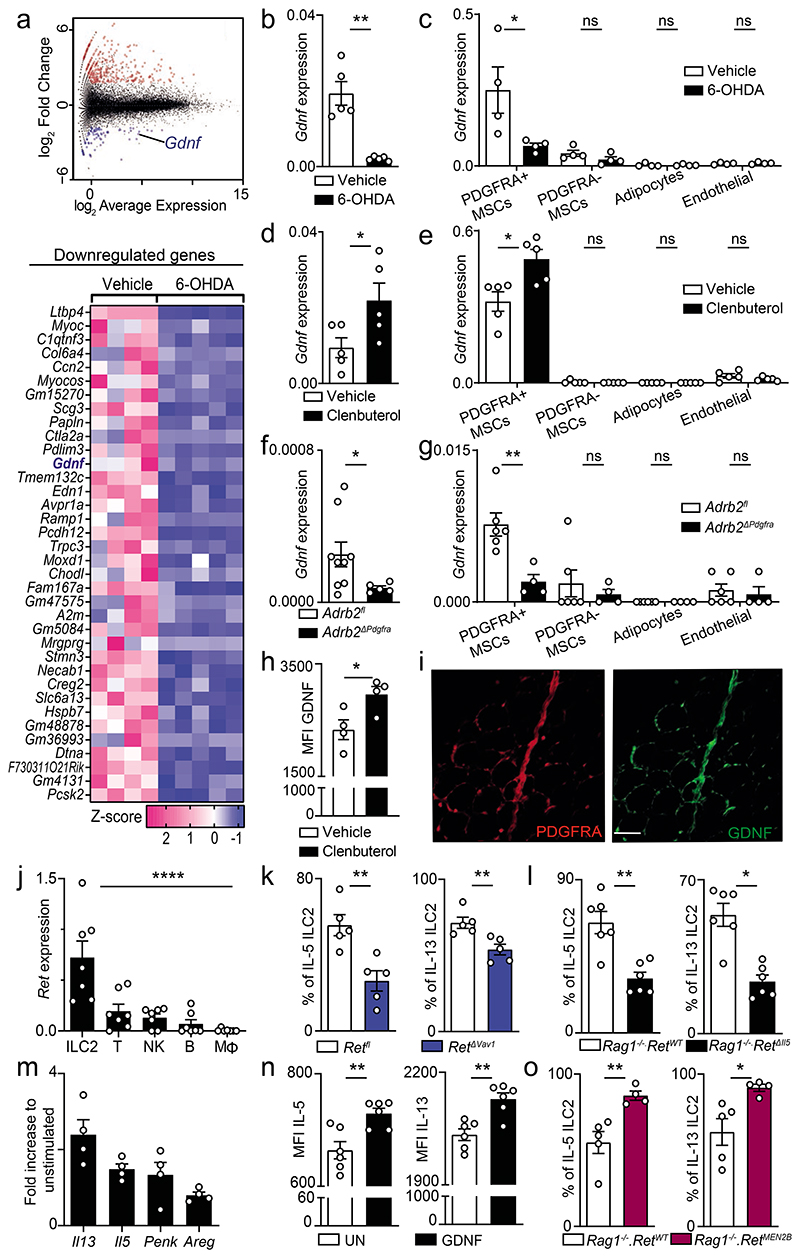
Figure 2. Sympathetic cues orchestrate mesenchyme-derived GDNF and innate type 2 cytokines.
6-OHDA treatment (a-c). a, RNAseq of PDGFRA+ MSC. Top: Mean-difference plot of vehicle versus 6-OHDA. Bottom: heatmap of downregulated genes. Vehicle n=4, 6-OHDA n=5. b, Total GAT RNA. n=5. c, GAT cell populations. n=4. (d,e) Clenbuterol administration. d, Total GAT RNA. n=5. e, GAT cell populations. n=5. f, Total GAT RNA. Adrb2fl n=9, Adrb2ΔPdgfra n=5. g, GAT cell populations. Adrb2fl n=6, Adrb2ΔPdgfra n=4. h, GDNF Median Flourescence Intensity (MFI) in MSC. n=4. i, GAT. Green: GDNF. Red: PDGFRA. Scale bar: 50μm. j, ILC2, T cells (T), natural killer cells (NK), B cells (B), macrophages (Mφ). n=7. k, GAT ILC2. n=5. l, GAT ILC2. n=6. (m, n) In vitro stimulation with GDNF. m, GAT ILC2. n=4. n, MFI innate type 2 cytokines. n=6. o, ILC2 from BM chimaeras. Rag1-/-.RetWT n=5, Rag1-/-.RetMEN2B n=4. Data are representative of 3 independent experiments. n represents biologically independent animals. Data are presented as mean values and error bars: SEM. Two-tailed unpaired Welch's t-test (b-h, k-o). One-way ANOVA (j). *P<0.05; **P<0.01; ***P<0.005; ns not significant.