Abstract
Metastasis is the primary cause of cancer-related mortality. Tumor cell interactions with cells of the vessel wall are decisive and potentially rate-limiting for metastasis. The molecular nature of this crosstalk is beyond candidate gene approaches hitherto poorly understood. Employing endothelial cell (EC) bulk and single-cell transcriptomics in combination with serum proteomics, we traced the evolution of the metastatic vascular niche in surgical models of lung metastasis. Temporal multiomics revealed that primary tumors systemically reprogram the body’s vascular endothelium to perturb homeostasis and to precondition the vascular niche for metastatic growth. The vasculature with its enormous surface thereby serves as amplifier of tumor-induced instructive signals. Comparative analysis of lung EC gene expression and secretome identified the Transforming growth factor-β (TGFβ) pathway specifier LRG1, Leucine-rich alpha-2-glycoprotein 1, as an early instructor of metastasis. In the presence of a primary tumor, ECs systemically upregulated LRG1 in a Signal transducer and activator of transcription 3 (STAT3)-dependent manner. A meta-analysis of retrospective clinical studies revealed a corresponding upregulation of LRG1 concentrations in the serum of cancer patients. Functionally, systemic upregulation of LRG1 promoted metastasis in mice by increasing the number of pro-metastatic NG2+ perivascular cells. In turn, genetic deletion of Lrg1 hampered growth of lung metastasis. Postsurgical adjuvant administration of an LRG1-neutralizing antibody delayed metastatic growth and increased overall survival. The study has thereby established the systems map of early primary tumor-induced vascular changes and identified LRG1 as a therapeutic target for metastasis.
Introduction
Metastasis is the primary cause of cancer-related mortality and the mechanistically least well-understood step of the tumor progression cascade (1–4). Successful metastasis relies on the close interaction of tumor cells with the metastatic vascular niche (5–7). The past decade has witnessed a fundamental change of paradigm from blood vessel wall-lining endothelial cells (ECs) being conceived as merely supportive of angiogenesis to active gatekeepers and modulators of the tumor microenvironment (8). Indeed, a growing body of evidence suggests that EC-derived angiocrine signals actively shape the metastatic niche to dictate the fate of single-seeded tumor cells (9–12).
Activated vascular niches were reported to promote metastatic colonization by inducing mesenchymal-to-epithelial transition in disseminated tumor cells and enhancing their stemness in a sex determining region Y box 2 (SOX2)/SOX9-dependent manner (10). Likewise, activated ECs release a plethora of protumorigenic cytokines including angiopoietin 2 (ANG2) (11). In turn, autocrine-acting ANG2 resulted in the amplification of endothelial inflammatory responses by promoting STAT3 signaling, and subsequently, increasing the expression of the chemoattractant C-C motif chemokine ligand 2 (CCL2) and the adhesion molecule intercellular adhesion molecule 1 (ICAM1). These molecular changes led to the selective infiltration of tumor-promoting myeloid-derived suppressor cells in the metastatic niche. Similarly, sustained activation of endothelial NOTCH signaling instigated a senescent phenotype in metastatic ECs that was accompanied by a strong increase in the expression of a wide-array of chemokines and adhesion molecules including vascular cell adhesion molecule 1 (VCAM1) (12). Quenching EC inflammatory responses either by genetic deletion of STAT3 or by pharmacological neutralization of ANG2 or VCAM1 suppressed metastatic colonization and prolonged survival of mice (11–14). Furthermore, therapeutic intervention with antibodies targeting ANG2 and/or vascular endothelial growth factor a (VEGFA) reprogramed the immune landscape to provide anti-tumor immunity, which was further potentiated by the addition of anti-PD1 therapy (15–18).
Despite numerous lines of evidence supporting the notion that vascular niches impose molecular checkpoints on metastatic progression, much of the research pursued so far has focused on well-described EC-specific signaling families in a candidate gene approach. To establish a dynamic systems map of the evolving vascular niche, we adopted an unbiased approach wherein complementary transcriptomics of the lung ECs and proteomics of sera were performed for each stage of metastatic progression. Employing postsurgical spontaneous metastasis mouse models, comparative multi-omics identified Leucine-rich alpha-2-glycoprotein 1 (LRG1) as an EC-specific angiocrine signal that assisted the process of metastasis. The expression of LRG1 was linked to systemic inflammation induced by the presence of a tumor. Elevated concentrations of circulating LRG1 promoted metastasis by increasing the number of perivascular cells in the lungs. Conversely, intervention with the LRG1-blocking antibody in clinically meaningful regimens, including postsurgical adjuvant administration, impeded metastatic growth and prolonged overall survival of mice. Beyond providing a resource for the identification of angiocrine regulators of the metastatic niche, the present study identifies and establishes LRG1 as an anti-metastatic target that warrants further clinical investigation.
Results
Temporal endothelial cell transcriptomics reveals pre-metastatic and metastatic changes in the lung vascular niche
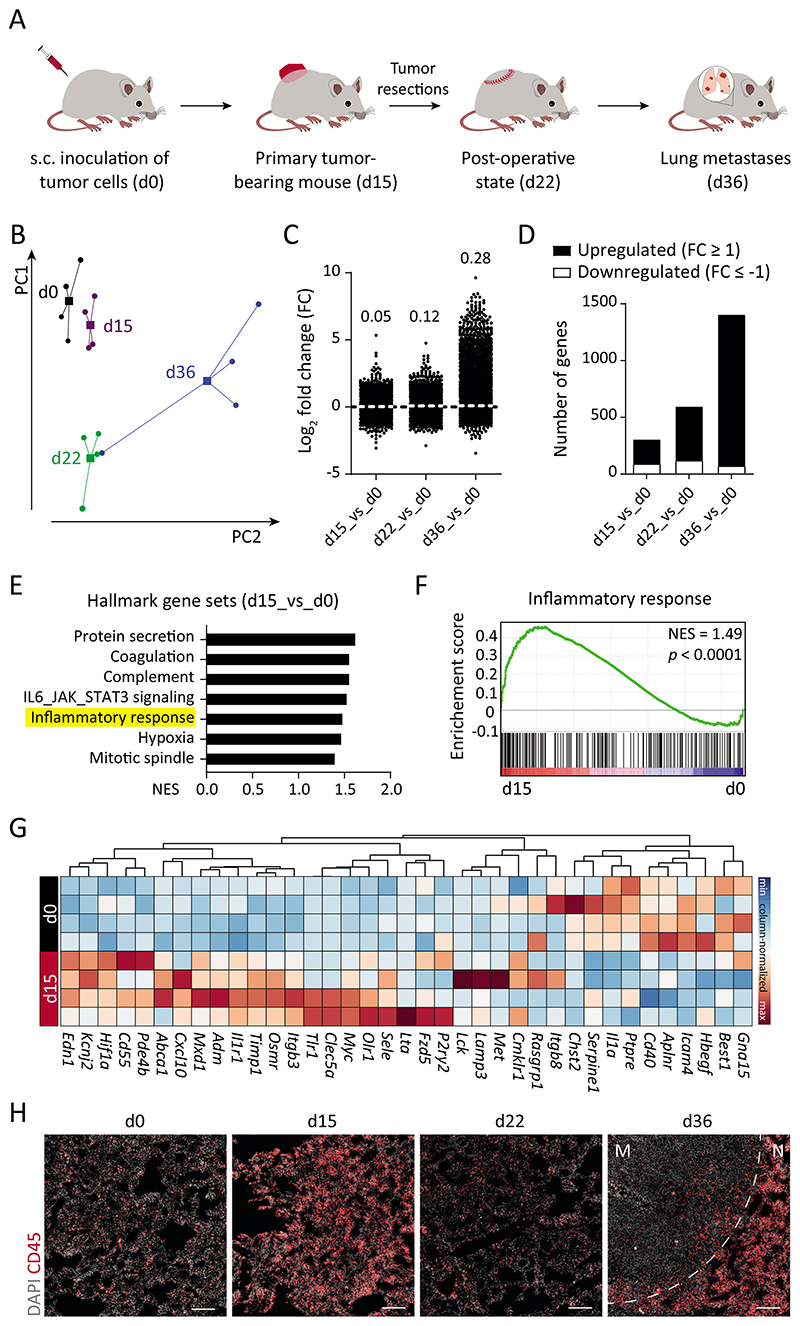
In order to identify molecular changes of ECs in the premetastatic and metastatic niche in an unbiased systems biology approach, we employed surgical metastasis models (19) and transcriptionally profiled target organ ECs over time. A primary screen was performed by subcutaneously inoculating lung metastasizing tumor cells (Lewis Lung Carcinoma, LLC) in C57BL/6N mice and analyzing lung ECs at sequential stages of tumor progression, including control (d0), small primary tumor-bearing (d15), 1 wk post-primary tumor resection (d22), and metastasis-bearing (d36) (Fig. 1A and fig. S1A). Lung ECs were isolated in high purity and used for global transcriptomic profiling (fig. S1, B to D). Differential gene expression analysis revealed transcriptional activation of ECs upon disease progression (Fig. 1, B to D and fig. S1E). Differentially altered genes at d15 and d36 were related to hallmark gene sets corresponding to protein secretion, inflammatory responses, hypoxia, and cellular proliferation (Fig. 1E and fig. S1F). The presence of a primary tumor is reported to evoke a systemic inflammation (20, 21), as evidenced by an inflammatory transcriptomic signature of lung ECs (Fig. 1, F and G). Concomitantly, strong immune cell infiltration, particularly of myeloid cells, was observed in d15 lung tissue as compared with d0 (Fig. 1H and fig. S2, A to E). A sharp decline in the expression of inflammatory genes and corresponding infiltrating immune cells was observed at d22 (Fig. 1H and figs. S1F and S2, A to E), suggesting subsided inflammation following primary tumor resection. Hence, the employed metastasis model truthfully captured the tumor cell-driven systemic alterations including initial myelopoiesis during primary tumor growth, rapid restoration of homeostasis following tumor resection, and finally myeloid cell expansion upon metastatic colonization. The immune cell infiltration in d36 lung tissue was largely restricted to the adjacent normal tissue rather than the metastatic nodules (Fig. 1H and fig. S2A). Together, the data show that the vascular and immune compartments within a metastatic organ exhibited a defined temporal signature that mirrored the kinetics of disease progression. Further comparative gene ontology analyses of disease and bio-functions supported the immune-phenotyping data and additionally identified disease stage-specific regulation of neovascularization-, cell viability- and metastasis-related gene sets (Fig. 2A).
Fig. 1. Transcriptomic evolution of lung ECs during metastasis.
(A) Schematic depiction of the LLC spontaneous metastasis model, in which C57BL/6N mice develop lung metastases around 3 weeks after primary tumor resection. (B) Principal component analysis of RNA-seq data of isolated lung ECs (n = 4 samples for each time point). Circles denote individual samples and squares denote the centroid of each group. (C) Dot plot showing Log2-fold change (FC) for genes with reads per kilobase of transcript, per million mapped reads (RPKM) ≥ 1 in at least one of the samples. The mean FC of all analyzed genes is indicated for each comparison. (D) Bar graph illustrating the number of significantly upregulated (↑) and downregulated (↓) genes in d15 (226 ↑, 89 ↓), d22 (480 ↑, 119 ↓), and d36 (1329 ↑, 71 ↓) lung ECs as compared with d0. (E) Gene Set Enrichment Analysis (GSEA) comparing d15 and d0 data sets. (F) The inflammatory response gene set was found to be positively correlated with d15 time point. (G) Heatmap highlighting genes in the inflammatory response gene set. (H) Immunofluorescence images showing infiltrating CD45+ immune cells in the lung tissue. Scale bars = 200 μm. M = metastatic nodule; N = normal adjacent tissue; NES = normalized enrichment score.
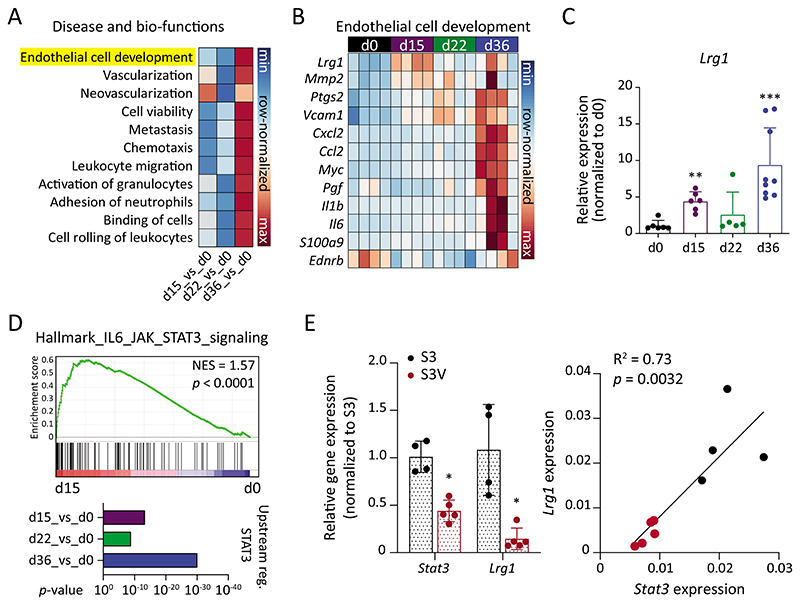
Fig. 2. Lrg1 expression closely reflects tumor progression.
(A) Comparison of disease and bio-functions was conducted using Ingenuity Pathway Analysis (IPA). Correlation scores (z-scores) are shown for the selected disease and bio-functions. (B) Genes involved in the EC development gene set are shown in row-normalized Log2-expression values. (C) qPCR quantitation of Lrg1 expression in lung ECs to validate RNA-seq data (mean ± SD, n = 5-9 mice). **, P<0.01; ***, P<0.001 (two-tailed Mann-Whitney U test, comparing to d0). (D) GSEA plot comparing IL6_JAK_STAT3 signaling between d15 and d0 (upper panel). IPA analysis to compare STAT3 as an upstream regulator during metastatic progression. (E) On the left, qPCR analysis of Stat3 and Lrg1 expression in lung ECs isolated from tumor-bearing Stat3fl/fl (S3) or Stat3fl/fl X VECadCreERT2 (S3V) mice (mean ± SD, n = 4-5 mice). *, P<0.05 (two-tailed Mann-Whitney U test, comparing to S3). On the right, Pearson’s correlation between Stat3 and Lrg1 expression.
Comparative lung transcriptomics and serum proteomics identify LRG1 as a key angiocrine factor during metastatic progression
Lrg1 was identified as one of the most differentially expressed EC-specific genes (Fig. 2, B and C and fig. S3A). LRG1 was reported to modulate endothelial TGFβ-signaling (22), promoting vascular activation and remodeling under pathological conditions. Lrg1 expression closely reflected the temporal pattern of systemic inflammation, thereby suggesting Lrg1 as an immediate endothelial response gene to tumor challenge. Endothelial STAT3 signaling has been described to actively orchestrate EC responses to inflammation and during metastasis (11, 14, 23). Concurrently, STAT3 signaling was enriched in a disease stage-specific manner in lung ECs (Fig. 2D and fig. S1F). To investigate whether STAT3 transcriptionally regulated Lrg1 expression, we employed a mouse model with EC-specific genetic deletion of Stat3 (fig. S4A). In line with previous in silico analysis of the Lrg1 promoter (24), Stat3 deletion strongly abrogated Lrg1 expression in lung ECs isolated from tumor-bearing mice (Fig. 2E). Further, primary tumor experiments in immunocompromised NSG mice manifested reduced expression of Lrg1 as compared with immunocompetent C57BL/6N mice (fig. S4B). Additionally, tumor cell-derived factors failed to directly induce Lrg1 expression in mouse lung ECs in in vitro Boyden chamber-based experiments (fig. S4C), thereby establishing LRG1 as an endothelial response factor to tumor-induced systemic inflammation.
We next performed proteomic analyses of serum specimens at sequential stages of LLC tumor progression. Consistent with the transcriptomic screen, LRG1 was one of the most abundant proteins differentially upregulated in d15 serum as compared with d0 specimens (Fig. 3A). Supporting the lung EC bulk RNA-seq data, the serum concentrations of circulating LRG1 closely reflected the temporal pattern of disease progression (Fig. 3B and fig. S4D).
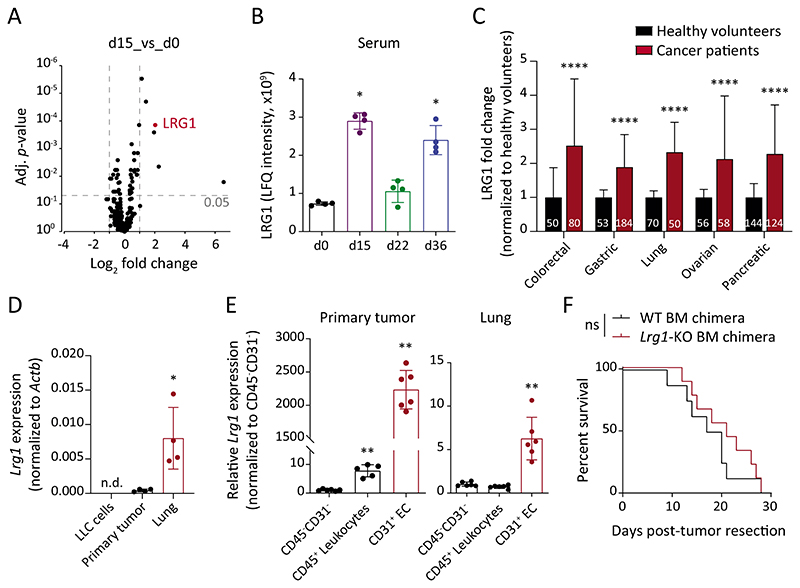
Fig. 3. LRG1 is systemically elevated during tumor progression.
(A) Volcano plot displaying FC and adjusted P-value for each identified protein in LC-MS analyses. The mean of 4 biological replicates is indicated. (B) Shown are LFQ intensities of LRG1 protein in serum samples (mean ± SD, n = 4 mice). *, P<0.05 (two-tailed Mann-Whitney U test, comparing to d0). (C) LRG1 protein amounts in sera of cancer patients and healthy volunteers were retrieved from previously published articles (25–29). The bar graph shows relative LRG1 abundance normalized to the corresponding healthy cohort. Data normalization removes differences originating due to varying measurement techniques employed in different studies. The size of each sample cohort is indicated in the graph. ****, P<0.0001 (multiple t-tests corrected with the Holm-Sidak method). (D) Comparison of Lrg1 expression between in vitro-cultured LLC cells, primary tumor and d15 lung tissue (mean ± SD, n = 4 mice). *, P<0.05 (two-tailed Mann-Whitney U test, comparing to primary tumor). (E) ECs, leukocytes, and CD31-CD45- cells were isolated from primary tumors and d15 lung tissues. Dot plots show relative Lrg1 expression in ECs and leukocytes as compared with CD31-CD45- cells (mean ± SD, n = 5-6 mice). **, P<0.01 (two-tailed Mann-Whitney U test, comparing to corresponding CD31-CD45- cells). (F) LLC tumors were implanted in WT or Lrg1-KO BM chimeras. Kaplan-Meier graph showing overall survival of mice after primary tumor resection (n = 8-9 mice). The comparison was rendered non-significant (ns) according to Log-rank (Mantel-Cox) test. n.d. = non-detectable.
To confirm the findings of the LLC screen in a second, less reductionist tumor model, we orthotopically implanted small bio-banked MMTV-PyMT breast tumor fragments in the mammary fat pads of syngeneic FVB/N mice (fig S5A) and traced spontaneous metastasis (fig. S5B). Similar to the LLC model, Lrg1 was upregulated in lung ECs and serum during metastatic progression in the MMTV-PyMT model (fig. S5, C and D). Likewise, Lrg1 expression in lung ECs was upregulated in nonobese diabetic scid gamma (NSG) mice with pancreatic PDX (PACO2) tumors when compared with wild-type littermates (fig. S5, E and F). Correspondingly, a meta-analysis of several retrospective clinical studies (25–29) revealed an upregulation of serum LRG1 concentrations for different human cancer entities, including colorectal, gastric, lung, ovarian, and pancreatic tumors, as compared with corresponding cohorts of healthy volunteers (Fig. 3C).
Endothelial cells serve as the primary source of elevated LRG1
To determine the primary source of circulating LRG1, we compared Lrg1 expression amongst in vitro cultured LLC cells, primary tumors, and d15 lung tissue. Whereas LLC cells did not express Lrg1, small amounts of Lrg1 were detectable in the primary tumor (Fig. 3D). However, lung tissue displayed substantially stronger Lrg1 expression when compared with primary tumor tissue (Fig. 3D). To further understand the cellular source of Lrg1, we isolated ECs, leukocytes, and CD31-CD45- cells (containing epithelial, mesenchymal and tumor cells) from both primary tumor and lung tissue. Lrg1 expression was enriched in the EC population (Fig. 3E and fig. S6A). In addition to ECs, hepatocyte and myeloid cells have previously been reported to express Lrg1 during homeostatic conditions (30–32). Whereas hepatocytes expressed much higher amounts of Lrg1 at steady-state (fig. S6B), only liver ECs manifested an upregulation of Lrg1 expression in the presence of a subcutaneous primary tumor (fig. S6C). As the pre-metastatic niche (d15) was found to be heavily infiltrated with myeloid cells (fig. S2, A to C), we investigated the functional relevance of leukocyte-derived LRG1 on tumor progression. To this end, bone marrow (BM)-chimeric mice were generated with either wild-type (WT) or Lrg1-KO BM cells (fig. S6, D to F). Lack of leukocyte-derived LRG1 neither affected primary tumor vasculature (fig. S6, G and H), nor did it impact overall survival of mice when compared with the WT BM-chimeras (Fig. 3F). Therefore, ECs appear to represent the major cellular source of elevated circulating LRG1 during tumor progression.
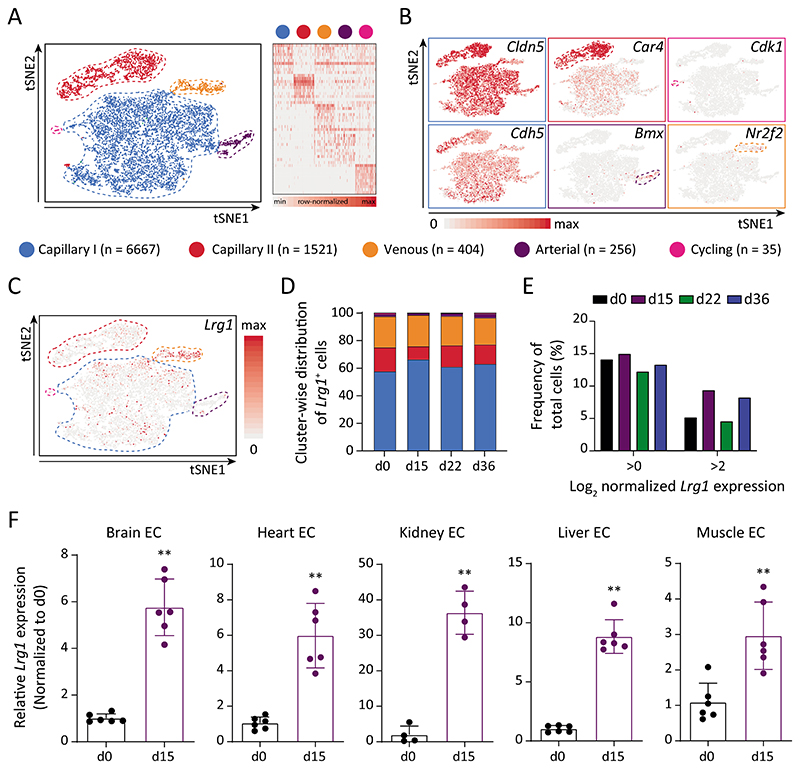
To gain insight into EC transcriptomic heterogeneity and to map Lrg1 expression across EC subpopulations, we performed single-cell RNA sequencing (scRNA-seq) of lung ECs isolated at sequential stages of tumor progression. The cellular heterogeneity was investigated both within and between the samples by applying t-distributed stochastic neighbor embedding (tSNE) and graph-based clustering. Following biologically-supervised filtering (fig. S7, A and B), 8,883 cells were annotated as capillary (sub-cluster I/II), arterial, venous, and cycling populations based on the top 10 differentially-expressed genes in each cluster (Fig. 4, A and B). The cluster annotation was in line with the current knowledge of prominent EC signaling families including Vegf-Vegfr, Ang-Tie, and Notch (fig. S7C), and corroborated with recently published single-cell data of homeostatic brain and lung ECs (33, 34). Lung ECs largely retained their arterio-venous identity as metastatic disease progressed (fig. S7D). Venous ECs were enriched for Lrg1 expression (Fig. 4C), attributing to the fact that LRG1 was initially identified as a marker for high-endothelial venules (31). Yet, Lrg1+ venous cells constituted merely 20% of total Lrg1+ cells, whereas the remaining 80% of Lrg1+ cells were uniformly dispersed amongst the other EC clusters (Fig. 4D). Although the frequency of cells expressing Lrg1 (Log2-normalized expression >0) remained largely unchanged between the samples, d15 and d36 witnessed a much higher fraction of total ECs with elevated expression of Lrg1 (Fig. 4E). In concordance with the bulk RNA-seq data, the single-cell data highlighted a systemic upregulation of Lrg1 expression in lung ECs in a tumor stage-specific pattern. Furthermore, ECs isolated from either metastatic tumors or adjacent lung tissue manifested upregulated Lrg1 expression as compared to control lung ECs (fig. S8, A and B), suggesting a uniform upregulation of Lrg1 across all lung ECs in metastasis-bearing mice. Additionally, we examined Lrg1 expression in different organ ECs and found them to be strongly upregulated at d15 across all examined vascular beds when compared with the resting vasculature (Fig. 4F). Enhanced Lrg1 expression across multiple vascular beds might have resulted in the observed increase in serum concentrations of LRG1, suggesting a corroborative role of multi-organ endothelium in systemic upregulation of tumor-induced angiocrine signals.
Fig. 4. Multi-organ vascular endothelial cells upregulate Lrg1 expression and serve as a signal amplifier.
(A) On the left, tSNE visualization of color-coded clusters of lung ECs (n = 8,883 cells). On the right, gene signature of the capillary I/II, arterial, venous, and cycling subpopulations based on 10 most-upregulated genes. (B) Feature plots indicating enriched genes for each identified subpopulation. EC-specific Cldn5 and Cdh5 were uniformly expressed by all subpopulations. (C) Feature plot displaying Lrg1 expression across all analyzed lung EC. (D) Shown is the cluster-wise spread of Lrg1-expressing cells for each sample. (E) The graph highlights the frequency of Lrg1-expressing cells (Log2-normalized expression >0 or >2) amongst the total number of cells per sample. (F) Lrg1 expression was analyzed in ECs isolated from multiple organs of d0 and d15 mice (mean ± SD, n = 4-6 mice). **, P<0.01 (two-tailed Mann-Whitney U test, comparing to d0).
Systemic upregulation of circulating LRG1 promotes metastasis
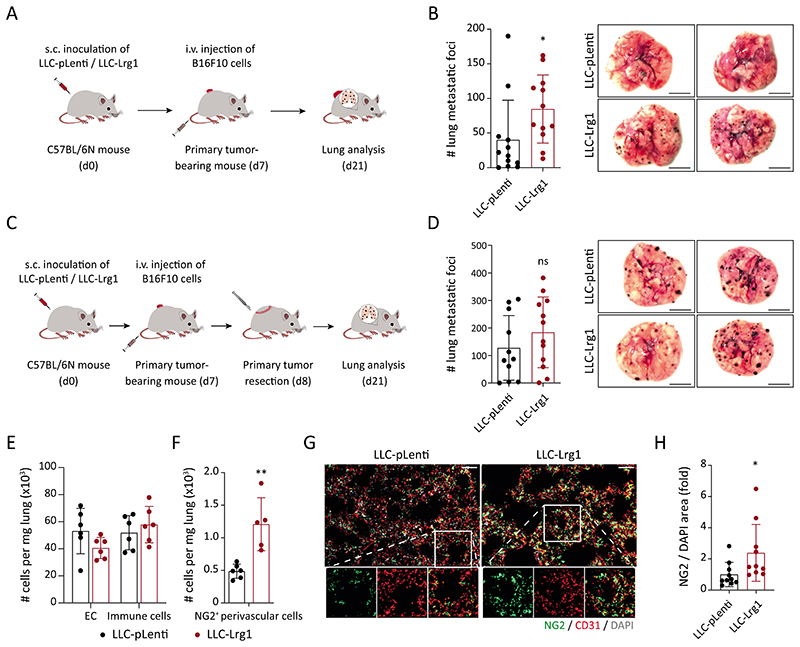
To dissect the function of LRG1 during metastatic progression, we established a systemic gain-of-function (GOF) experiment by ectopically expressing Lrg1 in LLC cells using lentiviral overexpression vectors (fig. S8, C to E). C57BL/6N mice were subcutaneously implanted with LLC-pLenti or LLC-Lrg1 tumors. Upon attaining an average tumor size of just 50 mm3, mice were intravenously injected with a second tumor cell line - melanoma (B16F10) cells to initiate an experimental metastasis assay (Fig. 5A). In this experiment setup, LLC tumors merely served as a source of secreted LRG1 and did not form metastatic colonies within the experiment’s duration. Mice with elevated concentrations of circulating LRG1 exhibited an increase in melanoma lung metastases (Fig. 5B), thereby establishing a pro-metastatic role of circulating LRG1. Likewise, in an experimental liver metastasis model, intravenous injection of WT31 cells resulted in a higher metastatic incidence in C57BL/6N mice with LRG1 GOF (fig. S8, F and G), suggesting that the pro-metastatic effects of LRG1 were not restricted to the lung. To further decipher the exact step of the metastatic cascade, LLC-pLenti and LLC-Lrg1 tumors were resected 24 h after intravenous injection of B16F10 cells (Fig. 5C). There were no differences observed between the two groups suggesting that the pro-metastatic effect of LRG1-GOF was rapidly lost upon withdrawal of the source of LRG1 during growth of metastases (Fig. 5D). To rule out any direct effect of LRG1 on tumor cell extravasation, we employed an LRG1-neutralizing antibody 15C4 (35, 36). Mice were preconditioned with a single injection of either 15C4 or control-IgG prior to intravenous injection of melanoma cells (fig. S9A). Consistent with the previous results, blocking LRG1 did not affect the extravasation of melanoma cells (fig. S9B).
Fig. 5. Systemic upregulation of LRG1 promotes metastasis.
(A) Lrg1-overexpressing LLC (LLC-Lrg1) or control-LLC (LLC-pLenti) cells were subcutaneously inoculated in C57BL/6N mice. 7 days after inoculation, melanoma (B16F10) cells were intravenously injected to initiate an experimental metastasis assay. (B) On the left, dot plot showing the number of melanoma metastases in the lung, and on the right, representative lung images (mean ± SD, n = 12 mice). Scale bars = 5 mm. *, P<0.05 (two-tailed Mann-Whitney U test, comparing to LLC-pLenti). (C) C57BL/6N mice were injected subcutaneously with LLC (pLenti/Lrg1) cells. On day 7, tumor-bearing mice were injected intravenously with melanoma (B16F10) cells. Primary tumors (the source of LRG1) were resected 24 hours after the intravenous injection of melanoma cells. (D) On the left, dot plot showing the number of melanoma metastases in the lung, and on the right, representative lung images (mean ± SD, n = 11-12 mice). The comparison was rendered non-significant (ns, P= 0.31) according to two-tailed Mann-Whitney U test. (E-H) WT or NG2-Cre X YFPfl/fl mice were injected with either LLC-Lrg1 or LLC-pLenti cells. FACS-based quantitation of ECs, immune cells, and NG2+ perivascular cells in the lung of tumor-bearing mice (E, F) (mean ± SD, n = 5-6 mice). **, P<0.01 (two-tailed Mann-Whitney U test, comparing to LLC-pLenti). Lung tissue sections were stained for NG2 (pericyte-specific). Representative images of lung sections (G). Scale bars = 50 μm. Quantitation of NG2/DAPI area is shown (H) (mean ± SD, n = 10 mice). *, P<0.05 (two-tailed Mann-Whitney U test, comparing to LLC-pLenti).
To investigate the functional impact of LRG1 on the metastatic niche, we quantitated different stromal populations in the lung (fig. S10, A and B). LRG1-GOF neither influenced EC proliferation nor did it affect the infiltration of different immune cells (Fig. 5E and fig. S10, C and D), thereby pointing towards an angiogenesis- and immune-independent role of LRG1 on the lung metastatic niche. We observed an increased number of NG2+ lung perivascular cells with upregulation of circulating LRG1 (Fig. 5F). Concomitant to the FACS analysis, immunostainings marked an increase in neural/glial antigen 2 (NG2) stained area in the lungs of mice with LRG1-GOF (Fig. 5, G and H). Further, we found an increase in normalized Desmin area in the lungs of LRG1-GOF mice, whereas alpha smooth muscle actin (aSMA), s100 calcium binding protein a4 (S100A4)/ fibroblast-specific protein-1 (FSP1) and platelet derived growth factor receptor alpha (PDGFRa) stained lung area remained largely unaltered (fig. S11A). These data suggest that LRG1-GOF resulted in an enhanced number of lung pericytes. Next, we corroborated these in vivo findings by conducting an in vitro experiment in which lung pericytes were stimulated with tumor conditioned media (CM). Indeed, incubation with CM derived from Lrg1-overexpressing LLC cells (Lrg1-CM) could enhance the proliferation of lung pericytes as compared with CM from control tumor cells (fig. S12A). Furthermore, global transcriptomic profiling of CM-treated lung pericytes revealed that Lrg1-CM led to an upregulation in the expression of MYC, a central orchestrator of cell cycle progression (37), and its downstream target genes (fig. S12, B and C). These NG2+ perivascular cells were recently described to establish a conducive metastatic niche and facilitate metastasis (38). Therefore, the data suggest that LRG1 facilitates the expansion of perivascular cells to support metastasis. Our findings are consistent with previous studies suggesting an upregulation of LRG1 to promote lung and cardiac fibrosis and to correlate with the expression of genes involved in glomerular fibrosis during kidney diseases (39–43).
To study the impact of LRG1-GOF on spontaneous metastasis, we traced metastatic progression in mice injected with either LLC-pLenti or LLC-Lrg1 cells. Analyzing lungs two weeks after primary tumor resection, we recorded an increase in metastatic burden in mice with Lrg1-GOF (fig. S13A), substantiating a pro-metastatic role of LRG1. Next, we employed Lrg1-KO mice to investigate metastatic progression in a loss-of-function (LOF) experiment. Following the resection of similar sized primary tumors (fig. S13B), Lrg1-KO mice manifested lower metastatic burden and reduced Desmin area in the lungs of Lrg1-KO mice when compared with control mice (fig. S13, C and D). Overall, both GOF and LOF experiments support a crucial role of LRG1 during metastasis.
LRG1-blocking antibody suppresses metastatic progression and prolongs survival
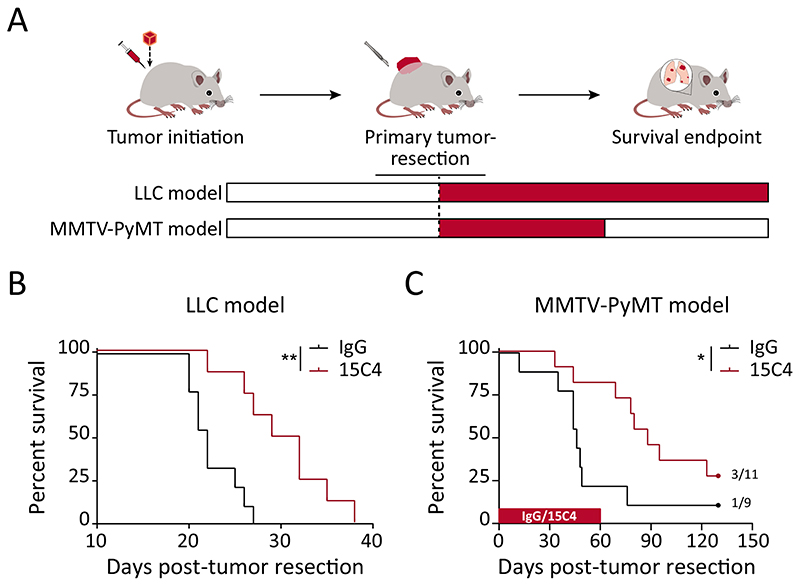
To assess the therapeutic potential of blocking LRG1 during metastatic progression, we administered an LRG1-neutralizing antibody 15C4 (35, 36) as postsurgical adjuvant therapy in two spontaneously metastasizing mouse models – the LLC and the MMTV-PyMT model (Fig. 6A). The administration of anti-LRG1 (15C4) or control-IgG was commenced 1-day post-primary tumor resection. In the LLC model, long-term adjuvant therapy resulted in reduced lung metastasis burden (fig. S14, A and B) and prolonged overall survival of mice by 8.5 days, which corresponded to an approximately 40% improvement over the control-IgG treated group (Fig. 6B and fig. S14C). Anti-LRG1 (15C4) as a monotherapy offered a substantial overall survival advantage in the LLC model that has previously been reported to be refractory to anti-VEGF therapy (44) and in which chemotherapy shows no effect on lung metastatic burden (11). Likewise, for the breast cancer (MMTV-PyMT) metastasis model, postsurgical adjuvant administration of anti-LRG1 (15C4) nearly doubled the median survival of mice when compared with IgG-treated mice (Fig. 6C and fig. S14C). A subset of mice developed lung metastases after the cessation of anti-LRG1 (15C4) treatment, suggesting LRG1-neutralization could delay the outgrowth of previously-seeded tumor cells in the lungs. Neutralizing LRG1 in a postsurgical adjuvant setting suppressed metastasis, thereby providing a substantial survival benefit in clinically-relevant metastasis models.
Fig. 6. LRG1 neutralization delays metastatic progression and prolongs overall survival.
(A) Therapeutic assessment of LRG1-blocking antibody 15C4 in LLC and MMTV-PyMT murine metastasis models using a postsurgical adjuvant strategy. (B) For the LLC tumor model, Kaplan-Meier graphs showing overall survival of mice when treated with control-IgG or anti-LRG1 (15C4) antibody in the postsurgical adjuvant (B; n = 8-9 mice) setting. **, P<0.01 [Log-rank (Mantel-Cox) test]. (C) For the MMTV-PyMT tumor model, Kaplan-Meier graph showing overall survival of mice when treated with control-IgG or anti-LRG1 (15C4) antibody in the postsurgical adjuvant setting (n = 9-11 mice). Mice were treated from day 1 until day 60 post-primary tumor resection with IgG or anti-LRG1 (15C4) antibody. 3 of 11 15C4-treated and 1 of 9 control-IgG-treated mice were healthy until the end of experiment. *, P<0.05 [Log-rank (Mantel-Cox) test]. For moribund mice (n = 8-9 for the LLC model and n = 8 per treatment group for the MMTV-PyMT model), fig. S14C shows the incidence of lung metastases.
Discussion
Exploiting a comparative systems biology approach, the present study captured the temporal evolution of vascular changes in the pre-metastatic and metastatic niches. In-depth bulk RNA-seq analysis of lung ECs complemented with serum proteomics served as a versatile tool for the identification of angiocrine molecules (https://augustinlab.dkfz.de/metastasis). The temporal approach with surgical removal of the primary tumor facilitated the discrimination between pre-metastatic and metastatic EC transcriptomic changes. Analyzing the temporal evolution of lung EC transcriptome uncovered gene signatures that are driven primarily by the presence of a primary tumor. The comparison between primary tumor-bearing (d15) and post-operative (d22, one week after surgery) mice illustrated that the lung ECs revert back to the homeostatic state following primary tumor resection. Likewise, d22 lungs had far lower number of infiltrating myeloid cells as compared with d15 time point, suggesting a quenching of systemic inflammation post-tumor resection.
Adult endothelial cells remain largely quiescent (45). However, blood vessels have been reported to orchestrate cytokine amplification and mitigate immune cell infiltration during pathological conditions such as viral infection and cancer (46–48). Lung ECs overexpressed a wide array of cytokines including Il1b, Il6, Ccl2, and Cxcl2 in the presence of a metastatic tumor. We data-mined for secreted angiocrine factors and identified an early EC-specific STAT3-dependent responsive signal, LRG1, that was tightly calibrated to the tumor-induced inflammation. In vitro Boyden chamber experiments suggested that tumor cell-derived factors failed to induce Lrg1 expression in mouse lung ECs, thereby hinting at tumor-associated inflammation as a potential intermediary factor that caused upregulation of Lrg1 expression in mouse lung EC. STAT3 has previously been reported to be a crucial mediator of EC responsiveness to an inflammatory challenge (11, 23, 49). Although the data presented here suggest that tumor-induced inflammation activates STAT3 signaling in ECs, thereby resulting in the upregulation of Lrg1 expression, future studies will need to identify circulating soluble factors that mediate systemic activation of STAT3 in distant endothelial cells. Likewise, the effect of anti-inflammatory drugs on STAT3 activation and subsequent expression of Lrg1 remains to be evaluated. Importantly, consistent induction, albeit of varying magnitudes, of Lrg1 expression in lung ECs of mice from different genetic backgrounds (C57BL/6N, FVB/N and NSG) in the presence of different cancers (lung - LLC, breast – MMTV-PyMT and pancreatic – PACO2) hints toward LRG1 as a common EC response signal to tumor-associated systemic alterations.
The analysis of multiple vascular beds unveiled a similar upregulation of Lrg1 expression during tumor progression. In the LLC postsurgical experiments, the lung remained the primary and the most frequent site for metastasis. We rarely observed metastases to the liver and never to the brain, heart, kidney or muscle. Despite not being frequent metastatic sites, brain, heart, kidney, liver and muscle ECs experienced a strong increase in Lrg1 expression in the presence of a distant primary tumor. The widespread regulation of Lrg1 expression throughout the vascular tree supports the notion that the vascular endothelium with its vast surface may serve as an amplifier of tumor-induced systemically-acting instructive signals. As such, multiorgan ECs secrete angiocrine signals that can proteomically be detected in the circulation. In line with our preclinical data, retrospective clinical studies analyzing sera from cancer patients have reported increased concentrations of LRG1 when compared with healthy volunteers as highlighted in the meta-analysis in figure 3C. Monitoring serum LRG1 concentrations allowed the discrimination of early-stage pancreatic cancer from chronic pancreatitis, and addition of LRG1 immunoassay to CA19-9 and TIMP1 led to robust detection of early-stage pancreatic cancers (50). These observations suggest that serum LRG1 may serve as a prognostic marker for monitoring tumor relapse.
Systemic upregulation of LRG1 was dispensable for extravasation but facilitated growth of tumor cells at distant metastatic sites. LRG1 was previously described to promote pathological angiogenesis in models of ocular neo-vascular diseases (22). However, LRG1 GOF did not enhance lung EC proliferation in our study. It is noteworthy that we observed a very small fraction (approximately 0.5%) of lung EC entering cell cycle. These data suggest that small lung metastases analyzed in the current study did not induce EC proliferation but potentially survived via vessel co-option. Our observations are in line with numerous published preclinical and clinical studies demonstrating that early lung and liver metastases rely on vessel co-option rather than sprouting angiogenesis (39, 51–55). Instead, we found that the elevated concentrations of LRG1 promoted proliferation of NG2+ perivascular cells in the lungs. These perivascular cells were found to assist in establishing a conducive niche for the colonization of disseminated tumor cells (38). Concomitantly, intervention with anti-LRG1 antibody (15C4) suppressed metastasis when administered in a postsurgical adjuvant regimen. Intervention with 15C4 seemed to keep seeded tumor cells in a dormancy-like state as lung metastases developed in MMTV-PyMT mice only following withdrawal of antibody therapy. Our preclinical data support a crucial role of LRG1 in tumor metastasis and warrant further translational studies of LRG1 as a therapeutic target for restricting metastasis.
The study’s limitations include that, in line with transcriptomic screens, subsequent mechanistic experiments were focused on the lung niche. Whereas Lrg1 expression was upregulated across all analyzed vascular beds in the presence of a primary tumor, the tumor models employed in the present study allowed reliable and quantitative readouts only in the lung and liver. Future studies, employing tumor models that preferentially metastasize to other organs, will need to investigate the effect of elevated LRG1 on the local tissue microenvironment and subsequent metastases. Moreover, our findings in preclinical mouse tumor models and meta-analysis of published clinical data need to be further validated with larger cohorts of human specimens, particularly in regards to LRG1 serving as a potential biomarker for early metastatic growth and disease relapse.
In conclusion, the comparative analysis of lung transcriptomics and serum proteomics offers a unique database to identify disease stage-specific angiocrine factors, such as LRG1, that not only mediate the locally confined cellular crosstalk within the metastatic niche but can also manifest a systemic response to body-wide alterations during metastatic progression.
Materials and Methods
Study design
The objective of this study was to investigate the role of EC-derived signals in orchestrating a conducive metastatic niche. Comparative analysis of EC transcriptomic and serum proteomic screens resulted in the identification of LRG1 as a key angiocrine molecule. Employing preclinical murine metastasis models, we assessed the effect of either genetic or pharmacological inhibition of LRG1 on metastatic progression. This was a hypothesis-driven study with unknown "effect magnitude". Therefore, statistical power was not computed prior to the experiments. The employed sample sizes were estimated based on the previous experience with the experimental models. For postsurgical metastasis experiments, mice with primary tumor regrowth were excluded for the survival analysis. For immuno-fluorescence image quantitation, images were excluded if artifacts were observed during automated signal thresholding. Individual mice served as biological replicates within an in vivo experiment. All in vitro experiments were repeated at least three independent times. Mice were randomized prior to initiating an in vivo experiment. In therapy experiments (Fig. 6), mice were randomly assigned by a blinded scientist into the cohorts of treatment. All enrolled mice for a therapy experiment were handled by animal caretakers. Mice were examined on daily basis for the experimental endpoint criteria. Animal caretakers had no knowledge about the experimental groups.
Mice
C57BL/6N, NSG and FVB/N mice were purchased from Charles River. NG2-Cre x YFPfl/fl and VECadCreERT2 X Stat3fl/fl were bred in-house at the DKFZ animal facility. Lrg1 transgenic mice were generated by the University of California Davies knockout mouse project (KOMP) repository (http://www.komp.org/) and bred at the Institute of Ophthalmology, University College London. 8-12 wks. old mice were used in this study unless otherwise indicated. All mice were housed on a 12 h light/dark cycle with free access to food and drinking water in specific pathogen-free animal facilities. All animal experiments were approved by the governmental (G164/16, G231/16, G254/18, G286/18, G9/19, G196/19, and G213/18 from Regierungspräsidium Karlsruhe, Germany,) and institutional (IRCBC-2018-006 to JH) Animal Care and Use Committees. All experiments were performed in accordance with the respective institutional guidelines for the care and use of laboratory animals.
Cells
LLC, bEnd3 and B16F10 cells (ATCC) were maintained according to ATCC standard culture instructions. WT31 cells were kindly provided by C. Géraud, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany. WT31 cells were cultured in RPMI-1640 media supplemented with 10% fetal calf serum (FCS) and 1% penicillin-streptomycin. Primary mouse lung microvascular ECs (mLECs) were purchased from Cell Biologics and were cultured in the manufacturer’s recommended media. Human lung pericytes (LPs) were kindly provided by B. Peault (University of Edinburgh, UK) and cultured in pericyte medium (ScienCell, #1201) supplemented with 2% FCS, 1% of the corresponding pericyte growth supplement and 1% penicillin-streptomycin. LLC cells were transduced with lentivirus to overexpress either Lrg1 or control vector pLenti. All cells were cultured at 37°C and 5% CO2 and routinely tested for mycoplasma by polymerase chain reaction (PCR).
Tumor models
All mice were routinely checked for the experimental endpoint criteria: (i) metastasis with a diameter of >1.5 cm, (ii) weight loss of 20% (compared with normal weight), (iii) lack of feed or water intake, (iv) noticeable respiratory symptoms, (v) abnormal posture or crouching, (vi) apathy or immobility, and (vii) pale extremities (13).
LLC tumor model
LLC cells (1x106 in phosphate-buffered saline (PBS)) were subcutaneously inoculated in C57BL/6N, NSG, or Lrg1-transgenic mice. Primary tumors were surgically resected at an average size of 300 mm3. For the therapy experiment, mice were administered with 50 mg/kg either anti-LRG1 antibody (UCL, clone 15C4) or control-IgG (BioXcell, clone MOPC-21) twice a week where therapy was initiated one day after tumor resections until the experimental endpoint criteria were reached. Image analysis for computing metastatic area was performed using FIJI software.
MMTV-PyMT tumor model
Bio-banked tumor fragments (˜10 mm3 in volume) were orthotopically implanted in the fourth mammary pad of syngeneic FVB/N mice. Primary tumors were surgically resected at an average size of 500 mm3. For adjuvant treatment, therapy was initiated one day after tumor resections until day 60 after tumor resection.
PACO2 pancreatic PDX model
The PACO2 tumor model was employed as previously described (56). Briefly, PACO2 cells (2x105 in Matrigel) were injected orthotopically into the pancreas. Engraftment of tumors and subsequent growth were monitored by regular palpation of the implantation site and in vivo bioluminescence. Mice were euthanized 60 days after injection. At the time of sacrifice, ex vivo bioluminescence imaging was performed on the lungs.
B16F10 experimental metastasis assay
C57BL/6N mice were pretreated with one shot of either anti-LRG1 or Control-IgG. 3 days later, B16F10 cells (2x105 in PBS) were injected into the tail vein. Lungs were collected two weeks after tumor cell inoculation, and metastatic foci were counted under a stereo-microscope.
LRG1 systemic GOF and B16F10 experimental metastasis assay
Lrg1-overexpressing or control LLC cells (1x106 in PBS) were inoculated subcutaneously in C57BL/6N mice. 7 days later, B16F10 melanoma cells (2x105 in PBS) were injected into the tail vein. Two weeks after B16F10 cell injection, lungs were collected, and melanoma metastatic foci were counted under a stereo-microscope. In a follow-up experiment (refer to Fig. 5C), primary LLC tumors were resected 24 h after intravenous injection of B16F10 cells.
LRG1 systemic GOF and WT31 experimental metastasis assay
Lrg1-overexpressing or control LLC cells (1x106 in PBS) were inoculated subcutaneously in C57BL/6N mice. 7 days later, WT31 cells (2.5x106 in PBS) were injected slowly into the tail vein as previously described (57). Two weeks later, livers were collected, and melanoma metastatic foci were analyzed under a stereo-microscope.
EC-specific Stat3 deletion
Stat3fl/fl mice were crossed with VECadCreERT2 mice to have tamoxifen-inducible EC-specific deletion of Stat3. For inducing Stat3 deletion, both Stat3fl/fl and VECadCreERT2 X Stat3fl/fl mice were injected intraperitoneally with 2mg tamoxifen (5x in two weeks). After induction, LLC cells were injected subcutaneously to initiate a tumor experiment.
Bone marrow chimeric mice
Bone marrow cells were isolated from Lrg1-KO or wild-type littermate mice. Genotyping PCR for LacZ (forward primer: TCCTGGTGGGAGAGGACTC; reverse primer: GTCTGTCCTAGCTTCCTCACTG) was performed to ensure the knockout of Lrg1. Bone marrow chimeras were generated as described previously (58). Briefly, 8-wk-old WT mice were lethally irradiated with a total dose of 9 Gy (split dose, 2 × 4.5 Gy). After a 2 h rest, mice were injected with Lrg1-KO/WT bone marrow cells, consisting of approximately 5000 LSK cells, via the tail vein. 2 months later, LLC metastasis experiment was performed in BM chimeras. Leukocyte chimerism of recipient animals was determined by analyzing Lrg1 gene expression in splenocytes after sacrificing animals.
Flow cytometry
EC isolation
Tissues were dissociated into single cell suspension with Liberase digestion enzyme mix (Roche). For brain, the single cell suspension was mixed with 22% bovine serum albumin in distilled water and centrifuged at 1300g for 15 mins to remove myelin. Following ACK lysis, ECs were enriched using CD31 microbeads (Miltenyi Biotec) according to the manufacturer’s instructions. Enriched ECs were further FACS-sorted for the surface marker profile CD45-LYVE1-PDPN-TER-119-CD31+ using fluorescence-conjugated antibodies [CD45 (30-F11), CD31 (MEC 13.3), PDPN (eBio8.1.1), TER-119 (TER-119), and LYVE1 (ALY7)].
EdU proliferation assay
Mice were injected 1mg of EdU (5-Ethynyl-2’-deoxyuridine) in PBS intraperitoneally. 16 h later, mice were sacrificed and lungs were dissected. Tissues were dissociated into single cell suspension with Liberase digestion enzyme mix (Roche). Following ACK lysis, single cell suspensions were stained with fluorescence-conjugated antibodies [CD45 (30-F11), CD31 (MEC 13.3), PDPN (eBio8.1.1), TER-119 (TER-119), and LYVE1 (ALY7)]. Thereafter, EdU staining were performed using Click-iT EdU assay kit (ThermoFisher Scientific) according to the manufacturer’s instructions.
FACS-based immunophenotyping
In the LLC metastasis model, lung and spleen tissues were collected at sequential stages of tumor progression and analyzed as described in (59). Briefly, lung tissue was dissociated into a single cell suspension with Liberase digestion enzyme mix (Roche). Spleen tissue was mechanically dissociated into single cell suspension. Following erythrocyte lysis, the remaining single cell solution was divided for lymphoid [CD45 (30-F11), CD3ε (17A2), CD4 (GK1.5), CD8a (53–6.7), CD45R-B220 (RA3-6B2), and NK- 1.1 (PK136)] and myeloid [CD45 (30-F11), CD11b (M1/70), Ly-6C (HK-1.4), Ly-6G (1A8), F4/80 (BM8), and CD11c (N418)] staining.
Dead cells were excluded by FxCycle Violet staining. Stained cells were analyzed using a BD bioscience Aria cell sorting platform, and frequency of individual cell populations was quantified with FlowJo software.
Bulk RNA-sequencing and data analysis
Lung EC transcriptomics
Lung ECs were isolated from four biological replicates at each stage of tumor progression and total RNA was isolated using Arcturus PicoPure RNA isolation kit (ThermoFisher Scientific) according to the manufacturer’s instructions. Quality control was performed by Bioanalyzer (Agilent) measurements. The sequencing library was generated with 10 ng of total RNA using the SMARTer Ultra Low RNA kit for Illumina sequencing (Clontech) according to the manufacturer’s protocol. Sequencing reads (100 bp paired-end) were generated on the HiSeq2000 platform (Illumina) with four samples per lane. The sequenced reads were aligned to the mouse reference genome mm10 using STAR aligner (60). Differential gene expression was computed using DEseq2 (61). Only transcripts with reads per kilobase of transcript, per million mapped reads (RPKM) ≥ 1 in at least one sample were considered for the downstream analysis. Gene Set Enrichment Analysis (GSEA) and Ingenuity Pathway Analysis (IPA) were undertaken to study regulated molecular pathways and corresponding biological functions.
Lung pericyte transcriptomics
Lung pericytes treated with tumor CM from four experimental repeats were used to isolate total RNA using GeneElute total mammalian RNA extraction kit (Merck-Sigma) according to the manufacturer’s instructions. Quality control was performed by Bioanalyzer (Agilent) measurements. The sequencing library was generated using the TruSeq Stranded mRNA Library Prep Kit (Illumina) according to the manufacturer’s protocol. Sequencing reads (100 bp single-end) were generated on the NovaSeq 6000 platform (Illumina). The sequenced reads were aligned to the human reference transcriptome (hg19) using Kallisto (version 0.43.1) (62). Only transcripts with transcripts per million reads (TPM) ≥ 1 were considered for the downstream gene set analysis using GSEA.
scRNA-seq
Lung ECs were isolated from four biological replicates at each stage of tumor progression. Cell suspensions, consisting of 10,000 randomly-selected cells, for each time point, were separately loaded on a Chromium Single Cell Instrument (10X Genomics). scRNA-seq libraries were prepared using Chromium Single cell 3’ Library (v2), Gel beads & Multiplex kit (10X Genomics). Multiplexed libraries were sequenced on the HiSeq4000 platform (Illumina).
scRNA-seq data processing
The sequenced data were aligned to the mouse reference genome (mm10) using Cell Ranger (version 2.1.1), thereby generating gene-barcode matrices. Low-quality cells, containing less than 200 detected genes or mitochondrial genes accounting for more than 10% of total transcripts, were filtered out. Outlying cells were identified by three median absolute deviations away from the median through scater R package (version 1.10.1). After quality control, Seurat R package (version 3.1.1) was used for further analysis (63). Count matrices for all quality-controlled cells per time point were read as individual Seurat objects (9,036 cells for 4 time-points). A set of anchors between four Seurat objects was calculated using FindIntegrationAnchors command (package version 3.1.1) using all default parameters. For the neighbor space search, 10, 20, 30, 40 and 50 dimensions were tested. Because there was no major difference with increasing dimensions, ten dimensions were used for final integration. The computed anchors from the previous step were used to further integrate Seurat objects from all time-points. The integrated data was scaled and further analysis was done on the scaled data.
Unsupervised clustering and visualization
We reduced dimensions of the merged data using principal component analysis. Initial unsupervised clustering with FindClusters function at resolution 0.25 yielded 8 clusters. Differentially expressed genes of each cluster were identified with FindAllMarkers function (marker genes for each cluster compared against all other clusters combined). t-Distributed Stochastic Neighbor Embedding (t-SNE) was employed for data visualization.
Biologically-supervised annotation and filtering
We manually annotated unsupervised clusters based on expression of previously-identified markers of different cell types in each cluster. Two clusters were identified as contaminants (mural cells and lymphatic ECs) and excluded from further analysis. Thereafter, 8,883 cells were reanalyzed for differential gene expression and re-clustered using resolution parameter set to 0.2. Two out of the six identified clusters that were weakly-distinguishable and embedded closely in t-SNE, were merged together. Altogether, we detected five clusters as indicated.
Serum proteomics
Sample and library preparation
Protein concentration in serum samples was measured with Bicinchoninic Acid (BCA) assay according to the manufacturer’s protocol (ThermoFisher Scientific). Samples were prepared according to previously published protocol (64). Briefly, 50 μg of protein were diluted in a total volume of 100 μL of 0.1% (w/v) RapiGest-SF (Waters) dissolved in 50 mM ammonium bicarbonate (ABC) (pH 8) and heated for 15 min at 95°C. Prior to protein digestion, disulfide bonds were reduced using dithiothreitol (DTT, 5 mM final concentration) for 30 min at 60°C and subsequently alkylated with 2-chloroacetamide (CAA, 15 mM final concentration) for 30 min at room temperature (RT). Proteolytic digestion was performed with trypsin (sequencing grade modified, Promega) in a protease-to-protein ratio of 1:50 (w/w) overnight at 37°C, shaking at 700 rpm. Following digestion, trifluoroacetic acid (TFA) was added (pH < 2) to a final concentration of 1% and incubated for 20 min at 37°C to stop digestion and break down RapiGest, followed by centrifugation at 20,000g for 10 min. The peptide-containing supernatants were collected in new tubes. MS injection-ready samples were stored at -20°C. For relative quantification, samples were subjected to liquid chromatography–mass spectrometry (LC-MS) analysis in single shots. Additionally, a library of protein identification was generated by deep fractionating a pool of all samples, using high pH reverse phase liquid chromatography.
The pooled library-sample was adjusted to pH 10 with ammonium formate at a final concentration of 20 mM. Peptide fractionation was performed on a 1200 Infinity HPLC system (Agilent) with a Gemini C18 column (3 μm, 110 Å, 100 × 1.0 mm; Phenomenex) using a linear 60 min gradient from 0% to 35% (v/v) ACN in 20 mM ammonium formate (pH 10) at a flow rate of 0.1 mL/min. Sixty 1-min fractions were collected, and pooled into twelve fractions,dried, and reconstituted in 0.1% formic acid (FA). MS injection-ready samples were stored at -20°C.
LC-MS analysis
The single shot samples and 12 library fractions were injected using an Easy-nLC 1200 nano-UPLC (ThermoFisher Scientific) onto a trap column (PepMap, 100 μm x 2cm, C18, 5 μm 100Å pores) at a constant flow of solvent A (0.1% FA in water) at a maximum pressure of 800 bar, and separated on an analytical column (PepMap RSLC 75 μm × 50 cm, C18, 2 μM, 100Å) at a constant flow of 0.3 μl/minute, at 55°C by applying a multistep gradient. During elution, the percentage of solvent B (0.1% FA, 80% ACN 19.9% water) was increased linearly from 3% to 8% in 4 minutes, then from 8% to 10% in 2 minutes, then from 10% to 32% in further 17 minutes, and then to 50% B in 3 minutes. Finally, the gradient was finished with 8 minutes at 100% solvent B, followed by 11 minutes 97% solvent A.
Eluting peptides were electro-sprayed by applying 2 kV on a 360 μm OD x 20μm ID; 10 μm Picotip coated emitter (New Objective) into a Q Exactive HF quadrupole mass spectrometer (ThermoFisher Scientific). The capillary temperature was set to 275°C. The mass spectrometer was operated in data dependent mode of acquisition. Briefly, for each cycle, one Full MS spectrum was acquired in the Orbitrap with a mass range of 350-1500 m/z and a resolution of 60,000 FWHM at 200 m/z. The automated gain control (AGC) target was set to 3 x 106 with a maximum injection time of 32 ms. Precursor ions were filtered according to charge state (required 2-7 z) and monoisotopic peak assignment. The top 20 most abundant ions per full scan were selected for an MS2 acquisition. Previously interrogated precursors were excluded using a dynamic exclusion window (40 s ± 10 ppm tolerance). For MS2 scans the resolution was set to 15,000 FWHM with an AGC of 1 x 105 ions and maximum fill time of 50 ms.
MS data processing and protein identification
The mass spectra were processed with MaxQuant (V1.5.1.2) using the Andromeda search engine against UniProtKB/Swiss-Prot databases of Mus musculus (13.09.2017), with the following search settings: digestion enzyme was set to trypsin/P, with a maximum of two missed cleavages allowed (65, 66). Precursor and product ion tolerances were set at 20 ppm and 0.5 Da, respectively. Carbamidomethylation of cysteine was set as a fixed modification, oxidation of methionine and acetylation (protein N-term) were set as variable modification. The match between run functions was enabled to match from the “library” to the “single shot samples” with a time window of 0.7 min and an alignment window of 20 min. A minimum of one unique peptide and a false discovery rate below 0.01 was set for peptide and protein identification. The protein quantification was performed using the label-free quantification algorithm of MaxQuant. As a decoy database, reversed sequences of the target database were used. If not stated otherwise, MaxQuant settings were left as default. For identification of differentially expressed proteins the LFQ values extracted from the protein groups table were used and Linear Models for Microarray Data (Limma, Version 3.36.2; Rstudio, Version 1.1.456), with a P-value below 0.05 was performed. The mass spectrometry data files have been deposited to the ProteomeXchange Consortium (67) under the accession number PXD013978.
In vitro Boyden chamber experiment. mLECs were seeded in 6-well plates. After forming monolayers, mLECs were co-cultured either alone or with a Boyden chamber (0.4 μm pore size) consisting of LLC cells for 48 h. Thereafter, total RNA was isolated from mLECs for gene expression analysis.
In vitro condition media (CM) experiment. 500,000 LLC cells (pLenti or Lrg1-overexpressing) were seeded in a 10 cm dish and allowed to form a monolayer. Thereafter, complete media was replaced by 5mL serum-free media (SFM, 0% Dulbecco's modified eagle medium (DMEM)) for 24 hours. Simultaneously, 100,000 lung pericytes per well were seeded in a 6-well dish. 24 hours later, complete pericyte media was replaced with 1:1 mixture of 1mL basal pericyte medium and 1mL of SFM, CM-pLenti, or CM-Lrg1 supplemented with 10μM EdU. 3 hours later, lung pericytes were processed either for isolating total RNA for gene expression analysis or for EdU staining using Click-iT Plus EdU Proliferation Kit (Thermo Fischer Scientific) according to the manufacturer’s protocol.
Immunofluorescence stainings and analysis
Primary tumors and lung tissues were stained and analyzed as described in (59). Briefly, tumor and lung tissues were embedded in Tissue-Tek OCT compound and were cut into 5 to 7 μm sections. Tissue sections were fixed in ice-cold methanol and were blocked using 10% ready-to-use normal goat serum (Thermo Fisher Scientific). The tissue sections were then incubated overnight at 4°C with primary antibodies [rat anti-CD31 (BD Biosciences, catalogue 550300); rabbit anti-Desmin (Abcam, catalogue Ab15200-1); rat anti-CD45 (Cedarlane labs, catalogue CL9446AP); rat anti-CD140a/PDGFRa (eBioscience, catalogue 14-1401-82); rabbit anti-NG2/CSPG4 (Merck-Millipore, catalogue AB5320); rabbit anti-FSP1/S100A4 (Abcam, catalogue ab27957)]. Staining with fluorescence-conjugated secondary antibodies (Thermo Fisher Scientific) was performed next day for 1 h at room temperature. Thereafter, tissue sections were incubated with PE-conjugated antibodies [CD3ε (17A2); CD4 (GK1.5); CD8a (53–6.7); CD11b (M1/70); Gr-1 (RB6-8C5)] for co-staining. Cell nuclei were stained with DAPI (Merck-Sigma). Images were taken using a Zeiss AxioScan slide scanner, and image analysis was performed using FIJI software.
Gene expression analysis
Total RNA was transcribed into cDNA using the Quantitect reverse transcription kit (Qiagen). Quantitative PCRs were performed with Taqman master mix (ThermoFisher Scientific). Taqman primers (Kdr - Mm01222421_m1; Flt1 - Mm00438980_m1; Icam2 - Mm00494862_m1; Cldn5 - Mm00727012_s1; Lrg1 - Mm01278767_m1; Mmp2 - Mm00439498_m1; Ptgs2 - Mm00478374_m1; Vcam1 - Mm01320970_m1; Cxcl2 - Mm00436450_m1; Ccl2 - Mm00441242_m1; Myc - Mm00487804_m1; Pgf - Mm00435613_m1; Il1b - Mm00434228_m1; Il6 - Mm01210733_m1; S100a9 - Mm00656925_m1; Ednrb - Mm00432989_m1; Actb - Mm00607939_S1; Pecam1 - Mm01242584_m1; MYC – Hs00153408_m1; ACTB - Hs01060665_g1) were ordered from Thermo Fisher Scientific. Gene expression was calculated based on the ΔΔCt relative quantification method. mRNA abundances were normalized to Actb or Pecam1 expression as indicated.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 8/9 (GraphPad Software). Data are expressed as the mean ± SD. Employed statistical tests are indicated in corresponding figure legends. A P-value of less than 0.05 was considered statistically significant.
Supplementary Material
One sentence summary.
A temporal systems biology screen of pre-metastatic endothelial cells identified LRG1 as a therapeutic target for restricting metastases.
Acknowledgements
The authors would like to thank J. Sleeman (ECAS, Medical Faculty Mannheim, Heidelberg University, Germany) for providing MMTV-PyMT tumors. We thank R. Adams (Max Planck Institute for Molecular Biomedicine, Muenster, Germany) for providing VECadCreERT2 mice. We thank F. Constantini (Columbia University, New York, NY) for providing Rosa-YFPfl/fl (YFPfl/fl) mice. We thank B. Peault (University of Edinburgh, UK) for kindly providing human lung pericytes. We thank J-P. Mallm, I. Heras Murillo and C. Previti for technical assistance. We are most grateful for the excellent technical support of the Flow Cytometry, the Light Microscopy, the Genomics and Proteomics, the Omics IT and Data Management, the Single-cell Open Lab and the Laboratory Animal Facilities of the DKFZ and IRCBC.
Funding
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) (project C5 within CRC1366 “Vascular control of organ function” [project number 39404578 to H.G.A. and C.M.] and projects A2 and Z4 within CRC1324 “Wnt signaling” [project number 331351713 to H.G.A. and J.K.]), the European Research Council Advanced Grant “AngioMature” [project 787181 to H.G.A.], the DFG-funded Research Training Group 2099 “Hallmarks of Skin Cancer” [project P8 to H.G.A.], and the Integrate-TN consortium funded by the Deutsche Krebshilfe and the Dietmar Hopp Foundation (to A.T.). J.H. is supported by the “The Thousand Young Talents Recruitment Program”. J.G. and S.E.M. are supported by the Wellcome Trust Investigator Award [206413/B/17/Z] and Medical Research Council UK [MR/N006410/1]. M.K. received a fellowship from the Naito Foundation and the Nakatomi Foundation.
Footnotes
Author contributions: MS, NG, AAAP, and HGA conceived and designed the study. MS, NG, AAAP, MK, CC, SFP, BL, EE, MH, MRS, AT, CM, and JH performed in vivo and in vitro experiments. BJK, and JK performed serum proteomics. LH, SRK, and MSc performed data analysis for bulk and single-cell RNAseq experiments. DMK, SEM, and JG provided reagents. EB, and DH provided technical support. MS, NG, AAAP, and HGA analyzed and interpreted data. MS and HGA supervised the project. MS, NG, AAAP, and HGA wrote the manuscript. All authors discussed the results and commented on the manuscript.
Competing interests: J.G. and S.E.M. are inventors on U.S. Pat. No. 10,556,007, entitled “Antibody which binds Lrg1 and methods of use”.
Data and materials availability
All data associated with this study are in the paper or the Supplementary Materials. NGS data have been deposited in the Gene Expression Omnibus under accession numbers GSE173482 (bulk LP RNA-seq), GSE131072 (bulk EC RNA-seq), GSE131110 (single-cell EC RNA-seq). The mass spectrometry data files have been deposited to the ProteomeXchange Consortium under the accession number PXD013978. The 15C4 antibody for non-commercial research purposes can be obtained from J.G. and S.E.M. under institutional material transfer agreement.
References
- 1.Abdul Pari AA, Singhal M, Augustin HG. Emerging paradigms in metastasis research. J Exp Med. 2021;218:e20190218. doi: 10.1084/jem.20190218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B, McGraw T, Mittal V. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer. 2019;19:9–31. doi: 10.1038/s41568-018-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steeg PS. Targeting metastasis. Nat Rev Cancer. 2016;16:201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Augustin HG, Koh GY. Organotypic vasculature: From descriptive heterogeneity to functional pathophysiology. Science. 2017;357:eaal2379. doi: 10.1126/science.aal2379. [DOI] [PubMed] [Google Scholar]
- 6.Butler JM, Kobayashi H, Rafii S. Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nat Rev Cancer. 2010;10:138–146. doi: 10.1038/nrc2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massague J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singhal M, Augustin HG. Beyond angiogenesis: Exploiting angiocrine factors to restrict tumor progression and metastasis. Cancer Res. 2020;80:659–662. doi: 10.1158/0008-5472.CAN-19-3351. [DOI] [PubMed] [Google Scholar]
- 9.Cao Z, Scandura JM, Inghirami GG, Shido K, Ding BS, Rafii S. Molecular checkpoint decisions made by subverted vascular niche transform indolent tumor cells into chemoresistant cancer stem cells. Cancer Cell. 2017;31:110–126. doi: 10.1016/j.ccell.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esposito M, Mondal N, Greco TM, Wei Y, Spadazzi C, Lin SC, Zheng H, Cheung C, Magnani JL, Lin SH, Cristea IM, et al. Bone vascular niche E-selectin induces mesenchymal-epithelial transition and Wnt activation in cancer cells to promote bone metastasis. Nat Cell Biol. 2019;21:627–639. doi: 10.1038/s41556-019-0309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srivastava K, Hu J, Korn C, Savant S, Teichert M, Kapel SS, Jugold M, Besemfelder E, Thomas M, Pasparakis M, Augustin HG. Postsurgical adjuvant tumor therapy by combining anti-angiopoietin-2 and metronomic chemotherapy limits metastatic growth. Cancer Cell. 2014;26:880–895. doi: 10.1016/j.ccell.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Wieland E, Rodriguez-Vita J, Liebler SS, Mogler C, Moll I, Herberich SE, Espinet E, Herpel E, Menuchin A, Chang-Claude J, Hoffmeister M, et al. Endothelial Notch1 activity facilitates metastasis. Cancer Cell. 2017;31:355–367. doi: 10.1016/j.ccell.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Gengenbacher N, Singhal M, Mogler C, Hai L, Milde L, Pari AAA, Besemfelder E, Fricke C, Baumann D, Gehrs S, Utikal J, et al. Timed Ang2-targeted therapy identifies the Angiopoietin-Tie pathway as key regulator of fatal lymphogenous metastasis. Cancer Discov. 2021;11:424–445. doi: 10.1158/2159-8290.CD-20-0122. [DOI] [PubMed] [Google Scholar]
- 14.Kim KJ, Kwon SH, Yun JH, Jeong HS, Kim HR, Lee EH, Ye SK, Cho CH. STAT3 activation in endothelial cells is important for tumor metastasis via increased cell adhesion molecule expression. Oncogene. 2017;36:5445–5459. doi: 10.1038/onc.2017.148. [DOI] [PubMed] [Google Scholar]
- 15.Schmittnaegel M, Rigamonti N, Kadioglu E, Cassara A, Wyser Rmili C, Kiialainen A, Kienast Y, Mueller HJ, Ooi CH, Laoui D, De Palma M. Dual angiopoietin-2 and VEGFA inhibition elicits antitumor immunity that is enhanced by PD-1 checkpoint blockade. Sci Transl Med. 2017;9:eaak9670. doi: 10.1126/scitranslmed.aak9670. [DOI] [PubMed] [Google Scholar]
- 16.Kloepper J, Riedemann L, Amoozgar Z, Seano G, Susek K, Yu V, Dalvie N, Amelung RL, Datta M, Song JW, Askoxylakis V, et al. Ang-2/VEGF bispecific antibody reprograms macrophages and resident microglia to anti-tumor phenotype and prolongs glioblastoma survival. Proc Natl Acad Sci U S A. 2016;113:4476–4481. doi: 10.1073/pnas.1525360113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterson TE, Kirkpatrick ND, Huang Y, Farrar CT, Marijt KA, Kloepper J, Datta M, Amoozgar Z, Seano G, Jung K, Kamoun WS, et al. Dual inhibition of Ang-2 and VEGF receptors normalizes tumor vasculature and prolongs survival in glioblastoma by altering macrophages. Proc Natl Acad Sci U S A. 2016;113:4470–4475. doi: 10.1073/pnas.1525349113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen E, Jabouille A, Rivera LB, Lodewijckx I, Missiaen R, Steri V, Feyen K, Tawney J, Hanahan D, Michael IP, Bergers G. Combined antiangiogenic and anti-PD-L1 therapy stimulates tumor immunity through HEV formation. Sci Transl Med. 2017;9:eaak9679. doi: 10.1126/scitranslmed.aak9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gengenbacher N, Singhal M, Augustin HG. Preclinical mouse solid tumour models: status quo, challenges and perspectives. Nat Rev Cancer. 2017;17:751–765. doi: 10.1038/nrc.2017.92. [DOI] [PubMed] [Google Scholar]
- 20.DePalma M, Biziato D, Petrova TV. Microenvironmental regulation of tumour angiogenesis. Nat Rev Cancer. 2017;17:457–474. doi: 10.1038/nrc.2017.51. [DOI] [PubMed] [Google Scholar]
- 21.Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, Psaila B, Kaplan RN, Bromberg JF, Kang Y, Bissell MJ, et al. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer. 2017;17:302–317. doi: 10.1038/nrc.2017.6. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Abraham S, McKenzie JAG, Jeffs N, Swire M, Tripathi VB, Luhmann UFO, Lange CAK, Zhai Z, Arthur HM, Bainbridge J, et al. LRG1 promotes angiogenesis by modulating endothelial TGF-beta signalling. Nature. 2013;499:306–311. doi: 10.1038/nature12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kano A, Wolfgang MJ, Gao Q, Jacoby J, Chai GX, Hansen W, Iwamoto Y, Pober JS, Flavell RA, Fu XY. Endothelial cells require STAT3 for protection against endotoxin-induced inflammation. J Exp Med. 2003;198:1517–1525. doi: 10.1084/jem.20030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naka T, Fujimoto M. LRG is a novel inflammatory marker clinically useful for the evaluation of disease activity in rheumatoid arthritis and inflammatory bowel disease. Immunol Med. 2018;41:62–67. doi: 10.1080/13497413.2018.1481582. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto M, Takahashi T, Serada S, Sugase T, Tanaka K, Miyazaki Y, Makino T, Kurokawa Y, Yamasaki M, Nakajima K, Takiguchi S, et al. Overexpression of leucine-rich alpha2-glycoprotein-1 is a prognostic marker and enhances tumor migration in gastric cancer. Cancer Sci. 2017;108:2052–2060. doi: 10.1111/cas.13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shinozaki E, Tanabe K, Akiyoshi T, Tsuchida T, Miyazaki Y, Kojima N, Igarashi M, Ueno M, Suenaga M, Mizunuma N, Yamaguchi K, et al. Serum leucine-rich alpha-2-glycoprotein-1 with fucosylated triantennary N-glycan: a novel colorectal cancer marker. BMC Cancer. 2018;18:406. doi: 10.1186/s12885-018-4252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu YS, Luo XY, Li QR, Li H, Li C, Ni H, Li RX, Wang R, Hu HC, Pan YJ, Chen HQ, et al. Shotgun and targeted proteomics reveal that pre-surgery serum levels of LRG1, SAA, and C4BP may refine prognosis of resected squamous cell lung cancer. J Mol Cell Biol. 2012;4:344–347. doi: 10.1093/jmcb/mjs050. [DOI] [PubMed] [Google Scholar]
- 28.Furukawa K, Kawamoto K, Eguchi H, Tanemura M, Tanida T, Tomimaru Y, Akita H, Hama N, Wada H, Kobayashi S, Nonaka Y, et al. Clinicopathological significance of Leucine-rich alpha2-glycoprotein-1 in sera of patients with pancreatic cancer. Pancreas. 2015;44:93–98. doi: 10.1097/MPA.0000000000000205. [DOI] [PubMed] [Google Scholar]
- 29.Andersen JD, Boylan KL, Jemmerson R, Geller MA, Misemer B, Harrington KM, Weivoda S, Witthuhn BA, Argenta P, Vogel RI, Skubitz AP. Leucine-rich alpha-2-glycoprotein-1 is upregulated in sera and tumors of ovarian cancer patients. J Ovarian Res. 2010;3:21. doi: 10.1186/1757-2215-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Donnell LC, Druhan LJ, Avalos BR. Molecular characterization and expression analysis of leucine-rich alpha2-glycoprotein, a novel marker of granulocytic differentiation. J Leukoc Biol. 2002;72:478–485. [PubMed] [Google Scholar]
- 31.Saito K, Tanaka T, Kanda H, Ebisuno Y, Izawa D, Kawamoto S, Okubo K, Miyasaka M. Gene expression profiling of mucosal addressin cell adhesion molecule-1+ high endothelial venule cells (HEV) and identification of a leucine-rich HEV glycoprotein as a HEV marker. J Immunol. 2002;168:1050–1059. doi: 10.4049/jimmunol.168.3.1050. [DOI] [PubMed] [Google Scholar]
- 32.Shirai R, Hirano F, Ohkura N, Ikeda K, Inoue S. Up-regulation of the expression of leucine-rich alpha(2)-glycoprotein in hepatocytes by the mediators of acute-phase response. Biochem Biophys Res Commun. 2009;382:776–779. doi: 10.1016/j.bbrc.2009.03.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanlandewijck M, He L, Mae MA, Andrae J, Ando K, DelGaudio F, Nahar K, Lebouvier T, Lavina B, Gouveia L, Sun Y, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554:475–480. doi: 10.1038/nature25739. [DOI] [PubMed] [Google Scholar]
- 34.Vila Ellis L, Cain MP, Hutchison V, Flodby P, Crandall ED, Borok Z, Zhou B, Ostrin EJ, Wythe JD, Chen J. Epithelial vegfa specifies a distinct endothelial population in the mouse lung. Dev Cell. 2020;52:617–630. doi: 10.1016/j.devcel.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moss S, Kallenberg D, Tripathi V, Davis S, George J, O’Connor M, Dowsett L, Greenwood J. Preclinical development and testing of a therapeutic antibody against LRG1. Cancer Res. 2018;78:13. [Google Scholar]
- 36.Kallenberg D, Tripathi V, Javaid F, Pilotti C, George J, Davis S, Blackburn JWD, O’Connor M, Dowsett L, Bowers CE, Liyanage S, et al. A humanized antibody against LRG1 that inhibits angiogenesis and reduces retinal vascular leakage. bioRxiv. 2020:2020.2007.2025.218149 [Google Scholar]
- 37.Bretones G, Delgado MD, Leon J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849:506–516. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 38.Murgai M, Ju W, Eason M, Kline J, Beury DW, Kaczanowska S, Miettinen MM, Kruhlak M, Lei H, Shern JF, Cherepanova OA, et al. KLF4-dependent perivascular cell plasticity mediates pre-metastatic niche formation and metastasis. Nat Med. 2017;23:1176–1190. doi: 10.1038/nm.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuczynski EA, Vermeulen PB, Pezzella F, Kerbel RS, Reynolds AR. Vessel co-option in cancer. Nat Rev Clin Oncol. 2019;16:469–493. doi: 10.1038/s41571-019-0181-9. [DOI] [PubMed] [Google Scholar]
- 40.Liu C, Lim ST, Teo MHY, Tan MSY, Kulkarni MD, Qiu B, Li A, Lal S, Dos Remedios CG, Tan NS, Wahli W, et al. Collaborative regulation of LRG1 by TGF-beta1 and PPAR-beta/delta modulates chronic pressure overload-induced cardiac fibrosis. Circ Heart Fail. 2019;12:e005962. doi: 10.1161/CIRCHEARTFAILURE.119.005962. [DOI] [PubMed] [Google Scholar]
- 41.Hong Q, Zhang L, Fu J, Verghese DA, Chauhan K, Nadkarni GN, Li Z, Ju W, Kretzler M, Cai GY, Chen XM, D’Agati VD, et al. LRG1 promotes diabetic kidney disease progression by enhancing TGF-beta-induced angiogenesis. J Am Soc Nephrol. 2019;30:546–562. doi: 10.1681/ASN.2018060599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haku S, Wakui H, Azushima K, Haruhara K, Kinguchi S, Ohki K, Uneda K, Kobayashi R, Matsuda M, Yamaji T, Yamada T, et al. Early enhanced leucine-rich alpha-2-glycoprotein-1 expression in glomerular endothelial cells of type 2 diabetic nephropathy model mice. Biomed Res Int. 2018;2018:2817045. doi: 10.1155/2018/2817045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Honda H, Fujimoto M, Serada S, Urushima H, Mishima T, Lee H, Ohkawara T, Kohno N, Hattori N, Yokoyama A, Naka T. Leucine-rich alpha-2 glycoprotein promotes lung fibrosis by modulating TGF-beta signaling in fibroblasts. Physiol Rep. 2017;5:e13556. doi: 10.14814/phy2.13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shojaei F, Wu X, Malik AK, Zhong C, Baldwin ME, Schanz S, Fuh G, Gerber HP, Ferrara N. Tumor refractoriness to anti-VEGF treatment is mediated by CD11b+Gr1+ myeloid cells. Nat Biotechnol. 2007;25:911–920. doi: 10.1038/nbt1323. [DOI] [PubMed] [Google Scholar]
- 45.Schlereth K, Weichenhan D, Bauer T, Heumann T, Giannakouri E, Lipka D, Jaeger S, Schlesner M, Aloy P, Eils R, Plass C, et al. The transcriptomic and epigenetic map of vascular quiescence in the continuous lung endothelium. Elife. 2018;7:e34423. doi: 10.7554/eLife.34423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pasquier J, Ghiabi P, Chouchane L, Razzouk K, Rafii S, Rafii A. Angiocrine endothelium: from physiology to cancer. J Transl Med. 2020;18:52. doi: 10.1186/s12967-020-02244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mantovani A, Bussolino F, Dejana E. Cytokine regulation of endothelial cell function. FASEB J. 1992;6:2591–2599. doi: 10.1096/fasebj.6.8.1592209. [DOI] [PubMed] [Google Scholar]
- 48.Teijaro JR, Walsh KB, Cahalan S, Fremgen DM, Roberts E, Scott F, Martinborough E, Peach R, Oldstone MB, Rosen H. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell. 2011;146:980–991. doi: 10.1016/j.cell.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dutzmann J, Daniel JM, Bauersachs J, Hilfiker-Kleiner D, Sedding DG. Emerging translational approaches to target STAT3 signalling and its impact on vascular disease. Cardiovasc Res. 2015;106:365–374. doi: 10.1093/cvr/cvv103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Capello M, Bantis LE, Scelo G, Zhao Y, Li P, Dhillon DS, Patel NJ, Kundnani DL, Wang H, Abbruzzese JL, Maitra A, et al. Sequential validation of blood-based protein biomarker candidates for early-stage pancreatic cancer. J Natl Cancer Inst. 2017;109:djw266. doi: 10.1093/jnci/djw266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bridgeman VL, Vermeulen PB, Foo S, Bilecz A, Daley F, Kostaras E, Nathan MR, Wan E, Frentzas S, Schweiger T, Hegedus B, et al. Vessel co-option is common in human lung metastases and mediates resistance to anti-angiogenic therapy in preclinical lung metastasis models. J Pathol. 2017;241:362–374. doi: 10.1002/path.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frentzas S, Simoneau E, Bridgeman VL, Vermeulen PB, Foo S, Kostaras E, Nathan M, Wotherspoon A, Gao ZH, Shi Y, Van den Eynden G, et al. Vessel co-option mediates resistance to anti-angiogenic therapy in liver metastases. Nat Med. 2016;22:1294–1302. doi: 10.1038/nm.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holash J, Maisonpierre PC, Compton D, Boland P, Alexander CR, Zagzag D, Yancopoulos GD, Wiegand SJ. Vessel cooption, regression, and growth in tumors mediated by angiopoietins and VEGF. Science. 1999;284:1994–1998. doi: 10.1126/science.284.5422.1994. [DOI] [PubMed] [Google Scholar]
- 54.Pezzella F, Pastorino U, Tagliabue E, Andreola S, Sozzi G, Gasparini G, Menard S, Gatter KC, Harris AL, Fox S, Buyse M, et al. Non-small-cell lung carcinoma tumor growth without morphological evidence of neo-angiogenesis. Am J Pathol. 1997;151:1417–1423. [PMC free article] [PubMed] [Google Scholar]
- 55.Szabo V, Bugyik E, Dezso K, Ecker N, Nagy P, Timar J, Tovari J, Laszlo V, Bridgeman VL, Wan E, Frentzas S, et al. Mechanism of tumour vascularization in experimental lung metastases. J Pathol. 2015;235:384–396. doi: 10.1002/path.4464. [DOI] [PubMed] [Google Scholar]
- 56.Noll EM, Eisen C, Stenzinger A, Espinet E, Muckenhuber A, Klein C, Vogel V, Klaus B, Nadler W, Rosli C, Lutz C, et al. CYP3A5 mediates basal and acquired therapy resistance in different subtypes of pancreatic ductal adenocarcinoma. Nat Med. 2016;22:278–287. doi: 10.1038/nm.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wohlfeil SA, Hafele V, Dietsch B, Schledzewski K, Winkler M, Zierow J, Leibing T, Mohammadi MM, Heineke J, Sticht C, Olsavszky V, et al. Hepatic endothelial Notch activation protects against liver metastasis by regulating endothelial-tumor cell adhesion independent of angiocrine signaling. Cancer Res. 2019;79:598–610. doi: 10.1158/0008-5472.CAN-18-1752. [DOI] [PubMed] [Google Scholar]
- 58.Singhal M, Liu X, Inverso D, Jiang K, Dai J, He H, Bartels S, Li W, Abdul Pari AA, Gengenbacher N, Besemfelder E, et al. Endothelial cell fitness dictates the source of regenerating liver vasculature. J Exp Med. 2018;215:2497–2508. doi: 10.1084/jem.20180008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singhal M, Gengenbacher N, La Porta S, Gehrs S, Shi J, Kamiyama M, Bodenmiller DM, Fischl A, Schieb B, Besemfelder E, Chintharlapalli S, et al. Preclinical validation of a novel metastasis-inhibiting Tie1 function-blocking antibody. EMBO Mol Med. 2020;12:e11164. doi: 10.15252/emmm.201911164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 63.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.:e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kramer G, Woolerton Y, van Straalen JP, Vissers JP, Dekker N, Langridge JI, Beynon RJ, Speijer D, Sturk A, Aerts JM. Accuracy and reproducibility in quantification of plasma protein concentrations by mass spectrometry without the use of isotopic standards. PLoS One. 2015;10:e0140097. doi: 10.1371/journal.pone.0140097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 66.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 67.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Perez E, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are in the paper or the Supplementary Materials. NGS data have been deposited in the Gene Expression Omnibus under accession numbers GSE173482 (bulk LP RNA-seq), GSE131072 (bulk EC RNA-seq), GSE131110 (single-cell EC RNA-seq). The mass spectrometry data files have been deposited to the ProteomeXchange Consortium under the accession number PXD013978. The 15C4 antibody for non-commercial research purposes can be obtained from J.G. and S.E.M. under institutional material transfer agreement.