Abstract
The hypothalamus contains the highest diversity of neurons in the brain, which use-dependently co-release neurotransmitters and neuropeptides. Investigators have hitherto relied on candidate protein-based tools to correlate behavioral, endocrine and gender traits with hypothalamic neuron identity. Here, we mapped neuronal identities in the hypothalamus by single-cell RNA-sequencing. We distinguish 62 neuronal subtypes producing glutamatergic, dopaminergic or GABAergic codes for synaptic neurotransmission and harboring the ability of task-dependent neurotransmitter switching. We identify dopamine neurons that uniquely co-express Onecut3 and Nmur2 genes, and place these in the periventricular nucleus with many synaptic afferents arising from neuromedin S+ neurons of the suprachiasmatic nucleus. These neuroendocrine dopamine cells may contribute to the dopaminergic inhibition of prolactin secretion diurnally because their neuromedin S+ inputs originate from pacemaker 2/3 (Per2/3)-expressing neurons and their tyrosine hydroxylase phosphorylation is circadian regulated. Overall, our catalogue of neuronal subclasses presents new understanding of hypothalamic organization and function.
Keywords: Connectivity, lightsheet microscopy, magnocellular, parvocellular, transcriptome
Introduction
In contrast to the cerebellum and cerebral cortex, the hypothalamus lacks distinct layering or other stringent anatomical, repetitive organizational principles. Since the discovery of vasopressin and oxytocin1, and of the releasing and inhibitory factors/hormones2 in the magno- and parvocellular hypothalamic systems, respectively, neuropeptides have been a basis for defining and understanding hypothalamic organization and functionality. Coordination of activity within the hypothalamic circuitry, including its functionally heterogeneous sub-networks composed of molecularly diverse and spatially segregated neuroendocrine neurons3, is the basis for maintaining vital adaptive responsiveness and homeostatic control of the body.
As an example of functional complexity within a circumscribed brain volume, eight subdivisons are distinguishable within the paraventricular nucleus (PVN) alone4. These contain both magno- and parvocellular neuroendocrine secretory motor neurons, as well as parvocellular neurons projecting to the brainstem and spinal cord. Alternatively, a continuum of dopamine neurons within the hypothalamus (A11, A12, A14, and A15 groups), centered around the arcuate nucleus (Arc) and adjacent hypothalamic/extrahypothalamic brain areas (e.g. preoptic region rostrally, zona incerta in dorsal extension and midbrain caudally, A8-11 and A13) is arranged such that spatial segregation codes for function determination5,6. However, the degree of molecular diversity amongst dopamine neurons remains unknown.
Here, we used single-cell RNA-seq in dissociated neurons from the medial hypothalamus to generate a comprehensive catalogue of neuronal identities. We reveal 62 neuronal subclasses in the conglomerate of hypothalamic nuclei sampled, many of which cluster uniquely through novel identity marks. We used a multiparametric approach aided by transgenic technologies to define four subtypes of dopaminergic neurons with one selectively expressing onecut-3 and neuromedin U receptor 2 (Nmur2). We then employed circuit mapping in intact tissues in combination with Ca2+ imaging to show that a neuromedin S-Nmur2 axis exists between glutamatergic suprachiasmatic nucleus (SCN) neurons and periventricular dopamine cells, thus resolving the long-standing debate about the identity of neurons and neuropeptide systems integrating dopamine output into the circadian circuitry7, 8.
Results
Diversity of cell types in the mouse hypothalamus
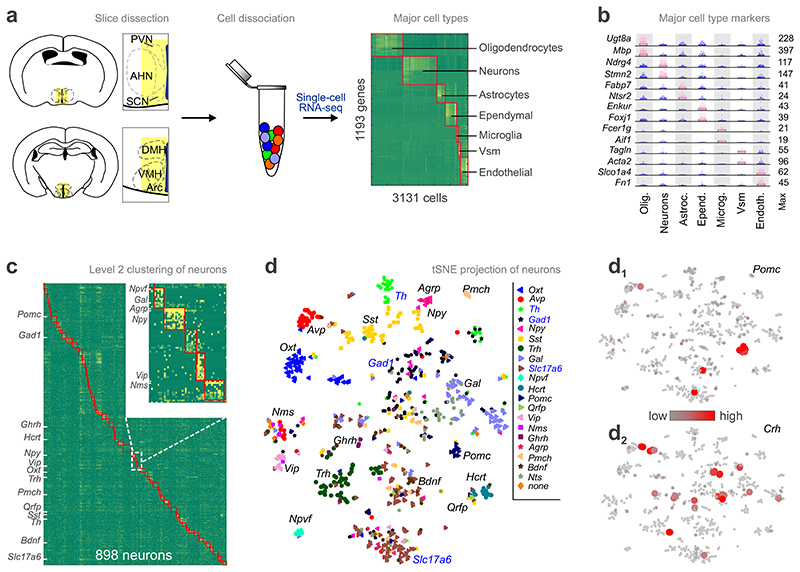
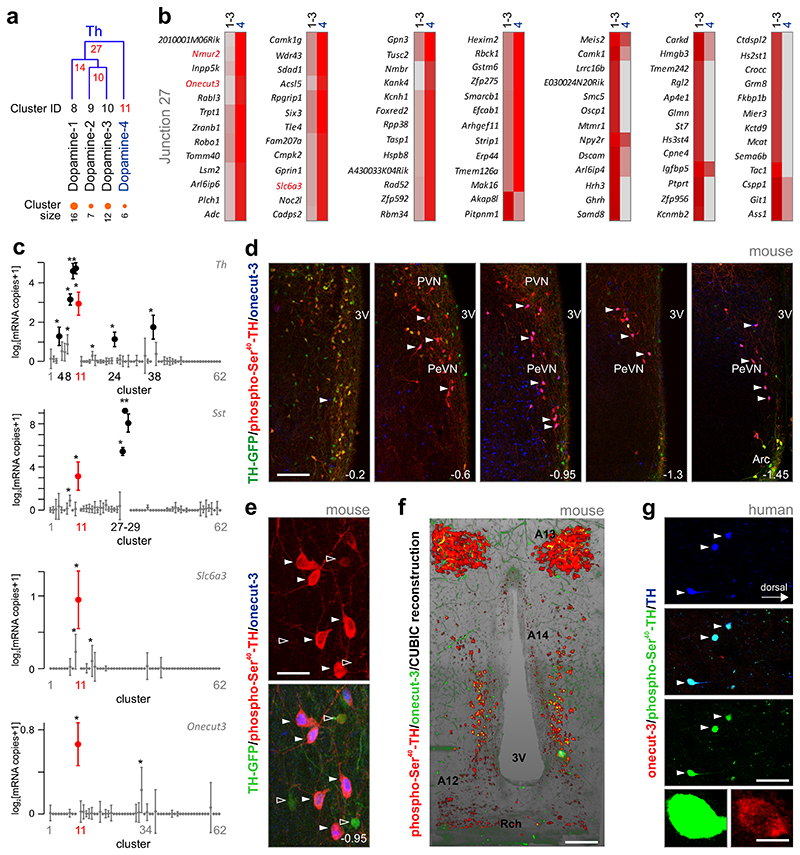
Single-cell RNA-seq data were obtained from 3,131 cells dissociated from a central column of the medial-ventral diencephalon (Fig. 1a), including in its rostrocaudal extent the posterior part of the preoptic area (Bregma = +0.5 mm) and in the caudal direction the Arc (Bregma = -2.1 mm). We relied on a divisive biclustering method using the BackSpinV2 algorithm9, which sorts cells into clusters of closest neighbors. Using lineage-specific known and new protogenes (see level 1 analysis, On-line Methods), this procedure allowed us to distinguish 7 major cell types (Fig. 1a,b). To determine neuronal subclass diversity, we performed 2nd level clustering on hypothalamic neurons (Fig. 1c). The analysis of 898 neurons classified these cells into 62 clusters with high-quality content (donors/cluster, detected genes/cell, total molecules detected, Supplementary Fig. 1a-c); prominently featuring neuropeptides (Fig. 1d-d2) and enzymes involved in neurotransmitter turnover.
Fig. 1. Cell-type diversity in the mouse hypothalamus.
(a) Workflow diagram to indiscriminately obtain intact single cells from the juvenile mouse hypothalamus and analyze them without selection. A central vertical column spanning the preoptic area and arcuate nucleus in its rostrocaudal extent (yellow shading) was dissected, enzymatically dissociated and processed for single-cell RNA-seq. Molecule counts from 3131 cells were first clustered to define main cell types. Heat-map shows blocks of genes enriched in each cell type. (b) mRNA transcripts specifying each of the 7 major cell types. Dot density plots depict means (bars) and individual values (circles). mRNA expression of selected known and novel markers. A marker pair is shown per class including a generally accepted and a lesser-known gene. Abbreviations: Astroc., astrocytes; Epend., ependymal cells; Microg., microglia; Olig., oligodendrocytes; Vsm, vascular and smooth muscle lineage; Endoth., endothelial cells. (c) Clustering of 898 neurons reveals unprecedented molecular diversity. Heat-map shows clustering results with blocks of clusters aligned along the diagonal axis and boxed in orange. Abbreviations in inset denote subtype-specific neuropeptides and hormones. (d) Visualization of hypothalamic neuron subtypes on a two-dimensional map using tSNE (1,194 genes, perplexity = 5, 200 principle components; see also Supplementary Figs. 3,4). Neurons were color-coded by highest expression of well-known, cluster-defining hypothalamic markers. (d1) Pomc mRNA expression is an example of phenotypic clustering on the same tSNE plot. (d2) In contrast, Crh is heterogeneously distributed on the tSNE scaffold, precluding its use to typify neurons. Abbreviations in (c,d): Agrp, agouti-related peptide; Avp, arginine-vasopressin; Bdnf, brain-derived neurotrophic factor; Gad1, glutamate decarboxylase 1; Gal, galanin; Ghrh, gonadotropin-releasing hormone; Hcrt, hypocretin; Nms, neuromedin S; Npvf, neuropeptide VF precursor; Npy, neuropeptide Y; Oxt, oxytocin; Pmch, pro-melanin-concentrating hormone; Pomc, pro-opiomelanocortin; Qrfp, RF(Arg-Phe)amide family 26 amino acid peptide; Slc17a6, vesicular glutamate transporter 2; Sst, somatostatin; Th, tyrosine hydroxylase; Trh, thyrotropin-releasing hormone; Vip, vasoactive intestinal polypeptide.
Cell-state changes and neuronal identity
The mammalian hypothalamus exhibits significant gender distinctions, including the number and content of neuronal subtypes10, 11. Previous genome-wide profiling studies aimed to resolve gender-related traits and linked those to specific behaviors12, 13. Even though we minimized the effects of sexual dimorphism by using mice before they reached the age of sexual maturation (postnatal days 21 ± 7), we used the molecular resolution of single-cell RNA-seq to test whether sex-determination exists in neuronal cohorts making up the mouse hypothalamus. Hierarchical clustering did not reveal major differences in neuronal heterogeneity between female and male hypothalamic counterparts (Supplementary Fig. 2a). This allowed us to pool data from both sexes. Nevertheless, several neuronal clusters were dominated by neurons from male (e.g., #55) or female (#6) subjects, suggesting, but not being causal to, gender-specific coding (Supplementary Fig. 2a).
Neuronal networks in the mammalian hypothalamus can change the predominance of their main fast neurotransmitter and/or neuropeptide signaling system within hours upon metabolic challenges14–16. For example, glutamate to GABA switches can occur due to the reorganization of synaptic circuits upon feeding16, 17. The ability of neurons to harbor mRNAs encoding enzymes and transporters indispensable for the production or release of multiple neurotransmitters may indicate an alternative mechanism of neuronal state-switches. Therefore, we asked whether acute stress by peripheral paraformaldehyde injection and sampling 6h later18 affected cluster dynamics. Our unbiased clustering rejected any change to cluster assignment, as this would argue for the rapid generation of novel neuronal subtypes upon stress (Supplementary Fig. 2b).
Neuronal heterogeneity in the hypothalamus
Classical views on neuronal diversity in the hypothalamus use neuropeptides19, and, less frequently, fast neurotransmitters as identifying marks for functionally specialized neuronal pools20. However, there is no consensus on, or direct correlation between, neurotransmitter-neuropeptide expression patterns.
Here, we first used t-dependent stochastic neighbor embedding (tSNE) to visualize our multi-dimensional dataset (Fig. 1d; Supplementary Information and Supplementary Fig. 3,4). By relying on a combination of neuropeptides (Supplementary Table 1), enzymes and neurotransmitter transporters that rate-limit synaptic signaling, we provide an unbiased list of neuropeptides and/or hormones that define neuronal subtypes by forming spatially segregated cell clusters (Fig. 1d). As such, proopiomelanocortin (Pomc gene) depicted a clustering example (Fig. 1d1). In contrast, the gene encoding corticotropin-releasing hormone (Crh) was not restricted to a neuronal subset (Fig. 1d2). Instead, our analysis suggest that many hypothalamic neurons, including both GABA and glutamate phenotypes (Supplementary Fig. 5a-d), can produce and secrete Crh under finite metabolic conditions (Fig. 1d2; Supplementary Fig. 5). Thus, singlecell RNA-seq allows to distinguish transcript patterns that encode homogeneity or, conversely, molecular differences.
Hierarchical clustering reveals neurotransmitter-neuropeptide relationships
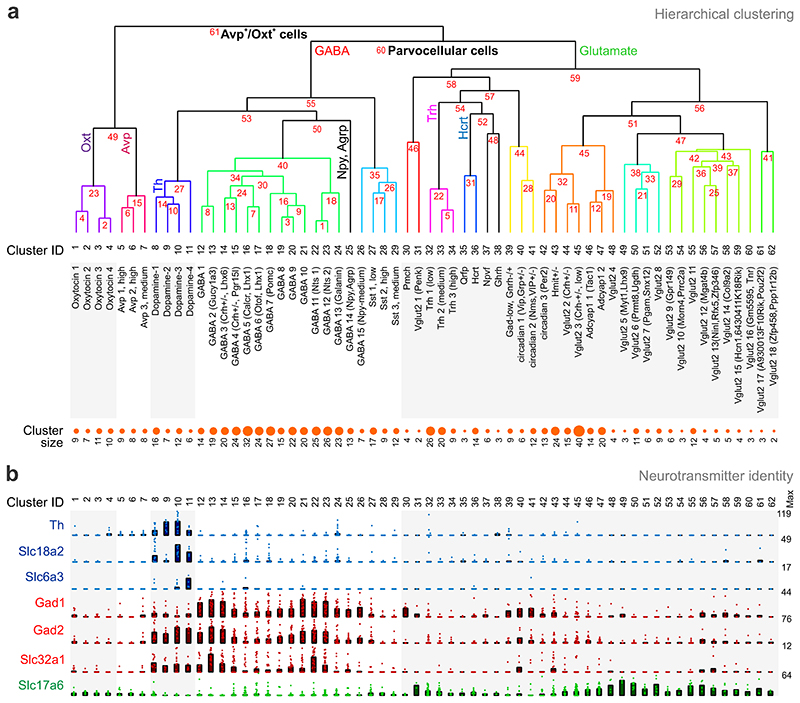
Next, we filtered our single-cell RNA-seq data 9 to distinguish stable neuronal clusters whose borders and marker gene homogeneities were manually confirmed (Supplementary Table 2). We produced a hierarchical dendrogram (Fig. 2a), which segregated neurotransmitter and neuropeptide-containing cells into subclasses (junction points and their specificities are listed in Supplementary Table 3). Based on the most abundant gene marks (“top five filtering”, Supplementary Table 3), we introduced a specific terminology that reflects differential gene expression related to neurotransmitters, neuropeptides and function determination (e.g. circadian rhythm) (Fig. 2a). Neurotransmitter identities included glutamatergic (clusters #1-7 and #30-62), dopaminergic (#8-11) and GABAergic (#12-29) cells. Notably, both the glutamate mark Slc17a6 and the dopamine mark Th were of low abundance in oxytocin (Oxt) and vasopressin (Avp)-containing neuroendocrine clusters (#1-7; Fig. 2b), suggesting that these cells under normal conditions may primarily act via their respective hormones. The significance of Th and tyrosine hydroxylase protein in these neurons remains unclear since they appear to lack the gene for aromatic amino-acid decarboxylase (AADC, for Ddc and other genes see Supplementary Table 2 and http://linnarssonlab.org/hypothalamus), the enzyme needed for dopamine synthesis. Subsequent differential mRNA expression analysis subdivided Oxt and Avp neurosecretory cells, which project to the posterior lobe of the pituitary4, into 3 and 4 subtypes, respectively (Supplementary Table 3).
Fig. 2. Hierarchical clustering of hypothalamic neuron subtypes.
(a) Classification of 62 neuronal subtypes as defined by unique molecular fingerprints (i.e. top five genes). Numbers in red indicate divergence points defined by the BackSpinV2 algorithm. We adopted a terminology that relies on neurotransmitter and subtype-specific gene sets. Cluster size indicates the number of neurons assigned to a specific subtype. (b) Neurotransmitter specificity in each neuronal subtype was defined by their expression of tyrosine hydroxylase (Th), vesicular monoamine transporter 2 (Slc18a2) and dopamine transporter 1 (Slc6a3) for dopaminergic neurons, glutamate decarboxylase 1 and 2 (Gad1/2) and vesicular GABA transporter (Slc32a1) for GABAergic neurons, and vesicular glutamate transporter 2 (Slc17a6) for glutamatergic neurons. Note the existence of dual neurotransmitter phenotypes. Dot density plots show mean expression per cluster × percentage of cells (bars), and individual values (circles). Numbers to the right indicate the maximum number of molecules for each gene, thus providing ranges from 0-to-maximum value.
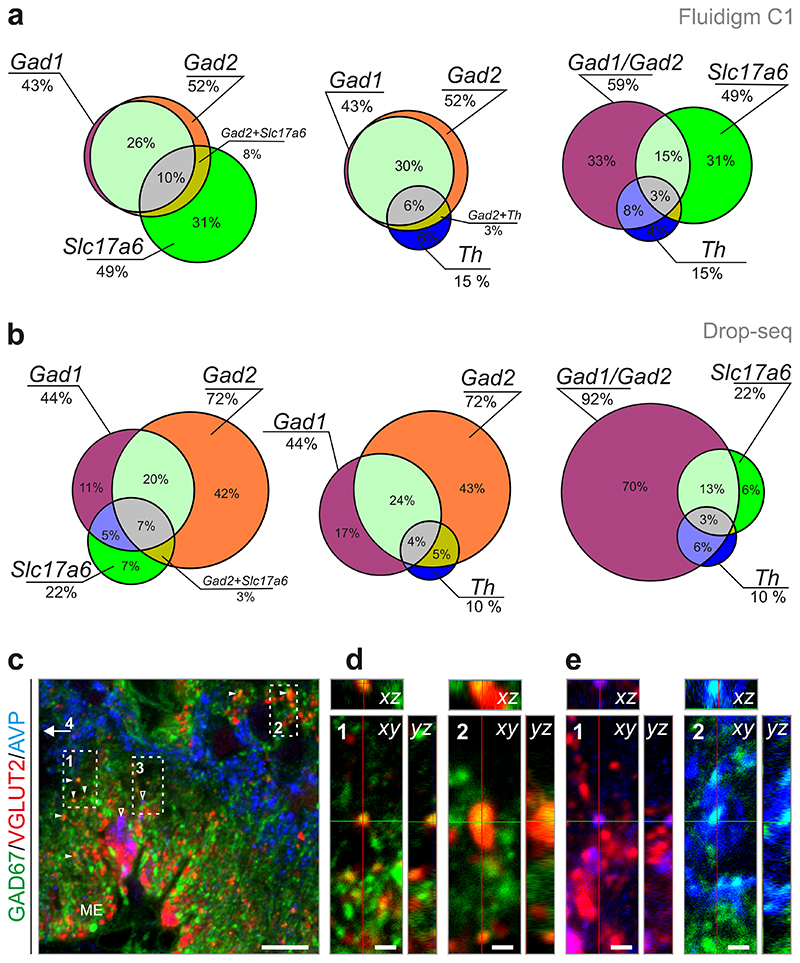
In comparison, non-magnocellular cells present a more heterogeneous trait, and can be separated into “GABAergic” (Gad1/Gad2/Slc32a1) and “glutamatergic” (Slc17a6) subtypes20. GABA neurons varied in their expression levels of Gad1 and Gad2, encoding GAD67 and GAD65, respectively (Fig. 2b). This was particularly notable for clusters #8-11 cells (with low Gad1 and high Gad2) which also harbored Th, Slc18a2 (vesicular monoamine transporter 2, VMAT2) and in some cases, Slc6a3 (encoding dopamine transporter 1, DAT), qualifying them as dopamine neurons and suggesting the existence of dual dopamine/GABA neurotransmitter phenotypes (Fig. 3a; see also 21). Our hierarchical clustering approach also separated 33 clusters of mostly “glutamatergic” modalities (#28-62). Some of these neuronal subtypes also contained Gad1/2 and/or Slc32al mRNAs. For further validation we performed an additional sequencing study using “Drop-seq” as another technical platform22. In 220 neurons, we recaptured subsets of GABA, glutamate and dopamine cells that contained genes for alternative neurotransmission (Fig. 3b). Importantly, and despite the different proportion of GABAergic, glutamatergic and dopaminergic cells in the pool of neurons sampled by “Drop-seq”, we found strikingly similar numbers of cells that exhibited the ability of dual neurotransmission (Fig. 3b). Lastly, we employed multiple immunofluorescence labeling to show the co-existence of VGLUT2 and GAD67 proteins in nerve endings at the median eminence (ME) (Fig. 3c,d). Likewise, we revealed that even minute mRNA copy numbers can translate into meaningful biological modalities by showing the co-existence of vasopressin and VGLUT2 (Fig. 3e) and GAD67 (Fig. 3e). Quantitatively, we confirmed by 3-dimensional laser-scanning microscopy that 36 terminals contained both VGLUT2 and GAD67 out of 790 VGLUT2+ (~4.6%) or 1,221 GAD67+ (~3.0%) nerve endings localized within identical surface areas in the ME. In sum, these data reinforce our conclusion that hypothalamic neurons might either co-release neurotransmitters, segregate them to subsets of synapses or switch between modes of neurotransmission in response to specific inputs, thus allowing fast output modulation 16, 17.
Fig. 3. Neurotransmitter phenotypes in hypothalamic neurons.
(a,b) Venn diagrams showing the proportion of dual and triple neuronal phenotypes in GABA/glutamate (left), GABA/dopamine (middle) and GABA/glutamate/dopamine (right) neurons after Fluidigm C1 (a) or Drop-seq (b) sequencing. Percentage values indicate the proportion of neurons falling into groups and intersections (“dual phenotype” categories). (c-e) Histochemical validation of neurotransmitter heterogeneity in axon terminals at the median eminence (ME) showing GAD67/VGLUT2 (d) and vasopressin (AVP)/GAD67, as well as AVP/VGLUT2 co-existence. Orthogonal image stacks are shown. Scale bars = 10 μM (c), 2 μM (d,e).
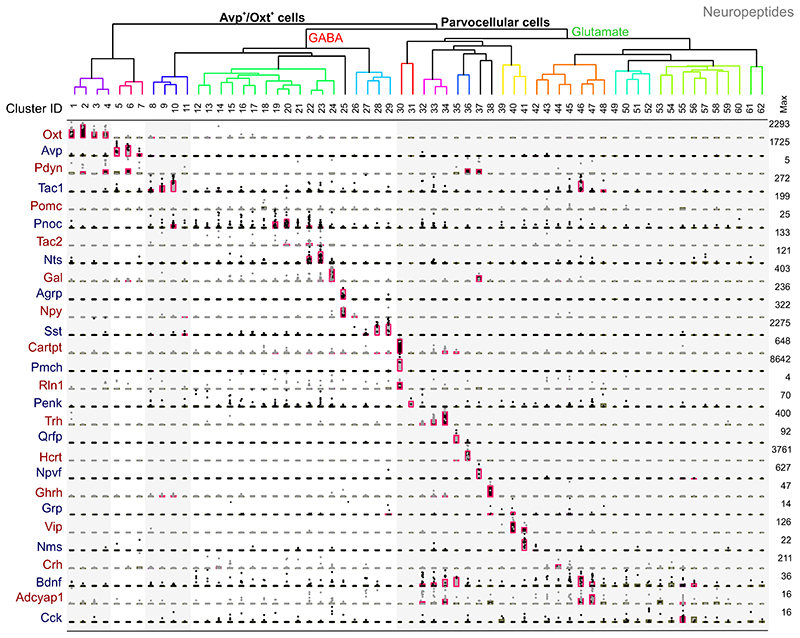
Owing to the hypothesis that neuropeptides can modulate the action of fast neurotransmitters, as well as being signal-competent on their own23, we established neurotransmitter/neuropeptide relationships. To this end, we determined the total number of mRNA molecules for the top 52 known neuropeptide “hits” (Supplementary Table 1), which ranged from 0 to >30,000 per neuronal subtype. As Fig. 4 shows, Oxt and Avp neurons frequently contained the Pdyn gene (encoding prodynorphin). Dopamine neurons chiefly co-expressed the genes for tachykinin 1 (Tac1) and nociceptin (Pnoc). For GABA neurons, clusters #12-17 (“GABA1-6 subtypes”) were relatively sparse in neuropeptide genes (in particular, Crh [clusters #14,15] and Tac2 [clusters #13,15]). In contrast, subclusters amongst #18-29 groups were separated by primary gene expression for neuropeptide contents, including Pomc (#18), Pnoc (#19,20), Tac2 (#20-24), Nts (#22,23), Gal (#24), Agrp (#25), Npy (#25,26) and Sst (#27-29) mRNAs (Fig. 4, Supplementary Table 4). Lastly, glutamatergic neurons showed an apparent variability in their gene expression for neuropeptides since clusters #33-35 contained Trh, Bdnf and Adcyap1 mRNAs. In contrast, clusters #57-62 exhibited only marginal (if any) expression of genes for polypeptides (particularly Cck) (Supplementary Table 4). However, even if clusters were closely placed, their differential expression profiles revealed specificity in individual and/or the combinatory expression of genes: #62-enriched genes included Zfp458, Ppp1r12b, whereas neurons in #61 were enriched in Pou2f2 (Supplementary Fig. 6a and Tables 2,3). For clusters #22,#23 we note the differential expression of the hypocretin receptor 1 and 2 genes (Hcrtr1, Hcrtr2), establishing their molecular divergence (http://linnarssonlab.org/hypothalamus). Cumulatively, these data advance existing knowledge on the molecular heterogeneity of dopamine, GABA and glutamate neurons in the hypothalamus and allow for inferences be made on the positioning, circuit recruitment and signal diversity of specific neuronal subtypes (for novel subtype-specific markers see Supplementary Fig. 6a).
Fig. 4. Neuropeptide associations to individual hypothalamic neuronal subtypes.
Neuronal subtypes were clustered as in Figure 2. Vertical shaded columns denote main neuropeptide mRNA contents per neuronal subclass as in Figure 2, including level of statistical significance at q < 0.05 (pink). Dot density plots show mRNA expression levels per cell. Abbreviations: Adcyap1, adenylate cyclase activating polypeptide 1; Agrp, agouti-related peptide; Avp, arginine-vasopressin; Bdnf, brain-derived neurotrophic factor; Cartpt, cocaine and amphetamine regulated transcript prepropeptide; Cck, cholecystokinin; Crh, corticotropin-releasing hormone; Gal, galanin; Ghrh, growth hormone-releasing hormone; Grp, gastrin-releasing peptide; Hcrt, orexin/hypocretin; Nms, neuromedin S; Npvf, Neuropeptide VF precursor; Npy, neuropeptide Y; Nts, neurotensin; Oxt, oxytocin; Pdyn, prodynorphin; Penk, proenkephalin; Pmch, pro-melanin-concentrating hormone; Pomc, pro-opiomelanocortin; Pnoc; prepronociceptin; Qrfp, pyroglutamylated RFamide peptide; Rin1, Ras and Rab interactor 1; Sst, somatostatin; Tac1, substance P (tachykinin 1); Tac2, tachykinin 2; Trh, thyrotropin-releasing hormone; Vip, vasoactive intestinal polypeptide.
Molecular validation through novel neuropeptide marks
Our single-cell RNA-seq data allowed the identification of many novel markers with a high degree of subclass selectivity (Supplementary Fig. 6a, Supplementary Tables 5,6). In addition, we also identified combinations of genes whose co-expression patterns predicted individual subclusters with a high degree of accuracy (e.g., Shf alone vs. Shf + Sfrp2 or Dlx1 alone vs. Dlx1 + Zfp865). Based on the cluster marks used (Supplementary Fig. 6, 7a), we identified two novel neuronal subtypes specifically expressing the gene for either pyroglutamylated RF amide (Qrfp; Supplementary Fig. 6b, b1) or neuropeptide VF (Npvf; Supplementary Fig. 6c, c1) 24, 25. Accordingly, Qrfp- and Npvf-expressing hypothalamic neurons are both glutamatergic and segregate into clusters #35 and #37, respectively (Supplementary Fig. 6b1, c1). Moreover, we resolved that Qrfp+ and Npvf hypothalamic neurons co-express the genes for orexin/hypocretin (Hcrt; Supplementary Fig. 6b) and galanin (Gal; Supplementary Fig. 6c), respectively. Thus, and also considering that both Qrfp and Npvf qualify as cell identity marks (Fig. 2a), we assign these neurons as bona-fide excitatory neuropeptidergic subtypes.
Expression-based prediction of a novel dopamine neuron subtype
Single-cell RNA-seq of dissociated tissues provides compelling information on the molecular make-up of particular cell lineages, and allows hypotheses to be formulated regarding functional divergence, particularly neuronal circuit wiring through e.g. transmitter-receptor relationships. However, the often small cluster size (< 10 cells) and the lack of positional information per se could render single-cell RNA-seq data liable to false positive outcomes. Therefore, we set out to validate our data by choosing the predicted 4th subtype of dopamine neurons (cluster #11; “dopamine-4 cells”), which despite its small sample size (6 neurons) segregated from other dopamine clusters in our predictive model (Fig. 5a).
Fig. 5. Molecular interrogation of dopamine neurons defines a onecut-3-expressing periventricular subtype.
(a) Segregation of tyrosine hydroxylase (Th)-expressing neurons into 4 subtypes, as defined by their divergent expression of top 5 genes (see also Figure 2). Note that dopamine neuron subtype #4 segregates early from the other subclasses. On the top, numbers in red indicate divergence points (junctions) defined by dendrogram construction. At the bottom, neuronal cluster sizes (number of cells) are shown. (b) Differential gene expression profile of dopamine neuron subtypes 1-3 (cumulative) vs. subtype 4. mRNA transcripts in red, including onecut-3 (Onecut3) define subclass identity. Incremental mRNA expression was color coded from grey (no detectable expression) to deep red. (c) Selective co-expression (in red) of Th, Slc6a3, onecut-3 and somatostatin in dopamine-4 subclass (cluster #11). Data are presented as ‘power 1 ‘ error-bar plots. Note that data exceeding >2x s.e.m. for Th and somatostatin in other neuronal clusters are shown in black. Statistics: *q < 0.05 (Wilcoxon rank-sum test corrected for multiple testing). (d) Distribution of phospho-Ser40-TH+/onecut-3+ neurons at select anterior-posterior coordinates (relative from Bregma 50) in the hypothalamus of Th-GFP reporter mice. Note that multiple-labeled neurons concentrate (arrowheads) in the periventricular hypothalamic nucleus (PeVN). (e) High-resolution photomicrograph depicts PeVN dopamine neurons co-expressing phospho-Ser40-TH and onecut-3 (solid arrowheads) with elaborate dendrite morphologies coursing in parallel with the ventricular wall. Open arrowheads denote Th-GFP neurons not containing onecut-3 signal (see also Supplementary Fig. 7d). (f) In-plane rendering of lightsheet microscopy reconstruction of the anterior-posterior distribution of dopamine subclass 4 (yellow label) along the wall of the third ventricle (see also Supplementary Video 1 and 2). Abbreviations: A13, zona incerta dopamine neurons, A14, PeVN dopamine neurons 27, A12, arcuate nucleus dopamine neurons 5; 3V, third ventricle; PVN, paraventricular nucleus, Rch, rethrochiasmatic area. (g) A molecularly equivalent dopamine neuron subclass exists in the human hypothalamus with co-existence of phospho-Ser40-TH and onecut-3. Scale bars = 300 μM (d), 160 μM (f), 70 μM (e,g), 7 μM (g, inset).
First, we compared fractional gene expression (Fig. 5b), as well as average amplitude expression (Supplementary Table 3) between dopamine subtypes 1-3 vs. 4 to reveal if any gene can be used to hallmark this predicted dopamine subclass. Differentially-expressed genes selectively enriched in dopamine-4 cells included those for the transcription factor onecut-3 (Onecut3), Nmur2, Nmbr, Robo1, Slc6a3, K+ channels (Kcnh1), secretory proteins (Cadps2), genes associated to intracellular signaling (Rabl3, Gpn3, Plch1), cytoskeletal dynamics (Kank4, Gprin1), un-annotated genes (2010001M06Rik, A430033K04Rik) and Sst (Supplementary Table 3). In turn, these cells lacked neuropeptides that were abundant in other dopamine subclasses like Ghrh1 and Tac1 (Fig. 5b), and partly Npy2r, the gene encoding neuropeptide Y receptor Y2. Subsequently, we broadened our specificity testing to all 62 predicted neuronal clusters and show that, as opposed to redundant Th and Sst expression, both Slc6a3 (DAT) and Onecut3 were predominantly found in this particular cell population (Fig. 5c). Thus, coincident DAT and onecut-3 localization could reveal if these neurons are spatially segregated from or intermingled with other dopamine cells.
Next, we explored the distribution of onecut-3 and DAT in hypothalamic dopamine neurons using a combination of immunohistochemistry (Fig. 5d) and mouse genetics, particularly in Th-GFP and Dat1-Cre::ROSA26SorGt:Tomato: mice, allowing life-long expression profiling (Supplementary Fig. 7b-d1). Coincident detection of onecut-3 and enzymatically-active TH phosphorylated at residue Ser40 26 on a Th- GFP background revealed that these dopamine neurons populate the periventricular nucleus of the hypothalamus (Fig. 5d), with their dendritic trees being primarily oriented vertically (Fig. 5e). By using serial sections (Fig. 5d) and reconstructive histochemistry in optically-cleared tissues (Fig. 5f, Supplementary Fig. 7e, Supplementary Movies 1-3), we show that onecut-3+ dopamine neurons form a cell continuum spanning the SCN-Arc domain with an increasing cell gradient caudally, thus qualifying as “A14” dopamine neurons6, 27. Lightsheet microscopy revealed 474 cells in an adult mouse brain (Fig. 5f, Supplementary Movies 1,2), while conventional neuroanatomy in serial sections projected 500-1,000 cells/brain. Notably, these neurons segregate dorsally from other dopamine cells residing in the Arc (Fig. 5d, f, Supplementary Movie 2). When quantifying GFP intensity in these TH+ cells, we find that their GFP signal is significantly lower than that of other TH+ cell groups (Supplementary Fig. 7c), while their phospho-Ser40 levels are distinctly elevated (Supplementary Fig. 7c). These data reinforce our single-cell RNA-seq results, which demonstrated low Th mRNA copy numbers in dopamine-4 neurons (Fig. 5c). Lastly, we studied post-mortem human hypothalami (Fig. 5g) to show that a neuronal cohort positioned proximal to the 3rd ventricle possesses a molecular make-up of onecut-3/phospho-Ser40-TH reminiscent of that in mouse, suggesting that our differential classification of dopamine neurons may be broadly relevant to defining neuronal diversity in the hypothalamus of various mammals.
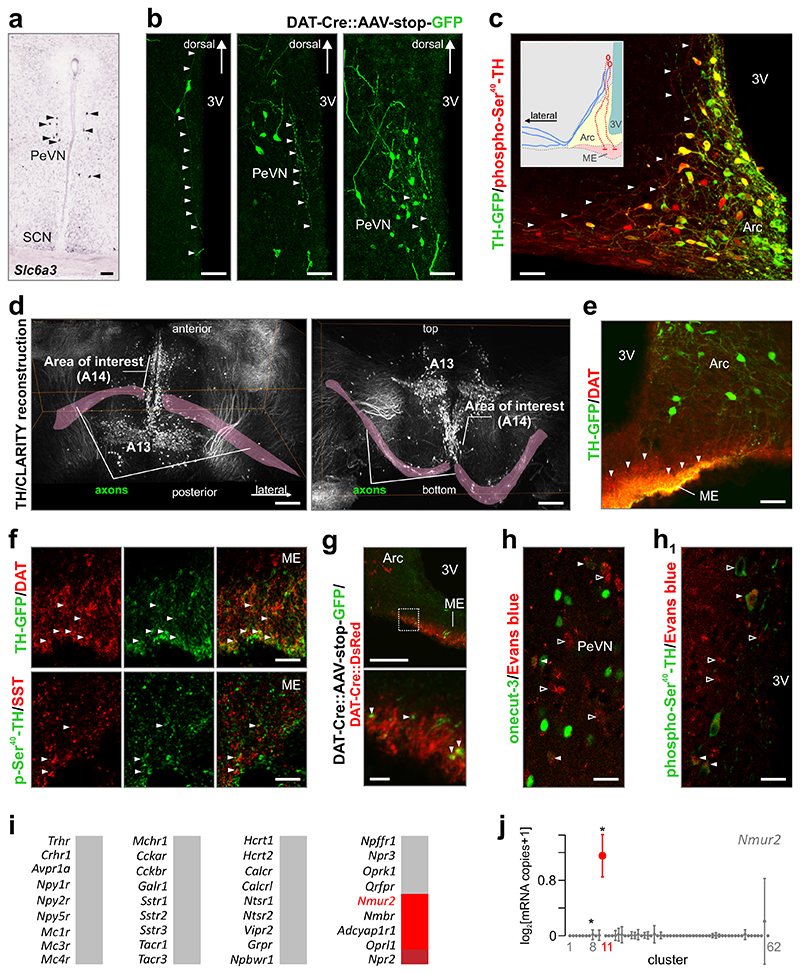
Morphological stratification of dopamine neurons in intact brains
We show that dopamine-4 neurons (A14, cluster #11) express the gene for DAT (Slc6a3; Fig. 6a), contrasting ventrolateral Arc/A12 neurons, but being in Slc6a3 expression similar to the dorsomedial A12 group 28. The latter neuronal pool also contains GAD21 and is a source of dopamine inhibiting prolactin release 29. The expression of DAT (Fig. 6a) could help A14, like dorsomedial Arc A12, neurons to regulate ambient levels of dopamine30. This feature also allowed us to combine advanced optical imaging and mouse genetics: we used AAV-driven and Cre-dependent GFP expression in Dat1 -Cre mice to address intrahypothalamic projection sites for dopamine-4 neurons (Fig. 6b). GFP distribution revealed ventrally-running fine processes with pearl-lace like appearance, likely axons. Most of these processes showed a detour around the Arc and coursed laterally as also suggested by the differential expression of phospho- Ser40-TH in processes putatively originating in A14 dopamine-4 neurons (Fig. 6c). Moreover, histochemical detection of TH using COLM technology demonstrated that most TH+ axons leaving the A14 area project laterally using the supraoptic decussation and upper lateral pathway and terminate outside the hypothalamus, likely in basolateral and central amygdaloid nuclei (Fig. 6d, Supplementary Fig. 7e, Supplementary Movie 3). Notably, the median eminence (ME) is a site where DAT immunoreactivity accumulates (Fig. 6e), including sporadic co-localization with Sst in terminal-like specializations (Fig. 6f), pinpointing the ME as a potential release site for A14 neurons. Indeed, viral GFP transduction in Dat1-Cre mice confirmed the presence of ME-oriented projections and likely terminals (Fig. 6g). These data cumulatively suggest that A14 dopamine neurons might, at least in part, be classified as “neuroendocrine cells” releasing their dopamine content directly into the portal circulation (not excluding projections to the intermediate lobe of the pituitary6, 31. We tested this hypothesis by peripheral Evans blue (EB) dye loading, since circulating EB does not cross the blood brain barrier and only accumulates in central neuronal somata that have access to the peripheral circulation. 24-48 h after dye-loading (i.p.), we found a subset of onecut-3+ or phospho-Ser40-TH+ neurons in the periventricular nucleus to accumulate EB (Fig. 6h,h1). Thus, we suggest that A14 dopamine neurons are bona fide neurosecretory cells (see also31), which are integrated into (extra-) hypothalamic neuronal networks. These data also recapitulate the known dichotomy of hypothalamic dopamine neurons releasing their contents either at the ME into the hypophyseal portal system 29 or using DAT to remove superfluous dopamine at central synapses.
Fig. 6. Efferent projections of periventricular onecut-3+ dopamine neurons.
(a) Dopamine transporter (Slc6a3) localization by in situ hybridization (www.brain-maps.org) reveals a Slc6a3+ cell cluster in the periventricular nucleus (arrows). (b) Microinjection of AAV-stop-GFP viruses into the periventricular region of Dat1-Cre mice reveals the morphology of periventricular Dat1+ neurons, including their ramifying dendrites and axons primarily running ventrally and, in some cases, laterally. (c) By exploiting the association of exceptionally high phospho-Ser40-TH expression and low GFP fluorescence to distinguish periventricular neurons (see also Supplementary Fig. 7c), their phospho-Ser40-TH+ axons (arrowheads) were confirmed to commute towards the median eminence (centrally, red in inset) as well as laterally (blue in inset). (d) Lightsheet microscopy optimized for CLARITY reconstruction 51 of TH immunoreactivity with a focus on axons (magenta overlay) emanating from A14/A12 cells. Note that lateral projections course towards amygdaloid nuclei. Horizontal (left) and vertical (right) views are shown, revealing an axonal detour around the ventromedial and lateral hypothalamic nuclei. (e) DAT immunoreactive terminals at the median eminence (ME) of adult mice (arrowheads). (f) Co-localization of DAT, but not somatostatin (SST), and GFP fluorescence (arrowheads) at the ME in Th-GFP mice. (g) Our Dat1 -Cre::AAV-stop-GFP viral labeling approach confirmed that DAT+ terminals (arrowheads) at the ME could originate, at least in part, in the periventricular nucleus (see bottom image for high-resolution) (h,h1) Systemic Evans blue administration led to its uptake by neurons residing in the periventricular nucleus and immunoreactive for onecut-3 (H) or phospho-Ser40-TH (h1). Solid arrowheads pinpoint co-localization, while open arrowheads denote neurosecretory cells whose identity was not pursued in this study. (i) Single-cell RNA-seq analysis of the receptor repertoire of onecut-3+ dopamine neurons. Incremental mRNA expression was color coded from grey (no significant expression) to red (q < 0.05). (j) Dopamine subclass 4 (cluster #11) predominantly expresses neuromedin U receptor 2 (Nmur2). *q < 0.05 (Wilcoxon rank-sum test corrected for multiple testing). Scale bars = 180 μm (d), 100 μm (a,b,g top), 70 μm (c), 40 μm (f,h, h1), 10 μm (g bottom). Abbreviations: 3V, third ventricle; Arc, arcuate nucleus; PeVN, periventricular nucleus; SCN, suprachiasmatic nucleus.
Dopamine-4 neurons are entrained by neuromedin S
Next, we explored whether dopamine-4 cells (cluster #11) receive specific afferentation that could contribute to their function. To this end, we mapped the receptor repertoire of dopamine-4 neurons by screening the expression of 36 neuropeptide receptors found by single-cell RNA-seq. We show that dopamine-4 cells abundantly expressed Nmur2 (neuromedin U receptor 2), Nmbr (neuromedin B receptor), Adcyap1r1 (adenylate cyclase activating polypeptide 1 receptor type I) and Oprl1 (opiate receptor-like 1) (Fig. 6i). Whereas Adcyap1r1 and Oprl1 are promiscuously expressed in most neuronal clusters (data not shown), Nmur2 and Nmbr are highly specific for dopamine-4 neurons (Fig. 5a, 6j).
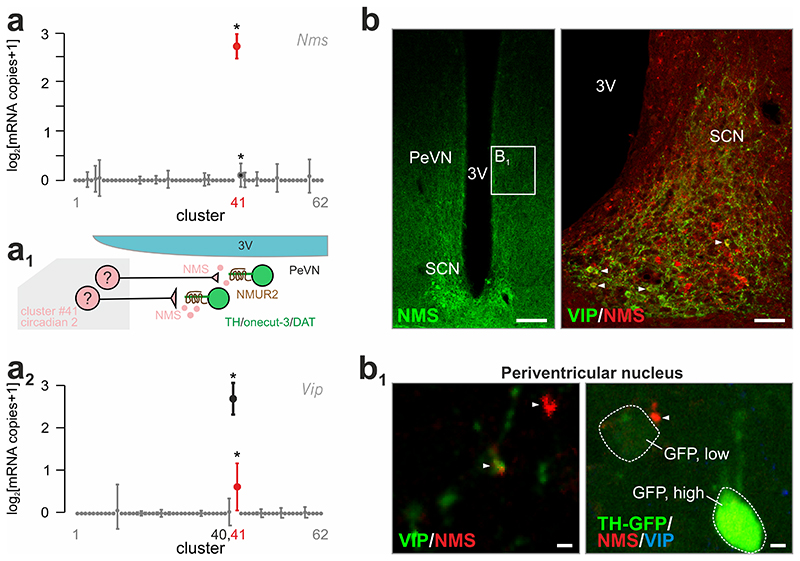
Neuromedin S is the endogenous agonist at neuromedin U receptor 2, with its production being concentrated in the SCN32. Indeed, our single-cell RNA-seq confirmed that neuromedin S (Nms) is present in all cells of hypothalamic glutamatergic neurons that formed cluster #41 (Fig. 7a). The combination of RNA and positional information allowed us to predict an intrahypothalamic network to tune periventricular dopamine-4 output through the release of neuromedin S (Fig. 7a1). It is noteworthy that neuromedin S-containing neurons in the SCN co-express Vip (42%) (Fig. 7a2). Meanwhile, other Vip+ neurons that, however, were Grp+/- and Nms- clustered separately (Fig. 7a,a2, Supplementary Fig. 8a,b). Subsequent histochemistry validated these data by spatially confining neuromedin S production to the SCN (Fig. 7b) 32. Moreover, the partial co-localization of neuromedin S with VIP (Fig. 7b,b1) and VIP and GRP within the SCN and in apposition to its periventricular targets (Supplementary Fig. 8c) lent further support to our RNA-seq-based prediction of neuronal network wiring. Finally, we used high-resolution laser-scanning microscopy to show that onecut-3+/TH+ neurons (note that we focused on particularly low TH-driven GFP levels in accord with Supplementary Fig. 7c) receive neuromedin S+ input (Fig. 7b1). Thus an SCN-periventricular nucleus intrahypothalamic network can rely on neuromedin S as neuropeptide modulator. Nevertheless, our histochemical data do not exclude the possibility that dopamine-4 neurons in the periventricular nucleus might receive neuromedin S-containing inputs of extrahypothalamic origin.
Fig. 7. Suprachiasmatic origin for neuromedin S inputs to onecut-3/Nmur2+ A14 dopamine neurons.
(a) Single-cell RNA-seq revealed highest neuromedin S (Nms) expression by cluster #41 (“circadian 2”) in the mouse hypothalamus. *q < 0.05 (Wilcoxon rank-sum test corrected for multiple testing). (a1) This ligand-receptor relationship allowed us to suggest a wiring diagram in which Nms- containing neurons of the suprachiasmatic nucleus can selectively innervate NMUR, TH, DAT and onecut-3-co-expressing periventricular dopamine neurons. (a2) Likewise, neurons in cluster #41 co-expressed vasoactive intestinal polypeptide (Vip). (b) Neuromedin S detection by histochemistry in the suprachiasmatic nucleus (SCN). Panel on the right shows neuromedin S and VIP co-existence in the SCN (solid arrowheads pointing to yellow composite color). Open rectangle denotes the general localization of images shown in B1. (b1) High-resolution analysis showed NMS+ boutons (solid arrowheads; left) in the periventricular area, which preferentially terminated in close apposition to dopamine neurons (arrowhead; right) marked by low GFP expression in Th-GFP reporter mice. Scale bars = 100 μm (b, left), 40 μm (b, right), 6 μm (b1). Abbreviations: 3V, third ventricle; PeVN, periventricular nucleus.
Neuromedin S+ neurons of the circadian pacemaker might modulate dopamine-4 cells
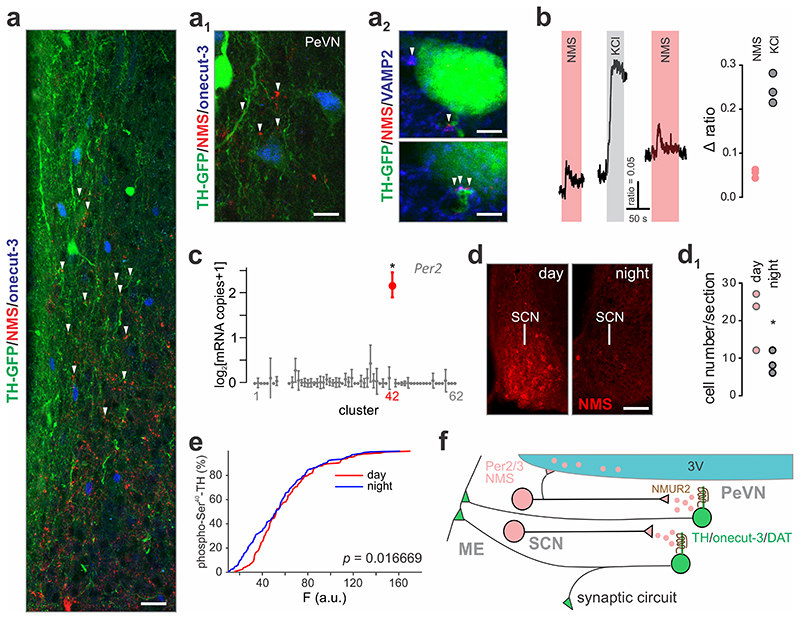
The presence of neuromedin S immunoreactivity in the periventricular nucleus prompted us to test whether this neuropeptide could affect dopamine-4 neurons through intercellular signaling. Therefore, we first mapped if neuromedin S co-exists with ubiquitous members of the presynaptic SNARE release machinery. Indeed, we find that neuromedin S co-localizes with vesicle-associated membrane protein 2 (VAMP2), marking presynapses, in close apposition to predominantly proximal dendrites of TH+/onecut-3+ periventricular neurons (Fig. 8a-a2). Subsequently, we deployed Ca2+ imaging with FURA2-AM to show that a subset (~7%) of periventricular dopamine (Th-GFP+) neurons exhibited resolvable Ca2+ responses upon acute wash-in of neuromedin S (500 nM) and using KCl as positive control (Fig. 8b).
Fig. 8. Periventricular onecut-3+ dopamine neurons respond to neuromedin S produced during light periods.
(a,a1) Immunohistochemical identification of synaptic contacts containing neuromedin S, which innervate onecut-3+ A14 neurons. (a2) VAMP2 was used as a ubiquitous presynaptic marker; perisomatic terminals are shown. (b) Bath application of 500 nM neuromedin S (NMS) leads to the generation of Ca2+ responses in a subset of periventricular TH+ cells, which was comparable to depolarization by 55 mM KCl. (c) The neuronal cluster containing neuromedin S (#42) also expresses the circadian pacemaker gene, Per2. Bold symbol denotes significant expression (> 2x s.e.m. from baseline); *q < 0.05. (d) Circadian fluctuations in neuromedin S content in the suprachiasmatic nucleus (SCN) as detected histochemically. (d1) Quantitative neuromedin S histochemistry using perisomatic fluorescence analysis on SCN neurons (n = 3 animals/group). *p < 0.05. (e) Circadian dependence of tyrosine hydroxylase phosphorylation at Ser40 as revealed by quantitative histochemistry (n = 4 animals/group). Cumulative distribution of function is shown. *p = 0.0167 (two-sample Kolmogorov-Smirnov test). (f) Synaptic wiring of a circadian pacemaker network regulating dopamine release from A14 neurons in the periventricular nucleus of the hypothalamus. Note that both synaptic and volume transmission mechanisms for neuromedin S modulation of dopaminergic output at the median eminence (ME) might exist. Scale bars = 20 μm (a), 10 μm (a1), 5 μm (a2, top), 3 μm (a2, bottom), 50 μm (d).
Even though the exact physiological role of neuromedin S is still debated, earlier data implicates it in circadian regulation 32. Indeed, our single-cell RNA-seq data showed that 42% of Nms+ cluster #41 coexpressed the period circadian clock-3, Per333 (Supplementary Fig. 8e). Likewise, 25% of neuromedin S+ neurons in cluster #41 co-expressed the “primary pacemaker” period circadian clock-2, Per234. Significantly, Per2 was also expressed in cluster #42 (Fig. 8c, Supplementary Fig. 8f), where 23% of Per2+ neurons express Nms (Supplementary Fig. 8d). These data predict that neuromedin S production might undergo circadian fluctuation. Therefore, we tested neuromedin S content in the SCN during day/night conditions (Fig. 8d), and confirmed circadian rhythm-driven oscillations in neuromedin S levels (Fig. 8d1).
If neuromedin S production is under circadian regulation and, as suggested by our data, in a position to act on dopamine-4 neurons, then the latter might be expected to show phase shifts. Therefore, we monitored TH phosphorylation at Ser40, known to mark increased enzymatic activity and dopamine production 26. This mark showed a distinct difference in the rising slope of cumulative distribution of function (CDF; p = 0.0167), suggesting different levels of active TH enzyme at day and night periods (Fig. 8e). Overall, these data suggest that periventricular dopamine output is coupled to the circadian clock through the recruitment of a neuromedin S-containing excitatory projection between the SCN and periventricular nucleus 7.
Neuromedin S release sites
Besides their release at central synapses, some neuropeptides undergo regulated release into the cerebrospinal fluid (CSF) to mediate volume transmission35, 36. We find neuromedin S+/VAMP2+ inputs to TH+/onecut-3+ neurons concentrated mostly in the rostral subdivision of the periventricular nucleus, positioned above the SCN. Since we could not find a similar gradient of Nmur2 expression (Allen Brain Atlas, experiment #81600118), neuromedin S release into the CSF would reconcile this difference and provide an alternative means of long-distance signaling. Unexpectedly, we identified a significant number of neuromedin S+ nerve endings along the wall of the third ventricle at the level of the anterior part of the median eminence (Supplementary Fig. 8g). These data, together with the recent identification of neuromedin S in human CSF 35, suggest that a release site might exist to allow volume transmission as an alternative to synaptic/non-synaptic communication within the periventricular nucleus (Fig. 8f).
Discussion
Our knowledge of neuronal subtypes in the neocortex, hippocampus and cerebellum, together with the origins and layer distribution of synaptic inputs 9, 37, 38 led to robust structure-function studies that uncovered key cellular substrates of higher cognitive and motor functions. In contrast, the study of brain regions that contain many functionally distinct, but positionally interspersed, neuronal subtypes in small volumes, such as the hypothalamus, remains a significant challenge. Here, we combined single-cell RNA-seq on hypothalamic neurons, spatially-resolved circuit mapping and functional assays to pioneer i) the molecular study of neuronal organization, ii) dual (or even triple) neurotransmitter phenotypes (Fig. 3) and their activity-dependent switching, iii) receptor repertoires for circuit prediction and iv) to resolve existing ambiguities in neuronal nomenclature by unifying neurotransmitter-neuropeptide relationships. Notably, we also detected many mRNAs previously localized to neurites39. Therefore, sequencing somatic contents will likely produce a representative catalogue of total mRNAs expressed by a single neuron.
The computational algorithms used here include tSNE and BackSPIN. tSNE is a pure dimensional reduction algorithm and was used only for the visualization. In turn, BackSPIN is not based on dimensional reduction but on a series of binary splits, which allow focusing on the most relevant set of genes while clustering a subset of cells. Taking into account the high neuronal diversity of the hypothalamus, as confirmed by both analysis routines, we consider these results (Supplementary Fig. 3,4) as a mutual cross-validation of both approaches.
Our molecular profiling focused on the central column of the hypothalamus, leaving its most lateral and caudal segments uncharted. Therefore, we presume that further novel neuronal subtypes, divergent either at the molecular or functional levels, might be revealed by sequencing tools that can process larger volumes of data, like “Drop-seq”22. Thus, one might suggest that the number of neuronal subtypes could reach even beyond 100 in the mammalian hypothalamus. During our effort to reconstruct neuronal networks through a combination of RNA-seq on dissociated cells and subsequent bioinformatics (“forward transcriptomics”), we discovered a unique subtype of dopamine(-4) neurons, situated in the A14 locus, and whose molecular phenotype challenges the concept of a “dopamine continuum” in the hypothalamus. We propose that adjacent blocks of phenotypically distinct dopamine cells assemble an anatomically continuous stream of cells that, when using known markers, such as TH, seems homogeneous. This principle might be applicable to other subtypes of hypothalamic neurons, as well, and therefore, the resource we offer can be informative to many structure-function studies addressing key outputs of hypothalamic neuronal circuits.
Any new method requires extensive validation. As an example, we studied the expression of some receptors regulating body metabolism through sensing ligand levels in the general circulation. In particular, our data reveal that pancreatic polypeptide receptor (Npy6r) mRNA is confined to Vip+/Nms+ circadian neurons (cluster #41) leaving Vip-only neurons of the neighboring circadian cluster (#40) unchecked. This result lends significant support to previous data 40 and advances knowledge in receptor-dependent function determination for pancreatic polypeptide. Similarly, our analysis of leptin receptor (Lepr) expression whose activity upon binding white fat-derived leptin 41 is associated with satiety, shows that Lepr is a low-expressed gene (1 mRNA copy/per cell) and it is only detected in ~3% of hypothalamic neurons. The low number of Lepr+ neurons we find is in contrast to the high numbers of cells in studies using Lepr-Cre driven GFP expression in mice 42. This discrepancy can be explained by the temporal “snap-shot” nature of our RNA-Seq strategy, compared to GFP-expressing animals that reflect the accumulated expression over the animal’s lifetime, as corroborated by our own Crh -IRES-Cre::stop-GFP (Supplementary Fig. 5a,a1) and Crh-EGFP experiments 18, showing that episodic expression of any gene during the lifetime of a neuron might be considered a common feature. At the level of neuronal identity, 30% of the Npy+/Agrp+ population (4 of 13 cells) contained Lepr mRNAs. Moreover, we characterized Lepr mRNA expression in some glutamatergic neurons, corroborating published reports42. An additional strength of our dataset is that it relies on both male and female animals kept under ad lib feeding conditions, suggesting that we resolved ground-state Lepr mRNA expression, relevant for behavioral output in both sexes (see also Supplementary Fig. 2a). Finally, our results on acute formalin injection-induced stress show that the formation of new neuronal clusters (that is, subtypes) does not occur under these circumstances, meaning that no novel neuronal subtype is generated over hour-long timescales.
By consensus definition, each neuron is expected to contain at least one fast neurotransmitter, which can be co-released with another neurotransmitter (e.g. dopamine), one or more peptide, gaseous or lipid neuromodulator20, 43–45. Using the abundance of mRNAs for neurotransmitter-producing enzymes and vesicular and reuptake transporters as a weighing factor (Fig. 2b), non-Oxt/non-Avp neurons were subdivided into primarily dopaminergic, GABAergic and glutamatergic subtypes. Yet, and in contrast to RNA-seq data on cortical neurons 9, we find unexpected combinations of neurotransmitter content in an estimated 18% of hypothalamic neurons. Therefore, nominal classification as “dopaminergic”, “GABAergic” or “glutamatergic” (“single neurotransmitter” criteria) must be used with care, because metabolic pathways for alternative neurotransmitters could co-exist (Fig. 3). These might be bioenergetically more favourable than rewiring strategies 16, 17. When comparing single-cell RNA-seq data from cortical/hippocampal vs. hypothalamic neurons, the following differences apply: i) in the cerebral cortex, higher mRNA transcript levels are seen in interneurons, which also contain both Gad1 and Gad2 transcripts, and ii) cortical neurons rarely, if at all, produce mixed excitatory/inhibitory phenotypes 9. Thus, the molecular make-up of hypothalamic neurons significantly differs from their cortical counterparts. Yet, and given its statistical prevalence, mixed neurotransmitter phenotypes are not an exception but rather may apply as a general principle. This arrangement could contribute to the flexibility of hypothalamic neuronal networks to adapt to ever-changing environmental stimuli.
Addressing the range of systemic consequences of hormone release at the ME has been at the forefront of neuroendocrinology research for decades 2. For dopamine, parvocellular dorsomedial neurosecretory cells (Th+) in the Arc (A12) give rise to large nerve endings in the ME, amassing 1/3 of all boutons in the lateral external layer 46, and release dopamine for the tonic inhibition of pituitary prolactin release29. For dopamine neurons in the periventricular nucleus (A14) a significant intrahypothalamic drive from the suprachiasmatic nucleus (SCN) has been established7, 8. This synaptic circuit is significant, since it integrates the circadian pacemaker (characterized by Per genes) 33, 34and hormonal output, particularly diurnally fluctuating dopamine levels in the general circulation 7. By histochemistry, we show that onecut- 3+ A14 dopamine neurons can terminate at the ME. Notably, our EB-loading experiments are also in agreement with a previously documented parallel projection to the intermediate lobe of the pituitary 31. Our data outline a potential cellular substrate that may participate in dopamine release into the hypophyseal portal system and/or the intermediate lobe for the chronospecific inhibition of pituitary prolactin secretion 29 (Fig. 6g,h). Such a role is in agreement with indications that periventricular dopamine cells are involved in feedback control of prolactin release 47. These newly distinguished neurons are exclusively situated at the medio-posterior subdivision of the periventricular area and receive Nms + synaptic afferents from SCN neurons regulating the circadian clock 32, 48, in addition to the expression of clock genes in the periventricular and Arc dopamine cells themselves 49. Thus, these onecut-3+/Th+ neurons can also serve as neurosecretory “hubs” to tune the release of many other hypothalamic neuropeptides through synaptic afferents. Cumulatively, we recognize onecut-3+ A14 dopamine neurons as distinctly different – at the level of molecular make-up and connectivity – from the dorsomedial A12 dopamine group; thus showcasing that single-cell RNA-seq in combination with neuroanatomy can produce a molecular protomap of hypothalamic neuronal organization and aid the future discovery of functionally-segregated neuronal subclasses.
Online Methods
Animals, tissue preparation and histochemistry
Single-cell RNA-seq and histochemistry were conducted in mice on a C57BL/6N background prior to sexual maturation during postnatal days (P)14-28. Transgenic animals were also on C57BL/6N backgrounds either from their generation or through backcrossing for multiple generations. The reporter lines GAD67+/gfp, GAD67+/gfp::CCKBAC/DsRedT3 52, 53, B6.B6D2-Tg(Th-EGFP)21-31Koba (developed by K. Kobayashi and available from Riken Bioresource Center), Crh-IRES-Cre::B6.Cg-Gt(ROSA)26Sortm6(CAG-ZsGreen 1)Hze/J (Ai6; both GFP reporters were from the Jackson laboratories, #012704 and #007906) and dopamine transporter (Dat1)-Cre::B6.Cg-Gt(ROSA)26Sortm14(CAG-dTomato)Hze/J mice were extensively validated and used for neuronal identity mapping by histochemistry (n = 3-5/group). Animals were housed conventionally (12/12-h light cycle, 55% humidity). Experimental protocols were in accordance with the European Communities Council Directive (86/609/EEC), approved by regional ethical committees and regulated by applicable local laws (Stockholms Norra Djurförsöksetiska Nämnd; N512/12 (Sweden) and Tierversuchsgesetz 2012, BGBI, Nr. 114/2012 (Austria)). Particular effort was directed towards minimizing the number of animals used and their suffering during experiments.
For histochemical evaluation, animals were transcardially perfused with a fixative composed of 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB; pH 7.4) that was preceded by a short pre-rinse with physiological saline (anesthesia: 5% isoflurane at 1 l/min flow rate). In experiments investigating circadian clock-related changes in neuromedin S production, transcardial perfusions were carried out during the periods of 10.00-11.00 (a.m./”day”) and 22.00-23.00 (p.m./”night”). After overnight post-fixation in the same fixative and cryoprotection in 30% sucrose for at least 48 h, 30-50-μm thick serial free-floating or 16-μm thick serial glass-mounted sections were cut on a cryostat microtome and processed for multiple immunofluorescence histochemistry according to published protocols 18. (E)GFP and DsRed immunofluorescence were not amplified in any of the transgenic models. Free-floating sections were rinsed in PB (pH 7.4) and pre-treated with 0.3% Triton X-100 (in PB) for 1 h at 22-24°C to enhance antibody penetration. Non-specific immunoreactivity was suppressed by incubating our specimens in a cocktail of 5% normal donkey serum (NDS; Jackson), 1% bovine serum albumin (BSA; Sigma) and 0.3% Triton X-100 (Sigma) in PB for 1 h at 22-24°C. Sections were then exposed to select combinations of primary antibodies diluted in PB containing 0.1% NDS and 0.3% Triton X-100 for 48 h at 4 °C. Primary antibodies were as follows: rabbit anti-oxytocin (1:5,000; Millipore, Cat. # Mab 5296), guinea pig anti-CRH (1:1,000; Peninsula, T-5007.0050), goat anti-neurophysin II (AVP; 1:50; Santa Cruz, Cat. # sc-27093), rabbit anti-tyrosine hydroxylase (TH) (1:1,000; Millipore, Cat. # AB152), mouse anti-TH (1:500; Millipore, Cat. # MAB5280), rabbit anti-phospho-Ser40-TH (1:1,000; Millipore, Cat. # AB5935), guinea pig anti- onecut-3 (1:5,000; 54), rat anti-somatostatin (1:250; Millipore, MAB354), rabbit anti-neuromedin S (1:1,000; Bachem, Cat. # T-4814.0400), mouse anti-vasoactive intestinal polypeptide (1:400; kindly provided by Dr. Helen Wong, David Geffen School of Medicine, UCLA), rabbit anti-GRP (1:200, ImmunoStar, Cat. # 20073), mouse anti-synaptobrevin 2 (VAMP2) (1:200; Synaptic Systems, Cat. # 104211), rabbit anti-dopamine transporter (DAT) (1:250; Synaptic Systems, Cat. # 284003) and guinea pig anti-DAT (1:250; Synaptic Systems, Cat. # 284005). After extensive rinsing in PB, immunoreactivities were revealed using carbocyanine (Cy) 2,3 or 5-tagged secondary antibodies raised in donkey (1:200; Jackson; 2 h at 22-24 °C incubation). Sections were mounted onto fluorescence-free glass slides and coverslipped with Entellan (in toluene; Merck).
Polyclonal (rabbit) antibodies against ARFGEF1 (1μg/ml, HPA023822), USP48 (1μg/ml, HPA030046), KIF5A (1μg/ml, HPA004469, all available from Atlas Antibodies), guinea pig anti-AVP (1:1,200; Peninsula), rabbit anti-vesicular glutamate transporter 2 (VGLUT2, 1:400; gift of Dr. M. Watanabe) and mouse anti-GAD67 (1:400; Millipore MAB5406) were diluted in PB to which 0.3% Triton X-100 had been added were applied to glass-mounted cryosections for 16-24 h at 4 °C. Immunoreactivity was visualized using the tyramide signal amplification method (1:100, PerkinElmer) 55. Sections were counterstained with DAPI as nuclear marker and whole slides were captured using a 10x (Plan-Apocromat 10x/0.45) primary objective on a Vslide slide-scanning microscope (Metasystems) equipped with appropriate filter sets. Individual field-of-view images were stitched to produce images of entire brain sections with high resolution. Extended image data on Atlas Antibodies are available in the most recent version (HPA14) of the human protein atlas (http://www.proteinatlas.org).
Systemic Evans blue administration
Neuroendocrine cells were identified by their uptake of systemically-administered Evans blue56. Briefly, mice (n = 7) were bolus-injected i.p. with 0.25 ml of Evans blue (3%), dissolved in physiological saline, and allowed to survive for an additional 24, 48 or 72 h. Brain slices containing the periventricular region were prepared on a vibratome (VT1200S, Leica), immersion-fixed in 4% PFA in 0.1M PB, and imaged on a Zeiss LSM780 confocal microscope. Upon successful Evans blue uptake, brain slices were processed for the co-localization of tyrosine hydroxylase, onecut-3 or phosphor-Ser40-tyrosine hydroxylase (as above).
Virus microinjection in vivo
Male Dat1 -Cre::B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J mice (2 months of age, n = 3) were used for the injection of AAV1/2-flex-GFP virus constructs as described 57, 58. Briefly, mice anaesthetized by isoflurane (5%, 1 l/min flow rate) and placed in a stereotaxic frame (Harvard) were injected with 250-450 nl of virus (6 x 108 particles/ml) bilaterally using a micropipette coupled to a Quintessential stereotaxic injector (Stoelting). Injections targeted the periventricular nucleus at coordinates: anterior-posterior: -1.0 mm, lateral: ± 0.2 mm and dorsoventral: -5.2 mm from dura mater. The micropipette was held in place for 5 min before slowly retracting it from the hypothalamus to limit virus spread. Buprenorphine (0.03 mg/kg) was applied for post-surgical analgesia. Procedures were approved by the regional authority (Stockholms Norra Djurförsöksetiska Nämnd; N166/15). After 14 days of survival, mice were re-anesthetized with pentobarbital (50 mg/kg, i.p.) and perfused with 4% PFA containing 0.1% picric acid in 0.1 M PB (pH7.4). Whole brains were post-fixed for 120 min in the same fixative 4 °C, followed by rinses in 10% sucrose (in 0.1M PB, pH 7.4) containing 0.01% sodium azide (Merck) and 0.02% bacitracin (Sigma) over 2-3 days at 4°C. Brains were then embedded in 4% agarose in 0.05 M phosphate-buffered saline and coronal sections (60 μm) were cut on a vibratome (VT1000, Leica). For immunohistochemistry, free-floating sections were washed in PBS, incubated in 1% H2O2 (in PBS) for 10 min, and “blocked” in PBS containing 0.3% Triton-X and 5% NDS (Jackson) for 1 hour at 22-24 °C. Specimens were then exposed to a chicken anti-GFP antibody (1:8,000; Abcam) at 4 °C overnight. Immunoreactivity was visualized using the TSA Plus kit (PerkinElmer) using a horseradish peroxidase-conjugated donkey anti-chicken antibody (1:200, 75 min, Jackson). Finally, sections were incubated in biotinyl tyramide-fluorescein (1:100 in amplification diluent) for 10 min, mounted onto SuperFrost Plus glass slides, air dried and coverslipped with 50% glycerol/PBS.
Laser-scanning and lightsheet microscopy by Lightsheet Z.1 in optically cleared hypothalami
Laser-scanning microscopy to obtain x,y (single plane) images was performed on a Zeiss 780LSM laserscanning microscope at 40x or 63x primary magnification and with maximal signal separation or spectral unmixing. Quantitative determination of the co-localization of VGLUT2 and GAD67 immunoreactivities at the ME was undertaken at 40x primary magnification and by using the ZEN2013 software package (Zeiss) to define signal coexistence in red/green channels.
For light-sheet microscopy, hypothalami from GAD67gfp/+ P23 mice were microdissected and cleared using a modified “CUBIC” protocol 59. In brief, blocks of 4% PFA-fixed tissues were incubated, under continuous agitation in CUBIC 1 solution (25% urea, 25% N,N,N’,N’-tetrakis-(2- hydroxypropyl)ethylenediamine and 15% Triton X-100) at 37 °C for 4 days. Immunostainings were carried out after CUBIC 1 clearing. Firstly, samples were washed 3 times for 30 min each in 0.1 M PB (pH 7.4) at 22-24 °C, during which tissues regained their opaque appearance. This was followed by a 6 h incubation in PB-based blocking solution containing 2.5% BSA, 5% NDS, 0.5% Triton X-100 and 10% DMSO. Sections were then incubated with rabbit anti-phospho-Ser40-TH (1:500; Millipore) and guinea pig anti- onecut-3 (1:1,000) primary antibodies diluted in PB containing 2% NDS, 0.1% BSA, 0.3% Triton X-100, 5% DMSO and 0.1% NaN3 for 96 h at 37 °C with gentle shaking. Following 3 washes of 30 min each in PB, immunoreactivities were revealed by incubation in Cy2 and Cy3-tagged secondary antibodies (1:200; Jackson Laboratories) diluted in PB containing 3% NDS and 0.1% NaN3 for 48 h at 22-24 °C. Samples subsequently underwent 4 washes of 60 min each in PB before immersion in CUBIC 2 solution (50% sucrose, 25% urea, 10% 2,2’,2’-nitrilotriethanol and 0.1% Triton X-100). Tissues were left to shake at 22-24 °C for 3 days before image acquisition. All samples were imaged in CUBIC 2 solution with a measured refractory index of 1.45. A sequence of fluorescence images was acquired on a Lightsheet Z.1 (Zeiss) microscope using a 5x detection objective, 5x illumination optics and laser excitation at 488 nm and 561 nm. Each plane was illuminated from a single side, and whole hypothalamic images were obtained through 3 x 4 tile scanning. All images were captured at 0.7x zoom, with z-stack intervals set at 3.0 μm with an exposure time of 250 ms and 600 ms for 561 nm (Cy3; phospho-Ser40-TH) and 488 nm (Cy2; onecut-3) laser lines, respectively. 3D-rendered images were visualized with Arivis Vision4D for Zeiss (v. 2.12). Brightness and contrast of the 3D-rendered images were manually adjusted to aid visual clarity. Composite figures were assembled in CorelDraw X7 (Corel Corp.).
Whole-brain mapping by CLARITY
Reconstruction of TH-positive neurons and their projections in the intact adult mouse brain were performed using passive CLARITY methods as described 51. Briefly, wildtype C57Bl6/N adult mouse brains were cleared using the passive CLARITY protocol (1%) hydrogel monomer solution followed by whole brain TH immunostaining using a polyclonal rabbit anti-TH antibody (1:50; Abcam). High-resolution whole brain imaging was performed using CLARITY Optimized Light-sheet Microscopy (COLM,51), and 3D volume renderings were generated using Amira (FEI).
Quantitative morphometry of periventricular onecut-3+/TH+ neurons
Male Dat1 -Cre::B6.Cg- Gt(ROSA)26Sortm14(ClAG-tdTomato)Hze/J mice (n = 6) were deeply anesthetized with Na-pentobarbital and perfused with Tyrode’s Ca2+-free solution (10 ml, 37°C) followed first by 10 ml 4% PFA and 0.2% picric acid (37°C) and 50 ml of the same fixative at 4°C. Brains were immersed in the same fixative for 90 min and cryoprotected in 15% sucrose in PBS (0.01M, pH 7.4). Fourteen μm-thick coronal sections were cut on a cryostat microtome and thaw-mounted onto glass slides. Sections rinsed in PBS were incubated with rabbit anti-TH (1:1,000; Millipore) diluted in PBS containing 0.3% Triton X-100 at 4°C for 16 h. After repeated rinses in PBS, sections were re-incubated with guinea pig anti-oncecut-3 antiserum (1:250) at 4°C for 16 h. Subsequently, tissues were simultaneously exposed to Alexa 488-conjugated goat-anti guinea pig and Alexa 647-conjugated goat anti-rabbit whole IgGs (Life Technologies). Sections were then washed for >30 min in PBS and mounted with ProLong® Gold (Life Technologies). Photomicrographs were captured on a Zeiss LSM780 (Zeiss) laser-scanning microscope. Three sections/animal, spaced at regular intervals across the periventricular region were selected for quantification of fluorescently-labeled neurons. The periventricular nucleus was divided into three levels: rostral/Bregma -0.22 mm (preoptic area), mid/Bregma -0.58 mm (at suprachiasmatic nucleus) and caudal/Bregma -0.94 mm (retrochiasmatic nucleus) levels according to Paxinos and Franklin 50. Confocal micrograph montages of single optical sections covering the entire periventricular nucleus of each section were acquired with a 20x objective utilizing an automatic stage controller. Occasionally, the brightness and contrast of the acquired images were adjusted. Cells within the periventricular region were counted manually off-line using the merged, as well as individual color channels. Only neurons situated ventral to the PVN and <120 μm from the lateral wall of the third ventricle were counted at both hemispheres (Fig. 5d, Supplementary Fig. 7d).
Cell capture, imaging, lysis and RNA-seq
C57BL6/N juvenile mice (P14-28) of both sexes from control (untreated) and acutely stressed (formalin stress induced by injection of 4% PFA into the left paw) 18 experimental groups were used for cell collection. We used a 6-h time point as global changes in RNA peak ~5-9 h after experimental manipulation 60. The processing of cells from male and female animals was random to avoid any methodological bias. Mice were deeply anesthetized (5% isoflurane) and transcardially perfused with 40 ml ice-cold pre-oxygenated (95% O2/5% CO2) cutting solution containing (in mM): 90 NaCl, 26 NaHCO3, 2.5 KCl, 1.2 NaH2PO4, 10 HEPES-NaOH, 5 Na-ascorbate, 5 Na-pyruvate, 0.5 CaCl2, 8 MgSO4 and 20 glucose. A central column of the mouse hypothalamus spanning the posterior preoptic area-Arc (rostrocaudal axis), paraventricular nucleus (dorsal) and the ventrolateral hypothalamic area (lateral; Fig. 1a) was microdissected from serial 300-μm thick coronal slices under microscopy guidance and then dissociated using the Papain Dissociation System (Worthington). Isolated single cells were concentrated by centrifugation to a density of 600-1,000 cells/μl. After mixing C1 suspension reagent (4 μl; “C1 reagents”; Fluidigm Inc.) with the cell suspension (7 μl), this mixture was loaded into a C1-AutoPrep IFC microfluidic chip designed for cells of 10-17-μm in diameter (Fluidigm), and processed on a Fluidigm C1 instrument using the ‘mRNA Seq: cell load (1772x/1773x)’ script (30 min at 4°C). The microfluidic plate was then transferred to an automated microscope (Nikon TE2000E) to acquire a bright- field image of each capture site at 20x magnification using μManager (www.micro-manager.org) in <15 min. Capture quality for exclusion of debris or doublets was performed after each capture experiment. Following lysis, cDNA synthesis, amplification and tagmentation, high-throughput RNA sequencing was performed on an Illumina HiSeq2000 sequencer9, 61.
BackSPIN algorithm
BackSPIN is a bi-clustering algorithm that works in parallel on cells and genes. BackSpin is not based on dimensional reduction but on a series of binary splits, which allow for focusing on the most relevant set of genes while clustering a subset of the cells. For each binary split, the points are sorted into one-dimensional order using the SPIN algorithm 62. Here, we used BackSpinV2, which is a more adaptive and scalable version of the original algorithm 9 as shown by Marques et al. 63. Conceptually, the V2 approach is identical to the BackSPIN algorithm 9 with its full source code available at https://github.com/linnarsson-lab/BackSPIN. The algorithm has by now been extensively validated using both computational (including comparisons to PCA, and independent-component analysis, tSNE, and GP-LVM) biological validation tools 9, 63–66.
Level 1 analysis
Data from all cells that passed visual quality control were imported as molecule counts (including all metadata annotations). Cells with more than 1,500 molecules/cell (excluding rRNA, mitochondrial RNA and repeats) were analyzed, and resulted in a total of 3,131 cells. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported for the cerebral cortex and hippocampus 9. Genes with <50 molecules in the whole dataset or expressed in >70% of the cells were excluded. Next, the BackSpinV2 algorithm was used with the following parameters: splitlev = 8; Nfeature = 500; N_to_backspin = 100; N_to_cells = 500; mean_tresh = 0.1; fdr_th = 0.2; min_gr_cells = 5; min_gr_genes = 2. Considering that our primary aim was to sort cells into molecularly-defined clusters that correspond to main tissue/lineage subtypes, we manually inspected the borders of the ensuing clusters and merged clusters when thought necessary for the “main” cell types: oligodendrocytes, neurons, astrocytes, ependymal cells, endothelial cell, vascular lineage and microglia (Fig. 1b). Fig. 1c shows a heat-map with equal representation of genes that are specific to each of the main cell lineages: i) oligodendrocytes (mRNAs for myelin basic protein (Mbp) and UDP glycosyltransferase-8 (Ugt8) involved in sphingolipid metabolism 67, 68); ii) astrocytes (genes Fabp7 and Ntsr2) 9, 69; iii) ependymal cells (genes associated with motile cilia, including Enkur9 and the transcription factor Foxj170); iv) microglia (mRNAs for macrophage lineage marker allograft inflammatory factor 1 (Aif1) and immunoglobulin E receptor (Fcer1g) 71); v) endothelial cells (genes coding fibronectin (Fn1) and organic anion transporter (Slco1a4)72); vi) vascular and smooth muscle lineage (mRNAs for α2-subunit of actin (Acta2) and transgelin (Tagln)73); and vii) neurons (genes Ndrg4 and Stmn2)9, 18, 74.
Level 2 analysis of neurons
This paper particularly addresses the diversity of hypothalamic neurons and therefore neuronal clusters (Fig. 1b) were subjected to in-depth analysis. Data were from 898 cells, which qualified as neurons based on level 1 clustering. To remove noise prior to clustering, we first used a primary (coarse) filter and selected for genes, which were distinguished by t-test to be enriched in neurons using a false discovery rate (FDR) of 5% (results in 9,013 neuronal genes in total). BackSpinV2 was then used with the following parameters: splitlev = 7; Nfeturel = 500; Nfeture = 200; N_to_backspin = 10; N_to_cells = 500; mean_tresh = 0.1; fdr_th = 0.3; min_gr_cells = 5; min_gr_genes = 3. This step distinguished 91 neuronal clusters (Fig. 1c). Next, we manually removed putative artifactual clusters that had formed due to low/variable quality of RNA, and merged clusters that showed a high percentage of similarity, i.e. none of the key markers differed between the two cell populations. Moreover, we omitted clusters that appeared thalamus specific because of the presence of known thalamus-specific homeobox genes and transcription factors. These refinements led to 62 neuronal sub-clusters being discerned.
Selection of cluster-enriched genes and markers
For each gene i and cluster j, we followed the following mathematical formula: ; , where Eik is the expression of gene i in cell k. Quantities represent molecule enrichment in a specific cluster and in the fraction of cells that expressed the gene within that cluster. Next, we combined the outcomes, weighed the fraction of positive cells and ranked the genes for each cluster by score: Si,j = enrichi,j × posfraci,jpower, where “power” sets the weight for the fraction of positive cells in the cluster. We used power = 0, 0.5, 1 to rank genes in every cluster and then use the top Xgenes as most enriched. Specifically, the dendrogram in Fig. 2a was built by using the top 5 genes per cluster. To identify new markers for each neuronal cluster (Supplementary Fig. 6, Supplementary Tables 2 and 4), we used power = 0.5 to identify the top hit in each cluster, excluding that a gene is amongst the “top 5 markers” for any other cluster.
Dendrogram construction and split point listing
For generation of a dendrogram (Fig. 2a), we first selected genes enriched within the cluster so that each cluster was represented in a balanced way (that is, the top enriched genes within and selection of the top 5 from every power parameter; see above). This returned 401 genes, all of which, after a log2+1 transformation were used to calculate the correlation between clusters. We used the “Ward method” to express correlation as distance for linkage to construct a hierarchical tree. To better understand the shape of the tree, we considered if specific genes existed to explain each of the tree junctions. To this end, we used two independent criteria: the difference in the average and the difference in the fraction of positive (>0) cells (Table S3). Thus, each junction defines a left and a right group by having the above score calculated at the single-cell level.
Error-bar plots
For the most comprehensive presentation of genes with meaningful expression over many clusters, we used the form of error-bar plots that show the means ± s.e.m. for each cluster. To construct relevant figures, we calculated the mean log expression for each gene in each cluster. First, we normalized expression such that the total number of molecules (sum of all genes detected; Supplementary Fig. 1c) in every cell was set at 10,000. Next, we transformed the results by log2(x+1) to calculate the means ± s.e.m. Likewise, we computed the fraction of positive cells per cluster. We present data in two forms: i) with power = 0, where there is no correction for the fraction of positive cells and ii) with power = 1, where the average is multiplied by the fraction of positive cells to avoid heterogeneous groups (cluster identity).
Calculating significance using the Wilcoxon rank-sum test
In this report, we use statistical analysis for the purpose of declaring i) that particular gene ‘i’ is expressed in group ‘j’ significantly higher than the basal (‘background’) level and ii) genes with hierarchical expression, e.g., which differentiate junctions of the cluster dendrogram. In order to obtain levels of significance we used the Wilcoxon rank-sum test, which is non-parametric and does not assume normality. We applied this test in a ‘one-tail’ fashion (using the MATLAB function ‘ranksum’ assuming that expression in the group tested is greater than in others). For the first case, the test is performed independently for each gene while sequentially analyzing the groups and testing each group against the rest of the cells. The process is subsequently repeated for each gene over the 62 groups, and for all genes. A similar process was carried out when testing dendrogram junctions. The q value was calculated as the p value corrected for multiple testing using horizontal correction across the 62 clusters with the Benjamini-Hochberg procedure 75 to control the false discovery rate (FDR). We consider gene expression significant if q < 0.05 (raw data for p and q values are shown in Supplementary Tables 5 and 6). To obtain the set of genes that distinguish any junction in the dendrogram (Fig. 2a), we tested each side of a particular junction against its other side using Wilcoxon’s rank-sum test, corrected for multiple testing since each gene was tested for all 61 junctions. In addition, since in junction analysis genes that are exclusive (or almost exclusive) are of particular interest, we tested the number of cells with positive (greater than zero) expression using binomial distribution. Here, the p value = binomial CDF (x,N,p), where x = number of positive cells, N = cluster size, p = fraction of positive cells in both groups. These results are shown in Supplementary Table 3 with the top genes relevant to differentiate each junction.
Statistical analysis of histochemical and imaging data
None of the experiments required data collection in a blinded fashion. Data were analyzed using SigmaPlot (Systat Software Inc.). Data were expressed as means ± s.e.m. A P-value of < 0.05 was considered statistically significant and calculated using Student’s t-test (on independent samples), one-way analysis of variance (ANOVA with Dunn’s post- hoc test) or Mann-Whitney’s U-test as appropriate. The non-parametric Kolmogorov-Smirnov test was used for the determination of diurnal variations in phospho-TH levels.
tSNE projections
We used t-distributed Stochastic Neighbor Embedding (tSNE)76 to visualize neuronal complexity in two dimensions (no effect on clustering). In order to present single-cell RNA-sequencing data, which is naturally of high dimensions, one needs to choose a dimensional reduction method that is either linear (principle component analysis) or non-linear (tSNE). We chose tSNE since many single-cell RNA-sequencing studies show this to be the most powerful in keeping multiple structures within the datasets 9, 64, 65. tSNE projections (Fig. 1d-d2) were calculated on neuronal data after selecting 1,194 cluster-enriched genes as used for dendrogram construction (see above). We used 200 principle components; perplexity = 5, 10 or 20, ε (initial learning rate) = 100 and correlation as a distance measure.
Drop-seq
Drop-seq was performed as previously published22. Briefly, single-cell suspensions were prepared in PBS/BSA at a concentration of 30.000 cells/ml. Barcoded beads (Chemgenes) were resuspended in lysis buffer containing DTT at a concentration of 100,000 beads/ml. Syringes containing oil, beads and cell suspension were connected to a Drop-seq microfluidics chip (FlowJEM), and individual flow rates were adjusted to achieve constant and productive flow. Droplets were collected in a 50 ml Falcon tube. Excess oil at the bottom of the tube below the droplet-containing phase was removed. Droplet breakage, subsequent reverse transcription and exonuclease treatment were conducted as described22. cDNA was PCR amplified by 4 initial cycles at 65 °C followed by 12 cycles at 67 °C annealing temperature. cDNAs were fragmented and amplified with the Nextera XT DNA sample prep kit (Illumina) using custom primers that enabled the specific amplification of only their 3’ ends. cDNA and library concentrations were assessed using the Qubit dsDNA HS assay (Life Technologies), and fragment distribution was determined with sensitivity DNA chips on a 2100 Bioanalyzer (Agilent). Sequencing was performed by using the 75bp paired-end configuration on an Illumina HiSeq 3000/4000 platform.
Drop-seq analysis
Drop-seq data processing of 220 neurons as output was performed using the Drop-seq Tools v1.12 software22. Briefly, each transcriptome Read 2 was tagged with the cell barcode (bases 1 to 12) and UMI barcode (bases 13 to 20) obtained from Read 1, trimmed for sequencing adapters and polyA sequences and aligned to the mouse genome (Ensembl GRCm38 release) using STAR v2.4.0 77. Reads aligning to exons were tagged with the respective gene name, and counts of unique UMIs per gene within each cell were used to build a digital gene expression matrix. Read counts for each gene were posteriorly normalized to the total coverage per cell.
Ca2+ imaging
Coronal hypothalamic slices containing the periventricular and Arc nuclei were prepared from male Th-GFP mice (2-4 weeks old). In brief, mice were deeply anesthetized (5% isoflurane), brains were rapidly removed, and immersed in ice-cold pre-oxygenated (95% O2/5% CO2) cutting solution containing (in mM): 90 NaCl, 26 NaHCO3, 2.5 KCl, 1.2 NaH2PO4, 10 Hepes-NaOH, 5 Na-ascorbate, 5 Na- pyruvate, 0.5 CaCl2, 8 MgSO4 and 20 glucose. Subsequently, 250 μm-thick coronal slices were cut on a vibratome (VT1200S, Leica). Slices encompassing the periventricular nucleus were selected and equilibrated in artificial cerebrospinal fluid (ACSF) containing (in mM): 124 NaCl, 26 NaHCO3, 2.5 KCl, 1.2 NaH2PO4, 2 CaCl2 and 2 MgSO4 at 22-24 °C for 1 - 4 h before recording. Prior to imaging, slices were transferred into ACSF containing 10-20 μM Fura2-AM for 40-90 min (loading). Recordings were done using a VisiChrome monochromator and VisiView software (Visitron Systems) on an AxioExaminer.D1 microscope (Zeiss) equipped with a CoolSnap HQ2 camera (Photometrics). Neuromedin S was from Tocris, 55 mM KCl solution contained (in mM): 71.5 NaCI, 26 NaHCO3, 55 KCl, 1.2 NaH2PO4, 2 CaCl2 and 2 MgSO4.
Human, tissue preparation and histochemistry
We have applied direct perfusion via the internal carotid and vertebral arteries, which facilitated the preservation of tissue integrity relative to alternative fixation methods. Human brains (n = 2, gender and age: female/83 years and male/79 years, ethical approval: TUKEB 84/2014, Hungary) were first perfused with physiological saline followed by a fixative containing 2% PFA and 0.1% glutaraldehyde in 0.1 M Tris-buffered saline (TBS, pH 7.4) 7 h or 11 h after death. The removal and subsequent preparation of human tissues were in accordance with ethical guidelines of Semmelweis University (1998, Budapest, Hungary). Hypothalami were dissected out, post-fixed in 2% PFA in TBS for 72 h, followed by immersion in cryoprotective 30% sucrose in 0.1M PB (pH 7.4) overnight. Coronal sections (50 μm) were cut on a cryostat microtome and processed for immunohistochemistry. Free-floating sections were rinsed in PB (pH 7.4) and pre-treated with 0.3% Triton-X 100 (in PB) for 1 h at 22-24 °C to enhance the penetration of antibodies. Non-specific immunoreactivity was suppressed by incubating our specimens in a cocktail of 5% NDS (Jackson), 10% BSA (Sigma) and 0.3% Triton X-100 (Sigma) in PB for 1 h at 22-24 °C. Sections were exposed for up to 72 h (at 4 °C) to the following mixture of primary antibodies diluted in PB to which 0.1% NDS and 0.3% Triton X-100 had been added: guinea pig anti-onecut-3 (1:5.00054), mouse anti-TH (Millipore, 1:500, Cat. # MAB5280), rabbit anti-phospho-Ser40-TH (Millipore, 1:500). After extensive rinsing in PB, immunoreactivities were revealed by Cy2, 3 or 5-tagged secondary antibodies raised in donkey (1:200 [Jackson], 2h at 22-24 °C). Lipofuscin autofluorescence was quenched by applying Sudan Black-B (1%, dissolved in 70% ethanol 78). Glass-mounted sections were coverslipped with Aquamount embedding medium (Dako). Sections were inspected and images acquired on a 710LSM confocal laser-scanning microscope (Zeiss) at 10x or 40x primary magnification, and pinhole settings limiting signal detection to 0.5-0.7 μm. Emission spectra for each dye were limited as follows: Cy2/505-530 nm, Cy3/560-610 nm, and Cy5/ 650-720 nm. Multi-panel figures were assembled in CorelDraw X7 (Corel Corp.).
In situ hybridization
In accord with applicable publication policies, images presented here are from publicly available resources of the Allen Brain Atlas. Particularly, images are from the Adult Mouse Atlas resource with experiments #75041585, 74882808, 79591637, 77371835, 793, 79591365 for the genes Qrfp, Npvf, Grp, Vip, Per2 and Slc6a3, respectively (www.alleninstitute.org).
Supplementary Material
Acknowledgements
The authors thank N.-G. Larsson and L. Olson for providing Dat1-Cre mice for the generation of reporter mice, H. Wong for antibodies and K. Meletis for his supervision of viral injections in Dat1-Cre mice. This work was supported by the Swedish Research Council (T.Ha., T.Hö., S.L., C.B.), Hjärnfonden (T.Ha.), the Petrus and Augusta Hedlunds Foundation (T. Ha.), the Novo Nordisk Foundation (T.Ha., T.Hö., C.B.), the National Brain Research Program of Hungary (MTA-SE NAP B, KTIA_NAP_13-2014-0013; A.A.), the European Commission (PAINCAGE grant, T.Ha., T.Hö.), the European Research Council (BRAINCELL; S.L., ENDOSWITCH; C.B. and SECRET-CELLS; T.Ha.), intramural funds of the Medical University of Vienna (T.Ha.) and an NIH Director’s Pioneer Award (T.L.H.). R.A.R. is an EMBO long-term research fellow (ALTF 596-2014) co-funded by the European Commission FP7 (Marie Curie Actions, EMBOCOFUND2012, GA-2012-600394). A.Z. received support from the Human Frontier Science Program. F.C. is a Research Associate of the Fonds de la Recherche Scientifique-FNRS, Belgium. The single-cell sequencing infrastructure at CeMM was supported by a New Frontiers Research Infrastructure grant from the Austrian Academy of Sciences.
Footnotes
Accession codes
Gene Expression Omnibus: GSE74672.
Author contributions
T.Ha. and R.A.R. conceived the general framework of this study. T.Ha., T.L.H., S.L., R.A.R., A.Z., T.Hö., C.B., K.D. designed experiments, T.Ha., T.L.H., S.L., T.Hö., C.B., K.D., A.A., J.M. and C.B. senior authors, sponsored research. R.A.R., A.Z., A.H., J.B., F.G., X.-B.G., Z.-W.L., A.A., E.K., R.T., B.H., A.K.C., D.C., M.Z., A.R. and M.F. performed research and analyzed data. H.M., C.S., D.C., Z.M., G.S., F.C., Y.Y., M.U., J.S.B. and P.W. provided unique reagents. R.A.R., A.Z., T.L.H. and T.Ha. wrote the paper. All authors reviewed the manuscript and approved its submission.
Competing Financial Interests Statement
T.Ha. declares support from GW Pharmaceuticals on projects unrelated to the focus of this report. All other authors report no conflict of interest.
Data Availability
Sequence data were deposited in Gene Expression Omnibus, accession number GSE74672. For each gene, data can be analyzed and visualized on-line at http://linnarssonlab.org/hypothalamus. All other data that support the findings of this study are available from Tibor Harkany upon reasonable request.
Data Availability Statement
All supplementary information and source data are available in the online version of the paper and at http://linnarssonlab.org/hypothalamus.
References
- 1.Du Vigneaud V. Hormones of the posterior pituitary gland: oxytocin and vasopressin. Harvey lectures. 1954;50:1–26. [PubMed] [Google Scholar]
- 2.Clarke IJ. Hypothalamus as an endocrine organ. Comprehensive Physiology. 2015;5:217–253. doi: 10.1002/cphy.c140019. [DOI] [PubMed] [Google Scholar]
- 3.Lantos TA, Gorcs TJ, Palkovits M. Immunohistochemical mapping of neuropeptides in the premamillary region of the hypothalamus in rats. Brain research Brain research reviews. 1995;20:209–249. doi: 10.1016/0165-0173(94)00013-f. [DOI] [PubMed] [Google Scholar]
- 4.Swanson LW, Kuypers HG. The paraventricular nucleus of the hypothalamus: cytoarchitectonic subdivisions and organization of projections to the pituitary, dorsal vagal complex, and spinal cord as demonstrated by retrograde fluorescence double-labeling methods. The Journal of comparative neurology. 1980;194:555–570. doi: 10.1002/cne.901940306. [DOI] [PubMed] [Google Scholar]
- 5.Dahlstrom A, Fuxe K. Evidence for Existence of Monoamine-Containing Neurons in Central Nervous System.I. Demonstration of Monoamines in Cell Bodies of Brain Stem Neurons. Acta Physiol Scand. 1964;62:1–55. [PubMed] [Google Scholar]
- 6.Bjorklund A, Moore RY, Nobin A, Stenevi U. The organization of tubero-hypophyseal and reticulo-infundibular catecholamine neuron systems in the rat brain. Brain research. 1973;51:171–191. doi: 10.1016/0006-8993(73)90371-5. [DOI] [PubMed] [Google Scholar]
- 7.Horvath TL. Suprachiasmatic efferents avoid phenestrated capillaries but innervate neuroendocrine cells, including those producing dopamine. Endocrinology. 1997;138:1312–1320. doi: 10.1210/endo.138.3.4976. [DOI] [PubMed] [Google Scholar]
- 8.Abizaid A, Horvath B, Keefe DL, Leranth C, Horvath TL. Direct visual and circadian pathways target neuroendocrine cells in primates. The European journal of neuroscience. 2004;20:2767–2776. doi: 10.1111/j.1460-9568.2004.03737.x. [DOI] [PubMed] [Google Scholar]
- 9.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 10.Scott N, Prigge M, Yizhar O, Kimchi T. A sexually dimorphic hypothalamic circuit controls maternal care and oxytocin secretion. Nature. 2015;525:519–522. doi: 10.1038/nature15378. [DOI] [PubMed] [Google Scholar]
- 11.Yang CF, et al. Sexually dimorphic neurons in the ventromedial hypothalamus govern mating in both sexes and aggression in males. Cell. 2013;153:896–909. doi: 10.1016/j.cell.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X, et al. Modular genetic control of sexually dimorphic behaviors. Cell. 2012;148:596–607. doi: 10.1016/j.cell.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome research. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dulcis D, Jamshidi P, Leutgeb S, Spitzer NC. Neurotransmitter switching in the adult brain regulates behavior. Science. 2013;340:449–453. doi: 10.1126/science.1234152. [DOI] [PubMed] [Google Scholar]
- 15.Meister B, Cortes R, Villar MJ, Schalling M, Hokfelt T. Peptides and transmitter enzymes in hypothalamic magnocellular neurons after administration of hyperosmotic stimuli: comparison between messenger RNA and peptide/protein levels. Cell and tissue research. 1990;260:279–297. doi: 10.1007/BF00318631. [DOI] [PubMed] [Google Scholar]
- 16.Pinto S, et al. Rapid rewiring of arcuate nucleus feeding circuits by leptin. Science. 2004;304:110–115. doi: 10.1126/science.1089459. [DOI] [PubMed] [Google Scholar]
- 17.Cristino L, et al. Obesity-driven synaptic remodeling affects endocannabinoid control of orexinergic neurons. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2229-2238. doi: 10.1073/pnas.1219485110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romanov RA, et al. A secretagogin locus of the mammalian hypothalamus controls stress hormone release. The EMBO journal. 2015;34:36–54. doi: 10.15252/embj.201488977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry FE, Sugino K, Tozer A, Branco T, Sternson SM. Cell type-specific transcriptomics of hypothalamic energy-sensing neuron responses to weight-loss. eLife. 2015;4 doi: 10.7554/eLife.09800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krashes MJ, et al. An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature. 2014;507:238–242. doi: 10.1038/nature12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everitt BJ, Hokfelt T, Wu JY, Goldstein M. Coexistence of tyrosine hydroxylase-like and gamma-aminobutyric acid-like immunoreactivities in neurons of the arcuate nucleus. Neuroendocrinology. 1984;39:189–191. doi: 10.1159/000123977. [DOI] [PubMed] [Google Scholar]
- 22.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SY, Foldy C, Szabadics J, Soltesz I. Cell-type-specific CCK2 receptor signaling underlies the cholecystokinin-mediated selective excitation of hippocampal parvalbumin-positive fast-spiking basket cells. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:10993–11002. doi: 10.1523/JNEUROSCI.1970-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moriya R, et al. RFamide peptide QRFP43 causes obesity with hyperphagia and reduced thermogenesis in mice. Endocrinology. 2006;147:2916–2922. doi: 10.1210/en.2005-1580. [DOI] [PubMed] [Google Scholar]
- 25.Zagoracz O, et al. Effects of direct QRFP-26 administration into the medial hypothalamic area on food intake in rats. Brain research bulletin. 2015;118:58–64. doi: 10.1016/j.brainresbull.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Tekin I, Roskoski R, Jr, Carkaci-Salli N, Vrana KE. Complex molecular regulation of tyrosine hydroxylase. Journal of neural transmission. 2014;121:1451–1481. doi: 10.1007/s00702-014-1238-7. [DOI] [PubMed] [Google Scholar]
- 27.Hokfelt T, Martensson R, Bjorklund A, Kleinau S, Goldstein M. In: Handbook of Chemical Neuroanatomy. Bjorklund A, Hokfelt T, editors. Elsevier Science Publishers; 1984. Distribution maps of tyrosine-hydroxylase-immunoreactive neurons in the rat brain; pp. 277–379. [Google Scholar]
- 28.Meister B, Elde R. Dopamine transporter mRNA in neurons of the rat hypothalamus. Neuroendocrinology. 1993;58:388–395. doi: 10.1159/000126568. [DOI] [PubMed] [Google Scholar]
- 29.Ben-Jonathan N, Hnasko R. Dopamine as a prolactin (PRL) inhibitor. Endocrine reviews. 2001;22:724–763. doi: 10.1210/edrv.22.6.0451. [DOI] [PubMed] [Google Scholar]
- 30.Stagkourakis S, Kim H, Lyons DJ, Broberger C. Dopamine Autoreceptor Regulation of a Hypothalamic Dopaminergic Network. Cell reports. 2016 doi: 10.1016/j.celrep.2016.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goudreau JL, Lindley SE, Lookingland KJ, Moore KE. Evidence that hypothalamic periventricular dopamine neurons innervate the intermediate lobe of the rat pituitary. Neuroendocrinology. 1992;56:100–105. doi: 10.1159/000126214. [DOI] [PubMed] [Google Scholar]
- 32.Mori K, et al. Identification of neuromedin S and its possible role in the mammalian circadian oscillator system. The EMBO journal. 2005;24:325–335. doi: 10.1038/sj.emboj.7600526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, et al. A PERIOD3 variant causes a circadian phenotype and is associated with a seasonal mood trait. Proceedings of the National Academy of Sciences of the United States of America. 2016 doi: 10.1073/pnas.1600039113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vitaterna MH, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holtta M, et al. An integrated workflow for multiplex CSF proteomics and peptidomics-identification of candidate cerebrospinal fluid biomarkers of Alzheimer’s disease. Journal of proteome research. 2015;14:654–663. doi: 10.1021/pr501076j. [DOI] [PubMed] [Google Scholar]
- 36.Irani DN. Cerebrospinal Fluid in Clinical Practice. W.B. Saunders; Philadelphia: 2009. CHAPTER 10 - Properties and Composition of Normal Cerebrospinal Fluid; pp. 69–89. [Google Scholar]
- 37.Bro S, Haycock JW. Visual cortex (translation of S. Ramon y Cajal) translated from L’Encorce Cerebrale Suivant les Regions L’Ecorce Visuelle, Chapter 25 in “Histologie du Systeme Nerveux de l’Homme et des Vertebres,” 1911. Behavioral biology. 1977;21:508–528. doi: 10.1016/s0091-6773(77)92165-4. [DOI] [PubMed] [Google Scholar]
- 38.Petilla Interneuron Nomenclature, G et al. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nature reviews Neuroscience. 2008;9:557–568. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zivraj KH, et al. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yulyaningsih E, et al. Pancreatic polypeptide controls energy homeostasis via Npy6r signaling in the suprachiasmatic nucleus in mice. Cell metabolism. 2014;19:58–72. doi: 10.1016/j.cmet.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 41.Friedman JM, Halaas JL. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 42.Scott MM, et al. Leptin targets in the mouse brain. The Journal of comparative neurology. 2009;514:518–532. doi: 10.1002/cne.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boulland JL, et al. Vesicular glutamate and GABA transporters sort to distinct sets of vesicles in a population of presynaptic terminals. Cerebral cortex. 2009;19:241–248. doi: 10.1093/cercor/bhn077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dawson TM, Snyder SH. Gases as biological messengers: nitric oxide and carbon monoxide in the brain. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1994;14:5147–5159. doi: 10.1523/JNEUROSCI.14-09-05147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jonas P, Bischofberger J, Sandkuhler J. Corelease of two fast neurotransmitters at a central synapse. Science. 1998;281:419–424. doi: 10.1126/science.281.5375.419. [DOI] [PubMed] [Google Scholar]
- 46.Ajika K, Hokfelt T. Ultrastructural identification of catecholamine neurones in the hypothalamic periventricular-arcuate nucleus-median eminence complex with special reference to quantitative aspects. Brain research. 1973;57:97–117. doi: 10.1016/0006-8993(73)90571-4. [DOI] [PubMed] [Google Scholar]
- 47.DeMaria JE, Lerant AA, Freeman ME. Prolactin activates all three populations of hypothalamic neuroendocrine dopaminergic neurons in ovariectomized rats. Brain research. 1999;837:236–241. doi: 10.1016/s0006-8993(99)01667-4. [DOI] [PubMed] [Google Scholar]
- 48.Lee IT, et al. Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron. 2015;85:1086–1102. doi: 10.1016/j.neuron.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sellix MT, et al. Anatomical and functional characterization of clock gene expression in neuroendocrine dopaminergic neurons. American journal of physiology Regulatory, integrative and comparative physiology. 2006;290:R1309-1323. doi: 10.1152/ajpregu.00555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. Academic Press; San Diego: 2001. [Google Scholar]
- 51.Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nature protocols. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamamaki N, et al. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. The Journal of comparative neurology. 2003;467:60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- 53.Mate Z, et al. Spatiotemporal expression pattern of DsRedT3/CCK gene construct during postnatal development of myenteric plexus in transgenic mice. Cell and tissue research. 2013;352:199–206. doi: 10.1007/s00441-013-1552-7. [DOI] [PubMed] [Google Scholar]
- 54.Pierreux CE, Vanhorenbeeck V, Jacquemin P, Lemaigre FP, Rousseau GG. The transcription factor hepatocyte nuclear factor-6/Onecut-1 controls the expression of its paralog Onecut-3 in developing mouse endoderm. The Journal of biological chemistry. 2004;279:51298–51304. doi: 10.1074/jbc.M409038200. [DOI] [PubMed] [Google Scholar]
- 55.Mulder J, et al. Secretagogin is a Ca2+-binding protein identifying prospective extended amygdala neurons in the developing mammalian telencephalon. The European journal of neuroscience. 2010;31:2166–2177. doi: 10.1111/j.1460-9568.2010.07275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weiss ML, Cobbett P. Intravenous injection of Evans Blue labels magnocellular neuroendocrine cells of the rat supraoptic nucleus in situ and after dissociation. Neuroscience. 1992;48:383–395. doi: 10.1016/0306-4522(92)90498-q. [DOI] [PubMed] [Google Scholar]
- 57.Murray AJ, et al. Parvalbumin-positive CA1 interneurons are required for spatial working but not for reference memory. Nature neuroscience. 2011;14:297–299. doi: 10.1038/nn.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pollak Dorocic I, et al. A whole-brain atlas of inputs to serotonergic neurons of the dorsal and median raphe nuclei. Neuron. 2014;83:663–678. doi: 10.1016/j.neuron.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 59.Susaki EA, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 60.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 61.Islam S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nature methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 62.Tsafrir D, et al. Sorting points into neighborhoods (SPIN): data analysis and visualization by ordering distance matrices. Bioinformatics. 2005;21:2301–2308. doi: 10.1093/bioinformatics/bti329. [DOI] [PubMed] [Google Scholar]
- 63.Marques S, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352:1326–1329. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fan J, et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nature methods. 2016;13:241–244. doi: 10.1038/nmeth.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tasic B, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nature neuroscience. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fuzik J, et al. Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nature biotechnology. 2016;34:175–183. doi: 10.1038/nbt.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sprong H, et al. UDP-galactose:ceramide galactosyltransferase is a class I integral membrane protein of the endoplasmic reticulum. The Journal of biological chemistry. 1998;273:25880–25888. doi: 10.1074/jbc.273.40.25880. [DOI] [PubMed] [Google Scholar]
- 68.Jahn O, Tenzer S, Werner HB. Myelin proteomics: molecular anatomy of an insulating sheath. Molecular neurobiology. 2009;40:55–72. doi: 10.1007/s12035-009-8071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ebrahimi M, et al. Astrocyte-expressed FABP7 regulates dendritic morphology and excitatory synaptic function of cortical neurons. Glia. 2015 doi: 10.1002/glia.22902. [DOI] [PubMed] [Google Scholar]
- 70.Roy S. The motile cilium in development and disease: emerging new insights. BioEssays : news and reviews in molecular, cellular and developmental biology. 2009;31:694–699. doi: 10.1002/bies.200900031. [DOI] [PubMed] [Google Scholar]
- 71.Liu G, Ma H, Jiang L, Zhao Y. Allograft inflammatory factor-1 and its immune regulation. Autoimmunity. 2007;40:95–102. doi: 10.1080/08916930601083946. [DOI] [PubMed] [Google Scholar]
- 72.Ose A, et al. Functional characterization of mouse organic anion transporting peptide 1a4 in the uptake and efflux of drugs across the blood-brain barrier. Drug metabolism and disposition: the biological fate of chemicals. 2010;38:168–176. doi: 10.1124/dmd.109.029454. [DOI] [PubMed] [Google Scholar]
- 73.Li L, Miano JM, Cserjesi P, Olson EN. SM22 alpha, a marker of adult smooth muscle, is expressed in multiple myogenic lineages during embryogenesis. Circulation research. 1996;78:188–195. doi: 10.1161/01.res.78.2.188. [DOI] [PubMed] [Google Scholar]
- 74.Tortoriello G, et al. Miswiring the brain: Delta9-tetrahydrocannabinol disrupts cortical development by inducing an SCG10/stathmin-2 degradation pathway. The EMBO journal. 2014;33:668–685. doi: 10.1002/embj.201386035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 76.van der Maaten L, Hinton G. Visualizing Data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 77.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schnell SA, Staines WA, Wessendorf MW. Reduction of lipofuscin-like autofluorescence in fluorescently labeled tissue. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1999;47:719–730. doi: 10.1177/002215549904700601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data were deposited in Gene Expression Omnibus, accession number GSE74672. For each gene, data can be analyzed and visualized on-line at http://linnarssonlab.org/hypothalamus. All other data that support the findings of this study are available from Tibor Harkany upon reasonable request.
All supplementary information and source data are available in the online version of the paper and at http://linnarssonlab.org/hypothalamus.