Abstract
Cerebrovascular dysregulation such as altered cerebral blood flow (CBF) can be observed in Alzheimer’s disease (AD) and may precede symptom onset. Genome wide association studies show that AD has a polygenic aetiology, providing a tool for studying AD susceptibility across the lifespan. Here, we ascertain whether the AD genetic risk effects on CBF previously observed (Chandler et al., 2019) are also present in later life. Consistent with our prior observations, AD genetic risk score (AD-GRS) was associated with reduced CBF in the ADNI sample. The regional association between AD-GRS and CBF were also spatially similar. Furthermore, CBF was related to the regional mRNA transcript expression of AD risk genes proximal to AD-GRS risk loci. These observations suggest that AD risk alleles may reduce neurovascular process such as CBF, potentially via mechanisms such as regional expression of proximal AD risk genes as an antecedent AD pathophysiology. Our observations help establish processes that underpin AD genetic risk-related reductions in CBF as a therapeutic target prior to the onset of neurodegeneration.
Keywords: Cerebral blood flow, Polygenic, Alzheimer’s disease, Gene expression, Lifespan
1. Introduction
Variability of cerebrovascular function is heritable and partly explained by additive effects of genetic factors that converge across several neurobiological processes (Ikram et al., 2018). In Alzheimer’s disease (AD), cerebrovascular dysregulation is a key concomitant factor (Kelleher and Soiza, 2013), and is one of the earliest markers of AD pathophysiology (Iturria-Medina et al., 2016; Kelleher and Soiza, 2013). Decreases in cerebrovascular function are observed both in patients with AD and young individuals with an increased risk of dementia (Chandler et al., 2019; Filippini et al., 2011; Montagne et al., 2020; Wolters et al., 2017) This broadly suggests that altered cerebrovascular function is a risk factor for AD, rather than a consequence of the disease, which may be present across an individual’s lifespan.
Genome-wide association studies (GWAS) demonstrate that AD is also highly polygenic, where potentially thousands of common risk alleles confer susceptibility for disease (Kunkle et al., 2019). Although polygenic analysis has shown utility in predicting AD (Escott-Price et al., 2017; Escott-Price et al., 2015), the neurobiological mechanisms by which these loci confer risk remains poorly understood, particularly in relation to cerebrovascular function. Furthermore, the impact of these risk alleles across the lifespan has been seldom explored. Several studies have suggested that the influence of AD risk alleles may be age-dependent (Matura et al., 2016), while other large studies demonstrate that the impact of AD risk alleles on risk factors such as cognition are influential across the entire lifespan (Hill et al., 2016). However, the impact of AD risk alleles on in-vivo measures of brain function has not been investigated across the lifespan.
In our previous work we used arterial spin labelling (ASL) with MRI to quantify regional cerebral perfusion in young healthy individuals (18–35 years) and observed negative associations between AD-polygenic risk and regional perfusion, as well as lower CBF in those who possess a copy of the APOE-ε4 allele. Our findings suggest that vascular alterations in those with a broad increased genetic risk for AD manifest decades prior to symptom onset (Chandler et al., 2019). While our prior work provided insight into the influence of genetic risk factors on the cerebrovasculature in early adulthood, it is not yet known whether the influence of AD genetic risk scores on grey matter cerebral blood flow (GM CBF) remains consistent across the lifespan.
In the current study, we aim to determine the impact of AD risk alleles on CBF in an older population (mean age = 70). We anticipate that the combined influence of AD risk alleles will be associated with a reduction in global CBF (similar to our findings in (Chandler et al., 2019). Here, one predicts that either (1) the effects of AD risk alleles on CBF remain consistent or (2) demonstrate a more pronounced influence later in life. As AD risk alleles are likely to confer susceptibility by influencing expression of proximal genes, we further anticipate that regional CBF is spatially related to the transcript expression of these AD risk alleles. Here, we aim to take advantage of the Allen Human Brain Atlas (AHBA), a gene expression atlas that has advanced the development of imaging transcriptomics; linking macroscale brain imaging data to molecular function (for overview, see (Arnatkevic Iute et al., 2019; Fornito et al., 2019)). Prior evidence has revealed regional variations in gene expression with both functional connectivity with resting-state MRI (Forest et al., 2017), and structural connectivity with tractography (Goel et al., 2014). Yet, the link between AD risk alleles, physiological MRI measures, including regional perfusion, and the regional co-expression of AD risk gene transcripts proximal to these AD risk loci remains to be examined.
In order to address this hypothesis, we probe the AHBA to understand the relationship between AD risk gene expression and regional CBF across the cortex to determine if the influence of AD risk alleles can be explained by the regional co-expression of gene transcripts proximal to these AD risk loci (Arnatkevic Iute et al., 2019). Specifically, we sought to investigate whether brain-wide AD-related gene expression correlates with regional variation in CBF. While prior investigations have determined associations between gene expression and MRI based markers of regional aging / AD – associated atrophy, this approach has not yet been considered for cerebrovascular architecture (Groot et al., 2021; Vidal-Pineiro et al., 2020; Zhang et al., 2021). These analyses will establish the regional cortical co-expression of AD risk genes and AD-risk gene related CBF reductions, providing a plausible mechanistic link between AD risk loci and a well-established pathophysiological process in AD.
2. Methods
2.1. Participants
2.1.1. ADNI sample
A total of 79 participants, classified as either healthy controls or having mild cognitive impairment took part in a series of MRI scans as part of their involvement in the ADNI protocol. To avoid population stratification issues between our AD GWAS - AD-GRS, and to compare to our prior sample (Chandler et al., 2019), we matched our test sample with demographically similar characteristics to the discovery sample (Kunkle et al., 2019), we included participants who self-reported as ‘White’ and ‘Not Hispanic or Latino’. Participants were further removed if they also contributed to discovery IGAP AD GWAS (NOVERLAP = 2). Some participants were scanned at several time points, where the final number of discrete data points NOBSERVATIONS = 127, where NPARTICIPANTS = 44 completed 1 scan and NPARTICIPANTS = 22 completed 2 scans and NPARTICIPANTS = 13 completed 3 scans in the final analysis. See Table 1 for further demographic information.
Table 1. Sample characteristics.
| Participants with baseline scan | Participants with 12-mo follow-up scan | Participants with 24-mofollow-up scan | Overall | |
|---|---|---|---|---|
| (N = 44) | (N = 22) | (N = 13) | (N TOTAL=127, NUNIQUE = 79) | |
| Age | ||||
| Mean (SD) | 71.0 (6.95) | 68.3 (5.26) | 67.2 (5.64) | 68.9 (6.11) |
| Median [Min, Max] | 70.3 [55.0, 83.4] | 68.3 [58.5, 78.9] | 65.5 [59.5, 78.5] | 68.2 [55.0, 83.4] |
| Sex | ||||
| Female | 19 (43.2%) | 10 (45.5%) | 6 (46.2%) | 57 (44.9%) |
| Male | 25 (56.8%) | 12 (54.5%) | 7 (53.8%) | 70 (55.1%) |
| Diagnosis | ||||
| CN | 13 (29.5%) | 5 (22.7%) | 5 (38.5%) | 38 (29.9%) |
| EMCI | 19 (43.2%) | 13 (59.1%) | 6 (46.2%) | 63 (49.6%) |
| LMCI | 12 (27.3%) | 4 (18.2%) | 2 (15.4%) | 26 (20.5%) |
| APOE4 | ||||
| 0 | 27 (61.4%) | 13 (59.1%) | 12 (92.3%) | 89 (70.1%) |
| 1 | 17 (38.6%) | 6 (27.3%) | 0 (0%) | 29 (22.8%) |
| 2 | 0 (0%) | 3 (13.6%) | 1 (7.7%) | 9 (7.1%) |
| Visit (Mos) | ||||
| 6 | 21 (47.7%) | 9 (40.9%) | 8 (61.5%) | 52 (40.9%) |
| 12 | 16 (36.4%) | 7 (31.8%) | 2 (15.4%) | 44 (34.6%) |
| 24 | 7 (15.9%) | 6 (27.3%) | 3 (23.1%) | 31 (24.4%) |
| HC > MCI (Conversion) | ||||
| 0 | 43 (97.7%) | 18 (81.8%) | 12 (92.3%) | 115 (90.6%) |
| 1 | 1 (2.3%) | 4 (18.2%) | 1 (7.7%) | 12 (9.4%) |
| MoCA | ||||
| Mean (SD) | 24.6 (2.98) | 25.3 (2.85) | 25.6 (1.98) | 25.2 (2.65) |
| Median [Min, Max] | 25.0 [16.0, 30.0] | 26.0 [18.0, 29.0] | 26.0 [22.0, 30.0] | 26.0 [16.0, 30.0] |
| Education (Yrs) | ||||
| Mean (SD) | 16.5 (2.64) | 17.4 (1.97) | 17.5 (2.88) | 17.1 (2.50) |
| Median [Min, Max] | 16.0 [12.0, 20.0] | 17.5 [14.0, 20.0] | 18.0 [12.0, 20.0] | 18.0 [12.0, 20.0] |
| Intracranial Volume (mm3) | ||||
| Mean (SD) | 1,510,000 (140,000) | 1,550,000 (163,000) | 1,570,000 (182,000) | 1,540,000 (160,000) |
| Median [Min, Max] | 1,500,000 [1,290,000, 1,830,000] | 1,550,000 [1,260,000, 2,070,000] | 1,550,000 [1,260,000, 1,830,000] | 1,530,000 [1,260,000, 2,070,000] |
2.1.2. Cardiff sample
our younger sample was identical to our previous sample (Chandler et al., 2019) and consisted of 75(NFEMALE = 47), righthanded individuals of western European descent, aged between 18–35, with at least 15 years of education. For further sample characterization (including ethics, exclusion criteria, genotyping methods, see Chandler et al., (2019).
2.2. Creation of polygenic scores
Polygenic score calculations were performed according to the procedure described by the International Schizophrenia Consortium, using the –score command in PLINK, via a wrapper function provided in the PRSice v1.25 software package (Euesden et al., 2015). Training data were from a recent AD GWAS (Kunkle et al., 2019), where SNPs were removed from summary statistics / geno-type data if they had a low minor allele frequency (p < 0.01) and data were pruned for linkage disequilibrium, removing SNPs within 500 kb and R2 > 0.1 with a more significantly associated SNP. For the creation of the AD-GRS, we considered SNPs that were associated with AD that surpassed the GWAS threshold (PT < 5 ×10–8), as performed in and to make comparable to our original study (Chandler et al., 2019). We also removed all SNPs from the APOE gene on chromosome 19, as previously recommended (Ware et al., 2020) and individual APOE ε4 status was independently modelled in all analyses. Twenty-three SNPs were considered in the final AD-GRS calculation (see Fig. 2). To minimize potential confounding from population stratification linked to AD-GRS, we included the first 5 principle components from a linkage-disequilibrium (LD) pruned version of the genotypes as covariates in all analysis (Choi et al., 2020).
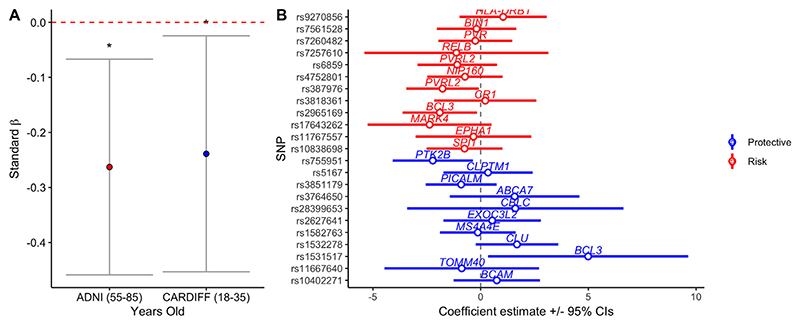
Fig. 2.
(A) Standardized AD-GRS effects on whole brain GM CBF for the young (18-35) and older (55-85) samples, * indicates p < 0.05, error bars represent 95% confidence intervals. (B) Diagnostic plot, demonstrating individual effects of AD risk (red) and protective (blue) SNPs on whole brain GMCBF, controlling for covariates in the older sample (55-85 years old). Circles / lines represent adjusted effect sizes and 95% confidence intervals. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.3. Imaging procedures and analysis of CBF
2.3.1. ADNI sample
A 3T siemens PICORE MRI sequence (Wong et al., 1997) with pulsed ASL (or Q2TIPS) (Luh et al., 1999). The sequence parameters include repetition time (TR) = 3400 ms, echo time (TE) = 12 ms, TI1 = 700 ms, TI2 = 1900 ms, field of view (FOV) = 256 mm × 256 mm, number of slices: 24 axial, slice thickness = 4 mm, and image matrix size = 64 × 64. Pre-processing steps were conducted in SPM8 and included motion correction of individual ASL frames by rigid body transformation and least squares fitting. To obtain perfusion weighted images, the ASL data were then split into tag and control images and the mean-untagged data were sub-tracted from the mean-tagged data. The first volume of the ASL scan was used in place of an M0 (providing fully relaxed signal) to estimate blood-water-density proxy and used for calibration. A 3D MPRAGE T1-weighted sequence was collected for registration with the following parameters: TR = 2300ms, TE = 2.98ms, TI = 900ms, 176 sagittal slices, FOV = 256 × 240mm2, voxel size = 1.1 × 1.1 ×1.2mm3, flip angle = 9°. The perfusion data were registered to T1 space and rescaled to obtain CBF in ml/100g/min. For both samples, GM CBF values were sampled in native space across 82 cortical and subcortical parcellations as segmented using a FreeSurfer template (Desikan et al., 2006; Potvin et al., 2017). Full analysis including details of distortion correction, registration and partial volume correction can be found via the ADNI web page (http://adni.loni.usc.edu).
2.3.2. Cardiff sample
Imaging data were collected on a 3T General Electric (GE) MRI scanner. Anatomical T1-weighted images were acquired with a 3D fast spoiled gradient echo sequence (FSPGR). Sequence parameters included: 172 contiguous sagittal slices with a slice thickness of 1 mm, TR = 7.9, TE = 3ms, inversion time of 450ms, flip angle = 20°, a FOV of 256 ×256 ×176 mm, matrix size 256 ×256 ×192 to yield 1 mm isotropic voxel resolution images. Resting CBF data were collected using a pseudo-continuous arterial spin labelling (PCASL) sequence. The study consisted of a single MRI session (which also comprised other functional and structural scans), and the PCASL sequence that lasted approximately 6 minutes. A PCASL sequence was acquired and included a 3D fast spin echo (FSE) spiral multi-slice readout. The sequence parameters included: number of excitations = 3, time to echo=32ms, echo time train length =64, TR =5.5seconds, matrix size =48 ×64 ×60, FOV =18 ×23 ×18cm, tag = 1500ms, PLD = 1500ms. The pcASL pre-processing for the sample can be found in (Chandler et al., 2019). Briefly, structural T1-weighted FSPGR images were registered to the M0 image acquired as part of the calibration of the CBF image acquisition, generating a transformation matrix. This transformation matrix was then applied to the skull stripped FSPGR (with reference/warping to the M0) using FSL’s Brain Extraction Tool. Next, linear registration via FSLs FLIRT registered the skull stripped anatomical image to the M0 transformation matrix (Montreal Neurological Institute (MNI) space) and the difference was calculated between this and the subject’s native space, providing data in the same space as the CBF data. The two transformation matrices for each participant were then concatenated to produce a matrix for the low resolution CBF image. All CBF images were then warped to standard MNI template using FSLs FLIRT. The priors for the grey matter were then registered to the skull stripped M0 image, generating a mask of grey matter from which CBF values was extracted.
2.4. Gene expression analysis
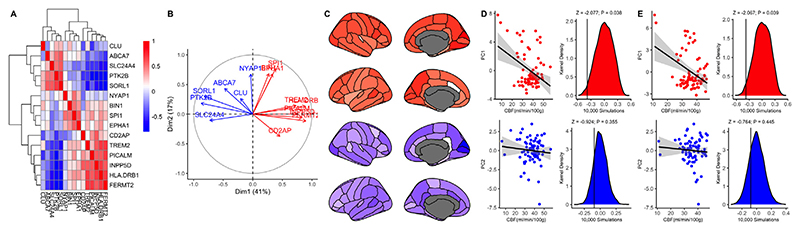
Publicly available human gene expression data from six post-mortem donors (NFEMALE = 1), aged 24–57 (42.5 ± 13.38) were obtained from the Allen Institute (Hawrylycz et al., 2012). Data reflect the microarray normalization pipeline implemented in March 2013 (http://human.brain-map.org) and analyses were conducted according to the guidelines of the Yale University Human Subjects Committee. Normalized brain-wide gene transcript expression was mapped to 82 cortical /subcortical regions of interest as defined by the Desikan-Killiany atlas in abagen v0.0.3 (Arnatkevic Iute et al., 2019). Comprehensive processing details for the gene expression pre-processing and analysis can be found at https://abagen.readthedocs.io/en/stable/user_guide/reporting.html. In order to quantify AD risk gene transcript expression, we performed principal component analysis (PCA) for regional transcript expression. We identified two principal modes of covariation (see Fig. 4A–B) which were then individually regressed against regional CBF for the Cardiff and ADNI samples.
Fig. 4.
(A) Correlation matrix showing expression of AD risk genes. (B) Principal component analysis identified two principal modes of covariation between expression of all AD risk genes across the brain. (C) PC1-2 mapped onto the cortical regions. (D-E). Scatter plots show relationship between regional AD gene expression for PC1 (upper) and PC2 (lower) and regional CBF for the (D) Cardiff sample and (E) the ADNI sample. (D-E) Density plots for the distribution of 10,000 randomly simulated regional values (scaled to CBF range) for PC1-2 for Cardiff (D) and ADNI (E) samples. Solid black vertical lines represent the actual, observed correlation between PC1/2 AD gene expression and regional CBF.
2.5. Statistical analysis
To maximize consistency of regression models across the samples, we included the same covariates for both sample analyses. Predictors were regressed against (1) whole GM CBF and (2) regional GM CBF for the 82 cortical / subcortical regions as defined by the Desikan-Killiany-Tourville (DKT) Atlas (Potvin et al., 2017). Regional CBF was z-normalized where each ROI was de-meaned and divided by the sample standard deviation to make comparison across ROIs and sample easier to interpret. The fixed effects of AD-GRS (PT < 5 ×10-8) and APOE (number of ε4 alleles (0/1/2)) were modelled while controlling for age, biological sex, education, ICV, and the first 5 genetic principal components, acquired via the LD-pruned datasets. For the ADNI sample, we further included fixed effect covariates for (1) diagnostic status (modelling both a) healthy control / mild cognitive impairment and b) progression from healthy control to CI); (2) years of education; (3) cognition (as measured via the Montreal Cognitive Assessment Score); (4) site; (5) visit code (1) 12 months; 2) 24 months), and nested random effects for both (3) visit code and (4) subject, modelled as repeated measures. We included multiple time points to maximize sample size / statistical power. We employed outlier labelling / detection (Hoaglin and Iglewicz, 1987), which defines outliers using the interquartile range outlier labelling rule (1.5 × interquartile range (Q3-Q1)). This approach dynamically removed data points for each GM CBF dependent variable to minimize the impact of outlier data points. We compared the (1) CBF and (2) beta coefficients for the AD-GRS fixed effect for each region of interest between the Cardiff and ADNI samples using simple pearson r correlation. To control for false positives in each analysis, we compare each correlation to (1) 10,000 generated regional gene expression profiles equaling the number of genes used to generate the average AD gene expression (NUNIQUE-GENES = 16) and (2) generated surrogate brain maps to simulate 10,000 null effects for CBF across 82 brain regions. To preserve potential autocorrelation across the brain maps and test for spatial specificity, we performed null spins of the brain parcellations to generate 10,000 surrogates for each map to create our null distributions (Alexander-Bloch et al., 2018; Burt et al., 2020; Wei et al., 2021). To assume that the strength of the gene expression – CBF covariation is more pronounced than expected by chance, the observed z-transformed correlation must surpass the alpha tail (Z > 1.96 / 95% CI: two-tailed) for the simulated distributions.
3. Results
3.1. Cerebral blood flow across the lifespan
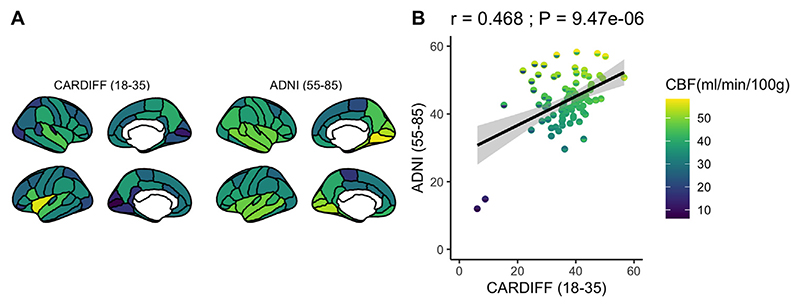
First, we observed that regional GM CBF showed a consistent pattern of positive covariation between the younger and older sample (Fig. 1A-B), where cortical regions that showed higher per-fusion (ml/100g/min) in the younger sample was also comparably higher in the older sample (r = 0.468: p < 0.001; Fig. 1C), suggesting a pattern of consistent, regional variation in GM CBF across the lifespan.
Fig. 1.
Regional GM CBF (ml/100g/min) in (A) the younger sample (aged: 18-35) previously described in Chandler et al., 2019 and (B) an older sample (aged: 55-85) and (C) Regional GM CBF (ml/100g/min) comparison for young (x-axis) and old (y-axis) across 82 cortical / subcortical regions.
3.2. AD-GRS effects on whole brain cerebral blood flow (ml/min/100g)
Similar to our original discovery (Chandler et al., 2019), we observed a significant negative association between whole brain GM-CBF and AD-GRS in the older (55 –85 years) ADNI sample (β = - 0.26; p = 0.011) after controlling for all covariates. Unlike our observation in younger individuals (Chandler et al., 2019) we did not observe a significant association between APOE ε4 absence / presence and whole brain GM CBF in the older sample (β = 0.28; p = 0.107). For all fixed effects and confidence intervals observed in the whole brain GM CBF analysis, see Table 2.
Table 2.
Fixed effect predictors (β estimate and 95% confidence intervals) regressed against whole brain GM CBF in the final sample of ADNI participants controlling for the top 5 principal components (PCs) as additional covariates of no interest and visit code / subject as nested random effects.
| FIXED EFFECTS | BETA | CI (2.5%) | CI (97.5%) | T-STATISTIC | DF | p-VALUE |
|---|---|---|---|---|---|---|
| (Intercept) | -1.19726 | -3.21405 | 0.819521 | -1.16353 | 76.11257 | 0.248248 |
| AD-GRS | -0.26292 | -0.45875 | -0.06709 | -2.63145 | 67.49825 | 0.010526 |
| APOE4 | 0.289192 | -0.05816 | 0.636544 | 1.631796 | 65.6209 | 0.107513 |
| MCI | 0.007101 | -0.31093 | 0.325133 | 0.043761 | 110.8895 | 0.965174 |
| MCI-CONVERSION | 0.357973 | -0.35038 | 1.066325 | 0.99049 | 61.00472 | 0.325847 |
| AGE | 0.018739 | -0.00964 | 0.047115 | 1.294349 | 72.57036 | 0.199648 |
| SEX(M) | -0.34147 | -0.78345 | 0.100503 | -1.51428 | 67.246 | 0.134642 |
| SITE | 0.000119 | -0.0009 | 0.001139 | 0.229399 | 11.33588 | 0.822645 |
| FOLLOW-UP (12M) | -0.49858 | -0.69474 | -0.30243 | -4.98176 | 62.90319 | 5.21E-06 |
| FOLLOW-UP (24M) | -0.82986 | -1.04813 | -0.6116 | -7.45195 | 56.49852 | 5.9E-10 |
| EDUCATION | 0.031833 | -0.16608 | 0.229751 | 0.315243 | 74.35791 | 0.75346 |
| MOCA | 0.062863 | -0.09299 | 0.218718 | 0.790535 | 99.64165 | 0.431093 |
| ICV | 0.260453 | 0.04815 | 0.472756 | 2.404479 | 66.19241 | 0.019001 |
Key: AD-GRS, alzheimer’s disease genetic risk score; MCI, mild cognitive impairment; MOCA, montreal Cognitive Assessment; ICV, intracranial volume
To assess the individual impact of each of the SNPs in our AD-GRS model, we performed a linear regression analysis where each individual SNP was regressed in an additive model against whole brain GM CBF, controlling for all aforementioned covariates. Consistent with broad polygenic modelling assumptions, we observed a general propensity for SNPs that increase risk for AD (odds ratio (OR) > 1) to associate with reduced whole GM CBF, while alleles that conferred relative protection (OR < 1) for AD where associated with an increase in whole GM CBF (Fig. 2; sign test for direction of effects: p = 0.041).
3.3. Comparing AD-GRS effects on cerebral blood flow in early adulthood and older age
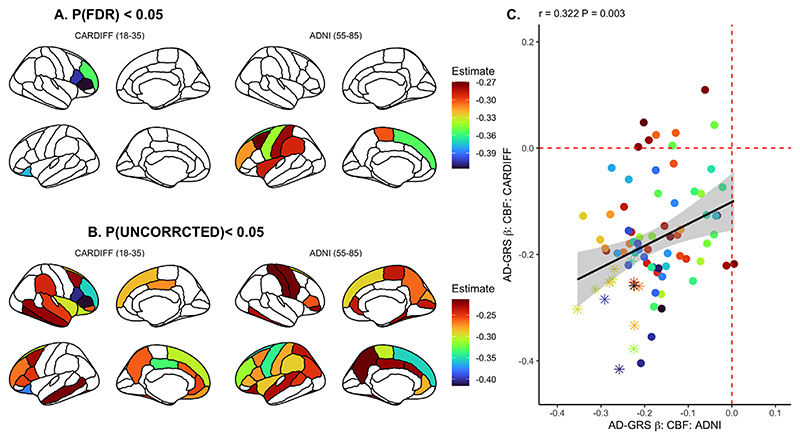
As we observed an association between AD-GRS and whole brain GM CBF for both younger and older samples (Fig. 3a), we proceeded to explore the association at a regional level. We repeated the linear mixed-model analysis across 82 cortical / subcortical regions. Building upon our initial analysis in the younger sample (Chandler et al., 2019 replotted here in Fig. 3b), we observed a relationship between regional effect sizes across the brain, where the most / least pronounced effects of AD-GRS were comparable between young and older samples (Fig. 3d). In the ADNI sample of older individuals (55-85 years old) we found several specific regions with significant effects after correcting for false discovery rate, specifically within left frontal cortices (see Fig. 3, Supplementary Table 2). We did not observe the influence of APOE ε4 status on (1) whole brain GM CBF in the older sample and (2) a regional effect of APOE ε4 on the younger sample, so did not proceed to investigate similarity between samples for APOE ε4 GM CBF effects at a regional level (see Supplementary Table 3). For additional comparability with the Cardiff sample, we repeated the ADNI sample analyses, restricting to a single time point per participant, reflecting their latest scan (N = 79). We observed comparable associations between whole brain CBF and AD-GRS (β = - 0.265; p = 0.034) and a similar profile of regional spatial similarity with the Cardiff sample (r = 0.22; p = 0.042). However, we did not observe any individual, regional associations after FDR correction (lowest PFDR = 0.054; left superior frontal gyrus).
Fig. 3.
Regional association between AD-GRS and CBF in both Cardiff (Chandler et al., 2019) and ADNI samples, corrected for false discovery rate (A) (PFDR < 0.05) and uncorrected (B) (PUNCORRCECTED < 0.05). (C) Linear relationship of effect sizes (standardized beta coefficients) across the brain when comparing all cortical regions between sample B & C, where data points represented as an asterisk reflect p < 0.05 in both samples. Each point in the scatter plot represents one cortical / subcortical region. Regression slope grey area represents 95% confidence intervals.
3.4. Regional AD risk gene expression overlap
We calculated the principal modes of covariation for transcript expression of AD risk genes proximal to the 23 SNPs used in AD-GRS model (NUNIQUE-GENES =16) for the 82 cortical / subcortical regions (Fig. 4A). We correlated AD risk gene expression with (1) regional CBF (ml/100g/min) for the younger/older samples. We observed that mean AD gene expression was negatively associated with regional GM CBF in the young (Z = -2.07, p = 0.038) and older sample (Z = -2.067, p = 0.039) with no difference in correlation strength (z = -0.02, p = 0.98), suggesting that AD risk gene expression is highest in cortical regions where GM CBF is generally lower. We observed a similar pattern of association for APOE transcript expression and CBF, but these were not significant following the permutation testing (Cardiff: Z = -1.69, p = 0.091; ADNI: Z = - 1.30, p = 0.194, see supplementary Fig. 1).
4. Discussion
We sought to further investigate the impact of common AD genetic risk alleles on cerebral perfusion. Critically, the negative association we observed between AD-GRS and whole brain GM CBF in our prior work (Chandler et al., 2019) was also evident in the older population. This observation was further supported by evidence that regional effects of AD-GRS on GM CBF were correlated across samples. This suggests that the regional impact of AD-GRS on CBF remains consistent across the lifespan, and preferentially influences specific cortical structures previously implicated in preclinical models of AD related pathophysiology. Together these observations show that the cumulative impact of AD risk loci on this hallmark feature of AD pathogenesis is consistent across the lifespan.
The mechanisms by which common (e.g. intronic and intergenic) SNPs identified via GWAS confer susceptibility are largely unknown. However, a growing body of work suggests that these SNPs act as expression quantitative trait locus (eQTLs) and influence the expression of AD risk genes. Here, we tested the hypothesis that the brain-wide CBF variability would be spatially convergent with the expression AD risk gene transcripts. We found consistent regional covariation between mean AD risk gene expression and regional CBF perfusion pattern in both young and old samples. These findings demonstrate that the impact of AD-GRS on perfusion may confer susceptibility via the altered expression of proximal AD genes.
Cerebral blood flow shows a gradual and steady decrease across the lifespan (Bertsch et al., 2009; Devous et al., 1986; Hagstadius and Risberg, 1989; Heo et al., 2010; Lu et al., 2011). Here, our correlation demonstrates some evidence for spatial convergence of CBF variation between the young and old samples, suggesting that regional variability in GM CBF across the cortex remains somewhat consistent across age. It is not entirely understood why there is variability in GM CBF at rest across the brain. However, prior evidence has shown that brain perfusion closely correlates with brain function and metabolism (Detre et al., 2009), suggesting that variability in regional perfusion may reflect differences in energy demand across the cortex at rest.
In the second analysis, we showed that the association between regional CBF and AD-GRS in the young sample correlated positively with the association between regional CBF and AD-GRS in the older sample. The most significant effects were mostly observed in the frontal and temporal cortical structures. Critically, this result demonstrates that the AD-GRS effects seen in the older sample are regionally congruent with those in the younger sample. Our findings suggest that SNPs included in the AD genetic/polygenic risk model have consistent negative effects on cerebral perfusion from young adulthood and throughout the lifespan. In addition to AD-GRS we also investigated the effects of APOE on CBF across the samples. We saw no influence of APOE ε4 status on whole brain or regional CBF in the ADNI sample (unlike in our prior study (Chandler et al., 2019). These findings are consistent with the large studies which have observed limited association between CBF and APOE in middle / later life (Moonen et al., 2022). Research into the association between CBF and APOE across the lifespan remains mixed, with some studies demonstrating CBF reductions in vulnerable medial temporal lobe structures in AD (Rubinski et al., 2021). We suggest that while the AD-GRS influence on CBF across the lifespan remains consistent, APOE status may have a more dynamic role in shaping CBF which may change across the lifespan (Wierenga et al., 2013) and requires further investigation.
In our third analysis we used gene expression data to identify how AD risk genes expression across the cortex correlates with regional CBF. Our results show a negative association between ADgene expression and regional CBF, suggesting that AD risk genes may spatially covary with regional cerebral perfusion. Moreover, our findings demonstrate that the regional covariation between cerebral perfusion and AD gene expression occurs throughout the lifespan.
While amyloid and tau-genic hypotheses provide important insight into preclinical AD models (Bloom, 2014; Gotz et al., 2001; Ittner and Gotz, 2011; Lewis et al., 2001), vascular dysregulation occurs prior to this AD pathophysiology (Iturria-Medina et al., 2016). We provide additional support for vascular dysregulation and hypoperfusion as early markers of AD risk that may be observed during young adulthood. Moreover, we suggest that cerebral perfusion is a potentially important AD related pathological feature and should be considered as a target for therapeutic intervention.
Our observations should be considered with the following limitations. First, while we observed consistent AD-GRS effects across the lifespan, we did not observe APOE related effects in our older sample, so we cannot infer that all AD genetic effects were consistent. This may be explained by dynamic APOE effects that have recently been discovered in recent MRI studies, where APOE effects manifest in early life, for example in the volume of medial temporal lobe structures such as the amygdala (Brouwer et al., 2020). We therefore suggest our study warrants further exploration of APOE / CBF in larger samples across the lifespan. Second, there are several differences within and between these samples, including non-equivalent scanner sequence (pCASL / pASL), site / scanner differences and pre-processing pathways which makes direct comparison or a combined analysis challenging, and observations about sample consistency should be interpreted with caution. Third, while we observed an association between whole brain CBF and AD-GRS in the ADNI sample and similar pattern of alterations to the Cardiff sample, the specific regions were not the same and the impact of AD-GRS on specific brain regions should be considered tentative until replicated in an independent, demographically similar sample. The negative association between AD-GRS and regional CBF were also weaker in the cross-sectional analysis of the ADNI, further suggesting larger samples will be beneficial in confirming the spatiotemporal relationship between genetic risk for AD and CBF. Last, we acknowledge potential confounding from other AD-related pathophysiology / cere-brovascular risk factors (Amen et al., 2020; Broce et al., 2019; Korte et al., 2020; Rubinski et al., 2021). We suggest that future research should incorporate further longitudinal molecular imaging such as glucose metabolism, amyloid, and tau imaging as well as known cerebrovascular risk factors. These would help to determine the trajectory of whole brain and regional perfusion changes across the lifespan and its association to further oxygenation changes and metabolic dysfunction in those at risk of AD.
To conclude, we demonstrate a consistent negative influence of additive genetic AD risk on cerebral perfusion across the lifespan, which was also related to regional expression of proximal AD risk genes across the cortex. Thus, reduced CBF may be a central, and proximal, process in the pathophysiology of AD, and a potential mechanism by which AD risk genes exert their adverse effects on brain structure and function.
Supplementary Material
Acknowledgements
HLC is funded by a Wellcome Strategic Award [104943/Z/14/Z].
KM is funded by the Wellcome Trust [WT200804].
TML acknowledges funding via a Sêr Cyrmu II Fellowship (East Wales European Regional Development Funds (PNU-80762-CU-14) and the UK Dementia Research Institute at Cardiff University (UK-DRI, which is supported by the Medical Research Council [UKDRI-3003], Alzheimer’s Research UK, and Alzheimer’s Society).
We are grateful to all professionals, patients and volunteers involved with the National Centre for Mental Health (NCMH) who took part in our Cardiff study. NCMH is funded by the National Institute for Social Care and Health Research (NISCHR), Welsh Government, Wales (Grant No. BR09).
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICOLtd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
CRediT authorship contribution statement
Hannah Chandler: Conceptualization, Methodology, Writing - original draft, Writing - review & editing. Richard Wise: Writing - review & editing, Supervision. David Linden: Writing - review & editing, Supervision, Funding acquisition. Julie Williams: Investigation, Writing - review & editing, Supervision. Kevin Murphy: Investigation, Writing - review & editing, Supervision, Funding acquisition. Thomas Matthew Lancaster: Conceptualization, Methodology, Writing - review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
All authors declare no conflict of interest with relevance to the current study.
Disclosure statement
The authors have no conflicts of interests or financial disclosures relevant to this article.
This work was supported by a Ser Cyrmu II Fellowship (European Regional Development Funds: CU149: “Imaging immunity in the genetic risk for Alzheimer’s disease) the Dementia Research Institute (DRI) for supporting this project. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.;Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.;Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
We currently have no related papers from this study under submission or submitted for publication.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at:http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
All authors has reviewed the contents of the manuscript being submitted and approve of its contents and validate the accuracy of the data.
References
- Alexander-Bloch AF, Shou H, Liu S, Satterthwaite TD, Glahn DC, Shinohara RT, Vandekar SN, Raznahan A. On testing for spatial correspondence between maps of human brain structure and function. Neuroimage. 2018;178:540–551. doi: 10.1016/j.neuroimage.2018.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amen DG, Wu J, George N, Newberg A. Patterns of Regional Cerebral Blood Flow as a Function of Obesity in Adults. J Alzheimers Dis. 2020;77(3):1331–1337. doi: 10.3233/JAD-200655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnatkevic Iute A, Fulcher BD, Fornito A. A practical guide to linking brain-wide gene expression and neuroimaging data. NeuroImage. 2019;189:353–367. doi: 10.1016/j.neuroimage.2019.01.011. [DOI] [PubMed] [Google Scholar]
- Bertsch K, Hagemann D, Hermes M, Walter C, Khan R, Naumann E. Resting cerebral blood flow, attention, and aging. Brain Res. 2009;1267:77–88. doi: 10.1016/j.brainres.2009.02.053. [DOI] [PubMed] [Google Scholar]
- Bloom GS. Amyloid-beta and tau: The trigger and bullet in Alzheimer disease pathogenesis. Jama Neurol. 2014;71(4):505–508. doi: 10.1001/jamaneurol.2013.5847. [DOI] [PubMed] [Google Scholar]
- Broce IJ, Tan CH, Fan CC, Jansen I, Savage JE, Witoelar A, Wen N, Hess CP, Dillon WP, Glastonbury CM, Glymour M. Dissecting the genetic relationship between cardiovascular risk factors and Alzheimer’s disease. Acta neuropathologica. 2019;137(2):209–226. doi: 10.1007/s00401-018-1928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer RM, Klein M, Grasby KL, Schnack HG, Jahanshad N, Teeuw J, Thomopoulos SI, Sprooten E, Franz CE, Gogtay N, Kremen WS. Dynamics of Brain Structure and its Genetic Architecture over the Lifespan. bioRxiv. 2020:2020.2004.2024.031138 [Google Scholar]
- Burt JB, Helmer M, Shinn M, Anticevic A, Murray JD. Generative modeling of brain maps with spatial autocorrelation. Neuroimage. 2020;220:117038. doi: 10.1016/j.neuroimage.2020.117038. [DOI] [PubMed] [Google Scholar]
- Chandler HL, Wise RG, Murphy K, Tansey KE, Linden DEJ, Lancaster TM. Polygenic impact of common genetic risk loci for Alzheimer’s disease on cerebral blood flow in young individuals. Sci Rep. 2019;9(1):467. doi: 10.1038/s41598-018-36820-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SW, Mak TS, O’Reilly PF. Tutorial: A guide to performing polygenic risk score analyses. Nature protocols. 2020 doi: 10.1038/s41596-020-0353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Detre JA, Wang J, Wang Z, Rao H. Arterial spin-labeled perfusion mri in basic and clinical neuroscience. Curr Opin Neurol. 2009;22(4):348–355. doi: 10.1097/WCO.0b013e32832d9505. [DOI] [PubMed] [Google Scholar]
- Devous MD, Sr, Stokely EM, Chehabi HH, Bonte FJ. Normal distribution of regional cerebral blood flow measured by dynamic single-photon emission tomography. J Cereb Blood Flow Metab. 1986;6(1):95–104. doi: 10.1038/jcbfm.1986.12. [DOI] [PubMed] [Google Scholar]
- Escott-Price V, Shoai M, Pither R, Williams J, Hardy J. Polygenic score prediction captures nearly all common genetic risk for Alzheimer’s disease. Neurobiol Aging. 2017;49:214 e217–214 e211. doi: 10.1016/j.neurobiolaging.2016.07.018. [DOI] [PubMed] [Google Scholar]
- Gerad/Perades, consortia, I. Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E, Badarinarayan N, Morgan K, Passmore P, Holmes C, Powell J. Common polygenic variation enhances risk prediction for alzheimer’s disease. Brain. 2015;138(Pt 12):3673–3684. doi: 10.1093/brain/awv268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31(9):1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N, Ebmeier KP, MacIntosh BJ, Trachtenberg AJ, Frisoni GB, Wilcock GK, Beckmann CF, Smith SM, Matthews PM, Mackay CE. Differential effects of the APOE genotype on brain function across the lifespan. NeuroImage. 2011;54(1):602–610. doi: 10.1016/j.neuroimage.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Forest M, Iturria-Medina Y, Goldman JS, Kleinman CL, Lovato A, Oros Klein K, Evans A, Ciampi A, Labbe A, Greenwood CMT. Gene networks show associations with seed region connectivity. Hum Brain Mapp. 2017;38(6):3126–3140. doi: 10.1002/hbm.23579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Arnatkeviciute A, Fulcher BD. Bridging the Gap between Connectome and Transcriptome. Trends in cognitive sciences. 2019;23(1):34–50. doi: 10.1016/j.tics.2018.10.005. [DOI] [PubMed] [Google Scholar]
- Goel P, Kuceyeski A, LoCastro E, Raj A. Spatial patterns of genome-wide expression profiles reflect anatomic and fiber connectivity architecture of healthy human brain. Hum Brain Mapp. 2014;35(8):4204–4218. doi: 10.1002/hbm.22471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz J, Chen F, van Dorpe J, Nitsch RM. Formation of neurofibrillary tangles in P301l tau transgenic mice induced by Abeta 42 fibrils. Science. 2001;293(5534):1491–1495. doi: 10.1126/science.1062097. [DOI] [PubMed] [Google Scholar]
- Groot C, Grothe MJ, Mukherjee S, Jelistratova I, Jansen I, van Loenhoud AC, Risacher SL, Saykin AJ, Mac Donald CL, Mez J, Trittschuh EH. Differential patterns of gray matter volumes and associated gene expression profiles in cognitively-defined Alzheimer’s disease subgroups. NeuroImage Clinical. 2021;30:102660. doi: 10.1016/j.nicl.2021.102660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstadius S, Risberg J. Regional cerebral blood flow characteristics and variations with age in resting normal subjects. Brain Cogn. 1989;10(1):28–43. doi: 10.1016/0278-2626(89)90073-0. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, Abajian C. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489(7416):391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo S, Prakash RS, Voss MW, Erickson KI, Ouyang C, Sutton BP, Kramer AF. Resting hippocampal blood flow, spatial memory and aging. Brain Res. 2010;1315:119–127. doi: 10.1016/j.brainres.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WD, Davies G, Group CCW, Liewald DC, McIntosh AM, Deary IJ. Age-Dependent Pleiotropy Between General Cognitive Function and Major Psychiatric Disorders. Biol Psychiatry. 2016;80(4):266–273. doi: 10.1016/j.biopsych.2015.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoaglin DC, Iglewicz B. Fine-Tuning Some Resistant Rules for Outlier Labeling. Journal of the American Statistical Association. 1987;82(400):1147–1149. [Google Scholar]
- Ikram MA, Zonneveld HI, Roshchupkin G, Smith AV, Franco OH, Sigurdsson S, van Duijn C, Uitterlinden AG, Launer LJ, Vernooij MW, Gudnason V. Heritability and genome-wide associations studies of cerebral blood flow in the general population. Journal of cerebral blood flow and metabolism: Official journal of the International Society of Cerebral Blood Flow and Metabolism. 2018;38(9):1598–1608. doi: 10.1177/0271678X17715861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittner LM, Gotz J. Amyloid-beta and tau–a toxic pas de deux in Alzheimer’s disease. Nat Rev Neurosci. 2011;12(2):65–72. doi: 10.1038/nrn2967. [DOI] [PubMed] [Google Scholar]
- Iturria-Medina Y, Sotero RC, Toussaint PJ, Mateos-Perez JM, Evans AC, Alzheimer’s Disease Neuroimaging, I. Early role of vascular dysregulation on late-onset Alzheimer’s disease based on multifactorial data-driven analysis. Nat Commun. 2016;7:11934. doi: 10.1038/ncomms11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ, Soiza RL. Evidence of endothelial dysfunction in the development of Alzheimer’s disease: Is Alzheimer’s a vascular disorder? Am J Cardiovasc Dis. 2013;3(4):197–226. [PMC free article] [PubMed] [Google Scholar]
- Korte N, Nortley R, Attwell D. Cerebral blood flow decrease as an early pathological mechanism in Alzheimer’s disease. Acta neuropathologica. 2020 doi: 10.1007/s00401-020-02215-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, Boland A, Vronskaya M, van der Lee SJ, Amlie-Wolf A, Bellenguez C, Alzheimer Disease Genetics, C., European Alzheimer’s Disease, I., Cohorts for, H., Aging Research in Genomic Epidemiology, C., Genetic, Environmental Risk in Ad/Defining Genetic, P., Environmental Risk for Alzheimer’s Disease, C. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51(3):414–430. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J, Dickson DW, Lin WL, Chisholm L, Corral A, Jones G, Yen SH, Sahara N, Skipper L, Yager D, Eckman C. Enhanced neurofibrillary degeneration in transgenic mice expressing mutant tau and APP. Science. 2001;293(5534):1487–1491. doi: 10.1126/science.1058189. [DOI] [PubMed] [Google Scholar]
- Lu H, Xu F, Rodrigue KM, Kennedy KM, Cheng Y, Flicker B, Hebrank AC, Uh J, Park DC. Alterations in cerebral metabolic rate and blood supply across the adult lifespan. Cereb Cortex. 2011;21(6):1426–1434. doi: 10.1093/cercor/bhq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luh WM, Wong EC, Bandettini PA, Hyde JS. QUIPSS II with thin-slice TI1 periodic saturation: A method for improving accuracy of quantitative perfusion imaging using pulsed arterial spin labeling. Magn Reson Med. 1999;41(6):1246–1254. doi: 10.1002/(sici)1522-2594(199906)41:6<1246::aid-mrm22>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Matura S, Prvulovic D, Hartmann D, Scheibe M, Sepanski B, Butz M, Oertel–Knochel V, Knochel C, Karakaya T, Fusser F, Hattingen E. Age-Related Effects of the Apolipoprotein E Gene on Brain Function. J Alzheimers Dis. 2016;52(1):317–331. doi: 10.3233/JAD-150990. [DOI] [PubMed] [Google Scholar]
- Montagne A, Nation DA, Sagare AP, Barisano G, Sweeney MD, Chakhoyan A, Pachicano M, Joe E, Nelson AR, D’Orazio LM, Buennagel DP. APOE4 leads to blood-brain barrier dysfunction predicting cognitive decline. Nature. 2020;581(7806):71–76. doi: 10.1038/s41586-020-2247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonen JEF, Nasrallah IM, Detre JA, Dolui S, Erus G, Davatzikos C, Meirelles O, Bryan RN, Launer LJ. Race, sex, and mid-life changes in brain health: Cardia MRI substudy. Alzheimers Dement. 2022 doi: 10.1002/alz.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potvin O, Dieumegarde L, Duchesne S, Alzheimer’s Disease Neuroimaging, I Freesurfer cortical normative data for adults using Desikan-Killiany-Tourville and ex vivo protocols. Neuroimage. 2017;156:43–64. doi: 10.1016/j.neuroimage.2017.04.035. [DOI] [PubMed] [Google Scholar]
- Rubinski A, Tosun D, Franzmeier N, Neitzel J, Frontzkowski L, Weiner M, Ewers M. Lower cerebral perfusion is associated with tau-PET in the entorhinal cortex across the Alzheimer’s continuum. Neurobiol Aging. 2021;102:111–118. doi: 10.1016/j.neurobiolaging.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Pineiro D, Parker N, Shin J, French L, Grydeland H, Jackowski AP, Mowinckel AM, Patel Y, Pausova Z, Salum G, Sørensen Ø. Cellular correlates of cortical thinning throughout the lifespan. Sci Rep. 2020;10(1):21803. doi: 10.1038/s41598-020-78471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware EB, Faul JD, Mitchell CM, Bakulski KM. Considering the APOE locus in Alzheimer’s disease polygenic scores in the Health and Retirement Study: A longitudinal panel study. BMC medical genomics. 2020;13(1):164. doi: 10.1186/s12920-020-00815-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, de Lange SC, Pijnenburg R, Scholtens LH, Ardesch DJ, Watanabe K, Posthuma D, van den Heuvel MP. Statistical testing in gene transcriptomic-neuroimaging associations: An evaluation of methods that assess spatial and gene specificity. bioRxiv. 2021:2021.2002.2022.432228. doi: 10.1002/hbm.25711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga CE, Clark LR, Dev SI, Shin DD, Jurick SM, Rissman RA, Liu TT, Bondi MW. Interaction of age and APOE genotype on cerebral blood flow at rest. Journal of Alzheimer’s disease: JAD. 2013;34(4):921–935. doi: 10.3233/JAD-121897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters FJ, Zonneveld HI, Hofman A, van der Lugt A, Koudstaal PJ, Vernooij MW, Ikram MA, Heart-Brain Connection Collaborative Research, G. Cerebral Perfusion and the Risk of Dementia: A Population-Based Study. Circulation. 2017;136(8):719–728. doi: 10.1161/CIRCULATIONAHA.117.027448. [DOI] [PubMed] [Google Scholar]
- Wong EC, Buxton RB, Frank LR. Implementation of quantitative perfusion imaging techniques for functional brain mapping using pulsed arterial spin labeling. Nmr Biomed. 1997;10(4-5):237–249. doi: 10.1002/(sici)1099-1492(199706/08)10:4/5<237::aid-nbm475>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ma M, Xie Z, Wu H, Zhang N, Shen J. Bridging the Gap Between Morphometric Similarity Mapping and Gene Transcription in Alzheimer’s Disease. Front Neurosci. 2021;15:731292. doi: 10.3389/fnins.2021.731292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.