Abstract
Social preference, the decision to interact with one member of the same species over another, is a key feature of optimizing social interactions. Thus, adult rodents favor interacting with novel conspecifics over familiar ones but whether this preference for social novelty stems from neural circuits facilitating interactions with novel conspecifics or suppressing interactions with familiar ones remains unknown. Here, we identify neurons in the infra-limbic area (ILA) of the mouse prefrontal cortex that express the neuropeptide corticotropin-releasing hormone (CRH) and project to the dorsal region of the rostral lateral septum (rLS). We show how release of CRH during familiar encounters disinhibits rLS neurons, thereby suppressing social interactions with familiar mice and contributing to social novelty preference. We further demonstrate how the maturation of CRH expression in ILA during the first two post-natal weeks enables the developmental shift from a preference for littermates in juveniles to a preference for novel mice in adults. Taken together, our findings suggest that the developmental maturation of CRH in ILA and its later release onto rLS is critical for controlling the preference for socially novel encounters exhibited by adult mice.
Keywords: social novelty preference, lateral septum, pre-frontal cortex, corticotropinreleasing hormone, CRF
Introduction
Social preference, the decision to interact with one conspecific over another, is a feature displayed by gregarious animals, which is critical to navigate their social space1,2. Adult rodents prefer to interact with their kin3,4, individuals from specific strains5 and members of the opposite sex6–9. In addition to innate factors (e.g., kin, strain, and sex), social preference is also influenced by social memory10, social hierarchy9,11,12 and the affective state of the conspecific13. Thus, adult rodents display social novelty preference (SNP), choosing to interact with novel individuals over familiar ones10. For the last two decades, social novelty preference has been used as a proxy to assess social memory14–16 but the neuronal circuits mediating social novelty preference remain elusive. In particular, it is unknown whether social novelty preference is due solely to a rewarding signal for novel social interactions17 or also involves the suppression of exploration of familiar individuals. We hypothesized the existence of neuronal circuits promoting the avoidance of familiar mice and therefore contributing to social novelty preference when novel and familiar mice are presented simultaneously.
Memory-based preferences, such as social novelty preference, also have a developmental window18 and can change during the life of altricial animals. For example, young mice prefer their mother to unfamiliar dams until weaning when they begin to prefer unfamiliar dams over their mother19. Similarly, rat pups display a preference for their familiar siblings during the first 2 postnatal weeks, after which the preference shifts toward novel pups3,4. Although the mechanisms that regulate these developmental shifts remain elusive, the lateral septum (LS), a brain region associated with the regulation of motivated behaviors including social interactions20, is necessary for kinship/familiarity preference in young rats3 as well as for social novelty preference in adult rodents20–22. Moreover, the ventral aspect of medial prefrontal cortex (mPFC), the infra-limbic area (ILA), is known for its involvement in decision-making, responds to social stimuli23–25 and is also necessary for social novelty preference26,27. The mPFC projects to LS to regulate food-seeking behavior28 but how these regions integrate social memory cues and communicate to regulate social interactions is still unclear.
Corticotropin-releasing hormone (CRH)29, a 41 amino acid peptide, regulates several processes including homeostatic and allostatic neuroendocrine mechanisms, memory30 and social behaviors in non-stressful context31,32. In humans, CRH is implicated in psychiatric disorders associated with social deficits such as depression33,34 and social phobia35. In rodents, systemic manipulations of the CRH system impair social interactions32,36–44. Given that CRH is expressed in ILA45 and CRHR1 is expressed in LS46, we hypothesized that CRH release from ILA to LS is involved in regulating social interactions and therefore social novelty preference.
We demonstrate through a combination of electrophysiological, chemogenetic, optogenetic, calcium recording and gene silencing techniques that the release of CRH from ILA neurons (ILACRH neurons) into the rostral region of LS (rLS) suppresses social interaction with familiar mice. This circuit therefore regulates familiarization (decrease in interaction as a novel rodent becomes familiar) and contributes to the social novelty preference exhibited by adult mice. In addition, we find that the increase in ILACRH neuron density during the second postnatal week is responsible for a developmental shift in the social preference of young mice from familiar to novel conspecifics.
Results
ILACRH cells project to the rostro-dorsal lateral septum
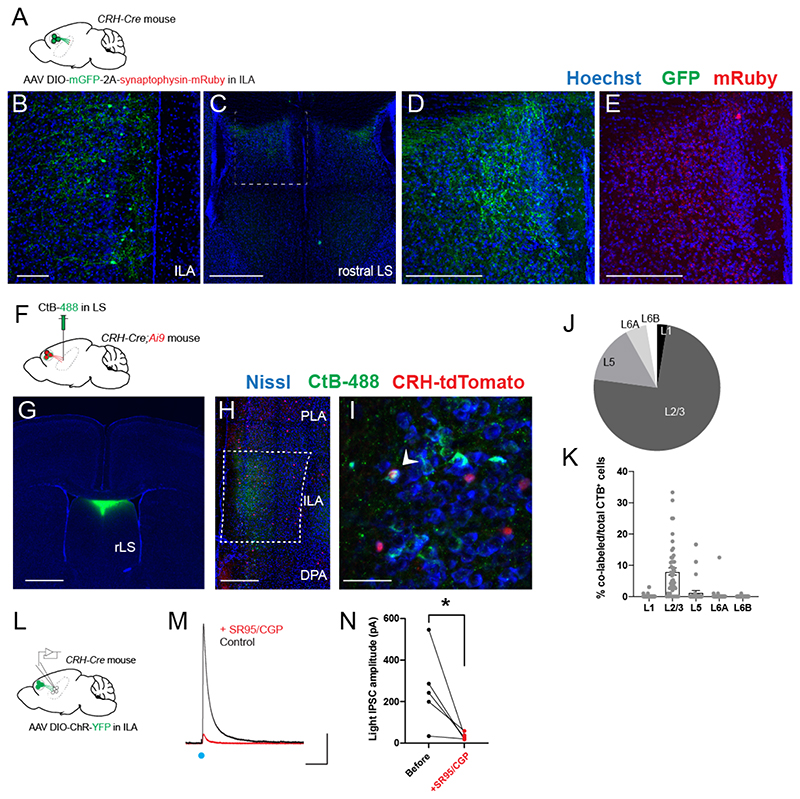
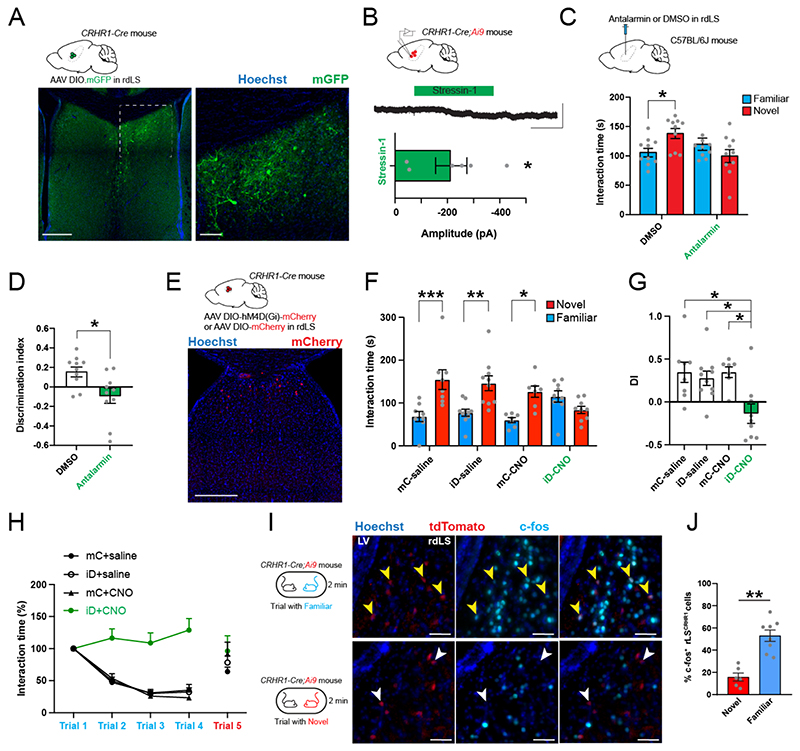
We injected CRH-Cre mice in ILA47 with a Cre-dependent adeno-associated virus (AAV) expressing membranous GFP and synaptophysin tagged with mRuby in order to visualize axons and synaptic terminals respectively (Fig. 1A-B). We observed GFP+ fibers in the rostral-dorsal region of the lateral septum (rdLS, (Fig. 1C-D). A closer examination confirmed the presence of mRuby-labeled axon terminals in this region (Fig. 1E). We did not observe fibers going to posterior LS or other brain regions (Fig. S1A-C). In addition, we did not observe fibers in rdLS following injections in nearby mPFC regions (pre-limbic or anterior cingulate area, Fig. S1D). Next, we injected the retrograde marker CtB-488 in rdLS of CRH-Cre mice crossed with a Cre-dependent tdTomato reporter line (CRH-Cre;Ai9 mice) to visualize CRH+ cells (Fig. 1F-G). tdTomato+ cells were distributed evenly throughout the rostro-caudal axis of ILA (Fig. S2). CtB retrogradely labelled many ILA neurons mostly located in layer 2/3 (Fig. 1H, J). Some CtB+ cells co-expressed tdTomato (Fig. 1I) and were mainly found in layer 2/3 of ILA (Fig. 1K). We confirmed this result by injecting a Cre-dependent retrograde monosynaptic herpes simplex virus expressing GFP in the rdLS of CRH-Cre mice (Fig. S3A). Consistent with our CtB injections, 79% of CRH/GFP+ cells were located in ILA with the rest being located in adjacent regions (Fig. S3B-C). Within ILA, 66% of GFP+ cells were located in layer 2/3 (Fig. S3D). Overall, these experiments show that ILACRH cells from layer 2/3 project to rdLS.
Figure 1. ILACRH cells project to rostro-dorsal LS.
A. CRH-Cre mice injected in ILA with AAV2/DJ hSyn.FLEX.mGFP.2A.Synatophysin-mRuby. B-E. Immunohistochemistry images of a ILA (B) and LS (C-E) sections labeled for GFP (B-D) or mRuby (E). Scale bars: 100 μm (B), 500 μm (C) and 200 μm (D-E). F. CRH-Cre;Ai9 mice injected in rdLS with CtB-488. G. Image of a coronal brain section containing the injection site. Scale bar: 1 mm. H-I. Images of coronal brain sections containing the mPFC. White arrowheads indicate CtB+/CRH+ cells. Scale bar: 400 μm (H), 50 μm (I). J. Distribution of CtB+ cells in ILA (6mice). K. Percentage of co-labeled tdTomato+ and CtB+ over the total number of CtB+ cells per ILA layer. Each point from a different section. 4 sections / mice, N = 6 mice. Bar graph represents mean ± S.E.M. Nested one-way ANOVA, F4,25 = 24.20, p < 0.0001. L. CRH-Cre mice injected in ILA with AAV2/9 EF1a.DIO.hChR2(E123T/T159C)-eYFP. M. Electrophysiological traces from a rdLS neuron recorded in voltage-clamp configuration at +10 mV. Blue dot, stimulation of CRH+ fibers expressing Channelrhodopsin using 1 ms pulse of blue light to elicit an IPSC before and after application of SR95 and CGP. Scale bars: 100 pA and 100 ms. N. IPSC amplitude before and after SR95 and CGP application (5 cells in 4 mice; paired t test, p = 0.04).
We labeled sections from the same mice for GABA and observed that 89% of GFP+ ILACRH cells are positive for GABA (Fig. S3E). Furthermore, using in situ hybridization markers for excitatory and inhibitory neurons, we found that 92% of ILACRH cells expressed the mRNA for glutamic acid decarboxylase 2 (Gad2) while only 3% expressed the mRNA for the vesicular glutamate transporter 1 (Slc17a7/VGlut1) (Fig. S3G-H), confirming the identity of these neurons as GABAergic. We also patched rdLS neurons in septal slices obtained from CRH-Cre mice injected with a Cre-dependent AAV expressing Channelrhodopsin in ILA (Fig. 1L). Stimulation with blue light elicited a large outward current when holding the neurons at + 10 mV (Fig. 1M) No inward currents were detected at -70 mV. The light-induced IPSCs following stimulation of CRH+ fibers from ILA were abolished upon application of 2 μM SR 95531 and 1 μM CGP 55845 which block GABAA and GABAB receptors, respectively (Fig. 1N). Overall, these results show that ILACRH neurons projecting to rdLS are a sub-population of GABAergic neurons.
ILACRH neurons suppress social interactions with familiar mice and support social novelty preference
Next, we used a chemogenetic approach to modulate the activity of ILACRH neurons and probe their behavioral function. We injected CRH-Cre mice in ILA with Cre-dependent AAVs expressing an inhibitory DREADD (designer receptor exclusively activated by designer drugs) tagged with mCherry (iDREADD) or mCherry only as a control (Fig. 2A-B). Three weeks later, mice were intra-peritoneally injected with the DREADD agonist clozapine N-oxide (CNO, 5 mg/kg) 30 min prior to conducting the behavioral tests. Since a previous study associated CRH+ cells in the pre-limbic area of the mPFC with anxiety45, we first tested the mice in the open-field to assess locomotion and anxiety (Fig. S4A). Silencing ILACRH cells had no effect on the distance travelled, the time spent in the center or the surround or the ratio of time spent in the center vs. surround (Fig. S4B-D). Next, we examined the effect of silencing in the elevated plus maze test of anxiety and found no effect on the number of entries or time spent in the open arms relative to the closed ones (Fig. S4E-I). Finally, since glutamatergic cells in the PFC projecting to LS have been reported to be involved in food-seeking behavior, we performed the anxiety-suppressed feeding behavior test, where a food-deprived mouse must venture into the center of an open-field in order to eat (Fig. S4J). Silencing ILACRH cells had no effect on the latency to feed, the time spent feeding or the number of entries into the food zone (Fig. S4K-M). These controls suggest that ILACRH cells are functionally distinct neurons without a prominent function in locomotion, anxiety or feeding-related behaviors.
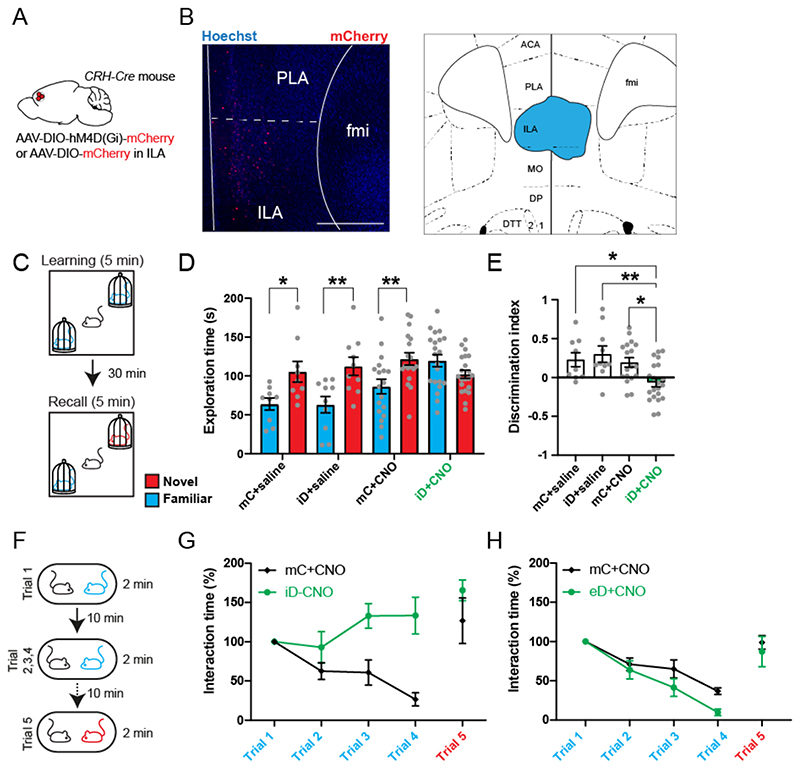
Figure 2. ILACRH cells support social novelty preference and familiarization.
A. CRH-Cre mice injected in ILA with AAV2/8 hSyn.DIO.hM4D(Gi)-mCherry (iDREADD) or AAV2/8 hSyn.DIO.mCherry. B. Left: immunohistochemistry image showing the extent of iDREADD expression. Scale bar: 500 μm. Right: maximal extent of iDREADD expression across several mPFC sections. C. Schematic of the social novelty preference test. D. Interaction time with novel (red) or familiar (blue) mouse during recall in mice expressing mCherry (mC) or hM4Di (iD). Both groups were injected with saline (left groups) or 5 mg/kg of the DREADD agonist CNO (right groups). Grey dots are different mice. 3-way ANOVA F(novelty x injection x virus)1,108 = 3.471, p = 0.02. Sidak’s multiple comparison tests novel vs. familiar: mC + saline, p = 0.04; iD + saline, p = 0.006; mC + CNO, p = 0.008; iD + CNO, p = 0.3. E. Discrimination indexes for social novelty preference of the four groups during recall trial. One-sample t tests compared to 0: mC + saline, p = 0.04; iD + saline, p = 0.02; mC + CNO, p = 0.006; iD + CNO, p =0.2. 2-way ANOVA: F(virus x injection)1, 54 = 4.7, p = 0.03; F(virus)1, 54 = 7.1, p = 0.01; F(injection)1, 54 = 1.5, p = 0.2. Tukey’s multiple comparison tests compared to the iD + CNO group: mC + saline, p = 0.04; iD + saline, p = 0.004; mC + CNO, p = 0.02. F. Schematic of the repetitive social presentation test. G. Normalized interaction times during social presentations (inhibitory DREADD-expressing mice and controls injected with CNO). 8 mice per group. Two-way ANOVA, F(triali1-4 x virus)3,55 = 5.44, p = 0.002; F(trial)3,55 = 1.28, p = 0.2; F(virus)3,55 = 26.82, p < 0.0001. Post-hoc Sidak’s multiple comparison tests between mC and iD groups, trial 2 p = 0.4, trial 3 p = 0.004 and trial 4 p < 0.0001. H. Normalized interaction times during repetitive social presentation test in CRH-Cre mice injected in ILA with AAV5 hSyn.DIO.hM3D(Gq)-mCherry (excitatory DREADD) or with AAV5 hSyn.DIO.mCherry as a control. 8 mice per group. Two-way ANOVA: F(triali1-4 x virus)3,56 = 1.36, p = 0.26; F(trial)3,56 = 33.05, p < 0.0001; F(virus)3,56 = 6.765, p = 0.012.
We next tested whether ILACRH cells regulate social interactions. The mPFC is known to regulate sociability, social preference, social hierarchy as well as emotion discrimination13,26,27 but it remains unclear whether specific sub-regions or populations control different facets of social interactions. First, we silenced ILACRH cells and assessed the sociability of the mice (preference for a mouse compared to an object, Fig. S5A)14. Both groups exhibited a strong preference for the mouse compared to the object (Fig. S5B-C). Next, we tested whether ILACRH cells regulate social novelty preference (Fig. 2C). A subject mouse was exposed to two novel stimulus mice inside wire cup cages in opposite corners of a squared open arena. After 5 min exploring both mice (learning trial), the subject mouse was removed from the arena, placed into an empty housing cage, and one of the two stimulus mice was replaced by a third (novel) mouse. After a 30 min inter-trial interval the subject mouse was reintroduced in the arena (recall trial). Social novelty preference is manifest when the subject mouse spends more time exploring the novel stimulus mouse compared to the familiar one during the recall trial. We also quantified the preference for exploring the novel mouse by calculating a discrimination index (DI), representing the percentage of extra time the subject mouse spent with the novel compared to the familiar stimulus (see Methods). We measured the social novelty preference of mice in which iDREADD-expressing ILACRH cells were silenced from the start of the task by injecting CNO systemically 30 min prior to the learning trial. To rule out off-target effects of CNO or of iDREADD expression alone, we examined three control groups of mice: 1. mice injected with CNO expressing mCherry in ILACRH cells, 2. Mice injected with saline expressing iDREADD in ILACRH cells; 3. Mice injected with saline expressing mCherry in ILACRH cells. During recall, the three control groups (mCherry + saline, iDREADD + saline, mCherry + CNO) exhibited a higher interaction time with the novel mouse compared to the familiar one (Fig. 2D), which translated into a high discrimination index preference for the novel mouse (Fig. 2E), indicating intact social novelty preference. However, in the test group in which ILACRH cells were silenced, the subject mice explored the novel and familiar mice to the same extent (Fig. 2D). As a result, the discrimination index for social novelty preference was not different from zero (Fig. 2E). During learning or recall, the total exploration time of the mice was similar across groups (Fig. S5D-E), suggesting that ILACRH cell silencing does not affect the motivation to explore. Overall, this experiment shows that ILACRH cells are necessary for social novelty preference.
How ILACRH cells regulate social novelty preference is however unclear. Are they regulating social memory or rather processes that utilize social memory cues such as social novelty preference? Because the mPFC is involved in executive functions27, we hypothesized that ILACRH cells leverage social memory cues to promote social novelty preference by regulating social interactions with novel and/or familiar mice. Specifically, do ILACRH cells support social novelty preference by promoting interactions with the novel mouse or by suppressing interactions with the familiar one? During the learning phase of the social novelty preference test, test mice explored each novel conspecific to the same extent than control mice (Fig. S5F), suggesting that silencing ILACRH cells does not impair social interactions with novel animals. We therefore tested the role of ILACRH cells during the repetitive social interaction test (also known as the habituation/dishabituation test), where a sex- and age-matched novel mouse is presented 4 times to the test mouse (Fig. 2F). This test offers the advantage of observing the evolution of social interaction with a single novel mouse becoming gradually familiar. Control mice showed a progressive decrease in interaction time with repeated presentations of the mouse (Fig. 2G). When a novel mouse was presented in the final fifth trial, the interaction time jumped back to its initial level, demonstrating that the decreased interaction was not due to fatigue or loss of engagement in the task. In contrast, mice expressing iDREADD showed no decrease in interaction time during the repeated presentations, suggesting that ILACRH cells are necessary for social familiarization (Fig. 2G). We repeated the experiments injecting saline instead of CNO and both groups exhibited a steady decrease in interaction (Fig. S5G). Next, we asked whether over-activating ILACRH cells could conversely promote social familiarization and repeated the repetitive social presentation test with mice expressing an excitatory DREADD in ILACRH neurons. Increasing the activity of ILACRH cells with CNO slightly facilitated the decrease in social interaction (Fig. 2H), which was not observed when mice were injected with saline (Fig. S5H), indicating that ILACRH cells can bidirectionally modulate the interaction time with familiar mice. Taken together, these experiments suggest that ILACRH cells repress social interaction with a familiar mouse and are necessary for social familiarization.
To confirm our chemogenetic approach, CRH-Cre mice expressing excitatory or inhibitory DREADD in ILA were injected with CNO or saline before presenting them with a familiar animal (Fig. S6A). We measured the overlap between c-fos and mCherry expression in ILA (Fig. S6B-C). iDREADD-expressing mice given CNO exhibited less c-fos/mCherry+ cells compared to the saline control, suggesting efficient ILACRH cells silencing. By contrast, eDREADD-expressing mice injected with CNO showed an increased c-fos/mCherry+ overlap, suggesting ILACRH cell excitation. Do ILACRH cells specifically control social interactions or does it extend to objects as well? We performed tests of novel object recognition, repetitive object presentation and familiar food preference while silencing the ILACRH and found no effect (Fig. S7), indicating that ILACRH neurons specifically regulate social preferences.
ILACRH neurons respond preferentially during familiar social interactions
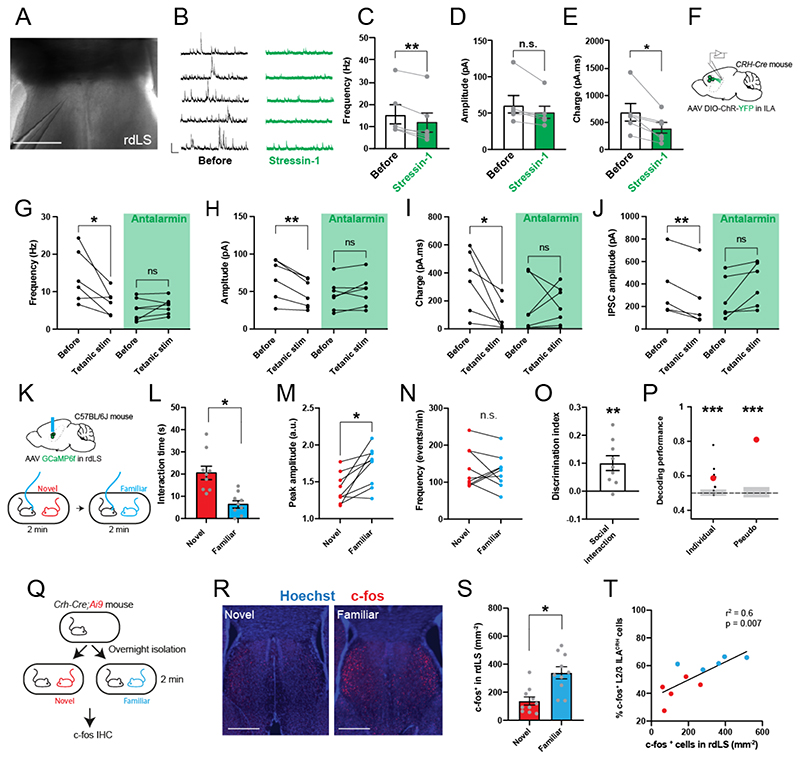
If ILACRH cells regulate social interactions with familiar mice, we can expect the cells to be more active when the mouse interacts with familiar mice than with novel ones. To test this prediction, we performed fiber-photometry of ILACRH cells. We injected the ILA of CRH-Cre mice with a Cre-dependent AAV expressing the calcium sensor GCaMP6f and implanted an optical ferrule above ILA (Fig. 3A and S8A). Subject mice were presented with novel then familiar mice meanwhile we recorded the calcium activity of the cell population (Fig. 3A-C). First, we calculated the peri-stimulus time histogram using the start of social interaction to synchronize traces (Fig. 3D) and found that familiar mouse presentation elicited a large increase of the calcium response while presentation of a novel mouse elicited a small decrease (Fig. 3E). Then, we automatically detected calcium transients and measured their average amplitude and frequency during each trial. The peaks were higher during familiar compared to novel mouse presentation (Fig. 3F), but we saw no difference in the frequency of events (Fig. 3G). We inverted the order of social presentation and obtained the same results (Fig. 3H). These observations suggest that ILACRH neuron activity is increased during familiar encounters compared to novel ones.
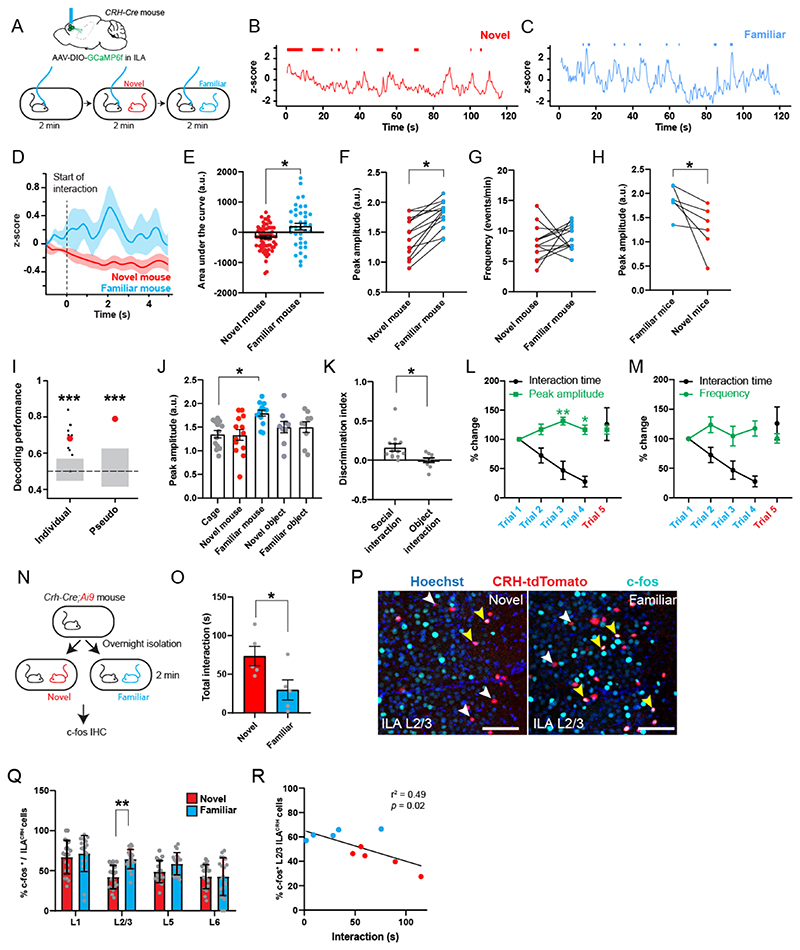
Figure 3. ILACRH cells respond preferentially to familiar mouse presentation.
A. CRH-Cre mice injected in ILA with AAV2/1 syn.FLEX.GCaMP6f and implanted with an optical ferrule over ILA (top). Schematic of the fiber-photometry recording experiment (bottom). B-C. Example traces of recording during presentation of a novel (B) or familiar (C) mouse to the same test mouse. Interaction bouts intervals are shown above each trace. D. Peri-stimulus time histogram during social interaction with novel or familiar mouse, 5 mice. E. Area under the curve during familiar and novel mouse interaction. Each point is an interaction, 5 mice. One-sample nested t tests: Familiar response vs. 0, p = 0.003, Novel responses vs. 0, p = 0.005. Nested t test between groups: p = 0.01. F. Average peak amplitude of the z-score during social presentations of a novel then familiar mouse. For panels F, G and J, each dot is a different recording session using 5 mice. Nested t test between groups: p = 0.02. G. Frequency of calcium events during presentation of a novel then familiar mouse. Nested t test: p = 0.4. H. Average peak amplitude of the z-score during inverted presentation of a familiar then a novel mouse. 3 mice per groups. 2 observations per mice. Nested t test, p = 0.03. I. Decoding performance for familiarity versus novelty from individual recordings or pseudo-simultaneous data. Small black dots on the left are the results from individual recording sessions (N = 5 mice), using 20 cross-validation iterations, large red dot is the average. Red dot on the right is the result of pseudo-population analysis from 100 cross-validation iterations. Grey areas denote chance level computed using permutation tests (2.5 – 97.5 percentiles in distribution of shuffled decoding performances). In both cases, statistical significance is determined by the probability of drawing the observed decoding performance from the distribution of shuffled decoding performances (null-hypothesis). p < 0.001 (two-tailed permutation test, see Methods). J. Average peak amplitude during each type of presentation (novel then familiar experiments only). Nested One-way ANOVA F4,16 = 24.20 followed by Tukey’s multiple comparison test: cage vs. novel mouse p = 0.9; cage vs. familiar mouse p = 0.03; cage vs. novel object p = 0.7; cage vs. familiar object p = 0.7. K. Discrimination indexes for familiarity preference calculated from z-scores during mouse or object presentation. Each point is one recording session. N = 5 mice. One-sample t tests compared to 0: p = 0.001 and p = 0.3 respectively. Unpaired t test between groups taking the average value per mice: p = 0.03. L. Fiber-photometry recording during repetitive social presentation test (10 sessions in 5 mice). One-sample t tests compared to: trial 1 p = 0.06; trial 3 p = 0.002 and trial 4 p = 0.04. M. Frequency of calcium events during repetitive social presentation test (10 sessions in 5 mice). N. CRH-Cre;Ai9 mice were presented with novel or familiar mice after overnight isolation before being processed for immunohistochemistry. O. Interaction times following 2 min social presentation. Unpaired t test novel vs. familiar interaction time, p = 0.04. P. Immunohistochemistry images of c-fos labelling in ILA layer 2/3. Yellow arrowheads: c-fos+ / tdTomato+ cells. White arrowheads: c-fos- / tdTomato+ cells. Scale bars: 100 μm. Q. Percentage of ILACRH cells positive for c-fos per layer. Each point corresponds to each side of 2 sections. 5 mice per group. Nested t test, p = 0.003. R. Percentage of layer 2/3 ILACRH cells positive for c-fos vs. interaction time during social interaction with novel (red) or familiar (blue) mouse. Each point represents one mouse. For the entire figure, bar graphs represent mean ± S.E.M.
To confirm whether ILACRH cell activity differs during novel and familiar mouse presentation, we trained linear classifiers to discriminate between interactions with a novel or familiar mouse using our fiberphotometry recordings. We implemented 2 classifiers using either individual recording sessions (individual) or a meta-session pooling all sessions (pseudo-simultaneous, Fig. 3I, see Methods)48. For each classifier, we also computed chance levels using permutation tests (grey bars). Most individual recording sessions yielded a decoding performance above chance with an average 68% accuracy. The pseudo-simultaneous data yielded a decoding performance even higher (79%). These results show that the ILACRH cell population can code for social familiarity.
We also presented novel and familiar objects and saw no change in activity compared to baseline (Fig. 3J) or between novel and familiar object (Fig. S8B-D). Using the peak amplitudes, we calculated the discrimination indexes (DI) for familiarity preference following object or social presentation (Fig. 3K). DI of social interaction showed a strong preference for social familiarity, unlike the one for object interaction. We then recorded ILACRH cells during the repetitive social presentation test and found the peak amplitude to increase during familiarization (Fig. 3L). The frequency of events however remained stable (Fig. 3M), similar to what was observed previously (Fig. 3G).
As a further assay of neuronal activity, we measured the expression of the immediate-early gene c-fos. CRH-Cre;Ai9 mice were presented with a novel or familiar mouse for 2 min (Fig. 3N). As expected, mice interacted more with novel than familiar mice (Fig. 3O). Mice were perfused 1 hour later and processed for c-fos immunohistochemistry in order to count the number of ILACRH neurons expressing c-fos (Fig. 3P). Despite shorter interactions, ILACRH cells in layer 2/3 exhibited higher c-fos expression following encounters with familiar mice compared to novel (Fig. 3Q), similar to what was already reported for the entire ILA cell population23. Indeed, the activation of layer 2/3 ILACRH cells negatively correlated with the amount of social interaction (Fig. 3R). Overall, our experiments demonstrate that ILACRH cells are more active during interaction with a familiar mouse than a novel one.
CRH release in rLS suppresses social interactions with familiar mice to promote social novelty preference
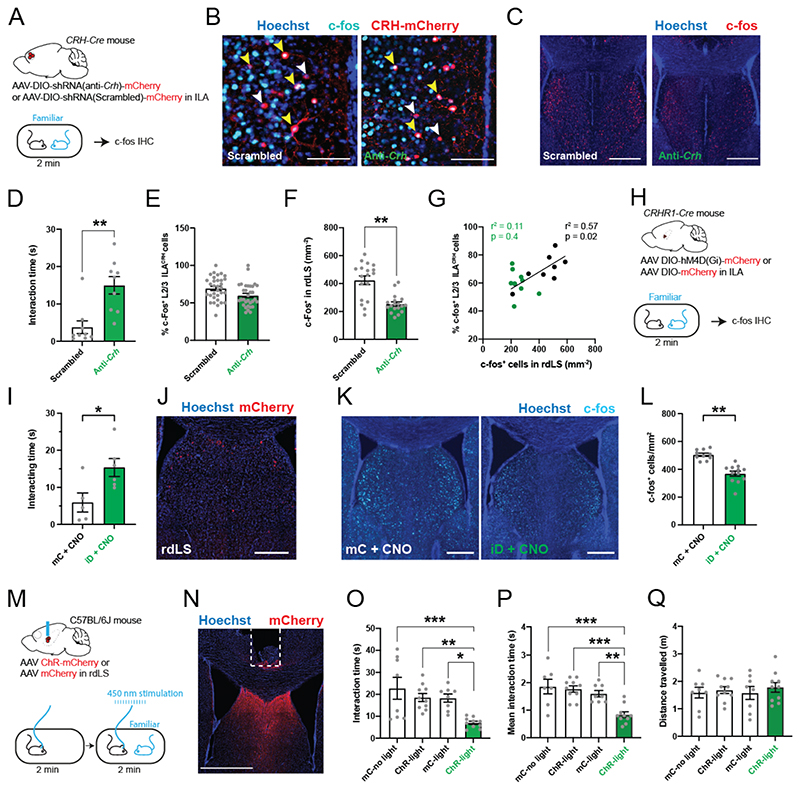
How could ILACRH cells suppress social interactions with a familiar mouse? ILACRH cells projects to rLS which expresses the CRH type-1 receptor (CRHR1)20 and regulates social interactions20, which led us to ask whether CRH release from ILA into rLS is necessary for familiarization. We designed a Cre-dependent shRNA against Crh (see Methods) and expressed it into ILACRH neurons (Fig. 4A, top). To assess the efficacy of Crh silencing, we quantified Crh and mCherry levels using in situ hybridization (Fig. 4A, bottom). ILACRH cells expressing the anti-Crh shRNA and mCherry showed a 4-fold decrease in the intensity of Crh labeling compared to nearby non-infected CRH+ cells that did not express mCherry (Fig. 4A-B). No change was seen in cells expressing the scrambled shRNA (Fig. 4A-B). These results indicate that our strategy to reduce Crh level in ILACRH neurons is both specific and efficient.
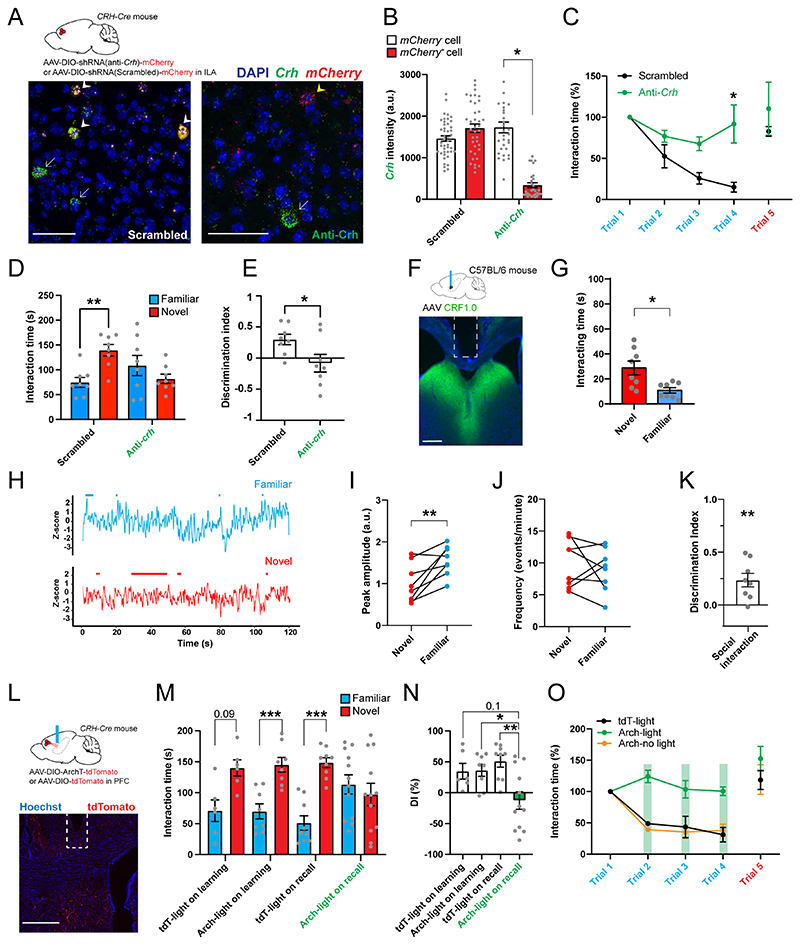
Figure 4. CRH release from ILA in rdLS suppresses social interactions with familiar mice and supports social novelty preference.
A. CRH-Cre mice injected in ILA with AAV2/9 CMV-DIO-(mCherry-U6)-shRNA(anti-Crh) to downregulate Crh or control AAV2/9 CMV-DIO-(mCherry-U6)-shRNA(scrambled) (top). In situ hybridization images of ILA slices expressing the scrambled shRNA (right) or the shRNA against Crh (left) labelled for mCherry and Crh. White arrows denote CRH+ neurons that do not express the virus. Yellow arrowhead denotes CRH+ cells expressing the anti-Crh shRNA, with reduced level of Crh. White arrowheads denote CRH+neurons expressing the scrambled shRNA, with intact Crh level. Scale bars: 50 μm. B. Quantification of Crh expression. In each slice neurons were classified as to whether they were uninfected or infected with virus based on mCherry expression (3 mice per group; each point is a different neuron). Nested one-way ANOVA F3,8 = 6.41, p = 0.016 followed by Tukey’s multiple comparison test, anti-Crh + mCherry+ vs. anti-Crh + mCherry-, p = 0.03. C. Normalized interaction time during the repetitive social presentation test in mice expressing scrambled or anti-Crh shRNAs. 4 mice per group. Two-way ANOVA; F(triali-4 x virus)3,24 = 4.4 p = 0.01; F(trial)3,24 = 9.6, p = 0.0002; F(virus)3,24 = 21.9, p < 0.0001 followed by Tukey’s multiple comparison test between scrambled and anti-Crh groups: trial 2, p = 0.8; trial 3, p = 0.2; trial 4, p = 0.009. D. Interaction time with familiar (blue) or novel (red) mouse during the recall trial of the social novelty preference test in mice expressing scrambled or anti-Crh shRNAs. Grey dots are different mice. 2-way ANOVA F(novelty x virus)1,28 = 11.53, p = 0.002. Sidak’s multiple comparison tests novel vs. familiar: scrambled, p = 0.004; anti-Crh, p = 0.3. Paired t test: p = 0.0095, p = 0.6. E. Discrimination indexes for social novelty preference during recall trial. Grey dots are different mice. Unpaired t test: p = 0.03. F. Top: C57BL/6J wild-type mice injected in rdLS with AAV2/9 Syn.CRF1.0 and implanted with an optical ferrule above rdLS. Bottom: immunohistochemistry image showing CRF1.0 expression in rdLS and the optical ferrule implanted above the injection site. Scale bar: 300 μm. G. Bar graph showing the interaction time with novel and familiar mice. 8 mice. Paired t test, p = 0.002. H. Trace of a representative fiberphotometry recording during interaction with a familiar mouse or a novel mouse. Interaction bouts are shown above each trace. I. Average peak amplitude of the z-score during presentation of a novel or familiar mouse. 8 mice. Paired t test: p = 0.008. J. Frequency of events during presentation of a novel or a familiar mouse. 8 mice. Paired t test: p = 0.5. K. Discrimination index for social familiarity preference calculated from z-scores. 8 mice. One-sample t tests compared to 0: p = 0.008. L. CRH-Cre mice injected in ILA with AAV2/2 CAG.FLEX.ArchT-tdTomato or control AAV2/2 CAG.FLEX.tdTomato. Optical ferrule implant is above rdLS. Scale bar: 500 μm. M. Interaction time with familiar (blue) or novel (red) mouse during the recall trial of the social novelty preference test in the same mice. Laser was on during the learning or recall trial. Each dot is a mouse. 3-way ANOVA F(novelty x light x virus)1,62=14.44, p=0.007. Sidak’s multiple comparison tests novel vs. familiar: p=0.03, 0.004, <0.0001 and 0.8. N. Discrimination index for social novelty preference during recall trial of the social novelty preference test. One-sample t-tests: p=0.04, 0.007, 0.0007 and 0.4. Two-way ANOVA: F(virus x light)1,3i=6.232, p=0.01. F(light)1,31=1.578, p=0.2; F(virus)1,3i=5.701, p=0.02. Sidak’s multiple comparison tests: p=0.09, 0.04 and 0.003.. O. Normalized interaction time during the repetitive social presentation test in the same mice. The laser was on during trials 1 to 4 of the Arch-light and mC-light groups (4 mice and 3 mice respectively). Laser was not on for the Arch-no light group (4 mice). Two-way ANOVA: F(triali-4 x virus)6,32 = 5.84, p = 0.0003; F(trial)3,32 = 14.35, p < 0.0001; F(group)2,32 = 49.32, p < 0.0001. For the entire figure, bar graphs represent mean ± S.E.M. Grey dots are different mice.
Next, we tested CRH-Cre mice expressing scrambled and anti-Crh shRNA during repetitive social presentations. Mice expressing the anti-Crh shRNA in ILACRH cells showed very little familiarization unlike mice expressing the scrambled shRNA (Fig. 4C). Then, we tested the mice for social novelty preference. Mice expressing anti-Crh shRNA showed no preference during the recall trial (Fig. 4D) and the discrimination index of this group was null (Fig. 4E). Total exploration during learning or recall trials was not different between groups (Fig. S9A-B). Given that ILACRH neurons co-express GABA (Fig. S3E-F), we repeated the same experiments using the shRNA against vGAT mRNA (Fig. S9C-D)49, which had been used previously to knock-down vGAT in hypothalamic CRH+ neurons50. Unlike knocking-down Crh, knocking-down vGAT expression in ILACRH cells failed to impair familiarization or social novelty preference (Fig. S9E-H).
Even though we did not observe ILACRH projections in any region other than rdLS (Fig. S1C), we sought to validate our previous results using a retrograde targeting approach. We injected a monosynaptic retrograde virus expressing Cre in rdLS (HSV-Cre) and the Cre-dependent viruses expressing anti-Crh shRNA or scrambled shRNA in the ILA of WT mice (Fig. S9I). Removing Crh from rdLS-projecting cells in the ILA impaired familiarization and social novelty preference (Fig. S9J-N), similar to our previous results.
We then asked whether CRH release from ILACRH in rLS cells was necessary to mediate familiarization and social novelty preference. First, we sought to confirm that CRH release in LS occurs preferentially during familiar social interaction and leveraged the recently developed CRH biosensor CRF1.051. WT mice were injected in rdLS with an AAV expressing CRF1.0 and an optical ferrule was implanted above it (Fig. 4F). We presented novel and familiar mice in a random order meanwhile recording CRH activity events (Fig. 4H). Presentation of a familiar mouse induced responses of larger amplitude compared to the presentation of a novel mouse (Fig. 4I), despite the mice interacting less (Fig. 4G). There was no change in the frequency of events (Fig. 4J). The discrimination index based on z-score peak amplitude (see Methods) showed a significant preference for the response for familiar mice (Fig. 4K). This experiment fits with our calcium recordings of ILACRH cells and demonstrates that CRH release in rLS is higher during familiar social interactions compared to novel social interactions.
Next, we used optogenetics to silence ILACRH cell terminals in rdLS. CRH-Cre mice were injected in ILA with Cre-dependent AAVs expressing Archaerhodopsin tagged with tdTomato (Arch) or tdTomato only as a control and an optical ferrule was implanted above rdLS (Fig. 4L). Light from a 561 nm laser was applied continuously during either the learning or recall trials of the social novelty preference test. When light was applied during the learning trial, both groups exhibited social novelty preference during the recall trial. However, when light was applied during the recall trial, mice expressing Arch failed to show a preference for the novel mouse while mice expressing tdTomato only did (Fig. 4M-N). All groups showed the same extent of total interaction during learning and recall (Fig. S9O-P). We also tested the mice during repetitive social presentations and applied the light stimulation during trials 2-4 or no light. Arch-expressing mice with light failed to familiarize to the novel mouse unlike tdTomato only -expressing mice with light or Arch-expressing mice with no light (Fig. 4O).
We tested the efficiency of our terminal-silencing approach combining Arch-mediated silencing and CRH recording. CRH-Cre mice were injected with a Cre-dependent virus expressing Arch in the ILA and a virus expressing the biosensor CRF1.0 in rdLS (Fig. S10A-B). Two optical ferrules were implanted above rdLS. Then, we exposed the mice to a familiar mouse for 3 sessions of 2 min separated by 10 min intervals. The 561 nm laser was turned on only during the middle presentation in order to stimulate Arch in ILACRH terminals in rdLS (Fig. SIOC). Activating Arch efficiently decreased the frequency of CRH-related transients (Fig. SIOD). The amplitude of the transients followed a similar trend (Fig. S10E). Importantly, turning on the laser increased the amount of social interaction with the familiar mouse, suggesting that ILACRH fibers regulate the amount of social interaction with familiar mice (Fig. S10F). Taken together, these experiments show that CRH release from ILACRH cells in rdLS during social encounters suppress social interactions with familiar mice to promote social novelty preference.
CRHR1 activation in rdLS suppresses social interaction with familiar mice and support social novelty preference
Since CRH receptor 1 (CRHR1) regulates social interaction and social novelty preference32,42, we looked whether it was expressed in the vicinity of the ILACRH cells terminals in LS. We injected CRHR1-Cre mice (courtesy of Jan Deussing)52 with a Cre-dependent AAV expressing GFP and labelled several neurons in rdLS (Fig. 5A), the same region targeted by terminals of ILACRH cells (Fig. 1C-D). Then, we crossed the CRHR1-Cre line with the Ai9 tdTomato reporter in order to visualize CRHR1+ neurons, prepared acute LS slices and obtained whole-cell patch-clamp recordings from rdLSCRHR1 neurons clamped at -60 mV. Application of 300 nM stressin-1, a CRHR1 agonist, induced an inward current (Fig. 5B), similar to previous results53. This experiment suggests that CRH release in rdLS depolarizes CRHR1+ neurons.
Figure 5. CRHR1+ neurons in rdLS are activated by social familiarity and regulate SNP and familiarization.
A. CRHR1-Cre mice injected in rdLS with AAV2/5 hSyn.DIO.mGFP. Scale bars: 300 μm (left), 50 μm (right). B. Whole-cell patch-clamp recording of CRHR1-tdTomato cells in rdLS of CRHR1-Cre;Ai9 mice (top). Voltage-clamp trace during bath application of 300 nM stressin-1 (middle). Scale bars: 100 pA and 2 min. Bar graph showing the amplitude of the decrease (bottom). C.C57BL/6J wild-type mice infused in rdLS with 2 μg of antalarmin dissolved in 0.6 μL of DMSO or DMSO as a control (top). Interaction time with familiar (blue) or novel (red) mouse during the recall trial of the social novelty preference test in mice infused with antalarmin or DMSO (bottom). Grey dots are different mice. 2-way ANOVA F(novelty x injection)1,36 = 7.699, p = 0.009. Sidak’s multiple comparison tests novel vs. familiar: DMSO, p = 0.04; antalarmin, p = 0.3. D. Discrimination index for social novelty preference during recall trial. Grey dots are different mice. One-sample t tests: p = 0.003 and p = 0.2. Unpaired t test:p = 0.01. E. CRHR1-Cre mice injected in rdLS with AAV2/8 hSyn.DIO.hM4D(Gi)-mCherry (iDREADD) or AAV2/8 hSyn.DIO.mCherry (top). Immunohistochemistry pictures of iD-mCherry expression in rdLS. Scale bar: 300 μm (bottom). F. Interaction time with novel (red) or familiar (blue) mouse during the recall trial of the social novelty preference test in mice expressing mCherry (mC) or hM4Di (iD). Grey dots are different mice. 3-way ANOVA F(novelty x injection x virus)1,60 = 3.845, p = 0.04. Sidak’s multiple comparison tests novel vs. familiar: mC + saline, p = 0.0003; iD + saline, p = 0.001; mC + CNO, p = 0.01; iD + CNO, p = 0.4. G. Discrimination indexes for social novelty preference of the four groups during recall trial. One-sample t tests compared to 0: mC + saline, p = 0.02; iD + saline, p = 0.009; mC + CNO, p = 0.001; iD + CNO, p = 0.3. 2-way ANOVA: F(virus x injection)1, 30 = 4.3, p = 0.04; F(virus)1, 30 = 7.654, p = 0.009; F(injection)1, 30 = 4.263, p = 0.05. Tukey’s multiple comparison tests compared to the iD + CNO group: mC + saline, p = 0.009; iD + saline, p = 0.02; mC + CNO, p = 0.01. H. Normalized interaction times during repetitive social presentations. 7-8 mice per group. Two-way ANOVA, F(triali1-4 x virus)9,104 = 6.612, p < 0.0001; F(trial)3,104 = 28.05, p < 0.0001; F(virus)3,104 = 52.74, p < 0.0001. I. Immunohistochemistry pictures against c-fos of CRHR1-tdTomato mouse rdLS of following interaction with a familiar or novel mice. Scale bars: 50 μm. J. Percentage of CRHR1+ neurons expressing c-fos. 3 and 4 mice. 2 observations per mice. Nested t test, p = 0.007.
Then, we implanted mice with a cannula in rdLS and infused them with the CRHR1 antagonist antalarmin diluted in DMSO54 or DMSO only before running them for the social novelty preference test (Fig. 5C). Mice infused with DMSO exhibited normal preference for the novel mouse whereas mice infused with antalarmin showed no novelty preference (Fig. 5C-D). Total exploration time during learning or recall was not different between groups (Fig. S11A-B).
Next, we performed chemogenetic silencing of CRHR1+ neurons in rdLS. We injected CRHR1-Cre mice with a Cre-dependent AAVs expressing inhibitory DREADD tagged with mCherry or mCherry only (Fig. 5E). Then, we tested the mice for social novelty preference and observed a preference for the novel mouse in the 3 control groups but not in the test group (Fig. 5F). Consequently, the discrimination index of the test group was not different from 0 unlike the ones of the control groups (Fig. 5G). The total interaction times during learning or recall or the interaction time with each novel mouse during learning were similar across all groups (Fig. S11C-E). We also ran the mice for the repetitive social presentation test and observed no familiarization in the test group (Fig. 5H). Overall, these experiment shows that activation of the CRHR1 receptor in rdLS is necessary for familiarization and social novelty preference.
Then, we tested whether CRHR1+ neurons in rdLS were preferentially activated during familiar encounters compared to novel ones. We presented CRHR1-Cre;Ai9 mice with either novel or familiar mice before perfusing them and labeling for c-fos (Fig. 5I). The percentage of rdLSCRHR1 neurons expressing c-fos during familiar interaction was 3-fold higher than the one during novel interaction (Fig. 5J). Overall, these experiments demonstrate that rdLSCRHR1 neurons are preferentially recruited during familiar encounters in order to regulate familiarization and social novelty preference.
CRHR1 activation disinhibits rLS to suppress social interactions with familiar mice
What is the effect of CRHR1 activation in rLS? We prepared acute LS slices from WT mice and recorded spontaneous inhibitory post-synaptic currents (IPSCs) from rdLS cells in whole-cell configuration before applying the CRHR1 agonist stressin-1 (300 nM, (Fig. 6A-B)53. Stressin-1 application for 15 min decreased the frequency and integrated charge of spontaneous IPSCs (Fig. 6B-C and E). IPSC amplitude also exhibited a trend toward a decrease (Fig. 6D). This effect was not seen when rLS neurons were recorded for 15 min without application of the agonist (Fig. S12A). In addition, application of stressin-1 while recording from vLS neurons in posterior LS slices had no effect (Fig. S12B), suggesting that the agonist effect was not generalized to the entire LS, consistent with the pattern of CRHR1 expression (Fig. 5A)20. Taken together, these results suggests that CRH release disinhibits rLS neurons.
Figure 6. CRH signaling from ILA and familiar social interaction disinhibit rdLS.
A. Differential interference contrast microscopy image of rdLS during patch-clamp recording. Scale bar: 500 μm. B. Example traces of IPSCs before or 15 min after application of 300 nM stressin-1. C. Frequency of IPSCs. D. Amplitude of IPSCs. E. IPSCs area under the curve. For C-E, points are obtained from individual cells recorded from separate slices in 6 mice. F. CRH-Cre mice injected with AAV2/9 EF1a.DIO.hChR2(E123T/T159C)-eYFP in ILA. G-I. Frequency (G), amplitude (H) and charge (I) of rdLS neuron spontaneous inhibitory events before and after tetanic light stimulation with or without 300 nM antalarmin. Each observation is from a different cell in separate brain slices obtained from 6 mice and 5 mice respectively. Paired t tests: p = 0.03, 0.2, 0.0003, 0.2, 0.03 and 0.9. J. Electrically evoked IPSC of rdLS neuron spontaneous inhibitory events before and after tetanic light stimulation with or without 300 nM antalarmin. Paired t tests: p = 0.006 and 0.07. K. C57BL/6J wild-type mice injected in rdLS with AAV2/1 Syn.GCaMP6f and implanted with an optical ferrule above rdLS. Implanted mice were presented with novel then familiar mice. L. Interaction time during social presentation (left). 9 recording sessions using 5 mice. Nested t test, p = 0.01. M. Average peak amplitude of the z-score during presentation of a novel or familiar mouse. Nested t test: p = 0.03. N. Frequency of events during presentation of a novel or a familiar mouse. Nested t test: p = 0.8. O. Discrimination index for social familiarity preference calculated from z-scores. Nested t tests compared to 0: p = 0.002. P. Decoding performance for familiarity versus novelty from individual recordings or pseudo-simultaneous data. Small black dots on the left are the results from each individual recording sessions using 20 cross-validation iterations, large red dot is the average. Red dot on the right is the result of pseudo-population analysis from 100 cross-validation iterations. Grey areas denote chance level computed using permutation tests (2.5 – 97.5 percentiles in distribution of shuffled decoding performances). In both cases, statistical significance is determined by the probability of drawing the observed decoding performance from the distribution of shuffled decoding performances (null-hypothesis). p < 0.001 (two-tailed permutation test, see Methods). Q. CRH-Cre;Ai9 mice were presented with novel or familiar mice after overnight isolation before being processed for immunohistochemistry against c-fos. R. Immunohistochemistry images of c-fos labelling in rdLS following social presentation with a novel or familiar mouse (same experiment than (Fig. 3N). Scale bars: 500 μm. S. Density of rdLS cells positive for c-fos. For each mouse, one observation on each side of a rLS section. 5 mice per group. Nested t test, p = 0.02. T. Percentage of layer 2/3 ILACRH cells positive for c-fos (cf. Fig. 3) vs. density of rdLS cells positive for c-fos following social interactions. Each point represents a mouse.
We thought of eliciting CRH release in rdLS from ILA fibers since CRH could originate from other regions. We expressed channelrhodopsin in ILACRH neurons and prepared rdLS slices (Fig. 6F). We obtained whole-cell recordings of rdLS neurons and recorded spontaneous inhibitory events (sIPSC) before and after applying a tetanic light stimulation. Light stimulation induced a decrease in sIPSC frequency, amplitude and charge (Fig. 6G-I), similar to stressin-1 application. In addition, we applied a local electrical stimulation to induce a large elicited IPSC.
The amplitude of which was also decreased following light stimulation (Fig. 6J). We repeated these recordings with 300 nM antalarmin in the bath to confirm that the decrease in inhibition following light stimulation was CRH-dependent. Indeed, antalarmin application blocked the disinhibition (Fig. 6G-J). Taken together these results demonstrate that CRH release from ILA disinhibits rLS neurons.
Is rLS disinhibited during social interactions with familiar mice and what is the effect of this disinhibition? To answer these questions, we recorded responses of rLS neurons during interaction with a novel or familiar mouse using fiber-photometry. C57BL/6J wild-type mice were injected in rLS with an AAV expressing GCaMP6f and an optical ferrule was implanted above it (Fig. 6K and S13A-C). We presented novel and familiar mice and measured the interaction time, average peak amplitude and peak frequency (Fig. 6L-N). Similar to our recordings of ILACRH neurons, presentation of a familiar mouse induced transients of larger amplitude compared to the presentation of a novel mouse (Fig. 6M), despite the mice interacting less (Fig. 6L). There was no change in the frequency of transients however (Fig. 6N). The discrimination index based on the peak amplitude (see Methods) showed a significant preference in response to a familiar mouse (Fig. 6O). To determine further whether rLS activity alone can differentiate between novel and familiar mouse presentation, we trained linear classifiers to discriminate between interactions with a novel or familiar mouse based on our fiberphotometry recordings. We implemented 2 classifiers using either individual recording sessions (individual) or creating a meta-session pooling all sessions (pseudo-simultaneous, (Fig. 6P). For each classifier, we also computed chance levels using permutation tests (grey bars). Most individual recording session yielded a decoding performance above chance with an average of 59% accuracy while the pseudo-simultaneous data yielded a decoding performance even higher (81%). This shows that rLS neurons can encode for social familiarity. We also measure rLS activity during the repetitive social presentation test and observed that the peak amplitude of calcium events increased from trial 1 to trial 3 when familiarization is taking place (Fig. S13D-E). Similar to the percentage of c-fos+ L2/3 ILACRH cells (Fig. 3R), the calcium activity anti-correlated with the amount of social interaction (Fig. S13F).
Which LS neurons are activated by familiar presentation? We examined c-fos expression in LS following novel or familiar social encounters (Fig. 6Q) in the same cohort of mice than the one used to look at c-fos expression in ILA (Fig. 3N). rLS responded preferentially to familiar mouse presentation (Fig. 6R-S), similar to layer 2/3 ILACRH neurons (Fig. 3Q). C-fos expression was specifically upregulated in a spatially defined band of rLS cells bordering the lateral ventricle (Fig. 6R, right) while exposure to a novel mouse failed to activate the same population (Fig. 6R, left). Taken together, the fiberphotometry recordings and c-fos labelling demonstrate that a population of rLS neurons is activated preferentially during a familiar encounter compared to a novel one. Similar to L2/3 ILACRH neurons, activation of rLS neurons tended to correlate negatively with the amount of social interaction (Fig. S14A). Interestingly, activation of layer 2/3 ILACRH cells plotted against rdLS activation demonstrated a strong positive correlation, suggesting that one population might control the other (Fig. 6T). We also quantified c-fos expression in posterior dorsal LS (dLS) and posterior ventral LS (vLS) and observed no preferential response to familiar presentation compared to novel nor correlation with the amount of interaction (Fig. S14B-G).
CRH release in rdLS disinhibits rLS to suppresses social interactions with familiar mice
Does the activation of rLS during familiar encounters depends on CRH release from ILACRH cells? We tested this hypothesis by measuring c-fos expression in mice where Crh was knocked down in ILA, similar to the approach used previously to disrupt memory retrieval55. CRH-Cre mice were injected in ILA with Cre-dependent AAVs expressing anti-Crh shRNA or a scrambled shRNA control, as described above. Mice were then presented with a familiar littermate and sections containing the ILA and rLS were labeled for c-fos (Fig. 7A-C). Importantly, mice expressing the anti-Crh shRNA interacted more with a familiar mouse than control mice, suggesting that Crh reduces social interaction with a familiar mouse (Fig. 7D). Loss of Crh did not alter c-fos expression in layer 2/3 ILACRH cells (Fig. 7E) but reduced the density of c-fos expression in rLS (Fig. 7F). As previously, control mice exhibited a correlation between c-fos levels in layer 2/3 ILACRH and rLS neurons but mice depleted of Crh in ILA did not (Fig. 7G).
Figure 7. CRH release from ILA and rdLSCRHR1 neurons regulate rdLS disinhibition and social interaction with a familiar mouse.
A. CRH-Cre mice injected in ILA with AAV2/9 CMV-DIO-(mCherry-U6)-shRNA(anti-Crh) or AAV2/9 CMV-DIO-(mCherry-U6)-shRNA(scrambled) presented with a familiar mouse for 2 min before being processed for immunohistochemistry against c-fos. B-C. Immunohistochemistry images of c-fos labelling in ILA (B) and rdLS (C). Yellow arrowheads: c-fos+ / tdTomato+ cells. White arrowheads: c-fos- / tdTomato+ cells. Scale bars: 100 μm and 300 μm. D. Duration of interaction during familiar presentation. Each point is one mouse. Unpaired t test, p = 0.001. E. Percentage of layer 2/3 ILACRH cells positive for c-fos in layer 2/3 of ILA. Each point corresponds to each side of 2 sections. 9 mice per group. F. Density of rdLS cells positive for c-fos. We made one observation on each side of a rLS section. 9 mice per group. Nested t test, p = 0.002. G. Percentage of layer 2/3 ILACRH cells positive for c-fos vs. density of rdLS cells positive for c-fos following social interaction with a familiar mouse. Each point represents one mouse. H. CRHR1-Cre mice injected in rdLS with AAV2/8 hSyn.DIO.hM4D(Gi)-mCherry (iDREADD) or AAV2/8 hSyn.DIO.mCherry presented with a familiar mouse for 2 min before being processed for immunohistochemistry against c-fos. I. Interaction time with familiar mouse. Each point is one mouse. Unpaired t test, p = 0.03. J. Immunohistochemistry picture of mCherry expression in rdLS. Scale bar: 400 μm. K. Immunohistochemistry pictures of c-fos expression in rdLS. Scale bar: 400 μm. L. Density of rdLS cells positive for c-fos. For each mouse, one observation on each side of a rLS section. 5 and 6 mice per group. Nested t test, p = 0.002. M. C57BL/6J wild-type mice were injected with AA2/2 hSyn1.hChR2(H134R)-mCherry or AA2/2 hSyn1.mCherry as control and an optical fiber was implanted above the injection site. Mice were then presented to a familiar mouse for 2 min meanwhile 450 nm light was applied (20 Hz, 1 ms). Mice were also run without light as additional controls. N. Immunohistochemistry picture of viral injection. Scale bar: 1 mm. O. Total interaction time with familiar mouse. Each point represents one mouse. One-way ANOVA: F3,32 = 7.01, p < 0.0001. Dunnett’s multiple comparison tests: ChR-light vs. YFP-no light p = 0.0005, ChR-light vs. ChR-no light p = 0.006, ChR-light vs. YFP-light p = 0.01. P. Average duration of each bout of social interaction. Each point represents one mouse. One-way ANOVA: F3,31 = 10.62, p < 0.0001. Dunnett’s multiple comparison tests: ChR-light vs. YFP-no light p = 0.0001, ChR-light vs. ChR-no light p = 0.0001, ChR-light vs. YFP-light p = 0.003. Q. Total distance travelled. Each point represents one mouse. For the entire figure, bar graphs represent mean ± S.E.M.
We next tested whether CRHR1+ cells in rdLS controls the disinhibition of rdLS following familiar social encounter. We expressed inhibitory DREADD in rdLSCRHR1 neurons and injected the mice with CNO before presenting them with a familiar mouse (Fig. 7H-I). We then perfused the mice and labeled rLS slices against c-fos (Fig. 7J-K). Silencing rdLSCRHR1 neurons decreased the density of c-fos labeled cells in rdLS (Fig. 7L) while increasing the amount of social interaction (Fig. 7I). Taken together, these experiments demonstrate that CRH release from ILA and activation of rdLSCRHR1 neurons during familiar encounters disinhibits a specific population of rLS cells bordering the lateral ventricles and suppress social interactions with a familiar mouse.
Our findings suggest that rLS disinhibition suppresses social interactions. To explore this further, we examined the effects of optogenetic activation of rLS neurons. We injected an AAV expressing Channelrhodopsin (ChR) tagged with mCherry or mCherry only in rLS of C57BL/6J wild-type mice and implanted an optical ferrule above it (Fig. 7M-N). We then presented a familiar mouse for 2 min while stimulating ChR using a 445 nm laser (1 ms stimulation at 20 Hz) and measured the interaction time as well as the mean duration of each interaction bout. As an additional control, we measured behavior in ChR-expressing mice without laser stimulation. Activation of rLS neurons decreased the amount of social interaction with familiar mice (Fig. 7O) due to shorter interaction bouts each time the mice met (Fig. 7P) without affecting locomotion (Fig. 7Q). These optogenetic experiments demonstrate that rLS is able to decrease social interactions. Altogether our study shows that ILACRH cells are activated during familiar mouse encounters, leading to release of CRH in rdLS causing its disinhibition. Disinhibition of rLS in turn suppress social interaction which leads to the decrease in social interaction observed during familiarization. Inhibition of social interaction with familiar mice promotes social novelty preference when novel and familiar mice are presented simultaneously.
Increased CRH expression in ILA supports a shift in social preference in young mice
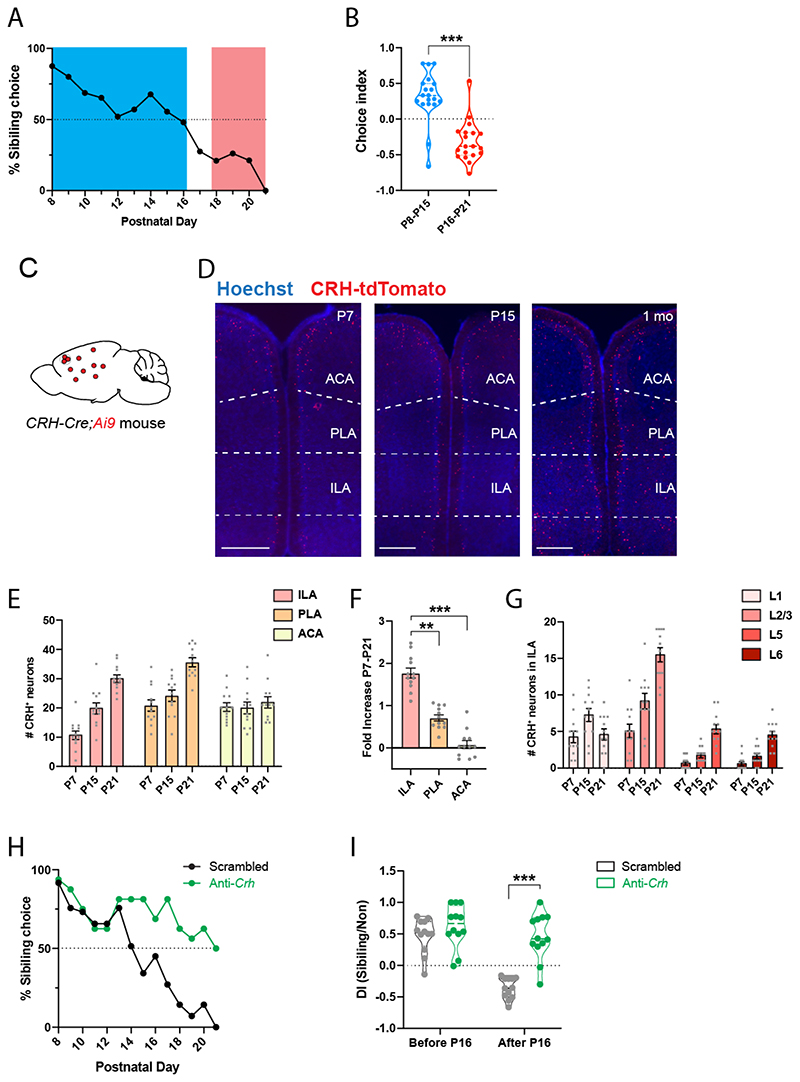
In contrast to adults, young rodents prefer to interact with their familiar siblings compared to novel pups3,4,56. We tested when the shift in social preference occurs in mice by giving young C57BL/6J wild-type mice the choice to interact with their familiar siblings or with unfamiliar non-siblings every day from P7 to P21. Between P7 and P15, mice preferred to interact with their siblings (Fig. 8A) and their discrimination index was strongly skewed toward familiarity (Fig. 8B). Preference then gradually shifted toward novel mice (Fig. 8A) and the index leaned toward social novelty after P16 (Fig. 8B).
Figure 8. Increased CRH expression in ILA supports a shift in social preference in young mice.
A. Percentage of familiar choice during development, 19 mice. B. Discrimination index for familiar kin before and after postnatal day 16. Each point represents a mouse, 19 mice.
Unpaired t test, p < 0.001. C. CRH-Cre;Ai9 mice. D. mPFC images of CRH-Cre;Ai9 mice at P7, P15 or P21. Scale bars: 500 μm. E. Number of CRH+ cells in ILA, PLA and ACA during development. Each point represents one observation made on each side of 2 section, 3 mice per group. Nested one-way ANOVA tests comparing CRH cells along postnatal day: F(ILA)2,6 = 18.64, p = 0.003; F(PL)2,6 = 11.47, p = 0.009; F(ACA)2,6 = 0.22, p = 0.8. F. Fold-increase of CHR+ cells between P7 and P21. P21 values compared to the average P7 value. Nested one-way ANOVA F1,6 = 63.03, p < 0.0001. Post-hoc Tukey’s multiple comparison test: ILA vs. PLA p = 0.001; ILA vs. ACA p < 0.0001. G. Number of CRH+ cells per ILA layers during development. Each point represents one observation made on each side of 2 section, 3 mice per group. H. Percentage of familiar choice during development in CRH-Cre mice injected in ILA with AAV2/9 CMV-DIO-(mCherry-U6)-shRNA(anti-Crh) to downregulate Crh or control AAV2/9 CMV-DIO-(mCherry-U6)-shRNA(scrambled). 12 pups per group. Chi-square test: p < 0.0001. I. Discrimination index for familiar kin before and after postnatal day 16. Each point represents a mouse, 12 pups per group. Unpaired t tests: p = 0.3 and p < 0.0001. For the entire figure, bar graphs represent mean ± S.E.M.
Both the PFC and LS have been shown to control social novelty preference and the LS is also involved in the preference young rats display for their own (familiar) kin compared to nonkin3. We then asked when ILA neurons begin to express CRH and whether the emergence of the ILA to rdLS circuit described above contributes to the shift in social preference. We counted CRH-tdTomato+ cells at P7, P15 and P21 in CRH-Cre;Ai9 mice (Fig. 8C-D) and observed a strong increase in the density of ILACRH cells from P7 to P21 (Fig. 8E). Interestingly, the increase was the strongest in ILA compared to other prefrontal regions (Fig. 8F). Within ILA, the increase of CRH+ cells proceeded mostly from an increase in CRH+ cells located in layer 2/3 (Fig. 8G), which is the layer containing ILACRH cells projecting to rdLS (Fig. 1K and S3D). Comparing the number of ILACRH cells per section to the retrogradely labeled ones (Fig. S3D), suggests that at least 60% of these cells project to rdLS. Closer inspection of CRH+ cells in PLA and ACA revealed less or no increase in layer 2/3 (Fig. S15A-B). In situ hybridization against Crh in C57BL/6 WT mice show a similar increase in Crh+ neuron density in mPFC, PLA and ILA (Fig. S15C-F). Overall, these experiments demonstrate that the strengthening of the ILACRH to rdLS circuit correlates with the shift from social familiarity to social novelty preference in young pups.
We next probed whether the emergence of the circuit causes the shift in preference. P5 pups were injected with AAVs expressing anti-Crh or scrambled shRNAs. We reasoned the AAVs would be taken up by many ILA neurons so that, as soon as CRH expression begins, so would the Cre recombinase expression and therefore shRNA expression under the control of the fast-expressing U6 promoter. Two days after the injection, we began testing the injected pups for social preference and observed preference shifted at P14 in the control group (Fig. 8H), similar to our previous experiment on wild-type mice (Fig. 8A). The test group lacking Crh however continued to exhibit familiar preference until P20 (Fig. 8H). Consistently, discrimination index for the control group inverted before and after P16 reflecting the shift in social preference while the index for the Crh-depleted group remained oriented toward familiar choice (Fig. 8I). We performed in situ hybridization against Crh and mCherry at the end of the behavioral testing (P21) in order to verify the efficacy of the shRNA-mediated Crhdepletion technique in young pups (Fig. S15G). Similar to our results in adults (Fig. 4B), cells expressing the anti-Crh shRNA exhibited a strong decrease in Crh labeling intensity (Fig. S15H). Overall, these experiments suggest that increased CRH expression in ILA is responsible for the shift in social preference displayed by young mice.
Discussion
We show that ILACRH cells respond to social interaction with familiar over novel mice and release CRH into rLS in order to suppress social interactions with familiar mice through LS disinhibition. During familiarization, increasingly responsive ILACRH cells control the decrease in interaction as a novel mouse becomes familiar. When given the choice between a familiar and a novel mouse, this circuit suppresses interaction with the familiar mouse to support social novelty preference.
We asked previously whether ILACRH cells control social memory or rather downstream processes, including social novelty preference that utilize social memory cues. Silencing ILACRH cell terminals to rdLS during the recall trial but not during the learning trial disrupts social novelty preference (Fig. 4L-M), suggesting that ILACRH cells do not contribute to social memory formation. This is in line with previous work showing that the CRH-binding protein is critical for the recall but not for the learning phase of social recognition32. Knocking-down Crh expression in ILA increases social interactions with familiar mice (Fig. 4C, (7D) while keeping c-fos expression in ILA intact (Fig. 7E), suggesting that ILACRH neurons integrate social familiarity cues before releasing CRH in order to regulate social interaction with familiar mice.
Previous hypotheses about the mechanisms underlying social novelty preference supposed the existence of a circuit promoting interaction with novel mice, perhaps under control of the rewarding properties of social novelty. In addition, the kin preference toward mothers or siblings displayed by young mice19,56 supposes the existence of other circuits controlling social preference. Very little is known however about the mechanisms supporting the rewarding properties of social cues. The lateral habenula, nucleus accumbens, dorsal raphe nucleus and ventral tegmental area modulate social reward57–61, some of them under the control of oxytocin57–59. Subsequent studies should aim to characterize how social novelty reward facilitates interaction with novel mice to regulate social preference. The lateral septum, which is heavily modulated by dopamine, vasopressin and oxytocin20, likely also integrate inputs promoting interaction with novel mice in order to regulate social preferences. Interestingly, Liu et al. (2022) showed that silencing of dorsal LS neurons (located posterior to rLS) suppresses social approach and facilitates avoidance with novel but not familiar mice. This confirms the importance of LS to modulate social interactions and suggests that different LS regions can regulate different types of social interactions and may work together to promote social novelty preference.
How specific is the regulation of social preference by ILACRH cells? We demonstrate that ILACRH cells control memory-based social novelty preference but not memory-based novel object preference. Since the three stimulus mice used during our social novelty preference test are siblings from the same cage (and thus also from the same strain, same age and same sex), mice must discriminate between novel and familiar individuals based on the idiosyncratic identity of each individual, that is, based on true individual recognition and not a more general class recognition. Whether ILACRH cells control other social preferences such as preferences based on sex, strain, kinship or anxiety (mice prefer to interact with non-stressed mice)13 remains to be determined. The associative nature of the mPFC and our rabies tracing experiment (data not shown) suggest that ILACRH cells integrate information from many brain regions which are likely to provide various social cues about the nature and identity of the stimulus mice. In this framework, we suppose that social cues of negative valence activate excitatory neurons projecting to ILACRH cells. For example, social memories of previous encounters are known to be stored in the pyramidal neurons of the ventral CA1 region of the hippocampus63 and vCA1 projection to mPFC is necessary for behaviors relying on social memory such as social novelty preference64,65. Consistently, rabies tracing demonstrates that ILACRH cells receive inputs from vCA1 (data not shown). However, whether the vCA1 neurons projecting to ILACRH cells carry social familiarly information remains to be confirmed.
Unlike adults, young rats and mice display kin preference for mother and siblings during the first weeks of life3,19. Here, we show how young mice reliably display social preference toward their siblings versus age-matched pups until CRH increase triggers a shift in preference toward the normotopic adult behavior. Indeed, while defenseless pups need to rely on the safety of their nest and company of their siblings, older and more able young mice will benefit from leaving their kin and venture out of the nest in order to sample resources (feeding behavior) and interact with novel conspecifics (reproductive behavior)66. Overall, orchestrating a wide range of sometimes antagonistic motivated behaviors including safety, feeding, novel social interactions and mating is essential and the lateral septum has been proposed to play a key role in setting up priorities between motivated behaviors67.
Unlike mice, monogamous prairie voles exhibit social novelty preference only during short-term tests but partner preference emerges during long-term tests68,69. This difference in social preference might be due to the fact that prairie vole mPFC does not express CRH70. Furthermore, CRH intra-cerebroventricular injections in prairie voles prior to short-term tests induces preference for a familiar vole over a novel one71. These experiments demonstrate the role of CRF in regulating social preferences in several rodent species and suggest that differences in mPFC CRH system can be responsible for novelty or partner preference.
Humans can suffer from social separation anxiety disorder, which manifests itself as an “unusually strong fear or anxiety to separating from people they feel a strong attachment to” 73. Patients present unusual distress at the discussion or experience of being parted from their attachment figure and a refusal to leave the attachment figure. Similarly, people affected with avoidant personality disorder avoids novel individual to favor familiar ones. In addition, CRH has been involved in various anxiety disorders, including social phobia35,74. Similar to our findings, familiarity cues activate the human PFC75,76 and septal77 regions, supporting the idea that the circuit we described in the mouse is conserved in humans. A potential cause for social anxiety disorders such as separation anxiety disorder or avoidant personality disorder could then be that patients exhibit low CRH level in the PFC, preventing them from seeking social novelty.
Technical limitations of the study
The exact demarcations of the rodent PFC are subjected to debate, including the demarcations of its sub-regions such as the ILA are unclear77. In this study we used the Paxinos atlas (4th edition) to delineate separations between brain regions47. Furthermore, even though shRNA-mediated silencing of vGAT decreased the intensity of vGAT labeling by 2-fold, we cannot exclude the possibility that enough vGAT remained to mediate some GABAergic transmission and perhaps participate in familiarization and social novelty preference. Finally, although it is technically challenging to over-express CRH in mouse pups to test whether it would accelerate the shift in preference, it would be interesting to over-express CRH in rodents exhibiting social familiarity preference.
STAR Methods
Key resources
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-c-fos antibody produced in rabbit | Abcam | #ab190289 RRID:AB_2737414 |
| Anti-GFP antibody produced in chicken | AVES Labs | #GFP-1020 RRID:AB_10000240 |
| Anti-GFP polyclonal antibody, Alexa Fluor 488 | Thermo-Fisher Scientific | #A21311 RRID:AB_221477 |
| Anti-RFP antibody produced in rabbit | Rockland Antibody | #600-401-379 RRID:AB_2209751 |
| Anti-mCherry antibody produced in goat | Biorbyt | #orb153320 |
| Anti-GABA antibody produced in guinea-pig | Abcam | #ab17413 RRID:AB_443865 |
| Donkey anti-goat IgG (H+L) secondary antibody, Alexa Fluor 568 conjugate | Thermo-Fisher Scientific | #A11057 RRID:AB_2534104 |
| Goat anti-rabbit IgG (H+L) secondary antibody, Alexa Fluor 568 conjugate | Thermo-Fisher Scientific | #A11011 RRID:AB_143157 |
| Goat anti-rabbit IgG (H+L) secondary antibody, Alexa Fluor 647 conjugate | Thermo-Fisher Scientific | #A32733 RRID: AB_2633282 |
| Goat anti-guinea-pig IgG (H+L) secondary antibody, Alexa Fluor 647 conjugate | Thermo-Fisher Scientific | #A21450 RRID:AB_2735091 |
| Goat anti-mouse IgG1 (H+L) secondary antibody, Alexa Fluor 647 conjugate | Thermo-Fisher Scientific | #A21240 RRID:AB_2536165 |
| Goat anti-rat IgG (H+L) secondary antibody, Alexa Fluor 488 conjugate | Thermo-Fisher Scientific | #A48262 RRID:AB_2896330 |
| Goat Anti-chicken IgG (H+L) secondary antibody, Alexa Fluor 488 Conjugate | Thermo-Fisher Scientific | #A11039 RRID:AB_142924 |
| In situ hybridization probes | ||
| Mm-Crh | ACD Bio | #316091 |
| mCherry-C2 | ACD Bio | #431201-C2 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Stressin-1 | Tocris | #1608 |
| Antalarmin | Tocris | #2778 |
| DMSO | Sigma-Aldrich | #D8418 |
| CNO | Cayman Chemical | #16882 |
| CGP55845 | Tocris | Cat# 1248 |
| SR 95531 | Tocris | Cat# 1262 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J Mus musculus | Jackson Laboratories | RRID:IMSR_JAX:000664 |
| B6(Cg)-Crhtm1<cre>ZJh/J | Jackson Laboratories | RRID:IMSR_JAX:012704 |
| B6-Crhr1tm4(cre>Jde | Jan Deussing | RRID:MGI:6281608 |
| B6.Cg-Gt(ROSA)26Sortm9lCAG’tdTomato>Hze/J | Jackson Laboratories | RRID:IMSR_JAX:007909 |
| Recombinant DNA | ||
| AAV2/DJ hSyn.FLEX.mGFP.2A.Synatophysin-mRuby | Stanford vector core | #1930 / Addgene #71760 |
| AAV2/9 EF1a.DIO.hChR2(E123T/T159C).eYFP | Addgene | #35505-AAV9 |
| HSV hEF1a.LSIL.GFP (HT) | Massachusetts General Hospital | #RN406 |
| HSV hEF1a-Cre (HT) | Massachusetts General Hospital | #RN242 |
| AAV2/8 hSyn.DIO.hM4D(Gi)-mCherry | Addgene | #44362-AAV8 |
| AAV2/8 hSyn.DIO.mCherry | Addgene | #50459-AAV8 |
| AAV2/5-hSyn-DIO-hM3D(Gq)-mCherry | Addgene | #44361-AAV5 |
| AAV2/5 hsyn.DIO.mCherry | Addgene | #50459-AAV5 |
| AAV2/1 syn.FLEX.GCaMP6f.WPRE.SV40 | Addgene | #100833-AAV1 |
| PX552 | Addgene | #60958 |
| rAAV-U6-shRNA1(CRH)-CMV-EGFP-SV40-polyA | Brain VTA | #PT-2784 |
| rAAV-U6-shRNA2(CRH)-CMV-EGFP-SV40-polyA | Brain VTA | #PT-2785 |
| rAAV-U6-shRNA3(CRH)-CMV-EGFP-SV40-polyA | Brain VTA | #PT-2786 |
| rAAV-U6-shRNA(scramble)-CMV-EGFP-SV40-polyA | Brain VTA | #PT-0916 |
| rAAV-CMV-CRH-P2A-EGFP-WPRE-hGH-polyA | Brain VTA | #PT-2827 |
| AAV2/9 CMV-DIO-(mCherry-U6)-shRNA(anti-Crh) | Brain VTA | #PT-2787 |
| AAV2/9 CMV-DIO-(mCherry-U6)-shRNA(scrambled) | Brain VTA | #PT-2788 |
| AAV2/9 hSyn.FLEX.dsRed-shRNA(vGAT) | Addgene | Addgene #67845 |
| AAV 2/9 hSyn-flex-dsRed-shRNA(scrambled) | Addgene | Addgene #71383 |
| AAV2/5 DIO.mGFP | University of North Carolina | #AV4310i |
| AAV2/2 CAG.FLEX.ArchT-tdTomato | University of North Carolina | Addgene #28305 |
| AAV2/2 CAG.FLEX.tdTomato | University of North Carolina | Addgene #28306 |
| AAV2/1 syn.GCaMP6f.WPRE.SV40 | Pennsylvania University | #AV-1-PV2822 |
| AAV2/9 syn.CRF1.0 | Yulong Li Lab | Ref. 51 |
| AA2/2 hSyn1.hChR2(H134R)-mCherry.WPRE | University of Zurich | #V-124 |
| AA2/2 hSyn1.mCherry.WPRE | Addgene | #114472-AAV2 |
| Software | ||
| AxoGraph | AxoGraph | 1.6.4 |
| PRISM 9 | Graphpad | 9.0.1 (128) |
| Microsoft Office Word | Microsoft | 2019 16.56 |
| Microsoft Office Exel | Microsoft | 2019 |
| Adobe Illustrator | Adobe | 2020 v24.1 |
| FIDJI | GPL v2 | 2.3.0/1.53f |
| MATLAB | Mathworks | 2018 |
| Python | 3.10.2 | |
| Guppy | Lerner Lab 78 | 1.1.4 |
| Leica Application Suite X | Leica | v3.7.4 |
| ANY-maze | Stoelting Co. | 4.99 |
| Doric Neuroscience Studio | Doric | 5.4.1.23 |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Félix Leroy (felxfel@aol.com).
Material availability
Plasmids generated in this study are available upon request to the lead contact and will be deposited to Addgene.
Experimental Models and Study Participants Details
All animal procedures were performed in accordance with the regulations of the UMH-CSIC lACUCs. We used P5 to 16-week-old C57BL6/J wild-type (Jackson Laboratories, #000664) mice as well as mice of the same age range from the following transgenic mouse lines: CRH-Cre mice (Jackson Laboratories, #012704) and CRHR1-Cre mice (courtesy of Jan Deussing). CHR-Cre were crossed to the Ai9 tdTomato Cre-reporter mice (Jackson Laboratories, #007909) in order to visualize CRH+ neurons. All transgenic mice were on the C57BL6/J background. During social interaction tests, stimulus mice were C57BL6/J wild-type mice of the same gender and age than the test mouse. We observed no difference related to sex and the results were pooled together. The table below summarizes the number of male and female mice used in each behavioral experiment.
Figures.
| Figure | EXPERIMENT | Male | Female |
|---|---|---|---|
| 2C-E | CRH-iD SNP | 30 | 28 |
| 2G | CRH-iD repetitive social presentation | 8 | 8 |
| 2H | CRH-eD repetitive social presentation | 8 | 8 |
| 3D-K | CRH fiberphotometry single presentation | 5 | 0 |
| 3L-M | CRH fiberphotometry repetitive social presentation | 5 | 0 |
| 3N-R | CRH-tdT c-fos of ILA and rdLS | 5 | 5 |
| 4C | shRNA anti-Crh repetitive social presentation | 4 | 4 |
| 4D-E | shRNA anti-Crh SNP | 8 | 8 |
| 4F-K | CRH biosensor | 8 | 0 |
| 4-L-N | CRH-Arch SNP | 15 | 0 |
| 4O | CRH-Arch repetitive social presentation | 11 | 0 |
| 5C | WT antalarmin infusion SNP | 10 | 10 |
| 5E-G | CRHR1-iD SNP | 15 | 10 |
| 5H | CRHR1-iD repetitive social presentation | 15 | 15 |
| 5I-J | CRHR1-iD c-fos of LS single presentation | 7 | 4 |
| 6K-O | WT Fiberphotometry in rLS single presentation | 5 | 0 |
| 7A-D | CRH-tdT c-fos of ILA and rdLS following single presentations | 5 | 5 |
| 7E-K | CRH-shRNA anti-Crh c-fos after familiar presentation | 14 | 4 |
| 7E-K | CRHR1-iD c-fos after familiar presentation | 7 | 5 |
| 7Q-U | WT ChR in LS | 17 | 0 |
| 8A-B | Kinship in WTs | 10 | 9 |
| 8H-I | Kinship in with anti-Crh shRNA | 15 | 9 |
Method Details
Anti-Crh shRNA design and in vitro validation
Three different shRNAs that target Crh (shRNA1: GCCCTTGAATTTCTTGCAGCC; shRNA2: GCATGGGTGAAGAATACTTCC; shRNA3: GGAAACTGATGGAGATTATCG) were cloned into the PX552 plasmid (Addgene #60958) to make rAAV-U6-shRNA1,2,3(CRH)-CMV-EGFP-SV40-polyA (Brain VTA #PT-2784, #PT-2785 and #PT-2786). In addition, a scrambled shRNA control was cloned into the same plasmid to make rAAV-U6-shRNA(scrambled)-CMV-EGFP-SV40-polyA (Brain VTA #PT-0916). To overexpress CRH mRNA, the sequence for mouse CRH was cloned into a plasmid to construct rAAV-CMV-CRH-P2A-EGFP-WPRE-hGH-polyA (Brain VTA #PT-2827). For validation, HEK293T cells were transfected, in triplicate, with one of the 3 plasmids containing anti-Crh shRNAs or the scrambled shRNA construct along with the overexpression plasmid for Crh (CMV-CRH-GFP). The cells were collected 48hr post-transfection and RNA was purified (MiniBEST Universal RNA Extraction Kit; Takara, 9767) and subjected to RT-PCR (One Step SYBR®PrimeScript™RT-PCR Kit II; Takara, RR086A) using primers for CRH (F: CCCCGCAGCCCTTGAATTTCTTG; R: GGGCGTGGAGTTGGGGGACAG) and GAPDH (F: GCAAATTCCATGGCACCGTCAAGG; R: CGCCAGCATCGCCCCACTTG) as a control. The RT-PCR results revealed that all three of the anti-Crh shRNAs showed robust decreases of Crh mRNA relative to the scrambled shRNA control. The anti-Crh shRNA-2 showed the highest knockdown efficiency and was selected for in vivo knockdown experiments. The anti-Crh shRNA-2 and the scrambled shRNA were cloned into a Cre-dependent plasmid 79 to make rAAV-CMV-DIO-(mCherry-U6)-shRNA(anti-Crh)-WPRE-hGH-polyA (Brain VTA, #PT-2787) and rAAV-CMV-DIO-(mCherry-U6)-shRNA(scrambled)-WPRE-hGH-polyA (Brain VTA, #PT-2788) which were subsequently packaged into AAV9.
Virus injections
For all injections, animals were anesthetized using isoflurane and given analgesics. A craniotomy was performed above the target region and a glass pipette was stereotaxically lowered down the desired depth. Injections were performed using a nano-inject II (Drummond Scientific). 23 nL were delivered 10 s apart until the total amount was reached. The pipette was retracted after 5 min. With homozygous animals (C57BL/6J wild-type or CRH-Cre mice), injection of the virus injection expressing DREADD, ArchT, shRNA(anti-Crh) and their control viruses (fluorophore only) was randomized within each cage.
AAVs injections in ILA
Injection coordinates were the following (in mm from Bregma): AP: 1.65, ML: ±0.1, DV: -2.6. Injections were done bilaterally with 100 nl injected per site. We injected AAV2/DJ hSyn.FLEX.mGFP.2A.Synatophysin-mRuby (Addgene #71760 prepared by the Stanford University vector core #1930), AAV2/8 hSyn.DIO.hM4D(Gi)-mCherry (Addgene #44362-AAV8), AAV2/8 hSyn.DIO.mCherry (Addgene #50459-AAV8), AAV2/5-hSyn-DIO-hM3D(Gq)-mCherry (Addgene, #44361-AAV5), AAV2/5 hsyn.DIO.mCherry (Addgene #50459-AAV5), AAV2/1 syn.FLEX.GCaMP6f.WPRE.SV40 (Addgene #100833-AAV1), AAV2/9 CMV-DIO-(mCherry-U6)-shRNA(anti-Crh) (VTA brain), AAV2/9 CMV-DIO-(mCherry-U6)-shRNA(scrambled) (VTA brain), AAV2/9 hSyn.FLEX.dsRed-shRNA(Vgat) (Addgene #67845), AAV 2/9 hSyn-flex-dsRed-shRNA(scrambled) (Addgene #71383), AAV2/2 CAG.FLEX.ArchT-tdTomato (Addgene #28305 prepared by the University of North Carolina vector core) and AAV2/2 CAG.FLEX.tdTomato (Addgene #28306 prepared by the University of North Carolina vector core) into the ILA of CRH-Cre mice. Viruses expressed for a minimum of 2 weeks.
ACA and PLA anterograde tracing injections
We injected AAV2/DJ hSyn.FLEX.mGFP.2A.Synatophysin-mRuby (Stanford vector core #1930 / Addgene #71760A) in ACA and PLA of CRH-Cre mice. Injections were done unilaterally with 100 nl injected per site. Injection coordinates were the following (in mm from Bregma): PLA: 1.65, ML: ±0.1, DV: -1.4 and ACA 1.65, ML: ±0.1, DV: -0.6. Viruses expressed for a minimum of 2 weeks.
AAV injections in rdLS
Unless specified otherwise, viruses were injected at the following coordinates in mm from Bregma: AP: 0.9, ML: ±0.2, DV: -2.8. We injected bilaterally 100 nL of HSV hEF1a.LSIL.mCherry, (Rachel Neve, Massachusetts General Hospital #RN406) into the rdLS of CRH-Cre mice. We injected 100 nL of AAV2/5 DIO.mGFP (University of North Carolina, #AV4310i) unilaterally into CRHR1-Cre mice. We injected unilaterally 200 nL of AAV2/1 syn.GCaMP6f.WPRE.SV40 (University of Pennsylvania, #AV-1-PV2822) or AAV2/9 syn.CRF1.0 (Yulong Li Lab, Wang et al., 2022) into C57BL/6J WT mice. We injected bilaterally 100 nL of AA2/2 hSyn1.hChR2(H134R)-mCherry.WPRE (University of Zurich #V-124) or AA2/2 hSyn1.mCherry.WPRE (Addgene #114472-AAV2) into C57BL/6J WT mice. We injected bilaterally 100 nL of AAV2/8 hSyn.DIO.hM4D(Gi)-mCherry (Addgene #44362) or AAV2/8 hSyn.DIO.mCherry (Addgene #50459) into the rdLS of CRHR1-Cre mice. All viruses expressed for a minimum of 2 weeks.
Dual injection in rdLS and ILA
We injected C57BL/6 mice with 200 nL of HSV hEF1a.Cre in rdLS (coordinates in mm from Bregma: AP: 0.9, ML: 0, DV: -2.8). We also injected bilaterally 100 nL of AAV2/9 CMV-DIO-(mCherry-U6)-shRNA(anti-Crh) or AAV2/9 CMV-DIO-(mCherry-U6)-shRNA(scrambled) in ILA (coordinates in mm from Bregma): AP: 1.65, ML: ±0.1, DV: -2.6). We waited 3 weeks for viral expression.
Optical ferrule implants
Animals were anesthetized using isoflurane and given analgesics. The scalp was removed and we applied Vetbond™ (3M™ #7000002814) along the cut. A craniotomy was performed above the target region and the optical ferrule was lowered until the desired depth. Superglue was applied to hold the lens in position and then dental cement (GC FujiCEM 2) was applied to cover the exposed skull and keep the optical ferrule in position. Animal were allowed to recovered for 5 days before being used.
-
-
For fiberphotometry recording of ILACRH cells in the right hemisphere we implanted the optical ferrule (B280-4419-3, Doric) at the following coordinates: AP: 1.65, ML: 0.1, DV: -2.4.
-
-
For fiberphotometry recording of rdLS cells we implanted the optical ferrule at the following coordinates: AP: 0.9, ML: 0, DV: -2.65.
-
-
For silencing of ILACRH cells fibers in rdLS cells we implanted optical ferrules (Thorlab # FT200UMT and # CFLC230-10) bilaterally at the following coordinates: AP: 0.9, ML: ±0, DV: - 2.65.
-
-
For excitation rdLS cells we implanted the optical ferrule (Thorlab # FT200UMT and # CFLC230-10) at the following coordinates: AP: 0.9, ML: 0, DV: -2.65
Cannula guide implantation and micro-infusion
The mouse scalp was removed and scored before a hole was drilled (AP +0.9, ML ±0). A cannula guide extending 2.4 mm below the pedestal (Plastics One #C315G 2-G11-SPC) was lowered slowly and kept in place using superglue. The skull was then covered with dental cement (GC FujiCEM 2) and dummy cannulas (Plastics One #C315DC-SPC) were inserted into the guides. The mice were returned to their home cages and left to recover for at least 1 week. For rdLS infusion, mice were placed under light isoflurane anesthesia (2%) and the dummy cannula was removed. A cannula (Plastics One #C315I-SPC) projecting 2.5 mm from the tip of the cannula guide was mounted. 0.6 μL of a solution containing 2 μg of antalarmin dissolved in DMSO or DMSO (Sigma-Aldrich # D8418) only were infused over 5 min using the Fusion 200 syringe pusher (Chemix Inc.) mounted with a 2-μl syringe (Hamilton #88511). The cannula was removed 2 min after the end of the micro-infusion. Mice typically recovered fully from the light anesthesia within 5 min. Mice were returned to their home cages 20 min before the test began.
Immunohistochemistry (IHC)
Mice were anesthetized using isoflurane then perfused in the heart with 10 mL saline and their brains were quickly extracted and incubated in 4% PFA overnight. After 1 h washing in PBS, 60 μm slices were prepared using a Leica VT1000S vibratome (Leica Biosystems). Unless indicated otherwise, slices were permeabilized for 2h in PBS with 0.5% Triton-X100 (T9284, Sigma-Aldrich) in PBS before being incubated overnight at 4°C with primary antibodies diluted in PBS with 0.5% Triton-X in PBS. The slices were washed in PBS for 1 h then incubated overnight at 4°C with secondary antibodies from Thermo-Fisher Scientific at a concentration of 1:500 diluted in PBS with 0.1% Triton-X. Hoechst counterstain was applied (Hoechst 33342 at 1:1000 for 30 min in PBS at RT) prior to mounting the slice using fluoromount (Sigma-Aldrich). Images were acquired using inverted confocal microscopes (lsm 900, Zeiss and SPII,Leica) or an epifluorescent microscope (Thunder, Leica). For post-hoc immunocytochemistry after patch-clamp recordings, slices were fixed for 1 h in PBS with 4% PFA and streptavidin was applied during secondary incubation.
Fig. 1B-E: For mRuby and GFP labelling, primary incubation was performed overnight at 4°C with rabbit anti-RFP (1:500, Rockland Antibody, #600-401-379) and chicken anti-GFP (1:1000, Aves, #GFP-1020) antibodies. Secondary incubation was performed with anti-rabbit antibody conjugated to Alexa 568 (#A11036) and anti-chicken antibody conjugated to 488 (#A11039).
Fig. 1M and supplementary fig. 1, 4, 11 and 14: For YFP, GFP, GCaMP or CRF1.0 labelling, incubation was performed overnight at 4°C with anti-GFP antibody conjugated to Alexa 488 (1:500, Thermo-Fisher Scientific #A21311). No immunohistochemistry was performed against mCherry.
Supplementary fig. 3E: For GABA labelling, incubation was performed during 2 days at 4°C with an anti-GABA antibody (1:200, Abcam, #ab17413). Secondary incubation was performed with anti-guinea-pig antibody conjugated to Alexa 647 (#A21450). No immunohistochemistry was performed against GFP.
Fig. 2B and 5E: For mCherry labelling, primary incubation was performed overnight at 4°C with an anti-RFP antibody (1:500, Rockland Antibody, #600-401-379). Secondary incubation was performed with anti-rabbit antibody conjugated to Alexa 568 (#A11036).
Fig. 3P, 7B, 7G and supplementary fig. 14B,E: For c-fos labelling, primary incubation was performed overnight at 4°C with anti-c-fos antibody (1:1000, Abcam, #ab190289). Secondary incubation was performed with an anti-rabbit antibody conjugated to Alexa 568 (#A11036).
Fig. 5I, 7F and supplementary fig. 6A: For c-fos labelling, primary incubation was performed overnight at 4°C with an anti-c-fos antibody (1:1000, Abcam, #ab190289). Secondary incubation was performed with anti-rabbit antibody conjugated to Alexa 647 (#A32733). No immunohistochemistry was performed against mCherry or tdTomato.
Fig. 4L, 7R and supplementary fig. S9I: For tdTomato or mCherry labelling, primary incubation was performed overnight at 4°C with an anti-RFP antibody (1:500, Rockland Antibody, 600-401-379). Secondary incubation was performed with anti-rabbit antibody conjugated to Alexa 568 (#A11036).
Supplementary fig. 10B: For tdTomato and CRF1.0 labelling, primary incubation was performed overnight at 4°C with an anti-RFP antibody (1:500, Rockland Antibody, 600-401-379). Secondary incubation was performed with anti-rabbit antibody conjugated to Alexa 568 (#A11036) and anti-GFP antibody conjugated to Alexa 488 (1:500, Thermo-Fisher Scientific #A21311).
Fig. 5A and supplementary fig. 8A & 13B-C: For GFP or GCaMP6f labelling, incubation was performed overnight at 4°C with an anti-GFP antibody conjugated to Alexa 488 (1:500, Thermo-Fisher Scientific, #A21311).
Fig. 7I-J: For c-fos and mCherry labelling, primary incubation was performed overnight at 4°C with an anti-c-fos antibody (1:1000, Abcam, #ab190289) and anti-mCherry antibody (1:1000, Biorbyt, #orb153320). Secondary incubation was performed with anti-rabbit antibody conjugated to Alexa 647 (#A32733) and anti-goat secondary antibody conjugated to Alexa 568 (#A11057).
Fluorescence quantification in CRH-Cre mice injected in the PFC with AAV2/DJ hSyn.FLEX.mGFP.2A.Synatophysin-mRuby
Images were acquired using an epifluorescent microscope (Thunder, Leica). Images at 8 bits (0 to 255 intensity units/pixel) were analyzed using the software Image J. For each picture, we measure the mean fluorescence value of the background and then subtracted it from the mean fluorescence value of the region of interest (ROI). Fluorescence intensity = (Mean fluorescence of ROI – Mean fluorescence of the background).
In situ hybridization (ISH)
Mice were anesthetized using isoflurane then decapitated and their brain quickly extracted. Brains were then immersed in dry-ice cold Butan X for 6 s before being stored at -80°C. 16 μm-thick slices were prepared using a Leica cryostat (CM3050 S, Leica Biosystems) and mounted on Superfrost Plus microscope slides (12-550-15, FisherBrand). For Crh and mCherry labelling (Fig. 4A), we processed the slices using the RNAscope Multiplex Fluorescent Detection kit v1 (ACD Bio, #320851) with the probes for Crh in C1 (#316091), mCherry in C2 (#431201-C2). We applied Protease IV for 30 s and used the Amp4 Alt-A color module. The first version of the kit was discontinued and we performed the Crh only labelling (Fig. S15C) using the RNAscope Multiplex Fluorescent Detection kit v2 (#323110) with Crh in C1(#316091) with 30 s of Protease IV for and used the TSA Vivid Dye 520 at 1:750. We also performed Vgat/dsRed (Fig. S9C) labelling using the RNAscope Multiplex Fluorescent Detection kit v2 (ACD Bio, #323110) vGAT in C1 (#319191) and dsRed in C2 (#481361-C2) with 2 min of Protease IV. We used fluorescent dyes TSA Vivid Dye 520 at 1:750 for vGAT and TSA Vivid Dye 650 at 1:750 for dsRed. DAPI was apply for 30 s prior to mounting using fluoromount. Images were acquired using a confocal microscope (SPII, Leica).
In vitro electrophysiological recordings
We prepared coronal brain slices from 8- to 12-week-old C57BL6/J mice. Animals were killed under isoflurane anesthesia by perfusion into the right ventricle of ice-cold solution containing the following (in mM): 125 NaCl, 2.5 KCl, 22.5 glucose, 25 NaHCO3, 1.25 NaH2PO4, 3 Na Pyruvate, 1 Ascorbic acid, 2 CaC2 and 1 MgC2). ACSF was saturated with 95% O2 and 5% CO2, pH 7.4. Brains were cut into 400 μm slices with a vibratome (VT1200S, Leica) in the same ice-cold dissection solution. Slices were then transferred to an incubation chamber containing the same ACSF solution. The chamber was kept at 34°C for 30 min and then at room temperature for at least 1 h before recording. All experiments were performed at room temperature. Slices were mounted in the recording chamber under a microscope. Recordings were acquired using the Multiclamp 700A amplifier (Molecular Device), data acquisition interface ITC-18 (Instrutech) and the Axograph software. We targeted CA2 PNs based somatic location and size in both deep and superficial layer. Whole-cell recordings were obtained from LS neurons in voltage-clamp mode at -70 mV patch pipette (3–5 MΩ) containing the following (in mM): 135 Cs-gluconate, 5 KCl, 0.2 EGTA-Na, 10 HEPES, 2 NaCl, 5 ATP, 0.4 GTP, 10 phosphocreatine, and 5 μM biocytin, pH 7.2 (280–290 mOsm). The liquid junction potential was 1.2 mV and was left uncorrected. Inhibitory currents were recorded in voltage-clamp configuration at +10 mV. We recorded rdLS neurons in septal slices from CRH-Cre mice expressing channelrhodopsin and applied 2 μM SR 95531 (Tocris, # 1262) and 1 μM CGP 55845 (Tocris, #1248) to block GABAA and GABAB receptors, respectively while monitoring the light induced IPSCs in rdLS neurons. We also applied a tetanic light stimulation (3 times 100 pulses of 1 ms at 100 Hz) with or without 300 nM antalarmin (Tocris, #2778) present in the bath. We also recorded LS neurons from WT mice. After 10 min of stable baseline recording, stressin-1 (300 nM, Tocris # 1608) was applied following a 1:1000 dilution from stock solution into the ACSF. We used the Axograph software for data acquisition, and Excel (Microsoft) and PRISM (Graphpad) for data analysis.
Behavioral tests
Based on our experience conducting similar social behavior experiments, we used group size of 10-15 animals. Animals that had viral expression outside of ILA or rdLS were excluded from analysis. This criterion was pre-established since we wanted to investigate the role of local neurons. The observer was blind to the identity of the mice while performing the behavioral experiments and the subsequent analyses.
Open arena test
Mice were introduced into a new arena (60 cm x 60 cm) and allowed to roam freely for 10 min. Using automatic tracking of the test mouse (Any-Maze 7, Stoelting), we quantified the total distance travelled as well as time spent in the surround (20% of the surface) or center (remaining 80% of the surface).
Elevated plus maze test
Mice were placed at the center of a maze consisting of a cross with two open arms and two closed arms. They were allowed to explore the maze freely for 5 min. We quantified the amount of time and number of entries in open or closed arms.
Sociability test
Test mice were introduced into a same open arena and allowed to roam freely for 10 min. Then, wire cup cages were introduced at opposite corners. One cup had a novel mouse from the same sex and age underneath it. The test mouse was allowed to explore each cup for 5 min. Using automatic tracking of the test mice (ANY-Maze, Stoelting), we quantified the time spent in the area surrounding each cup. Using the interaction times, we calculated a discrimination index for social preference as the following: DI = (time with mouse – time with empty cup) / total interaction time. DI represents the percentage of extra time spent with the mouse compared to the empty cup. The preference exhibited by the test mouse for the mouse compared to the empty cup was used as an index for sociability.
Social novelty preference test
This test was performed in the same arena as the open-field and sociability test. Test mouse was allowed to explore the arena for 10 min. Then, the test mouse explored for 5 min two wire cups on opposite corners (learning trial). Each cup contained a novel mouse. Stimulus mice were removed from the cups and the test mouse were placed in an empty cage, the size of its home cage for 30 min. Then, one of the now familiar mice was returned under its cup while a novel mouse was introduced under the other cup (recall trial). The test mouse was free to explore each cup again for 5 min. Importantly, all 3 stimulus mice were littermates housed in the same cage (thus preventing the test mice to use any cage-specific odor clue). The design of this test allows for better exploration of each cup than the classical 3-chamber test while still giving the test mouse freedom to explore the novel or familiar mice unlike the direct interaction test. Using automatic tracking of the test mice (ANY-Maze, Stoelting), we quantified the time the test mice spent in the immediate area surrounding each cup - and therefore, interacting with each stimulus mice - during learning and recall. Using the interaction time from the recall trial, we calculated a discrimination index for social novelty preference as the following: DI = (time with novel – time with familiar) / total interaction time. DI represents the percentage of extra time spent with novel compared to familiar. The preference normally exhibited by the test mouse for the novel compared to the familiar animal was used as an index for social memory. We also calculated the total time spent interacting with stimulus mice during each trial in order to verify that the sociability drive was not affected.
Novel object preference test
Same as above but with 3 different small toys of different shape about the size of the mouse. Objects were of the same color.
Repetitive social presentation test
Test mice were introduced in a clean cage the same size than their home cage and left to explore for 20 min. Then, a novel mouse of the same age and sex was introduced for four times during 2 min with 10 min inter-trial. For the 5th trial, a novel age- and sex-matched mouse housed in the same cage than the first one was presented. Films were scored offline for social interaction by a trained observer blind to the experimental condition. Sniffing of any part of the body, allo-grooming and close following counted as social interaction. Interaction times were normalized with respect to the first interaction.
Repetitive object presentation test
Same as above but with 2 different small toys of different shape about the size of the mouse. Objects were of the same color.
Food-seeking test
Mice were food-deprived overnight and then introduce into a large arena (60x60 cm) containing some crushed chow in the middle. We measured the latency to feed and the amount of time spent feeding.
Food preference test
Mice were food-deprived overnight and then introduce into a large arena (60x60 cm) containing their regular food (chow) in one corner, and chocolate, as a novel food, in the opposite corner of the arena. We measured the amount of time spent feeding as well as the duration spent in each food zone. Notice that mice are neophobic for food and prefer familiar chow even to more palatable food like chocolate.
One trial presentation of novel or familiar mice
Test mice were individually placed in a clean cage the night before the test. The next day a novel mouse or a littermate was introduced for 1 single trial during 2 min. The total time of interaction during the 2 min was simultaneously annotated during film recording. Social interaction was measure by a key-press using Any-Maze when sniffing of any part of the body or chasing occurred. Mice were perfused 1 hour after performing the test and processed for c-fos analysis.
Kinship preference test
For this test, we used two females with a positive plug from the same day in order to have two litters of the same developmental stage. After the pups of both litters were born, we started to test the percentage of sibling choice of each pup during consecutive days. From P5 to P14 the test was performed as following: each nest containing their corresponding litter was placed in each corner of the OA. Then we place each individual pup in a middle point equidistant from both nests. We recorded each pup during 2 min, then we annotated weather the pup ended reaching the sibling or non-sibling nest. Due to the natural increasing exploration behavior of the pups, after P14, we placed all pups of each litter under a pencil cup and placed them in opposite zones of the OA. Then we tested the percentage of sibling preference by measuring the time spent in each zone during 2 min. We only used litters of 5 pups or more, and with similar number of pups between the litters. We also measured the time spent near each nest to calculate a discrimination index for familiar kin similar to the one calculated previously.
Optogenetic stimulation during social interaction
Test mice were introduced in a clean cage the same size than their home cage and left to explore for 5 min. Then, 445 nm laser stimulation was applied and a littermate or a novel mouse of the same age and sex was introduced for 2 minutes. Films were scored offline for social interaction by a trained observer blind to the experimental condition. Sniffing of any part of the body, allo-grooming and close following counted as social interaction.
Fiberphotometry data acquisition and analysis
Fiber-photometry recording during social or object presentation
Test mice were individually placed in a clean cage the night before the test. The day of recording, mice were habituated to the optical fiber for three days by placing the fiber on the mouse head and letting it roam free for 15 min in their own cage before the test. Then, we recorded basal activity during two minutes. After 5 minutes, we performed 2 min social or object presentations with 5 minutes in between recordings. Novel and familiar mice or objects were introduced for 1 single trial during 2 min. The order of the familiar or novel presentations were randomized along all the sessions. Movies were annotated online to mark the beginning and end of the interactions.
Fiber-photometry recording during repetitive social presentation
Mice were habituated to the optical fiber for three days by placing the fiber on the mouse head and letting it roam free for 10 min. The test was conducted as described previously.
Optogenetic silencing of CRH-ILA terminals in rdLS and CRH-Biosensor recording in rdLS during interaction with familiar mice
Mice were injected with the Cre-dependent AAV2/2 CAG.FLEX.ArchT-tdTomato (Addgene #28305 prepared by the University of North Carolina vector core) in the ILA AP: 1.65, ML: ±0.1, DV: -2.6, and a with the CRH-Biosensor pAAV-hSyn-CRF1.0 in LS AP: 0.9, ML: 0, DV: -2.75. For fiber-photometry imaging of LS cells, we implanted the optical ferrule (B280-4419-3, Doric) at the following coordinates AP: 0.9, ML: 0, DV: -2.65. For CRH-ILA terminal silencing in rdLS, we implanted an optical ferrule of 200um using the following coordinates AP: 1.70 ML: 0.2 DV: 3.1, 30° angle. Test mice were introduced in a clean cage the same size as their home cage and left to explore for 15 min. Then, we perform three consecutive trials presenting a littermate for 2 minutes. Trials were developed within 10 min intervals. Light stimulation was developed using the Cobolt-Jive 561nm adjusted at 5mW and applied during Trial 2. CRH-Biosensor activity was recorded using a DORIC system (Basic FMC). Films were scored in real time for social interaction by a trained observer blind to the experimental condition. Sniffing of any part of the body, allo-grooming and close following counted as social interaction.
Fiberphotometry was conducted using a DORIC system (Basic FMC). Two LEDs (405 nm and 465 nm) were coupled to a fluorescence mini-cube (FMC) to deliver light into optical ferrules permanently implanted above the ILA or rdLS. We used 217 Hz as the oscillation frequency for the 465 nm calcium-dependent fluorescence channel, and 319 Hz as the frequency for the 405 nm isosbestic signal. For calcium recording, light was delivered at a final intensity of 2.24 mW (465nm) and 2.76 mW (405 nm) at the tip of the patch-cord before coupling with the implanted ferrule80. For the CRF biosensor recording, light was delivered at a final intensity of 1.1 mW (465nm) and 1.9 mW (405 nm) at the tip of the patch-cord in order to limit bleaching. Emitted light between 420 and 450 nm (with 405 nm excitation) and 500 and 540 nm (with 465 nm excitation) were collected through the FMC on separate fiber-coupled Newport 2151 photo-receiver modules. The collected fluorescent signals were collected in AC-low mode and converted to voltage via the formula V = PRG, where V is the collected voltage, P is the optical input power in watts, R is photodetector responsivity in amps/watts (0.2 – 0.4), and G is the trans-impedance gain of the amplifier. Raw signals for 473 nm excitation (GCaMP6f) and 405 nm excitation (isosbestic signal) were recorded using Doric Neuroscience Studio software.
We used the Guppy software for analysis78. After loading the data, we apply an artifact correction criterion consisting on the removal of the first second of the raw data. This allowed us to remove the artifacts that are usually generated right after the light sources turn on. Subtraction of the background fluorescence was calculated via a time-fitted running average of the 465 nm channel relative to the 405 nm control channel using a least squares polynomial fit of degree 1. ΔF/F is calculated by subtracting the fitted control channel from the signal channel and dividing by the fitted control channel using the formula (465 nm – 405 nm)/405 nm. A peak enveloping Fourier transform was applied to the ΔF/F signal across the entire trace to identify peaks in activity. Finally, we presented the data as the deviation of the ΔF/F from its mean (z-score) using the formula (ΔF/F – mean of AF/F/Standard deviation of AF/F).
For PSTH analysis, we analyzed the z-score for each interaction during the sessions based on the following time window (0 to 3 s after the timestamp).
Classifier analysis
Fiberphotometry recordings from ILACRH and rdLS were used to discriminate between novel or familiar interactions. On each session, we detected calcium transients and fit support vector machines (SVM) with linear kernel to discriminate whether each transient occurred during a novel or a familiar interaction. We used two different approaches to fit the SVMs: individual sessions and pseudo-populations81,82. For the individual analysis, we fitted an independent SVM on each individual session (10 sessions for ILACRH and 10 sessions for rdLS recordings) and evaluated the cross-validated decoding performance per session. Cross-validation was performed by training the classifier on 80% of the dataset and testing the performance on the remaining 20%. Since this partition was random, we repeated this process 20 times and reported the mean. To evaluate the significance of the obtained decoding performance, we built a null-distribution of decoding performances by randomly shuffling the labels for novel or familiar interaction. On each shuffle iteration, we first shuffled the labels and then evaluated the cross-validated performance as in the original dataset (80% train, 20% test, 20 iterations). We repeated this process 1000 times. The reported p-value was obtained by comparing the decoding performance of the original dataset with the null distribution. In all cases, the decoding performance was larger than the largest value of the null distribution (p < 0.001). For the pseudo-population analysis, for each session we randomly draw (with replacement) 100 trials from familiar and 100 trials from novel interactions. By combining all recording sessions from a particular brain region, we obtained one matrix for training and one matrix for testing (200 trials x 10 sessions) our linear classifiers (SVM). The matrices for training and testing were obtained by sampling with replacement from 80% and 20% of the trials from the original dataset, respectively. Our procedure ensured that there was no overlap between the train and test matrices. Since the procedure to build these two matrices was random, we repeated this process 100 times and reported the mean decoding performance. To evaluate the significance of the obtained decoding performance for the pseudo-population, we also built a null-distribution of decoding performances by randomly shuffling the labels for novel or familiar interaction on each recording session and proceeded as described above (1000 iterations). The reported p-value was obtained by comparing the decoding performance of the original dataset with the null distribution. As for the individual sessions analysis, the decoding performance was larger than the largest value of the null distribution. The shaded areas correspond to the 2.5 – 97.5 percentile of the null distribution. In all cases, the mean of the null distribution was very close to 50%. Code is available at https://github.com/ramonnogueira/decode_familiarity.
Statistics
Statistical tests were performed using PRISM (Graphpad). Results presented in the text, figures and figure legends are reported as the mean ± SEM. Unless specified otherwise all tests are two-tailed. * is for p < 0.05, ** is for p < 0.01, *** is for p < 0.001. When multiple observations were done in the same mouse, we used nested statistical tests to take into account the lower degree of freedom.
Supplementary Material
Acknowledgments
Our co-author Jay Shulkin sadly passed away during the final stage of the reviewing process. We would like to acknowledge his lifetime dedication to science, his good humor and boundless motivation to discuss experiments and results. We miss him dearly. We thank Christina Fregola and Georg Keller for providing viral vectors, Jan Deussing for sharing the CRHR1-Cre mouse line and Yulong Li for providing the CRF1.0 biosensor. We also thank Antoine Besnard, Claudio Elgueta, Azahara Oliva, Isabel Otaño-Perez, Francesco Papaleo and Steve Siegelbaum for critical reading of the manuscript and insightful comments. This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No 949652). F.L. also acknowledges support from a CIDEGENT grant from the Valencian Community, Severo Ochoa Foundation and a NARSAD young investigator grant from the Brain and Behavior research foundation, funded by the Osterhaus Family. R.N. is supported by NSF Neuronex (1707398) and the Gatsby Charitable Foundation (GAT3708).
Footnotes
Author Contributions
Conceptualization: N.S.L.R., J.S., A.A and F.L.; Intra-cellular recordings: N.S.L.R. and F.L.; Behavioral assays and viral injections: N.S.L.R., P.S.D., A.A. and F.L.; Immunohistochemistry and in situ hybridization: N.S.L.R., A.R, Y.N. and F.L.; shRNA design C.S., Fiberphotometry: N.S.L.R., Y.N.; Classifier analysis: R.N.; Writing – Original draft: N.S.L.R and F.L.; Writing – Review and editing: N.S.L.R., A.A. and F.L.; Visualization: N.S.L.R. and F.L.; Supervision: F.L.; Funding acquisition: F.L.
Declaration of interests
Christopher de Solis is currently an employee of Rejuvenate Bio, San Diego, USA. Arun Asok is currently an employee of Exponent Inc, Philadelphia, USA. Their current work is unrelated to the contents of this manuscript, and their contributions to this manuscript were made while previously employed at Columbia University.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. The code used to create the classifiers analysis of the fiber-photometry data is available at https://github.com/ramonnogueira/decode_familiarity. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
Bibliography
- 1.Amdam GV, Hovland AL. Measuring Animal Preferences and Choice Behavior. Nature Education Knowledge. 2012;3:1–9. [Google Scholar]
- 2.Blaustein AR, O’Hara RK. Genetic control for sibling recognition? Nature. 1981;290:246–248. doi: 10.1038/290246A0. [DOI] [PubMed] [Google Scholar]
- 3.Clemens AM, Wang H, Brecht M. The lateral septum mediates kinship behavior in the rat. Nat Commun. 2020;11:3161. doi: 10.1038/s41467-020-16489-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hepper PG. Sibling recognition in the rat. Anim Behav. 1983;31:1177–1191. doi: 10.1016/S0003-3472(83)80024-4. [DOI] [Google Scholar]
- 5.Kogo H, Kiyokawa Y, Takeuchi Y. Rats show a preference for unfamiliar strains of rats. bioRxiv. 2021:6. doi: 10.1101/2021.02.18.431764. [DOI] [PubMed] [Google Scholar]
- 6.Stowers L, Holy TE, Meister M, Dulac C, Koentges G. Loss of sex discrimination and male–male aggression in mice deficient for TRP2. Science (1979) 2002;295:1493–1500. doi: 10.1126/science.1069259. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Jiang YA, Si Y, Kim JY, Chen ZF, Rao Y. Molecular regulation of sexual preference revealed by genetic studies of 5-HT in the brains of male mice. Nature. 2011;472:95–99. doi: 10.1038/nature09822. 2011 472:7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kingsbury L, Huang S, Raam T, Ye LS, Wei D, Hu RK, Ye L, Hong W. Cortical Representations of Conspecific Sex Shape Social Behavior. Neuron. 2020;107:941–953.:e7. doi: 10.1016/J.NEURON.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mossman CA, Drickamer LC. Odor Preferences of Female House Mice (Mus domesticus) in Seminatural Enclosures. J Comp Psychol. 1996;110:131–138. doi: 10.1037/0735-7036.110.2.131. [DOI] [PubMed] [Google Scholar]
- 10.Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, Piven J, Crawley JN. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302. doi: 10.1111/J.1601-1848.2004.00076.X. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe S. The dominant/subordinate relationship between mice modifies the approach behavior toward a cage mate experiencing pain. Behavioural Processes. 2014;103:1–4. doi: 10.1016/J.BEPROC.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 12.van Loo PLP, de Groot AC, van Zutphen BFM, Baumans V. Do Male Mice Prefer or Avoid Each Other’s Company? Influence of Hierarchy, Kinship, and Familiarity. 2010;4:91–103. doi: 10.1207/S15327604JAWS0402_1. http://dx.doi.org/10.1207/S15327604JAWS0402_1. [DOI] [Google Scholar]
- 13.Scheggia D, Managò F, Maltese F, Bruni S, Nigro M, Dautan D, Latuske P, Contarini G, Gomez-Gonzalo M, Requie LM, et al. Somatostatin interneurons in the prefrontal cortex control affective state discrimination in mice. Nat Neurosci. 2020;23:47–60. doi: 10.1038/S41593-019-0551-8. [DOI] [PubMed] [Google Scholar]
- 14.Nadler JJ, Moy SS, Dold G, Trang D, Simmons N, Perez A, Young NB, Barbaro RP, Piven J, Magnuson TR, et al. Automated apparatus for quantitation of social approach behaviors in mice. Genes Brain Behav. 2004;3:303–314. doi: 10.1111/J.1601-183X.2004.00071.X. [DOI] [PubMed] [Google Scholar]
- 15.Oliva A, Fernandez-Ruiz A, Leroy F, Siegelbaum SA. Hippocampal CA2 sharp-wave ripples reactivate and promote social memory. Nature. 2020;587:264–269. doi: 10.1038/s41586-020-2758-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gheusi G, Bluthé RM, Goodall G, Dantzer R. Social and individual recognition in rodents: Methodological aspects and neurobiological bases. Behavioural Processes. 1994;33:59–87. doi: 10.1016/0376-6357(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 17.Manduca A, Carbone E, Schiavi S, Cacchione C, Buzzelli V, Campolongo P, Trezza V. The neurochemistry of social reward during development: What have we learned from rodent models? J Neurochem. 2021;757:1408–1435. doi: 10.1111/JNC.15321. [DOI] [PubMed] [Google Scholar]
- 18.Domínguez S, Rey CC, Therreau L, Fanton A, Massotte D, Verret L, Piskorowski RA, Chevaleyre V. Maturation of PNN and ErbB4 Signaling in Area CA2 during Adolescence Underlies the Emergence of PV Interneuron Plasticity and Social Memory. Cell Rep. 2019;29:1099–1112.:e4. doi: 10.1016/j.celrep.2019.09.044. [DOI] [PubMed] [Google Scholar]
- 19.Laham BJ, Diethorn EJ, Gould E. Newborn mice form lasting CA2-dependent memories of their mothers. Cell Rep. 2021;34:108668. doi: 10.1016/j.celrep.2020.108668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menon R, Süß T, de M Oliveira VE, Neumann ID, Bludau A. Neurobiology of the lateral septum: regulation of social behavior. Trends Neurosci. 2022;45:27–40. doi: 10.1016/j.tins.2021.10.010. [DOI] [PubMed] [Google Scholar]
- 21.Bielsky IF, Hu S-B, Ren X, Terwilliger EF, Young LJ. The V1a Vasopressin Receptor Is Necessary and Sufficient for Normal Social Recognition: A Gene Replacement Study. Neuron. 2005;47:503–513. doi: 10.1016/j.neuron.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 22.Lukas M, Toth I, Veenema AH, Neumann ID. Oxytocin mediates rodent social memory within the lateral septum and the medial amygdala depending on the relevance of the social stimulus: male juvenile versus female adult conspecifics. Psychoneuroendocrinology. 2013;38:916–926. doi: 10.1016/J.PSYNEUEN.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 23.Gutzeit VA, Ahuna K, Santos TL, Cunningham AM, Sadsad Rooney M, Muñoz Zamora A, Denny CA, Donaldson ZR. Optogenetic reactivation of prefrontal social neural ensembles mimics social buffering of fear. Neuropsychopharmacology. 2020;45:1068–1077. doi: 10.1038/s41386-020-0631-1. 2020 45:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy DR, Tamir T, Kaufman M, Parabucki A, Weissbrod A, Schneidman E, Yizhar O. Dynamics of social representation in the mouse prefrontal cortex. Nature Neuroscience. 2019;22:2013–2022. doi: 10.1038/s41593-019-0531-z. 2019 22:12. [DOI] [PubMed] [Google Scholar]
- 25.Kingsbury L, Huang S, Wang J, Gu K, Golshani P, Wu YE, Hong W. Correlated Neural Activity and Encoding of Behavior across Brains of Socially Interacting Animals. Cell. 2019;178:429–446.:e16. doi: 10.1016/J.CELL.2019.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park G, Ryu C, Kim S, Jeong SJ, Koo JW, Lee Y-S, Kim SJ. Social isolation impairs the prefrontal-nucleus accumbens circuit subserving social recognition in mice. Cell Rep. 2021;35:109104. doi: 10.1016/j.celrep.2021.109104. [DOI] [PubMed] [Google Scholar]
- 27.Yizhar O, Levy DR. The social dilemma: prefrontal control of mammalian sociability. Curr Opin Neurobiol. 2021;68:67–75. doi: 10.1016/j.conb.2021.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Carus-Cadavieco M, Gorbati M, Ye L, Bender F, van der Veldt S, Kosse C, Börgers C, Lee SY, Ramakrishnan C, Hu Y, et al. Gamma oscillations organize top-down signalling to hypothalamus and enable food seeking. Nature. 2017;542:232–236. doi: 10.1038/nature21066. [DOI] [PubMed] [Google Scholar]
- 29.Vale W, Spiess J, Rivier C, Rivier J. Characterization of a 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and betaendorphin. Science. 1981;273:1394–1397. doi: 10.1126/SCIENCE.6267699. [DOI] [PubMed] [Google Scholar]
- 30.Schulkin J. The CRF signal: Uncovering an information molecule. Oxford University Press; 2017. [DOI] [Google Scholar]
- 31.Hostetler CM, Ryabinin AE. The crf system and social behavior: A review. Front Neurosci. 2013:92. doi: 10.3389/FNINS.2013.00092/BIBTEX. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinrichs SC. Modulation of social learning in rats by brain corticotropin-releasing factor. Brain Res. 2003;994:107–114. doi: 10.1016/J.BRAINRES.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 33.Dinan TG, Lavelle E, Scott LV, Newell-Price J, Medbak S, Grossman AB. Desmopressin normalizes the blunted adrenocorticotropin response to corticotropin-releasing hormone in melancholic depression: evidence of enhanced vasopressinergic responsivity. J Clin Endocrinol Metab. 1999;84:2238–2240. doi: 10.1210/JCEM.84.6.5723. [DOI] [PubMed] [Google Scholar]
- 34.Raadsheer FC, Hoogendijk WJG, Stam FC, Tilders FJH, Swaab DF. Increased numbers of corticotropin-releasing hormone expressing neurons in the hypothalamic paraventricular nucleus of depressed patients. Neuroendocrinology. 1994;60:436–444. doi: 10.1159/000126778. [DOI] [PubMed] [Google Scholar]
- 35.Risbrough VB, Stein MB. Role of Corticotropin Releasing Factor in Anxiety Disorders: A Translational Research Perspective. Horm Behav. 2006;50:550. doi: 10.1016/J.YHBEH.2006.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dedic N, Kühne C, Gomes KS, Hartmann J, Ressler KJ, Schmidt Mv, Deussing JM. Deletion of CRH From GABAergic Forebrain Neurons Promotes Stress Resilience and Dampens Stress-Induced Changes in Neuronal Activity. Front Neurosci. 2019;13 doi: 10.3389/fnins.2019.00986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell BM, Morrison JL, Walker EL, Merchant KM. Differential regulation of behavioral, genomic, and neuroendocrine responses by CRF infusions in rats. Pharmacol Biochem Behav. 2004;77:447–455. doi: 10.1016/J.PBB.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 38.Dunn AJ, File SE. Corticotropin-releasing factor has an anxiogenic action in the social interaction test. Horm Behav. 1987;21:193–202. doi: 10.1016/0018-506X(87)90044-4. [DOI] [PubMed] [Google Scholar]
- 39.Elkabir DR, Wyatt ME, Vellucci Sv, Herbert J. The effects of separate or combined infusions of corticotrophin-releasing factor and vasopressin either intraventricularly or into the amygdala on aggressive and investigative behaviour in the rat. Regul Pept. 1990;28:199–214. doi: 10.1016/0167-0115(90)90018-R. [DOI] [PubMed] [Google Scholar]
- 40.Gehlert DR, Shekhar A, Morin SM, Hipskind PA, Zink C, Gackenheimer SL, Shaw J, Fitz SD, Sajdyk TJ. Stress and central Urocortin increase anxiety-like behavior in the social interaction test via the CRF1 receptor. Eur J Pharmacol. 2005;509:145–153. doi: 10.1016/J.EJPHAR.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 41.Mele A, Cabib S, Oliverio A, Melchiorri P, Puglisi-Allegra S. Effects of corticotropin releasing factor and sauvagine on social behavior of isolated mice. Peptides (NY) 1987;8:935–938. doi: 10.1016/0196-9781(87)90083-0. [DOI] [PubMed] [Google Scholar]
- 42.Zhao Y, Valdez GR, Fekete EM, Rivier JE, Vale WW, Rice KC, Weiss F, Zorrilla EP. Subtype-selective corticotropin-releasing factor receptor agonists exert contrasting, but not opposite, effects on anxiety-related behavior in rats. J Pharmacol Exp Ther. 2007;323:846–854. doi: 10.1124/JPET.107.123208. [DOI] [PubMed] [Google Scholar]
- 43.Bagosi Z, Czébely-Lénárt A, Karasz G, Csabafi K, Jaszberenyi M, Telegdy G. The effects of CRF and urocortins on the preference for social novelty of mice. Behavioural Brain Research. 2017;324:146–154. doi: 10.1016/J.BBR.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 44.Kasahara M, Groenink L, Kas MJH, Bijlsma EY, Olivier B, Sarnyai Z. Influence of transgenic corticotropin-releasing factor (CRF) over-expression on social recognition memory in mice. Behavioural brain research. 2011;218:357–362. doi: 10.1016/J.BBR.2010.12.029. [DOI] [PubMed] [Google Scholar]
- 45.Chen P, Lou S, Huang ZH, Wang Z, Shan QH, Wang Y, Yang Y, Li X, Gong H, Jin Y, et al. Prefrontal Cortex Corticotropin-Releasing Factor Neurons Control Behavioral Style Selection under Challenging Situations. Neuron. 2020;106:301–315.:e7. doi: 10.1016/J.NEURON.2020.01.033. [DOI] [PubMed] [Google Scholar]
- 46.van Pett K, Viau V, Bittencourt JC, Chan RKWK, Li HY, Arias C, Prins GS, Perrin M, Vale W, Sawchenko PE. Distribution of mRNAs encoding CRF receptors in brain and pituitary of rat and mouse. J Comp Neurol. 2000;428:191–212. doi: 10.1002/1096-9861(20001211)428:2<191::AID-CNE1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 47.Paxinos G, Franklin KBJ. Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates. 2012 doi: 10.1364/OE.20.020998. [DOI] [Google Scholar]
- 48.Bernardi S, Benna MK, Rigotti M, Munuera J, Fusi S, Salzman CD. The Geometry of Abstraction in the Hippocampus and Prefrontal Cortex. Cell. 2020;183:954–967.:e21. doi: 10.1016/J.CELL.2020.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu X, Ye Z, Houston CM, Zecharia AY, Ma Y, Zhang Z, Uygun DS, Parker S, Vyssotski AL, Yustos R, et al. Wakefulness Is Governed by GABA and Histamine Cotransmission. Neuron. 2015;87:164–178. doi: 10.1016/J.NEURON.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pomrenze MB, Giovanetti SM, Maiya R, Gordon AG, Kreeger LJ, Messing RO. Dissecting the Roles of GABA and Neuropeptides from Rat Central Amygdala CRF Neurons in Anxiety and Fear Learning. Cell Rep. 2019;29:13–21.:e4. doi: 10.1016/J.CELREP.2019.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H, Qian T, Zhao Y, Zhuo Y, Wu C, Osakada T, Chen P, Ren H, Yan Y, Geng L, et al. A toolkit of highly selective and sensitive genetically encoded neuropeptide sensors. bioRxiv. 2022:2022.03.26.485911. doi: 10.1101/2022.03.26.485911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dedic N, Kühne C, Jakovcevski M, Hartmann J, Genewsky AJ, Gomes KS, Anderzhanova E, Pöhlmann ML, Chang S, Kolarz A, et al. Chronic CRH depletion from GABAergic, long-range projection neurons in the extended amygdala reduces dopamine release and increases anxiety. Nat Neurosci. 2018;21:803–807. doi: 10.1038/s41593-018-0151-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Chen ZP, Zhuang QX, Zhang XY, Li HZ, Wang JJ, Zhu JN. Role of Corticotropin-Releasing Factor in Cerebellar Motor Control and Ataxia. Curr Biol. 2017;27:2661–2669.:e5. doi: 10.1016/J.CUB.2017.07.035. [DOI] [PubMed] [Google Scholar]
- 54.Asok A, Schulkin J, Rosen JB. Corticotropin releasing factor type-1 receptor antagonism in the dorsolateral bed nucleus of the stria terminalis disrupts contextually conditioned fear, but not unconditioned fear to a predator odor. Psychoneuroendocrinology. 2016;70:17–24. doi: 10.1016/j.psyneuen.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanaka KZ, Pevzner A, Hamidi AB, Nakazawa Y, Graham J, Wiltgen BJ. Cortical representations are reinstated by the hippocampus during memory retrieval. Neuron. 2014;84:347–354. doi: 10.1016/J.NEURON.2014.09.037. [DOI] [PubMed] [Google Scholar]
- 56.Clemens AM, Brecht M. Neural representations of kinship. Curr Opin Neurobiol. 2021;68:116–123. doi: 10.1016/j.conb.2021.02.007. [DOI] [PubMed] [Google Scholar]
- 57.Dölen G, Darvishzadeh A, Huang KW, Malenka RC. Social reward requires coordinated activity of nucleus accumbens oxytocin and serotonin. Nature. 2013;501:179–184. doi: 10.1038/NATURE12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hung LW, Neuner S, Polepalli JS, Beier KT, Wright M, Walsh JJ, Lewis EM, Luo L, Deisseroth K, Dölen G, et al. Gating of social reward by oxytocin in the ventral tegmental area. Science. 2017;357:1406–1411. doi: 10.1126/SCIENCE.AAN4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, Lammel S, Mirzabekov JJ, Airan RD, Zalocusky KA, et al. Natural Neural Projection Dynamics Underlying Social Behavior. Cell. 2014;157:1535–1551. doi: 10.1016/j.cell.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Zhong W, Wang D, Feng Q, Liu Z, Zhou J, Jia C, Hu F, Zeng J, Guo Q, et al. Serotonin neurons in the dorsal raphe nucleus encode reward signals. Nat Commun. 2016;7 doi: 10.1038/NCOMMS10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Golden SA, Heshmati M, Flanigan M, Christoffel DJ, Guise K, Pfau ML, Aleyasin H, Menard C, Zhang H, Hodes GE, et al. Basal forebrain projections to the lateral habenula modulate aggression reward. Nature. 2016;534:688–692. doi: 10.1038/nature18601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Y, Deng SL, Li LX, Zhou ZX, Lv Q, Wang ZY, Wang F, Chen JG. A circuit from dorsal hippocampal CA3 to parvafox nucleus mediates chronic social defeat stress-induced deficits in preference for social novelty. Sci Adv. 2022;8:eabe8828. doi: 10.1126/sciadv.abe8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okuyama T, Kitamura T, Roy DS, Itohara S, Tonegawa S. Ventral CA1 neurons store social memory. Science (1979) 2016;353:1536–1541. doi: 10.1126/science.aaf7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phillips ML, Robinson HA, Pozzo-Miller L. Ventral hippocampal projections to the medial prefrontal cortex regulate social memory. Elife. 2019;8 doi: 10.7554/eLife.44182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun Q, Li X, Li A, Zhang J, Ding Z, Gong H, Luo Q. Ventral Hippocampal-Prefrontal Interaction Affects Social Behavior via Parvalbumin Positive Neurons in the Medial Prefrontal Cortex. iScience. 2020;23 doi: 10.1016/J.ISCI.2020.100894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Castelhano-Carlos MJ, Sousa N, Ohl F, Baumans V. Identification methods in newborn C57BL/6 mice: A developmental and behavioural evaluation. Lab Anim. 2010;44:88–103. doi: 10.1258/la.2009.009044. [DOI] [PubMed] [Google Scholar]
- 67.Besnard A, Leroy F. Top-down regulation of motivated behaviors via lateral septum sub-circuits. Mol Psychiatry. 2022 doi: 10.1038/S41380-022-01599-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beery AK, Christensen JD, Lee NS, Blandino KL. Specificity in Sociality: Mice and Prairie Voles Exhibit Different Patterns of Peer Affiliation. Front Behav Neurosci. 2018;12 doi: 10.3389/FNBEH.2018.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee NS, Goodwin NL, Freitas KE, Beery AK. Affiliation, Aggression, and Selectivity of Peer Relationships in Meadow and Prairie Voles. Front Behav Neurosci. 2019;13:52. doi: 10.3389/fnbeh.2019.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lim MM, Tsivkovskaia NO, Bai Y, Young LJ, Ryabinin AE. Distribution of corticotropin-releasing factor and urocortin 1 in the vole brain. Brain Behav Evol. 2006;68:229–240. doi: 10.1159/000094360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DeVries AC, Guptaa T, Cardillo S, Cho M, Carter CS. Corticotropin-releasing factor induces social preferences in male prairie voles. Psychoneuroendocrinology. 2002;27:705–714. doi: 10.1016/S0306-4530(01)00073-7. [DOI] [PubMed] [Google Scholar]
- 72.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 5th Edition. American Psychiatric Publishing, Inc; 2013. Cautionary Statement for Forensic Use of DSM-5. [DOI] [Google Scholar]
- 73.Laryea G, Arnett MG, Muglia LJ. Behavioral Studies and Genetic Alterations in Corticotropin-Releasing Hormone (CRH) Neurocircuitry: Insights into Human Psychiatric Disorders. Behavioral sciences (Basel, Switzerland) 2012;2:135–171. doi: 10.3390/BS2020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aly M, Yonelinas AP, Kishiyama MM, Knight RT. Damage to the lateral prefrontal cortex impairs familiarity but not recollection. Behavioural Brain Research. 2011;225:297–304. doi: 10.1016/j.bbr.2011.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Horn M, Jardri R, D’Hondt F, Vaiva G, Thomas P, Pins D. The multiple neural networks of familiarity: A meta-analysis of functional imaging studies. Cogn Affect Behav Neurosci. 2016;16:176–190. doi: 10.3758/S13415-015-0392-1/TABLES/3. [DOI] [PubMed] [Google Scholar]
- 76.Moll J, Bado P, de Oliveira-Souza R, Bramati IE, Lima DO, Paiva FF, Sato JR, Tovar-Moll F, Zahn R. A neural signature of affiliative emotion in the human septohypothalamic area. J Neurosci. 2012;32:12499–12505. doi: 10.1523/JNEUROSCI.6508-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Laubach M, Amarante LM, Swanson K, White SR. What, If Anything, Is Rodent Prefrontal Cortex? eNeuro. 2018;5 doi: 10.1523/ENEURO.0315-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sherathiya VN, Schaid MD, Seiler JL, Lopez GC, Lerner TN. GuPPy, a Python toolbox for the analysis of fiber photometry data. Scientific Reports. 2021;11:1–9. doi: 10.1038/s41598-021-03626-9. 2021 11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Botta P, Demmou L, Kasugai Y, Markovic M, Xu C, Fadok JP, Lu T, Poe MM, Xu L, Cook JM, et al. Regulating anxiety with extrasynaptic inhibition. Nature Neuroscience. 2015;18:1493–1500. doi: 10.1038/nn.4102. 2015 18:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Owen SF, Kreitzer AC. An open-source control system for in vivo fluorescence measurements from deep-brain structures. J Neurosci Methods. 2019;311:170–177. doi: 10.1016/J.JNEUMETH.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boyle L, Posani L, Irfan S, Siegelbaum SA, Fusi S. The geometry of hippocampal CA2 representations enables abstract coding of social familiarity and identity. bioRxiv. 2022:2022.01.24.477361. doi: 10.1101/2022.01.24.477361. [DOI] [Google Scholar]
- 82.Nogueira R, Rodgers CC, Bruno RM, Fusi S. The geometry of cortical representations of touch in rodents. Nat Neurosci. 2023;26:239–250. doi: 10.1038/S41593-022-01237-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. The code used to create the classifiers analysis of the fiber-photometry data is available at https://github.com/ramonnogueira/decode_familiarity. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request