Abstract
Flaviviruses are arthropod-borne viruses, several of which represent emerging or re-emerging pathogens responsible for widespread infections with consequences ranging from asymptomatic seroconversion to severe clinical diseases and congenital developmental deficits. This variability is due to multiple factors including host genetic determinants the role of which has been investigated in mouse models and human genetic studies. In this review, we provide an overview of the host genes and variants which modify susceptibility or resistance to major mosquito-borne flaviviruses infections in mice and humans.
Keywords: Flavivirus, host genetics, mouse, human, innate immune response, HLA
Introduction
Flaviviruses constitute a large genus of arthropod-borne viruses, several of which represent emerging or re-emerging pathogens. Important members of this genus include West Nile (WNV), dengue (DENV), Zika (ZIKV), Japanese encephalitis (JEV) and yellow fever (YFV) viruses, all of which are transmitted by mosquitoes. In humans, flaviviruses infections can remain asymptomatic, trigger flu-like symptoms, or progress towards severe complications such as encephalitis, hemorrhagic fever or, in the case of ZIKV, Guillain-Barré syndrome and congenital brain developmental deficits. Because of the rapid progression of these infections, the innate immune response plays a key role in the quick control of viral multiplication and dissemination (Suthar et al. 2013). Flaviviruses cell entry and genome replication trigger multiple sensing events, activation of antiviral effectors through the type I interferon (IFN) pathway, cellular stress reaction and inflammation (Valadao et al. 2016). Like other viruses, they have evolved a variety of mechanisms to block the IFN pathway at different steps through interactions between their non-structural proteins (Cedillo-Barrón et al. 2018), in particular NS5 (Best 2017; Laurent-Rolle et al. 2010), and molecular components of this pathway (Cumberworth et al. 2017; Wu et al. 2017).
Mouse models have been developed to study the pathophysiology of mosquito-borne flavivirus infections, to model the complications observed in humans (in particular encephalitis and neuroinvasive disease, but also intrauterine growth restriction and fetal demise caused by ZIKV infection during pregnancy) and to test novel preventive and therapeutic countermeasures (Julander and Siddharthan 2017). However, while most laboratory strains of mice are naturally susceptible to WNV (Mashimo et al. 2002) and JEV (Wang and Deubel 2011), their infection with DENV fails to elicit overt signs of disease (Zellweger and Shresta 2014) and they are somewhat refractory to ZIKV with the exception of very young mice or genetically manipulated strains with immune deficit (Julander and Siddharthan 2017). In humans, DENV and ZIKV inhibit type I IFN response by STING cleavage (Aguirre et al. 2012; Ding et al. 2018) and by NS5-induced STAT2 degradation (Best 2017; Grant et al. 2016), but these restriction mechanisms are inefficient in mice (Best 2017; Ding et al. 2018; Miorin et al. 2017). Efficient infection is obtained in mice genetically deficient in the receptors for type I (encoded by the Ifnar1 and Ifnar2 genes) and/or type II (Ifngr1 and Ifngr2 genes) IFNs. Alternatively, the type I IFN receptor can be blocked pharmacologically by anti-IFNAR antibody injection prior to infection (Sheehan et al. 2006; Sheehan et al. 2015). Recently, an immunocompetent mouse model for ZIKV infection has been produced by infecting with a mouse-adapted ZIKV strain mice in which the Stat2 gene had been replaced with its human version (Gorman et al. 2018).
The mechanisms underlying the variable severity of symptoms in human patients and between mouse inbred strains remain poorly understood, although the viral strain and inoculum are obvious contributing factors. Host microbiota can also influence susceptibility to flaviviruses, as shown in mice by oral antibiotic treatment (Thackray et al. 2018). Host genetic determinants affect the susceptibility of humans or animal species to infections. In this review, we will summarize the evidence demonstrating the role of host genes in the susceptibility or resistance to flaviviruses in mice and humans, with emphasis on innate immunity. We will focus on infections caused by WNV, DENV, ZIKV, JEV and YFV. While many host genes have been shown to interfere with virus biology in cultured cells, we will consider only genes for which variants have been associated with differential clinical severity. Many mouse studies have focused on WNV, while human studies investigated mostly susceptibility to highly prevalent DENV, thus limiting comparisons between the two species.
Mouse models
In mice, the effect of specific genetic variants has been identified either through a genome-wide, forward genetic approach (from phenotypic variation to underlying gene variants), or by testing the consequences of genetic ablation of specific genes chosen from their function (reverse genetic approach). Table 1 presents a cross-compilation of all mouse genes which have been shown to influence clinical severity and/or lethality after infection with WNV, DENV, ZIKV, YFV or JEV. The corresponding experimental details, results and references are provided in Table 2.
Table 1.
List of mouse genes which have been shown to affect clinical outcome and/or mortality following infection with WNV, DENV, ZIKV, YFV and JEV. Details of supporting studies are given in Table 2.
| Gene (synonym) | WNV | DENV | ZIKV | YFV | JEV |
|---|---|---|---|---|---|
| Atg16l1 | X | ||||
| Ccr2 | X | X | |||
| Ccr4 | X | ||||
| Ccr7 | X | ||||
| Clec5a | X | ||||
| Cxcl10 | X | X | |||
| Cxcr3 | X | X | |||
| Ddx58 (RIG-I) | X | ||||
| Dhx58 (LGP2) | X | ||||
| Ifih1 (MDA5) | X | ||||
| Ifi27l2a | X | ||||
| Ifit2 | X | ||||
| Ifitm3 | X | ||||
| Ifna | X | ||||
| Ifnb | X | ||||
| Ifng | X | ||||
| Ifnar1 | X | X | X | X | |
| Ifngr1 | X | X | X | X | |
| Ifnlr1 | X | X | X | ||
| Il10 | X | ||||
| Il12a | X | ||||
| Il12b | X | ||||
| Il23a | X | ||||
| Irf1 | X | X | |||
| Irf3 | X | X | X | ||
| Irf5 | X | X | X | ||
| Irf7 | X | X | X | ||
| Isg15 | X | ||||
| Mavs (IPS-1) | X | ||||
| Mb21d1 (cGas) | X | ||||
| Myd88 | X | ||||
| Oas1b | X | ||||
| Rag1 | X | X | |||
| Rag2 | X | ||||
| Rnasel | X | ||||
| Rsad2 | X | ||||
| Sema7a | X | ||||
| Stat1 | X | X | X | X | |
| Stat2 | X | X | |||
| Tlr3 | X | ||||
| Tlr7 | X | ||||
| Tnfrsf9 | X |
Table 2. Detailed information on mouse genetic studies supporting the role of specific genes on susceptibility or resistance to flaviviruses.
| Genes identified by forward genetics approaches | ||||||
| Oas1b | Activation of RNA decay pathway by dsRNA recognition | WNV | Susceptible BALB/cByJ, C57BL/6J vs resistant MAI/Pas and MBT/Pas, backcrosses and congenic strains | WNV strain IS-98-ST1, 1000 ffu ip, 6-wk-old mice | Most laboratory inbred mouse strains carry a non-sense, loss of function mutation making them highly susceptible. | Mashimo et al. 2002 |
| WNV | MEFs from susceptible C3H/He and resistant C3H.PRI-Flyr congenic strain | WNV strain Eg101. In vitro infection of MEFs transfected with cDNAs of candidate genes | C3H.PRI-Flvr congenic strain carries a nonsense mutation resulting in higher viral replication in MEFs. | Perelygin et al. 2002 | ||
| WNV | Collaborative Cross (CC) and F1 crosses of CC mice | WNV strain TX-2002-HC, 102 pfu footpad, 8-10-wk-old mice | Oas1b plays a role in susceptibility to WNV but a wide range of WNV-associated phenotypes are observed for a given Oas1b allele. | Graham et al. 2015 | ||
| Statl | Promotes transcription of ISG and IFN subtypes | WNV | Backcross between C57BL/6-MHCII-Aα/β-/- and C3H | WNV strain Kunjin, 103 pfu footpad, 8-wk-old mice | Highly susceptible MHCII-Aα/β-/- mice have acquired a spontaneous amino-acid substitution mutation. | Larena et al. 2017 |
| Genes identified by their function in host immune responses | ||||||
| Ccr2 | Chemokine receptor expressed on Ly6chi inflammatory monocytes and other leukocyte subtypes | WNV | C57BL/6J WT and Ccr2 KO | WNV strain NY99-35262, 100 pfu sc, 8-12-wk-old mice | Increased mortality, large and selective reduction of Ly6chi monocyte accumulation in the brain, sustained monocytopenia. | Lim et al. 2011 |
| Ccr7 | Chemokine receptor expressed by numerous cell types, which regulates the homing of T cells into lymphoid organs | WNV | C57BL/6J WT and Ccr7 KO | WNV strain NY99, 104 pfu footpad, 8-12-wk-old mice | Increased mortality and CNS viral loads, leukocyte accumulation in the CNS with neuroinflammation and reduced viral clearance. | Bardina et al. 2017 |
| Cxcl10 | Chemokine of the CXC subfamily which binds to CXCR3 receptor | WNV | C57BL/6J WT and Cxcl10 KO | WNV strain NY99, 104 pfu footpad, 5-9-wk-old mice | Enhanced morbidity and mortality, increased viral burden in the brain, decreased CXCR3+CD8+ T-cell trafficking | Klein et al. 2005 |
| Cxcr3 | Receptor for chemokines CXCL9, CXCL10 and CXCL11 which stimulates leukocyte trafficking | WNV | C57BL/6J WT and Cxcr3 KO | WNV strain 3000.0259, 102 pfu footpad, 5-8-wk-old mice | Enhanced mortality with increased viral burden and reduced CD8+ T cell trafficking specifically in the cerebellum. | Zhang et al. 2008 |
| Ddx58 (RIG-I) | Cytoplasmic sensor of viral ssRNA and dsRNA | WNV | WT and Ddx58 KO (mixed genetic background) | WNV isolate TX 2002-HC, 100 pfu footpad, 6-12-wk-old mice | Increased mortality. Decreased innate immune signaling and virus control in MEFs. Ddx58-Ifih1 double KO are extremely susceptible. | Errett et al. 2013 |
| Dhx58 (LGP2) | Cytoplasmic sensor of viral RNA and inhibitor of antiviral RNAi by association with Dicer | WNV | C57BL/6J WT and Dhx58 KO | WNV isolate TX 2002-HC, 100 pfu footpad, 6-12-wk-old mice | Increased mortality despite activation of innate immune response. LGP2 regulates CD8+ T cell survival and effector functions. | Suthar et al. 2012 |
| C57BL/6J WT and Ifih1 KO | WNV strain 3000.0259, 102-104 pfu footpad, 9-12-wk-old mice | Reduced survival. Increased viral burden primarily in the CNS. | Lazear et al. 2013 | |||
| Ifi27l2a | ISG expressed at higher levels in granule cell neurons than in cortical neurons | WNV | C57BL/6J WT and Ifi27l2a KO | WNV strain 3000.0259, 100 pfu footpad, 8-12-wk-old mice | Increased susceptibility. Viral burden higher only in the CNS, associated with reduced cell death. | Lucas et al. 2016 |
| Ifit2 | Aka ISG54, member of the IFN-induced proteins with tetratricopeptide repeats | WNV | C57BL/6J WT and Ifit2 KO | WNV strain 3000.0259, 100 pfu footpad or 10 pfu ic, 8-10-wk-old mice | Increased susceptibility with higher replication in the brain. | Cho et al. 2013 |
| Ifitm3 | ISG inhibiting virus entry and preventing viral fusion and release of viral contents into the cytosol | WNV | C57BL/6J WT and Ifitm3 KO | WNV NY1999, 100 pfu sc, 8-9-wk-old mice | Increased susceptibility with higher viral burden in peripheral organs and CNS. Decreased B-cells, CD4+ T cells and Ag-specific CD8+ T cells. | Gorman et al. 2016 |
| Ifna | Interferon-α(type I) with multiple members | WNV | C57BL/6J treated with anti-IFN-αmAb | WNV strain 3000.0259, 100 pfu footpad or 10 pfu ic, 8-12-wk-old mice | Increased lethality when mAb injected one day prior and two days following WNV infection. | Sheehan et al. 2015 |
| Ifnb | Interferon-β(type I) | WNV | C57BL/6J WT and Ifnb KO | WNV strain 3000.0259, 100 pfu footpad or 10 pfu ic, 8-12-wk-old mice | Increased mortality with enhanced viral replication in peripheral tissues and the CNS. | Lazear et al. 2011 |
| Ifng | Interferon-γ(type II) | WNV | C57BL/6J WT and Ifng KO | WNV strain 3000.0259, 100 pfu footpad or 10 pfu ic, 8-12-wk-old mice | Increased mortality with enhanced viremia and viral replication in lymphoid tissues, earlier detection of WNV the CNS. | Shrestha et al. 2006 |
| Ifnar1 | Receptor of type I (αand β) interferons | WNV | C57BL/6J WT and Ifnar1KO 129Sv/Ev WT and Ifnar1 KO |
WNV strain 3000.0259, 100 pfu footpad or 10 pfu ic, 8-10-wk-old mice | High susceptibility with death in 4 days and increased infection in macrophages, B cells, and T cells in the spleen. | Samuel and Diamond 2005 |
| Ifngr1 | Receptor of type II (γ) interferon | WNV | C57BL/6J WT and Ifng KO | WNV strain 3000.0259, 100 pfu footpad or 10 pfu ic, 8-12-wk-old mice | Increased mortality with enhanced viremia and viral replication in lymphoid tissues, earlier detection of WNV the CNS. | Shrestha et al. 2006 |
| Ifnlr1 | Receptor of type III (λ) interferon | WNV | C57BL/6J WT and Ifnlr1 KO | WNV strain 3000.0259, 100 pfu footpad, 5-12-wk-old mice | Increased viral load in the brain due to enhanced blood-brain barrier permeability. | Lazear et al. 2015 |
| Il10 | Pleiotropic cytokine with immunosuppressive properties | WNV | C57BL/6J WT and Il10 KO | WNV strain 2741, 2000 pfu ip, 7-8wk-old mice | Markedly reduced infection with increased production of antiviral cytokines. | Bai et al. 2009 |
| Il12a | p35 subunit of IL12 which activates NK cells and induces differentiation of CD4+ T-cells into IFN-γproducing Th1 cells. | WNV | C57BL/6J WT and Il12a KO | WNV strain 2741, 2000 pfu ip, 8-12wk-old mice | No difference in susceptibility with wildtype controls. | Town et al. 2009 |
| Il12b | p40 subunit of IL12 | WNV | C57BL/6J WT and Il12b KO | WNV strain 2741, 2000 pfu ip, 8-12wk-old mice | Increased mortality. | Town et al. 2009 |
| Il23a | Associates with IL12B to form IL23, which can activate STAT4 and stimulate the production of IFN-γ | WNV | C57BL/6J WT and Il23a KO | WNV strain 2741, 2000 pfu ip, 8-12wk-old mice | Increased mortality. | Town et al. 2009 |
| Irf1 | Transcriptional regulator of interferon stimulated genes | WNV | C57BL/6J WT and Irf1 KO | WNV strain 3000.0259, 100 pfu footpad, 8-12-wk-old mice | Increased mortality, elevated viral burdens in peripheral tissues and the CNS. | Brien et al. 2011 |
| Irf3 | Transcriptional regulator of interferon stimulated genes | WNV | C57BL/6J WT and Irf3 KO | WNV strain 3000.0259, 100 pfu footpad, 8-12-wk-old mice | Increased mortality, elevated viral burdens in peripheral tissues and the CNS. | Daffis et al. 2007 |
| Irf5 | Transcriptional regulator of interferon stimulated genes | WNV | C57BL/6J WT and Irf5 KO | WNV strain 3000.0259, 100 pfu footpad, 9-10-wk-old mice | Increased lethality with elevated infection in peripheral organs and the CNS. | Thackray et al. 2014 |
| Irf7 | Transcriptional regulator of interferon stimulated genes | WNV | C57BL/6J WT and Irf7 KO | WNV strain 3000.0259, 100 pfu footpad, 8-12-wk-old mice | Increased lethality with blunted systemic type I IFN response. Increased viral titers in primary macrophages, fibroblasts, dendritic cells, and cortical neurons. | Daffis et al. 2008b |
| Mavs (IPS-1) | Adaptor molecule of RIG-I and MDA5 | WNV | C57BL/6J WT and Mavs KO | WNV isolate TX 2002-HC, 100 pfu footpad, 6-12-wk-old mice | High susceptibility in 8 days with enhanced viral replication and dissemination (similar to Ddx58-Ifih1 double KO). |
Suthar et al. 2010
Errett et al. 2013 |
| Mb21d1 (cGas) | Cytosolic DNA sensor, cGMP-AMP synthase, activator of Tmem173 (STING) | WNV | C57BL/6J WT and Mb21d1 KO | WNV NY1999, 100 pfu sc, 8-9-wk-old mice | Increased mortality but no increase in brain viral load | Schoggins et al. 2014 |
| C57BL/6J WT and Myd88 KO | WNV strain 3000.0259, 100 pfu footpad, 8-10-wk-old mice | Increased mortality. Increased viral burden primarily in the CNS. | Szretter et al. 2010 | |||
| Rnasel | Ribonuclease L (2’,5’-OAS-dependent) | WNV | C57BL/6J WT and Rnasel KO | WNV strain 3000.0259, 100 pfu footpad or 10 pfu ic, 8-10-wk-old mice | Increased mortality (even higher if combined with Eif2ak2 (PKR) knockout). Increased viral burden in tissues and CNS. | Samuel et al. 2006 |
| Rsad2 | Aka viperin : ISG expressed in response to type I and II interferons. Modulates cholesterol and isoprenoid biosynthesis, and lipid raft formation. | WNV | C57BL/6J WT and Rsad2 KO | WNV strain 3000.0259, 100 pfu footpad or 10 pfu ic, 8-10-wk-old mice | Increased lethality. Modest increase of viral replication in macrophages. No difference in cortical neurons. | Szretter et al. 2011 |
| Sema7a | A membrane-associated/secreted protein involved in connecting the neuronal and immune systems. | WNV | C57BL/6J WT and Sema7a KO | WNV strain 2741, 1000 pfu ip, 6-8wk-old mice | Increased survival, reduced viral burden and blood-brain barrier permeability. | Sultana et al. 2012 |
| WNV strain 3000.0259, 100 pfu footpad, 8-12-wk-old mice | Increased mortality with higher viral burden in the CNS. | Daffis et al. 2008a | ||||
| Tlr7 | Recognition of ssRNA | WNV | C57BL/6J WT and Tlr7 KO | WNV strain 2741, 2000 pfu ip, 8-12wk-old mice | Increased mortality. Increased viral burden systemically and in the CNS. | Town et al. 2009 |
| Ccr2 | Chemokine receptor expressed on Ly6chi inflammatory monocytes and other leukocyte subtypes | DENV | C57BL/6J WT and Ccr2 KO | Mouse-adapted DENV-2 strain P23085, 20 pfu ip, 8-10-wk-old male mice | Enhanced survival associated with decreased liver damage, decreased cell activation and decreased cytokine storm. | Guabiraba et al. 2010 |
| Ccr4 | Chemokine receptor which regulates leukocytes trafficking | DENV | C57BL/6J WT and Ccr4 KO | Mouse-adapted DENV-2 strain P23085, 20 pfu ip, 8-10-wk-old male mice | Enhanced survival associated with decreased hemoconcentration, thrombocytopenia, liver damage, systemic inflammation and leukocyte activation. | Guabiraba et al. 2010 |
| Clec5a | C-type lectin which regulates cell adhesion, cell-cell signaling during immune response | DENV | Stat1 KO (unspecified genetic background) treated with anti-CLEC5A mAb | Mouse-adapted DENV-2 strain New Guinea C-N, 105pfu ip and ic simultaneously, unspecified mouse age | Decreased plasma leakage and TNF-α serum level without suppression of viral replication and overall reduced lethality. | Chen et al. 2008 |
| Cxcl10 | Chemokine of the CXC subfamily which binds to CXCR3 receptor | DENV | C57BL/6J WT and Cxcl10 KO (unspecified genetic background) | Mouse-adapted DENV-2 strain New Guinea C-N, 2.105 pfu ic, 6-8-wk-old mice | Increased mortality rate but unchanged number of infiltrating T cells in the brain. Cxcl10 KO mice tend to be more susceptible than Cxcr3 KO mice. |
Hsieh et al. 2006
Ip and Liao 2010 |
| Cxcr3 | Receptor for chemokines CXCL9, CXCL10 and CXCL11 which stimulates leukocyte trafficking | DENV | C57BL/6J WT and Cxcr3 KO (unspecified genetic background) | Mouse-adapted DENV-2 strain New Guinea C-N, 2.105 pfu ic, 6-8-wk-old mice | Increased mortality rate with high viral load associated with a decrease in CD8+ T cells in the brain. | Hsieh et al. 2006 |
| 129Sv/Ev WT and Ifnar1KO (also called A129 mice) | DENV-2 S221 strain, 1011’12 genomic equivalents (GE) iv, 5-6-wk-old mice | 100% morbidity with observation of systemic disease but no sign of limb paralysis. Mortality rate varies between 0% and 100% according to the inoculum dose. Viral replication is observed in blood, spleen and blood marrow. | Prestwood et al. 2012 | |||
| Ifnar1 / Ifngr1 | Receptors of type I (αand β) and type II (γ) IFNs | DENV | 129Sv/Ev WT and Ifnar1-Ifngr1 double KO (also called AG129 mice) | DENV-2 strain PL046, 105-6-7-8 pfu iv or mouse adapted DENV-1 strain Mochizuki, 4.104 pfu iv, 5-6-wk-old mice | Highly susceptible to both viral strains. Mortality rate varies between 0 and 100% depending on DENV-2 infectious dose. Clinical signs include limb paralysis. Viral replication is observed in blood, spleen, lymph nodes and CNS. |
Shresta et al. 2004
Prestwood et al. 2012 |
| Irf1 / Irf3 / Irf5 / Irf7 | Transcriptional regulators of IFN stimulated genes | DENV | C57BL/6J WT and Irf3-Irf5-Irf7 triple KO (TKO) C57BL/6J WT and Irf1-Irf3-Irf5-Irf7 quadruple KO (QKO) | DENV-2 S221 strain, 5.106 ffu iv, under ADE conditions, 5-6-wk-old male and female mice | Both TKO and QKO mice sustain viral replication but only QKO mice succumb to infection. DENV infection of TKO mice results in minimal type I IFN production but a robust type II IFN response. | Carlin et al. 2017 |
| Rag1 | Protein involved in activation of immunoglobulin V(D)J recombination during B and T cell development | DENV | C57BL/6J WT and Rag1 KO (deficient in B and T lymphocytes) | DENV-2 strain PL046, 105-6-7-8 pfu iv or mouse adapted DENV-1 strain Mochizuki, 4.104 pfu iv, 5-6-wk-old mice | Increased mortality rate with mouse-adapted DENV-1 strain but no detection of viral replication in peripheral organs. | Shresta et al. 2004 |
| Rag2 | Protein involved in the initiation of V(D)J recombination during B and T cell development | DENV | 129Sv/Ev WT and Rag2 KO (deficient in B and T lymphocytes) | DENV-2 strain PL046, 105-6-7-8 pfu iv or mouse adapted DENV-1 strain Mochizuki, 4.104 pfu iv, 5-6-wk-old mice | Increased mortality rate with mouse-adapted DENV-1 strain but no detection of viral replication in peripheral organs. | Shresta et al. 2004 |
| Stat1 | Promotes transcription of ISG and IFN subtypes | DENV | 129Sv/Ev WT and Stat1 KO | DENV-2 strain PL046, 107-8 pfu iv or mouse adapted DENV-1 strain Mochizuki, 4.104 pfu iv, 5-6-wk-old mice | Increased mortality rate with both DENV strains but viral replication is only transient (DENV is undetected at day 3 p.i. in various tissues). |
Shresta et al. 2005
Perry et al. 2011 |
| Stat2 | Promotes transcription of ISG and IFN subtypes | DENV | C57BL/6J WT and Stat2 KO | Mouse adapted DENV-2 D2S10 strain, 106 ffu iv, under ADE conditions, unspecified mouse age | Extended viral detection in spleen and lymph nodes but no mortality. | Perry et al. 2011 |
| Stat1 / Stat2 | Promotes transcription of ISG and IFN subtypes | DENV | 129Sv/Ev WT and Stat1-Stat2 double KO | DENV-2 S221 strain, 2.105 pfu iv, 5-6-wk-old mice | Highly susceptible, 100% morbidity and lethality contrary to single-deficient mice. | Perry et al. 2011 |
| Atg16l1 | Part of a protein complex necessary for autophagy | ZIKV | C57BL/6J WT and Atg16l1Hm (hypomorphic) treated with anti-IFNAR mAb | ZIKV strain Paraiba 2015 (Asian), 103 ffu sc, 8-10-wk-old pregnant female mice | Restricted ZIKV vertical transmission and placental/fetal damage. Overall improved placental and fetal outcomes. | Cao et al. 2017 |
| 129Sv/Ev WT and Ifnar1 KO (also called A129 mice) | ZIKV strain MP1751 (African) or PRVABC59 (Asian), 10 or 106 ffu sc, 6-8-wk-old male mice ZIKV strain FSS13025 (Asian), 105 ffu ip, 3-5-11-wk-old mice | Highly susceptible to the African strain, 100% morbidity and lethality. Viral dissemination to the brain, spleen, liver and testis. Susceptibility to Asian strain is age-dependent, only mice under 6 weeks of age display morbidity and lethality. Viral replication is observed in all cases in blood, brain, spleen, liver and testis. |
Dowall et al. 2017
Rossi et al. 2016 |
|||
| Ifnar1 / Ifngr1 | Receptors of type I (α and β) and type II (γ) IFNs | ZIKV | 129Sv/Ev WT and Ifnar1-Ifngr1 double KO (also called AG129 mice) | ZIKV strain PF13 (Asian), 105 pfu sc, 3-4-8-wk-old mice | Highly susceptible regardless of mouse age or infectious dose. 100%morbidity and mortality. Viral replication is observed in blood and organs including spleen, liver and CNS but is associated with severe pathological findings only in the brain and muscle. |
Aliota et al. 2016 Other results reviewed in Winkler and Peterson 2017 |
| Ifnlr1 | Receptor of type III (λ) interferon | ZIKV | C57BL/6J WT and Ifnlr1 KO | ZIKV strain Paraiba 2015 (Asian), 103 pfu footpad | Increased ZIKV replication in the placenta and fetus | Jagger et al. 2017 |
| Irf3 / Irf5 / Irf7 | Transcriptional regulators of IFN stimulated genes | ZIKV | C57BL/6J WT and Irf3-Irf5-Irf7 triple KO (TKO) | ZIKV strain MR766 (African) or PF13 (Asian), 102 ffu sc, 5-6-wk-old mice | Highly susceptible to both ZIKV strains, 100% morbidity and lethality. | Lazear et al. 2016 |
| Isg15 | Ubiquitin-like protein activated by type I IFNs | ZIKV | C57BL/6J WT and Isg15 KO | ZIKV strain PRVABC59 (Asian), 104 pfu intravitreally in the right eye, 4-6-wk-old male and female mice | Severe chorioretinitis associated with increased retinal cell death and higher ZIKV RNA and protein levels. | Singh et al. 2017 |
| Rag1 | Protein involved in activation of immunoglobulin V(D)J recombination during B and T cell development | ZIKV | C57BL/6J WT and Rag1 KO (deficient in B and T lymphocytes) treated with anti-IFNAR mAb | Mouse-adapted ZIKV strain Dakar 41519 (African), 106 ffu sc, 7-wk-old male mice ZIKV strain Paraiba (Asian), 104 pfu ip, 8-10-wk-old male mice | ZIKV replication is observed in male reproductive organs. Enhanced morbidity with signs of severe disease. Viral replication is observed at high level in the brain and testes. |
Govero et al. 2016
Winkler et al. 2017 |
| Stat1 | Promotes transcription of ISG and IFN subtypes | ZIKV | 129Sv/Ev WT and Stat1KO (unspecified genetic background) | ZIKV strain MR766 (African), 104 pfu sc, 7-8-wk-old male mice | Highly susceptible, 100% morbidity and lethality. High viral load in the blood and in the brain. | Kamiyama et al. 2017 |
| Stat2 | Promotes transcription of ISG and IFN subtypes | ZIKV | C57BL/6J WT and Stat2 KO | ZIKV strain MR766 (African), 103 ffu sc, 5-6-wk-old female mice | Highly susceptible, 100% morbidity with neurological symptoms and 100% lethality. High viral loads in the CNS, gonads, spleen and liver. | Tripathi et al. 2017 |
| C57BL/6J and Ifnar1 KO | YFV 17D vaccine strain, 104 pfu ip, 3-4wk-old mice | Viscerotropic disease with mortality by ip but not by im, footpad or sc routes. | Erickson and Pfeiffer 2015 | |||
| Ifngr1 | Receptor of type III (γ) interferon | YFV | 129 and Ifngr1 KO | YFV Asibi and Angola wildtype strains, 104 pfu footpad and YFV 17D-204 vaccine strain, 106 pfu footpad, 3-4wk-old mice | Resistant to YFV wildtype and 17D vaccine strains. | Meier et al. 2009 |
| Ifnar1 / Ifngr1 | Receptors of type I (α and β) and type II (γ) interferons | YFV | 129 and Ifnar1-Ifngr1 double KO | 17D-204 vaccine strain, 106pfu footpad, 3-4wk-old 17D-204 vaccine strain, 2.105 pfu ip, 7-8-wk-old mice | Highly susceptible to 17D-204 vaccine strain. Highly susceptible to 17D-204 vaccine strain, with neurotropic and viscerotropic infection (high viral titers in brain and liver). |
Meier et al. 2009
Thibodeaux et al. 2012 |
| Ifnlr1 | Receptor of type III (λ) interferon | YFV | C57BL/6J WT and Ifnlr1 KO | 17D vaccine strain, 106-7 pfu iv, 2-6mo-old | KO resistant to vaccinal strain 17D like Ifnar1/2mice, but mice with double Ifnar1/2 and Inflr1KO highly susceptible. | Douam et al. 2017 |
| Stat1 | Promotes transcription of ISG and IFN-αsubtypes | YFV | 129 and Stat1 KO | YFV Asibi and Angola wildtype strains, 104 pfu footpad and YFV 17D-204 vaccine strain, 106 pfu footpad, 3-4wk-old mice | Highly susceptible to viscerotropic YFV strain but subclinical infection with 17D vaccine strain. | Meier et al. 2009 |
| Tnfrsf9 | Aka CD137, T cell costimulatory molecule | JEV | C57BL/6 WT and Tnfrsf9 KO | JEV Beijing-1 strain, 1.5-3.105 pfu ip, 4-5wk-old mice | Reduced mortality rate and reduced viral burden in extraneural tissues and the CNS. | Kim et al. 2015 |
Genome-wide search for genetic association
The forward genetic approach to identify flavivrus-resistance genes has been successfully applied so far only to WNV infection, taking advantage of the contrast between the high susceptibility of several laboratory inbred strains and the strong resistance of wild-derived inbred strains, one of the first examples of inherited resistance to a pathogen to be described in mice (Webster 1937; Webster and Clow 1936). Flavivirus resistance was found to be inherited in a monogenic, autosomal dominant manner (Darnell et al. 1974; Sangster et al. 1993) and was mapped to chromosome 5 (Urosevic et al. 1995). Two groups simultaneously identified a loss-of-function mutation in the 2’-5’ oligoadenylate synthetase 1b (Oas1b) mouse gene (Mashimo et al. 2002; Perelygin et al. 2002) and experimentally confirmed its causative role in knocked-in (Scherbik et al. 2007) and transgenic mice (Simon-Chazottes et al. 2011). As pointed out by Mashimo et al., the susceptibility to WNV of almost all laboratory strains most likely results from the shared inheritance of a haplotype carrying the Oas1b mutation from one of the very few progenitors at the origin of laboratory mice (Mashimo et al. 2002).
Oas1b is one of the hundreds of IFN-stimulated genes (ISG) which have antiviral and immune modulatory activity to limit viral replication and spread. The mouse Oas gene cluster includes Oas1, Oas2, Oas3, and OasL genes. The Oas1 gene has eight copies (Oas1a to Oas1h), compared to one copy (OAS1) in humans (Mashimo et al. 2003). Most Oas genes encode 2’-5’ oligoadenylate synthetases (OAS) which bind to double-stranded RNA (dsRNA) and polymerize ATP into 2’-5’-linked oligoadenylates (2-5A). 2-5A bind and activate ribonuclease L (RNase L), a latent endoribonuclease. Upon activation during viral infection, RNase L cleaves viral and cellular single-stranded RNAs (ssRNAs). RNase L contributes to host resistance to WNV since RNAse L-deficient mice showed increased mortality (Samuel et al. 2006). RNase L products of RNA degradation can also bind and activate RIG-I-like receptors (RLR), resulting in enhanced innate immune signaling (Choi et al. 2015). However, Oas1b lacks 2-5A activity (Elbahesh et al. 2011). Moreover, RNase L has an antiviral effect against WNV infection in mouse embryonic fibroblasts carrying either functional or deficient Oas1b alleles, indicating that Oas1b controls WNV infection through another, RNase L-independent, mechanism (Elbahesh et al. 2011; Scherbik et al. 2006). Further insight into this mechanism may come from the identification of molecular partners of Oas1b (Courtney et al. 2012) and from the analysis of innate immune gene signatures that correlate with variations in Oas1b gene dosage, in genetically diverse mouse populations such as the Collaborative Cross (Green et al. 2017). This collection of recombinant inbred strains with large genetic diversity (Churchill et al. 2004) is an ideal platform for modelling a large range of phenotypes and has led to the development of several new models for human WNV disease where Oas1b is not the sole determinant (Graham et al. 2016; Graham et al. 2015).
Despite differences in the genomic organization of the members of this gene family across mammals, the conservation of the OAS pathway and of its role in host response to WNV allowed to successfully identify a variant of the equine OAS1 gene associated with symptomatic forms of WNV disease in horses (Rios et al. 2010).
An amino-acid substitution in the Stat1 gene, resulting in partial inactivation of the IFN pathway, was recently identified in a backcross involving an MHC-II knockout mouse strain, through genome-wide SNP genotyping followed by sequence capture and sequencing of the candidate interval (Larena and Lobigs 2017).
Similar genetic association studies have not yet been reported with other flaviviruses. In the case of Dengue and Zika viruses, they are more difficult to perform due to the necessity to analyze mice with an abrogated type I IFN response.
Functional analysis of candidate genes
Host genetic factors involved in mouse susceptibility to flavivirus infections have also been identified through reverse genetic approaches, by evaluating the phenotype resulting from a specific genetic modification. Most candidate genes have been tested based on their role in immune responses and validated through functional analysis using mice carrying loss-of-function mutations. In particular, the IFN signaling pathways are crucial in the innate immune response against flaviviruses (Miorin et al. 2017), although adaptive immunity also plays a significant role, as illustrated by the enhanced susceptibility to DENV or ZIKV of mice lacking Rag1 or Rag2 (Shresta et al. 2004; Winkler et al. 2017), two genes critical for B and T cell development (Table 2). The importance of several mechanisms in the host susceptibility to these infections has been confirmed in vivo by reverse genetics (Figure 1). These host genetic determinants are reviewed hereafter according to their function in the immune responses to viral infection.
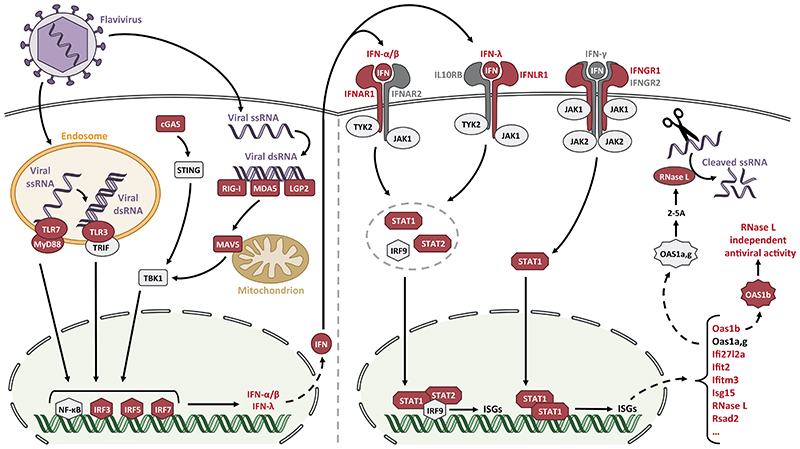
Figure 1. Innate immune response triggered by flavivirus infection, highlighting mouse genes involved in susceptibility or resistance.
Following infection by a flavivirus, viral RNA is detected by TLRs and RLRs which induce activation of several transcription factors such as NF-κB and IRFs which, in turn, promote the transcription of type I, type II and type III IFNs. Once secreted, each IFN subtype (α/β, γ, λ) binds to its specific receptor (IFNAR, IFNGR, IFNLR) which leads to the activation of the JAK/STAT transduction pathways. IFNAR and IFNLR both signal through STAT1, STAT2 and IRF9 whereas only STAT1 is activated by IFNGR. STAT proteins activate the transcription of hundreds of ISGs, including Oas genes, some of which sense viral dsRNA and further promote viral ssRNA cleavage by activating RNase L. Genes of the IFN signaling pathway identified in the mouse as host genetic factors of susceptibility to flaviviral infections are depicted in red.
Abbreviations: TLR, Toll-like receptor; RLR, RIG-I-like receptor; IFN, interferon; IRF, IFN regulatory factor; ISG, IFN stimulated gene; ssRNA, single-stranded RNA; dsRNA, double-stranded RNA.
Interferon responses
Type I IFNs are secreted by infected cells, induce an antiviral state and promote immune responses against viral pathogens in an autocrine and paracrine manner. These signaling pathways are finely regulated by host factors at multiple levels starting from viral sensing and recognition to transduction and regulation of transcription.
A variety of pattern recognition receptors (PRRs) are involved in the recognition of the virus, including Toll-like receptors (TLRs) and RLRs. Among the RLRs, Ddx58 and Ifih1 (which encode viral nucleic acid sensors known as RIG-I and MDA5, respectively), are essential PRR genes : mice deficient for either of them showed increased lethality after WNV infection, and mice lacking both genes were extremely susceptible (Errett et al. 2013; Lazear et al. 2013). Likewise, mice deficient for Mavs, the downstream adaptor molecule of these PRRs which coordinates pathways leading to the activation of NFκB, IFN regulatory factors (IRFs) 3 and 7, showed enhanced WNV replication and dissemination, with high mortality (Suthar et al. 2010). LGP2, another member of the RLR family, is not essential for induction of innate immune response but promotes antigen-specific CD8+ T cell survival, proliferation, and antiviral effector functions (Suthar et al. 2012). It was recently demonstrated that LGP2 associates with DICER and blocks the cleavage of viral dsRNAs, therefore inhibiting antiviral RNA interference (van der Veen et al. 2018). TLR7, a sensor for ssRNA, is another critical host sensor of WNV. Tlr7 inactivation in mice led to an ineffective viral clearance and resulted in increased susceptibility to lethal WNV encephalitis (Town et al. 2009). MYD88 is considered the exclusive adaptor molecule for TLR7 and is required for signal transduction after viral RNA sensing and for an effective IFN response against WNV (Wang et al. 2004). Accordingly, Myd88 KO mice displayed the same highly susceptible phenotype as Tlr7 KO mice following WNV infection (Szretter et al. 2010; Town et al. 2009). By contrast, the analysis of Tlr3-deficient mice has led to contradictory results. A first study found that Tlr3 KO mice were more resistant to WNV infection than controls, with decreased viral load in the brain and reduced blood-brain barrier permeability (Wang et al. 2004), while another study reported higher mortality and increased viral burden in the central nervous system (CNS) (Daffis et al. 2008a). Discrepancies were attributed to differences in passage history of the virus, and to dose or route of inoculation. Finally, MB21D1, also known as cGAS, is a cytosolic sensor of dsDNA and a cGMP-AMP synthase which plays a key role in restriction of DNA viruses (Ma and Damania 2016). Interestingly, it is also important for innate immunity against RNA viruses with no DNA intermediates in their life cycle, as demonstrated by the increased susceptibility of Mb21d1 KO mice to WNV (Schoggins et al. 2014). cGMP activates TMEM173, better known as STING (also MITA or MYPS). DENV protein NS2B3 is able to cleave human, but not mouse STING (Aguirre et al. 2012; Ma and Damania 2016; Stabell et al. 2018; Yu et al. 2012), a phenomenon which contributes to the natural resistance of mice to DENV. NS2B3 protein from WNV, ZIKV and JEV, but not YFV, showed the same property (Ding et al. 2018). However, Tmem173-deficient mice did not exhibit increased susceptibility to ZIKV (Ding et al. 2018).
Viral recognition by TLRs or RLRs activates multiple transcription factors including NF-κB and IRFs, directly or through TBK1. Irf3, Irf5 and Irf7 seem to have redundant functions since these three genes need to be inactivated to induce susceptibility to DENV or ZIKV (Carlin et al. 2017; Lazear et al. 2016). Although signaling through the same pathway, IRFs can influence host susceptibility through cell and tissue-specific processes. Mice deficient for either Irf1, Irf3, Irf5 or Irf7 showed increased mortality upon WNV infection with intact IFN-β production (Brien et al. 2011; Daffis et al. 2007; Daffis et al. 2008b; Thackray et al. 2014). IRFs stimulate in turn the transcription of type I IFNs. Mice deficient for the Ifnb gene, or injected with an antibody directed against IFN-α or/and IFN-β before and during WNV infection displayed increased lethality, highlighting the critical contribution of type I IFNs in antiviral responses against WNV (Lazear et al. 2011; Sheehan et al. 2015). Likewise, mice deficient for the type II IFN Ifng gene showed enhanced mortality with higher viremia and replication in lymphoid tissue (Shrestha et al. 2006).
IFN receptors are the central players of the IFN system and are activated upon binding of their subtype-specific IFN. Noteworthy, mice deficient in Ifnar1 and in both Ifnar1 and Ifngr1 genes (often referred to as A129 and AG129, respectively, when bred on the 129S2/SvPas background) are currently the most widely used models in YFV, DENV and ZIKV studies. Ifnar1 KO mice were more susceptible than WT controls to WNV (Samuel and Diamond 2005), ZIKV (Dowall et al. 2017; Lazear et al. 2016; Rossi et al. 2016; Tripathi et al. 2017), DENV (Orozco et al. 2012; Prestwood et al. 2012; Shresta et al. 2004), and YFV (Meier et al. 2009) infections, with high levels of viral replication and disease manifestations allowing for pathogenesis and mechanistic studies in vivo. Morbidity and lethality were very high with WNV, ZIKV and YFV, and were dose-dependent with DENV. By contrast, Ifngr1 KO mice were resistant to YFV (Meier et al. 2009), moderately susceptible to DENV (Shresta et al. 2004) but highly susceptible to WNV (Shrestha et al. 2006). Mice lacking the receptor for type III IFN (Ifnlr1 gene) were resistant to YFV but exhibited enhanced WNV neuroinvasion with increased blood-brain barrier permeability (Lazear et al. 2015) and enhanced ZIKV transplacental transmission (Jagger et al. 2017). For all the above viruses, Ifnar1/Ifngr1 double KO mice displayed very high susceptibility with 100% mortality (Aliota et al. 2016; Meier et al. 2009; Prestwood et al. 2012; Shresta et al. 2004; Thibodeaux et al. 2012). Likewise, Ifnar1/Ifnlr1 double KO mice were highly susceptible to YFV (Douam et al. 2017). These mouse models have allowed to decipher the roles of the different IFNs and IFN-receptors subtypes in response to flaviviruses. For example, in DENV infection, IFNAR signaling limits initial viral replication and controls its subsequent dissemination. By contrast, IFNGR-mediated responses appear to act at later stages of dengue disease by restricting viral replication in the periphery and eliminating virus from the CNS (Shresta et al. 2004).
IFN-receptors signal through different tyrosine kinases which recruit signal transducers and activators of transcription (STAT). STAT proteins are key mediators of the IFN response. IFNAR and IFNLR signal through both STAT1 and STAT2 whereas only STAT1 is activated after IFNGR stimulation. STAT2 is one of the targets of the NS5 protein of flaviviruses, a potent and specific antagonist of IFN signaling which acts through virus-specific mechanisms (Grant et al. 2016). ZIKV NS5 binds to and targets human (Kumar et al. 2016), but not mouse, STAT2 for proteasomal degradation, which provides the mechanism underlying the natural resistance of mice to ZIKV and DENV (Best 2017; Grant et al. 2016; Miorin et al. 2017). The difference between human and mouse STAT2 for the binding of DENV NS5 was mapped to the coiled-coil domain (Ashour et al. 2010). To bind STAT2, DENV and YFV require E3 ubiquitin ligases UBR4 (Best 2017; Grant et al. 2016) and TRIM23 (Laurent-Rolle et al. 2014), respectively. WNV NS5 binds to prolidase, a cellular peptidase, to suppress IFNAR maturation and cell surface expression (Laurent-Rolle et al. 2010; Lubick et al. 2015). By analyzing multiple crosses between mice inactivated for either Ifnar1, Ifngr1, Stat1 or Stat2 genes, Perry et al. have demonstrated the importance of STAT proteins in the host immune response against DENV. They have shown that the combined loss of STAT1 and STAT2 resulted in severe disease and death in mice challenged with DENV. They also showed, using high virus doses, that Stat1 KO mice succumbed to dengue disease unlike Stat2 KO mice, and concluded that STAT1 plays a more prominent role than STAT2 in anti-DENV responses (Perry et al. 2011). Stat1 KO mice were also highly susceptible to ZIKV and YFV infections although morbidity and mortality rates varied according to viral strains (Kamiyama et al. 2017; Meier et al. 2009). After ZIKV infection, Stat2 KO mice displayed neurological symptoms and viral dissemination to the brain and gonads. Interestingly, clinical signs of Zika fever were delayed and milder in Ifnar1 KO mice compared with Stat2 KO mice, pointing to a possible protective role of INF-λ response (Tripathi et al. 2017).
Once activated, STAT proteins stimulate the transcription of hundreds of ISGs. In addition to Oas1b, several ISGs have been shown to influence mouse susceptibility to WNV infection. Mice lacking Rsad2 (also known as viperin), Ifi27l2a, Ifitm3 or Ifit2 were all more vulnerable to lethal WNV challenge and allowed higher viral replication mainly in the CNS (Cho et al. 2013; Gorman et al. 2016; Lucas et al. 2015; Szretter et al. 2011). Rsad2 inhibits ZIKV replication by inducing proteasome-dependent degradation of ZIKV NS3 (Panayiotou et al. 2018) and the synthesis of a replication-chain terminator (Gizzi et al. 2018), and has cell-type specific activity in the CNS (Lindqvist et al. 2018). Ifitm proteins alter the properties of cell and viral membranes and can inhibit the replication of a wide range of pathogenic viruses (Perreira et al. 2013). They induce similar restriction of primary and antibody-dependent enhancement secondary DENV infections in human leukemia cells (Chan et al. 2012). Ifitm3 inhibits the early stages of Zika virus replication and can prevent Zika virus-induced cell death (Savidis et al. 2016). In a model of direct inoculation of ZIKV in the eye, Isg15 KO mice showed increased ocular tissue pathology, characterized by a severe chorioretinitis with enhanced retinal cell death (Singh et al. 2017).
Overall, many studies have emphasized the crucial role of IFN responses, and especially the type I IFN pathway, which constitute an essential line of defense for the host after infection by flaviviruses.
Cytokines and chemokines regulating the immune response
The role of cytokines and chemokines as important regulators of immune responses has also been investigated in flaviviral infections using mice deficient for cytokines, chemokines or chemokine receptors.
After WNV infection, Il10 KO mice had a decreased mortality rate, suggesting that IL-10, which has immunosuppressive properties, promotes WNV pathogenesis (Bai et al. 2009). Mice lacking IL-12b or IL-23a were more susceptible to WNV induced encephalitis, but not mice deficient for IL-12a, indicating that survival required intact IL-23 as opposed to IL-12 responses (Town et al. 2009).
Chemokines and chemokine receptors, which modulate leukocytes trafficking, play an important role in the regulation of immune responses. Several of them have been implicated in host susceptibility to flaviviruses, sometimes with opposing effects depending on the virus and its cell tropism. Deficiency in Cxcr3, the receptor for chemokines CXCL9, CXCL10 and CXCL11, resulted in increased lethality in mice infected by DENV or WNV, with a decrease in T lymphocytes in the brain, CD8+ T cells in particular (Hsieh et al. 2006; Zhang et al. 2008). Mice lacking CXCL10 also showed enhanced susceptibility to DENV infection with higher viral loads in the brain but unchanged number of infiltrating T cells, suggesting that CXCL10 protective effect in dengue might be due to its direct antiviral activity rather than its role in lymphocyte recruitment (Hsieh et al. 2006; Ip and Liao 2010). Cxcl10 KO mice showed enhanced susceptibility to WNV with increased viral burden in the brain and a decrease in CXCR3+ CD8+ T-cell trafficking, supporting a neuroprotective role for Cxcl10 in the brain (Klein et al. 2005). Ccr2 KO mice displayed increased survival time associated with decreased liver damage following DENV infection, whereas they showed a higher mortality rate combined with a reduction of monocyte accumulation in the brain after WNV infection (Guabiraba et al. 2010; Lim et al. 2011). Upon WNV infection, Ccr7-deficient mice exhibited enhanced mortality and CNS viral load, associated with marked leukocyte accumulation in the brain. These results indicate that CCR7 contributes to viral clearance and effectively modulates neuroinflammation in a model of WNV encephalitis (Bardina et al. 2017).
Other stimulatory molecules of immune cells have crucial functions in promoting or restricting flaviviral infections. CLEC5a is a C-type lectin which regulates cell adhesion and cell-cell signaling during the immune response. CLEC5a has been shown to act as a susceptibility factor in DENV infection. Stat1 KO mice treated with an anti-CLEC5a antibody showed increased survival associated with a reduction of plasma leakage and TNF-α serum level (Chen et al. 2008). Mice deficient in Tnfrsf9, a T cell co-stimulatory factor, displayed reduced mortality rate in a model of Japanese encephalitis, highlighting a detrimental role of this molecule in the immune response against JEV (Kim et al. 2015).
Other Mechanisms
Autophagy is an essential mechanism which targets cellular components for lysosomal degradation. In the immune system, autophagy has many functions in both innate and adaptive responses, such as intracellular pathogen detection, modulation of the inflammatory processes as well as regulation of lymphocytes homeostasis. Mice carrying a hypomorphic variant of Atg16l1, a key autophagy gene, and treated with an anti-IFNAR antibody showed reduced ZIKV vertical transmission and placental damage. This phenotype was shown to result from a placental cell-autonomous effect of autophagy activity indicating that autophagy promotes ZIKV pathogenesis during gestation (Cao et al. 2017).
Semaphorins constitute a group of proteins that are involved in connecting the neuronal and immune systems. Following WNV infection, Sema7A KO mice exhibited increased survival, correlated with a reduction of blood-brain barrier permeability. SEMA7A thus appears to play a deleterious role during WNV infection in vivo (Sultana et al. 2012).
Human vs mouse genetics
While mouse genetics studies improve our understanding of the function of genes and identify new susceptibility genes in genome wide screens, the main obstacle in studying flaviviruses using mouse models is the inherent resistance of mice to most mosquito-borne flaviviruses (such as DENV and ZIKV). Several mouse models frequently used in flaviviral research are constitutively deficient for the IFN type I and/or type II responses, and therefore do not reflect an intact functioning human immune system. Moreover, these models are generally more susceptible to mouse adapted viral strains, which are genetically different from human pathogens Therefore, human cohorts are important to understand the role of the genes in protection or pathogenesis of human diseases, although human genetic studies are highly dependent on patient numbers.
Human genetic studies
Multiple approaches have been taken to identify genetic variants in human populations associated with susceptibility or resistance to flaviviruses, in particular case-control studies to test candidate genes, genome-wide association studies, association with specific HLA alleles, and allelic selection in exposed populations or ethnicities. Many studies have focused on favorable or disadvantageous immunological mechanisms involved in the pathogeny of infection but few have identified underlying genetic variants.
West Nile virus
WNV is a neurotropic virus and is transmitted to humans, who are a dead end host, by Culex mosquitoes. WNV causes a self-limiting febrile illness in most individuals that occasionally progresses to severe neurological disease including meningitis and encephalitis (Colpitts et al. 2012). Currently, there is no vaccine licensed in humans. Although there were a few human genetic studies on WNV, the most significant data resulted from the confirmation of the findings of the mouse model. Two polymorphisms of human OAS1, a splicing variant (rs10774671) (Lim et al. 2009), and an intron 2 variant (rs34137742) (Bigham et al. 2011), were associated with symptomatic WNV seroconversion. Another study found a polymorphism of human OASL (rs3213545) associated with hospitalized WNV fever, meningitis and/or WNV encephalitis (Yakub et al. 2005). Two studies have identified associations between the clinical severity of WNV infection and HLA Class I and II alleles (Lanteri et al. 2011; Sarri et al. 2016).
Dengue virus
Classification of Dengue cases
DENV, which is the most common mosquito borne viral infection, is spreading worldwide. There is one dengue vaccine licensed. The strategic Advisory Group of Experts on Immunisation (SAGE) recommended limited usage of the vaccine in only seropositive individuals since April 2018 (http://www.who.int/immunization/diseases/dengue/revised_SAGE_recommendations_dengue_vaccines_apr2018/en/). There are four serotypes of DENV co-circulating (DENV-1 to DENV-4). Infection by one of the four can result in a spectrum of clinical outcomes ranging from asymptomatic to inapparent infection (patients developed mild disease but not enough to seek medical advice) to undifferentiated fever, classical dengue fever (DF) with or without hemorrhage, dengue hemorrhagic fever (DHF) with plasma leakage leading to shock (dengue shock syndrome (DSS)) and other organ involvement (such as hepatitis, encephalitis).
There are several difficulties in performing genetic studies of dengue even though there are a high number of cases. First, clinical case definition can be based on either WHO 1997 (WHO 1997) or WHO 2009 criteria (WHO 2009), that define severity of dengue in different ways. The WHO 1997 utilizes well defined criteria focused on plasma leakage to differentiate DF, DHF and DSS. The WHO 2009 criteria are designed for patient management and rely on more subjective criteria including several signs and symptoms of multiple organ involvement, hence complicating our understanding of disease pathogenesis. Therefore, most human genetic studies used WHO 1997 criteria, which is specific for plasma leakage.
Secondly, previous infection history and infecting dengue serotypes are the two most important confounding factors for genetic studies but are expensive, time consuming and not always possible to determine. Most studies, which are designed to distinguish primary from secondary infection and can identify the infecting dengue serotype indeed showed interaction of these variables and genetic factors (Simon-Loriere et al. 2015; Stephens et al. 2002).
Genetic study of Dengue
There were more human genetics studies of DENV compared to mouse genetic studies, which were more focused on candidate genes related to IFN pathways. There are several excellent recent reviews on human genetic susceptibility to dengue, including a meta-analysis (Xavier-Carvalho et al. 2017a). Associations could be replicated in at least two populations for only one locus and six genes (Xavier-Carvalho et al. 2017a). The region which was replicated in many populations is the HLA locus on chromosome 6 including HLA-A*24 (Loke et al. 2001; Malavige et al. 2011; Nguyen et al. 2008), MHC class I polypeptide-related sequence B MICB (rs3132468) (Dang et al. 2014; Khor et al. 2011; Whitehorn et al. 2013) and tumor necrosis factor TNF (-308, rs1800629) (Fernandez-Mestre et al. 2004; Fernando et al. 2015; Sam et al. 2015; Santos et al. 2017). The other six genes are C-type lectin, CD209 (rs4804803) (Sakuntabhai et al. 2005; Wang et al. 2011; Xavier-Carvalho et al. 2013), C-type lectin domain containing 5A CLEC5A (rs1285933) (Xavier-Carvalho et al. 2017b; Xavier-Carvalho et al. 2013), immunoglobulin heavy chain receptor FcgR IIA (Arg131His-rs1801274) (Garcia et al. 2010; Mohsin et al. 2015; Noecker et al. 2014), cytokine IL-10 (-1082/-819/-592) (Fernando et al. 2015; Perez et al. 2010), alpha tryptase 1 TPSAB1 (Velasquez et al. 2015) and phospholipase C epsilon 1 PLCE1 (rs3765524, and rs3740360) (Dang et al. 2014; Khor et al. 2011; Whitehorn et al. 2013). Among these genes, how CLEC5A contributes to pathology has been investigated in a mouse model for DENV (Chen et al. 2008).
Although there was a bias of patient selection among the studies (more studies on DHF/DSS than DF), there was evidence that different sets of genes are associated with DF (mild clinical dengue) and/or DHF/DSS (severe dengue). While the more severe form of dengue is associated with HLA class I, and genes associated with inflammation and immune response (CLEC5A) (Xavier-Carvalho et al. 2017b; Xavier-Carvalho et al. 2013), cytokine response (IL-10), NK cell (MICB), mast cell activity (TPSAB1) (Velasquez et al. 2015) and lipid metabolism (PLCE1, PLCB4, OSBPL10, RXRA) (Sierra et al. 2017), DF is associated with genes in xenobiotic pathway (CHST10, AHRR, PPP2R5E and GRIP1) (Oliveira et al. 2018) and HLA class II (LaFleur et al. 2002; Sierra et al. 2007; Stephens et al. 2002; Weiskopf et al. 2015). Genes involved in viral entry (CD209, FcgRII) and TNFα pathway were associated with both forms of diseases. In addition, a non-synonymous variant of OAS3 was associated with severe dengue caused by DENV serotype 2 (Simon-Loriere et al. 2015). Adaptive immunity could play a more important role in eliminating the virus and in the development of clinical and severe dengue disease. More human genetic studies with well-characterized patients are needed in order to understand protection and pathogenesis of mild clinical and severe dengue in humans.
Protective and enhancing HLA class I and class II alleles in dengue virus infections
An association between HLA class I alleles and DHF susceptibility was shown in a large cohort of Vietnamese patients. More specifically, it was found that children with HLA-A*33 were less likely to develop DHF, whereas children with HLA-A*24 were at increased risk of developing DHF (Loke et al. 2001). Analyses of NS3- and NS5-specific CD8 T cell responses in different donors suggest opposing roles for T cells in both protection and development of DHF. Likewise, a more recent study in Vietnam confirmed the HLA association between HLA-A*24 and DHF or DSS and showed that HLA-A*24 with histidine at codon 70 is a susceptible allele, whereas the HLA-DRB1*0901 class II allele is protective against development of DSS, in patients with DENV-2 infection (Nguyen et al. 2008).
A second and larger case-control study in ethnic Thai patients revealed a variety of HLA class I association with the severity of clinical disease during secondary DENV infections (Stephens et al. 2002). The HLA-A*0203 allele was in particular associated with less severe DF, regardless of the secondary infecting virus serotype. By contrast, HLA-A*0207 was associated with susceptibility to the more severe DHF in patients with secondary DENV-1 and DENV-2 infections. Conversely, HLA-B*51 was associated with the development of DHF in patients with secondary infections, and HLA-B*52 was associated with DF in patients with secondary DENV-1 and DENV-2 infections. Moreover, HLA-B44, B62, B76 and B77 also appeared to be protective against developing clinical disease after secondary dengue virus infection. Interestingly, at least for the HLA-A*02 class I alleles, the strong binding potential to viral peptides was suggested to enable T cell activation and protection observed in these patients with secondary DENV infections.
Another study of HLA polymorphism in the Cuban population revealed an increased frequency of HLA-A*31 and -B*15 class I alleles in symptomatic dengue virus infection compared with controls, and conversely, an elevated frequency of HLA-DRB1*07 and DRB1*04 class II alleles in control subjects compared with dengue case patients (Sierra et al. 2007). In a Mexican population, HLA-B*35 was negatively associated with symptomatic disease, whereas HLA-DQB1*0302 was positively associated with DHF, and HLA-DQB1*0202 was positively associated with DF only (Falcon-Lezama et al. 2009). Finally, in a Sri Lankan population, it was found that HLA-A*31 and HLA-DRB1*08 were associated with susceptibility to DSS, during secondary infection, and HLA-A*24 and HLA-DRB1*12 were strongly associated with DHF during primary dengue infection (Malavige et al. 2011).
One of the reasons for the association observed between certain HLA alleles and dengue disease severity is linked to the ability of these class I or class II alleles to induce a strong CD8+ or CD4+ T cell response against dengue epitopes. In that respect, it was found that T cell responses that were weak in magnitude, for example those observed against dengue epitopes restricted by HLA-A*0101 and HLA-A*2401, correlated with disease susceptibility, whereas strong and polyfunctional CD8+ T cell responses were observed in HLA-B*3501 individuals and were negatively associated with symptomatic disease (Weiskopf et al. 2013). Likewise, a higher resistance and susceptibility to severe dengue clinical outcome were shown to be associated with a more vigorous CD4+ T cell response in the context of DRB1*0401 and DRB1*0802, respectively (Weiskopf et al. 2015).
Protective or enhancing Killer immunoglobulin-like receptors (KIRs) in DENV infection
An association between KIR-ligands pairs and susceptibility to dengue in Southern Brazil was also detected, in the context of DENV3 infection (Beltrame et al. 2013). Although the exact role of NK cells expressing either activating or inhibitory KIRs has yet to be determined during DENV infection in humans, several studies strongly support the ability of DENV- or Flavivirus-derived conserved peptides, to stimulate or to inhibit KIR2DS2 or RIR3DL1 NK cells in vitro against NS1 or NS3 peptides, and in the context of HLA-C*0102 or HLA-B*57, respectively (Naiyer et al. 2017; Townsley et al. 2016).
The HLA transgenic mice as models to study the immune protection against DENV infection
To allow the identification of HLA-restricted peptides derived from the viral proteins, several HLA class I and class II transgenic mice have been developed to study the T cell response against the whole virus or the peptides derived from the viral proteins (Boucherma et al. 2013; Pascolo et al. 1997). Strikingly, most DENV-derived T cell epitopes inducing a T cell response in HLA class I mice correspond to the peptides identified from human individuals after DENV infection (Duan et al. 2015; Elong Ngono et al. 2017; Rivino et al. 2013a; Rivino et al. 2013b; Weiskopf et al. 2013; Weiskopf et al. 2011). Importantly, as the magnitude of the T cell response reflects the binding affinity of the different peptides to an HLA allele, analysis of the magnitude of T cell response in these transgenic mice against the different peptides covering the whole sequence should allow the identification of the most immunogenic peptides inducing a long lasting immunity.
An improvement of this mouse model involves a transient blockade of type I interferon signaling, after treatment with anti-IFNAR antibody. Commonly used in wild type mice to study the cellular effectors of the immune response to different viruses, such as the WNV or ZIKV (Lazear et al. 2016; Ng et al. 2015; Pinto et al. 2011; Sheehan et al. 2015; Zhao et al. 2016), this procedure is currently adapted to the HLA transgenic mice, with the objective to study the role of peptide-specific T cells in the induction of long lasting immunity against DENV and ZIKV infection.
Perspectives
A number of studies on flaviviruses have focused on the influence of viral virulence factors on viral multiplication, on the inhibition of immune responses of the infected host, and on disease severity. By contrast, the role of host genetic determinants on clinical severity of flavivirus infections remains elusive, with the notable exception of type I IFN responses. The few examples of genes identified which are not directly related to the type I IFN responses demonstrate that genome-wide, unbiased approaches will be essential in identifying novel actors. Despite differences in host-virus interactions, the mouse can successfully serve as an experimental model to assess the role of specific genes and to query the genome for host genetic determinants in genetic reference populations with vast genetic polymorphism, such as the Collaborative Cross (Aylor et al. 2011) and the Diversity Outbred mice (Recla et al. 2014). Their IFN response could be abrogated by antibodies directed against the type I IFN receptor (Lazear et al. 2016). These resources will be highly valuable to address the complex host-virus interactions through systems biology approaches.
Along with these innovative mouse model, well-characterized human cohorts of flaviviral infections should be collected. More DENV cohorts with well-characterized virological, immunological and clinical parameters from different countries are needed to understand human genetic basis of susceptibility to severe dengue, host-viral interaction and genetic susceptibility to ADE phenomenon. In addition, cohorts of asymptomatic DENV infected patients will help us understand protective immunity and will serve as the best control for symptomatic and severe dengue. Beyond DENV and WNV, ZIKV infection represents an ongoing public health challenge because of its complications. Collecting a sufficient number of samples from ZIKV-infected donors living in different endemic regions is a necessary step forward to be able to identify host genetic factors influencing persistent infection, susceptibility to neurological complications, mother-to-child transmission, and susceptibility to brain pathology.
Acknowledgments
C.M. was supported by a fellowship from the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (grant n°ANR-10-LABX-62-IBEID). T.C. was supported by a HHMI-Wellcome International Research Scholarship and Institut Pasteur International Network G4 grant.
Footnotes
Conflict of interest statement
On behalf of all authors, the corresponding authors state that there is no conflict of interest.
References
- Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, Rodriguez-Madoz JR, et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliota MT, Caine EA, Walker EC, Larkin KE, Camacho E, Osorio JE. Characterization of Lethal Zika Virus Infection in AG129 Mice. PLoS neglected tropical diseases. 2016;10:e0004682. doi: 10.1371/journal.pntd.0004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour J, Morrison J, Laurent-Rolle M, Belicha-Villanueva A, Plumlee CR, Bernal-Rubio D, Williams K, Harris E, Fernandez-Sesma A, Schindler C, García-Sastre A. Mouse STAT2 Restricts Early Dengue Virus Replication. Cell host & microbe. 2010;8:410–421. doi: 10.1016/j.chom.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, Ferris MT, Frelinger JA, Heise M, Frieman MB, Gralinski LE, et al. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 2011;21:1213–1222. doi: 10.1101/gr.111310.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Town T, Qian F, Wang P, Kamanaka M, Connolly TM, Gate D, Montgomery RR, Flavell RA, Fikrig E. IL-10 signaling blockade controls murine West Nile virus infection. PLoS Pathog. 2009;5:e1000610. doi: 10.1371/journal.ppat.1000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardina SV, Brown JA, Michlmayr D, Hoffman KW, Sum J, Pletnev AG, Lira SA, Lim JK. Chemokine Receptor Ccr7 Restricts Fatal West Nile Virus Encephalitis. J Virol. 2017;91:e02409-02416. doi: 10.1128/JVI.02409-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame LM, Sell AM, Moliterno RA, Clementino SL, Cardozo DM, Dalalio MM, Fonzar UJ, Visentainer JE. Influence of KIR genes and their HLA ligands in susceptibility to dengue in a population from southern Brazil. Tissue Antigens. 2013;82:397–404. doi: 10.1111/tan.12256. [DOI] [PubMed] [Google Scholar]
- Best SM. The Many Faces of the Flavivirus NS5 Protein in Antagonism of Type I Interferon Signaling. J Virol. 2017;91 doi: 10.1128/JVI.01970-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham AW, Buckingham KJ, Husain S, Emond MJ, Bofferding KM, Gildersleeve H, Rutherford A, Astakhova NM, Perelygin AA, Busch MP, Murray KO, et al. Host genetic risk factors for West Nile virus infection and disease progression. PLoS One. 2011;6:e24745. doi: 10.1371/journal.pone.0024745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucherma R, Kridane-Miledi H, Bouziat R, Rasmussen M, Gatard T, Langa-Vives F, Lemercier B, Lim A, Berard M, Benmohamed L, Buus S, et al. HLA-A*01:03, HLA-A*24:02, HLA-B*08:01, HLA-B*27:05, HLA-B*35:01, HLA-B*44:02, and HLA-C*07:01 Monochain Transgenic/H-2 Class I Null Mice: Novel Versatile Preclinical Models of Human T Cell Responses. J Immunol. 2013;191:583–593. doi: 10.4049/jimmunol.1300483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brien JD, Daffis S, Lazear HM, Cho H, Suthar MS, Gale M, Jr, Diamond MS. Interferon regulatory factor-1 (IRF-1) shapes both innate and CD8(+) T cell immune responses against West Nile virus infection. PLoS Pathog. 2011;7:e1002230. doi: 10.1371/journal.ppat.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B, Parnell LA, Diamond MS, Mysorekar IU. Inhibition of autophagy limits vertical transmission of Zika virus in pregnant mice. The Journal of experimental medicine. 2017;214:2303–2313. doi: 10.1084/jem.20170957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin AF, Plummer EM, Vizcarra EA, Sheets N, Joo Y, Tang W, Day J, Greenbaum J, Glass CK, Diamond MS, Shresta S. An IRF-3-, IRF-5-, and IRF-7-Independent Pathway of Dengue Viral Resistance Utilizes IRF-1 to Stimulate Type I and II Interferon Responses. Cell reports. 2017;21:1600–1612. doi: 10.1016/j.celrep.2017.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedillo-Barrón L, García-Cordero J, Shrivastava G, Carrillo-Halfon S, León-Juárez M, Bustos Arriaga J, León Valenzuela P, Gutiérrez Castañeda B. In: Virus Protein and Nucleoprotein Complexes. Harris JR, Bhella D, editors. Springer Singapore; Singapore: 2018. The Role of Flaviviral Proteins in the Induction of Innate Immunity; pp. 407–442. [DOI] [PubMed] [Google Scholar]
- Chan YK, Huang IC, Farzan M. IFITM proteins restrict antibody-dependent enhancement of dengue virus infection. PLoS One. 2012;7:e34508. doi: 10.1371/journal.pone.0034508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ST, Lin YL, Huang MT, Wu MF, Cheng SC, Lei HY, Lee CK, Chiou TW, Wong CH, Hsieh SL. CLEC5A is critical for dengue-virus-induced lethal disease. Nature. 2008;453:672–676. doi: 10.1038/nature07013. [DOI] [PubMed] [Google Scholar]
- Cho H, Shrestha B, Sen GC, Diamond MS. A role for Ifit2 in restricting West Nile virus infection in the brain. J Virol. 2013;87:8363–8371. doi: 10.1128/JVI.01097-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi UY, Kang JS, Hwang YS, Kim YJ. Oligoadenylate synthase-like (OASL) proteins: dual functions and associations with diseases. Exp Mol Med. 2015;47:e144. doi: 10.1038/emm.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, Bleich A, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Colpitts TM, Conway MJ, Montgomery RR, Fikrig E. West Nile Virus: biology, transmission, and human infection. Clin Microbiol Rev. 2012;25:635–648. doi: 10.1128/CMR.00045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney SC, Di H, Stockman BM, Liu H, Scherbik SV, Brinton MA. Identification of novel host cell binding partners of Oas1b, the protein conferring resistance to flavivirus-induced disease in mice. J Virol. 2012;86:7953–7963. doi: 10.1128/JVI.00333-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumberworth SL, Clark JJ, Kohl A, Donald CL. Inhibition of type I interferon induction and signalling by mosquito-borne flaviviruses. Cellular microbiology. 2017;19:e12737. doi: 10.1111/cmi.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S, Samuel MA, Keller BC, Gale M, Jr, Diamond MS. Cell-specific IRF-3 responses protect against West Nile virus infection by interferon-dependent and -independent mechanisms. PLoS Pathog. 2007;3:e106. doi: 10.1371/journal.ppat.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S, Samuel MA, Suthar MS, Gale M, Jr, Diamond MS. Toll-like receptor 3 has a protective role against West Nile virus infection. J Virol. 2008a;82:10349–10358. doi: 10.1128/JVI.00935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S, Samuel MA, Suthar MS, Keller BC, Gale M, Jr, Diamond MS. Interferon regulatory factor IRF-7 induces the antiviral alpha interferon response and protects against lethal West Nile virus infection. J Virol. 2008b;82:8465–8475. doi: 10.1128/JVI.00918-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang TN, Naka I, Sa-Ngasang A, Anantapreecha S, Chanama S, Wichukchinda N, Sawanpanyalert P, Patarapotikul J, Tsuchiya N, Ohashi J. A replication study confirms the association of GWAS-identified SNPs at MICB and PLCE1 in Thai patients with dengue shock syndrome. BMC Med Genet. 2014;15:58. doi: 10.1186/1471-2350-15-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell MB, Koprowski H, Lagerspetz K. Genetically determined resistance to infection with group B arboviruses I. Distribution of the resistance gene among various mouse populations and characteristics of gene expression in vivo. J Infect Dis. 1974;129:240–247. doi: 10.1093/infdis/129.3.240. [DOI] [PubMed] [Google Scholar]
- Ding Q, Gaska JM, Douam F, Wei L, Kim D, Balev M, Heller B, Ploss A. Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proceedings of the National Academy of Sciences of the United States of America. 2018 doi: 10.1073/pnas.1803406115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douam F, Soto Albrecht YE, Hrebikova G, Sadimin E, Davidson C, Kotenko SV, Ploss A. Type III Interferon-Mediated Signaling Is Critical for Controlling Live Attenuated Yellow Fever Virus Infection In Vivo. MBio. 2017;8:e12737. doi: 10.1128/mBio.00819-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowall SD, Graham VA, Rayner E, Hunter L, Atkinson B, Pearson G, Dennis M, Hewson R. Lineage-dependent differences in the disease progression of Zika virus infection in type-I interferon receptor knockout (A129) mice. PLoS neglected tropical diseases. 2017;11:e0005704. doi: 10.1371/journal.pntd.0005704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan ZL, Liu HF, Huang X, Wang SN, Yang JL, Chen XY, Li DZ, Zhong XZ, Chen BK, Wen JS. Identification of conserved and HLA-A*2402-restricted epitopes in Dengue virus serotype 2. Virus Res. 2015;196:5–12. doi: 10.1016/j.virusres.2014.10.022. [DOI] [PubMed] [Google Scholar]
- Elbahesh H, Jha BK, Silverman RH, Scherbik SV, Brinton MA. The Flvr-encoded murine oligoadenylate synthetase 1b (Oas1b) suppresses 2-5A synthesis in intact cells. Virology. 2011;409:262–270. doi: 10.1016/j.virol.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elong Ngono A, Vizcarra EA, Tang WW, Sheets N, Joo Y, Kim K, Gorman MJ, Diamond MS, Shresta S. Mapping and Role of the CD8+ T Cell Response During Primary Zika Virus Infection in Mice. Cell Host Microbe. 2017;21:35–46. doi: 10.1016/j.chom.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson AK, Pfeiffer JK. Spectrum of disease outcomes in mice infected with YFV-17D. J Gen Virol. 2015;96:1328–1339. doi: 10.1099/vir.0.000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errett JS, Suthar MS, McMillan A, Diamond MS, Gale M., Jr The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J Virol. 2013;87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon-Lezama JA, Ramos C, Zuniga J, Juarez-Palma L, Rangel-Flores H, Garcia-Trejo AR, Acunha-Alonzo V, Granados J, Vargas-Alarcon G. HLA class I and II polymorphisms in Mexican Mestizo patients with dengue fever. Acta Trop. 2009;112:193–197. doi: 10.1016/j.actatropica.2009.07.025. [DOI] [PubMed] [Google Scholar]
- Fernandez-Mestre MT, Gendzekhadze K, Rivas-Vetencourt P, Layrisse Z. TNF-alpha-308A allele, a possible severity risk factor of hemorrhagic manifestation in dengue fever patients. Tissue Antigens. 2004;64:469–472. doi: 10.1111/j.1399-0039.2004.00304.x. [DOI] [PubMed] [Google Scholar]
- Fernando AN, Malavige GN, Perera KL, Premawansa S, Ogg GS, De Silva AD. Polymorphisms of Transporter Associated with Antigen Presentation, Tumor Necrosis Factor-alpha and Interleukin-10 and their Implications for Protection and Susceptibility to Severe Forms of Dengue Fever in Patients in Sri Lanka. J Glob Infect Dis. 2015;7:157–164. doi: 10.4103/0974-777X.170501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G, Sierra B, Perez AB, Aguirre E, Rosado I, Gonzalez N, Izquierdo A, Pupo M, Danay Diaz DR, Sanchez L, Marcheco B, et al. Asymptomatic dengue infection in a Cuban population confirms the protective role of the RR variant of the FcgammaRIIa polymorphism. The American journal of tropical medicine and hygiene. 2010;82:1153–1156. doi: 10.4269/ajtmh.2010.09-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizzi AS, Grove TL, Arnold JJ, Jose J, Jangra RK, Garforth SJ, Du Q, Cahill SM, Dulyaninova NG, Love JD, Chandran K, et al. A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature. 2018;558:610–614. doi: 10.1038/s41586-018-0238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman MJ, Caine EA, Zaitsev K, Begley MC, Weger-Lucarelli J, Uccellini MB, Tripathi S, Morrison J, Yount BL, Dinnon KH, 3rd, Ruckert C, et al. An Immunocompetent Mouse Model of Zika Virus Infection. Cell Host Microbe. 2018;23:672–685.:e676. doi: 10.1016/j.chom.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman MJ, Poddar S, Farzan M, Diamond MS. The Interferon-Stimulated Gene Ifitm3 Restricts West Nile Virus Infection and Pathogenesis. J Virol. 2016;90:8212–8225. doi: 10.1128/JVI.00581-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govero J, Esakky P, Scheaffer SM, Fernandez E, Drury A, Platt DJ, Gorman MJ, Richner JM, Caine EA, Salazar V, Moley KH, et al. Zika virus infection damages the testes in mice. Nature. 2016;540:438–442. doi: 10.1038/nature20556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JB, Swarts JL, Wilkins C, Thomas S, Green R, Sekine A, Voss KM, Ireton RC, Mooney M, Choonoo G, Miller DR, et al. A Mouse Model of Chronic West Nile Virus Disease. PLoS Pathog. 2016;12:e1005996. doi: 10.1371/journal.ppat.1005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JB, Thomas S, Swarts J, McMillan AA, Ferris MT, Suthar MS, Treuting PM, Ireton R, Gale M, Jr, Lund JM. Genetic diversity in the collaborative cross model recapitulates human West Nile virus disease outcomes. MBio. 2015;6:e00493-00415. doi: 10.1128/mBio.00493-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sanchez-Seco MP, Evans MJ, Best SM, Garcia-Sastre A. Zika Virus Targets Human STAT2 to Inhibit Type I Interferon Signaling. Cell Host Microbe. 2016;19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R, Wilkins C, Thomas S, Sekine A, Hendrick DM, Voss K, Ireton RC, Mooney M, Go JT, Choonoo G, Jeng S, et al. Oas1b-dependent Immune Transcriptional Profiles of West Nile Virus Infection in the Collaborative Cross. G3 (Bethesda) 2017;7:1665–1682. doi: 10.1534/g3.117.041624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guabiraba R, Marques RE, Besnard AG, Fagundes CT, Souza DG, Ryffel B, Teixeira MM. Role of the chemokine receptors CCR1, CCR2 and CCR4 in the pathogenesis of experimental dengue infection in mice. PLoS One. 2010;5:e15680. doi: 10.1371/journal.pone.0015680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MF, Lai SL, Chen JP, Sung JM, Lin YL, Wu-Hsieh BA, Gerard C, Luster A, Liao F. Both CXCR3 and CXCL10/IFN-inducible protein 10 are required for resistance to primary infection by dengue virus. J Immunol. 2006;177:1855–1863. doi: 10.4049/jimmunol.177.3.1855. [DOI] [PubMed] [Google Scholar]
- Ip PP, Liao F. Resistance to dengue virus infection in mice is potentiated by CXCL10 and is independent of CXCL10-mediated leukocyte recruitment. J Immunol. 2010;184:5705–5714. doi: 10.4049/jimmunol.0903484. [DOI] [PubMed] [Google Scholar]
- Jagger BW, Miner JJ, Cao B, Arora N, Smith AM, Kovacs A, Mysorekar IU, Coyne CB, Diamond MS. Gestational Stage and IFN-lambda Signaling Regulate ZIKV Infection In Utero. Cell Host Microbe. 2017;22:366–376.:e363. doi: 10.1016/j.chom.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julander JG, Siddharthan V. Small-Animal Models of Zika Virus. J Infect Dis. 2017;216:S919–S927. doi: 10.1093/infdis/jix465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyama N, Soma R, Hidano S, Watanabe K, Umekita H, Fukuda C, Noguchi K, Gendo Y, Ozaki T, Sonoda A, Sachi N, et al. Ribavirin inhibits Zika virus (ZIKV) replication in vitro and suppresses viremia in ZIKV-infected STAT1-deficient mice. Antiviral research. 2017;146:1–11. doi: 10.1016/j.antiviral.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor CC, Chau TN, Pang J, Davila S, Long HT, Ong RT, Dunstan SJ, Wills B, Farrar J, Van Tram T, Gan TT, et al. Genome-wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nat Genet. 2011;43:1139–1141. doi: 10.1038/ng.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SB, Choi JY, Kim JH, Uyangaa E, Patil AM, Park SY, Lee JH, Kim K, Han YW, Eo SK. Amelioration of Japanese encephalitis by blockage of 4-1BB signaling is coupled to divergent enhancement of type I/II IFN responses and Ly-6C(hi) monocyte differentiation. J Neuroinflammation. 2015;12:216. doi: 10.1186/s12974-015-0438-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RS, Lin E, Zhang B, Luster AD, Tollett J, Samuel MA, Engle M, Diamond MS. Neuronal CXCL10 directs CD8+ T-cell recruitment and control of West Nile virus encephalitis. J Virol. 2005;79:11457–11466. doi: 10.1128/JVI.79.17.11457-11466.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, Branton W, Power C, Hobman TC. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep. 2016 doi: 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFleur C, Granados J, Vargas-Alarcon G, Ruiz-Morales J, Villarreal-Garza C, Higuera L, Hernandez-Pacheco G, Cutino-Moguel T, Rangel H, Figueroa R, Acosta M, et al. HLA-DR antigen frequencies in Mexican patients with dengue virus infection: HLA-DR4 as a possible genetic resistance factor for dengue hemorrhagic fever. Hum Immunol. 2002;63:1039–1044. doi: 10.1016/s0198-8859(02)00682-1. [DOI] [PubMed] [Google Scholar]
- Lanteri MC, Kaidarova Z, Peterson T, Cate S, Custer B, Wu S, Agapova M, Law JP, Bielawny T, Plummer F, Tobler LH, et al. Association between HLA class I and class II alleles and the outcome of West Nile virus infection: an exploratory study. PLoS One. 2011;6:e22948. doi: 10.1371/journal.pone.0022948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larena M, Lobigs M. Partial dysfunction of STAT1 profoundly reduces host resistance to flaviviral infection. Virology. 2017;506:1–6. doi: 10.1016/j.virol.2017.03.001. [DOI] [PubMed] [Google Scholar]
- Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, Liu W, Ashour J, Shupert WL, Holbrook MR, Barrett AD, et al. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J Virol. 2010;84:3503–3515. doi: 10.1128/JVI.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent-Rolle M, Morrison J, Rajsbaum R, Macleod JML, Pisanelli G, Pham A, Ayllon J, Miorin L, Martinez C, tenOever BR, Garcia-Sastre A. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe. 2014;16:314–327. doi: 10.1016/j.chom.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Daniels BP, Pinto AK, Huang AC, Vick SC, Doyle SE, Gale M, Jr, Klein RS, Diamond MS. Interferon-lambda restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Sci Transl Med. 2015;7:284ra259. doi: 10.1126/scitranslmed.aaa4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS. A Mouse Model of Zika Virus Pathogenesis. Cell Host Microbe. 2016;19:720–730. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Pinto AK, Ramos HJ, Vick SC, Shrestha B, Suthar MS, Gale M, Jr, Diamond MS. Pattern recognition receptor MDA5 modulates CD8+ T cell-dependent clearance of West Nile virus from the central nervous system. J Virol. 2013;87:11401–11415. doi: 10.1128/JVI.01403-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Pinto AK, Vogt MR, Gale M, Jr, Diamond MS. Beta interferon controls West Nile virus infection and pathogenesis in mice. J Virol. 2011;85:7186–7194. doi: 10.1128/JVI.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JK, Lisco A, McDermott DH, Huynh L, Ward JM, Johnson B, Johnson H, Pape J, Foster GA, Krysztof D, Follmann D, et al. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog. 2009;5:e1000321. doi: 10.1371/journal.ppat.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JK, Obara CJ, Rivollier A, Pletnev AG, Kelsall BL, Murphy PM. Chemokine receptor Ccr2 is critical for monocyte accumulation and survival in West Nile virus encephalitis. J Immunol. 2011;186:471–478. doi: 10.4049/jimmunol.1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist R, Kurhade C, Gilthorpe JD, Overby AK. Cell-type- and region-specific restriction of neurotropic flavivirus infection by viperin. J Neuroinflammation. 2018;15:80. doi: 10.1186/s12974-018-1119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke H, Bethell DB, Phuong CX, Dung M, Schneider J, White NJ, Day NP, Farrar J, Hill AV. Strong HLA class I--restricted T cell responses in dengue hemorrhagic fever: a double-edged sword? J Infect Dis. 2001;184:1369–1373. doi: 10.1086/324320. [DOI] [PubMed] [Google Scholar]
- Lubick Kirk J, Robertson Shelly J, McNally Kristin L, Freedman Brett A, Rasmussen Angela L, Taylor RT, Walts Avram D, Tsuruda S, Sakai M, Ishizuka M, Boer Elena F, et al. Flavivirus Antagonism of Type I Interferon Signaling Reveals Prolidase as a Regulator of IFNAR1 Surface Expression. Cell Host & Microbe. 2015;18:61–74. doi: 10.1016/j.chom.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas TM, Richner JM, Diamond MS. The Interferon-Stimulated Gene Ifi27l2a Restricts West Nile Virus Infection and Pathogenesis in a Cell-Type- and Region-Specific Manner. J Virol. 2015;90:2600–2615. doi: 10.1128/JVI.02463-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Damania B. The cGAS-STING Defense Pathway and Its Counteraction by Viruses. Cell Host Microbe. 2016;19:150–158. doi: 10.1016/j.chom.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malavige GN, Rostron T, Rohanachandra LT, Jayaratne SD, Fernando N, De Silva AD, Liyanage M, Ogg G. HLA class I and class II associations in dengue viral infections in a Sri Lankan population. PLoS One. 2011;6:e20581. doi: 10.1371/journal.pone.0020581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo T, Glaser P, Lucas M, Simon-Chazottes D, Ceccaldi PE, Montagutelli X, Despres P, Guenet JL. Structural and functional genomics and evolutionary relationships in the cluster of genes encoding murine 2’,5’-oligoadenylate synthetases. Genomics. 2003;82:537–552. doi: 10.1016/s0888-7543(03)00176-9. [DOI] [PubMed] [Google Scholar]
- Mashimo T, Lucas M, Simon-Chazottes D, Frenkiel MP, Montagutelli X, Ceccaldi PE, Deubel V, Guenet JL, Despres P. A nonsense mutation in the gene encoding 2’-5’-oligoadenylate synthetase/L1 isoform is associated with West Nile virus susceptibility in laboratory mice. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11311–11316. doi: 10.1073/pnas.172195399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier KC, Gardner CL, Khoretonenko MV, Klimstra WB, Ryman KD. A mouse model for studying viscerotropic disease caused by yellow fever virus infection. PLoS Pathog. 2009;5:e1000614. doi: 10.1371/journal.ppat.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miorin L, Maestre AM, Fernandez-Sesma A, Garcia-Sastre A. Antagonism of type I interferon by flaviviruses. Biochem Biophys Res Commun. 2017;492:587–596. doi: 10.1016/j.bbrc.2017.05.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsin SN, Mahmood S, Amar A, Ghafoor F, Raza SM, Saleem M. Association of FcgammaRIIa Polymorphism with Clinical Outcome of Dengue Infection: First Insight from Pakistan. The American journal of tropical medicine and hygiene. 2015;93:691–696. doi: 10.4269/ajtmh.15-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiyer MM, Cassidy SA, Magri A, Cowton V, Chen K, Mansour S, Kranidioti H, Mbirbindi B, Rettman P, Harris S, Fanning LJ, et al. KIR2DS2 recognizes conserved peptides derived from viral helicases in the context of HLA-C. Sci Immunol. 2017;2:eaal5296. doi: 10.1126/sciimmunol.aal5296. [DOI] [PubMed] [Google Scholar]
- Ng CT, Sullivan BM, Teijaro JR, Lee AM, Welch M, Rice S, Sheehan KC, Schreiber RD, Oldstone MB. Blockade of interferon Beta, but not interferon alpha, signaling controls persistent viral infection. Cell Host Microbe. 2015;17:653–661. doi: 10.1016/j.chom.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TP, Kikuchi M, Vu TQ, Do QH, Tran TT, Vo DT, Ha MT, Vo VT, Cao TP, Tran VD, Oyama T, et al. Protective and enhancing HLA alleles, HLA-DRB1*0901 and HLA-A*24, for severe forms of dengue virus infection, dengue hemorrhagic fever and dengue shock syndrome. PLoS neglected tropical diseases. 2008;2:e304. doi: 10.1371/journal.pntd.0000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noecker CA, Amaya-Larios IY, Galeana-Hernandez M, Ramos-Castaneda J, Martinez-Vega RA. Contrasting associations of polymorphisms in FcgammaRIIa and DC-SIGN with the clinical presentation of dengue infection in a Mexican population. Acta Trop. 2014;138:15–22. doi: 10.1016/j.actatropica.2014.05.021. [DOI] [PubMed] [Google Scholar]
- Oliveira M, Lert-Itthiporn W, Cavadas B, Fernandes V, Chuansumrit A, Anunciacao O, Casademont I, Koeth F, Penova M, Tangnararatchakit K, Khor CC, et al. Joint ancestry and association test indicate two distinct pathogenic pathways involved in classical dengue fever and dengue shock syndrome. PLoS neglected tropical diseases. 2018;12:e0006202. doi: 10.1371/journal.pntd.0006202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco S, Schmid MA, Parameswaran P, Lachica R, Henn MR, Beatty R, Harris E. Characterization of a model of lethal dengue virus 2 infection in C57BL/6 mice deficient in the alpha/beta interferon receptor. J Gen Virol. 2012;93:2152–2157. doi: 10.1099/vir.0.045088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayiotou C, Lindqvist R, Kurhade C, Vonderstein K, Pasto J, Edlund K, Upadhyay AS, Overby AK. Viperin Restricts Zika Virus and Tick-Borne Encephalitis Virus Replication by Targeting NS3 for Proteasomal Degradation. J Virol. 2018;92 doi: 10.1128/JVI.02054-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascolo S, Bervas N, Ure JM, Smith AG, Lemonnier FA, Perarnau B. HLA-A2.1-restricted education and cytolytic activity of CD8(+) T lymphocytes from beta2 microglobulin (beta2m) HLA-A2.1 monochain transgenic H-2Db beta2m double knockout mice. The Journal of experimental medicine. 1997;185:2043–2051. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelygin AA, Scherbik SV, Zhulin IB, Stockman BM, Li Y, Brinton MA. Positional cloning of the murine flavivirus resistance gene. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9322–9327. doi: 10.1073/pnas.142287799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez AB, Sierra B, Garcia G, Aguirre E, Babel N, Alvarez M, Sanchez L, Valdes L, Volk HD, Guzman MG. Tumor necrosis factor-alpha, transforming growth factor-beta1, and interleukin-10 gene polymorphisms: implication in protection or susceptibility to dengue hemorrhagic fever. Hum Immunol. 2010;71:1135–1140. doi: 10.1016/j.humimm.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Perreira JM, Chin CR, Feeley EM, Brass AL. IFITMs restrict the replication of multiple pathogenic viruses. Journal of molecular biology. 2013;425:4937–4955. doi: 10.1016/j.jmb.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry ST, Buck MD, Lada SM, Schindler C, Shresta S. STAT2 mediates innate immunity to Dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog. 2011;7:e1001297. doi: 10.1371/journal.ppat.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto AK, Daffis S, Brien JD, Gainey MD, Yokoyama WM, Sheehan KC, Murphy KM, Schreiber RD, Diamond MS. A temporal role of type I interferon signaling in CD8+ T cell maturation during acute West Nile virus infection. PLoS Pathog. 2011;7:e1002407. doi: 10.1371/journal.ppat.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestwood TR, Morar MM, Zellweger RM, Miller R, May MM, Yauch LE, Lada SM, Shresta S. Gamma interferon (IFN-gamma) receptor restricts systemic dengue virus replication and prevents paralysis in IFN-alpha/beta receptor-deficient mice. J Virol. 2012;86:12561–12570. doi: 10.1128/JVI.06743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recla JM, Robledo RF, Gatti DM, Bult CJ, Churchill GA, Chesler EJ. Precise genetic mapping and integrative bioinformatics in Diversity Outbred mice reveals Hydin as a novel pain gene. Mammalian genome : official journal of the International Mammalian Genome Society. 2014;25:211–222. doi: 10.1007/s00335-014-9508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios JJ, Fleming JG, Bryant UK, Carter CN, Huber JC, Long MT, Spencer TE, Adelson DL. OAS1 polymorphisms are associated with susceptibility to West Nile encephalitis in horses. PLoS One. 2010;5:e10537. doi: 10.1371/journal.pone.0010537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivino L, Kumaran EA, Jovanovic V, Nadua K, Teo EW, Pang SW, Teo GH, Gan VC, Lye DC, Leo YS, Hanson BJ, et al. Differential targeting of viral components by CD4+ versus CD8+ T lymphocytes in dengue virus infection. J Virol. 2013a;87:2693–2706. doi: 10.1128/JVI.02675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivino L, Tan AT, Chia A, Kumaran EA, Grotenbreg GM, MacAry PA, Bertoletti A. Defining CD8+ T cell determinants during human viral infection in populations of Asian ethnicity. J Immunol. 2013b;191:4010–4019. doi: 10.4049/jimmunol.1301507. [DOI] [PubMed] [Google Scholar]
- Rossi SL, Tesh RB, Azar SR, Muruato AE, Hanley KA, Auguste AJ, Langsjoen RM, Paessler S, Vasilakis N, Weaver SC. Characterization of a Novel Murine Model to Study Zika Virus. The American journal of tropical medicine and hygiene. 2016;94:1362–1369. doi: 10.4269/ajtmh.16-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuntabhai A, Turbpaiboon C, Casademont I, Chuansumrit A, Lowhnoo T, Kajaste-Rudnitski A, Kalayanarooj SM, Tangnararatchakit K, Tangthawornchaikul N, Vasanawathana S, Chaiyaratana W, et al. A variant in the CD209 promoter is associated with severity of dengue disease. Nat Genet. 2005;37:507–513. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sam SS, Teoh BT, Chinna K, AbuBakar S. High producing tumor necrosis factor alpha gene alleles in protection against severe manifestations of dengue. Int J Med Sci. 2015;12:177–186. doi: 10.7150/ijms.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Diamond MS. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J Virol. 2005;79:13350–13361. doi: 10.1128/JVI.79.21.13350-13361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Whitby K, Keller BC, Marri A, Barchet W, Williams BR, Silverman RH, Gale M, Jr, Diamond MS. PKR and RNase L contribute to protection against lethal West Nile Virus infection by controlling early viral spread in the periphery and replication in neurons. J Virol. 2006;80:7009–7019. doi: 10.1128/JVI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster MY, Heliams DB, MacKenzie JS, Shellam GR. Genetic studies of flavivirus resistance in inbred strains derived from wild mice: evidence for a new resistance allele at the flavivirus resistance locus (Flv) J Virol. 1993;67:340–347. doi: 10.1128/jvi.67.1.340-347.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AC, de Moura EL, Ferreira JM, Santos BR, Alves VM, de Farias KF, de Souza Figueiredo EV. Meta-Analysis of the Relationship between TNF-alpha (-308G/A) and IL-10 (-819C/T) Gene Polymorphisms and Susceptibility to Dengue. Immunol Invest. 2017;46:201–220. doi: 10.1080/08820139.2016.1248560. [DOI] [PubMed] [Google Scholar]
- Sarri CA, Markantoni M, Stamatis C, Papa A, Tsakris A, Pervanidou D, Baka A, Politis C, Billinis C, Hadjichristodoulou C, Mamuris Z, et al. Genetic Contribution of MHC Class II Genes in Susceptibility to West Nile Virus Infection. PLoS One. 2016;11:e0165952. doi: 10.1371/journal.pone.0165952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidis G, Perreira JM, Portmann JM, Meraner P, Guo Z, Green S, Brass AL. The IFITMs Inhibit Zika Virus Replication. Cell reports. 2016;15:2323–2330. doi: 10.1016/j.celrep.2016.05.074. [DOI] [PubMed] [Google Scholar]
- Scherbik SV, Kluetzman K, Perelygin AA, Brinton MA. Knock-in of the Oas1b(r) allele into a flavivirus-induced disease susceptible mouse generates the resistant phenotype. Virology. 2007;368:232–237. doi: 10.1016/j.virol.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Scherbik SV, Paranjape JM, Stockman BM, Silverman RH, Brinton MA. RNase L plays a role in the antiviral response to West Nile virus. J Virol. 2006;80:2987–2999. doi: 10.1128/JVI.80.6.2987-2999.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan KC, Lai KS, Dunn GP, Bruce AT, Diamond MS, Heutel JD, Dungo-Arthur C, Carrero JA, White JM, Hertzog PJ, Schreiber RD. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2006;26:804–819. doi: 10.1089/jir.2006.26.804. [DOI] [PubMed] [Google Scholar]
- Sheehan KC, Lazear HM, Diamond MS, Schreiber RD. Selective Blockade of Interferon-alpha and -beta Reveals Their Non-Redundant Functions in a Mouse Model of West Nile Virus Infection. PLoS One. 2015;10:e0128636. doi: 10.1371/journal.pone.0128636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shresta S, Kyle JL, Snider HM, Basavapatna M, Beatty PR, Harris E. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J Virol. 2004;78:2701–2710. doi: 10.1128/JVI.78.6.2701-2710.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shresta S, Sharar KL, Prigozhin DM, Snider HM, Beatty PR, Harris E. Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. J Immunol. 2005;175:3946–3954. doi: 10.4049/jimmunol.175.6.3946. [DOI] [PubMed] [Google Scholar]
- Shrestha B, Wang T, Samuel MA, Whitby K, Craft J, Fikrig E, Diamond MS. Gamma interferon plays a crucial early antiviral role in protection against West Nile virus infection. J Virol. 2006;80:5338–5348. doi: 10.1128/JVI.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra B, Alegre R, Perez AB, Garcia G, Sturn-Ramirez K, Obasanjo O, Aguirre E, Alvarez M, Rodriguez-Roche R, Valdes L, Kanki P, et al. HLA-A, -B, -C, and -DRB1 allele frequencies in Cuban individuals with antecedents of dengue 2 disease: advantages of the Cuban population for HLA studies of dengue virus infection. Hum Immunol. 2007;68:531–540. doi: 10.1016/j.humimm.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Sierra B, Triska P, Soares P, Garcia G, Perez AB, Aguirre E, Oliveira M, Cavadas B, Regnault B, Alvarez M, Ruiz D, et al. OSBPL10, RXRA and lipid metabolism confer African-ancestry protection against dengue haemorrhagic fever in admixed Cubans. PLoS Pathog. 2017;13:e1006220. doi: 10.1371/journal.ppat.1006220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Chazottes D, Frenkiel MP, Montagutelli X, Guenet JL, Despres P, Panthier JJ. Transgenic expression of full-length 2’,5’-oligoadenylate synthetase 1b confers to BALB/c mice resistance against West Nile virus-induced encephalitis. Virology. 2011;417:147–153. doi: 10.1016/j.virol.2011.05.018. [DOI] [PubMed] [Google Scholar]
- Simon-Loriere E, Lin RJ, Kalayanarooj SM, Chuansumrit A, Casademont I, Lin SY, Yu HP, Lert-Itthiporn W, Chaiyaratana W, Tangthawornchaikul N, Tangnararatchakit K, et al. High Anti-Dengue Virus Activity of the OAS Gene Family Is Associated With Increased Severity of Dengue. J Infect Dis. 2015;212:2011–2020. doi: 10.1093/infdis/jiv321. [DOI] [PubMed] [Google Scholar]
- Singh PK, Guest JM, Kanwar M, Boss J, Gao N, Juzych MS, Abrams GW, Yu FS, Kumar A. Zika virus infects cells lining the blood-retinal barrier and causes chorioretinal atrophy in mouse eyes. JCI insight. 2017;2:e92340. doi: 10.1172/jci.insight.92340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabell AC, Meyerson NR, Gullberg RC, Gilchrist AR, Webb KJ, Old WM, Perera R, Sawyer SL. Dengue viruses cleave STING in humans but not in nonhuman primates, their presumed natural reservoir. Elife. 2018;7 doi: 10.7554/eLife.31919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens HA, Klaythong R, Sirikong M, Vaughn DW, Green S, Kalayanarooj S, Endy TP, Libraty DH, Nisalak A, Innis BL, Rothman AL, et al. HLA-A and -B allele associations with secondary dengue virus infections correlate with disease severity and the infecting viral serotype in ethnic Thais. Tissue Antigens. 2002;60:309–318. doi: 10.1034/j.1399-0039.2002.600405.x. [DOI] [PubMed] [Google Scholar]
- Sultana H, Neelakanta G, Foellmer HG, Montgomery RR, Anderson JF, Koski RA, Medzhitov RM, Fikrig E. Semaphorin 7A contributes to West Nile virus pathogenesis through TGF-beta1/Smad6 signaling. J Immunol. 2012;189:3150–3158. doi: 10.4049/jimmunol.1201140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthar MS, Diamond MS, Gale M., Jr West Nile virus infection and immunity. Nat Rev Microbiol. 2013;11:115–128. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- Suthar MS, Ma DY, Thomas S, Lund JM, Zhang N, Daffis S, Rudensky AY, Bevan MJ, Clark EA, Kaja MK, Diamond MS, et al. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog. 2010;6:e1000757. doi: 10.1371/journal.ppat.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthar MS, Ramos HJ, Brassil MM, Netland J, Chappell CP, Blahnik G, McMillan A, Diamond MS, Clark EA, Bevan MJ, Gale M., Jr The RIG-I-like receptor LGP2 controls CD8(+) T cell survival and fitness. Immunity. 2012;37:235–248. doi: 10.1016/j.immuni.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szretter KJ, Brien JD, Thackray LB, Virgin HW, Cresswell P, Diamond MS. The interferon-inducible gene viperin restricts West Nile virus pathogenesis. J Virol. 2011;85:11557–11566. doi: 10.1128/JVI.05519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szretter KJ, Daffis S, Patel J, Suthar MS, Klein RS, Gale M, Jr, Diamond MS. The innate immune adaptor molecule MyD88 restricts West Nile virus replication and spread in neurons of the central nervous system. J Virol. 2010;84:12125–12138. doi: 10.1128/JVI.01026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackray LB, Handley SA, Gorman MJ, Poddar S, Bagadia P, Briseno CG, Theisen DJ, Tan Q, Hykes BL, Jr, Lin H, Lucas TM, et al. Oral Antibiotic Treatment of Mice Exacerbates the Disease Severity of Multiple Flavivirus Infections. Cell reports. 2018;22:3440–3453.:e3446. doi: 10.1016/j.celrep.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackray LB, Shrestha B, Richner JM, Miner JJ, Pinto AK, Lazear HM, Gale M, Jr, Diamond MS. Interferon regulatory factor 5-dependent immune responses in the draining lymph node protect against West Nile virus infection. J Virol. 2014;88:11007–11021. doi: 10.1128/JVI.01545-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeaux BA, Garbino NC, Liss NM, Piper J, Blair CD, Roehrig JT. A small animal peripheral challenge model of yellow fever using interferon-receptor deficient mice and the 17D-204 vaccine strain. Vaccine. 2012;30:3180–3187. doi: 10.1016/j.vaccine.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town T, Bai F, Wang T, Kaplan AT, Qian F, Montgomery RR, Anderson JF, Flavell RA, Fikrig E. Toll-like receptor 7 mitigates lethal West Nile encephalitis via interleukin 23-dependent immune cell infiltration and homing. Immunity. 2009;30:242–253. doi: 10.1016/j.immuni.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley E, O’Connor G, Cosgrove C, Woda M, Co M, Thomas SJ, Kalayanarooj S, Yoon IK, Nisalak A, Srikiatkhachorn A, Green S, et al. Interaction of a dengue virus NS1-derived peptide with the inhibitory receptor KIR3DL1 on natural killer cells. Clinical and experimental immunology. 2016;183:419–430. doi: 10.1111/cei.12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi S, Balasubramaniam VR, Brown JA, Mena I, Grant A, Bardina SV, Maringer K, Schwarz MC, Maestre AM, Sourisseau M, Albrecht RA, et al. A novel Zika virus mouse model reveals strain specific differences in virus pathogenesis and host inflammatory immune responses. PLoS Pathog. 2017;13:e1006258. doi: 10.1371/journal.ppat.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urosevic N, Mansfield JP, Mackenzie JS, Shellam GR. Low resolution mapping around the flavivirus resistance locus (Flv) on mouse chromosome 5. Mammalian genome : official journal of the International Mammalian Genome Society. 1995;6:454–458. doi: 10.1007/BF00360653. [DOI] [PubMed] [Google Scholar]
- Valadao AL, Aguiar RS, de Arruda LB. Interplay between Inflammation and Cellular Stress Triggered by Flaviviridae Viruses. Frontiers in microbiology. 2016;7:1233. doi: 10.3389/fmicb.2016.01233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen AG, Maillard PV, Schmidt JM, Lee SA, Deddouche-Grass S, Borg A, Kjaer S, Snijders AP, Reis ESC. The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long doublestranded RNA and blocks RNA interference in mammalian cells. The EMBO journal. 2018;37 doi: 10.15252/embj.201797479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez CV, Roman AD, Lan NT, Huy NT, Mercado ES, Espino FE, Perez ML, Huong VT, Thuy TT, Tham VD, Nga CT, et al. Alpha tryptase allele of Tryptase 1 (TPSAB1) gene associated with Dengue Hemorrhagic Fever (DHF) and Dengue Shock Syndrome (DSS) in Vietnam and Philippines. Hum Immunol. 2015;76:318–323. doi: 10.1016/j.humimm.2015.03.009. [DOI] [PubMed] [Google Scholar]
- Wang K, Deubel V. Mice with different susceptibility to Japanese encephalitis virus infection show selective neutralizing antibody response and myeloid cell infectivity. PLoS One. 2011;6:e24744. doi: 10.1371/journal.pone.0024744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Chen RF, Liu JW, Lee IK, Lee CP, Kuo HC, Huang SK, Yang KD. DC-SIGN (CD209) Promoter -336 A/G polymorphism is associated with dengue hemorrhagic fever and correlated to DC-SIGN expression and immune augmentation. PLoS neglected tropical diseases. 2011;5:e934. doi: 10.1371/journal.pntd.0000934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Town T, Alexopoulou L, Anderson JF, Fikrig E, Flavell RA. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat Med. 2004;10:1366–1373. doi: 10.1038/nm1140. [DOI] [PubMed] [Google Scholar]
- Webster LT. INHERITANCE OF RESISTANCE OF MICE TO ENTERIC BACTERIAL AND NEUROTROPIC VIRUS INFECTIONS. The Journal of experimental medicine. 1937;65:261–286. doi: 10.1084/jem.65.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster LT, Clow AD. EXPERIMENTAL ENCEPHALITIS (ST. LOUIS TYPE) IN MICE WITH HIGH INBORN RESISTANCE : A CHRONIC SUBCLINICAL INFECTION. The Journal of experimental medicine. 1936;63:827–845. doi: 10.1084/jem.63.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, Fernando AN, Broadwater A, Kolla RV, De Silva AD, de Silva AM, Mattia KA, et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2046–2053. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Bangs DJ, Sidney J, Kolla RV, De Silva AD, de Silva AM, Crotty S, Peters B, Sette A. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E4256–4263. doi: 10.1073/pnas.1505956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Yauch LE, Angelo MA, John DV, Greenbaum JA, Sidney J, Kolla RV, De Silva AD, de Silva AM, Grey H, Peters B, et al. Insights into HLA-restricted T cell responses in a novel mouse model of dengue virus infection point toward new implications for vaccine design. J Immunol. 2011;187:4268–4279. doi: 10.4049/jimmunol.1101970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehorn J, Chau TN, Nguyet NM, Kien DT, Quyen NT, Trung DT, Pang J, Wills B, Van Vinh Chau N, Farrar J, Hibberd ML, et al. Genetic variants of MICB and PLCE1 and associations with non-severe dengue. PLoS One. 2013;8:e59067. doi: 10.1371/journal.pone.0059067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Dengue haemorrhagic fever: diagnosis, treatment, prevention and control. World Health Organization; Geneva: 1997. [Google Scholar]
- WHO. Dengue: Guidelines for Diagnosis, treatment, prevention and control. Spec Program Res Train Trop Dis. 2009;147 [PubMed] [Google Scholar]
- Winkler CW, Myers LM, Woods TA, Messer RJ, Carmody AB, McNally KL, Scott DP, Hasenkrug KJ, Best SM, Peterson KE. Adaptive Immune Responses to Zika Virus Are Important for Controlling Virus Infection and Preventing Infection in Brain and Testes. J Immunol. 2017;198:3526–3535. doi: 10.4049/jimmunol.1601949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler CW, Peterson KE. Using immunocompromised mice to identify mechanisms of Zika virus transmission and pathogenesis. Immunology. 2018;153:443–454. doi: 10.1111/imm.12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Liu Q, Zhou J, Xie W, Chen C, Wang Z, Yang H, Cui J. Zika virus evades interferon-mediated antiviral response through the co-operation of multiple nonstructural proteins in vitro. Cell Discov. 2017;3:17006. doi: 10.1038/celldisc.2017.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier-Carvalho C, Cardoso CC, de Souza Kehdy F, Pacheco AG, Moraes MO. Host genetics and dengue fever. Infect Genet Evol. 2017a;56:99–110. doi: 10.1016/j.meegid.2017.11.009. [DOI] [PubMed] [Google Scholar]
- Xavier-Carvalho C, Cezar R, Freire NM, Vasconcelos CMM, Solorzano VEF, de Toledo-Pinto TG, Fialho LG, do Carmo RF, Vasconcelos LRS, Cordeiro MT, Baptista P, et al. Association of rs1285933 single nucleotide polymorphism in CLEC5A gene with dengue severity and its functional effects. Hum Immunol. 2017b;78:649–656. doi: 10.1016/j.humimm.2017.07.013. [DOI] [PubMed] [Google Scholar]
- Xavier-Carvalho C, Gibson G, Brasil P, Ferreira RX, de Souza Santos R, Goncalves Cruz O, de Oliveira SA, de Sa Carvalho M, Pacheco AG, Kubelka CF, Moraes MO. Single nucleotide polymorphisms in candidate genes and dengue severity in children: a case-control, functional and meta-analysis study. Infect Genet Evol. 2013;20:197–205. doi: 10.1016/j.meegid.2013.08.017. [DOI] [PubMed] [Google Scholar]
- Yakub I, Lillibridge KM, Moran A, Gonzalez OY, Belmont J, Gibbs RA, Tweardy DJ. Single nucleotide polymorphisms in genes for 2’-5’-oligoadenylate synthetase and RNase L inpatients hospitalized with West Nile virus infection. J Infect Dis. 2005;192:1741–1748. doi: 10.1086/497340. [DOI] [PubMed] [Google Scholar]
- Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, Lin YL. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 2012;8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger RM, Shresta S. Mouse models to study dengue virus immunology and pathogenesis. Front Immunol. 2014;5:151. doi: 10.3389/fimmu.2014.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Chan YK, Lu B, Diamond MS, Klein RS. CXCR3 mediates region-specific antiviral T cell trafficking within the central nervous system during West Nile virus encephalitis. J Immunol. 2008;180:2641–2649. doi: 10.4049/jimmunol.180.4.2641. [DOI] [PubMed] [Google Scholar]
- Zhao H, Fernandez E, Dowd KA, Speer SD, Platt DJ, Gorman MJ, Govero J, Nelson CA, Pierson TC, Diamond MS, Fremont DH. Structural Basis of Zika Virus-Specific Antibody Protection. Cell. 2016;166:1016–1027. doi: 10.1016/j.cell.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]