Summary
Most mitochondrial proteins are synthesized as precursors in the cytosol and post-translationally transported into mitochondria. The mitochondrial surface protein Tom70 acts at the interface of the cytosol and mitochondria. In vitro import experiments identified Tom70 as targeting receptor, particularly for hydrophobic carriers. Using in vivo methods and high-content screens, we revisit the question of Tom70 function and considerably expand the set of Tom70-dependent mitochondrial proteins. We demonstrate that the crucial activity of Tom70 is its ability to recruit cytosolic chaperones to the outer membrane. Indeed, tethering an unrelated chaperone-binding domain onto the mitochondrial surface complements most of the defects caused by Tom70 deletion. Tom70-mediated chaperone recruitment reduces the proteotoxicity of mitochondrial precursor proteins, particularly of hydrophobic inner membrane proteins. Thus, our work suggests that the predominant function of Tom70 is to tether cytosolic chaperones to the outer mitochondrial membrane, rather than to serve as a mitochondrion-specifying targeting receptor.
Graphical Abstract.
Introduction
With a concentration of 30,000 to 50,000 ribosomes per μm3, the eukaryotic cytosol is densely packed with molecular machines for protein synthesis that make up a considerable fraction of its total volume (Marini et al., 2020). Rapid and efficient protein folding in the cytosol is of pivotal importance for rapidly growing cells. Chaperones, particularly those of the Hsp70 and Hsp90 family, with the assistance of different co-chaperones and accessory factors, bind to nascent chains as soon as they emerge from the ribosome in order to facilitate their folding (Hartl et al., 2011; Kramer et al., 2019; Sontag et al., 2017) or hold them in a translocation-competent state for transport across membranes of organelles (Deshaies et al., 1988; Jores et al., 2018; Hoseini et al., 2016; Young et al., 2003).
Mitochondria are essential organelles of eukaryotic cells. They synthesize a handful of very hydrophobic polypeptides on mitochondrial ribosomes in the matrix. All other 900 (yeast) to 1,500 (humans) mitochondrial proteins are encoded by nuclear genes and synthesized in the cytosol (Vögtle et al., 2017; Morgenstern et al., 2017; Calvo et al., 2016). With the exception of a small number of inner membrane proteins (Williams et al., 2014; Tsuboi et al., 2020), the import of mitochondrial proteins occurs post-translationally, meaning that they are first synthesized in the cytosol and subsequently translocated into mitochondria; however, the spatiotemporal details of these processes are largely elusive (Jan et al., 2014; Gold et al., 2017).
Mitochondrial protein biogenesis strictly depends on the cytosolic chaperone capacity (Becker et al., 1996; Terada et al.,1996; Hoseini et al., 2016; Ben-Menachem et al., 2018; Deshaies et al., 1988; Döring et al., 2017; Pfanner et al., 1987; Stein et al.,2019). Presumably as a consequence of their strong tendency to sequester chaperones, precursor proteins accumulating in the cytosol induce a sudden growth arrest, trigger the heat shock response to increase components of the chaperone and proteasome system, and activate specific factors on the mitochondrial surface that clean off translocation intermediates (Mårtensson et al., 2019; Wrobel et al., 2015; Weidberg and Amon, 2018; Boos et al., 2019, 2020; Wang and Chen, 2015; Shakya et al.,2020).
Most mitochondrial proteins are synthesized with an N-terminal presequence that serves as a matrix-targeting signal (MTS) (Vögtle et al., 2009; von Heijne, 1986). Presequences are recognized by Tom20 and Tom22, two receptor proteins that are part of the translocase of the outer membrane (TOM) complex (Rimmer et al., 2011; Shiota et al., 2015; Araiso et al., 2019) before they lead the way into the matrix, across protein-conducting channels of the TOM complex and the presequence translocase (or TIM23 complex) in the inner membrane (Chacinska et al.,2009). Internal MTS-like (iMTS-L) sequences are frequently found in matrix proteins; although not sufficient as import signals, these patterns can strongly improve the import competence of precursors (Backes et al., 2018).
Many mitochondrial proteins, however, lack presequences and embark on other import routes into mitochondria. This is the case for all proteins of the outer membrane and most components of the intermembrane space (IMS) that use several distinct pathways (Drwesh and Rapaport, 2020; Wiedemann and Pfanner, 2017; Finger and Riemer, 2020; Edwards et al., 2020; Doan et al.,2020). In addition, many mitochondrial inner membrane proteins, in particular the members of the metabolite carrier family (carriers for short), lack presequences and are imported by a distinct “carrier pathway” (Horten et al., 2020; Rehling et al., 2004). From studies using the ATP/ADP carrier (i.e., Pet9 in yeast) as a model protein, it was proposed that the carrier pathway differs from the import route of matrix-destined proteins already on the surface of mitochondria, where carriers bind the “carrier receptor” Tom70 that would insert them into the universal protein-conducting channel of the TOM complex. In the IMS, a soluble chaperone complex consisting of small Tim proteins further transfers the carriers to a specific translocase of the inner membrane, the TIM22 complex, for membrane insertion. Although the mitochondrial steps of the carrier pathways were dissected in detail by powerful in vitro assays (Pfanner and Neupert, 1987; Ryan et al.,1999; Hasson et al., 2010), the early, i.e., cytosolic, steps remain unclear.
This lack of understanding is particularly obvious for the role of Tom70 (and its paralog Tom71), although this outer membrane protein was one of the first import components discovered (Söllner et al., 1990; Steger et al., 1990; Hines et al., 1990). In contrast to all other TOM subunits, Tom70 is presumably no stoichiometric component of the TOM complex but rather associates with the outer membrane translocase in a dynamic and transient fashion.
Tom70 offers dedicated binding sites for the recruitment of cytosolic Hsp70 and Hsp90 chaperones by their unique C-terminal EEVD tails (Young et al., 2003) and was found to interact with co-chaperones (Opaliński et al., 2018). It also directly binds mitochondrial precursor proteins (Papic et al., 2011; Brix et al., 1999, 2000; Iwata and Nakai, 1998; Melin et al., 2015) and prevents their aggregation (Yamamoto et al., 2009). However, the specific contribution of each of these properties in the context of mitochondrial protein import is not clear, particularly, because these functions of Tom70 cannot be reliably assessed with the in vitro import assays that were used in most studies.
In this study, we used a number of complementary in vivo approaches to elucidate the specific role of Tom70. Our assays demonstrate that Tom70 is not a specific receptor for carriers. Rather, Tom70 supports the biogenesis of some carriers (in particular, of Pet9) but also that of many other mitochondrial proteins. Many Tom70 clients contain hydrophobic transmembrane segments or other aggregation-prone regions. Most of these proteins are also sensitive to heat, which is in line with the temperature sensitivity of Tom70-deficient cells. Interestingly, the loss of Tom70 can be largely complemented in strains carrying an unrelated EEVD-binding protein on the mitochondrial surface. Thus, the predominant and crucial function of Tom70 is not that of a classical import receptor. Instead, Tom70 serves as cochaperone on the mitochondrial surface to suppress the toxic effects of mitochondrial precursor proteins.
Results
Tom70 is required for the biogenesis of many mitochondrial proteins
On the basis of in vitro experiments with a very small number of substrates, Tom70 was proposed to serve as a mitochondrial import receptor for precursors that are made without presequences (Papic et al., 2011; Becker et al., 2011; Brix et al., 1999; Steger et al., 1990), in particular of carriers such as Pet9 or Oac1 (Figure 1A). Tom70 also supports the in vitro import process of some presequence-containing proteins, such as Atp1, but not that of others, such as Hsp60 (Figure 1B). Hence, to date, there is no clear understanding of the substrate range of Tom70. To elucidate the in vivo substrate spectrum of Tom70, we hypothesized that proteins that are not properly imported into mitochondria will be degraded by cytosolic quality control and hence that we can use their steady-state cellular abundance as a proxy for their capacity to be properly targeted to mitochondria. Therefore, we measured, using quantitative mass spectrometry, the levels of all mitochondrial proteins in Δtom70/71 mutants lacking the genes for Tom70 and its barely expressed paralog Tom71 (Schlossmann et al., 1996;Figures 1C, S1A, and S1B; Tables S1 and S2). Cells were grown on galactose at 30°C, at which a loss of Tom70 does not result in reduced growth rates, to avoid secondary-growth-dependent effects (Figure S1C). Cells were harvested at exponential growth phase from which proteins were extracted and digested with trypsin. Peptides were labeled using a tandem mass tag (TMT) protocol and multiplexed to quantify proteins in combined sample runs (samples from additional mutants, described later, were also pooled).
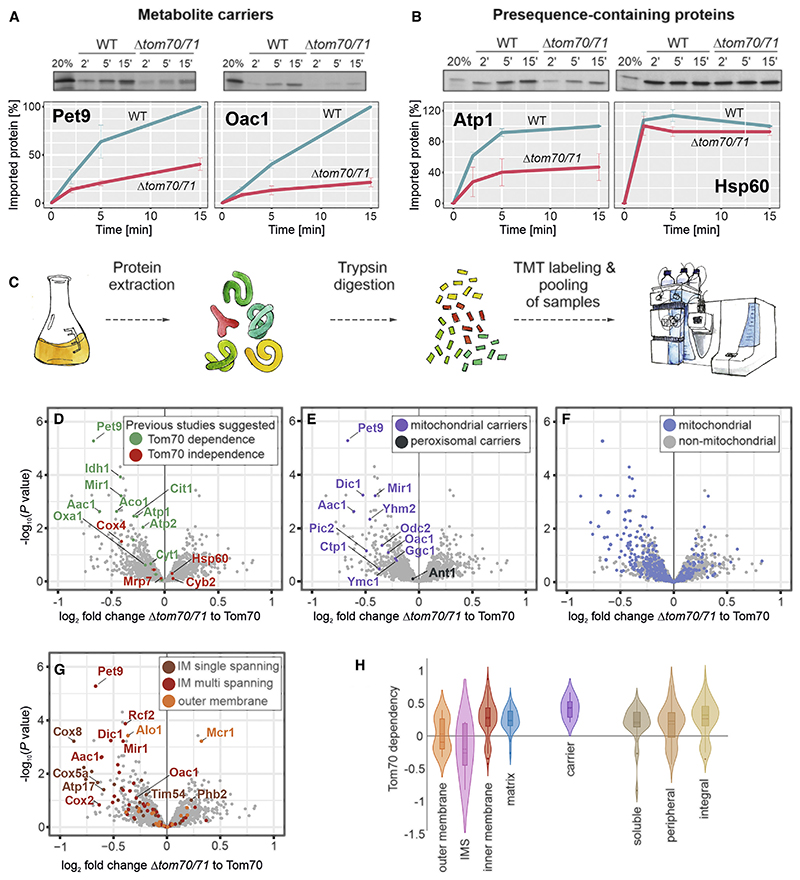
Figure 1. Identification of Tom70/71 clients.
(A and B) Radiolabeled Atp1, Hsp60, Pet9, and Oac1 were incubated with isolated wild-type and Δtom70/71 mitochondria for the times indicated at 25°C. Non-imported protein was removed by treatment with proteinase K, and samples were analyzed by SDS-PAGE and autoradiography. Graphs show mean values and standard deviations from three independent experiments.
(C) The proteomes of different mutants were compared using quantitative proteomics and multiplexing (see also Figure S1A).
(D–G) The proteomes of Δtom70/71 cells carrying either empty or Tom70-expressing plasmids (three biological replicates each) were measured by mass spectrometry. Shown are the mean values of the ratios obtained from Δtom70/71 (30°C) to Tom70-expressing cells (30°C) plotted against their statistical significances (p values). The points in the top left corner show the highest Tom70 dependence. The data point for Tom70 is shown in Figure S1B. Different groups of proteins are indicated in the same dataset. IM, inner membrane.
(H) The relative depletion of proteins in the Δtom70/71 to Tom70 comparison (log2 fold changes [FCs]) were taken as proxy for the Tom70 dependence of proteins. Shown are the distributions of these Tom70 dependence values for different groups of mitochondrial proteins (Morgenstern et al., 2017).
This experiment reliably confirmed previous reports about the Tom70-dependent or -independent nature of individual proteins (Figure 1D). The majority of mitochondrial carrier proteins, but not the peroxisomal carrier Ant1, were reduced in the Δtom70/71 mutant, albeit to different degrees (Figure 1E). In particular, Pet9 was confirmed as a Tom70/71-dependent mitochondrial protein, which is in line with the results from in vitro import assays. Interestingly, however, many other mitochondrial proteins were diminished in Δtom70/71 samples that were not previously reported to be dependent on Tom70 (Figure 1F). This observation points toward a much more general function of Tom70/71 in mitochondrial biogenesis. In particular, membrane proteins were found to be affected (Figure 1G). Many mitochondrial proteins belonging to all types of sub-mitochondrial compartments were consistently, but not strongly, reduced in the Δtom70/71 samples, and no proteins were depleted by more than 40%, explaining the efficient growth of the mutant even on respiratory media.
Taking protein levels in these strains as a proxy for their Tom70 dependence, we found that many inner membrane proteins, including carriers, as well as many proteins of the matrix use this outer membrane receptor (Figure 1H). Proteins of the outer membrane, on the other hand, showed a heterogeneous Tom70 dependence, and most IMS proteins did not show alterations in abundance in Tom70/71 mutants, which is consistent with previous studies (Lutz et al., 2003; Araiso et al., 2019; Gornicka et al., 2014). Many soluble proteins were affected in the Tom70 mutants, arguing against a role of Tom70 as a specific receptor for membrane proteins.
Mitochondrial proteins strongly differ in their dependence on Tom70
As a second, independent strategy to measure the Tom70 dependence of mitochondrial proteins under in vivo conditions, we selected a set of 113 MTS-independent mitochondrial proteins N-terminally fused with superfolder green fluorescent protein (sfGFP) and visualized them on the background of Tom70/71 mutants. Specifically, we picked the strains from the N-terminal SWAp-Tag (N-SWAT) library (Weill et al., 2018) in which each of these proteins is tagged with N-terminal sfGFP under the control of a NOP1 promoter (Figure 2A; see STAR Methods for details). We used fluorescence intensity as a proxy for protein abundance and, in addition, to determine whether the proteins localize correctly to mitochondria. Using automated approaches, we introduced the tagged strains into genetic backgrounds that lack either Tom70 (Δtom70), its paralog Tom71 (Δtom71), or both (Δtom70/71). High-content microscopy screening of these mutants showed three patterns (Figures 2B–2D and S2). The mitochondrial accumulation of some proteins, such as the ATP/ADP carrier Pet9, was considerably reduced in the absence of Tom70 and also affected to a certain degree in Δtom71 single mutants (Figure 2B). The degree of Pet9 reduction in this microscopy screen was very similar to that of the proteomics measurements, and both approaches showed that in the absence of Tom70/71, about one-half of the normal Pet9 levels accumulated in mitochondria.
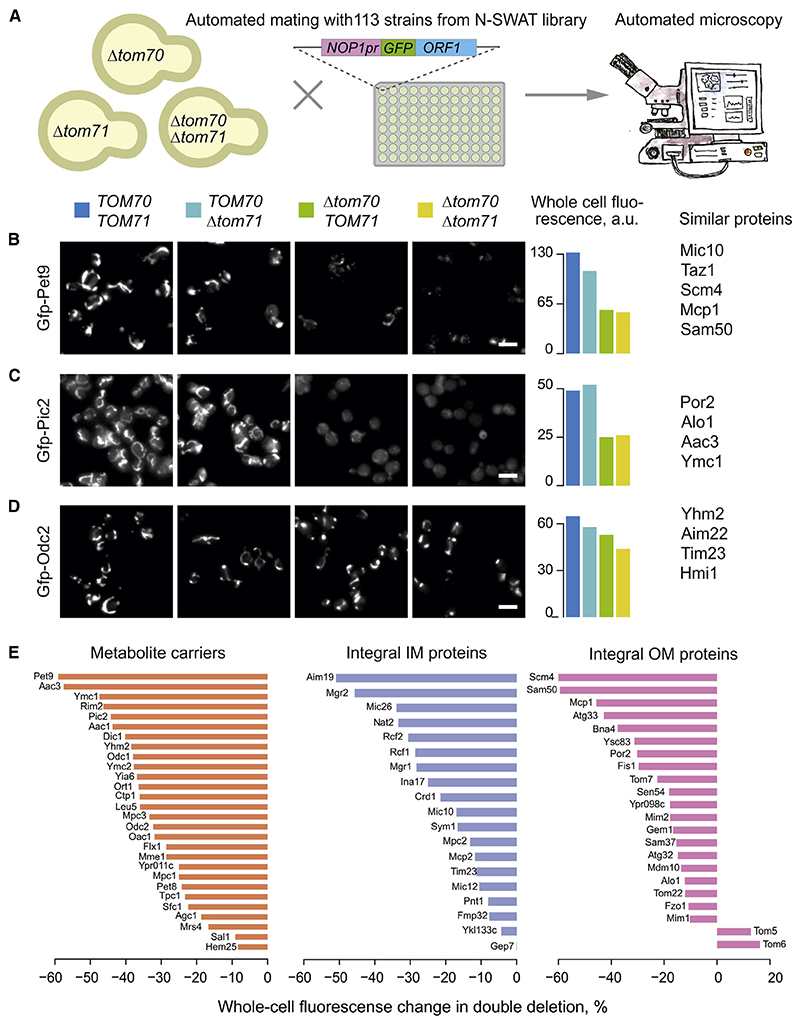
Figure 2. Mitochondrial proteins strongly differ in their Tom70 dependence.
(A) Scheme of the systematic visual screen of GFP-tagged mitochondrial proteins.
(B–D) The mitochondrial localization of 113 N-terminally GFP-tagged mitochondrial proteins (all lacking an MTS) were visualized. Proteins shown in (B) showed a strongly reduced mitochondrial localization in the absence of Tom70 and moderately reduced levels if Tom71 was deleted. Thus, these proteins depend to some degree on both receptors. Proteins shown in (C) were unaffected if Tom71 was deleted but still required Tom70. For proteins shown in (D), Tom70 and Tom71 were hardly, if at all, relevant.
(E) The whole-cell GFP signal change in Δtom70Δtom71 compared with wild-type cells measured for different mitochondrial protein classes. See Table S3 for details. Scale bars, 10 μm. OM, outer membrane.
Some proteins, such as the phosphate carrier Pic2, were Tom71 independent but required Tom70 for efficient accumulation in mitochondria (Figure 2C). A third group, including the dicarboxylate carrier Odc2, was only mildly affected in all mutants, even in the Δtom70/71 double deletion (Figure 2D). These results showed that Tom71 supports the biogenesis of some proteins but is not essential for any of them. But also, the relevance of Tom70 was surprisingly variable, and even many carrier proteins efficiently accumulated in mitochondria in Δtom70/71 mutants (Figures 2D and 2E).
By using two orthogonal approaches, we consistently found that, in vivo, carriers strongly vary in their Tom70 dependence (Figures S3A and S3B). The results of the image-based screen with GFP-tagged carriers thereby correlated well with the proteomic data (Figure S3C). Carriers are not per se more affected in Δtom70/71 mutants than other mitochondrial membrane proteins. These results challenge the long-standing notion that Tom70/71 is a specialized carrier receptor. Rather, Tom70 and (to a much lesser degree) Tom71 support mitochondrial biogenesis of many proteins. However, the individual dependence of mitochondrial proteins on these receptors varies considerably.
Tom70/71 supports biogenesis of aggregation-prone mitochondrial proteins
We mined our dataset of Tom70-dependent proteins for features that could determine whether a protein needs the assistance of Tom70 for its biogenesis. As expected, we found Tom70-dependent proteins to be enriched with iMTS-Ls, internal stretches that structurally mimic presequences (Figure 3A). They were reported before to be efficient binding sites for Tom70 (Backes et al.,2018). In addition, we found significantly higher aggregation propensities for Tom70-dependent proteins (Figure 3A). The large, hydrophobic carriers are predicted to be particularly aggregation prone, which might explain why many carriers depend more on Tom70; there is more heterogeneity for other inner membrane or matrix proteins (Figure 3B). The notion that Tom70 might be required for the correct biogenesis of proteins that are prone to misfolding and potentially aggregate is in line with the temperature-sensitive phenotype of the Δtom70/71 mutant (Figure 3C).
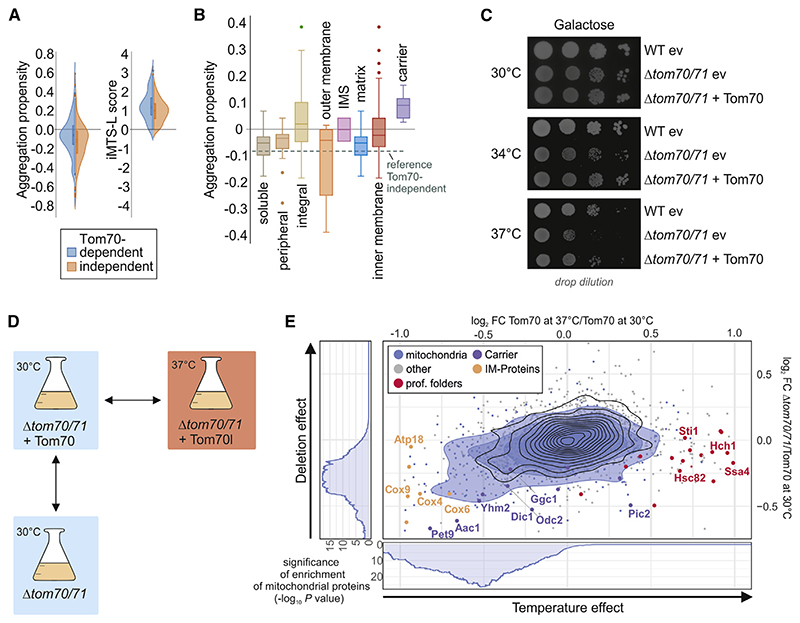
Figure 3. Tom70/71 supports biogenesis of aggregation-prone mitochondrial proteins.
(A) The aggregation propensities (Conchillo-Sole et al., 2007) and the presence of iMTS-L sequences in proteins (Boos et al., 2018) were calculated. Plotted are the distributions of these values for Tom70-dependent (log2 FC, <—0.2) and -independent (log2 FC, >0.2) proteins.
(B) Aggregation propensities were calculated for different groups of mitochondrial proteins. The dotted line shows the mean value of Tom70-independent proteins as a reference.
(C) The indicated strains were precultured in galactose-containing medium at 30°C and spotted on galactose medium, following 3 days of incubation at 30°C, 34°C, or 37°C. WT, wild-type; ev, empty vector.
(D and E) The influence of temperature (log2 FC of Tom70 37°C as compared to Tom70 30°C) and the absence of Tom70 (log2 FC of Δtom70/71 as compared to Tom70 at 30°C) were analyzed. Blue circles show the isobaric distribution of mitochondrial proteins, whereas black ones show the distribution of the entire proteome. Enrichment of mitochondrial proteins among proteins with a log2 FC below a certain threshold was calculated, and significance of this enrichment was plotted (side panels).
If Tom70 indeed predominantly supports the biogenesis of demanding, aggregation-prone proteins, one would expect that deletion of this stabilizing factor has similar effects on its clients as conditions that promote the general misfolding of proteins. To test this hypothesis, we compared the effect of the absence of Tom70 with the effect of a higher temperature. We grew cells on galactose at 30°C and 37°C and compared the temperature-dependent changes in proteome composition with those caused by deletion of TOM70/71 (Figures 3D and 3E). We observed that many chaperones were found at higher levels in 37°C-grown cells but were hardly influenced by the absence or presence of Tom70 (Figure 3E, marked in red), showing that Δtom70/71 cells do not suffer from generally perturbed protein homeostasis. Interestingly, levels of many mitochondrial proteins were reduced by high temperatures as well as by the loss of Tom70, and the effects in these conditions are remarkably similar (Figure 3E). This is most impressive for Pet9 that was strongly reduced under both conditions, again indicating its aggregation-prone nature.
Hence, these results suggest that its tendency to misfold or even aggregate determines whether a mitochondrial protein requires Tom70 for its biogenesis. Thus, in vivo, the predominant role of Tom70 may be not so much that of a receptor that confers directed or specific targeting of its cargo to the surface of mitochondria. Rather, Tom70 safeguards proteins that are intrinsically prone to acquire non-productive, presumably importincompetent or even toxic conformations. This function seems particularly relevant at elevated temperatures when proteostasis is in any way challenged.
Tom70 can be replaced by a chaperone-tether on the mitochondrial surface
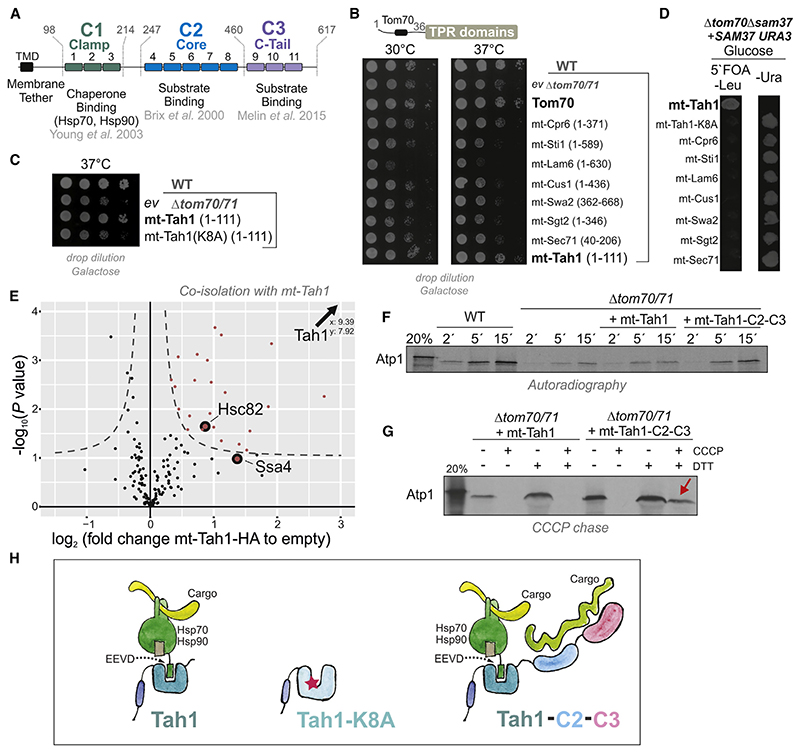
How does Tom70 support the biogenesis of proteins that are prone to misfolding and aggregation? The large size and the multi-domain structure of Tom70/71 indicate that it is not just a simple binder for precursor proteins, as is the case for the much smaller Tom20. Tom70 is tethered to the outer membrane by an N-terminal transmembrane domain and exposes three soluble domains to the cytosol, called Clamp, Core, and C-tail domains (Figure 4A; Young et al., 2003; Chan et al., 2006; Brix et al.,2000). For simplicity, we refer to them here as C1, C2, and C3. All three domains are formed by tetratricopeptide repeats (TPRs). C2 and C3 were reported to bind internal targeting signals of pre-cursor proteins, such as Pic2 (Brix et al., 2000) (C2) or Adh3 (Chan et al., 2006) and rat alcohol dehydrogenase (Melin et al.,2015) (C3). In contrast, C1 forms a binding groove to recruit the C-terminal EEVD tetrapeptides of Hsp70 and Hsp90 chaperones (Young et al., 2003; Li et al., 2009). We reasoned that it might be this chaperone-binding activity that contributes to the function of Tom70 as a stabilizer of aggregation-prone proteins. Expression of a mutant version of Tom70 that contains only the C1 domain proved to be impossible because it was unstable in vivo and never accumulated to detectable levels (Figure S4A). However, TPR domains are found in many cellular proteins where they serve as specific binding modules for different groups of ligands (Perez-Riba and Itzhaki, 2019).
Figure 4. Tom70 can be replaced by a chaperone tether on the mitochondrial surface.
(A) Schematic representation of the different domains of Tom70 formed by 11 TPR domains.
(B and C) The indicated sequences of yeast TPR proteins were fused to the membrane anchor of Tom70 and expressed in the Δtom70/71 mutant.
(D) A Δsam37 Δtom70 double mutant carrying SAM37 on a URA3-containing plasmid was transformed with plasmids for the expression of the indicated fusion proteins. Upon addition of 5-fluoroorotic acid (5°FOA), only cells that lost the URA3-containing SAM37 plasmid could grow.
(E) Cells of the Δtom70/71 mutant carrying the mt-Tah1 expression or an empty plasmid were grown in galactose medium to mid-log phase. Cells were washed, gently lysed with Triton X-100, and incubated with Sepharose beads carrying HA-specific antibodies (to pull out mt-Tah1). Samples from four independent replicates for each strain were analyzed by mass spectrometry. The full dataset can be found in Table S4.
(F) Radiolabeled Atp1 was incubated with mitochondria isolated from the indicated mutants. Non-imported Atp1 was removed by adding proteinase K after the times indicated. Mt-Tah1-C2-C3 is a fusion protein in which the C2 and C3 domains of Tom70 were fused to mt-Tah1.
(G) Radiolabeled Atp1 was incubated with mitochondria after the membrane potential was depleted by treatment with carbonyl cyanide m-chlorophenyl hydrazone (CCCP). When indicated, CCCP was quenched by dithiothreitol (DTT) to restore the membrane potential. The presence of the C2-C3 domains was essential to keep Atp1 bound to the mitochondria (indicated by the red arrow).
(H) Model of the chaperone binding property of the C1 domain of Tom70/71 and of mt-Tah1. The C2 and C3 domains facilitate direct substrate binding that is particularly relevant under the conditions of the in vitro import reaction.
Alignments of the first three TPR domains of Tom70 show sequence similarity to other TPR proteins of yeast (Figures S4B), which inspired us to test whether any of these other TPR domains can functionally replace Tom70. To this end, we fused six TPR domains to the transmembrane domain of Tom70 and expressed these sequences in the Δtom70/71 mutant (Figure S4C). Tethering the protein Tah1 to the outer membrane (mt-Tah1) suppressed the temperature-sensitive growth of the mutant, whereas the other TPR domains had no or even negative effects (Figures 4B and S4D).
Intriguingly, Tah1, just like Tom70, is an Hsp70/90-binding protein, although it functions in a completely different context as follows: Tah1 is a subunit of the cytosolic Rvb1-Rvb2-Tah1-Pih1 (R2TP) assembly complex that facilitates the biogenesis of RNA polymerases and other RNA-binding complexes (Back et al., 2013; Jiménez et al., 2012; Boulon et al., 2010). Its 111 residues form 1 globular domain comprising 2 TPR stretches and a capping helix that specifically bind the EEVD-tail of cytosolic Hsp90 (Millson et al., 2008). A point mutation in mt-Tah1 (K8A) that destroys its Hsp90-binding ability (Jiménez et al., 2012) was unable to suppress the phenotype of the Δtom70/71 mutant (Figure 4C). mt-Tah1, but not mt-Tah1(K8A), was even able to rescue the synthetic lethal Δtom70 Δsam37 double mutant, demonstrating that mt-Tah1 can specifically replace Tom70 and not only indirectly buffers the adverse effects of its absence (Figures 4D, S4E, and S4F).
Next, we purified the mt-Tah1 protein from whole-cell extracts by using its hemagglutinin (HA) tag and identified bound proteins by quantitative mass spectrometry (Figure 4E). A number of interaction partners of mt-Tah1 were identified, including the cytosolic chaperones Hsp82/Hsc82 (the yeast representatives of Hsp90) and the cytosolic Hsp70 Ssa4 (Table S4).
In contrast to the in vivo situation, in in vitro import experiments, mt-Tah1 was not able to replace Tom70 in its ability to facilitate the import of Tom70-dependent substrate proteins such as Atp1 (Figure 4F). Only when mt-Tah1 was extended with the C2 and C3 domains of Tom70, it was able to support the import of radiolabeled Atp1. This was particularly apparent in “CCCP chase experiments” (Haucke et al., 1995; Backes et al., 2018) in which Atp1 was initially bound to de-energized mitochondria and subsequently chased across the outer membrane upon restoration of the membrane potential (Figure 4G). In that setup, Tom70 is crucial to hold Atp1 on the mitochondrial surface, a function that obviously is not carried out by mt-Tah1. These results confirm that mt-Tah1 rescues the Δtom70/71 mutant by replacing it as a chaperone-recruitment factor on the outer membrane (Young et al., 2003), and not by direct binding to precursor proteins (Figure 4H). In summary, we conclude that the ability of Tom70 to bind substrates directly is largely dispensable under physiological in vivo conditions.
Chaperone binding by Tom70 is important for different cellular activities
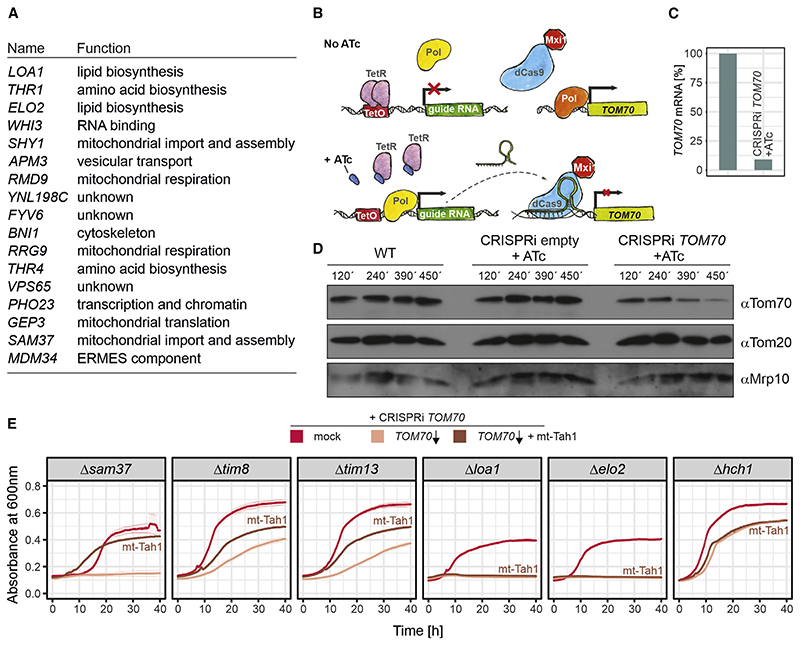
If indeed it is the co-chaperone activity of Tom70 that is essential for cellular and mitochondrial function, we would expect this to be evident from the cellular effects of its absence. To map such global effects, we set out to identify synthetic lethal or sick genetic interactions in Δtom70 mutants. We carried out a genome-wide genetic interaction screen (Baryshnikova et al.,2010; Tong and Boone, 2007) by crossing a Δtom70 query strain with the systematic yeast knockout library in which all non-essential genes were individually deleted. We measured the colony size of the resulting double mutants as a proxy for their fitness. The genes whose deletions led to considerable fitness reduction or death in the Δtom70 but not wild-type TOM70 background were regarded as synthetic sick or lethal genetic interactors of Tom70. We identified many genetic interactors, of which most are components relevant for mitochondrial biogenesis (Figure 5A; Table S5), such as Sam37, Tom71, Tom7, Tim8, Tim13, Pam17, Mip1, or Afg3. In addition, we found components relevant for peroxisome biogenesis (Pex17 and Pex18), for lipid metabolism (Loa1, Elo1, Elo2, Crd1, and Psd1), the ERMES complex (Mdm10, Mdm34, and Gem1), and cytosolic proteostasis (Pfd1 and Hch1). The fitness of cells lacking any of these components dropped considerably in the absence of Tom70.
Figure 5. Chaperone binding by Tom70 is important for different cellular activities.
(A) The Δtom70 allele was introduced into a systematic yeast deletion library by automated genetic manipulations. Colony sizes were measured, and the 100 most-affected deletion mutants (Table S5) were analyzed (see STAR Methods for details).
(B) Schematic illustration of the CRISPRi strategy used to knock down TOM70.
(C) TOM70 transcript levels were measured by qPCR 6.5 h after addition of anhydrotetracyclin (ATc). Shown are mean values of three replicates.
(D) Tom70 levels were analyzed by western blotting of the indicated strains at different time points after addition of ATc.
(E) Growth curves of the following strains: indicated single deletions without addition of ATc (mock); TOM70 is knocked down through the addition of 960 ng/μl ATc (TOM70 ↓); and TOM70 is knocked down through the addition of 960 ng/μl ATc, but mt-Tah1 rescues the synthetic growth defect of some mutants (TOM70 ↓ + mt-Tah1). Shown are mean values and standard deviations from three replicates.
The observation that Tom70 showed genetic interactions with these seemingly different functional groups of proteins could either point to several distinct functions of Tom70 or, alternatively, to the relevance of its chaperone-binding activity for multiple cellular activities. To study these genetic interactions further, despite the lethality of many double mutants, we used an improved, plasmid-based CRISPR interference (CRISPRi) system to specifically knock down transcription (Figure 5B). We expressed catalytically inactive Cas9 coupled to a potent transcriptional repressor (dCas9-Mxi1) together with a TOM70- specific guide RNA under the control of a tetracycline-inducible promoter (Smith et al., 2016). The concentrations of anhydrotetracycline (ATc) necessary to induce expression did not affect the fitness of yeast cells (Figure S4G). Within a few hours of induction, the levels of TOM70 mRNA and of the Tom70 protein dropped to about 10% (Figures 5C and 5D). Combining CRISPRi perturbations with several deletion backgrounds allowed us to verify genetic interaction partners of Tom70, including the genes encoding Tim8, Tim13, Sam37, Loa1, Elo2, and Hch1 (Figure 5E). Expression of mt-Tah1 in these strains partially rescued the synthetic defects of mitochondrial protein import mutants (Tim8, Tim13, and Sam37) but not those in lipid metabolism (Elo2 and Loa1). Interestingly, mutants lacking Elo2 or Loa1 could however be suppressed by expression of a mt-Tah1-C2-C3 fusion protein (Figure S4H), suggesting that the genetic interaction of Tom70 with other proteins can either be mediated by the C1 domain (with genes of the import components Tim8, Tim13, and Sam37) or by the C2-C3 domains for genes relevant for lipid biosynthesis (Figure S4I). Thus, the chaperone-recruiting function of Tom70 is of relevance especially in the context of protein biogenesis, and it might be dispensable for other roles.
Chaperone binding by Tom70 is crucial for the biogenesis of small inner membrane proteins
Artifcial chaperone recruitment to the outer mitochondrial membrane through mt-Tah1 relieves the temperature sensitivity of Δtom70/71 and its synthetic defects with mitochondrial biogenesis components. To analyze the mechanistic basis of this unexpected observation, we tested how mt-Tah1 influences the proteomic changes that we observed in the Δtom70/71 mutant.
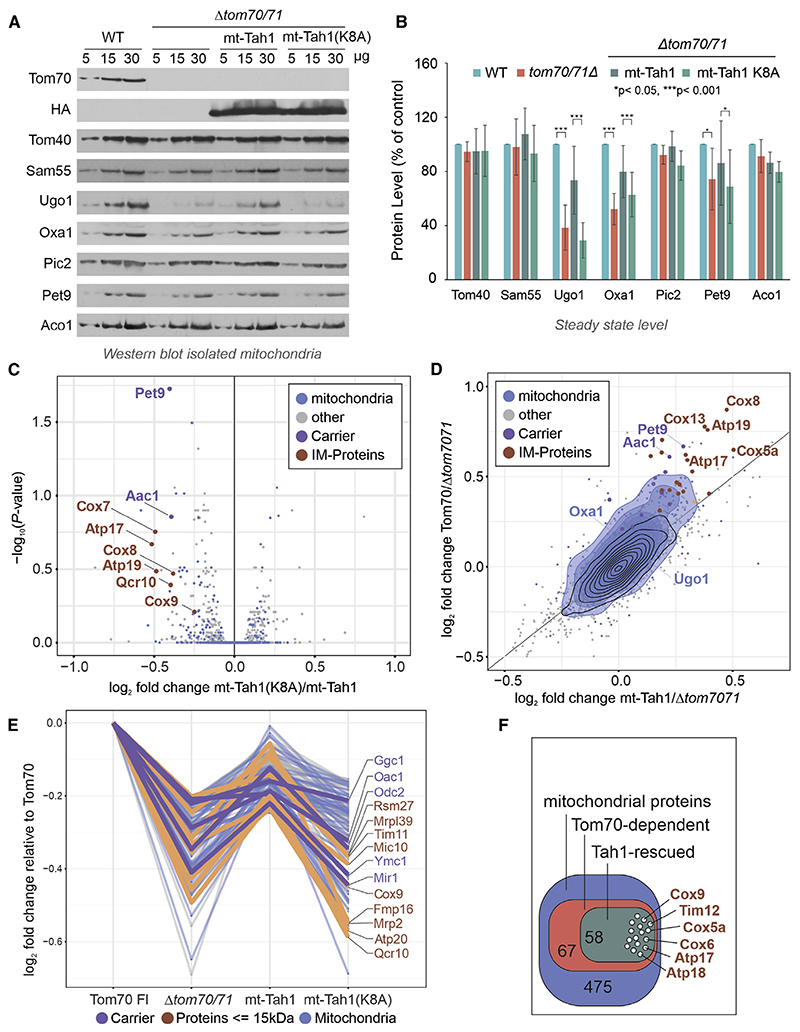
Western blots of isolated mitochondria showed that the expression of mt-Tah1, but not that of mt-Tah1(K8A), restores the levels of Tom70-dependent proteins, such as Ugo1, Oxa1, and Pet9 (Figures 6A, 6B, and S4J). Which other proteins rely on the chaperone-binding activity of Tom70? To address this question systematically, we compared the cellular proteomes of Δtom70/71 with the same strains expressing Tom70, mt-Tah1, or mt-Tah1(K8A). Indeed, many proteins that were depleted from Δtom70/71 cells were rescued by expression of mt-Tah1, but not mt-Tah1(K8A) (Figures 6C and 6D). Thereby, mt-Tah1 particularly supported the accumulation of many small proteins of the inner membrane, including many single-spanning subunits of the complexes of the respiratory chain. The levels of these small proteins were reduced in Δtom70/71 cells but restored if Tom70 or mt-Tah1 were expressed (Figures 6C–6F, protein names labeled in brown).
Figure 6. Chaperone binding by Tom70 is crucial for the biogenesis of small inner membrane proteins.
(A and B) Protein levels in mitochondria isolated from either wild-type cells or the indicated Δtom70/71 mutants were analyzed by western blotting. Six data points from three biological repeats were analyzed for each protein. The error bars refer to standard deviations. The p values were generated from the two-tailed paired t test.
(C) The volcano plot shows the comparison of the proteomes of Δtom70/71 cells that express the mt-Tah1(K8A) to those with mt-Tah1. The positions of several small inner membrane proteins (brown) and of carriers (purple), which are considerably stabilized by mt-Tah1 but not by mt-Tah1(K8A), are indicated.
(D) The effects by which Tom70 and mt-Tah1 influence the cellular proteomes are plotted against each other.
(E) Relative log2 FCs of Tom70-dependent mitochondrial proteins that are rescued by either mt-Tah1 or its variant.
(F) Graphical overview of the number of Tom70-dependent proteins that are rescued by expression of mt-Tah1 near to Tom70 full-length levels.
Chaperone binding by Tom70 prevents mitoprotein-induced toxicity
The inner membrane contains many proteins of less than 18 kDa, of which most are single-spanning non-catalytic subunits of respiratory chain complexes (Morgenstern et al., 2017). These small proteins lack iMTS-L sequences and thus predictable internal binding sites for the C2/C3 domains of Tom70, whereas iMTS-L sequences are ubiquitously found in carriers and otherTom70 clients (Figure S5A).
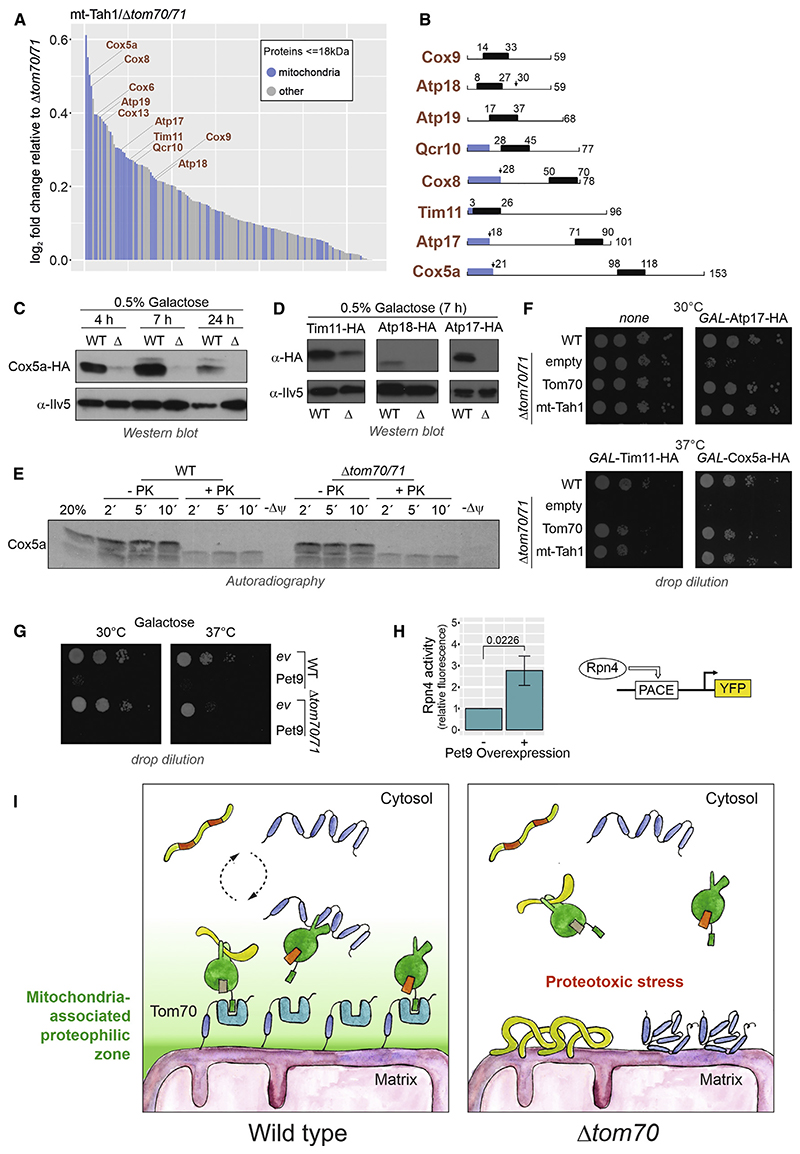
The presence of mt-Tah1 restored the levels of many small inner membrane proteins (Figure 7A). Only some of these proteins carry N-terminal presequences, but others do not (Figure 7B), and their import process was not studied in the past. We therefore chose some of these Tom70-dependent small inner membrane proteins (Cox5a, Tim11, Atp17, and Atp18) as model proteins for further follow-up.
Figure 7. Chaperone binding by Tom70 prevents mitoprotein-induced toxicity.
(A) Proteins that are enriched by the expression of mt-Tah1 compared with Δtom70/71 are shown. Only proteins with masses smaller than 18 kDa and positive enrichment factors were considered. Mitochondrial proteins are indicated in blue.
(B) Schematic representations of small inner membrane proteins for which information about their overall structure and targeting information exists. Blue regions show presequences, and black boxes indicate transmembrane domains.
(C and D) Cox5a-HA, Tim11-HA, Atp17-HA, and Atp18-HA were expressed under GAL1 control from multi-copy plasmids in wild-type and Δtom70/71 cells. The times indicate how long cells were shifted to 0.5% galactose-containing medium.
(E) Radiolabeled Cox5a was incubated with isolated mitochondria for the times indicated at 30°C. The membrane potential (Δψ) was depleted in control samples by addition of CCCP. Mitochondria were reisolated and incubated with or without proteinase K.
(F and G) The indicated strains were transformed with plasmids to express Atp17-HA, Tim11-HA, Cox5a-HA, and Pet9-HA under the control of the GAL1 promoter. All cultures were grown on lactate medium to mid-log phase, induced with 0.5% galactose for 4.5 h, and dropped onto galactose plates.
(H) Rpn4-driven gene expression was measured using a yellow fluorescent protein (YFP) reporter system (Boos et al., 2019).
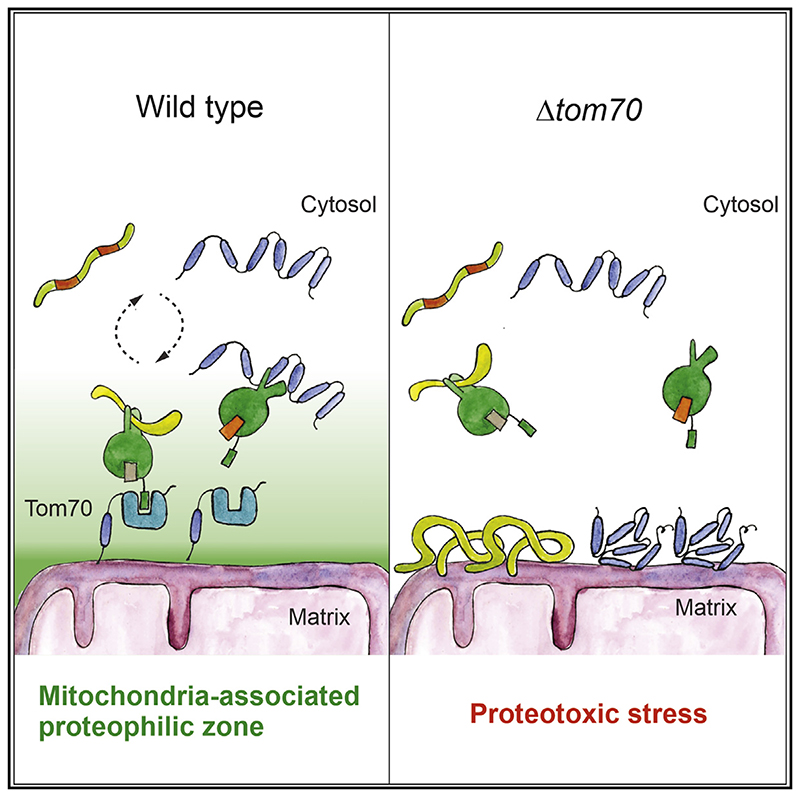
(I) Tom70 supports the biogenesis of aggregation-prone mitochondrial membrane proteins by recruiting cytosolic chaperones to the mitochondrial surface, thereby generating a “mitochondria-associated proteophilic zone.”
When expressed in vivo from a strong GAL1 promoter, Cox5a accumulated in cells only in the presence of Tom70/71 but not in a deletion mutant (Figure 7C). The same strong Tom70/71 dependence was observed when Tim11, Atp18, or Atp17 were expressed from GAL1 promoters (Figure 7D).
Cox5a was in vitro imported into wild-type and Δtom70/71 mitochondria with similar, rather low efficiency (Figure 7E). Even if the import was carried out at 37°C, the in vitro import of Cox5a did not depend on Tom70 (Figure S5D). This result indicates that the striking Tom70 dependence in vivo is not recapitulated in the in vitro import assay. Aggregation-prone Tom70 substrates could be toxic in the absence of Tom70/71, impeding their intracellular accumulation. Indeed, we observed a high toxicity of overexpressed Cox5a, Atp17, Tim11, and Pet9 (Figures 7F, 7G, and S5C) and moderate toxicity of Atp18 (Figure S5D) in the Δtom70/71 mutant, whereas wild-type cells were less sensitive. Regardless of whether Tom70 was present or absent, the overexpressed Cox5a co-isolated with mitochondria upon fractionation experiments (Figure S5E), and high temperature reduced the levels of cellular Cox5a to amounts similar to those found in Δtom70/71 cells (Figure S5F).
Why is the overexpression of Pet9 and many small inner membrane proteins so toxic? The overexpression of Pet9 strongly induced an Rpn4-driven reporter indicative for problems in cytosolic proteostasis (Figure 7H). An Rpn4-mediated gene induction was observed as a characteristic element of the mitoprotein-induced stress response in yeast (Boos et al., 2019) but is also triggered by other cytosolic proteotoxic stress conditions.
In summary, we conclude that many small inner membrane proteins as well as carrier proteins have the potential to be toxic to cells. Tom70/71 suppresses this toxicity and facilitates the accumulation of these proteins in mitochondria. This property of Tom70/71 depends on its chaperone-binding ability (and hence can be replaced by mt-Tah1). Thus, the primary function of Tom70/71 is that of a co-chaperone on the mitochondrial surface that is crucial to protect the cytosolic compartment against proteotoxicity arising from mitochondrial precursors.
Discussion
Traditionally, the problem of mitochondrial protein biogenesis is seen from the perspective of the organelle. From this direction, Tom70 was identified as “the carrier receptor,” owing to its ability to bind precursor proteins on the mitochondrial surface (by its C2 and C3 domains) and to promote the transfer of these proteins into the protein-conducting channel of the TOM complex. This function of Tom70 is well reflected in the results obtained from in vitro import experiments, in which Tom70 clearly supports the import of specific mitochondrial proteins, in particular of the ATP/ADP carrier Pet9 but also of other mitochondrial proteins (Papic et al., 2011; Becker et al., 2011; Söllner et al., 1990; Brix et al., 2000; Ryan et al., 1999; Yamamoto et al., 2009; Backes et al., 2018; Wiedemann et al., 2001). In this study, we used different approaches to elucidate the relevance of Tom70 in the physiological in vivo context. These approaches included the microscopic screens of collections of GFP-tagged proteins, quantitative mass-spectrometry-based proteomics of wholecell extracts, and the dominant-negative growth effects caused by excess precursor proteins. These three approaches clearly confirmed the particular relevance of Tom70/71 for Pet9 biogenesis. Nevertheless, even though the cellular levels of many mitochondrial proteins, and, in particular, those of some carriers, were clearly reduced in the absence of Tom70/71, this depletion was never severe. Even for Pet9, the levels in Δtom70/71 cells were about 40% (GFP signal intensity) to 60% (proteomics) of those in wild-type cells, and all other carriers were affected even less. Despite these rather moderate effects, Δtom70/71 cells are unable to grow under respiration conditions at elevated temperatures. The data presented here point at a crucial function of Tom70 to protect the cytosol against proteotoxic stress conditions for which mitochondrial precursors are presumably responsible. The rather abundant hydrophobic and aggregation-prone Pet9 protein is particularly problematic here. Previous studies already identified Pet9 and its human homolog ANT as proteins with high cytotoxic potential (Wang and Chen, 2015; Coyne and Chen, 2018; Liu et al., 2019; Hoshino et al., 2019; Wang et al., 2008). Mutations that even further increased their aggregation propensity are the cause of autosomal dominant progressive external ophthalmoplegia 2 in humans (Kaukonen et al., 2000) and trigger mitochondrial precursor over-accumulation stress (mPOS) in yeast (Wang and Chen, 2015).
The accumulation of mitochondrial precursor proteins in the cytosol was found to arrest cell growth (Wrobel et al., 2015; Boos et al., 2019; Labbadia et al., 2017; Melber and Haynes,2018), presumably due to their ability to sequester cytosolic chaperones. Obviously, mitochondrial biogenesis is a considerable challenge for eukaryotic cells (Labbadia et al., 2017; Bar-Ziv et al., 2020), and our observations indicate that Tom70 serves as a component to reduce these mitoprotein-induced stress conditions. In the context of the early stages of precursor targeting to mitochondria, Tom70 apparently serves as the interface between the cytosolic chaperone system and the mitochondrial import machinery (Papić et al., 2013; Hansen et al., 2018; Opaliński et al., 2018; Boos et al., 2020; Eliyahu et al., 2012).
How does Tom70 exhibit its cytoprotective activity? In this study, we observe that the artificial tethering of the chaperone-binding TPR protein Tah1 to the mitochondrial surface can almost fully replace Tom70/71 in its properties to promote cell growth at increased temperature to stabilize the mitochondrial proteome and provide resistance against overexpressed inner membrane proteins. This observation is astonishing as the function of endogenous Tah1 is completely unrelated to mitochondrial biogenesis. The profound suppression of the Δtom70/71 mutant by mt-Tah1 indicates that the physiologically relevant function of Tom70 is that of a chaperone-binding factor and that, in vivo, its direct substrate-binding properties and, hence, its receptor functions are of minor relevance. This is in line with the observation that an R171A mutation in the C1 domain completely wipes out Tom70 function in vivo (Young et al.,2003). This mutation is analogous to the K8A mutant in mt-Tah1 and prevents binding of the EEVD peptide. This again supports our conclusion that, in vivo, the chaperone-binding activity is of predominant relevance for the functionality of Tom70.
It appears likely that the recruitment of chaperones to the mitochondrial surface establishes a functionally important “mitochondria-associated proteophilic zone” at the organelle-cytosol interphase that facilitates protein biogenesis and suppresses the potential toxicity of precursor proteins (Figure 7I). The specific conditions of the in vitro import assay in which precursors and mitochondria are incubated in a relatively large volume of buffer might have overestimated the relevance of the high-affinity binding sites provided by the C2 and C3 domains of Tom70 for the import of hydrophobic mitochondrial precursor proteins such as carrier proteins.
The comparison of the proteome of the Δtom70/71 mutant with that of cells that express mt-Tah1 (chaperone-binding) or Tom70 (chaperone and substrate binding) showed that many proteins require the chaperone-binding activity of Tom70, especially those that are intrinsically aggregation prone. A particularly interesting group of proteins are small (6–18 kDa) inner membrane proteins. Overexpression of these proteins in the absence of Tom70 or mt-Tah1 is highly toxic and prevents cell growth. Apparently, chaperones play a crucial role in facilitating the productive translocation of these proteins to mitochondria. In vitro,Tom70 was either not required for these proteins (as for Cox5a) or these proteins could not be imported from reticulocyte lysates (not shown), which points toward a requirement of additional factors—potentially chaperones—that are not accurately reflected in the in vitro assay. Finally, we observed that the levels of some Tom70-dependent proteins were not rescued by mt-Tah1. They were inner membrane proteins with bipartite targeting signals such as Sco1 or Dld1. Potentially, these proteins require the direct binding to Tom70, and hence, mt-Tah1 does not support their biogenesis. Further studies will be required to study the mechanistic reactions of the Tom70 modules and of cytosolic chaperones in the translocation reactions of these protein classes in more detail.
Ribosomes synthesizing Pet9 and other inner membrane proteins were found to be enriched on the surface of mitochondria, potentially to avoid the accumulation of hydrophobic precursors in the cytosol (Jan et al., 2014; Williams et al., 2014). Interestingly, on the contrary, ribosomes for small proteins (less than 180 residues) were strongly depleted from the mitochondrial surface, including those synthesizing Cox5a and other small inner membrane proteins, for which Tom70 was found in this study to be highly relevant (Figure S5G). The profound post-translational mode of their targeting might explain why the chaperone-assisted targeting by Tom70 (or mt-Tah1) is of such high importance for small inner membrane proteins.
The cytosol contains many TPR proteins that are structurally similar to Tom70 and Tah1 (Perez-Riba and Itzhaki, 2019; Zeytuni and Zarivach, 2012). Examples are co-chaperones such as Sti1 that, like Tah1 or the C1 domain of Tom70, binds cytosolic chaperones (Hoseini et al., 2016; Schmid et al., 2012). However, TPR proteins apparently are generally used for the translocation of proteins across cellular membranes. Examples include Sec71 and Sec72 for secretory proteins, Sgt2 for tail-anchored proteins, Pex5 for peroxisomal proteins, or Toc64 for plastid proteins (Chartron et al., 2011; Tripathi et al., 2017; Graham et al., 2019; Schwenkert et al., 2018; Harano et al., 2001). It appears likely that early sorting intermediates in general pose a considerable threat for cytosolic proteostasis, which is countered by multiple TPR proteins on organellar membranes. It will be exciting to study the specific roles of this group of proteins more comparatively, not only in respect to their relevance for protein targeting to their respective home organelle but also for cellular proteostasis and fitness in general.
Star Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-Ilv5 | Johannes Herrmann lab | Peleh et al., 2017 |
| anti-Sod1 | Johannes Herrmann lab | Peleh et al., 2017 |
| anti-Tom70 | Nikolaus Pfanner lab | Söllner et al., 1990 |
| anti-Tom20 | Doron Rapaport lab | Papic et al., 2011 |
| anti-Oxa1 | Johannes Herrmann lab | Peleh et al., 2017 |
| anti-Aco1 | Ophry Pines Lab | Ben Menachem et al., 2018 |
| anti-Fum1 | Ophry Pines Lab | Ben Menachem et al., 2018 |
| anti-Sam55 | Doron Rapaport lab | Papic et al., 2011 |
| anti-Mim1 | Doron Rapaport lab | Papic et al., 2011 |
| anti-AAC | Doron Rapaport lab | Papic et al., 2011 |
| anti-Om14 | Doron Rapaport lab | Papic et al., 2011 |
| anti-Ugo1 | Doron Rapaport lab | Papic et al., 2011 |
| anti-Rabbit secondary antibody | BioRad | 172-1019 |
| anti-Mouse secondary antibody | BioRad | 172-1011 |
| anti-HA | Roche | 12013819001 |
| Chemicals, peptides, and recombinant proteins | ||
| Sera-Mag Beads | Thermo Scientific | 4515-2105-050250 |
| Water, HPLC grade | Chromanorm | 23595.294 |
| 0.2 M HEPES/NaOH pH 8.4 | Sigma Aldrich | H3375 |
| 100% Ethanol, HPLC grade | VWR | 153385E |
| Formic Acid, mass spectrometry grade | Sigma Aldrich | 94318 |
| Chloroacetamide | Sigma Aldrich | C0267 |
| Trypsin | Sigma Aldrich | T6567 |
| DMSO, HPLC grade | Sigma Aldrich | 42780.AK |
| TMT10plex isobaric label reagent set | Thermo Scientific | 90111 |
| Acetonitrile | Honeywell | 34967 |
| Hydroxylamine | Sigma Aldrich | 438227 |
| Dithiothreitol | BioChemica | A1101,0025 |
| Carbonyl cyanide 3-chlorophenylhydrazone | Sigma Aldrich | C2759-250MG |
| Anhydrotetracycline | Cayman chemical company | 10009542 |
| 5’Fluorootic acid Monohydrate | US Biological | F5050 |
| Critical commercial assays | ||
| Quick Coupled Transcription/Translation kit | Promega | L2080 |
| Turbo DNA free kit | Ambion | AM1907 |
| qScript cDNA Synthesis Kit | Quanta Biosciences | 95047 |
| iQ SYBR Green Supermix | BioRad | 1708886 |
| Pierce BCA Protein Assay Kit | Thermo Scientific | 23225 |
| TMT10plex isobaric label reagent set | Thermo Scientific | 90111 |
| Deposited data | ||
| Mass spectrometry proteomics data | This paper | ProteomeXchange PXD021173 |
| IP mass spectrometry data | This paper | ProteomeXchange PXD023149 |
| Experimental models: Organisms/strains | ||
| Yeast: YPH499 WT: MATa ade2-101 his3-200 leu2-1 ura3-52 trp1-63 lys2-80 | Herrmann lab | Woellhaf et al., 2016 |
| Yeast: YPH499 Δsam37: sam37Δ::HIS3 derivative of YPH499 WT | Doron Rapaport lab | N/A |
| Yeast: YPH499 Δsam37: sam37Δ::HIS3 (SAM37 URA3) derivative of YPH499 WT | Doron Rapaport lab | N/A |
| Yeast: YPH499 Δsam37Δtom70 shuffle strain: sam37Δ::HIS3 tom70Δ::NatNT2 (SAM37 URA3) derivative of YPH499 WT | This study | N/A |
| Yeast: BY4742 WT: MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 [ρ+] | Euroscarf | N/A |
| Yeast: BY4742 Δtim8: tim8Δ:: kanMX derivative of BY4742 WT | Euroscarf | N/A |
| Yeast: BY4742 Δelo2: elo2Δ:: kanMX derivative of BY4742 WT | Euroscarf | N/A |
| Yeast: BY4742 Δpex18: pex18Δ:: kanMX derivative of BY4742 WT | Euroscarf | N/A |
| Yeast: BY4742 Δpex17: pex17Δ:: kanMX derivative of BY4742 WT | Euroscarf | N/A |
| Yeast: BY4742 Δ pfd1: pfd1Δ:: kanMX derivative of BY4742 WT | Euroscarf | N/A |
| Yeast: BY4742 Δ hch1: hch1 Δ:: kanMX derivative of BY4742 WT | Euroscarf | N/A |
| Yeast: BY4742 Δloa1: loa1Δ:: kanMX derivative of BY4742 WT | Euroscarf | N/A |
| Yeast: BY4742 Δtim13: tim13Δ:: kanMX derivative of BY4742 WT | Euroscarf | N/A |
| Yeast: BY4742 Δsam37: sam37Δ:: kanMX derivative of BY4742 WT | Euroscarf | N/A |
| Yeast: W303 WT: MATa ura3-1 ade2-1 his3-11 leu2-3,112 trp1 Δ2 | Herrmann lab | Peleh et al., 2017 |
| Yeast: W303 Δtom70/71: tom70Δ::KanMX4 tom71 Δ::NatNT2 derivative of W303 WT | Doron Rapaport lab | Jores et al., 2018 |
| Yeast: yMS721 WT: MATα ura3Δ0 his3Δ1 leu2Δ0 his3Δ1 can1Δ::STE2pr-spHIS5; lyp 1ΔSTE3pr-LEU2; met15Δ0 | Maya Schuldiner lab | Tong and Boone, 2007 |
| Yeast: yMS721 Δtom70: tom70Δ::NAT2 derivative of yMS721 WT | This study | N/A |
| Yeast: yMS721 Δtom71: tom71 Δ::kanMX derivative of yMS721 WT | This study | N/A |
| Yeast: yMS721 Δtom70Δtom71: tom70Δ::NAT2 tom71Δ:: kanMX derivative of yMS721 WT | This study | N/A |
| Software and algorithms | ||
| Coral Photopaint X7 | Corel | N/A |
| BioFSharp | https://github.com/CSBiology/BioFSharp | 1.2.0 |
| Coral Draw X7 | Corel | N/A |
| R 3.6.3 | R Core Team | N/A |
| MaxQuant 1.6.10.43 | N/A | Tyanova et al., 2016 |
| Balony | Young and Loewen, 2013 | N/A |
| iMTS-L profiles | http://iMLP.bio.uni-kl.de/ | N/A |
| AIDA software | Elysia-raytest | N/A |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Prof. Dr. Johannes M. Herrmann (hannes.herrmann@biologie.uni-kl.de).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
Experimental Model and Subject Details
Yeast strains, plasmids and growth conditions
The yeast strains used in this study are either based on BY4742, W303 or YPH499 background. All strains used in this study are described in detail in the key resources table.
All strains were either grown on YP (1% yeast extract and 2% peptone) medium containing 2% glucose or galactose (Altmann et al., 2007) or on minimal synthetic medium (S) containing 0.67% (w/v) yeast nitrogen base and 2% glucose, galactose or lactate as carbon source. To express proteins from the GAL promoter, cells were shifted to 0.5% galactose containing medium for 4.5 h.
The shuffle strain for SAM37 was obtained by replacement of the SAM37 genomic open reading frame with a HIS3 cassette in a YPH499 WT. Afterward, a pRS426-TPI plasmid expressing SAM37 was transformed, following the subsequent replacement of the TOM70 genomic reading frame with a NAT2 cassette.
In order to anchor the TPR-containing proteins to the mitochondrial outer membrane, a pYX142-TPI vector containing the N-terminal Tom70-anchor (residues 1-98) was used to insert the various constructs (for residues see Figure 4B). The Tah1 mutation in the MEEVD binding site was achieved by PCR-based site-directed mutagenesis (Quick-Change method, Stratagene) using suitable primer sequences with the desired mutation. For construction of the mt-Tah1-C2-C3 variant, the sequences corresponding to the protein sequence of C2 and C3 domains of Tom70 (residues 247-460 and 461-617) were cloned into the mt-Tah1-containing pYX142 vector.
Yeast transformation was carried out by the lithium acetate method (Gietz et al., 1992). Empty vectors were also transformed in parallel to serve as negative controls.
Method Details
CRISPRI system construction
We employ an improved version of a previously generated single plasmid CRISPRi system (Smith et al., 2016) by making it compatible with Type IIS/Golden Gate Assembly, by employing an improved structural gRNA that reduces premature Polymerase III termination (Chen et al., 2013) and by addition of a KanMX resistance cassette. Briefly, we first generated the pKR297 plasmid, containing the RPR1(TetO) promoter, a BspQI-flanking gRNA cassette with an AscI site to remove uncut plasmid, the structural gRNA part and TetR. The pTef1-dCas9-Mxi1-tCyc1 fragment (from pRS416-dCas9-Mxi1: https://www.addgene.org/73796/) was then introduced to yield pKR359 (https://benchling.com/s/seq-gndJVnw6U1oisO0sL65k/edit), and KanMX was inserted to yield pKR366 (https://benchling.com/s/seq-Ymw9j7Wn3MM7g8N7Ny9K/edit).
To assemble the TOM70 gRNA, two oligonucleotides were annealed in CutSmart buffer (NEB) to form a double-stranded sticky end fragment. pKR366 was digested with BspQI (NEB), gel-purified, and the fragment inserted matching with the BspQI sites using T4 ligase (NEB), according to the supplier’s instructions.
Growth assays and viability tests
For spot analysis, the respective yeast strains were grown in liquid rich or synthetic media. Total yeast cells equivalent to 0.5/0.2 OD600 were harvested at exponential phase. The cells were washed in sterile water and subjected to ten-fold serial dilutions. Each dilution was spotted on rich or synthetic media followed by incubation at 30°C or 37°C. Pictures were taken after different days of the growth.
Growth curves were performed in a 96 well plate, using the automated ELx808 Absorbance Microplate Reader (BioTek®). The growth curves started at 0.1 OD600 and the OD600 was measured every 10 min for 72 h at 30°C. The mean of technical triplicates was calculated and plotted in R. For CRISPRi-mediated repression of TOM70, strains were incubated with 960 ng/ml Anhydrotetracycline for 6 h prior to growth curve analysis.
High-throughput screening of the GFP collection
To analyze the effect of TOM70 and TOM71 deletions on mitochondrial protein import we compiled a mini-library of 113 MTS-independent strains with mitochondrial GFP signal from the N-SWAT-library with NOP1 promotor and N-terminal sfGFP tag, but without a generic MTS inserted before the sfGFP (Yofe et al., 2016; Weill et al., 2018). This mini-library included many members of the metabolite carrier family, other inner membrane proteins, and outer membrane proteins (Table S3). We didn’t include any MTS-dependent strains in the mini-library since in the N-SWAT library they all carry a very strong generic MTS inserted before sfGFP and thus considerably influencing the import pathway taken by the protein. We constructed Δtom70, Δtom71, and double mutant Δtom70Δtom71 strains in the synthetic genetic array (SGA) compatible background (Tong and Boone, 2007) using standard yeast transformation techniques (Gietz and Woods, 2006; Janke et al., 2004) (See Key resources table). We mated these strains with the mitochondrial mini-library and selected for haploid cells harboring both the GFP tag and the required deletion using automated mating and selection approaches as described before (Tong and Boone, 2007; Cohen and Schuldiner, 2011). All mating and selection procedures were performed using RoToR high-density arrayer (Singer Instruments).
For imaging, the resulting haploid libraries with TOM70, TOM71, or double deletion were inoculated from agar plates into SD-URA liquid media (6.7 g l-1 yeast nitrogen base without amino acids, 20 g l-1 glucose, and optimized nutrient supplement without uracil supplemented with 200 μg ml-1 nourseothricin for Δtom70, 500 μg ml-1 geneticin for Δtom71, or both antibiotics for the Δtom17 Δtom71 double mutant in 384-well plates and grown overnight at 30°C with shaking. The donor mini-library was at the same time inoculated into SD-URA and grown in the same conditions. All liquid media operations and automated imaging were performed using a JANUS liquid handler (PelkinElmer) connected to an incubator (LiCONiC) and a microscope (Breker et al., 2013). The overnight cultures were diluted 20 times in SD-URA media without antibiotics. After 4 h of growth at 30°C the yeast cultures were transferred to Concanavalin A-coated (Sigma Aldrich) glass-bottom 384-well plates (Matrical Bioscience) and adhered for 20 minutes. Non-adhering cells were washed away with SD-URA-Riboflavin (same as regular SD-URA except yeast nitrogen base without riboflavin and without amino-acids is used to reduce media autofluorescence) that was also used as an imaging media. The plates were transferred to an automated ScanR miscroscopic system (Olympus) using a robotic swap arm (Hamilton). The cells were imaged in bright field and GFP (excitation filter 490/20 nm, emission filter 535/50 nm) channels with 60x air objective (NA 0.9) and the images were recorded on ORCA-ER charge-coupled device camera (Hammamatsu). The donor mini-library and the libraries crossed with TOM70 and TOM71 deletion strains were imaged on the same day.
For each strain the four images from the donor library, the library crossed with Δtom70, Δtom71, and Δtom70Δtom71 were displayed side by side and GFP signal localization and intensity were visually assessed (Table S3).
For quantitive analysis, the cells’ outlines were determined in bright field channel using a custom MATLAB script and median fluorescence intensity was calculated within these outlines. These values were averaged across all detected cells. The strain with the lowest mean cell intensity was taken as a cell background value and this value was subtracted from all other values to obtain background-corrected fluorescence intensities. The background fluorescence for each micrograph was also calculated using background subtraction procedure to assess illumination stability during the imaging process. The illumination was stable throughout the whole imaging period so we performed no additional corrections of the measured cell fluorescence intensities. Mean fluorescence for each strain of the donor mini-library was directly compared to the mean fluorescence of the same strain crossed to TOM70 and TOM71 deletions (Table S3).
For MitoTracker colocolazation experiments, yeast were adhered to Con A coated plates for 20 min and then stained with 50 nM MitoTracker CMTMRos in SD-URA without Riboflavin for 10 min. The staining solution was replaced with SD-URA without Riboflavin for imaging. The cells were imaged using VisiScope Confocal Cell explorer system consisting of Yokogawa spinning disk scanning unit attached to the Olympus IX83 microscope and equipped with PCO-Edge sCMOS detector controlled by VisView software. The imaging was performed with 60x oil objective. Representative areas of the micrographs were cropped and linearly adjusted for contrast using ImageJ.
TOM70 genetic interaction screen
To investigate genetic interactions of TOM70 we crossed the Δtom70 strain with the yeast full-genome knock-out collection (Giaever et al., 2002) and performed single and double mutant selection as described before (Tong and Boone, 2007). All mating and selection procedures were performed using RoToR high-density arrayer (Singer Instruments).
Briefly, after mating and sporulation all haploid MATα cells were selected on SD-LEU-LYS-ARG (6.7 g l-1 yeast nitrogen base without amino acids, 20 g l-1 glucose, complete set of supplements without leucine, lysine, and arginine) supplied with canavanine and thialysine. Then the haploids were plated on SD-LEU-LYS-ARG supplied with canavanine, thialysine, and geneticin (G418) to select all haploids that have the library knock-outs disregarding of the TOM70 allele. From these plates the strains were simultaneously replicated either on the same media (SD-LEU-LYS-ARG + canavanine + thialysine + G418) to measure the library knock out colony size or on the media additionally supplied with nourseothricin (NAT) to select both for the library knock out and Δtom70 allele and to measure the colony size of the double mutant. The plates were grown overnight and photographed the next day. Size of the colonies was determined and normalized for each plate using SGAtools (Wagih et al., 2013). For each library knock-out strain the colony size difference between single (library) mutant selection (SD-LEU-LYS-ARG + canavanine + thialysine + G418) and double mutant (library +Δtom70) selection media (SD-LEU-LYS-ARG + canavanine + thialysine + G418 + NAT) was calculated as a measure of genetic interaction with TOM70 (Table S5).
Overexpression assay
Yeast cells were inoculated in non-inducing medium. At mid-log phase (OD 0.6 – 0.8), cells were shifted to inducing conditions (0.5% galactose). At the indicated time points, 2 OD600 were harvested by centrifugation (20,000 g, 3 min, RT) and whole cell lysates were prepared. Whole cell lysates were prepared for the indicated time points to investigate the degradation behavior.
Fractionation assay
Overnight cultures of respective strains were diluted and induced with 0.5% Galactose. After 4.5 hours cells of 10 OD600 were harvested by centrifugation (5,000 g, 10min, RT). After washing with dH2O the pellet was resuspended in 1 ml MP1 buffer (100 mM Tris, 10 mM DTT) and incubated for 30 min at 30°C. Cells were pelleted by centrifugation (5,000 g, 3 min, RT) and washed with 1 ml 1.2 M Sorbitol. Afterwards pellets were resuspended in 1 ml MP2 buffer (1.2 M Sorbitol, 20 mM KPi pH 7.4, Zymolyase) and incubated 30 min at 30 °C. From now on, any step was done at 4°C or on ice. Pelleted cells were resuspended in 1 ml Homogenisation buffer (10 mM Tris pH7.4,1 mM EDTA, 0.6 M Sorbitol) and dounced in a cooled potter for 15 times. After centrifugation (5,000 g, 3 min, 4°C) the supernatant was taken and centrifuged again (5,000 g, 3 min, 4°C). The pellet (P1) was taken up in 100 μl Laemmli +DTT. To obtain the pellet (P2) the previous supernatant was centrifuged at 12,000 g for 5 min at 4°C and the pellet was taken up in 100 μl Laemmli +DTT. The supernatant was finally centrifuged at 30,000 g for 30min at 4°C. The pellet (P3) was resuspended in 100 μl Laemmli +DTT. The supernatant was transferred into a new tube and 200 μl 72% TCA was added. The samples were frozen at 20°C overnight. The next day the samples were thawed on ice and centrifuged at 30,000 g for 20 min at 4°C. The supernatant was discarded and the pellets were washed with ice-cold acetone. After centrifugation at 30,000 g for 20 min at 4°C the pellet (P4) was taken up in 100 μl Laemmli + DTT. To resuspend P1-P4, samples were incubated shaking for 30 min at 30°C. Per sample 25 μl were loaded on 16% SDS-Gel.
Overnight cultures of respective strains were diluted and induced with 0.5% galactose. After 4.5 hours cells of 10 OD600 were harvested by centrifugation (5,000 g, 10min, RT). After washing with dH2O the pellet was resuspended in 1 mL MP1 buffer (100 mM Tris, 10 mM DTT) and incubated for 30 min at 30°C. Cells were pelleted by centrifugation (5,000 g, 3 min, RT) and washed with 1 mL 1.2 M Sorbitol. Afterward pellets were resuspended in 1 mL MP2 buffer (1.2 M Sorbitol, 20 mM KPi pH 7.4, Zymolyase) and incubated 30 min at 30°C. From now on, any step was done at 4°C or on ice. Pelleted cells were resuspended in 1 mL homogenization buffer (10 mM Tris pH7.4,1 mM EDTA, 0.6 M Sorbitol) and dounced in a cooled potter 15 times. After centrifugation (5,000 g, 3 min, 4°C) the supernatant was taken and centrifuged again (5,000 g, 3 min, 4°C). The pellet (P1) was taken up in 100 μl Laemmli +DTT. To obtain the pellet (P2), the previous supernatant was centrifuged at 12,000 g for 5 min at 4°C and the pellet was taken up in 100 μl Laemmli +DTT. The supernatant was finally centrifuged at 30,000 g for 30min at 4°C. The pellet (P3) was resuspended in 100 μl Laemmli +DTT. The supernatant was transferred into a new tube and 200 μl 72% TCA was added. The samples were frozen at 20°C overnight. The next day the samples were thawed on ice and centrifuged at 30,000 g for 20 min at 4°C. The supernatant was discarded and the pellets were washed with ice-cold acetone. After centrifugation at 30,000 g for 20 min at 4°C the pellet (P4) was taken up in Laemmli + DTT. Samples were analyzed by SDS-Gel.
Heat-shock assay
Yeast cells were pre-grown in SD medium at 30°C. At mid-log phase (OD 0.6–0.8), 0.1 OD600 were exposed to a heat-shock at 50°C for the indicated time points. After each time point, 0.01 OD600 were equally plated on SD plates and incubated for two days at 30°C.
Sample preparation and mass spectrometric identification of proteins
For IP mass spectrometry, cells (Δtom70/71+ HA-tagged mt-Tah1 or Δtom70/71 + empty vector) were incubated in SGal-Leu medium and 20 OD of cells were harvested. Cell lysates were prepared in 1000 μl lysis buffer (10mM tris, 150mM NaCl, 0,5mM EDTA, 0.5% Triton, 1mM PMSF) using a FastPrep-24 5 G homogenizer (MP Biomedicals) with 3 cycles of 30 s, speed 6.0 m s–1, 120 s breaks, glass beads. Samples were centrifuged 10 min at 20.000 g. Cell lysates were used for an IP with protein-A-Sepharose beads (expedeon) and HA-serum. The tagged proteins were bound to the beads for 1 h at 4°C. After spinning the samples down the beads were washed 3x with 800 μl wash buffer I (150 mM NaCl, 50 mM Tris pH 7.5,5% Glycerol, 0.05% Tx-100) and afterward 2x with 500 μl wash buffer II (150 mM NaCl, 50 mM Trish pH 7.5,5% Glycerol). For elution and trypsin digestion 50 μl elution buffer I was added (2M Urea, 50 mM Tris pH 7.5,1 mM DTT, 5 ng/μl Trypsin) and incubated for 1 h at RT. Afterward 1 μl Trypsin was added and incubated for 10 min at RT. 50 μl elution buffer II (2M Urea, 50 mM Tris pH 7.5, 5mM CAA) were added. Samples were incubated ON in the dark at RT. pH of samples was adjusted to pH < 2 with Tri-flouracetic acid. Desalting/reversed-Phase cleanup with 3xC18 stage tips. Samples were dried down in speed-vac and resolubilized in 9 μl buffer A (0.1 % formic acid in MS grad water) and 1 μl buffer A* (0.1 % formic acid, 0.1 % TFA in MS grad water).
For mass spectrometry sample preparation of whole cell lysates, strains were pregrown in S-medium containing 2% galactose at 30°C and either shifted to 37°C or kept at 30°C for 16 h.
50 OD600 of cells were harvested at each time point by centrifugation (17,000 g, 3 min, 2°C), washed with prechilled water, snap-frozen in liquid nitrogen and stored at −80°C. Cells lysates were prepared in lysis buffer (50 mM Tris pH 7.6,5% (w/v)SDS) using a FastPrep-24 5G homogenizer (MP Biomedicals, Heidelberg, Germany) with 3 cycles of 30 s, speed 8.0 m/s, 120 s breaks, glass beads). Lysates were diluted to 2% (w/v) SDS and protein concentrations were determined using the Pierce BCA Protein Assay (Thermo Scientific, #23225). 20 μg of each lysate were subjected to an in-solution tryptic digest using a modified version of the Single-Pot Solid-Phase-enhanced Sample Preparation (SP3) protocol (Hughes et al., 2014,2019). Here, lysates were added to Sera-Mag Beads (Thermo Scientific, #4515-2105-050250,6515-2105-050250) in 10 μl 15% formic acid and 30 μl of ethanol. Binding of proteins was achieved by shaking for 15 min at room temperature. SDS was removed by four subsequent washes with 200 μl of 70% ethanol. Proteins were digested with 0.4 μg of sequencing grade modified trypsin (Promega, #V5111) in 40 μl HEPES/NaOH, pH 8.4 in the presence of 1.25 mM TCEP and 5 mM chloroacetamide (Sigma-Aldrich, #C0267) overnight at room temperature. Beads were separated, washed with 10 μl of an aqueous solution of 2% DMSO and the combined eluates were dried down. In total three biological replicates were prepared (n = 3). Each replicate included samples of all 5 strains at 30°C or 37°C (in total 10 samples per replicate). Peptides were reconstituted in 10 μl of H2O and reacted with 80 μg of TMT10plex (Thermo Scientific, #90111) (Werner et al., 2014) label reagent dissolved in 4 μl of acetonitrile for 1 h at room temperature. Excess TMT reagent was quenched by the addition of 4 μl of an aqueous solution of 5% hydroxylamine (Sigma, 438227). Peptides were mixed to achieve a 1:1 ratio across all TMT-channels. Mixed peptides were desalted on home-made StageTips containing Empore C18 disks (Rappsilber et al., 2007). The samples were then analyzed by LC-MS/MSon a Q Exactive HF instrument (Thermo Scientific) as previously described.
Briefly, peptides were separated using an Easy-nLC 1200 system (Thermo Scientific) coupled to a Q Exactive HF mass spectrometer via a Nanospray-Flex ion source. The analytical column (50 cm, 75 μm inner diameter (NewObjective) packed in-house with C18 resin ReproSilPur 120, 1.9 μm diameter Dr. Maisch) was operated at a constant flow rate of 250 nl/min. A 3 h gradient was used to elute peptides (Solvent A: aqueous 0.1% formic acid; Solvent B: 80 % acetonitrile, 0.1% formic acid). Peptides were analyzed in positive ion mode applying with a spray voltage of 2.3 kV and a capillary temperature of 250°C. MS spectra with a mass range of 375–1.400 m/z were acquired in profile mode using a resolution of 120.000 [maximum fill time of 80 ms or a maximum of 3e6 ions (automatic gain control, AGC)]. Fragmentation was triggered for the top 15 peaks with charge 2–8 on the MS scan (data-dependent acquisition) with a 30 s dynamic exclusion window (normalized collision energy was 32). Precursors were isolated with a 0.7 m/z window and MS/MS spectra were acquired in profile mode with a resolution of 60,000 (maximum fill time of 100 ms, AGC target of 2e5 ions, fixed first mass 100 m/z).
Analysis of mass spectrometry data
Peptide and protein identification and quantification was done using the MaxQuant software (version 1.6.10.43) (Cox and Mann,2008; Cox et al., 2011; Tyanova et al., 2016) and a Saccharomyces cerevisiae proteome database obtained from Uniprot. 10plex TMT was chosen in Reporter ion MS2 quantification, up to 2 tryptic miss-cleavages were allowed, protein N-terminal acetylation and Met oxidation were specified as variable modifications and Cys carbamidomethylation as fixed modification. The “Requantify” and “Second Peptides” options were deactivated. False discovery rate was set at 1% for peptides, proteins and sites, minimal peptide length was 7 amino acids.
The output files of MaxQuant were processed using the R programming language. Only proteins that were quantified with at least two unique peptides were considered for the analysis. Moreover, only proteins that were identified in at least two out of three MS runs were kept. A total of 2920 proteins passed the quality control filters. Raw signal sums were cleaned for batch effects using limma (Ritchie et al., 2015) and further normalized using variance stabilization normalization (Huber et al., 2002). Proteins were tested for differential expression using the limma package for the indicated comparison of strains.
A reference list of yeast mitochondrial proteins was obtained from (Morgenstern et al., 2017). Gene set enrichment analysis was performed using Fisher’s exact test. A Benjamini-Hochberg procedure was used to account for multiple testing, where this was performed (Benjamini and Hochberg, 1995).
Calculation of aggregation propensity
Aggregation propensity is determined from primary protein sequence using the “hot spot” approach according to Sanchez de Groot et al., 2005. A predictive model is based on the individual aggregation propensities of natural amino acids, which have already been experimentally validated in the literature and provide insights into the effect of disease-linked mutations in these polypeptides. Here, we used the average over a sliding window of 5, 7, 9 or 11 residues depending on total sequence length (≤75, ≤ 175, ≤ 300, or > 300). The resulting value is assigned to the central residue in the window and then averaged to obtain the aggregation-propensity of the respective protein (Conchillo-Solé et al., 2007). For convenient application the algorithm was implemented using BioFSharp 1.2.0 (https://github.com/CSBiology/BioFSharp).
Miscellaneous
The following methods were performed according to already published methods: Import into isolated mitochondria (Backes and Herrmann, 2017), CCCP chase experiment (Backes and Herrmann, 2017), iMTS-L profile generation (Backes and Herrmann, 2017), isolation of mitochondria (Saladi et al., 2020), whole cell lysates (Saladi et al., 2020), RNA-isolation (Boos et al., 2019), quantitative real-time PCR assays (Boos et al., 2019), PACE-YFP reporter assay (Boos et al., 2019).
Quantification and Statistical Analysis
Unless otherwise indicated, experiments were performed in n = 3 independent biological replicates and mean values and standard deviations are presented in the figures. Significance of results was assessed using standard statistical tests as detailed in the respective figure legends and descriptions in the STAR Methods section. In particular, statistical analysis of the mass spectrometry data was performed with the limma package within R.
Where multiple comparisons were analyzed, P values were adjusted for multiple hypothesis testing with the Benjamini-Hochberg procedure.
Western blot analyses were independently replicated with similar results, and representative data are shown in the figures. Quantification was performed with Fiji/ImageJ and significance testing was performed with Student’s t-Test.
Supplementary Material
In brief.
Backes et al. identify the spectrum of substrates of the mitochondrial Tom70 receptor. Many Tom70 clients are aggregation-prone and/or membrane proteins. In vivo, the critical function of Tom70 is the recruitment of chaperones to the mitochondrial surface, whereas its ability to directly bind precursor proteins is largely dispensable.
Highlight.
Tom70 supports the targeting of a wide range of precursor proteins to mitochondria
In vivo, the main function of Tom70 is to recruit chaperones to the outer membrane
Small inner membrane proteins are highly toxic in the absence of Tom70
Tom70 protects the cytosol against toxic effects of mitochondrial precursors
Acknowledgements
We thank Vera Nehr and Sabine Knaus for technical assistance. We thank Mikhail Savitski, Per Haberkant, and Frank Stein for help with setting up the mass spectrometry workflow. We thank Carina Groh and Carsten Balzer for the help with data analysis and Amir Fadel for help with the microscopy screens. Additionally, we thank Anna Mariam Schlagowski for providing the GAL1-Pet9 overexpression plasmid, Janina Laborenz for the Tom70-TMD plasmid, and Klaus Pfanner and Ophry Pines for antibodies. This study was supported by funding from the Deutsche Forschungsgemeinschaft (DIP MitoBalance to J.M.H., D.R., and M.S. and HE2803/10-1 to J.M.H.), the Joachim Herz Stiftung (to F.B.), and the Forschungsinitiative Rheinland-Pfalz BioComp (to J.M.H. and F.B.). Y.S.B. is supported by an EMBO postdoctoral fellowship. M.S. is an incumbent of the Dr. Gilbert Omenn and Martha Darling Professorial Chair in Molecular Genetics.
Footnotes
Author Contributions
S.B., Y.S.B., and T.F. designed, cloned, and verified the constructs and strains; J.M.H. conceived the project; S.B., L.K., F.B., M.R., and Z.S. carried out the mass-spectrometry-based proteomics; S.B., F.B., M.R., and T.M. performed the bioinformatical analysis of the mass spectrometry data; S.B., T.F., S.L., and J.Z. carried out the biochemical experiments to test the relevance of Tom70 for mitochondrial protein biogenesis; Y.S.B., C.B., and M.S. designed and performed genetic screens to identify Tom70-dependent proteins; C.J., J.D.S., and L.M.S. developed the CRISPR interference strategy and provided the tools that were used by S.B. and S.L.; S.B., D.R., M.S., F.B., and J.M.H. analyzed the data; J.M.H., S.B., and F.B. wrote the manuscript with the help and input of all authors.
Declaration of Interests
The authors declare no competing interests.
Data and code availability
The mass spectrometry proteomics data (see also Tables S1 and S2) have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD021173.
reviewer13653@ebi.ac.uk
Mass spectrometry data of the immunoprecipitation experiments (see also Table S4) are available via ProteomeXchange with identifier PXD023149.
reviewer_pxd023149@ebi.ac.uk
References
- Altmann K, Dürr M, Westermann B. In: Mitochondria Practical Protocols. Leister D, Herrmann JM, editors. Humana Press; 2007. Saccharomyces cerevisiae as a model organism to study mitochondrial biology. [DOI] [PubMed] [Google Scholar]
- Araiso Y, Tsutsumi A, Qiu J, Imai K, Shiota T, Song J, Lindau C, Wenz LS, Sakaue H, Yunoki K, et al. Structure of the mitochondrial import gate reveals distinct preprotein paths. Nature. 2019;575:395–401. doi: 10.1038/s41586-019-1680-7. [DOI] [PubMed] [Google Scholar]
- Back R, Dominguez C, Rothé B, Bobo C, Beaufils C, Moréra S, Meyer P, Charpentier B, Branlant C, Allain FH, Manival X. High-resolution structural analysis shows how Tah1 tethers Hsp90 to the R2TP complex. Structure. 2013;21:1834–1847. doi: 10.1016/j.str.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Backes S, Herrmann JM. Protein Translocation into the Intermembrane Space and Matrix of Mitochondria: Mechanisms and Driving Forces. Front Mol Biosci. 2017;4:83. doi: 10.3389/fmolb.2017.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes S, Hess S, Boos F, Woellhaf MW, Gödel S, Jung M, Mühlhaus T, Herrmann JM. Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. J Cell Biol. 2018;217:1369–1382. doi: 10.1083/jcb.201708044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Ziv R, Bolas T, Dillin A. Systemic effects of mitochondrial stress. EMBO Rep. 2020;21:e50094. doi: 10.15252/embr.202050094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryshnikova A, Costanzo M, Kim Y, Ding H, Koh J, Toufighi K, Youn JY, Ou J, San Luis BJ, Bandyopadhyay S, et al. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Methods. 2010;7:1017–1024. doi: 10.1038/nmeth.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Walter W, Yan W, Craig EA. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Wenz LS, Krüger V, Lehmann W, Müller JM, Goroncy L, Zufall N, Lithgow T, Guiard B, Chacinska A, et al. The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J Cell Biol. 2011;194:387–395. doi: 10.1083/jcb.201102044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Menachem R, Wang K, Marcu O, Yu Z, Lim TK, Lin Q, Schueler-Furman O, Pines O. Yeast aconitase mitochondrial import is modulated by interactions of its C and N terminal domains and Ssa1/2 (Hsp70) Sci Rep. 2018;8:5903. doi: 10.1038/s41598-018-24068-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Boos F, Mühlhaus T, Herrmann JM. Detection of Internal Matrix Targeting Signal-like Sequences (iMTS-Ls) in Mitochondrial Precursor Proteins Using the TargetP Prediction Tool. Biol-protocol. 2018;8:e2474. doi: 10.21769/BioProtoc.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos F, Krämer L, Groh C, Jung F, Haberkant P, Stein F, Wollweber F, Gackstatter A, Zöller E, van der Laan M, et al. Mitochondrial protein-induced stress triggers a global adaptive transcriptional programme. Nat Cell Biol. 2019;21:442–451. doi: 10.1038/s41556-019-0294-5. [DOI] [PubMed] [Google Scholar]
- Boos F, Labbadia J, Herrmann JM. How the Mitoprotein-Induced Stress Response Safeguards the Cytosol: A Unified View. Trends Cell Biol. 2020;30:241–254. doi: 10.1016/j.tcb.2019.12.003. [DOI] [PubMed] [Google Scholar]
- Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, Azzag K, Robert MC, Ahmad Y, Neel H, et al. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell. 2010;39:912–924. doi: 10.1016/j.molcel.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breker M, Gymrek M, Schuldiner M. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J Cell Biol. 2013;200:839–850. doi: 10.1083/jcb.201301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brix J, Rüdiger S, Bukau B, Schneider-Mergener J, Pfanner N. Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J Biol Chem. 1999;274:16522–16530. doi: 10.1074/jbc.274.23.16522. [DOI] [PubMed] [Google Scholar]
- Brix J, Ziegler GA, Dietmeier K, Schneider-Mergener J, Schulz GE, Pfanner N. The mitochondrial import receptor Tom70: identification of a 25 kDa core domain with a specific binding site for preproteins. J Mol Biol. 2000;303:479–488. doi: 10.1006/jmbi.2000.4120. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44(D1):D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NC, Likić VA, Waller RF, Mulhern TD, Lithgow T. The C-terminal TPR domain of Tom70 defines a family of mitochondrial protein import receptors found only in animals and fungi. J Mol Biol. 2006;358:1010–1022. doi: 10.1016/j.jmb.2006.02.062. [DOI] [PubMed] [Google Scholar]
- Chartron JW, Gonzalez GM, Clemons WM., Jr A structural model of the Sgt2 protein and its interactions with chaperones and the Get4/Get5 complex. J Biol Chem. 2011;286:34325–34334. doi: 10.1074/jbc.M111.277798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/ Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y, Schuldiner M. Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol Biol. 2011;781:127–159. doi: 10.1007/978-1-61779-276-2_8. [DOI] [PubMed] [Google Scholar]
- Conchillo-Solé O, de Groot NS, Avilés FX, Vendrell J, Daura X, Ventura S. AGGRESCAN: a server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinformatics. 2007;8:65. doi: 10.1186/1471-2105-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Coyne LP, Chen XJ. mPOS is a novel mitochondrial trigger of cell death - implications for neurodegeneration. FEBS Lett. 2018;592:759–775. doi: 10.1002/1873-3468.12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988;332:800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- Doan KN, Grevel A, Mårtensson CU, Ellenrieder L, Thornton N, Wenz LS, Opaliński Ł, Guiard B, Pfanner N, Becker T. The Mitochondrial Import Complex MIM Functions as Main Translocase for a-Helical Outer Membrane Proteins. Cell Rep. 2020;31:107567. doi: 10.1016/j.celrep.2020.107567. [DOI] [PubMed] [Google Scholar]
- Doring K, Ahmed N, Riemer T, Suresh HG, Vainshtein Y, Habich M, Riemer J, Mayer MP, O’Brien EP, Kramer G, Bukau B. Profiling Ssb-Nascent Chain Interactions Reveals Principles of Hsp70-Assisted Folding. Cell. 2017;170:298–311.:e20. doi: 10.1016/j.cell.2017.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drwesh L, Rapaport D. Biogenesis pathways of a-helical mitochondrial outer membrane proteins. Biol Chem. 2020;401:677–686. doi: 10.1515/hsz-2019-0440. [DOI] [PubMed] [Google Scholar]
- Edwards R, Gerlich S, Tokatlidis K. The biogenesis of mitochondrial intermembrane space proteins. Biol Chem. 2020;401:737–747. doi: 10.1515/hsz-2020-0114. [DOI] [PubMed] [Google Scholar]
- Eliyahu E, Lesnik C, Arava Y. The protein chaperone Ssa1 affects mRNA localization to the mitochondria. FEBS Lett. 2012;586:64–69. doi: 10.1016/j.febslet.2011.11.025. [DOI] [PubMed] [Google Scholar]
- Finger Y, Riemer J. Protein import by the mitochondrial disulfide relay in higher eukaryotes. Biol Chem. 2020;401:749–763. doi: 10.1515/hsz-2020-0108. [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. In: Yeast Protocol. Xiao W, editor. Humana Press; 2006. Yeast Transformation by the LiAc/SS Carrier DNA/PEG Method. [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold VA, Chroscicki P, Bragoszewski P, Chacinska A. Visualization of cytosolic ribosomes on the surface of mitochondria by electron cryotomography. EMBO Rep. 2017;18:1786–1800. doi: 10.15252/embr.201744261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornicka A, Bragoszewski P, Chroscicki P, Wenz LS, Schulz C, Rehling P, Chacinska A. Adiscrete pathway for the transfer of intermembrane space proteins across the outer membrane of mitochondria. Mol Biol Cell. 2014;25:3999–4009. doi: 10.1091/mbc.E14-06-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JB, Canniff NP, Hebert DN. TPR-containing proteins control protein organization and homeostasis for the endoplasmic reticulum. Crit Rev Biochem Mol Biol. 2019;54:103–118. doi: 10.1080/10409238.2019.1590305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KG, Aviram N, Laborenz J, Bibi C, Meyer M, Spang A, Schuldiner M, Herrmann JM. An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science. 2018;361:1118–1122. doi: 10.1126/science.aar8174. [DOI] [PubMed] [Google Scholar]
- Harano T, Nose S, Uezu R, Shimizu N, Fujiki Y. Hsp70 regulatesthe interaction between the peroxisome targeting signal type 1 (PTS1)-receptor Pex5p and PTS1. Biochem J. 2001;357:157–165. doi: 10.1042/0264-6021:3570157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hasson SA, Damoiseaux R, Glavin JD, Dabir DV, Walker SS, Koehler CM. Substrate specificity of the TIM22 mitochondrial import pathway revealed with small molecule inhibitor of protein translocation. Proc Natl Acad Sci USA. 2010;107:9578–9583. doi: 10.1073/pnas.0914387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haucke V, Lithgow T, Rospert S, Hahne K, Schatz G. The yeast mitochondrial protein import receptor Mas20p binds precursor proteins through electrostatic interaction with the positively charged presequence. J Biol Chem. 1995;270:5565–5570. doi: 10.1074/jbc.270.10.5565. [DOI] [PubMed] [Google Scholar]
- Hines V, Brandt A, Griffiths G, Horstmann H, Brütsch H, Schatz G. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 1990;9:3191–3200. doi: 10.1002/j.1460-2075.1990.tb07517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horten P, Colina-Tenorio L, Rampelt H. Biogenesis of Mitochondrial Metabolite Carriers. Biomolecules. 2020;10:1008. doi: 10.3390/biom10071008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoseini H, Pandey S, Jores T, Schmitt A, Franz-Wachtel M, Macek B, Buchner J, Dimmer KS, Rapaport D. The cytosolic cochaperone Sti1 is relevant for mitochondrial biogenesis and morphology. FEBS J. 2016;283:3338–3352. doi: 10.1111/febs.13813. [DOI] [PubMed] [Google Scholar]
- Hoshino A, Wang WJ, Wada S, McDermott-Roe C, Evans CS, Gosis B, Morley MP, Rathi KS, Li J, Li K, et al. The ADP/ATP translocase drives mitophagy independent of nucleotide exchange. Nature. 2019;575:375–379. doi: 10.1038/s41586-019-1667-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sültmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- Hughes CS, Foehr S, Garfield DA, Furlong EE, Steinmetz LM, Krijgsveld J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol Syst Biol. 2014;10:757. doi: 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CS, Moggridge S, Müller T, Sorensen PH, Morin GB, Krijgsveld J. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat Protoc. 2019;14:68–85. doi: 10.1038/s41596-018-0082-x. [DOI] [PubMed] [Google Scholar]
- Iwata K, Nakai M. Interaction between mitochondrial precursor proteins and cytosolic soluble domains of mitochondrial import receptors, Tom20 and Tom70, measured by surface plasmon resonance. Biochem Biophys Res Commun. 1998;253:648–652. doi: 10.1006/bbrc.1998.9769. [DOI] [PubMed] [Google Scholar]
- Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Jiménez B, Ugwu F, Zhao R, Ortí L, Makhnevych T, Pineda-Lucena A, Houry WA. Structure of minimal tetratricopeptide repeat domain protein Tah1 reveals mechanism of its interaction with Pih1 and Hsp90. J Biol Chem. 2012;287:5698–5709. doi: 10.1074/jbc.M111.287458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jores T, Lawatscheck J, Beke V, Franz-Wachtel M, Yunoki K, Fitzgerald JC, Macek B, Endo T, Kalbacher H, Buchner J, Rapaport D. Cytosolic Hsp70 and Hsp40 chaperones enable the biogenesis of mitochondrial β-barrel proteins. J Cell Biol. 2018;217:3091–3108. doi: 10.1083/jcb.201712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaukonen J, Juselius JK, Tiranti V, Kyttälä A, Zeviani M, Comi GP, Keränen S, Peltonen L, Suomalainen A. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science. 2000;289:782–785. doi: 10.1126/science.289.5480.782. [DOI] [PubMed] [Google Scholar]
- Kramer G, Shiber A, Bukau B. Mechanisms of Cotranslational Maturation of Newly Synthesized Proteins. Annu Rev Biochem. 2019;88:337–364. doi: 10.1146/annurev-biochem-013118-111717. [DOI] [PubMed] [Google Scholar]
- Labbadia J, Brielmann RM, Neto MF, Lin YF, Haynes CM, Morimoto RI. Mitochondrial Stress Restores the Heat Shock Response and Prevents Proteostasis Collapse during Aging. Cell Rep. 2017;21:1481–1494. doi: 10.1016/j.celrep.2017.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Qian X, Hu J, Sha B. Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J Biol Chem. 2009;284:23852–23859. doi: 10.1074/jbc.M109.023986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang X, Coyne LP, Yang Y, Qi Y, Middleton FA, Chen XJ. Mitochondrial carrier protein overloading and misfolding induce aggresomes and proteostatic adaptations in the cytosol. Mol Biol Cell. 2019;30:1272–1284. doi: 10.1091/mbc.E19-01-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz T, Neupert W, Herrmann JM. Import of small Tim proteins into the mitochondrial intermembrane space. EMBO J. 2003;22:4400–4408. doi: 10.1093/emboj/cdg421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini G, Nüske E, Leng W, Alberti S, Pigino G. Reorganization of budding yeast cytoplasm upon energy depletion. Mol Biol Cell. 2020;31:1232–1245. doi: 10.1091/mbc.E20-02-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mårtensson CU, Priesnitz C, Song J, Ellenrieder L, Doan KN, Boos F, Floerchinger A, Zufall N, Oeljeklaus S, Warscheid B, Becker T. Mitochondrial protein translocation-associated degradation. Nature. 2019;569:679–683. doi: 10.1038/s41586-019-1227-y. [DOI] [PubMed] [Google Scholar]
- Melber A, Haynes CM. UPRmt regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res. 2018;28:281–295. doi: 10.1038/cr.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin J, Kilisch M, Neumann P, Lytovchenko O, Gomkale R, Schendzielorz A, Schmidt B, Liepold T, Ficner R, Jahn O, et al. A presequence-binding groove in Tom70 supports import of Mdl1 into mitochondria. Biochim Biophys Acta. 2015;1853:1850–1859. doi: 10.1016/j.bbamcr.2015.04.021. [DOI] [PubMed] [Google Scholar]
- Millson SH, Vaughan CK, Zhai C, Ali MM, Panaretou B, Piper PW, Pearl LH, Prodromou C. Chaperone ligand-discrimination by the TPR-domain protein Tah1. Biochem J. 2008;413:261–268. doi: 10.1042/BJ20080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern M, Stiller SB, Lübbert P, Peikert CD, Dannenmaier S, Drepper F, Weill U, Höß P, Feuerstein R, Gebert M, et al. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 2017;19:2836–2852. doi: 10.1016/j.celrep.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opaliński Ł, Song J, Priesnitz C, Wenz LS, Oeljeklaus S, Warscheid B, Pfanner N, Becker T. Recruitment of Cytosolic J-Proteins by TOM Receptors Promotes Mitochondrial Protein Biogenesis. Cell Rep. 2018;25:2036–2043.:e5. doi: 10.1016/j.celrep.2018.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papic D, Krumpe K, Dukanovic J, Dimmer KS, Rapaport D. Multispan mitochondrial outer membrane protein Ugo1 follows a unique Mim1-dependent import pathway. J Cell Biol. 2011;194:397–405. doi: 10.1083/jcb.201102041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papić D, Elbaz-Alon Y, Koerdt SN, Leopold K, Worm D, Jung M, Schuldiner M, Rapaport D. The role of Djp1 in import of the mitochondrial protein Mim1 demonstrates specificity between a cochaperone and its substrate protein. Mol Cell Biol. 2013;33:4083–4094. doi: 10.1128/MCB.00227-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleh V, Zannini F, Backes S, Rouhier N, Herrmann JM. Erv1 of Arabidopsis thaliana can directly oxidize mitochondrial intermembrane space proteins in the absence of redox-active Mia40. BMC Biol. 2017;15:106. doi: 10.1186/s12915-017-0445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riba A, Itzhaki LS. The tetratricopeptide-repeat motif is a versatile platform that enables diverse modes of molecular recognition. Curr Opin Struct Biol. 2019;54:43–49. doi: 10.1016/j.sbi.2018.12.004. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, Neupert W. Distinct steps in the import of ADP/ATP carrier into mitochondria. J Biol Chem. 1987;262:7528–7536. [PubMed] [Google Scholar]
- Pfanner N, Tropschug M, Neupert W. Mitochondrial protein import: nucleoside triphosphates are involved in conferring import-competence to precursors. Cell. 1987;49:815–823. doi: 10.1016/0092-8674(87)90619-2. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Rehling P, Brandner K, Pfanner N. Mitochondrial import and the twin-pore translocase. Nat Rev Mol Cell Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- Rimmer KA, Foo JH, Ng A, Petrie EJ, Shilling PJ, Perry AJ, Mertens HD, Lithgow T, Mulhern TD, Gooley PR. Recognition of mitochondrial targeting sequences by the import receptors Tom20 and Tom22. J Mol Biol. 2011;405:804–818. doi: 10.1016/j.jmb.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MT, Müller H, Pfanner N. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J Biol Chem. 1999;274:20619–20627. doi: 10.1074/jbc.274.29.20619. [DOI] [PubMed] [Google Scholar]
- Saladi S, Boos F, Poglitsch M, Meyer H, Sommer F, Mühlhaus T, Schroda M, Schuldiner M, Madeo F, Herrmann JM. The NADH Dehydrogenase Nde1 Executes Cell Death after Integrating Signals from Metabolism and Proteostasis on the Mitochondrial Surface. Mol Cell. 2020;77:189–202.:e6. doi: 10.1016/j.molcel.2019.09.027. [DOI] [PubMed] [Google Scholar]
- Sanchez de Groot N, Pallares I, Vendrell J, Ventura S. Prediction of “hot spots” of aggregation in disease-linked polypeptides. BMC Struct Biol. 2005;5:18. doi: 10.1186/1472-6807-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlossmann J, Lill R, Neupert W, Court DA. Tom71, a novel homologue of the mitochondrial preprotein receptor Tom70. J Biol Chem. 1996;271:17890–17895. doi: 10.1074/jbc.271.30.17890. [DOI] [PubMed] [Google Scholar]
- Schmid AB, Lagleder S, Gräwert MA, Röhl A, Hagn F, Wandinger SK, Cox MB, Demmer O, Richter K, Groll M, et al. The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop. EMBO J. 2012;31:1506–1517. doi: 10.1038/emboj.2011.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenkert S, Dittmer S, Soll J. Structural components involved in plastid protein import. Essays Biochem. 2018;62:65–75. doi: 10.1042/EBC20170093. [DOI] [PubMed] [Google Scholar]
- Shakya VPS, Barbeau WA, Xiao T, Knutson CS, Hughes AL. The nucleus is a quality control center for non-imported mitochondrial proteins. bioRxiv. 2020 doi: 10.1101/2020.06.26.173781. [DOI] [Google Scholar]
- Shiota T, Imai K, Qiu J, Hewitt VL, Tan K, Shen HH, Sakiyama N, Fukasawa Y, Hayat S, Kamiya M, et al. Molecular architecture of the active mitochondrial protein gate. Science. 2015;349:1544–1548. doi: 10.1126/science.aac6428. [DOI] [PubMed] [Google Scholar]
- Smith JD, Suresh S, Schlecht U, Wu M, Wagih O, Peltz G, Davis RW, Steinmetz LM, Parts L, St Onge RP. Quantitative CRISPR interference screens in yeast identify chemical-genetic interactions and new rules for guide RNA design. Genome Biol. 2016;17:45. doi: 10.1186/s13059-016-0900-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T, Pfaller R, Griffiths G, Pfanner N, Neupert W. A mitochondrial import receptor for the ADP/ATP carrier. Cell. 1990;62:107–115. doi: 10.1016/0092-8674(90)90244-9. [DOI] [PubMed] [Google Scholar]
- Sontag EM, Samant RS, Frydman J. Mechanisms and Functions of Spatial Protein Quality Control. Annu Rev Biochem. 2017;86:97–122. doi: 10.1146/annurev-biochem-060815-014616. [DOI] [PubMed] [Google Scholar]
- Steger HF, Söllner T, Kiebler M, Dietmeier KA, Pfaller R, Trülzsch KS, Tropschug M, Neupert W, Pfanner N. Import of ADP/ATP carrier into mitochondria: two receptors act in parallel. J Cell Biol. 1990;111:2353–2363. doi: 10.1083/jcb.111.6.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein KC, Kriel A, Frydman J. Nascent Polypeptide Domain Topology and Elongation Rate Direct the Cotranslational Hierarchy of Hsp70 and TRiC/CCT. Mol Cell. 2019;75:1117–1130.:e5. doi: 10.1016/j.molcel.2019.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada K, Ueda I, Ohtsuka K, Oda T, Ichiyama A, Mori M. The requirement of heat shock cognate 70 protein for mitochondrial import varies among precursor proteins and depends on precursor length. Mol Cell Biol. 1996;16:6103–6109. doi: 10.1128/mcb.16.11.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AHY, Boone C. High-Throughput Strain Construction and Systematic Synthetic Lethal Screening in Saccharomyces cerevisiae. Methods Microbiol. 2007;36:369–386. [Google Scholar]
- Tripathi A, Mandon EC, Gilmore R, Rapoport TA. Two alternative binding mechanisms connect the protein translocation Sec71-Sec72 complex with heat shock proteins. J Biol Chem. 2017;292:8007–8018. doi: 10.1074/jbc.M116.761122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T, Viana MP, Xu F, Yu J, Chanchani R, Arceo XG, Tutucci E, Choi J, Chen YS, Singer RH, et al. Mitochondrial volume fraction and translation duration impact mitochondrial mRNA localization and protein synthesis. eLife. 2020;9:e57814. doi: 10.7554/eLife.57814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- Vögtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, Meisinger C. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- Vögtle FN, Burkhart JM, Gonczarowska-Jorge H, Kücükköse C, Taskin AA, Kopczynski D, Ahrends R, Mossmann D, Sickmann A, Zahedi RP, Meisinger C. Landscape of submitochondrial protein distribution. Nat Commun. 2017;8:290. doi: 10.1038/s41467-017-00359-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986;5:1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagih O, Usaj M, Baryshnikova A, VanderSluis B, Kuzmin E, Costanzo M, Myers CL, Andrews BJ, Boone CM, Parts L. SGAtools: one-stop analysis and visualization of array-based genetic interaction screens. Nucleic Acids Res. 2013;41:W591–W596. doi: 10.1093/nar/gkt400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen XJ. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature. 2015;524:481–484. doi: 10.1038/nature14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Salinas K, Zuo X, Kucejova B, Chen XJ. Dominant membrane uncoupling by mutant adenine nucleotide translocase in mitochondrial diseases. Hum Mol Genet. 2008;17:4036–4044. doi: 10.1093/hmg/ddn306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Amon A. MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. Science. 2018;360:eaan4146. doi: 10.1126/science.aan4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill U, Yofe I, Sass E, Stynen B, Davidi D, Natarajan J, Ben-Menachem R, Avihou Z, Goldman O, Harpaz N, et al. Genome-wide SWAp-Tag yeast libraries for proteome exploration. Nat Methods. 2018;15:617–622. doi: 10.1038/s41592-018-0044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Sweetman G, Savitski MF, Mathieson T, Bantscheff M, Savitski MM. Ion coalescence of neutron encoded TMT 10-plex reporter ions. Anal Chem. 2014;86:3594–3601. doi: 10.1021/ac500140s. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N. Mitochondrial Machineries for Protein Import and Assembly. Annu Rev Biochem. 2017;86:685–714. doi: 10.1146/annurev-biochem-060815-014352. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N, Ryan MT. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 2001;20:951–960. doi: 10.1093/emboj/20.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woellhaf MW, Sommer F, Schroda M, Herrmann JM. Proteomic profiling of the mitochondrial ribosome identifies Atp25 as a composite mitochondrial precursor protein. Mol Biol Cell. 2016;27:3031–3039. doi: 10.1091/mbc.E16-07-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L, Topf U, Bragoszewski P, Wiese S, Sztolsztener ME, Oeljeklaus S, Varabyova A, Lirski M, Chroscicki P, Mroczek S, et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature. 2015;524:485–488. doi: 10.1038/nature14951. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Fukui K, Takahashi H, Kitamura S, Shiota T, Terao K, Uchida M, Esaki M, Nishikawa S, Yoshihisa T, et al. Roles of Tom70 in import of presequence-containing mitochondrial proteins. J Biol Chem. 2009;284:31635–31646. doi: 10.1074/jbc.M109.041756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yofe I, Weill U, Meurer M, Chuartzman S, Zalckvar E, Goldman O, Ben-Dor S, Schütze C, Wiedemann N, Knop M, et al. One library to make them all: streamlining the creation of yeast libraries via a SWAp-Tag strategy. Nat Methods. 2016;13:371–378. doi: 10.1038/nmeth.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BP, Loewen CJ. Balony: a software package for analysis of data generated by synthetic genetic array experiments. BMC Bioinformatics. 2013;14:354. doi: 10.1186/1471-2105-14-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- Zeytuni N, Zarivach R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure. 2012;20:397–405. doi: 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data (see also Tables S1 and S2) have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD021173.
reviewer13653@ebi.ac.uk
Mass spectrometry data of the immunoprecipitation experiments (see also Table S4) are available via ProteomeXchange with identifier PXD023149.
reviewer_pxd023149@ebi.ac.uk