Abstract
Poxviruses comprise a group of large dsDNA viruses that include members relevant to human and animal health, such as variola virus, monkeypox virus, cowpox virus and vaccinia virus (VACV). Poxviruses are remarkable for their unique replication cycle, which is restricted to the cytoplasm of infected cells. The independence from the host nucleus requires poxviruses to encode most of the enzymes involved in DNA replication, transcription and processing. Here, we use the CRISPR/Cas9 genome engineering system to induce DNA damage to VACV (strain Western Reserve) genomes. We show that targeting CRISPR/Cas9 to essential viral genes limits virus replication efficiently. Although VACV is a strictly cytoplasmic pathogen, we observed extensive viral genome editing at the target site; this is reminiscent of a non-homologous end-joining DNA repair mechanism. This pathway was not dependent on the viral DNA ligase, but critically involved the cellular DNA ligase IV. Our data show that DNA ligase IV can act outside of the nucleus to allow repair of dsDNA breaks in poxvirus genomes. This pathway might contribute to the introduction of mutations within the genome of poxviruses and may thereby promote the evolution of these viruses.
Keywords: Poxvirus, viral genome repair, DNA ligase IV, CRISPR/Cas9, genome editing, mutagenesis
Introduction
Poxviruses are a group of large dsDNA viruses that are capable of infecting a wide range of host species [1]. Among these viruses are clinically and economically relevant species, including variola virus, the causative agent of smallpox, vaccinia virus (VACV), monkeypox virus and cowpox virus (CPXV). Although smallpox was declared eradicated in 1980, several variola virus strains are retained in two WHO-approved maximum security laboratories in USA and Russia. Variola virus is considered a bioterrorist threat. Monkeypox virus and CPXV are emerging pathogens that cause zoonotic infections in humans that are occasionally fatal [2]. VACV was used as the vaccine to eradicate smallpox and is both a promising gene delivery vector for vaccine development and a promising oncolytic agent [3–5]. VACV is considered to be a model poxvirus and is frequently used to study the orthopoxvirus life cycle.
In contrast to most DNA viruses, poxviruses replicate exclusively in the cytosol, where they form specialized replication sites known as virosomes or viral factories [6]. To allow virus replication outside of the nucleus, poxvirus genomes encode most of the factors necessary for DNA processing and transcription, including DNA polymerase, helicase–primase and ligase enzymes, as reviewed in [7]. Additionally, the virus encodes DNA-processing factors that aid the repair of damaged viral genomes [8, 9]. The FEN1-like nuclease encoded by the VACV G5R gene mediates double-stranded break (DSB) repair through homologous recombination [9]. The VACV DNA ligase is non-essential for virus replication [10, 11], but is reported to be part of the DNA replication machinery involved in the ligation of DNA fragments produced during lagging strand DNA synthesis [9, 12]. In addition, the viral DNA ligase plays a role in DNA repair upon damage [8]. Correspondingly, poxvirus mutants lacking the DNA ligase display enhanced sensitivity to DNA damage caused by UV irradiation and chemical inducers of DSBs [8]. However, the extent of this effect is cell line-dependent, suggesting that DNA repair may also involve host cell DNA ligases [8, 13, 14]. Other host cell factors may also be involved in VACV repair, as underscored by studies in cells derived from xeroderma pigmentosum (XP) patients [15]. These cells have mutations in their nucleotide-excision enzymes and consequently lack a functional DNA repair system [16]. The repair of VACV upon UV irradiation was highly reduced in XP cells, although this observation has also been disputed [14, 17]. Thus, the function of specific cellular and viral DNA repair enzymes in viral DNA repair is incompletely understood. In addition, the effect of repair pathways on the integrity of the viral genome remains unknown. In this respect, erroneous repair of damaged viral DNA may contribute to the generation of mutant viruses and thereby affect poxvirus evolution.
In the nucleus, DNA damage can be repaired through different pathways, including homologous recombination and non-homologous end-joining (NHEJ). Homologous recombination occurs generally at high fidelity and requires a DNA template that is complementary to the site of the DNA damage. The final ligation step of this pathway is mediated by cellular DNA ligase I or III [18]. In the absence of a donor template, DNA damage can be repaired through different error-prone non-homologous end-joining processes [19]. The DNA ligase IV-dependent canonical NHEJ pathway (C-NHEJ) initiates upon recognition of a DSB by Ku80/ Ku70 complexes [20]. Subsequently, additional factors are recruited, including the DNA-PKcs kinase, the endonuclease Artemis, which trims damaged DNA ends, and DNA polymerases that replace affected nucleotides. Ultimately, the DNA fragments are ligated by the XRCC4–DNA ligase IV complex. Although this process is error-prone and frequently induces small insertions or deletions (indels) at the site of the lesion, C-NHEJ is crucial in maintaining chromosomal integrity upon damage induced by e.g. DNA replication stress, irradiation or free oxygen radicals [21]. In the absensce of C-NHEJ, e.g. in cells lacking DNA ligase IV, DNA repair may involve alternative end-joining pathways (A-EJ). These pathways rely on DNA ligase I and III and induce extensive mutagenic rearrangements and chromosomal translocations [22].
The error-prone features of C-NHEJ are exploited by DSB-inducing genome editing techniques to induce site-specific alterations to cellular genomes, such as the clustered regularly interspaced short palindromic repeats (CRISPR)/ CRISPR-associated protein (Cas)9 (Cas proteins) system (CRISPR/Cas9) [23]. The Cas9 endonuclease is recruited to a desired genomic site by means of a guide RNA (gRNA) that consists of a CRISPR RNA (crRNA) that is complementary to a genomic target of interest, coupled to a trans-activating crRNA (tracrRNA). Cas9 subsequently recognizes and cleaves the genomic target DNA three nucleotides upstream of the protospacer-adjacent motif (PAM) site that is present immediately downstream of the targeted DNA sequence [24]. This DSB subsequently activates DNA repair pathways, including C-NHEJ [25]. When targeting protein-encoding genes, C-NHEJ may induce the disruption of the coding region, thereby knocking out the desired gene. In addition, the CRISPR/Cas9 system has been used to promote homologous recombination events by co-delivery of a donor template, thereby allowing specific mutagenesis at the target site [26]. The CRISPR/Cas9 system has become an indispensable tool for modifying genomes [27].
Recently, the CRISPR/Cas9 system has been adapted to target and manipulate the genomes of multiple human viruses [28]. CRISPR/Cas9-targeting of viral genomes efficiently generated desired mutations in adenovirus [29], herpes simplex virus type 1 (HSV-1) [29–31] and VACV [32]. In addition, the CRISPR/Cas9 system can efficiently limit and prevent infections of HIV-1 [33–39] human herpesviruses [40–42], human papillomavirus [43, 44], hepatitis B virus [45, 46], hepatitis C virus [47] and JC virus [48]. The ability to severely hamper virus replication makes the CRISPR/ Cas9 system a promising candidate for antiviral therapy.
In this study, we characterize the mechanisms of poxvirus DNA repair upon the introduction of DSBs in the VACV genome using the CRISPR/Cas9 system. The introduction of DSBs induces desired homologous recombination events in the presence of a template DNA. In the absence of a donor template, however, the introduction of DSB into the poxvirus genome severely limits virus replication. Nevertheless, error-prone NHEJ induced the generation of virus variants that bypass CRISPR/Cas9 targeting. These variants are selected quickly during the productive phase of the infection. Hence, although poxviruses replication occurs exclusively in the cytosol, we observed extensive viral genome editing via an NHEJ pathway. This process was not dependent on the viral DNA ligase, but critically involved the cellular DNA ligase IV-dependent C-NHEJ. Our data suggest that DNA ligase IV is recruited to the cytosolic viral factories to aid in poxvirus mutagenic DNA repair during virus infection.
Results
CRISPR/Cas9 promotes homologous recombination within the VACV genome
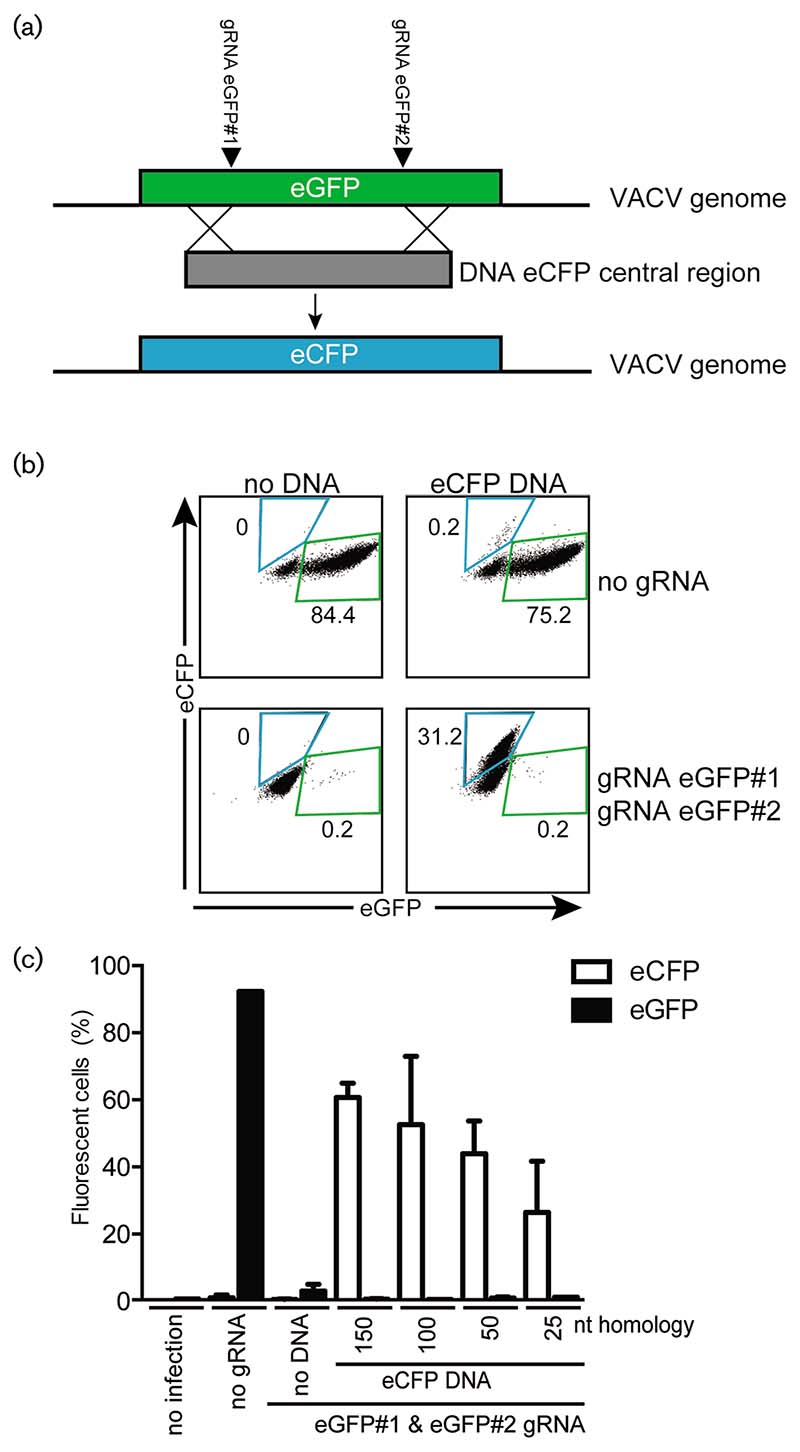
The protocols to generate specific dsDNA virus mutants rely predominantly on spontaneous homologous recombination events between a viral genome and a DNA template sharing sequence homology with a specific targeted region. This strategy is inefficient and time-consuming, with only 0.1% of the viruses containing the desired mutation. Recent studies, however, have indicated that homologous recombination events can be promoted effectively at sites of DSBs using the CRISPR/Cas9 genome editing system [49, 50]. Here, we used the CRISPR/Cas9 technology to direct the generation of specific VACV mutants in the human melanoma cell line MelJuSo (MJS). For this, we aimed to replace the eGFP marker gene from the recombinant VACV-eGFP virus with an eCFP gene. Two guide RNAs (gRNAs) targeting the regions 5' and 3' of the central part of the eGFP gene were expressed in cells. These cells were transfected with a PCR product encoding the central domain of eCFP and a flanking sequence homologous to eGFP. The homologous sequences between the viral eGFP and template eCFP would aid the homologous recombination event, thereby replacing eGFP for eCFP (Fig. 1a). Subsequently, the cells were infected with VACV-eGFP. Virus progeny were harvested in the supernatant and used to reinfect wild-type MJS cells. The production of eGFP- and eCFP-expressing virus was subsequently analysed by flow cytometry (Fig. 1b). Cells lacking gRNAs and a PCR template produced eGFP-expressing virus upon infection (Fig. 1b, top left panel). Upon the introduction of anti-eGFP gRNAs, however, the production of eGFP-expressing virus was efficiently abrogated (Fig. 1b, lower left panel). The transfection of a donor template comprising part of the eCFP gene with 5' and 3' sequences identical to eGFP (Fig. 1a) into cells not expressing gRNAs resulted in low-efficiency homologous recombination-induced production of eCFP-expressing virus (Fig. 1b, top right panel). In contrast, the transfection of the eCFP donor template in gRNA-expressing cells resulted in potent production of eCFP-, but not eGFP-, expressing virus (Fig. 1b, lower right panel). This indicates that the eCFP template efficiently replaced the eGFP sequence within the VACV-eGFP genome. Thus, the CRISPR/Cas9-mediated induction of DSBs increases the frequency of desired virus mutants in the presence of a homologous DNA template.
Fig. 1. CRISPR/Cas9 allows for efficient directed construction of VACV mutants.
(a) Schematic representation of the approach to generate VACV mutants by homologous recombination. Two anti-eGFP gRNAs introduce double-stranded breaks in the eGFP gene present on the genome of VACV-WR encoding eGFP (VACV-eGFP). Simultaneous administration of a repair PCR product comprising the eCFP gene with 5' and 3' sequence homology to the eGFP termini results in recombination and the production of VACV-eCFP. (b) Control cells (no gRNA) or cells expressing gRNAs targeting eGFP (eGFP#1 and eGFP#2) were transfected with the eCFP PCR product described in (a). MJS cells were subsequently infected with VACV-eGFP (m.o.i.: 0.1), and virus supernatant was harvested 8 days post-infection (p.i.) and used to infect new cells. Two days p.i., eGFP and eCFP expression was analysed by flow cytometry. The percentages of eGFP or eCFP-expressing cells are indicated. (c) PCR products with the indicated nucleotide overlap with the VACV genome at their termini were transfected into MJS cells. Subsequently, the cells were infected with VACV and analysed as in (b). The percentage of cells expressing the indicated fluorescent protein is presented.
Next, we assessed the impact of the length of the flanking sequence on the guiding of the homologous recombination event upon CRISPR targeting. For this, PCR products with different flanking sequence lengths were transfected into gRNA-expressing cells that were subsequently infected with VACV-eGFP. We observed an enhanced incorporation of template PCR products at the target region when longer flanking regions were used, although recombination occurred with flanking regions as short as 25 bps (Fig. 1c).
CRISPR/Cas9 targeting essential poxvirus genes limits virus replication
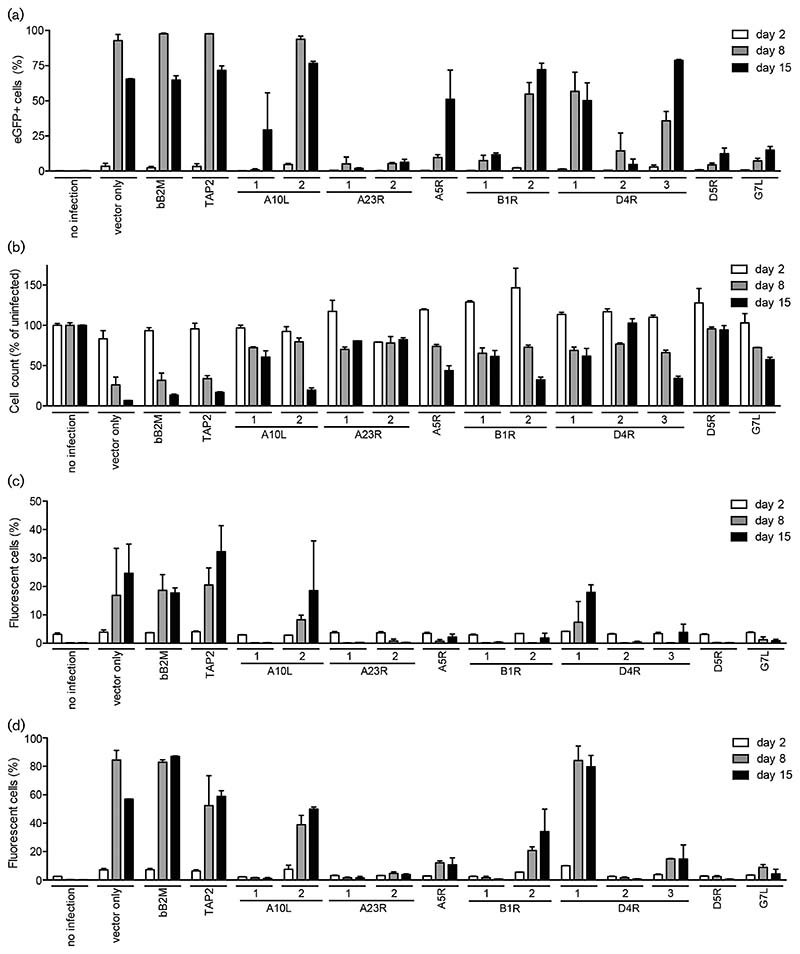
As CRISPR/Cas9 targeting of poxvirus genomes proved to be efficient in promoting homologous recombination events, we next asked whether targeting essential regions of the viral genome could limit virus replication. To test this hypothesis, gRNAs targeting poxvirus genes (see Table 1 in the Methods section) were co-expressed with Cas9 in MJS cells. Subsequently, the cells were infected with VACV-eGFP at an m.o.i. of 0.01 and monitored up to 15 days for eGFP expression (Fig. 2a). MJS cells expressing control vector only or gRNAs targeting human TAP2 or bB2M were readily infected with VACV-eGFP, resulting in potent eGFP expression in all cells at 8 days p.i. In contrast, VACV-eGFP infection was severely hampered in the presence of most gRNAs targeting VACV genes. Several of these gRNAs almost completely protected cells from productive VACV infection for up to 15 days, including A23R#1, gRNA A23R#2 and gRNA D5R. Some gRNAs showed a less pronounced effect, resulting in abrogated infection at early time points, but outgrowth of virus at 15 days post-infection (p.i.).
Table 1. CRISPR target sequences.
| CRISPR name | Target | Target sequence |
|---|---|---|
| A10L#1 | A10L | GTGAGATCGACGTTTGGTAA |
| A10L#2 | A10L | GTTTCCGAACTAATGTTAAT |
| A23R#1 | A23R | GAAAGAACGCATTTCCTCAG |
| A23R#2 | A23R | GTACAGTAGAGATATTTGAG |
| A5R | A5R | GTAAACGATTTTGACAAAGA |
| B1R#1 | B1R | GTTGGACCATTAATAGGAAA |
| B1R#2 | B1R | GAATCGATATTCCACATTAA |
| D4R#1 | D4R | GGTACACCAGTTCCATCTTT |
| D4R#2 | D4R | GGAACCAGTAATGAGTCAAT |
| D4R#3 | D4R | GTTAGTGTTCTTTATTGTTT |
| D5R | D5R | GTACACTATTCGAAAGTCTT |
| G7L | G7L | GGACAATCTTCGATGTCATT |
| bB2M | Bovine β-2M | GCTGCTGTCGCTGTCGGAC |
| TAP2 | Human TAP2 | GAAGAAGAAGGCGGCAACG |
| eGFP#1 | eGFP | GCTGAAGCACTGCACGCCGT |
| eGFP#2 | eGFP | GGAGCGCACCATCTTCTTCA |
| eGFP#3 | eGFP | GATGCCGTTCTTCTGCTTGT |
| LIGIV | Human DNA ligase IV | TAAACTACAGAACACCCAC |
Fig. 2. CRISPR/Cas9-targeting of poxvirus genes limit virus infection.
(a, b) MJS cells expressing the indicated gRNAs were infected with VACV-WR expressing eGFP (m.o.i. 0.01), and harvested 2, 8 and 15 days post-infection (p.i.). Subsequently, the cells were analysed for eGFP expression (a) and viability (b) by flow cytometry. The cell counts for infected cells were compared to those for uninfected cells (set at 100 %). Cells expressing Cas9 vector only, or in combination with gRNAs targeting bovine β-2M (bB2M) or TAP2, served as infection controls. (c, d) MJS cells expressing the indicated gRNAs were infected with cowpox virus strain Brighton Red expressing RFP and eGFP using an m.o.i. of 0.01 (c) or 0.1 (d). Fluorescence was analysed by flow cytometry at 5, 8 and 15 days p.i.
Under normal conditions, VACV infection results in cell death of infected MJS cells. To quantify cell survival upon VACV-eGFP infection in the presence or absence of anti-VACV gRNAs, we counted the percentage of viable cells at 2, 8 and 15 days p.i. (Fig. 2b). As expected, the percentage of viable cells was inversely correlated to the percentage of eGFP-expressing cells. Cell counts declined in control cells and cells expressing the non-protective gRNAs A10L#2, whereas all other anti-VACV gRNAs (partially) protected MJS cells from VACV-induced cell death up to 15 days p.i. (Fig. 2b).
The gRNAs used in this study target sequences that are highly conserved among orthopoxviruses. To assess whether anti-VACV gRNAs also protect against other members of the orthopoxvirus genus, we infected the gRNA-expressing MJS cells with eGFP-tagged CPXV. CPXV is a virus from rodents that can cause zoonotic infections in several mammals, including humans [2]. The progression of CPXV infection was monitored over time (Fig. 2c, d). Most anti-VACV gRNAs abrogated CPXV infection effectively.
CRISPR/Cas9-targeting of non-essential VACV genes triggers NHEJ to repair DSBs
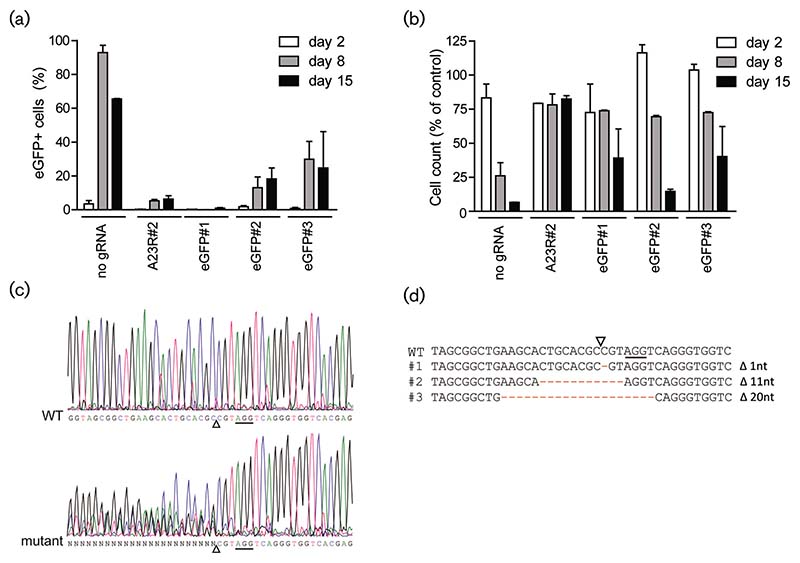
CRISPR/Cas9-mediated targeting of viral genes may abrogate infection by two different mechanisms: (1) a DSB induced by Cas9 results in a segmented poxvirus genome, or (2) the DSB is repaired but mutations are introduced that render the virus non-infectious. As the replication of poxviruses occurs outside of the nucleus [7], potential repair of DSBs might not involve the C-NHEJ response. We first assessed whether the CRISPR-Cas9 system is capable of inducing C-NHEJ-resembling mutagenesis at the target site, which would induce the generation of virus variants with small indels. For this, we studied the effect of anti-eGFP gRNAs on the eGFP gene present in the viral VACV-eGFP genome in more detail. Upon the infection of anti-eGFP gRNA-containing MJS cells with VACV-eGFP, expression of the fluorescent protein was significantly reduced in three out of three anti-eGFP gRNAs (Fig. 3a). gRNA eGFP#1 was especially potent at limiting eGFP expression. This is likely due to the location of the target site, which encodes amino acids that are crucial for eGFP fluorescence [51]. Interestingly, although no eGFP expression was observed, the anti-eGFP gRNA-containing cell cultures eventually showed cytopathic effects (CPE) and died (Fig. 3b). Apparently, upon CRISPR/Cas9 targeting of viral eGFP, the expression of eGFP is abrogated, but virus replication is retained. This suggests that VACV genomes may be repaired by an error-prone NHEJ pathway. Repair-induced mutations at the target site disrupt the eGFP gene, yet retain an intact infectious virus genome. Such mutations might be either indels or amino acid substitutions that disrupt the fluorescence of eGFP. Indeed, the eGFP gene was extensively edited in viral genomes produced by gRNA-expressing cells, but not control cells (Fig. 3c). Subsequent sequencing of individual virus clones confirmed that the eGFP gene was mutated, resulting in out-of-frame eGFP virus variants (Fig. 3d). In conclusion, our data indicate that the strictly cytosolic viral genome can be repaired at DSB sites, probably via NHEJ.
Fig. 3.
CRISPR/Cas9-targeting of the VACV genome triggers NHEJ to repair DSBs in the cytosol. Control cells (no gRNA), cells expressing a gRNA targeting the essential VACV gene A23R and cells expressing a gRNA targeting eGFP were infected with VACV-eGFP at an m.o.i. of 0.01. Subsequently, cells were analysed for eGFP expression (a) and survival (b) by flow cytometry. (c) Cells expressing gRNA eGFP#1 were infected with VACV and expression of the virus-encoded eGFP gene was subsequently assessed by Sanger sequencing. Sequence analysis of wild-type VACV (WT; upper panel) and polyclonal VACV mutants targeted by gRNA eGFP#1 (mutant; lower panel) is indicated. Solid lines correspond to the gRNA eGFP#1 PAM sequence; open triangles indicate the Cas9 cleavage site. (d) VACV genome sequences surrounding the gRNA eGFP#1 target site of wild-type virus and three isolated mutants.
CRISPR/Cas9-mediated VACV genome editing is partially mediated by the viral DNA ligase
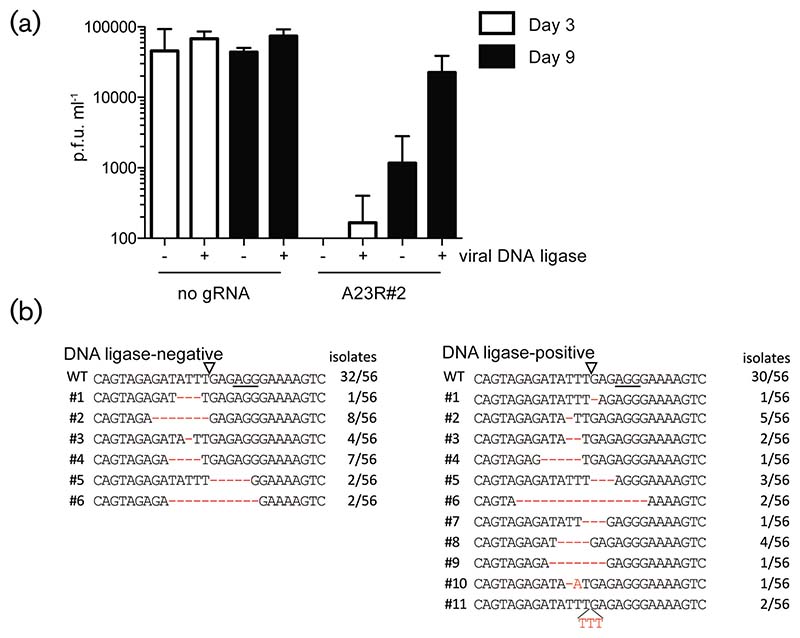
The occurrence of mutations at eGFP gRNA target sites in the VACV-eGFP genome suggests that mutagenic DSB repair can occur in the cytosol, which is the replication site for poxviruses [7]. Poxviruses encode several proteins that could be involved in this process, including the poxvirus DNA ligase encoded by the A50R gene. Indeed, an A50R-deletion virus (vSK20) is more sensitive to DNA-damaging agents than the revertant virus vSK21 that expresses the DNA ligase [8]. We used these viruses to assess whether the viral DNA ligase is involved in the observed editing of poxviruses upon CRISPR/Cas9 targeting. As expected, both vSK20 and vSK21 could readily infect and replicate in MJS cells, although the revertant virus propagated slightly faster than the ligase knockout mutant (Fig. 4a), as reported previously [8, 13]. Propagation of vSK20 and vSK21 was assessed in the presence of the anti-A23R#2 gRNA. The fact that this gRNA permitted some replication of VACV-eGFP after prolonged culture (see Fig. 2a) suggests that escape mutants may have formed that bypass CRISPR/Cas9 targeting. In the presence of the gRNA, virus production of vSK20 and vSK21 was severely reduced 3 days p.i. However, prolonged culture did give rise to virus replication, as the virus titres increased for the revertant vSK21 strain at 9 days p.i. Only a modest recovery was observed for the DNA ligase knockout virus vSK20 (Fig. 4a). To assess whether mutations arose at the gRNA target sites in these viruses, the gRNA target sites were sequenced (Fig. 4b). We observed frequent editing of the A23R#2 gRNA target site for both the knockout and revertant virus. Our data show that VACV lacking the A50R DNA ligase is more sensitive to CRISPR/Cas9-induced DNA damage, although mutagenic repair still occurs and results in the formation of escape variants.
Fig. 4.
Mutagenic DNA repair of VACV genomes occurs in the absence of viral DNA ligase. MJS cells expressing Cas9 only or in combination with the gRNA A23R#2 were infected with VACV DNA ligase mutant vSK20 (—) or revertant vSK21 (+). (a) The virus titres were determined in the cell supernatant 3 and 9 days p.i. using plaque assays. (b) VACV genomic regions surrounding gRNA target sites were amplified from virus supernatant and sequenced. Solid lines indicate the PAM sequences; open triangles indicate the Cas9 cleavage site. Numbers indicate the abundance of the given virus mutant within the 56 genomes per virus population. p.f.u., plaque-forming units.
Cellular DNA ligase IV mediates NHEJ of poxvirus genomes
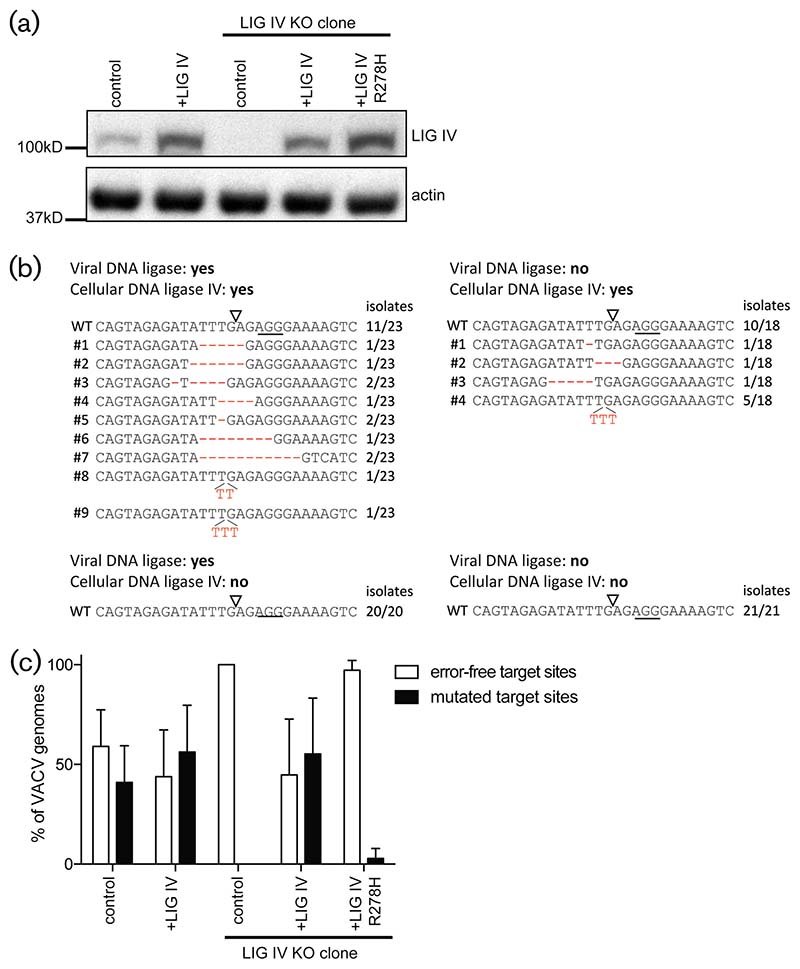
As the repair of DSBs still occurred in the absence of viral DNA ligase, editing at the gRNA target sites may be mediated by cellular DNA ligases that are recruited to the cytosol where poxvirus genomes are replicating. Cellular DNA ligase IV mediates C-NHEJ within the host cell nucleus [19]. To test whether DNA ligase IV is also involved in the repair of cytoplasmic poxvirus DNA, MJS cells lacking DNA ligase IV were generated using CRISPR/Cas9 (Fig. 4b). These KO cells also expressed the gRNA A23R#2 that has been used in the previous experiments. Cells were infected with the vSK20 virus lacking DNA ligase, or the revertant vSK21. Nine days after infection with VACV, cells were harvested and virus genomes were sequenced at the gRNA target site (Fig. 5b). For both the vSK20 and vSK21 reverant viruses, editing of the gRNA target site was readily observed in the parental line expressing cellular DNA ligase IV, as shown previously (Fig. 4). In cells lacking DNA ligase IV, however (Fig. 4b), no editing occurred, as only wild-type virus was identified (Fig. 5b, bottom conditions). In contrast, DNA ligase IV-expressing cells did allow editing of the viral genomes in both vSK20 and vSK21-infected cells (Fig. 5b, top conditions). This effect was not caused by clonal variation between the KO and parental cells, as the reintroduction of DNA ligase IV into the KO cell line rescued mutagenic repair of VACV upon CRISPR/Cas9 targeting (Fig. 5c). Introduction of the DNA ligase IVR278H, a variant lacking ligase activity, did not rescue the editing, showing that the ligase activity of DNA ligase IV is needed for the error-prone repair of disrupted poxvirus genomes. To conclude, C-NHEJ is capable of repairing poxvirus genomes upon the introduction of dsDNA breaks.
Fig. 5.
Cellular DNA ligase IV mediates NHEJ of VACV genomes upon CRISPR/Cas9 cleavage. (a) DNA ligase IV (upper panel) and actin (lower panel) protein expression levels in cells co-expressing Cas9 and gRNA A23R#2 (lanes 1–5). Lanes 1 and 2 represent a cell line expressing endogenous DNA ligase IV and lanes 3–5 represent a clonal DNA ligase IV knockout line. Cells were transduced with either a control lentiviral vector (control), a lentiviral vector encoding DNA ligase IV (+LIG IV), or the DNA ligase IV-R278H mutant (+LIG IV R278H). (b) Polyclonal cells expressing DNA ligase IV or the DNA ligase IV KO clone were infected with vSK20 virus lacking DNA ligase and the vSK21 revertant expressing DNA ligase. VACV genomic regions surrounding gRNA target sites were amplified from virus supernatant and sequenced. Solid lines indicate the PAM sequences; open triangles indicate the Cas9 cleavage site. Numbers indicate the abundance of the given virus mutant within the sequenced virus population. (c) Frequency of mutations observed at the A23R#2 CRISPR target site. The cells described in (a) were infected with VACV vSK21 in triplicate. Virus genomes were harvested 9 days p.i. and the A23R#2 target site of 24 isolates per triplicated infection was sequenced.
Discussion
Our data show that the CRISPR/Cas9 genome engineering system is highly effective in directing targeted mutagenesis of poxviral genomes as a means to generate defined virus variants. Upon the co-introduction of a virus-specific gRNA, Cas9, and a donor template sharing sequence homology with the target region, efficient exchange of genetic material can be obtained, resulting in the formation of specific virus mutants. As also shown by others [32], the proportion of virus mutants that underwent successful homologous recombination was increased upon DSB introduction via the CRISPR/Cas9 system. In the absence of DSB induction, successful homologous recombination events within the VACV genome are infrequent [52], whereas DSBs promote VACV-mediated recombination events [53, 54]. This is in line with our experiments, where template-guided recombination of VACV-eGFP in the absence of a gRNA proved inefficient (Fig. 1b, right top panel). Our results show the ease and potency of the CRISPR/Cas9 system in editing poxvirus genomes as an alternative to currently used approaches, such as spontaneous homologous recombination and the use of bacterial artificial chromosomes to induce desired mutations [52, 55–57]. We show that donor templates produced from PCR products readily recombine with the targeted VACV genome, even when short flanking sequences of 25 nt are used. A similar requirement for overlapping sequence homology has been observed previously, showing that 10-20nt homology is sufficient for vaccinia virus-induced homologous recombination [53, 54, 58, 59], although larger homologies may greatly enhance recombination events [54].
Altogether, CRISPR/Cas9-induced DSBs allow for the rapid generation of recombinant poxvirus strains to construct virus variants in a cell line of choice.
CRISPR/Cas9-targeting of viral genes severely hampered virus infection. The viral genes targeted in this study were highly conserved among poxviruses and were previously shown to be essential for CPXV [60] and VACV replication, including A10L [61] A23R [62], A5R [63], B1R [64], D4R [65], D5R [66] and G7L [67]. The majority of gRNAs targeting these genes severely limited infection at early time points. The loss of virus infectivity upon gRNA-targeting may be caused by different underlying mechanisms. A DSB may fragment the viral genome and thereby affect the incorporation of complete genomes into newly synthesized virus particles. In addition, gRNAs targeting genes that are essential for the virus life cycle may disrupt the expression of that gene, thereby stalling virus infection. Furthermore, repair of the DSB may alter amino acid sequences that are crucial for activity of the essential genes.
At prolonged times post-infection, virus replication was observed in the presence of VACV-specific gRNAs, suggesting that viruses escape targeting by CRISPR/Cas9. This may be due to C-NHEJ-mediated mutagenesis at the target region, resulting in loss of the gRNA recognition site but maintained functional gene expression. These viruses thus become resistant to subsequent CRISPR/Cas9-mediated cleavage and can propagate unrestricted [37, 41]. Some gRNAs, however, are capable of restricting poxvirus replication after prolonged infection; these gRNAs may target an Achilles’ heel of the virus.
The acquisition of viruses with mutations at the gRNA target site revealed that CRISPR/Cas9-induced DSBs are repaired by a mutagenic repair pathway. It was previously suggested that DSB repair of VACV genomes involves the viral DNA ligase, as VACV mutants lacking the enzyme were sensitive to DSBs induced by UV irradiation or the DNA-damaging drug bleomycin [8, 13]. In contrast, the DNA ligase of Shope fibroma virus did not mediate the repair of UV-damaged extragenomic DNA [13]. Unexpectedly, our results show that the viral DNA ligase is not crucial for error-prone VACV DNA repair induced by CRISPR/Cas9-mediated DNA breaks, as mutated target sites were still observed in the absence of viral DNA ligase. However, the lower titre of ligase-deficient viruses suggests that these viruses are more sensitive to DSBs, as observed previously [8]. This may imply that the viral ligase is involved in error-free repair through catalyzing e.g. homologous recombination of viral genomes [68]. Indeed, VACV DNA ligase is capable of complementing the functions of yeast DNA ligase, including catalysis of homologous recombination [8], although removal of the viral ligase may not always affect homologous recombination in infected cells [10, 13]. These contrasting findings may be explained by a complementary role of DNA ligase I, a host cell ligase involved in the final ligation step of homologous recombination [18, 69]. DNA ligase I is recruited to virus factories [12], and may partially mask the loss of viral DNA ligase in homology-directed repair.
Another factor that could contribute indirectly to the increased sensitivity of DNA ligase mutants to CRISPR/ Cas9-induced DSBs, is a slightly reduced replication of these viruses. Even in the absence of VACV-targeting gRNAs, DNA ligase-lacking viruses have smaller plaque sizes, and lower virus titres at prolonged infections (see Fig. 4a; [13]). This may allow for more efficient targeting by the CRISPR/ Cas9 system, and an increased sensitivity to DSBs. As observed previously, the quantity of virus particles directly affects the efficiency of the CRISPR/Cas9 system [29].
We identified DNA ligase IV as a crucial factor involved in mutagenic repair upon CRISPR/Cas9-mediated DSBs in VACV infection, as the absence of (functional) DNA ligase IV did not result in the formation of mutated target sites and all detected VACV genome copies contained the wild-type sequence. The occurrence of wild-type sequences was also apparent in the presence of DNA ligase IV, and may result from different processes within the infected cell. Potentially, not all cells may express (sufficient amounts of) Cas9 or the gRNA to efficiently cut all of the approximately 10 000 VACV genome copies per infected cell [70], thereby allowing the escape of unedited viral genomes. In addition, DSBs in viral genomes may be repaired through homologous recombination events. Recombination between viral genomes or transfected DNA is catalyzed in vivo and in vitro by the viral DNA polymerase E9 and the viral single-stranded DNA-binding protein I3 [58, 71]. The 3'-to 5' exonuclease activity of the DNA polymerase resects the DNA at the DSB, and the revealed single-stranded tail is targeted for recombination with a homologous template. The proofreading activity of the DNA polymerase-encoded exonuclease contributes to the high fidelity of such recombination events [53, 72]. Indeed, the great majority of homologous recombination events between poxvirus genomes show no sign of mutagenesis at recombination sites [73].
Homologous recombination events may even be further stimulated in the absence of a DNA ligase IV. In line with this, silencing genes essential for C-NHEJ, including DNA ligase IV [74] or targeting DNA ligase IV with the inhibitor Scr7 [75] was shown to enhance homologous recombination of Cas9-induced DSBs. In contrast, binding of DNA ligase IV to DSBs actively suppresses the initiation of homologous recombination by inhibiting DNA end resection, as shown in Saccharomyces cerevisiae [76].
The absence of DNA ligase IV may also induce alternative end-joining pathways (A-EJ) that rely on DNA ligase I and/ or DNA ligase III [77–81]. DNA ligase I has been found at cytosolic virus factories, where it plays a role in replication of the viral genome [12], and possibly homologous recombination [10, 68]. However, an additional role for DNA ligase I (or DNA ligase III) in A-EJ is unlikely, as this pathway is intrinsically mutagenic and induces extended insertions and deletions at the target site [82, 83]. As the VACV target sequences were exclusively wild-type, alternative end-joining pathways appear not to be involved in VACV genome repair in the absence of DNA ligase IV. In addition, DNA ligase IV-mediated C-NHEJ of some genomes may maintain the wild-type sequence. Indeed, DNA repair by C-NHEJ does not always result in mutagenesis at the target site [82, 84–86], and wild-type sequences have been observed frequently in viral genomes upon repair of CRISPR/Cas9-induced DSBs [29, 37].
DNA ligase IV is targeted to the nucleus via a bipartite nuclear localization signal in the C-terminal region of the protein [87]. DNA ligase IV binding to the cofactor XRCC4 further promotes nuclear localization [87]. In contrast, DNA ligase IV is absent from mitochondria, the other DNA-containing cellular compartment [88]. Upon DNA damage in the nucleus, DNA ligase IV forms repair foci together with other components of the C-NHEJ pathway [89]. The involvement of DNA ligase IV in DNA repair of VACV indicates that part of the C-NHEJ machinery gains access to the virus factories in the cytosol. However, we did not observe DNA ligase IV or XRCC4 in viral factories (data not shown). Nonetheless, it is possible that only small amounts of DNA ligase IV are present at the cytosolic sites of virus replication. Indeed, although the vast majority of DNA ligase IV localized to the nucleus, some DNA ligase IV was detected in the cytosol (data not shown). In line with this, cytosolic extracts of various cell lines do contain low levels of DNA ligase IV, and only small amounts of DNA ligase IV are sufficient for adequate C-NHEJ to occur [90, 91].
Poxviruses are renowned for their replication, independent of the nuclear environment, as illustrated by successful infections of enucleated cells [92, 93]. However, more recent work shows that some nuclear factors gain access to virus factories and are needed for poxvirus infection [94], including cellular DNA ligase I [12], topoisomerase II [95], HMG20A [96] and nuclear transcription factors [97]. The C-NHEJ factors Ku80 and DNA-PK have also been observed in association with viral factories, where they play a role in the activation of the host innate immune system [98] and are antagonized by the VACV protein C16 [99]. The results presented here indicate that DNA ligase IV can also access the poxvirus genome and repair CRISPR/Cas9-mediated DSBs, possibly with the aid of other enzymes that are crucial for C-NHEJ. Although DNA ligase IV is crucial for the repair of CRISPR/Cas9-induced damage, the role of DNA ligase IV in repairing naturally occurring DNA damage remains to be investigated.
Such damage may be caused by replication stress and subsequent replication fork collapse [100, 101]. In addition, DNA damage and subsequent repair by C-NHEJ may be induced by a multitude of other extrinsic and intrinsic stressors, including γ-irradiation, UV light and reactive oxygen species/reactive nitrogen species (ROS/RNS) [102–104].
The small indels introduced into the poxvirus genome upon C-NHEJ are reminiscent of the indels frequently observed throughout the genome of VACV and related members, including CPXV and variola virus [105–110]. These mutations are considered to be important factors that drive evolution and changes in virulence and host tropism after zoonosis [111]. The majority of these indels result from DNA replication errors, including replication stuttering and slippage, although a minority of these mutations cannot be explained by these erroneous pathways [105, 108–110]. C-NHEJ may be a reciprocal mechanism that is capable of inducing mutations, and through this driving evolution [112–114]. As shown for CPXV, even a small deletion in the protein-encoding gene CPXV012 is sufficient to establish a completely new function for the protein [115, 116].
Methods
Cells and viruses
The human melanoma cell line MelJuSo (MJS) [117] and T2 cells [118] were kindly provided by Professors Jacques Neefjes (LUMC, Leiden, the Netherlands) and Peter Cresswell (Yale University School of Medicine, USA), respectively. HEK-293T and Vero African Green monkey kidney epithelial cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). MJS and T2 cells were cultured in RPMI 1640 (Lonza AG, Switzerland) supplemented with 10% foetal bovine serum (FBS) (#S1810 Biowest Europe, France), 100 Uml−1 penicillin, 100 μg ml−1 streptomycin and 2mM l-glutamine (complete medium). HEK-293T and Vero cells were cultured in DMEM (Lonza AG, Switzerland) supplemented with 10% FBS, 100 Uml−1 penicillin, 100 μg ml−1 streptomycin and 2mM l-glutamine.
The VACV strain Western Reserve encoding eGFP from the p7.5 synthetic early/late promoter (VACV-eGFP) was a generous gift from Dr Jon Yewdell (NIH, Bethesda, USA). The CPXV strain Brighton Red (CPXV-BR) expressing RFP and eGFP was kindly provided by Dr Karsten Tischer (FU Berlin, Germany). VACV-eGFP and CPXV-BR were propagated and titrated on Vero cells using the standard methodology. VACV variants lacking viral DNA ligase (vSK20) and the revertant virus (vSK21) were described previously [8], and titrated on Vero cells.
Lentiviral CRISPR/Cas9 vector and other DNA vectors
A selectable lentiviral CRISPR/Cas9 vector based on the pSicoR vector (Addgene plasmid 11579, Tyler Jacks Lab, MIT, USA) was constructed as described previously [41, 119]. This lentiviral vector holds a human codon-optimized S. pyogenes Cas9 gene N-terminally fused to PuroR via a T2A ribosome-skipping sequence under control of the human EF1A promoter. Additionally, it contains a human U6 promoter, which drives the expression of a guideRNA (gRNA) consisting of a 18–20 bp target-specific CRISPR RNA (crRNA) fused to the trans-activating crRNA (tracrRNA) and a terminator sequence. Another variant of this vector was created by replacing the PuroR gene for a BlastR gene. These vectors are called pSicoR-CRISPR-PuroR and pSicoR-CRISPR-BlastR, respectively. For each virus gene target, crRNA target sequences were designed using an online CRISPR design tool (crispr.mit.edu; Zhang lab, MIT, USA). CRISPR gRNAs with the highest specificity and lowest off-target rate for the human genome were selected and cloned into the pSicoR-CRISPR-PuroR or pSicoR-CRISPR-BlastR vector. The CRISPR gRNA sequences used in this study are listed in Table 1.
For DNA ligase IV overexpression and rescue experiments, the human DNA ligase IV was amplified by PCR from genomic DNA isolated from MJS cells (Wizard genomic DNA isolation kit #A1125, Promega Benelux, The Netherlands) using primers 5'-TTCAGGTGTCGTGAGCTAGCAGTAT-TAATTAACCACCATGGCTGCCTCACAAACTTCAC-3' (LIG#1) and 5'-atgactaagctagtaccggttaggatgcaTgcttaaatcaaa-tactggttttcttcttg-3' (LIG#2). Subsequently, the PCR product was introduced into a dual-promoter lentiviral vector co-expressing ZeoR and the fluorescent mAmetrine gene [119] by means of Gibson assembly (#E2611L, Bioké, New England Biolabs, The Netherlands). The DNA ligase IVR278H mutation was introduced by PCR amplification of the DNA ligase IV template using primers LIG#1, and 5'-catttgca-taTgttcaccatc-3', and LIG#2 and 5'-GATGGTGAACA-TATGCAAATG-3'. The PCR products were introduced into the dual-promoter lentiviral vector using Gibson assembly.
Lentivirus production and transduction
Third-generation lentiviruses were produced in HEK-293T cells in a 24-well plate format using standard lentivirus production protocols. Lentiviruses were transduced into MJS cells using spin infection at 1000 g for 90 min at 33°C in the presence of 3.2 µg ml𢈒1 polybrene. After 3 days, transduced cells were selected using either puromycin (2 µg ml𢈒1), zeocin (400 µg ml𢈒1) or blasticidin (20 µg ml𢈒1). Cell lines expressing 2 gRNAs were generated by simultaneous transduction of pSico-CRISPR-PuroR and pSico-CRISPR-BlastR lentiviral vectors encoding different gRNAs followed by selection.
Homologous recombination assay
DNA templates for homologous recombination were obtained from IDT (gBLOCKs, Integrated DNA Technologies Inc.) comprising nucleotides (nt) 50–637 of eGFP or enhanced cyan fluorescent protein (eCFP). These gBlocks contained the silent mutations C198A and C465A to render the DNA resistant to CRISPR/Cas9 targeting by gRNA eGFP#1 and eGFP#3 (Table 1). The gBlocks were PCR-amplified to generate products with different lengths of homology flanking the gRNA target sites. Primers 5'-TCGAGCTGGACGGCG-3' and 5'-GGGGTCTTTGCTCA GGGC-3' were used for the PCR amplification of a product with a homology region of 150 nt flanking each gRNA target site; primers 5'-GGCGAGGGCGATGCC-3' and 5'-AGCAG-CACGGGGCCG-3' for 100 nt; primers 5'-CACCGG-CAAGCTGCC-3' and 5'-AGCTGCACGCTGCCG-3' for 50 nt; and primers 5'-CCCACCCTCGTGACCAC-3' and 5'-TGTGGCGGATCTTGAAGTTG-3' for a homology region of 25 nt.
To generate variants of VACV-eGFP by using homologous recombination, MJS cells expressing Cas9 and gRNAs eGFP#1 and eGFP#3 (Table 1) were plated in a 48-well plate at 104 cells per well. The following day, the cells were mock transfected, or transfected with the different DNA templates encoding eGFP or eCFP. After overnight incubation, the cells were infected with VACV-eGFP using an m.o.i. of 0.1. Eight days post-infection, the supernatant was collected and used for a secondary infection on MJS cells plated in a 48-well plate. Two days after the secondary infection, the cells were harvested and eCFP and eGFP expression was quantified by flow cytometry (FACS Canto II, BD Biosciences).
Primary infections of CRISPR-expressing cells with VACV-eGFP
MJS cells (104 cells/well) expressing different CRISPR gRNAs were plated in a 48-well plate and incubated over-night at 37°C. The next day, cells were infected at an m.o.i. of 0.1 or 0.01 in complete medium. Cells were harvested at 3, 5, 8, or 15 days p.i. and cell numbers and the percentage of infected (eGFP-positive) cells were quantified by flow cytometry. Prior to flow cytometric analysis, 5000 mCherry-positive T2 cells were mixed with the MJS cells to allow for normalization between the wells.
Plaque assays
MJS cells including supernatant were collected at 3 or 9 days p.i. To release viral particles, cells were freeze/thawed three times and sonicated for 10 min in a waterbath sonicator (M2 800-E, Branson Ultrasonics, The Netherlands). Samples were centrifuged for 3 min at 300 g, and various dilutions of the supernatant were incubated on 105 Vero cells seeded 1 day prior to infection in a 24-well format. After 1 h of incubation, the cells were overlaid with 0.5% agarose (Seakem LE Agarose, Lonza AG, Switzerland) in complete medium. After 7–9 days, the cells were fixed overnight at RT using 1% formaldehyde and cell monolayers were stained with 0.5% crystal violet solution. Plaques were counted by eye and virus titres were calculated as plaque-forming units (p.f.u.) ml−1.
Sequencing of CRISPR target sites
MJS cells including supernatant were collected 9 days p.i., freeze/thawed three times and sonicated for 10 min. The samples were centrifuged to remove cells and debris, and the supernatant was heated to 95 °C for 4 h to inactivate the virus particles. Heat-inactivated supernatant was used as a template to PCR-amplify the CRISPR target regions from the viral genomes, using primers 5'-acaagtt-cagcgtgtccg-3' (eGFP#1) and 5'-tgctcaggtagtggttgtcg-3' for the eGFP target region, and primers 5'-TGAAATAGAA-GATAGATATGCCAGAAC-3' and 5'-TTCGCCGTAATT-GACCTTTC-3' for the target region in A23R. The PCR products of the eGFP target region were sequenced directly using primer eGFP#1. To characterize individual mutations, PCR products were cloned into the pCR2.1 TOPO TA cloning vector (Thermo Fisher Scientific, Inc., USA) and transformed into OneShot TOP10 Escherichia coli cells (#K450001, Thermo Fisher Scientific Inc, USA). Positive colonies were selected using a β-galactosidase-dependent blue/white screen and the TA-cloned insert was PCR-amplified using the T7 forward primer 5'-TAATACGACTCAC-TATAGG-3' and the M13 reverse primer 5'-GGAAACAGCTATGACCATG-3', and sequenced (Macrogen Inc., USA) using the T7 primer. The sequences were analysed using SeqMan software (DNASTAR, Inc., USA).
Generation of DNA ligase IV KO cell lines and revertants
MJS cells were transfected with pSicoR-CRISPR-BlastR encoding the gRNA LIGIV targeting DNA ligase IV (Table 1). The transfected cells were transiently selected using blasticidin (20 µg ml−1) and subsequently cloned by limited dilution. DNA ligase IV knock-out was confirmed in selected clones by Western blot analysis.
DNA ligase IV was (re)introduced by transducing cells with the lentiviral vector encoding DNA ligase IV, or an inactive DNA ligase IVR278H mutant [120]. As a control, cells were transduced with an empty control vector. Cells were selected via zeocin (400 µg ml−1) treatment and DNA ligase IV expression was confirmed by Western blot analysis (see below).
SDS-PAGE and Western Blot analysis
Cells were lysed in 1 % Triton buffer [1 % Triton X-100, 20 mM 2-(N-morpholino)ethanesulfonic acid (MES), 100 mM NaCl, 30 mM Tris (pH7.5)] in the presence of 10 mM leupeptin and 1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride. To prepare samples for SDS-PAGE separation, whole-cell lysates were boiled in Laemmli sample buffer for 10 min at 70 C and subsequently sonicated using three short pulses of approximately 25 W to shear genomic DNA. After SDS-PAGE, proteins were transferred to PVDF membranes using the Trans-Blot Turbo Transfer system (#1703935, Bio-Rad, The Netherlands). Membranes were incubated with primary antibodies specific for actin (clone C4; Milipore #MAB1501R, The Netherlands), or DNA ligase IV (clone D-8; Santa Cruz biotechnology #sc271299, Germany), washed and incubated with the secondary HRP-conjugated goat anti-mouse IgG L-chain-specific antibody (#115-035-174, Jackson ImmunoResearch, Inc., USA). Bound antibodies were visualized by incubating membranes with ECL (Thermo Fisher Scientific Pierce, USA) and exposure to Amersham Hyperfilm (GE Healthcare, The Netherlands).
Acknowledgements
The authors would like to thank Ana I. Costa, Medical Microbiology, University Medical Center Utrecht, for critically reading and assisting with preparation of the manuscript.
Funding information
G. L. S. is a Wellcome Trust Principal Research Fellow. R. J. L. was supported by Marie Curie Career Integration Grant PCIG-GA-2011-294196 and I. D. was supported by the DFG funding GRK 1949.
Abbreviations
- A-EJ
alternative end joining
- bB2M
bovine β2 microglobulin
- BlastR
blasticidin resistance gene
- C-NHEJ
canonical NHEJ
- CPXV
cowpox virus
- CRISPR/Cas9
clustered regularly interspaced short palindromic repeats/CRISPR-associated protein-9 nuclease
- DNA-PK
DNA-dependent protein kinase
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
- DSB
double-stranded breaks
- eCFP
enhanced cyan fluorescent protein
- EF1A promoter
human elongation factor-1 alpha promoter
- FEN1
Flap endonuclease 1
- gRNA
guide RNA
- HEK-293T
human embryonic kidney 293-cell line expressing the SV40 large T antigen
- HIV-1
human immunodeficiency virus 1
- HMG20A
high-mobility group protein 20A
- NHEJ
non-homologous end joining
- p.i.
post-infection
- PuroR
puromycin resistance gene
- RFP
red fluorescent protein
- ROS/RNS
reactive oxygen species/reactive nitrogen species
- TAP2
transporter associated with antigen processing 2
- VACV
vaccinia virus
- XRCC4
X-ray repair cross-complementing protein 4
- ZeoR
zeocin resistance gene
Footnotes
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.McFadden G. Poxvirus tropism. Nat Rev Microbiol. 2005;3:201–213. doi: 10.1038/nrmicro1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Essbauer S, Pfeffer M, Meyer H. Zoonotic poxviruses. Vet Microbiol. 2010;140:229–236. doi: 10.1016/j.vetmic.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim M. Replicating poxviruses for human cancer therapy. J Microbiol. 2015;53:209–218. doi: 10.1007/s12275-015-5041-4. [DOI] [PubMed] [Google Scholar]
- 4.Gómez CE, N´ajera JL, Krupa M, Perdiguero B, Esteban M. MVA and NYVAC as vaccines against emergent infectious diseases and cancer. Curr Gene Ther. 2011;11:189–217. doi: 10.2174/156652311795684731. [DOI] [PubMed] [Google Scholar]
- 5.Volz A, Sutter G. Modified vaccinia virus Ankara: history, value in basic research, and current perspectives for vaccine development. Adv Virus Res. 2017;97:187–243. doi: 10.1016/bs.aivir.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katsafanas GC, Moss B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe. 2007;2:221–228. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moss B. Poxvirus DNA replication. Cold Spring Harb Perspect Biol. 2013;5:a010199. doi: 10.1101/cshperspect.a010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerr SM, Johnston LH, Odell M, Duncan SA, Law KM, et al. Vaccinia DNA ligase complements Saccharomyces cerevisiae cdc9, localizes in cytoplasmic factories and affects virulence and virus sensitivity to DNA damaging agents. EMBO J. 1991;10:4343–4350. doi: 10.1002/j.1460-2075.1991.tb05012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senkevich TG, Koonin EV, Moss B. Predicted poxvirus FEN1-like nuclease required for homologous recombination, double-strand break repair and full-size genome formation. Proc Natl Acad Sci USA. 2009;106:17921–17926. doi: 10.1073/pnas.0909529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colinas RJ, Goebel SJ, Davis SW, Johnson GP, Norton EK, et al. A DNA ligase gene in the Copenhagen strain of vaccinia virus is nonessential for viral replication and recombination. Virology. 1990;179:267–275. doi: 10.1016/0042-6822(90)90295-3. [DOI] [PubMed] [Google Scholar]
- 11.Kerr SM, Smith GL. Vaccinia virus DNA ligase is nonessential for virus replication: recovery of plasmids from virus-infected cells. Virology. 1991;180:625–632. doi: 10.1016/0042-6822(91)90076-n. [DOI] [PubMed] [Google Scholar]
- 12.Paran N, de Silva FS, Senkevich TG, Moss B. Cellular DNA ligase I is recruited to cytoplasmic vaccinia virus factories and masks the role of the vaccinia ligase in viral DNA replication. Cell Host Microbe. 2009;6:563–569. doi: 10.1016/j.chom.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parks RJ, Winchcombe-Forhan C, Delange AM, Xing X, Evans DH. DNA ligase gene disruptions can depress viral growth and replication in poxvirus-infected cells. Virus Res. 1998;56:135–147. doi: 10.1016/s0168-1702(98)00055-0. [DOI] [PubMed] [Google Scholar]
- 14.Klein B, Filon AR, van Zeeland AA, van der Eb AJ. Survival of UV-irradiated vaccinia virus in normal and xeroderma pigmentosum fibroblasts; evidence for repair of UV-damaged viral DNA. Mutat Res. 1994;307:25–32. doi: 10.1016/0027-5107(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 15.Z´avadova´ Z. Host-cell repair of vaccinia virus and of double stranded RNA of encephalomyocarditis virus. Nat New Biol. 1971;233:123. [PubMed] [Google Scholar]
- 16.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 17.Lytle CD, Aaronson SA, Harvey E. Host-cell reactivation in mammalian cells. II. Survival of herpes simplex virus and vaccinia virus in normal human and xeroderma pigmentosum cells. Int J Radiat Biol Relat Stud Phys Chem Med. 1972;22:159–165. [PubMed] [Google Scholar]
- 18.Timson DJ, Singleton MR, Wigley DB. DNA ligases in the repair and replication of DNA. Mutat Res. 2000;460:301–318. doi: 10.1016/s0921-8777(00)00033-1. [DOI] [PubMed] [Google Scholar]
- 19.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis AJ, Chen DJ. DNA double strand break repair via non-homologous end-joining. Transl Cancer Res. 2013;2:130–143. doi: 10.3978/j.issn.2218-676X.2013.04.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- 22.Ceccaldi R, Rondinelli B, D’Andrea AD. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll D. Genome editing by targeted chromosomal mutagenesis. Methods Mol Biol. 2015;1239:1–13. doi: 10.1007/978-1-4939-1862-1_1. [DOI] [PubMed] [Google Scholar]
- 24.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sternberg SH, Doudna JA. Expanding the biologist’s toolkit with CRISPR-Cas9. Mol Cell. 2015;58:568–574. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 28.Soppe JA, Lebbink RJ. Antiviral goes viral: harnessing CRISPR/ Cas9 to combat viruses in humans. Trends Microbiol. 2017;25:833–850. doi: 10.1016/j.tim.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Bi Y, Sun L, Gao D, Ding C, Li Z, et al. High-efficiency targeted editing of large viral genomes by RNA-guided nucleases. PLoS Pathog. 2014;10:e1004090. doi: 10.1371/journal.ppat.1004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell TA, Stefanovic T, Tscharke DC. Engineering herpes simplex viruses by infection-transfection methods including recombination site targeting by CRISPR/Cas9 nucleases. J Virol Methods. 2015;213:18–25. doi: 10.1016/j.jviromet.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Suenaga T, Kohyama M, Hirayasu K, Arase H. Engineering large viral DNA genomes using the CRISPR-Cas9 system. Microbiol Immunol. 2014;58:513–522. doi: 10.1111/1348-0421.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan M, Zhang W, Wang J, Al Yaghchi C, Ahmed J, et al. Efficiently editing the vaccinia virus genome by using the CRISPR-Cas9 system. J Virol. 2015;89:5176–5179. doi: 10.1128/JVI.00339-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci USA. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, et al. Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci Rep. 2016;6:22555. doi: 10.1038/srep22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao HK, Gu Y, Diaz A, Marlett J, Takahashi Y, et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun. 2015;6:6413. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Pan Q, Gendron P, Zhu W, Guo F, et al. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep. 2016;15:481–489. doi: 10.1016/j.celrep.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 38.Zhu W, Lei R, Le Duff Y, Li J, Guo F, et al. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology. 2015;12:22. doi: 10.1186/s12977-015-0150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lebbink RJ, de Jong DC, Wolters F, Kruse EM, van Ham PM, et al. A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Sci Rep. 2017;7:41968. doi: 10.1038/srep41968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Quake SR. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc Natl Acad Sci USA. 2014;111:13157–13162. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Diemen FR, Kruse EM, Hooykaas MJ, Bruggeling CE, Schürch AC, et al. CRISPR/Cas9-Mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 2016;12:e1005701. doi: 10.1371/journal.ppat.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roehm PC, Shekarabi M, Wollebo HS, Bellizzi A, He L, et al. Inhibition of HSV-1 replication by gene editing strategy. Sci Rep. 2016;6:23146. doi: 10.1038/srep23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kennedy EM, Kornepati AV, Goldstein M, Bogerd HP, Poling BC, et al. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J Virol. 2014;88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhen S, Hua L, Takahashi Y, Narita S, Liu YH, et al. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem Biophys Res Commun. 2014;450:1422–1426. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 45.Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, et al. The CRISPR/ Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo. Mol Ther Nucleic Acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kennedy EM, Bassit LC, Mueller H, Kornepati AV, Bogerd HP, et al. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology. 2015;476:196–205. doi: 10.1016/j.virol.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price AA, Sampson TR, Ratner HK, Grakoui A, Weiss DS. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc Natl Acad Sci USA. 2015;112:6164–6169. doi: 10.1073/pnas.1422340112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wollebo HS, Bellizzi A, Kaminski R, Hu W, White MK, et al. CRISPR/Cas9 System as an agent for eliminating polyomavirus JC infection. PLoS One. 2015;10:e0136046. doi: 10.1371/journal.pone.0136046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cong L, Ran FA, Cox D, Lin S, Barretto R, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 52.Nakano E, Panicali D, Paoletti E. Molecular genetics of vaccinia virus: demonstration of marker rescue. Proc Natl Acad Sci USA. 1982;79:1593–1596. doi: 10.1073/pnas.79.5.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao XD, Evans DH. Effects of DNA structure and homology length on vaccinia virus recombination. J Virol. 2001;75:6923–6932. doi: 10.1128/JVI.75.15.6923-6932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao XD, Evans DH. Construction of recombinant vaccinia viruses using leporipoxvirus-catalyzed recombination and reactivation of orthopoxvirus DNA. Methods Mol Biol. 2004;269:51–64. doi: 10.1385/1-59259-789-0:051. [DOI] [PubMed] [Google Scholar]
- 55.Domi A, Moss B. Cloning the vaccinia virus genome as a bacterial artificial chromosome in Escherichia coli and recovery of infectious virus in mammalian cells. Proc Natl Acad Sci USA. 2002;99:12415–12420. doi: 10.1073/pnas.192420599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Falkner FG, Moss B. Transient dominant selection of recombinant vaccinia viruses. J Virol. 1990;64:3108–3111. doi: 10.1128/jvi.64.6.3108-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mackett M, Smith GL, Moss B. General method for production and selection of infectious vaccinia virus recombinants expressing foreign genes. J Virol. 1984;49:857–864. doi: 10.1128/jvi.49.3.857-864.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Willer DO, Yao XD, Mann MJ, Evans DH. In vitro concatemer formation catalyzed by vaccinia virus DNA polymerase. Virology. 2000;278:562–569. doi: 10.1006/viro.2000.0686. [DOI] [PubMed] [Google Scholar]
- 59.Irwin CR, Farmer A, Willer DO, Evans DH. In-fusion® cloning with vaccinia virus DNA polymerase. Methods Mol Biol. 2012;890:23–35. doi: 10.1007/978-1-61779-876-4_2. [DOI] [PubMed] [Google Scholar]
- 60.Xu Z, Zikos D, Osterrieder N, Tischer BK. Generation of a complete single-gene knockout bacterial artificial chromosome library of cowpox virus and identification of its essential genes. J Virol. 2014;88:490–502. doi: 10.1128/JVI.02385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heljasvaara R, Rodríguez D, Risco C, Carrascosa JL, Esteban M, et al. The major core protein P4a (A10L gene) of vaccinia virus is essential for correct assembly of viral DNA into the nucleo-protein complex to form immature viral particles. J Virol. 2001;75:5778–5795. doi: 10.1128/JVI.75.13.5778-5795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Warren RD, Cotter CA, Moss B. Reverse genetics analysis of poxvirus intermediate transcription factors. J Virol. 2012;86:9514–9519. doi: 10.1128/JVI.06902-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seo D, Kim NY, Lee JA, Han KR, Hur GH, et al. Protection against lethal vaccinia virus infection in mice using an siRNA targeting the A5R gene. Antivir Ther. 2016;21:397–404. doi: 10.3851/IMP3022. [DOI] [PubMed] [Google Scholar]
- 64.Rempel RE, Traktman P. Vaccinia virus B1 kinase: phenotypic analysis of temperature-sensitive mutants and enzymatic characterization of recombinant proteins. J Virol. 1992;66:4413–4426. doi: 10.1128/jvi.66.7.4413-4426.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stuart DT, Upton C, Higman MA, Niles EG, McFadden G. A poxvirus-encoded uracil DNA glycosylase is essential for virus viability. J Virol. 1993;67:2503–2512. doi: 10.1128/jvi.67.5.2503-2512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Evans E, Traktman P. Characterization of vaccinia virus DNA replication mutants with lesions in the D5 gene. Chromosoma. 1992;102:S72–S82. doi: 10.1007/BF02451789. [DOI] [PubMed] [Google Scholar]
- 67.Szajner P, Jaffe H, Weisberg AS, Moss B. Vaccinia virus G7L protein Interacts with the A30L protein and is required for association of viral membranes with dense viroplasm to form immature virions. J Virol. 2003;77:3418–3429. doi: 10.1128/JVI.77.6.3418-3429.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hamilton MD, Evans DH. Enzymatic processing of replication and recombination intermediates by the vaccinia virus DNA polymerase. Nucleic Acids Res. 2005;33:2259–2268. doi: 10.1093/nar/gki525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goetz JD, Motycka TA, Han M, Jasin M, Tomkinson AE. Reduced repair of DNA double-strand breaks by homologous recombination in a DNA ligase I-deficient human cell line. DNA Repair. 2005;4:649–654. doi: 10.1016/j.dnarep.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 70.Salzman NP. The rate of formation of vaccinia deoxyribonucleic acid and vaccinia virus. Virology. 1960;10:150–152. doi: 10.1016/0042-6822(60)90015-5. [DOI] [PubMed] [Google Scholar]
- 71.Gammon DB, Evans DH. The 3'-to-5' exonuclease activity of vaccinia virus DNA polymerase is essential and plays a role in promoting virus genetic recombination. J Virol. 2009;83:4236–4250. doi: 10.1128/JVI.02255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ball LA. Fidelity of homologous recombination in vaccinia virus DNA. Virology. 1995;209:688–691. doi: 10.1006/viro.1995.1305. [DOI] [PubMed] [Google Scholar]
- 73.Qin L, Evans DH. Genome scale patterns of recombination between coinfecting vaccinia viruses. J Virol. 2014;88:5277–5286. doi: 10.1128/JVI.00022-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chu VT, Weber T, Wefers B, Wurst W, Sander S, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 75.Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, et al. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol. 2007;14:639–646. doi: 10.1038/nsmb1261. [DOI] [PubMed] [Google Scholar]
- 77.Liang L, Deng L, Nguyen SC, Zhao X, Maulion CD, et al. Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks. Nucleic Acids Res. 2008;36:3297–3310. doi: 10.1093/nar/gkn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu G, Duan J, Shu S, Wang X, Gao L, et al. Ligase I and ligase III mediate the DNA double-strand break ligation in alternative end-joining. Proc Natl Acad Sci USA. 2016;113:1256–1260. doi: 10.1073/pnas.1521597113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paul K, Wang M, Mladenov E, Bencsik-Theilen A, Bednar T, et al. DNA ligases I and III cooperate in alternative non-homologous end-joining in vertebrates. PLoS One. 2013;8:e59505. doi: 10.1371/journal.pone.0059505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soni A, Siemann M, Grabos M, Murmann T, Pantelias GE, et al. Requirement for Parp-1 and DNA ligases 1 or 3 but not of Xrcc1 in chromosomal translocation formation by backup end joining. Nucleic Acids Res. 2014;42:6380–6392. doi: 10.1093/nar/gku298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Be´termier M, Bertrand P, Lopez BS. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 2014;10:e1004086. doi: 10.1371/journal.pgen.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Frit P, Barboule N, Yuan Y, Gomez D, Calsou P. Alternative end-joining pathway(s): bricolage at DNA breaks. DNA Repair. 2014;17:81–97. doi: 10.1016/j.dnarep.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 84.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, et al. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 85.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal non-homologous ends. Proc Natl Acad Sci USA. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, et al. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 87.Girard PM, Kysela B, Härer CJ, Doherty AJ, Jeggo PA. Analysis of DNA ligase IV mutations found in LIG4 syndrome patients: the impact of two linked polymorphisms. Hum Mol Genet. 2004;13:2369–2376. doi: 10.1093/hmg/ddh274. [DOI] [PubMed] [Google Scholar]
- 88.Tadi SK, Sebastian R, Dahal S, Babu RK, Choudhary B, et al. Microhomology-mediated end joining is the principal mediator of double-strand break repair during mitochondrial DNA lesions. Mol Biol Cell. 2016;27:223–235. doi: 10.1091/mbc.E15-05-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Berg E, Christensen MO, Dalla Rosa I, Wannagat E, Jänicke RU, et al. XRCC4 controls nuclear import and distribution of ligase IV and exchanges faster at damaged DNA in complex with Ligase IV. DNA Repair. 2011;10:1232–1242. doi: 10.1016/j.dnarep.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 90.Francis DB, Kozlov M, Chavez J, Chu J, Malu S, et al. DNA ligase IV regulates XRCC4 nuclear localization. DNA Repair. 2014;21:36–42. doi: 10.1016/j.dnarep.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Windhofer F, Wu W, Iliakis G. Low levels of DNA ligases III and IV sufficient for effective NHEJ. J Cell Physiol. 2007;213:475–483. doi: 10.1002/jcp.21120. [DOI] [PubMed] [Google Scholar]
- 92.Prescott DM, Kates J, Kirkpatrick JB. Replication of vaccinia virus DNA in enucleated L-cells. J Mol Biol. 1971;59:505–508. doi: 10.1016/0022-2836(71)90313-5. [DOI] [PubMed] [Google Scholar]
- 93.Pennington TH, Follett EA. Vaccinia virus replication in enucleate BSC-1 cells: particle production and synthesis of viral DNA and proteins. J Virol. 1974;13:488–493. doi: 10.1128/jvi.13.2.488-493.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sivan G, Martin SE, Myers TG, Buehler E, Szymczyk KH, et al. Human genome-wide RNAi screen reveals a role for nuclear pore proteins in poxvirus morphogenesis. Proc Natl Acad Sci USA. 2013;110:3519–3524. doi: 10.1073/pnas.1300708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin YC, Li J, Irwin CR, Jenkins H, Delange L, et al. Vaccinia virus DNA ligase recruits cellular topoisomerase II to sites of viral replication and assembly. J Virol. 2008;82:5922–5932. doi: 10.1128/JVI.02723-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hsiao JC, Chao CC, Young MJ, Chang YT, Cho EC, et al. A poxvirus host range protein, CP77, binds to a cellular protein, HMG20A, and regulates its dissociation from the vaccinia virus genome in CHO-K1 cells. J Virol. 2006;80:7714–7728. doi: 10.1128/JVI.00207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Oh J, Broyles SS. Host cell nuclear proteins are recruited to cytoplasmic vaccinia virus replication complexes. J Virol. 2005;79:12852–12860. doi: 10.1128/JVI.79.20.12852-12860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife. 2012;1:e00047. doi: 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peters NE, Ferguson BJ, Mazzon M, Fahy AS, Krysztofinska E, et al. A mechanism for the inhibition of DNA-PK-mediated DNA sensing by a virus. PLoS Pathog. 2013;9:e1003649. doi: 10.1371/journal.ppat.1003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Allen C, Ashley AK, Hromas R, Nickoloff JA. More forks on the road to replication stress recovery. J Mol Cell Biol. 2011;3:4–12. doi: 10.1093/jmcb/mjq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hendriks G, Calléja F, Vrieling H, Mullenders LH, Jansen JG, et al. Gene transcription increases DNA damage-induced mutagenesis in mammalian stem cells. DNA Repair. 2008;7:1330–1339. doi: 10.1016/j.dnarep.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 102.Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2014;6:a016428. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kavanagh JN, Redmond KM, Schettino G, Prise KM. DNA double strand break repair: a radiation perspective. Antioxid Redox Signal. 2013;18:2458–2472. doi: 10.1089/ars.2012.5151. [DOI] [PubMed] [Google Scholar]
- 104.Sharma V, Collins LB, Chen TH, Herr N, Takeda S, et al. Oxidative stress at low levels can induce clustered DNA lesions leading to NHEJ mediated mutations. Oncotarget. 2016;7:25377–90. doi: 10.18632/oncotarget.8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Coulson D, Upton C. Characterization of indels in poxvirus genomes. Virus Genes. 2011;42:171–177. doi: 10.1007/s11262-010-0560-x. [DOI] [PubMed] [Google Scholar]
- 106.Elde NC, Child SJ, Eickbush MT, Kitzman JO, Rogers KS, et al. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell. 2012;150:831–841. doi: 10.1016/j.cell.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Esposito JJ, Sammons SA, Frace AM, Osborne JD, Olsen-Rasmussen M, et al. Genome sequence diversity and clues to the evolution of variola (smallpox) virus. Science. 2006;313:807–812. doi: 10.1126/science.1125134. [DOI] [PubMed] [Google Scholar]
- 108.Qin L, Upton C, Hazes B, Evans DH. Genomic analysis of the vaccinia virus strain variants found in Dryvax vaccine. J Virol. 2011;85:13049–13060. doi: 10.1128/JVI.05779-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Smith GL, Chan YS, Howard ST. Nucleotide sequence of 42 kbp of vaccinia virus strain WR from near the right inverted terminal repeat. J Gen Virol. 1991;72:1349–1376. doi: 10.1099/0022-1317-72-6-1349. [DOI] [PubMed] [Google Scholar]
- 110.Aguado B, Selmes IP, Smith GL. Nucleotide sequence of 21.8 kbp of variola major virus strain Harvey and comparison with vaccinia virus. J Gen Virol. 1992;73:2887–2902. doi: 10.1099/0022-1317-73-11-2887. [DOI] [PubMed] [Google Scholar]
- 111.Haller SL, Peng C, McFadden G, Rothenburg S. Poxviruses and the evolution of host range and virulence. Infect Genet Evol. 2014;21:15–40. doi: 10.1016/j.meegid.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Heidenreich E, Eisler H. Non-homologous end joining dependency of gamma-irradiation-induced adaptive frameshift mutation formation in cell cycle-arrested yeast cells. Mutat Res. 2004;556:201–208. doi: 10.1016/j.mrfmmm.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 113.Puig M, Castellano D, Pantano L, Giner-Delgado C, Izquierdo D, et al. Functional impact and evolution of a novel human polymorphic inversion that disrupts a gene and creates a fusion transcript. PLoS Genet. 2015;11:e1005495. doi: 10.1371/journal.pgen.1005495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goettel W, Messing J. Change of gene structure and function by non-homologous end-joining, homologous recombination, and transposition of DNA. PLoS Genet. 2009;5:e1000516. doi: 10.1371/journal.pgen.1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Luteijn RD, Hoelen H, Kruse E, van Leeuwen WF, Grootens J, et al. Cowpox virus protein CPXV012 eludes CTLs by blocking ATP binding to TAP. J Immunol. 2014;193:1578–1589. doi: 10.4049/jimmunol.1400964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lin J, Eggensperger S, Hank S, Wycisk AI, Wieneke R, et al. A negative feedback modulator of antigen processing evolved from a frameshift in the cowpox virus genome. PLoS Pathog. 2014;10:e1004554. doi: 10.1371/journal.ppat.1004554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van Ham SM, Tjin EP, Lillemeier BF, Grüneberg U, van Meijgaarden KE, et al. HLA-DO is a negative modulator of HLA-DM-mediated MHC class II peptide loading. Curr Biol. 1997;7:950–957. doi: 10.1016/s0960-9822(06)00414-3. [DOI] [PubMed] [Google Scholar]
- 118.Salter RD, Howell DN, Cresswell P. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics. 1985;21:235–246. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]
- 119.van de Weijer ML, Bassik MC, Luteijn RD, Voorburg CM, Lohuis MA, et al. A high-coverage shRNA screen identifies TMEM129 as an E3 ligase involved in ER-associated protein degradation. Nat Commun. 2014;5:3832. doi: 10.1038/ncomms4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Riballo E, Doherty AJ, Dai Y, Stiff T, Oettinger MA, et al. Cellular and biochemical impact of a mutation in DNA ligase IV conferring clinical radiosensitivity. J Biol Chem. 2001;276:31124–31132. doi: 10.1074/jbc.M103866200. [DOI] [PubMed] [Google Scholar]