Summary
Background
The clonal diversity underpinning trends in multi-drug resistant (MDR) Escherichia coli causing bloodstream infections (BSI) remains uncertain. We aimed to determine the contribution of individual clones to resistance over time, using large-scale genomics based molecular epidemiology.
Methods
We performed a longitudinal E. coli population genomic cohort study that sampled isolates from the 22,512 E. coli BSI included in the Norwegian surveillance program on resistant microbes (NORM) between 2002-2017. 15 out of 22 laboratories were able to share their isolates, and the first 22.5% (3500/15,552) from each year were requested. We used whole genome sequencing to infer the population structure (PopPUNK), and characterised the dominant MDR clonal complex (CC)131 using genetic markers of ST131 (Sequence Type), effective population size (BEAST) and presence of antimicrobial resistance determinants (ARIBA, PointFinder and ResFinder databases) over time and compared to a UK study by Kallonen et al.
Findings
Of the 3500 isolates requested, 3397 were received and 3254 successfully sequenced and included in the analysis. A significant increase (p<0.0001) of the MDR CC131, from 5.6% (71/1277) of isolates in 2002-2010 to 10.5% (207/1977) in 2011-2017, was the largest clonal expansion. CC131 dominated among ESBL positive (58.6%, 75/128) and fluoroquinolone non-susceptible isolates (39.3%, 148/377). Within CC131, clade A increased from 2002, whilst the global MDR clade C2 was not observed until 2007. Multiple de novo acquisitions of both blaCTX-M ESBL genes in clades A and C1, and of fluoroquinolone resistance in clade A were observed. We estimated that exponential increases in the effective population sizes of clades A, C1 and C2 occurred in the mid-2000’s, and in clade B a decade earlier. The rate of increase in the estimated effective population size of clade A (Ne=3147) was nearly ten-times that of C2 (Ne=345), with clade A over-represented in Norwegian CC131 isolates (27.0%, 75/278) compared to the UK study (5.4%, 8/147).
Interpretation
The early and sustained establishment of the predominantly susceptible CC131 clade A, relative to MDR clade C2, suggests that resistance is not necessary for clonal success. However, even in the low antibiotic use setting of Norway, resistance to important antimicrobial classes were quickly being selected for in this successful clone. This study demonstrates the importance of genomic surveillance in uncovering the complex ecology underlying MDR dissemination and competition which have implications for designing strategies and interventions to control the spread of high-risk MDR clones.
Funding
Trond Mohn Foundation, European Research Council and Marie Skłodowska-Curie Actions
Keywords: E. coli, ExPEC, ST131, Molecular epidemiology, antimicrobial resistance, multi-drug resistance
Introduction
Escherichia coli is a commensal of the gastrointestinal tract and a leading cause of bloodstream infections (BSI) worldwide, associated with considerable morbidity and mortality 1. E. coli associated with BSI are a subset of extraintestinal pathogenic E. coli (ExPEC), with BSI occurring as a result of underlying urinary tract infection (UTI), gastrointestinal colonisation or hepatobiliary infections 2. Though most ExPEC belong to phylogroup B2, they are not a monophyletic group of but a diverse collection of clones with the capacity to invade and cause disease through the acquisition of virulence factors 3. A limited set of globally dispersed ExPEC clones are responsible for the majority of infections 4 and increases in the incidence of E. coli-associated BSI have been reported in multiple countries 5–7. Increasing incidence is exacerbated by an increasing proportion of multi-drug resistant (MDR) ExPEC infections 5–7. One study estimated that E. coli resistant to third-generation cephalosporins caused 25-43 million cases of BSI and other serious infections globally in 2014 8.
In 2014, a single ExPEC clone, clonal complex (CC)131, was reported to predominate globally 9. A retrospective genomic study of BSI in the UK showed that CC131 had emerged quickly around 2003 10. Whole genome sequencing (WGS) has allowed the delineation of a number of phylogenetic sub-groups called clades within CC131, having different associations with antimicrobial resistance (AMR) 11–13. Clades C1 (alternatively referred to as H30-R) and C2 (H30-Rx) are characterised by fluoroquinolone resistance. Clade C2 is additionally associated with extended-spectrum β-lactamases (ESBL) of the CTX-M class conferring resistance to 3rd generation cephalosporins 11–13. Clades C1 and C2 of CC131 were reported to be the leading cause of MDR E. coli infections in the United States in 2011-2012 6. Additional CC131 clades; A, B, B0 and C0, have typically been reported to be minor clades with less resistance 11–13. Much of the literature proposes that AMR may have driven the emergence and success of CC131. There are, however, other successful E. coli BSI clones including CC73 and CC95 that have less extensive AMR10. Recent evidence suggests that success, through competition between E. coli strains, is a product of the relative frequency of the full complement of accessory genes, not just AMR, termed negative frequency-dependent selection (NFDS) 10,14.
Cases of E. coli BSI have been increasing in Norway, with the prevalence of ESBL-producing and fluoroquinolone non-susceptible E. coli causing BSI in Norway also increasing 5. Here we used WGS to analyse the clonal diversity of E. coli causing BSI in Norway over 16 years. This large nationwide longitudinal genomic collection allowed us to observe the emergence of clones and determine their contribution to rising BSI incidence and AMR prevalence.
Methods
Methods are expanded in the appendix p1-4.
Study design and bacterial isolates
A dataset of all 22,512 E. coli isolates 2002-2017, collected as part of the Norwegian surveillance program on resistant microbes (NORM) was collated5. Sample identifier, laboratory, year, sample date and antimicrobial susceptibility profile were reported for each isolate. We selected the first 22.5% of cases annually from 15 of the 22 laboratories participating in NORM (appendix p6) resulting in 3500/15,552 isolates for inclusion. The collection of data within NORM follows a standard protocol defined in the yearly surveillance reports (appendix p1) 5.
Isolates were plated on MacConkey agar no.3 (Oxoid Ltd., Thermo Fisher Scientific Inc., Waltham, MA, US), one colony was subsequently used to inoculate 1.6 mL Luria Bertani broth (Becton Dickinson, NJ, US) and incubated overnight at 37°C. Genomic DNA was extracted by QIAGEN (Hilden, Germany) using the DNeasy 96 Blood and Tissue kit (QIAGEN, Hilden, Germany). Samples were sequenced at the Wellcome Sanger Institute (Hinxton, UK) on the Illumina HiSeq platform (San Diego, CA, USA) using the NEB Ultra II custom kit (Emsworth, UK) with 392-plexing and a 150bp read length. Samples failed quality control if there was insufficient DNA to be sequenced, depth of coverage was <20, or there was evidence of mixed strain or species contamination Sequence data are available on the European Nucleotide Archive (ENA, Supplementary metadata).
Sequence analysis
Genome sequences were assembled and annotated using default parameters of a published pipeline 15,16 and the pangenome defined using Panaroo v1.0.217. Phylogenetic group18 and multilocus sequence type (MLST) were determined 19. PopPUNK v2.0.2 was used to cluster isolates with shared ancestry into clones 20. Clonal complex (CC) was defined here as the most prevalent sequence type (ST) within a PopPUNK grouping. ARIBA v2.12.2 was used with ResFinder and PointFinder (E. coli) databases21,22 to detect bacterial resistance determinants including blaCTX-M genes and fluoroquinolone resistance associated mutations in gyrA and/or parC/E 21–23. CTX-M genes served as a proxy for ESBL-production and ciprofloxacin efflux mutations were screened for in the Panaroo gene alignments 24–26.
PopPUNK4 (CC131) isolates were mapped to EC958 (GenBank HG941718.1), recombination detected and removed using GUBBINS v2.4.0, and a recombination-free phylogeny produced using RaxML v8.2.8 27,28. CC131 clades were assigned using clade-specific single nucleotide polymorphism (SNPs) and fimH alleles, and then corrected for the phylogenetic distribution of clades 11–13. Recombining K-mers were excluded before generating a reference-free alignment using SKA v1.0 for each CC131 clade 29. BEAST v1.10.4 was subsequently used with the GTR (generalised time reversible) substitution and gamma site heterogeneity models for 100,000,000 generations with 10% discarded as burn-in, sampled every 10,000 states 30. External collections of E. coli CC131 genomes used by McNally et al and a longitudinal BSI collection were used to compare to Norwegian isolates, the fastq data was processed using the above methods 10,14.
Statistical analysis
To determine if differences in proportions between two groups were significant, when any counts were <5 the 2-sided Fisher’s exact test was used, otherwise the chi-squared test was used using the p<0.05 threshold. We split the collection into two periods to assess differences in proportions to reduce any bias contained in any one year. The periods represented almost half the collection each 2002-2010 (39.4% 1277/3254) and 2011 (60.6%, 1977) but were split between 2010/11 after which many of the key changes of interest in the collection occurred, including a doubling of CTX-M prevalence observed between 2010 and 2011. Using these two periods we compared the prevalence of popPUNK4-CC131 and of each of the clades of CC131 as a proportion of the collection, and the prevalence of each of the CC131 clades within CC131. We also compared the prevalence of CTX-M positive, and ciprofloxacin non-susceptible isolates overall, that were CC131, and that belonged to each clade of CC131 as proportion of the collection, as a proportion of CC131 and as a proportion of each CC131 clade. We additionally compared the prevalence of CC131, and the prevalence of CTX-M positive and ciprofloxacin non-susceptible isolates between different health regions in Norway. Finally, we compared the prevalence of CC131, CC131 clades, CTX-M positive isolates and isolates in which gyrA and or parC were detected, between our collection and a UK collection by Kallonen et al 10. The BEAST Bayesian skyline model which uses phylogenetic history to provide estimates of the median effective population size over time with a 95% Highest Posterior Density (HPD), was used to identified the main exponential increase in population size and the slope representing the rate of increase in each clade. To assess the relative invasiveness of CC131 clades we used the ratio of effective population size and incidence in BSI in 2017 for CC131 clades. The incidence of each clade was estimated from the isolate counts adjusted for the proportion of total E. coli BSI in 2017 that was sampled in this study, reported per million population.
Ethical considerations
Data in NORM are disidentified. Ethical approval was not required after evaluation by the Regional Ethical Committee (REC North ref: 93528).
Role of the funding source
This work was supported by a Trond Mohn Foundation AMR grant TMS2019TMT04 (R.G., A.P., J.C., Ø.S., P.J.), Marie Skłodowska-Curie Actions 801133 (A.P.), and ERC grant 742158 (J.C.). The funders of the study played no role in study design, data collection, data analysis, data interpretation, or writing of the report. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
Of the 3500 isolates selected for inclusion, 3397 were received and sequenced. 3254 (95.8%) passed sequencing quality control criteria, and had a mean sequencing depth of 52.1 (SD 9.9). Excluded samples had no impact on the interpretation of the results (appendix p5).
Incidence/Prevalence
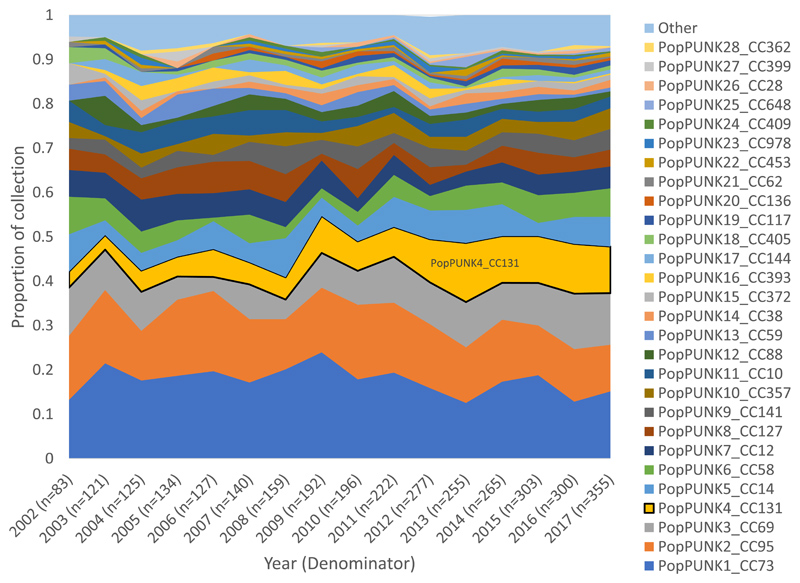
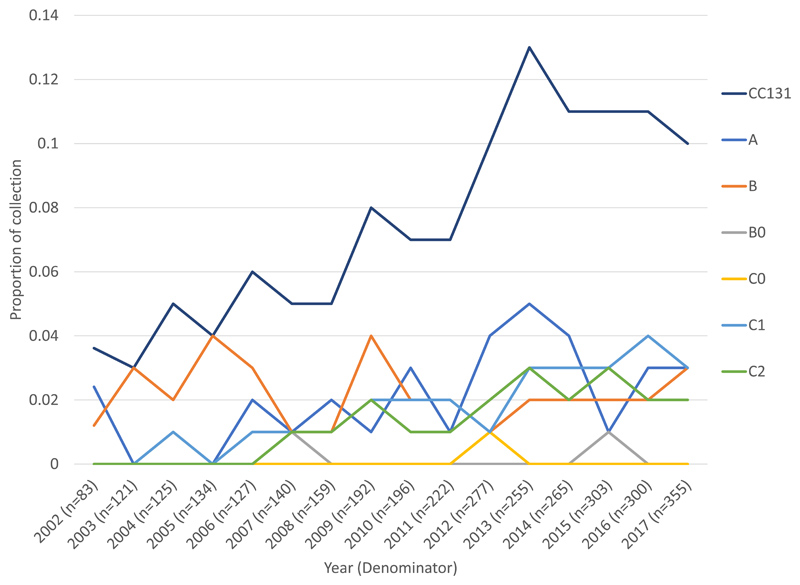
Sixty-seven percent of the collection (2180/3254) belonged to phylogroup B2. The collection was delineated into 136 clonal groups by PopPUNK (appendix p7). PopPUNK1 (CC73), 2 (CC95), 3 (CC69), 4 (CC131) and 5 (CC14) were the top five clones overall accounting for 54.1% (1761/3254) of the collection. PopPUNK4 (CC131) showed the largest proportional gain in the collection, occurring gradually from 2002 to its peak prevalence in 2013 (Figure 1, appendix p8), representing a significant increase between the periods 2002-2010 5.6% (71/1277) and 2011-2017 10.5% (207/1977, p<0.0001, Figure 1A). CC131 clades A (p=0.0043), C1 (p=0.0002) and C2 (p=0.0009) increased as a proportion of the collection between 2002-2010 and 2011-2017, whilst clade B remained stable (p=0.55, Figure 1B, appendix p18). Clades B0 and C0 combined represented only 2.9% (8/278) of CC131 (appendix p9). Within CC131 we only detected a change in proportion for clade B which significantly decreased as a proportion of CC131 (p=0.0001) between 2002-2010 (45.1%, 32/71) and 2011-2017 (20.3%,42/207).
Figure 1. Proportion of isolates by clone and by CC131 clades over time.
A) Clones as a proportion of the total collection (n=3254) by year. PopPUNK4 that represents clonal complex (CC)131 (yellow) had the largest gain of all lineages as proportion of the collection between 2002 and 2017.
B) CC131 (n=278) and its clades as a proportion of the total collection (n=3254) by year. Clades C1, C2 and A all contributed to the observed increase of CC131 as a proportion of collection.
Antimicrobial resistance
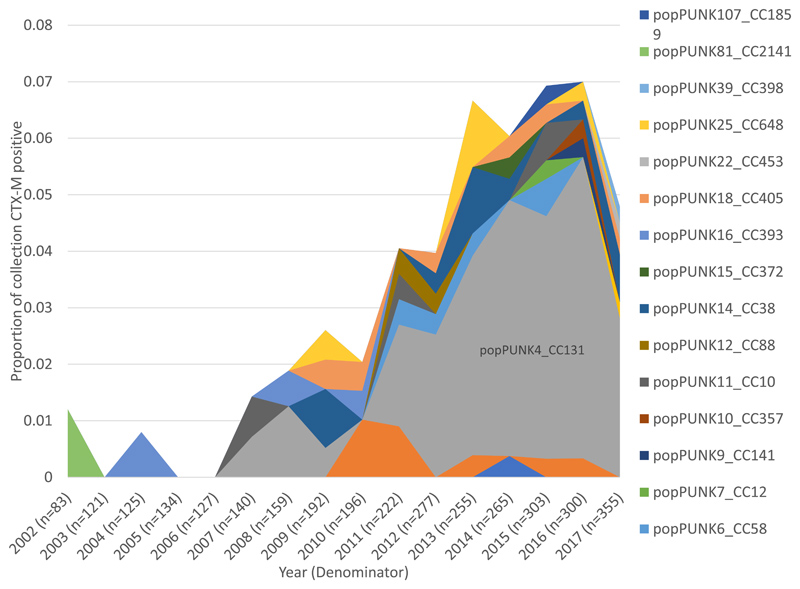
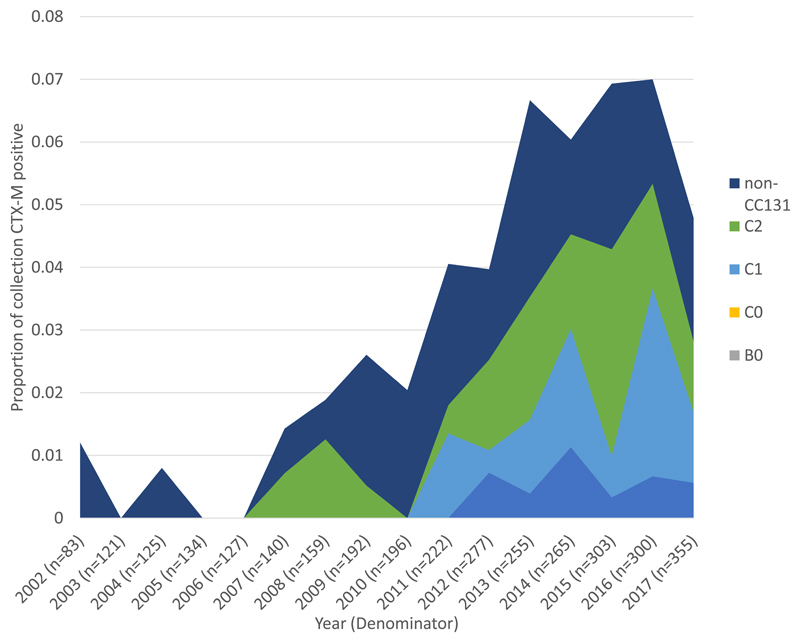
Using CTX-M as a marker for ESBL-production we detected 9 variants of CTX-M present in 3.9% (128/3254) of isolates across the whole collection (appendix p10). We did not detect a significant difference in CTX-M gene prevalence between 2002 1.2% (1/83) and the 2016 peak prevalence of 7.0% (21/300, p=0.058). CC131 was the single largest contributor of CTX-M positive isolates in the collection (58.6%, 75/128, Figure 2A). The remaining 41.4% (53/128) of CTX-M positive isolates belonged to 18 other PopPUNK clones. The number of clones with CTX-M positive isolates in 2017 was not significantly different to 2002 (p=0.19). Clades C2, C1 and A accounted for all CC131 CTX-M isolates and constituted 29.5% (33/112), 24.1% (27/112) and 9.8% (11/112), respectively, of all CTX-M isolates from 2011-2017 (Figure 2B). CTX-M isolates increased significantly overall, and CTX-M isolates that belonged to CC131, and the CC131 clades C1 and C2 as a proportion of the collection (appendix p19). C2 had the highest proportion of CTX-M isolates within it (76.7% 33/43) in 2011-2017.
Figure 2. Proportion of isolates that were CTX-M positive by clone and by CC131 clades over time.
A) CTX-M positive isolates (n=112) by clone as a proportion of collection year. In 2017 4.8% of isolates were CTX-M positive. PopPUNK 4 that represents clonal complex (CC)131 (grey) accounted for 59% of CTX-M positive isolates in 2017.
B) CTX-M positive isolates in CC131 (n=71) by clade as a proportion of the total collection (n=3254), over time. CC131 clades A and C1 start contributing CTX-M positive isolates in 2010 and 2011 respectively, corresponding to acquisitions of CTX-M in these clades.
CTX-M-15 was the most common CTX-M type in the entire collection (appendix p10). Within CC131, 70.6% (36/51) of clade C2 isolates were CTX-M-15 positive. The most common CTX-M type in clade C1 was CTX-M-27 (21.4%, 15/70) and CTX-M-15 (9.3%, 7/75) in clade A (appendix p20). All 74 clade B isolates were CTX-M negative. De novo CTX-M acquisitions were observed in clades A and C1 (appendix p11-13). Within C1 we estimated that there were 9 phylogenetically independent acquisitions of CTX-M contributing to the 27 CTX-M positive isolates. One acquisition of CTX-M-27, accounted for 14/27 isolates. However, the most acquisitions (5/9) were detected in a single isolate. Within clade A, we estimated that around 7 de novo acquisitions of CTX-M genes accounted for most of the CTX-M positive isolates, with most acquisitions (4/7) detected in only 1 isolate, and all acquisitions detected in a maximum of 2 isolates.
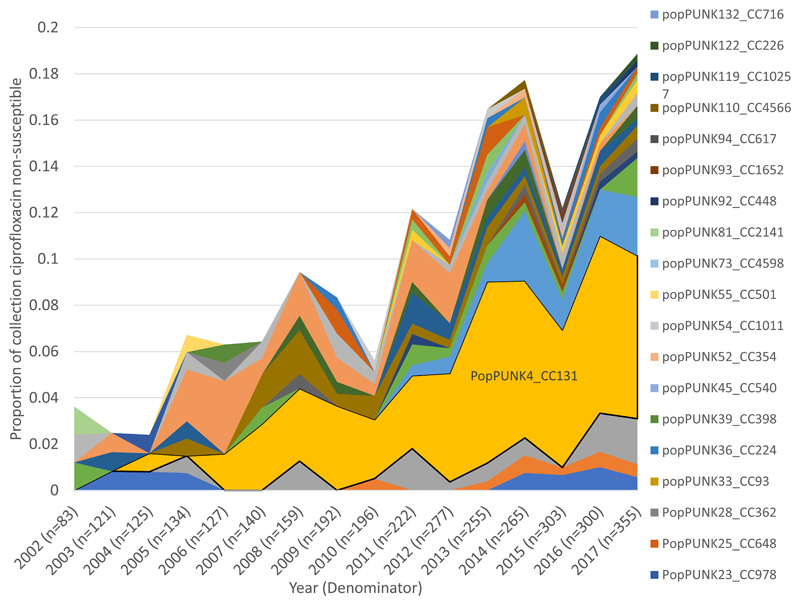
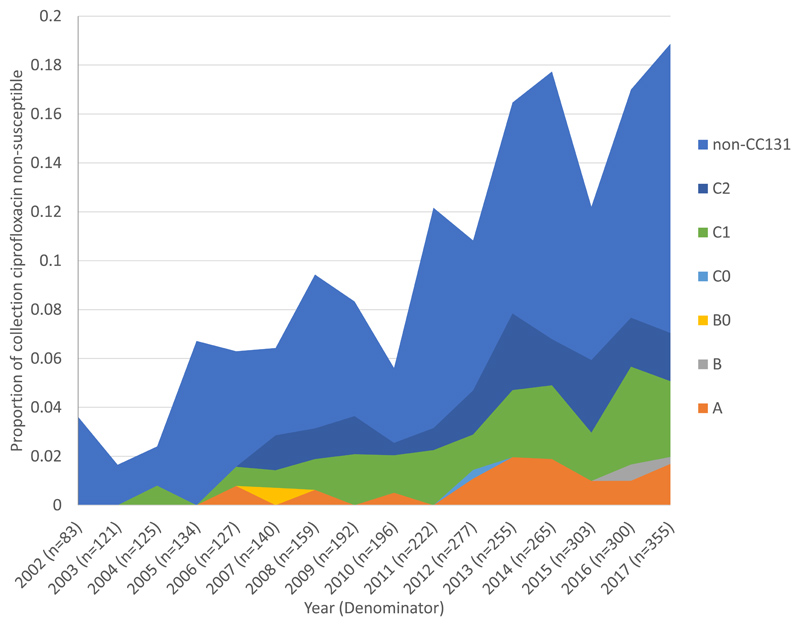
Ciprofloxacin non-susceptibility was observed in 11.6% (378/3254) of the isolates, significantly increasing from 3.6% (3/83) in 2002 to 18.9% (67/355) in 2017 (p=0.0006). Only 24.9% (94/378) of ciprofloxacin non-susceptible isolates were additionally CTX-M positive, whereas the majority (73.4%, 94/128) of CTX-M positive isolates were also ciprofloxacin non-susceptible. CC131 made the single largest contribution to ciprofloxacin non-susceptibility in the collection (39.2%, 148/378) and to isolates in the final year, 2017, (37.3%, 25/67). CC131, CC14 and CC69 combined accounted for 61.2% (41/67) of ciprofloxacin non-susceptibility in 2017. The remaining 38.8% (26/67) belonged to 34 other PopPUNK clones (Figure 3A). CC131 clades C1, C2 and A contributed 17.6% (53/301), 14.0% (42/301), and 8.3% (25/301), respectively, to 2011-2017 ciprofloxacin non-susceptibility. The proportion of isolates with ciprofloxacin non-susceptibility within CC131 significantly increased (p=0.0002), between 2002-2010 and 2011-2017 (Figure 3B, appendix p14,21). Clades C1 and C2 were near uniformly ciprofloxacin resistant (114/121) and all had the following mutations: GyrA S83L and D87N, and ParC S80I and E84V. The non-susceptible phenotype in clade A were represented by multiple independent phylogenetic clusters and were not well explained by detected gyrA, parC or parE mutations in the PointFinder (E. coli) resistance database (appendix p15). Eighty-two percent (23/28) of the ciprofloxacin non-susceptible isolates in clade A had resistance mutations in gyrA but not parC and most clade A isolates (71/75) had the ParE I529L mutation regardless of phenotype. Non-susceptible isolates in clade A did not have known efflux mutations or plasmid-mediated quinolone resistance genes.
Figure 3. Proportion of isolates that were ciprofloxacin non-susceptible by clone and by CC131 clades over time.
A) Ciprofloxacin non-susceptible isolates (n=377) by clone as a proportion of the total collection (n=3254) by year. PopPUNK4 that represents clonal complex (CC)131 (yellow) made the single largest contribution of any clone to ciprofloxacin nonsusceptibility. B) Ciprofloxacin non-susceptible isolates in CC131 (n=148) by clade as a proportion of the total collection (n=3254) by year. CC131 clades A, C1 and C2 all made considerable contributions to ciprofloxacin non-susceptible isolates in the collection overall.
Phylogenetic histories of CC131 clades
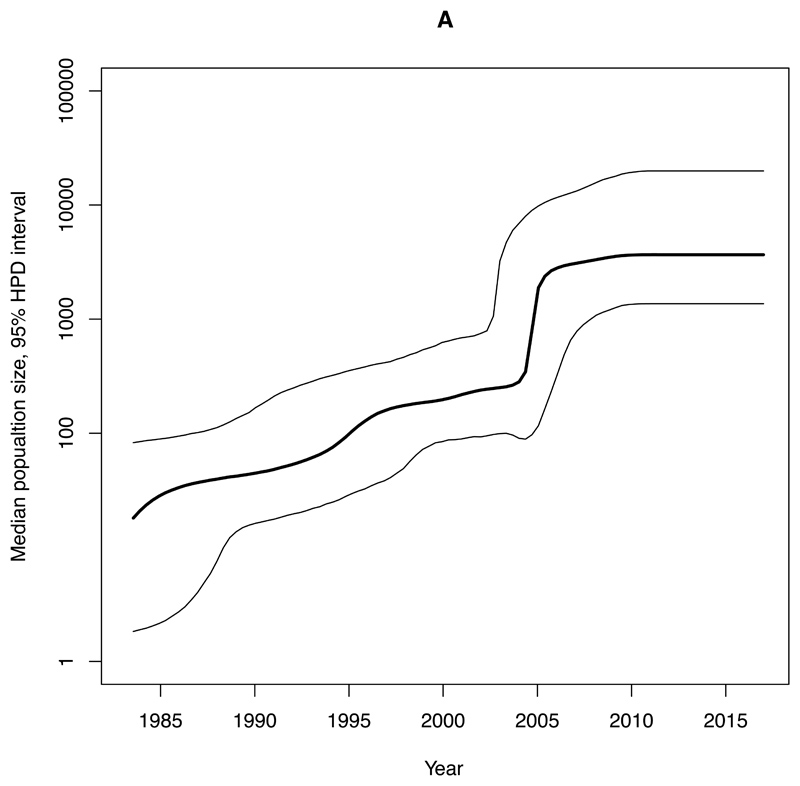
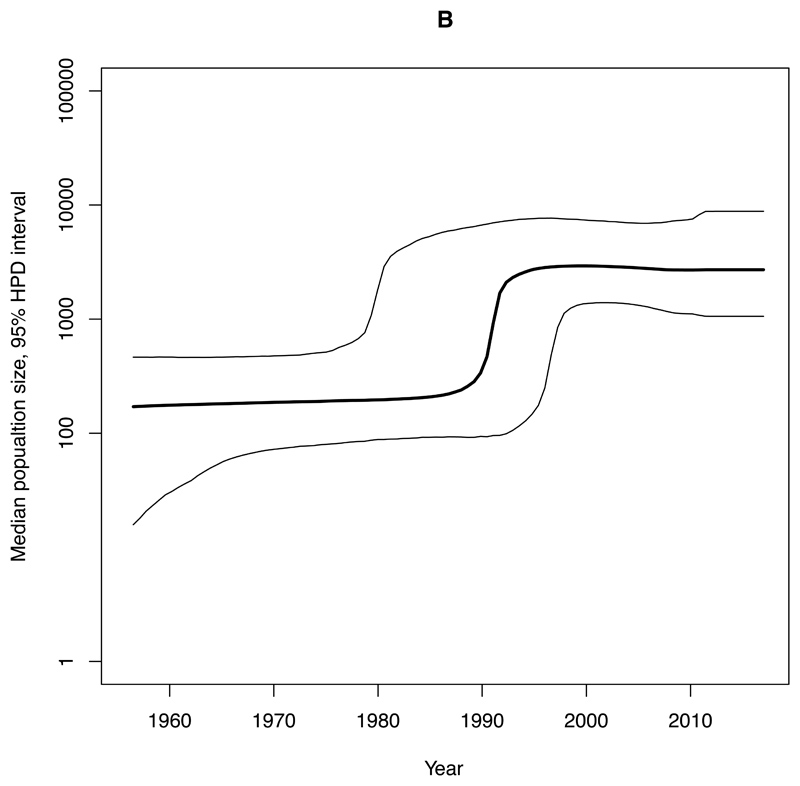
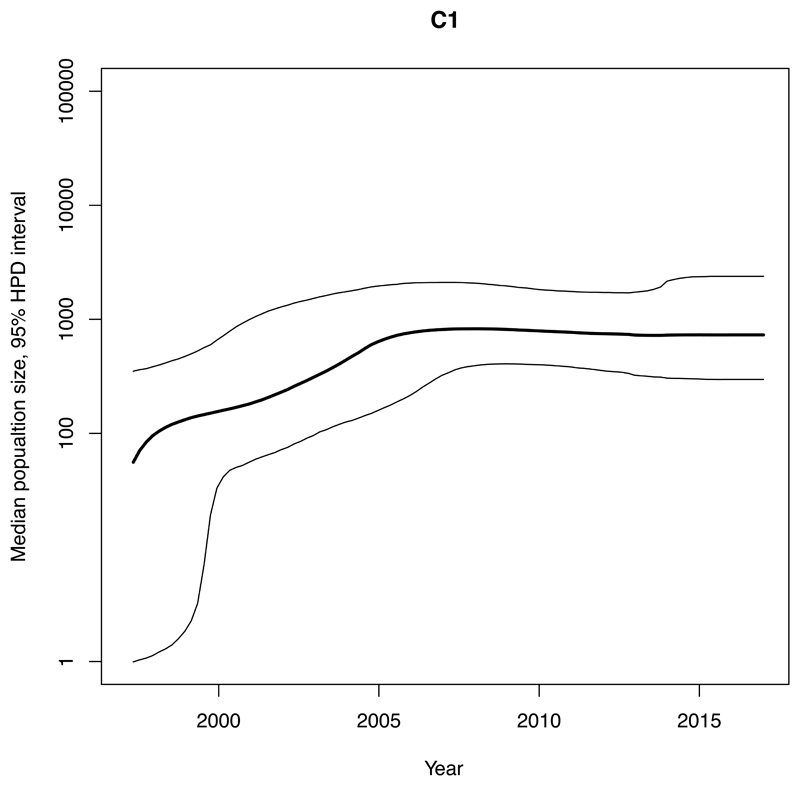
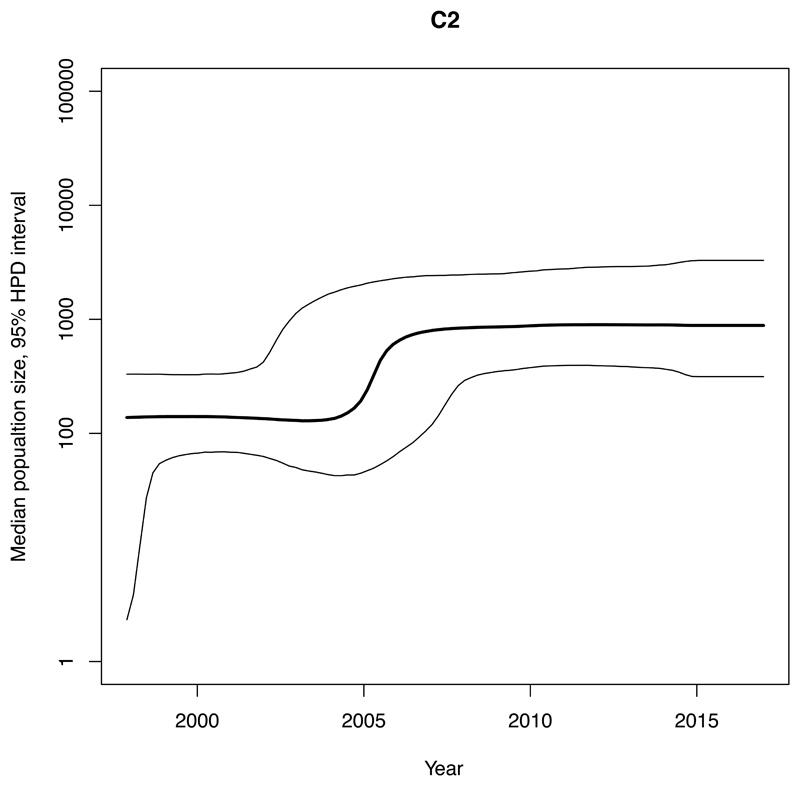
We estimated an exponential increase in the effective population sizes of clades A, C1 and C2, occurring in the mid 2000’s, whereas the increase in clade B occurred more than a decade earlier, around 1990 (Figure 4). The rate of expansion in the exponential phase of the effective population size over time (per year) for clade A (Ne=3147) was estimated to be nearly ten-times higher compared with C2 (Ne=345) and nearly twenty-five-times higher than for C1 (Ne=131) (Figure 4, Table 1).
Figure 4. Bayesian skyline plots of estimated median effective population size of CC131 clades over time.
95% Highest Posterior Density (HPD) intervals are plotted. Note that timescales on the x-axis differ. A) Clade A. B) Clade B. C) Clade C1. D) Clade C2.
Table 1. Median effective population size of CC131 clades.
Total cases were estimated for each CC131 clade by adjusting the number of genomes of a clade by the proportion of total 2017 cases that were sequenced, before calculating incidence per million population in Norway in 2017. tMRCA Time to most recent common ancestor. HPD 95% Highest Posterior Density.
| Clade | popSize 2017 [95% HPD] | Exponential time period | Growth rate [95% HPD] | Estimated incidence per million population 2017* | popSize/Incidence ratio 2017 | tMRCA [95% HPD] |
|---|---|---|---|---|---|---|
| A | 5488 [1365-19872] |
2004.6-2005.3 | 3147 [2909-3385] |
21.30 | 0.004 | 1997 [1968-1983] |
| B | 3341 [1058-8812] |
1989.8-1993.9 | 650 [608-692] |
23.67 | 0.007 | 1926 [1884-1965] |
| C1 | 1123 [314-3284] |
2002.8-2005.8 | 131 [126-137] |
26.03 | 0.023 | 1993 [1988-1997] |
| C2 | 889 [297-2382] |
2004.7-2006.7 | 345 [326-364] |
16.57 | 0.019 | 1992 [1985-1997] |
These four clades accounted for ~2-3% each of the collection in 2017 and were estimated (adjusting for sampling) to have similar BSI incidence per million population of Norway in 2017 (Table 1, appendix p4,22). The ratio of each clade’s estimated population size (used as a proxy for the carriage population) to incidence of disease (BSI) in the human population was used as a measure of invasiveness relative to the other clades. For clades C1 and C2 we estimated smaller population sizes and higher population size/incidence ratios, whilst clade A had the largest population size and lowest ratio. The time to the most recent common ancestors of our Norwegian isolates in CC131, C1 and C2 pre-date the study period by over a decade whilst clade A and B are estimated to be older (Table 1).
Spatio-temporal analysis
There was no discernable spatio-temporal spread or phylogenetic structure within Norway during the expansion of CC131 (appendix p16,23). In 2002, clade A was already observed in blood cultures in the North and West health regions. C2 was the last clade to be observed in BSI in 2007. CC131 (p=0.031) and CTX-M positive isolates (p=0.032) were significantly underrepresented in the Central health region compared to the rest of the collection. However, ciprofloxacin resistance was underrepresented in the North (p=0.041) and over-represented in the South-East (p=0.015, appendix p24).
We additionally compared CC131 in our collection with CC131 isolates from another longitudinal collection, from the UK, representing isolates from 2002-2011 published by Kallonen et al 10. Compared to the UK, a significantly larger proportion (p<0.0001) of Norwegian CC131 isolates belonged to clade A (5.4% 8/147 in the UK vs 27.0% 75/278 in Norway) and clade C1 (8.2% 12/147 vs 25.2% 70/278, appendix p25). The opposite was true for clade C2 (60.5% 89/147 vs 18.3% 51/278, p<0.0001). As such, resistant isolates within these clades made different contributions to resistance between the two countries. C2 made a significantly smaller contribution (p<0.0001), and C1 and clade A significantly larger contributions to both CC131 CTX-M positive isolates (p<0.0001, p=0.0028 respectively), and CC131 isolates with gyrA and/or parC mutations (p<0.0001, p<0.0001 respectively) in Norway than in the UK (appendix p26-27). Finally, we contextualised our Norwegian isolates within a global genomic collection 14. Aside from the overrepresentation of Norwegian isolates in clade A and underrepresentation in C2, Norwegian isolates were phylogenetically spread across the sampled genetic diversity from other locations within each of the CC131 clades (appendix p17).
Discussion
Cases of E. coli BSI have been increasing in Norway, the UK and elsewhere 5,7. It is even more concerning that the proportion of BSI caused by MDR isolates is also increasing. ESBL prevalence in E. coli BSI in Norway was reported to be 0.3% in 2002 increasing to 6.6% by 2017 5. Fluoroquinolone resistance in Norway has also continued to increase in recent years from 3.3% in 2002 to 18% in E. coli causing BSI in 2017 5. We observed similar prevalence and general increasing trends 5. It was reported that there was a clear correlation between the total usage of fluoroquinolones and non-susceptibility to these agents in Norway where antimicrobial usage in both animals and people is generally low and tightly-regulated 5.
We have shown the emergence of CC131 as the dominant contributor to the prevalence of ESBLs and fluoroquinolone resistance in E. coli causing BSI in Norway. The emergence of CC131 in our data appears to have been a gradual process spanning 2002-2013 compared to that reported for the UK where the expansion occurred quickly between 2002-2004 10. We additionally show that the prevalence of the individual CC131 clades are different with a higher prevalence and resistance for clade A and C1 in Norway than the UK, and lower prevalence and resistance in C2. Despite these differences we detected the same predominant CC131 CTX-M types reported by others of CTX-M-15 in C2, and CTX-M-27 and CTX-M-14 in C1 10,31. Additionally the CC131 diversity we observed overlapped with other datasets, and our estimated tMRCA for CC131 clades are in line with previous estimates 11–13,31,32.
It is striking that CC131 clades have different AMR prevalence within Norway, but also different estimated population sizes, growth rates, and potentially invasiveness. In vivo quantification of clade virulence combined with genomics could reveal underlying variation driving any differences in invasiveness. Evidence of varying evolutionary pressures also warrants a further elaboration of the NFDS model for ExPEC population evolution 14. Whilst harboring less resistance than the MDR clade C2, increasing resistance facilitated by multiple de novo acquisition events in other clades of CC131 is clinically concerning. Clade A, which was the most frequently observed clade during the study period, and clade B, are far less resistant than C2 suggesting resistance is not critical for their success. Furthermore, we show resistance being acquired multiple times sporadically within clades A and C1 with expansion of the clades rather than preferential expansion of resistant isolates also suggests resistance is being selected for even in a low usage setting subsequent to their establishment. Our data underscores that a longitudinal framework is pivotal in obtaining a thorough epidemiological understanding of the population dynamics of E. coli. Furthermore, clones and clades can differ significantly between European countries that are resource similar. Our findings thus highlight the need for country-specific intensified surveillance efforts to circumvent the threat posed by the current evolutionary trajectory of E. coli causing BSI. The dominance of CC131, also offers the possibility of targeted diagnostics in relation to infection control to limit further spread of this high-risk MDR clone.
Limitations/sources of bias
NORM isolates represent a subset of total BSI cases in Norway included for surveillance of AMR however they are unbiased in terms of resistance profiles and are considered to be representative5. We further sampled the first 22.5% annually, but as E. coli BSI does not exhibit seasonality, they should remain representative. Comparing two periods 2002-2010 and 2011-2017 or trends over time limits any bias in an individual year and the general trends in prevalence observed here concur with the NORM reports 5. For the analysis using phenotypic antimicrobial susceptibility testing (AST) data, we used reported categorical AST data reported in NORM based on the breakpoints for each year. Reported AST data were acquired by multiple methods during the time-period (appendix p1). Thus, the breakpoints could not be calibrated over the time-period. We used an earlier characterisation of ESBL-producers in the NORM surveillance that showed that in 2017, 100% of ESBL-producing E. coli BSI were CTX-M positive 5 to justify using CTX-M genes as a proxy for ESBL producers, as such we did not include isolates in which only SHV and/or TEM were detected in the ESBL/CTX-M analysis. Finally, the published UK collection only represents isolates up until 2011 whereas this collection continues until 2017; it is possible that the importance of clade A in the latter years may also be occurring in other locations warranting further surveillance.
Supplementary Material
Research in context.
Evidence before this study
Following initial reports in the early 2000’s, numerous surveillance and epidemiological studies have reported Escherichia coli clonal complex (CC)131 as the dominant cause of E. coli urinary tract (UTI) and bloodstream infections (BSI) across the world. We used Pubmed searches for (coli + ST131 + urinary + infection; and coli + ST131 + blood + infection; searches limited to English language primary research articles). This returned a total of 957 publications (May 22nd 2020) describing the isolation and relative frequency of CC131 among UTI and BSI cases. Of these, only one is a designed longitudinal survey of isolates, performed by Kallonen et al. on a UK collection of BSI E. coli isolates from 2001-2011 from the British Society for Antimicrobial Chemotherapy (BSAC). By performing a densely sampled longitudinal study they were able to determine that whilst CC131 did indeed increase in frequency in the early 2000’s, this increase was not as marked as suggested in single-site, single-time point studies. The study also suggested that MDR alone did not explain the increase in CC131 prevalence, a finding supported by a subsequent evolutionary genomics study by McNally et al. in 2019. Thus, a number of important questions remain regarding the epidemiology of E. coli from BSI including whether the results of the UK study are representative of the wider geographic picture and what has happened to the E. coli population from BSI post 2011.
Added value of this study
The study used a longitudinal sampling framework over an extended period of 16 years which provides a clearer picture of the underlying epidemiology of E. coli from BSI. As it was performed in a different country, with lower antimicrobial usage and resistance to previous studies, there was the opportunity to compare to a UK study and determine that findings in one setting were not generalisable. These data also allowed for comparisons of the growth and expansion of clones that cause BSI beyond the raw prevalence numbers available at any one time.
Implications of all the available evidence
A full picture of the emergence of E. coli as a major causative agent of BSI is required, driven by longitudinal understanding of the epidemiology and MDR of these organisms. Finding CC131 clade A as an expanding and increasing cause of MDR BSI was a novel finding not reported in previous studies and merits further surveillance for this clone world-wide. This study shows that the findings of individual, longitudinal surveys of E. coli epidemiology are not generalisable across countries, not even resource similar nations within Europe. The observation of both emerging MDR clones and the acquisition of MDR in established clones, has implications for our efforts to control MDR in this pathogen.
Acknowledgements
We are grateful for technical assistance from Ellen Josefsen, Miriam Nilsen, Lennart Maximillian van Ligtenberg and all those that prepared and shipped isolates for the study.
Footnotes
Declaration of interests
N.J.C. reports grants from GlaxoSmithKline and personal fees from Antigen Discovery Inc. outside the submitted work, and J.P. reports grants from Wellcome Trust during the conduct of the study. The remaining authors report no conflicts of interest.
Contributors
Authors’ contribution statement
RG: Methodology, Data Curation, Investigation, Formal Analysis, Visualisation, Writing – Original Draft Preparation,
AM: Conceptualisation, Funding, Methodology, Writing – Original Draft Preparation,
AP: Writing – Review and Editing
GT, JAL: Writing – Review and Editing, Software
KS: Data Curation, Writing – Review and Editing
FC, MOKC, BCH: Writing – Review and Editing, Investigation
KKB, KWG, RH, AK, HEL, PCL, IHL, ÅM, EN, MTN: Resources, Writing – Review and Editing
GSS: Resources, Writing – Review and Editing, Data curation
MS, ST, MV: Resources, Writing – Review and Editing
SDB, NJC, JP: Writing – Review and Editing,
PJJ: Conceptualisation, Funding, Methodology, Writing – Review and Editing,
ØS: Conceptualisation, Funding, Methodology, Verified underlying data of the study, Writing – Review and Editing, Data curation
JC: Conceptualisation, Funding, Methodology, Verified underlying data of the study, Formal Analysis, Writing – Original Draft Preparation
Data Sharing
Fastq sequence data are available on the European Nucleotide Archive (ENA). Metadata for samples including ENA accession (ERR) are included in the supplementary metadata. Phylogenetic trees overlaid with metadata are hosted in Microreact and URLs listed in appendix.
References
- 1.Kern WV, Rieg S. Burden of bacterial bloodstream infection-a brief update on epidemiology and significance of multidrug-resistant pathogens. Clin Microbiol Infect. 2020;26:151–7. doi: 10.1016/j.cmi.2019.10.031. [DOI] [PubMed] [Google Scholar]
- 2.Day MJ, Doumith M, Abernethy J, et al. Population structure of Escherichia coli causing bacteraemia in the UK and Ireland between 2001 and 2010. J Antimicrob Chemother. 2016;71:2139–42. doi: 10.1093/jac/dkw145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Köhler C-D, Dobrindt U. What defines extraintestinal pathogenic Escherichia coli? Int J Med Microbiol. 2011;301:642–7. doi: 10.1016/j.ijmm.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Manges AR, Geum HM, Guo A, Edens TJ, Fibke CD, Pitout JDD. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin Microbiol Rev. 2019;32 doi: 10.1128/CMR.00135-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.NORM-VET reports. [accessed Dec 22, 2020]. https://www.vetinst.no/en/surveillance-programmes/norm-norm-vet-report .
- 6.Johnson JR, Porter S, Thuras P, Castanheira M. The Pandemic H30 Subclone of Sequence Type 131 (ST131) as the Leading Cause of Multidrug-Resistant Escherichia coli Infections in the United States (2011-2012) Open Forum Infect Dis. 2017;4:ofx089. doi: 10.1093/ofid/ofx089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerver R, Mihalkova M, Abernethy J, et al. Annual epidemiological commentary: mandatory MRSA, MSSA and E. coli bacteraemia and C. difficile infection data, 2014/15. Public Health England, London. 2015:81. [Google Scholar]
- 8.Temkin E, Fallach N, Almagor J, et al. Estimating the number of infections caused by antibiotic-resistant Escherichia coli and Klebsiella pneumoniae in 2014: a modelling study. Lancet Glob Health. 2018;6:e969–79. doi: 10.1016/S2214-109X(18)30278-X. [DOI] [PubMed] [Google Scholar]
- 9.Nicolas-Chanoine M-H, Bertrand X, Madec J-Y. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014;27:543–74. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kallonen T, Brodrick HJ, Harris SR, et al. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res. 2017 doi: 10.1101/gr.216606.116. published online July 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roer L, Tchesnokova V, Allesøe R, et al. Development of a Web Tool for Escherichia coli Subtyping Based on fimH Alleles. J Clin Microbiol. 2017;55:2538–43. doi: 10.1128/JCM.00737-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben Zakour NL, Alsheikh-Hussain AS, Ashcroft MM, et al. Sequential Acquisition of Virulence and Fluoroquinolone Resistance Has Shaped the Evolution of Escherichia coli ST131. MBio. 2016;7:e00347-16. doi: 10.1128/mBio.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price LB, Johnson JR, Aziz M, et al. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. MBio. 2013;4:e00377–13. doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNally A, Kallonen T, Connor C, et al. Diversification of Colonization Factors in a Multidrug-Resistant Escherichia coli Lineage Evolving under Negative Frequency-Dependent Selection. MBio. 2019;10 doi: 10.1128/mBio.00644-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Page AJ, De Silva N, Hunt M, et al. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb Genom. 2016;2:e000083. doi: 10.1099/mgen.0.000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu153. published online March. [DOI] [PubMed] [Google Scholar]
- 17.Tonkin-Hill G, MacAlasdair N, Ruis C, et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020;21:180. doi: 10.1186/s13059-020-02090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waters NR, Abram F, Brennan F, Holmes A, Pritchard L. Easy phylotyping of Escherichia coli via the EzClermont web app and command-line tool. Access Microbiol. 2020;2:acmi000143. doi: 10.1099/acmi.0.000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inouye M, Dashnow H, Raven L-A, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lees JA, Harris SR, Tonkin-Hill G, et al. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res. 2019;29:304–16. doi: 10.1101/gr.241455.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother. 2017;72:2764–8. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–4. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunt M, Mather AE, Sánchez-Busó L, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom. 2017;3:e000131. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huseby DL, Pietsch F, Brandis G, Garoff L, Tegehall A, Hughes D. Mutation Supply and Relative Fitness Shape the Genotypes of Ciprofloxacin-Resistant Escherichia coli. Mol Biol Evol. 2017;34:1029–39. doi: 10.1093/molbev/msx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Podnecky NL, Fredheim EGA, Kloos J, et al. Conserved collateral antibiotic susceptibility networks in diverse clinical strains of Escherichia coli. Nat Commun. 2018;9:3673. doi: 10.1038/s41467-018-06143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pietsch F, Bergman JM, Brandis G, et al. Ciprofloxacin selects for RNA polymerase mutations with pleiotropic antibiotic resistance effects. J Antimicrob Chemother. 2017;72:75–84. doi: 10.1093/jac/dkw364. [DOI] [PubMed] [Google Scholar]
- 27.Croucher NJ, Page AJ, Connor TR, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 29.Harris SR. SKA: Split Kmer Analysis Toolkit for Bacterial Genomic Epidemiology. bioRxiv. 2018:453142 [Google Scholar]
- 30.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Decano AG, Downing T. An Escherichia coli ST131 pangenome atlas reveals population structure and evolution across 4,071 isolates. Sci Rep. 2019;9:17394. doi: 10.1038/s41598-019-54004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoesser N, Sheppard AE, Pankhurst L, et al. Evolutionary History of the Global Emergence of the Escherichia coli Epidemic Clone ST131. MBio. 2016;7:e02162. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fastq sequence data are available on the European Nucleotide Archive (ENA). Metadata for samples including ENA accession (ERR) are included in the supplementary metadata. Phylogenetic trees overlaid with metadata are hosted in Microreact and URLs listed in appendix.