Abstract
Recognition of the intron branch site (BS) by the U2 snRNP is a critical event during spliceosome assembly. In mammals, BS sequences are poorly conserved and unambiguous intron recognition cannot be achieved solely via a base-pairing mechanism. We isolated human 17S U2 snRNP and reconstituted in vitro its ATP-dependent remodelling and binding to the pre-mRNA substrate. We determined a series of high-resolution (2.0-2.2 Å) structures providing snapshots of the BS selection process. The substrate-bound U2 snRNP shows that SF3B6 stabilises the BS:U2 snRNA duplex, which could aid binding of introns with poor sequence complementarity. ATP-dependent remodelling uncoupled from substrate binding captures U2 snRNA in a conformation that competes with BS recognition, providing a selection mechanism based on branch helix stability.
Removal of introns from pre-mRNA is catalyzed by a large and dynamic RNA-protein complex known as the spliceosome. The spliceosome is assembled de novo on each pre-mRNA substrate from five small nuclear ribonucleoprotein particles (snRNPs) and several dozen protein factors. During spliceosome assembly, three conserved positions in the pre-mRNA, the 5’-splice site (5’-SS), branch-site (BS) and 3’-splice site (3’-SS), are specifically recognized by the components of the spliceosome allowing a two-step trans-esterification reaction to occur.
In mammalian cells, the BS is initially recognised by SF1 (mBBP) in cooperation with U2AF2 (U2AF65), which binds the polypyrimidine tract (PPT) sequence (1). Concomitantly, U1 snRNP binds to the 5’-SS and together they form the first, ATP-independent spliceosome assembly intermediate known as complex E (2). The U2 snRNP is loosely associated with complex E (3) and its stable incorporation into the prespliceosome (complex A) requires ATP and formation of base-pairing interactions between the BS and the U2 snRNA (4).
In yeast, several factors have been shown to facilitate complex A formation. Among them, Cus2 (human HTATSF1) (5) and the RNA-dependent DEAD-box ATPase, Prp5 (human DDX46), whose activity is required for Cus2 displacement (6–8) and fidelity control of branch site recognition (9, 10).
During BS recognition, an evolutionarily conserved branchpoint-interacting stem loop (BSL) presents U2 nucleotides to the intron BS for base-pairing (11, 12). Although branch helix formation is subject to a fidelity checkpoint, it is unclear how branch helix stability is sensed by the splicing machinery (9, 10).
Upon engagement of U2 snRNA with the pre-mRNA substrate, a 15 nt-long branch helix is formed, adopting helical geometry even in the absence of full complementarity. The length of the branch helix is conserved between yeast and human spliceosomes and maintained throughout different stages of splicing (13–15). In early splicing complexes, the branch helix is accommodated within a cavity formed by the heteroheptameric SF3b complex (7), which contacts the pre-mRNA around the BS and stabilises the U2 snRNA:BS base-pairing interaction (16, 17). The branchpoint adenosine (BP-A) is bulged out of the branch helix and binds into a pocket formed by SF3B1 and PHF5A (13, 14). Mutations in SF3B1 associated with myelodysplastic syndromes have been shown to modulate BS selection (18, 19).
SF3B6 (p14), which has no homologue in S. cerevisiae, was shown to crosslink to the BP-A in HeLa nuclear extract, indicating a potential role in BS recognition (20). However, the position of SF3B6 in human Bact spliceosomes (21) does not explain the crosslinking data or its role in splicing.
While parts of the U2 snRNP structure have been determined as a component of yeast and mammalian spliceosomes, there is no high-resolution structural information for the 17S U2 snRNP and early splicing complexes in humans (i.e. E and A). This is of particular interest as sequence conservation and base-pairing potential of human branch sites is weak compared to yeast (22) and the mechanism of branch site selection remains elusive.
Here, we isolated human 17S U2 snRNP and reconstituted in vitro its binding to a model BS and the remodelling leading to the dissociation of HTATSF1 from the complex. We determined a series of cryo-EM structures of the U2 snRNP in different conformational states, including two previously unknown assembly intermediates. Our high-resolution reconstructions provide unprecedented insight into the architecture and dynamics of the human U2 snRNP and pre-spliceosome formation. These new data point at the critical roles of HTATSF1 and SF3B6 in facilitating pre-mRNA recognition.
Results
Purification of the 17S U2 snRNP complex
Existing methods for the purification of the U2 snRNP use antibodies against SF3a or SF3b components (7, 12), which can in principle capture particles in multiple states. To specifically select a subset of U2 snRNPs representing a single functional state, we used CRISPR-Cas9-mediated genome editing to introduce a GFP-tag into the HTATSF1 genomic locus of HEK293F cells (Fig. S1). Affinity chromatography with anti-GFP nanobodies allowed isolation of an intact 17S U2 snRNP, containing U2 snRNA and 22 proteins accounting for a total estimated molecular weight of 1.08 MDa (Fig. S1).
High-resolution structure of the 17S U2 snRNP
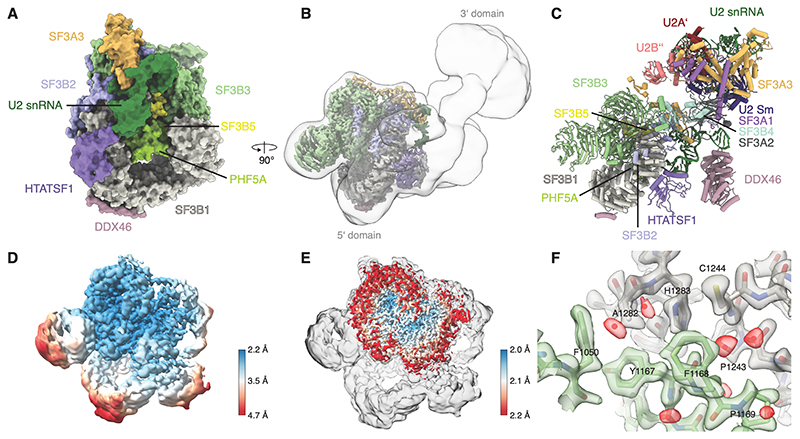
We determined the cryo-EM structure of the 5’-domain of the human 17S U2 snRNP at 2.2 Å resolution, which allowed accurate atomic modelling of the SF3b complex, SF3A3, HTATSF1RRM and the 5’-end of the U2 snRNA (Fig. 1, and Fig. S2-S4). The overall architecture of the complex agrees well with the previous studies (23, 24), including a recent low-resolution cryo-EM reconstruction (12). The low-pass filtered map reveals an unresolved density at the periphery, which likely corresponds to the U2 snRNP core (3’-domain) (Fig. 1B-C). This domain appears flexible relative to the resolved 5’-domain and could not be improved by further data processing.
Figure 1. High-resolution structure of the human 17S U2 snRNP.
(A) Surface representation of the 5’-domain of the 17S U2 snRNP model. (B) Experimental cryo-EM map for the 17S U2 snRNP showing the high-resolution 5’-domain (coloured by chain identity) embedded in a low-pass filtered map showing position of the 3’-domain. (C) Pseudo-atomic model for the fully assembled 17S U2 snRNP. 3’-domain was modelled by rigid-body docking of the previously reported coordinates (PDB:6Y5Q). (D) Cryo-EM map of the 17S U2 snRNP filtered and coloured by local resolution. (E) The cryo-EM map obtained by merging several U2 snRNP datasets overlaid with the map of the 17S U2 snRNP. (F) Atomic modelling into the highest-resolution region at an interface of SF3B1 and SF3B3. The map was coloured by chain identity, water molecules are coloured red.
Parts of SF3B2 and SF3A3 have been previously observed in cryo-EM maps of mammalian snRNPs and spliceosomes (21, 25), but due to limited resolution they were not interpreted with atomic coordinates. The high-resolution reconstruction provides atomic insights into several interfaces including HTATSF1RRM:SF3B1(Fig. 1A and C) and SF3B2:SF3A3 (Fig. 1 and S4), consistent with previous lower resolution structures (12, 26).
In vitro reconstitution of branch site recognition by the U2 snRNP
In order to obtain mechanistic insights into BS recognition via cryo-EM analysis, we reconstituted BS recognition in vitro with purified 17S U2 snRNP and a model BS oligonucleotide (BPS oligo). The BPS oligo is complementary to the positions 27-42 of the U2 snRNA and includes a bulged-out adenosine, which mimics the BP-A. Similar minimal substrates have been used previously (27, 28).
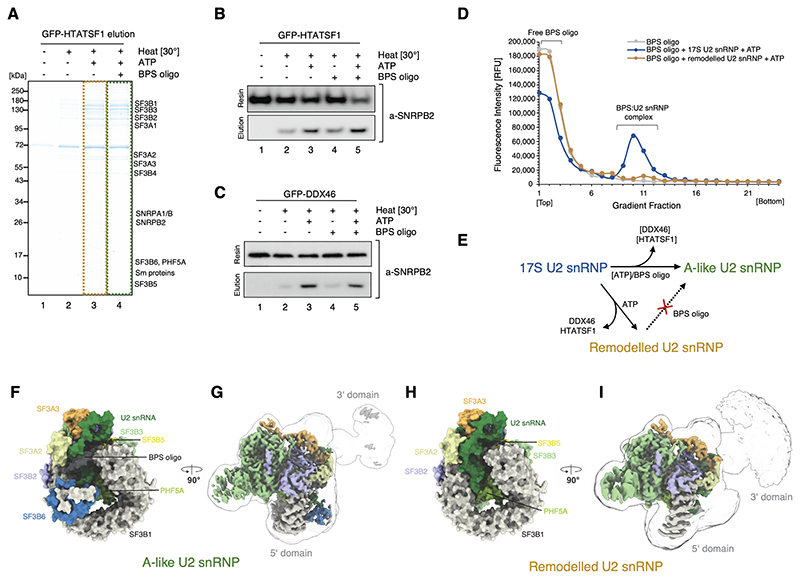
We immobilised 17S U2 snRNP on anti-GFP nanobody resin via a GFP-tag on HTATSF1 or DDX46 and incubated it under various conditions. In the presence of ATP and the BPS oligo, U2 snRNP is released from the resin (Fig.2A, lane 4) and remains bound to the BPS oligo when analysed by glycerol gradient centrifugation. These BPS oligonucleotide-bound complexes likely resemble the substrate-bound U2 snRNP within complex A. Thus, we refer to these complexes as A-like U2 snRNPs hereafter. Interestingly, addition of ATP alone, without the BPS oligo, also induces HTATSF1 dissociation at elevated temperature (Fig. 2A lane 3), suggesting that this remodelling can be functionally uncoupled from substrate binding leading to the formation of a remodelled U2 snRNP. We further investigated both reactions biochemically by probing eluates with antibodies specific to the core U2 snRNP component, SNRPB2 (Fig. 2B and C). Western blotting revealed that some HTATSF1 dissociation can occur spontaneously at elevated temperature (Fig. 2B, lane 2), but it is greatly stimulated by the presence of ATP (Fig. 2b, lanes 3 and 5), consistent with previous experiments (8). The same results were obtained regardless of whether the sample was tethered to the resin via HTATSF1 (Fig. 2B) or DDX46 (Fig. 2C).
Figure 2. Sample preparation and in vitro reconstitution of the branch site recognition by the U2 snRNP.
(A) SDS-PAGE analysis of the eluates from the GFP-HTATSF1-tagged 17S U2 snRNP immobilised on the GFP nanobody resin and incubated under various conditions. (B) Western blot analysis of the reconstitution reaction performed as in (A). Elution and resin fractions were probed with antibodies against SNRPB2, a core U2 snRNP component. (C) The same as in (B), but the 17S U2 snRNP sample was immobilised using GFP tag attached to DDX46. (D) Analysis of the Cy5-labelled BPS oligonucleotide binding to the 17S U2 snRNP or remodelled U2 snRNP by glycerol gradients. RFU, Relative Fluorescence Units. (E) Schematic summarising the outcome of the in vitro remodelling and substrate binding experiments. (F) and (H) Surface representation of the 5’-domains of the A-like and Remodelled U2 snRNPs models. (G) and (I) Experimental cryo-EM maps of A-like and Remodelled U2 snRNPs showing the high-resolution 5’-domain (coloured by chain identity) embedded in a low-pass filtered (5 Å) maps, showing positioning of their 3’-domains.
Next, we investigated the requirements for U2 snRNP engagement with a model substrate. 17S U2 snRNP engages efficiently with the BPS oligo (Fig. 2D) and the binding occurs in a wide range of conditions without requiring ATP (Fig. S5). Interestingly, no binding was observed when the remodelled (ATP-treated) U2 snRNP variant was used in this assay (Fig. 2D). This indicates that displacement of HTATSF1 and DDX46 uncoupled from substrate binding leads to the formation of an inhibited conformation of the U2 snRNP.
We determined high-resolution cryo-EM structures of these two newly identified U2 snRNP complexes (Fig. 2E-I and S2-4).
Structures of the minimal A-like and remodelled U2 snRNP complexes
The overall architecture of the A-like U2 snRNP is in good agreement with the lower-resolution descriptions of the U2 snRNP embedded within the fully assembled human B and Bact spliceosomes (21, 29) (Fig. 2F and G). However, the structure exhibits features that are incompatible with those later splicing complexes, indicating that the A-like U2 snRNP represents a distinct splicing intermediate. The remodelled U2 snRNP closely resembles A-like U2 snRNPs, except for the major differences in the 5’-end of the U2 snRNA and missing SF3B6 (Fig. 2F and H). Similar to the 17S U2 snRNP, both complexes can be divided into a well-resolved 5’-domain and a 3’-domain (Fig. 2G and I). The 3’-domains remain flexible, but occupy different positions compared to the 17S complex. HTATSF1 and the DDX46 helix are missing from these two reconstructions, consistent with the biochemical data and the sample preparation protocol.
Structure and dynamics of the U2 snRNA during branch site recognition
SLIIa of the U2 snRNA is resolved to nearly 2 Å in all our reconstructions, which allowed modelling of three additional, non-canonical base-pairs within this stem-loop and its interactions with the components of the SF3a and SF3b complexes (Fig. S4 and S6). These interactions remain unchanged during the transition from the 17S U2 snRNP to the A-like and remodelled U2 snRNP complexes.
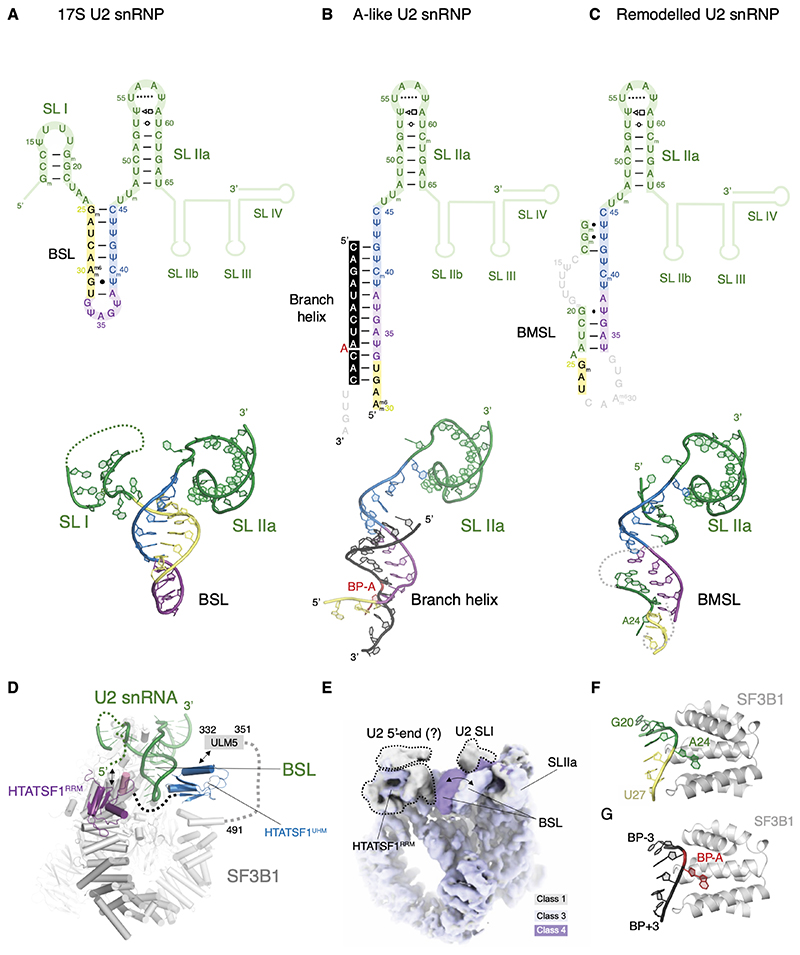
In the 17S U2 snRNP reconstruction, a helical density emerges from the 5’-end of SLIIa and points towards SF3B1 and PHF5A along the side of HTATSF1RRM. This density was interpreted as the BSL and modelled by rigid body docking of an idealised RNA helix with a base-pairing pattern based on previous predictions and the low-resolution reconstruction of the 17S U2 snRNP (12)(Fig. 3A). In contrast to the neighbouring U2 SLIIa, the BSL density is not well resolved in our map, pointing at the intrinsically dynamic nature of this structure. Indeed, a 3D classification focused on this region allows the separation of the ensemble structure into at least three distinct conformational states of the BSL (Fig. 3D and E), suggesting a dynamic probing mechanism for BS recognition.
Figure 3. Structure and dynamics of the U2 snRNA during branch site recognition.
Secondary and tertiary structure of the U2 snRNA in the (A) 17S, (B) A-like and (C) remodelled U2 snRNPs.
(D) BSL is stabilised directly by the two domains of HTATSF1 in the 17S U2 snRNP complex.
(E) Structural dynamics of the BSL visualised directly in the cryo-EM map (see also movie S2).
(F) Adenosine 24 of the U2 snRNA mimics BP-A in the remodelled U2 snRNP complex. (G) Environment of the BP-A in the A-like U2 snRNP in the same orientation as in (F).
During transition from 17S to A-like U2 snRNP the BSL sequence engages with the BPS oligo, forming a 12-nt U2 snRNA:BS duplex (Fig. 3B). This duplex forms interactions with SF3B1, PHF5A, SF3A2, and SF3A3. The SF3A2 zinc finger domain binds the branch helix as previously described (14, 21, 26). SF3A3 becomes more ordered in the A-like complex, likely stabilised by SF3A2 and its interaction with the branch helix (Fig. 4A). Most prominent of the contacts formed during this transition are the interactions of charged amino acids of SF3B1 (K1071, R1106, N1107, R1109, K1149) with the BPS RNA phosphate backbone, consistent with previous studies (14, 21). Interestingly, the SF3a complex is less well resolved in the A-like complex assembled in the presence of AMP-PCP, suggesting that the ATP-dependent remodelling may play a role in facilitating SF3a docking to the complex A (Fig. S7).
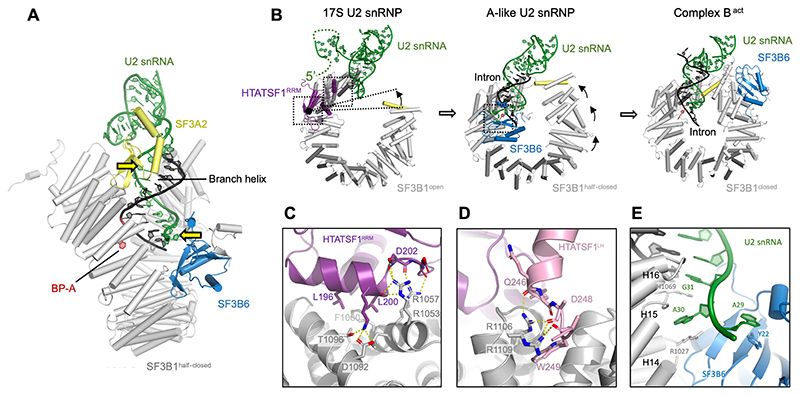
Figure 4. SF3B6 stabilises branch helix in the A-like U2 snRNP while SF3B1 HEAT repeats adopt a half-closed conformation.
(A) Side view of the A-like U2 snRNP showing positions of the branch helix and its stabilisation by the SF3A2 and SF3B6, yellow arrows indicate U2 snRNA contact points enforcing helical geometry of the branch helix. (B) Structure of the RNA and HEAT repeats in the 17S U2 snRNP, the A-like U2 snRNP and the Bact complex (PDB: 6FF7) showing two-step transition from open to close SF3B1 conformation. (C) and (D) Atomic model of the interfaces between SF3B1HEAT and HTATSF1RRM/LH. (E) Atomic model of the interface of SF3B6 with SF3B1HEAT and U2 snRNA.
5’-end of the U2 snRNA mimics pre-mRNA substrate in the absence of HTATSF1
Upon ATP-dependent remodelling and HTATSF1 dissociation, U2 snRNA nucleotides 11-44 form a novel bulged stem-loop structure in the remodelled U2 snRNP complex (Fig. 3C). Comparison with the A-like U2 snRNP reveals that this newly formed stem-loop closely mimics the branch duplex and its interactions with the U2 snRNP proteins (Fig. 3F and G), therefore we refer to this stem-loop as the Branch Helix-Mimicking Stem-Loop (BMSL). Formation of the BMSL is mutually exclusive with the pre-mRNA binding by the U2 snRNP, suggesting that the two structures could compete with one another during BS recognition. This finding provides a potential mechanism for how the branch helix stability can be selected by the spliceosome and could represent a novel BS fidelity checkpoint.
SF3B6 stabilises the branch helix in the A-like U2 snRNP
Upon BPS oligo binding to the 17S U2 snRNP, an additional density appears near H14 and H15 of SF3B1. This density could be unambiguously interpreted by rigid body docking of the SF3B6:SF3B1 crystal structure (PDB: 3LQV (30), Fig. S4). Although it is a stable component of the SF3b complex, SF3B6 has not been observed in any of previously reported structure of the SF3b complex or U2 snRNP (12, 23) and it differs dramatically from the SF3B6:SF3B1 interface in the Bact spliceosome (21). SF3B6 binds to the U2 snRNA at the 5’-end of the branch helix (Fig. 4A), therefore it defines the exact position of the bulged BP-A relative to the end of the branch helix. Such an interaction is supported by previous RNA-protein cross-linking (31, 32) and XL-MS data (12, 23). A29 of the U2 snRNA inserts into the same pocket where adenine was placed in the co-crystal structure (30) and stacks against residue Y22 of SF3B6 (Fig. 4E). Moreover, SF3B6 is oriented in such a way that its disordered N-terminus points towards BP-A and is close enough to explain previous cross-linking data (20, 33). Our data show that SF3B6 plays a previously unknown role in stabilising branch helix, which could be particularly relevant for branch sequences with poor complementarity to the U2 snRNA.
HTATSF1 stabilises the BSL in 17S U2 snRNP
Comparison of the 17S and A-like U2 snRNP shows that binding of SF3B6 and HTATSF1RRM to SF3B1HEAT are mutually exclusive and HTATSF1RRM needs to be displaced before stable docking of SF3B6 (Fig. 4B). Our reconstruction of the 17S U2 snRNP shows that HTATSF1RRM binds in a hydrophobic groove formed by HEAT repeats H15 and H16 of SF3B1 (Fig. 4C). The neighbouring H16 and H17 repeats form the interface for the C-terminus of the HTATSF1 linker helix (HTATSF1LH), comprising residues 239-251 (Fig. 4D). The C-terminus of HTATSF1LH points towards the BSL and a globular density nearby which likely belongs to the HTATSF1UHM domain that is known to bind the SF3B1ULM motif (Fig. 3d)(12, 34, 35). Therefore, the two domains of HTATSF1 form stable interfaces with SF3B1 and flank the U2 snRNA BSL from both sides suggesting a direct stabilization mechanism for this transient RNA secondary structure.
Interestingly, movement of the BSL correlates with the disappearance of the extra density on top of HTATSF1RRM and presumably the short variant of the U2 stem-loop I structure (Fig. 3C, Movie S1). Given the concerted movement with other U2 snRNA elements we speculate that at least part of this density could belong to the 5’-end of the U2 snRNA, especially that it occupies the surface that is typically involved in RRM-RNA binding. Indeed, the Y48D mutation (Y136 in HTATSF1) in the yeast homologue Cus2, abolishes U2 snRNA binding (5). Recombinant HTATSF1RRM exhibits some non-specific affinity for RNA (Fig. S8). This supports the hypothesis that interaction between HTATSF1RRM and the 5’-end of the U2 snRNA could additionally stabilise the BSL in an indirect manner by preventing BMSL formation.
Two-step conformational change in SF3B1 upon pre-mRNA binding
SF3B1 was previously reported to transition from an open to a closed conformation around a hinge between HEAT repeats H15 and H16 (36) Similar remodelling occurs in our in vitro system, with no extra factors needed, even when the BPS oligo is incubated on ice with the 17S U2 snRNP in the presence of AMP-PCP, a non-hydrolysable ATP analogue (Table 1, Fig. S7). This indicates that branch helix formation is the only driving force for the rearrangement around the first hinge region and that it does not depend on ATP hydrolysis.
Although the hinge-like movement of SF3B1 is reconstituted in our system, the conformation of the N-terminal part of the HEAT repeat differs significantly from any of the previously reported states. In the closed conformation, SF3B1 helix H1 (residues 509-523) inserts into the major groove of the branch helix providing additional stabilisation for the branch helix, while in the A-like complex it remains ~20 Å away from this binding site (Fig. 4B). We refer to this new SF3B1 conformation as half-closed, following the previous convention. The movement from half-closed to closed is different from the hinge-like closure and involves multiple small changes in the curvature of the HEAT repeats in its N-terminal part (H1-H12) (Fig. 4B). It is possible that binding of the intron sequence downstream from the BS could facilitate complete closure of the SF3B1.
Discussion
Recognition of the branch point sequences by the U2 snRNP is a critical step of spliceosome assembly. In this work, we used a minimal in vitro system to analyse the structure of the human U2 snRNP and its conformational changes upon ATP-dependent remodelling and engagement with the pre-mRNA substrate.
The 17S U2 snRNP structure shows that HTATSF1 and BSL stabilise each other in two distinct ways. Directly, through interactions between BSL and HTATSF1LH/UHM and indirectly, via a possible association of the 5’-end of the U2 snRNA with the HTATSF1RRM, which would prevent formation of RNA structures that compete with the BSL, i.e. the long variant of the SLI or the BMSL.
Our data show that a model BPS oligonucleotide can engage in base-pairing interaction with the U2 snRNP without requirement for prior remodelling. Although formation of complex A has been shown to be ATP-dependent in HeLa cell nuclear extract, Amin complex can form without ATP when the sequence upstream of the BS is missing (27, 37). This could be due to the absence of certain BS binding proteins (e.g. SF1) or lack of topological restraints for branch helix formation. To form the branch helix, U2 snRNA has to wind around the long pre-mRNA substrate and it is possible that ATP is required to liberate the 5’-end of the U2 snRNA from HTATSF1 to allow that. Indeed, in yeast, the ATPase activity of the DDX46 homologue Prp5 is required for complex A formation, but deletion of the HTATSF1 homologue Cus2 removes this dependence(8, 34). However, the ATP-dependent branch site fidelity control by Prp5 remains unchanged in the absence of Cus2, suggesting more complex function of this protein.
The structure of the A-like U2 snRNP captured SF3B6 interacting with the branch helix, which has two major implications. Firstly, it provides a specific binding site for the U2 snRNA in addition to SLIIa and SF3A2ZnF, which imposes helical geometry on the U2 snRNA within the branch helix binding pocket. This provides a mechanism for the stabilisation of weak branch point sequences, as those found in mammals, even in the absence of extensive complementarity. Secondly, SF3B6 binds at the junction of the branch helix duplex and the single stranded region of the U2 snRNA, therefore it defines the length of the branch helix and the exact position of the bulged BP-A relative to its end. It has been previously shown in an orthogonal yeast system that the position of the BP-A within the branch helix is critical for productive splicing (38). In budding yeast, the BS has evolved to be highly conserved and sequence complementarity between BS and U2 snRNA ensures proper positioning of the BP-A(39). Weak BS sequence conservation and base-pairing potential in other organisms, including mammals and fission yeast (22), require an additional BP-A positioning mechanism, which is fulfilled by SF3B6. Consequently, SF3B6 is conserved in many species with low BS conservation, but not in S. cerevisiae (Fig. S9).
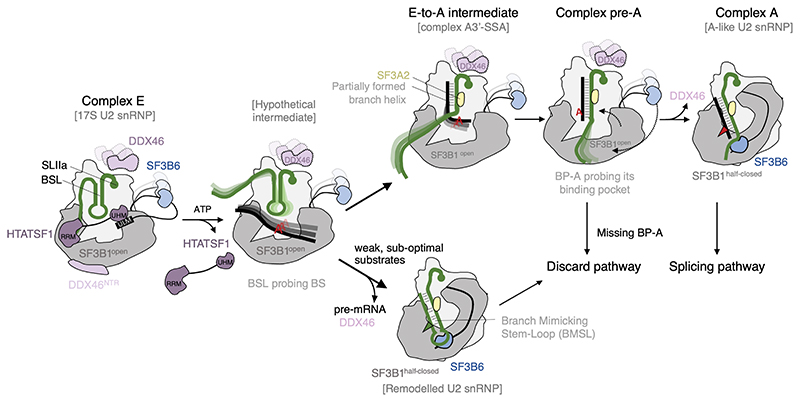
The emerging data suggest that the transition from E to A complex requires ATP-dependent displacement of HTATSF1, which destabilises the BSL and allows it to probe BS sequences(40) (Fig.5). The absence of HTATSF1 creates competition between the branch helix and the BMSL structure within U2 snRNA, providing a mechanism for the selection of the branch helix stability. Formation of the BMSL would mean rejection of the potential BS sequences. Therefore, the structure of the remodelled U2 snRNP likely represents an intermediate on the discard pathway after sub-optimal substrate rejection. Such a state was predicted to exist in the framework of the kinetic proofreading model (9).
Figure 5. Schematic model of branch site recognition by the U2 snRNP based on recent structural data.
U2 snRNP associated with spliceosomal complex E is likely structurally similar to the 17S U2 snRNP described by (12) and in this work. Dissociation of HTATSF1 creates competition between the formation of a branch helix and the BMSL. Rejection of weak, sub-optimal substrates results in the remodelled U2 snRNP, which is targeted to a discard pathway (this work). Stable substrates gradually form the branch helix as shown in the E-to-A (41) and pre-A (42) intermediates. In the absence of properly positioned, bulged out BP-A, the pre-A complex is targeted to a discard pathway. Productive engagement of the branch helix leads to the formation of complex A, wherein U2 snRNP is structurally similar to the A-like U2 snRNP (this work).
BS sequences that withstand competition with the BMSL would continue to progressively form the branch helix via a recently proposed toe-hold strand invasion mechanism (41). An intermediate state in this process (A3’-SSA complex) was captured by blocking spliceosome assembly with spliceostatin A (SSA) (41), which trapped U2 snRNP with a partially formed branch helix, missing bulged out BP-A. Consequently, the branch helix was not accommodated in its pocket and SF3B1 remains in the open conformation, resembling that found in the 17S U2 snRNP (Fig. 5).
Without inhibition by SSA, BS sequences would continue to fully form the branch helix. At this point another checkpoint would be reached. If a bulged out BP-A is present, it will bind the pocket in SF3B1, causing transition to the half-closed conformation and dissociation of DDX46, as shown in the A-like U2 snRNP. However, in the absence of a properly positioned BP-A, SF3B1HEAT remains in the open conformation and the spliceosome is stalled, as shown in the structure of the yeast pre-A complex (42). In this complex, Prp5 provides steric hindrance for the next step of spliceosome assembly, recruitment of the tri-snRNP(10). A prolonged block by Prp5 will likely initiate a discard pathway.
The remodelled U2 snRNP described in this work and the pre-A complex(42) are two distinct intermediates that direct sub-optimal substrates to the discard pathway. They represent different checkpoints in BS fidelity control, ensuring both, the formation of a stable branch helix, and the presence of properly positioned BP-A (Fig. 5).
Only properly positioned bulged out BP-A can bind the SF3B1-PHF5A pocket and cause the transition to the half-closed conformation of SF3B1, as observed in the A-like U2 snRNP. During this transition an extensive interaction surface forms between the branch helix, including the bulged BP-A and the HEAT repeats H15 to H19. Our minimal system shows that this interaction is the sole driving force for the SF3B1 hinge-like movement. It is not clear which factors are needed for the second phase of the transition from half-closed to closed SF3B1 conformation and when this conformational change occurs.
Upon A complex formation, poorly defined branch sequences would benefit from stabilisation by SF3B6, which enforces helical geometry of the U2 snRNA, even in the absence of extensive branch site complementarity. During subsequent steps of the spliceosome assembly, SF3B6 has to relocate to its binding site observed in the Bact complex (21), as its position in the A-like U2 snRNP would clash with Prp8 and prevent early Bact formation.
Our data provide several high-resolution snapshots of the complex process of branch site recognition by the U2 snRNP and contribute to a better understanding of the mechanism of pre-mRNA splicing in humans.
Materials and Methods
Generation of a knock-in cell line expressing endogenously GFP-tagged HTATSF1
FreeStyle 293-F Cells (Thermo Fisher Scientific) were edited to express EGFP-HTATSF1 fusion protein using a modification of a previously described CRISPR/Cas9 knock-in protocol (43). Two gRNAs were designed using the Benchling.com CRISPR gRNA design tool (Benchling; gRNA1: AAACATGAGCGGCACCAACT, gRNA2: TCATGTTTCCTACCTAGCTC) and were cloned into the plasmid pX335-U6-Chimeric_BB-CBh-hSpCas9n(D10A) (44), a gift from F. Zhang (Addgene plasmid #42335). The 800 bp sequences flanking the HTATSF1 start codon were obtained by PCR on genomic DNA obtained from FreeStyle 293-F cells using the PureLink™ Genomic DNA Mini Kit (Thermo Fisher Scientific) and subcloned into pUC19 (45), a gift from Joachim Messing (Addgene plasmid #50005; http://n2t.net/addgene:50005; RRID:Addgene_50005) digested with HindIII and EcoRI using Gibson Assembly (NEB). The homology-directed repair (HDR) donor plasmid was generated by cloning the sequence of the 3xHA-EGFP-3C protease site-SBP tag and the downstream flanking sequence into the pUC19-Upstream homology arm plasmid. FreeStyle 293-F Cells were grown adherently in DMEM medium supplemented with 10% FBS and GlutaMAX (all Thermo Fisher Scientific) and transfected with the HDR donor and the two pX335 plasmids using LipoD293 transfection agent (SignaGen Laboratories). 7 days post-transfection, after several passages, cells were suspended by treatment with trypsin-EDTA 0.05% (Thermo Fisher Scientific) and subjected to fluorescence-assisted cell sorting (FACS) using a BD FACSAria IIu (BD Biosciences). Cells expressing the EGFP-tag were sorted into 96-well plates. After approximately two weeks, wells with homogeneous fluorescence were transferred to 6-well plates and analyzed by western blotting for homozygous knock-in of the tag using an anti-HTATSF1 antibody (C-4; sc-514351; Santa Cruz) and an anti-HA tag antibody (F-7; sc-7392 HRP; Santa Cruz). A positive clone was selected and adapted to growth in FreeStyle suspension media (Thermo Fisher Scientific).
All adherent cells were incubated at 37°C in a humidified 5% CO2 atmosphere, all suspension cells in a non-humidified 8% CO2 atmosphere in shaker flasks.
Generation of a cell line for the inducible expression of GFP-DDX46
The ORF of DDX46 (NM_014829.4) with a C-terminal SBP-3C protease site-3xHA-GFP tag was cloned together with the Tet-One system (Takara Bio) into the pCMV6 plasmid using Gibson Assembly. Expi293F cells (Thermo Fisher Scientific) were transfected in adherent culture with this plasmid, split after two days and selected with G418 antibiotic (Thermo Fisher Scientific) for two weeks before single colonies were picked, checked for doxycycline-inducible expression using western blot with anti-DDX46 antibodies (B-6; sc-514071; Santa Cruz), and resuspended in ESF SFM Mammalian Cell Culture Medium (Expression Systems) for large-scale growth. Expression of DDX46-EGFP was induced with 2 μg/ml doxycycline (Sigma).
Purification of the 17S U2 snRNP
Nuclear extract was prepared from 1 liter of cells at 2x106 cells/ml as described(46). The nuclear extract was incubated with ~0.5 ml of home-made CNBr-activated sepharose resin coupled to GFP nanobody(47). The resin was washed four times with purification buffer (20 mM HEPES-KOH pH 7.9, 150 mM KCl, 2 mM MgCl2, 5% glycerol) and eluted by adding purification buffer supplemented with 1:50 molar ratio of HRV 3C protease and incubating for 1 h on ice. The A-like U2 snRNP was obtained by incubating ~0.5 ml anti-GFP resin loaded with GFP-HTATSF1 nuclear extract with 500 μl of the purification buffer supplemented with 2 mM ATP and 0.2 μM BPS oligo at 30 °C for 1 h. The remodelled U2 snRNP was obtained in the same way, but omitting the BPS oligo. The AMP-PCP-treated A-like U2 snRNP was obtained by treating the 3C protease elution with 20 μl 100 mM AMP-PCP (final 2 mM) and 2 μl 100 μM BPS oligo (final 0.2 uM) in 500 μl purification buffer for ~1 h on ice.
The samples were loaded onto 4 ml 10-30% glycerol gradients containing 20 mM HEPES-KOH pH 7.9, 150 mM KCl, 2 mM MgCl2. For cryo-EM studies, gradients also contained a 0-0.025% glutaraldehyde gradient, as in the GraFix protocol(48). Gradients were prepared using the Biocomp 108 Gradient Mixer (Biocomp). After 6 h centrifugation at 259,000g, gradients were fractionated into 24 fractions of 180 μl and the crosslinker was quenched with 40 mM (NH4)HCO3. Fractions containing the 17S U2 snRNP were identified by SDS-PAGE using the Novex Tris-Glycine system (Thermo Fisher Scientific), pooled and supplemented with an additional 100 mM KCl. The sample was concentrated in 50 kDa MWCO Amicon concentrators (Merck KGaA, Darmstadt) and then desalted to remove glycerol with Zeba Spin Desalting Columns, 7K MWCO (Thermo Fisher Scientific) that were equilibrated with 20 mM HEPES-KOH pH 7.9, 250 mM KCl, 2 mM MgCl2.
In vitro reconstitution reactions
300 μl 50% v/v Anti-GFP resin slurry was incubated with GFP-HTATSF1 or GFP-DDX46 nuclear extract as described above, but not eluted. To test the dissociation of U2 snRNP from HTATSF1 or DDX46, the loaded resin was incubated at 30 °C for 30 min under different conditions. For each reaction, 20 μl slurry was incubated with either 40 μl purification buffer or purification buffer supplemented with 2 mM ATP, 1 μM BPS oligo or both. The BPS RNA oligo Cy5-CAGAUACUAACACUUGA was synthesised and HPLC-purified by Eurofins Genomics. As a control reaction, resin was incubated with purification buffer on ice. The reaction was resuspended by flicking the tube every 5 min. After incubation, the resin was sedimented by centrifugation and the top 20 μl supernatant taken as the elution and mixed with SDS-PAGE sample buffer. The remaining suspension was washed twice with 1 ml purification buffer. Almost all supernatant was removed and SDS-PAGE loading buffer was added to the resin. Both elution and resin samples were analyzed by SDS-PAGE followed by Western blotting. Blots were incubated with primary antibodies mouse anti-SNRPB2 OTI3A7 (TA808139, OriGene) or rabbit anti-SNRPA1 (STJ114054, St John’s Laboratory) to visualise U2 snRNP in the elution.
Western blotting
A PVDF membrane (Merck KGaA, Darmstadt) was activated for 10 min in 100% EtOH and equilibrated for 5 min in transfer buffer (25 mM Tris, 192 mM glycine, 20% EtOH). A wet transfer was performed for 80 min at 40 V in an Invitrogen XCell II Blot Module (Thermo Fisher Scientific). The membrane was blocked with 5% milk in PBS supplemented with 0.1% Tween 20 (hereafter referred to as PBST) for 1 h at room temperature. Primary antibodies were added and incubated for 1 h at room temperature. The membrane was washed 3 times for 5 min with PBST and incubated for 1 h with secondary antibody (Goat Anti-Mouse IgG H&L (HRP), 31430, Thermo Fisher Scientific; Goat Anti-Rabbit IgG H&L (HRP), ab205718, Abcam). The membrane was washed 3 times for 5 min with PBST and developed with Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific), followed by imaging with a ChemiDoc MP imager (Bio-Rad).
RNA oligonucleotide binding assays
The 3C protease elution from anti-GFP resin loaded with GFP-HTATSF1 nuclear extract, i.e. 17S U2 snRNP, at A280=Ø.Ø8 was incubated with 0.5 μM BPS oligo at different conditions for 30 min. The BPS oligo binding was tested under the following conditions: buffer on ice, buffer at 30 °C, 1 mM AMP-PCP (Sigma) at 30 °C, 1 mM ATP disodium salt hydrate (Sigma) at 30 °C.
The same experiment was performed with the ATP-treated complex, i.e. the remodelled U2 snRNP. Because the ATP interfered with photometric concentration determination, the concentration was equalized to the 17S U2 snRNP by using the same input material, Coomassie-stain SDS-PAGE (InstantBlue Coomassie Protein Stain; ab119211; Abcam) and confirmation by Western blot. The BPS oligo binding was tested under the following conditions: buffer at 30 °C, 1 mM ATP at 30 °C, and 1 mM ATP and 2 μM HTATSF1-containing gradient fractions at 30 °C. The HTATSF1-containing gradient fractions were the fractions 5 and 6 of a glycerol gradient run with the 3C protease elution after ATP incubation of the resin. ATP incubation as described for the purification of the remodelled U2 snRNP removed most of the U2 snRNP, allowing the isolation of HTATSF1 in the low-molecular weight fractions of the gradient.
After the reaction, the mix was applied to 4 ml 10-30% glycerol gradient as described before. After fractionation the Cy5-fluorescence (excitation filter 610-30 nm, emission filter 675-50 nm) in each fraction was measured using the CLARIOstar Plus microplate reader (BMG LABTECH).
Electrophoretic Mobility Shift Assay
Human HTATSF1RRM domain (residues 1-248) with a N-terminal His6-GFP-3C protease site tag was cloned into the pEC-K expression vector, a gift from Eva Kowalinski. Protein was expressed in E. coli BL21(DE3)-RIL cells (Agilent). Cultures were grown in LB medium at 37°C, induced with 0.4 mM β-d-1-thiogalactopyranoside (IPTG) at OD600=0.8, grown overnight at 18°C and harvested by centrifugation. For protein purification, the bacterial pellet was lysed by sonication in buffer containing 50 mM Tris-HCl pH 7.5, 300 mM NaCl, 20 mM imidazole, 5 mM 2-mercaptoethanol, 5% glycerol (buffer A) and protease inhibitor cocktail (Roche cOmplete). Lysate was clarified by centrifugation at 185,700g and the supernatant was loaded on a HisTrap column (GE Healthcare) equilibrated in buffer A. After a wash with buffer A containing 60 mM imidazole, protein was eluted with buffer A containing 300 mM imidazole. After overnight incubation with 3C protease the eluted fraction was applied to 0.5 ml of sepharose resin coupled to GFP nanobody. Flow-through fractions that contained the tag-free HTATSF1RRM domain were collected and applied to Superdex 75 size-exclusion column (GE Healthcare) that was equilibrated in 20 mM Tris-HCl pH 7.5, 300 mM NaCl, 1 mM DTT and 5% glycerol. Peak fractions were concentrated on 10 kDa MWCO Amicon centrifugal filter (Merck KGaA, Darmstadt) and used for the electrophoretic mobility shift assay.
RNA substrates were synthesised and HPLC-purified by Dharmacon. Full-length U2 snRNA was obtained by phenol-chloroform extraction and ethanol precipitation of 17S U2 snRNP.
RNA substrates were adjusted to a final concentration of 20 nM and mixed with serial dilutions of HTATSF1RRM in binding buffer that contained 10 mM Tris-HCl pH 8.0, 50 mM KCl, 0.4 mM EDTA, 0.8 mM MgCl2, 0.6 mM DTT, 10 % (v/v) glycerol, 0.005 % (w/v) bromophenol blue.
After 30 minutes of incubation on ice, the samples were resolved by native 7% TBE-PAGE for 45 minutes at 100 V and 4°C in case of the 3’-Cy5 labelled U2 snRNA 5’-end fragments (residues 1-24) with or without naturally occurring modifications (pseudouridylation and 2’-O methylation). For the full-length U2 snRNA, the samples were resolved on native 4-12% TBE PAGE gels (Invitrogen) for 45 minutes at 140 V and 4°C. The gel was stained with SYBR Gold (Invitrogen) in TBE buffer for 30 minutes. The reaction products were visualised with a ChemiDoc MP imager (Bio-Rad).
Protein identification via LC-MS/MS
Complexes were purified as described using glycerol gradients without crosslinker and 40 μl of peak fractions were prepared for LC-MS/MS using the SP3 protocol (49). All reagents for LC-MS/MS were prepared in 50 mM HEPES pH 8.5. First, cysteines were reduced using 10 mM dithiothreitol at 56 °C for 30 minutes. Samples were kept at 24 °C and alkylated with 20 mM 2-chloroacetamide at room temperature in the dark for 30 minutes. They were digested with trypsin (Promega), and the peptides were cleaned up using OASIS HLB μElution Plate (Waters).
The outlet of an UltiMate 3000 RSLC nano LC system (Dionex) fitted with a trapping cartridge (μ-Precolumn C18 PepMap 100, 5μm, 300 μm i.d. x 5 mm, 100 Å) and an analytical column (nanoEase™ M/Z HSS T3 column 75 μm x 250 mm C18, 1.8 μm, 100 Å, Waters) was coupled directly to the Orbitrap Fusion Lumos (Thermo Fisher Scientific) mass spectrometer using the nanoFlex source. The mass spectrometer was operated in positive mode with the capillary temperature set at 275°C. The peptides were introduced into the mass spectrometer via a Pico-Tip Emitter (360 μm OD x 20 μm ID, 10 μm tip; New Objective) with an applied spray voltage of 2.4 kV.
With the Orbitrap mass spectrometer in profile mode, in the 300-1500 m/z mass range full mass scans were acquired with a resolution of 120000. The filling time was set to a maximum of 250 ms with a limit of 2e5 ions. The instrument was operated in data-dependent acquisition (DDA) mode and MSMS scans were acquired in the Iontrap with scan rate set to rapid and normal mass range, with a fill time of up to 35 ms. A normalized collision energy of 30 was applied. MS2 data was acquired in centroid mode.
To analyse the data, IsobarQuant5 (50) and Mascot v2.2.07 (Matrix Science) were used. Data were searched against the Uniprot Homo sapiens proteome database (UP000005640), which also contained common contaminants and reversed sequences. The following modifications were included into the search parameters: Carbamidomethylation (C) (fixed modification), Acetylation (Protein N-term) and Oxidation (M) (variable modifications). For the full scan (MS1) a mass error tolerance of 10 ppm and for MS/MS (MS2) spectra of 0.02 Da was set. Trypsin was set as protease with a maximum of two missed cleavages, the minimum peptide length was seven amino acids, and at least two unique peptides were required for a protein identification. The false discovery rate on peptide and protein level was set to 0.01.
Negative-stain EM
Negative-stain EM was used to check fractions of the GraFix gradient. For the preparation of negative-stain grids, CF300-Cu (Electron Microscopy Sciences) were glow-discharged for 30 s at 25 mA at 0.3 bar using the Pelco EasiGlow. 3 μl sample were applied and incubated for 30 s, then washed twice in a drop of water, once in 1.5% (w/v) solution of uranyl acetate and then incubated for 30s in uranyl acetate solution, before all liquid was blotted away and the grid was dried. The grid was imaged using a Tecnai G2 Spirit BT microscope (Thermo Fisher Scientific) operating at 120 kV.
Cryo-EM sample preparation
GraFix gradient crosslinked, desalted U2 snRNP samples were applied to EM grids glow-discharged on each side for 30 s at 25 mA at 0.3 bar using the Pelco EasiGlow. The sample concentration was adjusted to 0.25 A280nm with 20 mM HEPES-KOH pH 7.9, 250 mM KCl, 2 mM MgCl2. Grids were blotted and plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific) at 4°C, 100% humidity. For the 17S U2 snRNP, 2 μl sample was applied to each side of Quantifoil Au 300 mesh R1.2/1.3 grids (Quantifoil Micro Tools) before blotting 3 s at 10 Force. For the A-like U2 snRNP, 1 μl sample was applied to each side of Quantifoil Au 300 mesh R1.2/1.3 grids before blotting 1 s at 0 Force, and then a second 1 μl sample was applied to each side before blotting for 5 s 0 Force. For the remodelled U2 snRNP, 1 μl sample was applied to each side of UltrAufoil Au 300 mesh R1.2/1.3 grids (Quantifoil Micro Tools) before blotting 5 s at 5 Force. For the AMP-PCP-treated A-like U2 snRNP, 1 μl sample was applied to each side of UltrAufoil Au 300 mesh R1.2/1.3 grids (Quantifoil Micro Tools) before blotting 5 s at 5 Force.
Cryo-EM data collection and analysis
Grids were screened using a Glacios Cryo-TEM equipped with a Falcon 3EC Direct Electron Detector (Thermo Fisher Scientific). The 17S, A-like, and remodelled U2 snRNP data were collected on a Titan Krios TEM (Thermo Fisher Scientific) operated at 300 kV, equipped with a Quantum energy filter (Gatan) and a Gatan K3 Camera (51). A magnification of 130,000x was used, corresponding to a pixel size of 0.64 Å/pixel. Automated data acquisition was performed using SerialEM (52). Movies were recorded for 1 s, fractionated into 40 frames, with defocus values from -0.8 to -1.8 μm (0.1 μm steps). For the 17S U2 snRNP, 15,531 movies were recorded with 53.45 e/Å2 total dose. For the A-like U2 snRNP, 15,681 movies were recorded with 52.03 e/Å2 total dose. For the remodelled U2 snRNP, 7,809 movies were recorded with 50.76 e/Å2 total dose.
For the AMP-PCP-treated A-like U2 snRNP 2,230 movies were collected using a Glacios microscope with Falcon 3EC in electron-counting (EC) mode with a magnification of 150,000x (0.94 Å pixel size) and 40 e/Å2 total dose fractionated into 40 frames.
All image processing was performed within Relion 3.1 (53) unless otherwise stated. All three datasets were motion corrected using Relion’s implementation of MotionCorr2 1.4 (54) with a 5x5 patch model without binning followed by CTF estimation with CTFFIND 4.1.14 (55). Particles were picked in WARP (56) using a general model.
For the 17S U2 snRNP, 1,500,165 particles were extracted with two-fold binning (640 pixel original box size, 320 pixel binned box size, 1.28 Å/pixel). 784,879 of these particles were exported to cryoSPARC (Structura Biotechnology; (57)) to generate an initial model. In cryoSPARC, 326,150 particles were selected in 2D classification and used for ab initio reconstruction to generate 3 classes. These were heterogeneously refined to generate one class with 118,122 particles which was refined with Homogenous Refinement and then Local Refinement to a resolution of 3.3 Å. This reconstruction was low-pass filtered to 30 Å and used as a reference for 3D classification of all particles in Relion (performed in 2 batches). Each 3D classification led to one good class, which after merging yielded 697,430 particles. This dataset was auto-refined to 3 Å. After two rounds of bayesian polishing, including expansion of the box size to 640 pixel and of the pixel size to 0.64 Å/pixel, and two rounds of CTF refinement, a resolution of 2.45 Å was reached (Medium Resolution 17S U2 snRNP map). After one more round of bayesian polishing and CTF refinement, the high-resolution part of SF3b was 3D classified without image alignment and a T value of 100. One class with 225,943 particles showed good density. It was polished once more and refined with SIDESPLITTER (58) to a resolution of 2.35 Å. For the HEAT-focussed classification, the particles of the refinement that led to the medium-resolution 17S U2 snRNP map were 3D classified without image alignment and a T value of 100. One class with 152,253 particles was refined to 2.9 Å (HEAT-focussed map).
For the A-like U2 snRNP, 1,470,005 particles were extracted with two-fold binning (640 pixel original box size, 320 pixel binned box size, 1.28 Å/pixel) and were exported to cryoSPARC.
In cryoSPARC, 302,816 particles were selected after 2D classification and used for ab initio reconstruction to generate 3 references for heterogeneous refinement with all particles. One class containing 658,325 particles was refined with homogenous refinement and then local refinement to a resolution of 2.7 Å. The resulting volume and particle file including orientations were imported to Relion. The volume was low-pass filtered to 40 Å and used as a reference for 3D refinement. After two rounds of bayesian polishing, including expansion of the box size to 640 pixel and of the pixel size to 0.64 Å/pixel, and three rounds of CTF refinement, a resolution of 2.26 Å was reached.The high resolution part of SF3b was 3D classified without image alignment and a T value of 100. One class with 249,011 particles showed good density. It was polished and CTF refined once more and refined using SIDESPLITTER to a resolution of 2.13 Å, producing the A-like U2 snRNP map.
For the remodelled U2 snRNP, 800,711 particles were extracted with two-fold binning (640 pixel original box size, 320 pixel binned box size, 1.28 Å/pixel) and were exported to cryoSPARC. In cryoSPARC, 14,522 particles were selected in 2D classification and used for ab-initio reconstruction to generate four references (J8vol0, J8vol1, J8vol2, J8vol3). All 800,711 particles were used for heterogeneous refinement using as references the all four references and generating four classes (J9vol0, J9vol1, J9vol2, J9vol3). J9vol0 showed the structure of the U2 snRNP SF3b lobe and was thus refined to 3 Å (J12vol), followed by a heterogeneous refinement using J8vol1, J8vol2, J8vol3, and J12vol as references and generating four classes (J15vol0, J15vol1, J15vol2, J15vol3). J15vol2 showed the 17S U2 snRNP and was refined to 3.7 Å (J23vol), while J15vol3 showed the remodelled U2 snRNP and was refined to 2.9 Å (J24vol). J23vol converged on the wrong handedness and was flipped over the z-axis using cryoSPARC’s volume tools to generate J30vol. All 800,711 particles were also used for heterogeneous refinement using as references the good class J8vol0 and three times the noise class J8vol2. This generated J11vol0, which was dominated by noise containing stripes, likely generated by particles close to the micrograph edge. Combining the information that the dataset contained 17S U2 snRNP, remodelled U2 snRNP and many particles that cause a striped volume, a final heterogeneous refinement was performed. All 800,711 particles were used for heterogeneous refinement using as references J8vol1, J8vol2, J8vol3, twice J11vol0, J30vol, and J24vol and generating J29class0-6. J29class5 showed the 17S U2 snRNP and was refined to 4 Å (J47vol), then subjected to heterogeneous refinement using three times J47vol as reference, class 0 was homogenously refined to 3.5 Å (J51). J29class6 showed the remodelled U2 snRNP and was refined using local refinement to 2.7 Å (J32). The output was once again heterogeneously refined with the same references as J29 to remove the last impurities. Class 5 was refined to 2.6 Å. The resulting volume and particle file including orientations were imported to Relion. The volume was low-pass filtered to 20 Å and used as a reference for 3D refinement. After three rounds of bayesian polishing, including expansion of the box size to 640 pixel and of the pixel size to 0.64 Å/pixel, and three rounds of CTF refinement, a resolution of 2.21 Å was reached.
The refined datasets of 17S U2 snRNP, A-like U2 snRNP and remodelled U2 snRNP were independently refined to the A-like U2 snRNP as the reference and merged. The merged dataset was refined with a tight mask around the common structure of the U2 snRNP. After bayesian polishing of the particles, a 2.0 Å merged map was obtained. The particles were then separated into the three datasets again. The maps of 17S U2 snRNP and remodelled U2 snRNP were refined to their final 2.3 Å and 2.2 Å resolutions, respectively.
For the AMP-PCP-treated A-like U2 snRNP all processing was performed in cryoSPARC. 320,883 particles were extracted and subjected to two rounds of heterogeneous refinement. The selected 63,915 particles were refined to 3.3 Å using non-uniform refinement. All (local) resolutions of the AMP-PCP-treated A-like U2 snRNP were calculated in cryoSPARC.
Model building and validation
Templates for model building were acquired as described in Supplementary Table 2. The templates were rigid-body fitted into the maps in ChimeraX (59). The models were then refined into the maps using ISOLDE(60). Subsequently, Coot was used to add protein or RNA residues(61). The RNA part of each structure was refined in ISOLDE without noncanonical bases, modified nucleotides were added and real-space refined in Coot. The crystal structure of SF3B6:SF3B1(394-415; PDB: 3LQV) was fitted into density by rigid-body fitting and the residues at the interface with SF3B1HEAT and U2 snRNA were refined in real space in Coot.
Distance restraints for the protein part were generated using ProSMART(62) and using LIBG for the RNA(63). Models were refined to the high-resolution maps using REFMAC5(64) and validated using the wwPDB OneDep System. Additional validation statistics were obtained using MolProbity(65). Atomic models were visualised with ChimeraX or PyMol (Schrödinger).and the merged SF3b map are likely caused by the tight mask for local refinement. (B) Masked map-to-model FSC curves. (C) Unmasked map-to-model FSC curves. (D) Angular distribution of particles used in the reconstruction of the maps. (E)-(G), Overview of the modelled proteins and RNA in the structure of the 17S (E), remodelled (F), and A-like U2 snRNP (G). Filled in rectangles denote the sequences that were modelled, opaque rectangles were modelled by fitting a crystal structure, empty rectangles are parts of the proteins and RNA that could not be assigned to the reconstructed maps. Selected domains important for this work are indicated.
Supplementary Material
One-Sentence Summary.
High-resolution structures provide mechanistic insights into intron branch site recognition by the human spliceosome.
Footnotes
Author contributions:
Conceptualization: JT, WPG
Methodology: JT, MR, FW, WPG
Investigation: JT, MR
Visualization: JT, WPG
Funding acquisition: WPG
Project administration: JT, WPG
Supervision: WPG
Writing – original draft: JT, WPG
Writing – review & editing: JT, MR, FW, WPG
Competing interests: Authors declare that they have no competing interests.
Acknowledgments
We thank Moritz Pfleiderer and Daniel Peter for experimental advice; Angelique Fraudeau and Estelle Marchal for technical assistance; Sarah Schneider, Erika Pellegrini and Wim Hagen for ensuring smooth running of the EMBL cryo-EM facilities; EMBL Proteomic Core Facility for performing mass spectrometry experiments; Martin Pelosse and Alice Aubert (EMBL EEF facility) for their assistance with cell culture; Mylene Pezet (IAB Grenoble Flow Cytometry Platform) for cell sorting; Aymeric Peuch and the EMBL Grenoble IT team for the support with high-performance computing; Charles Query for an insightful discussion; Ra Pillai and Sebastian Fica for critical comments on the manuscript.
Funding
This project has received funding from the European Research Council (ERC) the European Union’s Horizon 2020 research and innovation programme (Grant agreeme 950278, awarded to W.P.G.)
Data and materials availability:
Cryo-EM maps were deposited in the EMDB with the following accession codes:
EMD-13793 (17S U2 snRNP core);
EMD-13810 (17S U2 snRNP HEAT repeats);
EMD-13811 (A-like U2 snRNP);
EMD-13813 (A-like U2 snRNP medium resolution/SF3B6 map);
EMD-13812 (Remodelled U2 snRNP);
EMD-13815 (Merged datasets – the highest resolution map);
EMD-13814 (AMP-PCP A-like U2 snRNP).
Atomic coordinates were deposited in the PDB database with the following accession codes: 7Q3L (17S U2 snRNP);
7Q4O (A-like U2 snRNP);
7Q4P (Remodelled U2 snRNP)
Materials generated in this study are available on request from the lead contact (wgalej @embl.fr)
References and Notes
- 1.Valcárcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA [corrected] Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 2.Michaud S, Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- 3.Das R, Zhou Z, Reed R. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol Cell. 2000;5:779–787. doi: 10.1016/s1097-2765(00)80318-4. [DOI] [PubMed] [Google Scholar]
- 4.Wu J, Manley JL. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989;3:1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- 5.Yan D, et al. CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol Cell Biol. 1998;18:5000–5009. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Day CL, Dalbadie-McFarland G, Abelson J. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J Biol Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- 7.Will CL, et al. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21:4978–4988. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perriman R, Barta I, Voeltz GK, Abelson J, Ares M. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc Natl Acad Sci USA. 2003;100:13857–13862. doi: 10.1073/pnas.2036312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y-Z, Query CC. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell. 2007;28:838–849. doi: 10.1016/j.molcel.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang W-W, Cheng S-C. A novel mechanism for Prp5 function in prespliceosome formation and proofreading the branch site sequence. Genes Dev. 2015;29:81–93. doi: 10.1101/gad.253708.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perriman R, Ares M. Invariant U2 snRNA nucleotides form a stem loop to recognize the intron early in splicing. Mol Cell. 2010;38:416–427. doi: 10.1016/j.molcel.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, et al. Molecular architecture of the human 17S U2 snRNP. Nature Publishing Group. 2020;583:310–313. doi: 10.1038/s41586-020-2344-3. [DOI] [PubMed] [Google Scholar]
- 13.Rauhut R, et al. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science. 2016;353:1399–1405. doi: 10.1126/science.aag1906. [DOI] [PubMed] [Google Scholar]
- 14.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science. 2016;353:904–911. doi: 10.1126/science.aag0291. [DOI] [PubMed] [Google Scholar]
- 15.Galej WP, et al. Cryo-EM structure of the spliceosome immediately after branching. Nature. 2016;537:197–201. doi: 10.1038/nature19316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- 17.Will CL, et al. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 2001;20:4536–4546. doi: 10.1093/emboj/20.16.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Q, et al. SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing. Genes Dev. 2016;30:2710–2723. doi: 10.1101/gad.291872.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrocci TJ, Zoerner DM, Paulson JC, Hoskins AA. SF3b1 mutations associated with myelodysplastic syndromes alter the fidelity of branchsite selection in yeast. Nucleic Acids Research. 2017;45:4837–4852. doi: 10.1093/nar/gkw1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacMillan AM, et al. Dynamic association of proteins with the pre-mRNA branch region. Genes Dev. 1994;8:3008–3020. doi: 10.1101/gad.8.24.3008. [DOI] [PubMed] [Google Scholar]
- 21.Haselbach D, et al. Structure and Conformational Dynamics of the Human Spliceosomal Bact Complex. Cell. 2018;172:454–464.:e11. doi: 10.1016/j.cell.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Taggart AJ, et al. Large-scale analysis of branchpoint usage across species and cell lines. Genome Research. 2017;27:639–649. doi: 10.1101/gr.202820.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cretu C, et al. Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations. Mol Cell. 2016;64:307–319. doi: 10.1016/j.molcel.2016.08.036. [DOI] [PubMed] [Google Scholar]
- 24.Krämer A, Grüter P, Gröning K, Kastner B. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J Cell Biol. 1999;145:1355–1368. doi: 10.1083/jcb.145.7.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, et al. Structure of the human activated spliceosome in three conformational states. Cell Res. 2018;28:307–322. doi: 10.1038/cr.2018.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plaschka C, Lin P-C, Nagai K. Structure of a pre-catalytic spliceosome. Nature Publishing Group. 2017;546:617–621. doi: 10.1038/nature22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Query CC, McCaw PS, Sharp PA. A minimal spliceosomal complex A recognizes the branch site and polypyrimidine tract. Mol Cell Biol. 1997;17:2944–2953. doi: 10.1128/mcb.17.5.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Folco EG, Coil KE, Reed R. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes Dev. 2011;25:440–444. doi: 10.1101/gad.2009411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhan X, Yan C, Zhang X, Lei J, Shi Y. Structures of the human pre-catalytic spliceosome and its precursor spliceosome. Cell Res. 2018;28:1129–1140. doi: 10.1038/s41422-018-0094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schellenberg MJ, Dul EL, MacMillan AM. Structural model of the p14/SF3b155 o branch duplex complex. RNA. 2011;17:155–165. doi: 10.1261/rna.2224411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dybkov O, et al. U2 snRNA-protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol Cell Biol. 2006;26:2803–2816. doi: 10.1128/MCB.26.7.2803-2816.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spadaccini R, et al. Biochemical and NMR analyses of an SF3b155-p14-U2AF-RNA interaction network involved in branch point definition during pre-mRNA splicing. RNA. 2006;12:410–425. doi: 10.1261/rna.2271406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schellenberg MJ, et al. Crystal structure of a core spliceosomal protein interface. Proc Natl Acad Sci USA. 2006;103:1266–1271. doi: 10.1073/pnas.0508048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talkish J, et al. Cus2 enforces the first ATP-dependent step of splicing by binding to yeast SF3b1 through a UHM-ULM interaction. RNA. 2019;25:1020–1037. doi: 10.1261/rna.070649.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loerch S, et al. The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface. J Biol Chem. 2019;294:2892–2902. doi: 10.1074/jbc.RA118.006764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cretu C, et al. Structural Basis of Splicing Modulation by Antitumor Macrolide Compounds. Mol Cell. 2018;70:265–273.:e8. doi: 10.1016/j.molcel.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 37.Newnham CM, Query CC. The ATP requirement for U2 snRNP addition is linked to the pre-mRNA region 5’ to the branch site. RNA. 2001;7:1298–1309. doi: 10.1017/s1355838201010561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith DJ, Konarska MM, Query CC. Insights into branch nucleophile positioning and activation from an orthogonal pre-mRNA splicing system in yeast. Mol Cell. 2009;34:333–343. doi: 10.1016/j.molcel.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sales-Lee J, et al. Coupling of spliceosome complexity to intron diversity. Curr Biol. 2021 doi: 10.1016/j.cub.2021.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kao C-Y, Cao E-C, Wai HL, Cheng S-C. Evidence for complex dynamics during U2 snRNP selection of the intron branchpoint. Nucleic Acids Research. 2021 doi: 10.1093/nar/gkab695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cretu C, et al. Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors. Nat Commun. 2021;12:4491–15. doi: 10.1038/s41467-021-24741-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, et al. Structural insights into how Prp5 proofreads the pre-mRNA branch site. Nature Publishing Group. 2021;596:296–300. doi: 10.1038/s41586-021-03789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koch B, et al. Generation and validation of homozygous fluorescent knock-in cells using CRISPR-Cas9 genome editing. Nat Protoc. 2018;13:1465–1487. doi: 10.1038/nprot.2018.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 46.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Research. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fridy PC, et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat Methods. 2014;11:1253–1260. doi: 10.1038/nmeth.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kastner B, et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5:53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 49.Hughes CS, et al. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol Syst Biol. 2014;10:757. doi: 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franken H, et al. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat Protoc. 2015;10:1567–1593. doi: 10.1038/nprot.2015.101. [DOI] [PubMed] [Google Scholar]
- 51.Weis F, Hagen WJH. Combining high throughput and high quality for cryo-electron microscopy data collection. Acta Crystallogr Sect D Struct Biol. 2020;76:724–728. doi: 10.1107/S2059798320008347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. Journal of Structural Biology. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Zivanov J, Nakane T, Scheres SHW. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ. 2020;7:253–267. doi: 10.1107/S2052252520000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. Journal of Structural Biology. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tegunov D, Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods. 2019;16:1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 58.Ramlaul K, Palmer CM, Nakane T, Aylett CHS. Mitigating local over-fitting during single particle reconstruction with SIDESPLITTER. Journal of Structural Biology. 2020;211:107545. doi: 10.1016/j.jsb.2020.107545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen EF, et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Croll TI. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr Sect D Struct Biol. 2018;74:519–530. doi: 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Emsley P, Cowtan K. IUCr, Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 62.Nicholls RA, Fischer M, McNicholas S, Murshudov GN. IUCr, Conformationindependent structural comparison of macromolecules with ProSMART. Acta Crystallogr Sect D Biol Crystallogr. 2014;70:2487–2499. doi: 10.1107/S1399004714016241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr Sect D Biol Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr Sect D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams CJ, et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altschul SF, Lipman DJ. Protein database searches for multiple alignments. Proc Natl Acad Sci USA. 1990;87:5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Tommaso P, et al. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Research. 2011;39:W13–7. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research. 2014;42:W320–4. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Irimia M, Roy SW. Evolutionary convergence on highly-conserved 3’ intron structures in intron-poor eukaryotes and insights into the ancestral eukaryotic genome. PLoS Genet. 2008;4:e1000148. doi: 10.1371/journal.pgen.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps were deposited in the EMDB with the following accession codes:
EMD-13793 (17S U2 snRNP core);
EMD-13810 (17S U2 snRNP HEAT repeats);
EMD-13811 (A-like U2 snRNP);
EMD-13813 (A-like U2 snRNP medium resolution/SF3B6 map);
EMD-13812 (Remodelled U2 snRNP);
EMD-13815 (Merged datasets – the highest resolution map);
EMD-13814 (AMP-PCP A-like U2 snRNP).
Atomic coordinates were deposited in the PDB database with the following accession codes: 7Q3L (17S U2 snRNP);
7Q4O (A-like U2 snRNP);
7Q4P (Remodelled U2 snRNP)
Materials generated in this study are available on request from the lead contact (wgalej @embl.fr)