Abstract
Adaptive features of natural killer (NK) cells have been reported in various species with different underlying mechanisms. It is unclear, however, which NK cell populations are capable of mounting antigen-specific recall responses, and how such functions are regulated at the molecular level. Here we identify and characterize a discrete population of CD49a+CD16- NK cells in the human liver that display increased epigenetic potential to elicit memory responses and has the functional properties to exert antigen-specific immunity on the skin as an effector site. Integrated chromatin-based epigenetic and transcriptomic profiling revealed unique characteristics of hepatic CD49a+CD16- NK cells when compared to conventional CD49a-CD16+ NK cells thereby defining active genomic regions and molecules underpinning distinct NK cell reactivity. In contrast to conventional NK cells, our results suggest adaptive CD49a+CD16- NK cells to be able to bypass the KIR receptor-ligand system upon antigen-specific stimulation. Furthermore, these cells were highly migratory towards chemokine gradients expressed in epicutaneous patch test lesions as an effector site of adaptive immune responses of the skin. These results define pathways operative in human antigen-specific memory NK cells and provide a roadmap for harnessing this NK cell subset for specific therapeutic or prophylactic vaccine strategies.
Introduction
Historically, NK cells are prototypic innate effector cells (1–4) regulated by activating and inhibitory germline-encoded receptors, but lacking the recombination machinery crucial for the generation of antigen receptor diversity in B and T cells (5). However, mounting evidence hints to the presence of NK cells that display long-lived antigen-specific memory akin to T cells.
A first demonstration of NK cell-mediated recall responses was reported in mice lacking T and B cells with the induction of antigen-specific contact hypersensitivity reaction that was mediated by NK cells (6). After the initial observation that RAG-independent antigen-specific memory reside in a subset of hepatic NK cells (6), murine CD49a+DX5- NK cells were demonstrated to exhibit antigen-specific features against various structurally different haptenized proteins (7), vesicular stomatitis virus (VSV), influenza virus and human immunodeficiency virus (HIV) (8). Subsequent studies identified a subset of Thy1+ murine NK cells to be relevant in antigen-specific responses against vaccinia virus (9) and bacteria (Salmonella typhimurium, Ehrlichia muris (10). In search of memory NK cells in humans, a recent work demonstrated antigen-specific recall responses of hepatic NK cells in a humanized mouse model, and revealed NK cells in blisters of individuals after re-exposure with the varicella zoster virus with a phenotype similar to murine memory NK cells (11), while another study found hepatitis B-specific NK cells in the peripheral blood (12). These observations are of fundamental biological and clinical relevance, but also open many questions about the regulation of antigen-specific NK cell memory in humans. Previous studies point to a heterogeneous composition of human hepatic NK cells (13, 14), but the precise functional repertoire of antigen-specific memory NK cells and the functional comparison of NK cells in various organs in humans have yet to be elucidated.
Here, we combine high-throughput single-cell and bulk RNA sequencing and flow cytometry approaches with functional ex vivo assays to identify a subset of liver-derived adaptive NK cells expressing CD49a, while lacking CD16. These NK cells are phenotypically and functionally different from a previously characterized NKG2C+ NK cell population in peripheral blood after cytomegalovirus infection in humans. Transcriptional as well as epigenetic profiling revealed factors and pathways of CD49a+CD16- liver NK cells underpinning their enhanced effector functions. In ex vivo recall responses we found that CD49a+CD16- NK cells exclusively recognized and lysed autologous target cells pulsed with hepatitis A and/or hepatitis B antigens or haptens patients had been sensitized with (nickel) via the skin. Furthermore, we suggest that CD49a+CD16- NK cells can bypass killer-cell immunoglobulin-like receptors (KIR) for cytotoxic activity of and migrate to inflamed skin upon contact hypersensitivity reactions. Our data uncover a functional specialization of human hepatic NK cells and imply the relevance of memory NK cells in adaptive immune responses.
Results
Human liver NK cells contain a CD49a+CD16- NK cell subset
Studies in mice have revealed the liver to be a reservoir for NK cells with memory activities towards different viral antigens and haptens. To characterize the phenotypic, functional and transcriptomic features of hepatic NK cells from humans we characterized CD45+ cells isolated from three human livers using droplet-based single-cell RNA sequencing. The combined data set yielded 16,113 cells passing quality control and collectively gave rise to a detailed cellular and molecular landscape of human hepatic immune cell types (Fig. 1a). Graph-based clustering on the tSNE projection identified 15 distinct clusters of cells. Cluster-specific gene expression profiles and known marker gene expression for immune cell types supported the assignment of clusters, including several myeloid subpopulations (all expressing CD14), clearly separated subtypes of B cells and plasma cells characterized by CD19, CD3-expressing T cell clusters subdivided into CD4- and CD8-expressing T cells as well as NCAM1+ (CD56), CD3+ and CD8+ putative NKT cells. Additionally, we recognized heterogeneous populations of NCAM1+ NK cells / innate lymphoid cells (ILCs) that comprise at least four clusters (Fig. 1a, Suppl. Fig. 1a). NK cells were defined as NCAM1+ cells lacking CD3 expression. One distinct NCAM1+ subpopulation was characterized by high expression of the transcription factors Eomes, TBX21 (T-bet) as well as KLRB1 (CD161), NKG2A, CXCR6, NKp46 and CD7, but low levels of the ITGA1 gene (CD49a) and FCGR3 (Fig. 1a, Suppl. Fig. 1a), which we classified as CD49a- CD16- NK cells. While most of the other NK cells expressed FCGR3, one major subpopulation of NK cells lacked FCGR3, but displayed high levels of the ITGA1 gene encoding CD49a (Fig. 1a). A subset of intrahepatic NK cells with CD49a expression that has been attributed to recall responses to allergens and viral infections in mice (7, 8, 15).
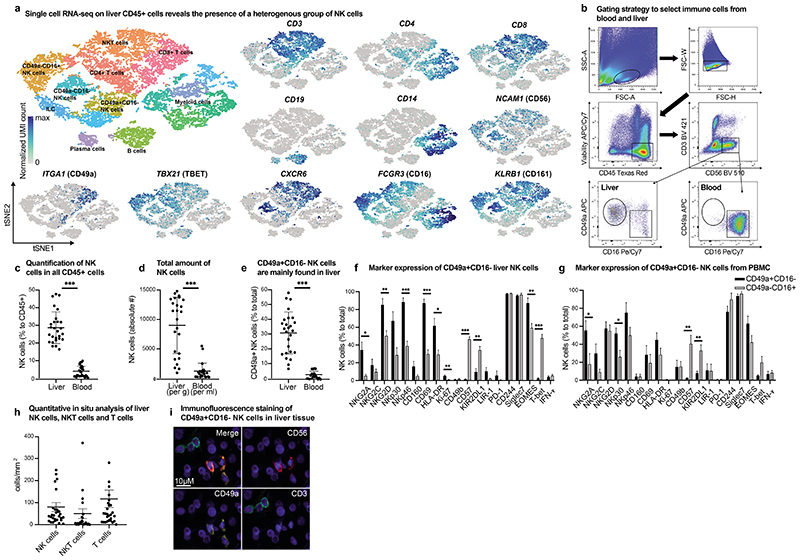
Figure 1. Human liver NK cells display a unique heterogeneity and contain a CD49a+ CD16- subpopulation.
a, tSNE projection of human hepatic CD45+ leukocytes. Each cell is grouped into one of 15 clusters (distinguished by their colors). UMI normalized expression of indicated genes within the depicted clusters. b, Gating strategy for CD49a+CD16- and CD49a- CD16+ NK cells from human liver mononuclear cells isolated of liver tissue and PBMCs. c, The percentage of total CD56+ NK cells in human liver and blood of total CD45+ leukocytes assessed by flow cytometry and presented as mean percentage of positive cells ± SD. n = 27. ***P < 0.0001. d, Mean numbers of NK cells in liver per gram and blood per ml assessed by flow cytometry and presented as mean absolute number of positive cells ± SD. n = 27. ***P < 0.0001. e, Mean percentage of CD49a+CD16- NK cells of total CD56+ NK cells in liver and blood. n = 27. ***P < 0.0001. f-g, Mean of intra- and extracellular markers of NK cells from the liver (f) and blood (g) according to their CD49a expression assessed by flow cytometry and depicted as percentage of positive cells ± SD. n = 5 (liver), 8 (blood). *P < 0.01, **P < 0.001, ***P < 0.0001. h, Quantitative in situ analysis of human livers of NK cells (CD56+CD3-), NKT cells (CD56+CD3+) and T cells (CD56-CD3+) by multicolor immunofluorescence staining. Data are given as absolute numbers of positive cells per mm2 ± SD. n = 30. i, Representative image of multicolor immunofluorescence staining of NK cells (CD56 PE/CD49a A647/ CD3 FITC) in human liver tissue.
Based on these data we prepared single-cell suspensions of blood and liver tissue from 27 patients to further phenotypically characterize CD49a+CD16- and CD49a-CD16+ NK cells by flow cytometry (Fig. 1b-g). NK cells accounted for almost one third of all CD45+ hepatic leukocytes (Fig. 1c, 1d). CD49a+CD16- NK cells were predominantly present in the liver (Fig. 1e, Suppl. Fig. 1b) and displayed a different phenotype compared to their CD49a-CD16+ counterparts with significantly elevated expression of NKG2A, NKG2D, NKp46, CD69 and HLA-DR (Figure 1f). Of note, the quantitative variability of CD49a+CD16- NK cells did not correlate with the underlying disease or tumor-derived inflammation (Suppl. Fig. 1c). Only a small number of CD49a+CD16- NK cells expressed CD57, NKG2C and KIR2DL1 (Fig. 1f, Suppl. Fig. 1d). While CD49a+CD16- NK cells were predominantly found in the human liver, NKG2C+CD16+NKp30- NK cells were present in the peripheral blood in comparable levels as in the liver, arguing for a different subset of NK cells (Suppl. Fig. 1e-g). Proliferation capacity of hepatic CD49a+CD16- NK cells was indicated by a significantly higher expression of Ki-67 compared to CD49a-CD16+ NK cells (Fig. 1f). Differences between CD49a+CD16- and CD49a-CD16+ were also reflected in the expression of the transcription factor Eomesodermin (EOMES) and T-box protein expressed in T cells (T-bet). CD49a+CD16- NK cells were T-betlow and EOMEShigh, whereas CD49a-CD16+ NK cells demonstrated moderate EOMES and T-bet expression. In general, CD49a+CD16- NK cells of the peripheral blood displayed a phenotype comparable to their hepatic CD49a+CD16- counterparts (Fig. 1g). Immunofluorescence staining and quantitative in situ analysis confirmed the distribution of T cells, NK cells and NKT cells in human liver tissue (Fig. 1h, 1i). Together these results show the heterogeneity of liver NK cells and indicate a subset of liver NK cells with a unique marker expression profile distinct from the majority of blood NK cells.
Transcriptomic and epigenomic profiling in liver NK cell subtypes reveal increased responsiveness of CD49a+CD16- NK cells towards IL-2/IL-15 stimulation
To test the activation properties of the two NK cell subsets, we exposed both NK cell subtypes to common activation signals for NK cells. In order to gain deeper insight into the regulatory factors responsible for genes specifically enriched in liver NK cells we in vitro stimulated FACS-purified CD49a+CD16- and CD49a-CD16+ liver NK cells with IL-2 and IL-15 for 4 hours. We then performed bulk Smart-seq2 RNA-sequencing and chromatin accessibility profiling using the assay for transposase accessible chromatin followed by sequencing (ATAC-seq) before and after stimulation. Principal component analysis (PCA) of the RNA-seq as well as the ATAC-seq data revealed differences between CD49a+CD16-and CD49a-CD16+ NK cells on gene expression levels (Suppl. Table 1) and on the level of accessible chromatin (Suppl. Table 2) in homeostatic/unstimulated conditions (Fig. 2a, b). Upon stimulation of CD49a+CD16- and CD49a-CD16+NK cells with IL-2/IL-15 the first principal component for our RNA-seq and ATAC-seq data (responsible for 42% and 67% of the observed variability, respectively) separated unstimulated from stimulated NK cells, although preserving the differences according to NK cell subtypes we had observed in the unstimulated/homeostatic conditions (Fig. 2a, b). Principal component 2 was able to separate the two tested NK cell types also when plotted against PC3 in case of RNA-seq and ATAC-seq (Suppl. Fig. 2a, b). To better understand the differences between the tested cell types, we performed differential gene expression and accessibility analysis of CD49a+CD16- and CD49a-CD16+ NK cells in unstimulated and stimulated conditions independent of each other (Fig. 2c, d). On gene expression level we observed a subset of genes that was expressed in CD49a+CD16- cells but was silent in CD49a-CD16+ cells and vice-versa. This was evident in both unstimulated and stimulated conditions. Importantly, a significant number of regions displayed open/accessible chromatin at promoter regions in unstimulated CD49a+CD16- cells, which were closed in CD49a-CD16+ NK cells (Fig. 2d). Pairwise comparison of the accessibility profiles of CD49a+CD16- and CD49a-CD16+ NK cells in either unstimulated or stimulated conditions revealed these promoter regions to remain accessible even after stimulation. This is suggestive of a pre-disposed chromatin state that primes the underlying genes for activation. The same regions were closed in CD49a-CD16+ NK cells and remained inaccessible even after stimulation with IL-2/IL-15, indicating a specific role of the genes associated with these promoters in CD49a+CD16- NK cells.
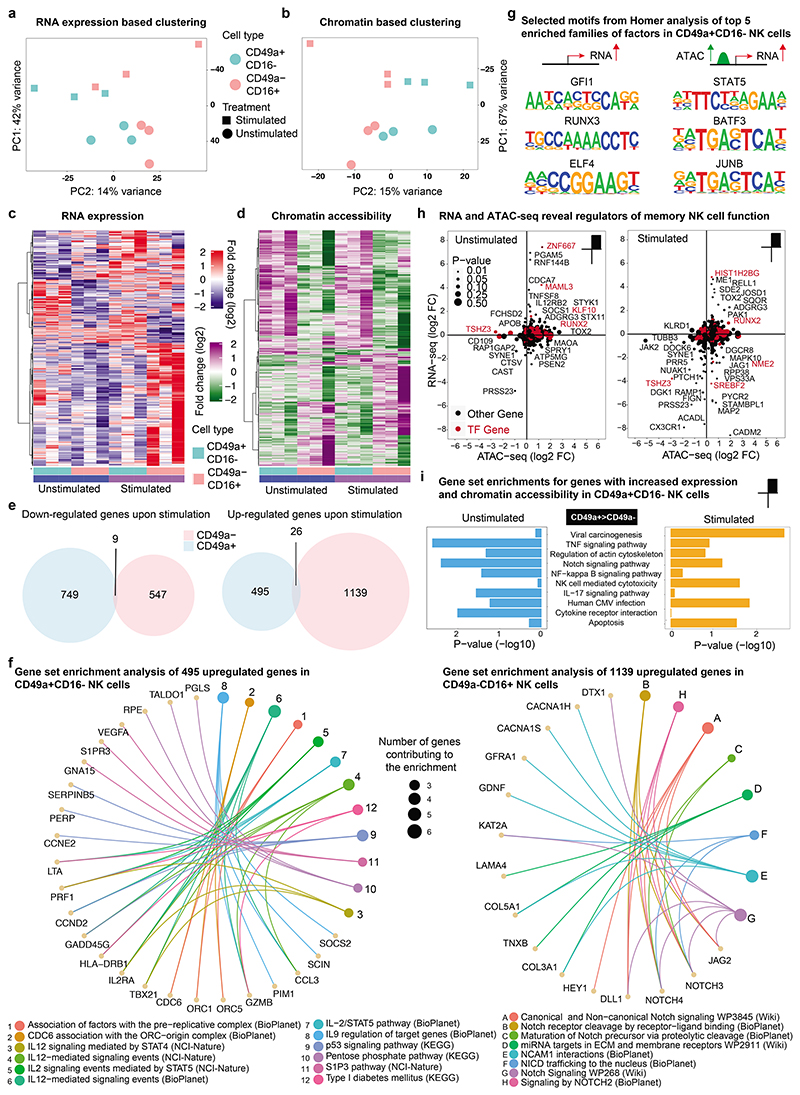
Figure 2. Transcriptomic and epigenomic differences in NK cell subtypes reveal priming of CD49a+CD16- NK cells for activation of genes with effector function.
a-i, CD49a+CD16- and CD49a-CD16+ NK cells were sorted from human liver from 3 donors by flow cytometry and stimulated with IL-2/IL-15. Smart-seq2 RNA sequencing and ATAC-seq was performed before and after stimulation. a, PCA analysis of bulk Smart-seq2 RNA-seq data on differentially expressed genes (P < 0.05) comparing unstimulated and IL-2/IL-15-stimulated CD49a+CD16- and CD49a-CD16+ NK cells. b, PCA analysis of ATAC-seq data on differentially accessible (DA) regions (P < 0.05) in CD49a+CD16- vs CD49a-CD16+ NK cells comparing unstimulated and IL-2/IL-15-stimulated cells. c, Heatmap of differentially expressed genes assessed by Smart-seq2 under stimulated and unstimulated conditions in CD49a+CD16- and CD49a-CD16+ subtypes. d, Heatmap of differentially accessible (DA) regions revealed by ATAC-seq between CD49a+CD16- vs CD49a-CD16+ NK cells under stimulated and unstimulated conditions. e, Venn diagram representing the number of up- and down-regulated genes in CD49a+CD16- and CD49a-CD16+ NK cells comparing stimulated versus unstimulated conditions. f, Functional enrichment (from BioPlanet 2019, KEGG 2019 Human, NCI-Nature 2016, and WikiPathways 2019 Human) analysis of 495 upregulated genes in CD49a+CD16- NK cells (left) and enrichment analysis of 1139 upregulated genes in CD49a-CD16+ NK cells (right). g, Selected consensus binding sequences resulting from HOMER motif enrichment analysis out of the top 5 enriched families of factors in CD49a+CD16- NK cells. h, Scatter plots of log2 fold changes from DA regions and differentially expressed genes, comparing CD49a+CD16- vs CD49a-CD16+ NK cells under stimulated and unstimulated conditions. Transcription factors are marked in red color. i, Enrichment of Top 10 selected KEGG pathways in stimulated and unstimulated conditions for genes with increased expression and accessibility profile.
To further investigate the differences in gene induction upon stimulation in the two NK cell subtypes we analyzed the number of differentially expressed genes upon stimulation and compared them between the two subtypes. We detected 749 genes to be downregulated specifically in CD49a+CD16- NK cells and 547 genes specifically downregulated in CD49a-CD16+ NK cells (Fig. 2e, left panel). Interestingly, only 9 genes displayed a downregulation to comparable extend in both NK cell subtypes. Furthermore, we detected 495 genes to be higher expressed in CD49a+CD16- NK cells and 1139 genes increased expression in CD49a-CD16+ NK cells with an overlap of only 26 genes upregulated in both entities (Fig. 2e, right panel). To understand the functional consequences of the genes induced in either of the two NK cell subtypes we performed enrichment analysis of the differentially upregulated genes in response to IL-2/IL-15 stimulation (Figure 2f). The gene set enrichments of the 495 genes with increased expression in CD49a+CD16- NK cells, using pathway enrichment with KEGG, NCI-Nature 2019 and BioPlanet 2019, revealed a predominant signature of at least four key pathways (Figure 2f, left panel). CD49a+CD16- NK cells displayed an increased metabolic activation centered around the pentose phosphate pathway including the genes PGLS, TALDO1 and RPE (16). The genes GNA15, SIPR3 and VEGFA gave rise to the enrichment of the S1P3 pathway, which is synonymous over an increased migratory potential of the CD49a+CD16- NK cell subtype upon stimulation (17). The KEGG-derived enrichment for p53 signaling is driven by a differential regulation of the genes CCNE2, PERP and SERPINB5, where the SERPIN protein family is known to play a critical role in protecting cells from granzyme-mediated apoptosis (18). Moreover, CD49a+CD16- NK cells displayed a greater responsiveness towards classical interleukin stimulation driven by STAT transcription factors as compared to their counterparts. The predominant factors driving the enrichment for IL-2, IL-9 and IL-12 signaling pathways were identified to be GZMB, the gene encoding granzyme B, SOCS2, a target gene and negative regulator of the JAK-STAT signal transduction, IL2RA, HLA-DRB1, LTA, PIM1, CCL3, TBX21, encoding T-bet, and PRF1, the gene encoding perforin, indicative of an increased potential for activation of factors contributing to classical NK cell killing, specification and proliferation.
In contrast, genes displaying increased expression in CD49a-CD16+ NK cells upon treatment with IL-2/IL-15 resulted in one dominant pathway to be significantly enriched in WikiPathways 2019 and BioPlanet 2019 – the Notch signaling pathway and Notch driven effectors (Fig. 2f, right panel). Factors contributing to these enrichments with known a link to and function in NK cells were: DTX1, a E3 ubiquitin ligase that acts as a positive regulator of Notch signaling and critical for NK cell development in the thymus; the Notch driven effector HEY1; the delta-like protein 1 – DLL1, as well as JAG2, which serve as ligands for the NOTCH receptors; as well as NOTCH3 and NOTCH4 – the receptor subunits itself showed higher expression.
Using the Homer motif enrichment tool, we next investigated the involvement of DNA binding factors potentially regulating the expression of the 495 genes we found upregulated in CD49a+CD16- NK cells after stimulation (Fig. 2g, left panel and Suppl. Table 3). Besides the consensus binding sequence of STAT5 we found the GFI, RUNX and ELF family of lineage determining transcription factors represented with highest significance at the promoters of the upregulated genes. Prompted by the discovery that a subset of transcriptionally induced genes also undergoes changes in chromatin accessibility we observed an even greater enrichment of interferon gamma activated sequences (GAS) known to bind STAT family members as well as the consensus for AP1 and/or BATF family members. All of the factors were also expressed in both NK cell subtypes in unstimulated and stimulated conditions, albeit to different levels (Suppl. Fig. 2c; Suppl. Table 1).
We conclude that distinct gene expression and chromatin profiles distinguish hepatic CD49a+CD16- NK cells from their CD49a-CD16+ counterparts. CD49a+CD16- NK cells respond more pronounced to the stimulation with IL-2/IL-15 leading to an upregulation of genes critical for NK cell effector functions mainly under the control of specific cell lineage determining factors. In contrast, CD49a-CD16+ NK cells predominantly upregulate genes in response to NOTCH signaling, but fail to express interleukin-inducible genes to a level comparable to CD49a+CD16- NK cells. Moreover, while lineage determining factors are associated with the upregulation of genes that display accessible chromatin profiles even in untreated conditions, inducible factors such as STATs, AP1 or BATF family members were associated with active reorganization of promoter chromatin in response to IL-2/IL-15 treatment.
Integrated chromatin profiling uncovers an increased epigenetic potential of CD49a+CD16- NK cells for activation of effector function genes
As indicated above the majority of ATAC-seq enriched and, thus, accessible regions did not change upon treatment in CD49a+CD16- NK cells (Fig. 2d). This provides support for the notion that the CD49a+CD16- NK cell type acquired an increased chromatin activation potential, as compared to their CD49a-CD16+ counterparts. To expand on distinguishing features of hepatic CD49a+CD16- and CD49a-CD16+ NK cells on the level of chromatin rearrangements we integrated RNA-seq data with ATAC-seq data and studied the changes across the genome/transcriptome directly comparing the two cell types either under unstimulated/homeostatic conditions or upon stimulation with IL-2/IL-15.
Our analysis revealed that in unstimulated CD49a+CD16- NK cells, 255 genes were expressed and displayed increased accessibility when compared to CD49a-CD16+ NK cells (Suppl. Fig. 2d). Among genes with highest significance we detected transcription-associated factors such as KLF10, RUNX2, MAML3, TOX2 or ZNF667 (Fig. 2h, left panel). RUNX1 and RUNX3 were targets of STAT4 during NK cell activation. Ablation of RUNX resulted in defective clonal expansion and memory formation during MCMV infection (19). Upregulation of TOX2 during NK cell differentiation is known to control T-bet expression (20). TBX21/T-bet was overexpressed in response to treatment of CD49a+CD16- NK cells with IL-2/IL-15 (Fig. 2f, left panel). Further we identified factors involved in secretion processes such as STX11 or secreted proteins such as TNFSF8, receptors such as IL12RB2, ADGRG2 and proteins involved in regulating intracellular signaling such as SOCS1, RNF144B and PGAM5. SOCS1 is known to mediate immunomodulation of NK cells through inhibition of JAK kinases (21).
A comparable number of 249 promoter regions displayed increased accessibility in CD49a+CD16- NK cells over CD49a-CD16+ NK cells without concomitant increase in gene expression (Suppl. Fig. 2d), including factors like PSEN2, MAOA and SPRY1 (Fig. 2h, left panel). 345 genes were found overexpressed with paralleled increase of promoter accessibility in CD49a+CD16- NK cells after stimulation with IL-2/IL-15 (Suppl. Fig. 2d), including an increase in the expression of transcription factors such as TOX2 and RUNX2 and the receptor-encoding gene ADGRG3 (Fig. 2h, right panel). In contrast, a total of 477 promoter regions displayed increased accessibility in CD49a+CD16- NK cells without concomitant gene expression changes (Suppl. Fig. 2d), indicative of a chromatin encoded potential for further activation. A significant number of signal transduction molecules and adhesion factors show active repression of expression and/or decreased accessibility in IL-2/IL-15-stimulated CD49a+CD16- NK cells, such as JAK2, DOCK6, RAMP1, CX3CR1, CADM2, NME2, JAG1 or MAPK10 (Figure 2h, right panel).
After the identification of the critical elements upregulated in CD49a+CD16- NK cells we set out to assess the factors involved in regulating the expression of genes in unstimulated CD49a+CD16- NK cells. To this end we conducted two complementary analytical approaches: First, we performed location overlap analysis (LOLA) enrichment (22) with the promoter regions from the 255 genes expressed and accessible in untreated CD49a+CD16- NK cells. On the one hand, several significant enrichments for data sets obtained after chromatin immune precipitation (ChIP) of polymerases in several cell lines and primary cells validated our promoter-centric approach. On the other hand, we were able to obtain significant enrichments for data obtained after IP of IRF1, E2F and ELF transcription factors (Suppl. Fig. 2e). Second, we performed Homer motif enrichment analysis for the consensus binding sequences within the promoters. We found differentially accessible binding sequences in CD49a+CD16- NK cells yielding enrichments for the consensus of factors, such as IRFs, ELF1, KLF6, KLF10 and RUNX (Suppl. Table 3). This validates the findings we obtained with LOLA and emphasizes the importance of those factors for actively regulating the gene expression program we specifically observed in CD49a+CD16- NK cells (Fig. 2a). Furthermore, all of the above-mentioned factors were significantly expressed in unstimulated and stimulated CD49a+CD16- NK cells with highest abundances observed for KLF6, RUNX1, IRF8 and ELF1 (Suppl. Fig. 2c). To identify the function of genes upregulated in unstimulated and stimulated CD49a+CD16- NK cells with chromatin accessibility we performed gene set enrichment analysis, yielding enrichments in untreated CD49a+CD16- NK cells for TNF signaling pathway, NFkB pathway, IL-17 signaling pathway, and cytokine-cytokine receptor interaction. This suggests the CD49a+CD16- NK cells to be hyper-responsive to pro-inflammatory cytokines under steady state conditions. Upon stimulation with IL-2/IL-15 we observed a significant enrichment for genes involved in viral carcinogenesis, apoptosis and natural killer cell-mediated cytotoxicity, such as ICAM1, KLRC1, PAK1, MAPK1, SOS2 and TNFRSF10A (Fig. 2i, Suppl. Table 1).
Taken together, our results are indicative of an increased epigenetic potential in CD49a+CD16- NK cells resulting in elevated inducibility towards stimuli triggering NK cell responses, a phenomenon describing the primed state of immune cells. However, the mechanistic basis maintaining the chromatin potential requires further verification in future functional studies.
Human hepatic CD49a+CD16- NK cells exhibit antigen-specific cytotoxicity
To validate the hypothesized specific functional repertoire of human CD49a+CD16- NK cells compared to CD49a-CD16+ NK cells, we performed killing assays with NK cells and T cells isolated by flow cytometry from the liver and blood. In a first set of experiments we used K562 as classical target cell line for NK cell-mediated cytotoxicity. Interestingly, similar to T cells, liver-derived CD49a+CD16- NK cells demonstrate a significant decreased cytotoxicity against K562 cells, while NK cells from peripheral blood and CD49a-CD16+ liver NK cells efficiently lysed K562 target cells (Fig. 3a), strengthening the concept of functionally different NK cells isolated from the human liver.
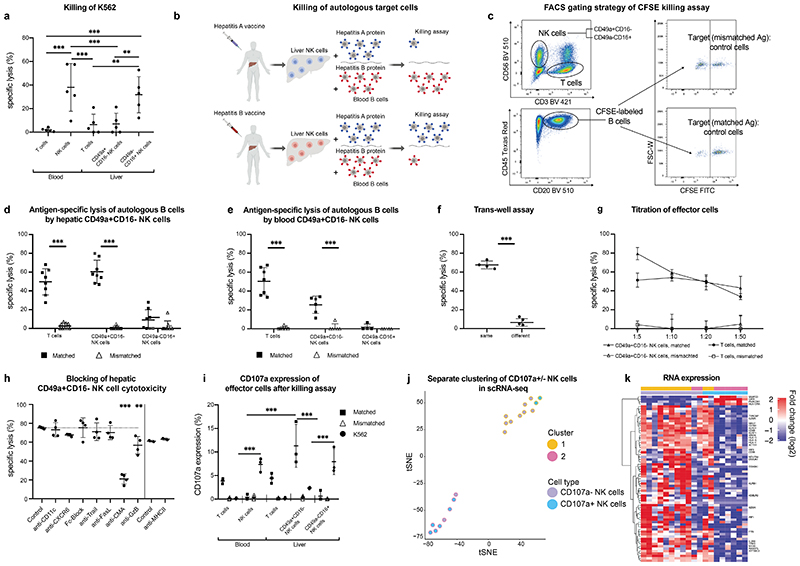
Figure 3. Human liver NK cells recognize and discriminate between matched and mismatched viral antigens.
a, Killing of K562 cells by hepatic CD49a+CD16- NK cells, CD49a-CD16+ NK cells and CD3+ T cells from blood and liver. Results are presented as percentage of specific lysis ± SD. n = 5. ** P < 0.01, *** P < 0.001. b, Graphical abstract of experiments assessing antigen-specific killing of hepatitis A/B-incubated autologous B cells by NK cells of vaccinated individuals c, Gating strategy for the cytotoxicity assay of hepatic NK cells and T cells. Autologous B cells of the patients pulsed with either hepatitis A or B and labelled with different concentrations of CFSE. Killing (“specific lysis”) is determined as the difference in the ratio of Ag-pulsed target B cells (CFSE low) and non-pulsed, intra-well control B cells (CFSE high). d-e, Killing of B cells pulsed with “Matched” or “Mismatched” antigens (either hepatitis A or B antigens according to hepatitis A or B serology) by hepatic NK cells (d) or blood derived NK cells (e) and CD8+ T cells as controls. The effector: target cell ratio was 1:10. Data represent the mean specific lysis ± SD of triplicates of each individual. Grey plots indicate individuals after hepatitis A/B infection. n = 6-8. ***P < 0.0001. f, Percentage of specific lysis of autologous B cells pulsed with matched antigens by NK cells in different and same wells in trans-well assays. n = 4. ***P < 0.0001. g, Titration of hepatic CD49a+CD16- NK cells, T cells and CD49a-CD16+ NK cells as effector cells and against target cells (B cells). Results are depicted as percentage of specific lysis ± SD. n = 4. h, Neutralizing antibodies or blocking agents were added to hepatic CD49a+CD16- NK cell cytotoxicity and compared to an isotype antibody control (dotted line). anti-FasL: anti-Fas Ligand. CMA: Concanamycin A, anti-GzB: anti-Granzyme B. n=4. **P < 0.01, ***P < 0.0001. i, Percentage of CD107a expression of depicted cells of blood and liver after killing assay of effector cells against autologous B cells incubated with matched, mismatched (according to serology status) and K562 cells. n=3, ***P < 0.001. j-k, Single-cell Smart-seq2 sequencing was performed on FACS-purified CD107a+CD49a+CD16- NK cells and CD107a-CD49a+CD16- NK cells after an antigen-specific cytotoxicity assay (shown in Fig.3i). j, Separate clustering of CD107a+/- NK cells after antigen-specific killing assay. k, Heat map of differentially expressed genes in the two identified clusters of CD107a+/- CD49a+CD16- NK cells.
NK cell killing towards K562 cells depends on the low expression levels of HLA on the surface of K562 cells. We speculated whether cytolytic effects of CD49a+CD16- NK cells required the detection of antigens presented via HLA. To assess antigen-specific memory functions of liver-derived NK cells based on their transcriptomic and epigenomic features and observations in mice (6), we performed ex vivo killing assays addressing their adaptive immune responsiveness (experimental outline depicted in Fig. 3b). We first classified patients according their hepatitis A and B vaccination/infection history and serology in four groups: i) hepatitis A positive, hepatitis B negative; ii) hepatitis A negative, hepatitis B positive; iii) hepatitis A positive, hepatitis B positive; iv) hepatitis A negative, hepatitis B negative. Autologous B cells pulsed with either hepatitis A or B proteins were used as target cells and non-pulsed B cells as control cells for the cytotoxicity assay (Fig. 3b). To differentiate target and control cells, we labeled autologous B cells with different concentrations of CFSE (Fig. 3c). T cells were used as positive controls for memory responses. After incubation with autologous hepatitis A or B pulsed B cells, cytotoxic activity of T cells was restricted to target cells that had been pulsed with the hepatitis antigen the patient had been pre-sensitized against (“matched antigens”), (Fig. 3d). In contrast, if patients had no positive serology against hepatitis A and/or hepatitis B, T cells did not lyse B cells that had been incubated with the respective antigens (“mismatched antigens”), arguing for antigen-specific killing of protein-pulsed target cells by T cells. CD49a-CD16+ or CD49a-CD16- NK cells did not show significant cytotoxic activity against autologous B cells in any condition. However, when incubating CD49a+CD16- liver NK cells with autologous B cells that had been pulsed with the matched antigen (hepatitis A and/or hepatitis B, depending on the vaccination/infection status), specific lysis of B cells pulsed with the matched antigen but not with the mismatched antigen was observed (Fig. 3d). Even though circulating in the peripheral blood in low numbers, CD49a+CD16- blood-derived NK cells lysed autologous B cells that had been incubated with matched antigens comparable to T cells (Fig. 3e). Notably, after incubation with the antigen, B cells did not differ in their activation state (Suppl. Fig. 3a). To functionally rule out a B cell-dependent effect of antigen-specific killing of NK cells, we performed cytotoxicity assays similar to above, but used autologous dendritic cells instead of B cells as target cells. We could fully replicate our results of antigen-specific NK cells killing autologous dendritic cells that had been pulsed with matched antigens (Suppl. Fig. 3b). To exclude the theoretical possibility of contaminating T cells exhibiting cytotoxic activity within our FACS-sorted NK cell population, we performed cytotoxicity assays with effector cells consisting of non-cytotoxic irrelevant filler cells (e.g., A431 cell line) containing titrated numbers of T cells. In a setting of 5% T cells titrated to A431 cells as potential killer cells, we observed a 20% cytotoxic activity against matched target cells (Suppl. Fig. 3c), which represented about one third of CD49a+CD16- NK cell-mediated killing. As T cells never accounted for >1% within purified NK populations, these experiments strongly argue for an antigen-specific cytolytic activity of CD49a+CD16- NK cells independent of the presence or absence of other leukocyte subsets (post-sort analysis Suppl. Fig. 3d). Hepatic CD49a-CD16- NK cells did not lyse autologous target cells incubated with matched antigens compared to CD49a+CD16- NK cells (Suppl. Fig. 3e).
Mechanisms of antigen-specific cytotoxicity of hepatic CD49a+CD16- NK cells
To further elucidate mechanisms of antigen-specific NK cell activity, we performed killing assays in trans-well plates and inhibited different cytotoxicity pathways. In trans-well assays, specific lysis of autologous target cells by CD49a+CD16- NK cells was achieved when target and effector cells were in direct contact within the same well, but not when separated in wells only connected via pores, indicating contact-dependent cytotoxic activity (Fig. 3f). To determine the potency of adaptive NK cells, we assessed various effector to target cell ratios. Decreasing the effector to target cell ratio resulted in reduced antigen-specific lysis by CD49a+CD16- NK cells comparable to T cells (Fig. 3g).
To unravel the mechanism underlying CD49a+CD16- NK cell-mediated antigen-specific cytotoxicity we blocked candidate receptors and pathways (Fig. 3h). We observed the strongest decrease in lysis of target cells by CD49a+CD16- NK cells when the perforin pathway was blocked with concanamycin A (CMA) and decreased specific lysis upon inhibition of granzyme B compared to controls; blockage of Fas ligand, TRAIL, Fcγ receptors, CD11c, CXCR6 and MHC-II had no effect on their cytotoxic activity. In line with our sequencing data this argues for the importance of the perforin and granzyme pathway for antigen-specific killing of CD49a+CD16- NK cells.
The diverse cytotoxic activity of CD49a+CD16- and CD49a-CD16+ NK cells from human livers was further demonstrated by assessing their degranulation by surface staining of the functional marker CD107a. In contrast to CD49a+CD16- NK cells and T cells, CD49a-CD16+ NK cells from liver tissue and blood-derived NK cells stained positive for CD107a after incubation with K562 (Fig. 3i). Importantly, CD49a+CD16- NK cells and T cells from the liver degranulated after incubation with autologous B cells that had been pulsed with matched antigens, reflecting their cytotoxic activity against target cells pulsed with antigens the patients had been vaccinated with.
Next, we assessed the transcriptomic features of antigen-specific NK cells after antigen recognition. We therefore isolated CD49a+CD16- NK cells after incubation with Ag-pulsed B cells and sorted them according their CD107a expression as a marker for degranulation to perform single-cell Smart-Seq2. We found that 167 genes displayed significant differences in expression in the two groups leading to clustering based on their transcriptomic profile (Fig. 3j). A heatmap of these genes separates CD107a+ and CD107a- CD49a+CD16- NK cells according to their transcriptomic differences (Fig. 3k, complete heatmap Suppl. Fig. 3f). Compared to CD107a+ after degranulation, CD107a- NK cells expressed high levels of IRF1 and IRF8 (Suppl. Table 4), in line with our transcriptome profiling obtained in bulk and a previous report indicating IRF8 of importance in adaptive NK cell responses in mice (23). Furthermore, CD107a- NK cells were enriched for genes encoding for surface and signaling receptors (IL2RB, CD160, TYROBP, CD69, CD96, HLA-C, HLA-E, HLA-A), various NK cell receptors (KLRB1, KLRD1, KLRF1), cytotoxic cytokines such as GZMK and GZMA as well as vesicle formation, traffic and storage of granules (SNAP23, NKG7, SRGN, KDELR2). Genes related to the upkeep of the cytoskeleton (TPM3, FYN, MYH9) and cell motility (RASSF5, ACTG1) were enhanced in CD107a- NK cells (Fig. 3k). These changes highlight that vesicle trafficking, cytoskeletal rearrangements and cellular mobility play essential roles for the recognition and response to antigen-specific stimulation of CD49a+CD16- NK cells. The high energy consumption of adaptive NK cell responses is reflected by differential expression of transcripts for genes of the respiratory chain for ATP production (NDUFA4, ATP5MC2, COX6A1). Only four transcripts of genes were expressed higher on degranulated CD49a+CD16- NK cells (CD107a+), namely CIITA, the mediator of MHC class II gene induction by interferon-γ, HLA-DRA, PHACTR1 and SNAP23 encoding for cell motility and transport vesicle docking and fusion (Fig. 3k).
In summary our data identify antigen-specificity of CD49a+CD16- hepatic NK cells in humans and reveal distinct patterns of cytotoxicity in NK cell subsets of human livers. Furthermore, our findings agree with the knowledge of degranulation and activation of NK cells and indicate that adaptive NK cells utilize a cytolytic machinery similar to conventional NK cells (24).
Antigen-specific killing by CD49a+CD16- hepatic NK cells bypasses KIR receptor-ligand patterns
To further unravel mechanistic principles of antigen-specific cytotoxicity of hepatic NK cells, we investigated if CD49a+CD16- hepatic NK cells are capable of antigen-specific lysis of allogeneic target cells. We phenotyped killer cell immunoglobulin like receptors (KIR) and their ligands (HLA-C and B allotypes) in patients and identified five donors with various constellations of KIR haplotypes, KIR ligands and serologic results for hepatitis A and hepatitis B (Fig. 4a). First, we isolated total hepatic NK cells from two donors with a different KIR repertoire (L21, L23) and performed killing assays against autologous and allogeneic B cells from L21, L23 and L27 resulting in allogeneic settings with matched (L23 versus L27) or mismatched (L21 versus L23/L27) KIR ligands (Fig. 4b). As expected, autologous B cells without further stimulation were not killed by NK cells (L21 versus L21, L23 versus L23, Fig. 4b). In allogeneic conditions, target cells were only killed, if the KIR ligands were mismatched (L21 NK cells versus L23 and L27 target cells, L23 NK cells versus L21 target cells, Fig. 4b). Of note, the KIR haplotypes did not determine the occurrence of killing but could be important for the extent of killing explaining the variation of cytotoxicity in this set of experiments (Fig. 4b).
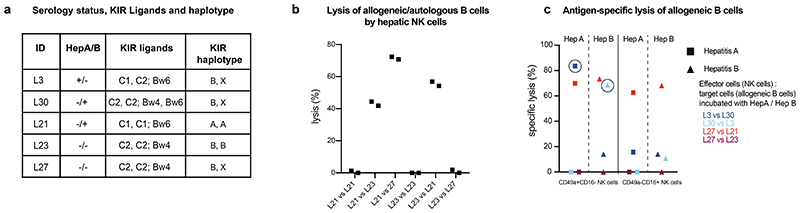
Figure 4. Antigen-specific killing by CD49a+CD16- hepatic NK cells of allogeneic target cells is dependent on KIR receptor-ligand match.
a, Serology status, KIR Ligands and KIR haplotype of five tested individuals. b, Percentage of lysis of allogeneic/autologous B cells by hepatic NK cells as indicated. B cells were not incubated with antigens. Data points depict two independent replicates of each condition. c, Percentage of specific lysis of allogeneic B cells after incubation with hepatitis A/B proteins by CD49a+CD16-/CD49a-CD16+ hepatic NK cells. Circled data points indicate NK cell killing that cannot be explained by their KIR pattern. Data are presented as mean specific lysis ± SD of triplicates of each individual.
Knowing the natural cytotoxic activity of NK cells in allogeneic settings based on the KIR repertoire, we addressed in a next step the antigen-specific activity of hepatic CD49a+CD16- and CD49a-CD16+ NK cells against allogeneic target cells that had been incubated with hepatitis A and hepatitis B proteins as described in figure 3. Hepatic CD49a+CD16- NK cells of L3 (dark blue) with a positive serology for hepatitis A, but negative for hepatitis B killed allogeneic B cells from L30 pulsed with the hepatitis A (matched antigen, dark blue circled square) but not hepatitis B (mismatched antigen). This killing took place despite of matched KIR ligands C2, which should in this allogeneic setting inhibit the response. This was mirrored by hepatic CD49a+CD16- NK cells from L30 (positive serology for hepatitis B, light blue) against B cells from L3, when killing was only observed upon incubation of allogeneic B cells with hepatitis B (matched antigen, light blue circled triangle) but not hepatitis A (mismatched antigen). Again, in this allogeneic setting, the KIR ligands C2 on the target cells of L3 should inhibit the cytotoxic response of CD49a+CD16- NK cells of L30. CD49a-CD16+ NK cells from livers of L3 and L30 did not exhibit cytotoxic activity of allogeneic target cells of these two donors. In contrast, L27 had a negative serology for hepatitis A and B. Consequently, CD49a+CD16- and CD49a-CD16+ NK cells from L27 did not exhibit antigen-specific cytotoxic activity against autologous B cells exposed to hepatitis A or B antigens (mismatched antigens). However, CD49a+CD16- and CD49a-CD16+ NK cells of L27 lysed allogeneic B cells from L21 with a KIR ligand mismatch (light red), while B cells from L23 with matched KIR ligands were not killed regardless prior incubation of B cells with hepatitis A or B antigens (dark red, Fig. 4c). Our data suggest that antigen-specific cytotoxicity of CD49a+CD16- NK cells may be able to bypass traditional NK cell biology upon antigen-specific recall responses, while both hepatic NK cell subsets can be activated by mismatched expression of KIR ligands. However further studies need to be undertaken to draw additional, significant conclusions.
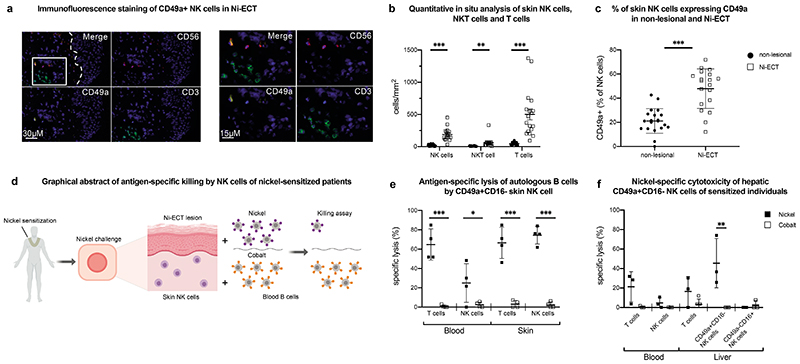
CD49a+CD16- NK cells of cutaneous contact hypersensitivity reactions mediate antigen-specific cytotoxicity
After defining the human liver as reservoir for antigen-specific NK cells, we examined the presence of NK cells at an effector site of antigen-specific memory reaction. As adaptive immune responses of liver NK cells were shown in a mouse model for skin contact hypersensitivity (7), we decided to use patients sensitized against nickel without a contact allergy against cobalt as an experimental model to test antigen specificity of skin-derived NK cells. By immunofluorescence staining of skin sections we observed significantly more NK cells in nickel-induced epicutaneous patch test lesions (Ni-ECT) compared to the non-lesional sites in parallel with an influx of CD3+ T cells (Fig. 5a, b). The majority of NK cells at lesional sites expressed CD49a (Fig. 5c). To elucidate direct antigen-specific effector functions of NK cells of skin lesions, we isolated T cells and NK cells from skin lesions and autologous blood-derived B cells as target cells and incubated B cells with either nickel or cobalt (as a negative control) (Fig. 5d). Similar to T cells from blood and Ni-ECT as positive controls, NK cells from Ni-ECT exhibited antigen-specific killing against autologous B cells that had been pulsed with nickel (matched) but not against target cells that had been incubated with cobalt (mismatched) (Fig. 5e). We obtained liver tissue from three patients with a medical history for contact allergy that could be confirmed by ECT testing for nickel, but not for cobalt. To gain insights into the potential connection of liver- and skin-derived NK cells we isolated liver-derived NK cells and T cells from those patients to perform killing assays with autologous B cells pulsed either with nickel or cobalt. Hepatic CD49a+CD16- but not hepatic CD49a-CD16+ or blood-derived NK cells exhibited antigen-specific killing against autologous B cells pulsed with nickel similar to CD8+ T cells (Fig. 5f). Although we have no direct evidence for a shared phenotype between liver- and skin-derived NK cells, these findings show the contribution of memory NK cells as effector cells to adaptive immune responses at the site of inflammation.
Figure 5. Human NK cells from nickel-induced epicutaneous patch test lesions exhibit antigen-specific memory responses and resemble liver NK cells.
a, Representative image of immunofluorescence multicolor staining of NK cells (CD56 PE/ CD49a A647/ CD3 FITC) in the upper dermis of cutaneous Ni-ECT lesions. Magnified image to the right. The dotted line in the merged picture indicates the dermal-epidermal junction and the rectangle displays the region of the magnified image. b, Quantitative in situ immunofluorescence analysis of NK cells (CD56+CD3-), NKT cells (CD56+CD3+) and T cells (CD56-CD3+) in the dermis of cutaneous Ni-ECT lesion. Data are given as absolute numbers of positive cells per mm2 ± SD. n = 20. **P < 0.001, ***P< 0.0001. c, Percentage of CD49a+ NK cells in non-lesional and lesional (Ni-ECT) skin to total NK cells. Data are presented as percentage of CD49a+ cells to total CD56+CD3- NK cells ± SD. n = 20. ***P < 0.0001. d, Graphical abstract of the experimental design with antigen-specific killing of nickel-incubated autologous B cells by NK cells of nickel sensitized individuals. e, Antigen-specific lysis of autologous B cells pulsed with “Matched” (nickel) or “Mismatched” (cobalt) antigens by isolated NK cells and T cells from Ni-ECT lesions and blood. Bars represent mean specific lysis ± SD of triplicates. n = 4. *P < 0.05, ***P < 0.0001. f, Nickel-specific cytotoxicity of hepatic NK cells of sensitized individuals. Antigen-specific lysis of autologous B cells pulsed with “Matched” (nickel) or “Mismatched” (cobalt) antigens by isolated NK cells and T cells from blood and liver of patients sensitized to nickel. Results represent mean specific lysis ± SD of triplicates. n=3. **P < 0.001.
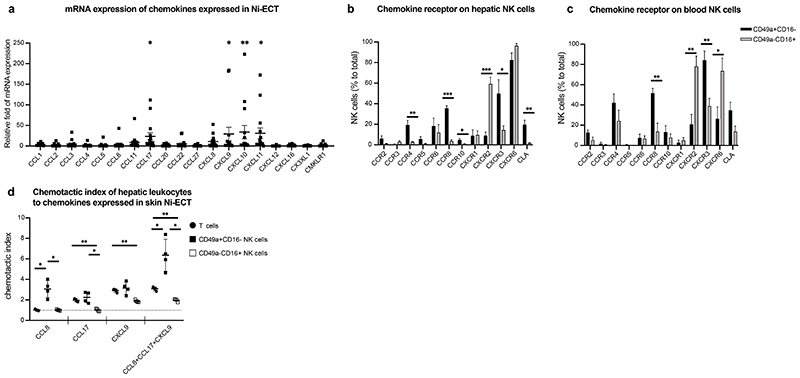
Specific migratory properties of CD49a+CD16- hepatic NK cells
To further address the possibility of a liver-skin axis with hepatic CD49a+CD16- NK cells leaving the liver and migrating towards effector sites such as the skin, we investigated the expression of chemokines in Ni-ECT potentially responsible to attract NK cells. CCL17, CXCL9, CXCL10 and CXCL11 were significantly increased in lesional skin of Ni-ECT compared to non-lesional skin (Fig. 6a). To associate the chemokine pattern of Ni-ECT with chemokine receptor expression of NK cells, we assessed the chemokine receptor profile of CD49a+CD16- and CD49a-CD16+ hepatic NK cells. Compared to CD49a-CD16+ NK cells, a large number of CD49a+CD16- NK cells expressed CCR8, CCR4 and CXCR3, while CXCR2 was mainly expressed by CD49a-CD16+ NK cells (Fig. 6b). The small number of CD49a+CD16- NK cells from peripheral blood showed a chemokine receptor repertoire similar to CD49a+CD16- NK cells from the liver (Fig. 6c). Interestingly, CXCR6 was expressed by CD49a+CD16- and CD49a-CD16+ liver-derived NK cells with no significant difference (Fig. 6b) and did not allow differentiation of hepatic NK cells subsets. To functionally assess the ability of hepatic NK cells to migrate towards chemokine gradients expressed in lesional skin, we performed migration assays of hepatic CD49a+CD16- and CD49a-CD16+ NK cells and T cells. While hepatic T cells showed moderate migration towards CCL17 and CXCL9 and CD49a-CD16+ NK cells towards CXCL9, CD49a+CD16- NK cells from liver tissue migrated towards CCL8, CCL17 and CXCL9 (Fig. 6d). Importantly, combination of these chemokines enhanced the chemotactic index of CD49a+CD16- NK cells, while we did not observe this effect on hepatic T cells and CD49a-CD16+ NK cells (Fig. 6d). We therefore conclude that CD49a+CD16- hepatic NK cells have an increased ability to migrate towards chemokines expressed in Ni-ECT compared to other effector cells. Appropriate stimulation may lead to emigration of CD49a+CD16- NK cells to effector sites, which needs to be further investigated. However, these findings are consistent with our sequencing data showing enrichment for components of the S1P3 pathway in CD49a+CD16- NK cells in response to IL-2/IL-15 (Fig. 2f), a common NK cell activation signal. The postulated increased migratory properties were recapitulated on functional level in context of CD49a+CD16- hepatic NK cells exposed to chemokines expressed in the skin of nickel allergic patients (Fig. 6d).
Figure 6. Human liver NK cells migrate towards cytokines expressed in nickel-induced epicutaneous patch test lesions.
a, Relative fold of mRNA expression of presented chemokines in Ni-ECT lesions. n = 15. *P < 0.05, **P < 0.001. b-c, Mean percentage of CD49a+CD16-/CD49a-CD16+ NK cells expressing depicted chemokine receptors ± SD of human livers (b) and blood (c). n = 5 (liver), 8 (blood). *P < 0.05, **P < 0.001, ***P < 0.0001. d, Chemotactic index of hepatic T cells and NK cells to the exposed chemokines measured by flow cytometry of trans-well assays. n = 4. *P < 0.05, **P < 0.001.
Discussion
In this study we characterize the immune repertoire of the human liver in an unbiased way at single-cell resolution with a focus on NK cells. We encountered a remarkable heterogeneity within the CD56+ cell population drawing our attention to a subtype of CD56+ NK cells, which expressed low levels of FCGR3 (CD16), but was positive for the ITGA1 (CD49a) gene and therefore different from hitherto described memory-like NK cells in the context of CMV infection. Linking their phenotypic characterization to chromatin accessibility profiling we found lineage determining factors such as the GFI, RUNX, KLF, IRF and ELF family of transcription factors to likely regulate gene expression of genes encoded in regions with a chromatin state of high epigenetic potential, primed for activation in CD49a+CD16- NK cells. Additionally, we identified factors of the STAT, AP1 and BATF family to be potential key-regulators orchestrating the response of CD49a+CD16- NK cells under steady state and after in vitro challenge with IL-2/IL-15. In CD49a+CD16- NK cells these factors predominantly function at promoter regions that require active chromatin reorganization to render the nearby encoded genes expressed. CD49+CD16- NK cells are hyper-responsive towards stimulation with IL-2/IL-15 and express high levels of genes involved in pro-inflammatory signal transduction cascades and display a burst in expression of genes involved in NK cell cytotoxic processes after stimulation with IL-2/IL-15. In accordance with interleukin driven genes CD49a+CD16- NK cells display elevated levels of genes functionally linked to the S1P3 pathway conferring cell mobility, a finding supported by functional tests assessing increased chemotactic properties of CD49a+CD16- NK cells towards chemokines secreted in the skin of nickel sensitive patients. We speculate that CD49+CD16- NK cells fulfill a surveillance function within the liver representing a first line of defense in the detection and response to blood-borne pathogens facilitated by cell type-specific chromatin architecture.
Innate immune memory or training has been discovered in recent years and was often attributed to myeloid cell populations (25). To our knowledge this is the first evidence of immune priming in human innate lymphocytes as indicated by our data obtained by chromatin accessibility profiling. CD49a as an integrin subunit is associated with lymphocytes binding to collagen IV (14). Since CD49a+ is a marker for tissue residency we cannot exclude that the origin of NK cells plays a role in the extent of lysis of K562 by CD49a-CD16+ or CD49a+CD16- NK cells. The lower killing activity towards K562 cells in our experiments suggests that triggering the activation of the primed CD49a+CD16- NK cells requires a different stimulus. Whether the primed genes for NK cell cytotoxicity are a consequence of dynamic immune responses and can be causally linked to antigen-specific immune responses or are the result of cell intrinsic properties remains to be tested.
Immunological memory of NK cells has potentially tremendous impact for vaccination strategies via utilizing their cytopathic effects in an antigen-specific fashion. Memory properties of NK cells were described in mice and non-human primates against a variety of haptens and viral antigens (6–10, 15, 26, 27). In our study we demonstrate the functional properties of antigen-specific NK cells derived from various human tissues under steady state and inflammatory conditions. CMV infection is known to induce long-lived NKG2C+ NK cells in humans and is associated with an expansion as well as differentiation of NK cells (28–30). However, the reported CMV driven maturation of a NKG2C+ NK cell population is distinct in function and phenotype from the antigen-specific NK cell subset we identified in this study. Our study goes way beyond the properties of antigen-specific NK cells described in recent years (11, 31, 32). The characterization of a discrete NK cell subset on a functional, transcriptomic and epigenomic level from various human tissues provides novel insights into their life cycle and regulation being essential for harnessing them in future. We confirmed that the liver harbors a unique lineage of liver resident CD49a+CD16- NK cells associated with a distinct phenotype (33, 34) and linked this subset to memory NK cells. In line with previous studies we observed a broad range in numbers of CD49a+CD16- liver NK cells (14, 34, 35), which was not related to clinical parameters of our study. It was previously shown that CD49a+CD16- hepatic NK cells express high levels of CXCR6 (8, 34), which we corroborated with our data. However, CXCR6 was expressed on CD49a-CD16+ hepatic NK cells in comparable levels, excluding it as a useful marker to define memory NK cells in humans.
Evidence for the existence of distinct developmental pathways of NK cells came from mouse studies demonstrating the importance of EOMES and T-bet for their development with complementary functions (36, 37). Liver-resident NK cells comprise various subsets with a variable set of markers used for their characterization (13, 14, 34, 38, 39). In accordance with previous results we show on protein and RNA level that human CD49a+CD16- hepatic NK cells, in contrast to CD49a-CD16+ NK cells from the liver, are EOMEShigh and T-betlow (13, 34, 38, 39), indicating different origins of NK cell subsets. The precursor of CD49a+CD16- NK cells that persist and potentially proliferate in the liver is unknown. Whether CD49a+CD16- NK cells are recruited from the circulating NK cell population into the liver tissue or are retained since fetal development (33, 40) still needs to be elucidated.
In this study we demonstrate a cellular antigen-specific immune response by T cells and NK cells in patients after vaccination against hepatitis A and B. Besides the induction of protective antibodies, T cell responses after hepatitis A and B vaccinations in humans are well described (41). In addition to T cell-mediated cellular immunity, it was recently shown for NK cells in people living in a malaria-endemic region that Plasmodium falciparum–infected red blood cells induced antibody-dependent NK cell degranulation after the addition of plasma from malaria-resistant individuals (42).
Many facets of memory NK cells are still unknown. The “career path” of naïve NK cells becoming memory cells was introduced in mice (43) and is further elucidated with this study. Here, we hypothesize that NK cells may constantly leave and re-circulate tissues. However, we do not have direct evidence that the increase of CD49a+CD16- NK cells of lesional skin are directly linked to hepatic CD49a+CD16- NK cells. Our work is supported by initial findings of Hunger et al., where NK cells have been detected in skin-draining lymph fluid from healthy volunteers and patients with contact dermatitis, which suggests that they are recruited from blood to skin during steady state and inflammatory conditions and return to the blood via draining lymphatics (44). Furthermore, epidermal application of contact sensitizers elicits the accumulation of NK cells in the exposed skin (7, 45, 46). This infiltration by antigen-specific NK cells in mouse CHS probably reflects recruitment of liver-derived NK cells from the blood as well as antigen-driven proliferation in situ (47). We observed blood CD49a+CD16- NK cells being more responsive to antigen-specific stimulation directly after in vivo challenge with haptens (Fig. 5e) compared to a situation without epicutaneous application of the antigen, which supports our concept of the mobilisation of CD49a+CD16- NK cells by antigen exposure and trafficing to effector sites via the peripheral blood. Although the detection of non-specific tissue injury by activating receptors could lead NK cells rapidly into the effector sites, the specific ways for antigen recognition by NK cells have yet to be found. It is not clear, if these mechanisms are distinct from adaptive NK cells that mediate recall responses against CMV infection in humans (29, 48) and mice (15, 49, 50). Notably, blood-derived expanded NK cells after infection of HCMV were shown to display increased transcription of CIITA and several genes related to the MHC class II pathway to present antigens to CD4+ T cells (51). The transcription factor IRF8 was essential in adaptive NK cells response in MCMV infection (23). We identified IRF8 and IRF1 as a prerequisite for memory NK cells. If these are stable features of NK cell-related memory in general still needs to be investigated.
In summary, our data prove human primary antigen-specific NK cells to reside under steady state in the liver. Even though we do not find the mechanism leading to specific antigen recognition by NK cells, their epigenetic and transcriptomic features combined with their cytotoxic and migratory functions shown in this study might form the basis for future work harnessing adaptive NK cells for the development of prophylactic and / or therapeutic vaccinations in various clinical settings.
Methods
Patients and tissue material
Patients with major liver resection for benign or malignant tumors were enrolled into the study after written consent. Liver tissue without apparent pathologies was obtained from the border of surgical resected tissue and 30ml peripheral blood were taken. Age, gender, liver disease, sensitization to nickel, the vaccination status including serologic testing and history on hepatitis A/B infection were documented (Suppl. Table 5). For skin samples, an epicutaneous patch test (ECT) was performed in nickel sensitized patients and a punch biopsy 72 hours after antigen challenge was taken from lesional and healthy-appearing skin (Suppl. Table 6). Studies involving patient material were performed according to the Declaration of Helsinki and are approved by the local ethics committee of the Medical University of Vienna (122/2006, 2032/2013). For cytotoxicity assays the K562 and A431 cell lines were purchased from ATCC.
Cell isolation and sorting
Human liver tissue was flushed with PBS, cut into small pieces with a sterile scalpel, passed through 70-μm mesh filters and rinsed with 10% FBS in PBS. Human skin samples were harvested as a 6-mm punch biopsy. Skin samples were minced and digested with Collagenase IV (10 units/ml) and DNase I (10mg/ml) for 90 min in RPMI supplemented with 5% FBS in PBS at 37°C. Leukocytes of the liver and blood were isolated by density gradient centrifugation using Ficoll gradients. For flow cytometry analysis or cell sorting, cells were stained with fluorescence antibodies listed in supplementary table 3. Samples were acquired and sorted on a FACSAria III (BD Biosciences) and analyzed with FlowJo software. Briefly, after doublet-exclusion we gated on viable CD45+ leukocytes. NK cells (CD45+CD56+CD3-) and T cells (CD45+CD56-CD3+) were characterized by a panel of markers depicted in Suppl. Table 7. Then NK cells (CD56+CD3-CD49a+CD16-, CD56+CD3-CD49a-CD16+ and/or CD56+CD3-CD49a-CD16-) and T cells (CD56-CD3+) were sorted for the killing assay (post-sorting analysis Suppl. Fig. 3d). B cells and dendritic cells from peripheral blood were isolated, sorted and used as target cells in an antigen-specific autologous (and allogeneic for B cells, Fig. 4) setting. A FACSAria III (BD Biosciences) was used for cell sorting with FACSDiva software and purity >98% was reported for all sorted populations.
In vitro killing assay
T cells (viable CD45+CD56-CD3+) and CD49a+CD16- and CD49a-CD16+ NK cells (viable CD45+CD56+CD3-) were sorted of human livers and peripheral blood as indicated above. Autologous B cells and dendritic cells as target cells from the peripheral blood were sorted as viable CD45+CD20+ cells and viable CD45+Lineage-HLA-DR+CD11c+ cells, respectively. From ECT lesions we sorted T cells and total NK cells due to smaller sample size and cell numbers. Hepatitis A or B (both ProSpec) proteins were added (final concentration: 0.5 mg/ml) and cells were incubated for 30 min at 37°C. B cells from the blood of nickel-sensitized patients were incubated with nickel or cobalt (both Sigma-Aldrich) for 30min at 25°C (final concentration: 0.5 mM). After extensive washing, target cells incubated with hepatitis A/B proteins or nickel/cobalt and control cells without protein/hapten incubation were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE, Thermo Fisher) in two concentration (0.5 or 5μM), resulting in CFSEhigh and CFSElow B cells that had been pulsed with various antigens or without antigen incubation of control cells (Fig 3d-i). After 8h of incubation at 37°C of NK cells or T cells with autologous or allogeneic B cells as target cells at effector to target cell ratio of 1:10 (except figure 3g, where different effector:target cell ratios are indicated), the proportion of CFSEhigh and CFSElow cells was assessed by flow cytometry. For inhibition of TRAIL–, Fas ligand–, Fc–, perforin and granzyme B–dependent lysis and to assess the contribution of CXCR6, CD11c or MHC-II to cytotoxicity of adaptive NK cells, azide-free neutralizing anti-TRAIL (R&D Systems), anti–Fas ligand (BD Biosciences), Fc blocking reagent (BD Biosciences), anti–granzyme B (Abcam), anti-CXCR6 (R&D Systems), anti-CD11c (BioLegend), concanamycin A (Sigma-Aldrich) and anti-MHC II (HLA-DR, -DP, -DQ; BD Biosciences), respectively, were added to effector cells 30 min before the addition of target cells. Anti-MHC II was added additionally 30 min prior to incubation of target cells with antigens. A 30-min preculture of effector cells with 5 μg/ml of an IgG1 isotype (Sigma-Aldrich) served as negative control (Fig. 3h).
The K562 cell line (ATCC) was cultured in RPMI 1640 media (Thermo Fisher), supplemented with 10% fetal bovine serum (Thermo Fisher), 2mM Glutamine (Thermo Fisher), 100IU/ml penicillin and 100ug/ml streptomycin (Thermo Fisher) at 37°C. FACS-sorted NK cells, T cells isolated from the blood and T cells as well as CD49a+CD16-/CD49a-CD16+ NK cells derived from the liver were cultured with K562 cells at effector to target cell ratio 1:10, respectively, in 96-well plates (Fig. 3a). Target cells were distinguished by CFSE labeling, resulting in CFSElow K562 cells. Unpulsed autologous B cells were used as CFSEhigh control cells. After 8h of incubation at 37°C, the proportion of CFSEhigh and CFSElow cells was assessed by flow cytometry.
Antigen-specific lysis was calculated as the following: (1-(CFSE high/CFSE low)no effector cells/(CFSE high/CFSE low)with effector cells) x 100.
For the assessment of degranulation of blood-derived NK cells and T cells as well as liver-derived CD49a+CD16-/CD49a-CD16+ NK cells and T cells (Fig. 3i), killing assays with autologous matched/mismatched B cells and K562 cells were performed as described above in the presence or absence of anti-CD107a antibody (Biolegend) and Monensin (Biolegend). CD107a expression was measured with flow cytometry.
Immunofluorescence
For immunofluorescence staining approximately 0.5cm3 of liver tissue and 4mm punch biopsies of skin were frozen and stored in Optimal Cutting Temperature (OCT) media.
Immunofluorescence stainings of 5μm liver cryosections for CD49a, CD56, CD3 and DAPI nuclear marker were performed with monoclonal antibodies listed in supplementary table 3. In brief, after incubation with the primary antibodies overnight at 4°C, an appropriate secondary fluorescence-labeled antibody was applied for 30 minutes at room temperature, followed by staining of DAPI nuclear marker. For evaluation of immunofluorescence results, images were acquired using a Z1 Axio Observer microscope equipped with a LD Plan-Neofluar 20x/0.4 objective (Zeiss) and quantified using TissueFAXS/TissueQUEST image analysis software (TissueGnostics GmbH).
RNA isolation and one-step qPCR
Skin biopsies of epicutaneous patch test lesions from patients sensitized to nickel frozen in TRI Reagent (Sigma-Aldrich) were thawed, cut into pieces and homogenized in the FastPrep-24™ classic instrument (mpbio) for RNA isolation according to the manufacturer’s instructions. The RNA concentration was determined with a Nanodrop 200c spectrophotometer (Thermo Fisher). A master mix containing the Luna Universal Probe One-Step Reaction Mix (New England Biolabs), Luna WarmStart RT Enzyme Mix (New England Biolabs) and the TaqMan Gene Expression Assay for the tested chemokines (Applied Biosystems) was prepared and pipetted into a MicroAmp Fast Optical 96-Well Reaction Plate (Applied Biosystems). One-step qPCR was conducted in the Step One Plus Real Time PCR System (Applied Biosystems) and fold-changes were calculated with the ΔΔCT method.
Migration Assay
Migration assays with sorted liver-derived NK Cells and T cells were performed using 24-transwell plates with 5μm pore size (CoStar). 5 x 104 CD49a+CD16-NK cells, CD49a-CD16+ NK cells and T cells were placed in l00μ1 in the upper chamber and 500μl medium alone or supplemented with CCL8 (50ng/ml), CCL17 (10ng/ml) or CXCL9 (2000ng/ml), respectively, in the lower chamber. Cells were incubated for 4 hours at 37°C. Triplicates were run for all conditions. Migrated cells were collected from the lower compartment and counted by flow cytometry. Migration was assessed as chemotactic index as the following: (viable cells lower chamber (with chemokine)/viable cells lower chamber (medium only)).
KIR genotyping and analysis
DNA was isolated either by the salting-out method or by using an automated DNA extraction device (GenoM-6; GenoVision). HLA class I and class II genes were typed to the high-resolution level by sequencing based typing (52). Sequence specific KIR primers (SSP-KIR) and amplification conditions used for typing of KIR genes as published (53, 54). Each typing included positive and negative controls for each gene. KIR genotype assignment of individuals and its confirmation was done as reported previously (55).
Following genotype assignment, 23 haplotypes (1–23) published (53) were used to identify haplotype pairs for each of our donors. Broader assignment of either A (1–2) or B (3–23) haplotypes was possible in each case. The associations of KIR haplotypes with NK cell cytotoxicity were analyzed by distinguishing between AA or BX haplotypes, where X stands for either A or B haplotype. Similarly, in the genotype HLA-C1CX, CX stands for either C1 or C2 allelic groups.
RNA sequencing and bioinformatic analysis
Droplet-based single-cell RNA-seq was performed using the 10X Genomics Chromium Single Cell Controller with the Chromium Single Cell 3’ V3 Kit following the manufacturer’s instructions. After quality control, libraries were sequenced on the Illumina HiSeq 4000 platform in 2x75bp paired-end mode. Raw sequencing data were processed with the CellRanger v1.3.0 software (10X Genomics) for demultiplexing and alignment to the GRCh38 human reference transcriptome. Processed data were analysed using pre-processed results from CellRanger using the Loop Cell Browser software tool (10X Genomics). Unless otherwise stated, we used default parameters throughout.
For single-cell SMART-seq2, we sorted CD49a+CD16- NK cells after antigen-specific stimulation according to their expression of CD107a in separate wells of 96-well plates. Sequencing of single-cells was performed using the SMART-seq2 protocol. Briefly, RNA isolation and cDNA synthesis were performed, followed by enrichment as previously described (56). ERCC (External RNA Controls Consortium) spike-in RNA controls were added to each sample and carried along through the entire library preparation for quality filtering. The libraries were sequenced using an Illumina HiSeq 3000/4000 platform and a 50 bp single-read configuration.
Single-end reads were trimmed for low quality bases and removal of low-quality reads using ReadTools (v.1.0.0) (57). Trimmed reads were mapped to the Homo sapiens genome (GRCh38.92 assembly) using STAR (v2.5.3a) mapper (58). The reads mapped in multiple genomic locations were eliminated by using Samtools (v1.4)(59). Read counts for features (exons) were generated using the featureCounts function from the Subread package (v1.22.1) (60).
For gene expression analysis with bulk RNA-seq data, lowly expressed genes (with < 10 reads) were eliminated from further analysis in all samples. Gene expression analysis was done with DESeq2 (v 3.22.3) (61) using the Ensembl Known Gene models (version GRCh38.92) as reference annotations. For differential gene expression analyses genes were considered differentially expressed, if they showed a P-value </= 0.05. Principal component analysis and hierarchical clustering were performed using the normalized gene expression from DESeq2 (v 3.22.3). Normalized count (log2) by DESeq2 was used to perform the principle component analysis (PCA), hierarchical clustering (HC) and to create heatmaps and scatter plots.
For single-cell RNA-seq data analysis, we had first removed the low-quality cells with log-library sizes that are more than 3 median absolute deviations (MADs) below the median log-library size and the log-transformed number of expressed genes is 3 MADs below the median. We also removed low expressed genes that were not present in at least 3 cells. After quality control we have normalized the count matrix by using the deconvolution method from Scater package (62). The technical bias was modelled using the “trendVar” and “decomposeVar” methods and removed by using the “denoisePCA” method from Scater package. Batch effect was removed by using the “removeBatchEffect” function from limma package (63). The clustering of cells was done with the t-stochastic neighbor embedding (t-SNE) method with “ncomponents=2, perplexicty=10 and ntop=500” parameters. The differentially expressed genes were identified with P-value < 0.01 and logFC > 2. The heatmaps for selected genes were created by using the pheatmap package (https://cran.r-project.org/web/packages/pheatmap/index.html).
KEGG pathway enrichment and Gene Ontology (GO) enrichment analysis were done by using the KEGGA and GOANA methods respectively from limma package (63). Top 20 KEGG pathways and GO terms were selected for each gene set at corrected p-value (p adj < 0.05).
Chromatin accessibility mapping
Upon stimulation of freshly isolated liver-derived CD49a+CD16- NK cells and CD49a-CD16+ NK cells with IL-2 (100Ui/ml) and IL-15 (10ng/ml) for 4 hours, we performed RNA sequencing and assay for transposase-accessible chromatin sequencing (ATAC-seq) to analyze the gene expression and chromatin accessibility changes. ATAC-seq was performed as described previously (64). Briefly, 10,000 to 20,000 cells were lysed in a buffer containing digitonin and Tn5 transposase enzyme (Illumina). After incubation at 37°C for 30 minutes, tagmented DNA was purified and enriched. After final purification, the libraries were quality checked using a 2100 Bioanalyzer (Agilent) with high-sensitivity DNA chips. DNA concentration was examined using Qubit Fluorometer, and the libraries were sequenced on the Illumina HiSeq 4000 platform in 1x50bp single-read mode. Raw sequencing data were trimmed for low quality bases and removal of low-quality reads using ReadTools (v.1.0.0) (57). Trimmed reads were mapped to the Homo sapiens genome (GRCh38.92 assembly) using STAR (v2.5.3a) mapper (58). The reads mapped in multiple genomic locations were eliminated by using Samtools (v1.4).(59) ATAC-seq peaks were called by using the “findPeaks” module from HOMER package (65) with parameter “-gsize 2.7e9 -style dnase -fdr 0.05”. After merging peaks across all ATAC-seq datasets and removing peaks that overlapped blacklisted regions from ENCODE consortia (ENCODE Project Consortium, 2012) were eliminated from the further analysis. We quantified for each input dataset the number of reads overlapping the retained peaks. Raw read counts were loaded into DESeq2 v1.22.257 for normalization and differential analysis (using a minimum absolute log2 fold change of 1 as threshold; n = 4 replicates per NK cell subset).
Enrichment analysis for genes and genomic regions
To understand the biological roles of differentially expressed genes and differentially active genomic regions, we employed KEGG and Gene Ontology analyses to test genes of T cells, CD49a+CD16- NK cells and CD49a-CD16+ NK cells. For the assessment of differentially expressed genes from bulk RNA-seq, we employed BioPlanet 2019, KEGG 2019 Human, NCI-Nature 2016, and WikiPathways 2019 Human functional enrichments by using “enrichR” R package (66). We selected top 5 enriched categories from each enrichment at corrected p-value (p adj < 0.05). For DA region we used Locus Overlap Analysis (LOLA; web version 1.3.1) with “activeDHS universe” as background to test regulatory regions identified in our ATAC-seq data for significant overlaps with experimentally determined TF-binding sites from publicly available ChIP-seq data. We have used “clusterProfiler” R package to organize the enriched terms into a network with edges connecting overlapping gene sets (67).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 (GraphPad Software). Statistical significance was determined by Student’s t-test when comparing two groups. The two-way analysis of variance (ANOVA) Tukey’s multiple comparison test was used when comparing three or more groups. Significance was set at a P value of less than 0.05.
Supplementary Material
One Sentence Summary.
CD49a+CD16- NK cells from the liver and contact dermatitis lesions specifically recognize antigens and contribute to adaptive immunity.
Acknowledgments
This work was supported by the Austrian Science Fund (FWF, P30972) and the Medical Scientific Fund of the Mayor of the City of Vienna (17121). M.F. was supported by the Innovation Fund of the Austrian Academy of Sciences (IF_2015_36) and the Austrian Science Fund (FWF) Special Research Program grant (FWF SFB F6102). We thank S. Wenda and I. Faé for performing KIR phenotyping. Graphical abstracts were designed with BioRender.
Footnotes
AUTHOR CONTRIBUTION
V.S., M.F. and G.S. designed the study. V.S., J.S., L.K., M.F. and G.S. performed the experiments. R.V.P. performed the bioinformatic analyses with input from C.B. and M.F. D.P., P.S. and G.S. provided access to human samples. G.F.F. provided results on KIR phenotyping. C.B. and M.F. supervised the high-throughput experiments. V.S. M.F. W.W. and G.S. wrote the manuscript with contributions of all authors.
COMPETING INTERESTS STATEMENT
The authors declare no competing interests.
Data availability
Processed single-cell RNA-seq, bulk RNA-seq and ATAC-seq data are openly available via Gene Expression Omnibus (GEO).
References
- 1.Herberman RB, Nunn ME, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic acid allogeneic tumors. I. Distribution of reactivity and specificity. Int J Cancer. 1975;16:216–229. doi: 10.1002/ijc.2910160204. [DOI] [PubMed] [Google Scholar]
- 2.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 3.Kiessling R, Klein E, Pross H, Wigzell H. “Natural” killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur J Immunol. 1975;5:117–121. doi: 10.1002/eji.1830050209. [DOI] [PubMed] [Google Scholar]
- 4.Thornthwaite JT, Leif RC. The plaque cytogram assay. I. Light and scanning electron microscopy of immunocompetent cells. J Immunol. 1974;113:1897–1908. [PubMed] [Google Scholar]
- 5.Lanier LL. NK cell receptors. Annu Rev Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 6.O’Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell-and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol. 2006;7:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- 7.Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, Sun R, Yokoyama WM, Tian Z. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J Clin Invest. 2013;123:1444–1456. doi: 10.1172/JCI66381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, Szczepanik M, Telenti A, Askenase PW, Compans RW, von Andrian UH. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol. 2010;11:1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillard GO, Bivas-Benita M, Hovav AH, Grandpre LE, Panas MW, Seaman MS, Haynes BF, Letvin NL. Thy1+ NK [corrected] cells from vaccinia virus-primed mice confer protection against vaccinia virus challenge in the absence of adaptive lymphocytes. PLoS Pathog. 2011;7:e1002141. doi: 10.1371/journal.ppat.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kupz A, Scott TA, Belz GT, Andrews DM, Greyer M, Lew AM, Brooks AGM, Smyth J, Curtiss R, 3rd, Bedoui S, Strugnell RA. Contribution of Thy1+ NK cells to protective IFN-gamma production during Salmonella typhimurium infections. Proc Natl Acad Sci U S A. 2013;110:2252–2257. doi: 10.1073/pnas.1222047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikzad R, Angelo LS, Aviles-Padilla K, Le DT, Singh VK, Bimler L, Vukmanovic-Stejic M, Vendrame E, Ranganath T, Simpson L, Haigwood NL, et al. Human natural killer cells mediate adaptive immunity to viral antigens. Sci Immunol. 2019;4 doi: 10.1126/sciimmunol.aat8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wijaya RS, Read SA, Truong NR, Han S, Chen D, Shahidipour H, Fewings NL, Schibeci S, Azardaryany MK, Parnell GP, Booth D, et al. HBV vaccination and HBV infection induces HBV-specific natural killer cell memory. Gut. 2020 doi: 10.1136/gutjnl-2019-319252. [DOI] [PubMed] [Google Scholar]
- 13.Filipovic I, Sonnerborg I, Strunz B, Friberg D, Cornillet M, Hertwig L, Ivarsson MA, Bjorkstrom NK. 29-Color Flow Cytometry: Unraveling Human Liver NK Cell Repertoire Diversity. Front Immunol. 2019;10:2692. doi: 10.3389/fimmu.2019.02692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marquardt N, Beziat V, Nystrom S, Hengst J, Ivarsson MA, Kekalainen E, Johansson H, Mjosberg J, Westgren M, Lankisch TO, Wedemeyer H, et al. Cutting edge: identification and characterization of human intrahepatic CD49a+ NK cells. J Immunol. 2015;194:2467–2471. doi: 10.4049/jimmunol.1402756. [DOI] [PubMed] [Google Scholar]
- 15.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardiner CM, Finlay DK. What Fuels Natural Killers? Metabolism and NK Cell Responses. Front Immunol. 2017;8:367. doi: 10.3389/fimmu.2017.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walzer T, Chiossone L, Chaix J, Calver A, Carozzo C, Garrigue-Antar L, Jacques Y, Baratin M, Tomasello E, Vivier E. Natural killer cell trafficking in vivo requires a dedicated sphingosine 1-phosphate receptor. Nat Immunol. 2007;8:1337–1344. doi: 10.1038/ni1523. [DOI] [PubMed] [Google Scholar]
- 18.Bird CH, Christensen ME, Mangan MS, Prakash MD, Sedelies KA, Smyth MJ, Harper I, Waterhouse NJ, Bird PI. The granzyme B-Serpinb9 axis controls the fate of lymphocytes after lysosomal stress. Cell Death Differ. 2014;21:876–887. doi: 10.1038/cdd.2014.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rapp M, Lau CM, Adams NM, Weizman OE, O’Sullivan TE, Geary CD, Sun JC. Core-binding factor beta and Runx transcription factors promote adaptive natural killer cell responses. Sci Immunol. 2017;2 doi: 10.1126/sciimmunol.aan3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vong QP, Leung WH, Houston J, Li Y, Rooney B, Holladay M, Oostendorp RA, Leung W. TOX2 regulates human natural killer cell development by controlling T-BET expression. Blood. 2014;124:3905–3913. doi: 10.1182/blood-2014-06-582965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endo TA, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, Miyazaki T, et al. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 22.Sheffield NC, Bock C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2016;32:587–589. doi: 10.1093/bioinformatics/btv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams NM, Lau CM, Fan X, Rapp M, Geary CD, Weizman OE, Diaz-Salazar C, Sun JC. Transcription Factor IRF8 Orchestrates the Adaptive Natural Killer Cell Response. Immunity. 2018;48:1172–1182.:e1176. doi: 10.1016/j.immuni.2018.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bezman NA, Kim CC, Sun JC, Min-Oo G, Hendricks DW, Kamimura Y, Best JA, Goldrath AW, Lanier LL, Immunological Genome Project C Molecular definition of the identity and activation of natural killer cells. Nat Immunol. 2012;13:1000–1009. doi: 10.1038/ni.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulder WJM, Ochando J, Joosten LAB, Fayad ZA, Netea MG. Therapeutic targeting of trained immunity. Nat Rev Drug Discov. 2019;18:553–566. doi: 10.1038/s41573-019-0025-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habib S, El Andaloussi A, Hisham A, Ismail N. NK Cell-Mediated Regulation of Protective Memory Responses against Intracellular Ehrlichial Pathogens. PLoS One. 2016;11:e0l53223. doi: 10.1371/journal.pone.0153223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reeves RK, Li H, Jost S, Blass E, Li H, Schafer JL, Varner V, Manickam C, Eslamizar L, Altfeld M, von Andrian UH, et al. Antigen-specific NK cell memory in rhesus macaques. Nat Immunol. 2015;16:927–932. doi: 10.1038/ni.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Verges S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, Houchins JP, Miller S, Kang SM, Norris PJ, Nixon DF, et al. Expansion of a unique CD57(+)NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci U S A. 2011;108:14725–14732. doi: 10.1073/pnas.1110900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, Scott JM, Kamimura Y, Lanier LL, Kim S. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity. 2015;42:431–442. doi: 10.1016/j.immuni.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM. Cytokine-induced memory-like natural killer cells. Proc Natl Acad Sci U S A. 2009;106:1915–1919. doi: 10.1073/pnas.0813192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu X, Liu Y, Li L, Li Q, Qiao D, Wang H, Lao S, Fan Y, Wu C. Human natural killer cells expressing the memory-associated marker CD45RO from tuberculous pleurisy respond more strongly and rapidly than CD45RO-natural killer cells following stimulation with interleukin-12. Immunology. 20ll;134:41–49. doi: 10.1111/j.1365-2567.2011.03464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martrus G, Kautz T, Lunemann S, Richert L, Glau L, Salzberger W, Goebels H, Langeneckert A, Hess L, Poch T, Schramm C, et al. Proliferative capacity exhibited by human liver-resident CD49a+CD25+ NK cells. PLoS One. 2017;12:e0182532. doi: 10.1371/journal.pone.0182532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stegmann KA, Robertson F, Hansi N, Gill U, Pallant C, Christophides T, Pallett LJ, Peppa D, Dunn C, Fusai G, Male V, et al. CXCR6 marks a novel subset of T-bet(lo)Eomes(hi) natural killer cells residing in human liver. Sci Rep. 2016;6:26157. doi: 10.1038/srep26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.AwYeang HX, Piersma SJ, Lin Y, Yang L, Malkova ON, Miner C, Krupnick AS, Chapman WC, Yokoyama WM. Cutting Edge: Human CD49e-NK Cells Are Tissue Resident in the Liver. J Immunol. 2017;198:1417–1422. doi: 10.4049/jimmunol.1601818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, Bienvenu J, Henry T, Debien E, Hasan UA, Marvel J, et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med. 2014;211:563–577. doi: 10.1084/jem.20131560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, Zhong C, Chase JM, Rothman PB, Yu J, Riley JK, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collins A, Rothman N, Liu K, Reiner SL. Eomesodermin and T-bet mark developmentally distinct human natural killer cells. JCI Insight. 2017;2:e90063. doi: 10.1172/jci.insight.90063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harmon C, Robinson MW, Fahey R, Whelan S, Houlihan DD, Geoghegan J, O’Farrelly C. Tissue-resident Eomes(hi) T-bet(lo) CD56(bright) NK cells with reduced proinflammatory potential are enriched in the adult human liver. Eur J Immunol. 2016;46:2111–2120. doi: 10.1002/eji.201646559. [DOI] [PubMed] [Google Scholar]
- 40.Popescu DM, Botting RA, Stephenson E, Green K, Webb S, Jardine L, Calderbank EF, Polanski K, Goh I, Efremova M, Acres M, et al. Decoding human fetal liver haematopoiesis. Nature. 2019;574:365–371. doi: 10.1038/s41586-019-1652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Penna A, Fowler P, Bertoletti A, Guilhot S, Moss B, Margolskee RF, Cavalli A, Valli A, Fiaccadori F, Chisari FV, et al. Hepatitis B virus (HBV)-specific cytotoxic T-cell (CTL) response in humans: characterization of HLA class II-restricted CTLs that recognize endogenously synthesized HBV envelope antigens. J Virol. 1992;66:1193–1198. doi: 10.1128/jvi.66.2.1193-1198.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hart GT, Tran TM, Theorell J, Schlums H, Arora G, Rajagopalan S, Sangala ADJ, Welsh KJ, Traore B, Pierce SK, Crompton PD, et al. Adaptive NK cells in people exposed to Plasmodium falciparum correlate with protection from malaria. J Exp Med. 2019 doi: 10.1084/jem.20181681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paust S, von Andrian UH. Natural killer cell memory. Nat Immunol. 2011;12:500–508. doi: 10.1038/ni.2032. [DOI] [PubMed] [Google Scholar]
- 44.Hunger RE, Yawalkar N, Braathen LR, Brand CU. The HECA-452 epitope is highly expressed on lymph cells derived from human skin. Br J Dermatol. 1999;141:565–569. doi: 10.1046/j.1365-2133.1999.03031.x. [DOI] [PubMed] [Google Scholar]
- 45.Bangert C, Friedl J, Stary G, Stingl G, Kopp T. Immunopathologic features of allergic contact dermatitis in humans: participation of plasmacytoid dendritic cells in the pathogenesis of the disease? J Invest Dermatol. 2003;121:1409–148. doi: 10.1111/j.1523-1747.2003.12623.x. [DOI] [PubMed] [Google Scholar]
- 46.Carbone T, Nasorri F, Pennino D, Eyerich K, Foerster S, Cifaldi L, Traidl-Hoffman C, Behrendt H, Cavani A. CD56highCD16-CD62L-NK cells accumulate in allergic contact dermatitis and contribute to the expression of allergic responses. J Immunol. 2010;184:1102–1110. doi: 10.4049/jimmunol.0902518. [DOI] [PubMed] [Google Scholar]
- 47.Buentke E, Heffler LC, Wilson JL, Wallin RP, Lofman C, Chambers BJ, Ljunggren HG, Scheynius A. Natural killer and dendritic cell contact in lesional atopic dermatitis skin--Malassezia-influenced cell interaction. J Invest Dermatol. 2002;119:850–857. doi: 10.1046/j.1523-1747.2002.00132.x. [DOI] [PubMed] [Google Scholar]
- 48.Guma M, Budt M, Saez A, Brckalo T, Hengel H, Angulo A, Lopez-Botet M. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood. 2006;107:3624–3631. doi: 10.1182/blood-2005-09-3682. [DOI] [PubMed] [Google Scholar]
- 49.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 50.Smith HR, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV, Iizuka K, Furukawa H, Beckman DL, Pingel JT, Scalzo AA, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci U S A. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costa-Garcia M, Ataya M, Moraru M, Vilches C, Lopez-Botet M, Muntasell A. Human Cytomegalovirus Antigen Presentation by HLA-DR+ NKG2C+ Adaptive NK Cells Specifically Activates Polyfunctional Effector Memory CD4+ T Lymphocytes. Front Immunol. 2019;10:687. doi: 10.3389/fimmu.2019.00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fischer GF, Fae I, Frey E, Mayr WR. HLA-A*02172* adds to the heterogeneity of HLA-A*02 alleles. Tissue Antigens. 1998;51:312–314. doi: 10.1111/j.1399-0039.1998.tb03109.x. [DOI] [PubMed] [Google Scholar]
- 53.Hsu KC, Liu XR, Selvakumar A, Mickelson E, O’Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. 2002;169:5118–5129. doi: 10.4049/jimmunol.169.9.5118. [DOI] [PubMed] [Google Scholar]
- 54.Velickovic M, Velickovic Z, Dunckley H. Diversity of killer cell immunoglobulin-like receptor genes in Pacific Islands populations. Immunogenetics. 2006;58:523–532. doi: 10.1007/s00251-006-0124-3. [DOI] [PubMed] [Google Scholar]
- 55.Ludajic K, Balavarca Y, Bickeboller H, Rosenmayr A, Fae I, Fischer GF, Kouba M, Pohlreich D, Kalhs P, Greinix HT. KIR genes and KIR ligands affect occurrence of acute GVHD after unrelated, 12/12 HLA matched, hematopoietic stem cell transplantation. Bone Marrow Transplant. 2009;44:97–103. doi: 10.1038/bmt.2008.432. [DOI] [PubMed] [Google Scholar]
- 56.Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 20l4;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 57.Gomez-Sanchez D, Schlotterer C. ReadTools: A universal toolkit for handling sequence data from different sequencing platforms. Mol Ecol Resour. 2018;18:676–680. doi: 10.1111/1755-0998.12741. [DOI] [PubMed] [Google Scholar]
- 58.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome S. Project Data Processing, The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 61.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCarthy DJ, Campbell KR, Lun AT, Wills QF. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, Kathiria A, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Processed single-cell RNA-seq, bulk RNA-seq and ATAC-seq data are openly available via Gene Expression Omnibus (GEO).