Summary
Listeria monocytogenes is a foodborne pathogen causing systemic infection with high mortality. To allow efficient tracing of outbreaks a clear definition of the genomic signature of a cluster of related isolates is required, but lineage specific characteristics call for a more detailed understanding of evolution. In our work we used core genome MLST (cgMLST) to identify new outbreaks combined to core genome SNP analysis to characterize the population structure and gene flow between lineages. Whilst analysing differences between the four lineages of L. monocytogenes we have detected differences in the recombination rate, and interestingly also divergence in the SNP differences between sub-lineages. In addition, the exchange of core genome variation between the lineages exhibited a distinct pattern, with lineage III being the best donor for horizontal gene transfer. Whilst attempting to link bacteriophage mediated transduction to observed gene transfer, we found an inverse correlation between phage presence in a lineage and the extent of recombination. Irrespective of the profound differences in recombination rates observed between sub-lineages and lineages we found that the previously proposed cut-off of 10 allelic differences in cgMLST can be still considered valid for the definition of a foodborne outbreak cluster of L. monocytogenes.
Introduction
Listeria monocytogenes, one of the most important foodborne pathogens, is the causative agent of Listeriosis, an infection that particularly affects the elderly, immuno-compromised individuals, and pregnant women (Lamond and Freitag 2018; Radoshevich and Cossart 2017). L. monocytogenes infections can progress to life threating conditions including septicaemia, meningitis, encephalitis and spontaneous abortion, resulting in a high mortality rate (EFSA and ECDC 2017). In the recent years a number of large listeriosis outbreaks have been seen worldwide, including the latest one reported in 2017-2018 in South Africa which was caused by the consumption of contaminated ready-to-eat processed meat products from one company (Smith et al. 2019; WHO 2018). This means that surveillance of outbreaks is a major concern for national and international authorities. With the advent of low-cost whole genome sequencing in the past few years outbreak analysis has shifted from the previous gold standard of PFGE analysis to core genome multi locus sequencing (cgMLST) (Lüth, Kleta, and Al Dahouk 2018). cgMLST is based upon just the genes present in the core genome of a species, or at least those present in the panel of genomes available for analysis (Ruppitsch et al. 2015). The 1701-gene cgMLST scheme (Ruppitsch et al. 2015) has a much higher capacity for resolution of isolates differences than the classical 7-locus MLST (Ragon et al. 2008; Salcedo et al. 2003). As such cgMLST is not only able to provide insight into overall population structure but is also able to contribute to short term nomenclature and the analysis of outbreaks. In the specific case of L. monocytogenes the cut off value for outbreak detection was fixed to ≤10 cgMLST alleles difference (Ruppitsch et al. 2015). More recently after analysis of isolates with known epidemiological link it was proposed that this cut off value should be reduced to ≤7 cgMLST allele differences (Alexandra Moura et al. 2016).
Reference laboratories favour cgMLST analysis over other genomic analyses due to the easier standardisation of the cgMLST scheme; however, analysis of population structure both at the outbreak and the lineage scales also benefits from the analysis of how SNPs and recombination contribute to bacterial evolution. This is of particular interest as L. monocytogenes in not a homogeneous species but contains four separate deep-branching lineages as is evident from both MLST work and more recent global genomic analyses (Haase et al. 2014; Lees et al. 2019; Alexandra Moura et al. 2016). When testing for point mutations and recombination using the 7-locus MLST data, it was found that the occurrence of recombination versus point mutations was about 6-times more frequent in lineage II than in lineage I (den Bakker et al. 2008). However, the knowledge is limited on population structure within sublineages and the gene flow between lineages at the finer scale.
Horizontal gene transfer mechanisms alter the core and accessory genome and are an important driving force in shaping bacterial population structures. With transformation not being documented for L. monocytogenes (Borezee et al. 2000), transduction, which is well documented in many bacterial species (Popa, Landan, and Dagan 2017), automatically gains relevance as a candidate mechanism for gene transfer. In this species interest in phages also has practical aspects due to the differences in lineage-specific (teichoic acids) and genus-specific (cell wall) phage adsorption targets (Dunne et al. 2018). Furthermore, there is significant research in the utilisation of phages as biocontrol in food industry (Carlton et al. 2005; Hagens and Loessner 2014). With respect to the actual contribution of bacteriophages to gene flow within Listeria, there is conflicting evidence. In addition to the relevance of phages in the shaping of the accessory genome of L. monocytogenes (Klumpp and Loessner 2013; Kuenne et al. 2013), there is clear evidence that phages are responsible for the transfer virulence associated genes from other species to Listeria (Chen and Novick 2009). With this, phages have been implicated with facilitating core-genome gene transfer by generalised transduction possibly with distinct impact in distinct lineages (Hodgson et al. 2000; Orsi, Bakker, and Wiedmann 2011). Contrary to this, the short size of homologous recombination events detected, do not support transduction as main transfer mechanism (den Bakker et al. 2008).
In the present work we identity new outbreak isolates from a panel of genome sequences from Swiss L. monocytogenes clinical and food isolates by using cgMLST scheme, and then we used core SNPs to characterise differences in the population structure and the gene flow at high resolution of the lineage I, II and III.
Methods
Bacterial isolates
The draft genome sequences of 158 L. monocytogenes isolates identified during surveillance by the Institute for Food Safety and Hygiene, the Swiss National Reference Laboratory, at the University of Zurich in the Swiss region of Europe mainly from 2011-2014 are presented in this work (Table S1). The addition of two lineage III isolates and six further outbreak isolates (Tasara et al. 2015, 2016; Weinmaier et al. 2013) creates a total database of 166 Swiss isolates. Ninety-six of the isolates were from human clinical cases (Althaus et al. 2014) and 66 isolates were isolated from a variety of food products (Ebner et al. 2015), one was from the environment and three were of unknown origin. Metadata for the isolates, including geographic location, collection data and source of the isolation, are listed in Table S1. For the comparative genomic analysis we included 414 isolates from Germany (Halbedel et al. 2018) and 128 Dutch isolates (Kremer et al. 2017; Lees et al. 2019) (Table S2) making a total of 708 genomes. We used the German isolates because it was our reference study for the outbreaks analysis and we included the Dutch isolates because it was our reference study for the PopPUNK analysis. In addition, for the population structure and gene flow analysis, we included 36 lineage III and four lineage IV isolates from public databases (Table S3).
Serotyping of the Swiss isolates was performed using the commercial set of Listeria O-factor and H-factor antisera from Denka Seiken (Pharma Consulting, Burgdorf, Switzerland) according to the manufacturer’s instructions.
Genome sequencing
DNA extraction for the 160 Swiss isolates was performed from pure cultures of each isolate grown in Brain Heart Infusion. In brief, 1.5 ml of overnight culture was pelleted, washed with 100% acetone and re-suspended in 400 μL TE (10 mM Tris-HCl, 10 mM EDTA, pH 7), 10 μL lysozyme (10 mg/mL, Sigma UK) and 10 μL RNAse A (10 mg/mL, Sigma UK) and incubated at 37°C for 30 minutes. Then 75 μL of 10% SDS (Sigma UK) and 10 μL proteinase K (10 mg/mL, Sigma UK) were added and incubated at 65°C for 30 minutes. DNA was then purified using the Genomic Clean and Concentrator kit (Zymo Research), using the manufacturer’s instructions. 150 bp paired-end DNA libraries, with an average insert size of 451 bp were produced using the NEB Ultra II custom kit and sequenced in pools on the HiSeq X10 platform; this resulted in an average of 2666434 reads per sample, corresponding to an average of 100X coverage per genome. Adapter and poor quality sequences were removed using Trimmomatic (Ver. 0.36) (Bolger, Lohse, and Usadel 2014) and the genomes were assembled using SPAdes (Ver. 3.9.0, default parameters) (Bankevich et al. 2012) and the assembly statistics for each isolate are found in the Table S1. For the downstream analysis assemblies were only included if they had a minimum depth coverage of 10-fold. Raw sequence files and draft genomes in contig level for all 160 Swiss isolates were submitted to the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under study accession number PRJNA486730 (Table S1). In silico MLST was performed using a BLAST-based tool (https://github.com/tseemann/mlst) on de novo genome assemblies (Jolley and Maiden 2010; Seemann 2018).
cgMLST and SNP analysis
A set of 1,701 genes well defined for cgMLST analysis (Halbedel et al. 2018; Ruppitsch et al. 2015) were used for the identification of cluster type (CT), sublineages (SL) and lineages in a total of 708 isolates (Table S1 and Table S2). The alleles for the 1,701-loci cgMLST were downloaded from the database https://www.cgmlst.org/ncs/schema/690488/ and used as query in BLASTn in order to identify and extract the target gene sequence. The parameters used in BLASTn were 100% of query coverage required, word size 11, mismatch penalty -3, match reward 1, gap open costs 5, and gap extension costs 2 and the hit with the highest % identity were kept. The sequences for each target gene per genome were extracted using bedtools v2.25.0 (https://bedtools.readthedocs.io), which creates a new FASTA file for each extracted sequence. Using a custom-made bash script, a multi-FASTA file was obtained for each target gene, and then the genes were aligned using Muscle v3.8.31 (Edgar 2004).
The evaluation of the cgMLST scheme was done using standard pairwise allelic mismatches and SNPs pairwise. The matrix of pairwise allelic mismatches per target gene was obtained from the previously aligned sequences using snp-dists v0.6 (https://github.com/tseemann/snp-dists), where any differences (SNPs and/or indels) was scored as 1 and no differences was scored as 0. Using a python custom script, the matrices of allele pairwise differences were combined obtaining the overall allele differences in 1,701-loci for each pairwise comparison. Gene absence was scored as a missing value. 1,596 (93.8%) target genes were present in all 708 isolates and this set of genes was used for the SNP pairwise analysis. 1,596 core genes from the gene by gene alignment were concatenated according to the order of the reference genome EGD-e (NC_003210.1) and then the matrix of SNPs pairwise differences was obtained using the snp-dists v0.6. The maximum-likelihood core genome phylogenetic tree was inferred from the concatenated alignment core genes using the GTR model in RAxML (Randomized Axelerated Maximum Likelihood) (version 8.2.12) (Stamatakis 2014). The visualization, manipulation and annotation of the phylogenetic tree was done using the ggtree R package v1.15.6 (Yu et al. 2017).
PopPUNK analysis
Clustering analysis of 708 isolates was performed using PopPUNK tool, which uses a K-mer approach to find genetic distances between isolates and then identify clusters of closely related isolates (Lees et al. 2019). PopPUNK was run on the genome assemblies using the default settings (lower k-mer length was 13). Through PopPUNK we identified clusters of isolates which were then compared with the clustering obtained from the phylogenetic analysis that was based on SNPs.
Chromosome painting and fineSTRUCTURE analysis
In order to detect genome-wide transfer of DNA sequence chunks through homologous recombination events between lineages, we expanded our initial data set (708 isolates) with additional genomes from Lineage III (36 isolates) and IV (4 isolates) taken from public databases (Table S2) making a total of 748 isolates. We inferred population structure at a finer scale among the isolates from the core-genome SNPs data by using ChromoPainter (version 0.04) and fineSTRUCTURE (version 2.1.1). (Lawson et al. 2012). Before executing the analysis, we removed isolates belonging to the same CC to generate a dataset with a single randomly selected genome for each CC. The genome collection for this analysis now contained 81 isolates with 15 isolates belonging to lineage I, 33 to lineage II, 29 to lineage III and 4 to lineage IV. In this genome collection the genetic distance was more than 3,000 SNPs between the single core-genomes (1,569 core genes). The genome sequences of the isolates were mapped against the complete reference genome N2306 (CP011004) through the SKA tool (Harris 2018), thus the alignment of the whole genome was obtained. ChromoPainter infers where DNA “chunks” have been donated from a donor to a recipient and reconstructs the chromosome haplotype of “recipient” individuals as a series of chunks from the other “donor” individuals in the sample. A “chunk” refers to a set of neighbouring/linked SNPs copied from a donor to a recipient. The output from ChromoPainter is a “co-ancestry matrix” which summarizes the expected number of chunks of DNA imported from a donor to a recipient genome. fineSTRUCTURE uses the output of ChromoPainter for clustering of individuals based on the co-ancestry matrix (Lawson et al. 2012). ChromoPainter and fineSTRUCTURE were used according to the procedure described in http://www.paintmychromosomes.com.
Recombination rate
The recombination rate in the core genome alignment was assessed with Gubbins Version: 2.3.1 (Croucher et al. 2015). Gubbins was run independently using separate data sets for lineage I (371 isolates), lineage II (335 isolates) and lineage III (38 isolates). The whole genome alignment was done by using SKA tool (Harris 2018) and complete reference genomes were used for the alignment; LL195 (HF558398) from Lineage I, Lm3136 (CP013723) from lineage II and M7 (CP002816) from lineage III. We did not estimate the recombination for lineage IV due to the small sample size of genomes deposited (4 isolates). Reference based alignments of the core genome were analysed using the default parameter of Gubbins, which identifies regions in the alignment with an atypically high density of SNPs, and that infers hypothetical recombination events. Two measures of the recombination rate were estimated: ρ/θ estimate the relative frequency of occurrence of recombination and mutation in the history of the lineage (Milkman and Bridges 1990), and r/m assess the relative impact of recombination and mutation in the genetic diversification of the lineage (Guttman and Dykhuizen 1994).
Transduction in silico analysis
L. monocytogenes prophages were identified in the set of 81 isolates (see above) using the PHASTER tool, which provides the phage sequence annotation as either intact, questionable or incomplete (Arndt et al. 2016; Zhou et al. 2011). The incomplete prophages were removed for further analysis. The intact and questionable prophages were validated against a well-characterised set of known prophages evaluated using MASH v2.1 (Ondov et al. 2016). This set drew on an additional 45 L. monocytogenes prophages available from a public database (http://millardlab.org/bioinformatics/bacteriophage-genomes). Through the Mash analysis we obtained a distance matrix to produce a heatmap of similarity between the prophages, and to determine whether questionable prophage were prophage or host chromosomal sequence. The same analysis allowed to identify those prophages from public database with no similarity with the prophage in our dataset. The mapping of the prophages to the set of 81 isolates was done by BLASTn. The identity threshold was 72% and the coverage value was recorded and plotted as a heatmap to quantify the length of the prophage region. To test in silico the effect of phages on competence, we mapped the known insertion of phages into the comK (lmo2270-lmo2233) in the set of 81 isolates by using BLASTn.
Results
The genome sequences of the 166 Swiss L. monocytogenes isolates
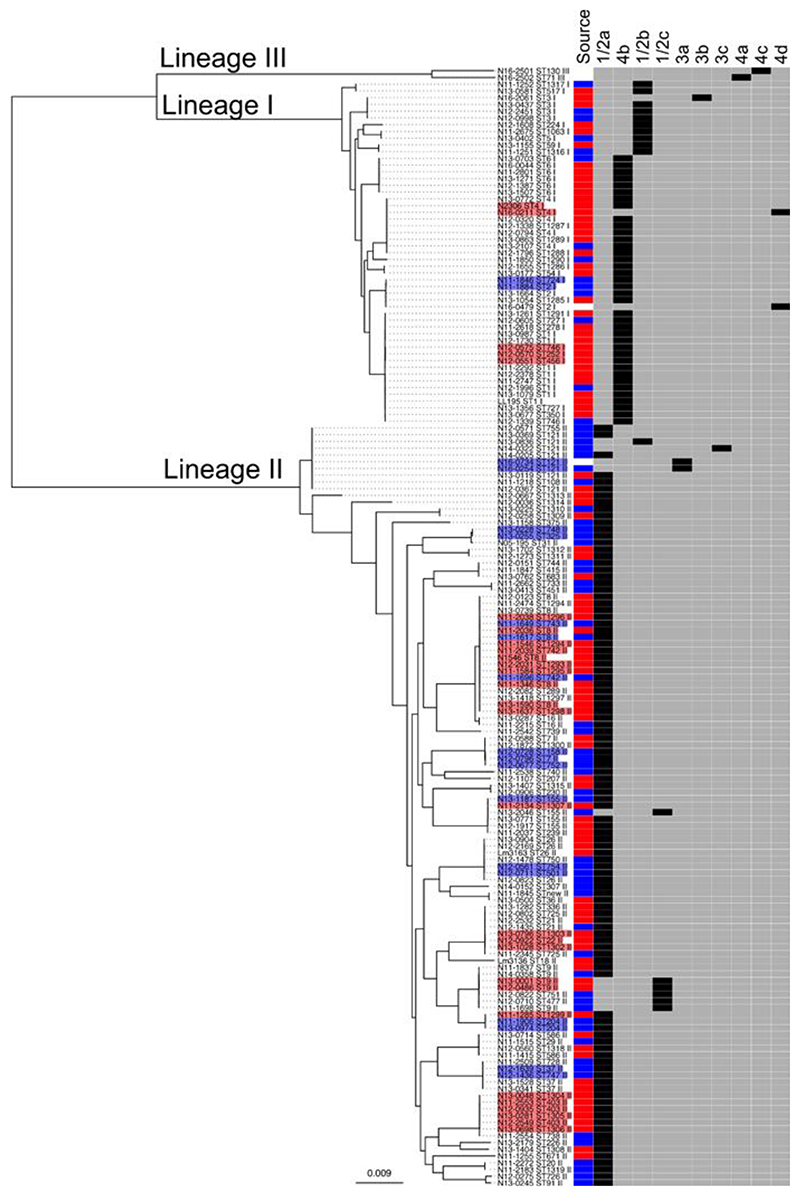
The species Listeria monocytogenes is subdivided into four phylogenetic lineages (I-IV) (Orsi, Bakker, and Wiedmann 2011; Ward et al. 2008). Here we present the draft genome sequences of L. monocytogenes isolates identified during surveillance by the Institute for Food Safety and Hygiene, the Swiss National Reference Laboratory, at the University of Zurich in the Swiss region of Europe mainly from 2011-2014 (Table S1). The mean length of the draft genomes was 2,948,113 bp with mean GC% content of 37.8%. The summary data of the assembly for each genome is reported in the Table S1. Core genome phylogenetic analysis of the 166 Swiss L. monocytogenes isolates found 51 isolates belonged to lineage I and 113 to lineage II (Figure 1, Table S1). No significant association of the two lineages was found with respect to the origin (clinical or food) of the isolates (clinical isolates: lineage I 36 isolates, lineage II 60 isolates; food isolates: lineage I 14 isolates and lineage II 52 isolates) (Figure 1, Table S1). Phenotypic testing was used to confirm the serotypes (1/2a, 1/2b, 1/2c, 3a, 3b, 3c, 4a, 4b, 4c, 4d) across the isolates. The two most prevalent serotypes were the lineage II serotype 1/2a (103 isolates) and the lineage I serotype 4b (38 isolates) (Figure 1). MLST sequence type (ST) information showed ST1 (8 isolates) to be the most prevalent ST in lineage I and ST8 and ST121 (with 7 and 8 isolates, respectively) in lineage II (Figure 1, Table S1).
Figure 1. Swiss L. monocytogenes core-genome phylogenetic tree.
A maximum likelihood phylogenetic SNP tree was constructed using 1596 core genes with the genome sequences of 166 Swiss isolates, which are associated with the lineage I, II and III. In the heatmap to the right of the tree the source of the isolates is shown (clinical in red, food in blue, other in white), and the serotype indicated (in black). Outbreak isolates are shaded in red and food clusters in blue.
Genome evolution in lineage I and II
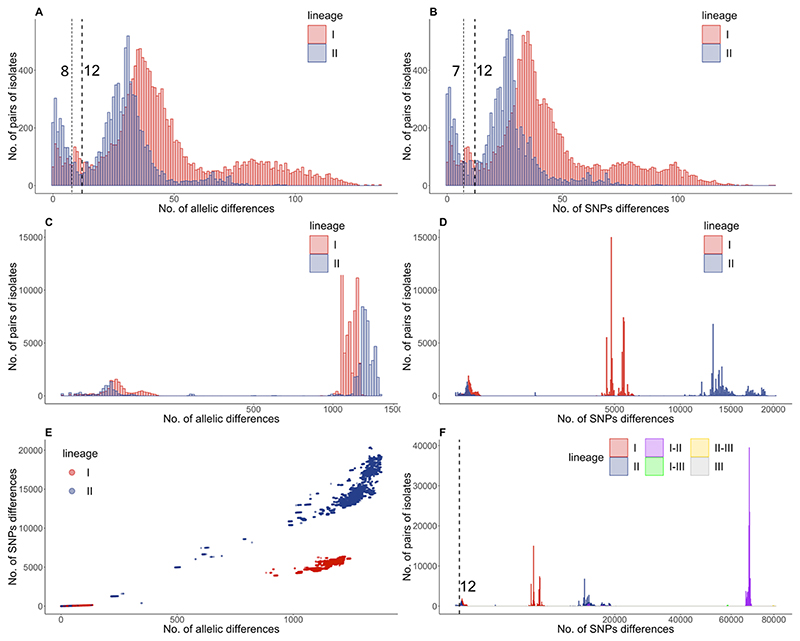
In recent years outbreak analysis has shifted from PFGE to core genome MLST (cgMLST) and a difference of ≤10 cgMLST alleles was identified as the most appropriate cut off (Ruppitsch et al. 2015). To verify if the proposed cut off for outbreaks of ≤10 cgMLST differences was appropriate in our data set, we included in the analysis recently published isolates from Germany (414 isolates) (Halbedel et al. 2018) and the Netherlands (128 isolates) (Kremer et al. 2017; Lees et al. 2019). To visualise the distribution and frequency of differences we constructed a pairwise matrix of either the core genome SNPs or the cgMLST allele differences and plotted the data (Figure 2). In the overall pairwise plots (Figure 2 E) we clearly identified differences that mapped to outbreaks, sublineages and lineages. When limiting the analysis to groups of isolates distinguished by fewer than 150 differences in cgMLST or SNPs (Figure 2 A-B) outbreaks were clearly identifiable on the plots both by analysing cgMLST allele difference and core genome SNPs. When choosing SNPs or cgMLST differences with the lowest frequency both approaches yielded the same cut off of ≤12 differences separating outbreak isolates but if being more stringent cut offs of ≤ 8 alleles and ≤ 7 SNP differences could also be considered (Figure 2 A-B). Pairwise analysis is plotted separately for diversity within sub-lineage (Figure 2 A-B), between sub-lineage within a lineage (Figure 2 C-D) and between lineages (Figure 2 E).
Figure 2. Lineage specific evolution of L. monocytogenes.
Pairwise cgMLST (A, B) and SNP (C, D, F) differences were analysed for 708 isolates form Switzerland, Germany and the Netherlands. Pairwise analyses are plotted separately for diversity within sub-lineage (A-B; up to 150 differences), between sub-lineage and within lineage (C-D; up to 1,500 cgMLST differences and 20,000 SNPs) and between lineages (F). The correlation between SNP and cgMLST differences are plotted in panel E. The pairwise SNP analysis considering the whole population (F) includes data for within lineage I differences (red), within lineage II differences (blue), within lineage III differences (grey) and the differences between lineage I and II (purple), between lineage I and III (green), and between lineage II and III (yellow).
When we plotted the data separately for lineage I and lineage II isolates (red and blue respectively in Figure 2) we noted differences in their genome diversity. Lineage I isolates were more diverse within sub-lineages than lineage II (Kruskal-Wallis test p-value < 0.01), with a broader distribution of pairwise distances, and a less clear peak corresponding to putative outbreaks. In both lineages the outbreak cut off was comparable, but the sub-lineages had different population structures with lineage I having peaks at 36 and at 80 cgMLST differences while lineage II showed tighter distribution with peaks at 31 and 65 cgMLST differences (Figure 2 A). A comparable distribution was given by plotting SNP differences where lineage I had more SNPs within sub-lineages than lineage II (Figure 2 B).
When looking instead at the differences between sublineages within a lineage (cut off 1,500 cgMLST alleles or 22,000 core genome SNPs) we observed the opposite arrangement with lineage I showing less genome diversity than lineage II (Kruskal-Wallis test p-value < 0.01) (Figure 2 C-D). cgMLST did not resolve this difference effectively as almost all the alleles were different; however SNP differences showed dramatic diversity with lineage I between-sublineages differences of about 5,000 SNPs and linage II of about 15,000 SNPs differences. These differences became more evident when plotting SNPs versus cgMLST differences (Figure 2 E). In conclusion, this suggests lineage II is composed of sub-lineages that are more highly divergent from one another than those within lineage I, but each lineage I sub-lineage contains slightly more diversity than its lineage II equivalent, which might complicate the inference of outbreaks in lineage I.
Genome diversity within sub-lineages
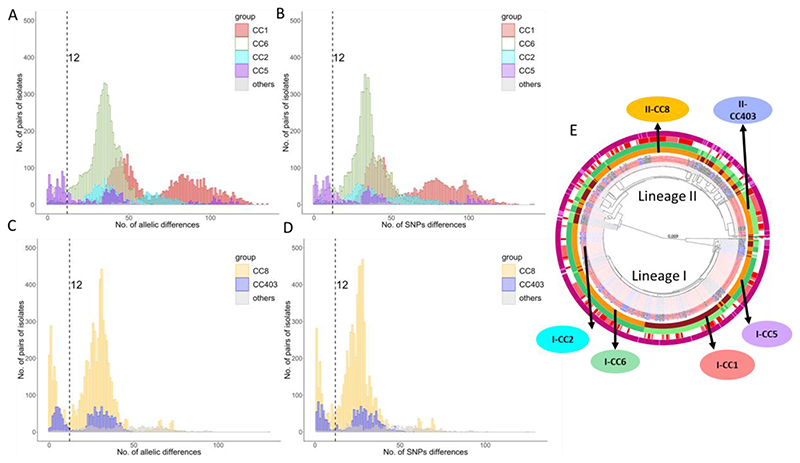
To better understand this potentially confounding within-lineage population structure, the genetic diversity of the collection was analysed using MLST and PopPUNK. This revealed a close correspondence between the cgMLST-identified sub-lineages and both MLST-defined clonal complexes (CCs) and PopPUNK clusters. The differences within sub-lineages were clearly defined by both cgMLST difference and SNPs but also mapped perfectly to clusters defined by PopPUNK (Lees et al. 2019) (Figure S1). To explore the rationale for the multiple peaks in the cgMLST and SNP analysis within sublineages, we plotted the values separately for the different CC/PopPUNK clusters/sub-lineages (Figure 3). Data show that the main sub-lineages and PopPUNK clusters match to clonal complexes (CC). In lineage I, CC1 included 100 isolates (89 isolates of which were ST1), CC2 included 60 isolates (58 of which were ST2), CC5 included 56 isolates (55 of which were ST5 and one ST1063) and CC6 included 111 isolates (110 of which were ST6) (Figure 3). Irrespective of the number of outbreak isolates (those with less than 12 cgMLST allele differences), there were different levels of SNP diversity within these CC/PopPUNK clusters/sub-lineages (Figure 3 A-B). For example, CC6 had one peak with cgMLST difference of 35 alleles and a comparable number of SNPs (Figure 3 A-B). In contrast both CC1 and CC2 showed two clearly distinct peaks, and CC5 had three peaks (Figure 3 A-B) indicating population structure at a finer level than sub-lineages. In CC1 there were several groups of isolates which differed by about 40 to 50 cgMLST alleles within the groups and which differed by 80 to 100 cgMLST alleles between the groups. Similarly, in CC2 there were a number of groups of isolates which differed by about 40 cgMLST alleles within the groups and differed by about 60 cgMLST alleles between the groups. The few isolates in clusters not belonging to either ST1 or ST2 could not account for this peculiar within ST population structure. In lineage II the two main two clusters were CC403, which included 54 isolates (50 of which ST403) and a large CC8 cluster with 118 isolates (102 of which ST8) (Figure 3 E). The two lineage II CC analysed (Figure 3 C-D) showed a similar population structure with a median cgMLST difference of 30 alleles for CC8 and 31 alleles for CC403. When analysing the k-mer distance using PopPUNK, data on pairwise distance could be generated in addition to the core genome, also for the accessory genome (Figure S2 A-B), with both sets of data showing quite a good overlap.
Figure 3. Genome diversity within clonal complexes within lineage I and II.
Pairwise cgMLST and SNP differences were analysed for the major clonal complexes for each lineage. For lineage I the distribution of the pairwise cgMLST (A) and SNP (B) differences are shown for clonal complexes CC1 (red), CC2 (cyan), CC5 (purple) and CC6 (green). For lineage II the distribution of the pairwise cgMLST (C) and SNP (D) differences are shown for CC8 (yellow) and CC403 (blue). The CCs shown in panels A-D are indicated on the core genome phylogenetic tree and the arrows indicate the respective PopPUNK clusters (orange and brown) plotted on the first ring (E). The complete legend and high-resolution image of this circular tree is in Figure S1.
Swiss outbreaks
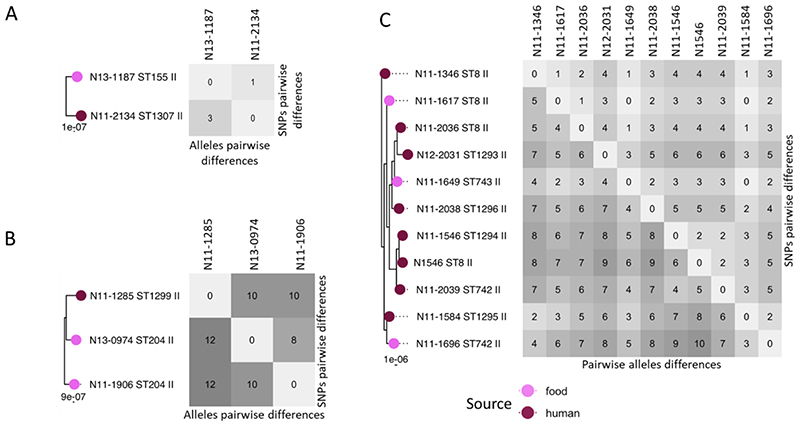
In the 166 Swiss isolates, from clinical and food sources, we could identify fifteen clusters of isolates with ≤12 cgMLST allele differences (Table 1). Nine cases were known outbreaks and six were clusters from food (Table 1). SNP differences correlated well to cgMLST data but often yielded fewer differences as our SNP data did not include indels and were calculated on just 1594 core genes, while cgMLST was performed on all 1701 genes. Two small clusters of two and three isolates respectively were from lineage I while all others mapped to lineage II with three clusters in CC8. In three instances isolates from human and food source are placed in the same cluster (Figure 4A-C). When we included the German and Dutch isolates, we observed that three “Swiss” outbreak clusters actually also contained isolates from Switzerland and/or Germany and/or the Netherlands.
Table 1. Swiss L. monocytogenes outbreaks and food clusters.
| Outbreak | Lineage | CC (ST) | No. isolates | cgMLST differences | SNP differences | Comment |
|---|---|---|---|---|---|---|
| Outbreak | ||||||
| 1 | I | 1 (252, 456, 746) | 3 | 4-6 | 3-4 | Human isolates |
| 2 | II | 8 (8, 742, 743, 1293-1296) | 11 | 2-10 | 1-6 | Food and human isolates |
| 3 | II | 8 (289) | 1 | 2 | 1 | Human isolates, international |
| 4 | II | 8 (8, 1298) | 2 | 2 | 1 | Human isolates, international |
| 5 | II | 9 (9) | 2 | 8 | 6 | Human isolates |
| 6 | II | 21 (22, 1302, 1303) | 3 | 5-7 | 4-6 | Human isolates |
| 7 | II | 155 (155, 1307) | 2 | 3 | 1 | Food and human isolate |
| 8 | II | 204 (204, 1299) | 3 | 10-12 | 8-10 | No conclusive epidemiological link |
| 9 | II | 403 (403, 1304-1306) | 6 | 2-11 | 1-10 | Human isolates |
| Food cluster | ||||||
| 1 | I | 2 (2, 724) | 2 | 3 | 2 | Food isolates |
| 2 | II | 7 (7, 158, 752) | 3 | 3-5 | 1-3 | Food isolates |
| 3 | II | 26 (501, 754) | 2 | 2 | 0 | Food isolates |
| 4 | II | 31 (325, 748) | 2 | 7 | 7 | Food isolates |
| 5 | II | 37 (37, 747) | 2 | 1 | 0 | Food isolates |
| 6 | II | 121 (121) | 2 | 3 | 1 | Food and environment isolates |
Figure 4. Swiss outbreaks with mix source origin.
Subset of three phylogenetic trees for three cases of Swiss outbreaks which included both human (dark purple) and food isolates (light purple). To the right of the trees is the respective matrix of the pairwise cgMLST and SNPs differences.
Recombination
The recombination rate of L. monocytogenes was analysed separately for lineages I, II and III using Gubbins on 371, 336 and 38 isolates respectively. The mean of the number of polymorphisms introduced via recombination with respect to mutation (r/m) and the number of recombination events with respect to polymorphisms introduced by mutation (ρ/θ) are reported for internal and terminal branches of the lineages (Table 2). There are differences in the mean recombination ratio between the internal and external branch for lineage I and III, but there are considerably fewer for lineage II, with recombination hot-spots (Kuenne et al. 2013) readily detectable in the core genome (Figure S3-S5). In agreement with our data above, recombination has occurred more frequently in lineage II and III where the r/m rate was 40 to 33 times higher when compared to the lineage I (Table 2). Graphical representations of the blocks of recombination clearly underlines the higher frequency of recombination and horizontal gene transfer in lineages II and III (Figure S3-S5).
Table 2. Recombination statistics among lineages.
| Mean r/ma | Mean ρ/θb | |||||
|---|---|---|---|---|---|---|
| internal | terminal | total | internal | terminal | total | |
| lineage I (n=371) | 0.0046 | 0.0091 | 0.0137 | 0.0002 | 0.0013 | 0.0015 |
| lineage II (n=336) | 0.3146 | 0.2324 | 0.5470 | 0.0149 | 0.0075 | 0.0224 |
| lineage III (n= 39) | 0.1100 | 0.3460 | 0.4559 | 0.0038 | 0.0114 | 0.0151 |
r/m is the ratio of base substitutions predicted to have been imported through recombination compared to those occurring through point mutations.
ρ/θ (rho/theta) is the ratio of the number of recombination events to point mutations, a measure of the relative rates of recombination and point mutation.
Sequence exchange between lineages
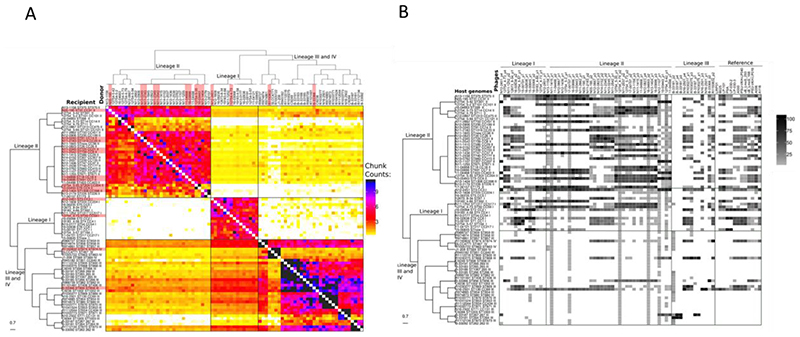
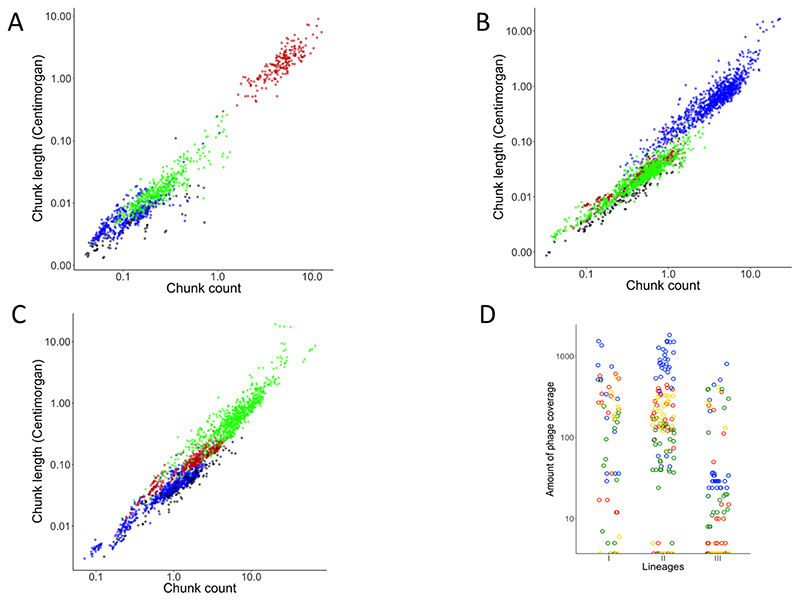
We characterized patterns of gene flow and population structure by using ChromoPainter and fineSTRUCTURE. To perform this analysis, we selected a panel of 81 isolates, with one isolate representing each CC, from all four lineages of L. monocytogenes. The co-ancestry matrix was visualized as a heatmap with each cell indicating the DNA ‘chunk’ counts that a recipient received from each donor (Figure 5A). We used all isolates as potential donors in order to characterize the gene flow among the isolates. The boxes in Figure 5A indicate evident signs of “gene flow”; we use the term “gene flow” when there is a horizontal transfer of DNA chunks between lineages. The co-ancestry matrix revealed asymmetries in the gene flow between lineages; i.e the four lineages imported higher numbers of chunks from other members of their own lineage. Both lineage I (p-value < 0.05) and lineage II (p-value=0.018) imported more chunk counts from Lineage III than from lineage II and I, respectively (Figure S6A). There is high genetic exchange from both lineages I and II to lineage III, with more chunk counts imported from lineage I (p-value < 0.05) (Figure S6A). Interestingly, despite the high frequency of exchanges observed overall, the analysis found little evidence for sequence exchange between lineages I and II (Figure 5A, Figure S6A). Lineage I and II donated more chunk counts to lineage III and lineage III donated more chunk counts to lineage II (p-value < 0.05) (Figure S6B). When we considered the total chunk counts of transferred DNA per isolate, we observed again that lineage I had fewer chunk counts from external lineage population, and that lineage II and III had more chunk numbers which matched well with the lower and higher respective rates of recombination observed with Gubbins (Figure 6A). Importantly the chunk counts also showed that each lineage predominantly imported DNA from the same lineage and that lineage III was the second “best” donor in both lineage I and II (Figure 6A). Here we did not analyse lineage IV due to the small sample size available.
Figure 5. The heatmap of ChromoPainter’s co-ancestry matrix and occurrence of the prophages.
A) The genome collection of 81 isolates for this analysis contained only a single isolate for each clonal complex including 15 isolates belonging to lineage I, 33 to lineage II, 29 to lineage III and 4 to lineage IV. The clustering of the genomes seen above the heatmap was done using FineStructure. Each row of the heat map represents the genomes of recipient isolates, with the number of chunks copied from each donor individuals as columns. Dark colouring denotes a high chunk count and light colour a low chunk count. The colour bar on the right indicates the number of chunks. B) Heatmap of phage alignment coverage in a set of 81 isolates. Each row of the heatmap represents the genomes of the host (bacteria) with the percentage of phage alignment coverage from each phage as columns. A square with dark colouring denotes high coverage and with light colouring low coverage.
Figure 6. Distribution of the total chunk count for lineages and total occurrence of the prophages.
The total chunk count was obtained for each genome from the co-ancestry matrix obtained from ChromoPainter and the distribution of this total count is show in the dot plot. Each circle represents one single genome for the lineage I (red), lineage II (blue), lineage III (green). Data for lineage I are in panel A, for lineage II in panel B and for lineage III in panel C. Panel D shows the distribution of the total occurrence of prophages (the total of the numerical values for the coverage by lineage is in Figure 5B) plotted as a dot plot for lineage I (red), lineage II (blue), lineage III (green) and prophages from public database (yellow). In these graphs we have not included the results for lineage IV due to small sample size.
Horizontal gene transfers by transduction and transformation
To test if bacteriophage were responsible for the horizontal gene transfer in our dataset, we mapped the prophages in our set of genomes, inferring that detection of similar phages or phage-remnants through the genomes of different CCs could be taken as a proxy for the transfer of DNA by transduction. We identified 41 intact and 47 questionable prophages across the 81 L. monocytogenes isolates (one representative for each CC) using PHASTER. After Mash analysis, through the heatmap of similarity we identified 28 of the questionable prophages as chromosomal sequence and thus they were removed, also 31 lytic phages from the public database (Denes et al. 2014; Dorscht et al. 2009) were removed as none had similarity with our genomes (Table S4). We then used the remaining 60 prophages plus 13 temperate phages from the public database as a query in a BLASTn analysis with all 81 isolates as the subject in order to predict sequence exchange between lineages mediated by prophage transduction. From BLASTn we obtained a phage coverage matrix which was plotted as a heatmap and linked to the phylogenetic tree (Figure 5B). Data showed that there is a high occurrence of prophages in both lineages I and II and lower occurrence of prophages in lineage III. Some genomes, particularly in lineage II, showed extensive homology to many phages or phage-remnants present in lineage I and II. The prophages identified in lineage III had greater similarity to prophage of lineage I and II than with others in lineage III. A similar pattern was observed with the prophages from the public databases (Figure 5B); however, for these we had the limitation that we did not have access to the lineage data from the isolates from which they were originally sourced. When we quantified the total coverage of an individual phage throughout all isolates of a given lineage, we observed that lineage II phages scored much higher than lineage I (p-value < 0.05), followed by lineage III (Figure 6D, Figure S7) indicating that phages from lineage II show much higher levels of sequence identity to the phages present in the other lineages. To check the impact of phage on competence for genetic transformation, we mapped the incidence of insertion of prophage into the competence regulator gene comK in the 81 isolates (Table S5). We detected a phage-interrupted comK gene in 2/15 isolates in lineage I, 10/33 isolates in lineage II, 1/29 in lineage III, and 1/4 in lineage IV, however none of these genomes showed any evidence of reduction in DNA exchange (Figure 5A).
When testing the gene flow between lineages using Chromopainter the data clearly showed a correlation between the number of chunks of exchanged DNA and their size (Figure 6 A-C). In each of the three lineages incoming DNA originating from the same lineage is detected more frequent (higher chunk counts) and larger (chunk size) (Figure 6 A-C). Of the three lineages, lineage III shows by far the highest numbers of recombination events (Figure 6C; note different X-axis scale). Interestingly when we compare incoming DNA from other lineages the data shows that lineage III is also the main donor to both lineages I and II (Figure 6 A-B, Figure S6A). Finally, through Pearson correlation we confirm an inverse correlation (p=0.023) between the prophage coverage and the chunk counts donated from lineage II and III to lineage I, and from I and III to lineage II.
Discussion
The core genome multi-locus sequence typing (cgMLST) scheme set up for L. monocytogenes typing is currently the most widely accepted approach for the identification of outbreaks (Halbedel et al. 2018; Alexandra Moura et al. 2016; Ruppitsch et al. 2015). This is because cgMLST has a greater discriminatory power when compared with MLST or PFGE (Lüth, Kleta, and Al Dahouk 2018). In this study, we explored the cut-off for outbreak identification by performing pairwise comparison with a 1,701-locus cgMLST scheme (Ruppitsch et al., 2015) against 708 de novo assembled genomes using BLASTN. Our work analysed 158 genomes of Swiss human and food L. monocytogenes isolates (this study) to which we added, for comparison, six known Swiss outbreak isolates (Tasara et al. 2015, 2016; Weinmaier et al. 2013), 414 German human isolates (Halbedel et al. 2018) and 128 Dutch human isolates (Kremer et al. 2017; Lees et al. 2019). Our results show a cut-off for the definition of outbreaks between 7 and 12 allelic or SNPs differences. Using the same set of cgMLST target genes the cut-off for the determination of outbreak isolates was previously identified as ≤10 alleles differences by analysing 42 and 424 genomes respectively (Halbedel et al. 2018; Ruppitsch et al. 2015). In contrast a lower threshold of ≤7 alleles was identified by Moura and colleagues using 1,696 genomes with a 1,748 gene cgMLST scheme (Alexandra Moura et al. 2016). The use of different sets of cgMLST has not been found by others to be critical for the general identification of clonal groups or outbreak isolates (Y. Chen et al. 2016). While we agree with this cut-off, especially when plotting all isolates on the same graph, it should be noted that there are also more subtle differences to be seen when mapping pairwise differences of different lineages and sub-lineages/clonal-complexes. Our data clearly show that, dependent upon the sub-lineage analysed, the most likely cut-off could be anywhere between 7 and 12 cgMLST differences when using the 1,701-locus cgMLST scheme (Figure 3). Plotting core genome SNP differences shows the same pattern of variation when the different lineages and sub-lineages are plotted separately. Our pairwise data analysis, in common with previous observations (Y. Chen et al. 2016), does not allow us propose defining a clonal-complex specific cut-off, however we would underline the necessity to consider the possibility of occurrence of outbreaks with a slightly higher cgMLST or SNP diversity. SNP analysis in core-genomes was used in previous work for the characterization of the genome diversity within sublineages but this was limited to CC1 and CC9 only (Alexandra Moura et al. 2016). In this study, we have greatly expanded this and characterized the genome diversity within lineages for CC1, CC2, CC5 and CC6 for lineage I and CC8 and CC403 for lineage II.
While the web-based identification of clonal complexes is straight forward, the definition of sublineages is not as obvious. To identify a computationally more approachable method, we tested the PopPUNK program for clustering clonal isolates (Lees et al. 2019). Our data show that PopPUNK clustering matches perfectly to isolates grouped into clonal complexes. Importantly this overlaps perfectly with the sublineages identified by cgMLST that are defined by having less than <150 alleles or SNPs differences (Alexandra Moura et al. 2016). One important aspect is that PopPUNK is much faster, more flexible and computationally less heavy for the definition of sublineages which confirms it as an ideal tool for the analysis of large datasets (Lees et al. 2019).
Nine cases of listeriosis were recorded in Switzerland in 2011 as due to imported cooked ham (Hächler et al. 2013), and in this study we have sequenced the genomes for the isolates from these cases. A follow-up study reported a genome sequence for the isolate N1546 which was associated with this listeriosis outbreak in 2011 (Tasara et al. 2015). In this study, we have identified a total of eleven isolates belonging to this outbreak; these include the seven isolates previously associated with outbreak, in addition four isolates newly reported in this study, three of which were from a food source (meat). Another isolate N2306, which is associated with the listeriosis outbreak in 2013-2014 (Tasara et al. 2016), also fell into a cluster with one of the newly sequenced isolates. Overall, using the cgMLST scheme and SNP approach, we have identified nine Swiss outbreaks of which, three included isolates with food and human sources. The fact that we have identified three outbreaks, which include isolates from Switzerland and Germany, shows that a posteriori genome analysis can highlight cross-border outbreaks which may fail to be noted by national surveillance programs. In this case inclusion of the large collection of French genomes (A Moura et al. 2017) may well have shown other cross-border outbreaks.
The population structure in L. monocytogenes had been previously evaluated using the STRUCTURE program to analyse seven house-keeping genes. These studies on MLST loci found that the genetic diversity between lineages was due to mutation rather than recombination (Haase et al. 2014; Ragon et al. 2008). In contrast previous work, which involved examination of two virulence genes, had found that the rate of recombination was six times higher than the mutation rate (den Bakker et al. 2008). In our work, which used 1,569 core genes, we have identified a higher recombination rate in lineage II and III, 40 and 33 times higher respectively, than in lineage I; these figures match well to the data of den Bakker (den Bakker et al. 2008). Our work is the first study to demonstrate that horizontal gene transfer occurs predominantly within a lineage, and that lineage III shows the highest rate of within-lineage exchange. In addition, lineage III was also observed to be the most proficient donor of DNA chunks. The surprisingly low gene flow between lineage I and II indicates a, so far, unexplained barrier to horizontal gene transfer between these lineages. As restriction modification systems in Listeria are specific to sequence types (P. Chen et al. 2017; De Ste Croix et al. 2017) these barriers to horizontal gene transfer cannot account for the observed lineage-specific DNA exchange. Bacteriophages have also been proposed to be responsible for DNA transfer in Listeria (Orsi, Bakker, and Wiedmann 2011). The preferential within-lineage transfer of DNA we have observed could have matched well to the lineage-specific serovars of wall teichoic acids to which Listeria phages adsorb and which therefore determine their serovar dependent host range (Dunne et al. 2018). In contrast, our results from the prophages mapping showed the opposite findings, with lineage III harbouring almost no bacteriophages whilst being the most proficient acceptor and donor of core genome chunks. Given that the detection of phage sequences was inversely proportional to the amount of recombination the hypothesis that horizontal gene transfer in Listeria is mainly due to transduction had to be excluded. The observation of the preferential gene flow to and from lineage III in absence of phage detection would exclude for Listeria phage-related mechanisms as mayor contributors to core genome evolutions, as also for example in E. coli (Tree et al. 2014). This observation would also exclude other phage related mechanisms of genome evolution in Listeria as for example the genome hypermobility through lateral gene transfer by transduction, as recently reported for the related species Staphylococcus aureus (Chen et al. 2018) for which phage-mediated gene transfer to Listeria had been documented (Chen and Novick 2009). The potential contribution of conjugation of plasmids and genomic islands to the horizontal gene flow was not evaluated here as their contribution to homologous recombination within the core genome is considered generally less significant. The other hypothesis for horizontal gene transfer, i.e. that it most likely occurs through transformation, has been examined previously by analysis of MLST locus recombination based on the relative short size of recombining DNA (den Bakker et al. 2008). The well described insertion of phages into the competence regulator comK (Kuenne et al. 2013; Loessner et al. 2000) was detected in a third of lineage II isolates and few other isolates however, this did not correlate with an appreciable reduction in horizontal gene transfer in these isolates. It is important to underline that the phage insertion into comK in the isolates tested for gene flow was generally representative of all isolates (as they were selected randomly) of those specific CCs, thus representing a stable trait that could have a potential impact on CC evolution. This does not rule out transformation as the mechanism for gene transfer in the core genome because the comK-integrated phages have been shown to excise efficiently during stress allowing comK transcription and ComK-mediated gene regulation in conditions like intracellular infection of macrophages (Molloy 2012; Rabinovich et al. 2012). The observation that the gene flow shows clearly that within lineage gene exchange is favoured over transfer of DNA to or from other lineages is consistent with homologous recombination, the main mechanism for horizontal gene transfer in the core genome, being facilitated by the lower diversity between isolates in the same lineage. While we have no direct evidence for the mechanism of core genome evolution, the fact that phage presence inversely correlates to recombination, indicates that transformation, even if it has never been experimentally tested in L. monocytogenes, should be viewed as the most likely mechanism.
In conclusion our data show important differences in the evolution of the four lineages of L. monocytogenes, with differences in recombination rate and flow of core genome genes. Furthermore, the finding that phage presence inversely correlates with core genome gene flow indicates that transformation and not transduction is the most likely mechanism for evolution of the core genome. From an epidemiological point of view, where the aim is for a clear definition of a cut-off for an outbreak, our work demonstrates that the proposed cut-offs of 10 cgMLST differences can still be considered valid irrespective of the different population structures found in the different L. monocytogenes lineages.
Supplementary Material
Originality-Significance Statement.
Listeria monocytogenes is an important foodborne pathogen, with reservoirs in the environment where it is often found in soil and water. L. monocytogenes is responsible for outbreaks with high mortality, therefore defining genomic cut offs for the identification of outbreaks and source tracking is of importance. In this work for the first time we use core genome MLST and core genome SNPs to characterize the population structure and gene flow in four lineages of L. monocytogenes and in 166 Swiss isolates.
Analysis of mutation frequency, recombination and the horizontal gene transfer between lineages revealed for the first time a clear pattern, with lineage III being the best donor in the transfer of genes to lineage I and II. Prophages are known mediators of horizontal gene transfer. Despite the presence of prophages in all lineages, phage mediated transduction could not be identified as the main driver of gene exchange, as might be expected. Phage exchange showed a negative correlation with the observed patterns of gene flow.
Overall, we demonstrated the exchange of the core genome between lineages and demonstrated that prophages were not the main mechanism responsible for transfer. This work revises our understanding of sub-lineages and lineage evolution in L. monocytogenes, but still allows to confirm the validity of the current genomic cut off for identification of outbreaks
Acknowledgements
The authors wish to acknowledge funding from BBSRC grant BB/N002903/1 to MRO, SDB and NJC. NJC is also supported by a Sir Henry Dale Fellowship, jointly funded by Wellcome and the Royal Society (Grant Number 104169/Z/14/Z). JDR is funded by a KTN CASE BBSRC studentship BB/P504737/1. The authors thank the work of the management team of the ALICE High Performance Computing Facility at the University of Leicester.
Footnotes
Competing interest
Authors declare no competing interest with respect to the work performed in the manuscript.
Data Availability
The genome sequences were deposited as reads at the European Nucleotide Archive ENA with Study accession PRJEB17888 and accession 4421STDY6601709 to 4421STDY6601879 and as FASTA files at GenBank as Bioproject PRJNA486730 with the accession numbers QYDA00000000 to RDTB00000000.
References
- Althaus Denise, et al. Characterization of Listeria Monocytogenes Strains Isolated during 2011-2013 from Human Infections in Switzerland. Foodborne pathogens and disease. 2014;11(10):753–58. doi: 10.1089/fpd.2014.1747. [DOI] [PubMed] [Google Scholar]
- Arndt David, et al. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic acids research. 2016;44(W1):W16-21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Bakker Henk C, et al. Lineage Specific Recombination Rates and Microevolution in Listeria Monocytogenes. BMC Evolutionary Biology. 2008;8(1):277. doi: 10.1186/1471-2148-8-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich Anton, et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. Journal of computational biology: a journal of computational molecular cell biology. 2012;19(5):455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger Anthony M, Lohse Marc, Usadel Bjoern. Trimmomatic: A Flexible Trimmer for Data Illumina Sequence. Bioinformatics (Oxford, England) 2014;30(15):2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borezee Elise, Msadek Tarek, Durant Lionel, Berche Patrick. Identification in Listeria Monocytogenes of MecA, a Homologue of the Bacillus Subtilis Competence Regulatory Protein. Journal of Bacteriology. 2000;182(20):5931–34. doi: 10.1128/jb.182.20.5931-5934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton RM, et al. Bacteriophage P100 for Control of Listeria Monocytogenes in Foods: Genome Sequence, Bioinformatic Analyses, Oral Toxicity Study, and Application. Regulatory Toxicology and Pharmacology. 2005;43(3):301–12. doi: 10.1016/j.yrtph.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Chen John, et al. Genome Hypermobility by Lateral Transduction. Science. 2018;362(6411):207–12. doi: 10.1126/science.aat5867. [DOI] [PubMed] [Google Scholar]
- Chen John, Novick RichardP. Phage-Mediated Intergeneric Transfer of Toxin Genes. Science. 2009;323(5910):139–41. doi: 10.1126/science.1164783. [DOI] [PubMed] [Google Scholar]
- Chen Poyin, et al. Comparative Genomics Reveals the Diversity of Restriction-Modification Systems and DNA Methylation Sites in Listeria Monocytogenes. Applied and environmental microbiology. 2017;83(3) doi: 10.1128/AEM.02091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Yi, et al. Dudley EG, editor. Core Genome Multilocus Sequence Typing for Identification of Globally Distributed Clonal Groups and Differentiation of Outbreak Strains of Listeria Monocytogenes. Applied and Environmental Microbiology. 2016;82(20):6258–72. doi: 10.1128/AEM.01532-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher Nicholas J, et al. Rapid Phylogenetic Analysis of Large Samples of Recombinant Bacterial Whole Genome Sequences Using Gubbins. Nucleic acids research. 2015;43(3):e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denes Thomas, et al. Björkroth J, editor. Comparative Genomic and Morphological Analyses of Listeria Phages Isolated from Farm Environments. Applied and Environmental Microbiology. 2014;80(15):4616–25. doi: 10.1128/AEM.00720-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorscht J, et al. Comparative Genome Analysis of Listeria Bacteriophages Reveals Extensive Mosaicism, Programmed Translational Frameshifting, and a Novel Prophage Insertion Site. Journal of Bacteriology. 2009;191(23):7206–15. doi: 10.1128/JB.01041-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne Matthew, Hupfeld Mario, Klumpp Jochen, Loessner Martin J. Molecular Basis of Bacterial Host Interactions by Gram-Positive Targeting Bacteriophages. Viruses. 2018;10(8) doi: 10.3390/v10080397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner Rebecca, et al. Phenotypic and Genotypic Characteristics of Listeria Monocytogenes Strains Isolated during 2011–2014 from Different Food Matrices in Switzerland. Food Control. 2015;57:321–26. [Google Scholar]
- Edgar Robert C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA and ECDC. The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2016. EFSA Journal. 2017;15(12) doi: 10.2903/j.efsa.2017.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman D, Dykhuizen D. Clonal Divergence in Escherichia Coli as a Result of Recombination, Not Mutation. Science. 1994;266(5189):1380–83. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- Haase Jana K, et al. The Ubiquitous Nature of Listeria Monocytogenes Clones: A Large-Scale Multilocus Sequence Typing Study. Environmental microbiology. 2014;16(2):405–16. doi: 10.1111/1462-2920.12342. [DOI] [PubMed] [Google Scholar]
- Hächler H, et al. Outbreak of Listerosis Due to Imported Cooked Ham, Switzerland 2011. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2013;18(18):20469. [PubMed] [Google Scholar]
- Hagens Steven, Loessner MartinJ. Phages of Listeria Offer Novel Tools for Diagnostics and Biocontrol. Frontiers in Microbiology. 2014;5:1–6. doi: 10.3389/fmicb.2014.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbedel Sven, et al. Whole-Genome Sequencing of Recent Listeria Monocytogenes Isolates from Germany Reveals Population Structure and Disease Clusters. Journal of clinical microbiology. 2018;56:6. doi: 10.1128/JCM.00119-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SR. SKA: Split Kmer Analysis Toolkit for Bacterial Genomic Epidemiology. bioRxiv preprint. 2018 [Google Scholar]
- Hodgson David A. Generalized Transduction of Serotype 1/2 and Serotype 4b Strains of Listeria Monocytogenes. Molecular Microbiology. 2000;35(2):312–23. doi: 10.1046/j.1365-2958.2000.01643.x. [DOI] [PubMed] [Google Scholar]
- Jolley Keith A, Maiden Martin CJ. BIGSdb: Scalable Analysis of Bacterial Genome Variation at the Population Level. BMC Bioinformatics. 2010;11(1):595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp Jochen, Loessner Martin J. Listeria Phages. Bacteriophage. 2013;3(3):e26861. doi: 10.4161/bact.26861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer PHC, et al. Benzalkonium Tolerance Genes and Outcome in Listeria Monocytogenes Meningitis. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2017;23(4):265.e1–265.e7. doi: 10.1016/j.cmi.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenne Carsten, et al. Reassessment of the Listeria Monocytogenes Pan-Genome Reveals Dynamic Integration Hotspots and Mobile Genetic Elements as Major Components of the Accessory Genome. BMC Genomics. 2013;14(1):47. doi: 10.1186/1471-2164-14-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond Nicole, Freitag Nancy. Vertical Transmission of Listeria Monocytogenes: Probing the Balance between Protection from Pathogens and Fetal Tolerance. Pathogens. 2018;7(2):52. doi: 10.3390/pathogens7020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson Daniel John, Hellenthal Garrett, Myers Simon, Falush Daniel. Copenhaver Gregory P. Inference of Population Structure Using Dense Haplotype Data. PLoS Genetics. 2012;8(1):e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees John A, et al. Fast and Flexible Bacterial Genomic Epidemiology with PopPUNK. Genome Research. 2019;29(2):304–16. doi: 10.1101/gr.241455.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loessner Martin J, Inman Ross B, Lauer Peter, Calendar Richard. Complete Nucleotide Sequence, Molecular Analysis and Genome Structure of Bacteriophage A118 of Listeria Monocytogenes: Implications for Phage Evolution. Molecular Microbiology. 2000;35(2):324–40. doi: 10.1046/j.1365-2958.2000.01720.x. [DOI] [PubMed] [Google Scholar]
- Lüth Stefanie, Kleta Sylvia, Al Dahouk Sascha. Whole Genome Sequencing as a Typing Tool for Foodborne Pathogens like Listeria Monocytogenes – The Way towards Global Harmonisation and Data Exchange. Trends in Food Science & Technology. 2018;73:67–75. [Google Scholar]
- Milkman R, Bridges MM. Molecular Evolution of the Escherichia Coli Chromosome. III. Clonal Frames. Genetics. 1990;126(4):505–5017. doi: 10.1093/genetics/126.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy Sheilagh. Bacterial Pathogenicity: A Competent Escape for Listeria. Nature reviews Microbiology. 2012;10(10):670. doi: 10.1038/nrmicro2885. [DOI] [PubMed] [Google Scholar]
- Moura A, et al. Real-Time Whole-Genome Sequencing for Surveillance of Listeria Monocytogenes France. Emerging infectious diseases. 2017;23(9):1462–70. doi: 10.3201/eid2309.170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura Alexandra, et al. Whole Genome-Based Population Biology and Epidemiological Surveillance of Listeria Monocytogenes. Nature microbiology. 2016;2:16185. doi: 10.1038/nmicrobiol.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov BrianD, et al. Mash: Fast Genome and Metagenome Distance Estimation Using MinHash. Genome Biology. 2016;17(1):132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi Renato H, den Bakker Henk C, Wiedmann Martin. Listeria Monocytogenes Lineages: Genomics, Evolution, Ecology, and Phenotypic Characteristics. International Journal of Medical Microbiology. 2011;301(2):79–96. doi: 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Popa Ovidiu, Landan Giddy, Dagan Tal. Phylogenomic Networks Reveal Limited Phylogenetic Range of Lateral Gene Transfer by Transduction. The ISME Journal. 2017;11(2):543–54. doi: 10.1038/ismej.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich Lev, et al. Prophage Excision Activates Listeria Competence Genes That Promote Phagosomal Escape and Virulence. Cell. 2012;150(4):792–802. doi: 10.1016/j.cell.2012.06.036. [DOI] [PubMed] [Google Scholar]
- Radoshevich Lilliana, Cossart Pascale. Listeria Monocytogenes Towards a Complete Picture of Its Physiology and Pathogenesis. Nature Reviews Microbiology. 2017;16(1):32–46. doi: 10.1038/nrmicro.2017.126. [DOI] [PubMed] [Google Scholar]
- Ragon Marie, et al. Dykhuizen Dan., editor. A New Perspective on Listeria Monocytogenes Evolution. PLoS Pathogens. 2008;4(9):e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppitsch Werner, et al. Defining and Evaluating a Core Genome Multilocus Sequence Typing Scheme for Whole-Genome Sequence-Based Typing of Listeria Monocytogenes. Journal of clinical microbiology. 2015;53(9):2869–76. doi: 10.1128/JCM.01193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo C, et al. Development of a Multilocus Sequence Typing Method for Analysis of Listeria Monocytogenes Clones. Journal of clinical microbiology. 2003;41(2):757–62. doi: 10.1128/JCM.41.2.757-762.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Mlst. 2018. https://github.com/tseemann/mlst .
- Smith Anthony M, et al. Outbreak of Listeria Monocytogenes in South Africa, 2017–2018: Laboratory Activities and Experiences Associated with Whole-Genome Sequencing Analysis of Isolates. Foodborne Pathogens and Disease. 2019;16(7):524–30. doi: 10.1089/fpd.2018.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis Alexandros. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics. 2014;30(9):1312–13. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ste Croix Megan, et al. Phase-Variable Methylation and Epigenetic Regulation by Type I Restriction-Modification Systems. FEMS microbiology reviews. 2017;41(Supp_1):S3–15. doi: 10.1093/femsre/fux025. [DOI] [PubMed] [Google Scholar]
- Tasara Taurai, Ebner Rebecca, Klumpp Jochen, Stephan Roger. Complete Genome Sequence of Listeria Monocytogenes N2306, a Strain Associated with the 2013-2014 Listeriosis Outbreak in Switzerland. Genome announcements. 2015;3(3) doi: 10.1128/genomeA.00553-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasara Taurai, Klumpp Jochen, Bille Jacques, Stephan Roger. Genome Sequences of Listeria Monocytogenes Strains Responsible for Cheese- and Cooked Ham Product-Associated Swiss Listeriosis Outbreaks in 2005 and 2011. Genome announcements. 2016;4(2) doi: 10.1128/genomeA.00106-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree Jai J, et al. Identification of Bacteriophage-Encoded Anti-SRNAs in Pathogenic Escherichia Coli. Molecular Cell. 2014;55(2):199–213. doi: 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward Todd J, et al. Multilocus Genotyping Assays for Single Nucleotide Polymorphism-Based Subtyping of Listeria Monocytogenes Isolates. Applied and environmental microbiology. 2008;74(24):7629–42. doi: 10.1128/AEM.01127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmaier Thomas, et al. Complete Genome Sequence of Listeria Monocytogenes LL195, a Serotype 4b Strain from the 1983-1987 Listeriosis Epidemic in Switzerland. Genome announcements. 2013;1(1) doi: 10.1128/genomeA.00152-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Listeriosis – South Africa. World Health Organization; 2018. https://www.who.int/csr/don/28-march-2018-listeriosis-south-africa/en/ [Google Scholar]
- Yu Guangchuang, et al. McInerny Greg. Ggtree: An R Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods in Ecology and Evolution. 2017;8(1):28–36. [Google Scholar]
- Zhou You, et al. PHAST: A Fast Phage Search Tool. Nucleic acids research. 2011;39(Web Server issue):W347-52. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences were deposited as reads at the European Nucleotide Archive ENA with Study accession PRJEB17888 and accession 4421STDY6601709 to 4421STDY6601879 and as FASTA files at GenBank as Bioproject PRJNA486730 with the accession numbers QYDA00000000 to RDTB00000000.