Abstract
The translocation of mechanosensitive transcription factors (TFs) across the nuclear envelope is a crucial step in cellular mechanotransduction. Yet the molecular mechanisms by which external mechanical cues control the nuclear shuttling dynamics of TFs through the nuclear pore complex (NPC) to activate gene expression are poorly understood. Here, we show that the nuclear import rate of myocardin-related transcription factor A (MRTFA) — a protein that regulates cytoskeletal dynamics via the activation of the TF serum response factor (SRF) — inversely correlates with the protein’s nanomechanical stability and does not relate to its thermodynamic stability. Tagging MRTFA with mechanically resistant proteins results in the downregulation of SRF-mediated myosin light-chain 9 (MYL9) gene expression and subsequent slowing down of cell migration. We conclude that the mechanical unfolding of proteins regulates their nuclear translocation rate through the NPC, and highlight the role of the NPC as a selective mechanosensor able to discriminate forces as low as ~10 pN. The modulation of the mechanical stability of TFs may represent a new strategy for the control of gene expression.
Introduction
Cellular mechanotransduction requires that cytoplasmic mechanical cues are relayed to the nucleus to activate gene expression1. Several transcription factors, such as YAP/TAZ2, β-catenin3 or zyxin4, have emerged as important mediators of mechanical signalling5. The myocardin related transcription factor A (MAL/MRTFA) stimulates the transcriptional activity of the serum response factor (SRF)6, which targets genes that encode cytoskeletal components7,8, including actin and other key proteins involved in cell motility, adhesion and differentiation9. Importantly, the Rho-dependent MRTFA/SRF activity is shown to control migration and invasion of metastatic cancer cell lines6. Thorough in-vitro cell-biology assays supported by structural-biology approaches have provided a seemingly complete picture of how MRTFA localization is regulated at the biochemical level10; in serum-starved cells, MRTFA is mostly cytoplasmic and bound to G-actin. By contrast, upon serum stimulation, Rho-GTPases trigger F-actin polymerization11 and the subsequent dissociation of G-actin from the MRTFA RPEL motif results in the exposure of a nuclear localization sequence (NLS)12 that signals MRTFA to translocate into the nucleus13. Consequently, the active remodelling of the cytoskeleton is tightly controlled by a Rho/actin-mediated mechanical feedback mechanism upstream of MRTFA/SRF transcriptional activity. Given that mechanical cues control both its cytoplasmic10 and nuclear14,15 activity, MRTFA is an excellent molecular platform to interrogate whether nuclear shuttling of transcription factors through the nuclear pore complex (NPC) — the primary transport gate for molecular exchange to and from the nucleus16 — is also per se a mechanically-activated process, with potential knock-on effects at the transcriptional and functional levels.
Recent single-molecule nanomechanical experiments on molecular pores showed that the translocation of polypeptides through the ClpX proteolytic E. coli machinery requires mechanical unfolding17,18, and that the pulling direction19 and mechanical stability of the degraded protein20 regulate its translocation kinetics. Similarly, proteins need to unfold before they cross nanopores upon electrical stimulation21. Unlike the narrow pores of the ClpX and α-hemolysin proteins, recent cryo-EM structures revealed that the central pore of the human NPC, such as that of U2OS cells22, is rather large (~40-50 nm width)23,24 in comparison to the size of the translocating cargo. Despite its considerable size and the absence of direct ATP consumption, transport across the NPC is extremely selective and finely regulated by a sophisticated set of intrinsically disordered25 and highly dynamic nucleoporin proteins (Nups) enriched in phenylalanine-glycine (FG) motifs that line the central NPC channel and extend filaments on both the cytoplasmic and nucleoplasmic faces26. These FG-Nups form a dense protein mesh that acts as an effective ‘molecular sieve’27. Several complementary models — including the virtual gating28, polymer brush29, selective gel phase30, forest30, bimodal structure31 and reduction of dimensionality32 models—have been proposed to explain the physical basis for the high selectivity and directionality observed during transport across the NPC, on the basis of the physicochemical properties of nucleoporins26,27. The general consensus is that translocating molecules need to overcome an entropic energy barrier created by the rapidly-fluctuating FG-Nups33. Given that mechanical unfolding drastically reduces the entropy of proteins34, it is tempting to speculate that a mechanically-extended protein cargo will traverse the repulsive mechanical gate more efficiently than the protein’s natively-folded conformation, exhibiting a much higher degree of steric hindrance. Early independent observations reported that ribonucleoprotein particles are straightened while translocating through the NPC35, and that transport of flexible cargos is faster than rigid ones of similar size26,36–38. More recently, we showed that nuclear translocation of the YAP transcription factor across the NPC is mechanoselective39, demonstrating that the physical deformation of the NPC resulted in enhanced YAP nuclear permeability. Motivated by these findings, we conjectured that the mechanical unfolding of proteins could accelerate their dynamics of nuclear import, having potential knock-on effects at the genetic and functional levels. Here we employed a multiscale experimental approach, using a combination of single-molecule force spectroscopy, single-cell mechanics,live-cell imaging and cellular-motility assays to quantitatively investigate the mechanosensor role of the NPC in regulating the dynamics of MRTFA mechanotransduction.
Results
To examine the dynamics of nuclear translocation in serum-starved U2OS cells after 15% serum stimulation —which significantly (>80%, Fig. S1) triggers nuclear shuttling, Fig. 1A—we used live-imaging confocal microscopy to track the fraction of nuclear MRTFA-GFP every 30 seconds for 20 minutes (Fig. 1B). Fitting the resulting time course with a single exponential yielded a total rate constant k = 3.91 ± 0.42 ×10-3 s-1 and an equilibrium nuclear accumulation ne = 78.4 ± 1.9 % (Fig. S2). Since MRTFA is known to be in dynamic equilibrium between the cytoplasm and the nucleus even under serum stimulation conditions10, we re-wrote the exponential term of our kinetic equation (see Supplementary Information) to decouple the contributions of the import and export processes, both being treated as pseudo-first order reactions, to yield both import kI = 3.07 ± 0.32 ×10-3 s-1 and export kE = 0.84 ± 0.14 × 10-3 s-1 rate constant. The dynamic equilibrium between cytoplasmic and nuclear MRTFA was further demonstrated by treating starved cells with leptomycin B, known to block nuclear export40, resulting in gradual nuclear accumulation of MRTFA-GFP (Fig. S3). To explore the effect of the mechanical stability of the translocating protein on its shuttling dynamics, the most common tactic would entail the rational modification of its mechanical stability via the introduction of well-defined point mutations into the force-bearing structural motif41. Unfortunately, our structural information of MRTFA is limited to its relatively small RPEL motif13. We hence used an alternative strategy that consisted of tagging fluorescent MRTFA constructs with different protein domains with varying mechanical stabilities. Specifically, as first candidates, we used 3 distinct immunoglobulin (Ig) domains – namely Ig1, Ig27 and Ig32 – from cardiac titin (Fig. 1C). This giant protein is responsible for the passive elasticity of muscle42 and its mechanical properties – increasing along the N-C termini direction43 – can be directly characterized in single molecule force spectroscopy experiments. To this goal, single polyproteins of the different Ig domains under study, namely (Ig1-Ig27C47A-C63A)4, (Ig27WT)8 and (Ig32)8, were individually stretched by an AFM tip at a constant velocity of 400 nm s-1 (Fig. 1D), resulting in the classic saw-tooth pattern of mechanical unfolding, whereby each force peak fingerprints the unfolding of an individual protein domain within the polyprotein chain (Fig. 1E). As expected, the mechanical stability of the Ig1 domain, located in the proximal region of titin’s elastic I-band, displays a lower mechanical stability (144 ± 27 pN) than the Ig27 (207 ± 29 pN) and the Ig32 (267 ± 33 pN) domains, which are placed further down into the distal I-band (Figs. 1F & Fig. S4). These results are all in general agreement with previous findings43–45.
Figure. 1. The kinetics of MRTFA nuclear import is regulated by its mechanical properties.
(a) Representative confocal image gallery showing MRTFA nuclear translocation, upon serum stimulation, in a U2OS cell stably expressing MRTFA-GFP. 10μm scale bar. (b) Individual (light grey lines) and averaged (green points, mean ± s.e.m.) time courses of nucleus/cell MRTFA. These translocation dynamics can be fit to n(t) = ne(1 – e–kt) (black line), yielding an equilibrium accumulation ne = 0.784 ± 0.019 and a total rate constant k = 3.91 ± 0.42 ×10-3 s-1, which can be decomposed into import and export rate constants kI = 3.07 ± 0.32 ×10-3 s-1 and kE = 0.84 ± 0.14 ×10-3 s-1, respectively. Data are from four independent experiments (n = 24). (c) Structural schematic of titin, showing the distribution of selected Ig domains along the N-C termini direction. (d) Schematics of a single molecule force spectroscopy experiment, whereby a polyprotein made of selected titin Ig domains is tethered between an AFM cantilever tip and a gold substrate. (e) Stretching individual (Ig1-Ig27C47A-C63A)4, (Ig27)8 (not shown here), and (Ig32)8 polyproteins at a constant velocity of 400 nm s-1 gives rise to unfolding trajectories exhibiting saw-tooth patterns, where each force peak corresponds to the unfolding of an individual Ig domain within the polyprotein chain. (f) Probability density histograms of unfolding forces for Ig1 (yellow), Ig27 (grey), and Ig32 (magenta) domains with the associated Gaussian probability density distributions (black) overlaid (unfolding force mean ± s.d.: Ig1, 144 ± 27 pN, n = 137; Ig27, 208 ± 28 pN, n = 186; Ig32, 267 ± 33 pN, n = 936). (g) Inserting Ig domains within MRTFA constructs enables one to probe the effect of mechanical stability on nuclear translocation. (h) Averaged (mean ± s.e.m.) time courses of nucleus/cell MRTFA in U2OS cells expressing MRTFA-Ig1-BFP, MRTFA-Ig27-BFP, or MRTFA-Ig32-BFP, after serum stimulation. The rate and extent of MRTFA nuclear translocation of the mechanically-labile Ig1 is higher than that of Ig27 and of the mechanically-stable Ig32. Data are from four independent experiments (Ig1, n = 34; Ig27, n = 37, Ig32, n = 44), only a representative error bar is shown per condition. (i) Total (squares), import (right-pointing triangles), and export (left-pointing triangles) rate constants associated with the nuclear translocation of Ig-domain-tagged MRTFA constructs plot against the mechanical stability (unfolding force) of the tagging Ig domain. The import rate constant displays exponential dependence (R2 = 0.999) with mechanical stability. By contrast, the export rate constant is largely independent of mechanical stability. Rate constants correspond to the fitting parameters from the averaged time courses in (h), associated error bars correspond to the s.e.m. of fitting parameters of the individual time courses. The unfolding forces correspond to the mean ± s.d. of unfolding forces as in (f). Dashed lines are a weighted linear fit (import) and average (export).
Having characterized the mechanical stability of each Ig domain independently, we measured the dynamics of nuclear translocation of each MRTFA-X-BFP (blue fluorescent protein) construct, with X = Ig1 (Fig. 1G), Ig27 and Ig32. Tagging MRTFA with the mechanically-labile Ig1 resulted in a much faster translocation (and a much higher degree of nuclear accumulation) than Ig27 and Ig32 (Figs. 1H & S5). The nuclear import rate constant exhibits a behaviour in line with an exponential dependence with the unfolding force measured by the AFM (Figs. 1I & S5). By contrast, the export rate constant is largely independent of the mechanical stability of the tagging domain. We then compared the kinetics of translocation when MRTFA was tagged with either of two domains with extreme mechanical properties (albeit with a different fold and molecular weights) such as the mechanically-labile R16 domain of spectrin46 and the mechanically-stable Spy0128 domain of pilin47, the complete unfolding of the latter being compromised by an internal isopeptide bond (Fig. S6). As expected, spectrin R16 exhibits a much higher import rate constant kI = 4.26 ± 0.45 ×10-3 s-1 and equilibrium nuclear accumulation ne = 72.2 ± 2.5% than Spy0128 (kI = 1.25 ± 0.19 ×10-3 s-1 and ne = 53.1 ± 2.8%, respectively). Taken together, these experiments suggest that the dynamics of MRTFA nuclear accumulation inversely follows the hierarchy of the mechanical stability of the tagging domains.
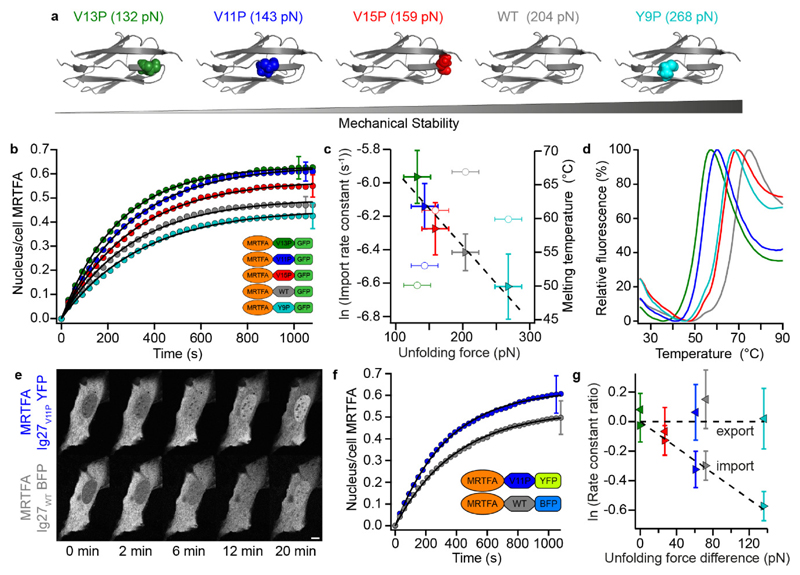
Although the different titin Ig domains share a similar Ig fold and are of comparable size, their sequences are different, and the differences in mechanical stability are rather large (~60 pN, Fig. 1F). An ideal comparison of the effect that a change in the protein’s mechanical stability has on the rate of nuclear translocation should be done, stricto sensu, by modulating the protein’s mechanical stability while keeping its sequence and structure largely unperturbed. With this purpose, we repeated the live-cell imaging experiments using MRTFA-Ig27X-GFP constructs, with the Ig27X domain either wild-type (X = WT) or harbouring a point mutation in well-localized position within the mechanical clamp, namely X = V13P, V11P, V15P or Y9P — all inducing a change in the mechanical stability with respect to the wild-type form in the order V13P < V11P < V15P < WT < Y9P41 (Fig. 2A). The dynamics of nuclear accumulation for each construct (Figs. 2B & S7) also followed the mechanical stability trend and, most strikingly, the import rate constant is again found to display an exponential dependence on the unfolding force measured under constant velocity conditions with the AFM41 (whereas the export rate is force-independent, Figs. 2C & S7). To learn whether the nuclear import rate also correlated with the thermodynamic stability of each protein, we conducted differential scanning fluorimetry48 and found no correlation between their melting temperature and their mechanical stability (Figs. 2C, 2D & S8) —notably, the Y9P mutant has an increased mechanical stability when compared to the wild-type, and yet exhibits a marked decrease in the thermal stability. These experiments unambiguously demonstrated that nuclear import is dictated by the mechanical —and not the thermal— stability of the translocating protein. Analogous experiments comparing the nuclear translocation for MRTFA-Ig27WT- GFP and MRTFA-Ig27V13P-GFP in MDA-MB-231 cells led to similar conclusions (Fig. S9), suggesting that the mechanical selectivity of the import mechanism might be general and independent of the cell line.
Figure. 2. The nuclear pore complex is highly mechanoselective.
(a) Selective introduction of point mutations in key residues in the mechanical-clamp region of the Ig27 structure result in subtle changes to its mechanical stability, in the order V13P < V11P < V15P < WT < Y9P, as measured elsewhere by single molecule force spectroscopy experiments41. (b) Averaged (mean ± s.e.m.) time courses of MRTFA nuclear translocation, upon serum stimulation, in U2OS cells expressing MRTFA-GFP tagged with different Ig27 mutants – the rate and extent of MRTFA nuclear accumulation is dependent on the tagging Ig27 variant. Data are from two (V11P, n = 26) or three (V13P, n = 22; V15P, n = 25; WT, n = 30; Y9P, n = 17) independent experiments, only a representative error bar is shown per condition. (c) The import rate constant (triangles) displays an exponential dependence (dashed line, linear fit, R2 = 0.988) with the mechanical stability of the tagging Ig27 mutant. By contrast, a weaker correlation is observed with their thermodynamic stability (circles). Import rate constants correspond to the fitting parameters from averaged time courses, associated error bars correspond to the s.e.m. of fitting parameters from individual time courses. Melting temperatures correspond to the mean of 12 replicates (with associated s.e.m. < 0.1 °C for each mutant, not shown). Unfolding forces correspond to the mean ± estimated s.d. (20 pN)41 as in (a). (d) Averaged melting curves of Ig27 variants (colour coded as in (a)) from differential scanning fluorimetry experiments. (e) Representative confocal image galleries showing MRTFA nuclear translocation, following serum stimulation, of a U2OS cell co-expressing MRTFA-I27WT-BFP and MRTFA-I27V11P-YFP. 10 μm scale bar. (f) Corresponding averaged (mean ± s.e.m.) time courses showing that, within the same cell, the rate and extent of nuclear accumulation of MRTFA tagged with the mechanically-labile Ig27V11P is higher than MRTFA tagged with the mechanically-stable Ig27WT. Data are from three independent experiments (n = 9), only a representative error bar is shown per condition. (g) Ratios (mean ± s.e.m.) of import (right-pointing triangles) and export (left-pointing triangles) rate constants plot against the unfolding force differences for co-expression experiments (full details in Fig. S10). Data are from three independent experiments (left to right, BFP/YFP: V13P/V13P, n = 9; V15P/V13P, n = 8; WT/V11P, n = 9; WT/V13P, n = 15; Y9P/V13P, n = 12). Ratios correspond to the mean ± s.e.m. of the ratios of the fitting parameters associated with translocation of the BFP construct, with respect to the YFP construct, from each individual cell. Dashed lines are a weighted linear fit (import) and average (export).
We next questioned whether the high mechanical selectivity in the nuclear accumulation that we observed —in some cases the AFM unfolding forces between two given mutations differ by only ~10 pN (Fig. 2A) — could be explained in terms of the intrinsic variability observed between individual cells (Fig. 1B). To rule out this possibility, we conducted control experiments whereby cells were co-transfected with MRTFA-Ig27X-BFP and MRTFA-Ig27Y-YFP (yellow fluorescent protein) constructs, where X, Y = V11P, V13P, V15P, WT or Y9P. The choice of BFP and YFP ensured that both emission wavelengths were sufficiently far apart to guarantee their individual detection without cross-talk, while presumably exhibiting very close (if not identical) mechanical stabilities49,50. This is confirmed as the translocation behaviour of MRTFA constructs differing only in their fluorescent tag (BFP or YFP) display near identical behaviour (Fig. S10). Dual fluorescence live-cell imaging to monitor the nuclear translocation of both MRTFA-Ig27WT-BFP and MRTFA-Ig27V11P-YFP constructs in an individual cell (Fig. 2E) showed that, at any given time, the degree of nuclear accumulation of the YFP construct (tagged with the mechanically-labile Ig27V11P) was always significantly higher than that of the BFP construct (tagged with the mechanically-stable Ig27WT), as observed in the overall kinetics of MRTFA nuclear localization (Fig. 2F). As expected, transport was completely blocked by overexpression of the dominant-negative vector of RanGTP, RanQ69L-mCherry (Fig. S11). Importantly, treatment of U2OS cells, co-expressing MRTFA-Ig27WT-BFP and MRTFA-Ig27V13P-YFP, with importazole, a small molecule inhibitor of importin-β51, noticeably inhibits their overall translocation, without affecting their selectivity (Fig. S11). Finally, the kinetic analysis of the dual fluorescence assays comparing MRTFA-Ig27X-BFP constructs benchmarked against MRTFA-Ig27V13P-YFP (or MRTFA-Ig27V11P-YFP) confirmed the mechanical hierarchy observed for the different Ig27 mutants when analysed independently (Fig. 2G). In conclusion, these experiments reiterate the high degree of mechanical sensitivity underpinning the nuclear translocation of MRTFA.
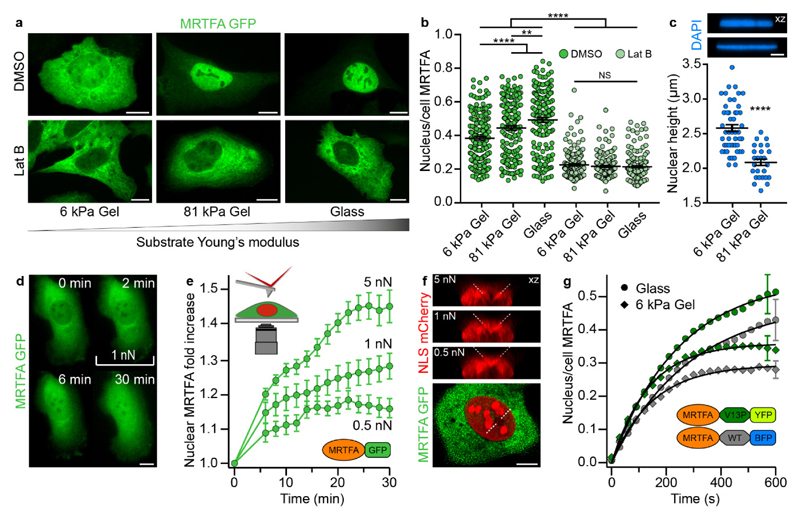
Such a novel mechanism for nuclear shuttling across the NPC, with the mechanical stability of the translocating protein regulating transport, is likely to complement an independent mechanism demonstrated for YAP39 whereby external forces directly applied to the NPC resulted in its physical deformation, triggering the subsequent increase in nuclear shuttling39. To probe whether this latter physical mechanism can also be extended to MRTFA, we first indirectly applied mechanical forces to the nuclear envelope by plating U2OS cells on polyacrylamide gels of different stiffnesses (Young’s moduli, 6 and 81 kPa) and glass (Fig. 3A). Nuclear localization of MRTFA was significantly increased on stiffer substrates (Fig. 3B), probably due to the combined effect of increasing actin polymerization (evidenced by an increase in the number of stress fibres, Fig. S12) resulting in a release of cytoplasmic MRTFA6 and the concomitant actin-mediated modification of the nuclear shape, which became significantly flattened as substrate stiffness increased (Fig. 3C)52. Control experiments using latrunculin B reversed the process, i.e. the number of actin stress fibres was reduced (Fig. S12) and MRTFA was mostly cytoplasmic, independently of substrate stiffness (Fig. 3A,B).
Figure. 3. Physical deformation of the nucleus is a complementary pathway to induce MRTFA nuclear translocation.
(a) Representative epifluorescence images of U2OS cells stably expressing MRTFA-GFP, plated on 6 or 81 kPa polyacrylamide gels or glass. 10 μm scale bars. (b) The nuclear accumulation of MRTFA-GFP increases significantly with substrate stiffness, a trend that is abolished when treating cells with latrunculin B. Data are from three independent experiments (DMSO: 6 kPa, n = 167; 81 kPa, n = 166; glass n = 162. Lat B: 6 kPa, n = 164; 81 kPa, n = 145; glass, n = 159). Two-way ANOVA, NS P > 0.05, ** P ≤ 0.01, and **** P ≤ 0.0001. (c) Analysis of nuclear heights via measuring the full-width half-maximum of fluorescent intensity on vertical sections through XZ sum projections of DAPI-stained nuclei. The inset shows representative XZ projections (top, 6 kPa; bottom, 81 kPa). Nuclear heights are significantly increased on 6 kPa gels compared to 81 kPa gels, concluding that the nuclear envelope is more deformed on stiffer substrates. 5 μm scale bar. Data are from two independent experiments (6 kPa, n = 49; 81 kPa, n = 26). Two-tailed t-test, **** P≤ 0.0001. (d) Representative epifluorescence image gallery of a U2OS cell stably expressing MRTFA-GFP, showing MRTFA nuclear translocation triggered by the application of 1 nN by a pyramidal AFM tip. (e) MRTFA nuclear translocation occurs in a force-dependent manner. Data points represent the mean ± s.e.m. of multiple experiments (0.5 nN, n = 7; 1 nN, n = 15; 5 nN, n = 10). (f) Localized nuclear deformation is induced by the AFM tip, as confirmed by vertical image slices of NLS-mCherry, dashed lines represent an estimation of the (diagonal) tip profile. (g) Averaged (mean ± s.e.m.) time courses of MRTFA nuclear translocation in U2OS cells, following serum stimulation, comparing the dynamics of MRTFA-Ig27WT-BFP and MRTFA-Ig27V11P-YFP when cells are plated on glass or 6 kPa gel substrates, revealing that, although nuclear accumulation is significantly decreased on 6 kPa gels (mostly due to a 4-fold increase in the export rate constant (Fig. S11)), the mechanical selectivity to the translocating protein remains unaltered. Data are from three independent experiments (glass, n = 15; 6 kPa gel, n = 13), only a representative error bar is shown per condition.
Hence, akin to the case of YAP39, it is very likely that mechanical deformation of the nuclear envelope induces a conformational change in the mechanically-stretched NPCs that favours nuclear import (or hinders nuclear export). We therefore speculated that nuclear transport could also be finely regulated by a more localized deformation of the nuclear envelope with a pyramidal AFM tip. To this end, we conducted experiments whereby a constant force was externally applied perpendicular to the cell nucleus. Application of 500 pN for 5 minutes resulted in a time-dependent accumulation of MRTFA (Fig. 3D). Increasing the pushing force up to 1 nN and 5 nN further promoted nuclear translocation (Fig. 3E). Image slices transecting the AFM tip (Fig. 3F) confirmed that larger forces resulted in an increasingly deformed nucleus, likely resulting in a mechanical perturbation of the NPCs. Even under high force conditions (5 nN) nuclei were not ruptured, as confirmed by the absence of cytoplasmic NLS-mCherry (data not shown). In contrast to YAP, we did not observe an increase of cytoplasmic accumulation of MRTFA upon withdrawal of the pushing force. Altogether, these experiments confirmed the presence of two complementary mechanical mechanisms that regulate MRTFA translocation across the NPC; in the first, external forces (directly or indirectly) applied to the nuclear envelope regulate nuclear entry upon elastically deforming the NPC (ref 39 and Fig. 3E), and a second mechanism characterised here where forces applied by the NPC modify nuclear entry according to the mechanical properties of the translocating protein (Figs. 1I & 2C). To further investigate the interplay between both mechanisms, we postulated that, if soft substrates result in rounder and less-distorted nuclei, their NPCs should be less stressed and feature a more ‘closed’ conformation, hence resulting in slower translocation. To test our hypothesis, we performed live-cell imaging experiments to measure the dynamics of nuclear translocation of the MRTFA-Ig27WT-BFP and MRTFA-Ig27V13P-YFP constructs in U2OS cells plated on 6 kPa soft gels (Fig. 3G). Comparison of the dynamics of nuclear translocation to that obtained for the same constructs when cells were plated on stiff (glass) substrates reveals that, on soft substrates nuclear accumulation is reduced, mainly due to an increase in the export rate constant (Fig. S13). However, the mechanical-stability-selectivity persisted in soft gels, suggesting that the combination of both complementary mechanosensitive roles of the NPC is likely to regulate, at any given time, both the rate and extent of MRTFA nuclear accumulation.
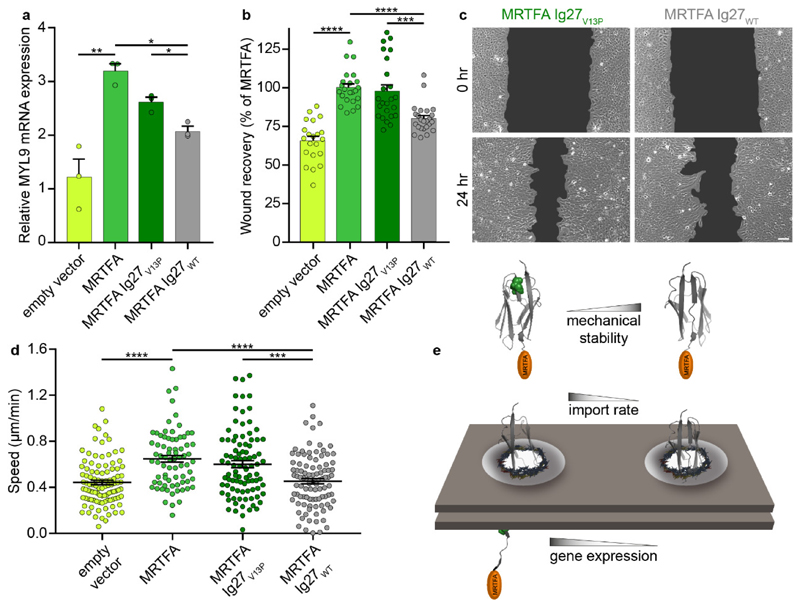
Given that MRTFA regulates serum response factor (SRF)-related genes6, we then posed the question of whether variations in the mechanical stability of the translocating MRTFA constructs had a direct impact on gene expression. Previous work reported that the myosin light chain 9 gene (MYL9), responsible for cell migration and cancer invasion, is regulated by MRTFA/SRF6,53. We conducted quantitative polymerase chain reaction (qPCR) experiments on stable U2OS cell lines expressing MRTFA-GFP and measured, 4 hours after serum stimulation, a large increase in the expression of MYL9 compared to cells stably expressing a GFP-empty vector (Fig. 4A). In addition, tagging MRTFA with GFP does not compromise SRF-mediated activity. Most importantly, cells stably expressing MRTFA-Ig27V13P-GFP exhibited increased levels of MYL9 mRNA expression compared to a MRTFA-Ig27WT-GFP stable cell line (Fig. 4A). Triggered by these observations, we then hypothesized that the variations in gene expression could potentially translate into functional consequences at the cellular level. Since MRTFA upregulates MYL9 and a variety of genes involved in cell migration, we postulated that the cells expressing MRTFA constructs tagged with mechanical-labile proteins —which feature an enhanced nuclear accumulation—would migrate faster than those cells where MRTFA was tagged with a mechanically-stable domain. To test this premise, we designed a wound-healing assay to compare, after 24 hours, the wound recovery of the U2OS stable cell lines described above. These experiments showed that the wound recovery of cells expressing the mechanically-labile MRTFA-Ig27V13P-GFP construct was significantly higher than that of cells expressing the mechanically-stable MRTFA-Ig27WT-GFP (Figs. 4B, 4C & S14). Finally, we conducted cell motility assays on the highly-metastatic MDA-MB-231 cells6, and tracked, during 13 hours, the migration speed of individual cells transfected with different MRTFA constructs (Fig. S14). In this case we also observed that cells transfected with the MRTFA construct harbouring the Ig27V13P domain moved significantly faster than those containing Ig27WT (Fig. 4D). Combined, these assays demonstrate that very subtle changes in the mechanical stability of translocating proteins have large effects at gene expression level, ultimately controlling the motility of two cancer cell lines (Fig. 4E).
Figure. 4. Mechanically-stable MRTFA constructs downregulate gene expression and cellular motility.
(a) Real-time quantitative PCR in U2OS cells stably expressing a GFP empty vector, MRTFA-GFP, MRTFA-Ig27WT-GFP or MRTFA-Ig27V13P-GFP, 4 hours after serum stimulation. MYL9 gene expression is significantly increased when MRTFA is tagged with the mechanically-labile Ig27V13P domain compared to the mechanically-stable I27WT. Data are from three independent experiments, bars show mean ± s.e.m. (b) Wound-healing assays on U2OS stable cell lines show that wound healing is significantly slower in cells expressing MRTFA-Ig27WT-GFP compared to MRTFA-Ig27V13P-GFP. Data are from three independent experiments (GFP empty vector, n = 21; MRTFA-GFP, n = 23; MRTFA-Ig27V13P-GFP, n = 24; MRTFA-Ig27WT-GFP, n = 24), bars show mean ± s.e.m. (c) Representative bright-field images of wound healing of MRTFA-Ig27V13P-GFP and MRTFA-Ig27WT-GFP stable cell lines. 100 μm scale bar. (d) Motility assays of MDA-MB-231 cells showing that the migration speed of cells transfected with MRTFA-Ig27V13P-GFP is significantly higher than those transfected with MRTFA-Ig27WT-GFP. Data are from three independent experiments (GFP empty vector, n = 101; MRTFA-GFP, n = 74; MRTFA-Ig27V13P-GFP, n = 92; MRTFA-Ig27WT-GFP, n = 98), bars show mean ± s.e.m. All statistical tests are two-tailed t-tests, * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, and **** P ≤ 0.0001. (e) Schematic representation of the proposed nuclear import mechanism by which the mechanical properties of the translocating protein regulate their nuclear shuttling dynamics.
Discussion
While recent structural advances have provided invaluable information on the composition and spatial arrangement of the multifaceted NPC machinery16,24,54, dynamic measurements of the NPC have proved challenging, especially due to the intrinsically-disordered and highly-mobile nature of the unstructured FG-Nup domains. Pioneering studies using AFM imaging measured the nanoscopic spatiotemporal dynamics of FG-Nups29, and probed the nanomechanical repulsion of the NPC central channel55. However, direct measurement of molecular transport at the nanoscale has lagged significantly behind. Our live-cell imaging experiments, inspired by our in-vitro single molecule nanomechanics results, have enabled new mechanistic insights into an elusive NPC-mediated mechanotransduction pathway. Our multiscale experiments revealed that transport across the NPC inversely scales with the mechanical properties of the shuttling protein.
This scenario is compatible with the current models of nucleocytoplasmic transport, both from a physical and chemical perspective. From a polymer physics view, mechanically-unfolded conformations might be able to surf the intricate and dense NPC FG-Nup ‘forest’ more efficiently than the sterically-bulkier folded counterpart. This picture would be compatible for example with the selective phase model27, whereby the ‘adaptive’ barrier would not need to dramatically open to let the ‘floppier’ mechanically-unfolded protein through. Similarly, within the FG-gel context27, it is easy to rationalise that unfolded proteins are likely to be more efficient at crossing smaller mesh sizes, thereby minimising transient kinetic traps. Considering the interactions established during nuclear shuttling, a very recent elegant report demonstrated that the nature of the surface chemical properties of proteins determines their passage rates across the NPC56; while negative residues and lysines hinder passage, hydrophobic residues, certain polar residues and arginines dramatically increase the passage efficiency as a result of the favourable interactions established with the FG-Nups. Although this rationale explains the high hydrophobic content of the nuclear transport receptors, it also applies to the nature of the translocating protein cargos56. In particular, folded globular proteins tend to bury their hydrophobic core from the solvent. Hence, we postulate that upon mechanical unfolding, the otherwise cryptic hydrophobic residues will be readily exposed, facilitating their interaction with the dynamic FG-Nups and resulting in an overall increase of their translocation rate. Of note, while our experiments show that those proteins with lower mechanical stability translocate faster, it is likely, as observed in the slow nuclear translocation of the Spy0128 domain of pilin (Fig. S6), that, while speeding up the process, mechanical unfolding might be not mandatory26 to cross the NPC. Lacking a specific ATP-dependent machinery, it remains unresolved what is effectively pulling on the translocating proteins. While the highly-dynamic Nups seem the first candidates to attach and pull from one of the protein’s termini, given the large (~30) number of different FG-Nup copies, it is difficult to anticipate the precise FG-Nup(s) responsible for the mechanical interaction. An educated structurally-based guess would suggest that those Nups forming the cytoplasmic ring, such as the Nup214–Nup88–Nup62 complex, or even the central Nup358, might play a crucial role24. Future work based on the systematic individual knock-down of the major Nup families will help shed direct light into this question.
A central discovery in our experiments is that the import rate constant of the different constructs (Figs. 1I & 2C) displays an exponential dependency with the unfolding force measured with the AFM when stretched at 400 nm s-1. Furthermore, the strong force dependency of the import process contrasts with the force independency observed in the export process. This implies that, regardless of any molecular consideration, import is barrier-activated whereas export is not. To further delve into this question, we measured the rate of nuclear localization of MRTFA-GFP at different temperatures, spanning 28-37°C (Fig. S15), confirming that, while import is barrier activated (with a considerable associated activation barrier of ~21 kBT at 37 °C), export is barrier-less. As a note of caution, it is possible that the binding of nuclear G-actin to MRTFA might affect the absolute value for the export kinetics14,57.
In conclusion, our experiments demonstrate that, in addition to protein size58,59 and surface properties56, mechanical stability emerges as an additional intrinsic property of proteins capable of regulating their dynamics of nuclear translocation. Our experimental approach integrating single molecule results into the cellular context, and with knock-on effects at the functional level, explicitly defines new functions of the NPC as an effective nuclear mechanosensor60 that actively senses mechanical stress. From a more applied perspective, our findings could be tested in vitro using artificial nuclear pore complexes61,62, and potentially used in the design of new molecular approaches aimed at externally modifying the mechanical stability of target transcription factors to selectively regulate nuclear localization and gene expression on demand.
Materials and methods
Plasmid constructs and (poly)protein engineering
All genes and consumables were obtained by Thermo Fisher Scientific unless stated otherwise. The MRTFA-GFP vector was kindly provided by Maria Vartiainen, and the pEBFP2-N1 and pEYFP-N1 vectors were kindly provided by Maddy Parsons. pEBFP2-C1, pmCherry-NLS, and pmCherry-RanQ69L were obtained from Addgene. Ig27 mutants were either created by site-directed mutagenesis by PCR or were ordered and subcloned into the pEGFP-MRTFA vector. The Spy0128 domain of pilin, R16 domain of spectrin, Ig27WT, and Ig27V11P were cloned using Kpnl restriction enzymes. Ig27V15P, Ig27V13P, and Ig27Y9P were cloned using Sall and Kpnl restriction enzymes. Ig1 was subcloned from pQE80L, using PCR amplification with the addition of SalI and KpnI restriction sites, into MRTFA-GFP. MRTFA-X-GFP (X= Ig27WT, Ig27V11P, Ig27V13P, Ig27V15P or Ig27Y9P) were subcloned into pEBFP2-N1 and pEYFP-N1 between HindIII and KpnI restriction sites by PCR amplification to incorporate an additional glycine residue in the coding sequence in order to remain in reading frame with the fluorescent tag. Ig32 was ordered with SalI and KpnI restriction sites for cloning into pEBFP2-N1 and pEYFP-N1 vectors and also incorporated an additional serine residue. Recombinant plasmids were transformed in XL1Blue competent cells (Agilent Technologies). Selected colonies were grown in Luria broth (LB) supplemented with 100 mg/ml kanamycin at 37 °C. Cells were lysed and plasmid DNA was purified prior to transfection using a Qiagen kit according to the manufacturer’s instructions. Immunoglobulin monomers for differential scanning fluorimetry (Ig27WT, Ig27V11P, Ig27V13P, Ig27V15P, and Ig27Y9P) were subcloned into pQE80L vector (Qiagen) between BamHI and KpnI restriction sites. Immunoglobulin polyproteins for single-molecule force spectroscopy, (Ig1-Ig27C47A-C63A)4, (Ig27WT)8, and (Ig32)8, all containing two additional cysteine residues, were constructed using BamHI, BglII and KpnI restriction sites. Constructs were expressed in E. coli BLR(D3) cells (Novagen). Cells were grown in LB supplemented with 100 mg/ml ampicillin at 37 °C. After reaching an OD600 of ~0.6, cultures were induced with 1 mM isopropyl-β-D-thiogalactopyranoside and grown at 25 °C for 16 hours. Cells were disrupted by French press and purified by Talon affinity resin (Clontech) using wash buffer (50 mM phosphate buffer, 300 mM NaCl, 20 mM imidazole, pH 7.0) and elution buffer supplemented with 250 mM imidazole. This was followed by gel filtration using a Superdex 200 10/300 GL column (GE Biosciences). Proteins were stored in PBS pH 7.3 at 4 °C.
Cell culture, stable and transient transfection, and drug treatment
U2OS (American Type Culture Collection) and MDA-MB-231 (kindly provided by Maddy Parsons) cells were grown in complete media – DMEM high glucose (Sigma-Aldrich) supplemented with 10% foetal bovine serum (FBS, Sigma-Aldrich), 100 U/ml penicillin, 100 mg/ml streptomycin, and 2 mM glutamine (Invitrogen). For transient expression, U2OS and MDA-MB-231 cells were transfected with 1 μg of constructs using Fugene (Promega) according to the manufacturer’s protocol. U2OS cells stably expressing GFP, MRTFA-GFP, MRTFA-Ig27WT-GFP or MRTFA-Ig27V13P-GFP were generated by transfection using Fugene, followed by selection with 500 μg/ml geneticin (Thermo Fisher Scientific). After FACS sorting, polyclonal cell lines were obtained and cultured in complete media supplemented with 500 μg/ml geneticin. Cells were starved by withdrawing serum and incubating in serum-starved medium containing 0.3% FBS for 24 hours. Starved cells were stimulated with 15% serum for the indicated time. Where indicated, U2OS cells were treated with 20 nM leptomycin B (Sigma-Aldrich) for 1 hour during live imaging experiments and for 4 hours for cells plated on polyacrylamide gels, 100 nM latrunculin B (Abcam) for 40 minutes, or 40 μM importazole (Abcam) for 1 hour.
Live cell imaging and quantification
Cells transfected with the indicated constructs were seeded and incubated overnight onto glass-bottom dishes. Cells were then serum starved for 24 hours. For live imaging acquisition, cells were stimulated with 15% FBS or treated with the drugs indicated. Images were acquired with a Nikon A1R confocal microscope with a 60× NA 1.40 oil objective and laser and emission filter wavelengths of 405 nm and 450/50 nm respectively for BFP, 488.2 nm and 540/30 nm respectively for GFP and YFP, and 561.9nm and 595/50 nm respectively for mCherry. The microscope was operated with the Nikon Perfect Focus System and controlled by NIS Elements software. Images were typically acquired every 30 seconds during 30 or 60 minutes. The microscope was enclosed in a Solent Scientific environmental chamber with temperature and CO2 control. Live cell images were processed to quantify MRTFA localisation using a custom-made MATLAB (MathWorks) script. First, the background was measured and subtracted. Next, the outlines of the whole cell and the nucleus were defined by the user to determine the total fluorescent intensities of the cell and nucleus. The nuclear intensity was divided by the cell intensity to provide a measure of the nuclear MRTFA fraction. This procedure was repeated for all frames in an imaging sequence to obtain the time-dependent behaviour of the nuclear MRTFA fraction.
Kinetic analysis
For each stimulated cell, the time-dependence of the nuclear MRTFA fraction, n(t), is fit to
where n0 and ne are the initial and equilibrium nuclear MRTFA fractions respectively, and k is the total rate constant (for simplicity we refer to ne as the equilibrium accumulation). For data from a stimulated cell to be considered ‘good’, fitting parameters must obey the criteria 0 ≤ n0 ≤ ne ≤ 1 and k ≥ 1 /T where T is the time duration over which the fit is applied. For U2OS cells, t = 0 is defined as 2 minutes after serum stimulation, such that the initial transient behaviour (before stimulation is fully activated) is not fitted. For MDA-MB-231 cells, which respond slower to serum, the definition of t = 0 rises to 5 minutes after stimulation. Before averaging the time courses of individual cells for a given condition, any variations in n0 between cells (which is not of interest) must be corrected for in a way that does not alter ne or k (which are of interest). This correction is performed by subtracting n0e−kt from the nuclear MRTFA fraction data to give a corrected nuclear MRTFA fraction, ñ(t). Assuming pseudo-first-order behaviour (see supplementary information), an equivalent mathematical description of the translocation kinetics is given by import and export rate constants, kI and kE respectively, which relate to the equilibrium accumulation and total rate constant via ne = k1/(kI + kE) and k = kI + kE. As such, there are two equivalent fittings to the corrected nuclear MRTFA fraction:
Final values of these parameters-of-interest are obtained from the fitting of averaged time courses. Errors in these values are given by the standard error of fitting parameters obtained from the fitting of the time courses from individual cells.
Single molecule force spectroscopy
Single molecule force spectroscopy experiments were conducted using both a home-made and a Luigs and Neumann force spectrometer operating at room temperature, as described previously63. Samples were prepared by depositing 0.5–5 μl of protein (at a concentration of 1–5 mg/ml in PBS pH 7.3) onto a gold-coated cover slide which had been plasma cleaned for 10 minutes prior to deposition. Each cantilever (MLCT-C, Bruker) was calibrated using the equipartition theorem, giving a typical spring constant of ~12–18 pN/nm. To pull/unfold an individual protein, the cantilever tip was first pushed against the surface at high force (2 nN) to promote non-specific adhesion between the protein and the tip. The tethered protein was subsequently pulled by retracting the cantilever at a constant velocity of 400 nm/s relative to the substrate. All data was recorded and analysed using custom-written software in Igor Pro (WaveMetrics). For the octomeric polyproteins (Ig27WT)8 and (Ig32)8 only traces showing the signature of at least 5 unfolding events were selected for analysis. For the tetrameric chimera polyprotein (Ig1–Ig27C47A-C63A)4 only trajectories displaying at least 2 Ig27C47A-C63A unfolding events (hallmarked by an increment of ~28 nm at ~200 pN) were analysed. There is no significant difference between the unfolding forces of Ig27WT and Ig27C47A-C63A (Fig. S4). Each protein unfolding event was fitted with the worm-like chain model of polymer elasticity to obtain the increment in contour length.
Differential Scanning Fluorimetry
Differential scanning fluorimetry (DSF) experiments were performed to find the melting temperatures of Ig27 mutants (Ig27V13P, Ig27V11P, Ig27V15P, Ig27WT, and Ig27Y9P) using a Stratagene Mx3005P Real-Time (RT) qPCR System. The process of thermally-induced unfolding was monitored by measuring the fluorescence of SYPRO Orange dye (Thermo Fisher Scientific) with the excitation and emission wavelengths of 587 nm and 607 nm respectively. Data were collected at 1 °C intervals from 25 °C through to 95 °C with a heating rate of 1 °C/min. Preliminary tests were carried out to find the optimal combination of protein concertation (Ig27WT used) and dye dilution. At the recommended concentrations, the fluorescence signal increase, which accompanied thermally-induced unfolding, was too low to produce reliable results. This was due to the small nature of Ig domains in question, and thus the sparsity of hydrophobic residues (to which the dye binds to become fluorescent) being exposed upon unfolding. An optimal signal-to-noise ratio was achieved with a 50 μM protein concentration and 1:500 dye dilution in 50 mM PBS pH 7.3 with 300 mM NaCl. As such, for each protein, 240 μl of 100 μM protein and 240 μl of 1:250 diluted dye were mixed and aliquoted into 12 wells (40 μl per well) of a 96-well, non-skirted, low-profile, 0.2 ml PCR plate (Thermo Fisher Scientific), sealed with an optical adhesive cover (Applied Biosystems). As such, for each Ig27 mutant, 12 DSF melting curves were obtained under identical conditions. To find the melting temperature of a given Ig27 mutant, each individual curve was fit to a sigmoidal function between its minimum and maximum fluorescence values and the temperature at the sigmoid midpoint was taken as the melting temperature for that curve. The quoted melting temperature is then the mean ± s.e.m. from these 12 measurements. For a given Ig27 mutant, averaged DSF melting curves were obtained after normalising each individual curve by setting the area under the curve to unity48.
Polyacrylamide gel preparation and characterisation
All reagents were obtained from Sigma-Aldrich unless specified otherwise. Polyacrylamide gels were prepared on 22 mm square glass coverslips or 35 mm glass-bottom dishes as previously described64. Glass surfaces were silanized by sequential coverage of 0.1 M sodium hydroxide for 5 minutes followed by 0.5% (3-aminopropyl)trimethoxysilane for 30 minutes. After washing for 30 minutes in running dH2O, glass surfaces were covered with 0.5% glutaraldehyde in PBS for 30 minutes then washed for a further 10 minutes. A solution containing 0.1% ammonium persulfate, 0.1% TEMED (N,N,N’,N’-tetramethylethylenediamine), and 10 mM HEPES pH 7.0 was then mixed with 4%/0.17% acrylamide/bis-acrylamide for 6 kPa gels or 10%/0.43% acrylamide/bis-acrylamide for 81 kPa gels, at which point gel polymerisation starts. Swiftly, 35 μl (for coverslips) or 20 μl (for dishes) from the generated solution was placed on the glass surface and covered with a 25 mm round glass coverslips. After 30 minutes polymerisation is complete and the round coverslips were removed and gels were washed with 50 mM HEPES. The gel surface was cross-linked by coverage of 0.5 mg/ml Sulfo-SANPAH (sulfosuccinimidyl 6-(4’-azido-2’-nitrophenylamino)hexanoate), Fisher Scientific) in 10 mM HEPES pH 7.0 and UV exposure (7500 μJ/cm2, 254 nm). Excess Sulfo-SANPAH was removed by washing with 50 mM HEPES. Cross-linked gels were incubated with 25 μg/ml fibronectin (FN) in PBS overnight at 4 °C or for 2 hours at 37 °C. The stiffness (Young’s modulus) of the gels was measured by nanoindentation using a Bruker BioScope Resolve AFM. Silicon nitride cantilevers, with a nominal spring constant of 0.03 nN/nm, with four-sided pyramidal tips, with an effective semi-included angle of 18°, nominal tip radius of 20 nm, and minimal tip height of 2.5 μm, were used (MLCT-D, Bruker). The actual spring constant was calibrated by thermal tuning in MilliQ water with the simple harmonic oscillator model fit in the NanoScope software. For each gel sample, four positions near the centre were selected, where 64 (8×8) or 256 (16×16) force-displacement curves were recorded in an area of 50×50 μm2, with a ramp size and speed of 6 μm and 6 μm/s respectively. To calculate the Young’s modulus, force-displacement data were fit to a linearized form of the force-indentation relationship for a four-sided pyramidal tip65:
where F is the force measured by the AFM, E is the Young’s modulus of the gel, θ is the semi-included angle of the AFM tip, v = 1 /2 is the Poisson’s ratio of the gel (assuming the gels are incompressible), and δ is the indentation of the AFM tip into the gel. For each acrylamide/bis-acrylamide-ratio gel, a fixed indentation range with a lower bound large enough to negate the finite tip radius and an upper bound small enough to not exceed the tip height was used. Ten gels, produced in six batches, were measured for each acrylamide/bis-acrylamide ratio. The Young’s modulus of each gel sample was obtained by taking the mean of all individual Young’s moduli obtained for that gel (one per each force-displacement curve). The final values represent the mean ± s.e.m. of these ten means. For 4%/0.17% acrylamide/bis-acrylamide gels the Young’s modulus was found to be 5.6 ± 0.4 kPa (hence we refer to these as 6 kPa gels). For 10%/0.43% acrylamide/bis-acrylamide gels the Young’s modulus was found to be 80.9 ± 6.8 kPa (hence we refer to these as 81 kPa gels).
Immunostaining and fixed-cell imaging and quantification
Gels and coverslips (both FN coated) were washed with PBS and incubated for 30 minutes with complete media at 37 °C prior cell seeding. Cells were left 4 hours to adhere and spread on the gels/coverslips. Cells were fixed with 4% paraformaldehyde (Alfa Aesar) for 10 minutes, then permeabilised with 0.1% Triton X-100 (Sigma-Aldrich) for 10 minutes. Cells were stained with 1:250 Alexa Fluor 546 Phalloidin (Thermo Fisher Scientific) in PBS for 1 hour at room temperature and 1:3000 DAPI (4’,6-diamidino-2-phenylindole, Sigma-Aldrich) in MilliQ water for 3 minutes before mounting on slides using ProLong Glass Antifade Mountant (Thermo Fisher Scientific). Fluorescent images were taken using either a Nikon Eclipse Ti inverted epifluorescence microscope or a Nikon A1 inverted confocal microscope with a 60x NA 1.40 oil immersion objective. Fluorescent images were analysed using ImageJ software. After background subtraction, the cell outline was user-defined from the GFP image whereas the nuclear outline was user-defined from the DAPI image. Then the nuclear GFP intensity was divided by the whole cell GFP intensity to provide a measure of the nuclear MRTFA fraction.
Mechanical perturbation of cells using an AFM
U2OS cells stably expressing MRTFA-GFP or co-expressing MRTFA-GFP and NLS-mCherry were plated on a glass-bottom dishes (WillCo Wells) and starved for 24 hours. Experiments were performed using a Bruker BioScope Resolve AFM. Silicon nitride cantilevers, with a nominal spring constant of 0.6 N/m, with four-sided pyramidal tips, with semi-included front, back, and side angles of 35° (and, as such, a semi-included diagonal angle of ~45°), nominal tip radius of 20 nm, and minimal tip height of 3.2 μm, were used (MLCT-Bio-DC Tip F, Bruker). These probes are designed to have a low thermal sensitivity to minimize the thermal influence of fluorescent excitation. The actual spring constant was calibrated by thermal tuning in MilliQ water with the simple harmonic oscillator model fit in the NanoScope. During acquisition, cells were maintained at 37 °C with a combination of stage and objective heaters, with the medium buffered with 50 mM HEPES. The Resolve AFM is incorporated with a Nikon Eclipse Ti inverted epifluorescence microscope with a 60x NA 1.40 oil immersion objective which allows the visualization of fluorescent cells. Cells with cytoplasmic MRTFA-GFP were identified and constant forces (0.5 nN to 5 nN, using z-feedback control) were applied perpendicular to the cell nucleus for 5 minutes. NLS-mCherry expression was used to monitor nuclear envelope integrity, which was confirmed by nuclear localization of this fluorescent signal. Images were acquired every 2 minutes for 30 minutes using an Andor iXon Ultra 888. For dual fluorescence acquisition, to avoid mechanical noise of filter cube turret rotation, an OptoSplit II Bypass Image Splitter (Cairn Research) was implemented, allowing the simultaneous acquisition of two different optical wavelengths images (here GFP and mCherry) on either side of the camera sensor. To assess the force-dependent nature of nuclear deformation by the AFM tip, image stacks were acquired with a tip applying different applied forces when positioned above a nucleus. GFP and mCherry image stacks were acquired with a z-step and range of 0.4 μm and 30 μm respectively. Deconvolution of these epifluorescence image stacks was accomplished using the DeconvolutionLab2 and PSF Generator plug-ins for ImageJ66,67. 200 iterations of the Richardson-Lucy algorithm were carried out on each stack using a Born and Wolf point spread function with n = 1.515, NA = 1.40, and λ = 519 nm (for GFP) and λ = 624 nm (for mCherry). Following deconvolution, image slices transecting the AFM tip perpendicular to the z-plane were obtained using the Volume Viewer plug-in for ImageJ.
RNA extraction and RT qPCR
RNA from U2OS cells was extracted on ice using RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions and treated with DNase (Invitrogen). RNA concentration was determined and purity was checked by measuring the A260/A280 ratio. Relative mRNA expression of genes was determined by RT qPCR assay using SYBR-Green detection chemistry (Agilent) and the ViiA7 RT qPCR System (Applied Biosystems). The primers used for MYL9 were CATCCATGAGGACCACCTCCG/CTGGGGTGGCCTAGTCGTC6. Relative abundances of template cDNA were calculated by the comparative CT (ΔΔCT) method. Each sample was normalized to the level of a GAPDH (PPH00150F-200 qPCR, Qiagen).
Wound healing assay
U2OS cells stably expressing a GFP empty vector, MRTFA-GFP, MRTFA-Ig27WT-GFP, or MRTFA-Ig27V13P-GFP were seeded onto 6-well plates and incubated overnight at 37 °C. After reaching 80% confluence, cells were first serum starved for 24 hours and then stimulated with 10% serum for other 24 hours. Scratch wounds were made by scraping a 10 μl pipette tip across each cell layer68. For each condition regions were imaged at time zero, to record the initial area of the wounds, and at 24 hours, using a Nikon Ti2 microscope with a 10x NA 0.3 and a Nikon DS-Qi2 camera, controlled by NIS Elements software. Wound recovery was calculated by measuring the change in the wound area from time zero to 24 hours, these areas were user-defined in ImageJ. Experiments were performed independently three times, evaluating eight regions within each scratch. Final values are normalised with respect to MRTFA-GFP.
Random migration assay
MDA-MB-231 transiently transfected with a GFP empty vector, MRTFA-GFP, MRTFA-Ig27WT-GFP, or MRTFA-Ig27V13P-GFP, were plated on FN-coated 6-well plates in serum-starved medium for 16 hours and the stimulated with 10% serum for other 24 hours. Cells were then imaged using a Nikon Ti2 microscope with a 10x NA 0.3 and a Nikon DS-Qi2 camera, controlled by NIS Elements software. In order to visualize and track successfully transfected cells, a GFP fluorescence image was acquired at time zero. Bright field images were then acquired every 5 minutes for 13 hours. Cells were tracked and their migration speed was determined using the Manual Tracking plug-in for ImageJ.
Supplementary Material
Acknowledgements
We thank Maria Vartiainen (University of Helsinki) for sharing the MRTFA-GFP plasmid and Maddy Parsons (King’s College London) for sharing the pEBFP2-N1 and pEYFP-N1 vectors and the MDA-MB-231 cell line. We thank Alexander Dulebo (Bruker) for assistance in setting up cell mechanics experiments. We thank the Nikon Centre at King’s College London for invaluable assistance in setting up the cell imaging experiments. We wish to thank Guang Yang for help in qPCR analysis, and Charlie Nichols and Sasi Conte (King’s College London) for help with differential scanning fluorimetry experiments. A.E.M.B. was funded by an EPSRC DTP fellowship. V.S.R. was funded by the BHF Centre for Research Excellence at King’s College London. This work was supported by the BHF grant (PG/13/50/30426), the European Commission (Mechanocontrol, grant agreement SEP-210342844), EPSRC Fellowship K00641X/1, EPSRC Strategic Equipment Grant (EP/M022536/1), the Leverhulme Trust Project Grant (RPG-2015-225) and by the Leverhulme Trust Research Leadership Award (RL-2016-015) to S.G-M.
Footnotes
Author contributions. S.G.-M. conceived the research. E.I. designed and performed cell biology, live-cell imaging experiments, motility assays and qPCR experiments. A.S. designed and performed live-cell imaging analysis and kinetic modelling. E.I., A.S. and S.G.-B. performed single-cell mechanical experiments. A.E.M.B. and Y.J.W. conducted single molecule nanomechanical experiments and A.E.M.B analysed data. P.R-L., S.J.B. and A.L. performed molecular biology work. E.R. with A.S. conducted and analysed differential scanning fluorimetry experiments. F.P. and V.S.R conducted polyacrylamide gel experiments. C.M.S.and P.R-C. participated in data discussion. S.G.-M. wrote the paper with contribution from E.I. and A.S.
Competing financial interests: The authors declare no competing financial interests.
Data Availability
Data supporting this research can be obtained from the corresponding author upon reasonable request.
References
- 1.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 2.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 3.Markiewicz E, et al. The inner nuclear membrane protein emerin regulates beta-catenin activity by restricting its accumulation in the nucleus. EMBO J. 2006;25:3275–3285. doi: 10.1038/sj.emboj.7601230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moon HS, Even-Ram S, Kleinman HK, Cha HJ. Zyxin is upregulated in the nucleus by thymosin beta4 in SiHa cells. Exp Cell Res. 2006;312:3425–3431. doi: 10.1016/j.yexcr.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 5.Fedorchak GR, Kaminski A, Lammerding J. Cellular mechanosensing: getting to the nucleus of it all. Prog Biophys Mol Biol. 2014;115:76–92. doi: 10.1016/j.pbiomolbio.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol. 2009;11:257–268. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain N, Iyer KV, Kumar A, Shivashankar GV. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc Natl Acad Sci U S A. 2013;110:11349–11354. doi: 10.1073/pnas.1300801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connelly JT, et al. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat Cell Biol. 2010;12:711–718. doi: 10.1038/ncb2074. [DOI] [PubMed] [Google Scholar]
- 10.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 11.Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature. 2013;497:507–511. doi: 10.1038/nature12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pawlowski R, Rajakyla EK, Vartiainen MK, Treisman R. An actin-regulated importin alpha/beta-dependent extended bipartite NLS directs nuclear import of MRTF-A. EMBO J. 2010;29:3448–3458. doi: 10.1038/emboj.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mouilleron S, Langer CA, Guettler S, McDonald NQ, Treisman R. Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator. Sci Signal. 2011;4:ra40. doi: 10.1126/scisignal.2001750. [DOI] [PubMed] [Google Scholar]
- 14.Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- 15.Baarlink C, Wang H, Grosse R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science. 2013;340:864–867. doi: 10.1126/science.1235038. [DOI] [PubMed] [Google Scholar]
- 16.Kim SJ, et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature. 2018;555:475–482. doi: 10.1038/nature26003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maillard RA, et al. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell. 2011;145:459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Singlemolecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olivares AO, Kotamarthi HC, Stein BJ, Sauer RT, Baker TA. Effect of directional pulling on mechanical protein degradation by ATP-dependent proteolytic machines. Proc Natl Acad Sci U S A. 2017 doi: 10.1073/pnas.1707794114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cordova JC, et al. Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell. 2014;158:647–658. doi: 10.1016/j.cell.2014.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez-Larrea D, Bayley H. Multistep protein unfolding during nanopore translocation. Nat Nanotechnol. 2013;8:288–295. doi: 10.1038/nnano.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maimon T, Elad N, Dahan I, Medalia O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure. 2012;20:998–1006. doi: 10.1016/j.str.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 23.Lin DH, et al. Architecture of the symmetric core of the nuclear pore. Science. 2016;352:aaf1015. doi: 10.1126/science.aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Appen A, et al. In situ structural analysis of the human nuclear pore complex. Nature. 2015;526:140–143. doi: 10.1038/nature15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemke EA. The Multiple Faces of Disordered Nucleoporins. J Mol Biol. 2016;428:2011–2024. doi: 10.1016/j.jmb.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grunwald D, Singer RH. Multiscale dynamics in nucleocytoplasmic transport. Curr Opin Cell Biol. 2012;24:100–106. doi: 10.1016/j.ceb.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt HB, Gorlich D. Transport Selectivity of Nuclear Pores, Phase Separation, and Membraneless Organelles. Trends Biochem Sci. 2016;41:46–61. doi: 10.1016/j.tibs.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Rout MP, Aitchison JD, Magnasco MO, Chait BT. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 2003;13:622–628. doi: 10.1016/j.tcb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 29.Lim RY, et al. Nanomechanical basis of selective gating by the nuclear pore complex. Science. 2007;318:640–643. doi: 10.1126/science.1145980. [DOI] [PubMed] [Google Scholar]
- 30.Frey S, Richter RP, Gorlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- 31.Yamada J, et al. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol Cell Proteomics. 2010;9:2205–2224. doi: 10.1074/mcp.M000035-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters R. Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality. Traffic. 2005;6:421–427. doi: 10.1111/j.1600-0854.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- 33.Lim RY, et al. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc Natl Acad Sci U S A. 2006;103:9512–9517. doi: 10.1073/pnas.0603521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher TE, Oberhauser AF, Carrion-Vazquez M, Marszalek PE, Fernandez JM. The study of protein mechanics with the atomic force microscope. Trends Biochem Sci. 1999;24:379–384. doi: 10.1016/s0968-0004(99)01453-x. [DOI] [PubMed] [Google Scholar]
- 35.Mehlin H, Daneholt B, Skoglund U. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell. 1992;69:605–613. doi: 10.1016/0092-8674(92)90224-z. [DOI] [PubMed] [Google Scholar]
- 36.Grunwald D, Singer RH. In vivo imaging of labelled endogenous beta-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467:604–607. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens BJ, Swift H. RNA transport from nucleus to cytoplasm in Chironomus salivary glands. J Cell Biol. 1966;31:55–77. doi: 10.1083/jcb.31.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowe AR, et al. Selectivity mechanism of the nuclear pore complex characterized by single cargo tracking. Nature. 2010;467:600–603. doi: 10.1038/nature09285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elosegui-Artola A, et al. Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell. 2017;171:1397–1410.:e1314. doi: 10.1016/j.cell.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 40.Kudo N, et al. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Carrion-Vazquez M, Oberhauser AF, Marszalek PE, Fernandez JM. Point mutations alter the mechanical stability of immunoglobulin modules. Nat Struct Biol. 2000;7:1117–1120. doi: 10.1038/81964. [DOI] [PubMed] [Google Scholar]
- 42.Kruger M, Linke WA. The giant protein titin: a regulatory node that integrates myocyte signaling pathways. J Biol Chem. 2011;286:9905–9912. doi: 10.1074/jbc.R110.173260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, et al. Reverse engineering of the giant muscle protein titin. Nature. 2002;418:998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- 44.Li H, Fernandez JM. Mechanical design of the first proximal Ig domain of human cardiac titin revealed by single molecule force spectroscopy. J Mol Biol. 2003;334:75–86. doi: 10.1016/j.jmb.2003.09.036. [DOI] [PubMed] [Google Scholar]
- 45.Carrion-Vazquez M, et al. Mechanical and chemical unfolding of a single protein: a comparison. Proc Natl Acad Sci U S A. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Randles LG, Rounsevell RW, Clarke J. Spectrin domains lose cooperativity in forced unfolding. Biophys J. 2007;92:571–577. doi: 10.1529/biophysj.106.093690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alegre-Cebollada J, Badilla CL, Fernandez JM. Isopeptide bonds block the mechanical extension of pili in pathogenic Streptococcus pyogenes. J Biol Chem. 2010;285:11235–11242. doi: 10.1074/jbc.M110.102962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosa N, et al. Meltdown: A Tool to Help in the Interpretation of Thermal Melt Curves Acquired by Differential Scanning Fluorimetry. J Biomol Screen. 2015;20:898–905. doi: 10.1177/1087057115584059. [DOI] [PubMed] [Google Scholar]
- 49.Dietz H, Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc Natl Acad Sci U S A. 2004;101:16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez-Jimenez R, Garcia-Manyes S, Ainavarapu SR, Fernandez JM. Mechanical unfolding pathways of the enhanced yellow fluorescent protein revealed by single molecule force spectroscopy. J Biol Chem. 2006;281:40010–40014. doi: 10.1074/jbc.M609890200. [DOI] [PubMed] [Google Scholar]
- 51.Soderholm JF, et al. Importazole, a small molecule inhibitor of the transport receptor importin-beta. ACS Chem Biol. 2011;6:700–708. doi: 10.1021/cb2000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lovett DB, Shekhar N, Nickerson JA, Roux KJ, Lele TP. Modulation of Nuclear Shape by Substrate Rigidity. Cell Mol Bioeng. 2013;6:230–238. doi: 10.1007/s12195-013-0270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Record J, et al. Immunodeficiency and severe susceptibility to bacterial infection associated with a loss-of-function homozygous mutation of MKL1. Blood. 2015;126:1527–1535. doi: 10.1182/blood-2014-12-611012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beck M, Hurt E. The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol. 2017;18:73–89. doi: 10.1038/nrm.2016.147. [DOI] [PubMed] [Google Scholar]
- 55.Bestembayeva A, et al. Nanoscale stiffness topography reveals structure and mechanics of the transport barrier in intact nuclear pore complexes. Nat Nanotechnol. 2015;10:60–64. doi: 10.1038/nnano.2014.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frey S, et al. Surface Properties Determining Passage Rates of Proteins through Nuclear Pores. Cell. 2018;174:202–217.:e209. doi: 10.1016/j.cell.2018.05.045. [DOI] [PubMed] [Google Scholar]
- 57.Plessner M, Grosse R. Extracellular signaling cues for nuclear actin polymerization. Eur J Cell Biol. 2015;94:359–362. doi: 10.1016/j.ejcb.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 58.Mohr D, Frey S, Fischer T, Guttler T, Gorlich D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009;28:2541–2553. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Timney BL, et al. Simple rules for passive diffusion through the nuclear pore complex. J Cell Biol. 2016;215:57–76. doi: 10.1083/jcb.201601004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kirby TJ, Lammerding J. Emerging views of the nucleus as a cellular mechanosensor. Nat Cell Biol. 2018;20:373–381. doi: 10.1038/s41556-018-0038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ketterer P, et al. DNA origami scaffold for studying intrinsically disordered proteins of the nuclear pore complex. Nat Commun. 2018;9:902. doi: 10.1038/s41467-018-03313-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fisher PDE, et al. A Programmable DNA Origami Platform for Organizing Intrinsically Disordered Nucleoporins within Nanopore Confinement. ACS Nano. 2018;12:1508–1518. doi: 10.1021/acsnano.7b08044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Popa I, Kosuri P, Alegre-Cebollada J, Garcia-Manyes S, Fernandez JM. Force dependency of biochemical reactions measured by single-molecule force-clamp spectroscopy. Nat Protoc. 2013;8:1261–1276. doi: 10.1038/nprot.2013.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fischer RS, Myers KA, Gardel ML, Waterman CM. Stiffness-controlled three-dimensional extracellular matrices for high-resolution imaging of cell behavior. Nat Protoc. 2012;7:2056–2066. doi: 10.1038/nprot.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rico F, et al. Probing mechanical properties of living cells by atomic force microscopy with blunted pyramidal cantilever tips. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;72:021914. doi: 10.1103/PhysRevE.72.021914. [DOI] [PubMed] [Google Scholar]
- 66.Sage D, et al. DeconvolutionLab2: An open-source software for deconvolution microscopy. Methods. 2017;115:28–41. doi: 10.1016/j.ymeth.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 67.Kirshner H, Aguet F, Sage D, Unser M. 3-D PSF fitting for fluorescence microscopy: implementation and localization application. J Microsc. 2013;249:13–25. doi: 10.1111/j.1365-2818.2012.03675.x. [DOI] [PubMed] [Google Scholar]
- 68.Rodriguez LG, Wu X, Guan JL. Wound-healing assay. Methods Mol Biol. 2005;294:23–29. doi: 10.1385/1-59259-860-9:023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting this research can be obtained from the corresponding author upon reasonable request.