Abstract
Infection of mammalian cells with viruses activates NF-κB to induce the expression of cytokines and chemokines and initiate an antiviral response. Here, we show that a vaccinia virus protein mimics the transactivation domain of the p65 subunit of NF-κB to inhibit selectively the expression of NF-κB-regulated genes. Using co-immunoprecipitation assays, we found that the vaccinia virus protein F14 associates with NF-κB co-activator CREB-binding protein (CBP) and disrupts the interaction between p65 and CBP. This abrogates CBP-mediated acetylation of p65, after which it reduces promoter recruitment of the transcriptional regulator BRD4 and diminishes stimulation of NF-κB-regulated genes CXCL10 and CCL2. Recruitment of BRD4 to the promoters of NFKBIA and CXCL8 remains unaffected by either F14 or JQ1 (a competitive inhibitor of BRD4 bromodomains), indicating that BRD4 recruitment is acetylation-independent. Unlike other viral proteins that are general antagonists of NF-κB, F14 is a selective inhibitor of NF-κB-dependent gene expression. An in vivo model of infection demonstrated that F14 promotes virulence. Molecular mimicry of NF-κB may be conserved because other orthopoxviruses, including variola, monkeypox and cowpox viruses, encode orthologues of F14.
An array of pattern recognition receptors (PRRs) recognizes viruses and activates a gene expression programme that initiates an antiviral response1. Engagement of PRRs and cytokine receptors by their cognate ligands (respectively, pathogen-associated molecular patterns (PAMPs) and cytokines, such as tumour necrosis factor (TNF)-α) activates multiple transcription factors, including nuclear factor kappa light-chain enhancer of activated B cells (NF-κB)2. NF-κB is a homodimer or heterodimer of Rel proteins, with the heterodimer of p50 (also known as NF-κB1 or NFKB1) and p65 (also known as RelA or RELA) being the prototypical form of NF-κB3. NF-κB recognizes and binds to a consensus DNA sequence in the promoter elements and enhancers of target genes, which include cytokines, chemokines and cell adhesion molecules, as well as proteins involved in other immune processes, such as MHC molecules, growth factors and regulators of apoptosis4–6. The p65 subunit is the transcriptionally active component of NF-κB and undergoes multiple post-translational modifications, in the cytoplasm or nucleus, that control its transcriptional activity through interactions with co-activators and basal transcription machinery3. Following stimulation with PAMPs or inflammatory cytokines, conserved residues S276 and S536 in p65 are phosphorylated, which promotes the interaction with co-activator CREB-binding protein (CBP) or its paralogue p300 (also known as CREBBP and EP300, respectively). These co-activators acetylate p65 to allow transcription initiation3,7,8. The bromodomain and extraterminal domain (BET) protein BRD4 docks onto acetylated K310 of p65 and recruits positive transcription elongation factor b (P-TEFb) to drive transcription of inflammatory genes9.
Targeting of the nuclear activity of NF-κB and its co-activator CBP by viral proteins has been reported. For example, human papillomavirus (HPV) E6 protein10 and herpes simplex virus (HSV) type 1 protein VP16 (ref. 11) antagonize NF-κB in the nucleus but the detailed mechanisms remain unclear. Poxviruses have large DNA genomes that encode multiple proteins that antagonize the host antiviral response. Vaccinia virus (VACV), the smallpox vaccine and prototypical poxvirus, encodes a family of proteins sharing structural similarity to cellular Bcl-2 proteins despite very limited sequence similarity. Viral Bcl-2-like proteins have evolved a wide range of functions, such as antagonism of NF-κB activation by VACV proteins A46, A49, A52, B14, K7 and N1 (refs. 12–14). Despite the expression of multiple inhibitors of NF-κB by VACV, strains lacking individual inhibitors have reduced virulence in mouse models, arguing against functional redundancy13,14. Previous work predicted that VACV encodes additional inhibitors of NF-κB because a mutant VACV strain (vv811ΔA49) lacking all known VACV inhibitors of NF-κB still suppressed NF-κB-dependent gene expression without preventing NF-κB translocation to the nucleus15. Here, we sought additional NF-κB antagonists and show that the VACV protein F14 inhibits NF-κB selectively and a VACV strain lacking F14 has reduced virulence in a mouse model.
Results
F14 is a virulence factor that inhibits NF-κB activation
The prediction that VACV encodes additional inhibitor(s) of NF-κB that function downstream of p65 translocation to the nucleus15 prompted a search for nuclear NF-κB inhibitors. VACV strain vv811ΔA49 lacks 56 open reading frames (ORFs) but retains the inhibitor(s)15,16 and so its encoded proteins were screened by bioinformatics for ones that met the following criteria: (1) early expression, based on previous VACV transcriptome studies17,18; (2) predicted not to be involved in replication and/or morphogenesis; (3) being poorly characterized; (4) the presence of putative nuclear localization signals (NLS) or predicted molecular mass <40 kDa that would allow diffusion into the nucleus19; and (5) the presence of domains indicative of function. The genomic position of the ORF and its conservation among orthopoxviruses were also considered given that VACV immunomodulatory genes are located towards the genome termini and often are less conserved than genes having functions in virus replication20.
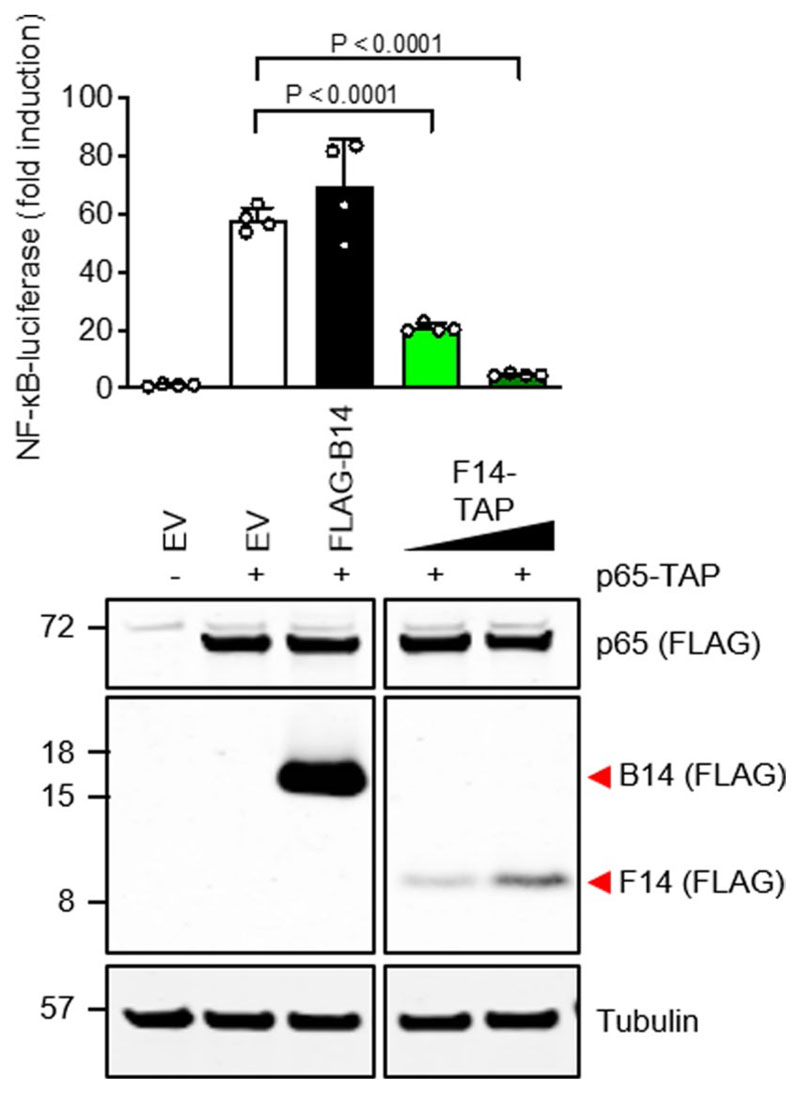
This approach yielded a list of seven ORFs, namely F6L, F7L, F14L, A47L, B6R, B11R and B12R. The proteins encoded by these ORFs were tested for inhibition of NF-κB activation in an NF-κB-luciferase reporter gene assay. GFP and VACV protein N2, an interferon regulatory factor (IRF) 3 inhibitor21, were used as negative controls, whereas VACV protein B14, a known inhibitor of NF-κB22, was used as positive control. Protein F14 inhibited TNF-α- and IL-1β-stimulated NF-κB activity in HEK 293T cells in a dose-dependent manner (Fig. 1a,b and Extended Data Fig. 1). This inhibitory activity was specific for the NF-κB pathway because F14 did not affect IFN-α-stimulated IFN-α/β receptor (IFNAR)/signal transducer and activator of transcription (STAT) or phorbol 12-myristate 13-acetate (PMA)-stimulated mitogen-activated protein kinase (MAPK)/AP-1 pathways (Fig. 1c,d). The inhibitory activity was exerted despite the lower levels of F14 when compared to protein B14 (Fig. 1e)22. Conversely, VACV protein C6 suppressed type I IFN signalling and B14 upregulated AP-1 activity, as observed previously (Fig. 1c,d)23,24.
Fig. 1. Vaccinia virus protein F14 inhibits NF-κB activation and contributes to virulence.
a-d, TNFR/NF-κB- (a), IL1R/NF-κB- (b), IFNAR/STAT- (c) or MAPK/AP-1- (d) dependent luciferase activities in HEK293T cells transfected with vectors expressing the indicated VACV proteins or EV and stimulated with TNF-α, IL-1β, IFN-α or PMA (as indicated). Means + s.d. (n = 4 per condition) are shown. e, Immunoblotting of whole cell lysates of transfected HEK 293T cells. f, C57BL/6 mice were infected intradermally in both ears with 104 p.f.u. of the indicated VACV strains and the lesions were measured daily. Means±s.e.m. (n = 10 mice from two independent experiments) are shown. g, Virus titres in the ears of mice infected as in f. Means±s.e.m. (n = 5 mice) are shown. h, Immunoblotting of protein extracts from HeLa cells infected with VF14-TAP (5 p.f.u. per cell) and treated with cytosine arabinoside (AraC, 40 μg ml-1) where indicated. i, NF-κB-dependent luciferase activity in A549 cells infected with VACV vv811 strains (5 p.f.u. per cell, 6h) and stimulated with TNF-α or IL-1β for additional 6 h. Means + s.d. (n = 4 per condition) are shown. j, Immunoblotting of protein extracts from A549 cells infected as in i. k, WT, wild type. Immunoblotting of protein extracts from T-REx-293 cells inducibly expressing F14, after doxycycline induction overnight and TNF-α stimulation. l, Quantification of NF-κB p65 localization after immunofluorescence of T-REx-293 cells inducibly expressing the EV or VACV proteins B14, C6 or F14, induced with doxycycline and stimulated with TNF-α for 15 min. The number of cells counted from two independent experiments (n) is stated below each bar. The positions of molecular mass markers in kDa are shown on the left of immunoblots. Immunoblots of tagged proteins are labelled with the protein name followed by the epitope tag antibody in parentheses. When multiple tagged proteins are shown in the same immunoblot, each protein is indicated by a red arrowhead. Statistical significance was determined by unpaired two-tailed Student’s f-test. Data shown in a-d, i, f and g, and e,h,j and k are representative of four, two or three separate experiments, respectively.
The virulence of VACV strains lacking specific genes has been tested mostly in intranasal or intradermal murine models13. Deletion of genes encoding VACV immunomodulatory proteins may give a phenotype in either, neither or both models. To evaluate if loss of F14 expression affects virulence, a recombinant VACV lacking F14 was generated, termed vΔF14. Intradermal injection of the ear pinnae of mice with vΔF14 produced smaller lesions (Fig. 1f) compared to wild-type virus (vF14) and a revertant strain (vF14-Rev), generated by reinserting F14 into vΔF14 at its natural locus (Fig. 1f). Attenuation of vΔF14 in the intradermal mouse model correlated with reduced viral titres in the infected ears 7 and 10 d post-infection (p.i.) but not 3 d p.i. (Fig. 1g). In contrast, in an intranasal mouse model, vΔF14 caused the same extent of body mass loss as wild type and revertant controls (Extended Data Fig. 2). In cell culture, vF14, vΔF14 and vF14-Rev displayed no differences in replication and plaque size (Extended Data Fig. 3). Altogether, these experiments showed that F14 is not essential for virus replication but contributes to virulence.
Previous analyses of the VACV transcriptome showed that the F14L ORF is transcribed and translated early during infection17,18,25. This was consistent with an upstream typical early promoter and a transcription termination motif T5NT downstream of the stop codon26. To investigate F14 expression during infection, a VACV strain was constructed in which F14 was tagged with a C-terminal TAP tag. The vF14-TAP strain replicated normally (Extended Data Fig. 3) and immunoblotting showed F14 protein expression was detected from 4 h p.i. and peaked by 8 h p.i., matching the accumulation of the early VACV protein C6 (Fig. 1h)27. F14 levels were notably low either when expressed ectopically (Fig. 1e) or during infection (Fig. 1h). This might explain why F14 was not detected in our recent quantitative proteomic analysis of VACV infection, which detected about 80% of the predicted VACV proteins28. Pharmacological inhibition of virus DNA replication with cytosine arabinoside (AraC) did not affect F14 protein levels, consistent with early expression, whereas late protein D8 was inhibited (Fig. 1h).
The existence of multiple VACV inhibitors of NF-κB that each contribute to virulence indicates that they are not redundant. To test if F14 affects NF-κB activation during infection, we deleted F14 from the vv811ΔA49 strain that lacks other known inhibitors of NF-κB15 and then infected an NF-κB reporter cell line15. As shown previously, vv811ΔA49 inhibited NF-κB to a reduced extent when compared to the parental vv811 strain (Fig. 1i)15 and deletion of F14L from vv811ΔA49 reduced NF-κB inhibition further (Fig. 1i). Immunoblotting confirmed equal infection with these viruses (Fig. 1j). Notably, vv811ΔA49ΔF14 still suppressed NF-κB activation considerably, which might be explained by: (1) the existence of additional virally encoded inhibitors that cooperate to inhibit NF-κB in the nucleus, or (2) the actions of viral decapping enzymes to reduce host messenger RNAs29.
To dissect how F14 functions, its impacts on three hallmarks of NF-κB signalling were studied: namely, degradation of IκBα, phosphorylation of p65 at S536 and p65 nuclear translocation2,3. A cell line that expresses F14 inducibly on addition of doxycycline was used to study the degradation of IκBα and phosphorylation and nuclear translocation of p65 following stimulation with TNF-α. IκBα degradation was evident 15 min after stimulation and its resynthesis had started by 30 min, but neither process was influenced by F14 (Fig. 1k). F14 also did not affect phosphorylation of p65 at S536 (Fig. 1k) or p65 translocation into the nucleus as measured by immunofluorescence (Fig. 1l and Extended Data Fig. 4). In contrast, VACV protein B14 inhibited translocation efficiently as reported22 and VACV protein C6, an IFN antagonist23,27, did not (Fig. 1l). Next, the NF-κB inhibitory activity of F14 was tested by reporter gene assay following pathway activation by p65 overexpression. In contrast to B14, F14 inhibited p65-mediated activation in a dose-dependent manner without affecting p65 levels (Extended Data Fig. 5). Altogether, these results showed that F14 blocks NF-κB in the nucleus at or downstream of p65. F14 thus fits the criteria described previously for the unknown inhibitor of NF-κB encoded by VACV and expressed by vv811ΔA49 (ref. 15).
F14 mimics p65 transactivation domain
Poxvirus immunomodulatory proteins are generally encoded in the variable genome termini, share lower sequence identity and show genus-specific distribution20. Even among orthopoxviruses, only a few of the immunomodulatory genes are present in all virus species20. Nonetheless, Viral Orthologous Clusters30 and protein BLAST searches found orthologues of VACV F14 in all orthopoxviruses, with 70.7–98.6% amino acid (aa) identity (Fig. 2a). The F14L orthologue from cowpox virus, CPXV062, is expressed during infection by two different strains of cowpox virus, as shown by RNA sequencing31. F14L orthologues from monkeypox and variola viruses are also likely to be expressed during infection because the nucleotide sequences surrounding the ORFs contained highly conserved transcriptional regulatory sequences; that is, canonical poxvirus early promoters and transcription termination signals26. Furthermore, historical strains of variola virus from the tenth and seventeenth centuries CE also encoded F14L orthologues with conserved flanking transcription regulatory sequences32,33.
Fig. 2. F14 binds to CBP and has transactivation activity.
a, The top section shows amino acid alignment of F14 orthologues of representative orthopoxviruses: VACV, Western Reserve (WR), VACV Copenhagen (Cop), modified vaccinia Ankara (MVA), horsepox virus (HSPV), monkeypox virus (MPXV), cowpox virus (CPXV), variola virus (VARV), camelpox virus (CMLV), taterapox virus (TATV), ectromelia virus (ECMV) and racoonpox virus (RCNV). Red, aa identical in all sequences; yellow, aa identical in at least 8/11 sequences. The percentage aa identities of F14 orthologues compared to the F14 protein of VACV-WR are shown on the right. The bottom of a shows alignment of the C termini of F14 and p65 highlighting their sequence similarity including the ΦXXΦΦ motif, above a schematic of p65 and its functional domains. Asterisks (*), identical aa; colons (:), conservative aa change; dots (.), non-conservative aa change. Nucleotide sequences used for this study are listed in the Data availability statement. b,c, Lysates from transfected HEK 293T cells were immunoprecipitated with anti-HA (b) or anti-FLAG (c). Immunoblots are representative of three independent experiments. d, HEK 293T and HeLa cells were infected with VACV strains vC6-TAP or vF14-TAP (5 p.f.u. per cell, 8 h) and lysates were immunoprecipitated with anti-FLAG. Immunoblots are representative of two independent experiments. e, NF-κB-dependent luciferase activity in HEK 293T cells transfected with vectors expressing p65, p65 mutants or EV. In the graph at the top, means + s.d. (n = 4 per condition) are shown. Statistical significance was determined by unpaired two-tailed Student’s t-test. The bottom panel shows immunoblotting. Protein molecular mass markers in kDa are shown on the left of the blots. Immunoblots of tagged proteins are labelled with the protein name followed the epitope tag antibody in parentheses. When multiple tagged proteins are shown in the same immunoblot, each protein is indicated by a red arrowhead. IB, immunoblotting.
The C-terminal half of F14 was more conserved and included a predicted coiled-coil region (aa 34–47), the only structural motif predicted via bioinformatic analyses. However, the Phyre2 algorithm34 predicted the C-terminal aa 55–71 to adopt an α-helical secondary structure similar to that of aa 534–546 of the p65 C terminus in complex with the PH domain of human general transcription factor Tfb1 or aa 534–546 of p65 in complex with the KIX domain of NF-κB co-activator CBP35. The C terminus of p65 harbours its transactivation domain (TAD), which is divided into two subdomains that have independent transcriptional activity: TA1 (aa 521–551) and TA2 (aa 428–521) (refs. 36,37). TA1 contributes at least 85% of p65 transcriptional activity and interacts directly with CBP35–37. The TAD domain of p65 belongs to the class of acidic activation domains, characterized by a preponderance of aspartic acid or glutamic acid residues surrounding hydrophobic motifs37. The F14 aa similarity was striking despite the low confidence of the Phyre2 model (41.4%). When aligned to p65 C-terminal aa 521–551, F14 shared 39% aa identity and 61% aa similarity, including the conservation of a ΦΧΧΦΦ motif and an upstream acidic residue, both essential for NF-κB transcriptional activity35,38,39. Notably, in F14 the position equivalent to S536 in p65, which is phosphorylated on NF-κB activation40, is occupied by the negatively charged residue D59 (Fig. 2a). The negative charge of F14-D59 closely resembles the negative charge conferred by phosphorylation of p65-S536 during NF-κB activation40.
These observations and the key role of CBP in NF-κB-dependent gene activation3 prompted investigation of whether F14 could interact with CBP. Immunoprecipitation (IP) of HA-tagged F14 coprecipitated CBP-FLAG from HEK 293T cells (Fig. 2b). Reciprocal IP experiments showed that ectopic CBP coprecipitated F14-HA, but not GFP-HA, with or without prior TNF-α stimulation (Fig. 2c). These interactions were also seen at endogenous levels in both HEK 293T cells and HeLa cells infected with vF14-TAP. F14, but not C6, coprecipitated endogenous CBP (Fig. 2d). To test whether the C terminus of F14 mediated transactivation via its binding to CBP, F14 aa 51–73 were fused to the C terminus of a p65 mutant lacking the TA1 subdomain of the TAD (ΔTA1) and the fusion protein was tested in a NF-κB reporter gene assay. Compared to wild-type p65, the p65ΔTA1 mutant was impaired in its transactivating activity, which was restored to wild-type levels on fusion to F1451-73 (Fig. 2e). This result argues strongly that the C terminus of F14 mimics the TA1 of p65 and this mimicry might explain how F14 inhibits NF-κB activation.
VP16 is a transcriptional activator from HSV-1 that bears a prototypical acidic TAD (Extended Data Fig. 6a) and inhibits the expression of virus-induced IFN-β by association with p65 and IRF3 (ref. 11). Although the VP16-mediated inhibition of the IFN-β promoter was independent of its TAD, we revisited this observation to investigate the effect of VP16 more specifically on NF-κB-dependent gene activation. VP16 inhibited NF-κB reporter gene expression in a dose-dependent manner and deletion of the TAD reduced NF-κB inhibitory activity of VP16 about twofold but some activity remained (Extended Data Fig. 6a). A search for other viral proteins that contain motifs resembling the ΦΧΧΦΦ motif present in acidic transactivation activation domains detected a divergent ΦΧΧΦΦ motif in protein E7 (aa 79–83) from HPV16, with acidic residues upstream (D75) or within (E80 and D81) the motif (Extended Data Fig. 6b). E7 has been reported to inhibit NF-κB activation, in addition to its role in promoting cell cycle progression10,41–43. We confirmed that HPV16 protein E7 inhibits NF-κB-dependent gene expression (Extended Data Fig. 6b). Furthermore, E7 mutants harbouring aa substitutions that inverted the charge of D75 (D75K) or added a positive charge to the otherwise hydrophobic L83 (L83R) were impaired in their capacity to inhibit NF-κB (Extended Data Fig. 6b). Lastly, the ability of VP16 and E7 to associate with CBP was assessed after ectopic expression in HEK 293T cells. Neither VP16 nor E7, like VACV protein C6 used as negative control, coprecipitated CBP under conditions in which F14 did (Extended Data Fig. 6c). These findings indicate that the mimicry of p65 TAD by F14 to target CBP is unique among known anti-NF-κB proteins from human pathogenic viruses.
F14 outcompetes NF-κB for binding to CBP
The similarity between the C termini of F14 and p65 led us to investigate if conserved aa residues contributed to the NF-κB inhibitory activity of F14. On the basis of the structure of CBP KIX domain in complex with p65 TA1 (ref. 35), residues of the F14 TAD-like domain corresponding to residues of p65 important for its transcriptional activity and binding to CBP were mutated. Three sites were altered by site-directed mutagenesis: the dipeptide D62/63 and the following L65 and L68 of the ΦΧΧΦΦ motif. F14 L65A or L68A still inhibited NF-κB (Fig. 3a), although the L65A mutant was slightly impaired. In contrast, mutation of D62/63 to either alanine (D62/63A) or lysine (D62/63K) abolished the inhibitory activity (Fig. 3a). Protein levels were comparable across the different F14 mutants (Fig. 3a). The loss of NF-κB inhibitory activity of D62/63A and D62/63K mutants correlated with their reduced capacity to coprecipitate CBP, whereas L65A and L68A mutants coprecipitated CBP to the same extent as wild-type F14 (Fig. 3b). The mutation of the negatively charged D62/63 to positively charged lysine residues was more efficient in disrupting the interaction between F14 and CBP than only abolishing the charge (Fig. 3b). Collectively, these results highlight the importance of the negatively charged dipeptide D62/63 within the TAD-like domain for NF-κB inhibition by F14.
Fig. 3. The dipeptide D62/63 of F14 is required for inhibition of NF-κB.
a, NF-κB-dependent luciferase activity in HEK 293T cells transfected with vectors expressing F14, F14 mutants or EV and stimulated with TNF-α for 8h. In the graph at the top, means + s.d. (n = 4 per condition) are shown. Statistical significance was determined by unpaired two-tailed Student’s t-test. The bottom panel shows immunoblotting. b, Lysates from transfected HEK 293T cells were affinity-purified with Strep-Tactin resin. DD/AA denotes D62/63 A mutant and DD/KK, D62/63 K mutant. AP, affinity purification. Protein molecular mass markers in kDa are shown on the left of the blots. Immunoblots of tagged proteins are labelled with the protein name followed the epitope tag antibody in parentheses. When multiple tagged proteins are shown in the same immunoblot, each protein is indicated by a red arrowhead. Data are representative of three independent experiments.
Next, we tested if F14 could disrupt the interaction of p65 with its co-activator CBP. HEK 293T cells were transfected with vectors expressing p65 and CBP or RIG-I (negative control) and VACV proteins F14 or C6. The amount of p65-HA immunoprecipitated by ectopic CBP was reduced by increasing amounts of F14 but not C6 (Fig. 4a,b). Quantitative analysis showed equivalent ectopic CBP immunoprecipitation with or without F14 (Fig. 4c). Furthermore, the mutation D62/63K diminished the capacity of F14 to disrupt the interaction of CBP and p65 (Fig. 4d). This observation correlated well with the reduced capacity of the D62/63K mutant to coprecipitate CBP (Fig. 3b).
Fig. 4. F14 outcompetes NF-κB for binding to CBP.
a, Lysates from transfected HEK 293T cells were Immunopreclpltated with anti-FLAG after TNF-α stimulation. b,c, Ratio of immunoprecipitate (IP) over input signal intensities from p65 (b) and CBP (c) immunoblots as in a. Means + s.d. (n = 3 independent experiments) are shown. Statistical significance was determined by unpaired two-tailed Student’s t-test. d, Lysates from transfected HEK 293T cells were immunoprecipitated with anti-HA after TNF-α stimulation. Protein molecular mass markers in kDa are shown on the left of the blots. Immunoblots of tagged proteins are labelled with the protein name followed the epitope tag antibody in parentheses. When multiple tagged proteins are shown in the same immunoblot, each protein is indicated by a red arrowhead.
F14 suppresses a subset of NF-κB-regulated genes
To address the impact of F14 on the stimulation of endogenous NF-κB-responsive genes by TNF-α, the cell line inducibly expressing F14 was used. NF-κB-responsive genes display different temporal kinetics on activation, with ‘early’ gene transcripts peaking between 30 and 60 min after stimulation before declining, whilst ‘late’ gene transcripts accumulate slowly and progressively, peaking 3 h post-stimulation4,5. When F14 expression was induced, mRNAs of NFKBIA and CXCL8 ‘early’ genes had equivalent accumulation kinetics compared to uninduced cells (Fig. 5a,b). The lack of inhibition of F14 on the expression of NFKBIA mRNA is in agreement with the previous finding that the resynthesis of IκBα (NFKBIA protein product) is unaffected by F14 after its proteasomal degradation stimulated by TNF-α (Fig. 1k). Conversely, F14 induction inhibited the accumulation of the mRNAs of CCL2 and CXCL10 ‘late’ genes in response to TNF-α (Fig. 5d,e). Similar results were observed when the F14-expressing cell line was compared to the cell line inducibly expressing C6 (Extended Data Fig. 7). CXCL8 and CXCL10 encode chemokines CXCL8 and CXCL10 (also known as IL-8 and IP-10, respectively). Following induction of VACV protein expression, the levels of these secreted chemokines were measured by ELISA and showed that the level of CXCL10, but not CXCL8, was inhibited by F14. In contrast, the secretion of both chemokines was inhibited, or unaffected, by VACV proteins B14 or C6, respectively, as expected (Fig. 5c,f,i and Extended Data Fig. 8). Thus, unlike other VACV NF-κB inhibitors, F14 is selective and inhibits only a subset of NF-κB-responsive genes.
Fig. 5. F14 suppresses expression of a subset of NF-κB-responsive genes.
a-f, T-REx-293 cells inducibly expressing the EV or VACV proteins F14, B14 or C6 in the absence (-F14 or -Dox) or in the presence (+F14 or +Dox) of doxycycline overnight and stimulated with TNF-α. qPCR with reverse transcription analysis of NF-κB-responsive genes NFKBIA (a), CXCL8 (b), CCL2 (d) and CXCL10 (e) in inducible T-REx-293-F14 cells. Means (n = 2 per condition) are shown. c,f, Quantification of secreted CXCL8 (c) and CXCL10 (f) by ELISA of culture supernatants from inducible T-REx-293 cells after 16 h of stimulation with TNF-α. Means + s.d. (n = 3 per condition) of the percentage of secretion in the presence of doxycycline (+Dox) versus in the absence of doxycycline (-Dox, equals 100%) are shown. g,h, Secretion of CXCL8 (g) and CXCL10 (h) as measured by ELISA of culture supernatants from T-REx-293-EV cells stimulated with TNF-α for 16 h in the absence or in the presence of JQ1. NS, non-stimulated. Means ± s.d. (n = 3 per condition) are shown. i, Immunoblotting of lysates of inducible T-REx-293 cell lines in the absence or in the presence of doxycycline overnight. Protein molecular masses in kDa are shown on the left of the blots. Immunoblots of tagged proteins are labelled with the protein name followed the epitope tag antibody in parentheses. When multiple tagged proteins are shown in the same immunoblot, each protein is indicated by a red arrowhead. Statistical significance was determined by unpaired two-tailed Student’s t-test.
Differential regulation of transcription activation downstream of NF-κB has been ascribed to the recruitment of BRD4 to some NF-κB-dependent inflammatory genes9. Via its bromodomains 1 and 2, BRD4 docks onto acetylated histones and non-histone proteins and recruits transcriptional regulatory complexes to chromatin44,45. The specific recognition of acetyl-lysine residues by BRD4 is competitively inhibited by small-molecule BET bromodomain inhibitors, such as JQ1 (ref. 46). Therefore, to gain more insight into the mechanism underpinning the selective inhibition of inflammatory genes by F14, the effect of JQ1 on the inducible expression of CXCL8 and CXCL10 was investigated. Following TNF-α stimulation, JQ1 inhibited the secretion of CXCL10, but not CXCL8, phenocopying the selective inhibition of inflammatory protein expression by F14 (Fig. 5g,h).
p65 acetylation and BRD4 recruitment are inhibited by F14
Post-translational modifications of p65 accompany NF-κB translocation to the nucleus and some, such as acetylation by acetyltransferases CBP and p300, are associated with increased transcriptional activity3,7,8,40. F14 did not interfere with the phosphorylation of p65 at S536 (Fig. 1k), so the acetylation of p65-K310 was investigated. Cell lines that express F14 inducibly or contain the empty vector (EV) control were transfected with plasmids expressing p65 and CBP in the presence of the inducer, doxycycline. Although both cell lines expressed equivalent amounts of ectopic p65 and CBP, the amount of p65 acetylated at K310 was greatly diminished by F14 (Fig. 6a). Quantitative analysis showed acetylated p65 was reduced 90% by F14 (Fig. 6b). This result, together with data in Fig. 5, indicated that the reduced acetylation of p65 was due to disruption of the interaction between p65 and CBP by F14.
Fig. 6. F14 antagonizes p65 acetylation and inducible recruitment of BRD4 to CCL2 and CXCL10 promoters.
a, Immunoblotting of protein lysates from T-REx-293 cells stably transfected with EV or inducibly expressing F14, induced with doxycycline and transfected with plasmids expressing p65 and CBP. Blots are representative of two independent experiments carried out with three biological replicates each. b, Ratio of acetylated (Ac) p65 over total ectopic p65 signal intensities from immunoblots as in a. Means + s.d. (n = 3 per condition) are shown. c-f, T-REx-293-F14 were left uninduced (-F14) or induced with doxycycline (+F14) and stimulated with TNF-α, T-REx-293-F14 were left uninduced (-F14) or induced with doxycycline (+F14) and stimulated with TNF-α. ChIP with anti-BRD4 or control IgG and qPCR for the promoters of NFKBIA (c), CXCL8 (d), CCL2 (e) and CXCL10 (f) genes. g, Immunoblotting from c to f. h-k, Alternatively, T-REx-293 cells were treated with JQ1 before TNF-α stimulation. ChIP with anti-BRD4 or control IgG and qPCR for the promoters of NFKBIA (h), CXCL8 (i), CCL2 (j) and CXCL10 (k) genes. Means + s.d. (n = 5 or 6 per condition from two independent experiments) are shown. Protein molecular mass markers in kDa are shown on the left of the blots. Immunoblots of tagged proteins are labelled with the protein name followed the epitope tag antibody in parentheses. Statistical significance was determined by unpaired two-tailed Student’s t-test.
Acetylated K310 on p65 serves as a docking site for the bromodomains 1 and 2 of BRD4, which then recruits P-TEFb to promote RNAP II elongation during transcription of some NF-κB-responsive genes9. The differential sensitivity of TNF-α-stimulated genes to the inhibition of NF-κB by F14 might reflect the differential requirement of p65 acetylated at K310 and the subsequent recruitment of BRD4, to activate the expression from NF-κB-responsive promoters9. This hypothesis was tested by chromatin immunoprecipitation (ChIP) with anti-BRD4 followed by quantitative polymerase chain reaction (qPCR) of the promoters of four representative genes: NFKBIA and CXCL8, resistant to F14 inhibition and CCL2 and CXCL10, sensitive to inhibition. BRD4 was recruited to these promoters after TNF-α stimulation, with BRD4 present on NFKBIA and CXCL8 promoters at 1 and 5 h post-stimulation, whereas BRD4 was more enriched on CCL2 and CXCL10 promoters only at 5 h post-stimulation, mirroring the kinetics of mRNA accumulation (Fig. 6b–f and Fig. 5a,b,d,e). In the presence of F14, the inducible recruitment of BRD4 to the NFKBIA and CXCL8 promoters remained unaffected, whilst its recruitment to CCL2 and CXCL10 was blocked (Fig. 6c). This strongly suggests that inhibition of acetylation of p65 at K310 by F14 is relayed downstream to the recruitment of BRD4 to the ‘F14-sensitive’ promoters but not to the ‘F14-resistant’ promoters.
The BRD4 recruitment to NFKBIA and CXCL8 promoters despite inhibition of p65-K310 acetylation prompted investigation of whether other acetyl-lysine residues are recognized. The bromodomain-mediated docking onto acetylated lysine residues is generally accepted as responsible for the recruitment of BRD4 to the chromatin44,45. For instance, histone 4 acetylated on K5, K8 and K12 (H4K5/K8/K12ac) is responsible for BRD4 recruitment to NF-κB-responsive genes on lipopolysaccharide stimulation47. The recruitment of BRD4 to the NFKBIA, CXCL8, CCL2 and CXCL10 promoters was tested in the presence of the bromodomain inhibitor JQ1, by ChIP–qPCR. BRD4 was still recruited to NFKBIA and CXCL8 promoters after TNF-α stimulation in the presence of JQ1, whilst inducible recruitment to CCL2 and CXCL10 promoters was abolished by JQ1 (Fig. 6h–k). As a control for JQ1 pharmacological activity, BRD4 recruitment to the CCND1 gene promoter was diminished by this small-molecule inhibitor (Extended Data Fig. 9). CCND1 is a BRD4 target gene that encodes the cell cycle regulator cyclin D1 and was used as a positive control48. Altogether, these results suggest that the inducible recruitment of BRD4 to some promoters is independent of the bromodomains.
Discussion
NF-κB couples the sensing of viral and inflammatory signals with the selective activation of target genes. Multiple immune evasion strategies of viruses have been reported, including interference with NF-κB activation. VACV encodes 15 proteins known to intercept NF-κB activation downstream of PRRs and cytokine receptors13,14,49,50. Nonetheless, a VACV strain lacking all these inhibitors still prevented NF-κB activation after p65 translocation into the nucleus15, indicating the existence of other inhibitor(s). Here, we report that VACV protein F14, which is conserved in all orthopoxviruses, including ancient variola viruses, inhibits NF-κB activation in the nucleus by disrupting the binding of p65 to its co-activator CBP (Figs. 4 and 5) and reducing acetylation of p65-K310. Downstream of reduced p65 acetylation, F14 inhibits the inducible recruitment of BRD4 to CCL2 and CXCL10 promoters but not to NFKBIA and CXCL8 promoters (Fig. 6). From the viral perspective, the selective inhibition of only a subset of NF-κB-responsive genes by F14 might represent an adaptation to counteract the host immune response more efficiently. If an NF-κB-activating signal reached the nucleus of an infected cell, maintaining expression of some NF-κB-dependent genes, particularly NFKBIA, might promote the signal termination by IκBα. Newly synthesized IκBα not only tethers cytoplasmic NF-κB but can also remove NF-κB from the DNA and cause its export from the nucleus2,51,52.
Recruitment of BRD4 to promoters and enhancers occurs via bromodomain-mediated docking onto acetyl-lysine residues on either histones or non-histone proteins and promotes chromatin remodelling and transcription44,45. For NF-κB-bound promoters, BRD4 recognizes p65 acetylated at K310 (ref. 9). This explains how F14 reduced inducible enrichment of BRD4 on the CCL2 and CXCL10 promoters following TNF-α stimulation: namely, reduced acetylation of p65 by CBP (Fig. 6a,b,e,f). Nonetheless, BRD4 enrichment on the NFKBIA and CXCL8 promoters remained unaffected in the presence of F14 (Fig. 6c,d). BRD4 enrichment on NFKBIA and CXCL8 promoters also remained unaffected in the presence of the bromodomain inhibitor JQ1 (Fig. 6h,i), indicating alternative mechanism(s) of BRD4 recruitment to some promoters. Downstream of p65, alternative recruitment via protein–protein interactions through the C-terminal domains of BRD4 might mediate BRD4 recruitment to some NF-κB-bound promoters independently of the recognition of acetyl-lysine residues by the N-terminal bromodomains, which is recognized as the main mechanism of BRD4 recruitment to chromatin44,45. Further investigation of acetylation-independent recruitment of BRD4 to inducible promoters observed here and elsewhere53 is warranted.
In the nucleus, p65 engages with multiple binding partners via its TADs, including the direct interactions between TA1 and TA2 and the KIX and transcriptional adaptor zinc finger (TAZ) 1 domains of CBP, respectively. These interactions are mediated by hydrophobic contacts of the ΦΧΧΦΦ motifs and complemented by electrostatic contacts by the acidic residues in the vicinity of the hydrophobic motifs35,54. Sequence analysis suggested that F14 mimics the p65 TA1 domain (Fig. 2a). Indeed, fusion of the TAD-like domain of F14 to a p65 mutant lacking the TA1 domain restored its transactivation activity to wild-type levels (Fig. 2e). Site-directed mutagenesis of F14 revealed that the dipeptide D62/63, but not L65 or L68 of the ΦΧΧΦΦ motif, is required for inhibition of NF-κB (Fig. 3a), for interaction with CBP (Fig. 3b) and for the efficient disruption of p65 binding to CBP (Fig. 4d). This contrasts with the molecular determinants of p65 TA1 function, that is, both hydrophobic (F542) and acidic (including D539 and D541) residues contribute to p65 TA1 transactivation activity38,39. Although the p65 TA1 binding to the KIX domain of CBP was shown to depend on F542, the importance of the electrostatic interactions by D539 and D541 is yet to be tested35. Of note, a recent high-throughput mutagenesis analysis of a model acidic activation domain provided useful insight into the relative contributions of hydrophobic and acidic residues for transcriptional activity. This analysis supports a model in which key hydrophobic residues require the acidic residues to keep them exposed to solvent where they can interact with co-activators55. We cannot rule out that F14 function depends on other C-terminal hydrophobic residues but our observation that F14-D62/63K (F14-D62 aligns with p65-D539) mutant is impaired in disrupting p65-KIX interaction in cells is in line with the hypothesis of how acidic activation domains work. Future elucidation of the structure of F14-KIX complex and its comparison with p65 TA1-KIX costructure will be necessary to address this apparent discrepancy. The ‘imperfect’ nature of F14 mimicry is not without precedent in poxviruses. VACV protein A49, a mimic of IκBα phosphodegron, contains an extra aa residue between the two phosphorylatable serine residues of the degron and requires the phosphorylation of just one of the two serines to interact with the E3 ligase β-TrCP and thus to prevent IκBα degradation56,57.
The abolished acetylation of p65-K310 is a direct consequence of the disruption of CBP and p65 interaction by F14. Other poxvirus proteins are reported to inhibit p65 acetylation. For instance, VACV protein K1 inhibits CBP-dependent p65 acetylation and NF-κB-dependent gene expression58. However, the vv811ΔA49 strain used to predict the existence of additional VACV inhibitors of NF-κB lacks K1 (ref. 15). The other poxviral protein that inhibits p65 acetylation is encoded by gene 002 of orf virus, a parapoxvirus that causes mucocutaneous infections in goats and sheep59. However, protein 002 differs from F14 in that it interacts with p65 to prevent acetylation by p300 (ref. 59). Our study adds VACV protein F14 to the list of viral binding partners of CBP and its paralogue p300, which includes adenovirus E1A protein60, human immunodeficiency virus (HIV) 1 Tat protein61 and high-risk HPV16 E6 protein62. Even though some of these proteins also inhibit NF-κB activation10,62,63, F14 is unique among them in mimicking p65 TA1 to bind to CBP and prevent its interaction with p65. HPV16 E6 also disrupts the interaction of CBP with p65 but, unlike F14, E6 lacks a ΦΧΧΦΦ motif surrounded by acidic residues and inhibits the expression of CXCL8 and therefore is mechanistically distinct10. After searching for additional viral proteins that might mimic p65 TAD, we focused on HPV16 E7 and HSV-1 VP16. The latter protein has a prototypical acidic TAD (Extended Data Fig. 6a), the former bears a motif resembling the ΦΧΧΦΦ motif (Extended Data Fig. 6b) and both proteins inhibit NF-κB activation10,11,41–43. Data presented here confirm that VP16 and E7 each inhibit NF-κB-dependent gene expression (Extended Data Fig. 6a,b). However, neither coprecipitated CBP under conditions in which F14 did (Extended Data Fig. 6c), suggesting VP16 and E7 inhibit NF-κB activation by a mechanism distinct from F14. The interaction between VP16 and CBP is contentious11,64 and data presented here suggest that these two proteins do not associate with each other under the conditions tested. Therefore, the molecular mimicry of F14 might be only rivalled by that of the avian reticuloendotheliosis virus, a retrovirus whose v-Rel gene was acquired from an avian host. A viral orthologue of c-Rel with weak transcriptional activity, v-Rel acts as dominant-negative protein to repress NF-κB-dependent gene activation in avian cells65. Overall, our search for additional inhibitors of NF-κB activation encoded by VACV unveiled a viral strategy to inhibit this transcription factor that is unique among known viral antagonists of NF-κB. By mimicking the TA1 domain of p65, F14 disrupts the interaction between p65 and its co-activator CBP, thus inhibiting the downstream molecular events that trigger the activation of a subset of inflammatory genes in response to cytokine stimulation.
Methods
Sequence analysis
Candidate ORFs encoding the unknown VACV inhibitor of NF-κB were first selected on the basis of VACV genomes available on the NCBI database (accession numbers NC_006998.1 for the Western Reserve strain and M35027.1 for the Copenhagen strain). The prediction of general properties of the candidate VACV gene products was done with ExPASy Compute pI/MW tool (https://web.expasy.org/compute_pi/) and SeqNLS (http://mleg.cse.sc.edu/seqNLS/)66, respectively. Domain searches were performed using InterPro (http://www.ebi.ac.uk/interpro/search/sequence/), UniProt (https://www.uniprot.org/uniprot/), HHpred (https://toolkit.tuebingen.mpg.de/tools/hhpred), PCOILS (https://toolkit.tuebingen.mpg.de/tools/pcoils) and Phobius (https://www.ebi.ac.uk/Tools/pfa/phobius/). Gene family searches were done within the Pfam database (https://pfam.xfam.org/) and conservation within the poxvirus family, with Viral Orthologous Clusters (https://4virology.net/virology-ca-tools/vocs/)30 and protein BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) searches. Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index)34 was used for the prediction of F14 protein structure. Multiple sequence alignments were performed using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) and ESPript 3.0 (http://espript.ibcp.fr/ESPript/ESPript/)67 was used for the visualization of protein sequence alignments. All poxvirus sequences referred to in this study are listed in the Data availability statement.
Expression vectors
The VACV F6L, F7L, F14L, A47L, B6R, B11R and B12R ORFs (strains Western Reserve and Copenhagen, if sequences diverged between strains) were codon-optimized for expression in human cells and synthesized by GeneArt (Thermo Fisher Scientific), with an optimal 5′ Kozak sequence and fused to an N-terminal FLAG epitope, with an (Ala)3 linker between the epitope tag and the protein. Nucleotide sequences encoding N-terminal FLAG-tagged VACV proteins were subcloned into the pcDNA4/TO vector (Invitrogen). Alternatively, codon-optimized F14 was also cloned with C-terminal HA, FLAG or TAP tags; the TAP tag consisted of two copies of the Strep-tag II epitope and one copy of the FLAG epitope68. The pcDNA4/TO-based expression vectors for VACV proteins C6 and B14 have been described24,69. The ORF encoding HPV16 E7 protein was amplified from a template provided by C. Kranjec and J. Doorbar (Department of Pathology, University of Cambridge, UK) and cloned into pcDNA4/TO with C-terminal TAP or HA tags. The ORF encoding HSV-1 VP16 and ΔTAD mutant (lacking aa 413–490) were amplified from the pEGFP-C2-VP16 plasmid provided by C. Crump (Department of Pathology, University of Cambridge, UK) and cloned into pcDNA4/TO with a C-terminal HA epitope. Mutant protein expression vectors were constructed with QuikChange II XL Site-Directed Mutagenesis kit (Agilent), using primers containing the desired mutations. The pcDNA4/TO plasmids encoding TAP- and HA-tagged p65 were described elsewhere49 and plasmids expressing FLAG-tagged (pCMV5-CBP-FLAG) and HA-tagged (pRcRSV-CBP-HA) mouse CBP were gifts from G. A. Blobel (University of Pennsylvania, USA) and T. Kouzarides (Department of Pathology and The Gurdon Institute, University of Cambridge, UK), respectively. Firefly luciferase reporter plasmids for NF-κB, STAT and AP-1, as well as the constitutively active TK-Renilla luciferase reporter plasmid were kind gifts from A. Bowie (Trinity College, Dublin, Republic of Ireland). The reporter plasmids encode firefly luciferase under the control of consensus NF-κB response element repeats ((GGGAATTTCC)5), AP-1 response element repeats ((TGACTAA)7) or IFN-stimulated response element (ISRE) ((TAGTTTCACTTTCCC)5). The oligonucleotide primers used for cloning and site-directed mutagenesis are listed in Supplementary Table 1. Nucleotide sequences of the inserts in all the plasmids were verified by Sanger DNA sequencing.
Cell lines
All cell lines were grown in medium supplemented with 10% foetal bovine serum (FBS, Pan Biotech), 100 units ml-1 of penicillin and 100 μg ml-1 of streptomycin (Gibco), at 37 °C in a humid atmosphere containing 5% CO2. Human embryo kidney (HEK) 293T cells (ATCC, CRL-11268) and monkey kidney BS-C-1 (ATCC, CCL-26) and CV-1 (ATCC, CCL-70) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco). Rabbit kidney RK13 cells (ATCC, CCL-37) were grown in minimum essential medium (MEM, Gibco) and human cervix HeLa cells (ATCC, CCL-2), in MEM supplemented with non-essential amino acids (Gibco). T-REx-293 cells (Invitrogen) were grown in DMEM supplemented with blasticidin (10 μg ml-1, InvivoGen), whilst the growth medium of T-REx-293-derived cell lines stably transfected with pcDNA4/TO-based plasmids was further supplemented with zeocin (100 μg ml-1, Gibco). The absence of mycoplasma contamination in the cell cultures was tested routinely with MycoAlert detection kit (Lonza), following the manufacturer’s recommendations.
Construction of recombinant viruses
A VACV Western Reserve (WR) strain lacking F14 (vΔF14) was constructed by introduction of a 137-base pair (bp) internal deletion in the F14L ORF by transient dominant selection70. A DNA fragment including the first 3 bp of F14L ORF and 297 bp upstream, intervening NotI and HindIII sites and the last 82 bp of the ORF and 218 bp downstream were generated by overlapping PCR and inserted into the PstI and BamHI sites of pUC13-Ecogpt-EGFP (pEE) plasmid, containing the Escherichia coli guanylphosphoribosyl transferase (Ecogpt) gene fused in-frame with the enhanced green fluorescent protein (EGFP) gene under the control of the VACV 7.5 K promoter27. The resulting plasmid contained an internal deletion of the F14L ORF (nucleotide positions 42,049–42,185 from VACV-WR reference genome, accession number NC_006998.1). The remaining sequence of F14L was out-of-frame and contained multiple stop codons, precluding the expression of a truncated version of F14. The derived plasmid was transfected into CV-1 cells that had been infected with VACV-WR at 0.1 plaque-forming units (p.f.u.) per cell for 1 h. After 48 h, progeny viruses that incorporated the plasmid by recombination and expressed the Ecogpt-EGFP were selected and plaque-purified three times on monolayers of BS-C-1 cells in the presence of mycophenolic acid (25 μg ml-1), supplemented with hypoxanthine (15 μg ml-1) and xanthine (250 μg ml-1). The intermediate recombinant virus was submitted to three additional rounds of plaque purification in the absence of the selecting drugs and GFP-negative plaques were selected. Under these conditions, progeny viruses can undergo a second recombination that results in loss of the Ecogpt-EGFP cassette concomitantly with either incorporation of the desired mutation (vΔF14) or reversal to wild-type genotype (vF14). Because vΔF14 and vF14 are sibling strains derived from the same intermediate virus, they are genetically identical except for the 137-bp deletion in the F14L locus. To restore F14 expression in vΔF14, the F14L locus was amplified by PCR, including about 250 bp upstream and downstream of the ORF. Alternatively, F14L ORF fused to the sequence coding a C-terminal TAP tag was also amplified by overlapping PCR, including the same flanking sequences described above. Both sequences were cloned into the PstI and BamHI sites of the pEE plasmid. These plasmids were used to generate two revertant strains derived from vΔF14: (1) vF14-Rev, in which F14 expression from its natural locus was restored and (2) vF14-TAP, expressing F14 fused to a C-terminal TAP tag. The vC6-TAP virus was described elsewhere69. A VACV vv811 strain lacking both A49 and F14 (vv811ΔA49ΔF14) was also constructed by transient dominant selection. The resultant virus contained the same 137-bp internal deletion in the F14L ORF within the vv811ΔA49 strain generated previously15. All viruses were analysed by PCR to identify recombinants by distinguishing wild-type and ΔF14 alleles and the presence or absence of the Ecogpt-EGFP cassette. The oligonucleotide primers used to generate the recombinant VACV strains are listed in Supplementary Table 1. To verify that all the final recombinant viruses harboured the correct sequences, PCR fragments spanning the F14L locus were sequenced.
Preparation of virus stocks
The stocks of virus strains derived from VACV-WR were prepared in RK13 cells. Infected cells were harvested by centrifugation, suspended in a small volume of DMEM supplemented with 2% FBS and submitted to multiple cycles of freezing/thawing and sonication to lyse the cells and disrupt aggregates of virus particles and cellular debris. These crude virus stocks were used for experiments in cultured cells. Crude stocks of vv811 and derived strains were prepared in the same way, except for the BS-C-1 cells used for infection. For the in vivo work, virus stocks were prepared by ultracentrifugation of the cytoplasmic fraction of infected-cell lysates through 36% (w/v) sucrose cushion and suspension of the virus samples in 10 mM Tris-HCl pH 9.0. The viral titres in the stocks were determined by plaque assay on BS-C-1 cells.
Virus growth, spread and infection assays
Virus single-step growth curve experiments were performed in HeLa cells. Cells were infected at 5 p.f.u. per cell in medium supplemented with 2% FBS. The inoculum was removed after 1 h of adsorption and the cells were replenished with fresh medium. At 1, 8 and 24 h p.i., infected-cell supernatants and monolayers were collected for determination of extracellular and cell-associated infectious virus titres, respectively, by plaque assay on BS-C-1 cells. Supernatants were clarified by centrifugation, whereas cell monolayers were scraped and disrupted by three cycles of freezing/thawing followed by sonication, to release intracellular virus particles. Virus spread in cell culture was assessed by plaque formation. Confluent monolayers of BS-C-1 cells in six-well plates were infected with 50 p.f.u. per well and overlaid with MEM supplemented with 2% FBS and 1.5% carboxymethylcellulose. After 48 h, infected-cell monolayers were stained with 0.5% crystal violet solution in 20% methanol and imaged. For immunoblotting and IP experiments, cells in six-well plates or 10-cm dishes, respectively, were infected at 5 p.f.u. per cell. Viral inocula were prepared in growth medium supplemented with 2% FBS. Viral adsorption was done at 37 °C for 1 h, after which the medium supplemented with 2% FBS was topped up to the appropriate volume and cells were incubated at 37 °C.
Construction of inducible F14-expressing T-REx-293 cell line
T-REx-293 cells (Invitrogen), which constitutively expresses the Tet repressor (TetR), were transfected with pcDNA4/TO-coF14-TAP plasmid, which encodes F14 under the control of the HCMV immediate early promoter and two tetracycline operator (TetO2) sites. Transfected cells were selected in the presence of blasticidin (10 μg ml-1) and zeocin (100 μg ml-1) and clonal cell lines were obtained by limiting dilution. Expression of protein F14 within these clones was analysed by immunoblotting and flow cytometry with anti-FLAG antibodies. T-REx-293-EV, T-REx-293-B14 and T-REx-293-C6 cell lines were described elsewhere21.
Reporter gene assays
HEK 293T cells in 96-well plates were transfected in quadruplicate with firefly luciferase reporter plasmid (NF-κB, ISRE or AP-1), TK-Renilla luciferase reporter plasmid (as an internal control) and the desired expression vectors or EV using TransIT-LT1 transfection reagent (Mirus Bio). On the following day, cells were stimulated with TNF-α (10 ng ml-1, PeproTech), IL-1β (20 ng ml-1, PeproTech) or IFN-a2 (1,000 U ml-1, PeproTech) for 8 h or phorbol 12-myristate 13-acetate (10 ng ml-1) for 24 h. Alternatively, NF-κB was activated by cotransfection of p65-overexpressing plasmid and cells were harvested 24 h after transfection. Where described, dilutions of the desired expression vectors were used (5 and 25 ng or 5, 10 and 25 ng, depending on the experiment). The total amount of transfected DNA was made equivalent by addition of EV. To measure NF-κB luciferase activation during infection, A549 cells transduced with a lentiviral vector expressing the firefly luciferase under the control of an NF-κB promoter (A549-NF-κB-Luc)15 were grown in 96-well plates and infected with VACV vv811 and derived strains at 5 p.f.u. per cell. After 6 h, cells were stimulated with TNF-α (10 ng ml-1, PeproTech) or IL-1β (20 ng ml-1, PeproTech) for additional 6 h. Lysates of A549-NF-κB-Luc cells grown in six-well plates and infected with the equivalent amount of virus for 12 h were analysed by immunoblotting. Cells were lysed using passive lysis buffer (Promega) and firefly and Renilla luciferase luminescence was measured using a FLUOstar luminometer (BMG), operated with FLUOstar Omega reader control software (BMG) and analysed with MARS data analysis software (BMG). During the measurement, the gain was adjusted to keep the reads within the dynamic range of the luminometer. Promoter activity was obtained by calculation of firefly/Renilla luciferase ratios and the promoter activity under pathway stimulation was normalized to the activity of the respective non-stimulated control of each protein under test. In parallel, aliquots of the replicas of each condition tested were combined, mixed with 5× SDS-gel loading buffer and immunoblotted to confirm the expression of the proteins tested.
In vivo experiments
All animal experiments were approved and conducted according to the Animals (Scientific Procedures) Act 1986 under the PPL 70/8524 issued by the UK Home Office. Mice were purchased from Envigo and housed in specific pathogen-free conditions in the Cambridge University Biomedical Services facility. For the intradermal model of infection, female C57BL/6 mice (6–8 weeks old) were inoculated with 104 p.f.u. in both ear pinnae and the diameter of the lesion was measured daily using a calliper71. For the intranasal model, female BALB/c mice (6–8 weeks old) were inoculated with 5 × 103 p.f.u. divided equally into each nostril and were weighed daily72. In both cases, viral inocula were prepared in phosphate-buffered saline (PBS) supplemented with 0.01% bovine serum albumin (BSA, Sigma-Aldrich) and the infectious titres in the administered inocula were confirmed by plaque assay. For quantification of virus replication after the intradermal infection, infected mice were killed 3, 7 and 10 d p.i. and ear tissues were collected, ground in a tissue homogenizer and passed through a 70-μm nylon mesh using DMEM containing 10% FBS. Samples were frozen, thawed and sonicated three times, to liberate cell-associated virus particles and the infectious titres present were determined by plaque assay on BS-C-1 cells.
Immunoblotting
Cells were washed with PBS and lysed on ice with cell lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 10% (v/v) glycerol, 1% (v/v) Triton X-100 and 0.05% (v/v) Nonidet P-40 (NP-40)), supplemented with protease (cOmplete Mini, Roche) and phosphatase (PhosSTOP, Roche) inhibitors, for 20 min. Lysed cells were scraped and lysates were clarified to remove insoluble material by centrifugation at 17,000g for 15 min at 4 °C. Protein concentration in the cell lysate was determined using a bicinchoninic acid protein assay kit (Pierce). After mixing with 5× SDS-gel loading buffer and boiling at 100 °C for 5 min, equivalent amounts of protein samples (15–50 μg per well) were loaded onto SDS-polyacrylamide gels or NuPAGE 4–12% Bis-Tris precast gels (Invitrogen), separated by electrophoresis and transferred onto nitrocellulose membranes (GE Healthcare). Membranes were blocked at room temperature with either 5% (w/v) non-fat milk or 3% (w/v) BSA (Sigma-Aldrich) in Tris-buffered saline (TBS) containing 0.1% (v/v) Tween-20. The membranes were incubated with specific primary antibodies diluted in blocking buffer at 4 °C overnight. After washing, membranes were probed with fluorophore-conjugated secondary antibodies (LI-COR Biosciences) diluted in 5% (w/v) non-fat milk at room temperature for 1 h. After washing, membranes were imaged using the Odyssey CLx imaging system (LI-COR Biosciences). The band intensities on the immunoblots were quantified using the Image Studio software (LI-COR Biosciences). The antibodies used for immunoblotting are listed in Supplementary Table 2.
Co-immunoprecipitation and pulldown assays
HEK 293T cells or HeLa cells in 10-cm dishes were infected at 5 p.f.u. per cell for 8 h or transfected overnight with the specified epitope-tagged plasmids using polyethylenimine (PEI, Polysciences, 2 μl of 1 mg ml-1 stock per μg of plasmid DNA). For the competition assays, cells were starved of FBS for 3 h and stimulated with TNF-α (40 ng ml-1, PeproTech) in FBS-free DMEM for 15 min before harvesting. Cells were washed with ice-cold PSB, scraped in IP buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5% (v/v) NP-40, 0.1 mM EDTA), supplemented with protease (cOmplete Mini, Roche) and phosphatase (PhosSTOP, Roche) inhibitors, on ice, transferred to 1.5-ml microcentrifuge tubes and rotated for 30 min at 4 °C. Cell lysates were centrifuged at 17,000g for 15 min at 4 °C and the soluble fractions were incubated with 20 μl of affinity resin equilibrated in IP buffer: (1) anti-FLAG M2 agarose (Sigma-Aldrich, catalogue no. A2220) for FLAG- or TAP-tagged proteins; (2) anti-HA agarose (Sigma-Aldrich, catalogue no. A2095) for HA-tagged proteins; or (3) Strep-Tactin Superflow agarose (IBA, catalogue no. 2-1206-025) for TAP-tagged proteins. After 2 h of rotation at 4 °C, the protein-bound resins were washed three times with ice-cold IP buffer. The bound proteins were eluted by incubation with 2× SDS-gel loading buffer and boiled at 100 °C for 5 min before analysis by SDS-polyacrylamide gel electrophoresis and immunoblotting, along with 10% input samples collected after clarification of cell lysates. The antibodies used for IP are listed in Supplementary Table 2.
Reverse transcription and quantitative PCR
T-REx-293-F14 in 12-well plates were left uninduced or induced overnight with 100 ng ml-1 doxycycline (Melford) to induce the expression of F14. Alternatively, T-REx-293-F14 and C6 in 12-well plates were induced overnight. The next day, cells were starved for 3 h by removal of serum from the medium and then stimulated in duplicate with TNF-α (40 ng ml-1, PeproTech) in FBS-free DMEM for 0, 1 or 6 h. RNA was extracted using RNeasy Mini Kit (Qiagen) and complementary DNA (cDNA) was synthesized using SuperScript III reverse transcriptase (Invitrogen) and oligo-dT primers (Thermo Scientific). The mRNA levels of target genes were quantified by quantitative PCR using gene-specific primer sets, fast SYBR green master mix (Applied Biosystems), the ViiA 7 real-time PCR system (Applied Biosystems) and QuantStudio software (Applied Biosystems). The primers used for the qPCR analysis of gene expression are listed in Supplementary Table 1. Fold induction was calculated by the 2-ΔΔCt method using non-induced and non-stimulated T-REx-293-F14 cells, or induced and non-stimulated T-REx-293-C6 cells, as the reference sample and GAPDH as the housekeeping control gene.
Enzyme-linked immunosorbent assay
The secretion of CXCL8 and CXCL10 was measured by enzyme-linked immunosorbent assay (ELISA). T-REx-293-EV, T-REx-293-B14, T-REx-293-C6 and T-REx-293-F14 cells in 12-well plates were incubated overnight in the presence or absence of 100 ng ml-1 doxycycline (Melford) to induce VACV protein expression. The next day, cells were stimulated in triplicate with TNF-α (40 ng ml-1, PeproTech) in DMEM supplemented with 2% FBS for 16 h. Alternatively, cells were treated with BRD4 bromodomain inhibitor (+)-JQ1 (5 μM, Abcam, catalogue ab141498, dissolved in DMSO) or the equivalent amount of DMSO (0.025% (v/v)) 30 min before TNF-α stimulation. The supernatants were assayed for human CXCL8 and CXCL10 using the respective DuoSet ELISA kits (R&D Biosystems).
Immunofluorescence
T-REx-293-EV, T-REx-293-B14, T-REx-293-C6 and T-REx-293-F14 cells were grown on poly-D-lysine-treated glass coverslips placed inside six-well plates. Following induction of protein expression with 100 ng ml-1 doxycycline (Melford) overnight, cells were starved of FBS for 3 h and then stimulated with 40 ng ml-1 TNF-α (PeproTech) in FBS-free DMEM for 15 min. Cells were washed twice with ice-cold PBS and fixed in 4% (v/v) paraformaldehyde for 10 min. After quenching of free formaldehyde with 150 mM ammonium chloride for 5 min, the fixed cells were permeabilized with 0.1% (v/v) Triton X-100 in PBS for 5 min and blocked with 10% (v/v) FBS in PBS for 30 min. Staining was carried out with primary antibodies for 1 h, followed by incubation with the appropriate AlexaFluor fluorophore-conjugated secondary antibodies (Invitrogen Molecular Probes) for 30 min and mounting onto glass slides with Mowiol 4-88 (Calbiochem) containing 0.5 μg ml-1 DAPI (4′,6-diamidino-2-phenylindole, Biotium). Images were acquired on an LSM 700 confocal microscope (Zeiss) using ZEN system software (Zeiss). Quantification of nuclear localization of p65 was done manually on the ZEN lite software (blue edition, Zeiss). The details about the antibodies used for immunofluorescence are listed in Supplementary Table 2.
Flow cytometry
T-REx-293-F14 cells were induced overnight with 100 ng ml-1 doxycycline (Melford) in the presence or absence of 10 μM MG132 (Abcam). Cells were detached with trypsin-EDTA (Gibco), washed in PBS and fixed with 4% paraformaldehyde in PBS for 10 min at room temperature with intermittent agitation by vortexing. After centrifugation, fixed cells were suspended in PBS containing 0.1% BSA (Sigma-Aldrich). Cells were permeabilized with 0.1% saponin (Sigma-Aldrich) in PBS and F14-TAP was stained with the mouse monoclonal antibody against the FLAG tag or isotype control, followed by PE goat anti-mouse IgG (Poly4053, BioLegend). Immunostained cells were fixed again with 1% paraformaldehyde in PBS. Data were acquired with a FACScan/Cytek DxP8-upgraded flow cytometry analyser FlowJo CE software and analysed with FlowJo software.
Chromatin immunoprecipitation and quantitative PCR
T-REx-F14 cells in 15-cm dishes were incubated overnight in the absence or presence of 100 ng ml-1 doxycycline (Melford) to induce F14 expression. The next day, cells were starved of FBS for 3 h and stimulated with TNF-α (40 ng ml-1, PeproTech) in FBS-free DMEM for 0, 1 or 5 h. Alternatively, T-REx-293-EV cells were treated with (+)-JQ1 (5 μM, Abcam, catalogue no. ab141498, dissolved in DMSO) or the equivalent amount of DMSO (0.025% (v/v)) 30 min before TNF-α stimulation. Cells were crosslinked with 1% (v/v) formaldehyde added directly to the growth medium. After 10 min at room temperature, crosslinking was stopped by the addition of 0.125 M glycine. Cells were then lysed in 0.2% NP-40, 10 mM Tris-HCl pH 8.0, 10 mM NaCl, supplemented with protease (cOmplete Mini, Roche), phosphatase (PhosSTOP, Roche) and histone deacetylase (10 mM sodium butyrate, Sigma-Aldrich) inhibitors and nuclei were recovered by centrifugation at 600g for 5 min at 4 °C. Nuclei were lysed in 1% (w/v) SDS, 50 mM Tris-HCl pH 8.0, 10 mM EDTA, plus protease/phosphatase/histone deacetylase inhibitors and lysates were sonicated in a Bioruptor Pico (Diagenode) to achieve DNA fragments of about 500 bp. After sonication, samples were centrifuged at 3,500g for 10 min at 4 °C and supernatants were diluted fourfold in IP dilution buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% (v/v) Triton X-100, 0.01% (w/v) SDS) supplemented with protease/phosphatase/histone deacetylase inhibitors. Protein G-conjugated agarose beads (GE Healthcare, catalogue no. 17-0618-02) equilibrated in IP dilution buffer were used to preclear the chromatin for 1 h at 4 °C with rotation. Immunoprecipitation was performed with 8 μg of anti-BRD4 antibody (Cell Signaling Technology, catalogue no. 13440) or anti-GFP (Abcam, catalogue no. ab290), used as negative IgG control, overnight at 4 °C with rotation; 20% of the precleared chromatin was kept as input control. Protein–DNA immunocomplexes were retrieved by incubation with 60 μl of equilibrated protein G-conjugated agarose beads (GE Healthcare), for 2 h at 4 °C, followed by centrifugation at 5,000g for 2 min at 4 °C. Immunocomplex-bound beads were then washed: (1) twice with IP wash I (20 mM Tris-HCl pH 8.0, 50 mM NaCl, 2 mM EDTA, 1% (v/v) Triton X-100, 0.1% (w/v) SDS); (2) once with IP wash buffer II (10 mM Tris-HCl pH 8.0, 250 mM LiCl, 1 mM EDTA, 1% (v/v) NP-40, 1% (w/v) sodium deoxycholate); and (3) twice with TE buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA). Antibody-bound chromatin was eluted with 1% SDS, 100 mM sodium bicarbonate for 15 min at room temperature. Formaldehyde crosslinks were reversed by incubation overnight at 67 °C in presence of 1 μg of RNase A and 300 mM NaCl, followed by proteinase K digestion for 2 h at 45 °C. Co-immunoprecipitated DNA fragments were purified using the QIAquick PCR purification kit (Qiagen) and analysed by quantitative PCR targeting the specific gene promoter elements. The primers used for the qPCR analysis of ChIP are listed in Supplementary Table 1 and target regions containing consensus κB sites (5′-GGGRNYYYCC-3′, in which R is a purine, Y is a pyrimidine and N is any nucleotide). Amplicons overlapping areas with histone modification often observed near active regulatory elements (H3K27ac), according to the UCSC Genome Browser (https://genome.ucsc.edu/index.html), were prioritized. Some primers have been described previously4,73,74. The chromatin immunoprecipitation and q PCR (ChIP–qPCR) data were analysed by the fold enrichment method. Briefly, the signals obtained from the ChIP with each antibody were first normalized to the signals obtained from the corresponding input sample (ΔCt = CtIP – CtInput). Next, the input-normalized signals ΔCt) were normalized to the corresponding 0 time-point control (that is, ΔΔCt = ΔCt – ΔCt0). The fold enrichment of each time-point was then calculated with the 2-ΔΔCt formula.
Statistical analysis
Experimental data are presented as means + s.d. or means ± s.e.m. for in vivo results, unless otherwise stated in figure legends. Sample size and number of repeats are indicated in the respective sublegend, when they apply to specific panels or in the end, when they apply to all above panels in the figure. Statistical significance was calculated by unpaired two-tailed Student’s t-test. In the figures, only P < 0.05 values are shown above horizontal brackets indicating the samples being compared. GraphPad Prism software was used for statistical analysis. No animals or data points were excluded from the analyses.
Biological materials
All unique materials are readily available from the corresponding authors on request. The availability of the antibodies recognizing VACV antigens and VACV proteins A49, C6 and D8 is limited.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. Screen of VACV strain WR ORFs for NF-κB inhibitory activity.
NF-κB-dependent luciferase activity in HEK 293T cells transfected with vectors expressing the indicated VACV proteins or empty vector (EV), and stimulated with TNF-α. Negative (EV, GFP, and N2) and positive (B14) controls are highlighted in the dashed black square, whilst F14 is highlighted in the dashed red square. Means + s.d. (n = 3-4 per condition) are shown.
Extended Data Fig. 2. Virulence of VACV mutant lacking F14 in the intranasal mouse model of infection.
BALB/c mice were infected intranasally with 5×103 p.f.u. of the indicated VACV strains and their body mass was measured daily. Body mass is expressed as the percentage ± s.e.m. of the mean of the same group of mice on day 0 (n = 5 mice).
Extended Data Fig. 3. Replication and spread of VACV mutant lacking F14 in cell culture.
(a,b) HeLa cells were Infected with the Indicated VACV strains (5 p.f.u./cell) and virus titres associated with the cells (a) and in the supernatants (b) were determined by plaque assay. Means (n = 2 per condition) are shown. (c) Plaque formation by the indicated VACV strains on BS-C-1 cells.
Extended Data Fig. 4. F14 does not inhibit the nuclear translocation of NF-κB subunit p65.
a, T-REx-293 cells inducibly expressing the empty vector (EV) or VACV proteins B14, C6, or F14, were induced with doxycycline and stimulated with TNF-α. Fixed and permeabilised cells were stained with anti-p65 antibody and DAPI, and analysed by confocal microscopy. Scale bars (50 μm) are shown in the bottom right of each micrograph. Representative micrographs of quantitative analysis shown in Fig. 1l. b, Flow cytometry analysis of T-REx-293-F14 induced with doxycycline in the absence and in the presence of the proteasome inhibitor MG132. F14 presence was detected by staining with an anti-FLAG antibody.
Extended Data Fig. 5. F14 inhibits NF-κB at or downstream of p65.
NF-κB activity in HEK 293 T cells transfected with vectors expressing p65, VACV proteins B14 or F14, or empty vector (EV). Top panel: Means + s.d. (n = 4 per condition) are shown. Statistical significance was determined by unpaired two-tailed Student’s t-test. Bottom panel: Immunoblotting. Protein molecular mass markers in kDa are shown on the left of the blots. Immunoblots of tagged proteins are labelled with the protein name followed by the epitope tag antibody in parentheses. When multiple tagged proteins are shown in the same immunoblot, each protein is indicated by a red arrowhead.
Extended Data Fig. 6. F14 is unique among known viral inhibitors of NF-κB.
(a) Top: Amino acid sequence of the TAD of HSV-1 VP16 with the acidic activation domain similar to p65 highlighted in red, and hydrophobic residues (Φ) are indicated. Middle: NF-κB-dependent luciferase activity in HEK 293T cells transfected with vectors expressing VP16, VP16 mutant, or empty vector (EV), and stimulated with TNF-α. Bottom: Immunoblotting. (b) Top: Amino acid residues 61-98 from HPV16 protein E7 encompassing a ΦΧΧΦΦ motif containing and preceded by negatively charged residues. Highlighted are two residues mutated to disrupt this motif. Middle: NF-κB-dependent luciferase activity in HEK 293T cells expressing E7 and two mutants as described in (a). Bottom: Immunoblotting. Means + s.d. (n = 4 per condition) are shown. c, Lysates from transfected HEK 293T cells were immunoprecipitated with anti-HA. Immunoblots are representative of two independent experiments. Protein molecular masses in kDa are shown on the left of the blots. Immunoblots of tagged proteins are labelled with the protein name followed the epitope tag antibody in parentheses. When multiple tagged proteins are shown in the same immunoblot, each protein is indicated a red arrowhead. Statistical significance was determined by the Student’s t-test.
Extended Data Fig. 7. F14 suppresses expression of a subset of NF-κB-responsive genes.
(a-d) RT-qPCR analysis of NF-κB-responsive gene expression in inducible T-REx-293 cells induced with doxycycline overnight to express VACV proteins F14 or C6, and stimulated with TNF-α. Means (n = 2 per condition) are shown. e, Immunoblotting of lysates of inducible T-REx-293 cell lines induced with doxycycline overnight. Data are representative of two independent experiments. Protein molecular masses in kDa are shown on the left of the blots.
Extended Data Fig. 8. F14 suppresses expression of CXCL10, but not CXCL8, after stimulation with TNF-α.
This shows data normalized for presentation in Fig. 5c,f. ELISA of CXCL8 (a) and CXCL10 (b) in culture supernatants from T-REx-293 cells inducibly expressing the empty vector (EV) or VACV proteins B14, C6, or F14, induced with doxycycline and stimulated with TNF-α. Means + s.d. (n = 3 per condition) are shown. Statistical significance was determined by the Student’s t-test.
Extended Data Fig. 9. JQ1 reduces BRD4 occupancy on CCND1 gene promoter.
Chromatin immunoprecipitation (ChIP) with anti-BRD4 antibody or control IgG, and qPCR for the promoters of CCND1 gene. T-REx-293 cells were treated with JQ1 and stimulated with TNF-α. Means + s.d. (n = 6 per condition from two independent experiments). Statistical significance was determined by the Student’s t-test.
Supplementary Material
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41564-021-01004-9.
Acknowledgements
We thank R. Seear, S. Macilwee and J. Milburn for technical support and F. Pfaff and M. Beer (Friedrich-Loeffler-Institut, Germany) for help with access to the cowpox RNA sequencing dataset. We also thank J. Doorbar and C. Crump (Department of Pathology, University of Cambridge, UK), T. Kouzarides (Department of Pathology and The Gurdon Institute, University of Cambridge, UK) and G. Blobel (University of Pennsylvania, Philadelphia, USA) for providing us with reagents. We are grateful to T. Kouzarides for helpful advice and to C. Talbot-Cooper for critical reading of the manuscript. This work was supported by grant no. 090315 from the Wellcome Trust (to G.L.S.). B.Y-W.C.’s laboratory is funded by the Medical Research Council (grant no. MR/R021821/1), the Biotechnology and Biological Sciences Research Council (grant no. BB/V017780.1) and Isaac Newton Trust (grant no. G101522). J.D.A. was a postdoctoral fellow of the Science without Borders programme from CNPq-Brazil (grant no. 235246/2014-0). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Author contributions
J.D.A., A.A.T. and G.L.S. conceived the idea. J.D.A., H.R., A.A.T., E.V.S., C.A.M., A.J.B., M.P.B. and B.Y-W.C. developed the methodology. J.D.A., H.R., A.A.T. and E.V.S. validated the results. J.D.A. and H.R. undertook the formal analysis. J.D.A., H.R., A.A.T. and E.V.S. undertook investigation. A.A.T., C.A.M., A.J.B., B.Y-W.C. and G.L.S. obtained resources. J.D.A. undertook data curation. J.D.A. wrote the original draft. J.D.A., H.R., A.A.T., C.A.M., A.J.B., B.Y-W.C. and G.L.S. reviewed and edited the manuscript. J.D.A. prepared visualization. J.D.A. and G.L.S. supervised the work. J.D.A. and G.L.S. were project administrators. J.D.A. and G.L.S. obtained funding. A.A.T., C.A.M., A.J.B., B.Y-W.C. and G.L.S. contributed resources.
Competing interests
The authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41564-021-01004-9.
Peer review information Nature Microbiology thanks the anonymous reviewers for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability
The authors declare that the main data supporting the findings of this study are available within the article and its Supplementary Information. Poxvirus nucleotide sequences mentioned in this study are publicly available on NCBI GenBank: VACV Western Reserve (NC_006998.1), VACV Copenhagen (M35027.1), VACV Modified Virus Ankara (AY603355.1), horsepox virus isolate MNR-76 (DQ792504.1), monkeypox virus Zaire-96-I-16 (NC_003310.10, cowpox virus Brighton Red (NC_003663.2), variola virus India-1967 (NC_001611.1), camelpox virus CMS (AY009089.1), taterapox virus Dahomey 1968 (DQ437594.1), ectromelia virus Moscow (AF012825.2), raccoonpox virus Herman (NC_027213.1), premodern variola virus (LR800247.1, LR800244.1, LR800245.1, LR800246.1) and variola virus VD21 (KY358055.1). Source data are provided with this paper.
References
- 1.Iwasaki A. A virological view of innate immune recognition. Annu Rev Microbiol. 2012;66:177–196. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 3.Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 4.Tian B, Nowak DE, Brasier AR. A TNF-induced gene expression program under oscillatory NF-kappaB control. BMC Genom. 2005;6:137. doi: 10.1186/1471-2164-6-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao M, et al. Transcriptional outcomes and kinetic patterning of gene expression in response to NF-kappaB activation. PLoS Biol. 2018;16:e2006347. doi: 10.1371/journal.pbio.2006347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaikkonen MU, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen LF, et al. NF-kappaB RelA phosphorylation regulates RelA acetylation. Mol Cell Biol. 2005;25:7966–7975. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 9.Huang B, et al. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang SM, McCance DJ. Down regulation of the interleukin-8 promoter by human papillomavirus type 16 E6 and E7 through effects on CREB binding protein/p300 and P/CAF. J Virol. 2002;76:8710–8721. doi: 10.1128/JVI.76.17.8710-8721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xing J, et al. Herpes simplex virus 1-encoded tegument protein VP16 abrogates the production of beta interferon (IFN) by inhibiting NF-kappaB activation and blocking IFN regulatory factor 3 to recruit its coactivator CBP. J Virol. 2013;87:9788–9801. doi: 10.1128/JVI.01440-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bahar MW, et al. How vaccinia virus has evolved to subvert the host immune response. J Struct Biol. 2011;175:127–134. doi: 10.1016/j.jsb.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith GL, et al. Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J Gen Virol. 2013;94:2367–2392. doi: 10.1099/vir.0.055921-0. [DOI] [PubMed] [Google Scholar]
- 14.Albarnaz JD, Torres AA, Smith GL. Modulating vaccinia virus immunomodulators to improve immunological memory. Viruses. 2018;10:101. doi: 10.3390/v10030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sumner RP, et al. Vaccinia virus inhibits NF-kappaB-dependent gene expression downstream of p65 translocation. J Virol. 2014;88:3092–3102. doi: 10.1128/JVI.02627-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perkus ME, et al. Deletion of 55 open reading frames from the termini of vaccinia virus. Virology. 1991;180:406–410. doi: 10.1016/0042-6822(91)90047-f. [DOI] [PubMed] [Google Scholar]
- 17.Assarsson E, et al. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc Natl Acad Sci USA. 2008;105:2140–2145. doi: 10.1073/pnas.0711573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z, et al. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc Natl Acad Sci USA. 2010;107:11513–11518. doi: 10.1073/pnas.1006594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gubser C, et al. Poxvirus genomes: a phylogenetic analysis. J Gen Virol. 2004;85:105–117. doi: 10.1099/vir.0.19565-0. [DOI] [PubMed] [Google Scholar]
- 21.Ferguson BJ, et al. Vaccinia virus protein N2 is a nuclear IRF3 inhibitor that promotes virulence. J Gen Virol. 2013;94:2070–2081. doi: 10.1099/vir.0.054114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen RA, et al. Inhibition of IkappaB kinase by vaccinia virus virulence factor B14. PLoS Pathog. 2008;4:e22. doi: 10.1371/journal.ppat.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuart JH, et al. Vaccinia virus protein C6 inhibits type I IFN signalling in the nucleus and binds to the transactivation domain of STAT2. PLoS Pathog. 2016;12:e1005955. doi: 10.1371/journal.ppat.1005955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torres AA, et al. Multiple Bcl-2 family immunomodulators from vaccinia virus regulate MAPK/AP-1 activation. J Gen Virol. 2016;97:2346–2351. doi: 10.1099/jgv.0.000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z, et al. Deciphering poxvirus gene expression by RNA sequencing and ribosome profiling. J Virol. 2015;89:6874–6886. doi: 10.1128/JVI.00528-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, et al. Genome-wide analysis of the 5′ and 3′ ends of vaccinia virus early mRNAs delineates regulatory sequences of annotated and anomalous transcripts. J Virol. 2011;85:5897–5909. doi: 10.1128/JVI.00428-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unterholzner L, et al. Vaccinia virus protein C6 is a virulence factor that binds TBK-1 adaptor proteins and inhibits activation of IRF3 and IRF7. PLoS Pathog. 2011;7:e1002247. doi: 10.1371/journal.ppat.1002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soday L, et al. Quantitative temporal proteomic analysis of vaccinia virus infection reveals regulation of histone deacetylases by an interferon antagonist. Cell Rep. 2019;27:1920–1933. doi: 10.1016/j.celrep.2019.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parrish S, Moss B. Characterization of a vaccinia virus mutant with a deletion of the D10R gene encoding a putative negative regulator of gene expression. J Virol. 2006;80:553–561. doi: 10.1128/JVI.80.2.553-561.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ehlers A, et al. Poxvirus orthologous clusters (POCs) Bioinformatics. 2002;18:1544–1545. doi: 10.1093/bioinformatics/18.11.1544. [DOI] [PubMed] [Google Scholar]
- 31.Tamosiunaite A, et al. What a difference a gene makes: identification of virulence factors of cowpox virus. J Virol. 2020;94:e01625–19. doi: 10.1128/JVI.01625-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muhlemann B, et al. Diverse variola virus (smallpox) strains were widespread in northern Europe in the Viking Age. Science. 2020;369:eaaw8977. doi: 10.1126/science.aaw8977. [DOI] [PubMed] [Google Scholar]
- 33.Duggan AT, et al. 17th century variola virus reveals the recent history of smallpox. Curr Biol. 2016;26:3407–3412. doi: 10.1016/j.cub.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelley LA, et al. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lecoq L, et al. Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappaB and transcription regulatory factors. Nucleic Acids Res. 2017;45:5564–5576. doi: 10.1093/nar/gkx146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitz ML, Baeuerle PA. The p65 subunit is responsible for the strong transcription activating potential of NF-kappa B. EMBO J. 1991;10:3805–3817. doi: 10.1002/j.1460-2075.1991.tb04950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitz ML, et al. Interaction of the COOH-terminal transactivation domain of p65 NF-kappa B with TATA-binding protein, transcription factor IIB, and coactivators. J Biol Chem. 1995;270:7219–7226. doi: 10.1074/jbc.270.13.7219. [DOI] [PubMed] [Google Scholar]
- 38.Schmitz ML, et al. Structural and functional analysis of the NF-kappa B p65 C terminus: an acidic and modular transactivation domain with the potential to adopt an alpha-helical conformation. J Biol Chem. 1994;269:25613–25620. [PubMed] [Google Scholar]
- 39.Blair WS, et al. Mutational analysis of the transcription activation domain of RelA: identification of a highly synergistic minimal acidic activation module. Mol Cell Biol. 1994;14:7226–7234. doi: 10.1128/mcb.14.11.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang F, et al. IKK beta plays an essential role in the phosphorylation of RelA/p65 on serine 536 induced by lipopolysaccharide. J Immunol. 2003;170:5630–5635. doi: 10.4049/jimmunol.170.11.5630. [DOI] [PubMed] [Google Scholar]
- 41.Richards KH, et al. The human papillomavirus (HPV) E7 protein antagonises an Imiquimod-induced inflammatory pathway in primary human keratinocytes. Sci Rep. 2015;5:12922. doi: 10.1038/srep12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spitkovsky D, et al. The human papillomavirus oncoprotein E7 attenuates NF-kappa B activation by targeting the Ikappa B kinase complex. J Biol Chem. 2002;277:25576–25582. doi: 10.1074/jbc.M201884200. [DOI] [PubMed] [Google Scholar]
- 43.Vandermark ER, et al. Human papillomavirus type 16 E6 and E 7 proteins alter NF-kB in cultured cervical epithelial cells and inhibition of NF-kB promotes cell growth and immortalization. Virology. 2012;425:53–60. doi: 10.1016/j.virol.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Devaiah BN, Gegonne A, Singer DS. Bromodomain 4: a cellular Swiss army knife. J Leukoc Biol. 2016;100:679–686. doi: 10.1189/jlb.2RI0616-250R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mochizuki K, et al. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J Biol Chem. 2008;283:9040–9048. doi: 10.1074/jbc.M707603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pallett MA, et al. Vaccinia Virus BBK E3 Ligase Adaptor A55 Targets Importin-Dependent NF-kappaB Activation and Inhibits CD8(+) T-Cell Memory. J Virol. 2019;93:e00051–19. doi: 10.1128/JVI.00051-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eaglesham JB, et al. Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS-STING signalling. Nature. 2019;566:259–263. doi: 10.1038/s41586-019-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arenzana-Seisdedos F, et al. Inducible nuclear expression of newly synthesized I kappa B alpha negatively regulates DNA-binding and transcriptional activities of NF-kappa B. Mol Cell Biol. 1995;15:2689–2696. doi: 10.1128/mcb.15.5.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arenzana-Seisdedos F, et al. Nuclear localization of I kappa B alpha promotes active transport of NF-kappa B from the nucleus to the cytoplasm. J Cell Sci. 1997;110:369–378. doi: 10.1242/jcs.110.3.369. [DOI] [PubMed] [Google Scholar]
- 53.Kanno T, et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol. 2014;21:1047–1057. doi: 10.1038/nsmb.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukherjee SP, et al. Analysis of the RelA:CBP/p300 interaction reveals its involvement in NF-kappaB-driven transcription. PLoS Biol. 2013;11:e1001647. doi: 10.1371/journal.pbio.1001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staller MV, et al. A high-throughput mutational scan of an intrinsically disordered acidic transcriptional activation domain. Cell Syst. 2018;6:444–455. doi: 10.1016/j.cels.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neidel S, et al. NF-kappaB activation is a turn on for vaccinia virus phosphoprotein A49 to turn off NF-kappaB activation. Proc Natl Acad Sci USA. 2019;116:5699–5704. doi: 10.1073/pnas.1813504116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mansur DS, et al. Poxvirus targeting of E3 ligase beta-TrCP by molecular mimicry: a mechanism to inhibit NF-kappaB activation and promote immune evasion and virulence. PLoS Pathog. 2013;9:e1003183. doi: 10.1371/journal.ppat.1003183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bravo Cruz AG, Shisler JL. Vaccinia virus K1 ankyrin repeat protein inhibits NF-kappaB activation by preventing RelA acetylation. J Gen Virol. 2016;97:2691–2702. doi: 10.1099/jgv.0.000576. [DOI] [PubMed] [Google Scholar]
- 59.Diel DG, et al. A nuclear inhibitor of NF-kappaB encoded by a poxvirus. J Virol. 2011;85:264–275. doi: 10.1128/JVI.01149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Connor MJ, et al. Characterization of an E1A-CBP interaction defines a novel transcriptional adapter motif (TRAM) in CBP/p300. J Virol. 1999;73:3574–3581. doi: 10.1128/jvi.73.5.3574-3581.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marzio G, et al. HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc Natl Acad Sci USA. 1998;95:13519–13524. doi: 10.1073/pnas.95.23.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patel D, et al. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999;18:5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cook JL, et al. Role of the E1A Rb-binding domain in repression of the NF-kappa B-dependent defense against tumor necrosis factor-alpha. Proc Natl Acad Sci USA. 2002;99:9966–9971. doi: 10.1073/pnas.162082999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naar AM, et al. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 65.Richardson PM, Gilmore TD. vRel is an inactive member of the Rel family of transcriptional activating proteins. J Virol. 1991;65:3122–3130. doi: 10.1128/jvi.65.6.3122-3130.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin JR, et al. Minimalist ensemble algorithms for genome-wide protein localization prediction. BMC Bioinf. 2012;13:157. doi: 10.1186/1471-2105-13-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gloeckner CJ, et al. A novel tandem affinity purification strategy for the efficient isolation and characterisation of native protein complexes. Proteomics. 2007;7:4228–4234. doi: 10.1002/pmic.200700038. [DOI] [PubMed] [Google Scholar]
- 69.Maluquer de Motes C, et al. Vaccinia virus virulence factor N1 can be ubiquitylated on multiple lysine residues. J Gen Virol. 2014;95:2038–2049. doi: 10.1099/vir.0.065664-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Falkner FG, Moss B. Transient dominant selection of recombinant vaccinia viruses. J Virol. 1990;64:3108–3111. doi: 10.1128/jvi.64.6.3108-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tscharke DC, Smith GL. A model for vaccinia virus pathogenesis and immunity based on intradermal injection of mouse ear pinnae. J Gen Virol. 1999;80:2751–2755. doi: 10.1099/0022-1317-80-10-2751. [DOI] [PubMed] [Google Scholar]
- 72.Williamson JD, et al. Biological characterization of recombinant vaccinia viruses in mice infected by the respiratory route. J Gen Virol. 1990;71:2761–2767. doi: 10.1099/0022-1317-71-11-2761. [DOI] [PubMed] [Google Scholar]
- 73.Harris DP, et al. Tumor necrosis factor (TNF)-alpha induction of CXCL10 in endothelial cells requires protein arginine methyltransferase 5 (PRMT5)-mediated nuclear factor (NF)-kappaB p65 methylation. J Biol Chem. 2014;289:15328–15339. doi: 10.1074/jbc.M114.547349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu X, et al. EVI1 acts as an inducible negative-feedback regulator of NF-kappaB by inhibiting p65 acetylation. J Immunol. 2012;188:6371–6380. doi: 10.4049/jimmunol.1103527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the main data supporting the findings of this study are available within the article and its Supplementary Information. Poxvirus nucleotide sequences mentioned in this study are publicly available on NCBI GenBank: VACV Western Reserve (NC_006998.1), VACV Copenhagen (M35027.1), VACV Modified Virus Ankara (AY603355.1), horsepox virus isolate MNR-76 (DQ792504.1), monkeypox virus Zaire-96-I-16 (NC_003310.10, cowpox virus Brighton Red (NC_003663.2), variola virus India-1967 (NC_001611.1), camelpox virus CMS (AY009089.1), taterapox virus Dahomey 1968 (DQ437594.1), ectromelia virus Moscow (AF012825.2), raccoonpox virus Herman (NC_027213.1), premodern variola virus (LR800247.1, LR800244.1, LR800245.1, LR800246.1) and variola virus VD21 (KY358055.1). Source data are provided with this paper.