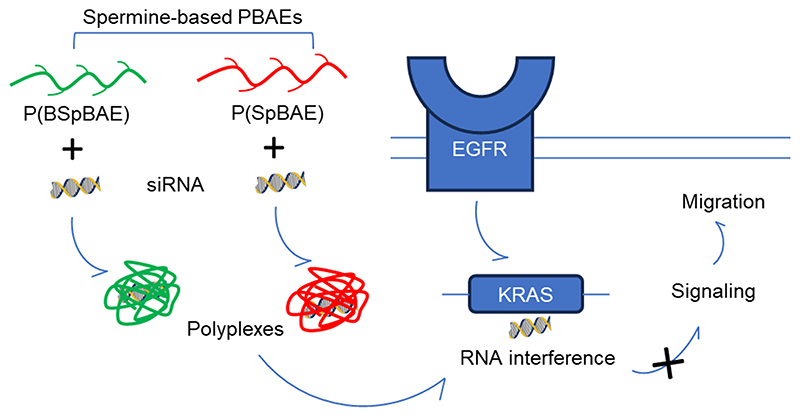
Abstract
Polyethylenimine (PEI) is a highly efficient cationic polymer for nucleic acid delivery, and although it is commonly used in preclinical studies, its clinical application is limited due to concerns regarding its cytotoxicity. Poly(β-amino ester)s are a new group of biodegradable and biocompatible cationic polymers which can be used for siRNA delivery. In this study, we synthesized Boc-protected and deprotected poly(β-amino ester)s, P(BSpBAE) and P(SpBAE) respectively, based on spermine and 1,4-butanediol diacrylate to deliver siRNA. The polymers were synthesized by Michael addition in a step-growth polymerization and characterized via 1H-NMR spectroscopy and size-exclusion chromatography (SEC). The polymers can encapsulate siRNA as determined by SYBR gold assays. Both polymers and polyplexes were biocompatible in vitro. Furthermore, cellular uptake of P(BSpBAE) and P(SpBAE)-polyplexes was higher than for branched PEI (25 kDa) polyplexes at same N/P ratios. P(BSpBAE) polyplexes achieved 60% eGFP knockdown in vitro, which indicates that the Boc-protection can improve siRNA delivery and gene silencing efficiency of PBAEs. P(BSpBAE) polyplexes and P(SpBAE) polyplexes showed different cellular uptake mechanisms, and P(BSpBAE) polyplexes showed lower endosomal entrapment, which could explain why P(BSpBAE) polyplexes more efficient mediated gene silencing than P(SpBAE) polyplexes.Furthermore, transfection of an siRNA against mutated KRAS in KRAS-mutated lung cancer cells led to around 35% P(BspBAE) to 45% P(SpBAE) inhibition of KRAS expression and around 33% P(SpBAE) to 55% P(BspBAE) decreased motility in a migration assay. These results suggest that the newly developed spermine-based poly(β-aminoester) sare promising materials for therapeutic siRNA delivery.
Keywords: poly(β-amino ester)s, spermine, siRNA delivery, PBAE, lung cancer, KRAS
1. Introduction
RNA interference (RNAi) is a promising strategy to silence gene expression by microRNA or small interfering RNA (siRNA)1. After the first discovery of RNAi by Fire and Mello in 1998, RNAi-based therapeutics are now progressing rapidly. siRNA is a double-stranded RNA of 21-25 nucleotides per strand, which can be used to mediate RNAi and downregulate overexpressed or pathologically expressed genes. 2 During the COVID-19 pandemic caused by the SARS-CoV-2 virus, potential siRNA sequences against the SARS-CoV-2 genome or accessory proteins or targeting host factors have been suggested provide alternative ways in the fight against SARS-CoV-2.3 However, the delivery of siRNA has remained a challenge. Due to its hydrophilicity, negative charges and high molecular weight (~13300 g/mol), siRNA duplexes cannot cross cell membrane passively, and thus their cellular uptake is limited. In addition, siRNA is rapidly degraded in blood and renally cleared.4
Numerous siRNA vectors have been developed in the past years, which can be mainly divided into viral vectors and non-viral vectors. Viral vectors can achieve high transduction but are associated with many safety problems such as immune responses and carcinogenesis.5 Non-viral vectors, such as polycationic polymers including polyethylenimine (PEI), have emerged as favorable candidates due to their good efficiency at a low commercial cost.6 However, low-molecular-weight PEI cannot deliver siRNA effectively, while high-molecular-weight (HMW) PEI is cytotoxic as a highly charged non-degradable macromolecule. HMW-PEI has been shown to induce cell necrosis and apoptosis in a variety of cell lines.7
Poly(β-amino ester)s abbreviated as PBAEs or PAEs refer to a polymer class synthesized from acrylatic compounds and amine-based monomers by Michael addition. The first representative of this class was synthesized based on linear poly(amido amines) by Ferruti et al. in the 1970s and named poly(β-amino ester) in 2000.8 Lynn et al. first used this polymer for DNA delivery, and since then, the interest in poly(β-amino ester) grew significantly.9 PBAEs are biocompatible, biodegradable and stimuli-responsive, which makes them very promising for siRNA delivery.10 PBAEs are biodegradable due to their hydrolysable ester bonds, which also decrease their cytotoxicity.11 The charge-reversible amine groups can be protonated at specific pH-values and interact with negatively charged cargos electrostatically to deliver them.12 In addition, PBAEs are compatible with various other polymers: for example, PEG, PCL, PLA, and other blocks can be conjugated to PBAEs to form block copolymers.13
Spermine is a naturally occurring tetraamine aiding in packaging cellular DNA into a compact state in eukaryotic cells14. However, spermine poorly condenses siRNA and shows a limited siRNA transfection efficiency because of its low molecular weight. Besides, the endosomal escape of spermine is also limited.15 Polymerization of spermine to increase its molecular weight has been shown to be beneficial for siRNA delivery.15, 16
In lung cancer, KRAS (Kirsten rat sarcoma viral oncogene homolog) is the most frequently mutated gene, and KRAS mutations are found in approximately 27% of all lung adenocarcinomas.17 The activating mutations in KRAS cause constitutive activation of the GTPase protein in the absence of growth factor signaling and result in a sustained proliferation signal within the cell which is related to the migration and invasion of cancer cells.18 The inhibition of mutant KRAS has been proven to be critical for the successful treatment of lung tumors.19, 20
In this study, we designed spermine-based poly(β-amino ester)s to deliver siRNA against mutated KRAS. We tested the encapsulation efficiency of the polymers, characterized the polyplexes, and evaluated the polyplexes in vitro using H1299 cells and eGFP expressing H1299/eGFP cells. The endosomal entrapment and cellular uptake pathway of the polyplexes were also investigated. To achieve a therapeutic relevant gene silencing, siRNA against mutated KRAS (siG12S) was delivered to KRAS mutated A549 lung adenocarcinoma cells using P(BSpBAE) and P(SpBAE) polyplexes, and therapeutic efficacy was evaluated by migration assays and western blot analysis.
2. Materials & Methods
2.1. Materials
Spermine (Fisher Scientific, Acros, USA), ethyl trifluoroacetate (Sigma Aldrich, Taufkirchen, Germany), 1,4-butanediol diacrylate (TCI, Japan), di-tert-butyl dicarbonate (Sigma Aldrich, Taufkirchen, Germany), chloroform D (Eurisotop, Germany), Deuterium oxide (Sigma Aldrich, Taufkirchen, Germany) were purchased from the suppliers indicated. Methanol, n-hexane, dichloromethane, isopropanol, dimethyl sulfoxide (DMSO), and dry ice are provided by Ludwig-Maximilians-University Munich.
Branched PEI (25 kDa), HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), thiazolyl blue tetrazolium bromide (MTT), chloroquine diphosphate, paraformaldehyde solution, 4’, 6-diamidino-2-phenylindole dihydrochloride (DAPI), FluorSave Reagent, RPMI-1640 medium, fetal bovine serum (FBS), Penicillin-Streptomycin solution, Dulbecco’s Phosphate Buffered Saline (PBS), trypsin-EDTA solution (0.05%), and Geneticin (G418) disulfate solution were purchased from Sigma-Aldrich (Taufkirchen, Germany). SYBR Gold Dye, Lipofectamine™ 2000, and Alexa Fluor 488 (AF488) were purchased from Life Technologies (Darmstadt, Germany).
Amine-modified eGFP siRNA (5’-pACCCUGAAGUUCAUCUGCACCACcg, 3’-ACUGGGACUUCAAGUAGACGGGUGGC) and scrambled siRNA (5’-pCGUUAAUCGCGUAUAAUACGCGUat,3’-CAGCAAUUAGCGCAUAUUAUGCGCAUAp) were purchased from Integrated DNA Technologies (Leuven, Belgium). “p” denotes a phosphate residue, lower case letters are 2’-deoxyribonucleotides, capital letters are ribonucleotides, and underlined capital letters are 2’-O-methylribonucleotides. siRNA against KRAS G12S (siG12S) was purchased from Eurofins Genomics (Ebersberg, Germany).
2.2. Synthesis and characterization of spermine-based poly(β-amino ester)s
2.2.1. Synthesis of tri-tert-Butyl Carbonyl Spermine (tri-Boc-spermine)
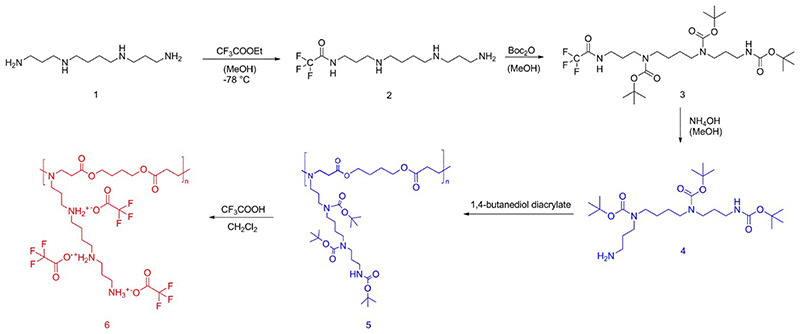
Tri-Boc-spermine was synthesized according to the literature with slight modifications.21 Spermine (1 eq.) was dissolved in methanol and stirred at -78°C, ethyl trifluoroacetate (1 eq.) was then added into the solution of spermine dropwise, kept stirring at -78°C for an hour and at 0°C for an hour to afford predominantly the mono-trifluoroacetate (2, Scheme 1). Without isolation, di-tert-butyl dicarbonate (4 eq.) in methanol was added dropwise into the solution and stirred at room temperature for 2 days to protect all other amines to afford the tri-Boc-protected mono-trifluoroacetate (3, Scheme 1). Finally, the pH of the solution was adjusted to be above 11 by 25% ammonia and stirred overnight to cleave the trifluoroacetamide protecting group.
Scheme 1. Synthesis route of spermine and 1,4-butanediol diacrylate based poly(β-amino ester)s.
The reaction mixture was concentrated in vacuum and the residue was diluted with dichloromethane (DCM) and then washed with water and saturated NaCl aqueous solution, before it was dried by MgSO4, filtered, and DCM was evaporated. After purification by column chromatography (CH2Cl2\MeOH\NH3, aq. 7:1:0.1, SiO2, KMnO4; Rf = 0.413), the desired product tri-Boc-spermine (4, Scheme 1) was isolated as a colorless oil. Yield: 37%.
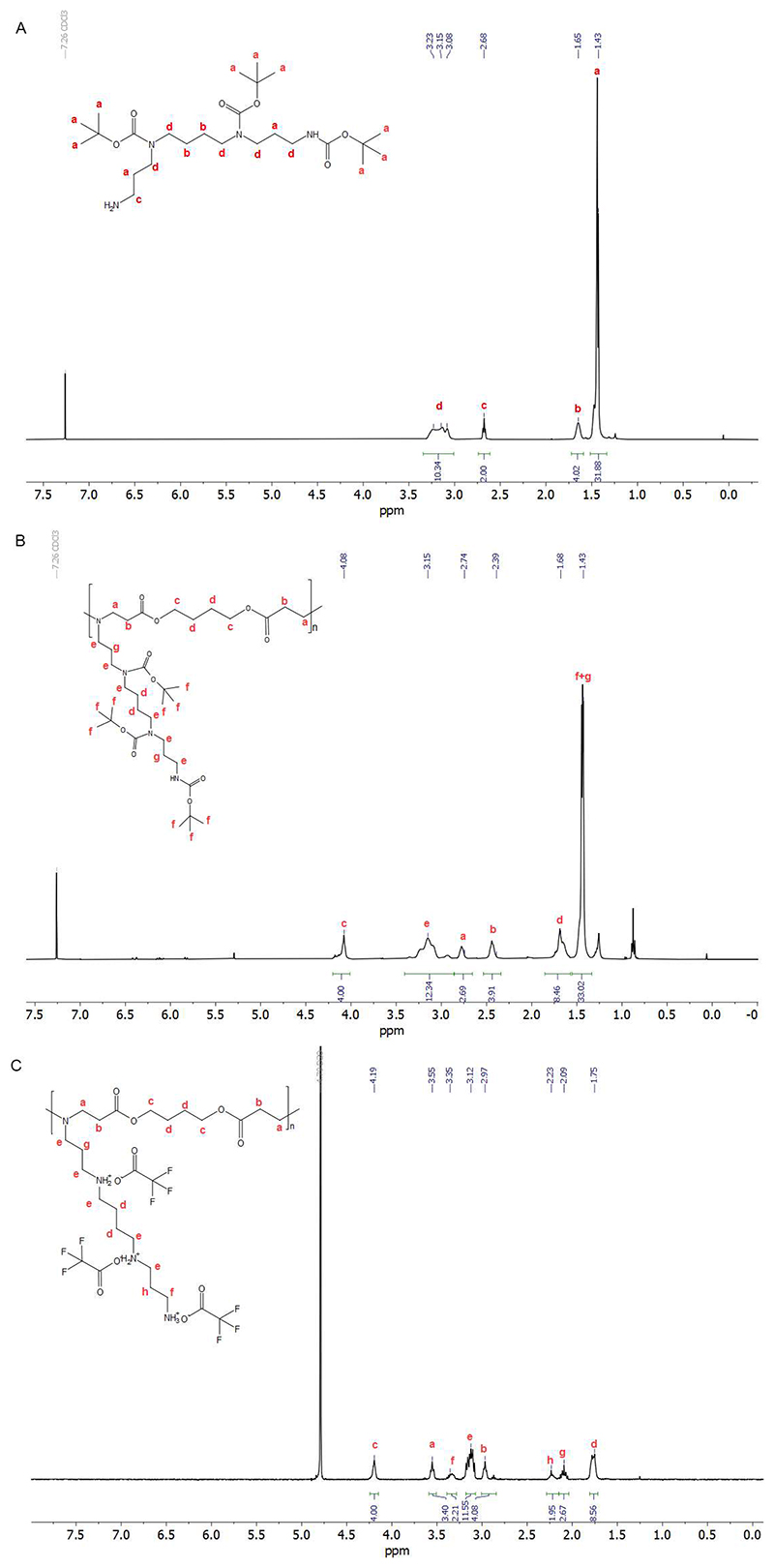
1H NMR, 400 MHz, CDCl3: 1.43–1.48 [m, 31H, 6-CH2, 7-CH2, O–C–(CH3)3×3, overlapping]; 1.65 (m, 6H, 2-CH2 11-CH2, NH2); 2.68 (t, 2H, 12-CH2); 3.08–3.23 (m, 10H, 1-CH2, 3-CH2, 5-CH2, 8-CH2, 10-CH2). ESI-MS: expected for (C25H50N4O6 + H)+: 503.3803, found: m/z = 503.3796.
2.2.2. Synthesis of spermine and 1,4-butanediol diacrylate-based poly(β-amino ester)s
Tri-Boc-spermine (4, Scheme 1) (1 eq.) was mixed with 1,4-butanediol diacrylate (1 eq.), and stirred at 120 °C overnight. The product was dissolved with DCM, precipitated in n-hexane, and dried in vacuum to afford polymeric tri-Boc-spermine- and 1,4-butanediol diacrylate-based poly(β-amino ester) abbreviated as P(BSpBAE) (5, Scheme 1), which was then characterized by 1H NMR spectroscopy and SEC (measurement relative to polystyrene and in chloroform at 30°C, 4 mg/mL). Yield: 83%.
1H NMR (400 MHz, CDCl3) δ 4.20 – 4.01 (m, 4H, 2CH2), 3.19 (d, 12H, 6CH2), 2.77 (s, 4H, 2CH2), 2.43 (d, 4H, 2CH2), 1.66 (d, 8H, 4CH2), 1.55 – 1.34 (m, 33H, 9CH3+4CH2).
P(BSpBAE) (5, Scheme 1) was then dissolved in a mixture of trifluoroacetic acid (TFA) and DCM (TFA: DCM 1:20 v/v) and stirred for 2 h at room temperature to deprotect amino functional groups. DCM and TFA were evaporated in vacuo, and the residue was dissolved in distilled water and lyophilized to obtain the final product: Spermine- and 1,4-butanediol diacrylate-based poly(β-amino ester) (6, Scheme 1), which will be termed as P(SpBAE) in short. Yield: 63%.
1H NMR (400 MHz, D2O) δ 4.19 (s, 4H, 2CH2), 3.55 (t, 4H, 2CH2), 3.36 (dd, 2H, CH2), 3.13 (dt, 10H, 5CH2), 2.96 (d, 4H, 2CH2), 2.23 (s, 2H, CH2), 2.09 (p, 2H, CH2), 1.77 (d, 8H, 4CH2).
2.3. Preparation and characterization of polyplexes
The polymer P(SpBAE) was dissolved in distilled water at a concentration of 1 mg/mL, and the polymer P(BSpBAE) was dissolved in acetone due to its limited solubility in water at a concentration of 10 mg/mL as stock solutions. To prepare polyplexes (the complexes of siRNA and polymer), various amounts of polymers calculated according to the respective N/P ratios were first diluted in 50 μL 10 mM HEPES buffer (pH 5.3). Subsequently, siRNA in 50 μL 10 mM HEPES buffer was mixed with polymer solution via pipetting. After incubation at room temperature for 30 min, the size and zeta potential of the polyplexes in 10 mM HEPES buffer were tested using a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK).
The amount of polymer was calculated according to the following equation:
where 52 is the number of nucleotides of the 25/27mer siRNA; N/P ratio is the molar ratio of the protonable polymer amino groups (N) and the siRNA phosphate groups (P); The protonable unit was calculated by dividing the molar mass of the repeating unit by the number of the protonable chemical groups in each repeating unit. Based on pKa calculations in MarvinSketch, two protonable amines/amides were considered for P(BSpBAE), while for P(SpBAE), 4 ionizable groups were counted.
2.4. siRNA encapsulation efficiency by SYBR gold assay
To evaluate the siRNA encapsulation capacity of P(BSpBAE) and P(SpBAE), SYBR Gold assays were carried out.22 SYBR Gold is a proprietary unsymmetrical cyanine dye which can bind with free RNA or DNA molecules and emits thousand-fold fluorescence upon excitation.23 In brief, polyplexes were prepared as described above at N/P ratios 0, 1, 3, 7, 10, 15, and 20. Subsequently, 100 μL of each polyplex solution was added to a black FluoroNunc 96-well pate (FisherScientific, Darmstadt, Germany). 4X SYBR Gold aqueous solution (30 μL per well) was added to each well and incubated for 10 min in the dark. The fluorescence intensity was determined using a fluorescence plate reader (Spark, TECAN, Männedorf, Switzerland, excitation: 485/20 nm, emission: 535/20 nm.). The fluorescence intensity of free siRNA (N/P = 0) was used as a control and set as 100% fluorescence.
2.5. In vitro compatibility assessment
The cytotoxicity of P(BSpBAE) and P(SpBAE) polymers and polyplexes was evaluated via MTT assays.24 In brief, H1299 cells were seeded in 96-well plates (5000 cells/well). After incubation in a CO2 incubator for 24 h, dilutions of P(BSpBAE) and P(SpBAE) in RMPI-1640 complete medium were added and incubated to reach a final concentration ranging from 1 to 100 μg/mL, and cells were grown in the CO2 incubator for another 24 h. Subsequently, the PBAEs solution was removed and MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide) in serum-free RPMI-1640 medium was added and incubated at 37 °C for 4 h. Afterwards, the MTT solution was replaced with DMSO and incubated at room temperature for 30 min. The optical density was finally determined at 570 nm and corrected with background measured at 680 nm using a microplate reader (TECAN, Männedorf, Switzerland). The cytotoxicity of polyplexes at N/P 10 was determined accordingly.
2.6. Quantification of cellular uptake
The cellular uptake of P(BSpBAE) and P(SpBAE) polyplexes was quantified by flow cytometry. The polyplexes were prepared as described above using Alexa Fluor 488-labeled siRNA (AF488-siRNA). PEI and Lipofectamine ™ 2000 were used as controls. H1299 cells were seeded in 24-well plates (50 000 cells/well). After incubation in the CO2 incubator (37°C, 5% CO2) for 24 h, polyplexes containing 50 pmol AF488-siRNA at N/P 10 in RPMI-1640 complete medium were added and incubated in the incubator for 24 h. Subsequently, the polyplexes solution was discarded, and the cells were rinsed with PBS and detached with 0.05% Trypsin-EDTA. The detached cells were then washed with PBS another 2 times and analyzed via flow cytometry (Attune NxT Acoustic Focusing Cytometer, Thermo Fisher Scientific, Darmstadt, Germany) with or without quenching by Trypan blue.
2.7. In vitro eGFP knockdown
To determine if the polyplexes can silence genes on the protein levels, silencing of the enhanced green fluorescent protein reporter gene (eGFP) was quantified by flow cytometry. H1299/eGFP cells were seeded in 24-well plates (25 000 cells in 500 μL medium/well). After growth in the CO2 incubator (37°C, 5% CO2) for 24 h, the cells were transfected with P(BSpBAE) polyplexes and P(SpBAE) polyplexes composed of scrambled siRNA (siNC, 50 pmol/well) or siRNA against GFP (siGFP, 50 pmol/well) with or without chloroquine (100 μM). Lipofectamine™ 2000 formulated with siNC and siGFP were used as respective control treatment. After 48h in the incubator, the cells were detached and washed with PBS for flow cytometry measurements (Attune NxT Acoustic Focusing Cytometer, Thermo Fisher Scientific, Darmstadt, Germany).
2.8. Endosomal entrapment
To visualize the endosomal entrapment behavior of the polyplexes, cells were imaged by confocal laser scanning microscopy (CLSM) after transfection with fluorescent siRNA. In brief, H1299 cells were seeded on 13-mm microscope cover glasses (VWR, Allison Park, PA, USA) in each well of a 24-well plate (50 000 cells/well). After growth for 24 h, the cells were further incubated with polyplexes formulated with 50 pmol of Alexa Fluor 488-labeled siRNA (AF488-siRNA) for 24 h. Subsequently, the polyplex containing medium was discarded and 75 nM Lysotracker Red™ DND 99 (Thermo Fisher Scientific, Darmstadt, Germany) solutions in full medium were added and incubated at 37°C for 1 h in the CO2 incubator. The cells were then washed with PBS twice and fixed with 4% paraformaldehyde (PFA) at room temperature for 15 min. After washing with PBS twice more, the cells were stained with DAPI (1 μg/mL) for 20 min. Finally, the cells were washed with PBS twice and mounted with FluorSave reagent (Sigma-Aldrich, Darmstadt, Germany). The fluorescence images were acquired using a laser scanning microscope (Leica SP8 inverted, software: LAS X, Leica microsystems GmbH, Wetzlar, Germany). A diode laser (405 nm), and argon laser (488 nm and 552 nm) were used for excitation. Emission in the blue channel for DAPI (627 nm – 750 nm), green channel for AF488 (750 nm – 755 nm), and red channel (755 nm – 760 nm) for lysotracker, were recorded respectively.
2.9. Cellular uptake pathway
To further understand the mechanism behind the different behavior of P(SpBAE) polyplexes and P(BSpBAE) polyplexes, we investigated the cellular uptake pathway of both types of polyplexes. KRAS mutated A549 cells were seeded in 24-well plates 24 h before use (50 000 cells/well). Various inhibitors of specific pathways, namely nystatin (10 μg/mL), wortmannin (12 ng/mL), chlorpromazine (10 μg/mL) and methyl-β-cyclodextrin (3 mg/mL) were added, and cells were incubated in the CO2 incubator at 37°C for 1 h. Polyplexes were subsequently added (50 pmol AF488-siRNA/well, N/P 10) and incubated for 24 h. After incubation, the polyplex-containing medium was discarded, and the cells were rinsed with PBS and detached with 0.05% Trypsin-EDTA. The detached cells were then washed with PBS, quenched with Trypan blue, and analyzed via flow cytometry (Attune NxT Acoustic Focusing Cytometer, Thermo Fisher Scientific, Darmstadt, Germany).
2.10. Migration assay
To determine therapeutically relevant gene silencing efficiency of the PBAEs, a migration assay was carried out to evaluate the potential of the polyplexes to treat lung cancer.19, 25 For this assay, Boyden chambers (VWR, 8 μm pore size) placed in 24 well plates were used. In brief, cells were seeded and transfected as described in section 2.7. Subsequently, 20 000 transfected A549 cells in 300 μL serum-free RPMI-1640 medium were added to the upper chambers, and 500 μL of complete RPMI-1640 medium was added to the lower compartment. The plates were incubated at 37°C and 5% CO2 in the incubator for 16 h, after which the cells were washed with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature. Cells on the upper surfaces of the membrane were removed with a cotton tip, and the membranes were washed 2 times with PBS. Subsequently, cells on the bottom surfaces of the membrane were stained with 0.1% crystal violet prepared in 20% ethanol for 30 min and washed with PBS 5 times to remove unbound crystal violet. The cells were then dried at 37°C and observed under the microscope (Keyence Biozero, Neu-Isenburg, Germany). Per sample, 3 fields were counted per chamber.
2.11. Western blot analysis of KRAS gene silencing
To quantify the ability of PBAE polyplexes to silence mutated KRAS, A549 cells were seeded in 6-well plates 24 h before use at a density of 2 × 105 cells/well. P(SpBAE) polyplexes and P(BSpBAE) polyplexes loaded with siNC and siG12S at a concentration of 50 nM were incubated with the cells at 37°C and 5% CO2 in a humidified incubator for 72 h. Cells were then washed with ice-cold PBS and then lysed in lysis buffer (800 μL RIPA buffer, 100 μL Phosphatase inhibitor, and 100 μL Protease inhibitor). The extracted protein was determined by Bradford reagent (Bio Rad, America), and equal amounts of protein were subjected to SDS-PAGE (Novex Wedgewell 10% Tris-Glycine Gel). Separated proteins were transferred to a nitrocellulose membrane (Amersham™ Protran®), which was then exposed to 5% nonfat milk TBST solution (Tris-buffered saline pH 8.0 + 1% Tween 20) for 1 h at room temperature and incubated with antibodies against KRAS (Proteintech, Germany) overnight at 4 °C. The membranes were washed 3 times with 1% TBST and then incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibodies (Proteintech, Germany) at 4 C for about 7 hours. Chemiluminescence reagents were finally applied to the membrane and photographed immediately with the ChemiDoc imager (Bio-Rad, Feldkirchen, Germany). The membrane was then treated with stripping buffer for 15 min, washed with TBST, and blocked with 5% nonfat milk TBST solution, stained with a primary antibody against histone 3 (Santa Cruz Biotechnologies, Heidelberg, Germany) at 4°C overnight, and then with an HRP-conjugated donkey anti-mouse antibody (Santa Cruz Biotechnologies, Heidelberg, Germany), before it was imaged in a GelDoc XR (Bio-Rad, Feldkirchen, Germany).
3. Results & Discussion
3.1. Synthesis and characterization of spermine-based poly(β-amino ester)s
To synthesize the target poly(β-amino ester)s, we first converted spermine to the monomer tri-Boc-spermine using a literature procedure. For the PBAE synthesis, widely used solvents are dichloromethane (DCM), tetrahydrofuran (THF) and dimethyl sulfoxide (DMSO). Lynn and Akinc et al. compared DCM and THF on the basis of the solubility of their monomers and found that the reaction in DCM yields higher molecular weight polymers.8, 26 DMSO is also common for PBAEs synthesis to avoid the cytotoxicity of halogenated solvents, especially when the resulting PBAE is used directly without further purification.13, 27 In addition to optimizing solvents, a high concentration of monomers in the absence of solvents can also lead to high molecular weight PBAEs, shorter reaction time, and less intramolecular cyclization.28 In contrast, here, we synthesized P(SpBAE) without any additional solvents, as the reactions in DCM and THF did not lead to desired polymers, which could be due to the low boiling point of the two solvents and low reaction temperature, and the yield in DMSO was too low in our case (data not shown).
Tri-Boc-spermine was then polymerized with 1,4-butanediol diacrylate. As shown in Figure 1B, the shift of the peak at 4.09 ppm (4.20 ppm in 1,4-butanediol diacrylate) and the new signal at 2.39 ppm indicated that tri-Boc-spermine was conjugated with 1,4-butanediol diacrylate. The peaks at 5.8 to 6.4 ppm belong to the terminal acrylate group. The peaks at 0.88 and 1.25 ppm belong to the residual n-hexane, which does not influence the next step and was further removed in vacuum before use. The product P(BSpBAE) was then analyzed by SEC, and a molar mass (Mn) of 3600 g/mol and a polydispersity of 1.73 were determined. The polymer was further treated with TFA to deprotect the Boc-protection groups, and the product P(SpBAE) as a TFA salt was obtained according to the 13C NMR (Figure S4). As shown in Figure 1C, the disappearance of the peak at 1.43 ppm indicated that N-Boc groups were removed.
Figure 1. 1H NMR of (A) tri-Boc-spermine in CDCl3; (B) P(BSpBAE) in CDCl3; (C) P(SpBAE) in D2O.
Polymer characteristics and results of the polymerization reactions in this study are summarized in Table 1.
Table 1. Step-growth polymerization (polyaddition) of tri-Boc spermine and 1,4-butanediol diacrylate and deprotection of the resulting polymer.
| Polymer | Solvent | Time (h) | Yield (%) | Mn(Da) | Ð | Protonable unit |
|---|---|---|---|---|---|---|
| P(BSpAE) 5[a] | neat | 15 | 83 | 3600[c] | 1.73[c] | 350 |
| P(SpBAE) 6[b] | DCM | 2 | 63 | 3800[d] | / | 186 |
reaction conditions: neat, 120 °C, overnight, the molar ratio of spermine and 1,4-butanediol diacrylate is 1:1.
reaction conditions: polymer 5, TFA, DCM, room temperature, 2 h.
Mn and Ð was assessed via SEC (measurement relative to polystyrene and in chloroform at 30 °C).
molecular weight was calculated on the basis of SEC of polymer 5 (Figure S3) and the theoretical chemical structure of polymer 6.
Poly(β-amino ester)s (P(BSpBAE) and P(SpBAE)) were synthesized via Michael-addition-based step-growth polymerization. To obtain a high molecular weight polymer, the monomers must be pure and the stoichiometric ratio of the monomers should be 1:1.29 Although different ratios of the monomers have been reported in the literature to obtain polymers with different terminal groups, according to the reaction kinetics, monomers with ratio 1:1 often lead to the highest molecular weight/conversion.30, 31 The reaction temperature of synthesizing PBAEs in the literature ranged from 45 °C to 120°C, and the reaction time ranged from 5 hours to 5 days. High reaction temperature usually corresponds to short reaction time.26, 28
3.3. siRNA encapsulation efficiency
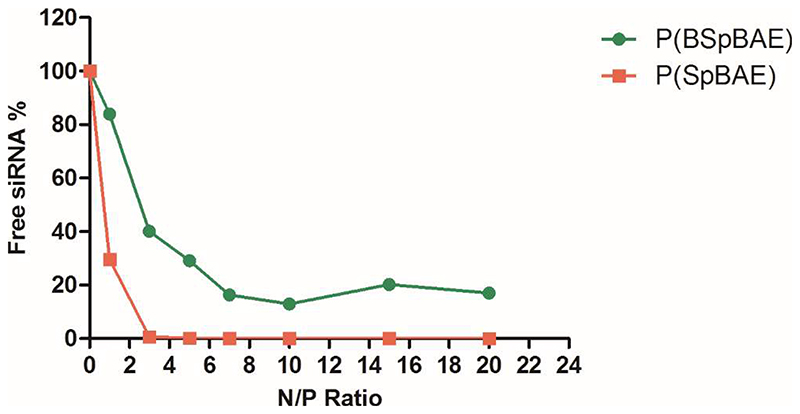
SYBR® Gold stain is a proprietary unsymmetrical cyanine dye that which can bind to double- or single-stranded DNA or RNA and exhibit high fluorescence intensity upon excitation. 23 Here we used the fluorescence of free siRNA as the negative control (100% free siRNA), and the free siRNA in other samples was calculated by dividing their fluorescence intensity by the negative control. As shown in Figure 2, in case of P(SpBAE) polyplexes, the siRNA was fully encapsulated from N/P 2 on, while P(BSpBAE) polyplexes encapsulated siRNA most efficiently at N/P 10. This observation can be explained by the primary amines and secondary amines in P(BSpBAE) being protected by the N-Boc groups, hence decreasing encapsulation efficacy on the basis of overall polymer weight.
Figure 2.
siRNA encapsulation profiles of polyplexes measured by SYBR Gold assays at various N/P ratios. 100% values (N/P = 0) are represented by the determined fluorescence of uncondensed free siRNA (data points indicate mean± SD, n = 3).
In fact, despite their decreased encapsulation efficiency due to the lack of primary and secondary amines, the overall performance of poly(β-amino ester)smay not be affected negatively. Professor Langer’s group published extensive research on the structure-function relationship of poly(β-amino ester)s. For instance, Anderson et al. found that the polymers synthesized from 1,4-butanediol diacrylate with 2-(piperidin-1-yl) ethan-1-amine and cyclohexane-1,4-diylbis(methylene) diacrylate with 4-aminol-1-butanol were most promising and had 4-8 times the transfection efficacy of poly(ethylene imine) (PEI).31 In those polymers, the amines are mainly tertiary amines. Other poly(β-amino ester)s containing mainly tertiary amines were also synthesized and widely used by different groups. For example, Kim et al. utilized 4-amino-1-butanol to synthesize PBAE. They also modified the PBAE with PEG to enhance the stability and transfection efficiency of the polymer.32 Kozielski et al. synthesized a bioreducible linear PBAE with 4-amino-1-butanol and bis(2-hydroxyethyl) disulfide reacted with acryloyl chloride. The repeating unit of the polymer contained only tertiary amines but achieved 92% GFP knockdown without measurable loss in metabolic activity in GBM 319 cells.33
3.2. Characterization of the polyplexes
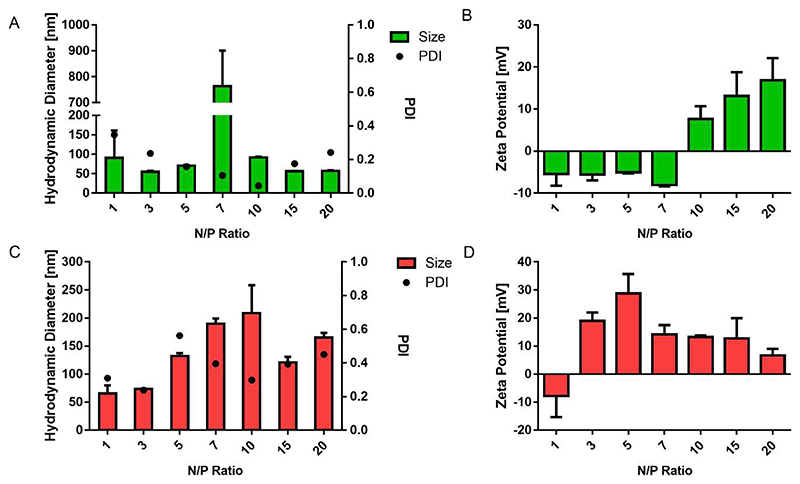
As shown in Figure 3, the size (hydrodynamic diameter) and zeta potential of the polyplexes were measured by dynamic light scattering and laser Doppler anemometry. The N/P ratio has a significant influence on the size and zeta potential of the polyplexes. For P(SpBAE) polyplexes, the size ranged from 65.6 nm to 209 nm; for P(BSpBAE) polyplexes, the size ranged from 55.1 nm to 92.2 nm except the formulation at N/P 7 with a size of 764 nm.
Figure 3.
(A) Hydrodynamic diameter of P(BSpBAE) polyplexes; (B) Zeta potential of P(BSpBAE) polyplexes; (C) Hydrodynamic diameter of P(SpBAE) polyplexes; (D) Zeta potential of P(SpBAE) polyplexes. (Size and zeta potential, mean ± SD, n = 3, PDI, mean, n = 3).
The size of the polyplexes depends on the property of the polymer and the amount of polymer used in a formulation. siRNA cannot be condensed efficiently at low N/P ratios, while when the N/P ratio is too high, the free polymer may cause toxicity, and aggregation may lead to faster sedimentation. For both P(BSpBAE) and P(SpBAE), the polyplexes with the lowest PDI formed at the N/P ratio where the siRNA was just most efficiently encapsulated. For instance, according to the SYBR gold assay of P(BSpBAE), approximately 90% of the siRNA is encapsulated at N/P 10, and the PDI of polyplexes at N/P 10 was 0.044, which is the lowest for these polyplexes among the different N/P ratios tested. Similarly, for P(SpBAE), the siRNA was fully encapsulated at N/P 3, and the polyplexes of N/P 3 showed the lowest PDI 0.238. The size of P(BSpBAE) polyplexes at N/P 7 was large, which could be caused by aggregation of negatively and positively charged polyplexes present at N/P 7. From N/P 7 to N/P 10, the zeta potential of the polyplexes switches from negative to positive. It is therefore also possible that neutral polyplexes are formed whose net charge is insufficient to stabilize the polyplexes, resulting in aggregation.
3.4. In vitro compatibility
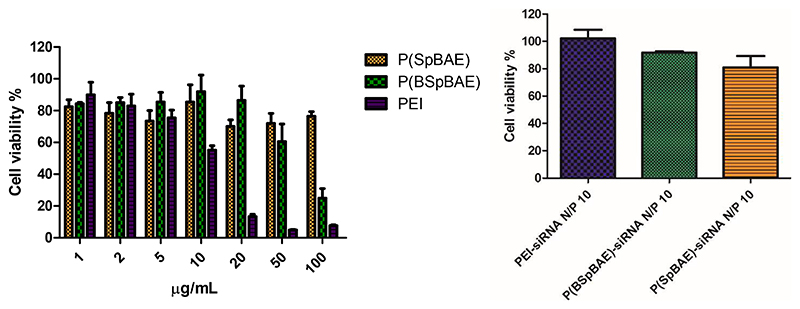
MTT assays were performed to investigate the polymers’ and polyplexes’ biocompatibility. As shown in Figure 4, PBAEs and PEI had comparable cell viability at low concentrations from 1 μg/mL to 5 μg/mL. However, above 10 μg/mL, PEI caused stronger cytotoxicity than the poly(β-amino ester)s P(SpBAE) and P(BSpBAE), confirming better biocompatibility of the poly(β-amino ester)s compared with PEI. All polyplexes mediated cell viability above 80% in H1299 cells, which indicated that the polyplexes were biocompatible. However, for the PBAE polyplexes, larger weight amounts of polymer are necessary, resulting in slight differences regarding polyplex biocompatibility.
Figure 4. Cell viability of polymers and polyplexes determined by diemthylthiazolyl blue diphenyltetrazolium bromide (MTT) assay in H1299 cells.
3.5. Cellular uptake
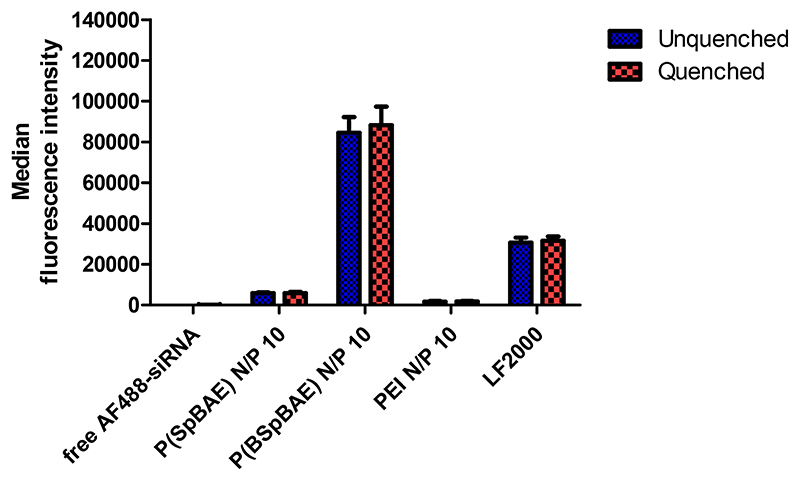
The cellular uptake of P(BSpBAE) polyplexes and P(SpBAE) polyplexes was determined by flow cytometry with or without trypan blue quenching. As shown in Figure 5, there is no significant difference between unquenched and quenched samples, which confirms cellular polyplex internalization, rather than adsorption on the surface of the cells. The uptake of P(SpBAE) polyplexes was 60 times higher than that of free AF488-siRNA (negative control), while the uptake of P(BSpBAE) polyplexes was 865 times higher than the negative control, 14 times higher than that of P(SpBAE) polyplexes, and 49 times higher than that of PEI polyplexes. The uptake of P(BSpBAE) polyplexes was more than 2 times higher than with Lipofectamine ™ 2000 with the same amount of siRNA (50 pmol AF488-siRNA) for each well.
Figure 5. Cellular uptake of polyplexes quantified by flow cytometry and presented as median fluorescence intensity corrected by autofluorescence of untreated blank cells (H1299 cells, AF488-siRNA).
The uptake of P(SpBAE) polyplexes at N/P 1 to 20 was also determined (Figure S1). The cellular uptake depended on the N/P ratios, and higher N/P ratios often lead to higher cellular uptake regardless of the high PDI. This observation can be explained by the positive charge of the polyplexes enhancing the interaction with the negatively charged cell membrane. However, too much positive charge also leads to cytotoxicity and thus a lower cellular uptake in viable cells. Besides charge, the amphiphilicity of a polymer and its resulting polyplexes is crucial and has a significant influence on their cellular uptake. In this experiment, P(BSpBAE) is hydrophobic, while P(SpBAE) is hydrophilic. Although the siRNA encapsulation efficiency of P(BSpBAE) is lower than that of P(SpBAE) according to the SYBR gold assays (Figure 3), the P(BSpBAE) polyplexes still achieved much higher cellular uptake.
3.6. In vitro eGFP knockdown
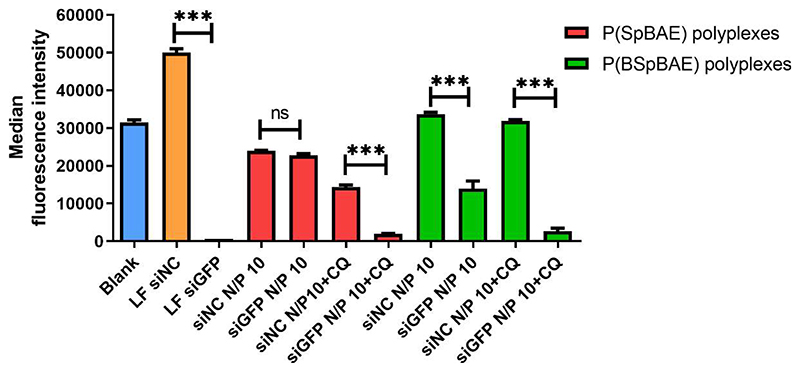
To evaluate the gene silencing efficiency of such hydrophobic and hydrophilic polyplexes on the protein level, we utilized H1299/eGFP cells that stably express the enhanced green fluorescent protein reporter gene (eGFP). Chloroquine (CQ) used as an anti-malaria drug has been a focus of research for its contribution to the endosomal escape process, and it has been proposed to increase endosome escape through several mechanisms.34 Chloroquine diffuses across the cell membrane and into endosomes passively, as the pH drops in endosomes, it becomes protonated and trapped inside the endosome resulting in a dramatic increase in its endosomal concentration. It is thought to insert a hydrophobic motif into the lipid bilayer and lyse the endosome at a critical concentration.35 As shown in Figure 6, Lipofectamine™ 2000 (LF) was used as a positive control. In transfection experiments without chloroquine, P(SpBAE) polyplexes mediated only 4.91% of knockdown which is not significant in statistics, while P(BSpBAE) polyplexes had a knockdown efficiency of 58.5%. After chloroquine treatment, the knockdown efficiency of P(SpBAE) polyplexes was 86.6%, which indicates that the P(SpBAE) polyplexes were entrapped in endosome/lysosomes. However, the fluorescence intensity of cells transfected with P(SpBAE)-siNC polyplexes also decreased after treatment with chloroquine which might be a sign of cytotoxicity. The knockdown efficiency of P(BSpBAE) polyplexes, on the other hand, increased to 91.6% with the treatment of chloroquine without visible signs of increased cytotoxicity.
Figure 6.
Enhanced green fluorescent protein (eGFP) knockdown of P(SpBAE) polyplexes and P(BSpBAE) polyplexes in H1299 cells expressing eGFP quantified by flow cytometry as median fluorescence intensity (Mean ± SD, n = 3, One-way ANOVA with Bonferroni multiple comparison test, ***p< 0.001, nsp> 0.05).
Chloroquine enhanced the knockdown of P(BSpBAE) and P(SpBAE) polyplexes, which means both of the polyplexes are not released from the endosome efficiently. However, the GFP knockdown efficiency of P(SpBAE) polyplexes with chloroquine was improved by a factor 17.6 times compared to the samples without chloroquine treatment. On the other hand, the knockdown efficiency of P(BSpBAE) polyplexes with chloroquine was only 1.57 times increased compared with samples without chloroquine treatment. This indicates that the P(BSpBAE) polyplexes may be less subject to endosomal entrapment than the P(SpBAE) polyplexes.
3.7. Endosome Entrapment
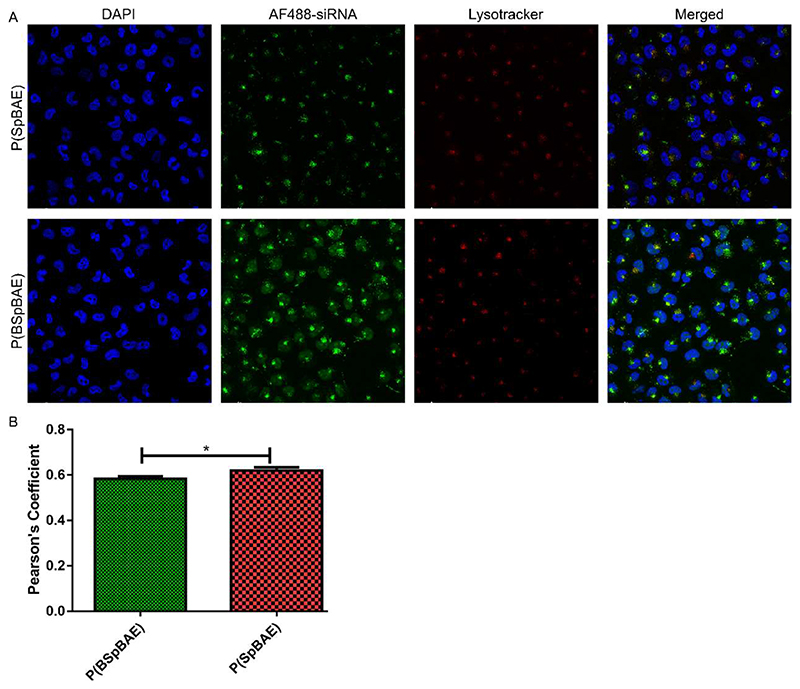
To find the mechanism behind the efficient knockdown of P(BSpBAE) polyplexes, the endosomal entrapment of the polyplexes was visualized via confocal laser scanning microscopy (CLSM). As shown in Figure 7, more AF488-siRNA P(BSpBAE) polyplexes seemed to reach the cytoplasm than P(SpBAE) polyplexes. This result is in line with the results obtained by flow cytometry. The P(BSpBAE) polyplexes were also distributed in other cell organelles. The colocalization of P(SpBAE) polyplexes with lysosomes (yellow dots in the merged figure) was more than that of P(BSpBAE) polyplexes, which indicates that the P(BSpBAE) polyplexes may have a better ability of endosomal escape. The colocalization coefficient was analyzed in Image J, which also confirmed that P(BSpBAE) polyplexes colocalized less with endosome/lysosomes than P(SpBAE) polyplexes.
Figure 7.
(A) Confocal images after treatment of H1299 cells with P(SpBAE) polyplexes and P(BSpBAE) polyplexes formulated with AF488-siRNA and stained with Lysotracker red™ DND 99 and DAPI. (B) Pearson’s coefficient analyzed via Image J (Mean ± SD, n = 3, Unpaired T test, *p < 0.05).
Endosomal escape is one of the critical hurdles to successful intracellular delivery of nucleic acid-based therapeutics. Nanoparticles internalization must be followed by endosomal escape. Otherwise, endosomal entrapment renders the nanocarriers and their cargo useless as it is degraded via the endo/lysosomal pathway.36 One of the most common strategies for endosomal escape is the hypothesized “proton sponge effect”. The sponges, which typically are polyamines, can absorb free protons in endosomes. As the absorbed protons accumulate, chloride will then begin to diffuse into the endosome as a compensation to restore the equilibrium potential which raises the osmotic pressure inside the vesicle. This causes the endosome/lysosome to swell and expand until a critical area strain is passed, and finally the lipid bilayer membrane ruptures and the polyplexes are released without degradation.37
Interestingly, the P(BSpBAE) polyplexes seem to have a better endosomal escape ability although lacking amines. We assume that this may be due to the partial removal of N-Boc groups in acidic endosome/lysosomes. The N-Boc group is a common amine protecting group, which is stable in neutral to basic environment but is unstable and removable in an acidic environment. When N-Boc groups are removed in endosome/lysosomes, the amines can be protonated and thus enhance the “proton sponge effect”. The P(BSpBAE) polyplexes might also achieve endosomal escape via other mechanisms. For example, the polyplex-mediated escape theory. In brief, the charged polyplexes interact directly with the anionic phospholipids of the inner membrane leaflet leading to membrane destabilization and nanoscale holes within the membrane and then vector escape from these holes.38, 39 Another similar theory is that the endosomal escape could be a result from free polymer-mediated membrane permeability and nanoscale hole formation. It was hypothesized that there is a dynamic equilibrium between dissociated polymer chains and polyplexes, and free polymer chains intercalate in the membrane of endo-lysosomes and cause membrane disintegration and hole formation.38, 40 It is also possible that due to the hydrophobicity of N-Boc groups, the polyplexes were internalized into the cells via a different pathway which results in less entrapment in endosome/lysosomes.
3.8. Cellular uptake pathway
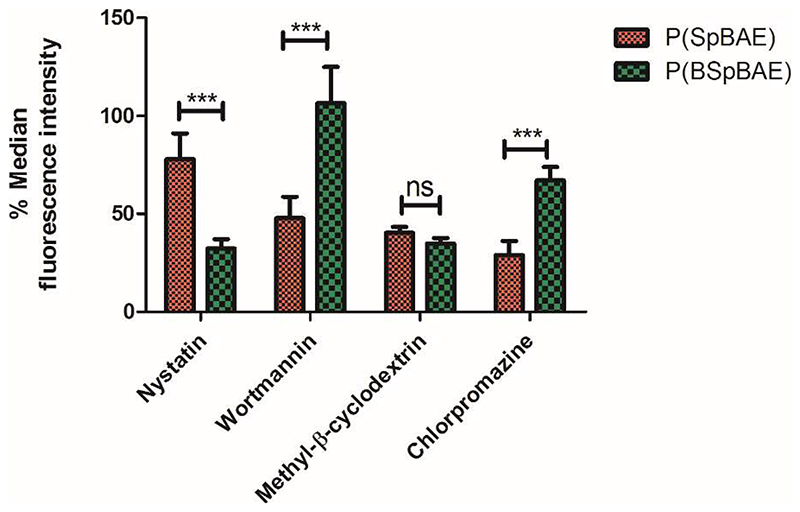
Four cellular uptake inhibitors were applied in this experiment to investigate the cellular uptake pathway. Specifically, nystatin can inhibit the internalization of caveolae and lipid rafts through the depletion of cholesterol in the cell membrane; wortmannin can block macropinocytosis; methyl-β-cyclodextrin inhibits the lipid raft-dependent endocytic pathway by interfering with cholesterol synthesis and decomposing cholesterol; chlorpromazine blocks clathrin-mediated endocytosis through destroying cell surface clathrin and the AP2 complex.22, 41 As shown in Figure 8, the P(SpBAE) polyplexes and P(BSpBAE) polyplexes showed different behavior after the treatment with these four inhibitors. Nystatin had almost no influence on the cellular uptake of P(SpBAE) polyplexes but decreased the uptake of P(BSpBAE) polyplexes’ by 70%; while wortmannin had no inhibition effect on the cellular uptake of P(BSpBAE) polyplexes, it inhibited the cellular uptake of P(SpBAE) polyplexes by about 50%; Methyl-β-cyclodextrin strongly inhibited the uptake of both P(SpBAE) polyplexes and P(BSpBAE) polyplexes; Chlorpromazine decreased the uptake of P(SpBAE) polyplexes by 71.1% but that of P(BSpBAE) polyplexes only by only 33.0%.
Figure 8. Cellular uptake of P(SpBAE) and P(BSpBAE) polyplexes (N/P = 10) after treatment with inhibitors nystatin, wortmannin, chlorpromazine, and methyl-β-cyclodextrin (M-β-CD).
(Results are shown as mean ± SD as a percentage of median fluorescence intensity normalized to non-inhibited samples, n = 3, two-way ANOVA with Bonferroni post-hoc test, ***p < 0.001, nsp > 0.05.)
These results indicate that P(SpBAE) polyplexes were mainly internalized via macropinocytosis, lipid raft-dependent, and clathrin-dependent endocytic pathways; P(BSpBAE) polyplexes, on the other hand were internalized mainly via caveolae and lipid raft-dependent endocytosis. Caveolae-dependent endocytosis is reported to have the potential to bypass lysosomes, and many pathogens including viruses and bacteria in fact select this pathway to avoid lysosomal degradation. Therefore, this route is believed to be beneficial for enhanced therapeutic effects.42 While clathrin-dependent endocytosis usually leads to endosomal/lysosomal localization.43 These differences in intracellular trafficking could explain why P(BSpBAE) polyplexes resulted in less endosomal entrapment and higher GFP knockdown efficiency.
3.9. Migration assay
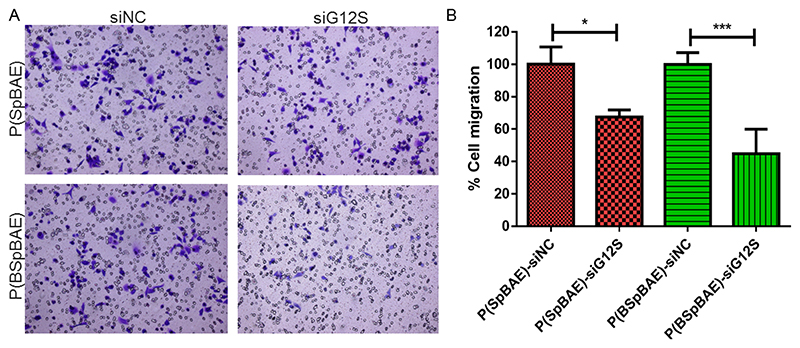
KRAS (Kirsten rat sarcoma virus) is a gene that provides instructions for a protein called K-Ras, which relays signals from outside the cell to the nucleus. The signals instruct the cell to grow and proliferate or to mature and differentiate. It is called KRAS because it was first identified as a viral oncogene in the Kirsten Rat Sarcoma Virus.44 KRAS is the most frequently mutated gene associated with lung cancer, and the activating mutations in KRAS result in constitutive activation of the GTPase protein in the absence of growth factor signaling, resulting in a sustained proliferation signal within the cell which is related to the migration and invasion of cancer cells.18 To achieve therapeutically relevant gene silencing, we used a siRNA sequence targeting a KRAS mutant, namely KRAS G12S. As shown in Figure 9, cells transfected with the siG12S encapsulated polyplexes showed less migration than cells transfected with the negative controls. Specifically, formulations with P(SpBAE)-siG12S inhibited about 33% of the migration, while P(BSpBAE)-siG12S inhibited about 55% of the migration.
Figure 9.
(A) The crystal violet staining of migrated A549 cells. (B) The percentage of the migrated cells was quantified by counting three fields per chamber and compared with controls (One way ANOVA, n = 3, *p < 0.05, ***p < 0.001).
Interestingly, P(SpBAE) polyplexes did not mediate a significant GFP knockdown but were able to inhibit the migration of A549 cells. It was hypothesized that although P(SpBAE) polyplexes did not achieve a knockdown on protein level, they were yet able to change the status of the cells, for example, the signal transduction, thus leading to an inhibition of the migration. It is also possible that transfection of A549 cells with P(SpBAE) polyplexes was in general more efficient than transfection of H1299 cells or that a smaller gene silencing effect of KRAS resulted in a more visible effect than eGFP silencing measured by flow cytomatry. The gene silencing efficiency is also related to the siRNA. It is thus possible that the siRNA against KRAS has a higher efficiency than the siRNA against GFP, thus the cytoplasmatically released siRNA delivered by P(SpBAE) polyplexes was able to achieved a KRAS knockdown and inhibited the migration of A549 cells. To observe the KRAS knockdown on protein level and investigate which assumption is correct, a western blot experiment was performed.
3.10. Western blot analysis
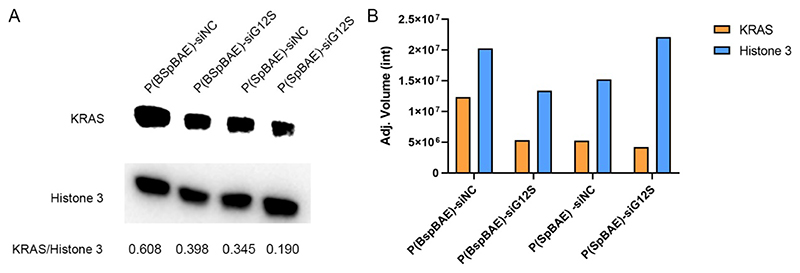
Western blot analysis was conducted to investigate if the P(SpBAE) polyplexes and P(BSpBAE) polyplexes can knock down KRAS on the protein level. As shown in Figure 10, both P(SpBAE) polyplexes and P(BSpBAE) polyplexes downregulated the expression of KRAS in relation to the housekeeping gene Histone 3 to about 60% of the controls which had been transfected with scrambled siRNA. Since cytotoxicity was negligible for both types of polyplexes, differences in the housekeeping gene expression were understood as loading differences, and gene silencing efficacy was in fact in line with the migration inhibition results in Figure 9. P(SpBAE) polyplexes did not knock down GFP significantly but inhibited the expression of KRAS. To understand the underlying principles of this phenomenon, we determined the cellular uptake of P(SpBAE) and P(BSpBAE) polyplexes in A549 cells. PEI and Lipofectamine™ 2000 were used as controls (Figure S2). The cellular uptake of P(BSpBAE) polyplexes was still higher than P(SpBAE) polyplexes. However, the cellular uptake of P(BSpBAE) polyplexes decreased compared with its behavior in H1299 cells, and the cellular uptake of P(SpBAE) polyplexes increased. This result could explain why P(SpBAE) polyplexes did not efficiently mediate GFP knockdown in H1299 cells but caused therapeutically relevant KRAS knockdown in A549 cells as shown in the migration assay. The phenomenon that nanoparticles show different behavior in different cell lines was also reported by other researchers. For example, Kim et al. investigated the transfection efficiency of DOTAP-liposomes with different DOTAP/DOPE ratios in different cell types including Huh7, AGS, COS7, and A549. They found that the formulation T1P0 and T3P1 showed higher luciferase activities than T1P1 or T1P3 in Huh7 and AGS cells. However, the transfection efficiencies of T3P1 and T1P1 were higher than those of T1P0 and T3P1 in COS7 cells. These results implied that specific cell lines can favor certain lipid compositions in gene delivery and the transfection efficiency is cell-line dependent.45 The good performance of P(SpBAE)-siG12S polyplexes could also be due to the higher knockdown efficiency of the siRNA sequence siG12S. The knockdown efficiency of siRNA is related to its length, nucleotide composition, and the distance of the target region to transcription start site.46
Figure 10. Western blot analysis of KRAS protein expression in A549 cells.
Histone 3 was used as the loading control. The ratio of KRAS/Histione 3 (A) was calcualted by the the adjusted volume corrected by the normalization factor (Adj. Volume (int)) generated by Image lab™ (B).
4. Conclusion
In this project, we have synthesized spermine-based poly(β-amino ester)s successfully. The PBAEs polymers were more biocompatible than PEI and were able to encapsulate siRNA and delivery siRNA efficiently. Interestingly, we observed that the PBAEs before removing the N-Boc protection groups P(BSpBAE) mediated much higher cellular siRNA uptake and GFP knockdown efficiency, benefiting from different cellular uptake mechanisms and lower endosomal entrapment. This indicates that the Boc-protection of the polymer could be beneficial for siRNA delivery and gene silencing efficiency. The PBAE polyplexes encapsulating siRNA against KRAS also showed promising results, which indicates the potential application of spermine-PBAEs for therapeutic purposes. Taken together, the spermine-based poly(β-amino ester)s are very promising for siRNA delivery and RNAi-based therapeutics.
Supplementary Material
Acknowledgments
Yao Jin appreciates the financial support from China Scholarship Council (CSC 201906010329). This research was funded by ERC-2014-StG – 637830 to Olivia Merkel.
References
- (1).Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101(1):25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- (2).McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nature reviews genetics. 2002;3(10):737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- (3).Uludağ H, Parent K, Aliabadi HM, Haddadi A. Prospects for RNAi Therapy of COVID-19. Frontiers in Bioengineering and Biotechnology. 2020;8(916) doi: 10.3389/fbioe.2020.00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Lam JK-W, Liang W, Chan H-K. Pulmonary delivery of therapeutic siRNA. Advanced drug delivery reviews. 2012;64(1):1–15. doi: 10.1016/j.addr.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Check E. Harmful potential of viral vectors fuels doubts over gene therapy. Nature. 2003;423(6940):573–575. doi: 10.1038/423573a. [DOI] [PubMed] [Google Scholar]
- (6).Lai W-F. In vivo nucleic acid delivery with PEI and its derivatives: current status and perspectives. Expert review of medical devices. 2011;8(2):173–185. doi: 10.1586/erd.10.83. [DOI] [PubMed] [Google Scholar]
- (7).Oh Y-K, Park TG. siRNA delivery systems for cancer treatment. Advanced drug delivery reviews. 2009;61(10):850–862. doi: 10.1016/j.addr.2009.04.018. [DOI] [PubMed] [Google Scholar]
- (8).Lynn DM, Langer R. Degradable poly (β-amino esters): synthesis, characterization, and self-assembly with plasmid DNA. Journal of the American Chemical Society. 2000;122(44):10761–10768. [Google Scholar]
- (9).Lynn DM, Langer R. Degradable Poly(β-amino esters): Synthesis, Characterization, Self-Assembly with Plasmid DNA. Journal of the American Chemical Society. 2000;122(44):10761–10768. doi: 10.1021/ja0015388. [DOI] [Google Scholar]
- (10).Green JJ, Langer R, Anderson DG. A Combinatorial Polymer Library Approach Yields Insight into Nonviral Gene Delivery. Accounts of Chemical Research. 2008;41(6):749–759. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Eltoukhy AA, Siegwart DJ, Alabi CA, Rajan JS, Langer R, Anderson DG. Effect of molecular weight of amine end-modified poly (β-amino ester) s on gene delivery efficiency and toxicity. Biomaterials. 2012;33(13):3594–3603. doi: 10.1016/j.biomaterials.2012.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Gao H, Cheng T, Liu J, Liu J, Yang C, Chu L, Zhang Y, Ma R, Shi L. Self-regulated multifunctional collaboration of targeted nanocarriers for enhanced tumor therapy. Biomacromolecules. 2014;15(10):3634–3642. doi: 10.1021/bm5009348. [DOI] [PubMed] [Google Scholar]
- (13).Liu Y, Li Y, Keskin D, Shi L. Poly(beta-Amino Esters): Synthesis, Formulations, and Their Biomedical Applications. Adv Healthc Mater. 2019;8(2):e1801359. doi: 10.1002/adhm.201801359. [DOI] [PubMed] [Google Scholar]
- (14).Estévez-Torres A, Baigl D. DNA compaction: fundamentals and applications. Soft Matter. 2011;7(15):6746. doi: 10.1039/c1sm05373f. [DOI] [Google Scholar]
- (15).Elsayed M, Corrand V, Kolhatkar V, Xie Y, Kim NH, Kolhatkar R, Merkel OM. Influence of oligospermines architecture on their suitability for siRNA delivery. Biomacromolecules. 2014;15(4):1299–1310. doi: 10.1021/bm401849d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lote AR, Kolhatkar VR, Insley T, Král P, Kolhatkar R. Oligospermines and Nucleic Acid Interaction: A Structure Property Relationship Study. ACS. Macro Letters. 2014;3(8):829–833. doi: 10.1021/mz500358w. [DOI] [PubMed] [Google Scholar]
- (17).El Osta B, Behera M, Kim S, Berry LD, Sica G, Pillai RN, Owonikoko TK, Kris MG, Johnson BE, Kwiatkowski DJ. Characteristics and outcomes of patients with metastatic KRAS-mutant lung adenocarcinomas: the lung cancer mutation consortium experience. Journal of Thoracic Oncology. 2019;14(5):876–889. doi: 10.1016/j.jtho.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Lievre A, Bachet J-B, Le Corre D, Boige V, Landi B, Emile J-F, Côté J-F, Tomasic G, Penna C, Ducreux M. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer research. 2006;66(8):3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- (19).Mehta A, Dalle Vedove E, Isert L, Merkel OM. Targeting KRAS Mutant Lung Cancer Cells with siRNA-Loaded Bovine Serum Albumin Nanoparticles. Pharm Res. 2019;36(9):133. doi: 10.1007/s11095-019-2665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Sotillo R, Schvartzman J-M, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464(7287):436–440. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Blagbrough IS, Geall AJ. Practical synthesis of unsymmetrical polyamine amides. Tetrahedron letters. 1998;39(5–6):439–442. Nalluri, S. K. M.; Voskuhl, J.; Bultema, J. B.; Boekema, E. J.; Ravoo, B. J. Light-responsive capture and release of DNA in a ternary supramolecular complex. Angewandte Chemie International Edition 2011, 50 (41), 9747-9751. [Google Scholar]
- (22).Hartl N, Adams F, Costabile G, Isert L, Doblinger M, Xiao X, Liu R, Merkel OM. The Impact of Nylon-3 Copolymer Composition on the Efficiency of siRNA Delivery to Glioblastoma Cells. Nanomaterials (Basel) 2019;9(7) doi: 10.3390/nano9070986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Tuma RS, Beaudet MP, Jin X, Jones LJ, Cheung C-Y, Yue S, Singer VL. Characterization of SYBR Gold nucleic acid gel stain: a dye optimized for use with 300-nm ultraviolet transilluminators. Analytical biochemistry. 1999;268(2):278–288. doi: 10.1006/abio.1998.3067. [DOI] [PubMed] [Google Scholar]
- (24).Jin Y, Liu Q, Zhou C, Hu X, Wang L, Han S, Zhou Y, Liu Y. Intestinal oligopeptide transporter PepT1-targeted polymeric micelles for further enhancing the oral absorption of water-insoluble agents. Nanoscale. 2019 doi: 10.1039/c9nr07029j. [DOI] [PubMed] [Google Scholar]
- (25).Zou Y, Zhou Y, Jin Y, He C, Deng Y, Han S, Zhou C, Li X, Zhou Y, Liu Y. Synergistically Enhanced Antimetastasis Effects by Honokiol-Loaded pH-Sensitive Polymer-Doxorubicin Conjugate Micelles. ACS Appl Mater Interfaces. 2018;10(22):18585–18600. doi: 10.1021/acsami.8b04854. [DOI] [PubMed] [Google Scholar]
- (26).Akinc A, Lynn DM, Anderson DG, Langer R. Parallel Synthesis and Biophysical Characterization of a Degradable Polymer Library for Gene Delivery. Journal of the American Chemical Society. 2003;125(18):5316–5323. doi: 10.1021/ja034429c. [DOI] [PubMed] [Google Scholar]
- (27).Lee J-S, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Gold, poly (β-amino ester) nanoparticles for small interfering RNA delivery. Nano letters. 2009;9(6):2402–2406. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Chen J, Huang S-W, Liu M, Zhuo R-X. Synthesis and degradation of poly(beta-aminoester) with pendant primary amine. Polymer. 2007;48(3):675–681. doi: 10.1016/j.polymer.2006.12.008. Akinc, A.; Anderson, D. G.; Lynn, D. M.; Langer, R. Synthesis of Poly(ß-amino ester)s Optimized for Highly Effective Gene Delivery. Bioconjugate Chemistry 2003, 14 (5), 979-988. DOI: 10.1021/bc034067y. [DOI] [Google Scholar]
- (29).Stille JK. Step-growth polymerization. ACS Publications; 1981. [Google Scholar]
- (30).Jere D, Kim TH, Arote RB, Jiang HL, Cho MH, Nah JW, Cho CS. Key Engineering Materials. Vol. 342. Trans Tech Publ; 2007. A poly (β-amino ester) of spermine and poly (ethylene glycol) diacrylate as a gene carrier; pp. 425–428. [Google Scholar]
- (31).Anderson DG, Akinc A, Hossain N, Langer R. Structure/property studies of polymeric gene delivery using a library of poly(β-amino esters) Molecular Therapy. 2005;11(3):426–434. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
- (32).Kim J, Kang Y, Tzeng SY, Green JJ. Synthesis and application of poly(ethylene glycol)-co-poly(β-amino ester) copolymers for small cell lung cancer gene therapy. Acta Biomaterialia. 2016;41:293–301. doi: 10.1016/j.actbio.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Kozielski KL, Tzeng SY, Green JJ. A bioreducible linear poly(beta-amino ester) for siRNA delivery. Chem Commun (Camb) 2013;49(46):5319–5321. doi: 10.1039/c3cc40718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Baradaran Eftekhari R, Maghsoudnia N, Dorkoosh FA. Chloroquine: a brand-new scenario for an old drug. Expert Opinion on Drug Delivery. 2020;17(3):275–277. doi: 10.1080/17425247.2020.1716729. [DOI] [PubMed] [Google Scholar]
- (35).Maxfield FR. Weak bases and ionophores rapidly and reversibly raise the pH of endocytic vesicles in cultured mouse fibroblasts. The Journal of cell biology. 1982;95(2):676–681. doi: 10.1083/jcb.95.2.676. Dowdy, S. F. Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol 2017, 35 (3), 222-229. DOI: 10.1038/nbt.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Smith SA, Selby LI, Johnston APR, Such GK. The Endosomal Escape of Nanoparticles: Toward More Efficient Cellular Delivery. Bioconjug Chem. 2019;30(2):263–272. doi: 10.1021/acs.bioconjchem.8b00732. Karlsson, J.; Tzeng, S. Y.; Hemmati, S.; Luly, K. M.; Choi, O.; Rui, Y.; Wilson, D. R.; Kozielski, K. L.; Quinones-Hinojosa, A.; Green, J. J. Photocrosslinked Bioreducible Polymeric Nanoparticles for Enhanced Systemic siRNA Delivery as Cancer Therapy. Advanced Functional Materials 2021, 31 (17), 2009768. DOI: 10.1002/adfm.202009768. [DOI] [PubMed] [Google Scholar]
- (37).Agirre M, Zarate J, Ojeda E, Puras G, Desbrieres J, Pedraz J. Low Molecular Weight Chitosan (LMWC)-based Polyplexes for pDNA Delivery: From Bench to Bedside. Polymers. 2014;6(6):1727–1755. doi: 10.3390/polym6061727. [DOI] [Google Scholar]
- (38).Bus T, Traeger A, Schubert US. The great escape: how cationic polyplexes overcome the endosomal barrier. Journal of Materials Chemistry B. 2018;6(43):6904–6918. doi: 10.1039/c8tb00967h. [DOI] [PubMed] [Google Scholar]
- (39).Rehman ZU, Hoekstra D, Zuhorn IS. Mechanism of polyplex-and lipoplex-mediated delivery of nucleic acids: real-time visualization of transient membrane destabilization without endosomal lysis. ACS. nano. 2013;7(5):3767–3777. doi: 10.1021/nn3049494. [DOI] [PubMed] [Google Scholar]
- (40).Bonner DK, Zhao X, Buss H, Langer R, Hammond PT. Crosslinked linear polyethylenimine enhances delivery of DNA to the cytoplasm. Journal of Controlled Release. 2013;167(1):101–107. doi: 10.1016/j.jconrel.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Park BG, Kim YJ, Min JH, Cheong T-C, Nam SH, Cho N-H, Kim YK, Lee KB. Assessment of Cellular Uptake Efficiency According to Multiple Inhibitors of Fe3O4-Au Core-Shell Nanoparticles: Possibility to Control Specific Endocytosis in Colorectal Cancer Cells. Nanoscale research letters. 2020;15(1):1–10. doi: 10.1186/s11671-020-03395-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Kou L, Sun J, Zhai Y, He Z. The endocytosis and intracellular fate of nanomedicines: Implication for rational design. Asian Journal of Pharmaceutical Sciences. 2013;8(1):1–10. [Google Scholar]
- (43).Sahay G, Alakhova DY, Kabanov AV. Endocytosis of nanomedicines. Journal of Controlled Release. 2010;145(3):182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Tsuchida N, Ryder T, Ohtsubo E. Nucleotide sequence of the oncogene encoding the p21 transforming protein of Kirsten murine sarcoma virus. Science. 1982;217(4563):937–939. doi: 10.1126/science.6287573FromNLM. [DOI] [PubMed] [Google Scholar]
- (45).Kim B-K, Hwang G-B, Seu Y-B, Choi J-S, Jin KS, Doh K-O. DOTAP/DOPE ratio and cell type determine transfection efficiency with DOTAP-liposomes. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2015;1848(10, Part A):1996–2001. doi: 10.1016/j.bbamem.2015.06.020. [DOI] [PubMed] [Google Scholar]
- (46).Fakhr E, Zare F, Teimoori-Toolabi L. Precise and efficient siRNA design: a key point in competent gene silencing. Cancer gene therapy. 2016;23(4):73–82. doi: 10.1038/cgt.2016.4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.