Abstract
The field of dental calculus research has exploded in recent years, predominantly due to the multitude of studies related to human genomes and oral pathogens. Despite having a subset of these studies devoted to non-human primates, little progress has been made in the distribution of oral pathogens across domestic and wild animal populations. This overlooked avenue of research is particularly important at present when many animal populations with the potentiality for zoonotic transmission continue to reside in close proximity to human groups due to reasons such as deforestation and climatic impacts on resource availability. Here, we analyze all previously available published oral microbiome data recovered from the skeletal remains of animals, all of which belong to the Mammalia class. Our genus level results emphasize the tremendous diversity of oral ecologies across mammals in spite of the clustering based primarily on host species. We also discuss the caveats and flaws in analyzing ancient animal oral microbiomes at the species level of classification. Lastly, we assess the benefits, challenges, and gaps in the current knowledge of dental calculus research within animals and postulate the future of the field as a whole.
Keywords: Dental calculus, Ancient DNA, Animal oral microbiomes, Metagenomics
1. Introduction
The calcified form of dental plaque, otherwise referred to as dental calculus, is not only considered to be ubiquitous across the fossil record, but contains a myriad of genetic, proteomic, metabolomic, and micro-fossil information related to both the host and the microorganisms therein. Although considered waste for decades by dental practitioners and a nuisance to many museum curators, it was only recently that the information nature of calculus was truly uncovered (Adler et al., 2013; Warinner et al., 2014). As the breadth of calculus studies grew, soon emerged an interest in mammalian dental calculus and the information they may hold regarding pathogen evolution and zoonotic reservoirs. Dental calculus is abundant within extant wild and captive animals considering those with high concentration of salivary calcium and phosphorus can exhibit calcified plaque within three days of biofilm development (Zander et al., 1960).
The earliest case of dental calculus present in non-human primates were found in Dryopithecus carinthiacus, fossil hominids from 12.5 mya (Fuss et al., 2018). The dental evidence suggests that these hominids consumed highly cariogenic fruits rich in sugars much more often than their extant primate cousins (Fuss et al., 2018). An additional case from Sivapithecus sivalensis dating to slightly more recently in the Miocene, 8.7–9.3 mya, did not find any conclusive dietary evidence (Hershkovitz et al., 1997). Although the preservation of DNA cannot extend back to the Miocene, recently, genetic material was successfully recovered from mammoths in Siberia dating back more than one million years ago (van der Valk et al., 2021). Proteins too have been shown to preserve beyond the one million year barrier; a huge benefit to both dental archaeologists and geneticists (Hendy, 2021). In recent decades, phytoliths (accumulation of silica within plant cells that can appear embedded within dental calculus) have been identified using traditional high resolution magnification (Armitage, 1975; Ciochon et al., 1990; Fox et al., 1994, 1996; Middleton and Rovner, 1994) and through scanning electron microscopy (Dobney and Brothwell, 1988; Hansen et al., 1991; Vandermeersch et al., 1994).
Studies revolving around non-human primate oral microbiomes, although few, have yielded fascinating insights into the health of our closest evolutionary relatives. Early studies focused on preventing plaque formation and thus, the progression of periodontal disease within captive primates (Beem et al., 1991; Willis et al., 1999). These studies paved the way for more in-depth analyses of periodontal disease present in macaques (Ebersole et al., 2019; Reynolds et al., 2009) and lorises (Plesker and Schulze, 2013), among others. Initial examinations of microremains explored two decades of chimpanzee occupation at Taï National Park, Côte d’Ivoire and uncovered the wealth of information about various dietary behaviors these assemblages collect, from plant consumption to age of weaning and even nut-cracking activities (Power et al., 2015).
Although microremains and phytoliths are incredibly informative, Weyrich and colleagues used DNA sequence information to infer dietary information along with microbial content in Neanderthal sites in Spain and Belgium (Weyrich et al., 2017). The authors found evidence of certain dietary sources (sheep, mushrooms) and self-medication due to health issues associated with a dental abscess and the presence of a gastrointestinal pathogen (Enterocytozoon bieneusi) by Neanderthals. However, other studies have suggested that caution must be exercised when inferring diet and behavior from DNA sequences alone (Charlier et al., 2018; Mann et al., 2018, 2020; Ottoni et al., 2019; Ozga et al., 2019). Subsequent primate calculus studies have investigated the oral pathogens in wild chimpanzees (Ozga et al., 2019) and microbial diversity within mummified Egyptian baboons (Ottoni et al., 2019). Ozga and colleagues showed a high abundance of Porphyromonas in historic wild chimpanzees from Gombe National Park compared to historic humans, while Ottoni and colleagues suggested unique oral signatures in captive Egyptian baboons compared to humans and wild primates (Ottoni et al., 2019; Ozga et al., 2019). Lastly, plaque studies have also ventured into living captive populations, as demonstrated in a study by Kirakodu and colleagues on rhesus monkeys from Puerto Rico that showed microbial differences across age associated with periodontal infections (2019).
In parallel to human investigations and primate studies, the veterinary community continued to be concerned with the modern manifestation of dental calculus across the animal kingdom, particularly within captive, endangered, domesticated, and those animals related to agriculture. There is a multitude of literature devoted to these topics, but they fell into several categories that are worth mentioning briefly. A bulk of the literature is associated with common household pets such as dogs (Carroll et al., 2020; Clarke et al., 2011; Quest, 2013) and cats (Bellows et al., 2012; Scherl et al., 2019) and the effort to combat plaque buildup through treats and chew toy interventions. Other common wild and domesticated animals that have been studied for oral pathologies and calculus deposition include horses (Earley and Rawlinson, 2013), camels (Eze et al., 2012), pandas (Jin et al., 2012), brown bears (Wenker et al., 1998), dolphins (Loch et al., 2011) and captive big cats (Kapoor et al., 2016). However, these studies rarely investigate the oral micro-biome, with those studies only involving dogs (Dewhirst et al., 2012; Oh et al., 2015), cats (Adler et al., 2016; Dewhirst et al., 2015), and most recently a number of ancient mammals (Brealey et al., 2020). The latter study in particular, demonstrated the potential of dental calculus as a tool to conduct comparative studies of oral microbiomes and pathogen evolution in a range of animal species, and also as a source of host DNA to reconstruct ancient genetic profiles (Brealey et al., 2020).
In this meta-analysis, we compare dental calculus metagenomic profiles from all previously published animal datasets along with several human datasets integral to the field of oral metagenomics, an effort to provide a more expansive view of oral microbial distribution across mammals. The dataset includes comparative modern, historic, and prehistoric human populations (Eisenhofer et al., 2020; Mann et al., 2018; Velsko et al., 2019; Warinner et al., 2014; Weyrich et al., 2017), along with non-human primate chimpanzees (Ozga et al., 2019) and baboons (Ottoni et al., 2019), and finally large mammals (Brealey et al., 2020). The human metagenomic data come from a variety of geographic locations and time periods (Eisenhofer et al., 2020; Warinner et al., 2014; Weyrich et al., 2017), and also include paired dentin and calculus data (Mann et al., 2018) along with microbial profiles from historic calculus, modern plaque, and modern calculus (Velsko et al., 2019). We have also included comparative data from human skin and soil (Table S1). Our meta-analysis of the DNA reads from these datasets suggest that microbiota belonging to animals of the same species predictably cluster together, but there is a tremendous breath of diversity across the animal kingdom. We will also address issues that come along with the comparison of animal microbiomes to a reference database largely composed of human derived microbial material. We maintain that further examination of both extant and extinct, captive and domestic mammalian oral microbiomes are necessary in order to fully understand oral microbial diversity within the host.
2. Materials and methods
Raw DNA reads of shotgun sequenced metagenomic samples from the literature (Table S1) were downloaded and pre-processed with AdapterRemoval (Schubert et al., 2016) (–minlength 30 –minquality 25 –trimns –trimqualities –collapse). Sequence duplicates were removed with Prinseq (Schmieder and Edwards, 2011). Taxonomic classification of pre-processed reads for each sample was done with a custom database of bacterial, viral, archaeal and organelle genomes from the NCBI RefSeq (https://www.ncbi.nlm.nih.gov/refseq/, as of November 2020). The reference genomes in the database were masked in Kraken2 for low-complexity regions with Dustmasker, to reduce the impact of potential spurious classifications (Wood et al., 2019). Read abundances for each species taxonomically identified in the Kraken2 output were estimated with Bracken (Lu et al., 2017). In the dental calculus samples from Egyptian baboons, bears, gorillas and reindeer, which were shown in the original studies to possess low proportions of oral taxa, environmental contaminants were filtered as previously described (Brealey et al., 2020; Ottoni et al., 2019). For the gorillas, we used the ‘washed’ samples, as reported in the study by Brealey and colleagues, as they showed a higher proportion of endogenous reads (Brealey et al., 2020).
Species abundance tables were normalized for genome length with a custom python script (https://github.com/claottoni). Namely, we divided the species reads abundance for the size (in Gb) of the species genome reported in the NCBI. To do that, we generated a table of genome lengths from the NCBI browser (https://www.ncbi.nlm.nih.gov/genome/browse/#!/overview/) for the taxa represented in our RefSeq custom database. This table was then used for normalization with a custom python script (https://github.com/claottoni). Read abundances were also normalized for library size through total sum scaling in R (Table S2). We used taxonomy-ranks (https://pypi.org/project/taxonomy-ranks/) to retrieve full taxonomic data of species abundances (from species up to phylum) and generate abundance tables at the genus-level (Table S3). Normalized species and genera abundance tables of bacteria and archaea were filtered to include only taxa with frequency >0.02% in the full dataset of samples.
Alignment to the reference genome of Olsenella sp. oral Taxon 807, Lawsonella clevelandensis, Acinetobacter johnsonii and A. lwoffii, which were found to be the most discriminant species among the animal groups investigated in DESeq2 (Table S4, see below), was conducted to screen the authenticity of the microbial species and their taxonomic assignation. Quality-filtered and adapter-trimmed reads were aligned against the “reference” or “representative” genomes available in the NCBI RefSeq database with bwa aln at high stringency (-n 0.1). Post-mortem damage in the form of cytosine deaminations was calculated with mapDamage (Jonsson et al., 2013).
We investigated the edit distance distribution by calculating the parameter -Δ% (negative difference proportion) in each sample (Table S6) (Hübler et al., 2019). To do so, we generated a bed file of edit distances to the reference of the mapped reads with bedtools bamtobed-tag NM, and we calculated the -Δ% in R with barplot. We regarded taxonomic assignations with -Δ% >0.8 as authentic, to account for the possibility of cross-alignments due to horizontal gene transfer and the presence of closely related microbial species in the sample, as recently proposed (Hübler et al., 2019). In the highly damaged ancient Egyptian baboon calculus samples, edit distances >0.8 were obtained after filtering the mapped reads with PMDtools (https://github.com/pontussk/PMDtools) for post-mortem damage (–threshold = 1) and mapping quality >30 (–requiremapq = 30).
Bray-Curtis dissimilarities from the species abundances were calculated for the microbiomes present in our comparative dataset with vegan and plotted using non-metric multidimensional scaling (Fig. 1). The genus-level Bray-Curtis dissimilarities of species abundances in the oral microbiomes were also visualized in a dendrogram with the unweighted-pair group method with arithmetic means (UPGMA) using the ape library in R (Fig. 2). Approximately Unbiased (AU) p-values computed by multiscale bootstrap resampling were estimated with pvclust in R. To detect the most differentially abundant species and genera among the animal groups (Table S4, Table S5) we used DESeq2 (Love et al., 2014), and generated heatmaps with pheatmap in R (Fig. 3).
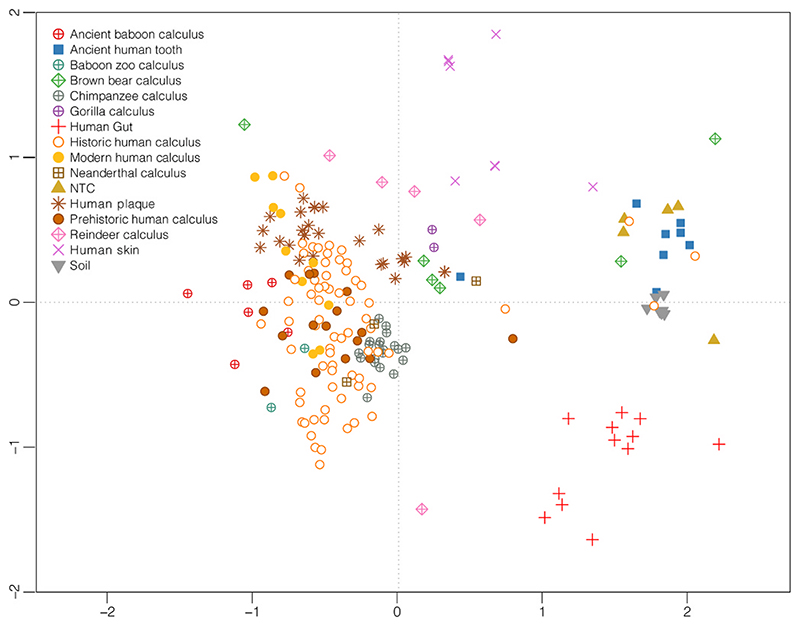
Fig. 1.
Non-metric Multidimensional scaling of Bray-Curtis bacterial and archaeal normalized species abundance obtained with Kraken2 of microbiomes analyzed. (see also Table S1). The stress value is 0.16.
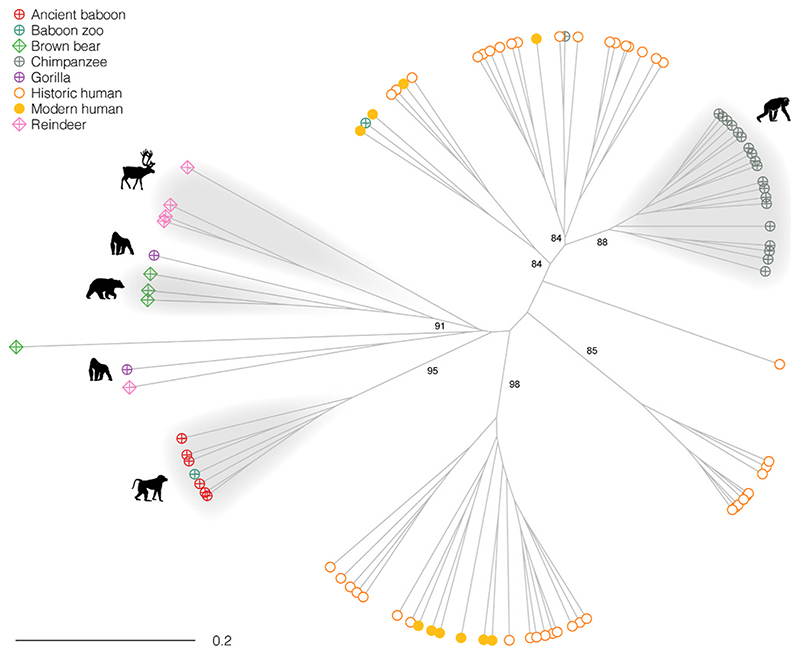
Fig. 2.
UPGMA of Bray-Curtis dissimilarities calculated from bacterial and archaeal species abundances of dental calculus samples analyzed. Approximate Unbiased (AU) p-values computed by multiscale bootstrap resampling in pvclust are indicated for the main clusters.
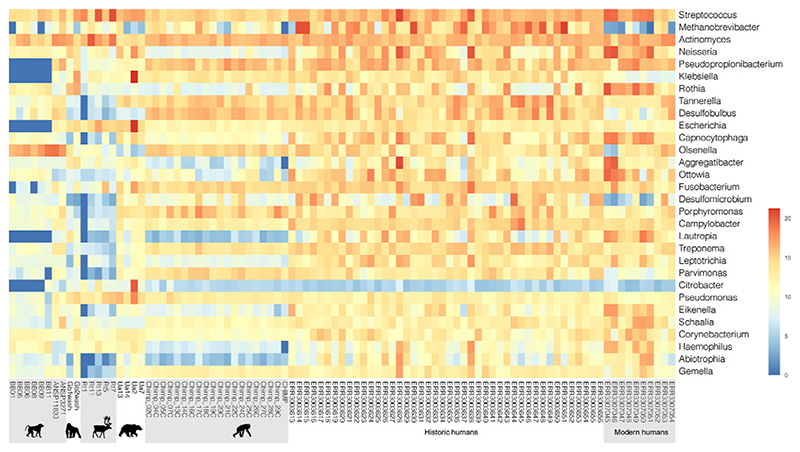
Fig. 3.
Heatmaps generated with DESeq2 of normalized abundances of the 40 most abundant and relevant bacterial and archaeal species detected by Kraken2 in the animal and human dental calculus samples analyzed.
3. Results and discussion
3.1. Caveats for taxonomic classification of animal shotgun sequencing data
Our first focus in the meta-analysis of previously published data was to determine the most accurate method for examining the presence of microbial communities across a variety of mammalian species, including primates. Originally, our taxonomic investigation of the animal dental calculus microbiomes was focused on the species-level classification (Table S2). In order to detect discriminant microbial species in groups of oral microbiomes of our comparative dataset (chimpanzees, baboons, bears, reindeers, modern and historic humans), we made pairwise comparisons between the dental calculus microbiomes in DESeq2 (Table S4) (Love et al., 2014) (gorillas were left out due to the low number of individuals (n = 2)). We found that the bacterial species Olsenella sp. oral Taxon 807 and Lawsonella clevelandensis were typical of non-human primates, baboons and chimpanzees, respectively. The latter, in particular, has been recently described as an emerging pathogenic bacterium isolated from human abscesses (Bell et al., 2016). As such, we opted to further investigate these microbiota at the species level in order to validate these results as authentic.
Then, we tested whether the use of microbial reference databases biased on human research, mostly originating from human-derived biological sources, could affect the species classification in animal-derived samples. To do that, we assessed the authenticity of Olsenella sp. oral Taxon 807 and Lawsonella clevelandensis by calculating the edit distance distribution and the -Δ% value. For Olsenella sp. oral Taxon 807 in the baboons, the -Δ% was compatible with the species assignation in the more recent historic samples (ANSP 11833 and ANSP 3271) and in the two ancient Egyptian samples BB01 and BB08 (Table S6). In the remaining four ancient baboons the Δ% was most likely affected by the high deamination rate, and after filtration of the damaged reads with PMDtools -Δ% increased to acceptable values above 0.8. For Lawsonella clevelandensis in the chimpanzees -Δ% was fairly low, ranging 0.56–0.61 (Table S6), even after filtration of reads carrying deaminated cytosines. This result suggests that a yet uncharacterized microbial species closely related to Lawsonella clevelandensis is discriminant for the chimpanzee oral microbiome that we reconstructed, and highlights the potential flaws associated with the use of human-biased microbial reference databases for metagenomic analysis of animal samples. For this reason we restricted the downstream analyses to the genus-level comparisons.
In addition to Lawsonella clevelandensis, we also found that species of the Acinetobacter genus (A. johnsonii and A. lwoffii) were distinct in bears and reindeers (Table S4), and to a lesser extent, chimpanzees. However, upon further examination, this genus has also been identified as a common contaminant for ancient samples (Eisenhofer et al., 2019; Salter et al., 2014). In order to check this species as a contaminant, we aligned the reads of the bear and reindeer samples to the reference sequences of the two most common Acinetobacter species detected in our species classification. We found low deamination levels (Table S6), corroborating the hypothesis that these taxa originate from a modern contaminant source that was not removed during the filtering process. Although members of the genus Acinetobacter are also ubiquitous free-living saprophytes isolated from soil, water, and various foods (Kämpfer, 2014), here they appear to be of modern origin and as such, cannot provide any additional information about the oral ecology of these animals. For this reason, this taxon was removed from the downstream genus-level analysis.
Finally, we tested the proportion of reads classified as bacteria in both the human and animal shotgun datasets in order to uncover any underlying database biases. Since taphonomic factors (including age and archaeological context) may drastically affect this value, we compared the unfiltered datasets of the chimpanzee (that lived over the past century) and the historic human samples (dated to the 18th-19th century AD, Table S1). We found that the chimps possessed significantly lower average percentages of bacterial classified reads than the historic humans from Radcliffe (14.8% vs 29.2%, p < 0.001, Pairwise Wilcoxon test). We believe this discrepancy can be explained by the scarcity of animal-derived microbial reference genomes, which then leads to a bias towards human-derived species. This poses questions about our ability to reconstruct full microbiomes in animal samples and highlights once more the need to characterize microbes more extensively in current databases.
3.2. Genus analysis of animal oral microbiomes
The nMDS of Bray-Curtis dissimilarities of species abundances that we conducted (Fig. 1) showed that most of the animal dental calculus microbiomes, in particular those from non-human primates (baboons, chimpanzees and gorillas), clustered closest to ancient and modern human calculus oral microbiomes. As expected, this larger cluster was more distant from microbiomes representing other non-oral sources such as soil and laboratory controls, along with other human body ecosystems such as the gut and skin. Samples from reindeer appeared to be more distantly related from the primate cluster and closer to modern human gut microbiomes. Similar to reindeer, brown bear samples showed a lack of central clustering, with only three samples (Ua13, Ua14, Ua7) out of six examined congregating near the oral microbiome samples. Those three samples from bears that remained unclustered could be partially due to unfiltered contamination (Ua6) or dysbiotic conditions associated with a caries lesion (Ua9) (Brealey et al., 2020). Interestingly, the microbiomes originating from ancient human teeth, clustered closer to soil samples and distant from those dental calculus samples, confirming previous observations that archaeological dental calculus is relatively less exposed to environmental contamination compared to ancient dentin samples (Mann et al., 2018, 2020). Additionally, it is worth mentioning that Neanderthal samples are clustered just outside of the traditional primate cluster, which mirrors previous publications (Ozga et al., 2019; Weyrich et al., 2017).
In order to remove extraneous non-oral taxa from these meta-analyses, we generated a UPGMA dendrogram examining animal dental calculus samples (Fig. 2), contrasted against a reduced sample of modern and historic dental calculus representative of the overall human oral microbiome variation. As expected, the animal samples clustered separately from humans. The chimpanzee and the baboon samples appear to be fairly distinctive and only in two instances do we lack clustering by species. In the first, a chimpanzee sample from Gombe National Park more closely aligns with human samples (Sample 17C). This could be the result of an overabundance of genus Methano-brevibacter in this chimpanzee (>11%, while it rangers from 0 to 4% in the other chimpanzees analyzed), a microbe which is a common human oral pathogen (found at 15% average frequency in the historic human sample). In the second instance, a historic baboon sample clustered with human dental calculus. All the baboon samples originated from animals held in captivity, in ancient Egypt (samples BB01 to BB11) and in a 19th century zoo (ANSP3271 and 11833). As previously observed in Ottoni and colleagues, data from wild specimens may help in the future to assess to what extent the clustering observed reflects the omnivorous foraging behavior of the baboons in natural conditions, or whether the oral microbiomes of the animals investigated were affected by captivity (Ottoni et al., 2019). The bear, gorilla and reindeer dental calculus microbiomes were not as distinctive as the others, clustering very close to each other. This could be due to the lack of resolution of the genus-level analysis and the low number of samples available.
In line with the observations at the species level, the analysis of differently abundant genera with DESeq2 (Table S5; Fig. 3) detected Olsenella and Lawsonella as typical for non-human primates. Compared to modern humans, the animal dental calculus samples were less represented by species of the genera Eikenella, Lautropia, Ottowia, and in particular for non-human primates, Neisseria. At present, it is not known whether this is due to a lack of data in the field of mammalian oral microbiomes, or whether these genera are more associated with primate oral disease. As addressed previously, a single chimpanzee sample was characterized by a high frequency (11.8%) of Methanobrevibacter, which was abundant in the historic human sample (15.7%). This taxon was found to be rare in other chimpanzees (0.6%) and may suggest a more serious oral health issue, as Methanobrevibacter species have been shown to promote secondary colonization by fermenters potentially increasing the likelihood of periodontitis (Lepp et al., 2004). However, the Methanobrevibacter genus has been poorly characterized in the current literature and may be more associated with mature calculus as opposed to biofilms (Velsko et al., 2019). Finally, the genus Porphyromonas, which was shown to be of high abundance in dental calculus recovered from chimpanzees (Ozga et al., 2019), is largely absent from the other animals examined aside from two bear samples and only present at an average of 2.9%. P. gingivalis, part of the Red Complex (Socransky et al., 1998), was initially tied to poor oral health, but also found to be common within healthy humans (Arora et al., 2014). As is the case in many microbiome studies, further population of databases and more extensive non-human oral examinations are necessary.
3.3. Conclusions and future directions
Our results suggest that many oral ecologies cluster based on host species, however the wide range of diversity, especially across non-primates such as bears and reindeers, leave many many unanswered questions in the field of animal oral microbiomes. Despite dental calculus generally considered to be ubiquitous across the fossil record (Velsko et al., 2019), little attention has been paid to animal micro-biomes outside of primates. Recently, more focus has been given to present-day animals in captivity and pets, in particular the effects of diet on oral health (e.g. industrial pet food for companion animals). For example, veterinary surveys showed that a ground meat diet provided to captive and domestic felids lacks the mechanical properties of food normally consumed in the wild, leading to severe dental calculus build-up and related periodontal disease, which on the other hand are rare in wild specimens (Kapoor et al., 2016). Another recent study demonstrated that diet has a strong influence on the oral bacterial composition of present-day domestic cats (Adler et al., 2016), showing differences in the microbiome of cats fed on dry-food (highly refined, cereal-based food typical of modern pet food industry) versus wet-food (canned and/or fresh meat-based). With this regard, dental calculus may represent a valuable diagnostic tool to investigate potential dietary shifts in animal oral microbiota within anthropogenic environments.
Continuing to investigate the oral ecosystems of animals in the fossil record along with living animals has multiple benefits. Examining a single mammal over extended time periods in wide ranging geographical contexts can aid in our understanding of oral diversity that may be related to diet, surrounding ecosystems, or captivity of these animals. As agricultural animals continue to exist in high numbers in close proximity to human populations, zoonotic transmission of diseases, especially through animal bites and consumption of contaminated meat, remains an utmost concern. Although zoonotically transmitted microbes cannot be parsed out from the samples examined in the meta-analysis, future studies may want to focus on specific pathogens that infect human populations originating from animal reservoirs. With this in mind, we also understand that we are limited in our analyses by only examining the oral microbes from the class Mammalia, as they are the only published shotgun datasets available. A complete understanding of microbial diversity is of critical importance in conservation efforts for all wildlife populations (Trevelline et al., 2019). Determining whether these common oral pathogens in non-primate mammals are vertically or horizontally transmitted may also give us a better understanding of the evolution of microbial taxa across multiple generations of species. Despite a low number of publications devoted to the characterization of oral microbiota within primates, a clearer understanding of similarities and differences across the animal kingdom is emerging. We are hopeful that continued interest in the field of animal microbiomes can only benefit our understanding of holobionts within our biosphere.
Supplementary Material
Acknowledgments
Bioinformatic analyses were performed on the Galileo super-computing cluster of Cineca, with support of Elixir-Italy and the HPC@CINECA program. This study has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 101002811 to CO, University of Rome Tor Vergata, and grant agreement no. 639286 to Emanuela Cristiani, Sapienza University of Rome).
Footnotes
Author contributions
The authors contributed equally to this work.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
All the data utilized for this meta-analysis came from previously published material (Table S1).
References
- Adler CJ, Dobney K, Weyrich LS, Kaidonis J, Walker AW, Haak W, Bradshaw CJ, Townsend G, Soltysiak A, Alt KW, Parkhill J, et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat Genet. 2013;45:450–455. doi: 10.1038/ng.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler CJ, Malik R, Browne GV, Norris JM. Diet may influence the oral microbiome composition in cats. Microbiome. 2016;4:23. doi: 10.1186/s40168-016-0169-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage PL. The extraction and identification of opal phytoliths from the teeth of ungulates. J Archaeol Sci. 1975;2:187–197. [Google Scholar]
- Arora N, Mishra A, Chugh S. Microbial role in periodontitis: have we reached the top? Some unsung bacteria other than red complex. J Indian Soc Periodontol. 2014;18:9–13. doi: 10.4103/0972-124X.128192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beem JE, Hurley CG, Magnusson I, McArthur WP, Clark WB. Subgingival microbiota in squirrel monkeys with naturally occurring periodontal diseases. Infect Immun. 1991;59:4034. doi: 10.1128/iai.59.11.4034-4041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell ME, Bernard KA, Harrington SM, Patel NB, Tucker T-A, Metcalfe MG, McQuiston JR. Lawsonella clevelandensis gen. nov., sp. nov., a new member of the suborder Corynebacterineae isolated from human abscesses. Int J Syst Evol Microbiol. 2016;66:2929–2935. doi: 10.1099/ijsem.0.001122. [DOI] [PubMed] [Google Scholar]
- Bellows J, Carithers DS, Gross SJ. Efficacy of a barrier gel for reducing the development of plaque, calculus, and gingivitis in cats. J Vet Dent. 2012;29:89–94. doi: 10.1177/089875641202900204. [DOI] [PubMed] [Google Scholar]
- Brealey JC, Leitao HG, van der Valk T, Xu W, Bougiouri K, Dalen L, Guschanski K. Dental calculus as a tool to study the evolution of the mammalian oral microbiome. Mol Biol Evol. 2020;37:3003–3022. doi: 10.1093/molbev/msaa135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll MQ, Oba PM, Sieja KM, Alexander C, Lye L, de Godoy MRC, He F, Somrak AJ, Keating SCJ, Sage AM, Swanson KS. Effects of novel dental chews on oral health outcomes and halitosis in adult dogs. Journal of animal science. 2020;98 doi: 10.1093/jas/skaa274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier P, Gaultier F, Hery-Arnaud G. Interbreeding between Neanderthals and modern humans: remarks and methodological dangers of a dental calculus microbiome analysis. J Hum Evol. 2018;126:124–126. doi: 10.1016/j.jhevol.2018.06.007. [DOI] [PubMed] [Google Scholar]
- Ciochon RL, Piperno DR, Thompson RG. Opal phytoliths found on the teeth of the extinct ape Gigantopithecus blacki: implications for paleodietary studies. Proc Natl Acad Sci Unit States Am. 1990;87:8120. doi: 10.1073/pnas.87.20.8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DE, Kelman M, Perkins N. Effectiveness of a vegetable dental chew on periodontal disease parameters in toy breed dogs. J Vet Dent. 2011;28:230–235. doi: 10.1177/089875641102800403. [DOI] [PubMed] [Google Scholar]
- Dewhirst FE, Klein EA, Bennett M-L, Croft JM, Harris SJ, Marshall-Jones ZV. The feline oral microbiome: a provisional 16S rRNA gene based taxonomy with full-length reference sequences. Vet Microbiol. 2015;175:294–303. doi: 10.1016/j.vetmic.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Klein EA, Thompson EC, Blanton JM, Chen T, Milella L, Buckley CMF, Davis IJ, Bennett M-L, Marshall-Jones ZV. The canine oral microbiome. PloS One. 2012;7:e36067. doi: 10.1371/journal.pone.0036067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobney K, Brothwell DR. A Scanning Electron Microscope Study of Archaeological Dental Calculus, Scanning Electron Microscopy in Archaeology. Archaeo press; Oxford: 1988. pp. 372–385. [Google Scholar]
- Earley E, Rawlinson JT. A new understanding of oral and dental disorders of the equine incisor and canine teeth. Vet Clin N Am Equine Pract. 2013;29:273–300. doi: 10.1016/j.cveq.2013.04.011. [DOI] [PubMed] [Google Scholar]
- Ebersole JL, Orraca L, Kensler TB, Gonzalez-Martinez J, Maldonado E, Gonzalez OA. Periodontal disease susceptible matrilines in the Cayo Santiago Macaca mulatta macaques. J Periodontal Res. 2019;54:134–142. doi: 10.1111/jre.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhofer R, Kanzawa-Kiriyama H, Shinoda K-i, Weyrich LS. Investigating the demographic history of Japan using ancient oral microbiota. Phil Trans Biol Sci. 2020;375:20190578. doi: 10.1098/rstb.2019.0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhofer R, Minich JJ, Marotz C, Cooper A, Knight R, Weyrich LS. Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol. 2019;27:105–117. doi: 10.1016/j.tim.2018.11.003. [DOI] [PubMed] [Google Scholar]
- Eze CA, Adamu SS, Bukar MM. Studies on dentition and oral disorders of camels in maiduguri abattoir, borno state, Nigeria. Trop Anim Health Prod. 2012;44:1953–1956. doi: 10.1007/s11250-012-0162-9. [DOI] [PubMed] [Google Scholar]
- Fox CL, Juan J, Albert RM. Phytolith analysis on dental calculus, enamel surface, and burial soil: information about diet and paleoenvironment. Am J Phys Anthropol. 1996;101:101–113. doi: 10.1002/(SICI)1096-8644(199609)101:1<101::AID-AJPA7>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Fox CL, Pérez-Perez A, Juan J. Dietary information through the examination of plant phytoliths on the enamel surface of human dentition. J Archaeol Sci. 1994;21:29–34. [Google Scholar]
- Fuss J, Uhlig G, Böhme M. Earliest evidence of caries lesion in hominids reveal sugar-rich diet for a Middle Miocene dryopithecine from Europe. PloS One. 2018;13:e0203307. doi: 10.1371/journal.pone.0203307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J, Meldgaard J, Nordqvist J. The Greenland Mummies. McGill-Queens University Press; Montreal: 1991. [Google Scholar]
- Hendy J. Ancient protein analysis in archaeology. Science Advances. 2021;7:eabb9314. doi: 10.1126/sciadv.abb9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershkovitz I, Kelly J, Latimer B, Rothschild BM, Simpson S, Polak J, Rosenberg M. Oral bacteria in MioceneSivapithecus. J Hum Evol. 1997;33:507–512. doi: 10.1006/jhev.1997.0149. [DOI] [PubMed] [Google Scholar]
- Hübler R, Key FM, Warinner C, Bos KI, Krause J, Herbig A. HOPS: automated detection and authentication of pathogen DNA in archaeological remains. Genome Biol. 2019;20:280. doi: 10.1186/s13059-019-1903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Lin W, Huang S, Zhang C, Pu T, Ma W, Lin D. Dental abnormalities in eight captive giant pandas (ailuropoda melanoleuca) in China. J Comp Pathol. 2012;146:357–364. doi: 10.1016/j.jcpa.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Jonsson H, Ginolhac A, Schubert M, Johnson PL, Orlando L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaömpfer P. In: Encyclopedia of Food Microbiology. second ed. Batt CA, Tortorello ML, editors. Academic Press; Oxford: 2014. Acinetobacter; pp. 11–17. [Google Scholar]
- Kapoor V, Antonelli T, Parkinson JA, Hartstone-Rose A. Oral health correlates of captivity. Res Vet Sci. 2016;107:213–219. doi: 10.1016/j.rvsc.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Kirakodu S, Chen J, Gonzalez Martinez J, Gonzalez OA, Ebersole J. Microbiome Profiles of Ligature-Induced Periodontitis in Nonhuman Primates across the Lifespan. Infection and Immunity. 2019 doi: 10.1128/IAI.00067-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA. Methanogenic Archaea and human periodontal disease. Proc Natl Acad Sci U S A. 2004;101:6176–6181. doi: 10.1073/pnas.0308766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loch C, Grando L, Kieser J, Simoes-Lopes P. Dental pathology in dolphins (Cetacea: delphinidae) from the southern coast of Brazil. Dis Aquat Org. 2011;94:225–234. doi: 10.3354/dao02339. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Breitwieser F, Thielen P, Salzberg SL. Bracken: estimating species abundance in metagenomics data. Peerj Computer Science. 2017;3 [Google Scholar]
- Mann AE, Fellows Yates JA, Fagernöas Z, Austin RM, Nelson EA, Hofman CA. Do I have something in my teeth? The trouble with genetic analyses of diet from archaeological dental calculus. Quat Int. 2020 [Google Scholar]
- Mann AE, Sabin S, Ziesemer K, Vågene ÄJ, Schroeder H, Ozga AT, Sankaranarayanan K, Hofman CA, Fellows Yates JA, Salazar-García DC, Frohlich B, et al. Differential preservation of endogenous human and microbial DNA in dental calculus and dentin. Sci Rep. 2018;8:9822. doi: 10.1038/s41598-018-28091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton WD, Rovner I. Extraction of opal phytoliths from herbivore dental calculus. J Archaeol Sci. 1994;21:469–473. [Google Scholar]
- Oh C, Lee K, Cheong Y, Lee S-W, Park S-Y, Song C-S, Choi I-S, Lee J-B. Comparison of the oral microbiomes of canines and their owners using nextgeneration sequencing. PloS One. 2015;10:e0131468. doi: 10.1371/journal.pone.0131468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottoni C, Guellil M, Ozga AT, Stone AC, Kersten O, Bramanti B, Porcier S, Van Neer W. Metagenomic analysis of dental calculus in ancient Egyptian baboons. Sci Rep. 2019;9:19637. doi: 10.1038/s41598-019-56074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga AT, Gilby I, Nockerts RS, Wilson ML, Pusey A, Stone AC. Oral microbiome diversity in chimpanzees from Gombe national Park. Sci Rep. 2019;9:17354. doi: 10.1038/s41598-019-53802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesker R, Schulze H. Dental disease in slender lorises (Loris tardigradus) Zoo Biol. 2013;32:571–574. doi: 10.1002/zoo.21072. [DOI] [PubMed] [Google Scholar]
- Power RC, Salazar-García DC, Wittig RM, Freiberg M, Henry AG. Dental calculus evidence of Taï Forest Chimpanzee plant consumption and life history transitions. Sci Rep. 2015;5:15161. doi: 10.1038/srep15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quest BW. Oral health benefits of a daily dental chew in dogs. J Vet Dent. 2013;30:84–87. doi: 10.1177/089875641303000203. [DOI] [PubMed] [Google Scholar]
- Reynolds MA, Dawson DR, Novak KF, Ebersole JL, Gunsolley JC, BranchMays GL, Holt SC, Mattison JA, Ingram DK, Novak MJ. Effects of caloric restriction on inflammatory periodontal disease. Nutrition. 2009;25:88–97. doi: 10.1016/j.nut.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherl DS, Coffman L, Davidson S, Stiers C. Two randomized trials demonstrate lactic acid supplementation in pet food inhibits dental plaque, calculus, and tooth stain in cats. J Vet Dent. 2019;36:129–134. doi: 10.1177/0898756419873986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Trevelline BK, Fontaine SS, Hartup BK, Kohl KD. Conservation biology needs a microbial renaissance: a call for the consideration of host-associated microbiota in wildlife management practices. Proc Biol Sci. 2019;286:20182448. doi: 10.1098/rspb.2018.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Valk T, Pecnerová P, Díez-del-Molino D, Bergstrom A, Oppenheimer J, Hartmann S, Xenikoudakis G, Thomas JA, Dehasque M, Saglican E, Fidan FR, et al. Millionyear-old DNA sheds light on the genomic history of mammoths. Nature. 2021;591:265–269. doi: 10.1038/s41586-021-03224-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermeersch B, Arensburg B, Tillier A, Rak Y, Weiner S, Spiers M, Aspillaga E. Middle paleolithic dental bacteria from kebara, Israel. Cr Acad Sci Ii. 1994;319:727–731. [Google Scholar]
- Velsko IM, Fellows Yates JA, Aron F, Hagan RW, Frantz LAF, Loe L, Martinez JBR, Chaves E, Gosden C, Larson G, Warinner C. Microbial differences between dental plaque and historic dental calculus are related to oral biofilm maturation stage. Microbiome. 2019;7:102. doi: 10.1186/s40168-019-0717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warinner C, Matias Rodrigues JF, Vyas R, Trachsel C, Shved N, Grossmann J, Radini A, Hancock Y, Tito RY, Fiddyment S, Speller C, et al. Pathogens and host immunity in the ancient human oral cavity. Nat Genet. 2014;46:336–344. doi: 10.1038/ng.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenker CJ, Müller M, Berger M, Heiniger S, Neiger-Aeschbacher G, Schawalder P, Lussi A. Dental health status and endodontic treatment of captive Brown bears (Ursus arctos ssp.) living in the bernese bear pit. J Vet Dent. 1998;15:27–34. doi: 10.1177/089875649801500104. [DOI] [PubMed] [Google Scholar]
- Weyrich LS, Duchene S, Soubrier J, Arriola L, Llamas B, Breen J, Morris AG, Alt KW, Caramelli D, Dresely V, Farrell M, et al. Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature. 2017;544:357–361. doi: 10.1038/nature21674. [DOI] [PubMed] [Google Scholar]
- Willis GP, Kapustin N, Warrick JM, Miller LL, Stookey GK, Hopkins DT, Doan EJ, Ross SR. Preventing dental calculus formation in lemurs (Lemur catta, Eulemur fulvus collaris) and baboons (Papio cynocephalus) J Zoo Wildl Med. 1999;30:377–382. [PubMed] [Google Scholar]
- Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander HA, Hazen SP, Scott DB. Mineralization of dental calculus. PSEBM (Proc Soc Exp Biol Med) 1960;103:257–260. doi: 10.3181/00379727-103-25479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data utilized for this meta-analysis came from previously published material (Table S1).