Abstract
GATA3 gene silencing in activated T cells displays a promising option to early-on undermine pathological pathways in the disease formation of allergic asthma. The central transcription factor of T helper 2 (Th2) cell cytokines IL-4, IL-5, and IL-13 plays a major role in immune and inflammatory cascades underlying asthmatic processes in the airways. Pulmonary delivery of small interfering RNAs (siRNA) to induce GATA3 knockdown within disease related T cells of asthmatic lungs via RNA interference (RNAi) presents an auspicious base to realize this strategy, however, still faces some major hurdles. Main obstacles for successful siRNA delivery in general comprise stability and targeting issues, while in addition the transfection of T cells presents a particularly challenging task itself. In previous studies, we have developed and advanced an eligible siRNA delivery system composed of polyethylenimine (PEI) as polycationic carrier, transferrin (Tf) as targeting ligand and melittin (Mel) as endosomolytic agent. Resulting Tf-Mel-PEI polyplexes exhibited ideal characteristics for targeted siRNA delivery to activated T cells and achieved efficient and sequence-specific gene knockdown in vitro. In this work, the therapeutic potential of this carrier system was evaluated in an optimized cellular model displaying the activated status of asthmatic T cells. Moreover, a suitable siRNA sequence combination was found for effective gene silencing of GATA3. To confirm the translatability of our findings, Tf-Mel-PEI polyplexes were additionally tested ex vivoin activated human precision-cut lung slices (PCLS). Here, the formulation showed a safe profile as well as successful delivery to the lung epithelium with 88% GATA3 silencing in lung explants. These findings support the feasibility of Tf-Mel-PEI as siRNA delivery system for targeted gene knockdown in activated T cells as a potential novel therapy for allergic asthma.
Keywords: siRNA therapy, pulmonary delivery, asthma, T cell targeting, cytokines, PCLS
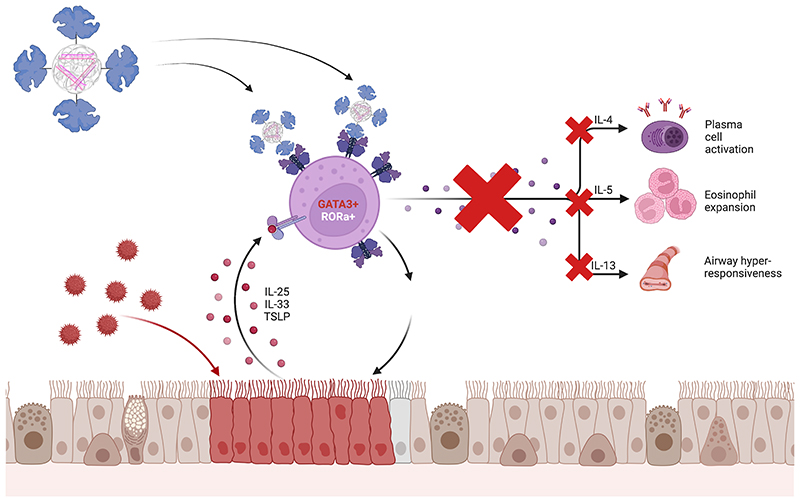
Graphical Abstract.
1. Introduction
Asthma is a major public health problem affecting 339 million people in all regions of the world and accounting for 1000 deaths per day. [1] The disease not only causes a high disability and death burden, but also raises tremendous socioeconomic issues. Although today in the majority of patients asthma symptoms can be controlled using standard therapies mainly based on corticosteroids as controller and beta-2-sympathomimetics as reliever medications, in a distinct portion of patients (5-10 %) symptoms can still not adequately be commanded.[2] This is particularly dangerous, as these patients experience worse asthma symptoms, more concomitant comorbidities and, moreover, a specifically high risk of mortality. It is therefore inevitable to find novel more precise treatment options for this group of patients. Asthma is a reversible disease of the airways characterized by inflammation resulting in bronchoconstriction, enhanced mucus production, hyperresponsiveness and ultimately remodeling of the airways. [3] The fact that current therapies only show limited benefits in certain subgroups of patients may be due to the heterogenicity of the disease resulting in several different phenotypes with distinct clinical, functional and pathological patterns. [4] The underlying pathological base is distinguished by different patterns of cytokine-based airway inflammation involving immune and inflammatory cell types such as T and B lymphocytes. These cytokines therefore display promising targets of novel therapies tailored for patients suffering from severe asthma who do not fully respond to convenient treatments. [5]
Allergic asthma plays a major role in this context, comprising all levels of disease severity and being involved in the disease pattern of 50-80 % of all severe asthma patients. [6] This asthma phenotype class is supposed to be actuated by an immune-inflammatory response driven by type 2 T helper (Th2) cells orchestrating a complex interplay between innate and adaptive immune system. [7] Prevalently, these processes already start in the childhood via sensitization to common inhaled allergens such as house dust mites or pollens. These allergens are taken up by dendritic cells (DCs) processing and presenting antigenic molecules to naïve T helper cells. Consequently, allergen-specific Th2 cells are activated and produce and secrete the major Th2 cytokines interleukin (IL)-4, IL-5, and IL-13, playing an essential role in the development of asthmatic inflammation. [8] Besides interfering with these single cytokines individually, silencing of their common transcription factor GATA3 [9] provides the option to catch all of them at once and therein early-on prevent the inflammatory cascade from commencing. However, transfection of T cells displays an especially challenging endeavor, since caveolae-mediated uptake, the commonly used strategy for non-viral vector-based transfection methods, cannot be applied to the non-caveolin expressing cells. [10]
RNA interference (RNAi) offers an auspicious base to even treat diseases that have so far thought to be “undruggable”, enabling the potential targeting of any chosen gene with an established sequence. [11] Small interfering RNA (siRNA) have therefore been subject to extensive research leading to the approval of currently four siRNA-products on the market. [12, 13] The main drawbacks, however, that are still holding back the potential of these novel treatment forms are the poor pharmacological properties of nucleic acids, which are easily degraded by nucleases ubiquitous in the bloodstream and rapidly excreted upon systemic injection. [14] In recent years, however, key improvements have been made in stable packaging of siRNA within nanoparticles mostly composed of polymers and/or lipid formulations. A further way to improve siRNA delivery and circumvent i.v. stability concerns, thereby also bypassing first pass metabolism, is to use local administration routes. In terms of asthma, pulmonary delivery obviously offers a suitable approach to effectively target disease related cells in asthmatic lungs. [15, 16]
In our previous work, we have developed and optimized an siRNA carrier system on the base of polyethylenimine (PEI), [17] the most extensively studied polymer for gene delivery. [18] The reason behind choosing PEI as delivery system for RNA is that formulations based on polyplexes can be easily processed or nebulized for direct administration to the lungs. Lipid-based delivery systems are instead more prone to instability problems once nebulized for inhalation. [19] The polycationic carrier was coupled to transferrin, a glycoprotein responsible for iron transport in biological fluids. Its receptor is therefore overexpressed on highly proliferating cell types such as activated T cells, [20] providing both an entry gate via receptor mediated endocytosis and the potential to specifically target these cells. Since naïve T cells only show a negligible expression of the transferrin receptor (TfR), they are not considerably transfected and a general immune suppression by the treatment can be avoided. We confirmed this by investigating the cellular distribution of Tf-PEI polyplexes in vivo in a murine model of asthma, where T cells were the cellular population showing the highest cellular uptake. Furthermore, T cell uptake was significantly higher for OVA-stimulated mice in comparison to the unstimulated group, confirming the transferrin-mediated targeted delivery of polyplexes to activated T cells.[17] In a follow-up study, the Tf-PEI polyplexes were then further advanced by blending them with melittin, a cationic peptide with lysosomal properties. Resulting in the Tf-Mel-PEI blend, these polyplexes were now able to efficiently be released from the endosomes after being taken up so that an optimized in vivo effect of the incorporated siRNA is expected. The optimized formulation was previously characterized in terms of size, PDI, zeta-potential. The results underlined optimal characteristics for pulmonary delivery, with sizes around 100 nm, low PDI and a slightly positive zeta-potential for Tf-Mel-PEI. Furthermore, a stability study in presence of mucus and lung surfactant suggested desirable stability of the formulation even in complex environments. [21]
The aim of this work was therefore to evaluate the therapeutic potential of these Tf-Mel-PEI/siRNA polyplexes in the context of targeted GATA3 knockdown in activated T cells in the lung for early interference with the inflammatory cascade in asthmatic patients. For this purpose, primary as well as continuous T cells were used to generate and optimize an appropriate model for the activated T cell status in inflamed lungs. A suitable GATA3 sequence combination was found for distinct gene silencing and downstream effects of this knockdown were investigated. To better understand the behavior of the formulation in a more relevant setup, an ex vivo model of inflammation was established. Human PCLS, which are living tissues from lung explants that retain the anatomical as well as cellular structure of the lung, were pre-treated in order to obtain activated T cells. Activated PCLS were then used to investigated the safety of the formulation as well as the delivery and the gene silencing activity.
Overall, the results gathered in this study give reason to assume that Tf-Mel-PEI polyplexes present a feasible approach to pointedly deliver siRNA for specific gene knockdown in disease-related T cells as a novel therapy for allergic asthma.
2. Experimental Section
Synthesis of Conjugates and Preparation of Polyplexes
Tf-PEI and Mel-PEI conjugates were prepared as previously described. [17] Briefly, Tf was coupled to 5 k PEI (Lupasol® G100, BASF, Ludwigshafen, Germany) via succinimidyl 3-(2-pyridyldithio) propionate (SPDP, Thermo Fisher Scientific, Waltham, USA) to yield the Tf-PEI conjugate with a reducible disulfide bond. Similarly, cysteine modified melittin (Mel, Pepmic, Suzhou, China) was coupled to PEI via 4-polyethylene glycol-N-succinimidyl 3-(2-pyridyldithio) propionate (PEG4-SPDP, Thermo Fisher Scientific, Waltham, USA) after masking of its lytic amine groups with 2,3-Dimethyl-maleic anhydride solution (DMMAn, Sigma-Aldrich, St. Louis, USA) to yield the pH-responsive Mel-PEI conjugate as described before.[21]
For Tf-Mel-PEI blend polyplexes, Tf-PEI and Mel-PEI were mixed in a 1:1 ratio and diluted in 5 % glucose. Defined amounts of siRNA were added to obtain specific amine to phosphate (N/P) ratios according to the following equation for calculating the mass of polymer required for 50 pmol siRNA: m (PEI in pg) = 50 pmol x 43.1 g/mol x N/P x 52 (protonable unit of PEI = 43.1 g/mol, number of nucleotides of 25/27mer siRNA = 52). Solutions were mixed by pipetting and incubated for 20 min at room temperature before proceeding with respective experiments.
Cell Culture
Jurkat cells, a human T lymphocyte cell line, were a kind gift from Prof. Dr. Heissmeyer (Institute for Immunology, Biomedical Center Munich). They were cultured in RPMI-1640 cell culture medium (Sigma-Aldrich) that was supplemented with 10 mM HEPES, 1 mM sodium pyruvate, 4500 mg/l glucose, 10 % (v/v) heat inactivated fetal bovine serum (FBS, Sigma-Aldrich) and 1 x penicillin/streptomycin (Pen/Strep, Sigma-Aldrich). Human and murine primary T cells were cultured in the same medium. MCF-7 cells were cultured in EMEM cell culture medium (Sigma-Aldrich) supplemented with 2 mM L-Glutamine, 1 % non-essential amino acids (NEAA), 10 % (v/v) heat inactivated fetal bovine serum (FBS) and 1 x penicillin/streptomycin (Pen/Strep, all supplements were obtained from Sigma-Aldrich). All cells were grown at a humidified atmosphere with 5 % CO2 at 37 °C.
T Cell Isolation and Activation
Primary human CD4+ T cells were isolated from freshly obtained buffy coats (DRK, Berlin, Germany). The isolation process was conducted via magnetic bead separation as described before. [22] CD4+ T cells were cultured in RPMI-1640 medium as described above. For activation with soluble antibodies, wells of a 96-well culture plate or 48-well culture plate (Thermo Fisher Scientific) were coated with anti-CD3 antibody (BD Biosciences, Franklin Lake, USA) at a concentration of 5 μg/ml in PBS and incubated for 3 h at 37 °C or overnight at 4 °C. After washing the wells 3 times with rising amounts of PBS, 100.000 (for TfR1 expression) or 500.000 (for gene expression) Jurkat cells or primary T cells, respectively, were added per well in fresh medium. Anti-CD28 antibody (BD Biosciences) was added at a final concentration of 1 μg/ml. For activation via beads, Dynabeads™ Human T-Activator CD3/CD28 for T Cell Expansion and Activation (Thermo Fisher Scientific) were washed and added to the cells in a bead:cell ratio of 1:1 according to the manufacturer’s protocol. Cells were activated for 24, 48, or 72 h at a humidified atmosphere with 5 % CO2 at 37 °C before further analysis.
Transferrin Receptor (TfR1) Expression
Jurkat cells and primary CD4+ T cells from the activation procedure were collected at different days post activation, washed with PBS and resuspended in 100 μl of PBS. Per sample, 10 μl of 1:10 diluted FcR binding inhibitor (eBioscience, Frankfurt, Germany) in PBS was added, mixed and incubated for 5 min at 4 °C. Subsequently, 1 μl of undiluted anti-CD71 antibody (eBioscience) was added to appropriate samples, while isotype controls were stained with IgG1 antibody (eBioscience) and blank samples were left unstained. All samples were vortexed and incubated for another 30 min at 4 °C protected from light. After washing with PBS 3 times, cells were resuspended in 400 μl PBS/2 mM EDTA and sample were analyzed using an Attune ® NxT flow cytometer (Thermo Fisher Scientific) with 488 excitation and 574/26 emission filter. The cells of all samples were gated according to morphology based on forward/sideward scattering, and 10.000 events were evaluated per sample.
GATA3 Expression
Jurkat cells and primary CD4+ T cells from the activation procedure were collected at different days post activation and total RNA was isolated with the PureLink RNA mini kit (Thermo Fisher Scientific) for Jurkat cells or the RNeasy micro kit (QIAGEN), respectively, according to the manufacturer’s protocol with additional DNAse I digestion. From the obtained RNA, cDNA was synthesized and amplified with the Brilliant III ultra-fast SYBR® green QRT-PCR master mix kit (Agilent Technologies, Santa Clara, USA), custom-synthesized GATA3 primers (Thermo Fisher Scientific) and QuantiTect® primer assay Hs_ACTB_2_SG (Qiagen, Venlo, Netherlands) on a qTOWER real-time PCR thermal cycler (Analytik Jena, Jena, Germany). Cycle threshold (Ct) values were calculated using the qPCRsoft software (Analytik Jena) and a 1:5 serial dilution of an untreated blank sample was prepared as a standard curve. Ct values were plotted against the assigned concentration of each point in the curve (1, 0.2, 0.004, 0.0008, and 0.00016). For each sample, gene expression of GATA3 was normalized by corresponding β-Actin expressionapplying theΔΔCt method.
ELISA
Primary CD4+ T cells from the activation procedure were harvested at different days post activation. Cells were centrifuged for 5 min at 350 x g and supernatants of the samples were collected and frozen at -80 °C. Interleukin concentrations were quantified with human ELISA kits for IL-4, IL-5, and IL-13 (Invitrogen, Carlsbad, USA), respectively, according to the manufacturer’s protocol. Absorbance was recorded at a wavelength of 450 nm using a FLUOstar Omega (BMG Labtech).
GATA3 Sequence Optimization and GATA3 Gene Knockdown
To find a suitable human siRNA sequence for efficient GATA3 knockdown, different sequences, namely Hs_GATA3_1, Hs_GATA3_7, Hs_GATA3_8, and Hs_GATA3_9 (QIAGEN) were screened. Therefore, 100.000 MCF-7 cells per sample were seeded in a 24-well plate (Thermo Fisher Scientific), transfected with lipoplexes containing 25 pmol siRNA either directed against GATA3 (siGATA3_1, siGATA3_7, siGATA3_8, siGATA3_9) or scrambled negative control siRNA (siNC), respectively, and incubated for 24 h. Cells were harvested and GATA3 gene expression was quantified as described above. In addition, an analogous experiment was performed with lipoplexes containing 25 pmol of a 1:1 mixture of sequences Hs_GATA3_8 and Hs_GATA3_9.
For GATA3 Gene Silencing experiments, 500.000 Jurkat cells per sample were seeded in a 96-well plate or 1x106 primary T cells were seeded in a 48-well plate (Thermo Fisher Scientific) and activated for 48 h as described above. Cells were transfected with polyplexes containing 25 pmol siRNA either directed against GATA3 (siGATA3_8+9, 1:1 mixture of sequences siGATA3_8 and siGATA3_9) or scrambled negative control siRNA (siNC), respectively, at an N/P ratio of 10. After 24 h of incubation, cells were harvested and GATA3 gene expression was quantified as described above.
Human donors and ethics statement
Human lung lobes were acquired from patients who underwent cancer lobe resection at Hannover Medical School (MHH, Hannover, Germany) or KRH Klinikum Siloah-Oststadt-Heidehaus (Hannover, Germany). Experiments were approved by the ethics committee of the Hannover Medical School (MHH, Hannover, Germany) and are in compliance with “The Code of Ethics of the World Medical Association” (renewed on 2015/04/22, number 2701–2015). All patients gave written informed consent for the use of their lung tissue for research.
Preparation of hPCLS
Human PCLS were prepared as previously described.[23] Briefly, a 1.5% agarose-DMEM solution was gently inflated into the lung explant and solidified at 4°C. Sections of 8 mm in diameter were sliced into approximately 250-300 μm slices using a Krumdieck tissue slicer. PCLS were cultured in DMEM medium (2 slices per 500 μl) under submerged conditions in a humidified atmosphere at 37 °C and 5% CO2.
Activation of hPCLS (Stimulation of PCLS for cytokine release)
Two PCLS per well were treated in duplicates with anti-CD3/CD28 Dynabeads™ (Human T-Activator CD3/CD28 for T Cell Expansion and Activation, Thermo Fisher Scientific), to activate tissue resident T cells at a bead:cells ratio of 1:1 in 500 μl DMEM medium for 48 h under standard cell culture conditions. Once the incubation time was completed, the supernatant was collected and stored for further analysis. Changes inT cell cytokineswere measured in PCLS supernatants using multiplex assays (Pro-Inflammatory Panel 1 Human V plex (IFN-γ, IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12p70, IL-13, TNF-α; Meso Scale Diagnostics) according to the manufacturer’s instructions. Graphs depicted in boxplots were created using GraphPad Prism 9 software. Negative controls consisted of unstimulated samples.
Cell viability and lactate dehydrogenase (LDH) release
Viability of the formulation following activation as well as incubation with Tf-PEI and Tf-Mel-PEI polyplexes containing 50 pmol siRNA was assessed via the metabolic WST-1 assay according to the manufacturer’s protocol and as described before.[23] After 48 h of stimulation with beads and 24 h of polyplex treatment, PCLS were incubated with 1:10 diluted WST-1 solution for 1 h at 37°C. Absorbance was then measured at 420-480 nm with a reference wavelength at 690 nm using a plate reader (Tecan, Männedorf, Switzerland). The effect of bead stimulation as well as polyplexes transfection on membrane integrity of PCLS and primary CD4+ T cells cells was evaluated by measuring the release of lactate dehydrogenase (LDH) using the Cytotoxicity Detection Kit (Th. Geyer, Renningen, Germany) according to the manufacturer’s protocol.For tissue toxicity studies, PCLS were stimulated for 5 days with beads in 24-well plates and incubated for 24 h with Tf-PEI and Tf-Mel-PEI polyplexes containing 50 pmol siRNA. For toxicity studies in primary CD4+ T cells 100,000 cells were seeded in a 96-well plate and incubated with Tf-PEI and Tf-Mel-PEI polyplexes for 24h. In brief, 50 μl of the supernatants were collected and incubated with 50 μl of the substrate mix for 20 min at room temperature protected from light. Absorbance was measured at 490 nm with a reference wavelength at 690 nm using a microplate reader (Tecan, Männedorf, Switzerland).
Microscopic assessment of siRNA delivery in hPCLS
PCLS were fixed overnight in 2% PFA and subsequently washed with PBS. DAPI staining was performed for 45 minutes, and the PCLS were embedded in ibidi mounting medium on two thin coverslips. PCLS were treated with 50 pmol of AF-647 labeled siRNA and fluorescence was detected in the Cy5 channel while autofluoresence of the tissue was recorded in the Cy3 channel. Z stacks of the tissue slide were imaged with a confocal microscope (Zeiss 880) and reconstructed in three dimensionsusing IMARIS® Software (Bitplane).
GATA3 gene silencing in hPCLS
Two PCLS/well were stimulated with anti-CD3/CD28 coupled magnetic beads in DMEM/F-12 to activate tissue resident T cells. PCLS were placed in 24-well-plates in 500 μl DMEM F-12 medium supplemented with 1 % penicillin/streptomycin and 0.1 % FBSand treated for 5 days with anti-CD3/CD28 coupled magnetic beads to activate tissue resident T-cells. Afterwards, PCLS were transfected with Tf-PEI and Tf-Mel-PEI polyplexes encapsulating 50 and 100 pmol siRNA against GATA3 or scrambled negative control. The tissue slices were incubated for 48 hours at 37°C and 5% CO2 in technical biological duplicates. Experiments were conducted with tissue from one human donor. RNA was isolated 48 hours post treatment according to an optimized protocol for PCLS.[24] Supernatants were frozen at -80°C, and cytokine level were later determined by LEGENDplex™ HU Th Cytokine Panel (Biolegend, USA). cDNA was synthesized from total RNA with a high-capacity cDNA synthesis kit (Applied Biosystems, Waltham, Massachusetts, USA). cDNA was then diluted 1:10 and qPCR was performed using the SYBR™ Green PCR master mix (Thermo Fisher Scientific) with primers for GATA3 (Qiagen, Hilden, Germany) and β-actin (Qiagen, Hilden, Germany) for normalization applying theΔΔCt method. For calibration to negative control, ΔΔCt values of samples treated with Tf-PEI and Tf-Mel-PEI polyplexes containing either 50 pmol or 100 pmolGATA3 siRNA were set in relation to the average ΔΔCt values of respective negative control samples treated with Tf-PEI and Tf-Mel-PEI containing either 50 pmol or 100 pmol negative control siRNA.Amplification and data analysis was performed using a QuantStudio 3 Real-Time PCR (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Cycle thresholds were acquired by autosetting with the qPCR software (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Values are given as mean values ± SEM.
Statistical Analysis
All experiments were performed in triplicates. All results are presented as mean value ± standard deviation (SD). One-way ANOVA and two-way ANOVA with Bonferroni posthoc post-test were conducted in GraphPad Prism (GraphPad Software, La Jolla, USA) to calculate p-values with 95 % confidence.
3. Results
3.1. Optimization of T Cell Activation
To optimize the T cell activation process, two different activation strategies were tested: soluble antibodies directed against CD3 and CD28 as well as so-called Dynabeads®, beads covalently coupled to anti-CD3 and anti-CD28 antibodies.Dynabeads® have a size of 4.5 μm to resemble the size of antigen presenting cells. They are chemically inert and supermagnetic, allowing them to be easily removed from culture medium with a magnet. Both strategies were applied to Jurkat cells, a human T lymphocyte cell line, as well as primary CD4+ T cells freshly isolated from buffy coats via magnetic bead separation. As parameters depicting the activation status of the cells, upregulation of TfR, gene expression of GATA3, and secretion of interleukins IL-4, IL-5, and IL-13, were chosen and analyzed.
Supplementary Figure 1 illustrates TfR(equivalent to CD71) expression in Jurkat cells (Figure S1A) and primary CD4+ T cells (Figure S1B) as measured at different time points post activation. As 100 % of Jurkat cells already express TfR constitutively, this number wasnot enhanced any further, but was steadily maintained for 3 days. The corresponding median fluorescence intensity (MFI) of CD71-PE labeled cells was, however, increased via bead activation at day 1, while treatment with soluble antibodies resulted in a consistent decrease of receptor expression. In contrast, naïve primary T cells only show a negligible TfR expression without activation stimuli and were therefore successfully activated, eventuating in a distinct increase in both CD71 positive cells and MFI of CD71-PE. Dynabead activation outperformed soluble antibodies, resulting in 45 % positive cells already after 24 h of activation and maintained for an additional 2 days, reflected in an increase of MFI from 1500 up to approximately 3800.
In Supplementary Figure 2, GATA3 expression of Jurkat cells (Figure S2A) and primary CD4+ T cells (Figure S2B) normalized to ß-Actin is depicted after the different activation procedures as quantified by qRT-PCR. In both cell types, GATA3 expression was upregulated via CD3/CD28 based activation compared to untreated blank cells. In Jurkat cells, beads and soluble antibodies worked almost comparably well, while in the primary T cells, the bead activation was again more effective, resulting in a twice as high expression after three days of activation compared to soluble antibodies and a three times higher expression compared to non-activated cells resulting in statistically significant upregulation after three days.
As a third activation parameter, the secretion of GATA3 related interleukins IL-4, 5, and 13, was chosen to be analyzed. Results of sandwich ELISAs from primary CD4+ T cells are shown in Supplementary Figure 3 after 2 days of activation with beads (Figure S3A) or soluble antibodies (Figure S3B) compared to non-stimulated naïve T cells. In general, all 3 interleukins were only secreted at a non-detectable level without activation. In marked contrast to this, secretion of all tested cytokines could considerably be upregulated by both activation processes, with IL-13 showing the strongest increase in each case. The bead based activation enhanced IL-13 concentration from about 20 to 200 pg/ml, composing a 10 fold increase. In general, trends achieved with beads and soluble antibodies are somehow comparable in activation effectiveness, while larger standard deviations for soluble antibodies suggest a more steady and reproducible procedure regarding bead activation resulting in statistically significant events.Interleukin measurements in activated Jurkat cells revealed irregular results (data not shown).
3.2. Optimization of GATA3 siRNA sequences
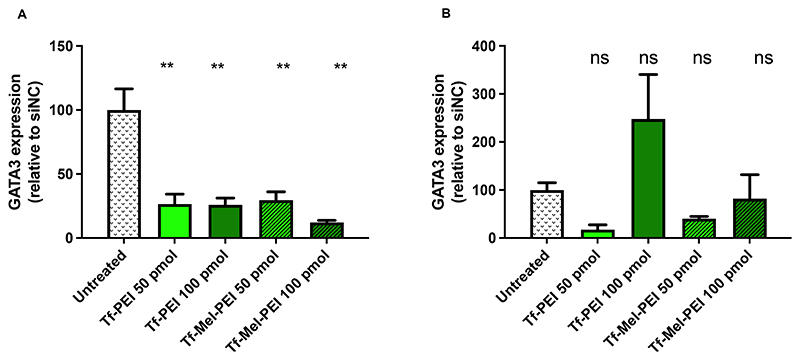
For optimization of siRNA sequences to downregulate GATA3, several different sequences were screened in preliminary experiments using lipofectamine as transfection reagent and MCF-7 cells as an easy-to-transfect model cell line. Supplementary Figure 4A depicts GATA3 expression 24 h after transfection with the four most promisingdifferent GATA3 sequences compared to untreated blank cells as well as cells treated with a scrambled negative control sequence. While sequences GATA3_1 and GATA3_7 did not result in a considerable knockdown effect, the two sequences GATA3_8 and GATA3_9 were found to induce significant gene silencing compared to the negative control sequence NC-siRNA.
Consequently, these two sequences were combined in a 1:1 mix and applied together, resulting in an even more distinct knockdown effect, showing significance compared to both the negative controls and the untreated blank samples (Figure S4B).
3.3. GATA3 Gene Silencing
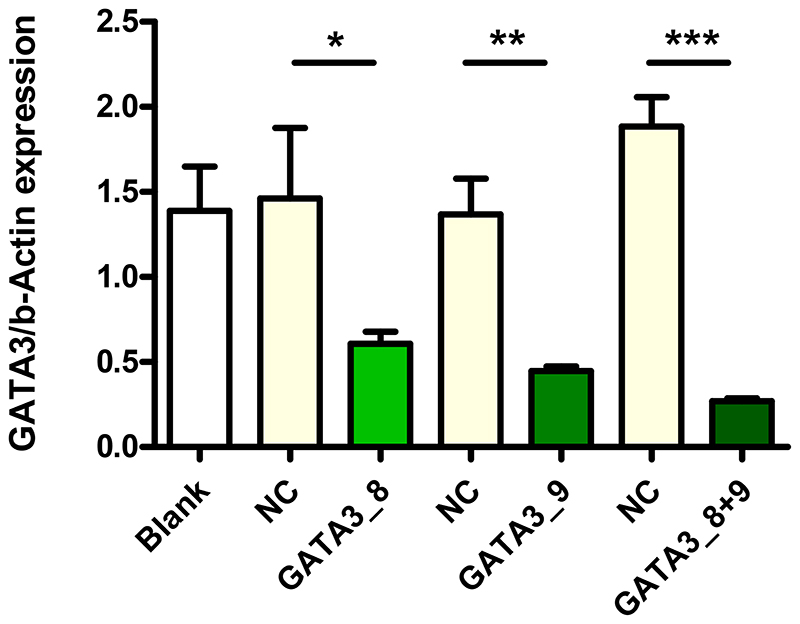
GATA3 gene silencing of the two sequences GATA3_8 and GATA3_9 within Tf-Mel-PEI polyplexes was confirmed in Jurkat cells. Figure 1 illustrates GATA3 expression normalized to ß-Actin 24 h after treatment with Tf-Mel-PEI polyplexes containing either only sequence GATA3_8 or GATA3_9 or a 1:1 mixture of both sequences, always formulated at an N/P ratio of 10. For comparison, again an untreated blank sample as well as the scrambled NC-siRNA were used. Here, both GATA3 sequences alone already resulted in a significant gene knockdown, while the 1:1 combination could even achieve an enhanced silencing effect. This mixture was consequently chosen to be used for further knockdown experiments.
Figure 1.
GATA3 knockdown in Jurkat cells transfected with Tf-Mel-PEI polyplexes containing 25 pmol of different GATA3 siRNA sequences at N/P 10 as measured by qRT-PCR and normalized against ß-Act in expression. Control cells were left untreated (blank) or transfected with respective polyplexes containing scrambled siRNA as negative control (NC). (Data points indicate mean ± SD, n = 3; One-way ANOVA, *, p < 0.05; **, p < 0.01; ***, p < 0.005).
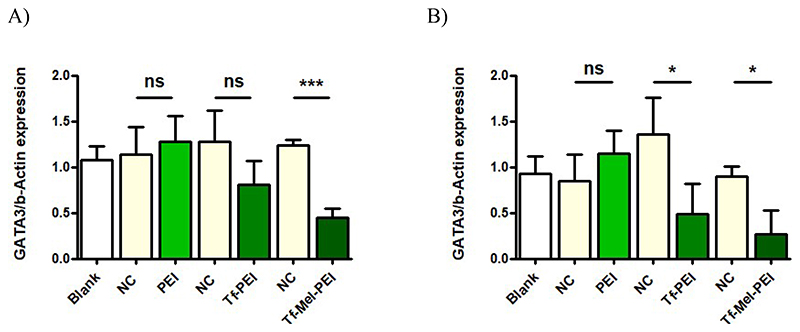
The optimized GATA3 sequences were then evaluated for gene knockdown in both Jurkat cells and primary CD4+ T cells within polyplexes prepared with unmodified PEI, the original Tf-PEI conjugate, as well as the Tf-Mel-PEI blend conjugate. Figure 2shows GATA3 expression normalized to ß-Actin after treatment with polyplexes at N/P 10 for 24 h. Neither in Jurkat cells (Figure 2A) nor in primary T cells (Figure 2B), PEI treatment could achieve an efficient gene silencing effect. While Tf-PEI polyplexes did result in a visible downregulation of GATA3 in Jurkat cells, this effect was, however, revealed to be not significant compared to treatment with scrambled negative control siRNA. In primary T cells, on the contrary, Tf-PEI in fact induced a significant gene knockdown. The Tf-Mel-PEI blend polyplexes were found to be the only treatment able to significantly and specifically silence GATA3 in both cell types, resulting in a knockdown efficiency of approximately 65 % in Jurkat cells and 70 % in human CD4+ primary T cells, compared to negative controls, respectively.
Figure 2.
GATA3 knockdown in A) Jurkat cells and B) human primary CD4+ T cells after transfection with 25 pmol of a 1:1 mixture of siRNA sequences GATA3_8 and GATA3_9 encapsulated within PEI, Tf-PEI, or Tf-Mel-PEI, respectively. Control cells were left untreated (blank) or transfected with respective polyplexes containing scrambled siRNA as negative control (NC). (Data points indicate mean ± SD, n = 3; One-way ANOVA, *, p < 0.05; **, p < 0.01; ***, p < 0.005).
3.4. Evaluation of Downstream Effects
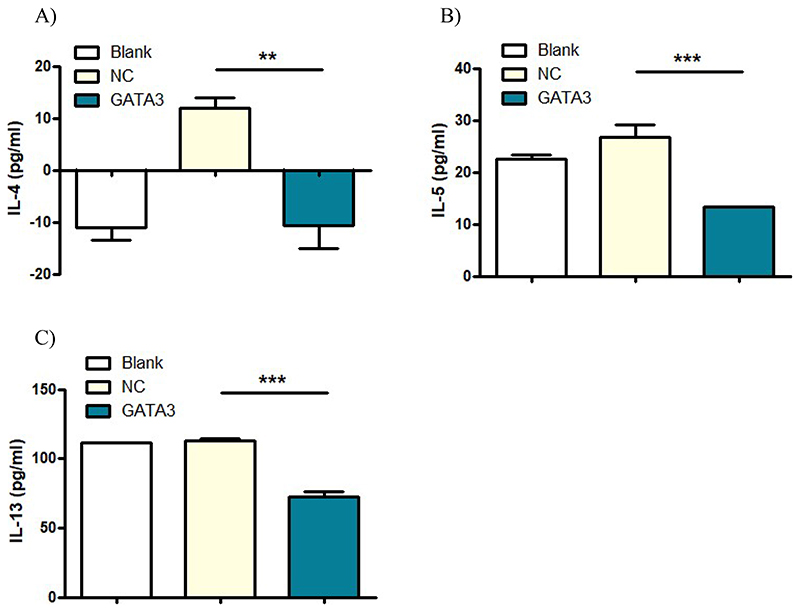
Downstream effects of GATA3 gene silencing on respective cytokine secretion of primary T cells was evaluated by ELISA. Concentrations of the three interleukins IL-4, IL-5, and IL-13 in the supernatant of treated cells were measured after 48 h of activation and subsequent transfection with Tf-Mel-PEI polyplexes containing either scrambled control NC-siRNA or siRNA directed against GATA3 (GATA3_8 and GATA3_9 in a 1:1 mixture) at N/P ratio 10. Figure 3 summarizes the results of respective sandwich ELISAs. Secretion of all three interleukins could significantly be downregulated via GATA3 targeted treatment compared to negative controls, indicating that GATA3 knockdown can indeed affect appropriate downstream signaling. The best results could be obtained for IL-5 and IL-13, were the GATA3 knockdown resulted in a consequent decrease of interleukin production of 50 % and 35 %, respectively, compared to negative controls. For IL-4, inconsistent and partly negative values were generated, nevertheless, a decrease in cytokine concentration could be observed for GATA3 treatment.
Figure 3.
Cytokine concentrations of A) IL-4, B) IL-5, and C) IL-13 in the supernatant of human primary CD4+ T cells after 48 h of activation and subsequent transfection with Tf-Mel-PEI polyplexes containing 25 pmol of a 1:1 mixture of siRNA sequences GATA3_8 and GATA3_9. Control cells were left untreated (blank) or transfected with respective polyplexes containing scrambled siRNA as negative control (NC). (Data points indicate mean ± SD, n = 3; One-way ANOVA, *, p < 0.05; **, p < 0.01; ***, p < 0.005).
3.5. T cell activation in hPCLS
Based on the T cell activation protocol optimized above for isolated primary T cells, tissueresident T cell activation was investigated in hPCLS after 3 days and 5 days of incubation with Dynabeads®. As shown in Supplementary Figure 5, each condition was compared to the respective non-stimulated PCLS. The incubation with Dynabeads® successfully induced an inflammatory state in PCLS. Cytokines typical of general inflammation (IL-12p70, IL-8, IL-6 and IL1b) as well as mediated by Th1 response (TNF-α, IFN-γ and IL-2) and Th2 cells (IL-4, IL-13 and IL-10) were upregulated following the activation process. The observation of activation and inflammation was confirmed by the upregulated expression of GATA3 in PCLS after 5 days of incubation with beads (Supplementary Figure 6). The expression of GATA3 was measured by qRT-PCR in PCLS obtained from three different patients.
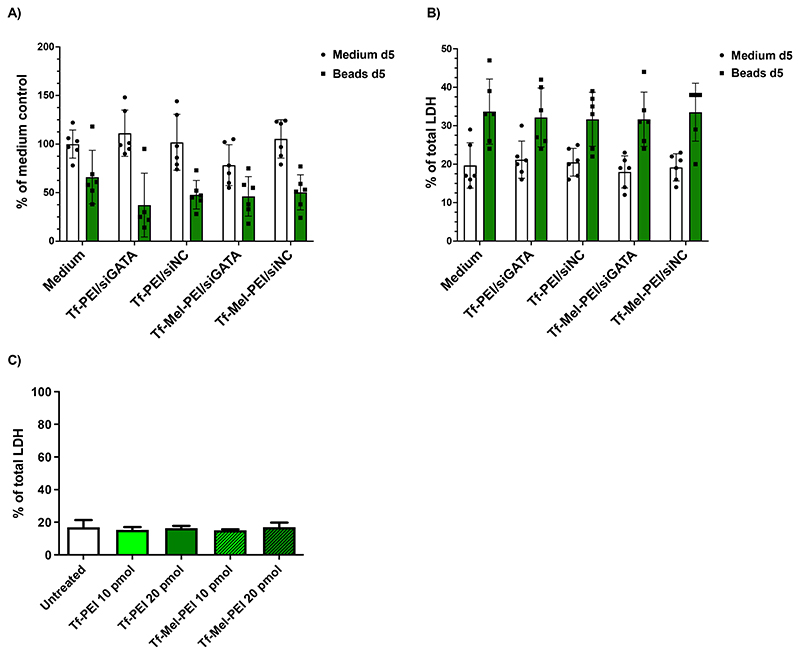
3.6. Effect of activation and transfection on metabolic activity and LDH release
The effects of incubation with Dynabeads™ as well as with polyplexes on metabolic activity was evaluated by WST-1 assays. Activated or non-activated PCLS were incubated for 5 days with Tf-PEI or Tf-Mel-PEI polyplexes encapsulating siGATA3 or a siNC negative control to observe if any of the different factors influenced cell viability. As it can be observed in Figure 4A, the treatment with Dynabeads® negatively affected metabolic activity, with a decrease of 40 to 50%. Nevertheless, the treatment with the Tf-PEI and Tf-Mel-PEI did not induce any relevant change in metabolic activity. To confirm the results, LDH assays were performed to detect any detrimental effect on membrane integrity (Figure 4B). In line with the metabolic activity results, non-activated samples showed only negligible membrane impairment, reflected in LDH release values below 20%. On the other hand, samples incubated with beads indicated an increase in membrane disturbance, with a maximum of 40% of LDH release. The transfection with the different formulation did not further increase the LDH release, in both stimulated and unstimulated samples. Toxicity studies in primary CD4+ T cells confirmed that no increase in membrane disturbance was observable after incubation with polyplexes (Figure 4C).
Figure 4.
Metabolic activity and plasma membrane integrity is not altered in activated and non-activated PCLS (three donors)as well as in primary CD4+ T cells after treatment with Tf-PEI and Tf-Mel-PEI polyplexes. (A) Evaluation of cellular viability by WST-1 assay in PCLS after transfection with 50 pmol siRNA. (B) Assessment of lactate dehydrogenase release by LDH assay following incubation with polyplexes in PCLS after transfection with 50 pmol siRNA and (C) primary CD4+ T cells after transfection with 10 pmol or 20 pmol siRNA. Data points indicate means ± SD.
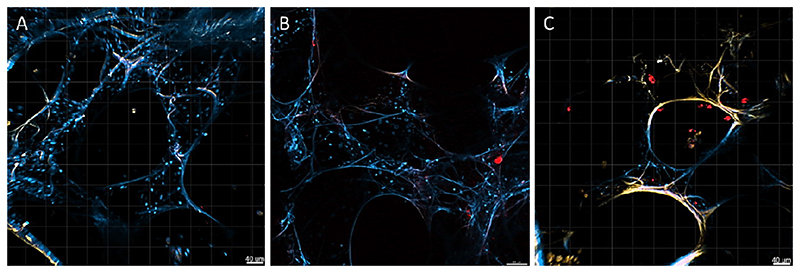
3.8. siRNA delivery to hPCLS
Confocal scanning laser microscopy was utilized to evaluate the ability of Tf-PEI and Tf-Mel-PEI polyplexes to deliver siRNA to PCLS. The encapsulated fluorescently labelled siRNA, AF647-siRNA, is presented in red, and nuclei were stained with DAPI, shown in blue. The autofluorescence observed in lung tissue is shown in yellow (Figure 5). From the qualitative assessment of this experiment, it appears that siRNA reaches the parenchyma and, more importantly, more siRNA shown as red dots seems to be delivered with Tf-Mel-PEI polyplexes compared to samples transfected with Tf-Mel-PEI.
Figure 5.
Delivery of Tf-PEI (B) and Tf-Mel-PEI (C) polyplexes loaded with50 pmol AF647-siRNA to human PCLS analyzed by confocal scanning laser microscopy. Red dots represent AF647-siRNA, blue corresponds to nuclei stained with DAPI and yellow reflects autofluorescence of PCLS. (A) represents a control PCLS receiving no treatment.
3.9. siRNA-mediated GATA3 gene silencing in hPCLS
The GATA3 gene silencing activity of the polyplexes was measured by qRT-PCR 7 days after activation and 2 days after transfection. For this experiment, a 50:50 blend of two siRNA sequences, GATA3_8 and GATA3_9 was selected, as they showed the best silencing activity in a preliminary experiment using Lipofectamine as transfection reagent. PCLSs were incubated with Tf-PEI or Tf-Mel-PEI polyplexes encapsulating different amounts of siGATA3. For comparison, untreated samples as well as samples transfected with scrambled siRNA were included as negative controls. Both Tf-PEI and Tf-Mel-PEI polyplexes significantly downregulated GATA3 expression with Tf-Mel-PEI mediating the strongest gene silencing efficacy (88%) after transfection with 100 pmol siRNA in comparison to 74% gene knockdown with Tf-PEI. Interestingly, statistically significant GATA3 downregulation was observed only in activated and not in non-activated samplesas shown in Figure 6.
Figure 6.
GATA3 knockdown in activated (A) and non-activated (B) human PCLS transfected with Tf-PEI and T-Mel-PEI polyplexes encapsulating 50 or 100 pmol of GATA3 siRNA as measured by qRT-PCR and normalized to β-actin expression. Each value is normalized to the GATA3/β-actin expression after treatment with negative control siRNA at the same conditions. Data points indicate mean ± SD. One way ANOVA, **p<0.01.
4. Discussion
The still not satisfactory controlled inflammatory cascades involved in allergic asthma reveal the need of novel more precise treatment options especially for severe cases. GATA3 as the central transcription factor in Th2 mediated asthmatic airway inflammation offers an auspicious target for respective medications, however, is challenging to reach. Transferrin was found to be a suitable option to achieve both the entry into otherwise hard-to-transfect T cells in general and the targeted delivery to activated disease-related T cells in particular. [17] As early as in 1999, the group of Wagner et al. described how coupling of transferrin to polycationic vectors such as PEI results in stable carriers for efficient and target-specific gene delivery to subcutaneously growing tumors. [25] This strategy was thenceforward constantly advanced and refined and applied for various cancer targeting approaches [26], but eventually also found suitable for further approaches other than tumor targeting. The transferrin receptor (TfR) as entry port for iron transport is not only overexpressed on different cancer cell types, but also on cells involved in inflammatory cascades, to meet their elevated iron requirements for enhanced cell proliferation and differentiation. It can therefore also be used to target disease-causing cell subsets in inflammatory diseases such as asthma. In our previous work, we developed a Tf-PEI based carrier system for specific delivery of siRNA to activated T cells as a potential novel asthma therapy. Despite showing optimal particle characteristics and being able to successfully and preferentially knock-down particular genes in vitro, [17] Tf-PEI polyplexes lacked efficient in vivo gene silencing capability.[22] In a follow-up study, the original Tf-PEI conjugate was therefore modulated with the endosomolytic peptide melittin in order to increase endosomal escape of the polyplexes which presumably hindered the full potential of in vivo effects. Indeed, the addition of melittin could distinctly improve polyplex transport from the endosomes to the cytosol, resulting in a significantly enhanced knockdown effect in vitro. [21] While this latter study concentrated on the characterization and optimization of the Tf-Mel-PEI polyplexes and their binding specificity towards the TfR, the focus of this work was put on resulting gene knockdown in activated T cells with an emphasis on GATA3 silencing and evaluation of respective downstream treatment effects.
The first step was to gain more precise insights into the T cell activation process and optimize respective processes to develop a suitable model mirroring the characteristics of activated, disease-related T cells in asthmatic patients. Therefore, two different activation procedures were tested, namely the use of soluble antibodies directed against CD3 and CD28 and the application of so-called Dynabeads®, magnetic ferric oxide beads covalently coupled to anti-CD3 and anti-CD28 antibodies. These beads have a diameter of 4,5 μm resembling the size of antigen presenting cells and, moreover, they are chemically inert and superparamagnetic, meaning that they can easily be removed from activated cell cultures with the help of a magnet. CD3 and CD28 were chosen as the typically used primary and costimulatory signals for T cell activation partially mimicking in vivo stimulation via antigen-presenting cells (APC). The activation process is generally initiated by recognition of peptide-loaded major histocompatibility complexes (MHCs) on APCs by T cell receptors (TCRs). These APCs provide at the minimum two signals required for T cell activation, one via the TCR/CD3 complex and one additional signal through one or more costimulatory cell interactions such as via CD28. [27] Exclusively in the presence of an appropriate costimulatory signal, the primed T cell gets capable of starting a productive immune responsive characterized by differentiation, proliferation, IL-2 production and following steps. [28]
To evaluate and compare activation processes in both continuous cell lines and primary cells, Jurkat cells, a human T lymphocyte cell line, were chosen as a model cell line, and human CD4+ primary T cells were isolated from freshly obtained buffy coats via magnetic bead separation. After preliminary studies for optimization of different parameters concerning cell viability during the activation process such as cell number and concentration or well-plate format (data not shown), three parameters were determined to optimize the activation procedure itself and its resulting outcome. As a first activation marker, expression of TfR was chosen and measured at different activation time points. The transferrin receptor plays a crucial role in this work, being both the targeting and entry port of Tf-Mel-PEI polyplexes for efficient T cell transfection. In our previous experiments, the targeting specificity of Tf-Mel-PEI towards the TfR was proven by surface Plasmon resonance spectroscopy (SPR) measurements and the preferential uptake in activated vs. naïve T cells was highlighted. [21] While in the Jurkat cell line nearly 100 % of all cells do already express the receptor constitutively, primary T cells only show a negligible TfR expression at naïve levels. Here, the receptor expression could distinctly be upregulated via both activation with soluble antibodies and antibody coated beads. The dynabeads, in particular, increased TfR expression resulting in a CD71 positive cell population accounting for 45 % compared to almost no positive cells at naïve status. This upregulation could furthermore be maintained for additional 48 h, indicating that the activation process and subsequent receptor expression proceeds in a sustained and reproducible way. In comparison, T cell activation with soluble CD3 and CD28 antibodies resulted in a lower, but still distinct upregulation of TfR expression, reaching a maximum of 30 % CD71 positive cells after 3 days. Concerning receptor expression, the beads can, therefore, be concluded as the more effective option.
Since the ultimate aim of this study is to generate a novel asthma therapy on the base of downregulation of the transcription factor GATA3 in activated T cells, GATA3 expression levels were determined as the second surrogate marker for the T cell activation status. Again, soluble antibodies and beads were compared in Jurkat as well as primary T cells, and again, both activation procedures led to a distinct activation with a subsequent increase of respective expression. While the maximally reached GATA3 expression in Jurkat cells was comparable for the two activation methods, in primary CD4+ T cells the beads clearly surpassed the soluble antibodies, resulting in a steady and activation time dependent increase in gene expression. A significant GATA3 expression was also found in mice by Ying et al. after naïve T cell activation, eventuating in Th2 differentiation. [29] Overall these results give reason to assume that the selected activation procedures are able to achieve increased GATA3 levels and thereby simulate respective inflammation processes orchestrated by activated T cells in the lungs of asthmatic patients.
To further evaluate this assumption, downstream effects of T cell activation and subsequent upregulated GATA3 expression on respective cytokine secretion was chosen as a third parameter to be investigated. Th2 related IL-4, IL-5, and IL-13 are all directly affected by GATA3 gene expression, as upon T cell receptor stimulation, the transcription factor is translocated to the nucleus [30] and involves in interleukin promoter activation and Th2 cell differentiation. The GATA3 protein supposedly docks on and interferes with promoter regions of the interleukin genes, in turn upregulating their expression, however, this mechanism is not yet fully clarified. [31] IL-4 mainly drives polarization of the CD4+ T cell response in the Th2 phenotype in combination with suppression of interferon (IFN)-γ-producing Th1 cells [32], having substantial impact on the Th1/Th2 imbalance in inflammatory cascades in asthma. Moreover, it directs differentiation of T cells into IL-4 producing effector T cells, as even naïve T cells are able to produce IL-4. In this way, a significant population of naïve CD4+ was stimulated to secrete IL-4 after primary activation. [29] The interleukin also controls the growth and differentiation of B cells [33] and the specificity of the immunoglobulin G (IgG) class switching, navigating further disease related processes. IL-5 is known to be key maturation and differentiation factor for eosinophils, leading to increased eosinophil numbers and antibody levels upon over-expression. It regulates expression of genes involved in cell survival, proliferation, and maturation of eosinophils as well as B cells and therefore plays a major role in both innate and acquired immune responses as well as eosinophilia. [34] IL-13, on the other hand, intersects with several biological activities of IL-4, both sharing the IL-4 receptor alpha-chain essential for signal transduction. As T cells, however, do not express functional IL-13 receptors, IL-13 is unlike IL-4 not able to induce Th2 cell differentiation. [35] The effects of the first-mentioned cytokine against this comprise goblet cell differentiation resulting in enhanced mucus production, increase of bronchial hyperresponsiveness, activation of fibroblasts as well as B cell antibody switch from IgM to IgE. [36] In this work, analogous to the first two tested factors TfR and GATA3 expression, a distinct increase could also be observed for the interleukin secretion upon soluble as well as bead based activation. As experiments in the Jurkat cell line unfortunately yielded inconsistent results, only data obtained from human primary CD4+ T cells are discussed here. The consistent enhancement of production of all three tested cytokines proves that both activation procedures do not only influence the insides of the T cells themselves, but furthermore also their surrounding environments and thereby presumably even further cell types and other downstream processes and cascades. As all three tested interleukins play key roles in the inflammation process underlying allergic asthma pathogenesis, it can hence be expected that this stage of the disease process can properly be illustrated in the activated T cell model. IL-13 was by far the strongest affected cytokine and will be discussed in more detail at a later point.
In summary it can be stated that both tested activation procedures, beads and soluble antibodies, achieved a distinct activation of continuous as well as primary T cells resulting in an upregulation of TfR, GATA3 as well as secretion of chosen interleukins. In comparison, CD3 and CD28 antibody coated beads were shown to be overall more effective. As the ferric oxide beads resemble antigen presenting cells in size, they can mimic these essential helpers in the activation process, thereby leading to a more precise signaling. Increasing the activation time from two to three days could not considerably increase activation parameters further, and rather resulted in elevated stress concerning cell viability. Consequently, dynabead activation for 48 h was chosen as the preferred T cell activation method to achieve a proper model for activated T cells mirroring the status in inflamed airways of asthmatic lungs.
For the knockdown of GATA3, a potent murine siRNA sequence had already been optimized by the group of Garn et al. and was shown to decrease the number of eosinophils and level of airway hyperresponsiveness in models of acute allergic airway inflammation after intranasal administration. [9] For humans, however, to the best of our knowledge, no comparably suitable sequence had been detected to this point. Therefore, the first step towards successful GATA3 knockdown in human cells was to discover an appropriate functional siRNA sequence resulting in significant gene silencing. After testing multiple sequences from different vendors in preliminary screening experiments with lipofectamine as transfection agent in easy-to-transfect MCF-7 cells, two sequences were found to be particularly eligible for this purpose, namely GATA3_8 and GATA3_9. It was then hypothesized that these sequences taken together could be even more effective as combining more than one siRNA sequence had been shown to have significant advantages over use of the corresponding individual siRNAs [37] and can lower off-target effects of single sequences due to smaller applied concentrations [38]. In fact, a 1:1 mixture of both functional siRNA sequences resulted in a GATA3 gene knockdown of almost 90 % compared to the negative control. This combination was subsequently chosen to be applied to all further gene silencing experiments.
Sequences GATA3_8 and GATA3_9 were then further validated in Jurkat cells and within the Tf-Mel-PEI blend polyplexes, proving the capability of both sequences alone, and all the more in combination, resulting in a significant silencing of the target gene. The knockdown potential of Tf-Mel-PEI was furthermore highlighted in comparative experiments in Jurkat cells as well as primary human CD4+ T cells involving unmodified PEI and the original Tf-PEI conjugate as controls. In both cell types, the superior gene silencing efficiency of the blend polyplexes could clearly be pointed out, again emphasizing the essential role of endosomal escape for effective siRNA therapy. Although Tf-PEI also resulted in a distinct knockdown effect compared to respective negative controls, only the addition of melittin aiding transport of the siRNA cargo from the endosomes to the cytosol could really maximize the treatment outcome.
To determine whether the achieved knockdown of GATA3 would also result in sufficient interference with respective parameters in downstream cascades of the transcription factors, concentrations of the three most relevant cytokines IL-4, IL-5, and IL-13 were measured in cell supernatants by sandwich ELISA. Even if unfortunately for IL-4, some negative values were obtained, a reduction of all three cytokines could be observed after treatment with GATA3 siRNA containing Tf-Mel-PEI polyplexes. The strongest decrease in secretion was achieved for IL-5, the predominant cytokine involved in eosinophilic activation, causing airway inflammation as a classic feature of severe allergic asthma. [39] IL-5 displays the main growth, differentiation, and activation factor for human eosinophils, highly expressing the IL-5 receptor on their surface membranes. Targeting of IL-5, therefore presents a promising base to develop treatments for patients with severe asthma, especially when suffering particularly from eosinophilic inflammation. [40] In fact, several monoclonal antibodies targeting IL-5, namely mepolizumab, reslizumab, and benralizumab have been developed to specifically bind and interfere with the IL-5 receptor on eosinophils and were recently approved for the treatment of adult patients with severe eosinophilic asthma. [40, 41]. Beyond IL-5, a significant reduction was also observed for IL-13 concentration. IL-13 is another central effector of asthmatic processes, most notably inducing goblet cell hyperplasia and mucus hypersecretion [2] and IL-13 signaling was shown to directly result in mucin secretion, airway hyperresponsiveness and pulmonary fibrosis in animal models of asthma [42]. Although glucocorticoids are effective and widely used maintenance therapies in acute and chronic asthma, some patients are non-responsive to steroids maintaining elevated IL-13 levels. [43] For these severe cases, antibodies targeting IL-13 are being under investigation as novel targeted add-on treatments. Despite of the benefit of these specific therapies concentrating on single interleukins, the aim of this work is to interfere with GATA3 as the central transcription factor of these different cytokines, thereby eradicating all of them simultaneously and being able to early-on undermine pathologic pathways. The discussed results give reason to assume that GATA3 knockdown via Tf-Mel-PEI polyplexes could indeed be a promising approach to downregulate specific pathways playing essential roles in the inflammation process in asthmatic patients and thereby ameliorate respective symptoms.
To further evaluate the behavior and treatment effects of Tf-Mel-PEI polyplexes under more in vivo like conditions, studies in precision cut lung slices were performed to investigate transfection efficacy not only in isolated T cells, but also in the complex environment of lung tissue. Activation of tissue resident T cells efficiently in a sophisticated ex vivo model of the lung efficiently mimicked the inflammatory pattern found in patients suffering from allergic asthma where a mixture of different inflammatory cytokines is found. Commonly, GATA3 related and non-related cytokines contribute to the pathogenesis of allergic asthma. Th2 related cytokines are directly affected by GATA3 expression, since it promotes and upregulates their secretion.[44] As increased expression of GATA3 and consistent secretion of the different inflammatory cytokines was observed after activation with Dynabeads®, a functional response at both the upstream and downstream level of the inflammatory cascade was successfully confirmed.
The observed decreased metabolic activity in the lung tissue was to be expected considering that sites with ongoing inflammation and triggered immune response present alterations of metabolic activity.[45] The increase in membrane leakiness, on the other hand can be linked to the inflammatory state induced by the beads. The transfection with the different formulation did not increase the LDH release, in both stimulated and unstimulated samples, confirming the safety of Tf-PEI and Tf-Mel-PEI as nanocarriers for siRNA. These results are in line with our previous studies in cell culture, where no significant cytotoxicity was observed for any of the different formulations neither in PCLS nor in primary CD4+ T cells, even at higher N/P ratios.[21]
Tf-PEI and Tf-Mel-PEI polyplexes successfully delivered siRNA to the lung tissue indicating more efficient delivery with Tf-Mel-PEI polyplexes compared to Tf-Mel-PEI. This observation is in line with our previous studies in immortalized and primary T cells, where the presence of melittin improved not only the endosomal escape, but also the uptake of siRNA.[21] Melittin is indeed a positively charged peptide that, in addition to endosmolytic activity, can also deliver nucleic acids. This specific property could explain the superior siRNA delivery observed for Tf-Mel-PEI polyplexes.[46]
Furthermore, the downregulation activity of Tf-PEI and Tf-Mel-PEI polyplexes against GATA3 was confirmed, with Tf-Mel-PEI showing the best activity results, in line with results obtained in isolated primary T cells. The fact that only in activated and not in non-activated lung samples statistically significant gene silencing was observed can be explained by the fact that TfR expression is negligible in non-activated T cells (Supplementary Figure 1B) and TfR-targeting is only beneficial for targeting activated T cells.[17] In line with our previous findings, the inclusion of melittin in the formulation improves the endosomal escape and consequently the RNA interference activity of the formulation, which is reflected in the results after transfection with 100 pmol siRNA. This experiment confirmed the potential of Tf-PEI and Tf-Mel-PEI polyplexes as delivery systems for siRNA. Interestingly, downstream effects on decreased secretion of IL-5 and IL-13, particularly, were more successfully obtained with Tf-PEI rather than Tf-Mel-PEI in activated PCLS samples (Supplementary Figure 7). To the best of our knowledge, we are the first to attempt GATA3 downregulation in human lung explants. In an unrelated study by Ruigrok et al., the downregulation of GAPDH was tested by qRT-PCR in mouse PCLS 48 h after transfection with siRNA. In that study, siRNA was delivered with a commercially available transfection reagent and achieved 50% reduction of mRNA expression.[47] Tf-Mel-PEI polyplexes here reachedgene silencing efficacy of up to88 %, thus confirming that our formulation represents a valid option for delivering siRNA to lung tissues and as a promising strategy to treat allergic asthma. However, the beneficial effects of melittin were less clear in lung tissue explants than in isolated T cells, reflecting potentially additional gene silencing in epithelial cells, which would nonetheless improve symptoms of asthma.
5. Conclusion
In conclusion, within this work, an optimized activation methods for continuous as well as primary T cells and T cells in lung tissue has been generated that is suitable to illustrate the inflammation status of activated T cells as a model for asthmatic processes. A functional sequence combination to silence GATA3 gene expression via siRNA mediated RNAi was found and applied for the specific knockdown in activated T cells. Tf-Mel-PEI polyplexes were proven to be an eligible carrier system for targeted GATA3 silencing in activated T cells, resulting in respective alleviation of downstream cytokine production. In an effort to continue experiments in models of the same species and to avoid switching to the murine version of the transferrin conjugate, human Tf-Mel-PEI was additionally tested under more in vivo like conditions in explanted lung tissue. The polyplexes efficiently mediated RNAi for GATA3 downregulation in human lung tissue without measurable toxic effects in the tissue. Overall, the blend particles display a promising way to deliver siRNA to activated T cells in the lung as a potential new therapy peculiarly for severe cases of allergic asthma.
Supplementary Material
Acknowledgements
This work was supported by ERC Starting Grant ERC-2014-StG – 637830 “Novel Asthma Therapy” to Olivia Merkel.
References
- [1].Addo-Yobo EOD. The global asthma network. The global asthma report. 2018 [Google Scholar]
- [2].Rael EL, Lockey RF. Interleukin-13 signaling and its role in asthma. World Allergy Organ J. 2011;4:54–64. doi: 10.1097/WOX.0b013e31821188e0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Holgate ST. Innate and adaptive immune responses in asthma. Nat Med. 2012;18:673–683. doi: 10.1038/nm.2731. [DOI] [PubMed] [Google Scholar]
- [4].Anderson GP. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet. 2008;372:1107–1119. doi: 10.1016/S0140-6736(08)61452-X. [DOI] [PubMed] [Google Scholar]
- [5].Pelaia G, Vatrella A, Maselli R. The potential of biologics for the treatment of asthma. Nat Rev Drug Discov. 2012;11:958–972. doi: 10.1038/nrd3792. [DOI] [PubMed] [Google Scholar]
- [6].Haselkorn T, Borish L, Miller DP, Weiss ST, Wong DA. High prevalence of skin test positivity in severe or difficult-to-treat asthma. J Asthma. 2006;43:745–752. doi: 10.1080/02770900601031540. [DOI] [PubMed] [Google Scholar]
- [7].Moore WC, Bleecker ER, Curran-Everett D, Erzurum SC, Ameredes BT, Bacharier L, Calhoun WJ, Castro M, Chung KF, Clark MP, Dweik RA, et al. L.B.I.s.S.A.R.P. National Heart Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119:405–413. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kudo M, Ishigatsubo Y, Aoki I. Pathology of asthma. Front Microbiol. 2013;4:263. doi: 10.3389/fmicb.2013.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sel S, Wegmann M, Dicke T, Sel S, Henke W, Yildirim AO, Renz H, Garn H. Effective prevention and therapy of experimental allergic asthma using a GATA-3-specific DNAzyme. J Allergy Clin Immunol. 2008;121:910–916.:e915. doi: 10.1016/j.jaci.2007.12.1175. [DOI] [PubMed] [Google Scholar]
- [10].Lamaze C, Dujeancourt A, Baba T, Lo CG, Benmerah A, Dautry-Varsat A. Interleukin 2 receptors and detergent-resistant membrane domains define a clathrin-independent endocytic pathway. Mol Cell. 2001;7:661–671. doi: 10.1016/s1097-2765(01)00212-x. [DOI] [PubMed] [Google Scholar]
- [11].Mocellin S, Provenzano M. RNA interference: learning gene knock-down from cell physiology. J Transl Med. 2004;2:39. doi: 10.1186/1479-5876-2-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Padda IS, Mahtani AU, Parmar M. Small Interfering RNA (siRNA) Based Therapy. StatPearls; Treasure Island (FL): 2022. [PubMed] [Google Scholar]
- [13].Hoy SM. Patisiran: First Global Approval, Drugs. 2018;78:1625–1631. doi: 10.1007/s40265-018-0983-6. [DOI] [PubMed] [Google Scholar]
- [14].Merkel OM, Kissel T. Nonviral pulmonary delivery of siRNA. Accounts of chemical research. 2012;45:961–970. doi: 10.1021/ar200110p. [DOI] [PubMed] [Google Scholar]
- [15].Kandil R, Merkel OM. Pulmonary delivery of siRNA as a novel treatment for lung diseases. Ther Deliv. 2019;10:203–206. doi: 10.4155/tde-2019-0009. [DOI] [PubMed] [Google Scholar]
- [16].Keil TWM, Baldassi D, Merkel OM. T-cell targeted pulmonary siRNA delivery for the treatment of asthma. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2020;12:e1634. doi: 10.1002/wnan.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xie Y, Kim NH, Nadithe V, Schalk D, Thakur A, Kilic A, Lum LG, Bassett DJ, Merkel OM. Targeted delivery of siRNA to activated T cells via transferrin-polyethylenimine (Tf-PEI) as a potential therapy of asthma. J Control Release. 2016;229:120–129. doi: 10.1016/j.jconrel.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Beyerle A, Braun A, Merkel O, Koch F, Kissel T, Stoeger T. Comparative in vivo study of poly(ethylene imine)/siRNA complexes for pulmonary delivery in mice. J Control Release. 2011;151:51–56. doi: 10.1016/j.jconrel.2010.12.017. [DOI] [PubMed] [Google Scholar]
- [19].Cipolla D, Gonda I, Chan HK. Liposomal formulations for inhalation. Ther Deliv. 2013;4:1047–1072. doi: 10.4155/tde.13.71. [DOI] [PubMed] [Google Scholar]
- [20].Kim NH, Nadithe V, Elsayed M, Merkel OM. Tracking and treating activated T cells. J Drug Deliv Sci Technol. 2013;23:17–21. doi: 10.1016/s1773-2247(13)50002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rima Kandil YX, Heermann Ralf, Isert Lorenz, Jung Kirsten, Mehta Aditi, Merkel Olivia M. Coming in and Finding Out: Blending Receptor-Targeted Delivery and Efficient Endosomal Escape in a Novel Bio-Responsive siRNA Delivery System for Gene Knockdown in Pulmonary T Cells. Advanced Therapeutics. 2019 doi: 10.1002/adtp.201900047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kandil R, Feldmann D, Xie Y, Merkel OM. Evaluating the Regulation of Cytokine Levels After siRNA Treatment in Antigen-Specific Target Cell Populations via Intracellular Staining. Methods Mol Biol. 2019;1943:323–331. doi: 10.1007/978-1-4939-9092-4_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Neuhaus V, Danov O, Konzok S, Obernolte H, Dehmel S, Braubach P, Jonigk D, Fieguth HG, Zardo P, Warnecke G, Martin C, et al. Assessment of the Cytotoxic and Immunomodulatory Effects of Substances in Human Precision-cut Lung Slices. J Vis Exp. 2018 doi: 10.3791/57042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Niehof M, Hildebrandt T, Danov O, Arndt K, Koschmann J, Dahlmann F, Hansen T, Sewald K. RNA isolation from precision-cut lung slices (PCLS) from different species. BMC Res Notes. 2017;10:121. doi: 10.1186/s13104-017-2447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kircheis R, Schuller S, Brunner S, Ogris M, Heider KH, Zauner W, Wagner E. Polycationbased DNA complexes for tumor-targeted gene delivery in vivo. J Gene Med. 1999;1:111–120. doi: 10.1002/(SICI)1521-2254(199903/04)1:2<111::AID-JGM22>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- [26].Xie Y, Killinger B, Moszczynska A, Merkel OM. Targeted Delivery of siRNA to Transferrin Receptor Overexpressing Tumor Cells via Peptide Modified Polyethylenimine. Molecules. 2016;21 doi: 10.3390/molecules21101334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kusmartsev SA, Li Y, Chen SH. Gr-1+ myeloid cells derived from tumor-bearing mice inhibit primary T cell activation induced through CD3/CD28 costimulation. J Immunol. 2000;165:779–785. doi: 10.4049/jimmunol.165.2.779. [DOI] [PubMed] [Google Scholar]
- [28].Lenschow DJ, Walunas TL, Bluestone JA. CD28/B7 system of T cell costimulation. Annu Rev Immunol. 1996;14:233–258. doi: 10.1146/annurev.immunol.14.1.233. [DOI] [PubMed] [Google Scholar]
- [29].Yagi R, Suzuki W, Seki N, Kohyama M, Inoue T, Arai T, Kubo M. The IL-4 production capability of different strains of naive CD4(+) T cells controls the direction of the T(h) cell response. Int Immunol. 2002;14:1–11. doi: 10.1093/intimm/14.1.1. [DOI] [PubMed] [Google Scholar]
- [30].Silva-Filho JL, Caruso-Neves C, Pinheiro AAS. IL-4: an important cytokine in determining the fate of T cells. Biophys Rev. 2014;6:111–118. doi: 10.1007/s12551-013-0133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhu J, Yamane H, Cote-Sierra J, Guo L, Paul WE. GATA-3 promotes Th2 responses through three different mechanisms: induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors. Cell Res. 2006;16:3–10. doi: 10.1038/sj.cr.7310002. [DOI] [PubMed] [Google Scholar]
- [32].Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, Bucher P. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem. 2001;276:6675–6688. doi: 10.1074/jbc.M001748200. [DOI] [PubMed] [Google Scholar]
- [33].Howard M. Interleukins for B lymphocytes. Surv Immunol Res. 1983;2:210–212. doi: 10.1007/BF02918413. [DOI] [PubMed] [Google Scholar]
- [34].Kouro T, Takatsu K. IL-5-and eosinophil-mediated inflammation: from discovery to therapy. Int Immunol. 2009;21:1303–1309. doi: 10.1093/intimm/dxp102. [DOI] [PubMed] [Google Scholar]
- [35].de Vries JE. The role of IL-13 and its receptor in allergy and inflammatory responses. J Allergy Clin Immunol. 1998;102:165–169. doi: 10.1016/s0091-6749(98)70080-6. [DOI] [PubMed] [Google Scholar]
- [36].Corren J. Role of interleukin-13 in asthma. Curr Allergy Asthma Rep. 2013;13:415–420. doi: 10.1007/s11882-013-0373-9. [DOI] [PubMed] [Google Scholar]
- [37].Parsons BD, Schindler A, Evans DH, Foley E. A direct phenotypic comparison of siRNA pools and multiple individual duplexes in a functional assay. PLoS One. 2009;4:e8471. doi: 10.1371/journal.pone.0008471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hannus M, Beitzinger M, Engelmann JC, Weickert MT, Spang R, Hannus S, Meister G. siPools: highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res. 2014;42:8049–8061. doi: 10.1093/nar/gku480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Varricchi G, Bagnasco D, Borriello F, Heffler E, Canonica GW. Interleukin-5 pathway inhibition in the treatment of eosinophilic respiratory disorders: evidence and unmet needs. Curr Opin Allergy Clin Immunol. 2016;16:186–200. doi: 10.1097/ACI.0000000000000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Varricchi G, Canonica GW. The role of interleukin 5 in asthma. Expert Rev Clin Immunol. 2016;12:903–905. doi: 10.1080/1744666X.2016.1208564. [DOI] [PubMed] [Google Scholar]
- [41].Markham A. Benralizumab: First Global Approval. Drugs. 2018;78:505–511. doi: 10.1007/s40265-018-0876-8. [DOI] [PubMed] [Google Scholar]
- [42].Munitz A, Brandt EB, Mingler M, Finkelman FD, Rothenberg ME. Distinct roles for IL-13 and IL-4 via IL-13 receptor alpha1 and the type II IL-4 receptor in asthma pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7240–7245. doi: 10.1073/pnas.0802465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ito K, Chung KF, Adcock IM. Update on glucocorticoid action and resistance. J Allergy Clin Immunol. 2006;117:522–543. doi: 10.1016/j.jaci.2006.01.032. [DOI] [PubMed] [Google Scholar]
- [44].Ray A, Cohn L. Th2 cells and GATA-3 in asthma: new insights into the regulation of airway inflammation. J Clin Invest. 1999;104:985–993. doi: 10.1172/JCI8204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kominsky DJ, Campbell EL, Colgan SP. Metabolic shifts in immunity and inflammation. J Immunol. 2010;184:4062–4068. doi: 10.4049/jimmunol.0903002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Paray BA, Ahmad A, Khan JM, Taufiq F, Pathan A, Malik A, Ahmed MZ. The role of the multifunctional antimicrobial peptide melittin in gene delivery. Drug Discov Today. 2021;26:1053–1059. doi: 10.1016/j.drudis.2021.01.004. [DOI] [PubMed] [Google Scholar]
- [47].Ruigrok MJR, Maggan N, Willaert D, Frijlink HW, Melgert BN, Olinga P, Hinrichs WLJ. siRNA-Mediated RNA Interference in Precision-Cut Tissue Slices Prepared from Mouse Lung and Kidney. AAPS J. 2017;19:1855–1863. doi: 10.1208/s12248-017-0136-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.