Summary
There is increasing recognition of the prognostic significance of tumor cell MHC class II (MHC-II) expression in anti-cancer immunity. Relapse of acute myeloid leukemia (AML) following allogeneic stem cell transplantation (alloSCT) has recently been linked to MHC-II silencing in leukemic blasts; however, the regulation of MHC-II expression remains incompletely understood. Utilizing unbiased CRISPR/Cas9 screens, we identify that the CtBP complex transcriptionally represses MHC-II pathway genes, while the E3 ubiquitin ligase complex component FBXO11 mediates degradation of CIITA, the principal transcription factor regulating MHC-II expression. Targeting these repressive mechanisms selectively induces MHC-II upregulation across a range of AML cell lines. Functionally, MHC-II+ leukemic blasts stimulate antigen-dependent CD4+ T cell activation and potent anti-tumor immune responses, providing fundamental insights into the graft-versus-leukemia effect. These findings establish the rationale for therapeutic strategies aimed at restoring tumor-specific MHC-II expression to salvage AML relapse post-alloSCT, and also potentially to enhance immunotherapy outcomes in non-myeloid malignancies.
Keywords: Acute myeloid leukemia, bone marrow transplantation, tumor immunology, cancer epigenetics, gene regulation
Introduction
Acute myeloid leukemia (AML) is an aggressive hematological malignancy that is often incurable with chemotherapy (Kantarjian et al., 2021). For fit and eligible patients with adverse prognostic features, allogeneic stem cell transplantation (alloSCT) from a related or genetically matched unrelated donor is the only consolidative intervention that offers the potential for durable disease remission, principally driven by an immunological graft-versus-leukemia (GVL) effect (Horowitz et al., 1990). Although alloSCT is the most established curative cellular immunotherapy (Copelan, 2006), post-transplantation relapses unfortunately remain common and are associated with a dismal prognosis, due to a paucity of effective salvage options (Bejanyan et al., 2015; Lim et al., 2018; Mielcarek et al., 2007). Although NK cell-mediated innate immunity has been shown to contribute to the GVL effect (Gill et al., 2009), it is likely that T cell-derived adaptive immune responses are predominantly responsible for driving sustained disease control. This is evidenced by the significantly higher relapse rates seen with T cell-depleted allografts (Marmont et al., 1991; Soiffer et al., 2011) and the ability of donor lymphocyte infusion, which mainly comprises T cells, to induce remissions in a minority of relapsing patients (Schmid et al., 2007). Adaptive immunity relies on activation of CD4+ and CD8+ T lymphocytes and B lymphocytes following recognition of tumor-specific antigens presented on major histocompatibility complex (MHC) molecules (Chaplin, 2010). Classically, CD8+ T lymphocytes recognize endogenous cellular peptides presented by MHC class I (MHC-I), whereas CD4+ T lymphocytes recognize exogenously-derived antigens presented by MHC-II on professional antigen-presenting cells (APCs). Following alloSCT, a therapeutic anti-leukemia immune response can be initiated by two different types of alloantigen recognition. In an HLA-matched setting, donor T cells recognize minor histocompatibility or leukemia-associated antigens processed and presented in the context of an MHC molecule that is shared by the donor and recipient. Alternatively, following haploidentical or HLA-mismatched alloSCT, donor T cells can directly recognize mismatched MHC molecules via molecular mimicry, whereby the mismatched MHC with bound peptide structurally mimics donor MHC-peptide complexes to trigger donor T cell activation (Macdonald et al., 2009).
Recently, downregulation of MHC-II was identified as a recurrent relapse phenomenon following alloSCT, which occurred in the absence of inactivating mutations in genes encoding components of the MHC-II pathway. Strikingly, selective MHC-II repression was observed in 39-50% of relapsing patients in independent cohorts (Christopher et al., 2018; Toffalori et al., 2019), while MHC-I expression was preserved. These findings potentially implicate CD4+ T cells in GVL-mediated immunosurveillance; however, the molecular mechanisms by which MHC-II is regulated to contribute to immune escape remain largely unknown. Given that epigenetic silencing of MHC-II may be targetable as a salvage strategy to re-establish immunological disease control, we sought to characterize the molecular mechanisms facilitating MHC-II repression in AML and its biological significance in the context of post-transplantation relapse.
Results
Genome-wide CRISPR screens identify key regulators of surface MHC-II expression in AML
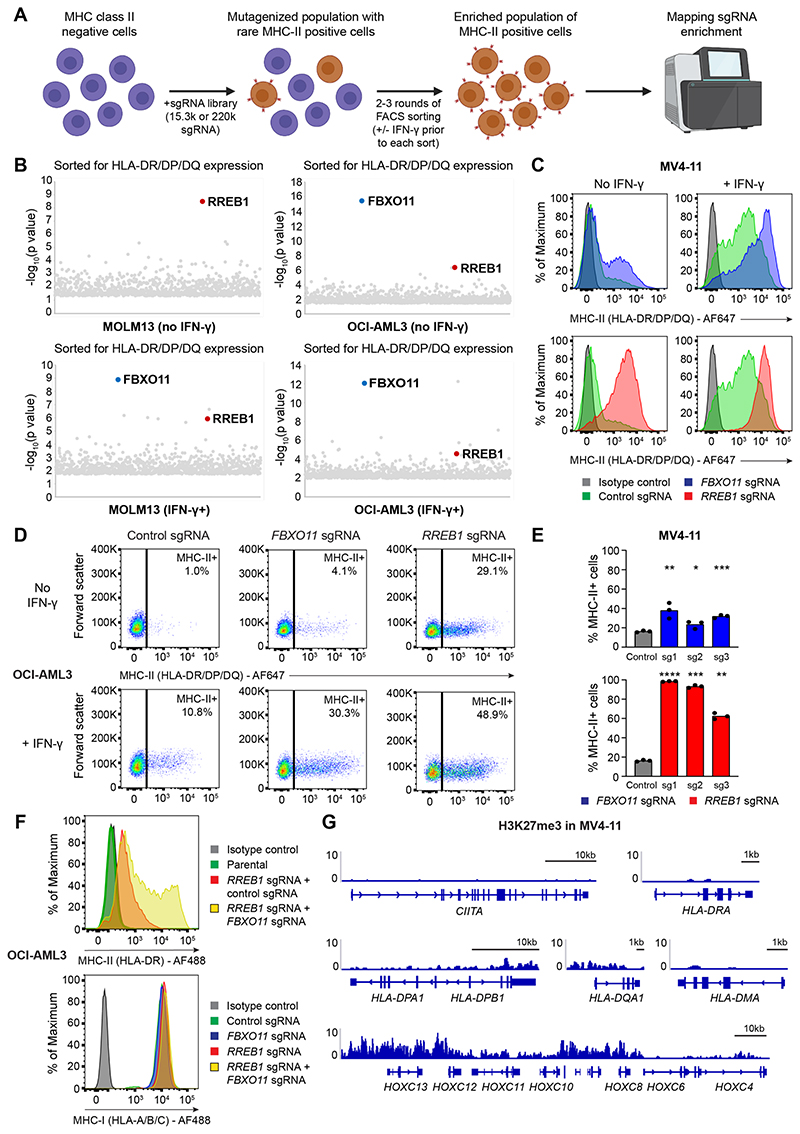
To identify negative regulators of MHC-II expression, we performed genome-wide positive selection CRISPR/Cas9 screens in MOLM13 and OCI-AML3 cells (Figure 1A) as these cell lines collectively encompass mutations in DNMT3A, NPM1 and FLT3, which are the most common oncogenic drivers in AML (Cancer Genome Atlas Research et al., 2013). Notably, these AML lines are of myelomonocytic lineage and have low or undetectable cell surface MHC-II in the absence of interferon-γ (IFN-γ) stimulation (Figure S1A). Two of the top candidate genes identified in both screens were F-box only protein 11 (FBXO11), a substrate recognition component of the SKP1-CUL1-F-box (SCF) E3 ubiquitin ligase complex, and Ras-responsive element binding protein 1 (RREB1), a zinc finger transcription factor (Figure 1B). These screen hits were also validated in MV4-11, another AML cell line with distinct driver mutations. RREB1 and FBXO11 knockout (KO) induced surface MHC-II expression in these cell lines, which was further potentiated by IFN-γ stimulation (Figures 1C-1E and S1B). Additionally, combined KO of RREB1 and FBXO11 further augmented MHC-II expression with no significant effect on surface MHC-I levels or transcription of key genes within the MHC-I antigen processing pathway (Figures 1F and S1C). Although CRISPR/Cas9-mediated disruption of RREB1 could not be confirmed by immunoblot due to a lack of antibodies that detect the endogenous protein, efficient on-target genomic editing was demonstrated by ICE analysis for each sgRNA (Figure S1D).
Figure 1. Genome-wide CRISPR screens identify key MHC-II regulators in AML.
(A)CRISPR screen workflow. MHC-II-negative cells (MOLM13 and OCI-AML3) were mutagenized with pooled lentiviral libraries comprising 220,000 sgRNA (whole-genome screen) or 15,300 sgRNA (targeted epigenetic screen) and MHC-II+ cells were enriched by FACS.
(B)Whole-genome CRISPR screen results. For IFN-γ+ sorts, cells were pulsed with IFN-γ 25 ng/mL for 48 hours prior to each sort. Bubble plots show the top 1,000 enriched genes identified. P values were calculated using the RSA algorithm (König et al., 2007).
(C and D) Cell surface MHC-II expression in MV4-11 Cas9 cells (C) and OCI-AML3 Cas9 cells (D) transduced with a pool of 3 sgRNAs targeting the indicated genes or a control sgRNA, in the presence or absence of IFN-γ 10 ng/mL (MV4-11) or 25 ng/mL (OCI-AML3) for 48 hours. Representative plots from 3 experiments.
(E)Cell surface MHC-II expression in MV4-11 Cas9 cells transduced with individual sgRNAs targeting the indicated genes or a control sgRNA. Bars depict the mean percentage of MHC-II+ cells from 3 independent experiments indicated by points. Statistical analysis by unpaired t test compared to control cells; p value * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001.
(F)Cell surface MHC-I and MHC-II expression in OCI-AML3 Cas9 cells transduced with sgRNAs targeting the indicated genes and/or a control sgRNA. Representative plots from 2 experiments.
(G)Chromatin immunoprecipitation (ChIP) sequencing showing H3K27me3 levels at MHC-II genes and the HoxC cluster. Data are from GEO: GSM1513828 (Göllner et al., 2017). Y axes indicate reads per million (rpm). See also Figure S1.
We and others have shown that Polycomb repressive complex 2 (PRC2) coordinates MHC-I silencing across multiple cancer types (Burr et al., 2019; Dersh et al., 2021; Ennishi et al., 2019). Although it has previously been proposed that PRC2 may contribute to MHC-II silencing in diffuse large B cell lymphoma (Ennishi et al., 2019) and AML (Gambacorta et al., 2022), no polycomb complex proteins were identified as hits within our genome-wide screens. We therefore sought to clarify whether PRC2 was a major negative regulator of MHC-II expression in AML. Treatment of human AML cell lines and a murine MLL-AF9-driven AML cell line with EPZ-011989, a potent and selective EZH2 inhibitor (Figure S1E), demonstrated only a minimal increase in baseline MHC-II expression and no substantial enhancement of MHC-II following IFN-γ stimulation (Figure S1F). Notably, previous studies implicating PRC2 in MHC-II repression in AML failed to demonstrate H3K27me3 at MHC-II genes (Gambacorta et al., 2022), raising the prospect that alternative mechanisms such as viral mimicry, a well-established consequence of PRC2 inhibition (Morel et al., 2021), may indirectly activate MHC-II through a cell-intrinsic interferon response. Consistent with this, chromatin immunoprecipitation sequencing (ChIP-seq) data for H3K27me3 in AML cells (Göllner et al., 2017) revealed minimal H3K27me3 deposition at MHC-II pathway genes compared to known polycomb target genes (Figure 1G). Collectively, these data indicate that PRC2 is not the dominant negative regulator of MHC-II expression in AML and that RREB1 and FBXO11 likely operate through PRC2-independent mechanisms.
The CtBP co-repressor complex catalyzes H3K9me2 at MHC-II pathway genes
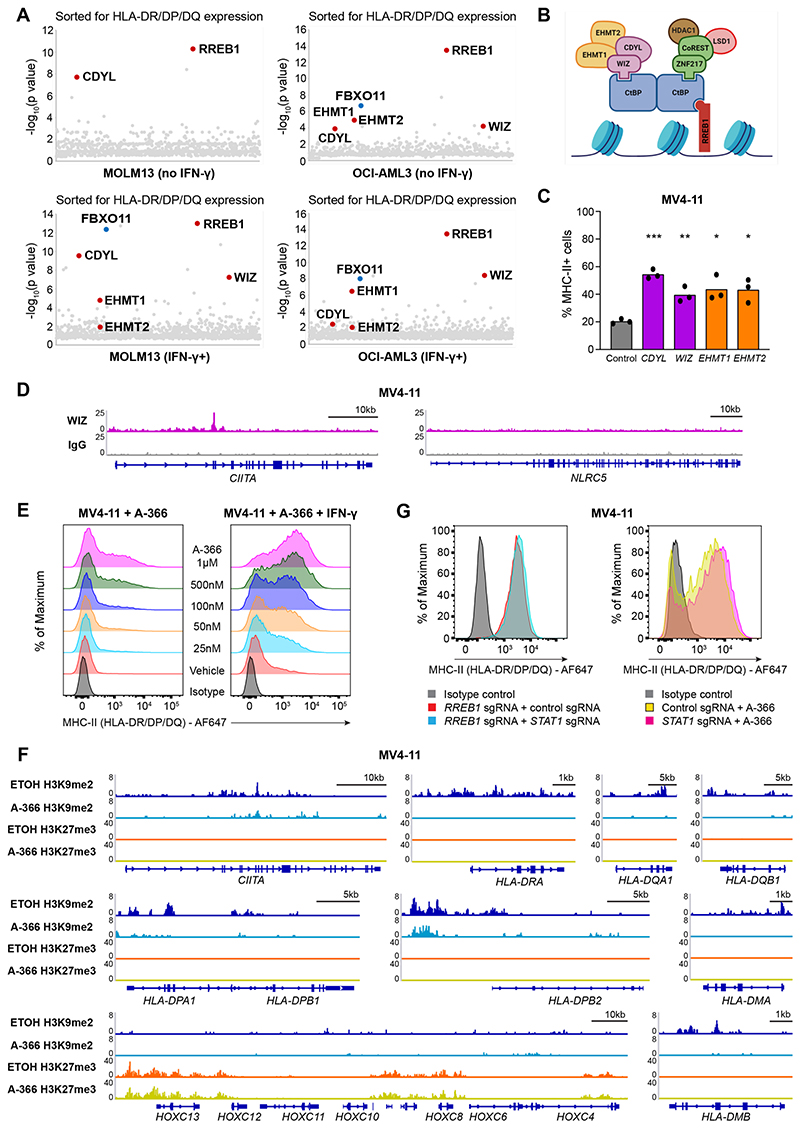
Whole-genome CRISPR screens are excellent at identifying selected members of major pathways required for a biological process; however, greater granularity is often achieved through more focused CRISPR screening approaches. We therefore repeated the positive-selection CRISPR screens in MOLM13 and OCI-AML3 cells, utilizing a library of 15,300 sgRNAs targeting epigenetic regulators and transcription factors. Validating the genome-wide screens, RREB1 and FBXO11 again emerged as top hits in these targeted screens. However, in addition, we now identified several components of the C-terminal binding protein (CtBP) co-repressor complex (Figure 2A). In vertebrates, the CtBP complex comprises a core homodimer or heterodimer of CtBP1 and CtBP2, which are highly homologous and often functionally redundant proteins, likely explaining why sgRNA targeting these genes were not enriched in our screens. Additionally, the combined loss of Ctbp1 and Ctbp2 results in embryonic lethality (Hildebrand and Soriano, 2002), illustrating their essential roles in gene regulation and development. CtBP1/2 associate with a broad range of DNA-binding proteins and accessory complex members involved in transcriptional regulation. The CtBP proteins directly interact with several nuclear proteins via two conserved binding motifs: (i) the PLDLS motif present on several DNA-binding transcription factors such as RREB1 or adapter proteins including WIZ and (ii) an RRT motif present in other DNA-binding and CtBP-interacting factors such as ZNF217 and RIZ1. Each CtBP dimer contains two PLDLS interaction clefts and two RRT-binding grooves and are consequently able to interact with DNA-bound transcription factors through one of these elements, whilst concurrently engaging adapter proteins that recruit the repressive enzymatic activity of histone methyltransferases (EHMT1/2), histone deacetylases (HDAC1/2) and histone demethylases (LSD1) (Chinnadurai, 2007). There are likely several CtBP complexes active within a cell, where specificity is conferred through the interaction with certain transcription factors and adapter proteins that enable the recruitment of chromatin-modifying enzymes to facilitate transcriptional repression.
Figure 2. Transcriptional silencing of MHC-II is mediated by the CtBP co-repressor complex.
(A)Targeted epigenetic CRISPR screen results. For IFN-γ+ sorts, cells were pulsed with IFN-γ 25 ng/mL for 48 hours prior to each sort. Bubble plots show the top 1,000 enriched genes identified, with components of the CtBP complex indicated in red. P values were calculated using the RSA algorithm (König et al., 2007).
(B)Schematic representation of the CtBP complex.
(C)Cell surface MHC-II expression in MV4-11 Cas9 cells transduced with a pool of 2 sgRNAs targeting the indicated genes or a control sgRNA. Bars depict the mean percentage of MHC-II+ cells from 3 independent experiments indicated by points. Statistical analysis by unpaired t test compared to control cells; p value * < 0.05, ** < 0.01, *** < 0.001.
(D)Chromatin immunoprecipitation sequencing (ChIP-seq) plots for WIZ in MV4-11 cells at indicated genes.
(E)Cell surface MHC-II expression in MV4-11 cells treated with EHMT1/2 inhibitor (A-366) at the indicated doses or vehicle control in the presence or absence of IFN-γ 10 ng/mL for 24 hours. Representative plots from 2 experiments.
(F)CUT&Tag chromatin profiling of H3K9me2 and H3K27me3 at MHC-II genes in MV4-11 cells treated with A-366 1μM or vehicle control (ETOH) for 7 days. The HoxC cluster is included as a positive H3K27me3 control.
(G)Cell surface MHC-II expression in MV4-11 RREB1 KO cells transduced with a pool of 2 sgRNAs targeting STAT1 or a control sgRNA, and MV4-11 STAT1 KO or control cells treated with A-366 1μM for 7 days. Representative plots from 2 experiments.
See also Figure S2.
Remarkably, the additional hits identified in our focused CRISPR screens functionally linked the transcription factor RREB1, the PLDLS-containing adapter protein WIZ along with the chromatin reader CDYL and the histone methyltransferases EHMT1 and EHMT2 as the specific CtBP complex that facilitates MHC-II repression (Figures 2A and 2B). In support of this contention, single gene KOs of these non-redundant CtBP complex members increased MHC-II expression even in the absence of stimulation with the inflammatory cytokine IFN-γ (Figures 2C and S2A) and ChIP-seq assays localized WIZ, a key CtBP complex member not known to associate with any other epigenetic complexes, to MHC-II pathway genes (Figures 2D and S2B-C) but not to NLRC5, the key transcriptional regulator of MHC-I expression (Meissner et al., 2010) (Figure 2D).
Having used unbiased genetic strategies to establish the requirement of the CtBP complex in MHC-II repression, we next wanted to address the role of selectively inhibiting the catalytic components of the complex. EHMT1 and EHMT2 form a heteromeric complex, whereby both proteins are cooperatively required for the in vivo catalysis of histone H3 lysine 9 monomethylation (H3K9me1) and dimethylation (H3K9me2) (Tachibana et al., 2005). As both were identified as the major chromatin-modifying enzymes involved in MHC-II repression via the CtBP complex, we utilized A-366 and UNC0638, which are two chemically distinct, potent and selective inhibitors of both EHMT1 and EHMT2 (Pappano et al., 2015; Vedadi et al., 2011). Treatment of AML cells with A-366 and UNC0638 resulted in dose-dependent MHC-II upregulation and increased IFN-γ sensitivity (Figures 2E and S2D-S2E), phenocopying the MHC-II upregulation observed with CRISPR-mediated KO of EHMT1/2 (Figure 2C). As H3K9me2 is associated with dynamic gene repression within facultative heterochromatin and euchromatin (Peters et al., 2003; Tachibana et al., 2002), we assessed H3K9me2 levels at MHC-II pathway genes, including the key transcriptional regulator CIITA. These data demonstrate that MHC-II pathway genes are marked by H3K9me2 at baseline and the re-expression of MHC-II after catalytic inhibition of EHMT1/2 is associated with depletion of H3K9me2 at these genes (Figure 2F). To distinguish whether MHC-II loci are also regulated by PRC2, we assessed H3K27me3 levels at these genes. Importantly, H3K27me3 is not seen at MHC-II genes, nor does A-366 alter the levels of H3K27me3 at known PRC2 target genes (Figure 2F). Taken together, these findings show that the EHMT1/2-containing CtBP co-repressor complex is the key transcriptional regulator of MHC-II expression.
It is increasingly recognized that therapies targeting epigenetic modifiers, particularly those involved in transcriptional repression, can reactivate endogenous retroviral elements (ERVs), triggering a type I interferon response and subsequent MHC upregulation (Chiappinelli et al., 2015; Roulois et al., 2015; Sheng et al., 2018). To assess whether MHC-II upregulation following CtBP complex disruption was principally due to enhanced interferon signaling, we generated MV4-11 cells with CRISPR-mediated disruption of STAT1 (Figure S2F), a transcription factor that is a key component of the canonical type I and type II interferon pathway (Durbin et al., 1996; Meraz et al., 1996). Loss of STAT1 had no effect on the level of MHC-II upregulation induced by both RREB1 KO and A-366 treatment (Figure 2G), confirming that targeting the CtBP complex derepresses MHC-II independent of interferon signaling due to ERV activation and viral mimicry.
The CtBP complex and FBXO11 are specific and synergistic repressors of surface MHC-II expression
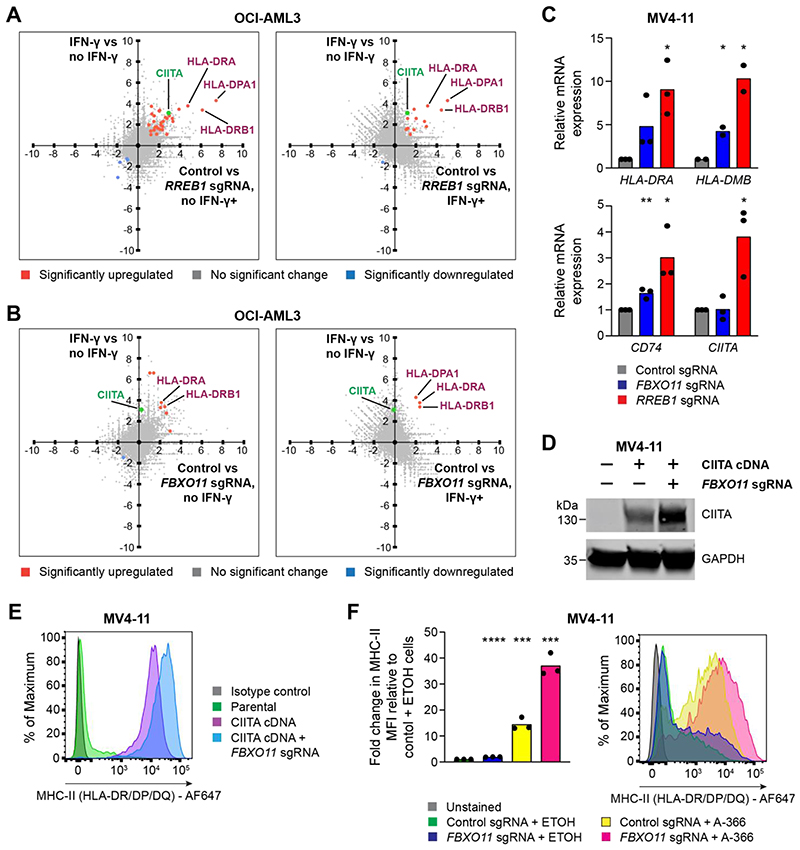
As loss of the RREB1-containing CtBP complex increases basal expression of MHC-II genes and potentiates the induction of these genes by inflammatory cytokines, we sought to investigate the specificity of the gene expression changes induced by functional disruption of this complex in the presence and absence of IFN-γ. Notably, we did not identify increased expression of any genes involved in the MHC-I antigen processing and presentation pathway following loss of RREB1 (Burr et al., 2019), including the non-classical MHC-I allele HLA-G (Figure S2G), of which RREB1 is a known regulator (Flajollet et al., 2009). Gene set enrichment analysis also predominantly highlighted terms driven by MHC-II gene upregulation, with no significant change in pathways relating to intracellular signaling, cell death or cell cycle progression (Figure S2H). Strikingly, the most highly upregulated genes following RREB1 KO in AML included multiple genes encoding MHC-II alleles,as well as the MHC-II transactivator CIITA (Figures 3A and S3A-S3B). CIITA is the master regulator of tissue-specific MHC-II expression and binds the proximal promoter of all classical MHC-II genes (encoding the HLA-DR, HLA-DP and HLA-DQ isotypes), as well as other MHC-II pathway genes including CD74 (which encodes the invariant chain), HLA-DM and HLA-DO. In conjunction with our WIZ ChIP-seq (Figures 2D and S2B) and H3K9me2 data (Figure 2F), these findings implicate the RREB1-containing CtBP complex as a major and selective transcriptional repressor of MHC-II genes.
Figure 3. The CtBP complex and FBXO11 synergistically regulate MHC-II in AML.
(A and B) RNA-seq correlation plots showing differential gene expression in OCI-AML3 cells in the presence or absence of IFN-γ 25 ng/mL for 48 hours (Y-axis) and OCI-AML3 Cas9 cells transduced with control and RREB1 (A) or FBXO11 (B) sgRNA in the presence or absence of IFN-γ 25 ng/mL for 48 hours (X-axis). Fold change > 1.0 and false discovery rate < 0.05 for highlighted genes (in blue and orange) with significant differential gene expression. CIITA is labelled separately in green and reaches significance only in the RREB1 sgRNA, no IFN-γ analysis.
(C)mRNA expression of MHC-II genes in MV4-11 Cas9 cells transduced with control, FBXO11 or RREB1 sgRNA. Bars depict mean fold change in expression from 3 independent experiments and points indicate the mean of technical triplicates from individual experiments. Statistical analysis by unpaired t test compared to control cells, with significant changes indicated; p value * < 0.05, ** < 0.01.
(D and E) Immunoblot (D) and cell surface MHC-II expression (E) in MV4-11 Cas9 cells transduced with a retroviral vector encoding CIITA cDNA and with or without FBXO11 sgRNA. Representative data from 2 experiments.
(F) Cell surface MHC-II expression and mean fold change in median fluorescence intensity (MFI) in MV4-11 Cas9 cells transduced with control or FBXO11 sgRNA and treated with EHMT1/2 inhibitor (A-366 1μM) or vehicle control (ETOH) for 7 days. Bars depict mean fold change in MFI from 3 independent experiments indicated by points. Statistical analysis by unpaired t test compared to control sgRNA + ETOH cells, with significant changes indicated; p value *** < 0.001, **** < 0.0001.
See also Figures S3 and S4.
FBXO11 is an F-box protein, which constitutes the specific substrate recognition component of Skp, Cullin, F-box-containing (SCF) multi-subunit E3 ubiquitin ligase complexes that catalyze polyubiquitination of proteins and target them for proteasomal degradation. Established targets of FBXO11 include BCL6 (Duan et al., 2012), through which it controls B cell development within lymph node germinal centers (Schneider et al., 2016), as well as BLIMP-1 (Horn et al., 2014), which is essential for terminal differentiation of B cells to plasma cells. Given that AML cells with low MHC-II expression exhibit silencing of multiple MHC-II genes, the protein target of FBXO11-mediated regulation was not immediately evident.
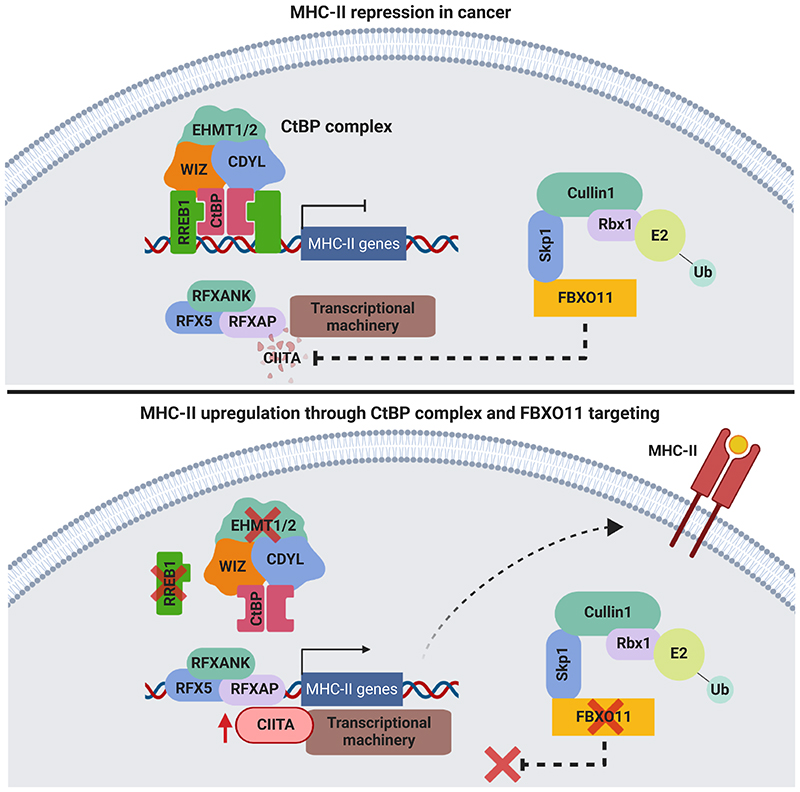
We initially performed RNA-sequencing (RNA-seq) to determine the transcriptional consequences of FBXO11 loss. Here we found that FBXO11 depletion in AML cells significantly increased expression of only a small subset of genes (30 genes in IFN-γ-treated cells, 45 genes in untreated cells). Importantly, these included multiple classical MHC-II genes but, in contrast to RREB1 KO, CIITA expression did not increase following FBXO11 KO (Figures 3B and S3C-S3D). Given the importance of CIITA in coordinating the transcriptional activation of MHC-II pathway genes, these data raised the possibility that FBXO11 may regulate CIITA protein stability. Consistent with this, the MHC-II upregulation that follows FBXO11 loss is most dramatic following IFN-γ stimulation (Figure 1D), which is known to induce CIITA gene expression. Validation of the RNA-seq findings by RT-qPCR in a different AML cell line further confirmed that, while loss of RREB1 led to activation of both CIITA and MHC-II gene expression, FBXO11 KO led to activation of downstream MHC-II genes with no change in CIITA mRNA levels (Figure 3C). To further investigate FBXO11-mediated post-translational regulation of CIITA, we expressed CIITA cDNA in AML cells under the control of a viral promoter, which drives stable MHC-II expression with no corresponding effect on MHC-I (Figure S4A). Depletion of FBXO11 in these cells led to a substantial accumulation of CIITA protein with a corresponding increase in cell surface MHC-II (Figures 3D and 3E). Together these data suggest that loss of FBXO11 increases cell surface MHC-II in leukemia cells due to stabilization of CIITA protein, which drives enhanced expression of multiple MHC-II genes.
Since our results had demonstrated a role for the CtBP complex in transcriptional repression of CIITA and FBXO11 as a specific regulator of CIITA protein stability, we predicted that combined targeting of these non-overlapping repressive mechanisms would synergistically induce high level MHC-II expression. As expected,depletion of FBXO11 further increased MHC-II expression in RREB1 KO cells (Figure 1F) and also enhanced the MHC-II upregulation induced by pharmacological EHMT1/2 inhibition (Figure 3F). Our data therefore establish that the CtBP complex and FBXO11 are highly selective and synergistic regulators of MHC-II expression in AML, and provide valuable insights into potential therapeutic strategies to facilitate re-expression of MHC-II in the setting of post-alloSCT relapse.
CtBP complex and FBXO11 targeting enhance MHC-II expression in non-hematological cancers
In an era defined by the widespread incorporation of cancer immunotherapies into clinical practice, there is increasing recognition of the prognostic importance of MHC-II expression on tumor cells, especially in terms of predicting therapeutic responses and improved patient survival outcomes (Axelrod et al., 2019). Interestingly, this observation is not limited to cancers arising from professional APCs that constitutively express MHC-II, such as diffuse large B cell lymphoma (Rimsza et al., 2004) and Hodgkin lymphoma (Roemer et al., 2018), but also includes epithelial and cutaneous malignancies where the cell of origin does not normally express MHC-II, such as non-small cell lung cancer (Johnson et al., 2020), breast cancer (Forero et al., 2016; Park et al., 2017) and melanoma (Johnson et al., 2016; Liu et al., 2019). This is illustrated through RNA-seq analysis of biopsy samples from cohorts of melanoma patients treated with immune checkpoint inhibitors (Gide et al., 2019) (Figure 4A), which demonstrates significant variability in MHC-II levels and a strong correlation of higher expression of MHC-II pathway genes with therapeutic response. Consequently, interventions that induce tumor cell MHC-II upregulation may not only potentiate the GVL effect in the setting of post-alloSCT relapse of AML, but could also help to improve immunotherapy responses in non-hematological cancers.
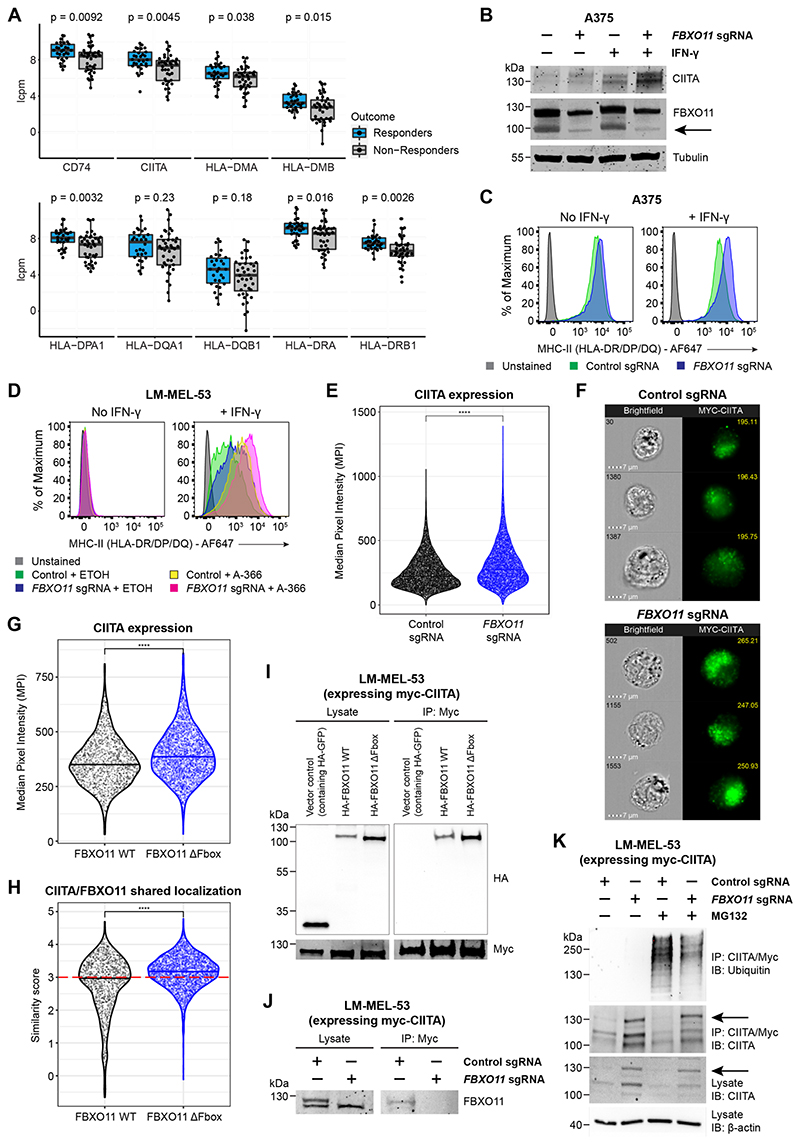
Figure 4. FBXO11 loss increases surface MHC-II expression through stabilization of CIITA.
(A)mRNA expression of MHC-II genes in tumor biopsy samples from melanoma patients treated with immune checkpoint inhibitors. The upper limit, center and lower limit of each box denotes the upper quartile, median and lower quartile of the data, respectively. P values were calculated using the Wilcoxon rank-sum test. Data are from ENA: PRJEB23709 (Gide et al., 2019). Y axes indicate log2 counts per million (lcpm).
(B and C) Immunoblot (B) and cell surface MHC-II expression (C) in A375 cells transduced with a retroviral vector encoding CIITA cDNA with or without FBXO11 sgRNA, in the presence or absence of IFN-γ 10 ng/mL for 48 hours. Representative data from 2 experiments.
(D)Cell surface MHC-II expression in LM-MEL-53 Cas9 cells transduced with control or FBXO11 sgRNA and treated with EHMT1/2 inhibitor (A-366 3μM) or vehicle control for 7 days, in the presence or absence of IFN-γ 10 ng/mL for 48 hours. Representative data from 2 experiments.
(E)Myc-CIITA expression measured as Median Pixel Intensity (MPI) in LM-MEL-53 Cas9 cells transduced with control or FBXO11 sgRNA. Each data point represents a single cell measured and bars represent the group median. Statistical significance was calculated using Dunn’s test with Bonferroni correction for multiple comparisons. **** = p < 0.0001. Representative data from 2 experiments.
(F)Representative images showing brightfield microscopy (cell number at top left) and Myc-CIITA expression (MPI value at top right) in LM-MEL-53 Cas9 cells transduced with control or FBXO11 sgRNA.
(G)Myc-CIITA expression measured as MPI in LM-MEL-53 cells expressing FBXO11 WT or FBXO11 ΔFbox. Each data point represents a single cell measured and bars represent the group median. Statistical significance was calculated using Dunn’s test with Bonferroni correction for multiple comparisons. **** = p < 0.0001. Representative data from 2 experiments.
(H)Summary of shared localization between CIITA and FBXO11 in LM-MEL-53 cells expressing FBXO11 WT or FBXO11 ΔFbox. High shared localization defined by similarity score ≥3 (red dotted line), as previously described (Beum et al., 2006; Erie et al., 2011; George et al., 2006). Statistical significance was calculated using Dunn’s test with Bonferroni correction for multiple comparisons. **** = p < 0.0001. Representative data from 2 experiments.
(I)Immunoprecipitation of myc-CIITA from lysates of LM-MEL-53 cells expressing FBXO11 WT or FBXO11 ΔFbox, analyzed by immunoblot. Lysate, 5% of input. Experiments performed twice.
(J)Immunoprecipitation of myc-CIITA from lysates of LM-MEL-53 Cas9 cells transduced with control or FBXO11 sgRNA, analyzed by immunoblot. Lysate, 5% of input. Experiments performed twice.
(K)Immunoprecipitation of myc-CIITA from lysates of LM-MEL-53 Cas9 cells transduced with control or FBXO11 sgRNA and with or without MG-132 treatment, analyzed by immunoblot. Lysate, 5% of input. Experiments performed twice.
See also Figures S4 and S5.
We therefore evaluated the effect of disrupting CtBP complex and FBXO11 function in melanoma cell lines with variable levels of cell surface MHC-II (Figure S4B), which is representative of the heterogeneity for MHC-II expression seen in this disease (Meyer et al., 2021). Mirroring our findings in AML, deletion of FBXO11 in melanoma cell lines with basal expression of MHC-II led to accumulation of endogenous CIITA and a corresponding increase in cell surface MHC-II (Figures 4B-4C and S4C-S4D). In contrast, in LM-MEL-53 cells where MHC-II is silenced, increased MHC-II upregulation following FBXO11 KO was only evident following induction of basal CIITA expression in response to IFN-γ treatment (Figure S4E). Similar to our observations in AML, targeting the catalytic activity of the EHMT1/2-containing CtBP complex with A-366 led to enhanced MHC-II expression in these cells, which was further augmented by concurrent FBXO11 KO (Figure 4D), with no effect on MHC-I levels (Figure S4F). Modest MHC-II upregulation was also seen in WM852 and A375 cells treated with A-366 (Figure S4G and S4H), corresponding with the lesser degree of basal MHC-II repression in these cells. These data reinforce the specificity of CtBP complex and FBXO11-mediated MHC-II repression, and broaden the applicability of treatments targeting these repressive mechanisms to upregulate tumor-specific MHC-II and potentially enhance melanoma responses to immune checkpoint inhibition.
FBXO11 interacts with CIITA and promotes CIITA ubiquitination
In order to further characterize the mechanism through which FBXO11 regulates CIITA protein stability, we generated LM-MEL-53 cells expressing epitope-tagged CIITA. As seen in MV4-11 cells (Figures 3D and 3E), genetic disruption of FBXO11 resulted in significant CIITA accumulation (Figures 4E and 4F). Increased CIITA levels were also observed following co-expression of an FBXO11 mutant (Figure 4G) in which the F-box domain (aa71-116) has been deleted, thereby abolishing binding to the remainder of the SCF ubiquitin ligase complex but leaving the substrate recognition domain intact (Horn et al., 2014). The FBXO11ΔFbox mutant showed increased nuclear localization compared to wildtype FBXO11 (Figure S5A), potentially reflecting impaired binding to SCF complex components within the cytoplasm. Expression of FBXO11ΔFbox increased the nuclear localization of co-expressed CIITA and dual staining revealed high CIITA and FBXO11ΔFbox colocalization (Figures 4H and S5A-B). These findings suggested that FBXO11ΔFbox may bind and stabilize CIITA by inhibiting its recruitment to the SCF ubiquitin ligase complex and subsequent degradation.
To more directly investigate whether CIITA and FBXO11 interact, we immunoprecipitated CIITA from CIITA-expressing LM-MEL-53 cells, which revealed co-immunoprecipitation of FBXO11 (Figure 4I). Consistent with the significant nuclear co-localization of CIITA and FBXO11ΔFbox, and the known function of the F-box in selectively binding Skp1 to target bound substrates for degradation, interaction between CIITA and FBXO11ΔFbox was readily detected. Supporting the specificity of this interaction, endogenous FBXO11 could also be detected in association with CIITA (Figure 4J). Incubation of cells with the proteasome inhibitor MG-132 led to accumulation of polyubiquitinated CIITA, indicating turnover of CIITA via proteasome-mediated degradation. Whilst total levels of CIITA were increased in the absence of FBXO11 (Figures 4B, 4E, 4F and 4K), knockout of FBXO11 led to a reduction in polyubiquitinated CIITA in MG-132-treated cells, supporting a role for FBXO11 in targeting CIITA for polyubiquitination and degradation by the proteasome (Figure 4K). These findings collectively establish FBXO11 as part of a key ubiquitin ligase complex regulating CIITA protein stability via proteasomal degradation. The stabilization of CIITA subsequently leads to enhanced downstream transcriptional activation of CIITA-regulated MHC-II genes and increased cell surface MHC-II expression.
MHC-II expression on leukemic blasts facilitates anti-cancer immune surveillance
To explore the contribution of tumor cell MHC-II expression to immunological anti-leukemia responses, we investigated whether MHC-II upregulation could be induced by CtBP complex and FBXO11 targeting in a mouse model of AML that does not express MHC-II. Interestingly, deletion of Fbxo11 and Rreb1 (Figure S5C) and A-366 treatment (Figure S5D) did not increase surface MHC-II levels, implying differences in the repressive mechanisms that regulate MHC-II expression in human and mouse. We therefore performed an unbiased CRISPR screen to determine the regulators of MHC-II in mouse AML cells, identifying Rcor1 (CoREST) and Zfp217 as the top hits (Figures S5E and S5F). RCOR1/CoREST and ZNF217 form a distinct CtBP complex to those that contain WIZ and CDYL, which we had shown are the major CtBP components involved in regulating MHC-II in human cells. Notably, RCOR1 and ZNF217 KO failed to significantly increase MHC-II levels in human AML cells (Figure S5G). Together these data highlight the fact that, although the CtBP complex regulates MHC-II expression in mouse and human, there are different facultative members involved in this process for each species.
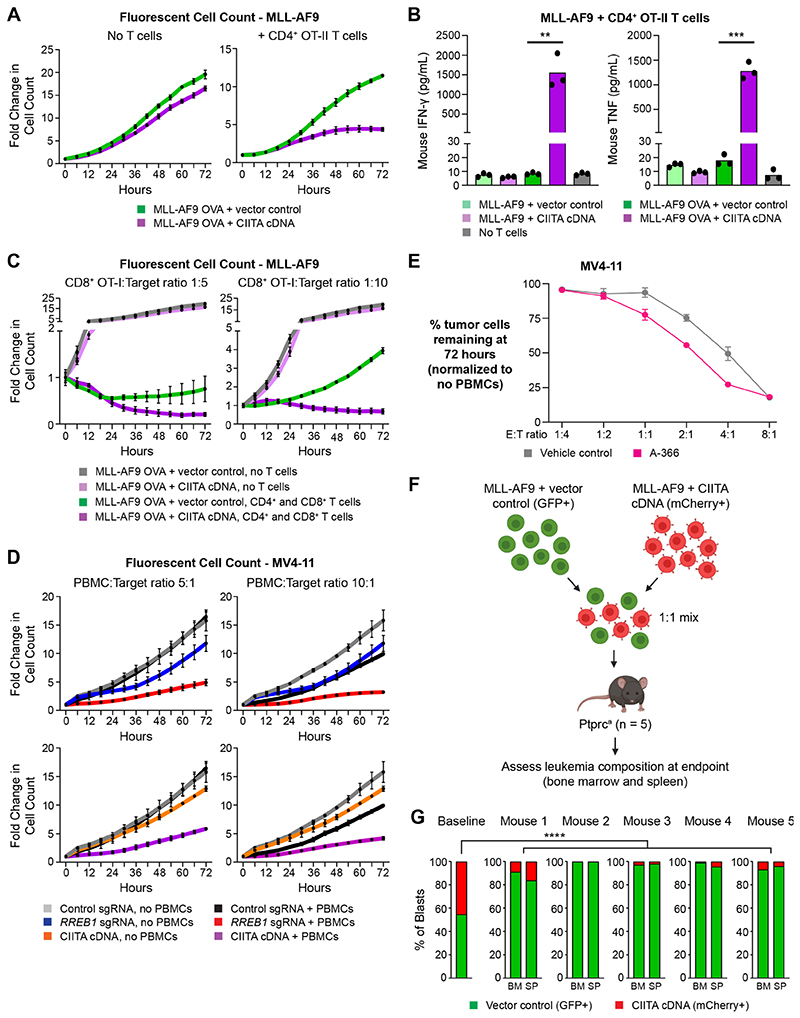
Similar to human AML cells (Figure S4A), expression of CIITA cDNA resulted in significant and selective MHC-II upregulation (Figure S6A and S6B) in mouse AML cells, enabling us to specifically test the functional effects of leukemia MHC-II expression using genetically matched immune effector cells. To investigate whether restoring MHC-II in MHC-II-negative AML cells is sufficient to stimulate antigen-dependent T cell responses, we engineered mouse leukemia cells to express full-length chicken ovalbumin (OVA) with a green fluorescent protein (GFP) reporter, as well as CIITA. These OVA-expressing MHC-II+ cells were then used for in vitro co-culture experiments on the Incucyte SX5 Live-Cell Analysis System, allowing objective real-time tracking of AML cell proliferation and killing using a combination of phase contrast imaging and detection of GFP fluorescence (Figure S6C). Co-culture with CD4+ OT-II T cells, which specifically recognize processed peptides derived from OVA protein and presented on mouse MHC-II, significantly reduced the number of OVA-expressing MHC-II+ MLL-AF9 cells compared to OVA-expressing cells that were negative for MHC-II (Figure 5A). High-level release of pro-inflammatory cytokines were also observed following co-culture of CD4+ OT-II T cells with OVA-expressing MHC-II+ leukemia cells (Figure 5B) compared to leukemia cells expressing either OVA or MHC-II alone, providing confirmation of antigen- and MHC-II-dependent T cell activation. Tumor cell-specific MHC-II expression also significantly potentiated CD8+ T cell killing in combined co-cultures of OVA-expressing leukemia cells with CD8+ OT-I T cells and CD4+ OT-II T cells (Figure 5C), suggesting a role for promotion of CD4+ T cell help in addition to direct CD4+ T cell cytotoxicity. These findings were validated using co-cultures of human peripheral blood mononuclear cells (PBMCs) with RREB1 KO, CIITA-expressing or A-366-treated MV4-11 cells, all of which resulted in clear reduction of MHC-II+ AML cells compared to control cells with low MHC-II expression (Figures 5D and 5E). Importantly, EHMT1/2 inhibition had no effect on human T cell subset composition (Figure S6D) or effector function (Figure S6E), reinforcing the specificity of CtBP complex targeting in augmenting anti-tumor immunity via upregulation of tumor cell MHC-II expression. Taken together, these data show that, despite their impaired capacity for differentiation, both human and murine leukemic blasts can process and present antigen via a functional MHC-II pathway, leading to antigendependent CD4+ T cell activation and potent anti-tumor immune effects. While our results demonstrate that either genetic or pharmacological CtBP complex disruption is sufficient to increase MHC-II expression and T cell-mediated killing, the potential role for targeting FBXO11 in isolation was comparatively more modest (Figures 1C-1E and 3C). This is consistent with its role in post-translational regulation of CIITA, which is absent or lowly expressed in MHC-II deficient cells and requires activation through disruption of the CtBP complex or stimulation with inflammatory cytokines.
Figure 5. MHC-II expression on leukemic blasts facilitates adaptive anti-cancer immune responses.
(A)Fold change of fluorescent cell count for MLL-AF9 OVA-expressing cells transduced with a retroviral vector encoding CIITA cDNA or vector control, following co-culture with CD4+ OT-II T cells at an effector:target (E:T) ratio of 4:1. Plots show mean ± SEM of technical triplicates from a representative experiment, performed twice.
(B)Cytometric bead array (CBA) assay for T cell cytokines following 24 hour co-culture of OT-II CD4+ T cells with MLL-AF9 cells expressing OVA peptide and/or CIITA cDNA or vector control, at an E:T ratio of 2:1. Bars depict the mean of technical triplicates from a representative experiment, performed twice. Statistical analysis by unpaired t test compared to respective vector control cells, with significant changes indicated; p value ** < 0.01, *** < 0.001.
(C)Fold change of fluorescent cell count for MLL-AF9 OVA-expressing cells transduced with a retroviral vector encoding CIITA cDNA or vector control, following co-culture with CD4+ OT-II T cells at an E:T ratio of 4:1 and CD8+ T cells at the indicated E:T ratios. Plots show mean ± SEM of technical triplicates from a representative experiment, performed twice.
(D)Fold change of fluorescent cell count for MV4-11 Cas9 cells transduced with RREB1 sgRNA, a retroviral vector encoding CIITA cDNA or vector control, following co-culture with human peripheral blood mononuclear cells (PBMC) at the indicated E:T ratios. Plots show mean ± SEM of technical triplicates from a representative experiment, performed twice.
(E)Percent remaining live MV4-11 cells following 72 hour co-culture with human CD4+ T cells at the indicated E:T ratios. Cells were treated with A366 1μM or vehicle control for the duration of the experiment. Plots show mean ± SEM of technical triplicates from a representative experiment, performed twice.
(F)Overview of in vivo competition assay, utilizing MLL-AF9 cells transduced with GFP vector control or a retroviral vector encoding mCherry and CIITA cDNA.
(G)Proportion of GFP+ control and mCherry+ CIITA-expressing cells in bone marrow (BM) and spleen (SP) at disease endpoint. Two-tailed binomial test was performed to compare expected and observed proportions in each sample compared to baseline. **** = p < 0.0001.
See also Figures S5 and S6.
The evaluation of immune responses in patient-derived or human cell line xenografts is clouded by the xenogenic immune environment. Moreover, when unmatched PBMCs are used as immune effectors in these assays, there is also likely a potent allogeneic effect that would mask the ability to evaluate the selective functional consequences of MHC-II expression. Therefore, to determine whether tumor cell MHC-II expression promotes immune-mediated clearance of MHC-matched AML in vivo, we chose to utilize the transplantation of mouse leukemia cells into genetically matched recipients with an identical MHC-I and MHC-II haplotype. As EHMT1/2 inhibitors do not regulate MHC-II expression in mice, we instead transplanted an equal ratio of control GFP+ leukemia cells (MHC-Ihigh) and mCherry+ CIITA-expressing leukemia cells (MHC-Ihigh + MHC-IIhigh) into immunocompetent mice (Figure 5F) and then assessed the relative proportion of GFP+ and mCherry+ cells contributing to resulting leukemia (Figure S6F). Remarkably, the leukemic composition in all five mice showed a marked predominance of GFP+ cells (Figure 5G), consistent with immune clearance of the mCherry+ MHC-II+ compartment. Although an important limitation of this in vivo model is an inability to fully replicate the GVL effect induced by alloSCT, it nevertheless underscores the biological relevance of MHC-II in driving immune-mediated anti-leukemia responses in an MHC-matched setting.
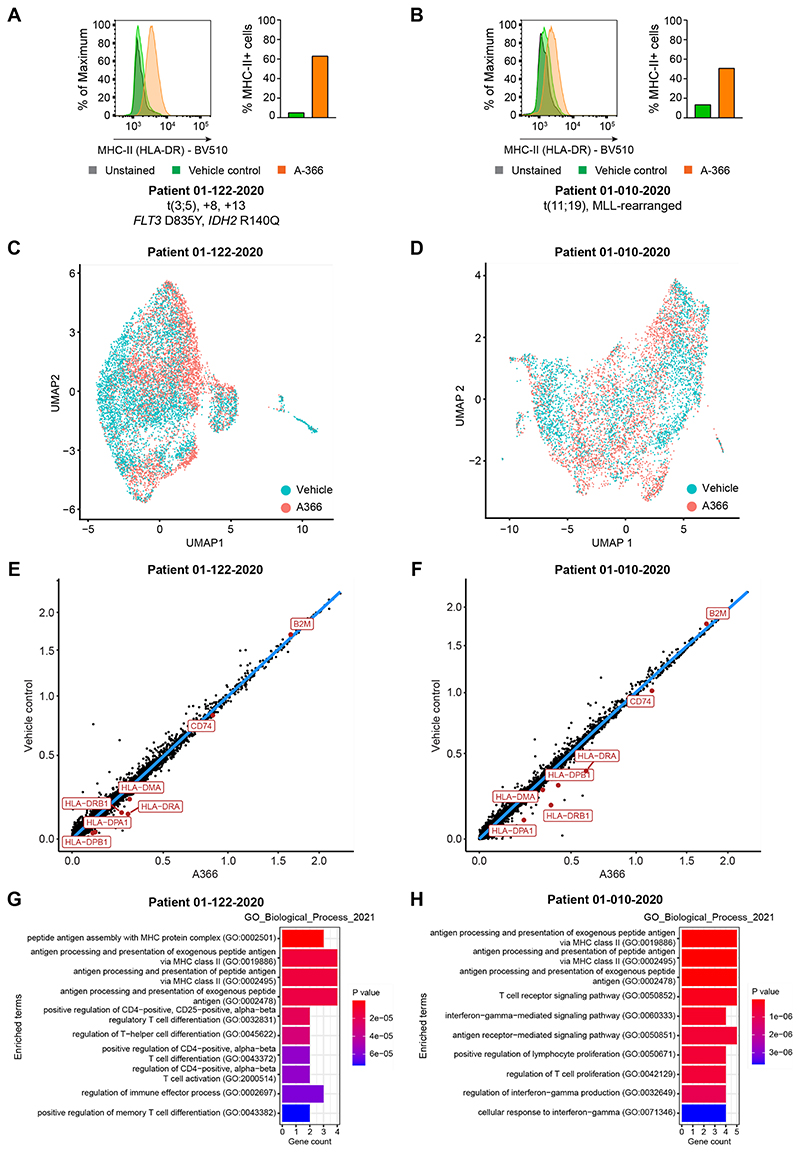
Finally, to illustrate the relevance of our findings to primary human leukemia samples, we assessed the effect of inhibiting EHMT1/2, the catalytic component of the CtBP complex, in primary AML cells with low basal levels of MHC-II expression. Consistent with our findings in human AML cell lines, A-366 treatment of leukemic blasts from two patients induced significant MHC-II upregulation, even in the absence of IFN-γ stimulation (Figures 6A and 6B). Although inhibition of epigenetic complexes is often associated with a diverse range of gene expression changes, we found that EHMT1/2 inhibition in human AML cells is remarkably specific. Despite increasing MHC-II cell surface expression, single-cell RNA sequencing following A-366 treatment did not reveal a global change in the transcriptional landscape of the treated cells, which clustered together with the control population (Figures 6C and 6D). Strikingly, when we assessed the global gene expression changes in both patient samples, we observed highly selective upregulation of only MHC-II pathway genes (Figures 6E-6H). These data confirm that CtBP-mediated repression is a targetable mechanism of MHC-II silencing in human AML. Moreover, the lack of widespread gene expression changes in the tumor cells alongside the preserved T cell function in the presence of EHMT1/2 inhibition (Figure S6E) provide the rationale for clinical implementation of this therapeutic strategy to circumvent leukemic immune escape.
Figure 6. Pharmacological EHMT1/2 inhibition selectively induces MHC-II upregulation in primary AML cells.
(A and B) Cell surface MHC-II expression in primary AML cells from patients treated with EHMT1/2 inhibitor (A-366 1μM) or vehicle control for 5-7 days. Data is shown from a representative experiment, performed twice.
(C and D) UMAP projections of primary AML cells treated with EHMT1/2 inhibitor (A-366 1μM) or vehicle control for 5-7 days. Cells are highlighted by treatment.
(E and F) Scatterplots showing normalized gene expression in primary AML cells treated with EHMT1/2 inhibitor (A-366 1μM) or vehicle control for 5-7 days. MHC-II pathway genes and B2M are highlighted.
(G and H) Gene set enrichment analysis performed on all genes differentially upregulated with A-366 treatment in primary AML cells.
Discussion
The ability of alloSCT to maintain durable remissions in a proportion of AML patients, even those with adverse-risk prognostic features, highlights the considerable power of the GVL effect. Consequently, for patients who relapse post-alloSCT, immunomodulatory therapies that attempt to reinstate and augment GVL responses represent a logical and promising salvage strategy. However, currently available interventions, such as donor lymphocyte infusion, only benefit a minority and carry a significant risk of exacerbating graft-versus-host disease with attendant treatment-related morbidity and/or mortality (Miyamoto et al., 2017; Schmid et al., 2007).
Our findings help to address this critical unmet need by identifying fundamental mechanisms of MHC-II repression in AML that can be exploited to evade the GVL effect. In contrast to prior work characterizing the key role of PRC2 in MHC-I silencing (Burr et al., 2019; Dersh et al., 2021; Ennishi et al., 2019), here we demonstrate that the CtBP co-repressor complex is the main regulator of MHC-II gene expression, while FBXO11 functions indirectly through post-translational control of CIITA. Interestingly, MHC-I levels are unaffected by CtBP complex targeting while MHC-II levels are unaffected by PRC2 inhibition, illustrating how these two distinct chromatin repressive complexes independently regulate MHC-I and MHC-II expression. The compartmentalization of these processes underscores the importance of delineating the mechanisms of immune evasion in a given cancer context in order to guide appropriate intervention.
From a translational perspective, we show that currently available small molecule inhibitors of EHMT1/2 efficiently reverse CtBP-mediated MHC-II silencing, supporting clinical trials of these inhibitors in post-alloSCT relapse particularly when MHC-II is lowly expressed on the emergent measurable residual disease (MRD). We elucidate the role of MHC-II in driving the adaptive immune responses underpinning the GVL effect, demonstrating that MHC-II+ leukemic blasts can activate CD4+ T cells in an antigen-dependent manner, stimulating CD4+ T cell-mediated cytotoxicity as well as potentiation of CD8+ effector T cell function. The ability to potently and selectively induce MHC-II re-expression in leukemic blasts provides a unique opportunity to enhance GVL responses. Targeting these specific repressive mechanisms also enables MHC-II upregulation in non-hematological cancer cells, broadening the scope of our findings and raising the prospect that these adjunctive therapeutic approaches to increase MHC-II expression could enhance immunotherapy responses, particularly with combinatorial inhibition of various immune checkpoints.
Star Methods
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mark Dawson (mark.dawson@petermac.org).
Materials availability
All unique reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental Model and Subject Details
Cell lines
MV4-11, LM-MEL-53, A375 and Drosophila S2 cells were obtained from ATCC. MOLM13 and OCI-AML3 cells were obtained from DSMZ. WM852 cells were obtained from ESTDAB. HEK-293ET cells were a gift from Dr. Felix Randow (MRC-LMB, Cambridge, UK). Mouse MLL-AF9 leukemic blasts were generated by magnetic bead selection (Miltenyi Biotec) of c-KIT+ cells from whole mouse bone marrow and subsequent retroviral transduction with an MSCV-MLL-AF9-IRES-YFP construct.
Human cell lines were cultured in RPMI-1640 or DMEM (HEK-293ET) supplemented with 2 mM Glutamax, 100 IU/mL Penicillin, 100 μg/mL Streptomycin and 10% HI-FCS. MLL-AF9 cells were cultured in RPMI-1640 supplemented with 2 mM Glutamax, 100 IU/mL Penicillin, 100 μg/mL Streptomycin, 10% HI-FCS, IL-3 10 ng/mL, IL-6 10 ng/mL and SCF 50 ng/mL (Peprotech). T cells isolated from mice were cultured in complete RPMI-1640 media (10% HI-FCS, 100 IU/mL Penicillin, 100 μg/mL Streptomycin, 1 mM sodium pyruvate, 2 mM Glutamax and 55 μM 2-mercaptoethanol). Drosophila S2 cells were cultured in Schneider’s Drosophila media (Life Technologies) supplemented with 100 IU/mL Penicillin, 100 μg/mL Streptomycin and 10% HI-FCS. All cell lines were cultured in 5% CO2 at 37°C with the exception of Drosophila S2, which was cultured at room temperature.
Cell lines were authenticated by STR profiling through the Australian Genome Research Facility (Melbourne, Victoria). Cell lines were regularly tested and verified to be mycoplasma-negative by in-house PCR testing.
Animal experiments
All mouse work was performed at the Peter MacCallum Cancer Centre animal facility, under approval E615 from the Peter MacCallum Cancer Centre Animal Ethics and Experimentation committee. All ethical obligations were met.
For the in vivo competition assay, MLL-AF9 cells were transduced with either pMSCV-IRES-GFP or pMSCV-IRES-mCherry-CIITA. A pool of 1 x 106 GFP+ and 1 x 106 mCherry+ cells were then transplanted into 6-8 week old female B6.SJL-Ptprc<a>Pepc<b>/BoyJ (Ptprca) mice. Leukemic blasts were harvested from bone marrow and spleen at disease endpoint, with the proportion of GFP+ and mCherry+ cells assessed by flow cytometry.
Patient samples
Samples were collected from patients with AML treated at the Alfred Hospital after obtaining informed consent. Studies were conducted in accordance with the approved protocols through the Human Research Ethics Committee of the Alfred Hospital and the Declaration of Helsinki for all subjects. Primary AML cells were cultured in StemSpan SFEM (STEMCELL Technologies), supplemented with 10 ng/mL IL-3, 10 ng/mL IL-6, 50 ng/mL SCF and 50 ng/mL FLT3L (all Peprotech) and 35 nM UM171 and 500 nM stemreginin (STEMCELL Technologies).
Method Details
Chemicals
EPZ-011989, A-366 and UNC0638 were purchased from Selleck Chemicals. For each inhibitor, titration experiments were performed to determine the maximally effective, tolerated dose in each cell line. Doses used and durations of treatment are indicated in figure legends. Cells were typically treated for 7-10 days prior to analysis and inhibitors were refreshed 3 times per week. For interferon induction of MHC-I and MHC-II expression, human cells were treated with recombinant human IFN-γ (Sigma-Aldrich) at a dose of 10-25 ng/mL for 24-48 hours, while mouse cells were treated with recombinant mouse IFN-γ (Abcam) at a dose of 10 ng/mL for 48 hours prior to analysis.
Plasmids
pcDNA3 myc CIITA (P#808) was a gift from Matija Peterlin (Addgene plasmid #14650) (Kanazawa et al., 2000). CIITA cDNA from pcDNA3 myc CIITA was subcloned into both pMSCV-IRES-mCherry FP (a gift from Paul Beavis) and pMSCV-IRES-GFP (modified from pMSCV-IRES-mCherry FP, a gift from Paul Beavis) via EcoRI and XhoI sites. FLAG-HA-Fbxo11-pcDNA3.1-(Addgene #52501) and FLAG-HA-Fbxo11-ΔFbox-pcDNA3.1-(Addgene #52502) were a gift from Adam Antebi (Horn et al., 2014). Wildtype FBXO11 and FBXO11 ΔFbox were PCR amplified from the above plasmids and subcloned into pHRSIN-PSFFV-eGFP-PPGK-Puro via BamHI and XhoI, replacing the GFP. An ovalbumin (OVA)-expressing retroviral vector was generated by PCR amplification of full-length chicken OVA from pcDNA3-OVA (Addgene #64599, a gift from Sandra Diebold & Martin Zenke) (Diebold et al., 2001) and subcloned into pMSCV-IRES-GFP via EcoRI and XhoI sites. All plasmids were verified by Sanger sequencing analysis through the Australian Genome Research Facility (Melbourne, Victoria).
CRISPR sgRNA libraries
The whole-genome CRISPR screens were performed using the Bassik Human CRISPR Knockout Library (a gift from Michael Bassik, Addgene #101296-101934). This 10-sgRNAs-per-gene CRISPR/Cas9 deletion library was designed to target all ~20,500 protein-coding human genes. The library contains two distinct classes of negative control gRNA: non-targeting control sgRNAs with no binding sites in the genome and safe-targeting sgRNAs targeting genomic locations with no annotated function. Further details are described in Morgens et al (Morgens et al., 2017). Library sgRNAs were expressed in the pMCB320 lentiviral sgRNA expression vector, which encodes puromycin and mCherry selection markers.
For the targeted CRISPR screens in human AML cell lines, we compiled a custom library of ~15,300 sgRNAs targeting ~ 2,400 recognized chromatin regulators (6 sgRNAs per gene), as well as non-targeting and safe-targeting control sgRNAs (guide sequences are provided in Table S1). The library was designed through a combination of searches for genes containing domains known to be enriched in transcriptional regulators and manual curation. The oligo pool was synthesized by CustomArray (GenScript). Library sgRNAs were PCR amplified and cloned into the pKLV-U6gRNA(BbsI)-PGKpuro2A BFP lentiviral sgRNA expression vector, which encodes puromycin and BFP selection markers. The ligated product was electroporated into electrocompetent cells (Lucigen) and grown in liquid culture overnight at 37°C before being extracted by Maxiprep (Macherey-Nagel). Low skewing of the library was confirmed by sequencing of the plasmid pool. For the CRISPR screens in mouse AML cells, we used a custom epigenetic library of ~ 14,000 sgRNAs (6 sgRNAs per gene) obtained as a gift from Johannes Zuber.
CRISPR screens
MOLM13, OCI-AML3 and MLL-AF9 cells were transduced with a lentiviral vector encoding Cas9 and selected with blasticidin. Cas9 cells (300 x 106 per cell line for whole-genome screens, 100 x 106 per cell line for targeted screens) were infected with the respective pooled lentiviral sgRNA libraries at a multiplicity of infection of 0.3. The percentage of cells infected was determined by flow cytometry-based evaluation of mCherry+ or BFP+ (sgRNA-expressing) cells 72 hours following library transduction. Infected cells were selected with 2 μg/mL puromycin for 72 hours, commencing 48 hours after transduction. Cells were pulsed with 25 ng/mL human IFN-γ or 10 ng/mL mouse IFN-γ for 48 hours prior to FACS sorting and enriched for MHC-II+ cells by 3 rounds of sorting at approximately days 12, 21 and 28 following transduction with the sgRNA libraries. Cells were sorted in parallel, stained with AF647-conjugated anti-human HLA-DR,DP,DQ antibody (Tü39, BioLegend) or AF647-conjugated anti-mouse I-A/I-E antibody (M5/114.15.2, BioLegend), incubated on ice for 20 minutes and washed with PBS plus 2% FCS prior to sorting for MHC-II+ and mCherry+ or BFP+ cells on the BD FACSAria Fusion cell sorter.
Genomic DNA was extracted (Puregene Core Kit A, Qiagen) from both the sorted cells and an unselected pool of mutagenized cells grown for the same amount of time. For the whole-genome screens and the mouse targeted screens, sgRNA sequences were amplified by two rounds of PCR, with the second round primers containing adaptors for Illumina sequencing. For the human targeted screens, one-step PCR was performed, containing adaptors for Illumina sequencing. Primers used for PCR amplification are listed in Table S2. Samples were sequenced with single-end 75 bp reads on an Illumina NextSeq500. The sequence reads were trimmed to remove the constant portion of the sgRNA sequences with fastx clipper (http://hannonlab.cshl.edu/fastx_toolkit/) or cutadapt v1.14 (Martin, 2011), then mapped to the reference sgRNA library with bowtie2 (Langmead and Salzberg, 2012). After filtering to remove multi-aligning reads, the read counts were computed for each sgRNA. The RSA algorithm (König et al., 2007) was used to rank the genes for which targeting sgRNAs were significantly enriched in the sorted populations compared to the control unsorted populations grown in parallel.
CRISPR/Cas9-mediated gene disruption
Single guide RNA (sgRNA) oligonucleotides (Integrated DNA Technologies) were cloned into lentiviral expression vectors pKLV-U6gRNA(BbsI)-PGKpuro2ABFP (Addgene #50946, a gift from Kosuke Yusa) (Koike-Yusa et al., 2014) or lentiGuide-Crimson (Addgene #70683, a gift from Daniel Bauer) (Canver et al., 2015) as described. Oligonucleotide sequences are listed in Table S2, with sources as indicated (Morgens et al., 2017; Wang et al., 2014). For CRISPR/Cas9-mediated gene disruption, cells were first transduced with the Cas9 expression vector pHRSIN-PSFFV-Cas9-PPGK-Blasticidin (Burr et al., 2017) or FUCas9Cherry (a gift from Marco Herold, Addgene #70182) (Aubrey et al., 2015), and selected with blasticidin or sorted for mCherry expression respectively. To generate polyclonal populations with targeted gene disruption, cells were subsequently transduced with pKLV-gRNA-PGKpuro2ABFP or lentiGuide-Crimson encoding either gene-specific sgRNAs or with a control sgRNA targeting a ‘safe’ genomic location with no annotated function (Morgens et al., 2017). Cells infected with pKLV-gRNA-PGKpuro2△BFP were selected with 2 μg/mL puromycin for 72 hours, commencing 48 hours after transduction, while cells infected with lentiGuide-Crimson were sorted for Crimson+ cells 5-7 days after transduction. Where possible, efficient functional CRISPR/Cas9-mediated disruption of target genes was confirmed by immunoblot. In cases where immunoblot was unsuccessful due to lack of working antibodies, Sanger sequencing of the expected DNA break site was performed, with insertion-deletion (indel) frequency estimated by online ICE analysis (https://ice.synthego.com).
Virus production and transduction
Lentivirus was produced by triple transfection of HEK-293ET cells with a lentiviral transfer vector and the packaging plasmids psPAX2 and pMD.G at a 0.5:0.35:0.15 ratio. Retrovirus was produced by triple transfection of HEK-293ET cells with a retroviral transfer vector, structural pMD1-gag-pol plasmid and pMD.G envelope plasmid at a 0.75:0.22:0.03 ratio. All transfections were performed using polyethylenimine (PEI). Viral supernatants were collected 48 hours following transfection, filtered through a 0.45 μm filter and added to target cells.
Flow cytometry
Cells were washed in PBS prior to Fc blocking with 5 μg human IgG (for human cells, Sigma-Aldrich) or 0.5 μg anti-mouse CD16/32 (for MLL-AF9 cells; clone 93, BioLegend) for 10 minutes at room temperature. Cells were then stained on ice for 20 minutes in PBS with 2% FCS. After washing in PBS/2% FCS, samples were resuspended in PBS with 2% FCS. Data were acquired on a BD LSRFortessa or BD LSR II and analyzed in FlowJo.
Immunoprecipitation
For immunoprecipitation, cells were lysed in 1% NP-40 in TBS pH 7.6 with Roche cOmplete EDTA-free protease inhibitor, 0.5 mM PMSF and 10 mM iodoacetamide (IAA) for 30 minutes on ice. Samples were incubated (rotating) with primary antibody and pre-washed Protein A Dynabeads (Thermo Fisher Scientific) overnight at 4°C. Samples were eluted in LDS sample buffer/DTT for 10 minutes at 70°C, then separated by SDS-PAGE and immunoblotted as described below. For ubiquitinated species, cells were incubated with 50 μM MG-132 for 4 hours. Cells were lysed in 1% NP-40 in 50 mM Tris-HCL with Roche cOmplete EDTA-free protease inhibitor, 0.5 mM PMSF, 10 mM IAA, and 20 μM DUB inhibitor (PR-619) for 10 minutes at room temperature. Incubation with primary antibody and Protein A Dynabeads, elution and immunoblotting procedures were performed as previously described.
Immunoblotting
Cells were lysed in 1% SDS in 100 mM Tris-HCl pH 8.0 with Roche cOmplete EDTA-free protease inhibitor at room temperature. DNA was fragmented by adding 1:100 Benzonase (Sigma). Lysates were heated to 70°C in SDS sample buffer with 50 mM DTT for 10 minutes, separated by SDS-PAGE and transferred to PVDF membrane (Millipore). Membranes were blocked in Intercept TBS blocking buffer (LI-COR), probed with the indicated antibodies and reactive bands visualized using the LI-COR Odyssey imaging system. For some experiments, membranes were blocked with 5% skim milk in TBS-T and visualized using iBright™ 1500 imaging system (Thermo Fisher Scientific). Prior to visualizing ubiquitinated proteins (VU-1), membranes were washed with PBS for 2 minutes x 3. Pre-treatment of membranes was performed with 0.5% glutaraldehyde/PBS pH 7.0 for 20 minutes at room temperature. Membranes were washed with PBS x 3. Standard blocking procedures were done using 5% skim milk in TBS-T.
Imaging flow cytometry
Cells were fixed and permeabilized with eBioscience Transcription Factor Staining Buffer Set (Thermo Fisher Scientific) according to manufacturer’s instructions. Cells were blocked using 2.5% BSA in PBS for 30 minutes at room temperature. Antibodies for intracellular staining were diluted in permeabilization wash buffer supplemented with 0.1% BSA. Cells were incubated with intracellular antibody cocktail at 4°C overnight. The next day, cells were stained with appropriate fluorochrome-conjugated secondary antibodies for 1 hour at room temperature. Cell nuclei were stained with DAPI (Sigma-Aldrich) at 20 ng/mL for 10 minutes at room temperature. Cells were washed with PBS and resuspended in PBS.
Data was acquired on an Amnis ImageStream X Mark II using 60X objective. At least 5000 images were collected for each sample. Post-acquisition spectral compensation and downstream analysis were performed using IDEAS® image analysis software (Amnis Corporation). To determine single cell populations and to separate from debris, doublets and aggregates, an initial gating strategy using Brightfield Area vs. Brightfield Aspect Ratio was performed. In-focus cells were selected using the Brightfield Gradient Root Mean Square (RMS) feature, which enables selection of high-quality images with RMS values > 50. The degree of shared localization and nuclear localization was calculated using the similarity feature, which is based on log-transformed Pearson’s correlation coefficient. This measures the degree to which the masked areas associated to the images of the markers of interest are linearly correlated. High similarity means that staining intensity of both images are high in the same pixels. Cells with a similarity score of ≥3 were classified as co-localized, as previously described (Beum et al., 2006; Erie et al., 2011; George et al., 2006). Expression analysis was calculated using Median Pixel Intensity (MPI). Violin plots for summary graphs were created using RStudio software (RStudio Team, 2022).
Isolation and in vitro activation of OT-I CD8+ T cells and OT-II CD4+ T cells
OT-I TCR transgenic mice were bred in-house and obtained from Joseph Trapani (Peter MacCallum Cancer Centre), while OT-II TCR transgenic mice were purchased from the Walter and Eliza Hall Institute (Melbourne, Australia). For expansion of CD8+ OT-I T cells, splenocytes were harvested from the spleen of OT-I mice and stimulated by incubation with 20 ng/mL of SIINFEKL (OVA peptide, Sigma-Aldrich) for 72 hours in complete RPMI-1640 media supplemented with IL-2 at 100 U/mL (NIH). For expansion of CD4+ OT-II T cells, splenocytes were harvested from the spleen of OT-II mice and stimulated by incubation with 300 nM OVA323-339 peptide (GenScript) for 72 hours in complete RPMI-1640 media supplemented with IL-2 at 100 U/mL (NIH). After washing to remove the peptide, cells were re-cultured in media supplemented with IL-2 for an additional 3 days. Successful enrichment of CD4+ and CD8+ T cell populations was confirmed by flow cytometry staining for CD3 (17A2, BioLegend), CD4 (GK1.5, BD Biosciences) and CD8 (53-6.7, BioLegend).
Isolation and in vitro activation of human peripheral blood mononuclear cells (PBMCs)
Human PBMCs were isolated from donor buffy coat preparations from the Australian Red Cross Lifeblood by Ficoll-Paque density gradient centrifugation and stimulated for 72 hours in complete RPMI-1640 media with 600 U/mL IL-2 (NIH), 30 ng/mL anti-CD3 (OKT3, eBioscience) and 0.5 μg/mL anti-CD28 (CD28.2, eBioscience). After washing, cells were re-cultured in media supplemented with IL-2 for 1-3 days before use in co-culture experiments. Successful enrichment of T cell populations was confirmed by flow cytometry staining for CD3 (UCHT1, BioLegend), CD4 (OKT4, BioLegend) and CD8 (SK1, BD Biosciences). CD4+ T cells were selected using the EasySep Human CD4+ T Cell Isolation Kit (STEMCELL Technologies).
T cell cytotoxicity assays
2 x 104 MLL-AF9 or MV4-11 cells expressing CIITA cDNA, OVA peptide and/or RREB1 sgRNA or corresponding vector controls (as indicated in figure legends) were plated in 96-well flat clear bottom black plates (Corning) pre-coated with 5 μg/mL fibronectin (Merck). For MLL-AF9 co-cultures, CD4+ OT-II T cells and/or CD8+ OT-I T cells were added at the indicated effector:target (E:T) ratios, with each treatment set up in triplicate wells. For MV4-11 co-cultures, human PBMCs were added at the indicated E:T ratios, with each treatment set up in triplicate wells. Plates were incubated in the Incucyte SX5 Live Cells Analysis System at 37°C for 72 hours and set to acquire images measuring the fluorescent cell count (green and/or red) every 6 hours. Proliferation or depletion of tumor cells in each treatment was calculated by fold change of the fluorescent cell count at each timepoint, normalized to baseline (T0).
Measurement of T cell cytokine secretion
1 x 105 MLL-AF9 cells, expressing CIITA cDNA and/or OVA peptide or corresponding vector controls (as indicated in figure legends), were co-cultured with 2 x 105 OT-II T cells in 96-well plates. Co-cultures were performed in triplicate for each condition. The cell culture media was collected after 24 hours of co-culture. Levels of T cell cytokines were measured using the BD Cytometric Bead Array Mouse IFN-γ and TNF Flex Sets, with data acquired on a BD FACSVerse and analyzed using FCAP Array software.
RT-qPCR
mRNA was prepared with a Qiagen RNeasy kit and cDNA synthesis was performed with a SuperScript VILO kit (Life Technologies), per the manufacturers’ instructions. Quantitative PCR analysis was performed on an Applied Biosystems StepOnePlus System or Roche LightCycler 480 Real-Time PCR System with SYBR Green reagents. All samples were assayed in triplicate. Relative expression levels were determined with the ΔCtmethod and normalized to GAPDH. RT-qPCR primers are listed in Table S2, with sources as indicated (Burr et al., 2019; Chang and Flavell, 1995; Chang et al., 1994; Ulbricht et al., 2012).
RNA-sequencing
RNA was extracted using the Qiagen RNeasy kit. RNA concentration was quantified using a Qubit Fluorometer (Thermo Fisher Scientific). Libraries were prepared using QuantSeq 3’ mRNA-seq Library Prep kit (Lexogen). Libraries were sequenced on the NextSeq500 using 75 bp single end chemistry.
RNA-sequencing analysis
Bcl2fastq (Illumina) was used to perform sample demultiplexing and to convert BCL files generated from the sequencing instrument into Fastq files. Reads were aligned to the human genome (G1k V37) using HiSAT2 (Kim et al., 2019) and reads were assigned to genes using htseq-count (Anders et al., 2014). Differential expression was calculated using DESeq2 (Love et al., 2014). Genes with a false discovery rate corrected for multiple testing using the method of Benjamini and Hochberg below 0.05 and a log fold change greater than 1 were considered significantly differentially expressed. Scatterplots depicting log fold change differences between conditions were generated in R using the ggplot2 package and custom R code.
Single-cell RNA sequencing (scRNA-seq)
scRNA-seq of all primary patient AML samples was conducted on the 10X Chromium system using the 10X Genomics Chromium Next GEM 5’ Single Cell Gene Expression Solution (10X Genomics). Cryopreserved samples (frozen after treatment with A-366 or vehicle control) were rapidly thawed at 37°C and viable cells were bulk sorted into Eppendorf tubes using a BD FACSAria Fusion cell sorter and propidium iodide viability dye. The cells were then washed twice with PBS + 1% BSA. All patient samples were pooled together and loaded onto a single lane of the 10X Chromium Single Cell Chip using TotalSeq-C anti-human Hashtag antibodies (BioLegend). Up to 1 million viable primary AML cells were resuspended in 100 μL of Hashing staining buffer (PBS + 2% BSA + 0.01% Tween). 1 μL of the relevant hashtag antibody was then added to the sample and stained on ice for 30 minutes. Cells were then washed 2 times with Hashing staining buffer and the cell concentration was determined. Hashtag-labelled samples were then pooled together at equal cell concentrations, spun down and resuspended in Suspension buffer (PBS + 0.04% BSA + 0.4 U/μL RNase Inhibitor (Roche)) at a final cell concentration suitable for superloading onto the 10X Chromium Single Cell Chip (>1500 cells per μL). The final cell suspension was put through a cell strainer and a final cell count was conducted for accuracy, with ~40,000 cells superloaded onto the 10X Chromium Single Cell Chip. Details of the Hashtag sequences and corresponding samples are located in Table S2.
Single cells were captured in droplet emulsions using the 10X Chromium Single-Cell Instrument and reverse transcription, cDNA amplification and library preparation were performed based on the manufacturer’s protocol using the Chromium Single Cell 3’ Library & Gel Bead Kit v3 (10X Genomics). An additional primer that was specific for the oligo sequence conjugated to the Total-seq antibody was added during the reverse transcription stage. Following this, cDNA was amplified and the fraction containing the amplified Hashtags were separated from the full-length mRNA from each cell using SPRI beads (10X Genomics). These two fractions were then processed as separate cDNA libraries. Quality of final libraries was checked using the 2100 Bioanalyser (Agilent) and they were stored at -20°C until sequencing. Libraries were pooled at equimolar ratios and sequenced on the Illumina NovaSeq 6000 system (PE 150 bp reads, ~30-50,000 reads per cell). Hashtag libraries were sequenced on the Illumina NextSeq or the Illumina MiSeq (PE 75 bp reads, 5000 reads per cell).
scRNA-seq data analysis
Count matrices were generated from demultiplexed scRNA-seq fastq files using the 10X Genomics Cell Ranger v3.1.0 count pipeline against the hg38/GRCh38 genome (v3.0.0). scRNA-seq quality control was performed using Seurat v4 in R (Butler et al., 2018; Stuart et al., 2019). Low quality cells were removed by filtering out cells that had fewer than 500 genes or 1000 unique molecular identifiers (UMIs). Cells with greater than 15% mitochondrial RNA content were also removed. Multiplexed scRNA-seq samples containing TotalSeq-C hashtag oligo information were de-multiplexed using the CITE-seq-count v1.4.3 software and HTODemux methods within Seurat v4 using the default settings (Stoeckius et al., 2018).
Where available, 10X scRNA-seq droplets containing doublet cells were identified based on hashtag oligo combinations with the HTODemux function in Seurat and were removed. Normalization of scRNA-seq datasets was performed using the SCTransform method (Hafemeister and Satija, 2019) with percentage mitochondrial reads, S phase cell cycle score and G2M cell cycle phase score variables included in the regression model as sources of technical variation to remove except where indicated. Dimensional reduction, k-nearest neighbor graph construction and clustering were performed using Seurat v4 using default settings. The first 50 principal components were used to compute a non-linear dimensional reduction using the Uniform Manifold Approximation and Projection (UMAP) method (Becht et al., 2018). Marker gene identification and differential gene expression analysis for clusters and groups of cells was performed using a Wilcoxon Rank-Sum test. Only groups that were comprised of at least 20 cells were considered for marker gene identification and DGE analysis.
Chromatin immunoprecipitation (ChIP)
For ChIP analysis, 20 x 106 cells were cross-linked with 1% formaldehyde (methanol-free, Thermo Fisher Scientific) for 15 minutes at room temperature. Cross-linking was stopped by the addition of 0.125 M glycine and washed twice with ice cold PBS. After fixation, pellets were flash frozen and stored at -80°C or directly lysed in 1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8.0 and Roche cOmplete EDTA-free protease inhibitor. For normalization, Drosophila S2 cells cross-linked and lysed under the same conditions were mixed with the target cell lysates at a ratio of 1:4 (Drosophila S2 cell number:target cell number). Lysates were sonicated in a Covaris ultrasonicator to achieve a mean DNA fragment size of 500 bp. Samples were diluted 1:10 in modified RIPA buffer (1% Triton X-100, 0.1% deoxycholate, 90 mM NaCl, 10 mM Tris-HCl pH 8.0 and protease inhibitors) and incubated rotating with antibody and Protein A Dynabeads (Life Technologies), to bind the antibody and associated chromatin, for a minimum of 12 hours at 4°C. Antibody used: rabbit WIZ (catalog NBP1-80586, Novus Biologicals).
After washing twice with low salt buffer (0.1% SDS, 1% Triton X-100, 20 mM Tris-HCl pH8.0, 2 mM EDTA, 150 mM NaCl), once with high salt buffer (0.1% SDS, 1% Triton X-100, 20 mM Tris-HCl pH 8.0, 2 mM EDTA, 500 mM NaCl) and once with TE buffer (10 mM Tris pH 8.0, 1 mM EDTA), samples were eluted from the beads for 30 minutes at 65°C in elution buffer (1% SDS, 0.1 M NaHCO3). Eluates were reverse cross-linked overnight by heating at 65°C with RNase A and 0.2 M NaCl and then purified using the QIAquick PCR purification kit (Qiagen). Sequencing libraries were prepared from eluted DNA using the Rubicon ThruPLEX DNA-seq kit. Libraries were size-selected between 200-700 bps and sequenced on the NextSeq500 using 75 bp single-end chemistry.
CUT&Tag
CUT&Tag was performed as previously described (Kaya-Okur et al., 2019). MV4-11 cells (0.375 x 106 per condition) were mixed with Drosophila S2 cells (0.125 x 106 per condition) and then serially washed with PBS and wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine, 1 x protease inhibitor cocktail) at room temperature. Concanavalin A beads were activated by washing twice with binding buffer (20 mM HEPES pH 7.9, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2), after which 10 μL of activated beads were added to each sample and incubated for 10 minutes at room temperature. Bead-bound cells were then resuspended in 150 μL of antibody buffer (wash buffer plus 0.05% digitonin, 2 mM EDTA, 0.1% BSA) with a 1:50 dilution of primary antibody and incubated for 2 hours at room temperature. Antibodies used were rabbit anti-H3K9me2 (Cat# 39239, Active Motif) and rabbit anti-H3K27me3 (C36B11, Cell Signaling Technology).
The supernatant was then removed and the beads were washed twice with digitonin buffer (wash buffer plus 0.1% digitonin) prior to re-suspension in 50 μL of Dig-300 buffer (20 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM spermidine, 1 x protease inhibitor cocktail, 0.01% digitonin) with pAG-Tn5 enzyme for 1 hour at room temperature. The supernatant was again aspirated and the beads washed twice with Dig-300 buffer, followed by re-suspension in 50 μL Tagmentation buffer (Dig-300 buffer plus 10 mM MgCl2) and incubation for 1 hour at 37°C. An equal volume of 2X stop buffer (Dig-300 buffer plus 40 mM EDTA, 0.2% SDS, 400 μg/mL proteinase K) was then added and the samples were incubated at 55°C for 2 hours, then held at 4°C overnight.
The following day, the supernatant was taken and purified using the MinElute PCR purification kit (Qiagen), with each sample eluted in 25 μL of elution buffer. To amplify libraries, 21 μL of eluted DNA was mixed with 2 μL of a universal i5 primer and 2 μL of a unique barcoded i7 primer, as well as 25 μL of NEBNext HiFi 2X PCR Master Mix. Following 14 cycles of PCR amplification, cleanup was performed using 1.1 x volume of AMPure XP beads (Beckman Coulter), analyzed on the Agilent 4200 TapeStation to determine size distribution of libraries and then sequenced on the NextSeq500 using 75 bp paired end chemistry.
ChIP and CUT&Tag sequencing analysis
Bcl2fastq (Illumina) was used to perform sample demultiplexing and to convert BCL files generated from the sequencing instrument into Fastq files. Adapters were trimmed from reads by trimmomatic (Bolger et al., 2014), then aligned to the human genome (GRCh37) combined with fly genome (BDGP5) with bowtie2 (Langmead and Salzberg, 2012) for CUT&Tag and BWA-MEM (Li, 2013) for ChIP-seq. Samtools (Danecek et al., 2021) was used to convert, sort and index bam files, and PCR duplicates were removed with picard. Peak calling was performed with MACS2 (Zhang et al., 2008) with default parameters, and deepTools (Ramírez et al., 2014) was used for normalization. Genome-browser images of ChIP-seq data was generated by converting files to TDF files with igvtools and viewing in IGV (Robinson et al., 2011).
Quantification and Statistical Analysis
Statistical analysis was carried out using GraphPad Prism 9. Details of statistical analysis performed are provided in Methods Details of STAR Methods and in the figure legends. Data were reported as mean ± SD, SEM or independent replicates shown as individual data points, as indicated in the figure legends. Significance was defined as p < 0.05.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa Fluor 647 mouse anti-human HLA-DR, DP, DQ antibody | BioLegend | Cat# 361704, clone Tü39, RRID: AB 2563169 |
| Alexa Fluor 488 mouse anti-human HLA-DR antibody | BioLegend | Cat# 307620, clone L243, RRID: AB 493175 |
| Brilliant Violet 510 mouse anti-human HLA-DR antibody | BioLegend | Cat# 307646, clone L243, RRID: AB 2561948 |
| Alexa Fluor 488 mouse anti-human HLA-A,B,C | BioLegend | Cat# 311413, clone W6/32; RRID: AB 493133 |
| Alexa Fluor 647 rat anti-mouse I-A/I-E antibody | BioLegend | Cat# 107618, clone M5/114.15.2, RRID: AB 493525 |
| Alexa Fluor 647 mouse anti-mouse H-2Kb antibody | BioLegend | Cat# 116512, clone AF6-88.5, RRID: AB 492917 |
| Brilliant Violet 510 mouse anti-human CD3 antibody | BioLegend | Cat# 300448, clone UCHT1, RRID: AB 2563468 |
| Brilliant Violet 605 mouse anti-human CD4 antibody | BioLegend | Cat# 317438, clone OKT4, RRID: AB 11218995 |
| BUV737 mouse anti-human CD8 antibody | BD Biosciences | Cat# 612754, clone SK1, RRID: AB 2870085 |
| PE anti-human CD45RO antibody | BioLegend | Cat# 304206, clone UCHL1, RRID: AB 314422 |
| Pacific Blue anti-human CD62L antibody | BioLegend | Cat# 304826, clone DREG-56, RRID: AB 2186977 |
| PE/Cyanine7 rat anti-mouse CD3 antibody | BioLegend | Cat# 100220, clone 17A2, RRID: AB 1732057 |
| BV650 rat anti-mouse CD4 antibody | BD Biosciences | Cat# 563232, clone GK1.5, RRID: AB 2738083 |
| PE rat anti-mouse CD8a antibody | BioLegend | Cat# 100708, clone 53 6.7, RRID: AB 312747 |
| CD3 monoclonal antibody (OKT3), functional grade | eBioscience | Cat# 16-0037-81, RRID: AB 468854 |
| CD28 monoclonal antibody (CD28.2), functional grade | eBioscience | Cat# 16-0289-81, RRID: AB 468926 |
| Purified rat anti-mouse CD16/32 antibody | BioLegend | Cat# 101302, clone 93, RRID: AB 312801 |
| Monoclonal rabbit anti-H3K27me3 antibody | Cell Signaling Technology |
Cat# 9733S, clone C36B11; RRID: AB 2616029 |
| Monoclonal mouse anti-HSP60 antibody | Santa Cruz | Cat# sc-376240, clone C-10, RRID: AB 10986282 |
| Polyclonal rabbit anti-H3K9me2 antibody | Active Motif | Cat# 39239, RRID: AB 2793199 |
| Polyclonal rabbit anti-STAT1 antibody | Merck-Millipore | Cat# 06-501, RRID: AB 310145 |
| Monoclonal mouse anti-alpha tubulin antibody | Invitrogen | Cat# 62204, clone DM1A, RRID: AB 1965960 |
| Rabbit anti-CIITA antibody | Cell Signaling Technology |
Cat# 3793S, RRID: AB 2080109 |
| Monoclonal rabbit anti-GAPDH antibody | Cell Signaling Technology |
Cat# 2118S, clone 14C10, RRID: AB 561053 |
| Polyclonal rabbit anti-WIZ antibody | Novus Biologicals | Cat# NBP1-80586, RRID: AB 11011659 |
| Polyclonal rabbit anti-FBXO11 antibody | Novus Biologicals | Cat# NB100-59826, RRID: AB 892469 |
| Myc-Tag mouse monoclonal antibody | Cell Signaling Technology |
Cat# 2276S, clone 9B11, RRID: AB 331783 |
| HA-Tag rabbit monoclonal antibody | Cell Signaling Technology |
Cat# 3724S, clone C29F4, RRID: AB 1549585 |
| β-Actin mouse monoclonal antibody | Cell Signaling Technology |
Cat# 3700S, clone 8H10D10, RRID: AB 2242334 |
| Anti-rabbit IgG (H+L), F(ab’)2 Fragment (PE conjugate) | Cell Signaling Technology |
Cat# 79408S, RRID: AB 2799931 |
| Anti-mouse IgG (H+L), F(ab’)2 Fragment (Alexa Fluor® 647 Conjugate) | Cell Signaling Technology |
Cat# 4410S, RRID: AB 1904023 |
| Anti-ubiquitin HRP-conjugated mouse monoclonal, VU-1 antibody | LifeSensors | Cat# VU101H, RRID: AB 2716558 |
| Secondary goat anti-mouse human ads-HRP | SouthernBiotech | Cat# 1010-05, RRID: AB 2728714 |
| Secondary goat anti-rabbit IgG (H+L) mouse/human ads-HRP | SouthernBiotech | Cat# 4050-05, RRID: AB 2795955 |
| Rabbit IgG, polyclonal – isotype control (ChIP Grade) | Abcam | Cat# ab171870, RRID: AB 2687657 |
| TotalSeq-C0251 anti-human Hashtag 1 antibody | BioLegend | Cat# 394661, RRID: AB 2801031 |
| TotalSeq-C0252 anti-human Hashtag 2 antibody | BioLegend | Cat# 394663, RRID: AB 2801032 |
| TotalSeq-C0253 anti-human Hashtag 3 antibody | BioLegend | Cat# 394665, RRID: AB 2801033 |
| TotalSeq-C0254 anti-human Hashtag 4 antibody | BioLegend | Cat# 394667, RRID: AB 2801034 |
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| EPZ011989 (EZH2 inhibitor) | Selleck Chemicals | Cat# S8075, CAS No. 2095432-26-9 |
| A-366 (EHMT1/2 inhibitor) | Selleck Chemicals | Cat# S7572, CAS No. 1527503-11-2 |
| UNC0638 (EHMT1/2 inhibitor) | Selleck Chemicals | Cat# S8071, CAS No. 1255580-76-7 |
| Recombinant human interferon gamma | Sigma-Aldrich | Cat# SRP3058 |
| Recombinant mouse interferon gamma | Abcam | Cat# ab123747 |
| Recombinant human interleukin-2 | NIH | Cat# Ro 23-6019 |
| Recombinant mouse interleukin-3 | Peprotech | Cat# 213-13 |
| Recombinant human interleukin-6 | Peprotech | Cat# 200-06 |
| Recombinant murine SCF | Peprotech | Cat# 250-03 |
| SIINFEKL (OVA peptide 257-264) | Sigma-Aldrich | Cat# S7951, CAS No 138831-86-4 |
| OVA peptide (323-339) | GenScript | Cat# RP10610 |
| MG-132 | MedChemExpress | Cat#HY-13259 |
| Phenylmethylsulfonyl fluoride (PMSF) | Sigma-Aldrich | Cat#11359061001 |
| Iodoacetamide (IAA) | Sigma-Aldrich | Cat#I6125 |
| Sodium dodecyl sulfate (SDS) | Sigma-Aldrich | Cat#L3771 |
| PR-619 (DUB inhibitor) | Sigma-Aldrich | Cat#SML0430 |
| cOmplete, EDTA free, protease inhibitor | Roche | Cat#05056489001 |
| NP-40 Surface-Amps Detergent Solution | Thermo Fisher | Cat#85124 |
| Dynabeads Protein A | Thermo Fisher | Cat#10002D |
| Critical commercial assays | ||
| Cytometric Bead Array Mouse Th1/Th2 Cytokine Kit | BD Biosciences | Cat# 551287 |
| EasySep Human CD4+ T Cell Isolation Kit | STEMCELL Technologies | Cat# 17952 |
| eBioscience Transcription Factor Staining Buffer Set | Thermo Fisher Scientific | Cat# 00-5523-00 |
| Deposited data | ||
| ChIP-seq: MV4-11 | Göllner et al, 2017 | GEO: GSM1513828 |
| Sequencing data (CRISPR screen, RNA-seq, scRNA-seq, ChIP-seq, CUT&Tag) | This paper | GEO: GSE189773 |
| Mendeley data (unprocessed immunoblot images) | This paper | http://dx.doi.org/10.17632/j5gt4fv75f.1 |
| Experimental models: Cell lines | ||
| Human: MV4-11 | ATCC | Cat# CRL-9591, RRID: CVCL 0064 |
| Human: LM-MEL-53 | ATCC | Cat# CRL-3337, RRID: CVCL UC56 |
| Human: A375 | ATCC | Cat# CRL-1619, RRID: CVCL 0132 |
| Drosophila: S2 | ATCC | Cat# CRL-1963, RRID: CVCL Z232 |
| Human: MOLM13 | DSMZ | Cat# ACC 554, RRID: CVCL 2119 |
| Human: OCI-AML3 | DSMZ | Cat# ACC 582, RRID: CVCL 1844 |
| Human: WM852 | ESTDAB | Cat# ESTDAB-084, RRID: CVCL 6804 |
| Human: HEK293-ET | Felix Randow (MRC-LMB, Cambridge, UK) | RRID: CVCL 6996 |
| Experimental models: Organisms/strains | ||
| Mouse: B6.SJL-Ptprc<a>Pepc<b>/BoyJ | Animal Resources Centre, Western Australia |
|
| Mouse: C57BL/6-Tg(OT-I) | Joseph Trapani (Peter MacCallum Cancer Centre) | |
| Mouse: C57BL/6-Tg(OT-II) | Walter and Eliza Hall Institute | |
| Oligonucleotides | ||
| See Table S1 and Table S2. | ||
| Recombinant DNA | ||
| pHRSIN-PSFFV-Cas9-PPGK-Blasticidin | Burr et al, 2017 | |
| FUCas9Cherry | Aubrey et al, 2015 | Addgene:70182 |
| Bassik Human CRISPR Knockout Library | Morgens et al, 2017 | Addgene:101296-101934 |
| Human targeted epigenetic CRISPR library | This paper | |
| Mouse targeted epigenetic CRISPR library | Johannes Zuber (IMP, Vienna, Austria) | |
| pKLV-U6gRNA(BbsI)-PGKPuro2ABFP | Koike-Yusa et al, 2014 | Addgene:50946 |
| lentiGuide-Crimson | Canver et al, 2015 | Addgene: 70683 |
| pcDNA3 myc CIITA | Kanazawa et al, 2000 | Addgene: 14650 |
| pMSCV-CIITA-IRES-mCherry | This paper | |
| pMSCV-CIITA-IRES-GFP | This paper | |
| FLAG-HA-Fbxo11 -pcDNA3.1 - | Horn et al, 2014 | Addgene: 52501 |
| FLAG-HA-Fbxo11 -ΔFbox-pcDNA3.1 - | Horn et al, 2014 | Addgene: 52502 |
| pHRSIN-PSFFV-Fbxo11-PPGK-Puro | This paper | |
| pHRSIN-PSFFV-Fbxo11-ΔFbox-PPGK-Puro | This paper | |
| pcDNA3-OVA | Diebold et al, 2001 | Addgene: 64599 |
| pMSCV-OVA-IRES-GFP | This paper | |
| Software and algorithms | ||
| Fastx Clipper | Hannon Laboratory | http://hannonlab.cshl.edu/fastxtoolkit/ |
| Cutadapt | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/ |
| Trimmomatic | Bolger et al, 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Redundant siRNA activity (RSA) Algorithm | König et al, 2007 | https://admin-ext.gnf.org/publications/RSA/ |
| Bcl2fastq | Illumina | https://support.illumina.com/downloads/bcl2fastq-conversion-software-v2-20.html |
| HiSAT2 | Kim et al, 2019 | http://www.ccb.jhu.edu/software/hisat/index.shtml |
| Htseq-count | Anders et al, 2014 | https://htseq.readthedocs.io/en/master/ |
| DESeq2 | Love et al, 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Picard | Broad Institute | https://broadinstitute.github.io/picard/ |
| MACS2 | Zhang et al, 2008 | https://github.com/taoliu/MACS |
| deepTools | Ramírez et al, 2014 | https://deeptools.readthedocs.io/en/develop/ |
| Samtools | Danecek et al, 2021 | http://www.htslib.org/ |
| igvtools | Broad Institute | https://software.broadinstitute.org/software/igv/igvtools |
| IGV | Robinson et al, 2011 | https://software.broadinstitute.org/software/igv/ |
| Seurat | Butler et al, 2018; Stuart et al, 2019 | https://satijalab.org/seurat/ |
| IDEAS | Amnis Corporation | https://www.luminexcorp.com/flowsight-imaging/#software |
| R Studio | RStudio Team (2022) | http://www.rstudio.com/ |
| Other | ||
Supplementary Material
Acknowledgments
We thank the Flow Cytometry facility and Molecular Genomics Core at the Peter MacCallum Cancer Centre, as well as S. Jackson, J. Schreuders, R. Walker, K. Warren, B. Blyth, H. Beetham and K. Simpson for their technical contributions to this project. We thank the following funders for fellowship, scholarship and grant support: NHMRC Postgraduate Scholarship and HSANZ New Investigator Scholarship (K.L.C); Snow Medical Research Foundation Fellowship, NHMRC Investigator Grant #1196598 and Cancer Research UK Clinician Scientist Fellowship C53779/A20097 (M.L.B); Cancer Council Victoria Sir Edward Dunlop Research Fellowship, NHMRC Investigator Grant #1196749 and Howard Hughes Medical Institute International Research Scholarship #55008729 (M.A.D); Peter MacCallum Postgraduate Scholarship (C.E.S); VCA Early Career Research Fellowship (I.G.H); VCA Mid-Career Research Fellowship (E.Y.N.L, P.A.B); US Cancer Research Institute Irvington Postdoctoral Fellowship (J.L); Nuovo-Soldati Foundation for Cancer Research and Fondation de France (S.G); NHMRC Investigator Grant #1176175 (F.C.B); Leukemia and Lymphoma Society, US, Specialized Center of Research grant 7015-18 and Medical Research Future Fund Fellowship #1141460 (A.H.W); CSL Centenary Fellowship and NHMRC Investigator Grant #1196755 (S-J.D); and NHMRC Grants #1164054 / #2010275 (M.L.B), #1085015 / #1106444 (M.A.D) and #1128984 (M.A.D, S-J.D). Schematics of experimental workflows were created with BioRender.com.
Footnotes
Author contributions
K.L.C, M.L.B and M.A.D designed the research, analyzed and interpreted the data and wrote the manuscript with helpful input from all of the authors. M.L.B and M.A.D supervised the research. K.L.C, J.G, C.C, N.K, C.E.S, J.A.K, D.M.O and E.N.W conducted experiments and analyzed data. K.L.C and S.G performed the CRISPR screens. E.Y.N.L led the analysis of the genomic data and CRISPR screens, with contribution from M.M.Y, Y-C.C, D.V and A.G. F.C.B, A.M, K.A.F, J.L, I.G.H, L.M, C-S.A, S-J.D, P.A.B and A.H.W provided critical expertise and/or reagents and contributed to manuscript preparation.
Declaration of interests
M.A.D has been a member of advisory boards for GlaxoSmithKline, CTx CRC, Storm Therapeutics, Celgene and Cambridge Epigenetix. S-J.D has been a member of advisory boards for AstraZeneca. The M.A.D and S-J.D laboratories have received research funding from CTx CRC and Pfizer. The S-J.D laboratory has received research funding from Genentech. P.A.B has received research funding from AstraZeneca, Bristol Myers Squibb and Gilead Sciences. A.H.W has been a member of advisory boards for Novartis, Janssen, Amgen, Roche, Pfizer, Abbvie, Servier, Gilead, BMS, Macrogenics and Agios; receives research funding to the Institution from Novartis, Abbvie, Servier, BMS, Astra Zeneca and Amgen; serves on speaker’s bureaus for Abbvie, Novartis and BMS. All other authors declare no competing interests.
Data and code availability
Unprocessed data (immunoblot images) are available at http://dx.doi.org/10.17632/j5gt4fv75f.1. Sequencing data has been deposited into the sequence read archive, hosted by the National Center for Biotechnology Information. The accession number for the sequencing data reported in this paper is NCBI sequence read archive: GSE189773. No custom programs were developed specifically for this manuscript.
References
- Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubrey BJ, Kelly Gemma L, Kueh Andrew J, Brennan Margs S, O’Connor L, Milla L, Wilcox S, Tai L, Strasser A, Herold Marco J. An Inducible Lentiviral Guide RNA Platform Enables the Identification of Tumor-Essential Genes and Tumor-Promoting Mutations In Vivo. Cell Rep. 2015;10:1422–1432. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Axelrod ML, Cook RS, Johnson DB, Balko JM. Biological Consequences of MHC-II Expression by Tumor Cells in Cancer. Clin Cancer Res. 2019;25:2392–2402. doi: 10.1158/1078-0432.CCR-18-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, Newell EW. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018 doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- Bejanyan N, Weisdorf DJ, Logan BR, Wang H-L, Devine SM, de Lima M, Bunjes DW, Zhang M-J. Survival of Patients with Acute Myeloid Leukemia Relapsing after Allogeneic Hematopoietic Cell Transplantation: A Center for International Blood and Marrow Transplant Research Study. Biol Blood Marrow Transplant. 2015;21:454–459. doi: 10.1016/j.bbmt.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beum PV, Lindorfer MA, Hall BE, George TC, Frost K, Morrissey PJ, Taylor RP. Quantitative analysis of protein co-localization on B cells opsonized with rituximab and complement using the ImageStream multispectral imaging flow cytometer. Journal of Immunological Methods. 2006;317:90–99. doi: 10.1016/j.jim.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr ML, Sparbier CE, Chan KL, Chan Y-C, Kersbergen A, Lam EYN, Azidis-Yates E, Vassiliadis D, Bell CC, Gilan O, et al. An Evolutionarily Conserved Function of Polycomb Silences the MHC Class I Antigen Presentation Pathway and Enables Immune Evasion in Cancer. Cancer Cell. 2019;36:385–401. doi: 10.1016/j.ccell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, Lam EYN, Henderson MA, Bell CC, Stolzenburg S, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–105. doi: 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ, Jr, Laird PW, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Flavell RA. Class II transactivator regulates the expression of multiple genes involved in antigen presentation. J Exp Med. 1995;181:765–767. doi: 10.1084/jem.181.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Fontes JD, Peterlin M, Flavell RA. Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J Exp Med. 1994;180:1367–1374. doi: 10.1084/jem.180.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin DD. Overview of the immune response. J Allergy Clin Immunol. 2010;125:S3–S23. doi: 10.1016/j.jaci.2009.12.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli KB, Strissel Pamela L, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote Neal S, Cope Leslie M, Snyder A, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol. 2007;39:1593–1607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Christopher MJ, Petti AA, Rettig MP, Miller CA, Chendamarai E, Duncavage EJ, Klco JM, Helton NM, O’Laughlin M, Fronick CC, et al. Immune Escape of Relapsed AML Cells after Allogeneic Transplantation. N Engl J Med. 2018;379:2330–2341. doi: 10.1056/NEJMoa1808777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copelan EA. Hematopoietic Stem-Cell Transplantation. N Engl J Med. 2006;354:1813–1826. doi: 10.1056/NEJMra052638. [DOI] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dersh D, Phelan JD, Gumina ME, Wang B, Arbuckle JH, Holly J, Kishton RJ, Markowitz TE, Seedhom MO, Fridlyand N, et al. Genome-wide Screens Identify Lineage- and Tumor-Specific Genes Modulating MHC-I- and MHC-II-Restricted Immunosurveillance of Human Lymphomas. Immunity. 2021;54:116–131. doi: 10.1016/j.immuni.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold SS, Cotten M, Koch N, Zenke M. MHC class II presentation of endogenously expressed antigens by transfected dendritic cells. Gene Ther. 2001;8:487–493. doi: 10.1038/sj.gt.3301433. [DOI] [PubMed] [Google Scholar]
- Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R, Pagano M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–93. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted Disruption of the Mouse Stat1 Gene Results in Compromised Innate Immunity to Viral Disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- Ennishi D, Takata K, Beguelin W, Duns G, Mottok A, Farinha P, Bashashati A, Saberi S, Boyle M, Meissner B, et al. Molecular and Genetic Characterization of MHC Deficiency Identifies EZH2 as Therapeutic Target for Enhancing Immune Recognition. Cancer Discov. 2019;9:546–563. doi: 10.1158/2159-8290.CD-18-1090. [DOI] [PubMed] [Google Scholar]
- Erie AJ, Samsel L, Takaku T, Desierto MJ, Keyvanfar K, McCoy JP, Young NS, Chen J. MHC class II upregulation and colocalization with Fas in experimental models of immune-mediated bone marrow failure. Experimental Hematology. 2011;39:837–849. doi: 10.1016/j.exphem.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajollet S, Poras I, Carosella ED, Moreau P. RREB-1 Is a Transcriptional Repressor of HLA-G. J Immunol. 2009;183:6948–6959. doi: 10.4049/jimmunol.0902053. [DOI] [PubMed] [Google Scholar]
- Forero A, Li Y, Chen D, Grizzle WE, Updike KL, Merz ND, Downs-Kelly E, Burwell TC, Vaklavas C, Buchsbaum DJ, et al. Expression of the MHC Class II Pathway in Triple-Negative Breast Cancer Tumor Cells is Associated with a Good Prognosis and Infiltrating Lymphocytes. Cancer Immunol Res. 2016;4:390–399. doi: 10.1158/2326-6066.CIR-15-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambacorta V, Beretta S, Ciccimarra M, Zito L, Giannetti K, Andrisani A, Gnani D, Zanotti L, Oliveira G, Carrabba MG, et al. Integrated Multiomic Profiling Identifies the Epigenetic Regulator PRC2 as a Therapeutic Target to Counteract Leukemia Immune Escape and Relapse. Cancer Discovery. 2022;12:1449–1461. doi: 10.1158/2159-8290.CD-21-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George TC, Fanning SL, Fitzgeral-Bocarsly P, Medeiros RB, Highfill S, Shimizu Y, Hall BE, Frost K, Basiji D, Ortyn WE, et al. Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. Journal of Immunological Methods. 2006;311:117–129. doi: 10.1016/j.jim.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Gide TN, Quek C, Menzies AM, Tasker AT, Shang P, Holst J, Madore J, Lim SY, Velickovic R, Wongchenko M, et al. Distinct Immune Cell Populations Define Response to Anti-PD-1 Monotherapy and Anti-PD-1/Anti-CTLA-4 Combined Therapy. Cancer Cell. 2019;35:238–255.:e236. doi: 10.1016/j.ccell.2019.01.003. [DOI] [PubMed] [Google Scholar]
- Gill S, Olson JA, Negrin RS. Natural Killer Cells in Allogeneic Transplantation: Effect on Engraftment, Graft-versus-Tumor, and Graft-versus-Host Responses. Biol Blood Marrow Transplant. 2009;15:765–776. doi: 10.1016/j.bbmt.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göllner S, Oellerich T, Agrawal-Singh S, Schenk T, Klein H-U, Rohde C, Pabst C, Sauer T, Lerdrup M, Tavor S, et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med. 2017;23:69–78. doi: 10.1038/nm.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand JD, Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol Cell Biol. 2002;22:5296–5307. doi: 10.1128/MCB.22.15.5296-5307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn M, Geisen C, Cermak L, Becker B, Nakamura S, Klein C, Pagano M, Antebi A. DRE-1/FBXO11-Dependent Degradation of BLMP-1/BLIMP-1 Governs C. elegans Developmental Timing and Maturation. Dev Cell. 2014;28:697–710. doi: 10.1016/j.devcel.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz M, Gale R, Sondel P, Goldman J, Kersey J, Kolb H, Rimm A, Ringden O, Rozman C, Speck B. Graft-versus-leukemia reactions after bone marrow transplantation. Blood. 1990;75:555–562. [PubMed] [Google Scholar]
- Johnson AM, Bullock BL, Neuwelt AJ, Poczobutt JM, Kaspar RE, Li HY, Kwak JW, Hopp K, Weiser-Evans MCM, Heasley LE, et al. Cancer Cell–Intrinsic Expression of MHC Class II Regulates the Immune Microenvironment and Response to Anti–PD-1 Therapy in Lung Adenocarcinoma. J Immunol. 2020;204:2295–2307. doi: 10.4049/jimmunol.1900778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DB, Estrada MV, Salgado R, Sanchez V, Doxie DB, Opalenik SR, Vilgelm AE, Feld E, Johnson AS, Greenplate AR, et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat Commun. 2016;7:10582. doi: 10.1038/ncomms10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa S, Okamoto T, Peterlin BM. Tat Competes with CIITA for the Binding to P-TEFb and Blocks the Expression of MHC Class II Genes in HIV Infection. Immunity. 2000;12:61–70. doi: 10.1016/s1074-7613(00)80159-4. [DOI] [PubMed] [Google Scholar]
- Kantarjian H, Kadia T, DiNardo C, Daver N, Borthakur G, Jabbour E, Garcia-Manero G, Konopleva M, Ravandi F. Acute myeloid leukemia: current progress and future directions. Blood Cancer J. 2021;11:41. doi: 10.1038/s41408-021-00425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019;10:1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MDC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- König R, Chiang C-y, Tu BP, Yan SF, DeJesus PD, Romero A, Bergauer T, Orth A, Krueger U, Zhou Y, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat Methods. 2007;4:847–849. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013:1303.3997 [Google Scholar]
- Lim ABM, Curley C, Fong CY, Bilmon I, Beligaswatte A, Purtill D, Getta B, Johnston AM, Armytage T, Collins M, et al. Acute myeloid leukaemia relapsing after allogeneic haemopoietic stem cell transplantation: prognostic factors and impact of initial therapy of relapse. Intern Med J. 2018;48:276–285. doi: 10.1111/imj.13522. [DOI] [PubMed] [Google Scholar]
- Liu D, Schilling B, Liu D, Sucker A, Livingstone E, Jerby-Arnon L, Zimmer L, Gutzmer R, Satzger I, Loquai C, et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med. 2019;25:1916–1927. doi: 10.1038/s41591-019-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald WA, Chen Z, Gras S, Archbold JK, Tynan FE, Clements CS, Bharadwaj M, Kjer-Nielsen L, Saunders PM, Wilce MCJ, et al. T Cell Allorecognition via Molecular Mimicry. Immunity. 2009;31:897–908. doi: 10.1016/j.immuni.2009.09.025. [DOI] [PubMed] [Google Scholar]
- Marmont A, Horowitz M, Gale R, Sobocinski K, Ash R, van Bekkum D, Champlin R, Dicke K, Goldman J, Good R. T-cell depletion of HLA-identical transplants in leukemia. Blood. 1991;78:2120–2130. [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal. 2011;17:10–12. [Google Scholar]
- Meissner TB, Li A, Biswas A, Lee K-H, Liu Y-J, Bayir E, Iliopoulos D, van den Elsen PJ, Kobayashi KS. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proceedings of the National Academy of Sciences. 2010;107:13794–13799. doi: 10.1073/pnas.1008684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraz MA, White JM, Sheehan KCF, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, et al. Targeted Disruption of the Stat1 Gene in Mice Reveals Unexpected Physiologic Specificity in the JAK-STAT Signaling Pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- Meyer S, Handke D, Mueller A, Biehl K, Kreuz M, Bukur J, Koehl U, Lazaridou M-F, Berneburg M, Steven A, et al. Distinct Molecular Mechanisms of Altered HLA Class II Expression in Malignant Melanoma. Cancers (Basel) 2021;13:3907. doi: 10.3390/cancers13153907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielcarek M, Storer BE, Flowers MED, Storb R, Sandmaier BM, Martin PJ. Outcomes among Patients with Recurrent High-Risk Hematologic Malignancies after Allogeneic Hematopoietic Cell Transplantation. Biol Blood Marrow Transplant. 2007;13:1160–1168. doi: 10.1016/j.bbmt.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Fukuda T, Nakashima M, Henzan T, Kusakabe S, Kobayashi N, Sugita J, Mori T, Kurokawa M, Mori S-i. Donor Lymphocyte Infusion for Relapsed Hematological Malignancies after Unrelated Allogeneic Bone Marrow Transplantation Facilitated by the Japan Marrow Donor Program. Biol Blood Marrow Transplant. 2017;23:938–944. doi: 10.1016/j.bbmt.2017.02.012. [DOI] [PubMed] [Google Scholar]
- Morel KL, Sheahan AV, Burkhart DL, Baca SC, Boufaied N, Liu Y, Qiu X, Cañadas I, Roehle K, Heckler M, et al. EZH2 inhibition activates a dsRNA–STING–interferon stress axis that potentiates response to PD-1 checkpoint blockade in prostate cancer. Nature Cancer. 2021;2:444–456. doi: 10.1038/s43018-021-00185-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgens DW, Wainberg M, Boyle EA, Ursu O, Araya CL, Tsui CK, Haney MS, Hess GT, Han K, Jeng EE, et al. Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat Commun. 2017;8:15178. doi: 10.1038/ncomms15178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappano WN, Guo J, He Y, Ferguson D, Jagadeeswaran S, Osterling DJ, Gao W, Spence JK, Pliushchev M, Sweis RF, et al. The Histone Methyltransferase Inhibitor A-366 Uncovers a Role for G9a/GLP in the Epigenetics of Leukemia. PLoS One. 2015;10:e0131716. doi: 10.1371/journal.pone.0131716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IA, Hwang S-H, Song IH, Heo S-H, Kim Y-A, Bang WS, Park HS, Lee M, Gong G, Lee HJ. Expression of the MHC class II in triple-negative breast cancer is associated with tumorinfiltrating lymphocytes and interferon signaling. PLoS One. 2017;12:e0182786. doi: 10.1371/journal.pone.0182786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AHFM, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AAHA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. Partitioning and Plasticity of Repressive Histone Methylation States in Mammalian Chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimsza LM, Roberts RA, Miller TP, Unger JM, LeBlanc M, Braziel RM, Weisenberger DD, Chan WC, Muller-Hermelink HK, Jaffe ES, et al. Loss of MHC class II gene and protein expression in diffuse large B-cell lymphoma is related to decreased tumor immunosurveillance and poor patient survival regardless of other prognostic factors: a follow-up study from the Leukemia and Lymphoma Molecular Profiling Project. Blood. 2004;103:4251–4258. doi: 10.1182/blood-2003-07-2365. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer MGM, Redd RA, Cader FZ, Pak CJ, Abdelrahman S, Ouyang J, Sasse S, Younes A, Fanale M, Santoro A, et al. Major Histocompatibility Complex Class II and Programmed Death Ligand 1 Expression Predict Outcome After Programmed Death 1 Blockade in Classic Hodgkin Lymphoma. J Clin Oncol. 2018;36:942–950. doi: 10.1200/JCO.2017.77.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen Shu Y, Han H, Liang G, Jones Peter A, Pugh Trevor J, et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C, Labopin M, Nagler A, Bornhäuser M, Finke J, Fassas A, Volin L, Gürman G, Maertens J, Bordigoni P, et al. Donor Lymphocyte Infusion in the Treatment of First Hematological Relapse After Allogeneic Stem-Cell Transplantation in Adults With Acute Myeloid Leukemia: A Retrospective Risk Factors Analysis and Comparison With Other Strategies by the EBMT Acute Leukemia Working Party. J Clin Oncol. 2007;25:4938–4945. doi: 10.1200/JCO.2007.11.6053. [DOI] [PubMed] [Google Scholar]
- Schneider C, Kon N, Amadori L, Shen Q, Schwartz FH, Tischler B, Bossennec M, Dominguez-Sola D, Bhagat G, Gu W, et al. FBXO11 inactivation leads to abnormal germinal-center formation and lymphoproliferative disease. Blood. 2016;128:660–666. doi: 10.1182/blood-2015-11-684357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng W, LaFleur MW, Nguyen TH, Chen S, Chakravarthy A, Conway JR, Li Y, Chen H, Yang H, Hsu PH, et al. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell. 2018;174:549–563.:e519. doi: 10.1016/j.cell.2018.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soiffer RJ, LeRademacher J, Ho V, Kan F, Artz A, Champlin RE, Devine S, Isola L, Lazarus HM, Marks DI, et al. Impact of immune modulation with anti–T-cell antibodies on the outcome of reduced-intensity allogeneic hematopoietic stem cell transplantation for hematologic malignancies. Blood. 2011;117:6963–6970. doi: 10.1182/blood-2011-01-332007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M, Zheng S, Houck-Loomis B, Hao S, Yeung BZ, Mauck WM, Smibert P, Satija R. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 2018;19:224. doi: 10.1186/s13059-018-1603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.:e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffalori C, Zito L, Gambacorta V, Riba M, Oliveira G, Bucci G, Barcella M, Spinelli O, Greco R, Crucitti L, et al. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat Med. 2019;25:603–611. doi: 10.1038/s41591-019-0400-z. [DOI] [PubMed] [Google Scholar]
- Ulbricht T, Alzrigat M, Horch A, Reuter N, von Mikecz A, Steimle V, Schmitt E, Krämer OH, Stamminger T, Hemmerich P. PML promotes MHC class II gene expression by stabilizing the class II transactivator. J Cell Biol. 2012;199:49–63. doi: 10.1083/jcb.201112015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, Wigle TJ, DiMaggio PA, Wasney GA, Siarheyeva A, et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Unprocessed data (immunoblot images) are available at http://dx.doi.org/10.17632/j5gt4fv75f.1. Sequencing data has been deposited into the sequence read archive, hosted by the National Center for Biotechnology Information. The accession number for the sequencing data reported in this paper is NCBI sequence read archive: GSE189773. No custom programs were developed specifically for this manuscript.