Abstract
Traditional approaches in the development of small-molecule drugs typically aim to inhibit the biochemical activity of functional protein domains. In contrast, targeted protein degradation aims to reduce overall levels of disease-relevant proteins. Mechanistically, this can be achieved via chemical ligands that induce molecular proximity between an E3 ubiquitin ligase and a protein of interest, leading to ubiquitination and degradation of the protein of interest. This paradigm-shifting pharmacology promises to address several limitations inherent to conventional inhibitor design. Most notably, targeted protein degradation has the potential not only to expand the druggable proteome beyond the reach of traditional competitive inhibitors but also to develop therapeutic strategies of unmatched selectivity. This review briefly summarizes key challenges that remain to be addressed to deliver on these promises and to realize the full therapeutic potential of pharmacologic modulation of protein degradation pathways.
Keywords: Targeted protein degradation, PROTACs, Molecular glues, Chemical biology, E3 ligase
The framework of chemically induced protein dimerization
Proximity and interactions between proteins underpin most cellular processes. This leaves their dynamic modulation as an important goal in chemical biology and ligand discovery. Several strategies to induce protein—protein interactions (PPIs) have been innovated and are summarized in an excellent recent review [1].
Historically, the concept of inducing novel PPIs through small molecules goes back to immunosuppressive natural products, such as rapamycin and FK506. Mechanistically, both bind the protein FKBP12, thus inducing novel interactions with the mechanistic target of rapamycin kinase (mTOR) and the phosphatase calcineurin, respectively. As a consequence of this drug-induced neoassociation, several activities of mTOR and calcineurin are modulated [2–4]. Thus, both small molecules act as ‘chemical neomorphs’ that achieve their cellular effect by inducing PPIs that otherwise do not occur in nature. Another example of such a chemical neomorph is the plant hormone indole-3-acetic acid (IAA, auxin). Auxin induces molecular proximity between the ubiquitin ligase substrate receptor TIR1 and a family of transcriptional regulators called Aux/IAAs [5,6]. As a consequence, IAAs get ubiquitinated and degraded by the proteasome. Thus, auxin modulates the function of the SCFTIR1 ligase, enabling molecular recognition and degradation of a protein it would not recognize in its absence. The notion that small molecules can artificially change the target spectrum of cellular effectors of the ubiquitin—proteasome system, in particular of E3 ubiquitin ligases, represents the core concept of targeted protein degradation (TPD) [7].
Monovalent molecular glues and heterobifunctional degraders
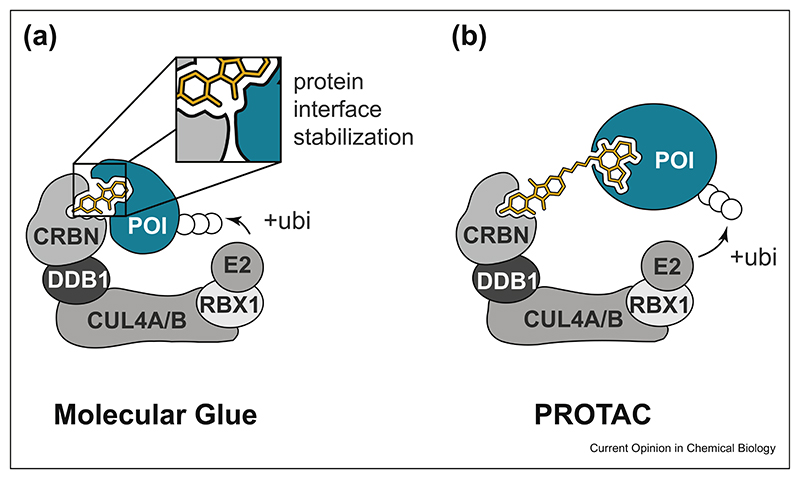
Conceptually, there are two chemical strategies to alter the substrate spectrum of an E3 ubiquitin ligase (Figure 1). First, E3 ligases can be reprogrammed by monovalent small molecules, often referred to as ‘molecular glues’ (MGs). Auxin is a prime example of how a MG connects an E3 ligase (SCFTIR1) and the target protein. Ligand binding to the E3 ligase alters protein interface properties, leading to dimerization with a neosubstrate. Similarly, the clinically approved drug thalidomide and its analogs, collectively called ‘immunomodulatory drugs’ or IMiDs, were found to act via a MG-type mechanism, namely, by reprogramming the target spectrum of the E3 ligase CRL4CRBN. After the initial identification of the substrate receptor Cereblon (CRBN) as the cellular target of thalidomide, seminal studies uncovered that IMiD binding to CRBN leads to recruitment and proteasomal degradation of zinc finger (ZF) proteins IZKF1 and IKZF3 [8–10]. In general, MGs such as IMiDs orchestrate molecular recognition between the E3 ligase and the neosubstrate in a highly cooperative manner (Figure 1A). This means that the MG typically induces novel PPIs between the E3 ligase and the target protein which contribute to the formation of a trimeric E3:MG:target protein complex. Notably, IMiDs per se have no measurable binding affinity to their degraded targets but hijack an entire surface patch on CRBN to induce productive dimerization [11]. While this outlines the exciting possibility of degrading unligandable proteins, the discovery of E3 modulating MGs has so far mostly been serendipitous and cannot be easily generalized for other targets and target classes.
Figure 1. Schematic comparison of molecular glues and PROTACs.
(a) Molecular glues are monovalent compounds that induce the dimerization of two proteins (here: an E3 ligase substrate receptor and a neosubstrate). Compound-induced proximity is often characterized by multiple interactions between the two proteins, resulting in high binding cooperativity. (b) PROTACs are heterobifunctional degraders that can individually bind to the E3 ligase and the protein of interest. Simultaneous binding induces molecular proximity and ensuing ubiquitination and degradation of the POI. In contrast to MGs, PROTAC-mediated dimerization is less dependent on compatible protein surfaces and associated binding cooperativity. PROTAC, proteolysis targeting chimeras; MG, molecular glue; POI, protein of interest.
The major alternative to MGs is heterobifunctional degraders, which consist of one ligand binding to a ubiquitin ligase and a second ligand designed to engage a protein of interest (POI). Both ligands are connected by a flexible linker of suitable length to allow simultaneous binding to the E3 and the POI, leading to molecular proximity (Figure 1B). Productive ternary complex formation ultimately leads to ubiquitination and degradation of the POI. Such heterobifunctional degraders are often referred to as proteolysis targeting chimeras (PROTACs) [12]. First PROTAC concepts relied on peptidic agents with limited cellular efficacy. However, over the past couple of years, significant progress has been made with nonpeptidic heterobifunctional molecules [13–15]. These efforts enabled the targeted degradation of a range of proteins with well-defined ligand binding sites, such as bromodomain proteins and kinases [16,17]. Among a small group of E3 ligases that can be harnessed for PROTAC development, CRBN and the von Hippel-Lindau tumor suppressor (VHL) stand out in terms of their versatility and in vivo compatibility [13,18,19]. Other accessible E3 ligases include MDM2 and cIAP1 [20,21]. The modular nature of PROTACs comes with tangible upsides but also considerable challenges. First, it allows a rational and straightforward design simply by exchanging the target-binding warhead. On the other hand, their molecular weight (typically above 800 Da) poses a challenge for pharmacokinetic optimization [22]. Furthermore, as PROTACs are required to individually bind both the target protein and the E3 ligase, the associated degradable space is limited to proteins that can efficiently be liganded with small molecules. In the following, we want to highlight some of the recent discoveries and trends in the field of PROTACs and MGs and put emphasis on some of the remaining key challenges.
Exploiting selectivity of TPD
Conventional pharmacologic inhibition of a protein typically depends on ligand binding at a functional site, which often is conserved throughout enzyme families. This poses a challenge for selective inhibitor design, as showcased, for instance, by the field of kinase inhibitors [23]. In contrast, TPD requires not only compound binding but also positioning of the E3 and the POI in a configuration conducive to ternary complex formation.
Finally, tripartite binding must occur in a manner that ensures accessibility of lysine residues within the ubiquitination zone of the ligase. Notably, recent studies have uncovered that these requirements can provide an avenue toward the design of highly selective degraders starting from more promiscuous targeting ligands. First observations of target discrimination through TPD where made with MZ1, a PROTAC that prompted preferential BRD4 degradation even though using a pan-bromodomain and extraterminal (BET) protein binding targeting ligand [14,24]. Moreover, when the multikinase inhibitor SNS-032 was conjugated to thalidomide to create a putative multikinase degrader, the resulting compound selectively degraded CDK9, despite retaining a multikinase binding spectrum [25]. Further studies have systematically addressed this phenomenon by generating CRBN- or VHL-based degraders based on additional multikinase inhibitor warheads [16,17]. Although the degraders would still bind to hundreds of kinases, degradation was achieved for only 10—20% of the targets across different cellular backgrounds. In addition, it was shown that the degradable target spectrum was dependent on the hijacked E3 ligase. Subtle changes in linker configuration were sufficient to generate isoform-specific degraders of the p38 mitogen-activated protein kinase (MAPK) family [26]. Together, these studies also highlighted that binding affinity to the POI was not correlated to its degradability. Further supporting the notion that degraders can elicit unprecedented selectivity, structural dissection of BET PROTACs led to the first rational development of a BRD4-selective degrader. Mechanistically, selectivity was dictated by protein— protein interface determinants outside of the ligand binding pocket on the POI [27,28]. Furthermore, subtle modification of the E3 ligase binder allows controlling for residual inhibitory consequences of the PROTAC [29].
Collectively, these studies have shown how selectivity of otherwise promiscuous scaffolds can be achieved through TPD and lead to highly specific degraders. This was ultimately exploited in a disease-relevant context by generating PROTACs capable of selectively degrading CDK6 over CDK4 [29,30]. Selective CDK6 degraders could emerge as valuable therapeutic options in hematologic malignancies and will enable addressing kinaseindependent scaffolding functions [31,32].
Expanding the scope of E3 ligases via covalent ligands
Rational identification of novel E3 ligands is a challenging task and most discoveries so far have happened serendipitously. The covalently acting natural product nimbolide is an example of such a finding. To elucidate the mechanism of action of nimbolide, activity-based proteomic profiling was applied, leading to the identification of the E3 ligase RNF114 as the primary target [33]. Other studies have explored the rational identification of covalent interactors of the E3 ligases. Gel-based activity-based proteomic profiling was used to screen for covalent binders of RNF4, identifying several probes with varying specificity for RNF4 over other proteins [34]. In another approach, broad cysteine-reactive probes were conjugated to an FKBP12 ligand and tested for their ability to degrade FKBP12 [35]. This led to the identification of a covalent ligand of the CRL4 substrate receptor DCAF16.
An important question was if the different covalent E3 binders could further be developed as E3 recruitment elements in a heterobifunctional degrader design. All of these studies assayed degradability of BET proteins by synthesizing PROTACs that connect the novel covalent E3 ligands to the known BET bromodomain antagonist JQ1 [24]. RNF114- and DCAF16-based degraders were selective for BRD4. The RNF4-based PROTAC however caused destabilization of a broader range of proteins including BRD4. Although effects of covalent binding of a POI through PROTACs have been studied previously, potential advantages and disadvantages of degraders that covalently engage the E3 ligase still remain to be experimentally validated [36]. Conceptually, these could include the reduction of the requirement for a ternary binding event to a binary interaction. Further ligand optimization will be required to develop second-generation, in vivo compatible degraders.
Exploring the limits of CRBN modulation with MGs
Although first PROTAC molecules recently entered clinical investigations, MG degraders, such as the aforementioned IMiDs, are already routine treatment options for different B-cell neoplasms. Generalizing and rationalizing the discovery of novel MGs thus marks a key future challenge. This could be achieved by further exploring the limits of the ‘degradable space’ in reach of the CRL4CRBN ligase complex or by chemically unlocking novel ligases. The first evidence that the CRL4CRBN ligase complex can be hijacked for the degradation of proteins other than the initially identified IKZF1/3 surfaced when CK1a was identified as an additional target of lenalidomide [37]. Soon, several additional IMiD targets such as GSPT1, ZFP91, and SALL4 were described [38–41]. This growing list motivated the first comprehensive assessment of the spectrum of neosubstrates that can be degraded by reprogramming CRL4CRBN with IMiD-like MGs [42]. Since the Cys2-His2 (C2H2)ZF domains of known substrates were sufficient for drug-induced recruitment, it was reasoned that IMiDs may degrade other proteins containing this feature. Indeed, four novel neosubstrates were identified using a scalable protein stability reporter assay. Structural and sequence homology analysis revealed that the overall ZF fold, rather than a particular linear amino acid sequence, is required for IMiD-based recruitment to the CRL4CRBN interface. This enabled a computational prediction of neosubstrates via a holistic docking approach, leading to the identification of several additional proteins that can biochemically be recruited to CRBN after IMiD treatment. Further chemistry efforts will be required to develop chemical matter that can expand on these efforts. Forthcoming studies will likely take advantage of these modern-day docking strategies to predict ‘glue-able’ interfaces between POIs and E3s, which might enable the prioritization of novel MG scaffolds.
MG approaches to other ubiquitin ligases
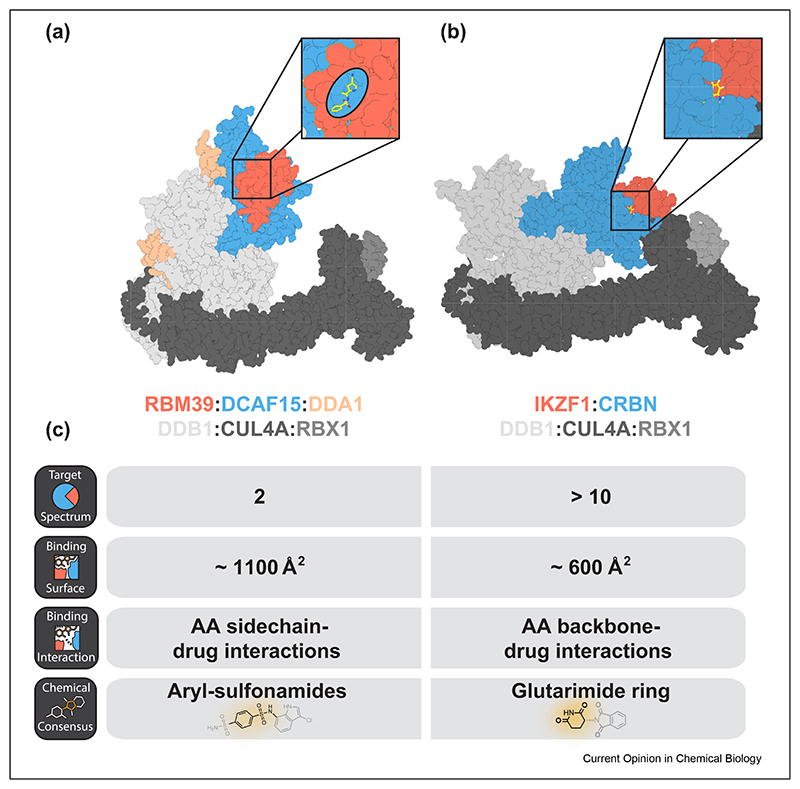
Recent research revealed that the molecular mechanism of IMiDs might not be a unique phenomenon. The arylsulfonamide indisulam and structurally related analogs were shown to induce the degradation of the splicing factor RBM39 by chemically reprogramming the substrate receptor DCAF15 [43,44]. Three recent independent studies have provided the structural and biochemical workup that unequivocally confirmed that indisulam and related aryl sulfonamides indeed function via a MG mechanism analogous to the IMiD pharmacology [45–47]. All three studies characterized a complex of DCAF15:DDB1:DDA1 together with an aryl sulfonamide recruiting RBM39. Of note, these compounds bind to DCAF15 with a significantly lower affinity than IMiDs bind to CRBN. Consistent with a model of high cooperativity, RBM39 binding greatly improves complex stability. In unbiased proteomics experiments, RBM23 is the only other destabilized protein after indisulam treatment [45]. Mechanistically, this is explained by high sequence conservation between their RRM domains involved in indisulam-induced molecular recognition. This remarkable selectivity likely stems from the buried surface area between DCAF15 and RBM39, which is approximately twice as large as that of CRBN and its neosubstrates (Figure 2). Furthermore, indisulam binding involves significant side chain interactions in the ternary complex resulting in increased specificity. This leads all three studies to the conclusion that the DCAF15:indisulam surface will likely not allow the same promiscuous target spectrum as the CRBN:IMiD interface. However, new chemical matter could potentially adopt some of the RBM39:DCAF15 interactions and thereby allow other targets to be degraded.
Figure 2. Comparison of DCAF15- and CRBN-based molecular glue degraders.
(a) A structure of DCAF15 (PDB: 6PAI) bound to indisulam, the RRM2 domain of RBM39 and DDB1 aligned with a structure of DDB1:CUL4A:RBX1 (PDB 6PAI). (b) A structure of CRBN (PDB: 6H0F) in complex with pomalidomide, the ZF2 domain of IKZF1 and DDB1 aligned with a structure of DDB1:CUL4A:RBX1 (PDB 6PAI). (c) Comparison of general properties of DCAF15- and CRBN-based molecular glues.
In addition to developing MGs that recruit true neosubstrates, another promising approach is to chemically reinforce known substrate—E3 interactions that are altered in the disease. One such example is β-catenin, a Wnt signaling effector protein. β-Catenin is often mutated at Ser33 and Ser37 [48]. This disrupts a phosphodegron recognized by the SCFβ-TrCP cullin-RING ligase, leading to stabilization of β-catenin and associated oncogenic consequences [49]. A recent study developed several biochemical screens coupled to structure-informed lead optimization to identify a compound that can re-enhance binding and ubiquitination of mutant β-catenin [50]. This marks the first successful study with the rationale of finding MGs for a specific ligase:substrate pair and provides a blueprint for similar future endeavors.
Collectively, the field of TPD has matured to a level where its generalizable nature is widely accepted. We will increasingly learn how to leverage some of the challenges that are associated with degrader design to our advantage to develop small molecules with unprecedented selectivity and potency. We believe that key challenges in the field will be to chemically unlock a larger number of ligases and to further rationalize the development of monovalent MGs. In addition, with the first heterobifunctional degraders in clinical trials, initial promising results on safety and druglikeness will have to be solidified (NCT03888612 and NCT04072952). This also leads to important considerations on anticipating, detecting, and circumventing associated resistance mechanisms. Along these lines, recent studies have shown that mutations in E3 ligase complexes or regulators thereof can lead to resistance to different types of degraders [51–53]. Similarly, CRBN mutations/loss of functions have been linked to the clinical mechanism of IMiD resistance, but their relevance have not fully been resolved [54,55]. Importantly, IMiDs are known to also act via a multiple indirect mechanisms, including T-cell and NK-cell modulation [56,57]. It thus remains to be seen how tumors will cope with the selective pressure ofTPD therapies that function primarily via cell-autonomous mechanisms.
Acknowledgements
The Center for Molecular Medicine (CeMM) and the Winter Laboratory are supported by the Austrian Academy of Sciences. This work was further supported by a FWF (Fonds zur Foerderung der Wissenschaftlichen Forschung) grant to GeorgWinter (project number P30271). We thank Matthias Brand for assistance with figure design. We want to apologize to colleagues whose work could not be included and highlighted in this review because of space limitations.
Footnotes
Conflict of interest statement
Nothing declared.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
* * of outstanding interest
- 1.Stanton BZ, Chory EJ, Crabtree GR. Chemically induced proximity in biology and medicine. Science. 2018;359:eaao5902–11. doi: 10.1126/science.aao5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J, Farmer JD, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 3.Griffith JP, Kim JL, Kim EE, Sintchak MD, Thomson JA, Fitzgibbon MJ, Fleming MA, Caron PR, Hsiao K, Navia MA. X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex. Cell. 1995;82:507–522. doi: 10.1016/0092-8674(95)90439-5. [DOI] [PubMed] [Google Scholar]
- 4.Stan R, McLaughlin MM, Cafferkey R, Johnson RK, Rosenberg M, Livi GP. Interaction between FKBP12-rapamycin and TOR involves a conserved serine residue. J Biol Chem. 1994;269:32027–32030. [PubMed] [Google Scholar]
- 5.Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 6.Kepinski S, Leyser O. Auxin-induced SCFTIR1 -Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc Natl Acad Sci. 2004;101:12381–12386. doi: 10.1073/pnas.0402868101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai AC, Crews CM. Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov. 2017;16:101–114. doi: 10.1038/nrd.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, Handa H. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [* This study shows uncovered CRBN as the cellular target of thalidomide] [DOI] [PubMed] [Google Scholar]
- 9.Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, Wong KK, Bradner JE, Kaelin WG. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [* Together with Krönke et al., this study highlights that lenalidomide modulates, rather than inhibits the function of the CRBN E3 ligase complex, leading to degradation of the neo-substrates IKZF1 and IKZF3.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kronke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M, Svinkina T, Heckl D, Comer E, Li X, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [* Together with Lu et al., this study highlights that lenalidomide modulates, rather than inhibits the function of the CRBN E3 ligase complex, leading to degradation of the neo-substrates IKZF1 and IKZF3] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chamberlain PP, Hamann LG. Development of targeted protein degradation therapeutics. Nat Chem Biol. 2019;86:129. doi: 10.1038/s41589-019-0362-y. [DOI] [PubMed] [Google Scholar]
- 12.Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, Bradner JE. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [** This study outlined the first all-chemical and in vivo compatible strategy for rational degrader design via CRBN reprogramming] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zengerle M, Chan K-H, Ciulli A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem Biol. 2015;10:1770–1777. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Qian Y, Altieri M, Dong H, Wang J, Raina K, Hines J, Winkler JD, Crew AP, Coleman K, et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol. 2015;22:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang H-T, Dobrovolsky D, Paulk J, Yang G, Weisberg EL, Doctor ZM, Buckley DL, Cho J-H, Ko E, Jang J, et al. A chemoproteomic approach to query the degradable kinome using a multi-kinase degrader. Cell Chem Biol. 2018;25:88–99.:e6. doi: 10.1016/j.chembiol.2017.10.005. [* Two comprehensive investigations of degrome of multi-kinase binding PROTACs including an assessment of how different ligases affect the spectrum of degradable targets] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bondeson DP, Smith BE, Burslem GM, Buhimschi AD, Hines J, Jaime-Figueroa S, Wang J, Hamman BD, Ishchenko A, Crews CM. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem Biol. 2018;25:78–87.:e5. doi: 10.1016/j.chembiol.2017.09.010. [* Two comprehensive investigations of degrome of multi-kinase binding PROTACs including an assessment of how different ligases affect the spectrum of degradable targets] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pettersson M, Crews CM. PROteolysis TArgeting Chimeras (PROTACs) - past, present and future. Drug Discov Today Technol. 2019;31:15–27. doi: 10.1016/j.ddtec.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bondeson DP, Mares A, Smith IED, Ko E, Campos S, Miah AH, Mulholland KE, Routly N, Buckley DL, Gustafson JL, et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol. 2015;11:611–617. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneekloth AR, Pucheault M, Tae HS, Crews CM. Targeted intracellular protein degradation induced by a small molecule: en route to chemical proteomics. Bioorg Med Chem Lett. 2008;18:5904–5908. doi: 10.1016/j.bmcl.2008.07.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh Y, Ishikawa M, Naito M, Hashimoto Y. Protein knockdown using methyl bestatin-ligand hybrid molecules: design and synthesis of inducers of ubiquitination-mediated degradation of cellular retinoic acid-binding proteins. J Am Chem Soc. 2010;132:5820–5826. doi: 10.1021/ja100691p. [DOI] [PubMed] [Google Scholar]
- 22.Edmondson SD, Yang B, Fallan C. Proteolysis targeting chimeras (PROTACs) in “beyond rule-of-five” chemical space: recent progress and future challenges. Bioorg Med Chem Lett. 2019;29:1555–1564. doi: 10.1016/j.bmcl.2019.04.030. [DOI] [PubMed] [Google Scholar]
- 23.Müller S, Chaikuad A, Gray NS, Knapp S. The ins and outs of selective kinase inhibitor development. Nat Chem Biol. 2015;11:818–821. doi: 10.1038/nchembio.1938. [DOI] [PubMed] [Google Scholar]
- 24.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olson CM, Jiang B, Erb MA, Liang Y, Doctor ZM, Zhang Z, Zhang T, Kwiatkowski N, Boukhali M, Green JL, et al. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol. 2017;14:1–14. doi: 10.1038/nchembio.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith BE, Wang SL, Jaime-Figueroa S, Harbin A, Wang J, Hamman BD, Crews CM. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat Commun. 2019;10:1–13. doi: 10.1038/s41467-018-08027-7. [* A structural and biochemical survey of PROTAC mediated target selectivity across isoforms of kinase family] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gadd MS, Testa A, Lucas X, Chan K-H, Chen W, Lamont DJ, Zengerle M, Ciulli A. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol. 2017;13:514–521. doi: 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nowak RP, DeAngelo SL, Buckley D, He Z, Donovan KA, An J, Safaee N, Jedrychowski MP, Ponthier CM, Ishoey M, et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat Chem Biol. 2018;14:706–714. doi: 10.1038/s41589-018-0055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brand M, Jiang B, Bauer S, Donovan KA, Liang Y, Wang ES, Nowak RP, Yuan JC, Zhang T, Kwiatkowski N, et al. Homolog-selective degradation as a strategy to probe the function of CDK6 in AML. Cell Chem Biol. 2019;26:300–306.:e9. doi: 10.1016/j.chembiol.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang B, Wang ES, Donovan KA, Liang Y, Fischer ES, Zhang T, Gray NS. Development of dual and selective degraders of cyclin-dependent kinases 4 and 6. Angew Chem Int Ed. 2019;58:6321–6326. doi: 10.1002/anie.201901336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kollmann K, Heller G, Schneckenleithner C, Warsch W, Scheicher R, Ott RG, Schäfer M, Fajmann S, Schlederer M, Schiefer A-I, et al. A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell. 2013;24:167–181. doi: 10.1016/j.ccr.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tigan A-S, Bellutti F, Kollmann K, Tebb G, Sexl V. CDK6-a review of the past and a glimpse into the future: from cellcycle control to transcriptional regulation. Oncogene. 2016;35:3083–3091. doi: 10.1038/onc.2015.407. [DOI] [PubMed] [Google Scholar]
- 33.Spradlin JN, Hu X, Ward CC, Brittain SM, Jones MD, Ou L, To M, Proudfoot A, Ornelas E, Woldegiorgis M, et al. Harnessing the anti-cancer natural product nimbolide for targeted protein degradation. Nat Chem Biol. 2019;15:747–755. doi: 10.1038/s41589-019-0304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward CC, Kleinman JI, Brittain SM, Lee PS, Chung CYS, Kim K, Petri Y, Thomas JR, Tallarico JA, McKenna JM, et al. Covalent ligand screening uncovers a RNF4 E3 ligase recruiter for targeted protein degradation applications. ACS Chem Biol. 2019 doi: 10.1021/acschembio.8b01083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Crowley VM, Wucherpfennig TG, Dix MM, Cravatt BF. Electrophilic PROTACs that degrade nuclear proteins by engaging DCAF16. Nat Chem Biol. 2019;15:737–746. doi: 10.1038/s41589-019-0279-5. [** This study outlines a rational approach towards the identification of novel covalent E3 ligands that are suited as recruitment elements for heterobifunctional degraders] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tinworth CP, Lithgow H, Dittus L, Bassi ZI, Hughes SE, Muelbaier M, Dai H, Smith IED, Kerr WJ, Burley GA, et al. PROTAC-mediated degradation of Bruton’s tyrosine kinase is inhibited by covalent binding. ACS Chem Biol. 2019;14:342–347. doi: 10.1021/acschembio.8b01094. [DOI] [PubMed] [Google Scholar]
- 37.Krönke J, Fink EC, Hollenbach PW, MacBeth KJ, Hurst SN, Udeshi ND, Chamberlain PP, Mani DR, Man HW, Gandhi AK, et al. Lenalidomide induces ubiquitination and degradation of CK1a in del(5q) MDS. Nature. 2015;523:183–188. doi: 10.1038/nature14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matyskiela ME, Lu G, Ito T, Pagarigan B, Lu C-C, Miller K, Fang W, Wang N-Y, Nguyen D, Houston J, et al. A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase. Nature. 2016;535:252–257. doi: 10.1038/nature18611. [DOI] [PubMed] [Google Scholar]
- 39.An J, Ponthier CM, Sack R, Seebacher J, Stadler MB, Donovan KA, Fischer ES. pSILAC mass spectrometry reveals ZFP91 as IMiD-dependent substrate of the CRL4CRBN ubiquitin ligase. Nat Commun. 2017;8:15398. doi: 10.1038/ncomms15398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matyskiela ME, Couto S, Zheng X, Lu G, Hui J, Stamp K, Drew C, Ren Y, Wang M, Carpenter A, et al. SALL4 mediates terato-genicity as a thalidomide-dependent cereblon substrate. Nat Chem Biol. 2018;14:981–987. doi: 10.1038/s41589-018-0129-x. [DOI] [PubMed] [Google Scholar]
- 41.Donovan KA, An J, Nowak RP, Yuan JC, Fink EC, Berry BC, Ebert BL, Fischer ES. Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane Radial Ray syndrome. eLife. 2018;7:154. doi: 10.7554/eLife.38430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sievers QL, Petzold G, Bunker RD, Renneville A, Sŀabicki M, Liddicoat BJ, Abdulrahman W, Mikkelsen T, Ebert BL, Thomä NH. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science. 2018;362:eaat0572. doi: 10.1126/science.aat0572. [** A comprehensive study investigating the the structural relationship between degradable zinc finger proteins and the IMiD:CRBN complex that also innovated computational methods to predict targets within the reach of IMiD-based CRBN reprogramming] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han T, Goralski M, Gaskill N, Capota E, Kim J, Ting TC, Xie Y, Williams NS, Nijhawan D. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science. 2017;356:eaal3755. doi: 10.1126/science.aal3755. [** This study describes the serendipitous finding that the anti-cancer activity of selected aryl sulfonamides is due to redirecting the activity of the E3 ligase DCAF15 to degrade the splicing factor RBM39] [DOI] [PubMed] [Google Scholar]
- 44.Uehara T, Minoshima Y, Sagane K, Sugi NH, Mitsuhashi KO, Yamamoto N, Kamiyama H, Takahashi K, Kotake Y, Uesugi M, et al. Selective degradation of splicing factor CAPERa by anticancer sulfonamides. Nat Chem Biol. 2017;13:675–680. doi: 10.1038/nchembio.2363. PubMed - NCBI. [DOI] [PubMed] [Google Scholar]
- 45.Bussiere DE, Xie L, Srinivas H, Shu W, Burke A, Be C, Zhao J, Godbole A, King D, Karki RG, et al. The structural basis of Indisulam-mediated recruitment of RBM39 to the DCAF15-DDB1-DDA1 E3 ligase complex. bioRxiv. 2019;44:737510 [Google Scholar]
- 46.Faust TB, Yoon H, Nowak RP, Donovan KA, Li Z, Cai Q, Eleuteri NA, Zhang T, Gray NS, Fischer ES. Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15. Nat Chem Biol. 2019;355:1–8. doi: 10.1038/s41589-019-0378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du X, Volkov OA, Czerwinski RM, Tan H, Huerta C, Morton ER, Rizzi JP, Wehn PM, Xu R, Nijhawan D, et al. Structural basis and kinetic pathway of RBM39 recruitment to DCAF15 by a sulfonamide molecular glue E7820. Structure. 2019;27:1625–1633.:e3. doi: 10.1016/j.str.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 48.Nusse R, Clevers H. Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 49.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 50.Simonetta KR, Taygerly J, Boyle K, Basham SE, Padovani C, Lou Y, Cummins TJ, Yung SL, Soly von SK, Kayser F, et al. Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction. Nat Commun. 2019;10:1–12. doi: 10.1038/s41467-019-09358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mayor-Ruiz C, Winter GE. Identification and characterization of cancer vulnerabilities via targeted protein degradation. Drug Discov Today Technol. 2019;31:81–90. doi: 10.1016/j.ddtec.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, Riley-Gillis B, Vijay P, Shen Y. Acquired resistance to BET-PROTACs (Proteolysis-Targeting chimeras) caused by genomic alterations in core components of E3 ligase complexes. Mol Cancer Ther. 2019;18:1302–1311. doi: 10.1158/1535-7163.MCT-18-1129. [DOI] [PubMed] [Google Scholar]
- 53.Ottis P, Palladino C, Thienger P, Britschgi A, Heichinger C, Berrera M, Julien-Laferriere A, Roudnicky F, Kam-Thong T, Bischoff JR, et al. Cellular resistance mechanisms to targeted protein degradation converge toward impairment of the engaged ubiquitin transfer pathway. ACS Chem Biol. 2019 doi: 10.1021/acschembio.9b00525. [DOI] [PubMed] [Google Scholar]
- 54.Kortüm KM, Mai EK, Hanafiah NH, Shi C-X, Zhu Y-X, Bruins L, Barrio S, Jedlowski P, Merz M, Xu J, et al. Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes. Blood. 2016;128:1226–1233. doi: 10.1182/blood-2016-02-698092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thakurta A, Gandhi AK, Waldman MF, Bjorklund C, Ning Y, Mendy D, Schafer P, Lopez-Girona A, Lentzsch S, Schey SA, et al. Absence of mutations in cereblon (CRBN) and DNA damage-binding protein 1 (DDB1) genes and significance for IMiD therapy. Leukemia. 2014;28:1129–1131. doi: 10.1038/leu.2013.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gandhi AK, Kang J, Havens CG, Conklin T, Ning Y, Wu L, Ito T, Ando H, Waldman MF, Thakurta A, et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.) Br J Haematol. 2014;164:811–821. doi: 10.1111/bjh.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hagner PR, Chiu H, Ortiz M, Apollonio B, Wang M, Couto S, Waldman MF, Flynt E, Ramsay AG, Trotter M, et al. Activity of lenalidomide in mantle cell lymphoma can be explained by NK cell-mediated cytotoxicity. Br J Haematol. 2017;179:399–409. doi: 10.1111/bjh.14866. [DOI] [PubMed] [Google Scholar]