Abstract
Prolonged drug residence times may result in longer-lasting drug efficacy, improved pharmacodynamic properties, and “kinetic selectivity” over off-targets with high drug dissociation rates. However, few strategies have been elaborated to rationally modulate drug residence time and thereby to integrate this key property into the drug development process. Herein, we show that the interaction between a halogen moiety on an inhibitor and an aromatic residue in the target protein can significantly increase inhibitor residence time. By using the interaction of the serine/threonine kinase haspin with 5-iodotubercidin (5-iTU) derivatives as a model for an archetypal active-state (type I) kinase–inhibitor binding mode, we demonstrate that inhibitor residence times markedly increase with the size and polarizability of the halogen atom. The halogen–aromatic π interactions in the haspin–inhibitor complexes were characterized by means of kinetic, thermodynamic, and structural measurements along with binding-energy calculations.
The kinetics of drug binding have emerged as important parameters in drug development. A long drug residence time will result in prolonged inhibition after the free drug concentration has dropped owing to in vivo clearance, potentially leading to improved drug efficiency and reduced off-target-mediated toxicity. In cases where slow off-rates are specific for the target, kinetic selectivity can be achieved over fast off-target dissociation despite similar binding constants.1 The kinetics of the interaction of a drug with its target are defined by the association rate (kon) and dissociation rate (koff) constants. For bimolecular interactions, the ratio of these two parameters defines the equilibrium dissociation constant (KD) of a drug, and hence the drug occupancy of the target. As on- and off-rates are coupled in simple rigid bimolecular interactions, more complicated binding modes are frequently evoked to explain high on- and low off-rates.
Kinases are particularly dynamic proteins and provide multiple opportunities for the development of inhibitors that target induced or allosteric binding sites.2 One of the first kinase inhibitors for which low dissociation rates have been described is the p38 inhibitor BIRB-796. This inhibitor binds to an inactive conformation in which the DFG motif is displaced in a so-called “DFG-out” conformation.3 Inhibitors that bind to this conformation are called type II inhibitors and often have prolonged residence times (τ). However, not all type II inhibitors show slow binding kinetics, suggesting that the DFG-out conformational change itself is not sufficient to explain the slow dissociation of BIRB-796.4 Indeed, more recent studies attributed the slow binding kinetics to efficient hydrophobic contacts rather than the kinetic dissociation barrier introduced by the DFG-out transition.5 However, conformational change also contributes to the slow off-rate of the breast cancer drug lapatinib, a type I inhibitor of the epidermal growth factor receptor.6
In addition to protein conformational changes, the rearrangement of water molecules has been discussed as a potential mechanism influencing inhibitor residence time.7 An example for the influence of water molecules on ligand binding kinetics is the type I CDK inhibitor roniciclib, whose slow off-rate is the result of changes in the hydration network coupled to conformational adaptation of the DFG motif.8 In some cases, the presence of water-shielded hydrogen bonds can also lead to slow dissociation.9 In addition, reversible covalent inhibitors provide an interesting strategy for prolonging target engagement by transient covalent-bond formation.10 However, the design of reversibly formed covalent interactions requires the presence of cysteine residues in the drug binding site. Many drug receptors, including a large number of kinases, contain cysteines in close proximity to their active sites,11 but the development of covalent inhibitors may not be feasible for all drug targets.
Herein, we present data that suggest that interactions mediated by halogen atoms, which are common in drugs, and aromatic residues, which are also typically found in drug binding sites on proteins,12 can be utilized to design ligands with slow off-rates. We used 5-iodotubercidin (5-iTU), a close analogue of ATP, as a model inhibitor for a canonical active-state kinase binding mode (type I). Screening against more than 100 diverse kinases showed that an aromatic gatekeeper residue that interacts with the halogen moiety of this inhibitor is required for high-affinity binding. We chose haspin, a serine/threonine kinase with a known three-dimensional structure,13 as a model system. Analysis of the ligand binding kinetics surprisingly showed that the binding of 5-iTU was slow. Mutation of the gatekeeper residue as well as removal or substitution of the iodide with other halogen atoms showed that the low inhibitor off-rates are due to a π-stacking interaction of the halogen with the aromatic gatekeeper. We present structural, biophysical, and computational data that strongly suggest that halogen interactions with aromatic residues can be exploited for the development of inhibitors with slow off-rates.
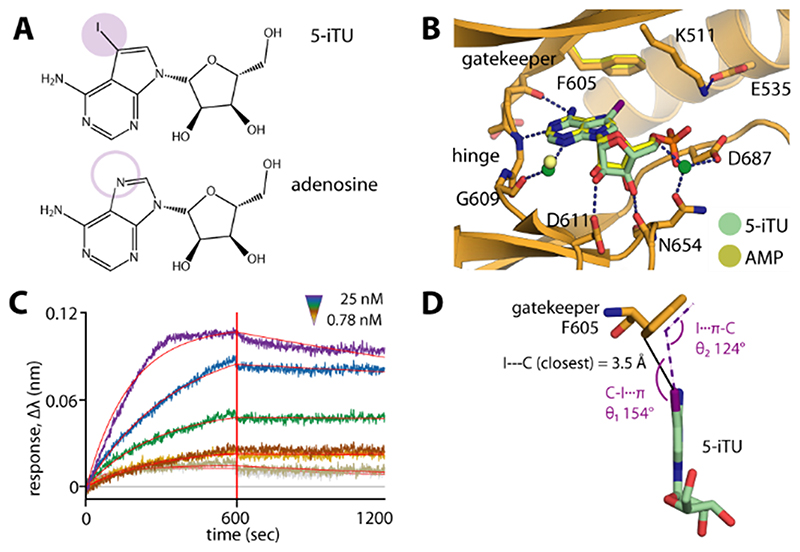
Comparative analysis of the high-resolution haspin structures revealed high conservation of the binding modes of both 5-iTU and the nucleoside adenosine (Figure 1 A, B). In contrast to adenosine or ATP, which binds with a KD value of about 180 μm,13b 5-iTU showed high affinity for haspin and an unexpectedly long target engagement time. We then further assessed the thermodynamics and kinetics of the binding by isothermal titration calorimetry (ITC), biolayer interferometry (BLI), and surface plasmon resonance (SPR) experiments, which consistently confirmed the tight binding with relatively slow binding kinetics, as measured for instance by BLI (Figure 1 C). Comparing the binding mode of 5-iTU with that of adenosine, the most striking structural difference between these highly similar molecules is the presence of the iodide moiety, which was positioned in close proximity to the F605 gatekeeper, forming a halogen–aromatic π interaction (Figure 1 D). We therefore hypothesized that this interaction might contribute most of the increase in the binding free energy (ΔG) and be responsible for the slow dissociation of 5-iTU from haspin.
Figure 1. The 5-iodotubercidin inhibitor (5-iTU) exhibits tight binding with slow dissociation kinetics from haspin.
A) Chemical structures of 5-iTU and adenosine. B) Superimposition of haspin–5-iTU and AMP (PDB ID: 4ouc) reveals similar binding modes for the two compounds. C) The BLI sensorgram suggests slow kinetics for the 5-iTU–haspin interaction. D) The iodide and benzene moieties of 5-iTU and F605, respectively, are located in close proximity, with a favorable geometry for a halogen–π bond.
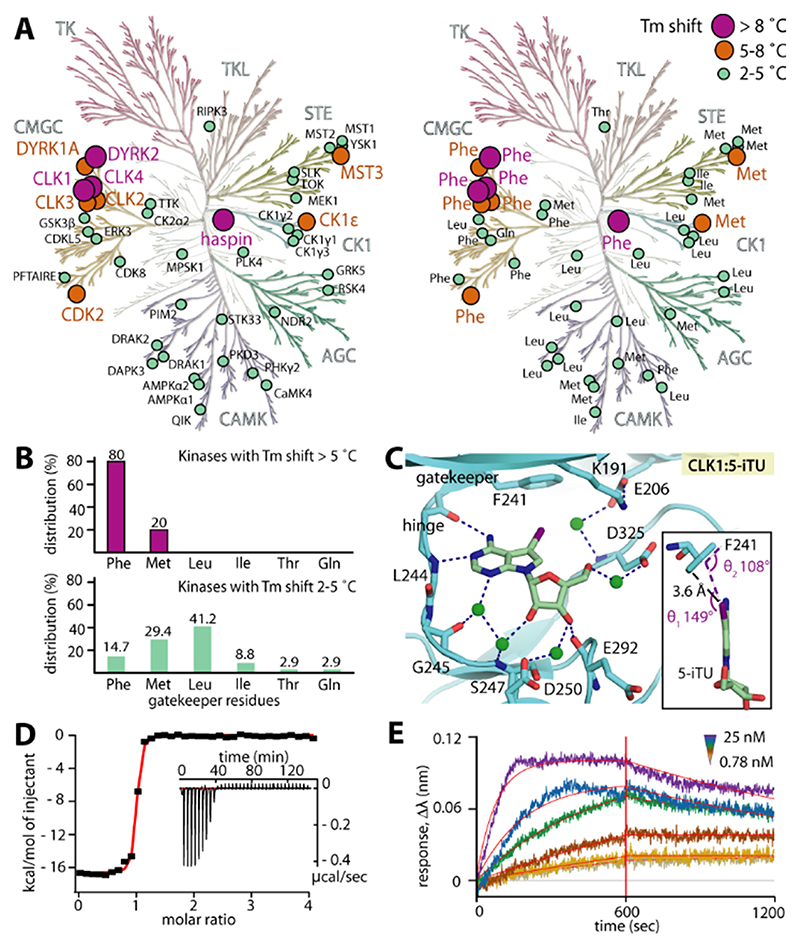
To address our hypothesis, we screened 5-iTU against 137 diverse kinases using temperature shift assays14 and observed unexpected selectivity of the inhibitor, with only ten kinases with ΔTm>5 °C (Figure 2 A and Table S1 in the Supporting Information). Interestingly, analyses of the gatekeeper residues of kinase targets that showed significant temperature shifts and therefore high affinity for 5-iTU revealed a strong preference for kinases harboring a phenylalanine (Phe) residue at this position whereas kinases that showed weak interactions (ΔTm=2–5 °C) revealed no preference for a certain residue. This analysis supported our hypothesis that the aromatic gatekeeper is important for high-affinity binding (Figure 2 B). To confirm these results, we determined the structure of 5-iTU bound to another high–affinity target that is structurally diverse from haspin, CLK1 (ΔTm=8.6 °C). As expected, the interaction of 5-iTU with CLK1 remarkably resembled that observed in haspin, including the conserved interaction geometry of the iodide with the CLK1 gatekeeper F241. 5-iTU bound CLK1 with high affinity (KD≈7 nm by ITC) and slow off-rates estimated to be about 50 min by BLI (Figure 2 C–E and Figure S1).
Figure 2. Interaction of 5-iTU with various human kinases.
A) Temperature shift assays show high ΔTm values preferentially for kinases harboring an aromatic gatekeeper. B) The distribution of gatekeeper residues reveals that the majority of high-ΔTm hits harbor a phenylalanine gatekeeper. C) The binding mode of 5-iTU in the off-target CLK1, including the iodide–gatekeeper interaction, is highly conserved. CLK1 has a high affinity for 5-iTU as measured by ITC (D) and indicated by a slow off-rate (E), as assessed by BLI.
The formation of a halogen–π interaction (C–X…π) is driven by the directional positive polarization along the halogen σ bond with the π molecular orbital of the aromatic system.15 However, the main focus of halogen–protein interactions has been on halogen–carbonyl/sulfonyl interactions (C–X…O/S bonds), with few examples that analyze halogen–aromatic interactions.16
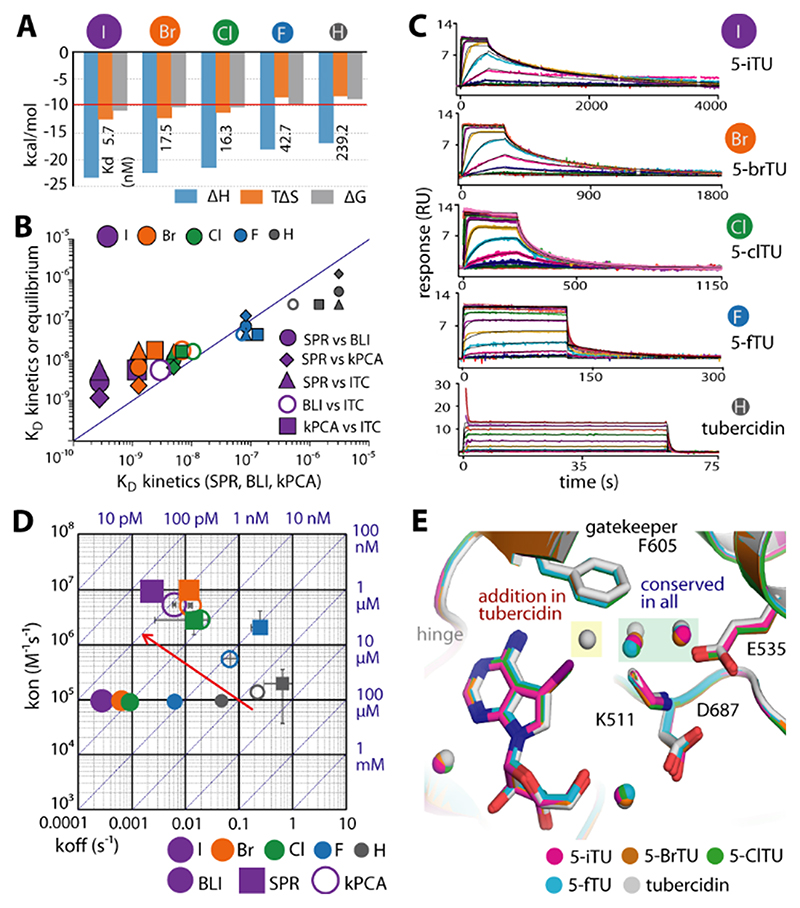
As the partial positive charge along the halogen σ bond diminishes with the size of the halogen atom, we next substituted the iodide by smaller halogen atoms and characterized the affinities and binding kinetics of these 5-iTU derivatives. Indeed, ITC experiments showed that the affinities of 5-iTU halogen derivatives were reduced with decreasing size of the halogen (Figure 3 and Table S2).
Figure 3. Binding kinetics of haspin with five tubercidin derivatives harboring halogen substituents at the 5-position.
A) ITC thermodynamic binding parameters. B) Comparison of dissociation constants (KD) measured by ITC, BLI, SPR, and kPCA shows good correlation for the measured equilibrium data. C) SPR sensorgrams demonstrating increasingly slow dissociation with increasing halogen size. D) Rate plot with isoaffinity diagonals (RaPID) of kon and koff constants measured by BLI, SPR, and kPCA. The red arrow indicates the trend to increasing kon and decreasing koff constants upon increasing the atomic radius of the halogen substituent. E) The crystal structures reveal the conserved binding modes of all five tubercidin derivatives, albeit with an additional water molecule adjacent to the inhibitor and F605 gatekeeper in tubercidin.
Removal of the halogen atom led to a 42-fold decrease in the potency of tubercidin (TU) compared to that of 5-iTU, and similarly an eightfold decrease was measured for 5-fluorotubercidin (5-fTU). Analyses of the binding kinetics of these five synthesized 5-tubercidin halogen derivatives with haspin were performed using three independent techniques: kinetic probe competition assays (kPCAs),17 BLI, and SPR. The binding affinities determined by these three independent methods correlated well with each other and also with the binding constants determined in solution by ITC (Figure 3 B). The dissociation rate constants from all experiments revealed the same behavior, with 5-iTU displaying the lowest off-rate. The off-rates increased with decreasing halogen size from the 5-iodo- to the 5-fluoro-substituted tubercidin, and the unsubstituted tubercidin showed the fastest off-rate (Figure 3 C, D). However, the absolute values differed somewhat between the different experimental methods used, with the residence time ranging from 60 min (BLI) to 7 min (SPR) for 5-iTU (Tables S2 and S3). We also observed lower on-rates using BLI but not in the SPR experiments. While the general trends were the same with both technologies, the differences in the on- and off-rates that have been observed might be due to differences in protein immobilization. As SPR is the more established technology, we used SPR kinetic data for quantitative analysis. The substitution from hydrogen to iodide at the 5-position of TU led to a 48-fold increase in the on-rates and a 274-fold decrease in the off-rates (Figure 3 D and Table S3).
Thus the fast dissociation kinetics observed for 5-fTU coincided with the lack of a pronounced σ hole in smaller halogen atoms, and hence an inability to form a polar halogen–π interaction with the aromatic gatekeeper. The increase in enthalpically favorable polar interactions with larger halogen atoms was also evident in the calorimetric data, which showed a steadily decreasing (more negative) binding enthalpy change from tubercidin to the larger halogens (H<F<Cl<Br<I). This effect of the halogen moieties was supported by the crystal structures, which showed that despite the highly conserved binding mode of all derivatives, the presence of an additional water molecule in the tubercidin complex within the space adjacent to the gatekeeper created by the removal of larger halogen substituents and the longer distance to the fluoro group presented suboptimal geometry for direct contacts with F605 in tubercidin and 5-fTU, respectively (Figure 3 E and Figure S2).
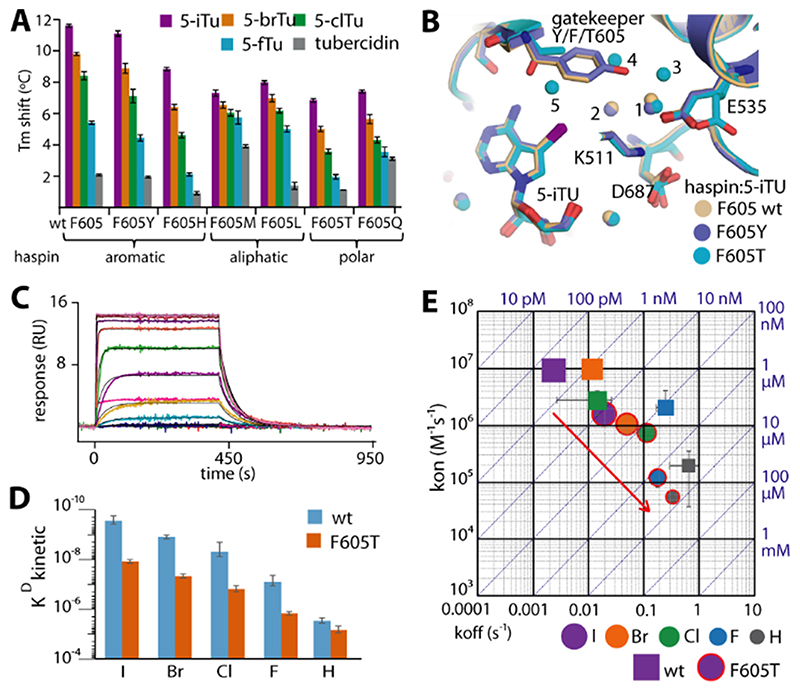
We next investigated the contributions of the Phe gatekeeper to the binding affinity and the observed slow dissociation kinetics of 5-iTU. Six haspin gatekeeper mutants using amino acids commonly found in kinases were generated, and their affinities against 5-iTU and its halogen derivatives were analyzed using DSF assays (Figure 4 A). As expected, the potency of 5-iTU with the aromatic tyrosine mutant was comparable to that of wild-type haspin.
Figure 4. The effect of gatekeeper mutation on the binding kinetics of haspin with tubercidin derivatives.
A) Tm shifts of six haspin mutants against five tubercidin derivatives. B) Superimposition of 5-iTU-complexed crystal structures of wild-type, F605Y, and F605T haspin reveals conserved binding modes for the inhibitor, but differences in the bound water molecules within the binding site. C) The SPR sensorgram demonstrates fast binding kinetics for the interaction between 5-iTU and the F605T mutant, which is accompanied by significant decreases in KD (D) and kon and generally increased koff constants (E).
For comparison, we analyzed the binding characteristics of 5-iTU with a representative mutant (F605T) in detail. Structural superimposition of the wild-type and the mutant structures (F605Y and F605T) showed that the gatekeeper mutation did not affect the binding mode of 5-iTU, yet led to a slight variation of the environment in the pocket. The substitution of the bulky aromatic residue F605 with the small threonine resulted in an extended water network, which filled the expanded binding site in the mutant (Figure 4 B and Figure S3). No direct contact was observed between the threonine side chain and the iodide of 5-iTU but interactions might be mediated through a water bridge. The absence of any strong contact was in agreement with the fast kinetics with an approximately 16-fold lower affinity as demonstrated by SPR (Figure 4 C, D). In comparison to the wild-type enzyme, the loss of the halogen–π contact in the F605T mutant led to a sixfold decrease and an eightfold increase in the association and dissociation rates, respectively, with the estimated residence time of 5-iTU dramatically dropping to less than one minute (Figure 4 E and Table S3).
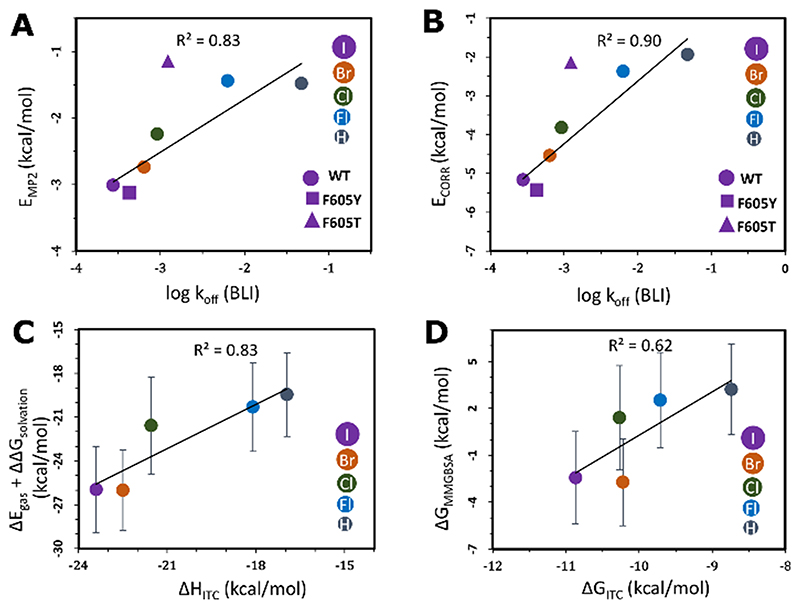
To assess the energetic contributions of the halogen–aromatic gatekeeper interaction, we calculated the energy of this interaction by ab initio quantum mechanics and classical methods. The second-order Møller–Plesset interaction energies (EMP2) between the inhibitor and the gatekeeper residue correlate well with the dissociation rate constants and equilibrium dissociation constants determined experimentally (Figure 5 A, Figure S5, and Tables S6–S9). Partitioning of EMP2 into its constituent energy components using a many-body interaction energy decomposition scheme showed that the major contribution to EMP2 is the correlation energy (ECORR), which describes second-order intermolecular dispersion interactions and the correlation corrections to the Hartree–Fock energy. ECORR increases in magnitude with an increase in the size of the halogen, which corresponds to the decreasing rate of dissociation measured experimentally (Figure 5 B), and indicates the importance of the halogen interaction with the aromatic gatekeeper for the prolongation of residence times as the halogen size increases. The computed ab initio energies also correlate for the interaction of 5-iTU with the F605Y mutant but the magnitude of the interaction energy of 5-iTU with the threonine mutant was underestimated. To account for the complete protein structure in the computation of the binding free energies of the haspin–ligand complexes, we used the classical MMGBSA approach with an implicit solvent model. The computed energies correlate well with calorimetric data measured by ITC, and are consistent with the increasingly favorable enthalpic contribution to binding as the halogen size increases (Figure 5 C, D and Table S10).
Figure 5. Correlation of the calculated binding free energies with experimental parameters for the halogen–gatekeeper interaction.
A) Second-order Møller–Plesset interaction energies (EMP2) and B) the second-order correlation correction energy terms (ECORR) between the TU derivatives and the gatekeeper residues versus the BLI koff values. The linear fits and correlation coefficients (R2) were computed by omitting the outlier F605T mutant. The experimental error bars are smaller than the size of the data points. Comparisons of C) MMGBSA internal and solvation contributions (ΔEgas+ΔΔGsolvation) versus the ITC enthalpies (ΔHITC) and D) the MMGBSA binding free energies (ΔGMMGBSA) versus the ITC binding free energies (ΔGITC) of the interactions between haspin and TU derivatives. Some ΔGMMGBSA values are positive as they only include translational and rotational entropic terms and do not include vibrational and conformational entropy contributions.
Halogen atoms are frequently found in approved drugs.12 A recent survey of the PDB archive for halogen–protein interactions reported that 33 % of all non-bonded interactions (excluding C–X…H contacts) of the heavier halogens in the protein database form aromatic stacking interactions of the C–X…π type.18 It is tempting to speculate that the presence of halogen atoms may lead to longer-lasting drug–protein interactions. Examples of the contribution of halogen atoms to prolonged off-rates in drug candidates have been demonstrated for the CDK inhibitor roniciclib,8 and it is likely that halogen atoms also contribute to the very slow off-rates observed for the ERK inhibitor VTX-11e.2b For the selected case of protein kinases, aromatic gatekeeper residues are the most frequently found residue type. Our proposed strategy of incorporating heavier halogen atoms into inhibitors to increase target residence time by designing interactions with aromatic residues is a general approach that may lead to active compounds with improved pharmacological properties. The biophysical, structural, and computational data presented herein on 5-halogen-substituted tubercidin derivatives provide a good basis for future studies on this exciting topic.
Supplementary Material
Acknowledgements
This study was supported by the European Union’s Seventh Framework Program (FP7/2007–2013) for the “Innovative Medicine Initiative” under Grant Agreement No. 115366. A.C. is supported by the SFB/DFG program “Autophagy”. G.K.G. and R.C.W. acknowledge financial support by the Klaus Tschira Foundation. P.B., A.C., and S.K. are grateful for support by the German cancer network DKTK and the SGC, a registered charity (no. 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, the Canada Foundation for Innovation, the Eshelman Institute for Innovation, Genome Canada through Ontario Genomics Institute [OGI-055], the Innovative Medicines Initiative (EU/EFPIA) [ULTRA-DD Grant No. 115766], Janssen, Merck KGaA, MSD, Novartis Pharma AG, the Ontario Ministry of Research, Innovation and Science (MRIS), Pfizer, the São Paulo Research Foundation-FAPESP, Takeda, and the Wellcome Trust. We thank Diamond Light Source for support.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Copeland RA, Pompliano DL, Meek TD. Nat Rev Drug Discovery. 2006;5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 2.2a Müller S, Chaikuad A, Gray NS, Knapp S. Nat Chem Biol. 2015;11:818–821. doi: 10.1038/nchembio.1938. [DOI] [PubMed] [Google Scholar]; 2b Chaikuad A, Tacconi EM, Zimmer J, Liang Y, Gray NS, Tarsounas M, Knapp S. Nat Chem Biol. 2014;10:853–860. doi: 10.1038/nchembio.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]; 2c Zhao Z, Wu H, Wang L, Liu Y, Knapp S, Liu Q, Gray NS. ACS Chem Biol. 2014;9:1230–1241. doi: 10.1021/cb500129t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pargellis C, Tong L, Churchill L, Cirillo PF, Gilmore T, Graham AG, Grob PM, Hickey ER, Moss N, Pav S, Regan J. Nat Struct Biol. 2002;9:268–272. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 4.Willemsen-Seegers N, Uitdehaag JC, Prinsen MB, de Vetter JR, de Man J, Sawa M, Kawase Y, Buijsman RC, Zaman GJ. J Mol Biol. 2017;429:574–586. doi: 10.1016/j.jmb.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 5.Schneider EV, Bottcher J, Huber R, Maskos K, Neumann L. Proc Natl Acad Sci USA. 2013;110:8081–8086. doi: 10.1073/pnas.1305378110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K, Alligood KJ, et al. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 7.Englert L, Biela A, Zayed M, Heine A, Hangauer D, Klebe G. Biochim Biophys Acta. 2010;1800:1192–1202. doi: 10.1016/j.bbagen.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Ayaz P, Andres D, Kwiatkowski DA, Kolbe CC, Lienau P, Siemeister G, Lucking U, Stegmann CM. ACS Chem Biol. 2016;11:1710–1719. doi: 10.1021/acschembio.6b00074. [DOI] [PubMed] [Google Scholar]
- 9.Schmidtke P, Luque FJ, Murray JB, Barril X. J Am Chem Soc. 2011;133:18903–18910. doi: 10.1021/ja207494u. [DOI] [PubMed] [Google Scholar]
- 10.10a Bradshaw JM, McFarland JM, Paavilainen VO, Bisconte A, Tam D, Phan VT, Romanov S, Finkle D, Shu J, Patel V, Ton T, et al. Nat Chem Biol. 2015;11:525–531. doi: 10.1038/nchembio.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]; 10b Miller RM, Paavilainen VO, Krishnan S, Serafimova IM, Taunton J. J Am Chem Soc. 2013;135:5298–5301. doi: 10.1021/ja401221b. [DOI] [PMC free article] [PubMed] [Google Scholar]; 10c Forster M, Chaikuad A, Bauer SM, Holstein J, Robers MB, Corona CR, Gehringer M, Pfaffenrot E, Ghoreschi K, Knapp S, Laufer SA. Cell Chem Biol. 2016;23:1335–1340. doi: 10.1016/j.chembiol.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; 10d Chaikuad A, Koch P, Laufer S, Knapp S. Angew Chem Int Ed. 2018;57:4372–4385. doi: 10.1002/anie.201707875. Angew. Chem. 2018, 130, 4456-4470. [DOI] [PubMed] [Google Scholar]
- 11.Liu Q, Sabnis Y, Zhao Z, Zhang T, Buhrlage SJ, Jones LH, Gray NS. Chem Biol. 2013;20:146–159. doi: 10.1016/j.chembiol.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z, Yang Z, Liu Y, Lu Y, Chen K, Zhu W. J Chem Inf Model. 2014;54:69–78. doi: 10.1021/ci400539q. [DOI] [PubMed] [Google Scholar]
- 13.13a Eswaran J, Patnaik D, Filippakopoulos P, Wang F, Stein RL, Murray JW, Higgins JM, Knapp S. Proc Natl Acad Sci USA. 2009;106:20198–20203. doi: 10.1073/pnas.0901989106. [DOI] [PMC free article] [PubMed] [Google Scholar]; 13b Villa F, Capasso P, Tortorici M, Forneris F, de Marco A, Mattevi A, Musacchio A. Proc Natl Acad Sci USA. 2009;106:20204–20209. doi: 10.1073/pnas.0908485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fedorov O, Niesen FH, Knapp S. Methods Mol Biol. 2012;795:109–118. doi: 10.1007/978-1-61779-337-0_7. [DOI] [PubMed] [Google Scholar]
- 15.Ho PS. Top Curr Chem. 2015;358:241–276. doi: 10.1007/128_2014_551. [DOI] [PubMed] [Google Scholar]
- 16.Tatko CD, Waters ML. Org Lett. 2004;6:3969–3972. doi: 10.1021/ol0483807. [DOI] [PubMed] [Google Scholar]
- 17.Schiele F, Ayaz P, Fernandez-Montalvan A. Anal Biochem. 2015;468:42–49. doi: 10.1016/j.ab.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Lu Y, Wang Y, Zhu W. Phys Chem Chem Phys. 2010;12:4543–4551. doi: 10.1039/b926326h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.