Abstract
Genetically encoded indicators engineered from G protein-coupled receptors (GPCRs) are important tools that enable high-resolution in vivo neuromodulator imaging. Here we introduce a family of sensitive multicolor norepinephrine indicators, which includes nLightG (green) and nLightR (red). These tools report endogenous norepinephrine (NE) release in vitro, ex vivo and in vivo with improved sensitivity, ligand selectivity and kinetics, as well as a distinct pharmacological profile compared to previous state-of-the-art GRABNE indicators. Using in vivo multi-site fiber photometry recordings of nLightG, we could simultaneously monitor optogenetically evoked NE release in the mouse locus coeruleus and hippocampus. Two-photon imaging of nLightG revealed locomotion and reward-related NE transients in the dorsal CA1 area of the hippocampus. Thus, the sensitive NE indicators introduced here represent an important addition to the current repertoire of indicators and provide the means for a thorough investigation of the norepinephrine system.
Introduction
The development of genetically-encoded fluorescent indicators based on engineering of circularly-permuted green fluorescent protein (cpGFP) into G protein-coupled receptors (GPCRs), hereafter called GPCR-based indicators, permits the in vivo optical detection of neuromodulators with high spatial and/or temporal resolution1,2. These indicators found widespread use by complementing previously-available techniques for monitoring neuromodulators in vivo (microdialysis, fast-scan cyclic voltammetry, CniFERs, iTango)1,2. The recent addition of red indicators to this class of tools further expanded potential applications, for example by enabling their combination with blue light-excited optogenetic tools or multiplexed imaging of neurotransmitters3,4. However, red indicators are so far only available for detecting dopamine.
Norepinephrine (NE) is a central neuromodulator that carries out important functions in the brain, including the regulation of wakefulness, alertness and the response to stress, among many others5,6. Despite the growing need for tools to directly probe NE in vivo their availability is currently limited, partly due to the labor-intensive screening efforts required for their development7. In fact, a large number of GPCR-based indicator variants (typically hundreds to thousands) needs to be screened in mammalian cells in order to identify candidate protein variants with suitable properties for in vivo applications 8,9,4,7. Current state-of-the-art NE indicators (GRABNE) enable optical detection of NE in vivo, but are solely based on an Alpha-2a adrenergic receptor (Alpha-2 AR) scaffold7. A diversification of the currently available toolkit would thus be highly desirable.
Here we set out to develop next-generation NE indicators by using a previously-unexplored receptor subtype. Adopting an Alpha-1a adrenergic receptor (Alpha-1 AR) as scaffold we engineered nLightG and nLightR, two highly sensitive green and red fluorescent norepinephrine (NE) indicators, respectively. We thoroughly benchmarked these tools in multiple experimental settings and demonstrated that they exhibit a distinct pharmacological profile, as well as improved sensitivity, ligand selectivity and kinetics compared to a state-of-the-art GRABNE indicator. Using these sensors we could accurately detect endogenous NE release both ex vivo and in vivo with a variety of techniques (widefield imaging, multi-site fiber photometry and two-photon microscopy). Our family of NE indicators offers ready-to-use tools for a more complete investigation of NE physiology, and has thus great potential for life science applications.
Results
Development of multicolor NE indicators
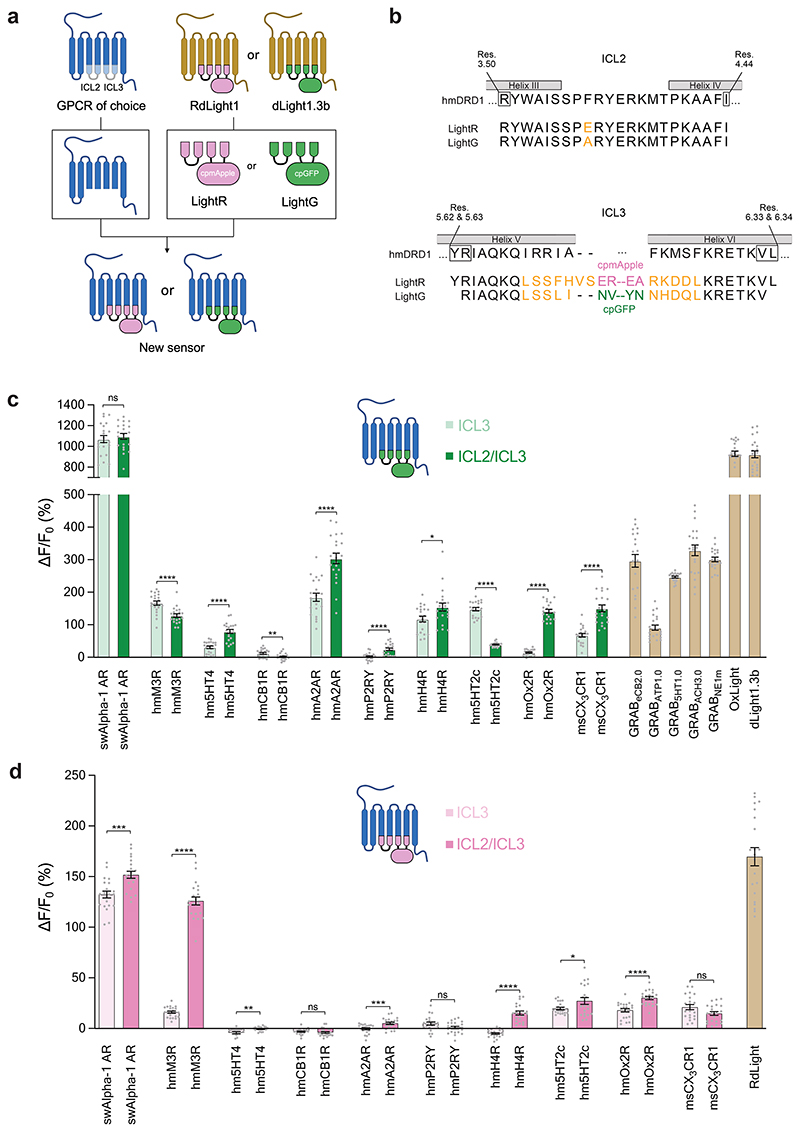
Despite the large sequence diversity of GPCRs, their structures and activation mechanisms are highly conserved10, and their seven transmembrane helices typically align well in space11. Thus, to expand the family of norepinephrine indicators we explored a grafting-based approach. Here, domains from the previously optimized green and red fluorescent dopamine (DA) indicators dLight1.3b8 and RdLight13, comprising the circularly-permuted fluorescent protein as well as parts of transmembrane helices V and VI were directly grafted onto target Alpha-1 ARs of choice.
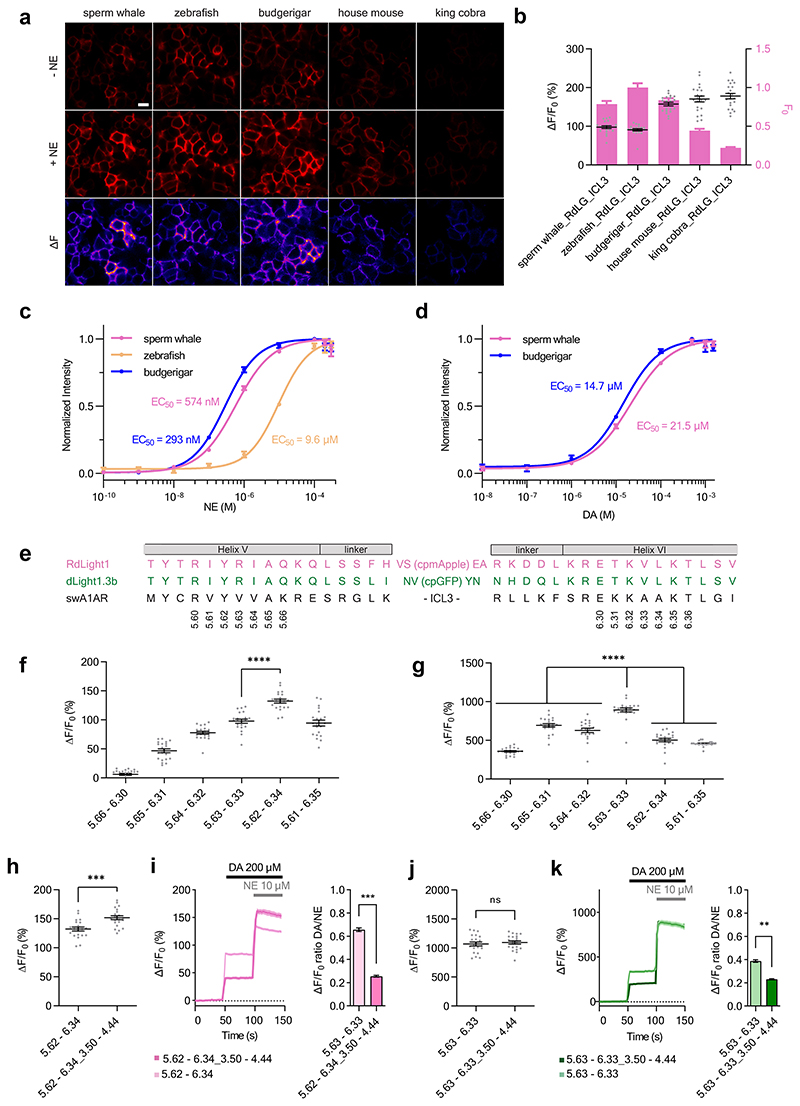
Given that all currently available NE indicators are green, we first set out to test our strategy by attempting the development of a red fluorescent NE indicator. We started by generating five indicator prototypes based on Alpha-1 ARs from different species (house mouse, budgerigar, king cobra, zebrafish and sperm whale). The fluorescent protein module was inserted according to a sequence alignment based on the Ballesteros-Weinstein (BW) numbering scheme12. Of note, structural alignment of the sperm whale Alpha-1 AR with the human DRD1 (Supplementary Figure 1) also indicated structural conservation at the insertion positions chosen based on BW numbering. All five prototype indicators were detectable on the membrane of transiently transfected HEK293T cells and gave a positive fluorescence response (ΔF/F0) in the range of 100% to 180% upon addition of 10 μM NE (Extended Data Fig. 1a-b). The indicators based on the sperm whale, budgerigar and zebrafish Alpha-1 ARs showed the highest basal brightness and were selected for the determination of the EC50 of NE and dopamine (DA). Out of these three indicators, the sperm whale-based indicator was selected for further development as it showed both high sensitivity and selectivity for NE (EC50 (NE) = 574 nM, EC50 (DA) = 21.5 μM) (Extended Data Fig. 1c-d). We then tested whether the same approach could work for the development of green fluorescent indicators by using an equivalent fluorescent protein module from dLight1.3b. The resulting NE indicator showed a ΔF/F0 response of approximately 1000%. To check whether the selected BW registries of the two modules were optimal, we systematically varied them in a parallel fashion. While the first prototype of the green fluorescent NE indicator showed the largest dynamic range, we identified an improved grafting registry for the red fluorescent NE indicator (Extended Data Fig. 1e-g).
In previous work, we discovered that mutations in the intracellular loop 2 (ICL2) can increase the fluorescence response of GPCR-based indicators3,8,9. Given the close proximity of the ICL2 region to transmembrane helices V and VI (Supplementary Fig. 1), we reasoned that also ICL2 grafting should be tested in the modular approach for engineering GPCR-based indicators. We grafted a module containing parts of helices III and IV and the entire ICL2 from dLight1.3b or RdLight1 onto the corresponding NE indicator prototypes. This led to an improvement in the fluorescent response for the red indicator, but not for the green indicator. Importantly, grafting the ICL2 significantly improved the selectivity for NE versus DA in both indicators (Extended Data Fig. 1h-k).
In vitro characterization of nLightG and nLightR
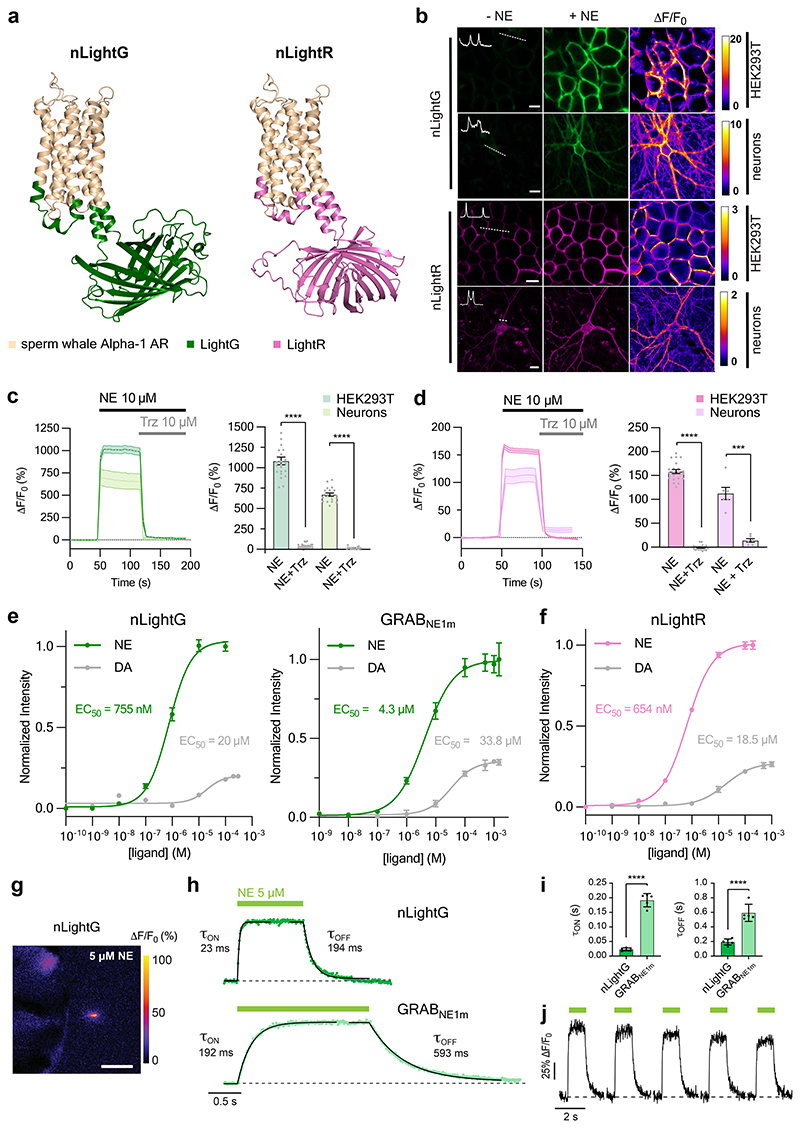
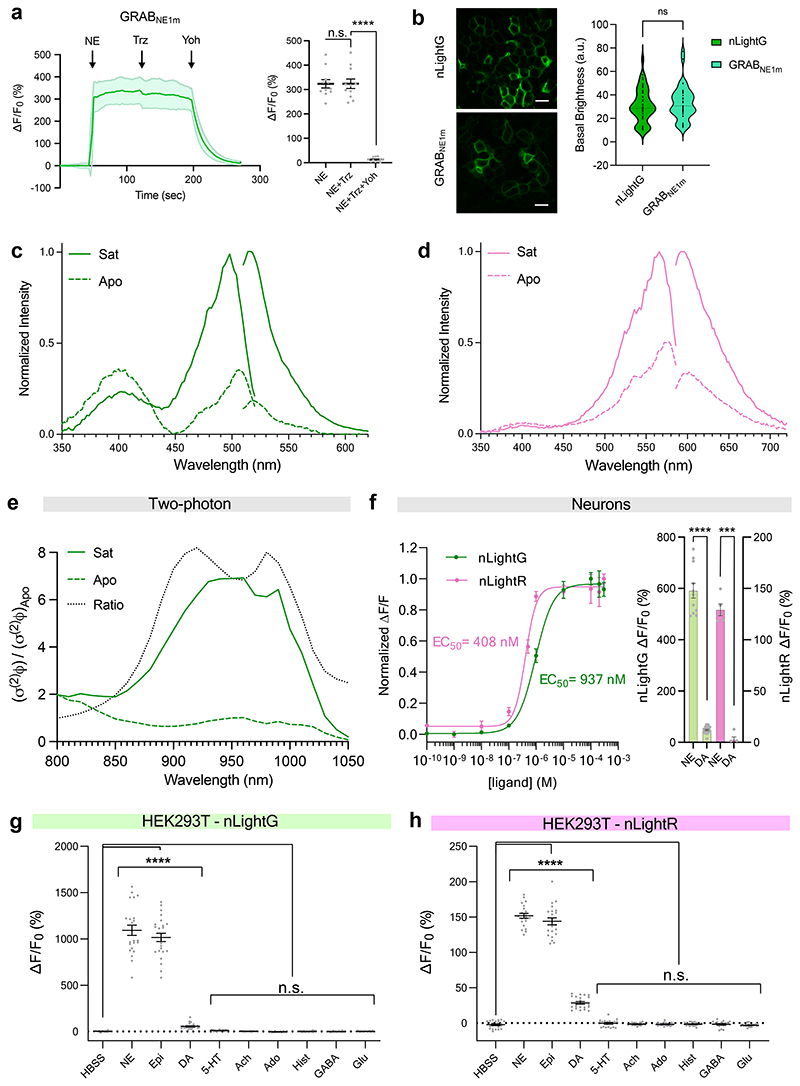
We next focused on characterizing the in vitro properties of our green and red fluorescent NE indicators (named nLightG and nLightR, respectively) (Fig. 1a). Both indicators localized well to the cell membrane and exhibited a similarly large fluorescence response upon stimulation with 10 μM NE in HEK293T cells and rat cortical neurons (ΔF/F0 HEK293T = 1083 ± 47%, ΔF/F0 neurons = 671 ± 21% for nLightG; ΔF/F0 HEK293T = 158 ± 4%, ΔF/F0 neurons = 113 ± 13% for nLightR; Fig. 1b-d). The activation of both indicators was fully reversed upon application of a small molecule Alpha-1 AR antagonist (trazodone, Fig. 1c,d), in contrast to the GRABNE1m indicator which was not affected by trazodone but was instead fully inactivated by the Alpha-2 AR antagonist yohimbine (Extended Data Fig. 2a), highlighting the distinct pharmacological profiles of the two indicator classes. Furthermore, a direct comparison between nLightG and GRABNE1m in HEK293T cells revealed that the indicators have similar basal brightness (Extended Data Fig. 2b). Spectral characterization of the indicators in HEK293T cells revealed peak one-photon excitations (λex) at 498 nm and 566 nm and peak emissions (λem) at 516 nm and 594 nm for nLightG and nLightR, respectively (Extended Data Fig. 2b-c). nLightG was also excitable through two-photon illumination, which showed peak performance at 920 nm (Extended Data Fig. 2d). A key feature of GPCR-based fluorescent indicators is their inherent, naturally-evolved molecular specificity. We tested a panel of eight neurotransmitters at a high concentration (10 μM) on nLightG and nLightR to check if the molecular specificity of the swAlpha-1 AR is conserved despite the changes introduced during the indicator-engineering process. As expected, only NE, epinephrine and (to a much lower extent) DA induced a significant fluorescence response (Extended Data Fig. 2e-g). The EC50 of NE was in a similar range for nLightG (EC50 HEK293T = 755 ± 85 nM, EC50 neurons = 937 ± 133 nM) and nLightR (EC50 HEK293T = 654 ± 47 nM, EC50 neurons = 408 ± 55 nM). In HEK293T cells the EC50 of DA was comparable between nLightG (EC50 = 20.0 ± 7.9 μM) and nLightR (EC50 = 18.5 ± 6.7 μM), and was approximately 30-fold higher than that of NE. Furthermore the maximal response to DA was only a small fraction of that induced by NE (23.0% for the nLightG and 25.6% for nLightR), indicating that DA only acts as a partial agonist at both indicators (Figure 1e,f). Similar measurements performed on GRABNE1m revealed an EC50 for DA which is only 8-fold higher than that for NE and that its maximal response to DA is 35% of the response to NE (Figure 1e). These results indicate that both nLightG and nLightR have higher selectivity for NE over DA compared to GRABNE1m. Furthermore, in cultured neurons the maximal response of both nLightG and nLightR to DA was further reduced and amounted to less than 10% of that induced by NE (Extended Data Fig. 2e). To establish the kinetic properties and reversibility of the fluorescence response for this family of indicators, we performed patch-clamp fluorometry on outside-out membrane patches from nLightG-transfected HEK293T cells (Fig. 1g, Supplementary Video 1). Using a piezo-controlled system for rapid solution switching between 0 and 5 μM NE in less than 1 ms13, we found that nLightG exhibited very rapid activation and deactivation kinetics (τon = 23 ± 5 ms and τoff = 194 ± 42 ms; mean ± SD, 6 patches, Fig. 1h). Similar experiments performed on membrane patches containing GRABNE1m revealed that this indicator has 8-fold slower activation kinetics and 3-fold slower deactivation kinetics (τon = 192 ± 23 ms and τoff = 593 ± 118 ms; mean ± SD, 5 patches) compared to nLightG (Fig. 1h-i). These experiments also confirmed the ability of nLightG to faithfully report multiple bidirectional changes in extracellular [NE] in rapid succession (Fig. 1j).
Figure 1. In vitro properties of nLightG and nLightR.
a, Structural models of nLightG (left) and nLightR (right) generated using ColabFold39. b, Representative images of HEK293T cells and neurons expressing nLightG or nLightR before/after application of NE (10 μM) and corresponding pixel-wise ΔF/F0 heatmaps. White insets indicate surface expression of the indicators. Scale bars, 10 μm (HEK293T), 20 μm (neurons). c, Left, timelapse of fluorescence of response (ΔF/F0) of nLightG in HEK293T (dark green) or neurons (light green) upon application of NE (10 μM) followed by application of trazodone (Trz, 10 μM). Right, quantification of maximal ΔF/F0 responses from left. Two-tailed Students t-test with Welch’s correction. n = 3 independent experiments with n=22 cells (HEK293T), n=18 cells (neurons). P = 2.979×10-16 (HEK293T), P = 8.558×10-17 (neurons). d, Same as in c for nLightR. n = 3 independent experiments with n = 21 cells (HEK293T), n=6 cells (neurons). P = 5.922×10-32 (HEK293T), P = 0.004 (neurons). e, Fluorescence dose-response curves of nLightG (left) and GRABNE1m (right) for NE and DA in HEK293T cells normalized to the maximum ΔF/F0 for NE for each indicator. Datapoints were fitted with four-parameter dose-response curves to determine EC50 values. n=6,5,3 wells for nLightG with NE, nLightG with DA, and GRABNE with NE/DA, respectively. f, Same as in e for nLightR with n=3 wells. All data are shown as mean ± SEM. g, Representative heatmap of nLightG fluorescence response (ΔF/F0 %) in an outside-out membrane patch from nLightG-expressing HEK293T cells (n = 6 independent experiments). Scale bar, 50 μm. h, ON and OFF kinetics of nLightG (top) and GRABNE1m (bottom) after ultrafast (<0.5 ms) switching of the perfusion pipette. Dots represent the average of 5 applications. Kinetic parameters were obtained using single-exponential fits on the average of all trials. i, Statistical comparison of kinetic parameters. Unpaired two-tailed Student’s t test. n=6 and 5 patches for nLightG and GRABNE1m, respectively. P = 3.0 ×10-8 (τON), P = 2.7 ×10-5 (τOFF). Mean ± standard deviation are shown. j, Five consecutive NE applications on nLightG (5 μM, 1 s). Images were acquired at 100 Hz. All experiments were repeated at least three times with similar results.
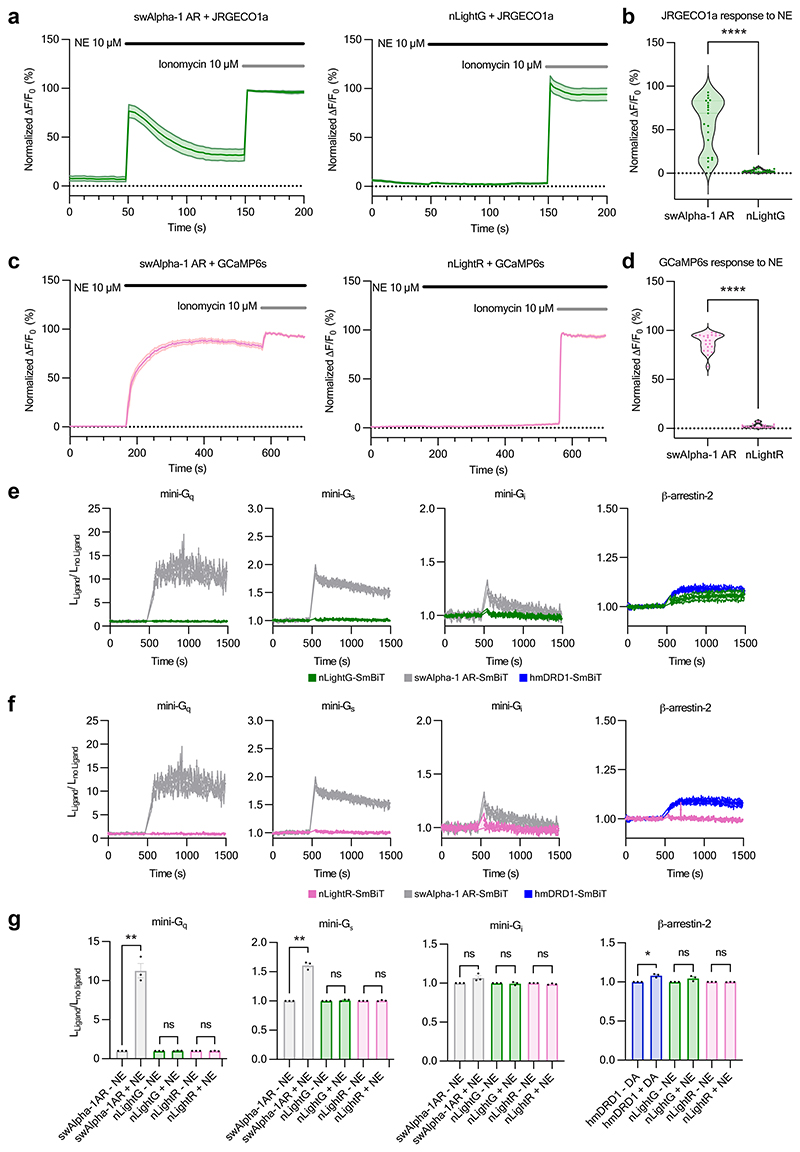
Intracellular signaling is the primary function of GPCRs, but should be avoided in GPCR-based fluorescent indicators. Using genetically-encoded calcium indicators14, we detected a significant increase of intracellular Ca2+ concentration in response to the activation of wild-type swAlpha-1 AR with NE, confirming the coupling to Gαq pathway expected for this adrenergic receptor subtype15. In the same experimental setup, activation of nLightG and nLightR did not induce a significant increase in the intracellular Ca2+ concentration, indicating that coupling to Gαq is disrupted (Extended Data Fig. 3a-d). To further investigate the downstream coupling capabilities of nLightG, nLightR, and swAlpha-1 AR, we characterized their direct interactions with mini-Gq, mini-Gs and mini-Gi16 upon agonist stimulation using a Nanoluciferase (NanoBiT)-based complementation assay17 (Extended Data Fig. 3e-g). Stimulation of the swAlpha-1 AR with NE showed significant coupling to mini-Gq and mini-Gs, but no significant coupling to mini-Gi. In contrast, stimulation of nLightG and nLightR with NE did not lead to a significant coupling to mini-Gq, mini-Gs or mini-Gi. Next, we investigated whether indicator activation induces recruitment of β-arrestin-2 using the same NanoBiT-based approach. Since the Alpha-1 AR subtype has not been consistently shown to couple to β-arrestins in previous studies18–20, we used the human DRD1 as positive control. Stimulation with DA induced significant recruitment of β-arrestin-2 to DRD1, while no recruitment was observed for nLightG or nLightR upon stimulation with NE (Extended Data Fig. 3e-g). In summary, the results presented here indicate that nLightG and nLightR are highly selective and sensitive NE indicators with minimal potential for interference with endogenous signaling pathways.
Ex vivo and in vivo benchmarking of nLightG and nLightR
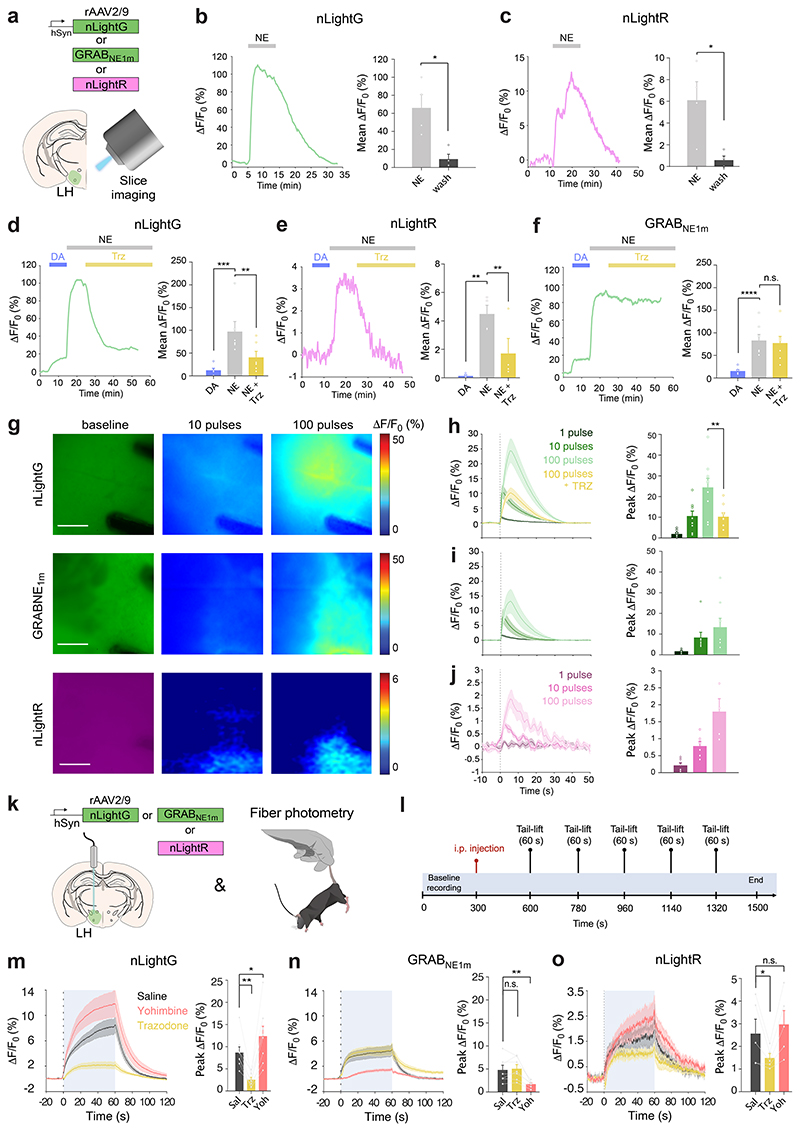
We then characterized the properties of nLightG and nLightR in brain slices and benchmarked them against those of the state-of-the-art NE indicator GRABNE1m. To this end, adeno-associated viruses (rAAVs) carrying either nLightG, nLightR or GRABNE1m driven by the human synapsin promoter were injected in the mouse Lateral Hypothalamus (LH) (rAAV2/9.hSynapsin1.nLightG/nLightR/GRABNE1m). After at least 4 weeks of expression, we prepared acute brain slices and imaged them under epifluorescence illumination (Fig. 2a). Perfusion of a high concentration of NE (50 μM) on the slices led to clearly detectable fluorescence responses from both nLightG and nLightR, which were reversible upon NE wash-out (Fig. 2b-c). Next we compared catecholamine-selectivity and response to an Alpha-1 AR-antagonist among all three NE indicators. Perfusion of a high concentration of DA (50 μM) elicited small, but detectable fluorescence responses from nLightG and nLightR, which amounted to about 12% and 3% of the responses to an identical concentration of NE, respectively (Fig. 2d-f). In comparison, the response to the same concentration of DA for GRABNE1m was larger and corresponded to about 19% of the response to NE. Furthermore, application of the Alpha-1 AR-antagonist trazodone (10 μM) largely reversed the NE-induced response from nLightG and nLightR while it had no significant effect on GRABNE1m, confirming that the two indicator families maintain distinct pharmacological profiles in brain tissue (Fig. 2d-f). Next, we investigated whether both nLightG and nLightR could be sensitive enough to detect endogenous NE release evoked by electrical pulses, as compared to GRABNE1m. We applied an increasing number of pulses to the slices (1, 10, 100 pulses) and quantified evoked fluorescence responses (Fig. 2g). Clear stimulus-evoked responses could be detected for both nLightG and GRABNE1m after application of a single pulse, while increasing the number of pulses was mirrored by a parallel increase in the responses (Fig. 2h-i). Fluorescence responses could also be clearly detected from nLightR-expressing slices upon application of 10 or 100 pulses, but not to a single pulse (Fig. 2j), indicating that also the red NE indicator can detect endogenous NE release.
Figure 2. Ex vivo and in vivo benchmarking of nLightG and nLightR.
a, Experiment schematics. b, Left, representative nLightG response to perfusion of NE (50 μM) and wash-out. Right, quantification of nLightG responses (two-sided paired t-test, P=0.0308, n=4 slices from 3 mice). c, Same as b, for nLightR (two-sided paired t-test, P = 0.0434, n=4 slices from 2 mice). d, Left, representative nLightG response to subsequent perfusions of DA (50 μM), NE (50 μM) and trazodone (Trz, 10 μM). Right, quantification of nLightG responses (repeated measures one-way ANOVA, P=0.0006 and Tukey’s multiple comparison test, P=0.0005 for DA against NE and P=0.0087 for NE against NE+Trz , n=6 slices from 2 mice). e, Same as d, for nLightR (repeated measures one-way ANOVA, P=0.0040 and Tukey’s multiple comparison test, P=0.0034 for DA against NE and P=0.0277 for NE against NE+Trz, n=4 slices from 2 mice). f, Same as e for GRABNE1m. (repeated measures oneway ANOVA, P=4.572 x 10-5 and Tukey’s multiple comparison test, P=8.501 x 10-5 for DA against NE, n=7 slices from 3 mice). g, Representative images of indicator expression and fluorescence responses. Scale bars, 100 μm. h, Left, average traces of nLightG responses to electrical stimulation using increasing numbers of pulses (1, 10, 100 pulses). Right, quantification of peak responses (two-tailed paired t-test, P=0.0018, n=9 slices from 2 mice). i, Same as in h for GRABNE1m. n=8 slices from 3 mice. j, Same as in i for nLightR. n=6 slices from 2 mice. k-l, Experiment schematics. m, Left, average traces of fluorescence response from nLightG-expressing animals after injection of different drugs (saline, Sal; trazodone, Trz; yohimbine, Yoh). Right, quantification of peak ΔF/F0 responses from left. Peak values were compared to the control (Sal). P=0.0013 for Trz, P=0.0345 for Yoh, n=8 mice. n, Same as m, for GRABNE1m. P=0.9010 for Trz, P=0.0015 for Yoh, n=7 mice. o, Same as m, for nLightR. P=0.0329 for Trz, P=0.457 for Yoh, n=5 mice. m-o, Analyses were performed with repeated measures one-way ANOVA with Dunnett’s multiple comparisons test. b-o, All data are mean ± S.E.M. All experiments were repeated at least three times with similar results.
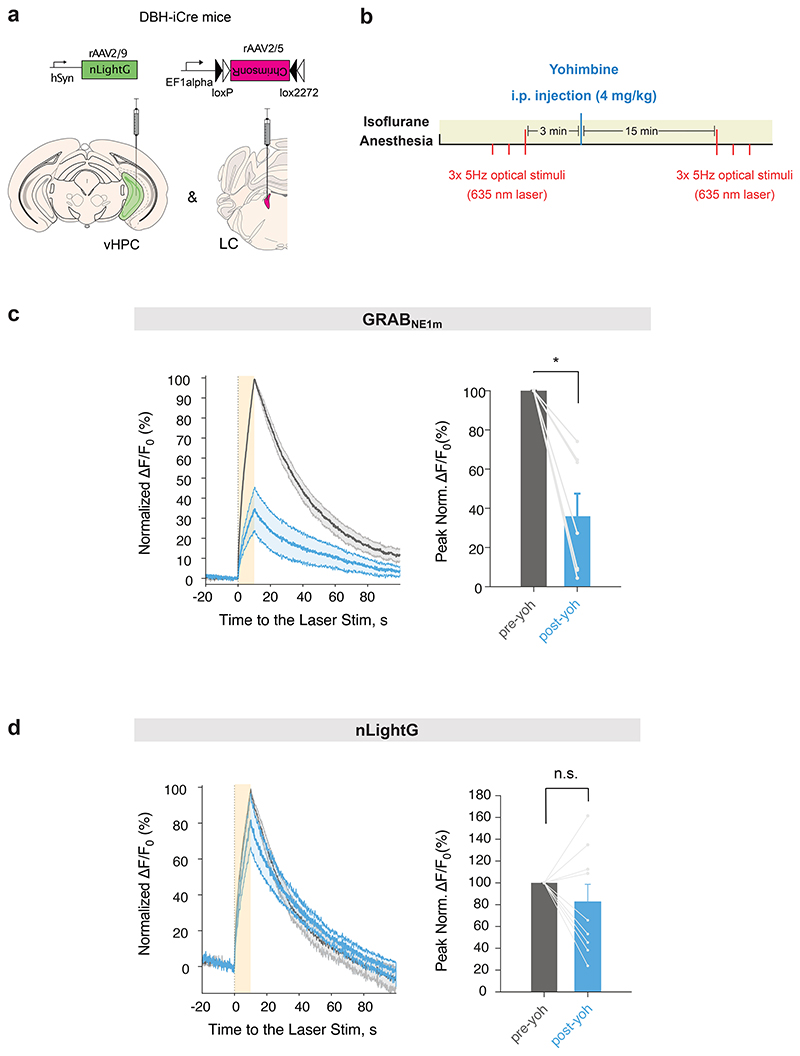
Alpha-2 ARs are densely expressed pre-synaptically and on the somata of locus coeruleus NE-producing neurons21,22, where they provide feedback inhibition in response to NE release. Accordingly, Alpha-2 AR antagonists, such as yohimbine, are known to increase natural LC excitability in response to external stimuli23, resulting in increased NE release24. Due to their intrinsic sensitivity to Alpha-2 AR-targeting drugs, Alpha-2 AR-based NE indicators (i.e. GRABNE1 family7) cannot be utilized to faithfully monitor the effects of these drugs on endogenous NE release. To determine whether nLightG and nLightR could overcome this limitation due to their orthogonal receptor pharmacology, we performed a side-by-side comparison in vivo with GRABNE1m using fiber photometry during systemic administration of subtype-selective adrenergic receptor antagonists. To reliably monitor NE release, we adopted a previously established tail-lifting assay7 (Fig. 2k). Mice received injections of saline, trazodone (1 mg/kg) or yohimbine (4 mg/kg) in separate trials followed by 5 consecutive tail-lifting events (Fig. 2l). Tail-lifting of the animals caused a stable and reproducible increase in nLightG signal (peak ΔF/F0 = 8.7%), which lasted for the entire duration of the event (Fig. 2m). Similar signals, although with smaller amplitudes, were detected in GRABNE1m (peak ΔF/F0 = 4.8%) and nLightR expressing animals (peak ΔF/F0 = 2.6%) (Fig. 2n-o). Administration of trazodone significantly reduced the tail-lift-induced response of nLightG and nLightR, while it had no effect on GRABNE1m signals. Conversely, yohimbine administration increased tail-lift induced responses from nLightG and nLightR, in line with the known disinhibitory effect of this drug on the LC system. In the case of GRABNE1m, yohimbine administration strongly attenuated the indicator response, an effect that had been previously reported7 (Fig. 2m-o). Finally, to determine whether the NE release-promoting effect was mediated by yohimbine via the Alpha-2 AR-feedback-loop, hence requiring conscious LC modulation by external stimuli, we also tested the effect of this drug on optogenetically-driven NE release in the ventral hippocampus (vHPC) during anesthesia (Extended Data Fig. 4a-b). Under these conditions, yohimbine significantly attenuated optogenetically-evoked NE signals monitored via GRABNE1m, while it did not have a significant effect on the NE signals reported by nLightG (Extended Data Fig. 4c-d). Overall, these results confirm that nLightG and nLightR have a pharmacological profile distinct to the GRABNE1m family indicators, which offers a valuable option to monitor the effects of Alpha-2 AR-targeting drugs on the endogenous NE system in vivo.
Optogenetic dissection of NE release across brain areas
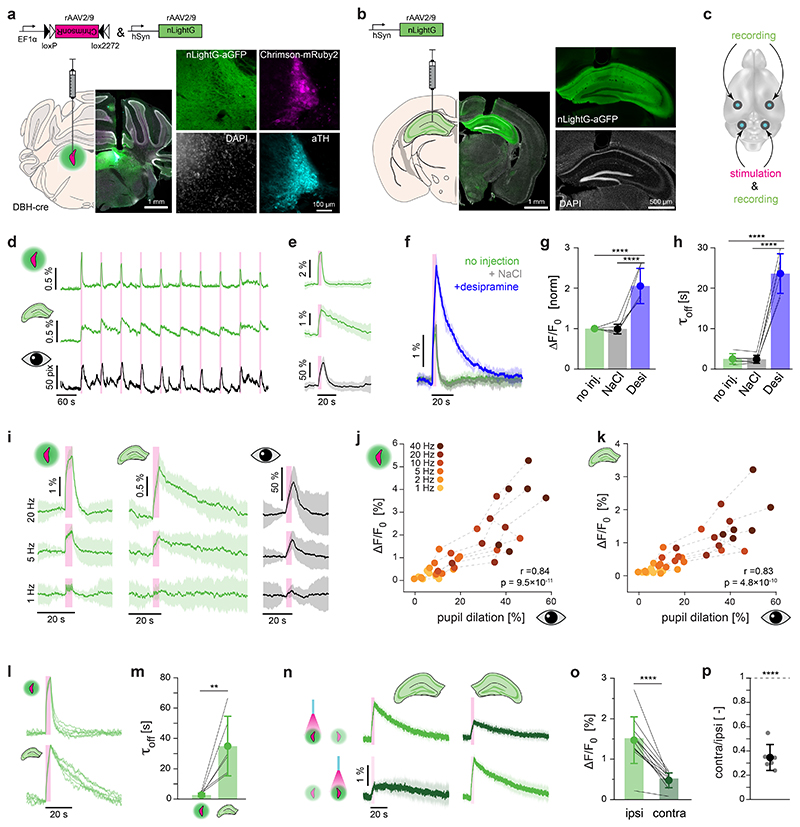
We next asked whether we could use the most sensitive of our newly developed NE indicators (nLightG) for detecting optogenetically-evoked NE release simultaneously in two brain areas with fiber photometry (Extended Data Fig 5). To do so, we bilaterally injected rAAV2/9.hSynapsin-1.nLightG in the dorsal hippocampus (dHPC) and rAAV2/9.hSynapsin-1.DIO.ChrimsonR-mRuby2 together with rAAV2/9.hSynapsin-1.nLightG in the locus coeruleus (LC) of DBH-cre mice (Fig. 3a-c). Optogenetic stimulation of the LC using 595 nm light (4 s pulse train, 20 ms pulses at 40 Hz, 10 mW at fiber tip), led to clearly detectable NE signals that simultaneously occurred in both LC and dHPC. As a second readout of LC activation, we measured pupil diameter 42 which, as expected, rapidly increased upon each optical stimulus (Fig. 3d,e). To establish the contribution of NE reuptake mechanisms on nLightG signals in the LC, we repeated the recordings after i.p. injections of saline or the norepinephrine transporter (NET) blocker desipramine. Saline injection did not alter the nLightG response profile, while desipramine led to a significant increase in both the magnitude and decay kinetics of the response (Fig. 3f-h), confirming the dependence of the nLightG response on extracellular norepinephrine. We then investigated the effects of different stimulation frequencies of optogenetic LC activation on nLightG responses in LC and dHPC and on pupil dilation. In the LC, nLightG responses could be detected at stimulation frequencies as low as 1 Hz (Fig. 3i). Upon increasing the frequency of optical stimuli, we found a strong, positive linear correlation between nLightG responses and pupil dilation in both recorded brain areas (Fig. 3j,k). Interestingly, the decay kinetics of nLightG responses to the same frequency of stimulation (40 Hz) were significantly slower in the dHPC (τoff = 37.3 ± 21 s) compared to the LC (τoff = 2.8 ± 1.3 s) (Fig. 3l,m), presumably reflecting differences in the density or functionality of local NE reuptake mechanisms. Finally, we used nLightG to explore the lateralization of noradrenergic innervation of the dHPC25,26: we found that unilateral LC stimulation most efficiently drove NE release in the ipsilateral dHPC, and that ipsilateral optogenetically-evoked dHPC-nLightG signals were approximately 3-fold larger than the contralateral ones (Fig. 3n-p).
Figure 3. In vivo dual-site recording of optogenetically-evoked NE release using nLightG.
a-c, Experimental schematics and histological verification for targeting LC (a) and dHPC (b). d, Example traces of pupil diameter (bottom) and simultaneously recorded nLightG fluorescence in the dHPC (center) and LC (top) upon optogenetic LC stimulation (4-second-long pulse train with 20 ms pulses at 40 Hz; 595 nm; 10 mW at fiber tip; pink bars). e, Mean ± SD of the recording shown in d (n=20 trials). f, Mean ± SD of nLightG fluorescence from LC in response to optogenetic stimulation (n=15 trials, 1 mouse) performed after indicated treatments. g, Peak ΔF/F0 of baseline-normalized averaged responses (One-way ANOVA, P=3.17×10-6 and Tukey’s test, P=1×10-4/1×10-5 for no injection/NaCl vs. desipramine; n=6 mice) and h,τoff of averaged responses (One-way ANOVA, P=1.89×10-9 and Tukey’s test, P=9.8×10-9/9.4×10-9 for no injection/NaCl vs. desipramine; n=6 mice) shown in f. i, Mean ± SD of simultaneously recorded nLightG fluorescence in the LC (left), dHPC (center), as well as pupil diameter (right) in an exemplary mouse, in response to pulses presented at 1, 5, and 20 Hz (n=20 trials each). j-k, Peak ΔF/F0 of averaged nLightG fluorescence in the LC (j) and dHPC (k) as a function of pupil dilation upon optogenetic stimulation (Pearson’s correlation coefficient r=0.84/0.83, P=9.5×10-11/4.8×10-1° for LC/dHPC; n=6 mice). l, Normalized responses of simultaneously recorded nLightG in the LC (top) and dHPC (bottom), during optogenetic LC stimulation (n = 6 mice). m, τoff of averaged responses shown in l. P10=0.0025, two-sided two-sample t-test, n=6 mice. (n) Mean ± SD of simultaneously recorded nLightG fluorescence from hippocampi in both hemispheres (columns) upon stimulation of either the left (top) or the right (bottom) LC (n=15 trials each). o, Peak ΔF/F0 of each dHPC upon ipsilateral (left) or contralateral (right) LC stimulation.. Pn=1.97×10-7, two-sided one-sample t-test, n=12 dHPC from n=6 mice. p, Contralateral divided by ipsilateral average response per mouse; 34.6±11 %; P5=2.4×10-5, two-sided one-sample t-test against 1; n=6 mice. n, o and p recorded under isoflurane anesthesia, all others in awake mice. All data are shown as mean ± standard deviation.
Two-photon imaging of NE release in the dorsal hippocampus
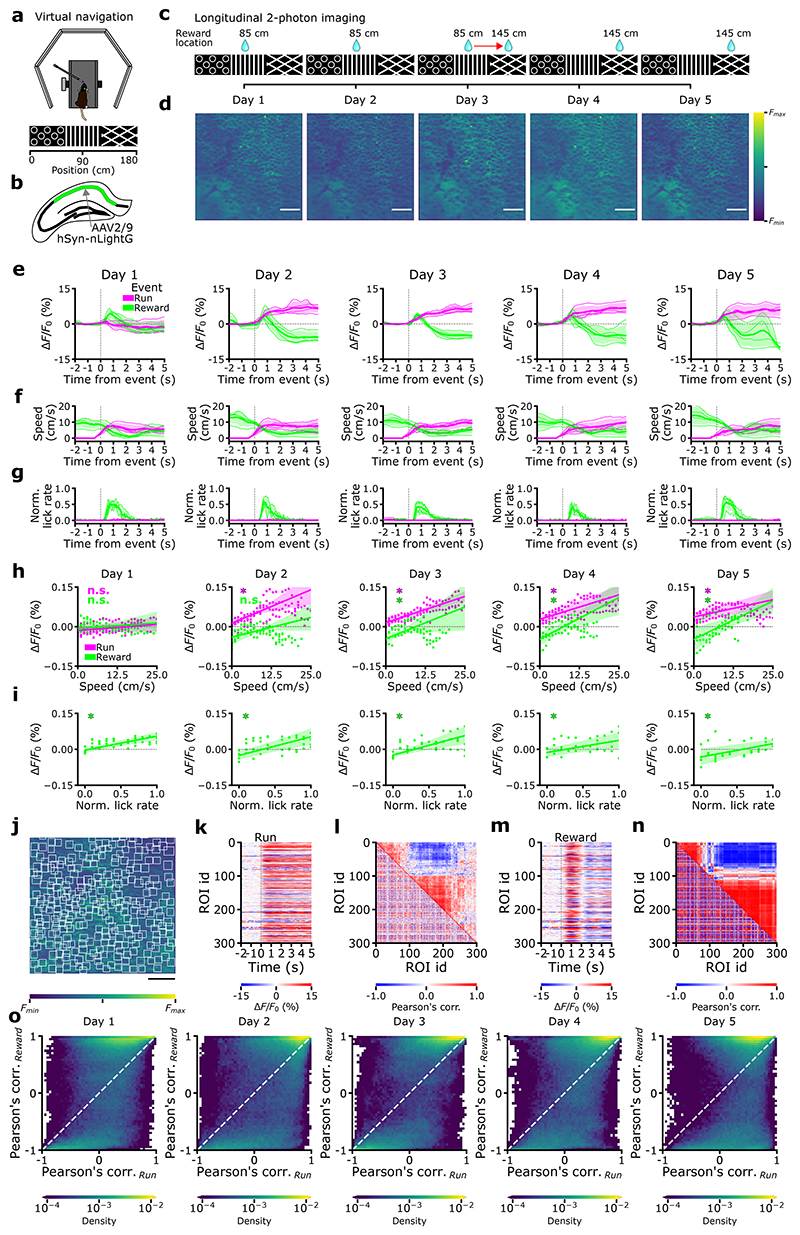
Next, we tested whether nLightG could be used in combination with two-photon excitation to image NE release in vivo with high spatial resolution. To this aim, we expressed nLightG in the CA1 region of the hippocampus and imaged nLightG signals through a chronic optical window using two-photon microscopy in awake head-fixed mice navigating in a virtual corridor (n=4 mice, Fig. 4a-b). Mice received a water reward positioned along the corridor and the same field-of-view (FOV) was imaged in five sessions on five consecutive days (one session for each imaging day, Fig. 4c-d).
Figure 4. Two-photon imaging of NE signals in awake behaving mice using nLightG.
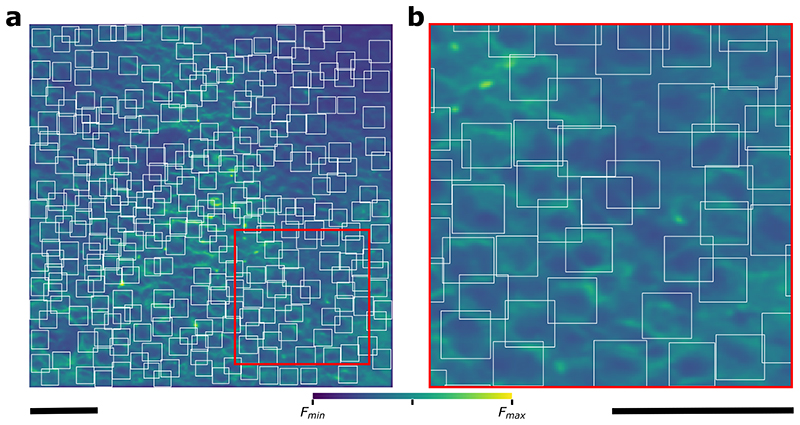
a-b, Experiment schematics. c, On day 1, 2, and 3, rewards were delivered at 85 cm from the start of the corridor. On day 3 at half of the recording session the reward delivery was repositioned to 145 cm, where it remained through days 4 and 5. d, Representative average time-projections of FOV, (images are scaled to their maximum, scale bar 50 μm). e, Event-triggered averages showing changes in nLightG signal amplitude over the whole field-of-view upon running (magenta) and reward position crossing (green) for the five days of recording. f-g, Event-triggered averages of animals’ speed (f) and lick rate (g) in the virtual corridor upon running initiation (magenta) and at reward position crossing (green). h-i, nLightG signal amplitude over the whole field-of-view as a function of running speed (h) and lick rate (i) for event-triggered averages upon running initiation (magenta) and reward position crossing (green, day 1-5). In h-i, * p < 0.05 Two-sided Rank-sum test, H0: slope of the linear model equals to 0 (for each test n=4 animals); n.s. not significant. In h p values are as follows, day 1: P(run)=2.48x10-1, P(reward)=2.48x10-1; day 2: P(run)=2.09x10-2, P(reward)=2.48x10-1; day 3: P(run)=2.09x10-2, p(reward)=2.09x10-2; day 4: P(run)=2.09x10-2, P(reward)=2.09x10-2; day 5: P(run)=2.09x10-2, P(reward)=2.09x10-2. In i, P value equals 2.09x10-2 for all sessions. In e-i, lines and shaded areas indicate (mean ± SD). j, Representative field-of-view showing nLightG-expressing hippocampal CA1 neurons. White boxes indicate regions-of-interest (ROIs) identified in the field-of-view using CITE-On29 (scale bar 50 μm). k, Event-triggered averages showing ΔF/F0 of nLightG signal when the mouse started running for all the ROIs identified in j. l, Lower-left triangle: cross-correlation matrix for all traces extracted from the ROIs displayed in j-k. Upper-right triangle: corresponding hierarchical clustering. m-n, Same as in k-l, but for event-triggered averages when the mouse crossed the reward position. o, Density map showing Pearson’s correlation value of nLightG signals from pairs of ROIs during reward position crossing as a function of that obtained during running. Data from 897,000 pairs from 6,000 ROIs in 4 mice over 5 imaging sessions.
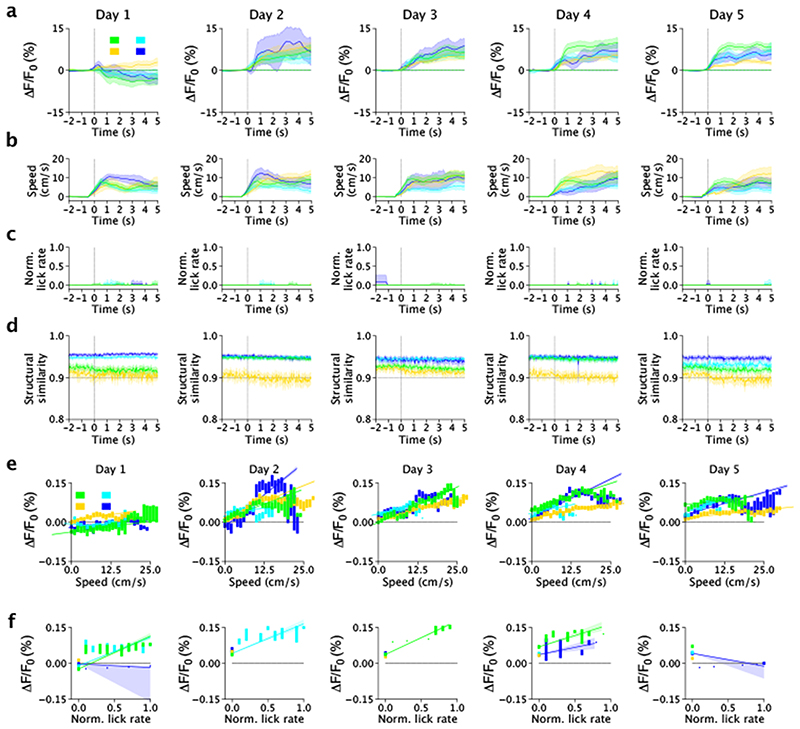
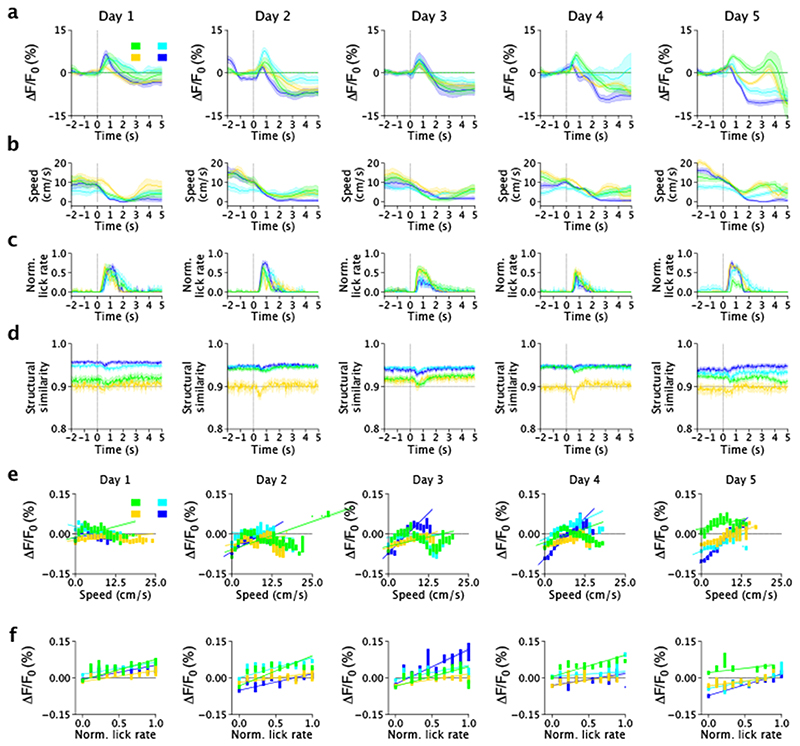
Since previous reports27,28 show that LC-projections activity is higher during locomotion and reward delivery, we sought to test the hypothesis that nLightG signals positively correlate with locomotion and reward. To this aim, we first computed event-triggered averages of nLightG signals over the whole field-of-view based on the time in which the mouse started to run (magenta color in Fig 4e-g, see Methods for details). While nLightG signal remained similar to baseline after the initial start of running in day 1 of recording, we found that after the mouse started to run nLightG signals displayed a clear increase with respect to baseline in all the following days (days 2-5, Fig. 4e, Extended Data Fig. 6a). Importantly, running speed dynamics were comparable across all five recording days. In day 2 to 5 (but not in day 1), the amplitude of nLightG signals positively correlated with the animal’s running speed (Fig. 4h; Extended Data Fig. 6b-c). Moreover, we investigated whether the nLightG signals that we recorded could be due to motion artifacts. We computed the structural similarity, a parameter evaluating the perceived change in structural information between individual frames with respect to the average projection of the whole imaging temporal series. We found structural similarity to be constant in the time window associated with the changes in nLightG signals described above (Extended Data Fig. 6d-f, see Methods for details). Moreover, we performed temporal shuffling of nLightG traces and found that the shuffling procedure destroyed the positive deflection of nLightG signals that we observed when traces were not shuffled (dashed horizontal line in Extended Data Fig. 6a).
We then built event-triggered averages of nLightG signals over the whole field-of-view based on the time in which the mouse crossed the reward position (green color in Fig. 4e-g, see Methods for details). We observed that nLightG signals showed a transient increase with respect to baseline followed by a decrease in all recording days (Fig. 4e, Extended Data Fig. 7a-c). In all five recording days, lick rate increased after the mouse crossed the reward position (Fig. 4g), while running speed decreased with respect to baseline after the mouse crossed the reward position (Fig. 4f). Importantly, the amplitude of nLightG signals increased with lick rate (Fig. 4i) across all recording days, and positively correlated with running speed across recording days 3 - 5 (Fig. 4h). The structural similarity in the time window associated with the changes in nLightG signals is shown in (Extended Data Fig. 7d-f, see Methods for details) and temporal shuffling of nLightG traces with respect to the timing in which the mouse crossed the reward position eliminated the changes of nLightG signals that we observed when traces were not shuffled (dashed horizontal line in Extended Data Fig. 7).
Finally, we segmented the field-of-view into anatomically identified regions-of-interest (ROIs) centered on putative cells using a recently developed machine learning algorithm29 (Figure 4j, Extended Data Fig. 8). We extracted event-triggered averages of nLightG signals for each identified ROIs based on the time at which the mouse started to run (Fig. 4k) and based on the time at which the mouse crossed the reward position (Fig. 4m). We computed the Pearson’s correlation value between nLightG signals of pairs of ROIs and built cross-correlation matrices (Fig. 4l,n). Across animals, we found that Pearson’s correlation values for the same pair of ROIs tended to differ when the mouse started to run (“Pearson’s corr.Run” in Fig. 4o) with respect to when it crossed the reward position (“Pearson’s corr.Reward” in Fig. 4o), suggesting that NE signaling in pairs of localized hippocampal regions is differentially correlated across behavioral conditions. Altogether, these results demonstrate that nLightG can be used to precisely image locomotion-induced and reward position-dependent NE dynamics in combination with two-photon microscopy in the hippocampus of awake head-fixed mice.
Rapid engineering of other GPCR-based indicators
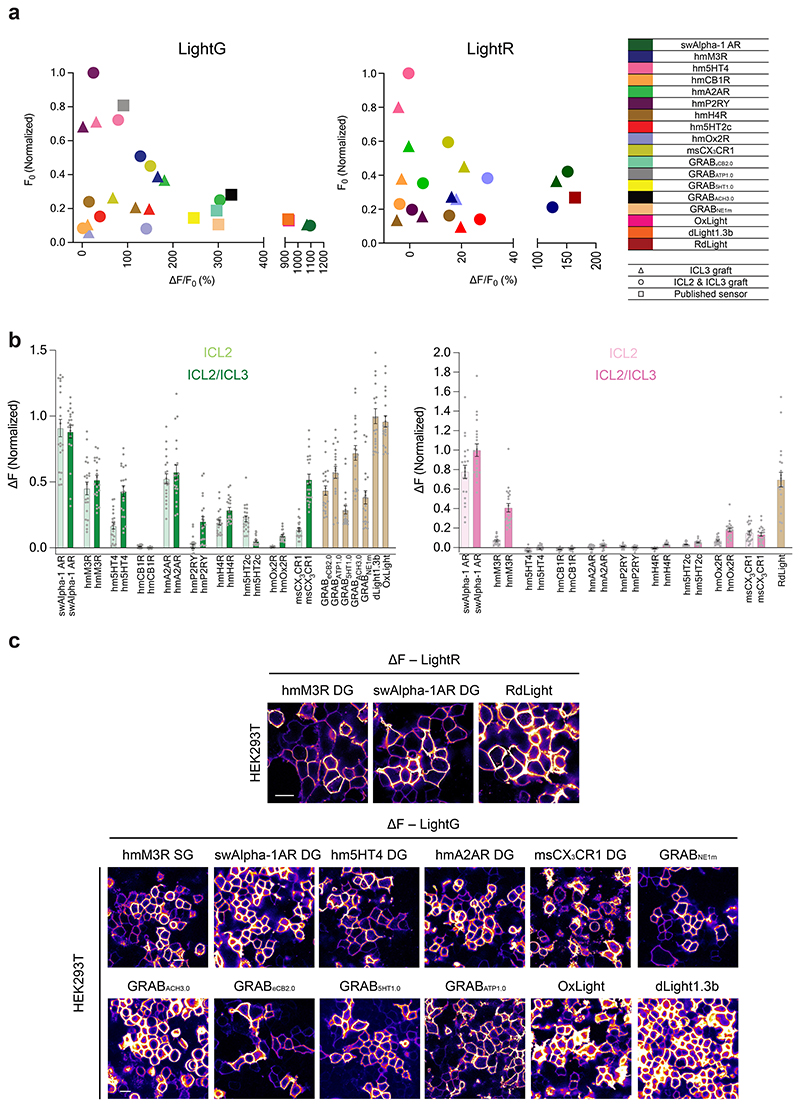
To test whether the two-step protein engineering strategy developed herein could be applied broadly, we tested it on 10 GPCRs in total. First, the region between BW residues 5.62/5.63 to 6.33/6.34 (ICL3, single-graft) was replaced, followed by the additional region between residues 3.50 and 4.44 (ICL2/ICL3, double-graft). Most of the 40 resulting indicators expressed well in HEK293T cells, with only a few exceptions. Eight out of ten GPCRs (80%) produced green indicators with a large response to ligand application (ΔF/F0 >50%) (Fig. 5c). On the other hand, two out of ten GPCRs (including Alpha-1 AR and the acetylcholine M3 receptor) produced red indicators with a large response to ligand application in the range (100 – 250%) of all previously in vivo-validated red fluorescent GPCR-based indicators3,4.
Figure 5. Rapid engineering of other multicolor GPCR-indicators.
a, Schematic illustration of a strategy for the two-step modular development of multicolor GPCR-based indicators using the LightR and LightG modules. b, Aminoacid sequence alignment of the LightG and LightR modules with portions of the human D1 receptor (hmDRD1). Individual residue differences between LightR and LightG are highlighted in orange. Helix-forming amino acids of hmDRD1 (according to the AlphaFold40 structural model of the receptor, Supplementary Figure 1) are indicated by grey boxes. The initial and terminal residues of the respective LightR and LightG building blocks (black frames) are numbered according to the Ballesteros-Weinstein numbering of hmDRD1. c, Quantification of the maximal ΔF/F0 response to bath-applied ligands (all tested at 10 μM concentration) of all generated green indicators. With the exception of msCX3CR1 DG (500 nM) and GRABATP1.0 (50 μM), for every experiment the ligand was added to a final concentration of 10 μM. The corresponding indicator-ligand pairs used were as follows: sperm whale Alpha-1 AR: swAlpha-1 AR, norepinephrine; human muscarinic M3 receptor: hmM3R, acetylcholine; human serotonin 5HT4 receptor: hm5HT4, serotonin; human adenosine 2A receptor: hmA2AR, adenosine; mouse CX3CR1 receptor: msCX3CR1, mouse fractalkine; GRABeCB2.0, 2-arachidonyl glycerol ether; GRABATP1.0, ATP; GrAB5HT1.0, 5HT; GRABACh3.0, acetylcholine; GRABNE1m, norepinephrine; OxLight1, Orexin-A; dLight1.3b, DA. Mean ΔF/F0 values of single-graft (only ICL3 replaced by grafting) and double-graft (ICL3 and ICL2 replaced by grafting) for each receptor were compared using a two-tailed Students t-test with Welch’s correction. P values (from left to right): 0.566; 7.342×10-5; 6.466×10-8; 4.143×10-3; 8.298×10-7; 1.355×10-5; 2.072×10-2; 3.629×10-16; 1.748×10-17; 8.324×10-8. d, Same as in c for red indicators. RdLight response was measured with 10 μM DA. P values (from left to right): 2.625×10-4; 2.910×10-19; 3.019×10-3; 0.477; 9.690×10-4; 5.954×10-2; 2.124×10-10; 4.593×10-2; 6.331×10-6; 6.032×10-2. c-d, n = 21 cells from 3 independent experiments for each indicator. Data are shown as mean ± S.E.M.
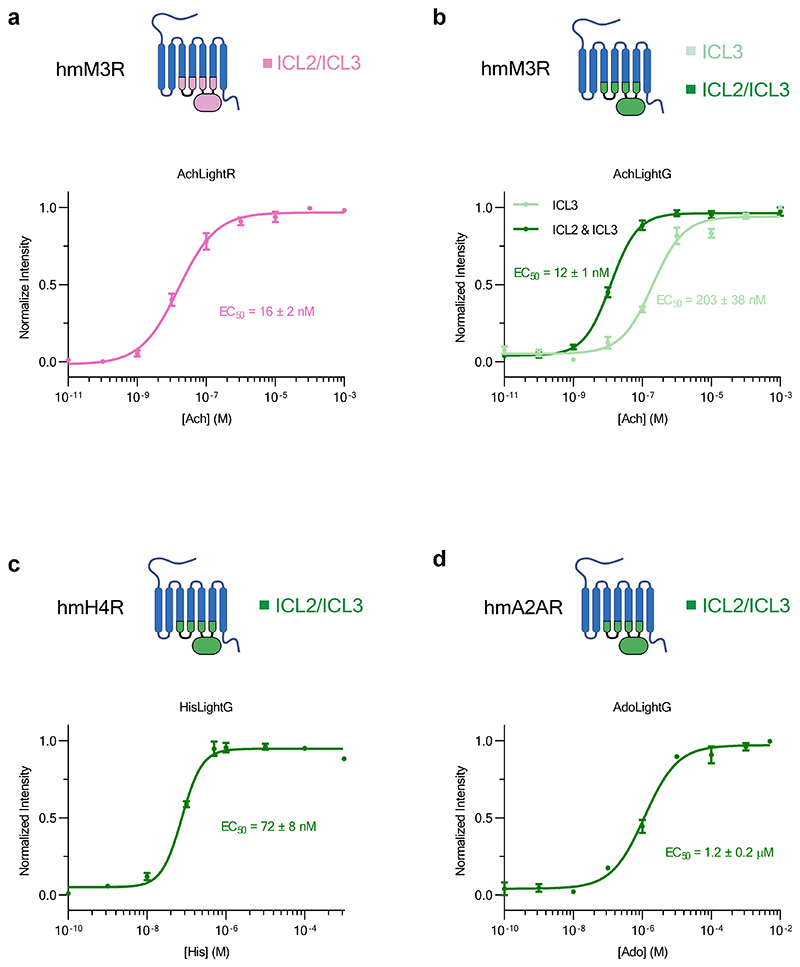
A comparison of the basal brightness (F0) revealed that the LightG- and LightR-based indicators often had a higher brightness in the absence of ligand compared to similar, previously developed indicators7,9,30,31 (Extended Data Fig. 9a-c). We also assessed whether grafting could affect the apparent affinity for the endogenous ligands by determining the half-maximal effective concentration (EC50) for a subset of the best performing indicators. We found the EC50 of the endogenous agonists was 203 nM for the green acetylcholine indicator (AchLightG, hmM3R single-graft), 16 nM for the red acetylcholine indicator (AchLightR, hmM3R double-graft), 1.2 μM for the green adenosine indicator (AdoLightG, hmA2AR double-graft) and 72 nM for the green histamine indicator (HisLightG, hmH4R double-graft) (Extended Data Fig. 10a-d). Interestingly, the affinity acetylcholine for AchLightR was approximately 100-fold higher than that reported for previously-developed indicators based on the same receptor subtype32. To check whether this effect stemmed from the additionally grafted ICL2 region, we titrated the double graft of AchLightG and measured an EC50 of 12 nM. This represents a 17-fold increase in affinity of acetylcholine compared to the single graft, with an EC50 similar to that of AchLightR (double graft), demonstrating that replacing the ICL2 with that of a different receptor can sometimes be used to tune the EC50 of an indicator. Overall, these data show that our two-step cloning strategy can in some cases, albeit not always, succeed in developing high-quality GPCR-based indicators.
Discussion
In addition to nLightG and nLightR, we developed indicators for several other neuromodulators. A recent study reported the use of a similar grafting approach for the generation of green GPCR-based indicators33. A direct comparison between these two approaches could be performed in the future to establish whether one is more successful than the other.
Our results show that nLightG and nLightR have improved selectivity for NE over DA compared to GRABNE1m. Nevertheless, both indicators showed detectable responses to perfusion of a high (50 μM) DA concentration in brain slices. This suggests that when planning the use of any of these sensors in areas of the brain with dense dopaminergic innervation (i.e. the basal ganglia)34,35, careful experimental design should be put in place to disambiguate the two catecholamines.
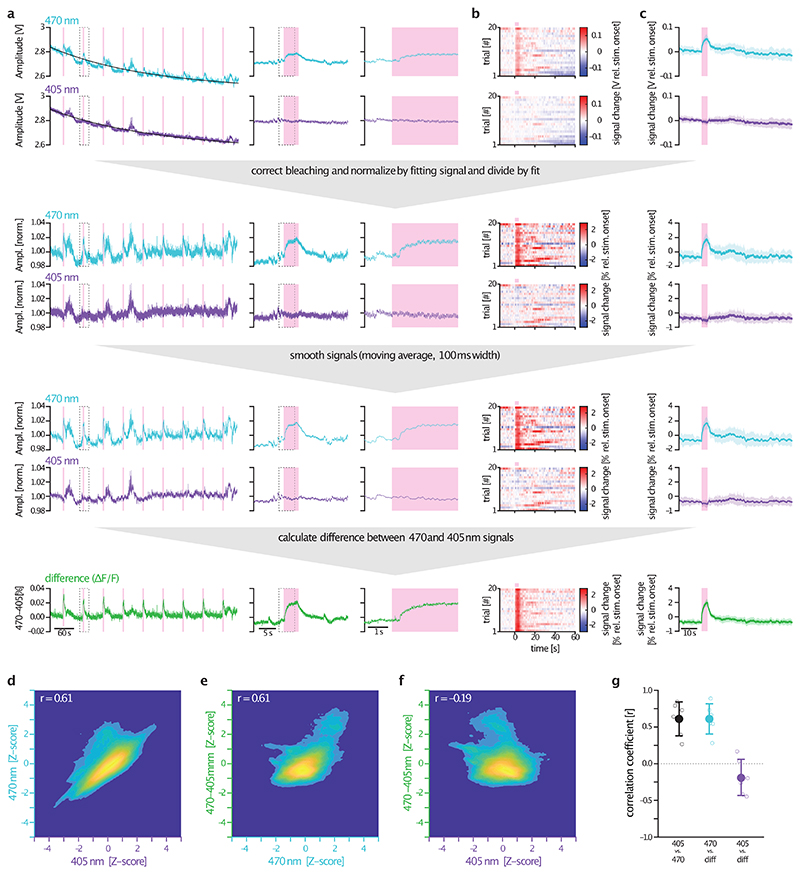
A potential limitation of fiber photometry recordings, which has recently gained attention, is the possible occurrence of artefacts caused by animal motion or hemodynamics36–38. These artefacts typically affect fluorescence signals excited at 405 nm or 470 nm uniformly37. To address this issue, our photometry recordings were conducted at both wavelengths, with 405 nm serving as a control channel. A comparison between 470 nm and 405 nm traces revealed that only the 470 nm channel exhibited optogenetically-evoked nLightG responses, while these were absent in the 405 nm channel. Therefore, the signals detected in fiber photometry experiments were specific to nLightG responses and were not influenced by hemodynamics or motion artefacts. In two-photon functional imaging experiments, we did not test a loss-of-function approach (e.g. a ligand-insensitive version of nLightG). However, structural similarity analysis and temporal shuffling of nLightG traces provided convincing evidence that the observed nLightG signals were not a result of motion-related artifacts.
Our demonstration of in vivo NE detection using nLightG led to several insights. First, we showed that nLightG can faithfully report the effects of an Alpha-2 AR antagonist on behaviorally-evoked NE release in vivo, while GRABNE1m fails to do so. Second, we could show functionally that the LC-dHPC system is highly lateralized, confirming previous evidence from neuroanatomical studies25,26, and demonstrated distinct NE signal kinetics across brain regions. Finally, via two-photon imaging of NE signaling in the dorsal hippocampus of awake mice navigating in a virtual corridor, we demonstrated differential correlation of localized NE signals associated with running and crossing reward position. Thus the next-generation NE indicators introduced here greatly advance the neurotechnological toolbox that is necessary to investigate the physiological functions of the norepinephrine system with high spatiotemporal resolution.
Methods
Molecular cloning
DNA sequences encoding individual GPCRs and GRAB indicators were either obtained from Addgene (GRABACh3.0, #121922; GRAB5HT1.0, #140552; GRABATP1.0, #167577; GRABeCB2.0, #164604) or directly ordered as gene fragments (Twist Bioscience, Thermo Fisher Scientific) and cloned into a mammalian expression vector downstream of a CMV-promoter (Addgene plasmid #60360) by restriction enzyme cloning using NotI and HindIII sites. Cloning of DNA sequences from dLight1.3b or RdLight1 (University of Zurich Viral Vector Facility, UZH-VVF) onto individual GPCRs for the generation of single graft and double graft constructs was performed by Gibson Assembly using the NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs). A list of primers used can be found in the Supplementary Data S2. Site-directed mutagenesis was performed by PCR using custom-designed primers (ThermoFisher Scientific) and PfuUltra II Hotstart PCR Master Mix (Agilent). To generate C-terminal fusions of SmBit onto DRD1, swAlpha-1 AR, nLightG, and nLightR, the SmBit sequence was PCR-amplified from the pCMV-Beta2AR-SmBit plasmid and cloned in place using Gibson assembly. All sequences were verified using Sanger sequencing (Microsynth). The DNA sequences encoding nLightG and nLightR were cloned into a viral vector under the control of a human Synapsin-1 promoter (UZH-VVF) by restriction enzyme cloning using BamHI and HindIII sites.
Cell culture, confocal imaging and quantification
HEK293T cells (ATCC #CRL-3216) were cultured as previously described41. The cell line was authenticated by the commercial provider. Cells were transfected at 40-50% confluency in glass-bottomed dishes or 6-well plates using PolyFect® Transfection Reagent (Qiagen) according to manufacturer instructions, and used for follow up experiments 24-48 h after transfection. Primary cortical neurons were prepared as follows: the cerebral cortex of 18 days old rat embryos were carefully dissected and washed with 5 ml sterile-filtered PBGA buffer (PBS containing 10 mM glucose, 1 mg/ml bovine serum albumin and antibiotic-antimycotic 1:100 (10,000 units/ml penicillin; 10,000 μg/ml streptomycin; 25 μg/ml amphotericin B)) (ThermoFisher Scientific). Cortices were cut into small pieces and digested in 5 ml sterile filtered papain solution for 15 min at 37°C. Tissues were then washed with complete DMEM medium containing 10% Fetal Calf Serum and penicillin/streptomycin (1:100), triturated and filtered through a 40 μm cell-strainer. Neurons were plated at a concentration of 40,000-50,000 cells per well onto poly-L-lysine (50 μg/ml in PBS, ThermoFisher Scientific) coated dishes and kept in NU-medium (Minimum Essential Medium (MEM) with 15% NU serum, 2% B27 supplement, 15 mM HEPES, 0.45% glucose, 1 mM sodium pyruvate, 2 mM GlutaMAX). The cultures were virally transduced after 4-6 days with adeno-associated viruses at a 1 x 109 GC/ml final titer and kept for 12-16 days in vitro. All cells were imaged at room temperature in Hank’s Balanced Salt Solution (HBSS) with CaCl2 and MgCl2 (ThermoFisher Scientific) in glass bottom dishes. For live-cell labeling cells were incubated with Alexa-647-anti-FLAG antibody (1:1000) for 10 minutes at room temperature and washed twice with PBS. Imaging was performed using Zen Blue software on an inverted Zeiss LSM 800 confocal microscope equipped with 488 nm laser (for green indicators) or a 561 nm laser (for red indicators), using either a 20X air-based or a 40X oil-based objective. During time-lapse imaging, ligands were manually applied using a pipette to reach the specified final concentrations. For quantification of the fluorescence response ΔF/F0, regions of interest (ROIs) that enclose isolated cell membranes were selected manually using the threshold function of Fiji (ImageJ). The fluorescence response of a indicator (ΔF/F0) was calculated as follows: (Ft - F0)/F0 with Ft being the ROI mean grey value at each time point t, and F0 being the mean grey value of the ten time points immediately prior to ligand addition.
Plate reader-based assays
For measuring dose-responses to titrations of ligands on the indicators, HEK293T cells were transfected in a T75 flask using polyethyleneimine hydrochloride (PEI, Sigma-Adlrich). For each flask 432 μL of PBS (ThermoFisher Scientific) supplemented with 150 mM of NaCl was mixed with 48 μL of PEI solution (1 mg/mL, pH 7.0 in ddH2O) and 9 μg of plasmid DNA was added prior to vortexing. The transfection mixture was incubated at RT for 45 min and added to the cells in a dropwise manner. The medium was replaced 24 h after transfection and the cells were used for titrations 48 h after transfection. The cells were detached using Versene (ThermoFisher Scientific), centrifuged (RT; 150g; 3 min), washed and resuspended in HBSS to a density of 6.66·106 cells/mL. The wells of a black, flat bottom, 96 well plate (Greiner Bio-One) were filled with 150 μL of cell suspension and to each well 150 μL of a 2-fold concentrated dilution series of ligand in HBSS was added. The fluorescence signal was detected at room temperature using a Tecan M200 Pro plate reader with an excitation wavelength of 480 nm or 560 nm (bandwidth of 9 nM) and an emission wavelength of 560 nm or 600 nM (bandwidth of 35 nM) for the green and the red indicators respectively. For each titration series three biological replicates were measured and the normalized means were fitted with a four-parameter dose-response curve to determine the EC50.
For measuring the one-photon excitation and emission spectra of the indicators, HEK293T cells were transfected using PolyFect® Transfection Reagent (Qiagen) according to manufacturer’s instructions and used for spectral measurements 48 h after transfection. The cells were plated in two wells of a black, flat bottom, 96 well plate as described above. To account for the autofluorescence of the cells a non-transfected control was measured at the exact same conditions. One-photon fluorescence excitation (λem = 560 nm or 620 nm for green and red indicators respectively) and emission (λexc = 470 nm or 550 nm for green and red indicators respectively) spectra were determined on a Tecan M200 Pro plate reader at room temperature.
To measure the recruitment of G-proteins and β-arrestin-2 we used a NanoBiT complementation-based assay17. For this we used the wild-type sperm whale Alpha-1 AR, a human DRD1 receptor, or the NE indicators carrying a C-terminally fused SmBiT fragment in combination with either mini-G protein probes (mini-Gs, mini-Gi and mini-Gq42) or β-arrestin-243 N-terminally fused to LgBiT, as specified. HEK293T cells were seeded in 6-well-plates (Techno Plastic Products) at a density of 250’000 cells/well and co-transfected using PolyFect® with the above-mentioned constructs at a 1:1 DNA ratio. Cells were detached using Versene 24 h after transfection, centrifuged (RT, 150 g, 3 min), washed and resuspended (0.5 x 106 cells/mL) in Fluorobrite™-DMEM (Thermo Fisher Scientific) complemented with 30 mM of HEPES (Thermo Fisher Scientific). For each interaction pair 200 μL of the cell suspension were gently mixed with 50 μL of a 20-fold dilution of Nano-Glo® reagent in LCS buffer (Promega) and distributed equally into two wells of a white flat-bottom 96-well plate OptiPlate (PerkinElmer) prior to incubation at 37°C for 45 min. The luminescence signal was measured at 37°C using a Tecan Spark® plate reader before and after the manual addition of 25 μL ligand solution (10 μM final concentration) in Fluorobrite®DMEM in one well or 25 μL of pure Fluorobrite®DMEM in the other well.
Patch-clamp fluorometry
To analyze the response kinetics of nLightG and GRABNE1m we used patch-clamp fluorometry similar to our previously described procedure13. Outside-out membrane patches from HEK cells expressing nLightG or GRABNE1m were placed in front of a double-barreled perfusion pipette that was rapidly moved by means of a piezo actuator44 while fluorescence was collected with an EMCCD camera. HEK293T cells were grown in DMEM with 7% FBS at 37 °C and 5% CO2 on poly-D-lysine/laminin-coated glass coverslips to minimize background fluorescence. Transfections were conducted after 24 h using polyethylenimine 25,000 (Sigma) with ~0.3 μg DNA per ml medium. After 72 h expression, coverslips with adherent cells were placed in external solution (138 mM NaCl, 1.5 mM KCl, 1.2 mM MgCl2, 2.5 mM CaCl2, 10 mM HEPES, pH 7.3) and membrane patches were pulled using borosilicate glass patch pipettes (G150TF-4, Warner Instruments; 4-6 MΩ resistance) filled with internal solution (135 mM K-gluconate, 10 mM NaCl, 2 mM MgCl2, 1 mM EGTA, 10 mM HEPES, pH 7.3). Cell-attached and then whole-cell configurations were established to excise outside-out patches using a Patchstar micro-manipulator (Scientifica) and Axopatch 200B patch-clamp amplifier (Molecular Devices). Experiments were performed at room temperature. pClamp 10.7 and the Digidata 1550 A/D converter (both Molecular Devices) were used for generating the voltage pulses for ligand application and triggering camera acquisition. Epifluorescence imaging of the excised patches was performed on an inverse DMi8 microscope (Leica) with a 20x objective (HC PL FLUOTAR L 20x/0.40 CORR PH1). Green fluorescence was excited using a 470 nm LED (Thorlabs) and a 470/40 nm excitation filter in combination with a 495 nm dichroic mirror and a 525/50 nm emission filter (all Chroma). The light intensity in the focal plane was ~2-10 mW/mm2. Images were acquired with an Evolve 512 delta EMCCD camera (Photometrics) using MicroManager2.045. The acquisition of individual frames (camera clearing mode: presequence; acquisition mode: strobed) was triggered by TTL pulses provided by pClamp. Imaging was performed with a frame rate of 100 fps (cropping to 256 x 256 pixels, 2x2 binning, 9 ms exposure, EM gain 200) or 200 fps (cropping to 256 x 256 pixels, 2x2 binning, 4.5 ms exposure, EM gain 500). Fast ligand application and removal was achieved by positioning the outside-out patches in front of a piezo-driven double-barreled Θ-glass pipette44. The perfusion pipette was pulled from borosilicate glass (OD 2.0 mm, ID 1.40 mm, septum 0.2 mm, Warner Instruments), broken to yield a ~250 μm diameter tip, and mounted to a piezo actuator (P842.20, Physik Instrumente). Lateral displacements were triggered with short voltage steps (3 V ramp in 0.7 ms), amplified by a power supply (E505.10, Physik Instrumente) and filtered at 1 kHz. Solutions were delivered using a syringe pump (0.3 ml/min per channel) with parallel bath perfusion at ~3-5 ml/min. NE (5 μM, Sigma 74480) in external solution (1.2-fold concentrated to obtain exchange currents) was applied in 1 s or 2 s pulses. Fiji46 (ImageJ) was used to calculate ΔF/F0 maps based on mean intensity projections of 100 frames before and 100 frames during NE application (Figure 2g). These maps were used to define the responsive patch region and placing of a rectangular ROI. The fluorescence intensity of this region was exported and normalized to the initial background fluorescence obtained from an adjacent region. Figure 2i shows 6 subsequent applications interrupted by 0.2 s breaks in the imaging/illumination sequence (white spaces). In this case a baseline correction was performed (dashed line) to account for minor signal loss in the patch region (0.72 e-t/40 s). However, consecutive applications showed full reversibility of the signal changes (see also Supplementary Video 1). To obtain estimates of the response times, data from 4-6 consecutive applications were averaged from each patch and single exponential fits were used to determine τon and τoff time constants by least-square fitting with ProFit 7.0 (Quantumsoft).
In vitro two-photon brightness measurements
Two-photon spectral characterizations of the nLightG indicator were performed on HEK293T cells before and after addition of NE (100 μM). Cells were transfected using Lipofectamine 3000 and imaged 24 hours post transfection. Prior to the spectral measurements the medium was switched to PBS to avoid DMEM autofluorescence. The two-photon spectra were acquired as described previously9.
Viral production
Adeno associated viruses (AAV) encoding for the indicators developed in this study were produced by the Viral Vector Facility at the University of Zurich (VVF). All other viruses were obtained either from the VVF or Addgene. The titers of the viruses used in this study were: rAAV2/9.hSynapsin1.nLightR, 1.4 x 1013 GC/mL; rAAV2/9.hSynapsin1.nLightG, 2.3 x 1013 GC/mL; rAAV2/9.hSynapsin1.GRABNE1m, 5.5 x 1012 GC/mL; rAAV2/5-hEF1α.DIO.ChrimsonR-tdTomato, 4.7 x 1012 GC/mL; rAAV2/9.EF1α.DIO.ChrimsonR-mRuby2, 4.5 x 1012 GC/mL.
Animals
Animal procedures were performed in accordance to the guidelines of the European Community Council Directive or the Animal Welfare Ordinance (TSchV 455.1) of the Swiss Federal Food Safety and Veterinary Office and were approved by the Zürich Cantonal Veterinary Office, the Hamburg state authority for animal welfare and the animal welfare officer of the University Medical Center Hamburg-Eppendorf, National Council on Animal Care of the Italian Ministry of Health. Rat embryos (E17) obtained from timed-pregnant Wistar rats (Envigo) were used for preparing primary cortical neuronal cultures. Wild-type C57BL/6 mice, heterozygous B6.Cg-Dbhtm3.2(cre)Pjen/J (DBH-Cre) mice47 and heterozygous C57BL/6-Tg(Dbh-iCre)1Gsc (DBH-iCre) mice48 of both sexes were used in this study. Mice were kept with ad libitum access to chow and water on a 12-h reversed light-dark cycle. Optogenetic and behavior experiments were performed during the dark phase. Two-photon imaging experiments were performed on animals older than 10 weeks which were housed at room temperature and in humidity-controlled rooms in groups of up to five littermates per cage with ad libitum access to food and water in a 12-hour light-dark cycle.
Animal surgeries and viral injections
Surgeries were conducted on adult anesthetized mice (males and females, age > 6 weeks). For slice imaging experiments, AAVs encoding nLightG, nLightR or GRABNE1m7 under the control of the human synapsin-1 promoter were unilaterally injected at a titer of ~5.5 x 1012 GC/mL into the LH (-1.4 mm AP, +1.1 mm ML, -5.2 mm DV, volume: 600 nL). For single-site in vivo photometry experiments, the following AAVs were unilaterally injected at a similar titer into either of the following regions: LH (-1.4 mm AP, +1.1 mm ML, -5.2 mm DV, volume: 250 nL), vHPC (-3.2 mm AP, -3.3 mm ML, -3.8 mm DV, volume: 250 nL) and commercial fiber optic cannulas were then implanted 0.1-0.2 mm above the injection sites (for LH, 400 μm core diameter, NA=0.57, Doric lenses; for vHPC, 200 μm core diameter, NA=0.37, Neurophotometrics). Mice used for optogenetic stimulation experiments also received an unilateral injection (1000 nL) in LC of undiluted rAAV2/5-hEF1α.DIO.ChrimsonR-tdTomato and were implanted an additional fiber optic cannula above the injection site. For dual-site in vivo photometry experiments, to bilaterally access the LC and dHPC, small craniotomies (~500 μm diameter) were drilled -5.4 mm posterior and ±1.1 mm lateral to Bregma (to access LC) as well as -2 mm posterior and ±1.5 mm lateral to Bregma (to access dHPC). 300 nL of viral suspension consisting of a mixture of rAAV2/9.hSynapsin-1.nLightG (~1.2 × 1013 GC/mL) and rAAV2/9.EF1α.DIO.ChrimsonR-mRuby2 (~4.5 × 1012 GC/mL) were injected into each LC at a depth of -3.6 mm. Subsequently, 500 nL of rAAV2/9.hSynapsin-1.nLightG (~1.2 × 1013 GC/mL) were injected into each dHPC at a depth of -1.6 mm. After each injection, the pipette was left in place for ~2 min, before it was slowly withdrawn. After pipette withdrawal, fiber optic cannulas (400 μm core diameter, NA=0.5, RWD) were slowly inserted at the injection coordinates to a depth of -3.5/-1.2 mm.
For two-photon imaging, neuronal-specific expression of nLightG was obtained by injecting rAAV2/9.hSynapsin-1.nLightG into the hippocampal CA1 region (-1.75 mm AP, +1.35 mm ML, -1.40 mm DV) through a craniotomy opened in the right hemisphere. 800 nL of AAV solution was injected using an injection apparatus (UltraMicroPump, WPI, Sarasota, FL). Following viral injection, a chronic hippocampal window was implanted following the procedures described in49–51. In brief, a 3 mm trephine-drill was used to open a craniotomy centered at coordinates 2.00 mm posterior and 1.80 mm lateral to bregma. Optical access to the hippocampus was obtained removing the overlying cortical tissue by careful aspiration. During the aspiration procedure, brain tissue was constantly irrigated with HEPES-buffered artificial cerebrospinal fluid. An optical window was fitted to the craniotomy above the external capsule and a thin layer of silicone elastomer (Kwik-Sil, World Precision Instruments, Sarasota, FL) was placed at the interface between the brain tissue and the optical window surface. A custom titanium headplate was attached to the skull using epoxy glue and implant components were further secured in place using dental cement (Super-Bond, Sun Medical, Japan). The scalp incision was finally sutured to adhere to the implant. Animals received an intraperitoneal bolus of antibiotic (BAYTRIL, Bayer, Germany) at the end of the surgery. Optical windows were built as previously described49 using a thin-walled stainless-steel cannula segment (OD, 3 mm; ID, 2.77 mm; height, 1.50 - 1.60 mm) and a 3.00 mm diameter round coverslip, which was secured to the lower end of the cannula using UV curable optical epoxy (Norland optical adhesive 63, Norland, Cranbury, NJ). All viruses were injected at a rate of ~100 nL/min.
Acute brain slice preparation, imaging and quantification
For acute brain slice preparation, at least 4 weeks after bilateral viral injections, mice were deeply anesthetized with an intraperitoneal injection of pentobarbital (200 mg/kg, 10mL/kg) and intracardially perfused with ice-cold artificial cerebrospinal fluid (aCSF) bubbled with 95/5% O2/CO2 containing, 120 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3, 5 mM HEPES, 1 mM MgCl2, 14.6 mM D-glucose and 2.5 mM CaCl2 (Osmolarity: 305-310 mOsm/kg). After perfusion, mice were decapitated and the brain was quickly extracted while submerged in the ice-cold aCSF. 250-300 μm thick coronal slices containing the LHA were obtained using a vibratome (HM 650V, Thermo Fisher Scientific). The slices were incubated at 34°C for 20 minutes in continuously oxygenated aCSF. Following incubation, brain slices were transferred at RT and kept until recording. Recordings were conducted in a slice chamber kept at 31°C perfused with aCSF. For electrical stimulation at 20 Hz, a bipolar electrode (Parallel Bipolar,30211, FHC) was positioned near the LHA. Electrical stimuli were synchronized by using Pulse Pal (Open Ephys). To visualize green or red norepinephrine indicators, slices were illuminated with either a blue (469 nm) or green (555 nm) LED (Colibri 7, Zeiss), respectively, on an upright Axio Examiner A1 microscope (Zeiss) using an N-Achroplan 10x/0.3 M27 objective (Zeiss). Images were collected at a sample rate of 5 Hz for nLightG and GRABNE1m of 2 Hz for nLightR (Live Acquisition, Thermo Fisher Scientific). The stimulation voltage was set at 2.5 V and the duration of each stimulation was 10 ms. The train duration of each session determined the number of pulses delivered to each slice. For pharmacological experiments, drugs were applied via a perfusion system at a flow rate at 2.5 mL/min and images were recorded at a 0.5 Hz sample rate for nLightG and GRABNE1m and at 0.2 Hz for nLightR. The analysis of slice imaging data was performed using a custom-written MATLAB vR2019b script. Regions of interest (ROIs) of equal size were selected for each slice using Fiji (Image J). For both electrical stimulation and ligand-perfusion experiments, the change in fluorescence ΔF/F0 %, defined as (F-F0)/F0 x 100 was calculated. F0 was defined using a 1 sec baseline for both the nLightG and GRABNE1m and a 2.5 sec baseline for nLightR. Depending on the rate and pattern of photobleaching, signals were fitted with either the MATLAB polyfit polynomial function of 4th degree or with the exp1 single-term exponential function and detrended using the detrend function. ΔF/F0 % signals for each indicator acquired from electrical stimulation experiments were averaged and smoothed with a 3-point moving mean filter. Peaks responses were calculated as maximum ΔF/F0 % response to each stimulation. For all the ligand-perfusion experiments 2-min bins were defined for each drug/condition (DA, NE, NE+Trz perfusions and NE washout) for mean ΔF/F0 % calculations. Representative traces were smoothed with a 3-point (for selectivity experiments) or a 5-point (for NE washout experiments) moving mean filter.
Fiber photometry
Recordings in lateral hypothalamus during tail-lifting
Fiber photometry recordings in the LH were performed using an iFMC6_IE(400-410)_E1(460-490)_F1(500-540)_E2(555-570)_F2(580-680)_S photometry system (Doric Lenses) controlled by the Doric Neuroscience Studio v6.1.2.0 software. A low-autofluorescence patch cord (400 μm, 0.57 N.A., Doric Lenses) was attached to the ferrule on mouse’s head and used to excite nLightG and GRABNE1m (465 nm) or nLightR (560nm) and collect the fluorescence emission, while 405 nm was used as a control fluorescence signal for all the indicators. Signals were sinusoidally modulated at 208 Hz and 572 Hz (405nm and 465nm, respectively) and at 333 Hz (560nm) via lock-in amplification, then demodulated on-line and low-passed filtered at 12 Hz. Mice were habituated to handling, injections, and tethering prior to all fiber photometry experiments. Experiments started 4 weeks after surgery to allow proper expression of the indicators. A baseline period of the fluorescent signal was recorded before drug administration and for 5 minutes after the injection of saline (0.9% NaCl, 10 μL/g bodyweight) or drug solution. Yohimbine (Y3125-1G; Sigma Aldrich) was diluted in saline to 4 μg/g bodyweight for i.p. injection (10 μL/g). trazodone (T6154-1G; Sigma Aldrich) was diluted in saline to 10 μg/g bodyweight for i.p. injection (10 μL/g). After i.p. injection, signals were recorded for additional 5 minutes before the tail-lifting task was performed. Each mouse was lifted by the tail 5 times of one-minute duration each, while indicator fluorescence was recorded. Between each tail lift mice had 2-minute intervals in the experimental cage.
Recordings in ventral hippocampus during optogenetic stimulation
The green fluorescence signal from NE indicators nLightG and GRABNE1m were recorded using a commercially available photometry system (Neurophotometrics, Model FP3002) controlled via the open-source software Bonsai (2.6.2 version). Two LEDs were used to deliver interleaved excitation light: a 470 nm LED for recording NE-dependent fluorescent signals (F470) and a 415 nm LED for control fluorescent signals (F415). The sampling rate was set at 120 Hz for both LEDs allowing 60 Hz for each channel individually. Excitation power at the fiber tip was set to 25-35 μW. Throughout the recording session mice were anesthetized (4% isoflurane during induction, 1.5-2% during maintenance) and the fiber implanted in the mouse brain was attached to a pre-bleached recording patch cord (200 μm, 0.39 numerical aperture; Doric Lenses). For yohimbine injection (10 μL/g, 4 mg/kg), a catheter was placed intraperitoneally. During the photometry recording session, an LC stimulation sequence was performed pre- and 15 min post-yohimbine injection. Each sequence consisted of three LC optical stimulations for 10 s each (5 Hz, 10 ms pulse width, 635 nm CNI laser at 10 mW output power) with 2 min interstimulation intervals.
Simultaneous dual-site recordings in locus coeruleus and dorsal hippocampus during optogenetic stimulation
3-4 weeks after surgery, animals were habituated to the experimenter by daily handling for a couple of days, before being habituated to the setup and to head-fixation on a linear treadmill (200-100 500 2100 and 700-100 100 0010; Luigs&Neumann). Fiber photometry recordings were realized using the recently developed PyPhotomety52 system: The PyPhotometry-board controlled a multi-color light source (pE-4000; CoolLED) connected to the 400-480 nm port of a dichroic cube (FMC5_E1(400-480)_F1(500-540)_E2(555-570)_F2(580-680)_S; Doric Lenses). Excitation light was band-pass filtered at 405/10 nm (#65-133, Edmund optics) and 470/10 nm (#7394, Alluxa) and delivered in a temporally interleaved manner52. Excitation power was adjusted such that the emission intensities of nLightG were similar when excited at 405 and 470 nm, respectively. Emission light was measured at the 500-540 nm port using a photodetector (DFD_FOA_FC; Doric Lenses), and digitized by the PyPhotometry-board at 130 Hz. Interfacing of the animal from the dichroic cube was achieved with a low autofluorescence patch cord (400 μm core diameter, 0.57 NA; MFP_400/430/3000-0.57_1m_FC-ZF1.25(F)_LAF, Doric Lenses), connected to the implanted cannula with a zirconia mating sleeve (SLEEVE_ZR_1.25; Doric Lenses), and covered with a black shrinking tube to minimize optical noise. For dual-site recordings, two of these systems were operated in parallel. Simultaneously, videographic images of the animal’s eye were taken using a monochrome camera (DMK 33UX249; The Imaging Source) equipped with a macro objective (TMN 1.0/50; The Imaging Source) and a 780 nm long-pass filter (FGL780; Thorlabs), while the eye was illuminated at 850 nm with an infrared spotlight. Optogenetic stimulation was achieved with custom scripts written in MATLAB (vR2019a, The MathWorks), actuating on a NI-DAQ-card (USB-6001; National Instruments), controlling a 594 nm diode laser (Obis 594 nm LS 100 mW; Coherent). Laser light was coupled into the 580-680 nm port of the dichroic cube for activation of ChrimsonR in noradrenergic neurons of the LC. Measurements were synchronized by custom scripts written in MATLAB (The MathWorks), actuating on a NI-DAQ card (PCIe-6323; National Instruments). For pharmacological experiments, data was first collected for the control condition, subsequently for the injection of saline (10 μL/g bodyweight, i.p., ~15 min. prior to the recording), and finally for the injection of desipramine (D3900-1G; Sigma Aldrich; 10 μg/g bodyweight, diluted in saline for injections of 10 μL/g bodyweight, i.p., ~15 min. prior to the recording), to avoid a bias in favor of the desipramine condition due to indicator bleaching. For experiments of LC lateralization, animals were anesthetized with isoflurane (1.5-2% in air) and placed on a heating pad to maintain body temperature. Recordings started 5-10 minutes after induction of anesthesia.
Data analysis for photometry and pupillometry recordings
For single-site photometry data, analysis of the raw photometry data was performed using a custom-written MATLAB script. First, to filter high frequency noise (above 1 Hz), the lowpass filter function was applied to both recorded signals: F560, F470/465 and control signal (Fcontrol, signal excited at either 405 nm or 415 nm).
Next, to correct for photobleaching of fluorescent signal, the baseline fluorescence Fcontrolbaseline fit was calculated as a linear fit applied to the filtered Fcontrol, the F470/465 or the F560 signals during the baseline 5 s window preceding each stimulus. The signal of the NE indicators were expressed as a percentage change in fluorescence: ΔF/F0 = 100*(F470/465/560(t) - Fcontrolbaseline fit(t))/Fcontrolbaseline fit(t), where F470/465/560(t) signifies the filtered fluorescence value at each time point t across the recording for either the 470 or 465 nm-excited signal or the 560 nm-excited signals, and Fcontrolbaseline fit(t) denotes the value of the fitted control signal at the time point t. The final ΔF/F0 signal was averaged over the multiple trials and smoothed with a 10-points moving mean filter. In the case of optogenetic experiments, all traces were normalized to the pre-yohimbine ΔF/F0 signal and plotted as percentage change.
For dual-site photometry data processing, the 405 and 470 nm excited signals were fitted with a polynomial function of 1st-4th degree (based on visual inspection) using MATLABs polyfit function, in order to capture the bleaching of each trace. Each signal was then divided by its fit, in order to correct for bleaching and normalize each signal. Traces were then smoothened with a moving average filter (100 ms window size). Finally, ΔF/F was calculated as the difference between the bleaching-corrected, normalized, smoothed signals excited at 405 and 470 nm (see also Extended Data Figure 7 for further details). To identify stimulus-evoked responses in pupillometric and photometry data, traces were cropped around each stimulation event, usually from 15 s before to 90 s after each event. Traces were then normalized by subtracting the median pupil diameter/fluorescence during the second before stimulus onset from each trace, before averaging all traces of a recording session. Finally, stimulus-evoked responses were defined as the median pupil diameter/fluorescence during the last second of stimulus presentation. Time constants of the decay of responses (τoff) were estimated by an exponential fit starting at stimulus offset. Processing of pupillometric data was performed as described before53: Briefly, the upper, lower, left, and right edges of the pupil were tracked using DeepLabCut54, samples with a certainty <0.5 were linearly interpolated between the next neighboring samples with a certainty of ≥0.5, and the pupil diameter was calculated as the maximum distance between two opposing points of the pupil. Eye opening was calculated by tracking the upper and lower edge of the eye, in order to remove blinking artefacts from pupil data by linearly interpolating regions in which the eye opening exceeds the moving median minus three times moving median absolute deviation with a sliding window of 30 s.
In vivo two-photon imaging
Two-photon imaging experiments during virtual navigation were carried out on a custom apparatus similar to that described in49. nLigthG signals were imaged using an Ultima Investigator scanhead (Bruker Corporation, Milan, Italy) equipped with galvo-resonant scanner, 16x/0.8 NA objective (Nikon, Milan, Italy), and multi-alkali photomultiplier tubes. Two-photon excitation was obtained using a Ti:Sapphire pulsed laser source tuned at 920 nm (Chameleon Ultra II, 80 MHz repetition rate, Coherent, Milan, Italy). Imaging average power at the objective focus was ~ 60 - 85 mW. Fluorescence emission was collected by a PMT detector downstream of a band-pass filter (525/70 nm) and Detector output signal was first amplified and then digitalized at 12 bits. Imaging sessions were acquired using resonant-scanning at ~ 30 Hz and 3x optical zooming factor. Images contained 512 pixels x 512 pixels (pixel size, 0.53 μm).
A custom virtual reality setup was implemented using the open-source 3D creation suite Blender (blender.org, version 2.78c). The virtual environment was a monodirectional linear corridor (length: 180 cm, width: 9 cm) with proximal walls characterized by three different white textures (vertical lines, mesh, and circles) on a black background. The character point of view (220° horizontal, 80° vertical) was rendered through a composite tiling of five thin-bezel led screens. Animal’s motion was rendered in 1:1 in the virtual corridor using a custom 3D printed wheel (radius 8 cm, width 9 cm) coupled with an optical rotary encoder (Avago AEDB-9140-A14, Broadcom Inc., San Jose, CA). Rotary encoder signals were acquired with a single board microcontroller (Arduino Uno R3, Arduino, Ivrea, Italy) and converted to USB-HID-compliant serial mouse input. Water rewards (~ 4 μl) were delivered through a custom lickport controlled by a solenoid valve (00431960, Christian Bürkert GmbH & Co., Ingelfingen, Germany) and licks were monitored using a capacitive indicator (MTCH102, Microchip Technology Inc., Chandler, AZ). Virtual reality rendering and two-photon imaging acquisition ran on asynchronous clocks while the start-of-frame TTL signal provided by the Ultima Investigator control system was used to synchronize imaging acquisitions with behavior.
Two weeks after surgery animals underwent water-scheduling, receiving approximately 1 ml of water per day. Weight was daily monitored, and it remained within 80-90% of the pre-scheduling weight throughout all procedures. Mice were first habituated to the operator and to the experimental rig for two sessions of few minutes duration. Animals were then habituated to head-tethering while receiving water rewards upon spontaneous running bouts. Head-tethering time was progressively increased reaching 45 minutes in five sessions. On average, mice underwent 8 training sessions where they were allowed to run spontaneously while receiving water rewards at pseudorandom times. Two-photon imaging experiments started on recording day 1, when mice were exposed to the virtual corridor for the first time and lasted for five consecutive days (day 1-5). Water rewards were available only at specific spatial locations in the virtual corridor. On recording day 1 and 2, the water reward was located at 85 cm from the beginning of the virtual corridor. At half of the recording session of day 3, the water reward was moved to a different position (145 cm) and was kept in that novel position for recordings performed on day 4-5. In each virtual corridor crossing, the mouse was teleported to the beginning of the corridor after reaching the end of the track (inter trial timeout interval 5 s). If the mouse did not reach the end of the corridor within 120 s, the trial was automatically terminated, and the mouse was teleported to the beginning of the corridor after an inter-trial timeout. On recording day 1, a set of reference images spanning different depths within the hippocampus was collected to identify an optical field-of-view and perform longitudinal imaging of the same population of CA1 neurons across experimental days 1 to 5. On each experimental session, three temporal series (12,500 frames/series; t-series duration, ~ 415 s), interleaved by three minutes breaks, were acquired during a ~ 45 minutes virtual navigation session. At the end of each imaging session, animals were returned to their home cage.
Data analysis for two-photon imaging experiments
Analysis was performed using custom code written in Python 3.6. t-series were pre-processed to correct motion artifacts using an open-source implementation of up-sampled phase cross-correlation55,56. Motion correction was first carried out for each day of recording, where each t-series was motion corrected using its average projection as reference frame. Average temporal projections of corrected t-series were then combined in an image stack and used to compute x-y drifts with respect to the first element of the image stack. Computed x-y drifts were applied to the corrected t-series, which were then concatenated in a single movie containing all imaging frames of an experimental session. The resulting movie was finally motion corrected using the global temporal average projection as a reference image. To maximize the alignment across longitudinal experimental sessions, we used the results of within-session motion correction procedures. For each session, we computed a global temporal average projection. Average projections were then combined in an image stack and subject to motion correction procedure using the average projection of the image stack as reference. This procedure yielded an array of across-session x-y drifts, and a stack of motion corrected average projections aligning each field-of-view across days. The resulting stack was used to compute a global average projection. The resulting x-y drifts were applied to all the frames of the relative imaging session, which was further motion corrected using the latest global average projection as a reference.
Fluorescence signals were computed either over the whole field-of-view as the average fluorescence intensity for each frame (Fig. 5a-i) or on identified ROIs (Fig. 5j-o). To identify potential axial displacement, for each session we computed the structural similarity index57 of each frame with the average projection of the whole session. To minimize the impact of this residual uncorrected motion artifacts, frames with a structural similarity index < 0.9 were excluded throughout all the following analysis. Importantly, the temporal dynamics of structural similarity values provide an internal control for the potential occurrence of stereotyped motion artifacts which would appear as repeated events in the temporal dynamics of structural similarity.
Fluorescence signals from ROIs (putative CA1 cells) were computed using CITE-On29 for both ROI detection and trace extraction. CITE-On algorithm was executed setting detection confidence threshold to 0.05 and upscaling factor to 1. Within detected ROIs, functional nLightG signals were extracted from pixels with intensity comprised within the 50th and the 95th percentile of the distribution of fluorescence intensity. For each session, putative running-epochs onset were identified as time points in which the mouse speed exceeded 1 cm/s. Runs were then selected when putative running-epochs onsets were preceded by 2 seconds of immobility (average speed < 1 cm/s) and followed by 5 seconds of sustained locomotion (average speed > 1 cm/s). Reward epochs were selected using the time-stamp of reward delivery provided by the virtual reality software. For both running- and reward-event triggered analysis fluorescence signals were analyzed within each epoch defining a time window of (-2, 5) seconds relative to the epoch onset. For whole field-of-view and ROI analysis, fluorescence F0 was defined as the average raw fluorescence intensity value observed in the interval (-2,0) seconds preceding the epoch onset. Within each epoch of (-2, 5) seconds fluorescence values F(t) were computed filtering raw traces with a uniform mean filter (width 500 ms) centered at t, and ΔF/F0 was calculated as (F(t)-F0)/F0. The shuffled traces reported in Extended data Fig. 8-9 were computed as the average of 1000 shuffling of the raw fluorescence traces with respect to behavioral event (either beginning of running or crossing the reward position). Least-squares linear models were fitted on each individual session for each animal using data recorded within event-triggered epochs intervals. Model fitting was performed using ordinary least-squares implementation in SciPy (v1.10.1)58. For running epochs data was selected within [0,5) seconds, while for reward epochs within [0,3) seconds. Average models were computed as averages of models fitted on individual animals’ data.
Pairwise correlation analysis of ROI signals, was conducted measuring the Pearson’s product-moment correlation coefficient for pairs of ROIs using data from the intervals [0,5) seconds or [0,3) seconds for running and reward epochs, respectively. Self-correlations were removed. Hierarchical clustering of correlation matrices was performed using the agglomerative unweighted pair group method with arithmetic mean (UPGMA) algorithm implemented in SciPy58 over the Euclidean distance.
Immunohistochemistry
For transcardial perfusions, mice were deeply anesthetized with intraperitoneal injections of Ketamine/Xylazine in saline (180/24 mg/kg), and deep anesthesia was confirmed by absence of the hind paw withdrawal reflex. Mice were initially perfused with ~50 ml of phosphate buffered saline (PBS), and subsequently with ~50 ml of paraformaldehyde (4% in PBS). Explanted brains were post-fixed in 4% PFA for at least 24h, embedded in 3% agarose, and coronal slices of ~50 μm thickness were obtained with a vibratome (VT100S; Leica Biosystems). Unspecific antibody binding sites in brain slices were blocked (10% normal goat serum (NGS) and 0.3% Triton X-100 in PBS, 2h, room temperature), before slices were incubated with primary antibodies against eGFP (chicken a-GFP, 1:750; A10262, Thermo Fisher Scientific) and tyrosine hydroxylase (LC only; rabbit a-TH, 1:1000; AB152, Merck Milipore) in carrier solution (2% NGS and, 0.3% Triton X-100 in PBS, 48h, 4°C). Slices were then washed in PBS (3 times for 5 minutes) before incubation with secondary antibodies (Alexa Fluor 488 goat anti-chicken; A11039, Thermo Fisher Scientific, and Alexa Fluor 647 goat anti-rabbit; A32733, Thermo Fisher Scientific; both diluted 1:1000 in carrier solution) over night at 4°C. Finally, slices were again washed in PBS (3 times for 5 minutes), incubated in DAPI in PBS for 5-10 minutes, and mounted on microscope slides using Fluoromount (Serva). Slices were then imaged using an epifluorescence microscope (AxioObserver, Zeiss) with a 20X/0.8 N.A. air objective and images were processed using ImageJ v1.52 (Fiji)46.
Statistical Analyses
For in vitro analysis of indicator variants, the statistical significance of their responses was determined using GraphPad Prism v9.0.0 on a case-by-case basis using either a two-tailed unpaired Student’s t-test with Welch’s correction or a Brown-Forsythe ANOVA test followed by Dunnett’s T3 multiple comparison. For in vivo single-site photometry experiments, we used one-sample, paired and unpaired t-tests for statistical analysis of parametric data, while Wilcoxon rank-sum or signed-rank tests for analysis of non-parametric data. For in vivo dual-site photometry experiments, one- and two-sample, two-tailed t-tests were performed to compare a single sample against a reference value or two samples against each other, after normality of the data was confirmed using a Jarque-Bera-Test. For comparison of three or more groups, one-way ANOVA was performed and - when significant - post-hoc comparisons between groups were performed using Tukey’s test. All P values are indicated in the figure legends. Data are displayed as mean with standard error of the mean (SEM) or standard deviation (SD), as noted in the figure legends. No statistical methods were used to predetermine sample size.
Extended Data
Extended Data Fig. 1. Development of nLightG and nLightR.
a, Representative images of indicators, generated by grafting LightR from residue 5.63 to residue 6.33 onto Alpha-1 ARs from five different species, expressed in HEK293T cells and imaged in the absence and presence of NE (10 μM). Heatmaps represent the pixelwise absolute change in fluorescence intensity upon addition of NE. b, Quantification of the basal brightness and the dynamic range upon addition of NE (10 μM) to HEK293T cells expressing the five prototype indicators. n = 21 cells. c, d, Normalized fluorescence intensity dose-response curves of nLightG and nLightR for NE (c) and DA (d) in HEK293T cells. Datapoints were fitted with four-parameter dose-response curves to determine the EC50 values. n = 3 wells per concentration for each ligand. e, Partial alignment of the amino acid sequence of RdLight1, dLight1.3b and the sperm whale Alpha-1 AR. BW numbering is shown for the residues involved in grafting registry optimization. f, g, Dynamic range of indicator variants generated by grafting of the cpmApple module of RdLight1 (f) or the cpGFP module of dLight1.3b (g) at different BW registries. The indicated BW numbers refer to the grafted part of RdLight1 or dLight1.3b replacing the equivalent part of the sperm whale Alpha-1 AR. The response was measured in HEK293T cells upon addition of NE (10 μM). n = 21 cells. P values were as follows: 1.585×10-8 (f); 5.66 - 6.30, 4.149×10-17; 5.65 - 6.31, 9.093×10-7; 5.64 - 6.32, 2.199×10-8; 5.62 - 6.34, 4.341×10-14; 5.61 - 6.35, 3.283×10-14 (g). h, Dynamic range of the red fluorescent swAlpha-1 AR-based indicator variants with the optimized grafting registry without or with additional grafting of the ICL2 from RdLight1. The grafted regions replacing the equivalent part of the swAlpha-1 AR are indicated with BW numbering. The response was measured in HEK293T cells upon addition of NE (10 μM). n = 21 cells. P = 2.625×10-5. i, Left, time trace of the relative fluorescent response of nLightR (with or without the ICL2 region grafted from RdLight1) transiently expressed in HEK293T. Saturating concentrations of DA (200 μM) and NE (10 μM) were added consecutively. n = 21 cells for each trace. Right, quantification of the ratio of the average fluorescent responses to DA versus NE from traces shown on left, for each indicator. P = 2.292× 10-4. j, Same as in (h) but for nLightG and using the ICL2 of dLight1.3b for grafting. P = 0.566. k, Same as in (i) but for nLightG (with or without the ICL2 region grafted from dLight1.3b). P = 2.103× 10-3. All data are shown as mean ± SEM and all experiments were repeated at least three times with similar results. The mean values were compared using a two-tailed Students t-test with Welch’s correction.
Extended Data Fig. 2. Additional in vitro characterization of nLightG and nLightR.
a, Time trace of fluorescent response of GRABNE1m in HEK293T, and quantification of mean values of GRABNE1m responses to different ligands. NE (10 μM), trazodone (Trz, 100 nM) and yohimbine (Yoh, 100 nM) were added consecutively. Two-tailed Students t-test with Welch’s correction. Error bars represent mean ± SEM. n = 11 cells from 3 independent experiments. n.s., P = 0.9902; **** P = 1.374×10-8. b, Left, representative images of cell expression for nLightG and GRABNE1m. Right, violin plot quantification and comparison of basal brightness between nLightG and GRABNE1m. Medians are represented by the thin dotted lines. n = 46, 37 cells for nLightG, and GRABNE1m, respectively, from 3 independent experiments. n.s., not significant. P = 0.4839. Two-tailed Students t-test with Welch’s correction. c, One-photon fluorescence excitation (λem = 560 nm) and emission (λex = 470 nm) spectra acquired from nLightG-expressing HEK293T cells in the presence (Sat) or absence (Apo) of NE (100 μM). Each trace is the average of four independent experiments. c, same as in (c) for nLightR (λem = 620 nm and λex = 550 nm). Each trace is the average of three independent experiments. Fluorescence excitation and emission were normalized to the respective maximal value in the absence of NE (Apo). e, Two-photon brightness of nLightG imaged in transfected HEK cells grown attached to a glass coverslip in the presence (Sat) or absence (Apo) of NE (100 μM). Spectra are normalized to the value of the Apo form at 950 nm. The ratio between Sat and Apo at all wavelengths is also shown as black dotted line. Each trace is the average of 3 independent experiments. f, Left, normalized fluorescence intensity dose-response curves of NE in nLightG- and nLightR-expressing primary rat cortical neurons. Each titration is normalized to the maximum ΔF/F0 response of the indicator. Datapoints were fitted with four-parameter dose-response curves to determine EC50 values. n = 11 cells for nLightG and n = 3 cells for nLightR from three independent experiments. Right, quantification of maximal ΔF/F0 response of nLightG- and nLightR-expressing primary rat cortical neurons to NE or DA. Both ligands were separately applied at 300 μM concentration on the cells. n = 11-24 cells for nLightG and n = 5 cells for nLightR. Average response to DA was compared to that to NE for each indicator, using a two-tailed Students t-test with Welch’s correction. P values: 3.421x10-9, nLightG; 2.240x10-6, nLightR. g, Maximal fluorescence response of nLightG in HEK293T cells upon addition of different small molecule neurotransmitters (in HBSS) at a final concentration of 10 μM. The mean values were compared to the control (pure HBSS) using Welch ANOVA with Dunett’s multiple comparison test. The data are shown as mean ± SEM. n ≥ 21 cells. P values were as follows: norepinephrine, <0.0001; epinephrine, <0.0001; dopamine, <0.0001; serotonin, 0.066; acetylcholine, 0.999; adenosine, 0.705; histamine, 0.999; γ-aminobutyric acid, 0.999; glutamic acid, 0.999. h, same as in (g) for nLightR. P values were as follows: norepinephrine,<0.0001; epinephrine, <0.0001; dopamine, <0.0001; serotonin, 0.887; acetylcholine, 0.998; adenosine, 0.999; histamine, 0.999; γ-aminobutyric acid, 0.999; glutamic acid, 0.998. All experiments were repeated at least three times with similar results.
Extended Data Fig. 3. Signaling characterization of nLightG and nLightR.
a, Intracellular calcium signaling recorded in HEK293T cells co-expressing nLightG (left) or the swAlpha-1 AR (right) along with the red fluorescent calcium indicator jRGECO1a. Fluorescence response of the calcium indicator was measured at baseline conditions, upon addition of NE (10 μM final concentration) and upon addition of ionomycin (10 μM final concentration), as indicated by the colored bars. Signals were normalized to the maximum response of the same cells after addition of 10 μM ionomycin. n= 19 and 22 cells for swAlpha-1 AR-jRGECO1a and nLightG-jRGECO1a. b, Statistical analysis of the responses shown in (a) for nLightG. Individual data points represent the mean ΔF/F0 response of jRGECO1a for each cell upon addition of NE. Violin plot represents the kernel density estimate of the probability density function for each sample. Mean values before and after addition of NE were compared using a two-tailed Students t-test with Welch’s correction. P = 2.887x10-7. c, Same as in a but for nLightR in combination with the green fluorescent calcium indicator GCaMP6s. n = 21 and 23 cells for swAlpha-1 AR-GCaMP6s and nLightR-GCaMP6s, respectively. d, Same as in b but for the data shown in c for nLightR in combination with GCaMP6s. P = 3.393x10-21. e, Time traces of the luminescence ratio of HEK293T cells co-expressing either nLightG (green traces), the swAlpha-1 AR (grey traces) or the hmDRD1 (blue traces) fused to SmBiT and a mini G-protein or β-arrestin-2 fused to LgBiT. NE (10 μM final concentration) or FluoroBrite DMEM was added to the cells between 490 s and 520 s. The luminescence ratio between stimulated and non-stimulated cells was calculated and normalized to the baseline luminescence ratio before the addition of NE. Each trace is the average of three independent experiments. f, Same as in e but for nLightR (magenta traces). g, Statistical analysis of data shown in e and f. The mean luminescence ratios before and after the addition of NE were calculated and compared using a two-tailed Students t-test with Welch’s correction. P values were as follows: mini-Gq, swAlpha-1 AR, 7.75×10-3; nLightG, 0.590; nLightR, 0.589; mini-Gs, swAlpha-1 AR, 3.88×10-3; nLightG, 0.281; nLightR, 0.410; mini-Gi, swAlpha-1 AR, 0.126; nLightG, 0.792; nLightR, 0.147; β-arrestin-2, swAlpha-1 AR, 8.97×10-2; nLightG, 0.134; nLightR, 0.377. All data are shown as mean ± SEM and all experiments were repeated three times with similar results.
Extended Data Fig. 4. Pharmacological characterization of nLightG and GRABNE1m in anesthetized mice.
a, Schematic representation of viral injections for photometry recordings of nLightG or GRABNE1m in vHPC during optogenetic stimulation of LC in anesthetized mice. b, Experimental protocol for optogenetic stimulation combined with drug injection during isoflurane anaesthesia. c, Left, average traces of normalized signal changes (ΔF/F0 %) of GRABNE1m photometry recordings in response to three LC optical stimulation protocols (5 Hz) pre- and post-yohimbine injection. Signals were normalized to the average peak value pre-yohimbine. The period of optogenetic stimulation is represented with an orange shade. Right, statistical comparison of peak normalized ΔF/F0 % responses to 5Hz LC between pre- and post-yohimbine injection. P = 0.0014, n = 7 mice, one-sample t-test. d, Same as in c for nLightG. n.s., non-significant (P = 0.31), n = 9 mice, one-sample t-test.
Extended Data Fig. 5. Processing of fiber photometry data presented in Figure 4.
a, Processing of an exemplary trace of fiber photometry data in response to optogenetic stimulation (magenta), stimulated and recorded in the same LC. nLightG was excited at wavelengths of 470/10 (“ligand-dependent”) and 405/10 nm (“control”), in a temporally interleaved manner. Note that excitation power was adjusted such that the emission power of both excitation modes was of comparable intensity. Data chunks of 10 minutes, 30 seconds, and 5 seconds duration are shown from left to right, dashed boxes indicate the magnified regions. In a first step, the raw data was fitted with a polynomial function of 1st-4th degree (based on visual inspection; black line) in order to capture the bleaching of each trace (top). Subsequently, each signal was divided by this fit, in order to correct for bleaching and normalize the signals to an amplitude of 1 (center top). In the next step, signals were smoothed with a moving average filter of a 100 ms window size (center bottom). Finally, ΔF/F0 was calculated as the difference between the bleaching-corrected, normalized, smoothed signals excited at 470/10 and 405/10 nm (bottom). While original signals contained substantial artefacts likely to originate from locomotion (e.g. large, slow peaks in the left column of the raw signal (top)) and hemodynamics (most obvious seen by the oscillations of 10-13 Hz likely to originate from the animal’s heartbeat 59; best seen in the bleaching-corrected, unsmoothed trace in the right column), these artefacts can be substantially reduced by subtracting the 405/10 nm excited signal from the 470/10 nm excited signal (bottom). b, Heatmap of 20 individual trials of optogenetic stimulation, corresponding to the processing steps shown in a. c, Mean ± standard deviation of the trials shown in b. d, Correlating the z-scored data across all animals (n = 6) excited at 405/10 nm vs 470/10 nm reveals a strong positive relationship of the functional and the isosbestic channel (Pearson’s correlation coefficient r = 0.61), likely to originate from motion and hemodynamic artefacts, which act in a similar manner on the light emitted by nLightG. e/f, Correlating the z-scored data across all animals (n = 6) excited at 470/10 nm e and 405/10 nm f against the calculated ΔF/F0 revealed strong positive (Pearson’s correlation coefficient r = 0.61) and moderate negative (Pearson’s correlation coefficient r = -0.19) relationships, as expected from the strong increase in emission light upon norepinephrine-binding when exciting nLightG at 470/10 nm, and the milder decrease in emission light upon norepinephrine-binding when exciting nLightG at 405/10 nm. g, Pearson’s correlation coefficients calculated for each animal (n = 6) individually, when correlating 405/10 nm vs 470/10 nm excited signals (left), 470/10 nm vs ΔF/F0 (center), and 405/10 nm vs ΔF/F0 (right). While the positive or negative sign of the correlations depend upon which signal is subtracted from the other (i.e. 470-405 nm vs. 405-470 nm), the strength of correlations as well as the opposing sign of the 470/10 nm vs ΔF/F0 as compared to the 405/10 nm vs ΔF/F0 agree with the expected modulations upon binding of norepinephrine by nLightG.
Extended Data Fig. 6. Running-induced nLightG signals in the mouse hippocampus.
a, Event-triggered averages for individual animals (n = 4) showing the changes in nLightG signal amplitude over the whole field-of-view when the mouse started running for the five consecutive recording days. Signals were recorded using two-photon microscopy. The colors indicate the different animals (subject 1-4). b, Event-triggered averages of running speed of individual animals in the virtual corridor when the mouse started running. Color code as in a. c, Same as in b for lick rate. d, Structural similarity in the time interval displayed in a-c. e, nLightG signal amplitude as a function of running speed for individual animals. f, nLightG signal amplitude as a function of lick rate for individual animals.
Extended Data Fig. 7. nLightG signals associated with reward position in the mouse hippocampus.
a, Event-triggered averages for individual animals (n = 4) showing the changes in nLightG signal amplitude over the whole field-of-view when the mouse crossed the reward position for the five consecutive days of recording. Signals were recorded using two-photon microscopy. The colors indicate the different animals (subject 1-4). b, Event-triggered averages of running speed of individual animals in the virtual corridor when the mouse crossed the reward position. Color code as in a. c, Same as in b for lick rate. d, Structural similarity in the time interval displayed in a-c. e, nLightG signal amplitude as a function of running speed for individual animals. f, nLightG signal amplitude as a function of lick rate for individual animals.
Extended Data Fig. 8. ROI selection for analysis of in vivo two-photon data.
a-b, Representative field-of-view showing hippocampal CA1 neurons expressing nLightG in vivo (same field-of-view as in Fig. 5j). The red solid line in (a) indicates the zoomed in area shown in (b). White boxes indicate regions-of-interest (ROIs) identified within the field-of-view and centered on putative cells using the machine learning algorithm CITE-On30. Scale bar, 50 μm.
Extended Data Fig. 9. Further comparison between newly-developed and published indicators.
a, Basal brightness of LightG and LightR indicator constructs plotted against their fluorescence response ΔF/F0 in HEK293T cells. Basal brightness values reflect the average brightness of indicator-expressing HEK293T cells in the ligand-free state. Grafts containing only the ICL3 module of LightG or LightR are represented as triangles. Grafts containing ICL2 and ICL3 modules of LightG or LightR are represented as circles. Previously published indicators are represented as rectangles. b, Absolute changes in fluorescence (ΔF) of LightG (green) and LightR (red) grafts measured in HEK293T cells for a set of ten GPCRs. n = 21 cells from three independent experiments. Data were obtained from the same imaging experiments shown in Figure 5. c, Heatmap of ΔF for a subset of indicators (those with ΔF > 0.3). Scale bars, 20 μm.
Extended Data Fig. 10. Ligand EC50 measurements for a subset of new indicators.
Normalized fluorescence intensity dose-response curves of AchLightR (AchLightR-DG, hmM3R double graft), AchLightG (AchLightG-SG, hmM3R single-graft, light green; AchLightG-DG, double graft, dark green), HisLightG (hmH4R, double-graft) and AdoLightG (hmA2AR, double-graft) expressed on HEK293T cells and titrated with their endogenous agonists. Datapoints were fitted with four-parameter dose-response curves to determine the EC50 values. n=3 wells per concentration for each ligand. All data are shown as mean ± SEM and all experiments were repeated three times with similar results.
Supplementary Material
Footnotes
AUTHOR CONTRIBUTIONS
T.P. led the study. T.P., V.R. and C.K. conceived the sensor engineering strategy. Z.K., V.R., C.K. performed molecular cloning, in vitro indicator screening and characterization in HEK293T cells and neurons, and analyzed data under the supervision of T.P. J.D. measured two-photon brightness under the supervision of L.R., B.W. and T.P. T.Z., A.R., L.M.W. and L.S. performed and T.Z. and A.R., analyzed patch clamp fluorometry experiments. M.A.B. prepared cortical neuronal cultures under the supervision of D.B. Z.K., M.W. and M.H. performed and analyzed imaging experiments in acute brain slices under the supervision of T.P. Z.K., M.W. and M.H. performed and analyzed in vivo tail-lift experiments under the supervision of T.P. S.N.D., M.W. performed and analyzed optogenetic experiments in ventral hippocampus under the supervision of J.B. and T.P. A.D. performed and analyzed optogenetic and photometry experiments in locus coeruleus and dorsal hippocampus mice under the supervision of J.S.W. S.C. performed and analyzed in vivo two-photon imaging experiments under the supervision of T.F. T.P., Z.K., V.R. and C.K. wrote the manuscript with contributions from all authors.
COMPETING FINANCIAL INTERESTS
T.P. is a co-inventor on a patent application related to the technology described in this article. All other authors have nothing to disclose.
Acknowledgements
The results are part of a project that has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement: 891959) (T.P.). We also acknowledge funding from H2020-ICT (grant agreement: 101016787) (T.F and T.P.), the University of Zürich and the Swiss National Science Foundation (grant agreement: 310030_196455) (T.P.), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, grant agreement: 178316478-B8) (J.S.W), and the NIH BRAIN Initiative (grant agreement: U19 NS107464) (T.F.). We would like to thank Jean-Charles Paterna and the Viral Vector Facility of the Neuroscience Center Zürich (ZNZ) for the kind help with virus production, Stefan Schillemeit for expert technical support with histology, and Kevin Assomou and Miriam Stoeber for cloning and providing the LgBit-mini-G protein fusions and for assistance with NanoBiT complementation assay. We thank Clémentine Lovato for help with perfusion pipette production and Silvia Schweer for cell culture assistance (both Ruhr University Bochum). The plasmids encoding Beta2AR-SmBit and LgBit-β-arrestin-2, as well as the Alexa-647 labelled M1 anti-FLAG antibody were a kind gift from Miriam Stoeber (University of Geneva). The rAAV2/9 carrying DNA encoding ChrimsonR-mRuby2 in a double-floxed inverted reading frame under control of the EF1α-promoter (Addgene viral prep #124603-AAV9) was kindly gifted by Christopher Harvey (Harvard University). The plasmid coding for DRD1-GFP was a kind gift from Dimitrios Stamou (University of Copenhagen).
Data Availability
DNA and protein sequences for indicators developed in this study were deposited on NCBI (accession numbers ON737776 - ON737782) or are available in Supplementary Data S1. DNA plasmids used for viral production have been deposited both on the UZH Viral Vector Facility (https://vvf.ethz.ch/) and on AddGene. Viral vectors can be obtained either from the Patriarchi laboratory, the UZH Viral Vector Facility, or AddGene. Source data are provided with the manuscript. Raw data can be made available upon reasonable request.
Code Availability
Custom MATLAB code is available on https://github.com/patriarchilab/nLightG.
References
- 1.Labouesse MA, Patriarchi T. A versatile GPCR toolkit to track in vivo neuromodulation: not a one-size-fits-all sensor. Neuropsychopharmacology. 2021:1–5. doi: 10.1038/s41386-021-00982-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Z, Lin D, Li Y. Pushing the frontiers: tools for monitoring neurotransmitters and neuromodulators. Nat Rev Neurosci. 2022;23:257–274. doi: 10.1038/s41583-022-00577-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patriarchi T, et al. An expanded palette of dopamine sensors for multiplex imaging in vivo. Nat Methods. 2020;17:1147–1155. doi: 10.1038/s41592-020-0936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun F, et al. Next-generation GRAB sensors for monitoring dopaminergic activity in vivo. Nature Methods. 2020;17:1156–1166. doi: 10.1038/s41592-020-00981-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sara SJ. The locus coeruleus and noradrenergic modulation of cognition. Nat Rev Neurosci. 2009;10:211–223. doi: 10.1038/nrn2573. [DOI] [PubMed] [Google Scholar]
- 6.Aston-Jones G, Cohen JD. An Integrative Theory Of Locus Coeruleus-Norepinephrine Function: Adaptive Gain and Optimal Performance. Annual Review of Neuroscience. 2005;28:403–450. doi: 10.1146/annurev.neuro.28.061604.135709. [DOI] [PubMed] [Google Scholar]
- 7.Feng J, et al. A Genetically Encoded Fluorescent Sensor for Rapid and Specific In Vivo Detection of Norepinephrine. Neuron. 2019;102:745–761.:e8. doi: 10.1016/j.neuron.2019.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patriarchi T, et al. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science. 2018;360:eaat4422. doi: 10.1126/science.aat4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffet L, et al. A genetically encoded sensor for in vivo imaging of orexin neuropeptides. Nat Methods. 2022;19:231–241. doi: 10.1038/s41592-021-01390-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauser AS, et al. GPCR activation mechanisms across classes and macro/microscales. Nat Struct Mol Biol. 2021;28:879–888. doi: 10.1038/s41594-021-00674-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cvicek V, Goddard WA, Abrol R. Structure-Based Sequence Alignment of the Transmembrane Domains of All Human GPCRs: Phylogenetic, Structural and Functional Implications. PLoS Comput Biol. 2016;12:e1004805. doi: 10.1371/journal.pcbi.1004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ballesteros JA, Weinstein H. In: Methods in Neurosciences. Sealfon SC, editor. Vol. 25. Academic Press; 1995. [19] Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors; pp. 366–428. [Google Scholar]
- 13.Kubitschke M, et al. sDarken: Next generation genetically encoded fluorescent sensors for serotonin. 2022:2022.03.10.483799. doi: 10.1101/2022.03.10.483799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen T-W, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu D, Katz A, Lee CH, Simon MI. Activation of phospholipase C by alpha 1-adrenergic receptors is mediated by the alpha subunits of Gq family. J Biol Chem. 1992;267:25798–25802. [PubMed] [Google Scholar]
- 16.Nehmé R, et al. Mini-G proteins: Novel tools for studying GPCRs in their active conformation. PLOS ONE. 2017;12:e0175642. doi: 10.1371/journal.pone.0175642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixon AS, et al. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem Biol. 2016;11:400–408. doi: 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- 18.Morris DP, Price RR, Smith MP, Lei B, Schwinn DA. Cellular Trafficking of Human α1a-Adrenergic Receptors Is Continuous and Primarily Agonist-Independent. Mol Pharmacol. 2004;66:843–854. doi: 10.1124/mol.104.000430. [DOI] [PubMed] [Google Scholar]
- 19.Chalothorn D, et al. Differences in the Cellular Localization and Agonist-Mediated Internalization Properties of the α1-Adrenoceptor Subtypes. Mol Pharmacol. 2002;61:1008–1016. doi: 10.1124/mol.61.5.1008. [DOI] [PubMed] [Google Scholar]
- 20.Stanasila L, Abuin L, Dey J, Cotecchia S. Different Internalization Properties of the α1a-and α1b-Adrenergic Receptor Subtypes: The Potential Role of Receptor Interaction with β-Arrestins and AP50. Mol Pharmacol. 2008;74:562–573. doi: 10.1124/mol.107.043422. [DOI] [PubMed] [Google Scholar]
- 21.Nörenberg W, Schöffel E, Szabo B, Starke K. Subtype determination of soma-dendritic alpha2-autoreceptors in slices of rat locus coeruleus. Naunyn Schmiedebergs Arch Pharmacol. 1997;356:159–165. doi: 10.1007/pl00005036. [DOI] [PubMed] [Google Scholar]
- 22.Starke K, Göthert M, Kilbinger H. Modulation of neurotransmitter release by presynaptic autoreceptors. Physiol Rev. 1989;69:864–989. doi: 10.1152/physrev.1989.69.3.864. [DOI] [PubMed] [Google Scholar]
- 23.Simson PE, Weiss JM. Alpha-2 receptor blockade increases responsiveness of locus coeruleus neurons to excitatory stimulation. J Neurosci. 1987;7:1732–1740. doi: 10.1523/JNEUROSCI.07-06-01732.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J, Kile BM, Mark Wightman R. In vivo voltammetric monitoring of norepinephrine release in the rat ventral bed nucleus of the stria terminalis and anteroventral thalamic nucleus. European Journal of Neuroscience. 2009;30:2121–2133. doi: 10.1111/j.1460-9568.2009.07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandler DJ, et al. Redefining Noradrenergic Neuromodulation of Behavior: Impacts of a Modular Locus Coeruleus Architecture. J Neurosci. 2019;39:8239–8249. doi: 10.1523/JNEUROSCI.1164-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones BE, Halaris AE, McIlhany M, Moore RY. Ascending projections of the locus coeruleus in the rat. I. axonal transport in central noradrenaline neurons. Brain Research. 1977;127:1–21. doi: 10.1016/0006-8993(77)90377-8. [DOI] [PubMed] [Google Scholar]
- 27.Kaufman AM, Geiller T, Losonczy A. A Role for the Locus Coeruleus in Hippocampal CA1 Place Cell Reorganization during Spatial Reward Learning. Neuron. 2020;105:1018–1026.:e4. doi: 10.1016/j.neuron.2019.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray SR, Ye L, Ye JY, Paukert M. Noradrenergic terminal short-term potentiation enables modality-selective integration of sensory input and vigilance state. Science Advances. 2021;7:eabk1378. doi: 10.1126/sciadv.abk1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sità L, et al. A deep-learning approach for online cell identification and trace extraction in functional two-photon calcium imaging. Nat Commun. 2022;13:1529. doi: 10.1038/s41467-022-29180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong A, et al. A fluorescent sensor for spatiotemporally resolved imaging of endocannabinoid dynamics in vivo. Nat Biotechnol. 2021:1–12. doi: 10.1038/s41587-021-01074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Z, et al. A sensitive GRAB sensor for detecting extracellular ATP in vitro and in vivo. Neuron. 2021:S0896627321009880. doi: 10.1016/j.neuron.2021.11.027. [DOI] [PubMed] [Google Scholar]
- 32.Jing M, et al. An optimized acetylcholine sensor for monitoring in vivo cholinergic activity. Nat Methods. 2020;17:1139–1146. doi: 10.1038/s41592-020-0953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ino D, Tanaka Y, Hibino H, Nishiyama M. A fluorescent sensor for real-time measurement of extracellular oxytocin dynamics in the brain. Nat Methods. 2022:1–9. doi: 10.1038/s41592-022-01597-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice ME, Patel JC, Cragg SJ. Dopamine release in the basal ganglia. Neuroscience. 2011;198:112–137. doi: 10.1016/j.neuroscience.2011.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C, Goel P, Kaeser PS. Spatial and temporal scales of dopamine transmission. Nat Rev Neurosci. 2021;22:345–358. doi: 10.1038/s41583-021-00455-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall JD, et al. Cell-Type-Specific Optical Recording of Membrane Voltage Dynamics in Freely Moving Mice. Cell. 2016;167:1650–1662.:e15. doi: 10.1016/j.cell.2016.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W-T, et al. Spectral fiber photometry derives hemoglobin concentration changes for accurate measurement of fluorescent sensor activity. Cell Rep Methods. 2022;2:100243. doi: 10.1016/j.crmeth.2022.100243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Formozov A, Dieter A, Wiegert JS. A flexible and versatile system for multi-color fiber photometry and optogenetic manipulation. Cell Rep Methods. 2023;3:100418. doi: 10.1016/j.crmeth.2023.100418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirdita M, et al. ColabFold: making protein folding accessible to all. Nat Methods. 2022:1–4. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jumper J, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duffet L, et al. A genetically encoded sensor for in vivo imaging of orexin neuropeptides. Nat Methods. 2022;19:231–241. doi: 10.1038/s41592-021-01390-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wan Q, et al. Mini G protein probes for active G protein-coupled receptors (GPCRs) in live cells. J Biol Chem. 2018;293:7466–7473. doi: 10.1074/jbc.RA118.001975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laschet C, Dupuis N, Hanson J. A dynamic and screening-compatible nanoluciferase-based complementation assay enables profiling of individual GPCR-G protein interactions. Journal of Biological Chemistry. 2019;294:4079–4090. doi: 10.1074/jbc.RA118.006231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pollok S, Reiner A. Subunit-selective iGluR antagonists can potentiate heteromeric receptor responses by blocking desensitization. Proceedings of the National Academy of Sciences. 2020;117:25851–25858. doi: 10.1073/pnas.2007471117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edelstein AD, et al. Advanced methods of microscope control using μManager software. Journal of Biological Methods. 2014;1:e10-e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tillage RP, et al. Elimination of galanin synthesis in noradrenergic neurons reduces galanin in select brain areas and promotes active coping behaviors. Brain Struct Funct. 2020;225:785–803. doi: 10.1007/s00429-020-02035-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parlato R, Otto C, Begus Y, Stotz S, Schütz G. Specific ablation of the transcription factor CREB in sympathetic neurons surprisingly protects against developmentally regulated apoptosis. Development. 2007;134:1663–1670. doi: 10.1242/dev.02838. [DOI] [PubMed] [Google Scholar]
- 49.Curreli S, Bonato J, Romanzi S, Panzeri S, Fellin T. Complementary encoding of spatial information in hippocampal astrocytes. PLOS Biology. 2022;20:e3001530. doi: 10.1371/journal.pbio.3001530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dombeck DA, Harvey CD, Tian L, Looger LL, Tank DW. Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat Neurosci. 2010;13:1433–1440. doi: 10.1038/nn.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheffield MEJ, Dombeck DA. Calcium transient prevalence across the dendritic arbour predicts place field properties. Nature. 2015;517:200–204. doi: 10.1038/nature13871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akam T, Walton ME. pyPhotometry: Open source Python based hardware and software for fiber photometry data acquisition. Sci Rep. 2019;9:3521. doi: 10.1038/s41598-019-39724-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang W, et al. Anesthetics fragment hippocampal network activity, alter spine dynamics, and affect memory consolidation. PLOS Biology. 2021;19:e3001146. doi: 10.1371/journal.pbio.3001146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mathis A, et al. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat Neurosci. 2018;21:1281–1289. doi: 10.1038/s41593-018-0209-y. [DOI] [PubMed] [Google Scholar]
- 55.Guizar-Sicairos M, Thurman ST, Fienup JR. Efficient subpixel image registration algorithms. Opt Lett, OL. 2008;33:156–158. doi: 10.1364/ol.33.000156. [DOI] [PubMed] [Google Scholar]
- 56.van der Walt S, et al. scikit-image: image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Avanaki AN. Exact global histogram specification optimized for structural similarity. OPT REV. 2009;16:613–621. [Google Scholar]
- 58.Virtanen P, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA and protein sequences for indicators developed in this study were deposited on NCBI (accession numbers ON737776 - ON737782) or are available in Supplementary Data S1. DNA plasmids used for viral production have been deposited both on the UZH Viral Vector Facility (https://vvf.ethz.ch/) and on AddGene. Viral vectors can be obtained either from the Patriarchi laboratory, the UZH Viral Vector Facility, or AddGene. Source data are provided with the manuscript. Raw data can be made available upon reasonable request.