Abstract
Primary cilia, antenna-like sensory organelles protruding from the surface of most vertebrate cell types, are essential for regulating signalling pathways during development and adult homeostasis. Mutations in genes affecting cilia cause an overlapping spectrum of >30 human diseases and syndromes, the ciliopathies. Given the immense structural and functional diversity of the mammalian cilia repertoire, there is a growing disconnect between patient genotype and associated phenotypes, with variable severity and expressivity characteristic of the ciliopathies as a group. Recent technological developments are rapidly advancing our understanding of the complex mechanisms that control biogenesis and function of primary cilia across a range of cell types and are starting to tackle this diversity. Here, we examine the structural and functional diversity of primary cilia, their dynamic regulation in different cellular and developmental contexts and their disruption in disease.
Introduction
Primary cilia are solitary, antenna-like sensory organelles protruding from the surface of most vertebrate cell types. They consist of a microtubule core, the axoneme [G] , which extends from a modified centriole [G] called basal body [G] and is surrounded by a lipid bilayer membrane that is continuous with, but compositionally distinct from, the cell body plasma membrane (Figure 1a). Although they are small, representing roughly 1/200th of the total surface of the cell, primary cilia are essential for development and homeostasis, as the cilium is highly enriched in receptors, ion channels and downstream effectors for several signalling pathways, including Hedgehog and G protein coupled receptor (GPCR) signalling; without this antenna-like organelle to localize to, these pathways fail to signal appropriately. Accordingly, mutations that impair ciliary biogenesis, structure or function cause deregulated signalling and lead to ciliopathies [G], an overlapping spectrum of >30 human diseases and syndromes, which affect most tissues or organs in the body, including the eye, kidney, liver, brain, skeleton and heart (Table 1).
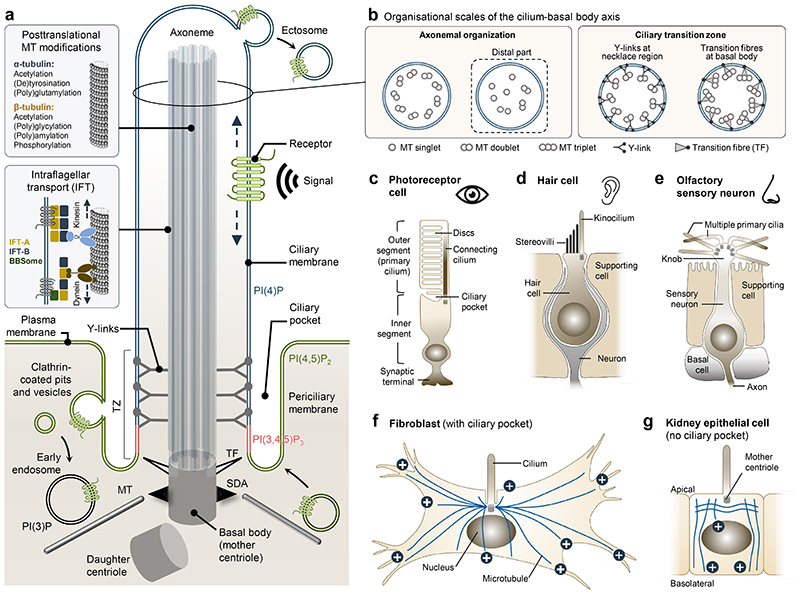
Figure 1. Architecture of primary cilia and main ciliary sub compartments.
a, Schematic of a generic primary cilium showing the main ciliary subcompartments. b, Cross-sectional views of the cilium-basal body axis (from tip to base) showing the changes in microtubule (MT) arrangement along the axis. The panel 1b right image presented in the box for axonemal organization illustrates the compromised 9+0 MT organization of unstructured bundles of MT singlets and doublets that can be observed at the distal region of the cilium in certain cell types. c, d, e, Diagrams of the indicated vertebrate cell types illustrating the diverse types of non-motile, sensory cilia found in these cells. f, g, Depictions of a fibroblast and polarized kidney epithelial cell showing the presence and absence, respectively, of a ciliary pocket, and the different cytoplasmic MT organisation (blue lines; MT plus ends are marked with “+”) in the two cell types. Abbreviations: IFT, intraflagellar transport; SDA, subdistal appendage; TF, transition fibre; TZ, transition zone.
Table 1. Established ciliopathies and associated disease genes.
| Namea | Commonly observed clinical featuresb | Known disease gene(s)c |
|---|---|---|
| Acrocallosal syndrome (ACLS) | Agenesis of corpus callosum, distal anomalies of limbs, minor craniofacial anomalies and intellectual disability. | KIF7, GLI3 |
| Alström syndrome (ALMS) | Vision and hearing abnormalities, childhood obesity, cardiomyopathy. Later in life diabetes mellitus, liver and kidney dysfunction may develop. | ALMS1 |
| Bardet-Biedl syndrome (BBS) | Cone-rod dystrophy, polydactyly, truncal obesity, hypogonadism, kidney abnormalities, learning difficulties, congenital heart defects, cardiomyopathy. | ARL6, BBPIP1, BBS1, BBS2, BBS4, BBS5, BBS7, BBS9, BBS10, BBS12, C8ORF37, CEP290, IFT27, IFT74, IFT172, LZTFL1, MKKS, MKS1, SDCCAG8, TRIM32, TTC8, WDPCP |
| Carpenter syndrome (CRPT) | Craniosynostosis, skeletal and dental abnormalities, vision and hearing loss, congenital heart defects, genital abnormalities, obesity, intellectual disability. | MEGF8, RAB23 |
| Cerebellar vermis defect, oligophrenia, ataxia, coloboma, hepatic fibrosis (COACH) syndrome | Intellectual disability, liver fibrosis, ataxia, ocular anomalies (coloboma, nystagmus). Considered a rare form of Joubert syndrome. | CC2D2A, RPGRIP1L, TMEM67 |
| Cranioectodermal dysplasia (CED; also known as Sensenbrenner syndrome) | Skeletal and ectodermal defects, nephronophthisis, liver fibrosis, ocular anomalies (mainly retinitis pigmentosa), congenital heart defects | IFT43, IFT122, WDR19 (IFT144), WDR35 (IFT121) |
| Curry–Jones syndrome (CRJS) | Syndromic craniosynostosis, agenesis of the corpus callosum, preaxial polysyndactyly and syndactyly of hands and/or feet, skin and intestinal abnormalities, ocular anomalies (colobomas, microphthalmia), occipital meningoceles, intellectual disability, tumours (smooth muscle hamartomas, desmoplastic medulloblastoma). | SMO |
| Ellis–Van Creveld (EVC) syndrome | Short stature, short arms and legs, narrow chest with short ribs, polydactyly, missing and/or malformed nails, dental abnormalities, congenital heart defects. | EVC, EVC2 |
| Endocrine-cerebroosteodysplasia (ECO) | Various anomalies of the endocrine, cerebral, and skeletal systems, neonatal mortality. | CILK1 |
| Greig cephalopolysyndactyly syndrome (GCPS) | Polydactyly, syndactyly, ocular hypertelorism, macrocephaly, intellectual disability. | GLI3 |
| Holoprosencephaly (HPE) | Abnormal brain development, cyclopia, proboscis, intellectual disability, pituitary gland anomalies. | CDON, FGF8, FOXH1, GLI2, NODAL, PTCH1, SHH, SIX3, TGIF1, ZIC2 |
| Hydrolethalus syndrome (HLS) | Severe foetal malformations including craniofacial dysmorphic features and abnormalities of central nervous system, heart, respiratory tract and limbs. | HYLS1, KIF7 |
| Jeune asphyxiating thoracic dystrophy (JATD)/ (also known as shortrib thoracic dysplasia (SRTD)) | Defective bone development, including small chest and short ribs causing impaired growth and expansion of the lungs and breathing difficulties; shortened bones in the arms and legs, polydactyly, unusually shaped pelvic bones. | CEP120, DYNC2H1, DYNC2I1 (WDR60), DYNC2I2 (WDR34), DYNC2LI1, DYNLT2B, IFT43, IFT52, IFT80, IFT81, IFT140, IFT172, INTU, KIAA0586 (TALPID3), KIAA0753 (MNR), NEK1, TCTEX1D2, TTC21B (IFT139), WDR19 (IFT144), WDR35 (IFT121) |
| Joubert syndrome (JBTS) | Defective brain development, including absence or underdevelopment of the cerebellar vermis and a malformed brain stem, which cause the characteristic molar tooth sign on MRI. Congenital heart defects. Other symptoms include hypotonia, abnormal breathing patterns and eye movements, ataxia, distinctive facial features, and intellectual disability. | AHI1, ARL13B, ARL3, ARMC9, B9D1, B9D2, CC2D2A, CEP41, CEP104, CEP120, CEP290, CPLANE1, CSPP1, FAM149B1 213,214, IFT74, INPP5E, KATNIP 215,KIAA0586 (TALPID3), KIAA0753 (MNR), KIF7, MKS1, NPHP1, OFD1, PDE6D, PIBF1, RPGRIP1L, SUFU, TCTN1, TCTN2, TCTN3, TMEM67, TMEM107, TMEM138, TMEM216, TMEM218 216, TMEM231, TMEM237, TOGARAM1, TTC21B (IFT139), ZNF423 |
| Kallmann syndrome (KS: central hypogonadism) | Hypogonadotropic hypogonadism leading to impaired sexual development; impaired sense of smell. | >50 genes; see Ref. 217 |
| Leber congenital amaurosis (LCA) | Retinal defects, causing severe visual impairment beginning in infancy. Other symptoms include photophobia, nystagmus, keratoconus and extreme farsightedness. | AIPL1, ALMS1, CEP290, CNGA3 218, CRB1, CRX, DTHD1, GDF6, GUCY2D, IDH3A, IMPDH1, IQCB1 (NPHP5), KCNJ13, LCA5, LRAT, MYO7A, NMNAT1, PHPH2, RD3, RDH12, RPE65, RPGRIP1, SPATA7, TUBB4B, TULP1, USP45 |
| McKusick–Kaufman syndrome (MKKS) | Genitourinary malformations, postaxial polydactyly, congenital heart defects, choanal atresia, pituitary dysplasia, esophageal atresia and distal tracheoesophageal fistula, Hirschsprung disease, vertebral anomalies, and hydrops fetalis. Syndrome is allelic with Bardet-Biedl. | MKKS |
| Meckel syndrome (MKS) | Multiple kidney cysts, occipital encephalocele, polydactyly, congenital heart defects. Affected children may also display anomalies of head, face, liver, lungs, genitals, and urinary tract. | B9D1, B9D2, CC2D2A, CEP290, KIF14, MKS1, NPHP3, RPGRIP1L, TCTN2, TMEM67, TMEM107, TMEM216, TMEM231, TXNDC15 |
| Mental retardation, truncal obesity, retinal dystrophy, and micropenis syndrome (MORMS) | Intellectual disability, truncal obesity, retinal dystrophy, and micropenis in males. Cataracts may occur later in life. | INPP5E |
| Morbid obesity and spermatogenic failure (MOSPGF) | Morbid obesity, hypertension, type 2 diabetes mellitus and dyslipidemia leading to early coronary disease, myocardial infarction and congestive heart failure; intellectual disability, decreased sperm counts or azoospermia. | CEP19 |
| Nephronophthisis (NPHP) | Renal dysfunction, chronic tubulointerstitial nephritis, renal cyst formation and progression to end stage renal disease. Congenital heart defects, cardiomyopathy. | ANKS6, CEP83, CEP164, CEP290, DCDC2, GLIS2, IFT172, INVS, IQCB1, MAPKBP1 219, NEK8, NPHP1, NPHP3, NPHP4, RPGRIP1L, SDCCAG8, TMEM67, TTC21B (IFT139), WDR19 (IFT144), ZNF423 |
| Oculocerebrorenal syndrome of Lowe (OCRL) | Defects in eyes, central nervous system and kidneys; hypotonia and feeding difficulties, developmental delay, intellectual disability, behavioural problems, seizures and short stature. Occurs almost exclusively in males. | OCRL |
| Oral-facial-digital syndrome (OFDS) | Defective development of brain, heart, face, limbs and kidneys; polycystic kidneys. | C2CD3, C5orf42, CPLANE1, DDX59, IFT57, INTU, KIAA0753 (MNR), NEK1, OFD1, SCLT1, SCNM1, TBC1D32, TCTN3, TMEM107, TMEM138, TMEM216, TMEM231, WDPCP |
| Pallister-Hall syndrome (PHS) | Polydactyly, syndactyly, hypothalamic hamartoma, and bifid epiglottis. Other symptoms include imperforate anus, abnormalities in the kidneys, cardiac defects, small genitalia, lack of fingers, nail problems, cleft palate, bifid uvula, and development delay and behavioural problems. | GLI3 |
| Pituitary stalk interruption syndrome (PSIS) | Congenital abnormality of the pituitary leading to pituitary deficiency. | CDON, IFT56 (TTC26) 220, GPR161, HESX1, LHX4, PROKR2, ROBO1, WDR11 |
| Polycystic kidney/liver disease (PKD) | Enlarged cystic and dysfunctional kidneys and/or livers. | ALG5, ALG8, DNAJB11, DZIP1L, GANAB, JAG1, LRP5, PKD1, PKD2, PKHD1, PRKCSH, SEC63, |
| Retinitis pigmentosa (RP) | Retinal defects leading to progressive vision loss. | >90 genes; see RetNet, the Retinal Information Network |
| RHYNS syndrome | Syndromic retinal disorder characterized by the association of retinitis pigmentosa, hypopituitarism, nephronophthisis, and skeletal dysplasia. | TMEM67 221 |
| Senior–Løken syndrome (SLSN) | Nephronophthisis (NPHP) associated with retinal dystrophy. | CEP164, CEP290, INV, IQCB1, NPHP1, NPHP3, NPHP4, SDCCAG8, TRAF3IP1, WDR19 (IFT144) |
| Stromme syndrome (STROMS) | Usually characterized by microcephaly, ocular anomalies, and apple-peel intestinal atresia. Other symptoms include facial dysmorphism, motor delay and intellectual disability, as well as heart, brain, kidney, and craniofacial abnormalities. | CENPF |
| Syndactyly-telecanthus-anogenital and renal malformations (STAR) syndrome | Syndactyly, telecanthus, anogenital and renal malformations. | CCNQ 222 |
| Usher (USH) syndrome | Sensorineural hearing loss or deafness and progressive vision loss due to retinitis pigmentosa. | ADGRV1, ARSG, CDH23, CEP78d, CEP250, CIB2, CLRN1, DFNB31, ESPN, HARS1, MYO7A, PCDH15, PDZD7, USH1C, USH1G, USH2A, WHRN |
| Von Hippel-Lindau (VHL) disease | Abnormal growth of both benign and cancerous tumours and cysts in many parts of the body, including central nervous system, kidney, pancreas, adrenal glands and endolymphatic sac. Anxiety disorders. | VHL |
| Weyer acrodental dysostosis (WAD) | Milder form of Ellis-Van Creveld syndrome without congenital heart defects. | EVC, EVC2 |
The list of ciliopathy disease genes was modified from Ref.7.
Adapted from Genetic and Rare Diseases Information Center (GARD) and Orphanet.
Unless otherwise indicated, the genes listed were obtained by searching Ref.7 and the OMIM database using the disease name as search entry. Genes indicated in bold encode proteins localizing to the cilium–centrosome axis according to the SYSCILIA gold standard version 2 223, or alternative studies as indicated for some genes, where supporting references are indicated.
Studies have suggested that patients with CEP78 mutations can present with atypical Usher syndrome or retinitis pigmentosa 224.
A major challenge for the field is deconvolving how genetic changes lead to the clinical phenotypes seen in patients with ciliopathies, and how defective primary cilia can give rise to isolated, single organ diseases or syndromic ciliopathies that affect multiple organs. While some types of ciliopathy-causing mutations, for example, in certain genes that cause Bardet–Biedl syndrome (BBS), can reliably predict patient phenotype1, in other cases a clear genotype–phenotype correlation can be challenging to establish. A prominent example is CEP290 (OMIM 610142), mutations in which can give rise to five distinct ciliopathies, including BBS, Joubert syndrome (JBTS), Meckel syndrome (MKS), nephronophthisis (NPHP) and the related Senior-Løken syndrome (SLSN), in addition to Leber congenital amaurosis (LCA), an early-onset vision loss (Table 1). Multiple factors account for the clinical heterogeneity in patients with ciliopathies, such as the type of alleles, overlapping and cell type-dependent functions of ciliary proteins, the specific timing of ciliary dysfunction during development, and modifier genes or genetic background 2–5.
Currently, 247 genes are known to give rise to ciliopathies when mutated, affecting either motile cilia, primary cilia or both2,6,7. Most of these established ciliopathy genes encode proteins that localize to the cilium–basal body axis and are associated with so-called first-order ciliopathies, whilst genes mutated in second-order ciliopathies encode proteins that are localized elsewhere in the cell and affect cilia indirectly2,7 (Table 1). Classic examples of first-order ciliopathy genes are those that encode components of the intraflagellar transport [G] (IFT) machinery, a highly conserved microtubule-based transport system operating within cilia, which is broadly required for ciliary assembly, maintenance and function. Importantly, the detailed 3D structure of protein complexes involved in IFT has now been solved (Box 1) 8, which is facilitating the structural modelling of pathogenic variants to predict patient phenotypes1. Examples of second-order ciliopathy genes include those encoding transcription factors that regulate the expression of ciliary genes, or proteins of the secretory pathway that affect, for example, glycosylation and trafficking of integral ciliary membrane proteins 2,7. In addition to these established ciliopathy genes, a recent survey of the Human Phenotype Ontology (HPO) database estimated that >300 additional human disorders that are not classified as ciliopathies may have a phenotypic spectrum that includes ciliary defects. These have been referred to as ‘disorders with ciliary contribution (DCC)’; the extent of ciliary dysfunction and how it arises in these diseases remain largely unknown7. Thus, the ciliopathies represent an expanding number of pleiotropic human diseases for which the underlying aetiology is not fully understood.
Box 1. Intraflagellar transport.
Intraflagellar transport (IFT) refers to the conserved microtubule-based motility system that moves bidirectionally along ciliary axonemes to transport cargoes into and out of the organelle. Originally identified in the green alga Chlamydomonas reinhardtii 225, IFT has subsequently been identified in all ciliated eukaryotic organisms, including protists, worms, and vertebrates 8,226,227.
Canonical IFT consists of anterograde (heterotrimeric kinesin-2) and retrograde (cytoplasmic dynein 2) microtubule motors that move trains of IFT-A and IFT-B complexes with associated cargoes from the ciliary base to the tip (kinesin-2) and back (dynein 2). Transport of axonemal building blocks, for example, tubulin, to the ciliary tip occurs through direct binding to specific IFT proteins 228–230, whereas IFT-mediated retrograde transport of ciliary integral or peripheral membrane proteins relies on the BBSome as cargo adaptor 25,231. As for anterograde IFT of transmembrane cargo, this is thought to occur by a mechanism involving both IFT-A and associated TUBBY-like proteins, which have also been implicated in ciliary import of lipidated cargoes 25,28–30,97. In some cell types and organisms, additional kinesins such as homodimeric kinesin-2 (OSM-3 in Caenorhabditis elegans, KIF17 in humans) and kinesin-3 (KLP-6 in C. elegans, KIF13B in humans) motors function in coordination with canonical IFT to modulate ciliary structure and function 232,233.
Numerous studies performed in different cell types and organisms have shown that IFT is required for assembly and function of almost all eukaryotic cilia and flagella 227,232,234. Fundamental studies on IFT were instrumental in providing the first link between primary cilia, developmental signalling and human disease 196–198, and mutations in IFT genes are now known to be causative of multiple ciliopathies 2,8.
Recent advances in imaging and structural biology, combined with molecular, genetic and biochemical approaches, have provided detailed insight into the 3D structure and molecular workings of the core IFT system 8,30,33,97,232,234–236, as well as the IFT-A-associated Tubby-like proteins and retrograde membrane cargo adaptor complex, the BBSome 29,30,237–239 (see the table for a list of IFT components).
| Complex | Protein name | Alternative name(s) | Gene | OMIM number |
|---|---|---|---|---|
| Heterotrimeric kinesin-2 (canonical anterograde motor) | KIF3A | NA | KIF3A | 604683 |
| KIF3B | KIAA0359 | KIF3B | 603754 | |
| KIF3AP | KAP3 | KIF3AP | 601836 | |
| Homodimeric kinesin 2 (accessory anterograde motor) | KIF17 | KIAA1405 | KIF17 | 605037 |
| Cytoplasmic dynein 2 (retrograde motor) | DYNC2H1 | DNCH2, DHC2, DHC1B | DYNC2H1 | 603297 |
| DYNC2I1 | WDR60 | DYNC2I1 | 615462 | |
| DYNC2I2 | WDR34 | DYNC2I2 | 613363 | |
| DYNC2LI1 | D2LIC, LIC3 | DYNC2LI1 | 617083 | |
| DYNLL1 | DNCL1, DLC1, PIN, LC8 | DYNLL1 | 601562 | |
| DYNLL2 | DLC2 | DYNLL2 | 608942 | |
| TCTEX1D2 | NA | DYNLT2B | 617353 | |
| DYNLT1 | TCTEL1 | DYNLT1 | 601554 | |
| DYNLT3 | TCTE1L | DYNLT3 | 300302 | |
| DYNLRB1 | DNCL2A, DNLC2A | DYNLRB1 | 607167 | |
| DYNLRB2 | DNLC2B | DYNLRB2 | 607168 | |
| IFT-B1 | IFT88 | TG737, Polaris, TTC10, DAF19 | IFT88 | 600595 |
| IFT81 | CDV1 | IFT81 | 605489 | |
| IFT74 | CCDC2, CMG1, BBS22, JBTS40, SPGF58 | IFT74 | 608040 | |
| IFT70 | TTC30B | TTC30B | NA | |
| IFT56 | TTC26, BRENS | TTC26 | 617453 | |
| IFT52 | SRTD16 | IFT52 | 617094 | |
| IFT46 | C11orf2 | IFT46 | NA | |
| IFT27 | RABL4, BBS19 | IFT27 | 615870 | |
| IFT25 | HSPB11 | HSPB11 | NA | |
| IFT22 | RABL5 | NA | NA | |
| IFT-B2 | IFT172 | SLB, KIAA1179, BBS20, RP71, SRTD10 | IFT172 | 607386 |
| IFT80 | KIAA1374, WDR56, SRTD2 | IFT80 | 611177 | |
| IFT57 | ESRRBL1, HIPPI, OFD18 | IFT57 | 606621 | |
| IFT54 | TRAF3IP1, MIPT3, SLSN9 | TRAF3IP1 | 616629 | |
| IFT38 | CLUAP1, FAP22, DYF3, QILIN | CLUAP1 | 616787 | |
| IFT20 | None | IFT20 | 614394 | |
| IFT-A1 | IFT144 | WDR19, SPGF72, CED4, NPHP13, SLSN8, SRTD5 | WDR19 | 608151 |
| IFT140 | KIAA0590, RP80, SRTD9 | IFT140 | 266920 | |
| IFT122 | WDR10, CED1 | IFT122 | 606045 | |
| IFT-A2 | IFT139 | TTC21B, THM1, NPHP12, SRTD4 | TTC21B | 612014 |
| IFT121 | WDR35, KIAA1336, CED2, SRTD7 | WDR35 | 613602 | |
| IFT43 | C14ORF179, CED3, RP81, SRTD18 | IFT43 | 614068 | |
| BBSome (retrograde IFT-B membrane cargo adaptor) | BBS1 | NA | BBS1 | 209901 |
| BBS2 | RP74 | BBS2 | 606151 | |
| BBS4 | NA | BBS4 | 600374 | |
| BBS5 | NA | BBS5 | 603650 | |
| BBS7 | FLJ10715 | BBS7 | 607590 | |
| BBS8 | TTC8, RP51 | TTC8 | 615985 | |
| BBS9 | PTHB1 | BBS9 | 615986 | |
| BBS18 | BBIP1 | BBIP1 | 615995 |
Names refer to human proteins/genes. NA, not available.
Related links
GARD https://rarediseases.info.nih.gov/
610142, 610937, 605446, 609863, 613846, 604730, 601197, 601313, 173910, 601309,
601500, 608922, 613037, 608002, 604695, 602676, 604011, 605413, 615519
604683, 603754, 601836, 605037, 603297, 615462, 613363, 617083, 601562, 608942,
617353, 601554, 300302, 607167, 607168, 600595, 605489, 608040, 617453, 617094,
615870, 607386, 611177, 606621, 616629, 616787, 614394, 608151, 266920, 606045,
612014, 613602, 614068, 209901, 606151, 600374, 603650, 607590, 615985, 615986,
615995
Addressing the knowledge gap between genotype and phenotype is essential to understand the aetiology of ciliopathies, improve diagnostics and develop treatments for these diseases. Recent work has demonstrated that primary cilia exhibit remarkable structural and compositional diversity across cell types, tissues and developmental scale, which allows cilia to function as versatile signalling hubs that can fine-tune their content and signalling function according to the needs of the cell and, ultimately, the organism. Such structural and functional diversity of cilia is regulated by a series of complex and dynamic mechanisms that may, at least in part, offer a plausible explanation for the genetic and phenotypic heterogeneity observed in ciliopathies.
Here, we discuss the architectural and functional diversity of primary cilia in different vertebrate cell and tissue types. We explore how ciliary signalling function is regulated by compartmentalization and dynamic changes in the ciliary proteome in response to cellular or environmental cues and highlight how new technological developments are advancing the field. Finally, we provide examples of how human disease genetics and patient-derived stem cells can be leveraged to dissect cilia diversity in vivo, pointing us towards both new genes and novel functions for known cilia proteins. We do not cover motile cilia and associated ciliopathies, which have been reviewed elsewhere6,9.
Architecture of primary cilia
All types of cilia consist of a membrane-covered, microtubule-based axoneme that extends directly from a basal body, which anchors the cilium to the cell body by means of transition fibres (derived from centriolar distal appendages) and subdistal appendages (Figure 1a) 10. At the interface between the basal body and cilium itself lies the transition zone [G] (TZ), a ciliary gating structure that connects the axonemal microtubules via Y-shaped assemblies to the ciliary membrane at a site known as the ciliary necklace [G] (Figure 1a).
TZ composition, structure and function
A proteomic analysis of purified TZs from Chlamydomonas identified 115 distinct proteins, several of which are known to be mutated in human ciliopathies 11, including components of the three main TZ protein modules, the MKS, NPHP and CEP290 modules 2,12–14. Super-resolution imaging and structural biology approaches have provided insight into the molecular architecture of the TZ 15,16 and its spatial arrangement of individual proteins 17–19 and lipids 20. The length, structure, and composition of the TZ can vary significantly between different cell types and species 21. For example, the connecting cilium of mouse photoreceptors, which is equivalent to the TZ in other cilia types, is 1-1.5 μm in length, whereas the TZ of primary cilia of a human retinal pigment epithelial cell line is about 0.3 μm in length 17,22. There is growing evidence supporting the existence of cell type-specific differences with respect to expression and function of TZ proteins, and how these could modulate bespoke ‘gating’ functions. For example, the related proteins RPGRIP1L (OMIM: 610937; also known as NPHP8 or MKS5) and RPGRIP1 (OMIM: 605446) are both highly expressed and contribute to TZ assembly in mouse embryonic fibroblasts and kidney cell lines, but in patients and mice only RPGRIP1L mutations contribute to syndromic phenotypes, whilst RPGRIP1 mutations cause non-syndromic inherited retinal dystrophy 23. Similarly, when the broadly expressed Tctn1 and Tctn2 genes, encoding components of the MKS complex of the TZ, are knocked out in mice, striking tissue-specific differences in ciliogenesis defects and ciliary content are observed, which could explain the spectrum of features found in patients with JBTS13, who harbour mutations in TCTN1 (OMIM: 609863) or patients with JBTS24, who harbour mutations in TCTN2 (OMIM: 613846) 24. It is still unclear why these tissue-specific sensitivities exist. Furthermore, in contrast to IFT-associated protein complexes (Box 1), the detailed 3D structure of ciliary TZ protein complexes remains largely unknown.
While the phenotypic consequences of TZ gene mutations vary from mild to severe, depending on the type of allele and gene affected, most lesions in such genes result in altered ciliary protein content, thereby affecting signalling 14. Studies in multiple organisms and cell types have shown that the TZ, together with the transition fibres, controls the ciliary entrance and exit of both soluble and membrane-bound proteins by forming a selective diffusion barrier between the cilium and cell body. The mechanisms that allow certain proteins to cross this barrier are still not clear, but two models, which are not mutually exclusive, have been proposed: the ‘open sesame’ and ‘motorized plow’ models. The open sesame model posits that cargo binding to ciliary import factors, such as the IFT-A subcomplex, induces their privileged crossing of the TZ independently of motor activity 14,25. This model is supported by studies indicating that several membrane-associated proteins enter cilia independently of IFT kinesin-2 motors 25,26, but in most cases require interaction with the IFT-A subcomplex and associated TUBBY domain proteins TULP3 (OMIM: 604730) and TUB (OMIM: 601197) for ciliary entrance 27–30. According to the motorized plow model, ciliary cargoes are dragged physically across the TZ diffusion barrier by the action of IFT motors 25. In support of this model, anterograde IFT trains with associated axonemal/soluble cargoes assemble at the TZ prior to ciliary entrance, and mutations in a kinesin-2 motor subunit impair IFT train localization at the TZ and prevent axoneme extension 31–34. Conversely, several studies showed accumulation of cargoes in cilia upon disruption of specific IFT dynein 2 subunits 35–38. Interestingly, in ciliated sensory neurons of Caenorhabditis elegans lacking the dynein 2 intermediate chain subunit WDR-60, such IFT accumulations were prevented by disruption of specific TZ modules, indicating that the underpowered dynein 2 motor is unable to breach the TZ barrier unless the latter is disrupted or weakened 38. Moreover, inactivation of the dynein 2 heavy chain (CHE-3) or IFT-A components in worms affected TZ assembly and gating function 39,40, indicating that the TZ and IFT machinery reciprocally affect each other.
Importantly, whilst many of these players were key to understanding functional compartmentalization of cilia, they are also human disease genes (Table 1). The next challenge is to understand how different patient variants in seemingly core components of ciliary protein modules operating at the TZ can result in tissue-specific dysfunction, such as reported for IFT140 or CEP290 variants causing non-syndromic inherited retinal diseases 41,42. In the case of CEP290 variants, tissue-specific isoforms seem to be at play and can be specifically targeted by therapeutics to halt or reverse visual decline43. Indeed, anti-sense oligonucleotide clinical trials targeting this cryptic intronic variant are underway 44, as was one for genome surgery which was recently halted but demonstrated proof-of-concept in patients with LCA 45.
The ciliary axoneme
While all cilia are microtubule-based, the configuration of the axonemal microtubules exhibits significant structural diversity. How this is genetically programmed and how it affects cilia signalling remain open questions. A reductionistic textbook view of a primary cilium [G] is one of 9 microtubule doublets lacking a central pair, termed a ‘9+0’ configuration versus the ‘9+2’ configuration of motile cilia axonemes (Figure 1a, b). However, as the detailed ultrastructure of primary cilia from more and more cell types is being uncovered, aided by advances in imaging technologies such as serial section electron tomography (SSET) 46, cryo-electron tomography (cryoET) 47 and focused ion beam scanning electron microscopy (FIB-SEM) 48,49, it becomes clear that substantial deviations from this canonical ciliary architecture exist. At one extreme are the vertebrate rod and cone photoreceptors whose outer segment is a specialized primary cilium composed of numerous opsin-containing membranous discs (rods) or lamellae (cones), which is bridged to the inner segment by a modified TZ called connecting cilium (Figure 1c). The distal end of the connecting cilium extends into an axoneme that terminates with an elongated microtubule singlet segment missing the B subfibre found in canonical axonemal outer doublet microtubules 50. Atypically shaped non-motile cilia with elaborate membranous appendages and/or distal axonemal singlet extensions are also found on sensory neuronal cells of nematodes 51, whereas the primary ciliary axoneme of mammalian kidney epithelial cell lines displays ‘9 + 0’ microtubule configuration only at the base, with the middle and distal regions being comprised largely by an unstructured bundle of microtubule singlets and actin filaments (Figure 1b) 46,47,52. Similar microtubule thinning towards the axoneme tip has been reported in zebrafish embryos using ultra-expansion microscopy (UExM) 53. Additional examples of vertebrate cilia with diverse axoneme structure include the ‘9+2’ structure of kinocilia of inner ear hair cells that retain outer dynein arms; these modified ‘primary’ cilia play key roles in hair cell morphogenesis and maintenance, hence act ‘passively’ to allow mechano-electrical transduction to sound (Figure 1d) 54,55. In mammals, olfactory sensory neuron cilia are also ‘9+2’ (Figure 1e), but are immotile as they lack dynein arms 56,57. The functional significance of maintaining a central pair remains unclear. There are also examples of ‘9+0’ sensory cilia with evidence of cilia motility, including multiciliated choroid plexus epithelial cells involved in sensing cerebral spinal fluid 58, and recently in glucose-dependent motility of β-cell cilia in pancreatic islets, which are necessary for insulin secretion 59. As long as there are rules, there will clearly be exceptions to this ‘two’ cilia state.
The molecular basis for the structural diversity of cilia is not fully understood, but studies in a range of model organisms are starting to provide some clues 60. First, employment of ‘accessory’ kinesins that work in concert with the canonical anterograde IFT motor, heterotrimeric kinesin-2, may promote assembly of extended distal axonemal singlet microtubules in specific subsets of cilia. This is exemplified by the C. elegans homodimeric kinesin-2 motor, OSM-3, which mediates assembly of distal axonemal singlet segments in amphid channel cilia but not amphid wing cilia 61. Similarly, the vertebrate OSM-3 homologue KIF17 seems to specifically modulate the assembly of olfactory cilia and photoreceptor outer segments, but not other cilia types 62,63. Second, confinement of IFT trafficking to specific axonemal doublet (or singlet) microtubules, as observed in Chlamydomonas reinhardtii 64 and Trypanosoma brucei 65, could allow for structural or molecular specialization of the microtubules not utilized as tracks for IFT 60. Such structural and functional diversification of microtubules within the same axoneme may be influenced by the ‘tubulin code’: either specific tubulin isotypes 66,67 or post-translational modifications that regulate microtubule dynamics (Figure 1a) 68,69. These reversible marks play critical roles in the timing of cilia assembly and disassembly as well as function of multiple cilia types, including vertebrate sperm flagella 70, photoreceptor outer segments 71, and subsets of C. elegans neuronal sensory cilia 72,73. Finally, cell type-specific expression of IFT cargoes 74 or of centriolar and TZ components 18,23, which provide the foundation for the axoneme and regulate ciliary protein content, could also influence ciliary structural diversity. Importantly, since the IFT system not only mediates intraciliary transport of axonemal building blocks, but also regulates ciliary transport of membrane-bound and soluble signalling molecules, as discussed below, the above-described mechanisms are likely to impinge on ciliary membrane composition and signalling function.
The ciliary membrane
The ciliary membrane is connected to the plasma membrane via the periciliary membrane [G], which in some cell types is invaginated, forming a ciliary pocket [G] that surrounds the proximal region of the cilium (Figure 1a). Other cilia types including vertebrate photoreceptor outer segments and a subset of C. elegans neuronal cilia feature elaborate membrane extensions at their distal end50,51. Despite being continuous with the cell body plasma membrane, the ciliary membrane has a different composition of proteins and lipids that function in signalling (Figure 2a), and which localize dynamically to cilia in response to cellular and environmental cues25,75. Indeed, morphology76 or length77 of cilia themselves can be altered by these signalling cues, underscoring the dynamic and highly responsive nature of these organelles.
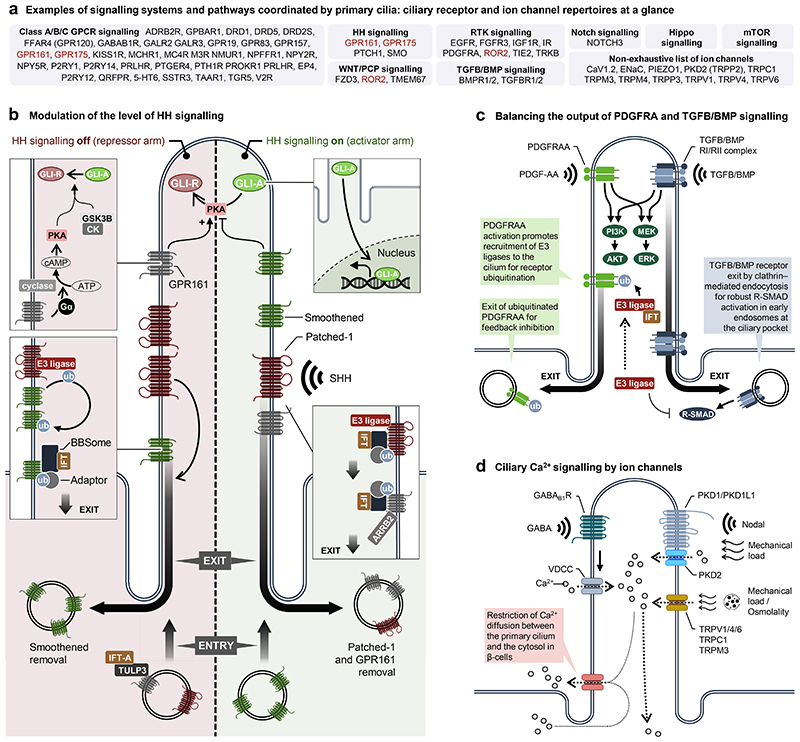
Figure 2. Overview of signalling pathways coordinated by primary cilia.
a, Overview of receptors and ion channels in ciliary signalling pathways. Receptors in red are listed twice, as they can be categorized in more than one signalling system. b, Overview of Hedgehog (HH) signalling. In the absence of sonic hedgehog (SHH) (in the repressor arm of HH signalling), the receptor patched-1 (PTCH1) is enriched in the ciliary membrane, preventing ciliary enrichment of smoothened (SMO) through WWP1 (E3-ligase)-mediated ubiquitination and ciliary exit by retrograde IFT. The class A GPCR, GPR161, is targeted to the cilium by tubby-like protein 3 (TULP3) and IFT-A to activate adenylate cyclases via G-proteins (Gα), leading to increased ciliary levels of cAMP. cAMP activates protein kinase A (PKA), which in complex with glycogen synthase kinase 3 β (GSK3β) and casein kinases (CK) promotes the limited proteolytic cleavage of full-length and activator versions of GLI2/3 transcription factors (GLI-A) into their repressor form (GLI-R). In the presence of SHH (in the activator arm of HH signalling), PTCH1 and GPR161 exit the cilium, allowing enrichment of ciliary SMO, which promotes formation of GLI-A. Exit of PTCH1 and GPR161 similarly relies on their ubiquitination and removal by BBSome-assisted retrograde IFT; GPR161 ubiquitination being controlled at the level of beta-arrestin 2 (ARRB2). Both GLI-A and GLI-R translocate from the cilium into the nucleus to induce and repress transcriptional activation of HH target genes, respectively. c, Overview of ciliary control of platelet-derived growth factor α (PDGFRα) and transforming growth factor β (TGF-β)/bone morphogenetic protein (BMP) signalling. Following activation of PDGFRα and downstream signalling via PI3K-AKT and MEK1/2-ERK1/2 pathways, E3 ligases of the CBL family ubiquitinate the receptor for internalization and feedback inhibition. TGFB/BMP signalling operates in the cilium via both canonical (R-SMAD) and non-canonical (e.g. PI3K-AKT and MEK1/2-ERK1/2 pathways). Robust canonical signalling relies on ciliary exit of activated TGFB receptors to activate R-SMADs, which are inhibited by E3-ligase SMURF1 at the ciliary pocket. d) Examples of stimulation modes (chemosensation, mechanosensation and osmolality) for ciliary Ca2+ signalling regulated by GPCRs and ion channels. Please see main text for further details. Abbreviations Ub: ubiquitination.
The ciliary membrane proteome
The best studied examples of ciliary membrane proteins include the autosomal dominant polycystic kidney disease (ADPKD) gene products polycystin-1 (PKD1; OMIM: 601313) and polycystin-2 (PKD2; OMIM: 173910), which form a cilium-localized heterodimeric receptor-cation channel complex essential for preventing cystogenesis 78–81, and the Sonic Hedgehog (SHH) co-receptor Patched-1 (PTCH1) (OMIM: 601309) and the class F GPCR, Smoothened (SMO) (OMIM: 601500), which accumulate in cilia in the absence and presence of SHH, respectively 82–84 (Figure 2b). Without cilia to localize to, while still being expressed normally, these signalling components no longer function properly, leading to characteristic fibrocystic features (in the case of ADPKD) or patterning defects (in the case of SHH signalling defects) observed in patients with ciliopathies. Intriguingly, even within the cilium itself, domains of protein organization may exist. Recent reports of microdomains of PKD2 localization enriched along the dorsal surface of immotile nodal cilia could explain how mechanosensory responses to directional flow exist 85,86.
Whilst less is known about how they signal downstream, numerous other receptors and ion channels have been reported to localize to the primary cilium in a dynamic and context-dependent manner, including a growing number of G protein coupled receptors (GPCRs) 87, receptor tyrosine kinases (RTKs) and TGFB/BMP receptors 88 (Figure 2a). Recent advances in cilia-targeted proximity-labelling and proteomics approaches have expanded the repertoire of cilia-localized receptors, ion channels and downstream effectors, and revealed how this may change in response to external signalling cues and/or mutations in specific ciliopathy disease genes, for example, BBS genes 89–94. However, to date the ciliary membrane/signalling proteome has been studied for only a handful of vertebrate cell types, including the IMCD3 kidney epithelial cell line 89,91, mouse fibroblast NIH3T3 cells 93 and mouse and zebrafish photoreceptor outer segments 90,94. In the future, it will be important to compare cilia proteomes across different cell types, both in vitro and in vivo, and during different cell differentiation and developmental stages.
The mechanisms controlling dynamic spatiotemporal localization of specific receptors and other signalling proteins to the cilium are complex and involve gating by the TZ (see: TZ composition, structure and function), a specialized ciliary import system for lipidated cargoes, and components of the IFT system (Box 1), which interacts with various adaptors to ferry specific integral or peripheral membrane proteins into or out of the organelle. During ciliary membrane protein import the IFT-A subcomplex binds to the TUBBY domain proteins TULP3 and TUB, which regulate the ciliary localization of a broad range of integral membrane proteins, including GPCRs and polycystins 27–30. TULP3 and TUB bind to phosphoinositides via their C-terminal TUBBY domain 27,28 and the IFT-A complex binds directly to phosphatidic acid and ceramide 95–97. Moreover, purified IFT-A was shown to bind to the ciliary targeting signal in SSTR3 98. Direct binding of TULP3/TUB to transmembrane cargoes remains undocumented, but a recent study indicated that TULP3 binds directly to ARL13B (OMIM: 608922) 29, an atypical GTPase that is highly enriched in cilia and mutated in JBTS 99,100. ARL13B is palmitoylated 101 and TULP3 is required for ciliary import of ARL13B as well as several other lipidated cargoes, including farnesylated INPP5E (OMIM: 613037) and myristoylated NPHP3 (OMIM: 608002) 29,102–104. In this context, ARL13B functions as a GEF for the small GTPase ARL3 (OMIM: 604695), and activated, ARL3-GTP promotes release of lipidated cargoes from their carrier proteins PDE6δ (OMIM: 602676) or UNC119/UNC119B (OMIM: 604011) causing their release into cilia 105.
In analogy with ARL3-dependent ciliary import of lipidated cargoes, the small GTPase RABL2B (OMIM: 605413) was shown to promote ciliary entrance of IFT trains in a GTP-dependent manner106. Shortly after IFT trains enter the ciliary compartment, GTP-bound RABL2 is inactivated by IFT81-IFT74, which function as a GAP to enhance GTP hydrolysis and inactivate RABL2 107. While a GDP-locked RABL2 variant failed to promote ciliogenesis, a GTP-locked variant caused aberrant accumulation of the BBSome [G] and associated cargoes in cilia 106,108, presumably owing to impaired dissociation of constitutively active RABL2 from the IFT machinery that prevents binding of the latter to BBSomes at the ciliary tip 107. Indeed, the BBSome is a well-described membrane cargo adaptor for the retrograde IFT machinery that binds to phospholipase D and various ubiquitinated transmembrane cargoes, including the GPCRs SSTR3 and SMO, to promote their exit from cilia 25,34,36,109–111. In this context, the BBSome seems to employ its own adaptor, the ancestral endosomal sorting complexes required for transport (ESCRT) protein TOM1L2 (OMIM: 615519), to facilitate interaction with ubiquitinated cargoes 111. Not surprisingly, mutations in BBS genes cause profound changes in the ciliary membrane proteome, as revealed using cilia-targeted proximity-labelling and proteomics approaches 36,89,90,94.
Ciliary lipid composition
Phosphoinositides (PIPs) are lipid signalling molecules that coordinate multiple membrane-associated molecular events. In primary cilia, high spatial organization of PIPs is observed; the ciliary membrane is enriched for PI(4)P, whereas the TZ and plasma membrane contain primarily PI(3,4,5)P3 and PI(4,5)P2, respectively (Figure 1a) 20,25. This boundary of distribution is highly regulated by specific cilia-localized lipid homeostatic enzymes, such as the ciliopathy gene product and PI(3,4,5)P3/PI(4,5)P2-specific phosphatase INPP5E 20,112-116, and is critical for regulating ciliary localization of various signalling receptors, including multiple GPCRs, which rely on the PI(4,5)P2- and IFT-A associated membrane adaptors TUB and TULP3, for recruitment to cilia 28. Other lipid species with critical functions in cilia include ceramides, which are essential for ciliogenesis in organisms ranging from Chlamydomonas to humans 96,117, and various sterols, which in vertebrates play essential roles in regulating GPCR-based signalling including Hedgehog118. Notably, while the lipid composition of the ciliary membrane affects its interaction with the IFT machinery thereby influencing ciliary protein import or export 28,96, perturbations in IFT or its associated membrane cargo adaptors, such as the BBSome, may conversely affect ciliary lipid composition. For example, a proteomics and lipidomics analysis of isolated photoreceptor outer segments from wild type and bbs1 mutant zebrafish showed enrichment of cholesterol and proteins involved in lipid homeostasis in outer segments of the mutants, indicating a critical role for the BBSome in regulating outer segment lipid homeostasis 94. Similarly, loss of BBS4 in Chlamydomonas caused altered composition of several lipid species within cilia, owing to abnormal accumulation of phospholipase D in the mutant cilia 36. Despite recent advances, the lipid composition of most vertebrate primary cilia types remains obscure owing to technical challenges associated with purifying primary cilia from such cells, combined with a paucity of reliable molecular probes for fluorescence imaging of most lipids 118.
Ciliary membrane biogenesis and homeostasis
Biogenesis of the ciliary membrane begins during initiation of ciliogenesis, which for vertebrate primary cilia can occur via two distinct pathways depending on the cell type. Mesenchymal cell types such as fibroblasts employ an intracellular pathway, in which ciliary membrane biogenesis is initiated by attachment and fusion of Golgi-derived vesicles to the mother centriole distal end before centriole docking at the plasma membrane 119. A similar intracellular pathway was reported for the biogenesis of vertebrate photoreceptor outer segments 120, whereas polarized kidney epithelial cells use an extracellular ciliogenesis pathway, whereby the mother centriole docks at the plasma membrane prior to ciliary membrane and axoneme extension 119. In the latter case, the midbody remnant and associated specialized membranes are believed to play a key role in initiating ciliary membrane outgrowth 121. For both pathways, proteins and lipids required for further ciliary membrane growth are transported in vesicles from the endoplasmic reticulum/Golgi towards the ciliary base, where vesicles are exocytosed and incorporated into the growing ciliary membrane. Many active players, for example, the small GTPases RAB8 and RAB11 and their associated effectors and regulators, are involved in such transport 119, some of which act in a cell type-dependent fashion. For example, the small GTPase RAB34 specifically promotes ciliary membrane biogenesis in the intracellular pathway but not the extracellular pathway 122,123. Considering the fundamentally different organization of cytoplasmic microtubules, along which vesicular transport of ciliary components occurs, in cell types that form cilia intracellularly versus extracellularly (Figure 1f, g), such cell type-specific requirement for ciliogenic factors is not surprising.
In addition to the enrichment processes described above, cilia also require a means to fine-tune concentration of these components to a ‘Goldilocks’ point for optimal signalling. It has been proposed that these enrichment mechanisms are counterbalanced by endocytosis at the ciliary pocket 124,125 and/or budding of ectosomes [G] or extracellular vesicles from the ciliary or periciliary membrane (Figure 1a) 126. While it is not yet clear whether cilia-associated endocytosis and ectosome shedding operate separately or in parallel, both processes would allow cilia appropriate content regulation by cell type and environmental conditions.
Cilia-dependent signalling
The dynamics of ciliary protein composition translate into a remarkable flexibility in the sensory capacity of primary cilia (Figure 2a), enabling the translation of quite diverse signalling inputs via elaborate signalling networks to process information in time and space 127. A prominent example includes that of the GPCR family, which probably functions in all types of primary cilia, albeit in a highly cell type-specific manner, to orchestrate diverse processes during development and in tissue homeostasis. GPCRs from different classes control light detection in photoreceptor cells; odorant sensation in olfactory sensory neurons; cognitive processes in the brain; and energy homeostasis and appetite via the concerted communication between multiple organs and tissue cell types, including, but not limited to, neurons in the arcuate nucleus of the hypothalamus, pancreatic islet cells, cholangiocytes and adipose tissue precursor cells 128–130.
Ciliary coordination of Hedgehog signalling
To ensure signalling flexibility, cells have evolved diverse mechanisms to monitor and fine-tune the temporal localization and interaction of regulatory proteins within the cilium-basal body axis that orchestrate cell type-specific signalling outputs in different developmental or environmental contexts 127. Perhaps the best described pathway system to be controlled by such mechanisms is Hedgehog signalling (Figure 2b), in which the concerted regulation of receptor trafficking and activity is orchestrated in part by unique lipid compositions of spatially distinct ciliary membrane domains 20,25,118.
In the absence of SHH, the cilium is enriched in PTCH1 and the class A GPCR GPR161 (OMIM 612250), which in combination form a Hedgehog signalling repression machinery that promotes the proteolytic cleavage of GLI2/3 transcription factors into their repressor forms (GLI-R), thereby preventing expression of Hedgehog target genes such as GLI1. In this scenario, GPR161 activates cAMP-dependent kinase (PKA), which in concert with GSK3B and CK phosphorylates GLI2/3 for their processing, while PTCH1 prevents ciliary accumulation of SMO, which counteracts proteolytic cleavage of GLI2/3. Binding of SHH to PTCH1 leads to ciliary removal of both PTCH1 and GPR161, allowing SMO to enter the ciliary compartment and inhibit PKA activity via its C-terminal PKA inhibitor (PKI) motif that functions as a decoy substrate sequence to physically block the active site of PKA and thereby switches off its enzymatic activity 131. This allows GLI2/3 to stay in their activator forms (GLI-A), which can traffic to the nucleus for target gene expression (Figure 2b). Furthermore, PKA inhibition in the activator arm of Hedgehog signalling was suggested to be controlled at the level of ciliary GPR175 (OMIM 608336) entry, which decreases cAMP production 132.
Regulation by ubiquitination
While molecular and mechanistic insights into the dynamics of ciliary signalling in different cellular, developmental or environmental contexts, and the implications thereof in disease, are not yet fully understood, emerging evidence points to a critical function of E3 ubiquitin ligases in coordinating ciliary signalling outputs 133. In the repressor arm of Hedgehog signalling, this is in part controlled at the level of SMO ubiquitination 110,134 by the WW domain E3 ligase, WWP1, which is brought into the cilium by PTCH1. This regulatory step allows SMO to interact with the BBSome for removal by retrograde IFT 135 (Figure 2b). Similarly, exit of PTCH1 and GPR161 in the activator arm of Hedgehog signalling relies on their ubiquitination. GPR161 ubiquitination is controlled by beta-arrestin 2 (ARRB2) 134, which is recruited to the cilium upon Hedgehog stimulation to remove GPR161 98,136 in concert with TOM1L2, an adaptor for BBSome-mediated retrieval of UbK63-tagged GPCRs from cilia 111. In these conditions, assembly and function of the BBSome complex rely on its own ubiquitination carried out by GPCR-cAMP-mediated mono-ubiquitination of BBSome subunits by the RING E3 ubiquitin ligase, PJA2 137. PTCH1 exit is co-regulated by the combined action of E3 ligases of the HECT domain family, SMURF1/2 138, and the endocytic adaptor protein NUMB, which when mutated results in attenuation of Hedgehog signalling and developmental defects, such as reduced size of the cerebellum 93. Furthermore, activation of Hedgehog signalling relies on ciliary localization of the CTLH E3 ubiquitin ligase complex, which when mutated in Xenopus laevis causes ciliopathy-like phenotypes, including, but not limited to, neural patterning defects 139. Similarly, ciliary modulation of downstream pathways in RTK and TGFB/BMP signalling (Figure 2c) 140–142 is coupled to the dynamic translocation and function of E3 ligases in the cilium-basal body axis. The timely feedback inhibition of PDGF-AA signalling proceeds through ciliary recruitment of CBL E3 ligases of the RING finger family, followed by ubiquitin-mediated internalization of its receptor, PDGFR-A (OMIM 173490) 143, while SMURF1 E3 ligase at the ciliary pocket region inhibits activation of R-SMAD transcription factors in TGF/BMP signalling during heart development 144 (Figure 2c).In addition, cilia assembly, length control and resorption are controlled by dynamic networks of E3 ubiquitin ligases and deubiquitinases 133,145,146, and proteomic approaches 147 have delineated ciliary ubiquitinomes, which mark cell type-specific signatures of ubiquitin-mediated processes, thereby contributing to the understanding of diversity in function of primary cilia across different cell types and tissues.
Ciliary Ca2+ signalling
Ciliary ion channels contribute to both osmo-, mechano- and chemosensory capabilities in various developmental and homeostatic contexts. In some cases, ion channel activity is controlled at the level of ciliary GCPRs130, such as in insulin-secreting β-cells of the pancreatic islet of Langerhans, where γ-aminobutyric acid-mediated activation of the class C GPCR GABAB1 receptor (GABAB1R) (OMIM 603540) promotes Ca2+ influx via L-type Ca2+ channel (VDCC) within the cilium proper148 (Figure 2d). In other cases, primary cilia sense mechanical loads, signalling molecules and/or changes in extracellular osmolality through various Ca2+ channels of the TRP family149–151 (Figure 1d). Through diverse mechanisms, primary cilia thus contribute to the orchestration of developmental processes, such as in left–right (LR) symmetry breaking during early embryogenesis 85,86,152, and in safeguarding function and remodelling of urinary, cardiovascular and musculoskeletal systems 153. While leftward fluid flow provided by rotating cilia in the pit of the LR organizer (LRO) during early embryogenesis is both necessary and sufficient to define the left side of the embryo in most vertebrates, including the mouse 154,155, the mechanisms by which this flow is interpreted into later asymmetrical placement and patterning of the internal organs and associated vasculature have been the subject of much debate over the past 25 years 156. Recent work using optical tweezers and advanced imaging techniques in mouse and zebrafish supports the conclusion that primary cilia at the left side of the LRO function as mechanosensors that convert the biomechanical forces of the flow into Ca2+ signals via PKD2 85,86, whereas the accumulation of the PKD1L1 polycystin channel at the left mouse LRO margin may provide a chemosensory channel for Nodal-mediated Ca2+ signalling in LR determination 152. It is therefore plausible that multiple mechanisms may act in concert via primary cilia to translate nodal flow, which when defective causes heterotaxy, including, but not limited to, cardiac laterality defects 157,158. Indeed, emerging evidence suggests that primary cilia combine chemosensory inputs with mechanical loads, such as in the detection of low-flow forces by primary cilia during development of the retinal vasculature, which sensitizes endothelial cells to BMP signalling 159.
Cilia dynamics in development and disease
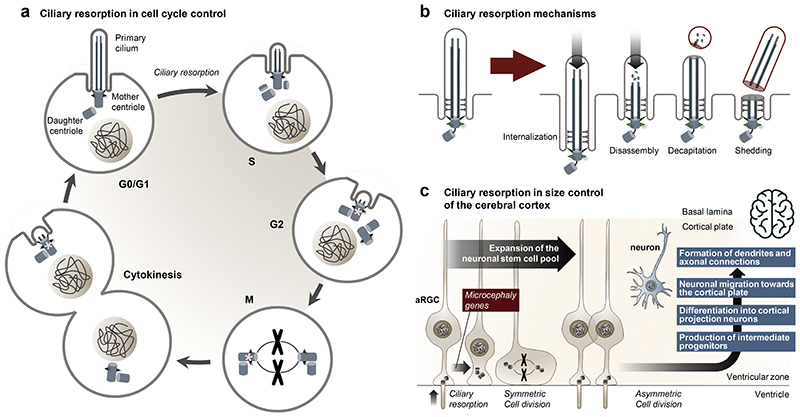
Unlike most organelles, cilia and the centrioles from which they are templated, are tightly regulated by the cell cycle. Cilia transit through a sequence of assembly, elongation and resorption coordinated by the centrosome [G] 160, with the latter being required to free up the centrioles for mitotic spindle pole formation (Figure 3). The molecular principles of orchestrating these transitions have been well defined in cell culture systems, generally using synchronization through serum starvation or addition 161. This reductionist approach has enabled the identification of cilia-linked growth factor pathways activated by serum, including RTKs 88 and lysophosphatidic acid receptors (LPAR) 162, the latter shown recently to trigger cilia disassembly and promote neurogenesis in vivo 163. In some cases, these mechanisms have not translated well in vivo. For example, the in vivo role of the second phase of the bi-phasic cilia resorption described in cell lines to depend on HDAC6-mediated axonemal tubulin de-acetylation 164 is unclear. Knock-out Hdac6 mice have no phenotype, but a mutation affecting HDAC6 post-transcriptional stability in humans (OMIM: 300272) could have a potentially ciliopathic phenotype 165. Nevertheless, defects in timely cilia resorption have been reported, for example, in patient fibroblasts, neuronal progenitors or brain organoids from patient induced pluripotent stem cells (iPSCs) with mutations in CPAP (OMIM: 609279), WDR62 (OMIM: 613583) or RRP7A (OMIM: 619449), resulting in depletion of the neuronal stem cell pool in the cerebral cortex leading to primary microcephaly (MCPH) 166–168 (Figure 3c). As such, not only the availability of a cilium to signal but also the ability to dismantle it, through gradual resorption, rapid excision or a combination of both 169, seems key to progenitor fate decisions and subsequent steps in tissue morphogenesis.
Figure 3. Ciliary dynamics.
a, Assembly and disassembly of primary cilia are tightly coordinated with the cell cycle. A single primary cilium assembles from the mother centriole in G0/G1 phase, and the cilium is resorbed during the G1/S and M phases to liberate duplicated centrioles for mitotic spindle pole formation. At the end of cytokinesis, the daughter cell inheriting the oldest mother centriole (marked with an asterisk), will begin forming a new primary cilium prior to the other daughter cells. b, Schematic illustration of different modes by which a cilium can be resorbed. c, During development of the cerebral cortex, apical radial glia cells (aRGC) primary cilia, which project into the ventricular lumen, are resorbed to allow expansion of the neural stem cell pool by symmetric cell divisions. By asymmetric cell divisions, aRGCs subsequently form intermediate progenitors that differentiate into cortical projecting neurons that migrate towards the cortical plate to form the dendrites and axonal connections. Consequently, dysfunction in the timely resorption of aRGC primary cilia is linked to proliferation–differentiation decision defects and reduced size of the cerebral cortex such as in microcephaly.
Cilia dynamics in proliferating tissue
Intricately linked with the cell cycle and cell differentiation state, primary cilia may not always be present on a cell in a growing tissue at a given moment. In a proliferative tissue, these dynamics would affect a cell’s signalling competence. Indeed, the daughter cell that inherits the ‘older’ mother centriole may become signalling competent first by assembling a cilium ahead of its sib 170–172. This cellular heterogeneity adds an extra layer of complexity as to how a transient organelle, which is required for mitogenic signalling including Hedgehog signalling, responds if it is only present for a portion of the cell cycle. Indeed, cilia have been shown to persist into S phase in a Hedgehog-responsive medulloblastoma cell line 173, in chick neural tube progenitors into late G2 in vivo 174 and on S/G2 cells in all embryonic cell types examined in Arl13b-Fucci2 mice 172. How Hedgehog can instruct robust developmental decisions may involve a ‘mitotic memory’ — a means to ‘remember’ signalling between ciliated phases in proliferating tissues —to deal with the variability of having a cilium in primary cerebellar granular precursors whose proliferation drives foliation of the developing cerebellum 173. As many developmental decisions are made in response to Hedgehog, determined by levels and length of time of exposure to ligand, this work connects the need for robust signalling with a dynamic organelle necessary to drive the growth of the developing brain, face and limb bud, systems often affected in ciliopathies 175.
More than just presence or absence, we do not understand how cilia content may change with the cell cycle, although evidence suggests this occurs for phosphoinositide content 176 and signalling competence in subsequent interphases 173. Moreover, specific and rapid remodelling of the cilia proteome occurs in the timescale of tens of minutes in response to Hedgehog ligand 92, but how the sensitivity and responsiveness may change with cell cycle stages remains unclear. These observations raise questions, particularly in rapidly dividing tissues or cancers, as to the physiological consequence of having a cilium at different stages of the cell cycle. Future studies, possibly combining organelle-specific proximity labelling techniques with cell cycle biosensors, will be needed to determine whether cilia can function as effective ‘signalling organelles’ if present regardless of cell cycle stage, or whether their contents and signalling competence are regulated in a cell cycle-specific manner.
Cilia dynamics during differentiation
Over longer periods, in this case developmental time, ciliation status or axoneme configuration may change within a tissue. Both olfactory 56 and choroid plexus 58 cilia transit from motile to sensory functions soon after their birth. Developing from a population of ciliated bipotential hepatoblast progenitor 177, the adult liver is composed mostly of hepatocytes, which are predominantly non-ciliated, and cholangiocytes of the biliary tract that are highly ciliated 178,179. Interestingly, mitogens, such as SHH, play key early roles in the expansion of the progenitors, inhibiting their hepatocyte lineage commitment 180,181. This finding suggests that temporal restriction of Hedgehog activation via cilia status is key for cell fate. In contrast to hepatocytes, cholangiocytes have very long cilia, which are proposed to function as mechanosensors (detecting PKD1/2-mediated flow) 182, osmosensors (sensing TRPV4-regulated hypotonicity) 183 and chemosensors (identifying purigenic receptor P2Y12 (OMIM: 600515) and bile acid receptor TGR5 (OMIM: 610147) signalling) 184 to regulate cholangiocyte homeostasis and the biophysical properties of bile. Consistent with roles for cilia across liver development and health, mutations in cilia genes are reported in congenital anomalies such as biliary atresia 185 and fibrocystic liver diseases 186, whilst cilia defects are reported during chronic inflammation and cholangiocarcinoma 187. In addition, primary cilia of pre-adipocytes are configured to orchestrate multiple signalling pathways that in different environmental contexts balance differentiation decisions of cells 129, including, but not limited to, IGF-1R (OMIM: 147370) 188 and class A GPCR signalling, the latter expanding white adipose tissue via omega-3 fatty acid-mediated activation of FFAR4 (GPR120, OMIM: 609044) 189. Understanding cilia states and their changing configurations and functions during differentiation remains a key challenge to interpreting disease phenotypes.
Rewiring of signalling caused by ciliary loss
Cilia loss as a result of mutation can result not only in dampening of signalling cascades that run through cilia but also misactivation of signalling responses normally held in check by an operational cilia axis. In cholangiocytes, ciliary loss drives activation of alternative signalling pathways either due to relocalization of or inhibited interaction with signalling molecules. This includes TGR5, a typically ciliary GPCR that when localized to the apical membrane alters the interaction with inhibitory to stimulatory Gα protein to drive cAMP production 190. Biliary exosomes in the bile interact with cilia to suppress ERK-mediated proliferation, whilst cholangiocytes without cilia do not respond to exosomes and proliferate 191. In the case of loss of Pkd1 or Pkd2 in the mouse, development of ADPKD depends on intact cilia; inappropriate signalling mediated by cilia upon disruption of PKD1/2 leading to cysts can be partially rescued by ablating cilia via Ift20 and/or Kif3a knockout 192. In postnatal mouse cholangiocytes, cilia loss leads to cystogenesis through an upregulation of a paracrine TGFB/R-SMAD proinflammatory axis, which remodels the cystic niche and biophysical properties of cystic ducts 193. Here, cilia seem to regulate the size, shape and mechanical properties of the committed bile ducts. From a translational perspective, remarkable recent work in mice has shown that similar aberrant changes in cell morphology, lumen size and extracellular matrix deposition of end-stage cystic kidneys can be reversed by restoring ADPKD gene function 194.
Conclusions and outlook
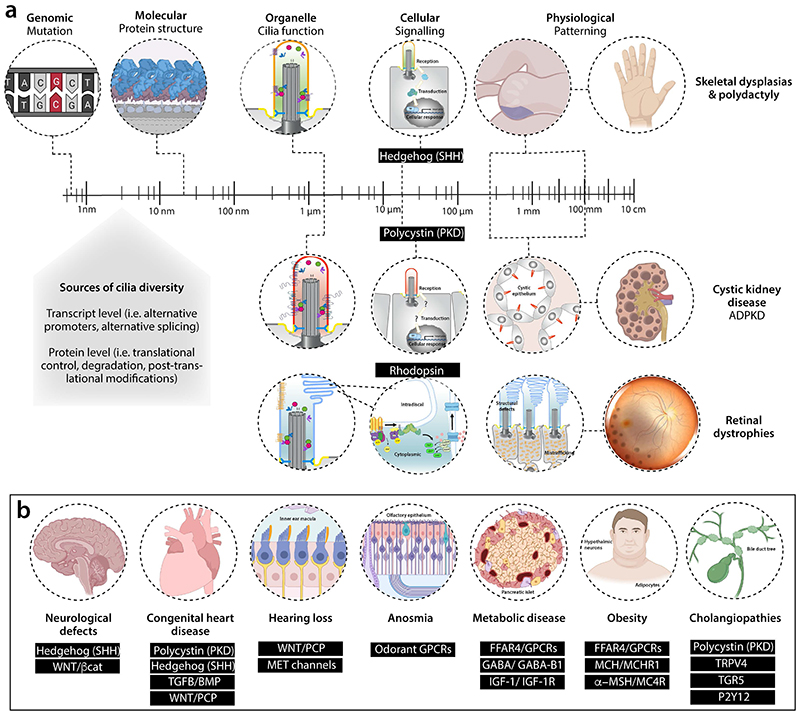
Once thought of as evolutionary remnants 195, primary cilia are known to play essential roles as coordinators of multiple signalling pathways that control key cellular processes during development and homeostasis of tissues and organs. Pioneered by early studies on IFT that provided the first links between primary cilia, vertebrate signalling and development and disease 196–198, a large body of literature has since substantiated the importance of primary cilia in human health and disease, with more than 30 distinct ciliopathies and 247 ciliopathy disease genes identified to date 2,6,7. Despite impressive recent progress in identifying genetic variation and dissecting the molecular aetiology of ciliopathies, understanding the correlation between genotype and clinical phenotypes of patients with ciliopathies remains an important challenge in the field — one of biology across scales (Figure 4). Moreover, how defective primary cilia can give rise to isolated, single organ diseases or syndromic ciliopathies affecting multiple organs remains unclear; it suggests not all cilia are ‘equal’, resulting in differences in sensitivity to dysfunction through mutation
Figure 4. Challenge of ciliopathies — biology across scales.
a, Schematic of how variant identification in a patient with ciliopathy is just the start of the challenge. Genetic changes are the same across all cell types. Understanding how these variants disturb different types of cilia structurally and functionally in terms of signalling readouts is our knowledge gap across cell types and developmental times, one that we need to address to understand patient phenotypes. Possible sources of tissue-specific phenotypes are highlighted. We focus on the best characterized cilia-dependent signalling defects resulting from ciliopathy mutations in different tissue types, including skeleton, kidney epithelia and photoreceptors resulting in ciliopathic disease. For the kidney, we have focused on autosomal dominant polycystic kidney disease through PKD1/2 signalling but other renal diseases include autosomal recessive polycystic kidney disease and nephronophthisis, the latter involving additional signaling defects (see Table 1). b, Examples of the signalling pathways disrupted in different tissue types in the ciliopathies. Abbreviations: ADPKD, autosomal dominant polycystic kidney disease; FFAR4, Free Fatty Acid Receptor 4; GABA, gamma-aminobutyric acid; GPCRs, G protein coupled receptors; IGF, insulin-like growth factor; MCH, melanin-concentrating hormone; MET, mechano-electrical transduction; MSH, melanocyte-stimulating hormone; P2Y12, purinergic receptor P2Y; PKD, Polycystin; SHH, Sonic Hedgehog; TGFB/BMP, Transforming Growth Factor Beta/ Bone Morphogenetic Protein; TGR5, Takeda G protein-coupled receptor 5; TRPV4, transient receptor potential vanilloid-type 4; WNT/ βcat, canonical Wingless/Integrated Beta catenin pathway; WNT/PCP, non-canonical Wingless/Integrated Planar Cell Polarity pathway.
Importantly, we need to tackle the complexity of cilia-mediated signalling in vivo at the intracellular level (for example, cross-talk between signalling pathways), at the level of specific cell types, and across tissues or centres to integrate complex physiological circuits (Figure 4a). To this end, we need better tools to track structural and functional diversity of primary cilia in biologically relevant cell types across developmental times (Figure 4b). Recent technological developments, including various omics approaches, have provided detailed insight into the ciliary signalling repertoire and its changes in response to extracellular signals or in specific mutant backgrounds 89,90,94,133. However, knowledge of differences in ciliary structure, composition and signalling function in vivo across cell types, tissues and environmental or developmental contexts remains scarce.
In this era of functional genomics, we need improved means to capture cell- and stage-type specific differences in expression of isoforms. At the mRNA level, alternative splicing or promoters could be captured via single-cell and spatial transcriptomics 199. The importance of post-transcriptional regulation is evidenced by human mutations in pre-mRNA processing factor genes (PRPF3/4/6/8/31 (OMIM: 607301, 607795, 613979, 607300, 606419), SNRNP200 (OMIM: 601664) and RP9 (OMIM: 607331)), which have been linked to 15–20% of autosomal dominant 200 retinitis pigmentosa. However, as these factors are ubiquitously expressed, most tissues seem to tolerate defective splicing, whilst the neuroretina is highly sensitive. We also need to tackle differences at the proteome level, to monitor differential translational and/or degradation programmes in different tissues, some of which have been shown to be regulated locally near cilia or centrosomes 201,202. Exciting advances in spatial proteomics, such as LOPIT-DC (Localisation of Organelle Proteins by Isotope Tagging after Differential ultracentrifugation) 203, which enables identification of isoform-specific localizations and assignment of proteins to suborganellar structures, may help to resolve differences in ciliary apparatus and centrosomes between cell types. Other new techniques for preserving tissue spatial context include DVP (Deep Visual Proteomics) 204, which combines a novel artificial intelligence-driven phenotype image analysis tool with single-cell laser microdissection and ultra-high-sensitivity mass spectrometry. Here, protein abundance is linked to complex cellular or subcellular phenotypes whilst preserving spatial context, say in adjacent cystic versus non-cystic epithelium in polycystic kidney disease, as well as associated pathological changes in the cystic niche, remodelled fibroblasts and inflammatory infiltrates.
A technical challenge remains in applying omics approaches more broadly to physiologically relevant tissues ex vivo or ideally in vivo. To maximally capitalize on the exquisite spatial and temporal resolution of proteomes, with APEX2 proximity labelling, for example, we need to overcome current limitations in cells with abundant endogenous peroxidases as well as sensitivity to H2O2 using synthetic biology and biorthogonal chemistry to evolve both better enzymes and specific substrates 205,206. In the future, extending current implementations of technologies such as proximity-labelling proteomics may allow for cell-, tissue- and developmental-stage-specific monitoring of cilia composition, providing insight into mechanisms underlying specific context-dependent roles for cilia and ciliary proteins in vivo. These studies may reveal differences in sensitivity to cilia dysfunction between tissues or stages, which could help to better understand pathomechanisms underlying ciliopathies.
In parallel, recent advances in imaging technologies 53 and optogenetics 207–209 can be harnessed to dissect ciliary structure and signalling function not only in vitro but also in vivo, such as in a recently reported, elegant study identifying a serotonergic axon–cilium synapse in the mouse brain 209. With respect to signalling, we need better imaging modalities that capture the highly dynamic (less than a second) nanoscale (tens of nanometers) events that underlie compartmentalized signalling, such as GPCR signalling within a cilium 210. Technical breakthroughs towards improved temporal resolution in single-molecule localization microscopy (SMLM) through development of faster, improved signal-to-noise, high-speed cameras than current EMCCD and sCMOS technology 211 as well as improved fluorophore photon yields and membrane permeability will be important for this field. New means to build better reporters using genome editing of endogenous loci and non-disruptive tagging approaches are also key. These could include expansion of the genetic code, permitting incorporation of synthetic amino acids into ciliary proteins, allowing functionalization through click chemistry 212. Together with new workflows, tools and biosensors, the field could begin to understand how signalling initiated from nanoscale complexes within cilia is propagated across the organelle, cell and tissue levels during development and how this flow is disrupted in disease.
Finally, adding another layer of complexity to our current view of the ciliary signalling repertoire, recent evidence based primarily on cultured cells of non-vertebrate model organisms has suggested that cilia may not only function as receivers of extracellular signals, but also emit signal to other cells in the form of extracellular vesicles 126. Determining the extent and physiological importance of this phenomenon in vivo will be an important goal for the future.
Glossary terms
- Axoneme
Core structure of cilia, which usually comprises 9 outer microtubule doublets and 0 or 2 central microtubule singlets in primary (9 + 0) and motile cilia (9 + 2), respectively.
- Basal body
Specialized centriole that provides the foundation of the axoneme and anchors the cilium to the cell body via distal and subdistal appendages or fibres.
- BBSome
A complex composed of eight Bardet–Biedl syndrome proteins, which is thought to function as a cargo adaptor for the retrograde IFT machinery during ciliary membrane protein export.
- Centriole
Barrel-shaped structure composed of 9 microtubule triplets that gives rise to the basal body and constitutes the core structure of the centrosome.
- Centrosome
Main microtubule organizing centre in animals. Composed of two centrioles (mother and daughter) surrounded by pericentriolar material.
- Ciliopathy
A genetic disorder characterized by ciliary dysfunction, and whose affected gene product localizes to (first-order ciliopathy) or indirectly affects (second-order ciliopathy) the cilium–centrosome axis.
- Ciliary necklace
Outer aspect of the ciliary membrane located at the transition zone region where Y-links connect the inside of the membrane to the axoneme.
- Ciliary pocket
An invagination of the periciliary membrane, which is a hotspot for endocytosis and exocytosis of vesicles derived from or destined to the ciliary membrane.
- Ectosomes
Extracellular vesicles that bud directly from the membrane, including the ciliary membrane.
- Intraflagellar transport
(IFT) Conserved intraciliary transport system in which kinesin-2 and cytoplasmic dynein 2 motors move trains of cargo-associated IFT-A/B complexes into and out of cilia. Required for ciliary assembly, maintenance and function.
- Periciliary membrane
Membrane region at the base of the cilium that connects the ciliary membrane with the plasma membrane. Sometimes invaginated to form a ciliary pocket.
- Phosphoinositides
(PIPs) Lipid signalling molecules that coordinate multiple membrane-associated molecular events.
- Primary cilium
Non-motile sensory organelle present on the cell surface. It typically has a 9 + 0 axoneme surrounded by a membrane enriched for specific receptors and ion channels involved in signalling.
- Transition zone (TZ)
Ciliary sub compartment located between the basal body and cilium itself, which functions as a barrier for selective transport of molecules into and out of the cilium.
Acknowledgements
P.M. is grateful for support from the MRC (MC_U_12018/26) and from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement n°866355). S.T.C. is grateful for support from the Lundbeck Foundation (R317-2019-889), and L.B.P. acknowledges funding from the Novo Nordisk Foundation (grant #NNF18OC0053024) and Independent Research Fund Denmark (grant #2032-00115B). The authors apologize to authors whose work was not cited due to space and reference limitations. The authors are thankful to Anthony Roberts and Esben Lorentzen for helpful discussions and appreciate the very helpful comments and suggestions by the reviewers, which greatly improved this review.
Footnotes
Author contributions
The authors contributed equally to all aspects of the manuscript.
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Genetics thanks Maxence V. Nachury, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
References
- 1.Niederlova V, Modrak M, Tsyklauri O, Huranova M, Stepanek O. Meta-analysis of genotype-phenotype associations in Bardet-Biedl syndrome uncovers differences among causative genes. Human Mutation. 2019;40:2068–2087. doi: 10.1002/humu.23862. [DOI] [PubMed] [Google Scholar]
- 2.Reiter JF, Leroux MR. Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol. 2017;18:533–547. doi: 10.1038/nrm.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wheway G, Mitchison HM, Genomics England Research, C Opportunities and Challenges for Molecular Understanding of Ciliopathies-The 100,000 Genomes Project. Front Genet. 2019;10:127. doi: 10.3389/fgene.2019.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandra B, Tung ML, Hsu Y, Scheetz T, Sheffield VC. Retinal ciliopathies through the lens of Bardet-Biedl Syndrome: Past, present and future. Prog Retin Eye Res. 2022;89:101035. doi: 10.1016/j.preteyeres.2021.101035. [DOI] [PubMed] [Google Scholar]
- 5.Van De Weghe JC, Gomez A, Doherty D. The Joubert-Meckel-Nephronophthisis Spectrum of Ciliopathies. Annu Rev Genomics Hum Genet. 2022;23:301–329. doi: 10.1146/annurev-genom-121321-093528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallmeier J, et al. Motile ciliopathies. Nat Rev Dis Primers. 2020;6:77. doi: 10.1038/s41572-020-0209-6. [DOI] [PubMed] [Google Scholar]
- 7.Lovera M, Luders J. The ciliary impact of nonciliary gene mutations. Trends Cell Biol. 2021;31:876–887. doi: 10.1016/j.tcb.2021.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Pigino G. Intraflagellar transport. Curr Biol. 2021;31:R530–R536. doi: 10.1016/j.cub.2021.03.081. [DOI] [PubMed] [Google Scholar]
- 9.Legendre M, Zaragosi LE, Mitchison HM. Motile cilia and airway disease. Semin Cell Dev Biol. 2021;110:19–33. doi: 10.1016/j.semcdb.2020.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Kumar D, et al. A ciliopathy complex builds distal appendages to initiate ciliogenesis. J Cell Biol. 2021;220 doi: 10.1083/jcb.202011133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diener DR, Lupetti P, Rosenbaum JL. Proteomic analysis of isolated ciliary transition zones reveals the presence of ESCRT proteins. Curr Biol. 2015;25:379–384. doi: 10.1016/j.cub.2014.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sang L, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–528. doi: 10.1016/j.cell.2011.04.019. Using high-confidence proteomics, this study identifies 850 interactors of nine NPHP/JBTS/MKS proteins and describes ATXN10 and TCTN2 as new NPHP-JBTS genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chih B, et al. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol. 2011;14:61–72. doi: 10.1038/ncb2410. Together with ref. 12 and ref. 24, this study identifies a complex of nine proteins (the MKS complex) at the ciliary transition zone that regulates ciliary assembly and composition, several of which are mutated in MKS or JBTS. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Gonzalo FR, Reiter JF. Open Sesame: How Transition Fibers and the Transition Zone Control Ciliary Composition. Cold Spring Harb Perspect Biol. 2017;9 doi: 10.1101/cshperspect.a028134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trepout S, Tassin AM, Marco S, Bastin P. STEM tomography analysis of the trypanosome transition zone. J Struct Biol. 2018;202:51–60. doi: 10.1016/j.jsb.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Greenan GA, Vale RD, Agard DA. Electron cryotomography of intact motile cilia defines the basal body to axoneme transition. J Cell Biol. 2020;219 doi: 10.1083/jcb.201907060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang TT, et al. Superresolution Pattern Recognition Reveals the Architectural Map of the Ciliary Transition Zone. Sci Rep. 2015;5:14096. doi: 10.1038/srep14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jana SC, et al. Differential regulation of transition zone and centriole proteins contributes to ciliary base diversity. Nat Cell Biol. 2018;20:928–941. doi: 10.1038/s41556-018-0132-1. [DOI] [PubMed] [Google Scholar]
- 19.Hazime KS, et al. STORM imaging reveals the spatial arrangement of transition zone components and IFT particles at the ciliary base in Tetrahymena. Sci Rep. 2021;11:7899. doi: 10.1038/s41598-021-86909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conduit SE, Vanhaesebroeck B. Phosphoinositide lipids in primary cilia biology. Biochem J. 2020;477:3541–3565. doi: 10.1042/BCJ20200277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisch C, Dupuis-Williams P. Ultrastructure of cilia and flagella - back to the future! Biol Cell. 2011;103:249–270. doi: 10.1042/BC20100139. [DOI] [PubMed] [Google Scholar]
- 22.Gilliam JC, et al. Three-dimensional architecture of the rod sensory cilium and its disruption in retinal neurodegeneration. Cell. 2012;151:1029–1041. doi: 10.1016/j.cell.2012.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiegering A, et al. Cell type-specific regulation of ciliary transition zone assembly in vertebrates. EMBO J. 2018;37 doi: 10.15252/embj.201797791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Gonzalo FR, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43:776–784. doi: 10.1038/ng.891. Together with ref. 12 and ref. 13, this study shows that the MKS complex at the ciliary transition zone regulates ciliary assembly and composition in a tissue-specific manner, and that loss of complex components causes MKS or JBTS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nachury MV, Mick DU. Establishing and regulating the composition of cilia for signal transduction. Nat Rev Mol Cell Biol. 2019;20:389–405. doi: 10.1038/s41580-019-0116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morthorst SK, Christensen ST, Pedersen LB. Regulation of ciliary membrane protein trafficking and signalling by kinesin motor proteins. The FEBS journal. 2018;285:4535–4564. doi: 10.1111/febs.14583. [DOI] [PubMed] [Google Scholar]
- 27.Mukhopadhyay S, et al. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 2010;24:2180–2193. doi: 10.1101/gad.1966210. This study identifies TULP3 as an interactor of the IFT-A complex and demonstrates a role for TULP3 and IFT-A in promoting ciliary trafficking of select GPCRs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Badgandi HB, Hwang S-h, Shimada IS, Loriot E, Mukhopadhyay S. Tubby family proteins are adapters for ciliary trafficking of integral membrane proteins. The Journal of Cell Biology. 2017;216:743–760. doi: 10.1083/jcb.201607095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palicharla VR, et al. Interactions between TULP3 tubby domain and ARL13B amphipathic helix promote lipidated protein transport to cilia. Mol Biol Cell. 2023:mbcE22100473. doi: 10.1091/mbc.E22-10-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hesketh SJ, Mukhopadhyay AG, Nakamura D, Toropova K, Roberts AJ. IFT-A structure reveals carriages for membrane protein transport into cilia. Cell. 2022;185:4971–4985.:e4916. doi: 10.1016/j.cell.2022.11.010. [DOI] [PubMed] [Google Scholar]
- 31.Cole DG, et al. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. First study to purify and characterize IFT-A and IFT-B polypeptides using Chlamydomonas flagella; it reveals that Chlamydomonas IFT polypeptides are homologous to proteins required for biogenesis of neuronal sensory cilia in C. elegans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuura K, Lefebvre PA, Kamiya R, Hirono M. Kinesin-II is not essential for mitosis and cell growth in Chlamydomonas. Cell Motil Cytoskeleton. 2002;52:195–201. doi: 10.1002/cm.10051. [DOI] [PubMed] [Google Scholar]
- 33.van den Hoek H, et al. In situ architecture of the ciliary base reveals the stepwise assembly of intraflagellar transport trains. Science. 2022;377:543–548. doi: 10.1126/science.abm6704. [DOI] [PubMed] [Google Scholar]
- 34.Lechtreck K. Cargo adapters expand the transport range of intraflagellar transport. J Cell Sci. 2022;135 doi: 10.1242/jcs.260408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ocbina PJ, Anderson KV. Intraflagellar transport, cilia, and mammalian Hedgehog signaling: analysis in mouse embryonic fibroblasts. Dev Dyn. 2008;237:2030–2038. doi: 10.1002/dvdy.21551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lechtreck KF, et al. Cycling of the signaling protein phospholipase D through cilia requires the BBSome only for the export phase. J Cell Biol. 2013;201:249–261. doi: 10.1083/jcb.201207139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goggolidou P, et al. ATMIN is a transcriptional regulator of both lung morphogenesis and ciliogenesis. Development. 2014;141:3966–3977. doi: 10.1242/dev.107755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De-Castro ARG, et al. WDR60-mediated dynein-2 loading into cilia powers retrograde IFT and transition zone crossing. J Cell Biol. 2022;221 doi: 10.1083/jcb.202010178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen VL, et al. Role for intraflagellar transport in building a functional transition zone. EMBO Rep. 2018;19 doi: 10.15252/embr.201845862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheidel N, Blacque OE. Intraflagellar Transport Complex A Genes Differentially Regulate Cilium Formation and Transition Zone Gating. Curr Biol. 2018;28:3279–3287.:e3272. doi: 10.1016/j.cub.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 41.den Hollander AI, Roepman R, Koenekoop RK, Cremers FP. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res. 2008;27:391–419. doi: 10.1016/j.preteyeres.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 42.Xu M, et al. Mutations in human IFT140 cause non-syndromic retinal degeneration. Hum Genet. 2015;134:1069–1078. doi: 10.1007/s00439-015-1586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parfitt DA, et al. Identification and Correction of Mechanisms Underlying Inherited Blindness in Human iPSC-Derived Optic Cups. Cell Stem Cell. 2016;18:769–781. doi: 10.1016/j.stem.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell SR, et al. Intravitreal antisense oligonucleotide sepofarsen in Leber congenital amaurosis type 10: a phase 1b/2 trial. Nature Medicine. 2022;28:1014–1021. doi: 10.1038/s41591-022-01755-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Editas Medicine, I. Single Ascending Dose Study in Participants With LCA10. U S National Library of Medicine; 2022. ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT03872479#moreinfo. [Google Scholar]
- 46.Sun S, Fisher RL, Bowser SS, Pentecost BT, Sui H. Three-dimensional architecture of epithelial primary cilia. Proc Natl Acad Sci U S A. 2019;116:9370–9379. doi: 10.1073/pnas.1821064116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiesel P, et al. The molecular structure of mammalian primary cilia revealed by cryo-electron tomography. Nat Struct Mol Biol. 2020;27:1115–1124. doi: 10.1038/s41594-020-0507-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Z, et al. Super-Resolution Microscopy and FIB-SEM Imaging Reveal Parental Centriole-Derived, Hybrid Cilium in Mammalian Multiciliated Cells. Dev Cell. 2020;55:224–236.:e226. doi: 10.1016/j.devcel.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu CS, et al. An open-access volume electron microscopy atlas of whole cells and tissues. Nature. 2021;599:147–151. doi: 10.1038/s41586-021-03992-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barnes CL, Malhotra H, Calvert PD. Compartmentalization of Photoreceptor Sensory Cilia. Front Cell Dev Biol. 2021;9:636737. doi: 10.3389/fcell.2021.636737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silverman MA, Leroux MR. Intraflagellar transport and the generation of dynamic, structurally and functionally diverse cilia. Trends Cell Biol. 2009;19:306–316. doi: 10.1016/j.tcb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 52.Gluenz E, et al. Beyond 9+0: noncanonical axoneme structures characterize sensory cilia from protists to humans. Faseb J. 2010;24:3117–3121. doi: 10.1096/fj.09-151381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steib E, et al. TissUExM enables quantitative ultrastructural analysis in whole vertebrate embryos by expansion microscopy. Cell Rep Methods. 2022;2:100311. doi: 10.1016/j.crmeth.2022.100311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flock A, Duvall AJ., 3rd The Ultrastructure of the Kinocilium of the Sensory Cells in the Inner Ear and Lateral Line Organs. J Cell Biol. 1965;25:1–8. doi: 10.1083/jcb.25.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones C, et al. Ciliary proteins link basal body polarization to planar cell polarity regulation. Nat Genet. 2008;40:69–77. doi: 10.1038/ng.2007.54. [DOI] [PubMed] [Google Scholar]
- 56.Jafek BW. Ultrastructure of human nasal mucosa. The Laryngoscope. 1983;93:1576–1599. doi: 10.1288/00005537-198312000-00011. [DOI] [PubMed] [Google Scholar]
- 57.Jenkins PM, McEwen DP, Martens JR. Olfactory Cilia: Linking Sensory Cilia Function and Human Disease. Chemical Senses. 2009;34:451–464. doi: 10.1093/chemse/bjp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nonami Y, Narita K, Nakamura H, Inoue T, Takeda S. Developmental changes in ciliary motility on choroid plexus epithelial cells during the perinatal period. Cytoskeleton. 2013;70:797–803. doi: 10.1002/cm.21132. [DOI] [PubMed] [Google Scholar]
- 59.Cho JH, et al. Islet primary cilia motility controls insulin secretion. Sci Adv. 2022;8:eabq8486. doi: 10.1126/sciadv.abq8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mallet A, Bastin P. Restriction of intraflagellar transport to some microtubule doublets: An opportunity for cilia diversification? Bioessays. 2022;44:e2200031. doi: 10.1002/bies.202200031. [DOI] [PubMed] [Google Scholar]
- 61.Evans JE, et al. Functional modulation of IFT kinesins extends the sensory repertoire of ciliated neurons in Caenorhabditis elegans . J Cell Biol. 2006;172:663–669. doi: 10.1083/jcb.200509115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pooranachandran N, Malicki JJ. Unexpected Roles for Ciliary Kinesins and Intraflagellar Transport Proteins. Genetics. 2016;203:771–785. doi: 10.1534/genetics.115.180943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lewis TR, et al. Cos2/Kif7 and Osm-3/Kif17 regulate onset of outer segment development in zebrafish photoreceptors through distinct mechanisms. Dev Biol. 2017;425:176–190. doi: 10.1016/j.ydbio.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stepanek L, Pigino G. Microtubule doublets are double-track railways for intraflagellar transport trains. Science. 2016;352:721–724. doi: 10.1126/science.aaf4594. [DOI] [PubMed] [Google Scholar]
- 65.Bertiaux E, et al. Bidirectional intraflagellar transport is restricted to two sets of microtubule doublets in the trypanosome flagellum. J Cell Biol. 2018;217:4284–4297. doi: 10.1083/jcb.201805030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silva M, et al. Cell-Specific alpha-Tubulin Isotype Regulates Ciliary Microtubule Ultrastructure, Intraflagellar Transport, and Extracellular Vesicle Biology. Curr Biol. 2017;27:968–980. doi: 10.1016/j.cub.2017.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mechaussier S, et al. <em>TUBB4B</em> variants specifically impact ciliary function, causing a ciliopathic spectrum. medRxiv. 2022:2022.2010.2019.22280748. doi: 10.1101/2022.10.19.22280748. [DOI] [Google Scholar]
- 68.Yu I, Garnham CP, Roll-Mecak A. Writing and Reading the Tubulin Code. J Biol Chem. 2015;290:17163–17172. doi: 10.1074/jbc.R115.637447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Magiera MM, Singh P, Gadadhar S, Janke C. Tubulin Posttranslational Modifications and Emerging Links to Human Disease. Cell. 2018;173:1323–1327. doi: 10.1016/j.cell.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 70.Gadadhar S, et al. Tubulin glycylation controls axonemal dynein activity, flagellar beat, and male fertility. Science. 2021;371 doi: 10.1126/science.abd4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bosch Grau M, et al. Alterations in the balance of tubulin glycylation and glutamylation in photoreceptors leads to retinal degeneration. J Cell Sci. 2017;130:938–949. doi: 10.1242/jcs.199091. [DOI] [PubMed] [Google Scholar]
- 72.O’Hagan R, et al. Glutamylation Regulates Transport, Specializes Function, and Sculpts the Structure of Cilia. Curr Biol. 2017;27:3430–3441.:e3436. doi: 10.1016/j.cub.2017.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Power KM, et al. Mutation of NEKL-4/NEK10 and TTLL genes suppress neuronal ciliary degeneration caused by loss of CCPP-1 deglutamylase function. PLoS Genet. 2020;16:e1009052. doi: 10.1371/journal.pgen.1009052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mukhopadhyay S, et al. Distinct IFT mechanisms contribute to the generation of ciliary structural diversity in C. elegans. EMBO J. 2007;26:2966–2980. doi: 10.1038/sj.emboj.7601717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garcia G, III, Raleigh DR, Reiter JF. How the Ciliary Membrane Is Organized Inside-Out to Communicate Outside-In. Current Biology. 2018;28:R421–R434. doi: 10.1016/j.cub.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mukhopadhyay S, Lu Y, Shaham S, Sengupta P. Sensory signaling-dependent remodeling of olfactory cilia architecture in C. elegans. Dev Cell. 2008;14:762–774. doi: 10.1016/j.devcel.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Besschetnova TY, et al. Identification of signaling pathways regulating primary cilium length and flow-mediated adaptation. Curr Biol. 2010;20:182–187. doi: 10.1016/j.cub.2009.11.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barr MM, Sternberg PW. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–389. doi: 10.1038/43913. By using C. elegans as a model, this study is the first to report cilia-associated functions for polycystins. [DOI] [PubMed] [Google Scholar]
- 79.Yoder BK, Hou X, Guay-Woodford LM. The Polycystic Kidney Disease Proteins, Polycystin-1, Polycystin-2, Polaris, and Cystin, Are Co-Localized in Renal Cilia. Journal of the American Society of Nephrology. 2002;13:2508–2516. doi: 10.1097/01.asn.0000029587.47950.25. Together with ref. 80 this study is the first to show localization of polycystins to primary cilia in mammalian kidney epithelial cells. [DOI] [PubMed] [Google Scholar]
- 80.Pazour GJ, San Agustin JT, Follit JA, Rosenbaum JL, Witman GB. Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr Biol. 2002;12:R378-380. doi: 10.1016/s0960-9822(02)00877-1. Together with ref. 79 this study is the first to show localization of polycystins to primary cilia in mammalian kidney epithelial cells. [DOI] [PubMed] [Google Scholar]
- 81.Ma M, Gallagher AR, Somlo S. Ciliary Mechanisms of Cyst Formation in Polycystic Kidney Disease. Cold Spring Harb Perspect Biol. 2017;9 doi: 10.1101/cshperspect.a028209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Corbit KC, et al. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018. doi: 10.1038/nature04117. First study showing that Smoothened localizes to and functions at the primary cilium to control Hedgehog signalling in vertebrates. [DOI] [PubMed] [Google Scholar]
- 83.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. First study showing that Patched-1 localizes to the primary cilium to prevent entry of Smoothened to the cilium. When Sonic Hedgehog binds to Patched-1, the receptor leaves the cilium, allowing ciliary accumulation of Smoothened to activate signalling. [DOI] [PubMed] [Google Scholar]
- 84.Bangs F, Anderson KV. Primary Cilia and Mammalian Hedgehog Signaling. Cold Spring Harb Perspect Biol. 2017;9 doi: 10.1101/cshperspect.a028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Katoh TA, et al. Immotile cilia mechanically sense the direction of fluid flow for left-right determination. Science. 2023;379:66–71. doi: 10.1126/science.abq8148. Using mouse embryos, this study, together with ref. 86, provides evidence that primary cilia of the left-right organizer are mechanosensors that translate extracellular fluid flow into Ca2+ signals to induce embryonic left-right asymmetry. [DOI] [PubMed] [Google Scholar]
- 86.Djenoune L, et al. Cilia function as calcium-mediated mechanosensors that instruct left-right asymmetry. Science. 2023;379:71–78. doi: 10.1126/science.abq7317. Using zebrafish embryos, this study, together with ref. 85, provides evidence that primary cilia of the left-right organizer are mechanosensors that translate extracellular fluid flow into Ca2+ signals to induce embryonic left-right asymmetry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hilgendorf KI, Johnson CT, Jackson PK. The primary cilium as a cellular receiver: organizing ciliary GPCR signaling. Curr Opin Cell Biol. 2016;39:84–92. doi: 10.1016/j.ceb.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Christensen ST, Morthorst SK, Mogensen JB, Pedersen LB. Primary Cilia and Coordination of Receptor Tyrosine Kinase (RTK) and Transforming Growth Factor beta (TGF-beta) Signaling. Cold Spring Harb Perspect Biol. 2017;9 doi: 10.1101/cshperspect.a028167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mick DU, et al. Proteomics of Primary Cilia by Proximity Labeling. Dev Cell. 2015;35:497–512. doi: 10.1016/j.devcel.2015.10.015. First study that uses cilium-targeted proximity-labelling proteomics to identify the proteome of primary cilia of cultured mammalian cells and reveal how it is altered in Ift27/Bbs19 mutant cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Datta P, et al. Accumulation of non-outer segment proteins in the outer segment underlies photoreceptor degeneration in Bardet-Biedl syndrome. Proc Natl Acad Sci U S A. 2015;112:E4400–4409. doi: 10.1073/pnas.1510111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kohli P, et al. The ciliary membrane-associated proteome reveals actin-binding proteins as key components of cilia. EMBO Rep. 2017;18:1521–1535. doi: 10.15252/embr.201643846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.May EA, et al. Time-resolved proteomics profiling of the ciliary Hedgehog response. J Cell Biol. 2021;220 doi: 10.1083/jcb.202007207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu X, et al. Cilium proteomics reveals Numb as a positive regulator of the Hedgehog signaling pathway. bioRxiv. 2022:2022.2010.2010.511655. doi: 10.1101/2022.10.10.511655. [DOI] [Google Scholar]
- 94.Masek M, et al. Loss of the Bardet-Biedl protein Bbs1 alters photoreceptor outer segment protein and lipid composition. Nat Commun. 2022;13:1282. doi: 10.1038/s41467-022-28982-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Quidwai T, et al. A WDR35-dependent coat protein complex transports ciliary membrane cargo vesicles to cilia. Elife. 2021;10 doi: 10.7554/eLife.69786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu D, et al. Ciliogenesis requires sphingolipid-dependent membrane and axoneme interaction. Proc Natl Acad Sci U S A. 2022;119:e2201096119. doi: 10.1073/pnas.2201096119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Meleppattu S, Zhou H, Dai J, Gui M, Brown A. Mechanism of IFT-A polymerization into trains for ciliary transport. Cell. 2022;185:4986–4998.:e4912. doi: 10.1016/j.cell.2022.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ye F, Nager AR, Nachury MV. BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. The Journal of Cell Biology. 2018 doi: 10.1083/jcb.201709041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 100.Cantagrel V, et al. Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am J Hum Genet. 2008;83:170–179. doi: 10.1016/j.ajhg.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cevik S, et al. Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J Cell Biol. 2010;188:953–969. doi: 10.1083/jcb.200908133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nozaki S, et al. Regulation of ciliary retrograde protein trafficking by the Joubert syndrome proteins ARL13B and INPP5E. J Cell Sci. 2017;130:563–576. doi: 10.1242/jcs.197004. [DOI] [PubMed] [Google Scholar]
- 103.Han S, et al. TULP3 is required for localization of membrane-associated proteins ARL13B and INPP5E to primary cilia. Biochem Biophys Res Commun. 2019;509:227–234. doi: 10.1016/j.bbrc.2018.12.109. [DOI] [PubMed] [Google Scholar]
- 104.Legue E, Liem KF., Jr Tulp3 Is a Ciliary Trafficking Gene that Regulates Polycystic Kidney Disease. Curr Biol. 2019;29:803–812.:e805. doi: 10.1016/j.cub.2019.01.054. [DOI] [PubMed] [Google Scholar]
- 105.Jensen VL, Leroux MR. Gates for soluble and membrane proteins, and two trafficking systems (IFT and LIFT), establish a dynamic ciliary signaling compartment. Curr Opin Cell Biol. 2017;47:83–91. doi: 10.1016/j.ceb.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 106.Kanie T, et al. The CEP19-RABL2 GTPase Complex Binds IFT-B to Initiate Intraflagellar Transport at the Ciliary Base. Dev Cell. 2017;42:22–36.:e12. doi: 10.1016/j.devcel.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boegholm N, et al. The IFT81-IFT74 complex enhances GTP hydrolysis to inactivate RabL2 during early steps of intraflagellar transport. bioRxiv. 2022:2022.2005.2031.494111. doi: 10.1101/2022.05.31.494111. [DOI] [Google Scholar]
- 108.Duan S, et al. Rabl2 GTP hydrolysis licenses BBSome-mediated export to fine-tune ciliary signaling. EMBO J. 2021;40:e105499. doi: 10.15252/embj.2020105499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. First study to identify the BBSome using cultured mammalian cells as model. [DOI] [PubMed] [Google Scholar]
- 110.Desai PB, Stuck MW, Lv B, Pazour GJ. Ubiquitin links smoothened to intraflagellar transport to regulate Hedgehog signaling. J Cell Biol. 2020;219 doi: 10.1083/jcb.201912104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shinde SR, et al. The ancestral ESCRT protein TOM1L2 selects ubiquitinated cargoes for retrieval from cilia. bioRxiv. 2022:2022.2009.2023.509287. doi: 10.1101/2022.09.23.509287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bielas SL, et al. Mutations in the inositol polyphosphate-5-phosphatase E gene link phosphatidyl inositol signaling to the ciliopathies. Nature genetics. 2009;41:1032–1036. doi: 10.1038/ng.423. Together with ref. 113 this study reports for the first time that mutations in INPP5E gene are causative of human ciliopathies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jacoby M, et al. INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nature Genetics. 2009;41:1027. doi: 10.1038/ng.427. Together with ref. 112 this study reports for the first time that mutations in INPP5E gene are causative of human ciliopathies. [DOI] [PubMed] [Google Scholar]
- 114.Chavez M, et al. Modulation of Ciliary Phosphoinositide Content Regulates Trafficking and Sonic Hedgehog Signaling Output. Dev Cell. 2015;34:338–350. doi: 10.1016/j.devcel.2015.06.016. Together with ref. 115 this study shows that INPP5E is required for enrichment of PI4P in the ciliary membrane, which in turn is needed for appropriate regulation of Hedgehog signalling. [DOI] [PubMed] [Google Scholar]
- 115.Garcia-Gonzalo FR, et al. Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling. Dev Cell. 2015;34:400–409. doi: 10.1016/j.devcel.2015.08.001. Together with ref. 114 this study shows that INPP5E is required for enrichment of PI4P in the ciliary membrane, which in turn is needed for appropriate regulation of Hedgehog signalling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dyson JM, et al. INPP5E regulates phosphoinositide-dependent cilia transition zone function. J Cell Biol. 2017;216:247–263. doi: 10.1083/jcb.201511055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tripathi P, et al. Palmitoylation of acetylated tubulin and association with ceramide-rich platforms is critical for ciliogenesis. J Lipid Res. 2021;62:100021. doi: 10.1194/jlr.RA120001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nguyen TD, Truong ME, Reiter JF. The Intimate Connection Between Lipids and Hedgehog Signaling. Front Cell Dev Biol. 2022;10:876815. doi: 10.3389/fcell.2022.876815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shakya S, Westlake CJ. Recent advances in understanding assembly of the primary cilium membrane. Fac Rev. 2021;10:16. doi: 10.12703/r/10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sedmak T, Wolfrum U. Intraflagellar transport proteins in ciliogenesis of photoreceptor cells. Biol Cell. 2011;103:449–466. doi: 10.1042/BC20110034. [DOI] [PubMed] [Google Scholar]
- 121.Labat-de-Hoz L, et al. A Model for Primary Cilium Biogenesis by Polarized Epithelial Cells: Role of the Midbody Remnant and Associated Specialized Membranes. Front Cell Dev Biol. 2020;8:622918. doi: 10.3389/fcell.2020.622918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stuck MW, Chong WM, Liao JC, Pazour GJ. Rab34 is necessary for early stages of intracellular ciliogenesis. Curr Biol. 2021;31:2887–2894.:e2884. doi: 10.1016/j.cub.2021.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ganga AK, et al. Rab34 GTPase mediates ciliary membrane formation in the intracellular ciliogenesis pathway. Curr Biol. 2021;31:2895–2905.:e2897. doi: 10.1016/j.cub.2021.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Benmerah A. The ciliary pocket. Curr Opin Cell Biol. 2013;25:78–84. doi: 10.1016/j.ceb.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 125.Pedersen LB, Mogensen JB, Christensen ST. Endocytic Control of Cellular Signaling at the Primary Cilium. Trends Biochem Sci. 2016;41:784–797. doi: 10.1016/j.tibs.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 126.Ojeda Naharros I, Nachury MV. Shedding of ciliary vesicles at a glance. J Cell Sci. 2022;135 doi: 10.1242/jcs.246553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Anvarian Z, Mykytyn K, Mukhopadhyay S, Pedersen LB, Christensen ST. Cellular signalling by primary cilia in development, organ function and disease. Nat Rev Nephrol. 2019;15:199–219. doi: 10.1038/s41581-019-0116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schou KB, Pedersen LB, Christensen ST. Ins and outs of GPCR signaling in primary cilia. EMBO Rep. 2015;16:1099–1113. doi: 10.15252/embr.201540530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hilgendorf KI. Primary Cilia Are Critical Regulators of White Adipose Tissue Expansion. Front Physiol. 2021;12:769367. doi: 10.3389/fphys.2021.769367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wachten D, Mick DU. Signal transduction in primary cilia - analyzing and manipulating GPCR and second messenger signaling. Pharmacol Ther. 2021;224:107836. doi: 10.1016/j.pharmthera.2021.107836. [DOI] [PubMed] [Google Scholar]
- 131.Happ JT, et al. A PKA inhibitor motif within SMOOTHENED controls Hedgehog signal transduction. Nat Struct Mol Biol. 2022;29:990–999. doi: 10.1038/s41594-022-00838-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Singh J, Wen X, Scales SJ. The Orphan G Protein-coupled Receptor Gpr175 (Tpra40) Enhances Hedgehog Signaling by Modulating cAMP Levels. The Journal of Biological Chemistry. 2015;290:29663–29675. doi: 10.1074/jbc.M115.665810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.May EA, Sroka TJ, Mick DU. Phosphorylation and Ubiquitylation Regulate Protein Trafficking, Signaling, and the Biogenesis of Primary Cilia. Front Cell Dev Biol. 2021;9:664279. doi: 10.3389/fcell.2021.664279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shinde SR, Nager AR, Nachury MV. Ubiquitin chains earmark GPCRs for BBSome-mediated removal from cilia. J Cell Biol. 2020;219 doi: 10.1083/jcb.202003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lv B, Stuck MW, Desai PB, Cabrera OA, Pazour GJ. E3 ubiquitin ligase Wwp1 regulates ciliary dynamics of the Hedgehog receptor Smoothened. J Cell Biol. 2021;220 doi: 10.1083/jcb.202010177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pal K, et al. Smoothened determines beta-arrestin-mediated removal of the G protein-coupled receptor Gpr161 from the primary cilium. J Cell Biol. 2016;212:861–875. doi: 10.1083/jcb.201506132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chiuso F, et al. Ubiquitylation of BBSome is required for ciliary assembly and signaling. EMBO Rep. 2023:e55571. doi: 10.15252/embr.202255571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yue S, et al. Requirement of Smurf-mediated endocytosis of Patched1 in sonic hedgehog signal reception. Elife. 2014;3 doi: 10.7554/eLife.02555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hantel F, et al. Cilia-localized GID/CTLH ubiquitin ligase complex regulates protein homeostasis of sonic hedgehog signaling components. J Cell Sci. 2022;135 doi: 10.1242/jcs.259209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schneider L, et al. PDGFRαα Signaling Is Regulated through the Primary Cilium in Fibroblasts. Current Biology. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. First study to show that PDGFRa localizes to and functions at the primary cilium in cultured vertebrate cells (mouse fibroblasts) [DOI] [PubMed] [Google Scholar]
- 141.Schneider L, et al. Directional Cell Migration and Chemotaxis in Wound Healing Response to PDGF-AA are Coordinated by the Primary Cilium in Fibroblasts. Cellular Physiology and Biochemistry. 2010;25:279–292. doi: 10.1159/000276562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Clement CA, et al. TGF-beta signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Rep. 2013;3:1806–1814. doi: 10.1016/j.celrep.2013.05.020. First study showing that TGFB signalling is regulated by receptors in the primary cilium and controlled at the level of clathrin-mediated endocytosis at the ciliary pocket in vertebrates. [DOI] [PubMed] [Google Scholar]
- 143.Schmid FM, et al. IFT20 modulates ciliary PDGFRalpha signaling by regulating the stability of Cbl E3 ubiquitin ligases. J Cell Biol. 2018;217:151–161. doi: 10.1083/jcb.201611050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Koefoed K, et al. The E3 ubiquitin ligase SMURF1 regulates cell-fate specification and outflow tract septation during mammalian heart development. Scientific Reports. 2018;8:9542. doi: 10.1038/s41598-018-27854-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hossain D, Tsang WY. The role of ubiquitination in the regulation of primary cilia assembly and disassembly. Semin Cell Dev Biol. 2019;93:145–152. doi: 10.1016/j.semcdb.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 146.Senatore E, et al. Pathophysiology of Primary Cilia: Signaling and Proteostasis Regulation. Front Cell Dev Biol. 2022;10:833086. doi: 10.3389/fcell.2022.833086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Aslanyan MG, et al. A targeted multi-proteomics approach generates a blueprint of the ciliary ubiquitinome. Frontiers in Cell and Developmental Biology. 2023;11 doi: 10.3389/fcell.2023.1113656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sanchez GM, et al. The beta-cell primary cilium is an autonomous Ca2+ compartment for paracrine GABA signaling. J Cell Biol. 2023;222 doi: 10.1083/jcb.202108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nauli SM, Pala R, Kleene SJ. Calcium channels in primary cilia. Curr Opin Nephrol Hypertens. 2016;25:452–458. doi: 10.1097/MNH.0000000000000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Saternos H, Ley S, AbouAlaiwi W. Primary Cilia and Calcium Signaling Interactions. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21197109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Spasic M, Jacobs CR. Primary cilia: Cell and molecular mechanosensors directing whole tissue function. Semin Cell Dev Biol. 2017;71:42–52. doi: 10.1016/j.semcdb.2017.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tanaka Y, Morozumi A, Hirokawa N. Leftward transfer of a chemosensory polycystin initiates left-dominant calcium signaling for lateralized embryonic development. bioRxiv. 2023:2023.2001.2012.523739. doi: 10.1101/2023.01.12.523739. [DOI] [Google Scholar]
- 153.Djenoune L, Berg K, Brueckner M, Yuan S. A change of heart: new roles for cilia in cardiac development and disease. Nat Rev Cardiol. 2022;19:211–227. doi: 10.1038/s41569-021-00635-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Okada Y, et al. Abnormal nodal flow precedes situs inversus in iv and inv mice. Mol Cell. 1999;4:459–468. doi: 10.1016/s1097-2765(00)80197-5. This study shows that abnormal nodal flow at the left-right organizer leads to laterality defects in mutant mouse embryos. [DOI] [PubMed] [Google Scholar]
- 155.Nonaka S, Shiratori H, Saijoh Y, Hamada H. Determination of left-right patterning of the mouse embryo by artificial nodal flow. Nature. 2002;418:96–99. doi: 10.1038/nature00849. Authors use artificial fluid flow at the node to control left-right patterning in mouse embryos, providing evidence for the role of mechanical fluid flow in embryonic symmetry breaking. [DOI] [PubMed] [Google Scholar]
- 156.Little RB, Norris DP. Right, left and cilia: How asymmetry is established. Semin Cell Dev Biol. 2021;110:11–18. doi: 10.1016/j.semcdb.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 157.Brueckner M. Heterotaxia, congenital heart disease, and primary ciliary dyskinesia. Circulation. 2007;115:2793–2795. doi: 10.1161/CIRCULATIONAHA.107.699256. [DOI] [PubMed] [Google Scholar]
- 158.Klena NT, Gibbs BC, Lo CW. Cilia and Ciliopathies in Congenital Heart Disease. Cold Spring Harb Perspect Biol. 2017;9 doi: 10.1101/cshperspect.a028266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vion A-C, et al. Primary cilia sensitize endothelial cells to BMP and prevent excessive vascular regression. The Journal of Cell Biology. 2018;217:1651–1665. doi: 10.1083/jcb.201706151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang L, Dynlacht BD. The regulation of cilium assembly and disassembly in development and disease. Development. 2018;145 doi: 10.1242/dev.151407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kasahara K, Inagaki M. Primary ciliary signaling: links with the cell cycle. Trends Cell Biol. 2021;31:954–964. doi: 10.1016/j.tcb.2021.07.009. [DOI] [PubMed] [Google Scholar]
- 162.Walia V, et al. Akt Regulates a Rab11-Effector Switch Required for Ciliogenesis. Dev Cell. 2019;50:229–246.:e227. doi: 10.1016/j.devcel.2019.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hu H-B, et al. LPA signaling acts as a cell-extrinsic mechanism to initiate cilia disassembly and promote neurogenesis. Nature Communications. 2021;12:662. doi: 10.1038/s41467-021-20986-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. This study shows that a HEF1-AURKA-HDAC6 signalling axis regulates primary cilia disassembly in cultured mammalian cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Simon D, et al. A mutation in the 3’-UTR of the HDAC6 gene abolishing the post-transcriptional regulation mediated by hsa-miR-433 is linked to a new form of dominant X-linked chondrodysplasia. Human Molecular Genetics. 2010;19:2015–2027. doi: 10.1093/hmg/ddq083. [DOI] [PubMed] [Google Scholar]
- 166.Gabriel E, et al. CPAP promotes timely cilium disassembly to maintain neural progenitor pool. The EMBO Journal. 2016;35:803–819. doi: 10.15252/embj.201593679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang W, et al. Modeling microcephaly with cerebral organoids reveals a WDR62-CEP170-KIF2A pathway promoting cilium disassembly in neural progenitors. Nat Commun. 2019;10:2612. doi: 10.1038/s41467-019-10497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Farooq M, et al. RRP7A links primary microcephaly to dysfunction of ribosome biogenesis, resorption of primary cilia, and neurogenesis. Nat Commun. 2020;11:5816. doi: 10.1038/s41467-020-19658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mirvis M, Siemers KA, Nelson WJ, Stearns TP. Primary cilium loss in mammalian cells occurs predominantly by whole-cilium shedding. PLoS Biol. 2019;17:e3000381. doi: 10.1371/journal.pbio.3000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Anderson CT, Stearns T. Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr Biol. 2009;19:1498–1502. doi: 10.1016/j.cub.2009.07.034. This paper which shows that following cell division, the daughter cell that inherits the older mother centriole ciliates first and is responsive for signalling faster. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Paridaen JT, Wilsch-Brauninger M, Huttner WB. Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell. 2013;155:333–344. doi: 10.1016/j.cell.2013.08.060. This study demonstrates that a ciliary membrane remnant remains associated with the centrosome around the mother centriole after cilia disassembly to allow asymmetric ciliation and signalling competency in the daughter that inherits it. [DOI] [PubMed] [Google Scholar]
- 172.Ford MJ, et al. A Cell/Cilia Cycle Biosensor for Single-Cell Kinetics Reveals Persistence of Cilia after G1/S Transition Is a General Property in Cells and Mice. Dev Cell. 2018;47:509–523.:e505. doi: 10.1016/j.devcel.2018.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ho EK, Tsai AE, Stearns T. Transient Primary Cilia Mediate Robust Hedgehog Pathway-Dependent Cell Cycle Control. Curr Biol. 2020;30:2829–2835.:e2825. doi: 10.1016/j.cub.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Spear PC, Erickson CA. Apical movement during interkinetic nuclear migration is a two-step process. Dev Biol. 2012;370:33–41. doi: 10.1016/j.ydbio.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ho EK, Stearns T. Hedgehog signaling and the primary cilium: implications for spatial and temporal constraints on signaling. Development. 2021;148 doi: 10.1242/dev.195552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Phua SC, et al. Dynamic Remodeling of Membrane Composition Drives Cell Cycle through Primary Cilia Excision. Cell. 2017;168:264–279.:e215. doi: 10.1016/j.cell.2016.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Airik M, et al. Loss of Anks6 leads to YAP deficiency and liver abnormalities. Hum Mol Genet. 2020;29:3064–3080. doi: 10.1093/hmg/ddaa197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Grisham JW. Ciliated Epithelial Cells in Normal Murine Intrahepatic Bile Ducts. Proc Soc Exp Biol Med. 1963;114:318–320. doi: 10.3181/00379727-114-28663. [DOI] [PubMed] [Google Scholar]
- 179.Tanuma Y, Ohata M. Occurrence of centrioles in interphasic hepatocytes of bat and chicken. Arch Histol Jpn. 1978;41:377–384. doi: 10.1679/aohc1950.41.377. [DOI] [PubMed] [Google Scholar]
- 180.Hirose Y, Itoh T, Miyajima A. Hedgehog signal activation coordinates proliferation and differentiation of fetal liver progenitor cells. Exp Cell Res. 2009;315:2648–2657. doi: 10.1016/j.yexcr.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 181.Grzelak CA, et al. The intrahepatic signalling niche of hedgehog is defined by primary cilia positive cells during chronic liver injury. J Hepatol. 2014;60:143–151. doi: 10.1016/j.jhep.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 182.Masyuk AI, et al. Cholangiocyte cilia detect changes in luminal fluid flow and transmit them into intracellular Ca2+ and cAMP signaling. Gastroenterology. 2006;131:911–920. doi: 10.1053/j.gastro.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gradilone SA, et al. Cholangiocyte cilia express TRPV4 and detect changes in luminal tonicity inducing bicarbonate secretion. Proc Natl Acad Sci U S A. 2007;104:19138–19143. doi: 10.1073/pnas.0705964104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Masyuk AI, et al. Cholangiocyte primary cilia are chemosensory organelles that detect biliary nucleotides via P2Y12 purinergic receptors. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2008;295:G725–G734. doi: 10.1152/ajpgi.90265.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lam WY, et al. Identification of a wide spectrum of ciliary gene mutations in nonsyndromic biliary atresia patients implicates ciliary dysfunction as a novel disease mechanism. EBioMedicine. 2021;71:103530. doi: 10.1016/j.ebiom.2021.103530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rock N, McLin V. Liver involvement in children with ciliopathies. Clin Res Hepatol Gastroenterol. 2014;38:407–414. doi: 10.1016/j.clinre.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 187.Gradilone SA, et al. HDAC6 Inhibition Restores Ciliary Expression and Decreases Tumor Growth. Cancer Research. 2013;73:2259–2270. doi: 10.1158/0008-5472.Can-12-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhu D, Shi S, Wang H, Liao K. Growth arrest induces primary-cilium formation and sensitizes IGF-1-receptor signaling during differentiation induction of 3T3-L1 preadipocytes. Journal of Cell Science. 2009;122:2760–2768. doi: 10.1242/jcs.046276. [DOI] [PubMed] [Google Scholar]
- 189.Hilgendorf KI, et al. Omega-3 Fatty Acids Activate Ciliary FFAR4 to Control Adipogenesis. Cell. 2019;179:1289–1305.:e1221. doi: 10.1016/j.cell.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Masyuk AI, et al. Ciliary subcellular localization of TGR5 determines the cholangiocyte functional response to bile acid signaling. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2013;304:G1013–G1024. doi: 10.1152/ajpgi.00383.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Masyuk AI, et al. Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2010;299:G990–G999. doi: 10.1152/ajpgi.00093.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ma M, Tian X, Igarashi P, Pazour GJ, Somlo S. Loss of cilia suppresses cyst growth in genetic models of autosomal dominant polycystic kidney disease. Nat Genet. 2013;45:1004–1012. doi: 10.1038/ng.2715. Using mouse genetics involving loss of polycystins and cilia in developing and adult kidney and liver, the authors discovered a novel pathway that inhibits cystic growth, which is inhibited by polycystins and activated by cilia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Waddell SH, et al. Primary cilia loss promotes reactivation of morphogenesis and cystfission through a deregulated TGF<em>β</em>-ECM-Integrin axis in polycystic liver disease. bioRxiv. 2022:2022.2004.2008.487546. doi: 10.1101/2022.04.08.487546. [DOI] [Google Scholar]
- 194.Dong K, et al. Renal plasticity revealed through reversal of polycystic kidney disease in mice. Nat Genet. 2021;53:1649–1663. doi: 10.1038/s41588-021-00946-4. Key paper about the reversability of ciliopathic disease phenotypes, which demonstrates that even late stage ADPKD can be rescued by restoring either PKD gene function, highlighting high plasticity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Pazour GJ, Witman GB. The vertebrate primary cilium is a sensory organelle. Current Opinion in Cell Biology. 2003;15:105–110. doi: 10.1016/S0955-0674(02)00012-1. [DOI] [PubMed] [Google Scholar]
- 196.Pazour GJ, et al. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. First study demonstrating that defects in IFT lead to aberrant assembly of primary cilia and cause ciliopathy (PKD) in vertebrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Haycraft CJ, Swoboda P, Taulman PD, Thomas JH, Yoder BK. The C. elegans homolog of the murine cystic kidney disease gene Tg737 functions in a ciliogenic pathway and is disrupted in osm-5 mutant worms. Development. 2001;128:1493–1505. doi: 10.1242/dev.128.9.1493. Together with ref. 196, this study is the first to show that defects in IFT lead to defective assembly of primary cilia and cause ciliopathy (PKD) in metazoans. [DOI] [PubMed] [Google Scholar]
- 198.Huangfu D, et al. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83. doi: 10.1038/nature02061. By performing a genetic screen in the mouse, this study shows for the first time that mutations in IFT genes lead to aberrant Hedgehog signalling and cause severe developmental defects in vertebrates. [DOI] [PubMed] [Google Scholar]
- 199.Olivieri JE, et al. RNA splicing programs define tissue compartments and cell types at single-cell resolution. Elife. 2021;10 doi: 10.7554/eLife.70692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yang C, et al. Pre-mRNA Processing Factors and Retinitis Pigmentosa: RNA Splicing and Beyond. Frontiers in Cell and Developmental Biology. 2021;9 doi: 10.3389/fcell.2021.700276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kwon OS, et al. Exon junction complex dependent mRNA localization is linked to centrosome organization during ciliogenesis. Nature Communications. 2021;12:1351. doi: 10.1038/s41467-021-21590-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Haward F, et al. Nucleo-cytoplasmic shuttling of splicing factor SRSF1 is required for development and cilia function. Elife. 2021;10 doi: 10.7554/eLife.65104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Geladaki A, et al. Combining LOPIT with differential ultracentrifugation for high-resolution spatial proteomics. Nature Communications. 2019;10:331. doi: 10.1038/s41467-018-08191-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mund A, et al. Deep Visual Proteomics defines single-cell identity and heterogeneity. Nature Biotechnology. 2022;40:1231–1240. doi: 10.1038/s41587-022-01302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Qin W, Cho KF, Cavanagh PE, Ting AY. Deciphering molecular interactions by proximity labeling. Nat Methods. 2021;18:133–143. doi: 10.1038/s41592-020-01010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhou Y, et al. Expanding APEX2 Substrates for Proximity-Dependent Labeling of Nucleic Acids and Proteins in Living Cells. Angewandte Chemie International Edition. 2019;58:11763–11767. doi: 10.1002/anie.201905949. [DOI] [PubMed] [Google Scholar]
- 207.Truong ME, et al. Vertebrate cells differentially interpret ciliary and extraciliary cAMP. Cell. 2021;184:2911–2926.:e2918. doi: 10.1016/j.cell.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hansen JN, et al. A cAMP signalosome in primary cilia drives gene expression and kidney cyst formation. EMBO Rep. 2022;23:e54315. doi: 10.15252/embr.202154315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Sheu SH, et al. A serotonergic axon-cilium synapse drives nuclear signaling to alter chromatin accessibility. Cell. 2022;185:3390–3407.:e3318. doi: 10.1016/j.cell.2022.07.026. Authors use focused ion beam-scanning electron microscopy, or FIB-SEM, of adult brain tissue to identify a novel ‘axociliary synapse’ where axons release serotonin directly onto cilia, acting as a shortcut for sending signals quickly and altering gene regulation in the nucleus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bock A, et al. Optical Mapping of cAMP Signaling at the Nanometer Scale. Cell. 2020;182:1519–1530.:e1517. doi: 10.1016/j.cell.2020.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lelek M, et al. Single-molecule localization microscopy. Nature Reviews Methods Primers. 2021;1:39. doi: 10.1038/s43586-021-00038-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Arsić A, Hagemann C, Stajković N, Schubert T, Nikić-Spiegel I. Minimal genetically encoded tags for fluorescent protein labeling in living neurons. Nature Communications. 2022;13:314. doi: 10.1038/s41467-022-27956-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Satoda Y, et al. BROMI/TBC1D32 together with CCRK/CDK20 and FAM149B1/JBTS36 contributes to intraflagellar transport turnaround involving ICK/CILK1. Mol Biol Cell. 2022;33:ar79. doi: 10.1091/mbc.E22-03-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Shaheen R, et al. Bi-allelic Mutations in FAM149B1 Cause Abnormal Primary Cilium and a Range of Ciliopathy Phenotypes in Humans. Am J Hum Genet. 2019;104:731–737. doi: 10.1016/j.ajhg.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sanders AA, et al. KIAA0556 is a novel ciliary basal body component mutated in Joubert syndrome. Genome Biol. 2015;16:293. doi: 10.1186/s13059-015-0858-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Epting D, et al. The ciliary transition zone protein TMEM218 synergistically interacts with the NPHP module and its reduced dosage leads to a wide range of syndromic ciliopathies. Hum Mol Genet. 2022;31:2295–2306. doi: 10.1093/hmg/ddac027. [DOI] [PubMed] [Google Scholar]
- 217.Grinspon RP. Genetics of congenital central hypogonadism. Best Pract Res Clin Endocrinol Metab. 2022;36:101599. doi: 10.1016/j.beem.2021.101599. [DOI] [PubMed] [Google Scholar]
- 218.Tanaka N, et al. Canine CNGA3 Gene Mutations Provide Novel Insights into Human Achromatopsia-Associated Channelopathies and Treatment. PLoS One. 2015;10:e0138943. doi: 10.1371/journal.pone.0138943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Schonauer R, et al. Novel nephronophthisis-associated variants reveal functional importance of MAPKBP1 dimerization for centriolar recruitment. Kidney Int. 2020;98:958–969. doi: 10.1016/j.kint.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.David O, et al. Pituitary stalk interruption syndrome broadens the clinical spectrum of the TTC26 ciliopathy. Clin Genet. 2020;98:303–307. doi: 10.1111/cge.13805. [DOI] [PubMed] [Google Scholar]
- 221.Brancati F, et al. Biallelic variants in the ciliary gene TMEM67 cause RHYNS syndrome. European Journal of Human Genetics. 2018;26:1266–1271. doi: 10.1038/s41431-018-0183-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Guen VJ, et al. STAR syndrome-associated CDK10/Cyclin M regulates actin network architecture and ciliogenesis. Cell Cycle. 2016;15:678–688. doi: 10.1080/15384101.2016.1147632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Vasquez SSV, van Dam J, Wheway G. An updated SYSCILIA gold standard (SCGSv2) of known ciliary genes, revealing the vast progress that has been made in the cilia research field. Mol Biol Cell. 2021;32:br13. doi: 10.1091/mbc.E21-05-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Lahteenoja L, et al. A novel frameshift variant in CEP78 associated with nonsyndromic retinitis pigmentosa, and a review of CEP78-related phenotypes. Ophthalmic Genet. 2022;43:152–158. doi: 10.1080/13816810.2022.2045511. [DOI] [PubMed] [Google Scholar]
- 225.Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci U S A. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. This paper reports the first discovery of IFT using digital interference contrast microscopy of Chlamydomonas cilia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.van Dam TJ, et al. Evolution of modular intraflagellar transport from a coatomer-like progenitor. Proc Natl Acad Sci U S A. 2013;110:6943–6948. doi: 10.1073/pnas.1221011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Taschner M, Lorentzen E. The Intraflagellar Transport Machinery. Cold Spring Harb Perspect Biol. 2016;8 doi: 10.1101/cshperspect.a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bhogaraju S, et al. Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science. 2013;341:1009–1012. doi: 10.1126/science.1240985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Craft JM, Harris JA, Hyman S, Kner P, Lechtreck KF. Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. The Journal of Cell Biology. 2015;208:223–237. doi: 10.1083/jcb.201409036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kubo T, et al. Together, the IFT81 and IFT74 N-termini form the main module for intraflagellar transport of tubulin. J Cell Sci. 2016;129:2106–2119. doi: 10.1242/jcs.187120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lechtreck KF, et al. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol. 2009;187:1117–1132. doi: 10.1083/jcb.200909183. By using Chlamydomonas as a model, this study demonstrates that the BBSome functions as a cargo adapter for certain membrane-associated and soluble ciliary proteins during retrograde IFT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Prevo B, Scholey JM, Peterman EJG. Intraflagellar transport: mechanisms of motor action, cooperation, and cargo delivery. The FEBS journal. 2017;284:2905–2931. doi: 10.1111/febs.14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Reilly ML, Benmerah A. Ciliary kinesins beyond IFT: Cilium length, disassembly, cargo transport and signalling. Biol Cell. 2019;111:79–94. doi: 10.1111/boc.201800074. [DOI] [PubMed] [Google Scholar]
- 234.Klena N, Pigino G. Structural Biology of Cilia and Intraflagellar Transport. Annu Rev Cell Dev Biol. 2022 doi: 10.1146/annurev-cellbio-120219-034238. [DOI] [PubMed] [Google Scholar]
- 235.Petriman NA, et al. Biochemically validated structural model of the 15-subunit intraflagellar transport complex IFT-B. EMBO J. 2022:e112440. doi: 10.15252/embj.2022112440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lacey SE, Foster HE, Pigino G. The molecular structure of IFT-A and IFT-B in anterograde intraflagellar transport trains. Nat Struct Mol Biol. 2023 doi: 10.1038/s41594-022-00905-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yang S, et al. Near-atomic structures of the BBSome reveal the basis for BBSome activation and binding to GPCR cargoes. Elife. 2020;9 doi: 10.7554/eLife.55954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Singh SK, Gui M, Koh F, Yip MC, Brown A. Structure and activation mechanism of the BBSome membrane protein trafficking complex. Elife. 2020;9 doi: 10.7554/eLife.53322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Klink BU, Gatsogiannis C, Hofnagel O, Wittinghofer A, Raunser S. Structure of the human BBSome core complex. Elife. 2020;9 doi: 10.7554/eLife.53910. [DOI] [PMC free article] [PubMed] [Google Scholar]