Abstract
Prime editing is a versatile genome editing tool but requires experimental optimization of the prime editing guide RNA (pegRNA) to achieve high editing efficiency. Here, we conducted a high-throughput screen to analyze prime editing outcomes of 92,423 pegRNAs on a highly diverse set of 13,349 human pathogenic mutations that include base substitutions, insertions and deletions. Based on this dataset, we identified sequence context features that influence prime editing and trained PRIDICT (PRIme editing guide preDICTion), an attention-based bi-directional recurrent neural network. PRIDICT reliably predicts editing rates for all small-sized genetic changes with a Spearman's R of 0.85 and 0.78 for intended and unintended edits, respectively. We validated PRIDICT on endogenous editing sites as well as an external dataset and showed that pegRNAs with high (>70) vs. low (<70) PRIDICT scores showed substantially increased prime editing efficiencies in different cell types in vitro (12-fold) and in hepatocytes in vivo (10-fold), highlighting the value of PRIDICT for basic- and translational research applications.
Introduction
Prime editing is a recently developed versatile genome editing technology that allows precise modifications of genomic DNA without introducing DNA double-strand breaks1. The prime editor (PE) is a Cas9 nickase fused to a reverse transcriptase, which copies novel genetic information encoded by the prime-editing guide RNA (pegRNA) into the target DNA locus. The pegRNA itself consists of three elements: a guide sequence (spacer) that directs the Cas9 nickase to the genomic target site (protospacer), a scaffold and the extension sequence (Fig. 1a). The latter is composed of the reverse transcriptase template (RTT) encoding the genetic modification and the primer binding sequence (PBS) for initiating reverse transcription.
Fig. 1. High-throughput screen for determinants of prime-editing efficiency.
(a) pegRNA visualization with different pegRNA domains. Example edit is depicted in red (A:T) at position 5 of the RTT domain, leading to a G to T base change in the target DNA. (b) Self-targeting construct with the promoter (hU6) and different pegRNA domains, target sequence, and primer location for NGS-PCR (Fw and Rv primer). (c) Visualization of the workflow of the self-targeting screen in HEK293T cells. (d) Effect of the maximum number of consecutive 'T' in spacer sequence or pegRNA extension on editing efficiency. (e-f) Comparison of edits with different insertion- and deletion-length on editing efficiency. (g) Effect of replacement bases in single base replacement edits on editing efficiency. (h) Heatmap visualizing editing efficiency of pegRNAs (single base replacements) with different RTT overhang lengths (3, 7, 10, 15 bp) and edit positions (1-15); PAM position highlighted with black rectangle. (i) Heatmap visualizing unintended editing rate of pegRNAs (single base replacements) with different RTT overhang lengths (3, 7, 10, 15 bp) and edit positions (1-15); PAM position highlighted with black rectangle. The number of analyzed pegRNA-target combinations are as follows: (d) n = 12,005, 42,611, 24,574, 9,223, 2,918, 1,092. (e) n = 21,882, 2,934, 1,103, 2,501. (f) n = 5,457, 459, 115, 52. (g) n = 4,482, 5,892, 6,576, 3,622, 5,252, 3,768, 4,868, 4,644, 4,864, 5,767, 4,832, 3,353. (h-i) n = 57,920. Boxplots in (d-g) represent the 25th, 50th, and 75th percentiles. Whiskers indicate 5 and 95 percentiles.
Achieving high prime editing rates typically requires the optimization of specific parts of the pegRNA by manually testing multiple pegRNA designs, a laborious and time-consuming process. Several web-based tools have therefore been developed to assist with the design process of pegRNAs by providing the sequence of oligos for cloning pegRNAs with different spacer, PBS, and RTT lengths2,3. In addition, Kim et al.4 and Li et al.5 developed models for in silico optimization of pegRNA designs by training machine learning algorithms on experimental prime editing datasets. However, as the number of diverse edits in their training data sets was limited, their models only reached high accuracy for a few edit types.
Here, we developed an attention-based bi-directional recurrent neural network and trained it on a large set of diverse human pathogenic mutations to predict pegRNA efficiencies. The resulting model, termed PRIDICT, is broadly applicable and can predict the efficiency of all edit types (all possible single base replacements as well as small insertions and deletions) with high accuracy (Spearman's R = 0.85 and Pearson's r = 0.86). PRIDICT can also predict the rates of unintended editing at the targeted loci with a correlation of R = 0.78 and r = 0.73, and has been validated in different cell lines in vitro and in mouse hepatocytes in vivo. PRIDICT is freely accessible at www.pridict.it.
Results
High-throughput screen for determinants of prime editing
To assess prime editing efficiencies in a high-throughput format, we generated a ‘self-targeting‘ lentiviral library in which each pegRNA is paired with its corresponding target site (library 1). The library consists of 119,701 pegRNAs targeting a diverse set of 14,238 human pathogenic mutations (ClinVar6), including all possible single base replacements as well as insertions (up to 24 bp) and deletions (up to 13 bp). To test the effect of different pegRNA designs, we targeted each disease mutation with multiple pegRNAs; these contained up to 3 different spacer sequences, a constant PBS of 13 bp, and 4 different RTTs. A constant 13 bp PBS was chosen because this length leads to high average editing efficiencies4, and because the diversity in PBS sequences already assures that the parameters relevant for primer binding vary within the library (GC content, melting temperature, minimum-free energy of extension template). In the RTT we varied the length of the RTT overhang, which corresponds to the sequence downstream of the edit and is complementary to the genomic target (Fig. 1a, Extended Data Fig. 1a-g). Since initial experiments revealed that a unidirectional orientation of pegRNA and target sequence on lentiviral vectors can lead to substantial background editing in cells that do not express the PE (Extended Data Fig. 1h,i), we inverted the direction of the target sequence in the library (Fig. 1b). After cloning synthesized oligonucleotides into the lentiviral backbone, the library was transduced into HEK293T cells (Fig. 1c), which were subsequently transfected with a construct expressing PE2. After 7 days of editing (6 days selection with Zeocin), we harvested the genomic DNA, and PCR amplified the integrated vectors for analysis by amplicon high-throughput sequencing (HTS).
To ensure the accuracy and sensitivity of the prime editing dataset, we excluded pegRNA/target site pairs with less than 100 reads per biological replicate and sequencing reads where the pegRNA spacer did not match the target sequence. Of the 92,423 pairs that passed the filtering steps, 57,920 represented single base replacements, 28,420 insertions, and 6,083 deletions. Analysis of editing outcomes revealed high consistency between the three independent biological replicates. We observed a correlation of R > 0.98 and r > 0.99 for the editing efficiency (percentage of reads with only the intended edit; Supplementary Fig. 1a-c) and R > 0.88, r > 0.94 for the unintended editing rate (percentage of reads that contained an unintended edit; Supplementary Fig. 1d-f). The median editing efficiency was 46%, and the median unintended editing rate was 7.4% (characteristics of unintended edits are further described in Supplementary Note 1 and Supplementary Fig. 2). We also observed a high variability in editing efficiency between pegRNAs targeting the same pathogenic locus (median difference of lowest vs. highest = 46%; Supplementary Fig. 1g) or protospacer (median difference of lowest vs. highest = 31%; Supplementary Fig. 1h), demonstrating the ability to improve correction efficiencies by optimizing pegRNA designs.
Analysis of pegRNA features influencing prime editing rates
First, we investigated whether we could identify pegRNA features that influenced the editing efficiency and unintended editing rate in our dataset (Fig. 1d-i, Supplementary Fig. 3-11). As PolyT sequences are known to cause catalytic inactivation and backtracking of RNA polymerase III7,8, we assumed that consecutive T bases in the spacer or extension sequence (RTT + PBS) of the pegRNA could have a strong negative influence on editing rates. Indeed, our dataset revealed that three, four, and five consecutive T bases led to a drop in the median editing efficiency from 55% to 43%, 10%, and 1%, respectively (Fig. 1d). Next, we assessed the impact of the different editing types. We found that single base replacements were installed more efficiently (median of 52%) than deletions and insertions (median of 31% and 31%, respectively), but were accompanied by similar rates of unintended editing (median of 6.8%) than deletions and insertions (median of 7.5% and 8.7%, respectively) (Supplementary Fig. 3a, Supplementary Fig. 4a). Analyzing the specific editing types more closely, we observed an inverse correlation between the length of insertions or deletions and the editing efficiency (Fig. 1e,f). Moreover, A-to-G conversions were more efficiently installed than other single base replacements (Fig. 1g), in line with a previous study that reported a strand-specific bias in repairing G:T mismatches towards the G:C base pair9. We also observed that bases directly flanking the targeted nucleotide influenced single base replacement editing rates, with G/C bases being more beneficial than A/T bases (Supplementary Fig. 3b). When we next assessed the impact of the edit position on the RTT on editing, we detected an increase in the editing efficiency and a decrease in the unintended editing rate when the target base was one of the G bases within the PAM (position 5 and 6 of the RTT) (Fig. 1h,i). These results suggest that PAM destruction prevents the PE from re-targeting the locus after the edit is installed. Further supporting this hypothesis, the editing efficiency of G-to-A conversions at these positions was lower compared to G-to-C or G-to-T conversions (SpCas9 is better in interacting with non-canonical NAG and NGA PAM sites compared to NCT/NTC/NCC/NTT PAM sites10) (Supplementary Fig. 3f). Next, we examined whether the length of the RTT and the RTT overhang influenced editing. While we only observed a weak positive correlation between the editing efficiency and the overall RTT length, a longer RTT overhang length and melting temperature substantially increased the editing efficiency (Fig. 1h, Supplementary Fig. 3j, Supplementary Fig. 7h) and reduced the unintended editing rate (Fig. 1i). Finally, we assessed if the PBS melting temperature had an influence on editing rates. Consistent with the temperature at which cells were cultured, we observed a reduced editing efficiency with PBS melting temperatures below 38 °C (Supplementary Fig. 7i).
Development of ML algorithms to predict prime editing rates
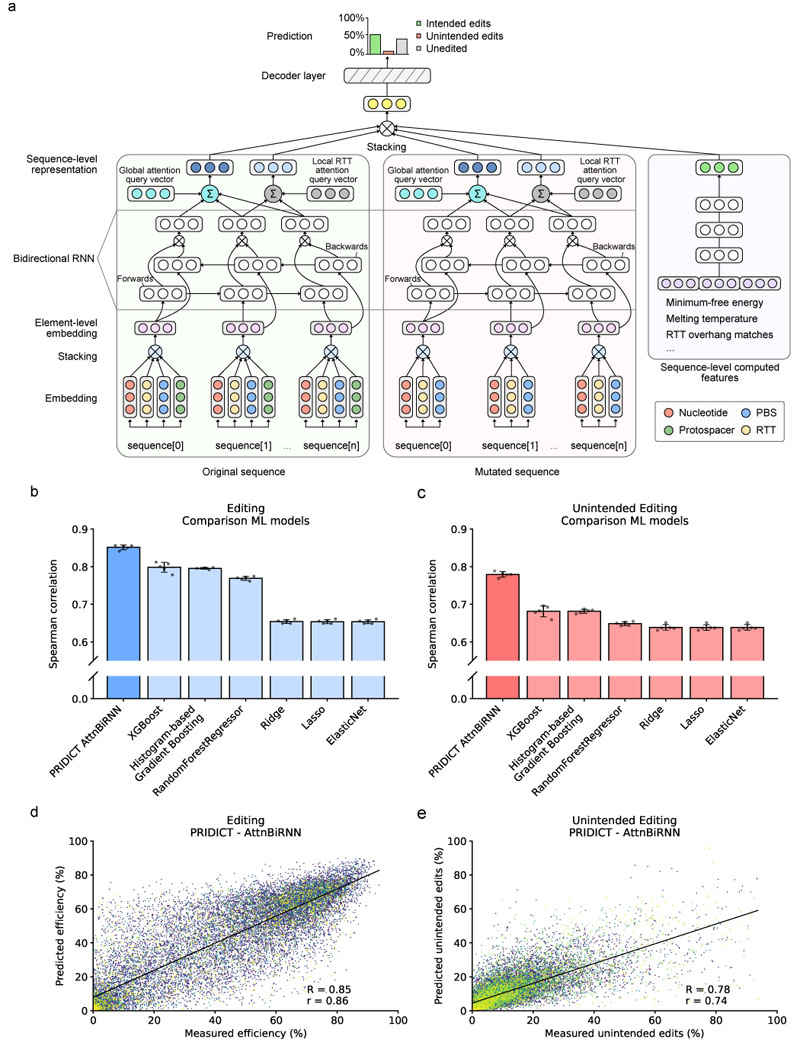
We next attempted to use our dataset of prime editing outcomes to develop machine learning models capable of predicting prime editing efficiencies and unintended editing rates. We therefore split the dataset into 80% training and 20% testing sequences, using a grouping in which pegRNAs for the same locus were not split. From the training sequences, we further split 10% as validation sequences that were used to optimize model hyperparameters during training. We then derived a set of 67 features for the pegRNA design and target sequence (Supplementary Table 1) to train linear regression based-models (Lasso, Ridge, and Elastic net) and tree-based models (Random Forest regressor, Histogram-Based Gradient Boosting, and extreme Gradient Boosting XGBoost) for predicting editing efficiencies and unintended editing rates of pegRNAs. Moreover, we developed an attention-based bi-directional recurrent neural network (AttnBiRNN), which in addition to learning from predefined pegRNA features is also capable of extracting and incorporating novel sequence features in an unbiased manner (i.e., data-driven feature representation). In short, the model consists of three encoder- and one decoder-neural network (Fig. 2a). While two separate encoder networks use bi-directional attention-based gated recurrent neural networks (RNN) to learn representations from given pairs of original- and edited sequences, the third encoder uses a feed-forward network to learn a representation from a set of pre-derived pegRNA features. The learned representations (i.e., fixed-length vectors) are then mapped using a decoder network to depict the probability distribution of the three possible outcomes: intended editing, unintended editing, or no editing (further information is available in Supplementary Methods 1). Training and validating the 6 baseline models on our dataset (five-fold cross-validation) revealed a higher Spearman (R) and Pearson (r) correlation between predicted and observed editing rates of pegRNAs with tree-based models compared to linear-based regression models, with XGBoost achieving the best average performance (R = 0.80 / r = 0.81 for intended editing and R = 0.68 / r = 0.63 for unintended editing; Fig. 2b,c). However, when we next trained the AttnBiRNN deep learning model on our dataset (trained model is termed PRIDICT for PRIme editing guide RNA preDICTion), we could further increase the prediction performance and obtained a correlation of R = 0.85 / r = 0.86 for the editing efficiency and R = 0.78 / r = 0.73 unintended editing rates (mean of five-fold cross-validation; Fig. 2b-e, Supplementary Fig. 12).
Fig. 2. Prediction of pegRNA editing rates by an attention-based bi-directional recurrent neural network.
(a) Illustration of the attention-based bi-directional recurrent neural network. Rectangles with circles represent vectors. (b) Comparison of ML model performances on editing efficiency prediction (mean of five-fold cross-validation). (c) Comparison of ML model performances on unintended editing rate prediction (five-fold cross-validation). (b,c) Each of the 5 cross-validations is visualized as individual data point (n = 5). Error bar = mean +/- SD (d) Evaluation of PRIDICT-AttnBiRNN model by comparing measured editing efficiency of one cross-validation data set with predicted efficiency. (e) Evaluation of PRIDICT-AttnBiRNN model by comparing measured unintended editing rate of one cross-validation data set with predicted unintended editing efficiency. (d-e) The black line corresponds to the least-squares polynomial fit. n = 18,485.
Effect of pegRNA features on machine learning models
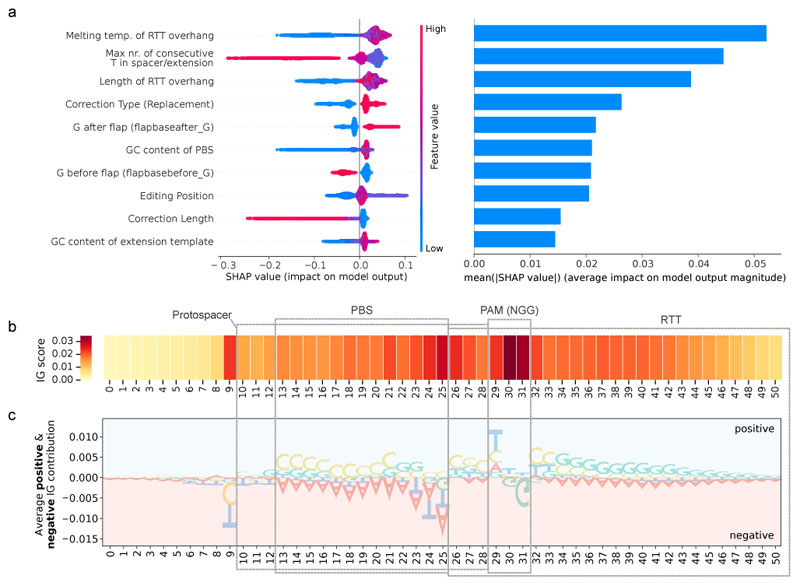
To identify the most important features for predicting pegRNA efficiencies, we first performed Shapley Additive exPlanations (SHAP) analysis11 on the XGBoost model (Fig. 3a, Supplementary Fig. 13). In line with our manual pegRNA feature analysis, we found that the melting temperature and length of the RTT overhang had a strong positive influence on predicted editing rates. Additional important features for the model were the occurrence of long polyT sequence stretches, the type (single base replacements vs. insertions and deletions), length and position of the edit, and the GC content of the extension sequence. The SHAP analysis also revealed that bases before- and after the end of the edited flap at the genomic locus influenced predicted prime editing efficiencies, with a G after the edited flap (flapbaseafter_G) leading to higher editing efficiency and a G before the edited flap end (flapbasebefore_G) leading to a lower editing efficiency (Fig. 3a, Supplementary Fig. 3c). As previously suggested by Anzalone et al.1, the latter phenomenon could be explained by a prolonged stem-loop in these pegRNAs, resulting from base pairing of the complementary C in the last position of the RTT with the guide scaffold. Finally, compared to previously established PE prediction tools4,5, the DeepSpCas9 score12 played a less prominent role in predicting prime editing efficiency of a pegRNA (Supplementary Fig. 3h).
Fig. 3. Feature importance overview for editing prediction.
(a) SHAP analysis on the test dataset of the XGBoost model visualizes the top 10 features influencing the outcome of the editing prediction. A high SHAP value associates with higher editing prediction. Feature values correspond to the values of each individual feature. Detailed list of all features is listed in Supplementary Table 1. (b) Analysis of the importance of individual sequence positions in the PRIDICT AttnBiRNN model. A high IG score associates with a high impact on editing prediction. (c) Average positive/negative IG contribution of individual positions. Large bases have a stronger effect on prediction.
We next implemented an integrated gradient (IG) posthoc explainability approach13 to weight the contributions of the different bases in the target site and pegRNA on the PRIDICT AttnBiRNN model (Fig. 3b; details of the IG method are reported in Supplementary Methods 1). The heatmap reflecting the importance of sequence positions suggests that the model puts the highest weights on the PAM site, followed by the bases flanking the nick position and the base 5’ to the 19-bp protospacer (Fig. 3b). To further assess the contribution of nucleotides at each position, we analyzed the effect of different bases at each position in the target sequence (Fig. 3c). This analysis confirmed findings from SHAP that a higher GC content in the extension sequence (PBS and RTT) leads to higher predicted editing efficiencies. In line with the observed higher Cas9 cleavage activity when the base after the PAM (position 32) is an A, C, or T or when the base before the nick (position 25) is a C14, these features also led to a higher prediction of prime editing efficiency.
Finally, we assessed the feature importance for unintended editing rates using SHAP (Supplementary Fig. 14) and IG (Supplementary Fig. 15). The majority of relevant features, such as Extension GC content, DeepSpCas9 value, or polyT values, influenced the prediction of intended- and unintended editing in the same direction, indicating that they generally affect the activity of the prime editor at that locus (e.g., by increasing PE binding to DNA). However, the overall RTT length and the feature flapbaseafter_G had an opposite effect on the prediction of intended- and unintended editing, demonstrating that they directly affect the accuracy of prime editing. We speculate that both features may determine the rates of unintended scaffold integration, with shorter RTTs facilitating read-through of the RT into the scaffold and a G base on the target DNA after the edited flap (flapbaseafter_G) protecting against 1 bp scaffold insertions (the scaffold ends with a complementary C).
PRIDICT validation on endogenous loci and external datasets
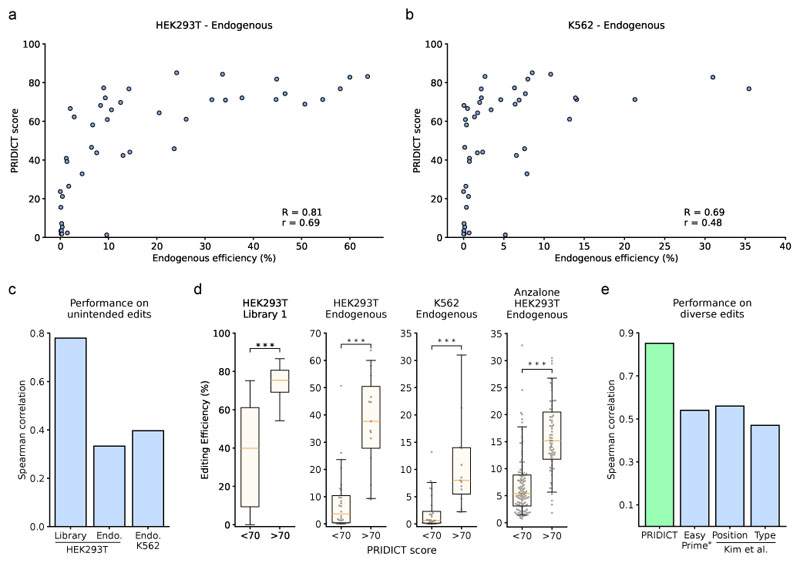
An important aspect of in silico prime editing prediction tools is their applicability to datasets generated under different experimental setups. Therefore, we next assessed the performance of PRIDICT on prime editing data generated i) on endogenous genomic loci, ii) in different cell lines, and iii) in other laboratories. Since experimental conditions strongly influence the absolute editing levels, the output of the PRIDICT model for these datasets was set to a score between 0 and 100 rather than the percentual editing rate. To test PRIDICT in an endogenous context, we targeted 15 endogenous loci for an arrayed validation (14 single base replacements, 1 deletion). Each locus was targeted by 3 pegRNAs covering low and high PRIDICT scores, and pegRNAs were transfected individually into HEK293T and K562 cells. Deep sequencing of the targeted loci revealed a correlation for intended editing of R = 0.81 in HEK293T cells and R = 0.69 in K562 cells (Fig. 4a,b) and for unintended editing of R = 0.33 in HEK293T cells and R = 0.40 in K562 cells (Fig. 4c). While we observed locus dependent variability of the editing efficiency, the variability between different pegRNAs within each locus was in a similar range (Supplementary Fig. 16a,b), further highlighting the potential for enhancing prime editing efficiencies at a given locus by optimizing the pegRNA design. Considering that certain pegRNAs showed low editing at endogenous loci despite a high PRIDICT score, we next investigated the influence of chromatin characteristics on editing efficiencies. While we observed a weak correlation of editing efficiency with open chromatin (ATAC-seq, DNAse-seq) as well as H3K4me3 and H3K27me3 marks (Supplementary Fig. 17a-f), these correlations alone were not sufficient to explain the low performance of certain pegRNAs (Supplementary Fig. 17g-j, Supplementary Note 2). Nevertheless, pegRNAs with a PRIDICT score above 70 resulted in consistent and substantially higher editing rates compared to pegRNAs with a PRIDICT score below 70 (10.2-fold increase in HEK293T and 12.2-fold increase in K562 cells - Fig. 4d), underlining the benefit of using PRIDICT to preselect pegRNA designs. Next, we tested PRIDICT on an external prime editing dataset of 181 pegRNAs1, generated under different experimental conditions (Supplementary Table 2). Despite differences in the scaffold, spacer length and selection period, pegRNAs with a PRIDICT score above 70 still led to a substantially higher median editing (15.2 %) compared to pegRNAs with a PRIDICT score below 70 (5.4 %) (Fig. 4d).
Fig. 4. Validation of PRIDICT on endogenous loci and external datasets.
(a,b) Editing on endogenous loci with 45 pegRNAs, targeting 15 loci (3 pegRNAs per locus). (a) Endogenous editing in HEK293T cells compared to PRIDICT score. (b) Endogenous editing in K562 cells compared to PRIDICT score. (c) PRIDICT prediction performance on unintended edits. (d) Comparison of pegRNAs (mean of 3 biological replicates) with PRIDICT score lower or higher than 70 in self-targeting library 1 test-dataset (n = 15,119 (<70), n = 2,059 (>70)), on endogenous edits in HEK293T/K562 (n = 30 (<70), n = 15 (>70)) and the Anzalone et al.1 dataset (n = 112 (<70), n = 69 (>70)). Mann-Whitney U rank test (two-sided): p < 0.001 (<1*10-16, 2.3*10-6, 5.6*10-6, 1.2*10-16 (from left to right)). Boxplots represent the 25th, 50th, and 75th percentiles. Whiskers indicate 5 and 95 percentiles. Individual data points are illustrated for endogenous experiments as grey dots. (e) Performance of the different prediction models on diverse edit types. The Spearman correlation for PRIDICT was calculated from data plotted in Fig. 2b (PRIDICT on test dataset of library 1). The Spearman correlations for the Position and Type models were retrieved from Kim et al.4 (Position model on the Position-test data and Type model on the Type-test data). The Spearman correlation of Easy-Prime PE2 was calculated by analyzing the model on the Easy-Prime test dataset5 filtered against edits at position 5 of the RTT to eliminate the strong bias towards this edit type in their dataset (see Extended Data Fig. 3).
Benchmarking PRIDICT to other PE prediction tools
Kim et al. have previously developed three different machine learning models for predicting prime editing efficiency4. The Position- and Type model, trained on nucleotide replacements, insertions, and deletions at several RTT positions, and DeepPE, trained on G-to-C edit at position 5 of the RTT (the second base of the targeted NGG PAM). While for the Position- and Type models relatively small training datasets were used (1,774 and 3,775 pegRNAs, respectively), leading to substantially lower performance than PRIDICT (R = 0.56 and r = 0.56 for the Position model and R = 0.47 and r = 0.48 for the Type model - Fig. 4e), DeepPE was trained on a library of 38,692 pegRNA and reached an accuracy in the range of PRIDICT (R = 0.80 and r = 0.75)4. This prompted us to directly benchmark the performance of PRIDICT to DeepPE. First, we specifically tested PRIDICT on G-to-C edits at position 5 of the RTT. We observed a performance of R = 0.78, r = 0.8 (Extended Data Fig. 2a), comparable to the performance of DeepPE. Next, we assessed the robustness of both models in tolerating differences in experimental conditions (Supplementary Table 2), and cross-compared PRIDICT on the DeepPE test dataset4 and DeepPE on our test data (library 1, filtered for G-to-C at position 5). For both models the prediction accuracy dropped markedly when tested on the external dataset (PRIDICT: R = 0.55 and r = 0.46; DeepPE: R = 0.2 and r = 0.17; Extended Data Fig. 2b,c). As the drop in accuracy for both models suggests that certain pegRNA features had a different influence on editing rates in the two experimental settings, we next performed SHAP analysis on XGBoost models trained and tested either on the DeepPE dataset (n = 43,149; Extended Data Fig. 2d) or on our library 1 filtered for G-to-C edits at position 5 of the RTT (n=540; Extended Data Fig. 2e). While most features had a similar influence on editing rates in both datasets and only showed differences in weighting, for RTT overhangs with low melting temperature we observed a negative effect on editing efficiency in our dataset, but a positive effect in the Kim et al. dataset, presumably due to the lack of RTT overhangs below 5 bp in their study (Extended Data Fig. 2f,g). In a final comparison we analyzed the performance of DeepPE on our endogenous loci. For the 18 pegRNAs with an edit that could be predicted by DeepPE, we observed a correlation of R = 0.57, r = 0.58 for HEK293T and R = 0.64, r = 0.84 for K562 cells (Extended Data Fig. 2h,i), which is again in the range of PRIDICT (Fig. 4a,b). Taken together, DeepPE performs with similar accuracy as PRIDICT, but has the limitation that it can only predict G-to-C edits at the second base of the targeted PAM.
Li et al. recently developed another tool for predicting pegRNA efficiency (Easy-Prime PE2-model)5. While their XGBoost-based model, similar to PRIDICT, allows the prediction of all small-sized edits, it was trained on a combination of datasets (from Kim et al. and Anzalone et al.)1,4 in which 87% of pegRNAs again encoded for a G-to-C edit at position 5 of the RTT (38,692 G-to-C at position 5 vs. 5748 diverse edits, Extended Data Fig. 3a). Therefore, we first tested the prediction accuracy of this model on diverse edits by filtering their test dataset against replacements at position 5 of the RTT. Using this approach, the model performance was lower than initially reported on their full test dataset (R = 0.54 and r = 0.56 vs. R = 0.67 and r = 0.63 on the full test dataset; Fig. 4e, Extended Data Fig. 3b). Furthermore, predicting prime editing efficiency with Easy-Prime PE2 on our test datasets (library 1, library 2, endogenous loci) only resulted in weak correlations (R = 0.23 - 0.37, r = 0.19 - 0.37; Extended Data Fig. 3c-g). The lower accuracy of Easy-Prime PE2 compared to PRIDICT was also confirmed when we analyzed how often the best-predicted pegRNA for a given locus overlaps with the best-performing pegRNA in library 1 and on endogenous loci (Extended Data Fig. 3h,i).
Model validation by MMR inhibition and pegRNA stabilization
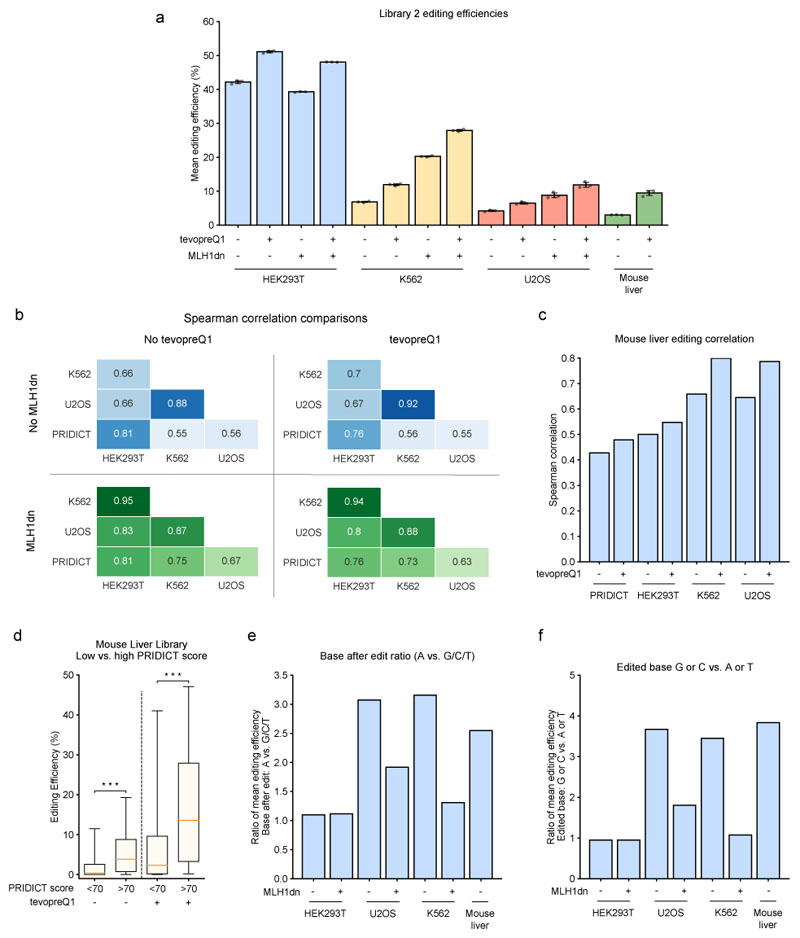
Two recent studies developed adaptations to the original prime editing method to enhance editing efficiency. Nelson et al.15 attached structured RNA motifs, such as tevopreQ1, to the 3' terminus of pegRNAs to prevent degradation by exonucleases. Chen et al.16 co-transfected a plasmid encoding a dominant-negative MLH1 variant to inhibit mismatch repair (MMR) to specifically increase the editing efficiency in MMR-proficient cell lines (HeLa, K562, U2OS). To assess the performance of PRIDICT on these prime editing approaches, we designed a ‘self-targeting’ library consisting of 1,938 pegRNAs (library 2) with and without the tevopreQ1 modification and tested it in HEK293T, K562, and U2OS cells with and without MMR inhibition. Verifying previous results, modification of pegRNAs with tevopreQ1 increased the average editing efficiency in all tested cell lines (1.2-fold in HEK293T, 1.7-fold in K562, and 1.5-fold in U2OS - Fig. 5a), and co-transfection of dominant-negative MLH1 (MLH1dn) increased the editing efficiency in K562 and U2OS cells (3-fold and 2.1-fold for unmodified pegRNAs and 2.3-fold and 1.8-fold for tevopreQ1 pegRNAs) but not in MMR-deficient HEK293T cells (Fig. 5a). We next correlated the editing efficiencies of the different datasets with each other and with PRIDICT predictions (Fig. 5b). This analysis revealed that PRIDICT performance on tevopreQ1 pegRNAs was comparable to the performance on unmodified pegRNAs (HEK293T R = 0.81 vs. 0.76, K562 R = 0.55 vs. 0.56 and U2OS R = 0.56 vs. 0.55). Moreover, inhibition of the MMR pathway led to a considerably higher PRIDICT performance in K562 and U2OS cells (from R = 0.55 to 0.75 in K562 cells and R = 0.56 to 0.67 in U2OS cells). This observation was expected due to the training of PRIDICT in MMR-deficient HEK293T cells, which show editing patterns more similar to MMR-inhibited K562 and U2OS cells (HEK293T vs. K562: R = 0.66, HEK293T-MLH1dn vs. K562-MLH1dn: R = 0.95 and HEK293T vs. U2OS: R = 0.66, HEK293T-MLH1dn vs. U2OS-MLH1dn: R = 0.83). Notably, we also performed the library 2 screen in K562 and U2OS cells with PEmax, a PE2 variant containing a codon-optimized RT domain and modifications in NLS sequences and the Cas9 domain16 (Extended Data Fig. 4a). PRIDICT performance in these screens was comparable to PE2 screens (K562: R = 0.75 vs. 0.73 and U2OS: R = 0.67 vs. 0.67 for PE2 vs. PEmax, both with MMR inhibition; Fig. 5b, Extended Data Fig. 4b,c), suggesting that PRIDICT can also be applied if pegRNAs are used in combination with optimized PE variants.
Fig. 5. Evaluation of MLH1dn and tevopreQ1 effect on PE2 editing efficiency and PRIDICT performance in library 2.
(a) Mean editing efficiencies of each replicate, including all pegRNAs in library 2 with different experimental conditions. Error bars indicate the mean +/- SD of three biologically independent replicates. n = 3. Mean editing of the library for each of the 3 replicates is based on the following number of pegRNAs for each data point (bars left to right) = 927, 925, 925, 926, 887, 887, 918, 926, 875, 866, 874, 866, 880, 928. (b) Spearman correlation between different experimental conditions and cell lines (HEK293T, K562, and U2OS). (c) Comparison of the editing efficiency in the mouse liver to other experimental conditions and the PRIDICT score. (d) Comparison of pegRNAs (mean of 3 biological replicates) with PRIDICT score lower or higher than 70 in the library 2 test-dataset (unmodified pegRNAs n(PRIDICT score < 70) = 771, n(PRIDICT score > 70) = 98, tevopreQ1 pegRNAs n(PRIDICT score < 70) = 803, n(PRIDICT score > 70) = 108). Mann-Whitney U rank test (two-sided): p = 3.7*10-11 and 2.7*10-13. Boxplots represent the 25th, 50th, and 75th percentiles. Whiskers indicate 5 and 95 percentiles. (e) Ratio of mean editing efficiency of pegRNAs in library 2 if base after the edit is an “A” vs. a “G”, “C”, or “G”. (f) Ratio of mean editing efficiency of pegRNAs in library 2 if the edited base is a “G” or “C” vs. an “A” or “T”.
To next investigate whether PRIDICT can also be used to predict prime editing efficiencies in vivo in living animals, we cloned library 2 into a lentiviral vector that expresses GFP under the liver-specific p3 promoter17 and systemically injected the lentiviral pool with an adenoviral PE2 vector into neonatal mice. After 7 weeks, we isolated GFP-positive hepatocytes from the liver and analyzed them by amplicon HTS for editing. Like in vitro in cell lines, also in vivo in hepatocytes the tevopreQ1 modification had a strong effect on the mean editing efficiency, leading to a 3.2-fold increase (from 3 to 9.5 % - Fig. 5a). As expected, editing efficiencies in the liver also correlated more closely to editing efficiencies in MMR-proficient cell lines, with the strongest correlation observed with tevopreQ1 pegRNAs in K562 cells (R = 0.80; Fig. 5c). Despite the lower correlation between editing efficiencies in hepatocytes and HEK293T cells, and the resulting drop in PRIDICT performance in vivo in hepatocytes (R = 0.43 for unmodified pegRNAs and R = 0.48 for tevopreQ1 pegRNAs - Fig. 5c), subdividing pegRNAs into PRIDICT scores below and above 70 led to a 5.9-fold (tevopreQ1 pegRNAs) and 9.6-fold (unmodified pegRNAs) increase in the median editing efficiency (Fig. 5d).
The observed variations in pegRNA efficiencies between cell types prompted us to assess feature contributions for prime editing in the different datasets based on SHAP (Supplementary Fig. 18). We observed that an A at the position after the edit (baseafter_A) had a major contribution on increased editing in K562, U2OS, and mouse hepatocytes, but not in HEK293T cells (Fig. 5e). Likewise, the introduction of a G or C rather than A or T as the edited base (edited_GC_content) only had a positive influence on the editing efficiency in K562, U2OS, and mouse hepatocytes (Fig. 5f). Suggesting that these features are associated with MMR proficiency, we observed a reduced contribution of these features in K562 and U2OS cells expressing MLH1dn (Fig. 5e,f).
Discussion
Prime editing is a powerful and versatile genome editing tool with great potential for correcting genetic diseases. Here, we conducted a high-throughput prime editing screen using ‘self-targeting’ pegRNA libraries to train an attention-based bi-directional RNN algorithm for predicting prime editing efficiency. Tested on diverse edits, PRIDICT performs with substantially higher accuracy than previously developed prediction tools4,5, and is capable of also predicting unintended editing rates (for further comparison to other prediction tools, see Supplementary Table 3). Furthermore, our model accurately predicts the efficiency of tevopreQ1 stabilized pegRNAs, and functions in various cell lines with and without functional MMR and in vivo in hepatocytes. PRIDICT may therefore find broad application to optimize pegRNA designs for installation/correction of specific mutations or to generate pegRNA libraries for high-throughput prime editing screens.
To enable easy access to PRIDICT, we also developed a web tool for designing and predicting the efficiency of pegRNAs (www.pridict.it – an example of the design process is shown in Supplementary Note 3). After submitting the desired sequence and edit, the tool provides a selection of potential pegRNAs together with their PRIDICT scores for editing efficiencies and unintended editing rates. In addition, the tool suggests sequences for potential nicking guide RNAs for the PE3 approach1 and provides oligo sequences for pegRNA cloning and for evaluating editing outcomes by deep sequencing (Primer3)18.
Methods
Cloning of pCMV-PE2-tagRFP-BleoR plasmid
pCMV-PE2 (Addgene #132775, a gift from David Liu1) was digested with EcoRI-HF (NEB). After digestion, the plasmid was dephosphorylated by rSAP (NEB), followed by gel extraction. tagRFP was PCR amplified from pLV312.3 (Addgene #119944)19. tagRFP was Gibson assembled into pCMV-PE2 using 50 ng backbone and 2:1 ratio of tagRFP amplicon. Assembly was performed with NEBuilder HiFi DNA Assembly Master Mix (NEB). Assembled plasmids were transformed into NEB Stable Competent E. coli, resulting in pCMV-PE2-tagRFP. P2A-BleoR (Zeocin resistance) was PCR amplified from an in-house plasmid that originated from pcDNA™3.1/Zeo (Invitrogen). pCMV-PE2-tagRFP was PCR amplified to get linearized plasmid, with amplicon end downstream of tagRFP. Gibson assembly was performed as described previously, resulting in pCMV-PE2-tagRFP-BleoR (Addgene #192508). Lenti-p3-eGFP plasmid was produced by replacing EF1a-PuroR (MluI-HF and ApaI) on Lenti-gRNA-puro (Addgene #84752, a gift from Hyongbum Kim20) with p3-eGFP sequence.
Oligo library design
The custom oligonucleotide pool containing pegRNAs and corresponding target sequence was ordered from Twist Bioscience. For library 1, target loci were selected via ClinVar database6 (December 2019; "Pathogenic" and "Likely Pathogenic" mutations) and the genomic sequences were downloaded from UCSC Genome Browser21. pegRNAs were designed based on the following criteria: “g” + 19bp protospacer, PAM motif (NGG) within 15bp window, PBS length 13bp, RTT Overhang Length 3/7/10/15 bp, 1bp replacements, Insertions Length 1-24bp, Deletions Length 1-13 bp. Oligos were designed with elements in the following order: 5' overhang assembly sequence, pegRNA (spacer, Esp3I spacer for cloning in scaffold, extension), polyT (7x), reverse target sequence, 3' overhang assembly sequence. For library 2, a subset of library 1 together with endogenous targets was used. Additionally, each pegRNA was also added with the tevopreQ1 modification15. Sequences are listed in Supplementary Table 4.
Library cloning and transformation
Step I: Amplification of oligo library
Library cloning protocol was adapted from Kim et al.22 as follows. The plasmid libraries containing the pegRNA and corresponding target sequence were prepared using a two-step cloning process. The oligonucleotide pool was PCR-amplified in 12 cycles (primers in Supplementary Table 5) with KAPA® HiFi HotStart Polymerase (Roche) following the manufacturer's protocol and resulting amplicons were gel extracted with a NucleoSpin Gel and PCR Clean-up Mini kit (Macherey-Nagel).
Step II: Cloning of oligo library into lentiviral vector
For libraries 1 and 2, we digested the Lenti-gRNA-Puro plasmid with Esp3I (NEB). For library 2 in vivo experiments, we digested the Lenti-p3-eGFP plasmid. After digestion, the plasmid was dephosphorylated by rSAP (NEB), followed by gel extraction. The oligo-pool amplicons were assembled into the linearized plasmid using NEBuilder HiFi DNA Assembly Master Mix (NEB). The product was precipitated by adding one volume of Isopropanol (99%) and 0.02 volumes of 5M NaCl solution. The mix was vortexed for 10 sec and incubated at room temperature for 15 min, followed by 15 min centrifugation (17,000 × g). The supernatant was discarded and replaced by four volumes of ice-cold ethanol (80%). Ethanol was removed immediately, the wash was repeated once, and the pellet was air-dried. Next, the pellet was dissolved in H2O. In 15 (library 1) or 2 (library 2) transformation replicates, 100 ng of plasmid library were transformed per 25 µl of Endura Competent Cells (Lucigen) using a Gene Pulser II device (Bio-Rad). Transformed cells were recovered in S.O.C. media for 1 h at 30 °C. Transformed cells were then spread on LB agar plates containing 100 µg/mL ampicillin. After incubation at 30 °C for 16 h, the colonies were scraped, and plasmids were purified using a Plasmid Maxi Kit (Qiagen).
Step III: Cloning of optimized guide scaffold into the plasmid library
We digested 50 mg of the purified plasmid library with Esp3I (NEB). After digestion, the plasmid was dephosphorylated by rSAP (NEB), followed by gel extraction. Oligo with optimized scaffold23 sequence was amplified by PCR with Q5 High-Fidelity DNA Polymerase (NEB) followed by gel extraction. The purified scaffold was digested with Esp3I (NEB), followed by gel extraction. The optimized scaffold was cloned into 500 ng of digested plasmid library via Golden Gate assembly (37 °C 30 min, 100 cycles: (37 °C 5 min, 20 °C 5 min), 37 °C 60 min, 65 °C 20 min) and electroporated and harvested on the following day as described in Step I.
Cell culture
HEK293T (ATCC CRL-3216) and U2OS cells (ATCC HTB-96) were maintained in DMEM++ (DMEM plus GlutaMAX (Thermo Fisher Scientific), supplemented with 10% (vol/vol) fetal bovine serum (FBS, Sigma-Aldrich) and 1% penicillin-streptomycin (Thermo Fisher Scientific)) at 37 °C and 5% CO2. TrypLE Express (Thermo Fisher Scientific) was used for splitting HEK293T cells. Trypsin-EDTA 0.25% (Thermo Fisher Scientific) was used for splitting U2OS cells. K562 cells (ATCC CCL-243) were maintained in RPMI++ (RPMI 1640 Medium with GlutaMAX Supplement (Thermo Fisher Scientific) supplemented with 10% (vol/vol) fetal bovine serum (FBS, Sigma-Aldrich) and 1% penicillin-streptomycin (Thermo Fisher Scientific)) at 37 °C and 5% CO2. Cells were maintained at confluency below 90% and were tested negative for Mycoplasma contamination. Cells were authenticated by the supplier by STR analysis.
Viral vector production
For lentiviral library production, transfection in 6x T175 cell culture flasks was conducted as follows: 3.5 µg pMD2.G, 10.4 µg psPAX2, and 13.8 µg target library plasmid were mixed in 1.83 ml serum-free Opti-MEM (Thermo Fisher Scientific), supplied with 138 µl of polyethylenimine (PEI, 1 mg/ml), vortexed for 10 s and incubated for 20 min. 1.6 ml of plasmid mix was added to 25 mL of DMEM++, and the medium-transfection mix was gently pipetted onto HEK293T cells at ~70% confluency (Day 0). The medium was changed on day 1. On day 3, supernatant-containing lentiviral particles were harvested and filtered using a Filtropur S 0.4 (Sarstedt) filter. The virus suspension was concentrated by ultracentrifugation at 20,000×g for 2 h (4 °C). Aliquots were frozen at −80 °C until use. Lentivirus for integrating reporter sequences of open- and closed-chromatin loci (Supplementary Note 2) was produced as described above (1x T175) but without ultracentrifugation. pMD2.G (Addgene plasmid #12259) and psPAX2 (Addgene plasmid #12260) were gifts from Didier Trono. AdV5-PE2ΔRnH vector24 was produced by ViraQuest Inc.
Animal studies
Animal experiments were performed in accordance with protocols approved by the “Veterinäramt Kanton Zürich” and in compliance with all relevant ethical regulations. C57BL/6J mice were housed in a pathogen-free animal facility at the Institute of Pharmacology and Toxicology of the University of Zurich. Mice were kept in a temperature- and humidity-controlled room (21 °C, 50% RH) on a 12-hour light-dark cycle. Newborn animals (P1, 3 mice, female) were injected with lentivirus (Lenti-p3-eGFP-library2) and 2.4 × 10^10 viral particles (vp) of human adenoviral vector 5 containing unsplit PE2ΔRnH (ViraQuest)24 via the temporal vein. Control animals (P1, 3 mice, male) received the lentiviral library without AdV5-PE2ΔRnH. Mice were euthanized 7 weeks after injection.
Primary hepatocyte isolation
Primary hepatocyte isolation was performed as previously described by Böck et al.24. Isolated hepatocytes were sorted by FACS (BD FACSAria III with BD FACS Diva 8.0.1) for expressing GFP (Supplementary Fig. 19).
Arrayed pegRNA cloning and transformation
Fifteen endogenous loci were selected for arrayed validation (11 from Kim et al. 20214 and 4 from Anzalone et al. 20191). For each locus, we additionally designed 2 pegRNAs introducing the same edit but with different PRIDICT scores to ensure a broad PRIDICT score spectrum (leading to a total of 45 pegRNAs). These pegRNAs were ordered as gene blocks from IDT (Supplementary Table 5). pegRNAs for targeting open and closed loci (G-to-C at position 5 edits using spacers previously used for Cas9 editing25 in open/closed loci) were designed by ordering 1 primer with backbone-spacer-scaffold (start) sequence and 1 primer with backbone-extension-scaffold(end) sequence followed by PCR on a plasmid containing the complete scaffold sequence. Then, the pU6-pegRNA-GG-acceptor for 45 arrayed pegRNAs was digested with BsaI (NEB) and BamHI (NEB). The pU6-pegRNA-GG-acceptor for open and closed loci cloning was digested with BsaI (NEB). After digestion, the plasmid was dephosphorylated by rSAP (NEB), followed by gel extraction. Next, gene blocks/PCR products were Gibson assembled into digested pU6-pegRNA-GG-acceptor using a 5:1 ratio of backbone (50 ng) and gene block or PCR DNA. Assembly was performed with NEBuilder HiFi DNA Assembly Master Mix (NEB), followed by transformation into NEB Stable Competent E. coli. Grown colonies were inoculated into LB (100 µg/ml Ampicillin) overnight, and plasmids were extracted via GeneJET Plasmid Miniprep Kit (ThermoFisher). Sequences were verified with Sanger sequencing. pU6-pegRNA-GG-acceptor (Addgene #132777) was a gift from David Liu1.
Pooled library screens
Coverage was kept above 500x for both libraries to prevent library skewing.
Library 1
HEK293T cells were cultured in 24x T175 flasks and transduced with lentivirus (MOI of 0.3) at a confluency of 75% in DMEM++ with 33 µg/ml Polybrene. After 1 day, cells were pooled, and 2/3 were split into 47x T175 flasks (1x control T175 flask) with 2.5 µg/ml puromycin for selection. The next day, the medium was replaced with fresh selection medium (2.5 µg/ml puromycin). After 3 days of selection, cells were frozen and kept in liquid nitrogen until use. For each replicate, cells were thawed separately and grown in 25 T175 flasks or 12 T175 flasks for untransfected control). At 70% confluency, cells were transfected with 18.5 µg of pCMV-PE2-tagRFP-BleoR plasmid in 508 µl Opti-MEM (Thermo Fisher Scientific) and 152 µl PEI (1 mg/ml) in 25 ml medium per flask (Day 0). On day 1, cells were split into 19 T300 flasks and selected with 750 ng/µl Zeocin (InvivoGen). Cells were maintained under Zeocin selection until harvest on day 7. Untransfected control replicates were maintained without selection for 7 days.
Library 2
HEK293T and U2OS cells were cultured in T175 flasks and transduced with library 2 lentivirus (MOI of 0.3) at 75% confluency. After 1 day, cells were split 1:2 into two T175 with 2.5 µg/ml puromycin for selection. Cells were expanded for 7 days and afterward frozen in liquid nitrogen. For each replicate, cells were thawed and seeded independently in T175 flasks. At 70% confluency, cells were transfected with either
-
(A)
18.5 µg of pCMV-PE2-tagRFP-BleoR plasmid (2.7 pmol)
-
(B)
14.4 µg pCMV-PE2-tagRFP-BleoR plasmid (2.1 pmol) together with 4.1 µg pEF1a-hMLH1dn (1.05 pmol)
-
(C)
Control (no DNA)
U2OS only:
-
(D)
17.2 µg pCMV-PEmax-P2A-BSD (2.7 pmol) with 1.3 µg pUC19 filler plasmid
-
(E)
13.4 µg pCMV-PEmax-P2A-BSD (2.1 pmol) with 4.1 µg pEF1a-hMLH1dn (1.05 pmol) and 1 µg pUC19 filler plasmid
in 508 µl Opti-MEM (Thermo Fisher Scientific) and 152 µl PEI (1 mg/ml) in 25 ml medium per flask (Day 0; 1x T-175 flask per replicate). On day 1, cells of conditions (A) and (B) were split into 2 T175 flasks with 750 ng/µl (HEK293T) or 200 ng/µl (U2OS) Zeocin (InvivoGen) added to the medium. Conditions (D) and (E) were split with 20 ng/µl Blasticidin (InvivoGen).
K562 cells were cultured in T175 flasks and transduced with library 2 lentivirus (MOI < 1) at 75% confluency. After 1 day, cells were split 1:2 into two T175 with 2.5 µg/ml puromycin for selection. Cells were expanded for 7 days and afterward frozen in liquid nitrogen. For each replicate, cells were thawed independently into T-175 flasks. On the day of the experiment, 100,000 K562 cells were seeded into 72 wells of 48-well plates. Cells were transfected with 1.5 µl Lipofectamine 2000 per well with the following plasmid concentration:
-
(A)
1000 ng of pCMV-PE2-tagRFP-BleoR plasmid (147 fmol)
-
(B)
777 ng pCMV-PE2-tagRFP-BleoR plasmid (114 fmol) together with 223 ng pEF1a-hMLH1dn (57 fmol)
-
(C)
932 ng pCMV-PEmax-P2A-BSD (147 fmol) with 68 ng pUC19 filler plasmid
-
(D)
724 ng pCMV-PEmax-P2A-BSD (114 fmol) with 223 ng pEF1a-hMLH1dn (57 fmol) and 53 ng pUC19 filler plasmid
-
(E)
Control (no DNA)
The next day (Day 1), the medium for (A) and (B) was replaced with RPMI++ with 200 ng/µl Zeocin (InvivoGen), whereas (C) and (D) were changed to RPMI++ with 20 ng/µl Blasticidin (InvivoGen).
After selection start, all cell lines were maintained under selection until harvest on day 7. Untransfected control replicates were maintained without selection for 7 days.
pCMV-PEmax-P2A-BSD (Addgene #174821) and pEF1a-hMLH1dn (Addgene #174824) were gifts from David Liu15,16.
Arrayed validation of pegRNAs on endogenous loci
For endogenous arrayed validation of pegRNAs in HEK293T, cells (130,000/well) were seeded into 48-well plates (Corning) and transfected 8 h after seeding with 750 ng of pCMV-PE2-tagRFP-BleoR, 250ng pegRNA, and 1 µl of Lipofectamine 2000 (Invitrogen) in Opti-MEM (Thermo Fisher Scientific) with a total volume of 50 µl (day 0). On day 1, the medium was removed, and cells were detached using 50 µl of TrypLE (Gibco) per well, resuspended in fresh medium containing 750 ng/µl Zeocin, and plated again into 48-well plates. K562 cells (100,000/well) were distributed into 48-well plates (Corning) and transfected 1 h after seeding with 750 ng of pCMV-PE2-tagRFP-BleoR, 250ng pegRNA, and 1.5 µl of Lipofectamine 2000 (Invitrogen) per well (day 0). On day 1, the medium was removed by centrifuging K562 cells and resuspending in fresh medium containing 200 ng/µl Zeocin. Cells were selected for 6 days and harvested on day 7. Experiments were performed with two biological replicates on separate days (each with two technical replicates).
Genomic DNA isolation and HTS
Genomic DNA from the library 1 screen was isolated by Blood & Cell Culture DNA Maxi Kit (Qiagen). DNA from endogenous editing experiments was isolated by direct lysis using direct lysis buffer: 10 µl of 4× lysis buffer (10 mM Tris-HCl pH 8, 2% Triton X-100, 1 mM EDTA, 1% freshly added proteinase K) was added to cells resuspended in 30 µl PBS and incubated at 60 °C for 60 min and 95 °C for 10 min. Genomic DNA from library 2 experiments was isolated by direct lysis followed by Phenol-Chloroform DNA purification and ethanol precipitation. Target sites of library screens were amplified by NEBNext Ultra II Q5 Master Mix (NEB; 26 cycles), and endogenous target sites were amplified by GoTaq G2 Hot Start Green Master Mix (Promega; 26 cycles). Library 2 was additionally amplified by Phusion High-Fidelity DNA Polymerase (NEB; 6 cycles) with an upstream reverse primer to sequence into the tevopreQ1 region. Amplicons from library DNA were purified with NucleoSpin Gel and PCR Clean-up Mini kit (Macherey-Nagel), and endogenous target amplicons were purified with Sera-Mag Select (Merck). Illumina sequencing adapters were added with NEBNext High-Fidelity 2X PCR Master Mix (endogenous target sites; NEB; 7 cycles) or Phusion High-Fidelity DNA Polymerase (library 1 and 2; NEB; 7 cycles) and purified by gel extraction. Final pools were quantified on a Qubit 3.0 (Invitrogen). Pooled library screens were sequenced paired-end on a Illumina NovaSeq 6000 using SP Reagent Kits (300 cycles). Endogenous targets were sequenced paired-end (2 × 150) on a Illumina MiSeq using MiSeq Reagent Nano Kit v2.
Library editing analysis
Editing levels were determined using a mix of in-house Python scripts and published sequence analysis tools. First, Cutadapt26 was used to trim sequences, and SeqKit27 was used for creating reverse complement sequences of R2 reads. Next, we analyzed trimmed sequences with an in-house Python script. Each sequencing read was assigned to the corresponding target sequence based on the spacer sequence, extension sequence, target sequence end, and barcode. Reads without a match for each region were excluded from further analysis (24% of total reads). Additionally, reads with recombination between the spacer and target/extension sequence (31% of reads with matches of all regions) were also excluded from further analysis. To calculate the editing rate, we compared the read sequence (2 bp upstream of nick position until 5 bp downstream of edited flap-end) with wild-type and edited sequence and assigned the labels' unedited', 'edited', or 'non-match'. The editing efficiency was calculated as previously described in Kim et al.4 with the following formula:
Background intended edit frequencies were determined by analyzing the control library pool, which was not transfected with the prime editor. Editing values were clamped to be within 0 and 100%. The unintended editing rate was calculated as described for intended editing but by using read counts of unintended editing ("non-match") instead. Target sequences were further filtered for having a minimum of 100 reads in every replicate (including control; discarding 20,963 sequences with low coverage). Target sequences where the corresponding control had > 20% unintended editing rate (6,245 sequences) or > 5% intended editing (70 sequences) were also discarded. This led to a total of 92,423 sequences that were used for machine learning. For each pegRNA, a selection of features was extracted for training statistical machine learning models and complementing deep learning models. These features included RTT-, PBS- and Correction-length, GC content, melting temperature, max. length of polyA/T/G/C sequence stretches and minimum free energy (MFE; ViennaRNA Package 2.028). DeepSpCas9 score12 was previously developed to represent cutting efficiency and was calculated for statistical machine learning models (linear regression- and tree-based models; using TensorFlow 2.4.1) but was not used in the final PRIDICT deep learning model. A more detailed description of features can be found in Supplementary Table 1, and Supplementary Table 7 contains Spearman correlations between each feature.
Analysis of unintended editing types
We used CRISPResso229 to analyze unintended editing in library 1 in more detail. Sequencing files from each replicate (+ control) were first demultiplexed into individual files containing reads with the target sequence of each pegRNA. Afterwards, we used CRISPRessoBatch with batch files containing the reference sequences (“amplicon_seq”), the edited sequences (“expected_hdr_amplicon_seq”), as well as the quantification windows (“quantification_window_coordinates”) spanning from 2bp before the nick until 5bp after the flap for each pegRNA. After running CRISPRessoBatch, we used the “Modification_count_vectors” files from each pegRNA to summarize the findings and generate position plots with mutations per position in the pegRNA (Supplementary Fig. 2).
Arrayed endogenous editing analysis
Arrayed editing experiments were analyzed with CRISPResso229 in batch mode. The original sequence (“amplicon_seq”), the expected sequence after editing (“expected_hdr_amplicon_seq”) and window of quantification (2 bp upstream of nick position until 5 bp downstream of edited flap-end; “quantification_window_coordinates”) were used for batch analysis. Editing efficiencies and unintended editing rates of technical replicates (transfection in separate wells but on the same day) were averaged and used as one independent biological replicate for the following analysis. Editing rates are listed in Supplementary Table 6.
Analysis of chromatin characteristics of endogenous loci
To analyze chromatin characteristics for endogenous loci in HEK293T and K562, we used publicly available datasets30–32 and mapped them to our loci. Of note, we used datasets from HEK293 for mapping to HEK293T loci due to the higher availability of high-quality chromatin accessibility datasets for HEK293. Bigwig files of ATAC-seq33 (K562 - GSM2902637, HEK293 - arithmetic mean of GSM2902624-7), DNAse-seq33 (K562 - GSM2902638, HEK293 - GSM2902639), and ChIP-seq results of H3K4me332 (K562 - ENCFF291SWG, HEK293 - ENCFF756EHF), H3K27me3 (K562 - ENCFF312LYO32, HEK293 - SRX636940634), H3K9me3 (K562 - SRX06752732, HEK293 - SRX89764135), H3K36me332 (K562 - ENCFF745HXR, HEK293 - ENCFF704SBO) were downloaded from public databases for the genome build hg19. The genomic coordinates of the edited sites were converted from the hg38 genome build to hg19 using liftOver36. Wiggletools37 was used to extract the fold change values from the bigwig file for the 2kb regions (1kb up and downstream from the middle of the loci) around the edited sites. Values from each dataset were normalized to the 45 arrayed endogenous pegRNAs (Fig. 4a) and the 10 open and closed chromatin loci (Supplementary Fig. 17d) of the respective cell line using z-score normalization. Normalized values are listed in Supplementary Table 6.
Machine learning model development
Machine learning models were developed based on the filtered dataset from library 1 (92,423 pegRNA design variants). We followed a grouped 5-fold cross-validation where the grouping was based on disease locus (pegRNAs for the same locus were not split between training-, test- and validation sequences). Each fold had 80% of the sequences for training and 20% for testing. A validation set for each fold was created by taking a 10% grouped random split (again, grouping was based on disease locus) from the fold’s training sequences which was used to optimize model hyperparameters. For the neural network model, we used a uniform random search strategy that randomly chose a set of hyperparameter configurations from the set of all possible configurations and trained corresponding models on one chosen random fold. Subsequently, the best model hyperparameters were determined based on the performance of the models on the validation set of the respective fold. Finally, these hyperparameters were used for the final training and testing of each model on all 5 folds. Notably, we observed that library 1 was large enough for creating PRIDICT since model performance (Spearman/Pearson) on test datasets only slightly increased when training on the whole training dataset compared to a 25% fraction of the training dataset (Supplementary Fig. 12g,h). For baseline models38,39, we used a random search strategy over each model’s specific hyperparameter space where the best hyperparameters were determined using 2-fold cross-validation on the combined training and validation set of each fold. Subsequently, the model achieving the best performance was retrained and tested (using the test set) on each corresponding fold of the five folds.
Features for baseline and neural network models are listed in Supplementary Table 1. Details of the attention-based bi-directional recurrent neural network model “PRIDICT” are described in Supplementary Methods 113,40–51.
Data analysis of external datasets
External validation datasets (Anzalone et al.1, Kim et al.4, Li et al.5) were analyzed with in-house Python scripts. Editing values of Anzalone et al.1 were analyzed and formatted by Li et al.5 and downloaded from their GitHub repository. In brief, sequences of external datasets were reformatted to fit the input of our prediction scripts (XXX(G/C)XXX), and the prediction of intended editing was compared with measured intended editing rates.
Statistics and reproducibility
Statistics were performed using Python 3.8 and SciPy 1.6.3. Mann-Whitney U rank test (two-sided) was used to compare editing efficiencies of pegRNAs with high vs. low PRIDICT scores. Pearson (r) and Spearman (R) correlations were determined to evaluate the correlation of predicted and measured pegRNA editing rates. Prime editing experiments were performed at least in independent biological duplicates. Sample sizes are described in figure legends.
Extended Data
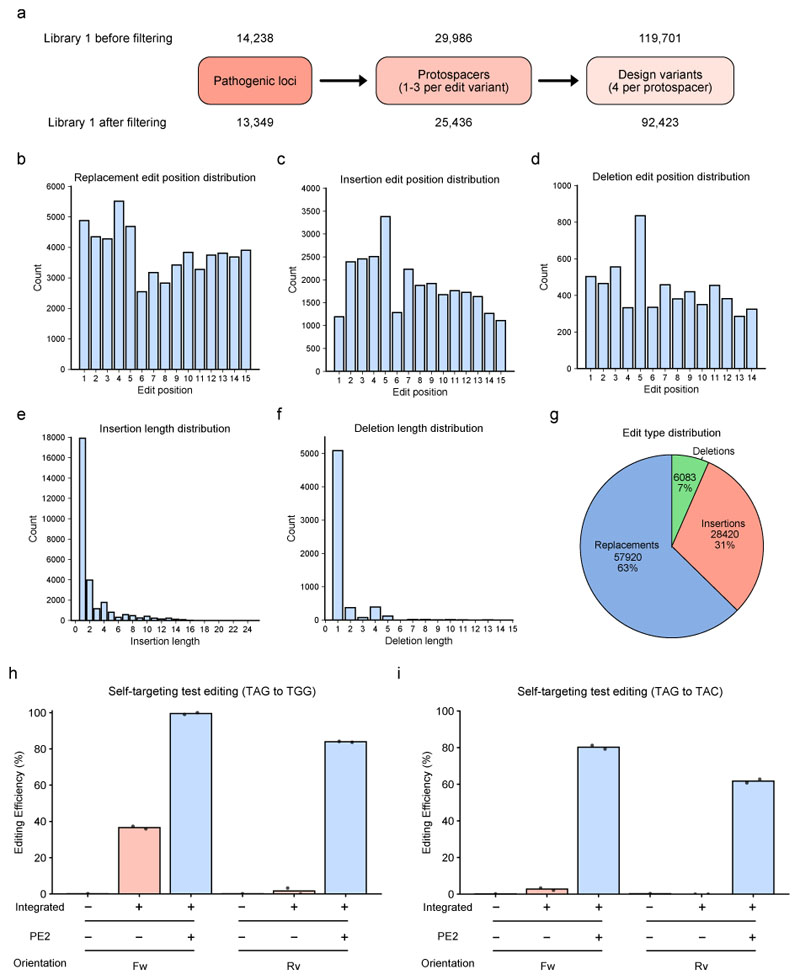
Extended Data Fig. 1. Self-targeting screen characteristics.
(a) Visualization of library design (library 1) and numbers before and after filtering results. (b) Distribution of edit positions for single base replacement edits in library 1. (c) Distribution of edit positions for insertion edits in library 1. (d) Distribution of edit positions for deletion edits in library 1. (e) Distribution of insertion lengths in library 1. (f) Distribution of deletion lengths in library 1. (g) Distribution of edit types in library 1 (number of design variants and percentage of the total library). (h-i) Editing rates of a test self-targeting locus with a forward (Fw) or reverse (Rv) orientation of the target sequence. Either on plasmid level or integrated by lentiviral transduction in HEK293T cells. Data points for bars (from left) 2,3 and 5,6 correspond to 2 technical replicates (simultaneous transfection of two separate wells). Only one data point was used for the plasmid controls (bar 1 and 4). (h) pegRNA with TAG to TGG edit. (i) pegRNA with TAG to TAC edit. The observed editing in the forward direction in the absence of PE2 could be caused by lentiviral reshuffling or ADAR-mediated A to I (G) RNA editing. The latter could occur during lentiviral packaging in HEK293T cells: HEK293T cells endogenously express ADAR and the target site is present as RNA on the lentiviral vector and targeted by the complementary pegRNA with a mismatch, providing an ideal template for ADAR-dependent RNA editing. The observation that primarily TAG to TGG (but not TAG to TAC) showed background editing is in line with this hypothesis, as previous studies showed ADAR preference for UAG sequences52.
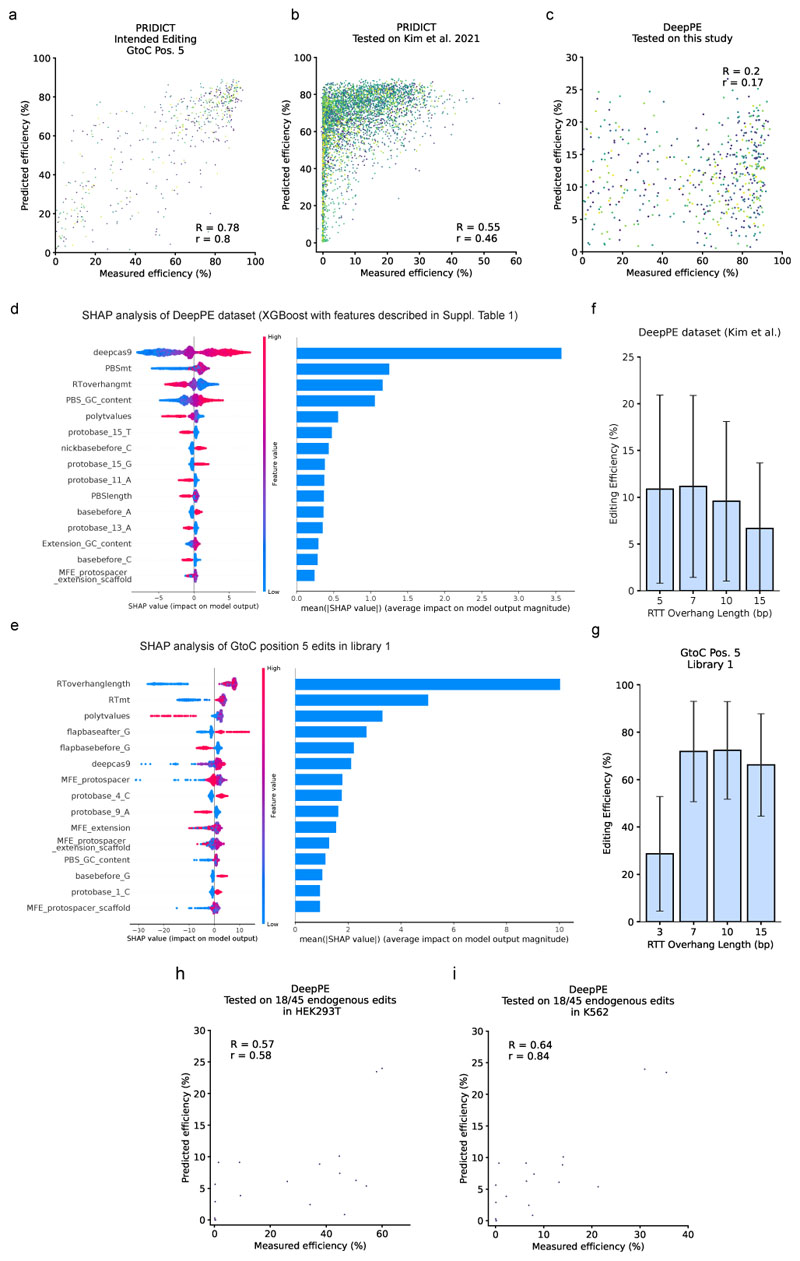
Extended Data Fig. 2. Additional validation of the DeepPE model.
(a) Predicted (PRIDICT) and measured intended editing efficiency for GtoC edits at position 5 of RTT in the dataset from this study. Data from all 5 test sets (five-fold cross-validation) were combined for this visualization. n = 540 (b) Evaluation of PRIDICT AttnBiRNN (trained on the dataset from this study) by testing on pegRNAs from Kim et al. 2021 HT dataset (only G to C at Position 5). n = 4,457. (c) Evaluation of DeepPE model (original, trained on Kim et al. 2021 HT dataset) by testing on the dataset from this study (only G to C at Position 5). n = 540. (d,e) SHAP analysis of XGBoost models trained and tested (d) on DeepPE dataset (n = 43,149) or (e) on G-to-C Position 5 edits from library 1. Feature descriptions are listed in Supplementary Table 1. (f) Editing efficiency with different RTT overhang lengths (5, 7, 10, 15 bp) in DeepPE (Kim et al.) dataset. n for each bar (left to right) = 10,746, 10,828, 10,921, 10,654. Error bars = mean +/- SD (g) Editing efficiency with different RTT overhang lengths (3, 7, 10, 15 bp) in GtoC Pos. 5 edits of library 1 for a direct comparison to identical edits in the DeepPE dataset. n for each bar (left to right) = 135, 135, 137, 133. (f,g) Error bars = mean +/- SD (h,i) Evaluation of DeepPE model (n = 18) on 18/45 endogenous edits from this study in HEK293T (h) and K562 (i).
Extended Data Fig. 3. Additional validation of the Easy-Prime PE2 model.
(a) Edit type count distribution in the original Easy-Prime test dataset. (b) Evaluation of Easy-Prime PE2 model by testing this XGBoost model on the original Easy-Prime test dataset5, filtered against 1bp edits at position 5 of the RTT to eliminate the bias towards this edit type. n = 585. (c-g) Evaluation of Easy-Prime PE2 by testing the model on datasets generated in this study. (c) Library 1 in HEK293T, n = 92,423. (d) Library 2 (editing with PE2 and pegRNAs without tevopreQ1) in HEK293T, n = 915. (e) Library 2 (editing with PE2 and pegRNAs without tevopreQ1) in K562, n = 876. (f,g) Endogenous loci from Fig. 4a,b in HEK293T (f) and K562 (g), n = 45. (h) Intended editing efficiency rank of the best-predicted pegRNA for each pathogenic locus in library 1 (PRIDICT and Easy-Prime). Pathogenic loci with multiple pegRNAs on rank 1 (identical efficiency) and loci with less than 3 pegRNAs were excluded from this analysis. Predictions from PRIDICT were taken from 5 different cross-validations to ensure none of the predictions are included in the training set. n = 12,189. (i) Intended editing efficiency rank of the best-predicted pegRNA for each endogenous locus (PRIDICT and Easy-Prime). n = 15.
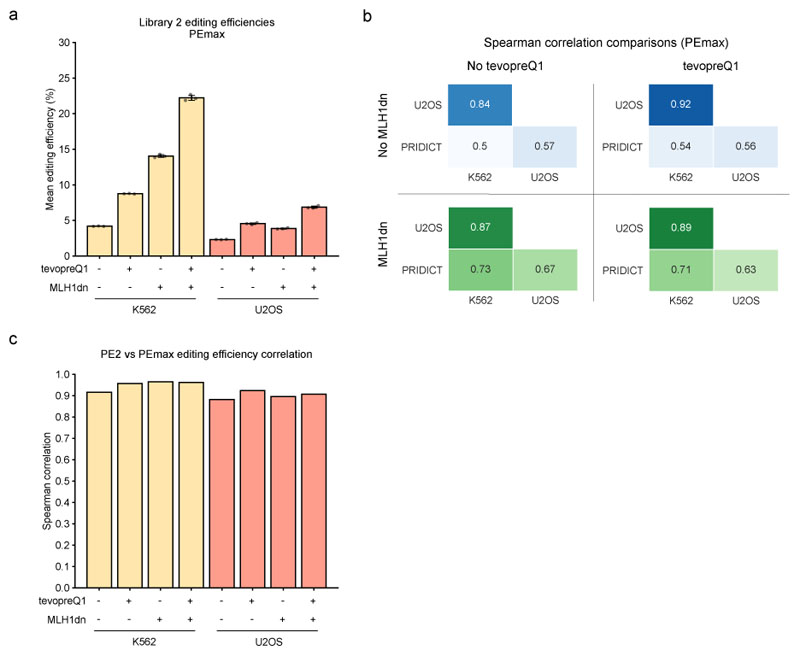
Extended Data Fig. 4. Additional library 2 evaluation with PEmax.
(a) Mean editing efficiencies of each replicate, including all pegRNAs in library 2 with different experimental conditions in U2OS and K562 cells. Error bars indicate the mean +/- SD of three biologically independent replicates. n = 3. Mean editing of library 2 for each of the 3 replicates is based on the following number of pegRNAs for each data point (bars left to right) = 916, 922, 917, 924, 879, 869, 877, 866. Note that absolute levels of editing efficiency for PEmax cannot be directly compared to PE2 in this study due to the use of different selection agents (Blasticidin for PEmax screens compared to Zeocin for PE2 screens). Previous studies showed that in identical setups, PEmax surpasses the performance of PE216. (b) Spearman correlation for PEmax editing efficiencies in library 2 between different experimental conditions (MLH1dn, tevopreQ1) and cell lines (K562, U2OS). (c) Editing efficiency rank correlations (Spearman) in library 2 between editing performed with PE2 vs. editing performed with PEmax.
Supplementary Material
Acknowledgments
We thank the Functional Genomics Center Zurich (FGCZ) for their help and support in next-generation sequencing; the Flow Cytometry Facility of the University of Zurich and especially Mario Wickert for performing liver hepatocyte sorting experiments; the Science IT team at the University of Zurich for providing infrastructure used for data analysis and especially Philip Shemella for helpful discussions about code performance optimizations; Ruben Schep for discussions about chromatin marks; Giuliana Affentranger for support in the design of figures; the members of the Schwank lab for fruitful discussions. This work was supported by the SNF (310030_185293 and 201184), the University Research Priority Program "Human Reproduction Reloaded" and “ITINERARE” of the University of Zurich. K.F.M. holds a PHRT iDoc Fellowship (PHRT_324).
Footnotes
Author contributions
N.M. designed the study, performed experiments, and analyzed data. A.A. designed and generated attention-based bi-directional recurrent neural networks (PRIDICT) and implemented feature extraction strategies. A.A. and N.M. built linear regression and tree-based machine learning models and performed feature extraction analysis. L.K. performed in vivo experiments. K.F.M and C.S. contributed to arrayed validation experiments. L.S. performed pegRNA and AdV cloning experiments. Z.B. performed the analysis of chromatin characteristics of endogenous loci. N.M., A.A., and G.S. wrote the manuscript. M.K. and G.S. designed and supervised the research. All authors revised the manuscript.
Competing interests
The authors declare no competing interests.
Data availability
Measured editing rates used for analysis and figures in this study are provided as Supplementary Tables and on GitHub (https://github.com/uzh-dqbm-cmi/PRIDICT). DNA-sequencing data is available via NCBI Sequence Read Archive (PRJNA825584). Target sequences of pathogenic mutations were based on the ClinVar database (accessed December 2019), and corresponding genomic sequences (flanking the edit) were acquired via UCSC Genome Browser (Table Browser, hg38). Plasmid encoding for pCMV-PE2-tagRFP-BleoR is available from Addgene (#192508).
Code availability
Custom Python code used in this study is provided on GitHub (https://github.com/uzh-dqbm-cmi/PRIDICT). Additional information on the PRIDICT algorithm can be found in Supplementary Methods 1.
References
- 1.Anzalone AV, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu JY, et al. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nat Commun. 2021;12:1034. doi: 10.1038/s41467-021-21337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hwang G-H, et al. PE-Designer and PE-Analyzer: web-based design and analysis tools for CRISPR prime editing. Nucleic Acids Res. 2021;49:W499–W504. doi: 10.1093/nar/gkab319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim HK, et al. Predicting the efficiency of prime editing guide RNAs in human cells. Nat Biotechnol. 2021;39:198–206. doi: 10.1038/s41587-020-0677-y. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Chen J, Tsai SQ, Cheng Y. Easy-Prime: a machine learning–based prime editor design tool. Genome Biol. 2021;22:235. doi: 10.1186/s13059-021-02458-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landrum MJ, et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen S, Yuzenkova Y, Zenkin N. Mechanism of Eukaryotic RNA Polymerase III Transcription Termination. Science. 2013;340:1577–1580. doi: 10.1126/science.1237934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Z, Herrera-Carrillo E, Berkhout B. Delineation of the Exact Transcription Termination Signal for Type 3 Polymerase III. Mol Ther Nucleic Acids. 2018;10:36–44. doi: 10.1016/j.omtn.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bill CA, Duran WA, Miselis NR, Nickoloff JA. Efficient repair of all types of single-base mismatches in recombination intermediates in Chinese hamster ovary cells: Competition between long-patch and G-T glycosylase-mediated repair of G-T mismatches. Genetics. 1998;149:1935–1943. doi: 10.1093/genetics/149.4.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walton RT, Christie KA, Whittaker MN, Kleinstiver BP. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368:290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lundberg SM, Lee SI. In: Advances in Neural Information Processing Systems. von Luxburg U, Guyon I, Bengio S, Wallach H, Fergus R, editors. 2017. A unified approach to interpreting model predictions; pp. 4766–4775. [Google Scholar]
- 12.Kim HK, et al. SpCas9 activity prediction by DeepSpCas9, a deep learning–based model with high generalization performance. Sci Adv. 2019;5:eaax9249. doi: 10.1126/sciadv.aax9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sundararajan M, Taly A, Yan Q. Axiomatic attribution for deep networks; 34th International Conference on Machine Learning, ICML 2017; 2017. pp. 5109–5118. [Google Scholar]
- 14.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson JW, et al. Engineered pegRNAs improve prime editing efficiency. Nat Biotechnol. 2022;40:402–410. doi: 10.1038/s41587-021-01039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen PJ, et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell. 2021;184:5635–5652.:e29. doi: 10.1016/j.cell.2021.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nair N, et al. Computationally designed liver-specific transcriptional modules and hyperactive factor IX improve hepatic gene therapy. Blood. 2014;123:3195–3199. doi: 10.1182/blood-2013-10-534032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Untergasser A, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115–e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villiger L, et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nature Medicine. 2018;24:1519–1525. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 20.Kim HK, et al. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat Methods. 2017;14:153–159. doi: 10.1038/nmeth.4104. [DOI] [PubMed] [Google Scholar]
- 21.Kent WJ, et al. The Human Genome Browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim N, et al. Prediction of the sequence-specific cleavage activity of Cas9 variants. Nat Biotechnol. 2020;38:1328–1336. doi: 10.1038/s41587-020-0537-9. [DOI] [PubMed] [Google Scholar]
- 23.Dang Y, et al. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 2015;16:1–10. doi: 10.1186/s13059-015-0846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Böck D, et al. In vivo prime editing of a metabolic liver disease in mice. Sci Transl Med. 2022;14 doi: 10.1126/scitranslmed.abl9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen KT, et al. Chromatin accessibility and guide sequence secondary structure affect CRISPR-Cas9 gene editing efficiency. FEBS Lett. 2017;591:1892–1901. doi: 10.1002/1873-3468.12707. [DOI] [PubMed] [Google Scholar]
- 26.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. [Google Scholar]
- 27.Shen W, Le S, Li Y, Hu F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS One. 2016;11:e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorenz R, et al. ViennaRNA Package 2.0. Algorithms for Molecular Biology. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clement K, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol. 2019;37:224–226. doi: 10.1038/s41587-019-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schep R, et al. Impact of chromatin context on Cas9-induced DNA double-strand break repair pathway balance. Mol Cell. 2021;81:2216–2230.:e10. doi: 10.1016/j.molcel.2021.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrett T, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2012;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Y, et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 2020;48:D882–D889. doi: 10.1093/nar/gkz1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karabacak Calviello A, Hirsekorn A, Wurmus R, Yusuf D, Ohler U. Reproducible inference of transcription factor footprints in ATAC-seq and DNase-seq datasets using protocol-specific bias modeling. Genome Biol. 2019;20:42. doi: 10.1186/s13059-019-1654-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamb KN, et al. Discovery and Characterization of a Cellular Potent Positive Allosteric Modulator of the Polycomb Repressive Complex 1 Chromodomain, CBX7. Cell Chem Biol. 2019;26:1365–1379.:e22. doi: 10.1016/j.chembiol.2019.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hattori T, et al. Antigen clasping by two antigen-binding sites of an exceptionally specific antibody for histone methylation. Proceedings of the National Academy of Sciences. 2016;113:2092–2097. doi: 10.1073/pnas.1522691113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee BT, et al. The UCSC Genome Browser database: 2022 update. Nucleic Acids Res. 2022;50:D1115–D1122. doi: 10.1093/nar/gkab959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zerbino DR, Johnson N, Juettemann T, Wilder SP, Flicek P. WiggleTools: parallel processing of large collections of genome-wide datasets for visualization and statistical analysis. Bioinformatics. 2014;30:1008–1009. doi: 10.1093/bioinformatics/btt737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 39.Chen T, Guestrin C. XGBoost; Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; August 13. 2016. pp. 785–794. [Google Scholar]
- 40.Marquart KF, et al. Predicting base editing outcomes with an attention-based deep learning algorithm trained on high-throughput target library screens. Nat Commun. 2020;12:1–25. doi: 10.1038/s41467-021-25375-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paszke A, et al. Automatic differentiation in pytorch; NIPS 2017; 2017. [Google Scholar]
- 42.Cho K, et al. Learning Phrase Representations using RNN Encoder–Decoder for Statistical Machine Translation; Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP); 2014. pp. 1724–1734. [DOI] [Google Scholar]
- 43.Chung J, Gulcehre C, Cho K, Bengio Y. Empirical Evaluation of Gated Recurrent Neural Networks on Sequence Modeling. ArXiv. 2014 [Google Scholar]
- 44.Hochreiter S, Schmidhuber J. Long Short-Term Memory. Neural Comput. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 45.Bengio Y, Simard P, Frasconi P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans Neural Netw. 1994;5:157–166. doi: 10.1109/72.279181. [DOI] [PubMed] [Google Scholar]
- 46.Graves A. Supervised Sequence Labelling with Recurrent Neural Networks. Vol. 385. Springer Berlin Heidelberg; 2012. [Google Scholar]
- 47.Luong T, Pham H, Manning CD. Effective Approaches to Attention-based Neural Machine Translation; Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing; 2015. pp. 1412–1421. [DOI] [Google Scholar]
- 48.Vaswani A, et al. Attention is all you need; NIPS 2017; 2017. [Google Scholar]
- 49.Ba JL, Kiros JR, Hinton GE. Layer Normalization. ArXiv. 2016 [Google Scholar]
- 50.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition; Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition; 2016. [Google Scholar]
- 51.Bergstra J, Bengio Y. Random Search for Hyper-Parameter Optimization. Journal of Machine Learning Research. 2012;13:281–305. [Google Scholar]
- 52.Eggington JM, Greene T, Bass BL. Predicting sites of ADAR editing in double-stranded RNA. Nat Commun. 2011;2:319. doi: 10.1038/ncomms1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Measured editing rates used for analysis and figures in this study are provided as Supplementary Tables and on GitHub (https://github.com/uzh-dqbm-cmi/PRIDICT). DNA-sequencing data is available via NCBI Sequence Read Archive (PRJNA825584). Target sequences of pathogenic mutations were based on the ClinVar database (accessed December 2019), and corresponding genomic sequences (flanking the edit) were acquired via UCSC Genome Browser (Table Browser, hg38). Plasmid encoding for pCMV-PE2-tagRFP-BleoR is available from Addgene (#192508).
Custom Python code used in this study is provided on GitHub (https://github.com/uzh-dqbm-cmi/PRIDICT). Additional information on the PRIDICT algorithm can be found in Supplementary Methods 1.