uptake that make possible biofilm formation in hypoxic niches is also dependent on Efg1. Biofilm formation allows C. albicans to generate foci for infections in oxygen-limiting microenvironments and is considered a major cause of persistent infections (Nobile & Mitchell, 2006). Collectively, all these studies emphasise the contribution of Efg1 in regulating hypoxic adaptation in C. albicans. Despite this knowledge, molecular components influencing the hypoxic regulation of Efg1 remain unidentified.
Treating candida infections is not only impeded by the ability of C. albicans to adapt to environmental changes by switching between the planktonic and biofilm mode of growth in host microenvironments but also due to the highly drug recalciterant nature of the biofilms. C. albicans develops resistance to existing azole antifungals due to the up-regulation of drug efflux pumps in both the planktonic as well as biofilm modes of growth. Induced expression of the drug efflux pumps, Cdr1, and Cdr2 in drug resistant C. albicans isolates is often accompanied by the simultaneous induction of a subset of genes (RTA3, IFU5, and HSP12) regulated by the transcription factor Tac1 (Coste, Karababa, Ischer, Bille, & Sanglard, 2004). The significance of these coregulated genes remains largely unexplored in C. albicans, which prompted us to ask if these proteins contribute to the development of azole resistance in C. albicans by affecting the function of Cdr1 and Cdr2. A recent study from our laboratory demonstrates that the 7-TM receptor protein Rta3 does not contribute to azole tolerance at least in a wild type C. albicans strain (SC5314) but is required for biofilm development in vivo (Srivastava et al., 2017). Thus, in order to investigate the relevance of other Tac1 coregulated genes, we characterised Ifu5, a WW domain-containing protein, in this pathogenic fungus.
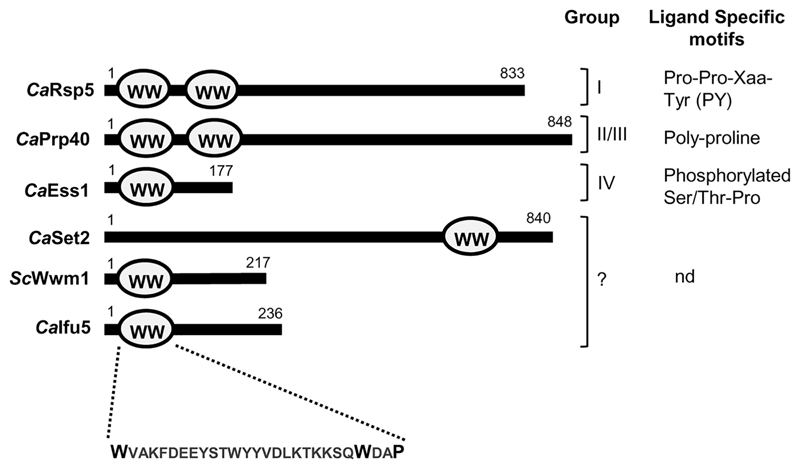
A large number of WW domain-containing proteins have been assigned relevance in Saccharomyces cerevisiae (Hesselberth et al., 2006), whereas they remain uncharacterised in C. albicans. WW domains are small modular domains that interact with peptide ligands that contain a core proline-rich sequence and are implicated in a plethora of cellular processes (Hesselberth et al., 2006). A typical WW domain consists of 35–40 amino acid residues with two highly conserved tryptophan residues separated by 20–23 amino acids followed by a proline residue in the protein. Ifu5 is a homolog of the S. cerevisiae WW domain-containing protein Wwm1 (73% similarity and 66% identity with Wwm1) that is implicated in apoptosis by means of interaction with a metacaspase(Figure S1; Szallies, Kubata, & Duszenko, 2002). Both Wwm1 and Ifu5 contain a single WW domain spanning 26 amino acid residues defined by two conserved tryptophan residues and a single proline residue(Figure 1). The WW domains are categorised into four groups based on the presence of signature residues within the domain and their ligand specificities(Figure 1; Hesselberth et al., 2006). In this study, we describe the role of WW domain-containing protein Ifu5 in mediating normoxic and hypoxic responses in C. albicans and demonstrate an alliance between Ifu5 and Efg1 that facilitates adaptation of this fungus to oxygen-limiting environments. Thus, our study uncovers a novel link between Ifu5 and Efg1 that is pivotal for modulating pathogenicity-related traits in C. albicans.
Figure 1.
Organisation of WW domain in different proteins. WW domain organisation of CaIfu5, ScWwm1 and members of different class of WW domain-containing proteins in Candida albicans. These proteins are classified into four groups based on their ligand specific motifs. nd, ligand specific motif not determined
2. Results
2.1. Tac1-regulated Ifu5 is involved in maintaining cell wall integrity
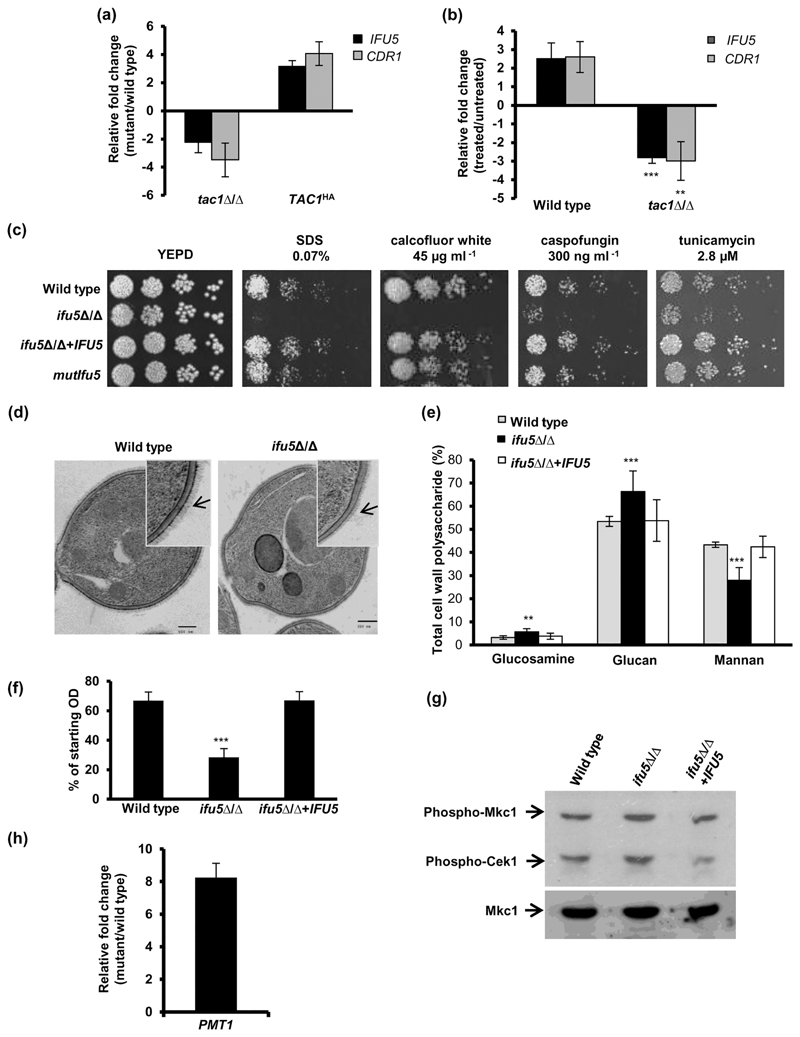
The up-regulation of the drug efflux proteins Cdr1 and Cdr2 by fluphenazine has been well documented (Coste et al., 2004). A genome wide transcriptome study showed that fluphenazine triggers the simultaneous up-regulation of IFU5 with CDR1 and CDR2 via the transcription factor Tac1 (Coste et al., 2004). In order to confirm Tac1-dependent co-ordinate regulation of CDR1 and IFU5, we first analysed their expression in tac1⍰/⍰ cells and in a strain carrying the TAC1HA (hyperactive Tac1) allele. Although the expression of IFU5 and CDR1 in tac1⍰/⍰ was two-fold and four-fold down-regulated, their expression was three-fold and four-fold up-regulated in a TAC1HA strain background, respectively (Figure 2a). Treatment of wild-type cells with fluphenazine resulted in an increase in expression of IFU5 and CDR1 by 2.5- and 2.6-fold, respectively(Figure 2b). On the contrary, tac1⍰/⍰ cells failed to enhance the expression of IFU5 and CDR1, upon fluphenazine treatment(Figure 2b). This set of data suggest that functional Tac1 is requisite for transcriptional regulation of IFU5 in both unstressed and stressed conditions, affirming that IFU5 is a target of Tac1.
Figure 2. Tac1-regulated Ifu5 is required for cell wall integrity.
(a) Expression analysis of IFU5 and CDR1 in tac1Δ/Δ and TAC1HA cells. (b) Expression analysis of IFU5 and CDR1 upon fluphenazine (20·μg·ml-1, 30 min) treatment in indicated strains. (c) Fivefold serial dilution of cell suspensions was spotted onto YEPD plates supplemented with indicated concentrations of drugs and incubated at 30°C for 48 hr. (d) Representative images of transmission electron microscopy analysis for the cell walls of wild type strain SC5314 and ifu5Δ/Δ. Arrow indicates the mannofibril layer. (e) Analysis of carbohydrate content of cell wall. Cell walls of the wild type, ifu5Δ/Δ, and the reconstituted strain were acid hydrolysed, and released monosaccharide was detected by HPAEC-PAD using a CarboPac PA10 analytical column. Results are expressed as a percentage of dried cell wall (μg°mg-1). Shown is the mole percentage average from four or five replicates analysed over two experiments and two-tailed, unpaired t test was used to determine the statistical relevance. **p < .01, ***p < .001. (f) Flocculation assay was done by measuring optical density of the cultures directly after vortex mixing followed by a resting period of 15 min. Values shown represent the ratio of the ODfinal/ODinitial. Values are means ± SD and are derived from three independent cultures. (g) Mitogen-activated protein kinase activation in indicated strains grown in YEPD medium at 37°C was performed. Anti-phospho-p44/p42 MAPK (Thr202/Tyr204) antibody was used to detect dually phosphorylated Mkc1 and Cek1 MAPKs. (h) Expression of PMT1 in ifu5Δ/Δ. Fold change in (a), (b), and (h; mutant/wild type or treated/untreated) is calculated by 2-ΔΔCT, normalised to ACT1 (endogenous control), with untreated strain as calibrator. Values are means ± SD and derived from three independent RNA preparations. Two-tailed, unpaired t test was used to determine the statistical relevance. **p < .01, ***p < .001
To determine the function of Ifu5 in C. albicans, ifu5⍰/⍰ and IFU5 reconstituted strains were constructed using the SAT1 flipper strategy and confirmed by Southern blot analysis(Figure S2). In an attempt to understand the importance of the WW domain in the function of Ifu5, we also mutagenised tryptophan and proline of this domain as these amino acids in other proteins are important for ligand (PPXY or PPLP motif) interactions(Figure 1; Hesselberth et al., 2006). We first examined ifu5⍰/⍰ cells and mutIfu5 for their ability to grow in presence of various antifungal compounds. Both the mutant strains displayed wild-type susceptibility to azole antifungals, suggesting that Ifu5 does not contribute to azole tolerance in the laboratory strain, SC5314(Figure S3). These mutants were then tested for their ability to grow in the presence of agents that target the membrane or the cell wall assembly such as caspofungin, calcofluor white, sodium dodecyl sulfate, and tunicamycin. Interestingly, ifu5⍰/⍰ cells, but not mutIfu5, exhibited a growth defect in the presence of these drugs(Figure 2c). The IFU5-reconstituted strain restored the growth defect of the ifu5⍰/⍰ cells to wild-type levels (Figure 2c).
Previous studies have correlated increased susceptibility to cell wall-damaging agents with alterations in cell wall composition and architecture (Lee et al., 2012). In order to assess for alterations in cell wall integrity, ifu5⍰/⍰ cells were subjected to transmission electron microscopy and cell wall composition analysis. Transmission electron microscopy revealed the presence of compromised fibrillar mannoprotein layer in the ifu5⍰/⍰ cells with no alteration in cell wall thickness compared with wild type(Figure 2d). Cell wall composition analysis showed significant changes in cell wall constituents in the mutant compared with wild type. Glucan constituted 66% of the total cell wall polysaccharide in ifu5⍰/⍰ cells, compared with 53% in the wild type. Consistent with the compromised fibrillar mannoprotein layer, there was a significant reduction in the mannan content in the ifu5⍰/⍰ cells (28% of total cell wall polysaccharide in mutant vs. 43% in wild type). This data also indicated a two-fold increase in the chitin content as determined by measuring the glucosamine content (6% vs. 3% in the wild type) in the ifu5⍰/⍰ cells(Figure 2e). Cell wall damage causes changes in cell surface hydrophobicity thus affecting cell–cell and cell-to-surface adhesion properties (Masuoka & Hazen, 1997). Hence, to investigate the effect of altered cell wall integrity on cell–cell adhesion, we performed flocculation assay with the mutant. The ifu5Δ/Δ cells flocculated extensively and aggregated at the bottom of the tube as indicated by a sharp decline in OD600 (>50% decline) within 15 min after the shaking was stopped in contrast to the wild type and the reconstituted strain(Figure 2f).
The cell wall integrity (CWI) pathway involving the Cek1 and Mkc1 mitogen-activated protein kinases (MAPKs) and the unfolded protein response pathway are requisite for cellular adaptation to cell wall stress (Román, Alonso-Monge, Miranda, & Pla, 2015). Given the strong cell wall phenotypes in ifu5⍰/⍰, we sought to analyse the impact of Ifu5 on the aforesaid adaptation processes and show that ifu5⍰/⍰ cells exhibit constitutive phosphorylation of Mkc1 and Cek1 (1.4- and 1.5-fold induction for Mkc1 and Cek1, respectively in mutant vs. wild type) under basal conditions (Figures 2g and S4). As activated Cek1 signalling pathway is equated to glycostress in C. albicans (van Wijlick, Swidergall, Brandt, & Ernst, 2016), we propose that absence of Ifu5 results in glycostress. The increased sensitivity of ifu5⍰/⍰ to tunicamycin (N-glycosylation inhibitor) and altered cell wall composition is consistent with this notion(Figure 2c,d). Cek1 pathway compensates for glycostress-induced cell wall damage by regulating the activity of O-mannosyltransferases (Pmt proteins), especially PMT1 (van Wijlick et al., 2016). PMT1 expression is repressed in cells with intact N-glycosylation, whereas the repression on PMT1 is relieved in cells with defective N-glycosylation via the Cek1 pathway (Cantero & Ernst, 2011). The ifu5⍰/⍰ mutant exhibited eightfold up-regulation in the transcript levels of PMT1(Figure 2h), concordont with glycostress-induced Cek1 phosphorylation. These findings suggest that Ifu5 impacts cell wall regulation by influencing glycosylation homeostasis in C. albicans.
2.2. EFG1 and biofilm-specific class of genes are differentially regulated in the absence of IFU5
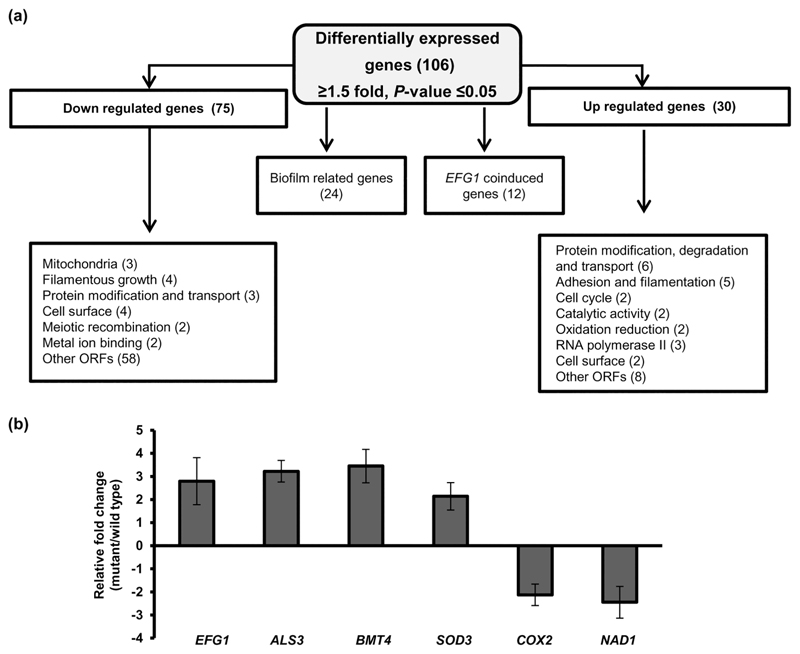
In order to obtain an insight into the genome-wide role of IFU5 in C. albicans, we compared the transcript profiles of wild-type and ifu5⍰/⍰ cells. After filtering, the entire data set resulted in a total of 106 statistically significant differentially regulated genes (p < .05, >1.5-fold up-regulated or down-regulated; Table S5). A total of 75 genes that were down-regulated were enriched for the GO annotation filamentous growth (FGR12, FGR18, NRG2, and HSP21), mitochondria (NAD1, COX2, CRD1, and COQ10), cell surface (PGA13, PGA16, and PGA46), and protein modification and transport(Figure 3a). The up-regulated genes (total of 30) were significantly enriched for the GO annotation protein modification and transport (PEX14, BMT4, ARC18, and RPT2), adhesion and filamentation (EFG1, ALS4, SSY1, and MHP1), oxidation–reduction (SOD3 and orf19.225), and cell surface (PGA39 and PGA22;Figure 2a). PEX14 (3.73-fold) and BMT4 (3.24-fold) were the highest up-regulated genes, whereas orf19.1557 (6.58-fold) was the highest down-regulated gene(Table 1). Out of the 106 differentially regulated genes, 24 genes were described as genes that are induced or repressed during biofilm formation(Figure 3a).
Figure 3. Transcriptional profiling of ifu5Δ/Δ cells.
(a) Flow chart indicating differential expression of genes in ifu5⍰/⍰. Boxes indicate list of functional categories according to GO annotation. Number of differentially expressed genes is indicated in brackets. (b) Quantitative polymerase chain reaction-based transcript analysis of Efg1 and its coregulated genes in ifu5Δ/Δ. Fold change (mutant/wild type) is calculated by 2-ΔΔCT, normalised to ACT1 (endogenous control) with untreated strain as calibrator. Values are means ± SD and are derived from three independent RNA preparations. A two-tailed, unpaired t test was used to determine the statistical relevance: ***p < .001
Table 1. Functional category of C. albicans genes whose transcript levels in the ifu5Δ/Δ strain are 1.5-fold up- and down-regulated (P≤ 0.05).
| Category and System Name | Gene | Function | Fold Change |
|---|---|---|---|
| Protein modification degradation and transport | |||
| orf19.1557 | RKM5 | S-adenosylmethionine-dependent methyltransferase activity | -7.34 |
| orf19.1805 | PEX14 | role in protein import into peroxisome matrix | 3.73 |
| orf19.5612 | BMT4 a,b,c | Beta-mannosyltransferase | 3.24 |
| orf19.5440 | RPT2 b | Putative ATPase of the 19S regulatory particle of the 26S proteasome | 1.96 |
| orf19.7106 | VPS70 | Ortholog(s) have role in protein targeting to vacuole | 1.76 |
| Cell Surface | |||
| orf19.3638 | PGA46 | GPI-anchored cell wall protein involved in cell wall synthesis | -2.62 |
| orf19.6420 | PGA13 b | GPI-anchored cell wall protein involved in cell wall synthesis | -1.88 |
| orf19.848 | PGA16 b | Putative GPI-anchored protein | -1.86 |
| orf19.6302 | PGA39 | GPI-anchored protein | 1.85 |
| orf19.3738 | PGA22 | Putative GPI-anchored protein | 1.54 |
| orf19.4555 | ALS4 b | GPI-anchored adhesion, Role in adhesion | 1.64 |
| orf19.1816 | ALS3 b,c | Cell wall adhesion, Promotes biofilm formation | 1.63 |
| Hypha formation and Virulence | |||
| orf19.5454 | FGR12 b | Filamentous growth | -5.17 |
| orf19.6339 | NRG2 | Transcription factor involved in biofilm | -3.60 |
| orf19.4067 | FGR18 | Filamentous growth | -2.61 |
| orf19.935 | AGA1 b | Spider biofilm induced | -2.47 |
| orf19.610 | EFG1 a,b | Transcription factor involved in biofilm | 2.73 |
| orf19.822 | HSP21 b | Small heat shock protein | -3.01 |
| Iron assimilation | |||
| orf19.1673 | PPT1 | Induced in high iron | -2.24 |
| orf19.6140 | FRE30 c | Protein with similarity to ferric reductases | 3.19 |
| Mitochondrial Function and Oxidation-Reduction Processes | |||
| CaalfMp01 | COX2 a | Subunit II of cytochrome c oxidase | -2.05 |
| orf19.6100 | CRD1 b | Cardiolipin synthase | -1.70 |
| orf19.6662 | COQ10 | Putative coenzyme Q (ubiquinone) binding protein | -1.50 |
| CaalfMp03 | NAD1 c | Subunit 6 of NADH:ubiquinone oxidoreductase (NADH:ubiquinone dehydrogenase) | -2.59 |
| orf19.7111.1 | SOD3 a,b,c | Cytosolic manganese-containing superoxide dismutase | 2.9 |
Genes that were validated by qPCR,
Genes that are annotated as flow model/RPMI/Spider/rat catheter biofilm induced or repressed in CGD.
Genes that are Efg1 regulated.
Interestingly, our data set shows two-fold transcriptional up-regulation of EFG1 in ifu5⍰/⍰ cells(Table 1). Previous transcriptional analysis shows that induced production of Efg1 under normoxia coinduces the expression of a subset of genes such as genes coding for cell wall proteins, superoxide dismutase, Fe3+ reductases, copper transport protein, and an O-mannosyltransferase (Stichternoth & Ernst, 2009). In accord with this, absence of IFU5 also results in a significant increase in expression of these aforesaid normoxia-dependent genes (Table 1). Genes such as ALS3, ALS4, PGA39, and PGA22 (cell wall proteins), BMT4 (beta-mannosyltransferase), FRE30, and SOD3 (redox homeostasis) are coinduced with EFG1 in ifu5⍰/⍰ cells. Furthermore, induced production of Efg1 also represses genes involved in oxidative metabolism (Doedt et al., 2004) in line with the reduced expression of genes such as NAD1, COX2, CRD1, and COQ10 observed in ifu5⍰/⍰ cells(Table 1). The expression of EFG1 and coinduced genes was validated by quantitative polymerase chain reaction (qPCR) analysis (Figure 3b). Coupled together, our data show that deletion of IFU5 results in altered expression of (a) biofilm associated genes and (b) genes that are induced in an Efg1-dependent manner under normoxia.
2.3. Ifu5 affects normoxic and hypoxic expression of EFG1
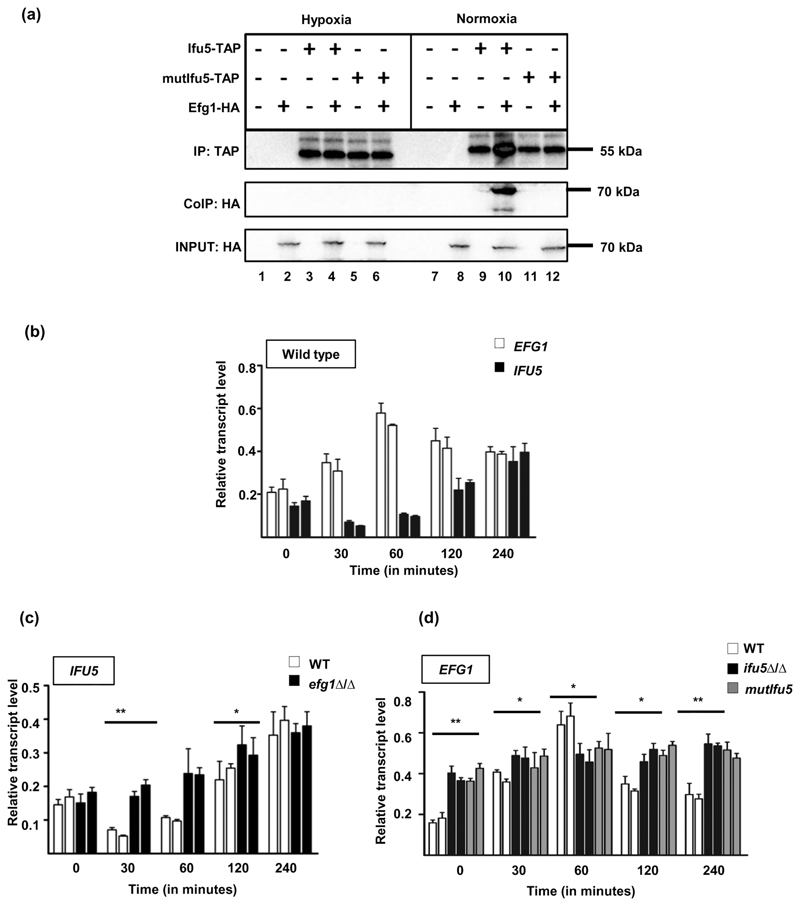
WW domains, present in structural and signalling proteins mediate protein–protein interactions by binding to specific motifs within their partners to coordinate cellular processes such as transcription, differentiation, and ubiquitination (Hesselberth et al., 2006). Considering that Ifu5 contains a WW domain and given the up-regulation of EFG1 in ifu5⍰/⍰ cells(Figure 3b), we predicted that Ifu5 could be negatively influencing the expression of Efg1 under normoxia, which may be due to direct interaction between these two proteins. Hypoxic conditions may relieve this repression resulting in disrupting the interaction between Ifu5 and Efg1; basis for the increased expression of Efg1 under hypoxia (Stichternoth & Ernst, 2009). Therefore, to assess for the interaction, strains producing hemagglutinin (HA)-tagged Efg1, tandem affinity purification (TAP)- tagged Ifu5 and mutIfu5 and a strain expressing both Ifu5-TAP and Efg1-HA were grown in yeast extract peptone dextrose (YEPD) at 30°C for 4 hr, under normoxia and hypoxia followed by cross-linking with formaldehyde to stabilise protein complexes. Thereafter, the total protein extracts were incubated with IgG-coated beads for immunoprecipitation of the Ifu5-TAP protein in the corresponding strain backgrounds. The Ifu5-TAP signal in beads incubated with extract from the strains carrying Ifu5-TAP allele was confirmed by immunoblotting with anti-TAP antibody, under both normoxia and hypoxia(Figure 4a, Anti-TAP panel). Interestingly, immunoblotting of the bound fractions with an anti-HA antibody (Co-IP, Anti-HA panel) allowed to detect Efg1-HA co-immunoprecipitation with Ifu5-TAP exclusively in normoxic conditions(Figure 4a). The Ifu5–Efg1 interaction was not detected with mutIfu5-TAP (Co-IP, Anti-HA panel), pointing to the essentiality of the WW domain in interacting with Efg1(Figure 4a). Our result therefore shows that Ifu5 protein via its WW domain physically associates with Efg1 protein solely during normoxia.
Figure 4. Role of Ifu5 in hypoxic adaptation.
(a) Strains expressing HA-Efg1 (AVL12, lanes 2 and 8), Ifu5-TAP (CLvW997, lanes 3 and 9), both HA-Efg1 and Ifu5-TAP (CLvW998, lanes 4 and 10), mutIfu5-TAP (CLvW990, lanes 5 and 11), HA-Efg1 and mutIfu5-TAP (CLvW989, lanes 6 and 12), and the control strain BWP17 (lanes 1 and 7) were grown in yeast extract peptone dextrose medium at 30°C in either hypoxic or normoxic conditions for 4 hr. The samples were immunoblotted and developed using anti-TAP (IP) or anti-HA (CoIP) antibodies (±). To verify presence of Efg1-HA, 1% of the total protein extracts was blotted and investigated with anti-HA. (b) Relative transcript levels of IFU5 and EFG1 were determined by quantitative polymerase chain reaction (qPCR). The indicated strains grown under normoxic conditions in yeast extract peptone dextrose medium for 4 hr at 30°C, were shifted under hypoxia (0.2% O2) and grown for additional 4 hr at 30°C, and 0.2% O2. RNA was isolated from cells at indicated time points. Time point 0 corresponds to 4-hr growth under normoxic conditions before shifting cells to hypoxic conditions. At each time point, two biological replicates and three technical replicates were assayed by qPCR. The ACT1 transcript was used as a calibrator to normalise IFU5 and EFG1 transcript levels. A two-tailed, unpaired t test was used to determine the statistical relevance: *p < .05, **p < .01. Relative transcript levels of (c) IFU5 transcript in wild type and efg1Δ/Δ and (d) EFG1 transcript in wild type, ifu5Δ/Δ, and mutifu5 was analysed upon shifting normoxia grown cells to hypoxic conditions. At each time point, two biological replicates and three technical replicates were assayed by qPCR. The ACT1 transcript was used as a calibrator to normalise IFU5 and EFG1 transcript levels, respectively. A two-tailed, unpaired t test was used to determine the statistical relevance:: *p < .05; **p < .01
Thereafter, based on the role of Efg1 in hypoxic adaptation in C. albicans, we were prompted to analyse the relevance of Ifu5–Efg1 interaction under hypoxia. First, we show that absence of Ifu5 does not affect growth under hypoxia suggesting that Ifu5 is not a determinant of hypoxic growth in C. albicans (data not shown). Second, we monitored transcript levels of EFG1 and IFU5 during the shift from normoxia to hypoxia in wild-type, efg1⍰/⍰, ifu5⍰/⍰, and mutIfu5 cells. We observed a decrease in the transcript levels of IFU5 after the shift to hypoxia with a simultaneous increase in the EFG1 transcript levels at the 30 and 60-min time points, compared with their transcript levels at the 0 time point in the wild type(Figure 4b). At the later time points of 120 and 240 min, there was an increase in the transcript level of IFU5 with a commensurate decrease in EFG1 transcript level, suggesting hypoxic repression of IFU5 during the early stages of hypoxia(Figure 4b). Additionally, transcript levels of IFU5 in efg1⍰/⍰ cells and of EFG1 in ifu5⍰/⍰ and mutIfu5 increased with time, pointing to the existence of reciprocal negative regulation between IFU5 and EFG1(Figure 4c,d). Thus, the result indicates that one of the functions of Ifu5 may be to effectively negatively regulate Efg1 expression by virtue of its WW domain under normoxia by physically interacting with this hypoxic regulator. Hypoxic growth conditions may result in the loss of this interaction, and negative reciprocal regulation coordinates the expression of IFU5 and EFG1 during various stages of hypoxia. Collectively, we surmise that Ifu5 is tied to regulating Efg1 expression and thus to hypoxic adaptation by means of its WW domain in C. albicans.
2.4. Ifu5 regulates filamentation and affects biofilm formation under hypoxia
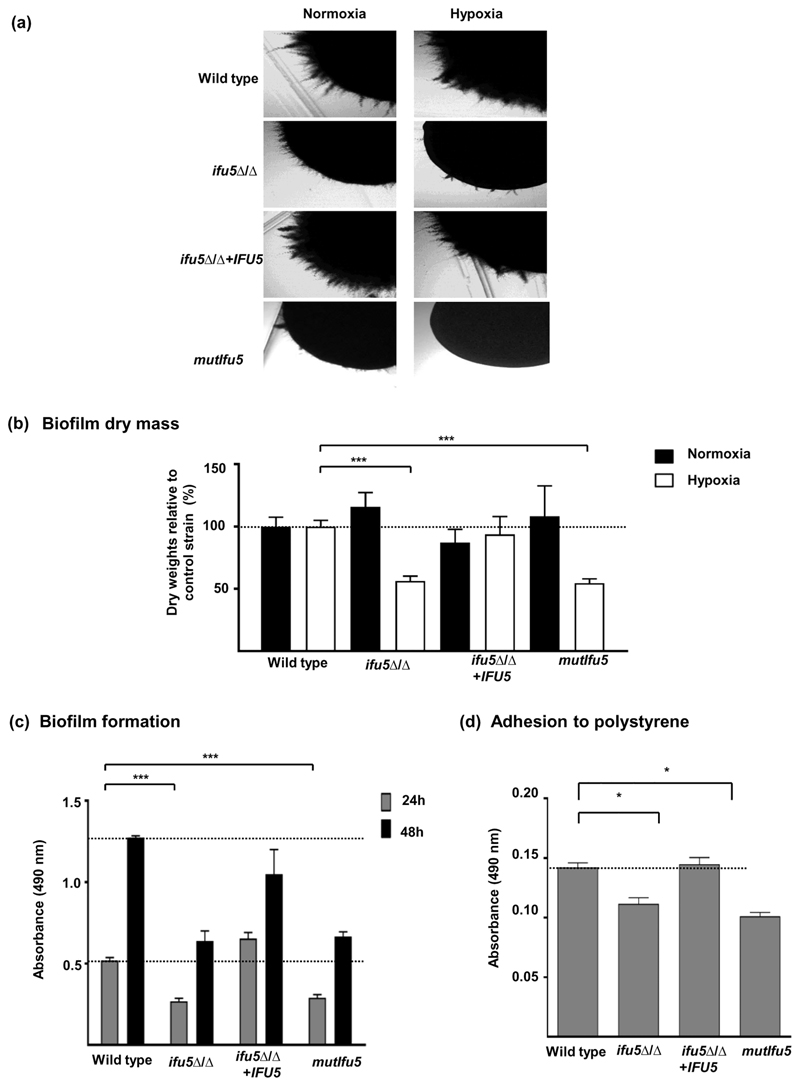
Hyphal morphogenesis is one of the process that is required by C. albicans to adapt to hypoxia, especially during its growth on agar surface (Setiadi et al., 2006). Therefore, given the role of Ifu5 in hypoxic adaptation, we were prompted to analyse the influence of Ifu5 on hyphal morphogenesis. Absence of IFU5 did not affect hyphal induction in liquid hyphal-inducing conditions such as Spider and serum-containing media. On the contrary, both ifu5⍰/⍰ and mutIfu5 exhibited filamentation (surface growth) defects on all the examined media in both normoxia and hypoxia, with the effect being more pronounced under hypoxia(Figure 5a).
Figure 5. Ifu5 influences filamentation and biofilm formation.
(a) Indicated strains were grown under hypoxia (0.2% O2) and normoxia (21% O2) on the surface of spider agar at 37°C for 3 days and photographed. (b) Biofilm mass (dry weight) after growth at 37°C under normoxia (21% O2, 6% CO2) and hypoxia (0.2% O2, 6% CO2) was estimated. Polystyrene wells were inoculated with 105 cells·ml-1 of the indicated strains and supplemented with Roswell Park Memorial Institute 1640 medium and incubated for 48 hr at 37°C under normoxia and hypoxia. Biofilms were washed twice with phosphate-buffered saline (PBS), and dry biomass of the biofilms were quantified and expressed as ratio of the indicated strain versus control strain (wild type). Means and standard deviations were determined from the results of three independent experiments. A t test was used to calculate the statistical relevance: ***p < .001. The dashed line indicates the relative mass of the control strain which was set to 100%. (c) Candida albicans wild type, ifu5Δ/Δ, IFU5 reconstituted, and mutifu5 strains were allowed to form biofilm under earlier performed gaseous conditions. Indicated strains were allowed to adhere to the surface of a polystyrene plate for 90 min in PBS at 37°C under hypoxic (0.2% O2) conditions. The wells were supplemented with Roswell Park Memorial Institute 1640 medium and incubated for 24 and 48 hr at 37°C under hypoxia. Biofilms were washed with PBS after the indicated time points and supplemented with XTT-menadione solution. After incubation in the dark for 30 min at 37°C, the supernatant was transferred to a new 96-well microtiter plate, and the colour change was measured at 490 nm. Dashed lines indicate absorbance of the control strain after 24 and 48 hr, respectively. A t test was used to calculate the statistical relevance: ***p < .001. Dashed lines indicate absorbance of the control strain after 24 and 48 hr, respectively. (d) Adhesion of C. albicans indicated strains to polystyrene wells was observed using XTT reduction assay. Strains were inoculated into polystyrene wells as they were for biofilm experiments. Following 90 min of incubation, non-adherent cells were removed by washing, and the numbers of cells were assessed by using an XTT reduction assay. A t test was used to calculate the statistical relevance: *p < .05. Dashed lines indicate absorbance of the control strain
Next, as cells in C. albicans biofilms go through Efg1-dependent hypoxic adaptation (Stichternoth & Ernst, 2009), we sought to assess the link between Ifu5 and this cellular process. As the ability of microbial cells to adhere to the substrate is central to the formation of fungal biofilms, we tested the strains of interest for their ability to adhere and form biofilm on 24-well polystyrene plate grown under normoxia and hypoxia. The dry mass of the biofilms formed by ifu5⍰/⍰ and mutIfu5 was similar to the wild type during normoxic growth (Figure 5b). Strikingly, although ifu5⍰/⍰ was able to form biofilm under hypoxia, the dry mass of the biofilm was two-fold less than that of the wild type, indicating that absence of Ifu5 impacts biofilm formation solely under hypoxia(Figure 5b). The difference in the extent of biofilm growth between the wild type, ifu5⍰/⍰ cells, mutIfu5, and the reconstituted strains in hypoxic condition was further assessed by the Cell Proliferation Kit II (XTT) reduction assay. In line with the decreased dry mass of biofilms, the mutants displayed a 50% reduction in the metabolic activity after 24 and 48 hr, compared with the wild type(Figure 5c). These data revealed that ifu5⍰/⍰ cells and mutIfu5 display a delay in mature biofilm formation. We then monitored the adherence ability of the mutant onto polystyrene plates under hypoxia. The adherence assay further confirmed the results of our metabolic assay as ifu5⍰/⍰ and mutIfu5 cells showed a significant 66% reduction in cells that had grown on the polystyrene surface compared with the wild type(Figure 5d). The extent of biofilm growth and adherence in the reconstituted strain was similar to wild type (Figure 5b–d). This set of data points to the role of Ifu5 in biofilm formation, specifically in oxygen-limiting conditions. Coupled together, we conclude that Ifu5 serves as a positive regulator of filamentation and hypoxic biofilm formation in C. albicans and that these processes are facilitated by the WW domain of Ifu5.
3. Discussion
In this study, we spotlight Ifu5, a Tac1-regulated WW domain-containing protein(Figure 1) that serves a role in hypoxic signalling and influences virulence related traits of C. albicans in oxygen-surfeit and oxygen-limiting niches. Despite being coregulated with the drug efflux pumps(Figure 2a,b), Ifu5 does not contribute to azole susceptibility in wild-type C. albicans(Figure S3). Through this study, we also demonstrate the significance of Ifu5 in maintaining cell wall integrity, one of the functions of this protein under normoxia (Figures 2 6). We propose that compromised Ifu5 function causes glycostress in unstressed cells, leading to constitutive high levels of phospho-Cek1 and consequentially up-regulated PMT1(Figure 2g,h). Thus, the ability of Ifu5 to contribute to cell wall regulation can be attributed to its role in maintaining glycosylation homeostasis in C. albicans. WW domain is a well-characterised, highly conserved protein domain implicated in variety of cellular processes. However, protein interaction partners of many WW domain-containing proteins are largely unknown (Figure 1). In this context, this study identifies the hypoxic regulator Efg1 as the interaction partner of Ifu5 and brings forward a relationship between Ifu5 and Efg1 that may be one of the underlying mechanisms for hypoxic adaptation in C. albicans(Figure 6).
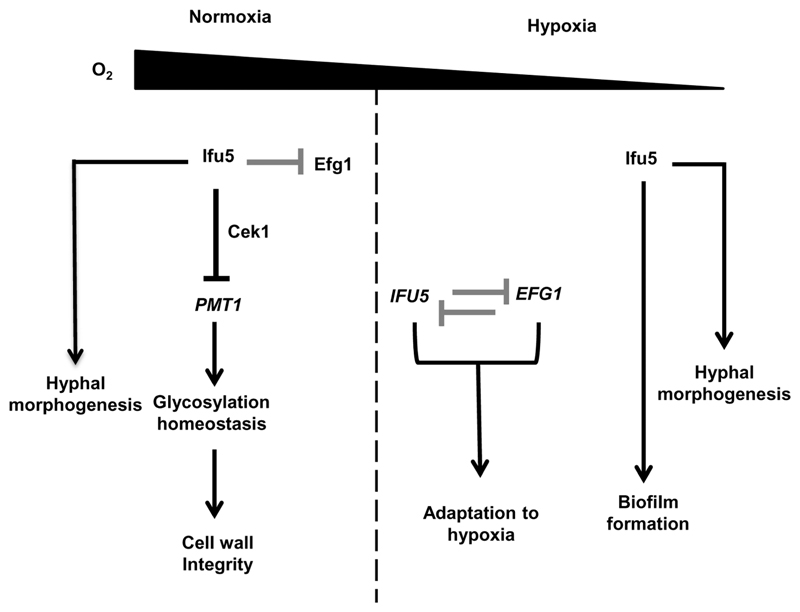
Figure 6.
Model depicting Ifu5 functions in Candida albicans. Ifu5 influences normoxic and hypoxic responses in C. albicans. Ifu5 represses PMT1 expression via the Cek1 MAP kinase pathway to maintain glycosylation homeostasis and thus cell wall integrity under normoxia. Direct binding of Ifu5 to Efg1 (grey blunt arrow) represses the function of this regulator. Hypoxia relieves this repression, and a negative reciprocal loop (grey blunt arrows) of regulation between IFU5 and EFG1 allows rapid hypoxic adaptation. Ifu5 positively regulates hypoxic biofilm formation and hyphal morphogenesis in both normoxic and hypoxic conditions
Transcriptome analysis of ifu5⍰/⍰ showed up-regulation of the hypoxic regulator EFG1, its normoxia-dependent target genes and biofilm-specific set of genes(Figure 3). We infer that Ifu5 not only regulates EFG1 expression but also Efg1 allied pathways involved in hypoxic adaptation. We propose that there is a complex molecular interaction between Ifu5 and Efg1 wherein Ifu5 regulates both normoxic and hypoxic expression of EFG1 transcript(Figure 4b–d) and coordinates with Efg1 to influence hypoxic responses in C. albicans. The repressive effect of Ifu5 on normoxic EFG1 expression and Efg1 activity is evident from transcriptional induction of EFG1 and its dependent genes in ifu5Δ/Δ cells(Figure 3b). This repression could be facilitated by WW domain-mediated direct interaction between Ifu5 and Efg1 under normoxia(Figure 4a). Considering that EFG1 negatively regulates its own transcription (Tebarth et al., 2003), we propose the possibility that Ifu5, by functioning as a corepressor of Efg1, may presumably allow this regulator to efficiently down-regulate its own expression. Thus, in ifu5Δ/Δ cells, the ability of Efg1 to autoregulate itself is compromised leading to its constitutive up-regulation(Figure 4d). This plausible explanation is justified by the established role of WW domain-containing proteins in binding to proline-rich sequences of transcription factors such as human PEBP2 (polyoma enhancer binding protein), NF-E2 (nuclear factor, eryt nuclear factor, erythroid 2) and BRN-2/POU3F2 (POU domain, class 3, transcription factor 2). The C-terminus of Efg1 contains a stretch of prolines (residues 332 to 338), located C-terminal to its APSES domain (Noffz, Liedschulte, Lengeler, & Ernst, 2008), which may serve as a putative binding site for Ifu5. The significance of the proline-rich stretch in mediating Ifu5–Efg1 interaction needs further investigations. During early hypoxic stress, it is likely that the repressive effect of Ifu5 on Efg1 is relieved, resulting in Efg1-mediated transcriptional repression of IFU5(Figure 4c). Increased IFU5 transcripts at later stages of hypoxic stress may be facilitated by low Efg1 levels, owing to the autoregulation of Efg1 that eventually causes a reduction in Efg1 protein levels (Lassak et al., 2011; Tebarth et al., 2003). Coupled together, these data suggest that Ifu5 in concert with Efg1 and unknown hypoxic regulators may ensure rapid adaptation to hypoxic environments and that Ifu5 and Efg1 may act as major points of integration for hypoxic adaptation in C. albicans(Figure 6).
Ifu5 also functions as a positive regulator of both filamentation and biofilm formation: two distinct hypoxia-dependent adaptation processes(Figure 6). Hyphal morphogenesis and hypoxic biofilm formation was blocked in ifu5⍰/⍰ cells despite the up-regulation of Efg1(Figure 5), suggesting that the requirement for Ifu5 during these processes cannot be bypassed by increased EFG1 expression in the mutant. This is in contrast to the established role of Efg1 as an inducer of morphogenesis and biofilm formation in both gas conditions (Desai et al., 2015; Setiadi et al., 2006). Our results thus argue that both Ifu5 and Efg1 induce filamentation and hypoxic biofilm development via similar factors and that the expression of these factors is largely dependent on functional Ifu5. It is likely that Efg1-dependent activation of biofilm-specific genes, especially under hypoxia, requires Ifu5-dependent expression of additional hypoxic regulators/factors that link to the Ifu5-Efg1 regulatory loop. Thus, the absence of Ifu5-dependent unknown hypoxic regulators may not allow Efg1 to fully exert its inducing effect on filamentation and biofilm formation in the mutant. The lack of normoxic biofilm phenotype(Figure 5b–d) for ifu5⍰/⍰ is surprising as the mutant shows reduced filamentation on agar surface. Likewise, mutants of ace2 and czf1 that are considered crucial for hypoxic filamentation are dispensable for hypoxic biofilm formation (Giusani, Vinces, & Kumamoto, 2002; Kelly et al., 2004). This indicates that regulators (factors) required for filamentation under specific environments may not have a role in biofilm development in similar environments. Collectively, our data reveal synergistic function for Ifu5 and Efg1 in promoting filamentation and hypoxic biofilm formation in C. albicans.
WW domain is the predominant interacting module within Ifu5 as mutagenising this domain resulted in not only loss of Ifu5–Efg1 interaction under normoxia but also filamentation and hypoxic biofilm formation (Figures 4a and 5). We posit that the WW domain of Ifu5 may serve as a platform linking Ifu5 with distinct proteins affiliated with multiple physiological networks. Although protein–protein interac-tions through WW domain may be crucial for Ifu5 to regulate Efg1 activity, its involvement in cell wall regulation could be a separate function independent of the WW domain(Figure 2c). The link between Ifu5 and cell wall regulation is reminiscent of the role of Sordaria macrospora protein PRO40 (protoperithecia) in modulating the CWI pathway (Teichert et al., 2014). PRO40 serves as a scaffold for the proteins of the CWI MAPK module, independent of the WW domain, linking them to the upstream activator Pkc1 (Teichert et al., 2014). It is plausible that similar to PRO40, Ifu5 may serve as the scaffold protein to regulate the activity of the C. albicans CWI pathway in a WW domain-independent manner, an area that ought to be explored. The mammalian tumor suppressor gene WWOX (WW domain-containing oxidoreductase) and the SO (SOFT) protein involved in hyphal fusion in Neurospora crassa are few additional examples that mediate certain functions in a WW domain-independent manner (Fleiβner & Glass, 2007; Jamous & Salah, 2018). It seems likely that Ifu5 may mediate its functions in C. albicans by interacting with and influencing the activities of different proteins in WW domain-dependent as well as WW domain-independent manner. Fungal hypoxic adaptation pathways are critical for virulence and exploiting them for antifungal therapy may improve the outcome of treating fungal infections. In vivo relevance of interfering with molecules involved in hypoxic adaptation may offer opportunities for prevention of fungal infections and prove to be favourable targets for drug development. We propose that Ifu5 is a promising component for potential therapeutic targeting of C. albicans associated infections under varying oxygen niches.
4. Experimental Procedures
4.1. Strains, chemicals, and growth conditions
Strains were maintained and propagated on YEPD medium (1% yeast extract, 2% Bacto peptone, 2% glucose, and 2% agar for solidification). YEPD medium supplemented with 200·μg·ml-1 of nourseothricin (Werner Bioagents, Jena, Germany) was used for selection of deletion mutants. To obtain nourseothricin-sensitive derivatives of transformants, strains were grown in yeast extract-peptone-maltose (1% yeast extract, 2% peptone, 2% maltose) for 8 hr and plated on 10-μg·ml-1 nourseothricin. For hyphal morphogenesis on solid media, cells were plated on Spider (1% nutrient broth, 1% D-mannitol and 0.2% K2HPO4) agar plate, kept under hypoxia (0.2% O2) and normoxia at 37°C for 3 days, and photographed. Cell surface stress was imposed with sodium dodecyl sulphate (Biobasic), caspofungin (Merck), Calcofluor White (Sigma Aldrich), and tunicamycin (Sigma Aldrich) at the concentrations specified in theFigure 2. For stress sensitivity assay strains grown overnight in YEPD media were diluted in 0.9% saline solution. Thereafter, 5-μl portions of four dilutions (5 × 103 to 5 × 105 cells) were spotted onto YEPD agar plates containing indicated drugs. Plates were photographed after incubation for 48 hr at 30°C.
4.1.1. Strain construction
Construction of deletion cassettes for IFU5
IFU5 was deleted by standard two-step disruptions using SAT1 flipper using the plasmid pSFS2B (Reuss, Vik, Kolter, & Morschhäuser, 2004). The generated strains, plasmids, and primers used for strain construction are listed in Tables S1, S2, and S3, respectively. Two different disruption cassettes were constructed for the two alleles of IFU5 in the disruption vector pSFS2B. For the IFU5 deletion construct, a 300-bp 5′ upstream noncoding region (NCR) of IFU5 (5′ IFU5NCR) was amplified from SC5314 genomic DNA with primers FP 1 and FP 2(Table S3), which introduced KpnI and XhoI restriction sites and cloned into 5′ end of the SAT1–FLP cassette in pSFS2B using the same enzymes. A 300-bp region of 3′ IFU5NCR was amplified with primers FP 3 and FP 4, which introduced SacII and SacI sites and was cloned in the 3′ end of the SAT1–FLP cassette (which already contains 5′ IFU5NCR). The plasmid thus constructed containing the disruption cassette for deletion of the first allele of IFU5 is referred to as pSR1(Table S2). For deleting the second allele, 5′ end of pSR1 was replaced by 300 bp of the IFU5NCR (amplified by primers FP 5 and FP 6) with KpnI and XhoI restriction sites, generating pSR2(Table S2). The IFU5 reconstitution construct was made by amplifying a 1.7-kb fragment containing the IFU5 ORF (0.7 kb) and the 5′ NCR (1.0 kb) by using primers FP 7 and FP 8 that introduced KpnI and XhoI sites. The fragment was then ligated into the KpnI and XhoI digested pSR1, which already contained the 3′ IFU5NCR. This procedure resulted in the IFU5 reconstitution plasmid (pSR3). The wild-type strain, SC5314, was electroporated with the first round disruption cassette, and deletion mutants were selected on 200-μg·ml-1 nourseothricin (SR11). To obtain nourseothricin-sensitive derivatives of transformants, strains were grown in yeast extract-peptone-maltose (1% yeast extract, 2% peptone, 2% maltose) and plated on 10-μg·ml-1 nourseothricin. These nourseothricin-sensitive heterozygous mutants (SR12) were then used for the second round of transformation generating the homozygous null mutant strain SR13. The CaSAT1 construct was flipped out from SR13, resulting in SR14. For reconstituted strain, 5.8-kb fragment from pSR3 digested with KpnI and SacI and was transformed in SR14 to yield, SR15. Proper integration at each step was confirmed by Southern hybridisation.
C-terminal Myc tagging
For constructing C-terminal 13X Myc-tagged IFU5, the vector pADH34 was used as described previously (Nobile et al., 2009), which introduces 13X Myc epitope upstream from stop codon of IFU5. Cassette carrying SAT1 marker, homology region of IFU5 and 13X Myc epitope was transformed in SC5314 and positive transformants were screened by polymerase chain reaction (PCR). SAT1 marker was recycled, and PCR product from nourseothricin sensitive colonies was sequenced to confirm proper integration of the cassette. Primers used for amplification of cassette and detection for integration are listed inTable S3.
Site-directed mutagenesis of the WW domain
For the construction of mutIfu5 strain, plasmid containing the gene IFU5 (pSR3) was used as template and tryptophan and proline substitution was performed by PCR (initial denaturation at 95°C for 5 min, annealing at 61°C for 1 min, extension at 68°C for 9 min, and final extension of 9 min) using primers listed inTable S3. The plasmid amplification products were then digested with DpnI at 37°C for 1 hr and then transformed in Escherichia coli. The mutations were confirmed by sequencing and the integration cassette was electroporated into SR16. Effective integration was confirmed by Southern blotting.
Construction of TAP modified Ifu5 and HA modified Efg1
For strains used for co-immunoprecipitation assay, hemagglutinin-epitope modified Efg1 (HA-Efg1) and TAP-epitope modified Ifu5 (Ifu5-TAP) were constructed. For C-terminal tagging of Ifu5, plasmid pFA-TAP-HIS (Lavoie, Sellam, Askew, Nantel, & Whiteway, 2008) was used to amplify sequences encoding the TAP and HIS1 cassette. The cassette was transformed in strain AVL12 that expresses HA-Efg1 from its native promoter (Noffz et al., 2008) and in BWP17 resulting in strains CLvW998 (HA-Efg1, Ifu5-TAP) and CLvW997 (Ifu5-TAP), respectively. Similarly, strains producing mutifu5-TAP were constructed by first replacing both IFU5 alleles with the mutated version encoded on plasmid pSR4 and subsequent transformation with the TAP-HIS cassette amplified with homologous-overhangs from plasmid pFA-TAP-HIS. The resulting strain CLvW990 (BWP17) produces mutifu5-TAP and strain CLvW989 produces (AVL12) HA-Efg1 and mutifu5-TAP. Chromosomal integration of the TAP-HIS1 cassette was confirmed by PCR using oligonucleotides colo-IFU5-for and colo-IFU5-rev.
Cell wall analysis
Transmission electron microscopy. Exponentially growing C. albicans yeast cells were snap-frozen in liquid nitrogen at high pressure using a Leica Empact high-pressure freezer (Leica, Milton Keynes, United Kingdom). The frozen samples were then fixed, warmed to −30°C, processed, and embedded in TAAB812 (TAAB Laboratories, Aldermaston, United Kingdom) epoxy resin as described previously (Walker et al., 2018). Ultrathin sections of 100 nm were cut on a Leica Ultracut E microtome and stained with uranyl acetate and lead citrate. Philips CM10 transmission microscope (FEI UK Ltd., Cambridge, United Kingdom) was used for viewing the samples, and a Gatan BioScan 792 camera system (Gatan UK, Abingdon, United Kingdom) was used for capturing the images.
Cell wall carbohydrate analysis. Yeast cells were grown in YEPD medium at 30°C, broken, and hydrolysed as described previously (Lee et al., 2012). Hydrolyzed samples were analysed by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) in a carbohydrate analyser system from Dionex. The total concentration of each cell wall component was expressed as microgram per milligram of dried cell wall, determined by calibration from the standard curves of glucosamine, glucose, and mannose monomers, and converted to a percentage of the total cell wall.
Flocculation assay. Overnight YEPD grown cells were harvested and resuspended in Roswell Park Memorial Institute (RPMI) medium with a starting OD600 of 0.3 and grown for additional 3 hr at 37°C with shaking at 220 r.p.m. Cells were then transferred to glass tubes and vortexed for 30 s. OD600 measured and cells were allowed to settle for 15 min; after which, OD600 was measured and an aliquot of cells were photographed. Experiment was done in triplicate and change in absorbance against each time point was plotted.
Microarray analysis
For transcriptional profiling, wild type and mutant cells were grown overnight in YEPD, subcultured from a starting OD600 of 0.3 in fresh YEPD and incubated at 30°C till OD600 reached 1.0. Thereafter, RNA was extracted from three biological replicates of both strains using an RNeasy minikit (Qiagen) and was used to perform the microarray experiments. The samples for gene expression were labelled using Agilent Quick-Amp labelling Kit (p/n5190-0442). The cDNA synthesis, in vitro transcription steps, and subsequent hybridisation on to genotypic-designed C. albicans_GXP_8X15k (AMADID No: 26377) array was carried out as described previously (Thomas et al., 2015). Data extraction from images was done using Feature Extraction software Version 11.5 of Agilent. The microarray data can be accessed under GEO accession number GSE110650.
Quantitative real-time PCR
The levels of specific transcripts were measured by qPCR in triplicate using independent biological replicates. RNA isolation was performed as described above and cDNA synthesis (Thermo scientific) carried out subsequently following manufacturer’s protocol. qPCR reactions were performed in a volume of 25 μl using Thermo Scientific Maxima SYBR Green mix in a 96-well plate. For measuring relative transcript levels of IFU5 and EFG1 under hypoxia, the indicated strains were first grown under normoxic conditions in YEPD medium for 4 hr at 30°C. Thereafter, a fraction of the cells was used to inoculate fresh YEPD medium (preincubated under hypoxia (0.2% O2), and cells were allowed to grow under hypoxic conditions for additional 4 hr at 30° C. After 30, 60, 120 and 240 min, a fraction of the cells was harvested, and total RNA was isolated. Time point 0 corresponds to 4hr growth under normoxic conditions before shifting cells to hypoxic conditions. At each time point, two biological replicates and three technical replicates were assayed by qPCR. ACT1 was used as the internal control and transcript level of the gene of interest was normalised to ACT1 levels. Results were statistically analysed using Student’s t test. The qPCR primers used in this study were designed by Primer Express 3.0 and are listed inTable S4.
Immunoblotting
To examine Mkc1 and Cek1 phosphorylation, protein extracts were prepared from exponentially grown C. albicans strains and these extracts were subjected to western blotting using previously described protocols (Martín, Arroyo, Sánchez, Molina, & Nombela, 1993). Anti-phospho-p44/p42 MAPK (Thr202/Tyr204) antibody (New England Biolabs) was used to detect dually phosphorylated Mkc1 and Cek1 MAPKs (indicated as Mkc1-P and Cek1-P inFigure 2g); anti-Mkc1 (Román, Nombela, & Pla, 2005) antibodies were used to detect total Mkc1 (loading control) as indicated in theFigure 2g. Blot imaging was done by using an Odyssey fluorescence imager (LI-COR) and quantified using Image Studio Lite (LI-COR).
Co-immunoprecipitation assay
Strains producing HA modified Efg1 (AVL12, lanes 2 and 8), TAP modified Ifu5 (CLvW997, lanes 3 and 9), both, HA modified Efg1 and TAP modified Ifu5 (CLvW998, lanes 4 and 10), mutIfu5 TAP modified (CLvW990, lanes 5 and 11), and a strain producing both HA modified Efg1 and TAP modified mutIfu5 (CLvW989, lanes 6 and 12) were grown in YEPD medium at 30°C in either hypoxic or normoxic (0.2% O2) conditions for 4 hr. A control strain BWP17 was used, expressing unmodified Efg1 and Ifu5 proteins (lanes 1 and 7). For cross-linking, cultures were treated with formaldehyde (1% v/v). Total protein extracts were incubated with Dynabeads, Mouse IgG (Invitrogen, Cat No: 11041) for immunoprecipitation of TAP modified Ifu5. The samples were immunoblotted and developed using anti-TAP (IP; Invitrogen, CAB1001, 1:1000) or anti-HA (CoIP; Santa Cruz Biotech, sc-7392, 1:1000) antibodies. To verify presence of HA-Efg1 1% of the total protein extracts was blotted and investigated with anti-HA.
Biofilm formation in polystyrene wells
Quantification of biofilm by dry weight of wells. To quantify the biofilm formation, dry weights of wells were used. Seven hundred microlitre of 105 cells ml-1 of the indicated strains were allowed to adhere to the surface of a polystyrene flat bottom 24-well plate (Corning Costar) for 90 min in phosphate-buffered saline (PBS) at 37°C under normoxic and hypoxic (0.2% O2) conditions. The medium and PBS used for hypoxic growth conditions were preincubated under hypoxia for 12 hr. Each well was washed twice with PBS to remove nonadherent cells. The wells were supplemented with RPMI 1640 medium and incubated for 48 hr at 37°C under normoxia and hypoxia. Biofilms were washed twice with PBS, and dry biomass of the biofilms were quantified and expressed as ratio of the indicated strain versus control strain (wild type). The experiment was performed twice, with two independent biological replicates, each assayed with three technical replicates.
Quantification of biofilm by XTT reduction assay. To determine the number of cells in the different biofilm stages, XTT reduction assay was performed. One hundred microlitre of 105 cells ml-1 of the indicated strains were allowed to adhere to the surface of a polystyrene flat bottom 96-well microtiter plate (Corning Costar) for 90 min in PBS at 37°C under hypoxic (0.2% O2) conditions. The medium and PBS buffer used was preincubated under hypoxic conditions for 12 hr. Each well was washed twice with PBS to remove nonadherent cells. The wells were supplemented with RPMI 1640 medium and incubated for 24 and 48 hr at 37°C under hypoxia. Biofilms were washed with PBS after the indicated time points and supplemented with XTT-menadione solution (1.5-mM XTT (2, 3-bis (2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide); 0.4-M menadione). After incubation in the dark for 30 min at 37°C, the supernatant was transferred to a new 96-well microtiter plate, and the colour change was measured at 490 nm. The experiment was performed twice, with two independent biological replicates, each assayed with three technical replicates.
Adhesion of C. albicans strains to polystyrene. To quantify adherence, XTT reduction assay was performed as described for quantification of biofilm formation except that after washing each well twice with PBS to remove nonadherent cells, XTT-menadione solution was added to the wells and incubated for 30 min at 37°C in dark. The supernatant was transferred to a new 96-well microtiter plate to monitor colour change at 490 nm. The experiment was performed twice, with two independent biological replicates, each assayed with three technical replicates. A t test was used to calculate the statistical relevance in all the aforesaid experiments.
Supplementary Material
Acknowledgements
We are grateful to Joachim Morschhauser and Suzanne Noble for providing us with the plasmids used in this study, to Patrick Van Dijck and Neeru Saini for performing initial experiments for this study and the entire lab for insightful discussions. We acknowledge Genotypic Technology Pvt., Ltd., Bangalore, India for microarray experiments. S. K. R., S. R., and P. A acknowledge the Department of Biotechnology, Council for Scientific and Industrial Research and women Scientist award from Department of Science & Technology, respectively for awarding Junior and Senior Research Fellowships. This work was funded by the Department of Biotechnology (BT/PR5347/MED/29/629/2012) to S. L. P. Umbrella grants from Jawaharlal Nehru University in the form of Capacity Build-up, UGC-Resource Networking and DST-PURSE are also acknowledged. Support from ERA-Net PathoGenoMics project OXYstress (www.pathogenomics-era.net/FundedProjects3rdJoint) and the Infect-ERA JTC2 project FunComPath (http://www.funcompath.eu/) to JFE, BIO2015-64777-P to JP and Wellcome Trust (101873, 086827, 075470, & 200208) and MRC Centre for Medical Mycology (N006364/1) to NG is acknowledged.
Funding information
Department of Biotechnology, Grant/Award Number: BT/PR5347/MED/29/629/2012
References
- Askew C, Sellam A, Epp E, Hogues H, Mullick A, Nantel A, Whiteway M. Transcriptional regulation of carbohydrate metabolism in the human pathogenCandida albicans. PLoS Pathogens. 2009;5(10):e1000612. doi: 10.1371/journal.ppat.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme J, Chauvel M, Goyard S, Roux P, Rossignol T, d’Enfert C. Contribution of the glycolytic flux and hypoxia adaptation to efficient biofilm formation by Candida albicans: Biofilm formation and adaptation to hypoxia inC. albicans. Molecular Microbiology. 2011;80(4):995–1013. doi: 10.1111/j.1365-2958.2011.07626.x. [DOI] [PubMed] [Google Scholar]
- Cantero PD, Ernst JF. Damage to the glycoshield activates PMT-directed O-mannosylation via the Msb2-Cek1 pathway inCandida albicans. Molecular Microbiology. 2011;80(3):715–725. doi: 10.1111/j.1365-2958.2011.07604.x. [DOI] [PubMed] [Google Scholar]
- Chang YC, Bien CM, Lee H, Espenshade PJ, Kwon-Chung KJ. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence inCryptococcus neoformans. Molecular Microbiology. 2007;64(3):614–629. doi: 10.1111/j.1365-2958.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation ofCandida albicans ABC transporters CDR1 and CDR2. Eukaryotic Cell. 2004;3(6):1639–1652. doi: 10.1128/EC.3.6.1639-1652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai PR, van Wijlick L, Kurtz D, Juchimiuk M, Ernst JF. Hypoxia and temperature regulated morphogenesis inCandida albicans . PLoS Genetics. 2015;11(8):e1005447. doi: 10.1371/journal.pgen.1005447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doedt T, Krishnamurthy S, Bockmühl DP, Tebarth B, Stempel C, Russell CL, Ernst JF. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Molecular Biology of the Cell. 2004;15(7):3167–3180. doi: 10.1091/mbc.e03-11-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst JF, Tielker D. Responses to hypoxia in fungal pathogens. Cellular Microbiology. 2009;11(2):183–190. doi: 10.1111/j.1462-5822.2008.01259.x. [DOI] [PubMed] [Google Scholar]
- Fleiβner A, Glass NL. SO,aproteininvolvedinhyphalfusionin Neurospora crassa, localizes to septal plugs. Eukaryotic Cell. 2007;6(1):84–94. doi: 10.1128/EC.00268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusani AD, Vinces M, Kumamoto CA. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics. 2002;160(4):1749–1753. doi: 10.1093/genetics/160.4.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahl N, Shepardson KM, Chung D, Cramer RA. Hypoxia and fungal pathogenesis: To air or not to air. Eukaryotic Cell. 2012;11(5):560–570. doi: 10.1128/EC.00031-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Miller JP, Golob A, Stajich JE, Michaud GA, Fields S. Comparative analysis of Saccharomyces cerevisiae WW domains and their interacting proteins. Genome Biology. 2006;7(4):R30. doi: 10.1186/gb-2006-7-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamous A, Salah Z. WW-domain containing protein poles in breast tumorigenesis. Frontiers in Oncology. 2018;8 doi: 10.3389/fonc.2018.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MT, MacCallum DM, Clancy SD, Odds FC, Brown AJP, Butler G. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence: C. albicans CaAce2 knock-out affects morphogenesis. Molecular Microbiology. 2004;53(3):969–983. doi: 10.1111/j.1365-2958.2004.04185.x. [DOI] [PubMed] [Google Scholar]
- Lassak T, Schneider E, Bussmann M, Kurtz D, Manak JR, Srikantha T, Ernst JF. Target specificity of the Candida albicans Efg1 regulator: Efg1 target recognition. Molecular Microbiology. 2011;82(3):602–618. doi: 10.1111/j.1365-2958.2011.07837.x. [DOI] [PubMed] [Google Scholar]
- Lavoie H, Sellam A, Askew C, Nantel A, Whiteway M. Atool-box for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics. 2008;9:578. doi: 10.1186/1471-2164-9-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, MacCallum DM, Jacobsen MD, Walker LA, Odds FC, Gow NAR, Munro CA. Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrobial Agents and Chemotherapy. 2012;56(1):208–217. doi: 10.1128/AAC.00683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes JP, Stylianou M, Backman E, Holmberg S, Jass J, Claesson R, Urban CF. Evasion of immune surveillance in low oxygen environments enhances Candida albicans virulence. MBio. 2018;9(6) doi: 10.1128/mBio.02120-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín H, Arroyo J, Sánchez M, Molina M, Nombela C. Activity of the yeast MAP kinase homologue Slt2 is critically required for cell integrity at 37 degrees C. Molecular General Genetics: MGG. 1993;241(1-2):177–184. doi: 10.1007/bf00280215. [DOI] [PubMed] [Google Scholar]
- Masuoka J, Hazen KC. Cell wall protein mannosylation determines Candida albicans cell surface hydrophobicity. Microbiology (Reading, England) 1997;143(Pt 9):3015–3021. doi: 10.1099/00221287-143-9-3015. [DOI] [PubMed] [Google Scholar]
- Nobile CJ, Mitchell AP. Genetics and genomics of Candida albicans biofilm formation. Cellular Microbiology. 2006;8(9):1382–1391. doi: 10.1111/j.1462-5822.2006.00761.x. [DOI] [PubMed] [Google Scholar]
- Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault J-S, Nantel A, Mitchell AP. Biofilm matrix regulation by Candida albicans zap1. PLoS Biology. 2009;7(6):e1000133. doi: 10.1371/journal.pbio.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noffz CS, Liedschulte V, Lengeler K, Ernst JF. Functional mapping of the Candida albicans Efg1 regulator. Eukaryotic Cell. 2008;7(5):881–893. doi: 10.1128/EC.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce JV, Kumamoto CA. Variation in Candida albicans EFG1 expression enables host-dependent changes in colonizing fungal populations. MBio. 2012;3(4):e00117–e00112. doi: 10.1128/mBio.00117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan A, Avelar GM, Bain JM, Childers DS, Larcombe DE, Netea MG, Brown AJP. Hypoxia promotes immune evasion by triggering β-glucan masking on the Candida albicans cell surface via mitochondrial and cAMP-protein kinase A signaling. MBio. 2018;9(6) doi: 10.1128/mBio.01318-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss O, Vik A, Kolter R, Morschhäuser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Román E, Alonso-Monge R, Miranda A, Pla J. The Mkk2 MAPKK regulates cell wall biogenesis in cooperation with the Cek1-pathway in Candida albicans. PloS One. 2015;10(7):e0133476. doi: 10.1371/journal.pone.0133476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román E, Nombela C, Pla J. The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Molecular and Cellular Biology. 2005;25(23):10611–10627. doi: 10.1128/MCB.25.23.10611-10627.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiadi ER, Doedt T, Cottier F, Noffz C, Ernst JF. Transcriptional response of Candida albicans to hypoxia: Linkage of oxygen sensing and Efg1p-regulatory networks. Journal of Molecular Biology. 2006;361(3):399–411. doi: 10.1016/j.jmb.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Sircaik S, Husain F, Thomas E, Ror S, Rastogi S, Panwar SL. Distinct roles of the 7-transmembrane receptor protein Rta3 in regulating the asymmetric distribution of phosphatidylcholine across the plasma membrane and biofilm formation in Candida albicans. Cellular Microbiology. 2017;19(12) doi: 10.1111/cmi.12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stichternoth C, Ernst JF. Hypoxic adaptation by Efg1 regulates biofilm formation by Candida albicans. Applied and Environmental Microbiology. 2009;75(11):3663–3672. doi: 10.1128/AEM.00098-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synnott JM, Guida A, Mulhern-Haughey S, Higgins DG, Butler G. Regulation ofthe hypoxic response in Candida albicans. Eukaryotic Cell. 2010;9(11):1734–1746. doi: 10.1128/EC.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szallies A, Kubata BK, Duszenko M. A metacaspase of Trypanosoma brucei causes loss of respiration competence and clonal death in the yeast Saccharomyces cerevisiae. FEBS Letters. 2002;517(1-3):144–150. doi: 10.1016/s0014-5793(02)02608-x. [DOI] [PubMed] [Google Scholar]
- Tebarth B, Doedt T, Krishnamurthy S, Weide M, Monterola F, Dominguez A, Ernst JF. Adaptation of the Efg1p morphogenetic pathway in Candida albicans by negative autoregulation and PKA-dependent repression ofthe EFG1 gene. Journal of Molecular Biology. 2003;329(5):949–962. doi: 10.1016/s0022-2836(03)00505-9. [DOI] [PubMed] [Google Scholar]
- Teichert I, Steffens EK, Schnaß N, Fränzel B, Krisp C, Wolters DA, Kück U. PRO40 is a scaffold protein of the cell wall integrity pathway, linking the MAP kinase module to the upstream activator protein kinase C. PLoS Genetics. 2014;10(9):e1004582. doi: 10.1371/journal.pgen.1004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas E, Sircaik S, Roman E, Brunel J-M, Johri AK, Pla J, Panwar SL. The activity of RTA2, a downstream effector of the calcineurin pathway, is required during tunicamycin-induced ER stress response in Candida albicans. FEMS Yeast Research. 2015:fov095. doi: 10.1093/femsyr/fov095. [DOI] [PubMed] [Google Scholar]
- van Wijlick L, Swidergall M, Brandt P, Ernst JF. Candida albicans responds to glycostructure damage by Ace2-mediated feedback regulation of Cek1 signaling. Molecular Microbiology. 2016;102(5):827–849. doi: 10.1111/mmi.13494. [DOI] [PubMed] [Google Scholar]
- Walker L, Sood P, Lenardon MD, Milne G, Olson J, Jensen G, Gow NAR. The viscoelastic properties of the fungal cell wall allow traffic of amBisome as intact liposome vesicles. MBio. 2018;9(1) doi: 10.1128/mBio.02383-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.