Abstract
Effector T-cells rely on integrins to drive adhesion and migration to facilitate their immune function. Heterodimeric transmembrane integrin LFA-1 (αLβ2) regulates adhesion and migration of effector T cells through linkage of the extracellular matrix with the intracellular actin treadmill machinery. We quantitated the velocity and direction of F-actin flow in migrating T-cells alongside single molecule localisation of transmembrane and intracellular LFA-1. Results showed that actin retrograde flow positively correlated and immobile actin negatively correlated with T-cell velocity. Plasma membrane localised LFA-1 forms unique nano-clustering patterns in the leading edge, compared to the mid-focal zone, of migrating T-cells. Deleting the cytosolic phosphatase PTPN22, loss-of-function mutations of which have been linked to autoimmune disease, increased T-cell velocity, and leading-edge co-clustering of pY397 FAK, pY416 Src family kinases and LFA-1. These data suggest that differential nanoclustering patterns of LFA-1 in migrating T-cells may instruct intracellular signalling. Our data presents a paradigm where T cells modulate the nanoscale organisation of adhesion and signalling molecules to fine tune their migration speed, with implications for the regulation of immune and inflammatory responses.
Keywords: Integrins, LFA-1, T cell migration, SMLM, dSTORM
Introduction
Integrin lymphocyte functional antigen-1 (LFA-1; αLβ2) is a transmembrane heterodimer highly expressed on T-cells. It is essential for T-cell function and is required for naïve lymphocyte antigenic activation, recruitment of immune cells to sites of infection and cytotoxic killing. LFA-1 expressed on effector T cells binds to intracellular adhesion molecule (ICAM)-1 to induce rapid migration in vivo and in vitro (Hons et al. 2018; Teijeira et al. 2017). The force required for migration is generated by actin polymerisation and myosin driven actin flow, and integrin adhesion molecules on the cell surface which couple the actin flow to the underlying substrate (Shannon et al. 2015). Actin and integrins are indirectly linked via a group of proteins collectively known as the molecular clutch, which regulates this mechanical coupling (Ishibashi et al. 2015; Chen et al. 2012; Case & Waterman 2015). The control of this linkage is integral to achieving high-speed migration (Hons et al. 2018), and is especially important during inflammation (Teijeira et al. 2017). It is also known that integrins can exist in low, intermediate and high affinity states that describe a specific pattern of localisation within migrating T cells. Low and intermediate affinity integrin are located in the leading edge, whereas high affinity LFA-1 is located in the mid cell body (focal zone)(Smith et al. 2005).
Control of adhesion is achieved through actin linkers (talin and vinculin) and regulatory kinases (FAK and Src family kinases)(Nordenfelt et al. 2016; Raab et al. 2017a), in common to other migrating cell types, that form large micron sized focal adhesions (Kanchanawong et al. 2010). These components were originally described in slow migrating adherent cells that characteristically form large micron sized focal adhesions (Kanchanawong et al. 2010). In T-cells, these complexes are small (nanoscale) (Burn et al. 2016), similar in size and composition to nascent adhesions (Changede et al. 2015) and their spatial organisation may represent a novel paradigm for nanoscale regulation of adhesion and migration.
In T cells, these complexes are small (nanoscale), containing only a few molecules (Burn et al. 2016) - similar in size and composition to nascent adhesions (Changede et al. 2015). Their spatial organisation therefore represents a novel paradigm for nanoscale regulation.
Conventional fluorescence microscopy is limited in resolution to around 200 nm. Recently, a number of super-resolution approaches have been developed which overcome this limitation. Single molecule localisation microscopy (SMLM), here implemented as stochastic optical reconstruction microscopy (STORM)(Rust et al. 2006), utilises fluorophore dark-states and sequential imaging to generate lists of molecular coordinates with mean 10 nm precision in fixed cells. Using Bayesian statistical cluster analysis(Griffié et al. 2016; Rubin-Delanchy et al. 2015), it is possible to accurately quantify protein clustering on the nanoscale. This includes clustering of intracellular molecular populations via 3D interferometric SMLM imaging featuring isotropic resolution (Shtengel et al. 2009; Griffié et al. 2017). Such populations of intracellular, membrane proximal LFA-1 and FAK may represent distinct nanoscale arrangements recruited to form new adhesions in the membrane.
We investigated the dynamics of retrograde actin flow alongside the nanoscale organisation of actively signalling LFA-1 nanoclusters in primary murine T blasts migrating on ICAM-1 coated coverslips. A range of conditions were used to modulate cell speed including manganese ions (which push integrins into the high affinity state)(Dransfield et al. 1992), high density ligand (to increase adhesion), the addition of chemokine CXCL12 (to induce signalling through CXCR4) and Cytochalasin-D (which inhibits actin polymerisation)(Goddette & Frieden 1986). We then included in our analysis cells lacking the phosphatase PTPN22, a negative regulator of migration (Burn et al. 2016) where a loss-of-function mutation (Svensson et al. 2011) causes a rapid migration phenotype in T cells and predisposes humans and mice to a range of autoimmune diseases (Dai et al. 2013; Burn et al. 2011; Brand et al. 2005; Bottini et al. 2006; Begovich et al. 2004). By characterising the nanoclustering of LFA-1 and its regulators as a previously uninvestigated aspect of the molecular clutch model, we are able to link changes in their membrane and submembrane arrangement to the functional output of cell speed and actin flow/engagement changes during rapid polarised migration.
Results
The speed of actin retrograde flow is positively correlated with T-cell velocity and negatively correlated with immobile actin
Inside-out and outside-in activation of LFA-1 regulates its affinity maturation and translates into functional effects on adhesion and migration behaviour. F-actin (filamentous actin) forms networks of actin that concurrently ‘slip and grip’ (Ponti et al. 2004; Jurado et al. 2005) to facilitate movement by linking highly regulated intracellular flowing actin to the extracellular substrate via transmembrane integrin adhesions (Wiseman et al. 2004; Lawson et al. 2012; Alexandrova et al. 2008; Goult et al. 2013; Changede et al. 2015; Sun et al. 2014). Transient engagement of the actin through adhesions is part of a mechanism that transduces force from flowing actin and is termed the molecular clutch (Chan & Odde 2008; Hu et al. 2007; Brown et al. 2006; Ishibashi et al. 2015; Chen et al. 2012; Case & Waterman 2015; Havrylenko et al. 2014). Flowing actin therefore represents a population that is unengaged with adhesions, whereas immobile actin (in the external reference frame) represents a population in contact with adhesions.
With these features in mind, we first investigated cell speed on a population level (1000s of cells measured) in polarised, migrating murine effector T-cells exposed to a range of stimuli, all in the presence of an ICAM-1 coating on a glass coverslip. Supplementary Figure 1 shows that cells slow down when exposed to MnCl2 or high surface concentrations of ICAM-1, and speed up in the presence of chemokine CXCL12 as well as in cells in which PTPN22 has been genetically deleted. To characterise the dynamics of actin flow in more detail, primary mouse T cells, expressing the Lifeact-GFP transgene were allowed to migrate on glass surfaces coated with ICAM-1 before being imaged by TIRF microscopy (Figure 1a). As a proxy for actin linked to the molecular clutch, Fourier analysis was used to determine the fraction of immobile actin in cells, since stationary parts of the actin cytoskeleton must necessarily be engaged with extracellular substrate while the cell migrates over that position. Subsequently, the centre of the cell was continually reset, shifting the frame of reference from the laboratory frame, to the cell frame (Figure 1b), while Spatio Temporal Image Correlation Spectroscopy (STICS) was used to measure the velocity and directionality of flowing (unengaged) actin relative to the cell, i.e. from the cell’s frame of reference (Figure 1c).
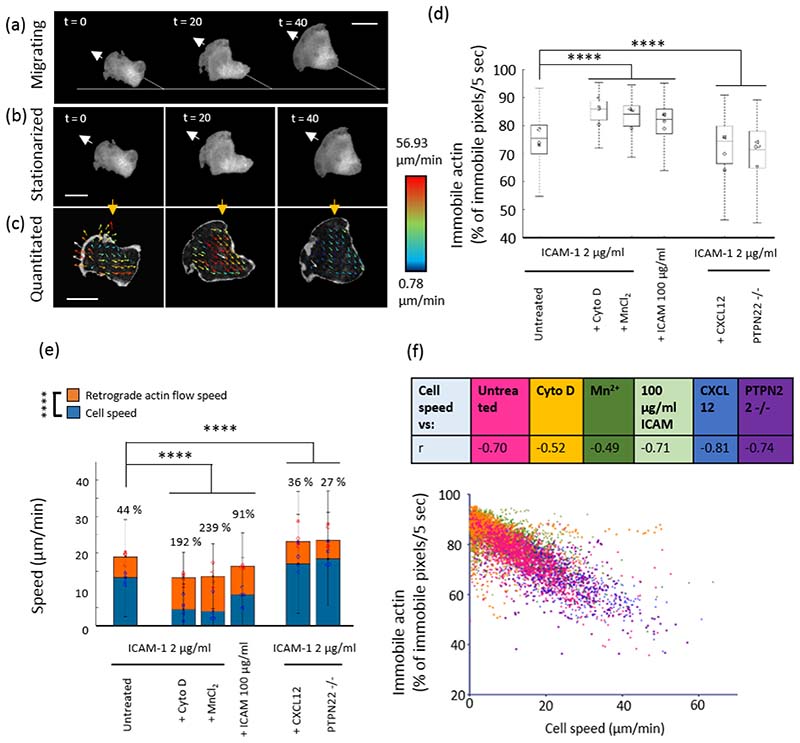
Figure 1. Fast cells exhibit decreased actin attachment to adhesions and increased flow speed.
Example frames from representative TIRF movies of actin in a migrating T cell (a). b) stationarised version of the same cell c) STICS vector maps from the stationarised cell. d) Box plot showing immobile actin - % of immobile pixels/5 seconds derived from the external reference frame, e) median cell speed (blue bars – blue symbols are median values for each mouse) and retrograde actin flow speed (orange bars and symbols) from the internal reference frame f) Plot showing negative correlation between % of immobile pixels/5 sec vs cell speed: table above shows r values for correlation in each condition. N = 4 mice, 50 cells per mouse, 200 cells per condition. Kruskal Wallis testing was used to compare pooled non-parametric data between each condition. P<0.0001 = ****.
For cells migrating on ICAM-1 (2 µg/ml), 75 ± 5% of pixels contained more than 50% immobile F-actin over 5 second time windows (Figure 1d). Slower cells, treated with cytochalasin D, manganese or a high ICAM concentration (100 µg/ml), displayed a significantly increased percentage of pixels displaying immobile actin p<0.0001 (Figure 1d). In contrast, faster migrating cells (treated with CXCL12 or deficient for PTPN22) had significantly lower percentages of pixels containing immobile actin p<0.0001.
STICS was then used to analyse the flow speeds of unengaged actin, relative to the cell (Figure 1e). T cells migrating on ICAM-1 (2 μg/ml) alone had an average cell speed of 13.19 μm/min (blue), with retrograde flowing actin moving faster at 18.95 μm/min (orange). Conditions which slowed cell migration also slowed actin retrograde flow speed. However, in all cases, actin flow speed always remained much higher than cell speed. The percentage values above the bars show that in slower cells, treated with Cyto D, Mn2+ or a high concentration of ICAM-1, actin speed was 192 %, 239 %, and 91% higher than the cell speed, respectively. Adding CXCL12 to cells or measuring cells deficient for PTPN22 led to faster cell migration which was also coupled to faster actin retrograde flow. Furthermore, Figure 1f shows that in all conditions, the fraction of pixels in which actin was immobile was inversely correlated to cell speed (r values for correlation shown in table, p < 0.0001). Therefore, we concluded that decreased actin engagement correlates with increased cell speed. Taken together, these data indicate that retrograde actin flow is always greater than the overall velocity of the T-cell and that the proportion of immobile actin is inversely related to cell speed.
The size and number of LFA-1 membrane nanoclusters increase with T-cell speed
When effector T-cells randomly migrate on ICAM-1 coated glass they adopt a polarised morphology (Hogg et al. 2004; Comrie et al. 2015; Valignat et al. 2013; Ridley et al. 2003). Previously, studies of integrin affinity have shown a specific pattern where LFA-1 within the leading edge exists predominantly in the intermediate affinity form, whereas high affinity LFA-1 is detected in the mid cell body/focal zone (Smith et al. 2007). It has been reported in slow moving cells that integrin-based adhesions change their size as they mature (Kanchanawong et al. 2010). We therefore tested whether T cells use differential membrane LFA-1 nano-clustering to regulate cell migration.
Integrin LFA-1 nanoscale clustering in conditions that speed up or slow T cell migration was investigated by dSTORM and Bayesian cluster analysis (Rubin-Delanchy et al. 2015; Griffié et al. 2016). Effector T-cells were allowed to migrate on ICAM-1 coated glass for 15min in the presence of conditioned/unconditioned media, and these were compared with cells deficient for PTPN22. Cells were fixed, surface LFA-1 detected with anti-LFA-1 AF647(clone 2D7), STORM acquisition performed and co-ordinate pointillist maps generated for Bayesian cluster analysis. Stationary effector T-cells were used as non-polarised T-cell controls (0 ICAM-1 on the coverglass). Representative STORM images and cluster maps of a 2 x 2 µm region of interest (ROI) within a stationary and migrating T-cell are shown (figure 2a and b). ROIs from round stationary cells were taken at random as they do not have a leading edge and focal zone. T-cell velocities were measured for each condition by automatic tracking before fixation for SMLM (as previously shown in Supplementary Figure 1).
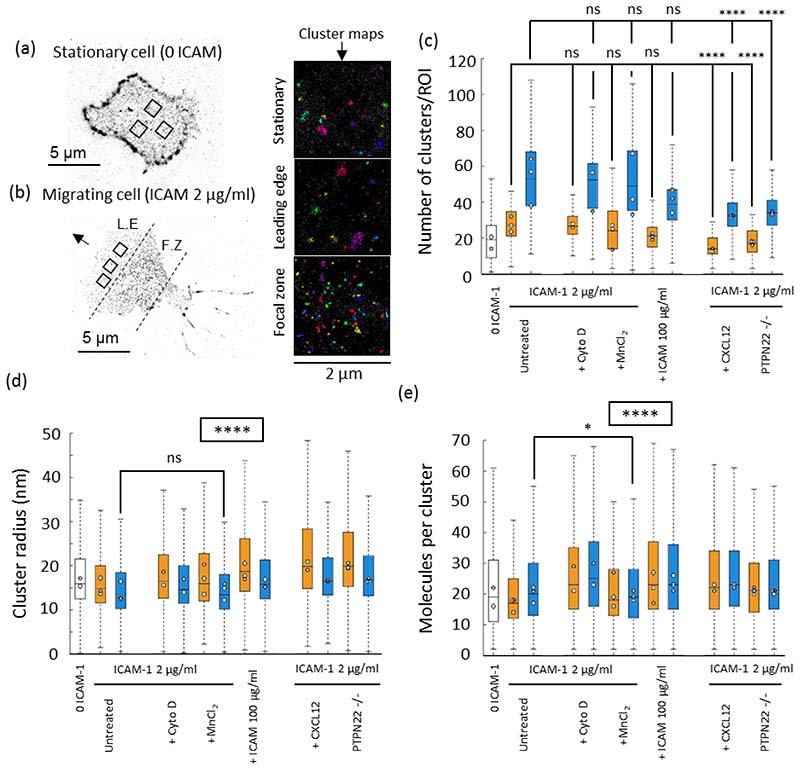
Figure 2. Integrin LFA-1 membrane nanoclusters increase in density and decrease in number in fast migrating cells.
Example pointillist maps derived from STORM imaging of LFA-1 in a stationary cells (a) and a migrating cell (b). Boxes in a and b denote 2 μm2 regions chosen for analysis and representative cluster maps on the right. In each plot, metrics extracted for stationary cells have clear bars, and migrating cells are split up into leading edge (orange bars) and focal zone (blue bars). From left to right conditions proceed: ICAM-1, 2 μg/ml ICAM-1 coated coverslip with added cytochalasin D, Mn2+ or a higher concentration of ICAM-1 (100 μg/ml), then, 2 μg/ml ICAM-1 coated coverslip with CXCL2 added at 150 ng/ml or using cells deficient for PTPN22. Metrics extracted consist of c) number of clusters per ROI, d) cluster radius (nm) and e) the molecules per cluster. (n=350 ROIs and 40 cells per condition from 3 separate mice). Kruskal Wallis testing was used to compare pooled non-parametric data between each condition. NS = not significant. P<0.05 = *. P<0.0001 = ****. Boxed **** in (d) and (e) denotes that p<0.0001 between every condition, apart from for untreated (FZ) versus MnCL2 (FZ).
In polarised T cells, the focal zone (FZ) displayed different LFA-1 clustering properties to the leading edge (LE). The number of LFA-1 clusters per ROI was consistently increased in the FZ (Figure 2c). Compared to the LE, the size of clusters was significantly decreased in the FZ (Figure 2d), whereas the number of molecules per cluster was significantly increased (Figure 2e). Together this represents a median increase in cluster density in FZ clusters (blue bars), compared to LE clusters (orange bars). This is achieved both through a size contraction and an increase in molecular content of the clusters. Compared to stationary cells, clusters in the cell membrane of polarised migrating T cells increase in number, in size and in molecular content. Together, this indicates that T cells adopt a regionally discriminated LFA-1 nanoclustering program to achieve polarised T cell migration.
The number of clusters significantly decreased in cells that migrated faster due to treatment with CXCL12 or deficiency for PTPN22 (Figure 2c) as compared to slower cells (Mn2+ treated, high ICAM-1 concentration and untreated controls). Slower moving cells had a similar number of LFA-1 nanoclusters to untreated control cells (Figure 2c). The size of clusters was significantly increased in the LE and FZ of T-cells treated with CXCL12 (32% LE and 22% FZ increase with CXCL12 compared to ICAM-1 alone (P < 0.0001)) and PTPN22-/- (33% LE and 23% FZ increase in PTPN22-/- compared to ICAM-1 alone (P < 0.0001)) (Figure 2d). The number of localisations per cluster was increased in faster moving T-cells to a greater extent in the LE than in the FZ (29% LE and 15% FZ increase with CXCL12 compared to ICAM-1 alone (P < 0.0001)); (24% LE and 5% FZ increase in PTPN22-/- compared to ICAM-1 alone (P < 0.0001)) (Figure 2e).
In slower moving T-cells treated with Cytochalasin D or MnCl2 we observed a minor increase in cluster size (< 9% LE and FZ),, and an intermediate increase of 25% and 17% in the LE and FZ of cells migrating on higher concentrations of ICAM-1 (100 μg/ml), respectively. In Cytochalasin D treated cells, the molecules per cluster increased by 35% and 25% in the LE and FZ respectively. In MnCl2 treated cells, the molecules per cluster increased by < 6 %, whereas in cells migrating on an increased concentration of ICAM-1, molecules per cluster was increased by 35% and 15% in the LE and FZ, respectively. Together, this indicates a more substantial increase in cluster size, and a concomitant increase in molecular content (representing maintained cluster density) in fast moving cells. Slow moving cells showed negligible changes in cluster size (Mn2+ or CytoD treated). Mn2+ treated clusters had negligible changes to molecular content, whereas Cytochalasin-D treated cells experienced an increase in cluster density. All changes were more substantial in the LE compared to the FZ. All changes are summarized in Supplementary Table 2.
These results suggest that integrin LFA-1 membrane clusters are modulated on the nanoscale and this influences the migration speed of the cell. Clusters adopt a small dense profile in the high affinity integrin rich focal zone, and a larger less dense profile in the reduced affinity leading edge. Clusters are larger and less dense in faster migrating cells. The data suggest that these zone-specific cluster patterns may be hardwired into polarised T-cells and indicate that the biophysical arrangement of LFA-1 is regulated in conjunction with integrin affinity to control cell speed. As there are fewer clusters in faster moving cells, we next investigated the 3D profile of nanoclusters of LFA-1, to determine whether this phenomenon relates to the recruitment of LFA-1 to the membrane from submembrane stores.
The size of 3D intracellular LFA-1 clusters increases above the focal zone as cell speed decreases
LFA-1 is constantly recycled to and from the cell membrane to facilitate cell migration. Accordingly, we investigated whether changes in the intracellular pool of membrane proximal LFA-1 was nanoclustered, and if so, was similarly zonally distributed to surface LFA-1 and whether that distribution was modulated with cell migration dynamics. Interferometric photoactivatable light microscopy (iPALM) (Shtengel et al. 2009) was used for the acquisition of 3D single molecule data with ˜20 nm z-precision to a depth of ˜500 nm from the height of the coverslip. The same battery of conditions were used to slow down and speed up migrating T cells, which were tracked to confirm their speed prior to fixation. Figure 3a illustrates a representative iPALM z-projection of cytosolic LFA-1 nanoclusters in a migrating T-cell (left) and the result of Bayesian 3D cluster analysis (Griffié et al. 2017) in a representative 2000 x 2000 x 400 nm ROI (right). Cluster descriptors for the size and number of molecules per cluster were then extracted from ROIs in the FZ or the LE and plotted as a function of z - height from the coverslip.
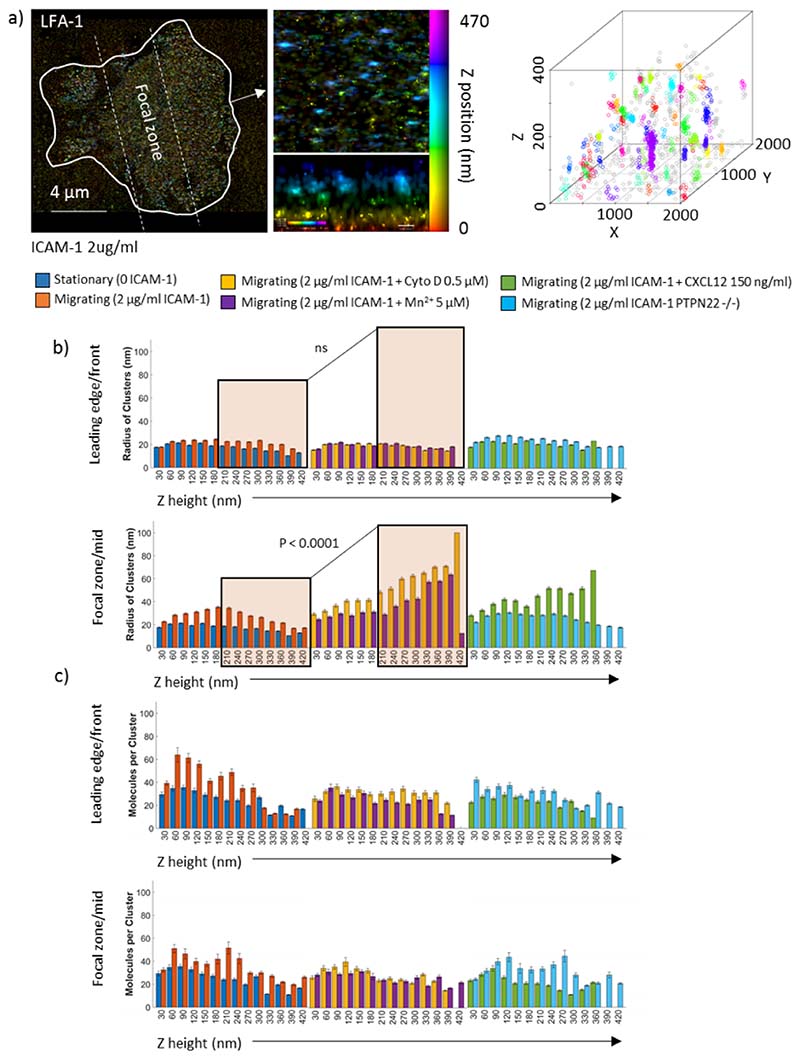
Figure 3. Intracellular LFA-1 nanoclusters are larger in slow moving cells above the focal zone.
A) Representative iPALM Z projection of LFA-1 in a migrating T cell (left panel), zoomed region in xy and xz side view (middle panel), and arbitrarily coloured clusters identified in a 2000 x 2000 nm region by Bayesian cluster analysis (right panel). The radius of clusters (b) and the molecules per cluster (c) were extracted from the cluster maps for stationary cells (0 ICAM-1) and migrating cells (2 μg/ml ICAM-1) (left hand plots), Cytochalasin D and MnCl2 treated (middle plots) and CXCL12 and PTPN22 -/- cells (right plots). Orange box in figure b denotes larger clusters higher above the membrane in slow cells, which are not present in untreated migrating cells. N = 3 mice, 20 cells and 200 ROIs per condition. Kruskal Wallis testing was used to compare pooled data from z heights 0 to 210 nm versus 240 to 420 nm.
Clustering in migrating cells was regionally distinct between the LE and FZ. In the leading edge, clusters in migrating cells compared to stationary cells exhibited almost no increase in size (figure 3b – top panel), but this was accompanied by an increase in the number of molecules per cluster (Figure 3c – top panel, p < 0.0001). Clusters in migrating cells also exhibited an increase in size, focused at a region 200 nm above the coverslip (Figure 3b – lower panel, p < 0.0001). The number of molecules per cluster was also increased in the FZ of migrating cells (Figure 3c, p < 0.0001). Together, this indicates that in the switch from stationary to migrating phenotype, 3D LFA-1 clustering becomes region specific, where clusters above the LE increase in density and clusters above the FZ increase in size and slightly increase their molecular content, concentrated close to the cell membrane.
In slowed cells (treated with Cyto D or Mn2+), effects on nanoclusters occur above the focal zone, whereas LE clusters remain constant (orange box, top panel figure 3b). FZ specific nanoclusters increase in size, especially in the range 210 to 410 nm above the cell membrane (orange box, lower panel figure 3b, p < 0.0001), with a similar number of molecules at all Z-heights in the range 0 to 420 nm. Thus, intracellular LFA-1 nanoclusters become larger and less dense in cells which migrate slower. In faster migrating PTPN22 deficient cells, the size of intracellular LFA-1 nanoclusters remains similar to those in PTPN22 proficient cells in both the LE and FZ, with a decrease in the number of molecules per cluster (figure 3b and c, orange compared to light blue bars). Activating T-cells with CXCL12 (figure 3b, orange compared to green) slightly increased the size of LFA-1 molecules per cluster high above the membrane (z = 210 to 420 nm) with an LE focused reduction the in molecules per cluster (figure 3d, orange to green). Intracellular LFA-1 clusters in PTPN22 deficient fast cells stay the same size, and remain the same density, and in CXCL12 treated cells become slightly larger higher up.
Deleting PTPN22 increases T-cell velocity, and the nanoscale colocalization of pY397FAK, pY416Src and LFA-1
Integrin based adhesions are regulated by an array of kinases and phosphatases. Phosphorylation of FAK is a feature of active ICAM-1/LFA-1 signalling (Zhang & Wang 2012; Sanchez-Martin et al. 2004), and is implicated in the first stages of nascent adhesion formation in non-leukocytes (Swaminathan et al. 2016). FAK is basally activated by Src-family kinase LCK in effector T-cells at Y576/577 and Y925. This primes FAK for full activation at the auto-phosphorylation site pY397 (Chapman & Houtman 2014) which in turn regulates LFA-1 adhesion and deadhesion(Raab et al. 2017b). Expression of dominant-negative mutants of FAK in T-cells reduce the speed of migration on ICAM-1 coated glass (Rose et al. 2003), and overexpression of a negative regulator of FAK phosphorylation impairs LFA-1 clustering and reduces ICAM-1/LFA-1 driven T-cell homo-aggregation (Giannoni et al. 2003). PTPN22 is a cytosolic phosphatase that negatively regulates LCK and ZAP-70 downstream of LFA-1 engagement and binds with C-terminal Src kinase (CSK) to function (Burn et al. 2016), a common target for FAK (Chapman & Houtman 2014). Auto-phosphorylated pY397 FAK has also described as being associated with active integrin in vesicles above the membrane(Nader et al. 2016; Kleinschmidt & Schlaepfer 2017).
We therefore employed iPALM and Bayesian cluster analysis to study 3D pY397 FAK localisation in stationary and migrating cells sufficient or deficient for PTPN22. An example pseudocoloured 3D localisation map is shown in figure 4a. ROIs were chosen from the cells, and Bayesian cluster analysis carried out to produce cluster maps (Figure 4b). pY397 FAK staining was verified using FAK inhibitor 14, which blocks Y397 phosphorylation (Supplementary Figure 2). ROIs from the LE or FZ regions were quantified in terms of their radius and molecules per cluster as a function of z (Figure 4c and d).
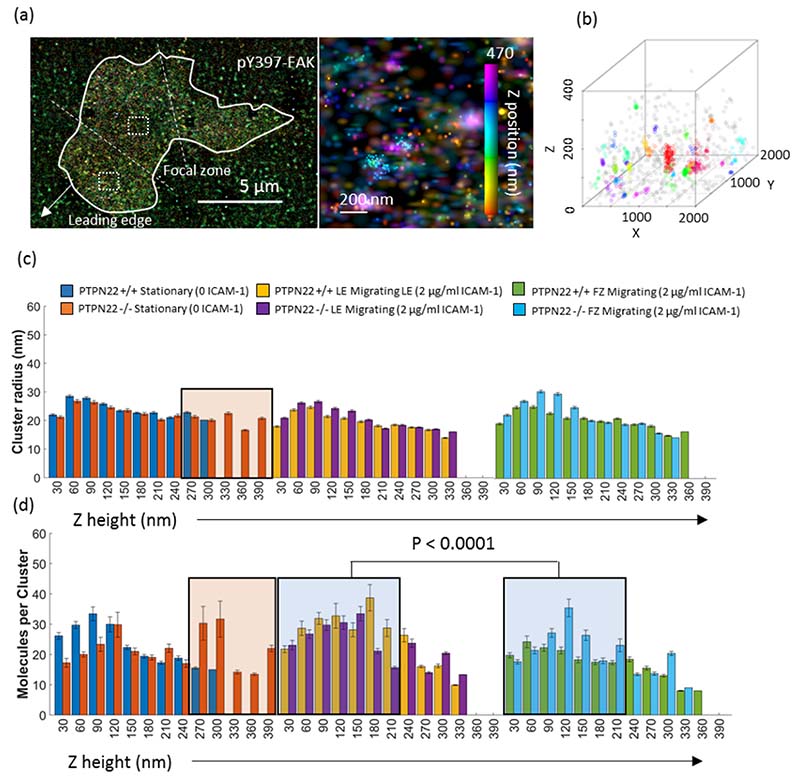
Figure 4. Intracellular 3D pY397 FAK nanoclusters are of similar size and density in migrating and stationary cells, while deleting PTPN22 increases membrane distal pools of pY397 FAK.
a) An example pseudocoloured cluster map (left) is shown alongside a zoomed image where clusters can be visualized at different height scales from 0 to 470 nm (middle panel with z colour bar). b) Shows an example cluster map with defined 3D clusters. For the radius of clusters (c) and the number of molecules per cluster (d), the leftmost plots show stationary PTPN22 +/+ or PTPN22 -/- cells (0 ICAM-1), the middle plots show the leading edge of migrating PTPN22 +/+ PTPN22 -/- cells (2 μg/ml ICAM-1) and the right hand plots show the focal zone of migrating PTPN22 +/+ PTPN22 -/- cells (2 μg/ml ICAM-1). N = 3 mice, 20 cells and 200 ROIs per condition. Error bars denote standard error of the mean. Orange boxes denote the population of clusters present in PTPN22 deficient cells and not in proficient cells. Blue boxes denote a reduction in the density of clusters in the FZ compared to the LE, close to the membrane. Kruskal Wallis testing was used to compare pooled data from z heights 0 to 210 nm versus 240 to 420 nm.
When comparing stationary cells to cells migrating on ICAM-1, pY397 FAK cluster size (Figure 4c) was largely unchanged above the leading edge and focal zone, in both PTPN22 proficient and deficient cells. The number of molecules per cluster was higher close to the cell membrane (Z = 0 to 210 nm) in the LE and FZ, meaning that such clusters retained their size but increased in density (figure 4d). The size of and number of molecules per cluster was similar between stationary and in the LE of migrating cells. Clusters became less dense (of similar size but with fewer molecules per cluster) close to the membrane in the FZ of migrating cells (blue translucent box, z = 0 to 210 nm, p < 0.0001).
PTPN22 deficient stationary cells (no ICAM-1) adopted pY397 FAK nanoclusters high above the membrane (orange box in figure 4c, comparing blue versus orange bars at z = 270 to 390 nm, p < 0.0001), which were not present in PTPN22 proficient cells. This population of high clusters present in PTPN22 deficient stationary cells has the same number of molecules per cluster as those lower down in z, representing a maintenance of density (Figure 4d). During migration, PTPN22 deficiency did not change the size or density of intracellular pY397 FAK clusters.
As nanoclusters resemble nascent adhesions in non-leukocytes, but occur throughout the cell, we next investigated how they are regulated in T cells. We tested whether the lack of PTPN22 causes changes in the colocalization of pFAK with LFA-1 and a second set of signalling intermediates: the Src family kinases, using multi-colour SMLM microscopy (Yi et al. 2016a; Yi et al. 2017). Wild type or PTPN22 deficient primary murine T cell blasts were plated on ICAM-1, allowed to migrate and tracked by live-cell imaging. They were then fixed, stained and imaged using madSTORM (Yi et al. 2016b) permitting the sequential imaging of pY397FAK, pY416Src family kinases and LFA-1. Supplementary Figure 3 shows the overall cell speeds where PTPN22 deficient cells were faster than PTPN22 proficient cells directly prior to fixation. Figure 5a shows a representative cell post fixation after three rounds of sequential staining and imaging.
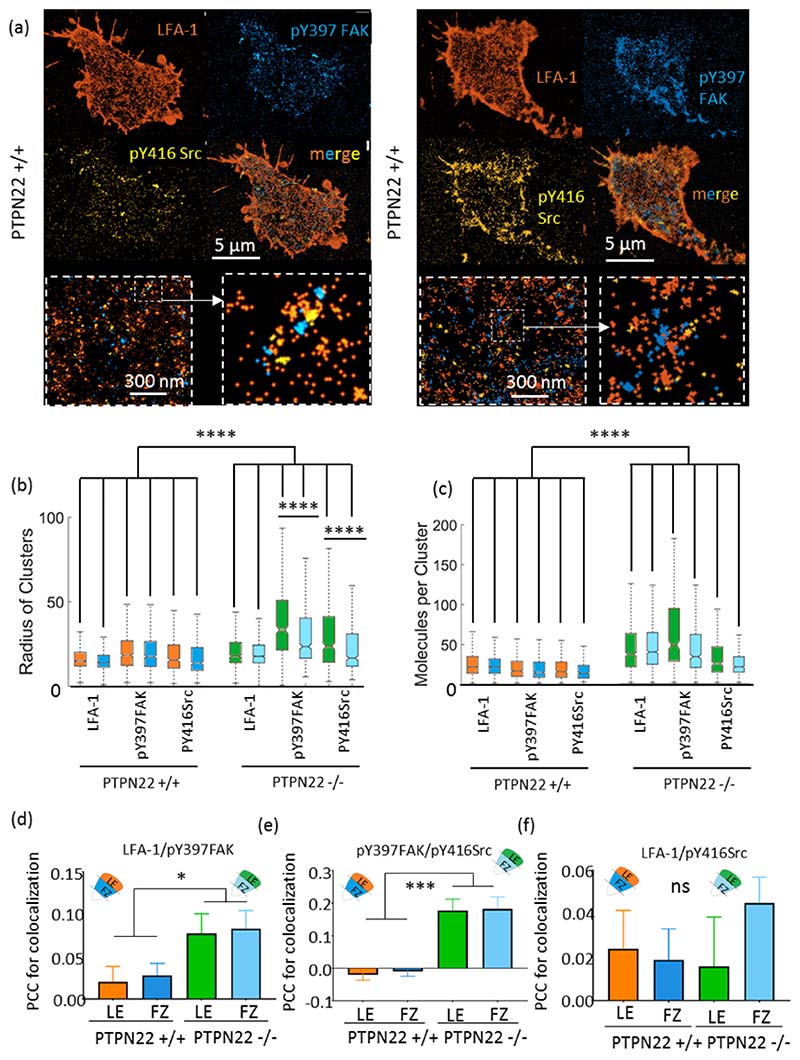
Figure 5. Cluster size and number of molecules per cluster increase in faster moving PTPN22 -/- cells in the case of LFA-1, pY397 FAK and pY416 Src, which are more colocalized.
Individual cells were sequentially stained and imaged by madSTORM: LFA-1. pY397 FAK and pY416 Src in the same cell are shown (a). LFA-1 clusters are larger (b) and denser (c) in faster moving PTPN22 -/- cells. pY397FAK and pY416 Src clusters are also larger and denser in PTPN22 -/- cells. The size of pY397 FAK and pY416 Src clusters in PTPN22 -/- are more strongly regionally discriminated: a) clusters in the leading edge are larger than those in the focal zone, though the molecular content stays the same. Pearson’s correlation coefficient for cluster colocalization: d) LFA-1 colocalized with pY397 FAK more in PTPN22 -/- cells, and pY397 FAK colocalized more with pY416 Src in PTPN22 -/- cells (e). LFA-1 and pY416 Src displayed similar levels of colocalization in PTPN22 +/+ and PTPN22 -/- cells (f). N = 21 PTPN22 +/+ cells and 13 PTPN22 -/- cells. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Boxplots in b and c denote the minimum and maximum values, the median and the IQR. Error bars in d, e and f denote standard error of the mean. Kruskal Wallis and Dunn’s post hoc testing was used to compare the clustering of each molecule, in each zone, in PTPN22 sufficient and deficient T cells.
In wild-type migrating T-cells, LFA-1, p Y397FAK and pY416Src clusters co-existed on a similar size scale and with similar numbers of molecules per cluster (Figure 5b and c). LFA-1, pY397FAK and pY416Src clusters were consistently larger with more molecules in the leading edge compared to the focal zone. PTPN22 deficient cells displayed significantly larger clusters of all three species in both the LE and FZ, combined with significantly more molecules per cluster. PTPN22 deficiency therefore increases the clustering of all three proteins, but especially pY397FAK and pY416Src family kinases. Extraction of Pearson’s Correlation Coefficient (PCC) for the colocalization of clusters of each species showed that LFA-1 clusters were more colocalized with pY397FAK clusters when PTPN22 was deficient (Figure 5d). pY397 FAK was also more colocalized with pY416Src family kinases in PTPN22 deficient cells (Figure 5e). The colocalization of LFA-1 clusters with pY416Src family kinase clusters was unchanged (Figure 5f). Taken together, faster migrating PTPN22 deficient cells display larger LFA-1, pFAK and pSrc clusters which are more often colocalized spatially on the nanoscale. The increase in the number of molecules per cluster in the case of pY416Src family kinases and pY397FAK supports existing literature describing the action of PTPN22 on Lck, a well characterized participant in early integrin adhesion signalling that itself upregulates FAK phosphorylation (Chapman & Houtman 2014). Additionally, we do not believe that the absence of PTPN22 is compensated for by upregulation of any other phosphatase activity, as the data show a clear increase in total phosphorylation of direct (Src family kinases) and indirect (FAK) PTPN22 substrates in PTPN22 deficient cells (Supplementary Figure 4).
Discussion
We employed advanced and super-resolved imaging methods to study the nanoscale migration machinery in T cells. Specifically, we quantitated the nanoscale distribution of membrane localised and intracellular LFA-1 alongside actin flow dynamics in migrating T-cells to understand the relationship between integrin affinity-clustering and engagement with flowing actin and the speed of T-cell propulsion. Our data indicate that 1) actin flow and engagement drive migration, where engagement is inversely correlated to cell speed and flow rate of unengaged actin is positively correlated, and that 2) T cells exploit differential clustering of hundreds of tiny nano-adhesions to translate force from the actin cytoskeleton to the substrate. 3) LFA-1 and pY397 FAK nano-adhesions adopt region specific clustering patterns both in 2D in the membrane and in 3D above the membrane. 4) Changes to cell speed induced by a battery of signals modulate adhesion nano-cluster size and density. 5) The deletion of PTPN22 phosphatase causes faster T cell migration, and modulates LFA-1 and pY397 FAK clustering in 2D and 3D. Importantly, lack of this phosphatase induced an increase in LFA-1-pYSrc family kinase-pY397FAK nanocluster colocalization coupled to actin flow speed increase, actin engagement decrease and cell migration speed increase.
The front(LE)/middle(FZ) trends in LFA-1 (2D and 3D), pYFAK (2D and 3D) and pYSrc (2D) indicated here are characterised for the first time. For LFA-1, reported trends for affinity occur in the same zones: high affinity integrin occurs in the FZ, whereas low/intermediate affinity integrin predominates in the LE (Persson et al. 2018; Evans et al. 2011; Smith et al. 2005). In addition, nano-adhesions here differ from adhesions in slower moving cells, which are founded as ‘nascent adhesions’ and grow as they mature into larger multi-molecular clutch assemblies towards the middle of the cell. The data here indicates a different system, where high affinity LFA-1 adopts smaller, denser more numerous cluster arrays in the FZ. Intracellular LFA-1 clusters are larger above the FZ, and intracellular pY397 FAK clusters are denser above the LE. Coupled with the actin work, this indicates that these clusters provide an area of increased adhesion in the FZ: it may be this adhesion imbalance between the LE and FZ that enables fast forward cell migration.
In conditions that increase cell speed, nano-adhesions become larger and are joined by larger clusters of phosphorylated kinases (pY397FAK and pYSrc family). The integration of multiple molecules into the same nanocluster may necessitate its increase in size in this way. That this effect is produced where PTPN22 is knocked-out of cells, which directly targets Lck, a Src family kinase that itself phosphorylates FAK, indicates that the size, density and number of LFA-1 clusters is modulated by such kinases and phosphatases on the nanoscale. Pools of intracellular LFA-1 decrease in density in rapidly migrating cells, and intracellular pY397 FAK clusters stay the same size, indicating a further level of control of adhesion based on the nature of LFA-1 undergoing recycling.
It is also important to note that T cells are capable of several modes of migration, depending on their environmental niche. Here, we study ICAM-1/LFA-1 dependent migration, coupled to actin treadmilling and engagement. High speed polarised T cell crawling is a phenomenon which occurs in the lymphatic capillaries, during lymph node entry, tissue entry and fast DC scanning2. Such integrin dependent migration is especially important during inflammation, and including in autoimmune disease2, and is a physiological requirement for high speed migration1. Integrin-free slower migration relies mainly on RhoGTPase mediated actin remodeling and predominates in the dense, fibrous interstitium (ICAM-1 blockade doesn’t affect cell migration here). In the lymph nodes, both integrin independent and integrin dependent fast migration occur, where ICAM-1 blockade reduces T cell migration speed considerably. In vitro, the presence of ICAM-1 potentiates migration speed by 50 to 30 %1 in confined 3D (under agarose), although in such a system the cells do not require integrin to migrate slowly. Previously, the simple unconfined ICAM-1 coated glass system used here has been used to characterise integrin affinity states. Here, we have used it to characterise the contribution of nanoclustering and its effect on actin engagement and cell speed.
Overall, we demonstrate that T cells use individually regulated integrin based, kinase/phosphatase coupled ‘nano-adhesions’ to achieve fast migration. Such nano-adhesions are similar to previously described nascent adhesions(Sun et al. 2014) in size and molecular composition, but differ in that they are regulated on a sub-diffraction length scale throughout the cell membrane, and not only in the leading edge. The differential inclusion of proxy markers for integrin activity (phosphorylated FAK and Src family kinases) indicates the highly spatial nature of such adhesion regulation. We posit that this nano-adhesion system, as opposed to a classical nascent > focal complex > focal adhesion system, represents a novel way for T cells to quickly turn over individual adhesions to achieve dynamic feats of migration depending on external or internal cues. Our results agree with the conventional understanding of how integrins associate with the actin treadmill as a ‘molecular clutch’ to facilitate adhesive traction, with the important difference being that T-cells utilise nanoscale ‘clutches’ of LFA-1 adhesive units at least 100-fold reduced in size and polarised in their cellular location. We suggest that these features potentiate speed and agility of T-cell migration.
The question of live cell dynamics and rare events in the regulation of nanoclusters will be key to understanding how cells use spatiotemporal molecular organisation to change their behaviour. New high throughput techniques (Holden et al. 2014; Gunkel et al. 2014; Yasui et al. 2018) to low power imaging (Chen et al. 2014), and new fluorescent proteins (Zhang et al. 2016; Chang et al. 2012; Tiwari et al. 2015) may soon enable these kind of investigations, and will be especially important in connecting nanoscale regulation to whole cell function in diverse settings (Shannon & Owen 2019), including in autoimmune disease.
Methods
Mice colonies
Mice were bred and maintained in the Biological Services Unit King’s College London in compliance with Home Office regulations and local ethically approved guidelines. Ptpn22-/- mice maintenance and breeding have been described elsewhere 52. Male β-actinCre/+; Lifeact-mEGFP/Y C57BL6 males were a gift from Dr Karen Liu’s lab, which were crossed with Ptpn22-/- C57BL6 females; offspring were backcrossed to generate Ptpn22+/+β-actinCre+Lifeact-mEGFP+ and Ptpn22-/-β-actinCre+Lifeact-mEGFP+ males for experiments. Tail blood was genotyped for EGFP expression by flow cytometry. Single cell suspensions of mouse lymph nodes and stimulated with 1 μg/ml of Concanavalin A. After 48 hrs T-blasts were re-suspended in medium supplemented with 20 ng/ml human recombinant IL-2 for an additional 3-5 days before being used for migration assays.
Primary murine T cell culture
Single cell suspensions were made from lymph nodes harvested from Ptpn22+/+; Ptpn22-/- and Ptpn22+/+ Lifeact-mEGFP EGFP+; Ptpn22-/-EGFP+ mice and stimulated with 1 μg/ml of Concanavalin A (Sigma L7647) at 3x 106 cells/ml for 48hrs in complete medium (RPMI 1640-Glutamax, 10% FBS; Penicillin, Streptomycin, 100 μM Sodium Pyruvate, 20 mM Hepes, 20 μM 2-Mercaptoethanol). After 48 hrs T-blasts were washed and re-suspended at 1-2 x 106 cells/ml in complete medium constantly supplemented with 20 ng/ml human recombinant IL-2 for an additional 3-5 days before being used in migration assays.
T cell migration assay
Migration media
RPMI 1640-Glutamax; 100μM Sodium Pyruvate, 25mM Hepes, 20μM 2ME; 2ug/ml hrIL-2; equilibrated overnight at 37C 5%CO2. Ibidi μ-Slide 8 well glass bottom chamber slides of #1.5H thickness were coated with 2μg/ml murine recombinant ICAM-1-Fc (R&D systems 796-IC-050) for 1.5 hours at 37C. Conditioned migration media: supplements were added to cells in migration media for 10min prior to addition of 2000,000 T cells to ICAM-1 coated slide chambers; 150 ng/ml CXCL12 (Peprotech 250-20A-10), 5μM Manganese Chloride (MnCl2+) (Sigma M1787) and 0.5μM Cytochalasin D (Sigma C2618).
Wide field live cell phase contrast time lapse microscopy and automatic tracking
Imaging was performed using a Nikon Eclipse TI-e microscope. 200 000 T cells were plated onto slide chambers, transferred to a pre-warmed microscope chamber and settled for 5-10min before acquisition of time lapse videos at 37C in 5 % CO2 under a 10x objective for 15min (50ms exposure; 25 seconds between frames). Automatic tracking to quantify T cell migration was performed using a custom ICY modular program. Image files were imported in the nd2 format with attached metadata to the ICY interface and cell tracking data was then exported to Graphpad prism or matlab for statistical analysis.
Analysis of actin engagement and flow
Analysis windows or ‘Time of Interest’ (TOI) for actin flow and engagement was 5 frames. Immobile actin was calculated in the external reference frame of the cell by taking the zero component of the temporal Fourier transform. A threshold of 50% was applied to each pixel for it to be counted as immobile. For STICS, we adapted a stationary cell program (Wilson & Theriot 2006), which keeps the cell centroid in the centre of the ROI. Actin flow analysis was then carried out using STICS(Hebert et al. 2005).
Fixation and labelling for STORM
Migrating T cells were fixed using 3% PFA + 0.1 % glutaraldehyde in kPIPES 80 mM, MgCl2 2mM, EGTA 0.5 mM and 2mM sucrose at pH6.8 for 10 min followed by 3%-PFA + 0.1 % glutaraldehyde in Borax 100 mM for 10 min. Slides were washed once PBS between each treatment. Immunofluorescence. T Cells were quenched for auto-fluorescence with 1mM NaBH4 for 15 minutes, L-Lysine 0.1M for 15 minutes, blocked with 10% normal goat serum (Thermo Fisher 50062Z) for 30 min at RT then stained with primary directly conjugated antibody at 1 in 100 dilution in 0.1% BSA for 1.5hr at RT. Antibodies. Anti-mouse CD11a (LFA-1) 2D7 clone (Biolegend, UK, 101002), anti-pTyr397-FAK (31H5L17), ABfinity Rabbit Monoclonal (Thermo Fisher, 700255) and anti-PTyr416-SRC (D49G4) Rabbit mAb #6943BF (Cell signalling technology, 6943BF). Immunofluorescence validation profiles for these antibodies are available at the company websites upon entering the product codes.
STORM and iPALM
T cells were fixed, blocked and stained with primary directly fluorophore conjugated (alexafluor 647) antibodies to CD11a (2D7 clone, Biolegend, catalogue number 101002), pY397 FAK (31H5L17 clone, Thermo Fisher, catalogue number 700255), and pY416 Src family kinases (D49G4 clone, Cell Signalling Technology, catalogue number 6943BF). Fixed and stained cells were placed in an oxygen scavenging buffer comprised of three parts which were stored separately. Part A was comprised of 4 mM Tris (2-carboxyethyl) phosphine hydrochloride (Sigma 51805-45-9), 50 % glycerol (Sigma 56-81-5), 25 mM KCl (Sigma 7447-40-7), 20 mM Tris HCl (Sigma 1185-53-1), 20 μg/ml catalase (Sigma 9001-5-2) and 1 mg/ml glucose oxidase (Sigma G2133) all at pH 7.5, made up to a total volume of 5 ml and stored in aliquots of 50 μl at – 20C. Part B contained 100 mg/ml glucose and 10 % glycerol and in practice was made to a total volume of 40 ml and stored in aliquots of 400 μl volume at – 20C. Part C contained 1 M Cysteamine-HCl, and in practice was made up directly prior to imaging to 1 ml volume. For imaging, an aliquot of each of Part A and Part B was thawed and added to 100 μl of freshly made Part C, supplemented with 450 μl PBS to a total volume of 1 ml. Imaging was performed on a Nikon N-STORM microscope using a Nikon Plan Apo 100 x NA oil immersion TIRF objective. Cells were imaged under TIRF illumination with a 15 mW 633 nm laser with the 405 nm laser set to maintain photo-blinking and transition to the triplet state by gradually increasing its power from 0.1 to 1 mW. Emitted fluorescence was collected at wavelengths between 640 and 790 nm, EM gain 300 on an Andor iXon EM-CCD camera at RT. Acquisition time was for ˜2.5 minutes: 10 000 frames were collected at 10 ms exposure time per frame and pixel size was 160 nm. Molecular coordinates were calculated using ThunderSTORM localisation software (Ovesný et al. 2014) with multi-emitter fitting analysis (Huang et al. 2011) enabled. Cluster analysis was performed using Bayesian analysis in 2D (Rubin-Delanchy et al. 2015; Griffié et al. 2016) or 3D (Griffié et al. 2017). Cluster analysis software is available for download from the articles listed above. For iPALM, custom gold nanorod fiducial embedded coverslips (Moore et al. 2018) were coated with ICAM-1. iPALM imaging was performed at the Advanced Imaging Centre at Janelia Research Campus. X, Y and Z coordinates were derived using custom Peakselector localisation software, which is available on request. It is important to note that clustering patterns in the first 60 nm of iPALM data closely matched those in the TIRF data. While detection efficiencies were different between the two techniques, reliable TIRF data closely matched iPALM data close to the coverslip.
Single cell correlative live tracking + multicolour dSTORM by multiplexed madSTORM
T cells were first tracked during migration. Cells were fixed, and their positions were recorded, and then multiplexed antibody madSTORM (Yi et al. 2016b; Yi et al. 2017) was used to achieve multicolour imaging. 3 colour pointillist maps were analysed channel per channel with Bayesian cluster analysis (Griffié et al. 2016), and colocalization was analysed (Rossy et al. 2014).
Statistics
D’Agostino and Pearson tests were used to check for normality in pooled data (from 3 or 4 mice in each condition, where leading edge and focal zone measurements were treated as separate conditions). Kruskal Wallis testing and Dunn’s post hoc testing was used to compare pooled, non-normal data between each condition. All statistical analyses were performed with Graphpad Prism software. It is important to note that due to high numbers of clusters being tested in the case of the radius and molecules per cluster, statistical testing gave very low P values between conditions, therefore these results are also discussed in terms of percentage changes. For multiple comparisons between conditions, one-way analysis of variance (ANOVA) and Kruskal Wallis testing was used. All statistical analyses were performed with Graphpad Prism software.
Supplementary Material
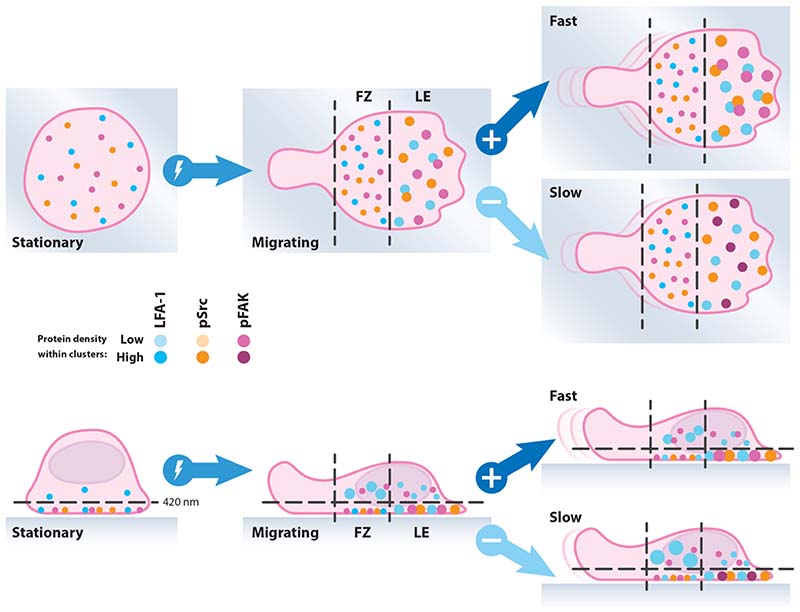
Figure 6. Schematic diagram of integrin nanoclustering in migrating T cells.
There is greater clustering of surface adhesion molecules at the front of migrating cells which become larger and more co-localised when those cells migrate rapidly. In contrast, a pool of large adhesion molecule clusters exists intracellularly above the focal zone, which become larger for slow moving cells.
Acknowledgements
This work was supported by ERC Starter Grant #337187, and Arthritis Research UK Programme grant 20525. We acknowledge use of the Nikon Imaging Centre (NIC), King’s College London. Male β-actinCreER/+; Lifeact-mEGFP/Y mice were a gift from Dr Karen Liu. For the STICS code and support, we acknowledge Paul W. Wiseman and Elvis Pandzic (McGill), with cell stationarity code written by Laurence Yolland and Brian Stramer (King’s College). iPALM imaging was done in collaboration with the Advanced Imaging Center at Janelia Research Campus, a facility jointly supported by the Gordon and Betty Moore Foundation, and the Howard Hughes Medical Institute. PTPN22 KO mice were a kind gift of Rose Zamoyska. Nanodiamonds were a kind gift of Keir Neumann, NIH. M.S. was supported by the King’s Bioscience Institute and the Guy’s and St Thomas’ Charity Prize PhD Programme in Biomedical and Translational Science.
References
- Alexandrova AY, et al. Comparative dynamics of retrograde actin flow and focal adhesions: formation of nascent adhesions triggers transition from fast to slow flow. [Accessed April 2, 2014];PloS one. 2008 3(9):e3234. doi: 10.1371/journal.pone.0003234. Available at: http://www.plosone.org/article/info:doi/10.1371/journal.pone.0003234#pone-0003234-g005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begovich AB, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. [Accessed May 31, 2014];American journal of human genetics. 2004 75(2):330–7. doi: 10.1086/422827. Available at: http://www.sciencedirect.com/science/article/pii/S0002929707624161 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottini N, et al. Role of PTPN22 in type 1 diabetes and other autoimmune diseases. [Accessed June 3, 2014];Seminars in immunology. 2006 18(4):207–13. doi: 10.1016/j.smim.2006.03.008. Available at: http://www.sciencedirect.com/science/article/pii/S1044532306000455 . [DOI] [PubMed] [Google Scholar]
- Brand O, Gough S, Heward J. <em>HLA</em>, <em>CTLA-4</em> and <em>PTPN22</em>: the shared genetic master-key to autoimmunity? [Accessed July 29, 2014];Expert Reviews in Molecular Medicine. 2005 7(23):1–15. doi: 10.1017/S1462399405009981. Available at: http://journals.cambridge.org/abstract_S1462399405009981 . [DOI] [PubMed] [Google Scholar]
- Brown CM, et al. Probing the integrin-actin linkage using high-resolution protein velocity mapping. [Accessed April 1, 2014];Journal of cell science. 2006 119(Pt 24):5204–14. doi: 10.1242/jcs.03321. Available at: http://jcs.biologists.org/content/119/24/5204.short . [DOI] [PubMed] [Google Scholar]
- Burn GL, et al. Superresolution imaging of the cytoplasmic phosphatase PTPN22 links integrin-mediated T cell adhesion with autoimmunity. [Accessed November 13, 2017];Science signaling. 2016 9(448):ra99. doi: 10.1126/scisignal.aaf2195. Available at: http://www.ncbi.nlm.nih.gov/pubmed/27703032 . [DOI] [PubMed] [Google Scholar]
- Burn GL, et al. Why is PTPN22 a good candidate susceptibility gene for autoimmune disease? [Accessed April 16, 2014];FEBS letters. 2011 585(23):3689–98. doi: 10.1016/j.febslet.2011.04.032. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21515266 . [DOI] [PubMed] [Google Scholar]
- Case LB, Waterman CM. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. [Accessed July 1, 2015];Nature cell biology. 2015 17(8):955–963. doi: 10.1038/ncb3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CE, Odde DJ. Traction dynamics of filopodia on compliant substrates. [Accessed December 17, 2014];Science (New York, N.Y.) 2008 322(5908):1687–91. doi: 10.1126/science.1163595. Available at: http://www.sciencemag.org/content/322/5908/1687.abstract . [DOI] [PubMed] [Google Scholar]
- Chang H, et al. A unique series of reversibly switchable fluorescent proteins with beneficial properties for various applications. [Accessed April 19, 2018];Proceedings of the National Academy of Sciences of the United States of America. 2012 109(12):4455–60. doi: 10.1073/pnas.1113770109. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22375034 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changede R, et al. Nascent Integrin Adhesions Form on All Matrix Rigidities after Integrin Activation. Developmental Cell. 2015;35(5):614–621. doi: 10.1016/j.devcel.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Chapman NM, Houtman JCD. Functions of the FAK family kinases in T cells: beyond actin cytoskeletal rearrangement. [Accessed July 24, 2017];Immunologic research. 2014 59(1–3):23–34. doi: 10.1007/s12026-014-8527-y. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24816556 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B-C, et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. [Accessed October 23, 2014];Science. 2014 346(6208):1257998. doi: 10.1126/science.1257998. Available at: http://www.sciencemag.org/content/346/6208/1257998.abstract . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, et al. The integrin-ligand interaction regulates adhesion and migration through a molecular clutch. [Accessed February 24, 2015];PloS one. 2012 7(7):e40202. doi: 10.1371/journal.pone.0040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comrie WA, Babich A, Burkhardt JK. F-actin flow drives affinity maturation and spatial organization of LFA-1 at the immunological synapse. [Accessed February 10, 2015];The Journal of Cell Biology. 2015 208(4):475–91. doi: 10.1083/jcb.201406121. Available at: http://www.ncbi.nlm.nih.gov/pubmed/25666810 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, et al. A disease-associated PTPN22 variant promotes systemic autoimmunity in murine models. [Accessed February 23, 2017];Journal of Clinical Investigation. 2013 123(5):2024–2036. doi: 10.1172/JCI66963. Available at: http://www.ncbi.nlm.nih.gov/pubmed/23619366 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dransfield I, et al. Divalent cation regulation of the function of the leukocyte integrin LFA-1. [Accessed June 26, 2015];The Journal of cell biology. 1992 116(1):219–26. doi: 10.1083/jcb.116.1.219. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2289255&tool=pmcentrez&rendertype=abstract . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R, et al. The integrin LFA-1 signals through ZAP-70 to regulate expression of high-affinity LFA-1 on T lymphocytes. [Accessed June 15, 2015];Blood. 2011 117(12):3331–42. doi: 10.1182/blood-2010-06-289140. Available at: http://www.bloodjournal.org/content/117/12/3331.abstract . [DOI] [PubMed] [Google Scholar]
- Giannoni E, et al. Lymphocyte function-associated antigen-1-mediated T cell adhesion is impaired by low molecular weight phosphotyrosine phosphatase-dependent inhibition of FAK activity. [Accessed March 1, 2019];The Journal of biological chemistry. 2003 278(38):36763–76. doi: 10.1074/jbc.M302686200. Available at: http://www.ncbi.nlm.nih.gov/pubmed/12815062 . [DOI] [PubMed] [Google Scholar]
- Goddette DW, Frieden C. Actin polymerization. The mechanism of action of cytochalasin D. [Accessed August 17, 2017];The Journal of biological chemistry. 1986 261(34):15974–80. Available at: http://www.ncbi.nlm.nih.gov/pubmed/3023337 . [PubMed] [Google Scholar]
- Goult BT, et al. RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover. [Accessed August 25, 2016];The Journal of biological chemistry. 2013 288(12):8238–49. doi: 10.1074/jbc.M112.438119. Available at: http://www.ncbi.nlm.nih.gov/pubmed/23389036 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffié J, et al. 3D Bayesian cluster analysis of super-resolution data reveals LAT recruitment to the T cell synapse. [Accessed September 7, 2017];Scientific reports. 2017 7(1):4077. doi: 10.1038/s41598-017-04450-w. Available at: http://www.nature.com/articles/s41598-017-04450-w . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffié J, et al. A Bayesian cluster analysis method for single-molecule localization microscopy data. [Accessed March 6, 2017];Nature Protocols. 2016 11(12):2499–2514. doi: 10.1038/nprot.2016.149. [DOI] [PubMed] [Google Scholar]
- Gunkel M, et al. Integrated and correlative high-throughput and super-resolution microscopy. [Accessed April 19, 2018];Histochemistry and Cell Biology. 2014 141(6):597–603. doi: 10.1007/s00418-014-1209-y. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24647616 . [DOI] [PubMed] [Google Scholar]
- Havrylenko S, et al. Extending the molecular clutch beyond actin-based cell motility. [Accessed February 25, 2015];New journal of physics. 2014 16(10):105012. doi: 10.1088/1367-2630/16/10/105012. Available at: http://stacks.iop.org/1367-2630/16/i=10/a=105012 . [DOI] [PubMed] [Google Scholar]
- Hebert B, Costantino S, Wiseman PW. Spatiotemporal image correlation spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. [Accessed March 26, 2014];Biophysical journal. 2005 88(5):3601–14. doi: 10.1529/biophysj.104.054874. Available at: http://www.sciencedirect.com/science/article/pii/S0006349505734093 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg N, et al. How T cells use LFA-1 to attach and migrate. [Accessed October 30, 2017];Immunology Letters. 2004 92(1–2):51–54. doi: 10.1016/j.imlet.2003.10.014. Available at: http://www.ncbi.nlm.nih.gov/pubmed/15081527 . [DOI] [PubMed] [Google Scholar]
- Holden SJ, et al. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. [Accessed April 19, 2018];Proceedings of the National Academy of Sciences of the United States of America. 2014 111(12):4566–71. doi: 10.1073/pnas.1313368111. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24616530 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hons M, et al. Chemokines and integrins independently tune actin flow and substrate friction during intranodal migration of T cells. [Accessed July 10, 2018];Nature Immunology. 2018 19(6):606–616. doi: 10.1038/s41590-018-0109-z. Available at: http://www.nature.com/articles/s41590-018-0109-z . [DOI] [PubMed] [Google Scholar]
- Hu K, et al. Differential transmission of actin motion within focal adhesions. [Accessed July 9, 2014];Science (New York, N.Y.) 2007 315(5808):111–5. doi: 10.1126/science.1135085. Available at: http://www.sciencemag.org/content/315/5808/111.full . [DOI] [PubMed] [Google Scholar]
- Huang F, et al. Simultaneous multiple-emitter fitting for single molecule super-resolution imaging. [Accessed July 18, 2017];Biomedical optics express. 2011 2(5):1377–93. doi: 10.1364/BOE.2.001377. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21559149 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi M, et al. Integrin LFA-1 regulates cell adhesion via transient clutch formation. [Accessed November 13, 2017];Biochemical and Biophysical Research Communications. 2015 464(2):459–466. doi: 10.1016/j.bbrc.2015.06.155. Available at: http://www.ncbi.nlm.nih.gov/pubmed/26143530 . [DOI] [PubMed] [Google Scholar]
- Jurado C, Haserick JR, Lee J. Slipping or gripping? Fluorescent speckle microscopy in fish keratocytes reveals two different mechanisms for generating a retrograde flow of actin. [Accessed March 20, 2014];Molecular biology of the cell. 2005 16(2):507–18. doi: 10.1091/mbc.E04-10-0860. Available at: http://www.molbiolcell.org/content/16/2/507.short . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. [Accessed August 9, 2017];Nature. 2010 468(7323):580–584. doi: 10.1038/nature09621. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21107430 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt EG, Schlaepfer DD. Focal adhesion kinase signaling in unexpected places. [Accessed May 16, 2017];Current Opinion in Cell Biology. 2017 45:24–30. doi: 10.1016/j.ceb.2017.01.003. Available at: http://www.sciencedirect.com/science/article/pii/S0955067417300145 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson C, et al. FAK promotes recruitment of talin to nascent adhesions to control cell motility. [Accessed May 16, 2017];Journal of Cell Biology. 2012 196(2):223–232. doi: 10.1083/jcb.201108078. Available at: http://jcb.rupress.org/content/196/2/223 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TI, et al. Measuring Integrin Conformational Change on the Cell Surface with Super-Resolution Microscopy. [Accessed May 7, 2018];Cell Reports. 2018 22(7):1903–1912. doi: 10.1016/j.celrep.2018.01.062. Available at: http://www.ncbi.nlm.nih.gov/pubmed/29444440 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader GPF, Ezratty EJ, Gundersen GG. FAK, talin and PIPKIγ regulate endocytosed integrin activation to polarize focal adhesion assembly. [Accessed September 4, 2017];Nature Cell Biology. 2016 18(5):491–503. doi: 10.1038/ncb3333. Available at: http://www.ncbi.nlm.nih.gov/pubmed/27043085 . [DOI] [PubMed] [Google Scholar]
- Nordenfelt P, Elliott HL, Springer TA. Coordinated integrin activation by actin-dependent force during T-cell migration. [Accessed February 8, 2018];Nature Communications. 2016 7:13119. doi: 10.1038/ncomms13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovesný M, et al. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. [Accessed June 3, 2015];Bioinformatics (Oxford, England) 2014 30(16):2389–90. doi: 10.1093/bioinformatics/btu202. Available at: http://bioinformatics.oxfordjournals.org/content/30/16/2389.short . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson H, et al. Spatial mapping of affinity changes for the integrin LFA-1 during cell migration using clusters identified based on local density. [Accessed October 9, 2018];Journal of Biophotonics. 2018 :e201800080. doi: 10.1002/jbio.201800080. Available at: http://www.ncbi.nlm.nih.gov/pubmed/30267470 . [DOI] [PubMed] [Google Scholar]
- Ponti A, et al. Two distinct actin networks drive the protrusion of migrating cells. [Accessed March 20, 2014];Science (New York, N.Y.) 2004 305(5691):1782–6. doi: 10.1126/science.1100533. Available at: http://www.ncbi.nlm.nih.gov/pubmed/15375270 . [DOI] [PubMed] [Google Scholar]
- Raab M, et al. LFA-1 activates focal adhesion kinases FAK1/PYK2 to generate LAT-GRB2-SKAP1 complexes that terminate T-cell conjugate formation. [Accessed February 8, 2018];Nature Communications. 2017a 8:16001. doi: 10.1038/ncomms16001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M, et al. LFA-1 activates focal adhesion kinases FAK1/PYK2 to generate LAT-GRB2-SKAP1 complexes that terminate T-cell conjugate formation. [Accessed July 24, 2017];Nature Communications. 2017b 8:16001. doi: 10.1038/ncomms16001. Available at: http://www.ncbi.nlm.nih.gov/pubmed/28699640 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, et al. Cell Migration: Integrating Signals from Front to Back. [Accessed August 17, 2017];Science. 2003 302(5651) doi: 10.1126/science.1092053. Available at: http://science.sciencemag.org/content/302/5651/1704.full . [DOI] [PubMed] [Google Scholar]
- Rose DM, et al. Paxillin Binding to the 4 Integrin Subunit Stimulates LFA-1 (Integrin L 2)-Dependent T Cell Migration by Augmenting the Activation of Focal Adhesion Kinase/Proline-Rich Tyrosine Kinase-2. [Accessed April 9, 2015];The Journal of Immunology. 2003 170(12):5912–5918. doi: 10.4049/jimmunol.170.12.5912. Available at: http://www.jimmunol.org/content/170/12/5912.full . [DOI] [PubMed] [Google Scholar]
- Rossy J, et al. Method for co-cluster analysis in multichannel single-molecule localisation data. [Accessed November 24, 2014];Histochemistry and cell biology. 2014 141(6):605–12. doi: 10.1007/s00418-014-1208-z. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24643361 . [DOI] [PubMed] [Google Scholar]
- Rubin-Delanchy P, et al. Bayesian cluster identification in single-molecule localization microscopy data. [Accessed November 13, 2017];Nature Methods. 2015 12(11):1072–1076. doi: 10.1038/nmeth.3612. [DOI] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) [Accessed August 9, 2017];Nature Methods. 2006 3(10):793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Martin L, et al. Signaling through the Leukocyte Integrin LFA-1 in T Cells Induces a Transient Activation of Rac-1 That Is Regulated by Vav and PI3K/Akt-1. [Accessed June 28, 2015];Journal of Biological Chemistry. 2004 279(16):16194–16205. doi: 10.1074/jbc.M400905200. Available at: http://www.ncbi.nlm.nih.gov/pubmed/14960575 . [DOI] [PubMed] [Google Scholar]
- Shannon M, Owen DM. Bridging the Nanoscopy-Immunology Gap. [Accessed February 18, 2019];Frontiers in Physics. 2019 6:157. doi: 10.3389/fphy.2018.00157/full. [DOI] [Google Scholar]
- Shannon MJ, et al. Protein clustering and spatial organization in T-cells. [Accessed May 31, 2015];Biochemical Society transactions. 2015 43(3):315–21. doi: 10.1042/BST20140316. Available at: http://www.biochemsoctrans.org/content/43/3/315.abstract . [DOI] [PubMed] [Google Scholar]
- Shtengel G, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. [Accessed July 13, 2017];Proceedings of the National Academy of Sciences of the United States of America. 2009 106(9):3125–30. doi: 10.1073/pnas.0813131106. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19202073 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, et al. A talin-dependent LFA-1 focal zone is formed by rapidly migrating T lymphocytes. [Accessed February 9, 2015];The Journal of cell biology. 2005 170(1):141–51. doi: 10.1083/jcb.200412032. Available at: http://jcb.rupress.org/content/170/1/141.short . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, et al. The role of the integrin LFA-1 in T-lymphocyte migration. [Accessed April 21, 2015];Immunological reviews. 2007 218:135–46. doi: 10.1111/j.1600-065X.2007.00537.x. Available at: http://www.ncbi.nlm.nih.gov/pubmed/17624950 . [DOI] [PubMed] [Google Scholar]
- Sun Z, Lambacher A, Fässler R. Nascent Adhesions: From Fluctuations to a Hierarchical Organization. Current Biology. 2014;24(17):R801–R803. doi: 10.1016/j.cub.2014.07.061. [DOI] [PubMed] [Google Scholar]
- Svensson L, et al. Lyp/PTPN22 is a negative regulator of integrin mediated T cell adhesion and migration; the disease associated PTPN22 allelic variant is a loss of function mutant that perturbs T cell migration. [Accessed July 4, 2014];Annals of the Rheumatic Diseases. 2011 70(Suppl 2):A7–A7. Available at: http://ard.bmj.com/content/70/Suppl_2/A7.1.short. [Google Scholar]
- Swaminathan V, Fischer RS, Waterman CM. The FAK-Arp2/3 interaction promotes leading edge advance and haptosensing by coupling nascent adhesions to lamellipodia actin. [Accessed August 30, 2017];Molecular biology of the cell. 2016 27(7):1085–100. doi: 10.1091/mbc.E15-08-0590. Available at: http://www.ncbi.nlm.nih.gov/pubmed/26842895 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijeira A, et al. T Cell Migration from Inflamed Skin to Draining Lymph Nodes Requires Intralymphatic Crawling Supported by ICAM-1/LFA-1 Interactions. [Accessed July 20, 2017];Cell Reports. 2017 18(4):857–865. doi: 10.1016/j.celrep.2016.12.078. Available at: http://linkinghub.elsevier.com/retrieve/pii/S2211124716318009 . [DOI] [PubMed] [Google Scholar]
- Tiwari DK, et al. A fast- and positively photoswitchable fluorescent protein for ultralow-laser-power RESOLFT nanoscopy. [Accessed September 14, 2017];Nature Methods. 2015 12(6):515–518. doi: 10.1038/nmeth.3362. Available at: http://www.ncbi.nlm.nih.gov/pubmed/25894946 . [DOI] [PubMed] [Google Scholar]
- Valignat M-P, et al. T lymphocytes orient against the direction of fluid flow during LFA-1-mediated migration. [Accessed May 30, 2014];Biophysical journal. 2013 104(2):322–31. doi: 10.1016/j.bpj.2012.12.007. Available at: http://www.sciencedirect.com/science/article/pii/S0006349512051181 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CA, Theriot JA. A correlation-based approach to calculate rotation and translation of moving cells. [Accessed March 6, 2017];IEEE Transactions on Image Processing. 2006 15(7):1939–1951. doi: 10.1109/tip.2006.873434. Available at: http://ieeexplore.ieee.org/document/1643701/ [DOI] [PubMed] [Google Scholar]
- Wiseman PW, et al. Spatial mapping of integrin interactions and dynamics during cell migration by image correlation microscopy. [Accessed March 29, 2014];Journal of cell science. 2004 117(Pt 23):5521–34. doi: 10.1242/jcs.01416. Available at: http://jcs.biologists.org/content/117/23/5521.long . [DOI] [PubMed] [Google Scholar]
- Yasui M, et al. Automated single-molecule imaging in living cells. [Accessed September 4, 2018];Nature Communications. 2018 9(1):3061. doi: 10.1038/s41467-018-05524-7. Available at: http://www.nature.com/articles/s41467-018-05524-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, et al. Highly Multiplexed, Super-resolution Imaging of T Cells Using madSTORM. [Accessed September 14, 2017];Journal of Visualized Experiments. 2017 (124):e55997–e55997. doi: 10.3791/55997. Available at: https://www.jove.com/video/55997/highly-multiplexed-super-resolution-imaging-of-t-cells-using-madstorm . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, et al. madSTORM: a superresolution technique for large-scale multiplexing at single-molecule accuracy. Molecular biology of the cell. 2016a;27(22):3591–3600. doi: 10.1091/mbc.E16-05-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, et al. madSTORM: a superresolution technique for large-scale multiplexing at single-molecule accuracy. [Accessed May 25, 2017];Molecular biology of the cell. 2016b 27(22):3591–3600. doi: 10.1091/mbc.E16-05-0330. Available at: http://www.ncbi.nlm.nih.gov/pubmed/27708141 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. Highly photostable, reversibly photoswitchable fluorescent protein with high contrast ratio for live-cell superresolution microscopy. [Accessed April 19, 2018];Proceedings of the National Academy of Sciences of the United States of America. 2016 113(37):10364–9. doi: 10.1073/pnas.1611038113. Available at: http://www.ncbi.nlm.nih.gov/pubmed/27562163 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang H. Integrin signalling and function in immune cells. [Accessed November 26, 2013];Immunology. 2012 135(4):268–75. doi: 10.1111/j.1365-2567.2011.03549.x. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3372743&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.