Abstract
Decreased antigen presentation contributes to the ability of cancer cells to evade the immune system. We used the minimal gene regulatory network of type 1 conventional dendritic cells (cDC1) to reprogram cancer cells into professional antigen presenting cells (tumor-APCs). Enforced expression of the transcription factors PU.1, IRF8 and BATF3 (PIB) was sufficient to induce cDC1 phenotype in 36 cell lines derived from human and mouse hematological and solid tumors. Within 9 days of reprogramming, tumor-APCs acquired transcriptional and epigenetic programs associated with cDC1 cells. Reprogramming restored the expression of antigen presentation complexes and costimulatory molecules on the surface of tumor cells, allowing the presentation of endogenous tumor antigens on MHC-I, and facilitating targeted killing by CD8+ T cells. Functionally, tumor-APCs engulfed and processed proteins and dead cells, secreted inflammatory cytokines and cross-presented antigens to naïve CD8+ T cells. Human primary tumor cells could also be reprogrammed to increase their capability to present antigen and to activate patient-specific tumor-infiltrating lymphocytes. In addition to acquiring improved antigen presentation, tumor-APCs had impaired tumorigenicity in vitro and in vivo. Injection of in vitro generated melanoma-derived tumor-APCs into subcutaneous melanoma tumors delayed tumor growth and increased survival in mice. Antitumor immunity elicited by tumor-APCs was synergistic with immune checkpoint inhibitors. Our approach serves as a platform for the development of immunotherapies that endow cancer cells with the capability to process and present endogenous tumor antigens.
Introduction
Cancer evades the immune system through various mechanisms, including exclusion of effector immune cells from the tumor microenvironment, immunosuppression, intratumor heterogeneity and downregulation of antigen presentation (1, 2). Immune checkpoint inhibitors (ICI) and adoptive T cell therapies (ACT) have revolutionized cancer treatment, although success is limited to a fraction of patients (1). Recent studies have highlighted the importance of tumor immunogenicity and antigen presentation to ensure efficient CD8+ T cell priming and response to ICI (3, 4). At the tumor cell-intrinsic level, immune evasion mechanisms include overexpression of checkpoint inhibitor molecules (5), editing of tumor neoantigens and downregulation of antigen presentation (2). These mechanisms are imposed by transcriptional and epigenetic downregulation of genes involved in antigen processing and presentation (6), posttranslational degradation of surface major histocompatibility complex-I (MHC-I) (7, 8), and modulation of interferon-γ (IFN-γ) signaling (9). Although the IFN-γ pathway (10, 11) and epigenetic factors (6, 12) have been implicated in the control of tumor immunogenicity, strategies to revert tumor cells’ intrinsic immune evasion mechanisms and drive immunity are lacking.
Cancer development is accompanied by modifications of the genome and epigenome. As for somatic cell types, cancer cells are endowed with epigenetic plasticity (13) that allows reversal or modification of cancer cell fate (14) by cellular reprogramming (15, 16). The extensive epigenetic remodeling elicited by cell fate reprogramming leads to the disruption of the oncogenic transcriptional network, resulting in reduced tumorigenicity (17). Previous efforts to reprogram cancer cells directly have aimed to decrease cancer cell oncogenic potential with transcription factor combinations to induce cancer cell differentiation (18) or conversion to an unrelated cell fate (19, 20). However, this approach requires the successful conversion of most tumor cells, which remains challenging due to the low efficiency of both reprogramming and the delivery approaches (21).
A reprogramming approach that would reduce oncogenic potential and at the same time impose immune activation mechanisms in tumor cells has the potential to be beneficial for cancer immunotherapy. The activation of immune surveillance strongly depends on type 1 conventional dendritic cells (cDC1s). Within the tumor microenvironment, immunogenic cDC1 cells cros-spresent tumor antigens to CD8+ T cells, a process that is critical to elicit anti-tumor immunity (22–24). In addition, cDC1 cells recruit immune effectors by secretion of pro-inflammatory cytokines and chemo-attractants, such as interleukin-12 (IL-12) and CXCL10 (25). The presence of cDC1 cells within a diverse array of human tumors positively correlates with better survival and responsiveness to ICI and ACT (25–27). We previously demonstrated direct reprogramming of mouse and human fibroblasts into immunogenic cDC1 cells with the transcription factors PU.1, IRF8, and BATF3 (termed PIB) (28, 29). Here, we hypothesize that PIB reprograms cancer cells into professional antigen-presenting cells (APCs), providing a strategy to counteract tumor immune evasion mechanisms and restore tumor cell immunogenicity. Findings that cDC1 reprogramming endows tumor cells with professional antigen-presenting capacity leading to improved antitumor immunity will support the development of a gene therapy strategy for cancer immunotherapy.
Results
Reprogramming induces cDC1 cell fate in human and mouse cancer cells
To assess whether antigen presentation could be imposed in cancer cells by direct cell reprogramming, we used a lentiviral polycistronic vector encoding mouse or human PIB followed by internal ribosomal entry site (IRES)-enhanced green fluorescent protein (eGFP) (PIB-eGFP, Fig. 1A, fig. S1A). For mouse cells, we used two poorly immunogenic murine tumor cell lines, Lewis Lung Carcinoma (LLC) and melanoma B16-F10 (B16), characterized by low MHC expression (10, 12, 30). First, we observed that ectopic expression of PIB induced surface expression of the pan-hematopoietic marker CD45 and the APC marker MHC-II in both cell lines within transduced cells (Fig. 1B-D, fig. S1B-C). The population of CD45+MHC-II+ cells was named tumor-APCs and used to quantify reprogramming efficiency, which was higher in LLC (15.8±10.3%) than in B16 (5.9±4.8%) cells (Fig. 1D, fig. S1D). We observed surface expression of the cDC1-specific marker CLEC9A (31) within CD45+MHC-II+ cells, suggesting the induction of a cDC1-like phenotype in mouse cancer cells (Fig. 1E, fig. S1D). For human cells, we used a panel of 28 human cancer cell lines derived from solid tumors (Fig. 1F-G, fig. S1E) and 5 cell lines from hematological malignancies (fig. S2A-B). Mirroring the reprogramming process in mouse cells, we observed emergence of a population of reprogrammed CD45+HLA-DR+ cells in all cell lines transduced with PIB, but not in eGFP transduced controls (Fig. 1F-G, fig. S1E), suggesting that cDC1 reprogramming is universally applicable to mouse and human cancer cells. In leukemic cell lines that already expressed CD45, reprogramming leads to activation of HLA-DR and increased CD45 median fluorescence intensity (MFI, fig. S2C). Furthermore, these data showed that cDC1 reprogramming efficiency ranged from 0.2±0.1% to 94.5±7.6% across human cancer cell lines, independently of transduction levels and proliferation rates (fig. S3A-B). Regarding the germ layer of origin, ectodermal and mesodermal-derived cells were more permissive to reprogramming than cells from endodermal origin (p<0.01) (fig. S3C), in line with low efficiency of inducing pluripotency and neural cell fate from hepatocytes (8, 32). Despite low reprogramming efficiency in cancer cell lines from lung and breast carcinoma, we detected large populations of cells acquiring either CD45 or HLA-DR expression, which represent partially reprogrammed cells that may have acquired dendritic cell features. Consistently with the mouse system, reprogrammed CD45+HLA-DR+ cells expressed the cDC1 surface markers CLEC9A (59.1±3.6%), CD226 (67.5±1.8%), and CD11c (54.4±3.6%) (Fig. 1H) and exhibited DC-like morphology (Fig. 1I).
Figure 1. Reprogramming induces cDC1 program in mouse and human cancer cells.
(A) Experimental strategy to restore immunogenicity in cancer cells with PIB encoded in a lentiviral vector as a polycistronic construction. Induced tumor-APCs were characterized in vitro and in vivo. (B) Reprogramming efficiency of murine LLC cells, analyzed by flow cytometry as the percentage of CD45+MHC-II+ cells (red) gated in eGFP+ cells at day 9, when transduced with PIB-eGFP or control eGFP lentiviruses. (C) Micrographs depicting expression of MHC-II in reprogrammed melanoma (B16) cells. Scale bars, 20 μm (D) Quantification of CD45+MHC-II+ cells (n=10-21) and (E) CLEC9A+ cells gated in CD45+MHC-II+ population (n=2-8). (F) Reprogramming efficiency of human glioblastoma (T98G) and melanoma (HMVII) cells and (G) quantification from 28 solid cancer cell lines. Reprogrammed (CD45+HLA-DR+) and partially reprogrammed populations (CD45+HLA-DR- and CD45-HLA-DR+) are shown (n=2-6). (H) Surface expression and quantification of cDC1 markers gated in CD45+HLA-DR+ (n=4). Fluorescence minus one (FMO) control is shown. (I) Scanning electron microscopy of tumor-APCs purified on day 9. Scale bars, 20 μm. (J) PCA for reprogrammed (day 9++) and partially reprogrammed (day 9+) tumor-APCs, eGFP transduced cells (day 0) and peripheral blood cDC1 (HLA-DR+CD11c+CD141+, grey) (n=3-8). Color code marks cell line of origin. Arrow highlights reprogramming trajectory. (K) Percentage of tumor-APC gene signature activation in reprogrammed (red) and partially reprogrammed cells (blue). (L) Top 6 Kyoto Encyclopedia of Genes and Genomes (KEGG) (mouse)/Reactome (human) pathways, and gene ontologies upregulated in tumor-APCs. Mean±SD is represented. n = biological replicates.
To map global gene expression changes in mouse and human cancer cells, we performed bulk RNA sequencing (RNA-seq) of 2 mouse and 17 human cell lines after 9 days of reprogramming. We purified reprogrammed (CD45+MHC-II+/CD45+HLA-DR+, day9++) and partially reprogrammed cells (expressing either CD45 or MHC-II/HLA-DR, day 9+) by fluorescence-activated cell sorting (FACS) and compared transcriptomes with eGFP-transduced cancer cells (day 0) and cDC1s from mouse spleens or from donor peripheral blood and induced DCs (iDC1) from mouse fibroblasts (28) (Fig. 1J, fig. 4A, Data file S1-S2). Principal component analysis (PCA) showed that reprogrammed cancer cells, regardless of their origin, showed a substantial shift in transcriptome mapping closely to natural cDC1 (PC1, 47% of variance) (Fig.1J, fig. S4B). Additionally, partially reprogrammed cells also showed major transcriptional rewiring. The lack of convergence in PC2 (8% of variance) indicated a maintenance of partial identity of parental cells, which may be important for retention and presentation of tumor antigens. In agreement, both partially and completely reprogrammed cells activated expression of human ZNF366 (DC-script), C1ORF54, XCR1 and CLEC9A (33) (fig.S5A) or mouse Clec9a, Xcr1, Cd24a and Itgax to levels comparable to cDC1 (fig.S4C). These data confirmed the conservation of the reprogramming process across multiple cell types (28, 29) including cancer cells. We detected endogenous expression of IRF8 and BATF3 in mouse and human reprogrammed cells, suggesting that a stable cDC1-like fate had been acquired (fig. S4D-E, fig. S5B). To determine reprogramming specificity, we integrated data with published gene signatures for cDC1, cDC2, and pDC subsets (34) and observed that reprogrammed cells specifically up-regulated cDC1 signatures (fig. S5C). Finally, we generated a human tumor-APC signature based on cDC1 genes commonly up-regulated during reprogramming. All tested lines scored >50% moderately correlating with reprogramming efficiency (R=0.4; Fig. 1K). Pathway, molecular function, and biological process analysis for mouse or human tumor-APC gene sets revealed terms associated with antigen processing and presentation, as well as immune interactions, suggesting that reprogrammed cells have established competence to prime T cells (Fig. 1L, fig. S5D). Collectively, our analysis showed that combined expression of cDC1 reprogramming transcription factors imposed a transcriptional and phenotypic remodeling towards cDC1 identity in a broad array of mouse and human cancer lines.
Figure 4. Tumor-APCs are endowed with cDC1 function.
(A) The APC function of mouse and human tumor-APCs was assessed at reprogramming day 9. (B) Secretion of IL12p70, IL-29, CXCL10, and TNFα by human reprogrammed CD45+HLA-DR+ cells (PIB, red) with or without Poly(I:C) or LPS stimulation. eGFP-transduced cells were used as controls (-) and enriched peripheral blood DCs as reference (n=3-8). (C) Micrographs depicting uptake of fluorescently labeled OVA (OVA-AF647, red) by reprogrammed CD45+MHC-II+ LLC cells. Flow cytometry histogram (middle) and quantification (right) at 4°C and 37°C with IFN-γ stimulation, where indicated (n=3-10). Scale bars, 25μm. (D) Micrographs showing processing of DQ-OVA+ (green) by reprogrammed LLC cells. Scale bars, 25μm. Flow cytometry quantification of the percentage of DQ-OVA+ cells (right) transduced with PIB (n=6-13). (E) PSMB10 and PSMB9 protein expression in tumor-APCs. BM-DCs were used as reference, and calnexin (CANX) as loading control. (F) Engulfment of fluorescently labeled dead cells quantified by flow cytometry (left) and visualized by time-lapse microscopy (right). Black arrows highlight a reprogrammed CD45+HLA-DR+ cell (green) engulfing a dead cell over time (red, white arrows). Scale bar, 100μm. (G) Quantification of CD44+CTVlow CD8+ OT-I T cells after a 3-day coculture period with BM-DCs (left, n=7-10) and reprogrammed LLC cells pre-incubated with OVA peptide (right, SINFEKL; n=4-12). (H) Quantification of CD44+CTVlow CD8+ OT-I T cells co-cultured with tumor-APCs pulsed with OVA protein (n=2-8). Mean±SD is represented. n= biological replicates. P value was calculated using Two-way ANOVA (B) or One-way ANOVA (C,D,H) followed by Tukey´s multiple comparison test. **P<0.01, *** P<0.001, ****P<0.0001.
Reprogramming imposes a stepwise cDC1 transcriptional and epigenetic remodeling in cancer cells
Human CD45+HLA-DR+ or mouse CD45+MHC-II+ cells emerged as early as 3 days after transduction with PIB and gradually increased over time, suggesting rapid reprogramming (Fig. 2A, fig. S6A). We detected increased apoptosis and cell death at early time points as previously described for iPSC reprogramming (35) (fig. S6B). To map the kinetics of human cancer cell reprogramming at the transcriptional and epigenetic levels, we profiled human reprogrammed (CD45+HLA-DR+, ++) and partially reprogrammed (CD45-HLA-DR+, +) T98G-derived tumor-APCs along a time-course using RNA-seq and assay for transposase-accessible chromatin (ATAC)-sequencing (Fig. 2A). PCA segregated all reprogramming stages (day 3, 5, 7, and 9) from parental cells (day 0, eGFP transduced cells), with day 7 and 9 mapping closer to peripheral blood cDC1s, indicating a progressive acquisition of a cDC1 transcriptional program. In agreement, partially reprogrammed cells lagged in the time course supporting the notion that these cells are on the way to successful reprogramming. Reprogramming of human embryonic fibroblasts (HEF) followed a similar reprogramming trajectory (Fig. 2B, Data file S3), indicating that reprogramming dynamics is conserved across malignant and noncancerous primary cells. PCA for differential open chromatin regions demonstrated that epigenetic remodeling occurred fast with major changes between day 0 and day 3 (62% variance), followed by a fine-tuning at later time points (day 3, 5, 7, 9), bringing cells closer to the open chromatin patterns of cDC1 (Fig. 2C, Data file S4). To confirm these observations, we used the tumor-APC gene signature and mapped changes along the time course. The signature was gradually imposed at the transcriptional level (Fig. 2D) and rapidly established at the chromatin level (Fig. 2E). These data showed that PIB-mediated reprogramming elicits rapid epigenetic remodeling followed by a gradual rewiring of the cDC1 transcriptional program. Correlating with this, PIB rapidly up-regulated expression of cDC1-specific genes including ZNF366, CADM1, C1ORF54, and CAMK2D, whereas the induction of CLEC9A, XCR1 and CXCR3 occurred at later stages of the reprogramming process (Fig. 2F, fig. S6C), highlighting the dynamic, stepwise unfolding of cDC1 cell fate. The endogenous expression of IRF8 and BATF3 reached cDC1 levels by day 5, indicating that a stable acquisition of cDC1 identity had been reached by this point (fig. S6D). This was in accordance with the rapid induction of transgene independence, which has been observed in B cell to macrophage reprogramming (35). Furthermore, pathway, molecular function and biological process analysis revealed enrichment in IFN-γ signaling, antigen presentation and processing, and immune cell activation as soon as day 3 (fig. S6E).
Figure 2. PIB induce rapid global transcriptional and epigenetic reprogramming.
(A) Experimental design to evaluate the kinetics of reprogramming at the transcriptional and epigenetic levels. The human glioblastoma cell line (T98G) was transduced with PIB-eGFP. Reprogrammed (CD45+HLA-DR+, ++) and partially reprogrammed (CD45-LA-DR+, +) cells purified and profiled by RNA-seq and ATAC-seq at day 3 (d3), 5 (d5), 7 (d7) and 9 (d9). eGFP transduced cells were used as control (d0) and peripheral blood cDC1 as reference (grey). (RNA-seq, n=4-8; ATAC-seq, n=2-3; biological replicates) (B) PCA of transcriptomes. Reprogramming of HEFs was included as a reference for the dynamics. Color code identifies time points. (C) PCA based on differentially accessible chromatin regions. Arrows highlight reprogramming trajectories. (D) Percentage of tumor-APC gene signature activation and (E) chromatin accessibility in reprogrammed and partially reprogrammed cells. (F) mRNA expression of the cDC1 specific genes, ZNF366 (DC-script), CLEC9A, and XCR1 (n=3-8). (G) Representative sequencing tracks for IFI16, ANPEP, and IRF8 loci showing ATAC-seq peaks during reprogramming. Grey boxes highlight relevant peaks. (H) Transcription factor (TF) binding motif enrichment analysis and (I) Gene Ontology biological processes enrichment analysis of differential ATAC-seq peaks. Circle size refers to the number of peaks intersecting with the respective database category. Color gradient depicts adjusted P values based on a binomial test.
We then investigated whether chromatin remodeling was a stepwise process at the level of individual loci. Genes associated with cDC1 function, which had closed chromatin in nonreprogrammed cells such as the cytoplasmic pathogen sensor IFI16, and the cDC1 genes ANPEP and IRF8, showed open chromatin regions at day 3, 7 and 9, respectively (Fig. 2G). Motif discovery on chromatin regions that became open in reprogrammed cells showed that they were enriched for PU.1 and composite PU.1:IRF8 motifs at day 3, confirming their dominant role in initiating cDC1 reprogramming (29) (Fig. 2H). Motif analysis during reprogramming progression (day 9 vs day 3) revealed recruitment of CTCF and BORIS, important regulators of chromatin architecture and macrophage function after transdifferentiation (36), suggesting a contribution for CTCF-mediated chromatin loops in fine tuning cDC1 identity during reprogramming (Fig. 2H). Pathway analysis revealed that chromatin regions that became accessible between day 3 and 9 were enriched for processes associated with cell migration and motility, reflecting functional maturation (Fig. 2I).
To characterize the differences between tumor-APCs and cDC1, we performed pathway analysis on up-regulated and down-regulated genes. Genes upregulated in cDC1 compared to tumor-APCs were enriched in general terms such as nuclear signaling and protein targeting and downregulated genes in extracellular matrix. In contrast, comparison of cDC1 to cancer cells showed terms associated with T cell activation (fig. S7A), suggesting that antigen presentation is properly established in tumor-APCs. When analyzing differential peak enrichment, we observed only a small number of up-regulated peaks in cDC1 compared to tumor-APCs (fig. S7B), validating the extensive epigenetic remodeling induced by PIB.
Tumor-APCs become immunogenic and present tumor-associated antigens
We then assessed whether cDC1 reprogramming increased intrinsic tumor cell immunogenicity. RNA-seq analysis showed a progressive activation of antigen processing and presentation machinery in mouse and human tumor cells (fig. S8A-B). Particularly, MHC-I and MHC-II master regulators Nlrc5 and Ciita were up-regulated by day 3, sustaining expression of downstream genes including B2m and Cd74 (2, 30) (fig. S8C). Indeed, in all 17 human cancer cell lines PIB activated an antigen presentation signature that correlated with reprogramming efficiency (R=0.5) (fig. S8D). Recent CRISPR screening approaches have highlighted the importance of IFN-γ signaling in unlocking anti-tumor immunity and cytotoxic T lymphocyte (CTL) sensitivity (3, 4, 12). Reprogramming induced IFN-γ and STING pathway gene signatures (fig. S9A-B), in agreement with increased immunogenicity of reprogrammed tumor cells. An enhanced immunogenic profile did not result in excessive upregulation of immune checkpoints as gene expression levels of Cd274 (that encodes PD-L1), Vsir, Icosl and Havcr2 in tumor-APCs were comparable to that in cDC1 cells (fig. S9C). Moreover, activation of IFN signatures, STING pathway, and Toll-like receptor (TLR)-induced maturation was validated in the human system using gene set enrichment analysis (fig. S9D-F).
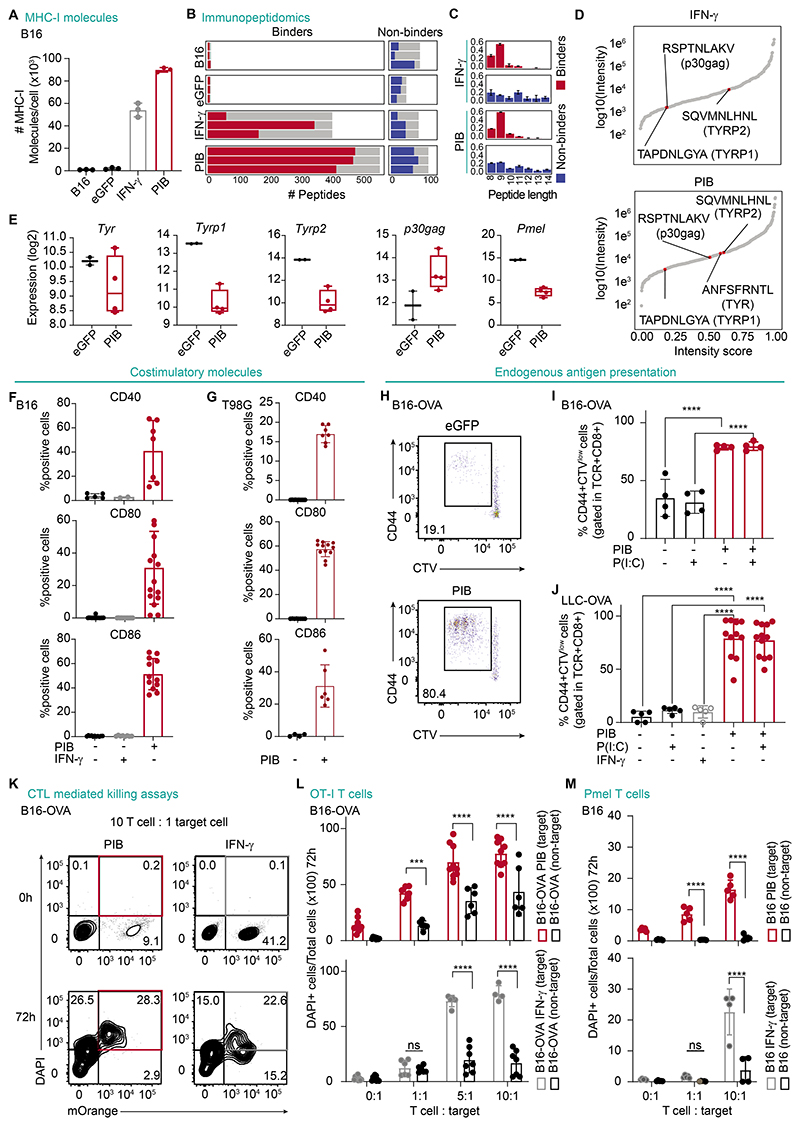
We detected increased surface expression of β-2-microglobulin (B2M) as well as MHC-I in mouse tumor-APCs (Fig. 3A, fig. S10A-D). In addition, surface expression of HLA-ABC and HLA-DR was up-regulated also in human T98G cells (fig. S10E), supporting that PIB gradually increases tumor immunogenicity. To investigate the ability of tumor-APCs to present tumor-associated antigens, we applied mass spectrometry-based immunopeptidomics on purified (CD45+) B16-derived tumor-APCs, eGFP-transduced, untransduced, and control cells treated with IFN-γ to induce MHC-I expression. We detected a higher number of peptides presented in reprogrammed cells compared with negative controls as well as IFN-γ-treated cells (Fig. 3B). These peptides were mainly predicted to bind to MHC-I and were in average of 9 amino acids long (Fig. 3C, Data file S5). B16-derived tumor-APCs showed increased presentation of peptides that originated from multiple known melanoma-associated antigens (Fig. 3D, fig. S10F). Peptides derived from TYR, TYRP1, TYRP2 and p30gag passed the binding affinity prediction threshold and were detected despite of reprogramming-induced transcriptional down-regulation (Fig. 3E). These data show that enhanced antigen presentation in tumor-APCs outweighs the transcriptional down-regulation of tumor-associated antigens.
Figure 3. Reprogrammed cancer cells become immunogenic.
(A) Flow cytometry quantification of the number of MHC-I molecules in B16 cells, tumor-APCs at day 9 (PIB) or B16 cells after eGFP transduction or stimulation with IFN-γ. (n=3). (B) Number of peptides predicted as binders and non-binders per biological replicate (n=3). Grey bars indicate the total number of distinct peptides per condition. (C) Peptide length distributions and (D) ranking by normalized intensity of predicted binders. Peptides derived from canonical melanoma tumor antigens are highlighted. (E) mRNA expression in B16-derived tumor-APCs on day 9. (F) Quantification of expression of the costimulatory molecules CD40, CD80 and CD86 by flow cytometry gated in CD45+MHC-II+ (mouse) (n=2-17) or (G) CD45+HLA-DR+ (human) (n=4-12). (H) Representative flow cytometry plots and (I) quantification of CD8+ T cell proliferation, measured by Cell Trace Violet (CTV) dilution, and activation (CD44+) after co-culture with purified B16-OVA cells at reprogramming day 3. Poly(I:C) (P(I:C)) stimulation overnight when indicated (n=4). (J) Quantification of CD44+CTVlow CD8+ T cells when co-cultured with tumor-APCs derived from LLC-OVA at day 9 (n=5-12). (K) T cell-mediated killing of B16-OVA target cells (mOrange+) that were either PIB-transduced or IFN-γ-treated, after 72 h of coculture. The percentage of target dead cells (mOrange+, DAPI+) is highlighted in red. (L) Quantification of cell killing of either target or nontarget tumor cells when cocultured with increasing ratios of OT-I T cells after 72 h (n=4-9). (M) Quantification of pmel-specific T cell killing of either target or nontarget B16 tumor cells after 72 h (n=4-6). Mean±SD is represented. n= biological replicates. P value was calculated using One-way ANOVA followed by Tukey´s multiple comparison test (I, J) or Two-way ANOVA followed by Sidak multiple comparison test. ***P<0.001, ****P<0.0001.
Exposure to IFN-γ has been employed to induce high levels of MHC-I, but T cell receptor (TCR) engagement by cancer cells in the absence of proper costimulation results in T cell anergy (37). Thus, we sought to assess the expression of the costimulatory molecules CD40, CD80 and CD86, as they represent a necessary second signal to ensure T cell activation (37). PIB induced the expression of costimulatory molecules in mouse and human tumor cells, whereas treatment of cancer cells with IFN-γ did not (Fig. 3F-G, fig. S10G-J). In human cells, we detected expression of costimulatory molecules starting at day 4 and increasing gradually until day 9. Tumor-APCs responded to TLR3/4 triggering, leading to an increased surface expression of CD40 (66.1±11.2%, vs 21.2±12.2%) (fig. S10G-J), implying that cDC1 reprogramming endows cancer cells with APC machinery to prime T cell responses.
To validate whether tumor-APCs present endogenously expressed antigens, we took advantage of the Ovalbumin (OVA)-expressing cell lines B16-OVA and LLC-OVA. First, we validated that these cell lines reprogram at similar efficiencies to parental cell lines (B16-OVA, 7.8±6.9%; LLC-OVA, 48.6±7.3%), confirming that OVA expression does not interfere with reprogramming (fig. S11A-B). To evaluate T cell priming ability of tumor-APCs, we cocultured magnetic-activated cell sorting (MACS)-enriched tumor-APCs with naïve OT-I CD8+ T cells which recognize OVA-derived SIINFEKL peptide loaded onto H-2Kb/B2M MHC-I complex (28). Whereas control eGFP-transduced B16-OVA and LLC-OVA cells showed low OVA antigen presentation capacity after IFN-γ or Polyinosinic-polycytidylic (Poly(I:C)) stimulation, tumor-APCs efficiently primed naïve OT-I CD8+ T cells independently of Poly(I:C) treatment (Fig. 3H-J). Next, to assess whether tumor-APCs became susceptible to CTL killing, we generated B16-OVA cells expressing the mOrange fluorescent protein. Tumor-APCs or B16-OVA cells treated with IFN-γ (target, mOrange+) were mixed with untreated B16-OVA cells (non-target, mOrange-) and cocultured for 3 days with increasing ratios of activated OT-I CD8+ T cells (3) (Fig. 3K-L). Importantly, we observed that tumor-APCs were more susceptible to CD8+ T cell-mediated killing compared to untreated B16-OVA cells. As expected, B16 cells that did not express OVA, were not targeted for T cell killing, even upon IFN-γ stimulation and the highest ratio of CTL (10:1), validating the specificity of the assay (fig. S11C). Notably, tumor-APCs were more efficiently killed by T cells than IFN-γ-stimulated B16-OVA cells at low (1:1) ratios (42.4±6.2% PIB vs 12.3±7.1% IFN-γ, Fig. 3L), and at early culture (24 h) time-points (12.9±12.1% PIB vs 2.7±3.1% IFN-γ, fig. S11D). Higher T cell to target cell ratios also resulted in killing of the nontarget population in tumor-APC co-cultures at later time-points (72 h; Fig. 3L). This may be explained by a bystander killing effect (38) which reflects sustained T cell activation by reprogrammed cells, increasing non-target cancer cell clearance. To evaluate whether T cells would effectively target endogenous tumor antigens in tumor-APCs, we tested the killing capacity of T cells against the gp100/pmel 17 melanoma antigen expressed in B16 cells (39). Accordingly, at 1:1 ratio, tumor-APCs were more efficiently killed by pmel-specific T cells than IFN-γ-stimulated cells after 72 h co-culture (8.6±2.8% PIB vs 1.6±0.5% IFN-γ, Fig. 3M, fig. S11E). Contrastingly, IFN-γ-stimulated B16 cells were only killed at high (10:1) T cell-cancer cell ratios. Together, our data indicate that cDC1 reprogramming promotes tumor antigen presentation resulting in enhanced immune recognition and elimination of cancer cells by CD8+ T cells.
Reprogramming generates functional APCs that elicit CD8+ T cell responses
Next, we sought to address whether tumor-APCs acquired cDC1 function by evaluating secretion of proinflammatory cytokines, antigen uptake, and presentation to CD8+ T cells (Fig. 4A). We first addressed tumor-APC capacity to secrete proinflammatory cytokines, the third signal required for T cell activation. We first observed expression of genes encoding inflammatory cytokines including Ifnb, Il6 and Cxcl10 (25) (fig. S12A) and confirmed secretion by mouse tumor-APCs of type I IFN (IFN-α and IFN-β), IL-1β, IL-6, IL-12p70 and CXCL10 after TLR3 stimulation with Poly(I:C) (fig. S12B). Similarly, in the human system tumor-APCs secreted IL12p70, IL-29 (IFN-λ), CXCL10 and TNFα in response to Poly(I:C) and LPS (26, 40) (Fig. 4B), showing that reprogramming endows cancer cells with the capacity to secrete cDC1-related cytokines and chemokines required for T cell recruitment and activation.
Furthermore, we sought to investigate whether tumor-APCs engulf protein and dead cells, a hallmark of cross-presenting DCs. Mouse and human tumor-APCs became fluorescently labelled with OVA-Alexa Fluor 647 (OVA-AF647) within 30 to 60 min of incubation (Fig. 4C, fig. S12C, Supplementary Movie S1). Antigen uptake by tumor-APCs was mediated by macropinocytosis, shown by Lucifer-yellow internalization, and by myeloid-specific mannose receptor-mediated endocytosis, as determined by treating tumor-APCs with mannan, a ligand for the mannose receptor (fig. S12D-E) (41). We next evaluated the capacity of tumor-APCs to process internalized antigens by fluorescence detection of cleaved DQ-OVA. Tumor-APCs efficiently processed OVA (Fig. 4D, Supplementary Movie S2), a process that was inhibited by Leupeptin and Lactacystin and was, therefore, dependent on protease and proteasome activity (fig. S12F). In DCs, efficient processing of antigens relies on the assembly and activity of the immunoproteasome (42). Accordingly, we observed an increase in the protein levels of the immunoproteasome subunits PSMB9 and PSMB10 in PIB-transduced LLC cells (Fig. 4E, fig. S12H), suggesting that enhanced antigen processing is mediated by the immunoproteasome. Moreover, human and mouse tumor-APCs engulfed fluorescently-labelled dead cells (Fig. 4F, fig. S12G and Supplementary Movie S3-S4), a hallmark of cross-presenting dendritic cells relying on CLEC9A engagement (31).
We evaluated T cell priming ability with OT-I CD8+ T cells. Reprogrammed LLC and B16 cells activated the proliferation of naïve CD8+ T cells after a pulse with the OVA peptide SIINFEKL, at comparable levels to CD103+ bone marrow-derived dendritic cells (BM-DCs) (Fig. 4G, fig. S12I, Supplementary Movie S5). Although eGFP-transduced cells failed to prime naïve CD8+ T cells, further stimulation of tumor-APCs with Poly(I:C) or IFN-γ improved T cell priming (76.8±15.8% PIB vs 91.1±2.1% Poly(I:C) and 82.4±11.9% IFN-γ), in accordance with enhanced IL-12 secretion by reprogrammed cells in the presence of Poly(I:C) (fig. S12I). Next, we evaluated cross-presentation of tumor-APCs after a pulse with OVA protein. We observed that tumor-APCs established competence to cross-present antigens to CD8+ T cells, which is further enhanced by TLR3 stimulation (63.5±8.5 vs 27.5±20.9%) (Fig. 4H). Consistently with an early establishment of global gene expression during reprogramming (fig. S8A-B), we demonstrated that tumor-APCs acquire cross-presenting ability by day 3 of reprogramming (Fig.4H, fig. S12J). Consistently, we found that reprogrammed cells pulsed with cytomegalovirus (CMV) peptides, with or without Poly(I:C) and LPS stimulation, efficiently elicited CMV+CD8+ T cell activation (fig. S12K).
We observed that reprogramming generates populations of partially reprogrammed cells (Fig.1G, fig. S13A-C) which acquired cDC1-like transcriptional signatures (Fig. 1K), suggesting that the process is either asynchronous or leads to heterogeneity. We FACS-purified CD45-HLA-DR+ partial reprogrammed cells at day 9 and showed that 68% of sorted cells became CD45+HLA-DR+ after 8 days of culture, showing that partial reprogrammed cells have potential to complete the reprogramming process with additional time (fig. S13D). We then sought to investigate whether partially reprogrammed cells acquire cDC1 functional capacity. Interestingly, partially reprogrammed cells also responded to stimulation by secreting IL-12p70, CXCL10, IL-29 and TNFα but at a lower extent than CD45+HLA-DR+ cells (fig. S13E). A similar pattern was observed for dead cell incorporation (fig. S13F). These data indicate that incompletely reprogrammed cells can, at least to certain extent act as tumor-APCs. Collectively, our results highlight that cDC1 reprogramming converts mouse and human cancer cells into tumor-APCs endowed with professional cDC1 function.
Primary patient-derived cancer cells can be reprogrammed towards a cDC1 fate
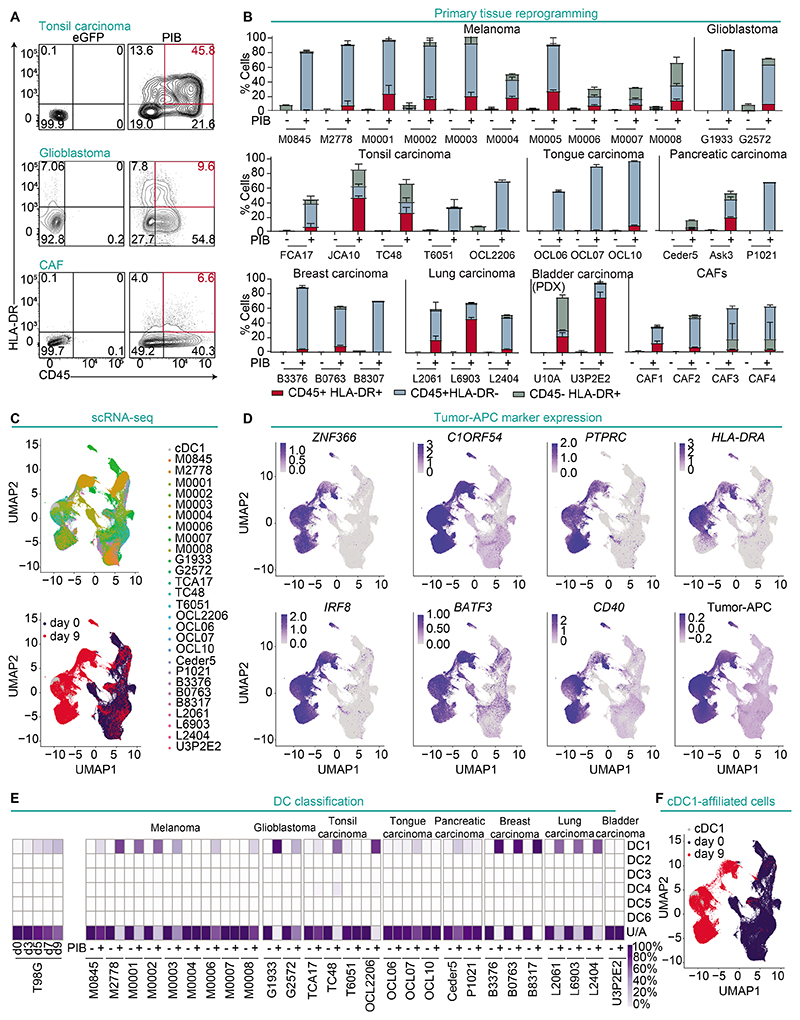
An important consideration for translation of tumor-APCs is whether reprogramming can be elicited in human primary cancer cells. To validate cDC1 reprogramming in primary cancer cells, 35 samples were collected from 7 different tumors obtained from patients with melanoma, lung, tonsil, tongue, pancreatic, breast and PDX-derived bladder carcinoma, as well as lung cancer associated fibroblasts (CAFs). Upon transduction with PIB, all primary cancer cells showed major phenotypic changes, initiating expression of CD45 and HLA-DR, reflecting reprogramming (Fig. 5A). Reprogramming efficiency ranged between 0.6±0.3% to 75.2%±6.8%. Samples from the same tumor types showed similar phenotypic profiles indicating relatively low variability across patients (Fig. 5B).
Figure 5. Human primary cancer cells are permissive to cDC1 reprogramming.
(A) Reprogramming efficiency of human primary tonsil carcinoma (JCA10), glioblastoma (G2572) and CAFs at reprogramming day 9 and (B) quantification from 35 patient samples. Reprogrammed (CD45+HLA-DR+) and partially reprogrammed populations (CD45+HLA-DR- and CD45-HLA-DR+) are shown. Individual patients are depicted by codes (n=2-6). Mean±SD is represented. (C) Reprogrammed and partially reprogrammed cells from 27 human primary tumor samples were purified and profiled by scRNA-seq without multiplexing. Peripheral blood cDC1 were used as reference, and eGFP-transduced cells as controls. UMAP analysis of single-cell transcriptomes showing 136,796 primary cancer cells according to their origin (upper panel) or treatment (bottom panel). (D) UMAP plots showing expression of cDC1 genes ZNF366 and C1ORF54, reprogramming markers PTPRC and HLA-DRA, endogenous expression of IRF8 and BATF3, costimulatory molecule CD40, and tumor-APC signature. (E) Integration of single-cell data from a reprogramming time-course of the T98G cell line (left) and primary tumor samples (right) with published DC subset data (GSE94820). Heatmap shows the percentage of single cells affiliated to individual cDC subsets or unaffiliated (U/A). (F) UMAP showing single-cell transcriptomes of cDC1-affiliated cells.
To verify that a cDC1 program had been established in single primary cells and uncover potential heterogeneity, we FACS-purified reprogrammed cells expressing at least one of the reprogramming markers derived from 27 patients and performed single-cell RNA-seq (scRNA-seq) (fig. S14A). Uniform manifold approximation and projection (UMAP) analysis revealed that regardless of parental origin, most reprogrammed cells mapped closely to donor peripheral blood cDC1, while clearly separating from eGFP transduced cells (day 0) (Fig. 5C, Data file S6). In agreement, reprogrammed cells mapping closer to cDC1s showed increased expression of cDC1 canonical markers (ZNF366, C1ORF54), reprogramming markers (PTPRC, HLA-DRA), costimulatory molecules (CD40), endogenous IRF8 and BATF3, as well as the tumor-APC signature at the single cell level (Fig. 5D), verifying successful reprogramming of human primary cancer cells.
To define the accuracy of cDC1 reprogramming in patient samples, we first generated a reference reprogramming trajectory with a single-cell transcriptomes time-course from reprogrammed T98G cells at day 3, 5, 7 and 9. T-distributed stochastic neighbor embedding (t-SNE) resolved the reprogramming process into the gradual acquisition of the cDC1 fate and expression of C1ORF54 and endogenous IRF8 and BATF3 (fig. S14B). We applied Monocle3 to recreate the reprogramming trajectory (fig. S14C). Analysis of gene modules activated during reprogramming showed gradual enrichment in modules 2 and 5 containing antigen processing and presentation and silencing of module 6 containing cell cycle progression genes, which is in line with reprogramming-induced antigen presentation and loss of tumorigenicity (fig. S14D-E). To develop a classification method of single reprogrammed cells, we used scPRED to integrate time-course single-cell data into six DC subsets (34). The percentage of cells classified as cDC1 gradually increased (from 3.7% at day 3 to 35.4% at day 9), whereas parental cancer cells were unaffiliated. Next, we applied the DC classification model to evaluate the fidelity of cDC1 reprogramming across all primary samples. We detected variable percentages of cDC1-affiliated cells (0.2% to 97.3%), demonstrating that PIB imposed the cDC1 signature with a degree of variability, but not crossing DC subset boundaries (Fig. 5E-F). Primary patient-derived samples are usually heterogenous. To ensure that reprogrammed cells originate from cancer cells, we employed CopyKAT algorithm to estimate genomic copy number variations in single cells (43) (fig. S15A). The percentages of malignant (aneuploidy) and normal (diploid) cells varied across samples before and after reprogramming (fig. S15B) but UMAP analysis showed transcriptional reprogramming in both cancerous and normal cells (fig. S15C), implying that cDC1 reprogramming in tumor derived cultures occurs regardless of transformation status. Reprogrammed malignant cells showed increased expression of cDC1 genes, the tumor-APC signature (fig. S15D), and were annotated to cDC1 subsets at frequencies comparable to reprogrammed diploid cells (fig. S15E). The exception was lung carcinoma that showed reduced efficiency from cancer cells in agreement with low reprogramming efficiency in lung cancer cell lines (fig. S15E-F). Together, our data show robust establishment of cDC1 fate in a wide spectrum of patient-derived tumor cell-types.
Patient-derived melanoma cells can be reprogrammed to functional cDC1-like antigen presenting cells
To evaluate functional capacity of tumor-APCs derived from primary cancer cells, we reprogrammed a panel of eight patient-derived melanoma cells. We validated reprogramming-induced phenotypic changes on day 3, 6, and 9. Consistent with previous data, PIB expression gradually induced reprogrammed CD45+HLA-DR+ populations that activate expression of CLEC9A, CD141, CD11c, HLA-ABC and the costimulatory molecules CD40, CD80 and CD86 (fig. S16A-C). The surface expression of the melanoma marker MSCP was down-regulated but still expressed by day 9, consistent with the presentation of tumor-associated antigens. Reprogrammed cells secreted the proinflammatory cytokines IL12p70, IL-29, CXCL10, TNFα, IL-28, IL1β, IL-6, and IL-8 with or without TLR engagement (fig. S16D), implying that reprogramming from primary cancer cells generated mature cDC1-like cells (fig. S16C). To validate the ability to activate memory and prime naïve T cells, we stimulated reprogrammed melanoma cells with HLA-A2 restricted CMV pp65 (NLVPMVATV) and MART-1 (ELAGIGLTV) peptides and co-cultured them with CD8+ T cells isolated from HLA-A2+ CMV+/MART-1+ donors. Eight-day cocultures with peptide-stimulated TLR-treated reprogrammed cells yielded higher percentages of both memory CMV+CD8+ T cells (24.8±12.1% to 5.7%±5.2%) and naïve CD8+ T cells (2.9±1.8% to 0.7±0.5%) to similar levels as monocyte-derived DCs (moDCs) used as reference (Fig. 6A-B, fig. S16E). Reprogrammed melanoma cells induced T cell activation after stimulation with long MART-1 peptide (44) (0.1±0.1% to 0.8±0.4%), demonstrating the ability of primary tumor-APCs to process and cross-present antigens (Fig. 6C). Consistent with a mature DC phenotype, reprogrammed cells could prime antigen-specific T cells even without TLR stimulation (Fig. 6A-C), confirming that cDC1 reprogramming endows primary cancer cells with professional antigen-presenting capacity.
Figure 6. cDC1 reprogramming imposes cDC1 function in primary melanoma cells.
(A) CD8+ T cells from CMV+ donors were co-cultured with day 9 PIB-transduced, eGFP-transduced melanoma cells primed with CMV peptide and LPS and Poly(I:C). Cocultures with moDCs were included as reference. Flow cytometry plots showing percentage of CMV+ CD8+ T cells (left, detected with two similar tetramers with PE and APC fluorophores) and quantification of CMV+ CD8+ T cells (middle) and TNFα+ IFN-γ+ CD8+ T cells after coculture (right, n=3-5). (B and C) Flow cytometry plots showing percentage and quantification of MART-1 specific CD8+ T cells after coculture with tumor-APCs primed with (B) MART-1 short peptide (right, n=6) and (C) long peptide (right, n=6). (D) PIB-reprogrammed and eGFP-transduced control melanoma cells were cocultured with autologous tumor infiltrating lymphocytes (TILs). Untransduced melanoma cells were included as additional controls. Quantification of the frequency of expression of the reactivity markers CD107a, CD137, TNFα, and IFN-γ by CD8+ T cells as a proportion of the total CD8+ T cell pool (right, n=2) and TIL-mediated lysis of melanoma cells (left, n=3). The lines in the left panel connect the mean values of cytolysis at individual timepoints. (E) Quantification of mean fluorescence intensity (MFI) for the exhaustion markers BTLA, LAG3, TIM3, PD-1 expressed on CD8+ TILs after co-culture with untransduced, eGFP-transduced, and reprogrammed cells. (n=4). Mean±SD is represented. n= biological replicates. P value was calculated using Two-Way ANOVA followed by Tukey´s multiple comparison test. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Finally, we sought to evaluate whether cDC1 reprogramming triggered killing of reprogrammed melanoma cells by tumor-infiltrating lymphocytes (TILs) isolated from the same patient. We observed that TILs that had been cocultured with reprogrammed melanoma cells for 8 hours had increased expression of the reactive markers CD107a, CD137, and had higher expression of the cytokines IFN-γ and TNFα compared to eGFP-transduced and untransduced melanoma cells (Fig. 6D). This correlated with enhanced cytotoxicity of the TILs against reprogrammed melanoma cells compared to nonreprogrammed melanoma cells (Fig. 6D). Additionally, we observed that TILs cocultured with reprogrammed melanoma cells had increased expression of the tumor experienced T cell markers, BTLA, TIM3, LAG3, PD-1, CD28 and CD69 (5) (Fig. 6E, fig. S16F). Addition of ICIs (anti-PD-1 and anti-CTLA4) during coculture resulted in a small increase in cytolysis of reprogrammed melanoma cells by the TILs (fig. S16G), suggesting that combining cDC1 reprogramming and immune checkpoint blockade could have the potential to be a synergistic immunotherapy approach.
Reprogramming reduces the tumorigenicity of cancer cells
In addition to altering the fate of tumor cells, it is possible that reprogramming cancer cells could reduce their proliferation and promote mitotic arrest. RNA-seq of cells along the reprogramming trajectory showed that genes associated with cell cycle progression were gradually down-regulated, suggesting that specifying a cDC1 fate reduces the tumorigenic drive of cancer cells (Fig. 7A). Genes associated with cell cycle progression, such as CCNA2, CDK1, MCM6, CDK2, PCNA, and MKI67, were repressed and the tumor suppressor genes, TP53, RB1 and CDKN1A, were activated with reprogramming (Fig. 7B). The gene signature associated with proliferation was decreased in the majority (16/17) of cancer cell lines profiled, pointing towards an induction of cell cycle arrest associated with reprogramming independently of cell type of origin (Fig. 7C). We then tested whether transcriptional patterns were reflected in reduced cell division in vitro. We labeled human tumor-APCs and control cells derived from 4 cancer lines with the membrane dye Cell Trace Violet (CTV) and quantified proliferation by dye dilution (Fig. 7D-E). Compared to control cells, both partially and completely reprogrammed cells slowed down cell division.
Figure 7. cDC1 reprogramming attenuates tumorigenicity in vitro and in vivo.
(A) Heatmap showing gene expression changes in cell cycle progression gene signatures in reprogramming. Reprogrammed (CD45+HLA-DR+; ++), partially reprogrammed (CD45-HLA-DR+; +), were profiled by bulk RNA-seq at day 3 (d3), 5 (d5), 7 (d7), and 9 (d9). eGFP-transduced cells were included as day 0 (d0). cDC1s served as reference. n=4-8; biological replicates. (B) mRNA expression of cell cycle progression (CCNA2, CDK1, MCM6, CDK2, PCNA, and MKI67) and cell cycle arrest genes (TP53, RB1, and CDKN1A) (n=3-8). (C) Changes in proliferation signature imposed by cDC1 reprogramming in 17 human cancer cell lines. (D) Flow cytometry analysis of cell proliferation by dye dilution. Reprogrammed T98G cells at day 9 were labelled with CTV, recultured and analyzed by flow cytometry after 1 (day 10), 3 (day 12), 4 (day 13), 6 (day 15), 8 (day 17), and 10 (day 19) days. (E) CTV MFI in reprogrammed CD45+HLA-DR+ (red) and eGFP+ (black) populations from 4 cell lines (n=2). (F) Anchorage-independent growth in soft agar of purified reprogrammed and partially reprogrammed cells. Colony formation after 4 to 6 weeks of culture and quantification for three cell lines (n=3-6). Relevant mutations are indicated. (G) Anchorage-independent growth of cells reprogrammed with DOX-inducible lentiviral PIB-expressing system. Colony formation after 5 weeks of culture after removal of DOX. (n=6). (H) Assessment of tumorigenic potential of tumor-APCs in vivo (top). Survival curves of NXG mice transplanted with reprogrammed T98G-derived CD45+HLA-DR+ cells (red, n=6), eGFP-transduced controls (black, n=9) and with 3P2C PDX-derived CD45+HLA-DR+ cells (red, n=4) and eGFP-transduced cells (black, n=5). For graphs in (A-G), mean±SD is presented. n= biological replicates. P values in (H) were calculated using Mantel-Cox test.
To assess loss of tumorigenicity, three cancer cell lines harboring mutations in the tumor-suppressor genes TP53, PTEN, and KRAS were selected and evaluated for their ability to show anchorage-independent growth and the ability to form colonies in soft agar. Although 582±43, 292±25, and 205±22.2 colonies were generated from eGFP-transduced cells, we did not observe colonies from purified completely or partially reprogrammed cells (Fig. 7F), indicating reduction of tumorigenic potential.
To determine the stability of the reprogrammed phenotype, we elicited reprogramming with doxycycline (DOX)-inducible lentiviral vectors which allow the controlled expression of reprogramming factors in transduced cells. We FACS-purified partially and completely reprogrammed cells on day 9 after lentiviral transduction, and plated them in soft agar for 5 weeks in the absence of DOX to stop PIB expression. We did not observe the formation of colonies from either partially or completely reprogrammed cells. These data demonstrate that sustained expression of exogenous PIB is not required to maintain reprogrammed tumor-APCs, and silencing does not result in reversal to tumorigenesis (Fig. 7G). To verify these findings in vivo, T98G-derived tumor-APCs or eGFP transduced cells were injected subcutaneously into the flank of NXG mice. In contrast to mice transplanted with control T98G cells, which all formed tumors and died during the first 5 months, animals implanted with tumor-APCs remained tumor-free and alive for the duration of the experiment. We confirmed these survival curves with reprogrammed PDX-derived bladder carcinoma (3P2C) cells (Fig. 7H). Altogether, our data show that cDC1 reprogramming enforces cell cycle arrest and reduces tumorigenicity of human cancer cells.
Reprogramming of cancer cells induces antitumor immunity in vivo
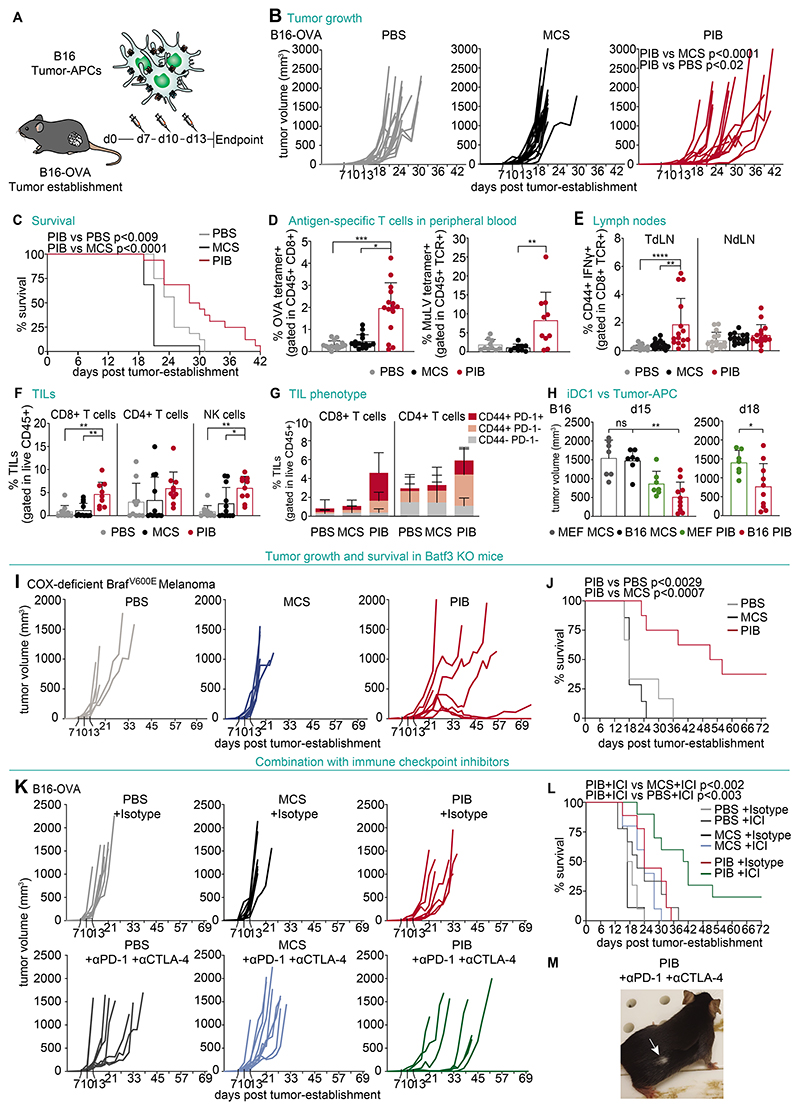
Finally, we tested the ability of tumor-APC that had been reprogrammed in vitro to drive antitumor immune responses in vivo. To do this, we established B16-OVA tumors in syngeneic mice for 7 days and generated B16-derived tumor-APCs, which were pulsed with OVA protein and stimulated with Poly(I:C). We injected the tumor-APCs intratumorally on days 7, 10 and 13 (Fig. 8A). Injections with B16-derived tumor-APC delayed tumor growth (Fig. 8B) and extended survival (Fig. 8C) compared with animals treated with phosphate-buffered saline (PBS) or injected with eGFP-transduced B16 cancer cells. On day 14, we detected increased frequencies of T cells specific for OVA-derived and tumor-associated antigen murine leukemia virus (MuLV)-derived MHC-I peptides in peripheral blood (Fig. 8D, fig. S17A). We also observed elevated levels of pmel-reactive CD8+ T cells in tumor-draining lymph nodes (TdLN) at day 18 (Fig. 8E), suggesting that local administration of reprogrammed cancer cells inside the tumor is sufficient to elicit expansion of antigen-specific T cells leading to the control of tumor growth. Immunophenotyping of lymphoid populations in treated tumors revealed a 5.5 and 7-fold increase of CD8+ T cells and NK cells, respectively (Fig. 8F). In contrast, we did not observe differences in myeloid populations including neutrophils, monocytes, CD64+ macrophages and CD11b+ dendritic cells (fig. S17B). Further characterization of infiltrating T cells revealed increased percentages of CD44+ PD-1+ and CD44+ PD-1- CD8+ and CD4+ T cells in tumor-APC-treated mice (Fig. 8G). These results indicate that tumor-APCs are sufficient to modify the tumor microenvironment promoting a hot tumor in the poorly immunogenic B16-OVA tumor model.
Figure 8. Tumor-APCs trigger anti-tumor immunity in vivo.
(A) B16-OVA tumors were injected intratumorally at day 7, 10 and 13 with B16-derived tumor-APCs pulsed with OVA protein and Poly(I:C). (B) Tumor growth and (C) survival of mice injected with tumor-APCs (PIB) compared with PBS and injection of control transduced cells (MCS) (n=16). (D) Flow cytometry quantification of peripheral blood T cells with OVA tetramer (left) or Murine Leukemia Virus (MuLV) tetramer (right), as a proportion of CD45+ CD8+ T cells, at day 14 after tumor establishment. (n=10-15) (E) Quantification of CD44+IFN-γ+ CD8+ T cells isolated from tumor-draining (TdLN) or non-draining lymph nodes (NdLN) after in vitro restimulation with pmel peptide at day 18. (n=15) (F) Quantification of tumor infiltrating lymphocytes (TILs) and (G) CD44+ and PD-1+ expression at day 18. (n=10) (H) Volumes of B16 tumors at day 15 (left) and 18 (right) treated with tumor-APCs (B16 PIB) compared with MEF-derived iDC1 (MEF PIB) after overnight stimulation with Poly(I:C) (n=7-10). (I) Tumor growth and (J) survival of BATF3-/- mice injected with Cox-deficient BRAFV600E melanoma tumor cells treated with tumor-APCs derived from the same cell line after overnight stimulation with Poly(I:C) (n=6-8). (K) Tumor growth and (L) survival of mice treated with ICIs (anti-PD-1 and anti-CTLA-4) or isotype controls (IgG2a and IgG2b) in combination with B16-derived tumor-APCs (PIB) after overnight incubation with OVA and Poly(I:C) (n=9-10). (M) Animal cured with combination therapy showing depigmentation (white arrow) on tumor regression site. Mean±SD is represented. n= biological replicates. P values were calculated using Kruskal Wallis (B, D-H), Mann-Whitney (H), and Mantel-Cox test (C, J, L). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
To test whether cDC1 reprogramming of tumor cells generated additional benefit over reprogramming mouse embryonic fibroblasts (MEF) into iDC1 (28), we compared the effect of injecting them intratumorally in B16 tumors. This comparison revealed smaller tumors in animals injected with B16-derived tumor-APCs on both day 15 and day 18 (Fig. 8H). Indeed, at day 14, tumor-APC injected animals displayed increased pmel and TRP2-reactive CD8+ T cells in peripheral blood (fig. S17C-D). To exclude that endogenous cDC1 would be indirectly responsible for the observed anti-tumor effects when injecting tumor-APCs, we used the cyclooxygenase (COX)-deficient BRAFV600E melanoma cell line that grows in BATF3-/- mice due to the lack of endogenous cDC1 (45). We observed reduced tumor growth, increased survival, and complete regression of tumor growth in 2 treated animals, showing that reprogramming was sufficient to elicit anti-tumor immunity in the absence of the cDC1 compartment (46) (Fig. 8I-J).
We next tested combination with the ICI (anti-PD-1 and anti-CTLA-4) using the low immunogenic B16-OVA model. The combination of tumor-APCs with ICI led to further reduction in tumor growth (Fig. 8K) compared with either treatment alone, and additionally extended the survival of tumor-bearing mice (Fig. 8L). We observed complete tumor regression in two animals, which showed skin and hair depigmentation at the tumor regression site (Fig. 8M) (47). These findings demonstrate synergism of tumor-APC mediated anti-tumor immunity with ICIs. Together, our results support that tumor cells reprogrammed in vitro acquired cDC1 function driving antitumor immunity against melanoma antigens in vivo.
Discussion
Here, we demonstrate that PIB-mediated direct reprogramming converts cancer cells into immunogenic cDC1-like cells endowed with professional antigen presentation and decreased tumorigenicity. We showed that enforced expression of PIB in cancer cells drives global transcriptional and epigenetic remodeling, establishment of cDC1 morphology, immunophenotype, and function, leading to tumor antigen presentation.
Conservation across cell types and species is a requirement to demonstrate the robustness of a reprogramming process, as shown for the induction of pluripotency (8, 48) and neural cell fate (32, 49). We show that cDC1 reprogramming is conserved across mouse and human cancer cell lines from hematologic cancers and solid tumors derived from the three germ layers. In addition, we show that patient-derived cancer cells, PDX-derived cancer cells and CAFs are amenable to cDC1 reprogramming, showing that tumor-APC generation is not limited by cell type of origin and can be induced across an unprecedented broad spectrum of cancer cell lines and primary cancer cells.
Direct cellular reprogramming is a gradual and asynchronous process. To recapitulate target cell identity with complete fidelity remains a challenge, mainly due to partial retention of parental identity (50). Partial reprogramming approaches aiming to rejuvenate aging hallmarks while retaining cell identity have been reported (51). Our data demonstrates that tumor-APC reprogramming is a stepwise process transiting through intermediate populations showing major global transcriptional and epigenetic remodeling and cDC1 functional features, which could be sufficient to elicit antitumor immunity. Although further refinement of the cDC1 reprogramming strategy may strengthen tumor-APC identity, partial reprogramming helps retaining the expression of tumor neoantigens and supports priming clonal expansion of T cells against the heterogeneous population of cancer cells. This highlights the advantage of utilizing partial cell fate reprogramming for cancer immunotherapy.
cDC1 reprogramming enables conversion of cancer cells into APCs restoring antigen-specific immune responses, unlike previous strategies utilizing direct reprogramming of cancer cells to halt tumorigenesis (16). Reprogramming of leukemic cells to macrophages was shown to inhibit tumorigenic potential and induce engulfment capacity (19, 20). Nevertheless, macrophages lack the molecular tools required for effective antigen cross-presentation and CD8+ T cell priming, and, within solid tumors, are largely pro-tumorigenic (52). cDC1 excel in CD8+ T cell stimulation and mediate tumor rejection at the tumor site (25, 53). Accordingly, we showed that tumor-APCs efficiently present endogenous antigens, cross-present exogenous antigens to naïve CD8+ T cells and induce antitumor immunity in vivo. cDC1 are rare and often impaired within tumors (54). The lack of methods allowing efficient generation of functional cDC1 cells has been a major hurdle in the development of DC-based vaccines (55). Because of easier isolation from peripheral blood, moDCs have been frequently used to produce DC vaccines. However, impaired priming capacity has limited clinical efficacy (56). Thus, reprogramming cancer cells to cDC1 offers a strategy to replenish the immune compartment that expresses endogenous tumor antigens.
Increased tumor cell immunogenicity and APC gene signatures are associated with better patient prognosis and response to ICIs (57). Strategies to increase tumor immunogenicity include targeting the IFN pathway and expressing cytokines or costimulatory molecules (58–60). Direct and indirect modulation of IFN-γ pathway through gene knockout or activation of the STING pathway has confirmed their importance for the recognition and killing of tumor cells by CTL (3, 4, 10, 12, 61), which has been attributed to increased antigen presentation through MHC-I. Moreover, recent efforts to promote MHC-I in IFN-deficient B16 was shown to restore sensitivity to ICI and ACT (30). Delivery of granulocyte-macrophage colony-stimulating factor (GM-CSF) and CD40L, as cancer vaccines or oncolytic viruses, stimulates existing immune populations within the tumor microenvironment (58, 59). However, application of these strategies has encountered challenges including systemic toxicity, protumorigenic effects, and failure to overcome resistance factors (1, 58, 62). Here, we show that reprogramming triggers a cascade of pathways (MHC-I and II, costimulatory molecules and immunoproteasome) that converge in restored immunogenicity, activating IFN gene signatures in mouse and human cancer cells and, in contrast to IFN-γ treatment, endows cancer cells with functional antigen presentation capacity. In addition, tumor-APCs express chemokine receptors (e.g. XCR1) and secrete chemokines (e.g. CXCL10) involved in chemo-attraction of lymphocytes (63). As a result, replenishing cDC1 within the tumor microenvironment recruited immune effectors to the tumor that work synergistically with ICI to slow down tumor growth and increased survival. In the future, it will be interesting to assess reprogramming and function elicited by IFN, STING, or chemokine-deficient cancer cells to further dissect the mechanisms of anti-tumor immunity mediated by tumor-APCs.
Tumor immunity was also shown to be elicited with autologous patient-derived iPSCs, an effect attributed to the expression of oncofetal antigens (64), with irradiated tumor cells engineered to express GM-CSF (GVAX) (55) or irradiated tumor cell-moDC fusions (65). For clinical translation and to prevent outgrowth of residual cancer cells, tumor-APCs may also be irradiated prior to reinfusion into patients. This may be particularly suitable for hematopoietic tumors, where hematopoietic blasts from AML or multiple myeloma patients would be isolated, reprogrammed and re-infused in the patient after irradiation. In the future, we envision that the cancer reprogramming technology described here can be further utilized in vivo to induce cDC1 fate and function in tumor cells in situ. The generation of tumor-APCs in vivo, by delivering PIB into tumors, would support ICI immunotherapy in an adjuvant or neoadjuvant manner (66), bringing cDC1 reprogramming one step closer to clinical translation. Exploring non-integrative viral vectors, nonviral delivery methods, cell-specific promoters for in vivo reprogramming will help advance cDC1 reprogramming into a safe and scalable cancer immunotherapy. In addition, the data presented in this manuscript independently supports the development of alternative methods for in vitro tumor-antigen specific TIL expansion and neoantigen discovery.
Overall, we have shown that cDC1 reprogramming enables the induction of antigen presentation directly in tumor cells, overcoming major immune evasion mechanisms, such as cancer heterogeneity, loss of antigen presentation, and lack of cDC1. Our study lays the foundation for the development of immunotherapies that would allow reprogramming of cancer cells to antigen-presenting cells in situ.
Materials and Methods
Study Design
Here, we aimed to evaluate whether direct reprogramming of cancer cells into cDC1 restores tumor immunogenicity and antitumor immune responses. To this end, we characterized mouse and human cancer cell lines and primary cancer cells transduced with PU.1, IRF8 and BATF3 at multiple timepoints at the phenotypic, transcriptional, epigenetic, and functional levels by flow cytometry, RNA-seq, scRNA-seq, and ATAC-seq. To evaluate the immunogenicity of tumor-APCs, we assessed endogenous antigen presentation by immunopeptidomics and evaluated their ability to stimulate CD8+ T cell killing. To evaluate cDC1 function, we tested the ability of tumor-APCs to secrete pro-inflammatory cytokines, uptake and process exogenous antigens, phagocyte dead cells, and prime antigen-specific naïve and memory T-cells. To evaluate loss of tumorigenic potential, we tested the ability for anchorage-independent growth and generation of tumors in vivo in immunodeficient mice. Finally, we addressed capacity of tumor-APCs to elicit anti-tumor immunity in syngeneic mouse models by injecting tumour-APCs intratumorally as monotherapy, or in combination with ICI, and assessed tumor growth, animal survival, and changes to tumor-infiltrating immune cells. For each experiment, the number of biological replicates and statistical significance are indicated in figure legends. All samples passing the quality controls were included in analyses. For in vivo experiments, all animals were randomized to treatment groups before and after tumor inoculation. Investigators were not blinded during experimental procedures nor during the assessment of outcomes. Numbers of mice for in vivo experiments were determined based on previous expertise, and power analysis was not performed.
Mice
Animal care and experimental procedures were performed in accordance with Swedish guidelines and regulations after approval from the local ethical committee. C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-I, The Jackson Laboratory), B6.Cg-Thy1a/Cy Tg(TcraTcrb)8Rest/J (pmel-1, The Jackson Laboratory), and B6.129S(C)-BATF3tm1Kmm (BATF3 KO, The Jackson Laboratory) mice were bred in-house and C57BL/6 females aged 6-10 weeks were purchased from Charles River. NOD-Prkdcscid-IL2rgTm1/Rj (NXG) females aged 6-7 weeks were purchased from Janvier. All mice were kept under a fixed 12-hour light/dark cycle with free access to food and water.
Cell culture
LLC-OVA were generated by stable expression of truncated cytoplasmic OVA (cOVA) with pHAGE-cOVA-IRES-Puro. B16-mOrange and B16-OVA-mOrange were generated by transduction with SFFV-mOrange, and mOrange+ cells were purified by FACS. Both cell lines were expanded from single cell sorted clones and used in experiments from passage 3-10 post-transduction. Cancer cell lines were maintained in Dulbecco´s Eagle Modified (DMEM), RPMI-1640, Ham´s Nutrient F10 Mixture (F10) and Ham´s Nutrient F12 Mixture (F12) growth medium supplemented with 10% (v/v) FBS, Glutamax and Penicillin/Streptomycin solution (100 U/ml penicillin, 100 mg/ml streptomycin. B16-OVA complete growth medium culture was supplemented with 0.4 mg mL-1 Geneticin (Gibco) whereas LLC-OVA with 1x10-3 mg mL-1 puromycin. Primary CD8+ OT-I T cells, CD8+ pmel-specific T cells and CD103+ BM-DCs were cultured in RPMI supplemented 10% FBS, 1% penicillin-streptomycin, 1% GlutaMAX, 1% sodium pyruvate and 50 mmol dm-3 2-mercaptoethanol (Gibco). Primary samples derived from lung tissue and head and neck carcinoma were cultured in Pneumacult Ex-Medium (Stem Cell Technologies) supplemented with hydrocortisone, amphotericin B, and penicillin/streptomycin. Human embryonic fibroblasts (HEF) and cancer-associated fibroblasts (CAF) were grown on plates coated with 0.1% gelatine and maintained in DMEM growth medium supplemented with 10% FBS, Glutamax and 1% penicillin/streptomycin. Early passage (3-6) HEF were used. Cell dissociation from tissue culture plates was done with TrypLE Express for 5-8 min at 37°C. All cells were cultured in a humid environment at 37°C and 5% CO2. All reagents used for cell culture were purchased from Thermo Fisher Scientific, Stem Cell Technologies and Nordic Biolabs. When mentioned, 1x10-9 mg mL-1 IFN-γ (Peprotech), 10x10-3 mg mL-1 Poly(I:C) (InvivoGen) and 1x10-4 mg mL-1 LPS (Enzo) were added 24 h prior to analysis. Detailed information of cells used in the study and culture conditions is provided in Data File S8.
Primary human tissue samples
Primary tumor tissue from head and neck, pancreatic, and bladder carcinoma was obtained according to the Helsinki Declaration and the European Network of Research Ethics Committees. Primary tissue-dissociated cancer cells derived from melanoma, breast and lung carcinoma, and cancer-associated fibroblast cultures were either purchased from Amsbio and BioIVT or provided by CCIT-DK. Tumor samples were cut into pieces and fat and muscle tissue removed. Fragments were digested following gentleMACS Octo Dissociator protocol (Miltenyi Biotec) according to the manufacturer’s recommendations using the 37°C_h_TDK_3 program. Dissociated cells were passed through a 70 μm cell strainer and seeded on 0.1% gelatine-treated plates and cultured. Alternatively, tissue was cut into small fragments and cells were grown under thin glass coverslips to promote cell attachment. Culture conditions are detailed in Data File S8.
Lentiviral production and cancer cell reprogramming
To induce reprogramming, we used the lentiviral vector pRRL.PPT.sf.PIB.i2eGFP expressing human or mouse PU.1, IRF8, and BATF3 (29) transcription factors in polycistronic constructions. Empty pRRL.PPT.sf.MCS.i2eGFP served as a mock control throughout reprogramming experiments. For defined experiments, lentiviral vectors lacking IRES2-eGFP were used. Lentiviral particles were produced in the Human Embryonic Kidney (HEK) 293T packaging cell line. A day before transfection, 7 million cells were seeded in a 15 cm dish to achieve ~80% confluence after 1 day. The next day, cells were transfected as follows: 10 μg lentiviral vector, 7.5 μg psPAX2.G lentiviral packaging vector and 2.5 μg pMD2 envelope vector was combined with 60 μl of 1 mg/ml Polyethylenimine (PEI, linear 25kDa, Polysciences) in 2 ml of OptiMEM (Thermo Fisher Scientific) and incubated for 15 minutes at room temperature. PEI-DMEM mixture was added dropwise to HEK293T cells and 12 hours later replaced with fresh medium. Viral supernatants were collected 48, 60, and 72 hours after transfection, filtered through a 0.45 μM cellulose-acetate filter and concentrated with Amicon centrifugal filters (Millipore) or with Lenti-X Concentrator (Takara). Lentiviral titers were estimated with Lenti-X qRT-PCR titration kit. Reprogramming was induced via transduction of cells with lentiviral particles in the presence of polybrene (8 μg/ml). For leukemic cell lines, polybrene was combined with spinfection (800 g, 60 minutes, room temperature). To assess reprogramming efficiency, cells were stained with anti-CD45 (hematopoietic marker) and anti-MHC-II/HLA-DR (antigen presentation marker) antibodies and analyzed by flow cytometry 9 days after transduction. Reprogramming efficiency was defined as the percentage of CD45+MHC-II/HLA-DR+ cells gated on live, transduced eGFP+ cells. LLC-OVA and B16-OVA were kept in antibiotic selection throughout reprogramming.
RNA-seq library preparation
For bulk RNA-seq, total RNA from 3,000–5,000 FACS-sorted mouse and human cancer cell lines or HEF was extracted using TRIzol Reagent (ThermoFischer Scientific) following the manufacturer’s protocol. cDNA was transcribed with SMARTSeq v4 Ultra low input RNA kit (TakaraBio), amplified with 8 PCR cycles, and purified with AMPure XP beads (Beckman). cDNA was quantified using a High sensitivity DNA kit on a Bioanalyzer 2100 (Agilent). Library preparation was performed using the Nextera XT DNA library preparation kit (Illumina) according to the manufacturer’s protocol. Briefly, cDNA was enzymatically tagmented, followed by indexing with 12 cycles of PCR. Libraries were normalized with beads, pooled, and sequenced on an Illumina NextSeq 500 platform (75bp paired-end reads).
ATAC-seq library preparation
To investigate epigenetic changes induced by reprogramming, 5,000–10,000 cells from defined populations were separated via FACS and processed to prepare bulk ATAC-seq libraries (68).
The quality was assessed using a High Sensitivity DNA Chip (Agilent Technologies), and the library was sequenced using a NextSeq 500/550 High Output Kit on a NextSeq 500 (Illumina).
Single-cell RNA sequencing
scRNA-seq was performed in T98G cells or patient-derived tumor cells transduced with PIB-eGFP or with eGFP lentiviral particles (day 0). Nine days after reprogramming, 5,000–10,000 transduced eGFP+ cells expressing at least one of the reprogramming markers, CD45 and HLA-DR, were FACS-purified and resuspended in PBS containing 0.04% bovine serum albumin (BSA). Day 0 controls were processed similarly. Cells were loaded on a 10x Chromium (10x Genomics) without multiplexing. Single-cell RNA libraries were prepared using a Chromium Single Cell 3+ v2 Reagent Kit (10x Genomics). Indexed sequencing libraries were quantified with a High Sensitivity DNA analysis kit (Agilent) and Agilent Bioanalyzer. Indexed libraries were pooled in an equimolar ratio and sequenced with an Illumina NextSeq 500 platform. Coverage was between 40,000 and 200,000 reads per single cell.
T cell priming and antigen cross-presentation assays
CD8+ T cells from spleen of OT-I mice were enriched using a naïve mouse CD8+ T cell isolation kit (Miltenyi Biotec). Enriched CD8+ T cells were labelled with CTV according to the manufacturer’s protocol. MACS-sorted reprogrammed cells, non-reprogrammed cancer cells, eGFP transduced cancer cells and CD103+ BM-DCs were incubated at 37 °C with OVA peptide (SIINFEKL, T cell priming assays) or protein (cross-presentation assays). OVA expressing cells were not incubated with exogenous OVA. Cells were incubated overnight in the presence of Poly(I:C) or IFN-γ where indicated. 5x103 antigen presenting cells were incubated with 1x105
CTV-labelled OT-I CD8+ T cells in 96-well round-bottom untreated-tissue culture plates. After 3-days of co-culture, T cells were collected, stained for viability (fixable viability dye eFluor-520, eBioscience), CD8α, TCR-β, and CD44 and analyzed by flow cytometry. T cell proliferation (dilution of CTV) and activation (CD44 expression) were determined by gating on live, single, TCR-β+ and CD8+ T cells. The threshold for data plotting was fixed at 1,000 events within live cell gating.
In vitro stimulation of MART-1 and CMV-specific CD8+ T cells
Day 9 reprogrammed melanoma cells (HLA-A2-positive) were harvested and plated in a 24-well plate in the presence or absence of LPS + Poly(I:C). Next day, cells were washed with PBS, resuspended in DMEM (10%FBS, 1%P/S; 0.5 x 106 cells/ml), and pulsed with MART-1 (1 μM) or CMV (1μM) peptides (Schafer-N) for 1 h at 37°C. Alternatively, day 9 reprogrammed cells were pulsed with long-MART-1 peptide (GHGHSYTTAEELAGIGILTVILGVL,10 μM) for 3 h in the presence of LPS and P(I:C). Afterwards, cells were washed and co-cultured with 0.5 x 106 CD8+ T cells (from MART-1+/CMV+ HLA-A2-positive donor) at a 1:10 ratio for 8 days in DMEM supplemented with 120 U/ml IL-2 and 10 μg/ml IL-7. On day 2 and 5 of co-culture, 50% of the medium was replaced with fresh DMEM. On day 8, cells were harvested and CMV and MART-1 responses were assessed by tetramer staining and intracellular cytokine staining. eGFP transduced cells served as controls. Monocyte-derived DCs (mo-DCs) served as reference.
Tumor establishment and treatment
To establish tumors, 2x105 B16OVA or 1x105 Cox-deficient BRAFV600E cells were injected subcutaneously into the right flank of recipient mice. For B16-OVA tumor challenges 6–10-week-old age-matched C57BL/6 females were used. Cox-deficient BRAFV600E melanoma tumors were established in 6-12-week-old BATF3 KO females and males. Tumor volumes were monitored with a digital caliper and calculated using the formula V=L x W x H x 1/2. Survival was determined by pre-defined endpoints (tumor size >1500 mm3, tumor ulceration, signs of animal suffering). For all treatments with tumor-APCs, cancer cells were transduced with SFFV-PIB and purified by MACS with anti-CD45 and anti-MHC-II antibodies at day 5. For iDC1 treatments, mouse embryonic fibroblasts (MEFs) from the C57BL/6 background were transduced with SFFV-PIB and MACS-purified with anti-CD45 and anti-MHC-II antibodies at day 5. PBS, cancer cells or MEFs transduced with empty vector (SFFV-MCS) served as controls. Intratumoral injections of tumor-APCs or iDC1 were performed with 1-2x105 cells at day 7, 10 and 13 post-tumor establishment. PBS or cancer cells transduced with empty lentiviruses (SFFV-MCS) were injected into tumors as controls. 24 h before injection, tumor-APCs, iDC1, or control cells were stimulated with Poly(I:C). For B16-OVA tumor experiments, B16-derived tumor-APCs were additionally pulsed with OVA protein and washed extensively. For combinatorial treatment with ICIs, mice received 200 μg of anti-PD1 (Bio X Cell, clone 29F.1A12) and 200 μg of anti-CTLA-4 (Bio X Cell, clone 9H10) or rat IgG2a (Bio X Cell, clone 2A3) and IgG2b (Bio X Cell, clone LTF-2) isotype control antibodies intraperitoneally at day 7, 10 and 13. Animals were randomized after tumor inoculation prior to further treatments.
Statistical Analysis
Data were subjected to a normality test before using One-way ANOVA, Two-Way ANOVA, Kruskall-Walis or Mann-Whitney test. Group comparisons were corrected using Sidak´s, Dunn’s, or Tukey´s multiple comparison test with GraphPad Prism version 9 (GraphPad).
Alternatively, a Wilcoxon rank-sum test was performed using the ´wilcox.test’ function in R software. The Spearman correlation coefficient was calculated using ‘cor’ function in R software. Survival curves were analyzed by the long-rank Mantel-Cox test. p-values are shown when relevant (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns – not significant).
Supplementary Material
One Sentence Summary.
A minimal cDC1 regulatory network reprograms cancer cells into tumor-APCs with low tumorigenicity that elicit anti-tumor immunity.
Acknowledgments
We thank the members of the Cell Reprogramming in Hematopoiesis and Immunity Laboratory for discussions and S. Pedreiro for technical assistance. We thank the Center for Translational Genomics, SciLifeLab Lund for providing sequencing services and Lund University Bioimaging Center for scanning electron microscopy assistance. We would also like to thank the Lund Stem Cell Center FACS facility for cell sorting assistance and the Centre for Comparative Medicine for animal facilities. We thank Francesca Granucci (University of Milano Bicocca, Italy) and Richard Marais (Cancer Research Manchester Institute, UK) for sharing mouse cell lines and Johan Bengzon (Lund University), Francesca Aguilo (Umeå University), and Göran B. Jönsson (Lund University) for providing human cancer cell lines. We thank Z. Kokaia, H. Ahlenius, and E. Quist (Lund University) for providing HEFs (ethical permit: 6.1.8-2887/2017). We also thank healthy donors and the center for Clinical Immunology and Transfusion Medicine at Skåne University Hospital for providing leukocyte concentrates (ethical permit: 2022:11).
Funding
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (Grant agreement No. 866448). This project was also funded by Cancerfonden (20 0939 PjF), the Swedish research council (2020-00615), NovoNordisk Fonden (0056527), Swelife and Medtech4Health (2020-04744), Eurostars-2 Joint Program (2021-03371), FCT (PTDC/MED-IMU/4520/2020) and Plano de Recuperação e Resiliência de Portugal pelo fundo NextGenerationEU (C644865576-00000005). The Knut and Alice Wallenberg foundation, the Medical Faculty at Lund University and Region Skåne are acknowledged for financial support. O.Z. is supported by a Cancerfonden postdoctoral fellowship (190008). A.G.F, I.C. and R.S.-A. are supported by FCT scholarships (SFRH/BD/133233/2017, PD/BD/146424/2019 and PD/BD/135725/2018). Expenses for salary and running costs for M.V.S. and I.M.S in connection with analyses performed at CCIT-DK were covered by Asgard.
Footnotes
Author contributions: O.Z., A.G.F., D.S.C., and E.A. conducted reprogramming experiments in vitro and analyzed the data. O.Z., A.G.F., E.A., C.F.P., and F.F.R. FACS purified tumor-APCs. I.K., O.Z., and A.G.F. performed RNA-seq and ATAC-seq analysis. O.Z., A.G.F, E.A., I.C. and R.S.A. performed immunofluorescence, live imaging and western blotting. K.M. and Y.N. provided human cancer cell lines. I.E.S., J.M., and M.B.S. performed and analyzed immunopeptidomic experiments. M.V.S., M.Ha., O.Z., F.F.R., C.F.P., Ö.M. and I.M.S designed and performed functional experiments with human primary melanoma cells. E.A., D.S.C., O.Z., M. Hu., S.H. and H.B. designed and performed in vivo experiments. M.B., R.A., C.B., M.Hö., D.G.J., L.G., M.L., Ö.M., and I.M.S. provided primary human cancer samples and discussed clinical applicability. C-F.P., F.F.R. and C.F.P. attracted funding. O.Z., A.G.F. and C-F.P. conceptualized the study and wrote the manuscript.
Competing interests: F.F.R., C.F.P. and C.-F.P. have equity interest and serve in management positions at Asgard Therapeutics AB, which develops cancer immunotherapies based on DC reprogramming technologies. F.F.R., C.F.P. and C.-F.P. are inventors on US patent 11,345,891, patent application WO 2018/185709 and patent application WO 2022/243448 (together with O.Z, A.G.F. and E.A.) held by Asgard Therapeutics that covers the cell reprogramming approach described here.
Data and materials availability
The sequencing data generated in this study is available from Gene Expression Omnibus (GEO) under accession codes GSE184527 (mouse RNA-seq), GSE200146 (human RNA-seq), GSE224941 (human scRNA-seq) and GSE200341 (human ATAC-seq). Published datasets re-analyzed in this study are available under accession codes GSE103618, GSE162650 and GSE94820. We have created a web application (cellreprolab.shinyapps.io/tumorAPC_atlas/) where visitors can explore processed RNA-seq and ATAC-seq data. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD039789. Constructs and vectors used for reprogramming are available from Asgard Therapeutics under a material transfer agreement with the company. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References
- 1.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jhunjhunwala S, Hammer C, Delamarre L. Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat Rev Cancer. 2021;21:298–312. doi: 10.1038/s41568-021-00339-z. [DOI] [PubMed] [Google Scholar]
- 3.Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, Collins NB, Bi K, La Fleur MW, Juneja VR, Weiss SA, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547:413–418. doi: 10.1038/nature23270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, Gartner JJ, Jia L, Steinberg SM, Yamamoto TN, Merchant AS, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537–542. doi: 10.1038/nature23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hashimoto M, Kamphorst AO, Im SJ, Kissick HT, Pillai RN, Ramalingam SS, Araki K, Ahmed R. CD8 T Cell Exhaustion in Chronic Infection and Cancer: Opportunities for Interventions. Annu Rev Med. 2018;69:301–318. doi: 10.1146/annurev-med-012017-043208. [DOI] [PubMed] [Google Scholar]
- 6.Guo W, Wang Y, Yang M, Wang Z, Wang Y, Chaurasia S, Wu Z, Zhang M, Yadav GS, Rathod S, Concha-Benavente F, et al. LincRNA-immunity landscape analysis identifies EPIC1 as a regulator of tumor immune evasion and immunotherapy resistance. Sci Adv. 2021;7 doi: 10.1126/sciadv.abb3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto K, Venida A, Yano J, Biancur DE, Kakiuchi M, Gupta S, Sohn ASW, Mukhopadhyay S, Lin EY, Parker SJ, Banh RS, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature. 2020;581:100–105. doi: 10.1038/s41586-020-2229-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 9.Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol. 2020;20:25–39. doi: 10.1038/s41577-019-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawson KA, Sousa CM, Zhang X, Kim E, Akthar R, Caumanns JJ, Yao Y, Mikolajewicz N, Ross C, Brown KR, Zid AA, et al. Functional genomic landscape of cancer-intrinsic evasion of killing by T cells. Nature. 2020;586:120–126. doi: 10.1038/s41586-020-2746-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen AM, Ge Y, Schuster MB, Pundhir S, Jakobsen JS, Kalvisa A, Tapia MC, Gordon S, Ambri F, Bagger FO, Pandey D, et al. H3K9 dimethylation safeguards cancer cells against activation of the interferon pathway. Sci Adv. 2022;8:eabf8627. doi: 10.1126/sciadv.abf8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin GK, Wu J, Iracheta-Vellve A, Patti JC, Hsu J, Davis T, Dele-Oni D, Du PP, Halawi AG, Ishizuka JJ, Kim SY, et al. Epigenetic silencing by SETDB1 suppresses tumour intrinsic immunogenicity. Nature. 2021;595:309–314. doi: 10.1038/s41586-021-03520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishay-Ronen D, Diepenbruck M, Kalathur RKR, Sugiyama N, Tiede S, Ivanek R, Bantug G, Morini MF, Wang J, Hess C, Christofori G. Gain Fat-Lose Metastasis: Converting Invasive Breast Cancer Cells into Adipocytes Inhibits Cancer Metastasis. Cancer Cell. 2019;35:17–32.:e16. doi: 10.1016/j.ccell.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Hochedlinger K, Blelloch R, Brennan C, Yamada Y, Kim M, Chin L, Jaenisch R. Reprogramming of a melanoma genome by nuclear transplantation. Genes Dev. 2004;18:1875–1885. doi: 10.1101/gad.1213504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmermannova O, Caiado I, Ferreira AG, Pereira CF. Cell Fate Reprogramming in the Era of Cancer Immunotherapy. Front Immunol. 2021;12:714–822. doi: 10.3389/fimmu.2021.714822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris H, Miller OJ, Klein G, Worst P, Tachibana T. Suppression of malignancy by cell fusion. Nature. 1969;223:363–368. doi: 10.1038/223363a0. [DOI] [PubMed] [Google Scholar]
- 18.Park NI, Guilhamon P, Desai K, McAdam RF, Langille E, O’Connor M, Lan X, Whetstone H, Coutinho FJ, Vanner RJ, Ling E, et al. ASCL1 Reorganizes Chromatin to Direct Neuronal Fate and Suppress Tumorigenicity of Glioblastoma Stem Cells. Cell Stem Cell. 2017;21:411. doi: 10.1016/j.stem.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Rapino F, Robles EF, Richter-Larrea JA, Kallin EM, Martinez-Climent JA, Graf T. C/EBPalpha induces highly efficient macrophage transdifferentiation of B lymphoma and leukemia cell lines and impairs their tumorigenicity. Cell Rep. 2013;3:1153–1163. doi: 10.1016/j.celrep.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 20.McClellan JS, Dove C, Gentles AJ, Ryan CE, Majeti R. Reprogramming of primary human Philadelphia chromosome-positive B cell acute lymphoblastic leukemia cells into nonleukemic macrophages. Proc Natl Acad Sci U S A. 2015;112:4074–4079. doi: 10.1073/pnas.1413383112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Yang Y, Liu J, Qian L. Direct cell reprogramming: approaches, mechanisms and progress. Nat Rev Mol Cell Biol. 2021;22:410–424. doi: 10.1038/s41580-021-00335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poulin LF, Reyal Y, Uronen-Hansson H, Schraml BU, Sancho D, Murphy KM, Hakansson UK, Moita LF, Agace WW, Bonnet D, Reis e Sousa C. DNGR-1 is a specific and universal marker of mouse and human Batf3-dependent dendritic cells in lymphoid and nonlymphoid tissues. Blood. 2012;119:6052–6062. doi: 10.1182/blood-2012-01-406967. [DOI] [PubMed] [Google Scholar]
- 23.Barry KC, Hsu J, Broz ML, Cueto FJ, Binnewies M, Combes AJ, Nelson AE, Loo K, Kumar R, Rosenblum MD, Alvarado MD, et al. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat Med. 2018;24:1178–1191. doi: 10.1038/s41591-018-0085-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kvedaraite E, Ginhoux F. Human dendritic cells in cancer. Sci Immunol. 2022;7:eabm9409. doi: 10.1126/sciimmunol.abm9409. [DOI] [PubMed] [Google Scholar]
- 25.Spranger S, Dai D, Horton B, Gajewski TF. Tumor-Residing Batf3 Dendritic Cells Are Required for Effector T Cell Trafficking and Adoptive T Cell Therapy. Cancer Cell. 2017;31:711–723.:e714. doi: 10.1016/j.ccell.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubert M, Gobbini E, Couillault C, Manh TV, Doffin AC, Berthet J, Rodriguez C, Ollion V, Kielbassa J, Sajous C, Treilleux I, et al. IFN-III is selectively produced by cDC1 and predicts good clinical outcome in breast cancer. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.aav3942. [DOI] [PubMed] [Google Scholar]
- 27.Mayoux M, Roller A, Pulko V, Sammicheli S, Chen S, Sum E, Jost C, Fransen MF, Buser RB, Kowanetz M, Rommel K, et al. Dendritic cells dictate responses to PD-L1 blockade cancer immunotherapy. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.aav7431. [DOI] [PubMed] [Google Scholar]
- 28.Rosa FF, Pires CF, Kurochkin I, Ferreira AG, Gomes AM, Palma LG, Shaiv K, Solanas L, Azenha C, Papatsenko D, Schulz O, et al. Direct reprogramming of fibroblasts into antigen-presenting dendritic cells. Sci Immunol. 2018;3 doi: 10.1126/sciimmunol.aau4292. [DOI] [PubMed] [Google Scholar]
- 29.Rosa FF, Pires CF, Kurochkin I, Halitzki E, Zahan T, Arh N, Zimmermannova O, Ferreira AG, Li H, Karlsson S, Scheding S, et al. Single-cell transcriptional profiling informs efficient reprogramming of human somatic cells to cross-presenting dendritic cells. Sci Immunol. 2022;7:eabg5539. doi: 10.1126/sciimmunol.abg5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalbasi A, Tariveranmoshabad M, Hakimi K, Kremer S, Campbell KM, Funes JM, Vega-Crespo A, Parisi G, Champekar A, Nguyen C, Torrejon D, et al. Uncoupling interferon signaling and antigen presentation to overcome immunotherapy resistance due to JAK1 loss in melanoma. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.abb0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sancho D, Joffre OP, Keller AM, Rogers NC, Martinez D, Hernanz-Falcon P, Rosewell I, Reis e Sousa C. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature. 2009;458:899–903. doi: 10.1038/nature07750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marro S, Pang ZP, Yang N, Tsai MC, Qu K, Chang HY, Sudhof TC, Wernig M. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell. 2011;9:374–382. doi: 10.1016/j.stem.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chopin M, Lun AT, Zhan Y, Schreuder J, Coughlan H, D’Amico A, Mielke LA, Almeida FF, Kueh AJ, Dickins RA, Belz GT, et al. Transcription Factor PU.1 Promotes Conventional Dendritic Cell Identity and Function via Induction of Transcriptional Regulator DC-SCRIPT. Immunity. 2019;50:77–90.:e75. doi: 10.1016/j.immuni.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 34.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, Jardine L, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356 doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Apostolou E, Stadtfeld M. Cellular trajectories and molecular mechanisms of iPSC reprogramming. Curr Opin Genet Dev. 2018;52:77–85. doi: 10.1016/j.gde.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stik G, Vidal E, Barrero M, Cuartero S, Vila-Casadesus M, Mendieta-Esteban J, Tian TV, Choi J, Berenguer C, Abad A, Borsari B, et al. CTCF is dispensable for immune cell transdifferentiation but facilitates an acute inflammatory response. Nat Genet. 2020;52:655–661. doi: 10.1038/s41588-020-0643-0. [DOI] [PubMed] [Google Scholar]
- 37.ElTanbouly MA, Noelle RJ. Rethinking peripheral T cell tolerance: checkpoints across a T cell’s journey. Nat Rev Immunol. 2021;21:257–267. doi: 10.1038/s41577-020-00454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Upadhyay R, Boiarsky JA, Pantsulaia G, Svensson-Arvelund J, Lin MJ, Wroblewska A, Bhalla S, Scholler N, Bot A, Rossi JM, Sadek N, et al. A Critical Role for Fas-Mediated Off-Target Tumor Killing in T-cell Immunotherapy. Cancer Discov. 2021;11:599–613. doi: 10.1158/2159-8290.CD-20-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Overwijk WW, Theoret MR, Finkelstein SE, Surman DR, de Jong LA, Vyth-Dreese FA, Dellemijn TA, Antony PA, Spiess PJ, Palmer DC, Heimann DM, et al. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J Exp Med. 2003;198:569–580. doi: 10.1084/jem.20030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bottcher JP, Reis e Sousa C. The Role of Type 1 Conventional Dendritic Cells in Cancer Immunity. Trends Cancer. 2018;4:784–792. doi: 10.1016/j.trecan.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olivar R, Luque A, Naranjo-Gomez M, Quer J, Garcia de Frutos P, Borras FE, Rodriguez de Cordoba S, Blom AM, Aran JM. The alpha7beta0 isoform of the complement regulator C4b-binding protein induces a semimature, anti-inflammatory state in dendritic cells. J Immunol. 2013;190:2857–2872. doi: 10.4049/jimmunol.1200503. [DOI] [PubMed] [Google Scholar]
- 42.de Verteuil DA, Rouette A, Hardy MP, Lavallee S, Trofimov A, Gaucher E, Perreault C. Immunoproteasomes shape the transcriptome and regulate the function of dendritic cells. J Immunol. 2014;193:1121–1132. doi: 10.4049/jimmunol.1400871. [DOI] [PubMed] [Google Scholar]
- 43.Gao R, Bai S, Henderson YC, Lin Y, Schalck A, Yan Y, Kumar T, Hu M, Sei E, Davis A, Wang F, et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat Biotechnol. 2021;39:599–608. doi: 10.1038/s41587-020-00795-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chauvin JM, Larrieu P, Sarrabayrouse G, Prevost-Blondel A, Lengagne R, Desfrancois J, Labarriere N, Jotereau F. HLA anchor optimization of the melan-A-HLA-A2 epitope within a long peptide is required for efficient cross-priming of human tumor-reactive T cells. J Immunol. 2012;188:2102–2110. doi: 10.4049/jimmunol.1101807. [DOI] [PubMed] [Google Scholar]
- 45.Zelenay S, van der Veen AG, Bottcher JP, Snelgrove KJ, Rogers N, Acton SE, Chakravarty P, Girotti MR, Marais R, Quezada SA, Sahai E, et al. Cyclooxygenase-Dependent Tumor Growth through Evasion of Immunity. Cell. 2015;162:1257–1270. doi: 10.1016/j.cell.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferris ST, Ohara RA, Ou F, Wu R, Huang X, Kim S, Chen J, Liu TT, Schreiber RD, Murphy TL, Murphy KM. cDC1 Vaccines Drive Tumor Rejection by Direct Presentation Independently of Host cDC1. Cancer Immunol Res. 2022;10:920–931. doi: 10.1158/2326-6066.CIR-21-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hotz C, Wagenaar TR, Gieseke F, Bangari DS, Callahan M, Cao H, Diekmann J, Diken M, Grunwitz C, Hebert A, Hsu K, et al. Local delivery of mRNA-encoded cytokines promotes antitumor immunity and tumor eradication across multiple preclinical tumor models. Sci Transl Med. 2021;13:eabc7804. doi: 10.1126/scitranslmed.abc7804. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 49.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 51.Lu Y, Brommer B, Tian X, Krishnan A, Meer M, Wang C, Vera DL, Zeng Q, Yu D, Bonkowski MS, Yang JH, et al. Reprogramming to recover youthful epigenetic information and restore vision. Nature. 2020;588:124–129. doi: 10.1038/s41586-020-2975-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borriello L, Coste A, Traub B, Sharma VP, Karagiannis GS, Lin Y, Wang Y, Ye X, Duran CL, Chen X, Friedman M, et al. Primary tumor associated macrophages activate programs of invasion and dormancy in disseminating tumor cells. Nat Commun. 2022;13:626. doi: 10.1038/s41467-022-28076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salmon H, Idoyaga J, Rahman A, Leboeuf M, Remark R, Jordan S, Casanova-Acebes M, Khudoynazarova M, Agudo J, Tung N, Chakarov S, et al. Expansion and Activation of CD103(+) Dendritic Cell Progenitors at the Tumor Site Enhances Tumor Responses to Therapeutic PD-L1 and BRAF Inhibition. Immunity. 2016;44:924–938. doi: 10.1016/j.immuni.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin JH, Huffman AP, Wattenberg MM, Walter DM, Carpenter EL, Feldser DM, Beatty GL, Furth EE, Vonderheide RH. Type 1 conventional dendritic cells are systemically dysregulated early in pancreatic carcinogenesis. J Exp Med. 2020;217 doi: 10.1084/jem.20190673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saxena M, van der Burg SH, Melief CJM, Bhardwaj N. Therapeutic cancer vaccines. Nature Reviews Cancer. 2021 doi: 10.1038/s41568-021-00346-0. [DOI] [PubMed] [Google Scholar]
- 56.Osugi Y, Vuckovic S, Hart DN. Myeloid blood CD11c(+) dendritic cells and monocyte-derived dendritic cells differ in their ability to stimulate T lymphocytes. Blood. 2002;100:2858–2866. doi: 10.1182/blood.V100.8.2858. [DOI] [PubMed] [Google Scholar]
- 57.Wang G, Chow RD, Bai Z, Zhu L, Errami Y, Dai X, Dong MB, Ye L, Zhang X, Renauer PA, Park JJ, et al. Multiplexed activation of endogenous genes by CRISPRa elicits potent antitumor immunity. Nature Immunology. 2019;20:1494–1505. doi: 10.1038/s41590-019-0500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lutz ER, Wu AA, Bigelow E, Sharma R, Mo G, Soares K, Solt S, Dorman A, Wamwea A, Yager A, Laheru D, et al. Immunotherapy converts nonimmunogenic pancreatic tumors into immunogenic foci of immune regulation. Cancer Immunol Res. 2014;2:616–631. doi: 10.1158/2326-6066.CIR-14-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eriksson E, Milenova I, Wenthe J, Stahle M, Leja-Jarblad J, Ullenhag G, Dimberg A, Moreno R, Alemany R, Loskog A. Shaping the Tumor Stroma and Sparking Immune Activation by CD40 and 4-1BB Signaling Induced by an Armed Oncolytic Virus. Clin Cancer Res. 2017;23:5846–5857. doi: 10.1158/1078-0432.CCR-17-0285. [DOI] [PubMed] [Google Scholar]
- 60.Wang S, He Z, Wang X, Li H, Liu XS. Antigen presentation and tumor immunogenicity in cancer immunotherapy response prediction. Elife. 2019;8 doi: 10.7554/eLife.49020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Falahat R, Perez-Villarroel P, Mailloux AW, Zhu G, Pilon-Thomas S, Barber GN, Mule JJ. STING Signaling in Melanoma Cells Shapes Antigenicity and Can Promote Antitumor T-cell Activity. Cancer Immunol Res. 2019;7:1837–1848. doi: 10.1158/2326-6066.CIR-19-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sabado RL, Balan S, Bhardwaj N. Dendritic cell-based immunotherapy. Cell Res. 2017;27:74–95. doi: 10.1038/cr.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harlin H, Meng Y, Peterson AC, Zha Y, Tretiakova M, Slingluff C, McKee M, Gajewski TF. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69:3077–3085. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kooreman NG, Kim Y, de Almeida PE, Termglinchan V, Diecke S, Shao NY, Wei TT, Yi H, Dey D, Nelakanti R, Brouwer TP, et al. Autologous iPSC-Based Vaccines Elicit Anti-tumor Responses In Vivo. Cell Stem Cell. 2018;22:501–513.:e507. doi: 10.1016/j.stem.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosenblatt J, Avivi I, Vasir B, Uhl L, Munshi NC, Katz T, Dey BR, Somaiya P, Mills H, Campigotto F, Weller E, et al. Vaccination with dendritic cell/tumor fusions following autologous stem cell transplant induces immunologic and clinical responses in multiple myeloma patients. Clin Cancer Res. 2013;19:3640–3648. doi: 10.1158/1078-0432.CCR-13-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Versluis JM, Long GV, Blank CU. Learning from clinical trials of neoadjuvant checkpoint blockade. Nat Med. 2020;26:475–484. doi: 10.1038/s41591-020-0829-0. [DOI] [PubMed] [Google Scholar]
- 67.Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015;109:21 29 21–21 29 29. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mayer CT, Ghorbani P, Nandan A, Dudek M, Arnold-Schrauf C, Hesse C, Berod L, Stuve P, Puttur F, Merad M, Sparwasser T. Selective and efficient generation of functional Batf3-dependent CD103+ dendritic cells from mouse bone marrow. Blood. 2014;124:3081–3091. doi: 10.1182/blood-2013-12-545772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 76.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby Ch, Zager M, Hoffman P, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.:e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, Trapnell C, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alquicira-Hernandez J, Sathe A, Ji HP, Nguyen Q, Powell JE. scPred: accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome Biol. 2019;20:264. doi: 10.1186/s13059-019-1862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reynisson B, Barra C, Kaabinejadian S, Hildebrand WH, Peters B, Nielsen M. Improved Prediction of MHC II Antigen Presentation through Integration and Motif Deconvolution of Mass Spectrometry MHC Eluted Ligand Data. J Proteome Res. 2020;19:2304–2315. doi: 10.1021/acs.jproteome.9b00874. [DOI] [PubMed] [Google Scholar]
- 82.Gfeller D, Schmidt J, Croce G, Guillaume P, Bobisse S, Genolet R, Queiroz L, Cesbron J, Racle J, Harari A. Predictions of immunogenicity reveal potent SARS-CoV-2 CD8+ T-cell epitopes. bioRxiv. 2022:2022.2005.2023.492800. doi: 10.1016/j.cels.2022.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Borowicz S, Van Scoyk M, Avasarala S, Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK, Winn RA. The soft agar colony formation assay. J Vis Exp. 2014:e51998. doi: 10.3791/51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data generated in this study is available from Gene Expression Omnibus (GEO) under accession codes GSE184527 (mouse RNA-seq), GSE200146 (human RNA-seq), GSE224941 (human scRNA-seq) and GSE200341 (human ATAC-seq). Published datasets re-analyzed in this study are available under accession codes GSE103618, GSE162650 and GSE94820. We have created a web application (cellreprolab.shinyapps.io/tumorAPC_atlas/) where visitors can explore processed RNA-seq and ATAC-seq data. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD039789. Constructs and vectors used for reprogramming are available from Asgard Therapeutics under a material transfer agreement with the company. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.